Abstract
Myrteae is the largest and most diverse tribe within Myrtaceae and represents the majority of its diversity in the Neotropics. Members of Myrteae hold ecological importance in tropical biomes for the provision of food sources for many animal species. Thus, due to its several roles, a growing interest has been addressed to this group. In this study, we report the sequencing and de novo assembly of the complete chloroplast (cp) genomes of six Myrteae species: Eugenia brasiliensis, E. pyriformis, E. nitida, Myrcianthes pungens, Plinia edulis and Psidium cattleianum. We characterized genome structure, gene content, and identified SSRs to detect variation within Neotropical Myrteae. The six newly sequenced plastomes exhibit a typical quadripartite structure, gene content and organization highly conserved among Myrtaceae species. Some differences in genome length, protein-coding genes and non-coding regions were found. Besides, IR boundaries present structural changes among species. Increased sequence diversity was observed in some intergenic regions, suggesting their suitability for investigating intraand interspecific genetic diversity in populational studies. These data also contribute to the improvement of taxa sampling in further phylogenetic investigations to understand Myrtaceae evolution.
Keywords: cpDNA, genomic resource, populational genetics, plastid, conservation
Myrtaceae encompasses over 6000 species of shrubs and trees, classified in 144 genera and subdivided into 17 tribes (Wilson et al., 2005; WCSP, 2019). This angiosperm family has a predominant Southern-Hemisphere distribution and is assumed to be of Gondwanan origin, being an important component in the forests of Southeast Asia, Australia, and South America (Wilson et al., 2005; Thornhill et al., 2015). In the Neotropical region, most of Myrtaceae is represented by the tribe Myrteae, which comprises over 50 genera and 2500 species, representing half of the diversity of the family (Wilson et al., 2005; WCSP, 2019). Myrteae species play an important ecological role in Neotropical environments as foraging resources to animals, especially to a variety of bee species (Fidalgo and Kleinert, 2009). Besides that, some studies focused on specific classes of compounds produced by Myrtaceae, such as terpenes, which present commercial uses (Guzman et al., 2014). Other studies have demonstrated the antifungal, antioxidant, antiinflammatory, gastroprotective, and other bioactivities of
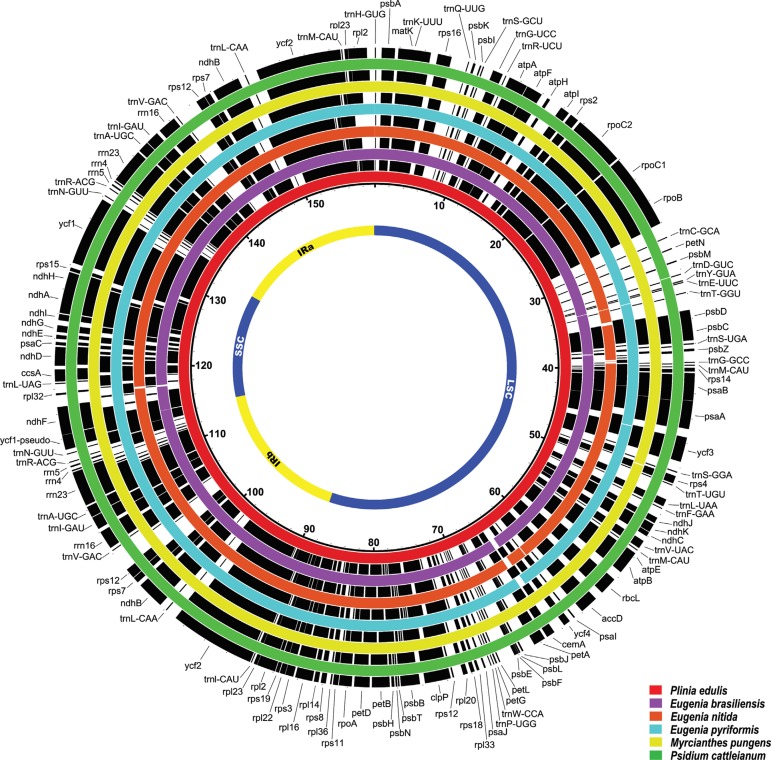
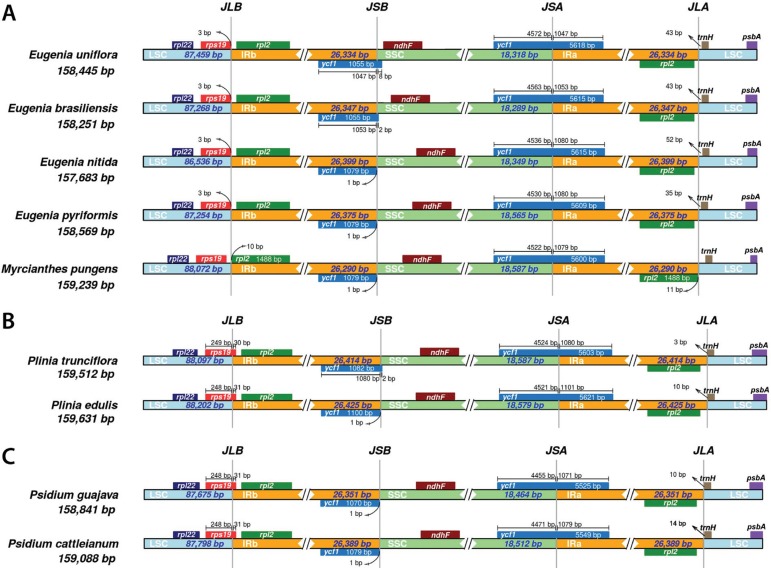
Myrteae species from Brazil (Salvador et al., 2011; Souza Moreira et al., 2019). Thus, due to its plethora of roles, a growing interest has been addressed to this group as a model for evolutionary, ecological and applied studies. For this study, leaves from Eugenia brasiliensis, Eugenia pyriformis, Eugenia nitida, Myrcianthes pungens, Plinia edulis and Psidium cattleianum trees were from a private in vivo collection in Gravataí, RS, Brazil (latitude (S): 29°51’52"; longitude (W): 50°53’53") and used to isolate chloroplasts by the modified high salt method, followed by cpDNA extraction with the CTAB method (Vieira et al., 2014). DNA quality was evaluated by electrophoresis in a 1% agarose gel, and DNA quantity was determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). For each species, one genomic paired-end library of 150 nt length was generated using an Illumina HiSeq2000 platform (Macrogen). The bases with quality below Q30 and adapter contamination were trimmed using Trim Galore!0.4.2, with a 50 bases minimum allowed. Paired-end sequence reads were filtered against 28 Myrtaceae plastomes (Table S1 (145.3KB, pdf) ) using BWA software (Li and Durbin, 2009) with two mismatches allowed. Mapped reads were used for the de novo assembly with ABySS software (Jackman et al., 2017). The plastome scaffolds were orientated by MUMmer (Delcher et al., 2003) using Eugenia uniflora (NC_026450.1), Plinia trunciflora (NC_034801.1) or Psidium guajava (NC_033355.1) as reference genomes for species of the same or closer genus. Genes were annotated using GeSeq (Tillich et al., 2017) by BLAST searches with 80% similarity. Circular plastome maps were drawn using the OGDRAW web toolkit (Lohse et al., 2013). IRa and IRb boundaries were analyzed using IRScope (Amiryousefi et al., 2018). For each species, local mVISTA (Frazer et al., 2004) was used to pairwise align plastomes with their respective reference. An overall genome comparison was performed with BLAST Ring Image Generator (BRIG) (Alikhan et al., 2011). Krait v0.11.4 (Du et al., 2018) was used to search and annotate perfect SSRs using the genomes and their annotated GFF3 file. The parameters for minimum repeat numbers were 8, 4, 3, 3, 3, 3 for mono-, di-, tri-, tetra-, penta- and hexanucleotide SSRs, respectively. DNA sequencing libraries were produced for each species, and these comprised 34.1-48.7 M raw Illumina paired-end reads (summarized in Table S2 (122.1KB, pdf) ). The percentage of removed reads due to trimming ranged from 1.17-1.45%. The number of filtered reads was 1.1-2.2 M reads. The obtained reads were de novo assembled into scaffolds that completely covered each plastome, without any gaps. The number of assembled scaffolds obtained ranged from four to eight. The minimum coverage ranged from 13 to 46 reads and the maximum coverage, 1,508-3,620 reads. The complete sequences were submitted to GenBank at accession numbers MN095407 to MN095411 and MN095413 (Table 1). The complete plastomes of six Myrteae have a narrow size range, from 157,683 bp in E. nitida to 159,631 bp in P. edulis, similar to the size of Myrtaceae species plastomes (Eguiluz et al., 2017 a,b; Machado et al., 2017). Figures S1 (252.8KB, pdf) , S2 (251.7KB, pdf) , S3 (252.6KB, pdf) , S4 (252.4KB, pdf) , S5 (251.2KB, pdf) , S6 (253KB, pdf) present the genome maps for each species. Four well-defined regions are present in all newly assembled genomes. Inverted regions (IR) ranged from ~26.3 to 26.4 kbp and had the smallest size variation, up to 78 bp. Short single copy (SSC) sections have ~18.2-18.5 kbp, while long single copy (LSC) sections have ~86.4-88.2 kbp (Table 1). Protein coding sequences comprise ~50% of the genome, rRNAs and tRNAs comprise ~7%, and non-coding regions, such as introns, pseudogenes and intergenic spacers correspond to ~43% (Table 1). Genome structure analysis showed a high degree of synteny among evaluated species (Figure 1). Genomes contained 129 genes in total, corresponding to 78 single-copy protein-coding genes, 30 transfer RNA (tRNA) genes, four ribosomal genes (rRNA) and one pseudogene (ycf1) (Figure 1, Table 1). In general, genomic features, such as size, structure, and gene abundance are similar to previously described Myrtaceae species (Eguiluz et al., 2017a,b). Despite the similarity in genomic features, the mVISTA comparison against each respective reference genome showed that some regions display lower similarity (Figures S7 (265.8KB, pdf) , S8 (257.7KB, pdf) , S9 (226.6KB, pdf) ). Non-coding regions, particularly the intergenic, had lower conservation and, therefore, more variation, such as psbI-trnS, trnT-psbD, trnS-psbZ-trnG, accD-psaI, and ndhF-rp132 in Eugenia and Myrcianthes; trnS-trnR, atpF-atpH, trnT-trnL, and rpl32-trnL in Plinia; and most intergenic regions of LSC in