Abstract
In addition to abiotic triggers, biotic factors such as microbial symbionts can alter development of multicellular organisms. Symbiont-mediated morphogenesis is well-investigated in plants and marine invertebrates but rarely in insects despite the enormous diversity of insect-microbe symbioses. The bean bug Riptortus pedestris is associated with Burkholderia insecticola which are acquired from the environmental soil and housed in midgut crypts. To sort symbionts from soil microbiota, the bean bug develops a specific organ called the “constricted region” (CR), a narrow and symbiont-selective channel, located in the midgut immediately upstream of the crypt-bearing region. In this study, inoculation of fluorescent protein-labeled symbionts followed by spatiotemporal microscopic observations revealed that after the initial passage of symbionts through the CR, it closes within 12–18 h, blocking any potential subsequent infection events. The “midgut closure” developmental response was irreversible, even after symbiont removal from the crypts by antibiotics. It never occurred in aposymbiotic insects, nor in insects infected with nonsymbiotic bacteria or B. insecticola mutants unable to cross the CR. However, species of the genus Burkholderia and its outgroup Pandoraea that can pass the CR and partially colonize the midgut crypts induce the morphological alteration, suggesting that the molecular trigger signaling the midgut closure is conserved in this bacterial lineage. We propose that this drastic and quick alteration of the midgut morphology in response to symbiont infection is a mechanism for stabilizing the insect-microbe gut symbiosis and contributes to host-symbiont specificity in a symbiosis without vertical transmission.
Subject terms: Microbial ecology, Symbiosis
Introduction
Phenotypic alteration in response to environmental factors is omnipresent in nature, and is thought to be a driving force for generation of evolutionary novelty [1]. In plants and animals that are intimately associated with microbes, the symbiont acquisition is integrated in the hosts’ development and sometimes triggers dramatic developmental responses. Well-known examples are found in the root nodule formation of the legume-Rhizobium symbiosis and the light organ morphogenesis of the squid-Vibrio symbiosis [2, 3]. Besides such drastic morphological changes in specific associations, recent studies on mammals and Drosophila gut microbiota have revealed that gut bacteria also play a significant role in proliferation and differentiation of gut epithelial cells and in metabolic and immunity homeostasis [4, 5]. Similarly, in the plant kingdom, rhizosphere microbiota modify the root and shoot architecture by modulating the phytohormone balance in the plant [6]. Even nonsymbiotic, environmental bacteria can have profound morphological effects on eukaryotes by the production of small molecules that act as essential developmental cues. The settlement and subsequent metamorphosis of free-swimming larvae of the marine tubeworm Hydroides elegans are trigged by biofilms of the bacterium Pseudoalteromonas luteoviolacea. These biofilms deliver a metamorphosis-triggering effector protein into the larvae via a contractile injection system [7–9]. Another striking example is the triggering of multicellularity from a single founder cell in the choanoflagellate Salpingoaeca rosetta by a sulfonolipid, derived from specific prey bacteria of the Bacteroidetes [10].
The class Insecta consists of over one million described species and is the most diverse animal group in the terrestrial ecosystem. Many insects harbor symbiotic bacteria in specialized cells called bacteriocytes or in gut crypts [11, 12]. In these symbiotic organs, bacterial symbionts play metabolic roles for the hosts, such as provision of essential nutrients and/or digestion of indigestible food materials [13–15]. Recent studies have revealed that, in addition, symbionts alter diverse aspects of host phenotypes, including heat tolerance, pathogen resistance, behavior, sex, cuticle hardening, body color, and tolerance to phytotoxins and insecticides [16–23]. Despite the accumulated knowledge about the diverse symbiosis-mediated phenotypes, symbiont-induced morphogenesis of the symbiotic organ has been poorly described in insects.
The bean bug Riptortus pedestris develops a number of crypts in a posterior region of the midgut, the lumen of which is densely colonized by the betaproteobacterial symbiont, Burkholderia insecticola [24–27]. Infection with B. insecticola improves growth and fecundity of the bean bug [28] probably because the symbiont recycles host metabolic wastes in the midgut crypts [29]. Unlike the typical insect-microbe symbioses in which symbionts are vertically transmitted from mother to offspring, the bean bug acquires B. insecticola from environmental soil every generation [30]. From hatching to adulthood, the bean bug development passes over five instar stages and the symbiont is acquired preferentially at the 2nd instar but also 3rd instar nymphs are compatible for infection [31].
Since diverse microorganisms abound in the environment, particularly in soils [32], hosts need to sort their specific partner from this microbial community. Furthermore, after symbiont colonization, they have to protect the symbiotic organ against invasion by superinfection to stabilize the mutualistic association. To sort the symbiont, the bean bug develops a specific organ called the “constricted region” (CR), a narrow passage filled with a mucus-like matrix, located in the midgut immediately upstream of the crypt-bearing region [33]. The CR sorts out contaminating microorganisms while B. insecticola can penetrate into the symbiotic region using an unusual, corkscrew-like flagellar motility [33–35]. However, the sorting mechanism is not perfectly tight and some allied nonsymbiont Burkholderia species as well as species of its outgroup Pandoraea can nevertheless penetrate the CR [36]. Interestingly, the non-native Burkholderia and Pandoraea species do not fully occupy the lumen of the midgut crypts and co-inoculation analyses revealed that B. insecticola always outcompetes these non-native bacteria inside the crypts, suggesting that the crypt’s luminal environment favors only the native symbiont [36]. Although the molecular basis remains unclear, the competition-based winnowing is therefore, thought of as a second-line mechanism ensuring the host-symbiont specificity. In addition to these partner choice mechanisms, here we discovered a third mechanism underpinning the specific gut symbiosis which we call “midgut closure”, wherein the CR is closed immediately after colonization of the crypt-bearing midgut region by the symbiont, preventing subsequent infections.
Materials and methods
Insects and bacterial strains
The strain of R. pedestris used in this study was originally collected from a soybean (Glycine max) field in Tsukuba, Ibaraki, Japan, and maintained in the laboratory over ten years. The insects were reared in petri dishes (90 mm in diameter and 20 mm high) at 25 °C under a long-day regimen (16 h light, 8 h dark) and fed with soybean seeds and distilled water containing 0.05% ascorbic acid (DWA). For inoculation tests of fluorescent-protein-labeled B. insecticola, a green fluorescent protein (GFP)-labeled strain, RPE225 [28], and a red fluorescent protein (RFP)-labeled strain, KT39 [36], were used. B. insecticola derivatives, including gene-deletion mutants, and nonsymbiotic bacteria used in this study are listed in Supplementary Table S1. They were labeled with GFP using the Tn7 minitransposon system, as previously described [28, 37].
Oral inoculation of cultured symbiont cells
The symbiont strains were grown to early log phase in YG medium (containing 30 µg/ml kanamycin for GFP-labeled RPE225 and RFP-labeled KT39) on a gyratory shaker (150 rpm) at 30 °C. Bacteria were harvested by centrifugation, resuspended in DWA, and adjusted to 104 cells/µl in DWA. Three types of inoculations were performed: (i) oral inoculation of the GFP strain at the 2nd instar, (ii) oral inoculation of the GFP strain at the 3rd instar, and (iii) oral inoculation of the RFP strain at the 2nd and then the GFP strain inoculated after a defined interval.
(i) Oral inoculation of the GFP strain at 2nd instar. Immediately after 1st instar nymphs molted to the 2nd instar, DWA was removed from the rearing containers so that the nymphs were kept without drinking water overnight. Then 1 µl DWA containing 104 cells/µl of the GFP-labeled RPE225 was supplied to the rearing containers, which the 2nd instar nymphs immediately exploited, leading to the acquisition of Burkholderia symbionts. After drinking, insects were supplied with DWA and the nymphs were further reared in the absence of bacteria for variable times before dissection to analyze their infection status as described in the Supplementary Information.
