Abstract
Scutellaria, or skullcaps, are medicinally important herbs in China, India, Japan, and elsewhere. Though Scutellaria is the second largest and one of the more taxonomically challenging genera within Lamiaceae, few molecular systematic studies have been undertaken within the genus; in part due to a paucity of available informative markers. The lack of informative molecular markers for Scutellaria hinders our ability to accurately and robustly reconstruct phylogenetic relationships, which hampers our understanding of the diversity, phylogeny, and evolutionary history of this cosmopolitan genus. Comparative analyses of 15 plastomes, representing 14 species of subfamily Scutellarioideae, indicate that plastomes within Scutellarioideae contain about 151,000 nucleotides, and possess a typical quadripartite structure. In total, 590 simple sequence repeats, 489 longer repeats, and 16 hyper-variable regions were identified from the 15 plastomes. Phylogenetic relationships among the 14 species representing four of the five genera of Scutellarioideae were resolved with high support values, but the current infrageneric classification of Scutellaria was not supported in all analyses. Complete plastome sequences provide better resolution at an interspecific level than using few to several plastid markers in phylogenetic reconstruction. The data presented here will serve as a foundation to facilitate DNA barcoding, species identification, and systematic research within Scutellaria, which is an important medicinal plant resource worldwide.
Introduction
Lamiaceae is the sixth largest angiosperm family and contains over 7000 species that are divided into 12 subfamilies [1, 2]. Scutellarioideae, while relatively small, is one of the most morphologically distinct subfamilies within Lamiaceae. As circumscribed in earlier classifications [3, 4], the subfamily contained only three genera, Scutellaria L., Perilomia Kunth, and Salazaria Torr., with the latter two genera synonymized with Scutellaria by Paton [5]. Subsequent studies based on morphological [6, 7] and molecular data [8, 9] expanded the subfamily to include Renschia Vatke, Tinnea Kotschy ex Hook. f., Holmskioldia Retz., and Wenchengia C. Y. Wu & S. Chow. Morphological synapomorphies for Scutellarioideae include pericarps with tuberculate or elongate processes [9], high densities of xylem fibers in the calyces [10], and racemose inflorescences (but most species of Tinnea and Holmskioldia have cymose inflorescences). The monophyly of the subfamily has also been supported by molecular phylogenetic studies [1, 8, 9, 11].
As currently defined, Scutellarioideae includes approximately 380 species in five genera [1]: Holmskioldia, Renschia, Wenchengia, Scutellaria, and Tinnea. The former three are monotypic genera. The genus Holmskioldia, comprising the single species H. sanguinea Retz., is native to the subtropical Himalayan region but is widely grown as an ornamental in warm climates and has become naturalized throughout the Old and New Worlds [12]. The monotypic Renschia, represented by R. heterotypica (S. Moore) Vatke, is narrowly endemic to the Ahl Mountains in northern Somalia [13], and its systematic position within Scutellarioideae remains unclear. The placement of Wenchengia in Scutellarioideae was resolved by Li et al. [9] based on the rediscovery of the extremely rare species, W. alternifolia C.Y. Wu & S. Chow. This genus was long thought to be endemic to Hainan Island in southern China [14, 15], but recently it was also reported from Vietnam [16]. With 19 species recognized to date, Tinnea is the second largest genus in Scutellarioideae, occurs mainly in fire-prone grassland, woodland, and scrub vegetation, and is endemic to Africa [17].
Scutellaria, containing approximately 360 species and commonly known as skullcaps, is the largest genus in Scutellarioideae [18]. The genus is distributed nearly worldwide and occurs in various habitats, but is mostly found in tropical montane and temperate regions [5, 19]. Most species are herbaceous perennials or small shrubs. The calyx of Scutellaria consists of two undivided lips and bears an appendage on the upper lip, which is described as a scutellum and is the most distinctive character of the genus; this feature is the basis for the common name skullcap. Many Scutellaria species possess medicinal uses, and some species are of economic importance. For example, S. baicalensis Georgi (baical skullcap or Chinese skullcap; ‘Huang-qin’ in Chinese) is a traditional Chinese medicinal herb that was first recorded in Shen Nong Ben Cao Jing in ca. 100 BC [20], and is widely used to treat hepatitis, jaundice, tumor, leukemia, hyperlipaemia, arteriosclerosis, diarrhea, and inflammatory diseases [21].
Due to tremendous diversity in habit, as well as calyx, corolla, inflorescence, and nutlet morphology, infrageneric boundaries within Scutellaria are poorly defined [3–5, 22–24]. Based on morphological data, Paton [5] subsumed Harlanlewisia Epling, Perilomia, and Salazaria into a broad Scutellaria as part of a global taxonomic revision, and divided Scutellaria into two subgenera: subg. Scutellaria and subg. Apeltanthus (Nevski ex Juz.) Juz. The former is further subdivided into five sections: Scutellaria, Salviifoliae (Boiss.) Edmondson, Salazaria (Torrey) Paton., Perilomia (Kunth) Epling, and Anaspi (Rech.f.) Paton. And the latter is divided into two sections: Apeltanthus and Lupulinaria A. Hamilt. As opposed to other large genera of Lamiaceae, such as Plectranthus L’Hér. [25–27], Salvia L. [28–34], and Isodon (Schrad. ex Benth.) Spach [35–38], molecular phylogenetic studies within Scutellaria are relatively scarce. Most previous work concentrated on genetic diversity and biogeography of taxonomic complexes (e.g. S. angustifolia Pursh [39, 40]), population genetics [21, 41], or species identification [42]. To date, only three phylogenetic studies have focused on Scutellaria [18, 41, 43]. Using both nuclear and chloroplast (cp) DNA markers, Chiang et al. [41] studied the relationships of Taiwanese Scutellaria and Safikhani et al. [18] focused on Iranian taxa. Similarly, when describing S. wuana C. L. Xiang & F. Zhao, only 41 taxa were involved in the phylogenetic analyses [43]. In total, only five DNA markers were used in these studies (nrITS, matK, ndhF-rpl32, rpl32-trnL, and trnL-trnF) and none generated phylogenetic trees with high resolution, ostensibly due to a lack of variability within these DNA markers among the sampled species.
The chloroplast is an essential organelle in angiosperms because it provides energy for plant cells [44]. This uniparentally inherited plastid is characterized by a circular double-stranded DNA molecule between 120,000–160,000 base pairs in length, multiple copies per cell, and a quadripartite structure that includes two identical regions in opposite orientations called the inverted repeat (IR), flanked by large single copy (LSC) and small single copy (SSC) regions [45]. With increasingly rapid and less expensive next generation sequencing (NGS) technologies continually developing, ever-increasing numbers of non-model species plastid genome are being sequenced and successfully used for resolving phylogenetic and taxonomic problems in flowering plants at various ranks [46–48]. However, using cp genomes to resolve phylogenetic questions within the mint family has been rare [49], and plastomes of only two species, Scutellaria baicalensis and S. indica L. var. coccinea S. Kim & S. Lee, have been published from Scutellarioideae [50, 51]. Sequences of S. insignis Nakai and S. lateriflora L. were uploaded to GenBank without any related publication or analyses. Consequently, little is known regarding plastome structure variation within Scutellaria.
In this study, we sequenced 12 plastomes from 11 species representing four of the five genera of Scutellarioideae. In addition, three previously released plastomes of Scutellaria (S. baicalensis, S. insignis and S. lateriflora) were downloaded from GenBank and included for comparative analyses. The species S. indica var. coccinea was exclude in this study because the sequence was unavailable. With these data, we aim to: 1) characterize and compare the structure and gene organization of plastid genomes within Scutellarioideae; 2) identify candidate molecular markers for future phylogenetic and/or population genetic studies within Scutellaria; and 3) reconstruct the phylogeny of Scutellarioideae using complete chloroplast genome sequences. The data presented in this study will provide abundant information for further studies about phylogeny, taxonomy, species identification, and population genetics of Scutellaria, and will also be helpful for exploration, utilization, and conservation of plant genetic resources of this important medicinal plant resources.
Materials and methods
Taxon sampling, DNA extraction, and sequencing
Plastomes of 12 samples, including eight species of Scutellaria, one species each of Holmskioldia and Tinnea, and two individuals of Wenchengia alternifolia, were newly generated for this study. Voucher information is listed in Table 1 and all voucher specimens were deposited at the Herbarium of Kunming Institute of Botany (KUN), Chinese Academy of Sciences. In addition, three complete plastomes of Scutellaria from GenBank, S. baicalensis (MF521633), S. insignis (KT750009), and S. lateriflora (KY085900), were included for comparative analyses (Table 1).
Table 1. Voucher information of the newly sequenced samples in this study.
| Species | Location | Vouchers | Coordinate |
|---|---|---|---|
| Wenchengia alternifolia C.Y. Wu & S. Chow HN | China, Hainan, Ding’an | Xiang et al. 1318 | E 110°17′50.19″, N 19°13′53.27″ |
| W. alternifolia C.Y. Wu & S. Chow VN | Vietnam, DaNang, Ba na Hill | Li et al. Lbo824 | E 107°59′17.91″, N 15°59′59.80″ |
| Holmskioldia sanguinea Retz. | China, Yunnan, XTBG* | Zhao et al. ZF014 | E 101°15′21.10″, N 21°55′38.06″ |
| Tinnea aethiopica Kotschy ex Hook. f. | Kenya, Kabarnet | Li et al. 4292 | E 35°44′36.30″, N 0°29′24.38″ |
| Scutellaria amoena var. amoena C.H. Wright | China, Yunnan, Kunming | Zhao et al. ZF034 | E 102°43′07.74″, N 25°07′19.91″ |
| S. calcarata C.Y. Wu & H.W. Li | China, Yunnan, Gongshan | Li et al. NJ023 | E 98°39′39.55″, N 27°44′26.32″ |
| S. mollifolia C.Y. Wu & H.W. Li | China, Sichuan, Emei | Chen et al. EM201 | E 103°20′01.25″, N 21°55′38.06″ |
| S. orthocalyx Hand.-Mazz. | China, Yunnan, KBG* | Zhao et al. ZF035 | E 102°44′38.26″, N 25°08′27.10″ |
| S. quadrilobulata Y.Z. Sun | China, Yunnan, Xinping | Li et al. XP965 | E 101°56′55.70″, N 23°56′51.32″ |
| S. kingiana Prain | China, Xizang, Cuona | Yang et al. ZJW-3890 | E 99°56′09.26″, N 28°05′18.95″ |
| S. altaica Fisch. ex Sweet | China, Xinjiang, Xinyuan | Zhang et al. 17CS16318 | E 84°02′23.18″, N 43°18′24.83″ |
| S. przewalskii Juz. | China, Xinjiang, Aletai | Chen et al. YC_ZX027 | E 88°02′42.31″, N 47°20′41.62″ |
*: XTBG: Xishaungbanna Tropical Botanical Garden; KBG: Kunming Botanical Garden.
