Abstract
Through the use of an unbiased, genome-scale CRISPR modifier screen, we identified NF1 suppression as a mechanism of resistance to EGFR inhibition in NRAS/KRAS/BRAFV600-wildtype colorectal cancer (CRC) cells. Reduced NF1 expression permitted sustained signalling through the MAPK (mitogen-activated protein kinase) pathway to promote cell proliferation in the presence of EGFR inhibition. Targeting of MEK in combination with EGFR inhibition lead to synergistic antiproliferative activity. Human KRAS/NRAS/BRAFV600-wildtype colorectal cancer cell lines with NF1 mutations displayed reduced NF1 mRNA or protein expression and were resistant to EGFR blockade by gefitinib or cetuximab. Co-occurring loss-of-function mutations in PTEN were associated with resistance to dual EGFR/MEK inhibition but co-treatment with a PI3 kinase inhibitor further suppressed proliferation. Loss of NF1 may be a useful biomarker to identify patients that are less likely to benefit from single agent anti-EGFR therapy in CRC and may direct potential combination strategies.
Introduction
Epidermal Growth Factor Receptor (EGFR) targeting monoclonal antibodies, cetuximab and panitumumab, have shown meaningful clinical efficacy as single agents and in combination with cytotoxic agents in patients with metastatic KRAS and NRAS exon 2-4 wildtype colorectal cancer (CRC)(1–3). In contrast, the 40% and 5% of CRC tumours that harbour activating KRAS and NRAS mutations respectively (3,4), exhibit primary resistance to anti-EGFR therapies, resulting in constitutive activation of the MAPK pathway and subsequent CRC cell survival and proliferation, despite upstream EGFR inhibition.
Nevertheless, even in patients with KRAS-wildtype tumours, the radiological response rate following treatment with anti-EGFR antibodies alone is limited to approximately 20%, with 25% of patients having progressive disease while on treatment (1). Other genetic aberrations may be implicated in anti-EGFR resistance, although these have not as yet been integrated in routine clinical practice. These include: activating mutations of the PIK3CA, BRAF, MAP2K1 genes, genetic and epigenetic loss of PTEN expression, acquired extracellular EGFR domain mutations and HER2 amplification (5–7). Aside from genetic aberrations, compensatory signalling via activation of other receptors (e.g. via other members of ERBB family) and increased expression of EGFR ligands have also been suggested to contribute to anti-EGFR resistance in KRAS/NRAS-wildtype CRC (6,8). Polyclonal resistance has also been reported to be a common feature in anti-EGFR–refractory CRC, with evolutionary changes typically leading to the selection of the “fittest” RAS-mutant resistant clones (9).
Targeted genome editing technologies, such as CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9, allow for the selective knock-out of genes and subsequent evaluation of the functional consequences of this. In the CRISPR/Cas9 system, the Cas9 nuclease is directed to the gene of interest through complementary binding of a single-guide RNA (sgRNA) to the target DNA. The Cas9 nuclease then cleaves DNA resulting in a double stranded DNA (dsDNA) break. This is then repaired by the error prone non-homologous end joining (NHEJ) leading to irreversible gene disruption, through insertion or deletion events. The CRISPR/Cas9 system has been shown to be superior to other commonly used high-throughput genetic approaches, such as pooled short hairpin RNA (shRNA)-based screens for performing functional genomic screens. In particular CRISPR/Cas9 has been shown to have lower noise and less off-target effects resulting in a low false-discovery rate and a better consistency across different cell lines and reagents (10). Pooled CRISPR knockout screens allow for the unbiased and simultaneous perturbation of multiple genes, at a whole genome scale, enabling the potential landscape of resistance mechanisms to be identified (11).
With the aim of improving our knowledge of mechanisms of resistance to anti-EGFR therapies, in KRAS/NRAS/BRAFV600-wildtype CRC we conducted an unbiased pooled, genome-wide CRISPR screen using an anti-EGFR sensitive CRC cell line under continuous exposure to the anti-EGFR tyrosine kinase inhibitor (TKI) gefitinib. Characterization of surviving cells and further validation identified inactivation of NF1, a known tumour suppressor gene that encodes for the RAS-GTPase–activating protein (RAS-GAP) neurofibromin, as a novel mechanism of resistance to both gefitinib and cetuximab. Our findings may aid personalised medicine approaches to direct therapeutic strategies for improved patient benefit.
Materials and Methods
Cell culture and reagents
Cell culture medium were obtained from Sigma Aldrich, UK and Life Technologies, UK. Fetal bovine serum (FBS Good) was from PAN Biotech UK Ltd. The HEK293T and LIM1215 cell lines were obtained from the American Tissue Culture Collection, HT115 cells were from Public Health England, DIFI cells were a kind gift from Professor Alberto Bardelli, NCIH508 cells were a kind gift from Dr Anguraj Sadanandam. SNUC4 and SNU1040 cells were obtained from the Korean Cell Line Bank. Cell lines were cultured in medium recommended by the suppliers for up to 3 months at a time, cell line authentication was not performed. Cell lines were tested for mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza). Small molecule inhibitors were purchased from Selleck Chemicals. Cetuximab was supplied under a material transfer agreement from Merck.
Immunoblotting
After the desired treatment, cells were washed with PBS and lysed in 1 % SDS lysis buffer (1 % SDS, 10 mM EDTA, 50 mM Tris, pH 8). Bicinchoninic acid (Sigma) was used to determine protein concentration. Equal amounts of protein were separated by gel electrophoresis, using NuPAGE polyacrylamide gels (Life Technologies). Proteins were transferred to a nitrocellulose membrane using the iBlot 2 system (Life Technologies) or by wet transfer using a Bio-Rad Critereon Blotter and then blocked with TBS Li-Cor blocking buffer (Li-Cor Biosciences). Membranes were incubated with the primary antibodies overnight at 4 °C, followed by IRdye-conjugated secondary antibodies (Li-Cor Biosciences) and detected using an Odyssey Fc imaging system (Li-Cor Biosciences). Quantification of Western blots was performed using Image Studio Lite (Li-Cor Biosciences). Details of the antibodies used can be found in Table S1.
Cell Proliferation assays
For GI50 determinations, cells were seeded in 96 well plates. The next day, cells were treated with increasing concentrations of inhibitor or with DMSO alone. After a 96 h incubation period, cell proliferation was quantified by fluorescent detection of the reduction of resazurin to resorufin by viable cells and normalised to DMSO treated wells. GI50 values were calculated using non-linear regression analysis in GraphPad Prism software. For longer-term colony assays, cells were seeded into 12 well plates (0.5-5x104 cells/well), treated with inhibitors the next day and incubated for 10-14 d. Cells were fixed with 4 % formaldehyde/PBS for 20 minutes, then stained with 0.5 % crystal violet in 70 % ethanol. Excess dye was removed by washing with water and plates were imaged with a GelCount (Oxford Optronix). Cell proliferation was quantified by solubilisation of crystal violet dye in 10 % acetic acid and absorbance measured at 595 nm using an EMax Plus plate reader (Molecular Devices), then expressed as a percentage of vehicle-treated cells.
Generation of Cas9-expressing cell line
DIFI cells were engineered to express Cas9 by centrifugation of 2x105 cells with pXPR101-Cas9 lentivirus (The Broad Institute), in the presence of 8 μg/ml polybrene for 1 h at 37 °C. Cells were incubated with fresh media overnight, before cells were trypsinized and pooled for selection with 10 μg/ml blasticidin for 7 d to select for successfully transduced cells.
