Abstract
Plants are now recognized as metaorganisms which are composed of a host plant associated with a multitude of microbes that provide the host plant with a variety of essential functions to adapt to the local environment. Recent research showed the remarkable importance and range of microbial partners for enhancing the growth and health of plants. However, plant–microbe holobionts are influenced by many different factors, generating complex interactive systems. In this review, we summarize insights from this emerging field, highlighting the factors that contribute to the recruitment, selection, enrichment, and dynamic interactions of plant-associated microbiota. We then propose a roadmap for synthetic community application with the aim of establishing sustainable agricultural systems that use microbial communities to enhance the productivity and health of plants independently of chemical fertilizers and pesticides. Considering global warming and climate change, we suggest that desert plants can serve as a suitable pool of potentially beneficial microbes to maintain plant growth under abiotic stress conditions. Finally, we propose a framework for advancing the application of microbial inoculants in agriculture.
Keywords: Abiotic and biotic stress, DARWIN21, desert bacteria, endophytes, plant growth-promoting rhizobacteria (PGPRs), plant microbiome, plant–microbe interaction, soil microbial community, synthetic community (SynCom)
Successful application of plant-associated microbial inoculants for sustainable agriculture requires a holistic approach of examining and understanding the soil environment, crop of interest, and associated microbial communities.
Introduction
According to the United Nations Organization, the current world population of 7.6 billion is expected to increase beyond 9.8 billion by the year 2050 (United Nations, 2017). Accompanying this dramatic growth in population is the anticipated increase in the demand for agricultural food and feed products and the evident rise in environmentally destructive human activities, such as deforestation and the overuse of chemical fertilizers and pesticides in agriculture. The continuous deforestation, industrialization, and excessive use of fossil fuels have escalated the rise of CO2 concentrations in the atmosphere, leading to higher greenhouse gas emissions and average global temperatures (Mgbemene et al., 2016). Subsequently, these activities and phenomena have led to reductions in cultivatable land and crop productivity. Furthermore, the scarcity of freshwater resources or its inaccessibility and the high costs of water treatment and desalination further present a challenge to meet water demand for the agriculture sector (Beltrán and Koo-Oshima, 2006; Rosegrant et al., 2009). The combination of all these problems and challenges poses a serious threat to global food security and stability of economies, especially in developing countries.
The solution to those challenges necessitates multiple approaches, including the use of plant growth-promoting microbes as biostimulants to increase crop productivity. The concept of using biostimulants in agriculture is not new, and application of microbial consortia or single microbes as inoculum was previously addressed (Kong et al., 2018; Woo and Pepe, 2018). However, the successful transfer of microbial inoculants from the lab to the field remains a challenge. This is primarily due to the presence of many crop species and crop varieties, variable environmental conditions between fields, and the exponential increase in the number of microbial isolates. Therefore, a holistic approach towards the use of ‘biostimulants’ is needed via ‘diagnostics’ of the field environment (e.g. soil) and desired crop (e.g. genotype), selecting best agricultural practices, screening for inoculants from available culture collections, increasing scientific research in the field of microbiomes, and, finally, incorporating all the latter into large-scale industrial production and field application (Mitter et al., 2019; Pascale et al., 2019).
In this review, we will shed light on the different biotic (e.g. plants or pathogens) and abiotic (e.g. soil or climate) factors shaping microbial communities in the soil, rhizosphere, and plant. The limitations and complexities of microbial community experiments and their applications in agriculture will also be discussed. Finally, a roadmap will be presented for the successful application of microbial inoculants in agriculture.
We are not alone: the concept of holobiont and plant-associated microbiota
Plants, animals, and almost all multicellular organisms are no longer considered as standalone individual organisms. Instead, they co-exist and are in constant interactions with their surrounding biota (Margulis and Fester, 1991; Turnbaugh et al., 2007; Bosch and McFall-Ngai, 2011). In the late 19th century, Karl Möbius named this interaction or co-existence as ‘biocenosis’ or ‘living community’ (Möbius, 1877). In 1991, Lynn Margulis proposed the term ‘Holobionts’—Holo is derived from the ancient Greek word ȍλος (hólos) for ‘whole’. Margulis described that any physical association between individuals of different species for a significant part of their life span is termed symbiosis and all participants in the symbiotic interaction are symbionts (Margulis and Fester, 1991; Bordenstein and Theis, 2015). A strictly microbe-dependent lifestyle has profound evolutionary consequences and suggests that the phenotype of a healthy host cannot be explained exclusively by its genome (Bosch and McFall-Ngai, 2011).
The advent of next-generation sequencing (NGS) opened up possibilities to study these close interactions between a host—human, animal, or plant—and its associated microbial community (Bosch and Miller, 2016; Greer et al., 2016; Sender et al., 2016). In addition, NGS can provide evidence for an active dialogue within the holobiont (host and associated microbiota) in coordinating and synchronizing signaling pathways and metabolic activities for maintaining a long-term, healthy co-existence (Rosenberg et al., 2010; Wier et al., 2010; Walter et al., 2011; Gilbert et al., 2012). Biological signals within the holobiont ecosystem could function as ‘Zeitgebers’ or time tuners (Leone et al., 2015; Lee et al., 2019). For example, signaling molecules produced by gut microbes were required for the functioning of the circadian clock in the host intestinal epithelial cells (Mukherji et al., 2013; Leone et al., 2015). Other living organisms, such as insects and plants, carry symbiotic microbes that provide defense against natural enemies (Arnold et al., 2003; Jaenike et al., 2010).
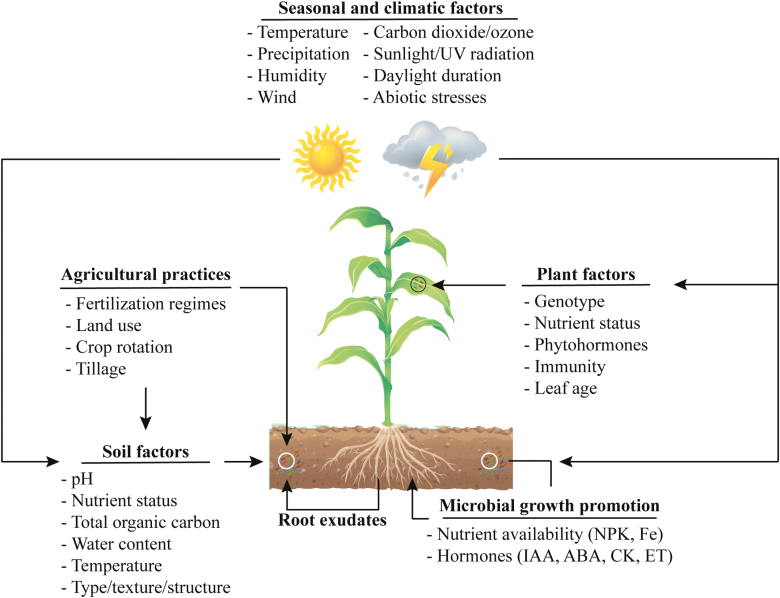
Advances in NGS and culture-independent methods demonstrated that terrestrial plants are heavily colonized by a wide diversity of microorganisms, including bacteria, fungi, oomycetes, and protozoa (Kemen, 2014; Bulgarelli et al., 2015; Hacquard et al., 2015). Plants accommodate and interact with different microbes (Fig. 1) within their tissues (endosphere); they also interact with the surrounding microbial community present in the narrow region of soil surrounding the root system (rhizosphere) and around the stems, leaves, flowers, and fruits (phyllosphere). It is also clear now that microbiota play a major role in plant health and fitness (Müller et al., 2016). These microbes can colonize different plant organs either inside (endophytic) or attached to the surface (ectophytic).
Fig. 1.
Microbial communities are shaped by several factors that must be considered in agricultural applications. Seasonal and climatic factors alter soil physicochemical properties and plant physiology. Microbial communities in the soil are affected by seasonal and climatic factors and soil factors. Plant factors alter microbial communities in the phyllosphere, endosphere, and rhizosphere, with the latter being via root exudates. Agricultural management practices can cause changes in the microbial communities in the soil either directly or via altering soil properties. Microbes associated with plants, in either the rhizosphere or the endosphere, are capable of promoting plant growth by making nutrients available or producing/modulating phytohormones.
The last two decades saw a steady increase in the number of studies investigating microbial communities of both above- and below-ground plants species. In the model plant Arabidopsis (Arabidopsis thaliana), a core microbial community was identified, where the bacterial community and function in the leaves overlapped with those in the roots (Lundberg et al., 2012; Bodenhausen et al., 2013; Bai et al., 2015). Similar studies were also shown for crop plants such as lettuce (Lactuca sativa) and tomato (Solanum lycopersicum) (Ottesen et al., 2013), wild and domesticated barley (Hordeum vulgare) (Bulgarelli et al., 2015), and maize (Zea mays) under field conditions (Peiffer et al., 2013) and in the greenhouse (Rastogi et al., 2012; Williams and Marco, 2014). Several pioneer desert plants such as Agave, Atriplex, Tribulus, Panicum, Euphorbia, and Zygophyllum were also studied (Kaplan et al., 2013; Coleman-Derr et al., 2016; Eida et al., 2018). All the aforementioned studies collectively agree that the plant root endosphere is dominated by a small number of bacterial lineages, with Actinobacteria, Bacteroidetes, and Proteobacteria being the dominant phyla when compared with soil and rhizosphere bacterial communities. Nevertheless, the relative abundances of individual phyla or genera are dependent on multiple physical, chemical, and biological factors.
In almost all ecosystems, multidimensional interactions exist between microbes and their hosts, and these are governed by biotic and abiotic factors. Biotic factors are the living components of an ecosystem, such as microbes, insects, plants, and animals. Abiotic factors are the non-living chemical and physical parts of the environment and are commonly affected by time (day/night) and seasonal or climate changes, such as soil chemical and physical properties, temperature, UV levels, precipitation (rainfall), and CO2 levels (Fig. 1). Abiotic factors, including stresses such as drought, soil salinity, or extreme temperatures, are very complex and affect the physiochemical properties of both the soil and plants, and their associated microbial communities.
The soil dictates which microbes are accommodated by a host plant
Soil represents a highly complex system comprising a variety of environments with different physical, chemical, and biological properties. It is one of the largest reservoirs of microbial biomass and diversity, and thus serves as a pool for recruitment of microbes and enrichment of root endophytic communities (Whitman et al., 1998; Hartman et al., 2008; Bulgarelli et al., 2012; Yeoh et al., 2017). The soil microbial community structures, functions, and compositions are susceptible to physical (e.g. soil structure), chemical (e.g. nutrient content), and biological (e.g. presence of pathogenic or beneficial microbes) changes in their surroundings (Fig. 1; Table 1) (Truog, 1947; de Vries et al., 2012; Fierer et al., 2012). High-throughput molecular techniques coupled with NGS have enhanced our ability to characterize prokaryotic (e.g. bacterial) and eukaryotic (e.g. fungal) communities in the soil in terms of taxonomic and phylogenetic structure, enzymatic activity, microbial function, and abundance and composition. Various factors influence soil microbial communities including pH, nutrient (e.g. carbon, nitrogen, phosphorus) content and availability, water/moisture content, temperature, and soil type, texture, and particle size.
Table 1.
Soil factors that are responsible for shaping microbial communities
| Factors | Summary | References |
|---|---|---|
| Soil and abiotic factors | pH can alter the solubility and availability of nutrients influencing microbial diversity and composition with stronger influence on bacteria than fungi. | Fierer and Jackson (2006); Lauber et al. (2008); Lauber et al. (2009); Shen et al. (2013); Maestre et al. (2015); Zhang et al. (2017); Shen et al. (2018) |
| Soil fertilization (e.g. NPK) and soil amendment (e.g. carbon) practices can affect nutrient status and influence bacterial and fungal communities in soil; C content is important for microbial growth and survival | Marschner et al. (2004); Eilers et al. (2010); Goldfarb et al. (2011); Kuramae et al. (2012); Siciliano et al. (2014); Maestre et al. (2015); Francioli et al. (2016) | |
| Changes in temperature and water content (or precipitation) can affect soil pH and nutrient status, and influence microbial community composition and function | Pettersson and Bååth (2003); Habekost et al. (2008); Bárcenas-Moreno et al. (2009); Bell et al. (2009); Castro et al. (2010); Koranda et al. (2013); Zhang et al. (2019) | |
| Soil type, texture, structure, and particle size can affect the flow and status of nutrients and water, and influence microbial communities in soil and rhizosphere | Gelsomino et al. (1999); Sessitsch et al. (2001); Girvan et al. (2003); Singh et al. (2007); Bach et al. (2010); Chau et al. (2011); Schreiter et al. (2014) | |
| Soil salinity can affect soil and plant-associated microbial communities | Lozupone and Knight (2007); Yaish et al. (2016); Thiem et al. (2018); Berens et al. (2019) | |
| Drought can affect soil and plant-associated microbial communities | Bachar et al. (2010); Hueso et al. (2012); Alster et al. (2013); Bogino et al. (2013); Naylor and Coleman-Derr (2018); Xu et al. (2018) | |
| Soil and biotic factors | Distinct microbial communities are correlated with the presence or occurrence of plant pathogens or diseases; suppressive soils could contain more microbes with antagonistic activity; initial differences in the soil microbiome composition can affect plant health | Sanguin et al. (2009); Mendes et al. (2011); Meng et al. (2012); Rosenzweig et al. (2012); Siegel-Hertz et al. (2018); Wei et al. (2019); Zhou et al. (2019) |
| Agricultural management practices and land use can cause changes in microbial community composition and diversity | Steenwerth et al. (2002); Garbeva et al. (2006); Lauber et al. (2008); Postma et al. (2008); Reeve et al. (2010); Carbonetto et al. (2014); Peralta et al. (2018); Sun et al. (2018); Le Guillou et al. (2019) |
Soil pH
Soil pH has a strong influence on the solubility and availability of nutrients (Cerozi and Fitzsimmons, 2016), such as carbon (C), nitrogen (N), phosphorus (P), potassium (K), iron (Fe), and zinc (Zn), which most living organisms need for survival and growth (Lindsay, 1995; Tack et al., 1996; Andersson et al., 2000; Wakelin et al., 2008; Griffiths et al., 2011). Accordingly, of all soil properties, the soil pH seems to be the most important factor in affecting soil bacterial diversity and community composition (Fierer and Jackson, 2006; Lauber et al., 2008). In agricultural soils, the bacterial and fungal community structure and catabolic function are also strongly correlated with soil pH (Lauber et al., 2009). Shen et al. (2013) further demonstrated that the bacterial community composition and diversity were strongly affected by pH, but no similar effects are observed for the fungal community. The authors suggest that the strong influence of pH on bacterial community composition but not on fungi may be due to the narrow range of pH for the optimal growth of bacteria as compared with fungi that tolerate a wider pH range. Similar findings indicated that soil pH was the best predictor for bacterial diversity (e.g. highest diversity at near-neutral pH), richness, and community composition, while the soil nutrient status was a stronger driver for the fungal community (Maestre et al., 2015; Zhang et al., 2017; Shen et al., 2018). However, a continental-scale study on dryland sites revealed that the soil pH does not correlate with bacterial diversity, possibly due to higher overall pH values (Li et al., 2017).
Fertilization and nutrient availability
Since soil pH controls nutrient availability and accessibility and, consequently, changes in bacterial and fungal community structure, then fertilization practices (e.g. chemical versus organic fertilization) and soil amendments must also play an important role. Francioli et al. (2016) showed that a combination of organic and inorganic fertilization led to increased total N and organic C, causing changes to the bacterial community composition, which correlated with taxa involved in organic matter decomposition and nutrient transformation. Goldfarb et al. (2011) found significant differences in the bacterial and fungal communities between mineral, organic, and mineral–organic combined fertilization. Organic fertilization (manure) increased bacterial diversity, stimulating microbial groups known to thrive in nutrient-rich environments, while soils without manure contained microbial groups adapted to nutrient-limited conditions (Eilers et al., 2010). In addition, fertilization altered the relative abundance of plant-beneficial and plant-pathogenic microbes. Overall, pH and total organic C were identified as the major factors driving the structure and activity of the soil microbial community.
One of the most essential nutrients required for cellular metabolism and growth of bacteria is C. Carbon soil amendments can also affect microbial communities (Maestre et al., 2015). Indeed, low molecular weight C amendments, particularly citric acid, of three soil types resulted in shifts in the bacterial communities (Siciliano et al., 2014). However, the responses in these shifts can vary depending on the soil type. Kuramae et al. (2012) concluded that organic C content had a direct positive effect on the diversity and abundance of bacteria and fungi. Further studies showed that other nutrients such as N and P were the major factors influencing bacterial and fungal community structures in the soil and rhizosphere (Marschner et al., 2004).
Soil water content and temperature
Seasonal cycles and, more importantly, global warming change the temperature, CO2 levels, daylight duration, wind, precipitation (rainfall), and/or humidity (Fig. 1). These changes can alter biological and chemical processes in living organisms, such as photosynthesis in plants, or nutrient recycling in the soil (Schuur and Matson, 2001; Borjigidai et al., 2006; Alvarez-Clare and Mack, 2011). Changes in precipitation, temperature, and vegetation as a result of seasonal changes caused shifts in the microbial community structure and function (Habekost et al., 2008; Bell et al., 2009; Koranda et al., 2013). For example, soil moisture had the highest impact on some microbial parameters (e.g. community structure, substrate activity) at the end of winter and the second highest impact at the end of summer (Bell et al., 2009). In addition, other parameters such as microbial biomass and fungal substrate activity highly correlated with temperature in different seasons (Table 1).
Temperature affects microbial growth and activity, and thus can cause shifts in community composition and function (Pettersson and Bååth, 2003; Bell et al., 2009). In some cases, temperature changes of 3 °C resulted in changes in the abundance of fungal and bacterial communities (Bárcenas-Moreno et al., 2009). In contrast, Castro et al. (2010) revealed undetectable effects on cyanobacterial abundance or bacterial community by a 2–3 °C increase in soil temperature, while altered precipitation had significant effects. Taking into account the different experimental settings, it is thus unclear which of the two—water content or temperature—has a larger effect on microbial communities in the soil.
