In this study, Ito et al. sought to understand the precise roles of MED1, and its various domains, at various stages of adipogenesis and in adipose tissue. Using multiple genetic approaches to assess requirements for MED1 in adipocyte formation and function in mice, they show that MED1 is indeed essential for the differentiation and/or function of both brown and white adipocytes, as its absence in these cells leads to, respectively, defective brown fat function and lipodystrophy.
Keywords: adipogenesis, MED1, Mediator complex, transcriptional regulation, coactivator, development, embryonic stem cell, lipodystrophy, thermogenesis
Abstract
The MED1 subunit has been shown to mediate ligand-dependent binding of the Mediator coactivator complex to multiple nuclear receptors, including the adipogenic PPARγ, and to play an essential role in ectopic PPARγ-induced adipogenesis of mouse embryonic fibroblasts. However, the precise roles of MED1, and its various domains, at various stages of adipogenesis and in adipose tissue have been unclear. Here, after establishing requirements for MED1, including specific domains, for differentiation of 3T3L1 cells and both primary white and brown preadipocytes, we used multiple genetic approaches to assess requirements for MED1 in adipocyte formation, maintenance, and function in mice. We show that MED1 is indeed essential for the differentiation and/or function of both brown and white adipocytes, as its absence in these cells leads to, respectively, defective brown fat function and lipodystrophy. This work establishes MED1 as an essential transcriptional coactivator that ensures homeostatic functions of adipocytes.
Adipose tissues play critical roles in energy homeostasis. The major role of white adipose tissue (WAT) is to store energy in the form of lipids, whereas brown adipose tissue (BAT) dissipates energy as heat and contributes to thermal regulation upon environmental challenges. Understanding the formation and function of these tissues is of critical importance as they play a fundamental role in metabolic diseases such as obesity and type II diabetes.
Brown adipocytes in the interscapular region originate from Myf5-expressing dermomyotomes, a population of cells that also gives rise to the skeletal muscle and dermis (Seale et al. 2008; Sanchez-Gurmaches and Guertin 2014; Wang et al. 2014). In contrast, the majority of white adipocytes originate from Myf5-negative mesodermal cell populations with different molecular characteristics depending on the depot to which they contribute (e.g., Prx1+ cells for subcutaneous adipocytes and WT1+ for a subpopulation of visceral adipocytes) (Chau et al. 2014; Sanchez-Gurmaches et al. 2015). Notably, one common essential transcription factor for all adipocytes is the peroxisome proliferator-activated receptor γ (PPARγ), which specifies the mesodermal precursors to the adipose fate and activates genes responsible for the function of mature adipocytes (e.g., Adiponectin, Fabp4, etc.).
PPARγ belongs to the nuclear receptor (NR) superfamily and is considered the master regulator of adipogenesis (Tontonoz and Spiegelman 2008). PPARγ exists in two distinct isoforms, γ1 and γ2, with the latter showing an adipocyte-specific expression pattern and a higher capacity for inducing adipogenesis (Mueller et al. 2002). Ectopic expression of PPARγ can convert nonadipogenic cells into lipid droplet-containing adipocytes in culture (Tontonoz et al. 1994b). PPARγ is essential for the formation and function of all adipose tissues, as both systemic- and adipocyte-specific loss of PPARγ results in severe postnatal lipoatrophy leading to metabolic disorders such as hepatic steatosis and insulin resistance (Barak et al. 1999; Rosen et al. 1999; Duan et al. 2007; Wang et al. 2013). Therefore, understanding the mechanisms underlying the expression and function of PPARγ is of critical importance for understanding adipose tissue physiology and how its alterations result in metabolic disorders.
DNA-binding transcription factors (TFs), including PPARγ, act in conjunction with transcriptional cofactors (coactivators and corepressors) to exert their functions in transcription. Coactivators include those that work to modify the chromatin structure through histone modifications and remodeling and those that function directly through the general transcriptional machinery (e.g., Mediator complex and TFIID) (Roeder 2005). While several coactivators have been demonstrated to bind to and function with PPARγ (Chen and Roeder 2011; Seale 2015; Gulyaeva et al. 2019; Lee et al. 2019), one key issue that remains to be addressed for many of them is whether they actually play essential roles in adipocyte differentiation and function. The transcriptional control of adipogenesis has been extensively studied using cell culture models (Green and Meuth 1974; Rosen and MacDougald 2006) that led to the elucidation of numerous activators (Cao et al. 1991; Tontonoz et al. 1994a; Kim and Spiegelman 1996), as well as their functions on specific genes (Wu et al. 1996; Siersbæk et al. 2011). However, increasing evidence shows that many of these factors are dispensable for adipogenesis in vivo (Shimano et al. 1997; Park and Ge 2017; Park et al. 2017), most likely due to the relatively artificial conditions used in culture and also to alternative mechanisms that include the action of tissue- or depot-selective factors (Rajakumari et al. 2013). Hence, it is crucial to examine the roles of factors both in culture, where molecular mechanisms are more easily elucidated, and in vivo.
The Mediator, of special interest here, is a large 30-subunit coactivator complex with a modular structure (comprised of head, middle, tail, and a dissociable kinase module), and physically and functionally links DNA-binding TFs to the general transcriptional machinery (RNA polymerase II and the general initiation factors) (Malik and Roeder 2010). MED1 was biochemically characterized as the Mediator subunit responsible for the strong ligand-dependent interaction between various nuclear receptors (including the adipogenic PPARγ) and the Mediator complex (Fondell et al. 1996; Zhu et al. 1997; Yuan et al. 1998; Chen and Roeder 2011). The two LxxLL motifs (NR boxes) present in MED1 are responsible for the strong ligand-dependent interaction and therefore for its strong transcriptional activation (Malik et al. 2004). Supporting this finding, and while generally dispensable for cell viability and general transcription (Ito et al. 2000), MED1 was found to be functionally required for ectopic PPARγ-mediated conversion of mouse embryonic fibroblasts (MEFs) into adipocytes (Ge et al. 2002), as well as forskolin- and ectopic PRDM16-mediated induction of BAT-specific genes in these cells (Iida et al. 2015). MED1 was thus proposed to play key roles for the formation and function of both BAT and WAT. Surprisingly, however, the NR boxes of MED1 appeared dispensable for most NR functions in a physiological setting (including adipogenesis) (Jiang et al. 2010) and also for the ectopic PPARγ-mediated adipogenic conversion of MEFs (Ge et al. 2008). Therefore, the requirement of MED1-mediated transcriptional control for adipose tissue formation and function in vivo, especially in relation to its presumed function as a critical PPARγ coactivator, has remained unclear.
Regarding the molecular function of MED1, recent reports proposed a model in which MED1 contributes to the formation of coactivator condensates in mouse embryonic stem cells (mESCs) through its phase-separation property within the disordered domain present in the C terminus of MED1 (Boija et al. 2018; Sabari et al. 2018). Although the relevance of this phenomenon has not been fully examined for its role in gene regulation, the results have raised the possibility of either an unanticipated role of MED1 in general transcription or its unique function specific to mESCs, both of which appear to challenge the previous finding that Med1 knockout embryos develop far beyond the preimplantation stages without obvious defects (Ito et al. 2000).
In this study, we addressed the requirement of MED1 for (1) the derivation and maintenance of mouse ESCs and (2) the formation and function of different mouse adipose tissues. Our data provide clear evidence that MED1 is not required for survival of ESCs or their proliferation. However, MED1 fulfills essential roles during the terminal phase of differentiation and hence for the function of mature adipose tissues.
Results
MED1 is dispensable for the derivation and maintenance of embryonic stem cells
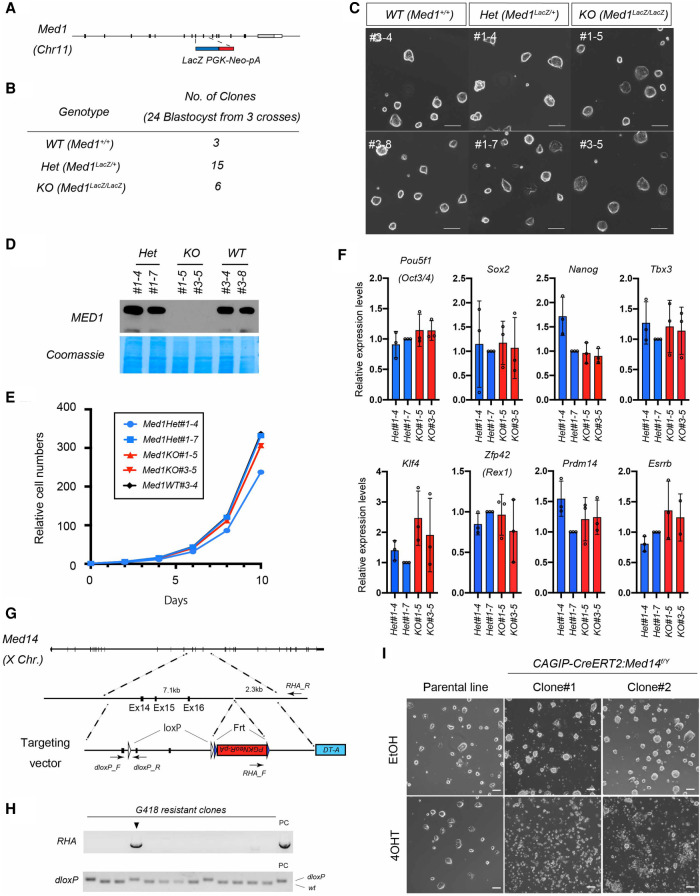
We have previously reported that Med1-null embryos develop relatively normally until embryonic days 10–11 (E10–E11) (Ito et al. 2000), which is beyond the preimplantation stage at which embryonic stem cells are derived. This allowed us to test whether embryonic stem cells can be derived from Med1-null blastocysts and whether MED1 is required for the derivation and maintenance of ESCs (Fig. 1A,B). To this end, we recovered blastocysts from Med1+/− heterozygous mice crosses and cultured them in vitro. Surprisingly, Med1-KO ES cells showed no obvious defects in morphology, growth rate, or expression of pluripotency-related transcription factors compared with control ESCs (derived from littermate blastocysts), at least when cultured in the presence of LIF and 2i (Fig. 1C–F). To confirm that this dispensability is limited to MED1, but not the Mediator, we also targeted Med14, encoding a central backbone subunit of the Mediator that physically tethers the head, middle, and tail modules (Cevher et al. 2014; Tsai et al. 2014), and generated conditional knockout ESCs. Subsequent to the gene targeting, a CreERT2-expressing construct was transduced to generate inducible Med14 knockout clones. We confirmed that MED14 is indeed essential for the maintenance of ESCs, as its conditional deletion resulted in complete loss of ESCs within 2 d of culture (Fig. 1G–I). These data strongly indicate that MED1, but not the Mediator as such, is dispensable for maintenance of ESCs in culture. This supports our previous view that the requirement of MED1 is context-dependent and is likely to function in an activator-specific manner rather than as an essential general component of the transcription machinery.
Figure 1.
MED1 is dispensable for the derivation and maintenance of mouse ESCs. (A) Design of Med1 conventional knockout allele. In the conventional knockout allele, Exon 9–10 is replaced by a LacZ-PGKNeo-pA cassette. (B) Derivation frequency of ESC lines from each Med1 genotype. Twenty-four blastocysts (from three Med1+− crosses) were individually plated in culture. (C) Morphology of established ESC lines in culture. Scale bars, 100 μm. (D) Immunoblot of ESC lysates showing the absence of MED1 only in the Med1−/− ESC clones. Two male lines for each genotype were used. (E) Cell growth curve of ESC lines in culture. Cells were counted and passaged every 2 d, and the numbers were recorded for 10 d. (F) Gene expression analysis by qRT-PCR for transcription factors involved in the pluripotency gene regulatory network (mean ± SD, n = 3 each, t-test all not significant). (G) Gene targeting strategy of the Med14 floxed allele. (dloxP) Distal loxP, (RHA) right homology arm. Short arrows indicate primers designed to detect correct homologous recombination by genomic PCR. (H) Validation of homologous recombination by genomic PCR flanking right homology arm and distal loxP site. (I) Morphology of Med14f/Y ESC lines carrying the CAGIPCreERT2 transgene treated either with vehicle (EtOH) or 4OHT for 48 h. All the CAGIPCreERT2:Med14f/Y ESC clones, but not the parental ESCs, treated with 4OHT died within 48 h. Scale bars, 100 μm.
