Abstract
Multiple myeloma (MM) is a currently incurable malignancy of antibody-secreting plasma cells. Long non-coding RNAs (lncRNAs) have been recognised as an important class of regulatory molecules which are increasingly implicated in tumorigenesis. While recent studies have demonstrated changes in expression of lncRNAs in MM, the functional significance and molecular pathways downstream of these changes remain poorly characterised. In this study, we have performed CRISPR-mediated deletion of the locus encoding the lncRNA Colorectal Neoplasia Differentially Expressed (CRNDE), a known oncogenic lncRNA that is overexpressed in plasma cells of MM patients and is a marker of poor prognosis. We found that CRISPR-mediated deletion of the CRNDE locus in MM cells decreases proliferation and adhesion properties, increases sensitivity to Dexamethasone and reduces tumour growth in an in vivo xenograft model. Transcriptomic profiling in CRNDE-deleted MM cells demonstrated that CRNDE activates expression of a number of genes previously implicated in the aetiology of MM, including IL6R. We further demonstrate that deletion of the CRNDE locus diminishes IL6 signalling and proliferative responses in MM cells. Altogether this study reveals the IL6 signalling pathway as a novel mechanism by which CRNDE impacts upon MM cell growth and disease progression.
Subject terms: Oncogenes, Myeloma, Cell signalling
Introduction
Multiple myeloma (MM), a clonal malignancy caused by the uncontrolled proliferation in the bone marrow of immunoglobulin-secreting plasma cells, is a disease of elderly adults which accounts for 10% of all haematological malignancies [1, 2]. The disease progresses from a pre-malignant precursor condition (monoclonal gammopathy of undetermined significance), through an indolent intermediate stage known as smouldering multiple myeloma, before evolving to full symptomatic myeloma, whose clinical features include immunosuppression, anaemia, lytic bone lesions, hypercalcaemia and renal failure [3, 4]. Despite significant therapeutic advances and improved survival times, disease relapse occurs frequently and at present MM remains essentially incurable [4, 5]. MM is characterised by a high degree of complexity, both in the primary genetic events driving initial transformation of plasma cells, as well as in the secondary mutations, copy number and gene expression changes, which control clonal evolution, disease progression and relapse [6–9]. Understanding the full range of secondary events implicated in this disease and the downstream molecular pathways by which they impact upon disease evolution should ultimately result in more tailored therapeutic strategies capable of achieving longer-term responses and eventually complete cure.
Over the last two decades, dysregulation of the activity of long non-coding RNAs (lncRNAs) has been increasingly implicated in the onset, aetiology and clinical heterogeneity of many solid and haematological tumours [10]. LncRNAs have been loosely defined as RNA molecules >200 nucleotides in length that have little or no protein-coding potential. There exist multiple sources of lncRNA annotations, with varying estimates of the number of lncRNA genes and proposed functional stratification [11]. Many lncRNAs act as regulators of gene transcription by recruiting chromatin-modifying proteins to influence the epigenetic state, by coordinating interactions between distal regulatory elements, or by establishing long-range chromosomal regulatory domains and nuclear bodies [12]. LncRNAs can also influence gene expression at the post-transcriptional level, by influencing mRNA splicing and stability, or by acting as molecular sponges which sequester and dampen the efficacy of microRNAs (miRNAs) [12, 13]. In MM, microarray and RNA-seq studies have identified large numbers of lncRNAs whose expression is deregulated in tumour plasma cells, and revealed prognostic lncRNA signatures [14–18]. Together these results indicate that lncRNAs exert an independent effect on disease outcome in MM and suggest the importance of characterising the molecular pathways by which specific dysregulated lncRNAs impact upon myeloma cell growth, disease progression and therapeutic response.
Here we report on CRISPR-mediated deletion of the locus encoding Colorectal Neoplasia Differentially Expressed (CRNDE), a lncRNA that is overexpressed in plasma cells of MM patients, and is a poor prognostic marker. We demonstrate that deletion of the CRNDE locus reduces expression of the interleukin 6 (IL6) receptor in MM cells, and that this is associated with decreased IL6 signalling and proliferative responses, together with reduced tumorigenic properties in vitro and in vivo. Altogether this study reveals a previously unrecognised mechanistic pathway by which CRNDE impacts upon MM growth and disease progression and indicates that this lncRNA may be a valuable therapeutic target.
Materials and methods
Culture of MM cell lines
MM cell lines KMS11 [19], RPMI 8226 [20], OPM2 [21] and NC1-H929 [22] as well as RAJI (Burkitt’s lymphoma) [23] cell lines were cultured in RPMI-1640 medium (ThermoFisher Scientific, Waltham, MA, USA) supplemented with 10% (or 20% for KMS11) foetal calf serum (FCS), 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM L-glutamine. The LP1 (MM) [24] and HeLa (cervical carcinoma) [25] cell lines were cultured in DMEM (ThermoFisher Scientific) supplemented with 10% FCS, penicillin, streptomycin and L-glutamine as above. Cells were maintained at 37 °C in a humidified incubator with 5% CO2. IL6 stimulation experiments were carried out as described previously [26] using 2 ng/ml human IL6 (Peprotech, Rocky Hill, NJ, USA) in serum-starved KMS11 cells.
Primary plasma cells
MM patients were treated at the APHP Department of Immuno-Hematology, Hôpital Saint Louis, between 1997 and 2017 (n = 70; mean age at diagnosis = 52.7 years; mean duration of treatment = 6.0 years; 23 patients with t(4;14); 24 patients with t(11;14)). Samples were obtained with full informed consent by the APHP Hematology Laboratory of Hôpital Saint Louis in accordance with the Declaration of Helsinki and French law. Mononuclear cells were purified from bone marrow samples of MM patients and healthy donors by density gradient centrifugation using Ficoll. The selection of plasma cells was performed using human CD138 microbeads and MACS separation (Miltenyi Biotec, Bergisch Gladbach, Germany).
Proliferation assays
In total, 1 × 106 KMS11 cells in PBS were pulse labelled with 5 µM CellTrace™ Violet Dye (ThermoFisher Scientific) for 20 min at 37 °C. Labelling was quenched with complete medium. Cells were cultured for a further 9 days and cultures were serially sampled at the timepoints indicated and analysed by flow cytometry on a Canto II flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) to monitor loss of the dye due to proliferation. For IL6 stimulation experiments, half the culture volume was harvested for flow cytometry analysis at the times indicated, and was replaced with fresh minimal medium with or without IL6 (2 ng/ml) as appropriate. For MTT assays, 2000 cells of each clone were plated in wells of a 96-well plate in complete medium (5 technical replicate wells per clone). After the number of days indicated, cells were incubated with complete culture medium supplemented with 0.5 mg/ml MTT (Methylthiazolyldiphenyl-tetrazolium bromide) (Merck, St. Louis, MO, USA) for 4 h at 37 °C. Cells were lysed overnight at 37 °C by adding an equal volume of 10% SDS/0.01 M HCl. Absorbance (590 nm) of lysates was read in a CLARIOStar Plus plate reader (BMG Labtech, Cary, NC, USA).
