Abstract
Vector population control using insecticides is a key element of current strategies to prevent malaria transmission in Africa. The introduction of effective insecticides, such as the organophosphate pirimiphos-methyl, is essential to overcome the recurrent emergence of resistance driven by the highly diverse Anopheles genomes. Here, we use a population genomic approach to investigate the basis of pirimiphos-methyl resistance in the major malaria vectors Anopheles gambiae and A. coluzzii. A combination of copy number variation and a single non-synonymous substitution in the acetylcholinesterase gene, Ace1, provides the key resistance diagnostic in an A. coluzzii population from Côte d’Ivoire that we used for sequence-based association mapping, with replication in other West African populations. The Ace1 substitution and duplications occur on a unique resistance haplotype that evolved in A. gambiae and introgressed into A. coluzzii, and is now common in West Africa primarily due to selection imposed by other organophosphate or carbamate insecticides. Our findings highlight the predictive value of this complex resistance haplotype for phenotypic resistance and clarify its evolutionary history, providing tools to for molecular surveillance of the current and future effectiveness of pirimiphos-methyl based interventions.
Author summary
Control of mosquito populations via insecticidal tools or interventions is a mainstay of campaigns to reduce malaria transmission. However, especially in sub-Saharan Africa, continued insecticidal selection pressure on the most important species of Anopheles malaria mosquitoes has favoured the evolutionary selection of increasingly effective resistance mechanisms. We investigate the genetic basis of resistance to the organophosphate pirimiphos-methyl, the dominant insecticide now used for indoor residual spraying campaigns in Africa. Genome-wide association analysis of a population from Cote d’Ivoire showed that resistant specimens share a unique combination of mutations in one gene, the acetylcholinesterase enzyme, which constitute the prime cause of pirimiphos-methyl resistance. Further testing of these mutations in diagnostic assays involving two major malaria vectors, A. coluzzii and A. gambiae, validate their use as informative predictors of pirimiphos-methyl resistance. Using data from a large collection of whole genome sequenced specimens from a broader range of locations (Burkina-Faso, Côte d’Ivoire, Ghana, and Guinea), our evolutionary analyses demonstrate that these mutations emerged in A. gambiae and transferred into A. coluzzii by inter-specific hybridisation. Our results show how resistance mechanisms in key malaria vectors have developed and spread, and provide validated tools for molecular surveillance to inform public health campaigns.
Introduction
Pirimiphos-methyl is an organophosphate insecticide that is widely used in control interventions against populations of the malaria vector Anopheles, especially in Africa [1,2]. Since 2013, the World Health Organization (WHO) has recommended the use of pirimiphos-methyl for indoor residual spraying (IRS) interventions, the major anti-vector strategy in malaria control after treated bednet distribution [1,3]. Strategic approaches to vector control often rely on the use of multiple insecticides to avoid or overcome the recurrent emergence of resistance in natural populations [4]. In that regard, various insecticide classes have been used in IRS, with pyrethroids—which target the voltage-gated sodium channel—being the dominant choice until recently [5,6]. However, the increase in pyrethroid resistance in Anopheles populations [7] has led to a progressive replacement with acetylcholinesterase-targeting insecticide classes, first the carbamate bendiocarb, and, latterly, the organophosphate pirimiphos-methyl [1]. Pirimiphos-methyl is the active ingredient in the most widely used insecticide for IRS in Africa, the spray formulation Actellic, which is highly effective and has strong residual performance [2,5,8,9]. However, resistance has recently been reported in several populations of African Anopheles s.l. [10,11], and though control failures have yet to be reported, it represents a clear threat to the efficacy of IRS strategies.
Mechanisms of resistance to pirimiphos-methyl are poorly understood, but organophosphates, as well as carbamates, all block the action of the acetylcholinesterase enzyme (ACE1, encoded by the Ace1 gene in mosquitoes) via competitive binding to its active site [12]. Studies in culicine and anopheline mosquitoes have found that non-synonymous mutations in Ace1 can result in resistance to carbamates and organophosphates other than pirimiphos-methyl [13–15]. The most common mutation in Anopheles is a glycine to serine mutation in codon 280 (G280S, also known as G119S after the codon numbering of a partial crystal structure from the electric ray Torpedo californica [13,15,16]), which is located near the active site gorge of ACE1 [17] and decreases its sensitivity to organophosphates and carbamates. This mutation results in reduced sensitivity to the acetylcholine neurotransmitter [18], thus carrying potential fitness costs, as demonstrated in Culex pipiens [19]. The resistance allele 280S has been regularly found in natural Anopheles populations: A. gambiae s.s. (henceforth, A. gambiae) and A. coluzzii from tropical West and Central Africa [13,20–25]; as well as in A. sinensis [26]. In addition, Ace1 is subject to duplication polymorphisms (copy number variants, or CNVs) that co-segregate with the 280S allele in both A. gambiae and A. coluzzii, and enhance resistance to carbamates and organophosphates, such as fenitrothion and chlorpyrifos-methyl [22,24,27–31].
In this study we provide an in-depth investigation of the relationship between Ace1 mutations and pirimiphos-methyl resistance in A. gambiae and A. coluzzii using whole-genome sequenced samples from the Anopheles gambiae 1000 Genomes project [32,33], and a wider testing of phenotyped specimens from across West Africa. In addition, we perform a first agnostic genome-wide scan for candidate regions contributing to pirimiphos-methyl resistance in a population of A. coluzzii from Côte d’Ivoire. Finally, we study the contemporary evolution of Ace1 to answer unresolved questions on the selective pressures and the pattern of inter-specific introgression associated with the spread of this resistance mechanism. Earlier studies provided support for the idea that a common resistance haplotype under positive selection might have introgressed between species [22,24,27], but they focused on a partial region of the Ace1 gene and did not address the relationship between introgression and the duplication, which extends beyond the gene [30,34]. Here we leverage population genomic resources from the Anopheles gambiae 1000 Genomes to overcome these limitations. Overall, our results demonstrate a widespread and dominant role for Ace1 mutations in pirimiphos-methyl resistance, provide critical insights into resistance diagnosis, and demonstrate that 280S alleles and Ace1 duplications co-occur on a single, swept, resistance haplotype that originated in West African A. gambiae and later introgressed into A. coluzzii.
Results
Conservation and distribution of Ace1 resistance mutations in Anopheles
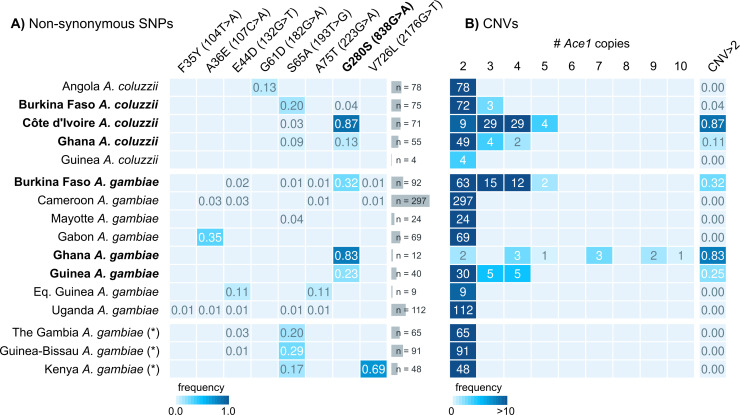
We examined the frequency and distribution of the two Ace1 mutations that have been associated with organophosphate and carbamate resistance in A. gambiae and A. coluzzii: the G280S non-synonymous single nucleotide polymorphism (SNP), and copy number variation (CNV) polymorphisms of Ace1 and the surrounding genomic region. G280S is sometimes known as G119S [15] based on its position in the truncated crystal structure of its homolog in the electric ray Torpedo californica, where ACE1 protein structure was first elucidated [16]. Due to a culicine-specific N-terminal insertion in ACE1, the exact position of this conserved codon differs among animal orthologs (S1 Data). We provide a list of homologous codon positions for Ace1 orthologs from selected animal species, including common insect vectors (S2 Data). Henceforth, we will use A. gambiae s.l.-based codon coordinates and refer to this SNP as G280S (wild-type allele, wt: 280G; resistant allele: 280S; gene accession number: AGAP001356-RA in AgamP4.12).
In the Anopheles 1000 Genomes cohort (Phase 2, n = 1142 genomes; Fig 1) [32], the 280S resistance allele is present across West African populations of A. coluzzii (Côte d’Ivoire, Burkina Faso, Ghana) and A. gambiae (Burkina Faso, Ghana, Guinea), with the highest frequencies observed in Ghanaian A. gambiae (83% of specimens carry 280S alleles) and Ivorian A. coluzzii (87%; Fig 1A), which is consistent with previous results [20–22,27]. According to the database of CNVs in the 1000 Genomes dataset generated by Lucas et al. [34] (which we have used in our analyses) Ace1 duplications have a similar distribution to the SNP in West African populations, as they overwhelmingly overlap with specimens also carrying 280S alleles (Figs 1B and 2; see below).
Fig 1. Ace1 mutations in African populations.
A) Frequency of non-synonymous SNPs in the Ace1 gene in African A. gambiae and A. coluzzii populations from Anopheles gambiae 1000 Genomes, Phase 2. For each SNP, we indicate peptide- and transcript-level coordinates and substitutions. B) Ace1 CNVs across African populations, including the frequency of specimens with >2 copies in each population. A diploid genome without duplications would have 2 copies. Populations where G280S and duplications are present are highlighted in bold text. Note: populations denoted with an asterisk (The Gambia, Guinea-Bissau and Kenya) have high frequency of hybridisation and/or unclear species identification.
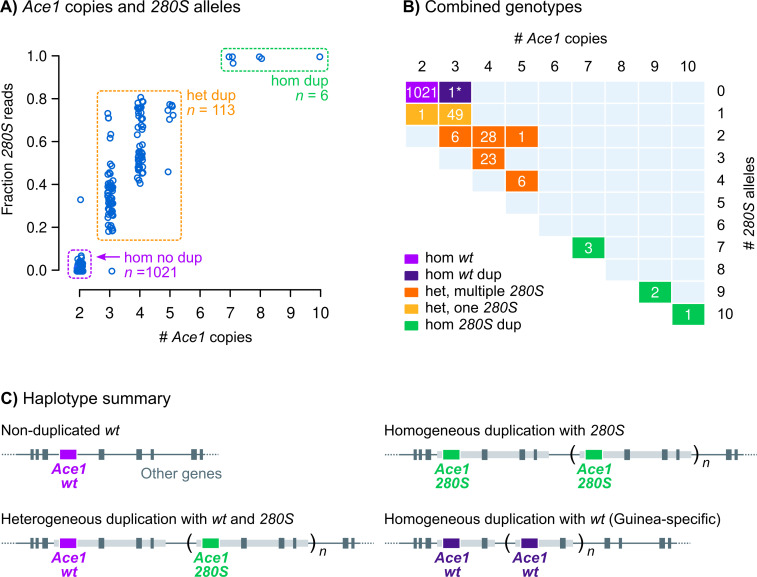
Fig 2. Combinations of Ace1 G280S and CNV genotypes.
A) Fraction of reads supporting 280S alleles and number of Ace1 copies (1000 Genomes dataset, n = 1142). Boxes highlight groups of haplotypes: non-duplicated wt (purple), duplications with both wt and 280S alleles (orange) and duplications with only 280S alleles (green). B) Breakdown of observed genotypes at the Ace1 locus according to Ace1 and 280S copy number. The asterisk (*) denotes a wt-homozygous specimen from Guinea that carries an independently evolved Ace1 duplication. C) Diagrammatic summary of Ace1 haplotypes and their surrounding flanks; light grey rectangles show duplicated regions.
The combination of CNVs and 280S alleles results in multiple genotypes being observed in the Ace1 locus, defined by a variable number of 280S copies. We used the fraction of sequencing reads supporting the wt and 280S alleles to estimate the number of 280S alleles in each sample (Fig 2A and 2B), which revealed that most specimens with duplications carry both wt and 280S alleles at various frequencies (n = 113, orange box in Fig 2A), whereas the vast majority of non-duplicated specimens only have wt alleles (n = 1021, purple box in Fig 2A). In addition, we identify six specimens with high Ace1 copy numbers that only carry 280S alleles, present in the Ghanaian A. gambiae population (Fig 1B).
Virtually all Ace1 CNVs identified in the 1000 Genomes cohort share the same duplication breakpoints [34], spanning a region ca. 200 kbp that includes a total of 11 genes (S5 Data). These breakpoints coincide with those of the strict tandem duplications identified by Assogba et al. in A. gambiae and A. coluzzii samples from the same region [30] (S17 Data). Fig 2C summarises the combinations of 280S and duplications that can be observed in this dataset: (i) non-duplicated loci with wt Ace1; (ii) chromosomes with Ace1 tandem duplications carrying wt and 280S alleles (heterogeneous duplications); and (iii) chromosomes containing multiple 280S alleles (homogeneous duplications). In addition to these three model haplotypes, one specimen from Guinea carries a distinct wt-homogeneous duplication (asterisk in Fig 2B). This Guinea-specific duplication is shorter than the one found in all other samples and has different breakpoints (ca. 70 kbp, including Ace1 and one other gene; S4 Data), implying an independent origin. Henceforth, all mentions of Ace1 CNVs will refer to the major duplication. The absence of specimens carrying the major duplication and lacking 280S alleles strongly implies that this CNV contains both 280S and wt alleles on the same chromosome.
In addition to G280S, we found seven non-synonymous SNPs in Ace1 with at least 1% frequency in at least one population (Fig 1A). None were in linkage disequilibrium with G280S (S3 Data), nor have any of them been previously associated with resistance [15]. The most salient of these additional mutations was a serine-to-alanine SNP in codon 65 (S65A), with frequencies of up to 30% in multiple West African populations. Codon 65 maps to the anopheline-specific N-terminal insertion of ACE1 proteins (S1) Data, which lacks predicted secondary structure and is far away from the active-site gorge of the enzyme [17]. This position is not conserved across ACE1 orthologs, being variously encoded as alanine, serine or threonine in different Anopheles species (S1 Data).
Pirimiphos-methyl resistance in Ivorian A. coluzzii is linked to Ace1 duplications and multiple 280S alleles
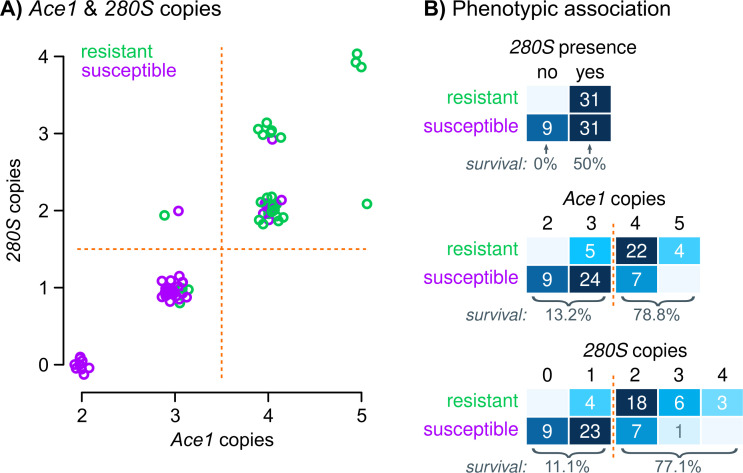
Next, we examined the association between Ace1 mutations and resistance to pirimiphos-methyl (Fig 3). We used 71 A. coluzzii female mosquitoes from Côte d’Ivoire, collected from rice fields in Tiassalé in 2012, that had been tested for resistance prior to genome sequencing. The G280S SNP and CNV co-occur at high frequencies in this population, with 87.3% of specimens carrying heterogeneous duplications and one or more 280S alleles (Fig 3A). Specimens with at least one 280S allele had a survival rate of 50%, as opposed to 0% in the wt homozygotes (Fig 3B), suggesting that 280S is linked to pirimiphos-methyl resistance in this population (p = 3.7 × 10−3 in Fisher’s exact test, Woolf-corrected odds ratio = 19.0 [95% confidence interval = 1.1–340.6]) but it does not fully explain the resistance phenotype. The only other non-synonymous SNP present in Ivorian A. coluzzii, S65A, was not associated with pirimiphos-methyl resistance (p = 0.605 Fisher’s exact test; S6A and S6B Data).
