Multidrug-resistant Shigella isolates with resistance to macrolides are an emerging public health threat. We define a plasmid/pathogen complex behind infections seen in the United States and globally in vulnerable patient populations and identify multiple outbreaks in the United States and evidence of intercontinental transmission.
KEYWORDS: genomics, Shigella, antimicrobial therapy, azithromycin, epidemiology, macrolide resistant, multidrug resistant, plasmid acquisition, public health, rapid diagnosis
ABSTRACT
Multidrug-resistant (MDR) Shigella infections have been identified globally among men who have sex with men (MSM). The highly drug-resistant phenotype often confounds initial antimicrobial therapy, placing patients at risk for adverse outcomes, the development of more drug-resistant strains, and additional treatment failures. New macrolide-resistant Shigella strains complicate treatment further as azithromycin is a next-in-line antibiotic for MDR strains, and an antibiotic-strain combination confounded by gaps in validated clinical breakpoints for clinical laboratories to interpret macrolide resistance in Shigella. We present the first high-resolution genomic analyses of 2,097 U.S. Shigella isolates, including those from MDR outbreaks. A sentinel shigellosis case in an MSM patient revealed a strain carrying 12 plasmids, of which two carried known resistance genes, the pKSR100-related plasmid pMHMC-004 and spA-related plasmid pMHMC-012. Genomic-epidemiologic analyses of isolates revealed high carriage rates of pMHMC-004 predominantly in U.S. isolates from men and not in other demographic groups. Isolates genetically related to the sentinel case further harbored elevated numbers of unique replicons, showing the receptivity of this Shigella lineage to plasmid acquisition. Findings from integrated genomic-epidemiologic analyses were leveraged to direct targeted clinical actions to improve rapid diagnosis and patient care and for public health efforts to further reduce spread.
INTRODUCTION
Sexually transmitted Shigella sonnei and S. flexneri have been reported among men who have sex with men (MSM) in the United States since 1974 (1–3). HIV-positive MSM are at increased risk for sporadic shigellosis, and infection with HIV is among a range of factors that can contribute to transmission of shigellosis among MSM (4). Multiple U.S. outbreaks of shigellosis among MSM have occurred over the past decade, resulting in calls for active additional testing, surveillance, and health education efforts focused on MSM, patients, and health care professionals to address this public health threat (5–8).
Antimicrobial resistance (AMR) in Shigella isolates is frequently mediated by plasmid-borne genes in addition to chromosomal determinants (9). Isolates of multidrug-resistant (MDR; defined as resistant to at least three classes of antibiotics [10]) Shigella from MSM in New York City have increasingly shown decreased susceptibility to the macrolide azithromycin (11), and a large proportion of Shigella isolates with decreased susceptibility to azithromycin have been shown among HIV-positive men (12). These findings are concerning given azithromycin’s role in empirical treatment of infectious diarrhea and use for Shigella resistant to fluoroquinolones, penicillins, and sulfamethoxazole/trimethoprim (13). However, in contrast to these other antibiotic classes, macrolides are not included in most automated clinical microbiology tests for drug resistance in Enterobacteriaceae. The lack of Clinical and Laboratory Standards Institute (CLSI) validated clinical breakpoints for interpretation of macrolide results in Shigella further confounds timely interpretation and reporting of resistance (14).
Azithromycin-resistant S. sonnei isolates carrying the mph(A) gene have been found in the United States since 1987 (15). Plasmid pKSR100, recently associated with S. sonnei and S. flexneri infections among MSM, carries mph(A) and has been identified as a primary driver of macrolide resistance in Shigella spp. and strongly associated with infections in MSM generally (16–19). Recent research on pKSR100 plasmids in Great Britain shows fluctuations in case numbers among different clades of Shigella, indicating that this plasmid/pathogen complex continues to evolve (20). Molecularly, insertion sequence 26 (IS26) has been identified as an important factor in the evolution of mph(A) within this plasmid family, with significant plasmid sequence diversity seen in Canada (19).
We identified an MSM patient with MDR S. sonnei that demonstrated resistance to multiple antibiotic classes, including macrolides (10). Integrated genomic and epidemiologic analyses of 2,097 U.S.-based Shigella cases, with functional in vitro studies, identified strain- and plasmid-specific drivers of drug resistance, risks for spread, and potential for further acquisition of resistance. The clear identification of strain- and plasmid-level drivers of resistance and spread directed targeted clinical actions to improve rapid diagnosis and to support public health efforts to further reduce spread.
RESULTS
Clinical case.
A 50- to 60-year-old HIV-positive self-reported MSM reported 2 weeks of watery, blood-streaked diarrhea, left-lower quadrant abdominal pain, and 25 pounds of weight loss over a month, without fever or chills. He had been off antiretroviral therapy for 4 years, had traveled to Central America 1 year prior, and had recent same-sex sexual activity prior to the onset of symptoms. The patient’s white blood cell count was 6,000/µl with 19% eosinophils and a CD4 cell count of <50/µl. His HIV viral load was >150,00 copies/ml, and he tested positive for cytomegalovirus viremia and Strongyloides IgG. An abdominal computerized tomography scan showed proctocolitis.
S. sonnei SBJ-9962 was isolated from stool culture. The patient was initially treated with ciprofloxacin but switched to intravenous (i.v.) ceftriaxone after susceptibility testing showed resistance to ciprofloxacin (>32 µg/ml), ampicillin (>32 µg/ml), and sulfamethoxazole-trimethoprim (>320 µg/ml), with susceptibility to ceftriaxone (<1 µg/ml). Upon request, an azithromycin Etest showed a MIC of >256 µg/ml. Symptoms resolved after 4 days, and he was transitioned to oral cefpodoxime. Antiretroviral therapy was resumed.
The patient was discharged to home on cefpodoxime, but several days later he developed a recurrence of diarrhea and abdominal pain. Repeat stool cultures again isolated S. sonnei with the same susceptibilities, producing the isolate SBJ-9961. He was continued on oral cefpodoxime, but symptoms persisted. At that time, he was restarted on 2 g of ceftriaxone i.v. daily. Symptoms improved after 1 week of therapy, and repeat stool cultures were negative. After two additional weeks of intravenous ceftriaxone, stool cultures remained negative.
National infrastructure for pathogen genomic surveillance identifies diverse plasmid replicons in MSM Shigella.
The isolates SBJ-9962 and SBJ-9961 were submitted to the FDA’s GenomeTrakr Network (21) and to CDC’s PulseNet network (22), distributed networks of laboratories in the United States and abroad that utilize whole-genome sequencing for pathogen detection and analysis. Deidentified genomic data from isolates were deposited into NCBI from both sources for access by local hospital, public health, and governmental personnel to support surveillance and active outbreak investigations from local through international levels.
