Abstract
Single cells are the building blocks of tissue systems that determine organ phenotypes, behaviors, and function. Understanding the differences between cell types and their activities might provide us with insights into normal tissue functions, development of disease, and new therapeutic strategies. Although -omic level single cell technologies are a relatively recent development that been used only in laboratory studies, these approaches might eventually be used in the clinic. We review the prospects of applying single cell genome, transcriptome, epigenome, proteome, and metabolome analyses to gastroenterology and hepatology research. Combining data from multi-omic platforms and rapid technological developments could lead to new diagnostic, prognostic, and therapeutic approaches.
Keywords: heterogeneity, transcriptomics, proteomics, metabolomics, epigenomics
During the last decade, significant technological advances occurred in the biomedical sciences due to rapid development of next-generation sequencing and mass spectrometry (MS) technologies. Today, we can easily generate comprehensive data on the genomic, epigenome, metabolome/lipidome, and proteome from patient specimens.1-3 This information, generated at sample level, enables studies of pathogenesis and identification of disease subtypes.1, 2, 4 Although bulk approaches have enabled discoveries based on inter-patient or inter-sample variation, intra-sample heterogeneities carried by individual cells over time are lost.5 This information is critical in understanding disease mechanisms; different tissue functions are carried out by specific cell types, and disease progression, such as in cancer relapse or metastasis, are usually mediate by small populations of cells.6, 7
To address cellular level heterogeneities, researchers developed new methods and techniques to generate high-throughput and multi-dimensional data at the single cell resolution (Fig. 1). The emerging single cell biology field is revolutionizing basic sciences by addressing sophisticated biological questions in developmental biology.8 In the clinic, single cell level metrics such as the Immunoscore9 can affect decisions about management of patients with cancer. Furthermore, in patients with gastrointestinal malignancies, tumors with increased cellular heterogeneity tend to respond poorly to treatment.10, 11 Increasing our understanding of these phenomena and obtaining higher resolution single cell data, especially from patients, might improve care. We review single cell technologies and how they can be used to study digestive diseases. However, we emphasize that single cell -omics level technologies have only been applied in a research setting, including translational research, and cannot yet be used to make actionable clinical decisions.
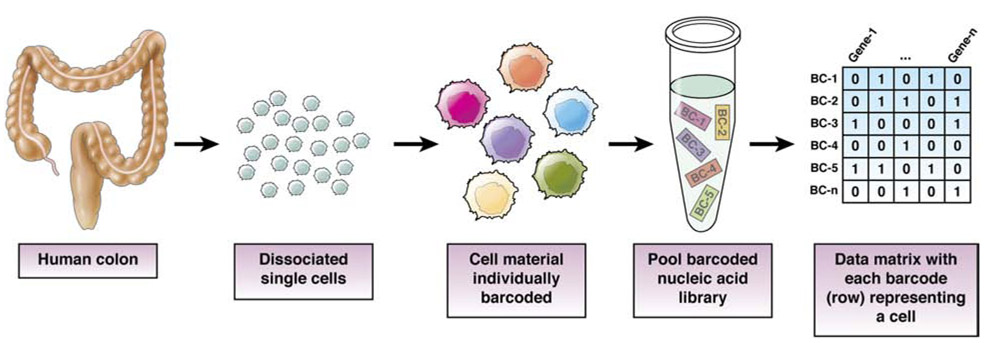
Figure 1.
Barcoding Scheme for High-throughput Genomics Analysis of Single Cells
Candidate-based Approaches
Analyses of molecular features of individual cells are not new to clinical research—pathologists have been using microscopy, immunohistochemistry, and in situ hybridization for decades to identify cells, such as infiltrating lymphocytes, in human tissues. Flow cytometry is used to perform immunophenotype analyses of blood cells in patients with immune-mediated and other diseases. These techniques are known as candidate-based approaches, because they require probes to detect specific molecules.
Advances have mainly focused on exceeding the limitations of spectral overlap in fluorescence to enable highly multiplexed (such as multi-analyte) analyses at the single cell level, such that machines can potentially identify cells from multivariate measurements in lieu of a human expert.12 There are 2 main ways by which multiplexing is achieved. First, elemental isotope labeling of antibodies enable mass spectrometric deconvolution of probes with minimal spill over. Metal antibody labeling results in flow cytometry (mass cytometry, also known as CyTOF)13 and microscopy modalities (imaging mass cytometry, multiplex ion beam imaging)14, 15 capable of analyzing 30–40 analytes at single cell resolution.
Mass cytometry has been optimized to study signaling within the intestinal epithelium16, and immune cells in the lamina propria.17 For example, Chuang et al studied immune cells infiltration in lamina propria samples from patients with Crohn’s disease using CyTOF; they found monocytes that carried a frameshift mutation in CSF2RB had reduced responses to granulocyte-macrophage colony stimulating factor.18 Martin et al also used CyTOF to identify unique fibroblasts and immune cells in ileal tissues from patients with Crohn’s disease that contribute to resistance to tumor necrosis factor antagonists.19
Iterative schemes can be used with microscopy, such that different probes are applied in different imaging rounds even though the same detection labels are used. With advanced registration approaches20, multi-channel images, analogous to standard 3- or 4- channel fluorescence images, are produced. Iterative schemes can be used to detect protein21-25 or transcripts12, 26, with differences between these schemes being the detection method (for example, direct conjugate vs tagged nucleic acids) affecting iteration times. Gerdes et al stained 61 protein antigens in tissue microarrays comprising 747 samples from patients with colorectal cancer (CRC); they found extensive heterogeneity among tumors that corresponds to specific signaling pathways.27 Using multiplex protein imaging, Ligorio et al found that cancer-associated fibroblasts contribute to heterogeneity within pancreatic tumors.28 Algorithms to analyze multi-dimensional images can be segmentation-based to produce single cell resolution data similar to mass cytometry29, 30, or pixel based31. The unifying theme behind these studies revolves around the discovery of new, unexpected cell populations that associate with disease processes. These efforts are made possible by labeling tissues with multiple probes that can detect a wide variety of cell types, and for combinatorial labeling to define unexpected cell types or states. The experiment therefore acts a screen for unexpected cell populations that correlate to phenotype, which fulfills a fundamentally different goal than single-target approaches, such as immunohistochemistry, which a single marker to query a single cell population.
Candidate-based approaches are most ready to be adopted for human research due to their congruence with traditional pathology analyses, and machine learning on large sets of histological images has already been explored.32, 33 However, candidate-based approaches require accurate probes—it is a challenge to determine specificity and sensitivity of detection for probes (such as antibodies) in human tissues, due to lack of control samples. Furthermore, measurements of preselected markers provide only a narrow scope of new insights into factors that contribute to disease development.
Single-Cell Analysis of Transcriptomes from Cell Suspensions
Untargeted sequencing does not depend on candidate probe quality, because sequencing data are mostly categorical in nature (4 nucleotides- ACGT). Single-cell RNA sequencing (scRNA-seq) is one of the most mature single cell genomics tools for studying cell heterogeneity.34 Since 2016, there has been a large increase in studies using scRNA-seq analysis, due to the advent of droplet-based techniques that enable high-throughput analyses of thousands of cells instead of hundreds.35-37 Different protocols provide different aspects of transcriptomic information (Supplemental Table 1), such as isoform information from full-length analysis38 and differential gene expression based on 3’ poly(A) capture transcript counting.39 For a review on scRNA-seq, see refs34, 40. We focus on aspects most relevant to human research.
