Abstract
Nucleosomes are a significant barrier to the repair of UV damage because they impede damage recognition by nucleotide excision repair (NER). The RSC and SWI/SNF chromatin remodelers function in cells to promote DNA access by moving or evicting nucleosomes, and both have been linked to NER in yeast. Here, we report genome-wide repair maps of UV-induced cyclobutane pyrimidine dimers (CPDs) in yeast cells lacking RSC or SWI/SNF activity. Our data indicate that SWI/SNF is not generally required for NER but instead promotes repair of CPD lesions at specific yeast genes. In contrast, mutation or depletion of RSC subunits causes a general defect in NER across the yeast genome. Our data indicate that RSC is required for repair not only in nucleosomal DNA but also in neighboring linker DNA and nucleosome-free regions (NFRs). Although depletion of the RSC catalytic subunit also affects base excision repair (BER) of N-methylpurine (NMP) lesions, RSC activity is less important for BER in linker DNA and NFRs. Furthermore, our data indicate that RSC plays a direct role in transcription-coupled NER (TC-NER) of transcribed DNA. These findings help to define the specific genomic and chromatin contexts in which each chromatin remodeler functions in DNA repair, and indicate that RSC plays a unique function in facilitating repair by both NER subpathways.
The nucleotide excision repair (NER) pathway plays a critical role in removing bulky, helix-distorting lesions from genomic DNA (Scharer 2013). Individuals with inherited genetic defects in the NER pathway have a more than 1000-fold higher risk of skin cancer (DiGiovanna and Kraemer 2012), owing to their inability to repair helix-distorting UV damage, primarily cyclobutane pyrimidine dimers (CPDs). NER consists of two subpathways that differ in the mechanism by which helix-distorting DNA damage is recognized. The transcription-coupled nucleotide excision repair (TC-NER) pathway recognizes and repairs DNA damage that stalls RNA polymerase II (Pol II) transcription, and therefore is confined to repairing damage residing in transcribed DNA (Hanawalt and Spivak 2008; Lans et al. 2019). In contrast, the global-genomic nucleotide excision repair (GG-NER) pathway uses a damage sensor consisting of Rad4 and Rad23 in yeast, in conjunction with accessory proteins such as the Rad7–Rad16 complex, to recognize DNA damage throughout the genome (Prakash and Prakash 2000; Friedberg et al. 2006). Although GG-NER can efficiently repair UV damage in naked DNA, the packaging of DNA into chromatin represents a significant barrier to GG-NER (Mao and Wyrick 2019). Even the nucleosome, which represents the primary packaging unit of chromatin, inhibits GG-NER both in vitro (Hara et al. 2000; Liu and Smerdon 2000) and in cells (Smerdon and Thoma 1990; Wellinger and Thoma 1997; Tijsterman et al. 1999; Mao et al. 2016, 2020; Brown et al. 2018).
ATP-dependent chromatin remodeling complexes (ACRs) reorganize chromatin structure to promote transcription but have also been implicated in promoting repair by the GG-NER and base excision repair (BER) pathways (Czaja et al. 2012; Hinz and Czaja 2015). ACRs use energy from ATP hydrolysis to alter nucleosome positions or even evict the histone octamer. For example, the Remodels Structure of Chromatin (RSC) complex is required to expand nucleosome-free regions (NFRs) in yeast promoters by shifting the positions of the flanking +1 and −1 nucleosomes (Badis et al. 2008; Hartley and Madhani 2009; Krietenstein et al. 2016; Kubik et al. 2018), whereas both the RSC and Switch/Sucrose Non-Fermentable (SWI/SNF) complexes can also evict histones (Lorch et al. 2006; Liu and Hayes 2010; Rando and Winston 2012). In yeast, both RSC and SWI/SNF have been linked to the NER pathway (Gong et al. 2006; Srivas et al. 2013). SWI/SNF interacts with the Rad4–Rad23 DNA damage sensor and is required for repair of CPD lesions in the heterochromatic HML locus (Gong et al. 2006) but not the repressed MFA2 locus (Yu et al. 2005), indicating it may play a loci-specific role in GG-NER. However, the specific genomic or chromatin contexts in which SWI/SNF stimulates NER are not known. RSC is also recruited to chromatin by Rad4 in response to UV damage, and deletion of the gene encoding a nonessential RSC subunit (rsc2Δ) confers UV sensitivity and affects the repair of the RPB2 gene in yeast (Srivas et al. 2013). RSC has also been linked to efficient BER of methyl methanesulfonate (MMS)–induced DNA base lesions at the yeast GAL1 gene (Czaja et al. 2014). Whether RSC is required for repair of UV- and MMS-induced lesions only in nucleosomes or potentially in other chromatin contexts (i.e., linker DNA, NFR) is unknown.
There have been hints that ACRs may also function in TC-NER. Mutations in both SWI/SNF and RSC complexes enhance the UV sensitivity of a rad16 mutant, in which GG-NER is eliminated, indicating that these ACR mutations may affect TC-NER (Gong et al. 2006; Srivas et al. 2013). Moreover, deletion of the gene encoding the Rsc2 subunit affects the repair of UV damage in the transcribed strand (TS) of the RPB2 gene (Srivas et al. 2013). Because the TS can be repaired by both NER subpathways, this could mean that RSC also functions in the TC-NER pathway or, alternatively, that RSC only affects the GG-NER of the RPB2 TS. Studies in other organisms (Caenorhabditis elegans) have potentially implicated ACRs in the TC-NER pathway (Lans et al. 2010), but definitive evidence linking ACRs to the TC-NER pathway has been lacking.
The advent of sequencing technologies such as CPD-seq to measure the repair of UV damage across the genome (Mao et al. 2016; Mao and Wyrick 2019) provides a unique opportunity to assess the specific roles of ACRs in each NER subpathway and in different genomic and chromatin contexts. Here, we report genome-wide maps of UV-induced CPD repair in yeast cells lacking RSC or SWI/SNF, which indicate that the RSC and SWI/SNF complexes play distinct roles in promoting NER across the genome.
Results
RSC is generally required for NER in yeast, whereas SWI/SNF is not
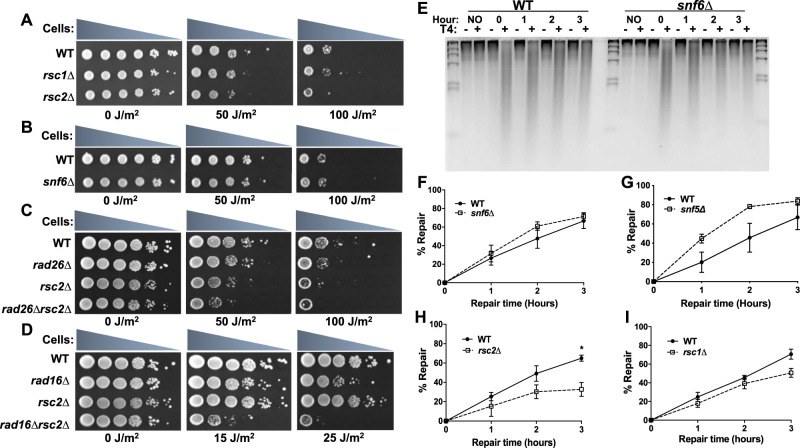
Although previous studies have implicated the yeast SWI/SNF and RSC complexes in NER at specific yeast loci (Gong et al. 2006; Srivas et al. 2013), it is not known if either complex is generally required for NER across the yeast genome. To investigate the roles of these ACR complexes in NER, we characterized the impact of SWI/SNF- or RSC-deletion mutants on the repair of UV-induced CPD lesions in yeast. Consistent with a previous report (Srivas et al. 2013), deletion of the Rsc2 subunit (rsc2Δ) imparts UV sensitivity in yeast (Fig. 1A), whereas deletion of Rsc1, which is incorporated in place of Rsc2 in a subset of RSC complexes (Cairns et al. 1999), does not increase UV sensitivity. Neither the snf6Δ nor snf5Δ mutants cause UV sensitivity under normal growth conditions (Fig. 1B; Supplemental Fig. S1A), but prior growth on galactose-containing medium rendered the snf6Δ mutant UV sensitive (Supplemental Fig. S1B), as previously reported (Gong et al. 2006). The rsc2Δ mutant enhances the UV sensitivity of both the GG-NER-defective rad16Δ and TC-NER-defective rad26Δ strains (Fig. 1C,D; Supplemental Fig. S1C), potentially implicating Rsc2 in both NER subpathways (Srivas et al. 2013).
Figure 1.
RSC is generally required for repair of CPD lesions in yeast, whereas SWI/SNF is not. (A,B) UV sensitivity of yeast mutants in the RSC (rsc1Δ and rsc2Δ) or SWI/SNF complex (snf6Δ). (C,D) UV sensitivity of rad26Δrsc2Δ and rad16Δrsc2Δ double mutants compared with single mutants. (E) Representative alkaline gel of CPD repair in WT and snf6Δ-mutant cells. Genomic DNA was isolated at the indicated time following damage induction with 100 J/m2 UVC light and treated with or without (+/−) T4 endonuclease V. (F–I) Quantification of CPD repair in RSC or SWI/SNF mutants based on alkaline gel analysis. The percentage of CPDs repaired is plotted as the mean ± SEM of a minimum of three replicates. P-values were calculated using an unpaired t-test with Holm–Sidak correction for multiple hypothesis testing. (*) P ≤ 0.05.