(ii) Oral inoculation of the GFP strain at 3rd instar. Nymphs were kept with soybean seeds and symbiont-free DWA until the 2nd instar. Immediately after 2nd instar nymphs molted to the 3rd instar, the DWA was removed from the rearing containers so that the nymphs were kept without drinking water overnight. Then 1 µl DWA containing 104 cells/µl of the GFP-labeled RPE225 was supplied to the rearing containers, which the 3rd instar nymphs immediately exploited, leading to the acquisition of Burkholderia symbionts. After drinking, insects were supplied with DWA and the nymphs were further reared in the absence of bacteria for variable times before dissection to analyze their infection status as described in the Supplementary Information.
(iii) Oral inoculation of the RFP strain, and re-inoculation of the GFP strain after an interval. After 2nd instar molting, DWA was removed so that nymphs became thirsty, as described above. Then 1 µl DWA containing 104 cells/µl of the RFP-labeled strain, KT39, was orally inoculated to the 2nd instar nymphs. After drinking the bacteria-containing DWA, insects were kept with soybean seeds for 6, 12, 15, 18 h, or 3 days. After the interval, 1 µl DWA containing 104 cells/µl of the GFP-labeled RPE225 was supplied to the rearing containers. As for the first infection, insects were deprived of water before the second infection. These nymphs were further reared for 2 days with symbiont-free DWA, and then dissected to check their infection status of the RFP and GFP strains. In the 3-day-interval group, insects became 3rd instar during the interval, and the molted nymphs were subjected to the re-inoculation test.
Experimental removal of crypt-colonizing symbionts by antibiotic treatment
To investigate the plasticity of the midgut closure, experimental removal of crypt-colonized B. insecticola by antibiotic treatment followed by re-inoculation of the symbiont was performed. For this experiment, a chloramphenicol (Cm)-susceptible GFP-labeled strain, KT33, and a Cm-resistant RFP-labeled strain, RPE744, were used (Supplementary Table S1). The GFP-labeled Cm-susceptible KT33 was constructed by the Tn7 minitransposon system [37], while the RFP-labeled Cm-resistant RPE744 strain carried the stable plasmid pIN29 [38]. Immediately after 1st instar nymphs molted to the 2nd instar, DWA was removed from the rearing containers and the nymphs were kept without drinking water overnight. Then DWA containing 104 cells/µl of the GFP-labeled Cm-susceptible strain KT33 was supplied to the rearing containers for 24 h. After 24 h, the symbiont-containing DWA was replaced by DWA containing 100 µg/ml chloramphenicol to remove the symbiont. Insects were treated with the antibiotic during the 2nd instar stage, and after molting to the 3rd instar, the Cm-resistant RFP-labeled strain RPE744 was inoculated as described above. These nymphs were further reared for 2 days with symbiont-free DWA, and then dissected to check their infection status with the GFP or RFP strains. To verify the first treatments in the 2nd instar stage, half of the treated insects (10 insects/20 in total) were sacrificed in the second instar, dissected and their infection status was verified. In addition, two types of control experiments were performed: first, insects were treated with chloramphenicol but without inoculation of the GFP strain in the 2nd instar stage, and then the RFP-labeled RPE744 strain was inoculated after molting to the 3rd instar; second, insects were inoculated with the GFP-labeled symbiont at the 2nd instar stage without chloramphenicol treatment, and the insects were inoculated with the RFP-labeled RPE744 at the 3rd instar stage.
Methods for “microscopic observations of dissected symbiotic organs”, “oral inoculation of colonization-deficient mutants and nonsymbiotic bacteria” and “bottleneck estimation” are described in the Supplementary Information.
Results
Closure of the constricted region during symbiont colonization
The midgut of R. pedestris consists of four distinct sections called M1, M2, M3, and M4 (Supplementary Fig. S1). The symbiotic Burkholderia specifically colonizes the crypt-bearing M4 section. In front of M4, a distinct, bulbous part called the M4 bulb (M4B) is developed, wherein symbionts flow from the M4 section and are digested [29, 39]. The M3 and M4B are connected by the CR. Our previous study using a GFP-labeled Burkholderia symbiont revealed that in the 2nd instar, colonization of the symbiotic M4B-M4 region of the midgut starts around 6 h post inoculation (hpi) [28]. Around 24 hpi, the symbiont cells appear in the main tract of the M4 region and start the colonization of the crypts. At 48 hpi most of the crypts are colonized and around 72 hpi, crypts are entirely filled by the symbiont cells. This colonization pattern is then stably maintained during the subsequent nymphal stages.
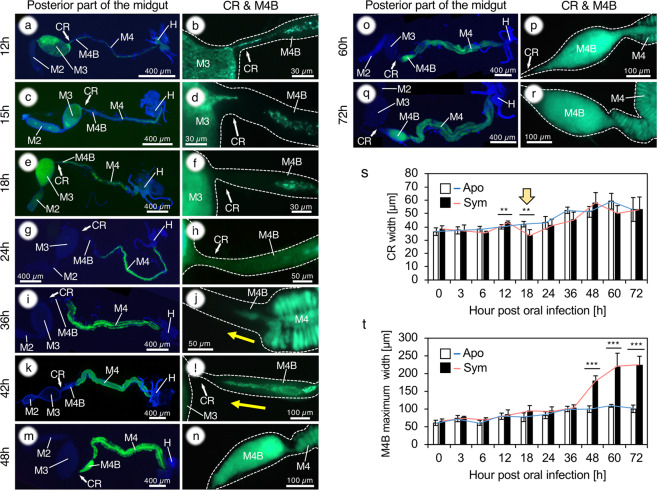
In order to further detail the dynamics of the infection process, we performed time course observations focusing on the CR and M4B regions of the midgut. As described before, the infection from M3 to M4B through the CR started around 6 hpi and a continuous migration of GFP-labeled bacteria was observed (Fig. 1a, b; Supplementary Fig. S2). However, around 15 hpi the connection was abruptly interrupted, despite the continuous presence of a dense population of symbiont cells in the M3 (Fig. 1c, d; Supplementary Fig. S2). Subsequently, the M4B was further closed from the side of the CR, and the closure was accomplished around 24 hpi (Fig. 1e–h; Supplementary Fig. S2). Laser scanning microscopy (LSM) observations revealed that after symbiont colonization, at 24 hpi, the epithelial cells of M4B are bulged at the luminal side and are arranged like interlocking teeth, closed like a zipper (Fig. 2a, b).
Fig. 1. Morphological alteration of the constricted region and M4B during symbiont colonization in 2nd instar nymphs of R. pedestris.
a–r A GFP-labeled Burkholderia symbiont was orally administered, and the posterior part of the midgut (M2-M4) was observed by epifluorescence microscopy, starting from the time of symbiont inoculation to 72 h after inoculation, just before the 3rd instar molting. Host nuclear DNA was stained by DAPI. The M4B is closed during symbiont infection (a–h) and reopened by backflow of symbiont cells during the colonization of M4 (i–r). M2, midgut second section; M3, midgut third section; M4, midgut fourth section with crypts (symbiotic organ); M4B, M4 bulb; H, hindgut. s Change of the CR width. While the CR width continuously increases in aposymbiotic insects, the CR becomes thinner during 12–18 hpi (arrow). This timing corresponds to the midgut closure event. t Change of the maximum M4B width. After the flow back of symbiont cells from M4 to M4B, occurring around 36–48 hpi, the M4B becomes bulbous. n = 5. Welch’s t test was performed between aposymbiotic and symbiotic insects (**P < 0.05; ***P < 0.005).
