Abstract
Detection of lymphocytes that target tumor-specific mutant neoantigens—derived from products encoded by mutated genes in the tumor—is mostly limited to tumor-resident lymphocytes1,2, but whether these lymphocytes often occur in the circulation is unclear. We recently reported that intratumoral expression of the programmed cell death 1 (PD-1) receptor can guide the identification of the patient-specific repertoire of tumor-reactive CD8+ lymphocytes that reside in the tumor3. In view of these findings, we investigated whether PD-1 expression on peripheral blood lymphocytes could be used as a biomarker to detect T cells that target neoantigens. By using a high-throughput personalized screening approach, we identified neoantigen-specific lymphocytes in the peripheral blood of three of four melanoma patients. Despite their low frequency in the circulation, we found that CD8+PD-1+, but not CD8+PD-1−, cell populations had lymphocytes that targeted 3, 3 and 1 unique, patient-specific neoantigens, respectively. We show that neoantigen-specific T cells and gene-engineered lymphocytes expressing neoantigen-specific T cell receptors (TCRs) isolated from peripheral blood recognized autologous tumors. Notably, the tumor-antigen specificities and TCR repertoires of the circulating and tumor-infiltrating CD8+PD-1+ cells appeared similar, implying that the circulating CD8+PD-1+ lymphocytes could provide a window into the tumor-resident antitumor lymphocytes. Thus, expression of PD-1 identifies a diverse and patient-specific antitumor T cell response in peripheral blood, providing a novel noninvasive strategy to develop personalized therapies using neoantigen-reactive lymphocytes or TCRs to treat cancer.
Cancer immunotherapies—including high-dose IL-2 (ref. 4), adoptive transfer of tumor-infiltrating lymphocytes (TILs)5 and immune-checkpoint inhibitors targeting the cell surface receptors PD-1 and cytotoxic T lymphocyte–associated protein (CTLA)-4 (refs. 6–9)—can mediate antitumor responses in patients with metastatic melanoma. Melanomas exhibit a high prevalence of mutations10,11, and accumulating correlative data suggest that T cell reactivity to neoantigens has a critical role in the clinical activity of cancer immunotherapies12–15. This has stimulated attempts to develop vaccines or T cell–based therapies targeting unique, patient-specific neoantigens16–19. We reported that adoptive transfer of selected CD4+ lymphocytes that recognize a mutant tumor-specific neo-epitope in ERBB2-interacting protein (encoded by ERBB2IP) into a patient with cholangiocarcinoma induced sustained tumor regression16. However, frequent detection of CD8+ and CD4+ lymphocytes that target neoantigens is currently limited to T cells isolated from tumor deposits1,2,15,16, which are often not available. Given the increasing evidence supporting their importance in cancer immunotherapy, we explored whether lymphocytes that target tumor-specific neoantigens frequently occur in peripheral blood and could thus be more easily exploited therapeutically.
Evidence of circulating neoantigen-specific lymphocytes in cancer patients is scarce, although rare clonotypes have been identified12,20 and isolated21 using human leukocyte antigen (HLA) multimers22, which are available for a limited number of HLA restriction elements and depend on the accuracy of peptide-prediction algorithms. Alternatively, mutant peptide–specific lymphocytes have been detected after multiple rounds of in vitro sensitization23. Thus, a biomarker that could prospectively identify a potentially diverse neoantigen-specific T cell response in peripheral blood, without previous knowledge of the specific neo-epitope targeted or the HLA restriction element, would be highly advantageous. Tumor-reactive and neoantigen-specific lymphocytes that infiltrate melanomas coexpress multiple inhibitory and co-stimulatory receptors3,24,25, and we found that expression of PD-1 on CD8+ TILs can identify the patient-specific repertoire of antitumor T cells3,26. Here we investigated whether PD-1 could be used as a biomarker to identify circulating neoantigen-specific lymphocytes in patients with melanoma.
We first compared the expression of PD-1 on tumor-resident and circulating CD8+ lymphocytes. PD-1 expression accounted for ~36% of the CD8+ TIL population, but peripheral blood samples from the same individuals contained only a median of 4.1% CD8+PD-1+ cells (Fig. 1a). Moreover, circulating CD8+ lymphocytes had limited coexpression of the inhibitory and co-stimulatory cell surface receptors PD-1, TIM-3, LAG-3 and 4–1BB as compared to tumor-resident CD8+ lymphocytes (Fig. 1b). A more detailed phenotypic characterization of the circulating CD8+PD-1+ lymphocytes is summarized in Supplementary Figure 1. Thus, few PD-1–expressing circulating CD8+ lymphocytes are present in patients with melanoma, and their significance is unknown.
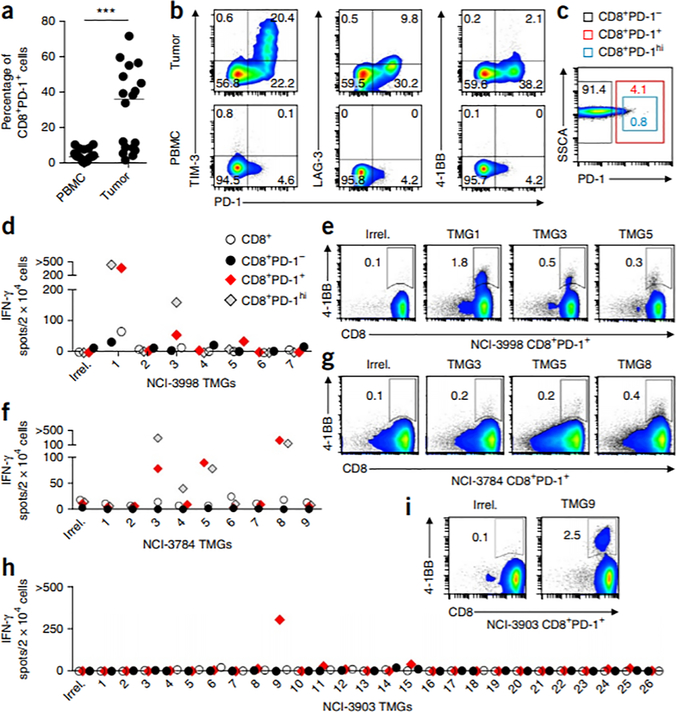
Figure 1.
