Abstract
Purpose:
Existing cell-free DNA (cfDNA) methods lack the sensitivity needed for detecting minimal residual disease (MRD) following therapy. We developed a test for tracking hundreds of patient-specific mutations to detect MRD with a 1000-fold lower error rate than conventional sequencing.
Experimental Design:
We compared the sensitivity of our approach to digital droplet PCR (ddPCR) in a dilution series, then retrospectively identified two cohorts of patients who had undergone prospective plasma sampling and clinical data collection: 16 patients with ER+/HER2− metastatic breast cancer (MBC) sampled within six months following metastatic diagnosis and 142 patients with stage 0-III breast cancer who received curative-intent treatment with most sampled at surgery and one year post-op. We performed whole-exome sequencing of tumors and designed individualized MRD tests, which we applied to serial cfDNA samples.
Results:
Our approach was 100-fold more sensitive than ddPCR when tracking 488 mutations, but most patients had fewer identifiable tumor mutations to track in cfDNA (median 57, range 2–346). Clinical sensitivity was 81% (n=13/16) in newly diagnosed MBC, 23% (n=7/30) at post-op and 19% (n=6/32) at one year in early-stage disease, and highest in patients with the most tumor mutations available to track. MRD detection at one year was strongly associated with distant recurrence (HR=20.8 [95%CI: 7.3–58.9]). Median lead time from first positive sample to recurrence was 18.9 months (range: 3.4–39.2 months).
Conclusions:
Tracking large numbers of individualized tumor mutations in cfDNA can improve MRD detection, but its sensitivity is driven by the number of tumor mutations available to track.
Keywords: minimal residual disease, breast cancer, circulating tumor DNA (ctDNA), molecular profiling, liquid biopsy
INTRODUCTION
Each year, more than 600,000 people worldwide die of breast cancer, largely due to metastatic recurrence. Although these recurrences can occur early, the majority in patients with hormone receptor-positive breast cancer occur more than 5 years after initial diagnosis, with risk extending for decades.1,2 Systemic recurrence likely arises from micrometastatic disease present at initial diagnosis, but undetectable by imaging or conventional blood tests. Once overt metastatic breast cancer develops, it is generally incurable. Appropriate adjuvant systemic therapy can substantially reduce the risk of cancer recurrence3,4; however, current clinical tools are imperfect in identifying which patients require adjuvant systemic therapy and assessing in real-time which therapies are achieving their intended therapeutic effects. The current American Society of Clinical Oncology guidelines recommend 10 years of adjuvant endocrine therapy for women presenting with stage II or III, estrogen receptor-positive (ER+) breast cancer, based upon randomized trial data demonstrating, on average, an absolute improvement in distant disease-free survival of 1.4–1.9% compared to 5 years of endocrine therapy.5–7 More sensitive methods to detect micrometastatic disease may allow therapy to be escalated in a subset of higher risk patients, while sparing other patients potentially adverse effects of unnecessary treatments.
Recent studies have shown that liquid biopsies can detect minimal residual disease (MRD) in multiple cancer types8–12 including breast,13–15 by tracking tumor mutations in circulating cell-free DNA (cfDNA). However, in these previous studies, clinical sensitivity has been limited at the early post-operative time points, at which treatment decisions are typically made, and the lead time prior to clinical presentation of overt metastatic disease has been relatively short. There is a need for more sensitive liquid biopsies, with greater dynamic range, to identify patients with MRD sooner, with sufficient lead time prior to overt metastatic disease such that additional curative-intent therapy could be effective.
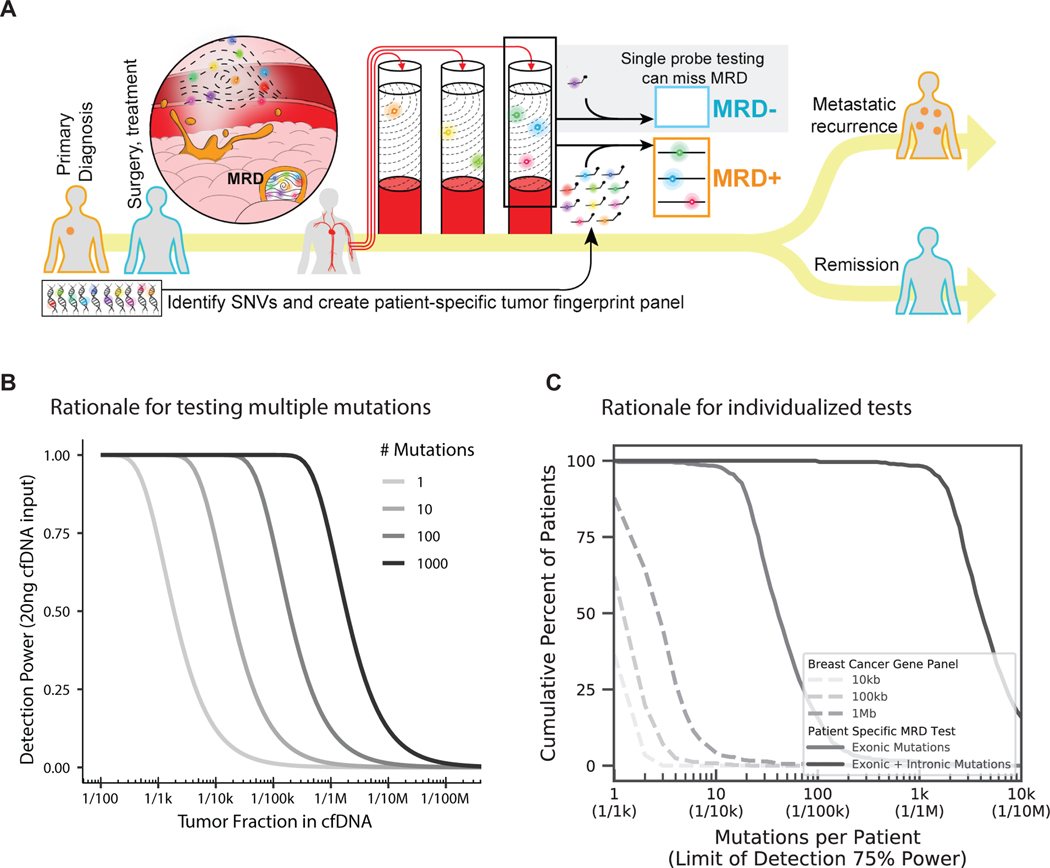
One major challenge in detecting MRD from peripheral blood is that a typical blood draw samples only a few thousand copies of each gene.16 This means that existing methods, which track one or few mutations, will be unable to detect MRD when the fraction of cancerous cfDNA in the bloodstream is lower than twice the inverse of the number of copies of each gene in a given sample, which we call the genomic equivalent (GE) limit (Fig. 1A). Collecting more blood is not always feasible; however, tracking many mutations per patient may increase the likelihood that cfDNA fragments containing the desired targeted mutations are captured when tumor fraction (i.e. the proportion of cfDNA derived from tumor rather than non-cancerous tissues)17–19 in blood is lower than the GE limit (Figs. 1A,B).20
Figure 1.