Lentiviral production
To generate the lentiviral particles for the Brunello whole genome library (Addgene #73178) containing 77,440 sgRNAs targeting 19,110 genes, (12,13), HEK293T cells were seeded at 40 % confluence in T225 flasks. After 24 h, medium was replaced with 13 ml OptiMEM 1 h before transfection with plasmids encoding the library. 20 μg of Brunello plasmid, 10 μg of pMD2.G and 15 μg of psPAX2 were added to 4 ml of OptiMEM. 100 μl of Lipofectamine 2000 were added in another 4 ml of OptiMEM. After 5 min these were mixed and added to the HEK293T cells. After 6 h the media was aspirated and replaced with 30 ml DMEM, 10 % FBS. After 60 h medium was harvested and centrifuged. The supernatant containing lentiviral particles was then filtered through a 0.45 μm low protein binding membrane and was stored in 1 ml aliquots at -80 ˚C. Viral titre was determined by transducing target cells with increasing volumes of virus (50, 100, 200, 300, 500 μl/well) and centrifugation at 2000 rpm for 2 h at 30 ˚C with 8 μg/ml polybrene. 2 ml fresh media were then added to each well. The next day the cells were trypsinised and re-plated in 6 well plates at a density of 3 x 105 cells per well. Each virus dilution was performed in duplicate, with one well treated with 1 μg/ml puromycin. After 72 h, cells were trypsinised and counted to determine the infection efficiency. This was calculated by dividing the cell number of puromycin treated cells by the number for untreated cells for each of the virus dilutions. For individual sgRNAs (Table S2), HEK293T cells were seeded at a density of 2.4x106 cells/plate in 10 cm plates. The next day cells were transfected with sgRNA plasmid (3 μg) and the packaging plasmids psPAX2 (2.1 μg) and pmD2.G (0.9 μg) using 30 μl lipofectamine per transfection. Cells were incubated for 72 h at 37 °C, after which the supernatant was collected and stored in 0.5 ml aliquots at -80 °C for future experiments. Each batch of lentivirus was titrated on cells to determine concentration needed for 100 % infection efficiency. To generate cell populations expressing each sgRNA, cells were transduced with the appropriate lentivirus and selected for using 1 μg/ml puromycin. Knockout/suppression of the target gene was confirmed by Western blotting.
Genome-wide synthetic lethal screen protocol
DIFI-Cas9 cells were seeded in 12 well plates at a density of 1.5x106 cells/well in 2 ml medium. Cells were transduced with the Brunello pooled lentiviral library with a predicted representation of 750 cells/sgRNA at an infection efficiency of 50 %. Cells were centrifuged at 2000 rpm for 2 h at 30 °C in the presence of lentivirus and 8 μg/ml polybrene, followed by incubation in fresh medium overnight. Cells were pooled and seeded into T175 flasks at a density of 5x106 cells/flask for selection with 1 μg/ml puromycin for 7 d and passaged as necessary. In parallel, cells were seeded in 6 well plates to determine infection efficiency. After 7 d of selection, DIFI-Cas9 cells were split into three arms and treated with either 0.1 % DMSO or 240 nM gefitinib. Whilst we initially intended to use cetuximab in the screen, we found that cetuximab treatment did not produce the expected antiproliferative effects in the large-scale, longer-term cell culture format required for the screen. We hypothesise that cetuximab was binding to the plastic of the triple layer flasks and the free-concentration of cetuximab in the culture medium was reduced, lessening the antiproliferative activity. We therefore elected to use gefitinib for the CRISPR screen, which behaved as expected under these culture conditions. We then validated our gefitinib screen results using both cetuximab and gefitinib.
Throughout the screen cells were passaged as necessary, maintaining a total representation of 750 cells/sgRNA in each replicate (three per condition). After 8 population doublings, cells from each arm were collected and cell pellets stored at -80 °C. Genomic DNA was extracted using the QIAamp DNA Blood Maxi Kit (Qiagen). PCR amplification of sgRNA sequences and next generation sequencing was carried out by Cellecta, catalogue no: LNGS-900. The representation of sgRNA in each sample was quantified by counting the number of specific reads generated by NGS on an Illumina Instrument (e.g. NextSeq 500 or HiSeq) (12).
CRISPR screen analysis
The abundance of each sgRNA in each replicate was quantified by calculating the Log2(sequencing reads/million) (RPM), according to the formula below.
The log2 fold change (LFC) from the pDNA sample was calculated by normalising RPM for each sgRNA in each replicate to that in the pDNA sample. The LFC between the DMSO and drug treated arms was calculated as the difference in average LFC across 3 replicates. This was used to rank individual sgRNAs according to their selective depletion or enrichment in the drug treated arms. Top scoring genes were ranked according to the number of independent high scoring sgRNAs targeting the same gene, according to the STARS gene-ranking algorithm (12). In order to assess depletion of essential genes from the population, as a positive control for successful gene editing, the RPM for each sgRNA was normalised to the plasmid DNA to calculate the LFC from baseline. The list of 885 core essential genes was kindly provided by Dr Marco Licciardello (The Institute of Cancer Research) and is compiled from the genes that were consistently and significantly depleted in all cell lines tested from three previous publications (14–16).
Results
A genome-scale CRISPR screen to identify drivers of resistance to EGFR inhibition
We identified the DIFI cell line as a suitable candidate for use in a genome-scale CRISPR screen to discover genes that when suppressed, could promote resistance to EGFR-targeting therapies. The DIFI cell line harbours amplification of the EGFR gene, which is reflected in elevated protein expression and exquisite sensitivity to gefitinib, a small molecule kinase inhibitor of EGFR and to cetuximab, an antibody that targets the extracellular domain of EGFR. In order to conduct a CRISPR/Cas9 screen, DIFI cells were transduced with a lentiviral expression vector for the RNA-guided nuclease Cas9. Both parental DIFI cells and the DIFI-Cas9 cells had near-identical GI50 values for both gefitinib and cetuximab and the phosphorylation of EGFR, ERK and AKT was suppressed to a similar extent by both treatments (Figure S1, Table S3).
For the whole-genome CRISPR screen, we elected to use the Brunello lentiviral sgRNA library which is composed of 76,441 sgRNAs targeting 19,114 genes and 1000 control, non-targeting sgRNAs. DIFI-Cas9 cells were transduced with the Brunello library and after selection and expansion of the cell population, cells were cultured in the presence of either dimethyl sulfoxide (DMSO) or 240 nM gefitinib (Figure 1A), a concentration that inhibits EGFR and ERK phosphorylation and results in durable suppression of cell proliferation (Figure S2). Initially, gefitinib-treated cells did not proliferate but after 3-4 weeks, proliferation resumed at a rate similar to the DMSO-treated cells (Figure 1B). Cells were passaged for up to 8 population doublings then genomic DNA was purified, sgRNAs were amplified by PCR and the abundance of each sgRNA was determined by next generation sequencing (NGS). The abundance of each sgRNA was determined relative to the Brunello plasmid DNA library as a reference. Comparison of two DMSO-treated replicates demonstrated good concordance between the two replicates (Person correlation 0.722), that non-targeting control (NTC) sgRNAs were largely unaltered in representation in the population and that sgRNAs targeting known essential genes were depleted from the population (Figure 1C).
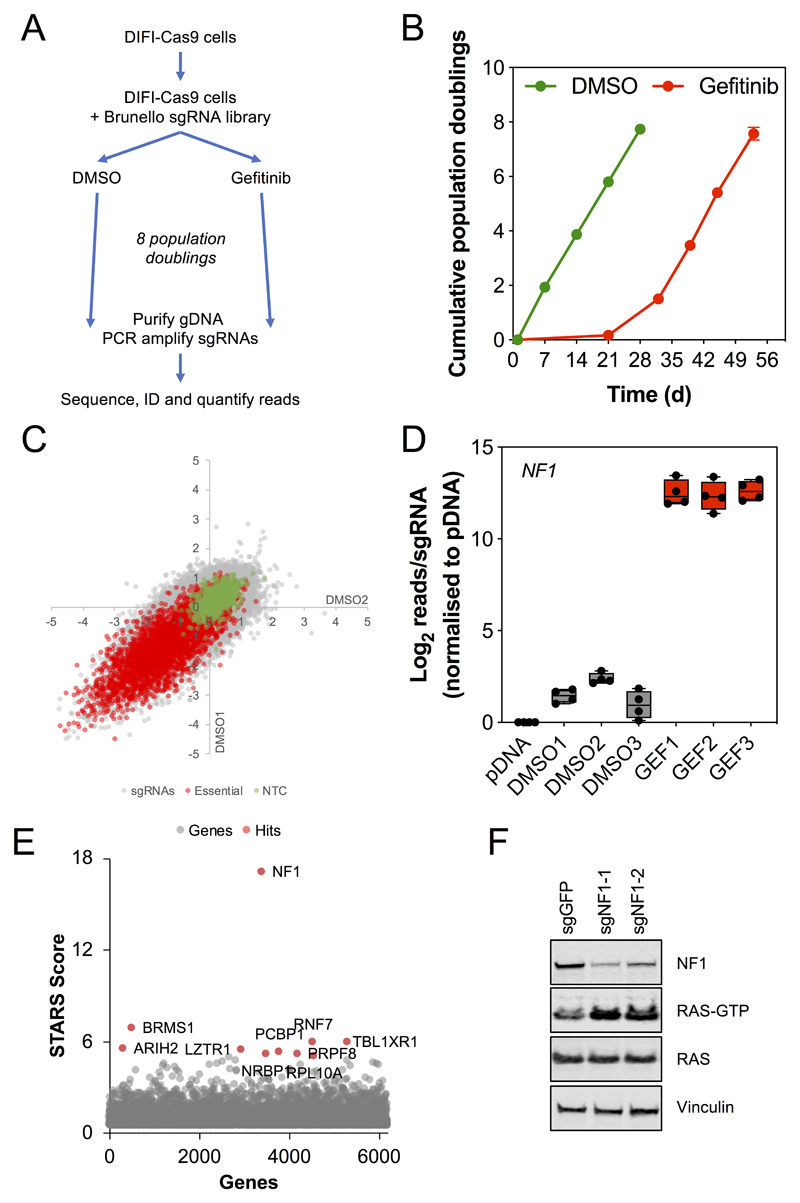
Figure 1. A genome-scale CRISPR screen identifies NF1 suppression as a driver of resistance to EGFR inhibition.