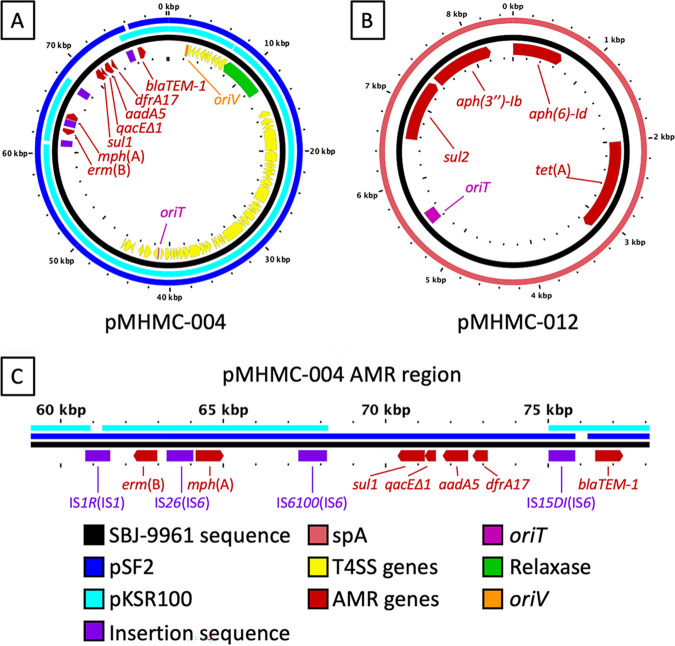
A high-resolution, closed genome of strain SBJ-9962 identified 12 distinct plasmids, including pMHMC-004, which showed >99% sequence similarity to the original pKSR100 draft sequence (GenBank accession no. LN624486.1) but with three gaps: one in the replication machinery, one at IS1R, and one between IS6100 and IS15DI. pMHMC-004 was nearly identical to pSF2, a pKSR100 family member seen first in Canada, where a gap in alignment exists next to IS15DI (Fig. 1; see also Fig. S1 in the supplemental material) (19). The second MDR plasmid, pMHMC-012, a plasmid with widespread distribution among S. sonnei isolates, showed sequence similarity to plasmid spA from S. sonnei strain Ss046 (23, 24). Six plasmids, including the MDR plasmids pMHMC-004 and pMHMC-012, contained known origins of transfer (Fig. S1). Both pMHMC-003 and pMHMC-004 encoded putatively intact type IV secretion systems. Genomic single-nucleotide polymorphism (SNP) comparison and clinical phenotypes showed that the patient’s two isolates, SBJ-9962 and SBJ-9961, are clonally related, with no clinical antibiotic resistance differences found and only 3 SNPs found in chromosomal coding regions.
FIG 1.
pMHMC-004 and pMHMC-012 are related to known multi-AMR gene plasmids. Plasmid BLAST identity over 80% with a minimum length of 100 bp is shown by colored bars exterior to the plasmid backbone in black. Genes and genetic features are labeled as directional arrows and boxes, respectively, in the inner circle. (A) pMHMC-004. (B) pMHMC-012. (C) Detail of the AMR gene region of pMHMC-004 with IS elements labeled; the IS element family is in parentheses. There were no putatively intact IS sequences found outside this region, and none were found in pMHMC-012.
Twelve plasmids of S. sonnei SBJ-9962. Genes and genetic features are labeled as directional arrows and boxes, respectively, in the inner circle for selected gene categories. Only pMHMC-003 and pMHMC-004 encode a putative T4SS, while those plus pMHMC-006, pMHMC-009, pMHMC-011, and pMHMC-012 encode a known origin of transfer. Two plasmids carry known AMR genes, pMHMC-004 and pMHMC-012. Most plasmids encoded a typical replicon, with the larger plasmids having Inc type replicons and the smaller plasmids having Col type replicons. Download FIG S1, PDF file, 0.5 MB (479.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Functional studies of the host range for the two MDR plasmids showed that pMHMC-012 could be transferred by conjugation to S. sonnei and Escherichia coli, while pMHMC-004 could only be transformed into several S. sonnei strains, including recently isolated clinical strains, by electroporation, but not E. coli DH5α or E. coli J53 (Data Set S2).
Antibiotic resistance profile.
S. sonnei SBJ-9961 demonstrated resistance to seven antibiotic classes: aminoglycosides, fluoroquinolones, macrolides, penicillins, sulfonamides, and tetracyclines (Table S1). Based on plasmid electroporation into the putatively pan-susceptible strain S. sonnei ATCC 25931, pMHMC-004 conferred resistance to macrolides, penicillins, sulfonamides, and sulfamethoxazole-trimethoprim, while pMHMC-012 conferred resistance to tetracycline, streptomycin, and sulfonamides. Both plasmids alone confer an MDR phenotype, defined as resistant to antimicrobials in at least three antibiotic classes. All strains remained susceptible to cephalosporins, colistin, beta-lactam combination agents, and several other classes of antimicrobials (Table S1) (10).
Antibiotic susceptibility testing on strains carrying plasmids pMHMC-004 and pMHMC-012. Antibiotic resistance testing results for SBJ-9961, ATCC 25931, ATCC 25931 pMHMC-004, and ATCC 25931 pMHMC-012. Symbols: *, intermediate range for resistance phenotypes, determined by CLSI; †, intermediate; ‡, resistant. These tests were done for comprehensive antimicrobial resistance testing. Not all drugs listed should be considered for treatment of shigellosis. Download Table S1, DOCX file, 0.02 MB (16.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Antimicrobial resistance determinants.
SBJ-9962 carried chromosomal dfrA1 and sat2 AMR genes in a class 2 integron in Tn7 downstream of glmS, which promote resistance to trimethoprim and streptothricin, respectively, but without aadA1, which is usually present in this integron (25). The chromosome also carries the fluoroquinolone resistance mutations gyrA S83L and D87G and parC S80I, mutations also identified in California outbreak strains in 2014 (15). Two of the 12 plasmids carried known resistance genes: pMHMC-004, which encodes erm(B), mph(A), sul1, qacEΔ1, aadA5, dfrA17, and blaTEM-1; and pMHMC-012, which encodes aph(6)-Id, tet(A), sul2, and aph(3″)-Ib (Fig. 1). pMHMC-004 harbored four intact ISs, three of which belong to the IS6 family and one to the IS1 family (Fig. 1C). IS26 and IS6100, both IS6 family members, flank mph(A) (Fig. 1C).
SBJ-9962 belongs to an S. sonnei clade that harbors high plasmid replicon counts.
The NCBI Pathogen Detection program (https://www.ncbi.nlm.nih.gov/pathogens/) calculates groups of genetically related isolates by genomic SNP content. The genome sequences enabled improved analyses of S. sonnei outbreak strains in the United States and abroad, particularly of SNP cluster PDS000033428. Members within this cluster averaged 5.6 unique replicons among their plasmids (Fig. 2). However, the subclade within PDS000033428 that includes SBJ-9962, referred to here as clade A, showed 6.9 different replicons on average (n = 718), including the 8 unique replicons found in SBJ-9962, versus 4.1 different replicons on average for those not in clade A (n = 598, P < 0.001). The distribution of unique replicons within the overall cluster is bimodal with respect to this phylogenetic distinction. The patient isolates contain an ISSfl4 insertion in cas6e, an insertion seen in other Shigella isolates (26).
FIG 2.