Fig. 2. Closing of the constricted region.
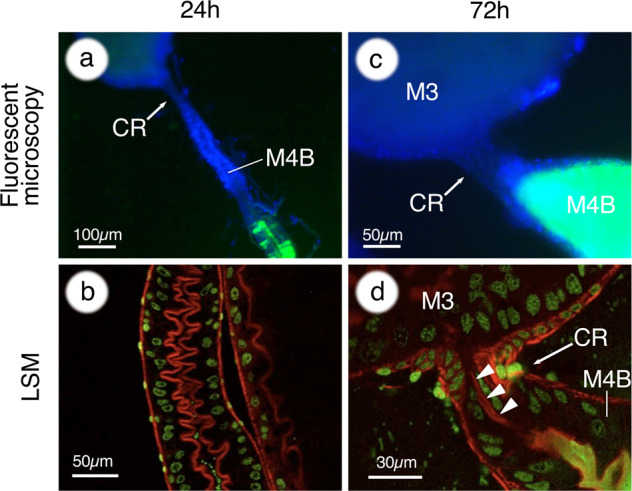
a At 24 h after inoculation, the CR and the M4B region are completely closed. b LSM observations reveal that epithelial cells of M4B are bulged at the luminal side and closed like a zipper during symbiont colonization. c At 72 h after inoculation, symbiont cells flow back into M4B, but never flow over into M3. d LSM observations indicate that the CR (arrowheads) is kept closed after the reopening of M4B. In a and c, host nuclei are stained by DAPI, shown in blue. In b and d, host nuclei and cytoskeleton are stained with SYTOX green (green) and phalloidin (red), respectively.
In the M4 crypts, the Burkholderia symbionts started colonization from 12 hpi, and then proliferated in them until they fully occupied the crypts by 48 hpi (Supplementary Fig. S3). Notably, in parallel with the filling of crypts, symbiont cells started to flow backwards from M4 into M4B around 36–42 hpi (Fig. 1i–l; Supplementary Fig. S2). M4B was entirely filled by symbiont cells and became bulbous and maturated around 48 hpi (Fig. 1m–r; Supplementary Fig. S2). However, no overflow was observed from M4B to M3, which at this stage became free of symbionts. LSM observations of tissues stained with phalloidin showed that the CR is kept tightly closed (Fig. 2c, d). The closure and reopening of M4B were not observed in aposymbiotic insects (Fig. 3a–g). Serial measurements of the CR and M4B (Supplementary Fig. S4) confirmed the marked morphological changes of the CR (Fig. 1s) and M4B (Fig. 1t) after symbiont infection.
Fig. 3. The closure of the constricted region and M4B during symbiont colonization in 3rd instar nymphs of R. pedestris.
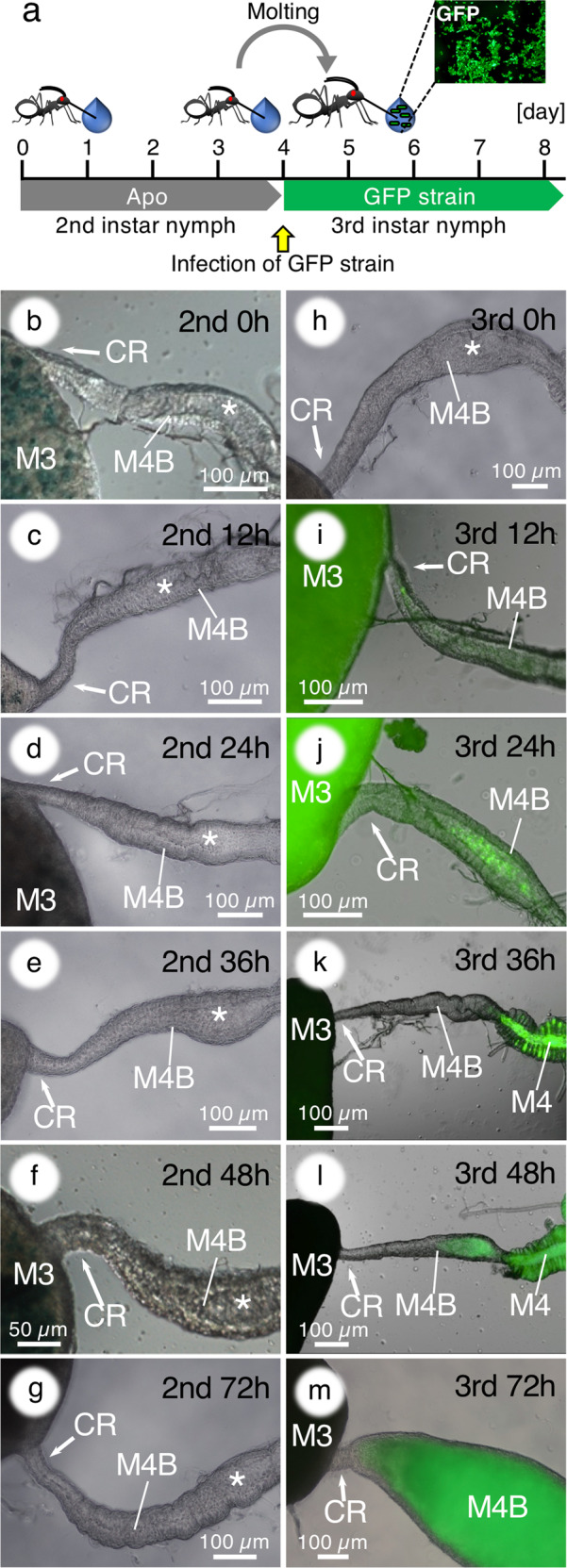
a Infection scheme. b–g No morphological alteration in the M4B of aposymbiotic 2nd instar insects. Asterisks indicate the lumen of M4B. h–m A GFP-labeled Burkholderia symbiont was orally administered to aposymbiotic 3rd instar nymphs and their M4B morphogenesis was investigated. Merged fluorescence and DIC images are shown. Abbreviations are as in Fig. 1.
Symbiont infection of 3rd instar nymphs triggers midgut closure
Our previous study reported that the symbiont acquisition occurs mainly at the 2nd instar but 3rd instar nymphs are still competent to acquire the symbiont [31]. To demonstrate the symbiont-dependency of the gut morphological alteration, we kept insects aposymbiotic during the 2nd instar, and then inoculated symbiont cells after molting and entry into the 3rd instar (Fig. 3a). While the closure and reopening of M4B were not observed in the aposymbiotic 2nd instar stage (Fig. 3b–g), the insects that ingested the symbionts at the 3rd instar stage showed the closure and reopening of M4B after symbiont infection in a similar way as during 2nd instar infection and colonization (Fig. 3h–m). However, the response time in the 3rd instar stage was delayed compared with the 2nd instar stage (Fig. 1). These results clearly demonstrated that the drastic morphological alteration of the midgut is triggered by the symbiont.
Midgut closure blocks superinfection of the M4 crypts
To confirm the physical closure of the CR after the symbiont colonization, a re-inoculation test of the symbiont was performed, in which a GFP-labeled symbiont is inoculated to nymphs that already acquired an RFP-labeled symbiont (Supplementary Fig. S5a). Insects accepted GFP-labeled symbionts when the symbionts were re-inoculated after short, 6–12 h, intervals (Supplementary Figs. S5b, S6). However, insects only rarely acquired the GFP-labeled symbionts when the bacteria were re-inoculated after a 15 h interval (Supplementary Figs. S5b, S6), which strikingly corresponded to the start of the midgut closure observed by microscopy (Fig. 1), and it never happened when the interval was longer (Supplementary Figs. S5b, S6).