Frequency of PD-1 expression on circulating and tumor-resident CD8+ lymphocytes and prospective identification of circulating neoantigen-reactive cells in melanoma patients. (a) Expression of PD-1 on CD8+ lymphocytes in matched PBMC and tumors (n = 18). The median is plotted. ***P < 0.001; by Mann-Whitney U test. (b) Representative flow cytometry analysis for coexpression of PD-1 and TIM-3 (left), LAG-3 (middle) or 4–1BB (right) on CD8+ lymphocytes in the tumor (top) and PBMC (bottom) from one patient (of n = 18). The percentage of cells expressing each combination of receptors is shown. (c) Image showing gates used for flow cytometry–based sorting of the circulating CD8+ lymphocytes. SSCA, side-scatter area. (d–i) IFN-γ ELISPOT assays (d,f,h) and flow cytometry analysis for 4–1BB expression (e,g,i) showing reactivity of in vitro–expanded subsets (CD8+, CD8+PD-1hi, CD8+PD-1− and CD8+PD-1+) sorted from pretreatment PBMC from patients NCI-3998 (d,e), NCI-3784 (f,g) and NCI-3903 (h,i) to autologous DCs transfected with RNAs encoding an irrelevant TMG (Irrel.) or the indicated TMGs. Representative flow cytometry plots show the percentage of 4–1BB+ lymphocytes after coculture of circulating CD8+PD-1+ cells with the TMGs specified. Plotted cells were gated on live CD3+ lymphocytes. ‘>’ denotes greater than 500 spots/2 × 104 cells. Experiments were performed without duplicates. All data are representative of at least two experiments.
Next we examined whether selection of circulating CD8+PD-1+ lymphocytes was able to prospectively identify neoantigen-specific CD8+ T cells in the blood of four individuals with melanoma. We used a high-throughput personalized screening strategy capable of evaluating T cell reactivity to neoantigens presented on all of the HLA restriction elements of the individual (depicted in Supplementary Fig. 2). Briefly, mutations selected on the basis of tumor-exome and transcriptome analyses (see Online Methods) were incorporated into oligonucleotides (minigenes) that encoded a 25-residue peptide (25-mer), and these oligonucleotides were then concatenated to yield tandem minigenes (TMGs; designated in numerical order and for each patient), as previously described15,16. Each TMG encoded up to 16 minigenes, and we constructed the requisite number of TMGs that allowed for the expression of all of the mutant 25-mers that were identified. In parallel, CD8+ lymphocytes were separated from peripheral blood mononuclear cells (PBMC) before treatment (hereafter referred to as pretreatment PBMC), using flow cytometry–based cell sorting, into CD8+, CD8+PD-1−, CD8+PD-1+ and CD8+PD-1hi cells (defined as the top 20% of PD-1–expressing CD8+ T cells; Fig. 1c), and these were expanded for 15 d. In vitro–transcribed TMG RNA was electroporated into immature autologous dendritic cells (DCs), which were used as targets in a T cell coculture assay. By using this approach, we screened the circulating in vitro–expanded CD8+ subsets from four individuals with metastatic melanoma (patients NCI-3998, NCI-3784, NCI-3903 and NCI-3926; see Supplementary Table 1) for recognition of 115, 140, 308 and 128 mutant 25-mers, respectively (Supplementary Table 2).
Although the unseparated peripheral blood CD8+ cells, as well as the CD8+PD-1− lymphocytes, from NCI-3998 showed limited recognition of the mutant 25-mers encoded by TMG1(hereafter referred to as recognition of TMG1 or TMG1 reactive), the circulating CD8+PD-1+ lymphocyte subset showed enhanced TMG1 reactivity and low, but reproducible, reactivity to TMG3 and TMG5 (Fig. 1d). Based on upregulation of the activation marker 4–1BB, the frequency of CD8+PD-1+ cells that were reactive to DCs expressing these TMG-encoded peptides was 1.8% for TMG1, 0.5% for TMG3 and 0.3% for TMG5 (Fig. 1e). Additionally, recognition of TMG1 and TMG3 by the CD8+PD-1hi subset was also observed (Fig. 1d). Similarly, CD8+PD-1+ and CD8+PD-1hi, but not CD8+ or CD8+PD-1−, lymphocytes from the peripheral blood of subjects NCI-3784 and NCI-3903 showed T cell reactivity to neoantigens (Fig. 1f–i). Circulating CD8+PD-1+ cells from subject NCI-3784 recognized at least three neoantigens encoded by TMG3, TMG5 and TMG8 (Fig. 1f,g), whereas those from subject NCI-3903 detected at least one neoantigen expressed by TMG9 (Fig. 1h,i). Peripheral blood lymphocytes from subject NCI-3926 did not show T cell reactivity to any of the neoantigens screened (data not shown). Overall, circulating neoantigen-reactive lymphocytes were prospectively identified in three of the four melanoma patients evaluated, and these cells were consistently detected within the CD8+PD-1+ lymphocyte subpopulation. Notably, with the exception of subject NCI-3998—from whom the unseparated population of circulating CD8+ T cells showed low-level recognition of TMG1—selection of CD8+PD-1+ or PD-1hi lymphocytes from the blood of the patients was necessary to expose CD8+ T cell reactivity to neoantigens.