A) Overview of MRD testing including why tracking many mutations is important, particularly when there is a low fraction of cancerous cfDNA in the bloodstream, B) Theoretical detection power for varied numbers of mutations tracked per patient, C) Cumulative fraction of patients harboring varied numbers of mutations (and in turn, the predicted detection limits based on Fig. 1B) using breast cancer gene sequencing panels versus individualized MRD tests. See methods.
One practical limitation for tracking more mutations is that each patient’s cancer is unique and harbors few mutations in common with other patients.16 Either a substantial number of digital droplet PCR (ddPCR) assays or deep sequencing of very large gene panels would be required to afford broad patient coverage, and even then, would only cover a limited number of mutations per patient (Fig. 1C, Supplementary Fig. 1). Individualized assays are therefore needed to track many tumor mutations in most patients, and groups are beginning to do this, but existing methods lack sufficient specificity10,21. We estimate that error rates need to be 100-fold lower (e.g. ~1 × 10−6) to reliably track hundreds of individualized mutations, because this involves scanning hundreds of thousands to millions of bases for single mutations.
Beyond limiting the false detection of MRD, we reason that it will be important to calibrate differences in detection power among individualized MRD tests, particularly when the test result is negative for MRD. Existing liquid biopsy assays do not compute detection power, which obscures whether the test was powered to detect MRD in a patient if the tumor fraction in cfDNA is low. As multiple biological and technical factors affect detection power from a liquid biopsy, we hypothesized that creating a single metric for detection power, which encompasses the potential variation, would help to better understand the significance of a negative MRD test.
In this study, we established that tracking hundreds of patient-specific tumor mutations enables reliable detection of MRD at tumor fractions significantly lower than the GE limit. We also developed a power calculation for each individual MRD test. We hypothesized that tracking multiple mutations would enable earlier detection of metastatic disease. In the first part of our study, we applied our newly developed assay to patients with estrogen receptor-positive (ER+) breast cancer within six months following metastatic diagnosis. The primary objective was to determine the sensitivity of the assay in patients at time of metastatic recurrence. In the second part of our study, we applied the assay to track mutation fingerprints in plasma and identified breast cancer recurrence following surgery and systemic therapy. The primary objectives were to determine the clinical sensitivity at post-op and one year following surgery (which reflect key points in patient care at which treatment decisions are made) and the lead interval between detection of ctDNA in plasma and clinical detection of overt metastatic disease.
METHODS
Patients and Samples
All patients provided written informed consent to allow the collection of blood and/or tumor tissue and analysis of clinical and genetic data for research purposes. Patients with MBC were prospectively identified for enrollment into tissue analysis and banking cohorts (Dana-Farber Cancer Institute [DFCI] protocol identifiers 05–246, 09–204). From within this cohort we retrospectively identified patients with ER-positive, HER2-negative MBC with plasma isolated from 20cc blood in Streck tubes and tissue sampling within six months of diagnosis. In the curative-intent cohort, we included patients participating in two clinical studies (DFCI IRB-approved protocols 05–055 and 06–169) given receipt of curative-intent treatment and available biospecimens (Supplementary Figure 2B). Plasma was derived from 10–20cc whole blood in EDTA tubes. From protocol 06–169, we included the first 99 patients enrolled with breast cancer diagnosis at age <35. From 05–055, all study patients were included, if tissue and plasma samples were available. At the time of analysis and data cut off, all patients had completed adjuvant local and systemic therapy aside from hormonal therapy, and median follow up had reached 7.1 years. Fresh whole blood (10–20 cc) from appropriately consented healthy donors was obtained through Research Blood Components and Boston Biosciences. HapMap cells, NA12878 and NA19238 were purchased from Coriell. This research was conducted in accordance with the provisions of the Declaration of Helsinki, and the U.S. Common Rule.
Sample Processing
Collection and processing of blood samples followed the same protocol as previously described.17 Germline DNA (gDNA) was extracted from buffy coat or whole blood and sheared using a Covaris Ultrasonicator. cfDNA and gDNA libraries were constructed using duplex UMI adapters (IDT) with up to 20ng of DNA input. Hybrid selection (HS) was performed using patient specific probe panels (Twist Biosciences and IDT, Supplementary Methods).
Sequencing and Data Analysis
Samples were sequenced on either a HiSeq 4000 or HiSeq X (Illumina). Exome sequencing was used to identify patient-specific single nucleotide variants (SNVs) as previously described.17 Patient specific SNVs were used to design custom MRD tests, which were subsequently applied to cfDNA and gDNA libraries. MRD detection, including detection limit and tumor fraction, was determined using a custom pipeline (Supplementary Methods).
