Abstract
This paper reviews how statistical tests of neutrality have been used to address questions in molecular ecology are reviewed. The work consists of four major parts: a brief review of the current status of the neutral theory; a review of several particularly interesting examples of how statistical tests of neutrality have led to insight into ecological problems; a brief discussion of the pitfalls of assuming a strictly neutral model if it is false; and a discussion of some of the opportunities and problems that molecular ecologists face when using neutrality tests to study natural selection.
Keywords: neutral theory, neutrality test, selection
Introduction
The fields of evolutionary biology and population genetics have been dominated by molecular studies for the last quarter century (Lewontin 1974; 1991). Over much of that time, there has been a persistent debate about whether natural selection or random drift is the dominant force in molecular evolution (Kimura 1983; Gillespie 1991). In recent years, considerable progress has been made in the ability to detect natural selection from patterns of DNA sequence variation, and the ‘selectionist/neutralist debate’ has matured into an effort to estimate the distribution of selective effects on genetic variation (Kreitman 1996; Hey 1999). The strict neutral theory, on the other hand, has become the standard null hypothesis used to approach the study of molecular evolution, even if it is often rejected.
In this work, how molecular inferences about natural selection have been and can be used to address questions in ecology and conservation biology are reviewed. The fields of conservation genetics and molecular ecology make heavy use of molecular techniques, but generally do so under the assumption that essentially all observed variation is selectively neutral. For example, of the papers published in Molecular Ecology over the last 2 years, only a handful made use of statistical tests of neutrality. The primary goal of this review is to convince molecular ecologists that molecular population genetics can be a fruitful way to study natural selection and adaptation.
There have been several recent reviews of the statistical techniques used to test for selection on DNA sequences (Kreitman & Akashi 1995; Hughes 1999; Kreitman 2000). Therefore, these techniques are reviewed in sufficient detail only to make this paper reasonably self‐contained (Information box), and interested readers are referred to the reviews cited above and the primary literature for more detailed treatments. Similarly, there have been numerous reviews of selection on protein electrophoretic variation (e.g. Gillespie 1991; Mitton 1997); in this review I therefore focus exclusively on DNA sequence variation and, in particular, on those aspects of adaptive molecular variation likely to be of most interest to molecular ecologists.
This review consists of four parts. First, the status of the neutral theory after 15 years of DNA sequence surveys is reviewed briefly. Second, I review how neutrality tests have been used to study ecological questions, focusing on what types of genes have been found to be subject to selection resulting in rates of evolution or levels of diversity greater than expected under neutrality (positive selection) and discussing in detail several examples likely to be of particular interest to molecular ecologists. Third, the review briefly discusses how failure to test the neutrality hypothesis adequately can produce misleading results focusing particularly on recent developments such as the use of microsatellites. Finally, the review concludes with a discussion of the positive implications of using DNA sequence data to study natural selection, and discusses some of the obstacles that will limit its near‐term utility.
The current status of the neutral theory
Studies that use molecular markers to address questions in ecology and conservation biology often assume a strictly neutral model of molecular evolution as the basis for analysing and interpreting results. It is therefore worthwhile to review briefly the status of the neutral theory after nearly 20 years of studies of DNA sequence variation within and among species. This has been the topic of several recent reviews (Kreitman 1996; Hey 1999; Hughes 1999), so this section will concentrate only on a few key findings.
The strictly neutral theory proposes that the vast majority of new mutations fall into one of two categories: deleterious or selectively neutral (Kimura 1983). Deleterious mutations are expected to be eliminated rapidly due to natural selection against them and therefore presumably contribute little to variation within or among species. Mutations that are selectively equivalent to the allele(s) already present in the population, on the other hand, are expected to have dynamics governed by genetic drift and to make up the vast majority of the observed variation both within and among species. Beneficial mutations are expected to be extremely rare and to contribute little to observed patterns of DNA sequence variation.
Overall, patterns of DNA sequence variation generally support one key aspect of the neutral theory — that many of the observed differences within and between species are nonadaptive (Kimura 1983; Hughes 1999). For example, there is a broad negative correlation between a nucleotide site's functional importance and its substitution rate. Sites that involve a change in a protein or a change in gene regulation are on average more conserved both within and among species than nonfunctional sites, exactly as predicted by the neutral theory. Additionally, vast parts of the genomes of many organisms contain noncoding DNA that serves no known purpose, and most mutations in these areas are presumably neutral.
A second key aspect of the neutral theory, that most variation has dynamics governed predominantly by genetic drift, is not supported by observed patterns of DNA variation. An important general finding from studies in Drosophila is that there is a positive correlation between a gene's average recombination rate and its level of variability within species (Begun & Aquadro 1992; Aquadro et al. 1994). Similar correlations have been found in humans (2001, 1998), sea beets (Kraft et al. 1998) and maize (Tenaillon et al. 2001). The difference cannot be explained by differences in the neutral mutation rate because there is no correlation between a gene's interspecific divergence and its recombination rate (Begun & Aquadro 1992). The explanation for the correlation appears to be due to the ‘hitchhiking’ of putatively neutral variation with sites that are under selection. One hitchhiking hypothesis that fits fairly comfortably within the paradigm of the neutral theory is that hitchhiking is due mainly to selection against new deleterious mutations that never reach high frequency (the background selection hypothesis —Charlesworth et al. 1993; Charlesworth 1996). The other hypothesis is that the hitchhiking is due mainly to selection of new beneficial mutations that, because of their rapid fixation, ‘selectively sweep’ away large swaths of neutral variation in regions of low recombination (Begun & Aquadro 1992; Aquadro et al. 1994; Hudson 1994; Stephan 1995). The relative degree to which selective sweeps and background selection shape patterns of variation across the genome remains unknown, and distinguishing between the two modes is active area of research (e.g. Begun & Whitley 2000; Fay & Wu 2000; Kim & Stephan 2000; Carr et al. 2001; Kauer et al. 2002; Wang et al. 2002).
The correlation between recombination and variation has two important implications for the neutral theory as it is used typically by molecular ecologists. First, regardless of whether the background selection or selective sweep hypothesis is more important, it means that a gene's recombinational environment is a large contributing factor in determining patterns of variation within species. Second, if the selective sweep hypothesis emerges as a major factor in creating the correlation, this would mean that even if much observed variation is selectively neutral, the dynamics of this neutral variation could well be governed more by linkage to selected sites than genetic drift. This is almost certainly true of genes in regions of low recombination, but linkage to selected sites can in theory be important even in regions of high recombination (Gillespie 2000).
Another aspect of the neutral theory that is not well supported by patterns of DNA sequence data is the hypothesis that adaptive DNA variation is expected to be exceedingly rare. Strictly speaking, this may be true on a per nucleotide site basis, considering that many changes in third codon position, introns and noncoding regions may often be selectively equivalent. However, the fraction of genes for which there is evidence of positive selection may be larger than postulated by the neutral theory. The only published study to attempt to estimate systematically the proportion of genes subject to positive selection is that of Endo et al. (1996), who estimated nonsynonymous/synonymous substitution rate ratios (d n /d s) for 3595 groups of genes extracted from public databases. A d n /d s ratio > 1 provides evidence for positive selection (see Information box). Of the 3595 homologous groups of genes, Endo et al. (1996) found only 17 with evidence for positive selection — certainly a small fraction. However, their criteria for positive selection, d n /d s > 1 averaged over entire genes, was extremely stringent, considering that in most cases only a small fraction of the sites in a gene may be subject to positive selection (Yang & Bielawski 2000). Simple d n /d s ratios are also far from the only method of detecting adaptive evolution at a gene (Information box). Other methods of detecting selection, particularly those that compare d n /d s ratios within and between species, are expected to be more powerful (Charlesworth et al. 2001; Fay et al. 2001), and have been used to find selection in many genes with simple d n /d s ratios < 1 (Table 1). In addition, there are several examples of selection on noncoding sequences (e.g. Schulte et al. 1997; Wang et al. 1999) which would not be detected with any sort of d n /d s comparison.
Table 1.
