Abstract
Modern health care requires a proactive and individualized response to diseases, combining precision diagnosis and personalized treatment. Accordingly, the approach to patients with allergic diseases encompasses novel developments in the area of personalized medicine, disease phenotyping and endotyping, and the development and application of reliable biomarkers. A detailed clinical history and physical examination followed by the detection of IgE immunoreactivity against specific allergens still represents the state of the art. However, nowadays, further emphasis focuses on the optimization of diagnostic and therapeutic standards and a large number of studies have been investigating the biomarkers of allergic diseases, including asthma, atopic dermatitis, allergic rhinitis, food allergy, urticaria and anaphylaxis. Various biomarkers have been developed by omics technologies, some of which lead to a better classification of distinct phenotypes or endotypes. The introduction of biologicals to clinical practice increases the need for biomarkers for patient selection, prediction of outcomes and monitoring, to allow for an adequate choice of the duration of these costly and long‐lasting therapies. Escalating healthcare costs together with questions about the efficacy of the current management of allergic diseases require further development of a biomarker‐driven approach. Here, we review biomarkers in diagnosis and treatment of asthma, atopic dermatitis, allergic rhinitis, viral infections, chronic rhinosinusitis, food allergy, drug hypersensitivity and allergen immunotherapy with a special emphasis on specific IgE, the microbiome and the epithelial barrier. In addition, EAACI guidelines on biologicals are discussed within the perspective of biomarkers.
Keywords: allergen immunotherapy, allergic rhinitis, asthma phenotypes and endotypes, biomarkers, food allergy
1. INTRODUCTION
Allergic diseases represent a group of conditions caused by hypersensitivity of the immune system to allergens present in the environment. 1 These diseases include food allergy, asthma, atopic dermatitis (AD), allergic rhinitis (AR), conjunctivitis and chronic rhinosinusitis with or without nasal polyposis (CRSwNP). 2 , 3 , 4 The 100‐year‐old personalized allergen‐specific management of allergic diseases has been a particular advantage in our specialty contributing to the early awareness of personalized approaches and precision medicine. The use of multiple omics, big data and systems biology has demonstrated a profound complexity and dynamic variability and enabled the discovery of novel biomarkers. 5
Generally, a biomarker is a measurable indicator of the presence and severity of diseases or their response to a treatment with clear cutoff points. Regarding the prediction, diagnosis or monitoring of diseases, biomarkers are gaining importance in clinical practice as they provide an objective and measurable way to characterize a disease. However, it is challenging to identify convincing biomarkers as the genetic and regulatory networks for individual patients differ significantly. Biomarkers represent measurable indicators linking an underlying pathway to a phenotype or endotype of a disease. 6 , 7 , 8 Regrettably, current biomarkers are not precise in selecting the specific endotype that will respond to a targeted treatment. A good example is the observation that blood eosinophilia predicts therapeutic responses to all currently available or future‐targeted interventions in severe asthma (i.e, anti‐IL‐5, IL‐4/IL‐13, CRTH2 antagonists). 8 , 9 Precision medicine in allergic diseases demands accurate diagnoses, 10 which mostly rely on the combination of the clinical history and respective gold standards, which are all subject to the operator, observer and interpretation variability. 11 , 12 Some of the approaches are time‐consuming, and in vivo challenges may result in severe side‐effects and, in rare cases, even death. Therefore, the discovery, validation and clinical applicability of molecular biomarkers become increasingly important. 13
The cellular, biochemical or molecular changes in allergic patients which are measurable in blood, sputum or nasal secretions can be considered as biomarkers. 14 These biomarkers are used for disease diagnosis, selection of targeted therapy, disease monitoring and prediction of prognosis. 15 Except for the well‐known biomarkers (e.g, IgE, blood or sputum eosinophilia, fractional exhaled nitric oxide [FeNO]), 16 , 17 , 18 research focusing on pro‐inflammatory mediators, genes, the epithelial barrier and microbiomes is now emerging, which highlights more potential biomarkers for allergic diseases. 19 , 20 Some of the biomarkers showing a strong ability to identify disease endotypes or phenotypes may also act as therapeutic targets. 21 , 22 This article reviews the biomarkers identified to date and potential targeted therapies in allergy. In addition, it briefly reviews the biomarkers included in EAACI guidelines.
2. BIOMARKERS IN ASTHMA
In the past decades, it has been increasingly recognized that asthma is a highly heterogeneous disorder with different underlying mechanisms and pathways translating into variable responses to standard treatment across the different subsets or clinical phenotypes. 23 , 24 Unbiased approaches and cluster analyses identified four major clinical phenotypes: (a) early‐onset allergic asthma, (b) early‐onset allergic moderate‐to‐severe asthma, (c) late‐onset nonallergic eosinophilic asthma and (d) late‐onset nonallergic noneosinophilic asthma. 25 The late‐onset subsets tend to present as more severe or more difficult to treat than early‐onset asthma. To promote an adequate treatment strategy, asthma can be subdivided into Type 2 (high) and non‐Type 2 (or Type 2 low) endotypes based on their underlying inflammatory pathways. 26 As part of a more general syndrome often including nasal polyps with or without NSAID‐Exacerbated Respiratory Disease (NERD), 27 , 28 Type 2 asthma currently comprises the best defined asthma subset(s) in terms of underlying immunopathology, corresponding biomarkers 8 and targeted treatment options with biologicals and small molecules. 13 , 29 , 30
In parallel with the available (targeted) treatment options, biomarkers have been validated along the corresponding inflammatory pathways aimed for pheno‐/endotyping and to guide treatment for Type 2 asthma. 8 Clinically applicable point‐of‐care biomarkers include blood eosinophils, or whenever feasible, sputum eosinophil counts, serum‐specific IgE and FeNO. 8 , 31 Although overlapping in Type 2 biomarkers may occur within patients, all biomarkers represent different aspects of the Type 2 inflammatory pathways with IgE associating with allergy, while FeNO is linked to the IL‐13 pathway and epithelium‐derived inflammation. 8 Based on these point‐of‐care biomarkers in combination with clinical characteristics (age of onset, comorbidities, exacerbations, need for maintenance systemic corticosteroids) and physiological parameters (lung function, airway hyperresponsiveness, etc), current guidelines have now adapted algorithms which can help to predict a response to (targeted) treatments and/or can be used to monitor the subsequent treatment response. 23 , 32 , 33 In this context, some confounders have been recognized for the existing point‐of‐care biomarkers, i.e, for FeNO mainly related to ICS use, smoking, dietary nitrate intake, virus infections and bronchoconstriction, while for blood eosinophils circadian variation, parasites and systemic corticosteroids were found to be the most common perturbing factors. 23 , 34 In parallel with FeNO, oxidative stress can also be caused by an excess of reactive oxygen and nitrogen species. Many direct or indirect markers of oxidative stress such as bromotyrosine, malondialdehyde, isoprostane, thiobarbituric acid, glutathione disulfide have been detected in urine, plasma, sputum and BAL fluids of patients with asthma, and the level of these markers correlated with the clinical output and severity of the disease. 35 , 36 , 37 A noninvasive way of analysis, exhaled breath condensate collection, has allowed direct measurements of pH changes, H2O2 and the measurement of several indirect by‐products of oxidation like 8‐isoprostane and ethane. 38 , 39 Currently, the presence of high levels of urinary bromotyrosine is a promising noninvasive biomarker of oxidative stress for clinical use in asthma patients. In this context, a clinically relevant issue has been raised, i.e, whether “true” non‐Type 2 (noneosinophilic) asthma really exists among patients with severe asthma, given the fact that high‐dose inhaled and oral corticosteroids may potentially mask preexisting Type 2 inflammation interfering with its biomarkers, especially blood eosinophils and FeNO. 40 , 41 Currently ongoing corticosteroid‐tapering studies (RASP‐UK) in patients with non‐Type 2 severe asthma should answer this question. Alternatively, airway neutrophilia (“neutrophilic asthma”) may often reflect (subclinical) airway infection. 41 , 42 , 43
In contrast, for non‐Type 2 asthma which is by default defined as asthma without Type 2 biomarkers, underlying pathways and, hence, clinically applicable biomarkers and targeted treatment options are still largely under exploration. 41 , 44 Apart from most patients with mild clinically stable asthma, 26 , 45 clinical phenotypes frequently associated with non‐Type 2 asthma include very late‐onset asthma (women), obesity‐associated asthma, smoking‐associated neutrophilic asthma and paucigranulocytic asthma. Although generally based on increased sputum neutrophils or absence of normal levels of (sputum) eosinophils and neutrophils (paucigranulocytic) with normal levels of other Type 2 markers, the diagnosis of non‐Type 2 asthma is difficult to establish as often based on cross‐sectional data potentially affected by confounders including respiratory infections or anti‐inflammatory therapies. 41 In the absence of targeted biologicals, in non‐Type 2 asthma treatable traits should be targeted, 46 , 47 , 48 e.g, obesity, smoking habits, psychological aspects, neutrophilia as a potential indicator of respiratory infection and airway narrowing or airway hyperresponsiveness as an indicator of ASM dysfunction, while corticosteroids may not be effective and should be tapered off (Figure 1). 23
FIGURE 1.
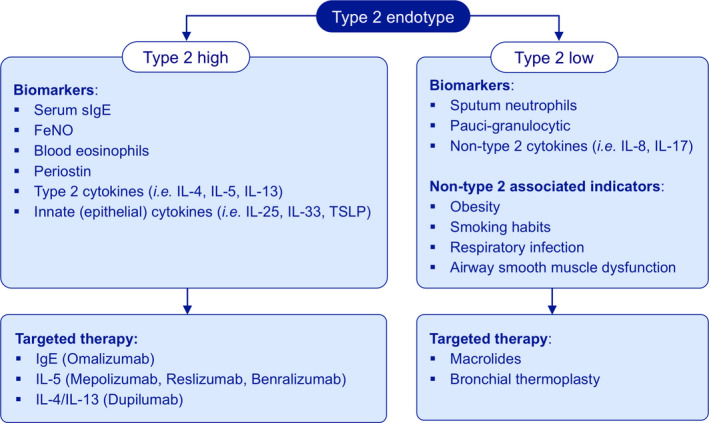
Treatment based on molecular biomarkers for endotypes in asthma. Asthma can be subdivided into Type 2 (high) and non‐type 2 (or type 2 low) endotypes based on their underlying inflammatory pathways. For Type 2 high asthma, potential biomarkers could be serum‐specific IgE (sIgE), fractional exhaled nitric oxide (FeNO) and blood or sputum eosinophils, and in some more specialized centers periostin. Moreover, Type 2 cytokines (IL‐4, IL‐5 and IL‐13) and innate (epithelial) cytokines (IL‐25, IL‐33 and TSLP) can also be important biomarkers. The options to treat with biologicals emphasizing biomarkers of Type 2 high endotype have entered the market: IgE (omalizumab), IL‐5 (mepolizumab, reslizumab, benralizumab) and IL‐4/IL‐13 (dupilumab). In contrast, the diagnosis of Type 2 low asthma is difficult to establish as generally based on increased sputum neutrophils or paucigranulocytic with normal levels of other Type 2 markers, and non‐Type 2 cytokines (IL‐8 or IL‐17). There are still some associated indicators including obesity, smoking habits and psychological aspects. Therefore, therapeutic strategies for patients with Type 2 low asthma could be macrolides and bronchial thermoplasty
In conclusion, despite substantial progress in our understanding, applicable biomarkers and targeted treatment options for Type 2 asthma, further characterization of molecular pathways by omics technologies, 49 , 50 , 51 sophisticated imaging 52 and innovative anatomical approaches 53 should help to further unravel the complexity of asthma and to define reliable (composite) biomarkers and therapeutic strategies for patients nonresponsive to currently available (targeted) treatment options including non‐Type 2 asthma.
3. BIOMARKERS LINKED TO MICROBIOME AND ASTHMA
An enormous variety of microbes colonize mucosal body surfaces, and these microbes are organized within complex community structures, utilizing nutrients from other microbes, host secretions and the diet. Modern lifestyles, medications and social interactions have fundamentally altered and disrupted the human microbiome metacommunity and, as a consequence, risk of immune‐mediated diseases such as allergy and asthma. 54 , 55 The mechanisms that mediate host–microbe communication are highly sophisticated and are being intensely investigated by many research groups across the world. However, there is accumulating evidence that microbiome composition and metabolic activities within the gut and the airways can influence asthma pathogenesis. 56 , 57 , 58 Here, we summarize some of the key recent findings that identify specific microbes or associated metabolites that may be useful as biomarkers to predict asthma risk, asthma severity or guide existing or novel therapies.
Alterations in the gut microbiota within the first year of life have been associated with asthma risk in multiple birth cohort studies. 19 The lower relative abundance of genera including Lachnospira, Veillonella, Faecalibacterium, Rothia, Bifidobacterium and Akkermansia in the gut during early life has been associated with the development of asthma. 59 , 60 While fewer studies have examined preschool children (2‐4 years of age), a recent study in this age group demonstrated that certain bacterial genera within the gut were still associated with wheezing (Collinsella and Dorea) or subsequent development of asthma (Gemmiger and Escherichia). 61 In addition to the gut microbiota, studies are also showing changes in the microbial populations of the airways. Microbial diversity and the relative abundances of Veillonella and Prevotella in the airways at age 1 month were associated with asthma by age 6 years. 62 Interestingly, higher relative abundance of these bacteria was associated with reduced TNF‐α and IL‐1β and increased CCL2 and CCL17 within the airways. A switch from a Corynebacterium and Dolosigranulum cluster in the upper airways to a Moraxella cluster was associated with a higher risk of severe asthma exacerbation in children with asthma. 63 In adults, increased relative abundance of the phylum Proteobacteria (including Haemophilus, Comamonadaceae, Sphingomonadaceae, Nitrosomonadaceae, Oxalobacteraceae and Pseudomonadaceae) is often associated with asthma or with worse asthma control. 64 Microbial changes within the gut, upper and lower airways of adult asthma patients are magnified in obese asthma patients and in those with severe disease. 65 Bronchoalveolar lavage levels of IL‐5 and eosinophils correlated with a variety of microbes within the airways. Of note, severe asthma negatively correlated with fecal Akkermansia levels and oral administration of Akkermansia to murine models significantly reduced airway hyper‐reactivity and airway inflammation (Figure 2).
FIGURE 2.
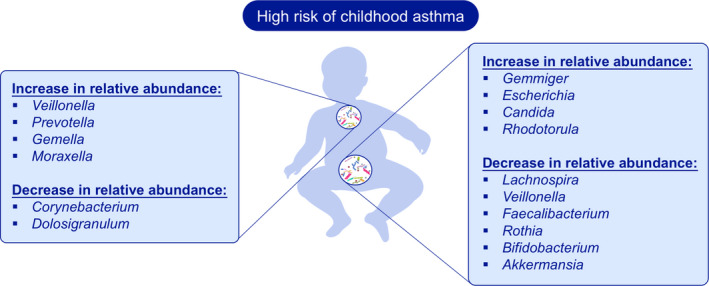
Microbiome Biomarkers in Asthma. Alterations in the gut and airway microbiota during childhood have been associated with asthma risk. The higher relative abundance of Veillonella and Prevotella and a switch from a Corynebacterium and Dolosigranulum cluster to a Moraxella cluster in the upper‐airways were associated with a higher risk of severe asthma exacerbation in children with asthma. The lower relative abundance of genera including Lachnospira, Veillonella, Faecalibacterium, Rothia, Bifidobacterium and Akkermansia in the gut during early life has been associated with the development of asthma. The increases in relative abundance of Gemmiger, Escherichia, Candida and Rhodotorula within the gut were also associated with the subsequent development of asthma
In addition to microbiota composition, microbial metabolites may also be useful biomarkers in asthma. The fecal metabolome of children at increased risk of asthma contained increased levels of pro‐inflammatory metabolites, among which 12, 13 DiHOME was able to induce IL‐4 production in CD4+T cells and decreased the abundance of Tregs. 60 High levels of short‐chain fatty acids (SCFAs), such as butyrate and propionate, at 1 year of age were associated with reduced risk of atopic sensitization and asthma by school age. 66 Multiple immune modulatory effects have been described in murine models for SCFAs, which include the promotion of Treg development and the inhibition of pulmonary ILC2 functions and subsequent development of airway hyper‐reactivity. 67 In adults, an increased abundance of histamine‐secreting bacteria was observed within the gut of patients with asthma, while disease severity correlated with high levels of the histamine‐secreting microbe Morganella morganii. 68 Murine models have demonstrated that bacterial‐derived histamine within the gut can influence inflammatory responses within the lungs. 69
In the future, the application of recent advances in metagenomic sequencing technologies and bioinformatics will likely lead to the identification of novel functional traits and metabolites within the gut and airway microbiome of asthma patients. 70 In addition, future asthma studies should include the microbiome as potential biomarkers that predict or associate with responses to biologics, as already observed for Faecalibacterium, Bifidobacterium and Akkermansia that associate with immunotherapy responses in certain groups of cancer patients. 71
4. SKIN BACTERIAL MICROBIOME AS CLINICAL BIOMARKER IN ATOPIC ECZEMA
Diagnosis of atopic eczema (AE) severity is still today a semiquantitative clinical score based on subjective information from the patients together with a doctor's subjective estimation of the severity of skin lesions and patient's history of itching and sleep loss. 72 , 73 In the era of targeted therapy, and thus more complex therapy management requirements, more objective criteria are urgently needed. The serum thymus and activation‐regulated chemokine (TARC) level has been reported as the most reliable biomarker for disease severity with strong pooled correlation coefficients with AD. Additional biomarkers that could prove useful but require additional research include serum cutaneous T‐cell‐attracting chemokine (CTACK), sE‐selectin, macrophage‐derived chemokine (MDC), lactate dehydrogenase (LDH) and IL‐18. 74 A diagnostic biomarker to distinguish between the different subgroups of AE is still needed. AE, likewise, lacks a prognostic biomarker: AE 75 affects 30% of children but only 5% of adults—thus, the question remains who keeps the disease, who emerges from it and who embarks on the full career of an atopic individual. Skin microbiome dysbiosis, measured either as microbiome diversity or more reliably as abundance of S aureus, was shown to correlate with both the AE clinical score and the expression of skin barrier molecules. 76 It is still a matter of scientific debate whether the relative frequency of various bacteria (e.g, S aureus frequency as obtained from 16S‐based NGS) is an adequate biomarker or rather the absolute microbial load (e.g, as obtained from qPCR) is better. Furthermore, is it enough to quantify the DNA abundance from nonstandardized amounts of skin samples, or rather is the absolute microbial load of standardized skin samples needed?
S aureus is important for AE pathogenesis even though it is still a matter of debate whether overgrowth of S aureus is a cause or a result of barrier disruption. 77 Thus, microbiome analysis, at least on the species level, but ideally on the strain level, would enable us to identify personalized biomarkers. This highlights a methodological drawback, as currently tools for annotation on species level are not reliable. Furthermore, the current methods for skin microbiome measurement are not standardized; testing the same material in different laboratories is prone to give different results. For skin microbiome to be used clinically as a biomarker, standardized methodology needs to be developed and validated so it can be reliably used across different laboratories. 78 Combinatory biomarkers between skin microbiome and biomarkers of Type 2 immunity would also be of great potential. 79 Recently, biofilm propensity of S aureus skin isolates as a cause and possible target has become more and more of a central issue. 80 Thus, resolving the enigma of skin–microbe interaction as a function of skin homeostasis has to take more players into the sight. 81
In conclusion, skin bacterial microbiome shows great potential to be used as a clinically important biomarker for atopic eczema. To reach this aim, we need to perform prospective clinical trials and large longitudinal registries that include skin microbiome testing. Furthermore, it is critical to advance standardized and foremost quantitative methodologies for skin bacterial microbiome analysis. New technologies, such as single‐molecule real‐time (SMRT) sequencing , need to be further developed and tested in order to improve skin microbiome analysis with higher accuracy and/or longer sequencing length. Collaboration between large academic consortia and pharmaceutical companies is essential for such endeavors.
5. BIOMARKERS IN DIAGNOSIS OF ALLERGIC RHINITIS
With deeper insights into mechanisms of AR, novel biomarkers have recently been identified in its diagnosis. Furthermore, several immune cells and mediators, genes and metabolites have been studied to explore their potential utilization in diagnosis of AR.
5.1. Immune cells and mediators
Several potential immune cells (granulocytes, lymphocytes, etc) and mediators might serve as diagnostic biomarkers of AR. 14 , 82 Izuhara and colleagues have reported that induction and increased expression of periostin reflect Type 2 inflammation and remodeling and could be regarded as an emerging biomarker for allergic diseases. 83 One study has demonstrated that allergen‐induced surface CD203c expression on basophils exhibits a time‐of‐day‐dependent variation, and allergen‐specific basophil reactivity shows daily variations depending on circadian clock activity in basophils, which could partly be responsible for temporal symptomatic variations in AR. 84 One recent study has suggested that circulating group 2 innate lymphoid cells (ILC2s) may play an important role in the pathology of AR, particularly as increased levels of ILC2s correlated with symptom scores and IL‐13 levels in house dust mite (HDM)‐sensitized AR patients, 85 and these cells produce large amounts of proinflammatory mediators in response to Th2 cytokines. 86 , 87 Indeed, a more recent study by Tojima and colleagues found that prostaglandin D2 (PGD2) and cysteinyl leukotriene (cysLTs) might induce ILC2s to produce Th2 cytokines such as IL‐5 and IL‐13. 88 Similarly, ST2‐expressing pathogenic memory T helper (Th) 2 cells, producing substantial amounts of IL‐33‐induced IL‐5 and IL‐13, have been shown to be linked to sensitization and the onset and progression of AR (Figure 3). 89
FIGURE 3.