Single-cell RNA sequencing involves isolation, containment, and processing of single cells into nucleic acid libraries for sequencing. An important pre-analytical variable lies in tissue collection and handling. Ischemic conditions and dissociation of single cells can introduce significant artefacts to cell transcriptomes. Fresh specimens collected from accessible surgical or biopsy samples, immediately delivered for processing, yield the highest quality data. Single cells are dissociated by exposure to enzymes such as trypsin or collagenase, followed by mechanical trituration. Optimization in fresh tissue processing, such as cold protease dissociation, enables minimized perturbation during single cell isolation.41 For tissue types where dissociation is challenging or situations in which times of ischemia cannot be controlled, tissues can be flash frozen, so quality of nucleic acids can be preserved and nuclei can be isolated for single nuclei (sn) RNA-seq.42, 43 snRNA-seq has the advantage of being amendable to retrospectively collected frozen archive specimens, but it should be noted that nuclei possess less material compared with what is in an entire cell, with marked differences in representation in transcripts.44 Regardless of protocols used, relative abundances of particular cell types might be artificially changed due to selective filtering and retrieval during tissue preparation steps.
A variety of methods, from plates to microdroplets35, 36, exist to contain single cells such that cDNA libraries from individual cells can be prepared (Table 1). The most technically amenable methods are droplet based35-37 or nanowell based.45-47 These are high-throughput approaches that require less manual handling due to barcoding (Fig. 1), miniaturization, and automation, and commercial counterparts exist for turnkey applications. Although there are nuances to library preparation downstream that results in different coverage48-51, all scRNA-seq libraries require amplification, due to the low amount of starting material. From small amounts of RNA, technical noise arises due to Poisson sampling, especially for less abundant transcripts.39 Researchers should balance the cost benefit of using exponential amplification (such as PCR) and linear amplification (such as in vitro transcription).51-53 Exponential amplification is easy and requires less technical skills and awareness, but results in amplification bias of highly abundant transcripts that requires more sequencing (and therefore, higher costs) to overcome. Linear amplification allows low abundance transcripts to be represented but is a multi-day process requiring multiple steps in handling RNA. Ziegenhain et al and Wang et al performed comparative analyses of different scRNAseq methods that could be informative for choosing particular methods for particular applications.54, 55
Table 1:
Cell Capture and Barcoding Methods for Genomic Analyses of Single Cells
| Micropipetting | Laser Capture Microdissection | Flow Sorting | Droplet | Nanowell | Combinatorial Indexing | |
|---|---|---|---|---|---|---|
| Input Material | tissue in situ | tissue in situ | suspension | suspension | suspension | suspension |
| Input Cell Requirement | No lower bound | No lower bound | ~10^5 | ~10^4 | ~10^2 | ~10^6 |
| Cell Output | < 10^2 | < 10^2 | ~10^3 | ~10^3 | ~10^3 | ~10^5 |
| Cell Capture | cell by cell picking | cell by cell picking | automated | automated | deposition by gravity | combinatorial scheme |
| Downstream Steps | well by well | well by well | well by well | pooled | pooled | pooled |
| Ease of Use | hard | hard | moderate | easy | moderate | moderate |
There are several alternatives for downstream processing and analysis of sequencing data (see ref56). Mapping of FASTQ data can be achieved via a variety of mapping algorithms including TopHat, STAR, and Kallisto Bustools57. Popular data analysis packages such as Seurat58 or Scanpy59 can be used for downstream analyses. These software tools already incorporate dimension reduction algorithms for visualization of multi-dimensional data in lower dimensional space, such as t-distributed stochastic neighbor embedding (t-SNE)60, and uniform manifold approximation and projection (UMAP).61
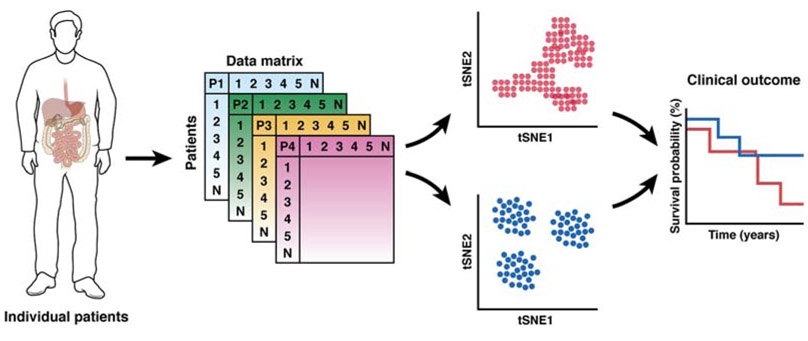
These tools allow data points, and therefore cells with similar expression profiles, to be grouped, enabling identification and analysis of cell types and states. One significant challenge, however, is the quality of single cell level data that results from incomplete sampling of the transcriptome. These noisy data require additional attention paid to quality control regarding the filtering of low quality cells 62, as well as non-reliant and irrelevant gene features63. Of note for scRNA-seq applications in human studies is the eventual acquisition and analysis of datasets from large cohort of patients (Fig. 2). Researchers have developed tools64, 65 that can compare entire single cell landscapes that can be used for subtyping samples and patients.
Figure 2. Potential Clinical Application of Single-cell -omic Technologies.
Single-cell -omics profiles of digestive organs from cohort of patients may reveal single cell landscapes that predict clinical outcome.
Slide-based Spatial Transcriptome Analyses
Transcriptomic analysis is routinely performed on single cell suspensions where spatial context is lost. The location of cells determines their interactions with other cells, via short- and long-range signaling mechanisms66, and their identities and behaviors as a function of environment (such as hypoxia or normoxia).67 Furthermore, cells in tissues, especially in patients with diseases, are spatially heterogeneous. For instance, Crohn’s disease intestinal tissues contain pathological skip lesions that are interspersed amongst uninflamed areas.68 Suspension-based scRNA-seq in this case produces data on admixtures of healthy and affected tissues, blending out disease-specific signals. In situ analysis does not require dissociation nor single cell retrieval, thus should maintain accurate representation of cell types of the native tissue. To produce spatially resolved analyses, researchers rely on candidate-based approaches for targeted analyses. To scale these approaches to the genome level, seqFISH+12 and MERFISH26 use temporal fluorophore barcoding with hamming distance correction to extend the number of mRNA measured to the scale of 10,000. For temporal barcoding to work, individual molecules of mRNA must be resolved, which limits the volume of tissue that can be profiled. These approaches are therefore best for high-, sub-cellular resolution imaging with high-end microscopes that are not always accessible outside of research settings. As such, recent studies have combined seqFISH+ with expansion microscopy to visualize gene expression at deeper, organelle levels.69
At the other end of the spectrum, genome-scale data can be obtained when single cell resolution is not needed. For instance, laser capture microdissection can be used in a low-throughput manner, even with fixed specimens, to profile transcriptomes of a few to 10s of cells.70, 71 For lower-resolution tissue profiling, multiple groups have developed techniques that involve spatial barcoding in lieu of single cell barcoding.72, 73 In general, these approaches immobilize barcoded oligonucleotides73 or oligonucleotide-conjugated beads onto slides72; tissue sections are then cut and permeabilized, enabling mRNA capture by the underlying oligos. The oligos are then released, pooled, and prepared as a library in a similar manner to scRNA-seq. The spatial barcode that was incorporated into the cDNA during capture could be mapped back to the original position in the tissue section.
These methods couple spatial information for analysis of tissue architecture and global transcriptome profiling; they have been used to characterize infiltration of immune cells into tumors during immunotherapy.73 The resolution of these approaches is on the order of 50–100 microns, such that each barcoded pixel contains information about dozens of cells. Improved protocols with fewer than 10 micron resolution are being developed, although data quality, in terms of number of transcripts detected, is compromised.74 Nevertheless, slide-based digital pathology and scRNA-seq have become more common, so these combined approaches could be rapidly advanced for analysis of tissues from patients.
Application of Single-cell Transcriptome Analyses to Gastroenterology
scRNA-seq can be used to evaluate tissue heterogeneity, and the cell types responsible for a particular function or disease in the gastrointestinal tract75-77 and liver78, 79 is a direct application of scRNA-seq. In a basic science setting, scRNA-seq has been used for finding new populations of endocrine80, 81 and tuft cells75, 82 in the gut epithelium, as well as immune83 and stromal cells.84 Because scRNA-seq is an endpoint assay where insights are derived from a snapshot in time when tissue is collected, it can applied to range of specimens from model organisms to human patients.85 scRNA-seq forms the basis of the next stepping stone towards precision medicine, akin to prior efforts such as TGCA and CPTAC. Large national and international consortia are being formed, utilizing scRNA-seq to characterize large cohorts of healthy and diseased tissues. Here, we will highlight studies utilizing scRNA-seq on human patient specimens.