Genes with evidence for positive selection
| Genes | Taxa | Tests a | References |
|---|---|---|---|
| Host defence genes | |||
| Alpha 1‐proteinase inhibitors | mammals | dn > ds | Goodwin et al. (1996) |
| CD45 | Bos sp. (cattle) | dn > ds | Ballingal et al. 2000 |
| Chitinases | Arabis sp. and other dicots | dn > ds | Bishop et al. (2000) |
| Interleukin‐2 gene (IL‐2) | mammals | dn > ds | Zelus et al. (2000); Zhang & Nei (2000) |
| MHC genes | vertebrates | dn > ds | Hughes & Nei (1988); Hughes et al. (1990); Mikko & Andersson (1995); Miller & Withler (1996); Miller et al. (1997); Hedrick et al. (2000) |
| nef | HIV virus | dn > ds | Zanotto et al. (1999) |
| Polygalacturonase inhibitor | various plants | dn > ds | Stotz et al. (2000) |
| protein genes | |||
| ref(2)p | Drosophila melanogaster | other | Wayne et al. (1996) |
| RGC2 complex | Lactuca sativa (lettuce) | dn > ds | Meyers et al. (1998) |
| RHAG | primates | dn > ds | Huang et al. (2000) |
| Ribonuclease genes | primates, rodents | dn > ds | Zhang et al. (1998); Zhang et al. (2000); Zhang & Rosenberg (2000) |
| Rpm1 | Arabidopsis thalians | other | Stahl et al. (1999) |
| RPP1 complex | Arabidopsis thaliana | dn > ds | Botella et al. (1998) |
| RPP13 | Arabidopsis thaliana | dn > ds | Bittner‐Eddy et al. (2000) |
| RPP8 complex | Arabidopsis thaliana | dn > ds | McDowell et al. (1998) |
| Transferrin | Salmonids | dn > ds | Ford et al. (1999); Ford 2000; Ford (2001) |
| Pyrin gene | primates | dn > ds | Schaner et al. (2001) |
| Colicin genes | Escherichia coli | dn > ds | Riley (1993); (Yang 1998) |
| Defensin genes | rodents | dn > ds | Hughes & Yeager (1997) |
| Fv1 | Mus | dn > ds | Qi et al. (1998) |
| Immunoglobulin VH genes | mammals | dn > ds | Tanaka & Nei (1989) |
| Type I interferon‐ω gene | mammals | dn > ds | Hughes (1995) |
| Parasite response genes | |||
| 10 Plasmodium genes | Plasmodium sp. | dn > ds | Hughes (1991); Hughes (1992); Hughes & Hughes (1995) |
| Envelope gene (env) | HIV | dn > ds | Nielsen & Yang (1998) |
| Envelope gene (env) | HTLV/STLV | dn > ds | Salemi et al. (2000) |
| HA1 | human flu virus | dn > ds | 1997, 2000 |
| Polygalacturonases | various fungi | dn > ds | Stotz et al. (2000) |
| porB | Neisseria gonorrhoeae | dn > ds | Posada et al. (2000) |
| wsp | Wolbachia sp. | dn > ds | Schulenburg et al. (2000) |
| Capsid gene | FMD virus | dn > ds | Haydon et al. (2001) |
| Delta‐antigen coding region | hepatitis D virus | dn > ds | Wu et al. 1999 |
| E gene | Phages G4, φX174 and S13 | dn > ds | Endo et al. (1996) |
| msp 1α | Anaplasma falciparum | dn > ds | Endo et al. (1996) |
| Invasion plasmid antigen‐1 gene | Shigella | dn > ds | Endo et al. (1996) |
| gH glycoprotein gene | Pseudorabies virus | dn > ds | Endo et al. (1996) |
| Outer membrane protein gene | Clamydia | dn > ds | Endo et al. (1996) |
| S and HE glycoprotein genes | Murine coronavirus | dn > ds | Baric et al. (1997) |
| Sigma‐1 protein gene | reovirus | dn > ds | Endo et al. (1996) |
| Virulence determinant gene | Yersinia | dn > ds | Endo et al. (1996) |
| Detoxification genes | |||
| Alcohol genes (Adh) | Drosophila melanogaster | HKA, MK | Hudson et al. (1987); Kreitman & Hudson (1991); McDonald & Kreitman (1991); Berry & Kreitman (1993) |
| Sod | Drosophila melanogaster | other | Hudson (1994); Hudson et al. (1997) |
| Developmental genes | |||
| CAULIFLOWER gene (BoCal) | Brassica sp.; Arabidopsis thaliana | TD, other | Purugganan & Suddith (1998); Purugganan et al. (2000) |
| Teosinte branched 1 (tb1) | Zea mays | HKA | Wang et al. (1999) |
| Genes involves in digestion | |||
| Lysozyme | primates | dn > ds | Swanson et al. (1991); Messier & Stewart (1997) |
| K‐casein gene | Bovidae | dn > ds | Ward et al. (1997) |
| Genes involved in energy metabolism | |||
| 6‐phosophogluconate dehydrogenase (Pgd) | Drosophila melanogaster | HKA | Begun & Aquadro (1994) |
| AGT gene | primates | dn > ds | Holbrook et al. (2000) |
| Amylase (Amy) | Drosophila melanogaster | HKA, MK | Araki et al. (2001) |
| COII gene | primates | other | Andrews & Easteal (2000) |
| G6pd | Drosophila melanogaster | HKA, MK | Eanes et al. (1993); Eanes et al. (1996) |
| G6pd | Homo sapiens | other | Tishkoff et al. (2001) |
| Gld | Drosophila melanogaster | other | Hamblin & Aquadro (1997) |
| Lactate dehydrogenase – B (Ldh‐B) | Fundulus heteroclitus (killifish) | HKA | Schulte et al. (1997); Powers & Schulte (1998); Schulte et al. (2000) |
| PGI gene | Gryllus sp. (field crickets) | other | Katz & Harrison (1997) |
| PgiC | Leavenworthia sp. | other | Liu et al. (1999) |
| PgiC | Arabidopsis thaliana | MK, TD, other | Kawabe et al. (2000) |
| Pgm | Drosophila melanogaster | MK | Verrelli & Eanes (2000, 2001) |
| Tpi | Drosophila melanogaster; D. simulans | HKA | Hasson et al. (1998) |
| ATP synthase F0 subunit gene | Escherichia coli | dn > ds | Endo et al. (1996) |
| COX7A isoform genes | primates | dn > ds | Schmidt et al. (1999) |
| Odour receptors | |||
| Odour receptor genes | Ictalurus punctatus (channel catfish) | dn > ds | Ngai et al. (1993) |
| Odour receptor genes | humans | HKA, MK | Gilad et al. (2000) |
| Pigmentation genes | |||
| White (w) | Drosophila melanogaster | other | Kirby & Stephan (1995, 1996) |
| Melanocortin 1 receptor gene | primates | dn > ds | Rana et al. (1999) |
| (MC1R) | |||
| Genes involved in reproduction | |||
| Odysseus (ods) | Drosophila simulans; D. mauritiana | dn > ds | Ting et al. (1998) |
| pem protein gene | rodents | dn > ds | Sutton & Wilkinson (1997) |
| het‐c | Sordariaceae (filamentous fungi) | dn > ds | Wu et al. (1998) |
| S locus | various plants | dn > ds | Clark & Kao (1991) |
| 19 seminal protein genes | Drosophila melanogaster; D. simulans | dn > ds | Swanson et al. (2001c) |
| Acp26Aa | Drosophila melanogaster; D. mauritiana | dn > ds | Tsaur & Wu (1997); Tsaur et al. (1998), (2001) |
| Acp29AB | Drosophila melanogaster | MK | Aguade (1999) |
| Acp29AB and Acp36DE | Drosophila melanogaster; D. simulans | MK | Begun et al. (2000) |
| Acp70A | Drosophila melogaster | other | Cirera & Aguade (1997) |
| Androgen‐binding protein gene (Abpa) | Mus sp. (house mice) | dn > ds, HKA | Karn & Nachman (1999) |
| Bindin | Echinoida (sea urchins) | dn > ds | Metz & Palumbi (1996); Beirmann (1998); Palumbi (1999) |
| Gastropod sperm proteins (lysin and others) | Haliotis sp. (abalone); Tegula sp. (sea snails) | dn > ds | Lee & Vacquier (1992); Vacquier et al. (1997); Metz et al. (1998); Hellberg & Vacquier (1999); Hellberg et al. (2000); Swanson et al. (2001a) |
| Protamine P1 | primates | dn > ds | Rooney & Zhang (1999); Rooney, Zhang & Nei (2000) |
| ZP3 | mammals | dn > ds | Swanson et al. (2001b) |
| Egg‐laying hormone genes | Aplysia californica | dn > ds | Endo et al. (1996) |
| S‐Rnase gene | Rosaceae | dn > ds | Ishimizu et al. (1998) |
| Other genes | |||
| desat2 | Drosophila melanogaster | other | Takahashi et al. (2001) |
| methuselah (mth) | Drosophila melanogaster | MK, other | Schmidt et al. (2000) |
| Mylin proteolipid protein gene | vertebrates | dn > ds | Kurihaha et al. (1997) |
| Pregnancy associated glycoproteins | artiodactyls | dn > ds | Xie et al. (1997) |
| pantophysin gene (pan 1) | Gadus morhua (Atlantic cod) | MK | Pogsen (2001) |
| CDC6 | Saccharomyces cerevisiae | dn > ds | Endo et al. (1996) |
| Alpha 1 Na/K‐Atpase gene | Artemia fanciscana | HKA, TD | Saez et al. (2000) |
| Toxin genes | Conus sp.; reptiles | dn > ds | Duda & Palumbi (1999); Kordiš& Gubenšek (2000) |
| Jingwei | Drosophila | dn > ds | Long & Langley (1993) |
| Growth hormone gene | vertebrates | dn > ds | Wallis (1996) (76) |
| Prostatic steriod binding protein | Rat | dn > ds | Endo (1996) |
| Haemoglobin | Nototenioids (antarctic fishes) | dn > ds | Bargelloni et al. (1998) |
| a HKA refers to the test of Hudson et al. (1987). TD refers to Tajima's (1989) test. MK refers to contingency test of McDonald & Kreitman (1991). See Information box for additional information. | |||
The proportion of genes subject to positive selection therefore remains unknown, but it is sufficiently high that it has become relatively common to find evidence for positive selection. For example, in a literature search for papers documenting positive selection using DNA sequence data (Table 1), three studies were found published in the 1980s, 57 published in the 1990s and 45 published in 2000 and the first half of 2001. The rapid increase in the number of papers documenting positive selection is due both to advances in DNA sequencing techniques that allow more rapid acquisition of data and advances in statistical methods for detecting positive selection (Kreitman & Akashi 1995; Yang & Bielawski 2000).
In summary, the ‘neutralist/selectionist’ debate of the 1970s and 1980s has died away (Kreitman 1996; Hey 1999). It has been replaced by a more complicated view in which selection, both positive and negative, appears to leave deep footprints in the genome and evidence for positive selection at many specific genes is accumulating. The strictly neutral theory, rather than being a simple explanation for patterns of genetic diversity, has instead become the primary null hypothesis used to test for the effects of natural selection.
Using inferences about selection to study ecology
The field of ecology encompasses a broad range of questions about why species and populations are where they are, and how they interact with other organisms and their environment. The fields of molecular ecology and molecular conservation genetics have involved primarily the application of molecular data to questions of biogeography, gene flow, hybridization and relatedness among individuals (Avise 1994, 1995). These questions are important and can be addressed to greater or lesser degrees within the framework of the neutral theory. Other ecological questions address adaptation and natural selection, however. If inferences about natural selection could be made readily from molecular data, this would be of enormous importance to the field of molecular ecology.
What types of genes are subject to positive selection?
To address this question, a literature search was conducted for studies documenting positive selection. Only studies that used statistical tests on DNA sequence variation, to infer the action of natural selection at particular genes, were included; direct functional studies, studies documenting hitchhiking over broad genomic areas and studies based solely on protein variation were not included. The search resulted in over 100 studies published in the last 15 years documenting statistical evidence for positive selection on over 119 genes or gene groups (Table 1). Table 1 is not an exhaustive list of all studies documenting positive selection, but certainly includes the bulk of studies published prior to early 2001. The only types of genes that are probably systematically underrepresented in Table 1 are microbial and viral genes, at which observations of high d n /d s ratios are common and therefore difficult to compile comprehensively. Studies documenting positive selection are being published at an increasing rate (over half of the studies cited in Table 1 are from 2000 and 2001), and the list of positively selected genes can be expected to grow dramatically in the coming years.
Of the 119 positively selected genes or gene groups, 92 had d n /d s significantly > 1 for all or part of the gene, nine had significant McDonald–Kreitman tests and 25 had statistically non‐neutral patterns of overall polymorphism or divergence indicative of natural selection (e.g. significant HKA, Tajima’s, or similar tests — Information box I).
Of the genes that show evidence for positive selection, 47 have been shown or hypothesized to be involved in host–parasite interactions, either as part of the host's defence system or the parasite's system for evading host defences (Table 1). Examples of host immune response genes with evidence for positive selection include multiple loci in the major histocampatibility complex (MHC) in various mammals and fishes; interleukin‐2, ribonucleases and CD45 in mammals, transferrin in salmonids; and chitinases, a polygalacturonase inhibitor, and various other disease resistance genes in plants (Table 1). With regard to pathogens, there is evidence for positive selection in multiple viral, bacterial, fungal and protist genes (Table 1).
A second large group of genes with evidence for positive selection consists of genes that are, or have been hypothesized to be, involved in sexual reproduction. In particular, there is evidence for positive selection in at least 35 reproductive genes (Table 1). Positive selection on sex‐related genes has been reported across a wide variety of taxa, including three sperm proteins from several marine invertebrates (sea urchins; abalone; sea snails); a large number of Drosophila ejaculatory proteins; a mouse androgen binding protein; genes related to interspecific mating isolation in Drosophila; mammalian egg surface proteins; and genes involved in self‐mating avoidance in plants and fungi (Table 1). The specific functions of these genes vary dramatically, and selective mechanisms have been documented directly in only a few cases (e.g. S‐loci in plants, Clark & Kao 1991).