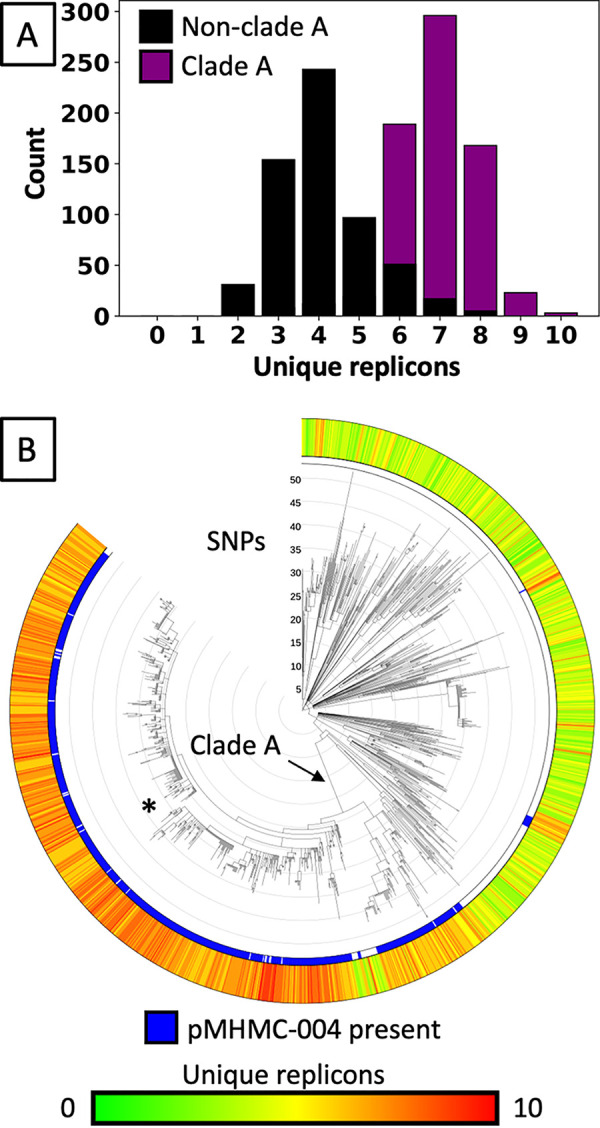
Unique replicon counts within PDS000033428. (A) Histogram of the number of different replicons detected in each isolate in PDS000033428.133 using the PlasmidFinder database. Isolates from a phylogenetically defined subgroup, clade A, average more distinct replicons. (B) The SNP cluster PDS000033428.133, which includes SBJ-9962 (*) with 8 different replicons, contains clade A with an elevated number of plasmid replicons. Clade A has 718 isolates.
Distribution of pMHMC-004 among S. sonnei, S. flexneri, and E. coli.
Approximately 20% of all S. flexneri and S. sonnei isolate genomes listed in the Pathogen Detection Isolates Browser showed carriage of pMHMC-004 or pKSR100 homologs (Table S2). In contrast, only 2 of 42,465 E. coli isolates harbored any of these plasmids, one of which was adherent-invasive E. coli (AIEC) LF82 (NC_011993.1), and the other was cultured from an asymptomatic case of bacteriuria (ABU), E. coli 83972 (NC_017631.1) (27, 28).
pMHMC-004 carriage in strains of Shigella and Escherichia coli. Prevalence of pKSR100 and pMHMC-004 across S. sonnei and S. flexneri genomes in NCBI. Download Table S2, DOCX file, 0.01 MB (13.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Among Shigella, pMHMC-004 occurred in all Shigella clades (Fig. S2), with multiple instances of plasmid acquisition and loss. Additional strains across clades carried gene regions with homology to portions of pMHMC-004 but at thresholds below those for calling pMHMC-004 or pKSR100 presence. Given the known sequence diversity for the pKSR100-related plasmids, heterogenous plasmid populations exist with broadly shared genetic cassettes (18, 19).
Large SNP-defined groups of S. sonnei and S. flexneri intercontinental transmission and plasmid acquisition and loss events. pMHMC-004 and pKSR100 presence is indicated in the exterior two rings. Total plasmid coverage is shown by a gradient in the first ring outside the dendrogram. Country of origin is shown for the United States, United Kingdom, and Australia. Other countries of origin and those without specified origins are grouped in a fourth category. Download FIG S2, PDF file, 1.1 MB (1.1MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Intercontinental strain and plasmid transmission.
Shigella isolates among all SNP clusters defined by NCBI at the time of access originated primarily from the United Kingdom (2,130 isolates), the United States (2,097 isolates), and Australia (321 isolates), with 132 isolates from other countries or that were of unspecified origin (Fig. S2). Isolates within each SNP cluster occurred across the three predominant countries, and subclades within these clusters showed strong geographical biases, suggesting localized spread with genetic divergence from a common ancestor. Cluster PDS000033428, which includes SBJ-9962, is particularly diverse in origin, with the repeated occurrence of the strain between the United States and the United Kingdom. These findings illustrate the occurrence of plasmid/strain complexes across continents and demonstrate putative instances of intercontinental transmission.
pMHMC-004 and related plasmids have association with adult males in the United States.
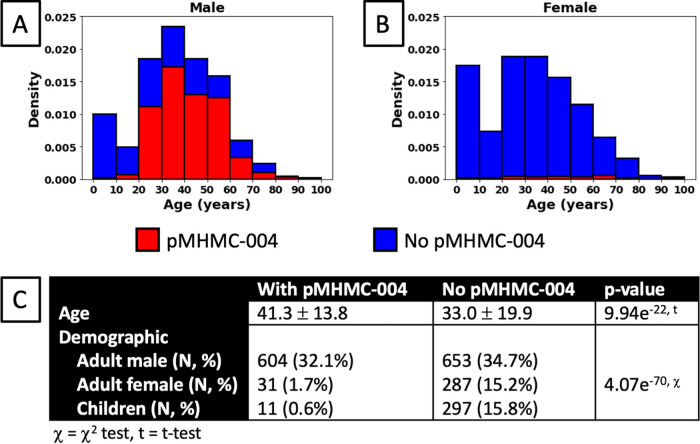
Epidemiologic analyses of 1,883 U.S.-based Shigella cases with known demographic data identified strong association of the MDR S. sonnei strain and plasmids with adult males (Fig. 3). The average age of patients linked to isolates with pMHMC-004 was 41.3 ± 13.8, while that of patients linked to isolates without the plasmid averaged 33.0 ± 19.9 (P = 9.9 × 10−22). The discrepancy in age was largely due to higher numbers of children (ages 0 to 18) in the latter category, where only 11 of 308 (3.6%) isolates carried the plasmid. Similarly, of cases involving adult females, only 31 of 318 (9.7%) isolates carried pMHMC-004. However, among isolates from adult males, 604 of 1,257 (52%) cases carried the plasmid. In total, of all cases involving the plasmid, 604 out of 646 cases (93%) with both age and sex recorded involved adult males. The observed difference in this distribution was highly significant (χ2 = 320, df = 2, P = 4.1e−70).