Total genomic DNA was extracted from 150 mg fresh or silica-gel dried leaves using the CTAB method [52]. The DNA samples were sheared into fragments of about 300 bp to construct libraries according to manufacturer’s instructions (Illumina, San Diego, CA, USA). Paired-end (PE) sequencing of 150 bp was conducted on an Illumina Hiseq-2500 platform (Illumina Inc.) at BGI-Wuhan.
Quality control of raw sequence reads was carried out using FastQC toolkit (http://www.bioinformatics.babraham.ac.uk/projects/fastqc; [53]) with the parameter set as Q ≥ 25 to acquire high-quality clean reads for downstream analyses. De novo assembling of the plastomes was implemented in the GetOrganelle pipeline [54]. The filtered de Bruijn graphs file “gfa” was visualized in Bandage v. 0.8.1 [55] and the complete chloroplast sequence paths were manually selected, with the minimum depth of contigs above 100 × and the minimum length > 300 bp. Then all PE reads were mapped to the assembled plastomes using the Bowite2 [56] plugin in Geneious v.11.0.4 [57] to verify quality and correct assembly errors.
Plastome annotation was first performed using the online programs Dual Organellar Genome Annotator (DOGMA) [58] and Ge-seq [59]. We then inspected and curated all annotation manually with comparisons to the published plastome of S. baicalensis (MF521633) in Geneious v.11.0.4 [57]. The tRNAs were verified using the online tRNAscan-SE service with default parameters [60]. The resulting circular plastome maps were drawn using the OrganellarGenomeDRAW tool [61].
Characterization of simple sequence repeats and repeat structure
The simple sequence repeats (SSRs) in plastomes were identified using MISA perl script (http://pgrc.ipk-gatersleben.de/misa). Thresholds for the minimum repeated size were set as follows: ≥ 10 for mono-nucleotide, ≥ 5 for di-nucleotide, ≥ 4 for tri-nucleotide, and ≥ 3 for tetra-nucleotide, penta-nucleotide, and hexa-nucleotide repeats. The location and size of the repeating sequences (forward, reverse, palindromic and complement) were visualized in REPuter [62] with the parameter set as with a hamming distance of 3 and a minimum repeat size of 30 bp following the procedure outlined in Jiang et al. [50].
Comparative plastome and sequence divergence analysis
Comparative analyses of 15 plastomes of Scutellarioideae were carried out using the Mauve v.2.3.1 [63] plugin in Geneious v.11.0.4 [57]. We applied mVISTA [64] to visualize the results and evaluate the similarity among different plastomes, using default parameters to align plastomes under the LAGAN model and the annotations of S. baicalensis (MF521633) as a reference. In order to investigate the IR contraction or expansion, we also compared the boundaries between IR and SC regions in Geneious v.11.0.4 [57]. Two data sets (alignments of all 15 samples from Scutellarioideae and 11 species of Scutellaria) were used for the sliding window analysis to evaluate the intergeneric and intrageneric nucleotide sequence variabilities (Pi). Sequences were aligned using MAFFT v.7.221 [65] and misaligned regions were manually adjusted in Geneious v.11.0.4. [57]. DnaSP v.6 [66] was then used to calculate the Pi. The step size was set to 200 bp, with a 600 bp window length.
Phylogenetic analysis based on complete plastome sequences
In addition to the previously published plastomes of Scutellaria, plastomes of 31 species from within other subfamilies of Lamiaceae (12 Nepetoideae, 15 Lamioideae, two Ajugoideae, and one each from Premnoideae and Tectonoideae) were also included in the analyses to evaluate the utility of complete plastome sequences for resolving broad relationships within Scutellarioideae. Based on previous studies [1], Callicarpa americana (assembly from the WGS data under the SRR6940059) from Callicarpoideae was selected as the outgroup. GenBank accession numbers are provided in S1 Table.
Alignments were initially performed using MAFFT v.7.221 [65] with default settings, and subsequently manually adjusted in Geneious v.11.0.4 [57]. Ambiguously aligned regions (e.g. characters of uncertain homology among taxa and single-taxon insertions) were excluded before phylogenetic analyses. Since the plastid genome is uniparentally inherited and does not undergo recombination [67], we combined all sequences and constructed three matrices: (i) combined coding regions (dataset CR); (ii) combined non-coding regions (dataset NCR); (iii) combined whole plastome sequences (dataset CPG). In order to reduce the overrepresentation of duplicated sequences, only the IRa region was included in all data sets. In addition, in order to evaluate the efficacy of the complete plastome sequences for phylogeny reconstruction within Scutellarioideae, we also created two additional datasets for phylogenetic analyses and comparison. One was a combined dataset of hyper-variable regions (16VAR) detected in this study, the other dataset consisted of six commonly used DNA regions (6CP) from previous studies [9, 41, 68].
Maximum likelihood (ML) and Bayesian inference (BI) analyses were performed on the Cyberinfrastructure for Phylogenetic Research Science (CIPRES) Gateway (http://www.phylo.org/; [69]. ML analyses were conducted using RAxML HPC2 v.8.2.9.0 [70] with the general time reversible (GTR) + G model and 1000 bootstrap replicates. BI analyses were carried out using MrBayes v.3.2.6 [71]. The best substitution model for each data set was determined using jModelTest2 [72] on the CIPRES Gateway, under the Bayesian information criterion (BIC) [73]. Four Markov Chain Monte Carlo (MCMC) chains (one cold and three heated) were run for 20 million generations. Convergence of the MCMC runs and estimated sample size (ESS) were analyzed by Tracer v.1.7.0 [74]. The first 25% of trees discarded as burn-in, and the remaining trees were summarized to construct the 50% majority-rule consensus tree.
Results
Genome assembly, features, and gene content across scutellarioideae
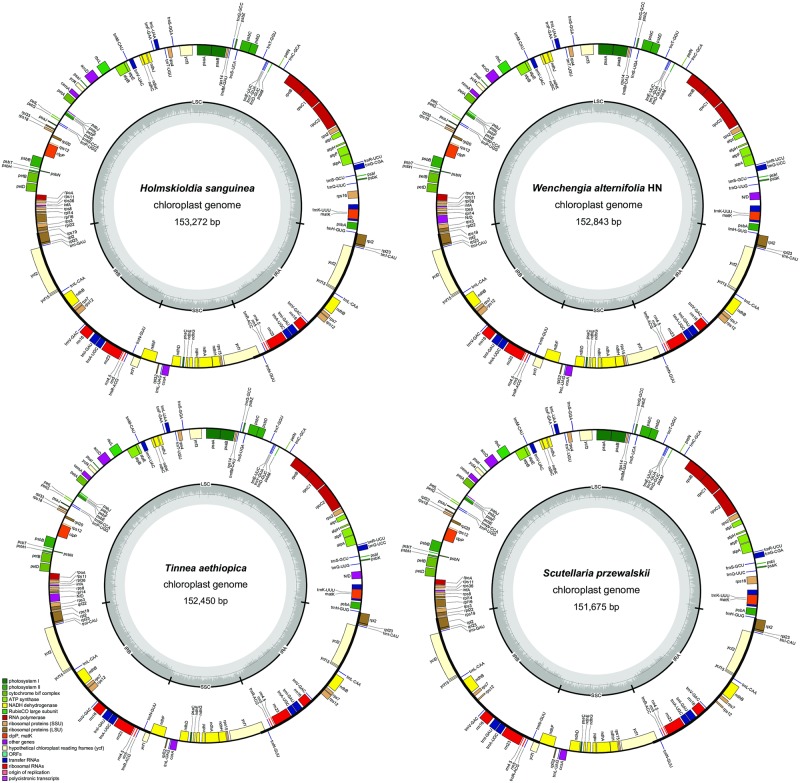
Illumina paired-end sequencing generated 16,687,912–27,007,418 clean reads for the 12 newly sequenced samples, with the mean coverage ranging from 618× in Scutellaria altaica Fisch. ex Sweet to 4510× in S. kingiana Prain. The genome size ranged from 151,675 bp in S. przewalskii Juz. to 153,272 bp in Holmskioldia sanguinea (Table 2). All 15 plastomes of Scutellarioideae displayed the typical quadripartite structure consisting of a pair of IR regions (25,208–25,634 bp) separated by the LSC (83,891–84,807bp) and SSC (16,750–17,569 bp) regions (Table 2). The GC content was similar among different species of Scutellarioideae and the average GC content was 38.3% (Table 2). In general, the GC content in the IR regions (43.4–43.6%) was higher than in the LSC (36.3–36.5%) and SSC (32.4–32.8%) regions, and the GC content within non-coding regions (35.0%) was lower than within coding regions (40.5%).
Table 2. Features of the complete plastomes of 15 species of Scutellarioideae.