Fig 3. Genotype-phenotype association in Ivorian A. coluzzii.
A) Number of Ace1 copies compared to the estimated number of 280S copies in Ivorian A. coluzzii (n = 71), color-coded according to resistance phenotypes. Random jitter has been added for clarity. B) Cross-tabulation of pirimiphos-methyl resistance and three Ace1 mutations in Ivorian A. coluzzii: 280S allele presence, number of Ace1 copies, and number of 280S alleles. Orange lines denote ad hoc groups of genotypes where we identify changes in survival rates (included at the bottom of each table). Source data available in S6.
We found a strong bias towards higher Ace1 copy numbers in resistant specimens, all of which had at least 3 copies of the Ace1 locus and most of them had 4 or more (Fig 3B; odds ratio from a binomial generalised linear model [GLM] = 16.9 [5.1–56.6]; p = 6.4 × 10−10 in a χ2 test compared to a null model). Concordantly, the estimated number of 280S alleles was also associated with resistance (GLM odds ratio = 10.7 [3.6–31.7]; p = 8.9 × 10−10 in χ2 test): survival rates increased with higher number of 280S copies (Fig 3B), with a threshold apparent between individuals with zero or one copies of 280S (11.1% survival) and those with two or more (77% survival). Crucially, the group of duplicated specimens with one 280S and two wt alleles (n = 36), did not show a statistically significant increase in survival compared to non-duplicated wt specimens (Fisher’s exact test p = 0.553; Woolf-corrected odds ratio = 3.6 [0.2–74.3]).
We examined the combined effects of these mutations (280S presence, CNVs and number of 280S alleles) using binomial GLMs and a step-wise variable addition procedure (S6B Data). Starting from a basic model based on the presence of 280S, we found that adding the number of 280S copies improved its predictive ability (at p = 2.56 × 10−7 in a χ2 ANOVA test), and so did the addition of CNVs (at p = 1.96 × 10−7). On the other hand, a full model with all three mutations was not significantly better than simpler models excluding either of the two duplication-related variables (at p = 0.16–0.22 in χ2 tests), and it was also not significantly better than a minimal model with CNVs as the only remaining variable, obtained according to the Bayesian Information Criterion (p = 0.44). Together, these results suggest that duplication-related mutations are necessary to explain the observed levels of resistance in this population, although it is difficult to disentangle their effects due to the fact that they co-occur in the same specimens in this dataset.
Pirimiphos-methyl resistance diagnostics are repeatable across A. gambiae and A. coluzzii populations
We investigated the extent to which the role of Ace1 and 280S copy numbers in pirimiphos-methyl resistance could be generalised to other populations beyond the A. coluzzii from Tiassalé. Specifically, we surveyed Ace1 mutations and phenotypic resistance in nine West African populations of A. gambiae and A. coluzzii from additional locations in Côte d’Ivoire, Togo, and Ghana (Table 1 and Table 2; list of specimens in S6C and S6D Data).
Table 1. Association of resistance genotypes with pirimiphos-methyl resistance in nine West African populations of A. coluzzii and A. gambiae.
For each population, we report number of sampled specimens (n), the number of specimens with only wt, only 280S, and both alleles; survival frequency among mutated groups, and the odds ratios of survival in specimens with 280S alleles (if available) in a binomial generalised linear model, and the p-value of this model in a χ2 ANOVA comparison with a null model. Odds ratios (OR) are reported with 95% confidence intervals. Dagger signs (†) denote survival rates in genotypes with fewer than 5 specimens. Samples listed in S6C Data; statistical analysis in S6D Data. Country abbreviations: CI, Côte d’Ivoire; GH, Ghana; TG, Togo.
| Population | n | Frequencies wt / wt+280S / 280S | % survival wt+280S | OR wt+280S | % survival 280S | OR 280S | p |
|---|---|---|---|---|---|---|---|
| coluzzii Aboisso (CI) | 55 | 42 / 13 / 0 | 46.1% | 35.1 (3.6–338.0) |
- | - | 1.3 × 10−4 |
| coluzzii Korle-Bu (GH) | 214 | 25 / 165 / 24 | 49.6% | 4.2 × 107 (0 –Inf) |
95.8% | 9.8 × 108 (0 –Inf) |
1.2 × 10−13 |
| coluzzii Madina (GH) | 42 | 2 / 37 / 3 | 37.0% | 0.4 (0.02–6.5) |
100% (†) | 4.3 × 107 (0 –Inf) |
0.023 |
| coluzzii Weija (GH) | 131 | 60 / 70 / 1 | 54.1% | 7.3 (3.0–17.6) |
100% (†) | 3.7 × 107 (0 –Inf) |
2.4 × 10−6 |
| gambiae Aboisso (CI) | 82 | 3 / 45 / 34 | 77.8% | 7.0 (0.6–85.4) |
97% | 66.0 (2.9–1491.2) |
3.7 × 10−3 |
| gambiae Madina (GH) | 172 | 19 / 112 / 41 | 58.9% | 12.2 (2.7–55.4) |
100% | 9.8 × 108 (0 –Inf) |
4.4 × 10−14 |
| gambiae Obuasi (GH) | 140 | 0 / 28 / 112 | 17.8% | - | 58.9% | 6.6 (2.3–18.6) |
6.0 × 10−5 |
| gambiae Weija (GH) | 13 | 7 / 4 / 2 | 75% (†) | 7.3 × 1017 (0 –Inf) |
100% (†) | 2.6 × 109 (0 –Inf) |
1.6 × 10−3 |
| gambiae Baguida (TG) | 102 | 0 / 14 / 88 | 57.1% | - | 56.8% | 0.9 (0.3–3.1) |
0.982 |
Table 2. Association of Ace1 duplications with pirimiphos-methyl resistance in specimens carrying both 280G and 280S alleles, from nine West African populations of A. coluzzii and A. gambiae.
For each population, we report number of sampled specimens (n), and the results of three binomial generalised linear models (GLM): (i) using the number of Ace1 copies as the predictor variable, (ii) using the 280S-to-280G allelic ratio, and (iii) a minimal model obtained with step-wise reduction, according to the Bayesian Information Criterion (BIC). For each model, we report the p-value from a comparison with a null model (ANOVA χ2 test) and odds ratios (OR) with 95% confidence intervals for the variables included in the model. Sample phenotypes and genotypes listed in S6E Data; statistical analysis in S6F Data. Country abbreviations: CI, Côte d’Ivoire; GH, Ghana; TG, Togo. Note that the Weija A. gambiae population was excluded from this analysis due to small sample size.
| Population | n | GLM Ace1 copies (single variable) | GLM 280S/280G allelic ratio (single variable) | Minimal GLM (BIC) | ||||
|---|---|---|---|---|---|---|---|---|
| OR | p | OR | p | OR Ace1 copies | OR allelic ratio | p | ||
| coluzzii Aboisso (CI) | 11 | 1.3 (0.7–2.3) |
0.361 | 52.9 (0.01 – 2.2 × 105) |
1.3 × 10−3 | - | 52.9 (0.01 – 2.2 × 105) |
1.3 × 10−3 |
| coluzzii Korle-Bu (GH) | 46 | 1.8 (1.2–2.6) |
1.0 × 10−4 | 2.8 (1.6–5.1) |
4.1 × 10−7 | 1.5 (0.9–2.3) |
2.3 (1.3–4.0) |
3.0 × 10−7 |
| coluzzii Madina (GH) | 34 | 1.4 (1.04–1.8) |
0.013 | 71.5 (3.3–1539.8) |
9.9 × 10−4 | - | 71.5 (3.3–1539.8) |
9.9 × 10−4 |
| coluzzii Weija (GH) | 21 | 0.5 (0.3–1.0) |
0.013 | 3.4 (0.4–28.6) |
0.021 | 0.4 (0.1–1.1) |
50 (0.3–9505.0) |
2.9 × 10−3 |
| gambiae Aboisso (CI) | 31 | 1.0 (0.6–1.5) |
0.933 | 2.6 (0.3–24.9) |
0.382 | - | - | - |
| gambiae Madina (GH) | 19 | 5.2 (1.1–23.9) |
7.7 × 10−3 | 6.5 × 1085 (0 –Inf) |
2.1 × 10−6 | 8.7 × 10−20 (0 –Inf) |
5.5 × 1045 (0 –Inf) |
2.6 × 10−9 |
| gambiae Obuasi (GH) | 26 | 1.5 (0.9–2.3) |
0.038 | 3.1 (0.85–10.8) |
0.038 | 1.5 (0.9–2.3) |
- | 0.038 |
| gambiae Weija (GH) | 0 | - | - | - | - | - | - | - |
| gambiae Baguida (TG) | 13 | 1.4 (0.7–2.6) |
0.350 | 2.9 (0.1–69.4) |
0.498 | - | - | - |
Pirimiphos-methyl resistance was associated with G280S mutations in seven out of nine populations (p < 0.01 in a χ2 test comparing GLM of each genotype to a null model; Table 1). However, whilst 280S homozygotes were strongly associated with resistance in populations where they were present in significant numbers (e.g. 95.8% survival rate in A. coluzzii from Korle-Bu), specimens with both 280S and wt alleles often exhibited lower, intermediate survival rates (e.g. 49.6% in A. coluzzii from Korle-Bu; six out of nine populations had survival rates <60%; Table 1) similar to that of the A. coluzzii population from Tiassalé (Côte d’Ivoire) mentioned above (Fig 3A).
Following our findings for the Tiassalé population (Fig 3), we further investigated the phenotypic variation in specimens with both 280S and wt alleles using two CNV-related variables: (i) the ratio of 280S to 280G alleles (measured as the ratio of FAM-to-HEX dye signal in a TaqMan qPCR assay) [35]; and (ii) the estimated number of Ace1 copies, normalised to two single-copy references genes, and assessed relative to the wt non-duplicated Kisumu A. gambiae colony [28] (see Methods).
Pirimiphos-methyl resistance was associated with CNV-related measures in specimens carrying both wt and 280S alleles from A. gambiae and A. coluzzii populations, according to generalised linear models that included 280S allelic ratios and/or Ace1 copy number as potential informative markers (Table 2). In A. coluzzii from Korle-Bu and Weija, as well as A. gambiae from Madina, both the 280S allelic ratio and the Ace1 copy number were included in the minimal model (at p < 0.01 in a χ2 comparison with a null model); whereas in A. coluzzii from Aboisso and Madina the allelic ratio was the only variable associated with resistance (at p < 10−3 in χ2 tests). In Obuasi A. gambiae specimens with both wt and 280S alleles, the minimal model only included Ace1 copy number (at p = 0.038) but neither measure was strongly predictive, as was the case in A. gambiae from Aboisso and Baguida. Overall, these results indicate that the combination of 280S allelic ratios and CNVs provides a similar pirimiphos-methyl resistance mechanism in both species. However, diagnostic capacity appears to vary among populations, suggesting the possible existence of additional resistance mechanisms.
Genome-wide identification of pirimiphos-methyl resistance variants in Ivorian A. coluzzii
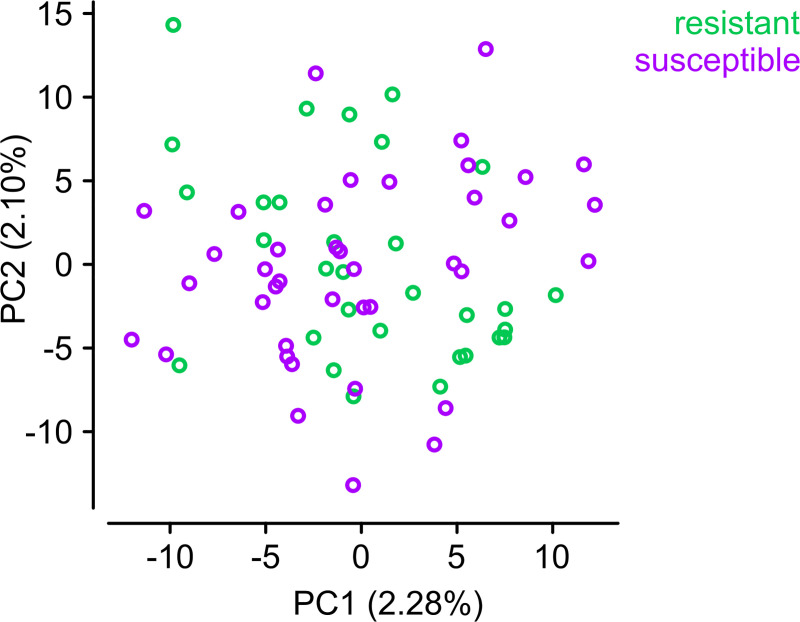
We examined the genomes of the 71 A. coluzzii specimens from Côte d’Ivoire to identify additional genetic variants—other than Ace1 G280S and duplications—that could be linked to pirimiphos-methyl resistance. All Ivorian specimens had been collected in the same location and time (Tiassalé, 2012), before the widespread adoption of pirimiphos-methyl in IRS interventions [1,3]. The absence of a long period of pirimiphos-methyl use prior to collection implies that adaptation to this insecticide would presumably be caused by cross-resistance with previously employed insecticides and/or standing variation, rather than novel selective sweeps. In accordance with this expectation of low differentiation, a principal component analysis of genetically unlinked variants (see Methods) did not reveal population stratification between resistant and susceptible mosquitoes (Fig 4), and average Hudson’s FST between them was low along all chromosomal arms (FST ≈ 0%).
Fig 4. Principal component analysis of Côte d’Ivoire A. coluzzii.
PCA constructed using genotypes of 791 unlinked variants from chromosome 3.
We thus aimed to identify signals of selection in resistant A. coluzzii using the population branch statistic (PBS, Fig 5A), which identifies regions with an excess of genetic differentiation in a focal population (here, resistant Ivorian mosquitoes) relative to a basal level of differentiation between a closely related population (susceptible Ivorian mosquitoes) and a more distant control (a population of A. coluzzii from Angola; see Methods). PBS is a powerful test to detect recent selection acting on incomplete sweeps and standing variation [36,37], and is therefore well-suited to investigate a scenario—like ours—in which the population of interest has not yet diverged.
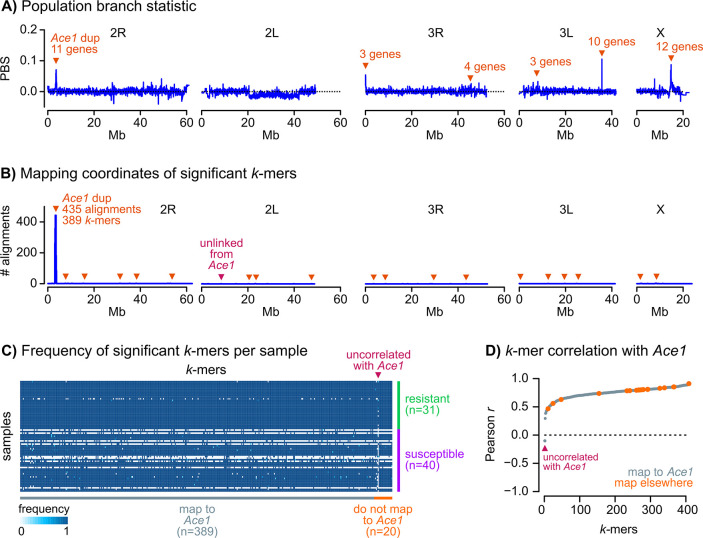
Fig 5. Genome-wide scan of variants associated with pirimiphos-methyl resistance in Ivorian A. coluzzii.