Accordingly, when the GFP-labeled symbiont was inoculated to 3rd instar nymphs that already acquired the RFP-labeled symbiont during the 2nd instar stage (Fig. 4a), insects never acquired the GFP-labeled symbiont (Supplementary Figs. S5b, S6). LSM observations revealed that, after colonization of the symbiotic region by an RFP-labeled symbiont, no GFP-labeled symbionts were able to enter the CR, although the GFP-labeled symbiont cells were readily detected in the M3 region (Fig. 4b, c). These results confirmed that the morphological closure of the CR after symbiont colonization in M4 crypts and maturation of M4B, effectively prevents subsequent entry of bacteria in the symbiotic organ.
Fig. 4. The midgut closure prevents subsequent infections.
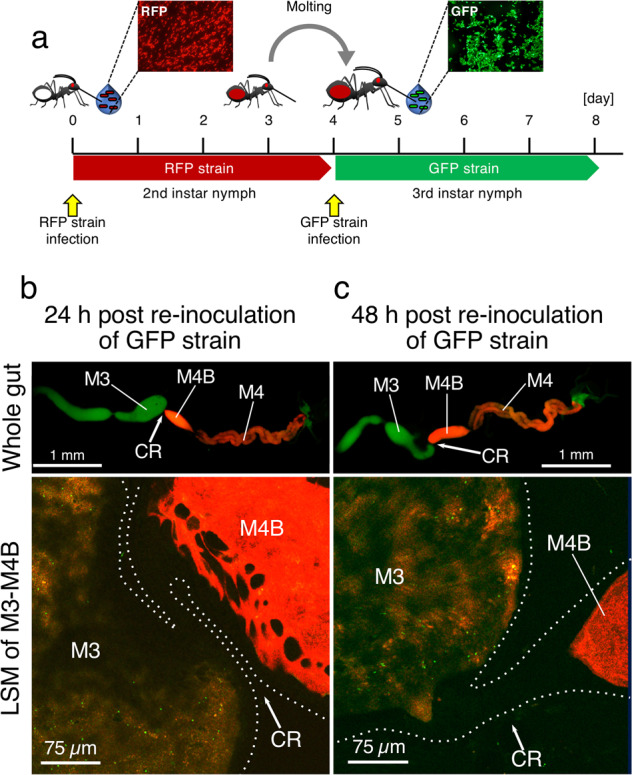
a Experimental set-up of re-inoculation of the Burkholderia symbiont. An RFP-labeled symbiont was orally administered at the 2nd instar stage. After the symbiont colonized the gut symbiotic organ and the nymphs were molted into the 3rd instar, a GFP-labeled symbiont was fed to the insects. b, c GFP-labeled symbionts are not able to enter the M4B and M4 regions anymore after the midgut closure following the initial infection. Observations were made 24 h (b) and 48 h (c) after inoculation of the GFP-labeled symbiont. Whole gut images and LSM images of M3-M4B region are shown. While in the LSM images, GFP signals are detected in the lumen of the M3, no GFP is detected behind the CR. Abbreviations are as in Fig. 1.
The midgut closure is correlated with a bottleneck of symbiont infection
The rapid closing of the CR, together with its narrow diameter of a few micrometers [33] could constrict the number of bacteria that can transit from M3 into M4 and thus cause a bottleneck of symbiont infection. Then, how many symbiont cells can enter the symbiotic region before the closing? To determine the bottleneck size, GFP-labeled symbiont cells were co-inoculated with non-labeled wild-type symbiont cells in different ratios ranging from one in ten to one in 2 × 105. The total inoculum per insect was adjusted to 105 CFU throughout the experiments. The rationale of the experiment was that if the dilution factor of the GFP-labeled strain is below the bottleneck, the marked strain should readily colonize the crypts while it should not when the dilution factor is higher than the bottleneck. If the dilution factor is around the bottleneck size, some insects should be infected and others not with the GFP-marked strain. Based on the detection rate of GFP-labeled symbiont cells, we concluded that the bottleneck of symbiont infection is 3950 cells under the used experimental conditions (Supplementary Fig. S7).
Midgut closure is permanent
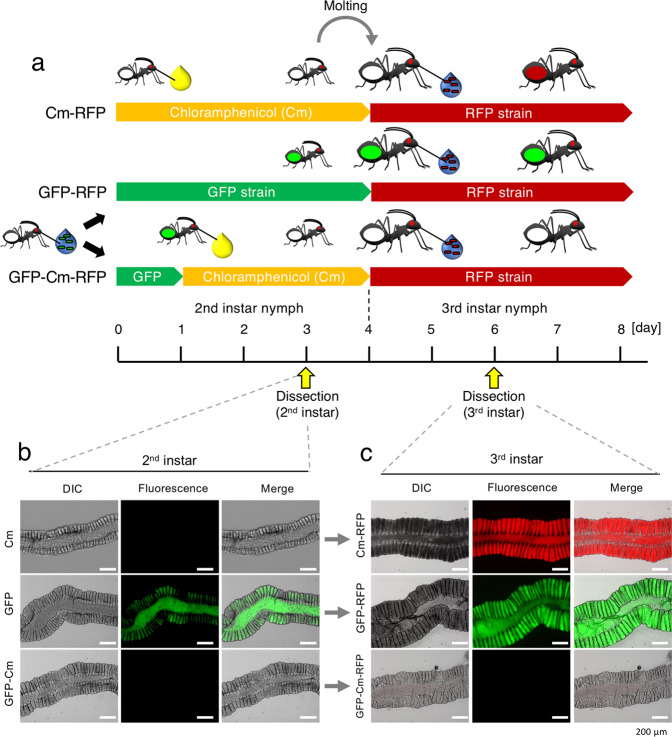
To investigate the plasticity of the midgut closure, an experimental symbiont removal and re-inoculation test was performed. During the 2nd instar stage, a GFP-labeled and chloramphenicol sensitive symbiont was allowed to establish in the M4 crypts and was subsequently removed by chloramphenicol treatment. An RFP-labeled and chloramphenicol resistant symbiont was then re-inoculated at the 3rd instar stage (Fig. 5a). The chloramphenicol treatment during the 2nd instar stage efficiently removed the GFP-labeled symbiont cells from the crypts and the insects became aposymbiotic (GFP-Cm in Fig. 5b). The removal was complete since the GFP-labeled symbionts did not reappear after the stop of the chloramphenicol treatment in the 3rd instar insects (GFP-Cm-RFP in Fig. 5c). Despite this complete elimination of the GFP-labeled symbionts of the first inoculum, no RFP-labeled symbiont was detected in 3rd instar nymphs when the RFP-labeled symbiont was re-inoculated to the cured insects (GFP-Cm-RFP in Fig. 5c). In control experiments, the re-inoculated RFP-labeled symbiont was accepted when insects were treated with chloramphenicol without inoculation with the GFP-labeled symbiont during the 2nd instar stage (Cm-RFP in Fig. 5c). On the other hand, the re-inoculated RFP-labeled symbiont was rejected when a GFP-labeled symbiont was inoculated but not cured by chloramphenicol during the 2nd instar stage (GFP-RFP in Fig. 5c). These results indicate that there is no phenotypic plasticity in the midgut closure (i.e., no reopening of the closed region) and that the closure is maintained once it is triggered, even in the absence of colonizing symbiont cells in the M4 region.
