Abstract
Nucleic acid aptamers can be chemically modified to enhance function, but modifying previously selected aptamers can have nontrivial structural and functional consequences. We present a reselection strategy to evaluate the impact of several modifications on preexisting aptamer pools. RNA aptamer libraries with affinity to HIV-1 reverse transcriptase (RT) were retranscribed with 2′-F, 2′-OMe, or 2′-NH2 pyrimidines and subjected to three additional selection cycles. RT inhibition was observed for representative aptamers from several structural families identified by high-throughput sequencing when transcribed with their corresponding modifications. Thus, reselection identified specialized subsets of aptamers that tolerated chemical modifications from unmodified preenriched libraries. Inhibition was the strongest with the 2′-F-pyrimidine (2′-FY) RNAs, as compared to inhibition by the 2′-OMeY and 2′-NH2Y RNAs. Unexpectedly, a diverse panel of retroviral RTs were strongly inhibited by all 2′-FY-modified transcripts, including sequences that do not inhibit those RTs as unmodified RNA. The magnitude of promiscuous RT inhibition was proportional to mole fraction 2′-FY in the transcript. RT binding affinity by 2′-FY transcripts was more sensitive to salt concentration than binding by unmodified transcripts, indicating that interaction with retroviral RTs is more ionic in character for 2′-FY RNA than for unmodified 2′-OH RNA. These surprising features of 2′-FY-modified RNA may have general implications for applied aptamer technologies.
Keywords: SELEX, in vitro selection, 2′ modifications, 2′-fluoro pyrimidines
INTRODUCTION
Aptamers are structured nucleic acids that bind to molecular targets, often with high affinity and specificity. They are generated through an iterative selection process termed systematic evolution of ligands by exponential enrichment (SELEX) (Ellington and Szostak 1990; Tuerk and Gold 1990). In some cases, chemically modified nucleotide building blocks within an aptamer can improve the binding affinity of an aptamer to its target of interest. For example, very high affinity aptamers against the human immunodeficiency virus type 1 reverse transcriptase (HIV-1 RT) and HIV-1 integrase were recently selected using the 2′-deoxy-2′-fluoroarabinonucleotide (FANA) modification (Alves Ferreira-Bravo et al. 2015; Rose et al. 2019), and various classes of modifications can improve the biostability of aptamers and enable reaction chemistries that are inaccessible to unmodified nucleic acids (Kuwahara and Sugimoto 2010; Stovall et al. 2014; Lapa et al. 2016). To stabilize RNA aptamers against rapid degradation by serum endonucleases for therapeutic and diagnostic applications, one strategy is to modify the 2′ position on the ribose ring of pyrimidines (Adler et al. 2008; Keefe et al. 2010; Dellafiore et al. 2016). Common 2′-modifications such as 2′-fluoro (-F), 2′-amino (-NH2), and 2′-O-methyl (-OMe) significantly increase RNA serum half-life, but they also can have structural and functional consequences for the aptamer by perturbing the binding interface, binding affinity, three-dimensional structure, and overall pharmacokinetic and pharmacodynamic properties (Burnett and Rossi 2012). Among the above-mentioned modifications, the 2′-F-pyrimidine (2′-FY) modification has been extensively used in selecting aptamers for use in vivo or in serum, such as cell-targeted or cell-internalizing aptamers (Zhou and Rossi 2016; Tawiah et al. 2017). Modifications at the 2′-position of the pentose sugar in nucleic acids can affect sugar puckering and the types of hydrogen bond patterns (Shigdar et al. 2013) and alters electronic distribution throughout the nucleotide (Guschlbauer and Jankowski 1980). The high electronegativity of the fluorine makes it a weak hydrogen bond acceptor relative to 2′-OH, and enhances the C3′-endo character of the sugar pucker relative to unmodified ribose to reinforce the canonical A-form helix (Guschlbauer and Jankowski 1980; Zhou et al. 2009; Patra et al. 2012). In contrast, the 2′-NH2 substitution more strongly prefers the C2′-endo sugar pucker found in DNA-like B-form helices (Guschlbauer and Jankowski 1980). 2′-OMe and unmodified 2′-OH nucleotides are intermediate between the two extremes (2′-F and 2′-NH2), favoring the C3′-endo sugar pucker to approximately the same degree (Guschlbauer and Jankowski 1980). Many other nucleotide analogs are available with modifications on the sugar, nucleobase, or internucleotide linkages, each associated with their own potential for modulating nucleic acid function.
Two approaches are conventionally used to generate chemically modified aptamers. In the first approach de novo selection is performed with chemically modified random libraries from the beginning. An advantage of this in-SELEX strategy is that it leverages the full sequence diversity of the random library to investigate a large number of modified sequences simultaneously (Meek et al. 2016). Many types of modified nucleotides can be incorporated into selections to enhance aptamer functionality and stability, such as modifications within the nucleobases, phosphate backbone, or other sugar modifications (Dellafiore et al. 2016). However, the modified nucleotides must be recognized and/or incorporated by relevant polymerases (Lauridsen et al. 2012), and many modified nucleotides are expensive or require specialized synthetic expertise to produce them in-house (Darmostuk et al. 2015). Polymerase engineering has yielded many new polymerases that incorporate diverse modified nucleotides efficiently and greatly expanded the alphabet of accessible nucleotide analogs for in vitro selection (Pinheiro et al. 2012; Larsen et al. 2016; Kimoto et al. 2017). For example, the Y639F mutation in T7 RNA polymerase allows efficient incorporation of 2′-F-, 2′-NH2-, and 2′-H nucleotides into RNA during in vitro transcription (Sousa and Padilla 1995; Huang et al. 1997), and the Y639F/H784A double mutant T7 RNA polymerase incorporates bulkier 2′-modified nucleotides such as 2′-OMe and 2′-azido nucleotide triphosphates (NTPs) (Padilla and Sousa 2002). Another example is the DNA polymerase I from Geobacillus stearthermophilus (Bst), as it is able to reverse transcribe RNA and xeno nucleic acids (XNAs) into DNA (Jackson et al. 2019), including FANA and threose nucleic acid (TNA), and has been used in in vitro selections (Mei et al. 2017, 2018; Wang et al. 2018). In the second approach chemical modifications are incorporated through post-SELEX chemical optimization of aptamers that were originally selected as native DNA, RNA, or other polymers by systematically introducing modified nucleotides into preexisting aptamers in various combinations and determining their impact on binding affinity, serum stability, and function through systematic screens (Eaton et al. 1997; Aaldering et al. 2015; Darmostuk et al. 2015; Meek et al. 2016). This process is repeated until the modified aptamer has the desired properties, which typically requires screening a large number of variants (Lapa et al. 2016), which can be laborious, expensive, and hit-or-miss.
We developed a reselection strategy that identifies aptamers that can either accommodate or adapt to nucleotide chemical modifications and that reduces many of the obstacles commonly faced with the de novo modified SELEX and post-SELEX optimization approaches. We began with a preenriched library of RNA aptamers (2′-OH) that had previously undergone 14 rounds of affinity selection against HIV-1 RT (Burke et al. 1996). This library was retranscribed with either 2′-F, 2′-OMe, or 2′-NH2 pyrimidines, and each new set was subjected to three additional rounds of selection followed by high-throughput sequencing and bioinformatics analyses. From the resulting enrichment data, we chose candidate aptamers to further characterize their ability to bind and inhibit HIV-1 RT enzymatic activity in primer extension assays. This approach identified 2′-modified RNA aptamers that maintained their originally selected function of binding to HIV-1 RT despite the 2′ modification. Unexpectedly, all transcripts carrying 2′-FY modifications—including nonbinding control sequences—were found to bind and inhibit several retroviral RTs, but either did not inhibit or only very weakly inhibited bacterial and human DNA polymerases. The enhanced inhibition by the 2′-FY-modified transcripts is proportional to the mole fraction of 2′-FY and is due, in part, to increased ionic interactions of the 2′-FY transcripts with HIV-1 RT in comparison with 2′-OH transcripts. These observed properties of 2′-FY-modified RNA may provide new insights on the binding interactions of 2′-FY aptamers to their respective targets.
