Summary
Available 3D structures of bacteriophage modules combined with predictive bioinformatic algorithms enabled the identification of adhesion modules in 57 siphophages infecting Streptococcus thermophilus (St). We identified several carbohydrate‐binding modules (CBMs) in so‐called evolved distal tail (Dit) and tail‐associated lysozyme (Tal) proteins of St phage baseplates. We examined the open reading frame (ORF) downstream of the Tal‐encoding ORF and uncovered the presence of a putative p2‐like receptor‐binding protein (RBP). A 21 Å resolution electron microscopy structure of the baseplate of cos‐phage STP1 revealed the presence of six elongated electron densities, surrounding the core of the baseplate, that harbour the p2‐like RBPs at their tip. To verify the functionality of these modules, we expressed GFP‐ or mCherry‐coupled Tal and putative RBP CBMs and observed by fluorescence microscopy that both modules bind to their corresponding St host, the putative RBP CBM with higher affinity than the Tal‐associated one. The large number of CBM functional domains in St phages suggests that they play a contributory role in the infection process, a feature that we previously described in lactococcal phages and beyond, possibly representing a universal feature of the siphophage host‐recognition apparatus.
We found that for each streptococcal phage several CBMs are present in the virions and are involved in phage adhesion. Moreover, we uncovered an additional ORF that harbours a predicted Receptor Binding Protein (RBP) at its C‐terminus. To confirm these predictions, we determined by electron microscopy the structure of streptococcal phage STP1 baseplate and assessed the binding capability of the CBM and the RBP‐like domain to their specific host using fluorescent binding assays.
Introduction
The dairy fermentation industry depends on robust starter cultures of lactic acid bacteria (LAB). Strains of Lactococcus lactis and Streptococcus thermophilus are the most widely utilized in dairy starter cultures and are consistently under the threat of bacteriophage (or phage) infection (Garneau and Moineau, 2011; Lavelle, et al., 2018). These phages may cause fermentation disruptions and product inconsistencies that are among the primary causes of economic losses in the associated dairy fermentation industry. This phage‐mediated financial threat has been a constant catalyst for very substantial scientific efforts to isolate and characterize LAB phages, which as a result, are among the most intensely studied phage groups (Brüssow, 2018).
The first step in phage infection is the recognition and binding of the phage to its bacterial host. Tailed phages (Caudovirales order) recognize and bind to their host utilizing a unique adhesion device at the distal end of their tail, referred to as the baseplate (Veesler, et al., 2012; Spinelli, et al., 2014; Mahony, et al., 2017; Dowah and Clokie, 2018; Dunne et al., 2018) or tail tip (Plisson, et al., 2007; Chaban, et al., 2015; Arnaud, et al., 2017). In addition to extensive studies on host binding devices and mechanisms of Myoviridae and Podoviridae phages (Xiang, et al., 2009; Taylor, et al., 2016), large adhesion structures of certain Siphoviridae phages have also been resolved in recent years. Among these are the distinct baseplates of lactococcal P335 group phage TP901‐1 (Veesler, et al., 2012) and that of the 936‐group phage p2 (Sciara, et al., 2010), both of which are located at the tip of their tail as a hetero‐oligomeric association of at least three proteins, that are typically encoded in the following genetic order: Dit (Distal tail protein) (Veesler, et al., 2010; Flayhan, et al., 2014), Tal (tail‐associated lysin) and RBP (receptor‐binding protein) (Spinelli, et al., 2006; Spinelli, et al., 2006; Sciara, et al., 2010; Veesler, et al., 2012), together with a variable number of accessory proteins. These devices recognize and bind to very particular host cell surface‐located polysaccharide (Mahony, et al., 2013; Farenc, et al., 2014). In contrast, other siphophages including lactococcal phage c2 (Valyasevi, et al., 1991), Bacillus phage SPP1 (Vinga, et al., 2012) and coliphage T5 bind to membrane‐embedded protein receptors (Breyton, et al., 2013).
Phage binding to a host is presumed to involve a unique protein, the so‐called RBP. This dogma, however, has recently been challenged: a Lactobacillus phage (Dieterle, et al., 2016; Dieterle, et al., 2017) and a large number of lactococcal 936 phages incorporate so‐called ‘evolved’ Dit proteins in their tail tip that specifically bind to the host surface (Hayes, et al., 2018). These extended Dits possess one or two internal insertion(s) that exhibit structural similarity to the BppA carbohydrate‐binding modules (CBMs) (accessory baseplate protein; PDB ID: 5E7T), which represents an ancillary baseplate protein of the baseplate of lactococcal phage Tuc2009, being distinct from the RBP, yet involved in host binding (Collins, et al., 2013; Legrand, et al., 2016). Using HHpred analysis and subsequent experimental verification, we then determined that the vast majority of lactococcal 936 phages incorporate, in addition to the aforementioned Dit proteins, further CBMs into their major tail proteins (MTP), forming MTP extensions, and/or in their neck passage proteins (NPS) (Hayes, et al., 2019). All these CBMs are involved in host adhesion, together with the bona fide RBP, and exhibit a binding activity that supports the same host specificity (Hayes, et al., 2018; Hayes, et al., 2019). On the other hand, just a few 936‐group phages (e.g. p2 and sk1) do not contain additional CBMs, while phage MP1 is unique as it contains all three CBM decorations in addition to the bona fide RBP (Hayes, et al., 2019).
Streptococcus thermophilus phages represent an industrially significant group of phages that have been thoroughly studied in recent years (Mahony, et al., 2012; McDonnell, et al., 2016; McDonnell, et al., 2017; Lavelle, et al., 2018). As more genome sequences have become available, it has become clear that various phage–host interactions may be in operation that warrant closer inspection. In the current work, we sought to determine whether adhesion mechanisms similar to those observed in the 936‐group lactococcal phages are operational in S. thermophilus phages. To this end, we examined in all four currently described groups of streptococcal phages (the so‐called cos, pac (Brussowvirus genus), 5093 and 987 group phages) the occurrence of CBMs in the amino acid sequences of deduced baseplate components, namely Dit and Tal. We also analysed the product of the open reading frame (ORF) immediately following the tal gene in these genomes and found that it encodes a p2/TP901‐1 RBP‐like module, making it a putative RBP. It was identified that, in most cases, Dit and Tal from a given S. thermophilus phage incorporate CBMs, beyond the putative RBP. We determined the 21 Å resolution electron microscopy (EM) structure of cos phage STP1 in order to localize the various components of the baseplate and identify for the first time a peripheral structure that we assigned to a putative RBP, with a p2 RBP‐like domain at its tip. To verify the functionality of these domains, we expressed GFP‐ or mCherry‐coupled Tal CBM and RBP module and observed by fluorescence microscopy that both modules bind to their corresponding streptococcal host, though the RBP module does this with a higher affinity than the Tal‐associated CBM.
Results
Bioinformatic analyses
We selected a number of ORFs that encode products corresponding to tail structural proteins from available genome sequences of representatives of the four known phage groups that infect Streptococcus thermophilus, namely cos, pac, 987 and 5093 (Lavelle, et al., 2018; Lavelle, et al., 2018). These ORFs correspond to genes located downstream of the gene encoding the tape measure protein (TMP), namely those specifying the Dit and Tal proteins, which will be discussed below (Fig. 1A) (Sciara, et al., 2010; Veesler, et al., 2010; Veesler and Cambillau, 2011). Additionally, we assessed other ORFs within the morphogenesis module of the investigated streptococcal phage genomes, in particular those that encode the MTP. However, no CBM decorations were identified, being in contrast to the situation in various members of the lactococcal 936 family which typically do contain a CBM in their MTP proteins (Hayes, et al., 2019).
Fig. 1.