Fig. 5. The midgut closure is irreversible.
a Infection schemes for three series of experiments. In the Cm-RFP control experiment, 2nd instar insects are kept aposymbiotic and fed with chloramphenicol (Cm) in the drinking water; after molting, these insects are infected with the RFP strain. In the GFP-RFP control experiment, 2nd instar insects are infected with the GFP strain present in the drinking water and after molting, the 3rd instars are exposed to the RFP strain present in the drinking water. In the GFP-Cm-RFP experiment, 2nd instar insects are infected with the GFP strain present in the drinking water; after 24 h, this drinking water is replaced by chloramphenicol-containing drinking water; the 3rd instars are then exposed to the RFP strain. In all infection schemes, the infection status was analyzed by dissection and microscopy in 3 dpi 2nd instar insects and in 3 dpi 3rd instar insects. b Infection status in 2nd instar insects. c Infection status in 3rd instar insects. In b, c, red and green fluorescence, DIC and merged images are shown. Note the “fluorescence” panels are merged images of red and green.
Triggering of midgut closure by colonization-deficient mutants and nonsymbiotic bacteria
The rapid midgut closure after infection with the symbiont and its absence in aposymbiotic insects suggest that the passage of bacteria through the CR and/or the colonization of the crypts are required to trigger this developmental response. To test this hypothesis directly, a re-inoculation test of the symbiont was performed, wherein an RFP-labeled B. insecticola was inoculated to nymphs that already acquired a GFP-labeled strain of either colonization-deficient mutants of B. insecticola or nonsymbiotic bacteria (Fig. 6a). The following colonization-defective gene-deletion mutants were used: the flagellar mutant fliC::Tn5 which can reach M3 but is completely blocked by the CR [33] and the purine biosynthesis mutant purL::Tn5 [40], the LPS biosynthesis mutant ΔwaaF [41] and the PHA biosynthesis mutant ΔphaB [42], which can each reach the crypts but cannot proliferate well leading to poor crypt colonization. The insects inoculated with the fliC::Tn5 mutant were colonized by wild-type B. insecticola after re-inoculation indicating no midgut closure by this mutant (Fig. 6c). On the contrary, the purL::Tn5, ΔwaaF, and ΔphaB mutants induced the midgut closure as evidenced by the complete block of infection by the re-inoculated wild type (Fig. 6d–f).
Fig. 6. Midgut closure requires passage through the constricted region and crypt colonization.
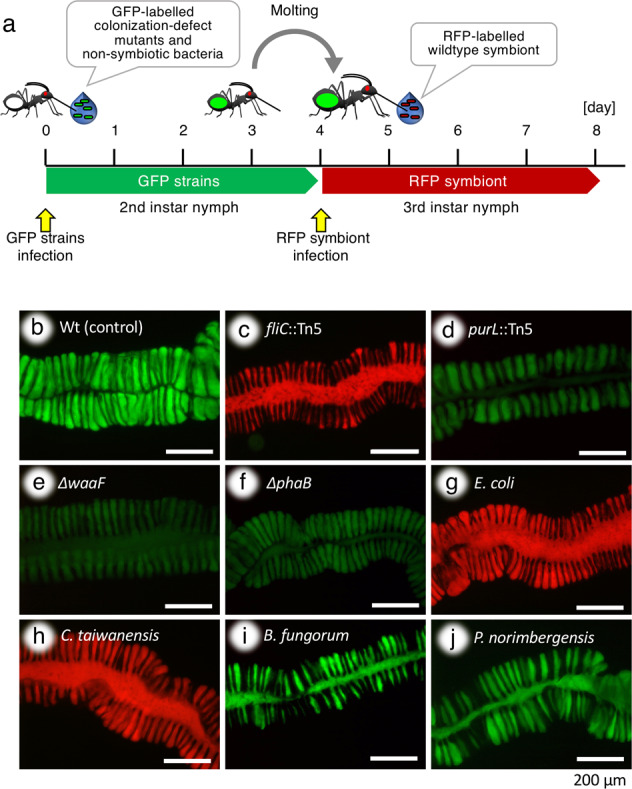
a Infection scheme: 2nd instar insects were infected with different GFP-labeled strains and after molting, 3rd instars were infected with the wild-type RFP symbiont to verify the occurrence or not of midgut closure triggered by the first inoculation. b–j Infection status of 3 dpi 3rd instar insects, analyzed by dissection and microscopy, after infection in the 2nd instar with the wild type (b), the fliC::Tn5 (c), purL::Tn5 (d), ΔwaaF (e) and ΔphaB (f) mutants of B. insecticola, with E. coli (g) and C taiwanensis (h) that cannot pass through the CR, and with B. fungorum (i) and P. norimbergensis (j) that can pass through the CR and partially colonize the M4 crypts. In b–j, fluorescence images are shown. Note the fluorescence panels are merged images of red and green.
As nonsymbiotic bacteria, the following four species were inoculated: Escherichia coli and Cupriavidus taiwanensis which are both blocked at the CR; Burkholderia fungorum and Pandoraea norimbergensis which can both pass through the CR and partially colonize the midgut crypts [36]. Insects inoculated with E. coli or C. taiwanensis did not induce the midgut closure and were colonized by wild-type B. insecticola after re-inoculation (Fig. 6g, h). In contrast, B. fungorum and P. norimbergensis induced the midgut closure and completely blocked the re-inoculated wild type (Fig. 6i, j). Together, these experiments confirm that the midgut closure is triggered only when bacteria have entered the symbiotic organ while their presence in the M3 is not sufficient.
Discussion
Our previous study reported that the bean bug R. pedestris develops in the midgut a mucus-filled sorting organ, the CR. This unusual organ selects the symbiotic Burkholderia from the enormously diverse soil microorganisms that can penetrate the intestine during feeding and drinking [33]. Through the detailed observations of the symbiont infection process in the 2nd instar stage, we revealed here additional important features of this selection mechanism. Our observations showed that: (i) the CR as well as the M4B are closed in response to symbiont infection (Figs. 1, 2; Supplementary Fig. S2); the morphological alteration of the M4B and the CR did not occur in aposymbiotic nymphs, but was reproduced in 3rd instar nymphs when insects were reared in the absence of bacteria in the 2nd instar and then inoculated after molting (Fig. 3); (ii) after the closure, the symbiont cells proliferating in the M4 crypts flow back into M4B, which opens again and becomes bulbous and filled with symbiont cells (Fig. 1; Supplementary Fig. S2); (iii) in spite of the M4B opening, the CR is kept closed (Fig. 2), blocking any further infection events (Fig. 4); (iv) once triggered, the CR is kept closed permanently, even when symbionts are removed from the crypts by antibiotic treatment (Fig. 5); (v) to triggered the closure, the passage of bacteria—symbionts or other species capable to do so—through the CR and their installation in the crypts is required (Fig. 6) and (vi) the midgut closure imposes an infection bottleneck on the symbiont population, limiting the founding population that will populate the symbiotic organ to a maximum of a few thousand cells (Supplementary Fig. S7). Together, these results demonstrate that the establishment of bacteria in the midgut crypts provokes a marked and rapid morphogenic response in the symbiotic organ, which is already largely formed before the establishment of the symbiosis. To our knowledge, this is a rare example in the insects of symbiotic microorganisms that trigger a developmental alteration.