Statistical Analysis
This study was designed to assess primarily the association between MRD status and distant disease recurrence and secondarily the time between MRD detection and clinical metastasis. We determined to identify at least 120 patients to give 80% power to detect a hazard ratio of 3 for distant recurrence-free survival (DRFS). The associations between MRD overall status and demographic/clinical factors were evaluated via Chi-square test for categorical variables and Wilcoxon rank-sum test for continuous variables. Both the univariate and multivariate analysis (MVA) of DRFS were conducted using the Cox proportional hazard model. MRD status was measured at 3 time points post-operatively. To cope with the time-variant nature of MRD status (a patient’s MRD status may vary across time), univariate analysis of DRFS was done in the frame of landmark analysis at post-operative and year 1 blood sample draw. Patients who had distant recurrence prior to the landmark time point, post-operation or year 1, were removed from the analysis. Multivariate analysis of DRFS was conducted using MRD status as a time-dependent covariate. A patient was coded as in the MRD-negative group until the patient tested positive at one time point and then was switched to the positive group. MRD status, stage, grade, and HR status were included in the MVA. All analyses were conducted using SAS 9.2. Kaplan Meier curves were made using the Survival package of R (version R2.15).
RESULTS
Establishment of a novel, scalable assay for detection of circulating tumor DNA
We have developed an ultrasensitive blood test for MRD. Our approach involved tracking hundreds of individualized tumor mutations, identified from a patient’s tumor tissue, in the cfDNA from a blood draw. We applied whole-exome sequencing to define up to several hundred mutations from each patient’s tumor. To limit potential errors, we employed strict criteria to select somatic single nucleotide variants (SNVs) to track using duplex sequencing22 in cfDNA. We further required detection of ≥ 2 mutations for a cfDNA sample to be called MRD-positive and excluded any mutations also found in a patient’s own germline DNA (Supplementary Methods, Supplementary Fig. 3).
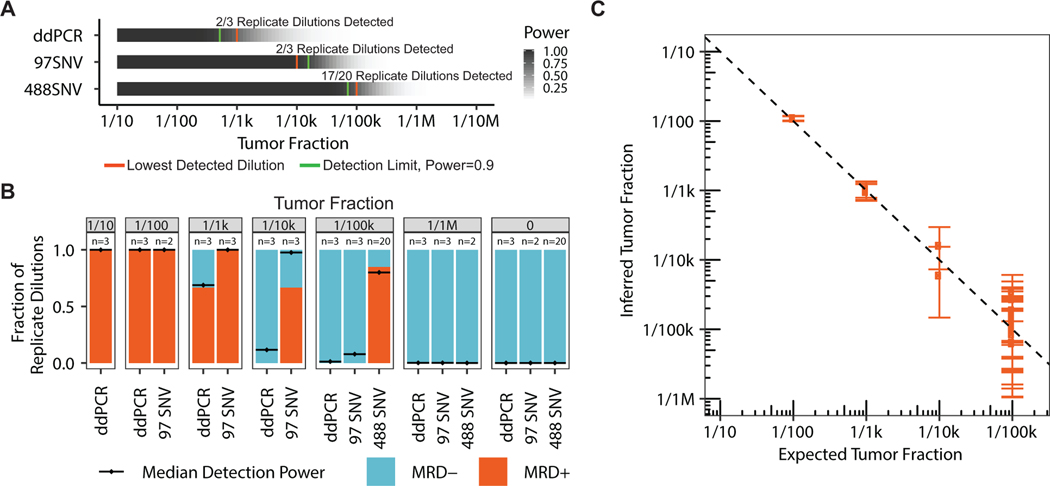
We first investigated whether tracking many mutations enabled detection of MRD at tumor fractions lower than the GE limit. To model blood draws from cancer patients, we created large volume dilutions of sheared genomic DNA from two well-characterized cancer cell lines and obtained 20 ng DNA samples comprising ~6k GEs. We applied a single mutation ddPCR test and observed detection down to tumor fraction 1/1k as expected for a GE limit of 1/3k (Fig. 2A, Supplementary Fig. 4). Whereas, MRD testing using a fingerprint comprised of 97 and 488 mutations exhibited reliable detection down to tumor fraction 1/10k and 1/100k, respectively, with all lower dilutions and negative controls being MRD-negative. The proportion of MRD-positive samples at each dilution aligned with the expected MRD-positive samples based on calculated detection power. We computed the median detection power for the 1/100k samples with 488 mutation fingerprints to be 82% and found 17/20 (85%) to be MRD-positive (Fig. 2A–B, Supplementary Fig. 5). Observed tumor fractions also correlated with expected values (r2=0.94, Fig. 2C, Supplementary Fig. 6, see Methods), including for another dilution series of cancer patient and healthy donor cfDNA (Supplementary Fig. 7), and further testing confirmed high specificity in healthy donors (Supplementary Fig. 8). These results demonstrate that tracking of many mutations enables reliable detection of MRD at tumor fractions that are up to 100-fold lower than the GE limit.
Figure 2.
Serial dilution benchmarking of our MRD test for 100 and 500 mutations, versus digital droplet PCR for a single mutation, using 20ng admixtures of sheared genomic DNA from two well-characterized cell lines. A) Detection power for a range of tumor fractions using our MRD assay and an equivalent ddPCR single mutation assay. B) Fraction of replicate dilutions detected by our MRD assay and an equivalent ddPCR assay as well as the median detection power for those samples. C) Tumor fraction estimation benchmarking for several replicates across a range of tumor fractions.
We then evaluated the reproducibility and cost effectiveness of patient-specific assays by examining the performance of 142 patient-specific panels applied to 370 samples. We found patient-specific assays with duplex sequencing technology provide high efficiency (87.4% +/− 15.5% bases on bait) and uniformity (coefficient of variation [CV]=0.23 +/− 0.12) in target capture; exhibit error rates, which are on average 1000-fold lower than conventional sequencing (1.3E-06 +/− 2.3E-06); detect the same DNA duplexes in replicate capture (92% shared) and sequencing (89% shared); and saturate recovery of DNA duplexes in many samples with a median of 1.46M reads per mutation site (Supplementary Figs. 9, 10). Of note, we found our MRD test results to be reproducible in benchmarking data sets (Fig. 2, Supplementary Fig. 7, Supplementary Fig. 10) but did not perform repeated testing of patient samples in our study. Considering these parameters and the mutational diversity in breast cancer, de novo assays are feasible for most patients and require less sequencing than predefined panels of recurrently mutated genes in breast cancer (due to less genomic territory targeted) to achieve higher detection power for MRD (Fig. 1C).