We performed a T4 endonuclease V alkaline gel assay to measure the repair of CPD lesions in yeast genomic DNA (Fig. 1E). Neither the snf5Δ nor snf6Δ deletion mutant showed a defect in bulk repair of UV-induced CPD lesions relative to the WT (Fig. 1F,G), indicating that the SWI/SNF complex is not generally required for NER. Indeed, CPD repair appeared to be marginally faster in the snf5Δ mutant relative to the WT (Fig. 1G). In contrast, the rsc2Δ mutant causes a general defect in CPD repair relative to the WT (Fig. 1H), particularly at the 3-h time point. This combined with the increased UV sensitivity of the rsc2Δ-mutant strain indicates that Rsc2 is generally important for repair of CPD lesions across the yeast genome. Previous analysis indicates that the rsc2Δ mutant does not affect the expression of any known NER genes (Srivas et al. 2013), indicating that the NER defect is likely a direct consequence of the rsc2Δ mutant. A subset of RSC complexes contains the nonessential Rsc1 subunit instead of Rsc2 (Cairns et al. 1999). Our data reveal that there is relatively little difference in CPD repair in the rsc1Δ mutant relative to the WT (Fig. 1I), confirming that unlike Rsc2, Rsc1 is largely dispensable for repair (Srivas et al. 2013).
SWI/SNF affects NER at a subset of yeast genes
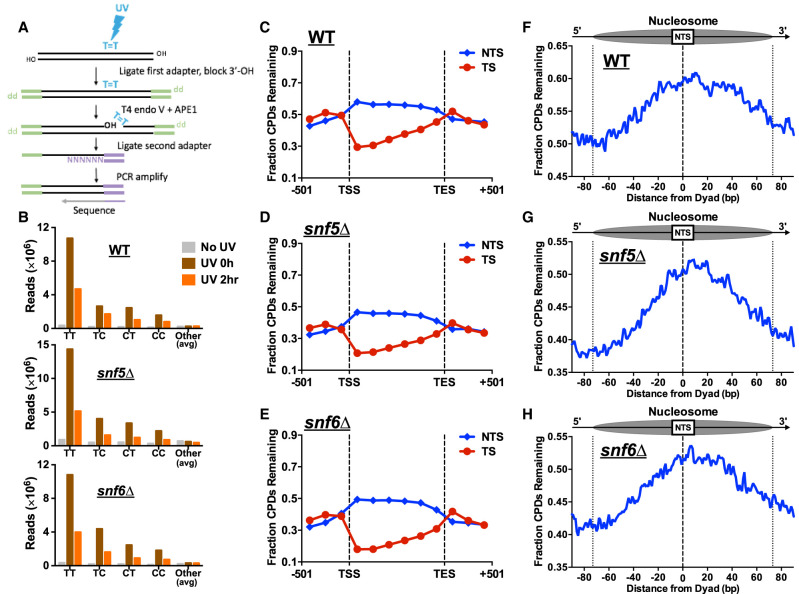
Although SWI/SNF has been reported to facilitate repair of UV damage at specific yeast genes (e.g., HML), our data indicate that it is not generally required for NER in yeast (Fig. 1). To better understand the genomic or chromatin contexts in which SWI/SNF facilitates repair, we used the CPD-seq method (Fig. 2A) to analyze the change in repair of UV-induced CPD lesions across the yeast genome at single-nucleotide resolution (Mao et al. 2016) in snf5Δ- or snf6Δ-mutant cells relative to WT. Initial analysis of the CPD-seq data confirmed that sequencing reads were primarily associated with lesion-forming dipyrimidine sequences (e.g., TT, TC, CT, CC) immediately after UV irradiation (0 h) relative to the “No UV” controls (Fig. 2B), consistent with previous reports (Mao et al. 2016, 2018). The number of lesion-associated CPD-seq reads was lower following a 2-h repair incubation (Fig. 2B), reflecting cellular repair of CPD lesions. Analysis of CPD repair across approximately 5000 yeast genes revealed faster repair of TS relative to the NTS in WT cells (Fig. 2C), owing to rapid removal of CPD lesions from the TS by TC-NER. In the snf5Δ- or snf6Δ-mutant cells, the fraction of unrepaired CPDs was somewhat lower than the WT along both the TS and NTS of yeast genes (Fig. 2C–E), consistent with the results of the alkaline gel analysis (Fig. 1). This analysis confirms that SWI/SNF (or at least the Snf5 and Snf6 subunits) is not generally required for NER, and instead suggests that repair proceeds slightly faster in SWI/SNF deficient cells.
Figure 2.
CPD-seq analysis indicates that SWI/SNF is dispensable for repair at most genomic and chromatin contexts. (A) Schematic of the CPD-seq method for mapping CPD lesions at single-nucleotide resolution. Adapted from Mao et al. (2016). (B) Enrichment of CPD-seq reads associated with putative CPD lesions at dipyrimidine sequences in UV-irradiated WT, snf5Δ, and snf6Δ cells but not the “No UV” control. (C–E) The fraction of CPDs remaining after 2-h repair was plotted for CPD-seq data for both the transcribed strand (TS) and nontranscribed strand (NTS) between the transcription start site (TSS) and transcription end site (TES) of approximately 5000 yeast genes. (F–H) Analysis of the fraction of CPDs remaining following 2-h repair within nucleosomes. Repair of the NTS oriented in the 5′ to 3′ orientation for the WT, snf5Δ, and snf6Δ cells was examined in aggregate for the first three nucleosomes (+1 to +3) downstream from the TSS of approximately 5200 genes. Dotted lines at positions −73 and +73 bp from the dyad center indicate the boundary of the nucleosome core particle.
Because the SWI/SNF ACR plays a role in altering nucleosome positions and evicting nucleosomes, we used the CPD-seq data to test whether the SWI/SNF complex is specifically required for repair in nucleosomes. High-resolution analysis of repair in WT cells near the transcription start sites (TSSs) of approximately 5200 yeast genes revealed peaks of unrepaired damage on the NTS coinciding with positioned nucleosomes in gene coding regions (Supplemental Fig. S2A). These results are consistent with our recent report that GG-NER is inhibited along the NTS of nucleosomes in coding regions (Mao et al. 2020). Deletion of snf5 or snf6 did not significantly alter this nucleosome-modulated repair pattern along the NTS (Supplemental Fig. S2B,C). We further analyzed CPD repair along the NTS in the first three nucleosomes downstream from the TSS (e.g., the +1, +2, and +3 nucleosomes). In WT cells, there was a higher fraction of unrepaired CPDs near the central dyad axis of the nucleosome relative to the nucleosome edges or adjacent linker DNA (Fig. 2F), owing to GG-NER inhibition in nucleosomes. Moreover, repair inhibition was asymmetric, with more unrepaired damage accumulating on the slower-repairing 3′ side of the nucleosomal DNA (Fig. 2F), consistent with our previous report (Mao et al. 2020). In the snf5Δ and snf6Δ mutants, there was relatively little change in the pattern of repair in nucleosomes (Fig. 2G,H). In summary, this analysis indicates that the SWI/SNF complex is largely dispensable for repair both across the genome and in positioned nucleosomes.
Although our genome-wide data averaged over all yeast genes showed no repair defects in SWI/SNF mutants, it is possible that repair at a subset of genes is affected. To identify genes that require SWI/SNF for efficient NER, we used the CPD-seq data to screen for genes with more unrepaired CPDs along either the NTS or TS in the snf5Δ or snf6Δ mutants relative to the WT (see Methods). We identified 227 genes in which repair of the NTS or TS was slower in one or both of the SWI/SNF mutants, reflected by an elevated fraction of unrepaired CPDs relative to the WT control (Supplemental Fig. S3). The set of genes with slower repair of the NTS was significantly enriched for ribosomal protein genes (P < 1 × 10−14) (Supplemental Fig. S3A), indicating that GG-NER of these genes may specifically require SWI/SNF activity. Repair of the TS for these ribosomal protein genes was not impeded in the snf5Δ and snf6Δ mutants (Supplemental Fig. S3B), consistent with a GG-NER defect. Genes with slower repair of the TS were significantly enriched for genes involved in glycolysis and gluconeogenesis (P < 1 × 10−6) and fungal-type cell wall (P < 1 × 10−9). The majority of these genes were significantly down-regulated in expression in snf2Δ, snf5Δ, or snf6Δ mutants (P < 1 × 10−5) (Supplemental Fig. S3C). Because TC-NER efficiency is correlated with transcription rate (Li et al. 2018), this finding suggests that the accumulation of unrepaired CPDs along the TS (Supplemental Fig. S3C) is a consequence of reduced transcription of these genes in SWI/SNF mutants. In contrast, expression of the set of slower repairing NTS genes was not significantly altered in the snf5Δ or snf6Δ mutants (P > 0.01), indicating that ribosomal protein genes may specifically require SWI/SNF to efficiently repair UV damage along the NTS.