A) Profile of population branching statistics along all chromosomal arms, calculated in consecutive blocks of 1000 segregating variants, using resistant and susceptible Ivorian A. coluzzii as populations A and B, and Angolan A. coluzzii as outgroup. Orange triangles indicate windows with extreme PBS values (p-values derived from a standardised distribution of PBS along each chromosomal arm, and FDR < 0.001), and the number of genes therein. B) Mapping coordinates of k-mers significantly associated with pirimiphos-methyl (n = 439). Most k-mers map to the Ace1 duplication region (n = 414) or, despite mapping elsewhere in the genome (n = 24), are correlated with Ace1 copy number (orange triangles). Only one k-mer mapping outside of the Ace1 duplication is not correlated with Ace1 copy number (purple triangle). C) Normalised frequency of each significant k-mer (n = 439, horizontal axis) in each genome (n = 71, vertical axis). k-mers are sorted according to their mapping location (in Ace1 or elsewhere), and genomes are sorted according to their phenotype (resistant/susceptible). D) Pearson’s correlation coefficients (r) between k-mer frequency and number of Ace1 copies in each genome (n = 439 significant k-mers). k-mers are coloured according to their mapping location (in Ace1 or elsewhere) and sorted by the values of r.
We estimated PBS along genomic windows to identify regions in which resistant Ivorian specimens had an excess of genetic differentiation (see Methods). In total, we identified 43 genes within six regions with excess differentiation in resistant A. coluzzii (PBS > 0, FDR < 0.001; see Methods; Fig 5A). This set of candidates contained eleven mostly uncharacterised genes located downstream of Ace1 and within the duplicated region, two ionotropic receptors (GLURIIb and GLURIIa), and several proteases, among others (S7 Data). However, a detailed examination of haplotypes in these regions did not identify selection signatures specific to the resistant specimens, either as swept haplotypes or increased Garud H12 values (S16 Data). In fact, genetic differentiation between resistant and susceptible specimens in each of these genes was low (FST < 6% in all cases, including coding and non-coding variants; S7 Data). These results suggest that they are unlikely to be directly associated with the resistance phenotype. The Ace1 gene itself was on the verge of the significance threshold (FDR = 3.2 × 10−3, PBS = 0.031) and exhibited low differentiation (FST = 1.2%). This apparent contradiction regarding the Ace1 phenotypic association can be explained by the fact that pirimiphos-methyl resistance is caused by a combination of mutations—a SNP within a heterogeneous duplication—that is not captured by the diploid allelic frequencies used in PBS calculations [36].
To overcome this limitation, we performed a k-mer association study comparing the resistant and susceptible Ivorian A. coluzzii. k-mer association studies test for differential frequencies of tracts of nucleotides of length k in different groups of genomes, and are able to identify genetic variation patterns linked to both SNPs and structural variants, such as CNVs, without requiring a reference annotation [38]. We identified ca. 767 million different k-mers of length k = 31 bp across all 71 samples (see Methods). Among these, 9,603 k-mers were significantly enriched in resistant specimens (Spearman correlation test, FDR < 0.001). The 9,603 significant k-mers were assembled into 446 unique sequences composed of more than one k-mer (median length = 54 bp), which we then aligned to the A. gambiae reference genome. We retained sequences that could be mapped to chromosomes 2, 3 or X (409 out of 446) for further analysis (listed in S8 Data). Among these, the vast majority (n = 389, 95.1%) aligned to the Ace1 duplicated region, while the remaining significant sequences aligned in scattered regions across the rest of the genome (n = 20, 4.9%; Fig 5B). Yet, 19 out of 20 k-mers that did not map to Ace1 had a very similar frequency profile in resistant and susceptible samples than the 389 Ace1-linked k-mers, being absent and present in the same genomes (Fig 5C). In fact, the k-mer frequencies of these 19 sequences was strongly correlated with Ace1 copy number (Pearson correlation r > 0 and p < 1 × 10−4; Fig 5D). This suggests that these 19 k-mers represent a non-independent signal that also reflects the association of Ace1 mutations with resistance, and can be parsimoniously explained by non-specific sequence alignments (see Discussion). Only one remaining sequence did not correlate with Ace1 copy number (Pearson correlation r < 0.0, maroon arrow in Fig 5C and 5D), indicating that it may represent another variant involved in resistance. The primary alignment of this sequence mapped to a non-coding region of chromosomal arm 2L (from 2L:8,662,023), with no proximity to any gene of known function, and did not exhibit signals of selection (S16 Data). Altogether, these results support the conclusion that Ace1 mutations are the primary driver of pirimiphos-methyl resistance in this A. coluzzii population.
Selection and introgression of Ace1 duplications in A. gambiae and A. coluzzii
A high degree of geographical and phylogenetic overlap is evident between Ace1 duplications and 280S alleles across four countries (Côte d’Ivoire, Ghana, Burkina Faso and Guinea) and two different species (A. coluzzii and A. gambiae; Fig 2A). Previous studies provide two key insights to understand this pattern. First, Ace1 duplications across West African populations have concordant breakpoints [30,34], which suggests they share a common evolutionary origin despite their multi-species distribution. Second, partial sequencing of Ace1 has shown that 280S alleles share highly similar haplotypic backgrounds in both West African A. gambiae and A. coluzzii [22,24,27], which is suggestive of inter-specific introgression and a selective sweep. The most parsimonious hypothesis linking our results and these observations would posit that (i) the high similarity of 280S-carrying haplotypes across the A. gambiae–A. coluzzii species boundary is shared along the entire duplicated region (ca. 200 kbp); and (ii) this similarity is due to a selective sweep linked to 280S and the duplication, which was followed by introgression between A. gambiae and A. coluzzii.
We tested this hypothesis by examining the profile of haplotype conservation and signatures of selection around the Ace1 duplication. To this end, we first had to phase the Ace1 duplication. However, accurate variant phasing within the duplication is confounded by the fact that this region is effectively polyploid in multiple samples, which results in uneven sequencing depth along this region (S17B Data) and a low density of phased variants (S9A and S9B Data). To circumvent these limitations, we scanned the duplication region to find tagging variants that (i) were in strong linkage disequilibrium with G280S and the duplication (Huff and Rogers’ r > 0.95; S9B Data, C); and (ii) belonged to diploid regions within the duplication (S9D–S9F Data). We found three tagging variants embedded in regions that fitted this definition, located within −26 kbp and +12 kbp of G280S (see Methods for details). We used these tagging variants to phase the duplication, building micro-haplotype networks (minimum spanning trees; Fig 6) and identifying sets of haplotypes in linkage disequilibrium with the Ace1 mutations.
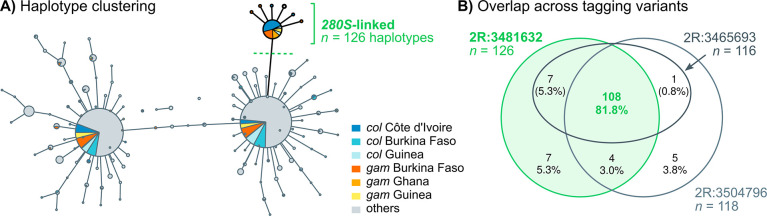
Fig 6. Haplotype clustering network around Ace1.
A) Minimum spanning tree of haplotypes around the 280S-linked variant 2R:3481632 (± 300 bp, n = 104 phased variants). Node size reflects number of haplotypes belonging to each cluster, which are color-coded according to species and geographical origin. Edges link haplotype clusters separated by one substitution. Singleton clusters have been removed from this view (see S10 Data). A cluster of identical haplotypes linked to the 280S-tagging variant is highlighted in green. B) Venn diagram representing the overlap of samples belonging to the 280S-linked haplotype clusters identified around each of the three tagging variants (S10 Data).
The minimum spanning tree built from haplotypes located around the first tagging variant (Fig 6A) identified a cluster of 126 identical haplotypes carrying the 280S-linked allele (Fig 6A, labelled with green text) and two larger haplotype clusters linked to the wt 280G allele (Fig 6A). The 280S cluster contains identical haplotypes from West African populations (Côte d’Ivoire, Burkina Faso, Ghana and Guinea) of both A. coluzzii and A. gambiae. On the other hand, wt-linked clusters have wider geographical distributions (including samples from West, Central and Eastern sub-Saharan Africa). The other three tagging variants show haplotype networks with a similar structure, with clusters of ca. 120 280S-linked identical haplotypes originating in the same A. gambiae and A. coluzzii specimens from West Africa (Fig 6B, S10 Data). The consistent clustering among the three tagging variants implies that these micro-haplotype networks reflect genetic signatures common across the duplicated region, and are therefore robust to the difficulties in phasing variants within a heterogeneous duplication (S9 Data). Thus, this result suggests that haplotype similarity between A. gambiae and A. coluzzii extends beyond the Ace1 gene.
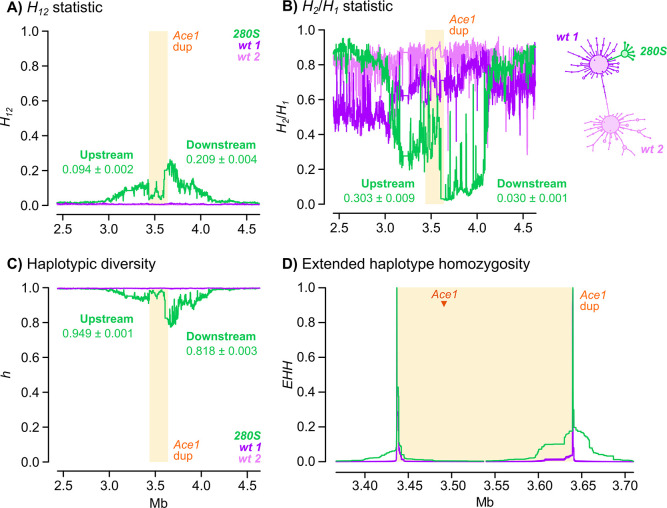
We also investigated possible signals of positive selection upstream and downstream of the Ace1 duplication breakpoints using Garud’s H statistics, haplotypic diversity, and extended haplotype homozygosity (Fig 7, S11 and S12 Data). The profile of Garud’s H12 statistic in 280S-linked haplotypes showed peaks both upstream and downstream of the Ace1 duplication (Fig 7A), which coincided with a low H2/H1 ratio (Fig 7B) and low haplotypic diversity (Fig 7C), and is thus indicative of a hard selective sweep in this region [39,40]. Notably, the region of extended haplotype homozygosity associated with this sweep was still apparent upstream and downstream of the duplication breakpoints, i.e., far away from the focal tagging variant used to define 280S-linked haplotypes (Fig 7D).
Fig 7. Positive selection around the Ace1 duplication.
A-C) Profile of Garud’s H12, H2/H1 and haplotypic diversity around the Ace1 duplication, for haplotypes carrying the 280S- or wt-tagging variants at 2R:3481632. Statistics are calculated in blocks of 500 variants with 20% block overlap. Includes averages of each statistic outside the duplication, upstream and downstream of the breakpoints (with standard errors from jackknife haplotype resampling). D) Extended haplotype homozygosity (EHH) at the duplication breakpoints for haplotypes carrying the 280S- or wt-tagging variants at the 2R:3481632 locus. Additional plots for all tagging variants are available in S11 and S12 Data. Note that Garud’s H statistics and EHH assume a diploid genome, and estimates from within the duplication can thus be biased.
Next, we tested whether 280S alleles and the duplication spread jointly between A. coluzzii and A. gambiae via introgression (Fig 8). We examined the chromosome-wide profile of introgression between A. gambiae and A. coluzzii populations using Patterson’s D statistic [41,42], which compares allele frequencies between three putatively admixing populations (A, B and C) and one outgroup (O); and can identify introgression between populations A and C (D > 0) or B and C (D < 0). Specifically, we tested whether duplicated A. coluzzii from West African populations (population A) introgressed with A. gambiae populations from West and Central Africa (C), using wt Angolan A. coluzzii as a control (B) and A. arabiensis as an outgroup (O).
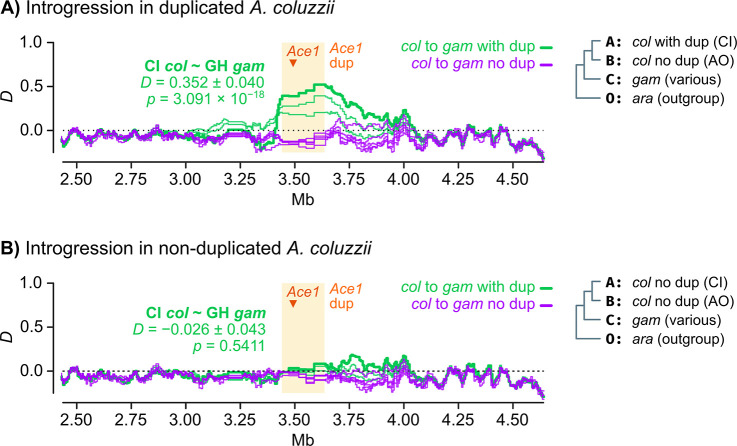
Fig 8. Introgression of the Ace1 duplication.
A) Profile of Patterson’s D statistic around the Ace1 duplication, testing evidence of introgression between duplicated A. coluzzii from Côte d’Ivoire (population A) and various A. gambiae populations (population C) with or without duplications (green and purple lines, respectively). We used non-duplicated Angolan A. coluzzii as a contrast (population B) and A. arabiensis as outgroup (O). D is calculated in windows of 5,000 variants with 20% overlap, and the average value of D along the duplication region is shown for the Côte d’Ivoire A. coluzzii / Ghana A. gambiae comparison, with standard errors derived from block-jackknife. B) Id., but using non-duplicated A. coluzzii from Côte d’Ivoire as population A. Detailed lists of all statistical tests and replicate analyses with additional populations are available in S13 Data. Species abbreviations: gam, A. gambiae; col, A. coluzzii; ara, A. arabiensis. Country abbreviations: AO, Angola; CI, Côte d’Ivoire; GH, Ghana.
We evaluated whether the Ace1 duplication introgressed between A. gambiae and A. coluzzii in two scenarios: (i) in specimens with CNVs from populations with Ace1 resistance mutations (Fig 8A), and (ii) in non-duplicated specimens from the same populations (Fig 8B). We only find evidence of introgression in the first case, i.e. between West African A. gambiae and A. coluzzii specimens carrying Ace1 duplications (Fig 8A). For example, we found evidence of introgression between duplicated Ghanaian A. gambiae and duplicated Ivorian A. coluzzii (D = 0.352 +/- 0.040, p = 3.1 × 10−18 from a standard distribution of block-jackknifed estimates; green lines in Fig 8A); but not with non-duplicated Ivorian A. coluzzii (D = -0.026 +/- 0.043, p = 0.54; green lines in Fig 8B). In all comparisons where introgression was apparent, the genomic region of increased D values extended along the entire duplicated region. On the other hand, there was no evidence of introgression in any comparison involving non-duplicated A. gambiae (D ≈ 0; purple lines in Fig 8A and 8B).