MRD detection in newly diagnosed, hormone receptor-positive MBC
Given our intent to detect micrometastatic disease, we first explored the clinical sensitivity of our MRD assay in patients with radiologically-overt newly diagnosed MBC. We focused on patients with hormone receptor-positive (HR+) and human epidermal growth factor receptor 2-negative (HER2-) breast cancer, in which recurrences may occur years to decades after initial treatment. We reasoned that understanding the detectability of MRD and corresponding tumor fraction in cfDNA would help to establish upper bounds for detection of MRD at earlier stages in care.
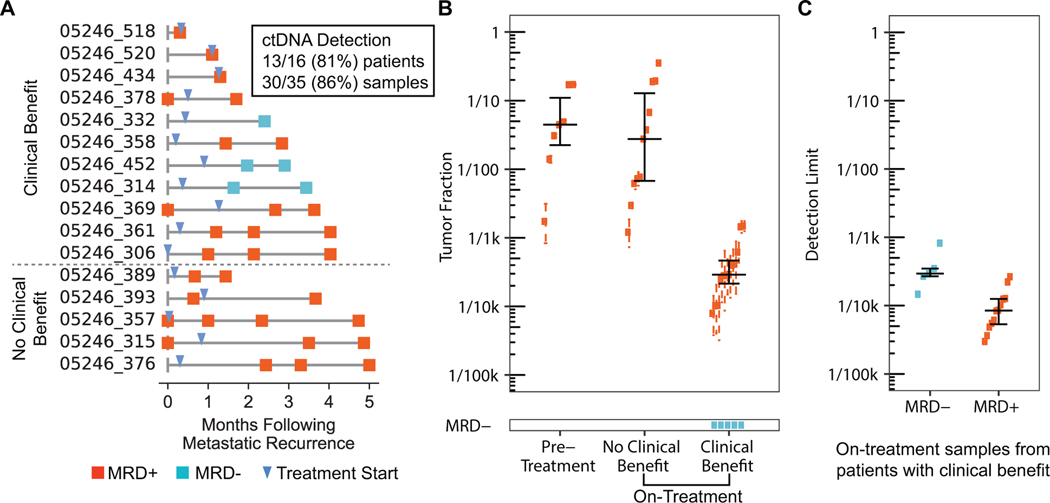
We applied our assay to 35 samples (range 1–4 per patient) from 16 patients with newly diagnosed, HR+ MBC, who had undergone tumor biopsy and cfDNA blood draw within six months following diagnosis (Supplementary Fig. 11A). Of note, we used patients’ metastatic tumor biopsies to define mutation fingerprints, tracking a median of 51.5 SNVs (range 6 – 120). Overall, 30/35 (86%) samples from 13/16 (81%) patients had detectable cancer cfDNA (or MRD-positive), including 7/7 (100%) samples taken prior to initiation of therapy, 11/11 (100%) samples taken from patients not benefiting from therapy, and 12/17 (69%) samples taken on therapy from patients experiencing clinical benefit, defined as continuation of treatment for at least 6 months from time of initiation (Fig. 3A–B, Supplementary Fig. 11B). Application of the same MRD tests to cfDNA from healthy donors yielded MRD-negative results in all cases (Supplementary Fig. 11A). Tumor fractions were highest when patients were sampled prior to therapy (median 0.044, range 0.002–0.17) and remained high when patients were on ineffective therapy (median 0.028, range 0.0012–0.35, Supplementary Fig. 11B). By contrast, tumor fractions were lower in patients sampled when on effective therapy (median 0.00025, range 0–0.0015). Notably, we observed significant declines in tumor fraction when patients were sampled prior to, and then again while on effective therapy (Supplementary Fig. 11C) as has previously been seen in metastatic prostate cancer.2
Figure 3.
MRD testing in patients with newly diagnosed, HR+ MBC. A) Patient timelines with samples collected less than six months after metastatic recurrence. B) Number of MRD-positive and MRD-negative samples including corresponding tumor fractions. Many patients had multiple blood draws, both pre-treatment and on-treatment. C) Comparison of detection limit for MRD− vs MRD+ samples from patients on treatment and experiencing clinical benefit.
We also investigated whether we could have tracked fewer mutations and found that MRD could have been detected using fewer mutations in the cfDNA samples which had the highest tumor fractions: those which were taken prior to therapy or on ineffective therapy. However, tracking fewer mutations would have failed to detect MRD in many of the cfDNA samples which had the lowest tumor fractions: those which were collected on effective therapy (Supplementary Fig. 11D). This suggests that our large fingerprint approach is most useful when seeking to detect trace levels of ctDNA, at tumor fractions below the GE limit.
Our assay failed to detect MRD in 5 samples from 3 patients with known metastatic disease. As these samples were taken while on effective therapy, when tumor fraction is expected to be low, we explored whether limited detection power could explain these results. We computed the tumor fraction at which detection power is 90% (which we call the detection limit) for all samples from patients experiencing clinical benefit (Fig. 3B, Methods). We found the 5 false-negative samples to exhibit a median detection limit of 0.00029 (range 0.00014–0.00082), which is 3-fold higher – i.e. worse – than the 12 MRD-positive samples from the rest of patients experiencing clinical benefit (median 0.0000848, range 0.0000299–0.000265). We cannot rule out the possibility that these samples actually had no tumor DNA in them, but our calculated detection limits suggest that it would have been less likely to detect MRD below ~1/1k in these 5 samples. In all, our results indicate that detection limits should be in the range of 1/10k or better to detect MRD prior to clinical metastatic recurrence.