Genes whose products code for enzymes involved in energy metabolism form a third group of genes with statistical evidence for positive selection (reviewed by Mitton 1997; Eanes 1999). There is statistical evidence for positive selection in least 15 metabolic genes (Table 1), most of which have been surveyed extensively in many species using protein electrophoresis. Most of the studies documenting positive selection at these genes have focused on variation within and among closely related species (Table 1), and violations of the neutral model at these genes generally involve non‐neutral patterns of overall polymorphism and divergence or significant McDonald–Kreitman tests (see Information box). Many of the genes targeted for DNA studies had been hypothesized previously to be subject to selection based on either clinal patterns of protein variation or function studies of protein variants (Mitton 1997; Eanes 1999).
The remaining genes with strong evidence for positive selection do not fall into any obvious functional category. These genes include lysozyme and K‐casein, which are involved in digestion; genes for various animal toxins; a melanocortin receptor gene involved in hair and skin colour; regulatory genes involved in plant morphology; several genes expressed in artiodactyl placentas; haemoglobin in Antarctic fishes; the cod pantophysin gene; odour receptors in humans and catfish, and the pyrin gene in primates (Table 1).
What are the selective mechanisms?
The number of reported cases of positive selection has grown remarkably over the last several years, but in most cases the actual source of selection remains unknown. In some cases, even the functions of the genes in question are obscure. This situation has come about because it has become relatively easy to collect and analyse DNA sequences but it is much more difficult to elucidate a gene's functions, let alone determine how variation in those functions was influenced by natural selection in current or past populations. In some cases, particularly when a gene's function(s) are at least partially understood, it is possible to postulate a plausible adaptive hypothesis for why a gene has been subject to positive selection. These hypotheses can then suggest further experiments or observations that could be used to confirm or refute them as explanations for observed patterns of genetic variation. On the other hand, some of the cases of positive selection reported in the literature are inferred by comparing sequences from species that have diverged tens or sometimes even hundreds of millions of years ago. In most of these cases, a detailed understanding of the selective mechanisms leading to observed patterns of variation may well be unattainable (Hughes 1999).
Even in cases where there is little information on the ecological or functional mechanisms leading to patterns of DNA variation indicative of positive selection, some useful information can be gleaned directly from the DNA sequence data. For example, patterns of DNA sequence polymorphism can be used to determine if selection acting on a gene is directional (e.g. G6pd: Eanes et al. 1993), balancing (e.g. PgiC: Liu et al. 1999) or both (e.g. Adh: Kreitman & Hudson 1991). High d n /d s ratios among genes that make up a multigene family provide strong evidence that there has been selection for new functions after gene duplication (reviewed by Hughes 1999). The observation of strong directional or balancing selection at a gene or gene region can be a useful tool for determining whether the gene is a good candidate for future functional studies. This approach is particularly powerful if specific nucleotide sites that have been the target of positive selection can be identified. Molecular biologists have long made use of a key paradigm of the neutral theory — that functionally important parts of a gene will be conserved — to identify candidate gene regions for further functional study. A similar approach can now be taken by identifying specific variable sites that show statistical evidence for adaptive evolution (Nielsen & Yang 1998; Suzuki & Gojobori 1999; Fig. 1).
Figure 1.
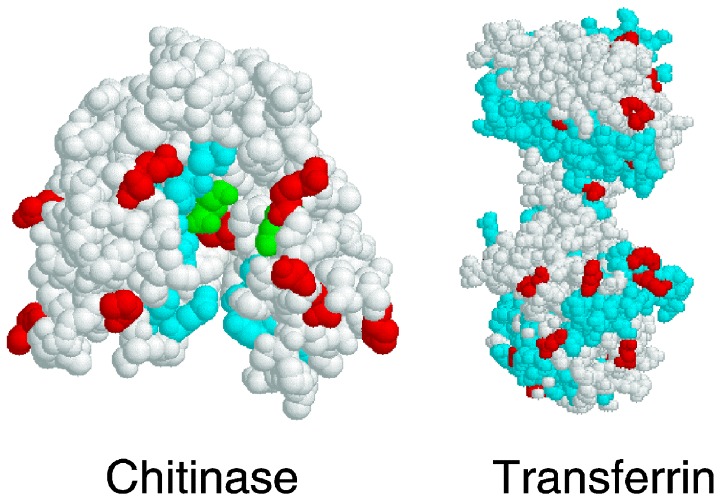
Examples of the identification of codons with high probability of being subject to positive selection. (right) Model of transferrin with sites subject to positive selection in salmonids coloured red and sites contacted by bacterial transferrin‐binding proteins in humans coloured blue (adapted from Ford 2001). (left) Model of class I plant chitinase (A chain only), with positively selected sites coloured red, substrate binding sites coloured blue and catalytic sites coloured green (adapted from Bishop et al. 2000).
Examples of ecologically interesting positive selection
Selection on allozyme loci
A basic goal of physiological ecology is understanding how an organism's metabolism allows it to live in its environment. Many of the proteins that can be studied electrophoretically are involved in energy metabolism. After high levels of allozyme variation were discovered in the late 1960s (Harris 1966; Lewontin & Hubby 1966), many studies were aimed at determining the functional and adaptive significance, if any, of observed variation (reviewed by Mitton 1997). Functional studies have documented that variation in metabolic proteins can affect ecologically important traits such as flying or swimming performance and mating ability (see Powers et al. 1991; Mitton 1997; Watt & Dean 2000 for reviews), and DNA sequence studies have found that many metabolic genes have patterns of variation inconsistent with neutrality (Table 1). Studying allozyme loci at the DNA level is attractive because many of the genes code for proteins whose biochemical roles are well understood and which may contain variation important for understanding the distribution or behaviour of organisms (Mitton 1997; Eanes 1999). Studies of DNA sequence variation at alloyzme genes are particularly useful for acquiring additional insight into the evolutionary history of previously documented allozyme variation, although the inferred histories can complicated (reviewed by Eanes 1999). For example, the first allozyme locus to be studied extensively at the DNA level, alcohol dehydrogenase in Drosophila (Adh), has a peak of synonymous site polymorphism centred on the site that causes the allozyme polymorphism (Fig. 2: Kreitman & Hudson 1991). This pattern of variation is inconsistent with a simple neutral model, and consistent with a balancing selection model (Kreitman & Hudson 1991). In a more recent analysis of additional Adh sequences, however, Begun et al. (1999) suggest that the evidence of an old balanced polymorphism at Adh is not compelling, in part because the peak of synonymous polymorphism appears to predate the alloyzme polymorphism. Other Drosophila protein polymorphisms also have quite complicated inferred histories based on patterns of DNA sequence variation. For example, Sod, G6pd, Pgm, Tpi, Pgd and Amy all have patterns of variation that are inconsistent with neutrality but that are not suggestive of old balanced polymorphisms (see Table 1 for references). The type of selection occurring at these loci remains obscure, but may involve transient or regionally varying selection or evolutionarily young balanced polymorphisms. One unifying factor in studies of selection in D. melanogaster is its relatively recent migration from Africa, which may have created new selection pressures in non‐African populations (Begun & Aquadro 1995; Takahashi et al. 2001; Kauer et al. 2002). Outside Drosophila, other cases in which DNA sequence studies have found or confirmed evidence of positive selection at allozyme loci include Ldh‐B in Fundulus (Powers et al. 1991; Schulte et al. 1997; Powers & Schulte 1998), and PgiC in crickets (Katz & Harrison 1997) and several plant species (Arabidopsis thaliana: Kawabe et al. 2000; Leavenworthia sp.: Liu et al. 1999).
Figure 2.
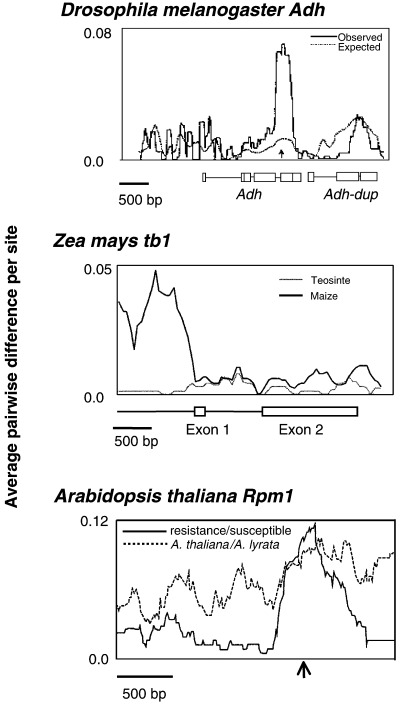
Three examples of sliding window analyses showing non‐neutral patterns polymorphism and divergence. (a) Drosophila Adh, showing a peak of intraspecific variation associated with the only amino acid polymorphism in the population (adapted from Hudson & Kreitman 1991; see also Begun et al. 1999). (b) Maize and teosinte tb1, showing reduction of diversity in maize in the 5′ untranslated region of the gene (adapted from Wang et al. 1999). (c) Arabidopsis thaliana Rpm1 region (adapted from Stahl et al. 1999). Dark line shows divergence between A. thaliana resistant (Rpm1 present) and susceptible (Rpm1 deleted) alleles, and the dotted line shows divergence between A. thaliana and A. lyrata. The arrow marks the location of the Rpm1 insertion/deletion.
Domestication selection in agricultural systems
Understanding the ecology and evolution of agricultural systems is important for solving many applied agricultural problems. For example, concerns have been raised that modern agricultural plants and animals are becoming increasingly depauperate of genetic diversity and therefore vulnerable to environmental or biological insults their wild ancestors would have resisted (e.g. Notter 1999). Understanding the genetic architecture and history of the domestication process provides useful information for the feasibility and utility of introducing new genetic material from a crop's wild ancestors (Tanksley & McCouch 1997). Agricultural crops have also provided a useful experimental context for addressing fundamental ecological and evolutionary questions applicable to natural systems, including the genetic basis of morphological variation, genetic and ecological interactions between hosts and pathogens and the evolution of phenotypic plasticity. Finally, many agricultural crops are amenable to extensive transmission genetic analysis such that genes of interest can be located in the genome and cloned. This makes agricultural systems particularly attractive for studying the population genetics of genes that control or influence complex traits (Tanksley & McCouch 1997).
The domestication of maize (Zea mays) from its wild grass ancestor teosinte (Z. mays parviglumis) provides a superb example of how molecular inferences about natural selection provided insight into an important ecological and evolutionary question. Recently, Doebley and colleagues used genetic mapping techniques to show that the teosinte branched‐1 (tb1) locus is primarily responsible for one of the key morphological differences between maize and teosinte (1995, 1997). They then used a prediction of the neutral theory — that loci that have recently experienced strong directional selection are expected to have low levels of intraspecific variation — to see if there was evidence for selection at tb1 during the domestiction of maize (Wang et al. 1999). Wang et al. (1999) tested this expectation by sequencing a sample of tb1 alleles from maize and teosinte. They found that the 5′ untranslated region of tb1 had much lower levels of variability in maize than in teosinte, consistent with a selective sweep of that gene region in maize (Fig. 2). An HKA test ( Information box) confirmed that the reduction in variability in maize was inconsistent statistically with neutral evolution.
