Abstract
MAD7 is an engineered class 2 type V-A CRISPR-Cas (Cas12a/Cpf1) system isolated from Eubacterium rectale. Analogous to Cas9, it is an RNA-guided nuclease with demonstrated gene editing activity in Escherichia coli and yeast cells. Here, we report that MAD7 is capable of generating indels and fluorescent gene tagging of endogenous genes in human HCT116 and U2OS cancer cell lines, respectively. In addition, MAD7 is highly proficient in generating indels, small DNA insertions (23 bases), and larger integrations ranging from 1 to 14 kb in size in mouse and rat embryos, resulting in live-born transgenic animals. Due to the different protospacer adjacent motif requirement, small-guide RNA, and highly efficient targeted gene disruption and insertions, MAD7 can expand the CRISPR toolbox for genome enginnering across different systems and model organisms.
Introduction
The field of gene editing has been revolutionized by the application of CRISPR systems discovered in bacteria and archaea. To date, the most widely used system for gene editing is the class 2 effector nuclease Cas9 from Streptococcus pyogenes (SpCas9).1–4 Unlike multicomponent class 1 CRISPR effectors, this nuclease is a single protein that can easily be reprogrammed to target 20-nucleotide recognition sequences in double-stranded mammalian DNA located next to a short protospacer adjacent motif (PAM) when loaded with a targeting crRNA and accessory tracrRNA (or a chimeric version containing both elements: single guide RNA [sgRNA]). This ease of programmability gives CRISPR gene targeting an advantage over previous gene editing methods such as zinc finger nucleases and TALENs, for which the entire protein must be re-engineered for each target site.5
Gene editing nucleases such as CRISPR-Cas9 make a targeted double-stranded break (DSB) in the genome, whereby in mammalian systems, the cut is repaired by endogenous repair mechanisms, primarily non-homologous end joining (NHEJ), which often result in small insertions/deletions (indels), and homology-directed repair (HDR), which can effect precise gene edits through the use of an exogenous DNA template donor sequence.4,6,7 Although CRISPR-Cas9 currently dominates applications of CRISPR systems for gene editing, recently, a wealth of additional CRISPR systems have been discovered with potential to be harnessed for targeted DNA manipulation.8,9 A diversity of CRISPR-Cas nucleases can be envisioned to address issues including increased functionality, increased specificity, smaller protein size for packaging into viral delivery systems, an increased targeting range with various PAM sequence and the potential for PAM-orthogonal, and multimodal gene regulation (e.g., one deactivated Cas protein targeting a sequence for gene activation or repression, and another Cas protein targeting DNA for gene disruption or tagging).10
An example of alternative DNA-targeting CRISPR-Cas systems discovered so far includes the class 2 type V Cas12a/Cpf1 enzymes, originally reported from Acidaminococcus and Lachnospiraceae species.11 Like Cas9, these are single effector nucleases that have evolved independently but within pathways similar to Cas9. However, they differ in several critical aspects.8,12 For example, instead of recognizing a guanine-rich PAM sequence (5′-NGG-3′ for SpCas9) immediately downstream of crRNA, Cas12a recognizes a thymidine-rich PAM sequence (5′-NTTV-3′) upstream of crRNA. Whereas Cas9 uses two nuclease domains (HNH and RuvC) to cut each DNA strand and form a near-simultaneous blunt-ended DSB, Cas12a enzymes use a single RuvC-like endonuclease domain to cut the DNA in a staggered fashion, leaving 4-nucleotide 5′ overhangs. Unlike Cas9, Cas12a nucleases use a shorter (42- or 56-nucleotide) guide RNA (gRNA) and does not require a tracrRNA.12 In addition, since Cas12a has both DNA and RNA nuclease activity, it could autonomously process pre-crRNA into mature crRNA, which makes it more friendly for targeting multiple genomic loci with polycistronic crRNA.13
MAD7 is an engineered Cas12a variant originating from the bacterium Eubacterium rectale (refseq WP_055225123.1) found on the island of Madagascar, which shares 76% identical nucleotides with the native form. It encodes a monomeric 147.9 kDa polypeptide consisting of 1,263 amino acids and shows preference for 5′-TTTN-3′ and 5′-CTTN-3′ PAM sites. Notably, this engineered variant only shares 31% homology with the canonical AsCpf1 from Acidominococcus species at the amino acid level and has evolved further away from Cas9 compared to AsCpf1.14 MAD7 has been shown to have significant gene editing activity in the Escherichia coli bacterium and Saccharomyces cerevisiae yeast (unpublished data from Inscripta, Inc.), but data on its activity in other systems, including mammalian cells, remain scarce (https://www.inscripta.com/uploads/downloads/2-mad7-mammalian.pdf).14
Here, we report that we achieved high efficiencies of targeted gene disruption and knock-ins in both human tumor cell lines and rodent embryos. The knock-ins range from small restriction site (using DNA oligo as donor) to mid-sized Cre recombinase and fluorescent protein tags to a large (14 kb) multiple-protein expression cassette, demonstrating a large range of CRISPR-MAD7 functionality in different species. Our work demonstrates the practical applications of the MAD7 enzyme to mammalian genome engineering. Combined with a small gRNA and orthogonal PAM to SpCas9, we believe MAD7 offers a robust extension to the use of CRISPR-Cas systems in genome manipulation, from generation of model systems to potentially human therapy.
Methods
MAD7 expression plasmid, mRNA, and protein
MAD7 expression plasmid was obtained from Inscripta (Boulder, CO). For generation of MAD7 mRNA, the plasmid was first digested with XbaI restriction enzyme (New England Biolabs, Ipswich, MA) and then purified. Capped MAD7 mRNAs were generated with a MessageMAX™ T7 ARCA-Capped Message Transcription Kit (CELLSCRIPT, Madison, WI) using purified plasmid. The capped MAD7 mRNAs were then polyadenylated using the Poly(A) Polymerase Tailing Kit (Epicentre (An Ilumina Company), Madison, WI) and then purified according to the manufacturer's protocol. The integrity of the MAD7 mRNAs was verified by gel analysis and quantitated by Nanodrop and Qubit fluorometer. MAD7 protein containing a C-terminal NLS was manufactured by Feldan Therapeutics (Quebec, Canada).
gRNA design and synthesis
Expressed MAD7 and SpCas9 gRNAs were cloned into hU6-containing mammalian expression vectors with synthesized cloning DNA oligonucleotide sequences (Eurofins; Supplementary File S1). For MAD7, we used a 35-nucleotide direct repeat (GUCAAAAGACCUUUUUAA UUUCUACUCUUGUAGAU) 5′ of the 21- nucleotide targeting sequence, making the expressed gRNA a 56-mer. Synthetic MAD7 and SpCas9 gRNAs were generated by Horizon Discovery/Dharmacon (Lafayette, CO). MAD7 crRNAs were synthesized as 56-mers with the above 35-nucleotide direct repeat sequence (Supplementary File S1).