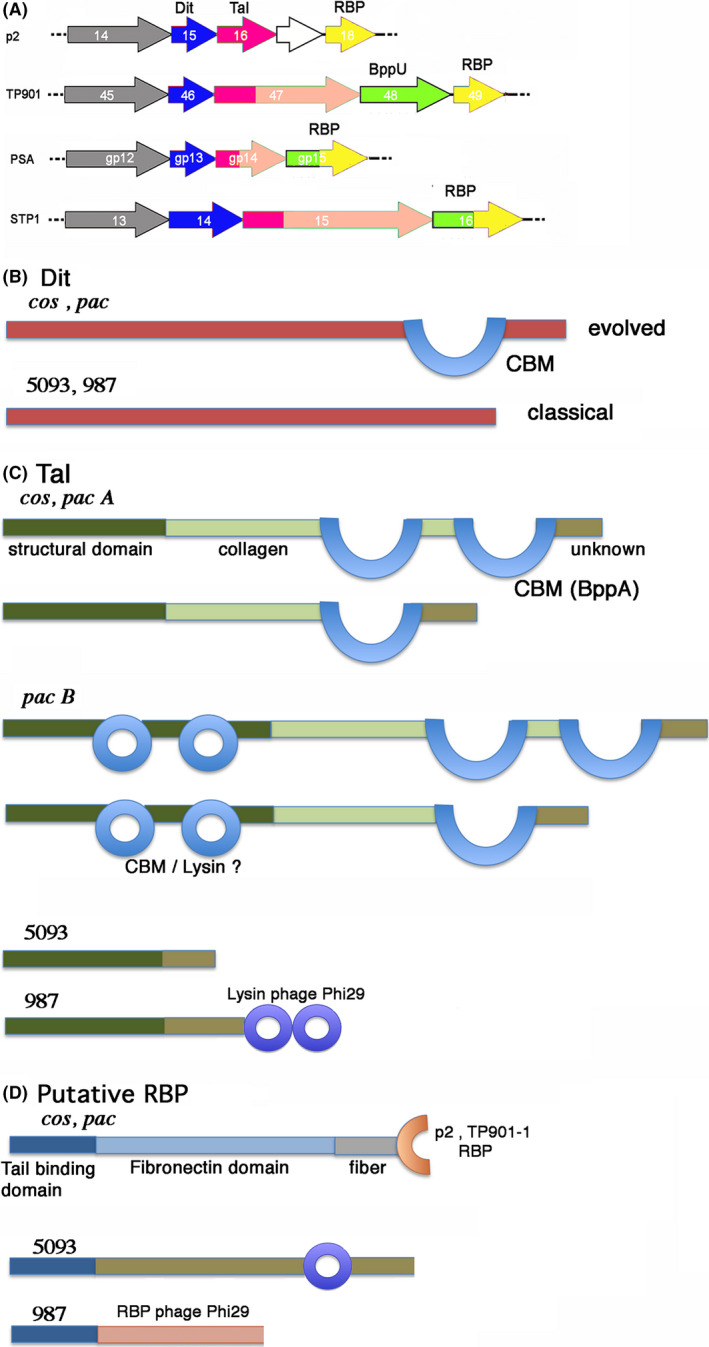
Schematic depiction of the Dit, Tal and putative RBP of S. thermophilus phages based on HHpred analysis.
A. Schematic representation of genes coding for the adhesion device of representative phages: p2, TP901‐1, PSA and STP1. TMPs are colour‐coded grey; Dits are colour‐coded blue; Tals are colour‐coded dark pink (Nt‐structural domain), light pink (Tal extension), RBPs are colour coded yellow; ancillary proteins and tail fibres are colour‐coded green. White genes are non‐structural. PSA and STP1 have chimeric RBPs with a BppU like N‐terminus.
B. The insertions observed by HHpred in cos and pac phages 'evolved' Dits correspond to Lactobacillus phage J1 inserted CBM domain, in blue (Dieterle, et al., 2017). The 5093 and 987 Dits do not possess any insertion and are hence 'classical'. B – The topology observed by HHpred in Tals: the structural N‐terminal domain is dark green; the collagen extension is light green; the BppA‐like CBM is blue, and the C‐terminal unknown domain is brown. The cos and pac phages Tals have variable lengths and bear 1 or 2 CBM insertions. The pac N‐terminal domain (dark green) harbours two insertions with lysin‐like folds identified by HHpred, casting doubt that it might not be a CBM. The 5093 phages possess only an unknown domain after the N‐terminal domain. The 987 phages possess a C‐terminal Lysin domain.
C. The putative RBPs HHpred analysis in cos and pac phages identified a N‐terminal tail binding domain (dark blue), a fibronectin‐like extension (light blue) and a C‐terminal domain resembling the RBPs of lactococcal phages p2 or TP901‐1. 5093 and 987 phages possess also a N‐terminal tail binding domain (dark blue), followed by an unknown linker and a lysin doman (5093) or a phage Phi29 RBP (987).
Distal tail proteins (Dits)
Sequence analysis of the Dits from cos and pac phages revealed that they in both cases represent evolved Dits as their length, 526 and 520 amino acids, respectively, exceed those of classical Dits (< 300 aa; Fig. S1A,B). Conversely, those specified by the 5093 and 987 phages are truly classical Dits based on their length of 239 and 253 amino acids, respectively (Fig. S1C). HHpred analysis of Dits encoded by cos and pac phages indeed revealed that they contain an insertion (residues 206–454) and that this insertion possesses the fold of a CBM similar to that present in Lactobacillus casei phage J1 (5LY8; probability > 99%; 18% identity) (Dieterle, et al., 2017), while HHpred analysis of 5093 and 987 Dits suggests that they do not contain any insertion (Fig. 1B, Fig. S2).
Tail‐associated lysins (Tals)
The Tals are the most complex and heterogeneous elements of streptococcal virions and are described by group below.
The cos group
The Tals encoded by cos phages range in size from 860 to 1450 residues (Fig. S3A). HHpred analysis showed that they all comprise the ~400 residues long classical gp27‐like structural domain within their N‐terminal region, similar to the situation in lactococcal phages TP901‐1 and p2, among others (Fig. 1C, Fig. S4A) (Bebeacua, et al., 2010; Sciara, et al., 2010; Veesler and Cambillau, 2011). This gp27‐like domain is followed by an all β‐stranded domain, likely representing a triple helix collagen‐like domain varying in length between ~100 and 350–400 residues followed by insertions of one or two CBM domains (Fig. S4A, Fig. 1C). The ~180–200 residues at the C‐terminus can be dissected in a probable collagen‐like linker (100 aa) and an unknown domain of ~80–100 aa (Fig. S4A, Fig. 1C). The CBM domains are identified by HHpred mainly as folds that resemble the BppA CBM of lactococcal phage Tuc2009 (PDB ID: 5E7T) (Legrand, et al., 2016).
The pac group
The Tals encoded by pac phages vary in size between 890 and 2020 residues (Fig. S3B). They can be divided into two sub‐groups, based on the structure of their N‐terminal domain (Fig. S4B). The first group (designated here as pac Tal group A) comprises Tals with the classical ~400 residues N‐terminus gp27‐like structural domain (Fig. S4Ba, Fig. 1C) followed by a 100 to 500 aa long collagen‐like domain and the insertion of one or two BppA‐like CBMs (or related CBMs). This is then followed by a short collagen linker and a ~100 aa long C‐terminal domain of unknown fold. The topology of these Tals resembles that of the cos group (Fig. 1C).
In the second pac Tal group, termed here the pac Tal group B, the N‐terminal domain of these Tals exhibits a to‐date unique feature: the N‐terminus structural domain exhibits two CBM domains inserted within two loops of its classical gp27 fold, leading to a completely ‘decorated’ N‐terminal domain of ~800 residues (Fig. 1C, Figs S3B and S4B). These insertions do not possess the BppA fold, but folds that resemble that found in two hydrolases from Sphingomonas and Streptococcus (PDB IDs 2zyc and 4f88). It remains to be determined, however, whether these domains are hydrolases or CBMs. This decorated structural domain is followed by a long stretch similar to that found in the cos Tals and pac Tal group A (see above): a 100 to 500 aa long collagen‐like domain which is followed by two BppA‐like domains, a short collagen linker and a ~100 aa C‐terminal domain of unknown fold (Fig. 1C).
Of note, three Tals do not follow this scheme, i.e. those of phages O1205, Sfi11 and TPJ‐34. Three unassigned ORFs lie downstream of these Tals. When the sequences of these ORFs are assembled together with that of the annotated Tal, HHpred reports a structural pattern that is similar to those of members of the pac Tal group B. This observation suggests that sequencing errors may have occurred or that introns localized in the Tal sequences were not taken into consideration.
The 5093 and 987 groups
Tals from the 5093 group are 508 residues long and exhibit 97.4% identity to each other (Fig. S5), yet substantially differ from the cos and pac Tals (Fig. 1C). HHpred returns a ~400 residues N‐terminal gp27‐like domain followed by ~100 residues of an unknown fold. In the case of the 987 phages, the Tal N‐terminal structural domain is followed by a ~200 amino acid long domain of unknown fold that links it to a ~300 amino acid domain resembling the cell wall degrading enzyme found in the tail of phage Phi29 (Xiang, et al., 2008).
Putative RBPs encoded by the ORF located downstream of tal
The components that constitute the baseplate of lactococcal phages are encoded in the order Dit ‐ Tal ‐ RBP, with occasionally additional ORFs inserted between the genes encoding Tal and RBP, for example orf17 in phage p2, bppU (upper baseplate protein) in phages TP901‐1 (Veesler, et al., 2012) and bppU + bppA (accessory baseplate protein) in phage Tuc2009 (Fig. 1A) (Legrand, et al., 2016). We therefore analysed the ORFs that are located downstream of the Tal‐encoding gene as their location in the phage genome may follow functional synteny and therefore may encode a component of the baseplate, possibly the RBP. Of note, the topology and possible function of these genes have not been examined previously.
The cos and pac groups
The proteins encoded by the ORFs after Tal, the putative RBPs, from members of the cos and pac phage groups are very similar as shown by sequence alignments (Fig. S6A) and HHpred analyses (Fig. S7A, Fig. 1D). HHpred could not identify a hit for the ~140 N‐terminal amino acids. We hypothesize that this N‐terminal domain may possess a fold that is close to that of the BppU N‐terminal domain, as observed for 987 phages (see below). Following this N‐terminus, a ~300 residue domain is identified by HHpred as a series of ß‐stranded domains (eg fibronectin3‐like domains). This is then followed by a ~70 amino acids β‐stranded linker domain and a ~100 amino acids C‐terminal domain resembling the RBP head domain of listeria phage PSA (Dunne, et al., 2019) and of lactococcal phages TP901‐1 and p2 (Spinelli, et al., 2006; Spinelli, et al., 2006).