A major application of scRNAseq is in cancer biology where intra- and inter-tumor heterogeneity is a major challenge in rational therapy design.1, 11, 86 Our descriptions of cancer phenotypes and subtypes rely mostly on bulk sequencing or proteomic assays that mask the potentially important cell populations, such as those that are metastasis-capable, whereas scRNA-seq can be used to deconvolve cellular heterogeneity. Furthermore, a comprehensive characterization of tumor landscape by scRNA-seq, including immune and fibroblast infiltration within the microenvironment, can possibly be applied to describe different disease subtypes and predict patient prognosis .28 Some recent studies have already shown successful application of scRNA-seq to pre-cancers and cancers of the digestive system, where cancer cell genotypes, phenotypes, and the microenvironment are systematically dissected. Zhang et al discovered a dynamic relationship of T cells in CRC and identified subgroup of CRC that are likely going to show favorable responses to immune-checkpoint blockade.87 Bian et al dissected 10 CRC using multi-omics approaches that provide fundamental insights for understanding the molecular alterations that occur during primary CRC progression and metastasis.88 Another study examined the transcriptional dynamics of different gastric cancer subtypes using scRNA-seq where they identified OR51E1 as a marker for unique endocrine cells in the early-malignant lesion in gastric cancer.89 Owen et al studied Barrett’s esophagus, that progress to esophageal cancer, using scRNA-seq and found transcriptional similarity between Barrett’s and normal esophagus.90 A recent liver cell atlas identified the cellular composition of liver and unraveled the phenotypic changes that occur in hepatocellular carcinoma.78 Using scRNAseq, Zheng et al identified infiltrating T cells with distinct functions in liver cancer that provided valuable insights to understand the immune landscape and prognostic potential in this cancer.91 Immunotherapy is gaining traction in providing sustained responses in many cancer types. However, only a fraction of patients will respond to immunotherapy, as partly determined by the degree of immune cell infiltration into the intra-tumoral space of the naive tumor.92, 93 scRNA-seq can contribute to immunotherapy by deeply characterizing the immune microenvironment as to predict patient response.
Inflammatory bowel disease are characterized by the chronic inflammation of the intestines.94 Complex conditions such as IBD where shifts in cellular ecosystem is correlated to disease phenotype can be dissected by scRNA-seq. Parikh et al performed a comparative study between IBD and normal colon to identify specific cell type that drives IBD77 They found a pH sensing and proton channel OTOP2-expressing colonic absorptive cells at the top of the crypt that are often dysregulated in IBD as well as cancer. Using scRNAseq, they also dissect cell type-specific responses to colonic inflammation and find that WFDC2 secretion is required for mucus barrier integrity. scRNA-seq has also been used to characterize a network of IgG plasma cells, inflammatory mononuclear phagocytes, activated T cells, and stromal cells in patients with Crohn’s disease resistant to tumor necrosis factor antagonists.19 Smillie et al generated a single cell atlas of ulcerative colitis to map the GWAS risk variants precisely to cell types and pathways where they characterized specific cell subsets responsible for inflammation and therapy resistance.95 Kinchen et al performed single cell profiling of colonic mesenchymal cells that revealed unexpected heterogeneity and mesenchymal remodeling associated with inflammation and barrier dysfunction in IBD.96 Single-cell analyses can be used to study microenvironment communication mechanisms and increase our understanding of complex gastrointestinal disorders.
Although clinical decisions cannot yet be made based on scRNA-seq data, they can be used to determine patient prognoses and disease subtypes.97, 98 scRNA-seq data might also be used as a reference to deconvolve cell compositions from existing bulk RNA-seq data, with algorithms such as MuSiC, xCell, and CIBERSORTx.99-101 The promise here is for new patient categories to be defined with finer resolution, such that possible outcomes of new and formerly diagnosed patients can be predicted from a retrospective resource. Despite the promise and wide accessibility of single cell transcriptome analysis methods, there are limitations that preclude their use in the clinic. There are still many places in transcriptome analysis protocols where artefacts can be introduced, reducing reproducibility and interpretation—the technical and computational expertise required is quite high. The cost and turn-around required for interpretable results might be unacceptably high for clinical decision making. Lastly, the utmost need for accessible fresh tissue to generate the best quality scRNA-seq data is a challenge for many clinics. Combination of all these factors results in a high barrier of adoption, but many groups are moving toward solving these problems.
Single-cell Epigenetic Analyses
Layers of epigenetic information connect the genome to the transcriptome. 102, 103 Microfluidic paradigms used to determine single cell transcriptomes have been expanded for studies of single cell epigenomes. These technologies have led to methods for investigating chromatin accessibility, protein–DNA interactions, chromosome conformation, and DNA methylation in single cells.
Single-cell assays for transposase-accessible chromatin using sequencing (scATAC-seq) target accessible genomic regions by exploiting the kinetic favorability of Tn5-mediated transposition reactions with DNA unincorporated by nucleosomes. These captured genome sequences can be cis-acting DNA elements poised for transcription or regulation by transcription factors. Borrowing the microfluidic platforms of single cell transcriptomics, several scATAC-seq methods have been established (see studies by Cusanovich et al, Buenrostro et al, and Lareau et al104-106). These methods isolate individual cells using plated micro-wells, integrated fluidic circuits, and encapsulation into nanoliter droplets, respectively. Each nucleus therefore produces a single barcoded library of genomic fragments enriched for regions of accessible genomic loci.
Single-cell chromatin conformation capture methods can be used to identify cis-acting DNA elements and determine their physical proximities to potential regulators. Topologically associated domains and long-range chromatin interactions mediated by loop structures can be probed by 3C methods; more recently, at single cell resolution. Hi-C, and its single cell variants like sc-Hi-C and sci-Hi-C, developed by Nagano et al and Ramani et al, combine chromatin crosslinking, restriction digestion, and proximity-based ligation to create libraries that capture spatially proximal DNA fragments.107, 108 sci-Hi-C, in particular, isolates nuclei in microwells and incorporates combinatorial indexing. These methods result in a single library per cell, containing fragments that represent pairs of proximally adjacent genomic loci. Although it is not exactly a single cell chromatin conformation capture method, a modified form of ATAC-seq, called ATAC-See, developed by Chen et al, permits covalent tagging of accessible chromatin with visualizable fluorophores. This allows for visualization by microscopy and subsequent high-throughput sequencing.109
Single-cell chromatin immunoprecipitation methods target protein–DNA interactions within single, isolated cells. These methods retain the same strategy as their bulk approaches, relying on specific antibody-protein interactions. Drop-ChIP, a method developed by Rotem et al, takes protein-associated genomic fragments generated from droplet-isolated single cells and tags them with unique DNA barcodes.110 These nanoliter droplets, which contain the contents of a single cell, are broken and aggregated for immunoprecipitation and library generation. This information can also be obtained using CUT&Tag, described by Kaya-Okur et al. This method uses protein-A-tethered Tn5 transposons to localize these elements to protein-bound antibodies.111 Target-localized transposons fragment the genome in a way that enriches target protein-associated loci. These reactions are amenable to nanowell-based single cell isolation systems, because antibody binding and transposon introduction can be performed at a bulk level before isolation. Both of these methods produce sequencing libraries that contain cis-acting regulatory elements associated with the antibody-targeted protein.