Cell line culturing and transfection
HCT116 (HD PAR-073) cells were maintained in HyClone™ RPMI-1640 medium (SH3096; GE Healthcare, Marlborough, MA), supplemented with 2 mM HyClone l-glutamine and 10% HyClone fetal bovine serum (FBS). HCT116 cells were seeded on 96-well plates at 10,000 cells per well 1 day prior to transfections. Cells were transfected with 100 ng nuclease plasmid and 100 ng gRNA plasmid using 0.8 μL/well of DharmaFECT™ kb transfection reagent (cat. # T-2006-01; Dharmacon), followed by 24 h of selection with 20 μg/mL blasticidin applied 24 h post transfection. For RNP transfections, cells were transfected with 25 nM MAD7 or Edit-R Cas9 protein nuclease (cat. # CAS11201; Dharmacon) and 100 nM crRNA (or 100 nM crRNA:tracrRNA for Cas9 nuclease) using 0.4 μL/well of DharmaFECT™ Duo transfection reagent (cat. # T-2010-03; Dharmacon). U2OS (cat. # HTB-96; ATCC, Manassas, VA) cells were maintained in standard growth medium per the manufacturer's recommendations. U2OS cells were seeded on a 96-well plate at 10,000 cells per well 1 day prior to transfection. Cells were transfected with 200 ng nuclease plasmid and 200 ng gRNA plasmid using 0.3 μL/well of DharmaFECT™ Duo transfection reagent (cat. # T-2010-03; Dharmacon). For HDR transfections, 200 ng repair plasmids were used with 200 ng nuclease plasmid and 200 ng gRNA plasmid.
Mouse Neuro-2A (cat. # CCL-131; ATCC) and rat C6 glioma (cat. # CCL-107; ATCC) cells were cultured and nucleofected as previously reported with CRISPR-Cas9 reagents.15 Briefly, cells were nucleofected with 2 μg each MAD7 mRNA and gRNA, cultured for 48 h, and then screened for MAD7 cutting activity. Green fluorescent protein (GFP) control plasmid was used regardless of cell line separately to determine nucleofection efficiency.
HDR repair template design and construction
An enhanced GFP (eGFP) plasmid repair template was designed with Edit-R HDR Donor Designer (Horizon Discovery), and polymerase chain reaction (PCR) primers were selected to give the following homology arm lengths: 5′ homology arm—605 bps, forward primer AAGAAGTTTGCTGTGAAGGCCA, reverse primer CATAGTGCCCGCCAGCTTT; 3′ homology arm—718 bps, forward primer GGGAAAAAACAAAACAAGAAGAAAGTGG, reverse primer AATTCTTGGGGAATGGG GAAGC.
The mouse Rosa26 CAG-SpCas9-2A-eYFP-2A-LbCpf1 and rat Calb2 CRE-tagging donor plasmids with corresponding homology arms were generated by traditional cloning. The mouse Rosa26 oligo donor repair template (Supplementary File S1) was synthesized by Integrated DNA Technologies (IDT; Coralville, IA).
Animal husbandry and micro-injection
Mouse and rat work in this study was performed at Horizon Discovery's Association for Assessment and Accreditation of Laboratory Animal Care–accredited facility (St. Louis, MO), which operated under approved animal protocols overseen by Horizon's Institutional Animal Care and Use Committee.
C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME), and Sprague Dawley (SD) rats purchased from Taconic Biosciences (Rensselaer, NY) were housed in standard cages and maintained on a 12 h light/dark cycle with ad libitum access to food and water. For rats, 3- to 4-week-old donors were injected with 20 IU of pregnant mare serum (PMS) followed by 50 IU of human chorionic gonadotropin (hCG) 48 h later just prior to mating. For mice, 5 IU of PMS and 5 IU of hCG were injected into 3- to 4-week-old mice 48 h apart prior to mating. In both cases, fertilized eggs were harvested 1 day later for micro-injections. MAD7 and Cas9 mRNAs, protein, and gRNAs were injected into the pronuclei of fertilized eggs. The range of final concentrations of MAD7 or Cas9 mRNA was 50–100 ng/μL, MAD7 or Cas9 protein 90–150 ng/μL, gRNAs 80–100 ng/μL, oligo-donor 50–200 ng/μL, and plasmid donors 0.5–3.5 ng/μL. Recipient female rats were injected with 40 μg luteinizing hormone-releasing hormone 96 h prior to mating to vasectomized males, while recipient female mice were determined to be in estrus by physical examination and were mated the same day. The presence of vaginal plugs the morning after mating was evidence that both mice and rats were in estrus and had therefore ovulated and released their unfertilized eggs. Micro-injected eggs were transferred to these pseudo-pregnant recipients.
DNA mismatch detection assay using T7EI and Cel-I
DNA mismatch detection assays were performed on human cell extracts using T7EI endonuclease (New England Biolabs).16 The Surveyor Mutation Detection Kit (IDT) with Cel-I enzyme were used on crude genomic DNA samples from mouse or rat transfected cells, micro-injected embryos, or tissue clips from live-born animals.15 Briefly, MAD7 target regions were PCR amplified (for PCR primer sequences, see Supplementary File S1) using the extracted DNA as template and primers flanking the cleavage site of each gene. The PCR amplification products were used in a DNA mismatch assay (T7EI or Cel-I; listed within the figure legends) to determine indels and in BamHI restriction digest assays for the mouse Rosa26 oligo-donor integration. The percent editing for each sample, when listed, was estimated using described calculations.1
Detection of eGFP integration by flow cytometry
U2OS cells were analyzed on an Accuri C6 Plus flow cytometer (BD Biosciences, San Jose, CA) 3 days post transfection using the manufacturer's protocols in order to determine the percentage of the population expressing eGFP. Briefly, after cell dissociation using trypsin, cells were washed with phosphate-buffered saline (PBS) and then re-suspended in a cell sorting buffer consisting of Ca/Mg++ free PBS supplemented with 1 mM EDTA, 25 mM HEPES pH 7.0, and 1% FBS.
Mouse and rat junction PCR genotyping and sequencing
Junction PCRs for the mouse and rat targeted integrations were performed using standard amplification procedures (for PCR primer sequences, see Supplementary File S1), with the only exception being for the mouse Rosa26 large integration, where the upstream amplification product is re-amplified with nested PCR oligonucleotide primers to visualize positive junction PCR reactions.
Sanger sequencing, when needed, was performed by Elim Biopharma (Hayward, CA). Next-generation sequencing (NGS) of PCR amplified products was performed by the Genome Engineering and iPSC Center (GEiC) at Washington University (St. Louis, MO). Data found within Supplementary File S2 give details about the various indels found in each sample based on length compared to the wild-type sequence using NGS.