Among a range of climate change drivers (CO2, temperature, and precipitation), precipitation had the largest effect on bacterial and fungal community composition (Johnson et al., 2012). By testing the effects of wetting events, Castro et al. (2010) found that the amount of water added had a much greater impact than the irrigation frequency on shaping the bacterial and fungal community structures. In another study, the abundance and community structure of fungi was unaffected by extreme precipitation events compared with more frequent moderate events, which increased bacterial abundance (Frossard et al., 2015). These effects may be due to changes in soil pH or availability of nutrients upon precipitation. For example, the continental-scale study of Zhang et al. (2019) on dryland sites revealed that aridity indirectly affected soil pH and organic C content, leading to reduced diversity and abundance of soil bacteria and fungi.
Soil type, texture, and structure
Soil type can be a primary determinant of microbial communities because soils comprise a range of characteristics, such as nutrient and water content, cation exchange capacity, or texture and structure. A variety of studies have shown that the soil type can have a strong influence on the soil microbial communities (Girvan et al., 2003; Maestre et al., 2015), as observed in the rhizosphere of grass (Lolium perenne) and lettuce (Gelsomino et al., 1999; Singh et al., 2007; Schreiter et al., 2014). The soil texture/structure can affect the size and distribution of particles and pore spaces (Table 1), influencing the flow of water and nutrients and, consequently, lead to changes in the soil microbial community (Girvan et al., 2003; Lauber et al., 2008). Bach et al. (2010) showed that the microbial community differed between silty clay loam and loamy fine sand soil. Chau et al. (2011) observed that soil texture affected bacterial species richness but not bacterial diversity. The microbial community structure was also significantly affected by particle size, whereby higher microbial diversity was attributed to smaller silt and clay particle size than coarse sand fractions (Sessitsch et al., 2001). Moreover, particle size fraction affected the bacterial community structure more than the type of organic soil amendment (Sessitsch et al., 2001).
Soil salinity and drought: abiotic stresses affecting microbial communities in plants
Plant microbiome studies showed the complex relationship between environmental factors and bacterial community structures, especially in open field conditions, emphasizing the possible bias in laboratory experiments due to the absence of the variability of environmental changes. Extreme environmental changes or abiotic stresses, especially in light of climate change, can cause changes in microbial communities. The soil microbiome can be affected by abiotic stresses both directly (e.g. survival of drought-, salt- or heat-tolerant taxa) (Martiny et al., 2017; Naylor and Coleman-Derr, 2017) or indirectly (e.g. through altered soil chemistry or diffusion rates) (Schimel et al., 2007; Liptzin et al., 2011). Soil salinity and drought are arguably the biggest threats to global food security, and are clearly important factors affecting the structure and dynamics of soil microbiomes and, in turn, the plant microbiota, especially root endophytes. Recently, Berens et al. (2019) demonstrated that salinity treatment, along with leaf age, were crucial factors in determining the microbial community composition in Arabidopsis leaves. The study also identified a leaf age/developmental stage-dependent response to biotic and abiotic stress.
A meta-analysis of soil microbial communities revealed that the global microbial composition in saline soils is more affected by salinity than by any other abiotic factor (Lozupone and Knight, 2007). A significant difference in the endophytic microbial community composition was observed in black alder (Alnus glutinosa Gaertn.) roots grown in saline soil, with a decrease in the bacterial diversity and species richness and evenness (Thiem et al., 2018). In date palm (Phoenix dactylifera), the endophytic bacterial community in salinity-treated plants contained a higher number of total operational taxonomic units (OTUs) and higher species evenness and diversity, compared with control plants (Yaish et al., 2016). The root microbiome under drought stress conditions is determined by how the stress shapes both the host plant and the surrounding soils, where the total bacterial biomass is reduced (Hueso et al., 2012; Alster et al., 2013). Bogino et al. (2013) demonstrated that the rhizosphere of alfalfa (Medicago sativa) plants exposed to differing water-limiting conditions harbor distinct bacterial communities with different abilities to develop biofilms, and thus to establish themselves in this microenvironment. A recent study on the root microbiome of sorghum (Sorghum bicolor) demonstrated that drought causes enrichment of a distinct set of roots microbes. The discovery of this drought-induced enrichment and associated shifts in metabolite exchange between the plant and the microbes revealed a potential blueprint for manipulating plant microbiomes for improved crop fitness (Xu et al., 2018).
Suppressive soil: effects of soil biotic factors on plant health and microbial communities
Soil microbiome studies increasingly focus on improving soil health, quality, and fertility by promoting growth of beneficial while suppressing pathogenic microbes (Schlatter et al., 2017). This is particularly evident when discussing suppressive soils, which are soils that possess the ability to limit the growth and survival of plant pathogens (Baker and Cook, 1974). Suppressive soils fall into two general categories: general disease suppression is attributed to the soil’s total microbiome antagonistic activity against a broad range of soil-borne pathogens, while specific suppression is attributed to an individual taxon or group of microbes and is transferrable by adding pure cultures or small amounts of suppressive soil to conducive (non-suppressive) soil (Weller et al., 2002; Schlatter et al., 2017). The microbial community composition and diversity in soil is important for pathogen suppression, as demonstrated by the lower suppression rates of sterilized or semi-sterilized soil compared with unsterile soil (Garbeva et al., 2006; Mendes et al., 2011; Meng et al., 2012; Svenningsen et al., 2018). Studies have revealed changes in the rhizosphere microbial community upon pathogen infection and identified key taxa that may be involved in suppression of plant pathogens or diseases (Mendes et al., 2011). A 10 year long wheat (Triticum aestivum) field study revealed that the outbreak, decline, and suppression of the take-all disease, caused by the pathogenic fungus Gaeumannomyces graminis var. tritici, was correlated with changes in the rhizosphere bacterial communities (Sanguin et al., 2009). Distinct microbial communities also exist in potato (Solanum tuberosum) when comparing common scab-conducive soil, caused by pathogenic bacteria Streptomyces scabies, with suppressive soil (Meng et al., 2012; Rosenzweig et al., 2012).
Meta-barcoding analysis of Fusarium wilt-suppressive and conducive soils demonstrated that specific genera of fungi were exclusively present, and some bacterial genera were more abundant in suppressive soils (Siegel-Hertz et al., 2018). Zhou et al. (2019) also observed a higher fungal and bacterial richness and diversity in Fusarium wilt-conducive than suppressive soils. Furthermore, the type of soil pathogen and soil properties may also play a role in the suppressive potential. Postma et al. (2008) found that, depending on the pathogen, soil suppression correlated not only with specific antagonistic microbes but also with different soil properties. More importantly, a recent study revealed that small initial differences in the soil microbiome composition can affect plant–pathogen interactions and, therefore, plant health under natural field conditions (Wei et al., 2019).
Additionally, agricultural management practices can affect the suppressive potential of soils, primarily due to changes in the microbial communities. Garbeva et al. (2006) revealed a correlation between agricultural management practices on soil microbial community structure and its effect on soil suppression of the pathogenic fungus Rhizoctonia solani AG3. Based on crop rotational diversity practices, Peralta et al. (2018) suggested that microbial community composition might be more crucial than microbial diversity in disease suppression.
Plant-associated microbial communities
Since plant phenotype and fitness depend on the associated microbiome, plants try to recruit the best microbial community under given conditions (e.g. nutrient availability, pathogenic infection, and abiotic stresses). These factors not only affect the plant microbiota but also shape the soil microbiome and, more specifically, the rhizosphere. Hereafter, we will describe different plant-related factors responsible for defining the selected microbial communities (bacteria and fungi) in different plant organs (e.g. seeds, roots, or shoots) (Fig. 1; Table 2).
Table 2.
Plant factors that are responsible for shaping microbial communities
Plant seeds harbor their own microbial community
Seed coating is an efficient tool to deliver beneficial microbes for agricultural applications (Rocha et al., 2019). Interestingly, the seeds of native plants harbor a more specific microbiota than that reported for crop plants, allowing plant populations to survive, persist, and germinate under harsh natural conditions (Fenner et al., 2005; Wassermann et al., 2019). Different studies have investigated the dynamics of the seed microbiota during germination and emergence. Eight plant genotypes mostly affiliated to Brassicaceae were evaluated at three physiological stages: seed, germinating seed, and seedling states (Barret et al., 2015). Similar to bacterial and fungal taxa associated with the rhizosphere and the phyllosphere of various plant species (Toju et al., 2019), the seed microbiota was shown to be composed of three major bacterial phyla, Actinobacteria, Firmicutes, and Proteobacteria, and two fungal classes, the Dothideomycetes and Tremellomycetes (Barret et al., 2015). This suggests that the seeds might serve as a microbial bank for other plant compartments (van Overbeek et al., 2011) where the plants can select for beneficial microbes, which explains the high content of β-Proteobacteria, γ-Proteobacteria, and Firmicutes to suppress diseases (Berendsen et al., 2012). Indeed, bacterial endophytes of maize and rice (Oryza sativa) seeds were also found in the root endosphere and rhizosphere of these plants (Johnston-Monje and Raizada, 2011; Hardoim et al., 2012). Notably, Wassermann et al. (2019) conducted a study on seeds of eight native alpine plant species and highlighted the importance of the plant genotype as the main driver of the seed microbial community composition and diversity.
Plant genotype determines microbiome composition
In the past 20 years, evidence has accumulated that plant genotypes dictate the development of plant phenotypes and influence the microbial community composition of roots, leaves, and seeds (Table 2) (Adams and Kloepper, 2002; Bálint et al., 2013; Sapkota et al., 2015; Müller et al., 2016; Wagner et al., 2016; Adam et al., 2018). For example, Delmotte et al. (2009) investigated the microbiome of different plants of the Fabaceae and Brassicaceae families (e.g. clover, soybean, and Arabidopsis) and highlighted that, despite the ~130 million years of evolutionary divergence between the families (Hyung et al., 2014; Johnston-Monje et al., 2016), ~70% of the phyllosphere microbiota were conserved. This indicates the presence of a large core microbiome with minor host-specific functions of the microbiota. Balint-Kurti et al. (2010) revealed consistent differences among maize genotypes in the diversity of the epiphytic microbial population and identified UV-B-specific loci that genetically correlated with resistance to fungal pathogen infection. The microbes inhabiting the phyllosphere and rhizosphere are affected by the plant species to different degrees due to different plant phenotypic characteristics (Delmotte et al., 2009; Jones et al., 2019). The phyllosphere microbial community is affected by time (day/night) and exclusively by the plant genotype because the compounds secreted in the leaves are limited (Lindow and Brandl, 2003). Sapkota et al. (2015) observed that plant genotype at the species level of cereals (wheat, barley, oat, rye, and triticale) provides 43% of the variance in the total fungal community. By investigating the phyllosphere of five dominant temperate forest tree species (Acer saccharum, Acer rubrum, Betula papyrifera, Abies balsamea, and Picea glauca), Laforest-Lapointe et al. (2016) demonstrated that host species features, such as wood density and leaf N content, drive the bacterial community structure. Similar results were found for fungal communities in European beech (Fagus sylvatica), which were more impacted by leaf physiological characteristics (Unterseher et al., 2016). Moreover, Li et al. (2018) showed that the leaf and root microbiomes of spruce trees grown in a common garden are affected by host genotype, with differences found between the phyllosphere and soil and between bacteria and fungi. Therefore, phenotypic characteristics of the host plant shape the composition of its associated microbial community (Li et al., 2018; Jones et al., 2019).
Root exudates and their interactions with root-associated microbes
Interactions between plants and their microbial communities are not unidirectional. The host plant provides novel metabolic capabilities to its microbial associates, leading to the adaptation of niche-specialized inhabitants that can have either a positive (mutualist), neutral (commensal), or deleterious (pathogen) impact on plant fitness (Thrall et al., 2007). The rhizosphere is a complex habitat that is surrounded by a soil matrix where the plant roots constantly produce and secrete a diverse suite of metabolites and compounds called root exudates (Knief et al., 2012; Zhalnina et al., 2018). Root exudates are commonly produced with great variation in the chemical composition which is under genetic control of the host (Inderjit and Weston, 2003; Canarini et al., 2019). Root exudates are mainly comprised of primary metabolites such as sugars, amino acids, and carboxylic acids, as well as a diverse set of secondary metabolites (Cesco et al., 2010; Hu et al., 2018).
Root exudates, which represent up to 20% and 15% of fixed C and N, respectively (Haichar et al., 2016; Venturi and Keel, 2016), enrich the soil and rhizosphere and lead to changes in the microbial communities. The rhizosphere community is influenced by both the soil and plant genotype due to differences in root exudate quality and quantity secreted in the soil (Berg and Smalla, 2009; Aira et al., 2010; Gomes et al., 2018). Typically, the quality and quantity are determined by the size, age, and physiological condition of the plant root system. Abiotic stresses can also affect plant root exudates and the microbial community, as shown for citrus plants under salinity and temperature stress (Vives-Peris et al., 2018). Exudates from Macrophylla salt-stressed plants were able to promote the growth of Pseudomonas putida KT2440 and Novosphingobium sp. HR1a, whereas exudates from Carrizo salt-stressed plants did not promote bacterial growth. Moreover, in the presence of exudates from Macrophylla salt-stressed plants, growth promotion by Novosphingobium sp. HR1a was higher than with P. putida KT2440, which could be due to the higher tolerance of this strain to salinity stress (Vives-Peris et al., 2018).
Root exudates can also play a role as signaling molecules, attractants, or stimulants in establishing a symbiotic relationships with different microbes and, additionally, function in defense against pathogens (Perret et al., 2000; Kobayashi et al., 2004; Cesco et al., 2010; Baetz and Martinoia, 2014). The growth of soil microbes is usually C limited and the high amounts of sugars, amino acids, and organic acids that plants deposit into the rhizosphere represent a valuable nutrition source for microbial growth (Bais et al., 2006). However, depositing C will attract both pathogenic and beneficial microbes, suggesting that plants not only evolved recognition mechanisms to discriminate between beneficial and pathogenic microorganisms (Passera et al., 2019), but can also change root exudate composition to serve such selective mechanisms. Clear examples are the secretion of communication molecules/attractants such as flavonoids, strigolactones (SLs), or terpenoids (Bais et al., 2006; Venturi and Fuqua, 2013; Massalha et al., 2017). Flavonoids (2-phenyl-1,4-benzopyrone derivatives) are the most important molecules from the symbiotic perspective. Although found throughout the plant kingdom, flavonoids specifically trigger the expression of the rhizobial genes (nod, nol, and noe) required for nodulation and efficient N2 fixation of different legume members (Kobayashi et al., 2004; Zgadzaj et al., 2016; Saad et al., 2019). The nodulation capacity varies with flavonoids and rhizobia; and, in some cases, flavonoids may inhibit nodulation (Cooper, 2007; Hassan and Mathesius, 2012). Interestingly, plant fitness determines exudate quality, as seen in non-infected healthy Arabidopsis and rice plants that constitutively produce and release metabolites such as antimicrobial diterpene rhizathalene A or momilactone A to protect plants against infection (Vaughan et al., 2013).
The role of root exudates in defense responses
Upon pathogen infection, plants produce low molecular weight antimicrobial compounds, called phytoalexins, that are not detectable in healthy plants (VanEtten et al., 1994). Clear evidence for this comes from Fusarium graminearum-infected barley roots, where the infected plant induced the production of antifungal compounds (Lanoue et al., 2010). Glucosinolates are another group of plant metabolites with antimicrobial activities that are specifically produced by Brassicaceae. An Arabidopsis CYP79A1 transgenic line, which produces exogenous glucosinolates, altered the bacterial and fungal communities in the rhizosphere and root tissues (Bressan et al., 2009). The synthetic SL analog GR24 inhibits the growth of an array of phytopathogenic fungi when present in the growth medium (Dor et al., 2011), indicating that secreted SLs can affect natural enemies directly or indirectly by modulating hormonal defense pathways and contribute to below-ground plant biotic stress responses (Torres-Vera et al., 2014). Triterpenes are another group of plant metabolites that possess antifungal and antibacterial activities, suggesting potential roles in shaping the plant microbes (Brown et al., 1963; Papadopoulou et al., 1999; Augustin et al., 2011). Recently, Huang et al. (2019) demonstrated that Arabidopsis produces a range of specialized triterpenes that direct the assembly and maintenance of an Arabidopsis-specific microbiota, enabling it to shape and tailor the microbial community within and around its roots for its own purposes.
Root exudates shape the rhizosphere microbiota
Plants also use root exudates to alter the root microbial (bacterial and fungal) communities and exploit them for their own benefits. Maize plants were found to produce and release a mixture of metabolites from the roots, including benzoxazinoids (BXs) such as DIMBOA, which influence the composition of the root-associated microbiota (Hu et al., 2018; Cotton et al., 2019). DIMBOA is relatively short lived and is rapidly converted to the more stable MBOA that accumulates in the soil. As a result, MBOA triggers changes in the structure of the root-associated microbiota in the next plant generation. The microbiota-mediated BX-dependent effects on plant growth and defense were strongly associated with changes in the bacterial, rather than the fungal, rhizosphere community. These changes resulted in increased leaf defense, suppression of herbivore growth, and decreased plant growth, and the latter depended on the plant genetic background (Hu et al., 2018).
Stringlis et al. (2018b) provided direct evidence of how a specialized root exudate, the antimicrobial coumarin scopoletin, can cause changes in the microbial community structure and diversity in the rhizosphere. Scopoletin inhibits the fungal pathogens Fusarium oxysporum and Verticillium dahliae but not the growth-promoting rhizobacteria Pseudomonas simiae WCS417 and P. capeferrum WCS358. Voges et al. (2019) showed that the lack of coumarin biosynthesis in ‘f6′h1’ mutant lines caused a shift in the root microbial community specifically under Fe deficiency, demonstrating a potential role for Fe-mobilizing coumarins in shaping the Arabidopsis root bacterial community by inhibiting the proliferation of a relatively abundant Pseudomonas species via a redox-mediated mechanism.
Overall, the secretion of the root exudates (genotype) leads to chemical changes in the soil composition, soil properties, available nutrients (see below), and toxic elements in the rhizosphere (Neumann and Römheld, 2000; Marschner et al., 2004). All the above studies suggest that molecules derived from these specialized metabolites may play a role in the local adaptation of the plant to the soil environment and microbial ecology. Therefore, the exudation of bioactive compounds in root exudates probably defines the assembly of the plant-specific root and rhizosphere microbial communities for the benefit of the plant.