In contrast to the intracellular symbioses that are typically found in insects, the gut symbiotic association of R. pedestris and related stinkbugs is an open system and the M4 symbiotic organ is in direct contact with the digestive tract, located upstream in the intestine (the M1 to M3 midgut regions). Therefore, the symbiotic organ faces a constant risk of invasion by other microorganisms, including cheaters and potential pathogens. From this point of view, the midgut closure found in the bean bug is one of the ultimate mechanisms to limit contamination of the gut symbiotic organ by unwanted microbes. Indeed, the re-inoculation test of the Burkholderia symbiont clearly demonstrated that the closure effectively prevents entrance of bacteria once the symbiont is established in the M4B-M4 region (Fig. 4; Supplementary Fig. S5). Moreover, the midgut closure operates within an intestinal organ that has a very distinctive organization and functioning. The bean bug externally digests the content of seeds of leguminous plants and sucks up liquid food [43]. By use of food colorings, we showed before that ingested liquid food is completely digested in M3 and absorbed into the hemolymph [33] and never enters the M4B and M4 region. Thus the symbiotic organ is isolated from the digestive flow in the intestine and this disconnection is independent of the symbiosis since it is established already in aposymbiotic animals [33]. Metabolic wastes are removed by the malpighian tubes and excreted via the hindgut, although transcriptome analyses suggest that some metabolic wastes are supplied to the M4 crypts and recycled by the symbiotic Burkholderia [29]. We propose that the symbiont-mediated midgut closure, in conjunction with the CR and the unique digestive system, plays a significant role to stabilize the specific host-symbiont interaction in the bean bug.
The inoculation tests with colonization-defective mutants (Fig. 6c–f) suggest how the midgut closure is triggered by the symbiont. First, the absence of the midgut closure by the fliC::Tn5 mutant that cannot pass through the CR, indicates that passage through this channel and/or M4 colonization is necessary to trigger the closure but that it cannot be triggered by bacteria that are located at the M3-side of the CR. Second, the results of the colonization-defective mutants purL::Tn5, ΔwaaF, and ΔphaB suggest that a weak colonization of the crypts is sufficient to trigger the midgut closure. Although the molecular mechanism of the midgut closure remains totally unknown, it appears likely that the bean bug recognizes the Burkholderia symbiont either in the CR or in the M4 by epithelial cells in a dose-dependent manner. In the legume-Rhizobium system, a lipochito-oligosaccharide (Nod factor) triggers the nodule formation [44]. In the case of the bobtail squid, a peptidoglycan fragment, tracheal cytotoxin (TCT), derived from Vibrio fischeri alters morphogenesis of the light organ [45, 46]. It is notable that in both cases, the host organisms recognize symbiont-associated molecular patterns via immune-related receptors to trigger a developmental response [44, 47]. Gut immunity genes have been reported in R. pedestris [48, 49], which may be involved in the here described symbiont-dependent morphogenesis in this insect. The bean bug is an ideal experimental system, where reverse genetic approaches have been established in both the host and in the symbiotic Burkholderia [25]. Transcriptomic analyses of the CR during the closure, followed by RNAi screening, and identification of bacterial compounds triggering midgut closure in the absence of live bacteria, could be a strategy leading towards the understanding of the molecular basis underlying the symbiont-triggered morphogenesis in the bean bug.
We recently demonstrated that some nonsymbiotic species of the genus Burkholderia and its outgroup Pandoraea can pass through the CR and stably but partially accommodate in the midgut crypts. Moreover, this colonization is beneficial for the host survival and development [36]. Notably, while E. coli and C. taiwanensis showed neither M4 colonization nor induction of the midgut closure (Fig. 6g, h), B. fungorum and P. norimbergensis colonized the midgut crypts and induced the closure (Fig. 6i, j), suggesting that the microbial factors triggering the morphological alteration are conserved in this bacterial lineage. This reveals a potential drawback of the midgut closure mechanism because when induced by nonsymbiotic species, it can restrict the access of the native symbiont to the M4 region if the symbiont is not present in the first infection event. However, these nonsymbionts are rarely detected in natural populations of the bean bug [24, 30, 50]. Considering that the symbiotic Burkholderia, nonsymbiotic Burkholderia and Pandoraea are very common bacterial groups in soil environments and frequently detected together [51–53], it is plausible that, in natural environments, these nonsymbiotic species are usually ingested together with symbiotic Burkholderia and are then outcompeted by the native symbiont inside the midgut crypts [36], which ensures the specific association in natural populations of the bean bug.
Bottlenecks imposed on symbiont populations during their transmission or acquisition by the host are thought to strongly affect the population structure and evolution of symbionts [54]. Such bottlenecks have been well described in insect symbioses with vertical transmission. These bottlenecks are large but vary from hundreds or thousands to several millions of symbiont cells transferred from mother to offspring, depending on the insect host and the specific transmission mode [54–57]. On the contrary, in the few previously characterized symbioses without vertical transmission, symbionts seem to be selected from the environment in much smaller numbers; experimental studies on the legume-Rhizobium and squid-Vibrio model systems revealed that only a few symbiont cells (mostly only one bacterial cell) initiate the colonization, regardless of symbiont density in the environment [58, 59], suggesting a drastic partner choice in the initial stage of the infection. Relative large bottlenecks during vertical transmission seem to be compatible with the fact that the symbiont lineage has been preselected efficiently in the previous generations of the host while the very narrow bottlenecks in environmental transmission modes might be required to cope with the extreme richness of microbes in the environment. However, unlike the legume and squid model systems, we determined a symbiont infection bottleneck of about 4000 cells in R. pedestris, indicating a milder sorting, at least in the initial infection stage. Thus, the bean bug seems to adopt a different strategy than legumes and the squid and enables a range of bacteria, including some nonsymbiont species, to pass the gate of the symbiotic organ (i.e., the CR). However, it applies then a severe selection inside the crypts of the symbiotic organ where nonsymbiotic bacteria are very efficiently outcompeted by symbiont species [36].
The CR is widely conserved among diverse stinkbug species that harbor gut symbiotic bacteria in midgut crypts [33], including the Bukholderia-associated coreoid and lygaeid species and members of the superfamily Pentatomoidea that vertically transmit gamma-proteobacterial symbionts. As the risk of contamination is a common issue in these gut symbiotic systems, it is plausible that the midgut closure after symbiont colonization is prevalent among the stinkbug species. In the case of members of the Plataspidae and Urostylididae families belonging to the Pentatomoidea superfamily, the M3 and M4B are only joint by a thin connective tissue in adult insects [33, 60–62], suggesting that the CR is indeed closed and completely disconnected from the M3 after symbiont infection in these species. However, in case of the brown-winged green bug Plautia stali, the CR becomes looser after the 5th instar stage is reached and a food coloring can pass through from M3 to M4 [63]. Thus, a morphological diversity of the CR exists among stinkbug species and may reflect a functional difference of this organ. This potential divergence requires comprehensive comparative analyses between different stinkbug species.
Recent microbiome analyses have revealed that a number of animals possess diverse but highly specialized microbiota in their gut, which are maintained by various physiochemical, molecular, and structural mechanisms. In human gut microbiota, mucus layers containing specific polysaccharides and antimicrobial peptides (AMPs) control the microbiota and facilitate specific colonization of Bacteroides and Acinatobacter in colon crypts [64]. In termites and honey bees, the hypoxia condition of the gut lumen remarkably contributes to shape their specific microbiota [15, 65, 66]. In Drosophila, immune-related AMPs and reactive oxygen species limit the growth of pathogenic bacteria and play a significant role to establish its indigenous gut microbiota [67, 68]. A more extreme case is found in the Sonoran Desert turtle ant, wherein a proventriculus valve located between the foregut and midgut filters out bacteria and keeps an exclusively specific microbiota in the midgut [69]. Among these diverse mechanisms, the midgut closure reported here is an additional and unique mechanism to maintain the population of a specific bacterial partner and prevent chronic contamination with unwanted microbes. Our findings expand the understanding of how animals sustain specific gut symbiosis.