Single-cell methylation and hydroxymethylation (sc-5mc and sc-5hmc) assays measure covalent modifications on genomic cytosine residues. Often, these modifications are enriched in CpG islands—high concentrations can result in silencing or reversible downregulation of gene expression.112 These methods are classified by their sodium bisulfite dependence, where dependent methods convert cytosine residues into sequencing-detectable uracil. Single-cell genome-wide and reduced-representation sequencing methods, which depend on bisulfite conversion, have been developed to capture varying breadths of the methylome.113, 114 In contrast, single cell CpG island methylation sequencing combines methylation-sensitive restriction digestions with multiple displacement amplification to generate a sequencing library enriched for loci associated with methylated CpG islands, while avoiding destructive bisulfite conversions.115 Other new methods include scAba-seq, which targets 5hmc and retains strand-specific information through bisulfite-independent, but glucosylation-dependent enzymatic reactions.116
Like single cell transcriptomes, epigenetic data from single cells can be used in human research, possibly to determine patient prognoses or select therapy. Bormann et al examined the CpG island methylator phenotype along with cell of origin signatures in colorectal tumor tissues and identified epigenetically defined subtypes of tumors that correlated with patient survival.117 Other tumor types have epigenetic heterogeneity along with functional heterogeneity. Litzenburger et al used scATAC-seq to demonstrate that differences in chromatin accessibility associated with sensitivity of cancer cell lines to drugs.118
Despite promise of single cell epigenomics, there is no integrated epigenomic method that could capture all levels of epigenetic modifications simultaneously. Nevertheless, as approaches for epigenomic analyses of single cells develop, we expect applications for gastrointestinal tissues and diseases.
Single-cell Metabolomics
Metabolites are small molecules (typically fewer than 2000 Da) including amino acids, sugars, lipids, small peptides, and break down products of drugs. These molecules can regulate cell structure, fuel, signaling, enzyme regulation, and pathogenesis. 119 It is a challenge to analyze metabolites because they are highly dynamic, 120 with vast structural diversity and many isomers.121 Single-cell analysis introduces additional demands because metabolites cannot be amplified, necessitating ultrasensitive analytical assays.
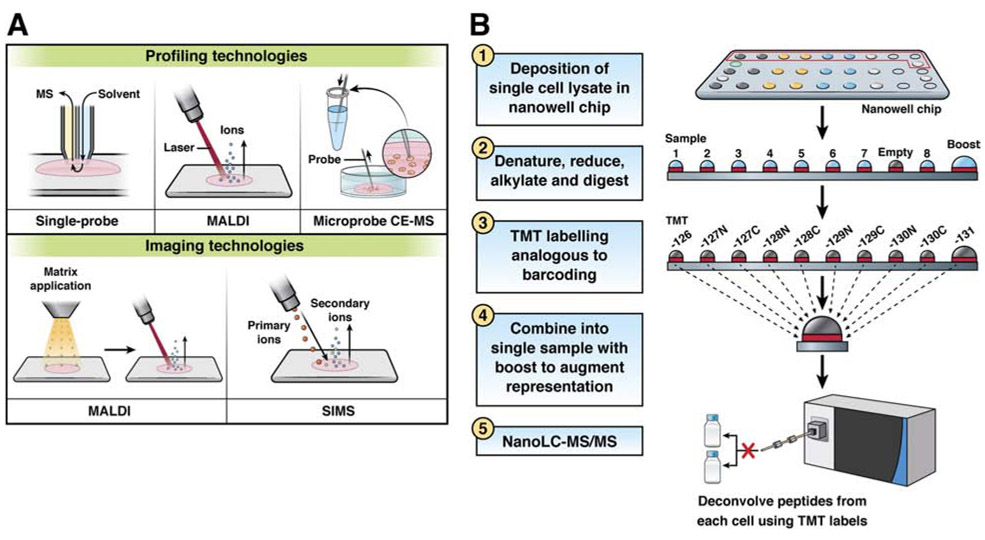
MS is the most widely used technology for analysis of single cell metabolomes.122 MS is label-free, untargeted (it can detect thousands of molecules concurrently), sensitive (fM limits of detection), and specific (it can discriminate molecules that differ by less than 0.001 Da).123 Single-cell MS analyses of metabolomes are typically performed as either profiling experiments, in which a single spectral signature is collected from each cell type, or as an imaging MS experiment to visualize metabolite distributions within a cell or across a tissue microenvironment (Figure 3A). In all cases, special care needs to be taken to ensure the metabolic profile of the cell is not perturbed during sample preparation because of the relatively high rate of turnover of some metabolites.124
Figure 3. MS-based Technologies for Analysis of Single Cells.
(A) Schematics of common single cell metabolomics profiling and imaging technologies.
(B) Workflow of the nanoPOTS-TMT-based single cell proteomics platform.
Most metabolomic profiling experiments of single cell types are performed on cell cultures or isolates from tissue. Targeted cell populations are separated by manual manipulation,125 fluorescence-activated cell sorting, microarray cell printing,126 or using microfluidic devices127 prior to MS analysis. There have been significant advances in development of technologies for rapid MS analysis of single cells in suspensions and on surfaces. For example, a single-probe MS sampling technology was developed to analyze live single cancer stem cells.128 The single-probe MS system uses a dual-bore capillary to deliver the lysis and extraction solvent to the cell surface and carry the molecular extract to the mass spectrometer for analysis by electrospray ionization. By bringing the single-probe tip directly in contact with individual cells, dispersed onto a glass slide, researchers were able to perform single cell metabolomic analyses of living cells with no sample preparation.
In a significant advancement in throughput, Neumann et al were able to perform MS lipid analysis on 30,000 individual cells using matrix-assisted laser desorption/ionization (MALDI).125 They achieved this by dissociating cells onto a slide and using a multimodal imaging approach to identify cells for MS analysis. Fluorescence microscopy was used to determine locations of intact cells for MALDI data acquisition. Using this technology, researchers detected more than 500 lipid features and were able to determine 101 significantly different cell clusters, using a combination of t-SNE and Louvain-Jaccard clustering. Others have been working to extend single cell metabolome profiling to more complex cell systems and tissues. For example, Portero et al coupled microsampling with capillary electrophoresis and electrospray ionization MS to enable in situ metabolite quantification in live cells and tissues.129
Recent advances in MS instrumentation and data processing have enabled imaging MS while maintaining spatial resolution.130, 131 The imaging MS experiment is typically performed on tissue sections, which were thawed and mounted onto MS compatible glass slides. Ions are produced at each position (pixel) within a designated region of the tissue resulting in the generation of a mass spectrum for each pixel. In a single imaging experiment, hundreds of metabolites are detected and are visualized as images by plotting the intensities for each molecule over array of pixels.
Although all imaging MS technologies are conceptually similar, in that they generate spatial molecular data, MALDI132 and secondary ion MS (SIMS)133, 134 are most commonly used for high spatial resolution of metabolomes. MALDI imaging is a laser-based surface analysis method in which the sample is coated with a light-absorbing matrix that assists in desorption and ionization of endogenous molecules. Spatial resolution is defined by the diameter of the focused laser beam at the sample surface. MALDI imaging is routinely performed at higher than 5 μm resolution; specialized setups can achieve 1 μm pixel sizes with good molecular coverage. SIMS imaging is similarly performed except that no matrix is used, and a tightly focused ion beam is used for surface sampling, allowing for pixel sizes down to 100 nm. However, SIMS typically induces molecular fragmentation during the sampling process, significantly complicating data interpretation.
Prentice et al used MALDI imaging MS to identify specific phospholipid and glycolipid isoforms in pancreatic islets.135 They performed serial imaging MS and immunofluorescence microscopy to correlate metabolite signals with specific cell types. Multimodal imaging studies have also been performed to combine MALDI and SIMS imaging for spatial metabolomics.136 This workflow was used to generate complimentary high spatial resolution SIMS and high molecular content MALDI images of human colon cancer tissue. The combination of SIMS and MALDI data can be used to determine localizations of lipids and cholesterol esters in regions of tumor cells, stroma, and necrosis.
Imaging and single cell metabolomic profiling are developing fields with rapidly advancing technologies focused on improving spatial resolution and molecular specificity. These innovations are leading to finer spatial fidelity, more complete molecular coverage, and ultimately a deeper understanding of the molecular characteristics and interactions between cells in tissue microenvironments.