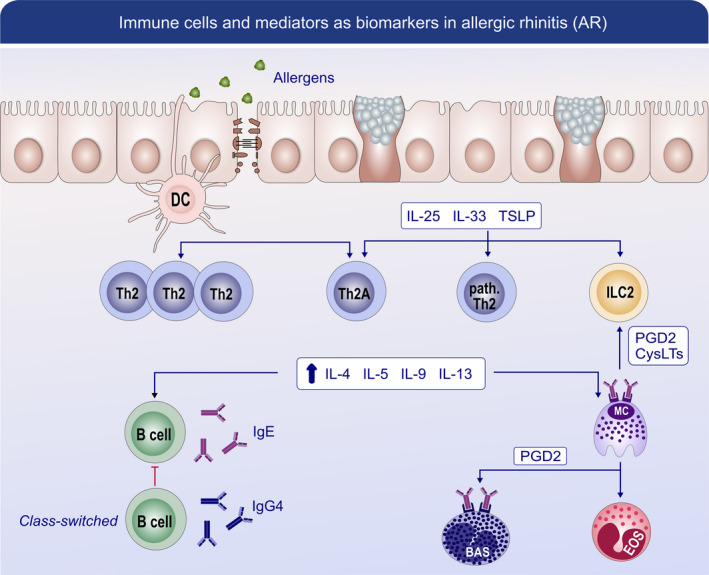
Immune cells and mediators as biomarkers in allergic rhinitis (AR). AR is associated with abnormalities in epithelial barrier function which is caused by exposure to exogenous proteases from allergens bacteria and viruses. These changes in epithelial barrier could contribute to the allergen absorption and disruption of epithelial tight junction. Activated dendritic cells (DCs) present allergen peptides to naive T cells and drive them to differentiate into Th2 cells and also allergen‐specific Th2A cells. Damaged epithelial cells release a high level of alarmin (TSLP, IL‐25, and IL‐33), which activate the group 2 innate lymphoid cells (ILC2s) as well as pathogenic memory T helper (Th) 2 cells. All these cells produce large amounts of proinflammatory mediators including IL‐4, IL‐5, IL‐9, and IL‐13. Besides, IL‐4 and IL‐13 are involved in IgE class switch in B cells. IgE binding to mast cells can trigger the release of mast cell‐associated mediators, such as prostaglandin D2 and leukotrienes, which could also activate the function of ILC2. PGD2 signaling could be a promising biomarker, as it can also activate eosinophils and basophils. Moreover, CD203c expression on basophils exhibits a time‐of‐day‐dependent variation, which could partly be responsible for temporal symptomatic variations in AR. IgG4 increased during allergen immunotherapy (AIT) is purported to be a blocking antibody by competing for allergen binding with IgE bound to Fcε receptors on mast cells and basophils. cysLT, leukotrienes; PGD2, prostaglandin D2
5.2. Genes
Epigenetic modifications, particularly DNA methylation and microRNAs (miRNAs), might have the potential to identify AR patients. One recent study has demonstrated changes in DNA methylation of tryptase gamma 1 (TPSG1), schlafen (SLFN12) and mucin 4 (MUC4) genes, following controlled allergen challenge, and suggested that baseline epigenetic status may act as a potential biomarker for AR symptom severity. 90 Another recent study has indicated that the nasal epigenome associated with asthma, FeNO and IgE may serve as a sensitive biomarker of asthma, allergy and airway inflammation in children. 91 Other studies have reported that subsets of circulating miRNAs are solely expressed in the blood of patients with AR and asthmatics and may therefore be used as noninvasive biomarkers for diagnosis and characterization of these diseases. 92 , 93
5.3. Metabolites
Metabolites have also been proposed as biomarkers for AR. A very recent study of serum metabolomics has demonstrated that at least nine metabolites (13(S)‐HPODE, bilirubin, leukotriene D4, hypoxanthine, L‐stercobilinogen, N‐succinyl‐L‐diaminopimelic acid, chlorophyll b, 15‐hydroxyeicosatetraenoic acid and urate) were significantly altered in the serum of AR patients and therefore may provide a better understanding of the metabolic pathways involved in the etiology of AR. 94 Additionally, a decreased serum lactoferrin level has been reported to be associated with the phenotype of Dermatophagoides pteronyssinus (Der p 1)‐sensitive AR and, in combination with serum Der p 1‐specific IgE levels, may serve as a serologic biomarker for early detection of AR. 95
5.4. Clinical biomarkers of allergic rhinitis
Clinically, rhinitis phenotypes include nonallergic rhinitis (NAR), AR, local allergic rhinitis (LAR) with a localized allergic response but no systemic atopy. In some cases, basic SPT and an sIgE test are not efficient to discriminate between these phenotypes. Recently, a retrospective study was conducted to investigate the safety and reproducibility of the nasal allergen challenge (NAC) carried out over 12 years in Spain. It was shown that 99.97% NACs were well tolerated without delayed, local severe or systemic adverse events in allergic patients for both children and adults. Moreover, there were no significant differences in three consecutive NAC with a single allergen per session (NAC‐S) proving the reproducibility of NACs. 96 Regarding the monitoring of the NAC, it was shown that the %Vol2‐6 cm by acoustic rhinometry (AcRh) displayed an optimal discriminative power for AR patients from both NAR and HC subjects. 97
Although the nasal provocation test (NPT) is considered a key tool to diagnose LAR, it requires well‐trained personnel and is time‐consuming. In this regard, the basophil activation test (BAT) should be helpful for supporting the diagnosis of LAR. BAT shows 50%‐66.6% sensitivity and 90%‐100% specificity of LAR, which is more sensitive than an sIgE test and less time‐consuming than NACs as an in vitro test. 98 , 99 , 100 There is a new AR phenotype named dual allergic rhinitis (DAR), where patients show SPT positivity to seasonal allergens only, but suffer from perennial symptoms and react to both perennial and seasonal allergens. For these patients, BAT displays 100% positivity with seasonal allergens and 60% positivity with perennial allergens. 101
Overall, NAC can act as a gold standard in distinguishing the AR phenotype. And BAT could also be useful as an in vitro tool for LAR/DAR diagnosis in the daily practice.
5.5. Biomarkers in therapy of allergic rhinitis
Currently, optional therapeutic measures for AR involve patient education, environmental control, pharmacotherapy, allergen immunotherapy (AIT) and surgery. 102 , 103 Traditional medications include nasal corticosteroids, antihistamines, mast cell stabilizers, decongestants, etc MP29‐02, a combination of nasal corticosteroid and antihistamine, is a novel topical medication which has proved to be effective in reducing nasal hyperreactivity and nasal mediators such as substance P, in patients with AR. 104 As ILC2s have been shown to produce significant amounts of proinflammatory mediators in response to epithelium‐derived cytokines 86 , 87 and PGD2 and cysLTs 88 in AR patients, agents targeting the ILC2s and the mediators activating these cells have become targets for therapy. Rittchen and Heinemann have recently reviewed the central role of hematopoietic PGD2 synthase in allergic inflammation and indicated that PGD2 signaling might be a promising therapeutic target for AR, as PGD2 can activate Th2 cells, eosinophils and basophils. 105 Indeed, a randomized controlled phase II clinical trial has recently demonstrated that ONO‐4053, a novel prostaglandin D receptor 1 antagonist, was more effective than pranlukast, a leukotriene receptor antagonist, in treating patients with seasonal AR. 106 Most recently, emerging studies have focused on biologics for treating allergic diseases; especially severe, uncontrolled asthma and AD, as well as AR. 107 , 108 To date, a high number of specific biologics targeting markers of Th1/2/17 inflammation have been introduced; with more underdevelopment. 107 , 109 In particular, targeting IgE by omalizumab, a recombinant humanized anti‐IgE antibody, has been shown to significantly improve symptoms in patients with inadequately controlled AR. 110 Furthermore, combining omalizumab with subcutaneous immune therapy (SCIT) in patients with SAR and comorbid seasonal allergic asthma has been shown to lead to greater clinical improvements in AR and lung function than SCIT alone. 111 Similarly, dupilumab, a biologic which targets IL‐4Rα to block the activity of both IL‐4 and IL‐13, has been shown to provide nasal symptom relief in patients with uncontrolled asthma and comorbid AR. 112
6. BIOMARKERS OF VIRAL INFECTIONS IN EXACERBATION OF ALLERGIC RHINITIS AND ASTHMA
Respiratory viral infections may exacerbate chronic airway inflammatory diseases, including allergic inflammation through both Type 2 (e.g, IL‐25, IL‐33 and TSLP) and non‐Type 2 (e.g, IFN types I and III, RIP3, OSM, MCIDAS) mechanisms.
Over the past decade, our understanding of immunological mechanisms underlying allergic diseases such as AR has substantially increased through the discovery of T helper (Th) subsets and their importance in allergic inflammation. Emerging data now provide new insights on the Type 2 immune response that is an immune response to allergens and involves Th2 cells, Type 2 B cells, ILC2s, Type 2 macrophages, a small fraction of IL‐4‐secreting NK cells, IL‐4‐secreting NK‐T cells, basophils, eosinophils and mast cells. 113 At the same time, it has also been established that viral infection synergizes with allergic inflammation causing more severe exacerbations and symptoms compared to both conditions alone. 114 , 115 There are increasing evidences that most respiratory viral infections could trigger or exacerbate chronic Type 2 inflammatory responses via excessive release of chemokines and cytokines into the airways. 116 , 117 , 118 While much of these studies focus on lower airway inflammatory diseases instead of AR, insights from these studies can be applied to ongoing studies of virus‐induced AR exacerbations and the search for its associated markers.
Like other chronic airway inflammatory diseases, AR patients also suffer from altered responses and potentially increased susceptibility toward viral infection. 119 , 120 , 121 This is similarly due to the reduced Type 3 interferon response, which is crucial against incoming viral infection in the upper airway. 119 , 120 , 121 Hence, markers for virus‐induced AR exacerbation may have a significant overlap with findings from other inflammatory airway diseases. Proinflammatory cytokines such as TNF‐α, IL‐4, IL‐5, IL‐13, RANTES, Eotaxin, TSLP, IL‐25 and IL‐33 are usually expressed at higher concentrations in chronically inflamed airways, some of which are also found in AR. 113 , 122 , 123 These cytokines can be further triggered directly or indirectly by virus‐induced IFNs, cytokines and chemokines. Infections such as RSV can even further shunt antiviral responses toward a more Type 2‐centric response. 124 , 125 , 126 , 127 , 128 In addition, the discovery of ILC2s, a group of lymphoid cells, further emphasized the role of epithelial alarmins IL‐25, IL‐33 and TSLP in viral‐induced exacerbation. 129 During viral infection, these three cytokines were secreted in response to epithelial injury. 130 , 131 , 132 It has been reported that IL‐5, IL‐13 and IL‐33 levels were elevated in both the BAL and nasal fluid of asthmatics after RV16 infection compared to healthy subjects. The nasal IL‐33 level was significantly and positively correlated with the total lower respiratory symptom score. Moreover, IL‐33 secreted by RV‐infected BECs directly induced IL‐5 and IL‐13 production by human blood ILC2s. 133 Together, it indicated that activated ILC2s in the upper and lower airways could cooperate to aggravate a Type 2 inflammation resulting in acute viral exacerbation. However, there are higher ILC2 levels in the blood of allergic asthmatics compared to patients with allergic rhinitis and even higher levels in patients with combined asthma and AR. 87 , 134 Furthermore, ILC2s from allergic asthmatics were more responsive to IL‐33 and IL‐2 treatment compared to patients with allergic rhinitis. 134 These differences may cause the diverse severity of allergic airway diseases (Figure 4).
FIGURE 4.
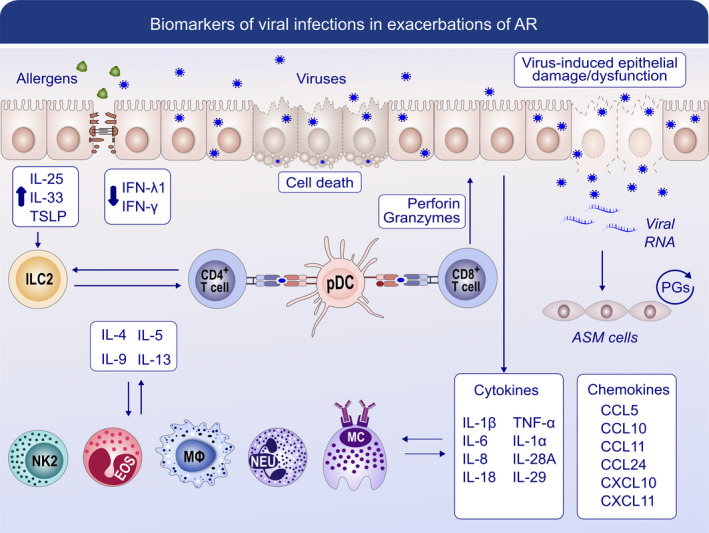
Biomarkers of viral infections in the exacerbation of AR. After the epithelial cells are infected with viruses, the replicating virus can cause cell lysis and direct damage to the epithelium, which causes deficiency in the production of antiviral interferon (IFN)‐β and IFN‐λ1. Together with the allergen‐induced cytokines IL‐25, IL‐33 and TSLP, ILC2s are activated and produce more Type 2 cytokines. Subepithelial plasmacytoid dendritic cells (pDCs) recognize virus antigens and present them to CD4+ T cells and CD8+ T cells through MHC class Ⅱ or Ⅰ and drive them toward a more Type 2 centric response. Excessive release of chemokines and cytokines can be triggered by infections such as respiratory–syncytial virus (RSV). Together with Type 2 cytokines, they could further promote the function of Type 2 macrophages, a small fraction of IL‐4‐secreting NK cells, IL‐4‐secreting NK‐T cells, neutrophils, eosinophils and mast cells and augment Type 2 responses in chronically inflamed airways. With the production of perforin and granzymes, CD8+ T cells can show cytotoxicity to virus‐infected epithelial cells and induce apoptosis. The viral RNA is released and detected by airway smooth muscle cells and stimulates the production of prostaglandins (PGs) in an autocrine manner
In addition, respiratory viral infections may also exacerbate chronic airway inflammatory diseases, including allergic inflammation through other non‐Type 2 mechanisms, in which other markers can also be used as an indicator of these exacerbations. Viral infections can lead to the destruction of epithelial barrier and disruption of mucociliary function due in part to cell death in the virus‐infected epithelium. Hence, markers for cell death (e.g, RIP3) and mucociliary dysfunction (e.g, MCIDAS) constitute part of the viral exacerbation repertoire. 124 Viral infection also causes increase in factors such as OSM and ANGPTL4 which disrupts tight junctions leading to increased allergen invasion and their contact with immune cells in the sub‐epithelium region, thereby exacerbating allergic symptoms. 135 , 136 In addition, miRNAs are increasingly implicated in the mis‐regulation of inflammatory responses and several of them are found to be dysregulated in an inflamed airway. For example, expressional changes of miRNAs such as miR‐21 may coincide with viral infection responses and hence linked to virus‐induced exacerbations. 137 Finally, an emerging field of bioenergetics and mitochondrial function may also contribute to the mechanism of virus‐induced exacerbation in AR. Oxidative stress and mitochondrial dysfunction from viral infection may induce increased inflammation, and thus ROS and its associated markers may potentially serve as key markers for viral exacerbation. 138 , 139
7. BIOMARKERS IN CHRONIC RHINOSINUSITIS
CRS can be divided into different pheno‐ and endotypes. The mostly used phenotype is the division into CRS with and without nasal polyps (CRSwNP and CRSsNP), although many other pheno‐ and endotypes are known. 28 , 140 However, recently, the options to treat with biologicals have put more emphasis on markers of Th2 disease irrespective of the presence of nasal polyps. The first Type 2 targeting biologic anti‐IL4Rα (Dupilumab) has entered the market for CRSwNP patients, and others like anti‐IgE, anti‐IL5 and anti‐IL5Rα may follow shortly. 141 , 142 , 143 Cluster analysis of CRS has shown that CRSsNP and CRSwNP are not dichotomous but instead have overlapping inflammatory signatures with Type 2 inflammation as the predominant endotype mainly in CRSwNP but also CRSsNP, especially in western parts of the world. Interestingly, some patients with CRS express a mixture of two or more inflammatory endotypes. 22 , 144 , 145 The recently published EPOS2020 proposes a new clinical classification based on the disease being localized (often unilateral) or diffuse (always bilateral). Both these groups can be further divided into Th2 or non‐Th2 disease. 4
In the very near future, it may be possible to offer personalized medicine for CRS patients where treatment is based on molecular biomarkers for the endotype or subendotype activated in an individual patient. 27 , 146 The major challenge is to find reliable biomarkers that define Th2 inflammation and predict reaction to treatment. Ideally, these biomarkers should be supported by a body of evidence clarifying the biological significance, be quantifiable in a cost‐efficient way and be easily measurable, preferably in blood or nasal secretion. 14 Potential biomarkers could be eosinophils, neutrophils, 147 , 148 IgE, 149 Th2 cytokines, 150 innate (epithelial) cytokines, 123 , 149 , 151 but also phenotypical phenomena like smell loss, 152 asthma and response to systemic corticosteroids. 146 Contrary to FeNO in asthma, nasal NO has not been shown to be helpful to identify the T2 endotype because the main source of production of nasal NO is the sinuses that are closed off when CRS occurs. 153 The main biomarkers used at the moment to define Th2 disease are eosinophils, IgE levels and in some more specialized centers periostin and/or IL‐5. There is quite some evidence showing that tissue and blood eosinophils are a reasonable surrogate marker for Th2 disease and that blood eosinophils are a reasonable biomarker to predict eosinophilic CRS with or without nasal polyps. 145 On the other hand, a lack of tissue eosinophilia, lower serum eosinophilia and absence of tissue squamous metaplasia may predict a CRS phenotype suitable for a trial of long‐term macrolide therapy when surgery and topical therapy have failed. 154 Unfortunately, recent large studies with monoclonal antibodies directed to Type 2 endotypes have not found reliable biomarkers to predict response to treatment. 141 , 142 , 155 , 156 , 157 As in asthma, 8 we need large, maybe real‐life studies to find better predictors to identify responders to biological treatments. For now, our treatment decisions still heavily rely on phenotypical characteristics such as smell loss, asthma and response to surgery and systemic corticosteroids. 4 , 27
8. BIOMARKERS IN FOOD ALLERGY
Apart from clinical determinants of food allergy and the respective gold standard, the oral food challenge, biomarkers represent an area of extensive research. In food allergy, the focus is on genetic risk factors, allergen‐specific and nonspecific humoral and cellular biomarkers. 158 , 159 , 160
Although genetic markers for food allergy are not yet at the level of clinical relevance, genes linked to HLA‐genes, and more importantly to epithelial integrity and consequently reduced barrier function like filaggrin, SPINK5 161 and SERPINB7, are linked to eczema development and also food allergy. 162 , 163 , 164 , 165 , 166 , 167 , 168 Given the importance of the exposome in allergy development, epigenetics may even play a more important role. Promising results in the context of peanut allergy await replication in larger cohorts. 169 , 170 Regulation at another level has been linked to the microRNA 193a‐5p. It is involved in the posttranscriptional regulation of IL‐4 and downregulated in PBMCs from milk allergic children. 171 Due to the importance of barrier (dys)function in atopic diseases, 172 , 173 , 174 measurement of skin integrity may be a very important tool to identify high‐risk populations. Electrical impedance spectroscopy, successfully tested in rodents, may be capable of assessing this biomarker also in humans. 175
Allergen extract‐based testing methodologies like skin prick test (SPT) and/or specific IgE (sIgE) tend to be less efficient for the diagnosis of food allergy. Thus, more specific approaches focusing on specific allergens (see section on allergens) and epitope‐specific antibody response patterns are explored. 176 Diversity of IgE binding to linear epitopes correlated with the severity of peanut and milk allergy, 177 , 178 , 179 and IgG4 and IgE antibody binding to specific milk epitopes was stronger and more diverse in children who do not outgrow their milk allergy. 178 By measuring IgE and IgG4 responses with bead‐immobilized milk epitopes and applying machine learning approaches, nonreactivity to baked milk could be predicted twice as successful as by conventional approaches. 180 , 181 , 182 The soluble high‐affinity IgE receptor (FcεRI) may also act as a biomarker for IgE‐mediated pathologies in a less allergen‐independent way. 183
Although allergen‐specific T‐cells are extremely rare, they display a pronounced Th2 type in allergic individuals. 184 , 185 A subset of allergen‐specific memory Th2 cells called TH2a cells, which are CD45+CD27−CD45RB−CRTH2+CD161+CD49+, has been discovered. They are almost exclusively found in allergic individuals, secrete IL‐5 and IL‐9, and within that group, the percentage of Type 2 cytokine double, triple, or quadruple positive cells is higher compared to Th2 cells. Moreover, mRNA expression of IL‐25, IL‐33 and TSLP receptors is higher. 186
Our understanding of B‐cell regulation has significantly evolved over the last few years. 187 Evidence is pointing toward an extreme rarity of IgE memory B‐cells in peripheral blood of allergic individuals, which may be absent in nonpeanut allergic individuals. 188 New therapeutic and diagnostic options opened up from data on allergen‐specific monoclonal antibodies that were generated via single‐cell sorting of allergen‐specific memory B‐cells. 189 , 190 B‐cell IgE antibody mutational maturation has been associated with barrier dysfunction. 191 Recently, the co‐emergence of short‐lived IgE plasmablasts and IgG memory B‐cells early in grass AIT in the absence of memory IgE + B‐cells has been reported. 192 , 193 Both subsets shared clonotypes supporting the existence of pools of specific B‐cell subsets, e.g, from IgG1‐positive allergen‐specific B‐cells upon switch factors and stimulation as demonstrated in mice. 192 , 194 Yet many questions on the emergence of IgE‐producing cells and their regulation have to be answered, and new biomarkers in this context will arise.
Functional tests that simulate allergen exposure in vitro like the basophil activation test (BAT) offer the possibility to assess allergen‐induced IgE cross‐linking. The BAT suggests adding significant diagnostic value to IgE‐based test methods. 195 , 196 , 197 Promising results on the usage of passive sensitization strategies, mast cell lines 198 or precursors 199 have been reported.
There is still a significant need to develop biomarkers to diagnose and predict anaphylaxis to prevent near fatalities and fatalities. 200 , 201 Beyond tryptase, which can be a very good marker in the emergency setting when baseline values exist, 82 , 202 predictors of life‐threatening reactions which can be measured on a routine basis or in multicenter trials are still missing.
There is the hope that the expanding array of novel mechanistic and diagnostic biomarkers provide the toolkit to develop algorithms or machine learning approaches to diagnose food allergy and predict treatment outcomes (Figure 5).
FIGURE 5.