Rapid depletion of the Sth1 subunit of RSC also causes a global defect in NER
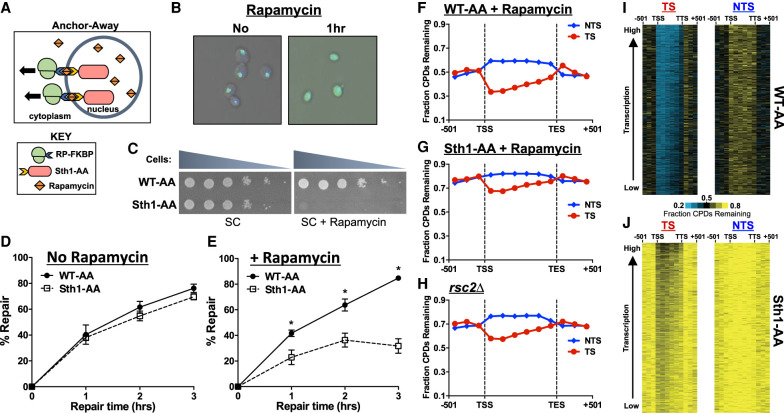
The data so far indicate that SWI/SNF is required for repair of specific genes and/or chromatin states, whereas the Rsc2 subunit of RSC plays a more general role in NER. To confirm that the RSC complex is generally required for NER, we used the anchor-away method (Haruki et al. 2008) to rapidly deplete the essential Sth1 subunit, which forms the catalytic core of RSC, from the nucleus of cells (Fig. 3A). Microscopy analysis confirmed that the addition of rapamycin caused rapid depletion of the GFP-tagged Sth1–anchor-away strain (denoted Sth1-AA) from the nucleus into the cytoplasm (Fig. 3B). Moreover, growth of the Sth1-AA strain on rapamycin-containing plates resulted in loss of cell viability (Fig. 3C), confirming that nuclear localization of Sth1 is required for cell viability. In contrast, rapamycin treatment had no effect on growth of the wild-type–anchor-away (WT-AA) control (Fig. 3C), in which Sth1 is not anchor-away-tagged.
Figure 3.
Depletion of RSC results in a genome-wide defect in NER. (A) Schematic of the anchor-away method. Ribosomal protein is tagged with a FKBP12 tag, whereas the nuclear protein of interest (Sth1) is tagged with a FRB tag. Upon treatment with rapamycin, the two tags rapidly dimerize, effectively depleting the nucleus of the protein of interest. (B) Microscopic confirmation of Sth1 depletion. The Sth1–anchor-away (Sth1-AA) protein is tagged with GFP, allowing for visualization of Sth1 trafficking from the nucleus to the cytoplasm following 1 h of rapamycin treatment. (C) Confirmation of Sth1 depletion via plating assay. Dilutions of WT-AA and Sth1-AA yeast were plated on synthetic complete (SC) and SC + rapamycin plates. The essential Sth1 subunit is successfully depleted on SC + rapamycin plates, resulting in cell death. (D,E) Quantification of T4 endonuclease V alkaline gel analysis of CPD repair for WT-AA and Sth1-AA with or without rapamycin treatment. The percentage of CPDs repaired at each time point is plotted as the mean ± SEM of a minimum of three replicates. P-values were calculated using an unpaired t-test with Holm–Sidak correction for multiple hypothesis testing. (*) P ≤ 0.05. (F–H) CPD-seq analysis examining the repair of CPDs on both DNA strands across approximately 5000 transcribed regions of the genome in WT-AA and Sth1-AA cells treated with rapamycin or in a rsc2Δ mutant. The fraction of CPDs remaining is calculated as the ratio of damage after 2 h of repair compared with the damage immediately following UV irradiation (0 h). (I,J) Gene plot analysis of WT-AA and Sth1-AA cells to examine CPD repair within individual genes on each DNA strand. The fraction of CPDs remaining following 2-h repair is plotted. Genes ordered based on transcription frequency (Holstege et al. 1998).
We measured bulk repair in the WT-AA and Sth1-AA strains using the T4 endonuclease V alkaline gel assay. There was no difference in repair of UV-induced CPD lesions between the WT-AA and Sth1-AA cells when not treated with rapamycin (Fig. 3D). However, depletion of Sth1 by incubation of Sth1-AA cells with rapamycin (see Supplemental Methods) caused a large repair defect at all repair time points relative to the WT-AA control (Fig. 3E), in which only about a third of the CPDs are removed in Sth1-AA-depleted cells relative to WT-AA after 3 h of repair. These data are consistent with our rsc2Δ-mutant data and indicate that RSC is generally required for NER. Because of the loss of viability of the Sth1-AA cells upon rapamycin treatment, we were unable to test the UV sensitivity following anchor-away depletion. However, repression of Sth1 expression using the TET-off system (Czaja et al. 2014) caused increased UV sensitivity (Supplemental Fig. S4A), consistent with a role for Sth1 in NER of UV-induced DNA lesions.
RSC is required for efficient NER throughout the yeast genome
Our data indicate that deletion or depletion of RSC subunits causes a defect in repair in bulk genomic DNA; however, it is unclear whether RSC is required for repair throughout the genome or primarily in particular genomic or chromatin contexts. To address this question, we used the CPD-seq method to measure repair of UV-induced CPD lesions in rapamycin-treated Sth1-AA and control WT-AA cells. CPD-seq reads at lesion-forming dipyrimidine sequences decreased by ∼50% after 2 h of repair incubation in WT-AA cells (Supplemental Fig. S4B), but significantly more of these lesions were present in the Sth1-AA cells after 2-h repair (Supplemental Fig. S4C), consistent with the NER defect observed in Sth1-depleted cells measured via alkaline gel electrophoresis (Fig. 3E).
Analysis of CPD repair within and adjacent to approximately 5000 yeast genes revealed that Sth1 depletion caused a general defect in repair relative to the WT-AA control (Fig. 3F,G). In rapamycin-treated Sth1-AA cells, the fraction of unrepaired CPDs following 2-h repair was significantly higher not only on the NTS of yeast genes but also on the TS and adjacent intergenic regions (Fig. 3G; Supplemental Fig. S4D,F). There was still somewhat faster repair of the TS relative to the NTS in Sth1-depleted cells, but overall repair of the TS was significantly slower than the WT-AA control. CPD-seq analysis of the rsc2Δ strain also showed a general defect in repair in both the TS and NTS of yeast genes (Fig. 3H; Supplemental Fig. S4E,G). The magnitude of the repair defect in the rsc2Δ strain appeared to be less severe than that in the Sth1-AA-depleted strain, likely because a minor fraction of Rsc1-containing RSC complexes remained functional in this mutant strain. Gene plot analysis confirmed that repair was significantly diminished at nearly all genes in either the Sth1-AA- or rsc2Δ-mutant cells (Fig. 3I,J; Supplemental Fig. S5). Taken together, our CPD-seq data indicate that the RSC complex is required for efficient NER throughout the yeast genome.
RSC plays a direct role in promoting TC-NER
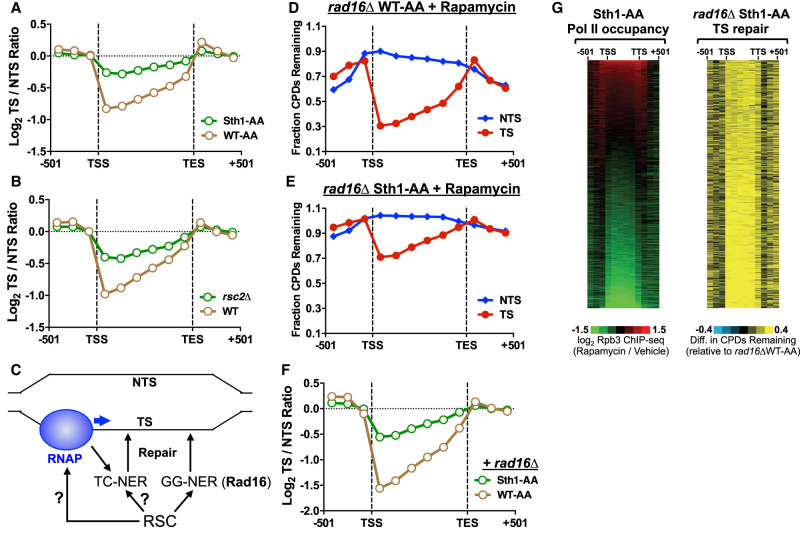
In Sth1-depleted cells, repair of the TS is significantly reduced relative to the WT-AA control (Fig. 3, cf. F and G), suggesting the RSC complex may play a direct role in TC-NER. To test this hypothesis, we analyzed the log ratio of unrepaired CPDs on the TS relative to the NTS for approximately 5000 yeast genes following 2 h of repair, as a measure of transcriptional strand asymmetry in repair owing to TC-NER. In the WT-AA control, the log2 TS/NTS ratio was significantly less than zero between the TSS and transcription end site (TES) (Fig. 4A), reflecting fewer unrepaired CPDs on the TS owing to TC-NER. Repair asymmetry was highest at the transcribed bin nearest the TSS, where there were nearly twofold fewer unrepaired CPDs on the TS relative to the NTS (i.e., log2 TS/NTS ratio ∼−1) (see Fig. 4A). This is consistent with previous reports indicating that TC-NER is more robust near the TSS (Li et al. 2018; Duan et al. 2020). In Sth1-depleted cells, repair asymmetry was significantly reduced (i.e., log2 TS/NTS ratio was closer to zero) (Fig. 4A), indicating that TC-NER is less efficient in removing CPDs from the TS. Consistently, repair asymmetry was also reduced in rsc2Δ cells relative to the WT control (Fig. 4B). Taken together, these results suggest that RSC is required for efficient TC-NER.