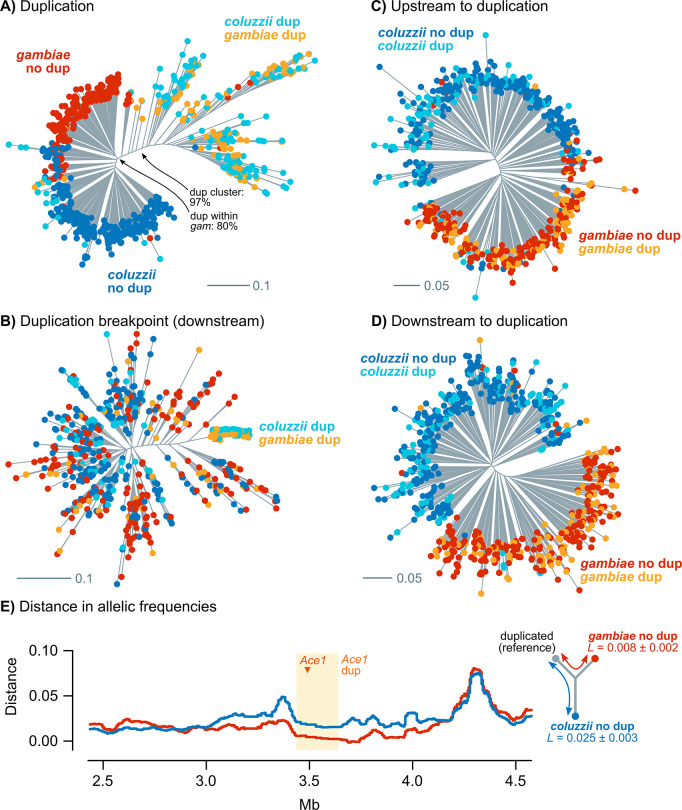
To establish the direction of introgression, we performed a phylogenomic analysis of haplotypes from the duplicated region (Fig 9). Firstly, haplotypes with Ace1 duplications formed a single clade containing A. gambiae and A. coluzzii sequences to the exclusion of all non-duplicated sequences from both species. This clustering was also found in haplotypes located immediately downstream of the duplication breakpoint (Fig 9B, coinciding with the region of D > 0 in Fig 8A); but not in control phylogenies built from non-introgressed regions further upstream and downstream of the duplication (1 Mb away), which exhibited a topology with species-specific clades for both duplicated and non-duplicated specimens (Fig 9C and 9D). Secondly, we found that duplicated sequences were phylogenetically closer to the wt A. gambiae clade than to wt A. coluzzii (Fig 9A). Thirdly, an analysis of distance in allelic frequencies between wt A. gambiae, wt A. coluzzii and duplicated haplotypes also indicated that the duplicated haplotypes are more similar to wt A. gambiae than to wt A. coluzzii: the branch leading to wt A. coluzzii since the divergence from the duplicated specimens was longer (L = 0.026 ± 0.003 standard error) than the branch leading to wt A. gambiae (L = 0.008 ± 0.002, Fig 9E). This difference was also apparent when we analysed duplicated A. coluzzii and A. gambiae specimens separately (S18 Data), and extended immediately upstream and downstream to the duplication breakpoints (Fig 9E). Together, these results indicate that the duplicated haplotypes emerged from a A. gambiae wt background and later introgressed into A. coluzzii.
Fig 9. Phylogenetic analysis of introgression in Ace1.
A) Maximum-Likelihood phylogenetic analysis of 690 Western African haplotypes from the Ace1 duplicated region (2,787 phased variants), using a GTR model with ascertainment bias correction, empirical state frequencies and four Γ rate categories. B-D) Id., using variants beyond the downstream duplication breakpoint (512 variants) and 1Mb upstream and downstream of the duplication (3,935 and 3,302). Tips are color-coded according to species and duplication presence. Source alignments and complete phylogenies with supports in all nodes are available in S14 and S15 Data. E) Distance in allelic frequencies between groups of duplicated specimens and wt A. coluzzii and A. gambiae, calculated using the three-population branch statistic in windows of 5,000 variants along 2R (see Methods). Includes estimated branch lengths (L) from within the duplication region.
Altogether, these results show that the 280S resistance alleles present across West Africa appear in similar genetic backgrounds irrespective of the sampled population and species, and that this similarity extends beyond the Ace1 gene to encompass the entire duplicated region (Fig 6). The low haplotypic diversity is due to a hard selective sweep, detectable at the duplication breakpoints (Fig 7). This resistance haplotype, which spans ca. 200 kbp and includes Ace1 and ten other genes (S3 Data) emerged in A. gambiae and later spread to A. coluzzii (Figs 8 and 9).
Discussion
Evolutionary history of G280S and Ace1 duplications
Our results show that pirimiphos-methyl resistance is associated with a combination of two mutations in Ace1: the G280S SNP and CNVs. In West Africa, virtually all CNVs are found as duplications of a wide region (ca. 200 kbp) that includes Ace1 and 10 other genes [30,34]. This duplication has a unique evolutionary origin in the A. gambiae/A. coluzzii species pair, as its breakpoints are consistent in all populations from both species studied so far [30,34]. The distribution of duplication-280S resistance haplotypes observed in the 1000 Genomes cohort can be explained by three evolutionary events: the G280S mutation, an internally heterogeneous duplication, and inter-specific introgression. Furthermore, recent sampling efforts have identified internal deletions within the duplication in both A. gambiae and A. coluzzii, which appear to be increasing in frequency [31]. Homogeneous duplications carrying only 280S alleles are rare in the 1000 Genomes dataset but appear to be quite common in some A. gambiae populations, though less so in A. coluzzii (Table 2; [24,31]).
At present, we cannot establish the relative order of occurrence of the SNP and CNV in Ace1 because these mutations are tightly linked in the 1000 Genomes dataset (Fig 2A). Nevertheless, the detection of wt homogeneous duplications in samples collected in 2002 in southern Ghana [24]—a country where 280S alleles have more recently been found at high frequencies (Fig 1)—raises the possibility that the SNP might have occurred on an already duplicated background. This order of events would have initially resulted in heterogeneous duplications, providing resistance and reducing the fitness costs associated with impaired acetylcholinesterase activity in the absence of insecticide [19,29]. If that were the case, the emergence of 280S-homogeneous duplications could have occurred via any of the following mechanisms: (i) a secondary loss of a wt copy; (ii) a parallel G280S mutation replacing the wt nucleotide in the initially heterogeneous duplication; (iii) or double recombination or gene conversion events involving the 280S-carrying copy.
After these sequential mutations, the joint duplication-280S resistance haplotype introgressed from A. gambiae into A. coluzzii (Figs 8 and 9). This A. gambiae origin is consistent with earlier studies of Ace1 variation in West African locations, which generally reported higher frequencies of 280S [20,22,27] and CNVs [31] in A. gambiae than in A. coluzzii, as one would expect if they had a longer evolutionary history in the former. Whilst previous studies had suggested that the similarity of 280S haplotypes in A. coluzzii and A. gambiae was due to inter-specific introgression [22,24,27], they were focused on sequencing of the Ace1 gene and could not assess the relationship between introgression and the wider duplicated region (ca. 30 times longer than the Ace1 coding region). Furthermore, analyses focused on the Ace1 gene body are confounded by the lack of sequence variation in this region (S9 Data). By leveraging variation in linkage disequilibrium beyond the gene, we are able identify clear signatures of introgression along the entire Ace1 duplication region (Fig 8).
Genetic basis of pirimiphos-methyl resistance in A. coluzzii and A. gambiae
We have uncovered a strong association between resistance to pirimiphos-methyl and the possession of two or more copies of the Ace1 280S allele in A. coluzzii from Côte d’Ivoire (Fig 3). While the G280S mutation alone has been previously linked to resistance to carbamates and organophosphates in multiple insects [13–15,19,27,43–45], our results indicate that resistance to pirimiphos-methyl specifically relies on the presence of multiple 280S alleles.
Across the 1000 Genomes cohort, specimens with 280S alleles can be grouped into two categories according to the number of Ace1 copies (Fig 2C): (i) heterogeneous duplications with multiple 280S alleles and/or multiple wt, and (ii) high-copy, 280S-homogeneous duplications. Heterogeneous duplications are the most frequent combination of Ace1 mutations (113 out of 119 samples, Fig 2B) and the only one identified in the A. coluzzii population from Côte d’Ivoire (Tiassalé). We next examined the contribution of 280S and duplications to pirimiphos-methyl resistance in a wider array of A. gambiae and A. coluzzii populations from Ghana, Côte d’Ivoire, and Togo (Tables 1 and 2). We identified a common theme in most populations surveyed here: (i) a combination of wt and 280S alleles was the most common resistant genotype in seven out of nine populations analysed, including all of the A. coluzzii sampling locations and three out of five in A. gambiae (Table 1); and (ii) the ratio of 280S alleles and Ace1 copy number were significantly associated with resistance in six out of eight tested populations (Table 2; note that low sample size precluded this analysis in the ninth population). This provides a wider demonstration that the combined effect of G280S and Ace1 duplications is significantly associated with pirimiphos-methyl resistance across multiple West African populations of both species; consistent with the requirement for multiple copies of 280S—shown in the our Tiassalé population analysis (Fig 3B)—to produce a resistant phenotype.
We also identified six genomes with high-copy, 280S-homogeneous loci sampled from the Ghanaian A. gambiae population in the 1000 Genomes dataset (Figs 1 and 2). These samples had not been assayed for pirimiphos-methyl resistance prior to sequencing. However, our extended genotype-phenotype analysis in nine West African populations (Table 1) showed that (i) 280S homozygotes exhibited increased resistance to pirimiphos-methyl, and that, in fact, they were the most abundant resistance allele in two A. gambiae populations (Baguida and Obuasi). It is worth mentioning that survival rates among 280S homozygotes were noticeably lower these two locations (56.8% - 58.9%) than in populations where specimens with both wt and 280S alleles were more common (> 95%; Table 1). Our analyses cannot fully capture the resistance landscape in these populations, because we have focused on the role of 280S-heterogeneous duplications (as they were the most abundant resistant type in the 1000 Genomes cohort; Fig 1). Thus, we cannot presently explain these differences, which may reflect an effect of different phenotyping strategies employed in each population (see Methods), or additional resistance mechanisms. A detailed examination of 280S-homogeneous duplications—e.g. effects on gene dosage or fitness, as performed on heterogeneous duplications in Culex pipiens [46]—could shed light onto the phenotypic variability observed in populations with high penetrance of Ace1 mutations.
Finally, we have performed a genome-wide scan in a population of A. coluzzii from Côte d’Ivoire (Tiassalé) to identify loci associated with pirimiphos-methyl resistance (Fig 5). Our investigation of the association between k-mer frequencies and resistance confirmed the overwhelming concentration of phenotypic association in the Ace1 duplication (Fig 5B), supporting the conclusion that it is the primary driver of resistance in this population. The correlations between Ace1 duplication and all but one of the k-mers significantly associated with resistance, but mapping elsewhere in the genome strongly suggests non-independence of the signals, but the exact cause of this is unclear. Whilst we cannot entirely discount linkage disequilibrium arising from epistasis, a simpler and parsimonious explanation would be that this result is caused by non-specific or incorrect sequence alignments, possibly associated with currently unrecognised variation in the Ace1 duplication.
The lack of clear signals of pirimiphos-methyl adaptation other than Ace1 in this population may reflect its collection at an early stage of the development of pirimiphos-methyl resistance in West Africa [47]. Therefore, our genome-wide analyses would likely only capture pre-existing variants that were enriched in samples resistant to other insecticides. Ace1 mutations and duplications, which cause cross-resistance with previously employed pesticides [22,24,27–31,48], fit this definition and are sufficient to produce significant resistance. However, future analyses on recent collections from populations subjected to regular pirimiphos-methyl-based IRS may reveal additional mechanisms more specifically selected by this insecticide.
Implications for insecticide intervention programmes
Our results demonstrate that the duplication-280S haplotype represents a powerful marker to diagnose resistance to pirimiphos-methyl in A. coluzzii and A. gambiae, permitting the spread of resistance to be tracked from any preserved sample from which DNA can be extracted. Our initial survey of an A. coluzzii population from Tiassalé (Côte d’Ivoire) shows that discrimination between susceptible and resistant specimens can be attained with high accuracy in this population according to the number of 280S alleles. Specifically, following the resistance thresholds apparent in Fig 3B, we can (i) classify wt homozygous samples as susceptible (100% predictive value) and (ii) separate individuals with alleles between those with more wt than 280S alleles (susceptible) and those with equal or more 280S than wt alleles (resistant). This would yield a positive predictive value of 77% for resistance, and a negative predictive value of 85% for susceptibility.
In practice, precise quantification of the number of resistant 280S alleles will often be difficult. Nevertheless, we show that key variation in Ace1 can be captured using either of two qPCR genotyping assays that predict pirimiphos-methyl resistance in populations of A. gambiae and A. coluzzii (Table 2): (i) a measurement of the ratio of 280S to 280G alleles [35], and (ii) the number of Ace1 copies [28]. Crucially, measurements of the 280S allelic ratio can be obtained from the first assay, which is currently widely used for genotyping of the G280S mutation [35]. This provides good diagnostic capacity not only in specimens with 280S-homogeneous duplications, but also in those with heterogeneous duplications, by scoring of the 280G/S allele balance. The phenotypic assays we employed were intended to provide more accurate separation than standard dose approaches [49], but the strong effect of Ace1 CNVs and G280S mutations on pirimiphos-methyl resistance is sufficiently robust for resistance diagnosis in standard discriminating dose assays as well–as used in the Tiassalé A. coluzzii specimens from the 1000 Genomes cohort.
Furthermore, monitoring studies must note that this resistance haplotype is still evolving, especially in the light of the fitness trade-offs associated with Ace1 resistance alleles–namely, that heterogeneous duplications offset the fitness cost of 280S alleles in the absence of insecticide exposure [19,29], and that variable copy number can result in gene dosage disturbances [30]. A recent survey has shown that internal deletions within the duplicated region, downstream to Ace1, are spreading in both A. gambiae and A. coluzzii [31]. The authors have proposed that these deletions reduce the fitness costs of duplications by ameliorating deleterious changes in gene expression [31]. These small deletions are absent from the Ivorian A. coluzzii genomes analysed here [34], but future genome monitoring studies could investigate their spread and identify signals of selection associated with their proposed selective advantage. Likewise, the independently evolved Ace1 CNV that we have identified in one A. gambiae specimen from Guinea (Fig 2B, S4 Data) might merit further attention in the future. Finally, as noted above, we cannot rule out the possibility that, following several years of widespread use of pirimiphos-methyl for IRS, additional non-target site-based resistance mechanisms might have emerged.
The duplication-280S haplotype pre-dates the widespread adoption of pirimiphos-methyl in 2013 [1], which indicates that it likely spread due to its adaptive value with respect to previously employed insecticides used in disease control or agriculture, such as carbamates (e.g. bendiocarb [28,30]) or other organophosphates (e.g. fenitrothion, chlorpyrifos-methyl [22,30]). In that regard, we show that the evolution of the pirimiphos-methyl resistance haplotype is deeply intertwined with that of previous insecticide adaptations, and reveal how continued genomic monitoring studies can help us understand the influence of previous interventions on future population control efforts. An unsuccessful small-scale trial of prototype pirimiphos-methyl wall hangings—interestingly also conducted in Tiassalé at a similar time to collection of our samples—documented significantly greater survival of Ace1 280S carriers (primarily heterozygotes) than wt [23]. However, operational impacts of the Ace1 mutations have yet to be demonstrated and investigation represents a high priority.
Whilst efficacy of pirimiphos-methyl in Actellic IRS formulations appears to have been retained, at least until 2017 [5], its sustainability could be limited in West African Anopheles populations where resistance is already common (Figs 1 and 2). This emphasises the crucial importance of currently-commencing resistance management strategies rotating pirimiphos-methyl spraying with additional IRS insecticides with alternative modes of action [50]. Our demonstration of the utility of Ace1-based diagnostics should aid in molecular surveillance to support these insecticide resistance management programmes.
Methods
Data collection
We used genomic variation data from individual mosquitoes, obtained from the Phase 2 (AR1) of the Anopheles gambiae 1000 Genomes project, as described in [32,33]. This dataset consists of 1,142 wild-caught mosquitoes (1,058 females and 84 males) from 33 sampling sites located in 13 sub-Saharan African countries (listed in S4 Data), and collected at different times between 2009 and 2012 (with the exception of samples from Gabon and Equatorial Guinea, collected in 2000 and 2002 respectively).
A detailed explanation of the methods used in whole-genome sequencing and variant calling can be found in the original publications [32,33]. Briefly, DNA from each specimen was extracted with a Qiagen DNeasy Blood and Tissue Kit (Qiagen Science, US) and sequenced with the Illumina HiSeq 2000 platform (Wellcome Sanger Institute, UK) using paired-end libraries (100 bp reads, 100–200 bp insert sizes), aiming at a 30× coverage per specimen. Variant calling was performed using bwa [51] 0.6.2 and the GATK 2.7.4 UnifiedGenotyper [52]. Haplotype phasing was estimated with SHAPEIT2 [53], and variant effects were predicted using SnpEff 4.1b [54]. We downloaded the genotype calls, SNP effect predictions, and haplotype phasing as available in Anopheles gambiae 1000 Genomes Phase 2 online archive [55].