Drug sensitivity
Cells were cultured in complete medium containing Bortezomib (Selleck Chemicals, Houston, TX, USA) or Dexamethasone (Mylan, Canonsburg, PA, USA) at the indicated concentrations for 72 h. Apoptosis was measured using the FITC Annexin V Apoptosis Detection kit with 7AAD (Biolegend, San Diego, CA, USA) according to the manufacturer’s instructions and flow cytometry analysis was carried out on a Canto II flow cytometer using DIVA software v8.0.1 (Becton Dickinson).
Adhesion assay
KMS11 clones were harvested using Enzyme-free Cell Dissociation Buffer (ThermoFisher Scientific) and washed with PBS. Twenty-four-well tissue culture plates were coated with Matrigel (Corning LifeSciences, Corning, NY, USA) (100 µg/ml in PBS) for 1 h at room temperature (RT). In total, 2 × 105 KMS11 cells were plated per well in 1 ml complete medium and incubated for 6 h at 37 °C. Non-adherent cells were removed by washing with PBS/1 mM CaCl2/1 mM MgCl2, and remaining adherent cells were fixed in 10% acetic acid/10% methanol for 15 min at RT. Cells were stained with Crystal Violet dye (5 mg/mL in 2% ethanol) for 10 min at RT, washed in distilled water and dried overnight. Stained cells were lysed in 200 µl 10% acetic acid and absorbance at 590 nm was measured using a CLARIOStar Plus plate reader (BMG Labtech).
In vivo tumorigenicity assay
Female NOD-SCID IL2Rγ−/− immunodeficient mice (8–12 weeks) were injected intra-caudally with 3 × 106 CRNDE+/+ or CRNDEΔ/Δ KMS11 cells. At 6 weeks post injection, bone marrows were harvested and stained with mouse-α-human CD38-FITC antibody (555459; Becton Dickinson) and analysed by flow cytometry on a Canto II flow cytometer (Becton Dickinson). All animal experiments were performed in accordance with the guidelines of the Institutional Review Board of the Institut de Recherche Saint Louis, Paris.
Results
The lncRNA CRNDE is upregulated in plasma cells of MM patients and is associated with poor prognosis
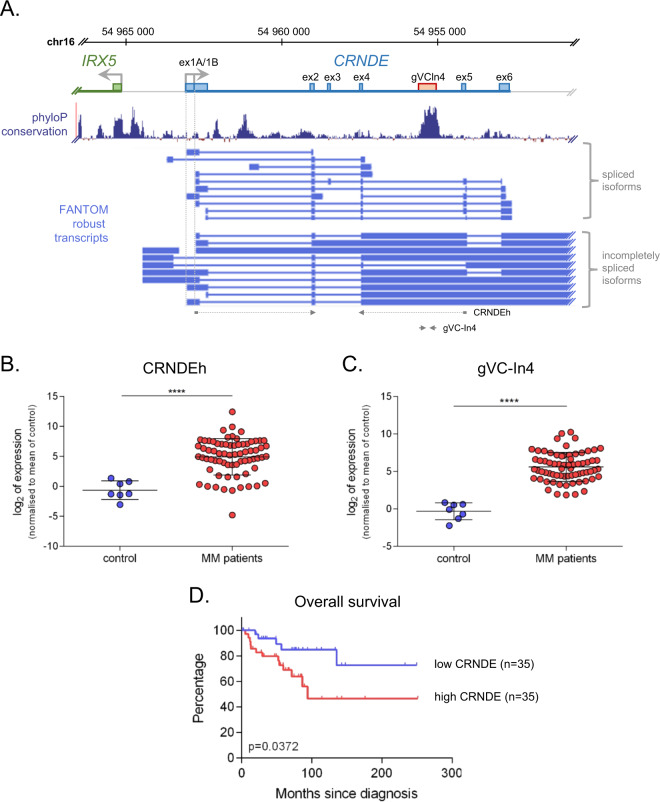
At the outset of this study, we carried out data-mining using a published transcriptomic dataset [27] in order to identify lncRNAs differentially expressed in CD138+ plasma cells from newly diagnosed MM patients relative to control subjects (Supplementary Fig. 1). Among the 105 dysregulated lncRNA genes identified using this approach (60 upregulated and 45 down-regulated in MM patients (Supplementary Fig. 1B and Supplementary Tables 1 and 2)), we observed the lncRNA CRNDE to be upregulated in patient plasma cells (Supplementary Fig. 1C). CRNDE is a known oncogenic lncRNA originally identified in colorectal adenomas and carcinomas, which has also been reported to be upregulated in diverse solid and haematological tumours [28]. Upregulation of CRNDE had previously been reported in MM [29], but was not detected in a more recent RNA-seq profiling of MM tumour cells [15]. To investigate CRNDE further, we analysed expression of CRNDE by real time reverse-transcription PCR (RT-PCR) in CD138+ plasma cells isolated from 70 newly diagnosed MM patients and 7 control subjects using primers detecting the major spliced and intron-retaining CRNDE isoforms (Fig. 1a) [30, 31]. We found that CRNDE was significantly overexpressed in plasma cells of MM patients in this cohort, with upregulation observed for both spliced transcript isoforms (CRNDEh, Fig. 1b), as well as 3′ intron-retaining transcripts containing the conserved gVC-In4 element (Fig. 1c). Broad cytogenetic stratification of samples revealed that CRNDE was upregulated in groups bearing the t(4;14) or t(11;14) translocations as well as in a group containing neither of these major cytogenetic events (Supplementary Fig. 2A). Further, analysis of a second transcriptomic dataset [32] revealed that the level of CRNDE in plasma cells of MM patients increases with disease progression (Supplementary Fig. 2B). Kaplan–Meier analysis showed that patients in our cohort with higher expression of CRNDE at diagnosis had significantly shorter overall survival than those expressing lower CRNDE levels (Fig. 1d). Altogether these findings confirm upregulation of multiple isoforms of the lncRNA CRNDE independent of primary cytogenetic translocation in a novel cohort of MM patients, where it is associated with poor prognosis.
Fig. 1. CRNDE is a marker of poor prognosis in multiple myeloma.
a The CRNDE locus at human chromosome 16p. The CRNDE gene and upstream antisense gene IRX5 are indicated in blue and green respectively, with annotated core exons (RefSeq) shown as boxes. A highly conserved element in intron 4 of CRNDE (gVC-In4) is shown in pink. Underneath is shown phyloP basewise conservation (http://compgen.cshl.edu/phast/) and FANTOM-annotated robust CRNDE transcripts (https://fantom.gsc.riken.jp/), indicating fully-spliced and incompletely spliced transcript isoforms. At bottom is shown the location of PCR primers for RT-PCR amplicons (CRNDEh, gVC-In4) used in this study (see Supplementary Table 6). Analysis of CRNDE expression in CD138+ plasma cells of control individuals (n = 7) and MM patients at diagnosis (n = 70) by real time RT-PCR using amplicons CRNDEh (b) and gVC-In4 (c) as shown in (a). Expression (relative to ACTB) was normalised to the mean value observed in healthy controls and is shown on a log scale (****p < 0.0001, Mann–Whitney). d Kaplan–Meier curves showing overall survival in MM patients with high (>median; n = 35) or low (<median; n = 35) expression of CRNDE (assessed with the CRNDEh amplicon). The log-rank statistic p value is indicated.