Cycling of nutrients between the soil, plant, and associated microbes
Plants are dependent on the growth of soil microbes which possess the metabolic machinery to depolymerize and mineralize organic forms of N, P, K, S, and Fe. In soil, most compounds are bound to organic molecules and are, therefore, minimally bioavailable for plants. To access these nutrients, plants adopt different strategies to interact with their environment for the solubilization and acquisition of nutrients (Lambers et al., 2008; Orwin et al., 2010; Grigulis et al., 2013). These strategies strongly influence plant–microbiota interactions due to the competition between plants and microorganisms for soil nutrients. The impact of plant nutrient resource strategies, plant functional traits, and the diversity of active microbiota through root exudation was studied extensively in the last decade (Guyonnet et al., 2018).
Nitrogen, phosphorus, and potassium (NPK)
The relationships between the plant and soil microbiome are governed by the trade-off theory where the plant provides C and, in return, can benefit from essential nutrients provided or facilitated by microbes, such as N, P, and K. For example, different studies highlighted the involvement of N2-fixing microbes (free-living ‘non-symbiotic’ or mutualistic ‘symbiotic’) in promoting plant growth (Vitousek et al., 2002; Graham and Vance, 2003; Bahulikar et al., 2014; Gaby and Buckley, 2015). Some bacteria and fungi can solubilize inorganic P or mineralize organic P (Eida et al., 2017; Nehls and Plassard, 2018). Many of these P-mobilizing strains are growth-promoting microbes which can promote plant growth via a wide variety of mechanisms. Thus, it is difficult to correlate P-mobilization mechanisms to the observed growth promotion elicited by these strains (Richardson and Simpson, 2011). However, under P-deficient conditions, plants respond by shaping the root microbial community (Castrillo et al., 2017; Finkel et al., 2019), which could enrich P-solubilizing/mobilizing microbes. Another vital nutrient considered as a key parameter of soil fertility and plant growth is K. As described by Sheng and He (2006), the inoculation of plants by Bacillus edaphicus NBT strains increased the production of citric, oxalic, tartaric, succinic, and α-ketogluconic acids, leading to K mobilization from K-containing minerals (e.g. mica and biotite) and chelation of silicon.
Iron
Fe is another essential element needed by all living organisms and is considered as a key micronutrient for soil fertility. The combination of the low concentration of Fe3+ together with high demand from both plants and microbes leads to a competition for Fe3+ in the rhizosphere (Guerinot and Yi, 1994). Bacteria and plants employ different strategies to overcome Fe limitations. For example, different groups of bacteria (e.g. Pseudomonas, Azotobacter, Bacillus, Enterobacter, Serratia, Azospirillum, and Rhizobium) produce low molecular weight proteins called siderophores with high affinity to chelate Fe from the soil (Loper and Buyer, 1991; Glick, 2014). Depending on the genotype, plants have adapted different strategies for Fe acquisition, such as the secretion of protons (Guerinot and Yi, 1994), the plasmalemma transport of Fe2+ by transporters (Eide et al., 1996; Vert et al., 2002), and/or the reduction of Fe3+ to the more stable Fe2+ by an NADPH-ferric chelate reductase (Yi and Guerinot, 1996). On the other hand, grasses can synthesize phytosiderophores to form complexes with Fe3+ complexes for uptake by specific transporters (von Wirén et al., 2000).
As a part of the adaptive responses to Fe deficiency, plants such as Arabidopsis can produce coumarins: active metabolites that change microbial dynamics by limiting the growth of a plant pathogenic Pseudomonas strain (Voges et al., 2019). Moreover, plant iron homeostasis is not only affected upon pathogen infection (Aznar et al., 2015), but also upon root colonization by plant growth-promoting rhizobacteria (PGPRs) (Zamioudis et al., 2015; Verbon et al., 2017). PGPRs are known to trigger induced systemic resistance (ISR) that primes plant tissues for enhanced defense against a broad spectrum of pathogens (Lugtenberg and Kamilova, 2009). A clear connection between ISR and iron homeostasis was demonstrated by Leeman et al. (1996), where the elicitation of ISR against Fusarium wilt in radish (Raphanus sativus) by beneficial Pseudomonas spp. was shown to be more effective under low-iron conditions. Siderophores secreted by Pseudomonas spp. were subsequently shown to act as elicitors of ISR in tomato (Meziane et al., 2005) and rice (De Vleesschauwer et al., 2008).
Phytohormones and their roles in shaping plant microbiota
Plant hormones (phytohormones) play diverse roles in plant physiological processes including mutualistic interactions with soil microbiota (Shigenaga and Argueso, 2016). The well-studied phytohormones are jasmonic acid (JA), salicylic acid (SA) (Boatwright and Pajerowska-Mukhtar, 2013), ethylene (ET) (Ju et al., 2015), abscisic acid (ABA) (Finkelstein, 2013), auxin (Austin et al., 2002), gibberellins (GAs) (Binenbaum et al., 2018), cytokinins (CKs) (Jiang et al., 2013), brassinosteroids (BRs) (Nolan et al., 2017), and SLs (Zwanenburg et al., 2016).
Auxin
Indole acetic acid (IAA) plays a role in shaping the microbiome because it regulates the development of lateral and secondary roots, which represent the preferential sites for microbial colonization (Kaldorf and Ludwig-Müller, 2000; Contreras-Cornejo et al., 2009; Zamioudis et al., 2013; Stringlis et al., 2018a). Applications of various forms of auxins (IAA, indole-3-butyric acid, 2,4-dichlorophenoxyacetic acid, and 1-naphthaleneacetic acid) promoted the spread of arbuscular mycorrhizal (AM) fungi and arbuscular abundance (J. Liu et al., 2016). The auxin (IAA)-deficient bushy mutant (Symons et al., 1999) showed reduced AM colonization but did not alter AM fungal structures inside the roots (Foo, 2013). Moreover, the tomato auxin-resistant diageotropica (dgt) mutant showed lower AM fungal development in both monoxenic and ex vitro conditions (Hanlon and Coenen, 2011). On the other hand, different soil microbes, either free-living or plant-associated, produce IAA themselves. Interestingly, 60% of phyllosphere bacteria and 80% of epiphytic bacteria can produce IAA (Spaepen et al., 2007; Kim et al., 2011; Spaepen and Vanderleyden, 2011). The synthesis of IAA and its derivatives was reported for Acidovorax, Agrobacterium, Arthrobacter, Bacillus, Chryseobacterium, Enterobacter, Pseudomonas, Ochrobactrum, Mycobacterium, Methylobacterium, and Stenotrophomonas species (Omer et al., 2004; Egamberdieva, 2009; Egamberdieva et al., 2015; Eida et al., 2018; Tsolakidou et al., 2019). This large number of bacterial IAA producers suggests that IAA synthesis might be a trait that contributes to survival in the plant environment (Kim et al., 2011). This idea is supported by several reports of different bacteria: IAA mutants of Erwinia herbicola (Brandl and Lindow, 1998; Manulis et al., 1998) and Pseudomonas savastanoi (Spaepen et al., 2007) showed reduced bacterial proliferation on leaves. Together with the plant endogenous IAA pool, bacterial auxin stimulates plant cell growth and proliferation, as well as plant tolerance to abiotic stresses (Panwar et al., 2016; Sorty et al., 2016; Barnawal et al., 2017).
Abscisic acid
Among other functions, ABA is a key regulator of abiotic stress responses. Therefore, ABA-producing bacteria could be selected by plants to promote abiotic stress tolerance. Different soil microorganisms can produce ABA, including several phytopathogenic fungi, such as Cercospora rosicola, C. cruenta and Botrytis cinerea (Zeevaart and Creelman, 1988; Sharon et al., 2007), or bacteria, such as Azospirillum (Forchetti et al., 2007; Cohen et al., 2008). Interestingly, bacteria commonly found in the human body, which can live in soil and in water (Proteus mirabilis, P. vulgaris, Bacillus megaterium, B. cereus, Klebsiella pneumoniae, and Escherichia coli), are also capable of producing ABA (Karadeniz et al., 2006).
Cytokinin
A number of bacteria produce CKs, such as Arthrobacter, Bacillus, Azospirillum, Pseudomonas, and Methylobacterium (Naz et al., 2009; Jorge et al., 2019). The CK-producing B. subtilis strain IB-22 enhances growth of lettuce and wheat, with high colonization rates throughout the vegetative period and increased wheat productivity (Arkhipova et al., 2006, 2007). Other B. subtilis isolates stimulated root biomass of Platycladus orientalis by 14% and increased CK levels in leaves by 47% under water stress conditions (Liu et al., 2013). Similar increases of shoot and root biomass were observed in soybean (Glycine max) inoculated with Pseudomonas and Arthrobacter spp. under salinity stress (Naz et al., 2009). Pseudomonas aurantiaca TSAU22 and P. extremorientalis TSAU6 and TSAU20 enhanced the growth of wheat under salinity stress (Egamberdieva, 2009). Moreover, different members of the Methylobacterium genus produce high levels of CKs and increase the tolerance of plants to abiotic stresses (e.g. salt and drought stress) (Knief et al., 2012; Lee et al., 2015; Chanratana et al., 2017; Jorge et al., 2019).
Ethylene
A variety of plant processes involve the olefin hydrocarbon ET, including nodulation of legumes by rhizobia (Tamimi and Timko, 2003) and mycorrhizal root interaction (Gamalero et al., 2008). Plants use ET as a regulator of stress responses, such as extreme temperatures, water, UV light, and insect and nematode damage and wounding, as well as in interactions with fungi and bacteria (Abeles et al., 1992). Plant genotype, organ, developmental stage, and the associated microbiota are major determinants of ET signaling and responses (Pierik et al., 2007; Dugardeyn and Van Der Straeten, 2008). Interestingly, more than one-third of all cultivable soil bacteria can produce ET via different pathways (Nagahama et al., 1992). Several plant-associated microbes can increase plant ET levels by 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS) activity (Suganuma et al., 1995) or produce intermediates, such as KMBA, that cav n be converted to ET in planta (de Zélicourt et al., 2018). Plant-associated microbes can also decrease ET levels by producing ACC deaminase, an enzyme responsible for the cleavage of the plant ET precursor ACC into ammonia and α-ketobutyrate. Engineering bacteria with ACC deaminase activity promoted resistance of banana (Musa spp.) to Fusarium (Liu et al., 2019). ACC deaminase-containing bacteria are relatively common in soil, possibly providing these bacteria with a competitive advantage over other rhizosphere microorganisms by using ACC as an N source (Glick, 2014).
Jasmonic acid
JA and its volatile methyl ester, MeJA, play crucial roles in plant defense responses against insects and microbial pathogens (Bari and Jones, 2009). Interestingly, JAs also act as signaling molecules that facilitate interactions between plants and root-associated microorganisms (Pieterse et al., 2009). Current evidence indicates that JA influences the composition of the Arabidopsis root-associated microbiome (Carvalhais et al., 2017). Induction of JA signaling increased the relative abundance of bacterial populations closely related to taxa that are reported to suppress phytopathogens and insects (Schlaeppi and Bulgarelli, 2015). Interestingly, the host genotype determines the effect of JA signaling. For example, JA signaling in rice restricts endophytic colonization by certain N2-fixing Azoarcus bacterial strains when the host–bacterium interaction is less compatible (Miché et al., 2006) and suppresses nodule formation in the legume Lotus japonicus (Nakagawa and Kawaguchi, 2006). On the other hand, JA signaling does not impact the structure of the phyllosphere and root microbiomes of wild tobacco (Nicotiana attenuate) (Santhanam et al., 2014). Carvalhais et al. (2013) reported that JA signaling pathways affected the composition of root exudates and rhizosphere bacterial and archaeal communities, and these changes significantly correlated with each other. D cFurthermore, a correlation between root exudate content and the abundance of the bacterial communities was reported by Liu et al. (2017). The authors demonstrated that activation of JA signaling in wheat reduced the diversity and changed the composition of bacterial communities in the root endosphere but not in the shoots or rhizosphere. All this evidence suggests that the changes in root endophyte communities in response to JA signaling may reflect a co-evolved mechanism by which plants recruit microbial symbionts that enhance host biotic stress tolerance (Carvalhais et al., 2015, 2017).
Salicylic acid
SA mediates plant defense responses against pathogens (Loake and Grant, 2007; An and Mou, 2014) and establishes beneficial symbioses in legume–rhizobia interactions (Martinez-Abarca et al., 1998). SA has also been shown to modulate the composition of the root microbiota at the family level in Arabidopsis (Lebeis et al., 2015). Depending on the host plant species, different responses of the microbial community were reported for SA. For example, activation of the SA signaling pathway in wheat had no significant impact on the diversity of root-associated microbiomes (Liu et al., 2018). A comparison of the bacterial root microbiome of wild-type Arabidopsis with a set of mutants lacking biosynthesis and/or signaling of SA, JA, and ET (Lebeis et al., 2015) demonstrated clear microbial compositional changes of the root microbiome. Moreover, it was shown that certain bacterial endophytic families may require SA-related processes to colonize the root system. Exogenous application of SA altered the microbial community profile composition in both bulk soil and endophytic compartment samples, indicating SA-mediated selection for microbial families. Moreover, different bacterial strains can use SA in different ways, whether as a growth signal or as a C source. Thus, SA may influence the microbial community structure of the root by ‘gating’ bacterial taxa via a homeostatic control of immune system outputs (Lebeis et al., 2015).
The plant immune system during beneficial microbe interactions
The plant immune system is a prime microbial target to establish beneficial or pathogenic interactions. Plants can detect both beneficial and pathogenic microbes via pattern recognition receptors that bind microbe-associated molecular patterns (MAMPs), such as chitin for fungi or flagellin for bacteria, triggering a basal defense system to halt the growth of most microbes. This defense mechanism is known as MAMP-triggered immunity (MTI) (Boller and Felix, 2009). Some microbes secrete effector proteins to suppress MTI, allowing successful plant infection via effector-triggered susceptibility (ETS). Another plant defense system relies on plant resistance proteins that can recognize microbial effector proteins to activate effector-triggered immunity (ETI). ETI activates local and systemic responses, such as the SA signaling and expression of pathogen-related (PR) proteins. Activation of systemic acquired resistance (SAR) confers a l ong-lasting protection against a wide variety of pathogens (Glazebrook, 2005).
Rhizobial PGPRs: manipulation of the host immune system
Beneficial microbes evolved different strategies to modulate the plant immune system for beneficial association/symbiosis. In legumes, rhizobia evolved different mechanisms to avoid pathogen recognition (Cao et al., 2017). For example, rhizobial flagellin appears to lack the flg22 epitope required for flagellin sensing-2 (FLS2)-mediated MAMP activity (Lopez-Gomez et al., 2012). In contrast to pathogens, the identified rhizobial MAMPs, including flagellin, lipopolysaccharides, peptidoglycans, and K-antigen-type polysaccharides, appear to lack MAMP activity and do not trigger MTI in their hosts (Lopez-Gomez et al., 2012). However, rhizobia induce MTI in the early stages of the infection process in legume roots (Libault et al., 2010). Similar changes in defense-related gene expression patterns were also reported in two other legumes (i.e. L. japonicus and Medicago truncatula) when inoculated with Rhizobia species (Stacey et al., 2006; Jones et al., 2008). For successful invasion and nodule formation, rhizobia can also modulate plant SA levels. Interestingly, when alfalfa plants were inoculated with a non-compatible rhizobium strain, the plants showed increased levels of endogenous SA. However, no changes of SA levels were detected upon inoculation with a compatible rhizobium strain (Martinez-Abarca et al., 1998). Similar results were obtained using a Nod factor synthesis-impaired mutant, indicating the role of Nod factors (host-determined compatibility) in suppressing MTI and SA-triggered responses (Liang et al., 2013).
Similar to pathogens, rhizobium strains manipulate the plant immune system by using effector proteins secreted via type 3 secretion systems (T3SSs), collectively named Nodulation outer protein ‘Nops’ (Marie et al., 2003; Saad et al., 2005; Songwattana et al., 2017). In Sinorhizobium fredii strain NGR234, NopL acts as a virulence factor when ectopically expressed in tobacco plants, down-regulating virus-induced PR protein accumulation (Bartsev et al., 2004). Similar to NopL, NopM is involved in the inhibition of plant immunity through misregulation of host mitogen-activated protein kinase (MAPK) activation and by inhibiting reactive oxygen species (ROS) production (Bartsev et al., 2004; Xin et al., 2012). Another effector protein, NopT, induces immune responses and cell death, suggesting the presence of a cognate resistance protein (Dai et al., 2008). The same is likely to be true for the rhizobial effector NopP, as nopP mutants in NGR234 showed enhanced nodule formation and lower Pathogenesis-Related 1 (PR1) gene expression when inoculated in soybean (Skorpil et al., 2005; López-Baena et al., 2009). T3SS effector suppression of MTI responses is most probably superimposed on the dominant suppressive functions of exopolysaccharides (EPSs) and Nod factors (Zamioudis and Pieterse, 2012).
Strategies employed by non-rhizobia PGPRs
Non-rhizobial PGPRs also evolved different strategies to overcome the plant immune system. The presence of T3SSs was also reported in a number of plant-associated PGPR (non-rhizobia) strains including different species of Pseudomonas with a potential to synthesize and deliver effector proteins (Loper et al., 2012). Pseudomonas fluorescens strains SBW25 and Q8r1-96 have a complete T3SS machinery, and SBW25 secretes multiple effectors including members of the AvrE family (e.g. RopE) (Preston et al., 2001), while Q8r1-96 secretes RopAA of the HopAA1-1 effector family. All the T3SS effectors can suppress typical innate immune responses when ectopically expressed in tobacco (Nicotiana benthamiana) (Mavrodi et al., 2011). The supramolecular structure of the T3SS in P. fluorescens strain 2P24 was resolved and shown to have retained the ability to secrete effector proteins (P. Liu et al., 2016). The presence of T3SS and potential effector proteins of other beneficial Pseudomonas spp. (e.g. P. simiae WCS417 and P. defensor WCS374) was reported by Stringlis et al. (2019). Effector delivery via T3SS may be one mechanism by which PGPRs can either assist in the suppression of MTI responses or manipulate certain host metabolic processes (Zamioudis and Pieterse, 2012).