Psidium. Regarding protein-coding genes, we observed a conservation decrease in accD and ccsA in Eugenia species and M. pungens; and rpoC2 in Plinia and Psidium. Protein-coding genes matK, ndhF and ycf1, showed more nucleotide diversity (4.6 to 6.1%) in all analyzed species (Figures S7 (265.8KB, pdf) , S8 (257.7KB, pdf) , S9 (226.6KB, pdf) , S10 (321.5KB, pdf) ). This diversity corroborates previous studies based on plastidial genes and non-coding regions with contrasting substitution rates (Thornhill et al., 2015; Machado et al., 2017). Some structural changes in the IRa and IRb boundaries were found for the evaluated species (Figure 2). Within the IRb-LSC boundaries, the boundaries of the rps19 gene were located on the left side. In the IRb region, except for M. pungens, the IRb-LSC boundary was embedded in rps19 and had a length of three bp in Eugenia (Figure 2A), ~30 bp in Plinia (Figure 2B), and 31 bp in Psidium, contained in the IRb (Figure 2C). The IRb-SSC boundaries were embedded in the ycf1 pseudogene, ranging from one to eight bp in Eugenia/Myrcianthes, one and two bp in Plinia, and only one bp in Psidium species. The ndhF gene was located on the right side of the IRb-SSC at a distance from the boundary of 10 bp in E. uniflora, 36 to 121 bp in other Eugenia, 72 bp in M. pungens, 109 bp and 120 bp in Plinia, and 111 bp and 124 bp in Psidium. The SSC-IRa boundary was embedded in ycf1, with a length of 1047 to 1080 bp in Eugenia/Myrcianthes, 1080 and 1011 bp in Plinia, and 1071 and 1079 bp in Psidium in the IRa region. The trnH-GUG gene was located on the right side of the IRa-LSC boundary ranging from 11 to 52 bp in Eugenia/Myrcianthes, from 3 to 10 bp in Plinia and 10 to 14 bp in Psidium. The contraction and expansion of IR regions are measurable events of plastome evolution. These results demonstrate a genus-specific IR conservation, which can be considered one of the reasons for genome size variation among species. In this work, we present, for the first time, a characterization of IR boundaries from species of the same genus in Myrteae because they compared different genera, previous studies could not report a significant variation in IRb-SSC border within Myrteae species (Eguiluz et al., 2017a,b; Machado et al., 2017). All plastomes presented a similar number of SSRs. In total, over 315 SSRs were identified for each species (Table 2). The mononucleotide SSRs of A/T were the most frequent, varying in number from 85/98 in E. brasiliensis/E. pyriformis to 93/103 in P. edulis/P. cattleianum (Table S3 (163.4KB, pdf) ). This AT richness was already demonstrated in previous studies and reflects the lower GC content in these plastid genomes (Eguiluz et al., 2017a,b). The secondmost common were the trinucleotide SSRs, ranging in number from 61 in E. nitida to 71 in P. edulis. In addition, the number of SSRs located in different regions were similar: in intergenic regions ranging from 171 in E. nitida to 185 in P. edulis; in genes, ranging from 96 in E. brasiliensis, E. pyriformis, M. pungens, and P. cattleianum, and 98 in E. nitida and P. edulis; and in introns, ranging from 43 in E. brasiliensis to 47 in E. nitida, P. edulis, and P. cattleianum (Table 2). Hexanucleotide SSRs could not be found in the genomes. All found SRRs are listed in Table S4 (1.7MB, pdf) . These SSR results provide more information on molecular markers that could be used to evaluate intra- and interspecific diversity. This work provides reference genomes for six Neotropical Myrtaceae species, increasing the genetic information available for the Myrteae tribe, and allowing the improvement of taxa sampling in further investigations into Myrtaceae evolution.