The genealogy of the tb1 gene also provided insight into the biogeography of maize domestication. The consistent clustering between maize tb1 5′ alleles and teosinte alleles sampled from a specific teosinte subspecies native to particular river valley provided evidence for the location in which maize was initially domesticated (Wang et al. 1999). In this case, genealogical analysis of a gene under strong selection provided a much clearer biogeographical signal than genes evolving neutrally (Hilton & Gaut 1998). The analysis of genetic variation at the CAULIFLOWER locus (BoCal) in wild and domesticated Brassica oleracea provides another good example of how neutrality tests were used to gain insight into selection at a gene involved in plant domestication (Purugganan et al. 2000).
Cuticular hydrocarbon variation in D. melanogaster
Understanding the reasons for intraspecific variation in behaviour, morphology or physiology is an important goal of ecology. For example, populations of D. melanogaster vary geographically in the components of their cuticles, with African and Caribbean populations having a high ratio of two major hydrocarbon isomers and all other populations worldwide having a low ratio of the same isomers. The functional significance of these differences is not well understood, but sexual selection or adaptation for desiccation resistance have been hypothesized as possible explanations (Ferveur et al. 1996; Takahashi et al. 2001).
Recently, two groups determined independently that the variation in the isomer ratios was due to a difference at a single gene, desat2 (Coyne et al. 1999; Dallerac et al. 2000; Takahashi et al. 2001). Based on patterns of variation at desat2, Takahashi et al. (2001) found that the cosmopolitan low ratio alleles were evolutionarily derived from an African/Caribbean high ratio allele. The low ratio alleles displayed little intra‐allelic diversity and had a frequency spectrum inconsistent with an equilibrium neutral model but qualitatively consistent with a recent selective sweep. Takahashi et al. (2001) concluded that the most probable explanation for their data was that the low ratio allele originated recently and increased in frequency in non‐African populations due to natural selection either during or subsequent to D. melanogaster’s recent emigration from Africa.
Host–pathogen coevolution
Understanding interactions among species is another important goal of ecology. One type of interspecific interaction that appears to lend itself readily to molecular analysis is the relationship between hosts and parasites. Recently, several investigators have used DNA sequence variation in plant genes involved in resistance to pathogens to gain insight into host/pathogen coevolution (reviewed by Bergelson et al. 2001; Holub 2001). These studies have found that positive natural selection has played a role in the evolution of plant resistance genes in a variety of ways, including diversifying selection among members of resistance gene families (e.g. Botella et al. 1998); directional selection on genes among different species (e.g. Bishop et al. 2000); and balancing selection among alleles within species (Fig. 2; Stahl et al. 1999; Bittner‐Eddy et al. 2000).
Stotz et al.'s (2000) study of fungal polygalacturonases (PGs) and their plant inhibitors (PGIPs) is a particularly good example of how neutrality tests can contribute to the understanding of interactions between hosts and pathogens. Stotz et al. (2000) analysed interspecific variation among 19 fungal PGs and 22 plant PGIPs, and found that many comparisons within each group had d n /d s significantly > 1, indicating that both the pathogen proteins and plant inhibitors were evolving by positive selection. More detailed analysis of d n /d s ratios using Nielsen & Yang's (1998) maximum likelihood method allowed Stotz et al. (2000) to identify nine codons in each group of genes with high posterior probabilities of being subject to positive selection.
One strength of the Stotz et al. (2000) study is that the molecular structures of both PGs and PGIPs are known, and the physical locations of sites identified as statistically likely to be subject to positive selection can be visualized three‐dimensionally. This three‐dimensional physical information can provide additional insight into the mechanism of natural selection and can generate hypotheses for further experimental study (Nielsen & Yang 1998, Fig. 1).
Using neutrality tests to study natural selection
The conclusion that variation at many genes is influenced by positive selection, either directly on the genes themselves or indirectly through the effects of linkage, has important implications for molecular ecology and conservation genetics. For example, the finding of pervasive effects of positive selection suggests that molecular ecologists must be cautious about analyses that rely upon untested assumptions of neutrality (Rand 1995). For example, molecular ecologists and conservation geneticists use molecular genetics routinely to help address questions such as defining conservation units, studying biogeography and estimating effective population sizes or rates and patterns of gene flow. In most cases the genetic variants that are analysed are of no particular interest in themselves, but are used instead as putatively neutral markers of demographic or evolutionary histories (Avise 1994). If a marker, or even an entire group of markers, that is assumed to be evolving neutrally is actually subject to selection, conclusions based on patterns of variation at the marker could be misleading. On the other hand, the ability to make strong inferences about the selective history of a gene opens up exciting new research areas for molecular ecologists to explore, although for most organisms there are several roadblocks standing in the way of research programmes based on studying natural selection at the molecular level. In what follows, the implications are discussed for molecular ecology if many putatively neutral markers turn out to be non‐neutral.
Implications for ignoring selection
Rand (1995) warned conservation geneticists that assuming selective neutrality for allozyme and mtDNA variation routinely was problematic due to considerable evidence for the effects of natural selection at these classes of genetic markers. Assuming strict neutrality when in fact variation is affected by selection is particularly worth avoiding when conducting quantitative analyses, such as estimating divergence times, rates of gene flow or effective population sizes. In the 6 years since Rand's review, the number of published examples of positive selection has increased dramatically (Table 1), so the need to examine carefully the consequences of false assumptions of neutrality appears even more justified now than it did in 1995.
One development that has come about after Rand's review is the now ubiquitous use of microsatellites in molecular ecological studies. Variation at these loci is usually assumed to be selectively neutral, in part because microsatellites are often located in noncoding regions. Several recent results suggest that assumptions of strict selective neutrality at microsatellite loci must be viewed with caution, however. For example, some microsatellites are located in coding or promoter regions and may be under direct selection (e.g. microsatellites in the promoter region of Ldh‐B in Fundulus heteroclitus: Schulte et al. 1997).
Even if variation at a specific microsatellite locus is selectively equivalent, it may not fit a strict neutral model due to selection at linked sites. Furthermore, because microsatellite mutation rates vary across taxa, the power to detect the effects of hitchhiking selection on equilibrium levels of variation will vary as well (Schug et al. 1998; Wiehe 1998). For example, in D. melanogaster there is a strong correlation between recombination rate and microsatellite variation similar to that observed for single nucleotide variation and due presumably to the combined effects of background selection and positive hitchhiking (Schug et al. 1998). In contrast, in humans there is no evidence of a correlation between microsatellite variation and recombination, but there is a positive correlation between recombination and single nucleotide polymorphism (Nachman et al. 1998; Payseur & Nachman 2000; Nachman 2001). The lack of a correlation between microsatellite variation and recombination in humans is due apparently to the high mutation rate of human microsatellites (∼10−4, references in Payseur & Nachman 2000). Microsatellites in regions of low recombination will have the same reduced coalscent times as other sequences in the same regions, but may not have reduced levels of variation in humans due to mutational saturation (Wiehe 1998). In contrast, in Drosophila microsatellite mutation rates are much lower (∼10−6), take longer to become mutationally saturated and therefore have patterns of variation that are influenced more readily by background selection or selective sweeps (Schug et al. 1998).
The effects of violations of selective neutrality will vary from study to study depending on the specific questions being addressed, and it is beyond the scope of this review to address the topic in any detail (see Rand 1995 for a comprehensive treatment, particularly with regard to mtDNA). The point is that it is important to test assumptions about selective neutrality before using ‘neutral’ markers to make inferences about populations.
Studying natural selection at the molecular level
The ability to make strong inferences about the action of selection from patterns of DNA sequence variation is a potentially powerful way of studying adaptation. For example, one might be interested in knowing if geographical patterns of variation in a particular trait are adaptations that arose through natural selection. Conceptually, a way to answer this question would be to survey DNA sequence variation at a gene or genes with major effects on the trait and determine if the null hypothesis of selective neutrality can be rejected (i.e. a variation of Endler's (1986) model fitting method of studying natural selection). If the patterns of variation are statistically inconsistent with the neutral theory, but statistically consistent with positive natural selection, this would provide evidence that the variation in the trait of interest arose in response to natural selection. It may also be possible to make inferences about how, when and perhaps even where the selection occurred. Several of the studies described above used successfully this approach of studying the history of natural selection on specific traits.
Using DNA sequence data to study natural selection is useful but there are several obstacles, both practical and conceptual, that will limit its near‐term applicability. The primary practical limitation for most molecular ecologists studying nonmodel organisms will be finding appropriate genes to study. Many of the traits that ecologists find interesting are complex or quantitative traits of morphology, physiology or behaviour. For most organisms, the genetic basis of these traits is unknown and likely to be influenced by many genes. Finding the genes that influence variation in quantitative (or for that matter even simple) traits is difficult and time‐consuming even in ‘model’ genetic organisms such as Drosophila and maize (Flint & Mott 2001; Mackay 2001), and is not currently feasible for many of the less genetically studied organisms that are usually of interest to ecologists or conservation biologists.
In addition to the practical problem of isolating genes, a related conceptual problem is that genetic variation in traits of interest may not always involve variation at a gene of large effect. If a trait of interest is influenced only by many genes of small effect, it is more likely that the effects of selection on the trait would also be dispersed widely around the genome and therefore more difficult to detect from patterns of DNA sequence variation. In Drosophila, where considerable progress has been made dissecting the genetic basis of quantitative traits, the distribution of genic effects for several traits is roughly exponential with a few genes of large effect and many genes of small effect (Mackay 2001). If this patterns holds for other traits and other species, it would mean that most traits will be influenced by some genes of large effect. Genetic mapping experiments in other species do generally find quantitative trait loci (QTL) of large effect (e.g. Schemske & Bradshaw 1999; Peichel et al. 2001), but in these studies individual QTL almost always encompass large chromosomal regions containing hundreds of genes, so the effects of individual genes cannot be evaluated without extensive additional genetic analysis (Flint & Mott 2001).
If isolating genes of interest is a struggle even in model genetic organisms, is there any hope for the ecologist studying an organism with no genetic map, no sophisticated genetic tools and no probable prospect for a government‐funded genome project? There are several potential approaches. First, for many organisms it will be more fruitful to examine variation in genes whose functions are already well understood than to start with a phenotypic trait of interest and try to find the genes that influence it. A place to start might be genes whose products are involved in basic energy metabolism, such as the enzymes in the glycolytic pathway and the Krebs cycle. These genes code for products with well‐understood biochemical functions that are similar across many organisms. Variation in at least some of these genes has been shown to be subject to positive natural selection and to contribute to ecological interesting variation in a variety of species (see above), making them good candidates for studying adaptive variation. Cloning these genes using heterologous probes or degenerate PCR primers is relatively straightforward (e.g. Katz & Harrison 1997) and will become more so as more sequences become available. For a great many organisms, variation at these genes has been surveyed using protein electrophoresis, and in some cases patterns of electrophoretic variation may suggest good candidates to study at the DNA level.