Off-target analysis for mouse and rat gRNAs
gRNAs were run against mouse and rat genomes using Benchling software.17 The top 10 off-target sites plus gene encoding regions were selected according to the score (Supplementary File S3). Briefly, gRNAs and PAM sequences were entered into the program and analyzed for on- and off-target scores. Flanking oligonucleotide primers (for PCR primer sequences, see Supplementary File S1) were designed to PCR amplify 300–600 bp fragments (Supplementary File S4) of the predicted off-target sites. These amplified fragments were analyzed by DNA mismatch assays using Cel-I assays, and the results were screened for the predicted cutting patterns. A wild type, an individual indel sample or pool of all the indel samples within the mouse Rosa26 and rat Calb2 target sites, was used within the assay.
Results
MAD7 editing in mammalian cancer cells
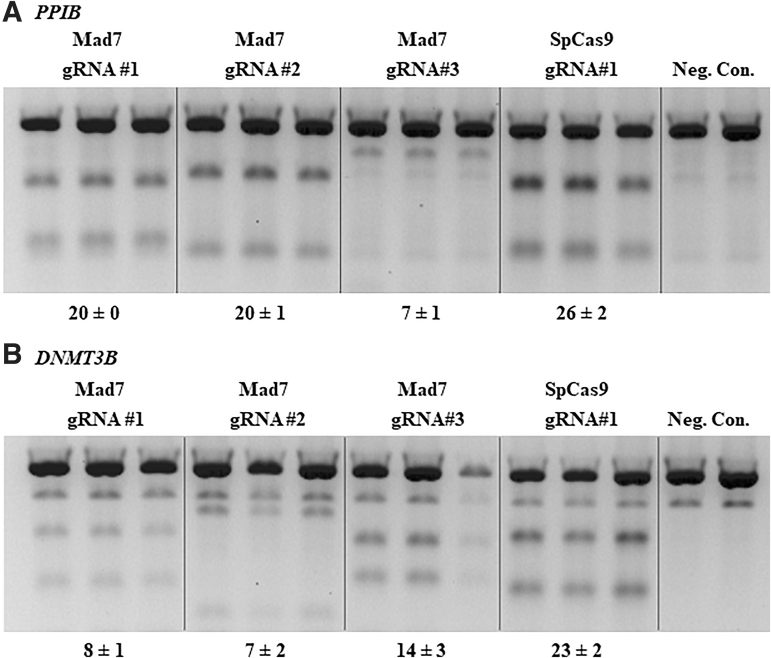
We first evaluated the activity of expressed MAD7 nuclease in NHEJ-mediated genome editing in cultured HCT116 mammalian cells using expressed gRNAs targeting human PPIB and DNMT3B genes (Fig. 1A and B) and compared it to Cas9 controls targeting the same amplicons but at non-overlapping target sites. The editing efficiency was estimated using a DNA mismatch assay. We found that three different expressed gRNAs targeting PPIB and DNMT3B and MAD7 created 7–20% and 7–14% indels, while the Cas9 editing efficiency with gene specific control gRNAs was 26% and 23%, respectively. We then sought to expand our evaluation to other gene targets, and test whether gRNAs with identical target sites or gRNAs with overlapping seeds will produce comparable results between MAD7 and Cas9. We used an established framework and selected a number of gRNAs with fully overlapping target sequences within the genes NF1, STAG2, and ALK, or with overlapping seeds for genes CACNA1D and PPP1R12C.18 The gRNAs we chose were reported to have various functional activities when tested with a related Cas12a enzyme.18 We found that for some gene targets (NF1 and STAG2) MAD7 was 5% less active when compared to SpCas9, while other gene targets (ALK, CACNA1D, and PPP1R12C) displayed much lower gene editing activity (Table 1 and Supplementary Fig. S1), which suggests that MAD7 and SpCas9 most likely have different gRNA design parameters that need to be better understood.
FIG. 1.
A DNA mismatch detection assay using T7EI enzyme for non-overlapping MAD7 and Cas9 targets using expressed enzyme and guide RNAs (gRNAs) in HCT116 cells. MAD7 or Cas9 enzyme and corresponding gRNAs to gene target PPIB (A) or DNMT3B (B) listed above each gel. Polymerase chain reaction (PCR) amplicons for each gene editing target site were produced from cell lysate generated from each individual transfection. PCR amplicons from untransfected cells for each gRNA target site were used as negative controls (Neg. Con.) in the T7EI assay. Percent indel formation is shown at the bottom of the gels plus or minus standard deviation.
Table 1.
Comparison of expressed MAD7 and Cas9 activity in human HCT116 cells
| Gene target | Guide RNA | MAD7 indels (%) | Cas9 indels (%) |
|---|---|---|---|
| PPIBa | 1 | 20 ± 0 | 26 ± 2 |
| 2 | 20 ± 1 | ||
| 3 | 7 ± 1 | ||
| DNMT3Ba | 1 | 8 ± 1 | 23 ± 2 |
| 2 | 7 ± 2 | ||
| 3 | 14 ± 3 | ||
| NF1b | 1 | 15 ± 2 | 20 ± 2 |
| STAG2b | 1 | 23 ± 2 | 28 ± 2 |
| ALK2b | 1 | 4 ± 0 | 14 ± 1 |
| CACNA1Dc | 1 | 14 ± 2 | 29 ± 1 |
| PPP1R12Cc | 1 | 6 ± 1 | 15 ± 1 |
Non-overlapping targets.
Fully overlapping targets.
Overlapping seed.