Single-cell Proteomics
Cell structure and function are largely determined by the proteome—the complement of proteins expressed within the cell at a given time.137 Yet, a corresponding technology for unbiased and broad profiling of the protein expression at the single cell level has been lacking. Single cells contain only picogram total amounts of protein, and as no amplification strategy is available, effectively analyzing these trace samples poses an enormous challenge. Consistent improvements in instrumentation (such as liquid chromatography, electrospray ionization, and MS) have extended protein detection limits to the single cell level138, but conventional sample processing is incompatible with single cells due to physical challenges, such as nonspecific adsorption of proteins to the surfaces of well plates and inefficient digestion kinetics.
Recent innovations have helped us overcome this bottleneck, such than in-depth proteome profiling of single cells is now feasible. For example, a microfluidic approach to sample processing, called nanodroplet processing in 1 pot for trace samples (nanoPOTS),139 has reduced sample preparation volumes from 10s or 100s of microliters to approximately 200 nanoliters, greatly reducing surface exposure and corresponding protein losses and increasing protein concentrations for efficient digestion kinetics. This is accomplished using a robotic nanopipetting system for liquid handling, which is capable of subnanoliter dispensing accuracy and submicrometer positioning control. The nanopipettor interfaces with a microfabricated glass nanowell chip, which replaces the conventional well plate and reduces surface exposure in each well to approximately 1 mm2 Coupled with evaporation controlling mechanisms, highly sensitive and miniaturized nanoflow liquid chromatography, and latest-generation MS instruments, the nanoPOTS platform enables several hundred to more than 1000 protein groups to be identified in label-free, single cell analyses.
In addition to miniaturizing sample preparation and increasing the sensitivity of analytical instrumentation, researchers have analyzed single cells using tandem mass tag (TMT) barcoding, which allows multiple single cell proteomes to be determined in 1 liquid chromatography-MS analysis. This approach can increase measurement throughput and proteome coverage. Budnik et al first applied TMT labeling to the analysis of single mammalian cells with the SCoPE-MS workflow,140 in which isobarically labeled single cells were analyzed in the presence of a larger carrier sample comprising hundreds of cells. The peptide signals from the combined carrier and single cell samples facilitated identification, and peptides from each cell were differentiated based on their corresponding reporter ion intensities following MS/MS fragmentation. Importantly, TMT workflows have recently been combined with miniaturized sample processing to take advantage of improved sample processing efficiency and multiplexing (Fig. 3B). These advances have been applied to studies of single cells140, 141 and trace samples139, 142, including those relevant to the digestive system. 139, 142 For example, liver tissues were profiled with 50 μm resolution142, and 10-μm sections of individual pancreatic islets were studied from patients with and without type 1 diabetes 139. Researchers were able to compare the 2 proteomes at the single-islet level. Of the nearly 3000 proteins profiled from the islets, approximately 300 were found to differ significantly in level between the two groups.
Compared with single cell immunoblotting143, which is an antibody-based assay designed to analyze a small number of proteins (approximately 10) per cell, MS-based single cell proteomics can quantify nearly 1000 proteins/cell in an unbiased, label-free analysis. However, every cell essentially occupies its own liquid chromatography-MS run, so single cell proteomic analysis requires more time, coupled to automation for analysis. This is one of the barriers to MS-based methods, and keeps them from being widely adopted.
Analyses of metabolomes and proteomes of single cells can identify differences in functions among seemingly identical cells; this information might be used in development of therapeutic agents. However, technologies for broad proteome and metabolome profiling at and near the single cell level are in their infancy; improvements in coverage, measurement throughput, and automation are needed before they can used routinely in human research.
Integration of Multi-omic Technologies
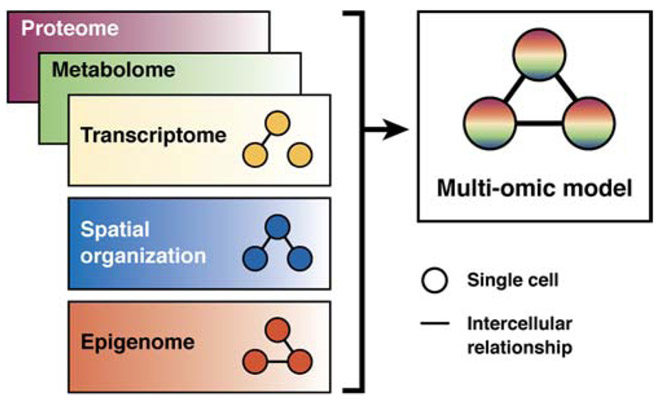
For a comprehensive picture of a cell, researchers require information on the genome, transcriptome, epigenome, and metabolome to be integrated (Fig. 4). Thus, comprehensive multi-omic profiling of single cells is the next frontier of development. Examples of current technologies include simultaneous profiling the transcriptome and chromatin-accessibility landscape144, 145, and the transcriptome plus a set of candidate protein markers.146 On the computational side, in silico multi-modal analytical frameworks permit exploration of phenomena indescribable by a single type of data alone and can often operate outside of the restrictions of multi-omic experimental methods. Frameworks such as Seurat v3 use distributions of cells collected by separate scRNA-seq and scATAC-seq experiments to co-embed cells into the same low-dimensional space. Further examples borrow other principles of multi-modal analysis, based on, but are not limited to, Bayesian modeling, graph theory, or deep learning. Respective examples include BREM-SC147, iOmics-PASS148, and SAUCIE149. Combinatorial multi- omics approaches could reveal different layers of heterogeneities that govern biology and disease phenotypes, but their prospect of being implemented in human research remains in question.
Figure 4.
Integration of Multi-omic Data
Future Directions
Although none of technologies for -omic analysis of single cells are ready for routine use in the clinic, many are used in human research and provide information about disease development and treatment. The degree by which each technology is adopted into clinical studies depends on its ease of use, precision, and reproducibility in generating data from minute amounts of material (Fig. 5). Some technologies, such as single cell proteomics, require sophisticated instruments and will probably be limited to specialized use in near term. Others, such as droplet-based scRNA-seq, is already available widely. A recent case study reported use of data from scRNA-seq analysis to guide treatment of drug-induced hypersensitivity syndrome.150 The next few years will be an exciting time as various atlasing efforts will test the limit of these technologies in generating data that can illuminate basic scientific knowledge and inform biomedical decisions.
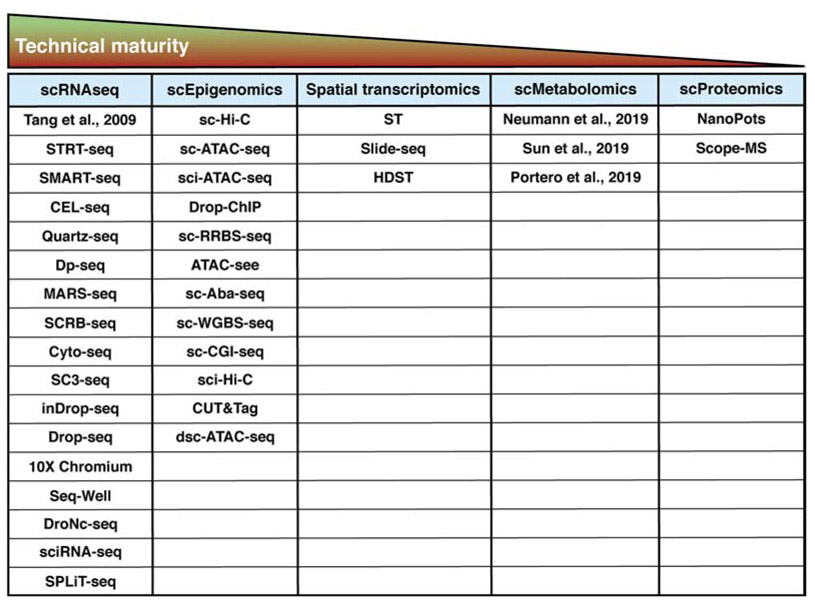
Figure 5. Technical Maturity of Single-cell -omic Technologies.