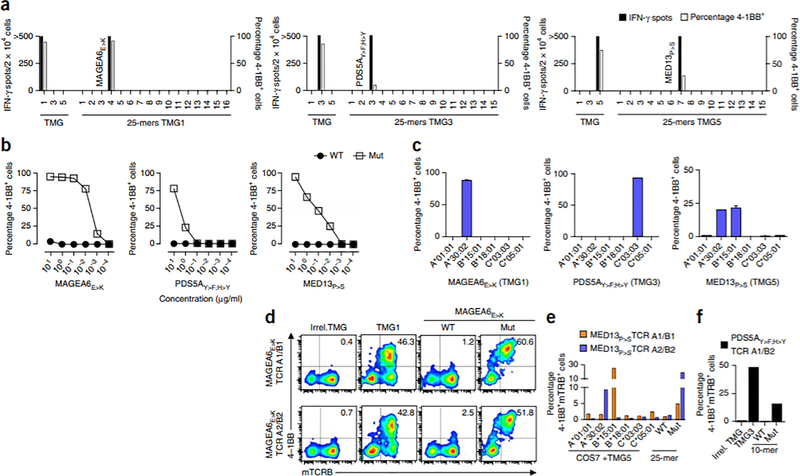
We next analyzed the specific neoantigens targeted by the TMG-reactive lymphocytes that we identified. Given the low frequency of some of the reactivities and the polyclonal nature of the circulating PD-1+ subset, we first enriched for TMG-reactive cells by selecting 4–1BB+ lymphocytes after coculturing circulating CD8+PD-1+ cells with DCs transfected with RNAs for the specific TMGs, expanded them in vitro and co-incubated them with DCs individually pulsed with the mutated 25-mers encoded by the corresponding TMG (Supplementary Table 2). In a representative example, TMG1-, TMG3- and TMG5-reactive cells isolated from the circulating CD8+PD-1+ subset of subject NCI-3998 showed reactivity to neoantigens derived from mutations in the genes MAGE family member A6 (MAGEA6), PDS5 cohesin–associated factor A (PDS5A) and mediator complex subunit 13 (MED13) (which we refer to as MAGEA6E>K, PDS5AY>F;H>Y and MED13P>S, respectively; Fig. 2a). The minimal predicted epitopes were determined, synthesized and tested (see Supplementary Table 3), and the TMG–reactive cells demonstrated specific recognition of the neo-epitopes, as compared to the wild-type counterparts (Fig. 2b). We also interrogated which HLA alleles presented the neoantigens identified. Although MAGEA6E>K and PDS5AY>F;H>Y were presented by the alleles encoding HLA-A*30:02 and HLA-C*03:03, respectively, recognition of the MED13P>S neo-epitope was restricted to alleles encoding HLA-A*30:02 and HLA-B*15:01 (Fig. 2c). Deep-sequencing analyses of the variable V-J or V-D-J region of the TRA and TRB genes (which encode the hypervariable regions of the TCR-α and TCR-β chains that are important for peptide recognition by the TCR) of the enriched populations of neoantigen-specific CD8+ T cells revealed multiple dominant TRA and TRB sequences that were unique for each of the T cell populations. To study the specificity of the neoantigen-specific cells at the clonal level, we constructed TCRs by pairing the sequences encoding the two most-dominant TRA and TRB sequences27 from the MAGEA6E>K, PDS5AY>F;H>Y or the MED13P>S neoantigen–specific lymphocytes (Supplementary Table 4) and cloning them into retroviral vectors that we then used to transduce autologous PBMC. The two TCRs constructed by pairing the most dominant and the second most dominant TRA and TRB sequences (which we refer to as TCR A1/B1 and TCR A2/B2) from the MAGEA6E>K-reactive population showed MAGEA6E>K recognition, as determined by 4–1BB upregulation following co-incubation with the mutant MAGEA6E>K minimal epitope (Fig. 2d). Four TCRs (TCR A1/B1, TCR A1/B2, TCR A2/B1 and TCR A2/B2) were assembled for each of the remaining MED13P>S- and PDS5AY>F;H>Y-specific lymphocyte populations. Two of the four potential MED13P>S-specific TCR–expressing lymphocytes tested (TCR A1/B1 and TCR A2/B2) recognized the mutant MED13P>S 25-mer peptide, and recognition of MED13P>S was restricted to HLA-B*15:01 and HLA-A*30:02, respectively (Fig. 2e). Finally, of four PDS5AY>F;H>Y-specific TCRs constructed and screened, only one showed specific recognition of TMG3 and the PDS5AY>F;H>Y neo-epitope (Fig. 2f).
Figure 2.
Characterization of neoantigen-specific lymphocytes isolated from the circulating CD8+PD1+ subset of subject NCI-3998. (a–c) Reactivity of TMG1- (left), TMG3- (middle) and TMG5-reactive (right) lymphocytes to the indicated TMGs or to the individual mutant 25-mers encoded by the indicated TMG (a), serial dilutions of the wild-type (WT) or mutant (Mut) MAGEA6E>K, PDS5AY>F;H>Y and MED13P>S minimal peptides (b), and COS7 cells cotransfected with the corresponding constructs expressing the indicated TMG and the individual HLA alleles that encode the HLA class I molecules indicated on the x axis (such as HLA-A*01:01; denoted A*01:01) (c). In c, the mean ± s.d. is plotted. (d–f) Reactivity of autologous PBMC that were transduced with retroviruses encoding neoantigen-specific TCRs. The T cell population of origin and the rank of the TRA and TRB sequences used to construct each TCR is denoted (as ‘TCR (A rank number)/(B rank number)’). The constructed TCRs expressed mouse constant regions, enabling the detection of the TCR with antibodies specific for the mouse TRB constant region (mTRB). Analyses showing reactivity of two distinct MAGEA6E>K-specific TCRs to TMG1 and the WT and Mut MAGEA6E>K minimal epitopes (CD3+CD8+ cells are plotted and the percentage of mTRB+4–1BB+ cells is shown) (d), mutant neo-epitope recognition and HLA restriction of two different MED13P>S-specific TCRs (e), and reactivity of a PDS5AY>F;H>Y-specific TCR to TMG3 and to the WT and Mut PDS5AY>F;H>Y peptides (f). Unless otherwise specified, experiments were performed without duplicates. All data are representative of at least two experiments.
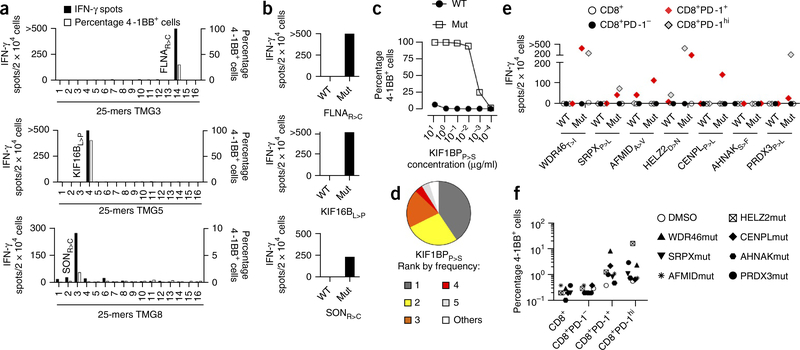
For subject NCI-3784, we identified neoantigen-specific responses in peripheral blood cells to three mutant neo-epitopes derived from filamin A (FLNAR>C), kinesin family member 16B (KIF16BL>P) and SON DNA-binding protein (SONR>C), all of which were presented by HLA-B*07:02 (Fig. 3a,b, Supplementary Fig. 3 and Supplementary Table 3). Moreover, circulating CD8+PD-1+ lymphocytes from subject NCI-3903 that were reactive to TMG9 showed mutant peptide–specific recognition of a KIF1BPP>S 8-mer presented by HLA-B*38:01, and this population contained three dominant TRB clonotypes (Fig. 3c,d, Supplementary Fig. 3 and Supplementary Table 3). Thus, selection of circulating CD8+PD-1+ lymphocytes led to the prospective identification of a diverse neoantigen-specific T cell response in three of four individuals with melanoma that we tested, with recognition of one or three unique tumor-specific neoantigens.
Figure 3.