We further tested the activity of MAD7 using transient ribonucleoprotein (RNP) complexes with synthetic gRNAs designed to target a few genes mentioned above. For PPIB and DNMT3B, we chose the two most active MAD7 gRNA sequences from the expressed experiments (PPIB gRNA#1 and gRNA#2; DNMT3B gRNA#1 and gRNA#3) and added an additional one (gRNA #4) that was targeting a sequence in close proximity to the Cas9 gRNA binding site (Supplementary Fig. S2A and B). We show that MAD7 RNPs can have activity within 5% to Cas9 RNP, therefore enabling DNA-free gene editing that can be beneficial in biological systems sensitive to DNA or that require a short editing time frame to lower off-targeting.19,20
MAD7-assisted eGFP tagging using plasmid repair templates
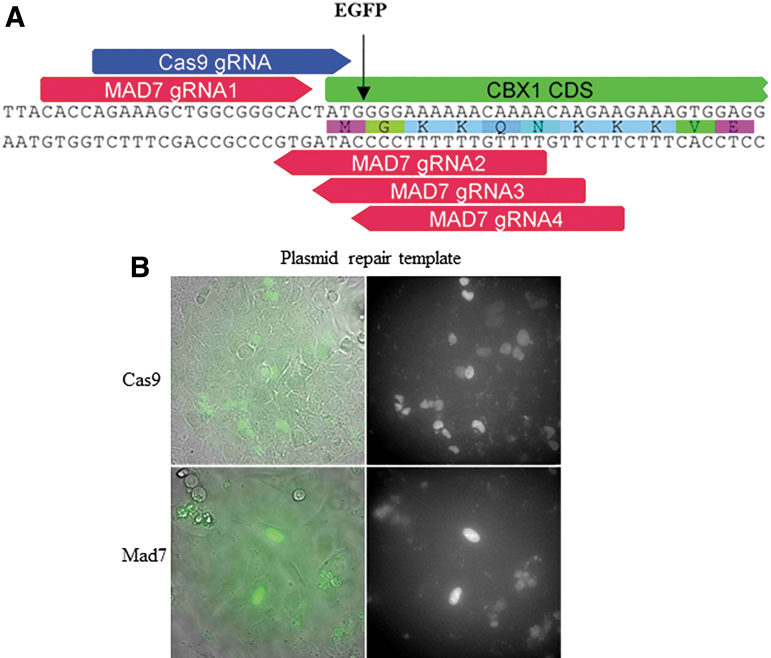
Gene editing enzymes have been used to make a variety of precise modifications in cell lines and other model systems via the HDR pathway. Similarly, we sought to determine whether we could tag eGFP to the N-terminus of the human CBX1 gene with MAD7 cleavage-induced HDR. CBX1 is a highly conserved non-histone nuclear localized protein, and tagging has previously been demonstrated with CRISPR-Cas9.21 We identified four different MAD7 gRNAs and a single Cas9 gRNA target site near the start codon of CBX1 (Fig. 2A). We tested the cleavage potential of all four MAD7 gRNAs and the one for Cas9 via the DNA mismatch detection assay (data not shown), and we found that MAD7 gRNA#1 (2% indel formation) and Cas9 (5% indel formation) had detectable cleavage activity. We subsequently transfected each nuclease and its cognate, active gRNAs each in a separate expression plasmid, into U2OS cells with an eGFP repair template in the form of a circularized plasmid. The repair template was designed to direct eGFP insertion at the N-terminus of CBX1, just after the start codon (Fig. 2A). Both MAD7 and Cas9 transfected U2OS cells were able to produce nuclear localized eGFP cells (Fig. 2B). Consistent with the higher DNA mismatch activity of Cas9/gRNA, we observed a larger eGFP expressing cell population with Cas9 compared to MAD7 gRNA #1 (3.8% to 1%, respectively; cell sort data not shown).
FIG. 2.
Enhanced green fluorescent protein (eGFP) N-terminal tagging of the human CBX1 gene. (A) Four MAD7 gRNAs (red) and a Cas9 gRNA (blue) each designed to direct cleavage of the genomic gene target around the start codon of CBX1 denoted by the black arrow. (B) U2OS cells co-transfected with plasmids expressing either MAD7 or Cas9 nuclease and the corresponding gRNA, with a repair plasmid to direct homology-directed repair–mediated insertion of the eGFP gene. eGFP expression was detected via epifluorescence microscopy.
To confirm the HDR-related tagging further, we performed junction PCR across the CBX1 eGFP integration site on genomic DNA isolated from MAD7 transfected cells, followed by blunt end cloning and Sanger sequencing of individual E. coli colonies. We were able to identify clones that displayed a precise integration of eGFP into the intended integration site with no additional genomic modifications either upstream or downstream of the eGFP (Supplementary Fig. S3), validating the presence of precise MAD7-induced HDRs. Our data show that MAD7 can be used for precise insertion of fluorescent reporters.
Generation of indels, small and large fragment knock-ins at the mouse Rosa26 locus
The mouse Rosa26 locus has been targeted successfully for generating indel mutations and gene integrations with ZFNs, TALENs, and CRISPR-Cas9.22–25 To assess the gene-targeting potential of the MAD7 enzyme in mice, we designed four gRNAs within the first intron of the mouse Rosa26 locus (Supplementary Fig. S4A). These gRNAs along with MAD7 were transfected into mouse Neuro-2A cells and then screened by DNA mismatch detection assays. All four gRNAs induced mutations at the Rosa26 locus (Supplementary Fig. S4B). To test its activity in vivo further, we selected Rosa26 gRNA #1 and micro-injected it with MAD7 mRNA into the pronucleus of fertilized C57BL/6J mouse embryos, transferred the embryos to pseudo-pregnant females, and collected embryos that had developed to at least embryonic day 14 (E14). Sixty percent of the E14 embryos had mutations at the Rosa26 locus as determined by NGS analysis of DNA mismatch PCR products (25/42 embryos, summarized in Table 2; Supplementary File S2). One out of the 25 embryos were homozygous for the same deletion (#20; Supplementary File S2), while several other embryos (15 in total) were mosaic, which is inherent in transgenic mouse production (NGS of PCR products; Supplementary File S2).26
Table 2.
MAD7 gene editing efficiency in mouse and rat embryos
| Targeting gRNA | MAD7 form | Donor | No. of injected zygotes | No. of surviving and transferred | No. of ∼E14 embryos (%)a | No. of newborns | No. of mutations (%)b | No. of integrations (%)b |
|---|---|---|---|---|---|---|---|---|
| Mouse Rosa26 #1 | mRNA | — | 90 | ND | 42 | — | 25 (60) | - |
| Mouse Rosa26 #1 | mRNA | ssODN | 55 | 50 | 10 (20) | — | 3 (30) | 2 (20) |
| Mouse Rosa26 #1 | Protein | ssODN | 80 | 60 | 5 (8) | — | 3 (60) | 1 (20) |
| Mouse Rosa26 #1 | mRNA | ssODN | 100 | 48 | — | 3 | 1 (33) | 1 (33) |
| Mouse Rosa26 #2 | mRNA + Protein | Plasmid | 528 | 296 | — | 48 | 39 (81) | 1 (2) |
| Rat Calb2 #1 | mRNA + Protein | Plasmid | 351 | 273 | — | 49 | 10 (20) | 12 (24) |
% calculated as number of E14 embryos divided by the number of surviving and transferred embryos.
% calculated as number of mutations or integrations divided by the number of E14 embryos or newborns.
E14, embryonic day 14.