Technical maturity is the time it takes to run the samples, the equipment requirements for these techniques, and the personnel expertise necessary for successful application. Abbreviations and citations: STRT-seq (Single-cell Tagged Reverse Transcription, Islam et al (2011)); SMART-seq (Switching mechanism at 5′ end of the RNA transcript, Ramskold et al (2012)); CEL-seq (Cell expression by linear amplification and sequencing, Hashimshony et al (2012)); Quartz-seq (Sasagawa et al (2013)); Dp-seq (Designed Primer-based RNA-sequencing strategy, Bhargava et al (2013)); MARS-seq (Massively parallel RNA single cell sequencing, Jaitin et al (2014)); SCRB-seq (single cell RNA barcoding and sequencing, Soumillon et al (2014)); Cyto-seq (Gene expression cytometry, Fan et al (2015)); SC3-seq (single cell mRNA 3-prime end sequencing, Nakamura et al (2015)); inDrop-seq (Indexing Droplets sequencing, Klein et al (2015)); Drop-seq (Individual cell in Droplets, Macosko et al (2015)); 10X Chromium (Zheng et al (2017)); Seq-well (Sequencing in nanowell, Gierahn et al (2017)); DroNc-seq (Droplet based single-nuclear sequencing, Habib et al(2017)); sciRNA-seq (single cell combinatorial indexing RNA sequencing, Cao et al (2017)); SpLiT-seq (split-pool ligation-based transcriptome sequencing, Rosenberg et al (2017)); sc-Hi-C (Single-cell Hi-C, Nagano et al (2013)); sc-ATAC-seq (Single-cell assay for transposase-accessible chromatin using sequencing, Buenrostro et al (2015)); sci-ATAC-seq (single cell indexing ATAC-seq, Cusanovich et al (2015)); Drop-ChIP (Droplet based single cell ChIP sequencing, Rotem et al (2015)); sc-RRBS-seq (Single-cell reduced-representation bisulfite sequencing, Guo et al (2015)); ATAC-see (Assay for transposase accessible chromatin with visualization, Chen et al (2016)); sc-Aba-seq (Single-cell 5hmC sequencing by AbaSI nuclease, Mooijman et al (2016)); sc-WGBS-seq (Single-cell whole genome bisulfide sequencing, Gravina et al (2016)); sc-CGI-seq (CpG island (CGI) methylation sequencing for single cells, Han et al (2017)); sci-Hi-C (single cell combinatorial indexed Hi-C, Ramani et al (2019)); CUT&Tag (Cleavage Under Targets and Tagmentation, Kaya-Okur et al (2019)); dsc-ATAC-seq (droplet single cell assay for transposase-accessible chromatin using sequencing, Lareau et al (2019)); ST (Spatial Transcriptomics, Stahl et al (2016)); Slide-seq (Slide based spatial sequencing, Rodriques et al (2019)); HDST (High-Density Spatial Transcriptomics, Vickovic et al (2019)); NanoPOTS (nanodroplet processing in one pot for trace samples, Zhu et al (2018)); Scope-MS (Single Cell ProtEomics by Mass Spectrometry, Budnik et al (2018)).
Supplementary Material
Supplemental Table 1. RNA Library Preparation Methods for Analyses of Single Cells
Acknowledgements
K.S.L is funded by R01DK103831, the Human Tumor Atlas Network U2CCA233291, and the Leona M. and Harry B. Helmsley Charitable Trust Gut Cell Atlas G-1903-03793. K.S.L. and J.M.S are funded by HuBMAP U54DK120058. M.I. is supported by P50CA236733. B.C. is supported by T32LM012412. R.T.K. is funded by R33CA225248.
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu Y, Sethi NS, Hinoue T, et al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer cell 2018;33:721–735. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasaikar S, Huang C, Wang X, et al. Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities. Cell 2019;177:1035–1049. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 2008;135:216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature 2016;529:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim C, Gao R, Sei E, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 2018;173:879–893. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potter SS. Single-cell RNA sequencing for the study of development, physiology and disease. Nature Reviews Nephrology 2018;14:479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. The Lancet 2018;391:2128–2139. [DOI] [PubMed] [Google Scholar]
- 10.Greenbaum A, Martin DR, Bocklage T, et al. Tumor Heterogeneity as a Predictor of Response to Neoadjuvant Chemotherapy in Locally Advanced Rectal Cancer. Clinical colorectal cancer 2019;18:102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pectasides E, Stachler MD, Derks S, et al. Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer discovery 2018;8:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eng C-HL, Lawson M, Zhu Q, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 2019;568:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011;332:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodenmiller B, Zunder ER, Finck R, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nature biotechnology 2012;30:858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angelo M, Bendall SC, Finck R, et al. Multiplexed ion beam imaging of human breast tumors. Nature medicine 2014;20:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmons AJ, Banerjee A, McKinley ET, et al. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-α-induced apoptosis in vivo. Molecular systems biology 2015;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyler CJ, Pérez-Jeldres T, Ehinger E, et al. Implementation of mass cytometry as a tool for mechanism of action studies in inflammatory bowel disease. Inflammatory bowel diseases 2018;24:2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang L-S, Villaverde N, Hui KY, et al. A frameshift in CSF2RB predominant among Ashkenazi jews increases risk for Crohn's disease and reduces monocyte signaling via GM-CSF. Gastroenterology 2016;151:710–723. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin JC, Chang C, Boschetti G, et al. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 2019;178:1493–1508. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Y, Gao K, Zhang T, et al. A novel image registration approach via combining local features and geometric invariants. PloS one 2018;13:e0190383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goltsev Y, Samusik N, Kennedy-Darling J, et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 2018;174:968–981. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsujikawa T, Kumar S, Borkar RN, et al. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell reports 2017;19:203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gut G, Herrmann MD, Pelkmans L. Multiplexed protein maps link subcellular organization to cellular states. Science 2018;361:eaar7042. [DOI] [PubMed] [Google Scholar]
- 24.Remark R, Merghoub T, Grabe N, et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immunol 2016;1:aaf6925–aaf6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jungmann R, Avendaño MS, Woehrstein JB, et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nature methods 2014;11:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen KH, Boettiger AN, Moffitt JR, et al. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015;348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerdes MJ, Sevinsky CJ, Sood A, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proceedings of the National Academy of Sciences 2013;110:11982–11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ligorio M, Sil S, Malagon-Lopez J, et al. Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer. Cell 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinley ET, Sui Y, Al-Kofahi Y, et al. Optimized multiplex immunofluorescence single-cell analysis reveals tuft cell heterogeneity. JCI insight 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinley ET, Roland JT, Franklin JL, et al. Machine and deep learning single-cell segmentation and quantification of multi-dimensional tissue images. bioRxiv 2019:790162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J, Choi W, Tiesmeyer S, et al. Segmentation-free inference of cell types from in situ transcriptomics data. bioRxiv 2019:800748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Science translational medicine 2011;3:108ra113–108ra113. [DOI] [PubMed] [Google Scholar]
- 33.Fu Y, Jung AW, Torne RV, et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. bioRxiv 2019:813543. [DOI] [PubMed] [Google Scholar]
- 34.Baslan T, Hicks J. Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nature Reviews Cancer 2017;17:557. [DOI] [PubMed] [Google Scholar]
- 35.Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015;161:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nature communications 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picelli S, Faridani OR, Björklund ÅK, et al. Full-length RNA-seq from single cells using Smart-seq2. Nature protocols 2014;9:171. [DOI] [PubMed] [Google Scholar]
- 39.Islam S, Zeisel A, Joost S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nature methods 2014;11:163. [DOI] [PubMed] [Google Scholar]
- 40.Kolodziejczyk AA, Kim JK, Svensson V, et al. The technology and biology of single-cell RNA sequencing. Molecular cell 2015;58:610–620. [DOI] [PubMed] [Google Scholar]
