Abstract
Eukaryotic histone deacetylation, critical for maintaining nucleosome structure and regulating gene expression, is mediated by histone deacetylases (HDACs). Although nucleosomes have been reported to regulate mRNA polyadenylation in humans, the role of HDACs in regulating polyadenylation has not been uncovered. Taking advantage of phenotypic studies on Arabidopsis, HDA6 (one of HDACs) was found to be a critical part of many biological processes. Here, we report that HDA6 affects mRNA polyadenylation in Arabidopsis. Poly(A) sites of up-regulated transcripts are closer to the histone acetylation peaks in hda6 compared to the wild-type Col-0. HDA6 is required for the deacetylation of histones around DNA on nucleosomes, which solely coincides with up-regulated or uniquely presented poly(A) sites in hda6. Furthermore, defective HDA6 results in an overrepresentation of the canonical poly(A) signal (AAUAAA) usage. Chromatin loci for generating AAUAAA-type transcripts have a comparatively low H3K9K14ac around poly(A) sites when compared to other noncanonical poly(A) signal–containing transcripts. These results indicate that HDA6 regulates polyadenylation in a histone deacetylation–dependent manner in Arabidopsis.
As one of the post-transcriptional modifications of mRNA, the polyadenylation process can critically regulate the gene expression in eukaryotes (Tian and Manley 2017). This universally required 3′ end processing for RNA polymerase II (Pol II)-dependent transcripts is mainly mediated by a protein complex which recognizes a poly(A) signal, cleavages the pre-mRNA, and synthesizes the poly(A) tail (Clerici et al. 2018). Recent studies revealed that alternative polyadenylation (APA) sites can be positioned at either the 3′ untranslated region (UTR) or outside of the 3′ UTR (Deng and Cao 2017; Tian and Manley 2017). APA results in increased diversity of transcripts and may affect mRNA stability, localization, and exportation. Consequently, the expression of these genes may be regulated by APA. By using high throughput sequencing approaches, it was found that over 50% of genes contain more than two poly(A) sites in eukaryotes (Xing and Li 2011; Tian and Manley 2017).
Poly(A) signal is mainly recognized by cleavage and polyadenylation specificity factors (CPSFs) (Clerici et al. 2018). Although CPSFs are highly conserved between mammals and plants (Zhang et al. 2008a; Hunt et al. 2012), the poly(A) signal compositions are quite different. More than 50% of transcripts from mammals use the canonical poly(A) signal AAUAAA for polyadenylation (Tian and Manley 2017), whereas the transcripts using AAUAAA only account for about 10% of plant transcripts (Wu et al. 2011b; Xing and Li 2011). The other 40% and 50% of plant transcripts contain 1-nt variations of AAUAAA and other poly(A) signals (Loke et al. 2005), respectively. This distinguished characteristic of poly(A) signal reflects a more complex polyadenylation mechanism in plants that is not yet understood.
Nucleosomes are formed when histone proteins are wrapped with DNA. These nucleosomes are then packed into highly condensed chromatins in the eukaryotic nucleus. It was reported that the nucleosome position affects the usage of proximal or distal poly(A) sites, suggesting the regulatory role of chromatin status in orchestrating mRNA polyadenylation (Huang et al. 2013; Lee and Chen 2013). Overall, the compactness of chromatin is determined by histone modifications such as methylation and acetylation. Within these modifications, histone acetylation is known to be associated with the accessibility of chromatin and the recruitment of DNA-binding proteins to activate gene expression. However, acetylation of the histones can be erased by histone deacetylases (HDACs), resulting in structural changes to the chromatin. Thus, the dynamic acetylation of a histone is an epigenetic hallmark to gate transcription activation. HDA6 (encoded by AT5G63110), one of the HDACs, is associated with heterochromatin formation and gene repression (Tan et al. 2018). In addition, HDA6 is also involved in other biological processes, such as stress tolerance, flowering time, and circadian rhythm in Arabidopsis (Yu et al. 2011; Kim et al. 2017; Wang et al. 2017; Hung et al. 2018). A recent study has revealed that nuclear retention of HDA6 results in gene silencing (Zhu et al. 2019). Essentially, HDA6 deficiency could also result in a significant increase of histone H3K9ac, H3K14ac, (To et al. 2011), and gene activation (Earley et al. 2006; Tanaka et al. 2008; Yu et al. 2017). It was reported that H3K9ac is required for the maximal Pol II pausing release to initiate and elongate transcription (Gates et al. 2017). The hyperacetylation of H3K9ac in coding and intergenic regions is correlated with transcription, reflecting the global regulatory role of histone acetylation on gene expression (Kurdistani et al. 2004). By facing the challenge of environmental stress, H3K9ac is associated with the expression of induced stress response genes (Kim et al. 2008; Chinnusamy and Zhu 2009). The other type of histone acetylation, H3K14ac, co-occurs with H3K9ac at gene regulatory elements, suggesting their coordinated regulatory role in gene expression (Karmodiya et al. 2012). Furthermore, H3K14ac can be rewritten by acetyltransferases during stress responses which affect the transcriptional elongation of target genes (Johnsson et al. 2009). Moreover, H3K14ac can crosstalk with H3K4 dimethylation during stress responses in soybeans (Wu et al. 2011a). Together, these results indicate that H3K9ac and H3K14ac are important histone epigenetic hallmarks for regulating gene expression. However, their roles in essential RNA maturation processes, polyadenylation, and APA are unknown.
Thus, we carried out poly(A) tag sequencing (PAT-seq) and ChIP-seq of histone acetylation on wild type and HDA6 mutants of Arabidopsis to uncover the crosstalk between histone deacetylation and mRNA 3′ end processing.
Results
HDA6 affects mRNA 3′ end processing
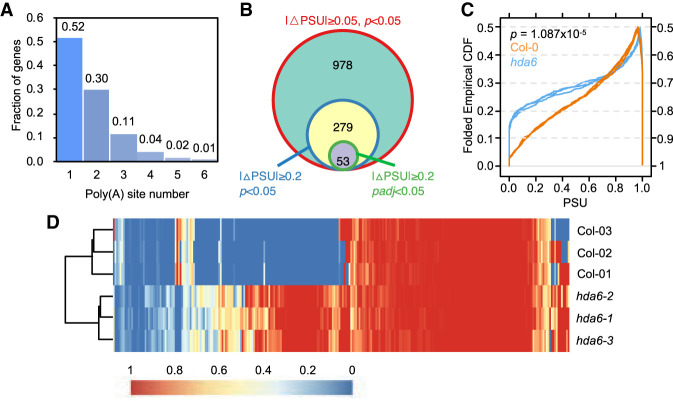
PAT-seq, which uniquely sequences the 3′ end of mRNA, was applied for Col-0 and HDA6 mutants to uncover the effect of HDA6 on polyadenylation of Arabidopsis. An overview of the experimental and computational analysis scheme is summarized in Supplemental Figure S1. It has been shown that mRNA polyadenylation sites are not exclusively positioned in the 3′ UTR (Wu et al. 2011b). They can be located anywhere with a suitable poly(A) signal, resulting in truncated or prolonged mRNA isoforms (Supplemental Fig. S2A). About 50% of genes and intergenic loci were identified with more than two poly(A) sites (Fig. 1A). Principle component analysis of PAT-seq data shows a good repeatability of this PAT-seq experiment (Supplemental Fig. S2B). Furthermore, the percentage of reads clustered into each poly(A) site in a gene is calculated to represent the poly(A) site usage (PSU). By excluding intergenic poly(A) sites, we identified 10,681 genes with ≥2 poly(A) sites. DESeq2 was used to calculate the p and adjusted P value (padj) of each poly(A) site. To explore the effect of HDA6 on APA, different thresholds were used. According to the published threshold of absolute value of PSU changes (|△PSU|≥0.05, P < 0.05) (Zheng et al. 2018), 1310 (978 + 279 + 53) genes were identified with significant APA events (Fig. 1B, red circle). Within these, about 25.34% (279 + 53) of genes have a larger (|△PSU|≥0.2) shift of poly(A) site usage (Fig. 1B, blue circle). However, if the threshold was set to |△PSU|≥0.2 and padj < 0.05, only 53 genes were identified with significant APA events (Fig. 1B, green circle; Supplemental Table S1). These results indicate that HDA6 affects mRNA polyadenylation and APA of genes by different measures.
Figure 1.
Mutation of HDA6 affects mRNA polyadenylation. (A) Fraction of genes with different number of poly(A) sites. (B) Venn diagram of identified APA genes by different threshold values. A total of 10,681 genes with at least two poly(A) sites were used for selections. PSU indicates the frequency of poly(A) site usage. (C) Folded cumulative distribution frequency (CDF) of PSUs of Col-0 and hda6. Only poly(A) sites with padj < 0.05 were selected for plotting. Three replicates were plotted separately. The y-axis indicates the fraction of poly(A) sites. (D) Heat map of PSUs of Col-0 and hda6. Only poly(A) sites with padj < 0.05 were selected for plotting. The scale bar is on the bottom.
The cumulative distribution curves present genome-wide polyadenylation profiles of hda6 and Col-0 and are folded at 50% cumulative frequency to show the median PSU (Supplemental Fig. S2C). Although there is no large difference between the cumulative distribution curves of hda6 and Col-0, a Kolmogorov–Smirnov test (K-S test) shows a significant difference in the PSU profiles of hda6 and Col-0, reflecting that HDA6 has an impact on polyadenylation in Arabidopsis. These results were further confirmed by the PAT-seq of another HDA6 mutant allele, sil1 (Liu et al. 2012) with a K-S test P value < 2.2 × 10−16 (Supplemental Fig. S2D). Furthermore, by CDF plot again, we found that the PSU profile of DE poly(A) sites (n = 1387; threshold: padj < 0.05) has a significant difference between hda6 and Col-0 (Fig. 1C). A heat map of these PSU profiles also illustrates that the PSUs of most DE poly(A) sites were varied between hda6 and Col-0, and this was consistent among replicates (Fig. 1D). These trends are also observed between sil1 and Col-0 (Supplemental Fig. S2E,F).
To evaluate the potential that DE poly(A) sites resulted from the total gene expression variation, RT-qPCR was carried out with a pair of primers which quantified the DE poly(A) sites and a pair of primers located on the 5′ end of genes which quantified the total gene expression levels (Supplemental Fig. S3; Supplemental Table S2). Results showed that DE poly(A) sites identified by DESeq2 were validated by RT-qPCR, and variation of their corresponding total gene expression levels was neither significant (Supplemental Fig. S3A–D) nor in an opposite direction (Supplemental Fig. S3E–H).
As a control, we selected AT1G35680 and AT2G38310 genes with non-DE poly(A) sites in the PAT-seq experiment. As shown in Supplemental Figure S3I, RT-qPCR confirmed that AT1G35680 was indeed not a DE poly(A) site gene. For the second control gene AT2G38310, while its 3′ UTR was significantly down-regulated in hda6 by the RT-qPCR experiment, the total gene expression level also had the same trend (Supplemental Fig. S3J). It indicates that the variation of the 3′ UTR of AT2G38310 may be induced by the repression of gene expression in hda6, and it was not identified as a DE poly(A) site in the PAT-seq analysis.
Taken together, DE poly(A) sites identified by PAT-seq were mainly contributed by the change in polyadenylation activity in hda6, and these results suggest that HDA6 affects mRNA polyadenylation.
Histone acetyltransferase inhibitor minimizes the molecular effect of HDA6 mutation
It was demonstrated that defective HDA6 results in an increased gene expression (Earley et al. 2006; Tanaka et al. 2008; Yu et al. 2017). Thus, we asked whether defective HDA6 causes differential expression of transcript isoforms. We found that more differentially expressed transcripts are up-regulated in hda6 (Fig. 2A). Within the 1387 transcripts with DE poly(A) sites, 882 are up-regulated and 505 are down-regulated. Since HDA6 functions in deacetylation of chromatins, we asked whether the effects of HDA6 on polyadenylation can be minimized under a low histone acetylation level. A histone acetyltransferase inhibitor, MB-3, was used to reduce the histone acetylation levels of hda6, sil1, and Col-0. PAT-seq of MB-3-treated seedlings shows that the variances of those 1387 HDA6-dependent poly(A) sites are minimized (Fig. 2B). Within those 1387 poly(A) sites, 929 show no significant difference after MB-3 treatment. The numbers of both up- and down-regulated poly(A) sites are reduced a lot.
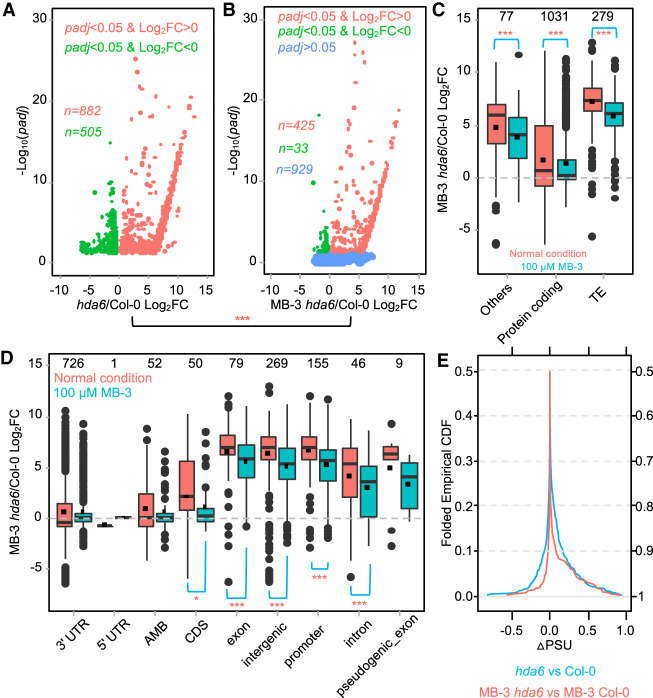
Figure 2.
The histone acetyltransferase inhibitor partially restored the effect of hda6 on polyadenylation. DE poly(A) sites (padj < 0.05) between hda6 and Col-0 are used for plotting. (***) Wilcoxon signed-rank test P < 0.001; (*) P < 0.05, when compared to nontreated groups. (A) More poly(A) transcripts are up-regulated in hda6. Log2FC indicates log2 of fold change of gene expression level. (B) MB-3 treatment minimizes the DE counts between hda6 and Col-0. (C,D) Distribution of DE poly(A) sites. Counts of poly(A) sites in each category are shown on the top of boxes. (TE) Transposon elements, (AMB) ambiguous regions which have overlapped features. Lines and dots in the boxes indicate the medians and means, respectively. (E) MB-3 treatment reduces the difference of PSUs between hda6 and Col-0. ΔPSU represents the difference of PSU of a poly(A) site in hda6 and Col-0.
Under normal conditions, the majority of DE poly(A) sites resided in protein coding genes (Fig. 2C, red boxes) and are mainly located in the 3′ UTR (Fig. 2D, red boxes). The interquartile ranges of boxplots and median values are minimized under MB-3 treatment, indicating that MB-3 treatment significantly reduces the variance of transcription expression levels among these three different gene types (Fig. 2C). The median expression level (dashed lines in red boxes) of DE poly(A) sites is slightly down in the 3′ UTR (canonical poly(A) site), whereas it is up-regulated in most other noncanonical poly(A) sites, such as the intergenic regions and introns (Fig. 2D, red boxes). The up-regulating pattern is also observed in sil1, although the expression pattern of CDS poly(A) transcripts is different from hda6 (Supplemental Fig. S2G).
MB-3 treatment also reduces the ΔPSU values of 1387 DE poly(A) sites (Fig. 2E), which reflects the assertion that MB-3 partially patches the effect of mutated HDA6. This trend was also found in MB-3 treatment on sil1 (Supplemental Fig. S2H). Together, these results show that the effect of HDA6 on polyadenylation is associated with histone acetylation dynamics.
Gene Ontology enrichment analyses show that genes with significant up- and down- regulated poly(A) sites are both enriched in terms related to stress response (Supplemental Fig. S4A,B, green dashed box). Genes with up-regulated poly(A) sites are also significantly enriched in translation-associated terms (Supplemental Fig. S4A, red dashed box), whereas genes with down-regulated poly(A) sites are also enriched in light response-associated terms (Supplemental Fig. S4B, orange dashed box). By checking the PAT-seq profiles, we found distinct APA on AT2G29500 encoding a HSP20-like protein, AT5G67300 encoding MYB DOMAIN PROTEIN 44 (MYB44), AT5G52882 encoding a P-loop protein in the nucleoside triphosphate hydrolases superfamily, and AT4G26090 encoding a leucine-rich repeat protein RPS2 (Supplemental Fig. S4C). Under MB-3 treatment, poly(A) sites of these genes in hda6 and sil1 are similar to Col-0 but rather different in the nontreated samples, reflecting that the mutation effect of HDA6 is abolished under low chromatin acetylation level. These distinct APA genes between hda6 and Col-0 may contribute to the phenotypic outcomes of hda6, such as affecting abiotic stress tolerance by AT5G52882 and MYB44 (Ali et al. 2013; Kim et al. 2017), altering heat shock tolerance by HSP20-like gene (Popova et al. 2013), and changing sensitivity to pathogen infection by RPS2 (Bhandari et al. 2019).
HDA6 is associated with mRNA 3′-UTR length
One of the important effects of APA is changing the tandem 3′-UTR length of mRNA (Tian and Manley 2017; Chen et al. 2018; Srivastava et al. 2018). This molecular effect connects to various post-transcriptional regulations, including alteration of mRNA stability, the gain or loss of miRNA, and/or RNA-binding protein targets (Mayr 2017). By genome-wide calculation of the weighted 3′-UTR length of genes, we found that 3′ UTR lengthening events (1157) are more abundant than 3′ UTR shortening events (373) in hda6 compared to Col-0 (Fig. 3A). These results suggest that HDA6 may play a role in ensuring proper 3′-UTR length for gene expression, indicating a connection between histone acetylation and 3′-UTR length control. The length of the 3′ UTR was reported to be correlated with total gene expression (not at the individual transcript level) under certain conditions or in specific tissues (Zheng et al. 2018; Fu et al. 2019). RNA-seq data of the seedlings of hda6 and Col-0 from the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) database were downloaded to obtain complete gene expression profiles. We did not observe a clear relationship between 3′-UTR length and gene expression. However, 3′ UTR lengthening and shortening events coincide with those of stable genes that are not significantly affected by the mutation of HDA6 (Fig. 3B). Figure 3C shows that decreased poly(A) site usage is coupled with an increase of another poly(A) site usage on the 3′ UTR of AT2G30720 and AT3G15518, which may explain the minor variance in total gene expression. Again, MB-3 treatment abolished the difference of poly(A) profile alteration of AT2G30720 and AT3G15518 between mutants and Col-0, reflecting that the 3′ UTR APA may depend on deacetylation by HDA6. In general, these evidences indicate that defective HDA6 results in both gene expression activation and 3′-UTR length variance. However, 3′ UTR APA in hda6 is independent of the transcription activity but associated with histone acetylation status.
Figure 3.
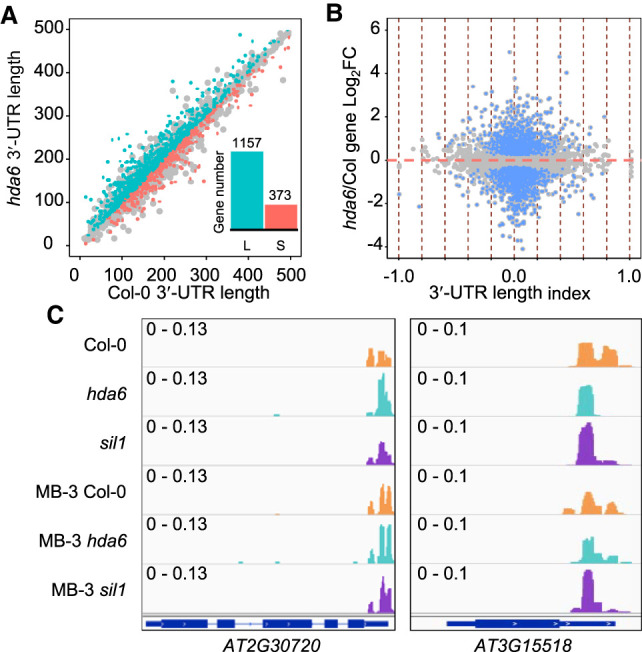
3′-UTR length variation is independent of gene expression activity in hda6. (A) Scatter-plot of weighted 3′-UTR lengths of hda6 and Col-0. (L) 3′-UTR lengthening event, (S) 3′ UTR shortening event. (B) Effect of HDA6 on 3′-UTR length and gene expression are independent. Blue dots indicate DE genes (padj < 0.05) identified from RNA-seq profiles; red vertical dashed lines indicate 0.2 intervals of 3′-UTR length index (>0, lengthening; <0, shortening). (C) Examples of a 3′ UTR lengthening event (left panel) and a 3′ UTR shortening event (right panel), respectively. Scale of each sample is indicated by data range.
Furthermore, tandem 3′ UTR sequences of genes with a switching of 3′-UTR length were submitted to the psRNATarget webserver (Dai et al. 2018) for identifying potential small RNA targets. For lengthening events, psRNATarget reported 832 pairs of small RNAs interacting with said 3′ UTRs that may support cleavage inhibition, and 203 pairs of interactions that may be subject to translation inhibition (Supplemental Table S3). For shortening events, psRNATarget predicted 233 cleavage inhibition and 43 translation inhibition pairs (Supplemental Table S4). These data suggest that the lengthening of the 3′ UTR in hda6 may increase possibilities of small RNA targeting for cleavage or translation inhibition, whereas the shortening of the 3′ UTR in hda6 may result in transcripts escaping from small RNA targeting. Thus, mRNA stability may be affected by the switch of 3′-UTR length.
Defective HDA6 alters poly(A) signal usage
Poly(A) signal recognition is a crucial step for polyadenylation (Clerici et al. 2018; Sun et al. 2018). Disruption of the poly(A) signal binding factor, CPSF30 (encoded by AT1G30460), results in genome-wide APA (Thomas et al. 2012; Chan et al. 2014). We performed a single nucleotide usage analysis around the DE poly(A) sites (Supplemental Fig. S5). The single nucleotide usage plots do not show any obvious differences between DE and non-DE poly(A) sites. The most common and important poly(A) signal is AAUAAA, which resides between −35 ∼ −10 nt upstream of the poly(A) site and is designated as the near upstream element (NUE) in plants (Loke et al. 2005; Xing and Li 2011). Thus, we calculated the preference of poly(A) signal usage within the NUE of DE poly(A) sites. Our results show that up-regulated poly(A) sites have a much higher AAUAAA cis-element frequency, whereas down-regulated poly(A) sites have almost the same AAUAAA cis-element frequency as non-DE poly(A) sites (Fig. 4A). The frequency of 1-nt variation of the AAUAAA poly(A) signal is depleted in both up- and down-regulated poly(A) sites. Moreover, down-regulated poly(A) site clusters were found with a high frequency of nonobvious poly(A) signal (designated as others). A χ2 test shows that the usage of NUE signals among three transcript groups has a significant difference (P = 3.64 × 10−8). These differences reflect that changes of polyadenylation induced by defective HDA6 may be associated with NUE poly(A) signal choices.
Figure 4.
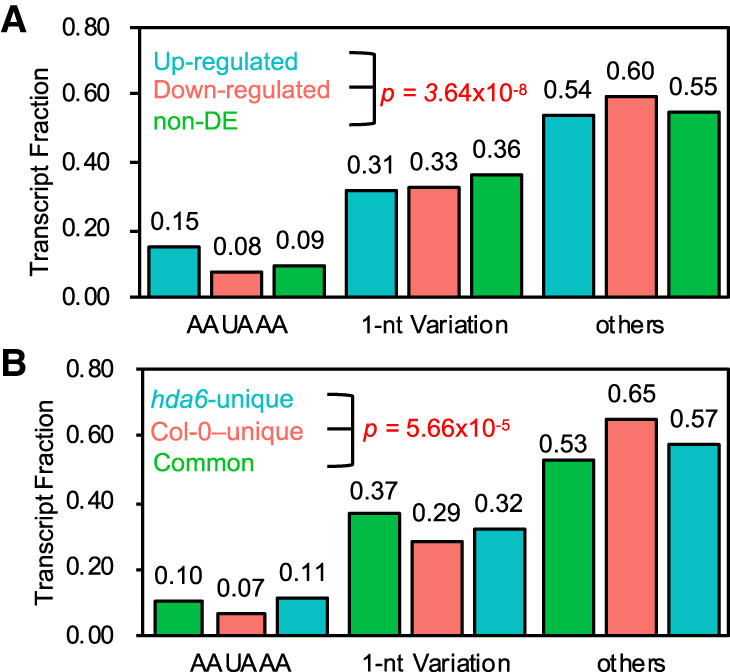
Defective HDA6 alters poly(A) signal usage. (A) NUE poly(A) signal usage of up-regulated, down-regulated, and non-DE poly(A) sites. (B) NUE poly(A) signal usage of common, Col-0–unique, and hda6-unique poly(A) sites.
Unique poly(A) sites can also be an indication of different poly(A) signal usage between the mutant and wild type (Thomas et al. 2012). Using a strict threshold (total reads ≥ 15, of which each replicate > 0 in one Arabidopsis strain, each replicate = 0 in the other strain), we found 870 and 259 unique poly(A) sites in hda6 and Col-0, respectively (Supplemental Fig. S6A,B). For instance, FLOWERING LOCUS C (FLC, AT5G10140) contains both hda6-unique (also unique in sil1) and Col-0–unique poly(A) sites on its antisense strand (Supplemental Fig. S6C). MB-3 treatment almost eliminated the unique poly(A) sites of hda6 and Col-0 on antisense of FLC but came out with new proximal antisense poly(A) sites. These suggests that unique poly(A) sites of hda6 are associated with the acetylation dynamic. We further found that hda6-unique poly(A) sites have almost the same single nucleotide usage profile as common poly(A) sites (Supplemental Fig. S6D). However, unique poly(A) sites in Col-0 show a lower A and U usage within −35 to 0 and −150 to −35 nt upstream of poly(A) sites (Supplemental Fig. S6D, red arrow). This result indicates that the poly(A) signal usage of unique transcripts within Col-0 may be different from that unique in hda6 and common poly(A) sites. Scanning of NUE poly(A) signals also confirmed this assumption (Fig. 4B). In Col-0, AAUAAA only accounts for 7% of unique poly(A) sites, and the frequency of other poly(A) signals (65%) is higher than hda6-unique and common poly(A) sites. However, AAUAAA accounts for 10% in hda6-unique poly(A) sites and common poly(A) sites. The differences of NUE signal usage among three transcript groups are also significant (χ2 test, P = 5.66 × 10−5). This result indicates that an increase of histone acetylation may eliminate the expression of those transcripts without an AAUAAA-like poly(A) signal.
Differentially expressed poly(A) sites are associated with the distance to acetylation peaks
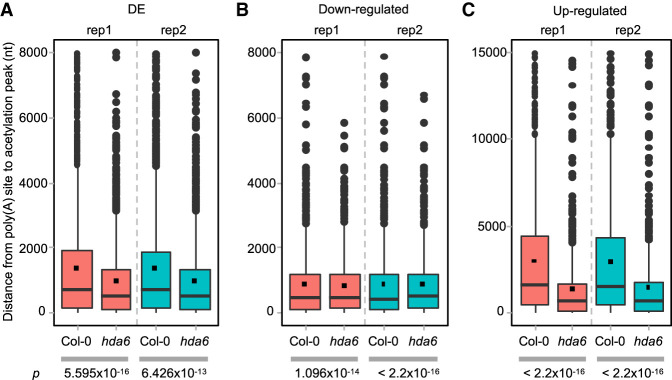
To elucidate the relationship between acetylation and polyadenylation, ChIP-seq of H3K9ac and H3K14ac (designated as H3K9K14ac) was also performed with an antibody targeting both histone acetylation markers. Peaks were called by MACS2 (Zhang et al. 2008b) to find the enriched acetylation regions, and the distances between DE poly(A) sites and their nearest upstream acetylation peaks were calculated. We found that the distance between DE poly(A) sites and acetylation peaks in hda6 is much shorter than that in Col-0 (Fig. 5A). Furthermore, we separated the up- and down-regulated poly(A) sites and re-analyzed the distance distribution. Wilcoxon signed-rank tests show that the distances between acetylation peaks and DE poly(A) sites, either up- or down-regulated groups, are significantly shorter in hda6 than that in Col-0. It appears that there is no large difference between the distances in down-regulated poly(A) sites and acetylation peaks between hda6 and Col-0 (Fig. 5B). However, there is a difference in the distances between up-regulated poly(A) sites and acetylation peaks between hda6 and Col-0, as well as all mixed DE poly(A) sites (Fig. 5C). This evidence reflects that up-regulated poly(A) sites are more eligible to be affected by the histone H3K9K14ac level. To confirm this finding, ChIP-seq of H3K9ac was performed with an antibody only targeting to H3K9ac. The result also shows that up-regulated poly(A) sites are significantly closer to H3K9ac peaks in hda6 than those in Col-0 (Supplemental Fig. S7). Taken together, the alteration of H3K9K14ac induced by defective HDA6 may contribute to mRNA polyadenylation disturbance in hda6.
Figure 5.
Expression of mature transcripts is associated with the distance between acetylation peaks to poly(A) sites. (A) DE poly(A) sites. (B) Down-regulated poly(A) sites. (C) Up-regulated poly(A) sites. The Wilcoxon signed-rank test was applied to evaluate the statistical significance of difference between two samples, and P values are indicated below.
H3K9ac and H3K14ac around poly(A) sites are associated with differential expression of poly(A) sites
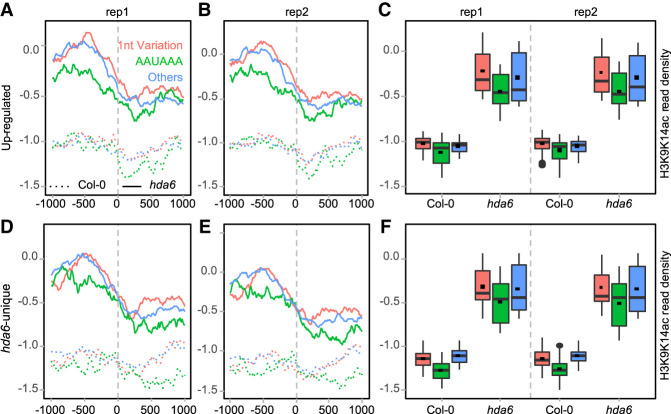
To further study the effects of acetylation on polyadenylation, the levels of H3K9K14ac around poly(A) sites were investigated (Fig. 6). Generally, H3K9K14ac levels at or near the poly(A) sites are lower than other locations within the −1 kb ∼ 1 kb region around poly(A) sites (Fig. 6AB,D,E), indicating that H3K9K14ac may be involved in the positioning of poly(A) sites. Moreover, the H3K9K14ac level upstream of poly(A) sites is higher than that downstream from poly(A) sites, including both hda6 preferred poly(A) sites (up-regulated and hda6-unique poly(A) sites) and Col-0 preferred poly(A) sites (down-regulated and Col-0–unique poly(A) sites) (Fig. 6A,B,D,E). These results indicate that the H3K9K14ac decrease is associated with polyadenylation.
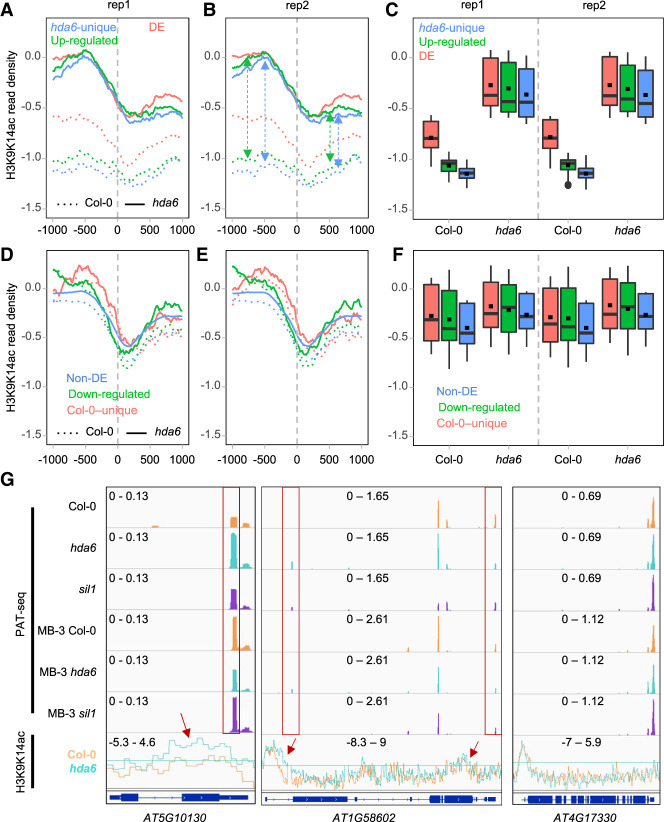
Figure 6.
H3K9K14ac profiles across the poly(A) sites. (A–C) H3K9K14ac profiles around DE, up-regulated, and hda6-unique poly(A) sites, respectively. (D–F) H3K9K14ac profiles around non-DE, down-regulated, and Col-0–unique poly(A) sites, respectively. (G) H3K9K14ac and APA of AT5G10130, AT1G58620 (RPP7), and non-DE poly(A) sites gene AT4G17330. Red arrows indicate H3K9K14ac is higher in hda6 than Col-0. Red rectangles indicate the potential DE poly(A) sites affected by deacetylation abortion.
In Col-0, H3K9K14ac profiles around DE, up-regulated, and hda6-unique poly(A) sites are much lower (ranging from −1.5 to −0.5) than that around non-DE, down-regulated, and Col-0–unique poly(A) sites (ranging from −1.0 to 0 or higher) (Fig. 6A,B,D,E). These differences are also shown as boxplots of the H3K9K14ac levels around corresponding poly(A) sites (Fig. 6C,F; Supplemental Table S5). Since a higher H3K9K14ac level presents in down-regulated poly(A) sites of Col-0 (green dotted lines in Fig. 6D,E), the H3K9K14ac profiles of DE poly(A) sites (red dotted lines in Fig. 6A,B) are diverged from that of up-regulated and hda6-unique poly(A) sites (blue and green dotted lines in Fig. 6A,B) in Col-0. Meanwhile, mutation of HDA6 results in an up-regulation of H3K9K14ac around up-regulated and hda6-unique poly(A) sites, whereas H3K9K14ac around down-regulated and Col-0–unique poly(A) sites do not change too much (Fig. 6A–F), although their statistical differences are significant (Supplemental Table S5). Thus, the poly(A) site usage of up-regulated and hda6-unique poly(A) sites is much more susceptible to be regulated through histone H3K9K14ac modification by HDA6.
Additionally, H3K9K14ac is enhanced within 1 kb upstream of up-regulated and hda6-unique poly(A) sites, whereas 1 kb downstream from those two poly(A) sites groups has a comparatively small increase of H3K9K14ac (Fig. 6A,B, green and blue dashed double-headed arrows marked on Fig. 6B). These large differences are not observed in down-regulated and Col-0–unique poly(A) sites (Fig. 6D,E). It suggests that HDA6 is more eligible to deacetylate upstream of poly(A) sites.
Individual gene examples show that changes of acetylation levels in hda6 are associated with APA dynamics. AT5G10130 and AT1G58602 contain both acetylation level change and APA dynamics, whereas AT4G17330 presents neither acetylation level shifting nor significant APA dynamics (Fig. 6G). Under MB-3 treatment, the poly(A) profiles of AT5G10130 and AT1G58620 in all samples were changed compared to nontreated Col-0, reflecting that polyadenylation of these two genes are acetylation-sensitive. However, this is not seen in AT4G17330. Again, after MB-3 treatment, the poly(A) profiles of AT5G10130 and AT1G58620 in hda6 and sil1 appeared similar to that of Col-0. It indicates that the deacetylation role of HDA6 is associated with polyadenylation in Arabidopsis.
In hda6, H3K9ac profiles upstream of poly(A) sites across DE, up-regulated, and hda6-unique poly(A) sites are also enhanced when compared to Col-0 (Supplemental Fig. S8A,B; Supplemental Table S6), whereas in non-DE, down-regulated, and Col-0–unique poly(A) sites, they are not (Supplemental Fig. S8C,D).
Taken together, these results strongly suggest that up-regulated and hda6-unique poly(A) sites are targets of HDA6, and deacetylation of H3K9K14ac or H3K9ac level upstream of poly(A) sites is associated with polyadenylation.
HDA6 differentially regulates H3K9K14ac upstream of poly(A) sites among different poly(A) signal types
Defective HDA6 varies in the usage of poly(A) signals (Fig. 4), and the H3K9K14ac profiles are different around poly(A) sites between up- and down-regulated poly(A) sites (Fig. 6). Thus, we speculated that the H3K9K14ac profiles around poly(A) sites are different among three types of NUE poly(A) signals including AAUAAA, 1-nt variations of AAUAAA, and other unclassified poly(A) signals. We mainly focused on the up-regulated and hda6-unique poly(A) sites. Results show that the H3K9K14ac profiles of up-regulated and hda6-unique poly(A) sites across all three types of poly(A) signals are increased in hda6 compared to Col-0 (Fig. 7A–F). This result further supports the conclusion that H3K9K14ac may mainly contribute to up-regulated and newly activated poly(A) sites.
Figure 7.
Different poly(A) signals are associated with different H3K9K14ac profiles. The Wilcoxon signed-rank test was used to evaluate the statistical significance of the acetylation level differences, and P values are listed in Supplemental Table S7. (A–C) Up-regulated poly(A) sites. (D–F) hda6-unique poly(A) sites.
By comparing the H3K9K14 profiles of up-regulated poly(A) sites classified by the three types of poly(A) signals, no wide deviants are found in Col-0 (Fig. 7A–C), although they are statistically different (Supplemental Table S7). However, in hda6, AAUAAA containing up-regulated poly(A) sites appears to have a lower level of H3K9K14ac compared to the categories of 1-nt variation of AAUAAA and other unclassified poly(A) signals, especially within the upstream area of poly(A) sites (Fig. 7A–C). In hda6-unique poly(A) sites group, the AAUAAA category also presents a lower H3K9K14ac level in hda6, especially around −500 bp (Fig. 7D–F). Although H3K9ac profiles between hda6 and Col-0 are different among DE, up-regulated, and hda6-unique poly(A) sites, we did not find large differences among the three types of poly(A) signals (Supplemental Fig. S9; Supplemental Table S8). This result indicates that the poly(A) signal choices may not solely be affected by H3K9ac.
In conclusion, H3K9K14ac mediation by HDA6 may play an important role in poly(A) signal choices in guiding polyadenylation during mRNA 3′ end processing. Canonical AAUAAA NUE signals containing transcripts require a low amount of H3K9K14ac upstream of poly(A) sites for polyadenylation, compared to 1-nt variation AAUAAA and other unclassified poly(A) signals containing transcripts.
Discussion
Epigenetic modifications play an important role in regulating gene expression (Kurdistani et al. 2004; Chinnusamy and Zhu 2009; Lawrence et al. 2016; Yan et al. 2018). In addition to transcription initiation and elongation, epigenetic modifications also function in alternative splicing (Luco et al. 2011; Jimeno-González and Reyes 2016; Zhu et al. 2018). Recently, epigenetic markers on genomic DNA contexts were reported to affect polyadenylation through the EDM2-AIPP1-ASI1 complex (Lei et al. 2014; Duan et al. 2017). The flowering time regulator, FPA, is also involved in this process (Deremetz et al. 2019). These findings indicate that chromatin status is sufficient in coordinating 3′ end processing during transcription. However, the effect of histone epigenetic modifications on transcription polyadenylation is rarely studied. It was reported that nucleosome structure is associated with the choice of proximal and distal polyadenylation sites (Huang et al. 2013; Jimeno-González and Reyes 2016). In addition, polyadenylation sites are less sensitive to DNase I digestion and prefer to position in less nucleosome structured regions in human cells (Lee and Chen 2013). However, the role of histone modification in polyadenylation is unclear.
In this study, we employed PAT-seq to uncover the effects of HDA6 on polyadenylation in Arabidopsis. Data show that HDA6 is able to affect polyadenylation genome-wide. As expected, defective HDA6 results in transcription activation and the emergence of unique transcripts in the mutant, especially in the intergenic regions. A histone acetyltransferase inhibitor assay confirmed that the effect of HDA6 mutation on polyadenylation is in an acetylation-dependent manner. HDA6 is associated with the regulation of 3′-UTR lengths of transcripts. Since transcription stability and small RNA abundance are sensitive to environmental conditions (Sidaway-Lee et al. 2014; Ma et al. 2018), the lengthening of the 3′ UTR induced by defective HDA6 may result in the alteration of mRNA stability or small RNA targeting, especially under certain stress conditions. Thus, the transcriptome homeostasis is shifted by APA and results in phenotypic outcomes. Moreover, the larger the variance in 3′-UTR lengths, the less total gene expression difference was observed. It suggests that HDA6 functions in coordinating 3′-UTR length and gene expression separately. However, this may also implicate that a 3′ UTR APA mechanism occurs to maintain a temporary homeostasis of gene expression in hda6. This strategy may be effective for the survival and normal phenotypic outcomes of hda6 under normal growth conditions, but further large scale studies on different growth conditions and different tissues are required.
To directly elucidate the relationship between histone acetylation and polyadenylation, H3K9K14ac profiles of hda6 and Col-0 were analyzed by ChIP-seq. An increase of H3K9K14ac specifically supports the overrepresented transcription of both up-regulated and unique poly(A) site usage in hda6 (Fig. 6). Since the acetylation peaks are much closer to up-regulated poly(A) sites in hda6 than in Col-0 (Fig. 5), we conclude that H3K9K14ac promotes polyadenylation in certain poly(A) site usage affected by HDA6. H3K9ac profiles of poly(A) site upstream contexts are also significantly different between DE, up-regulated, and hda6-unique poly(A) sites, although H3K9ac variations between hda6 and Col-0 are smaller than H3K9K14ac (Fig. 6; Supplemental Fig. S8). These results indicate that both H3K9ac and H3K14ac may function in the polyadenylation of those activated genes. Besides, H3K9ac profiles of poly(A) site downstream contexts are insensitive to HDA6 modulating, which reflects that the roles of H3K9ac and H3K14ac in transcriptional polyadenylation may be partially different. The difference between H3K9K14ac and H3K9ac is mainly found downstream from poly(A) sites in DE, up-regulated, and hda6-unique poly(A) sites. It appears that H3K9ac is suppressed in the downstream from poly(A) sites in hda6. These data suggest that H3K9ac may exclusively affect histone acetylation in gene bodies. However, the footprint of H3K14ac may be more general or even genome-wide than H3K9ac. Collectively, these results indicate that up-regulated and unique poly(A) sites in hda6 are potential deacetylation targets of HDA6, and the usage of these poly(A) sites is associated with H3K9K14ac marker. Further studies should be carried out to elucidate the roles of different histone acetylation markers in RNA processing.
AAUAAA is a common canonical poly(A) signal in eukaryotes (Clerici et al. 2018). It presents in ∼50% of mRNA in mammals (Clerici et al. 2017, 2018; Tian and Manley 2017), whereas the frequency in plants is only ∼10% (Wu et al. 2011b; Xing and Li 2011; Thomas et al. 2012). One-nt variation and other unclassified poly(A) signals account for the rest of poly(A) signals. However, how plants distinguish these three types of poly(A) signals is still unclear. Here, we found that the H3K9K14ac of activated transcripts (up-regulated and hda6-unique poly(A) sites) is enhanced by HDA6 defects (Fig. 6). Moreover, transcripts containing AAUAAA are associated with a lower amount of H3K9K14ac upstream of the poly(A) sites (Fig. 7). We speculate that the strong cis-element AAUAAA is sufficient to recruit the polyadenylation machinery for transcription termination, whereas weak poly(A) signals require other mechanisms such as histone modifications to ensure transcription polyadenylation.
In addition, we observed MB-3 has significant impact on polyadenylation in Col-0. It was reported that MB-3 does not solely reduce H3K9K14ac but also affects the genome-wide H3K27ac level (Chen et al. 2017). Hence, the impact of MB-3 on polyadenylation should be carefully considered and required for mass further studies to reach a conclusion. Moreover, we observed the 3′-UTR length of AT2G01930 is not lengthened in sil1 as in hda6 (Supplemental Fig. S10). It was reported that hda6 and sil1 have different strengths on deacetylating H3K9K14 and regulation on gene expression (Liu et al. 2012). Thus, the genetic difference may mediate the different APA of AT2G01930 between hda6 and sil1. This also requires further studies to elucidate the underlying mechanism.
To conclude, our studies provide direct evidence that histone deacetylation orchestrates polyadenylation in Arabidopsis to regulate gene expression. Moreover, the histone epigenetic marker H3K9K14ac is involved in distinguishing different poly(A) signals during cotranscriptional RNA processing.
Methods
Plant materials and growth conditions
Col-0, hda6 (axe1-5), and sil1 were surface-deterged and synchronized in 4°C with soaking in ddH2O for 3 d. The synchronized seeds were grown vertically for 14 d on 1/2MS with 1% sucrose and 0.8% phytagel. For the histone acetyltransferase inhibitor assay, synchronized seeds were geminated on normal 1/2MS with 1% sucrose and 0.8% phytagel plates for 3 d, transferred to 1/2MS plates containing 100 μM MB-3 (Sigma-Aldrich M2449), and kept in vertical growth for 14 d (Chen et al. 2017). The light period was set at 16 h light and 8 h dark in a growth chamber.
PAT-seq
The 14-d nontreated or MB-3-treated seedlings were collected and ground in liquid nitrogen. Fine powders of samples were collected in a 1.5-mL RNase-free tube for total RNA extraction by TRIzol (Thermo Fisher Scientific 15596018), followed by removal of putative DNA contamination. Two micrograms of each replicate of clean total RNA were used for PAT-seq library preparation. PAT-seq was performed following the protocol of a previous publication, with minor modifications (Ma et al. 2014). Briefly, clean RNA was sheared by 5× first strand buffer (supplied with transcriptase) at 94°C for 4 min. The poly(A) tags were immediately enriched by oligo d(T)25 magnetic beads. Enriched poly(A) tags were reverse-transcribed with barcoded and anchored oligo d(T)18 Illumina adaptors by SMARTScribe Reverse Transcriptase (Clontech 639538) for 2 h at 42°C. Locked Nucleic Acid (LNA) modified 5′ adaptor was added with 1 μL more of SMARTScribe for 2 h template switching at 42°C. cDNA was purified by HighPrep PCR beads (Magbio MGAC60050) and amplified by a pair of primers with Illumina adaptors. The products were separated on a 2% agarose gel and bands between 300–500 bp were gel-purified for sequencing. The purified products were qualified by Qubit and Aglient Bioanalyzer, then quantified by qPCR. Single-end 74-nt mode without index sequencing was chosen for sequencing on HiSeq 2500.
Chromatin immunoprecipitation sequencing assay
Chromatin extracts were prepared from seedlings treated with 1% formaldehyde. The chromatin was sheared to the mean length of 500 bp by sonication; proteins and DNA fragments were then immunoprecipitated using antibodies against acetylated histone H3K9K14 (Millipore-Sigma 06-599) or acetylated histone H3K9 (Millipore-Sigma 07-352). The DNA cross-linked to immunoprecipitated proteins was reversed and then participated by 70% EtOH. A total of 5 ng of DNA from at least five ChIPs was pooled to ensure that there was enough starting DNA for library construction. Two biological replicates of H3K9K14ac were prepared and sequenced for each ChIP-seq experiment. The ChIP DNA was first tested by RT-qPCR and then used to prepare ChIP-seq libraries. End repair, adaptor ligation, and amplification were carried out using the Illumina Genomic DNA Sample Prep kit according to the manufacturer's protocol. An Illumina HiSeq 2500 instrument was used for the high-throughput sequencing of the ChIP-seq libraries.
PAT-seq data processing
Raw reads quality filtering was performed using FASTX-Toolkit with a threshold q = 20. Barcodes and poly(T) stretches of high-quality reads were trimmed. Clean reads were then mapped to reference genome TAIR10 by Bowtie 2 for end-to-end alignment (Langmead and Salzberg 2012). Poly(A) sites, which are the cleavage sites for adding poly(A) tails, were extracted from uniquely mapped reads, and internal priming events were filtered out using published customized scripts (Wu et al. 2015b). Because of the microheterogeneity of polyadenylation, the location of qualified poly(A) tags were clustered by a sliding 24-nt interval according to our previous publications, and the locations supported by the most reads were adopted as reference coordinates of poly(A) sites (Wu et al. 2011b, 2015b). The BAM files of mapping results were converted into bedGraph format and normalized by “counts per million” for visualization in Integrative Genomics Viewer (IGV) (Robinson et al. 2011). Tracks from replicates were summed up into one track.
For removing low-confident sites, poly(A) sites with a total tag number of all nontreatment samples < 15 were filtered out. Poly(A) site usage was calculated as , in which i indicates the ith poly(A) site of a gene and n indicates the total number of poly(A) sites of the gene. Folded empirical cumulative distribution plots were generated by Moutainplot (Xue and Titterington 2011) in R (R Core Team 2019). The preserved poly(A) sites were subjected to DE analysis using DESeq2 (Love et al. 2014). A threshold of padj < 0.05 was chosen to control the false discovery rate of identification of DE poly(A) sites. Gene Ontology enrichment analysis was performed using the agriGO v2.0 webserver (Tian et al. 2017). For 3′ UTR variation analysis, a mean length of the 3′ UTR of a target gene was calculated as (leni represents the 3′-UTR length of the ith poly(A) site), and a 3′-UTR length index r was calculated by a Pearson product moment correlation coefficient that indicates the strength of 3′ UTR shortening (r < 0) or 3′ UTR lengthening (r > 0) between two conditions. The tandem 3′ UTR sequencing of the shortening or lengthening transcripts was submitted to the psRNATarget webserver (Dai et al. 2018) for small RNA targets alignment.
For profiling the single nucleotide composition around the poly(A) sites, nucleotide sequences 300 nt upstream of and 100 nt downstream from poly(A) sites were extracted and the corresponding frequency of each position was calculated. The poly(A) site was designated as the “0” point in figures. Poly(A) signal usage was calculated by customized Perl scripts published previously (Wu et al. 2015a), and the χ2 test was applied for testing the NUE signal usage among different transcript groups.
For RNA-seq data processing, raw sequencing data were downloaded from the NCBI SRA database under BioProject accession PRJNA314484. Reads were mapped to reference genome TAIR10 by Bowtie 2. Unique mapped reads were extracted for featureCounts (Liao et al. 2014) to quantify the gene expression levels. DE gene analysis was done by the DESeq2 package in R.
ChIP-seq data processing
Raw reads were quality-controlled by FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) with q ≥ 20. Clean reads were mapped to the TAIR10 genome by Bowtie 2 (Langmead and Salzberg 2012). Uniquely mapped reads were reserved, sorted, and indexed by SAMtools (Li et al. 2009). The output BAM files of samples were used for peak calling by MACS2 (Zhang et al. 2008b). The distance from DE poly(A) sites to identified peaks were calculated and statistically tested by a paired t-test in R. The output BAM files of samples were normalized to input sequencing BAM files and scaled with the “counts per million” method by deepTools (Ramírez et al. 2016). A metaplot of acetylation profiles around poly(A) sites of interest were performed at a resolution with bin size of 10 nt in R. The Wilcoxon signed-rank test was used to evaluate the statistical significance of the acetylation profile differences between each two compared groups.
Data access
The sequencing data sets generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA552176.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
The authors thank Haidong Qu, Xiuxiu Wang, Xiaoxuan Zhou, and Wenjia Lu for technical assistance. We also thank Dr. Zhicheng Dong for helpful discussions, Taylor L. Li for language editing, and Xu Pan for data mining assistance. We thank Technology Commons, College of Life Science, National Taiwan University for the convenient use of instruments. This work was supported in part by a grant from the National Key R&D Project of China (2016YFE0108800) to Q.Q.L.; by grants (2017M620274, 2018T110649) from the China Postdoctoral Science Foundation to J.L.; and a grant (61802323) from the National Natural Science Foundation of China to C.Y. This work was also supported by a grant from the Ministry of Science and Technology of Taiwan (105-2311-B-002-012-MY3 and 107-2313-B-002-001) and an NTU-Academia Sinica joint grant (NTU-AS-108L104310) to K.W.
Author contributions: Q.Q.L. and K.W. designed the research. J.L. and F.-Y.H. performed experiments and data analysis. C.Y. supported with bioinformatics codes and data analysis. L.H. and Y.-H.S. were involved in parts of experiments. J.L., F.-Y.H., C.Y., K.W., and Q.Q.L. wrote and revised the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.255232.119.
References
- Ali MA, Plattner S, Radakovic Z, Wieczorek K, Elashry A, Grundler FMW, Ammelburg M, Siddique S, Bohlmann H. 2013. An Arabidopsis ATPase gene involved in nematode-induced syncytium development and abiotic stress responses. Plant J 74: 852–866. 10.1111/tpj.12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari DD, Lapin D, Kracher B, von Born P, Bautor J, Niefind K, Parker JE. 2019. An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis. Nat Commun 10: 772 10.1038/s41467-019-08783-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR, Ule J, Manley JL, Shi Y. 2014. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 28: 2370–2380. 10.1101/gad.250993.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li C, Wang Y, Renaud J, Tian G, Kambhampati S, Saatian B, Nguyen V, Hannoufa A, Marsolais F, et al. 2017. Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat Plants 3: 814–824. 10.1038/s41477-017-0023-7 [DOI] [PubMed] [Google Scholar]
- Chen M, Lyu G, Han M, Nie H, Shen T, Chen W, Niu Y, Song Y, Li X, Li H, et al. 2018. 3′ UTR lengthening as a novel mechanism in regulating cellular senescence. Genome Res 28: 285–294. 10.1101/gr.224451.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK. 2009. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12: 133–139. 10.1016/j.pbi.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Faini M, Aebersold R, Jinek M. 2017. Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex. eLife 6: e33111 10.7554/eLife.33111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Faini M, Muckenfuss LM, Aebersold R, Jinek M. 2018. Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex. Nat Struct Mol Biol 25: 135–138. 10.1038/s41594-017-0020-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX. 2018. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46: W49–W54. 10.1093/nar/gky316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Cao X. 2017. Roles of pre-mRNA splicing and polyadenylation in plant development. Curr Opin Plant Biol 35: 45–53. 10.1016/j.pbi.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Deremetz A, Le Roux C, Idir Y, Brousse C, Agorio A, Gy I, Parker JE, Bouché N. 2019. Antagonistic actions of FPA and IBM2 regulate transcript processing from genes containing heterochromatin. Plant Physiol 180: 392–403. 10.1104/pp.18.01106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C-G, Wang X, Zhang L, Xiong X, Zhang Z, Tang K, Pan L, Hsu C-C, Xu H, Tao WA, et al. 2017. A protein complex regulates RNA processing of intronic heterochromatin-containing genes in Arabidopsis. Proc Natl Acad Sci 114: E7377–E7384. 10.1073/pnas.1710683114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. 2006. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev 20: 1283–1293. 10.1101/gad.1417706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Wang P, Wu X, Zhou X, Ji G, Shen Y, Gao Y, Li QQ, Liang J. 2019. Distinct genome-wide alternative polyadenylation during the response to silicon availability in the marine diatom Thalassiosira pseudonana. Plant J 99: 67–80. 10.1111/tpj.14309 [DOI] [PubMed] [Google Scholar]
- Gates LA, Shi J, Rohira AD, Feng Q, Zhu B, Bedford MT, Sagum CA, Jung SY, Qin J, Tsai M-J, et al. 2017. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem 292: 14456–14472. 10.1074/jbc.M117.802074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Chen J, Liu H, Sun X. 2013. The nucleosome regulates the usage of polyadenylation sites in the human genome. BMC Genomics 14: 912 10.1186/1471-2164-14-912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung F-Y, Chen F-F, Li C, Chen C, Lai Y-C, Chen J-H, Cui Y, Wu K. 2018. The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res 46: 10669–10681. 10.1093/nar/gky749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AG, Xing D, Li QQ. 2012. Plant polyadenylation factors: conservation and variety in the polyadenylation complex in plants. BMC Genomics 13: 641 10.1186/1471-2164-13-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno-González S, Reyes JC. 2016. Chromatin structure and pre-mRNA processing work together. Transcription 7: 63–68. 10.1080/21541264.2016.1168507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson A, Durand-Dubief M, Xue-Franzén Y, Rönnerblad M, Ekwall K, Wright A. 2009. HAT–HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep 10: 1009–1014. 10.1038/embor.2009.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. 2012. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13: 424 10.1186/1471-2164-13-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-M, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A, Toyoda T, Kimura H, Shinozaki K, Seki M. 2008. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol 49: 1580–1588. 10.1093/pcp/pcn133 [DOI] [PubMed] [Google Scholar]
- Kim J-M, To TK, Matsui A, Tanoi K, Kobayashi NI, Matsuda F, Habu Y, Ogawa D, Sakamoto T, Matsunaga S, et al. 2017. Acetate-mediated novel survival strategy against drought in plants. Nat Plants 3: 17097 10.1038/nplants.2017.97 [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M. 2004. Mapping global histone acetylation patterns to gene expression. Cell 117: 721–733. 10.1016/j.cell.2004.05.023 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Daujat S, Schneider R. 2016. Lateral thinking: how histone modifications regulate gene expression. Trends Genet 32: 42–56. 10.1016/j.tig.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Lee C-y, Chen L. 2013. Alternative polyadenylation sites reveal distinct chromatin accessibility and histone modification in human cell lines. Bioinformatics 29: 1713–1717. 10.1093/bioinformatics/btt288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, La H, Lu K, Wang P, Miki D, Ren Z, Duan CG, Wang X, Tang K, Zeng L, et al. 2014. Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns. Proc Natl Acad Sci 111: 527–532. 10.1073/pnas.1320106110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu X, Yu CW, Duan J, Luo M, Wang K, Tian G, Cui Y, Wu K. 2012. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol 158: 119–129. 10.1104/pp.111.184275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke JC, Stahlberg EA, Strenski DG, Haas BJ, Wood PC, Li QQ. 2005. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol 138: 1457–1468. 10.1104/pp.105.060541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550–550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144: 16–26. 10.1016/j.cell.2010.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Pati PK, Liu M, Li QQ, Hunt AG. 2014. High throughput characterizations of poly(A) site choice in plants. Methods 67: 74–83. 10.1016/j.ymeth.2013.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Chen C, Liu Y, Zeng M, Meyers BC, Li J, Xia R. 2018. Coupling of microRNA-directed phased small interfering RNA generation from long noncoding genes with alternative splicing and alternative polyadenylation in small RNA-mediated gene silencing. New Phytol 217: 1535–1550. 10.1111/nph.14934 [DOI] [PubMed] [Google Scholar]
- Mayr C. 2017. Regulation by 3′-untranslated regions. Annu Rev Genet 51: 171–194. 10.1146/annurev-genet-120116-024704 [DOI] [PubMed] [Google Scholar]
- Popova OV, Dinh HQ, Aufsatz W, Jonak C. 2013. The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol Plant 6: 396–410. 10.1093/mp/sst023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidaway-Lee K, Costa MJ, Rand DA, Finkenstadt B, Penfield S. 2014. Direct measurement of transcription rates reveals multiple mechanisms for configuration of the Arabidopsis ambient temperature response. Genome Biol 15: R45 10.1186/gb-2014-15-3-r45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu JK. 2018. UTR-dependent control of gene expression in plants. Trends Plant Sci 23: 248–259. 10.1016/j.tplants.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhang Y, Hamilton K, Manley JL, Shi Y, Walz T, Tong L. 2018. Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc Natl Acad Sci 115: E1419–E1428. 10.1073/pnas.1718723115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan LM, Zhang CJ, Hou XM, Shao CR, Lu YJ, Zhou JX, Li YQ, Li L, Chen S, He XJ. 2018. The PEAT protein complexes are required for histone deacetylation and heterochromatin silencing. EMBO J 37: e98770 10.15252/embj.201798770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kikuchi A, Kamada H. 2008. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146: 149–161. 10.1104/pp.107.111674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PE, Wu X, Liu M, Gaffney B, Ji G, Li QQ, Hunt AG. 2012. Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell 24: 4376–4388. 10.1105/tpc.112.096107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z. 2017. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45: W122–W129. 10.1093/nar/gkx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TK, Kim J-M, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T. 2011. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet 7: e1002055 10.1371/journal.pgen.1002055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hu Q, Wu Z, Wang H, Han S, Jin Y, Zhou J, Zhang Z, Jiang J, Shen Y, et al. 2017. HISTONE DEACETYLASE 6 represses pathogen defence responses in Arabidopsis thaliana. Plant Cell Environ 40: 2972–2986. 10.1111/pce.13047 [DOI] [PubMed] [Google Scholar]
- Wu T, Pi E-X, Tsai S-N, Lam H-M, Sun S-M, Kwan YW, Ngai S-M. 2011a. GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant Biol 11: 178 10.1186/1471-2229-11-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ, Hunt AG. 2011b. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci 108: 12533–12538. 10.1073/pnas.1019732108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Ji G, Li QQ. 2015a. Computational analysis of plant polyadenylation signals. In Polyadenylation in plants: methods and protocols (ed. Hunt AG, Li QQ), pp. 3–11. Springer New York, New York: 10.1007/978-1-4939-2175-1_1 [DOI] [PubMed] [Google Scholar]
- Wu X, Ji G, Li QQ. 2015b. Poly(A)-Tag deep sequencing data processing to extract Poly(A) sites. In Polyadenylation in plants: methods and protocols (ed. Hunt AG, Li QQ), pp. 39-48. Springer, New York: 10.1007/978-1-4939-2175-1_4 [DOI] [PubMed] [Google Scholar]
- Xing D, Li QQ. 2011. Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip Rev RNA 2: 445–458. 10.1002/wrna.59 [DOI] [PubMed] [Google Scholar]
- Xue J-H, Titterington DM. 2011. The p-folded cumulative distribution function and the mean absolute deviation from the p-quantile. Stat Probab Lett 81: 1179–1182. 10.1016/j.spl.2011.03.014 [DOI] [Google Scholar]
- Yan J, Chen S-AA, Local A, Liu T, Qiu Y, Dorighi KM, Preissl S, Rivera CM, Wang C, Ye Z, et al. 2018. Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res 28: 204–220. 10.1038/cr.2018.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-W, Liu X, Luo M, Chen C, Lin X, Tian G, Lu Q, Cui Y, Wu K. 2011. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol 156: 173–184. 10.1104/pp.111.174417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-W, Tai R, Wang S-C, Yang P, Luo M, Yang S, Cheng K, Wang W-C, Cheng Y-S, Wu K. 2017. HISTONE DEACETYLASE6 acts in concert with histone methyltransferases SUVH4, SUVH5, and SUVH6 to regulate transposon silencing. Plant Cell 29: 1970–1983. 10.1105/tpc.16.00570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Addepalli B, Yun K-Y, Hunt AG, Xu R, Rao S, Li QQ, Falcone DL. 2008a. A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS One 3: e2410 10.1371/journal.pone.0002410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W. 2008b. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Wang R, Ding Q, Wang T, Xie B, Wei L, Zhong Z, Tian B. 2018. Cellular stress alters 3'UTR landscape through alternative polyadenylation and isoform-specific degradation. Nat Commun 9: 2268–2268. 10.1038/s41467-018-04730-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L-Y, Zhu Y-R, Dai D-J, Wang X, Jin H-C. 2018. Epigenetic regulation of alternative splicing. Am J Cancer Res 8: 2346–2358. [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Chang Y, Xu X, Tang K, Chen C, Lei M, Zhu JK, Duan CG. 2019. EXPORTIN 1A prevents transgene silencing in Arabidopsis by modulating nucleo-cytoplasmic partitioning of HDA6. J Integr Plant Biol 61: 1243–1254. 10.1111/jipb.12787 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.