MRD detection after definitive treatment for breast cancer
To determine the predictive power of MRD testing and associated lead time to recurrence in patients treated for early-stage breast cancer, we identified a patient cohort representative of the major breast cancer subtypes (HR+/HER2−, HER2+, and HR−/HER2− [triple-negative breast cancer, TNBC]), with appropriately long-term follow up given the potential for late recurrence (Supplementary Table 1). Our cohort included 142 patients who had been treated for stage 0-III breast cancer, had postoperative blood and plasma samples available, and were monitored for distant recurrences for up to 13 years (Supplementary Fig. 2A–B). All patients had biopsy-proven breast cancer: 86 (61%) with HR+/HER2− disease, 31 (22%) with TNBC, and 25 (18%) with HER2+ disease. Overall, 130 (92%) received either neoadjuvant or adjuvant chemotherapy, and 108 (76%) received adjuvant endocrine therapy (Supplementary Table 1); 104 (73%) received adjuvant radiation treatment. In terms of stage, 3 (2%) patients presented with stage 0 disease, 32 (23%) with stage I, 68 (48%) with stage II, and 39 (27%) with stage III breast cancer at diagnosis. All patients were treated with curative intent surgery. Thirty-seven (26%) patients (24 [65%] ER+/HER2−, 7 [19%] HER2+, 6 [16%] TNBC) experienced distant recurrence, at a median follow up across the cohort from surgery to time of death or last contact of 7.1 years. Median time to distant recurrence was 32 months for those patients with recurrent disease.
From these 142 patients’ primary tumors, we selected a median of 53 SNVs (range 2 – 346) to track. We identified 271 postoperative plasma samples from these patients, collected at a median of 3.53 months (range 0.23–8.43) after surgery and at approximately year 1 (median 14.2 months, range 6.77–21.7) and year 4 (median 54.97 months, range 42.43–71.27) from surgery (Supplementary Table 2). Most patients had a postoperative (111, 78%) or year 1 (122, 86%) sample available but far fewer had a year 4 (38, 27%) sample. Forty-seven (33%) and 20 (14%) patients with postoperative and year 1 samples, respectively, had their samples collected while on active chemotherapy. Of the 37 patients who experienced distant recurrence, 30 and 32 had a postoperative and year 1 sample available, respectively, but only 3 patients had a year 4 sample. Median plasma volume was 3.4 mL (range 0.4–5.3), and a median of 19.9 ng cfDNA (range 2.6–21.6) was allocated for MRD testing. We then created individualized assays targeting a median of 57 mutations (range 2–346) per patient, identified via whole-exome sequencing of primary tumor tissue and germline DNA from whole blood. Of note, the number of identifiable tumor mutations to track in each patient’s cfDNA was much lower than we had used in our benchmarking (n=488 mutations) to achieve 1/100k detection limit from 20ng DNA (Fig 2A, Supplementary Fig. 2C). Nonetheless, we applied the assays to all plasma, and germline DNA samples when available.
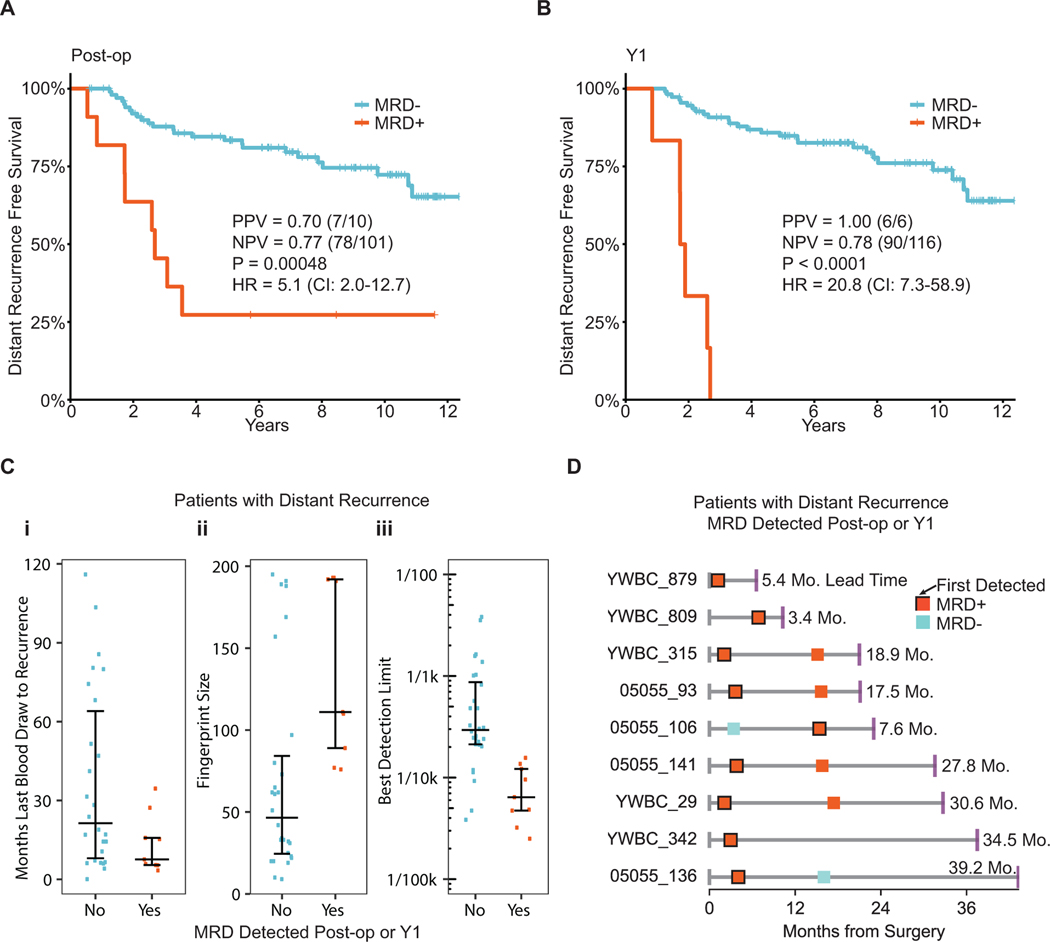
We first examined the relationship between postoperative MRD status and distant recurrence via a post-operative landmark analysis, and found that detection of MRD soon after curative surgery was highly predictive of distant relapse (hazard ratio=5.1 [95% confidence interval, CI: 2.0–12.7], Fig. 4A, Supplementary Fig. 12). However, we also hypothesized that because adjuvant systemic therapy reduces the risk of breast cancer recurrence,3 we would expect to detect MRD in some patients in whom cancer did not recur due to effective adjuvant treatment. Indeed, 3 patients had a single, postoperative MRD-positive sample, but remained recurrence free at last follow up. Two of these patients were undergoing adjuvant therapy at the time of blood sampling. The third patient sample was drawn 7 days after surgery. In all 3 of these patients, subsequent plasma samples were MRD-negative (Supplementary Fig. 13). Based on the post-op blood test, the positive predictive value (PPV) for distant recurrence was 0.70 (n=7/10) and clinical specificity was 0.96 (n=78/81). The negative predictive value (NPV) was 0.77 (n=78/101) and clinical sensitivity was 0.23 (n=7/30). Of note, MRD was not detected in the 6 patients who experienced local-only recurrence (Supplementary Fig. 14).