CRISPR/Cas9-mediated deletion of the CRNDE locus impairs MM cell tumorigenic properties in vitro and in vivo
In order to investigate the functional effects of CRNDE in MM cells, we adopted a CRISPR/Cas9 deletion approach. CRNDE is expressed from a gene on human chr16q12.2 which is situated in a head-to-head antisense orientation with the coding gene IRX5, with whom its expression is strongly correlated (Fig. 1a) [30]. The RefSeq-annotated gene contains five core exons (including an alternative first exon 1A/1B), as well as an additional annotated exon which is less utilised (exon 3) (Fig. 1a). Because the CRNDE promoter is likely to be bi-directionally shared with the neighbouring coding gene IRX5, rather than deleting the promoter region, CRISPR/Cas9 was used to delete a 5.1 kb region spanning exons 4–6 of the CRNDE gene (including the gVC-In4 intronic element) (Supplementary Fig. 3A). This CRISPR deletion was initially carried out in two MM cell lines, RPMI 8226 and KMS11, which express high and intermediate levels of CRNDE, respectively (Supplementary Fig. 3B). In both of these cell lines, sub-cellular fractionation studies revealed distinct compartmentalisation of the major CRNDE isoforms, with the spliced CRNDEh transcripts being primarily cytoplasmic and gVC-In4 retaining transcripts being primarily nuclear (Supplementary Fig. 3C), consistent with previous observation in colorectal carcinoma cell lines [31]. CRISPR-mediated deletion in KMS11 generated heterozygous- (CRNDE+/Δ) and homozygous- (CRNDEΔ/Δ) deleted clones at a frequency of 13% and 6%, respectively, while in RPMI 8226, heterozygous clones were obtained less frequently (5%) and no homozygous deleted clones were recovered (Supplementary Fig. 3D and Supplementary Methods). Subsequent experiments therefore focused on KMS11 as a cellular model. Real-time RT-PCR confirmed no detectable expression of the CRNDEh and gVC-In4 isoforms in CRNDEΔ/Δ KMS11 clones (Supplementary Fig. 3E).
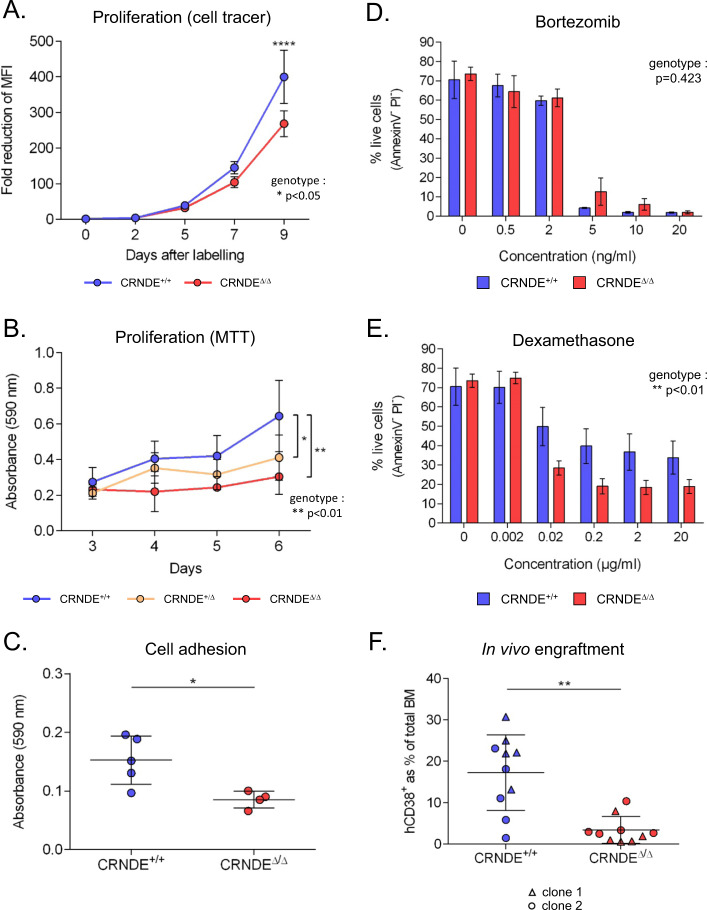
Although CRNDEΔ/Δ KMS11 clones could be maintained and expanded in culture, they proliferated more slowly than CRNDE+/+ clones, as assessed both by monitoring the reduction of fluorescent cell tracing dye during cell division (Fig. 2a) and using the MTT assay of cellular metabolic activity (Fig. 2b). In MTT assays, a more subtle but significant growth defect was also observed in heterozygous CRNDE+/Δ clones, indicating a CRNDE allele-dose-dependent effect on MM cell growth (Fig. 2b). These findings were confirmed in another CRNDE-expressing myeloma cell line NC1-H929 (Supplementary Fig. 3B) using a CRISPRi knockdown approach (Supplementary Fig. 4A). These experiments again showed that depletion of CRNDE led to a significant decrease in in vitro proliferation (Supplementary Fig. 4B).
Fig. 2. CRISPR-mediated deletion of the CRNDE locus impairs MM cell tumorigenic properties.
a CRNDE+/+ and CRNDEΔ/Δ KMS11 clones (4 per genotype) were pulse labelled with CellTracer dye, and dilution of the dye due to cell division was monitored by flow cytometry. Numbers indicate fold decrease of mean fluorescence intensity (MFI) relative to day 0 (p < 0.05 for genotype as a significant variable; for individual comparisons, ****p < 0.0001; 2-way ANOVA). b MTT proliferation assay of CRNDE+/+ clones (n = 4), CRNDE+/Δ clones (n = 4) and CRNDEΔ/Δ clones (n = 2). Absorbance (590 nm) was measured over 6 days (p < 0.01 for genotype as a significant variable; for individual comparisons *p < 0.05 and **p < 0.01; 2-way ANOVA). c Cell adhesion of CRNDE+/+ and CRNDEΔ/Δ clones. Points represent the mean of duplicate experiments for 5 CRNDE+/+ clones and 4 CRNDEΔ/Δ clones (*p < 0.05; Mann–Whitney). CRNDE+/+ and CRNDEΔ/Δ clones (replicate cultures on 3 clones per genotype) were cultured in complete medium containing Bortezomib (d) or Dexamethasone (e) at the concentrations indicated for 72 h and assessed for apoptosis by FACS. Shown is the percentage of live cells (AnnexinV− Propidium Iodide-) (p values for genotype as a significant variable are shown; 2-way ANOVA). f Percentage of KMS11 cells (hCD38+) in bone marrows of recipient NSG mice at 6 weeks after injection with CRNDE+/+ or CRNDEΔ/Δ clones (2 clones per genotype, each injected into 5 mice (**p < 0.01; Mann–Whitney)).