Identification of neoantigens targeted by circulating CD8+PD-1+ cells isolated from subjects NCI-3784, NCI-3903 and NCI-3713. (a,b) Reactivity of NCI-3784 TMG3- (top), TMG5- (middle) and TMG8-reactive (bottom) CD3+CD8+ lymphocytes to autologous DCs pulsed with the individual mutant 25-mers encoded by the indicated TMG (a) or to autologous DCs pulsed with FLNAR>C, KIF16BL>P or SONR>C 25-mers, or their WT counterparts (b). (c) Recognition of B cells pulsed with serial dilutions of the Mut KIF1PBP>S minimal peptide or its WT counterpart by NCI-3903 TMG9-reactive CD3+CD8+ lymphocytes. (d) Frequency of the top five TRB clonotypes, as determined by deep-sequencing analysis of TRB from NCI-3903 TMG9-reactive lymphocytes. (e,f) IFN-γ ELISPOT assays (e) and frequency of 4–1BB expression (f) of NCI-3713 pretreatment PBMC that were sorted into CD8+, CD8+PD-1−, CD8+PD-1+ and CD8+PD-1hi cells, expanded in vitro and cocultured with B cells pulsed with either DMSO as a control or the WT and Mut epitopes from the indicated proteins. Experiments were performed without duplicates. All data are representative of at least two experiments.
To validate these findings, we studied patient NCI-3713, who experienced complete tumor regression following administration of TIL-3713, autologous TILs derived from a lung metastasis of the same patient. Previous studies showed that TIL-3713 cells recognize multiple mutant neo-epitopes, including WDR46T>I, SRPXP>L, AFMIDA>V, HELZ2D>N, CENPLP>L, AHNAKS>F and PRDX3P>L (T.D.P., J.S.C., C.J. Cohen (Bar-Ilan University), J.G., X. Yao (NCI) et al., personal communication). Analysis of the pretreatment PBMC from this patient demonstrated the recognition of six of seven neo-epitopes tested (Fig. 3e). Reactivity was uniquely identified within the circulating CD8+PD-1+ and CD8+PD-1hi, but not the CD8+ or the CD8+PD-1−, lymphocytes. T cell reactivities observed were mutant-peptide specific, as they showed preferential recognition of the mutant versus the wild-type peptides (Fig. 3e), and the percentage of neoantigen-specific cells, on the basis of 4–1BB upregulation, ranged from 0.5% to up to 21% of the CD8+PD-1hi cells (Fig. 3f). Thus, selection of circulating CD8+PD-1+ lymphocytes revealed that the T cell response to mutant antigens displayed by TIL-3713 also existed in the blood of this patient before TIL therapy.
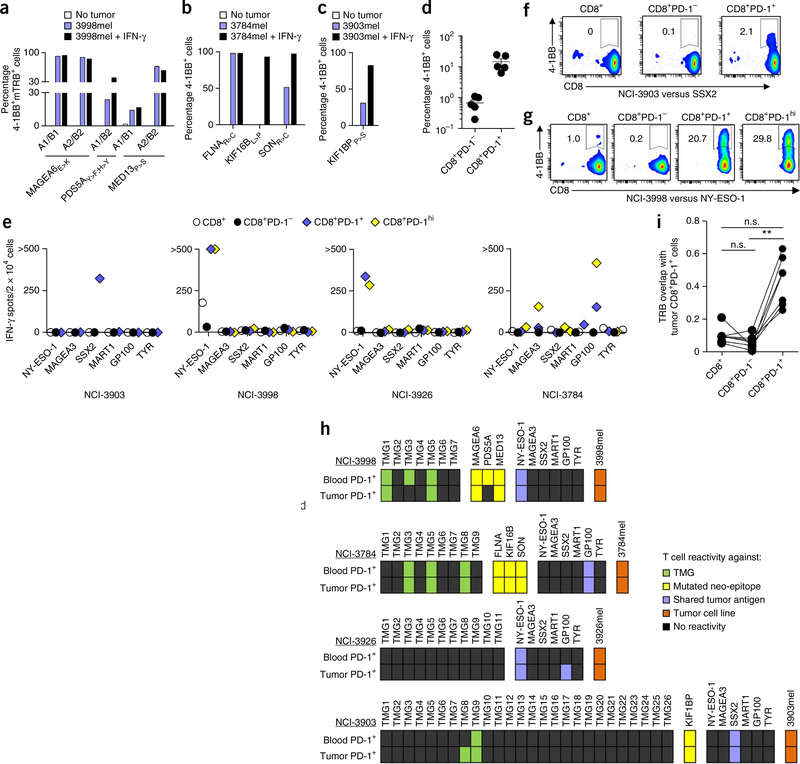
In view of their potential use in treating cancer, we next examined the recognition of autologous tumors by enriched populations of circulating neoantigen-specific T cells and by lymphocytes transduced with retroviruses expressing neoantigen-specific TCRs. Gene-engineered T cells from subject NCI-3998 that expressed MAGEA6E>K-, PDS5AY>F;H>Y- or MED13P>S-specific TCRs, and neoantigen-specific CD8+ T cells derived from the blood of subjects NCI-3784 and NCI-3903, recognized their corresponding autologous tumor cell lines at variable levels (Fig. 4a–c), either with or without pretreatment of the autologous tumor cell lines with interferon (IFN)-γ, which enhances processing and presentation of epitopes on HLA molecules. Furthermore, in all five individuals studied, the circulating CD8+PD-1+, but not CD8+PD-1−, lymphocytes showed direct tumor recognition, as evidenced by detection of 4–1BB upregulation (Fig. 4d) and IFN-γ release (data not shown). The frequency of tumor-reactive cells within the circulating CD8+PD-1+ lymphocytes ranged from 6.3%–24.6%. Notably, circulating CD8+PD-1+ cells from subject NCI-3926 did not recognize any of the mutant antigens tested, but they did recognize the autologous tumor. Additionally, the percentages of tumor-reactive CD8+PD-1+ lymphocytes from subjects NCI-3998 and NCI-3784 (9.5%, and 24.6%, respectively) exceeded those observed for the neoantigens evaluated (Fig. 1e,g), suggesting that either additional neoantigens or nonmutant tumor antigens may be recognized by the circulating CD8+PD-1+ subset. Indeed, in all four patients that were evaluated, the circulating CD8+PD-1+ and or CD8+PD-1hi cells also showed recognition of one or more cancer–germ line antigens or melanoma differentiation antigens that we tested, including cancer/testis antigen 1B (CTAG1B; also known as NY-ESO-1), MAGEA3, synovial sarcoma, X breakpoint 2 (SSX2), melan-A (MLANA; also known as MART1), premelanosome protein (PMEL; also known as GP100) and tyrosinase (TYR) (Fig. 4e). Whereas the peripheral blood CD8+PD-1+ T cells from subject NCI-3903 recognized SSX2 (Fig. 4f), circulating CD8+PD-1+ T cell subsets derived from subjects NCI-3926 and NCI-3998 recognized NY-ESO-1 (Fig. 4g), and the CD8+PD-1hi lymphocytes from subject NCI-3784 were reactive to MAGEA3 and GP100. MART1 and TYR were not recognized by any of the CD8+ T cell subsets tested. The relative frequency of circulating CD8+PD-1+ T cells targeting mutant antigens and self-antigens was highly variable between subjects (Supplementary Table 5).
Figure 4.
Recognition of tumors and self-antigens by TCRs or CD8+ T cells isolated from peripheral blood, and comparison of the specificity and TCR repertoire between circulating and tumor-infiltrating CD8+ T cell subsets. (a–c) Reactivity (as determined by 4–1BB upregulation on CD3+CD8+ cells) of retrovirally transduced lymphocytes from subject NCI-3998 expressing MAGEA6E>K-, PDS5AY>F;H>Y- or MED13P>S-specific TCRs (a), circulating FLNAR>C-, KIF16BL>P- or SONR>C-specific lymphocytes from subject NCI-3784 (b) and KIF1BPP>S-specific lymphocytes from subject NCI-3903 (c) that were cocultured with their corresponding autologous tumor cell lines (3998mel, 3784mel and 3903mel, respectively) pretreated with or without IFN-γ. (d) Reactivity of the circulating CD8+PD-1− and CD8+PD-1+ lymphocytes from subjects NCI-3998, NCI-3784, NCI-3903, NCI-3926 and NCI-3713 to their corresponding autologous tumor cell line. Each dot represents the frequency of 4–1BB upregulation for one patient sample (n = 5). Mean ± s.e.m. is shown. (e–g) IFN-γ ELISPOT assays (e) and analysis of 4–1BB upregulation by flow cytometry (representative plots shown, gated on CD3+ cells) (f,g) of pretreatment PBMC CD8+ subsets from subjects (indicated below each graph) that were screened for recognition of the shared tumor antigens indicated. Experiments were performed without duplicates. All data are representative of at least two experiments. (h) Antigens recognized by circulating and tumor-infiltrating CD8+PD-1+ lymphocytes. Each rectangle represents a target antigen screened. (i) Deep-sequencing analysis of TRB from intratumoral CD8+ PD-1+ cells and matched pretreatment PBMC CD8+, CD8+PD-1− and CD8+PD-1+ cells (n = 7) was used to determine TRB sequence overlap between the tumor-resident CD8+PD-1+ cells and the blood-derived CD8+, CD8+PD-1− and CD8+PD-1+ cells (see Online Methods for calculation methodology). A TRB sequence overlap of 1 indicates 100% similarity between two populations. **P < 0.01 using Dunn’s test for multiple comparisons; n.s., not significant.
Our findings indicated that circulating CD8+PD-1+ lymphocytes were enriched for cancer neoantigen–specific cells, as well as for other tumor-specific T cells. Additionally, simultaneous screening of matched circulating and tumor-resident CD8+PD-1+ lymphocytes from four patients revealed a high degree of similarity in the tumor antigens targeted by both populations (Fig. 4h, Supplementary Figs. 4 and 5, and Supplementary Table 5). In concordance, deep-sequencing analyses of the variable V-D-J region of the TRB gene from matched tumor-resident and circulating lymphocytes in the absence of in vitro expansion manifested a relatively high degree of overlap between TRB repertoires of the tumor-infiltrating and circulating CD8+PD-1+ subsets, but there was far less of an overlap with the circulating CD8+ or CD8+PD-1− cells (Fig. 4i and Supplementary Table 6). The specific antigens recognized by the circulating CD8+PD-1+ lymphocytes and the TIL infusion product these patients received were also similar (Supplementary Table 7).
Our work demonstrates the presence of antitumor T cells in the peripheral blood of melanoma patients and provides a novel noninvasive strategy to dissect, study and potentially exploit these reactivities therapeutically. Selection of circulating CD8+PD-1+ T cells, which account for <5% of all peripheral blood CD8+ lymphocytes, revealed the existence of CD8+ lymphocytes that target mutant and/or shared tumor antigens in all the melanoma patients studied. This data supports the idea of isolating circulating CD8+PD-1+ cells or neoantigen-specific T cell receptors from this population for the development of personalized T cell–based therapies. Moreover, these cells are probably targeted by PD-1–blocking antibodies, and their contribution to the antitumor efficacy of PD-1–specific antibody therapy warrants further investigation. The findings reported here—along with the advances that have made whole-exome sequencing possible from paraffin-fixed sections of the original tumor28, tumor-needle biopsies29 or from liquid-tumor biopsies30,31—suggest that a relatively noninvasive approach to T cell–based therapies that target neoantigens is possible.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Subjects, tumor biopsies and PBMC
Leukapheresis products and tumor samples were obtained from individuals with stage 4 melanoma enrolled on a clinical protocol (03-C-0277) approved by the institutional-review board (IRB) of the National Cancer Institute (NCI). Informed consent was obtained from all subjects, and they all had progressive disease at the time of sample acquisition. The five individuals studied in detail were chosen on the basis of availability of pretreatment leukapheresis and matched frozen-fresh tumor to perform whole-exome sequencing and transcriptome analysis. Patients were either treatment naive (NCI-3998), or had undergone prior therapies including surgery, chemotherapy and immunotherapy (NCI-3713, 3784, 3903 and 3926). The patient characteristics are provided in Supplementary Table 1. The patients that received prior therapies had been last treated from 7–55 months before the leukapheresis product was obtained. A summary of the individuals included in the phenotypic characterization of circulating and tumor-infiltrating lymphocytes is provided in Supplementary Table 8. Melanoma specimens were surgically resected and digested into single-cell suspensions using the gentleMACS Dissociator (Miltenyi Biotec, Gladbach, Germany), as previously described3, and cryopreserved. Pretreatment peripheral blood mononuclear cells (PBMC) were obtained by leukapheresis, prepared over a Ficoll-Hypaque gradient (LSM; MP Biomedicals, Santa Ana, CA) and cryopreserved until analysis. Melanoma cell lines were established from enzymatically separated tumor cells cultured in RPMI 1640 medium supplemented with 10% FBS (HyClone Defined, Logan, UT) at 37 °C and 5% CO2. Melanoma cell lines were mycoplasma negative and were authenticated based on the identification of patient-specific somatic mutations and HLA molecules.