PGPRs can activate ISR, which involves both JA and ET signaling pathways, leading to the expression of defense-related genes. Both SAR and ISR are activated for different responses, and, although ISR-mediated protection is less effective, SAR and ISR can also work together to provide the best protection and resistance against pathogens (van Wees et al., 2000). Induction of ISR against a broad range of pathogens through activation of SA-, JA-, or ET-responsive defense-related genes in plants w shown for Bacillus spp., Serratia liquefaciens, Penicillium spp., and Trichoderma spp. (Djonović et al., 2006; Hossain et al., 2007; Ongena et al., 2007). In Arabidopsis, the activation of ISR in roots by Pseudomonas simiae WCS417r (PGPR) is not accompanied by SA-responsive PR protein gene expression, indicating that WCS417r-mediated ISR functions independently of SA (Pieterse et al., 1996). WCS417 is able to suppress flagellin-triggered MTI responses in Arabidopsis roots via apoplastic secretion of low molecular weight molecules (Millet et al., 2010; Stringlis et al., 2018a). By using large-scale transcriptomic analysis and reverse genetics approaches, several components, such as MYB72, β-glucosidase U42 (BGLU42), and MYC2, were shown to be involved in rhizobacteria-mediated ISR (Van der Ent et al., 2009; Zamioudis et al., 2014). Recently, it was shown that the root-specific transcription factor MYB72 plays an important role in rhizobacteria-induced secretion of coumarins that shape the assembly of the microbiome in the rhizosphere, potentially optimizing the association with ISR-inducing rhizobacteria (Stringlis et al., 2018b). Other strategies employed by different PGPRs to suppress the root immune system were reported by Yu et al. (2019). For example, P. capeferrum WCS358 and P. simiae WCS417 quench local Arabidopsis root immune responses by lowering the environmental pH via bacterial gluconic acid (Yu et al., 2019).
Another strategy to modulate the plant immune system by PGPRs is phenotypic variation, where bacteria switch between different morphologies (flagella, lipopolysaccharides, pigmentation, etc.) or change their genetic make-up (Davidson and Surette, 2008; Wisniewski-Dyé and Vial, 2008). A clear example of such a process was observed with P. brassicacearum NFM421 (Achouak et al., 2004), which was isolated as a major root-colonizing population from Arabidopsis. NFM421 showed morphological phase variation during root colonization of Arabidopsis, resulting in different colony appearances on agar surfaces. Phase II cells localized to the surface of young roots and root tips, whereas phase I cells localized to root basal parts. The ability of phase II cells to spread and colonize new sites on the root surface correlated with the overproduction of flagellin. Phenotypic variation on plant roots is likely to be a colonization strategy that may explain the high colonization power of P. brassicacearum (Achouak et al., 2004) which, similarly to animal pathogens, employs phase variation to avoid detection by the immune system (Kingsley and Bäumler, 2000). Phase variation could also be a strategy of PGPRs to prime the plant immune system. The idea of priming could be explained by the plant–microbiota interaction, where plants discriminate friend from foe and respond by either ignoring, supporting, or eliminating microbes. Furthermore, the response pattern to non-pathogenic bacteria can be determined by the plant genotype (Ofek-Lalzar et al., 2014) and can differ across accessions in their recruitment of P. fluorescens (Haney et al., 2015). All these factors must be considered when studying plant-associated microbial communities and selecting individuals for inoculants.
Synthetic holobiont communities
Recent culture-independent analyses and culture collections have paved the way for developing artificially constructed communities, called synthetic communities (SynComs), for studying plant–microbe interactions and promoting plant growth and health (Lundberg et al., 2012; Bulgarelli et al., 2013; Bodenhausen et al., 2014; Bai et al., 2015; Helfrich et al., 2018; Carlström et al., 2019). SynComs can be assembled by rational bottom-up principles by co-culturing several individual microbes. For example, using a culture-dependent collection from sugarcane (Saccharum sp.), Armanhi et al. (2018) designed a SynCom comprised of highly abundant bacterial groups and successfully exploited the SynCom for promoting plant growth (increased biomass) in maize. By genome sequencing and comparative genomic analysis of this SynCom, coupled with colonization experiments, de Souza et al. (2019) found that functions related to nutrient acquisition were enriched in robust colonizers. Lebeis et al. (2015) revealed the importance of SA in shaping the root microbiota. Durán et al. (2018) demonstrated the importance of bacterial root commensals for Arabidopsis survival and biocontrol against filamentous eukaryotes and the importance of bacteria–fungi–oomycete consortia for plant growth promotion. Tsolakidou et al. (2019) used tomato rhizosphere bacteria for designing SynComs that were able to promote tomato growth and suppress Fusarium wilt symptoms. Using a bacterial SynCom composed of 185 members, Finkel et al. (2019) showed that excluding Bulkholderia isolates from the SynCom resulted in the accumulation of higher phosphate shoot levels in plants under P starvation compared with the full SynCom. Finally, drop-out and late introduction experiments using a SynCom made up of 62 leaf bacterial strains by Carlström et al. (2019) revealed that established microbiota are subject to change by late colonizers. The authors also showed that keystone taxa could play important roles in shaping the community structure, especially of strains that are present at very low relative abundance.
It is important to note that in previously mentioned experiments, the assembly of SynComs was performed by choosing either the most abundant taxa or whole collections based on what was cultured. However, we suggest that the selection should be based on the functional traits and abilities of each SynCom member (e.g. hormone production/modulation, nutrient solubilization, volatile production, colonization abilities, and production of antimicrobial compounds) (Fig. 1). In this way, unique traits of each member can complement each other. Furthermore, functional redundancy of SynCom members can increase the resilience of the inoculants, especially in a complex field system. It is also crucial to determine if the SynCom members are compatible with each other or with the plant and environment.
Desert plants and endophytic bacteria: a model approach for application of microbial inoculants
Hyper-arid deserts and semi-arid grasslands represent two of the harshest terrestrial environments and occupy >20% of the land surface of Earth. Agriculture in these areas faces many challenges, especially considering climate change-driven increases in temperature and aridity and the detrimental effects of abiotic stresses on crop productivity (Boyer, 1982; Bray et al., 2000). Here, we propose that microbial stimulants, whether single isolates or SynComs, should be selected on the basis of their target environment (e.g. bacteria isolated from salinity-stressed environments to promote salinity stress tolerance in plants). For example, pioneer desert plants or crops grown in semi-arid conditions could serve as a target source for isolating bacterial inoculants or SynComs, which can be exploited for semi-arid agriculture to increase the yield of cash crops (Marasco et al., 2012; Eida et al., 2018). The rhizosphere of drought-sensitive pepper (Capsicum annuum), cultivated in the North-Western desert region of Egypt, was enriched in PGPRs with growth-promoting abilities on pepper under drought stress (Marasco et al., 2012). Daur et al. (2018) and de Zélicourt et al. (2018) showed that bacterial strains isolated from the rhizosphere and endosphere of desert plants, respectively, in Saudi Arabia were able to boost the yield of alfalfa plants under desert agricultural conditions. The endophytic bacterium Enterobacter sp. SA187 from one of these collections survives under abiotic stresses and has PGPR traits (Andrés-Barrao et al., 2017). Application of SA187 was successful in field trials with alfalfa using low and high saline irrigation under desert conditions (de Zélicourt et al., 2018). The success of transferring beneficial microbe-induced abiotic stress tolerance from the lab to the field was probably because the field trials were performed in a similar environment to that from which the bacteria were isolated.
In an effort to achieve sustainable agriculture on semi-arid land, the DARWIN21 project (http://www.darwin21.org/) provides a database of bacterial strains isolated from pioneer desert plants native to the Middle East deserts (e.g. Jordan, Saudi Arabia, and Pakistan). Specific strains showed a great potential for desert agriculture (Bang et al., 2018; de Zélicourt et al., 2018; Bokhari et al., 2019), and draft genome sequences of some of these bacterial isolates have been released (Lafi et al., 2016a, b, c, 2017a, b, c, d), in addition to complete genome sequence analyses (Andrés-Barrao et al., 2017; Eida et al., 2020). We suggest that root-associated microbiota isolated from plants living in extreme conditions, possibly due to evolutionary selection, are ideal for obtaining plant growth-promoting microbes with traits for plant growth and promotion of abiotic or biotic stress tolerance.
Limitations of microbial community experiments and their applications in agriculture
Plant beneficial microbes become increasingly important for application in agriculture, primarily due to the significant effects of indigenous microbial communities on plant growth and health and the possibility of engineering microbiomes to control plant traits and produce antimicrobial compounds (Mueller and Sachs, 2015; Gopal and Gupta, 2016; Helfrich et al., 2018; Herrera Paredes et al., 2018). However, to understand the molecular and ecological functions of individual members in host-associated microbiomes is a major scientific challenge. This is due to the high complexity and genetic diversity at the species level in microbial communities, including the changing abiotic and biotic factors that dramatically structure microbial communities and the limitations in culturability of many microbes and in nucleic acid-based ‘omic’ approaches (Curtis et al., 2002; Morales and Holben, 2011). Furthermore, the lack of both systematic and comprehensive microbial culture collections for reconstruction experiments and model organisms for understanding plant–microbe interactions limits the progress in this field.
Culture-dependent community analysis and culture collections
Many studies demonstrate the limitations of culture-dependent community analysis when compared with culture-independent approaches. Two main problems arise when comparing these two methods: culturability and presence of rare taxa. Only a small fraction of the bacterial community can be cultured and those microbes often occur at very low abundance (Sogin et al., 2006; Pereira et al., 2011; Yashiro et al., 2011; Shade et al., 2012; Lee et al., 2016; Eida et al., 2018). The detection of rare species in culture-independent approaches depends on the sequencing technology or, more specifically, the sequencing depth and quality, the amplicon size, and primer pairs (Hiergeist et al., 2015; Beckers et al., 2016). Furthermore, DNA extraction and marker gene sequencing often do not discriminate between intracellular DNA from intact cells and extracellular DNA from lysed or dead cells (Nielsen et al., 2007). Challenges in culturability arise due to several reasons: (i) different species require different growth media and/or fastidious growth conditions; (ii) some microbes are obligate endophytes and need a host to survive; (iii) fast-growing or antagonistic microbes can constrain or inhibit growth of slow-growing strains; and (iv) growth or dominance of some species relies on the presence of others (Vartoukian et al., 2010; Yashiro et al., 2011; Niu et al., 2017; Sarhan et al., 2019).
Microbial community studies under natural, field, and laboratory settings
There are also other limitations in understanding community changes under natural or field settings. First, natural or field settings contain multiple interdependent factors that cannot be controlled. The soil properties, biological components, and climate all converge, giving rise to a complex environment where a certain microbial community structure is formed and where the root microbiota’s function could be affected (Fig. 1). Any change in one factor could affect all others, leading to false correlations as to which determinant factor caused changes in the community. For example, Bárcenas-Moreno et al. (2009) reasoned that changing soil pH would introduce changes in several other factors, making it difficult to separate pH from the other effects on soil. Often, comparing one factor (e.g. soil pH) from different natural soils can introduce further problems due to the presence of other factors (e.g. soil nutrients) which may play important roles in shaping the microbial community. Experimenting on microbial communities using single factors is only possible under laboratory conditions allowing an understanding of how each component of the soil environment plays a role in changing the microbial communities.
Technical aspects of community experiments
The sampling method, such as taking soil samples from different depths, can also lead to variable conclusions. For example, the bacterial and fungal communities differ depending on soil depth (topsoil versus subsoil) over long-term fertilization studies (Gu et al., 2017). Many studies have shown that the microbial diversity typically decreases with soil depth, probably owing to the decreased exposure to fertilizers from topsoil to subsoil (Li et al., 2014; Feng et al., 2019). Sampling of bulk soil can introduce high variability and, therefore, it is important to take into consideration sampling strategies to account for this variability (Ogram et al., 2007). Similarly, sampling plant tissue (e.g. leaves) of different age or developmental stage could introduce variability (Fig. 1).
The efficiency of genomic DNA extraction and the number of 16S rRNA copies per cell can vary depending on the bacteria (Frostegård et al., 1999; Klappenbach et al., 2000; Shrestha et al., 2007; Ketchum et al., 2018). Therefore, obtaining accurate abundances of each bacterial strain without knowing the number of 16S rRNA copies within their genomes is an additional limitation. Furthermore, contamination of samples, whether during sampling or during library preparation, can give rise to sequences not representative of the reality (Tanner et al., 1998). Finally, human factors such as agricultural management practices and land use can also affect microbial communities and soil health, and thus should also be considered when performing community experiments (Fig. 1) (Steenwerth et al., 2002; Lauber et al., 2008; Reeve et al., 2010; Carbonetto et al., 2014; Peralta et al., 2018; Sun et al., 2018; Le Guillou et al., 2019).
A roadmap for successful applications of plant-associated microbial inoculants
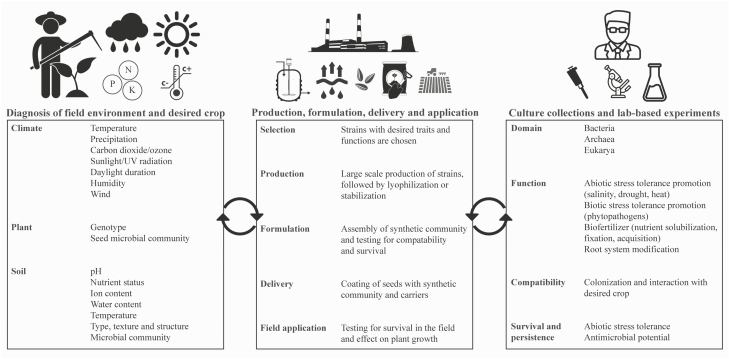
The construction and application of customized inoculants serve an important purpose for enhancing sustainable agriculture by increasing crop health and productivity. Microbiome studies and application of SynComs would greatly advance our knowledge of plant–microbe interactions when complemented with efforts to study and develop model systems from these synthetic communities. As discussed earlier, microbial community structure, function, and composition largely depend on the plant host/genotype, soil properties, the indigenous microbial community, and abiotic factors (Fig. 1; Tables 1, 2). Thus, there are many limitations and challenges of applying microbial inoculants in real, large-scale agricultural field settings. Here, we propose a framework in which the farmers, scientific community, and agricultural technology companies collectively contribute to reach the goal of successful microbial inoculant applications (Fig. 2).
Fig. 2.
Proposed framework for the successful application of microbial inoculants in agriculture. A framework in which the farmers/farming industry, scientific community, and research and agricultural technology companies collectively contribute to reach the goal of successful microbial inoculant applications. Microbial inoculants must be customized for the target crop, climate, and soil properties (left panel). An increase in scientific research of plant microbiomes, culture collections, and functional characterization of potential microbial inoculants paves the way for meeting farmers’ requirements (right panel). The integration of available microbial inoculants with farmers’ requirements and the large-scale production and formulation (especially for SynComs) is performed by agricultural technology companies (center panel). Collaboration and constant feedback between all three entities is required for the success of field application.
The first aspect in this framework is the thorough analysis of the target field environment and crop of interest (Fig. 2, left panel). Due to the presence of many factors that could affect microbial communities, the inoculants have to be customized to the target crop, field, environmental conditions, and agricultural management practices. Here, a characterization and solid understanding of the climate of the geographical location of the field (e.g. temperature, annual precipitation and humidity levels, and wind speeds) and soil properties (e.g. pH, nutrient status, moisture content, temperature, and microbial community) is performed. Then, the choice of crop plant and its genotype/variety are determined, specifically based on compatibility with climate/soil and economic feasibility. This also requires analysis of the indigenous seed microbial community as it may interfere with the applied inoculants.
The second aspect is a cornerstone in this framework and is pivotal for the success of inoculant application over a wide geographical context. This step requires an increase in scientific research on plant microbiomes and lab-based experiments as well as more culture-dependent isolations. Culturability of environmental microbes can be challenging, therefore its increase will require clever integration of omics (e.g. meta-transcriptomics) and novel culturomics techniques (e.g. plant-based media) (Bomar et al., 2011; Kwak et al., 2018; Sarhan et al., 2019). More importantly, characterization of the single isolates’ functions, survival abilities, plant growth-promoting traits, growth/stress tolerance-promoting mechanisms. and their compatibility with the desired crop is crucial (Fig. 2, right panel). Efforts to achieve this require the increase of whole-genome sequencing, functional characterization of isolates, plant phenotyping, and application of meta-omics (genomics, transcriptomics, proteomics, and metabolomics) approaches, in addition to developing sequencing technologies, bioinformatics models, and tools (Grosskopf and Soyer, 2014; Levy et al., 2018; Sergaki et al., 2018; Marco and Abram, 2019). For example, Rodrigues et al. (2018) recently developed a user-friendly web tool that makes use of the large amounts of microbiome data sets to identify the core microbiome associated with different habitats. Cross et al. (2019) used a reverse genetics approach to isolate and cultivate previously uncultured bacteria. Carper et al. (2019, Preprint) developed advanced computational programs that could overcome the limitations of amplicon sequencing in distinguishing members of a diverse community while maintaining desired member attributes.
The formation of systematic culture collections that cover a broad range of microbial domains (e.g. bacteria, archaea, and eukaryotes) must be considered (Fig. 2, right panel). Although most microbiome studies take into account bacteria and fungi, other microorganisms are also present in soil and could interact, symbiotically or antagonistically, with plants. For example, a recently isolated ammonia-oxidizing archaeon can promote the growth of Arabidopsis and induce systemic resistance against necrotrophic and biotrophic bacteria (Jung et al., 2016; Song et al., 2019). Indeed, archaea are important players in plants (e.g. rice), and their community composition responds to changes (e.g. plant aging and development) or stresses (e.g. drought) (Erkel et al., 2006; Edwards et al., 2018). Interestingly, bacteriophages have been recently shown to control soil-borne pathogens and thus should not be disregarded as a factor in selecting inoculants (Wang et al., 2019). Therefore, future community experiments and culture-dependent isolation should not disregard the presence of other microbial players, which could provide a clearer picture of the complex nature of microbiomes and improve their field application.
The third aspect of our proposed framework is the integration of the field data from farmers and available microbial resources from scientific research by agricultural technology companies in order to customize a suitable inoculant for the prescribed purpose (Fig. 2, center panel). First, the strain(s) are selected from the culture collections based on the desired traits and function, which are determined by field analysis. Large-scale production of the strains is then needed, followed by other processes, such as lyophilization for long-term storage and transport. For formulation of inoculants, the assembly of SynComs and testing the compatibility and survival of each SynCom member with each other is necessary. The single strain or SynComs inoculants can then be delivered for field testing either as lyophilized powder or by coating of seeds of the target crop. Finally, the performance of the inoculant in a field setting similar to the target field/environment is evaluated.
Finally, the framework relies on the constant feedback between all three aspects. Additionally, the increase in soil and plant microbiome data and development of models could assist in predicting how SynComs respond, adapt, and/or survive in the target environment and crop plant. The increase in funding for plant–microbiome research and formation of policies for the use of microbial inoculants in different countries are also needed to be considered for the overall success of achieving global food security in the future.
Acknowledgements
We would like to thank all members of the Hirt lab and the CDA management team for technical assistance and for their help in many aspects of this work. We would also like to thank Khaled Abd El Gawad and Wiam F. Alsharif for preparing the illustrations. The authors would like to thanks the reviewers for the great positive input in improving the review article.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This publication is supported by the King Abdullah University of Science and Technology (KAUST) to HH no. BAS/1/1062-01-01.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abeles FB, Morgan PW, Saltveit ME. 1992. Roles and physiological effects of ethylene in plant physiology: dormancy, growth, and development. In: Abeles FB, Morgan PW, Saltveit ME, eds. Ethylene in plant biology, 2nd edn New York: Academic Press, 120–181. [Google Scholar]
- Achouak W, Conrod S, Cohen V, Heulin T. 2004. Phenotypic variation of Pseudomonas brassicacearum as a plant root-colonization strategy. Molecular Plant-Microbe Interactions 17, 872–879. [DOI] [PubMed] [Google Scholar]
- Adam E, Bernhart M, Müller H, Winkler J, Berg G. 2018. The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant and Soil 422, 35–49. [Google Scholar]
- Adams PD, Kloepper JW. 2002. Effect of host genotype on indigenous bacterial endophytes of cotton (Gossypium hirsutum L.). Plant and Soil 240, 181–189. [Google Scholar]
- Aira M, Gómez-Brandón M, Lazcano C, Bååth E, Domínguez J. 2010. Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biology and Biochemistry 42, 2276–2281. [Google Scholar]
- Alster CJ, German DP, Lu Y, Allison SD. 2013. Microbial enzymatic responses to drought and to nitrogen addition in a southern California grassland. Soil Biology and Biochemistry 64, 68–79. [Google Scholar]
- Alvarez-Clare S, Mack MC. 2011. Influence of precipitation on soil and foliar nutrients across nine Costa Rican forests. Biotropica 43, 433–441. [Google Scholar]
- An C, Mou Z. 2014. Salicylic acid and defense responses in plants. In: Tran L-SP, Pal S, eds. Phytohormones: a window to metabolism, signaling and biotechnological applications. New York: Springer New York, 191–219. [Google Scholar]
- Andersson S, Nilsson SI, Saetre P. 2000. Leaching of dissolved organic carbon (DOC) and dissolved organic nitrogen (DON) in mor humus as affected by temperature and pH. Soil Biology and Biochemistry 32, 1–10. [Google Scholar]
- Andrés-Barrao C, Lafi FF, Alam I, de Zélicourt A, Eida AA, Bokhari A, Alzubaidy H, Bajic VB, Hirt H, Saad MM. 2017. Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium. Frontiers in Microbiology 8, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova TN, Prinsen E, Veselov SU, Martinenko EV, Melentiev AI, Kudoyarova GR. 2007. Cytokinin producing bacteria enhance plant growth in drying soil. Plant and Soil 292, 305–315. [Google Scholar]
- Arkhipova TN, Veselov SY, Melent’ev AI, Martynenko EV, Kudoyarova GR. 2006. Comparison of effects of bacterial strains differing in their ability to synthesize cytokinins on growth and cytokinin content in wheat plants. Russian Journal of Plant Physiology 53, 507–513. [Google Scholar]
- Armanhi JSL, de Souza RSC, Damasceno NB, de Araújo LM, Imperial J, Arruda P. 2018. A community-based culture collection for targeting novel plant growth-promoting bacteria from the sugarcane microbiome. Frontiers in Plant Science 8, 2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AE, Mejía LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA. 2003. Fungal endophytes limit pathogen damage in a tropical tree. Proceedings of the National Academy of Sciences, USA 100, 15649–15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin JM, Kuzina V, Andersen SB, Bak S. 2011. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72, 435–457. [DOI] [PubMed] [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE. 2002. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Aznar A, Chen NW, Thomine S, Dellagi A. 2015. Immunity to plant pathogens and iron homeostasis. Plant Science 240, 90–97. [DOI] [PubMed] [Google Scholar]
- Bach EM, Baer SG, Meyer CK, Six J. 2010. Soil texture affects soil microbial and structural recovery during grassland restoration. Soil Biology and Biochemistry 42, 2182–2191. [Google Scholar]
- Bachar A, Al-Ashhab A, Soares MI, Sklarz MY, Angel R, Ungar ED, Gillor O. 2010. Soil microbial abundance and diversity along a low precipitation gradient. Microbial Ecology 60, 453–461. [DOI] [PubMed] [Google Scholar]
- Baetz U, Martinoia E. 2014. Root exudates: the hidden part of plant defense. Trends in Plant Science 19, 90–98. [DOI] [PubMed] [Google Scholar]
- Bahulikar RA, Torres-Jerez I, Worley E, Craven K, Udvardi MK. 2014. Diversity of nitrogen-fixing bacteria associated with switchgrass in the native tallgrass prairie of northern Oklahoma. Applied and Environmental Microbiology 80, 5636–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Müller DB, Srinivas G, et al.. 2015. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annual Review of Plant Biology 57, 233–266. [DOI] [PubMed] [Google Scholar]
- Baker K, Cook RJ. 1974. Biological control of plant pathogens. San Francisco: W.H. Freeman and Company. [Google Scholar]
- Bálint M, Tiffin P, Hallström B, O’Hara RB, Olson MS, Fankhauser JD, Piepenbring M, Schmitt I. 2013. Host genotype shapes the foliar fungal microbiome of balsam poplar (Populus balsamifera). PLoS One 8, e53987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint-Kurti P, Simmons SJ, Blum JE, Ballaré CL, Stapleton AE. 2010. Maize leaf epiphytic bacteria diversity patterns are genetically correlated with resistance to fungal pathogen infection. Molecular Plant-Microbe Interactions 23, 473–484. [DOI] [PubMed] [Google Scholar]
- Bang C, Dagan T, Deines P, et al.. 2018. Metaorganisms in extreme environments: do microbes play a role in organismal adaptation? Zoology 127, 1–19. [DOI] [PubMed] [Google Scholar]
- Bárcenas-Moreno G, Gómez-Brandón M, Rousk J, Bååth E. 2009. Adaptation of soil microbial communities to temperature: comparison of fungi and bacteria in a laboratory experiment. Global Change Biology 15, 2950–2957. [Google Scholar]
- Bari R, Jones JD. 2009. Role of plant hormones in plant defence responses. Plant Molecular Biology 69, 473–488. [DOI] [PubMed] [Google Scholar]
- Barnawal D, Bharti N, Pandey SS, Pandey A, Chanotiya CS, Kalra A. 2017. Plant growth-promoting rhizobacteria enhance wheat salt and drought stress tolerance by altering endogenous phytohormone levels and TaCTR1/TaDREB2 expression. Physiologia Plantarum 161, 502–514. [DOI] [PubMed] [Google Scholar]
- Barret M, Briand M, Bonneau S, Préveaux A, Valière S, Bouchez O, Hunault G, Simoneau P, Jacquesa MA. 2015. Emergence shapes the structure of the seed microbiota. Applied and Environmental Microbiology 81, 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsev AV, Deakin WJ, Boukli NM, McAlvin CB, Stacey G, Malnoë P, Broughton WJ, Staehelin C. 2004. NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiology 134, 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers B, Op De Beeck M, Thijs S, Truyens S, Weyens N, Boerjan W, Vangronsveld J. 2016. Performance of 16s rDNA primer pairs in the study of rhizosphere and endosphere bacterial microbiomes in metabarcoding studies. Frontiers in Microbiology 7, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CW, Acosta-Martinez V, McIntyre NE, Cox S, Tissue DT, Zak JC. 2009. Linking microbial community structure and function to seasonal differences in soil moisture and temperature in a Chihuahuan desert grassland. Microbial Ecology 58, 827–842. [DOI] [PubMed] [Google Scholar]
- Beltrán JM, Koo-Oshima S. 2006. Water desalination for agricultural applications. Rome: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Berendsen RL, Pieterse CM, Bakker PA. 2012. The rhizosphere microbiome and plant health. Trends in Plant Science 17, 478–486. [DOI] [PubMed] [Google Scholar]
- Berens ML, Wolinska KW, Spaepen S, et al.. 2019. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proceedings of the National Academy of Sciences, USA 116, 2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Smalla K. 2009. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiology Ecology 68, 1–13. [DOI] [PubMed] [Google Scholar]
- Binenbaum J, Weinstain R, Shani E. 2018. Gibberellin localization and transport in plants. Trends in Plant Science 23, 410–421. [DOI] [PubMed] [Google Scholar]
- Boatwright JL, Pajerowska-Mukhtar K. 2013. Salicylic acid: an old hormone up to new tricks. Molecular Plant Pathology 14, 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA. 2014. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genetics 10, e1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen N, Horton MW, Bergelson J. 2013. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8, e56329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogino CP, Oliva DM, Sorroche GF, Giordano W. 2013. The role of bacterial biofilms and surface components in plant–bacterial associations. International Journal of Molecular Sciences 14, 15838–15859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhari A, Essack M, Lafi FF, et al.. 2019. Bioprospecting desert plant Bacillus endophytic strains for their potential to enhance plant stress tolerance. Scientific Reports 9, 18154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annual Review of Plant Biology 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bomar L, Maltz M, Colston S, Graf J. 2011. Directed culturing of microorganisms using metatranscriptomics. MBio 2, e00012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Theis KR. 2015. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biology 13, e1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borjigidai A, Hikosaka K, Hirose T, Hasegawa T, Okada M, Kobayashi K. 2006. Seasonal changes in temperature dependence of photosynthetic rate in rice under a free-air CO2 enrichment. Annals of Botany 97, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TC, McFall-Ngai MJ. 2011. Metaorganisms as the new frontier. Zoology 114, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TC, Miller DJ. 2016. The holobiont imperative. Vienna: Springer-Verlag. [Google Scholar]
- Boyer JS. 1982. Plant productivity and environment. Science 218, 443–448. [DOI] [PubMed] [Google Scholar]
- Brandl MT, Lindow SE. 1998. Contribution of indole-3-acetic acid production to the epiphytic fitness of Erwinia herbicola. Applied and Environmental Microbiology 64, 3256–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA, Bailey-Serres J, Weretilnyk E. 2000. Responses to abiotic stresses. In: Buchanan BB, Gruissem W, Jones RL, eds. Biochemistry and molecular biology of plants. Rockville, MD: American Society of Plant Physiologists, 1158–1203. [Google Scholar]
- Bressan M, Roncato MA, Bellvert F, Comte G, Haichar FZ, Achouak W, Berge O. 2009. Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. The ISME Journal 3, 1243–1257. [DOI] [PubMed] [Google Scholar]
- Brown JMM, Rimington C, Sawyer BC. 1963. Studies on biliary excretion in the rabbit II. The relationship between the chemical structure of certain natural or synthetic pentacyclic triterpenes and their icterogenic activity. Part 1: the substituents on carbon atoms 3, 17, 22 and 24. Proceedings of the Royal Society B: Biological Sciences 157, 473–491. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P. 2015. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host & Microbe 17, 392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, et al.. 2012. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annual Review of Plant Biology 64, 807–838. [DOI] [PubMed] [Google Scholar]
- Canarini A, Kaiser C, Merchant A, Richter A, Wanek W. 2019. Root exudation of primary metabolites: mechanisms and their roles in plant responses to environmental stimuli. Frontiers in Plant Science 10, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Halane MK, Gassmann W, Stacey G. 2017. The role of plant innate immunity in the legume–rhizobium symbiosis. Annual Review of Plant Biology 68, 535–561. [DOI] [PubMed] [Google Scholar]
- Carbonetto B, Rascovan N, Álvarez R, Mentaberry A, Vázquez MP. 2014. Structure, composition and metagenomic profile of soil microbiomes associated to agricultural land use and tillage systems in Argentine Pampas. PLoS One 9, e99949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlström CI, Field CM, Bortfeld-Miller M, Müller B, Sunagawa S, Vorholt JA. 2019. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nature Ecology & Evolution 3, 1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carper DL, Lawrence TJ, Carrell AA, Pelletier DA, Weston DJ. 2019. DISCo-microbe: design of an identifiable synthetic community of microbes. PeerJ Preprints 7, e27898–27891. [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhais LC, Dennis PG, Badri DV, Kidd BN, Vivanco JM, Schenk PM. 2015. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Molecular Plant-Microbe Interactions 28, 1049–1058. [DOI] [PubMed] [Google Scholar]
- Carvalhais LC, Dennis PG, Badri DV, Tyson GW, Vivanco JM, Schenk PM. 2013. Activation of the jasmonic acid plant defence pathway alters the composition of rhizosphere bacterial communities. PLoS One 8, e56457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhais LC, Schenk PM, Dennis PG. 2017. Jasmonic acid signalling and the plant holobiont. Current Opinion in Microbiology 37, 42–47. [DOI] [PubMed] [Google Scholar]
- Castrillo G, Teixeira PJ, Paredes SH, et al.. 2017. Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW. 2010. Soil microbial community responses to multiple experimental climate change drivers. Applied and Environmental Microbiology 76, 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerozi BdS, Fitzsimmons K. 2016. The effect of pH on phosphorus availability and speciation in an aquaponics nutrient solution. Bioresource Technology 219, 778–781. [DOI] [PubMed] [Google Scholar]
- Cesco S, Neumann G, Tomasi N, Pinton R, Weisskopf L. 2010. Release of plant-borne flavonoids into the rhizosphere and their role in plant nutrition. Plant and Soil 329, 1–25. [Google Scholar]
- Chanratana M, Han GH, Roy Choudhury A, Sundaram S, Halim MA, Krishnamoorthy R, Kang Y, Sa T. 2017. Assessment of Methylobacterium oryzae CBMB20 aggregates for salt tolerance and plant growth promoting characteristics for bio-inoculant development. AMB Express 7, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau JF, Bagtzoglou AC, Willig MR. 2011. The effect of soil texture on richness and diversity of bacterial communities. Environmental Forensics 12, 333–341. [Google Scholar]
- Cohen AC, Bottini R, Piccoli PN. 2008. Azospirillum brasilense Sp 245 produces ABA in chemically-defined culture medium and increases ABA content in arabidopsis plants. Plant Growth Regulation 54, 97–103. [Google Scholar]
- Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, North G, Visel A, Partida-Martinez LP, Tringe SG. 2016. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytologist 209, 798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Cornejo HA, Macías-Rodríguez L, Cortés-Penagos C, López-Bucio J. 2009. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiology 149, 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JE. 2007. Early interactions between legumes and rhizobia: disclosing complexity in a molecular dialogue. Journal of Applied Microbiology 103, 1355–1365. [DOI] [PubMed] [Google Scholar]
- Cotton TEA, Pétriacq P, Cameron DD, Meselmani MA, Schwarzenbacher R, Rolfe SA, Ton J. 2019. Metabolic regulation of the maize rhizobiome by benzoxazinoids. The ISME Journal 13, 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross KL, Campbell JH, Balachandran M, et al.. 2019. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nature Biotechnology 37, 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT, Scannell JW. 2002. Estimating prokaryotic diversity and its limits. Proceedings of the National Academy of Sciences, USA 99, 10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Wei X, Alfonso AA, Pei L, Duque UG, Zhang Z, Babb GM, Beachy RN. 2008. Transgenic rice plants that overexpress transcription factors RF2a and RF2b are tolerant to rice tungro virus replication and disease. Proceedings of the National Academy of Sciences, USA 105, 21012–21016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daur I, Saad MM, Eida AA, Ahmad S, Shah ZH, Ihsan MZ, Muhammad Y, Sohrab SS, Hirt H. 2018. Boosting Alfalfa (Medicago sativa L.) production with rhizobacteria from various plants in Saudi Arabia. Frontiers in Microbiology 9, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson CJ, Surette MG. 2008. Individuality in bacteria. Annual Review of Genetics 42, 253–268. [DOI] [PubMed] [Google Scholar]
- Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA. 2009. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proceedings of the National Academy of Sciences, USA 106, 16428–16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza RSC, Armanhi JSL, Damasceno NB, Imperial J, Arruda P. 2019. Genome sequences of a plant beneficial synthetic bacterial community reveal genetic features for successful plant colonization. Frontiers in Microbiology 10, 1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Djavaheri M, Bakker PA, Höfte M. 2008. Pseudomonas fluorescens WCS374r-induced systemic resistance in rice against Magnaporthe oryzae is based on pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiology 148, 1996–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries FT, Manning P, Tallowin JRB, et al.. 2012. Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecology Letters 15, 1230–1239. [DOI] [PubMed] [Google Scholar]
- de Zélicourt A, Synek L, Saad MM, et al.. 2018. Ethylene induced plant stress tolerance by Enterobacter sp. SA187 is mediated by 2-keto-4-methylthiobutyric acid production. PLoS Genetics 14, e1007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djonović S, Pozo MJ, Dangott LJ, Howell CR, Kenerley CM. 2006. Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Molecular Plant-Microbe Interactions 19, 838–853. [DOI] [PubMed] [Google Scholar]