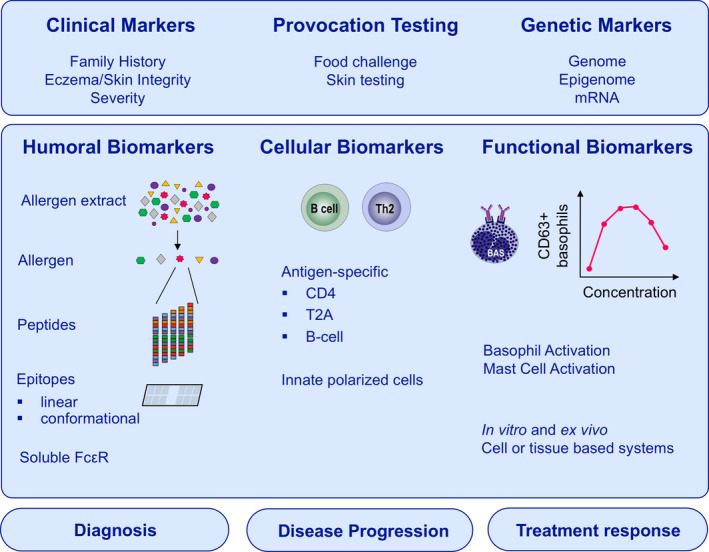
Biomarkers in food allergy diagnosis and treatment outcomes prediction. Conventional clinical approaches to diagnose food allergy include family history, skin integrity and the oral food challenge. Nowadays, expanded approaches focusing on genetic risk factors, allergen‐specific and nonspecific humoral and cellular biomarkers were explored. Genome, epigenome and mRNA linked to epithelial integrity and barrier (dys)function are linked to the development of food allergy. The measurement of IgE and IgG4 binding to linear or conformational epitopes could be more powerful to diagnose food allergy than conventional approaches. The soluble high‐affinity IgE receptor (FcεRI) may also act as a biomarker for IgE‐mediated pathologies in a less allergen‐independent way. Moreover, allergen‐specific Th2A cells and memory B cells have been discovered as new cellular biomarkers. Functional tests that simulate allergen exposure in vitro or ex vivo like the basophil activation test (BAT) and mast cell activation test (MAT) offer the possibility to assess allergen‐induced IgE cross‐linking
9. sIGE AS BIOMARKER IN DIAGNOSIS OF FOOD ALLERGY
Accurate diagnoses are essential for the management of food allergy. 159 They depend on a detailed clinical history, objective markers of sensitization and double‐blind placebo‐controlled food challenges (DBPCFC). 203 These are time‐consuming and require specialized medical facilities, and side‐effects may occur. Consequently, molecular allergy diagnosis aims to reveal significant associations between sIgE and clinical phenotypes.
9.1. Peanut
A retrospective study of 205 peanut‐challenged Danish patients found the best correlation between sIgE and clinical thresholds for the 2S albumin Ara h 2. 204 A diagnostic model for peanut allergy predicted the food challenge outcome with 100% accuracy in 59% versus Ara h 2 in 50% of 100 Danish peanut‐allergic patients. 205 Co‐sensitization to Ara h 2 and 6 was associated with severe allergy in peanut‐challenged Finnish patients. 206 A French study of 48 peanut‐allergic children found that Ara h 2 sIgE titers could predict the risk of anaphylaxis. 207
9.2. Soy
The cupins Gly m 5 and 6 were suggested as markers for severe allergic reactions in 30 soy‐allergic European patients. 208 A high diagnostic value of the 2S albumin Gly m 8 was reported in soybean‐sensitized Japanese children with and without symptoms. 209 Gly m 8 had an AUC = 0.75 for soy allergy, while the values for Gly m 5 and 6 were 0.69 and 0.64, respectively. In a study on soy allergy diagnostics, Gly m 8 had the highest AUC (0.79), comparable to skin prick test (0.76) and sIgE to soy extract (0.77). 210 In this study, the cupins Gly m 5 and 6 were related to mild symptoms.
9.3. Hazelnut
sIgE to the cupin Cor a 9 and the 2S albumin Cor a 14 was strongly associated with clinical symptoms in 161 Dutch hazelnut‐sensitized patients. 211 sIgE to Cor a 9 and 14 allowed correct diagnosis of 90% of severely hazelnut‐allergic Belgian children. 212 In 423 hazelnut‐allergic patients, Cor a 9 and 14 were associated with severe symptoms (AUC = 0.70). 213 A model combining clinical symptoms and sIgE to Cor a 14 and walnut increased the AUC to 0.91. In a prospective multicenter study of 90 peanut‐ and 44 hazelnut‐allergic German children, a 90% probability for a positive food challenge was calculated for Ara h 2‐specific IgE at 14.4 kU/L and for Cor a 14‐specific IgE at 47.8 kU/L. 214
9.4. Walnut
sIgE to the 2S albumin Jug r 1, the cupin Jug r 2 and the nsLTP1 Jug r 3 (AUC = 0.79, 0.70, 0.62, resp.) predicted anaphylaxis in 45 walnut‐allergic children. 215 In 91 walnut‐allergic subjects from Switzerland, Germany and Spain, severe reactions correlated with higher sIgE levels to Jug r 1 and the cupin Jug r 4. 216 sIgE to Jug r 1 (AUC = 0.79) from 32 walnut‐allergic Korean children better discriminated clinical walnut allergy from tolerance than sIgE to walnut extract (AUC = 0.56). 217 In 34 peanut‐, hazelnut‐ or walnut‐allergic Italian children, sIgE to Ara h 1 and Ara h 2, Cor a 9 and particularly Cor a 14 or Jug r 1, 2 and 3 was associated with anaphylaxis. 218
9.5. Cashew
In 63 cashew‐allergic Greek children, sIgE to the 2S albumin Ana o 3 (AUC = 0.97) performed better than extracts for predicting cashew allergy. 219 A markedly greater risk of a positive food challenge was observed for higher levels of sIgE to the cupins Ana o 1 and 2, and to Ana o 3 in 173 Dutch children with suspected cashew nut allergy. 220 Ana o 3 discriminated between allergic and tolerant children better than extract‐specific IgE with an AUC = 0.94 versus 0.78. A 95% probability for a positive cashew challenge was estimated for Ana o 3‐sIgE at 2.0 kU/L. 221
9.6. Egg and shrimp
sIgE to the ovomucoid Gal d 1 correlated with an increased risk of persistent egg allergy. 222 Sensitization to tropomyosin and sarcoplasmic calcium‐binding protein was associated with clinical reactivity in 58 shrimp‐allergic patients. 223
9.7. Cow's milk
Caseins (Bos d 8), the major protein fraction of cow's milk (80%), comprise four different proteins, αS1‐casein (Bos d 9, 32%), αS2‐casein (Bos d 10, 10%), β‐casein (Bos d 11, 28%) and κ‐casein (Bos d 12, 10%). α‐lactalbumins (Bos d 4) and β‐lactoglobulins (Bos d 5) make up the whey proteins in cow's milk. 224 An Italian study including 79 children found that Bos d 8 could differentiate children at risk for cow's milk anaphylaxis (AUC = 0.718) compared to Bos d 4 (AUC = 0.491) and Bos d 5 (AUC = 0.634). The levels of Bos d 8 sIgE reflected the severity of the milk allergy. 225 Additionally, low or undetectable levels of Bos d 8‐sIgE indicated tolerance to baked milk products. 226
In conclusion, severe reactions to legume seeds and tree nuts are predominantly caused by sensitization to storage proteins rather than by pollen‐related allergens such as Bet v 1 or profilin homologues, or nsLTPs (Table 1).
Table 1.
Specific IgE to these allergens is associated with severe symptoms
| Allergen source | 2S albumin |
Cupin *vicilin type **legumin type |
nsLTP1 | Ovomucoid | Tropomyosin | Caseins | References |
|---|---|---|---|---|---|---|---|
| Peanut |
Ara h 2 Ara h 6 |
Ara h 1* | 204, 205, 206, 207, 218 | ||||
| Soy | Gly m 8 |
Gly m 5* Gly m 6** |
208, 209, 210 | ||||
| Hazelnut | Cora 14 | Cor a 9** | 211, 212, 213, 214, 218 | ||||
| Walnut | Jug r 1 |
Jug r 2* Jug r 4** |
Jug r 3 | 215, 216, 218 | |||
| Cashew | Ana o 3 |
Ana o 1* Ana o 2** |
219, 220, 221 | ||||
| Egg | Gal d 1 | 222 | |||||
| Shrimp | Lit v 4 | 223 | |||||
| Cow's milk | Bos d 8 | 225 |
However, biomarkers for food allergy are also affected by geographical variations and can be age‐related. In Mediterranean pediatric patients, Ara h 6 and Ara h 2 are the best predictors of peanut allergy with the prevalence of 64% and 63%, respectively. 227 In birch‐endemic regions, preschool and school‐aged children with systemic reactions to hazelnut are mostly sensitized to Cor a 9. However, adults in these regions are highly sensitized to Cor a 1.04. Therefore, it is important to take regional and age variations into account when working on sIgE for food allergy. 228
10. BIOMARKERS IN DRUG HYPERSENSITIVITY
Drug hypersensitivity reactions include those mediated by a specific immunological mechanism and those nonspecific immune mediated (Figure 6). 229
FIGURE 6.
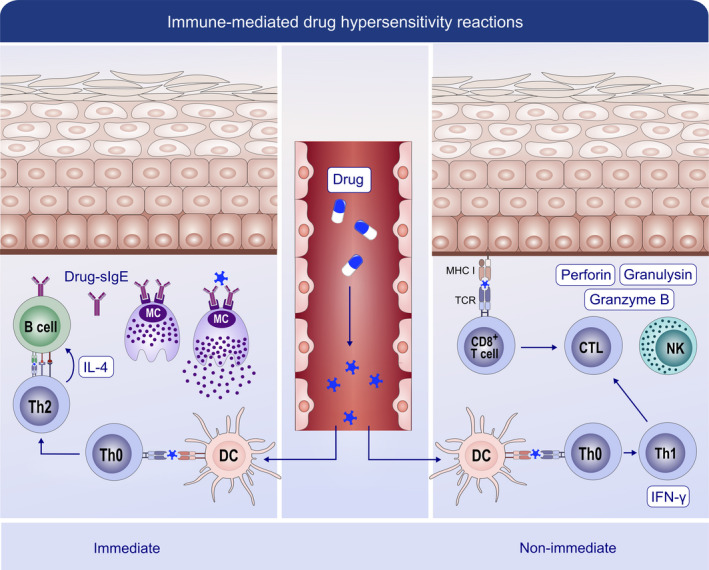
Mechanisms of immune‐mediated reactions to drugs. These reactions encompass immediate reactions (mediated by IgE) and nonimmediate reactions (mediated by T cells). In immediate reactions, drug‐induced polarization of Th2 cells from Th0 cells promotes B cells to produce specific IgE (sIgE). This sIgE binds to the FcεRI receptor on mast cells. In subsequent drug contacts, the simultaneous recognition by at least two sIgE initiates the degranulation and release of mediators. Nonimmediate reactions are generally characterized by a Th1 response with the increased secretion of IFN‐γ from Th1 cells and granulysin from NK cells
10.1. Immunologically mediated specific reactions
These reactions are classified into immediate reactions (IR) and nonimmediate reactions (NIRs) depending on whether they occur within 1‐6 hours or later after the drug intake. The first group is mostly associated with an IgE‐mediated mechanism and the latter with a T‐cell‐dependent type. 229 , 230
Skin tests (STs) are useful biomarkers for IRs to beta‐lactam (BL). 14 , 229 , 230 , 231 , 232 However, their sensitivity based on the classical antigenic determinants has decreased over the last decades 233 possibly due to the changing patterns of consumption (e.g, amoxicillin/clavulanic acid is replacing penicillin). 231 Interestingly, one antigenic determinant recognized by most patients with confirmed reactions to clavulanic acid has been recently identified. 234 Therefore, amoxicillin and any suspected BL must be included when performing STs. 230 , 233 , 235 , 236 For other drugs and for NIRs, the value of STs is very limited. 14 , 230 , 231 , 237
Regarding in vitro tests, during the acute phase of the reaction, serum tryptase is the most valuable biomarker for confirming mast cell activation in IR. 229 The expression of granzyme B and granulysin in blood cells may be useful for detecting lymphocyte activation in severe NIR. 238 At the resolution phase, immunoassays are used in IRs to determine sIgE, although the sensitivity is lower than for STs 229 , 239 , 240 and only commercially available for limited drugs. For BLs, it shows a variable sensitivity (0%‐50%), 229 , 230 , 239 with the possibility of inducing false‐positive results when testing for Penicillin V. 241 The value of BAT in IR has been proven for BL and quinolones, giving a sensitivity up to 55% 242 , 243 and 83%, 237 , 244 respectively. The sensitivity of both sIgE and BAT correlates with severity reaction, 239 decreases with time, 243 , 245 and depends on the activation basophil marker assessed, e.g, the detection of CD203c increases BAT sensitivity although remaining low (36.4%). 244 , 246
The value of lymphocyte transformation test (LTT) in NIRs has shown to be unsatisfactory. 14 , 229 The Tim3/galectin‐9 axis functions as a checkpoint inhibitor for Th1 cells. Interestingly, Th1 cells and dendritic cells of patients with drug‐induced maculopapular exanthema expressed lower levels of Tim3/galectin‐9 at baseline compared with tolerant individuals. 247 This observation might help identify subjects at risk of NIRs.
10.2. Immunologically mediated nonspecific reactions
The most important group in this type of reactions is cross‐intolerance to NSAIDs (CI), 248 , 249 in which patients react to NSAIDs from different pharmacological groups related to its COX‐1 inhibitory activity. 229 , 248 CI has been classically divided into phenotypes with exclusive skin involvement (NSAIDs‐exacerbated cutaneous disease (NECD) and NSAIDs‐induced urticarial angioedema (NIUA)) or with exclusive respiratory involvement (NSAIDs‐exacerbated respiratory disease, NERD). 28 Nevertheless, novel data indicate that almost 30% of patients with CI can experience blended reactions, especially involving skin and airways. 250 Genetic predisposition might account for the burden of some CI phenotypes (e.g, variants of GNAI2 in NIUA) (Figure 7). 251
FIGURE 7.
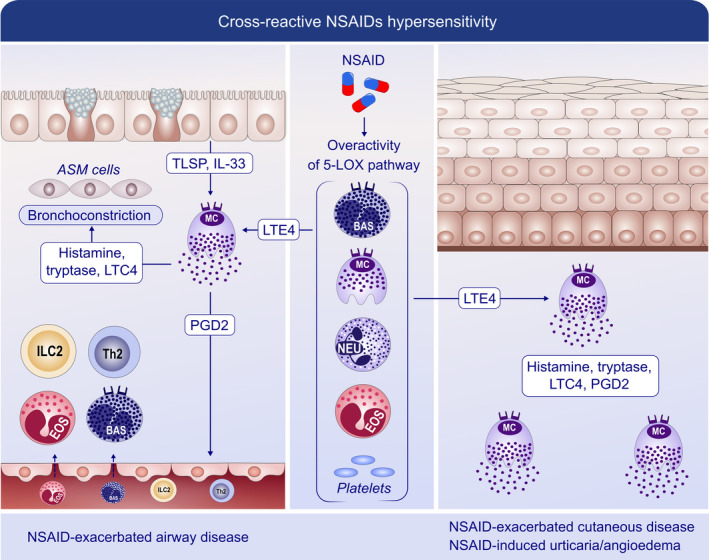
Mechanisms of cross‐reactive hypersensitivity reactions to NSAIDs. NSAIDs induce reactions relying on their COX‐1 inhibitory activity, i.e, activation of mast cells and other immune cells without involvement of adaptive immunity. During NSAID‐exacerbated respiratory disease (NERD), the administration of NSAIDs permits strong 5‐lipoxygenase (5LOX) activation and further generation of leukotriene E4 (LTE4). LTE4 induces the release of IL‐33 and TSLP, and consequent mast‐cell activation, with bronchoconstriction occurring as a result of the direct effects of leukotriene C4 (LTC4), prostaglandin D2 (PGD2) and other mast cell‐derived products. PGD2 recruits effector cells such as Th2 cells, group 2 innate cells (ILC2s), basophils and eosinophils to the airway. Consistently, in NSAIDs‐exacerbated cutaneous disease (NECD) and NSAIDs‐induced urticarial angioedema (NIUA), increased PGD2 can act on the skin epidermis. In addition, cross‐reactive hypersensitivity to NSAIDs may involve additional sources of inflammatory mediators, such as eosinophils and platelets. ILC2: innate lymphoid cells 2; LTE4/C4: leukotriene E4/C4; NSAID: nonsteroidal anti‐inflammatory drugs; PGD2: prostaglandin D2; TSLP: thymic stromal lymphopoietin; and 5LOX: 5‐lipooxygenase
As the underlying mechanism is related to arachidonic acid metabolism, potential biomarkers focus on determining leukotriene and prostaglandin metabolites. Therefore, urinary LTE4 has been reported as a useful biomarker to distinguish between different subphenotypes in NERD, 252 between NERD and aspirin‐tolerant asthma 252 and phenotypes with skin involvement (NIUA and NECD). 21 Sensitivity and specificity of urinary LTE4 for identifying NERD have been reported to range from 0.55 to 0.81 and 0.77 to 0.82, respectively, which depends on the detection method used. 253 Serum LTE4 and LTE4/PGF2 ratio have also been reported as potential in vitro biomarkers for NERD 254 and urinary PGF2 for NIUA and NECD (Table 2). 21
Table 2.
Biomarkers for drug hypersensitivity
| Biomarker | Disease | References |
|---|---|---|
| LTE4 | Aspirin sensitivity in asthma | 253, 254 |
| LTE4/PGF2 ratio | Aspirin sensitivity in asthma | 254 |
| LTE4 and PGF2 | NIUA and NECD | 21 |
| Tryptase | Acute phase of IgE‐mediated reactions | 229, 239 |
| Skin tests | Immediate and nonimmediate reactions to drugs | 231 |
| Specific IgE | Immediate reactions to betalactams | 240 |
| Basophil activation |
Immediate reactions to betalactams Immediate reactions to FQ |
242, 244 |
| Lymphocyte transformation test | Nonimmediate reactions | 239 |
| Granzyme B and granulysin | Stevens–Johnson syndrome | 238 |
11. BIOMARKERS IN IMMUNE MONITORING OF ALLERGEN‐SPECIFIC IMMUNOTHERAPY
Allergen immunotherapy (AIT) is the only therapy that can alter the natural course of immune responses to specific allergens, directing it toward desensitization and perhaps, toward tolerance. Routes of AIT include subcutaneous (SCIT), sublingual (SLIT), oral (OIT) or epicutaneous (EPIT). 255 , 256 Licensed products are available for therapy of AR, allergic asthma, stinging insect hypersensitivity, AD. 257 , 258 , 259 , 260 , 261 , 262 AIT for food allergy is currently investigational; however, phase 3 trials are promising. 263 Current evidence indicates that immunological changes associated with AIT differ from permanent tolerance as in healthy nonallergic individuals. AIT‐acquired desensitization is often temporary, waning with time, but can be maintained with regular exposure to the allergen. 256
Biomarkers that can identify responders, monitor treatment, predict durability of desensitization and determine adverse event risk would aid clinical decisions and delivery of targeted and effective treatment. With the advent of data‐driven “‐omic” technologies, more potential biomarkers have been identified. 264 Here, we briefly review the more promising candidates that are being evaluated for immune monitoring with an expanded number in Figure 8. 265
FIGURE 8.
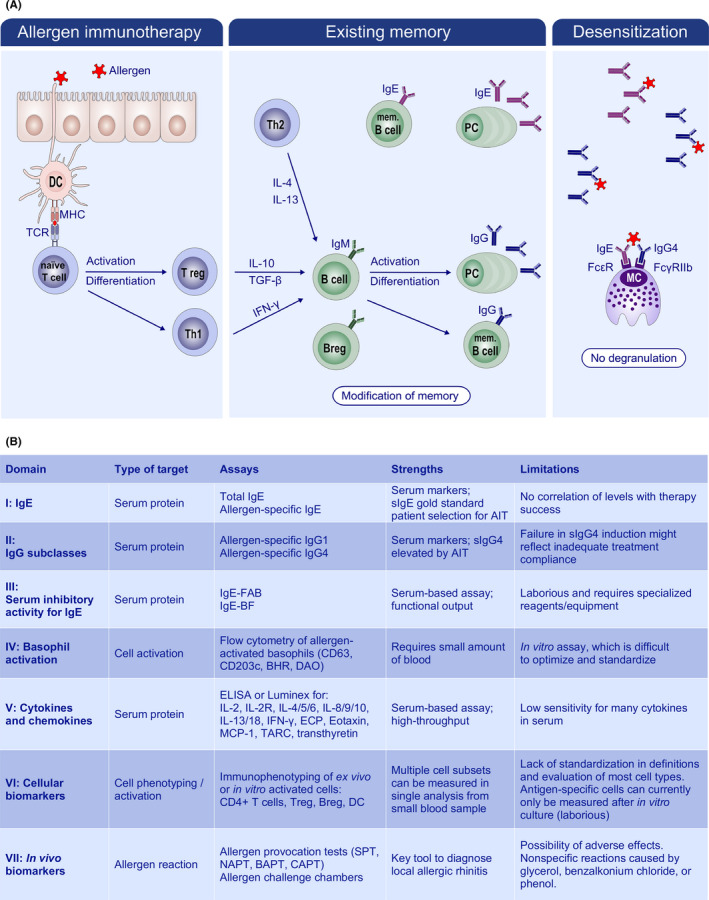
Current view on mechanism and biomarkers in use to monitor AIT. A, Scheme of immune modulation by AIT, where low‐dose, repeated exposure to allergen is thought to occur with limited to no inflammation. As a result, Th skewing is balanced toward Th1 and Treg, which subsequently modify the B‐cell response. In particular, the production of IL‐10 is thought to drive IgG4 class switching. Thus, local and systemic memory is rebalanced, in both the T‐cell and the B‐cell compartments, and there is a strong increase in allergen‐specific IgG4 antibodies. Upon allergen challenge, IgG4 and potentially other soluble factors are thought to inhibit IgE‐mediated degranulation of target cells, i.e, desensitization. Together with the loss of Th2 skewing, this underlies the observed clinical tolerance. B, Laboratory biomarkers utilized in diagnostics and clinical trials for AIT (adapted from 267 ). Abbreviations: IgE‐FAB, IgE‐facilitated allergen binding; IgE‐BF, IgE‐blocking factor; BHR, basophil histamine release; DAO, diamine oxidase; Treg, regulatory T cell; Breg, regulatory B‐cell; DC, dendritic cell. Figure reproduced from 265
11.1. Specific IgE and IgG
With the discovery of IgE in the 1960s, measurements of IgE have been a first step in diagnosis for atopic diseases. Both allergen‐specific IgE (sIgE) and total IgE levels increase during the initial stages of AIT and subsequently decrease 266 ; however, decreases may not accompany a positive clinical outcome. 267 Numerous studies indicate IgG1 and IgG4 levels increase with therapy but do not always differentiate between responders and nonresponders. IgG4 increases during AIT may reflect compliance not clinical efficacy. 267 sIgG4 is purported to be a blocking antibody by competing for allergen binding with IgE bound to Fcε receptors on mast cells and basophils, preventing degranulation. IgA and other subclasses of IgG may have similar blocking function. 268 Recent data support skewing toward IgG2 and IgG4 subclasses after SLIT for temperate grass pollen. 193 The IgG4/IgE ratio may monitor AIT progress and outcome, but has demonstrated conflicting utility. 269 , 270 , 271 Flow cytometry‐based assay (IgE‐FAB) and a solid‐phase assay enzyme‐linked immunosorbent‐facilitated antigen binding (ELIFAB) assay can determine IgE‐inhibitory activity. Although robust with good clinical efficacy correlation for AIT, these technologies are complex with usage limited to specialized centers. 267 , 272 , 273
11.2. Immunophenotyping of immune cells
Allergic sensitization is associated with multiple changes in blood immune cells, 187 and high‐dimensional immunophenotyping using flow and mass spectrometry (i.e, Cytometry Time of Flight; CyTOF) has contributed greatly to this identification. 274 A study on bee venom AIT provided detailed characterization of allergen‐specific B cells before and after bee venom tolerance in allergic patients and bee‐keepers with Api m 1‐specific B cells showing increased CCR5 expression after high‐dose allergen exposure. 275 In a study of OIT for milk allergy, researchers found, using flow cytometry, a significant increase in blood invariant natural killer cells and a shift from a Type 2 T helper (Th‐2; i.e, IL‐4, IL‐13) to a Th‐1 (ie, IFN‐γ) cytokine profile. 276 Similar studies in other immunotherapy models can assist with determining potential biomarkers for monitoring AIT.