Supplementary information
Acknowledgements
We thank H. Ooi (AIST) for technical assistance. Part of the fluorescence microscopy was performed at the Imagerie-Gif facilities (http://www.i2bc.paris-saclay.fr/spip.php?rubrique184). This study was supported by the JSPS-CNRS Bilateral Open Partnership Joint Research Project to YK and PM, the Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI to YK (18KK0211), and the JSPS Research Fellowship for Young Scientists to TO (20170267 and 19J01106) and SJ (201911493).
Author contributions
YK, TO, and PM designed the study. YK, TO, and SJ performed inoculation tests, antibiotic treatments, and microscopy observations. TO and PM measured morphological change of gut parts and bottleneck of symbiont infection. YK and PM wrote the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-0633-3) contains supplementary material, which is available to authorized users.
References
- 1.Gilbert SF, Epel D. Ecological developmental biology: integrating epigenetics, medicine, and evolution. Sunderland, MA: Sinauer Associates Inc; 2009. [Google Scholar]
- 2.Nyholm SV, McFall-Ngai M. The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–42. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 3.Kondorosi E, Mergaert P, Kereszt A. A paradigm for endosymbiotic life: cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu Rev Microbiol. 2013;67:611–28. doi: 10.1146/annurev-micro-092412-155630. [DOI] [PubMed] [Google Scholar]
- 4.Sommer F, Backhed F. The gut microbiota-masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–38. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 5.Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio. 2014;5:e01117–14. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vacheron J, Desbrosses G, Bouffaud M-L, Touraine B, Moënne-Loccoz Y, Muller D, et al. Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci. 2013;4:356. doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, Newman DK. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science. 2014;343:529–33. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shikuma NJ, Antoshechkin I, Medeiros JM, Pilhofer M, Newman DK. Stepwise metamorphosis of the tubeworm Hydroides elegans is mediated by a bacterial inducer and MAPK signaling. Proc Natl Acad Sci USA. 2016;113:10097–102. doi: 10.1073/pnas.1603142113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ericson CF, Eisenstein F, Medeiros JM, Malter KE, Cavalcanti GS, Zeller RW, et al. A contractile injection system stimulates tubeworm metamorphosis by translocating a proteinaceous effector. eLife. 2019;8:e46845. doi: 10.7554/eLife.46845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alegado RA, Brown LW, Cao S, Dermenjian RK, Zuzow R, Fairclough SR, et al. A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. eLife. 2012;1:e00013. doi: 10.7554/eLife.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchner P. Endosymbiosis of animals with plant microorganism. New York: Interscience; 1965. [Google Scholar]
- 12.Kikuchi Y. Endosymbiotic bacteria in insects: their diversity and culturability. Microbes Environ. 2009;24:195–204. doi: 10.1264/jsme2.me09140s. [DOI] [PubMed] [Google Scholar]
- 13.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–90. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 14.Douglas AE. Lessons from studying insect symbioses. Cell Host Microbe. 2011;10:359–67. doi: 10.1016/j.chom.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Utami YD, Kuwahara H, Murakami T, Morikawa T, Sugaya K, Kihara K, et al. Phylogenetic diversity and single-cell genome analysis of “Melainabacteria”, a non-photosynthetic cyanobacterial group, in the termite gut. Microbes Environ. 2018;33:50–7. doi: 10.1264/jsme2.ME17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T. Symbiont acquisition alters behaviour of stinkbug nymphs. Biol Lett. 2008;4:45–8. doi: 10.1098/rsbl.2007.0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 18.Feldhaar H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol. 2011;36:533–43. [Google Scholar]
- 19.Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. Symbiont-mediated insecticide resistance. Proc Natl Acad Sci USA. 2012;109:8618–22. doi: 10.1073/pnas.1200231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flórez LV, Biedermann PH, Engl T, Kaltenpoth M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep. 2015;32:904–36. doi: 10.1039/c5np00010f. [DOI] [PubMed] [Google Scholar]
- 21.Tsuchida T. Molecular basis and ecological relevance of aphid body colors. Curr Opin Insect Sci. 2016;17:74–80. doi: 10.1016/j.cois.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Anbutsu H, Moriyama M, Nikoh N, Hosokawa T, Futahashi R, Tanahashi M, et al. Small genome symbiont underlies cuticle hardness in beetles. Proc Natl Acad Sci USA. 2017;114:E8382–91. doi: 10.1073/pnas.1712857114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh H, Tago K, Hayatsu M, Kikuchi Y. Detoxifying symbiosis: microbe-mediated detoxification of phytotoxins and pesticides in insects. Nat Prod Rep. 2018;35:434–54. doi: 10.1039/c7np00051k. [DOI] [PubMed] [Google Scholar]
- 24.Kikuchi Y, Meng XY, Fukatsu T. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae) Appl Environ Microbiol. 2005;71:4035–43. doi: 10.1128/AEM.71.7.4035-4043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeshita K, Kikuchi Y. Riptortus pedestris and Burkholderia symbiont: an ideal model system for insect-microbe symbiotic associations. Res Microbiol. 2016;168:175–87. doi: 10.1016/j.resmic.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Takeshita K, Tamaki H, Ohbayashi T, Meng XY, Sone T, Mitani Y, et al. Burkholderia insecticola sp. nov., a gut symbiotic bacterium of the bean bug Riptortus pedestris. Int J Syst Evol Microbiol. 2018;68:2370–4. doi: 10.1099/ijsem.0.002848. [DOI] [PubMed] [Google Scholar]
- 27.Kaltenpoth M, Flórez LV. Versatile and dynamic symbioses between insects and Burkholderia bacteria. Annu Rev Entomol. 2020;65:145–70. doi: 10.1146/annurev-ento-011019-025025. [DOI] [PubMed] [Google Scholar]
- 28.Kikuchi Y, Fukatsu T. Live imaging of symbiosis: spatiotemporal infection dynamics of a GFP-labelled Burkholderia symbiont in the bean bug Riptortus pedestris. Mol Ecol. 2014;23:1445–56. doi: 10.1111/mec.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohbayashi T, Futahashi R, Terashima M, Barrière Q, Lamouche F, Takeshita K, et al. Comparative cytology, physiology and transcriptomics of Burkholderia insecticola in symbiosis with the bean bug Riptortus pedestris and in culture. ISME J. 2019;13:1469–83. doi: 10.1038/s41396-019-0361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikuchi Y, Hosokawa T, Fukatsu T. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl Environ Microbiol. 2007;73:4308–16. doi: 10.1128/AEM.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kikuchi Y, Hosokawa T, Fukatsu T. Specific developmental window for establishment of an insect-microbe gut symbiosis. Appl Environ Microbiol. 2011;77:4075–81. doi: 10.1128/AEM.00358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahram M, Hildebrand F, Forslund SK, Anderson JL, Soudzilovskaia NA, Bodegom PM, et al. Structure and function of the global topsoil microbiome. Nature. 2018;560:233–7. doi: 10.1038/s41586-018-0386-6. [DOI] [PubMed] [Google Scholar]