RESULTS
Reselection strategy to identify aptamers that can tolerate chemical modifications
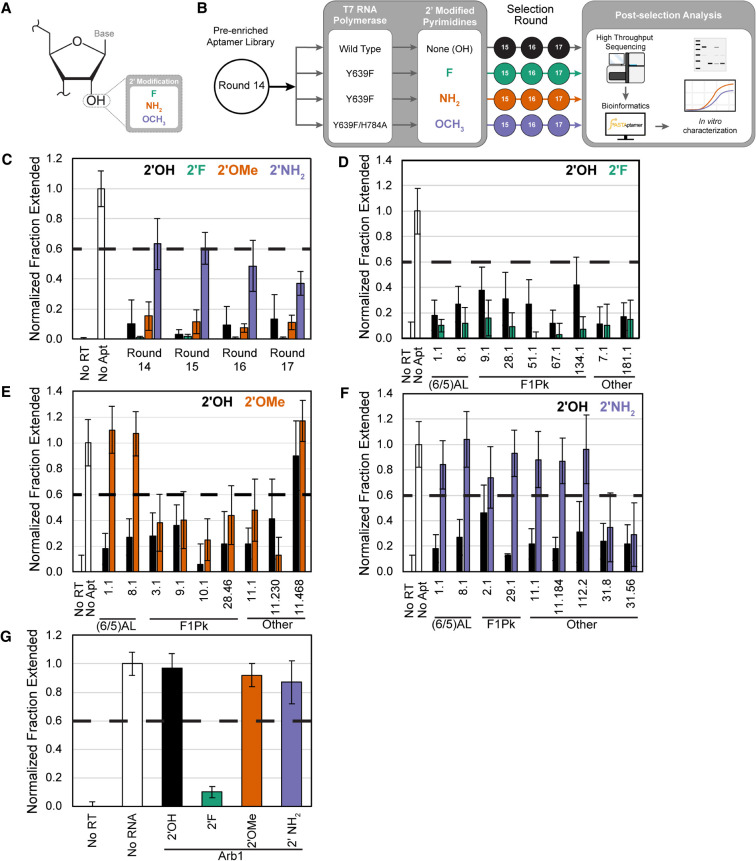
To rapidly identify aptamers that can productively accommodate or adapt to alternative nucleotide alphabets, a preenriched population of RNA aptamers that emerged from 14 rounds of selection for binding to HIV-1 RT (Burke et al. 1996) was subjected to three additional rounds of selection for binding to HIV-1 RT in which the populations carried either the original (2′-OH) or 2′ modified pyrimidines (2′-FY, 2′-OMeY, or 2′-NH2Y) (Fig. 1A,B). Each reselection trajectory was done in duplicate. Output libraries for each round were screened for RT inhibition, defined as a reduction in the “normalized fraction extended” value from 1.0 in the absence of aptamer to less than 0.6 (Fig. 1C; Nguyen et al. 2020). The results shown in Figure 1 demonstrate that the 2′-FY and 2′-OMeY preenriched libraries strongly inhibited RT. In contrast, the preenriched library was noninhibitory upon substitution to 2′-NH2Y but regained some RT inhibition after several rounds of reselection with 2′-NH2Y.
FIGURE 1.
Effect of 2′-pyrimdine modifications on RT inhibition by candidate aptamers. (A) Structure of ribose in RNA highlighting the 2′ position and modifications tested in this study. (B) Schematic of the reselection strategy. A preenriched aptamer library that had previously undergone 14 rounds of selection for affinity to HIV-1 RT was transcribed with either 2′-OH, 2′-F, 2′-OMe, or 2′-NH2 pyrimidines and reselected for three additional rounds, with each trajectory performed in duplicate. (C) Quantification of primer extension assays showing fraction of primer converted into full-length product in control reactions in the absence of RT (No RT) or aptamer (No Apt) and in reactions containing selected aptamer libraries from the 2′-OH (black), 2′-F (green), 2′-OMe (orange), or 2′-NH2 (purple) trajectory after each reselection round. (D–F) Quantification of primer extension assays showing fraction of primer converted into full-length product in control reactions in the absence of RT (No RT) or aptamer (No Apt) or in reactions containing aptamers transcribed with either 2′-OH or (D) 2′-F, (E) 2′-OMe, or (F) 2′-NH2 pyrimidines. Aptamers are grouped together by structural class: (6/5) asymmetric loop family [(6/5)AL] or family 1 pseudoknot (F1Pk). Aptamers that did not contain the consensus sequence features of any of the characterized structural motifs were grouped as Other. (G) Evaluation of primer extension assays in the presence of 2′-modified Arb1. Plotted values and vertical error bars represent the means and standard deviations of fraction primer extended to full-length product normalized to the no aptamer (or no RNA) control (set to 1) and to the no RT control (set to 0) of four independent replicates (n = 4). RNAs are considered inhibitory if the normalized fraction extended value is below 0.6, which is marked by a dashed horizontal line.
High-throughput sequencing data for aptamer populations from rounds 14 and 17 were analyzed using FASTAptamer, a bioinformatics toolkit that counts the number of times an individual sequence is present in each population, clusters highly similar sequences within a population, and calculates enrichment of sequences between populations (Alam et al. 2015). The total reads and number of unique sequences for each aptamer population is given in Supplemental Table S1. Aptamers that coenriched at least twofold in each of the duplicate trajectories for a given modification were prioritized for further characterization (Supplemental Tables S2, S3). Most of these belonged to the characterized family 1 pseudoknot (F1Pk) motif (Tuerk et al. 1992; Burke et al. 1996; Ditzler et al. 2013) or (6/5) asymmetric loop [(6/5)AL] motif (Ditzler et al. 2013) aptamer families (Supplemental Fig. S1A). (6/5)AL aptamers are broad-spectrum inhibitors of reverse transcriptases across multiple strains of HIV-1, while F1Pk aptamers are potent inhibitors of HIV-1 reverse transcriptases from Group M:Subtype B, which is the target against which they were originally selected. A single point mutation R277K confers resistance to inhibition by F1Pk aptamers. For comparison, additional sequences were chosen that codepleted at least twofold in each of the duplicate trajectories. Aptamers from the different structural families demonstrated different sensitivities to the three modifications tested, and similar results were observed with a subset of previously identified HIV-1 RT aptamers (Supplemental Fig. S1). 2′-FY versions of aptamers from the 2′-FY trajectories retained ability to inhibit RT to at least the same degree as the unmodified (2′-OH) versions of these same aptamers (Fig. 1D). 2′-OMeY versions of aptamers from the 2′-OMe trajectory exhibited different effects on RT inhibition depending on the structural class of the aptamer (Fig. 1E). The two (6/5)AL aptamers no longer inhibited RT as 2′-OMeY transcripts, while the F1Pk aptamers were only moderately affected (see Supplemental Text and Supplemental Fig. S2 for more analysis on the effects of the 2′-OMeY modification on these structural classes). 2′-NH2Y versions of aptamers from the 2′-NH2 trajectory were generally unable to inhibit HIV-1 RT (Fig. 1F), although each remained inhibitory as 2′-OH transcripts. Only two aptamers, both from cluster 31, inhibited RT to the same degree as their unmodified RNA versions when generated as 2′-NH2Y transcripts (Fig. 1F; also see Supplemental Text and Supplemental Fig. S3 for initial structural evaluation). Over the course of the selections, relative enrichment of the F1Pk and (6/5)AL structural families as a whole followed the trends predicted from screens of individual exemplars of these families. The F1Pk structural motif did not enrich or deplete more than twofold in any of the trajectories (Supplemental Table S4). In contrast, the (6/5)AL motif was much more strongly depleted in the 2′-OMeY and 2′-NH2Y trajectories (Supplemental Table S5), consistent with its sensitivity to these modifications. These results demonstrate the feasibility of the reselection strategy for identifying the subset of aptamers in a preenriched library that can function with modified nucleotides, and they highlight the variable degree to which 2′-modified pyrimidines impact various structural families of RNA aptamers with affinity for HIV-1 RT.
2′-FY transcripts as general inhibitors of retroviral RTs
In many of the RT enzymatic inhibition assays above, 2′-FY aptamer transcripts were more potent than 2′-OH or other forms in inhibiting HIV-1 RT. We therefore evaluated the impact of 2′-FY modifications on RT inhibition for Arb1, an arbitrary control RNA that contains 70 nucleotides from the luciferase gene mRNA, flanked by the same constant regions as the aptamers. This sequence has previously been shown not to bind RT in vitro or to inhibit virus replication in cells (Lange et al. 2012, 2017). As expected, a 2′-OH version of Arb1 did not inhibit HIV-1 RT. 2′-OMeY and 2′-NH2Y versions were similarly noninhibitory. In contrast, the 2′-FY version of Arb1 strongly inhibited HIV-1 RT (Fig. 1G). In other words, substituting the 2′-FY in place of 2′-OH converted a noninhibitory RNA transcript into an inhibitor for HIV-1 RT.
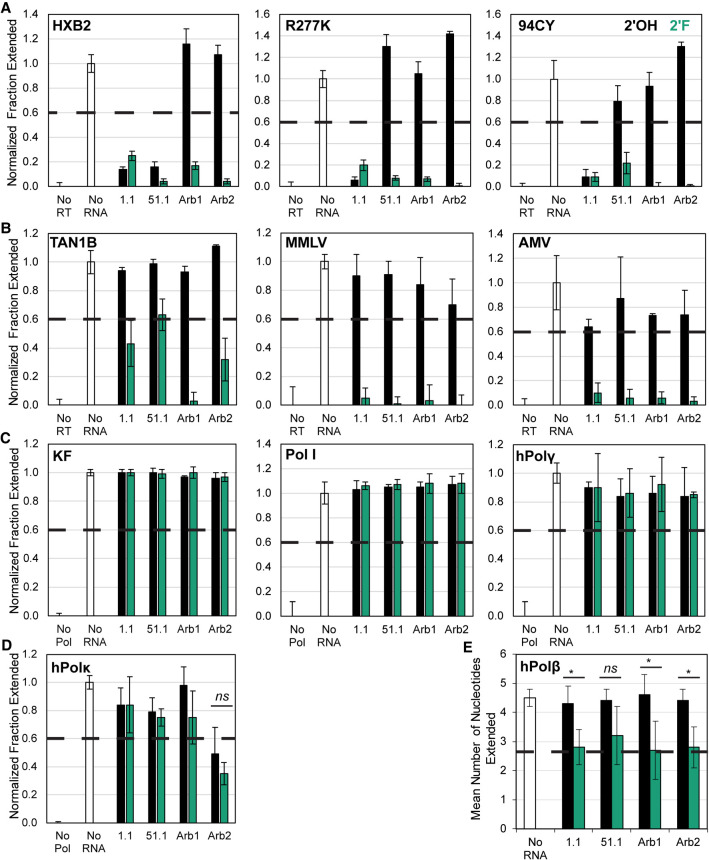
To establish the generalizability of this unexpected result, unmodified and 2′-FY modified transcripts were generated for a broad-spectrum (6/5)AL aptamer (70N 1.1) and for an F1Pk aptamer (70N 51.1), as well as for control RNAs Arb1 described above and Arb2 (an RNA aptamer selected to bind an unrelated protein). These eight transcripts (four sequences, each ± 2′-FY modifications) were screened for their abilities to inhibit a panel of 11 DNA polymerases that included RTs from phylogenetically diverse HIV-1, RTs from other retroviruses, and nonviral DNA polymerases. In unmodified form, both anti-RT aptamers inhibited the HIV-1 RT from strain HIV-1 HXB2 (Group M:Subtype B), but only the (6/5)AL aptamer inhibited the R277K point mutant of HIV-1 HXB2 RT and the RT from HIV-1 strain 94CY (Group M:Subtype A), consistent with previously observed specificities for these structural families (Held et al. 2007; Ditzler et al. 2013; Alam et al. 2018). Neither Arb1 nor Arb2 inhibited these three RTs as 2′-OH transcripts. In contrast, the 2′-FY versions of all four RNAs strongly inhibited these Group M RTs (Fig. 2A). The RT from the chimpanzee strain of simian immunodeficiency virus (SIVcpz), TAN1B, was not inhibited by either anti-RT aptamer as 2′-OH transcripts, consistent with previous observations (Held et al. 2007; Alam et al. 2018), while the 2′-FY-modified versions of these aptamers were moderately better inhibitors of TAN1B RT. Arb1 and Arb2 were both noninhibitory as 2′-OH transcripts but strongly inhibited TAN1B RT as 2′-FY transcripts (Fig. 2B, left panel).
FIGURE 2.
2′-FY RNA sequences inhibit retroviral reverse transcriptases. Primer extension assays (n = 4) were used to monitor polymerase inhibition by unmodified RNA (black) and 2′-FY RNA (green) transcripts using (A) RT from HIV-1 Group M strains and an F1Pk-resistant point mutant; (B) RT from SIVcpz and other retroviruses; (C) Klenow Fragment and holoenzyme of DNA Polymerase I and human DNA polymerase γ; (D) human DNA polymerase κ; and (E) human DNA polymerase β. In E, the dashed horizontal line marks 60% of the No RNA value. Inhibition is observed if the mean number of nucleotides incorporated is below this value. ns (P > 0.05), (*) (P < 0.05).
RTs from two other retroviruses were also screened. Moloney murine leukemia virus (MMLV) is a gammaretrovirus for which the 75 kDa RT is monomeric in crystal structures (Das and Georgiadis 2004). Avian myeloblastosis virus (AMV) is an alpharetrovirus with a heterodimeric RT consisting of a 63 kDa α subunit and a 95 kDa β subunit (Konishi et al. 2011). The 2′-FY RNAs strongly inhibited both retroviral RTs, but none of the unmodified RNAs inhibited either MMLV or AMV RT (Fig. 2B, middle and right panels). Finally, nonviral DNA-dependent DNA polymerases were screened. The Klenow fragment of Escherichia coli DNA polymerase I (KF), E. coli DNA polymerase I holoenzyme, and human DNA polymerase γ (hPolγ) were not inhibited by 2′-OH nor 2′-FY RNAs (Fig. 2C). Human DNA polymerase κ (hPolκ), a Y-family DNA polymerase (Sale et al. 2012), was not inhibited by the 2′-OH or by the 2′-FY versions of the RT aptamers and Arb1 (Fig. 2D). Although it was modestly inhibited to equivalent extents by both the 2′-OH and 2′-FY versions of Arb2, there is no clear 2′-FY-mediated enhancement of this inhibition. Human DNA polymerase β (hPolβ) is an X-family DNA polymerase that is typically involved in the base excision repair pathway (Yamtich and Sweasy 2010). hPolβ is typically processive for short gaps (1–6 nt) (Yamtich and Sweasy 2010), and its low processivity was observed here, compared to the other polymerases tested in this panel. Therefore, we quantified the mean number of nucleotides extended from the 18 nt primer that is annealed to the 31 nt template (maximum of 13 nt extension). A representative gel from this primer extension assay is shown in Supplemental Fig. S4. The mean number of nucleotides incorporated in the absence of aptamer or when 2′-OH RNAs were present was approximately 4, while the mean number of nucleotides incorporated when 2′-FY RNAs were present was approximately 3 (Fig. 2E). Therefore, while there is a difference between most unmodified and 2′-FY RNAs, the mean number of nucleotides incorporated was not reduced below 60% of the No RNA value. We conclude that 2′-FY RNAs bind and inhibit RTs from HIV-1 and other retroviruses with little or no specificity, but that this generalized inhibition does not extend to the nonreverse transcribing DNA polymerases tested here.