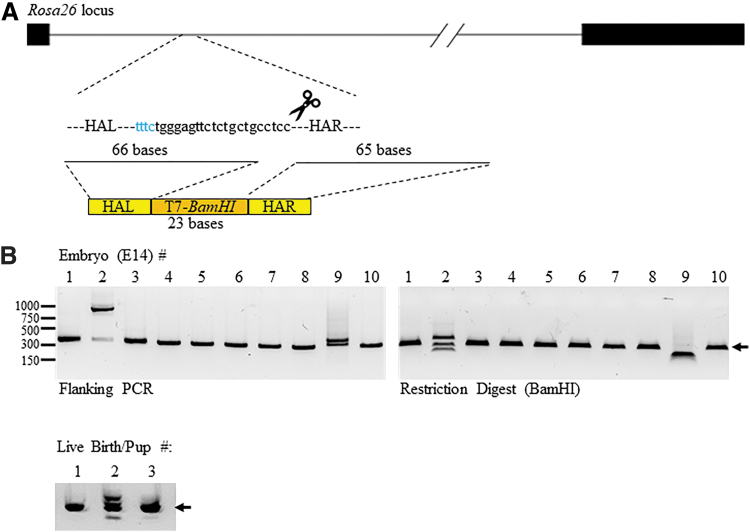
To test whether we could use MAD7 to target small DNA fragments into the genome of mice, we designed an oligo donor across the target site for gRNA #1 above to integrate a 23 bp DNA sequence containing a T7-promoter and BamHI restriction site and minus 2 bp into the Rosa26 locus (Fig. 3A and Supplementary Fig. S4A). We again micro-injected mouse embryos with MAD7 mRNA and Rosa26 gRNA #1 as well as the oligo donor and collected E14 embryos. In addition, we also let more embryos go to full term and screened the live-born pups for mutations and integration of the oligo donor. Analysis of the PCR amplicons flanking the target site (Fig. 3B, left panel) and BamHI restriction digests (Fig. 3B, right panel) via agarose gels found that 20% of the E14s had oligo donor integration at the expected Rosa26 locus (#2 and #9) as evident by multiple or larger bands within the PCR amplicon compared to the expected and smaller or multiple bands after BamHI restriction digest. NGS analysis of the PCR amplicons revealed that #9 embryo had the correct integration (+21; 60.5% of the reads) in addition to a 64 bp sequence (+64; 32.2% of the reads) integrated downstream of the PAM site and upstream of the inserted T7-BamHI sequence (Supplementary File S2 and Supplementary Fig. S5A). The larger amplification product observed for embryo #2 was not detected with NGS, since the size was out of the range, exposing a potential limitation of NGS-based gene targeting analysis methods. Therefore, we performed direct Sanger sequencing of the amplification product. The resulting sequences revealed that the E14 embryo #2 had a 13 bp insertion, a spliced mRNA sequence (529 bp) from the mouse Socs7 gene and the oligo donor +1 base (Supplementary Fig. S5B). The extra base (A) in this case is more than likely from the oligo donor synthesis. We repeated the BamHI restriction digests of the PCR amplicons and resolved the reactions via acrylamide gels to visualize the predicted banding pattern of the integration better (202 and 121 bp). For E14 embryo #9, we observed the predicted bands and an additional band of 165 bp corresponding to the 64 bp integration detected via NGS (Supplementary Fig. S5C). We also observed the expected banding patterns for the larger integration within E14 embryo #2 (376, 284, and 205 bp), since the Socs7 cDNA contained an additional BamHI restriction site (Supplementary Fig. S5C). In addition to the two integrations, it was determined by DNA mismatch assay and NGS analysis of the other E14 embryo PCR amplicons that the 50% total had cutting activity at the mouse Rosa26 locus (two integrations and three mutations/10 E14 embryos, summarized in Table 2, Supplementary Fig. S5D, and Supplementary File S2). Analysis of the three newborn pups showed that #2 had the correct integration and indel mutations (summarized in Table 2, Fig. 3B, lower panel, and NGS analysis, Supplementary File S2).
FIG. 3.
Mouse Rosa26 target with oligo donor integrations. (A) Schematic representation of the Rosa26 locus and the targeting gRNA. Dark bars represent exons and the solid line intron 1. The blue text represents the protospacer adjacent motif site and the remaining targeting sequence. The size of the homology arms (HA; bases) within the oligo donor is listed and the lines represent where it starts within the target region. The bottom schematic represents the HA (R, right; L, left) in yellow and the T7-BamHI integration sequence in orange. (B) PCR amplification products that flank the Rosa26 locus from purified DNA of isolated oligo-donor micro-injected embryonic day 14 (E14) embryos (left panel), BamHI restriction digests of the amplification products (right panel), and PCR amplification products for live-born pups (bottom panel). The dark arrow on the right of each gel represents the expected size (302 bp) of the PCR amplification product. The numbers to the left represent the location of the DNA size markers in base pairs.
We further tested MAD7 activity within mouse embryos using transient RNP complexes. Mouse embryos were micro-injected with MAD7 protein and Rosa26 gRNA #1 as well as the oligo donor above, and E14 embryos were collected. NGS analysis of the PCR amplicons flanking the target site revealed that we had correct integration within 20% (1/5) of the E14s and 80% total had cutting activity at the mouse Rosa26 locus (one integration and three mutations/five E14 embryos; summarized in Table 2, NGS analysis, Supplementary File S2). The activities of MAD7 being delivered as RNP or mRNA are comparable. However, the percentage of E14s collected out of the total number of embryos transferred was considerable lower for RNP when compared to mRNA delivery (8% and 20%, respectively; summarized in Table 2). Further studies will be necessary to determine if the observed lower number of E14s is due to protein toxicity within the mouse embryos.