We next investigated effects of CRNDE on cell adhesion, a critical determinant of various aspects of MM disease evolution [33]. Adhesion assays showed that CRNDEΔ/Δ KMS11 clones were less effective than wild type clones at adhering to wells coated with the extra-cellular matrix Matrigel (Fig. 2c). Adhesion of myeloma cells stimulates bone marrow stromal cells to secrete IL6 [34]. Consistent with decreased adhesive properties observed on matrigel, we found that CRNDEΔ/Δ KMS11 cells also induced lower levels of IL6 secretion by bone marrow mesenchymal stromal cells in co-culture assays, suggesting a perturbed interaction between the CRNDE-null KMS11 cells and the stroma (Supplementary Fig. 5). We further investigated the effects of CRNDE on the sensitivity of KMS11 MM cells to Bortezomib and Dexamethasone, two conventional drugs used in the treatment of MM. While deletion of the CRNDE locus did not affect the Bortezomib response (Fig. 2d), CRNDEΔ/Δ KMS11 clones showed increased sensitivity to Dexamethasone (Fig. 2e). Finally, we assessed the effect of the CRNDE deletion on the in vivo tumorigenicity of KMS11 cells using an in vivo xenograft mouse model. At 6 weeks post injection in NOD-SCID IL2Rγ−/− (NSG) mice, FACS analysis of KMS11 cells (hCD38+) in the bone marrow revealed a reduced infiltration by clones bearing the CRNDEΔ/Δ deletion (Fig. 2f). Interestingly, deletion of the CRNDE locus also decreased both the frequency and KMS11-infiltration of extra-medullary tumours observed in this xenograft model (Supplementary Fig. 6). Taken together, these experiments indicate that CRNDE impacts on proliferation, drug sensitivity, cell adhesion and in vivo tumorigenic properties of MM cells.
CRNDE regulates the expression of genes associated with signalling and adhesion pathways implicated in MM
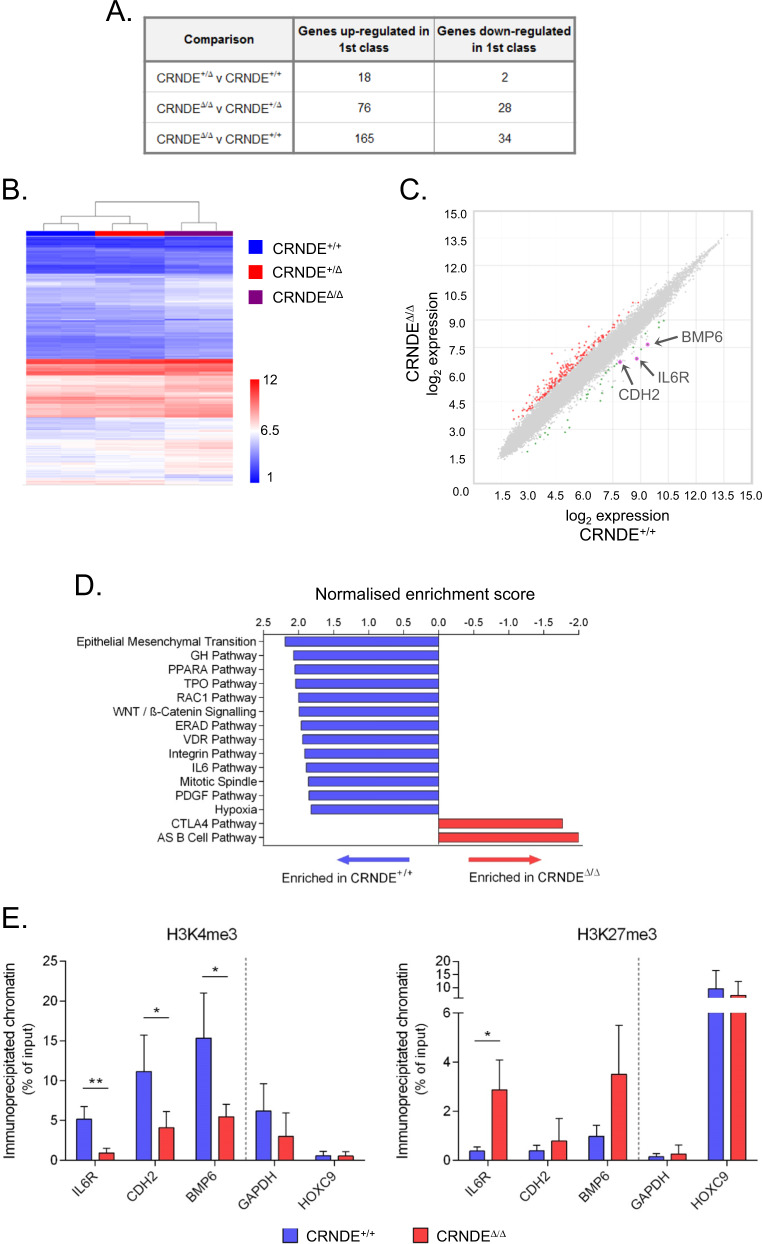
In order to characterise the molecular pathways affected by CRNDE in KMS11 myeloma cells in an unbiased fashion, we carried out microarray expression profiling of CRNDE+/+, CRNDE+/Δ and CRNDEΔ/Δ KMS11 clones. Exon-level analysis confirmed truncation of exons 4–6 of the CRNDE transcript in the CRNDEΔ allele, while exons 1–3 were expressed normally, suggesting that the 3′ end of the CRNDE locus is of particular functional importance (Supplementary Fig. 7A). Gene-level analysis identified 199 genes differentially expressed between CRNDEΔ/Δ and CRNDE+/+ KMS11 clones (Fig. 3a–c and Supplementary Table 3). In contrast, relatively few gene expression changes were observed between CRNDE+/Δ and CRNDE+/+ clones (Fig. 3a, b). Gene Set Enrichment Analysis (GSEA) [35] using Hallmark and BioCarta genesets revealed that relative to wild type cells, CRNDEΔ/Δ cells showed depletion of pathways previously linked to MM cell proliferation, survival and homing to the bone marrow including the WNT/β-catenin [36], IL6 [37] and Integrin [38] signalling pathways (Fig. 3d, Supplementary Fig. 8A and Supplementary Table 4). Leading edge analysis revealed that a cluster of genes enriched in wild type cells reflected activation of canonical pathways of MM survival and proliferation, including JAK/STAT (STAT3, STAT5A/5B), PI3K-AKT (PIK3CG) and MAPK (MAP2K1, JUN, SRF, SHC1) (Supplementary Fig. 8B, C). GSEA also revealed strong enrichment of pathways associated with both cell-substrate and cell–cell adhesion in wild type cells relative to CRNDEΔ/Δ cells (Supplementary Fig. 8D and Supplementary Table 5) consistent with the decreased adhesive properties of the deleted clones observed in vitro (Fig. 2c).