A. Schematic outline of the CRISPR modifier screen. DIFI colorectal cancer cells were transduced with lentiviral particles encoding for the expression of the Cas9 gene. DIFI-Cas9 cells were then transduced with lentiviral particles for the Brunello library of 77,440 sgRNAs targeting 19,110 genes. Cells were cultured in the presence of either DMSO or 240 nM gefitinib for up to 8 population doublings, genomic DNA was then purified and sgRNAs amplified by PCR and sequenced. sgRNAs were identified, mapped to their target genes and reads quantified.
B. Proliferation of DIFI-Cas9 cells transduced with the Brunello library and cultured in the presence of DMSO (0.1%) or 240 nM gefitinib for up to 8 population doublings.
C. Comparison of the LFC for two DMSO-treated replicates normalised to the pDNA library sample shows good correlation between replicates. Non-targeting control sgRNAs are indicated in green and sgRNAs targeting essential genes are indicated in red.
D. Abundance of sgRNAs targeting NF1 in the pDNA library, DMSO- and gefitinib-treated DIFI-Cas9 cells for each experimental replicate.
E. STARs analysis of sgRNA abundance in gefitinib-versus DMSO-treated DIFI-Cas9 cells. Top-ranking genes are shown in red.
F. DIFI-Cas9 cells were transduced with two independent sgRNAs targeting NF1 and a control sgRNA targeting GFP. Transduced cells were selected in the presence of puromycin for 7 d to establish stable cell line pools. The expression of the indicated proteins was determined by Western blotting. Levels of RAS-GTP were determined using a RAS-GTP pulldown assay. Data are representative of 3 independent experiments.
The log-fold change (LFC) of the gefitinib-treated replicates was compared to the DMSO-treated replicates to identify sgRNAs that were enriched in the presence of gefitinib. Strikingly, all four sgRNAs targeting NF1 were the most highly enriched (ranked 1-4) in the presence of gefitinib relative to DMSO (Figure 1D). STARS analysis was then used to rank genes for positive enrichment in the presence of gefitinib based on the sgRNA rankings, NF1 was the most highly ranked gene (Figure 1E, Table S4). In comparison, the next three most highly ranked genes BRMS1, TBL1XR1 and RNF7 showed inconsistent and relatively weak enrichment in the presence of gefitinib for individual sgRNAs and also across the three replicates (Figure S3). NF1 encodes the protein neurofibromin, a RAS-GTPase activating protein (RAS-GAP) which stimulates RAS proteins to hydrolyse bound GTP to GDP and consequently switch off RAS (17). Suppression of NF1 would therefore maintain RAS in the active, GTP-bound conformation and promote downstream signalling. We confirmed that targeting of NF1 with two independent sgRNAs not used in the genome-scale screen suppressed NF1 protein expression and resulted in increased levels of RAS-GTP in the DIFI-Cas9 cells (Figure 1F).
Consequences of NF1 suppression on cellular signalling changes induced by EGFR inhibition
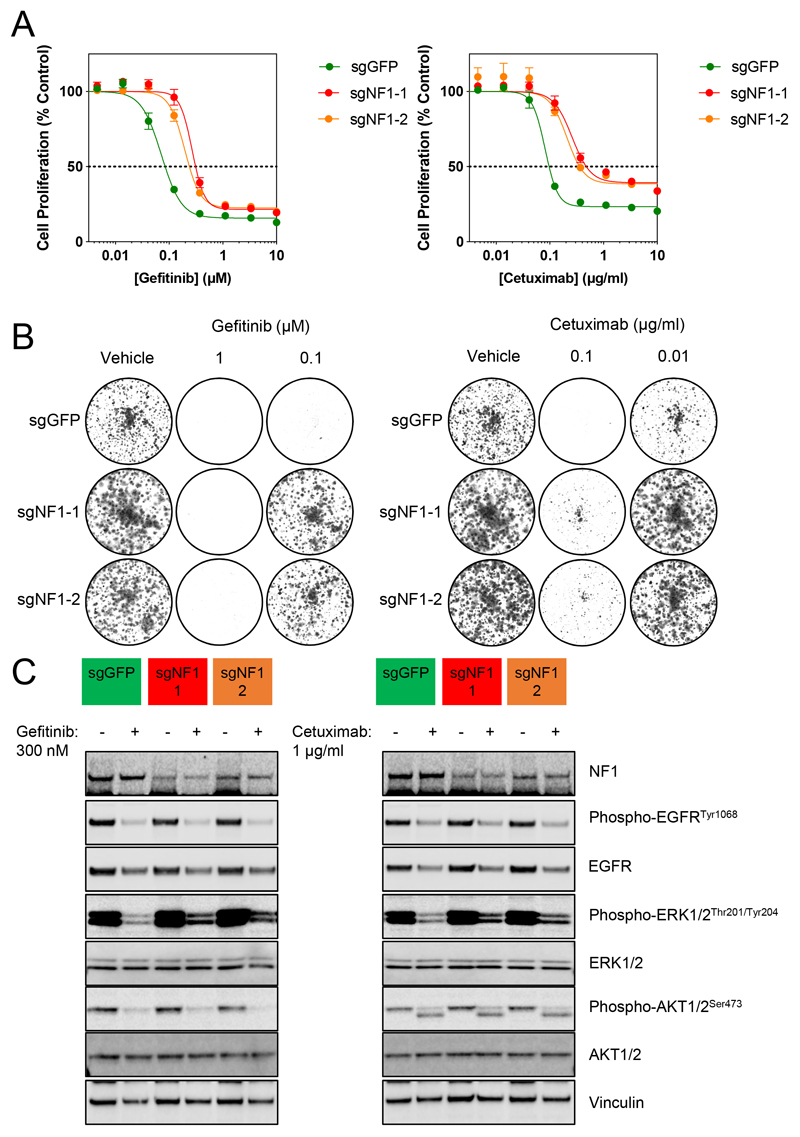
We generated stable pools of DIFI cells which expressed either an sgRNA targeting GFP or two different sgRNAs targeting NF1. To validate the CRISPR screen findings, these cells were treated with increasing concentrations of gefitinib or cetuximab for 96 h. Cell proliferation was determined by fluorescent detection of the reduction of resazurin to resorufin (Figure 2A). Suppression of NF1 was associated with an increase in the GI50 for gefitinib or cetuximab (Table S3). Furthermore, longer-term colony formation assays demonstrated resistance to gefitinib and cetuximab in cells expressing NF1-targeting sgRNAs (Figure 2B, Figure S4). Western blotting of lysates from cells treated with 300 nM gefitinib or 1 μg/ml cetuximab demonstrated impaired inhibition of ERK phosphorylation in the NF1-targeted cells compared to the GFP-targeted controls, despite comparable inhibition of EGFR phosphorylation. In contrast, inhibition of phospho-AKT was unaffected by suppression of NF1 (Figure 2C). Therefore, suppression of NF1 results in sufficient MAPK pathway activity to sustain cell proliferation in the presence of gefitinib or cetuximab, consistent with activation of RAS proteins.
Figure 2. Resistance to gefitinib and cetuximab through loss of NF1 is associated with incomplete inhibition of the MAPK pathway.