Figure 4.
RSC plays a direct role in transcription-coupled NER. (A) TC-NER efficiency is reduced in Sth1-AA cells relative to the WT-AA control, as reflected by the log ratio of unrepaired CPDs on the TS relative to the NTS being closer to zero (i.e., no repair asymmetry owing to TC-NER). CPD-seq data are for the 2-h repair time point relative to the 0-h control for approximately 5000 genes. (B) Same as panel A, except for rsc2Δ-mutant cells. (C) Schematic of RSC's potential roles in TC-NER. It is possible RSC may promote repair of the TS through GG-NER or may affect TC-NER indirectly (by altering RNA polymerase II [RNAP] transcription) or directly. (D,E) CPD-seq analysis examining the repair of CPDs on both DNA strands across approximately 5000 genes of the genome in GG-NER-deficient rad16Δ WT-AA and rad16Δ Sth1-AA cells treated with rapamycin. The fraction of CPDs remaining is calculated as the ratio of damage after 2 h of repair compared with the damage immediately following UV irradiation (0 h). (F) Same as panel A, except for GG-NER-deficient rad16Δ WT-AA and rad16Δ Sth1-AA cells. (G) Gene plot analysis of RNA polymerase II (Pol II) occupancy in Sth1-AA cells (left panel). Values calculated from log ratio of Rpb3 ChIP-seq data from Sth1-AA cells treated with rapamycin relative to the vehicle control (Kubik et al. 2018). Gene plot analysis of CPD repair on the TS of rad16Δ Sth1-AA cells. Left and right display relationship between RNA Pol II occupancy and unrepaired CPDs in each of the approximately 5000 genes mapped, ordered by decreasing Pol II occupancy in Sth1-AA-depleted cells.
The TS of yeast genes is repaired by both the TC-NER and GG-NER pathways (Fig. 4C), so it is theoretically possible that the decrease in repair of the TS in Sth1-depleted cells is owing to defective GG-NER and not to TC-NER. To test this possibility, we deleted the RAD16 gene, which is essential for GG-NER in yeast (Prakash and Prakash 2000), in each anchor-away strain (Sth1-AA and WT-AA) and repeated the CPD-seq experiments in rapamycin-treated cells. In the rad16Δ WT-AA strain, more unrepaired CPDs remained after 2 h along the NTS and flanking DNA (relative to the RAD16 WT-AA control) (cf. Figs. 3F and 4D), but repair of the TS was largely unaffected, owing to fast repair by TC-NER. However, in the rad16Δ Sth1-AA strain, the fraction of unrepaired CPDs on the TS was much higher than in the rad16Δ WT-AA control (Fig. 4D,E), suggesting that Sth1 affects repair of the TS independently of GG-NER. Loss of GG-NER because of the rad16Δ mutant resulted in greater repair asymmetry (i.e., a more negative log2 TS/NTS ratio) (Fig. 4F) in the WT-AA strain. In contrast, repair asymmetry was still significantly diminished in the rad16Δ Sth1-depleted cells (Fig. 4F). High-resolution analysis of repair around the TSS revealed that repair of the TS was reduced throughout the gene in the rad16Δ Sth1-AA strain (Supplemental Fig. S6A,B), including the TSS-proximal region, which is repaired independently of Rad26 (Supplemental Fig. S6C; see Duan et al. 2020). These findings show that RSC is required for efficient repair throughout the TS even in the absence of GG-NER (i.e., rad16Δ), indicating that RSC plays a role in both Rad26-dependent and Rad26-independent TC-NER.
Although our data suggest RSC plays a direct role in TC-NER, it is also possible that RSC affects TC-NER indirectly by regulating RNA Pol II transcription (Fig. 4C). To test this possibility, we stratified yeast genes by the change in RNA Pol II occupancy (measured by Rpb3 ChIP-seq) following anchor-away depletion of Sth1 (Kubik et al. 2018). Pol II occupancy is decreased at many genes following Sth1 depletion (Fig. 4G, left panel), consistent with the known role of RSC in promoting Pol II transcription (Ganguli et al. 2014; Spain et al. 2014). The fraction of unrepaired CPDs on the TS was significantly elevated in the rad16Δ Sth1-AA strain relative to the control (rad16Δ WT-AA), not only for genes with decreased Pol II occupancy (Fig. 4G, bottom) but also for genes with no change or even elevated Pol II occupancy following Sth1 depletion (Fig. 4G, top). To confirm this conclusion, we stratified yeast genes into three distinct groups based on whether Pol II occupancy was higher, unchanged, or diminished following Sth1 depletion (Supplemental Fig. S7A). Repair asymmetry was significantly diminished in each group (Supplemental Fig. S7B–D). This analysis indicates that repair of the TS is significantly diminished in Sth1-AA cells, independent of Sth1-mediated changes in Pol II occupancy. Taken together, these data suggest that RSC plays a direct role of facilitating TC-NER in chromatin.
RSC is required for repair in linker DNA and NFRs
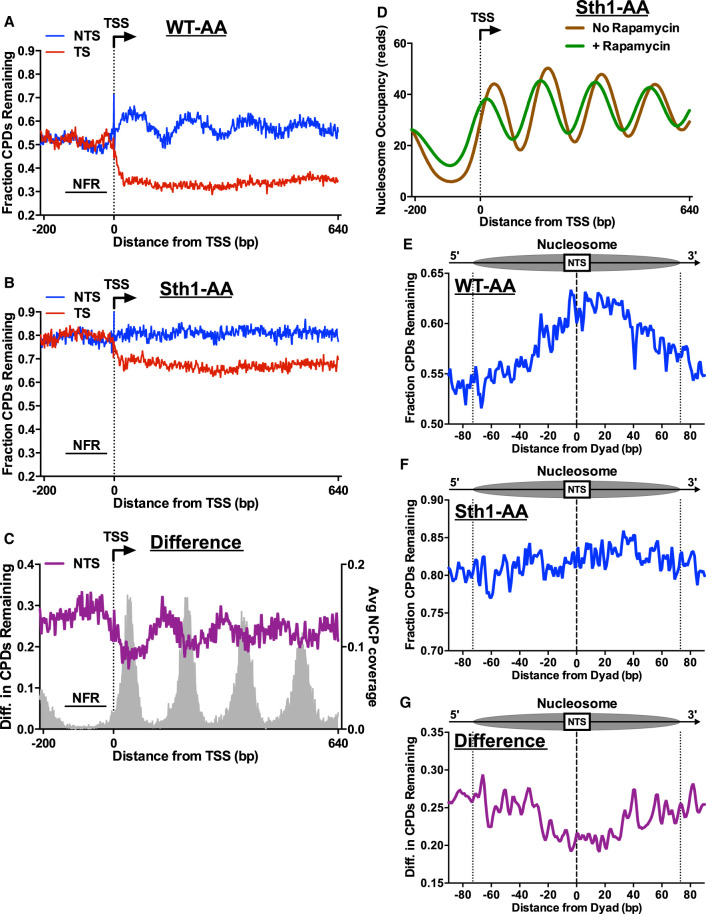
Because RSC is involved in nucleosome eviction and remodeling, we wondered whether the repair defect in the NTS and intergenic regions in Sth1-depleted cells was specifically associated with decreased repair in nucleosomes. To investigate this possibility, we plotted the CPD-seq data at single-nucleotide resolution around the TSS of approximately 5200 yeast genes. In WT-AA cells, there is a clear periodicity in repair along the NTS (Fig. 5A), with peaks of unrepaired CPDs associated with positioned nucleosomes in gene coding regions (Supplemental Fig. S8A). These peaks disappear in the GG-NER-defective rad16Δ mutant (Supplemental Fig. S6A), indicating they represent slow GG-NER in positioned nucleosomes, consistent with our previous report (Mao et al. 2020). Because RSC facilitates access to nucleosomal DNA (Clapier and Cairns 2009), we hypothesized that Sth1 depletion would cause an exaggerated repair periodicity, owing to even slower repair of CPDs in positioned nucleosomes relative to adjacent linker DNA or NFRs. However, in the Sth1-depleted strain, the nucleosome-associated periodicity in CPD repair is lost (Fig. 5B; Supplemental Fig. S8B). Similar analysis of the rsc2Δ mutant also showed little to no repair periodicity along the NTS (Supplemental Fig. S8C,D), consistent with the Sth1-AA results. Comparison between Sth1-AA and the WT-AA control revealed that Sth1 depletion causes elevated levels of unrepaired CPDs, particularly in linker DNA and NFRs (Fig. 5C).