Fig. 3. Gene expression changes in CRNDEΔ/Δ KMS11 multiple myeloma cells.
a Microarray analysis of CRNDE KMS11 CRISPR clones (2 clones per genotype). Shown is the number of genes up- and down-regulated in each comparison (cutoffs of FC < −2 or FC > 2 and ANOVA p ≤ 0.01). b Unsupervised clustering based on genes differentially expressed (F test p ≤ 0.01). c Scatter plot representation of comparison of CRNDE+/+ and CRNDEΔ/Δ KMS11 cells. Genes up- or down-regulated in CRNDEΔ/Δ cells relative to wild type are shown in red and green respectively. The probes for the IL6R, CDH2 and BMP6 genes are indicated. d GSEA analysis showing the Normalised enrichment score for Hallmark and Biocarta genesets significantly enriched (cut-off FDR q < 0.05) in CRNDE+/+ (blue) or CRNDEΔ/Δ (red) KMS11 clones. e Analysis by ChIP of enrichment of H3K4me3 (left panel) and H3K27me3 (right panel) at the promoters of the IL6R, CDH2 and BMP6 genes in CRNDE+/+ and CRNDEΔ/Δ KMS11 clones. The GAPDH (expressed) and HOXC9 (not expressed in KMS11) promoters were used as controls. Replicate ChIP experiments were performed on two clones per genotype (*p < 0.05, **p < 0.01; student’s t test).
We found that the majority of changing genes were upregulated in the CRNDE-deleted clones, suggesting that in MM cells CRNDE may act primarily as a repressor of gene expression (Fig. 3c). Nevertheless, the expression of several genes implicated in MM survival, proliferation and adhesion was decreased in CRNDEΔ/Δ clones, suggesting direct or indirect activation by CRNDE. This was the case for IL6R (encoding the gp80 subunit of the IL6 receptor complex), BMP6 (encoding a member of the TGFβ superfamily) and CDH2 (encoding the cell adhesion molecule N-cadherin) (Fig. 3c and Supplementary Table 3) [37, 39, 40]. Decreased expression of these genes following deletion of CRNDE was validated by real time RT-PCR (Supplementary Fig. 7B). A common mechanism of action of lncRNAs is to recruit chromatin-modifying factors to target genes to alter their epigenetic state and transcriptional activity [13]. Chromatin immunoprecipitation (ChIP) experiments revealed a depletion of the activating histone modification H3K4me3 at the promoters of IL6R, BMP6 and CDH2 in CRNDEΔ/Δ KMS11 clones (Fig. 3e). We also observed a tendency towards increased levels of the repressive histone modification H3K27me3 at these promoters in CRNDEΔ/Δ clones, although this change only reached significance at the promoter of the IL6R gene (Fig. 3e). Taken together these results implicate CRNDE in the regulation of key signalling and adhesion pathways that contribute to MM cell survival, proliferation and disease progression, and suggest that CRNDE increases the expression in MM cells of important target genes such as IL6R, BMP6 and CDH2 by affecting their epigenetic state either directly or indirectly.
CRNDE plays a critical role in the IL6 signalling pathway in MM cells
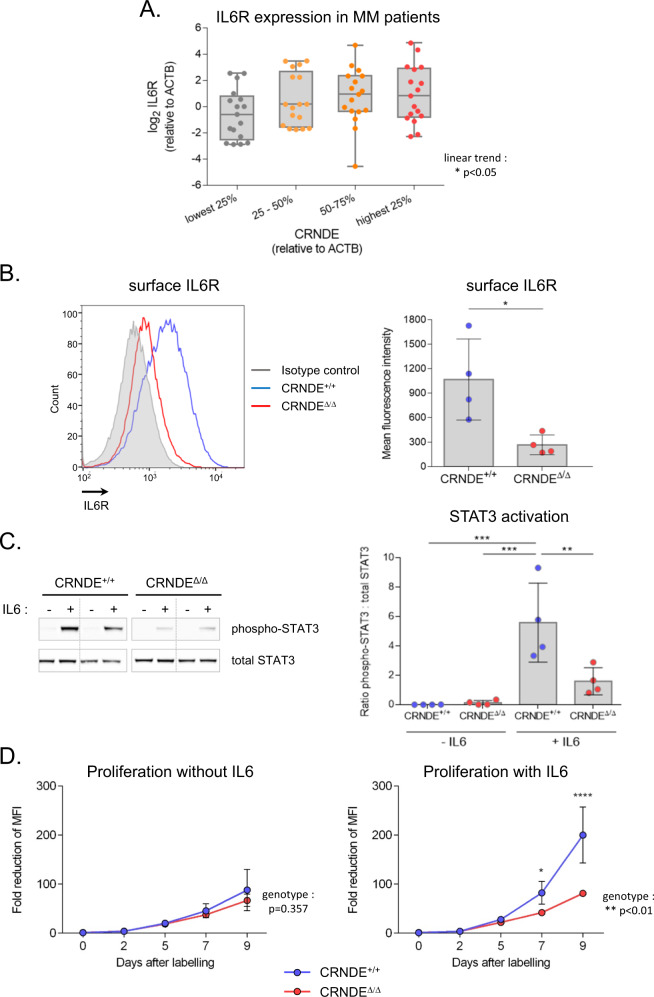
As shown above, IL6R was among the genes down-regulated in CRNDEΔ/Δ KMS11 clones, and these cells also showed transcriptomic changes consistent with decreased signalling through the IL6 pathway (Supplementary Fig. 8A,B). Consistent with our finding that CRNDE activates expression of IL6R in the KMS11 cell line, we observed a significant correlation between CRNDE and IL6R expression by real time RT-PCR in our own cohort of MM patients (Fig. 4a) as well as in a previously published microarray dataset [41] (Supplementary Fig. 9). Since activation of IL6 signalling drives proliferation of MM cells and limits apoptosis in response to chemotherapy [42], we further investigated the role of CRNDE in the regulation of IL6 signalling in MM. Consistent with decreased expression of the IL6R gene, flow cytometry revealed reduced levels of surface IL6R in CRNDEΔ/Δ KMS11 clones (Fig. 4b). Further, by Western blot, we demonstrated decreased phosphorylation of STAT3 (Tyr705) in response to IL6 in CRNDEΔ/Δ clones (Fig. 4c). To assess the contribution of diminished IL6 signalling to the proliferation defect observed in the absence of CRNDE, serum-starved cells were labelled with a fluorescent tracer dye and then cultured for 9 days in minimal medium supplemented or not with IL6 and proliferation (dilution of tracer dye) was monitored by flow cytometry. While no difference was observed between the proliferation of CRNDE+/+ and CRNDEΔ/Δ clones under minimal conditions, CRNDEΔ/Δ clones exhibited a proliferative defect in conditions of IL6 stimulation (Fig. 4d). Taken together these findings demonstrate a defect in the IL6 signalling pathway in CRNDE-deleted KMS11 MM cells and suggest that perturbed IL6 signalling is a major determinant of the proliferative defect observed in these cells.