FIG 3.
Demographic distribution of pMHMC-004-containing isolates. (A) Normalized histogram showing the proportion of isolates with or without pMHMC-004 by age for male patients. (B) Normalized histogram showing the proportion of isolates with or without pMHMC-004 by age for female patients. (C) Univariate analysis of plasmid carriage likelihood by age and demographic groups.
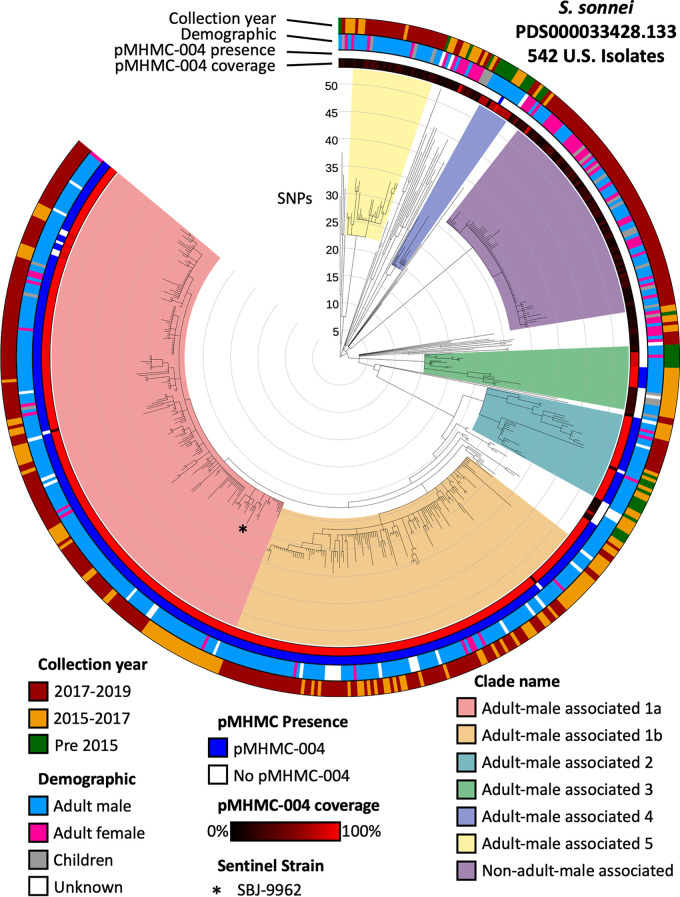
The largest SNP cluster, PDS000033428, which includes SBJ-9962, has five large clades that are primarily associated with adult males (>80%). Adult male-associated clades 1, 2, 3, and 4 demonstrated high prevalence of pMHMC-004-related sequence, a trait that adult male-associated clade 5 does not demonstrate (Fig. 4, Fig. S3, Data Set S3). The clade of non-adult male-associated strains does not have strong pKSR100 family sequence representation.
FIG 4.
SNP cluster PDS000033428.133 includes isolates from a recent and large clade of MDR Shigella sonnei in the United States. The rings, from interior to exterior, represent pMHMC-004 coverage, pMHMC-004 presence, demographic group, and year of isolation. Clades of interest within this SNP cluster are color coded directly on the tree. Adult male-associated clades have >80% of isolates from adult males.
SNP cluster PDS000033428.133 has highly variable AMR gene content driven partially by ISs in family IS6. The rings, from interior to exterior, represent pMHMC-004 coverage, pMHMC-004 presence, and then, in larger groups of rings, pMHMC-004 AMR gene content and pMHMC-012 AMR gene content. Clades of interest within this SNP cluster are color coded directly on the tree and are the same as those in Fig. 6. Download FIG S3, PDF file, 0.7 MB (686.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
pMHMC-004 and pMHMC-012 show variable carriage of AMR genes over time.
Significant variation occurred in the AMR genes seen in clades associated with cluster PDS000033428 (Fig. 4, Fig. S3, Data Set S4). After points of acquisition of macrolide resistance genes, sporadic isolates lacking mph(A) or erm(B) occurred, suggesting genetic excision events involving the three IS6 family ISs. Several strains lacked some or all genes bounded by these elements, indicative of multiple possible rearrangements involving these IS6 elements. The variability in MDR genes illustrates the degree of heterogeneity in the pKSR100 family of plasmids, attributes that are not unique to this or other Shigella plasmids, including spA/pMHMC-012.
DISCUSSION
We present the first functional and genomic epidemiologic analyses of Shigella strains seen predominantly among men in the United States, suggesting transmission among MSM, and we further define how these components fit more broadly within Shigella cases globally (29, 30). Shigella spp. can quickly acquire and develop antibiotic resistance (31). Notably, strains seen among men can carry many different plasmids, as highlighted by the 12 unique plasmids identified in SBJ-9961. The patient isolates contain an insertion in cas6e, a component of the CRISPR-Cas system that may play a role in its capacity to maintain a high plasmid count (26). pMHMC-004 belongs to an emerging family of MDR plasmids that includes pKSR100 and has shown frequent acquisition and loss of antimicrobial resistance genes commonly mediated by recombination events among homologous insertion sequences and other acquired repetitive elements (17, 19).
pMHMC-004 showed significant association in Shigella cases of adult males, including the sentinel MSM case that started our investigation, and not in other patient populations. This combined plasmid/pathogen complex significantly complicated our patient’s treatment. Chromosomal resistance to fluoroquinolones combined with plasmid-mediated resistance to ampicillin, sulfamethoxazole-trimethoprim, and azithromycin rendered the usual oral therapies ineffective and required intravenous treatment. The receptivity of Shigella to plasmid acquisition raises further concerns for the acquisition of additional resistance genes, including those for extended-spectrum and other beta-lactamase enzymes, that could further limit treatment options (32–35).
While pMHMC-004 encodes a putative type IV secretion system, we were unable to conjugatively transfer the plasmid to susceptible recipients under laboratory conditions. However, pMHMC-012 was transferrable by conjugation, demonstrating the existence of functioning conjugative machinery in this isolate. Electroporation experiments demonstrated a primary host range limited to other Shigella species and not to other E. coli strains, a finding further supported by genomic epidemiologic analyses that identified only two instances of pMHMC-004 homologs in >40,000 E. coli genomes, strengthening the idea that this plasmid has a limited host range (Table S2).
Relatives of the pMHMC-004 plasmid (18, 19) demonstrate variation in their antibiotic resistance gene profiles, suggesting that this family of plasmids provides an adaptable backbone for the transmission of antibiotic resistance across Shigella. Clinical microbiologic phenotypic testing for antibiotic resistance in strains infecting vulnerable patient populations is paramount to ensure proper treatment and, when appropriate during outbreaks, the implementation of measures for prevention. Furthermore, ongoing national and international efforts for genomic surveillance provide means to actively monitor AMR gene acquisition to inform clinical, public health, and food safety agencies for appropriate monitoring, diagnostic, and therapeutic options to ensure optimal and timely treatment. As part of surveillance activities, the collection of demographic and behavioral data is essential to further characterize populations disproportionately impacted by MDR Shigella.