The catalog of Ace1 CNVs in the 1000 Genomes dataset (Phase 2) was obtained from [34]. There, the authors calculated the coverage (sequencing depth) of each whole-genome sequenced sample in non-overlapping windows along the genome (300 bp) and normalised this value so as to obtain an expected average value of coverage = 2 in non-duplicated, diploid regions. A Gaussian HMM procedure was then applied to the normalised windowed coverage data so as to call windows with heightened normalised coverage (>2). A detailed account of these methods can be found in the original publication [34]. The duplication calls, breakpoint sequences and per-window coverage calculations at the Ace1 region are available in S4 Data and S17 Data.
We retrieved the reference genome information for A. gambiae from VectorBase [56], including the genome assembly (PEST, AgamP4 version), gene annotation coordinates in GFF format (AgamP4.9) and gene names and descriptions.
Genotype-phenotype association in Côte d’Ivoire A. coluzzii genomes
As part of Anopheles gambiae 1000 Genomes Phase 2, 71 A. coluzzii specimens from Côte d’Ivoire were collected and phenotyped for pirimiphos-methyl resistance before whole-genome sequencing [32]. These samples were collected as larvae in rice fields near Tiassalé (coordinates: -4.823, 5.898) between May and September 2012, and were identified as A. coluzzii using a PCR assay [57]. The 71 larvae were tested for resistance to pirimiphos-methyl resistance using a WHO tube assay with 0.25% impregnated papers [58], which led to the identification of 31 resistant (live) and 40 susceptible (dead) specimens.
We tested the association of these resistance phenotypes with various genetic variants present in Ace1 in this population. This includes the non-synonymous mutations G280S and S65A, the number of Ace1 copies, and the number of 280S alleles in each individual sample (listed in S6A Data).
For the non-synonymous mutations, we assessed genotype-phenotype associations for each variant independently using Fisher’s exact test (gmodels R library [59]) and estimated odds ratio and 95% confidence intervals using the Woolf method with Haldane-Anscombe correction (Prop.or function in the pairwiseCI R library [60]). For the Ace1 and 280S copy numbers, we used used binomial generalised linear models (glm function in R stats library, family binomial) to obtain odds ratio estimates for each of the four variables.
We also built a binomial generalised linear models with all four variables (G280S and A65S genotypes; Ace1 and 280S copy number) and a step-wise procedure to remove non-informative variants from the model (step function in R stats using k = log(n) as the degree of freedom, as required by the Bayesian Information Criterion; n represents the number of samples in our dataset). The number of Ace1 copies and the number of mutated alleles were encoded as continuous variables, and the genotypes of G280S and S65A were encoded as categorical variables. The performance of all generalised linear models was assessed relative to a null model (intercept as the only variable) using a χ2 test in an analysis of variance (anova function in the R stats library). A detailed statistical analysis of all comparisons mentioned above can be found in S6A and S6B Data.
The number of Ace1 copies in each genome of Anopheles gambiae 1000 Genomes Phase 2 was retrieved from [34]. In that study, copy number states of multiple CNVs were inferred using a HMM-based predictive model that took as input the normalised sequencing depth calculated along non-overlapping 300 bp genomic windows.
The estimated number of 280S alleles in each genome was estimated in the following manner: (i) we retrieved the copy numbers of Ace1 in each genome (C; see above); (ii) we calculated the fraction of reads supporting the reference and alternate alleles; and (iii) assigned the number of 280S alleles (S) as the value that minimised the difference between the fraction of alternate alleles and S/C, for all discrete S values between 0 and C. For example, a genome with three Ace1 copies (C = 3) and 30% of reads supporting 280S would carry one 280S allele (S = 1), given that S/C = 1/3 ≈ 30%.
Genotype-phenotype association in additional West African populations
We collected 1080 female specimens of two species (A. coluzzii and A. gambiae) from six different locations across West Africa (Côte d’Ivoire: Aboisso; Ghana: Madina, Korle-Bu, Weija and Obuasi; Togo: Baguida). Species identity was determined using two methods designed to discriminate between A. gambiae, A. coluzzii and A. arabiensis: a PCR of species-specific SINE insertion polymorphisms as described in [61], and a high-resolution melt curve analysis [62]. Details for each of these methods, including primer sequences, are available in S6G Data. The complete list of specimens, sampling times and locations, and species identification are available in S6C Data and S6E Data.
All 1080 specimens were phenotyped for pirimiphos-methyl resistance using a custom dose-response assay with WHO standard tubes [58]. For the Aboisso (Côte d’Ivoire), Korle-Bu, Weija and Madina samples, resistant specimens were determined as surviving a threshold concentration of pirimiphos-methyl after exposure during the larval stage, and susceptible specimens were identified as dead after a lower threshold dose (constant exposure time of 60 minutes). The exact concentration thresholds for the dose-response assay were determined on a per-site basis, tailored to the observed levels of resistance in each population and collection logistics, and are listed in S6C Data. Out of 1080 specimens, 951 could be assigned to one of the following populations, defined as combinations of species and collection locations with determined resistance phenotypes: A. coluzzii from Aboisso, Côte d’Ivoire (n = 55); A. coluzzii from Korle-Bu (n = 214), Madina (n = 42), and Weija (n = 131), Ghana; A. gambiae from Aboisso, Côte d’Ivoire (n = 82); A. gambiae from Baguida, Togo (n = 102); A. gambiae from Madina (n = 172), Obuasi (n = 140) and Weija (n = 13), Ghana.
For each sample, we determined the G280S genotype using a qPCR TaqMan assay as described by Bass et al. [35] (list of samples in S6C Data), in which 280S- and 280G-specific primers where labeled with different fluorescent dyes (FAM and HEX, respectively). We also calculated the ratio of FAM-to-HEX fluorescent dye signal in hetrerozygotes, which label 280S and 280G alleles respectively, as an index of the fraction of 280S allele copies [28,48]. Detailed methods for these two genotyping assays are available in S6G Data.
We assessed the association of G280S mutations with resistance for each of the populations listed above using generalised linear models (glm function in R stats library, binomial family). The results from these tests are available in S6D Data.
A sub-set of these specimens (n = 201; listed in S6E Data) were also genotyped for CNV polymorphisms in the Ace1 locus using a qPCR assay and a combination of primers for Ace1 and two non-duplicated control genes (the CYP4G16 gene AGAP001076 and the elongation factor AGAP005128), and were measured relative to values in A. gambiae specimens from the wt Kisumu colony (non-duplicated). Detailed methods for CNV genotyping assay are available in S6G Data and [28]. These samples were selected from the subset of specimens with both 280S and wt alleles, in order to investigate the effect of both CNVs and fraction of 280S alleles on resistance.
We used generalised linear models (glm function in R stats library, binomial family) to assess the effect of these two variables within each of the eight populations with available CNV data (four from A. coluzzii, four from A. gambiae; listed in Table 2 and S6F Data). Then, we obtained the minimal significant model for each population using a stepwise reduction procedure and the BIC criterion (step function in the R stats library), and assessed their fit against a null model (intercept as the only variable) with an analysis of variance and a χ2 test (anova function in R stats). The results from these tests are available in S6F Data.
Haplotype clustering and analysis of selection signals in Ace1
Haplotypes along chromosome arm 2R were reconstructed using the dataset of phased variants from the Anopheles gambiae 1000 Genomes Phase 2 data. Specifically, we retained phased variants that were biallelic, non-singletons and segregating in at least one population. It must be noted that phased variants are scarce in the Ace1 region (S9A and S9B Data) because the phasing procedure used in the original dataset can only be applied to diploid genomic regions [32,33], which is generally not the case along the Ace1 duplication. In particular, phased variants had to be (i) biallelic and (ii) belong to an accessible genomic region, defined as having even sequencing coverage across samples and not having ambiguous alignments in more than one specimen. Yet, a duplicated region can still encompass short segments of diploid sequences, either due to internal deletions, as found in Ace1 [31]); or to high divergence in one of the paralogous sequences, which results in only reads from the non-diverged (wt) paralog mapping to the reference genome and being phased as diploid.
We identified chromosomes carrying Ace1 duplications that could be used for further analyses of selection signatures. To do so, we calculated the linkage disequilibrium between position G280S (itself tightly linked to the duplication) and all variants located between positions 3416800 and 3659600 along 2R (i.e., 20kbp upstream and downstream of the duplication breakpoints [3436800–3639600]). Linkage disequilibrium was estimated from genotype counts in each sample using Rogers’ and Huff r correlation statistic [63] as implemented in scikit-allel v1.1.10 library (rogers_huff_r) [64]. Then, we retrieved all variants with r > 0.95 that were (i) present in the subset of phased variants and (ii) were embedded within genomic regions that were, on average, represented by two haplotypes across samples (i.e. diploid). Three tagging variants fitted this definition (S9 Data): they were located in intergenic regions −26 kbp and +12 kbp of G280S, had minor allele frequencies similar to that of 280S in Côte d’Ivoire (43%), were embedded in genomic windows that were effectively diploid (haplotype score ≈ 2; S9D–S9E Data, calculated with GATK Unified Genotyper in [32]), and had lower normalised sequencing depths across samples than the Ace1 gene locus (S9F Data and S4B Data; coverage ≈ 2; data from [34]). These properties imply that it is possible to recover diploid haplotypes around the tagging variants.
We retrieved haplotypes surrounding each of the three tagging variants and clustered them by similarity using minimum spanning trees. Specifically, we retrieved phased variants ± 300 bp of each tagging variant (retaining variants that were biallelic, non-singletons and segregating in at least one population; these regions contained between 67 and 164 phased variants depending on the analysis; S10 Data). We used allele presence/absence data from each haplotype to build minimum spanning trees of haplotypes based on Hamming distances (breaking edges for distances >1). This distance matrix was then used to build medium spanning trees (minimum_spanning_tree function in SciPy 1.1.0 Python library [65], from the sparse.csgraph submodule). Tree visualizations were produced using the graphviz 2.38.0 Python library [66] and the hapclust library [67,68], and clusters of haplotypes were color-coded according to the population of origin or the presence/absence of the alternate allele (S10 Data).
We used these trees to identify groups of highly similar haplotypes linked to the 280S or wt alleles in Ace1 (S11 Data). We calculated the profile of Garud’s H statistics (moving_garud_h function, scikit-allel) [39,40] and haplotype diversity (moving_haplotype_diversity in scikit-allel) around the Ace1 duplication region in windows of consecutive variants (500 variants with 20% overlap). These calculations were performed separately for the main groups of identical haplotypes (280S-linked or wt-linked) as identified around each of the tagging variants (minimum size = 100 haplotypes). In addition, we calculated the averages of these same statistics in the regions immediately upstream and downstream of the duplication breakpoints (2R:3436800–5000 bp and 2R:3639600 + 5000 bp, respectively), and we calculated standard errors of these estimates using sample jack-knifing.
Finally, we calculated the rates of extended haplotype homozygosity decay for each cluster (S12 Data), using 10,000 phased variants upstream and downstream of the duplication breakpoints (ehh_decay function in scikit-allel).
Introgression analysis
We used Patterson’s D statistic [41,42] to detect introgression between A. coluzzii and A. gambiae. We retrieved the allele frequencies of all biallelic, non-singleton variants from chromosome arm 2R that were segregating in West African populations where we had identified Ace1 duplications (namely: 75 A. coluzzii from Burkina Faso, 55 from Ghana, and 71 from Côte d’Ivoire; and 92 A. gambiae from Burkina Faso, 12 from Ghana and 40 from Guinea; total n = 345 genomes) [34], as well as Central African populations that we used as non-admixed controls (78 A. coluzzii from Angola; 69 A. gambiae from Gabon and 297 from Cameroon). In addition, we retrieved the genotypes at the same variant positions for the populations of A. arabiensis (12 genomes), A. quadriannulatus (10), A. merus (10) and A. melas (4) populations analysed in [69], which we used as outgroups in the calculation of Patterson’s D. In total, we retained ca. 12% of the 47,817,813 variants in 2R in each analysis.
To determine whether duplicated and non-duplicated populations had different introgression patterns in Ace1, we calculated windowed means of Patterson’s D (length 5,000 variants, 20% step; moving_patterson_d in scikit-allel) using various combinations of populations with and without the Ace1 duplication (Fig 5A). This test requires genotype frequencies in four populations (two ingroups A and B; one candidate donor/receptor C with which A or B might have introgressed; and one unadmixed outgroup O) branching in a (((A,B),C),O) topology (Fig 5A). We observed the following convention: (i) we used West African A. coluzzii populations as A, discriminating between duplicated and non-duplicated subpopulations; (ii) we used non-duplicated A. coluzzii from Angola as control B; (iii) we used various A. gambiae populations as C, again analysing duplicated and non-duplicated specimens within each population separately; and (iv) A. arabiensis, A. quadriannulatus, A. merus and A. melas as outgroup O.
For each combination of (((A,B),C),O) populations, we calculated the average D statistic within the duplicated (position 3,436,800 to 3,639,600, spanning 202.8 kbp around Ace1), and estimated its deviation from the null expectation (no introgression: D = 0) with a block-jackknife procedure (block length = 100 variants; average_patterson_d function in scikit-allel), which we used to estimate the standard error, Z-score and the corresponding p value from the two-sided normal distribution (S13 Data).
In parallel, we performed a phylogenomic analysis of the reconstructed haplotypes of the Ace1 CNV region (position 3436800 to 3639600) in the West African populations where duplications had been identified (75 A. coluzzii from Burkina Faso, 55 from Ghana, and 71 from Côte d’Ivoire; and 92 A. gambiae from Burkina Faso, 12 from Ghana and 40 from Guinea; total n = 345 genomes, 690 haplotypes). Specifically, we built an alignment of phased variants along the duplication (2,787 positions) and computed a Maximum-Likelihood phylogenetic tree using IQ-TREE 1.6.10 [70]. We used the GTR nucleotide substitution model with ascertainment bias correction, empirical state frequencies observed from each alignment, and four gamma distribution (Γ) categories (GTR+F+ASC+G4 model in IQ-TREE). This model was selected by the IQ-TREE implementation of ModelFinder [71] (TEST mode) as the best-fitting model according to the BIC criterion. The best-fitting tree was found after 22,000 iterations (correlation coefficient threshold = 0.99). We estimated node statistical supports from 1,000 UF bootstraps [72,73]. Using the same approach, we built three additional phylogenies from genomic regions located immediately downstream of the duplication breakpoint (5 kbp, 512 variants), as well as upstream and downstream of the duplication breakpoints (3,935 and 3,302 variants extracted from 50 kbp segments located −1Mb and +1Mb), finding the best-fitting trees after 5,500, 7,659 and 27,400 iterations, respectively (correlation coefficient threshold = 0.99). The resulting phylogenetic trees were plotted, unrooted, using the phytools 0.6–60 [74] and ape 5.3 libraries (plot.phylo) [75]. Phylogenies and source alignments are available as S14 Data and S15 Data.
We also used allelic frequencies within the inversion to estimate the divergence times between specimens carrying the duplication (both A. gambiae and A. coluzzii), wt A. coluzzii and wt A. gambiae. This three-way calculation of divergence times reflects the amount of allele frequency change between one of the three groups relative to the other two, and is therefore analogous to the population branch statistic [36,76]. Following this logic, we estimated separately (i) the branch length of wt A. coluzzii relative to the separation of wt A. gambiae and the duplicated sequences, and (ii) the branch length of wt A. gambiae relative to the separation of wt A. coluzzii and the duplicated sequences. We repeated these analyses using only duplicated specimens from either species. We performed these calculations in non-overlapping windows of 100 variants with the pbs function in scikit-allel, and estimated standard errors using a block-jackknifing procedure. We used windows of 5,000 variants at 10% steps for visualisation (Fig 9E and S18 Data).
Genetic differentiation in A. coluzzii from Côte d’Ivoire
We assessed the degree of genetic differentiation between resistant and susceptible A. coluzzii from Côte d’Ivoire. We focused on non-singleton variants that were segregating in this population. In total, 8,431,869 out of 57,837,885 (14.57%) variants from chromosomes 2, 3 and X were retained for further analysis.