Comprehensive analysis of the subunit composition of MED1-deficient Mediator complex
We previously demonstrated that the basal function of the Mediator complex remains active in the absence of MED1 and does not lead to the loss of other tested subunits as individually examined by immunoblotting (Malik et al. 2004). In order to confirm more rigorously that loss of MED1 does not dislodge any subunits from the complex, we examined the subunit composition of the Mediator complex in the absence or presence of MED1 by mass spectrometry. In order to obtain MED1-deficient Mediator, we first generated HEK293 cells solely expressing an endogenously N-terminal FLAG-tagged Mediator core subunit MED10 (Supplemental Fig. S1A–D) and then knocked out MED1 in this cell line (Supplemental Fig. S1E–G) using CRISPR/Cas9 (Ran et al. 2013). We also generated HEK293 cell lines in which the endogenous MED1 subunit is FLAG-V5 epitope-tagged (either at the N or the C terminus) (Supplemental Fig. S1H–K). Immunoprecipitation of the Mediator complex through MED1 assures the presence of MED1 within the complex, thus bypassing the concern that MED1 might exist in a subpopulation of the Mediator complex (Zhang et al. 2005). Mediator complexes were purified from nuclear extracts prepared from the engineered cell lines, and their subunit compositions were analyzed by mass spectrometry. We confirmed that, except for the expected loss of MED1 in MED1-deficient Mediator, the subunit composition and stoichiometry were essentially unchanged between MED1-containing and MED1-deficient Mediator complexes (Supplemental Fig. S1L). Although we cannot rule out a conformational change of the Mediator complex in the absence of MED1, this data allows us to attribute the phenotypes resulting from MED1-deletion to the absence of MED1 itself, rather than the dislodgement of other subunits from the complex.
MED1 is required for adipogenesis in cultured cells
Since MED1 seems not to be required for general transcription in cell culture systems, we next asked whether this subunit is required for specific processes and re-examined the MED1 requirement for adipogenesis in cell culture. Our previous work used nonadipogenic immortalized mouse embryonic fibroblasts derived from Med1-null embryos and ectopically expressed PPARγ2 to force differentiation (Ge et al. 2002, 2008). In order to examine the MED1 requirement in an adipogenic context, we first chose to use adipogenic 3T3-L1 fibroblasts due to their well-characterized molecular features (Siersbæk et al. 2011, 2014). In this classical system, adipogenesis relies on the induction and activation of transcription factors such as C/EBPβ, C/EBPδ, and glucocorticoid receptor (GR) by the use of artificial hormonal cocktails (superphysiological dose of steroid, insulin, and the nonphysiological agent IBMX) added to cell culture medium to initiate the expression of C/EBPα and PPARγ (Farmer 2006). These factors ultimately determine the adipogenic fate within the first 2 d of differentiation (Supplemental Fig. S2A). While C/EBPα and PPARγ induce genes responsible for adipocyte function, they also show a positive cross regulation that creates a feed-forward loop and reinforces a stable adipocyte phenotype (Wu et al. 1999; Rosen et al. 2002). This system, although nonphysiological, has been classically used as a reproducible method to achieve an adipocyte-like state in culture that enables more amenable mechanistic studies of adipogenesis. Addition of rosiglitazone (Rosi), a potent synthetic full agonist specific for PPARγ, can efficiently enhance the differentiation process and, conversely, can be used to confirm the presence and functionality of PPARγ.
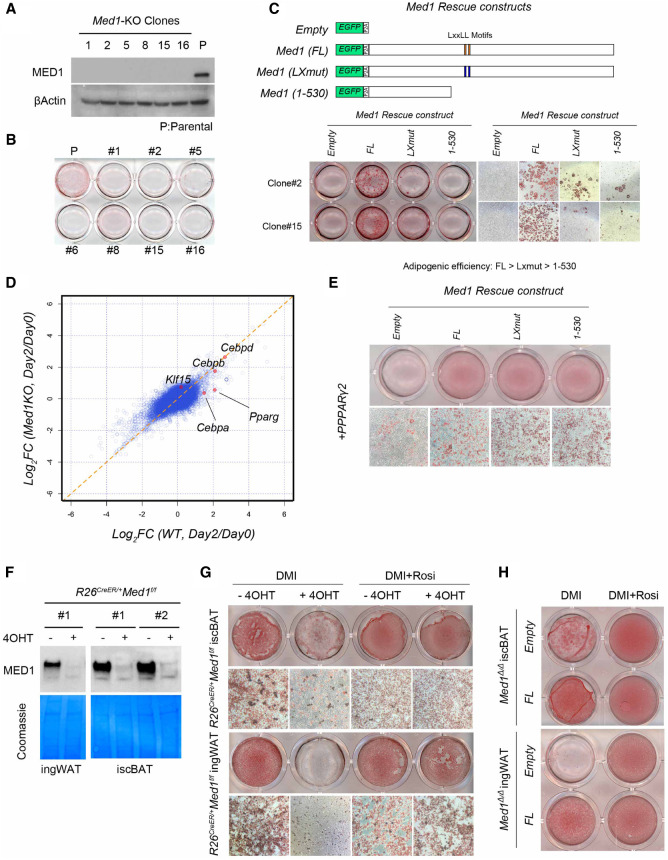
Med1-deficient 3T3-L1 clones were generated via CRISPR/Cas9 (Fig. 2A; Supplemental Fig. S2B), and their ability to undergo adipogenesis was assessed on day 6 by Oil Red O staining of lipids and neutral triglycerides (Proescher 1927). We found that none of the Med1-deficient clones were able to accumulate lipid droplets, even in the presence of Rosi (Fig. 2B). Reintroduction of intact MED1 rescued this adipogenesis defect in two independent clones even in the absence of Rosi, strongly confirming that this defect is MED1-dependent rather than a clonal defect (Fig. 2C). Interestingly, in this context (no Rosi), the MED1(LX) and C-terminal-deleted MED1 (1–530) mutants (Fig. 2C, top panel) were inefficient in inducing lipid droplet accumulation in the Med1-deficient 3T3-L1 cells by day 14 (Fig. 2C, bottom panel). In contrast, in the presence of Rosi, both WT and MED1(LXmut) forms of MED1 fully rescued the adipogenesis defect, whereas the MED1(1–530) form was still unable to do so (Supplemental Fig. S2C). Given that Rosi serves to activate PPARγ, the dependence of the responsiveness to Rosi on full length MED1 suggested that the early phase induction of PPARγ in 3T3-L1 cells relies, at least in part, on MED1 sequences beyond those present in the N-terminal (1–530) fragment. In this regard, a transcriptomic analysis in Med1-deficient 3T3-L1 cells at day 0 and day 2 of differentiation revealed defects in the induction of key transcription factors PPARγ and C/EBPα, whereas C/EBPβ and C/EBPδ were induced at comparable levels (Fig. 2D). Quantitative RT-PCR analysis confirmed this finding and, as expected, clones expressing MED1 or MED1(LXmut) showed higher levels of PPARγ (especially PPARγ2) compared with clones expressing MED1(1–530) or lacking MED1 entirely (Supplemental Fig. S2D). These data suggested that 3T3-L1 cells require MED1 to acquire an adipogenic (PPARγ-induced) fate, and that the N-terminal region of MED1 alone is insufficient for this process.
Figure 2.
MED1 is required for adipocyte differentiation in culture. (A) Immunoblot of lysates prepared from established 3T3-L1 clones, showing loss of MED1. β-Actin was used as a loading control. Parental 3T3-L1 cells were used as control. (B) Plate image of Oil Red O staining after 6 d of differentiation of parental and Med1-KO 3T3-L1 cells. None of the KO clones showed successful differentiation. (C) Lentiviral transduction of empty (eGFP only) vector or different forms of MED1 with the eGFP-P2A-construct. Note that only the full length restores efficient differentiation in the absence of Rosi. (D) Analysis of cDNA microarray data obtained from parental (WT) and pooled Med1-KO clones at day 0 and day 2 of differentiation. Ratio of expression levels at day 2 over day 0 is plotted for each condition. Classical adipogenic transcription factors are plotted in red dots. Note that Pparg and Cebpa are uninduced in KO conditions. (E) Ectopic PPARγ2 (introduced by retrovirus) rescue of Med1-KO 3T3-L1 clones with different forms of MED1. Note that efficient lipid accumulation does not occur in the absence of MED1. (F) Immunoblot of preadipocyte lysates. Efficient depletion of MED1 is seen only in 4OHT-treated (+) and not in vehicle-treated (−) cultures. Coomassie staining of corresponding membrane is shown as a loading control. (G) Oil Red O staining of differentiated brown and white preadipocytes pretreated with vehicle or 4OHT. (H) Oil Red O staining of differentiated brown and white preadipocytes pretreated with 4OHT and then transduced with lentivirus expressing empty (eGFP) or eGFP-P2A-MED1 constructs.
Given these observations, we then asked whether forced expression of PPARγ, which bypasses the initial induction phase, could rescue this adipogenesis defect in Med1-deficient 3T3-L1 cells. To this end, PPARγ2 was retrovirally expressed in Med1-deficient 3T3-L1 cells expressing different forms of MED1 in the absence of Rosi (Fig. 2E). Consistent with our previous results with Med1-null MEFs (Ge et al. 2008), ectopic expression of PPARγ2 failed to efficiently induce lipid droplet accumulation in the absence of MED1 but, in contrast, fully restored adipogenesis when coexpressed with WT MED1, MED1(LXmut), or MED1(1–530). These results indicate that, apart from promoting adipogenesis per se, abnormally high levels of ectopic PPARγ can also bypass otherwise normal requirements for C-terminal domains of MED1, including the NR boxes that mediate strong MED1 interactions with PPARγ upon ligand binding. In relation to NR box functions, we note that, whereas the MED1(LXmut) is inefficient relative to MED1 WT in facilitating adipogenesis in the absence of ectopic PPARγ (Fig. 2C), it nonetheless (like MED1 WT) promotes a considerable level of PPARγ2 induction in the early phase of differentiation under these conditions (Supplemental Fig. S2D). This observation, plus the above-described ability of ectopic PPARγ2 to efficiently promote adipogenesis in the absence of intact MED1 NR boxes (Fig. 2E), suggests that under classical conditions (no ectopic PPARγ expression), the NR boxes likely participate in the maintenance of endogenous PPARγ expression levels after the PPARγ-induction phase of 3T3-L1 differentiation.
Overall, these data suggest that MED1 is normally required to achieve efficient adipocyte differentiation in 3T3-L1 cells, in vitro, both before and after acquisition of the adipogenic fate (PPARγ and C/EBPα expressed state). These results further show that manipulation of PPARγ (expression levels or activity) can bypass some of the normal MED1 domain requirements in this in vitro model of adipogenesis.
To further examine the requirement of MED1 in a more physiological model of adipogenesis, we established immortalized brown and white preadipocyte cell lines from interscapular (isc) BAT or inguinal (ing) WAT, respectively, of newborn mice carrying a homozygous Med1-floxed allele and a Rosa-CreER allele (Med1f/f; R26CreER/+ mice). These cell lines retain features of their in vivo counterparts and have been reported to show differentiation cocktail responses similar to that observed for 3T3-L1 cells, although the effects of depot-selective factors are not fully understood (Morganstein et al. 2008; Siersbæk et al. 2012; Lai et al. 2017). Efficient depletion of MED1 after continuous exposure to 4-hydroxyl tamoxifen (4OHT) was confirmed (Fig. 2F), and adipogenic capability was assessed in the presence and absence of Rosi. In contrast to 3T3-L1 cells that completely fail to accumulate lipid droplets in the absence of MED1 and Rosi, the MED1-deficient brown preadipocyte cell lines showed modest accumulation of lipid droplets in the absence of Rosi, although to a drastically lower degree than the control cells (Fig. 2G), thereby confirming the requirement of MED1 for the optimal differentiation of these cells. On the other hand, the MED1-deficient white preadipocyte cell lines completely failed to accumulate lipid droplets in the absence of Rosi. Surprisingly, the observed defects in both cell lines were completely rescued by the addition of Rosi (Fig. 2G). Lentiviral transduction of MED1, but not the control lentiviral empty vectors, into 4OHT-treated cells allowed recovery of lipid droplet accumulation, demonstrating that the phenotypes observed were indeed MED1-dependent and were not caused by the adverse effects of 4OHT (Fig. 2H). These data suggested (1) that the degree of the MED1 requirement for the lipid accumulation phenotype differs between different types of preadipocytes; (2) that PPARγ is expressed in the absence of MED1 in cell lines derived from adipose tissues, as indicated by their responsiveness to Rosi; and (3) that hyperactivation of PPARγ by the potent, synthetic full agonist Rosi can bypass the normal MED1 requirement in these cell lines. Indeed, like these preadipocytes, even 3T3-L1 cells overexpressing ectopic PPARγ2 in the absence of MED1 showed efficient lipid droplet accumulation when Rosi was added during differentiation (Supplemental Fig. S2E).
In summary, whereas the manipulation of PPARγ-levels (via overexpression) or PPARγ-activity (via Rosi-mediated hyperactivation) can promote MED1-independent adipogenesis, our cell-based analyses (1) have established a MED1 requirement for adipogenesis in both 3T3-L1 cells and brown/white preadipocytes under classical, in vitro, and adipogenic conditions (summarized in Supplemental Fig. S2F) and (2) provided clues about requirements for specific MED1 domains. These results have prompted further tests of the MED1 requirement for adipose tissue formation and function in vivo, as well as tests of its physiological relevance for whole body metabolism.
MED1 is not critical for the formation of BAT, but is required for BAT function in newborn mice
In order to examine the MED1 requirement for the formation and function of adipose tissues, we first focused on BAT. Brown adipogenesis occurs in the interscapular region of the embryo, originating from dermomyotomes expressing Myf5 at E10.5 (Wang et al. 2014). These cells adopt an adipogenic fate (PPARγ+) by E12.5 and subsequently acquire brown adipocyte features around E17.5 (Mayeuf-Louchart et al. 2019). We used the Myf5Cre knock-in mouse (Tallquist et al. 2000) and a Med1-floxed allele (Med1f/f) to delete Med1 in the Myf5+ lineage to assess the formation and function of BAT in the absence of MED1. The Myf5Cre conditional Med1 knockout (Myf5CreMed1f/f) mice were born at the expected Mendelian ratio but demonstrated severe growth retardation after birth (Supplemental Fig. S3A,B) and died perinatally around weaning age (Supplemental Fig. S3C). We speculate that the lethality phenotype was caused by defects either in skeletal muscles or in a minor region of the brain (Gensch et al. 2008), as brown fat deficiency itself does not lead to lethality in mice (Yoneshiro et al. 2019).
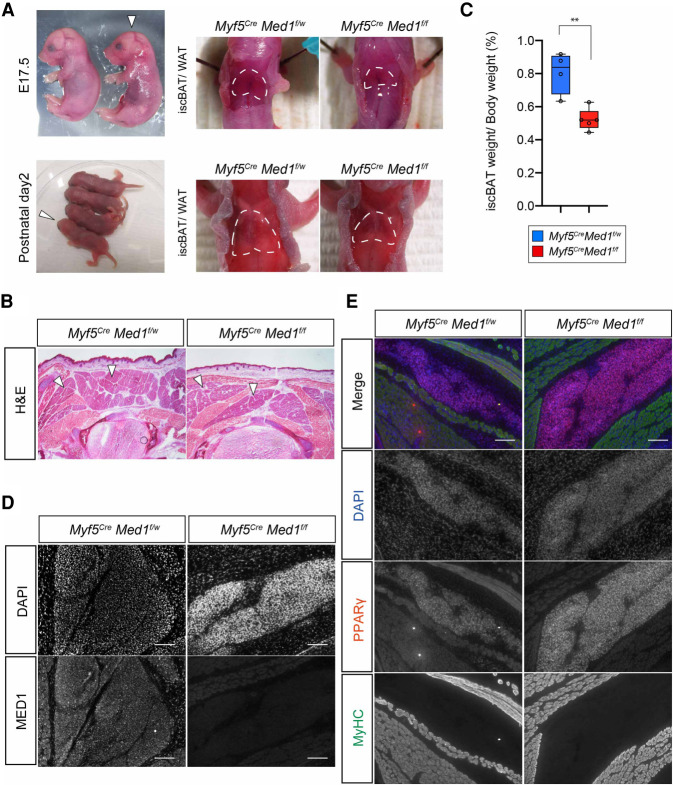
We therefore focused our analysis on the perinatal stages (E17.5 and P1) when Myf5CreMed1f/f animals were grossly indistinguishable from littermate controls (Fig. 3A). In these mice, the BAT was macroscopically visible, and histological examination confirmed the presence of BAT in the interscapular region (Fig. 3A,B), but with a reduction in mass compared with control littermates (Fig. 3C). Immunohistochemical analysis of Myf5CreMed1f/f embryos showed efficient loss of MED1 in BAT and skeletal muscle by E17.5 (Fig. 3D), whereas markers of adipocytes (PPARγ) and skeletal muscles (MyHC) were expressed at levels comparable with that of their littermate controls, indicating the successful formation of embryonic adipocytes and skeletal muscle in the absence of MED1 (Fig. 3E). These data suggest that cells of the Myf5+ dermomyotome can undergo fate determination into the (PPARγ+) adipocyte lineage in the absence of MED1 but require MED1 for optimal growth of the BAT. However, whether this is due to an intrinsic phenotype of MED1-deficient BAT, or due to extrinsic effects through MED1 deficient nonadipose cells, is unknown.
Figure 3.
Brown adipose tissue is formed in Myf5CreMed1f/f mice. (A) Macroscopic examination of BAT of Myf5CreMed1f/w and Myf5CreMed1f/f mice at perinatal (E17.5 and P1) stages. (Left panel) White arrows indicate the Myf5CreMed1f/f mice. BAT is present at the upper back of the Myf5CreMed1f/f mice. (B) H&E staining of thoracic transversal cryosection of E17.5 Myf5CreMed1f/w and Myf5CreMed1f/f mice. Arrowheads indicate the location of BAT. (C) Percentage of BAT weight over body weight of Myf5CreMed1f/w (n = 4) and Myf5CreMed1f/f (n = 5) mice at P1 (mean ± SD, unpaired t-test, P < 0.005). (D) Immunohistochemistry of MED1 in E17.5 BAT regions. Absence of MED1 in Myf5CreMed1f/f mice is shown. Scale bars, 100 μm. (E) Immunohistochemistry of PPARγ (BAT marker) and MyHC (skeletal muscle) in E17.5 embryos. Scale bars, 100 μm.
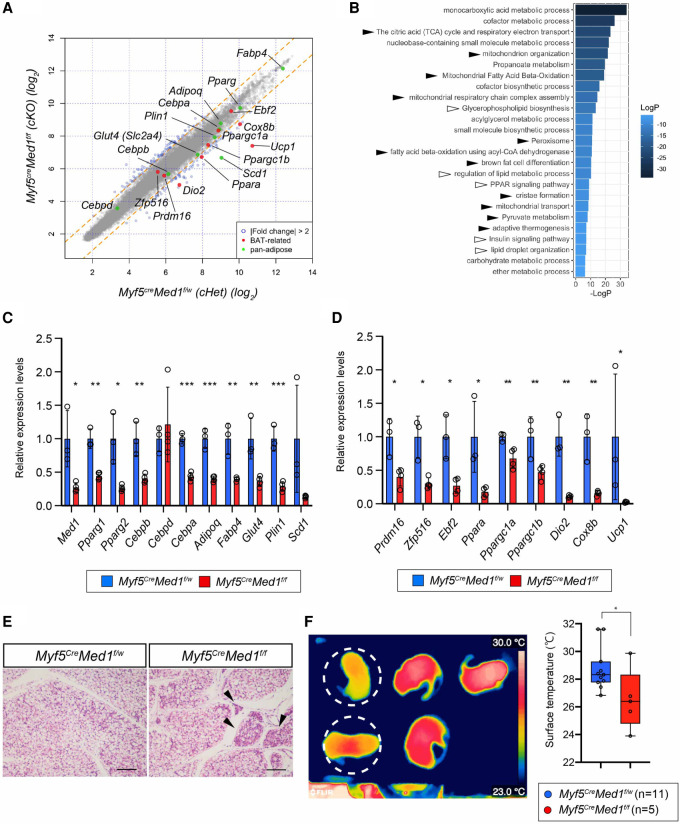
To molecularly characterize the BAT of Myf5CreMed1f/f embryos, we carried out a microarray-based gene expression analysis of BAT dissected from E17.5 male embryos (Myf5CreMed1f/f vs. Myf5CreMed1f/w). This analysis revealed a general decrease in pan-adipocyte gene expression. The most dramatically down-regulated genes in the Myf5CreMed1f/f BAT were those, such as Ucp1, Dio2, and Cox8b, known to be involved in the thermogenic activity of brown adipocytes (Fig. 4A). Gene ontology analysis of the down-regulated genes (greater than twofold) showed a strong enrichment of biological processes related to adipocyte function (such as lipid metabolism, PPAR-signaling) and brown fat functions (mitochondria functions, fatty acid oxidation, and thermogenesis) (Fig. 4B). RT-qPCR confirmed the depletion of Med1, as well as a decrease of various pan-adipocyte genes (i.e., Pparγ, C/ebpα, Adipoq, Plin1, Fabp4, etc.) (Fig. 4C) and a severe down-regulation of brown adipose-specific regulators and brown adipose functional genes (Fig. 4D). These data suggested that the BAT of Myf5CreMed1f/f mice is impaired in functions that are common to white and brown adipocytes (lipid accumulation) as well as those unique to BAT (energy expenditure and thermogenesis). Indeed, BAT of postnatal Myf5CreMed1f/f mice exhibited cells with less lipid droplet accumulation in newborn pups (Fig. 4E); and this phenotype became more pronounced during postnatal development (Supplemental Fig. S4). Although we could not rule out a systemic effect due to the growth retardation of the Myf5CreMed1f/f mice, we assume that this is a cell-intrinsic effect as brown preadipocyte cell lines lacking MED1 also presented a decreased lipid droplet accumulation phenotype upon differentiation (Fig. 2G).
Figure 4.
Characterization of MED1-deficient BAT at perinatal stages. (A) Microarray analysis of E17.5 iBAT from Myf5CreMed1f/w (n = 3) and Myf5CreMed1f/f (n = 4) male embryos. Array signals are plotted, and genes expressed differentially over twofold are plotted in blue. Genes common to all adipocytes are marked in green, BAT-specific genes are labeled in red. (B) Gene ontology enrichment analysis of genes down-regulated in Myf5CreMed1f/f mice using Metascape. Enriched GO terms (top 25) are displayed. White arrowheads correspond to pan-adipose tissue-related terms and black arrowheads correspond to BAT-related terms. (C) qRT-PCR validation of microarray results. Pan-adipose-related genes and Med1 (for confirmation) are presented. (D) qRT-PCR validation of BAT-related genes. In C and D, 36B4 (RPLP0) mRNA is used for an internal control. mean ± SD; unpaired t-test, (*) P < 0.05, (**) P < 0.005, (***) P < 0.0005. (E) H&E staining of P5 BAT from Myf5CreMed1f/w and Myf5CreMed1f/f mice. Scale bars, 100 μm. (F, left) Representative image taken by thermography camera of surface temperature of newborn (P4–P8) mice 10 min after isolation from the mother and nest. Pups circularized with white dashed lines are Myf5CreMed1f/f mice. (Right) Measurements of body surface temperature are summarized (mean ± SD; unpaired t-test). (*) P < 0.05.
It is believed that there is a high demand for BAT-mediated thermogenesis in newborn mice, especially when the pups are separated individually from the mother, which causes a drastic heat loss in the absence of fully grown hair and an unfavorable surface-to-volume ratio (Vinter et al. 1982; Cannon and Nedergaard 2004). Therefore, we examined the thermogenic capacity of BAT in Myf5CreMed1f/f mice by challenging the newborn pups to generate heat following separation from the mother and the nest. We found that, when isolated from their mother, the Myf5CreMed1f/f mice showed lower body surface temperature compared with their littermate controls (Fig. 4F). This strongly suggests that Myf5CreMed1f/f BAT has a defect in heat production that leads to whole-body hypothermia. Collectively, these data suggest a critical role for MED1 in the functional maturation of brown adipocytes during perinatal stages.
MED1 is required for BAT functions in adult mice
While the Myf5Cre model clearly indicates a functional defect in BAT in the absence of MED1, the postnatal growth defect and lethality prevented us from precisely addressing the function of MED1-deficient BAT during adulthood. Therefore, we generated Ucp1CreMed1f/f and AdipoqCreMed1f/f mice that lack MED1 either in differentiated brown (and potentially beige) adipocytes or in all adipocytes (both white and brown adipocytes), respectively. These mice were viable and fertile and allowed us to examine the role of MED1 in adult BAT.
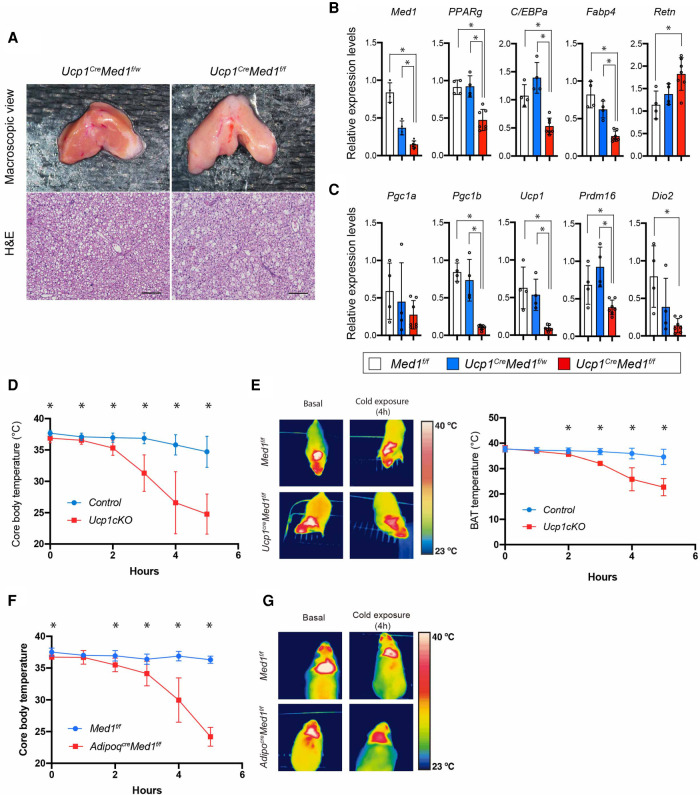
Ucp1CreMed1f/f mice did not show a significant difference in body weight, although there appeared to be a tendency toward an increase in body weight compared with control littermates in young female adults (Supplemental Fig. S5A). Histological examination of the Ucp1CreMed1f/f BAT showed less lipid droplet accumulation compared with WT animals (Fig. 5A), a feature that was also observed in postnatal BAT of Myf5CreMed1f/f mice (Supplemental Fig. S4). While the Ucp1CreMed1f/f animals showed a decrease in BAT weight, we observed an inverse increase in WAT weight (both subcutaneous and visceral WAT) and liver (Supplemental Fig. S5B), suggestive of a decrease in whole-body energy expenditure or an ectopic mobilization of lipids to non-BAT organs. Gene expression analysis of BAT under basal conditions showed a significant decrease in pan-adipocyte genes (Fig. 5B) and a severe down-regulation of BAT functional genes (Ucp1 and Dio2) specifically in the Ucp1CreMed1f/f mice (Fig. 5C). In order to examine the thermogenic capacity of the BAT in Ucp1CreMed1f/f adult mice, we challenged the animals with acute cold stress (22°C–4°C) in which BAT thermogenesis plays critical roles in maintaining the core body temperature. The Ucp1CreMed1f/f mice were cold-intolerant, as their core body temperature, as well as their local interscapular temperature, dropped rapidly after acute cold challenge in 4°C (Fig. 5D,E).
Figure 5.
MED1 is required for BAT function (thermogenesis) in adult mice. (A, top panel) Representative macroscopic images of BAT isolated from Ucp1CreMed1f/w and Ucp1CreMed1f/f mice. (Bottom panel) H&E staining of paraffin sections of BAT are shown. Scale bars, 100 μm. (B,C) qRT-PCR analysis of Med1 as well as pan-adipose gene (B) and BAT-related gene transcripts (C) in BAT isolated from Med1f/f (n = 4), Ucp1CreMed1f/w (n = 4), and Ucp1CreMed1f/f (n = 6) mice. Note that the majority of genes do not show a difference between Med1f/f and Ucp1CreMed1f/w mice. 36B4 mouse mRNA is used as an internal control (mean, ±SD; unpaired t-test) (*) P < 0.05. (D) Core body temperature changes in control and Ucp1CreMed1f/f mice upon acute cold challenge (room temperature to 4°C). Both Med1f/f and Ucp1CreMed1f/w are used as controls. Data from both males and females are combined and presented together. (E, left) Thermography image of BAT region of control and Ucp1CreMed1f/f mice before and after 4 h of cold challenge. (Right) Measurement of BAT temperature of control and Ucp1CreMed1f/f mice by wireless probe during the cold exposure. (F) Core body temperature changes of Med1f/f and AdipoqCreMed1f/f mice upon acute cold challenge (room temperature to 4°C). In D–F, mean, ±SD; unpaired t-test. (*) P < 0.05. (G) Thermographic images of BAT regions of Med1f/f and AdipoqCreMed1f/f mice before and after 4 h of cold challenge.
Consistent with the BAT phenotypes in Myf5CreMed1f/f mice and Ucp1CreMed1f/f mice, histological examination of postnatal BAT of AdipoqCreMed1f/f showed decreased lipid accumulation with animal growth (Supplemental Fig. S5C). In all the conditional knockout models, abnormally large-sized lipid-containing areas appeared in patches within the BAT. This phenomenon has also been observed in other models of adipocyte cell death as well as in lipodystrophy (Pajvani et al. 2005; Cautivo et al. 2016; Wang et al. 2018), and we consider that it is likely to be a consequence of coalescence of lipid droplets from spontaneously dying brown adipocytes. Adipocyte death is known to trigger an immune response, and immunohistochemical analysis of F4/80 antigen indeed suggested a dramatic increase in infiltration of macrophages (Supplemental Fig. S5D). We also performed microarray-based gene expression analysis of BAT isolated from AdipoqCreMed1f/f and control littermates (Supplemental Fig. S5E). Supporting the histology data, gene ontology analysis of the up-regulated genes in AdipoqCreMed1f/f BAT showed enrichment of genes related to the inflammatory response. On the other hand, and consistent with the Ucp1CreMed1f/f mice, AdipoqCreMed1f/f BAT showed down-regulation of pan-adipocyte genes as well as those involved in BAT functions (mitochondrial biogenesis, fatty acid oxidation, and thermogenesis). Finally, upon acute cold challenge, AdipoqcreMed1f/f mice demonstrated cold intolerance at lower BAT temperature compared with control littermates (Fig. 5F,G). Overall, these data suggest that MED1 is essential for BAT to maintain its function both at perinatal stages and in adults, with its absence leading to whole-body metabolic and thermogenic defects.
The role of MED1 in the formation of white adipose tissue
During the course of our analysis of the Myf5CreMed1f/f mice, we also observed that the amount of subcutaneous WAT in the upper back was significantly reduced in newborn pups (Supplemental Fig. S6A). As 40%–50% of isc/anterior subcutaneous (asc) WAT originates from Myf5+ cells (Sanchez-Gurmaches et al. 2012; Wang et al. 2014), there is a possibility of a strong MED1 requirement for either the formation or the expansion of a subpopulation of adipocytes in isc/asc WAT. However, as Myf5cre mice displayed growth defects after birth, we were unable to rule out potential systemic effects contributing to this phenotype. Moreover, as there are currently no ideal Cre driver mouse strains that specifically label the origin of the white preadipocytes while not affecting the other lineages, we were also unable to precisely determine the role of MED1 for formation of the 50%–60% of isc/asc white adipose tissue that is not of Myf5+ cell origin.
MED1 is required for the maintenance of adipose tissues in adult mice
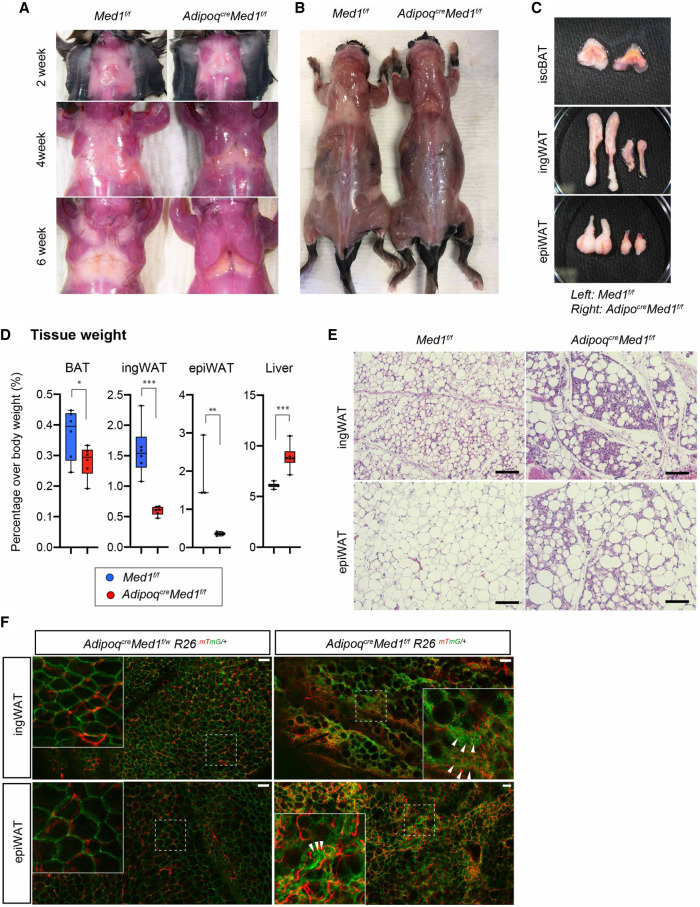
Finally, in order to address a MED1 requirement for WAT function, we took advantage of AdipoqCre mice to delete Med1 in committed preadipocytes (Hong et al. 2015). Macroscopic examination of AdipoqCreMed1f/f mice revealed a drastic loss of white adipose tissues around 3–4 wk of age (at the time of weaning) and a near complete absence of WAT by 6 wk of age (Fig. 6A; Supplemental Fig. S6B–E). This post-weaning lipodystrophy phenotype persisted throughout the lifetime of the knockout mice (Fig. 6B). In contrast, and consistent with previous BAT-specific MED1 deletion models (Fig. 5A), BAT was macroscopically visible with a relatively minor change compared with that of WAT (Fig. 6A–C). The AdipoqCre Med1f/f mice further showed a slight increase in adult body weight, similar to observations with other lipodystrophic mouse models (Supplemental Fig. S6E; Moitra et al. 1998), which correlates with the observed progression of WAT loss (Fig. 6A; Supplemental Fig. S6B–E). Supporting these macroscopic changes, WAT weight was severely reduced while the BAT weight showed a relatively mild (but significant) decrease in AdipoqCreMed1f/f mice. However, these mice also presented a significant increase in liver weight, a phenomenon that might partially explain the increase in whole-body weight and that also indicates systemic metabolic abnormalities (Fig. 6D).
Figure 6.
MED1 is required for WAT function and maintenance. (A) Representative macroscopic images of upper back regions (isc/ascWAT) of postnatal Med1f/f and AdipoqCreMed1f/f mice at 2, 4, and 6 wk of age. Results show the onset of lipodystrophy before 4 wk of age. (B) Macroscopic image of 10-wk-old Med1f/f and AdipoqCreMed1f/f mice with subcutaneous WAT exposed. (C) Representative images of dissected adipose tissues of Med1f/f and AdipoqCreMed1f/f male mice at 6 wk of age. (D) Percentage of tissue weight over body weight in 2- to 3-mo-old mice is presented. Severe decrease in WAT weight and a mild decrease in BAT weight is observed, while the liver shows a significant increase in weight. mean ± SD; unpaired t-test, (*) P < 0.05, (**) P < 0.005, (***) P < 0.0005. (E) H&E staining of WAT tissues from 10-wk-old Med1f/f and AdipoqCreMed1f/f male mice. Scale bars, 100 μm. (F) Whole-mount fluorescence confocal images of ingWAT and epiWAT from AdipoqCreMed1f/w and AdipoqCreMed1f/f male mice harboring a Rosa-mTmG allele. Magnified images of areas surrounded by dotted squares are shown in the top left corners. White arrowheads indicate examples of clustered mGFP-labeled non-lipid-containing cells. Scale bars, 100 µm.
Histological examination revealed that the WAT in the AdipoqCreMed1f/f mice consisted of non-lipid-containing cells along with large abnormally shaped lipid droplet-containing areas scattered within the tissue (Fig. 6E). Similar to the case of BAT, we consider that this is also a consequence of the coalescence of lipid droplets that result from spontaneous death of adipocytes. In order to assess the AdipoqCre Med1f/f WAT in more detail, we generated AdipoqCreMed1f/f mice harboring a Rosa26-mTmG reporter allele. In this mouse, the Adipoq+ lineage was labeled with a membrane-targeted green fluorescent protein (mGFP) while cells of the nonadipose lineage remained labeled with membrane-targeted tdTomato. Macroscopic examination confirmed the specific labeling of adipose tissue by mGFP and loss of mTomato in the same area (Supplemental Fig. S6G). Whole-mount imaging of WAT from these mice showed that the thick space between the large lipid-containing area consisted of clustered mGFP-expressing cells intercalated by mTomato-expressing nonadipose cells (Fig. 6F; Supplemental Fig. S6H). These data suggest that non-lipid-containing areas consisted of cells of the adipose lineage (MED1-depleted) as well as cells of the nonadipocyte lineage. As observed for the BAT tissues, we observed an increase in monocytes/macrophages (CD11b+ cells) (Supplemental Fig. S6I) as well as progression of fibrosis (Masson's Trichrome staining) (Supplemental Fig. S6J) within the WAT of AdipoqCreMed1f/f mice. These results confirm that MED1 is essential for the maintenance and proper function of the adult WAT.
As lipodystrophy is linked to metabolic disorders such as hepatic steatosis and insulin resistance in other organs, we also assessed the metabolic status of AdipoqCreMed1f/f animals. Consistent with a significant increase in weight (Fig. 6D), AdipoqCreMed1f/f livers were macroscopically larger and displayed a pale color (Supplemental Fig. S7A). Histological examination revealed ectopic fat deposition specifically in the livers of AdipoqCreMed1f/f mice, confirming the development of hepatic steatosis (Supplemental Fig. S7A). Next, we also examined the pancreas of the AdipoqCreMed1f/f mice. We observed a dramatic increase in the size of pancreatic islets (Supplemental Fig. S7B) that is indicative of β-cell compensation, a hallmark of insulin resistance. Indeed, these mice showed higher levels of circulating glucose, insulin, and triglycerides in the nonfasting state, indicating that the animals had defects in handling glucose as a consequence of lipid storage defects (Supplemental Fig. S7C–E). Finally, we tested the ability of the AdipoqCreMed1f/f mice to regulate glucose metabolism and also to respond to insulin by performing glucose and insulin tolerance tests. The AdipoqCreMed1f/f mice were glucose-intolerant (Supplemental Fig. S7F). Furthermore, compared with wild-type mice, AdipoqCreMed1f/f mice appeared to be unresponsive to insulin injections, suggesting that they were insulin-resistant (Supplemental Fig. S7G). These results suggest that MED1 plays critical roles in the homeostatic functions of adipose tissues in adults—such that its absence leads to severe lipodystrophy associated with hepatic steatosis and systemic glucose intolerance and insulin resistance.
Discussion
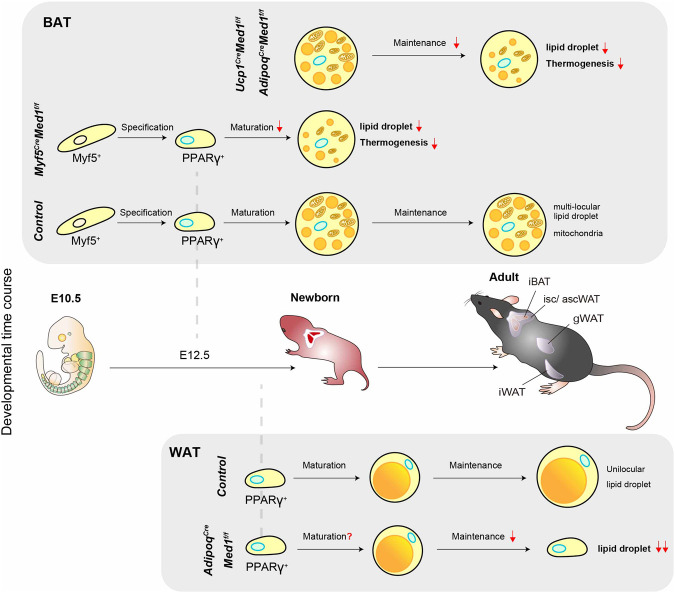
While it has been assumed from cell-based adipogenesis assays that MED1, a potent nuclear receptor coactivator, plays key roles in adipogenesis (Ge et al. 2002, 2008; Harms et al. 2015; Iida et al. 2015; LeBlanc et al. 2016), this has never been formally validated in vivo; and the contribution of relevant domains in MED1 are poorly understood. Here, we show (1) that, in the absence of ectopic PPARγ overexpression and/or hyperactivation by the synthetic agonist rosiglitazone, MED1 is required for adipogenesis in all the conventional in vitro cell culture models tested and shows variable mutant phenotypes and domain requirements at different stages of differentiation and between different assays; (2) that, in vivo, MED1 is not necessarily required for the formation of BAT, as might have been anticipated, but clearly is essential for the establishment and maintenance of BAT functions, such that its absence leads to lipid storage and thermogenesis defects; and (3) that, in vivo, MED1 is essential for the maintenance of adipocytes in WAT, as its loss in white adipocytes leads to severe lipodystrophy associated with systemic metabolic abnormalities postweaning. These in vivo findings are summarized in Figure 7. We also confirm that MED1 is not universally required for gene regulation, with MED1 depletion resulting in no obvious phenotypic abnormalities during ESC derivation and maintenance, whereas the Mediator complex itself (based on core subunit MED14 depletion) is indispensable for ESC maintenance. These results firmly establish the physiological importance of MED1, specifically as a unique and essential component of the adipocyte gene expression program.
Figure 7.
Model and summary of the phenotypes. Summary of the BAT and WAT phenotypes of different Med1 conditional knockout models used in this study. The top panel describes the effect of the loss of MED1 in BAT. The bottom panel describes the effect of the loss of MED1 in WAT.
MED1 requirements in cell culture models of adipogenesis
Using the conventional 3T3-L1 cell differentiation assay, we first demonstrated that MED1 is required at multiple steps of the adipogenic differentiation program. Further analyses revealed that C-terminal sequences beyond the conserved N terminus are required for optimal induction of the adipogenic transcription factors PPARγ and C/EBPα during the initial phase of differentiation, while the nuclear receptor-interacting NR boxes appear to play a role in the post-PPARγ induction phase. Given such a role for the NR boxes, attenuated activity of later stage-acting nuclear receptors such as PPARγ or LXRα (Seo et al. 2004), both of which function to control endogenous PPARγ levels, is one possible mechanism underlying the NR box mutant phenotype in 3T3-L1. The results from 3T3-L1 cells were surprising as our previous work had shown (1) that the N-terminal MED1 fragment (1–530) lacking the NR boxes is necessary and sufficient for ectopic (overexpressed) PPARγ2-mediated adipogenesis of MEFs (220) (Ge et al. 2008), as confirmed here for 3T3-L1 cells expressing ectopic PPARγ2, and (2) that the NR-boxes of MED1 appeared to be dispensable for the formation of adipose tissue in vivo (Jiang et al. 2010). However, given the now-established essentiality of MED1 for adipocyte gene expression in vivo (discussed below) and that cell-based assays do not necessarily recapitulate full aspects of normal adipogenesis (in vivo), future mouse genetic approaches must assess possible functions of MED1 C-terminal sequences and, especially, whether the classical model of robust PPARγ function through strong ligand- and MED1 NR box-dependent MED1/Mediator interactions plays a role in any aspect of adipocyte function in vivo (for example, under variable metabolic conditions).
Toward more physiological, albeit still artificial, cell-based assays with Med1-deficient brown and white preadipocyte cell lines that are widely believed to retain substantial in vivo features, we demonstrated that MED1 is critical for efficient lipid droplet accumulation, although brown preadipocytes showed a milder phenotype with MED1 loss than did white preadipocytes. However, these defects were rescued by the PPARγ-specific ligand Rosi, indicating that both of these cell types express levels of PPARγ at early phases of differentiation that are sufficient for Rosi-mediated adipogenesis even in the absence of MED1, which is consistent with the BAT developmental phenotypes observed in vivo (Fig. 3E). The difference between 3T3-L1 cells and brown or white preadipocytes in Rosi responsiveness (i.e., PPARγ expression) in the absence of MED1 likely reflects the presence of depot- or tissue-specific factors that allow MED1-independent PPARγ expression. The identity and mechanism of action of these factors require further study.
Finally, we showed that efficient adipogenesis could be achieved in Med1-KO 3T3-L1 cells by Rosi addition, as long as PPARγ2 was ectopically overexpressed. This is consistent with previous reports of MED1-independent PPARγ2 activation by Rosi in MEFs (Ge et al. 2008; Grøntved et al. 2010), although differentiation (lipid droplet accumulation) was not observed under these conditions in the previous studies. These results indicate differences in adipogenic competence between cell types used (Med1-KO 3T3-L1 vs. immortalized Med1-KO MEFs). The precise mechanisms of MED1-independent, Rosi-dependent PPARγ activation remain an interesting topic for further exploration, especially since PPARγ agonists that include the thiazolidinedione-class of drugs (such as Rosi) have been used as insulin-sensitizing drugs (Soccio et al. 2014).
Notably, these analyses have shown that MED1 is indeed required for efficient adipogenesis under conventional differentiation conditions in all the cellular models tested. However, the variable requirements for MED1 (and its domains) among different cell types (MEFs, 3T3-L1, and brown and white preadipocytes), differentiation stages (early vs. late), and culture conditions (with or without Rosi) further highlight the importance of our complementary in vivo genetic analysis for a better understanding of the role of MED1 in adipogenesis.
Essential roles of MED1 in adipose tissue formation and function in vivo
Using the Myf5Cre driver, we showed that MED1 is not necessarily required for the formation of a significant amount of BAT but is required for normal BAT mass and for functional maturation of BAT at perinatal stages. Besides the defective BAT phenotype, Myf5CreMed1f/f mice presented postnatal growth retardation and lethality. Since defects in BAT do not usually lead to lethality, the most likely explanation of this phenotype is defects in non-BAT tissues such as the skeletal muscle or a minor region of the brain that also originate from Myf5-expressing cells. The cause of this lethality will be addressed elsewhere. Deletion of Med1 in differentiated brown adipocytes through Ucp1Cre or AdipoqCre drivers also resulted in defective BAT, indicating that MED1 is also essential for the maintenance of gene expression programs in brown adipocytes in adult mice. The use of AdipoqCre also allowed us to address the role of MED1 in WAT. We showed that loss of MED1 in WAT leads to a striking loss of WAT around weaning, indicating a critical role of MED1 for the maintenance of white adipocytes. It is known that genetic regulation of DNA binding factors (especially C/EBPs and PPARγ) is essential for the adipocyte gene expression program, with either loss-of-function or dominant-negative alleles leading to lipodystrophy (Tanaka et al. 1997; Moitra et al. 1998; Linhart et al. 2001; Wang et al. 2013). One could speculate that ablation of coactivators required for the function of these activators would also phenocopy these models. However, heretofore, besides those cofactors (e.g., CBP/p300) that are universally required across various cell types (Lee et al. 2013; Namwanje et al. 2019), there has been only one very recent report of a mouse model with a specific coactivator mutation (MED19) that presents lipodystrophy (Dean et al. 2020) (discussed below). Although the MED1-null phenotypes were comparatively less severe than that for the PPARγ deficiency model (Wang et al. 2013), the post-weaning lipodystrophy in mice with adipocyte-specific loss of MED1 (AdipoqCreMed1f/f model) strongly implicates MED1 as an essential coactivator for one or more adipogenic transcriptional activators in vivo.