Figure 4.
MRD testing in early stage breast cancer. A-B) Kaplan-Meier curves showing distant recurrence-free survival for patients treated for early stage breast cancer and in whom MRD was detected or not at the post-op and year 1 (Y1) time points. Post-op and Y1 blood samples were drawn at a median of 3.6 months (0.23 – 8.43) and 14.3 months (6.77 – 21.70) after surgery respectively. C) Comparison of patients with distant recurrence in which MRD was detected or not at the post-op or Y1 time points. D) Timelines for patients with distant recurrence that were detected at post-op or Y1 time points. End points represent distant recurrence.
Reasoning that MRD status after completion of all local therapy and chemotherapy might better predict for distant recurrence, we conducted a landmark analysis at one year following surgery and found all patients (6/6) with detectable MRD experienced recurrence (hazard ratio=20.8 [95% CI: 7.3–58.9]) at a median of 21.7 months (range 10.1–32.2) (Fig. 4B, Supplementary Fig. 12). Based on the year 1 sample, PPV for distant recurrence was 1.00 (n=6/6) and clinical specificity was 1.0 (n=90/90). NPV was 0.78 (n=90/116) and clinical sensitivity was 0.19 (n=6/32). We then evaluated the subset of patients with available year 4 samples (38, 27%) to assess for MRD status and risk of late recurrence. Of the 38 patients who underwent year 4 sampling, only 3 went on to experience recurrence. We detected MRD in one of three patients with recurrence 18.9 months prior to clinical metastatic diagnosis (the other two samples were underpowered with poor detection limits of 0.0015 and 0.0023), and in no patients without breast cancer recurrence. Finally, in a multivariate model, MRD remained highly statistically significant for risk of distant recurrence (Supplementary Table 2), independent of stage, subtype, and age at diagnosis.
We next explored potential factors which could have affected our clinical sensitivity for MRD detection across the cohort. We first reasoned that duration of time between MRD test and time of recurrence could impact MRD detection, as patients in our study experienced recurrence up to 10 years later. Indeed, the median interval from last blood test to recurrence was 11.5 months for patients in whom we detected MRD, versus 23.8 months for those in whom we failed to detect MRD (Fig. 4C–i).
Given that the technical sensitivity of our approach was driven by number of mutations tracked (Fig 2A, Supplementary Fig. 15A), and that most patients in our study had limited mutations identified from whole-exome sequencing of their tumor biopsies, we also explored the association between number of mutations tracked and clinical sensitivity. Expectedly, clinical sensitivity was highest among patients in whom we tracked the most mutations (Fig 4C–ii) and whose MRD tests yielded the best detection limits (Fig 4C–iii). Reassuringly, tracking more mutations was not associated with increased MRD detection in the 105 patients who did not experience distant recurrence (Supplementary Fig. 15B). We were also more likely to detect mutations in the cfDNA that had higher allele fractions and cancer cell fractions (CCF) in the tumor (Supplementary Fig. 16A–B). Had we tracked fewer mutations per patient, we would have failed to detect MRD in 36% (n=5/14) of samples (Supplementary Fig. 15C). However, tumor fractions in cfDNA of patients who experienced recurrence (median 0.00042, range 0.000056 – 0.041) were close to the detection limits, suggesting that we may need to track even more mutations per patient in future studies.
Interestingly, among the patients with most mutations tracked, we observed a long median lead time to clinical recurrence of 18.9 months (range 3.4 – 39.2, Fig 4D, Supplementary Table 3). Such lead times could enable early intervention, long before patients develop metastatic recurrence.
DISCUSSION
In this study we present a novel approach to MRD tracking, based on the premise that tracking many, rather than one or a handful of mutations, inherently increases assay sensitivity, and assay personalization maximizes sensitivity in a disease without multiple recurrent mutations. We applied the assay to a group of breast cancer patients treated with curative intent, and detected MRD in many patients prior to presentation of metastatic disease.
The concept of tracking more mutations to improve sensitivity is not new, but existing methods have had insufficient specificity to track many individualized mutations. To our knowledge, this is the first study to track hundreds of personalized mutations simultaneously in patients with cancer, using an approach with sufficient analytical specificity (~1×10−6). Our approach enables reliable detection well below the GE limit, as compared to following a single mutation via ddPCR. Tracking MRD from blood in cancer patients is challenging from a diagnostic perspective, as cancer recurrence happens in the future, and no current standard metric exists against which to measure a true positive or negative test. Here, we quantify two key aspects: the detection limit, and tumor fraction. Detection limit determination allows for a clear understanding of whether a specific test is more likely negative because of assay performance or as a result of a lack of MRD.