(NA means not available).
| Taxa | Accession number | Complete | LSC | SSC | IR | Assembly Reads | Mean coverage (×) | Genes number | Protein coding genes | tRNA genes | rRNA genes | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | GC content (%) | Length (bp) | GC content (%) | Length (bp) | GC content (%) | Length (bp) | GC content (%) | ||||||||
| Holmskioldia sanguinea | MN128389 | 153,272 | 38.20 | 84,688 | 36.30 | 17,330 | 32.50 | 25,627 | 43.40 | 27,007,418 | 1655 | 114 | 80 | 30 | 4 |
| Wenchengia alternifolia HN | MN128379 | 152,843 | 38.30 | 84,807 | 36.30 | 16,768 | 32.80 | 25,634 | 43.40 | 21,196,548 | 1943 | 114 | 80 | 30 | 4 |
| W. alternifolia VN | MN128378 | 152,171 | 38.30 | 84,329 | 36.30 | 16,750 | 32.70 | 25,546 | 43.40 | 24,616,880 | 3068 | 114 | 80 | 30 | 4 |
| Tinnea aethiopica | MN128380 | 152,450 | 38.40 | 84,414 | 36.40 | 17,482 | 32.60 | 25,277 | 43.60 | 25,813,343 | 1612 | 114 | 80 | 30 | 4 |
| Scutellaria altaica | MN128387 | 151,779 | 38.30 | 83,984 | 36.30 | 17,327 | 32.60 | 25,234 | 43.60 | 24,309,408 | 618 | 114 | 80 | 30 | 4 |
| S. amoena var. amoena | MN128386 | 151,833 | 38.30 | 84,001 | 36.30 | 17,340 | 32.70 | 25,246 | 43.60 | 21,404,966 | 2893 | 114 | 80 | 30 | 4 |
| S. calcarata | MN128385 | 152,033 | 38.40 | 84,023 | 36.40 | 17,532 | 32.60 | 25,239 | 43.60 | 24,676,179 | 3257 | 114 | 80 | 30 | 4 |
| S. kingiana | MN128388 | 152,395 | 38.30 | 84,608 | 36.30 | 17,305 | 32.40 | 25,241 | 43.60 | 22,854,411 | 4510 | 114 | 80 | 30 | 4 |
| S. mollifolia | MN128384 | 152,417 | 38.30 | 84,432 | 36.40 | 17,569 | 32.60 | 25,208 | 43.60 | 25,602,286 | 1755 | 114 | 80 | 30 | 4 |
| S. orthocalyx | MN128383 | 152,071 | 38.40 | 84,072 | 36.40 | 17,519 | 32.60 | 25,240 | 43.60 | 16,687,912 | 1577 | 114 | 80 | 30 | 4 |
| S. przewalskii | MN128382 | 151,675 | 38.30 | 83,891 | 36.40 | 17,320 | 32.60 | 25,232 | 43.60 | 24,156,080 | 1496 | 114 | 80 | 30 | 4 |
| S. quadrilobulata | MN128381 | 152,066 | 38.30 | 84,052 | 36.40 | 17,544 | 32.50 | 25,235 | 43.60 | 25,241,479 | 2740 | 114 | 80 | 30 | 4 |
| S. baicalensis | MF521633 | 151,817 | 38.30 | 83,960 | 36.30 | 17,331 | 32.70 | 25,263 | 43.60 | NA | NA | 114 | 80 | 30 | 4 |
| S. insignis | KT750009 | 151,908 | 38.40 | 83,913 | 36.50 | 17,517 | 32.60 | 25,239 | 43.60 | NA | NA | 114 | 80 | 30 | 4 |
| S. lateriflora | KY085900 | 152,283 | 38.30 | 84,340 | 36.30 | 17,465 | 32.50 | 25,239 | 43.60 | NA | NA | 114 | 80 | 30 | 4 |
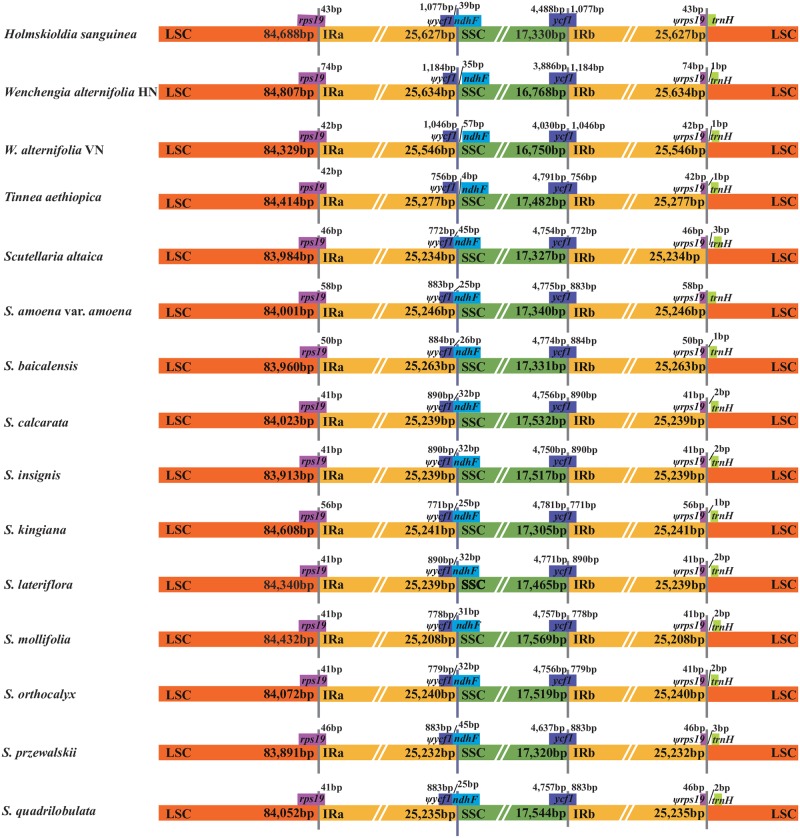
Intraspecific plastome polymorphisms can be evaluated among multiple individuals from the same species. The sequence identity between the two samples of Wenchengia alternifolia was 98.6%, with only two large indels (> 100 bp), within the intergenic psbE-petL (344 bp) and psbM-trnD (GUC) (226 bp) regions, detected. The plastome maps of Holmskioldia sanguinea, W. alternifolia HN, Tinnea aethiopica, and Scutellaria przewalskii are presented as representatives of Scutellarioideae (Fig 1), while maps of the remaining species are provided in supplementary materials (S1 Fig). All newly sequenced and annotated plastomes were submitted to the National Center for Biotechnology Information (NCBI) database under accession numbers MN128378–MN128389 (Table 2).
Fig 1. Complete plastome maps of Holmskioldia sanguinea, Wenchengia alternifolia, Tinnea aethiopica, and Scutellaria przewalskii.
When duplicated genes in IR regions were counted only once, each of the plastomes included 114 unique genes (80 protein-coding genes, 30 tRNAs and four rRNAs; Table 2) that were arranged in the same order. A total of 18 genes exist in duplication within the IR region, including seven protein-coding genes, seven tRNAs and four rRNAs (Table 3). Ten of the protein-coding genes and six of the tRNA genes contained one intron, and two genes (ycf3 and clpP) contained two introns. Among those newly sequenced samples, protein-coding regions accounted for 52.1–53.5% of the length of the whole genome, while tRNA and rRNA regions accounted for 1.78–1.92% and 5.9–5.96%, respectively (S2 Table). The remaining regions were non-coding sequences, including intergenic spacers, introns, and pseudogenes. All of the gene functions and groups were shown in Table 3.
Table 3. The gene functions of the plastomes of 15 species of Scutellarioideae.
| Category for genes | Group of genes | Name of genes |
|---|---|---|
| Photosynthesis | Subunits of NADH-dehydrogenase | ndhA*, ndhB*(2x), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3** | |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| Large chain of rubisco | rbcL | |
| Self-replication | Ribosomal RNA genes | rrn16 (2x), rrn23 (2x), rrn4.5 (2x), rrn5 (2x) |
| Transfer RNA genes 30 tRNA genes | (6 contain one intron, 7 are duplicated in the IR region) | |
|
trnA-UGC*(2x), trnfM-CAU, trnI-GAU*(2x), trnM-CAU, trnR-ACG(2x), trnS-UGA, trnC-GCA, trnG-GCC*, trnK-UUU*, trnN-GUU(2x), trnW-CCA, trnT-GGU, trnD-GUC, trnG-UCC, trnL-CAA(2x), trnY-GUA, trnR-UCU, trnT-UGU, trnE-UUC, trnH-GUG, trnL-UAA*, trnP-UGG, trnS-GCU, trnV-GAC(2x), trnF-GAA, trnI-CAU(2x), trnL-UAG, trnQ-UUG, trnS-GGA, trnV-UAC* |
||
| Small subunit of ribosome | rps2, rps3, rps4, rps7 (2x), rps8, rps11, rps12, rps14, rps15, rps16*, rps18, rps19 | |
| Large subunit of ribosome | rpl2* (2x), rpl14, rpl16*, rpl20, rpl22, rpl23 (2x), rpl32, rpl33, rpl36 | |
| RNA polymerase subunits | rpoA, rpoB, rpoC1*, rpoC2 | |
| Other genes | Translation initiation factor | infA |
| Maturase | matK | |
| Protease | clpP** | |
| Envelope membrane protein | cemA | |
| Subunit of acetyl-CoA-carboxylase | accD | |
| cytochrome c biogenesis protein | ccsA | |
| Component of TIC complex | ycf1 | |
| Genes of unknown function | ycf2, ycf4, ycf15 (2x) | |
*gene with a single intron,
**gene with two introns, (2x) duplicated gene.
SSRs and repeat structure
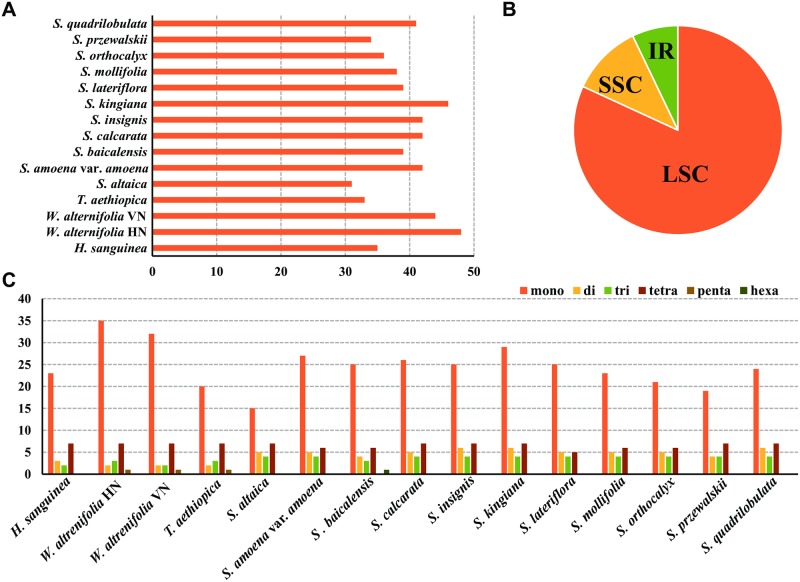
In total, 590 SSRs were identified in the 15 plastomes of Scutellarioideae, of which 483 SSRs (81.86%) were in the LSC region, 65 SSRs (11.02%) were in the SSC region, and 42 SSRs (7.12%) were in the IR region (Fig 2, S3 Table). The number of SSRs (or microsatellite loci) ranged from 31 (Scutellaria altaica) to 48 (Wenchengia alternifolia HN) among species of Scutellarioideae (Fig 2). The mononucleotide represents the highest variability with the repeat number ranging from 15 (S. altaica) to 35 (W. alternifolia HN), while the number of dinucleotide, trinucleotide, and tetranucleotide repeats showed no significant difference among the 15 samples. The number and frequency of each repeat type within the 15 plastomes of Scutellarioideae is shown in Fig 2 and S3 Table.