Sequence and compositional determinants of the 2′-FY effect for RT inhibition
In principle, RNA structure, length, and composition could all contribute to the strong RT inhibition observed for 2′-FY transcripts that were otherwise noninhibitory as unmodified 2′-OH transcripts. The Arb1 control RNA is predicted by mfold (Zuker 2003) to be fairly unstructured, with 30%–50% of the nucleotides unpaired. When the full-length 2′-FY version of Arb1 was annealed with its unmodified reverse complement, Arb1 alone and the 2′-FY/2′-OH duplex, but not the unmodified reverse complement strand, strongly inhibited HXB2 RT (Supplemental Fig. S5). Next, Arb1 RNA was truncated from the 3′ end to generate 100, 80, 64, and 45 nt versions, each of which had similar ratios of pyrimidines to purines as the full-length version. All truncations strongly inhibited HXB2 RT (Supplemental Fig. S5). The 2′-fluoro moieties must be part of an oligonucleotide to observe RT inhibition, as adding monomeric 2′-FY NTPs to the extension reactions did not inhibit HXB2 RT (Supplemental Fig. S6). To determine how extensively the pyrimidines need to be 2′-F-modified in order to observe strong inhibition of HIV-1 RT, four RNAs were transcribed in vitro with different percentages of 2′-FY NTP substitutions, ranging from 0% to 100%, and each RNA was tested for inhibition of RTs that they would not normally inhibit. The F1Pk aptamer was tested against the R277K point mutant of HXB2 RT since this mutation confers resistance to inhibition by F1Pk aptamers, as noted above. The other three RNAs (Arb1, Arb2, and a random-sequence library with 56 random positions) were evaluated against both HXB2 RT and the R277K point mutant. For all seven RT-RNA combinations, no inhibition was observed for fully unmodified transcripts (0% 2′-FY) and strong inhibition was observed for fully modified transcripts (100% 2′-FY), with half-maximal inhibition being observed at ∼40%–60% 2′-FY for most RNAs and slightly higher for the F1Pk aptamer against the R277K point mutant (Fig. 3). The progressive increase in RT inhibition with increasing mole fraction 2′-FY, observed across multiple unrelated transcripts with difference sequences and structures, suggests that the RT binding mode of 2′-FY RNAs is likely due to physico-chemical contributions of the 2'F groups to aptamer interactions with RT.
FIGURE 3.
Effect of 2′-FY composition on RT inhibition. RNA sequences indicated in the upper left of each panel were transcribed with the indicated percentages of 2′-FY NTPs present in the transcription reactions and were then tested for RT inhibition (n = 4) of the RT indicated in the upper right of each panel. Data are plotted for (A) F1Pk family aptamer 70N 51.1, (B) Arb1 control, (C) Arb2 control, and (D) 56N random RNA library.
The 2′-FY modification increases the ionic character of the interaction between RNA and HIV-1 RT
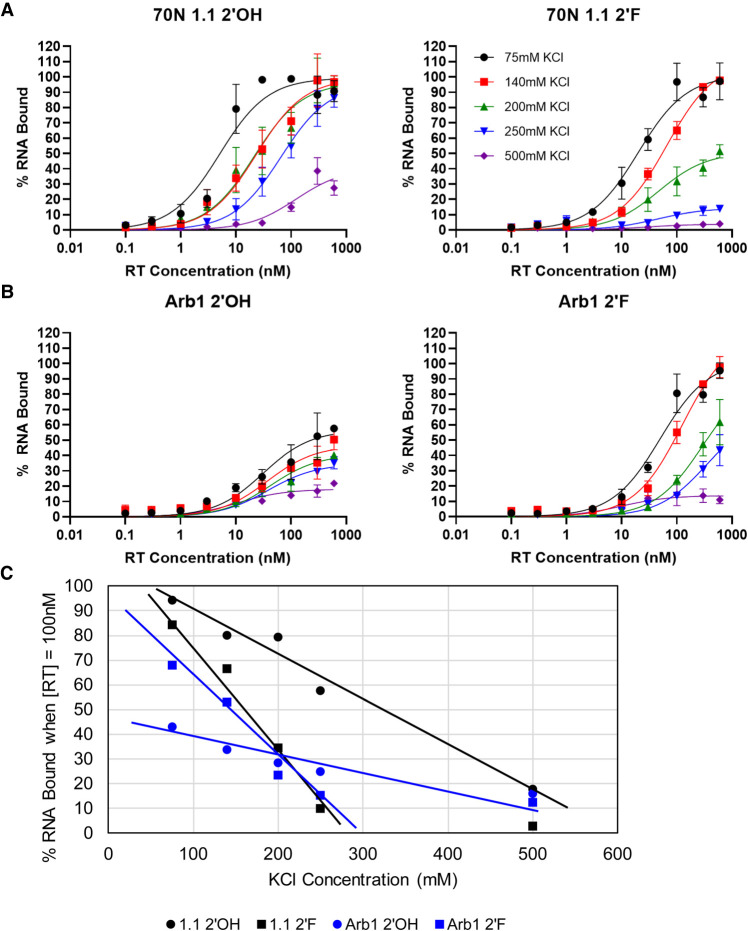
The enzymatic inhibition assays above indirectly monitor binding of RT by the RNA transcripts because the transcripts compete with primer-template for access to RT. Therefore, nitrocellulose filter binding assays were performed to measure RT binding directly and to compare apparent binding affinities of the modified and unmodified transcripts. When generated as unmodified transcripts, the (6/5)AL aptamer 70N 1.1 and the F1Pk aptamer 70N 51.1 had apparent dissociation constants (KD) in the low nanomolar range, as expected, while arbitrary sequences Arb1 and Arb2 did not strongly bind RT (Table 1; Supplemental Fig. S7). As 2′-FY RNA transcripts, aptamers 70N 1.1 and 70N 51.1 bound RT with a similar or slightly stronger affinity as the unmodified version. For the 2′-FY versions of arbitrary control RNAs, Arb1 bound RT to a similar degree as 70N 1.1 while Arb2 bound approximately twofold less strongly.
TABLE 1.
Apparent binding affinity (KD) of unmodified and 2′-FY RNAs to HXB2 RT

To evaluate the ionic character of the binding interaction between RT and 2′-FY RNA, we investigated how salt concentration affected the binding of RT to the 2′-FY and unmodified versions of 70N 1.1 and Arb1. Nitrocellulose filter binding assays were done using binding buffers in which the potassium chloride (KCl) concentration ranged from 75 to 500 mM. In all cases, affinity decreased with increasing KCl, as expected for a protein-nucleic acid complex. However, the effect appears to be stronger for the 2′-FY transcripts (Fig. 4A,B). By plotting the percent RNA bound from each of the fitted curves for the four RNAs when RT concentration is equal to 100 nM versus the KCl concentration present in the binding buffer, there was a nearly linear decrease in binding as salt concentration increased for the four RNAs (Fig. 4C). However, the binding of 2′-FY RNAs to RT decreased more rapidly as salt concentrations increased, in comparison to the slower drop-off observed for their corresponding unmodified versions, indicating that the 2′-FY modification increases the ionic character of the interaction between the modified RNAs and HIV-1 RT.
FIGURE 4.
Effect of increasing salt concentrations on binding of RNA sequences to HXB2 RT. Binding of unmodified and 2′-FY modified (A) 70N 1.1 and (B) Arb1 RNAs to HXB2 RT at increasing salt concentrations. Binding was measured using a nitrocellulose filter binding assay with alkaline-treated filters (n = 3). (C) Plot of the percent RNA bound when RT concentration is equal to 100 nM from each of the fitted curves for the four data sets in A,B. A line-of-best-fit is plotted for the first four data points of each set to illustrate the general trend of decreasing binding as salt concentration increases for each sequence.
DISCUSSION
This study demonstrated a reselection strategy for the identification of chemically modified aptamers and provided new insights on the impact of 2′-FY modifications on target recognition. A preenriched RNA library with affinity for HIV-1 RT was retranscribed with 2′ modifications, and three additional rounds of selection with the modified libraries were performed. High-throughput sequencing and bioinformatics identified aptamer sequences that productively accommodated 2′ pyrimidine modifications and that still bound HIV-1 RT in vitro, and coenrichment in the parallel trajectories helped identify which sequences to test. Structurally distinct aptamer families exhibited different inhibition profiles to the 2′ modifications. All aptamers tested strongly inhibited RT when 2′-F pyrimidines were incorporated. None of the HIV-1 RT aptamers from previously identified structural families inhibited RT when 2′-NH2 pyrimidines were incorporated, likely due in part to the fact that the 2′-NH2 modifications favor the C2′-endo sugar pucker and destabilize RNA duplexes (Aurup et al. 1994). The reselection identified two new aptamers from a single cluster (cluster 31) that were able to inhibit HIV-1 RT when transcribed with 2′-NH2 pyrimidines. The impact of 2′-OMe pyrimidines was structure-specific, as F1Pk aptamers still inhibited RT, but (6/5)AL and UCAA motif aptamers no longer inhibited RT.
The reselection strategy described here is conceptually similar to the recently described “Poly-Target” selection approach, in which the preenriched RNA aptamer library with affinity for RT from the BH10 strain of HIV-1 was subjected to additional cycles of selection for affinity to RTs from phylogenetically diverse lentiviruses (Alam et al. 2018). Both approaches sought to find specialized aptamer subsets from within preenriched libraries. While the present study looked to find aptamers that tolerate or productively accommodate various 2′ pyrimidine modifications, the goal of the Poly-Target selection approach was to identify aptamers with broad-spectrum target recognition (Alam et al. 2018). An analogous approach could be used to find aptamer subsets that are compatible with alternative alphabets from among preenriched aptamer libraries with other initial compositions and with affinity for other targets. In addition, while the reselection phase of the strategy described here introduced 2′-pyrimidine modifications, the strategy can easily be generalized to other chemical modifications, so long as there are reverse transcriptases and/or polymerases that recognize and incorporate the modified nucleotides. The “preenrichment + reselection” approach requires that there be overlap in the fitness landscapes in which the same sequence (or a close relative) is active for target recognition when generated from two different monomer alphabets. Given appropriately matched monomer sets for the two phases of the selection, this approach could enable exploration of nucleotide chemistries that are too expensive or technically challenging to carry out with the modified nucleotides from the beginning.
Promiscuous recognition of viral reverse transcriptases by generic 2′-FY-modified RNAs has not been described previously and was unexpected. The 2′-F moiety is relatively small and favors the same sugar pucker as unmodified RNA. Previous studies suggested that 2′-F-modified aptamers often have higher affinities than unmodified aptamers and attributed this to formation of more thermodynamically stable secondary structures (Aurup et al. 1994; Cummins et al. 1995; Pagratis et al. 1997). Additional studies determined that 2′-F-modified RNA duplexes are less hydrated than unmodified RNA duplexes, resulting in stronger Watson–Crick hydrogen bonding and π–π stacking interactions between nucleobases (Pallan et al. 2011; Patra et al. 2012). A combination of these effects may partially explain our observation that RT inhibition in primer extension assays by bona fide RT aptamers was equivalent or more potent for 2′-FY transcripts than for the corresponding 2′-OH versions. Nevertheless, it was surprising that the negative control 2′-FY arbitrary RNA sequences also strongly inhibited RT (Figs. 1G, 2) and that the 2′-FY RNA sequences inhibited diverse retroviral RTs but generally did not inhibit nonviral DNA polymerases (Fig. 2). For multiple unrelated transcripts, inhibition by 2′-FY RNAs was proportional to the mole fraction of 2′-F pyrimidines present in the transcript (Fig. 3), thereby establishing that no specific position or subset of positions can be responsible for the observed effects, and further suggesting that the 2′-F moieties may bind directly with the RTs. We also observed that the binding of 2′-FY RNAs to HIV-1 RT was more sensitive to increasing salt concentrations than were their unmodified versions (Fig. 4), suggesting that the 2′-F modification may increase the ionic character of the interaction between the RNA transcripts and reverse transcriptases. The precise physico-chemical basis for the increased ionic character is not immediately apparent. An important contributing factor may be loss of intramolecular interactions involving the proton of the 2′-hydroxyl. Specifically, in unmodified RNA, the 2′-hydroxyl proton can partially neutralize the negative charge of the phosphate backbone, but when replaced with the more electronegative fluorine group, this partial neutralization no longer occurs, and the phosphate retains its full negative charge. The additional negative charge on the phosphate may then be responsible for the enhanced ionic character of the interaction with RT.
2′-FY modifications are commonly used in aptamer selections because 2′-FY incorporation confers serum stability and because the nucleotide triphosphates are commercially available and are efficiently incorporated by modified T7 RNA polymerases. Our findings raise the possibility that fluoro modifications could contribute directly to the affinities observed for other aptamer-target pairs by various mechanisms, including via contributions to ionic interactions. 2′-FY RNA aptamers with affinity for HIV-1 RT have not been previously selected directly from random libraries; however, Alves Ferreira-Bravo et al. recently reported selection of FANA aptamers with enhanced affinity against HIV-1 RT (Alves Ferreira-Bravo et al. 2015). FANA is structurally distinct from 2′-F NTPs, as the fluoro substituent is in a β conformation for FANA, as compared to an α conformation for 2′-F ribose, and the FANA sugar pucker is in a C2′/O4′-endo conformation, as compared to C3′-endo in 2′-F NTPs (Berger et al. 1998; Trempe et al. 2001). Those authors found that some of the selected FANA aptamers, such as “FA1,” appear to bind RT with KD values in the single-digit pM range, while the FANA random pools bound HIV-1, MMLV, and AMV RTs at low nanomolar affinities. Although they observed that FA1 affinity for RT decreased with increasing ionic strength, it is not yet known whether the salt sensitivity of FANA transcripts is greater than that of RNA or DNA, or whether an increased ionic character may account for some of the enhanced affinity observed for FANA aptamers to HIV-1 RT.
Increased ionic character may be an important feature of other 2′-FY aptamers that are in development for use as theranostics and biosensors. In addition, our observation that the 2′-FY modifications can convert noninhibitory RNA transcripts into RT inhibitors raises the possibility that 2′-FY aptamers for other targets may be more prone to off-target binding than aptamers with other compositions when presented with highly basic alternative targets, especially under low salt conditions. In general, in vitro assessment of aptamer function should carefully control for ionic composition, particularly in comparing results from one lab or experimental setting to another. Finally, the inferences made in this study regarding potential contributions of fluoro moieties to target recognition were only made possible by observing a gain of function when unmodified aptamer or arbitrary sequences were retranscribed with 2′-FY nucleotides. Many aptamers that were initially selected with 2′-F pyrimidines have been shown to lose affinity for their targets when transcribed with unmodified pyrimidines. The root cause of this loss of affinity is rarely delineated with precision and could often simply reflect structural or other constraints associated with having evolved with the 2′-F modification. It remains to be seen whether fluoro moieties in 2′-FY or FANA aptamers contribute directly to binding affinities of other fluoro-substituted aptamers to their respective targets and/or to unwanted interactions with alternative molecular targets.
MATERIALS AND METHODS
RT expression and purification
Enzymatically active p66/p51 heterodimer RTs from different viral strains of HIV were purified and validated as described previously (Alam et al. 2018). Protein concentration was calculated from UV absorbance at 280 nm using an estimated extinction coefficient of 260,120 M−1cm−1 based on the amino acid sequence. RT preps were validated for purity and size by SDS-PAGE and for activity by comparison of primer extension assays against previous preps. Proteins were stored at −80°C after addition of glycerol to 50% (v/v).
Modified aptamer reselection, high-throughput sequencing, and bioinformatics
The starting library for this work was the round 14 70N library described previously (Burke et al. 1996). Double-stranded DNA from this library was transcribed in vitro to generate unmodified (2′-OH) and 2′-pyrimidine-modified input RNA libraries for the reselection. 2′-modified pyrimidines were purchased from TriLink Biotechnologies. Run-off transcription reactions of 2′-OH, 2′-FY, and 2′-NH2Y transcripts were performed using the Y639F mutant T7 RNA polymerase (Sousa and Padilla 1995), in vitro transcription buffer (50 mM Tris-HCl pH 7.5, 15 mM MgCl2, 5 mM DTT, and 2 mM spermidine), and 2 mM each of ATP, GTP, and the corresponding 2′-modified CTP and UTP. Transcription reactions for 2′-OMe RNAs utilized the same components as above, except that a Y639F/H784 double mutant T7 RNA polymerase (Padilla and Sousa 2002) was used, and 2 mM each 2′-OMe-CTP and 2′-OMe-UTP, 6 mM MnCl2, 2 mM GMP, 10% PEG 6000, and 1 U 10U/µL inorganic pyrophosphatase (Thermo Fisher Scientific) were added to the reaction. Reactions were incubated at 37°C for a minimum of 4 h and halted with the addition of denaturing gel loading buffer (90% formamide, 50 mM EDTA and trace amounts of xylene cyanol and bromophenol blue). RNAs were purified by denaturing polyacrylamide gel electrophoresis (6% TBE-PAGE, 8 M urea), and bands corresponding to the expected product sizes were excised from the gel and eluted while tumbling overnight in 300 mM sodium acetate pH 5.4. Eluates were ethanol precipitated, resuspended in buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA), and stored at −20°C until further use. RNA concentrations were determined on a NanoDropOne spectrophotometer (Thermo Fisher Scientific). For each trajectory and round of selection, 200 pmol of transcribed libraries (∼1.2 × 1014 molecules) were resuspended in 100 µL binding buffer (50 mM Tris-HCl [pH 7.5], 150 mM KCl, and 10 mM MgCl2) and renatured by heating to 65°C and cooling on ice. An amount of 40 pmol HXB2 RT was then added to a final concentration of 400 nM, and the mixture was incubated on ice for an additional 20 min. The bound RNA species were partitioned from the unbound species using nitrocellulose filters and recovered as previously described (Alam et al. 2018). The recovered RNA was reverse-transcribed using ImProm-II Reverse Transcriptase (Promega) and PCR-amplified for the next round of selection or for HTS. Round 14 and Round 17 libraries were prepared for sequencing using a series of PCR steps to add Illumina adapters and sequencing indices for multiplexing of the 70N libraries as previously described (Ditzler et al. 2013). The primers used to append the Illumina adapters and sequencing indices can be found in Supplemental Table S6. Sequencing was performed on an Illumina HiSeq2000 (University of Missouri DNA Core Facility). Populations were demultiplexed to identify and parse the 5′ and 3′ constant regions. Data preprocessing was performed using cutadapt (Martin 2011) to trim 5′ and 3′ constant regions from sequences and to discard any uncut sequences or sequences not within ±3 nt of the expected size (70 nt) after trimming. Trimmed sequences were then filtered for high-quality reads using FASTQ quality filter from the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx-toolkit/). Quality filtering eliminated a sequence if a single position had a Phred quality score of less than 20. Trimmed and quality filtered sequences were then processed using the FASTAptamer toolkit (Alam et al. 2015) to count and normalize sequence reads (FASTAptamer-Count), calculate fold enrichment from round 14 to round 17 (FASTAptamer-Enrich), and group related sequences into clusters (FASTAptamer-Cluster). Aptamers were named according to the cluster in which they were found in round 14 and their rank in terms of abundance within the cluster. For example, aptamer 70N 31.56 was part of cluster 31 in round 14 and was the 56th most abundant sequence within that cluster. To determine motif enrichment values, FASTAptamer-Search was used to identify sequences from the libraries that matched sequence patterns for the F1Pk [TCCG and GCCC] and (6/5)AL [RCGTY and RARAC] structural motifs, and a custom Perl script was used to count the total number of motif reads, normalize motif reads, and calculate motif enrichment values.
DNA templates and RNA transcription
For each aptamer to be transcribed, DNA oligonucleotides were purchased as “left” and “right” halves from Integrated DNA Technologies. Oligonucleotides corresponding to the “right” half were 5′-phosphorylated using T4 polynucleotide kinase (New England Biolabs). Equimolar amounts of “right” strand, “left” strand, and a complementary bridge oligo were annealed and ligated using T4 DNA ligase (New England Biolabs). Ligated templates were PCR amplified using Pfu DNA polymerase, a forward primer to append the T7 promoter, and a reverse primer complimentary to the 3′ constant region. Amplified products were verified for size using agarose gel electrophoresis. The double-stranded DNA templates were then transcribed in vitro and purified as above. The RNA sequences used in this study are in Supplemental Table S2. The Arb2 sequence was kindly provided by Dr. Margaret Lange (University of Missouri). Two guanine nucleotides were added to the 5′ end of the reverse complement of Arb1 to aid transcription, resulting in a 2-nt 5′ overhang in the annealed duplex. For transcriptions to generate mixed 2′-OH/2′-FY polymers, NTPs were adjusted to maintain a constant total NTP concentration. For example, transcription of an RNA containing 40% 2′-FY NTPs utilized 0.8 mM each of 2′-F-CTP and 2′-F-UTP and 1.2 mM each of CTP and UTP, in addition to 2 mM each ATP and GTP.
Primer extension assays
Inhibition of the DNA-dependent DNA polymerase activities of HIV-1 RT and other DNA polymerases was monitored using the primer extension assay as previously described (Ditzler et al. 2013). Briefly, 5′-Cy3-end-labeled 18 nt DNA primer corresponding to the 3′ end of tRNALys3 was annealed with a 31 nt DNA template. 20 nM RT was preassembled with excess (100 nM) RNA in extension buffer (50 mM Tris-HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2) for 10 min. Note that the ionic conditions of the extension buffer were different than those of the binding buffer used in the binding studies. The lower monovalent and higher divalent ion concentrations in the extension buffer, relative to the binding buffer, may affect binding affinity of 2′-FY RNAs to RT. Polymerization reactions were initiated by the addition of preannealed 10 nM primer and 20 nM template and 100 µM of each dNTP (final concentrations). After 10 min at 37°C, reactions were stopped by adding an equal volume of denaturing gel loading buffer, then heated to 90°C for 2 min immediately prior to loading onto a 10% polyacrylamide, 8 M urea denaturing gel. Gels were scanned for Cy3 fluorescence using a Typhoon FLA 9000 phosphoimager (GE Healthcare Life Sciences). The fraction of primer extended to full-length product was quantified by measuring the intensity of the bands using Multigauge software (Fujifilm) and normalized by setting the fraction of primer extended when no RNA was present to 1. Extension reactions using the Klenow fragment of E. coli DNA polymerase I (New England Biolabs) were done using 20 nM enzyme per reaction for 5 min in extension buffer containing 10 mM Tris-HCl [pH 7.9], 50 mM NaCl, 10 mM MgCl2, and 1 mM DTT. MMLV RT (Promega) extension reactions were done using 20 nM enzyme per reaction for 10 min in extension buffer containing 50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2, and 10 mM DTT. Human DNA polymerase γ was a gift from Dr. Whitney Yin (University of Texas Medical Center), human DNA polymerase κ was a gift from Dr. Robert Eoff (University of Arkansas Medical Center), and human DNA polymerase β, E. coli DNA polymerase I holoenzyme, and AMV RT were gifts from Dr. Mukund J. Modak (Rutgers University). Extension time for AMV RT and human DNA polymerase γ was 60 min, and the final concentration for these two proteins was 100 nM. The AMV RT reactions were done in buffer containing 50 mM Tris-HCl [pH 8.3], 50 mM KCl, 10 mM MgCl2, and 10 mM DTT. The human polymerase γ reactions were done in buffer containing 25 mM Tris-HCl [pH 8], 100 mM KCl, 10 mM MgCl2, 1 mM DTT, and 1 mg/mL BSA. The final concentration of the DNA polymerase I holoenzyme was 20 nM, and the extension time was 15 min. The buffer used for this enzyme was 50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM MgCl2, and 1 mM DTT. For human DNA polymerase κ, the final concentration was 100 nM, and extension time was 30 min. The buffer used for hPolκ was 40 mM Tris-HCl [pH 8], 60 mM KCl, 5 mM MgCl2, and 10 mM DTT. For human DNA polymerase β, the final concentration was 200 nM, and the extension time was 60 min. The buffer used for hPolβ was the same used for the DNA polymerase I holoenzyme. To quantify the largely distributive extension observed due to the low processivity by hPolβ, the mean number of nucleotides extended was determined. First, the intensity of each individual band within a given lane (up to 14 bands total, 0–13 nt extended onto the primer) was quantified. Values were weighted by multiplying each band's intensity by the number of nucleotides that were extended to produce that DNA product. The sum of the weighted intensities was then divided by the sum of the unweighted intensities within the lane to calculate the mean number of nucleotides extended. P values were calculated using an unpaired t-test computed by GraphPad Prism.