A. DIFI-Cas9 cells expressing sgRNAs targeting GFP or NF1 were exposed to increasing concentrations of gefitinib or cetuximab for 4 d. Cell proliferation was assessed by fluorescent detection of the reduction of resazurin to resorufin by viable cells and expressed as a percentage of vehicle-treated cells. GI50 values were determined by non-linear regression in GraphPad Prism. Mean values are shown ± standard error (n=4), data are representative of 3 independent experiments.
B. DIFI-sgGFP and -sgNF1 cells were treated with 1 or 0.1 μM gefitinib, 0.1 or 0.01 μg/ml cetuximab for 10 d and colonies were stained with crystal violet prior to imaging. Data are representative of 3 independent experiments.
C. DIFI-Cas9 cells expressing sgRNAs targeting GFP or NF1 were exposed to vehicle, 300 nM gefitinib or 1 μg/ml cetuximab for 24 h. Cell lysates were analysed for the indicated proteins by Western blotting. Data are representative of 3 independent experiments.
The effect of MEK inhibition in DIFI cells with suppression of NF1
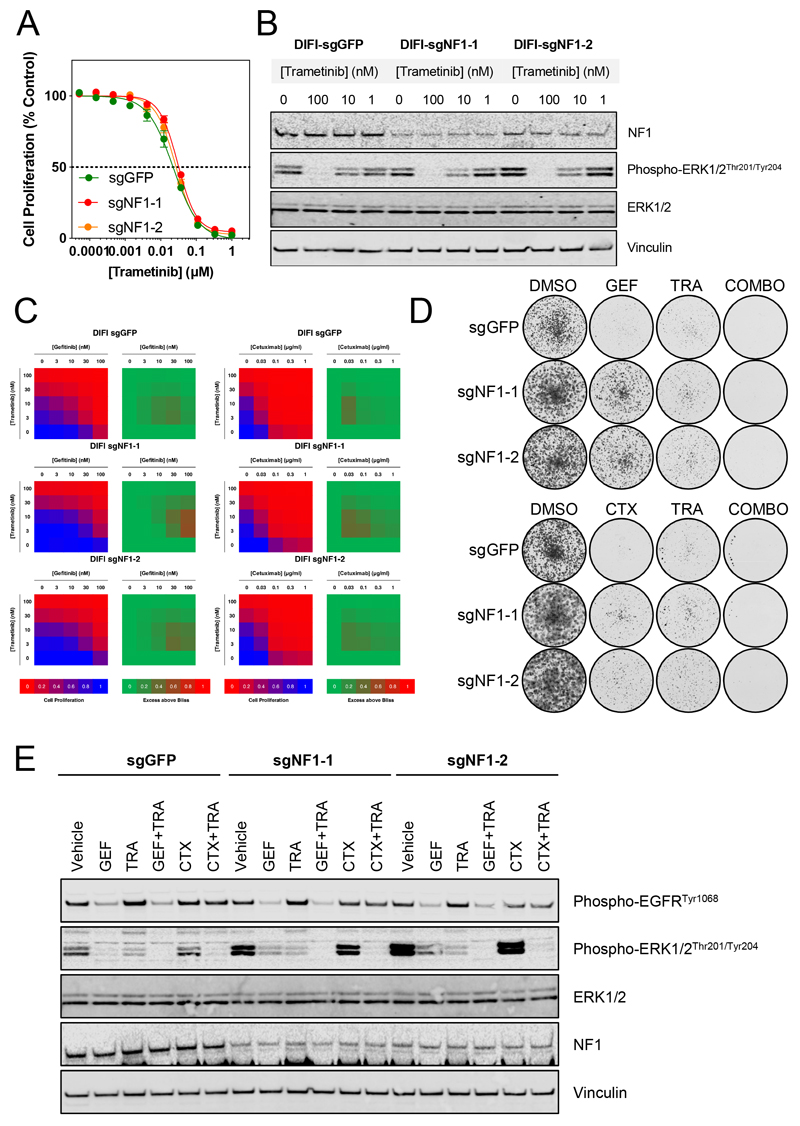
We hypothesised that inhibition of MEK may be necessary to overcome loss of NF1-mediated resistance to EGFR inhibition. Hence, DIFI cells expressing NF1-targeting sgRNAs were treated with increasing concentrations of the MEK inhibitor trametinib for 96 h and cell proliferation determined. As anticipated, cells with decreased NF1 expression were equally sensitive to trametinib compared to the GFP-targeting control (Figure 3A, Table S3). Western blotting of lysates from cells treated with increasing concentrations of trametinib for 24 h demonstrated comparable inhibition of ERK phosphorylation relative to the GFP-targeting control (Figure 3B). Prior reports suggest that the combination of EGFR and MEK inhibitors elicits synergistic activity in EGFR-inhibitor-resistant models. We therefore tested the combination of gefitinib or cetuximab with trametinib. DIFI cells with NF1 suppression were treated with a matrix of gefitinib and trametinib or cetuximab and trametinib for 96 h and cell proliferation was determined. Synergy was assessed using the Bliss independence model and demonstrated synergistic activity across both DIFI-sgGFP cells and DIFI-sgNF1 cells, affirming that this combination has potential therapeutic utility in NF1-suppressed models (Figure 3C)(18). In longer-term colony formation assays, modest resistance to trametinib was observed in DIFI-sgNF1 cells compared to DIFI-sgGFP cells but the combination of both gefitinib and trametinib or cetuximab and trametinib resulted in near-complete inhibition of colony formation (Figure 3D, Figure S5). Western blotting of lysates from cells treated with the combination of gefitinib or cetuximab with trametinib for 24 h demonstrated higher basal levels of ERK phosphorylation in DIFI-sgNF1 cells and incomplete inhibition of ERK phosphorylation following gefitinib or cetuximab treatment (Figure 3E). The combination of trametinib with either gefitinib or cetuximab was required to achieve near-complete suppression of ERK phosphorylation. Therefore, inhibition of the MAPK pathway appears to be required for a robust antiproliferative effect in cells lacking NF1 expression.
Figure 3. MEK inhibition is effective in DIFI cells with loss of NF1 and synergises with EGFR inhibitors.
A. DIFI-Cas9 cells expressing sgRNAs targeting GFP or NF1 were exposed to increasing concentrations of trametinib for 4 d. Cell proliferation was assessed by fluorescent detection of reduction of resazurin to resorufin by viable cells and normalised to vehicle-treated cells. GI50 values were determined by non-linear regression in GraphPad Prism. Mean values are shown ± standard error (n=4), data are representative of 3 independent experiments.
B. DIFI-Cas9 cells expressing sgRNAs targeting GFP or NF1 were treated with increasing concentrations of trametinib for 24 h. Cell lysates were analysed by Western blotting for the indicated proteins. Data are representative of 3 independent experiments.
C. DIFI-Cas9 cells expressing sgRNAs targeting GFP or NF1 were exposed to a matrix of increasing concentrations of trametinib and gefitinib or cetuximab. Cell proliferation was assessed by fluorescent detection of the reduction of resazurin to resorufin by viable cells and expressed relative to vehicle-treated cells (blue to red heatmap). Synergy was assessed by the Bliss assay, values > 0 are indicative of synergy (green to red heatmap). Data are representative of 3 independent experiments.
D. DIFI-Cas9 cells expressing sgRNAs targeting GFP or NF1 were exposed to 100 nM gefitinib, 30 nM trametinib or their combination for 10 d. Cells were fixed and stained with crystal violet prior to imaging. Data are representative of 3 independent experiments.
E. DIFI-Cas9 cells expressing sgRNAs targeting GFP or NF1 were exposed to 300 nM gefitinib, 30 nM trametinib, 0.3 μg/ml cetuximab or the indicated combinations for 24 h. Cell lysates were analysed by Western blotting for the indicated proteins. Data are representative of 2 independent experiments.