Fig 2. Comparisons of the simple sequence repeats (SSR) among the 15 plastomes of Scutellarioideae.
(A) Number of SSRs detected in each plastome; (B) Frequencies of identified SSRs in LSC, IR, and SSC regions; (C) Number of SSR types detected in each plastome.
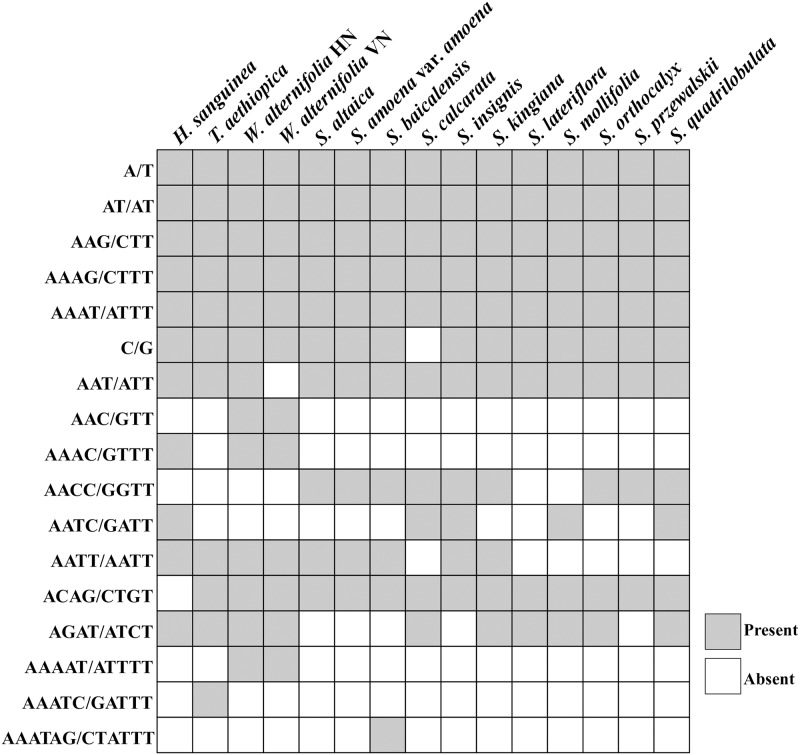
When the cyclic queues and reverse complements were regarded as the same SSRs, the 590 SSRs can be classified into 17 different repeat types. The mononucleotide repeat unit (A/T); dinucleotide repeat unit (AT/AT), trinucleotide repeats unit (AAG/CTT) and tetranucleotide repeat unit (AAAG/CTTT, AAAT/ATTT) were shared in all the 15 samples (Fig 3). The mononucleotide repeat unit (G/C) was absent in Scutellaria calcarata. Within the trinucleotide repeat, the repeat unit (AAC/GTT) was unique to Wenchengia, and the repeat unit (AAT/ATT) was shared by the other samples except the Wenchengia alternifolia VN accession. The tetranucleotide repeats showed the most polymorphisms, the repeat unit (AAAC/GTTT) were shared with Holmskioldia and the two samples of Wenchengia; the repeat unit (AACC/GGTT) was detected in nine species from Scutellaria; the repeat unit (AATC/GATT) was found in Holmskioldia and four Scutellaria species (S. calcarata, S. insignis, S. mollifolia and S. quadrilobulata); the repeat unit (AATT/AATT) was not found in S. calcarata, S. lateriflora, S. mollifolia, S. orthocalyx and S. quadrilobulata. The repeat unit (ACAG/CTGT) was shared by other species excluding the Holmskioldia, and repeat unit (AGAT/ATCT) didn’t present in S. altaica, S. amoena var. amoena, S. baicalensis, S. insignis and S. przewalskii. The pentanucleotide repeats were detected in both individuals of W. alternifolia and in Tinnea aethiopica, while the hexanucleotide repeats were only found in S. baicalensis. The distribution of the 17 repeat types among the 15 plastomes and their relationships is shown in Fig 3.
Fig 3. Distribution of the 17 types of SSR repeat units among 15 plastomes of Scutellarioideae and their relationships.
The horizontal axis indicates the species name and the Y-scale indicates the type of repeat unit.
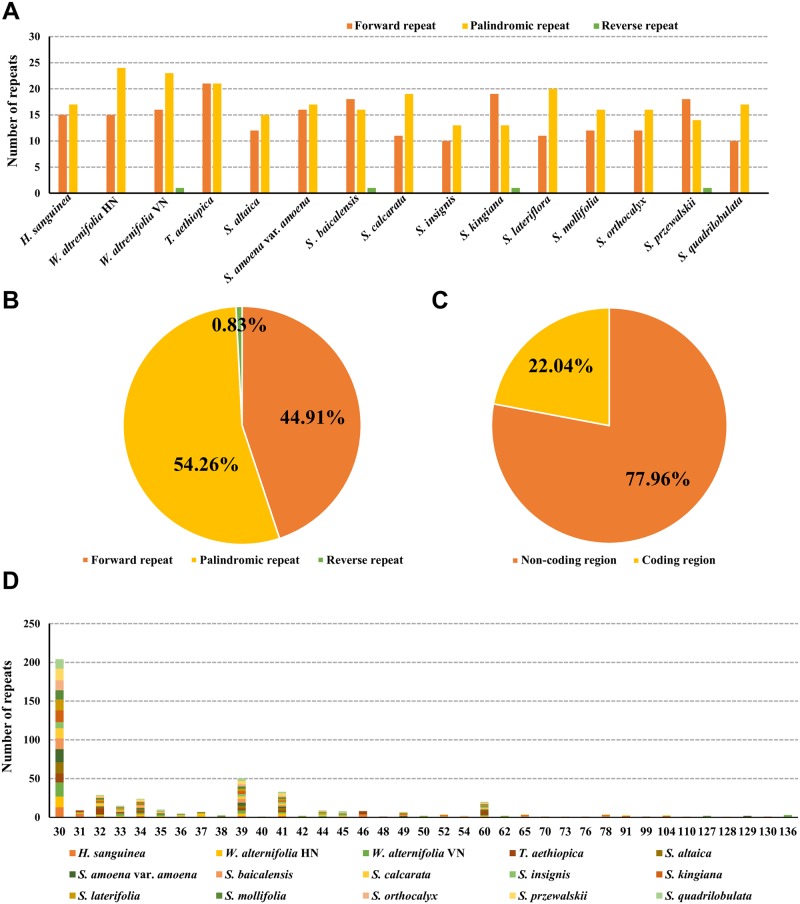
In total, 489 long repeats including forward, reverse, and palindromic were detected in the 15 plastomes (Fig 4). The most abundant type were the palindromic repeats, which accounted for 54.26% of the total repeats, followed by forward repeats (44.91%). The reverse repeats were rare and accounted for only 0.83% of the total repeats (Fig 4). Most repeats were located in the non-coding regions (77.96%; Fig 4). The length of the repeats ranged from 30 bp to 136 bp, and most of the repeat sequences were 30 bp, 32 bp, 39 bp, 41 bp, and 60 bp long (Fig 4, S4 Table).
Fig 4. Long repeat sequences in the complete plastomes of 15 taxa of Scutellarioideae.
(A) Number of repeat types detected in each plastome; (B) Frequency of each repeat type; (C) Percentages of repeat type loci in the non-coding and coding regions; (D) Frequencies of repeats longer than 30 bp.
Comparative analysis of plastomes of Scutellarioideae
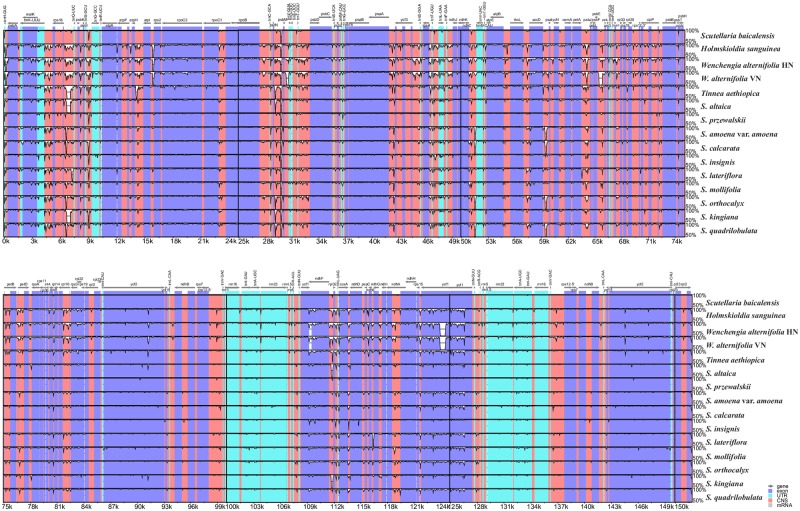
The Mauve results showed that the organization of the plastomes in Scutellarioideae is highly conserved; neither translocations nor inversions were detected. However, differences in the size of the plastomes were detected. For example, the plastome of Scutellaria przewalskii was the shortest (151,675 bp), while that of Holmskioldia sanguinea (153,272 bp) was longer than the other species (S2 Fig). Results from the analyses by mVISTA showed that the two IR regions were less divergent than the LSC and SSC regions. Moreover, the non-coding regions and the intergenic spacers exhibited a higher divergence than the coding regions (Fig 5). In all species, the IRa/LSC junctions were located within the rps19 gene, with a 41–74 bp protrusion of the rps19 gene into the IRa region that resulted in a part of the rps19 gene (ψrps19) present in the IRb region. In Wenchengia alternifolia and Tinnea aethiopica, the ndhF gene was completely located in the SSC region while in H. sanguinea and all species of Scutellaria a small fragment of the ndhF gene extended into the IRa region with (29 bp in H. sanguinea and 25–45 bp among species of Scutellaria). The IRb/SSC boundary was within the ycf1 gene, with between 771 and 1,184 bp in the IRb region. An equal length ycf1 pseudogene (ψycf1) was detected in the IRa region. The IRb/LSC boundary was located between the pseudogene rps19 (ψrps19) and trnH-GUG across the 15 plastomes. The distance between trnH-GUG and the IRb/LSC boundary for all species varied from 0 to 3 bp (Fig 6).
Fig 5. Sequence alignment of the whole plastomes of 15 taxa of Scutellarioideae using the LAGAN alignment algorithm in mVISTA, with Scutellaria baicalensis as the reference.
The horizontal axis indicates the coordinates within the plastomes. The Y-scale indicates the percentage of identity, ranging from 50% to 100%. Genome regions are color coded as protein coding, trnA gene, rrnA gene, intron, mRNA, and conserved non-coding sequences.