Binding affinity determination
Dissociation constant (KD) values were determined using a nitrocellulose filter binding assay. Approximately 20,000 counts-per-minute of 5′-radiolabeled and refolded RNA was incubated with varying concentrations of RT (0.1 to 800 nM or without RT to determine background binding) in binding buffer (50 mM Tris-HCl [pH 7.5], 140 mM KCl, 1 mM MgCl2, and 0.1 µg/mL BSA, unless otherwise noted) and allowed to equilibrate at room temperature for 15 min. RNA:RT complexes were then partitioned from unbound RNA by passing samples through a prewet nitrocellulose filter under vacuum, as described above for the partitioning step of the selection, and immediately washing with 500 µL binding buffer. Radioactivity retained on the filter was counted by placing filters into scintillation vials, adding 4 mL liquid scintillation fluid, and counting using a liquid scintillation counter. An unfiltered “No Wash” sample was counted to determine the total amount of radioactivity (“100%”) that was added to each binding reaction. The percent RNA bound was calculated by dividing the radioactivity retained on the filter by the radioactivity present in the “No Wash” sample multiplied by 100%. The percent RNA bound values were fit to a one-site, specific binding curve (Y = Bmax*X/[KD + X]) using GraphPad Prism 6.2. In the equation, Bmax is the maximum specific binding, KD is the dissociation constant, X is the RT concentration, and Y is the percent RNA bound. Binding assays were done in triplicate. To decrease background binding to nitrocellulose filters for Arb2 RNA and during salt titrations, nitrocellulose filters were incubated in 0.5 M KOH for 20 min, washed extensively with MilliQ water, and then incubated in the appropriate binding buffer for at least 45 min. Such alkaline treatment has been previously shown to reduce nonspecific binding to nitrocellulose filters (McEntee et al. 1980).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the members of the Burke laboratory for their helpful discussions. This work was supported by the National Institutes of Health (R01AI074389).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.077008.120.
Freely available online through the RNA Open Access option.
REFERENCES
- Aaldering LJ, Tayeb H, Krishnan S, Fletcher S, Wilton SD, Veedu RN. 2015. Smart functional nucleic acid chimeras: enabling tissue specific RNA targeting therapy. RNA Biol 12: 412–425. 10.1080/15476286.2015.1017234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler A, Forster N, Homann M, Goringer HU. 2008. Post-SELEX chemical optimization of a Trypanosome-specific RNA aptamer. Comb Chem High Throughput Screen 11: 16–23. 10.2174/138620708783398331 [DOI] [PubMed] [Google Scholar]
- Alam KK, Chang JL, Burke DH. 2015. FASTAptamer: a bioinformatic toolkit for high-throughput sequence analysis of combinatorial selections. Mol Ther Nucleic Acids 4: e230 10.1038/mtna.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam KK, Chang JL, Lange MJ, Nguyen PDM, Sawyer AW, Burke DH. 2018. Poly-target selection identifies broad-spectrum RNA aptamers. Mol Ther Nucleic Acids 13: 605–619. 10.1016/j.omtn.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves Ferreira-Bravo I, Cozens C, Holliger P, DeStefano JJ. 2015. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res 43: 9587–9599. 10.1093/nar/gkv1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurup H, Tuschl T, Benseler F, Ludwig J, Eckstein F. 1994. Oligonucleotide duplexes containing 2′-amino-2′-deoxycytidines: thermal stability and chemical reactivity. Nucleic Acids Res 22: 20–24. 10.1093/nar/22.1.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger I, Tereshko V, Ikeda H, Marquez VE, Egli M. 1998. Crystal structures of B-DNA with incorporated 2′-deoxy-2′-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability. Nucleic Acids Res 26: 2473–2480. 10.1093/nar/26.10.2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke DH, Scates L, Andrews K, Gold L. 1996. Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J Mol Biol 264: 650–666. 10.1006/jmbi.1996.0667 [DOI] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ. 2012. RNA-based therapeutics: current progress and future prospects. Chem Biol 19: 60–71. 10.1016/j.chembiol.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins LL, Owens SR, Risen LM, Lesnik EA, Freier SM, McGee D, Guinosso CJ, Cook PD. 1995. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res 23: 2019–2024. 10.1093/nar/23.11.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. 2015. Current approaches in SELEX: an update to aptamer selection technology. Biotechnol Adv 33: 1141–1161. 10.1016/j.biotechadv.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Das D, Georgiadis MM. 2004. The crystal structure of the monomeric reverse transcriptase from Moloney murine leukemia virus. Structure 12: 819–829. 10.1016/j.str.2004.02.032 [DOI] [PubMed] [Google Scholar]
- Dellafiore MA, Montserrat JM, Iribarren AM. 2016. Modified nucleoside triphosphates for in-vitro selection techniques. Front Chem 4: 18 10.3389/fchem.2016.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzler MA, Lange MJ, Bose D, Bottoms CA, Virkler KF, Sawyer AW, Whatley AS, Spollen W, Givan SA, Burke DH. 2013. High-throughput sequence analysis reveals structural diversity and improved potency among RNA inhibitors of HIV reverse transcriptase. Nucleic Acids Res 41: 1873–1884. 10.1093/nar/gks1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton BE, Gold L, Hicke BJ, Janjic N, Jucker FM, Sebesta DP, Tarasow TM, Willis MC, Zichi DA. 1997. Post-SELEX combinatorial optimization of aptamers. Bioorg Med Chem 5: 1087–1096. 10.1016/S0968-0896(97)00044-8 [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- Guschlbauer W, Jankowski K. 1980. Nucleoside conformation is determined by the electronegativity of the sugar substituent. Nucleic Acids Res 8: 1421–1433. 10.1093/nar/8.6.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held DM, Kissel JD, Thacker SJ, Michalowski D, Saran D, Ji J, Hardy RW, Rossi JJ, Burke DH. 2007. Cross-clade inhibition of recombinant human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus SIVcpz reverse transcriptases by RNA pseudoknot aptamers. J Virol 81: 5375–5384. 10.1128/JVI.01923-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Eckstein F, Padilla R, Sousa R. 1997. Mechanism of ribose 2′-group discrimination by an RNA polymerase. Biochemistry 36: 8231–8242. 10.1021/bi962674l [DOI] [PubMed] [Google Scholar]
- Jackson LN, Chim N, Shi C, Chaput JC. 2019. Crystal structures of a natural DNA polymerase that functions as an XNA reverse transcriptase. Nucleic Acids Res 47: 6973–6983. 10.1093/nar/gkz513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe AD, Pai S, Ellington A. 2010. Aptamers as therapeutics. Nat Rev Drug Discov 9: 537–550. 10.1038/nrd3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto M, Meyer AJ, Hirao I, Ellington AD. 2017. Genetic alphabet expansion transcription generating functional RNA molecules containing a five-letter alphabet including modified unnatural and natural base nucleotides by thermostable T7 RNA polymerase variants. Chem Commun 53: 12309–12312. 10.1039/C7CC06661A [DOI] [PubMed] [Google Scholar]
- Konishi A, Nemoto D, Yasukawa K, Inouye K. 2011. Comparison of the thermal stabilities of the αβ heterodimer and the α subunit of avian myeloblastosis virus reverse transcriptase. Biosci Biotech Biochem 75: 1618–1620. 10.1271/bbb.110238 [DOI] [PubMed] [Google Scholar]
- Kuwahara M, Sugimoto N. 2010. Molecular evolution of functional nucleic acids with chemical modifications. Molecules 15: 5423–5444. 10.3390/molecules15085423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange MJ, Sharma TK, Whatley AS, Landon LA, Tempesta MA, Johnson MC, Burke DH. 2012. Robust suppression of HIV replication by intracellularly expressed reverse transcriptase aptamers is independent of ribozyme processing. Mol Ther 20: 2304–2314. 10.1038/mt.2012.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange MJ, Nguyen PDM, Callaway MK, Johnson MC, Burke DH. 2017. RNA–protein interactions govern antiviral specificity and encapsidation of broad spectrum anti-HIV reverse transcriptase aptamers. Nucleic Acids Res 45: 6087–6097. 10.1093/nar/gkx155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapa SA, Chudinov AV, Timofeev EN. 2016. The toolbox for modified aptamers. Mol Biotechnol 58: 79–92. 10.1007/s12033-015-9907-9 [DOI] [PubMed] [Google Scholar]
- Larsen AC, Dunn MR, Hatch A, Sau SP, Youngbull C, Chaput JC. 2016. A general strategy for expanding polymerase function by droplet microfluidics. Nat Commun 7: 11235 10.1038/ncomms11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauridsen LH, Rothnagel JA, Veedu RN. 2012. Enzymatic recognition of 2′-modified ribonucleoside 5′-triphosphates: towards the evolution of versatile aptamers. ChemBioChem 13: 19–25. 10.1002/cbic.201100648 [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17: 3 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McEntee K, Weinstock GM, Lehman IR. 1980. recA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc Natl Acad Sci 77: 857–861. 10.1073/pnas.77.2.857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek KN, Rangel AE, Heemstra JM. 2016. Enhancing aptamer function and stability via in vitro selection using modified nucleic acids. Methods 106: 29–36. 10.1016/j.ymeth.2016.03.008 [DOI] [PubMed] [Google Scholar]
- Mei H, Shi C, Jimenez RM, Wang Y, Kardouh M, Chaput JC. 2017. Synthesis and polymerase activity of a fluorescent cytidine TNA triphosphate analogue. Nucleic Acids Res 45: 5629–5638. 10.1093/nar/gkx368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei H, Liao JY, Jimenez RM, Wang Y, Bala S, McCloskey C, Switzer C, Chaput JC. 2018. Synthesis and evolution of a threose nucleic acid aptamer bearing 7-deaza-7-substituted guanosine residues. J Am Chem Soc 140: 5706–5713. 10.1021/jacs.7b13031 [DOI] [PubMed] [Google Scholar]
- Nguyen PDM, Zheng J, Gremminger TJ, Qiu L, Zhang D, Tuske S, Lange MJ, Griffin PR, Arnold E, Chen S-J, et al. 2020. Binding interface and impact on protease cleavage for an RNA aptamer to HIV-1 reverse transcriptase. Nucleic Acids Res 48: 2709–2722. 10.1093/nar/gkz1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla R, Sousa R. 2002. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res 30: e138 10.1093/nar/gnf138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagratis NC, Bell C, Chang Y-F, Jennings S, Fitzwater T, Jellinek D, Dang C. 1997. Potent 2′-amino-, and 2′-fluoro-2′- deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat Biotechnol 15: 68–73. 10.1038/nbt0197-68 [DOI] [PubMed] [Google Scholar]
- Pallan PS, Greene EM, Jicman PA, Pandey RK, Manoharan M, Rozners E, Egli M. 2011. Unexpected origins of the enhanced pairing affinity of 2′-fluoro-modified RNA. Nucleic Acids Res 39: 3482–3495. 10.1093/nar/gkq1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra A, Paolillo M, Charisse K, Manoharan M, Rozners E, Egli M. 2012. 2′-fluoro RNA shows increased Watson-Crick H-bonding strength and stacking relative to RNA: evidence from NMR and thermodynamic data. Angew Chem Int Ed Engl 51: 11863–11866. 10.1002/anie.201204946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, Chaput JC, Wengel J, Peak-Chew S-Y, McLaughlin SH, et al. 2012. Synthetic genetic polymers capable of heredity and evolution. Science 336: 341–344. 10.1126/science.1217622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose KM, Alves Ferreira-Bravo I, Li M, Craigie R, Ditzler MA, Holliger P, DeStefano JJ. 2019. Selection of 2′-deoxy-2′-fluoroarabino nucleic acid (FANA) aptamers that bind HIV-1 integrase with picomolar affinity. ACS Chem Biol 14: 2166–2175. 10.1021/acschembio.9b00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 13: 141–152. 10.1038/nrm3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigdar S, Macdonald J, Connor M, Wang T, Xiang D, Al Shamaileh H, Qiao L, Wei M, Zhou S-F, Zhu Y, et al. 2013. Aptamers as theranostic agents: modifications, serum stability and functionalisation. Sensors 13: 13624–13637. 10.3390/s131013624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R, Padilla R. 1995. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J 14: 4609–4621. 10.1002/j.1460-2075.1995.tb00140.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovall GM, Bedenbaugh RS, Singh S, Meyer AJ, Hatala PJ, Ellington AD, Hall B. 2014. In vitro selection using modified or unnatural nucleotides. Curr Protoc Nucleic Acid Chem 56: 9.6.1–9.6.33. 10.1002/0471142700.nc0906s56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawiah KD, Porciani D, Burke DH. 2017. Toward the selection of cell targeting aptamers with extended biological functionalities to facilitate endosomal escape of cargoes. Biomedicines 5: 51 10.3390/biomedicines5030051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Wilds CJ, Denisov AY, Pon RT, Damha MJ, Gehring K. 2001. NMR solution structure of an oligonucleotide hairpin with a 2′F-ANA/RNA stem: implications for RNase H specificity toward DNA/RNA hybrid duplexes. J Am Chem Soc 123: 4896–4903. 10.1021/ja003859p [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- Tuerk C, MacDougal S, Gold L. 1992. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci 89: 6988–6992. 10.1073/pnas.89.15.6988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ngor AK, Nikoomanzar A, Chaput JC. 2018. Evolution of a general RNA-cleaving FANA enzyme. Nat Commun 9: 5067 10.1038/s41467-018-07611-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamtich J, Sweasy JB. 2010. DNA polymerase family X: function, structure, and cellular roles. Biochim Biophys Acta 1804: 1136–1150. 10.1016/j.bbapap.2009.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Rossi J. 2016. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov 16: 181–202. 10.1038/nrd.2016.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Zou J, Tian F, Shang Z. 2009. Fluorine bonding—How does it work in protein−ligand interactions? J Chem Inf Model 49: 2344–2355. 10.1021/ci9002393 [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.