Sensitivity of NF1-mutant colorectal cancer cell lines to EGFR inhibitors
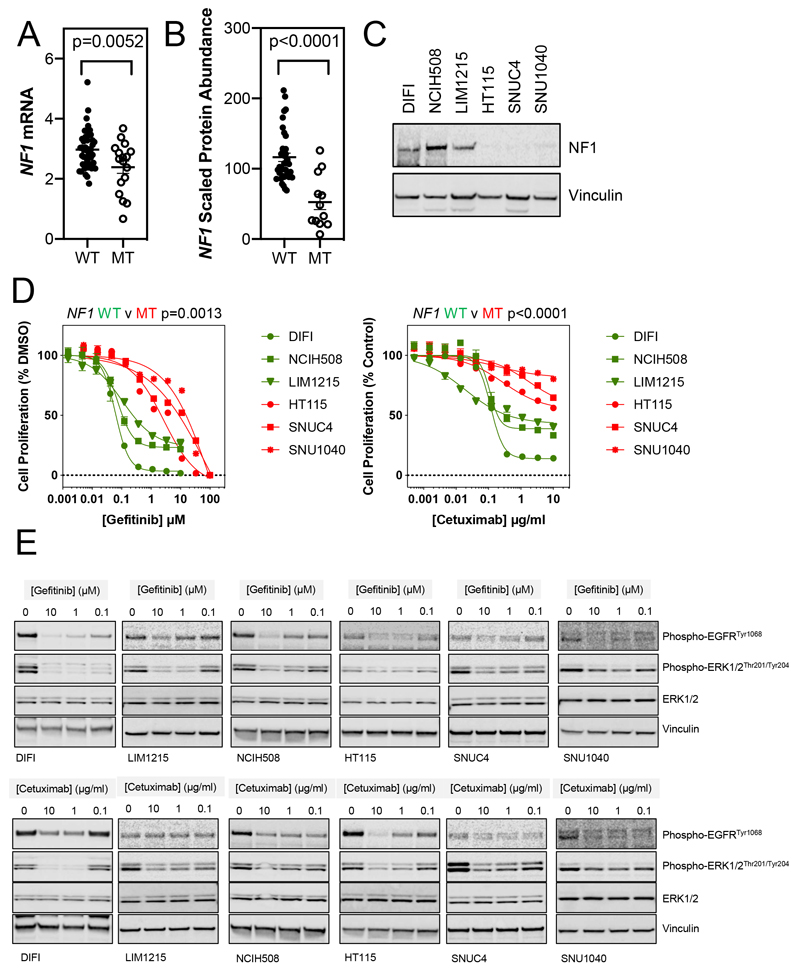
To translate the findings from our genetic models of NF1-suppression to established cell lines, we consulted the Cancer Dependency Map for CRC cell lines that harboured mutations in NF1. Of 60 CRC cell lines, 17 (28%) were NF1-mutant. NF1 mutation was associated with a significant reduction in NF1 mRNA expression (Figure 4A). Moreover, we utilised a recent proteomics-based profiling of 50 CRC cell lines, in which 12 (24%) were NF1-mutant (19). NF1 mutation was associated with significantly reduced expression of NF1 protein (Figure 4B). Therefore, we selected three NF1-wildtype and three NF1-mutant CRC cell lines (all KRAS/NRAS/BRAFV600-wildtype) and assessed NF1 expression by Western blotting. The NF1-mutant cell lines HT115 (NF1R1241*), SNUC4 (NF1T676fs) and SNU1040 (NF1R461*) had little or no expression of NF1 protein (Figure 4C).
Figure 4. NF1-mutant colorectal cancer cell lines are resistant to EGFR inhibitors.
A. Data for NF1 mutation and mRNA expression was obtained from DepMap.org. Of 60 colorectal cancer cell lines, 17 (28%) harboured mutations in the NF1 gene. The expression of NF1 mRNA was compared between wildtype and mutant cell lines (two-tailed T-test). WT = wildtype, MT = mutant.
B. Data for NF1 mutation and protein expression was obtained from Roumeliotis et al (19). Of 50 colorectal cancer cell lines, 12 (24%) harboured mutations in the NF1 gene. The expression of NF1 protein was compared between wildtype and mutant cell lines (two-tailed T-test). WT = wildtype, MT = mutant.
C. Lysates from NF1-wildtype (DIFI, NCIH508 and LIM1215) and NF1-mutant (HT115, SNUC4 and SNU1040) colorectal cancer cell lines were analysed by Western blotting for the indicated proteins. Data are representative of 3 independent experiments.
D. NF1-wildtype (DIFI, NCIH508 and LIM1215) and NF1-mutant (HT115, SNUC4 and SNU1040) colorectal cancer cell lines were exposed to increasing concentrations of gefitinib or cetuximab for 4 d. Cell proliferation was assessed by fluorescent detection of reduction of resazurin to resorufin by viable cells and normalised to vehicle-treated cells. GI50 values were determined by non-linear regression in GraphPad Prism. Mean values are shown ± standard error (n=4), data are representative of 3 independent experiments. The GI50 values for NF1-wildtype versus NF1-mutant cell lines were compared (two-tailed T-test).
E. Cell lines as in D were exposed to the indicated concentrations of gefitinib or cetuximab for 3 d. Cell lysates were analysed for the indicated proteins by Western blotting. Data are representative of 3 independent experiments.
The sensitivity of this panel of CRC cell lines to gefitinib and cetuximab was assessed and NF1 mutation was associated with reduced sensitivity to both agents (Figure 4D, Table S3). Inhibition of EGFR and ERK phosphorylation was also assessed in these cell lines following a 72 h exposure to increasing concentrations of gefitinib or cetuximab (Figure 4E). While inhibition of EGFR phosphorylation was generally observed across all cell lines treated with gefitinib, inhibition of ERK phosphorylation was less complete in NF1-mutant cell lines. Cetuximab treatment also resulted in decreased EGFR and ERK phosphorylation although interestingly, the association with cetuximab sensitivity was less pronounced.
The effect of combined gefitinib and trametinib or gefitinib and BYL719 on NF1-mutant colorectal cancer cell proliferation
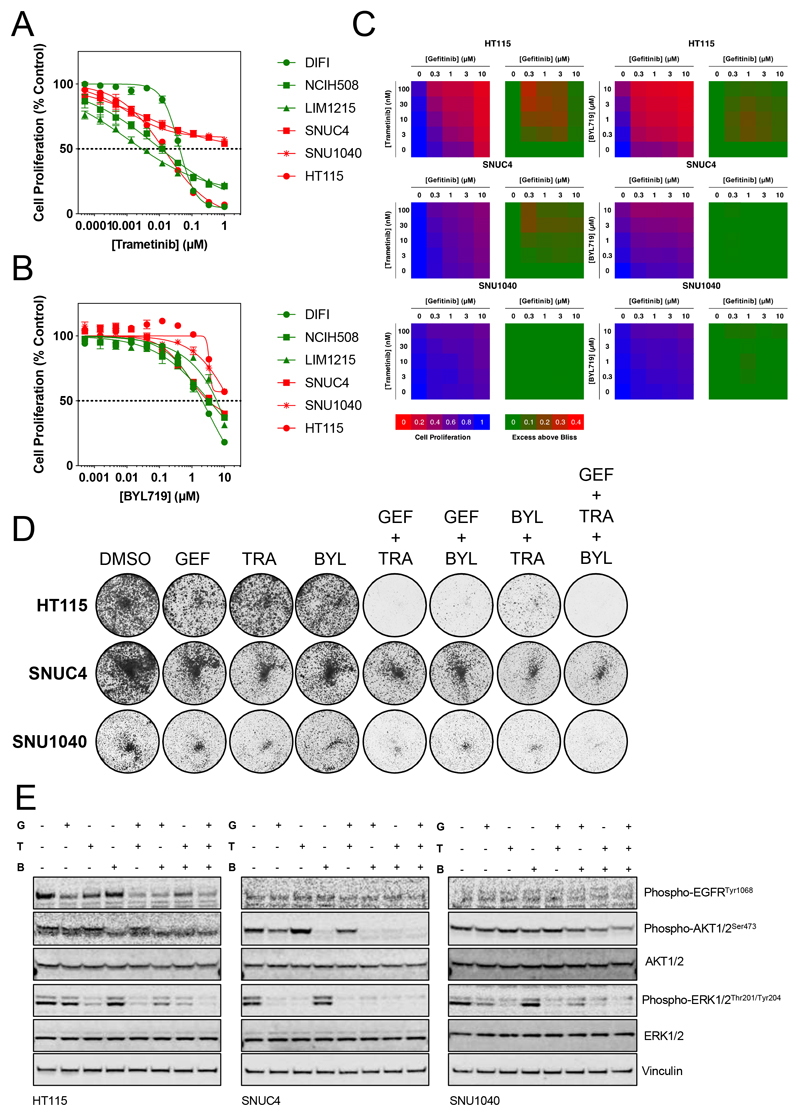
As we have shown that DIFI cells with NF1 suppression were equally sensitive to the MEK inhibitor trametinib, the panel of CRC cell lines was assessed for sensitivity to trametinib (Figure 5A, Table S3). Only the HT115 NF1-mutant cell line showed sensitivity to trametinib similar to the NF1-wildtype cell lines DIFI, LIM1215 and NCIH508. In order to determine a suitable therapeutic strategy for those cell lines not sensitive to trametinib we studied the mutational profile of the SNUC4 and SNU1040 cells for events that may drive resistance to trametinib. Both cell lines have damaging mutations in PTEN, which could promote resistance via activation of the PI3K pathway (www.DepMap.org). Notably, HT115 cells have mutations in PIK3CA, resulting in a triple mutant (PIK3CAE321D, R770Q, R88Q), although this did not appear to be sufficient to drive resistance to trametinib. We first tested the cell line panel for sensitivity to the p110α-specific PI3K inhibitor BYL719 and all cell lines appeared to be resistant (Figure 5B, Table S3). We therefore tested the combination of gefitinib and trametinib or gefitinib and BYL719 for synergy as assessed by the Bliss independence model (Figure 5C). Some synergy was observed in the HT115 cell line between gefitinib and trametinib or gefitinib and BYL719 but no synergy was seen with this combination in SNUC4 or SNU1040 cells. As both SNUC4 and SNU1040 cell lines lack expression of PTEN (20), it was possible that activation of the PI3K pathway via PTEN loss was sufficient to confer resistance to the dual EGFR/MEK inhibitor combination. We confirmed that both SNUC4 and SNU1040 cells exhibited high levels of AKT phosphorylation, relative to the other PTEN-wildtype cell lines in our panel, indicative of PTEN loss (Figure S6). Despite mutations in PIK3CA, HT115 cells did not exhibit high levels of AKT phosphorylation, which may explain their relative sensitivity to MEK inhibition.