- 41.Adam M, Potter AS, Potter SS. Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development 2017;144:3625–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Habib N, Avraham-Davidi I, Basu A, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nature methods 2017;14:955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slyper M, Porter CB, Ashenberg O, et al. A single-cell and single-nucleus RNA-seq toolbox for fresh and frozen human tumors. bioRxiv 2019:761429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bakken TE, Hodge RD, Miller JA, et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PloS one 2018;13:e0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gierahn TM, Wadsworth MH II, Hughes TK, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nature methods 2017;14:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han X, Wang R, Zhou Y, et al. Mapping the mouse cell atlas by microwell-seq. Cell 2018;172:1091–1107. e17. [DOI] [PubMed] [Google Scholar]
- 47.Goldstein LD, Chen Y-JJ, Dunne J, et al. Massively parallel nanowell-based single-cell gene expression profiling. BMC genomics 2017;18:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L, Duff MO, Graveley BR, et al. Genomewide characterization of non-polyadenylated RNAs. Genome biology 2011;12:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Islam S, Kjällquist U, Moliner A, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome research 2011;21:1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phipson B, Zappia L, Oshlack A. Gene length and detection bias in single cell RNA sequencing protocols. F1000Research 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashimshony T, Wagner F, Sher N, et al. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell reports 2012;2:666–673. [DOI] [PubMed] [Google Scholar]
- 52.Islam S, Kjällquist U, Moliner A, et al. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nature protocols 2012;7:813. [DOI] [PubMed] [Google Scholar]
- 53.Jaitin DA, Kenigsberg E, Keren-Shaul H, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 2014;343:776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziegenhain C, Vieth B, Parekh S, et al. Comparative analysis of single-cell RNA sequencing methods. Molecular cell 2017;65:631–643. e4. [DOI] [PubMed] [Google Scholar]
- 55.Wang YJ, Schug J, Lin J, et al. Comparative analysis of commercially available single-cell RNA sequencing platforms for their performance in complex human tissues. bioRxiv 2019:541433. [Google Scholar]
- 56.Herring CA, Chen B, McKinley ET, et al. Single-cell computational strategies for lineage reconstruction in tissue systems. Cellular and molecular gastroenterology and hepatology 2018;5:539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bray NL, Pimentel H, Melsted P, et al. Near-optimal probabilistic RNA-seq quantification. Nature biotechnology 2016;34:525. [DOI] [PubMed] [Google Scholar]
- 58.Satija R, Farrell JA, Gennert D, et al. Spatial reconstruction of single-cell gene expression data. Nature biotechnology 2015;33:495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome biology 2018;19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lvd Maaten, Hinton G. Visualizing data using t-SNE. Journal of machine learning research 2008;9:2579–2605. [Google Scholar]
- 61.McInnes L, Healy J, Melville J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426 2018. [Google Scholar]
- 62.Liu Q, Sheng Q, Ping J, et al. scRNABatchQC: multi-samples quality control for single cell RNA-seq data. Bioinformatics 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen B, Herring CA, Lau KS. pyNVR: investigating factors affecting feature selection from scRNA-seq data for lineage reconstruction. Bioinformatics 2018;35:2335–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Q, Herring CA, Sheng Q, et al. Quantitative assessment of cell population diversity in single-cell landscapes. PLoS biology 2018;16:e2006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Büttner M, Miao Z, Wolf FA, et al. A test metric for assessing single-cell RNA-seq batch correction. Nature methods 2019;16:43. [DOI] [PubMed] [Google Scholar]
- 66.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell 2012;149:1192–1205. [DOI] [PubMed] [Google Scholar]
- 67.Crosetto N, Bienko M, Van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nature Reviews Genetics 2015;16:57. [DOI] [PubMed] [Google Scholar]
- 68.Odze R Diagnostic problems and advances in inflammatory bowel disease. Modern pathology 2003;16:347. [DOI] [PubMed] [Google Scholar]
- 69.Alon S, Huynh GH, Boyden ES. Expansion microscopy: enabling single cell analysis in intact biological systems. The FEBS journal 2019;286:1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nichterwitz S, Chen G, Benitez JA, et al. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nature communications 2016;7:12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh S, Wang L, Schaff DL, et al. In situ 10-cell RNA sequencing in tissue and tumor biopsy samples. Scientific reports 2019;9:4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodriques SG, Stickels RR, Goeva A, et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019;363:1463–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ståhl PL, Salmén F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016;353:78–82. [DOI] [PubMed] [Google Scholar]
- 74.Vickovic S, Eraslan G, Klughammer J, et al. High-density spatial transcriptomics arrays for in situ tissue profiling. bioRxiv 2019:563338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haber AL, Biton M, Rogel N, et al. A single-cell survey of the small intestinal epithelium. Nature 2017;551:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hockley JR, Taylor TS, Callejo G, et al. Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut 2019;68:633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parikh K, Antanaviciute A, Fawkner-Corbett D, et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 2019;567:49. [DOI] [PubMed] [Google Scholar]
- 78.Aizarani N, Saviano A, Mailly L, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 2019;572:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.MacParland SA, Liu JC, Ma X-Z, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nature communications 2018;9:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gehart H, van Es JH, Hamer K, et al. Identification of enteroendocrine regulators by real-time single-cell differentiation mapping. Cell 2019;176:1158–1173. e16. [DOI] [PubMed] [Google Scholar]
- 81.Grün D, Lyubimova A, Kester L, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 2015;525:251. [DOI] [PubMed] [Google Scholar]
- 82.Herring CA, Banerjee A, McKinley ET, et al. Unsupervised trajectory analysis of single-cell RNA-seq and imaging data reveals alternative tuft cell origins in the gut. Cell systems 2018;6:37–51. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biton M, Haber AL, Rogel N, et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 2018;175:1307–1320. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu N, Sun H, Zhao X, et al. Map3k2-Regulated Intestinal Stromal Cells (MRISC) Define a Distinct Sub-cryptic Stem Cell Niche for Damage Induced Wnt Agonist R-spondin1 Production. bioRxiv 2019:723221. [Google Scholar]
- 85.Venema WTU, Voskuil MD, Vila AV, et al. Single-Cell RNA sequencing of blood and ileal T cells from patients with crohn's disease reveals tissue-specific characteristics and drug targets. Gastroenterology 2019;156:812–815. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roerink SF, Sasaki N, Lee-Six H, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 2018;556:457. [DOI] [PubMed] [Google Scholar]
- 87.Zhang L, Yu X, Zheng L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018;564:268. [DOI] [PubMed] [Google Scholar]
- 88.Bian S, Hou Y, Zhou X, et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science 2018;362:1060–1063. [DOI] [PubMed] [Google Scholar]
- 89.Zhang P, Yang M, Zhang Y, et al. Dissecting the Single-Cell Transcriptome Network Underlying Gastric Premalignant Lesions and Early Gastric Cancer. Cell reports 2019;27:1934–1947. e5. [DOI] [PubMed] [Google Scholar]
- 90.Owen RP, White MJ, Severson DT, et al. Single cell RNA-seq reveals profound transcriptional similarity between Barrett’s oesophagus and oesophageal submucosal glands. Nature communications 2018;9:4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng C, Zheng L, Yoo J-K, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017;169:1342–1356. e16. [DOI] [PubMed] [Google Scholar]
- 92.Galon J, Mlecnik B, Bindea G, et al. Towards the introduction of the ‘Immunoscore’in the classification of malignant tumours. The Journal of pathology 2014;232:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mlecnik B, Bindea G, Angell HK, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016;44:698–711. [DOI] [PubMed] [Google Scholar]
- 94.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology 2008;134:577–594. [DOI] [PubMed] [Google Scholar]
- 95.Smillie CS, Biton M, Ordovas-Montanes J, et al. Intra-and Inter-cellular rewiring of the human colon during ulcerative colitis. Cell 2019;178:714–730. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kinchen J, Chen HH, Parikh K, et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 2018;175:372–386. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li H, Courtois ET, Sengupta D, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nature genetics 2017;49:708. [DOI] [PubMed] [Google Scholar]
- 98.Sarobe P, Corrales F. Getting insights into hepatocellular carcinoma tumour heterogeneity by multiomics dissection. Gut 2019;68:1913–1914. [DOI] [PubMed] [Google Scholar]