Table 1. Myrteae chloroplast genome features.
| Feature |
Eugenia
brasiliensis |
Eugenia nitida |
Eugenia
pyriformis |
Myrcianthes
pungens |
Plinia edulis |
Psidium
cattleianum |
|---|---|---|---|---|---|---|
| GenBank accession | MN095407 | MN095411 | MN095410 | MN095409 | MN095413 | MN095408 |
| Total cpDNA size (bp) | 158,251 | 157,683 | 158,569 | 159,239 | 159,631 | 159,088 |
| LSC size (bp) | 87,201 | 86,436 | 87,189 | 87,910 | 88,202 | 87,798 |
| SSC size (bp) | 18,290 | 18,349 | 18,566 | 18,587 | 18,579 | 18,512 |
| IR size (bp) | 26,380 | 26,449 | 26,407 | 26,371 | 26,425 | 26,390 |
| Protein coding regions (%) | 50.01 | 50.22 | 49.90 | 49.67 | 49.48 | 49.65 |
| rRNA and tRNA (%) | 7.48 | 7.51 | 7.46 | 7.43 | 7.43 | 7.44 |
| Introns size (% total) | 13.02 | 13.09 | 13.02 | 12.97 | 12.90 | 12.92 |
| Intergenic sequences (%) | 31.21 | 30.85 | 31.32 | 31.63 | 31.79 | 31.58 |
| Number of genes | 129 | 129 | 129 | 129 | 129 | 129 |
| Number of different protein coding genes | 78 | 78 | 78 | 78 | 78 | 78 |
| Number of different tRNA genes | 30 | 30 | 30 | 30 | 30 | 30 |
| Number of different rRNA genes | 4 | 4 | 4 | 4 | 4 | 4 |
| Number of duplicated genes | 16 | 16 | 16 | 16 | 16 | 16 |
| Pseudogenes | 1 | 1 | 1 | 1 | 1 | 1 |
| GC content (%) | 36.95 | 37.04 | 37.01 | 36.94 | 36.93 | 37.05 |
Figure 1. Complete gene map of six Myrteae plastomes. Gene annotations are in black. The plastomes are in red (P. edulis), purple (E. brasiliensis), orange (E. nitida), blue (E. pyriformis), yellow (M. pungens), green (P. cattleianum). LSC: large single-copy region; SSC: small single-copy region; IR: inverted repeat. The numbers near P. edulis (red circle) represent the nucleotide positions (in kbp).
Figure 2. Comparison ofborder positions ofLSC, SSC and IRamong (A) Eugenia uniflora, (B) Psidium guajava and (C) Plinia trunciflora andrelated new species. JSA/JSB, junction of SSC-IRa/IRb; JLA/JLB, junction of LSC-IRa/IRb. Boxes above or under the main line indicate the predicted genes; ycfl pseudogenes are at JSB and their lengths are displayed in the corresponding regions. The figure is not to scaled, and shows relative changes at or near the IR-SC borders.