For each chromosome arm (2R, 2L, 3R and X), we calculated genotype counts in each subpopulation (resistant and susceptible), and calculated their genetic differentiation using the Hudson’s FST statistic [77,78] along non-overlapping windows of 1,000 variants (moving_hudson_fst function in the scikit-allel v1.1.10 library [64] from Python 3.4). We also calculated the average FST in each chromosomal arm, with standard errors obtained from a block-jackknife procedure (using non-overlapping windows of 1,000 variants; average_hudson_fst function).
We calculated the normalised population branching statistic (PBS) [36,37] in non-overlapping windows of 1,000 variants along each chromosomal arm (pbs function in scikit-allel). using resistant and susceptible Ivorian A. coluzzii as test populations, and A. coluzzii from Angola as an outgroup. Angolan A. coluzzii (n = 78) where selected as outgroup due to their relative isolation relative to West African A. coluzzii populations [32,33] and their putatively naive profile of organophosphate resistance [1,4]. The distribution of PBS estimates along each chromosomal arm was standardised to unit variance (standardize function in scikit-allel), and the resulting distribution of Z-scores was used to derive a two-sided p-value that reflected extreme values of PBS (highly positive or negative). We corrected for multiple testing using local estimation of false discovery rates (fdrtool function in the fdrtool 1.2.15 R library [79]). Finally, we selected genomic windows with high differentiation in resistant specimens (standardised PBS > 0, significance threshold FDR < 0.001). To further characterise these PBS peaks, we (i) calculated Hudson’s FST in each genomic window (S7 Data); (ii) built neighbour joining phylogenies from the phased variants within each peak, to identify swept haplotypes (S16A Data; nj tool in the ape R package [75]); and (iii) calculated the profile of Garud’s H12 in the neighbouring regions, for resistant and susceptible specimens separately (S16B Data).
We performed a principal component analysis of resistant and susceptible Ivorian A. coluzzii. We first obtained a set of genetically unlinked variants from chromosomal arms 3R and 3L (so as to avoid the confounding effects of chromosomal inversions in arms 2R, 2L and X). We discarded linked variants within 500 bp consecutive windows (using a 200 bp step), using a Rogers and Huff’s r threshold value of 0.1 (locate_unlinked function in scikit-allel), and repeated this process for ten iterations for each chromosome arm. This filtering procedure resulted in 791 genetically unlinked variants from both chromosomal arms. We used the alternate allele count in each variant to construct a PCA using singular value decomposition (pca function from scikit-allel), and scaling the resulting coordinates using Patterson’s procedure [80].
k-mer enrichment analysis in A. coluzzii from Côte d’Ivoire
We obtained k-mer counts for each of the 71 A. coluzzii samples using the count function in jellyfish v. 2.2.10 [81], using a k = 31 bp (parameters: -C -m 31—out-counter-len 2 -s 300M —bf-size 10G). To reduce the computer memory footprint of the k-mer count tables, the k-mer strings were recoded as integers, split into separate lexicographical groups according to the leading nucleotides (i.e. k-mers beginning with AAA, AAC, etc. were assigned to different groups), and, within each set, sorted lexicographically again. In total, we recorded the frequency of 1,734,834,987 k-mers across 71 samples. The resulting count tables were filtered to retain k-mers showing variation in the population: we discarded k-mers present in fewer than 3 samples, or absent in fewer than 3 samples. These filters removed 967,274,879 k-mers, leaving 767,560,108 k-mers for further analysis.
To test whether the frequencies of any k-mers were associated with pirimiphos-methyl resistance, normalised k-mer frequencies were obtained by dividing the k-mer counts by the total number of variant k-mers in each sample. We calculated the Spearman’s rank correlation of each k-mer frequency with the resistance phenotype for each of the 767,560,108 k-mers (cor.test function in the R stats library [82]), correcting for multiple testing using local estimation of false discovery rates (fdrtool function in fdrtool library [79]), and using a significance threshold of FDR < 0.001.
Given that multiple k-mers can overlap a single mutation, it was likely that many of the k-mers identified as significant were overlapping. We therefore took the k-mers that showed a significant association with pirimiphos-methyl resistance and assembled them by joining any k-mers that overlapped perfectly over at least 10 bp. The resulting assembled k-mers were aligned against the A. gambiae reference genome using bwa mem version 0.7.12-r1034 (parameters: -T 0) [51]. The mapping coordinates and sequences of the significant assembled k-mers are available in S8 Data.
A minority of assembled k-mers aligned in regions other than the Ace1 duplication. To determine whether this was due to mis-alignment, we correlated the frequency of these assembled k-mers in each sample against the Ace1 copy number. Frequency for each assembled k-mer was calculated as the mean normalised frequency of all the k-mers that were used in its assembly. Because these k-mers and the Ace1 copy number share the property of being correlated with pirimiphos-methyl resistance phenotype, they are statistically likely to have correlated frequencies even if physically unlinked. To control for this, the residuals of k-mer frequency and Ace1 copy number were calculated within each of the resistant and susceptible groups of samples. Pearson’s correlation was then calculated on these residuals for each of the assembled k-mers (available in S8C Data).
Scripts to reproduce the k-mer analysis steps described above (counting, filtering, significance testing, assembly, mapping, and Ace1 correlation analysis) are available on Github (see Availability of data and materials).
Alignment and phylogenetic analysis of ACE proteins in animals
To obtain a candidate list of homologs of the Ace1 gene, we retrieved all genes belonging to the orthogroup 339133at33208 from OrthoDB [83] (682 genes in total). We aligned these candidates to a curated database of predicted proteomes from 89 complete animal genomes (listed in S2 Data, including data sources) using Diamond 0.9.22.123 [84], and retained all alignments with an identity >95% to any of the candidate queries (130 genes). Next, we performed a multi-sequence alignment with MAFFT 7.310 (1,000 rounds of iterative refinement, L-INS-i algorithm) [85] of these 130 genes, together with the truncated sequence of the Pacific electric ray Torpedo californica protein used for its crystal structure analysis (PDB accession number: 1W75) and the full-sequence of the ortholog from its close relative T. marmorata (Uniprot accession number: P07692). This alignment (n = 132 genes) was trimmed position-wise using trimAL 1.4 [86] (automated1 mode), and 277 conserved columns were retained. A maximum-likelihood phylogenetic analysis of the trimmed alignment was then performed using IQ-TREE 1.6.10 [70], using a LG substitution matrix [87] with four Γ categories and accounting for invariant sites (LG+I+G4 model). IQ-TREE was run for 469 iterations until convergence was attained (correlation coefficient ≥ 0.99). Node statistical supports were calculated using the UF bootstrap procedure (1,000 replicates) [72,73]. Alignment visualizations (S1 Data) were obtained from Geneious 11.1.4 [88]. Complete alignments are available as S2 Data.
Supporting information
A) Alignment of ACE homologs (protein sequences) in selected species (A. gambiae, Culex quinquefasciatus, Aedes aegypti, Homo sapiens, and Torpedo californica), used to determine the homology of non-synonymous mutations in this gene (A65S and G280S are highlighted). B) Alignment of ACE protein homologs in 20 culicine species, focusing on the vicinity of codon 280 (highlighted). C) Maximum-Likelihood phylogenetic analysis of ACE homologs from 89 animals (listed in S2 Data, including data sources and accession numbers).
(PDF)
A) Peptide coordinates of A. gambiae Ace1 codon 280 in orthologs of Ace1 from 89 animal genomes. B) Alignment of Ace1 homologs from 89 animal genomes. C) Data sources, accession numbers, species abbreviation and taxonomy of the 89 animal genomes used in the analysis of ACE homology.
(XLSX)
Coordinates of non-synonymous mutations in Ace1 and frequencies in each population of the Anopheles gambiae 1000 Genomes Phase 2 dataset. Species codes: A. gambiae, gam, A. coluzzii, col. Population codes: Angola, AOcol; Burkina-Faso, BFcol and BFgam; Côte d’Ivoire, CIcol; Cameroon, CMgam; Mayotte, FRgam; Gabon, GAgam; Ghana, GHcol and GHgam; The Gambia, GM; Guinea, GNcol and GNgam; Equatorial Guinea, GQgam; Guinea-Bissau, GW; Kenya, KE; Uganda, UGgam.
(XLSX)
List of samples from the Anopheles gambiae 1000 Genomes Phase 2 dataset with accession numbers, sample metadata (population and region of origin, collection date, species, sex), and summary of the main Ace1 mutations in each sample (genotypes in the G280S mutation, number of reads supporting 280G and 280S alleles, number of CNVs). CNV coordinates and copy number from Lucas et al. (2019).
(XLSB)
List of genes in the Ace1 duplication region, with genomic coordinates along the 2R chromosomal arm.
(XLSX)
A) Genotypes of G280S and A65S mutations in A. coluzzii samples from Côte d’Ivoire (Anopheles gambiae 1000 Genomes Phase 2), and resistance phenotype to pirimiphos-methyl. B) Phenotype-genotype association tests for the 71 Ivorian A. coluzzii samples. Includes summaries of single-variable GLM models for the following variables: 280S presence (including a subset of samples with only one 280S allele), 65S, CNV and number of 280S alleles. Also includes the minimal model obtained using stepwise reduction of a starting multi-variable model (BIC criterion). Model significance is measured with ANOVA comparison with null model (no variables) and a χ2 test. C) G280S genotypes and pirimiphos-methyl resistance phenotype for 1080 mosquitoes from West African populations of A. gambiae and A. coluzzii collected from six locations. For each sample, we also report collection dates, geographical locations, and details of the species identification, genotyping, and phenotyping (concentrations, exposure time). D) Phenotype-genotype association tests for G280S alleles in nine West African populations. Includes summaries of GLM models for each population. Model significance is measured with ANOVA comparison with null model (no variables) and a χ2 test. E) Number of Ace1 copies and pirimiphos-methyl resistance phenotypes for 167 mosquitoes from the same West African populations. F) Phenotype-genotype association tests for CNV mutations (Ace1 copies [CNV] and 280S-to-280G allele ratio [ratio_FAM_HEX]) in West African populations. Includes summaries of GLM models for each population and each variable separately, and a minimal GLM obtained using the BIC criterion from an initial model with both variables. Model significance is measured with ANOVA comparison with null model (no variables) and a χ2 test. G) Protocols for species identification and Ace1 mutation genotyping, including primer sequences.
(XLSX)
A) Genetic differentiation statistics (Hudson’s FST and PBS) between resistant and susceptible A. coluzzii from Côte d’Ivoire, in windows of 10,000 variants along the genome. PBS values are calculated using Angolan A. coluzzii as outgroup, and we also report a Z-score and p-value derived from a two-sided normal distribution. B) Genes overlapping the regions of high PBS (FDR < 0.001 and standardised PBS > 0).
(XLSX)
A) Alignment coordinates of k-mers that are significantly associated with pirimiphos-methyl resistance in Ivorian A. coluzzii samples from the Anopheles gambiae 1000 Genomes Phase 2. For each k-mer, we report the alignment sequence and coordinates, and whether it overlaps the Ace1 gene or the Ace1 duplication. This table includes both primary and secondary alignments as reported by bwa mem, as well as non-aligned k-mers (chromosome “NA”). B) Correlation of alignment frequencies and Ace1 copy number in each sample, for each of the 409 k-mers that mapped to chromosomal arms 2R, 2L, 3R, 3L or X. We report Pearson’s correlation coefficients (r) and p-values. C) Sequences of the 446 assembled k-mers that are significantly associated with pirimiphos-methyl resistance. D) Frequencies of the 9,603 k-mers (before assembly) in each of the Côte d’Ivoire A. coluzzii samples (Anopheles gambiae 1000 Genomes Phase 2, n = 71).
(XLSX)
A) Density of phased variants in the Ace1 duplication region, from Anopheles gambiae 1000 Genomes Phase 2. B) Density of phased variants within the Ace1 duplication, focusing on the Ace1 gene body. C) Linkage disequilibrium between the G280S alleles and nearby phased variants (Huff and Roger’s r), highlighting the coordinates of nearby variants tightly linked to 280S alleles that could be used as tagging variants for 280S (in green), or variants that were later discarded by other filtering steps (in grey), and their frequencies in Côte d’Ivoire A. coluzzii. D) Haplotype score values in the vicinity of the Ace1 locus, calculated from the Phase 2 1000 Genomes cohort. Variants within low-score regions can be used to tag 280S (in green), variants within peaks were discarded (grey). E) Distribution of haplotype score values for variants within the regions used to build haplotype networks (green, tagging variant ± 300 bp) and in the Ace1 gene body (magenta). Distributions are shown as cumulative frequency distributions and boxplots (box lines represent the first, second and third quartiles). F) Distribution of the sequencing coverage of each sample with Ace1 duplications, for the genomic window containing each tagging variant (green) and Ace1 (magenta). A normalised coverage of two implies diploidy. Tagging variants are located in regions of lower sequencing coverage than Ace1, even though they all fall within the duplicated region.
(PDF)
Minimum spanning tree networks built from phased variants located around each of the three tagging variants (panels A to C). Each node in the network is colored according to its population composition (left panels) or linkage to the 280S or wt alleles in Ace1 (right panels).
(PDF)
Garud H statistics and haplotype diversity in 280S-linked and wt-linked haplotypes in the genomic window around the Ace1 duplication breakpoints, calculated for each of the main haplotype clusters defined around each of the three tagging variants (panels A to C; haplotype clusters from S10). For each tagging variant and duplication breakpoint (upstream/downstream), we report the average value of each statistic and standard errors from sample jack-knifing.
(PDF)
Extended haplotype homozygosity (EHH) of 280S-linked and wt-linked haplotypes in the genomic window around the Ace1 duplication breakpoints, calculated for each of the main haplotype clusters defined around each of the four tagging variants (panels A to D; haplotype clusters from S10). For each tagging variant and duplication breakpoint (upstream/downstream), we report the area under the EHH curve (a) and the distance around the duplication breakpoint where EHH>0.05 and EHH>0.95.
(PDF)
Patterson D statistics to test introgression of the Ace1 duplication region between various populations of A. coluzzii (populations A/B), A. gambiae (populations C) and multiple outgroup species (population O). Specifically, we use A. coluzzii from Côte d’Ivoire (CIcol), Ghana (GHcol) and Burkina-Faso (BFcol) with and without duplications (labelled as TRUE and FALSE respectively) as populations A; A. coluzzii from Angola as population B (always labelled as FALSE); and various A. gambiae populations C from Ghana (GHgam), Burkina-Faso (BFgam), Guinea (GNgam), Gabon (GAgam) and Cameroon (CMgam) with and without duplications (TRUE and FALSE labels, respectively); and four different outgroup species (panels A to D: A. arabiensis, A. melas, A. merus and A. quadriannulatus). For each comparison, we report the average D statistic from the Ace1 duplicated region with standard errors, and Z-scores and p-values derived from standardised D values (unit variance).
(PDF)
A) Alignment of phased variants from within the Ace1 duplication region (2R:3,436,800–3,639,600), using 345 samples from West African populations (Anopheles gambiae 1000 Genomes) with Ace1 duplications (Guinea A. gambiae, Côte d’Ivoire A. coluzzii, Ghana A. gambiae and A. coluzzii, Burkina Faso A. gambiae and A. coluzzii). B) Id., from a region downstream of the duplication (5 kbp starting at 3,639,600). C) Id., from a region upstream of the duplication (50 kbp starting at 3,436,800–1 Mb). D) Id., from a region downstream of the duplication (50 kbp starting at 3,639,600 + 1 Mb).
(XLSX)
Maximum-Likelihood phylogenetic analyses of haplotypes from within the Ace1 duplication (A), downstream breakpoint (B), upstream (C) and downstream (D) regions. Trees are unrooted. Tips are color-coded according to duplication presence/absence and species. UF bootstrap supports indicated in each node.