Figure 5.
RSC promotes repair of both linker and nucleosomal DNA. CPD-seq analysis examining the repair of CPDs on both the TS and NTS of approximately 5200 yeast genes in WT-AA (A) and Sth1-AA (B). Data plotted for each nucleotide position spanning −200 bp upstream of and +640 bp downstream from the TSS. The fraction of CPDs remaining is calculated as the ratio of damage after 2 h of repair compared with the damage immediately following UV irradiation (0 h). (C) Plot depicting the difference in unrepaired CPDs in Sth1-AA cells compared with WT-AA cells. Difference in unrepaired CPDs was smoothed using a 3-nt window. Gray peaks correspond to the sequencing coverage of dyad positions for position nucleosomes (Weiner et al. 2015). (D) Nucleosome occupancy plot of Sth1-AA cells with and without rapamycin treatment to visualize nucleosome shifting that is known to occur in RSC-depleted cells. Data taken from Kubik et al. (2018). (E,F) Analysis of CPD repair within nucleosomes. Repair of the NTS of WT-AA and Sth1-AA cells was examined for nucleosomes in yeast genes to examine potential GG-NER defects in RSC-depleted cells. Dotted lines at positions −73 and +73 bp from the dyad center indicate the boundary of the nucleosome core particle. Nucleosome positions for WT-AA are from vehicle-treated Sth1-AA cells, whereas those for Sth1-AA are from rapamycin-treated Sth1-AA cells (Kubik et al. 2018). (G) Plot depicting the difference in unrepaired CPDs within nucleosomes in Sth1-AA cells compared with WT-AA cells. Difference in unrepaired CPDs was smoothed using a 3-nt window.
One possible explanation for these findings is that Sth1 depletion (or rsc2Δ) disrupts phasing of coding nucleosomes, thereby suppressing the nucleosome-mediated repair periodicity relative to the TSS (Kubik et al. 2018). To test this hypothesis, we analyzed published nucleosome positioning data from WT-AA and Sth1-AA yeast (Kubik et al. 2018). This analysis confirmed that nucleosome positioning relative to the TSS is somewhat altered in the Sth1-depleted cells, with many nucleosome positions shifted toward the NFR (Fig. 5D). However, nucleosomes in the Sth1-AA still show strong phasing relative to the TSS, even though our data indicate that this phasing is not reflected in repair along the NTS in Sth1-depleted cells.
To further test how nucleosomes modulate repair in Sth1-depleted cells, we analyzed CPD repair in strongly positioned nucleosomes in transcribed genes, using nucleosome dyad coordinates derived from untreated or Sth1-depleted yeast (Kubik et al. 2018). In WT-AA cells, unrepaired CPDs accumulate near nucleosome centers, whereas CPDs are more rapidly repaired near the nucleosomal DNA ends and in adjacent linker DNA (Fig. 5E). However, in Sth1-AA cells, the levels of unrepaired CPDs are elevated both within the nucleosome and in adjacent linker DNA (Fig. 5F, linker DNA shown outside of the dashed lines), even after accounting for Sth1-AA-dependent changes in nucleosome positioning (Kubik et al. 2018). Indeed, the difference in unrepaired CPDs between Sth1-depleted cells and the WT-AA control was greatest near the nucleosomal DNA ends and adjacent linker DNA (Fig. 5G). This analysis indicates that RSC is more important for repair in DNA regions distal from the nucleosomal dyad, including adjacent linker DNA, relative to the dyad-proximal region.
RSC is not required for repair of linker DNA and NFRs by the BER pathway
Because a previous study indicated that RSC is also required for BER in yeast (Czaja et al. 2014), we wondered whether Sth1 depletion would cause a similar genome-wide defect in BER, particularly in linker DNA or NFRs. To test this hypothesis, we used the NMP-seq method (Fig. 6A) to map the repair of MMS-induced N-methylpurine (NMP) lesions (Mao et al. 2017), primarily consisting of 7-methylguanine (7meG) and 3-methyladenine (3meA). Treatment of yeast cells with 0.4% MMS for 10 min resulted in a significant enrichment of NMP-seq reads associated with lesions at the G and A nucleotides, relative to the “No MMS” control (Supplemental Fig. S9A). This result is consistent with MMS inducing 7meG and, to a lesser extent, 3meA lesions. Following a 2-h repair incubation, there were reduced numbers of NMP-seq reads at the G and A nucleotides (Supplemental Fig. S9A), reflecting ongoing BER. In Sth1-depleted cells, there was a higher proportion of G reads following 2 h of repair (Supplemental Fig. S9B), consistent with the published role of RSC in promoting repair of NMP lesions (Czaja et al. 2014). For these experiments, isogenic strains lacking the methyladenine DNA glycosylase (mag1Δ) were used as the 0-h control in order to eliminate repair during the 10-min exposure to MMS (Mao et al. 2017), and cells were arrested in the cell cycle using nocodazole in order to prevent replication-mediated dilution of NMP lesions.
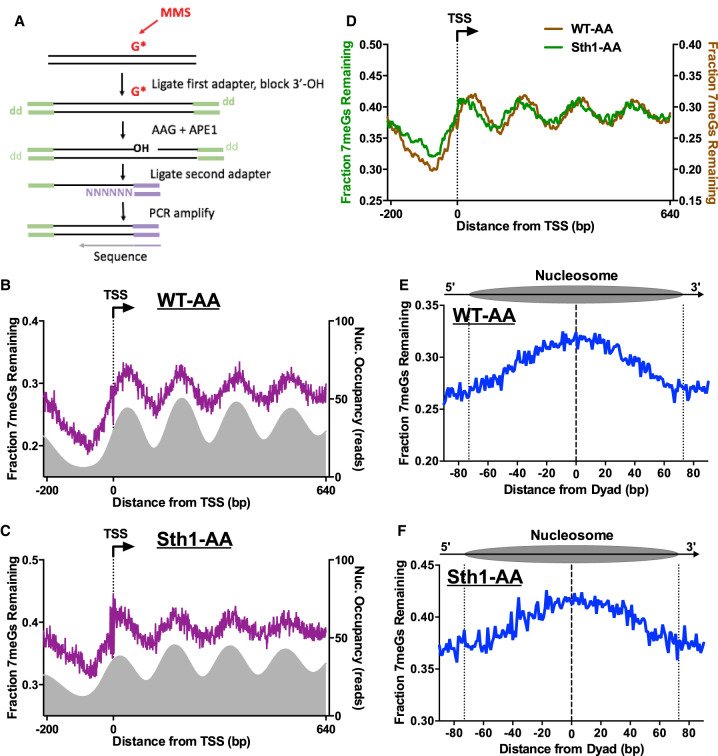
Figure 6.
RSC regulates BER in chromatin. (A) Schematic of the NMP-seq method. Yeast was exposed to 0.4% MMS to induce alkylation damage (primarily 7-methylguanine [7meG], represented by G*), and the resulting N-methylpurine (NMP) lesions are mapped using the indicated method. Adapted from Mao et al. (2017). (B,C) NMP-seq analysis of unrepaired 7meG lesions remaining after 2 h of repair of WT-AA (B) and Sth1-AA cells treated with rapamycin (C). Because 7meG lesions are not thought to be repaired by TC-NER, both DNA strands are combined and plotted in aggregate. Data are shown for positions −200 bp upstream of and +640 bp downstream from the TSS. Nucleosome positions for WT-AA are from vehicle-treated Sth1-AA cells, whereas those for Sth1-AA are from rapamycin-treated Sth1-AA cells (Kubik et al. 2018). (D) Overlay of unrepaired 7meG lesions following 2-h repair in WT-AA and Sth1-AA cells treated with rapamycin to highlight that the shift in mapped lesions corresponds to the shift in nucleosomes toward the TSS that occurs in RSC-depleted cells. NMP-seq data are smoothed in GraphPad Prism using a second-order polynomial and 7 bp on each side. (E,F) NMP-seq analysis of 7meG repair on both DNA strands of nucleosomes in WT-AA and Sth1-AA cells. Nucleosome positions for WT-AA are from vehicle-treated Sth1-AA cells, whereas those for Sth1-AA are from rapamycin-treated Sth1-AA cells (Kubik et al. 2018). Dotted lines at positions −73 and +73 bp from the dyad center indicate the boundary of the nucleosome core particle.