Fig 6. Comparisons of the LSC, IR, and SSC borders of plastomes of Scutellaria and related genera.
Sequence divergence and nucleotide diversity
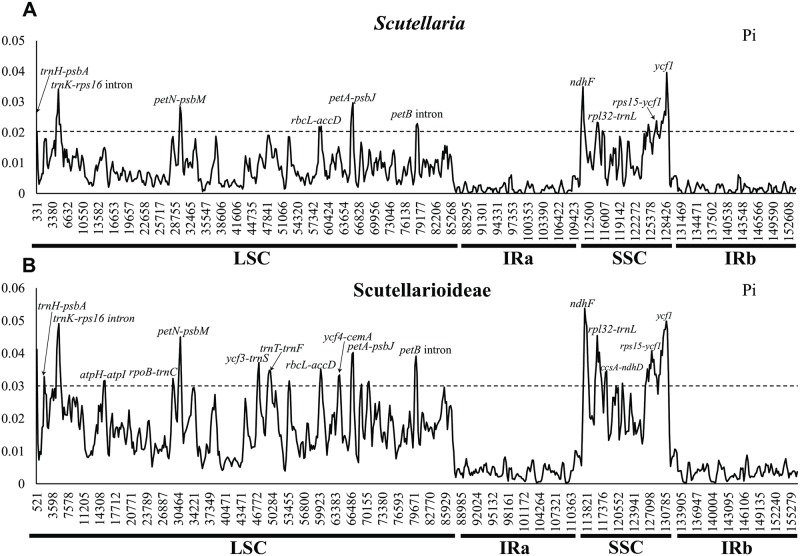
The average nucleotide variability (Pi) of plastomes was estimated to be 0.004 in Scutellaria (Fig 7). The SSC region showed the highest average nucleotide diversity (Pi = 0.0148), followed by the LSC region (Pi = 0.0087) and the IR region (Pi = 0.0019). Among the 11 species of Scutellaria, ten hyper-variable regions were identified, including two genes (ndhF, ycf1) and eight intergenic spacers (psbA-trnH, trnK-rps16 intron, petN-psbM, rbcL-accD, petA-psbJ, petB-petD intron, rpl32-trnL, and rps15-ycf1), with the variation exceeding 2.0%.
Fig 7. Sliding window analysis of the whole chloroplast genomes.
(A) the 11 species of Scutellaria; (B) the 15 samples of Scutellarioideae.
As for the 15 samples of Scutellarioideae, the average nucleotide variability (Pi) of the whole plastome was 0.014, while that of the LSC, SSC, and IR regions were 0.0178, 0.028, and 0.003, respectively. In the LSC region, we found 11 hyper-variable loci with Pi values > 0.03 (psbA-trnH, trnK-rps16 intron, atpH-atpI, rpoB-trnC, petN-psbM, ycf3-trnS, trnT-trnF, rbcL-accD, ycf4-cemA, petA-psbJ, and petB-petD intron), while in the SSC region, only five hyper-variable loci with Pi values > 0.03 (ndhF, rpl32-trnL, ccsA-ndhD, rps15-ycf1, and ycf1) were detected (Fig 7).
Characteristics of the datasets and phylogenetic relationships within Scutellarioideae
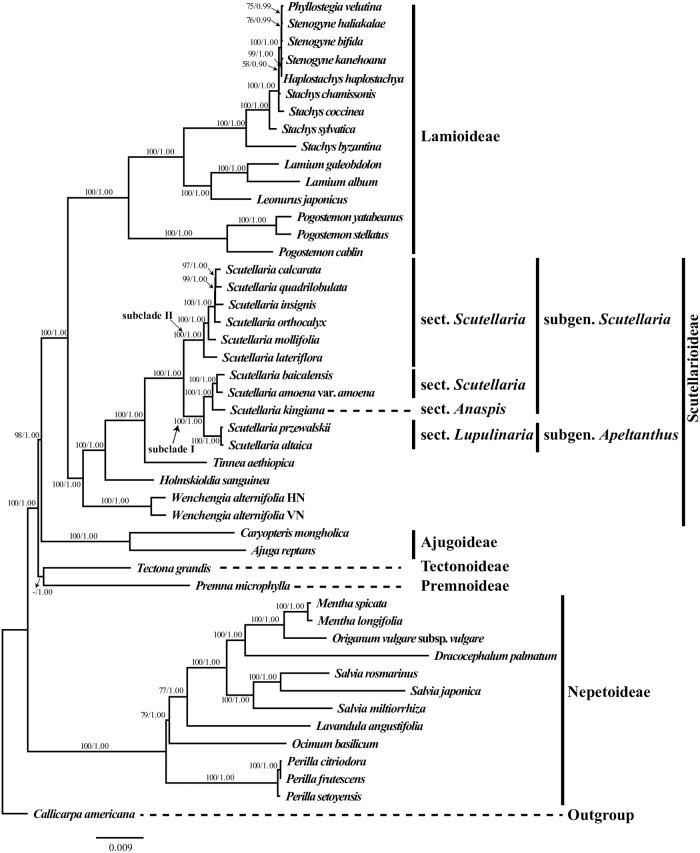
After the exclusion of ambiguously aligned sites, the total length of the complete aligned dataset (CPG) was 144,120 bp, of which 36,934 bp were variable (25.63%). The length of the CR dataset was 70,046 bp, of which 14,288 bp (20.4%) were variable. The noncoding dataset (NCR) was 72,624 bp, of which 23,032 bp (31.71%) were variable. The hyper-variable dataset (16VAR) was 24,090 bp, of which 9,953 bp (39.8%) were variable. The six commonly used cpDNA regions (6CP) was 8,346 bp, of which 2,820 bp (33.6%) were variable. Data characteristics with models selected for each dataset used for Bayesian phylogenetic analyses are list in Table 4. Topologies obtained from both ML and BI analyses for all three datasets were identical, thus the ML topology resulting from the analysis of the CPG dataset (Fig 8) is presented here for subsequent discussion of phylogenetic relationships.
Table 4. The number of parsimony-informative sites and the best fit model for each data set.
| Data set* | Aligned length [bp] | GC content (%) | No. of variable sites [bp] | No. of parsimony-informative sites [bp] | Best fit model (BIC) |
|---|---|---|---|---|---|
| CR | 70,046 | 38.20 | 14,288 (20.4%) | 8,695 (12.41%) | GTR+I+Γ |
| NCR | 72,624 | 33.60 | 23,032 (31.71%) | 13,208 (18.19%) | GTR+I+Γ |
| CPG | 144,120 | 37.10 | 36,934 (25.63%) | 21,763 (15.10%) | GTR+I+Γ |
| 16VAR | 24,090 | 31.60 | 9,953 (39.8%) | 5,865 (24.30%) | GTR+I+Γ |
| 6CP | 8,346 | 35.70 | 2,820 (33.6%) | 1,812 (21.71%) | GTR+I+Γ |
*: CPG, complete plastome sequences; CR, coding regions; NCR, non-coding regions; 16VAR: 16 hyper-variable regions; 6CP: six commonly cpDNA regions.
Fig 8. The best-score tree from maximum likelihood analysis of Scutellarioideae based on the complete plastome sequences.
Support values BS ≥ 50% or PP ≥ 0.90 are displayed on the branches follow the order MLBS/BIPP (“-” indicates a support value BS < 50%). Scale bar denotes the expected number of substitutions per site in maximum likelihood analysis.
In all our analyses, the Scutellarioideae was supported as monophyletic (ML/BS 100%, BI/PP 1.00) [all values follow this order hereafter] (Fig 8, S3–S8 Figs). The two samples of the monotypic genus Wenchengia formed a well-supported clade (100%, 1.00) sister to remaining genera of Scutellarioideae. All species of Scutellaria were recovered in a strongly supported clade (100%, 1.00), in which two subclades were recognized. Subclade I (100%, 1.00) comprised five species from three sections: sect. Lupulinaria (S. altaica and S. przewalskii, sect. Scutellaria (S. baicalensis and S. amoena var. amoena), and sect. Anaspis (S. kingiana). Subclade II (100%, 1.00) consist of six species from sect. Scutellaria.
Discussion
General characteristics of the plastomes of Scutellarioideae
Prior to this study, three plastomes of Scutellaria were available on GenBank, but two of them were without any related publication or analysis; only S. baicalensis was formally published [50]. The species S. indica var. coccinea has since been published, but the sequences were not yet available [51]. Here, we report on 12 complete plastomes representing 11 species from four genera of Scutellarioideae for the first time. In total, 15 plastomes were included for comparative analysis.
The length of plastomes of the 15 taxa from Scutellarioideae ranged from 151,675 bp to 153,272 bp, with the variation mainly caused by large indels (insertions/deletions) in the noncoding regions. The plastomes of Scutellarioideae are highly conserved in structure, gene order, and content. All the 15 plastomes encode 114 unique genes in the same gene order and display the typical quadripartite structure, including a pair of IR regions separated by the LSC and SSC regions (Fig 1 and S1 Fig). Lee and Kim [51] have recently identified 115 genes from the plastome of S. indica var. coccinea. In comparison with the present study, one extra tRNA gene was identified. Because sequences and annotation information of this plastome have not been released, we could not include it for comparative analysis. The average GC content of Scutellarioideae plastomes in our study was38.3%, very similar to other species in Lamiaceae [50, 51, 75–77].
The complete aligned sequences indicate that the 15 plastomes of Scutellarioideae are conserved, with the sequence identity among genera higher than 95% and no major structural rearrangements or gene losses discovered. The location of the IR boundaries, especially as this pertains to IR contraction and expansion, can be exploited for phylogenetic purposes as small expansions or contractions tend to have similar endpoints in closely related species [78]. We find that the variation in the IR boundaries in Scutellarioideae, however, is not as extensive as reported in previous studies [79].
Chen et al. [79] reported that the LSC/IR regions within Lamiales can be divided into four different types: type I, with the LSC/IR regions being located in the intergenic rpl2-rps19; type II, with the rps19 pseudogene at the LSC/IR border; type III, with the ycf2 pseudogene at the IR/LSC border; and type IV, with the IR extending to include the trnH gene and a truncated psbA pseudogene at the IR/LSC border. Subsequently, Gao et al. [48] detected a new type where the IR/LSC border was found in the intergenic rpl2-rps19. In our study, the LSC/IR junction of all 15 species of Scutellarioideae belongs to type II, and the boundary of the SSC and IRa regions in Wenchengia alternifolia and Tinnea aethiopica is aberrant, with an expansion that involved the complete ndhF gene being included in the SSC region (Fig 6).