The 987 and 5093 groups
The putative RBPs of members of the 987 group also encode very similar proteins (98.5% identity; Fig. S6B). HHpred analysis reports that the 128 first residues possess the fold of the Tuc2009 BppU N‐terminal domain (Fig. 1D, Fig. S7B). It is followed by a short linker of ~100 residues and by a domain of ~450 residues resembling the phage Phi29 RBP (Fig. S7B). Noteworthy, we have reported above that the 987 Tals resemble the phage Phi29 tail tip.
The putative RBPs ORFs are highly conserved in the 5093 group and significantly longer that those present in members of the cos, pac and 987 phage groups (Fig. 1D, Fig. S6B). Those from phages 0091‐0094 are almost identical (Fig. S6B). An equivalent ORF could not be identified in phage 5093. However, when grouping the sequences of its ORFs 30, 31 and 32, the resulting sequence is found to be very close to those of the other members of the group (Fig. S6B). However, HHpred analysis returned very limited data, as only an esterase fold is identified between residues 513 and 736 (Fig. S7B).
Schematic models of the baseplates of S. thermophilus phages
The general topology and stoichiometry of Siphoviridae infecting LAB is now well documented (Sciara, et al., 2010; Veesler, et al., 2010; Veesler and Cambillau, 2011; Veesler, et al., 2012; Dieterle, et al., 2017). Dit is a hexamer bound to a tail‐associated MTP hexamer on one side and to a Tal trimer on the other side. The RBPs and other decorating proteins are trimeric and peripheral. Based on this knowledge and our bioinformatic analyses, schematic models of the Dit‐Tal‐RBP assembly can be put forward. For cos and pac phages, Dits are evolved and insert one (Cos, Pac‐A) or two (Pac‐B) CBMs, thereby resembling the Dit of Lactobacillus casei phage J1 (Dieterle, et al., 2017) (Fig. 2A). In contrast, Dits from the 5093 and 987 groups are classical (Fig. 2B,C). In Siphoviridae, Tals are attached to and positioned ‘below’ the Dits (i.e. distal from the capsid) (Sciara, et al., 2010; Veesler and Cambillau, 2011; Spinelli, et al., 2014). The Tal protein attaches to the Dit hexameric ring as a trimer through their conical N‐terminal domain which is highly conserved among siphophages and beyond (e.g. Myoviridae T4 (Taylor, et al., 2016) and Mu (Buttner, et al., 2016)) (Fig. 2A). Noteworthy, this Tal‐associated N‐terminal domain is ‘evolved’ in the Pac Tal B group, which appears to be a unique feature to date among siphophages (Fig. 2A‐insert). The remainder of the protein forms an elongated fibre‐like domain containing one or two CBM domains, with a fold resembling that of Tuc2009 BppA (Legrand, et al., 2016) (Fig. 2A). In comparison, the Tals of the 5093 and 987 groups are simpler: they assemble the N‐terminal structural domain followed by a short, unknown domain (5093) or by a Phi29 phage hydrolase‐like domain (987) (Xiang, et al., 2008) (Fig. 2B,C).
Fig. 2.
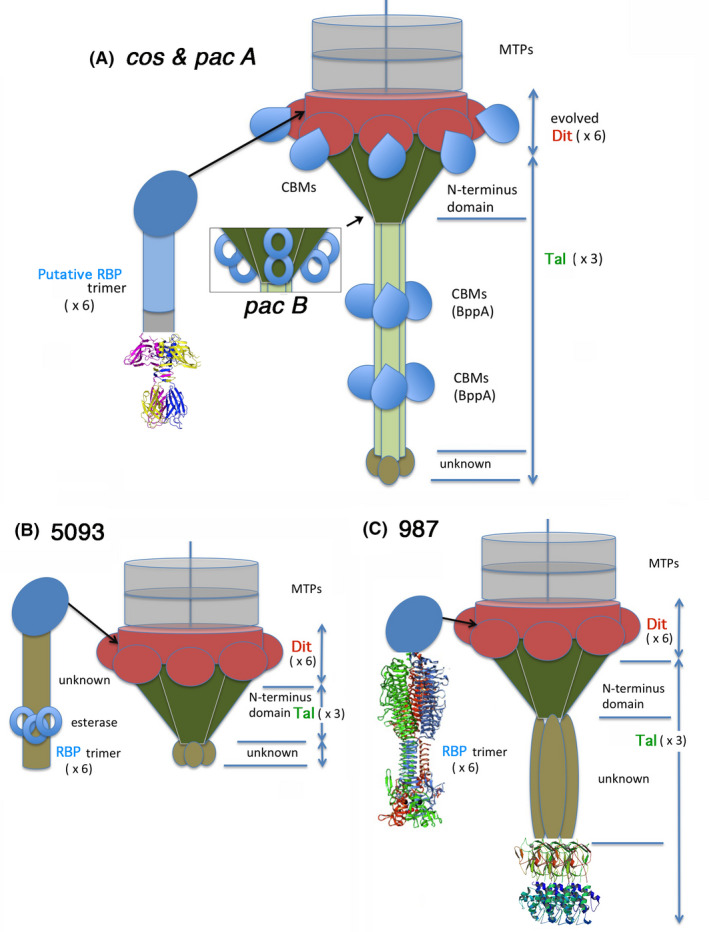
Putative schematic model of the baseplate structure of S. thermophilus phages.
A. The cos and pac phages. The Dit hexameric structural ring and galectin domains (top) are coloured red. The Dit CBM insertions are coloured blue. The trimeric Tal structural N‐terminal domain is coloured dark green. The trimeric Tal extensions are light green and the BppA insertions are blue. The unknown terminal domain is brown. In pac phages (insert), two insertions are observes in the Tal structural domain (blue rings). The putative RBP N‐terminal domain is depicted as a dark blue oval. A fibronectin extension (ligh blue) joins it to the p2‐like C‐terminal domain depicted with ribbons.
B. The 5083 phages. The Dit hexameric structural ring and galectin domains (top) are coloured red and do not exhibit insertions. The trimeric Tal structural domain, dark green, has no insertions. It is followed by a unknown domain (brown). The putative RBP N‐terminal domain is depicted as a dark blue oval is followed by an unknown domain harbouring esterases.
C. The 987 phages. The Dit hexameric structural ring and galectin domains (top) are coloured red and do not exhibit insertions. The trimeric Tal structural domain, dark green, has no insertions. It is followed by a unknown domain (brown) and a hydrolase tandem at the C‐terminus. The putative RBP N‐terminal domain is depicted as a dark blue oval is followed by a receptor‐binding domain.
In comparison to the Dit/Tal complexes, modelling of the putative RBPs within the baseplates of cos and pac group phages proved to be challenging. In cos and pac phages, the ~140 amino acid N‐terminus was not identified by HHpred. It is followed by a ß‐stranded domain and a TP901‐1/p2 RBP‐head like domain. Jpred secondary structure predictor reports an all ß‐stranded ORF. These ORFs have been shown to be present in the virion and should be attached close to the baseplate Dit/Tal core. The previously reported EM pictures reveal, however, a bulky baseplate deprived of any lateral features (see figure 7 in reference (Lavelle, et al., 2018)). Since more experimental details are needed to allow a more accurate and reliable topological assignment, we embarked on the low‐resolution 3D EM reconstruction of cos phage STP1 by negative staining (see below).
HHpred data concerning the 5093 group are scarce, with only an esterase fold identified at the C‐terminus of the ORF following Tal. However, EM pictures (Fig. S8) (Mills et al., 2011; McDonnell, et al., 2016; McDonnell, et al., 2017) reveal interesting features: six long hexameric arms are projected from the baseplate core with large bulbs at their extremities. This suggests i) the presence of a structure bound to the baseplate and ii) the association of long linker modules to the baseplate. We may assume that its N‐terminus plays the same role as BppU‐Nt to attach the rest of the protein to the central baseplate. From the attachment module, six long ß‐stranded linkers would attach the globular esterase‐like domain, which may represent the putative RBP module (Fig. S8) (Mills et al., 2011; McDonnell, et al., 2016; McDonnell, et al., 2017).
Phages belonging to the 987 group exhibit an N‐terminal BppU domain followed by a short linker and a large globular domain resembling the RBP of phage Phi29. Examination of EM images (Fig. S8) (Mills et al., 2011; McDonnell, et al., 2016; McDonnell, et al., 2017) confirms the presence of large bulbs following a six‐mer (C6) symmetry, attached to the baseplate core. The presence of an N‐terminal BppU‐like domain suggests that this ORF may form a 18‐mer ring above the Dit as observed in phages TP901‐1 and Tuc2009 (Veesler, et al., 2012; Legrand, et al., 2016) and Staphylococcus aureus phage 80α (Kizziah et al., 2020). This Dit ring would then project six trimeric linkers to each of which a trimeric RBP is attached (Fig. 2C).
Structure of the cos phage STP1 baseplate by electron microscopy
Preliminary tests indicated that phage STP1 was most amenable to negative staining EM (NS EM) studies. We collected images from STP1 samples prepared with 2, 1 and 0.5% (w/v) of uranyl acetate. The images of the phages at 1 and 2% resembled those reported in the literature (Lavelle, et al., 2018). However, at concentrations at or below 0.5% uranyl acetate, additional features appeared at the sides of the tail extremity.