We used NMP-seq to analyze BER in both WT and Sth1-depleted cells. We focused on repair of 7meG lesions, because these are the predominant lesions induced by MMS and their repair is modulated by nucleosomes (Mao et al. 2017). Analysis of BER in WT-AA cells near the TSS for approximately 5200 genes revealed peaks of unrepaired 7meGs on both DNA strands (Fig. 6B), corresponding to the locations of strongly positioned nucleosomes (Fig. 6B, gray peaks). In Sth1-depleted cells, there were also peaks of unrepaired 7meGs associated with positioned nucleosomes in yeast coding regions (Fig. 6C), although the locations of these peaks were shifted toward the TSS relative to the WT control (Fig. 6D). This is consistent with the shift in nucleosome positions toward the NFR in Sth1-depleted cells (cf. Figs. 6D and 5D). In both WT and Sth1-depleted cells, BER is relatively faster in NFRs upstream of the TSS compared with gene coding regions (Fig. 6B,C), although BER is generally slower in the Sth1-depleted cells, including at NFRs. This analysis indicates that in contrast to repair of CPD lesions, a nucleosome-modulated repair pattern for NMP lesions is clearly evident in Sth1-depleted cells.
Analysis of repair in strongly positioned nucleosomes revealed more unrepaired 7meGs located in nucleosome centers relative to the nucleosomal DNA ends or adjacent linker DNA in the WT-AA control (Fig. 6E). These results indicate that BER is inhibited in nucleosomes relative to flanking linker DNA, consistent with our previous findings (Mao et al. 2017). A similar repair pattern was observed in positioned nucleosomes in Sth1-depleted cells (Fig. 6E,F) after accounting for Sth1-mediated changes in nucleosome positioning (Kubik et al. 2018). Taken together, these results indicate that although BER of 7meG lesions is generally slower in Sth1-depleted cells, Sth1 is not especially required for BER of 7meG lesions in NFRs or adjacent linker DNA, as is the case for NER of CPDs. Hence, our data reveal that the NER and BER pathways have differing requirements for RSC in promoting repair of lesions in linker DNA and NFRs.
Discussion
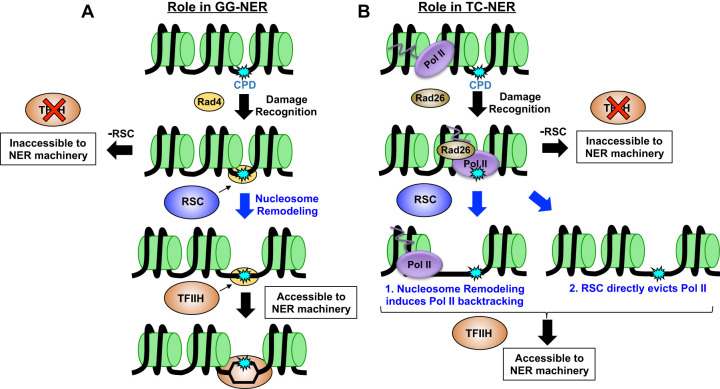
Although previous studies linked both SWI/SNF and RSC to NER, the specific contributions of these ACR complexes to the repair of UV damage across the genome were unclear. Here, we used the high-resolution CPD-seq method to define the specific genomic and chromatin contexts in which each ACR functions in NER. We have shown that SWI/SNF is largely dispensable for NER across the genome but does promote NER at specific genes, including those encoding ribosomal proteins. In contrast, our data indicate that RSC has a general role in promoting NER across the genome, being required for both the GG-NER and TC-NER subpathways. RSC not only is important for GG-NER of CPD lesions in nucleosomal DNA but also is especially important for efficient repair in NFRs and linker DNA. However, these RSC-dependent effects are unique to NER, as Sth1 depletion does not have the same effect on repair of 7meG lesions by the BER pathway in these chromatin contexts. We have further shown that RSC directly promotes TC-NER, independent of its effects on GG-NER or RNA Pol II transcription. Taken together, our data suggest a model in which RSC promotes NER by facilitating steps after lesion recognition for both NER subpathways in a diverse array of chromatin and genomic contexts (Fig. 7).
Figure 7.
Working model of role of RSC in NER. Our data suggest RSC plays a role in both subpathways of NER. (A) In GG-NER, we propose RSC is recruited to UV damage through Rad4 and is subsequently tasked with remodeling nucleosomes in order to increase accessibility to TFIIH and/or other downstream NER proteins. (B) In TC-NER, RSC may be recruited to remodel nucleosomes to induce RNA Pol II backtracking, or may directly act on RNA Pol II and aid in its eviction from the CPD lesion to promote subsequent repair.
Repair analysis, using both bulk T4 endonuclease V digestion coupled to alkaline gel electrophoresis and our CPD-seq method, indicates that the Snf5 and Snf6 subunits of the SWI/SNF complex are not generally required for NER. This is consistent with the observation that these mutants lack a UV-sensitivity phenotype under normal growth conditions (e.g., rich media containing glucose). A previous study found that the snf6Δ mutant caused a defect in CPD repair at the silent HML locus (Gong et al. 2006). Analysis of the snf6Δ CPD-seq data revealed marginally slower repair of at least part of the HML locus (Supplemental Fig. S10), roughly consistent with this prior study. It is possible that SWI/SNF mutants may cause a more severe repair defect in the differing growth conditions used in a previous study (e.g., media containing galactose) (Gong et al. 2006), as SWI/SNF is important for the regulation of galactose metabolism (Peterson et al. 1998), which could affect repair efficiency. Our data indicate that even under standard growth conditions, SWI/SNF is important for efficient repair of specific subsets of yeast genes. Many of these genes (e.g., ribosomal protein genes, genes involved in glycolysis, gluconeogenesis, and cell wall) are known regulatory targets of the SWI/SNF complex (Dutta et al. 2017) and have chromatin modifications conducive to SWI/SNF recruitment (Supplemental Fig. S11), indicating that SWI/SNF targeting to these genes may not only affect their expression but also facilitate their repair. In some cases (e.g., genes involved in glycolysis, gluconeogenesis, and cell wall), SWI/SNF facilitates TC-NER by promoting transcription of these target genes. However, for other target genes (e.g., ribosomal protein genes), SWI/SNF likely promotes repair of the NTS by facilitating DNA access to NER factors. Elsewhere in the genome, repair of CPD lesions is marginally faster in SWI/SNF-mutant strains, particularly snf5Δ. It is possible that in accessible chromatin contexts, in which SWI/SNF chromatin remodeling is dispensable, the physical interaction between SWI/SNF and Rad4 following UV irradiation (Gong et al. 2006) may actually inhibit repair, perhaps by interfering with the subsequent binding and recruitment of TFIIH. Alternatively, SWI/SNF may direct repair toward specific gene subsets and hard-to-repair heterochromatic regions such as the HML locus (Gong et al. 2006), thereby causing a delay in NER throughout the remainder of the genome. Future studies will be needed to test these hypotheses.
In contrast to the more localized role of SWI/SNF in repair, our data indicate that the RSC complex is generally required for NER throughout the yeast genome. Cells lacking either the Rsc2 or Sth1 subunits show a similar, significant defect in repair in both the TS and NTS of yeast genes, as well as nontranscribed intergenic DNA. These findings are consistent with a previous report (Srivas et al. 2013), which showed that the rsc2Δ mutant caused defective repair of both the TS and NTS of the RPB2 gene. These findings are also consistent with the UV sensitivity of yeast strains lacking Rsc2 (Fig. 1A). One possible explanation for its broad role in NER is that RSC may regulate the expression of one or more key NER genes. Indeed, depletion of the SMARCA4 (also known as BRG1) homolog of Sth1 causes an NER defect in human cells owing to down-regulation of the gene encoding the subunit 1 of general transcription factor IIH, GTF2H1 (also known as P62) (Ribeiro-Silva et al. 2018). However, a previous study revealed that the rsc2Δ mutant in yeast does not affect the expression of any known NER genes (Srivas et al. 2013), despite its broad effect on NER activity. Moreover, RT-qPCR analysis of the TFB1 gene, which is the yeast homolog of the gene encoding the p62 subunit (i.e., GTF2H1), shows no change in expression in the rsc2Δ mutant (Supplemental Fig. S12), consistent with published expression data (Hu et al. 2007). Furthermore, rapid depletion of the essential Sth1 subunit using the anchor-away method, which should mitigate indirect effects on gene expression, also caused a similar general defect in repair. Taken together, these results indicate that the repair defects in RSC-depleted cells are unlikely to be caused by expression changes in NER genes but are likely a direct consequence of RSC inactivation.
Given the primary function of RSC as an ATP-dependent nucleosome remodeler, we hypothesized that RSC would primarily be required for repair in nucleosomes. However, our data indicate that RSC is important for repair in linker DNA and NFRs, to the extent that repair of CPD lesions in rsc2Δ- or Sth1-depleted cells is no longer modulated by nucleosomes (i.e., slower in nucleosome centers relative to adjacent linker DNA). Our analysis indicates that this is not simply owing to disrupted nucleosome phasing in cells lacking RSC, as nucleosomes still show strong positioning in Sth1-depleted cells (Kubik et al. 2018), and NMP-seq analysis of the repair of MMS lesions shows significant nucleosome modulation of BER activity (Fig. 6). Rather, this analysis suggests that RSC is required for efficient repair of CPD lesions located in accessible regions of the genome, such as linker DNA or NFRs.