SSRs are widely used in molecular identification, genetic diversity, and population genetics studies [80]. Studies have shown that A/T mononucleotides are often very rich in SSRs [50, 76, 77]. Our analyses also show that SSRs in Scutellarioideae are generally composed of short polyadenine (poly A) or polythymine (poly T) repeats and rarely contain tandem guanine (G) and/or cytosine (C). In this study, a total of 455 SSRs are made up of A or T bases, accounting for approximately 77% of the total SSRs. In addition, most mononucleotide repeats were detected in the non-coding regions (S3 Table). A potential reason for the higher frequencies of the AT repeats is the strand separation for ATs is relatively easier than GCs during plastome replication, which increases slipped-strand mispairing. There is a tendency for SSRs to occur in the non-coding region of the chloroplast genome of higher plants [81]. The molecular processes that give rise to repeats are more likely to be preserved in non-coding regions because there is strong selection against them in coding regions. In addition, because the non-coding regions are so AT rich, there is an expectation that repeats will be biased towards AT content, especially in the single copy regions. In general, the structure and organization of plastomes is conserved and SSRs primers are transferable across species or genera. Thus, the new SSRs detected in this study are potential resources for estimating the genetic diversity of some important medicinal species of Scutellaria, and for phylogenetic study among species and genera.
It has been demonstrated that short dispersed repeats are a major factor promoting plastome rearrangements in land plants [82], but within the unrearranged plastid sequence the function of these repeats remains unknown [76]. Our study reveals three types of repeats (forward, reverse, and palindromic) in the 15 plastomes of Scutellarioideae. As has been reported in other species of Lamiales [79, 83], most of these repeats are located in the intergenic spacers and introns, but several also occur in the coding regions. In total, 22.04% of the repeats occur in four protein coding regions (psaB, psaA, ycf1, and ycf2; S4 Table). The genes ycf1 and ycf2 have been demonstrated to be associated with repeat events [84]. In our study, the richest repeats are found in the ycf2 gene, similar to other studies [48, 79, 83]. However, only one palindromic repeat, in the ycf1 gene of Wenchengia alternifolia VN was detected. The absence of the dispersed repeats from the ycf1 gene in this study is partially because the plastomes from closely related species are highly similar and lack of variation.
Potential DNA barcodes for Scutellaria
Genomic comparative analyses of complete plastome sequences have become necessary for developing variable DNA barcodes, especially for finding mutation “hotspot” regions for novel DNA barcodes in addition to the set of widely used DNA markers (matK, rbcL, psbA-trnH, and nrITS [85–87]).
Though Scutellaria is the second largest genus within Lamiaceae and has medicinally important [88], DNA barcoding research within the genus is wanting. Guo et al. [68] attempted to distinguish the most widely used medicinal species, S. baicalensis, from its congeners, S. amoena, S. rehderiana Diels, and S. viscidula Bunge. However, this study had sparse sampling and only three DNA regions were used (matK, rbcL, and psbA-trnH). In previous studies, the cpDNA markers rps16 (as part of the trnK-rps16 intron), ndhF, rps15-ycf1, and ycf1 were used to resolve the systematic position of some genera within Lamiaceae [89, 90], and fragments of psbA-trnH, rpl32-trnL, rps15-ycf1, and ycf1 were applied to infer the intrageneric relationships [91, 92]. Some fragments, such as petN-psbM and petA-psbJ have been commonly used in seed plant phylogenetic studies [93, 94], but never have been used to resolve phylogenetic relationships in Lamiaceae. The intergenic spacer rbcL-accD and petB-petD intron have been identified as highly variable regions in other plants [95, 96]. The 10 highly variable regions (psbA-trnH, trnK-rps16 intron, petN-psbM, rbcL-accD, petA-psbJ, petB-petD intron, ndhF, rpl32-trnL, rps15-ycf1, and ycf1; Fig 7) identified here could be used as potential barcodes for species identification and phylogenetic study of Scutellaria. Although further research is needed to investigate the reliability and effectiveness of using these regions and/or complete plastome sequences for DNA barcodes in Scutellaria, the results obtained here could be a reference for future studies on global genetic diversity assessment, phylogeny, and population genetics.
Phylogenetic relationships within Scutellarioideae
Our study is the first to use complete plastome sequences to reconstruct the phylogeny of Scutellarioideae. The phylogenetic tree obtained here is largely consistent with previous studies based on the plastid DNA markers [1, 9, 97, 98]. However, some phylogenetic relationships within Lamiaceae differ from recent nuclear trees [99]. Such incongruence between plastid and nuclear phylogenies emphasizes a need for phylogenetic inferences based on both plastome sequences and nuclear data, which can together both robustly resolve relationships and point to potential ancient hybridization events.
The monophyly of Scutellarioideae is confirmed based on the analyses of all datasets (Fig 8, S3–S8 Figs), and the major splits determined in this study for Scutellarioideae agree with previous studies [1, 9]. This study confirmed that the monotypic genus Wenchengia is sister to the remainder of Scutellarioideae (Fig 8). This relationship has been reported in a previous study using two DNA markers (i.e. rbcL and ndhF; [9]). The accession of W. alternifolia from Vietnam was recovered in a clade with an accession of W. alternifolia from Hainan, China in our analyses. The genus has long been thought to be endemic to Hainan Island in China and was only recently reported from Vietnam. As suggested by Paton et al. [16], the distribution of Wenchengia in Vietnam indicates that the Hainan populations are probably relicts of a once more widely distributed W. alternifolia. The discovery of living plants in Vietnam offers the opportunity for population genetic and biogeographic studies of Wenchengia in future.
The African genus Tinnea is sister to Scutellaria, as reported by Wagstaff et al. [8] and Li et al. [1, 9]. Although Renschia has never been included in a molecular analysis, morphological characters, e.g. ciliate anthers, well-developed nectar disk, bilabiate calyx with entire, rounded lips, and the closing of the calyx during fruit maturation [6]), suggest a close relationship among Renschia, Tinnea, and Scutellaria. Renschia is probably most closely related to Tinnea based on distribution (both genera are distributed in Africa; Renschia is endemic to North Somalia and Tinnea to tropical Africa) and morphology. Vatke [100] established Renschia based on Tinnea heterotypica S. Moore, and distinguished Renschia from Tinnea by its protruding stamens, the short and basal areoles of nutlets, and the indistinct nervation of calyces.
A total of 11 species of Scutellaria were sampled from both subgenera sensu Paton [5]. The monophyly of Scutellaria is supported here as in other studies [1, 9, 18, 43], but the infrageneric classification of Scutellaria as proposed by Paton [5] is not supported by the present study (Fig 8). As shown in Fig 8, in our sampling Scutellaria is comprised of two subclades: Subclade I included five taxa from subg. Scutellaria and two taxa from subg. Apeltanthus; Subclade II consists of six species from subg. Scutellaria sect. Scutellaria. Species from sect. Scutellaria are recovered in both subclades, thus the monophyly of subgenus Scutellaria and sect. Scutellaria is not supported by the plastome sequences in this study or nuclear ribosomal sequences in previous studies [18, 43]. With only one species of sect. Anaspis sampled here, it is premature to assess its monophyly. Though a recent study by Safikhani et al. [18] revealed that sect. Anaspis is a well-supported group, only four representatives of the section from Iran were included in their study. Subgenus Apeltanthus is well supported in all studies [18, 43]. The two sections of subg. Apeltanthus, sect. Apeltanthus and sect. Lupulinaria, are shown to be monophyletic in our study as in Zhao et al. [43]. However, based on a broader sampling, Safikhani et al. [18] revealed that neither of the two sections is supported. Further phylogenetic study of subg. Apeltanthus is needed based on a more comprehensive sampling and more DNA markers.
Despite the limited sampling, our study, based on complete plastomes, presents a more resolved and better supported phylogeny of Scutellarioideae than previous studies [1, 9, 18, 43, 98]. All the phylogenetic trees inferred from the complete plastome sequences have higher resolution (Fig 8) than trees based on the six commonly used chloroplast DNA regions (matK, ndhF, rbcL, rpL32-trnL, rps16-intron, and trnL-F; S7 Fig) in previous studies [9, 41, 68] and 16 hyper-variable chloroplast regions (S8 Fig), demonstrating that complete plastome sequences can markedly improve phylogenetic resolution, at least within Scutellarioideae and Lamiaceae.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
(PDF)
Bootstrap values > 50% are indicated at individual branches.
(PDF)
Bayesian posterior probabilities ≥ 0.95 are indicated at individual branches.
(PDF)
Bootstrap values > 50% are indicated at individual branches.
(PDF)
Bayesian posterior probabilities ≥ 0.95 are indicated at individual branches.
(PDF)
Support values BS ≥ 50% or PP ≥ 0.90 are displayed on the branches follow the order MLBS/BIPP (“-” indicates a support value BS < 50%). Scale bar denotes the expected number of substitutions per site in maximum likelihood analysis.
(PDF)
Support values BS ≥ 50% or PP ≥ 0.90 are displayed on the branches follow the order MLBS/BIPP (“-” indicates a support value BS < 50%). Scale bar denotes the expected number of substitutions per site in maximum likelihood analysis.
(PDF)
Acknowledgments
The authors are grateful to Prof. Shi-Xiao Luo, Prof. Shen-Zhuo Huang, Mr. Hong-Liang Chen, Mr. Yi Yang, Miss Yuan-Yuan Li, and Miss Qiao-Rong Zhang for their assistance in sample collection. We also thank Dr. Richard Olmstead and another anonymous reviewer for their constructive suggestions that greatly improved the paper.
Data Availability
All sequences used in this study are available from the National Center for Biotechnology Information (NCBI) MN128378–MN128389.
Funding Statement
This study was funded by the “Ten Thousand Talents Program of Yunnan (Top-notch Young Talents)” (No. YNWR-QNBJ-2018-279), CAS “Light of West China” Program and the “Excellent Youth Fund Project” (No. 2019FI009) of Yunnan Provincial Science and Technology Department to CLX, and the National Natural Science Foundation of China (No. 31870181) to QW.
References
- 1.Li B, Cantino PD, Olmstead RG, Bramley GL, Xiang CL, Ma ZH, et al. A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci Rep. 2016; 6: 34343 10.1038/srep34343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li B, Olmstead RG. Two new subfamilies in Lamiaceae. Phytotaxa. 2017; 313(2): 222–226. 10.11646/phytotaxa.313.2.9 [DOI] [Google Scholar]
- 3.Benthem G. Scutellaria, Salazria and Perilomia In: Bentham G, Hooker JD, editors. Genera Plantarum. 2 London: Lovell Reeve & Co; 1876. pp. 1201–1203. [Google Scholar]
- 4.Briquet J. Scutellaria, Salazaria and Perilomia In: Engler, A & Prantl, K. A. E. Die, editors. Naturlichen Pflanzenfamilien ed.1. 1896;4(3a):[Scutellaria and Salzaria: 224–227;Perilomia: 232–233;Position of seed: 199].