Model genetic organisms provide a second potential source of candidate genes for molecular adaptation studies (reviewed by Haag & True 2001). Ford & Aquadro (1996), for example, in their study of speciation in D. athabasca, surveyed variation in candidate mating song genes isolated originally from D. melanogaster. Lukens & Doebley (2001) studied patterns of variation in the tb1 gene among members of the grass family to determine if this gene, which influences morphology in maize, had also been evolving adaptively in other grass species. A coat colour gene, Mc1r, isolated originally in mice, has been shown to affect coat colour in a variety of farm animals (Andersson 2001), and there is evidence for positive selection acting on the same gene during primate evolution (Rana et al. 1999). There is even weak evidence that the gene is subject to positive selection in humans, where it may influence skin and hair colour (Rana et al. 1999). The CAULIFLOWER gene, isolated originally in Arabidopsis, was also found to affect floral morphology in domesticated Brassica sp. (Purugganan & Suddith 1998; Purugganan et al. 2000). MHC genes have been isolated from, and found to be subject to positive selection in, a variety of species (Table 1). As the functions of more and more genes are elucidated in a few ‘model’ organisms, molecular ecologists will have a large pool of candidate genes to draw upon for population genetic study. Confirming that candidate genes actually influence the trait of interest in the organism studied is rarely a trivial task, however (Flint & Mott 2001).
A third potential source of genes to study could come from randomly or systematically sampling a species’ genome for genes that show evidence for positive selection. Pogson et al. (1995) took this approach in their study of genetic variation in Atlantic cod. They surveyed variation initially within and among cod populations at a group of randomly cloned cDNA fragments, and found that as a class the random clones had greater levels of diversity among populations than did allozyme loci, suggesting the action of natural selection on one or both classes of loci. Pogson (2001) conducted a detailed analysis of sequence variation at one of the random clones and found that the gene (pantophysin) had d n /d s > 1 within cod populations. That observation provided additional evidence for positive selection, although the form of selection and even the gene's function remain obscure (Pogson 2001). There is no way to know how often such random searches can be expected to find genes of interest, but this example suggests that developing methods of scanning genomes for positively selected genes would be worthwhile.
Finally, the availability of complete genome sequences combined with well‐developed genetic tools provides a compelling reason to study the ecology of genetically well‐characterized organisms. In plants, Arabidopsis thaliana is rapidly becoming a model plant for ecological genetic studies (Pennisi 2000). D. melanogaster is one of the best understood animals genetically and developmentally, and studies in the species have led to fundamental advances in population genetics (Aquadro et al. 1994; Hey 1999). Despite this wealth of knowledge, D. melanogaster has been the subject of relatively few ecological studies and the ecological factors causing selection on even the famous Adh polymorphism are not well understood (Hughes 1999). More intensive ecological study of D. melanogaster, and other genetically well‐characterized organisms, would clearly be beneficial.
In summary, statistical analysis of DNA sequence diversity within and among species is a powerful way to study natural selection and adaptation. In large part, this is because DNA sequences can provide an informative record of their own evolutionary history. Over the last 20 years, investigators have made substantial progress in their ability to read and make sense of the historical information stored in DNA sequences. As more genes are sequenced and their allelic diversity analysed within and among species, the challenge will be not so much to find genes that have evolved under positive selection but to understand causes of the selective pressures.
Michael J. Ford heads the Genetics and Evolution Program in the Conservation Biology Division of the Northwest Fisheries Science Center. The Conservation Biology Division studies the processes maintaining biological diversity so that diversity can be sustained in the face of human impacts. The purpose of the Genetics and Evolution Program is to understand how genetic processes contribute to species viability, and to develop and use molecular genetic tools for addressing evolutionary, ecological and resource management questions.
Information box
Methods of detecting positive selection using DNA sequence data
Soon after protein electrophoretic studies began demonstrating that natural populations were polymorphic at enzyme loci, investigators developed methods of testing the fit of observed patterns of variation to expectations under neutrality (e.g. Ewens 1972; Lewontin & Krakauer 1973; Watterson 1978). These tests can suffer from low power and their assumptions are frequently violated, making strong inferences about selection difficult. Many of the statistical tests of the fit of DNA sequence data to neutral models have been more successful, due largely to the greater information content available in the data (Kreitman & Akashi 1995; Hey 1999; Kreitman 2000). A plethora of neutrality tests have been developed for DNA sequence data, and four important and representative ones are illustrated briefly.
The HKA test. Hudson et al. (1987) developed the first statistical test of neutrality to take advantage of the greater information content available from DNA sequence data compared to protein electrophoretic data. The test is based on the expectation that under an infinite sites neutral model (appropriate for DNA sequence variation within and among closely related species) the level of polymorphism at a gene within a species is proportional to the amount of divergence at that gene between species. This expectation can be tested by comparing levels of polymorphism and divergence at two or more genes using Hudson et al.′s (1987) test statistic. The HKA test can be applied to both DNA sequence and RFLP data, and has been used commonly to test for departures from neutral expectations (Table 1). The HKA and related tests, like the earlier tests designed for allozyme data, makes specific assumptions about population structure which, if violated, can lead to rejection of the neutral model for reasons other than positive selection (Ford & Aquadro 1996; Ford 1998). Sliding window graphs (Fig. 2) are a method of visualizing the HKA concept, and identifying specific regions of a gene responsible for the departure from neutral expectations. 1996, 1998) has developed a formal statistical test using the sliding window approach.
Tajima's D‐test. Tajima (1989) developed a statistical test of neutrality that uses only polymorphism data within a population. The test statistic, D, is based on the difference between two estimators of the neutral polymorphism parameter 4N e µ, one based on the total number of polymorphic nucleotide sites observed and the other based on the average number of differences between all pairs of sequences sampled. Under neutrality, both methods are expected to produce the same value, and Tajima's D statistic is designed to test whether the difference between them is unlikely to be observed by chance alone. The test is essentially a method of testing the fit of the data to the expected neutral site frequency distribution, with negative D‐values indicating an excess of rare variants and positive values indicating an excess of intermediate frequency variants. Like the HKA test, Tajima's test can be used on both RFLP and full sequence data. The test is sensitive to violations of its demographic assumptions and is not particlarly powerful (Tajima 1989;, Simonsen et al. 1995), so interpretation of test results can be uncertain. Fu (1997) discusses a number of similar tests.
Dn/ds tests. Hill & Hastie (1987) and Hughes & Nei (1988) were the first to use the ratio of nonsynonymous differences to synonymous differences among DNA sequences as a test for positive selection. The idea behind the test is that if synonymous mutations are essentially neutral because they do not result in a change in a protein, the rate of synonymous site evolution (d s) will equal the mutation rate (Kimura 1983). Nonsynonymous mutations, because they result in a change in a protein product, are more likely to be subject to natural selection. If most nonsynonymous mutations are deleterious, then the rate of nonsynonymous evolution (d n) will be lower than neutral rate, resulting in d n /d s < 1. If a substantial fraction of nonsynonymous mutations are beneficial, however, the average rate of nonsynonymous evolution can be higher than the neutral rate, resulting in d n /d s > 1. Conceptually, all that is involved in performing a d n /d s ratio test is estimating the number of synonymous and nonsynonymous substitutions between two sequences and the number of synonymous and nonsynonymous sites in the sequences and then performing a statistical test to determine if the difference between the two ratios is greater than expected by chance alone. In practice, complications such as accounting for multiple substitutions at the same site, estimating the number synonymous and nonsynonymous sites, and accounting for transition/transversion ratios add considerable complexity to the problem, especially for highly divergent sequences (Nielsen 1997; Yang & Nielsen 2000).
An exciting recent development in the application of d n /d s tests has been the development of maximum likelihood approaches for estimating d n /d s and related parameters (Goldman & Yang 1994; Nielsen & Yang 1998; Yang & Bielawski 2000). In addition to providing potentially more accurate d n /d s estimates (Yang & Nielsen 2000), the likelihood approaches allow estimation of d n /d s for individual branches of a phylogenetic tree, estimation of the proportion of codons in a sequence that are subject to positive selection, and identification of specific codon sites that are likely to have been subject to positive selection (Nielsen & Yang 1998, Fig. 1). Suzuki & Gojobori (1999) have developed an different method of detecting positive selection at single codons that uses a parsimony approach (see Suzuki & Nei 2001 for an initial comparison of the two methods).
The d n /d s ratio tests do not require any assumptions about population structure or equilibrium conditions, and therefore can provide particularly compelling evidence for positive selection. Various versions of the d n /d s ratio test do make assumptions about codon usage bias, transition/transition transversion rate ratios and selective neutrality of synonymous mutations. If these assumptions are violated the tests can produce misleading results, but these problems can be potentially avoided by using appropriate evolutionary models when conducting the tests (Yang & Bielawski 2000; Yang & Nielsen 2000).
McDonald–Kreitman test. McDonald & Kreitman (1991) proposed a two‐by‐two contingency test using the numbers of nonsynonymous and synonymous polymorphisms polymorphic within species and the numbers of nonsynonymous and synonymous differences between species. Under neutrality, the ratio of nonsynonymous to synonymous polymorphisms with species is expected to be the same as the ratio of nonsynonymous to synonymous differences between species (Sawyer & Hartl 1992). One striking finding that has emerged from using the test is that animal mtDNA genes generally have a greater number of nonsynonymous polymorphisms within species than expected compared to nonsynonymous divergence among species (e.g. Rand & Kann 1996), apparently due to weak selection against slightly deleterious nonsynonymous mutations (Nielsen & Weinreich 1999).
Acknowledgements
The author thanks Paul Moran, Robin Waples, Peter Kareiva and two anonymous reviewers for helpful comments on previous versions of the paper.
References
- Aguade M (1999) Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila . Genetics, 152, 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L (2001) Genetic dissection of phenotypic diversity in farm animals. Nature Genetics Reviews, 2, 130–138. [DOI] [PubMed] [Google Scholar]
- Andrews TD, Easteal S (2000) Evolutionary rate acceleration of cytochrome c oxidase subunit I in simian primates. Journal of Molecular Evolution, 50, 562–568. [DOI] [PubMed] [Google Scholar]
- Aquadro CF, Begun DJ, Kindahl EC (1994) Selection, recombination, and DNA polymorphism in Drosophila In: Non‐Neutral Evolution: Theories and Molecular Data (ed. Golding B.), pp. 46–56. Chapman & Hall, New York. [Google Scholar]
- Araki H, Inomata N, Yamazaki T (2001) Molecular evolution of duplicated amylase gene regions in Drosophila melanogaster: evidence of positive selection in the coding regions and selective constraints in the cis‐regulatory regions. Genetics, 157, 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC (1994) Molecular Markers, Natural History and Evolution. Chapman & Hall, New York. [Google Scholar]
- Avise JC (1995) Mitochondrial DNA polymorphism and a connection between genetics and demography of relevance to conservation. Conservation Biology, 9, 686–690. [Google Scholar]
- Ballingall KT, Waibochi L, Holmes EC et al. (2001) The CD45 locus in cattle: allelic polymorphism and evidence for exceptional positive natural selection. Immunogenetics, 52, 276–283. [DOI] [PubMed] [Google Scholar]
- Bargelloni L, Marcato S, Patarnello T (1998) Antarctic fish hemoglobins: evidence for adaptive evolution at subzero temperature. Proceedings of the National Academy of Science USA, 95, 8670–8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric RS, Yount B, Hensley L, Peel SA, Chen W (1997) Episodic evolution mediates interspecies transfer of a murine coronavirus. Journal of Virology, 71, 1946–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF (1992) Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster . Nature, 356, 519–520. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF (1994) Evolutionary inferences from DNA variation at the 6‐phosphogluconate dehydrogenase locus in natural populations of Drosophila: selection and geographic differentiation. Genetics, 136, 155–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF (1995) Molecular variation at the vermilion locus in geographically diverse populations of Drosophila melanogaster and D. simulans . Genetics, 140, 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Betancourt AJ, Langley CH, Stephan W (1999) Is the fast/slow allozyme variation at the Adh locus of Drosophila melanogaster an ancient balanced polymorphism? Molecular Biology and Evolution, 16, 1816–1819. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Whitley P (2000) Reduced X‐linked nucleotide polymorphism in Drosophila simulans . Proceedings of the National Academy of Science USA, 97, 5960–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D, Whitley P, Todd BL, Waldrip‐Dail HM, Clark AG (2000) Molecular population genetics of male accessory gland proteins in Drosophila. Genetics, 156, 1979–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson JB, Kreitman M, Stahl EA, Tian D (2001) Evolutionary dynamics of plant R‐genes. Science, 292, 2281–2285. [DOI] [PubMed] [Google Scholar]
- Berry A, Kreitman M (1993) Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics, 134, 869–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann CH (1998) The molecular evolution of sperm bindin in six species of sea urchins (Echinoida: Strongylocentrotidae). Molecular Biology and Evolution, 15, 1761–1771. [DOI] [PubMed] [Google Scholar]
- Bishop JG, Dean AM, Mitchell‐Olds T (2000) Rapid evolution in plant chitinases: molecular targets of natural selection in plant–pathogen coevolution. Proceedings of the National Academy of Sciences USA, 97, 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner‐Eddy PD, Crute IR, Holub EB, Beynon JL (2000) RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant Journal, 21, 177–188. [DOI] [PubMed] [Google Scholar]
- Botella MA, Parker JE, Frost LN et al. (1998) Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell, 10, 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr M, Soloway JR, Robinson TE, Brookfield JFY (2001) An investigation of the cause of low variability on the fourth chromosome of Drosophila melanogaster . Molecular Biology and Evolution, 18, 2260–2269. [DOI] [PubMed] [Google Scholar]
- Charlesworth B (1996) Background selection and patterns of Genetic diversity in Drosophilla melanogaster . Genetical Research, 68, 131–149. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, McVean GAT (2001) Genome sequences and evolutionary biology, a two‐way interaction. Trends in Ecology and Evolution, 16, 235–242. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D (1993) The effect of deleterious mutations on neutral molecular variation. Genetics, 134, 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirera S, Aguade M (1997) Evolutionary history of the sex‐peptide (Acp70A) gene region in Drosophila melanogaster . Genetics, 147, 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Kao T‐H (1991) Excess nonsynonymous substitution at shared polymorphic sites among self‐incompatibility alleles of Solanaceae. Proceedings of the National Academy of Sciences USA, 88, 9823–9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Wicker‐Thomas C, Jallon JM (1999) A gene responsible for a cuticular hydrocarbon polymorphism in Drosophila melanogaster . Genetical Research, 73, 189–203. [DOI] [PubMed] [Google Scholar]
- Dallerac R, Labeur C, Jallon JM, Knipple DC, Roelofs WL, Wicker‐Thomas C (2000) A delta 9 desaturase gene with a different substrate specificity is responsible for the cuticular diene hydrocarbon polymorphism in Drosophila melanogaster . Proceedings of the National Academy of Science USA, 17, 9449–9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Gustus C (1995) Teosinte branched 1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics, 141, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature, 386, 485–488. [DOI] [PubMed] [Google Scholar]
- Duda TF Jr, Palumbi SR (1999) Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus . Proceedings of the National Academy of Science USA, 96, 6820–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes WF (1999) Analysis of selection on enzyme polymorphisms. Annual Review of Ecology and Systematics, 30, 301–326. [Google Scholar]
- Eanes WF, Kirchner M, Yoon J (1993) Evidence for adaptive evolution of the G6pd gene in the Drosophila melanogaster and Drosophila simulans lineages. Proceedings of the National Academy of Sciences USA, 90, 7475–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes WF, Kirchner M, Yoon J et al. (1996) Historical selection, amino acid polymorphism and lineage‐specific divergence at the G6pd locus in Drosophila melanogaster and D. simulans . Genetics, 144, 1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler JA (1986) Natural Selection in the Wild. Princeton University Press, Princeton, New Jersey. [Google Scholar]
- Endo T, Ikeo K, Gojobori T (1996) Large‐scale search for genes on which positive selection may operate. Molecular Biology and Evolution, 13, 685–690. [DOI] [PubMed] [Google Scholar]
- Ewens WJ (1972) The sampling theory of selectively neutral alleles. Theoretical Population Biology, 3, 87–112. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu C‐I (2000) Hitchhiking under positive Darwinian selection. Genetics, 155, 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu C‐I (2001) Positive and negative selection on the human genome. Genetics, 158, 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferveur JF, Cobb M, Boukella H, Jallon JM (1996) World‐wide variation in Drosophila melanogaster sex pheromone: behavioral effects, genetic bases and potential evolutionary consequences. Genetica, 97, 73–80. [DOI] [PubMed] [Google Scholar]
- Fitch WM, Bush RM, Bender CA, Cox NJ (1997) Long term trends in the evolution of H (3) HA1 human influenza type A. Proceedings of the National Academy of Sciences USA, 94, 7712–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM, Bush RM, Bender CA, Subbarao K, Cox NJ (2000) Predicting the evolution of human influenza A. The Journal of Heredity, 91, 183–185. [DOI] [PubMed] [Google Scholar]
- Flint J, Mott R (2001) Finding the molecular basis of quantitative traits: successes and pitfalls. Nature Genetics Reviews, 2, 437–445. [DOI] [PubMed] [Google Scholar]
- Ford MJ (1998) Testing models of migration and isolation among populations of chinook salmon (Oncorhynchus tschawytscha). Evolution, 52, 539–557. [DOI] [PubMed] [Google Scholar]
- Ford MJ (2000) Effects of natural selection on patterns of DNA sequence variation at the transferrin, somatolactin, and p53 genes within and among chinook salmon (Oncorhynchus tshawytscha) populations. Molecular Ecology, 9, 843–855. [DOI] [PubMed] [Google Scholar]
- Ford MJ (2001) Molecular evolution of transferrin: evidence for positive selection in Salmonids. Molecular Biology and Evolution, 18, 639–647. [DOI] [PubMed] [Google Scholar]
- Ford MJ, Aquadro CF (1996) Selection on X‐linked genes during speciation in the Drosophila athabasca complex. Genetics, 144, 689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MJ, Thornton PJ, Park LK (1999) Natural selection promotes divergence of transferrin among salmonid species. Molecular Ecology, 8, 1055–1061. [DOI] [PubMed] [Google Scholar]
- Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147, 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Segre D, Skorecki K, Nachman MW, Lancet D, Sharon D (2000) Dichotomy of single‐nucleotide polymorphism haplotypes in olfactory receptor genes and pseudogenes. Nature Genetics, 26, 221–224. [DOI] [PubMed] [Google Scholar]
- Gillespie JH (1991) The Causes of Molecular Evolution. Oxford University Press, New York. [Google Scholar]
- Gillespie JH (2000) The neutral theory in an infinite population. Gene, 261, 11–18. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z (1994) A codon‐based model of nucleotide substitution for protein‐coding DNA sequences. Molecular Biology and Evolution, 11, 725–736. [DOI] [PubMed] [Google Scholar]
- Goodwin RL, Baumann H, Berger FG (1996) Patterns of divergence during evolution of alpha 1‐proteinase inhibitors in mammals. Molecular Biology and Evolution, 13, 346–358. [DOI] [PubMed] [Google Scholar]
- Haag ES, True JR (2001) Perspective: from mutants to mechanisms? Assessing the candidate gene paradigm in evolutionary biology. Evolution, 55, 1077–1084. [DOI] [PubMed] [Google Scholar]
- Hamblin MT, Aquadro CF (1997) Contrasting patterns of nucleotide sequence variation at the glucose dehydrogenase (Gld) locus in different populations of Drosophila melanogaster . Genetics, 145, 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H (1966) Enzyme polymorphism in man. Proceedings of the Royal Society Series B, 164, 298–310. [DOI] [PubMed] [Google Scholar]
- Hasson E, Wang I‐N, Zeng L‐W, Kreitman M, Eanes WF (1998) Nucleotide variation in the triosephosphate isomerase (Tpi) locus of Drosophila melanogaster and Drosophila simulans . Molecular Biology and Evolution, 15, 756–769. [DOI] [PubMed] [Google Scholar]
- Haydon DT, Bastos AD, Knowles NJ, Samuel AR (2001) Evidence for positive selection in foot‐and‐mouth disease virus capsid genes from field isolates. Genetics, 157, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW, Lee RN, Parker KM (2000) Major histocompatibility complex (MHC) variation in the endangered Mexican wolf and related canids. Heredity, 85, 617–624. [DOI] [PubMed] [Google Scholar]
- Hellberg ME, Moy GW, Vacquier VD (2000) Positive selection and propeptide repeats promote rapid interspecific divergence of a gastropod sperm protein. Molecular Biology and Evolution, 17, 458–466. [DOI] [PubMed] [Google Scholar]
- Hellberg ME, Vacquier VD (1997) Rapid evolution of fertilization selectivity and lysin cDNA sequences in teguline gastropods. Molecular Biology and Evolution, 16, 839–848. [DOI] [PubMed] [Google Scholar]
- Hey J (1999) The neutralist, the fly and the selectionist. Trends in Ecology and Evolution, 14, 35–38. [DOI] [PubMed] [Google Scholar]
- Hill RE, Hastie ND (1987) Accelerated evolution in the reactive centre regions of serince protease inhibitors. Nature, 326, 96–99. [DOI] [PubMed] [Google Scholar]
- Hilton H, Gaut B (1998) Speciation and domestication in maize and its wild relatives. Evidence from the globulin‐1 gene. Genetics, 150, 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook JD, Birdsey GM, Yang Z, Bruford MW, Danpure CJ (2000) Molecular adaptation of alanine: glyoxylate aminotransferase targeting in primates. Molecular Biology and Evolution, 17, 387–400. [DOI] [PubMed] [Google Scholar]
- Holub EB (2001) The arms race is ancient history in Arabidopsis, the wildflower. Nature Genetics Reviews, 2, 516–527. [DOI] [PubMed] [Google Scholar]
- Huang C‐H, Liu Z, Apoil P‐A, Blancher A (2000) Sequence, organization, and evolution of Rh50 glycoprotein genes in nonhuman primates. Journal of Molecular Evolution, 51, 76–87. [DOI] [PubMed] [Google Scholar]
- Hudson RR (1994) How can the low levels of DNA sequence variation in regions of the Drosophila genome with low recombination rates be explained? Proceedings of the National Academy of Science USA, 91, 6815–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics, 116, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Saez AG, Ayala FJ (1997) DNA variation at the Sod locus of Drosophila melanogaster: an unfolding story of natural selection. Proceedings of the National Academy of Sciences USA, 94, 7725–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL (1991) Circumsporozoite protein genes of malaria parasites (Plasmodium spp.): evidence for positive selection on immunogenic regions. Genetics, 127, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL (1992) Positive selection and interallelic recombination at the merozoite surface antigen‐1 (MSA‐1) locus of Plasmodium falciparum . Molecular Biology and Evolution, 9, 381–393. [DOI] [PubMed] [Google Scholar]
- Hughes AL (1995) The evolution of the type I interferon gene family in mammals. Journal of Molecular Evolution, 41, 539–548. [DOI] [PubMed] [Google Scholar]
- Hughes AL (1999) Adaptive Evolution of Genes and Genomes. Oxford University Press, New York. [Google Scholar]
- Hughes MK, Hughes AL (1995) Natural selection on Plasmodium surface proteins. Molecular and Biochemical Parasitology, 71, 99–113. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Nei M (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature, 335, 167–170. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Ota T, Nei M (1990) Positive Darwinian selection promotes charge profile diversity in the antigen‐binding cleft of class I major‐histocompatibility‐complex molecules. Molecular Biology and Evolution, 7, 515–524. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Yeager M (1997) Coordinated amino acid changes in the evolution of mammalian defensins. Journal of Molecular Evolution, 44, 675–682. [DOI] [PubMed] [Google Scholar]
- Ishimizu T, Endo T, Yamaguchi‐Kabata Y, Nakamura KT, Sakiyama F, Norioka S (1998) Identification of regions in which positive selection may operate in S‐RNase of Rosaceae: implication for S‐allele‐specific recognition sites in S‐RNase. FEBS Letters, 440, 337–342. [DOI] [PubMed] [Google Scholar]
- Karn RC, Nachman MW (1999) Reduced nucleotide variability at an androgen‐binding protein locus (Abpa) in house mice: evidence for positive natural selection. Molecular Biology and Evolution, 16, 1192–1197. [DOI] [PubMed] [Google Scholar]
- Katz LA, Harrison RG (1997) Balancing selection on electrophoretic variation of phosphoglucose isomerase in two species of field cricket Gryllus veletis and G. pennsylvanicus . Genetics, 147, 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer M, Zangerl B, Dieringer D, Schlötterer C (2002) Chromosomal patterns of microsatellite variability contrast sharply in African and non‐African populations of Drosophila melanogaster . Genetics, 160, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A, Yamane K, Miyashita N (2000) DNA polymorphism at the cytosolic phosphoglucose isomerase (PgiC) locus of the wild plant Arapidopsis thaliana . Genetics, 156, 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Stephan W (2000) Joint effects of genetics hitchhiking and background selection on neutral variation. Genetics, 155, 1415–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M (1983) The Neutral Theory of Molecular Evolution. Cambridge University Press, New York. [Google Scholar]
- Kirby DA, Stephan W (1995) Haplotype test reveals departure from neutrality in a segment of the white gene of Drosophila melanogaster . Genetics, 14, 1483–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby DA, Stephan W (1996) Multi‐locus selection and the structure of variation at the white gene of Drosophila melanogaster . Genetics, 144, 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordiš D, Gubenšek F (2000) Adaptive evolution of animal toxin multigene families. Gene, 261, 43–52. [DOI] [PubMed] [Google Scholar]
- Kraft T, Säll T, Magnusson‐Rading IM, Nilsson N‐O, Halldén C (1998) Positive correlation between recombination rates and levels of genetics variation in the natural populations of sea beet (Beta vulgaris subsp. maritima). Genetics, 150, 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M (1996) The neutral theory is dead. Long live the neutral theory. Bioessays, 18, 678–683. [DOI] [PubMed] [Google Scholar]
- Kreitman M (2000) Methods to detect selection in populations with applications to the human. Annual Review of Genomics and Human Genetics, 1, 539–559. [DOI] [PubMed] [Google Scholar]
- Kreitman M, Akashi H (1995) Molecular evidence for natural selection. Annual Review of Ecology and Systematics, 26, 403–422. [Google Scholar]
- Kreitman M, Hudson RR (1991) Inferring the evolutionary histories of Adh and Adh‐dup loci in Drosophila melanogaster from patterns of polymorphism and divergence. Genetics, 127, 565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara T, Sakuma M, Gojobori T (1997) Molecular evolution of myelin proteolipid protein. Biochemical and Biophysical Research Communications, 237, 559–561. [DOI] [PubMed] [Google Scholar]
- Lee YH, Vacquier VD (1992) The divergence of species‐specific abalone sperm lysins is promoted by positive Darwinian selection. Biological Bulletin, 182, 97–104. [DOI] [PubMed] [Google Scholar]
- Lewontin RC (1974) The Genetic Basis of Evolutionary Change. Columbia University Press, New York. [Google Scholar]
- Lewontin RC (1991) Electrophoresis in the development of evolutionary genetics: milestone or millstone? Genetics, 128, 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, Hubby JL (1966) A molecular approach to the study of genetic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura . Genetics, 54, 595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, Krakauer J (1973) Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics, 75, 175–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Charlesworth D, Kreitman M (1999) The effect of mating system differences on nucleotide diversity at the phosphoglucose isomerase locus in the plant genus Leavenworthia. Genetics, 151, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Langley CH (1993) Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila . Science, 260, 91–95. [DOI] [PubMed] [Google Scholar]
- Lukens L, Doebley J (2001) Molecular evolution of the teosinte branched gene among maize and related grasses. Molecular Biology and Evolution, 18, 627–638. [DOI] [PubMed] [Google Scholar]
- Mackay TFC (2001) Quantitative trait loci in Drosophila . Nature Genetics Reviews, 2, 11–20. [DOI] [PubMed] [Google Scholar]
- McDonald JH (1996) Detecting non‐neutral heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Molecular Biology and Evolution, 13, 253–260. [DOI] [PubMed] [Google Scholar]
- McDonald JH (1998) Improved test for heterogeneity across a region of DNA sequence in the ratio of polymorphism to divergence. Molecular Biology and Evolution, 15, 377–384. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila . Nature, 354, 114–116. [DOI] [PubMed] [Google Scholar]
- McDowell JM, Dhandaydham M, Long TA et al. (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis . Plant Cell, 10, 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier W, Stewart CB (1997) Episodic adaptive evolution of primate lysozymes. Nature, 385, 151–154. [DOI] [PubMed] [Google Scholar]
- Metz EC, Palumbi SR (1996) Positive selection and sequence rearrangements generate extensive polymorphism in the gamete recognition protein bindin. Molecular Biology and Evolution, 13, 397–406. [DOI] [PubMed] [Google Scholar]
- Metz EC, Robles‐Sikisaka R, Vacquier VD (1998) Nonsynonymous substitution in abalone sperm fertilization genes exceeds substitution in introns and mitochondrial DNA. Proceedings of the National Academy of Science USA, 95, 10676–10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW (1998) Receptor‐like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell, 11, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikko S, Andersson L (1995) Low major histocompatibility complex class II diversity in European and North American moose. Proceedings of the National Academy of Sciences USA, 92, 4259–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Withler RE (1996) Sequence analysis of a polymorphic Mhc class II gene in Pacific salmon. Immunogenetics, 43, 337–351. [DOI] [PubMed] [Google Scholar]
- Miller KM, Withler RE, Beacham TD (1997) Molecular evolution at Mhc genes in two populations of chinook salmon Oncorhynchus tshawytscha . Molecular Ecology, 6, 937–954. [DOI] [PubMed] [Google Scholar]
- Mitton JB (1997) Selection in Natural Populations. Oxford University Press, New York. [Google Scholar]
- Nachman MW (2001) Single nucleotide polymorphisms and recombination rate in humans. Trends in Genetics, 17, 481–485. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Bauer VL, Crowell SL, Aquadro CF (1998) DNA variability and recombination rates at X‐linked loci in humans. Genetics, 150, 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngai J, Dowling MM, Buck L, Axel R, Chess A (1993) The family of genes encoding odorant receptors in the channel catfish. Cell, 72, 657–666. [DOI] [PubMed] [Google Scholar]
- Nielsen R (1997) The ratio of replacement to silent divergence and tests of neutrality. Journal of Evolutionary Biology, 10, 217–231. [Google Scholar]
- Nielsen R, Weinreich DM (1999) The age of nonsynonymous and synonymous mutations in animal mtDNA and implications for the mildly deleterious theory. Genetics, 153, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Yang Z (1998) Likelihood models for detecting positively selected amino acid sites and applications to the HIV‐1 envelope gene. Genetics, 148, 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notter DR (1999) The importance of genetic diversity in livestock populations of the future. Journal of Animal Science, 77, 61–69. [DOI] [PubMed] [Google Scholar]
- Palumbi SR (1999) All males are not created equal: fertility differences depend on gamete recognition polymorphisms in sea urchins. Proceedings of the National Academy of Sciences USA, 96, 12632–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Nachman MW (2000) Microsatellite variation and recombination rate in the human genome. Genetics, 156, 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel CL, Nereng KS, Ohgi KA et al. (2001) The genetic architecture of divergence between threespine stickleback species. Nature, 414, 901–905. [DOI] [PubMed] [Google Scholar]
- Pennisi E (2000) Stalking the wild mustard. Science, 290, 2055–2057. [DOI] [PubMed] [Google Scholar]
- Pogson GH (2001) Nucleotide polymorphism and natural selection at the Pantophysin (Pan I) locus in the Atlantic cod, Gadus morhua (L.). Genetics, 157, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson GH, Mesa KA, Boutilier RG (1995) Genetic population structure and gene flow in the Atlantic cod, Gadus morhua: a comparison of allozyme and nuclear RFLP loci. Genetics, 139, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA, Nguyen M, Demma JC, Viscidi RP (2000) Population genetics of the porB gene of Neisseria gonorrhoeae: different dynamics in different homology groups. Molecular Biology and Evolution, 17, 423–436. [DOI] [PubMed] [Google Scholar]
- Powers DA, Lauerman T, Crawford D, DiMichele L (1991) Genetic mechanisms for adapting to a changing environment. Annual Review of Genetics, 25, 629–659. [DOI] [PubMed] [Google Scholar]
- Powers DA, Schulte PM (1998) Evolutionary adaptation of gene structure and expression in natural populations in relation to a changing environment: a multidisciplinary approach to address the million‐year saga of a small fish. Journal of Experimental Zoology, 282, 71–94. [PubMed] [Google Scholar]
- Purugganan MD, Boyles AL, Suddith JI (2000) Variation and selection at the CAULIFLOWER floral homeotic gene accompanying the evolution of domesticated Brassica oleracea . Genetics, 155, 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD, Suddith JI (1998) Molecular population genetics of the Arabidopsis CAULIFLOWER regulatory gene: nonneutral evolution and naturally occurring variation in floral homeotic function. Proceedings of the Natural Academy of Science USA, 95, 8130–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi CF, Bonhomme F, Buckler‐white A et al. (1998) Molecular phylogeny of Fv1 . Mammalian Genome, 9, 1049–1055. [DOI] [PubMed] [Google Scholar]
- Rana BK, Hewett‐Emmett D, Jin L et al. (1999) High polymorphism at the human melanocortin 1 receptor locus. Genetics, 151, 1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM (1995) Neutrality tests of molecular markers and the connection between DNA polymorphism, demography, and conservation biology. Conservation Biology, 10, 665–671. [Google Scholar]
- Rand DM, Kann LM (1996) Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Molecular Biology and Evolution, 13, 735–748. [DOI] [PubMed] [Google Scholar]
- Riley MA (1993) Positive selection for colicin diversity in bacteria. Molecular Biology and Evolution, 10, 1048–1059. [DOI] [PubMed] [Google Scholar]
- Rooney AP, Zhang J (1999) Rapid evolution of a primate sperm protein: relaxation of functional constraint or positive Darwinian selection? Molecular Biology and Evolution, 16, 706–710. [DOI] [PubMed] [Google Scholar]
- Rooney AP, Zhang J, Nei M (2000) An unusual form of purifying selection in a sperm protein. Molecular Biology and Evolution, 17, 278–283. [DOI] [PubMed] [Google Scholar]
- Saez AG, Escalante R, Sastre L (2000) High DNA sequence variability at the α1 Na/K‐ATPase locus of Artemia franciscana (brine shrimp): polymorphism in a gene for salt‐resistance in a salt‐resistant organism. Molecular Biology and Evolution, 17, 235–250. [DOI] [PubMed] [Google Scholar]
- Salemi M, Desmyter J, Vandamme A‐M (2000) Temp and mode of human and simian T‐lymphotropic virus (HTLV/STLV) evolution revealed by analyses of full‐genome sequences. Molecular Biology and Evolution, 17, 374–386. [DOI] [PubMed] [Google Scholar]
- Sawyer SA, Hartl DL (1992) Population genetics of polymorphism and divergence. Genetics, 132, 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner P, Richards N, Wadhwa A et al. (2001) Episodic evolution of pyrin in primates: human mutations recapitulate ancestral amino acid states. Nature Genetics, 27, 318–321. [DOI] [PubMed] [Google Scholar]
- Schemske DW, Bradshaw HDJ (1999) Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus). Proceedings of the National Academy of Science USA, 96, 11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS, Duvernell DD, Eanes WF (2000) Adaptive evolution of a candidate gene for aging in Drosophila . Proceedings of the National Academy of Science USA, 97, 10861–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TR, Goodman M, Grossman LI (1999) Molecular evolution of the COX7A gene family in primates. Molecular Biology and Evolution, 16, 619–626. [DOI] [PubMed] [Google Scholar]
- Schug MD, Hutter CM, Noor MA, Aquadro CF (1998) Mutation and evolution of microsatellites in Drosophila melanogaster . Genetica, 102–103, 359–367. [PubMed] [Google Scholar]
- Schulenburg JHG, Hurst GDD, Huigens TME, Van Meer MMM, Jiggins FM, Majerus MEN (2000) Molecular evolution and phylogenetic utility of Wolbachia ftsZ and wsp gene sequences with special reference to the origin of male‐killing. Molecular Biology and Evolution, 17, 584–600. [DOI] [PubMed] [Google Scholar]
- Schulte PM, Glemet HE, Fiebig AA, Powers DA (2000) Adaptive variation in lactate dehydrogenase‐B gene expression: role of a stress‐responsive regulatory element. Proceedings of the National Academy of Science USA, 97, 6597–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte PM, Gomez‐Chiarri M, Powers DA (1997) Structural and functional differences in the promoter and 5′ flanking region of Ldh‐B within and between populations of the teleost Fundulus heteroclitus . Genetics, 145, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen KL, Churchill GA, Aquadro CF (1995) Properties of statistical tests of neutrality for DNA polymorphism data. Genetics, 141, 413–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J (1999) Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis . Nature, 400, 667–671. [DOI] [PubMed] [Google Scholar]
- Stephan W (1995) An improved method for estimating the rate of fixation of favourable mutations based on DNA polymorphism data. Molecular Biology and Evolution, 12, 959–962. [DOI] [PubMed] [Google Scholar]
- Stotz HU, Bishop JG, Bergmann CW et al. (2000) Identification of target amino acids that affect interactions of fungal polygalacturonases and the plant inhibitors. Physiological and Molecular Plant Pathology, 56, 117–130. [Google Scholar]
- Sutton KA, Wilkinson MF (1997) Rapid evolution of a homeodomain: evidence for positive selection. Journal of Molecular Evolution, 45, 579–588. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Gojobori T (1999) A method for detecting positive selection at single amino acid sites. Molecular Biology and Evolution, 16, 1315–1328. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nei M (2001) Reliabilities of parsimony‐based and likelihood‐based methods for detecting positive selection at single amino acid sites. Molecular Biology and Evolution, 18, 2179–2185. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Aquadro CF, Vacquir VD (2001a) Polymorphism in abalone fertilization proteins is consistent with the neutral evolution of the egg's receptor for lysin (VERL) and positive darwinian selection of sperm lysin. Molecular Biology and Evolution, 18, 376–383. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Clark AG, Waldrip‐Dail HM, Wolfner MF, Aquadro CF (2001c) Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila . Proceedings of the National Academy of Sciences USA, 98, 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KW, Irwin DM, Wilson AC (1991) Stomach lysozyme gene of the langur monkey: tests for convergence and positive selection. Journal of Molecular Evolution, 33, 418–425. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Yang Z, Wolfner MF, Aquadro CF (2001b) Positive darwinian selection drives the evolution of several female reproductive proteins in mammals. Proceedings of the National Academy of Science USA, 98, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical methods for testing the neutral mutation hypothesis by DNA polymorphisms. Genetics, 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Tsaur S‐C, Coyne JA, Wu C‐I (2001) The nucleotide changes governing cuticular hydrocarbon variation and their evolution in Drosophila melanogaster . Proceedings of the National Academy of Science USA, 98, 3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Nei M (1989) Positive Darwinian selection observed at the variable‐region genes of immunoglobulins. Molecular Biology and Evolution, 6, 447–459. [DOI] [PubMed] [Google Scholar]
- Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science, 277, 1063–1066. [DOI] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proceedings of the National Academy of Science USA, 98, 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C‐T, Tsaur SC, Wu M‐L, Wu C‐I (1998) A rapidly evolving homeobox at the site of a hybrid sterility gene. Science, 282, 1501–1504. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Varkonyi R, Cahinhinan N et al. (2001. ) Haplotype diversity and linkage diequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science, 293, 455–462 [DOI] [PubMed] [Google Scholar]
- Tsaur SC, Ting C‐T, Wu C‐I (1998) Positive selection driving the evolution of a gene of male reproduction, Acp26Aa, of Drosophila. II. Divergence versus polymorphism. Molecular Biology and Evolution, 15, 1040–1046. [DOI] [PubMed] [Google Scholar]
- Tsaur SC, Ting C‐T, Wu C‐I (2001) Sex in Drosophila mauritiana: a very high level of amino acid polymorphism in a male reproductive protein gene Acp26Aa . Molecular Biology and Evolution, 18, 22–26. [DOI] [PubMed] [Google Scholar]
- Tsaur SC, Wu C‐I (1997) Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila . Molecular Biology and Evolution, 14, 544–549. [DOI] [PubMed] [Google Scholar]
- Vacquier VD, Swanson WJ, Lee YH (1997) Positive Darwinian selection on two homologous fertilization proteins: what is the selective pressure driving their divergence? Journal of Molecular Evolution, 44, S15–S22. [DOI] [PubMed] [Google Scholar]
- Verrelli BC, Eanes WF (2000) Extensive amino acid polymorphism at the Pgm locus is consistent with adaptive protein evolution in Drosophila melanogaster . Genetics, 156, 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli BC, Eanes WF (2001) Clinal variation for amino acid polymorphisms at the Pgm locus in Drosophila melanogaster . Genetics, 157, 1649–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis M (1996) The molecular evolution of vertebrate growth hormones: a pattern of near‐stasis interrupted by sustained bursts of rapid change. Journal of Molecular Evolution, 43, 93–100. [DOI] [PubMed] [Google Scholar]
- Wang R‐L, Stec A, Hey J, Lukens L, Doebley J (1999) The limits of selection during maize domestication. Nature, 398, 236–239[correction, 410, 718]. [DOI] [PubMed] [Google Scholar]
- Wang W, Thornton K, Berry A, Long M (2002) Nucleotide variation along the Drosophila melanogaster fourth chromosome. Science, 295, 134–137. [DOI] [PubMed] [Google Scholar]
- Ward TJ, Honeycutt RL, Derr JN (1997) Nucleotide sequence evolution at the kappa‐casein locus: evidence for positive selection within the family Bovidae. Genetics, 147, 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt WB, Dean AM (2000) Molecular‐functional studies of adaptive genetic variation in prokaryotes and eukaryotes. Annual Review of Genetics, 34, 593–622. [DOI] [PubMed] [Google Scholar]
- Watterson GA (1978) The homozygosity test of neutrality. Genetics, 88, 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne ML, Contamine D, Kreitman M (1996) Molecular population genetics of ref(2)P, a locus which confers viral resistance in Drosophila . Molecular Biology and Evolution, 13, 191–199. [DOI] [PubMed] [Google Scholar]
- Wiehe T (1998) The effect of selective sweeps on the variance of the allele distribution of a linked multiallele locus: hitchhiking of microsatellites. Theoretical Population Biology, 53, 272–283. [DOI] [PubMed] [Google Scholar]
- Wu JC, Chiang TY, Shiue WK, Wang SY, Sheen IJ, Huang YH, Syu WJ (1999) Recombination of hepatitis D virus RNA sequences and its implications. Molecular Biology and Evolution, 16, 1622–1632. [DOI] [PubMed] [Google Scholar]
- Wu J, Saupe SJ, Glass NL (1998) Evidence for balancing selection operating at the het‐c heterokaryon incompatibility locus in a group of filamentous fungi. Proceedings of the National Academy of Sciences USA, 95, 12398–12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Green J, Bixby JB, Szafranska B, DeMartini JC, Hect S, Roberts RM (1997) The diversity and evolutionary relationships of the pregnancy‐associated glycoproteins, and aspartic proteinase subfamily consisting of many trophoblast‐expressed genes. Proceedings of the National Academy of Science USA, 94, 12809–12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular Biology and Evolution, 15, 568–573. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. Trends in Ecology and Evolution, 15, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Molecular Biology and Evolution, 17, 32–43. [DOI] [PubMed] [Google Scholar]
- Zanotto PMA, Kallas EG, Souza RF, Holmes EC (1999) Genealogical evidence for positive selection in the nef gene of HIV‐1. Genetics, 153, 1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelus D, Robinson‐Rechavi M, Delacre M, Auriault C, Laudet V (2000) Fast evolution of interleukin‐2 in mammals and positive selection in ruminants. Journal of Molecular Evolution, 51, 234–244. [DOI] [PubMed] [Google Scholar]
- Zhang J, Dyer KD, Rosenberg HF (2000) Evolution of the rodent eosinophil‐associated RNase gene family by rapid gene sorting and positive selection. Proceedings of the National Academy of Sciences USA, 97, 4701–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nei M (2000) Positive selection in the evolution of mammalian interleukin‐2 genes. Molecular Biology and Evolution, 17, 1413–1416. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF (2000) Sequence variation at two eosinophil‐associated ribonuclease loci in humans. Genetics, 156, 1949–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M (1998) Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proceedings of the National Academy of Sciences USA, 95, 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]