Although PPARγ is a prime candidate for the essential coactivation function of MED1, in view of the strong MED1 selectivity for nuclear receptors, the previous biochemical demonstrations of physical/functional PPARγ-MED1 interactions (discussed below), and the MED1 requirement for PPARγ-mediated adipogenesis in preadipocytes and 3T3L1 cells (in the absence of ectopic PPARγ or Rosi), it remains unknown whether MED1 is directly required for the coactivation of PPARγ in vivo or, if so, whether through an unknown natural ligand that facilitates a direct PPARγ-MED1 interaction or through the action of an intermediary factor. In this regard, our studies indicate potential MED1 N-terminal domain-dependent/NR box-independent functions based on the 3T3L1 adipogenesis studies (with ectopic PPARγ). However, since a direct interaction between the N-terminal domain of MED1 and PPARγ has never been demonstrated, the results lead to the prediction of a MED1 (N terminus) interacting factor(s) that either (1) binds DNA to act cooperatively with PPARγ, or (2) acts as a bridge between PPARγ and MED1. Indeed, there are DNA binding activators (e.g., C/EBPβ) and cofactors (e.g., CCAR1) that fulfill these criteria and also appear to be important during adipogenesis in culture (Li et al. 2008; Mizuta et al. 2014; Ou et al. 2014). In addition to such MED1-interacting factors that may be involved in the lipodystrophy phenotype, the PRDM16 and PGC-1α coactivators that are involved in thermogenic gene regulation in brown adipocytes are known to bind to and function through MED1 (Wallberg et al. 2003; Chen et al. 2009; Harms et al. 2015; Iida et al. 2015). The severe down-regulation of brown fat-specific thermogenic genes in MED1-deficient BAT is consistent with these reports and further validates the conclusions in a more physiological setting.
Future work on the identification of genes whose expression is directly under the control of MED1 should give us a more detailed understanding of the mechanisms underlying these phenotypes. In particular, a kinetic approach that allows the measurement of immediate transcriptional changes upon rapid (degron-based) removal of MED1 should be ideal to identify genes directly regulated by MED1. Of note, ChIP-seq studies of MED1 during adipogenesis in culture and in adult brown adipose tissues have been reported previously (Harms et al. 2015; Lai et al. 2017; Siersbæk et al. 2017). These studies led to the identification of clusters of enhancers whose target genes are known to dictate the identity of preadipocytes and adipocytes. However, as the mechanisms underlying MED1-dependent gene regulation may be operative at various steps of transcriptional activation (including, for example, Mediator recruitment or function at transcription initiation or post-initiation steps), these data sets need to be analyzed together with additional transcriptomic and biochemical data for the proper establishment of causal relationships to function and underlying mechanisms.
Another issue that deserves mention, especially in light of the variable requirements for MED1, relates to potential redundancy (for biological robustness) between MED1 and other coactivators. Thus, since all of the factors/cofactors that contribute to various aspects of adipogenesis are still unknown, it could well be that the loss of another implicated (co)factor would elicit an essential function for MED1 during a given developmental/functional phase where it is not normally essential based on single knockout analysis. Moreover, as mentioned earlier, MED1 requirements/functions not evident in normal adipocyte development/function, or in the vitro assays, could well be relevant in other (e.g., pathological) situations.
Transcriptional regulation via the MED1/Mediator
The classical model of transcriptional regulation through the Mediator involves Mediator recruitment through interactions of DNA-binding activators with specific subunits of the Mediator (mainly in the tail module) followed by effector functions involving interactions of core subunits (head and middle modules) with the general transcriptional machinery at the promoters (Cevher et al. 2014; Jeronimo and Robert 2017). MED1, despite being annotated to the middle module, locates close to the tail module (Khattabi et al. 2019) and has been found to interact only with a select few transcriptional regulators, most notably nuclear receptors that include PPARγ, to facilitate Mediator recruitment and function (Chen and Roeder 2011). In this regard, rigorous biochemical analyses have established a role for direct PPARγ-MED1/Mediator interactions (involving the PPARγ ligand-binding [activation] domain and MED1 NR boxes) in optimal PPARγ-dependent transcription (Ge et al. 2002, 2008). At the same time, and although we have demonstrated that MED1 loss does not lead to dissociation of additional subunits from the core Mediator, reports of conformational changes in Mediator upon activator binding (Taatjes et al. 2004; Meyer et al. 2010; Tsai et al. 2014) raise the possibility that MED1 loss may lead to Mediator conformation changes that prevent optimal interactions with specific activators. Whether a common mechanism (including a potential conformational change of the middle module) underlies the phenotypic similarities between our adipocyte-specific MED1 knockout mice and the recently reported adipocyte-specific MED19 knockout mice (Dean et al. 2020) remains an interesting topic to be explored.
Last, recent studies of gene regulation have shed light on potentially related “coactivator condensates” (Hnisz et al. 2017; Sabari et al. 2018) and further suggested that observed MED1 condensates in ESCs may be of functional significance through inclusion of the activation domains of various transcription factors (Boija et al. 2018). While MED1-containing condensates may occur within cells, we demonstrated that MED1 itself (and hence its phase separation property) is dispensable for ESCs. A recent screen to identify essential Mediator subunits in several mammalian cell lines also suggested that this is the case (Khattabi et al. 2019). Consequently, the role of the phase separation properties of MED1 in gene regulation is unclear and requires further functional studies to identify relevant cellular contexts in which it might play a role. One such possibility is in the early phase of 3T3-L1 adipogenesis, which requires the large MED1 C-terminal region that contains (among other sequences) the large disordered region that is responsible for the condensate properties of MED1.
Materials and methods
Mice
All animal experiments were performed following the guidelines and protocols approved by the Institutional Animal Care at The Rockefeller University. Mice were group-housed with enrichment in a temperature- and humidity-controlled, specific pathogen-free animal facility at 22°C under a 12:12-h light:dark cycle with free access to standard chow and water. The Med1 conventional knockout (LacZ) mouse was generated in house (Ito et al. 2000). Med1f/f mice were kindly provided by Dr. Janardan Reddy (Jia et al. 2004) (Northwestern University). The conditional knockout mice were backcrossed at least three times to C57BL6 mice (Charles River 475) upon arrival and used for breeding. Med1f/f mice were then crossed to Myf5-Cre (Jackson Laboratory 007893), Adipoq-Cre (Jackson Laboratory 010803), Ucp1-Cre (Jackson Laboratory 024670), or Rosa26-CreERT2 (Jackson Laboratory 008463) mice. AdipoqCreMed1f/f mice were crossed to Rosa26-mTmG mice (Jackson Laboratory 007676).
Cell culture
Embryonic stem cells were cultured in ESC medium (Glasgow minimum essential medium [SIGMA G6148], 10% ESC grade FBS [Thermo Fisher 10439024], 1× nonessential amino acid [Thermo Fisher 11140-050], 1× sodium pyruvate [Thermo Fisher 11360-070], 1× penicillin-streptomycin-glutamine [PSG] [Thermo Fisher 10378016], β-mercaptoethanol [Gibco 21985-023], 1000 U/mL leukemina inhibitory factor [ESGRO, ESG1107], 0.4 µM PD0329501 [BioVision 1990-1], 3 µM CHIR99021 [BioVision 1677-5]). The Cy2.4 ESC used for Med14 gene targeting was kindly provided by Dr. Chingwen Yang (The Rockefeller University). HEK293 (ATCC CRL-1573) and derived cell lines, 293gp cell line (a gift from Dr. Nikunj Somia, University of Minnesota) (Somia et al. 2000), and 3T3-L1-derived clones (ATCC CL-173) were cultured in DMEM (Thermo Fisher 11995040) supplemented with 10% FBS and 1× PSG. 293gpg cells (a gift from Dr. Richard Mulligan, Harvard Medical School) (Ory et al. 1996) were maintained in the above-mentioned DMEM-based media supplemented with 2 µg/mL puromycin (Sigma P7255), 0.3 mg/mL G418 (Takara Bio 631308), and 1 µg/mL tetracycline (Sigma T7660) until the induction of virus production. Brown and white preadipocyte cell lines were cultured in the presence of DMEM/F12 medium (Thermo Fisher 11330-032) supplemented with10% FBS and 100 U/mL pen-strep-glutamine. Generation of Med1-null ESCs, Med14 conditional knockout ESCs, Med1-KO 3T3-L1, immortalized brown and white preadipocyte lines, and endogenous Mediator subunit-tagged HEK293 cell lines and the modification of these cell lines via various transgenesis approaches are described in Supplemental Material.
RNA isolation, cDNA preparation, and real-time qPCR analysis
Total RNA was prepared using TRIzol reagent (Thermo Fisher 15596018) as described in Gerber et al. (2020). Tissues were minced and homogenized in TRIzol, and after centrifugation of the homogenate, the supernatant was used for RNA extraction. Cultured cells were harvested directly in TRizol after removal of the culture medium. RNA was reverse-transcribed using a SuperScript III first strand synthesis kit (Invitrogen). Real-time PCR was performed on an Applied Biosystems 7300 real-time PCR system using QuantiTect SYBR Green mix. Changes in mRNA expression were calculated using the DD Ct method and are presented as fold change relative to expression of the GAPDH mRNA for studies of ESCs, 36B4 mRNA for 3T3-L1 and adipose tissues. Primer pairs used for the experiments are listed in Supplemental Table S1.
Protein extraction and immunoblotting
Cell lysates were prepared by suspending cells in RIPA buffer (25 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). Extracts were separated using 4%–20% gradient SDS-PAGE gels (Bio-Rad), and immunoblotting was performed according to standard protocols. Antibodies used were MED1 (Bethyl Laboratories A300-793A) and β-Actin (Santa Cruz Biotechnology sc-47778).
Biochemical purification and mass spectrometric analysis of MED1-deficient Mediator complex
Parental (unmodified), Flag-MED10#5, and two clones of Flag-MED10#5/MED1-KO (#2 and #5) HEK293 cell lines were expanded in up to 10 15-cm culture dishes. Cells were transferred to large flasks and adapted to large scale (4-liter) suspension culture using DMEPO4 medium (prepared at the Memorial Sloan Kettering Cancer Center) supplemented with 5% newborn calf serum (Gemini Bio). Nuclear extracts were prepared as described (Dignam et al. 1983) with slight modifications. Briefly, harvested cells were swollen in a hypotonic buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl), and nuclei were released by 10 strokes of dounce homogenization. Pelleted nuclei were suspended in a low-salt buffer (20 mM HEPES, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 20 mM KCl), and a high-salt buffer (20 mM HEPES, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1.2 M KCl) was then added in a dropwise manner to reach 420 mM KCl. Nuclear extracts were separated from nuclear pellets by centrifugation, dialyzed against buffer C (20 mM HEPES at pH 7.9 at 4°C, 20% glycerol, 0.2 mM EDTA) containing 100 mM KCl and loaded onto a P11 phospho-cellulose (Whatman 753807) column (Bio-Rad 737-2522). After washing, bound components were eluted in a stepwise manner with buffer C containing 100 mM, 300 mM, and 850 mM KCl. The 850 mM KCl fractions were collected and dialyzed against 300 mM KCl. Dialyzed extracts were incubated with an anti-FLAG M2 affinity gel (Sigma A-2220) overnight in the presence of 0.1% NP40. The gel was then washed five times with buffer C containing 300 mM KCl and 0.1% NP40, and bound proteins were eluted with buffer C containing 166 ng/mL 3xFLAG peptide (Sigma F4799). Eluted proteins were processed at the Proteomics Core Laboratory at The Rockefeller University.
Mass spectrometry was performed at the Proteomics Laboratory at The Rockefeller University, as previously described (Zhu et al. 2019). Briefly, following immunoprecipitation, proteins were cleaned up by ice-cold acetone precipitation. Samples were reduced in solution containing 8 M urea, 0.1 M ammonium bicarbonate (ABC), and 10 mM dithiothreitol. After ultrasound sonication for 1 h, samples were then alkylated by adding iodoacetamide and incubated in the dark for 45 min. Samples were digested with Endoproteinase LysC (Wako Chemicals) and trypsin (Promega) and analyzed by reversed phase nano LC-MS/MS using Fusion Lumos (ThermoFisher). Data were quantified and searched against a Uniprot human database (March 2016) using ProteomeDiscoverer v. 1.4.0.288 (Thermo Scientific) combined with Mascot v. 2.5.1 (Matrix Science).
Peptides from the parental (untagged) sample were used as a negative control to confirm the specific enrichment of the Mediator complex in tagged cell line-derived samples. Individual preparations of FlagV5-MED1 or MED1-FlagV5 Mediator and individual preparations of MED1-deficient Mediator from KO clones #2 and # 5 were compared. Peptide abundance was determined using the three most abundant peptides detected and calculated as area signals. Samples were normalized to the abundance of MED14 (core Mediator subunit). Data were analyzed and visualized using R Studio (ver.1.0.153). Mass spectrometry analysis (normalized to MED14) is listed in Supplemental Table S2.
Histology and immunohistochemistry
Tissues and embryos were fixed in 4% paraformaldehyde overnight. Buffer solution was exchanged to 10% sucrose solution for 12 h followed by 12 h incubation in 30% sucrose solution. For preparation of cryosections, fixed tissues or embryos were embedded in Tissue-Tek O.C.T. compound (Sakura) and kept frozen at −80°C. A Leica cryostat was used to prepare 16- µm-thick cryosections that were subsequently used for immunofluorescence analysis, H&E staining (Abcam ab245880), or Oil Red O staining. Antibodies used for immunofluorescence analysis were anti-MED1 (M-255, Santa Cruz Biotechnology sc-8998), anti-PPARγ (81B8, Cell Signaling Technology 2443), and anti-myosin heavy chain (DSFB MF-20). 4′,6-diamidino-2-phenylindole (DAPI) (Roche 10236276001) was use to stain the nuclei. Paraffin sections were prepared at the Laboratory of Comparative Pathologies (LCP) of the Memorial Sloan Kettering Cancer Center. H&E staining, Masson's Trichrome staining, and immunohistochemical staining (F4/80 and CD11b) on paraffin sections were also performed at the LCP.
Microscopy imaging
Images of cultured cells and cells used for immunocytochemistry were collected using an Eclipse Ti-S microscope (Nikon). Differentiated adipocytes stained with Oil Red O were scanned in 24-well format using an EPSON scanner, and magnified images were taken using an adaptor mounted on an iPhone6S (LabCam). Images of specimens were collected by a BZ-X700 (Keyence) microscope or Nikon ECLIPSE E400 microscope. Dissections of embryos and postnatal mice were performed under stereomicroscope (Nikon SMZ 745T) attached to a camera (Nikon DS-Fi2). Dissections of mice carrying the Rosa26-mTmG allele were performed under a SteREO Discovery.V12 fluorescent microscope (Zeiss) to confirm correct labeling of the adipose tissues with GFP. Dissected adipose tissues were placed on slides with chambers filled with PBS and then mounted with coverslips. Whole-mount fluorescent images were taken using the Caliber ID scanning confocal microscope at the Bio-imaging Resource Center at The Rockefeller University. Obtained images were analyzed using IMARIS software.
Microarray analysis
RNA quality was assessed using an Agilent Bioanalyzer. Microarray analysis was performed at the Genomics Resource Center at The Rockefeller University using the Affymetrix GeneChip mouse Transcriptiome 2.0 array (Thermo Fisher 902162) according to the manufacturer's instructions. The data expressed as CEL files were normalized by the robust multiarray average method with transcriptome analysis console (TAC) 4.0.1 (Thermo Fisher) and have been deposited in the Gene Expression Omnibus database as GSE158083.
GO analysis was performed using the Metascape website (Zhou et al. 2019), with genes with signals >2.0 in any of the conditions included as the background (total expressed) gene lists. Data visualization was performed using R Studio (ver.1.0.153).
Thermogenesis assays in newborn mice
For thermogenesis assays of Myf5CreMed1f/f mice, iBAT temperatures of newborn pups (between P4–8) were measured with a thermography camera (FLIR systems). An average of three thermal images per litter were taken at 10 min after pup isolation from the mother. Images were taken by placing the newborn pups into six-well plates at the time of recording.
Cold tolerance test
Animals housed at room temperature were transferred to 4°C in cages without food and free access to water for a maximum of 5 h. Measurements of core and iBAT temperatures were obtained using, respectively, an anal probe (Braintree Scientific) and a wireless implantable temperature probe (Bio Medic Data Systems IPTT-300) in tandem with a portable reader. iscBAT temperatures was also measured using a thermography camera (FLIR systems). Individual mice were removed from the cold if their core body temperatures dropped >10°C from the baseline.
Serum chemistry tests
Whole blood from mice was collected via retro-orbital puncture, incubated for 30 min at room temperature, and subjected to centrifugation at 1200g for 5 min. Glucose, insulin, and triglycerides were measured at the Laboratory of Comparative Pathology at the Memorial Sloan Kettering Cancer Center.
Glucose tolerance and insulin tolerance tests
Animals (3–4 mo old) were fasted for 6 h and then received intraperitoneal glucose (2 g/kg). Insulin tolerance tests were also done on 3- to 4-mo-old) animals. Animals were fasted for 6 h and then received intraperitoneal humulin R insulin (0.5 U/kg; NovolinR). Note that, in a preliminary test using 8-wk-old mice, only the control group showed a severe hypoglycemic response and died when 1 U/kg insulin was injected. Based on this preliminary experiment, we decided to reduce the dosage and also to use older mice. Blood glucose levels were measured at 0, 20, 40, 60, 90, 120, and 150 min following treatment using Novamax plus a glucose meter and glucose strips.
Quantification and statistical analysis
Unpaired two-tailed t-test was used to analyze differences between two groups. P-values < 0.05 were considered statistically significant. The analysis was performed using GraphPad Prism8 (GraphPad software)
Supplementary Material
Acknowledgments
We thank Sohail Malik and Mitsuhiro Ito for comments on the manuscript; Satoshi Iida, Tomoyoshi Nakadai, Wei Chen, and Sicong Zhang for discussion of the work; and Yoko Tajima for assistance with data presentation using R. We thank Dr. Janardan Reddy for kindly providing Med1 conditional knockout mouse. We thank the Rockefeller University Flow Cytometry Resource Center, Henrik Molina, and the Proteomics Core Facility at the Rockefeller University for mass spectrometry; Christina Pyrgaki and the Bioimaging Resource Center at the Rockefeller University for adipose tissue imaging; Rada Norinsky, Roxana Cubias, and the Rockefeller University Transgenic and Reproductive Technology Center for the derivation of the Med1-null mouse from frozen sperm; the Rockefeller University Genomics Core Facility for microarray analysis; and the Laboratory of Comparative Pathology at the Memorial Sloan Kettering Cancer Center for histology studies. This work was supported by National Institutes of Health grants DK071900 and CA234575 to R.G.R. K.I. was supported by National Cancer Institute T32 grant CA009673 and by a Japan Society for the Promotion of Science postdoctoral fellowship for research abroad.
Author contributions: K.I. designed and performed experiments, analyzed and interpreted data, and wrote the manuscript. M.S. assisted with the cold tolerance test. A.G. designed and assisted with the cell culture assays. M.J. assisted with the Mediator purification and analysis. F.M. assisted with the analysis of brown fat functions. A.V.M. assisted with primary culture assays and insulin and glucose tolerance tests. P.C. and J.M.F. supervised associated projects. R.G.R. supervised the overall project and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.346791.120.
Competing interest statement
The authors declare no competing interests.
References
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. 1999. PPARγ is required for placental, cardiac, and adipose tissue development. Mol Cell 4: 585–595. 10.1016/S1097-2765(00)80209-9 [DOI] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. 2018. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175: 1842–1855.e16. 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. 2004. Brown adipose tissue: function and physiological significance. Physiol Rev 84: 277–359. 10.1152/physrev.00015.2003 [DOI] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev 5: 1538–1552. 10.1101/gad.5.9.1538 [DOI] [PubMed] [Google Scholar]
- Cautivo KM, Lizama CO, Tapia PJ, Agarwal AK, Garg A, Horton JD, Cortés VA. 2016. AGPAT2 is essential for postnatal development and maintenance of white and brown adipose tissue. Mol Metab 5: 491–505. 10.1016/j.molmet.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevher MA, Shi Y, Li D, Chait BT, Malik S, Roeder RG. 2014. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat Struct Mol Biol 21: 1028–1034. 10.1038/nsmb.2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau Y-Y, Bandiera R, Serrels A, Martínez-Estrada OM, Qing W, Lee M, Slight J, Thornburn A, Berry R, McHaffie S, et al. 2014. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat Cell Biol 16: 367–375. 10.1038/ncb2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Roeder RG. 2011. Mediator-dependent nuclear receptor function. Semin Cell Dev Biol 22: 749–758. 10.1016/j.semcdb.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Yang Q, Roeder RG. 2009. Dynamic interactions and cooperative functions of PGC-1α and MED1 in TRα-mediated activation of the brown fat-specific UCP-1 gene. Mol Cell 35: 755–768. 10.1016/j.molcel.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JM, He A, Tan M, Wang J, Lu D, Razani B, Lodhi IJ. 2020. MED19 regulates adipogenesis and maintenance of white adipose tissue mass by mediating PPARγ-dependent gene expression. Cell Rep 33: 108228. 10.1016/j.celrep.2020.108228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489. 10.1093/nar/11.5.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan SZ, Ivashchenko CY, Whitesall SE, D'Alecy LG, Duquaine DC, Brosius FC, Gonzalez FJ, Vinson C, Pierre MA, Milstone DS, et al. 2007. Hypotension, lipodystrophy, and insulin resistance in generalized PPARγ-deficient mice rescued from embryonic lethality. J Clin Invest 117: 812–822. 10.1172/JCI28859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR. 2006. Transcriptional control of adipocyte formation. Cell Metab 4: 263–273. 10.1016/j.cmet.2006.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. PNAS 93: 8329–8333. 10.1073/pnas.93.16.8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Guermah M, Yuan C-X, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. 2002. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 417: 563–567. 10.1038/417563a [DOI] [PubMed] [Google Scholar]
- Ge K, Cho Y-W, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, Roeder RG. 2008. Alternative mechanisms by which Mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor γ-stimulated adipogenesis and target gene expression. Mol Cell Biol 28: 1081–1091. 10.1128/MCB.00967-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. 2008. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development 135: 1597–1604. 10.1242/dev.019331 [DOI] [PubMed] [Google Scholar]
- Gerber A, Ito K, Chu C-S, Roeder RG. 2020. Gene-specific control of tRNA expression by RNA polymerase II. Mol Cell 78: 765–778.E7. 10.1016/j.molcel.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Meuth M. 1974. An established pre-adipose cell line and its differentiation in culture. Cell 3: 127–133. 10.1016/0092-8674(74)90116-0 [DOI] [PubMed] [Google Scholar]
- Grøntved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. 2010. MED14 tethers Mediator to the N-terminal domain of peroxisome proliferator-activated receptor γ and is required for full transcriptional activity and adipogenesis. Mol Cell Biol 30: 2155–2169. 10.1128/MCB.01238-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyaeva O, Dempersmier J, Sul HS. 2019. Genetic and epigenetic control of adipose development. Biochim Biophys Acta Mol Cell Biol Lipids 1864: 3–12. 10.1016/j.bbalip.2018.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MJ, Lim H-W, Ho Y, Shapira SN, Ishibashi J, Rajakumari S, Steger DJ, Lazar MA, Won K-J, Seale P. 2015. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev 29: 298–307. 10.1101/gad.252734.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A phase separation model for transcriptional control. Cell 169: 13–23. 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KY, Bae H, Park I, Park D-Y, Kim KH, Kubota Y, Cho E-S, Kim H, Adams RH, Yoo O-J, et al. 2015. Perilipin+ embryonic preadipocytes actively proliferate along growing vasculatures for adipose expansion. Development 142: 2623–2632. 10.1242/dev.125336 [DOI] [PubMed] [Google Scholar]
- Iida S, Chen W, Nakadai T, Ohkuma Y, Roeder RG. 2015. PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with Mediator subunit MED1. Genes Dev 29: 308–321. 10.1101/gad.252809.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Yuan C-X, Okano HJ, Darnell RB, Roeder RG. 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell 5: 683–693. 10.1016/S1097-2765(00)80247-6 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Robert F. 2017. The Mediator complex: at the nexus of RNA polymerase II transcription. Trends Cell Biol 27: 765–783. 10.1016/j.tcb.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Jia Y, Qi C, Kashireddi P, Surapureddi S, Zhu Y-J, Rao MS, Roith DL, Chambon P, Gonzalez FJ, Reddy JK. 2004. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARα-regulated gene expression in liver. J Biol Chem 279: 24427–24434. 10.1074/jbc.M402391200 [DOI] [PubMed] [Google Scholar]