(PDF)
A) Neighbour joining trees built from phased variants located within the PBS peaks identified between resistant and susceptible Ivorian A. coluzzii (1000 Genomes dataset). B) Profile of Garud’s H12 in these same regions.
(PDF)
A) Sequence of the duplication breakpoints described by Assogba et al. (2016) with their coordinates in the A. gambiae genome assembly (AgamP4) and the corresponding genome window in the CNV database by Lucas et al. (2019). B) Normalised sequencing coverage along the Ace1 duplication locus in each sample of the Anopheles gambiae 1000 Genomes dataset (grouped by population of origin). Detailed coverage of each sample available in S4B Data.
(PDF)
A) Distance in allelic frequencies between A. coluzzii specimens with duplications and wt A. coluzzii and A. gambiae, calculated using the three-population branch statistic in windows of 5,000 variants along the genome. Includes estimated distance (L) from within the duplication region. B) Id., using A. gambiae specimens with duplications.
(PDF)
Acknowledgments
We thank Sean Tomlinson (LSTM) and Chris Clarkson (Wellcome Sanger Institute) for fruitful discussions on the analyses. ¶ Anopheles gambiae 1000 genomes members: Jorge Edouardo Amaya-Romero, Diego Ayala, C.J. Battey, Philip Bejon, Nora J. Besansky, Austin Burt, Jorge Cano, Beniamino Caputo, Edi Constant, Carlo Costantini, Boubacar Coulibaly, Alessandra della Torre, Abdoulaye Diabaté, João Dinis, Martin J. Donnelly, Eleanor Drury, Jorge Edouardo, Nohal Elissa, John Essandoh, Michael C. Fontaine, H. Charles J. Godfray, Matthew W. Hahn, Nicholas J. Harding, Christa Henrichs, Christina Hubbart, Alison T. Isaacs, Musa Jawara, Anna E. Jeffreys, Dushyanth Jyothi, Maryam Kamali, Andrew D. Kern, Dominic P. Kwiatkowski, Chris S. Clarkson, Mara K.N. Lawniczak, Gilbert Le Goff, Eric R. Lucas, Cinzia Malangone, Henry D. Mawejje, Charles Mbogo, Daniel Mead, Janet Midega, Alistair Miles, Davis C. Nwakanma, Samantha O’Loughlin, João Pinto, Michelle M. Riehle, Vincent Robert, Kirk A. Rockett, Kyanne R. Rohatgi, Kate Rowlands, Daniel R. Schrider, Igor Sharakhov, Victoria Simpson, Jim Stalker, Arlete D. Troco, Kenneth D. Vernick, David Weetman, Bradley J. White, Craig S. Wilding. Affiliations and contact details are available at www.malariagen.net/projects/ag1000g.
Data Availability
Python 3.4.7 and R 3.6.2 scripts to reproduce the main analyses from the paper are available in the following Github repository: https://github.com/xgrau/ace1-anopheles-report. All genome variation data has been obtained from the publicly available repositories of the Anopheles gambiae 1000 Genomes project (Phase 2 AR1): https://www.malariagen.net/data/ag1000g-phase-2-ar1 (download instructions can be found in the Github repository).
Funding Statement
This work was supported by the National Institute of Allergy and Infectious Diseases (R01-AI116811), the Wellcome Trust (090770/Z/09/Z; 090532/Z/09/Z; 098051), the Medical Research Council and the Department for International Development (MR/M006212/1), and the Medical Research Council (MR/P02520X/1 and MR/T001070). This UK funded award is part of the EDCTP2 programme supported by the European Union. These fellowships were awarded to MJD and DW. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Oxborough RM. Trends in US President’s Malaria Initiative-funded indoor residual spray coverage and insecticide choice in sub-Saharan Africa (2008–2015): urgent need for affordable, long-lasting insecticides. Malar J. 2016;15: 146 10.1186/s12936-016-1201-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dengela D, Seyoum A, Lucas B, Johns B, George K, Belemvire A, et al. Multi-country assessment of residual bio-efficacy of insecticides used for indoor residual spraying in malaria control on different surface types: results from program monitoring in 17 PMI/USAID-supported IRS countries. Parasit Vectors. 2018;11: 71 10.1186/s13071-017-2608-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Report of the Sixteenth WHOPES Working Group Meeting. Genèva; 2013.
- 4.World Health Organization. Global report on insecticide resistance in malaria vectors: 2010–2016. Genèva; 2018.
- 5.Sherrard-Smith E, Griffin JT, Winskill P, Corbel V, Pennetier C, Djénontin A, et al. Systematic review of indoor residual spray efficacy and effectiveness against Plasmodium falciparum in Africa. Nat Commun. 2018;9: 4982 10.1038/s41467-018-07357-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Berg H, Zaim M, Yadav RS, Soares A, Ameneshewa B, Mnzava A, et al. Global Trends in the Use of Insecticides to Control Vector-Borne Diseases. Environ Health Perspect. 2012;120: 577–582. 10.1289/ehp.1104340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranson H, N’Guessan R, Lines J, Moiroux N, Nkuni Z, Corbel V. Pyrethroid resistance in African anopheline mosquitoes: what are the implications for malaria control? Trends Parasitol. 2011;27: 91–98. 10.1016/j.pt.2010.08.004 [DOI] [PubMed] [Google Scholar]
- 8.Wagman J, Gogue C, Tynuv K, Mihigo J, Bankineza E, Bah M, et al. An observational analysis of the impact of indoor residual spraying with non-pyrethroid insecticides on the incidence of malaria in Ségou Region, Mali: 2012–2015. Malar J. 2018;17 10.1186/s12936-017-2160-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abong’o B, Gimnig JE, Torr SJ, Longman B, Omoke D, Muchoki M, et al. Impact of indoor residual spraying with pirimiphos-methyl (Actellic 300CS) on entomological indicators of transmission and malaria case burden in Migori County, western Kenya. Sci Rep. 2020;10: 1–14. 10.1038/s41598-020-61350-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kisinza WN, Nkya TE, Kabula B, Overgaard HJ, Massue DJ, Mageni Z, et al. Multiple insecticide resistance in Anopheles gambiae from Tanzania: A major concern for malaria vector control. Malar J. 2017;16: 439 10.1186/s12936-017-2087-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chukwuekezie O, Nwosu E, Nwangwu U, Dogunro F, Onwude C, Agashi N, et al. Resistance status of Anopheles gambiae (s.l.) to four commonly used insecticides for malaria vector control in South-East Nigeria. Parasites and Vectors. 2020;13 10.1186/s13071-020-3896-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oakeshott JG, Devonshire AL, Claudianos C, Sutherland TD, Horne I, Campbell PM, et al. Comparing the organophosphorus and carbamate insecticide resistance mutations in cholin- and carboxyl-esterases. Chem Biol Interact. 2005;157–158: 269–275. 10.1016/j.cbi.2005.10.041 [DOI] [PubMed] [Google Scholar]
- 13.Weill M, Lutfalla G, Mogensen K, Chandre F, Berthomieu A, Berticat C, et al. Insecticide resistance in mosquito vectors. Nature. 2003;423: 136–137. 10.1038/423136b [DOI] [PubMed] [Google Scholar]
- 14.Weill M, Malcolm C, Chandre F, Mogensen K, Berthomieu A, Marquine M, et al. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol Biol. 2004;13: 1–7. 10.1111/j.1365-2583.2004.00452.x [DOI] [PubMed] [Google Scholar]
- 15.Feyereisen R, Dermauw W, Van Leeuwen T. Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pestic Biochem Physiol. 2015;121: 61–77. 10.1016/j.pestbp.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 16.Greenblatt HM, Guillou C, Guénard D, Argaman A, Botti S, Badet B, et al. The complex of a bivalent derivative of galanthamine with torpedo acetylcholinesterase displays drastic deformation of the active-site gorge: implications for structure-based drug design. J Am Chem Soc. 2004;126: 15405–11. 10.1021/ja0466154 [DOI] [PubMed] [Google Scholar]
- 17.Cheung J, Mahmood A, Kalathur R, Liu L, Carlier PR. Structure of the G119S Mutant Acetylcholinesterase of the Malaria Vector Anopheles gambiae Reveals Basis of Insecticide Resistance. Structure. 2018;26: 130–136.e2. 10.1016/j.str.2017.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourguet D, Roig A, Toutant JP, Arpagaus M. Analysis of molecular forms and pharmacological properties of acetylcholinesterase in several mosquito species. Neurochem Int. 1997;31: 65–72. 10.1016/s0197-0186(96)00118-0 [DOI] [PubMed] [Google Scholar]
- 19.Labbé P, Berthomieu A, Berticat C, Alout H, Raymond M, Lenormand T, et al. Independent Duplications of the Acetylcholinesterase Gene Conferring Insecticide Resistance in the Mosquito Culex pipiens. Mol Biol Evol. 2007;24: 1056–1067. 10.1093/molbev/msm025 [DOI] [PubMed] [Google Scholar]
- 20.Dabiré KR, Diabaté A, Namontougou M, Djogbenou L, Kengne P, Simard F, et al. Distribution of insensitive acetylcholinesterase (ace-1R) in Anopheles gambiae s.l. populations from Burkina Faso (West Africa). Trop Med Int Heal. 2009;14: 396–403. 10.1111/j.1365-3156.2009.02243.x [DOI] [PubMed] [Google Scholar]
- 21.Ahoua Alou LP, Koffi AA, Adja MA, Tia E, Kouassi PK, Koné M, et al. Distribution of ace-1R and resistance to carbamates and organophosphates in Anopheles gambiae s.s. populations from Côte d’Ivoire. Malar J. 2010;9: 167 10.1186/1475-2875-9-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Essandoh J, Yawson AE, Weetman D. Acetylcholinesterase (Ace-1) target site mutation 119S is strongly diagnostic of carbamate and organophosphate resistance in Anopheles gambiae s.s. and Anopheles coluzzii across southern Ghana. Malar J. 2013;12: 404 10.1186/1475-2875-12-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngufor C, Chouaïbou M, Tchicaya E, Loukou B, Kesse N, N’Guessan R, et al. Combining organophosphate-Treated wall linings and long-lasting insecticidal nets fails to provide additional control over long-lasting insecticidal nets alone against multiple insecticide-resistant Anopheles gambiae in Côte d’Ivoire: An experimental hut. Malar J. 2014;13: 1–10. 10.1186/1475-2875-13-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weetman D, Mitchell SN, Wilding CS, Birks DP, Yawson AE, Essandoh J, et al. Contemporary evolution of resistance at the major insecticide target site gene Ace-1 by mutation and copy number variation in the malaria mosquito Anopheles gambiae. Mol Ecol. 2015;24: 2656–2672. 10.1111/mec.13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elanga-Ndille Nouage, Ndo Binyang, Assatse Nguiffo-Nguete, et al. The G119S Acetylcholinesterase (Ace-1) Target Site Mutation Confers Carbamate Resistance in the Major Malaria Vector Anopheles gambiae from Cameroon: A Challenge for the Coming IRS Implementation. Genes (Basel). 2019;10: 790 10.3390/genes10100790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng X, Yang C, Yang Y, Li J, Lin K, Li M, et al. Distribution and frequency of G119S mutation in ace-1 gene within Anopheles sinensis populations from Guangxi, China. Malar J. 2015;14: 470 10.1186/s12936-015-1000-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Djogbénou L, Chandre F, Berthomieu A, Dabiré R, Koffi A, Alout H, et al. Evidence of Introgression of the ace-1R Mutation and of the ace-1 Duplication in West African Anopheles gambiae s. s. Carter DA, editor. PLoS One. 2008;3: e2172 10.1371/journal.pone.0002172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edi C V., Djogbénou L, Jenkins AM, Regna K, Muskavitch MAT, Poupardin R, et al. CYP6 P450 Enzymes and ACE-1 Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito Anopheles gambiae. Zhang J, editor. PLoS Genet. 2014;10: e1004236 10.1371/journal.pgen.1004236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Assogba BS, Djogbénou LS, Milesi P, Berthomieu A, Perez J, Ayala D, et al. An ace-1 gene duplication resorbs the fitness cost associated with resistance in Anopheles gambiae, the main malaria mosquito. Sci Rep. 2015;5: 14529 10.1038/srep14529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Assogba BS, Milesi P, Djogbénou LS, Berthomieu A, Makoundou P, Baba-Moussa LS, et al. The ace-1 Locus Is Amplified in All Resistant Anopheles gambiae Mosquitoes: Fitness Consequences of Homogeneous and Heterogeneous Duplications. Barton N, editor. PLOS Biol. 2016;14: e2000618 10.1371/journal.pbio.2000618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Assogba BS, Alout H, Koffi A, Penetier C, Djogbénou LS, Makoundou P, et al. Adaptive deletion in resistance gene duplications in the malaria vector Anopheles gambiae. Evol Appl. 2018;11: 1245–1256. 10.1111/eva.12619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Anopheles gambiae 1000 Genomes Consortium. Genome variation and population structure among 1142 mosquitoes of the African malaria vector species Anopheles gambiae and Anopheles coluzzii. Genome Res. 2020;30: 1533–1546. 10.1101/gr.262790.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miles A, Harding NJ, Bottà G, Clarkson CS, Antão T, Kozak K, et al. Genetic diversity of the African malaria vector Anopheles gambiae. Nature. 2017;552: 96–100. 10.1038/nature24995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucas ER, Miles A, Harding NJ, Clarkson CS, Lawniczak MKN, Kwiatkowski DP, et al. Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes. Genome Res. 2019;29: 1250–1261. 10.1101/gr.245795.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bass C, Nikou D, Vontas J, Williamson MS, Field LM. Development of high-throughput real-time PCR assays for the identification of insensitive acetylcholinesterase (ace-1R) in Anopheles gambiae. Pestic Biochem Physiol. 2010;96: 80–85. 10.1016/J.PESTBP.2009.09.004 [DOI] [Google Scholar]
- 36.Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, et al. Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude. Science. 2010;329: 75–78. 10.1126/science.1190371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malaspinas AS, Westaway MC, Muller C, Sousa VC, Lao O, Alves I, et al. A genomic history of Aboriginal Australia Nature. Nature Publishing Group; 2016. pp. 207–214. 10.1038/nature18299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman A, Hallgrímsdóttir I, Eisen M, Pachter L. Association mapping from sequencing reads using k-mers. Elife. 2018;7 10.7554/eLife.32920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garud NR, Messer PW, Buzbas EO, Petrov DA. Recent Selective Sweeps in North American Drosophila melanogaster Show Signatures of Soft Sweeps. Copenhaver GP, editor. PLOS Genet. 2015;11: e1005004 10.1371/journal.pgen.1005004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messer PW, Petrov DA. Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol Evol. 2013;28: 659–669. 10.1016/j.tree.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durand EY, Patterson N, Reich D, Slatkin M. Testing for Ancient Admixture between Closely Related Populations. Mol Biol Evol. 2011;28: 2239–2252. 10.1093/molbev/msr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, et al. Ancient Admixture in Human History. Genetics. 2012;192: 1065–1093. 10.1534/genetics.112.145037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alout H, Berthomieu A, Cui F, Tan Y, Berticat C, Qiao C, et al. Different Amino-Acid Substitutions Confer Insecticide Resistance Through Acetylcholinesterase 1 Insensitivity in Culex vishnui and Culex tritaeniorhynchus (Diptera: Culicidae) from China. J Med Entomol. 2007;44: 463–469. 10.1603/0022-2585(2007)44[463:dascir]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- 44.Dabiré RK, Namountougou M, Diabaté A, Soma DD, Bado J, Toé HK, et al. Distribution and Frequency of kdr Mutations within Anopheles gambiae s.l. Populations and First Report of the Ace.1G119S Mutation in Anopheles arabiensis from Burkina Faso (West Africa). Brooke B, editor. PLoS One. 2014;9: e101484 10.1371/journal.pone.0101484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liebman KA, Pinto J, Valle J, Palomino M, Vizcaino L, Brogdon W, et al. Novel mutations on the ace-1 gene of the malaria vector Anopheles albimanus provide evidence for balancing selection in an area of high insecticide resistance in Peru. Malar J. 2015;14: 74 10.1186/s12936-015-0599-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Labbé P, Milesi P, Yébakima A, Pasteur N, Weill M, Lenormand T. Gene-dosage effects on fitness in recent adaptive duplications: ACE-1 in the mosquito culex pipiens. Evolution (N Y). 2014;68: 2092–2101. 10.1111/evo.12372 [DOI] [PubMed] [Google Scholar]
- 47.Edi CA V, Koudou BG, Bellai L, Adja AM, Chouaibou M, Bonfoh B, et al. Long-term trends in Anopheles gambiae insecticide resistance in Côte d’Ivoire. Parasit Vectors. 2014;7: 500 10.1186/s13071-014-0500-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Djogbénou LS, Assogba B, Essandoh J, Constant EA V., Makoutodé M, Akogbéto M, et al. Estimation of allele-specific Ace-1 duplication in insecticide-resistant Anopheles mosquitoes from West Africa. Malar J. 2015;14: 507 10.1186/s12936-015-1026-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weetman D, Donnelly MJ. Evolution of insecticide resistance diagnostics in malaria vectors. Trans R Soc Trop Med Hyg. 2015;109: 291–3. 10.1093/trstmh/trv017 [DOI] [PubMed] [Google Scholar]