- 99.Wang X, Park J, Susztak K, et al. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nature communications 2019;10:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome biology 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nature biotechnology 2019;37:773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nature Reviews Genetics 2015;16:716. [DOI] [PubMed] [Google Scholar]
- 103.D’Urso A, Brickner JH. Mechanisms of epigenetic memory. Trends in genetics 2014;30:230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cusanovich DA, Daza R, Adey A, et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015;348:910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Buenrostro JD, Wu B, Litzenburger UM, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015;523:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lareau CA, Duarte FM, Chew JG, et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nature Biotechnology 2019:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nagano T, Lubling Y, Stevens TJ, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013;502:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ramani V, Deng X, Qiu R, et al. Sci-Hi-C: a single-cell Hi-C method for mapping 3D genome organization in large number of single cells. Methods 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen X, Shen Y, Draper W, et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. nAture methods 2016;13:1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rotem A, Ram O, Shoresh N, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nature biotechnology 2015;33:1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kaya-Okur HS, Wu SJ, Codomo CA, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nature communications 2019;10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Karemaker ID, Vermeulen M. Single-cell DNA methylation profiling: technologies and biological applications. Trends in biotechnology 2018;36:952–965. [DOI] [PubMed] [Google Scholar]
- 113.Gravina S, Dong X, Yu B, et al. Single-cell genome-wide bisulfite sequencing uncovers extensive heterogeneity in the mouse liver methylome. Genome biology 2016;17:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guo H, Zhu P, Guo F, et al. Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nature protocols 2015;10:645. [DOI] [PubMed] [Google Scholar]
- 115.Han L, Wu H-J, Zhu H, et al. Bisulfite-independent analysis of CpG island methylation enables genome-scale stratification of single cells. Nucleic acids research 2017;45:e77–e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mooijman D, Dey SS, Boisset J-C, et al. Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nature biotechnology 2016;34:852. [DOI] [PubMed] [Google Scholar]
- 117.Bormann F, Rodríguez-Paredes M, Lasitschka F, et al. Cell-of-origin DNA methylation signatures are maintained during colorectal carcinogenesis. Cell reports 2018;23:3407–3418. [DOI] [PubMed] [Google Scholar]
- 118.Litzenburger UM, Buenrostro JD, Wu B, et al. Single-cell epigenomic variability reveals functional cancer heterogeneity. Genome biology 2017;18:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 2016;17:451–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weibel KE, Mor JR, Fiechter A. Rapid sampling of yeast cells and automated assays of adenylate, citrate, pyruvate and glucose-6-phosphate pools. Anal Biochem 1974;58:208–16. [DOI] [PubMed] [Google Scholar]
- 121.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proc Natl Acad Sci U S A 2015;112:12549–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Duncan KD, Fyrestam J, Lanekoff I. Advances in mass spectrometry based single-cell metabolomics. Analyst 2019;144:782–793. [DOI] [PubMed] [Google Scholar]
- 123.Cornett DS, Frappier SL, Caprioli RM. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal Chem 2008;80:5648–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lu W, Su X, Klein MS, et al. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annu Rev Biochem 2017;86:277–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neumann EK, Ellis JF, Triplett AE, et al. Lipid Analysis of 30000 Individual Rodent Cerebellar Cells Using High-Resolution Mass Spectrometry. Anal Chem 2019;91:7871–7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ellis SR, Ferris CJ, Gilmore KJ, et al. Direct lipid profiling of single cells from inkjet printed microarrays. Anal Chem 2012;84:9679–83. [DOI] [PubMed] [Google Scholar]
- 127.Tan WH, Takeuchi S. A trap-and-release integrated microfluidic system for dynamic microarray applications. Proc Natl Acad Sci U S A 2007;104:1146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun M, Yang Z. Metabolomic Studies of Live Single Cancer Stem Cells Using Mass Spectrometry. Anal Chem 2019;91:2384–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Portero EP, Nemes P. Dual cationic-anionic profiling of metabolites in a single identified cell in a live Xenopus laevis embryo by microprobe CE-ESI-MS. Analyst 2019;144:892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Spraggins JM, Djambazova KV, Rivera ES, et al. High-Performance Molecular Imaging with MALDI Trapped Ion-Mobility Time-of-Flight (timsTOF) Mass Spectrometry. Anal Chem 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Niehaus M, Soltwisch J, Belov ME, et al. Transmission-mode MALDI-2 mass spectrometry imaging of cells and tissues at subcellular resolution. Nat Methods 2019;16:925–931. [DOI] [PubMed] [Google Scholar]
- 132.Berry KA, Hankin JA, Barkley RM, et al. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem Rev 2011;111:6491–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Heien ML, Piehowski PD, Winograd N, et al. Lipid detection, identification, and imaging single cells with SIMS. Methods Mol Biol 2010;656:85–97. [DOI] [PubMed] [Google Scholar]
- 134.Bruinen AL, Fisher GL, Balez R, et al. Identification and High-Resolution Imaging of alpha-Tocopherol from Human Cells to Whole Animals by TOF-SIMS Tandem Mass Spectrometry. J Am Soc Mass Spectrom 2018;29:1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Prentice BM, Hart NJ, Phillips N, et al. Imaging mass spectrometry enables molecular profiling of mouse and human pancreatic tissue. Diabetologia 2019;62:1036–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Desbenoit N, Walch A, Spengler B, et al. Correlative mass spectrometry imaging, applying time-of-flight secondary ion mass spectrometry and atmospheric pressure matrix-assisted laser desorption/ionization to a single tissue section. Rapid Commun Mass Spectrom 2018;32:159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Saghatelian A, Cravatt BF. Assignment of protein function in the postgenomic era. Nature Chemical Biology 2005;1:130–142. [DOI] [PubMed] [Google Scholar]
- 138.Li SY, Plouffe BD, Belov AM, et al. An Integrated Platform for Isolation, Processing, and Mass Spectrometry-based Proteomic Profiling of Rare Cells in Whole Blood. Molecular & Cellular Proteomics 2015;14:1672–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhu Y, Piehowski PD, Zhao R, et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10-100 mammalian cells. Nature Communications 2018;9:882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Budnik B, Levy E, Harmange G, et al. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol 2018;19:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhu Y, Clair G, Chrisler WB, et al. Proteomic Analysis of Single Mammalian Cells Enabled by Microfluidic Nanodroplet Sample Preparation and Ultrasensitive NanoLC-MS. Angewandte Chemie-International Edition 2018;57:12370–12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu KR, Liang YR, Piehowski PD, et al. Benchtop-compatible sample processing workflow for proteome profiling of < 100 mammalian cells. Analytical and Bioanalytical Chemistry 2019;411:4587–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hughes AJ, Spelke DP, Xu Z, et al. Single-cell western blotting. Nature methods 2014;11:749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen S, Lake BB, Zhang K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nature biotechnology 2019;37:1452–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu L, Liu C, Quintero A, et al. Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity. Nature communications 2019;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mimitou EP, Cheng A, Montalbano A, et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nature methods 2019;16:409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang X, Sun Z, Zhang Y, et al. BREM-SC: A Bayesian Random Effects Mixture Model for Joint Clustering Single Cell Multi-omics Data. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Koh HW, Fermin D, Vogel C, et al. iOmicsPASS: network-based integration of multiomics data for predictive subnetwork discovery. NPJ systems biology and applications 2019;5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Amodio M, Van Dijk D, Srinivasan K, et al. Exploring single-cell data with deep multitasking neural networks. Nature methods 2019:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim D, Kobayashi T, Voisin B, et al. Targeted therapy guided by single-cell transcriptomic analysis in drug-induced hypersensitivity syndrome: a case report. Nature Medicine 2020:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. RNA Library Preparation Methods for Analyses of Single Cells