Figure 5. Combinations targeting EGFR, MEK and p110α show additive to synergistic antiproliferative activity in NF1-mutant colorectal cancer cell lines.
A. NF1-wildtype (DIFI, NCIH508 and LIM1215) and NF1-mutant (HT115, SNUC4 and SNU1040) colorectal cancer cell lines were exposed to increasing concentrations of trametinib for 4 d. Cell proliferation was assessed by fluorescent detection of reduction of resazurin to resorufin by viable cells and normalised to vehicle-treated cells. GI50 values were determined by non-linear regression in GraphPad Prism. Mean values are shown ± standard error (n=4), data are representative of 3 independent experiments.
B. NF1-mutant (HT115, SNUC4 and SNU1040) colorectal cancer cell lines were exposed to increasing concentrations of BYL719 for 4 d. Cell proliferation was assessed by fluorescent detection of reduction of resazurin to resorufin by viable cells and normalised to vehicle-treated cells. GI50 values were determined by non-linear regression in GraphPad Prism. Mean values are shown ± standard error (n=4), data are representative of 3 independent experiments.
C. NF1-mutant HT115, SNUC4 and SNU1040 cell lines were exposed to a matrix of increasing concentrations of geftinib and trametinib or gefitinib and BYL719. Cell proliferation was assessed by fluorescent detection of the reduction of resazurin to resorufin by viable cells and expressed relative to vehicle-treated cells (blue to red heatmap). Synergy was assessed by the Bliss assay, values > 0 are indicative of synergy (green to red heatmap). Data are representative of 3 independent experiments.
D. HT115, SNUC4 and SNU1040 NF1-mutant cell lines were treated with 300 nM gefitinib (GEF), 1 μM BYL719 (BYL), 30 nM trametinib (TRA) or combinations of these agents for 10 d. Cells were fixed and stained with crystal violet prior to imaging. Data are representative of 3 independent experiments.
E. HT115, SNUC4 and SNU1040 NF1-mutant cell lines were treated with 300 nM gefitinib (G), 30 nM trametinib (T), 1 μM BYL719 (B), or combinations of these agents for 24 h. Cell lysates were analysed for the indicated proteins by Western blotting. Data are representative of 2 independent experiments.
Antiproliferative activity of combinations targeting EGFR, MEK and p110α in NF1-mutant colorectal cancer cell lines
In light of the above data, we hypothesised that the triple combination of gefitinib, trametinib and BYL719 may inhibit cell proliferation more effectively than single agents or paired combinations. HT115, SNUC4 and SNU1040 cells were exposed to pathway-inhibitory concentrations of gefitinib, trametinib and BYL719 alone or in combination (Figure 5D). In HT115 cells, the combination of gefitinib and trametinib reduced colony formation by >90% and the combination of gefitinib and BYL719 inhibited colony formation to a similar extent. Therefore, dual inhibition of EGFR and MEK or EGFR and p110α appears to be synthetic lethal in HT115 cells. In contrast, the SNUC4 and SNU1040 cell lines were more resistant to the combination of gefitinib and trametinib but the addition of BYL719 further decreased colony formation (Figure 5D, Figure S7). Analysis of lysates from cell lines treated with gefitinib, trametinib or BYL719 for 24 h indicated that dual suppression of both AKT and ERK phosphorylation by the triple combination was associated with greater antiproliferative activity (Figure 5E). The above data suggest that the triple combination of gefitinib, trametinib and BYL719 may yield greater antiproliferative activity than single agents or dual combinations. Given the high frequency of PI3K pathway mutations in KRAS/NRAS/BRAFV600-wildtype CRC cell lines, these data raise the tantalising prospect that the addition of a PI3K inhibitor to cetuximab and trametinib may enhance antitumor activity, as has been observed in BRAF-mutant colorectal cancers with the combination of BRAF, EGFR and PI3K inhibitors.
Discussion
The advent of targeted therapies has enabled the concept of precision medicine, whereby drugs are administered to a select group of patients based on the expression (or lack of expression) of evidence-based molecular biomarkers. This approach not only maximises efficacy and cost-effectiveness, it also prevents unwarranted toxicity. However, drug resistance whether through selection of sub-clonal populations that harbour pre-existing resistant clones or by genetic or non-genetic adaptions to treatment, limit the duration of response and lead to disease progression. Understanding potential mechanisms of resistance to targeted therapies may therefore enable alternate therapeutic strategies such as combinations of drugs to overcome bypass mechanisms or targeting points of convergence of the primary target and resistance pathways.
CRISPR/Cas9 screens enable parallel assessment of the loss of thousands of genes on sensitivity to drug treatment. Typically, a synthetic-lethal approach is taken whereby loss of a particular gene may sensitise to treatment and sgRNAs targeting the sensitising gene are depleted from the population. However, these ‘drop-out’ screens require transduction of a greater number of cells to ensure sufficient sgRNA representation and greater sequencing depth, resulting in a reduced signal to noise ratio and potentially reduced confidence in hit calling. In this case, we sought genes that when lost, confer resistance to treatment and sgRNAs that target such genes are enriched in the population. The benefits of this approach are that sgRNA representation can be lower, requiring fewer cells for the initial sgRNA library viral transduction step, a lower sequencing depth and greater signal to noise ratio, permitting more statistically robust hit calling.
Our genome-scale CRISPR screen identified suppression of NF1 expression as the most highly-ranked and robust driver of resistance to EGFR inhibition. Because NF1 acts as a RAS-GAP, loss of this protein results in the activation of RAS, as it is stabilised in its GTP-bound form (17). KRAS and NRAS mutations are a well-established driver of resistance to EGFR-targeting therapy in CRC, therefore, reduced expression of NF1 may well phenocopy genetic alterations that promote RAS activity.
There have been recent publications that further support the notion that impairment of NF1 function may be implicated in anti-EGFR resistance in CRC. A study that analysed the mutation profiles of 33 patients with metastatic CRC using next-generation sequencing before starting cetuximab suggested that patients with NF1 mutations (n=4) had significantly shorter progression free survival, following treatment with cetuximab when compared to those with NF1-wildtype tumours (21). In our own institution, a separate study investigated anti-EGFR therapy resistance in RAS-wildtype CRC by carrying out exome and RNA sequencing from biopsies taken at baseline and upon progression to cetuximab. Tumors from two of the 35 patients with primary anti-EGFR resistance had inactivating mutations of NF1 (22). Recently it was reported that in a limited study of CRISPR-mediated knockout of RAS-GAPs in human colorectal cancer organoids, NF1 knockout conferred resistance to the EGFR TKI afatinib (23). Our genome-scale approach further demonstrated that NF1 was the most highly-ranked loss of function event to drive resistance. Compelling evidence in support of NF1 loss as a possible resistance mechanism to anti-EGFR therapies was also reported by a study that used genome-wide siRNA screen in EGFR-mutant non-small cell lung cancer (NSCLC) cells that were exposed to another anti-EGFR TKI, erlotinib. The authors demonstrated that NF1 mRNA expression is downregulated in an EGFR-driven, erlotinib-resistant inducible model of NSCLC, potentially as an alternative resistance mechanism to mutation of the gatekeeper residue of EGFR itself. Furthermore, knockdown of NF1 expression by shRNA also conferred resistance to EGFR inhibition in the PC9 NSCLC xenograft model, demonstrating this effect translated from in vitro models to in vivo models. Reduced expression of NF1 was also associated with resistance to EGFR-directed treatment and lower survival in NSCLC patients (24).
Our findings strongly demonstrate that relief of negative regulation of RAS via NF1 loss is a mechanism of resistance in NRAS/KRAS/BRAFV600-wildtype CRC. Mutation of NF1 has been reported to occur in approximately 5% of CRC tumours (25). Aside genomic aberrations, loss of NF1 expression may occur via epigenetic mechanisms such as gene promoter methylation, therefore, loss of NF1 function may account for > 5% of anti-EGFR resistance in KRAS/NRAS/BRAFV600-wildtype CRC. Use of this information in clinical decision-making may spare these patients from unnecessary treatment.
Interestingly, our data point to activation of the MAPK pathway as being a major driver of resistance, as targeting of MEK overcomes resistance associated with suppression of NF1. This is in line with previous studies that suggested that escape routes from EGFR blockade in CRC biochemically converge on MAPK activation and that vertical suppression with concomitant blockade of EGFR and MEK could overcome primary and acquired resistance to anti-EGFR agents in KRAS-mutant CRC cells (26,27). However, in our initial validation we identified 2 cells lines that harboured NF1 mutations, SNUC4 and SNU1040, that were relatively resistant to trametinib or the combination of trametinib and gefitinib. These cell lines have co-occurring mutations in PTEN which may contribute to the resistant phenotype, as combined inhibition of EGFR, MEK and p110α suppressed cell proliferation as depicted in our simplified model in Figure 6. Aside from PTEN inactivation, compensatory PI3K pathway activation may also arise from other mechanisms including: activating PIK3CA mutations and non-genetic compensatory activation of the PI3K signalling pathway. The notion of compensatory PI3K pathway activation in cells with constitutive activation of the MAPK pathway is strongly supported by two studies that utilised KRAS-mutant CRC cell lines. In one study KRAS-mutant CRC cells were rendered resistant to the combination of cetuximab and refametinib (a selective MEK inhibitor) after continuous exposure to increasing concentration of both drugs. It was suggested that resistance to this combination was due to secondary PI3K activation, following cooperative activation of multiple receptor tyrosine kinases: HER2, HER3 and IGF1R (28). Similarly, in another study shRNA knockdown of KRAS expression in KRAS-mutant CRC cell lines led to ERK suppression but was not sufficient to downregulate the PI3K pathway. Instead PI3K activation was regulated via activation of other receptor tyrosine kinases rather than KRAS itself (29). Similar to KRAS-mutant CRC, we showed that in CRC cells with NF1 suppression and co-occurring PTEN mutation, bypass PI3K pathway activation may also occur. Our data agree with studies in CRC or other EGFR-driven cancers where the combination of EGFR inhibitors and MEK or PI3K inhibitors leads to greater antiproliferative activity, including in the context of RAS-driven resistance in both in vivo models (30–32) and early phase clinical trials (33).
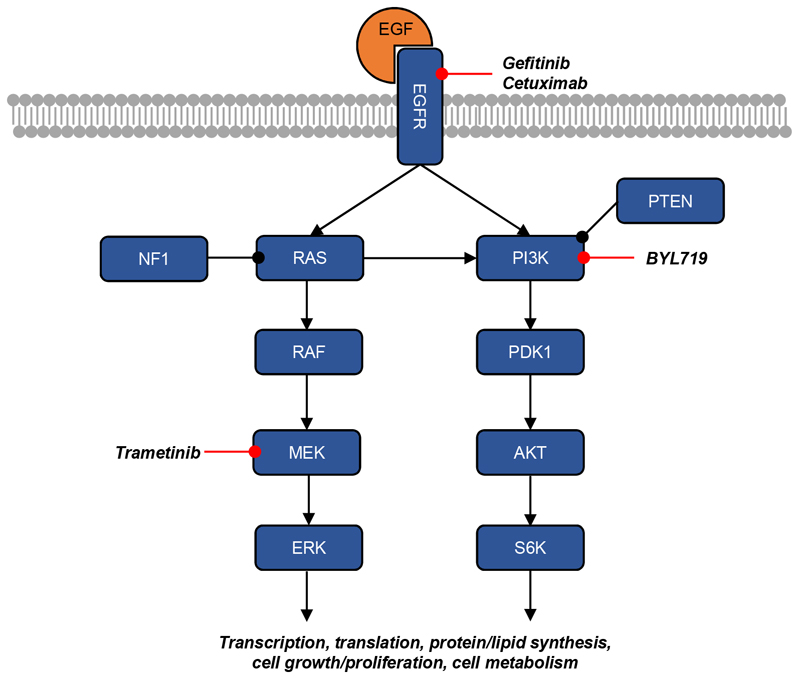
Figure 6. Cartoon of EGFR-mediated signalling pathways and their regulation by NF1.
EGFR engages the MAPK and PI3K pathways in response to EGF ligand. NF1 negatively regulates RAS such that NF1 loss leads to activation of RAS. In DIFI cells, loss of NF1 results in moderate activation of the MAPK pathway but has no effect on the PI3K pathway. Combined inhibition of EGFR and MEK is synergistic in NF1-wildtype and NF1-targeted DIFI cells. However, SNUC4 and SNU1040 cells are less sensitive to EGFR/MEK inhibition and require inhibition of EGFR, MEK and PI3K. As observed in BRAF-mutant colorectal cancer, a triple combination of EGFR, MEK and PI3K inhibitors may warrant investigation in BRAF/KRAS/BRAFV600-wildtype colorectal cancer.
In conclusion, our results suggest that loss of NF1 function may be a predictive biomarker of response to anti-EGFR therapy in CRC and that in CRC cells with NF1 suppression concomitant inhibition of the MAPK and PI3K pathways may be required to overcome anti-EGFR resistance. Our data support the concept that higher-order drug combinations may be required to elicit robust antiproliferative effects in CRC (34). Further clinical validation of NF1 status as a predictor of response to anti-EGFR targeting antibodies in CRC patients with KRAS/NRAS/BRAFV600-wildtype tumours is warranted.
Supplementary Material
Implications.
This study suggests that further clinical validation of NF1 status as predictor of response to anti-EGFR targeting antibodies in CRC patients with KRAS/NRAS/BRAFV600-wildtype tumours is warranted.
Acknowledgements
We thank Dr Marco Licciardello, Institute of Cancer Research, for assistance with generating the Brunello pooled lentiviral library.
Funding
This work was supported by a Cancer Research UK grant awarded for A.G clinical research fellowship at The Institute of Cancer Research. Authors acknowledge funding from the National Institute of Health (NIHR) Biomedical Research Centre (BRC) and Experimental Cancer Medicine Centre (ECMC) initiatives at The Institute of Cancer Research and The Royal Marsden NHS Foundation trust. U.B. is a recipient of a NIHR research professorship award (RP-2016-07-028).
Footnotes
Disclosure of Potential Conflicts of Interest
A.G, A.S and S.W declare no conflicts of interest. D.C has received grants from 4SC, Amgen, AstraZeneca, Bayer, Celgene, Clovis, Eli Lilly, Janssen, Medimmune, Merck, Merrimack and Sanofi outside the submitted work. U.B has received honoraria from Astellas, Novartis, Karus Therapeutics, Phoenix Solutions, Eli Lilly, Astex, Vernalis, Janssen and Boehringer Ingelheim. His institutional financial interests include funding for Phase I investigator-initiated trials from Onyx Pharmaceuticals, BTG International, Carrick Therapeutics, Chugai, AstraZeneca and Verastem. He is an employee of the Institute of Cancer Research, London, which is involved in the development of PI3K, HSP90, HDAC, AKT, ROCK, RAF, CHK1, and HSF1 inhibitors.