- 50.Oxborough RM, Seyoum A, Yihdego Y, Dabire R, Gnanguenon V, Wat’Senga F, et al. Susceptibility testing of Anopheles malaria vectors with the neonicotinoid insecticide clothianidin; Results from 16 African countries, in preparation for indoor residual spraying with new insecticide formulations. Malar J. 2019;18: 264 10.1186/s12936-019-2888-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;11: 11.10.1 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J. Haplotype estimation using sequencing reads. Am J Hum Genet. 2013;93: 687–696. 10.1016/j.ajhg.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.The Anopheles gambiae 1000 Genomes Consortium. Ag1000G Phase 2 AR1 data release In: MalariaGEN; [Internet]. 2017. Available: https://www.malariagen.net/data/ag1000g-phase-2-ar1 [Google Scholar]
- 56.Giraldo-Calderón GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43: D707–D713. 10.1093/nar/gku1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fanello C, Santolamazza F, Della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002;16: 461–464. 10.1046/j.1365-2915.2002.00393.x [DOI] [PubMed] [Google Scholar]
- 58.World Health Organization. Test procedures for insecticide resistance monitoring in malaria vector mosquitoes 2nd ed WHO; Genèva: World Health Organization; 2018. [Google Scholar]
- 59.Warnes GR, Bolker B, Lumley T, from Randall C. Johnson are Copyright SAIC-Frederick RCJC, by the Intramural Research Program IF, of the NIH, et al. gmodels: Various R Programming Tools for Model Fitting; 2018. [Google Scholar]
- 60.Schaarschmidt F, Gerhard D. pairwiseCI: Confidence Intervals for Two Sample Comparisons. 2019. [Google Scholar]
- 61.Santolamazza F, Mancini E, Simard F, Qi Y, Tu Z, della Torre A. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7: 163 10.1186/1475-2875-7-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chabi J, Van’t Hof A, N’dri LK, Datsomor A, Okyere D, Njoroge H, et al. Rapid high throughput SYBR green assay for identifying the malaria vectors Anopheles arabiensis, Anopheles coluzzii and Anopheles gambiae s.s. Giles. PLoS One. 2019;14: e0215669 10.1371/journal.pone.0215669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogers AR, Huff C. Linkage disequilibrium between loci with unknown phase. Genetics. 2009;182: 839–44. 10.1534/genetics.108.093153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miles A, Harding N. scikit-allel. 2017. 10.5281/zenodo.822784 [DOI] [Google Scholar]
- 65.Jones E, Oliphant T, Peterson P, others. SciPy: Open source scientific tools for Python. 2019. [Google Scholar]
- 66.Ellson J, Gansner E, Hu Y, Janssen E, North S. Graphviz—Graph Visualization Software. Available: https://www.graphviz.org/about/ [Google Scholar]
- 67.Clarkson CS, Miles A, Harding NJ, Weetman D, Kwiatkowski D, Donnelly M, et al. The genetic architecture of target-site resistance to pyrethroid insecticides in the African malaria vectors Anopheles gambiae and Anopheles coluzzii. BioRxiv. 2018. 10.1101/323980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clarkson C, Miles A. Hapclust. 2018. Available: https://github.com/malariagen/agam-vgsc-report
- 69.Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov I V., et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347: 1258524 10.1126/science.1258524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen L-TT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32: 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14: 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol. 2018;35: 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30: 1188–95. 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 2012;3: 217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- 75.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Schwartz R, editor. Bioinformatics. 2019;35: 526–528. 10.1093/bioinformatics/bty633 [DOI] [PubMed] [Google Scholar]
- 76.Cavalli-Sforza L. Human Diversity. Proceedings of the 12th International Congress of Genetics. 1969. pp. 405–416.
- 77.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132: 583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: The impact of rare variants. Genome Res. 2013;23: 1514–1521. 10.1101/gr.154831.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klaus B, Strimmer. K. fdrtool: Estimation of (Local) False Discovery Rates and Higher Criticism. 2015.
- 80.Patterson N, Price AL, Reich D. Population Structure and Eigenanalysis. PLoS Genet. 2006;2: e190 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27: 764–770. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria; 2017. Available: https://www.r-project.org/
- 83.Kriventseva E V, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, Simão FA, et al. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019;47: D807–D811. 10.1093/nar/gky1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12: 59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 85.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30: 772–80. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25: 1972–3. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25: 1307–20. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- 88.Geneious. Geneious. 2019. Available: www.geneious.com
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Alignment of ACE homologs (protein sequences) in selected species (A. gambiae, Culex quinquefasciatus, Aedes aegypti, Homo sapiens, and Torpedo californica), used to determine the homology of non-synonymous mutations in this gene (A65S and G280S are highlighted). B) Alignment of ACE protein homologs in 20 culicine species, focusing on the vicinity of codon 280 (highlighted). C) Maximum-Likelihood phylogenetic analysis of ACE homologs from 89 animals (listed in S2 Data, including data sources and accession numbers).
(PDF)
A) Peptide coordinates of A. gambiae Ace1 codon 280 in orthologs of Ace1 from 89 animal genomes. B) Alignment of Ace1 homologs from 89 animal genomes. C) Data sources, accession numbers, species abbreviation and taxonomy of the 89 animal genomes used in the analysis of ACE homology.
(XLSX)
Coordinates of non-synonymous mutations in Ace1 and frequencies in each population of the Anopheles gambiae 1000 Genomes Phase 2 dataset. Species codes: A. gambiae, gam, A. coluzzii, col. Population codes: Angola, AOcol; Burkina-Faso, BFcol and BFgam; Côte d’Ivoire, CIcol; Cameroon, CMgam; Mayotte, FRgam; Gabon, GAgam; Ghana, GHcol and GHgam; The Gambia, GM; Guinea, GNcol and GNgam; Equatorial Guinea, GQgam; Guinea-Bissau, GW; Kenya, KE; Uganda, UGgam.
(XLSX)
List of samples from the Anopheles gambiae 1000 Genomes Phase 2 dataset with accession numbers, sample metadata (population and region of origin, collection date, species, sex), and summary of the main Ace1 mutations in each sample (genotypes in the G280S mutation, number of reads supporting 280G and 280S alleles, number of CNVs). CNV coordinates and copy number from Lucas et al. (2019).
(XLSB)
List of genes in the Ace1 duplication region, with genomic coordinates along the 2R chromosomal arm.
(XLSX)
A) Genotypes of G280S and A65S mutations in A. coluzzii samples from Côte d’Ivoire (Anopheles gambiae 1000 Genomes Phase 2), and resistance phenotype to pirimiphos-methyl. B) Phenotype-genotype association tests for the 71 Ivorian A. coluzzii samples. Includes summaries of single-variable GLM models for the following variables: 280S presence (including a subset of samples with only one 280S allele), 65S, CNV and number of 280S alleles. Also includes the minimal model obtained using stepwise reduction of a starting multi-variable model (BIC criterion). Model significance is measured with ANOVA comparison with null model (no variables) and a χ2 test. C) G280S genotypes and pirimiphos-methyl resistance phenotype for 1080 mosquitoes from West African populations of A. gambiae and A. coluzzii collected from six locations. For each sample, we also report collection dates, geographical locations, and details of the species identification, genotyping, and phenotyping (concentrations, exposure time). D) Phenotype-genotype association tests for G280S alleles in nine West African populations. Includes summaries of GLM models for each population. Model significance is measured with ANOVA comparison with null model (no variables) and a χ2 test. E) Number of Ace1 copies and pirimiphos-methyl resistance phenotypes for 167 mosquitoes from the same West African populations. F) Phenotype-genotype association tests for CNV mutations (Ace1 copies [CNV] and 280S-to-280G allele ratio [ratio_FAM_HEX]) in West African populations. Includes summaries of GLM models for each population and each variable separately, and a minimal GLM obtained using the BIC criterion from an initial model with both variables. Model significance is measured with ANOVA comparison with null model (no variables) and a χ2 test. G) Protocols for species identification and Ace1 mutation genotyping, including primer sequences.
(XLSX)
A) Genetic differentiation statistics (Hudson’s FST and PBS) between resistant and susceptible A. coluzzii from Côte d’Ivoire, in windows of 10,000 variants along the genome. PBS values are calculated using Angolan A. coluzzii as outgroup, and we also report a Z-score and p-value derived from a two-sided normal distribution. B) Genes overlapping the regions of high PBS (FDR < 0.001 and standardised PBS > 0).
(XLSX)
A) Alignment coordinates of k-mers that are significantly associated with pirimiphos-methyl resistance in Ivorian A. coluzzii samples from the Anopheles gambiae 1000 Genomes Phase 2. For each k-mer, we report the alignment sequence and coordinates, and whether it overlaps the Ace1 gene or the Ace1 duplication. This table includes both primary and secondary alignments as reported by bwa mem, as well as non-aligned k-mers (chromosome “NA”). B) Correlation of alignment frequencies and Ace1 copy number in each sample, for each of the 409 k-mers that mapped to chromosomal arms 2R, 2L, 3R, 3L or X. We report Pearson’s correlation coefficients (r) and p-values. C) Sequences of the 446 assembled k-mers that are significantly associated with pirimiphos-methyl resistance. D) Frequencies of the 9,603 k-mers (before assembly) in each of the Côte d’Ivoire A. coluzzii samples (Anopheles gambiae 1000 Genomes Phase 2, n = 71).
(XLSX)
A) Density of phased variants in the Ace1 duplication region, from Anopheles gambiae 1000 Genomes Phase 2. B) Density of phased variants within the Ace1 duplication, focusing on the Ace1 gene body. C) Linkage disequilibrium between the G280S alleles and nearby phased variants (Huff and Roger’s r), highlighting the coordinates of nearby variants tightly linked to 280S alleles that could be used as tagging variants for 280S (in green), or variants that were later discarded by other filtering steps (in grey), and their frequencies in Côte d’Ivoire A. coluzzii. D) Haplotype score values in the vicinity of the Ace1 locus, calculated from the Phase 2 1000 Genomes cohort. Variants within low-score regions can be used to tag 280S (in green), variants within peaks were discarded (grey). E) Distribution of haplotype score values for variants within the regions used to build haplotype networks (green, tagging variant ± 300 bp) and in the Ace1 gene body (magenta). Distributions are shown as cumulative frequency distributions and boxplots (box lines represent the first, second and third quartiles). F) Distribution of the sequencing coverage of each sample with Ace1 duplications, for the genomic window containing each tagging variant (green) and Ace1 (magenta). A normalised coverage of two implies diploidy. Tagging variants are located in regions of lower sequencing coverage than Ace1, even though they all fall within the duplicated region.
(PDF)
Minimum spanning tree networks built from phased variants located around each of the three tagging variants (panels A to C). Each node in the network is colored according to its population composition (left panels) or linkage to the 280S or wt alleles in Ace1 (right panels).
(PDF)
Garud H statistics and haplotype diversity in 280S-linked and wt-linked haplotypes in the genomic window around the Ace1 duplication breakpoints, calculated for each of the main haplotype clusters defined around each of the three tagging variants (panels A to C; haplotype clusters from S10). For each tagging variant and duplication breakpoint (upstream/downstream), we report the average value of each statistic and standard errors from sample jack-knifing.
(PDF)
Extended haplotype homozygosity (EHH) of 280S-linked and wt-linked haplotypes in the genomic window around the Ace1 duplication breakpoints, calculated for each of the main haplotype clusters defined around each of the four tagging variants (panels A to D; haplotype clusters from S10). For each tagging variant and duplication breakpoint (upstream/downstream), we report the area under the EHH curve (a) and the distance around the duplication breakpoint where EHH>0.05 and EHH>0.95.
(PDF)
Patterson D statistics to test introgression of the Ace1 duplication region between various populations of A. coluzzii (populations A/B), A. gambiae (populations C) and multiple outgroup species (population O). Specifically, we use A. coluzzii from Côte d’Ivoire (CIcol), Ghana (GHcol) and Burkina-Faso (BFcol) with and without duplications (labelled as TRUE and FALSE respectively) as populations A; A. coluzzii from Angola as population B (always labelled as FALSE); and various A. gambiae populations C from Ghana (GHgam), Burkina-Faso (BFgam), Guinea (GNgam), Gabon (GAgam) and Cameroon (CMgam) with and without duplications (TRUE and FALSE labels, respectively); and four different outgroup species (panels A to D: A. arabiensis, A. melas, A. merus and A. quadriannulatus). For each comparison, we report the average D statistic from the Ace1 duplicated region with standard errors, and Z-scores and p-values derived from standardised D values (unit variance).
(PDF)
A) Alignment of phased variants from within the Ace1 duplication region (2R:3,436,800–3,639,600), using 345 samples from West African populations (Anopheles gambiae 1000 Genomes) with Ace1 duplications (Guinea A. gambiae, Côte d’Ivoire A. coluzzii, Ghana A. gambiae and A. coluzzii, Burkina Faso A. gambiae and A. coluzzii). B) Id., from a region downstream of the duplication (5 kbp starting at 3,639,600). C) Id., from a region upstream of the duplication (50 kbp starting at 3,436,800–1 Mb). D) Id., from a region downstream of the duplication (50 kbp starting at 3,639,600 + 1 Mb).
(XLSX)
Maximum-Likelihood phylogenetic analyses of haplotypes from within the Ace1 duplication (A), downstream breakpoint (B), upstream (C) and downstream (D) regions. Trees are unrooted. Tips are color-coded according to duplication presence/absence and species. UF bootstrap supports indicated in each node.
(PDF)
A) Neighbour joining trees built from phased variants located within the PBS peaks identified between resistant and susceptible Ivorian A. coluzzii (1000 Genomes dataset). B) Profile of Garud’s H12 in these same regions.
(PDF)
A) Sequence of the duplication breakpoints described by Assogba et al. (2016) with their coordinates in the A. gambiae genome assembly (AgamP4) and the corresponding genome window in the CNV database by Lucas et al. (2019). B) Normalised sequencing coverage along the Ace1 duplication locus in each sample of the Anopheles gambiae 1000 Genomes dataset (grouped by population of origin). Detailed coverage of each sample available in S4B Data.
(PDF)
A) Distance in allelic frequencies between A. coluzzii specimens with duplications and wt A. coluzzii and A. gambiae, calculated using the three-population branch statistic in windows of 5,000 variants along the genome. Includes estimated distance (L) from within the duplication region. B) Id., using A. gambiae specimens with duplications.
(PDF)
Data Availability Statement
Python 3.4.7 and R 3.6.2 scripts to reproduce the main analyses from the paper are available in the following Github repository: https://github.com/xgrau/ace1-anopheles-report. All genome variation data has been obtained from the publicly available repositories of the Anopheles gambiae 1000 Genomes project (Phase 2 AR1): https://www.malariagen.net/data/ag1000g-phase-2-ar1 (download instructions can be found in the Github repository).