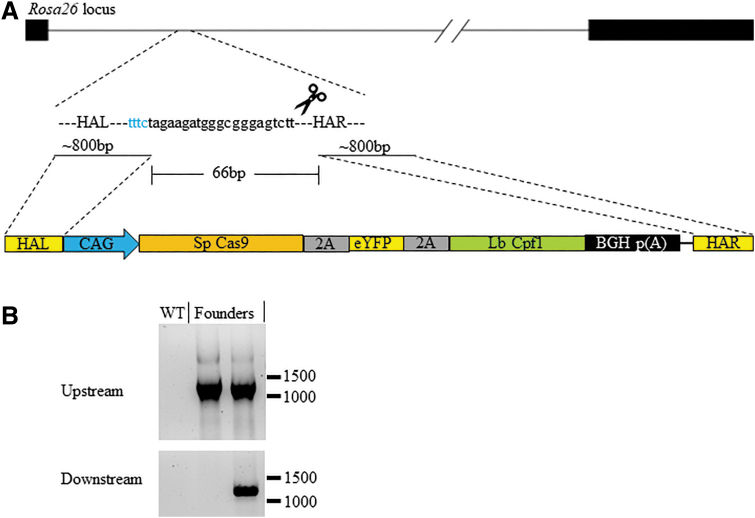
We have previously succeeded in integrating a large 14 kb (14,027 bp) construct with CRISPR-Cas9 into the mouse Rosa26 locus and obtained live pups (15%, three integrations/20 newborns; Supplementary Fig. S6A and B). The construct contains an expression cassette of SpCas9-2A-eYFP-2A-LbCpf1 (Fig. 4A) driven by the CAG promoter and 800 bp of flanking homology arms. This CRISPR-Cas9 target site overlaps with the MAD7 Rosa26 gRNA #2 that was active in Neuro-2A cells (Supplemental Fig. S6A). Therefore, we tested whether we could target the same large DNA fragment into the mouse Rosa26 locus with MAD7 (Fig. 4A). We co-injected this gene-targeting vector along with MAD7 mRNA and protein to increase efficiency and Rosa26 gRNA#2 into embryos, transferred it, and then screened live-born pups.27 Two percent of the pups were positive for integration (one integration/48 newborns; Table 2), as evident by a positive PCR amplification at the upstream and downstream junctions of the Rosa26 locus (Fig. 4B). One other pup was positive for the upstream junction PCR but not for the downstream junction. In addition, and including the targeted integration results, 83% (40/48) of the live births had indels at the Rosa26 locus (data not shown; summarized in Table 2). Since the construct includes both SpCas9 and LbCpf1 that are under the control of constitutive CAG promoter and the Rosa26 gRNA#2 co-injected with MAD7 could also be utilized by LbCpf1, it is possible that the expressed Cas9 and LbCpf1 from the construct itself may contribute to its own integration within the embryo.11 Therefore, we set out to determine if the SpCas9 and LbCpf1 genes were active by nucleofecting mouse Neuro-2A cells and micro-injecting B6 mouse embryos with the construct and Cas9 or MAD7 gRNAs. By DNA mismatch assays, we observed cutting activity at the mouse Rosa26 locus with the inclusion of the Cas9 gRNA but not with the MAD7 gRNA (Neuro-2A, Supplementary Fig. S7A, MAD7 RNP used as a control; E14 B6 embryos, Supplemental Fig. 7B and C, MAD7 and Cas9, respectively). In addition, no eYFP was visible within the Neuro-2A cells or in the knock-in mouse (data not shown). These data suggest that the only active gene within the construct is the SpCas9, and due to unknown reasons, neither eYFP nor LbCpf1 is active. Therefore, although it is possible that sustained expression of SpCas9 from the targeting construct may have helped the integration of the construct when Cas9/gRNA is used, it is unlikely that LbCpf1 has served a similar role when MAD7 is used. This may explain the differences in integration efficiency between MAD7 (2%) and Cas9 (15%).
FIG. 4.
Mouse Rosa26 target with large plasmid donor integrations. (A) Schematic representation of the Rosa26 locus, targeting gRNA and the large targeting donor plasmid. The size of the HA within the plasmid donor and the lines to where they start within the target region. The middle 66 bp listed represents the distance between where the HA start within the Rosa26 locus. The bottom schematic represents the HA (R, right; L, left) in yellow, CAG promoter in blue, poly(A) signal in black, and the remaining Cas9-eYFP-Cpf1 gene fusion. (B) Upstream (top panel) and downstream (bottom panel) junction PCR amplification products from purified DNA from wild-type (WT) and founder mice. The numbers on the right in each panel represent DNA size markers in base pairs.
To determine the specificity of MAD7 in gene targeting in vivo, we examined potential off-targets for the Rosa26 gRNAs. The top 10 possible off-target sites plus any gene encoding regions within the mouse genome with similarity to the target sequences in Rosa26 (Supplementary Files S3 and S4) were selected and analyzed by DNA mismatch assay of wild type, an individual indel sample, and the pool of all the positive indel samples. We found no notable off-target cutting at the 13 potential sites for gRNA #1 with E14 gDNA samples (top 10 plus three gene coding regions; Supplementary Fig. S8A) and the top 10 for gRNA #2 (Supplementary Fig. S8B).
Cre targeting downstream of the rat Calb2 gene
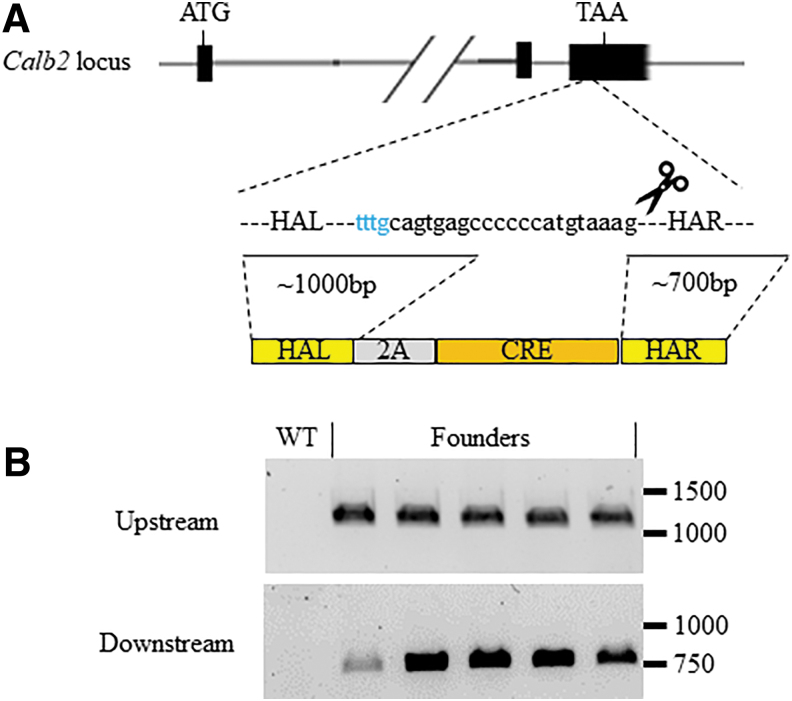
We previously generated Cre knock-in rat lines for use in conditional knockout and tissue specific expression studies via ZFNs and CRISPR-Cas9.28,29 While we had a good success rate with CRISPR/Cas9-based methods to knock-in Cre recombinase into the immediate downstream of various neuron-specific genes, we had trouble inserting the T2A-Cre cassette downstream of the Calb2 gene for unknown reasons. Encouraged by the activity of MAD7 in mouse embryos thus far, we were interested to know whether we could obtain Calb2-T2A-Cre knock-in rats with MAD7. We designed two gRNAs for MAD7 and validated that both were active in rat C6 glioma cells (Supplementary Fig. S9). We then micro-injected embryos from SD rats with a mixture of gRNA#2, MAD7 mRNA and protein, and the same gene targeting vector used with Cas9 previously (1,119 bp with flanking homology arms; Fig. 5A), transferred the embryos to pseudo-pregnant female rats, and then screened live-born pups. We performed upstream and downstream junction PCR and found that approximately 25% of the newborn pups were positive for the correct targeting of the Cre recombinase (12 integrations/49 newborns; Fig. 5B and Table 2). In addition, another 10 pups bear small indels in the Calb2 locus (data not shown). Therefore, 45% of the live-born pups have a modified Calb2 locus (22/49; summarized in Table 2).