The NS EM reconstruction of the baseplate at 21 Å resolution (Fig. 3, Fig. S9) assembles the MTP tube, followed by the Dit ring and the N‐terminal Tal structural domain (the long and thin Tal extensions are cancelled out by averaging due to their flexibility). Strikingly, it reveals the presence of six large extra‐domains at the periphery of the tube, attached at the level of the Dit and pointing upwards (i.e. towards the capsid). As the putative RBP is the only one that has the proper genomic localization to belong to the baseplate, we suggest that these lateral extra‐domains are the putative RBPs that detach in the presence of 1–2% of uranyl acetate.
Fig. 3.
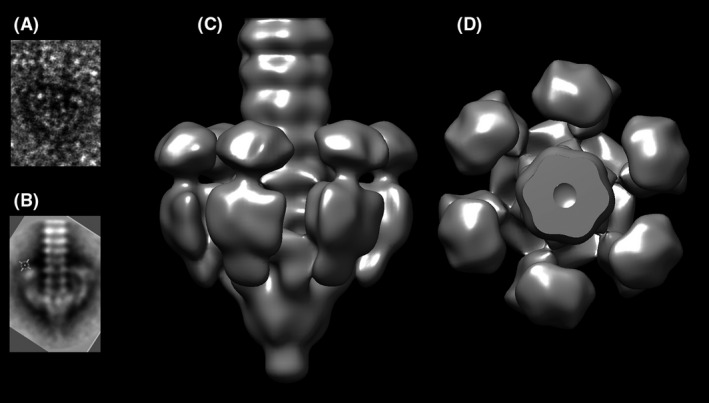
The 21 Å resolution nsEM structure of the tail terminal part of cos phage STP1.
A. View of a single image.
B. View of a 2D class.
C. Single particle reconstruction.
D. Same view as C, rotated 90°.
The proteins identified by HHpred were fitted as rigid bodies into the NS EM map (Fig. 4). The Dit hexamer from Bacillus subtilis phage SPP1 (Veesler, et al., 2010) (PDB ID: 2X8K) and the Tal trimer from lactococcal phage p2 (Sciara, et al., 2010) (PDB ID: 2WZP) were positioned using Chimera ‘fit in map’ option (Pettersen, et al., 2004) at the distal part of the map. The resolution of the map did not allow, however, to position the inserted CBM model. The tail MTPs were modelled and placed above Dit using three Hcp1 hexamers from type VI secretion system (Douzi, et al., 2014) (PDB ID: 4HKH). Of note, the extremity of the peripheral part of the map, assigned to the RBP module, respects a quasi threefold (C3) symmetry. We therefore could readily fit the trimeric RBP of phage p2 (PDB ID: 2BSD) at the extremity of the lateral extension of the map (Fig. 4).
Fig. 4.
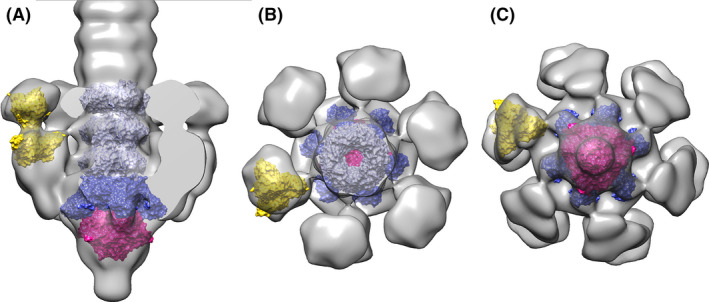
Fit of the proteins identified by HHpred into the EM map of STP1 tail.
A. Slabbed lateral view displaying, from top to bottom, three MTP hexamers (Hcp1, violet), the Dit hexamer (Dit SPP1, blue), the Tal trimer (Phage p2 Tal, red) and p2 RBP (yellow) at the tip of the RBP assigned density.
B. View rotated by −90° relative to A, showing the top of the model and C – view rotated by +90° relative to A, showing the view from the bottom of the model.
Binding studies of phage STP1 predicted adhesion modules
The Tal and putative RBP (encoded by ORFs 15 and 16, respectively) of phage STP1 were subject to a detailed bioinformatic analysis using HHpred (Zimmermann, et al., 2018) (Figs S4A and S6A). Tal harbours a CBM with significant structural similarity (HHpred 99.9% probability) to that of BppA (PDB ID: 5E7T), the accessory binding protein of L. lactis phage Tuc2009 (Legrand, et al., 2016). To note, this CBM corresponds to the previously reported VR1 region of DT1 phage (Duplessis and Moineau, 2001; Duplessis, et al., 2006), which also possesses a BppA fold according to HHpred (Fig. S4A). The putative RBP of STP1 was predicted to possess a p2‐like C‐terminal domain (Fig. S6A) (Spinelli, et al., 2006). Hence, the regions that were predicted by HHpred to encode CBMs of Tal (ORF 15, residues 505–765) and the RBP module (ORF 16, residues 482–682) were amplified, cloned into the GFP fusion vector pHTP9 and pHTP1‐mCherry, expressed and purified.
Fluorescent binding studies
STP1 is a cos type phage that infects a limited number of S. thermophilus strains (including UCCSt102 and UCCSt87). The host strains of STP1, St102 and St87, in addition to three non‐host strains (St95, St89, St12), were used as test strains in fluorescent labelling trials with the CBMs of STP1 Tal and the RBP module at protein concentrations in the range of 5 – 100 µg/ml. STP1 Tal CBM binding to the primary host strain St102 was clearly observed in the range of 20–100 µg/ml, whereas STP1 RBP module was observed to bind to St102 cells at lower concentrations of 5 µg/ml (Fig. 5).
Fig. 5.
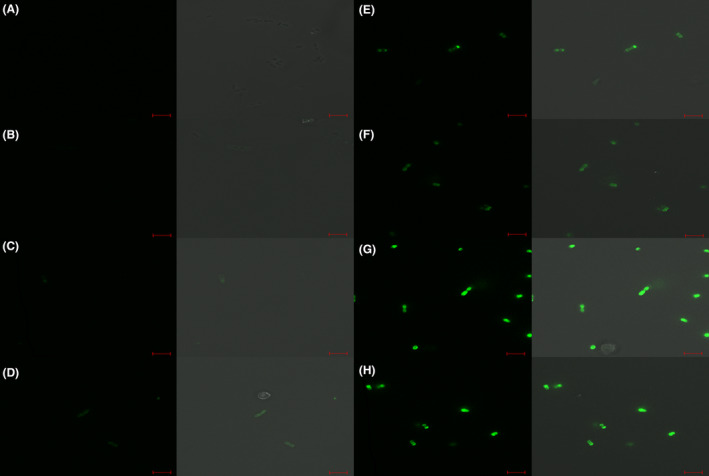
Fluorescent labelling of host strain St102 by Tal–CBM‐GFP (panels A‐D) and RBP‐module‐GFP (panels E‐H) at respective concentrations of 5, 20, 50 and 100 µg/ml. Cells were visualized using differential interference contrast (DIC) microscopy (panels on the right) and fluorescent confocal microscopy (panels on the left) at the GFP excitation wavelength of 488 nm. Scale bars correspond to 5µm.
Labelling assays using the secondary host strain, St87, returned a comparable result with fluorescent labelling observed at similar concentrations (Fig. 6). In contrast, no labelling was observed for non‐host strains highlighting the specificity of the CBMs for host strains of the phage from which they are derived. Of note, a deleterious effect was observed on the cell pellets of both St102 and St87 following incubation with Tal CBM at concentrations of 20 µg/ml and above. Similarly, less chain formation was visible under the microscopic field of view compared to that of negative control cells or cells incubated with a lower concentration of protein. The reason for this effect is unclear. It may be hypothesized that the addition of Tal‐CBM (in excess) results in cell surface modifications that alter pellet formation.
Fig. 6.
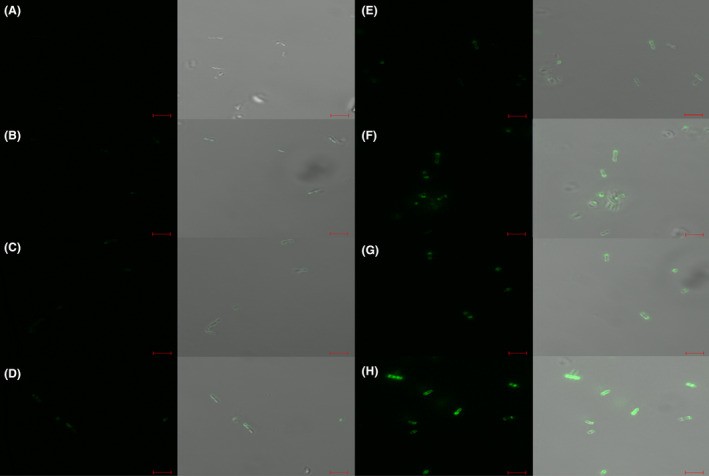
Fluorescent labelling of secondary host St87 by Tal‐CBM‐GFP (panels A‐D) and RBP‐module‐GFP (panels E‐H) at respective concentrations of 5, 20, 50 and 100 µg/ml. Cells were visualized using differential interference contrast (DIC) microscopy (panels on the right) and fluorescent confocal microscopy (panels on the left) at the GFP excitation wavelength of 488 nm. Scale bars correspond to 5µm.
Competition Assays between the CBMs of Tal and the putative RBP
In order to determine whether the CBMs of Tal and the RBP module have different binding affinities for the host cell receptor, competition assays were performed in which equal concentrations (100 µg/ml) of both CBMs, representing 11.27µM of Tal and 14.36 µM of RBP module, were incubated with host cells for 12.5 minutes at 37 °C. In these assays, both proteins were labelled separately with the GFP and mCherry labels to ascertain if the fluorescent label was responsible for seemingly preferential binding. Binding assays with Tal‐GFP/RBP‐module‐mCherry demonstrated a higher dominance of the RBP‐module‐mCherry than Tal‐GFP with reverse tagging assays (i.e. Tal‐mCherry/ RBP‐module‐GFP) supporting the initial findings of a dominance for RBP module over that of Tal CBM, thus eliminating any potential bias from fluorophore intensity (Fig. 7). The dominance of RBP module over that of Tal CBM can be assigned to a competition between both binders to the same site, reducing therefore the apparent affinity.
Fig. 7.
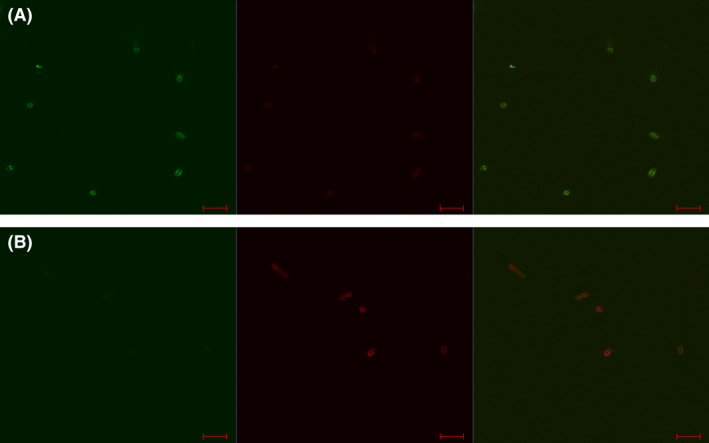
Competition assays performed on host strain St102 using an equal concentration (100 µg/ml) of Tal CBM and RBP module fusion constructs. Reverse tagging confirmed a dominance of RBP module over that of Tal CBM at the cell surface. Panels A: Tal‐CBM‐mCherry/ RBP‐module‐GFP. Panels B: Tal‐CBM‐Gfp/ RBP‐module‐mCherry. Panels on the left: cells were visualized using fluorescent confocal microscopy at the GFP excitation wavelength of 488 nm. Centre panels: cells were visualized using fluorescent confocal microscopy at the mCherry excitement wavelength of 514 nm. Panels on the right: cells were visualized using fluorescent confocal microscopy at the GFP excitation wavelength of 488 nm and at the mCherry excitement wavelength of 514 nm. Scale bars correspond to 5 µm. Scale bar represents 5 µm.
Discussion
Using HHpred, we identified multiple CBMs, putatively acting as receptor‐binding domains in all currently known groups of S. thermophilus siphophages. The cos phages possess a CBM insertion in their Dit and one or two insertions in the long C‐terminal extension of their Tal. It is noteworthy that the identified Tal insertions share the same fold as the CBM of phage Tuc2009 BppA (Legrand, et al., 2016). We analysed for the first time the protein encoded by the gene downstream of tal. We identified a CBM at its extremity, with a fold resembling that of the RBP of lactococcal phages p2 and TP901‐1 (Spinelli, et al., 2006; Spinelli, et al., 2006).
The pac phage analysis yielded results similar to those obtained for cos phages. Their Dit exhibits the same extension, while the Tal of some of the pac phages (the Tal A group) resembles those of cos, containing one or two CBM extensions. However, Tals of group B exhibit two extra CBM insertions in the N‐terminal structural domain of Tal, and the Tal extension bears two additional CBM insertions. As for cos phages, the protein following Tal possesses a p2/TP901‐1 RBP‐like fold. Hence, while the Tal of cos phages contains three to four CBM domains, this number increases to three to six in the Tal for certain pac phages. All in all, cos and pac phages share many identical topological features.
Phages from the 5093 and 987 phage groups in contrast differ strongly from cos and pac phages and from each other. Their Dits are classical, i.e. they do not harbour any extension. The Tals of the 5093 phages possess the characteristic N‐terminal structural domain followed by a short domain of unknown fold. The proteins encoded by the gene after tal constitute a long domain of unknown fold followed by an esterase domain and by a C‐terminus of unknown fold. In the phages from the 987 group, the Tal N‐terminal structural domain is followed by a long domain of unknown fold that links it to a domain resembling the cell wall degrading enzyme of phage Phi29 tail (Xiang, et al., 2008). The N‐terminus of the phage 987 protein specified by the gene after tal gene possesses the fold of the N‐terminal BppU domain of phage TP901‐1 that attaches to the Dit protein. Here, it is followed by a linker of ~200 amino acids and a large domain of ~600 amino acids with the fold of phage Phi29 RBP (Xiang and Rossmann, 2011). This suggests that baseplates of S. thermophilus 987 siphophages exhibit some similarities to that of podophage Phi29 from Bacillus subtilis (Xiang, et al., 2008; Xiang and Rossmann, 2011). The negative stained EM images of phages 5093 and 987 reported previously (McDonnell, et al., 2016; McDonnell, et al., 2017) support our analysis of short Tal extensions and ‘puffy’ lateral arms attributed to the protein encoded by the gene following tal, the putative RBP. In both cases, it is most probable that the proteins coded by the gene after tal correspond to bona fide RBPs.
In contrast, the structure–function relationship of the phages belonging to the cos or pac groups proved to be more elusive. Negatively stained EM images of cos and pac phages exhibit a thin and long extension at the tip of the tail tube containing one or two pearl‐like features, identified previously on negative stained EM images (Lavelle, et al., 2018). These features may represent CBM decorations of Tal.
Although the cos and pac proteins encoded by the putative rbp gene are similarly sized to those of the 5093 or 987 groups, their previously reported low‐resolution negatively stained EM (nsEM) images do not exhibit the same puffy features. Our nsEM structure of St phage STP1 reveals the presence of six peripheral structures, possibly attached to or around the Dit, pointing towards the capsid. We assigned these structures to the putative RBPs, the putative BppU domain being the attachment device to the tail (as in phages TP901‐1 (Veesler, et al., 2012), A112 (Cambillau, 2015) and S. aureus phage 80α (Kizziah et al., 2020)), while the p2/TP901‐1 RBP‐like domain would point upwards, towards the capsid, opposite to the host's cell wall. This suggests that, as in the case of lactococcal phage p2, the putative RBP changes conformation by rotating ~180° towards the host's cell wall (Sciara, et al., 2010).
The Tal domains have been analysed functionally, as it has been previously reported that gene exchange of Tal (called RBP in these papers) between phages DT1 and MD4 results in a host specificity exchange (Duplessis and Moineau, 2001). Furthermore, the authors identified two islands of variable sequence VR1 and VR2 within an otherwise conserved Tal that they attribute to the RBP (Duplessis, et al., 2006). More recently, fluorescent derivatives of the Streptococcus phage CHPC951 were expressed and were found to bind to the corresponding host (Szymczak, et al., 2019). These studies definitively assess an important role of Tal‐associated CBMs in host binding.
The STP1 host specificity that we observed in our binding studies of both CBM Tal and putative RBP module expands on the current knowledge and understanding of S. thermophilus phage–host interactions. The fluorescent labelling assays performed within this study have now established a functional role for the STP1 C‐term RBP module in the phage host‐recognition process. The higher binding dominance of RBP‐module‐mCherry than CBM‐Tal‐GFP is similar to that of 936 phages bona fide RBPs over the ancillary CBMs in Dit, MTP or NPS (Hayes, et al., 2018; Hayes, et al., 2019). Furthermore, the interplay of multiple proteins is common among phages in possession of a peripheral baseplate structure that mediates the recognition of a host‐encoded saccharidic receptor (Stockdale, et al., 2013; Hayes, et al., 2018; Hayes, et al., 2019).
The STP1 baseplate structure indicates that the RBPs may have a crucial role to ensure the proper orientation of cos and pac phages before DNA injection. In line with this hypothesis, we know that Myoviridae, Podoviridae and Siphoviridae phages recognize their saccharidic receptor using RBPs at the baseplate periphery, as this topology ensures a favourable DNA injection topology, perpendicular to the host cell wall (Bebeacua, et al., 2013; Hu, et al., 2013; Taylor, et al., 2016). Therefore, a variety of receptor‐binding domains at various exposed positions on the virion (e.g. on capsid, NPS, MTP, Dit, Tal and other ancillary proteins) can be employed for host recognition and binding, while the bona fide RBP may be the only one to place the phage in a correct, i.e. infection‐competent, position, with respect to its host.
Experimental procedures
Bioinformatical analysis
Protein sequence and structure analyses were performed using HHpred (Homology detection & structure prediction by HMM‐HMM comparison) with standard mode (Zimmermann, et al., 2018). The secondary structure prediction was performed with JJpred (Drozdetskiy, et al., 2015). Sequence alignments were performed with Multalin (Corpet, 1988). Model visualization and modelling were performed with Coot (Emsley, et al., 2010) and Chimera (Pettersen, et al., 2004).
Preparation and clarification of phage STP1 for TEM analysis
Phage STP1 was propagated on its host strain, S. thermophilus P1, at 42 °C in LM17 broth (Sigma‐Aldrich) supplemented with 10 mM CaCl2. Upon lysis of the culture, the supernatant was filtered through a 0.45 µm membrane filter and centrifuged at 71 124 × g for 2 h at 4 °C to concentrate phage particles. The pellet was resuspended in 3 ml of SM buffer (50 mM Tris‐HCL pH 7.5, 100 mM NaCl, 10 mM MgSO4) and clarified though the addition of 10% chloroform (Sigma‐Aldrich) followed by centrifugation at 16 000 × g for 10 min. The clarification step was repeated twice, after which the aqueous layer was filtered through a 0.22 µm membrane filter followed by phage enumeration using the standard double layer plaque assay method (Lillehaug, 1997) to determine the titre as PFU/mL. The purified phage lysate was stored at 4 °C prior to TEM imaging.
Electron microscopy
Negative staining grid preparation
A 10‐μl droplet of CsCl‐purified phage sample at a final concentration of 109 pfu was applied onto a 300‐mesh copper‐carbon‐coated grid (Agar Scientific), glow‐discharged for 2 min at 15 mA using a PELCO easiGlow cleaning system (Ted Pella, Inc). After 4 min incubation, the excess of sample was removed by side blotting. The grid was washed twice with water and stained with 0.5% (w/v) uranyl acetate solution for 15 s. The grid was blotted dry and imaged on a T12 Spirit transmission electron microscope operating at 120 kV (Thermo Fischer Scientific).
Negative staining data collection and image processing
Images were acquired using a Veleta CCD camera (Olympus) at nominal magnification of 55,000x, corresponding to a pixel size of 4.83 Å/pixel at the specimen level, with an applied defocus values between −0.9 and −1.5 μm. A total of 739 images were recorded and imported in RELION‐3.0.8 for all subsequent image processing tasks (Zivanov, et al., 2018). Estimation of the contrast transfer function (CTF) was performed using CTFFIND4 (Rohou and Grigorieff, 2015). Particles were manually picked and extracted in Relion. The dataset was subjected to several rounds of refrence‐free 2D classsification, and junk particles were removed. An initial 3D reference was generated using a Stochastic Gradient Descent algorithm, and subsequent rounds of 3D classification and refinement were performed imposing C6 symmetry. The overall estimate resolution was calculated from Fourier shell correlations at 0.143 (Fig. S14). The tail reconstruction has been deposited at the Electron Microscopy Data Bank (EMDB) with accession code EMD‐10913.
Bacterial strains and phage propagation
The bacterial strains used in this study, namely S. thermophilus UCCst102, S. thermophilus UCCSt87 (previously named P1 and P3 in), S. thermophilus UCCSt95, S. thermophilus UCCSt13 and S. thermophilus UCCSt12, were grown in M17 broth (Oxoid, UK) supplemented with 5% lactose as 37 °C. Phage STP1 was propagated on its primary host S. thermophilus UCCSt102 in LM17 supplemented with 10 mM CaCl2. Following complete lysis, the lysate was filtered (0.45µm) and the titre determined using the standard double agar layer technique, before storing at 4 °C.
Fusion protein production
The regions that are predicted to encode CBMs of Tal (ORF 15) and putative RBP module (ORF 16) were amplified from a fresh lysate of phage STP1 with a titre of 107 PFU/ml using the primers listed in Table S1. A tobacco etch virus (TEV) protease specific cleavage site (ENLYFQ\G) was incorporated to the forward cloning primer to allow for downstream cleavage of the fluorescent tag if required. An in‐frame stop codon was added to the reverse primer to prevent the inclusion of a C‐terminal His‐tag. The purified CBMs of Tal (ORF −15) and the RBP module (ORF16) were cloned into the GFP fusion vector pHTP9 and pHTP1‐mCherry (NZYTech, Portugal) according the manufacturer’s instructions. Ligation mixtures were transformed into competent E.coli BL21 (DE3) via heat shock at 42 °C × 45 s and plated on LB agar plates supplemented with 50 µg/ml kanamycin (Sigma, USA). The integrity of the recombinant plasmid sequences was verified by Sanger sequencing (Eurofins Genomics, Germany) using vector‐specific or insert‐specific primers.
Protein expression
About 1 ml of E. coli BL21 (DE3) harbouring the plasmid of interest was inoculated to 100 ml of auto‐induction media (NZYTech, Portugal) supplemented with 1% glycerol and 50 µg/ml kanamycin. Incubation was performed with agitation (300 rpm) at 24 °C for 24 hours. Cells were harvested at 4620 × g for 30 min before suspending in lysis buffer (50 mM Tris‐HCl pH7.5, 500 mM NaCl, 5% glycerol, 1% triton X, 30 mM imidazole, 50 mg/ml lysozyme) and freezing at −80 °C for a minimum of 24 hours. After thawing, cells were subject to five cycles of sonication (MSE Soniprep, Sanyo, Japan) at maximum amplitude for 30 s followed by a 30 s rest, with a holding step on ice for at least 2 minutes after the third round of sonication. Recombinant proteins were separated from cellular debris by centrifugation prior to purification via a standard Ni‐NTA agarose column according to the manufacturer’s instructions (Qiagen, UK). An elution gradient of 50 mM to 250 mM imidazole was applied across the column, with fusion proteins eluting primarily in the 200 mM fraction.
Fluorescent labelling with STP1 Tal‐CBM and RBP module
Labelling assays were performed as previously described (Dieterle, et al., 2017) with the following modifications; for all labelling assays, LM17 was supplemented with 10 mM CaCl2 before fluorescently tagged STP1 Tal CBM and RBP module were added to 300 µl of cells in the early exponential phase (OD600nm 0.3–0.4) at a final concentration of 5, 10, 20, 50 or 100 µg/ ml. In the case of competitive inhibition assays, both fluorescently tagged Tal CBM and RBP module were added at a final concentration of 100 µg/ ml each. Incubation took place at 37 °C for 12.5 min before washing in 120 µl of SM buffer (50 mM Tris‐HCl pH 7.5, 100 mM NaCl, 10 mM MgSO4). Fluorescent labelling of cells was visualized using an LSM 5 Exciter (Zeiss, Germany). Final images were analysed using the Zen 2.3 SP1 software package (Zeiss, Germany).
Conflict of interest
The authors declare no competing interests.
Author contributions
D.vS., J.M. and C.C. designed and conceived the experiments. KL, AG, SS and C.C. performed experiments (KL: Binding studies; AG and SS: electron microscopy; C.C.: bioinformatic analysis of phages CBMs). C.C. wrote the paper with contributions from all other authors. All authors revised and approved the manuscript.
Supporting information
Fig. S1. Sequence alignment of the cos (A), pac (Brussowvirus) (B), 5093 and 987 (C) Dit proteins. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black.
Fig. S2. HHpred analysis of the Dit proteins from cos, pac, 5083 and 987 representative phages. The horizontal bars are proportional to the proteins lengthes, and the labels within the bars correspond to the PDB identification of the structurally closest protein. The insertions observed in Dits found in cos and pac phages correspond to Lactobacillus phage J1 inserted CBM domain (25).
Fig. S3. (A) Sequence alignment of the cos Tal proteins. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black. To note, the excellent conservation of the N‐terminal gp27‐like domain, as well as that of the C‐terminus of unknown fold. (B) Sequence alignment of the pac (Brussowvirus) Tal proteins. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black. To note, the excellent conservation of the N‐terminal gp27‐like domain, as well as that of the BppA2 and of the C‐terminus of unknown fold.
Fig. S4. A) HHpred analysis of the Tal proteins from cos representative phages. The horizontal bars are proportional to the protein lengths, and the labels within the bars correspond to the PDB identification of the structurally closest protein. The gp27‐like domain correspond to the conserved N‐terminal structural domain of Tal. The 1 or 2 insertions in the Tal extensions correspond to the inserted CBM domain of the Tuc2009 BppA protein (28). B). HHpred analysis of the Tal proteins from pac (Brussowvirus) representative phages. A‐ The pac A sub‐group does not exhibit extensions in the conserved N‐terminal structural domain. B‐ The pac B sub‐group exhibits two extensions in the conserved N‐terminal structural domain. C‐ The pac ouliers. The horizontal bars are proportional to the proteins lengthes, and the labels within the bars correspond to the PDB identification of the structurally closest protein. The gp27‐like domain correspond to the conserved N‐terminal structural domain of Tal. The 1 or 2 insertions in the Tal extensions correspond to the inserted CBM domain of the Tuc2009 BppA protein (28).
Fig. S5. Sequence alignment and HHpred analysis of the Tal proteins from 5093 and 987 representative phages. A‐ Sequence alignment of the Tal proteins. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black. B‐ The horizontal bars are proportional to the proteins lengthes, and the labels within the bars correspond to the PDB identification of the structurally closest protein.
Fig. S6. (A) Sequence alignment of the cos and pac (Brussowvirus) putative RBPs. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black. (B) Sequence alignment of the 5093 and 987 putative RBPs. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black.