We followed patients for up to 13 years from definitive surgery. This is specifically important in the context of HR+/HER2− breast cancer, where recurrence may occur years to decades from initial treatment. Interestingly, our assay had a median lead time (the time from a positive test to metastatic diagnosis) of 18.9 months, and up to 39 months, without compromising clinical specificity. This is significantly longer than what has been seen in prior studies such as Garcia-Murillas et al (median 10.7 months) and Coombes et al (median 8.9 months) which tracked 1 – 16 mutations per patient, but is based on a small number of patients and could be a function of longer follow up.13,14 If additional prospective evaluation confirms a lead time of this magnitude, our assay could offer an opportunity to detect and treat MRD long before the development of overt metastatic disease, with the goal of offering curative treatment to patients for whom it is not currently an option. Testing whether treatment of MRD can, in fact, be curative will be an important next step.
One limitation of our study was related to the infrequent sampling of blood relative to the duration of clinical follow up. Whereas prior studies have collected blood samples every 6 months (or more frequently) and reported high rates of MRD detection based on any of many time points tested over a multi-year period being positive13,14, we sampled patients’ blood relatively infrequently and focused MRD testing on key time points at which treatment decisions are typically made. For instance, most patients in our study had a single post-op and year one blood sample following completion of all local therapy and adjuvant chemotherapy, and only 3 patients who experienced recurrence had a year 4 time point. When restricted to the first post-op time point, our clinical sensitivity (23%, n=7/30) was similar to Garcia-Murillas et al 2015 (19%, n=7/37).14 Garcia-Murillas et al 2019 did not compute clinical sensitivity at post-op but stated that most patients who experienced recurrence did not have an initial MRD+ test at post-op.15 Meanwhile, Coombes et al reported higher sensitivity in the first post-op sample tested per patient but this was later for most patients, up to 3 years after completion of adjuvant chemotherapy.13 Comparison of clinical sensitivity and lead time among studies is challenging, however, due to numerous potential confounders such as differences in patient populations, treatments, timing of sampling, and duration of follow-up.
Another limitation of our study was that most patients had too few mutations identified via whole-exome sequencing of their tumor biopsies, to fully leverage the large fingerprint approach in cfDNA. Whereas our benchmarking had shown that tracking 488 mutations achieved reliable detection of 1/100k dilution from 20ng DNA (Fig 2), in contrast to 1/1k for ddPCR, our detection limits for most MRD tests were constrained to 1/1k – 1/10k due to insufficient numbers of mutations having been identified from patients’ tumor biopsies (Fig 3C, 4C–iii). Indeed, clinical sensitivity was highest in patients with most mutations tracked (Fig 4C–ii). We reason that future efforts will require whole-genome sequencing (instead of whole-exome sequencing) to identify more mutations to track in all patients.
Other limitations of our study were that (i) most patients in our cohort had HR+/HER2− disease, the most common breast cancer subtype, which meant we were unable to draw meaningful conclusions about the variations of MRD detection by subtype, (ii) for some patients, the last blood sample was taken nearly a decade before metastatic recurrence, (iii) volumes of plasma processed were small, meaning the number of GEs sampled was lower, and (iv) blood samples were collected and processed for plasma many years ago, before cfDNA-specific protocols existed, and variable processing may have led to cfDNA degradation.
Breast cancer is a uniquely challenging disease. Widespread use of screening and effective systemic therapies have led to improved rates of cure. However, most breast cancer-related deaths are due to recurrence of breast cancers that arise from microscopic residual disease. Current clinico-pathologic risk assessments for patients with breast cancer are imperfect and result in both undertreatment of patients in whom MRD exists but is not detected, and overtreatment of patients who may have been cured with more limited therapy. An accurate MRD test could offer clinicians a more precise method for differentiating between patients who need further treatment and those who do not. Furthermore, clinicians treating patients with other solid tumors face similar challenges, and our findings might be generalizable beyond breast cancer.
Of note, modern adjuvant breast cancer clinical trials often enroll thousands of patients, are expensive, and take many years to complete. An accurate MRD test could potentially transform this approach. A validated MRD test could be used to select eligible participants for clinical trials by identifying patients at highest risk of recurrence, in whom the study of new therapies would be appropriate. This approach would have a number of benefits. It would limit toxicity of additional therapies to those truly at risk. Trials could include substantially fewer participants, putting fewer patients at risk and costing much less to complete. MRD clearance could be tested as a surrogate clinical trial endpoint. If validated, clinical trials could be completed in a shorter time. Finally, an MRD test could potentially address overtreatment, though we acknowledge that doing this would require an assay with exceptional sensitivity. Otherwise, an insufficiently sensitive assay could lead to inappropriate treatment de-escalation in patients who remain at risk of recurrence.
In conclusion, we found that tracking a customized fingerprint of up to hundreds of mutations is a promising approach to identifying MRD in breast cancer patients, but leveraging the power of this approach requires identifying enough tumor mutations to track in cfDNA. We anticipate that tracking even more mutations may increase assay sensitivity without compromising specificity, with studies currently underway. We look forward to applying this assay to prospective studies in breast cancer to prove clinical utility of MRD testing.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
Optimizing systemic therapy for patients with early-stage breast cancer remains challenging. An ultrasensitive blood test for minimal residual disease (MRD) could identify survivors who might benefit from additional systemic treatment versus de-escalation. Here we present an ultrasensitive blood test for MRD and evaluate its clinical validity in breast cancer. We show that it is 100-fold more sensitive than ddPCR at limiting dilution when tracking 488 mutations but its sensitivity depends on the number of mutations available to track in cfDNA. We examined MRD detection in 142 patients treated for early-stage breast cancer at post-op and one year after surgery. MRD detection was strongly associated with distant recurrence and provided significant lead time to recurrence. Clinical sensitivity was highest in patients who had most mutations identified from their tumor biopsies to track in cfDNA. Our approach could inform future testing of earlier therapeutic intervention in patients who may otherwise develop metastatic recurrence.