- 5.Paton A. A global taxonomic investigation of Scutellaria (Labiatae). Kew Bull. 1990;45(3): 399–450. [Google Scholar]
- 6.Cantino PD. Towa rd a phylogenetic classification of the Labiatae In: Hartley R, Reynolds T, editors. Advances in Labiatae science: Royal Botanic Gardens, Kew Richmond; 1992. pp. 27–37. [Google Scholar]
- 7.Cantino PD. Evidence for a Polyphyletic origin of the Labiatae. Ann Mo Bot Gard. 1992; 79(2): 361–379. 10.2307/2399774 [DOI] [Google Scholar]
- 8.Wagstaff SJ, Hickerson L, Spangler R, Reeves PA, Olmstead RG. Phylogeny in Labiatae s.l., inferred from cpDNA sequences. Plant Syst Evol. 1998; 209: 265–274. [Google Scholar]
- 9.Li B, Xu WX, Tu TY, Wang ZS, Olmstead RG, Peng H, et al. Phylogenetic position of Wenchengia (Lamiaceae): A taxonomically enigmatic and critically endangered genus. Taxon. 2012; 61(2): 392–401. [Google Scholar]
- 10.Ryding O. Amount of calyx fibres in Lamiaceae, relation to calyx structure, phylogeny and ecology. Plant Syst Evol. 2007; 268(1): 45–58. [Google Scholar]
- 11.Wagstaff SJ, Olmstead RG. Phylogeny of Labiatae and Verbenaceae inferred from rbcL sequences. Syst Bot. 1997; 22(1): 165–179. 10.2307/2419684 [DOI] [Google Scholar]
- 12.Moldenke HN. Notes on the genus Holmskioldia. Phytologia. 1981; 48: 313–356. [Google Scholar]
- 13.Lovett J, Friis I. Patterns of endemism in the woody flora of north-east and east Africa In: van der Maesen LJG, van der Burgt XM, van Medenbach de Rooy JM, editors. The biodiversity of African plants. Dordrecht: Springer; 1996. pp. 582–601. [Google Scholar]
- 14.Wu CY, Chow S. Duo Taxa Nova Labiatarum. Acta Phytotaxon Sin. 1965; 10(3): 249–257. [Google Scholar]
- 15.Francisco-Ortega J, Wang ZS, Wang FG, Xing FW, Liu H, Xu H, et al. Seed plant endemism on Hainan Island: A framework for conservation actions. Bot Rev. 2010; 76(3): 346–376. 10.1007/s12229-010-9055-7 [DOI] [Google Scholar]
- 16.Paton A, Phillipson PB, Suddee S. Records of Wenchengia (Lamiaceae) from Vietnam. Biodivers Data J. 2016; (4): e9596 10.3897/BDJ.4.e9596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harley RM, Atkins S, Budantsev AL, Cantino PD, Conn BJ, Grayer R, et al. Labiatae In: Kadereit JW, editor. The Families and Genera of Vascular Plants 7: Springer; 2004. pp. 167–275. [Google Scholar]
- 18.Safikhani K, Jamzad Z, Saeidi H. Phylogenetic relationships in Iranian Scutellaria (Lamiaceae) based on nuclear ribosomal ITS and chloroplast trnL-F DNA data. Plant Syst Evol. 2018; 304(9): 1077–1089. 10.1007/s00606-018-1533-0. [DOI] [Google Scholar]
- 19.Paton A. The Phytogeography of Scutellaria L. Notes Roy Bot Gard Edinburgh. 1990; 46(3): 345–359. [Google Scholar]
- 20.Huang S. Shen Nong Ben Cao Jing (The Divine Farmer’s Materia Medica): Beijing: Traditional Chinese Medicine Ancient Books Press; 1982. [Google Scholar]
- 21.Yuan QJ, Zhang ZY, Hu J, Guo LP, Shao AJ, Huang LQ. Impacts of recent cultivation on genetic diversity pattern of a medicinal plant, Scutellaria baicalensis (Lamiaceae). BMC Genet. 2010; 11: 29 10.1186/1471-2156-11-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton A. Esquisse d’une monographie du genre Scutellaria, ou Toque. Lyon: Louis Perrin; 1832. [Google Scholar]
- 23.Epling C. The American species of Scutellaria: University of California Press; 1942. pp. 1–146. [Google Scholar]
- 24.Juzepczuk SV. Scutellaria L In: Shishkin BK, Juzepczuk SV, editors. Flora of the USSR. 20 Russer: Izdatel’stvo Akademii NaukSSSR Moskva‐Leningrad; 1954. pp. 50–150. [Google Scholar]
- 25.Paton AJ, Springate D, Suddee S, Otieno D, Grayer RJ, Harley MM, et al. Phylogeny and evolution of basils and allies (Ocimeae, Labiatae) based on three plastid DNA regions. Mol Phylogenet Evol. 2004; 31(1): 277–299. 10.1016/j.ympev.2003.08.002 [DOI] [PubMed] [Google Scholar]
- 26.Paton A, Mwanyambo M, Culham A. Phylogenetic study of Plectranthus, Coleus and allies (Lamiaceae): taxonomy, distribution and medicinal use. Bot J Linn Soc. 2018; 188(4): 355–376. 10.1093/botlinnean/boy064 [DOI] [Google Scholar]
- 27.Heckenhauer J, Large D, Samuel R, Barfuss MHJ, Prins PDH. Molecular phylogeny helps to delimit Plectranthus hadiensis from its related morph occurring in Sri Lanka. Ceylon J Sci. 2019; 48(2): 133–141. 10.4038/cjs.v48i2.7617 [DOI] [Google Scholar]
- 28.Drew BT, Gonzalez-Gallegos JG, Xiang CL, Kriebel R, Drummond CP, Walker JB, et al. Salvia united: The greatest good for the greatest number. Taxon. 2017; 66(1): 133–145. 10.12705/661.7 [DOI] [Google Scholar]
- 29.Fragoso-Martínez I, Martínez-Gordillo M, Salazar GA, Sazatornil F, Jenks AA, García-Peña MR, et al. Phylogeny of the neotropical sages (Salvia subg. Calosphace; Lamiaceae) and insights into pollinator and area shifts. Plant Syst Evol. 2018; 304(1): 43–55. [Google Scholar]
- 30.Hu GX, Takano A, Drew BT, Liu ED, Soltis DE, Soltis PS, et al. Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Ann Bot. 2018; 122(4): 649–668. 10.1093/aob/mcy104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takano A, Okada H. Phylogenetic relationships among subgenera, species, and varieties of Japanese Salvia L. (Lamiaceae). J Plant Res. 2011; 124(2): 245–252. 10.1007/s10265-010-0367-9 [DOI] [PubMed] [Google Scholar]
- 32.Walker JB, Sytsma KJ. Staminal evolution in the genus Salvia (Lamiaceae): molecular phylogenetic evidence for multiple origins of the staminal lever. Ann Bot. 2007; 100(2): 375–391. 10.1093/aob/mcl176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker JB, Sytsma KJ, Treutlein J, Wink M. Salvia (Lamiaceae) is not monophyletic: implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am J Bot. 2004; 91(7): 1115–1125. 10.3732/ajb.91.7.1115 [DOI] [PubMed] [Google Scholar]
- 34.Will M, Claßen-Bockhoff R. Why Africa matters: evolution of Old World Salvia (Lamiaceae) in Africa. Ann Bot. 2014; 114(1): 61–83. 10.1093/aob/mcu081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen YP, Wilson TC, Zhou YD, Wang ZH, Liu ED, Peng H, et al. Isodon hsiwenii (Lamiaceae: Nepetoideae), a new species from Yunnan, China. Syst Bot. 2019; 44(4): 913–922. 10.1600/036364419x15710776741486 [DOI] [Google Scholar]
- 36.Dohzono I, Suzuki K. Morphological and genetic differentiation in Isodon umbrosus by altitudinal variation in bumblebee pollinator assemblages. Plant Spec Biol. 2010; 25(1): 20–9. 10.1111/j.1442-1984.2010.00268.x [DOI] [Google Scholar]
- 37.Yu XQ, Maki M, Drew BT, Paton AJ, Li HW, Zhao JL, et al. Phylogeny and historical biogeography of Isodon (Lamiaceae): rapid radiation in south-west China and Miocene overland dispersal into Africa. Mol Phylogenet Evol. 2014; 77: 183–94. 10.1016/j.ympev.2014.04.017 [DOI] [PubMed] [Google Scholar]
- 38.Zhong JS, Li J, Li L, Conran JG, Li HW. Phylogeny of Isodon (Schrad. ex Benth.) Spach (Lamiaceae) and related genera inferred from nuclear ribosomal ITS, trnL–trnF region, and rps16 intron sequences and morphology. Syst Bot. 2010; 35(1): 207–219. 10.1600/036364410790862614 [DOI] [Google Scholar]
- 39.Olmstead RG. Phylogeny, Phenotypic evolution, and biogeography of the Scutellaria angustifolia complex (Lamiaceae) inference from morphological and molecular data. Syst Bot. 1989; 14(3): 320–338. 10.2307/2418923 [DOI] [Google Scholar]
- 40.Olmstead RG. Biological and Historical factors influencing genetic diversity in the Scutellaria angustifolia complex (Labiatae). Evolution. 1990; 44(1): 54–70. 10.2307/2409524 [DOI] [PubMed] [Google Scholar]
- 41.Chiang YC, Huang BH, Liao PC. Diversification, biogeographic pattern, and demographic history of Taiwanese Scutellaria species inferred from nuclear and chloroplast DNA. PLoS ONE. 2012; 7(11): e50844 10.1371/journal.pone.0050844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosokawa K, Minami M, Nakamura I, Hishida A, Shibata T. The sequences of the plastid gene rpl16 and the rpl16-rpl14 spacer region allow discrimination among six species of Scutellaria. J Ethnopharmacol. 2005; 99(1): 105–108. 10.1016/j.jep.2004.02.0311 [DOI] [PubMed] [Google Scholar]
- 43.Zhao F, Liu ED, Peng H, Xiang CL. A new species of Scutellaria (Scutellarioideae, Lamiaceae) from Sichuan Province in southwest China. PeerJ. 2017; 5: e3624 10.7717/peerj.3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016; 17: 29 10.1186/s13059-016-1004-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruhlman TA, Jansen RK. The plastid genomes of flowering plants. Methods Mol Biol. 2014; 1132: 3–38. 10.1007/978-1-62703-995-6_1 [DOI] [PubMed] [Google Scholar]
- 46.Fu CN, Li HT, Milne R, Zhang T, Ma PF, Yang J, et al. Comparative analyses of plastid genomes from fourteen Cornales species: inferences for phylogenetic relationships and genome evolution. BMC Genomics. 2017; 18(1): 956 10.1186/s12864-017-4319-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, et al. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 2017; 214(3): 1355–1367. 10.1111/nph.14461 [DOI] [PubMed] [Google Scholar]
- 48.Gao CM, Wang J, Deng YF. The complete chloroplast genomes of Echinacanthus species (Acanthaceae): phylogenetic relationships, adaptive evolution, and screening of molecular markers. Front Plant Sci. 2018; 9: 1989 10.3389/fpls.2018.01989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welch AJ, Collins K, Ratan A, Drautz-Moses DI, Schuster SC, Lindqvist C. The quest to resolve recent radiations: plastid phylogenomics of extinct and endangered Hawaiian endemic mints (Lamiaceae). Mol Phylogenet Evol. 2016; 99: 16–33. 10.1016/j.ympev.2016.02.024 [DOI] [PubMed] [Google Scholar]