Fig. S7. HHpred analysis of the proteins following Tal (putative RBPs) from cos, pac, 5083 and 987 representative phages. A‐ phages cos and pac putative RBPs. The N‐terminus (blue, residues 1‐~140) was putatively assigned to a BppU‐like N‐terminal domain. The last ~140 residues share the fold of p2/TP901‐1 phages RBPs. B‐ phages 5083 and 987 putative RBPs. The N‐terminus (blue, residues 1‐~140) was putatively assigned to a BppU‐like Nterminal domain. The horizontal bars are proportional to the proteins lengthes, and the labels within the bars correspond to the PDB identification of the structurally closest protein.
Fig. S8. Electron microscopy views of phage 5093 (40) and 987 (33).
Fig. S9. Fourier shell correlation (FSC) curves of the final three‐dimensional reconstruction. This curve was obtained by correlation of two different three‐dimensional images created by splitting the particle set into two subsets. The resolutions was at 21 Å for the tail tip.
Table S1. Primers used in this study. TEV cleavage sites are represented in bold while incorporated stop codons are underlined.
Acknowledgements
Dr. Alain Roussel (AFMB, Marseille) is greatly acknowledged for his constant support. JM is the recipient of a Starting Investigator Research Grant (Ref. No. 15/SIRG/3430) funded by Science Foundation Ireland (SFI). D.vS is supported by a Principal Investigator award (Ref. No. 13/IA/1953) through SFI. This work was supported by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR‐10‐INSB‐05‐01.
Microbial Biotechnology (2020) 13(6), 1765–1779
Funding information
JM is the recipient of a Starting Investigator Research Grant (Ref. No. 15/SIRG/3430) funded by Science Foundation Ireland (SFI). D.vS is supported by a Principal Investigator award (Ref. No. 13/IA/1953) through SFI. This work was supported by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR‐10‐INSB‐05‐01.
Contributor Information
Jennifer Mahony, Email: j.mahony@ucc.ie.
Christian Cambillau, Email: cambillau@afmb.univ-mrs.fr.
References
- Arnaud, C.A. , Effantin, G. , Vives, C. , Engilberge, S. , Bacia, M. , Boulanger, P. , et al (2017) Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection. Nat Commun 8: 1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebeacua, C. , Bron, P. , Lai, L. , Vegge, C.S. , Brondsted, L. , Spinelli, S. , et al (2010) Structure and molecular assignment of lactococcal phage TP901‐1 baseplate. J Biol Chem 285: 39079–39086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebeacua, C. , Tremblay, D. , Farenc, C. , Chapot‐Chartier, M.P. , Sadovskaya, I. , van Heel, M. , et al (2013) Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. J Virol 87: 12302–12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyton, C. , Flayhan, A. , Gabel, F. , Lethier, M. , Durand, G. , Boulanger, P. , et al (2013) Assessing the conformational changes of pb5, the receptor‐binding protein of phage T5, upon binding to its Escherichia coli receptor FhuA. J Biol Chem 288: 30763–30772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow, H. (2018) Population genomics of bacteriophages In Population Genomics: Microorganisms Population Genomics. Polz M.F., and Rajora O.P. (eds). pp. 297–334. [Google Scholar]
- Buttner, C.R. , Wu, Y. , Maxwell, K.L. , and Davidson, A.R. (2016) Baseplate assembly of phage Mu: Defining the conserved core components of contractile‐tailed phages and related bacterial systems. Proc Natl Acad Sci USA 113: 10174–10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambillau, C. (2015) Bacteriophage module reshuffling results in adaptive host range as exemplified by the baseplate model of listerial phage A118. Virology 484: 86–92. [DOI] [PubMed] [Google Scholar]
- Chaban, Y. , Lurz, R. , Brasiles, S. , Cornilleau, C. , Karreman, M. , Zinn‐Justin, S. , et al (2015) Structural rearrangements in the phage head‐to‐tail interface during assembly and infection. Proc Natl Acad Sci USA 112: 7009–7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, B. , Bebeacua, C. , Mahony, J. , Blangy, S. , Douillard, F.P. , Veesler, D. , et al (2013) Structure and functional analysis of the host recognition device of lactococcal phage tuc2009. J Virol 87: 8429–8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet, F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle, M.E. , Fina Martin, J. , Duran, R. , Nemirovsky, S.I. , Sanchez Rivas, C. , Bowman, C. , et al (2016) Characterization of prophages containing "evolved" Dit/Tal modules in the genome of Lactobacillus casei BL23. Appl Microbiol Biotechnol 100: 9201–9215. [DOI] [PubMed] [Google Scholar]
- Dieterle, M.E. , Spinelli, S. , Sadovskaya, I. , Piuri, M. , and Cambillau, C. (2017) Evolved distal tail carbohydrate binding modules of Lactobacillus phage J‐1: a novel type of anti‐receptor widespread among lactic acid bacteria phages. Mol Microbiol 104: 608–620. [DOI] [PubMed] [Google Scholar]
- Douzi, B. , Spinelli, S. , Blangy, S. , Roussel, A. , Durand, E. , Brunet, Y.R. , et al (2014) Crystal structure and self‐interaction of the type VI secretion tail‐tube protein from enteroaggregative Escherichia coli. PLoS ONE 9: e86918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowah, A.S.A. , and Clokie, M.R.J. (2018) Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram‐positive bacteria. Biophys Rev 10: 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy, A. , Cole, C. , Procter, J. , and Barton, G.J. (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43: W389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne, M. , Hupfeld, M. , Klumpp, J. , and Loessner, M.J. (2018) Molecular basis of bacterial host interactions by gram‐positive targeting bacteriophages. Viruses 10: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne, M. , Rupf, B. , Tala, M. , Qabrati, X. , Ernst, P. , Shen, Y. , et al (2019) Reprogramming bacteriophage host range through structure‐guided design of chimeric receptor binding proteins. Cell Rep 29: 1336–1350.e1334. [DOI] [PubMed] [Google Scholar]
- Duplessis, M. , and Moineau, S. (2001) Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol Microbiol 41: 325–336. [DOI] [PubMed] [Google Scholar]
- Duplessis, M. , Levesque, C.M. , and Moineau, S. (2006) Characterization of Streptococcus thermophilus host range phage mutants. Appl Environ Microbiol 72: 3036–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P. , Lohkamp, B. , Scott, W.G. , and Cowtan, K. (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farenc, C. , Spinelli, S. , Vinogradov, E. , Tremblay, D. , Blangy, S. , Sadovskaya, I. , et al (2014) Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein. J Virol 88: 7005–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flayhan, A. , Vellieux, F.M. , Lurz, R. , Maury, O. , Contreras‐Martel, C. , Girard, E. , et al (2014) Crystal structure of pb9, the distal tail protein of bacteriophage T5: a conserved structural motif among all siphophages. J Virol 88: 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau, J.E. , and Moineau, S. (2011) Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb Cell Fact 10(Suppl 1): S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, S. , Vincentelli, R. , Mahony, J. , Nauta, A. , Ramond, L. , Lugli, G.A. , et al (2018) Functional carbohydrate binding modules identified in evolved dits from siphophages infecting various Gram‐positive bacteria. Mol Microbiol 110: 777–795. [DOI] [PubMed] [Google Scholar]
- Hayes, S. , Mahony, J. , Vincentelli, R. , Ramond, L. , Nauta, A. , van Sinderen, D. , and Cambillau, C. (2019) Ubiquitous carbohydrate binding modules decorate 936 lactococcal siphophage virions. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Margolin, W. , Molineux, I.J. , and Liu, J. (2013) The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science 339: 576–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizziah, J.L. , Manning, K.A. , Dearborn, A.D. , and Dokland, T. (2020) Structure of the host cell recognition and penetration machinery of a Staphylococcus aureus bacteriophage. PLoS Pathog. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle, K. , Murphy, J. , Fitzgerald, B. , Lugli, G.A. , Zomer, A. , Neve, H. , et al (2018) A decade of Streptococcus thermophilus phage evolution in an Irish dairy plant. Appl Environ Microbiol 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle, K. , Martinez, I. , Neve, H. , Lugli, G.A. , Franz, C. , Ventura, M. , et al (2018) Biodiversity of Streptococcus thermophilus phages in global dairy fermentations. Viruses 10: 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand, P. , Collins, B. , Blangy, S. , Murphy, J. , Spinelli, S. , Gutierrez, C. , et al (2016) The atomic structure of the phage Tuc 2009 baseplate tripod suggests that host recognition involves two different carbohydrate binding modules. MBio 7: e01781‐01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehaug, D. (1997) An improved plaque assay for poor plaque‐producing temperate lactococcal bacteriophages. J Appl Microbiol 83: 85–90. [DOI] [PubMed] [Google Scholar]
- Mahony, J. , Ainsworth, S. , Stockdale, S. , and van Sinderen, D. (2012) Phages of lactic acid bacteria: the role of genetics in understanding phage‐host interactions and their co‐evolutionary processes. Virology 434: 143–150. [DOI] [PubMed] [Google Scholar]
- Mahony, J. , Kot, W. , Murphy, J. , Ainsworth, S. , Neve, H. , Hansen, L.H. , et al (2013) Investigation of the relationship between lactococcal host cell wall polysaccharide genotype and 936 phage receptor binding protein phylogeny. Appl Environ Microbiol 79: 4385–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony, J. , Cambillau, C. , and van Sinderen, D. (2017) Host recognition by lactic acid bacterial phages. FEMS Microbiol Rev 41: S16–S26. [DOI] [PubMed] [Google Scholar]