- Dor E, Joel DM, Kapulnik Y, Koltai H, Hershenhorn J. 2011. The synthetic strigolactone GR24 influences the growth pattern of phytopathogenic fungi. Planta 234, 419–427. [DOI] [PubMed] [Google Scholar]
- Dugardeyn J, Van Der Straeten D. 2008. Ethylene: fine-tuning plant growth and development by stimulation and inhibition of elongation. Plant Science 175, 59–70. [Google Scholar]
- Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, Hacquard S. 2018. Microbial interkingdom interactions in roots promote arabidopsis survival. Cell 175, 973–983.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JA, Santos-Medellín CM, Liechty ZS, Nguyen B, Lurie E, Eason S, Phillips G, Sundaresan V. 2018. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biology 16, e2003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egamberdieva D. 2009. Alleviation of salt stress by plant growth regulators and IAA producing bacteria in wheat. Acta Physiologiae Plantarum 31, 861–864. [Google Scholar]
- Egamberdieva D, Wirth S, Alqarawi AA, Abd Allah EF. 2015. Salt tolerant Methylobacterium mesophilicum showed viable colonization abilities in the plant rhizosphere. Saudi Journal of Biological Sciences 22, 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eida AA, Bougouffa S, Alam I, Hirt H, Saad MM. 2020. Complete genome sequence of Paenibacillus sp. JZ16, a plant growth promoting root endophytic bacterium of the desert halophyte Zygophyllum simplex. Current Microbiology (in press). [DOI] [PubMed] [Google Scholar]
- Eida AA, Hirt H, Saad MM. 2017. Challenges faced in field application of phosphate-solubilizing bacteria. In: Mehnaz S, ed. Rhizotrophs: plant growth promotion to bioremediation, Vol. 2 Singapore: Springer Singapore, 125–143. [Google Scholar]
- Eida AA, Ziegler M, Lafi FF, Michell CT, Voolstra CR, Hirt H, Saad MM. 2018. Desert plant bacteria reveal host influence and beneficial plant growth properties. PLoS One 13, e0208223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. 1996. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences, USA 93, 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers KG, Lauber CL, Knight R, Fierer N. 2010. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biology and Biochemistry 42, 896–903. [Google Scholar]
- Erkel C, Kube M, Reinhardt R, Liesack W. 2006. Genome of rice cluster I archaea—the key methane producers in the rice rhizosphere. Science 313, 370–372. [DOI] [PubMed] [Google Scholar]
- Feng H, Guo J, Wang W, Song X, Yu S. 2019. Soil depth determines the composition and diversity of bacterial and archaeal communities in a Poplar plantation. Forests 10, 550. [Google Scholar]
- Fenner N, Freeman C, Reynolds B. 2005. Observations of a seasonally shifting thermal optimum in peatland carbon-cycling processes; implications for the global carbon cycle and soil enzyme methodologies. Soil Biology and Biochemistry 37, 1814–1821. [Google Scholar]
- Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proceedings of the National Academy of Sciences, USA 103, 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proceedings of the National Academy of Sciences, USA 109, 21390–21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel OM, Salas-González I, Castrillo G, Spaepen S, Law TF, Teixeira PJPL, Jones CD, Dangl JL. 2019. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biology 17, e3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. 2013. Abscisic acid synthesis and response. The Arabidopsis Book 11, e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E. 2013. Auxin influences strigolactones in pea mycorrhizal symbiosis. Journal of Plant Physiology 170, 523–528. [DOI] [PubMed] [Google Scholar]
- Forchetti G, Masciarelli O, Alemano S, Alvarez D, Abdala G. 2007. Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Applied Microbiology and Biotechnology 76, 1145–1152. [DOI] [PubMed] [Google Scholar]
- Francioli D, Schulz E, Lentendu G, Wubet T, Buscot F, Reitz T. 2016. Mineral vs. organic amendments: microbial community structure, activity and abundance of agriculturally relevant microbes are driven by long-term fertilization strategies. Frontiers in Microbiology 7, 1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frossard A, Ramond JB, Seely M, Cowan DA. 2015. Water regime history drives responses of soil Namib Desert microbial communities to wetting events. Scientific Reports 5, 12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frostegård A, Courtois S, Ramisse V, Clerc S, Bernillon D, Le Gall F, Jeannin P, Nesme X, Simonet P. 1999. Quantification of bias related to the extraction of DNA directly from soils. Applied and Environmental Microbiology 65, 5409–5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaby JC, Buckley DH. 2015. Assessment of nitrogenase diversity in the environment. In: Bruijn FJd, ed. Biological nitrogen fixation. Chichester: John Wiley & Sons, 209–216. [Google Scholar]
- Gamalero E, Berta G, Massa N, Glick BR, Lingua G. 2008. Synergistic interactions between the ACC deaminase-producing bacterium Pseudomonas putida UW4 and the AM fungus Gigaspora rosea positively affect cucumber plant growth. FEMS Microbiology Ecology 64, 459–467. [DOI] [PubMed] [Google Scholar]
- Garbeva P, Postma J, van Veen JA, van Elsas JD. 2006. Effect of above-ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3. Environmental Microbiology 8, 233–246. [DOI] [PubMed] [Google Scholar]
- Gelsomino A, Keijzer-Wolters AC, Cacco G, van Elsas JD. 1999. Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. Journal of Microbiological Methods 38, 1–15. [DOI] [PubMed] [Google Scholar]
- Gilbert SF, Sapp J, Tauber AI. 2012. A symbiotic view of life: we have never been individuals. Quarterly Review of Biology 87, 325–341. [DOI] [PubMed] [Google Scholar]
- Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS. 2003. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Applied and Environmental Microbiology 69, 1800–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Glick BR. 2014. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiological Research 169, 30–39. [DOI] [PubMed] [Google Scholar]
- Goldfarb KC, Karaoz U, Hanson CA, Santee CA, Bradford MA, Treseder KK, Wallenstein MD, Brodie EL. 2011. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Frontiers in Microbiology 2, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes EA, Lana UGP, Quensen JF, de Sousa SM, Oliveira CA, Guo J, Guimarães LJM, Tiedje JM. 2018. Root-associated microbiome of maize genotypes with contrasting phosphorus use efficiency. Phytobiomes Journal 2, 129–137. [Google Scholar]
- Gopal M, Gupta A. 2016. Microbiome selection could spur next-generation plant breeding strategies. Frontiers in Microbiology 7, 1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH, Vance CP. 2003. Legumes: importance and constraints to greater use. Plant Physiology 131, 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer AH, Mills G, Yin H. 2016. Amplicon-based NGS detects targeted variants in paired tissue and ctDNA samples. Cancer Research 76. [Google Scholar]
- Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS. 2011. The bacterial biogeography of British soils. Environmental Microbiology 13, 1642–1654. [DOI] [PubMed] [Google Scholar]
- Grigulis K, Lavorel S, Krainer U, et al.. 2013. Relative contributions of plant traits and soil microbial properties to mountain grassland ecosystem services. Journal of Ecology 101, 47–57. [Google Scholar]
- Grosskopf T, Soyer OS. 2014. Synthetic microbial communities. Current Opinion in Microbiology 18, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Wang Y, Lu S, Xiang Q, Yu X, Zhao K, Zou L, Chen Q, Tu S, Zhang X. 2017. Long-term fertilization structures bacterial and archaeal communities along soil depth gradient in a paddy soil. Frontiers in Microbiology 8, 1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y. 1994. Iron: nutritious, noxious, and not readily available. Plant Physiology 104, 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyonnet JP, Guillemet M, Dubost A, Simon L, Ortet P, Barakat M, Heulin T, Achouak W, Haichar FEZ. 2018. Plant nutrient resource use strategies shape active rhizosphere microbiota through root exudation. Frontiers in Plant Science 9, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habekost M, Eisenhauer N, Scheu S, Steinbeiss S, Weigelt A, Gleixner G. 2008. Seasonal changes in the soil microbial community in a grassland plant diversity gradient four years after establishment. Soil Biology and Biochemistry 40, 2588–2595. [Google Scholar]
- Hacquard S, Garrido-Oter R, González A, et al.. 2015. Microbiota and host nutrition across plant and animal kingdoms. Cell Host & Microbe 17, 603–616. [DOI] [PubMed] [Google Scholar]
- Haichar FEZ, Heulin T, Guyonnet JP, Achouak W. 2016. Stable isotope probing of carbon flow in the plant holobiont. Current Opinion in Biotechnology 41, 9–13. [DOI] [PubMed] [Google Scholar]
- Haney CH, Samuel BS, Bush J, Ausubel FM. 2015. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nature Plants 1, 15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon MT, Coenen C. 2011. Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytologist 189, 701–709. [DOI] [PubMed] [Google Scholar]
- Hardoim PR, Hardoim CC, van Overbeek LS, van Elsas JD. 2012. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One 7, e30438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman WH, Richardson CJ, Vilgalys R, Bruland GL. 2008. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proceedings of the National Academy of Sciences, USA 105, 17842–17847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S, Mathesius U. 2012. The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant–microbe interactions. Journal of Experimental Botany 63, 3429–3444. [DOI] [PubMed] [Google Scholar]
- Helfrich EJN, Vogel CM, Ueoka R, Schäfer M, Ryffel F, Müller DB, Probst S, Kreuzer M, Piel J, Vorholt JA. 2018. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nature Microbiology 3, 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera Paredes S, Gao T, Law TF, et al.. 2018. Design of synthetic bacterial communities for predictable plant phenotypes. PLoS Biology 16, e2003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiergeist A, Gläsner J, Reischl U, Gessner A. 2015. Analyses of intestinal microbiota: culture versus sequencing. ILAR Journal 56, 228–240. [DOI] [PubMed] [Google Scholar]
- Hossain MM, Sultana F, Kubota M, Koyama H, Hyakumachi M. 2007. The plant growth-promoting fungus Penicillium simplicissimum GP17-2 induces resistance in Arabidopsis thaliana by activation of multiple defense signals. Plant & Cell Physiology 48, 1724–1736. [DOI] [PubMed] [Google Scholar]
- Hu L, Robert CAM, Cadot S, et al.. 2018. Root exudate metabolites drive plant–soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nature Communications 9, 2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ACC, Jiang T, Liu YX, Bai YC, Reed J, Qu BY, Goossens A, Nutzmann HW, Bai Y, Osbourn A. 2019. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364, 546. [DOI] [PubMed] [Google Scholar]
- Hueso S, García C, Hernández T. 2012. Severe drought conditions modify the microbial community structure, size and activity in amended and unamended soils. Soil Biology and Biochemistry 50, 167–173. [Google Scholar]
- Hyung D, Lee C, Kim JH, et al.. 2014. Cross-family translational genomics of abiotic stress-responsive genes between Arabidopsis and Medicago truncatula. PLoS One 9, e91721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderjit, Weston LA. 2003. Root exudates: an overview. In: de Kroon H, Visser EJW, eds. Root ecology, Vol. 168 Berlin, Heidelberg: Springer Berlin Heidelberg, 235–255. [Google Scholar]
- Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. 2010. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329, 212–215. [DOI] [PubMed] [Google Scholar]
- Jiang CJ, Shimono M, Sugano S, Kojima M, Liu X, Inoue H, Sakakibara H, Takatsuji H. 2013. Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Molecular Plant-Microbe Interactions 26, 287–296. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Kuske CR, Carney TD, Housman DC, Gallegos-Graves LV, Belnap J. 2012. Increased temperature and altered summer precipitation have differential effects on biological soil crusts in a dryland ecosystem. Global Change Biology 18, 2583–2593. [Google Scholar]
- Johnston-Monje D, Lundberg DS, Lazarovits G, Reis VM, Raizada MN. 2016. Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant and Soil 405, 337–355. [Google Scholar]
- Johnston-Monje D, Raizada MN. 2011. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS One 6, e20396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC. 2008. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proceedings of the National Academy of Sciences, USA 105, 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Garcia BJ, Furches A, Tuskan GA, Jacobson D. 2019. Plant host-associated mechanisms for microbial selection. Frontiers in Plant Science 10, 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge GL, Kisiala A, Morrison E, Aoki M, Nogueira APO, Emery RJN. 2019. Endosymbiotic Methylobacterium oryzae mitigates the impact of limited water availability in lentil (Lens culinaris Medik.) by increasing plant cytokinin levels. Environmental and Experimental Botany 162, 525–540. [Google Scholar]
- Ju CL, Van de Poel B, Cooper ED, Thierer JH, Gibbons TR, Delwiche CF, Chang CR. 2015. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nature Plants 1, 14004. [DOI] [PubMed] [Google Scholar]
- Jung MY, Kim JG, Sinninghe Damsté JS, et al.. 2016. A hydrophobic ammonia-oxidizing archaeon of the Nitrosocosmicus clade isolated from coal tar-contaminated sediment. Environmental Microbiology Reports 8, 983–992. [DOI] [PubMed] [Google Scholar]
- Kaldorf M, Ludwig-Müller J. 2000. AM fungi might affect the root morphology of maize by increasing indole-3-butyric acid biosynthesis. Physiologia Plantarum 109, 58–67. [Google Scholar]
- Kaplan D, Maymon M, Agapakis CM, Lee A, Wang A, Prigge BA, Volkogon M, Hirsch AM. 2013. A survey of the microbial community in the rhizosphere of two dominant shrubs of the Negev Desert highlands, Zygophyllum dumosum (Zygophyllaceae) and Atriplex halimus (Amaranthaceae), using cultivation-dependent and cultivation-independent methods. American Journal of Botany 100, 1713–1725. [DOI] [PubMed] [Google Scholar]
- Karadeniz A, Topcuoğlu Ş, İnan S. 2006. Auxin, gibberellin, cytokinin and abscisic acid production in some bacteria. World Journal of Microbiology & Biotechnology 22, 1061–1064. [Google Scholar]
- Kemen E. 2014. Microbe–microbe interactions determine oomycete and fungal host colonization. Current Opinion in Plant Biology 20, 75–81. [DOI] [PubMed] [Google Scholar]
- Ketchum RN, Smith EG, Vaughan GO, Phippen BL, McParland D, Al-Mansoori N, Carrier TJ, Burt JA, Reitzel AM. 2018. DNA extraction method plays a significant role when defining bacterial community composition in the marine invertebrate Echinometra mathaei. Frontiers in Marine Science 5, 255. doi: 10.3389/fmars.2018.00255 [Google Scholar]
- Kim YC, Leveau J, McSpadden Gardener BB, Pierson EA, Pierson LS 3rd, Ryu CM. 2011. The multifactorial basis for plant health promotion by plant-associated bacteria. Applied and Environmental Microbiology 77, 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Bäumler AJ. 2000. Host adaptation and the emergence of infectious disease: the Salmonella paradigm. Molecular Microbiology 36, 1006–1014. [DOI] [PubMed] [Google Scholar]
- Klappenbach JA, Dunbar JM, Schmidt TM. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Applied and Environmental Microbiology 66, 1328–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, von Mering C, Vorholt JA. 2012. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. The ISME Journal 6, 1378–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Naciri-Graven Y, Broughton WJ, Perret X. 2004. Flavonoids induce temporal shifts in gene-expression of nod-box controlled loci in Rhizobium sp. NGR234. Molecular Microbiology 51, 335–347. [DOI] [PubMed] [Google Scholar]
- Kong Z, Hart M, Liu H. 2018. Paving the way from the lab to the field: using synthetic microbial consortia to produce high-quality crops. Frontiers in Plant Science 9, 1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koranda M, Kaiser C, Fuchslueger L, Kitzler B, Sessitsch A, Zechmeister-Boltenstern S, Richter A. 2013. Seasonal variation in functional properties of microbial communities in beech forest soil. Soil Biology & Biochemistry 60, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramae EE, Yergeau E, Wong LC, Pijl AS, van Veen JA, Kowalchuk GA. 2012. Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiology Ecology 79, 12–24. [DOI] [PubMed] [Google Scholar]
- Kwak MJ, Kong HG, Choi K, et al.. 2018. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nature Biotechnology 36, 1100–1109. [DOI] [PubMed] [Google Scholar]
- Lafi FF, Alam I, Bisseling T, Geurts R, Bajic VB, Hirt H, Saad MM. 2017a Draft genome sequence of the plant growth-promoting rhizobacterium Acinetobacter radioresistens strain SA188 isolated from the desert plant Indigofera argentea. Genome Announcements 5, e01708-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafi FF, Alam I, Geurts R, Bisseling T, Bajic VB, Hirt H, Saad MM. 2016a Draft genome sequence of the phosphate-solubilizing bacterium Pseudomonas argentinensis strain SA190 isolated from the desert plant Indigofera argentea. Genome Announcements 4, e01431-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafi FF, Alam I, Geurts R, Bisseling T, Bajic VB, Hirt H, Saad MM. 2017b Draft genome sequence of Ochrobactrum intermedium strain SA148, a plant growth-promoting desert rhizobacterium. Genome Announcements 5, e01707-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafi FF, AlBladi ML, Salem NM, Al-Banna L, Alam I, Bajic VB, Hirt H, Saad MM. 2017c Draft genome sequence of the plant growth-promoting Pseudomonas punonensis strain D1-6 isolated from the desert plant Erodium hirtum in Jordan. Genome Announcements 5, e01437-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafi FF, Bokhari A, Alam I, Bajic VB, Hirt H, Saad MM. 2016b Draft genome sequence of the plant growth-promoting Cupriavidus gilardii strain JZ4 isolated from the desert plant Tribulus terrestris. Genome Announcements 4, e00678-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafi FF, Ramirez-Prado JS, Alam I, Bajic VB, Hirt H, Saad MM. 2016c Draft genome sequence of Halomonas elongata strain K4, an endophytic growth-promoting bacterium enhancing salinity tolerance in planta. Genome Announcements 4, e01214-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafi FF, Ramirez-Prado JS, Alam I, Bajic VB, Hirt H, Saad MM. 2017d Draft genome sequence of plant growth–promoting Micrococcus luteus strain K39 isolated from Cyperus conglomeratus in Saudi Arabia. Genome Announcements 5, e01520-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest-Lapointe I, Messier C, Kembel SW. 2016. Tree phyllosphere bacterial communities: exploring the magnitude of intra- and inter-individual variation among host species. PeerJ 4, e2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Raven JA, Shaver GR, Smith SE. 2008. Plant nutrient-acquisition strategies change with soil age. Trends in Ecology & Evolution 23, 95–103. [DOI] [PubMed] [Google Scholar]