Based on these findings, we propose that RSC does not primarily facilitate lesion access and recognition by Rad4/Rad23 during GG-NER but instead promotes subsequent steps of the NER pathway (Fig. 7A). Following lesion detection, Rad4 recruits TFIIH and Rad14 to verify the lesion and unwind ∼20–30 nucleotides (nt) of adjacent DNA. Hence, although lesions located in ∼10–15 nt of linker DNA between yeast nucleosomes should be readily detectable by Rad4, subsequent recruitment and DNA unwinding by TFIIH likely would be impeded by adjacent nucleosomes (Fig. 7A). We propose that RSC functions to promote DNA access to TFIIH and other downstream NER factors after lesion recognition by Rad4 via nucleosome remodeling and/or eviction (Fig. 7A; Supplemental Fig. S13). This model would explain our data indicating RSC depletion causes repair defects in otherwise accessible DNA (e.g., linker, NFR). Moreover, this model is consistent with a report indicating that Rad4 is required to recruit RSC to UV damage in yeast (Srivas et al. 2013), indicating RSC is recruited after lesion recognition. This model is also consistent with a prior study showing that TFIIH recruitment to UV lesions requires ATP-dependent chromatin remodeling in mammalian cells (Rüthemann et al. 2017).
Our data indicate that RSC plays a direct role in promoting TC-NER. Previous studies have hinted that ACRs like RSC may function in TC-NER (Lans et al. 2010), but definitive evidence for this role has been lacking. Chromatin remodelers also influence RNA Pol II transcription and repair by the GG-NER pathway, both of which could cause defects in repair of the TS of yeast genes. Here, we have shown that RSC directly affects TC-NER even in GG-NER mutants (rad16Δ), and this repair defect is independent of changes in RNA Pol II transcription. We hypothesize that RSC may promote a shared downstream step of both NER subpathways, such as TFIIH recruitment (Fig. 7A,B). This could occur by RSC remodeling neighboring nucleosomes to facilitate TFIIH loading or RNA Pol II backtracking. Alternatively, RSC could directly stimulate RNA Pol II backtracking or eviction during TC-NER. Such a possibility has been hinted at in a previous report, which suggested that the Ino80 chromatin remodeling complex promotes RNA Pol II eviction and degradation (Lafon et al. 2015). Future studies will be required to test these hypotheses to better define the mechanism by which RSC functions in TC-NER.
In summary, our study has defined the distinct roles for the SWI/SNF and RSC chromatin remodeling complexes in repairing UV damage across the genome of a model eukaryote. Although RSC is generally required for NER, SWI/SNF facilitates the repair of specific subsets of genes, particularly those to which it is targeted during transcription regulation. Our data further suggest that RSC may function in a shared step downstream from lesion recognition in both the GG-NER and TC-NER pathways. It is tempting to speculate that RSC may be required to remove DNA-bound obstacles (e.g., nucleosomes or stalled RNA Pol II) that would otherwise impede recruitment of TFIIH and other core NER factors following lesion recognition. By analogy, RSC plays a similar role in removing obstacles that otherwise prevent preinitiation complex formation during transcription initiation (Kubik et al. 2018), which includes TFIIH recruitment and DNA unwinding. In contrast, BER factors require a much smaller DNA footprint, which can explain why BER is less affected by RSC depletion. As both XPC/Rad4 and accessory factors (e.g., UV-DDB) can bind UV damage resident in nucleosomes (Fei et al. 2011; Puumalainen et al. 2016; Matsumoto et al. 2019), this model suggests that chromatin remodeling is primarily required for the subsequent lesion verification and incision steps, presumably because such an extensive region of DNA must be bound by repair factors and unwound during these NER steps. Because RSC and SWI/SNF homologs (e.g., SMARCA4 and SMARCA2 [also known as BRM]) are mutated in a variety of cancers (Moloney et al. 2009; Weissman and Knudsen 2009), including UV-exposed skin cancers such as melanoma (Reisman et al. 2009), the role of these ACRs in NER may have important ramifications for carcinogenesis. It will be important to investigate in future studies to what extent ACR dysregulation in cancer contributes to genome instability and chemotherapy resistance.
Methods
Yeast strains
Strains used in this study are listed in Supplemental Table S1. Details of the yeast strain construction are provided in the Supplemental Methods.
Alkaline gel assay of CPD repair
Alkaline gel analysis to measure CPD repair in bulk genomic DNA isolated from both WT and mutant yeast was performed essentially as previously described (Hodges et al. 2019). Details are provided in the Supplemental Methods.
CPD-seq
The CPD-seq protocol was performed as previously described (Mao et al. 2016; Mao and Wyrick 2020). CPD levels were mapped in both WT and mutant yeast either immediately after UV irradiation (0 h) or after 2 h of repair. Analysis of strand-specific CPD repair in yeast genes and nucleosomes was performed following published methods (Duan et al. 2020; Mao et al. 2020), with modifications. For additional details regarding the CPD-seq experiments, data processing, and data analysis, see the Supplemental Methods.
SWI/SNF gene set analysis
Genes with decreased repair of either the NTS or TS in the snf5Δ and/or snf6Δ mutants were identified by calculating the average change in fraction CPDs remaining in the snf5- or snf6-mutant data relative to WT across the six coding region bins (e.g., between the TSS and NTS). Genes with an average change in fraction CPDs remaining greater than or equal to the chosen threshold (0.03 for the TS and 0.04 for the NTS) in either mutant were included in each gene set (62 genes for TS and 170 for the NTS). Gene set enrichment analysis was performed using the FunSpec tool (Robinson et al. 2002). Gene expression analysis was performed using RegulatorDB (Choi and Wyrick 2017).
NMP-seq
NMP-seq was used to map the repair of alkylation damage in WT-AA or Sth1-AA yeast following treatment with MMS using our previously published protocol (Mao et al. 2017). Additional details about the NMP-seq experiments and data processing are provided in the Supplemental Methods.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE161929, GSE161930, and GSE168369. Code for processing and analyzing the CPD-seq and NMP-seq data is available in the Supplemental Code.
Supplementary Material
Acknowledgments
We thank Mark Wildung and Wei Wei Du for technical assistance with Ion Proton sequencing. We thank Dr. Frank Holstege for providing yeast strains and Dr. David Shore for providing nucleosome coordinates. We thank Dr. Steven Roberts for providing purified T4 endonuclease V and Angelica Washington for helping to construct yeast strains. This research was supported by National Institute of Environmental Health Sciences grants R21ES027937 (to J.J.W.), R03ES027945 (to P.M.), R21ES029302 (to P.M. and J.J.W.), and R01ES028698 (to J.J.W. and M.J.S.). K.A.B. was funded by a National Institute of General Medical Sciences training grant (T32GM008336).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.274373.120.
Competing interest statement
The authors declare no competing interests.