FIG. 5.
Cre targeting of the Rat Calb2 gene. (A) Schematic representation of the Calb2 locus, targeting gRNA and the Cre-targeting donor plasmid. The locations of start (ATG) and stop (TAA) codons are listed above and the dotted lines to where the targeting vector will integrate upstream of the stop codon. The Calb2 locus is shortened for simplicity and to include the relevant exons for Cre-targeting. The size of the HA within the plasmid donor and the lines to where they start within the target region. The bottom schematic represents the HA (R, right; L, left) in yellow and the Cre gene in orange. (B) Upstream (top panel) and downstream (bottom panel) junction PCR amplification products from purified DNA from WT and founder rats. The numbers on the right in each panel represent DNA size markers in base pairs.
As with the mouse target sites, we screened for the top 10 potential off-targets within the rat genome with similarity to the target sequences in Calb2 (Supplementary Files S3 and S4 and Supplementary Fig. S8C). However, we observed two PCR amplicons for the individual and pool indel samples in comparison to the wild-type sample for off-target #5. No cutting activity was observed, but further analysis of the two amplicons is currently under investigation to determine if there is MAD7-mediated modification at this intergenic region of the genome.
Discussion
CRISPR-Cas9 is a gold-standard of RNA-guided endonuclease capable of inducing a DSB in a wide variety of model systems and cell lines. While creating specific genomic edits via CRISPR-Cas9 is starting to become a routine practice, the limitation of the Cas9 NGG PAM can be problematic in genomic regions that are AT rich. Cas12a enzymes are a class of RNA-guided endonucleases that are able to target genomic sites using an AT-rich TTTN and CTTN PAM. In this study, we show that the Cas12a enzyme, MAD7, is capable of generating indels and knock-ins in human tumor cell lines and mouse and rat embryos.
When co-expressing gRNAs with the MAD7 or Cas9 enzyme in HCT116 cells, indel formation was higher with Cas9 at the majority of the targets tested, regardless of gRNA location (non- or fully overlapping targets, overlapping seed) when compared to MAD7 (Fig. 1 and Supplementary Fig. S1). However, efficiencies can be different for different sequences, even for the same enzyme, and while there are multiple developed algorithms for predicting CRISPR-Cas9 gRNAs with high functionalities, Cas12a algorithms have not been developed. Nonetheless, the MAD7 is active within an expressed or transient system but will need further optimization to identify parameters for the identification of MAD7 gRNAs with higher efficiency.
CRISPR-Cas9 is an established system to generate targeted, blunt DNA cuts using a G-rich NGG PAM. This G-rich PAM requirement can be problematic in AT-rich regions. While Cas9's PAM has been mutated to recognize alternate bases, the Cas12a enzymes have a T-rich PAM requirement, and their ability to make a staggered cut has intrigued the field in terms of HDR mechanism or targeted insertions.30 Here, we wanted to compare Cas9 and MAD7 directly and their ability to induce HDR at the same gene target, CBX1. We identified active gRNAs for each nuclease and supplied repair template, each designed to knock-in an eGFP tag at the N-terminus of CBX1. While Cas9 and MAD7 were both able to induce HDR in the same CBX1 target gene, the eGFP-positive cell population initiated by Cas9 cleavage exceeded that of MAD7. This observation is similar to a previous study comparing SpCas9 and Cas12a/Cpf1 enzymes.31
In previous reports, knock-out mice and rats had been generated with Cas12a/Cpf1 enzymes.32–34 In addition, previous studies restored dystrophin in muscular dystrophy mice with oligo-donors using Cas12a/Cpf1.35 Here, we report comparable indel frequencies and oligo donor integration in mice with Cas12a/MAD7 as reported with Cas12a/Cpf1. Of course, targeted cutting and integration efficiencies can vary by genomic loci, but the data suggest that the MAD7 enzyme is adaptable to mouse genome editing. Larger (kb) integrations have not been reported with Cas12a enzymes in mice, and here we were successful in the integration of a 14 kb construct using the MAD7 enzyme at the mouse Rosa26 site, which, to our knowledge, is the largest fragment that has been knocked into the mouse genome with a CRISPR-based system, including Cas9. Our construct does contain SpCas9, which we show does utilize the provided Cas9 gRNA for targeted cutting. However, we have not detected any expression of eYFP or LbCpf1 cutting activity in cells nor the successive generations from the founder animals for unknown reasons (data not shown). Therefore, the MAD7 14 kb founder animal we generated was ascribed to the activity of the MAD7 enzyme and its gRNA. Although the sustained expression of SpCas9 from the targeting plasmid construct may explain why the integration efficiencies were higher when compared to MAD7, how much this had contributed may largely depend on the available gRNA and would require further testing. Clearly, further optimization to the MAD7 enzyme and/or modifications to the gRNA is necessary for increased efficiencies of >10 kB integrations. Regardless, our data suggest that there is another CRISPR editing enzyme capable of generating larger knock-ins within the mouse genome.
We previously had difficulty in targeting the 2A-Cre expression cassette downstream of the Calb2 gene with CRISPR/Cas9 in the rats. In great contrast, we obtained an approximately 25% integration rate with MAD7 in live-born pups (12/49). This HDR efficiency is comparable to other similar-sized (∼1 kB) targeted integrations performed in the rat with Cas9 (Liu et al., unpublished). Whether this high HDR efficiency is locus specific or species specific remains to be tested. Nevertheless, to our knowledge, this is the first knock-in rat that was created with a Cas12a enzyme. It is also worth noting that we observed a relatively high frequency of indels in the rest of the pups (10/49; ∼20%), which was comparable to previous observations with CRISPR/Cpf1 while generating Apoe- and Ldlr-deficient rats (combined modifications to the Calb2 locus 45%, compared to 39, 55%, respectively).34 This adds yet another species as reported here, in addition to zebrafish, of being adaptable to gene editing (knock-out and knock-ins) with MAD7.14 It also suggests there should be no limitations to applying CRISPR-MAD7 within other organisms.
In summary, we successfully generated knockout and knock-in human cell lines, mice, and rats with MAD7. As mentioned before, Cas12a enzymes such as MAD7 have many unique features when compared to Cas9, and they help to open up more regions of the genome to further targeting. Our work therefore adds yet another tool to the gene editing toolbox and proves the utility of the MAD7 not only for knockout alleles, but also for larger targeted integrations.