Exome and RNA sequencing
Tumor biopsies and normal PBMC were subjected to DNA extraction, library construction, exome capture of approximately 20,000 coding genes and next-generation sequencing by Macrogen (Rockville, MD), Personal Genome Diagnostics (PGDX, Baltimore, MD) or the Broad Institute (Cambridge, MA). The average number of distinct high-quality sequences at each base ranged between 100 and 150 for the individual exome libraries. Alignments and variant calling were performed as previously described16. The total number of putative nonsynonymous mutations (see Supplementary Table 1) was determined by using filters consisting of >2 exome-variant reads, ≥10% variant-allele frequency (VAF) in the tumor exome, >10 normal reads, tumor/normal variant frequency ≥5 and by filtering out single-nucleotide polymorphisms in dbSNP build 138. An mRNA-sequencing library was also prepared from a tumor biopsy using Illumina TruSeq RNA library prep kit. RNA alignment was performed using STAR32 duplicates and marked using Picard’s MarkDuplicate tools, and fragments per kb per million mapped reads (FPKM) values were calculated using cufflinks33. The levels of transcripts encoding putative nonsynonymous variants, calculated as FPKM, were used to assess expression of candidate mutations identified using whole-exome data.
We used the following criteria to prioritize mutations for immunological screening (Supplementary Table 2). Initially, we selected mutations with a VAF >10% in the tumor exome, as well as mutations that were identified in both transcriptome and exome analysis without any additional filters. For some samples (NCI-3903), the mutations selected based on exome analysis only were prioritized by selecting those with >10 variant reads to increase the confidence of mutation calling. All raw fastq files for samples used in this study can be accessed through the NCBI BioProject database with accession number PRJNA298310. For each of the immunogenic antigens detected, the amino acid changes are specified.
Antibodies and phenotypic characterization of T cells
We used fluorescently labeled antibodies purchased from BD Biosciences, San Jose, CA (UCHT1, 1.6:100, CD3 PE-CF594; SK7, 1:100, CD3 APC-Cy7; SK1, 0.5:100, CD8 PE-Cy7; 4B4–1, 1.25:100, CD137 APC; NK-1, 3:100, CD57 FITC; J168–540, 1.2:100, BTLA PE), eBioscience, San Diego, CA (H57–597, 0.5:100, mTRB FITC; O323, 2:100, CD27 BV605), Biolegend, San Diego, CA (EH12.2H7, 0.7:100, PD-1 BV421), R&D Systems, Minneapolis, MN (344823, 2.6:100, TIM-3 PE and APC), Enzo Life Sciences, Farmingdale, NY (17B4. 1:100, LAG-3 FITC) and Miltenyi Biotec (4B4–1, 2.6:100, 4–1BB PE). Anti–PD-1 antibody was kindly provided by L. Liu from Amplimmune (Gaithersburg, MD, AMP-514, 1:300, PD-1 Alexa Fluor 647). Cell-sorting experiments were carried out using anti–PD-1 AMP-514 antibody.
To perform the phenotypic characterization, PBMC and tumor single-cell suspensions were thawed into T cell medium (1:1 mix of AIMV medium (Life Technologies, Waltham, MA) and RPMI 1640 medium (Lonza, Walkersville, MD), 5% in-house human serum, 100 U/ml penicillin and 100 μg/ml streptomycin (Life Technologies), 2 mM l-glutamine (Life Technologies), 10 μg/ml gentamicin (Quality Biological Inc., Gaithersburg, MD), 12.5 mM HEPES (Life Technologies)) supplemented with DNase (Genentech Inc., San Francisco, CA, 1:1,000), centrifuged and plated at 2 × 106 cells/well in a 24-well plate in the absence of cytokines. After resting the cells overnight at 37 °C and 5% CO2, the cells were harvested, and 2 × 106 cells were resuspended in 50 μl of staining buffer (PBS, 0.5% BSA, 2 mM EDTA) containing antibodies. Cells were incubated for 30 min at 4 °C and washed twice before acquisition. We carried out flow cytometry acquisition on a modified Fortessa, equipped to detect 18 fluorescence parameters, or a Canto II flow cytometer (BD Biosciences). Flow cytometry data were analyzed using FlowJo software (Ashland, OR). Data were gated on live cells (propidium iodide (PI) negative) and single cells. Gates were set based on fluorescence minus one (FMO) controls (see Supplementary Fig. 6).
T cell sorting and in vitro expansion
Cell sorting was carried out using the BD jazz (BD Biosciences). For all experiments requiring cell sorting from PBMC, CD8+ cells were first enriched using CD8 microbeads (Miltenyi Biotec), and stained as described above. When sorting T cells from fresh-tumor single-cell suspensions, this pre-enrichment step was not performed. Cells were gated on live (PI negative), single cells, CD3+ and CD8+ cells, and on the population of interest. The gating strategy used to sort circulating lymphocyte subsets based on PD-1 expression is shown in detail in Supplementary Figure 6. Half of the T cells isolated were spun down and snap-frozen to perform deep sequencing of the variable V-D-J region of the TRB gene, and the other half was expanded in vitro. T cell yields ranged from 3 × 103 to 3 × 105. A similar sorting strategy was used to sort the 4–1BB+ lymphocytes, following a 20-h coculture.
T cells were expanded in vitro using an excess of irradiated allogeneic feeder cells (5,000 rad) pooled from three donors in T cell medium supplemented with 30 ng/ml anti-CD3 (OKT3, Miltenyi Biotec) and 3,000 IU of interleukin (IL)-2 (Aldesleukin, Chiron). After day 6, half of the medium was replaced with fresh T cell medium containing IL-2 every other day. At day 15, T cells were either used in coculture assays or cryopreserved until analysis. Of note, enrichment of neoantigen-specific T cells was consistent between replicate CD8+PD-1+ T cell cultures, but stochastic outgrowth or loss of T cell reactivities can be observed and become more apparent when starting with less than 3 × 103 CD8+PD-1+ T cells (Supplementary Table 9). The minimum material required to sort 3 × 103 CD8+PD-1+ cells is ~1 × 107 PBMC.
Generation of autologous antigen-presenting cells (APCs)
Immature dendritic cells (CD11c+CD14−CD80lowCD86+HLA-DR+) were generated from PBMC using the plastic-adherence method, as previously described16. On day 3, DC medium (RPMI supplemented with 5% human serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine (Life Technologies), 800 IU/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) and 200 U/ml IL-4 (Peprotech, Rocky Hill, NJ)) was added, and at day 5 or 6 DCs were harvested and used in electroporation experiments or cryopreserved. When used after cryopreservation, cells were thawed into DC medium, spun at 1,000 r.p.m. for 10 min, resuspended in DC medium at 2 × 106 cells/ml and incubated at 37 °C and 5% CO2 for 2 h before electroporation or peptide pulsing.
Autologous B cells were isolated from autologous PBMC by positive selection using CD19+ microbeads (Miltenyi Biotec) and expanded using irradiated NIH3T3 CD40L cells and IL-4 (Peprotech), as previously described16. NIH3T3 CD40L cells were established by transducing NIH3T3 cells (obtained from the American Type Culture Collection) with a retrovirus encoding CD40L. B cells were harvested at day 5 or 6 after the initial stimulation and were restimulated, cryopreserved or used in coculture assays. When used after cryopreservation, B cells were thawed into B cell medium 16–24 h before using them in coculture assays. B cell medium comprised of Iscove’s modified Dulbecco’s medium (IMDM) (Quality Biological Inc., Gaithersburg, MD) supplemented with 10% human serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 2mM l-glutamine and 200 U/ml IL-4 (Peprotech, Rocky Hill, NJ).
Construction of TMGs and in vitro transcription of TMG RNA
Tandem minigenes (TMGs) were constructed as previously described15,16. Briefly, a minigene was constructed for each nonsynonymous variant identified and consisted of the mutant amino acid flanked by 12 amino acids of the wild-type protein sequence. Up to 16 minigenes were strung together to generate a tandem minigene (TMG) construct. These TMG constructs were codon-optimized and cloned in frame into pcRNA2SL using EcoRI and BamHI. pcRNA2SL is based on the pcDNA3.1 and was modified to include a signal sequence and a DC-LAMP trafficking sequence to enhance processing and presentation34. The sequences were verified by Sanger sequencing. Following linearization of the constructs, we performed phenol-chloroform extraction and precipitated the DNA with sodium acetate and ethanol. Next we used 1 μg of linearized DNA to generate in vitro–transcribed (IVT) RNA using the Mmessage Mmachine T7 Ultra kit (Life Technologies) as instructed by the manufacturer. RNA was precipitated using LiCl2 and resuspended at 1 μg/μl. To screen for recognition of the cancer–germ line antigens NY-ESO-1, MAGEA3 and SSX2, and the melanoma differentiation antigens MART1, GP100 and TYR, DNA sequences encoding the full-length amino acid sequences were cloned individually into pcRNA2SL using EcoRI and BamHI, and these constructs were used to generate IVT RNA, as described above.
Transfection of RNA or DNA
DCs were resuspended in Opti-MEM (Life Technologies) at 10 × 106 to 40 × 106 cells/ml. 8 μg of IVT RNA was aliquoted into the bottom of an electroporation cuvette with a 2-mm gap, and 100 μl of DCs was added. DCs were electroporated at 150 V for 10 ms for one pulse, using a BTX-830 square-wave electroporator (Holliston, MA). Cells were gently resuspended into DC medium and transferred into ultra-low–attachment polysterene 24-well plates (Corning) at approximately 1 × 106 DCs/ml and rested overnight at 37 °C, 5% CO2. Transfection efficiencies were routinely between 70–90%, as assessed with a control green fluorescent protein (GFP)-encoding RNA (data not shown). In coculture assays, the ‘irrelevant TMG RNA’ control was a random TMG derived from a different patient.
HLA alleles were cloned into pcDNA3.1. To interrogate which HLA alleles presented the neoantigens identified, COS7 cells were cotransfected with TMG DNA constructs and plasmids encoding the individual HLA molecules using Lipofectamine 2000 (Life Technologies). After 16 h, cells were harvested and used as targets in coculture assays.
HLA class I (HLA-I) alleles, peptide prediction and pulsing
HLA was determined from next-generation sequencing data using the algorithm PHLAT35 (NCI-3713: HLA-A*02:01, A*29:02, B*44:03, B*51:01, C*15:02, C*16:01; NCI-3998: HLA-A*01:01, A*30:02, B*15:01, B*18:01, C*03:03, C*05:01; NCI-3784: HLA-A*01:01, A*03:01, B*07:02, C*07:02; NCI-3903: HLA-A*02:01, A*24:02, B*27:02, B*38:01, C*02:02, C*12:03; NCI-3926: HLA-A*01:01, A*02:01, B*08:01, B*13:02, C*06:02, C*07:01).
Candidate 8- to 11-mers containing the mutant residues that were predicted to bind with high affinity to the patients’ HLA-I molecules were identified using the immune epitope database (IEDB)36. Crude and HPLC-purified peptides were synthesized by GenScript (Piscataway, NJ), resuspended in DMSO at 10 mg/ml and stored at −20 °C.
For experiments requiring peptide pulsing, DCs or B cells were resuspended in DC or B cell medium, respectively, at 1 × 106 cells/ml. DCs were incubated overnight at 37 °C and 5% CO2 with wild-type or mutant 25-mers at a concentration of 10 μg/ml in DC medium. B cells were pulsed with 1 μg/ml or with tenfold serial dilutions of minimal epitopes, starting at 10 μg/ml, for 2 h at 37 °C and 5% CO2. DCs or B cells were washed once with PBS before co-incubation with T cells.
Coculture assays: IFN-γ enzyme-linked immunospot (ELISPOT) assays and detection of activation marker 4–1BB using flow cytometry
Both IFN-γ ELISPOT assays and 4–1BB upregulation at 20 h after the coculture were used to measure target cell recognition by T cells. After 15 d of T cell expansion, or following overnight rest of cryopreserved T cells in T cell medium supplemented with 3,000 IU/ml IL-2, T cells were washed to remove excess cytokines. In the ELISPOT assays, 2 × 104 T cells were added per well in a 96-well plate. When DCs electroporated with IVT RNAs encoding TMGs or shared antigens were used as targets, approximately 3 × 104 to 7 × 104 cells/well were used in a 96-well plate. When peptide-pulsed B cells were used, 8 × 104 to 1.5 × 105 cells were added per well. All cocultures were carried out in T cell medium in the absence of exogenously added cytokines. T cells cultured alone or stimulated with plate bound anti-CD3 (OKT3 antibody) were used as controls in all assays. RNA encoding epitopes derived from CMV, EBV, and Flu (CEF RNA) were included as controls in all the immunological screening assays37.
IFN-γ ELISPOT assays were done as previously described16. The raw data were plotted without subtracting the background. Greater than 40 spots and greater than twice the background was considered positive T cell reactivity. Prior to processing the ELISPOT assay, cells were harvested for flow cytometric detection of 4–1BB upregulation, as previously described16.