References
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887. 10.1016/j.molcel.2008.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Mao P, Smerdon MJ, Wyrick JJ, Roberts SA. 2018. Nucleosome positions establish an extended mutation signature in melanoma. PLoS Genet 14: e1007823. 10.1371/journal.pgen.1007823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell 4: 715–723. 10.1016/S1097-2765(00)80382-2 [DOI] [PubMed] [Google Scholar]
- Choi JA, Wyrick JJ. 2017. RegulatorDB: a resource for the analysis of yeast transcriptional regulation. Database (Oxford) 2017: bax058. 10.1093/database/bax058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Czaja W, Mao P, Smerdon MJ. 2012. The emerging roles of ATP-dependent chromatin remodeling enzymes in nucleotide excision repair. Int J Mol Sci 13: 11954–11973. 10.3390/ijms130911954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja W, Mao P, Smerdon MJ. 2014. Chromatin remodelling complex RSC promotes base excision repair in chromatin of Saccharomyces cerevisiae. DNA Repair (Amst) 16: 35–43. 10.1016/j.dnarep.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiovanna JJ, Kraemer KH. 2012. Shining a light on xeroderma pigmentosum. J Invest Dermatol 132: 785–796. 10.1038/jid.2011.426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M, Selvam K, Wyrick JJ, Mao P. 2020. Genome-wide role of Rad26 in promoting transcription-coupled nucleotide excision repair in yeast chromatin. Proc Natl Acad Sci 117: 18608–18616. 10.1073/pnas.2003868117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Sardiu M, Gogol M, Gilmore J, Zhang D, Florens L, Abmayr SM, Washburn MP, Workman JL. 2017. Composition and function of mutant Swi/Snf complexes. Cell Rep 18: 2124–2134. 10.1016/j.celrep.2017.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Kaczmarek N, Luch A, Glas A, Carell T, Naegeli H. 2011. Regulation of nucleotide excision repair by UV-DDB: prioritization of damage recognition to internucleosomal DNA. PLoS Biol 9: e1001183. 10.1371/journal.pbio.1001183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. 2006. DNA repair and mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- Ganguli D, Chereji RV, Iben JR, Cole HA, Clark DJ. 2014. RSC-dependent constructive and destructive interference between opposing arrays of phased nucleosomes in yeast. Genome Res 24: 1637–1649. 10.1101/gr.177014.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Fahy D, Smerdon MJ. 2006. Rad4–Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol 13: 902–907. 10.1038/nsmb1152 [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. 2008. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol 9: 958–970. 10.1038/nrm2549 [DOI] [PubMed] [Google Scholar]
- Hara R, Mo J, Sancar A. 2000. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol Cell Biol 20: 9173–9181. 10.1128/MCB.20.24.9173-9181.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458. 10.1016/j.cell.2009.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31: 925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Hinz JM, Czaja W. 2015. Facilitation of base excision repair by chromatin remodeling. DNA Repair (Amst) 36: 91–97. 10.1016/j.dnarep.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges AJ, Plummer DA, Wyrick JJ. 2019. NuA4 acetyltransferase is required for efficient nucleotide excision repair in yeast. DNA Repair (Amst) 73: 91–98. 10.1016/j.dnarep.2018.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. 10.1016/S0092-8674(00)81641-4 [DOI] [PubMed] [Google Scholar]
- Hu Z, Killion PJ, Iyer VR. 2007. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet 39: 683–687. 10.1038/ng2012 [DOI] [PubMed] [Google Scholar]
- Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, Korber P. 2016. Genomic nucleosome organization reconstituted with pure proteins. Cell 167: 709–721.e12. 10.1016/j.cell.2016.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubik S, O'Duibhir E, de Jonge WJ, Mattarocci S, Albert B, Falcone JL, Bruzzone MJ, Holstege FCP, Shore D. 2018. Sequence-directed action of RSC remodeler and general regulatory factors modulates +1 nucleosome position to facilitate transcription. Mol Cell 71: 89–102.e5. 10.1016/j.molcel.2018.05.030 [DOI] [PubMed] [Google Scholar]
- Lafon A, Taranum S, Pietrocola F, Dingli F, Loew D, Brahma S, Bartholomew B, Papamichos-Chronakis M. 2015. INO80 chromatin remodeler facilitates release of RNA Polymerase II from chromatin for ubiquitin-mediated proteasomal degradation. Mol Cell 60: 784–796. 10.1016/j.molcel.2015.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Marteijn JA, Schumacher B, Hoeijmakers JH, Jansen G, Vermeulen W. 2010. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet 6: e1000941. 10.1371/journal.pgen.1000941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Hoeijmakers JHJ, Vermeulen W, Marteijn JA. 2019. The DNA damage response to transcription stress. Nat Rev Mol Cell Biol 20: 766–784. 10.1038/s41580-019-0169-4 [DOI] [PubMed] [Google Scholar]
- Li W, Adebali O, Yang Y, Selby CP, Sancar A. 2018. Single-nucleotide resolution dynamic repair maps of UV damage in Saccharomyces cerevisiae genome. Proc Natl Acad Sci U S A 115: E3408–E3415. 10.1073/pnas.1801687115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Hayes JJ. 2010. When push comes to shove: SWI/SNF uses a nucleosome to get rid of a nucleosome. Mol Cell 38: 484–486. 10.1016/j.molcel.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Smerdon MJ. 2000. Nucleotide excision repair of the 5 S ribosomal RNA gene assembled into a nucleosome. J Biol Chem 275: 23729–23735. 10.1074/jbc.M002206200 [DOI] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. 2006. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci 103: 3090–3093. 10.1073/pnas.0511050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Wyrick JJ. 2019. Organization of DNA damage, excision repair, and mutagenesis in chromatin: a genomic perspective. DNA Repair (Amst) 81: 102645. 10.1016/j.dnarep.2019.102645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Wyrick JJ. 2020. Genome-wide mapping of UV-induced DNA damage with CPD-seq. Methods Mol Biol 2175: 79–94. 10.1007/978-1-0716-0763-3_7 [DOI] [PubMed] [Google Scholar]
- Mao P, Smerdon MJ, Roberts SA, Wyrick JJ. 2016. Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc Natl Acad Sci 113: 9057–9062. 10.1073/pnas.1606667113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Brown AJ, Malc EP, Mieczkowski PA, Smerdon MJ, Roberts SA, Wyrick JJ. 2017. Genome-wide maps of alkylation damage, repair, and mutagenesis in yeast reveal mechanisms of mutational heterogeneity. Genome Res 27: 1674–1684. 10.1101/gr.225771.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Brown AJ, Esaki S, Lockwood S, Poon GMK, Smerdon MJ, Roberts SA, Wyrick JJ. 2018. ETS transcription factors induce a unique UV damage signature that drives recurrent mutagenesis in melanoma. Nat Commun 9: 2626. 10.1038/s41467-018-05064-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Smerdon MJ, Roberts SA, Wyrick JJ. 2020. Asymmetric repair of UV damage in nucleosomes imposes a DNA strand polarity on somatic mutations in skin cancer. Genome Res 30: 12–21. 10.1101/gr.253146.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Cavadini S, Bunker RD, Grand RS, Potenza A, Rabl J, Yamamoto J, Schenk AD, Schubeler D, Iwai S, et al. 2019. DNA damage detection in nucleosomes involves DNA register shifting. Nature 571: 79–84. 10.1038/s41586-019-1259-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney FJ, Lyons JG, Bock VL, Huang XX, Bugeja MJ, Halliday GM. 2009. Hotspot mutation of Brahma in non-melanoma skin cancer. J Invest Dermatol 129: 1012–1015. 10.1038/jid.2008.319 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Zhao Y, Chait BT. 1998. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J Biol Chem 273: 23641–23644. 10.1074/jbc.273.37.23641 [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. 2000. Nucleotide excision repair in yeast. Mutat Res 451: 13–24. 10.1016/S0027-5107(00)00037-3 [DOI] [PubMed] [Google Scholar]
- Puumalainen MR, Rüthemann P, Min JH, Naegeli H. 2016. Xeroderma pigmentosum group C sensor: unprecedented recognition strategy and tight spatiotemporal regulation. Cell Mol Life Sci 73: 547–566. 10.1007/s00018-015-2075-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Winston F. 2012. Chromatin and transcription in yeast. Genetics 190: 351–387. 10.1534/genetics.111.132266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D, Glaros S, Thompson EA. 2009. The SWI/SNF complex and cancer. Oncogene 28: 1653–1668. 10.1038/onc.2009.4 [DOI] [PubMed] [Google Scholar]
- Ribeiro-Silva C, Aydin ÖZ, Mesquita-Ribeiro R, Slyskova J, Helfricht A, Marteijn JA, Hoeijmakers JHJ, Lans H, Vermeulen W. 2018. DNA damage sensitivity of SWI/SNF-deficient cells depends on TFIIH subunit p62/GTF2H1. Nat Commun 9: 4067. 10.1038/s41467-018-06402-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Grigull J, Mohammad N, Hughes TR. 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35. 10.1186/1471-2105-3-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüthemann P, Balbo Pogliano C, Codilupi T, Garajovà Z, Naegeli H. 2017. Chromatin remodeler CHD1 promotes XPC-to-TFIIH handover of nucleosomal UV lesions in nucleotide excision repair. EMBO J 36: 3372–3386. 10.15252/embj.201695742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer OD. 2013. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 5: a012609. 10.1101/cshperspect.a012609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdon MJ, Thoma F. 1990. Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell 61: 675–684. 10.1016/0092-8674(90)90479-X [DOI] [PubMed] [Google Scholar]
- Spain MM, Ansari SA, Pathak R, Palumbo MJ, Morse RH, Govind CK. 2014. The RSC complex localizes to coding sequences to regulate Pol II and histone occupancy. Mol Cell 56: 653–666. 10.1016/j.molcel.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivas R, Costelloe T, Carvunis AR, Sarkar S, Malta E, Sun SM, Pool M, Licon K, van Welsem T, van Leeuwen F, et al. 2013. A UV-induced genetic network links the RSC complex to nucleotide excision repair and shows dose-dependent rewiring. Cell Rep 5: 1714–1724. 10.1016/j.celrep.2013.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, de Pril R, Tasseron-de Jong JG, Brouwer J. 1999. RNA polymerase II transcription suppresses nucleosomal modulation of UV-induced (6-4) photoproduct and cyclobutane pyrimidine dimer repair in yeast. Mol Cell Biol 19: 934–940. 10.1128/MCB.19.1.934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hsieh TH, Appleboim A, Chen HV, Rahat A, Amit I, Rando OJ, Friedman N. 2015. High-resolution chromatin dynamics during a yeast stress response. Mol Cell 58: 371–386. 10.1016/j.molcel.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman B, Knudsen KE. 2009. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res 69: 8223–8230. 10.1158/0008-5472.CAN-09-2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Thoma F. 1997. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J 16: 5046–5056. 10.1093/emboj/16.16.5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Teng Y, Liu H, Reed SH, Waters R. 2005. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc Natl Acad Sci 102: 8650–8655. 10.1073/pnas.0501458102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.