Our sentinel strain and others in the United States among adult males had putative macrolide resistance mediated via mph(A) (17). Given the current lack of CLSI clinical breakpoints to call macrolide resistance in Shigella species, laboratories may opt to proactively test Shigella with MIC-based methods to provide a minimum antibiotic concentration to direct clinical therapy. Longer term, understanding the current drivers of antimicrobial resistance is best incorporated in ongoing national and international efforts to provide timely frameworks for diagnostic laboratories in support of patient care. These issues also highlight the need for reflex cultures if culture-independent diagnostic testing (CIDT) is used as a primary modality to diagnose Shigella infections, particularly for patients who do not clear infection or who may be at higher risk for infection with MDR strains.
We note that drug-resistant cases of Shigellosis are not unique to MSM patient populations (5, 7, 17, 18). Healthcare providers should consider the potential for drug-resistant Shigella in gastroenteritis patients who are immunocompromised and have not recovered or in any case are failing to respond to antimicrobial therapy. Education for patients with MDR shigellosis to prevent transmission through a range of modes, including food, water, and both sexual and nonsexual person-to-person contact, are important to further limit pathogen spread. Among MSM, where the disease appears to be more prevalent, physicians should take sexual histories into account in providing prevention information. While multiple pathogens and clinical diseases can present with such symptoms, standard clinical microbiologic methods for identifying enteric pathogens and for susceptibility testing of identified Shigella are needed to provide appropriate diagnostic information to guide patient care and management.
MATERIALS AND METHODS
Clinical case.
An MDR S. sonnei infection was identified in the Brigham and Women’s Hospital Clinical Microbiology Laboratory and flagged by Infection Control for genomic analyses through the Partner’s Pathogen Genomic Surveillance program (IRB protocol 2011-P002883; L. Bry) to identify drivers of antimicrobial resistance and relatedness with outbreak Shigella seen in the Northeast (36, 37). Details about this case and the two isolates from it are presented in Results.
Bacterial isolation and maintenance.
Shigella isolates were isolated by stool culture on Hektoen enteric agar (Remel, Lenexa, KS) and species determined by Vitek 2 (bioMérieux, Durham, NC). Rifampin-resistant mutants of clinical strains, used for functional studies, were created by plating a 10-µl loopful of cells from LB agar (Becton, Dickinson and Company, Sparks, MD) onto LB agar with rifampin overnight at 37°C. The following antibiotics were used for selections: ampicillin (MilliporeSigma, Saint Louis, MO), 200 µg/ml; rifampin (G-Biosciences, Saint Louis, MO), 100 µg/ml; and tetracycline (MilliporeSigma, Saint Louis, MO), 15 µg/ml. Bacterial strains used in this study are listed in Table S3 in the supplemental material.
Isolates used in this study. A table of isolates, NCBI genome accession numbers, and isolate information. Strains without a reference originate from this study. Download Table S3, DOCX file, 0.02 MB (20.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Plasmid transfer analyses.
Bacterial transformations were done as previously described (38), with additional screening done on CHROMagar MH orientation agar (CHROMagar, Paris, France) and MacConkey enteric agar (Remel, Lenexa, KS) to confirm species and strain phenotypes.
For conjugation and electroporation studies of plasmid transfer, donor and recipient strains were grown overnight on LB agar at 37°C with selective antibiotics whenever appropriate (Table S3). Cells were suspended in sterile LB to an optical density at 600 nm (OD600) of 0.1, and then donor and recipient were mixed 1:1 and 50 µl was plated onto LB agar. The reaction mixtures were incubated 16 to 20 h at 37°C, 30°C, or room temperature to evaluate the temperature dependence of conjugation. Reaction mixtures were resuspended into 5 ml sterile LB using a sterile loop and diluted before plating. Thirty microliters of dilution was plated onto half a selective or nonselective agar plate for determining cell concentrations.
Electroporation was performed as described previously, with the following modifications (39). Cells were grown on solid media and resuspended in 300 mM sucrose (MilliporeSigma, Saint Louis, MO), and 2-mm cuvettes (Thermo Fisher Scientific, Waltham, MA) were used with a 2.5-kV, 25-mF, 200-Ω program and 100 µl of prepped cells. DNA for electroporation was purified using a QIAfilter plasmid midi kit (Qiagen, Germantown, MD) with 100 ml of late-log-phase-growth culture grown in dual selection with ampicillin and tetracycline at 37°C. One microliter of plasmid DNA solution (400 ng) was used in electroporation reactions.
Antibiotic susceptibility testing.
Kirby-Bauer, Etest, and broth microdilution studies were performed within Clinical and Laboratory Standards Institute (CLSI) guidelines (14, 40, 41). Azithromycin resistance levels were determined by Etest (bioMérieux, Durham, NC).
Genomic analyses.
The Partners Pathogen Genomic Surveillance program is a node on the FDA’s GenomeTrakr Network and submitted the strain for genomic analysis. DNA isolation and Illumina MiSeq sequencing were performed as described previously (36). PacBio sequencing was performed as previously described (42). Sequence accession numbers used in genomic analyses are listed in Data Set S1.
NCBI accession numbers and plasmid presence data for strains analyzed in this study. Download Data Set S1, XLSX file, 0.7 MB (216.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Electroporation and transconjugation experiments reveal a limited host range for pMHMC-004. Individual experiments are recorded on separate sheets, and a comparison is provided on the Summary sheet. Download Data Set S2, XLSX file, 0.7 MB (32.8KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
SNP clusters are variable in their carriage of pMHMC-004 and pKSR-100. The data set contains dendrograms similar to those shown in Fig. S2 but for all clusters. Download Data Set S3, PDF file, 0.7 MB (2.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Demographic information, collection time, and antimicrobial resistance plasmid presence for U.S. Shigella isolates across SNP clusters. The data set contains dendrograms similar to those in Fig. 4 but for all clusters and without named clades. Download Data Set S4, PDF file, 0.7 MB (3.4MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Sequences were analyzed for resistance genes using the Bacterial Antimicrobial Resistance Reference Gene Database (PRJNA313047) and BLAST (43). AMR genes were screened for using the thresholds of 80% protein sequence identity and 60% gene length, keeping the best hit per genome locus. All hits had >95% protein sequence identity and length. Replicons were identified using the PlasmidFinder reference sequences and with homologous plasmids called at 80% nucleotide identity and 60% coverage (44). An independent t test was used to determine if differences in averages of the number of different replicons in isolates between two groups were significant. In the case of pMHMC-011, which had 92% sequence identity and 53% replicon coverage for Col(MG828), the replicon is interrupted by the beginning of the sequence. Plasmid-encoded type IV secretion system elements were identified using oriTfinder (45). GenBank files with all features annotated were created using Biopython (46).