Fig. 4. Deletion of CRNDE impairs IL6 signalling in multiple myeloma cells.
a Expression of CRNDEh and IL6R analysed by real time RT-PCR in CD138+ plasma cells isolated from bone marrow of newly diagnosed MM patients (n = 68). Expression (relative to ACTB) was normalised to the mean level observed in a control group of healthy individuals. Samples were stratified into quartiles based on expression of CRNDE (p < 0.05; ANOVA with post-test for linear trend). b Surface IL6R expression. Left panel: Representative flow cytometry staining of a CRNDE+/+ (blue) and CRNDEΔ/Δ (red) KMS11 clone with anti-IL6R-PE antibody or isotype control (grey). Right panel: Quantitation of mean fluorescence intensity of IL6R staining (n = 4 clones for each genotype) (*p < 0.05; Mann–Whitney). c IL6-mediated activation of STAT3. Left panel: Representative Western blot analysis of phosphorylated (Tyr705)-STAT3 and total STAT3 in two CRNDE+/+ and CRNDEΔ/Δ clones (serum starved) following treatment with or without IL6 (2 ng/ml) for 90 min. Right panel: Quantitation of Western blots. Shown is the ratio of phospho-STAT3: total STAT3 (n = 4 clones for each genotype) (**p < 0.01; ***p < 0.001; ANOVA). d IL6-mediated proliferation. Following pulse labelling with CellTracer dye, KMS11 clones (4 clones per genotype) were grown in minimum medium without (left) or with (right) 2 ng/ml IL6. Numbers indicate fold decrease in MFI relative to day 0. The p values for genotype as a significant factor are shown. For comparisons at each time point, *p < 0.05, ****p < 0.0001 (2-way ANOVA).
Discussion
CRNDE is a known oncogenic lncRNA which has previously been implicated in diverse solid tumours and haematological malignancies [28]. All these studies have revealed a variety of mechanisms by which CRNDE can affect tumour growth, suggesting that the molecular pathways involved are likely to be disease- and context-dependent. Consistent with the results of Meng et al. [29], here we show that CRNDE is also upregulated in tumour plasma cells of MM patients, where high expression correlates with poor prognosis. LncRNAs present unique challenges to functional assessment by classical siRNA/shRNA knockdown approaches (due to their frequent localisation in the nuclear space [43]), or by “single-cut” CRISPR-approaches, (since their biological activity is not dependent on maintenance of an open reading frame). Here, we used CRISPR/Cas9 to truncate both the major cytoplasmic and nuclear isoforms of CRNDE, as well as CRISPRi to reduce their expression in MM cell lines, allowing a thorough functional and molecular characterisation of the effects of this lncRNA in MM. Depletion of CRNDE led to a decrease in MM cell adhesion, proliferation and survival in vitro as well as reduced bone-marrow engraftment and extra-medullary tumours in a mouse tumorigenesis model. Taken together, these in vitro and in vivo findings suggest that the CRNDE locus plays an important role in the aetiology and progression of MM.
Despite being implicated in a variety of malignant diseases, the effect of CRNDE on genome-wide expression has only been investigated in the context of colorectal cancer and not in haematological malignancies [31, 44, 45]. In this study, we found that the deletion of CRNDE was largely associated with upregulation of gene expression, suggesting that CRNDE acts primarily as a repressor of gene expression in MM cells. It has been shown previously that CRNDE (also called lincIRX5) can interact with components of Polycomb repressive complex 2 (PRC2) and a significant overlap was observed between genes whose expression is upregulated in human fibroblasts following knockdown of CRNDE and knockdown of PRC2 components [44, 46, 47]. Interestingly, the histone methyltransferase EZH2, a core PRC2 component, is frequently overexpressed in MM where, like CRNDE, its expression correlates with disease progression and poor prognosis [48]. Taken together these observations suggest that the overexpression of CRNDE could co-operate with and exacerbate the effects of EZH2 upregulation during MM disease progression.
Mechanistically, many studies have reported that CRNDE can function as a molecular sponge to dampen the activity of miRNAs, and thereby regulate expression of miRNA target genes in a post-transcriptional manner [49–54]. In particular, it has recently been reported that CRNDE controls proliferation and survival of the U266 MM cell line by acting as a molecular sponge for miR-451 [29]. Further, another microRNA, miR-136-5p, which is observed to be sponged by CRNDE in several tumours [49, 54–56] has been reported to target the IL6R transcript in rat microglial cells [57]. However, miR-451 and miR-136-5p were not detectable by real time RT-PCR in either wild type or CRNDEΔ/Δ KMS11 cells, or in the NC1 CRISPRi cell lines used in the current study (data not shown), indicating that the effects of CRNDE in these models was not mediated via these miRNAs. Further, ChIP experiments revealed that the down-regulation of IL6R, CDH2 and BMP6 in CRNDEΔ/Δ cells was associated with epigenetic changes at their gene promoters, suggesting that CRNDE regulates expression of these target genes at the level of transcription, rather than a post-transcriptional effect via a microRNA. Further studies will be required to characterise the molecular mechanisms of transcriptional activation of these genes by CRNDE, and in particular the partner proteins with which this lncRNA interacts in MM cells.
Signalling by IL6 and the activation of downstream pathways (STAT, RAS, MAPK and PI3K/AKT) promotes MM cell proliferation and protects against apoptotic death induced by therapeutic agents, contributing to drug resistance and disease relapse [42]. Given the importance of this pathway in the aetiology of MM, IL6 signalling has long been recognised as a target for anti-MM therapies [58]. Here, we identified IL6R, encoding the gp80 subunit of the IL6 receptor complex, as a downstream activational target of CRNDE in KMS11 cells, and observed a correlation between IL6R and CRNDE expression in patient plasma cells. CRNDE-deleted KMS11 cells demonstrate impaired IL6 signalling and proliferative responses together with transcriptional changes indicating disruption of JAK/STAT, PI3K-AKT and MAPK pathways. In pre-clinical models and some clinical trials, inhibition of IL6 signalling has been shown to increase response to therapeutic drugs, and in particular to the effects of glucocorticoids, an important first line component of chemotherapy regimes [59–62]. Interestingly, we found that deletion of CRNDE increased the sensivity of KMS11 cells to Dexamethasone, consistent with impairment of IL6-dependent resistance mechanisms in these cells. Since resistance to glucocorticoid drugs is frequently observed in relapsed and refactory MM, our findings suggest that CRNDE may present an interesting novel target to attenuate IL6 signalling and improve or prolong drug sensitivity in some MM patients. Further, since IL6 signalling plays an important tumour cell-intrinsic role in diverse malignant diseases [63], it will be of interest to investigate the effect of CRNDE on this signalling pathway in other tumour cells where the lncRNA is upregulated.