- Jiang P, Hu Q, Ito M, Meyer S, Waltz S, Khan S, Roeder RG, Zhang X. 2010. Key roles for MED1 LxxLL motifs in pubertal mammary gland development and luminal-cell differentiation. Proc Natl Acad Sci 107: 6765–6770. 10.1073/pnas.1001814107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattabi LE, Zhao H, Kalchschmidt J, Young N, Jung S, Blerkom PV, Kieffer-Kwon P, Kieffer-Kwon K-R, Park S, Wang X, et al. 2019. A pliable Mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell 178: 1145–1158.e20. 10.1016/j.cell.2019.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Spiegelman BM. 1996. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev 10: 1096–1107. 10.1101/gad.10.9.1096 [DOI] [PubMed] [Google Scholar]
- Lai B, Lee J-E, Jang Y, Wang L, Peng W, Ge K. 2017. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res 45: 6388–6403. 10.1093/nar/gkx234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc SE, Wu Q, Lamba P, Sif S, Imbalzano AN. 2016. Promoter–enhancer looping at the PPARγ2 locus during adipogenic differentiation requires the Prmt5 methyltransferase. Nucleic Acids Res 44: 5133–5147. 10.1093/nar/gkw129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-E, Wang C, Xu S, Cho Y-W, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, et al. 2013. H3k4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation s.eLife 2: e01503. 10.7554/eLife.01503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-E, Schmidt H, Lai B, Ge K. 2019. Transcriptional and epigenomic regulation of adipogenesis. Mol Cell Biol 39: e00601-18. 10.1128/MCB.00601-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gade P, Nallar SC, Raha A, Roy SK, Karra S, Reddy JK, Reddy SP, Kalvakolanu DV. 2008. The Med1 subunit of transcriptional Mediator plays a central role in regulating CCAAT/enhancer-binding protein-β-driven transcription in response to interferon-γ. J Biol Chem 283: 13077–13086. 10.1074/jbc.M800604200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart HG, Ishimura-Oka K, DeMayo F, Kibe T, Repka D, Poindexter B, Bick RJ, Darlington GJ. 2001. C/EBPα is required for differentiation of white, but not brown, adipose tissue. PNAS 98: 12532–12537. 10.1073/pnas.211416898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772. 10.1038/nrg2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Guermah M, Yuan C-X, Wu W, Yamamura S, Roeder RG. 2004. Structural and functional organization of TRAP220, the TRAP/Mediator subunit that is targeted by nuclear receptors. Mol Cell Biol 24: 8244–8254. 10.1128/MCB.24.18.8244-8254.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeuf-Louchart A, Lancel S, Sebti Y, Pourcet B, Loyens A, Delhaye S, Duhem C, Beauchamp J, Ferri L, Thorel Q, et al. 2019. Glycogen dynamics drives lipid droplet biogenesis during brown adipocyte differentiation. Cell Rep 29: 1410–1418.e6. 10.1016/j.celrep.2019.09.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Lin S, Bernecky C, Gao Y, Taatjes DJ. 2010. P53 activates transcription by directing structural shifts in Mediator. Nat Struct Mol Biol 17: 753–760. 10.1038/nsmb.1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta S, Minami T, Fujita H, Kaminaga C, Matsui K, Ishino R, Fujita A, Oda K, Kawai A, Hasegawa N, et al. 2014. CCAR1/CoCoA pair-mediated recruitment of the Mediator defines a novel pathway for GATA1 function. Genes Cells 19: 28–51. 10.1111/gtc.12104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra J, Mason MM, Olive M, Krylov D, Gavrilova O, Marcus-Samuels B, Feigenbaum L, Lee E, Aoyama T, Eckhaus M, et al. 1998. Life without white fat: a transgenic mouse. Genes Dev 12: 3168–3181. 10.1101/gad.12.20.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morganstein DL, Christian M, Turner JJO, Parker MG, White R. 2008. Conditionally immortalized white preadipocytes: a novel adipocyte model. J Lipid Res 49: 679–685. 10.1194/jlr.D700029-JLR200 [DOI] [PubMed] [Google Scholar]
- Mueller E, Drori S, Aiyer A, Yie J, Sarraf P, Chen H, Hauser S, Rosen ED, Ge K, Roeder RG, et al. 2002. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor γ isoforms. J Biol Chem 277: 41925–41930. 10.1074/jbc.M206950200 [DOI] [PubMed] [Google Scholar]
- Namwanje M, Liu L, Chan M, Aaron N, Kraakman MJ, Qiang L. 2019. The depot-specific and essential roles of CBP/p300 in regulating adipose plasticity. J Endocrinol 240: 257–269. 10.1530/JOE-18-0361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. PNAS 93: 11400–11406. 10.1073/pnas.93.21.11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou C-Y, Chen T-C, Lee JV, Wang J-C, Stallcup MR. 2014. Coregulator cell cycle and apoptosis regulator 1 (CCAR1) positively regulates adipocyte differentiation through the glucocorticoid signaling pathway. J Biol Chem 289: 17078–17086. 10.1074/jbc.M114.548081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajvani UB, Trujillo ME, Combs TP, Iyengar P, Jelicks L, Roth KA, Kitsis RN, Scherer PE. 2005. Fat apoptosis through targeted activation of caspase 8: a new mouse model of inducible and reversible lipoatrophy. Nat Med 11: 797–803. 10.1038/nm1262 [DOI] [PubMed] [Google Scholar]
- Park Y-K, Ge K. 2017. Glucocorticoid receptor accelerates, but is dispensable for, adipogenesis. Mol Cell Biol 37: e00260-16. 10.1128/MCB.00260-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y-K, Wang L, Giampietro A, Lai B, Lee J-E, Ge K. 2017. Distinct roles of transcription factors KLF4, Krox20, and peroxisome proliferator-activated receptor γ in adipogenesis. Mol Cell Biol 37: e00554-16. 10.1128/MCB.00554-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proescher F. 1927. Oil Red O pyridin, a rapid fat stain. Stain Technol 2: 60–61. 10.3109/10520292709115655 [DOI] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim H-W, Giang A-H, Won K-J, Reed RR, Seale P. 2013. EBF2 determines and maintains brown adipocyte identity. Cell Metab 17: 562–574. 10.1016/j.cmet.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. 2005. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett 579: 909–915. 10.1016/j.febslet.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. 2006. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7: 885–896. 10.1038/nrm2066 [DOI] [PubMed] [Google Scholar]
- Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. 1999. PPARγ Is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell 4: 611–617. 10.1016/S1097-2765(00)80211-7 [DOI] [PubMed] [Google Scholar]
- Rosen ED, Hsu C-H, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM. 2002. C/EBPα induces adipogenesis through PPARγ: a unified pathway. Genes Dev 16: 22–26. 10.1101/gad.948702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361: eaar3958. 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Guertin DA. 2014. Adipocyte lineages: tracing back the origins of fat. Biochim Biophys Acta 1842: 340–351. 10.1016/j.bbadis.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Hung C-M, Sparks CA, Tang Y, Li H, Guertin DA. 2012. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab 16: 348–362. 10.1016/j.cmet.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Hsiao W-Y, Guertin DA. 2015. Highly selective in vivo labeling of subcutaneous white adipocyte precursors with Prx1-Cre. Stem Cell Reports 4: 541–550. 10.1016/j.stemcr.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P. 2015. Transcriptional regulatory circuits controlling brown fat development and activation. Diabetes 64: 2369–2375. 10.2337/db15-0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454: 961–967. 10.1038/nature07182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JB, Moon HM, Kim WS, Lee YS, Jeong HW, Yoo EJ, Ham J, Kang H, Park M-G, Steffensen KR, et al. 2004. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor γ expression. Mol Cell Biol 24: 3430–3444. 10.1128/MCB.24.8.3430-3444.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD. 1997. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest 100: 2115–2124. 10.1172/JCI119746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk R, Nielsen R, John S, Sung M-H, Baek S, Loft A, Hager GL, Mandrup S. 2011. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J 30: 1459–1472. 10.1038/emboj.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk MS, Loft A, Aagaard MM, Nielsen R, Schmidt SF, Petrovic N, Nedergaard J, Mandrup S. 2012. Genome-wide profiling of peroxisome proliferator-activated receptor γ in primary epididymal, inguinal, and brown adipocytes reveals depot-selective binding correlated with gene expression. Mol Cell Biol 32: 3452–3463. 10.1128/MCB.00526-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, Poulsen LLC, Rogowska-Wrzesinska A, Jensen ON, Mandrup S. 2014. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep 7: 1443–1455. 10.1016/j.celrep.2014.04.042 [DOI] [PubMed] [Google Scholar]
- Siersbæk R, Madsen JGS, Javierre BM, Nielsen R, Bagge EK, Cairns J, Wingett SW, Traynor S, Spivakov M, Fraser P, et al. 2017. Dynamic rewiring of promoter-anchored chromatin loops during adipocyte differentiation. Mol Cell 66: 420–435.e5. 10.1016/j.molcel.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Soccio RE, Chen ER, Lazar MA. 2014. Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metab 20: 573–591. 10.1016/j.cmet.2014.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somia NV, Miyoshi H, Schmitt MJ, Verma IM. 2000. Retroviral vector targeting to human immunodeficiency virus type 1-infected cells by receptor pseudotyping. J Virol 74: 4420–4424. 10.1128/JVI.74.9.4420-4424.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ, Schneider-Poetsch T, Tjian R. 2004. Distinct conformational states of nuclear receptor–bound CRSP–Med complexes. Nat Struct Mol Biol 11: 664–671. 10.1038/nsmb789 [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellstrom M, Soriano P. 2000. Early myotome specification regulates PDGFA expression and axial skeleton development. Development 127: 5059–5070. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Yoshida N, Kishimoto T, Akira S. 1997. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J 16: 7432–7443. 10.1093/emboj/16.24.7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM. 2008. Fat and beyond: the diverse biology of PPARγ. Annu Rev Biochem 77: 289–312. 10.1146/annurev.biochem.77.061307.091829 [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. 1994a. mPPARγ2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 8: 1224–1234. 10.1101/gad.8.10.1224 [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. 1994b. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79: 1147–1156. 10.1016/0092-8674(94)90006-X [DOI] [PubMed] [Google Scholar]
- Tsai K-L, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. 2014. Subunit architecture and functional modular rearrangements of the transcriptional Mediator complex. Cell 157: 1430–1444. 10.1016/j.cell.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinter J, Hull D, Elphick MC. 1982. Onset of thermogenesis in response to cold in newborn mice. NEO 42: 145–151. [DOI] [PubMed] [Google Scholar]
- Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. 2003. Coordination of p300-mediated chromatin remodeling and TRAP/Mediator function through coactivator PGC-1α. Mol Cell 12: 1137–1149. 10.1016/S1097-2765(03)00391-5 [DOI] [PubMed] [Google Scholar]
- Wang F, Mullican SE, DiSpirito JR, Peed LC, Lazar MA. 2013. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARγ. Proc Natl Acad Sci 110: 18656–18661. 10.1073/pnas.1314863110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kissig M, Rajakumari S, Huang L, Lim H, Won K-J, Seale P. 2014. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci 111: 14466–14471. 10.1073/pnas.1412685111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QA, Zhang F, Jiang L, Ye R, An Y, Shao M, Tao C, Gupta RK, Scherer PE. 2018. Peroxisome proliferator-activated receptor γ and its role in adipocyte homeostasis and thiazolidinedione-mediated insulin sensitization. Mol Cell Biol 38: e00677-17. 10.1128/MCB.00677-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Bucher NL, Farmer SR. 1996. Induction of peroxisome proliferator-activated receptor γ during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPβ, C/EBPδ, and glucocorticoids. Mol Cell Biol 16: 4128–4136. 10.1128/MCB.16.8.4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. 1999. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 3: 151–158. 10.1016/S1097-2765(00)80306-8 [DOI] [PubMed] [Google Scholar]
- Yoneshiro T, Wang Q, Tajima K, Matsushita M, Maki H, Igarashi K, Dai Z, White PJ, McGarrah RW, Ilkayeva OR, et al. 2019. BCAA catabolism in brown fat controls energy homeostasis through SLC25A44. Nature 572: 614–619. 10.1038/s41586-019-1503-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C-X, Ito M, Fondell JD, Fu Z-Y, Roeder RG. 1998. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. PNAS 95: 7939–7944. 10.1073/pnas.95.14.7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG. 2005. MED1/TRAP220 exists predominantly in a TRAP/ Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell 19: 89–100. 10.1016/j.molcel.2005.05.015 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10: 1523. 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. 1997. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem 272: 25500–25506. 10.1074/jbc.272.41.25500 [DOI] [PubMed] [Google Scholar]
- Zhu XG, Puthenveedu SN, Shen Y, La K, Ozlu C, Wang T, Klompstra D, Gultekin Y, Chi J, Fidelin J, et al. 2019. CHP1 regulates compartmentalized glycerolipid synthesis by activating GPAT4. Mol Cell 74: 45–58.e7. 10.1016/j.molcel.2019.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.