- Lanoue A, Burlat V, Henkes GJ, Koch I, Schurr U, Röse US. 2010. De novo biosynthesis of defense root exudates in response to Fusarium attack in barley. New Phytologist 185, 577–588. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology 75, 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Strickland MS, Bradford MA, Fierer N. 2008. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biology and Biochemistry 40, 2407–2415. [Google Scholar]
- Le Guillou C, Chemidlin Prévost-Bouré N, Karimi B, et al.. 2019. Tillage intensity and pasture in rotation effectively shape soil microbial communities at a landscape scale. MicrobiologyOpen 8, e00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeis SL, Paredes SH, Lundberg DS, Glavina T, Jones CD. 2015. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. [DOI] [PubMed] [Google Scholar]
- Lee SA, Park J, Chu B, Kim JM, Joa JH, Sang MK, Song J, Weon HY. 2016. Comparative analysis of bacterial diversity in the rhizosphere of tomato by culture-dependent and -independent approaches. Journal of Microbiology 54, 823–831. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Morse D, Hijri M. 2019. Holobiont chronobiology: mycorrhiza may be a key to linking aboveground and underground rhythms. Mycorrhiza 29, 403–412. [DOI] [PubMed] [Google Scholar]
- Lee Y, Krishnamoorthy R, Selvakumar G, Kim K, Sa T. 2015. Alleviation of salt stress in maize plant by co-inoculation of arbuscular mycorrhizal fungi and Methylobacterium oryzae CBMB20. Journal of the Korean Society for Applied Biological Chemistry 58, 533–540. [Google Scholar]
- Leeman M, denOuden EM, vanPelt JA, Dirkx FPM, Steijl H, Bakker P, Schippers B. 1996. Iron availability affects induction of systemic resistance to Fusarium wilt of radish by Pseudomonas fluorescens. Phytopathology 86, 149–155. [Google Scholar]
- Leone V, Gibbons SM, Martinez K, et al.. 2015. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host & Microbe 17, 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Conway JM, Dangl JL, Woyke T. 2018. Elucidating bacterial gene functions in the plant microbiome. Cell Host & Microbe 24, 475–485. [DOI] [PubMed] [Google Scholar]
- Li C, Yan K, Tang L, Jia Z, Li Y. 2014. Change in deep soil microbial communities due to long-term fertilization. Soil Biology and Biochemistry 75, 264–272. [Google Scholar]
- Li F, Chen L, Zhang J, Yin J, Huang S. 2017. Bacterial community structure after long-term organic and inorganic fertilization reveals important associations between soil nutrients and specific taxa involved in nutrient transformations. Frontiers in Microbiology 8, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wu X, Chen T, et al.. 2018. Plant phenotypic traits eventually shape its microbiota: a common garden test. Frontiers in Microbiology 9, 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Cao Y, Tanaka K, Thibivilliers S, Wan J, Choi J, Kang Ch, Qiu J, Stacey G. 2013. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341, 1384–1387. [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Brechenmacher L, et al.. 2010. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiology 152, 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Applied and Environmental Microbiology 69, 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay WL. 1995. Chemical reactions in soils that affect iron availability to plants. A quantative approach. In: Abadía J, ed. Iron nutrition in soils and plants. Dordrecht: Springer Netherlands, 7–14. [Google Scholar]
- Liptzin D, Silver WL, Detto M. 2011. Temporal dynamics in soil oxygen and greenhouse gases in two humid tropical forests. Ecosystems 14, 171–182. [Google Scholar]
- Liu F, Xing S, Ma H, Du Z, Ma B. 2013. Cytokinin-producing, plant growth-promoting rhizobacteria that confer resistance to drought stress in Platycladus orientalis container seedlings. Applied Microbiology and Biotechnology 97, 9155–9164. [DOI] [PubMed] [Google Scholar]
- Liu H, Carvalhais LC, Schenk PM, Dennis PG. 2017. Effects of jasmonic acid signalling on the wheat microbiome differ between body sites. Scientific Reports 7, 41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Carvalhais LC, Schenk PM, Dennis PG. 2018. Activation of the salicylic acid signalling pathway in wheat had no significant short-term impact on the diversity of root-associated microbiomes. Pedobiologia 70, 6–11. [Google Scholar]
- Liu J, Guo C, Chen ZL, He JD, Zou YN. 2016. Mycorrhizal inoculation modulates root morphology and root phytohormone responses in trifoliate orange under drought stress. Emirates Journal of Food and Agriculture 28, 251–256. [Google Scholar]
- Liu P, Zhang W, Zhang LQ, Liu X, Wei HL. 2016. Supramolecular structure and functional analysis of the type III secretion system in Pseudomonas fluorescens 2P24. Frontiers in Plant Science 6, 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhu A, Tan H, Cao L, Zhang R. 2019. Engineering banana endosphere microbiome to improve Fusarium wilt resistance in banana. Microbiome 7, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loake G, Grant M. 2007. Salicylic acid in plant defence—the players and protagonists. Current Opinion in Plant Biology 10, 466–472. [DOI] [PubMed] [Google Scholar]
- Loper JE, Buyer JS. 1991. Siderophores in microbial interactions on plant surfaces. Molecular Plant-Microbe Interactions 180, 108–117. [Google Scholar]
- Loper JE, Hassan KA, Mavrodi DV, et al.. 2012. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genetics 8, e1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Baena FJ, Monreal JA, Pérez-Montaño F, Guasch-Vidal B, Bellogín RA, Vinardell JM, Ollero FJ. 2009. The absence of Nops secretion in Sinorhizobium fredii HH103 increases GmPR1 expression in Williams soybean. Molecular Plant-Microbe Interactions 22, 1445–1454. [DOI] [PubMed] [Google Scholar]
- Lopez-Gomez M, Sandal N, Stougaard J, Boller T. 2012. Interplay of flg22-induced defence responses and nodulation in Lotus japonicus. Journal of Experimental Botany 63, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. 2007. Global patterns in bacterial diversity. Proceedings of the National Academy of Sciences, USA 104, 11436–11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F. 2009. Plant-growth-promoting rhizobacteria. Annual Review of Microbiology 63, 541–556. [DOI] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, et al.. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestre FT, Delgado-Baquerizo M, Jeffries TC, et al.. 2015. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proceedings of the National Academy of Sciences, USA 112, 15684–15689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manulis S, Haviv-Chesner A, Brandl MT, Lindow SE, Barash I. 1998. Differential involvement of indole-3-acetic acid biosynthetic pathways in pathogenicity and epiphytic fitness of Erwinia herbicola pv. gypsophilae. Molecular Plant-Microbe Interactions 11, 634–642. [DOI] [PubMed] [Google Scholar]
- Marasco R, Rolli E, Ettoumi B, et al.. 2012. A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS One 7, e48479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco DE, Abram F. 2019. Editorial: using genomics, metagenomics and other ‘Omics’ to assess valuable microbial ecosystem services and novel biotechnological applications. Frontiers in Microbiology 10, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L, Fester R. 1991. Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. Cambridge, MA: MIT Press. [PubMed] [Google Scholar]
- Marie C, Deakin WJ, Viprey V, Kopciñska J, Golinowski W, Krishnan HB, Perret X, Broughton WJ. 2003. Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Molecular Plant-Microbe Interactions 16, 743–751. [DOI] [PubMed] [Google Scholar]
- Marschner P, Crowley D, Yang CH. 2004. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant and Soil 261, 199–208. [Google Scholar]
- Martinez-Abarca F, Herrera-Cervara JA, Bueno P, Sanjuan J, Bisseling T, Olivares J. 1998. Involvement of salicylic acid in the establishment of the Rhizobium meliloti–alfalfa symbiosis. Molecular Plant-Microbe Interactions 11, 153–155. [Google Scholar]
- Martiny JB, Martiny AC, Weihe C, Lu Y, Berlemont R, Brodie EL, Goulden ML, Treseder KK, Allison SD. 2017. Microbial legacies alter decomposition in response to simulated global change. The ISME Journal 11, 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massalha H, Korenblum E, Tholl D, Aharoni A. 2017. Small molecules below-ground: the role of specialized metabolites in the rhizosphere. The Plant Journal 90, 788–807. [DOI] [PubMed] [Google Scholar]
- Mavrodi DV, Joe A, Mavrodi OV, Hassan KA, Weller DM, Paulsen IT, Loper JE, Alfano JR, Thomashow LS. 2011. Structural and functional analysis of the type III secretion system from Pseudomonas fluorescens Q8r1-96. Journal of Bacteriology 193, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, et al.. 2011. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. [DOI] [PubMed] [Google Scholar]
- Meng Q, Yin J, Rosenzweig N, Douches D, Hao JJ. 2012. Culture-based assessment of microbial communities in soil suppressive to potato common scab. Plant Disease 96, 712–717. [DOI] [PubMed] [Google Scholar]
- Meziane H, Van der Sluis I, Van Loon LC, Höfte M, Bakker PA. 2005. Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Molecular Plant Pathology 6, 177–185. [DOI] [PubMed] [Google Scholar]
- Mgbemene CA, Nnaji CC, Nwozor C. 2016. Industrialization and its backlash: focus on climate change and its consequences. Journal of Environmental Science and Technology 9, 301–316. [Google Scholar]
- Miché L, Battistoni F, Gemmer S, Belghazi M, Reinhold-Hurek B. 2006. Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Molecular Plant-Microbe Interactions 19, 502–511. [DOI] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM. 2010. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. The Plant Cell 22, 973–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter B, Brader G, Pfaffenbichler N, Sessitsch A. 2019. Next generation microbiome applications for crop production—limitations and the need of knowledge-based solutions. Current Opinion in Microbiology 49, 59–65. [DOI] [PubMed] [Google Scholar]
- Möbius KA. 1877. Die auster und die austernwirthschaft. Berlin: Wiegandt, Hempel & Parey. [Google Scholar]
- Morales SE, Holben WE. 2011. Linking bacterial identities and ecosystem processes: can ‘omic’ analyses be more than the sum of their parts? FEMS Microbiology Ecology 75, 2–16. [DOI] [PubMed] [Google Scholar]
- Mueller UG, Sachs JL. 2015. Engineering microbiomes to improve plant and animal health. Trends in Microbiology 23, 606–617. [DOI] [PubMed] [Google Scholar]
- Mukherji A, Kobiita A, Ye T, Chambon P. 2013. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153, 812–827. [DOI] [PubMed] [Google Scholar]
- Müller DB, Vogel C, Bai Y, Vorholt JA. 2016. The plant microbiota: systems-level insights and perspectives. Annual Review of Genetics 50, 211–234. [DOI] [PubMed] [Google Scholar]
- Nagahama K, Ogawa T, Fujii T, Fukuda H. 1992. Classification of ethylene-producing bacteria in terms of biosynthetic pathways to ethylene. Journal of Fermentation and Bioengineering 73, 1–5. [Google Scholar]
- Nakagawa T, Kawaguchi M. 2006. Shoot-applied MeJA suppresses root nodulation in Lotus japonicus. Plant & Cell Physiology 47, 176–180. [DOI] [PubMed] [Google Scholar]
- Naylor D, Coleman-Derr D. 2017. Drought stress and root-associated bacterial communities. Frontiers in Plant Science 8, 2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz I, Bano A, Tamoor-ul-Hassan. 2009. Isolation of phytohormones producing plant growth promoting rhizobacteria from weeds growing in Khewra salt range, Pakistan and their implication in providing salt tolerance to Glycine max L. African Journal of Biotechnology 8, 5762–5766. [Google Scholar]
- Nehls U, Plassard C. 2018. Nitrogen and phosphate metabolism in ectomycorrhizas. New Phytologist 220, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Neumann G, Römheld V. 2000. The release of root exudates as affected by the plant physiological status. In: Pinton R, Varanini Z, Nanipieri P, eds. The rhizosphere; biochemistry and organic substances at the soil–plant interface. New York: Marcel Dekker Inc., 41–98. [Google Scholar]
- Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D. 2007. Release and persistence of extracellular DNA in the environment. Environmental Biosafety Research 6, 37–53. [DOI] [PubMed] [Google Scholar]
- Niu B, Paulson JN, Zheng X, Kolter R. 2017. Simplified and representative bacterial community of maize roots. Proceedings of the National Academy of Sciences, USA 114, E2450–E2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T, Chen J, Yin Y. 2017. Cross-talk of brassinosteroid signaling in controlling growth and stress responses. The Biochemical Journal 474, 2641–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, Minz D. 2014. Niche and host-associated functional signatures of the root surface microbiome. Nature Communications 5, 4950. [DOI] [PubMed] [Google Scholar]
- Ogram A, Castro H, Chauhan A. 2007. Methods of soil microbial community analysis. Manual of environmental microbiology, 3rd edn Washington, DC: American Society of Microbiology. [Google Scholar]
- Omer ZS, Tombolini R, Broberg A, Gerhardson B. 2004. Indole-3-acetic acid production by pink-pigmented facultative methylotrophic bacteria. Plant Growth Regulation 43, 93–96. [Google Scholar]
- Ongena M, Jourdan E, Adam A, Paquot M, Brans A, Joris B, Arpigny JL, Thonart P. 2007. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environmental Microbiology 9, 1084–1090. [DOI] [PubMed] [Google Scholar]
- Orwin KH, Buckland SM, Johnson D, Turner BL, Smart S, Oakley S, Bardgett RD. 2010. Linkages of plant traits to soil properties and the functioning of temperate grassland. Journal of Ecology 98, 1074–1083. [Google Scholar]
- Ottesen AR, González Peña A, White JR, et al.. 2013. Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiology 13, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar M, Tewari R, Gulati A, Nayyar H. 2016. Indigenous salt-tolerant rhizobacterium Pantoea dispersa (PSB3) reduces sodium uptake and mitigates the effects of salt stress on growth and yield of chickpea. Acta Physiologiae Plantarum 38, 278. [Google Scholar]
- Papadopoulou K, Melton RE, Leggett M, Daniels MJ, Osbourn AE. 1999. Compromised disease resistance in saponin-deficient plants. Proceedings of the National Academy of Sciences, USA 96, 12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes SH, Gao T, Law TF, et al.. 2018. Design of synthetic bacterial communities for predictable plant phenotypes. PLoS Biology 16, e2003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale A, Proietti S, Pantelides IS, Stringlis IA. 2019. Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Frontiers in Plant Science 10, 1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passera A, Compant S, Casati P, et al.. 2019. Not just a pathogen? Description of a plant-beneficial Pseudomonas syringae strain. Frontiers in Microbiology 10, 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. 2013. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proceedings of the National Academy of Sciences, USA 110, 6548–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta AL, Sun Y, McDaniel MD, Lennon JT. 2018. Crop rotational diversity increases disease suppressive capacity of soil microbiomes. Ecosphere 9, e02235. [Google Scholar]
- Pereira P, Ibáñez F, Rosenblueth M, Etcheverry M, Martínez-Romero E. 2011. Analysis of the bacterial diversity associated with the roots of maize (Zea mays L.) through culture-dependent and culture-independent methods. ISRN Ecology 2011, 10. [Google Scholar]
- Perret X, Staehelin C, Broughton WJ. 2000. Molecular basis of symbiotic promiscuity. Microbiology and Molecular Biology Reviews 64, 180–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M, Bååth E. 2003. Temperature-dependent changes in the soil bacterial community in limed and unlimed soil. FEMS Microbiology Ecology 45, 13–21. [DOI] [PubMed] [Google Scholar]
- Pierik R, Sasidharan R, Voesenek LACJ. 2007. Growth control by ethylene: adjusting phenotypes to the environment. Journal of Plant Growth Regulation 26, 188–200. [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. 2009. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, van Wees SC, Hoffland E, van Pelt JA, van Loon LC. 1996. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. The Plant Cell 8, 1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma J, Schilder MT, Bloem J, van Leeuwen-Haagsma WK. 2008. Soil suppressiveness and functional diversity of the soil microflora in organic farming systems. Soil Biology and Biochemistry 40, 2394–2406. [Google Scholar]
- Preston GM, Bertrand N, Rainey PB. 2001. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Molecular Microbiology 41, 999–1014. [DOI] [PubMed] [Google Scholar]
- Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau JH. 2012. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. The ISME Journal 6, 1812–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve JR, Schadt CW, Carpenter-Boggs L, Kang S, Zhou J, Reganold JP. 2010. Effects of soil type and farm management on soil ecological functional genes and microbial activities. The ISME Journal 4, 1099–1107. [DOI] [PubMed] [Google Scholar]
- Richardson AE, Simpson RJ. 2011. Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiology 156, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha I, Ma Y, Souza-Alonso P, Vosátka M, Freitas H, Oliveira RS. 2019. Seed coating: a tool for delivering beneficial microbes to agricultural crops. Frontiers in Plant Science 10, 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues RR, Rodgers NC, Wu X, Williams MA. 2018. COREMIC: a web-tool to search for a niche associated CORE MICrobiome. PeerJ 6, e4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosegrant MW, Ringler C, Zhu T. 2009. Water for agriculture: maintaining food security under growing scarcity. Annual Review of Environment and Resources 34, 205–222. [Google Scholar]
- Rosenberg E, Sharon G, Atad I, Zilber-Rosenberg I. 2010. The evolution of animals and plants via symbiosis with microorganisms. Environmental Microbiology Reports 2, 500–506. [DOI] [PubMed] [Google Scholar]
- Rosenzweig N, Tiedje JM, Quensen JF 3rd, Meng Q, Hao JJ. 2012. Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Disease 96, 718–725. [DOI] [PubMed] [Google Scholar]
- Saad MM, Kobayashi H, Marie C, Brown IR, Mansfield JW, Broughton WJ, Deakin WJ. 2005. NopB, a type III secreted protein of Rhizobium sp. strain NGR234, is associated with pilus-like surface appendages. Journal of Bacteriology 187, 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad MM, Michalet S, Fossou R, Putnik-Delić M, Crèvecoeur M, Meyer J, de Malézieux C, Hopfgartner G, Maksimović I, Perret X. 2019. Loss of NifQ leads to accumulation of porphyrins and altered metal-homeostasis in nitrogen-fixing symbioses. Molecular Plant-Microbe Interactions 32, 208–216. [DOI] [PubMed] [Google Scholar]
- Sanguin H, Sarniguet A, Gazengel K, Moënne-Loccoz Y, Grundmann GL. 2009. Rhizosphere bacterial communities associated with disease suppressiveness stages of take-all decline in wheat monoculture. New Phytologist 184, 694–707. [DOI] [PubMed] [Google Scholar]