- 33.Ohbayashi T, Takeshita K, Kitagawa W, Nikoh N, Koga R, Meng XY, et al. Insect’s intestinal organ for symbiont sorting. Proc Natl Acad Sci USA. 2015;112:E5179–88. doi: 10.1073/pnas.1511454112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinosita Y, Kikuchi Y, Mikami N, Nakane D, Nishizaka T. Unforeseen swimming and gliding mode of an insect gut symbiont, Burkholderia sp. RPE64, with wrapping of the flagella around its cell body. ISME J. 2018;12:838–48. doi: 10.1038/s41396-017-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raina JB, Fernandez V, Lambert B, Stocker R, Seymour JR. The role of microbial motility and chemotaxis in symbiosis. Nat Rev Microbiol. 2019;17:284–94. doi: 10.1038/s41579-019-0182-9. [DOI] [PubMed] [Google Scholar]
- 36.Itoh H, Jang S, Takeshita K, Ohbayashi T, Ohnishi N, Meng XY, et al. Host-symbiont specificity determined by microbe-microbe competition in an insect gut. Proc Natl Acad Sci USA. 2019;116:22673–82. doi: 10.1073/pnas.1912397116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norris MH, Kang Y, Wilcox B, Hoang TT. Stable, site-specific fluorescent tagging constructs optimized for Burkholderia species. Appl Environ Microbiol. 2010;76:7635–40. doi: 10.1128/AEM.01188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vergunst AC, Meijer AH, Renshaw SA, O’Callaghan D. Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infect Immun. 2010;78:1495–508. doi: 10.1128/IAI.00743-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JK, Kim NH, Jang HA, Kikuchi Y, Kim CH, Fukatsu T, et al. Specific midgut region controlling the symbiont population in an insect-microbe gut symbiotic association. Appl Environ Microbiol. 2013;79:7229–33. doi: 10.1128/AEM.02152-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim JK, Jang HA, Won YJ, Kikuchi Y, Heum Han S, Kim CH, et al. Purine biosynthesis-deficient Burkholderia mutants are incapable of symbiotic accommodation in the stinkbug. ISME J. 2014;8:552–63. doi: 10.1038/ismej.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JK, Jang HA, Kim MS, Cho JH, Lee JB, Lorenzo FD, et al. The lipopolysaccharide core oligosaccharide of Burkholderia plays a critical role in maintaining a proper gut symbiosis with the bean bug Riptortus pedestris. J Biol Chem. 2017;292:19226–37. doi: 10.1074/jbc.M117.813832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim JK, Won YJ, Nikoh N, Nakayama H, Han SH, Kikuchi Y, et al. Polyester synthesis genes associated with stress resistance are involved in an insect-bacterium symbiosis. Proc Natl Acad Sci USA. 2013;110:E2381–9. doi: 10.1073/pnas.1303228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasunaga T, Takai M, Yamashita I, Kawamura M, Kawasawa T. A field guide to Japanese bugs. Tokyo, Japan: Zenkoku Nouson Kyouiku Kyoukai; 1993. [Google Scholar]
- 44.Zipfel C, Oldroyd GE. Plant signalling in symbiosis and immunity. Nature. 2017;543:328–36. doi: 10.1038/nature22009. [DOI] [PubMed] [Google Scholar]
- 45.Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–8. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 46.Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol. 2009;191:2012–22. doi: 10.1128/JB.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14:668–75. doi: 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim JK, Lee JB, Jang HA, Han YS, Fukatsu T, Lee BL. Understanding regulation of the host-mediated gut symbiont population and the symbiont-mediated host immunity in the Riptortus-Burkholderia symbiosis system. Dev Comp Immunol. 2016;64:75–81. doi: 10.1016/j.dci.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 49.Park KE, Jang SH, Lee J, Lee SA, Kikuchi Y, Seo YS, et al. The roles of antimicrobial peptide, rip-thanatin, in the midgut of Riptortus pedestris. Dev Comp Immunol. 2017;78:83–90. doi: 10.1016/j.dci.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 50.Kikuchi Y, Hosokawa T, Fukatsu T. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 2011;5:446–60. doi: 10.1038/ismej.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Itoh H, Navarro R, Takeshita K, Tago K, Hayatsu M, Hori T, et al. Bacterial population succession and adaptation affected by insecticide application and soil spraying history. Front Microbiol. 2014;5:457. doi: 10.3389/fmicb.2014.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Itoh H, Hori T, Sato Y, Nagayama A, Tago K, Hayatsu M, et al. Infection dynamics of insecticide-degrading symbionts from soil to insects in response to insecticide spraying. ISME J. 2018;12:909–20. doi: 10.1038/s41396-017-0021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stopnisek N, Bodenhausen N, Frey B, Fierer N, Eberl L, Weisskopf L. Genus-wide acid tolerance accounts for the biogeographical distribution of soil Burkholderia populations. Environ Microbiol. 2014;16:1503–12.. doi: 10.1111/1462-2920.12211. [DOI] [PubMed] [Google Scholar]
- 54.Campbell MA, Łukasik P, Meyer MC, Buckner M, Simon C, Veloso C, et al. Changes in endosymbiont complexity drive host-level compensatory adaptations in cicadas. mBio. 2018;9:e02104–18.. doi: 10.1128/mBio.02104-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mira A, Moran NA. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Micro Ecol. 2002;44:137–43. doi: 10.1007/s00248-002-0012-9. [DOI] [PubMed] [Google Scholar]
- 56.Kaltenpoth M, Goettler W, Koehler S, Strohm E. Life cycle and population dynamics of a protective insect symbiont reveal severe bottlenecks during vertical transmission. Evol Ecol. 2010;24:463–77. [Google Scholar]
- 57.Hosokawa T, Kikuchi Y, Fukatsu T. How many symbionts are provided by mothers, acquired by offspring, and needed for successful vertical transmission in an obligate insect-bacterium mutualism? Mol Ecol. 2007;16:5316–25. doi: 10.1111/j.1365-294X.2007.03592.x. [DOI] [PubMed] [Google Scholar]
- 58.Wollenberg MS, Ruby EG. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol. 2009;75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gage DJ. Analysis of infection thread development using GFP- and DsRed-expressing Sinorhizobium meliloti. J Bacteriol. 2002;184:7042–6. doi: 10.1128/JB.184.24.7042-7046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider G. Beiträge zur Kenntnis der symbiontischen Einrichtungen der Heteropteren. Z Morphol O'kol Tiere. 1940;36:565–644. [Google Scholar]
- 61.Fukatsu T, Hosokawa T. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl Environ Microbiol. 2002;68:389–96. doi: 10.1128/AEM.68.1.389-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaiwa N, Hosokawa T, Nikoh N, Tanahashi M, Moriyama M, Meng XY, et al. Symbiont-supplemented maternal investment underpinning host’s ecological adaptation. Curr Biol. 2014;24:2465–70. doi: 10.1016/j.cub.2014.08.065. [DOI] [PubMed] [Google Scholar]
- 63.Oishi S, Moriyama M, Koga R, Fukatsu T. Morphogenesis and development of midgut symbiotic organ of the stinkbug Plautia stali (Hemiptera: Pentatomidae) Zool Lett. 2019;5:16. doi: 10.1186/s40851-019-0134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brune A, Emerson D, Breznak JA. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl Environ Microbiol. 1995;61:2681–7. doi: 10.1128/aem.61.7.2681-2687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc Natl Acad Sci USA. 2017;114:4775–80. doi: 10.1073/pnas.1701819114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buchon N, Broderick NA, Lemaitre B. Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat Rev Microbiol. 2013;11:615–26. doi: 10.1038/nrmicro3074. [DOI] [PubMed] [Google Scholar]
- 68.Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol. 2014;10:416–24. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 69.Lanan MC, Rodrigues PA, Agellon A, Jansma P, Wheeler DE. A bacterial filter protects and structures the gut microbiome of an insect. ISME J. 2016;10:1866–76. doi: 10.1038/ismej.2015.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.