Authors’ Contributions
Conceptualization: A.G, U.B and S.W; methodology: A.G, U.B and S.W; data collection: A.G, A.S and S.W; data analysis: A.G and S.W; writing—original draft preparation: A.G and S.W; writing—review and editing: A.G, A.S, D.C and S.W; supervision: U.B and S.W
References
- 1.Price TJ, Peeters M, Kim TW, Li J, Cascinu S, Ruff P, et al. Panitumumab versus cetuximab in patients with chemotherapy-refractory wild-type KRAS exon 2 metastatic colorectal cancer (ASPECCT): a randomised, multicentre, open-label, non-inferiority phase 3 study. The Lancet Oncology. 2014;15(6):569–79. doi: 10.1016/S1470-2045(14)70118-4. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet (London, England) 2010;375(9719):1030–47. doi: 10.1016/s0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E, Lenz HJ, Kohne CH, Heinemann V, Tejpar S, Melezinek I, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(7):692–700. doi: 10.1200/jco.2014.59.4812. [DOI] [PubMed] [Google Scholar]
- 4.Schirripa M, Cremolini C, Loupakis F, Morvillo M, Bergamo F, Zoratto F, et al. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. International journal of cancer. 2015;136(1):83–90. doi: 10.1002/ijc.28955. [DOI] [PubMed] [Google Scholar]
- 5.Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nature reviews Cancer. 2017;17(4):268. doi: 10.1038/nrc.2017.24. [DOI] [PubMed] [Google Scholar]
- 6.Dienstmann R, Salazar R, Tabernero J. Overcoming Resistance to Anti-EGFR Therapy in Colorectal Cancer. American Society of Clinical Oncology educational book / ASCO American Society of Clinical Oncology Meeting. 2015;35:e149–56. doi: 10.14694/EdBook_AM.2015.35.e149. [DOI] [PubMed] [Google Scholar]
- 7.Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. The Lancet Oncology. 2016;17(6):738–46. doi: 10.1016/s1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 8.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25(22):3230–7. doi: 10.1200/jco.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 9.Khan KH, Cunningham D, Werner B, Vlachogiannis G, Spiteri I, Heide T, et al. Longitudinal Liquid Biopsy and Mathematical Modeling of Clonal Evolution Forecast Time to Treatment Failure in the PROSPECT-C Phase II Colorectal Cancer Clinical Trial. Cancer discovery. 2018;8(10):1270–85. doi: 10.1158/2159-8290.Cd-17-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evers B, Jastrzebski K, Heijmans JP, Grernrum W, Beijersbergen RL, Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nature biotechnology. 2016;34(6):631–3. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 11.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science (New York, NY) 2014;343(6166):84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature biotechnology. 2016;34(2):184–91. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piccioni F, Younger ST, Root DE. Pooled Lentiviral-Delivery Genetic Screens. Curr Protoc Mol Biol. 2018;121:32 1 1–1 21. doi: 10.1002/cpmb.52. [DOI] [PubMed] [Google Scholar]
- 14.Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, et al. Gene essentiality and synthetic lethality in haploid human cells. Science (New York, NY) 2015;350(6264):1092–6. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 15.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163(6):1515–26. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. Identification and characterization of essential genes in the human genome. Science (New York, NY) 2015;350(6264):1096–101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu GF, Lin B, Tanaka K, Dunn D, Wood D, Gesteland R, et al. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990;63(4):835–41. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- 18.Bliss CI. THE TOXICITY OF POISONS APPLIED JOINTLY1. Annals of Applied Biology. 1939;26(3):585–615. doi: 10.1111/j.1744-7348.1939.tb06990.x. [DOI] [Google Scholar]
- 19.Roumeliotis TI, Williams SP, Goncalves E, Alsinet C, Del Castillo Velasco-Herrera M, Aben N, et al. Genomic Determinants of Protein Abundance Variation in Colorectal Cancer Cells. Cell reports. 2017;20(9):2201–14. doi: 10.1016/j.celrep.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nature communications. 2015;6:7002. doi: 10.1038/ncomms8002. [DOI] [PubMed] [Google Scholar]
- 21.Mei Z, Shao YW, Lin P, Cai X, Wang B, Ding Y, et al. SMAD4 and NF1 mutations as potential biomarkers for poor prognosis to cetuximab-based therapy in Chinese metastatic colorectal cancer patients. BMC cancer. 2018;18(1):479. doi: 10.1186/s12885-018-4298-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woolston A, Khan K, Spain G, Barber LJ, Griffiths B, Gonzalez-Exposito R, et al. Genomic and Transcriptomic Determinants of Therapy Resistance and Immune Landscape Evolution during Anti-EGFR Treatment in Colorectal Cancer. Cancer cell. 2019;36(1):35–50.:e9. doi: 10.1016/j.ccell.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Post JB, Hami N, Mertens AEE, Elfrink S, Bos JL, Snippert HJG. CRISPR-induced RASGAP deficiencies in colorectal cancer organoids reveal that only loss of NF1 promotes resistance to EGFR inhibition. Oncotarget. 2019;10(14):1440–57. doi: 10.18632/oncotarget.26677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bruin EC, Cowell C, Warne PH, Jiang M, Saunders RE, Melnick MA, et al. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer discovery. 2014;4(5):606–19. doi: 10.1158/2159-8290.Cd-13-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philpott C, Tovell H, Frayling IM, Cooper DN, Upadhyaya M. The NF1 somatic mutational landscape in sporadic human cancers. Human genomics. 2017;11(1):13. doi: 10.1186/s40246-017-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misale S, Arena S, Lamba S, Siravegna G, Lallo A, Hobor S, et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Science translational medicine. 2014;6(224):224ra26. doi: 10.1126/scitranslmed.3007947. [DOI] [PubMed] [Google Scholar]
- 27.Troiani T, Napolitano S, Vitagliano D, Morgillo F, Capasso A, Sforza V, et al. Primary and acquired resistance of colorectal cancer cells to anti-EGFR antibodies converge on MEK/ERK pathway activation and can be overcome by combined MEK/EGFR inhibition. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(14):3775–86. doi: 10.1158/1078-0432.Ccr-13-2181. [DOI] [PubMed] [Google Scholar]
- 28.Vitiello PP, Cardone C, Martini G, Ciardiello D, Belli V, Matrone N, et al. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. Journal of experimental & clinical cancer research : CR. 2019;38(1):41. doi: 10.1186/s13046-019-1035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. The Journal of clinical investigation. 2011;121(11):4311–21. doi: 10.1172/jci57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spoerke JM, O’Brien C, Huw L, Koeppen H, Fridlyand J, Brachmann RK, et al. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(24):6771–83. doi: 10.1158/1078-0432.CCR-12-2347. [DOI] [PubMed] [Google Scholar]
- 31.Misale S, Bozic I, Tong J, Peraza-Penton A, Lallo A, Baldi F, et al. Vertical suppression of the EGFR pathway prevents onset of resistance in colorectal cancers. Nature communications. 2015;6:8305. doi: 10.1038/ncomms9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupo B, Gagliardi P, Sassi F, Zanella ER, Cottino F, Bertotti A, et al. Abstract LB-293: Targeting the PI3K pathway to intercept cetuximab tolerance in metastatic colorectal cancer. Cancer research. 2017;77(13 Supplement):LB-293-LB. doi: 10.1158/1538-7445.Am2017-lb-293. [DOI] [Google Scholar]
- 33.D’Amato V, Rosa R, D’Amato C, Formisano L, Marciano R, Nappi L, et al. The dual PI3K/mTOR inhibitor PKI-587 enhances sensitivity to cetuximab in EGFR-resistant human head and neck cancer models. British journal of cancer. 2014;110(12):2887–95. doi: 10.1038/bjc.2014.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horn T, Ferretti S, Ebel N, Tam A, Ho S, Harbinski F, et al. High-Order Drug Combinations Are Required to Effectively Kill Colorectal Cancer Cells. Cancer research. 2016;76(23):6950–63. doi: 10.1158/0008-5472.Can-15-3425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.