Deep sequencing of TCR-encoding genes and analysis
Deep sequencing of the variable V-J or V-D-J regions of the TRA and TRB genes, which encode for the hypervariable amino acid sequence of the TCR-α and the TCR-β chains responsible for contact with the cognate peptide (i.e., complementarity-determining region 3 (CDR3)), was performed on genomic DNA by Adaptive Biotechnologies (Seattle, WA). For the enriched populations of TMG-reactive cells, DNA was extracted from 1 × 106 lymphocytes. The number of circulating and tumor-resident CD8+ lymphocytes that were sequenced ranged from 3 × 103 to 3 × 105. The coverage per sample was >10×. Only productive rearrangements were used in the calculations of TRB clonotype frequencies and TRB overlap. Analysis of TRB overlap of nucleotide sequences encoding CDR3 between two given populations (A and B) was calculated using immunoSEQ (Adaptive Biotechnologies, Seattle, WA), using the following formula: sample TRB overlap = ((shared sequence reads in A + shared sequence reads in B) / (Σsequence reads in A + B)). Weighing in the frequency of the shared sequences rather than the total number of shared sequences helped account for potentially different-sized samples. A TRB overlap of 1 represents 100% overlap between two populations.
Cloning of TCRs into retroviral vectors, retrovirus production and transduction of T cells
For NCI-3998, we constructed TCRs by pairing the TRA and TRB sequences encoding the dominant chains in the TMG1 (MAGEA6E>K)-, TMG3 (PDS5AY>F;H>Y)- or TMG5 (MED13P>S)-reactive populations. The rank of the TRA and TRB sequences used to construct each TCR is denoted as ‘TCR (A rank number)/(B rank number)’. In total, two TCRs were assembled from the TMG1 (MAGEA6E>K)-reactive population (TCR A1/B1 and TCR A1/B2), and four TCRs were assembled from the TMG3 (PDS5AY>F;H>Y)-reactive, as well as the TMG5 (MED13P>S)-reactive, populations (TCR A1/B1, TCR A1/B2, TCR A2/B1 and TCR A2/B2). Briefly, TRA V-J–encoding sequences and TRB V-D-J–encoding sequences were fused to sequences encoding the mouse constant TRA and TRB chains38, respectively. Mouse constant regions were modified, as previously described39,40. The full-length TRB and TRA chains were cloned, in this orientation and separated by a furin SGSG P2A linker, into pMSGV1 retroviral vector (GenScript).
Transient retroviral supernatants were generated, and autologous pretreatment PBMC were transduced as previously described16. Transduced T cells were used at day 15 or cryopreserved until used. T cells either mock-transduced or transduced with a construct encoding GFP were used as controls in all transduction experiments.
Statistical analysis
Data were reported as the median, mean ± s.e.m. or mean ± s.d., as specified. We used the Mann-Whitney U test to compare the percentage of PD-1 expression between PBMC and fresh-tumor single-cell suspensions. Dunn’s test for multiple comparisons was used to analyze the statistical differences in TRB overlap. Statistical analysis was carried out using Prism program 6.0 (GraphPad Software Inc., La Jolla, CA). Unless otherwise specified, experiments were performed without duplicates. All data are representative of at least two experiments.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Center for Cancer Research intramural research program of the NCI, US National Institutes of Health (NIH). We thank the members of the Surgery branch for helpful discussions, R. Somerville and members of the tumor-infiltrating lymphocytes (TIL) lab for technical support, and L. Liu for kindly providing the PD-1–specific AMP-514 antibody. This work used the computational resources of the NIH High-Performance Computing (HPC) Biowulf cluster (http://hpc.nih.gov).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession codes. All raw fastq files for the samples used in this study can be accessed via the National Center for Biotechnology Information’s BioProject database using accession number PRJNA298310.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Robbins PF et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med 19, 747–752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linnemann C et al. High-throughput epitope discovery reveals frequent recognition of neoantigens by CD4+ T cells in human melanoma. Nat. Med 21, 81–85 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Gros A et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J. Clin. Invest 124, 2246–2259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg SA et al. Treatment of 283 consecutive patients with metastatic melanoma or renal cell cancer using high-dose bolus interleukin 2. J. Am. Med. Assoc 271, 907–913 (1994). [PubMed] [Google Scholar]
- 5.Rosenberg SA et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T cell–transfer immunotherapy. Clin. Cancer Res 17, 4550–4557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamid O et al. Safety and tumor responses with lambrolizumab (anti–PD-1) in melanoma. N. Engl. J. Med 369, 134–144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larkin J et al. Combined nivolumab and ipilimumab, or monotherapy, in untreated melanoma. N. Engl. J. Med 373, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Postow MA et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med 372, 2006–2017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Topalian SL et al. Survival, durable tumor remission and long-term safety in patients with advanced melanoma receiving nivolumab. J. Clin. Oncol 32, 1020–1030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizvi NA et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small-cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snyder A et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le DT et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu YC et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin. Cancer Res 20, 3401–3410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran E et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carreno BM et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg SA & Restifo NP Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rooij N et al. Tumor-exome analysis reveals neoantigen-specific T cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol 31, e439–e442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen CJ et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Invest 125, 3981–3991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altman JD et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996). [PubMed] [Google Scholar]
- 23.Lennerz V et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc. Natl. Acad. Sci. USA 102, 16013–16018 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmadzadeh M et al. Tumor antigen–specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 114, 1537–1544 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baitsch L et al. Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. J. Clin. Invest 121, 2350–2360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inozume T et al. Selection of CD8+PD-1+ lymphocytes in fresh human melanomas enriches for tumor-reactive T cells. J. Immunother 33, 956–964 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linnemann C et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat. Med 19, 1534–1541 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Van Allen EM et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med 20, 682–688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee HB et al. The use of FNA samples for whole-exome sequencing and detection of somatic mutations in breast cancer surgical specimens. Cancer Cytopathol. 123, 669–677 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Murtaza M et al. Noninvasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Lohr JG et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol 32, 479–484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu TC et al. Engineering an intracellular pathway for major histocompatibility complex class II presentation of antigens. Proc. Natl. Acad. Sci. USA 92, 11671–11675 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai Y, Ni M, Cooper B, Wei Y & Fury W Inference of high-resolution HLA types using genome-wide RNA- or DNA-sequencing reads. BMC Genomics 15, 325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vita R et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 43, D405–D412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen JS, Wick DA, Tran E, Nelson BH & Webb JR An in vitro–transcribed mRNA polyepitope construct encoding 32 distinct HLA class I–restricted epitopes from CMV, EBV and influenza for use as a functional control in human immune-monitoring studies. J. Immunol. Methods 360, 149–156 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA & Morgan RA Enhanced antitumor activity of murine-human hybrid T cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 66, 8878–8886 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen CJ et al. Enhanced antitumor activity of T cells engineered to express T cell receptors with a second disulfide bond. Cancer Res. 67, 3898–3903 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haga-Friedman A, Horovitz-Fried M & Cohen CJ Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J. Immunol 188, 5538–5546 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.