- McDonnell, B. , Mahony, J. , Neve, H. , Hanemaaijer, L. , Noben, J.P. , Kouwen, T. , and van Sinderen, D. (2016) Identification and analysis of a novel group of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus . Appl Environ Microbiol 82: 5153–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell, B. , Mahony, J. , Hanemaaijer, L. , Neve, H. , Noben, J.P. , Lugli, G.A. , et al (2017) Global survey and genome exploration of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus . Front Microbiol 8: 1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, S. , Griffin, C. , O’Sullivan, O. , Coffey, A. , McAuliffe, O.E. , Meijer, W.C. , et al (2011) A new phage on the ‘Mozzarella’ block: Bacteriophage 5093 shares a low level of homology with other Streptococcus thermophilus phages. Int Dairy J 21: 963–969. [Google Scholar]
- Pettersen, E.F. , Goddard, T.D. , Huang, C.C. , Couch, G.S. , Greenblatt, D.M. , Meng, E.C. , and Ferrin, T.E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Plisson, C. , White, H.E. , Auzat, I. , Zafarani, A. , Sao‐Jose, C. , Lhuillier, S. , et al (2007) Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J 26: 3720–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou, A. , and Grigorieff, N. (2015) CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciara, G. , Bebeacua, C. , Bron, P. , Tremblay, D. , Ortiz‐Lombardia, M. , Lichiere, J. , et al (2010) Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc Natl Acad Sci U S A 107: 6852–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli, S. , Campanacci, V. , Blangy, S. , Moineau, S. , Tegoni, M. , and Cambillau, C. (2006) Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901‐1. J Biol Chem 281: 14256–14262. [DOI] [PubMed] [Google Scholar]
- Spinelli, S. , Desmyter, A. , Verrips, C.T. , de Haard, H.J. , Moineau, S. , and Cambillau, C. (2006) Lactococcal bacteriophage p2 receptor‐binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat Struct Mol Biol 13: 85–89. [DOI] [PubMed] [Google Scholar]
- Spinelli, S. , Veesler, D. , Bebeacua, C. , and Cambillau, C. (2014) Structures and host‐adhesion mechanisms of lactococcal siphophages. Front Microbiol 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli, S. , Bebeacua, C. , Orlov, I. , Tremblay, D. , Klaholz, B.P. , Moineau, S. , and Cambillau, C. (2014) Cryo‐electron microscopy structure of lactococcal siphophage 1358 virion. J Virol 88: 8900–8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale, S.R. , Mahony, J. , Courtin, P. , Chapot‐Chartier, M.P. , van Pijkeren, J.P. , Britton, R.A. , et al (2013) The lactococcal phages Tuc 2009 and TP901‐1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J Biol Chem 288: 5581–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak, P. , Rau, M.H. , Monteiro, J.M. , Pinho, M.G. , Filipe, S.R. , Vogensen, F.K. , et al (2019) A comparative genomics approach for identifying host‐range determinants in Streptococcus thermophilus bacteriophages. Sci Rep 9: 7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.M. , Prokhorov, N.S. , Guerrero‐Ferreira, R.C. , Shneider, M.M. , Browning, C. , Goldie, K.N. , et al (2016) Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533: 346–352. [DOI] [PubMed] [Google Scholar]
- Valyasevi, R. , Sandine, W.E. , and Geller, B.L. (1991) A membrane protein is required for bacteriophage c2 infection of Lactococcus lactis subsp. lactis C2. J Bacteriol 173: 6095–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler, D. , and Cambillau, C. (2011) A common evolutionary origin for tailed‐bacteriophage functional modules and bacterial machineries. Microbiology and molecular biology reviews : MMBR 75: 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler, D. , Robin, G. , Lichiere, J. , Auzat, I. , Tavares, P. , Bron, P. , et al (2010) Crystal structure of bacteriophage SPP1 distal tail protein (gp19.1): A baseplate hub paradigm in gram‐positive infecting phages. J Biol Chem 285: 36666–36673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesler, D. , Spinelli, S. , Mahony, J. , Lichiere, J. , Blangy, S. , Bricogne, G. , et al (2012) Structure of the phage TP901‐1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc Natl Acad Sci USA 109: 8954–8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinga, I. , Baptista, C. , Auzat, I. , Petipas, I. , Lurz, R. , Tavares, P. , et al (2012) Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol Microbiol 83: 289–303. [DOI] [PubMed] [Google Scholar]
- Xiang, Y. , and Rossmann, M.G. (2011) Structure of bacteriophage phi29 head fibers has a supercoiled triple repeating helix‐turn‐helix motif. Proc Natl Acad Sci USA 108: 4806–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Y. , Morais, M.C. , Cohen, D.N. , Bowman, V.D. , Anderson, D.L. , and Rossmann, M.G. (2008) Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage phi29 tail. Proc Natl Acad Sci USA 105: 9552–9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Y. , Leiman, P.G. , Li, L. , Grimes, S. , Anderson, D.L. , and Rossmann, M.G. (2009) Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol Cell 34: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, L. , Stephens, A. , Nam, S.Z. , Rau, D. , Kubler, J. , Lozajic, M. , et al (2018) A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430: 2237–2243. [DOI] [PubMed] [Google Scholar]
- Zivanov, J. , Nakane, T. , Forsberg, B.O. , Kimanius, D. , Hagen, W.J. , Lindahl, E. , and Scheres, S.H. (2018) New tools for automated high‐resolution cryo‐EM structure determination in RELION‐3. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Sequence alignment of the cos (A), pac (Brussowvirus) (B), 5093 and 987 (C) Dit proteins. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black.
Fig. S2. HHpred analysis of the Dit proteins from cos, pac, 5083 and 987 representative phages. The horizontal bars are proportional to the proteins lengthes, and the labels within the bars correspond to the PDB identification of the structurally closest protein. The insertions observed in Dits found in cos and pac phages correspond to Lactobacillus phage J1 inserted CBM domain (25).
Fig. S3. (A) Sequence alignment of the cos Tal proteins. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black. To note, the excellent conservation of the N‐terminal gp27‐like domain, as well as that of the C‐terminus of unknown fold. (B) Sequence alignment of the pac (Brussowvirus) Tal proteins. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black. To note, the excellent conservation of the N‐terminal gp27‐like domain, as well as that of the BppA2 and of the C‐terminus of unknown fold.
Fig. S4. A) HHpred analysis of the Tal proteins from cos representative phages. The horizontal bars are proportional to the protein lengths, and the labels within the bars correspond to the PDB identification of the structurally closest protein. The gp27‐like domain correspond to the conserved N‐terminal structural domain of Tal. The 1 or 2 insertions in the Tal extensions correspond to the inserted CBM domain of the Tuc2009 BppA protein (28). B). HHpred analysis of the Tal proteins from pac (Brussowvirus) representative phages. A‐ The pac A sub‐group does not exhibit extensions in the conserved N‐terminal structural domain. B‐ The pac B sub‐group exhibits two extensions in the conserved N‐terminal structural domain. C‐ The pac ouliers. The horizontal bars are proportional to the proteins lengthes, and the labels within the bars correspond to the PDB identification of the structurally closest protein. The gp27‐like domain correspond to the conserved N‐terminal structural domain of Tal. The 1 or 2 insertions in the Tal extensions correspond to the inserted CBM domain of the Tuc2009 BppA protein (28).
Fig. S5. Sequence alignment and HHpred analysis of the Tal proteins from 5093 and 987 representative phages. A‐ Sequence alignment of the Tal proteins. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black. B‐ The horizontal bars are proportional to the proteins lengthes, and the labels within the bars correspond to the PDB identification of the structurally closest protein.
Fig. S6. (A) Sequence alignment of the cos and pac (Brussowvirus) putative RBPs. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black. (B) Sequence alignment of the 5093 and 987 putative RBPs. The conserved amino acids are in red and the partially conserved in blue. Otherwise in black.
Fig. S7. HHpred analysis of the proteins following Tal (putative RBPs) from cos, pac, 5083 and 987 representative phages. A‐ phages cos and pac putative RBPs. The N‐terminus (blue, residues 1‐~140) was putatively assigned to a BppU‐like N‐terminal domain. The last ~140 residues share the fold of p2/TP901‐1 phages RBPs. B‐ phages 5083 and 987 putative RBPs. The N‐terminus (blue, residues 1‐~140) was putatively assigned to a BppU‐like Nterminal domain. The horizontal bars are proportional to the proteins lengthes, and the labels within the bars correspond to the PDB identification of the structurally closest protein.
Fig. S8. Electron microscopy views of phage 5093 (40) and 987 (33).
Fig. S9. Fourier shell correlation (FSC) curves of the final three‐dimensional reconstruction. This curve was obtained by correlation of two different three‐dimensional images created by splitting the particle set into two subsets. The resolutions was at 21 Å for the tail tip.
Table S1. Primers used in this study. TEV cleavage sites are represented in bold while incorporated stop codons are underlined.