Sequencing reads from the E. coli and Shigella isolate genomes in the NCBI Pathogen Detection Isolates Browser (https://www.ncbi.nlm.nih.gov/pathogens) were downloaded from the Sequence Reads Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra) in May 2019. There are tools available for comparing these to known clusters, including those previously seen in the United Kingdom (K. Holt, J. Hawkey, and K. Paranagama, Sonneityping, https://github.com/katholt/sonneityping) (20, 23). The reads were de novo assembled using SPAdes with the –careful –cov-cutoff auto options to generate draft-level genome assemblies (47). Putative plasmids were evaluated using BLASTn comparisons between the complete plasmid sequences and the assembled genomes. Eighty percent sequence-level identity was used as a cutoff for mapping reads to plasmids in calculating the coverage of reference plasmid sequence in draft genomes. Due to the nature of similarity between the plasmids pMHMC-004 and pKSR100, a minimum plasmid coverage of 95% was used. The dfrA17-sul1 resistance region was also used to differentiate pMHMC-004 and pKSR100 type plasmids (Fig. 1C) (17). Kraken2 was used to confirm strain species assignments (48).
For comparisons of pMHMC-004 to related plasmids pKSR100 and pSF2, GView was used on default settings of greater than 80% nucleotide sequence identity and an E value of less than 1e−10 to determine overall sequence alignment and coverage of pMHMC-004 to the two reference plasmids (49). Plasmid sequence composition comparisons were done using BLASTn (43). Intact ISs were detected using ISfinder’s BLAST function using 80% reference sequence coverage and 80% nucleotide identity as cutoffs for pMHMC-004 and pMHMC-012 (50).
Genomic SNP comparison of the patient isolates was performed using BLASTn with nucleotide features of SBJ-9962 (CP053751.1) used to search the NCBI SBJ-9961 assembly AAVBEV000000000.1 (43).
Phylogenetic analyses.
Phylogenetic relationships were obtained from the NCBI Pathogen Detection Isolates Browser (https://www.ncbi.nlm.nih.gov/pathogens/isolates/). Visualizations with plasmid sequence alignment were made with the Interactive Tree of Life (51).
Epidemiological data management and statistics.
U.S. isolates had year of isolation, patient age, and patient sex information collected for analysis when available. Sexual orientation or sexual behavior data were not available for U.S. isolates. Patients were classified into one of three demographic groups: adult males are patients who were of male sex and at least 18 years of age, adult females are patients who were of female sex and at least 18 years of age, and children are all patients under 18 years of age. The proportion of adult males was used as an indicator to suggest MSM transmission. Previous work used gender distributions to identify excess cases of shigellosis among men, which can suggest MSM transmission (29, 30). For determination of the significance of patient age by isolate pMHMC-004 carriage, a t test was used. A χ2 test was used to evaluate the significance of demographic group between patient isolate pMHMC-004 carriage levels. t test and χ2 tests were performed using SciPy (52).
Data accessibility.
The sequences determined in the course of this work have been deposited in NCBI under BioSample accession numbers SAMN11948688 (S. sonnei 726), SAMN07450846 (S. sonnei SBJ-12001), SAMN05510450 (S. sonnei SBJ-13001), SAMN07662568 (S. sonnei SBJ-9961), SAMN07663113 (S. sonnei SBJ-9962).
ACKNOWLEDGMENTS
Analyses were supported by P30 DK034854 and a capital grant from the Massachusetts Life Sciences Center to the Massachusetts Host-Microbiome Center. The work of J.W. was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health. Kristen Hysell was supported by an NIH T32 grant (T32AI007433).
We acknowledge and thank the PulseNet participating laboratories whose data were used for the creation of this publication.
Jay Worley performed biological experiments, performed bioinformatic and statistical analyses, crafted figures, and wrote the manuscript. Kiran Javkar, Errol Strain, and Mihai Pop performed bioinformatic analyses and provided computational support. Maria Hoffmann and Marc Allard sequenced the bacteria and the plasmids. Kristen Hysell provided the case report. Amanda Garcia-Williams, Kaitlin Tagg, and Louise Francois Watkins provided epidemiological data and analysis. Louis Francois Watkins and Amanda Garcia-Williams provided public health information and messaging. Sanjat Kanjilal provided case data and research guidance. Lynn Bry wrote and edited the manuscript and provided direction.
We have no competing interests to declare.
Footnotes
This article is a direct contribution from Marc Allard, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Kate Baker, University of Liverpool, and François-Xavier Weill, Institut Pasteur.
Citation Worley JN, Javkar K, Hoffmann M, Hysell K, Garcia-Williams A, Tagg K, Kanjilal S, Strain E, Pop M, Allard M, Francois Watkins L, Bry L. 2021. Genomic drivers of multidrug-resistant Shigella affecting vulnerable patient populations in the United States and abroad. mBio 12:e03188-20. https://doi.org/10.1128/mBio.03188-20.
REFERENCES
- 1.Dritz SK, Back AF. 1974. Letter: Shigella enteritis venereally transmitted. N Engl J Med 291:1194. [PubMed] [Google Scholar]
- 2.Bader M, Pedersen AH, Williams R, Spearman J, Anderson H. 1977. Venereal transmission of shigellosis in Seattle-King county. Sex Transm Dis 4:89–91. doi: 10.1097/00007435-197707000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Drusin LM, Genvert G, Topf-Olstein B, Levy-Zombek E. 1976. Shigellosis. Another sexually transmitted disease? Br J Vener Dis 52:348–350. doi: 10.1136/sti.52.5.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aragón TJ, Vugia DJ, Shallow S, Samuel MC, Reingold A, Angulo FJ, Bradford WZ. 2007. Case-control study of shigellosis in San Francisco: the role of sexual transmission and HIV infection. Clin Infect Dis 44:327–334. doi: 10.1086/510593. [DOI] [PubMed] [Google Scholar]
- 5.Bowen A, Eikmeier D, Talley P, Siston A, Smith S, Hurd J, Smith K, Leano F, Bicknese A, Norton JC, Campbell D, Centers for Disease Control and Prevention (CDC) . 2015. Notes from the field: outbreaks of Shigella sonnei infection with decreased susceptibility to azithromycin among men who have sex with men—Chicago and metropolitan Minneapolis-St. Paul, 2014. MMWR Morb Mortal Wkly Rep 64:597–598. [PMC free article] [PubMed] [Google Scholar]
- 6.Hines JZ, Pinsent T, Rees K, Vines J, Bowen A, Hurd J, Leman RF, Hedberg K. 2016. Notes from the field: shigellosis outbreak among men who have sex with men and homeless persons—Oregon, 2015-2016. MMWR Morb Mortal Wkly Rep 65:812–813. doi: 10.15585/mmwr.mm6531a5. [DOI] [PubMed] [Google Scholar]
- 7.Bowen A, Grass J, Bicknese A, Campbell D, Hurd J, Kirkcaldy RD. 2016. Elevated risk for antimicrobial drug–resistant Shigella infection among men who have sex with men, United States, 2011–2015. Emerg Infect Dis 22:1613–1616. doi: 10.3201/eid2209.160624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danila RN, Eikmeier DL, Robinson TJ, La Pointe A, DeVries AS. 2014. Two concurrent enteric disease outbreaks among men who have sex with men, Minneapolis-St Paul area. Clin Infect Dis 59:987–989. doi: 10.1093/cid/ciu478. [DOI] [PubMed] [Google Scholar]
- 9.Chang H-H, Cohen T, Grad YH, Hanage WP, O'Brien TF, Lipsitch M. 2015. Origin and proliferation of multiple-drug resistance in bacterial pathogens. Microbiol Mol Biol Rev 79:101–116. doi: 10.1128/MMBR.00039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 11.Murray K, Reddy V, Kornblum JS, Waechter H, Chicaiza LF, Rubinstein I, Balter S, Greene SK, Braunstein SL, Rakeman JL, Dentinger CM. 2017. Increasing antibiotic resistance in Shigella spp. from infected New York City residents, New York, USA. Emerg Infect Dis 23:332–335. doi: 10.3201/eid2302.161203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heiman KE, Karlsson M, Grass J, Howie B, Kirkcaldy RD, Mahon B, Brooks JT, Bowen A, Centers for Disease Control and Prevention (CDC) . 2014. Shigella with decreased susceptibility to azithromycin among men who have sex with men—United States, 2002–2013. MMWR Morb Mortal Wkly Rep 63:132–133. [PMC free article] [PubMed] [Google Scholar]
- 13.Klontz KC, Singh N. 2015. Treatment of drug-resistant Shigella infections. Expert Rev Anti Infect Ther 13:69–80. doi: 10.1586/14787210.2015.983902. [DOI] [PubMed] [Google Scholar]
- 14.CLSI. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed. Clinical and Laboratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 15.Kozyreva VK, Jospin G, Greninger AL, Watt JP, Eisen JA, Chaturvedi V. 2016. Recent outbreaks of shigellosis in California caused by two distinct populations of Shigella sonnei with either increased virulence or fluoroquinolone resistance. mSphere 1:e00344-16. doi: 10.1128/mSphere.00344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker KS, Dallman TJ, Field N, Childs T, Mitchell H, Day M, Weill F-X, Lefèvre S, Tourdjman M, Hughes G, Jenkins C, Thomson N. 2018. Horizontal antimicrobial resistance transfer drives epidemics of multiple Shigella species. Nat Commun 9:1462. doi: 10.1038/s41467-018-03949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker KS, Dallman TJ, Ashton PM, Day M, Hughes G, Crook PD, Gilbart VL, Zittermann S, Allen VG, Howden BP, Tomita T, Valcanis M, Harris SR, Connor TR, Sintchenko V, Howard P, Brown JD, Petty NK, Gouali M, Thanh DP, Keddy KH, Smith AM, Talukder KA, Faruque SM, Parkhill J, Baker S, Weill F-X, Jenkins C, Thomson NR. 2015. Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect Dis 15:913–921. doi: 10.1016/S1473-3099(15)00002-X. [DOI] [PubMed] [Google Scholar]
- 18.Ingle DJ, Easton M, Valcanis M, Seemann T, Kwong JC, Stephens N, Carter GP, Gonçalves da Silva A, Adamopoulos J, Baines SL, Holt KE, Chow EPF, Fairley CK, Chen MY, Kirk M, Howden BP, Williamson DA. 2019. Co-circulation of multidrug-resistant Shigella among men who have sex with men, Australia. Clin Infect Dis 69:1535–1544. doi: 10.1093/cid/ciz005. [DOI] [PubMed] [Google Scholar]
- 19.Yousfi K, Gaudreau C, Pilon PA, Lefebvre B, Walker M, Fournier É, Doualla Bell F, Martineau C, Longtin J, Bekal S. 2018. Genetic mechanisms behind the spread of reduced susceptibility to azithromycin in Shigella strains isolated from men who have sex with men in Québec, Canada. Antimicrob Agents Chemother 63:e01679-18. doi: 10.1128/AAC.01679-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bardsley M, Jenkins C, Mitchell HD, Mikhail AFW, Baker KS, Foster K, Hughes G, Dallman TJ. 2020. Persistent transmission of shigellosis in England Is associated with a recently emerged multidrug-resistant strain of Shigella sonnei. J Clin Microbiol 58:e01692-19. doi: 10.1128/JCM.01692-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allard MW, Strain E, Melka D, Bunning K, Musser SM, Brown EW, Timme R. 2016. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol 54:1975–1983. doi: 10.1128/JCM.00081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV, CDC PulseNet Task Force . 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis 7:382–389. doi: 10.3201/eid0703.017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker KS, Dallman TJ, Field N, Childs T, Mitchell H, Day M, Weill F-X, Lefèvre S, Tourdjman M, Hughes G, Jenkins C, Thomson N. 2018. Genomic epidemiology of Shigella in the United Kingdom shows transmission of pathogen sublineages and determinants of antimicrobial resistance. Sci Rep 8:7389. doi: 10.1038/s41598-018-25764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F, Yang J, Zhang X, Chen L, Jiang Y, Yan Y, Tang X, Wang J, Xiong Z, Dong J, Xue Y, Zhu Y, Xu X, Sun L, Chen S, Nie H, Peng J, Xu J, Wang Y, Yuan Z, Wen Y, Yao Z, Shen Y, Qiang B, Hou Y, Yu J, Jin Q. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res 33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramírez MS, Piñeiro S, Group AIS, Centrón D, Argentinian Integron Study Group . 2010. Novel insights about class 2 integrons from experimental and genomic epidemiology. Antimicrob Agents Chemother 54:699–706. doi: 10.1128/AAC.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almendros C, Mojica FJM, Díez-Villaseñor C, Guzmán NM, García-Martínez J. 2014. CRISPR-Cas functional module exchange in Escherichia coli. mBio 5:e00767-13. doi: 10.1128/mBio.00767-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zdziarski J, Brzuszkiewicz E, Wullt B, Liesegang H, Biran D, Voigt B, Grönberg-Hernandez J, Ragnarsdottir B, Hecker M, Ron EZ, Daniel R, Gottschalk G, Hacker J, Svanborg C, Dobrindt U. 2010. Host imprints on bacterial genomes—rapid, divergent evolution in individual patients. PLoS Pathog 6:e1001078. doi: 10.1371/journal.ppat.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miquel S, Peyretaillade E, Claret L, de Vallée A, Dossat C, Vacherie B, Zineb EH, Segurens B, Barbe V, Sauvanet P, Neut C, Colombel J-F, Medigue C, Mojica FJM, Peyret P, Bonnet R, Darfeuille-Michaud A. 2010. Complete genome sequence of Crohn’s disease-associated adherent-invasive E. coli strain LF82. PLoS One 5:e12714. doi: 10.1371/journal.pone.0012714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beltrami JF, Shouse RL, Blake PA. 2005. Trends in infectious diseases and the male to female ratio: possible clues to changes in behavior among men who have sex with men. AIDS Educ Prev 17:49–59. doi: 10.1521/aeap.2005.17.Supplement_B.49. [DOI] [PubMed] [Google Scholar]
- 30.Mook P, Gardiner D, Kanagarajah S, Kerac M, Hughes G, Field N, McCarthy N, Rawlings C, Simms I, Lane C, Crook PD. 2018. Use of gender distribution in routine surveillance data to detect potential transmission of gastrointestinal infections among men who have sex with men in England. Epidemiol Infect 146:1468–1477. doi: 10.1017/S0950268818001681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker S, The HC. 2018. Recent insights into Shigella. Curr Opin Infect Dis 31:449–454. doi: 10.1097/QCO.0000000000000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman M, Shoma S, Rashid H, Siddique AK, Nair GB, Sack DA. 2004. Extended-spectrum β-lactamase-mediated third-generation cephalosporin resistance in Shigella isolates in Bangladesh. J Antimicrob Chemother 54:846–847. doi: 10.1093/jac/dkh413. [DOI] [PubMed] [Google Scholar]
- 33.Taneja N, Mewara A, Kumar A, Verma G, Sharma M. 2012. Cephalosporin-resistant Shigella flexneri over 9 years (2001-09) in India. J Antimicrob Chemother 67:1347–1353. doi: 10.1093/jac/dks061. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Luo Y, Li J, Lin L, Ma Y, Hu C, Jin S, Ran L, Cui S. 2011. Wide dissemination of multidrug-resistant Shigella isolates in China. J Antimicrob Chemother 66:2527–2535. doi: 10.1093/jac/dkr341. [DOI] [PubMed] [Google Scholar]
- 35.Huang I-F, Chiu C-H, Wang M-H, Wu C-Y, Hsieh K-S, Chiou CC. 2005. Outbreak of dysentery associated with ceftriaxone-resistant Shigella sonnei: first report of plasmid-mediated CMY-2-type AmpC beta-lactamase resistance in S. sonnei. J Clin Microbiol 43:2608–2612. doi: 10.1128/JCM.43.6.2608-2612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pecora ND, Li N, Allard M, Li C, Albano E, Delaney M, Dubois A, Onderdonk AB, Bry L. 2015. Genomically informed surveillance for carbapenem-resistant Enterobacteriaceae in a health care system. mBio 6:e01030. doi: 10.1128/mBio.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nudel K, Zhao X, Basu S, Dong X, Hoffmann M, Feldgarden M, Allard M, Klompas M, Bry L. 2018. Genomics of Corynebacterium striatum, an emerging multi-drug resistant pathogen of immunocompromised patients. Clin Microbiol Infect 24:1016.e7–1016.e13. doi: 10.1016/j.cmi.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertani G 1951. Studies on lysogenesis I. J Bacteriol 62:293–300. doi: 10.1128/JB.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi K-H, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 40.CLSI. 2018. Performance standards for antimicrobial disk susceptibility tests, 13th ed. Clinical and Laboratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 41.CLSI. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed. Clinical and Laboratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 42.Pecora N, Zhao X, Nudel K, Hoffmann M, Li N, Onderdonk AB, Yokoe D, Brown E, Allard M, Bry L. 2018. Diverse vectors and mechanisms spread New Delhi metallo-β-lactamases among carbapenem-resistant Enterobacteriaceae in the greater Boston area. Antimicrob Agents Chemother 63:e02040-18. doi: 10.1128/AAC.02040-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Xie Y, Liu M, Tai C, Sun J, Deng Z, Ou H-Y. 2018. oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res 46:W229–W234. doi: 10.1093/nar/gky352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. 2010. Interactive microbial genome visualization with GView. Bioinformatics 26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0 Contributors . 2020. SciPy 1.0–fundamental algorithms for scientific computing in Python. Nat Methods 17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Twelve plasmids of S. sonnei SBJ-9962. Genes and genetic features are labeled as directional arrows and boxes, respectively, in the inner circle for selected gene categories. Only pMHMC-003 and pMHMC-004 encode a putative T4SS, while those plus pMHMC-006, pMHMC-009, pMHMC-011, and pMHMC-012 encode a known origin of transfer. Two plasmids carry known AMR genes, pMHMC-004 and pMHMC-012. Most plasmids encoded a typical replicon, with the larger plasmids having Inc type replicons and the smaller plasmids having Col type replicons. Download FIG S1, PDF file, 0.5 MB (479.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Antibiotic susceptibility testing on strains carrying plasmids pMHMC-004 and pMHMC-012. Antibiotic resistance testing results for SBJ-9961, ATCC 25931, ATCC 25931 pMHMC-004, and ATCC 25931 pMHMC-012. Symbols: *, intermediate range for resistance phenotypes, determined by CLSI; †, intermediate; ‡, resistant. These tests were done for comprehensive antimicrobial resistance testing. Not all drugs listed should be considered for treatment of shigellosis. Download Table S1, DOCX file, 0.02 MB (16.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
pMHMC-004 carriage in strains of Shigella and Escherichia coli. Prevalence of pKSR100 and pMHMC-004 across S. sonnei and S. flexneri genomes in NCBI. Download Table S2, DOCX file, 0.01 MB (13.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Large SNP-defined groups of S. sonnei and S. flexneri intercontinental transmission and plasmid acquisition and loss events. pMHMC-004 and pKSR100 presence is indicated in the exterior two rings. Total plasmid coverage is shown by a gradient in the first ring outside the dendrogram. Country of origin is shown for the United States, United Kingdom, and Australia. Other countries of origin and those without specified origins are grouped in a fourth category. Download FIG S2, PDF file, 1.1 MB (1.1MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
SNP cluster PDS000033428.133 has highly variable AMR gene content driven partially by ISs in family IS6. The rings, from interior to exterior, represent pMHMC-004 coverage, pMHMC-004 presence, and then, in larger groups of rings, pMHMC-004 AMR gene content and pMHMC-012 AMR gene content. Clades of interest within this SNP cluster are color coded directly on the tree and are the same as those in Fig. 6. Download FIG S3, PDF file, 0.7 MB (686.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Isolates used in this study. A table of isolates, NCBI genome accession numbers, and isolate information. Strains without a reference originate from this study. Download Table S3, DOCX file, 0.02 MB (20.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
NCBI accession numbers and plasmid presence data for strains analyzed in this study. Download Data Set S1, XLSX file, 0.7 MB (216.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Electroporation and transconjugation experiments reveal a limited host range for pMHMC-004. Individual experiments are recorded on separate sheets, and a comparison is provided on the Summary sheet. Download Data Set S2, XLSX file, 0.7 MB (32.8KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
SNP clusters are variable in their carriage of pMHMC-004 and pKSR-100. The data set contains dendrograms similar to those shown in Fig. S2 but for all clusters. Download Data Set S3, PDF file, 0.7 MB (2.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Demographic information, collection time, and antimicrobial resistance plasmid presence for U.S. Shigella isolates across SNP clusters. The data set contains dendrograms similar to those in Fig. 4 but for all clusters and without named clades. Download Data Set S4, PDF file, 0.7 MB (3.4MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The sequences determined in the course of this work have been deposited in NCBI under BioSample accession numbers SAMN11948688 (S. sonnei 726), SAMN07450846 (S. sonnei SBJ-12001), SAMN05510450 (S. sonnei SBJ-13001), SAMN07662568 (S. sonnei SBJ-9961), SAMN07663113 (S. sonnei SBJ-9962).