- Santhanam R, Groten K, Meldau DG, Baldwin IT. 2014. Analysis of plant–bacteria interactions in their native habitat: bacterial communities associated with wild tobacco are independent of endogenous jasmonic acid levels and developmental stages. PLoS One 9, e94710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota R, Knorr K, Jørgensen LN, O’Hanlon KA, Nicolaisen M. 2015. Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytologist 207, 1134–1144. [DOI] [PubMed] [Google Scholar]
- Sarhan MS, Hamza MA, Youssef HH, et al.. 2019. Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media—a review. Journal of Advanced Research 19, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse J, Martinoia E, Northen T. 2018. Feed your friends: do plant exudates shape the root microbiome? Trends in Plant Science 23, 25–41. [DOI] [PubMed] [Google Scholar]
- Schimel J, Balser TC, Wallenstein M. 2007. Microbial stress-response physiology and its implications for ecosystem function. Ecology 88, 1386–1394. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K, Bulgarelli D. 2015. The plant microbiome at work. Molecular Plant-Microbe Interactions 28, 212–217. [DOI] [PubMed] [Google Scholar]
- Schlatter D, Kinkel L, Thomashow L, Weller D, Paulitz T. 2017. Disease suppressive soils: new insights from the soil microbiome. Phytopathology 107, 1284–1297. [DOI] [PubMed] [Google Scholar]
- Schreiter S, Ding GC, Heuer H, Neumann G, Sandmann M, Grosch R, Kropf S, Smalla K. 2014. Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Frontiers in Microbiology 5, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuur EA, Matson PA. 2001. Net primary productivity and nutrient cycling across a mesic to wet precipitation gradient in Hawaiian montane forest. Oecologia 128, 431–442. [DOI] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biology 14, e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergaki C, Lagunas B, Lidbury I, Gifford ML, Schäfer P. 2018. Challenges and approaches in microbiome research: from fundamental to applied. Frontiers in Plant Science 9, 1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessitsch A, Weilharter A, Gerzabek MH, Kirchmann H, Kandeler E. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Applied and Environmental Microbiology 67, 4215–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Hogan CS, Klimowicz AK, Linske M, McManus PS, Handelsman J. 2012. Culturing captures members of the soil rare biosphere. Environmental Microbiology 14, 2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon A, Elad Y, Barakat R, Tudzynski P. 2007. Phytohormones in Botrytis–plant interactions. In: Elad Y, Williamson B, Tudzynski P, Delen N, eds. Botrytis: biology, pathology and control. Dordrecht: Springer Netherlands, 163–179. [Google Scholar]
- Shen C, Xiong J, Zhang H, Feng Y, Lin X, Li X, Liang W, Chu H. 2013. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biology and Biochemistry 57, 204–211. [Google Scholar]
- Shen G, Zhang S, Liu X, Jiang Q, Ding W. 2018. Soil acidification amendments change the rhizosphere bacterial community of tobacco in a bacterial wilt affected field. Applied Microbiology and Biotechnology 102, 9781–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng XF, He LY. 2006. Solubilization of potassium-bearing minerals by a wild-type strain of Bacillus edaphicus and its mutants and increased potassium uptake by wheat. Canadian Journal of Microbiology 52, 66–72. [DOI] [PubMed] [Google Scholar]
- Shigenaga AM, Argueso CT. 2016. No hormone to rule them all: interactions of plant hormones during the responses of plants to pathogens. Seminars in Cell & Developmental Biology 56, 174–189. [DOI] [PubMed] [Google Scholar]
- Shrestha PM, Noll M, Liesack W. 2007. Phylogenetic identity, growth–response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environmental Microbiology 9, 2464–2474. [DOI] [PubMed] [Google Scholar]
- Siciliano SD, Palmer AS, Winsley T, et al.. 2014. Soil fertility is associated with fungal and bacterial richness, whereas pH is associated with community composition in polar soil microbial communities. Soil Biology and Biochemistry 78, 10–20. [Google Scholar]
- Siegel-Hertz K, Edel-Hermann V, Chapelle E, Terrat S, Raaijmakers JM, Steinberg C. 2018. Comparative microbiome analysis of a fusarium wilt suppressive soil and a fusarium wilt conducive soil from the Châteaurenard Region. Frontiers in Microbiology 9, 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Munro S, Potts JM, Millard P. 2007. Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Applied Soil Ecology 36, 147–155. [Google Scholar]
- Skorpil P, Saad MM, Boukli NM, Kobayashi H, Ares-Orpel F, Broughton WJ, Deakin WJ. 2005. NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Molecular Microbiology 57, 1304–1317. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proceedings of the National Academy of Sciences, USA 103, 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song GC, Im H, Jung J, Lee S, Jung MY, Rhee SK, Ryu CM. 2019. Plant growth-promoting archaea trigger induced systemic resistance in Arabidopsis thaliana against Pectobacterium carotovorum and Pseudomonas syringae. Environmental Microbiology 21, 940–948. [DOI] [PubMed] [Google Scholar]
- Songwattana P, Noisangiam R, Teamtisong K, Prakamhang J, Teulet A, Tittabutr P, Piromyou P, Boonkerd N, Giraud E, Teaumroong N. 2017. Type 3 secretion system (T3SS) of Bradyrhizobium sp. DOA9 and its roles in legume symbiosis and rice endophytic association. Frontiers in Microbiology 8, 1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorty AM, Meena KK, Choudhary K, Bitla UM, Minhas PS, Krishnani KK. 2016. Effect of plant growth promoting bacteria associated with halophytic weed (Psoralea corylifolia L) on germination and seedling growth of wheat under saline conditions. Applied Biochemistry and Biotechnology 180, 872–882. [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J. 2011. Auxin and plant–microbe interactions. Cold Spring Harbor Perspectives in Biology 3, a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. 2007. Indole-3-acetic acid in microbial and microorganism–plant signaling. FEMS Microbiology Reviews 31, 425–448. [DOI] [PubMed] [Google Scholar]
- Stacey G, McAlvin CB, Kim SY, Olivares J, Soto MJ. 2006. Effects of endogenous salicylic acid on nodulation in the model legumes Lotus japonicus and Medicago truncatula. Plant Physiology 141, 1473–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwerth KL, Jackson LE, Calderón FJ, Stromberg MR, Scow KM. 2002. Soil microbial community composition and land use history in cultivated and grassland ecosystems of coastal California. Soil Biology and Biochemistry 34, 1599–1611. [Google Scholar]
- Stringlis IA, Proietti S, Hickman R, Van Verk MC, Zamioudis C, Pieterse CMJ. 2018a Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. The Plant Journal 93, 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis IA, Yu K, Feussner K, de Jonge R, Van Bentum S, Van Verk MC, Berendsen RL, Bakker PAHM, Feussner I, Pieterse CMJ. 2018b MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proceedings of the National Academy of Sciences, USA 115, E5213–E5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis IA, Zamioudis C, Berendsen RL, Bakker PAHM, Pieterse CMJ. 2019. Type III secretion system of beneficial rhizobacteria Pseudomonas simiae WCS417 and Pseudomonas defensor WCS374. Frontiers in Microbiology 10, 1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma N, Yamauchi H, Yamamoto K. 1995. Enhanced production of ethylene by soybean roots after inoculation with Bradyrhizobium japonicum. Plant Science 111, 163–168. [Google Scholar]
- Sun R, Li W, Dong W, Tian Y, Hu C, Liu B. 2018. Tillage changes vertical distribution of soil bacterial and fungal communities. Frontiers in Microbiology 9, 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen NB, Watts-Williams SJ, Joner EJ, Battini F, Efthymiou A, Cruz-Paredes C, Nybroe O, Jakobsen I. 2018. Suppression of the activity of arbuscular mycorrhizal fungi by the soil microbiota. The ISME Journal 12, 1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Murfet IC, Ross JJ, Sherriff LJ, Warkentin TD. 1999. Bushy, a dominant pea mutant characterised by short, thin stems, tiny leaves and a major reduction in apical dominance. Physiologia Plantarum 107, 346–352. [Google Scholar]
- Tack FM, Callewaert OW, Verloo MG. 1996. Metal solubility as a function of pH in a contaminated, dredged sediment affected by oxidation. Environmental Pollution 91, 199–208. [DOI] [PubMed] [Google Scholar]
- Tamimi SM, Timko MP. 2003. Effects of ethylene and inhibitors of ethylene synthesis and action on nodulation in common bean (Phaseolus vulgaris L.). Plant and Soil 257, 125–131. [Google Scholar]
- Tanner MA, Goebel BM, Dojka MA, Pace NR. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Applied and Environmental Microbiology 64, 3110–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiem D, Gołębiewski M, Hulisz P, Piernik A, Hrynkiewicz K. 2018. How does salinity shape bacterial and fungal microbiomes of Alnus glutinosa roots? Frontiers in Microbiology 9, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall PH, Hochberg ME, Burdon JJ, Bever JD. 2007. Coevolution of symbiotic mutualists and parasites in a community context. Trends in Ecology & Evolution 22, 120–126. [DOI] [PubMed] [Google Scholar]
- Toju H, Kurokawa H, Kenta T. 2019. Factors influencing leaf- and root-associated communities of bacteria and fungi across 33 plant orders in a grassland. Frontiers in Microbiology 10, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Vera R, García JM, Pozo MJ, López-Ráez JA. 2014. Do strigolactones contribute to plant defence? Molecular Plant Pathology 15, 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truog E. 1947. Soil reaction influence on availability of plant nutrients1. Soil Science Society of America Journal 11, 305–308. [Google Scholar]
- Tsolakidou M-D, Stringlis IA, Fanega-Sleziak N, Papageorgiou S, Tsalakou A, Pantelides IS. 2019. Rhizosphere-enriched microbes as a pool to design synthetic communities for reproducible beneficial outputs. FEMS Microbiology Ecology 95, fiz138. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature 449, 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations 2017. World population prospects: the 2017 revision. Geneva: United Nations Department of Economic and Social Affairs. [Google Scholar]
- Unterseher M, Siddique AB, Brachmann A, Peršoh D. 2016. Diversity and composition of the leaf mycobiome of beech (Fagus sylvatica) are affected by local habitat conditions and leaf biochemistry. PLoS One 11, e0152878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent S, Van Hulten M, Pozo MJ, Czechowski T, Udvardi MK, Pieterse CM, Ton J. 2009. Priming of plant innate immunity by rhizobacteria and beta-aminobutyric acid: differences and similarities in regulation. New Phytologist 183, 419–431. [DOI] [PubMed] [Google Scholar]
- VanEtten HD, Mansfield JW, Bailey JA, Farmer EE. 1994. Two classes of plant antibiotics: phytoalexins versus ‘Phytoanticipins’. The Plant Cell 6, 1191–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek LS, Franke AC, Nijhuis EH, Groeneveld RM, da Rocha UN, Lotz LA. 2011. Bacterial communities associated with Chenopodium album and Stellaria media seeds from arable soils. Microbial Ecology 62, 257–264. [DOI] [PubMed] [Google Scholar]
- van Wees SC, de Swart EA, van Pelt JA, van Loon LC, Pieterse CM. 2000. Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 97, 8711–8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian SR, Palmer RM, Wade WG. 2010. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiology Letters 309, 1–7. [DOI] [PubMed] [Google Scholar]
- Vaughan MM, Wang Q, Webster FX, et al.. 2013. Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. The Plant Cell 25, 1108–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V, Fuqua C. 2013. Chemical signaling between plants and plant-pathogenic bacteria. Annual Review of Phytopathology 51, 17–37. [DOI] [PubMed] [Google Scholar]
- Venturi V, Keel C. 2016. Signaling in the rhizosphere. Trends in Plant Science 21, 187–198. [DOI] [PubMed] [Google Scholar]
- Verbon EH, Trapet PL, Stringlis IA, Kruijs S, Bakker PAHM, Pieterse CMJ. 2017. Iron and immunity. Annual Review of Phytopathology 55, 355–375. [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. 2002. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. The Plant Cell 14, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitousek PM, Cassman K, Cleveland C, et al.. 2002. Towards an ecological understanding of biological nitrogen fixation. Biogeochemistry 57, 1–45. [Google Scholar]
- Vives-Peris V, Molina L, Segura A, Gómez-Cadenas A, Pérez-Clemente RM. 2018. Root exudates from citrus plants subjected to abiotic stress conditions have a positive effect on rhizobacteria. Journal of Plant Physiology 228, 208–217. [DOI] [PubMed] [Google Scholar]
- Voges MJEEE, Bai Y, Schulze-Lefert P, Sattely ES. 2019. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proceedings of the National Academy of Sciences, USA 116, 12558–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wirén N, Khodr H, Hider RC. 2000. Hydroxylated phytosiderophore species possess an enhanced chelate stability and affinity for iron(III). Plant Physiology 124, 1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MR, Lundberg DS, Del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T. 2016. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nature Communications 7, 12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakelin SA, Macdonald LM, Rogers SL, Gregg AL, Bolger TP, Baldock JA. 2008. Habitat selective factors influencing the structural composition and functional capacity of microbial communities in agricultural soils. Soil Biology and Biochemistry 40, 803–813. [Google Scholar]
- Walter J, Britton RA, Roos S. 2011. Host–microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proceedings of the National Academy of Sciences, USA 108 Suppl 1, 4645–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wei Z, Yang K, Wang J, Jousset A, Xu Y, Shen Q, Friman VP. 2019. Phage combination therapies for bacterial wilt disease in tomato. Nature Biotechnology 37, 1513–1520. [DOI] [PubMed] [Google Scholar]
- Wassermann B, Cernava T, Müller H, Berg C, Berg G. 2019. Seeds of native alpine plants host unique microbial communities embedded in cross-kingdom networks. Microbiome 7, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Gu Y, Friman VP, Kowalchuk GA, Xu Y, Shen Q, Jousset A. 2019. Initial soil microbiome composition and functioning predetermine future plant health. Science Advances 5, eaaw0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller DM, Raaijmakers JM, Gardener BB, Thomashow LS. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annual Review of Phytopathology 40, 309–348. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proceedings of the National Academy of Sciences, USA 95, 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, et al.. 2010. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proceedings of the National Academy of Sciences, USA 107, 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TR, Marco ML. 2014. Phyllosphere microbiota composition and microbial community transplantation on lettuce plants grown indoors. MBio 5, e01564-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski-Dyé F, Vial L. 2008. Phase and antigenic variation mediated by genome modifications. Antonie Van Leeuwenhoek 94, 493–515. [DOI] [PubMed] [Google Scholar]
- Woo SL, Pepe O. 2018. Microbial consortia: promising probiotics as plant biostimulants for sustainable agriculture. Frontiers in Plant Science 9, 1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin DW, Liao S, Xie ZP, Hann DR, Steinle L, Boller T, Staehelin C. 2012. Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathogens 8, e1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Naylor D, Dong Z, et al.. 2018. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proceedings of the National Academy of Sciences, USA 115, E4284–E4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaish MW, Al-Lawati A, Jana GA, Vishwas Patankar H, Glick BR. 2016. Impact of soil salinity on the structure of the bacterial endophytic community identified from the roots of Caliph Medic (Medicago truncatula). PLoS One 11, e0159007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro E, Spear RN, McManus PS. 2011. Culture-dependent and culture-independent assessment of bacteria in the apple phyllosphere. Journal of Applied Microbiology 110, 1284–1296. [DOI] [PubMed] [Google Scholar]
- Yeoh YK, Dennis PG, Paungfoo-Lonhienne C, Weber L, Brackin R, Ragan MA, Schmidt S, Hugenholtz P. 2017. Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nature Communications 8, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML. 1996. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. The Plant Journal 10, 835–844. [DOI] [PubMed] [Google Scholar]
- Yu K, Liu Y, Tichelaar R, et al.. 2019. Rhizosphere-associated Pseudomonas suppress local root immune responses by gluconic acid-mediated lowering of environmental pH. Current Biology 29, 3913–3920.e4. [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Hanson J, Pieterse CM. 2014. β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytologist 204, 368–379. [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Korteland J, Van Pelt JA, et al.. 2015. Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB72 expression in Arabidopsis roots during onset of induced systemic resistance and iron-deficiency responses. The Plant Journal 84, 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis C, Mastranesti P, Dhonukshe P, Blilou I, Pieterse CM. 2013. Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiology 162, 304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis C, Pieterse CM. 2012. Modulation of host immunity by beneficial microbes. Molecular Plant-Microbe Interactions 25, 139–150. [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA. 1988. Metabolism and physiology of abscisic acid. Annual Review of Plant Physiology and Plant Molecular Biology 39, 439–473. [Google Scholar]
- Zgadzaj R, Garrido-Oter R, Jensen DB, Koprivova A, Schulze-Lefert P, Radutoiu S. 2016. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proceedings of the National Academy of Sciences, USA 113, E7996–E8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhalnina K, Louie KB, Hao Z, et al.. 2018. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nature Microbiology 3, 470–480. [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu W, Kang X, Cui X, Wang Y, Zhao H, Qian X, Hao Y. 2019. Changes in soil microbial community response to precipitation events in a semi-arid steppe of the Xilin River Basin, China. Journal of Arid Land 11, 97–110. [Google Scholar]
- Zhang Y, Shen H, He X, Thomas BW, Lupwayi NZ, Hao X, Thomas MC, Shi X. 2017. Fertilization shapes bacterial community structure by alteration of soil pH. Frontiers in Microbiology 8, 1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Jing T, Chen Y, Wang F, Qi D, Feng R, Xie J, Li H. 2019. Deciphering microbial diversity associated with Fusarium wilt-diseased and disease-free banana rhizosphere soil. BMC Microbiology 19, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwanenburg B, Pospíšil T, Ćavar Zeljković S. 2016. Strigolactones: new plant hormones in action. Planta 243, 1311–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]