Recent evidence shows that AIT modulates the balance between circulating T follicular helper (Tfh) and regulatory cells (Tfr), with Tfr as a potential biomarker for AIT efficacy. 277 , 278 Upregulation of the activated allergen‐specific Tregs and downregulation of dysfunctional allergen‐specific Treg cell subset, associated with improved clinical response, were recently described. 279 Responder status was shown to be associated with increased frequency of IgA‐ and IgG4‐expressing allergen‐specific B cells, plasmablasts and IL‐10+ and/or IL‐1RA + Breg cells. 280
AIT‐induced T regulatory cells secreting IL‐35 (iTR35) cells promote production of IL‐10 from CD19 + B cells, Breg subsets and Tfr cells. 281 Circulating Tfr cells share properties of memory cells and are distinct from their lymph nodes (LN) counterpart as they suppress B and Tfh cells with a much lower capacity, while circulating memory‐like Tfh cells are more potent than LN effector Tfh cells. 282 In addition, circulating memory‐like Tfr cells persist for long periods; thus, they could support the long‐term immunomodulatory effect of peptide AIT. Besides the accumulative effects, AIT also activated Treg functions. Treg cells isolated from Dermatophagoides pteronyssinus (DP) AR patients treated with DP‐SCIT for 12 months could suppress the function and proliferation of Th2 and Th9 cells, but increased the proportion of IL‐10‐producing cells. 283 Functional evaluation of T regulatory cells via expression of glycoprotein A repetitions predominant (GARP) and special AT‐rich sequence binding protein 1 (SATB1) is also interesting as a future potential biomarker. Tregs uniquely express GARP on their cell surface, and GARP functions as a delivery system for latent TGF‐β, which might augment the immunosuppressive role of Tregs on effector cells. 284 GARP expression was described as an activation marker of parasitic infection‐induced Tregs that strongly suppress allergic inflammation; thus, it is a novel potential mechanistic pathway for AIT. SATB1 is a genome organizer protein expressed in a lineage‐specific manner in CD4 + T‐cells. During the early Th2 cell differentiation, IL‐5 expression is repressed through direct binding of SATB1 to the IL‐5 promoter. Thus, SATB1 modulation might expand the impact of AIT on eosinophilic inflammation. In addition, SATB1‐dependent Treg‐cell‐specific super‐enhancers activation is crucial for Treg cell lineage specification in the thymus.
11.3. Basophil activation test
Blood basophils express high levels of FcεRI that have bound IgE. To determine sensitization, basophils are incubated in vitro with allergen, followed by examination of degranulation. Fusion of granules to the cell membrane leads to upregulation of surface markers CD63 and CD203c, which are indicative of specific IgE functional activity. In patients with AR undergoing SLIT, basophil activation decreased with clinical efficacy. 285 Similar decreases occurred in patients undergoing peanut immunotherapy. 271 However, in grass pollen SLIT, basophil activation did not correlate with clinical efficacy (Table 3). 286
Table 3.
Identification of changes in immune cells associated with allergic sensitization
| Immune cell type | Biomarkers | Assay | References |
|---|---|---|---|
| Basophils | Decrease in basophil activation post‐allergen‐specific OIT (CD63, CD203, histamine release) | BAT, iBAT | 287, 288, 289 |
| B cells | Induction of Breg cells on AIT, serological readouts: food, and allergen‐specific IgE, IgG4 epitope‐specific IgE and IgG4 | Flow cytometry, ImmunoCAP, epitope mapping | 182, 271, 280, 290 |
| T cells | Functional changes in tetramer positive/CFSElo/CD69 + CD40L+ allergen‐specific CD4 + T‐cells, tracking Th2A cells Decrease in T‐cell‐derived Type 2 cytokines Epigenetic changes in T‐cell subsets (Tregs) | Flow and mass cytometry, Luminex/ELISA, pyrosequencing | 291, 292, 293, 294, 295, 296 |
| Dendritic cells | Changes in costimulatory potential (CD80, CD86) | Flow cytometry, DC‐T cell co‐culture | 297, 302 |
11.4. Soluble cytokines
With immunotherapy, there is redirection of Th2 phenotype toward a Th1 and Treg phenotype with decreases in Th2 cytokines (eg, IL‐4, IL‐19 13, IL‐9) and upregulation of Th1 (eg, IFNγ) and regulatory cytokines (eg, TGFβ and IL‐10). 193 , 298 However, serum cytokine measurements are challenging due to low levels, which are often below the detection limit of current methods. 267 Changes in cytokine production by CD4 T cells following AIT are assessable through in vitro stimulation of PBMC from patients using allergen extracts, at both the protein and the transcript levels. In T‐cell of patients with HDM allergy, high levels of IL10 transcripts predicted immunotherapy success. 299
11.5. Epigenetic biomarkers
Beyond cells, proteins and transcripts, epigenetic biomarkers may suggest prognosis. 300 , 301 DNA methylation of promoter regions is associated with gene silencing, and FOXP3 gene expression is in part controlled through this. FoxP3 is the master regulator of Treg cells, and Syed et al found that peanut OIT resulted in hypomethylation of FOXP3, indicating increased gene expression with immunotherapy. 302
In conclusion, technological advances inform novel methods for monitoring immune responses with increased sensitivity and specificity. Studies indicate dysregulation of a number of molecular markers with AIT. Detection method optimization continues and should facilitate precision immunotherapy.
12. BIOMARKERS OF THE EPITHELIAL BARRIER
Environmental factors, microbiome, epithelial cells and immune cells show a dynamic cross talk at the skin and mucosal barriers in the development of AD, AR, CRS, eosinophilic esophagitis and asthma. 174 , 303 , 304 , 305 , 306 , 307 , 308 Studies on the pathogenesis of these diseases have clearly demonstrated a barrier defect in the skin and involved mucosas and a systemic inflammation. 307 , 308 , 309 , 310 Defects in the epithelial barriers, caused by both environmental risk factors and a genetic predisposition, may represent the starting point of a chronic inflammation and allergen sensitization. 307 , 308 , 309 , 310 A significant number of studies have reported that environmental factors directly affect the barrier function of epithelium. 162 , 311 , 312 In addition, T helper 2 cells, ILC2s and their cytokine IL‐13 damage skin and lung barriers. 307 , 313 In addition, mast cells and their enzyme chymase damage the epithelial barrier. 304 , 314 The effects of environmental factors may, at least in part, be mediated by epigenetic mechanisms. Histone deacetylase activation by Type 2 immune response has a major effect on leaky barriers, and blocking of histone deacetylase activity corrects the defective barrier in human air–liquid interface cultures and mouse models of allergic asthma with rhinitis. 107 , 307 , 315 The assessment of the barrier function of the skin and mucosas in vivo has an extremely high value in the clinics to identify barrier leakiness for an individual patient and requires the discovery of biomarkers.
The value of barrier detection in allergic diseases.
Early prediction of atopic dermatitis development in babies
Patient selection for certain therapies, endotyping and personalized medicine
Early prediction and monitoring response to treatments
Early and rapid analyses of environmental hazards to skin and mucosas before severe lesions start
To date, there are a few noninvasive methods to assess the skin epidermal barrier function in vivo. The quantification of transepidermal water loss (TEWL) in the skin across the stratum corneum has received some interest for early prediction of atopy prone children and detection of skin barrier. 316 Although TEWL increases in proportion to the level of damage, it is also affected by environmental factors such as humidity, temperature, season and moisture content of the skin. 317 The noninvasive and rapid measurement of natural moisturizing factor by Raman spectroscopy provides a method suitable for use in children. The association of natural moisturizing factor, filaggrin null mutations and AD suggests Raman spectroscopy as a promising approach for stratification of endotypes in AD in the clinics. Other noninvasive methods currently used include assessment of the stratum corneum hydration, colorimetry, skin surface pH and sebometry, but they only provide information on different characteristics and/or the condition of the skin and do not directly measure the barrier function. 318
Recently, electrical impedance spectroscopy (EIS) has been identified as an effective and stable tool for the detection of epidermal barrier. 175 This method works by transmitting a harmless electrical signal through the skin at several depths and frequencies and measuring the electrical resistance and impedance response of the tissue. EIS reflects particularly the tissue barrier status by collecting the electric impedance information from extracellular and intracellular tissue. 319 Recently, Rinaldi et al have studied the impairment of the epidermal barrier in mice by the epicutaneous administration of barrier‐damaging proteases, such as papain, trypsin, Vibrio cholera toxin or by tape stripping. 175 They showed the barrier‐damaging effects of these substances which correlate with the electric conductivity of the skin, causing a decrease in electrical impedance. According to these results, EIS shows a broad range of possible clinical applications in AD and atopic march, including early prediction of atopy‐prone children, early diagnosis of the disease, stratification and endotyping of patients, evaluation of the overall therapy response as well as single lesions and assessment of disease severity.
A critical feature of the gastrointestinal epithelium is intestinal barrier permeability as it must allow an efficient passage of nutrients while restricting the entry of larger molecules to avoid food allergy development. Multiple autoimmune diseases have been identified to arise or be exacerbated by a leaky gut, such as in inflammatory bowel disease, celiac disease, Type 1 diabetes, systemic lupus erythematosus, multiple sclerosis. 320 , 321 , 322 , 323 Recent studies point to a leaky gut as the initiator of Type 1 diabetes because a subclinical intestinal barrier dysfunction was already detected before clinical onset of Type 1 diabetes. 324 Zonulin is a prehaptoglobulin protein and a biomarker for gut barrier leakiness that downregulates TJ function, and it has been proposed to play a role in several autoimmune diseases. 325
Translocation of bacterial endotoxin (lipopolysaccharide, LPS) from the gut microbiota to blood circulation stimulates systemic inflammatory responses. 326 Measurement of intestinal permeability is often used in the examination of inflammatory gastrointestinal disorders. It can be assessed by measurement of urinary recovery of ingested nonmetabolizable lactulose and mannitol. Urine L/M ratio measured by 1 H NMR spectroscopy showed high correlation with the standard measurement of the urinary recoveries by enzymatic assays. In conclusion, NMR metabolomics enables simultaneous intestinal permeability testing and discovery of biomarkers associated with an impaired intestinal permeability. 327 In conclusion, identification of clinically reliable biomarkers for skin, respiratory and intestinal barrier measurements represents an important future research area.
13. IN VIVO BIOMARKERS IN AIT DEVELOPMENT PROGRAMS: EUROPEAN IMPLICATIONS
An increasing number of clinical trials in AIT have been published underlining both efficacy and safety of AIT as the only disease‐modifying treatment option for patients with IgE‐mediated respiratory allergic diseases. 328 With country‐specific exceptions, AIT products are regulated by authorities such as the European Medicines Agency 329 and others on the basis of methodological guidelines.
In its guideline “Clinical Development of Products for Specific Immunotherapy for The Treatment of Allergic Diseases”, 329 the European regulatory authority (European Medicine Agency (EMA)) has provided strict guidance for designing and performing clinical development programs in AIT. 113 , 330 In Germany, these principles were followed in the “Therapy Allergen Ordinance (TAV)“ for future registration of allergens and allergen mixtures (derived from grass pollen, early flowering trees pollen, house dust mites, and bee and wasp venom) based on the main prevalent respiratory allergies in Germany. 331 , 332 , 333 First registrations have been granted fulfilling the TAV demands by the German Paul Ehrlich Institut (PEI). 334 Besides, a sufficient body of evidence exists in the clinical documentation of both SLIT and SCIT AIT products. 335 , 336 , 337
An important unmet need for further harmonization of methodological principles in AIT study design 338 has been followed in a series of Task‐Force initiatives of the European Academy of Allergy and Clinical Immunology (EAACI) 339 as overviewed in the 2019 report of our group. 113 As such, position papers and guidelines on allergen‐challenge procedures including clear standardization of procedures (SOPs) through the nasal 340 or conjunctival 341 route have been provided by the EAACI. As aforementioned, the safety, reproducibility and cutoff points for NAC have also been proven recently. 96 , 97
In addition, the combined symptom and medication score (CSMS) as defined by the EAACI as standard primary endpoint for future (pivotal) trials in AIT 342 has been recently used recently in an increasing number of key trials in AIT. 343 , 344 , 345 However, further formal validation and amendments especially for the pediatric population are needed. 334 , 346 As another example, a further Task Force initiative aimed to provide clear clinically and aerobiologically justified definitions of pollen counts for onset, peak and duration of pollen seasons 347 and they have been confirmed to be robust 348 and clinically relevant as reflecting patients’ symptom load in different countries in Europe in recent reports. 348 , 349 Recently, the EAACI has published a Position Paper reporting the impact of the placebo effect in AIT from different methodological perspectives and outlining possible strategies to minimize this bias in clinical trials. 350
Taken together, further emphasis should be put on international collaborations of clinical experts, methodologists and regulatory authorities to optimize methodological standards for AIT clinical development programs aimed to increase the level of evidence of AIT as the only disease‐modifying therapy available. 334 , 337 , 351
14. BIOMARKERS ON THE CLINICAL USAGE OF BIOLOGICALS IN THE EAACI
The EAACI recently published the Guidelines on the use of biologicals in severe asthma. Recommendations were formulated following the GRADE approach 352 , 353 , 354 for each biological (benralizumab, dupilumab, mepolizumab, omalizumab, reslizumab) and each outcome (decrease in asthma exacerbations, oral and inhaled corticosteroids and rescue medication use; improvement in asthma control, quality of life and lung function; and safety). 33 The recommendations were informed by three systematic reviews. 355 , 356 , 357 FeNO, sputum and blood eosinophils were scored as outcomes with low importance; however, the evidence was analyzed for subgroups based on biomarkers and comorbidities. The higher the blood eosinophils, the higher the expected impact of benralizumab, dupilumab and mepolizumab in reducing severe asthma exacerbations. The improvement in asthma control was significantly better in the high‐eosinophil subgroup for benralizumab and reslizumab. Lung function increased significantly more in the high eosinophil subgroup for benralizumab, dupilumab and reslizumab. Benralizumab improved QoL better in the high‐eosinophil subgroup. The effect of omalizumab on exacerbations did not depend on blood eosinophils. Neither the atopic status nor total IgE predicts the magnitude of effect of benralizumab, and serum IgE thresholds (within regulatory limits) did not influence the response to omalizumab (Table 4).
Table 4.
Current biologicals, biomarkers, asthma outcomes and GRADE recommendation
| Biological | Asthma‐related outcome | Biomarkers | Recommendation as per GRADE methodology |
|---|---|---|---|
|
Benralizumab Dupilumab Reslizumab |
Decrease in severe asthma exacerbations | The higher the blood eosinophils, the higher the expected impact | Conditional recommendation |
| Omalizumab | Decrease in severe asthma exacerbations | The effect does not depend on blood eosinophils | Conditional recommendation |
|
Benralizumab Reslizumab |
Improvement in asthma control | The higher the blood eosinophils, the higher the expected impact | Conditional recommendation |
| Benralizumab | Improvement in asthma‐related QoL | The higher the blood eosinophils, the higher the expected impact | Conditional recommendation |
| Dupilumab | Improvement in lung function (FEV1) | Clinically significant effect in asthmatic and adolescent adults with blood eosinophils > 300 cells/μL and/or with FeNO levels > 50 ppb | Strong recommendation |
|
Benralizumab Reslizumab |
Improvement in lung function (FEV1) | The higher the blood eosinophils, the higher the expected impact | Conditional recommendation |
| Benralizumab | All asthma‐related outcomes | Neither the atopic status or total IgE predicts the magnitude of effect | Conditional recommendation |
| Omalizumab | All asthma‐related outcomes | Serum IgE thresholds (within regulatory limits) do not influence the response | |
| HDM immunotherapy | All asthma‐related outcomes | No biomarkers sufficiently predict response to HDM‐AIT in HDM‐driven allergic asthma | Conditional recommendation |
In addition, biomarkers were used to characterize the population where a strong recommendation is made instead of a conditional one, for example for dupilumab in improving lung function for asthmatic and adolescent adults with blood eosinophils > 300 cells/μL and/or with FeNO levels > 50 ppb. This upgrade was possible as in this subgroup the addition of dupilumab to standard of care improved FEV1 above the minimal important difference threshold.
The Guideline Development Group (GDG) formulated a three‐pillar decision tree aiming to help the clinician in reaching the decision to start on a particular biological based on the combination between phenotypic traits, biomarkers (blood and sputum eosinophils, FeNO) and clinically relevant asthma‐related endpoints. For suboptimal response, the GDG recommends to re‐assess airway inflammation and airway hyperresponsiveness (conditional recommendation, expert opinion based). Induced sputum is the preferred option to re‐assess airway inflammation (conditional recommendation, expert opinion based) as a noninvasive validated tool. The GDG strongly recommends joint efforts from academia, industry and healthcare systems to develop both educational tools and resources supporting a wider use of induced sputum evaluation in severe asthma. If at re‐evaluation for suboptimal response there is no airway eosinophilia and neutrophilic inflammation is present, the biological should be interrupted and measures addressing non‐T2 asthma such as macrolides should be considered (conditional recommendation, expert opinion based). In cases with no airway inflammation, addressing airway hyperresponsiveness (LABA/LAMA combinations or bifunctional drugs) or airway remodeling (bronchial thermoplasty in selected cases) is recommended (conditional recommendation, expert opinion based).
Identification of clinically relevant biomarkers in order to select responders to the currently available biologicals was identified as high priority, and proof‐of‐concept prospective studies evaluating patient selection based on biomarkers were proposed as a tool to overcome this barrier in severe asthma research.
Another recently published EAACI Guideline formulated recommendations for the use of AIT with HDM for HDM‐driven allergic asthma. To date, there are no biomarkers that sufficiently predict response to HDM‐AIT that can be used to decide on initiation or cessation of HDM AIT in HDM‐driven allergic asthma. 358
15. CONCLUSIONS
In our discipline, a huge amount of omics research has been employed during recent years that needs to be analyzed in depth. Allergic diseases are a group of complex and heterogeneous diseases. Specific, sensitive and reliable (point‐of‐care) biomarkers are critical for selecting the proper treatment for the proper patient and enabling precision medicine. Precision endotyping is linking pathobiological mechanisms with visible properties via specific biomarkers. It is expected to be translated into pathway‐specific diagnostic tests and opens a pathway to accurate disease classification and individualized targeted treatment options. We are expecting to change the diagnostic and therapeutic landscape in our specialty as well as of all medical disciplines. However, endotype‐driven treatment still needs to face multiple challenges before its implementation in daily practice in allergic diseases and asthma. Most of the disease endotyping research is conducted in severe forms of allergic diseases nowadays. Over the past few years, the biomarker discovery field has gained great advances, which facilitates the identification of novel methods for disease diagnosis and therapy. With these fast growing efforts, we can imagine that in the near future, we will be able to find better ways to treat allergic diseases, with a therapeutic approach targeted to the individual patient.
CONFLICTS OF INTEREST
Author CA reports grants from Allergopharma, Idorsia, Swiss National Science Foundation, Christine Kühne‐Center for Allergy Research and Education, European Commission's Horizon's 2020 Framework Programme, Cure, Novartis Research Institutes, Astra Zeneca, Scibase and others from Sanofi/Regeneron are outside the submitted work. Author ZD reports having acted in the past 3 years as Executive and Scientific Medical Director at a phase I/II pharmacological unit (QPS‐NL), which performs clinical studies for pharmaceutical companies. In addition, ZD received honoraria, consultancy and speaker fees from Acucort, Astrazeneca, ALK, Aquilon, Boehringer Ingelheim, CSL, HAL Allergy, MSD, Sanofi‐Genzyme. Author TE reports grants or others from DBV, Innovation fund Denmark, and Regeneron are outside the submitted work. TE is the Co‐I or scientific leader in three investigator‐initiated oral immunotherapy trials supported by the Allergy and Anaphylaxis Program Sickkids, and he is on the local advisory board for ALK. Author KN reports grants and/or personal fees from Allergenis, Ukko Pharma, EAT, NIAID, Novartis, and FARE, Sanofi, Astellas, Nestle, BeforeBrands, Alladapt, ForTra, Genentech, AImmune Therapeutics, DBV Technologies, AnaptysBio, Adare Pharmaceuticals, Stallergenes‐Greer, NHLBI, NIEHS, EPA, WAO Center of Excellence, Iggenix, Probio, Vedanta, Centecor, Seed, Immune Tolerance Network, NIH, Regeneron, Astrazeneca, ImmuneWorks and Cour Pharmaceuticals are outside the submitted work. In addition, KN has patents for Inhibition of Allergic Reaction to Peanut Allergen using an IL‐33 Inhibitor pending, Special Oral Formula for Decreasing Food Allergy Risk and Treatment for Food Allergy pending, Basophil Activation Based Diagnostic Allergy Test pending, Granulocyte‐based methods for detecting and monitoring immune system disorders pending, Methods and Assays for Detecting and Quantifying Pure Subpopulations of White Blood Cells in Immune System Disorders pending, Mixed Allergen Compositions and Methods for Using the Same pending, and Microfluidic Device and Diagnostic Methods for Allergy Testing Based on Detection of Basophil Activation pending. Author LO’M reports grants and/or personal fees from AHL and GSK are outside the submitted work. Author OP reports grants and/or personal fees from ALK‐Abelló, Allergopharma, Stallergenes Greer, HAL Allergy Holding BV/HAL Allergie GmbH, Bencard Allergie GmbH/Allergy Therapeutics, Lofarma, ASIT Biotech Tools SA, Laboratorios LETI/LETI Pharma, Anergis SA, Biomay, Circassia, Glaxo Smith Kline, MEDA Pharma/MYLAN, Mobile Chamber Experts (a GA2LEN Partner), Indoor Biotechnologies, Astellas Pharma Global, EUFOREA, ROXALL, Novartis, Sanofi Aventis, Med Update Europe GmbH, streamedup! GmbH are outside the submitted work. Author WF reports grants and personal fees from Sanofi, Novartis, GSK during the conduct of the study; grants from Mylan, ALK, Allergy Therapeutics, Chordate are outside the submitted work. Author CTH reports personal fees from Novartis, Sanofi, Lilly Pharma, Töpfer GmbH, Bencard, Danone Nutricia, Lancôme and L’Oréal are outside the submitted work. Authors DW, HB, IA, LZ, MJT, RO’H and YP report no conflicts of interest in relation to this work. Authors CA, CTH, DW, HB, IA, KN, LO’M, LZ, MJT, OP, RO’H, TE, WF, ZD are editors of Allergy.
ACKNOWLEDGMENTS
The authors thank Anna Globinska for expertly preparing the figures.
Breiteneder H, Peng Y‐Q, Agache I, et al. Biomarkers for diagnosis and prediction of therapy responses in allergic diseases and asthma. Allergy. 2020;75:3039–3068. 10.1111/all.14582
REFERENCES
- 1. Galli SJ, Tsai M, Piliponsky AM. The development of allergic inflammation. Nature. 2008;454(7203):445‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Samitas K, Carter A, Kariyawasam HH, Xanthou G. Upper and lower airway remodelling mechanisms in asthma, allergic rhinitis and chronic rhinosinusitis: the one airway concept revisited. Allergy. 2018;73(5):993‐1002. [DOI] [PubMed] [Google Scholar]
- 3. Barbarot S, Auziere S, Gadkari A, et al. Epidemiology of atopic dermatitis in adults: results from an international survey. Allergy. 2018;73(6):1284‐1293. [DOI] [PubMed] [Google Scholar]
- 4. Fokkens WJ, Lund VJ, Hopkins C, et al. European position paper on rhinosinusitis and nasal polyps 2020. Rhinology. 2020;58(Suppl S29):1‐464. [DOI] [PubMed] [Google Scholar]
- 5. De Meulder B, Lefaudeux D, Bansal AT, et al. A computational framework for complex disease stratification from multiple large‐scale datasets. BMC Syst Biol. 2018;12(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agache I, Sugita K, Morita H, Akdis M, Akdis CA. The complex type 2 endotype in allergy and asthma: from laboratory to bedside. Curr Allergy Asthma Rep. 2015;15(6):29. [DOI] [PubMed] [Google Scholar]
- 7. Agache I, Rogozea L. Asthma biomarkers: do they bring precision medicine closer to the clinic? Allergy Asthma Immunol Res. 2017;9(6):466‐476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diamant Z, Vijverberg S, Alving K, et al. Toward clinically applicable biomarkers for asthma: an EAACI position paper. Allergy. 2019;74(10):1835‐1851. [DOI] [PubMed] [Google Scholar]
- 9. Agache I, Strasser DS, Klenk A, et al. Serum IL‐5 and IL‐13 consistently serve as the best predictors for the blood eosinophilia phenotype in adult asthmatics. Allergy. 2016;71(8):1192‐1202. [DOI] [PubMed] [Google Scholar]
- 10. Yii ACA, Tay TR, Choo XN, Koh MSY, Tee AKH, Wang DY. Precision medicine in united airways disease: a "treatable traits" approach. Allergy. 2018;73(10):1964‐1978. [DOI] [PubMed] [Google Scholar]
- 11. Bachert C, Zhang N. Medical algorithm: diagnosis and treatment of chronic rhinosinusitis. Allergy. 2020;75(1):240‐242. [DOI] [PubMed] [Google Scholar]
- 12. Cardona V, Demoly P, Dreborg S, et al. Current practice of allergy diagnosis and the potential impact of regulation in Europe. Allergy. 2018;73(2):323‐327. [DOI] [PubMed] [Google Scholar]
- 13. Diamant Z, Vijverberg SJ, Agache I, et al. Much ado about biologicals: highlights of the master class on biologicals, Prague, 2018. Allergy. 2019;74(4):837‐840. [DOI] [PubMed] [Google Scholar]
- 14. Eguiluz‐Gracia I, Tay TR, Hew M, et al. Recent developments and highlights in biomarkers in allergic diseases and asthma. Allergy. 2018;73(12):2290‐2305. [DOI] [PubMed] [Google Scholar]
- 15. Guerra ENS, Acevedo AC, de Toledo IP, Combes A, Chardin H. Do mucosal biomarkers reveal the immunological state associated with food allergy? Allergy. 2018;73(12):2392‐2394. [DOI] [PubMed] [Google Scholar]
- 16. Chen LC, Tseng HM, Kuo ML, et al. A composite of exhaled LTB(4), LXA(4), FeNO, and FEV(1) as an "asthma classification ratio" characterizes childhood asthma. Allergy. 2018;73(3):627‐634. [DOI] [PubMed] [Google Scholar]
- 17. Rodrigo‐Muñoz JM, Cañas JA, Sastre B, et al. Asthma diagnosis using integrated analysis of eosinophil microRNAs. Allergy. 2019;74(3):507‐517. [DOI] [PubMed] [Google Scholar]
- 18. Siroux V, Boudier A, Nadif R, Lupinek C, Valenta R, Bousquet J. Association between asthma, rhinitis, and conjunctivitis multimorbidities with molecular IgE sensitization in adults. Allergy. 2019;74(4):824‐827. [DOI] [PubMed] [Google Scholar]
- 19. Lunjani N, Satitsuksanoa P, Lukasik Z, Sokolowska M, Eiwegger T, O'Mahony L. Recent developments and highlights in mechanisms of allergic diseases: microbiome. Allergy. 2018;73(12):2314‐2327. [DOI] [PubMed] [Google Scholar]
- 20. Su MW, Lin WC, Tsai CH, et al. Childhood asthma clusters reveal neutrophil‐predominant phenotype with distinct gene expression. Allergy. 2018;73(10):2024‐2032. [DOI] [PubMed] [Google Scholar]
- 21. Dona I, Jurado‐Escobar R, Perkins JR, et al. Eicosanoid mediator profiles in different phenotypes of nonsteroidal anti‐inflammatory drug‐induced urticaria. Allergy. 2019;74(6):1135‐1144. [DOI] [PubMed] [Google Scholar]
- 22. Liao B, Liu JX, Li ZY, et al. Multidimensional endotypes of chronic rhinosinusitis and their association with treatment outcomes. Allergy. 2018;73(7):1459‐1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Asthma GIf. https://ginasthma.org/. accessed June 30, 2020
- 24. Seys SF, Quirce S, Agache I, et al. Severe asthma: Entering an era of new concepts and emerging therapies: highlights of the 4th international severe asthma forum, Madrid, 2018. Allergy. 2019;74(11):2244‐2248. [DOI] [PubMed] [Google Scholar]
- 25. Kaur R, Chupp G. Phenotypes and endotypes of adult asthma: moving toward precision medicine. J Allergy Clin Immunol. 2019;144(1):1‐12. [DOI] [PubMed] [Google Scholar]
- 26. Fahy JV. Type 2 inflammation in asthma–present in most, absent in many. Nat Rev Immunol. 2015;15(1):57‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fokkens WJ, Lund V, Bachert C, et al. EUFOREA consensus on biologics for CRSwNP with or without asthma. Allergy. 2019;74(12):2312‐2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kowalski ML, Agache I, Bavbek S, et al. Diagnosis and management of NSAID‐exacerbated respiratory disease (N‐ERD)‐a EAACI position paper. Allergy. 2019;74(1):28‐39. [DOI] [PubMed] [Google Scholar]
- 29. Roth‐Walter F, Adcock IM, Benito‐Villalvilla C, et al. Comparing biologicals and small molecule drug therapies for chronic respiratory diseases: An EAACI taskforce on immunopharmacology position paper. Allergy. 2019;74(3):432‐448. [DOI] [PubMed] [Google Scholar]
- 30. Woodruff PG, Boushey HA, Dolganov GM, et al. Genome‐wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA. 2007;104(40):15858‐15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alving K, Diamant Z, Lucas S, et al. Point‐of‐care biomarkers in asthma management: time to move forward. Allergy. 2020;75(4):995‐997. [DOI] [PubMed] [Google Scholar]
- 32. Holguin F, Cardet JC, Chung KF, et al. Management of severe asthma: a European respiratory society/American thoracic society guideline. Eur Respir J. 2020;55(1):1900588 10.1183/13993003.00588-2019 [DOI] [PubMed] [Google Scholar]
- 33. Agache I, Akdis C, Akdis M, et al. EAACI Biologicals Guidelines ‐ Recommendations for severe asthma. Allergy. 2020. 10.1111/all.14425 [DOI] [PubMed] [Google Scholar]
- 34. Diamant Z, Boot JD, Mantzouranis E, Flohr R, Sterk PJ, Gerth van Wijk R. Biomarkers in asthma and allergic rhinitis. Pulm Pharmacol Ther. 2010;23(6):468‐481. [DOI] [PubMed] [Google Scholar]
- 35. Calhoun WJ, Reed HE, Moest DR, Stevens CA. Enhanced superoxide production by alveolar macrophages and air‐space cells, airway inflammation, and alveolar macrophage density changes after segmental antigen bronchoprovocation in allergic subjects. Am Rev Respir Dis. 1992;145(2 Pt 1):317‐325. [DOI] [PubMed] [Google Scholar]
- 36. Comhair SA, Bhathena PR, Dweik RA, Kavuru M, Erzurum SC. Rapid loss of superoxide dismutase activity during antigen‐induced asthmatic response. Lancet. 2000;355(9204):624. [DOI] [PubMed] [Google Scholar]
- 37. Comhair SAA, Erzurum SC. Antioxidant responses to oxidant‐mediated lung diseases. Am J Physiol Lung Cell Mol Physiol. 2002;283(2):L246‐L255. [DOI] [PubMed] [Google Scholar]
- 38. Aldakheel FM, Thomas PS, Bourke JE, Matheson MC, Dharmage SC, Lowe AJ. Relationships between adult asthma and oxidative stress markers and pH in exhaled breath condensate: a systematic review. Allergy. 2016;71(6):741‐757. [DOI] [PubMed] [Google Scholar]
- 39. Thomas PS, Lowe AJ, Samarasinghe P, et al. Exhaled breath condensate in pediatric asthma: promising new advance or pouring cold water on a lot of hot air? A systematic review. Pediatr Pulmonol. 2013;48(5):419‐442. [DOI] [PubMed] [Google Scholar]
- 40. McDowell PJ, Heaney LG. Different endotypes and phenotypes drive the heterogeneity in severe asthma. Allergy. 2020;75(2):302‐310. [DOI] [PubMed] [Google Scholar]
- 41. Sze E, Bhalla A, Nair P. Mechanisms and therapeutic strategies for non‐T2 asthma. Allergy. 2020;75(2):311‐325. [DOI] [PubMed] [Google Scholar]
- 42. Taylor SL, Leong LEX, Choo JM, et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J Allergy Clin Immunol. 2018;141(1):94‐103. [DOI] [PubMed] [Google Scholar]
- 43. Green BJ, Wiriyachaiporn S, Grainge C, et al. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One. 2014;9(6):e100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tliba O, Panettieri RA Jr. Paucigranulocytic asthma: uncoupling of airway obstruction from inflammation. J Allergy Clin Immunol. 2019;143(4):1287‐1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18(5):716‐725. [DOI] [PubMed] [Google Scholar]
- 46. Agusti A, Bafadhel M, Beasley R, et al. Precision medicine in airway diseases: moving to clinical practice. Eur Respir J. 2017;50(4):1701655. [DOI] [PubMed] [Google Scholar]
- 47. Chung KF, Adcock IM. Precision medicine for the discovery of treatable mechanisms in severe asthma. Allergy. 2019;74(9):1649‐1659. [DOI] [PubMed] [Google Scholar]
- 48. Simpson AJ, Hekking PP, Shaw DE, et al. Treatable traits in the European U‐BIOPRED adult asthma cohorts. Allergy. 2019;74(2):406‐411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lefaudeux D, De Meulder B, Loza MJ, et al. U‐BIOPRED clinical adult asthma clusters linked to a subset of sputum omics. J Allergy Clin Immunol. 2017;139(6):1797‐1807. [DOI] [PubMed] [Google Scholar]
- 50. Schofield JPR, Burg D, Nicholas B, et al. Stratification of asthma phenotypes by airway proteomic signatures. J Allergy Clin Immunol. 2019;144(1):70‐82. [DOI] [PubMed] [Google Scholar]
- 51. Ivanova O, Richards LB, Vijverberg SJ, et al. What did we learn from multiple omics studies in asthma? Allergy. 2019;74(11):2129‐2145. [DOI] [PubMed] [Google Scholar]
- 52. Dunican EM, Elicker BM, Gierada DS, et al. Mucus plugs in patients with asthma linked to eosinophilia and airflow obstruction. J Clin Investig. 2018;128(3):997‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Erjefalt JS. Unravelling the complexity of tissue inflammation in uncontrolled and severe asthma. Curr Opin Pulm Med. 2019;25(1):79‐86. [DOI] [PubMed] [Google Scholar]
- 54. Walter J, O'Mahony L. The importance of social networks‐An ecological and evolutionary framework to explain the role of microbes in the aetiology of allergy and asthma. Allergy. 2019;74(11):2248‐2251. [DOI] [PubMed] [Google Scholar]
- 55. Savage JH, Lee‐Sarwar KA, Sordillo J, et al. A prospective microbiome‐wide association study of food sensitization and food allergy in early childhood. Allergy. 2018;73(1):145‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kozik A, Huang YJ. Ecological interactions in asthma: from environment to microbiota and immune responses. Curr Opin Pulm Med. 2020;26(1):27‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sokolowska M, Frei R, Lunjani N, Akdis CA, O'Mahony L. Microbiome and asthma. Asthma Res Pract. 2018;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sbihi H, Boutin RC, Cutler C, Suen M, Finlay BB, Turvey SE. Thinking bigger: How early‐life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy. 2019;74(11):2103‐2115. [DOI] [PubMed] [Google Scholar]
- 59. Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. [DOI] [PubMed] [Google Scholar]
- 60. Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22(10):1187‐1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bannier M, van Best N, Bervoets L, et al. Gut microbiota in wheezing preschool children and the association with childhood asthma. Allergy. 2020;75(6):1473‐1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thorsen J, Rasmussen MA, Waage J, et al. Infant airway microbiota and topical immune perturbations in the origins of childhood asthma. Nat Commun. 2019;10(1):5001 10.1038/s41467-019-12989-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhou Y, Jackson D, Bacharier LB, et al. The upper‐airway microbiota and loss of asthma control among asthmatic children. Nat Commun. 2019;10(1):5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang YJ, Nariya S, Harris JM, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. 2015;136(4):874‐884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Michalovich D, Rodriguez‐Perez N, Smolinska S, et al. Obesity and disease severity magnify disturbed microbiome‐immune interactions in asthma patients. Nat Commun. 2019;10(1):5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roduit C, Frei R, Ferstl R, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. 2019;74(4):799‐809. [DOI] [PubMed] [Google Scholar]
- 67. Lewis G, Wang B, Shafiei Jahani P, et al. Dietary fiber‐induced microbial short chain fatty acids suppress ILC2‐dependent airway inflammation. Front Immunol. 2019;10:2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Barcik W, Pugin B, Westermann P, et al. Histamine‐secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol. 2016;138(5):1491‐1494. [DOI] [PubMed] [Google Scholar]
- 69. Barcik W, Pugin B, Bresco MS, et al. Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy. 2019;74(5):899‐909. [DOI] [PubMed] [Google Scholar]
- 70. Lee JJ, Kim SH, Lee MJ, et al. Different upper airway microbiome and their functional genes associated with asthma in young adults and elderly individuals. Allergy. 2019;74(4):709‐719. [DOI] [PubMed] [Google Scholar]
- 71. Jobin C. Precision medicine using microbiota. Science. 2018;359(6371):32‐34. [DOI] [PubMed] [Google Scholar]
- 72. Maurer M, Hawro T, Krause K, et al. Diagnosis and treatment of chronic inducible urticaria. Allergy. 2019;74(12):2550‐2553. [DOI] [PubMed] [Google Scholar]
- 73. Bieber T, Traidl‐Hoffmann C, Schäppi G, Lauener R, Akdis C, Schmid‐Grendlmeier P. Unraveling the complexity of atopic dermatitis: The CK‐CARE approach toward precision medicine. Allergy. 2020;75(11):2936‐2938. [DOI] [PubMed] [Google Scholar]
- 74. Thijs J, Krastev T, Weidinger S, et al. Biomarkers for atopic dermatitis: a systematic review and meta‐analysis. Curr Opin Allergy Clin Immunol. 2015;15(5):453‐460. [DOI] [PubMed] [Google Scholar]
- 75. Venter C, Meyer RW, Nwaru BI, et al. EAACI position paper: Influence of dietary fatty acids on asthma, food allergy, and atopic dermatitis. Allergy. 2019;74(8):1429‐1444. [DOI] [PubMed] [Google Scholar]
- 76. Altunbulakli C, Reiger M, Neumann AU, et al. Relations between epidermal barrier dysregulation and Staphylococcus species‐dominated microbiome dysbiosis in patients with atopic dermatitis. J Allergy Clin Immunol. 2018;142(5):1643‐1647. [DOI] [PubMed] [Google Scholar]
- 77. Moriwaki M, Iwamoto K, Niitsu Y, et al. Staphylococcus aureus from atopic dermatitis skin accumulates in the lysosomes of keratinocytes with induction of IL‐1α secretion via TLR9. Allergy. 2019;74(3):560‐571. [DOI] [PubMed] [Google Scholar]
- 78. Reiger M, Traidl‐Hoffmann C, Neumann AU. The skin microbiome as a clinical biomarker in atopic eczema: promises, navigation, and pitfalls. J Allergy Clin Immunol. 2020;145(1):93‐96. [DOI] [PubMed] [Google Scholar]
- 79. Gokkaya M, Damialis A, Nussbaumer T, et al. Defining biomarkers to predict symptoms in subjects with and without allergy under natural pollen exposure. J Allergy Clin Immunol. 2020;146(3):583‐594. [DOI] [PubMed] [Google Scholar]
- 80. Gonzalez T, Stevens ML, Baatrebek Kyzy A, et al. Biofilm propensity of Staphylococcus aureus skin isolates is associated with increased atopic dermatitis severity and barrier dysfunction in the MPAACH pediatric cohort. Allergy. 2020. 10.1111/all.14489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hülpüsch C, Tremmel K, Hammel G, et al. Skin pH‐dependent Staphylococcus aureus abundance as predictor for increasing atopic dermatitis severity. Allergy. 2020. 10.1111/all.14461 [DOI] [PubMed] [Google Scholar]
- 82. Vitte J, Amadei L, Gouitaa M, et al. Paired acute‐baseline serum tryptase levels in perioperative anaphylaxis: an observational study. Allergy. 2019;74(6):1157‐1165. [DOI] [PubMed] [Google Scholar]
- 83. Izuhara K, Nunomura S, Nanri Y, Ono J, Takai M, Kawaguchi A. Periostin: an emerging biomarker for allergic diseases. Allergy. 2019;74(11):2116‐2128. [DOI] [PubMed] [Google Scholar]
- 84. Ando N, Nakamura Y, Ishimaru K, et al. Allergen‐specific basophil reactivity exhibits daily variations in seasonal allergic rhinitis. Allergy. 2015;70(3):319‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhong H, Fan XL, Yu QN, et al. Increased innate type 2 immune response in house dust mite‐allergic patients with allergic rhinitis. Clin Immunol. 2017;183:293‐299. [DOI] [PubMed] [Google Scholar]
- 86. Dhariwal J, Cameron A, Trujillo‐Torralbo MB, et al. Mucosal type 2 innate lymphoid cells are a key component of the allergic response to aeroallergens. Am J Respir Crit Care Med. 2017;195(12):1586‐1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yu QN, Guo YB, Li X, et al. ILC2 frequency and activity are inhibited by glucocorticoid treatment via STAT pathway in patients with asthma. Allergy. 2018;73(9):1860‐1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tojima I, Matsumoto K, Kikuoka H, et al. Evidence for the induction of Th2 inflammation by group 2 innate lymphoid cells in response to prostaglandin D2 and cysteinyl leukotrienes in allergic rhinitis. Allergy. 2019;74(12):2417‐2426. [DOI] [PubMed] [Google Scholar]
- 89. Iinuma T, Okamoto Y, Morimoto Y, et al. Pathogenicity of memory Th2 cells is linked to stage of allergic rhinitis. Allergy. 2018;73(2):479‐489. [DOI] [PubMed] [Google Scholar]
- 90. North ML, Jones MJ, MacIsaac JL, et al. Blood and nasal epigenetics correlate with allergic rhinitis symptom development in the environmental exposure unit. Allergy. 2018;73(1):196‐205. [DOI] [PubMed] [Google Scholar]
- 91. Cardenas A, Sordillo JE, Rifas‐Shiman SL, et al. The nasal methylome as a biomarker of asthma and airway inflammation in children. Nat Commun. 2019;10(1):3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Panganiban RP, Wang Y, Howrylak J, et al. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol. 2016;137(5):1423‐1432. [DOI] [PubMed] [Google Scholar]
- 93. Panganiban RP, Lambert KA, Hsu MH, Laryea Z, Ishmael FT. Isolation and profiling of plasma microRNAs: biomarkers for asthma and allergic rhinitis. Methods. 2019;152:48‐54. [DOI] [PubMed] [Google Scholar]
- 94. Ma GC, Wang TS, Wang J, Ma ZJ, Pu SB. Serum metabolomics study of patients with allergic rhinitis. Biomed Chromatogr. 2020;34(3):e4739 10.1002/bmc.4739 [DOI] [PubMed] [Google Scholar]
- 95. Choi GS, Shin SY, Kim JH, et al. Serum lactoferrin level as a serologic biomarker for allergic rhinitis. Clin Exp Allergy. 2010;40(3):403‐410. [DOI] [PubMed] [Google Scholar]
- 96. Eguiluz‐Gracia I, Testera‐Montes A, Gonzalez M, et al. Safety and reproducibility of nasal allergen challenge. Allergy. 2019;74(6):1125‐1134. [DOI] [PubMed] [Google Scholar]
- 97. Eguiluz‐Gracia I, Testera‐Montes A, Salas M, et al. Comparison of diagnostic accuracy of acoustic rhinometry and symptoms score for nasal allergen challenge monitoring. Allergy. 2020. 10.1111/all.14499 [DOI] [PubMed] [Google Scholar]
- 98. Gomez E, Campo P, Rondon C, et al. Role of the basophil activation test in the diagnosis of local allergic rhinitis. J Allergy Clin Immunol. 2013;132(4):975‐976. [DOI] [PubMed] [Google Scholar]
- 99. Campo P, Rondon C, Gould HJ, Barrionuevo E, Gevaert P, Blanca M. Local IgE in non‐allergic rhinitis. Clin Exp Allergy. 2015;45(5):872‐881. [DOI] [PubMed] [Google Scholar]
- 100. Duarte Ferreira R, Ornelas C, Silva S, et al. Contribution of in vivo and in vitro testing for the diagnosis of local allergic rhinitis. J Investig Allergol Clin Immunol. 2019;29(1):46‐48. [DOI] [PubMed] [Google Scholar]
- 101. Eguiluz‐Gracia I, Fernandez‐Santamaria R, Testera‐Montes A, et al. Coexistence of nasal reactivity to allergens with and without IgE sensitization in patients with allergic rhinitis. Allergy. 2020;75(7):1689‐1698. [DOI] [PubMed] [Google Scholar]
- 102. Bousquet J, Pfaar O, Togias A, et al. 2019 ARIA Care pathways for allergen immunotherapy. Allergy. 2019;74(11):2087‐2102. [DOI] [PubMed] [Google Scholar]
- 103. Reitsma S, Subramaniam S, Fokkens WWJ, Wang Y. Recent developments and highlights in rhinitis and allergen immunotherapy. Allergy. 2018;73(12):2306‐2313. [DOI] [PubMed] [Google Scholar]
- 104. Kortekaas Krohn I, Callebaut I, Alpizar YA, et al. MP29‐02 reduces nasal hyperreactivity and nasal mediators in patients with house dust mite‐allergic rhinitis. Allergy. 2018;73(5):1084‐1093. [DOI] [PubMed] [Google Scholar]
- 105. Rittchen S, Heinemann A. Therapeutic potential of hematopoietic prostaglandin D2 synthase in allergic inflammation. Cells. 2019;8(6):619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Okubo K, Hashiguchi K, Takeda T, et al. A randomized controlled phase II clinical trial comparing ONO‐4053, a novel DP1 antagonist, with a leukotriene receptor antagonist pranlukast in patients with seasonal allergic rhinitis. Allergy. 2017;72(10):1565‐1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Meng Y, Wang C, Zhang L. Recent developments and highlights in allergic rhinitis. Allergy. 2019;74(12):2320‐2328. [DOI] [PubMed] [Google Scholar]
- 108. Eyerich S, Metz M, Bossios A, Eyerich K. New biological treatments for asthma and skin allergies. Allergy. 2020;75(3):546‐560. [DOI] [PubMed] [Google Scholar]
- 109. Mukherjee M, Bakakos P, Loukides S. New paradigm in asthma management: switching between biologics! Allergy. 2020;75(4):743‐745. [DOI] [PubMed] [Google Scholar]
- 110. Tsabouri S, Tseretopoulou X, Priftis K, Ntzani EE. Omalizumab for the treatment of inadequately controlled allergic rhinitis: a systematic review and meta‐analysis of randomized clinical trials. J Allergy Clin Immunol Pract. 2014;2(3):332‐340. [DOI] [PubMed] [Google Scholar]
- 111. Kopp MV, Hamelmann E, Bendiks M, et al. Transient impact of omalizumab in pollen allergic patients undergoing specific immunotherapy. Pediatr Allergy Immunol. 2013;24(5):427‐433. [DOI] [PubMed] [Google Scholar]
- 112. Weinstein SF, Katial R, Jayawardena S, et al. Efficacy and safety of dupilumab in perennial allergic rhinitis and comorbid asthma. J Allergy Clin Immunol. 2018;142(1):171‐177. [DOI] [PubMed] [Google Scholar]
- 113. Breiteneder H, Diamant Z, Eiwegger T, et al. Future research trends in understanding the mechanisms underlying allergic diseases for improved patient care. Allergy. 2019;74(12):2293‐2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Murray CS, Poletti G, Kebadze T, et al. Study of modifiable risk factors for asthma exacerbations: virus infection and allergen exposure increase the risk of asthma hospital admissions in children. Thorax. 2006;61(5):376‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Akbarshahi H, Menzel M, Ramu S, Mahmutovic Persson I, Bjermer L, Uller L. House dust mite impairs antiviral response in asthma exacerbation models through its effects on TLR3. Allergy. 2018;73(5):1053‐1063. [DOI] [PubMed] [Google Scholar]
- 116. Oliver BG, Robinson P, Peters M, Black J. Viral infections and asthma: an inflammatory interface? Eur Respir J. 2014;44(6):1666‐1681. [DOI] [PubMed] [Google Scholar]
- 117. Johnston NW, Johnston SL, Duncan JM, et al. The September epidemic of asthma exacerbations in children: a search for etiology. J Allergy Clin Immunol. 2005;115(1):132‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hasegawa K, Hoptay CE, Harmon B, et al. Association of type 2 cytokines in severe rhinovirus bronchiolitis during infancy with risk of developing asthma: a multicenter prospective study. Allergy. 2019;74(7):1374‐1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Globinska A, Pawelczyk M, Piechota‐Polanczyk A, et al. Impaired virus replication and decreased innate immune responses to viral infections in nasal epithelial cells from patients with allergic rhinitis. Clin Exp Immunol. 2017;187(1):100‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Jeon YJ, Lim JH, An S, et al. Type III interferons are critical host factors that determine susceptibility to Influenza A viral infection in allergic nasal mucosa. Clin Exp Allergy. 2018;48(3):253‐265. [DOI] [PubMed] [Google Scholar]
- 121. Gilles S, Blume C, Wimmer M, et al. Pollen exposure weakens innate defense against respiratory viruses. Allergy. 2020;75(3):576‐587. [DOI] [PubMed] [Google Scholar]
- 122. Flayer CH, Haczku A. The Th2 gene cluster unraveled: role of RHS6. Allergy. 2017;72(5):679‐681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hong HY, Chen FH, Sun YQ, et al. Local IL‐25 contributes to Th2‐biased inflammatory profiles in nasal polyps. Allergy. 2018;73(2):459‐469. [DOI] [PubMed] [Google Scholar]
- 124. Tan KS, Andiappan AK, Lee B, et al. RNA sequencing of H3N2 influenza virus‐infected human nasal epithelial cells from multiple subjects reveals molecular pathways associated with tissue injury and complications. Cells. 2019;8(9):986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Roan F, Obata‐Ninomiya K, Ziegler SF. Epithelial cell‐derived cytokines: more than just signaling the alarm. J Clin Investig. 2019;129(4):1441‐1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tan KS, Ong HH, Yan Y, et al. In vitro model of fully differentiated human nasal epithelial cells infected with rhinovirus reveals epithelium‐initiated immune responses. J Infect Dis. 2018;217(6):906‐915. [DOI] [PubMed] [Google Scholar]
- 127. Becker Y. Respiratory syncytial virus (RSV) evades the human adaptive immune system by skewing the Th1/Th2 cytokine balance toward increased levels of Th2 cytokines and IgE, markers of allergy–a review. Virus Genes. 2006;33(2):235‐252. [DOI] [PubMed] [Google Scholar]
- 128. Malinczak CA, Rasky AJ, Fonseca W, et al. Upregulation of H3K27 demethylase KDM6 during respiratory syncytial virus infection enhances proinflammatory responses and immunopathology. J Immunol. 2020;204(1):159‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Scanlon ST, McKenzie AN. Type 2 innate lymphoid cells: new players in asthma and allergy. Curr Opin Immunol. 2012;24(6):707‐712. [DOI] [PubMed] [Google Scholar]
- 130. Han JJ, Goldsmith AM, Hong JY, Sajjan U, Hershenson MB. Rhinovirus induces the expression of thymic stromal lymphopoietin in human airway epithelial cells. Am J Respir Crit Care Med. 2012;185:A6875. [Google Scholar]
- 131. Beale J, Jayaraman A, Jackson DJ, et al. Rhinovirus‐induced IL‐25 in asthma exacerbation drives type 2 immunity and allergic pulmonary inflammation. Sci Transl Med. 2014;6(256):256ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Shaw JL, Fakhri S, Citardi MJ, et al. IL‐33‐responsive innate lymphoid cells are an important source of IL‐13 in chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med. 2013;188(4):432‐439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Jackson DJ, Makrinioti H, Rana BM, et al. IL‐33‐dependent type 2 inflammation during rhinovirus‐induced asthma exacerbations in vivo. Am J Respir Crit Care Med. 2014;190(12):1373‐1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Bartemes KR, Kephart GM, Fox SJ, Kita H. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J Allergy Clin Immunol. 2014;134(3):671‐678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Tian T, Zi X, Peng Y, et al. H3N2 influenza virus infection enhances oncostatin M expression in human nasal epithelium. Exp Cell Res. 2018;371(2):322‐329. [DOI] [PubMed] [Google Scholar]
- 136. Li L, Chong HC, Ng SY, et al. Angiopoietin‐like 4 increases pulmonary tissue leakiness and damage during influenza pneumonia. Cell Rep. 2015;10(5):654‐663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Taka S, Tzani‐Tzanopoulou P, Wanstall H, Papadopoulos NG. MicroRNAs in asthma and respiratory infections: identifying common pathways. Allergy Asthma Immunol Res. 2020;12(1):4‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Tiwari BS, Belenghi B, Levine A. Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol. 2002;128(4):1271‐1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Aizawa H, Koarai A, Shishikura Y, et al. Oxidative stress enhances the expression of IL‐33 in human airway epithelial cells. Respir Res. 2018;19(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Manji J, Thamboo A, Tacey M, Garnis C, Chadha NK. The presence of Interleukin‐13 in nasal lavage may be a predictor of nasal polyposis in pediatric patients with cystic fibrosis. Rhinology. 2018;56(3):261‐267. [DOI] [PubMed] [Google Scholar]
- 141. Bachert C, Han JK, Desrosiers M, et al. Efficacy and safety of dupilumab in patients with severe chronic rhinosinusitis with nasal polyps (LIBERTY NP SINUS‐24 and LIBERTY NP SINUS‐52): results from two multicentre, randomised, double‐blind, placebo‐controlled, parallel‐group phase 3 trials. Lancet. 2019;394(10209):1638‐1650. [DOI] [PubMed] [Google Scholar]
- 142. Bachert C, Sousa AR, Lund VJ, et al. Reduced need for surgery in severe nasal polyposis with mepolizumab: randomized trial. J Allergy Clin Immunol. 2017;140(4):1024‐1031. [DOI] [PubMed] [Google Scholar]
- 143. Bachert C, Zinreich SJ, Hellings PW, et al. Dupilumab reduces opacification across all sinuses and related symptoms in patients with CRSwNP. Rhinology. 2020;58(1):10‐17. [DOI] [PubMed] [Google Scholar]
- 144. Tomassen P, Vandeplas G, Van Zele T, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. 2016;137(5):1449‐1456. [DOI] [PubMed] [Google Scholar]
- 145. Fokkens WJ, Reitsma S. Medical algorithms: management of chronic rhinosinusitis. Allergy. 2019;74(7):1415‐1416. [DOI] [PubMed] [Google Scholar]
- 146. Xu X, Ong YK, Wang Y. Novel findings in immunopathophysiology of chronic rhinosinusitis and their role in a model of precision medicine. Allergy. 2020;75(4):769‐780. [DOI] [PubMed] [Google Scholar]
- 147. Arebro J, Drakskog C, Winqvist O, Bachert C, Kumlien Georen S, Cardell LO. Subsetting reveals CD16(high) CD62L(dim) neutrophils in chronic rhinosinusitis with nasal polyps. Allergy. 2019;74(12):2499‐2501. [DOI] [PubMed] [Google Scholar]
- 148. Succar EF, Li P, Ely KA, Chowdhury NI, Chandra RK, Turner JH. Neutrophils are underrecognized contributors to inflammatory burden and quality of life in chronic rhinosinusitis. Allergy. 2020;75(3):713‐716. [DOI] [PubMed] [Google Scholar]
- 149. Jonstam K, Westman M, Holtappels G, Holweg CTJ, Bachert C. Serum periostin, IgE, and SE‐IgE can be used as biomarkers to identify moderate to severe chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2017;140(6):1705‐1708. [DOI] [PubMed] [Google Scholar]
- 150. Zhang Y, Derycke L, Holtappels G, et al. Th2 cytokines orchestrate the secretion of MUC5AC and MUC5B in IL‐5‐positive chronic rhinosinusitis with nasal polyps. Allergy. 2019;74(1):131‐140. [DOI] [PubMed] [Google Scholar]
- 151. Ogasawara N, Klingler AI, Tan BK, et al. Epithelial activators of type 2 inflammation: Elevation of thymic stromal lymphopoietin, but not IL‐25 or IL‐33, in chronic rhinosinusitis with nasal polyps in Chicago. Illinois. Allergy. 2018;73(11):2251‐2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Rouyar A, Classe M, Gorski R, et al. Type 2/Th2‐driven inflammation impairs olfactory sensory neurogenesis in mouse chronic rhinosinusitis model. Allergy. 2019;74(3):549‐559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Rimmer J, Hellings P, Lund VJ, et al. European position paper on diagnostic tools in rhinology. Rhinology. 2019;57(Suppl S28):1‐41. [DOI] [PubMed] [Google Scholar]
- 154. Oakley GM, Christensen JM, Sacks R, Earls P, Harvey RJ. Characteristics of macrolide responders in persistent post‐surgical rhinosinusitis. Rhinology. 2018;56(2):111‐117. [DOI] [PubMed] [Google Scholar]
- 155. Bidder T, Sahota J, Rennie C, Lund VJ, Robinson DS, Kariyawasam HH. Omalizumab treats chronic rhinosinusitis with nasal polyps and asthma together‐a real life study. Rhinology. 2018;56(1):42‐45. [DOI] [PubMed] [Google Scholar]
- 156. Jonstam K, Swanson BN, Mannent LP, et al. Dupilumab reduces local type 2 pro‐inflammatory biomarkers in chronic rhinosinusitis with nasal polyposis. Allergy. 2019;74(4):743‐752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Tsetsos N, Goudakos JK, Daskalakis D, Konstantinidis I, Markou K. Monoclonal antibodies for the treatment of chronic rhinosinusitis with nasal polyposis: a systematic review. Rhinology. 2018;56(1):11‐21. [DOI] [PubMed] [Google Scholar]
- 158. Castan L, Bogh KL, Maryniak NZ, et al. Overview of in vivo and ex vivo endpoints in murine food allergy models: suitable for evaluation of the sensitizing capacity of novel proteins? Allergy. 2020;75(2):289‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Eiwegger T, Hung L, San Diego KE, O'Mahony L, Upton J. Recent developments and highlights in food allergy. Allergy. 2019;74(12):2355‐2367. [DOI] [PubMed] [Google Scholar]
- 160. Ponce M, Diesner SC, Szepfalusi Z, Eiwegger T. Markers of tolerance development to food allergens. Allergy. 2016;71(10):1393‐1404. [DOI] [PubMed] [Google Scholar]
- 161. Ashley SE, Tan HT, Vuillermin P, et al. The skin barrier function gene SPINK5 is associated with challenge‐proven IgE‐mediated food allergy in infants. Allergy. 2017;72(9):1356‐1364. [DOI] [PubMed] [Google Scholar]
- 162. Tan HT, Hagner S, Ruchti F, et al. Tight junction, mucin, and inflammasome‐related molecules are differentially expressed in eosinophilic, mixed, and neutrophilic experimental asthma in mice. Allergy. 2019;74(2):294‐307. [DOI] [PubMed] [Google Scholar]
- 163. Leung DYM, Calatroni A, Zaramela LS, et al. The nonlesional skin surface distinguishes atopic dermatitis with food allergy as a unique endotype. Sci Transl Med. 2019;11(480):eaav2685 10.1126/scitranslmed.aav2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Goleva E, Berdyshev E, Leung DY. Epithelial barrier repair and prevention of allergy. J Clin Investig. 2019;129(4):1463‐1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. van Ginkel CD, Flokstra‐de Blok BM, Kollen BJ, Kukler J, Koppelman GH, Dubois AE. Loss‐of‐function variants of the filaggrin gene are associated with clinical reactivity to foods. Allergy. 2015;70(4):461‐464. [DOI] [PubMed] [Google Scholar]
- 166. Suaini NHA, Wang Y, Soriano VX, et al. Genetic determinants of paediatric food allergy: a systematic review. Allergy. 2019;74(9):1631‐1648. [DOI] [PubMed] [Google Scholar]
- 167. Kivisto JE, Clarke A, Dery A, et al. Genetic and environmental susceptibility to food allergy in a registry of twins. J Allergy Clin Immunol Pract. 2019;7(8):2916‐2918. [DOI] [PubMed] [Google Scholar]
- 168. Marenholz I, Grosche S, Kalb B, et al. Genome‐wide association study identifies the SERPINB gene cluster as a susceptibility locus for food allergy. Nat Commun. 2017;8(1):1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Do AN, Watson CT, Cohain AT, et al. Dual transcriptomic and epigenomic study of reaction severity in peanut‐allergic children. J Allergy Clin Immunol. 2020;145(4):1219‐1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Mondoulet L, Dioszeghy V, Busato F, et al. Gata3 hypermethylation and Foxp3 hypomethylation are associated with sustained protection and bystander effect following epicutaneous immunotherapy in peanut‐sensitized mice. Allergy. 2019;74(1):152‐164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. D'Argenio V, Del Monaco V, Paparo L, et al. Altered miR‐193a‐5p expression in children with cow's milk allergy. Allergy. 2018;73(2):379‐386. [DOI] [PubMed] [Google Scholar]
- 172. Ruffner MA, Song L, Maurer K, et al. Toll‐like receptor 2 stimulation augments esophageal barrier integrity. Allergy. 2019;74(12):2449‐2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Rahrig S, Dettmann JM, Brauns B, et al. Transient epidermal barrier deficiency and lowered allergic threshold in filaggrin‐hornerin (FlgHrnr(‐/‐) ) double‐deficient mice. Allergy. 2019;74(7):1327‐1339. [DOI] [PubMed] [Google Scholar]
- 174. Mitamura Y, Nunomura S, Nanri Y, et al. The IL‐13/periostin/IL‐24 pathway causes epidermal barrier dysfunction in allergic skin inflammation. Allergy. 2018;73(9):1881‐1891. [DOI] [PubMed] [Google Scholar]
- 175. Rinaldi AO, Morita H, Wawrzyniak P, et al. Direct assessment of skin epithelial barrier by electrical impedance spectroscopy. Allergy. 2019;74(10):1934‐1944. [DOI] [PubMed] [Google Scholar]
- 176. Chauveau A, Dalphin ML, Mauny F, et al. Skin prick tests and specific IgE in 10‐year‐old children: agreement and association with allergic diseases. Allergy. 2017;72(9):1365‐1373. [DOI] [PubMed] [Google Scholar]
- 177. Flinterman AE, Knol EF, Lencer DA, et al. Peanut epitopes for IgE and IgG4 in peanut‐sensitized children in relation to severity of peanut allergy. J Allergy Clin Immunol. 2008;121(3):737‐743. [DOI] [PubMed] [Google Scholar]
- 178. Caubet JC, Lin J, Ahrens B, et al. Natural tolerance development in cow's milk allergic children: IgE and IgG4 epitope binding. Allergy. 2017;72(11):1677‐1685. [DOI] [PubMed] [Google Scholar]
- 179. Cerecedo I, Zamora J, Shreffler WG, et al. Mapping of the IgE and IgG4 sequential epitopes of milk allergens with a peptide microarray‐based immunoassay. J Allergy Clin Immunol. 2008;122(3):589‐594. [DOI] [PubMed] [Google Scholar]
- 180. Sackesen C, Suarez‐Farinas M, Silva R, et al. A new Luminex‐based peptide assay to identify reactivity to baked, fermented, and whole milk. Allergy. 2019;74(2):327‐336. [DOI] [PubMed] [Google Scholar]
- 181. Suprun M, Getts R, Raghunathan R, et al. Novel bead‐based epitope assay is a sensitive and reliable tool for profiling epitope‐specific antibody repertoire in food allergy. Sci Rep. 2019;9(1):18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Suarez‐Farinas M, Suprun M, Chang HL, et al. Predicting development of sustained unresponsiveness to milk oral immunotherapy using epitope‐specific antibody binding profiles. J Allergy Clin Immunol. 2019;143(3):1038‐1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Monino‐Romero S, Lexmond WS, Singer J, et al. Soluble FcvarepsilonRI: a biomarker for IgE‐mediated diseases. Allergy. 2019;74(7):1381‐1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Saidova A, Hershkop AM, Ponce M, Eiwegger T. Allergen‐specific T cells in IgE‐mediated food allergy. Arch Immunol Ther Exp (Warsz). 2018;66(3):161‐170. [DOI] [PubMed] [Google Scholar]
- 185. Chiang D, Chen X, Jones SM, et al. Single‐cell profiling of peanut‐responsive T cells in patients with peanut allergy reveals heterogeneous effector TH2 subsets. J Allergy Clin Immunol. 2018;141(6):2107‐2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wambre E, Bajzik V, DeLong JH, et al. A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med. 2017;9(401):eaam9171 10.1126/scitranslmed.aam9171.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Heeringa JJ, Rijvers L, Arends NJ, et al. IgE‐expressing memory B cells and plasmablasts are increased in blood of children with asthma, food allergy, and atopic dermatitis. Allergy. 2018;73(6):1331‐1336. [DOI] [PubMed] [Google Scholar]
- 188. Jimenez‐Saiz R, Ellenbogen Y, Bruton K, et al. Human BCR analysis of single‐sorted, putative IgE(+) memory B cells in food allergy. J Allergy Clin Immunol. 2019;144(1):336‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Nielsen SCA, Boyd SD. New technologies and applications in infant B cell immunology. Curr Opin Immunol. 2019;57:53‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Croote D, Darmanis S, Nadeau KC, Quake SR. High‐affinity allergen‐specific human antibodies cloned from single IgE B cell transcriptomes. Science. 2018;362(6420):1306‐1309. [DOI] [PubMed] [Google Scholar]
- 191. Nielsen SCA, Roskin KM, Jackson KJL, et al. Shaping of infant B cell receptor repertoires by environmental factors and infectious disease. Sci Transl Med. 2019;11(481):eaat2004 10.1126/scitranslmed.aat2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Hoof I, Schulten V, Layhadi JA, et al. Allergen‐specific IgG(+) memory B cells are temporally linked to IgE memory responses. J Allergy Clin Immunol. 2020;146(1):180‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Heeringa JJ, McKenzie CI, Varese N, et al. Induction of IgG2 and IgG4 B‐cell memory following sublingual immunotherapy for ryegrass pollen allergy. Allergy. 2020;75(5):1121‐1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Jimenez‐Saiz R, Ellenbogen Y, Koenig JFE, et al. IgG1(+) B‐cell immunity predates IgE responses in epicutaneous sensitization to foods. Allergy. 2019;74(1):165‐175. [DOI] [PubMed] [Google Scholar]
- 195. Hoffmann HJ, Santos AF, Mayorga C, et al. The clinical utility of basophil activation testing in diagnosis and monitoring of allergic disease. Allergy. 2015;70(11):1393‐1405. [DOI] [PubMed] [Google Scholar]
- 196. Hemmings O, Kwok M, McKendry R, Santos AF. Basophil activation test: old and new applications in allergy. Curr Allergy Asthma Rep. 2018;18(12):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Hung L, Obernolte H, Sewald K, Eiwegger T. Human ex vivo and in vitro disease models to study food allergy. Asia Pac Allergy. 2019;9(1):e4 10.5415/apallergy.2019.9.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Santos AF, Couto‐Francisco N, Becares N, Kwok M, Bahnson HT, Lack G. A novel human mast cell activation test for peanut allergy. J Allergy Clin Immunol. 2018;142(2):689‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Bahri R, Custovic A, Korosec P, et al. Mast cell activation test in the diagnosis of allergic disease and anaphylaxis. J Allergy Clin Immunol. 2018;142(2):485‐496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Pouessel G, Beaudouin E, Tanno LK, et al. Food‐related anaphylaxis fatalities: analysis of the allergy vigilance network((R)) database. Allergy. 2019;74(6):1193‐1196. [DOI] [PubMed] [Google Scholar]
- 201. Pouessel G, Turner PJ, Worm M, et al. Food‐induced fatal anaphylaxis: from epidemiological data to general prevention strategies. Clin Exp Allergy. 2018;48(12):1584‐1593. [DOI] [PubMed] [Google Scholar]
- 202. De Schryver S, Halbrich M, Clarke A, et al. Tryptase levels in children presenting with anaphylaxis: temporal trends and associated factors. J Allergy Clin Immunol. 2016;137(4):1138‐1142. [DOI] [PubMed] [Google Scholar]
- 203. Flores Kim J, McCleary N, Nwaru BI, Stoddart A, Sheikh A. Diagnostic accuracy, risk assessment, and cost‐effectiveness of component‐resolved diagnostics for food allergy: a systematic review. Allergy. 2018;73(8):1609‐1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Eller E, Bindslev‐Jensen C. Clinical value of component‐resolved diagnostics in peanut‐allergic patients. Allergy. 2013;68(2):190‐194. [DOI] [PubMed] [Google Scholar]
- 205. Klemans RJ, Otte D, Knol M, et al. The diagnostic value of specific IgE to Ara h 2 to predict peanut allergy in children is comparable to a validated and updated diagnostic prediction model. J Allergy Clin Immunol. 2013;131(1):157‐163. [DOI] [PubMed] [Google Scholar]
- 206. Kukkonen AK, Pelkonen AS, Makinen‐Kiljunen S, Voutilainen H, Makela MJ. Ara h 2 and Ara 6 are the best predictors of severe peanut allergy: a double‐blind placebo‐controlled study. Allergy. 2015;70(10):1239‐1245. [DOI] [PubMed] [Google Scholar]
- 207. Martinet J, Couderc L, Renosi F, Bobée V, Marguet C, Boyer O. Diagnostic value of antigen‐specific immunoglobulin E immunoassays against Ara h 2 and Ara h 8 peanut components in child food allergy. Int Arch Allergy Immunol. 2016;169(4):216‐222. [DOI] [PubMed] [Google Scholar]
- 208. Holzhauser T, Wackermann O, Ballmer‐Weber BK, et al. Soybean (Glycine max) allergy in Europe: Gly m 5 (beta‐conglycinin) and Gly m 6 (glycinin) are potential diagnostic markers for severe allergic reactions to soy. J Allergy Clin Immunol. 2009;123(2):452‐458. [DOI] [PubMed] [Google Scholar]
- 209. Ebisawa M, Brostedt P, Sjolander S, Sato S, Borres MP, Ito K. Gly m 2S albumin is a major allergen with a high diagnostic value in soybean‐allergic children. J Allergy Clin Immunol. 2013;132(4):976‐978. [DOI] [PubMed] [Google Scholar]
- 210. Klemans RJ, Knol EF, Michelsen‐Huisman A, et al. Components in soy allergy diagnostics: Gly m 2S albumin has the best diagnostic value in adults. Allergy. 2013;68(11):1396‐1402. [DOI] [PubMed] [Google Scholar]
- 211. Masthoff LJ, Mattsson L, Zuidmeer‐Jongejan L, et al. Sensitization to Cor a 9 and Cor a 14 is highly specific for a hazelnut allergy with objective symptoms in Dutch children and adults. J Allergy Clin Immunol. 2013;132(2):393‐399. [DOI] [PubMed] [Google Scholar]
- 212. Faber MA, De Graag M, Van Der Heijden C, et al. Cor a 14: missing link in the molecular diagnosis of hazelnut allergy? Int Arch Allergy Immunol. 2014;164(3):200‐206. [DOI] [PubMed] [Google Scholar]
- 213. Datema MR, van Ree R, Asero R, et al. Component‐resolved diagnosis and beyond: multivariable regression models to predict severity of hazelnut allergy. Allergy. 2018;73(3):549‐559. [DOI] [PubMed] [Google Scholar]
- 214. Beyer K, Grabenhenrich L, Hartl M, et al. Predictive values of component‐specific IgE for the outcome of peanut and hazelnut food challenges in children. Allergy. 2015;70(1):90‐98. [DOI] [PubMed] [Google Scholar]
- 215. Ciprandi G, Pistorio A, Silvestri M, Rossi GA, Tosca MA. Walnut anaphylaxis: the usefulness of molecular‐based allergy diagnostics. Immunol Lett. 2014;161(1):138‐139. [DOI] [PubMed] [Google Scholar]
- 216. Ballmer‐Weber BK, Lidholm J, Lange L, et al. Allergen recognition patterns in walnut allergy are age dependent and correlate with the severity of allergic reactions. J Allergy Clin Immunol Pract. 2019;7(5):1560‐1567. [DOI] [PubMed] [Google Scholar]
- 217. Lee J, Jeong K, Jeon SA, Lee S. Component resolved diagnosis of walnut allergy in young children: Jug r 1 as a major walnut allergen. Asian Pac J Allergy Immunol. 2019. 10.12932/AP-161118-0443 [DOI] [PubMed] [Google Scholar]
- 218. Giovannini M, Comberiati P, Piazza M, et al. Retrospective definition of reaction risk in Italian children with peanut, hazelnut and walnut allergy through component‐resolved diagnosis. Allergol Immunopathol (Madr). 2019;47(1):73‐78. [DOI] [PubMed] [Google Scholar]
- 219. Savvatianos S, Konstantinopoulos AP, Borga A, et al. Sensitization to cashew nut 2S albumin, Ana o 3, is highly predictive of cashew and pistachio allergy in Greek children. J Allergy Clin Immunol. 2015;136(1):192‐194. [DOI] [PubMed] [Google Scholar]
- 220. van der Valk JP, Gerth van Wijk R, Vergouwe Y, et al. sIgE Ana o 1, 2 and 3 accurately distinguish tolerant from allergic children sensitized to cashew nuts. Clin Exp Allergy. 2017;47(1):113‐120. [DOI] [PubMed] [Google Scholar]
- 221. Lange L, Lasota L, Finger A, et al. Ana o 3‐specific IgE is a good predictor for clinically relevant cashew allergy in children. Allergy. 2017;72(4):598‐603. [DOI] [PubMed] [Google Scholar]
- 222. Dang TD, Peters RL, Koplin JJ, et al. Egg allergen specific IgE diversity predicts resolution of egg allergy in the population cohort HealthNuts. Allergy. 2019;74(2):318‐326. [DOI] [PubMed] [Google Scholar]
- 223. Pascal M, Grishina G, Yang AC, et al. Molecular diagnosis of shrimp allergy: efficiency of several allergens to predict clinical reactivity. J Allergy Clin Immunol Pract. 2015;3(4):521‐529. [DOI] [PubMed] [Google Scholar]
- 224. Fiocchi A, Brozek J, Schunemann H, et al. World allergy organization (WAO) diagnosis and rationale for action against cow's milk allergy (DRACMA) guidelines. Pediatr Allergy Immunol. 2010;21(Suppl 21):1‐125. [DOI] [PubMed] [Google Scholar]
- 225. Cingolani A, Di Pillo S, Cerasa M, et al. Usefulness of nBos d 4, 5 and nBos d 8 specific IgE antibodies in cow's milk allergic children. Allergy Asthma Immunol Res. 2014;6(2):121‐125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Caubet J‐C, Nowak‐Węgrzyn A, Moshier E, Godbold J, Wang J, Sampson HA. Utility of casein‐specific IgE levels in predicting reactivity to baked milk. J Allergy Clin Immunol. 2013;131(1):222‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Agabriel C, Ghazouani O, Birnbaum J, et al. Ara h 2 and Ara h 6 sensitization predicts peanut allergy in Mediterranean pediatric patients. Pediatr Allergy Immunol. 2014;25(7):662‐667. [DOI] [PubMed] [Google Scholar]
- 228. De Knop KJ, Verweij MM, Grimmelikhuijsen M, et al. Age‐related sensitization profiles for hazelnut (Corylus avellana) in a birch‐endemic region. Pediatr Allergy Immunol. 2011;22(1 Pt 2):e139‐e149. 10.1111/j.1399-3038.2011.01112.x [DOI] [PubMed] [Google Scholar]
- 229. Mayorga C, Fernandez TD, Montanez MI, Moreno E, Torres MJ. Recent developments and highlights in drug hypersensitivity. Allergy. 2019;74(12):2368‐2381. [DOI] [PubMed] [Google Scholar]
- 230. Romano A, Atanaskovic‐Markovic M, Barbaud A, et al. Towards a more precise diagnosis of hypersensitivity to beta‐lactams ‐ an EAACI position paper. Allergy. 2020;75(6):1300‐1315. [DOI] [PubMed] [Google Scholar]
- 231. Brockow K, Garvey LH, Aberer W, et al. Skin test concentrations for systemically administered drugs – an ENDA/EAACI Drug Allergy Interest Group position paper. Allergy. 2013;68(6):702‐712. [DOI] [PubMed] [Google Scholar]
- 232. Torres MJ, Celik GE, Whitaker P, et al. A EAACI drug allergy interest group survey on how European allergy specialists deal with β‐lactam allergy. Allergy. 2019;74(6):1052‐1062. [DOI] [PubMed] [Google Scholar]
- 233. Dona I, Romano A, Torres MJ. Algorithm for betalactam allergy diagnosis. Allergy. 2019;74(9):1817‐1819. [DOI] [PubMed] [Google Scholar]
- 234. Barbero N, Fernandez‐Santamaria R, Mayorga C, et al. Identification of an antigenic determinant of clavulanic acid responsible for IgE‐mediated reactions. Allergy. 2019;74(8):1490‐1501. [DOI] [PubMed] [Google Scholar]
- 235. Torres MJ, Celik GE, Whitaker P, et al. A EAACI drug allergy interest group survey on how European allergy specialists deal with beta‐lactam allergy. Allergy. 2019;74(6):1052‐1062. [DOI] [PubMed] [Google Scholar]
- 236. Yang MS, Kang DY, Seo B, et al. Incidence of cephalosporin‐induced anaphylaxis and clinical efficacy of screening intradermal tests with cephalosporins: a large multicenter retrospective cohort study. Allergy. 2018;73(9):1833‐1841. [DOI] [PubMed] [Google Scholar]
- 237. Dona I, Perez‐Sanchez N, Salas M, et al. Clinical characterization and diagnostic approaches for patients reporting hypersensitivity reactions to quinolones. J Allergy Clin Immunol Pract. 2020;8(8):2707‐2714. [DOI] [PubMed] [Google Scholar]
- 238. Porebski G, Pecaric‐Petkovic T, Groux‐Keller M, Bosak M, Kawabata TT, Pichler WJ. In vitro drug causality assessment in Stevens‐Johnson syndrome ‐ alternatives for lymphocyte transformation test. Clin Exp Allergy. 2013;43(9):1027‐1037. [DOI] [PubMed] [Google Scholar]
- 239. Mayorga C, Celik G, Rouzaire P, et al. In vitro tests for drug hypersensitivity reactions: an ENDA/EAACI drug allergy interest group position paper. Allergy. 2016;71(8):1103‐1134. [DOI] [PubMed] [Google Scholar]
- 240. Fontaine C, Mayorga C, Bousquet PJ, et al. Relevance of the determination of serum‐specific IgE antibodies in the diagnosis of immediate beta‐lactam allergy. Allergy. 2007;62(1):47‐52. [DOI] [PubMed] [Google Scholar]
- 241. Johansson SG, Adedoyin J, van Hage M, Gronneberg R, Nopp A. False‐positive penicillin immunoassay: an unnoticed common problem. J Allergy Clin Immunol. 2013;132(1):235‐237. [DOI] [PubMed] [Google Scholar]
- 242. Torres MJ, Padial A, Mayorga C, et al. The diagnostic interpretation of basophil activation test in immediate allergic reactions to betalactams. Clin Exp Allergy. 2004;34(11):1768‐1775. [DOI] [PubMed] [Google Scholar]
- 243. Torres MJ, Ariza A, Mayorga C, et al. Clavulanic acid can be the component in amoxicillin‐clavulanic acid responsible for immediate hypersensitivity reactions. J Allergy Clin Immunol. 2010;125(2):502‐505. [DOI] [PubMed] [Google Scholar]
- 244. Fernandez TD, Ariza A, Palomares F, et al. Hypersensitivity to fluoroquinolones: the expression of basophil activation markers depends on the clinical entity and the culprit fluoroquinolone. Medicine (Baltimore). 2016;95(23):e3679 10.1097/MD.0000000000003679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Fernandez TD, Torres MJ, Blanca‐Lopez N, et al. Negativization rates of IgE radioimmunoassay and basophil activation test in immediate reactions to penicillins. Allergy. 2009;64(2):242‐248. [DOI] [PubMed] [Google Scholar]
- 246. Van Gasse AL, Sabato V, Uyttebroek AP, et al. Immediate moxifloxacin hypersensitivity: is there more than currently meets the eye? Allergy. 2017;72(12):2039‐2043. [DOI] [PubMed] [Google Scholar]
- 247. Fernandez‐Santamaria R, Palomares F, Salas M, et al. Expression of the Tim3‐galectin‐9 axis is altered in drug‐induced maculopapular exanthema. Allergy. 2019;74(9):1769‐1779. [DOI] [PubMed] [Google Scholar]
- 248. Dona I, Perez‐Sanchez N, Eguiluz‐Gracia I, et al. Progress in understanding hypersensitivity reactions to nonsteroidal anti‐inflammatory drugs. Allergy. 2020;75(3):561‐575. [DOI] [PubMed] [Google Scholar]
- 249. Dona I, Perez‐Sanchez N, Bogas G, Moreno E, Salas M, Torres MJ. Medical algorithm: Diagnosis and treatment of nonsteroidal antiinflammatory drugs hypersensitivityity. Allergy. 2020;75(4):1003‐1005. [DOI] [PubMed] [Google Scholar]
- 250. Dona I, Barrionuevo E, Salas M, et al. NSAIDs‐hypersensitivity often induces a blended reaction pattern involving multiple organs. Sci Rep. 2018;8(1):16710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Blanca M, Oussalah A, Cornejo‐Garcia JA, et al. GNAI2 variants predict nonsteroidal anti‐inflammatory drug hypersensitivity in a genome‐wide study. Allergy. 2020;75(5):1250‐1253. [DOI] [PubMed] [Google Scholar]
- 252. Lee HY, Ye YM, Kim SH, et al. Identification of phenotypic clusters of nonsteroidal anti‐inflammatory drugs exacerbated respiratory disease. Allergy. 2017;72(4):616‐626. [DOI] [PubMed] [Google Scholar]
- 253. Hagan JB, Laidlaw TM, Divekar R, et al. Urinary leukotriene E4 to determine aspirin intolerance in asthma: a systematic review and meta‐analysis. J Allergy Clin Immunol Pract. 2017;5(4):990‐997. [DOI] [PubMed] [Google Scholar]
- 254. Ban GY, Cho K, Kim SH, et al. Metabolomic analysis identifies potential diagnostic biomarkers for aspirin‐exacerbated respiratory disease. Clin Exp Allergy. 2017;47(1):37‐47. [DOI] [PubMed] [Google Scholar]
- 255. Lei DK, Saltoun C. Allergen immunotherapy: definition, indications, and reactions. Allergy Asthma Proc. 2019;40(6):369‐371. [DOI] [PubMed] [Google Scholar]
- 256. Miller JM, Davis CM, Anvari S. The clinical and immune outcomes after food allergen immunotherapy emphasizing the development of tolerance. Curr Opin Pediatr. 2019;31(6):821‐827. [DOI] [PubMed] [Google Scholar]
- 257. Sindher SB, Long A, Acharya S, Sampath V, Nadeau KC. The use of biomarkers to predict aero‐allergen and food immunotherapy responses. Clin Rev Allergy Immunol. 2018;55(2):190‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Couroux P, Ipsen H, Stage BS, et al. A birch sublingual allergy immunotherapy tablet reduces rhinoconjunctivitis symptoms when exposed to birch and oak and induces IgG4 to allergens from all trees in the birch homologous group. Allergy. 2019;74(2):361‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Huang Y, Wang C, Wang X, Zhang L, Lou H. Efficacy and safety of subcutaneous immunotherapy with house dust mite for allergic rhinitis: a meta‐analysis of randomized controlled trials. Allergy. 2019;74(1):189‐192. [DOI] [PubMed] [Google Scholar]
- 260. Schmitt J, Wustenberg E, Kuster D, Mucke V, Serup‐Hansen N, Tesch F. The moderating role of allergy immunotherapy in asthma progression: results of a population‐based cohort study. Allergy. 2020;75(3):596‐602. [DOI] [PubMed] [Google Scholar]
- 261. Varona R, Ramos T, Escribese MM, et al. Persistent regulatory T‐cell response 2 years after 3 years of grass tablet SLIT: links to reduced eosinophil counts, sIgE levels, and clinical benefit. Allergy. 2019;74(2):349‐360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Wahn U, Bachert C, Heinrich J, Richter H, Zielen S. Real‐world benefits of allergen immunotherapy for birch pollen‐associated allergic rhinitis and asthma. Allergy. 2019;74(3):594‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Investigators PGoC , Vickery BP, Vereda A, et al. AR101 oral immunotherapy for peanut allergy. N Engl J Med. 2018;379(21):1991‐2001. [DOI] [PubMed] [Google Scholar]
- 264. Virkud YV, Kelly RS, Wood C, Lasky‐Su JA. The nuts and bolts of omics for the clinical allergist. Ann Allergy Asthma Immunol. 2019;123(6):558‐563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. van Zelm MC, McKenzie CI, Varese N, Rolland JM, O'Hehir RE. Recent developments and highlights in immune monitoring of allergen immunotherapy. Allergy. 2019;74(12):2342‐2354. [DOI] [PubMed] [Google Scholar]
- 266. Kim EH, Yang L, Ye P, et al. Long‐term sublingual immunotherapy for peanut allergy in children: clinical and immunologic evidence of desensitization. J Allergy Clin Immunol. 2019;144(5):1320‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Shamji MH, Kappen JH, Akdis M, et al. Biomarkers for monitoring clinical efficacy of allergen immunotherapy for allergic rhinoconjunctivitis and allergic asthma: an EAACI Position Paper. Allergy. 2017;72(8):1156‐1173. [DOI] [PubMed] [Google Scholar]