ACKNOWLEDGEMENTS
The authors would like to first and foremost acknowledge the courageous patients and their families for their contributions to this study. The authors acknowledge the Gerstner Family Foundation for its generous support. This study was also supported in part by the NBTII Foundation and The Bridge Project, a partnership between the Koch Institute for Integrative Cancer Research at MIT and the Dana-Farber/Harvard Cancer Center (DF/HCC), The Terri Brodeur Breast Cancer Foundation, the Friends of Dana-Farber, the Cheryl Tessler Solit Foundation, Breast Cancer Research Foundation (to N.U.L.), National Comprehensive Cancer Network/Pfizer Collaborative Grant Program (to N.U.L.), Fashion Footwear Association of New York, National Cancer Institute Specialized Program of Research Excellence in Breast Cancer (NCI P50CA168504). DFCI trial 05-055 was supported by Genentech/Roche. This study used cell line samples from the NINDS Repository; NINDS Repository sample numbers corresponding to the samples used are: NA12878 and NA19238.
Financial support: The authors acknowledge the Gerstner Family Foundation for its generous support. This study was also supported in part by the NBTII Foundation and The Bridge Project, a partnership between the Koch Institute for Integrative Cancer Research at MIT and the Dana-Farber/Harvard Cancer Center (DF/HCC), The Terri Brodeur Breast Cancer Foundation, the Friends of Dana-Farber, the Cheryl Tessler Solit Foundation, Breast Cancer Research Foundation (to N.U.L.), National Comprehensive Cancer Network/Pfizer Collaborative Grant Program (to N.U.L.), Fashion Footwear Association of New York, National Cancer Institute Specialized Program of Research Excellence in Breast Cancer (NCI P50CA168504), and R01CA221874 (to GMM). DFCI trial 05-055 was supported by Genentech/Roche.
Conflict of Interest statement: H.A. Parsons reports advisory role for Foundation Medicine and institutional research support from Puma Biotechnology. D. Rotem reports a consulting role and equity with Celsuis Pharmaceuticals (spouse) and has filed patents (US2018 0051319A1, WO2017136508A1, WO2018183908, WO2018183921) with Dana-Farber Cancer Institute and Broad Institute (spouse). L. Trippa reports consulting for Galera Therapeutics. D. Neuberg reports stock ownership in Madrigal Pharmaceuticals; research support from Pharmacyclics; and consulting (no fee) for Clear Creek Bio. S. Freeman reports a patent application filed with Broad Institute. N.J. Lennon reports advisory roles at Genturi, Inc. and New England Biolabs, Inc.. G. Ha reports a patent application filed with Broad Institute. A.D. Choudhury reports research funding from Bayer. M. Meyerson reports consulting for OrigiMed; royalties from LabCorp; research support and royalties from Bayer; and research support from Ono and Janssen. N.U. Lin reports research funding from Genentech, Seattle Genetics, Pfizer and Merck; and consulting/advisory board roles at Seattle Genetics, Puma and Daichii. E.L. Mayer reports consulting for Eisai, Pfizer, Lilly, Novartis, Roche, and Context Therapeutics; and institutional research support from Pfizer and Myriad. T.R. Golub reports advisor role (paid) at Foundation Medicine, GlaxoSmithKline and Sherlock Biosciences. V. Adalsteinsson reports a patent application filed with Broad Institute and is a member of the scientific advisory board of AGCT GmbH, which was not involved in this study. The remaining authors report no conflicts of interest.
Footnotes
DATA AVAILABILITY STATEMENT
Data have been deposited into dbGaP under accession code phs001977.v1.p1.
The prevalence of mutations in various cancer types was determined using data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
REFERENCES
- 1.Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–1953. [DOI] [PubMed] [Google Scholar]
- 2.Pan H, Gray R, Braybrooke J, et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N Engl J Med. 2017;377(19):1836–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Early Breast Cancer Trialists’ Collaborative G, Peto R, Davies C, et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379(9814):432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Early Breast Cancer Trialists’ Collaborative G, Dowsett M, Forbes JF, et al. Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet. 2015;386(10001):1341–1352. [DOI] [PubMed] [Google Scholar]
- 5.Goss PE, Ingle JN, Pritchard KI, et al. Extending Aromatase-Inhibitor Adjuvant Therapy to 10 Years. N Engl J Med. 2016;375(3):209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamounas EP, Bandos H, Lembersky BC, et al. Abstract S1–05: A randomized, double-blinded, placebo-controlled clinical trial of extended adjuvant endocrine therapy (tx) with letrozole (L) in postmenopausal women with hormone-receptor (+) breast cancer (BC) who have completed previous adjuvant tx with an aromatase inhibitor (AI): Results from NRG Oncology/NSABP B-42. Cancer Res. 2017;77(4_Suppl):S1–05. [Google Scholar]
- 7.Burstein HJ, Lacchetti C, Anderson H, et al. Adjuvant Endocrine Therapy for Women With Hormone Receptor-Positive Breast Cancer: ASCO Clinical Practice Guideline Focused Update. J Clin Oncol. 2019;37(5):423–438. [DOI] [PubMed] [Google Scholar]
- 8.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017;7(12):1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545(7655):446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen E, Birkenkamp-Demtroder K, Sethi H, et al. Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma. J Clin Oncol. 2019;37(18):1547–1557. [DOI] [PubMed] [Google Scholar]
- 12.Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol. 2019;5(8):1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coombes RC, Page K, Salari R, et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin Cancer Res. 2019;25(14):4255–4263. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Murillas I, Chopra N, Comino-Mendez I, et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019. August 1 [Epub ahead of print]. DOI: 10.1001/jamaoncol.2019.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volik S, Alcaide M, Morin RD, Collins C. Cell-free DNA (cfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies. Mol Cancer Res. 2016;14(10):898–908. [DOI] [PubMed] [Google Scholar]
- 17.Adalsteinsson VA, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8(1):1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choudhury AD, Werner L, Francini E, et al. Tumor fraction in cell-free DNA as a biomarker in prostate cancer. JCI Insight. 2018;3(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stover DG, Parsons HA, Ha G, et al. Association of Cell-Free DNA Tumor Fraction and Somatic Copy Number Alterations With Survival in Metastatic Triple-Negative Breast Cancer. J Clin Oncol. 2018;36(6):543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald BR, Contente-Cuomo T, Sammut SJ, et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med. 2019;11(504). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109(36):14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.