Supplementary Material
Acknowledgments
We would like to thank Inscripta, Inc. (Boulder, CO) for their gift of the MAD7 plasmid clone. We also thank Evguenia Kouranova and Xiaoxia Cui at the GEiC at Washington University (St. Louis, MO) for their assistance with NGS. Lastly, we thank our Horizon Discovery colleagues, Hidevaldo Machado, Michael D'Angelo, Philippe Collin, Annaleen Vermeulen, Joseph Abbott, Ryan Donnelly, and Andrew Brown, for their helpful discussion and insight.
Author Disclosure Statement
All of the authors are or were full-time employees of ENVIGO Research Model Services or Horizon Discovery.
Funding Information
Funding for the project was provided by Horizon Discovery Group.
Supplementary Material
References
- 1. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823. DOI: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337:816–821. DOI: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jinek M, East A, Cheng A, et al. RNA-programmed genome editing in human cells. Elife 2013;2:e00471 DOI: 10.7554/eLife.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823–826. DOI: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaj T, Gersbach CA, Barbas CF III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013;31:397–405. DOI: 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ratz M, Testa I, Hell SW, et al. CRISPR/Cas9-mediated endogenous protein tagging for RESOLFT super-resolution microscopy of living human cells. Sci Rep 2015;5:9592 DOI: 10.1038/srep09592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013;154:1370–1379. DOI: 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 2019;18:67–83. DOI: 10.1038/s41579-019-0299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Makarova KS, Wolf YI, Koonin EV. Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J 2018;1:325–336. DOI: 10.1089/crispr.2018.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klompe SE, Sternberg SH. Harnessing “a billion years of experimentation”: the ongoing exploration and exploitation of CRISPR-Cas immune systems. CRISPR J 2018;1:141–158. DOI: 10.1089/crispr.2018.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015;163:759–771. DOI: 10.1016/j.cell.2015.09.038.Cpf1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swarts DC, Jinek M.. Cas9 versus Cas12a/Cpf1: structure–function comparisons and implications for genome editing. Wiley Interdiscip Rev RNA 2018:e1481 DOI: 10.1002/wrna.1481 [DOI] [PubMed] [Google Scholar]
- 13. Zetsche B, Heidenreich M, Mohanraju P, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol 2017;35:31–34. DOI: 10.1038/nbt.3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wierson WA, Simone BW, WareJoncas Z, et al. Expanding the CRISPR toolbox with ErCas12a in zebrafish and human cells. CRISPR J 2019;2:417–433. DOI: 10.1089/crispr.2019.0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kouranova E, Forbes K, Zhao G, et al. CRISPRs for optimal targeting: delivery of CRISPR components as DNA, RNA, and protein into cultured cells and single-cell embryos. Hum Gene Ther 2016;27:464–476. DOI: 10.1089/hum.2016.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson EM, McClelland S, Maksimova E, et al. Lactobacillus gasseri CRISPR-Cas9 characterization in vitro reveals a flexible mode of protospacer-adjacent motif recognition. PLoS One 2018;13:1–14. DOI: 10.1371/journal.pone.0192181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benchling-Bilogy Software. Available online at https://benchling.com (last accessed 2019)
- 18. Wang Y, Liu KI, Sutrisnoh NAB, et al. Systematic evaluation of CRISPR-Cas systems reveals design principles for genome editing in human cells. Genome Biol 2018;19:1–16. DOI: 10.1186/s13059-018-1445-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liang X, Potter J, Kumar S, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 2015;208:44–53. DOI: 10.1016/j.jbiotec.2015.04.024 [DOI] [PubMed] [Google Scholar]
- 20. Hultquist JF, Schumann K, Woo JM, et al. A Cas9 ribonucleoprotein platform for functional genetic studies of HIV–host interactions in primary human T cells. Cell Rep 2016;17:1438–1452. DOI: 10.1016/j.celrep.2016.09.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamiyama D, Sekine S, Barsi-Rhyne B, et al. Versatile protein tagging in cells with split fluorescent protein. Nat Commun 2016;7:11046 DOI: 10.1038/ncomms11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujii W, Kawasaki K, Sugiura K, et al. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res 2013;41 DOI: 10.1093/nar/gkt772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kasparek P, Krausova M, Haneckova R, et al. Efficient gene targeting of the Rosa26 locus in mouse zygotes using TALE nucleases. FEBS Lett 2014;588:3982–3988. DOI: 10.1016/j.febslet.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 24. Meyer M, de Angelis MH, Wurst W, et al. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci U S A 2010;107:15022–15026. DOI: 10.1073/pnas.1009424107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perez-pinera P, Ousterout DG, Brown MT, et al. Gene targeting to the ROSA26 locus directed by engineered zinc finger nucleases. Nucleic Acids Res 2012;40:3741–3752. DOI: 10.1093/nar/gkr1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yen S, Zhang M, Min J, et al. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev Biol 2014;393:3–9. DOI: 10.1016/j.ydbio.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chu VT, Weber T, Graf R, et al. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol 2016;16:4 DOI: 10.1186/s12896-016-0234-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brown AJ, Fisher DA, Kouranova E, et al. Whole-rat conditional gene knockout via genome editing. Nat Methods 2013;10:638–640. DOI: 10.1038/nmeth.2516 [DOI] [PubMed] [Google Scholar]
- 29. Liu Z, Brown A, Fisher D, et al. Tissue specific expression of Cre in rat tyrosine hydroxylase and dopamine active transporter-positive neurons. PLoS One 2016;11:1–19. DOI: 10.1371/journal.pone.0149379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kleinstiver BP, Sousa AA, Walton RT, et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol 2019;37:276–282. DOI: 10.1038/s41587-018-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tóth E, Czene BC, Kulcsár PI, et al. Mb- and FnCpf1 nucleases are active in mammalian cells: activities and PAM preferences of four wild-type Cpf1 nucleases and of their altered PAM specificity variants. Nucleic Acids Res 2018;46:10272–10285. DOI: 10.1093/nar/gky815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim Y, Cheong S-A, Lee JG, et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol 2016; 34: 808–810. DOI: 10.1038/nbt.3614 [DOI] [PubMed] [Google Scholar]
- 33. Watkins-Chow DE, Varshney GK, Garrett LJ, et al. Highly efficient Cpf1-mediated gene targeting in mice following high concentration pronuclear injection. G3 (Bethesda) 2017;7:719–722. DOI: 10.1534/g3.116.038091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee JG, Ha CH, Yoon B, et al. Knockout rat models mimicking human atherosclerosis created by Cpf1-mediated gene targeting. Sci Rep 2019;9:1–9. DOI: 10.1038/s41598-019-38732-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y, Baskin KK, Bassel-Duby R, et al. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci Adv 2017;3:e1602814 DOI: 10.1126/sciadv.1602814 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.