- 268. Viswanathan RK, Busse WW. Allergen immunotherapy in allergic respiratory diseases: from mechanisms to meta‐analyses. Chest. 2012;141(5):1303‐1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Datema MR, Eller E, Zwinderman AH, et al. Ratios of specific IgG4 over IgE antibodies do not improve prediction of peanut allergy nor of its severity compared to specific IgE alone. Clin Exp Allergy. 2019;49(2):216‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Kulis M, Yue X, Guo R, et al. High‐ and low‐dose oral immunotherapy similarly suppress pro‐allergic cytokines and basophil activation in young children. Clin Exp Allergy. 2019;49(2):180‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Chinthrajah RS, Purington N, Andorf S, et al. Sustained outcomes in oral immunotherapy for peanut allergy (POISED study): a large, randomised, double‐blind, placebo‐controlled, phase 2 study. Lancet. 2019;394(10207):1437‐1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Feng M, Su Q, Lai X, et al. Functional and immunoreactive levels of IgG4 correlate with clinical responses during the maintenance phase of house dust mite immunotherapy. J Immunol. 2018;200(12):3897‐3904. [DOI] [PubMed] [Google Scholar]
- 273. Fukano C, Ohashi‐Doi K, Lund K, Nakao A, Masuyama K, Matsuoka T. Establishment of enzyme‐linked immunosorbent facilitated antigen binding as a biomarker assay for Japanese cedar pollen allergy immunotherapy. J Pharmacol Sci. 2019;140(3):223‐227. [DOI] [PubMed] [Google Scholar]
- 274. Chinthrajah RS, Purington N, Sampath V, et al. High dimensional immune biomarkers demonstrate differences in phenotypes and endotypes in food allergy and asthma. Ann Allergy Asthma Immunol. 2018;121(1):117‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Boonpiyathad T, Meyer N, Moniuszko M, et al. High‐dose bee venom exposure induces similar tolerogenic B‐cell responses in allergic patients and healthy beekeepers. Allergy. 2017;72(3):407‐415. [DOI] [PubMed] [Google Scholar]
- 276. Cianferoni A, Saltzman R, Saretta F, et al. Invariant natural killer cells change after an oral allergy desensitization protocol for cow's milk. Clin Exp Allergy. 2017;47(11):1390‐1397. [DOI] [PubMed] [Google Scholar]
- 277. Schulten V, Tripple V, Seumois G, et al. Allergen‐specific immunotherapy modulates the balance of circulating Tfh and Tfr cells. J Allergy Clin Immunol. 2018;141(2):775‐777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Yao Y, Wang ZC, Wang N, et al. Allergen immunotherapy improves defective follicular regulatory T cells in patients with allergic rhinitis. J Allergy Clin Immunol. 2019;144(1):118‐128. [DOI] [PubMed] [Google Scholar]
- 279. Boonpiyathad T, Sokolowska M, Morita H, et al. Der p 1‐specific regulatory T‐cell response during house dust mite allergen immunotherapy. Allergy. 2019;74(5):976‐985. [DOI] [PubMed] [Google Scholar]
- 280. Boonpiyathad T, van de Veen W, Wirz O, et al. Role of Der p 1‐specific B cells in immune tolerance during 2 years of house dust mite‐specific immunotherapy. J Allergy Clin Immunol. 2019;143(3):1077‐1086. [DOI] [PubMed] [Google Scholar]
- 281. Sharif H, Singh I, Kouser L, et al. Immunologic mechanisms of a short‐course of Lolium perenne peptide immunotherapy: a randomized, double‐blind, placebo‐controlled trial. J Allergy Clin Immunol. 2019;144(3):738‐749. [DOI] [PubMed] [Google Scholar]
- 282. Sage PT, Alvarez D, Godec J, von Andrian UH, Sharpe AH. Circulating T follicular regulatory and helper cells have memory‐like properties. J Clin Investig. 2014;124(12):5191‐5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Gonzalez M, Dona I, Palomares F, et al. Dermatophagoides pteronyssinus immunotherapy changes the T‐regulatory cell activity. Sci Rep. 2017;7(1):11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Sun L, Jin H, Li H. GARP: a surface molecule of regulatory T cells that is involved in the regulatory function and TGF‐β releasing. Oncotarget. 2016;7(27):42826‐42836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Caruso M, Cibella F, Emma R, et al. Basophil biomarkers as useful predictors for sublingual immunotherapy in allergic rhinitis. Int Immunopharmacol. 2018;60:50‐58. [DOI] [PubMed] [Google Scholar]
- 286. Van Overtvelt L, Baron‐Bodo V, Horiot S, et al. Changes in basophil activation during grass‐pollen sublingual immunotherapy do not correlate with clinical efficacy. Allergy. 2011;66(12):1530‐1537. [DOI] [PubMed] [Google Scholar]
- 287. Tsai M, Mukai K, Chinthrajah RS, Nadeau KC, Galli SJ. Sustained successful peanut oral immunotherapy associated with low basophil activation and peanut‐specific IgE. J Allergy Clin Immunol. 2020;145(3):885‐896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Burton OT, Logsdon SL, Zhou JS, et al. Oral immunotherapy induces IgG antibodies that act through FcgammaRIIb to suppress IgE‐mediated hypersensitivity. J Allergy Clin Immunol. 2014;134(6):1310‐1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Frischmeyer‐Guerrerio PA, Masilamani M, Gu W, et al. Mechanistic correlates of clinical responses to omalizumab in the setting of oral immunotherapy for milk allergy. J Allergy Clin Immunol. 2017;140(4):1043‐1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Vickery BP, Berglund JP, Burk CM, et al. Early oral immunotherapy in peanut‐allergic preschool children is safe and highly effective. J Allergy Clin Immunol. 2017;139(1):173‐181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Ryan JF, Hovde R, Glanville J, et al. Successful immunotherapy induces previously unidentified allergen‐specific CD4+ T‐cell subsets. Proc Natl Acad Sci USA. 2016;113(9):E1286‐E1295. 10.1073/pnas.1520180113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Bedoret D, Singh AK, Shaw V, et al. Changes in antigen‐specific T‐cell number and function during oral desensitization in cow's milk allergy enabled with omalizumab. Mucosal Immunol. 2012;5(3):267‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Abdel‐Gadir A, Schneider L, Casini A, et al. Oral immunotherapy with omalizumab reverses the Th2 cell‐like programme of regulatory T cells and restores their function. Clin Exp Allergy. 2018;48(7):825‐836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Blumchen K, Trendelenburg V, Ahrens F, et al. Safety, and quality of life in a multicenter, randomized, placebo‐controlled trial of low‐dose peanut oral immunotherapy in children with peanut allergy. J Allergy Clin Immunol Pract. 2019;7(2):479‐491. [DOI] [PubMed] [Google Scholar]
- 295. Syed IA, Sulaiman SA, Hassali MA, Syed SH, Shan LH, Lee CK. Factors associated with poor CD4 and viral load outcomes in patients with HIV/AIDS. J Med Virol. 2016;88(5):790‐797. [DOI] [PubMed] [Google Scholar]
- 296. Berin MC, Grishin A, Masilamani M, et al. Egg‐specific IgE and basophil activation but not egg‐specific T‐cell counts correlate with phenotypes of clinical egg allergy. J Allergy Clin Immunol. 2018;142(1):149‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Gorelik M, Narisety SD, Guerrerio AL, et al. Suppression of the immunologic response to peanut during immunotherapy is often transient. J Allergy Clin Immunol. 2015;135(5):1283‐1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Ihara F, Sakurai D, Yonekura S, et al. Identification of specifically reduced Th2 cell subsets in allergic rhinitis patients after sublingual immunotherapy. Allergy. 2018;73(9):1823‐1832. [DOI] [PubMed] [Google Scholar]
- 299. Gueguen C, Luce S, Lombardi V, Baron‐Bodo V, Moingeon P, Mascarell L. IL‐10 mRNA levels in whole blood cells correlate with house dust mite allergen immunotherapy efficacy. Allergy. 2019;74(11):2223‐2226. [DOI] [PubMed] [Google Scholar]
- 300. Potaczek DP, Harb H, Michel S, Alhamwe BA, Renz H, Tost J. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics. 2017;9(4):539‐571. [DOI] [PubMed] [Google Scholar]
- 301. Zhang H, Kaushal A, Merid SK, et al. DNA methylation and allergic sensitizations: a genome‐scale longitudinal study during adolescence. Allergy. 2019;74(6):1166‐1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Syed A, Garcia MA, Lyu SC, et al. Peanut oral immunotherapy results in increased antigen‐induced regulatory T‐cell function and hypomethylation of forkhead box protein 3 (FOXP3). J Allergy Clin Immunol. 2014;133(2):500‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Simon D, Page B, Vogel M, et al. Evidence of an abnormal epithelial barrier in active, untreated and corticosteroid‐treated eosinophilic esophagitis. Allergy. 2018;73(1):239‐247. [DOI] [PubMed] [Google Scholar]
- 304. Akdis CA, Arkwright PD, Bruggen MC, et al. Type 2 immunity in the skin and lungs. Allergy. 2020;75(7):1582‐1605. [DOI] [PubMed] [Google Scholar]
- 305. Kubo T, Wawrzyniak P, Morita H, et al. CpG‐DNA enhances the tight junction integrity of the bronchial epithelial cell barrier. J Allergy Clin Immunol. 2015;136(5):1413‐1416. [DOI] [PubMed] [Google Scholar]
- 306. Soyka MB, Wawrzyniak P, Eiwegger T, et al. Defective epithelial barrier in chronic rhinosinusitis: the regulation of tight junctions by IFN‐gamma and IL‐4. J Allergy Clin Immunol. 2012;130(5):1087‐1096. [DOI] [PubMed] [Google Scholar]
- 307. Wawrzyniak P, Wawrzyniak M, Wanke K, et al. Regulation of bronchial epithelial barrier integrity by type 2 cytokines and histone deacetylases in asthmatic patients. J Allergy Clin Immunol. 2017;139(1):93‐103. [DOI] [PubMed] [Google Scholar]
- 308. Kortekaas Krohn I, Seys SF, Lund G, et al. Nasal epithelial barrier dysfunction increases sensitization and mast cell degranulation in the absence of allergic inflammation. Allergy. 2020;75(5):1155‐1164. [DOI] [PubMed] [Google Scholar]
- 309. Werfel T, Allam JP, Biedermann T, et al. Cellular and molecular immunologic mechanisms in patients with atopic dermatitis. J Allergy Clin Immunol. 2016;138(2):336‐349. [DOI] [PubMed] [Google Scholar]
- 310. McAleer MA, Irvine AD. The multifunctional role of filaggrin in allergic skin disease. J Allergy Clin Immunol. 2013;131(2):280‐291. [DOI] [PubMed] [Google Scholar]
- 311. Jin Y, Lu L, Tu W, Luo T, Fu Z. Impacts of polystyrene microplastic on the gut barrier, microbiota and metabolism of mice. Sci Total Environ. 2019;649:308‐317. [DOI] [PubMed] [Google Scholar]
- 312. Hole AM, Draper A, Jolliffe G, Cullinan P, Jones M, Taylor AJ. Occupational asthma caused by bacillary amylase used in the detergent industry. Occup Environ Med. 2000;57(12):840‐842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Sugita K, Altunbulakli C, Morita H, et al. Human type 2 innate lymphoid cells disrupt skin keratinocyte tight junction barrier by IL‐13. Allergy. 2019;74(12):2534‐2537. [DOI] [PubMed] [Google Scholar]
- 314. Zhou X, Wei T, Cox CW, Jiang Y, Roche WR, Walls AF. Mast cell chymase impairs bronchial epithelium integrity by degrading cell junction molecules of epithelial cells. Allergy. 2019;74(7):1266‐1276. [DOI] [PubMed] [Google Scholar]
- 315. Steelant B, Wawrzyniak P, Martens K, et al. Blocking histone deacetylase activity as a novel target for epithelial barrier defects in patients with allergic rhinitis. J Allergy Clin Immunol. 2019;144(5):1242‐1253. [DOI] [PubMed] [Google Scholar]
- 316. Kelleher MM, Dunn‐Galvin A, Gray C, et al. Skin barrier impairment at birth predicts food allergy at 2 years of age. J Allergy Clin Immunol. 2016;137(4):1111‐1116. [DOI] [PubMed] [Google Scholar]
- 317. Sindher S, Alkotob SS, Shojinaga MN, et al. Pilot study measuring transepidermal water loss (TEWL) in children suggests trilipid cream is more effective than a paraffin‐based emollient. Allergy. 2020. 10.1111/all.14275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Antonov D, Schliemann S, Elsner P. Methods for the assessment of barrier function. Curr Probl Dermatol. 2016;49:61‐70. [DOI] [PubMed] [Google Scholar]
- 319. Birgersson U, Birgersson E, Aberg P, Nicander I, Ollmar S. Non‐invasive bioimpedance of intact skin: mathematical modeling and experiments. Physiol Meas. 2011;32(1):1‐18. [DOI] [PubMed] [Google Scholar]
- 320. Fasano A, Shea‐Donohue T. Mechanisms of disease: the role of intestinal barrier function in the pathogenesis of gastrointestinal autoimmune diseases. Nat Clin Pract Gastroenterol Hepatol. 2005;2(9):416‐422. [DOI] [PubMed] [Google Scholar]
- 321. Mu Q, Kirby J, Reilly CM, Luo XM. Leaky gut as a danger signal for autoimmune diseases. Front Immunol. 2017;8:598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Jiminez JA, Uwiera TC, Douglas Inglis G, Uwiera RR. Animal models to study acute and chronic intestinal inflammation in mammals. Gut Pathog. 2015;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Camara‐Lemarroy CR, Silva C, Greenfield J, Liu WQ, Metz LM, Yong VW. Biomarkers of intestinal barrier function in multiple sclerosis are associated with disease activity. Mult Scler. 2019;1352458519863133 10.1177/1352458519863133 [DOI] [PubMed] [Google Scholar]
- 324. Bosi E, Molteni L, Radaelli MG, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49(12):2824‐2827. [DOI] [PubMed] [Google Scholar]
- 325. Fasano A. Zonulin, regulation of tight junctions, and autoimmune diseases. Ann N Y Acad Sci. 2012;1258:25‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Kiecolt‐Glaser JK, Wilson SJ, Bailey ML, et al. Marital distress, depression, and a leaky gut: translocation of bacterial endotoxin as a pathway to inflammation. Psychoneuroendocrinology. 2018;98:52‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Alinaghi M, Nguyen DN, Sangild PT, Bertram HC. Direct implementation of intestinal permeability test in NMR metabolomics for simultaneous biomarker discovery‐a feasibility study in a preterm piglet model. Metabolites. 2020;10(1):22 10.3390/metabo10010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Muraro A, Roberts G, Halken S, et al. EAACI guidelines on allergen immunotherapy: executive statement. Allergy. 2018;73(4):739‐743. [DOI] [PubMed] [Google Scholar]
- 329. European Medicines Agency . Committee for medicinal products for human use (CHMP): guideline on the clinical development of products for specific immunotherapy for the treatment of allergic diseases (CHMP/EWP/18504/2006). http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009/09/WC500003605.pdf; 2008.
- 330. Bonertz A, Roberts G, Slater JE, et al. Allergen manufacturing and quality aspects for allergen immunotherapy in Europe and the United States: an analysis from the EAACI AIT Guidelines Project. Allergy. 2018;73(4):816‐826. [DOI] [PubMed] [Google Scholar]
- 331. Kaul S, Englert L, May S, Vieths S. Regulatory aspects of specific immunotherapy in Europe. Curr Opin Allergy Clin Immunol. 2010;10(6):594‐602. [DOI] [PubMed] [Google Scholar]
- 332. Englert L, May S, Kaul S, Vieths S. The therapy allergens ordinance ("Therapieallergene‐Verordnung"). Background and effects. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012;55(3):351‐357. [DOI] [PubMed] [Google Scholar]
- 333. Mahler V, Esch RE, Kleine‐Tebbe J, et al. Understanding differences in allergen immunotherapy products and practices in North America and Europe. J Allergy Clin Immunol. 2019;143(3):813‐828. [DOI] [PubMed] [Google Scholar]
- 334. Pfaar O, Agache I, de Blay F, et al. Perspectives in allergen immunotherapy: 2019 and beyond. Allergy. 2019;74(Suppl 108):3‐25. [DOI] [PubMed] [Google Scholar]
- 335. Dhami S, Nurmatov U, Arasi S, et al. Allergen immunotherapy for allergic rhinoconjunctivitis: a systematic review and meta‐analysis. Allergy. 2017;72(11):1597‐1631. [DOI] [PubMed] [Google Scholar]
- 336. German Society for Allergology and Clinical Immunology (DGAKI) . http://www.dgaki.de/leitlinien/s2k‐leitlinie‐sit/ (accessed on 07 Dec 2018).
- 337. Roberts G, Pfaar O, Akdis CA, et al. EAACI guidelines on allergen immunotherapy: allergic rhinoconjunctivitis. Allergy. 2018;73(4):765‐798. [DOI] [PubMed] [Google Scholar]
- 338. Pfaar O, Alvaro M, Cardona V, Hamelmann E, Mosges R, Kleine‐Tebbe J. Clinical trials in allergen immunotherapy: current concepts and future needs. Allergy. 2018;73(9):1775‐1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. https://www.clinicaltrialsregister.eu/ctr‐search/search?query=2016‐000051‐27
- 340. Auge J, Vent J, Agache I, et al. EAACI Position paper on the standardization of nasal allergen challenges. Allergy. 2018;73(8):1597‐1608. [DOI] [PubMed] [Google Scholar]
- 341. Fauquert JL, Jedrzejczak‐Czechowicz M, Rondon C, et al. Conjunctival allergen provocation test: guidelines for daily practice. Allergy. 2017;72(1):43‐54. [DOI] [PubMed] [Google Scholar]
- 342. Pfaar O, Demoly P, Gerth van Wijk R, et al. Recommendations for the standardization of clinical outcomes used in allergen immunotherapy trials for allergic rhinoconjunctivitis: an EAACI Position Paper. Allergy. 2014;69(7):854‐867. [DOI] [PubMed] [Google Scholar]
- 343. Mosges R, Bachert C, Panzner P, et al. Short course of grass allergen peptides immunotherapy over 3 weeks reduces seasonal symptoms in allergic rhinoconjunctivitis with/without asthma: a randomized, multicenter, double‐blind, placebo‐controlled trial. Allergy. 2018;73(9):1842‐1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Vély F, Barlogis V, Vallentin B, et al. Evidence of innate lymphoid cell redundancy in humans. Nat Immunol. 2016;17(11):1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Pfaar O, Bachert C, Kuna P, et al. Sublingual allergen immunotherapy with a liquid birch pollen product in patients with seasonal allergic rhinoconjunctivitis with or without asthma. J Allergy Clin Immunol. 2019;143(3):970‐977. [DOI] [PubMed] [Google Scholar]
- 346. Pfaar O, Gerth van Wijk R, Klimek L, Bousquet J, Creticos P. Clinical trials in allergen immunotherapy in the age group of children and adolescents: current concepts and future needs. Clin Transl Allergy. 2020;10:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Pfaar O, Bastl K, Berger U, et al. Defining pollen exposure times for clinical trials of allergen immunotherapy for pollen‐induced rhinoconjunctivitis ‐ an EAACI position paper. Allergy. 2017;72(5):713‐722. [DOI] [PubMed] [Google Scholar]
- 348. Karatzas K, Riga M, Berger U, Werchan M, Pfaar O, Bergmann KC. Computational validation of the recently proposed pollen season definition criteria. Allergy. 2018;73(1):5‐7. [DOI] [PubMed] [Google Scholar]
- 349. Pfaar O, Karatzas K, Bastl K, et al. Pollen season is reflected on symptom load for grass and birch pollen‐induced allergic rhinitis in different geographic areas‐An EAACI task force report. Allergy. 2020;75(5):1099–1106. [DOI] [PubMed] [Google Scholar]
- 350. Pfaar O, Agache I, Bergmann K, et al. Placebo effects in allergen immunotherapy–an EAACI task force position paper. Allergy. 2020. 10.1111/all.14331 [DOI] [PubMed] [Google Scholar]
- 351. Pfaar O, Bonini S, Cardona V, et al. Perspectives in allergen immunotherapy: 2017 and beyond. Allergy. 2018;73(Suppl 104):5‐23. [DOI] [PubMed] [Google Scholar]
- 352. Guyatt GH, Oxman AD, Vist GE, et al. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ. 2008;336(7650):924‐926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Guyatt G, Oxman AD, Akl EA, et al. GRADE guidelines: 1. Introduction—GRADE evidence profiles and summary of findings tables. J Clin Epidemiol. 2011;64(4):383‐394. [DOI] [PubMed] [Google Scholar]
- 354. Santesso N, Glenton C, Dahm P, et al. GRADE guidelines 26: Informative statements to communicate the findings of systematic reviews of interventions. J Clin Epidemiol. 2020;119:126‐135. [DOI] [PubMed] [Google Scholar]
- 355. Agache I, Beltran J, Akdis C, et al. Efficacy and safety of treatment with biologicals (benralizumab, dupilumab, mepolizumab, omalizumab and reslizumab) for severe eosinophilic asthma. A systematic review for the EAACI Guidelines‐recommendations on the use of biologicals in severe asthma. Allergy. 2020;75(5):1023‐1042. [DOI] [PubMed] [Google Scholar]
- 356. Agache I, Rocha C, Beltran J, et al. Efficacy and safety of treatment with biologicals (benralizumab, dupilumab and omalizumab) for severe allergic asthma: a systematic review for the EAACI Guidelines ‐ recommendations on the use of biologicals in severe asthma. Allergy. 2020;75(5):1043‐1057. [DOI] [PubMed] [Google Scholar]
- 357. Agache I, Song Y, Rocha C, et al. Efficacy and safety of treatment with dupilumab for severe asthma: a systematic review of the EAACI guidelines‐Recommendations on the use of biologicals in severe asthma. Allergy. 2020;75(5):1058‐1068. [DOI] [PubMed] [Google Scholar]
- 358. Agache I, Lau S, Akdis CA, et al. EAACI Guidelines on Allergen Immunotherapy: house dust mite‐driven allergic asthma. Allergy. 2019;74(5):855‐873. [DOI] [PubMed] [Google Scholar]