Supplementary information
Acknowledgements
This study was funded by grants from INSERM, the Fondation ARC (Association pour la Recherche sur le Cancer), and the Fondation Française pour la Recherche contre le Myélome et les Gammapathies (FFRMG). AD was supported by a fellowship from the FFRMG. SZ was supported by a Université de Paris IDEX doctoral bourse. We are grateful to Jérôme Moreaux and Christelle Freitas for helpful scientific discussions. We thank Adrienne Anginot, Karl Balabanian, Marion Espeli and Valeria Bisio for assistance with stromal cell co-culture experiments. We thank members of the Plateforme Technologique of the Institut de Recherche Saint Louis for support and technical assistance with cytometry and microarrays and the Centre de Ressources Biologiques (CRB)—Cellulothèque Saint-Louis Laboratoire d’Hématologie for patient sample conservation.
Data availability
The microarray datasets generated in this study are available under accession number GSE155091 in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/7/2021
A Correction to this paper has been published: 10.1038/s41375-021-01305-2
Supplementary information
The online version of this article (10.1038/s41375-020-01034-y) contains supplementary material, which is available to authorised users.
References
- 1.Network THMR. 2016. www.hmrn.org/statistics/incidence.
- 2.Turesson I, Bjorkholm M, Blimark CH, Kristinsson S, Velez R, Landgren O. Rapidly changing myeloma epidemiology in the general population: Increased incidence, older patients, and longer survival. Eur J Haematol. 2018. 10.1111/ejh.13083. [DOI] [PMC free article] [PubMed]
- 3.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–60. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 4.Kumar SK, Rajkumar V, Kyle RA, van Duin M, Sonneveld P, Mateos MV, et al. Multiple myeloma. Nat Rev Dis Prim. 2017;3:17046. doi: 10.1038/nrdp.2017.46. [DOI] [PubMed] [Google Scholar]
- 5.Ribatti D. A historical perspective on milestones in multiple myeloma research. Eur J Haematol. 2018;100:221–8. doi: 10.1111/ejh.13003. [DOI] [PubMed] [Google Scholar]
- 6.Manier S, Salem KZ, Park J, Landau DA, Getz G, Ghobrial IM. Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol. 2017;14:100–13.. doi: 10.1038/nrclinonc.2016.122. [DOI] [PubMed] [Google Scholar]
- 7.Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33:3911–20. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broyl A, Hose D, Lokhorst H, de Knegt Y, Peeters J, Jauch A, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116:2543–53. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 9.Jang JS, Li Y, Mitra AK, Bi L, Abyzov A, van Wijnen AJ, et al. Molecular signatures of multiple myeloma progression through single cell RNA-Seq. Blood Cancer J. 2019;9:2. doi: 10.1038/s41408-018-0160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Investig. 2016;126:2775–82. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao H, Wahlestedt C, Kapranov P. Strategies to annotate and characterize long noncoding RNAs: advantages and pitfalls. Trends Genet. 2018;34:704–21.. doi: 10.1016/j.tig.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21:542–51.. doi: 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- 13.Sun Q, Hao Q, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34:142–57.. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronchetti D, Agnelli L, Pietrelli A, Todoerti K, Manzoni M, Taiana E, et al. A compendium of long non-coding RNAs transcriptional fingerprint in multiple myeloma. Sci Rep. 2018;8:6557. doi: 10.1038/s41598-018-24701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samur MK, Minvielle S, Gulla A, Fulciniti M, Cleynen A, Aktas Samur A, et al. Long intergenic non-coding RNAs have an independent impact on survival in multiple myeloma. Leukemia. 2018;32:2626–35.. doi: 10.1038/s41375-018-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu AX, Huang ZY, Zhang L, Shen J. Potential prognostic long non-coding RNA identification and their validation in predicting survival of patients with multiple myeloma. Tumour Biol. 2017;39:1010428317694563. doi: 10.1177/1010428317694563. [DOI] [PubMed] [Google Scholar]
- 17.Ronchetti D, Agnelli L, Taiana E, Galletti S, Manzoni M, Todoerti K, et al. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget. 2016;7:14814–30. doi: 10.18632/oncotarget.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou M, Zhao H, Wang Z, Cheng L, Yang L, Shi H, et al. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J Exp Clin Cancer Res. 2015;34:102. doi: 10.1186/s13046-015-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Namba M, Ohtsuki T, Mori M, Togawa A, Wada H, Sugihara T, et al. Establishment of five human myeloma cell lines. Vitr Cell Dev Biol. 1989;25:723–9. doi: 10.1007/BF02623725. [DOI] [PubMed] [Google Scholar]
- 20.Moore GE, Kitamura H. Cell line derived from patient with myeloma. NY State J Med. 1968;68:2054–60. [PubMed] [Google Scholar]
- 21.Katagiri S, Yonezawa T, Kuyama J, Kanayama Y, Nishida K, Abe T, et al. Two distinct human myeloma cell lines originating from one patient with myeloma. Int J Cancer. 1985;36:241–6. doi: 10.1002/ijc.2910360217. [DOI] [PubMed] [Google Scholar]
- 22.Gazdar AF, Oie HK, Kirsch IR, Hollis GF. Establishment and characterization of a human plasma cell myeloma culture having a rearranged cellular myc proto-oncogene. Blood. 1986;67:1542–9. [PubMed] [Google Scholar]
- 23.Epstein MA, Achong BG, Barr YM, Zajac B, Henle G, Henle W. Morphological and virological investigations on cultured Burkitt tumor lymphoblasts (strain Raji) J Natl Cancer Inst. 1966;37:547–59. [PubMed] [Google Scholar]
- 24.Pegoraro L, Malavasi F, Bellone G, Massaia M, Boccadoro M, Saglio G, et al. The human myeloma cell line LP-1: a versatile model in which to study early plasma-cell differentiation and c-myc activation. Blood. 1989;73:1020–7. [PubMed] [Google Scholar]
- 25.Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 1953;97:695–710. doi: 10.1084/jem.97.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abroun S, Ishikawa H, Tsuyama N, Liu S, Li FJ, Otsuyama K, et al. Receptor synergy of interleukin-6 (IL-6) and insulin-like growth factor-I in myeloma cells that highly express IL-6 receptor alpha [corrected] Blood. 2004;103:2291–8. doi: 10.1182/blood-2003-07-2187. [DOI] [PubMed] [Google Scholar]