- 50.Jiang D, Zhao ZY, Zhang T, Zhong WH, Liu CS, Yuan QJ, et al. The chloroplast genome sequence of Scutellaria baicalensis provides insight into intraspecific and interspecific chloroplast genome diversity in Scutellaria. Genes. 2017; 8(9): 227–240. 10.3390/genes8090227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y, Kim S. The complete chloroplast genome of Scutellaria indica var. coccinea (Lamiaceae), an endemic taxon in Korea. Mitochondrial DNA Part B. 2019; 4(2): 2539–2540. 10.1080/23802359.2019.1640649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doyle J, Doyle J. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull. 1987; 19(11): 11–15. [Google Scholar]
- 53.FastQC: a quality control tool for high throughput sequence data [Internet]. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 54.Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 2018:256479 10.1101/256479 [DOI] [Google Scholar]
- 55.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015; 31(20): 3350–3352. 10.1093/bioinformatics/btv383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012; 9(4): 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28(12): 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004; 20(17):3252–3255. 10.1093/bioinformatics/bth352 [DOI] [PubMed] [Google Scholar]
- 59.Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, et al. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017; 45(W1): W6–W11. Epub 2017/05/10. 10.1093/nar/gkx391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016; 44(W1): W54–W57. 10.1093/nar/gkw413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013; 41(W1): W575–W581. 10.1093/nar/gkt289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001; 29(22): 4633–4642. 10.1093/nar/29.22.4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004; 14(7): 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004; 32: W273–W279. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013; 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017; 34(12): 3299–3302. 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- 67.Olmstead RG, Sweere JA. Combining data in phylogenetic systematics: an empirical-approach using 3 molecular-data sets in the Solanaceae. Syst Biol. 1994; 43(4): 467–481. 10.2307/2413546 [DOI] [Google Scholar]
- 68.Guo XR, Wang XG, Su WH, Zhang GF, Zhou R. DNA barcodes for discriminating the medicinal plant Scutellaria baicalensis (Lamiaceae) and its adulterants. Biol Pharm Bull. 2011; 34(8): 1198–1203. 10.1248/bpb.34.1198 [DOI] [PubMed] [Google Scholar]
- 69.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 gateway computing environments workshop (GCE); 2010: 1–8. Ieee.
- 70.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, et al. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012; 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012; 9(8): 772 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978; 6(2): 461–464. [Google Scholar]
- 74.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018; 67(5): 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lukas B, Novak J. The complete chloroplast genome of Origanum vulgare L. (Lamiaceae). Gene. 2013; 528(2): 163–169. 10.1016/j.gene.2013.07.026 [DOI] [PubMed] [Google Scholar]
- 76.Qian J, Song J, Gao H, Zhu Y, Xu J, Pang X, et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE. 2013; 8(2): e57607 10.1371/journal.pone.0057607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yi DK, Kim KJ. The complete chloroplast genome sequences of Pogostemon stellatus and Pogostemon yatabeanus (Lamiaceae). Mitochondrial DNA Part B. 2016; 1(1): 571–573. 10.1080/23802359.2016.1192509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Plunkett GM, Downie SR. Expansion and contraction of the chloroplast inverted repeat in Apiaceae Subfamily Apioideae. Syst Bot. 2000; 25(4): 648–667. 10.2307/2666726 [DOI] [Google Scholar]
- 79.Chen H, Shao J, Zhang H, Jiang M, Huang L, Zhang Z, et al. Sequencing and analysis of Strobilanthes cusia (Nees) Kuntze chloroplast genome revealed the rare simultaneous contraction and expansion of the inverted repeat region in angiosperm. Front Plant Sci. 2018; 9: 324 10.3389/fpls.2018.00324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He SL, Wang YS, Volis S, Li DZ, Yi TS. Genetic diversity and population structure: implications for conservation of wild soybean (Glycine soja Sieb. et Zucc) based on nuclear and chloroplast microsatellite variation. Int J Mol Sci. 2012; 13(10): 12608–12628. 10.3390/ijms131012608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Menezes AP, Resende-Moreira LC, Buzatti RS, Nazareno AG, Carlsen M, Lobo FP, et al. Chloroplast genomes of Byrsonima species (Malpighiaceae): comparative analysis and screening of high divergence sequences. Sci Rep. 2018; 8(1): 2210 10.1038/s41598-018-20189-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang RJ, Cheng CL, Chang CC, Wu CL, Su TM, Chaw SM. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol Biol. 2008; 8: 36 10.1186/1471-2148-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nazareno AG, Carlsen M, Lohmann LG. Complete chloroplast genome of Tanaecium tetragonolobum: the first Bignoniaceae plastome. PLoS ONE. 2015; 10(6): e0129930 10.1371/journal.pone.0129930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao XY, Zhang X, Meng HH, Li J, Zhang D, Liu CN. Comparative chloroplast genomes of Paris Sect. Marmorata: insights into repeat regions and evolutionary implications. BMC Genomics. 2018; 19(10): 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE. 2007; 2(6): e508 10.1371/journal.pone.0000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hollingsworth ML, Clark AA, Forrest LL, Richardson J, Pennington RT, Long DG, et al. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour. 2009; 9(2): 439–457. 10.1111/j.1755-0998.2008.02439.x [DOI] [PubMed] [Google Scholar]
- 87.Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. P Natl Acad Sci USA. 2011; 108(49): 19641–19646. 10.1073/pnas.1104551108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Irvin L, Jackson C, Hill AL, Bajaj R, Mahmoudi C, Vaidya BN, et al. Skullcaps (Scutellaria spp.): Ethnobotany and Current Research In: Joshee N, Dhekney S, Parajuli P, editors. Medicinal Plants: Springer; 2019. pp. 141–168. [Google Scholar]
- 89.Drew BT, Sytsma KJ. Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am J Bot. 2012; 99(5): 933–953. 10.3732/ajb.1100549 [DOI] [PubMed] [Google Scholar]
- 90.Chen YP, Li B, Olmstead RG, Cantino PD, Liu ED, Xiang CL. Phylogenetic placement of the enigmatic genus Holocheila (Lamiaceae) inferred from plastid DNA sequences. Taxon. 2014; 63(2): 355–366. 10.12705/632.8 [DOI] [Google Scholar]
- 91.Salmaki Y, Kattari S, Heubl G, Bräuchler C. Phylogeny of non-monophyletic Teucrium (Lamiaceae: Ajugoideae): implications for character evolution and taxonomy. Taxon. 2016; 65(4): 805–822. 10.12705/654.8. [DOI] [Google Scholar]
- 92.Yao G, Drew BT, Yi TS, Yan HF, Yuan YM, Ge XJ. Phylogenetic relationships, character evolution and biogeographic diversification of Pogostemon s.l. (Lamiaceae). Mol Phylogenet Evol. 2016; 98: 184–200. 10.1016/j.ympev.2016.01.020 [DOI] [PubMed] [Google Scholar]
- 93.Adams RP, Farzaliyev V, Gucel S, Leschner HV, Mataraci T, Tashev AN, et al. nrDNA and petN-psbM sequencing reveals putative Juniperus oxycedrus L. from Azerbaijan, Bulgaria, Cyprus and Israel to be J. deltoides R.P. Adams. Phytologia. 2015; 97(4): 286–290. [Google Scholar]
- 94.Latvis M, Mortimer SM, Morales‐Briones DF, Torpey S, Uribe‐Convers S, Jacobs SJ, et al. Primers for Castilleja and their utility across Orobanchaceae: I Chloroplast primers. Appl Plant Sci. 2017; 5(9): 1700020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khatfan A, Riablershirun N, Sathornviriyapong V, Juntawong N. Analysis of phylogenetic relationship between sacred lotus and woody plants using nucleotide sequences of rbcL and its adjacent genes. Thai J For. 2015; 34(3): 56–68. [Google Scholar]
- 96.Liu HR, Gao QB, Zhang FQ, Khan G, Chen SL. Westwards and northwards dispersal of Triosteum himalayanum (Caprifoliaceae) from the Hengduan Mountains region based on chloroplast DNA phylogeography. PeerJ. 2018; 6: e4748 10.7717/peerj.4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bendiksby M, Thorbek L, Scheen AC, Lindqvist C, Ryding O. An updated phylogeny and classification of Lamiaceae subfamily Lamioideae. Taxon. 2011; 60(2): 471–484. [Google Scholar]
- 98.Xiang CL, Zhao F, Cantino PD, Drew BT, Li B, Liu ED, et al. Molecular systematics of Caryopteris (Lamiaceae) and its allies with reference to the molecular phylogeny of subfamily Ajugoideae. Taxon. 2018; 67(2): 376–394. 10.12705/672.7 [DOI] [Google Scholar]
- 99.Mint Evolutionary Genomics Consortium. Electronic address bme, Mint Evolutionary Genomics C. Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Mol Plant. 2018; 11(8): 1084–1096. 10.1016/j.molp.2018.06.002 [DOI] [PubMed] [Google Scholar]
- 100.Vatke W. Plantas in itinere africano ab J. M. Hildebrandt collectas determinare pergit. Linnaea. 1880; 43: 83–112. [Google Scholar]