Table 2. Types, locations, and numbers of SSRs in the chloroplast genomes of six Myrteae species.
| Feature | Eugenia brasiliensis | Eugenia nitida | Eugenia pyriformis | Myrcianthes punges | Plinia edulis | Psidium cattleianum | |
|---|---|---|---|---|---|---|---|
| SSR type | Mono | 188 | 193 | 189 | 194 | 197 | 198 |
| Di | 50 | 49 | 49 | 52 | 49 | 45 | |
| Tri | 65 | 61 | 65 | 63 | 71 | 64 | |
| Tetra | 12 | 12 | 13 | 13 | 13 | 13 | |
| Penta | 0 | 1 | 1 | 0 | 0 | 1 | |
| Location | Intergenic | 176 | 171 | 175 | 181 | 185 | 178 |
| Genes | 96 | 98 | 96 | 96 | 98 | 96 | |
| Introns | 43 | 47 | 46 | 45 | 47 | 47 | |
| Total | 315 | 316 | 317 | 322 | 330 | 321 |
Acknowledgments
This study was carried out with financial fellowship supports from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasil - Finance code 001) and Fundação de Amparo à Pesquisa do Rio Grande do Sul (FAPERGS) with PRONEX grant number 16/2551-0000491-9.
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Ana Tereza Vasconcelos
References
- Alikhan NF, Petty NK, Ben Zakour. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402–402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiryousefi A, Hyvonen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinform. 2003;10(10):3–3. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- Du L, Zhang C, Liu Q, Zhang X, Yue B. Krait: An ultrafast tool for genome-wide survey of micro satellites, and primer design. Bioinformatics. 2018;34:681–683. doi: 10.1093/bioinformatics/btx665. [DOI] [PubMed] [Google Scholar]
- Eguiluz M, Rodrigues NF, Guzman F, Yuyama P, Margis R. The chloroplast genome sequence from Eugenia uniflora, a Myrtaceae from Neotropics. Plant Syst Evol. 2017a;303:1199–1212. [Google Scholar]
- Eguiluz M, Yuyama PM, Guzman F, Rodrigues NF, Margis R. Complete sequence, and comparative analysis of the chloroplast genome of Plinia trunciflora . Genet Mol Biol. 2017b;40:871–876. doi: 10.1590/1678-4685-GMB-2017-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo ADO, Kleinert ADMP. Reproductive biology of six Brazilian Myrtaceae: Is there a syndrome associated with buzz-pollination? New Zeal J Bot. 2009;47:355–365. [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman F, Kulcheski FR, Turchetto-Zolet AC, Margis R. De novo assembly of Eugenia uniflora L. transcriptome, and identification of genes from the terpenoid biosynthesis pathway. Plant Sci. 2014;229:238–246. doi: 10.1016/j.plantsci.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Jackman SD, Vervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, et al. ABySS 2 . 0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 2017;27:768–777. doi: 10.1101/gr.214346.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast, and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid, and mitochondrial genomes, and visualizing expression data sets. Nucleic Acids Res. 2013;41:W575–W581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado LO, Vieira LD, Stefenon VM, Oliveira Pedrosa, Souza EM, Guerra MP, Nodari RO. Phylogenomic relationship of feijoa (Acca sellowiana (O.Berg) Burret) with other Myrtaceae based on complete chloroplast genome sequences. Genetica. 2017;145:163–174. doi: 10.1007/s10709-017-9954-1. [DOI] [PubMed] [Google Scholar]
- Salvador MJ, de Lourenfo, Andreazza NL, Pascoal AC, Stefanello ME. Antioxidant capacity, and phenolic content of four myrtaceae plants of the South of Brazil. Nat Prod Commun. 2011;6:977–982. [PubMed] [Google Scholar]
- Souza-Moreira TM, Severi JA, Rodrigues ER, de Paula, Freitas JA, Vilegas W, Pietro RCLR. Flavonoids from Plinia cauliflora (Mart.) Kausel (Myrtaceae) with antifungal activity. Nat Prod Res. 2019;33:2579–2582. doi: 10.1080/14786419.2018.1460827. [DOI] [PubMed] [Google Scholar]
- Thornhill AH, Ho SYW, Kulheim C, Crisp MD. Interpreting the modern distribution of Myrtaceae using a dated molecular phylogeny. Mol Phylogenet Evol. 2015;93:29–43. doi: 10.1016/j.ympev.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. GeSeq Versatile, and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira LN, Faoro H, Fraga HP, Rogalski M, de Souza, de Oliveira, Nodari RO, Guerra MP. An improved protocol for intact chloroplasts, and cpDNA isolation in conifers. PLoS One. 2014;9:e84792. doi: 10.1371/journal.pone.0084792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WCSP World checklist of selected plant families. 2019. http://wcsp.science.kew.org/
- Wilson PG, O’Brien MM, Heslewood MM, Quinn CJ. Relationships within Myrtaceae sensu lato based on a matK phylogeny. Plant Syst Evol. 2005;251:3–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.