- 27.Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–58. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai M, Li S, Qin X. Colorectal neoplasia differentially expressed: a long noncoding RNA with an imperative role in cancer. Onco Targets Ther. 2018;11:3755–63.. doi: 10.2147/OTT.S162754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng YB, He X, Huang YF, Wu QN, Zhou YC, Hao DJ. Long Noncoding RNA CRNDE Promotes Multiple Myeloma Cell Growth by Suppressing miR-451. Oncol Res. 2017;25:1207–14.. doi: 10.3727/096504017X14886679715637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis BC, Molloy PL, Graham LD. CRNDE: a long non-coding RNA involved in cancer, neurobiology, and development. Front Genet. 2012;3:270. doi: 10.3389/fgene.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellis BC, Graham LD, Molloy PL. CRNDE a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta. 2014;1843:372–86. doi: 10.1016/j.bbamcr.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 32.Zhan F, Barlogie B, Arzoumanian V, Huang Y, Williams DR, Hollmig K, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109:1692–700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz BZ. Adhesion molecules-The lifelines of multiple myeloma cells. Semin Cancer Biol. 2010;20:186–95. doi: 10.1016/j.semcancer.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Uchiyama H, Barut BA, Mohrbacher AF, Chauhan D, Anderson KC. Adhesion of human myeloma-derived cell lines to bone marrow stromal cells stimulates interleukin-6 secretion. Blood. 1993;82:3712–20.. [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derksen PW, Tjin E, Meijer HP, Klok MD, MacGillavry HD, van Oers MH, et al. Illegitimate WNT signaling promotes proliferation of multiple myeloma cells. Proc Natl Acad Sci USA. 2004;101:6122–7. doi: 10.1073/pnas.0305855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gado K, Domjan G, Hegyesi H, Falus A. Role of INTERLEUKIN-6 in the pathogenesis of multiple myeloma. Cell Biol Int. 2000;24:195–209. doi: 10.1006/cbir.2000.0497. [DOI] [PubMed] [Google Scholar]
- 38.Hosen N. Integrins in multiple myeloma. Inflamm Regen. 2020;40:4. doi: 10.1186/s41232-020-00113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grcevic D, Kusec R, Kovacic N, Lukic A, Lukic IK, Ivcevic S, et al. Bone morphogenetic proteins and receptors are over-expressed in bone-marrow cells of multiple myeloma patients and support myeloma cells by inducing ID genes. Leuk Res. 2010;34:742–51. doi: 10.1016/j.leukres.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 40.Mrozik KM, Cheong CM, Hewett D, Chow AW, Blaschuk OW, Zannettino AC, et al. Therapeutic targeting of N-cadherin is an effective treatment for multiple myeloma. Br J Haematol. 2015;171:387–99. doi: 10.1111/bjh.13596. [DOI] [PubMed] [Google Scholar]
- 41.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosean TR, Tompkins VS, Tricot G, Holman CJ, Olivier AK, Zhan F, et al. Preclinical validation of interleukin 6 as a therapeutic target in multiple myeloma. Immunol Res. 2014;59:188–202. doi: 10.1007/s12026-014-8528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–77. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding J, Li J, Wang H, Tian Y, Xie M, He X, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8:e2997. doi: 10.1038/cddis.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang H, Wang Y, Ai M, Wang H, Duan Z, Wang H, et al. Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways. Cell Death Dis. 2017;8:e2862. doi: 10.1038/cddis.2017.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang M, Gao C, Yang Y, Li G, Dong J, Ai Y, et al. Long noncoding RNA CRNDE/PRC2 participated in the radiotherapy resistance of human lung adenocarcinoma through targeting p21 expression. Oncol Res. 2018;26:1245–55.. doi: 10.3727/096504017X14944585873668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alzrigat M, Jernberg-Wiklund H, Licht JD. Targeting EZH2 in multiple myeloma-multifaceted anti-tumor activity. Epigenomes. 2018;2:16. doi: 10.3390/epigenomes2030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao H, Song X, Kang T, Yan B, Feng L, Gao L, et al. Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer. Onco Targets Ther. 2017;10:205–16.. doi: 10.2147/OTT.S116178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu B, Ye X, Du Q, Zhu B, Zhai Q, Li XX. The long non-coding RNA CRNDE promotes colorectal carcinoma progression by competitively binding miR-217 with TCF7L2 and enhancing the Wnt/beta-catenin signaling pathway. Cell Physiol Biochem. 2017;41:2489–502.. doi: 10.1159/000475941. [DOI] [PubMed] [Google Scholar]
- 52.Chen Z, Yu C, Zhan L, Pan Y, Chen L, Sun C. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am J Cancer Res. 2016;6:2299–309.. [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C, et al. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24:1199–215. [DOI] [PMC free article] [PubMed] [Retracted]
- 54.Huan J, Xing L, Lin Q, Xui H, Qin X. Long noncoding RNA CRNDE activates Wnt/beta-catenin signaling pathway through acting as a molecular sponge of microRNA-136 in human breast cancer. Am J Transl Res. 2017;9:1977–89.. [PMC free article] [PubMed] [Google Scholar]
- 55.Li DX, Fei XR, Dong YF, Cheng CD, Yang Y, Deng XF, et al. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget. 2017;8:88163–78.. doi: 10.18632/oncotarget.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu L, Liu Y, Chen Q, Yu G, Chen J, Chen K, et al. Long-noncoding RNA colorectal neoplasia differentially expressed gene as a potential target to upregulate the expression of IRX5 by miR-136-5P to promote oncogenic properties in hepatocellular carcinoma. Cell Physiol Biochem. 2018;50:2229–48.. doi: 10.1159/000495084. [DOI] [PubMed] [Google Scholar]
- 57.Zhang D, Mou JY, Wang F, Liu J, Hu X. CRNDE enhances neuropathic pain via modulating miR-136/IL6R axis in CCI rat models. J Cell Physiol. 2019;234:22234–41. doi: 10.1002/jcp.28790. [DOI] [PubMed] [Google Scholar]
- 58.Matthes T, Manfroi B, Huard B. Revisiting IL-6 antagonism in multiple myeloma. Crit Rev Oncol Hematol. 2016;105:1–4. doi: 10.1016/j.critrevonc.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 59.Honemann D, Chatterjee M, Savino R, Bommert K, Burger R, Gramatzki M, et al. The IL-6 receptor antagonist SANT-7 overcomes bone marrow stromal cell-mediated drug resistance of multiple myeloma cells. Int J Cancer. 2001;93:674–80. doi: 10.1002/ijc.1388. [DOI] [PubMed] [Google Scholar]
- 60.Fulciniti M, Hideshima T, Vermot-Desroches C, Pozzi S, Nanjappa P, Shen Z, et al. A high-affinity fully human anti-IL-6 mAb, 1339, for the treatment of multiple myeloma. Clin Cancer Res. 2009;15:7144–52. doi: 10.1158/1078-0432.CCR-09-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voorhees PM, Chen Q, Small GW, Kuhn DJ, Hunsucker SA, Nemeth JA, et al. Targeted inhibition of interleukin-6 with CNTO 328 sensitizes pre-clinical models of multiple myeloma to dexamethasone-mediated cell death. Br J Haematol. 2009;145:481–90. doi: 10.1111/j.1365-2141.2009.07647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voorhees PM, Manges RF, Sonneveld P, Jagannath S, Somlo G, Krishnan A, et al. A phase 2 multicentre study of siltuximab, an anti-interleukin-6 monoclonal antibody, in patients with relapsed or refractory multiple myeloma. Br J Haematol. 2013;161:357–66. doi: 10.1111/bjh.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumari N, Dwarakanath BS, Das A, Bhatt AN. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. 2016;37:11553–72.. doi: 10.1007/s13277-016-5098-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray datasets generated in this study are available under accession number GSE155091 in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo).