Abstract
Although we have made significant progress, we still possess a limited understanding of how genomic and epigenomic information directs gene expression programs through sequence-specific transcription factors (TFs). Extensive research has settled on three general classes of TF targets in metazoans: promoter accessibility via chromatin regulation (e.g., SAGA), assembly of the general transcription factors on promoter DNA (e.g., TFIID) , and recruitment of RNA polymerase (Pol) II (e.g., Mediator) to establish transcription pre-initiation complex (PIC). Here we discuss TFs and their targets. We also place this in the context of our current work with Saccharomyces (yeast), where we find that promoters typically lack an architecture that supports TF function. Moreover, yeast promoters that support TF binding also display interactions with cofactors like SAGA and Mediator, but not TFIID. It is unknown to what extent all genes in metazoans require TFs and their cofactors.
Keywords: Gene regulation models, Transcription mechanisms, Enhancers, Promoters
Graphical Abstract. Promoter activation.
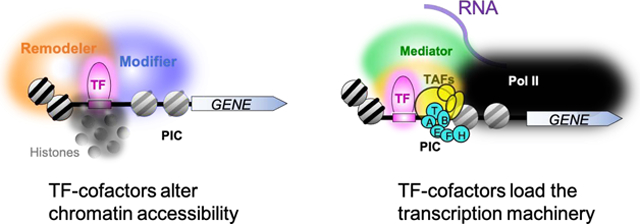
TFs bind to their cognate sequences in enhancers and promoters, where they coalesce cofactors to increase chromatin accessibility and assembly of the transcription machinery.
Enhancer–Promoter Communication
Logic of gene regulation
Cells need to constantly produce more biomolecules to maintain homeostasis. Maintenance of homeostasis also requires cells to reprogram themselves in response to environmental changes [1]. This is achieved by regulating gene expression. In maintaining the basic constituents of life that exist in all cells, each gene often needs to be expressed at a characteristic level relative to all other genes. The promoters that drive such constitutive expression may have a fundamentally common architecture. If they do not change in a gene-specific manner in response to signaling events, then they would not have evolved gene-specific control. Reprogramming in response to environmental and cellular signals is expected to incur additional regulatory architectures that tie the signal(s) to increased or decreased gene expression from its prior state [2, 3]. This creates a large dynamic range of expression that is focused in part on the initiation of transcription as one of many points of gene regulation.
To achieve these systems requirements, each gene may be under the control of a basic core promoter having a limited range of output [4]. Cis-regulatory elements (enhancers) can augment output as needed [5]. Whether all aspects of transcription initiation control can be classified into these two broad groups (core promoters and enhancers) remains to be determined. In mammals, it is unknown whether all genes are defined by enhancers, or just those that require a greater dynamic range of control than can be offered by a core promoter architecture. How enhancers communicate with core promoters remains an active area of investigation with many outstanding questions. For example, what exactly are the protein constituents of enhancers, and their function? How diverse is their composition? What are the protein-protein interactions that define enhancer-promoter connectivity? How pervasive and diverse are condensates that bring multiple promoters together into hubs of greater activity? Here we review some of the literature that has helped elucidate our understanding of gene control through regulated transcription initiation.
Core promoter and enhancer basics
Gene regulatory elements are highly prevalent in mammalian genomes, albeit promoter elements are not always all present in every promoter and the biological functions of some elements are unclear [6]. Among those gene regulatory elements are individual and clusters of sequence motifs that act on core promoters locally or from afar [7] (Fig. 1). Long-range interactions by promoter-enhancer loops fundamentally contribute to the control of transcription [8]. A core promoter with a transcription start site (TSS) assembles RNA polymerase (Pol) II transcription machinery with general transcription factors (GTFs) TFIIA, -B, -D, -E, -F and -H to establish a pre-initiation-complex (PIC) [9]. However, transcription driven only by a core promoter which lacks gene-specific enhancer elements is usually at a low level [9]. Core promoter sequences that would drive specificity seem to be largely undefined in mammalian and yeast systems, although a small fraction of all core promoters have a variety of site-specific DNA regulatory elements [6]. These range from TATA boxes [10] that help define the transcription start site (TSS) to dispersed sequence-specific transcription factor (TF) binding sites (in mammals) with less-defined positioning [11]. The TATA box binds the TATA-binding protein (TBP) to position the PIC [12], whereas surrounding TF binding sites help recruit the transcription machinery. Invertebrates, as modeled through Drosophila, appear to have much more defined core promoter elements [13].
Fig. 1. Simplified model of enhancer-core promoter interactions.
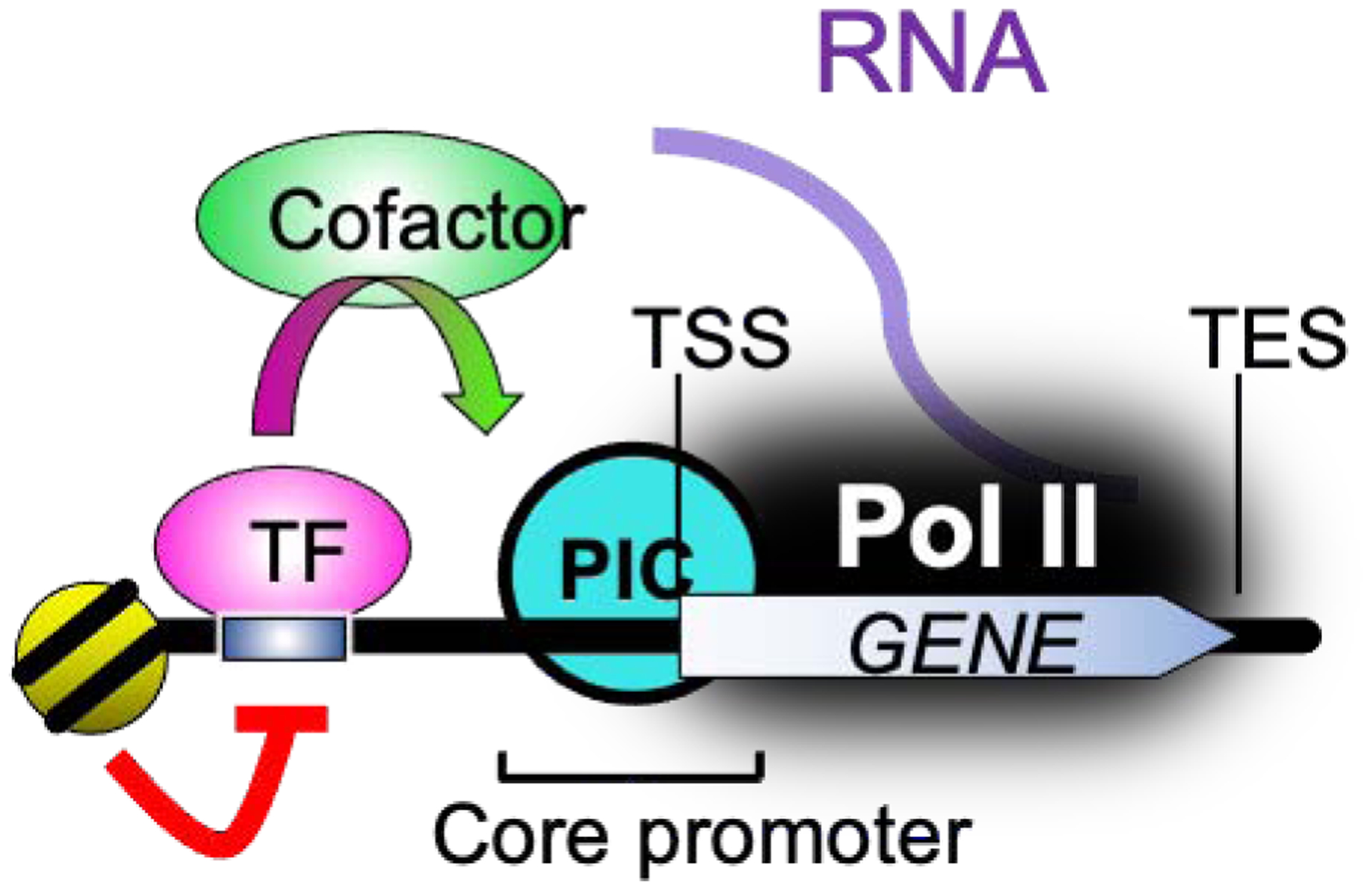
Enhancers contain specific sequence motifs that are bound by TFs . They activate core promoters by regulating promoter/enhancer interactions, PIC assembly, and Pol II recruitment through a wide variety of cofactors.
High levels of transcription at genes is achieved by enhancers acting on core promoters, often in a tissue-specific manner [14]. Active transcription at enhancers also appears to be a critical but poorly understood phenomenon [15]. Enhancers are bound by transcription factors (TFs) that recognize 6–12 bp-long DNA sequences. The traditional classification of gene regulatory elements define promoter and enhancers as having distinct molecular features that are placed far apart. However it has been shown that regulatory elements can have both promoter and enhancer activities [16]. Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) [17, 18] has identified the bound locations of TFs genome-wide. TF binding can increase or decrease at diverse sets of enhancers at different cell developmental stages or conditions [19–22]. Such dynamic and context-dependent TF occupancy often corresponds to combinatorial interplay of multiple cofactors and regional deposition of epigenetic markers like histone acetylation or DNA methylation.
CTCF and 3D architecture
Enhancer-promoter communication has traditionally been observed to occur in the context of chromosome folding. Eukaryotic genome organization is constructed in part for the formation of self-associating domains and enhancer-promoter communication. Current models of eukaryotic genomes depict a hierarchical series of folding steps, bringing genomic regions that are relatively distal from each other into close proximity, particularly within “topologically associating domains” (TADs) [23]. These steps may be driven by TFs with the consequence of orchestrating developmental programs [1, 24, 25]. TADs allow distal enhancer factors and their target promoters to coalesce [26].
How TFs interact with or maintain their interactions with targets in the context of TAD formation is unclear. In mammalian nuclei, cohesin directs TAD formation through a ‘loop extrusion’ process. CTCF anchored at looped barriers interacts directly with cohesin to protect it from loop release [27]. CTCF occupancy does not always coincide with TAD insulation that restricts enhancer activity to within the TAD. In one view, enhancers might scan throughout a TAD for its intended target promoters. This might contribute to the formation of genome topology that is cell-type specific [28].
Condensates
Eukaryotic cellular processes require the coalescence of molecules to facilitate their interactions and reactions [29]. During induced transcription, the concentration of Pol II is increased locally in membrane-less compartments, within which its unstructured and highly repetitive C-terminal domain (CTD) undergoes cooperative liquid phase separation [30]. The transcription events led by Pol II clustering are often related to super enhancers, that compartmentalize and concentrate large quantities of TFs, cofactors (like Mediator and BRD4), and the transcription machinery [31–33]. Super enhancers are clusters of smaller enhancers that are located close to each other, with the effect of compartmentalizing and concentrating the transcription apparatus [32, 34, 35]. This model is supported by a large body of molecular genetic evidence and advanced live-cell imaging methods. A series of in vitro reconstitution experiments validated the role of intrinsically disordered regions of TFs and Mediator subunits in formation of liquid droplet condensates. They typically assemble together through multivalent interactions among proteins and nucleic acids (Fig. 2) [31, 33, 36]. However, not all observations of condensates may be physiologically real [37].
Fig. 2. Condensate-based model of enhancer-promoter control.
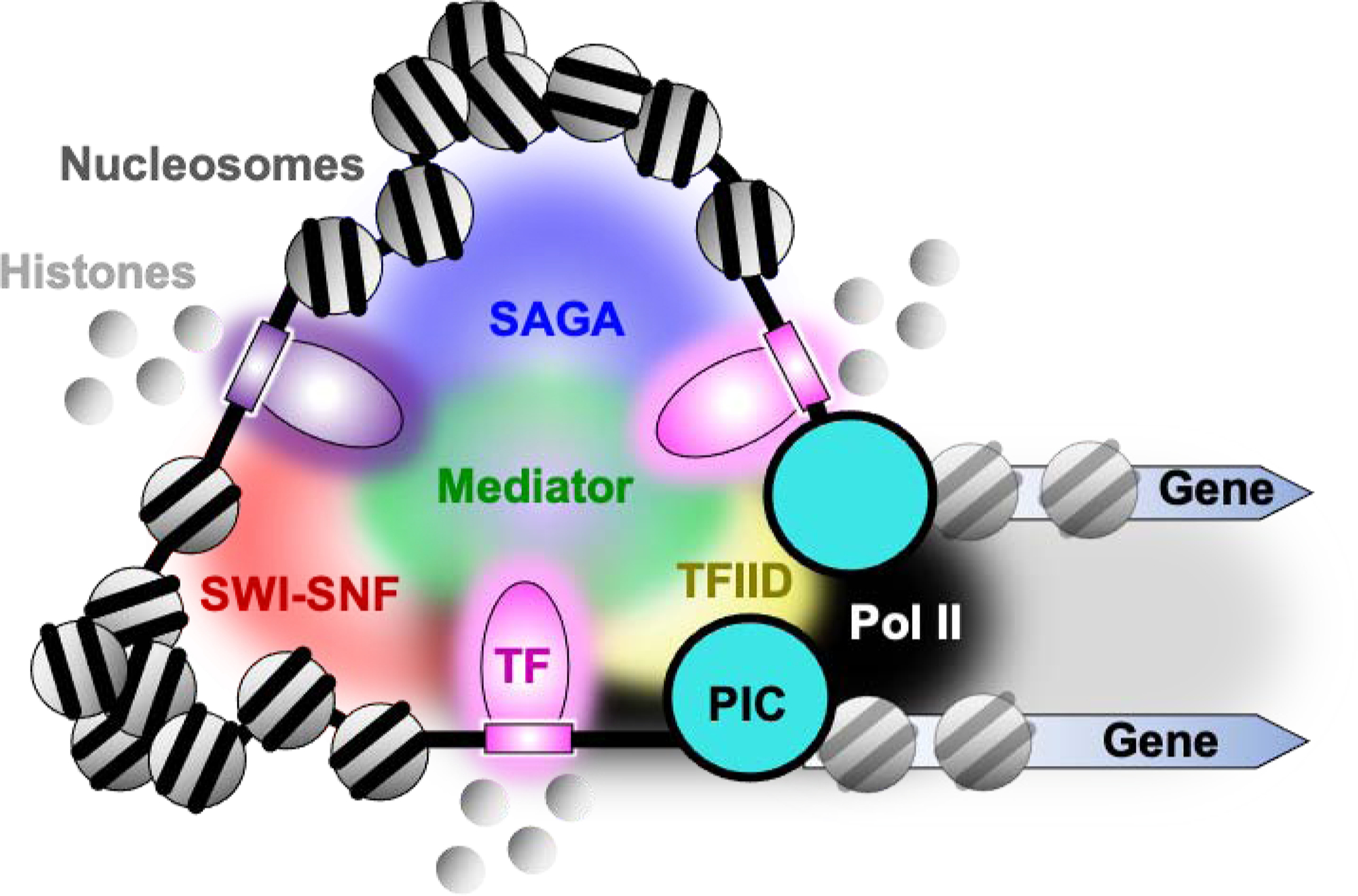
TFs with their intrinsically disordered regions (IDRs) along with other IDRs in cofactors coalesce and demix, thereby dramatically increasing the local concentration of components of the transcription machinery. Potential consequences include increased nucleosome dynamics affecting enhancer/promoter accessibility, more frequent PIC assembly, and a steady stream of Pol II loading and firing into an elongation complex.
Condensate/enhancer composition
The gene expression driven by super enhancers and molecular condensates are likely fundamental for cell identity or fate-determined processes [38]. For example, in mouse embryonic stem cells, the key regulatory transcription factors that will maintain ESC state and control gene expression programs are OCT4, SOX2, and NANOG [39]. Those master transcription factors that define the cell identity are highly co-occupied at super enhancers [31, 32]. Apart from TFs, signaling factors for the WNT, TGF-β, and JAK/STAT pathways can also be recruited to the super-enhancer-associated condensates [40]. A recent study found that kidney injury lead to enhancers/super enhancers activation in responding to cell injury and repair [41]. Pharmacological inhibition of BRD4 to block enhancer activation impaired recovery of injured kidney cells. Transcription factors HNF4A, GR, STAT3 and STAT5 were identified at those elements, indicating their roles in shaping the enhancer landscape in the regulation of kidney repair.
Cofactors
One broad category of TF cofactors includes complexes that chemically modify (acetylated, methylate, etc.) histones or conformationally remodel nucleosomes [42]. They endow TFs with the ability to alter the chromatin accessibility that is critical for PIC assembly. Another broad category of TF cofactors regulates genes via their ability to connect TFs to the transcription machinery. Mediator is one cofactor complex that links TFs to Pol II. At least in metazoans, TFIID may represent another cofactor complex that links TFs to GTFs through TFIID TAF subunits. Together they stimulate the assembly and function of PICs independent of TF distance and orientation from the core promoter [43–46].
Activation by recruitment is one of the best-studied regulatory mechanisms and there is overwhelming evidence to suggest that this is a major means of transcription stimulation in eukaryotes [47, 48]. Evidence for interactions between TFs and cofactors has been accumulated by in vivo and in vitro assays. In vivo cell environments provide the natural context in which protein-protein interactions reside. However, ascertaining direct interactions which help define mechanistic relationships is confounded by non-native artificial experimental reconstitutions and by co-localization events that are not necessarily direct interactions. Biochemical reconstitutions help delineate direct versus indirect interactions. However, they typically do not recapitulate the native context of properly organized and folded chromatin, along with appropriate condensates.
A yeast model for enhancer-promoter organization.
We have recently mapped the binding location at near-bp resolution of most classes of genome-interacting proteins in the budding yeast Saccharomyces cerevisiae by ultra-high resolution chromatin immunoprecipitation followed by exonuclease (ChIP-exo assay) [3, 49–54]. The 5’-3’ single-stranded exonuclease is stopped by an immunoprecipitated protein-DNA crosslink, thereby marking the location of the crosslink for a particular protein via the digested DNA 5’ end. Not only does its near-bp resolution allow the positional organization of chromatin proteins to be precisely determined, but their characteristic exonuclease stoppage patterns in bulk cells provides a view of potential interacting partners. These partners may not bind DNA directly, but crosslink through DNA-bound TF, thereby producing the same ChIP-exo pattern as the underlying TF. From this, and the bulk of related experimental literature in Saccharomyces, we have gained some fundamental overarching principles of enhancer and core promoter organization that guide our discussion.
We find that most promoters are active in yeast and have a PIC assembled near its TSS. Two fundamental gene classes arise based on their molecular architectures at promoter regions: constitutive and induced. Most promoters evolved an architecture that allows for constitutive gene expression only (Fig. 3). No TFs or their binding sites or cofactors are found at these genes beyond a subset having sequence-specific nucleosome organizing factors. They position the −1 and +1 nucleosomes that flank an intrinsically nucleosome-free promoter region (NFR). Underlying sequences like poly(dA:dT) tracts, along with chromatin remodelers, drive constitutive nucleosome exclusion in these NFRs [55]. The combination of an NFR and +1 nucleosome may promote a constitutively low level of PIC assembly. This limited architecture appears to suffice for many yeast genes.
Fig. 3. Binary architecture of yeast promoters.
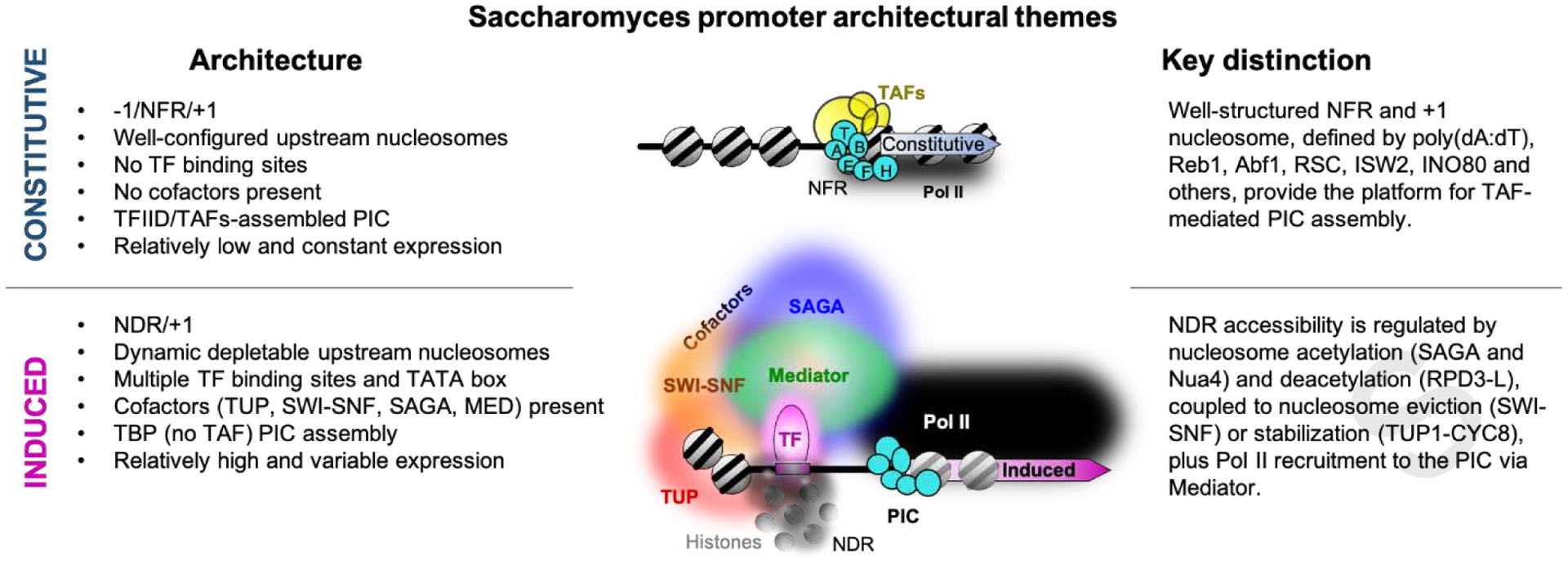
Summary schematic of yeast chromatin structures with key architectural arrangements at constitutive (upper) and induced (lower) promoter classes. T, A, B, E, F, H denote TBP, TFIIA, TFIIB, TFIIE, TFIIF, and TFIIH, respectively. NFR and NDR denote nucleosome-free region and nucleosome-depleted region, respectively.
In contrast, ~20% of yeast genes have their expression directly controlled by inducible sequence-specific regulatory TFs and their cognate cofactors. This inducibility involves TFs creating local nucleosome depleted regions (NDRs). NDRs are distinct from NFRs in being intrinsically nucleosomal and having regulated depletion rather than a constitutive absence of nucleosomes. At inducible promoters, regulated depletion occurs through TFs and their cofactors, whereas at constitutive promoters, nucleosome positioning rather than depletion is a key characteristic.
At inducible promoters, nucleosomal dynamics are critical. There, TFs directly engage with chromatin cofactors like SAGA and NuA4 for histone acetylation, and RPD3-L for deacetylation. They also interact with SWI/SNF for nucleosome eviction or Ssn6/Cyc8-Tup1 for nucleosome stabilization (Fig. 3). TFs then drive PIC assembly through TBP and Mediator recruitment. Remarkably, this PIC assembly appears to involve TBP without the TAF subunits of TFIID (although do involve TAFs that are part of SAGA). TFIID appears to provide PIC assembly at promoters that lack a relationship with TFs and the other cofactors (i.e., constitutive promoters). However, TFIID may contribute to basal transcription at inducible promoters in yeast, independent of TF-cofactor relationships. Beyond perhaps Rap1, we find little or no stable interactions between TFs and TAFs. Since TFIID in general may not be a stable TF target in yeast, it raises an interesting contrast to metazoan systems, where there is more abundant evidence for such interactions.
TF-cofactor regulation of chromatin
Eukaryotic cells employ a wide range of histone modifying enzymes and chromatin remodelers to regulate DNA accessibility [56]. They are targeted to specific promoters through sequence-specific interactions of TFs that interact with these chromatin regulators. We focus here on a few examples of their interactions with TFs.
TF interactions with nucleosome modifiers.
CREB-binding protein (CBP) and its homologue p300 catalyze the acetylation of histone H3 at amino acid K27 specifically at nucleosomes located within active enhancers [57, 58]. While p300/CBP bind chromatin at both active and poised enhancers, the acetylation on H3K27 at the poised enhancer cannot be established by CBP/p300 until it is recruited [59]. Then the transcriptional network is triggered. CBP/p300 are TF cofactors that interact with more than 400 binding partners [60]. Through a series of biochemistry assays including pull-down (using immobilized protein to capture diffusible proteins) and electrophoretic mobility shift (EMSA, which measures the slower migration of protein-DNA interactions in an electric field), p300 was found to interact with STAT1 homodimers that are bound to DNA [61]. Structural analysis of the interactions revealed that activated and dimerized TFs IRF3 or STAT1 can bind p300 and enhance the HAT activity of p300 [62–64].
TF interactions with nucleosome remodelers.
ATP-dependent chromatin remodeling complexes can reorganize nucleosomes to regulate chromatin accessibility. At specific genes, selective co-occupancy of Pol II with chromatin remodeler SWI/SNF in yeast and mammalian cells indicate that remodelers could be recruited through direct interactions with gene-specific activator [65]. A well-studied example of TF-cofactor interactions in mammalian cells that drive nucleosome depletion or alterations are the TFs OCT4, SOX2, and NANOG [39]. As a pioneer transcription factor, OCT4 may bind to nucleosomal DNA, leading to chromatin opening that is necessary for gene expression [66, 67]. OCT4 physically interacts with the BRG1 subunit of SWI/SNF chromatin-remodeling complexes [68]. In OCT4-depleted stem cells, chromatin accessibility examined by ATAC-seq is significantly reduced at OCT4 targets. OCT4-dependent ATAC-seq accessible regions also coincide with BRG1 occupancy detected by ChIP-seq, suggesting that local inaccessible regions are opened with BRG1 [66, 69]. OCT4 recruits P300/CBP to acetylate nucleosomes through chromatin reader BRD4 [70]. BRD4 directly interacts with OCT4 to determine pluripotent states [71].
SAGA cofactor
The histone acetyltransferase (HAT) GCN5 is part of different chromatin-modifying multiprotein complexes, most of which are conserved from yeast to human [72]. GCN5 is part of SAGA, SAGA-like complex (SLIK/ SALSA), Spt3-TAF9-GCN5L acetylase (STAGA), TBP-free TAFII-containing complex (TFTC), and p300/CBP-associated factor (PCAF) [73]. The SAGA complex in yeast is a well-studied GCN5-containing complex and its functions serve as a model for other SAGA like complexes in metazoan. The SAGA complex functions through its GCN5 histone acetyltransferase (HAT) [74] and deubiquitinase (DUB) activities [75], as well as assisting the recruitment of TATA box binding protein (TBP) to inducible (SAGA-dominated) genes through TF interactions [76, 77]. SAGA has been variously reported to be gene-selective but also general to all genes [78, 79]. It may be that its role in TBP delivery is gene-specific, whereas its HAT and DUB activities may be more general [80].
TF-SAGA interactions
Tra1 is the largest subunit of yeast SAGA and a major target of TFs. The direct protein-protein interaction between Tra1 and the yeast Gal4 TF was first detected by fluorescence resonance energy transfer (FRET) assay in living cells [81]. Further, Gal4-Tra1 interactions are important for the SAGA recruitment. Photo-crosslinkers on the Gal4 activating domain also identified Tra1 as a Gal4 interacting target in vitro [82]. Also based on ChIP and other biochemical studies, Spt3 and Spt8 subunits of SAGA stimulate TBP recruitment to promoters [76, 77, 81]. There are two PIC assembly pathways in yeast, depending on the coactivators TFIID or SAGA to recruit TBP at promoters, and the genes that are dominated relatively more by one or the other are labeled as TFIID-dominated and SAGA-dominated [77]. The majority of yeast genes (~85%) are TFIID dominated with these promoters being relatively depleted of a TATA box and also being constitutively active. In contrast, ~15% of genes have relatively greater enrichment of TATA boxes, are SAGA-dominated, and stress-induced. Gene-selective recruitment of SAGA through gene-specific TFs creates an opportunity to locally augment nucleosome acetylation and promoter activation.
In metazoan systems, TFs also interactions with SAGA-related complexes. TFs like MYC, E2F, E1A and p53 bind the homolog of Tra1, called TRRAP (transformation-transactivation domain-associated protein) [83–85]. These TFs recruit SAGA and related TRRAP-containing complexes to DNA. The recruited GCN5 can then acetylate nucleosomes, commonly on histone H3 [74]. Tra1 and TRRAP are also part of the NuA4 HAT complex, which commonly acetylates histones H4 and H2A [86]. This allows TFs to recruit multiple HATs that prepare nucleosomes for remodeling. More specifically, the association between MYC and human STAGA and TFTC complexes were validated based on co-immunoprecipitation and pull-down assays. The direct physical interaction between MYC and TRRAP is necessary to recruit GCN5 and other components of the STAGA complex [87]. Protein crosslinking assays further identified interaction between GCN5 and MYC [88]. Based on protein crosslinking, P53 was found to interact with the STAGA subunits TAF9, GCN5 and ADA2b [89]. These results indicate multiple interactions, beyond TRRAP may be involved in STAGA/TFTC recruitment to promoters.
TF interactions with TAFs shared between SAGA and TFIID
One challenge in identifying the complexes that TFs target in vivo is where subunits are shared among multiple complexes. For example, TAF10, as a subunit of both TFIID and SAGA, physically interacts with GATA1 to mediate TFIID and/or SAGA formation for erythroid cell lineage [90]. In another example, TAF9 is present in TFIID and TFTC, SAGA, SLIK, and STAGA [73]. TAF9 interacts with the TF EKLF and downstream promoter elements to activate transcription of erythroid genes [91]. Whether this involves TFIID or other complexes is unclear. Through a conserved consensus recognition motif, the TFs GLI1, GLI2, viral VP16, and TP53 bind TAF9 [92–94]. The interactions between GLI1 or GLI2 with TAF9 are important in small cell lung cancers. TP53 interacts with TAF9 with higher affinity than GLI1. Consequently, TP53 might contribute to inhibition of GLI1 activity in cancer treatments through sequestering TAF9. Whether TAF9 in other complexes could also be recruited by TFs or how discrimination among these complexes is achieved has yet to be explored.
TF-cofactor regulation of PIC assembly.
TF-TFIID interactions in yeast
TFIID serves as a central scaffold for PIC formation. TFIID is composed of TBP and 13 TAFs that are evolutionarily conserved from yeast to human. In yeast, the promoters of ribosomal protein (RP) genes are often TATA-less (TBP binds to TATA elements). The activation of RP genes involves recruitment of TFIIA and TFIID through the TF Rap1 [95]. Direct interactions between Rap1 and TAFs have been determined biochemically and genetically [96]. Furthermore, a Cryo-EM structure of TFIID-TFIIA-Rap1 indicates that activating signals could be transduced by the formation of a protein bridge [97]. Rap1 represents the only clear example in yeast where a TF interacts with TAFs. This case may be unique because yeast RP genes are expressed at very high constitutive levels, far beyond the norms of most constitutive gene expression. We examined 78 TFs in yeast for genome-wide interaction with TFIID and find little or no evidence for TF-TFIID interactions beyond Rap1 [98].
TF-TFIID interactions in metazoans
Bulk evidence by protein interaction analysis in metazoan systems provide evidence for TFs directly targeting TFIID prior to and/or after DNA binding, depending on the context and conditions at specific promoters (Table 1). TFs may also direct distinct tissue-specific isoforms of TFIID (containing paralogous TBP and TAF subunits) [99]. What distinctive purpose they play apart from canonical TFIID is unclear.
Table 1:
Experimental basis for TF-TFIID interactions
| Interaction | Regulation | in-vivo | in-vitro | Complex reconstructi on | Ref |
|---|---|---|---|---|---|
| SP1 and TAF4 | Intrinsically disordered proteins (IDPs) of SP1 and TAF4, have weak but specific association. | Sp1 DNA affinity columns Yeast Two-Hybrid | NMR spectroscopy, EM and single-particle 3D reconstructions | [101] [100] [102] | |
| TAFH domain of TAF4 and E proteins | TAFH-AD3 interaction enhances TFIID binding to the core promoter of E protein target gene. | IP, mass spectrometry, Immunoblot, ChIP, RNA-seq | in-vitro transcription assays, immobilized template assays | [105] | |
| TP53 and TAF1 | TAF1 directly phosphorylates TP53 at Thr-55, TAF1 induces G1 progression in a partly TP53-dependent manner. | IP, endogenous domain mutation . | Far-Western analysis, In-gel phosphorylation assay. | [108], | |
| Diacetyl K373/K382 P53 and TAF1 bromodomain | Recruitment of TAF1 to the p21 promoter is mediated through acetylated p53 in response to DNA Damage | IP, ChIP, RT-PCR assays | Pulldown, TAF1 binding competition experiments | Protein recruitment on Immobilized promoter. | [109] |
| Mdm2 and TAF1 | TAF1 downregulates Mdm2 auto-ubiquitylation, leading to Mdm2 stabilization, and promotes p53-Mdm2 association and turnover of p53 | IP, ectopic expression and in vivo ubiquitylation assay | Pulldown | [110] | |
| TP53 and TFIID | P53 facilitates TFIID recruitment onto the TP53 target promoter, TP53 helps conversion of canonical TFIID to the structurally rearranged form with DNA. | Single-molecule fluorescence microscopy, Single-particle cryo-electron microscopy. | [111] | ||
| EKLF and TAF9 | TAF9 on the β-Globin Promoter is EKLF dependent and the interaction is mediated via the zinc finger domain of EKLF and the CR domain of TAF9. | IP, ChIP, RNAi, RT-PCR and transactivation assays | Pulldown | [91] | |
| VP16, GLI1, GLI2, TP53 and TAF9 | A conserved consensus recognition motif FXXΦΦ serves to interact with TAF9 and binding competition between GLI and p53 are involved in cancer | IP, proximity ligation, cell transformation assays | Pulldown, transcriptional activity assays, Two-hybrid assays | NMR | [92] [93] [94] |
| GATA1 and TAF10 | TAF10 with GATA1 is important to facilitate the recruitment of TFIID and/or SAGA to GATA1-responsive promoters | IP, mass spectrometry, ChIP | Pulldown | [90] | |
| MYC 98–111 and TBP-TAF1TAND1 | Promote TBP-TAF1 interaction and prevent TBP-DNA binding. | BioID-MS, Proximity ligation | Pulldown | Crystal structure, Biolayer Interferometry, NMR | [112] |
| MYC115–124 and TBP | MYC competes out TAF1TAND2 on TBP to release TAF1. |
TF-TAF4 interactions in metazoans
Human TAF4 was the first TAF subunit demonstrated to interact with glutamine (Q)-rich TFs like SP1 and CREB. Most direct evidence of interactions were found through in vitro reconstitution [100, 101]. The two Q-rich domains of SP1 and the four Q-rich domains at the N-terminus of TAF4 are intrinsically disordered, which mediate interactions with no measurable conformation changes [102]. Overexpression of the Q1-Q4 domains of TAF4 in HeLa cells specifically inhibits Sp1-dependent transcriptional activation [103]. These findings are consistent with previously reported interactions between Drosophila TAF4 (TAFII110) and the activation domain of SP1 [103]. Based on these interactions, TAF4 was proposed to help TFIID recruitment to promoters, and promote TBP engagement by stabilizing a rearranged state of TFIID.
Apart from four Q-domains, there is a TAFH domain of human TAF4 that is highly conserved. It too might serve as a potential target of transcriptional regulators. Crystal structure analysis of the human TAFH domain found a binding surface for potential TAF4 interacting factors [104]. Biochemical studies have also shown that direct TFIID–E-protein interactions are mediated through this domain. E proteins are widely expressed basic helix-loop-helix TFs. Certain E proteins interact with p300/CBP to regulate cell growth, differentiation, and apoptosis. This interaction enhances TFIID binding to the core promoters and is critical for activation of E protein target genes [105]. In the model proposed, the enhancer-bound E protein recruits the coactivator p300/CBP, then p300/CBP-mediated acetylation of nucleosomes leads to increased promoter accessibility to GTFs like TFIID and TFIIA. The E protein then recruits TFIID, whose binding is further stabilized with core promoter elements. Perhaps related to TAFH domain function, transcriptional corepressor ETO has a related TAFH domain. It can interact with E proteins, to inhibits it activation potential, by blocking p300/CBP interactions [70]. This may negatively impact TFIID recruitment, although its mechanism remains unclear.
TF-TAF enzymatic crosstalk
Human TAF1 of TFIID possesses intrinsic protein kinase, putative histone acetyltransferase (HAT), and ubiquitin conjugating activities [106, 107]. TAF1 phosphorylates tumor suppressor TF TP53 at amino acid T55, contributing to the stabilization of TP53 [108]. Additionally, TAF1 contains a double bromodomain that can recognize TP53 that is acetylated at amino acids K373 and K382. In response to DNA damage, TAFs are recruited to the human p21 gene for transcription. During the process, TAF1 is recruited through interactions between TP53 diacetyl lysines and TAF1 bromodomains [108, 109]. TAF1 also stimulates the ubiquitination and degradation of TP53 in a MDM2 dependent manner. This process is achieved by a two-step process. First, the interaction between MDM2 and TAF1 increases MDM2 stabilization, and then promotes association between MDM2 and TP53. Subsequently, MDM2 stimulates ubiquitination and degradation of TP53 [110]. By single-molecule fluorescence microscopy, it was found that TP53 facilitates TFIID assembly on the synthetic core promoters of target genes [111]. Further, cryo-EM structures revealed a dynamic process of TFIID subunit reorganization that is facilitated by TP53 and DNA. This ultimately disengages TP53, thereby allowing it to participate in additional recruitment steps.
MYC-TFIID interactions
MYC interacts with human TFIID in a multipartite manner at promoters [112]. In the MYC-TFIID crystal structure, MYC98–111 interacts with the TAND1 domain of TAF1, whereas MYC115–124 interacted with TBP independently of TAND1. TAND1 is an N-terminal portion of TAF1 that also interacts with TBP to inhibit its binding [113, 114]. The evidence for MYC-TFIID interaction has been validated by both in vivo and in vitro assays. Mutations on these two TFIID-anchoring regions of MYC significantly decreases the cell proliferation and transformation potential of MYC. Activator-TFIID interaction is proposed to energetically modulate the conformational landscape of TFIID from the canonical to a rearranged state [115]. With the presence of promoter DNA, TAF1 can bind to downstream DNA and neighboring +1 nucleosome and release TBP upstream of the TSS for local DNA scanning [115]. During that process, MYC possibly extends the coverage of the TBP-DNA interacting region while obscuring TBP DNA binding [112]. Then, the interactive interface of MYC-TBP changes to the regulatory region of TBP with MYC115–124, after which the TFIID complex transitions to the extended state that favors release of TAF1 TAND2 (a second TBP-interacting domain). After DNA and TFIIA are involved, the interactions between MYC-TFIID are proposed to be excluded.
TF-Mediator regulation of Pol II recruitment.
Mediator-core promoter interactions
Mediator physically bridges enhancer-bound TFs to Pol II, so that functional information can be transmitted from enhancers to the transcription machinery [46]. The Med14 subunit works as a scaffold to hold together the head, middle, and tail modules of the Mediator. Generally, the head and middle contact the PIC and Pol II, while the tail interacts with TFs. Interactions between TFs and Mediator subunits have been observed in both human and yeast cells [116, 117]. Remarkable progress has been made in revealing the structure and function of RNA polymerase II–Mediator core initiation complex with the development of cryo-EM [118]. Structures of Pol II, TFIIB, TBP/TFIID, and TFIIF that fit into the holo-PIC complex, as well as protein crosslinking map help us understand how Mediator cooperates with PIC components [119]. This TF-Mediator-Pol II trifecta along with its tendency to form condensates that coalesce many promoters into hubs, makes Mediator a focal point for transcriptional regulation.
TF- Med1 interactions
About 75 known and novel Mediator-interacting partners have been discovered in neural stem cells using mass spectrometry [120]. They include TFs, coactivators/corepressors and chromatin modifiers. Thus, we may have only scratched the surface of direct and indirect targets of TFs and cofactors. Different TFs bind to different Mediator subunits, although those with similar activation domain properties typically target the same Mediator subunit. Mammalian Mediator was identified in part through its association with the thyroid hormone nuclear receptor (TR) in a ligand-dependent manner [121]. When nuclear receptors are triggered by their cognate ligands, they bind to the regulatory elements of target genes, orchestrating the assembly of the transcription machinery through Mediator [122]. TR interactions with Med1 illustrate one example of TF-Mediator specificity [123]. Co-immunoprecipitation, pull-down, and surface plasmon resonance helped to identify the LXXLL domains of Med1 as being important for nuclear receptor interactions [124]. Nuclear receptors include retinoid X receptor, thyroid hormone receptor, vitamin D3 receptor, and peroxisome proliferator-activated receptor [125].
TF-MED23 interactions
MED23 is the largest subunit of the tail module and interacts with several TFs. MED23 is involved in development, for example, of osteoblasts by interacting with osteogenic factor RUNX2 [126]. MED23 knockdown in mesenchymal stem cells (MSCs) reduce the expression of genes governed by RUNX2. Mouse MSCs that are deficient in MED23 exhibit impaired bone ossification that phenocopies the skeletal defects in RUNX2 +/− . In the mitogen-activated protein kinase (MAPK) signaling pathway, the activator domain of transcription factor ELK1 is phosphorylated by MAP kinase. MED23 interacts with the phosphorylated ELK1 and they colocalize at ELK1 target genes [127, 128] . Moreover, in mouse embryonic fibroblasts, MED23 deficiency downregulates a set of ELK1 target genes that control differentiation into adipocytes, but up-regulates a set of RhoA target genes for smooth muscle cells (SMCs) [129]. These data suggest that MED23 controls a cell fate determination program between SMCs and adipocytes. Other TF-MED23 interactions have been suggested in glucose and lipid metabolism [130], lung cancers [131] and and T-cell activation [132]. However, the evidence of those interactions was largely based on in vivo colocalization assays like ChIP and immunofluorescence, which does not necessarily equate to direct interactions.
Mediator-CDK module
Mediator can reversibly associate with its kinase module, called CDK–Mediator. The kinase module includes the kinases CDK8 or CDK19 together with three subunits: CCNC, MED12 and MED13 [133]. Treating cells with cortistatin A that specifically inhibits CDK8/CDK19, revealed TFs as target proteins along with other proteins involved in chromatin and DNA repair [134]. TF phosphorylation by CDK8 regulates TF activity including bidirectional enhancer-associated eRNA transcription and formation of enhancer-promoter loops [133]. For example, the Mediator kinases phosphorylate STAT1 on amino acid S727 upon IFN- γ treatment, and this leads to eRNA transcription [135, 136]. eRNAs, together with Mediator subunits, participate in enhancer-promoter loop formation. Knockdown of the eRNA or Mediator results in decreased looping and decreased occupancy of Mediator at target promoters [137, 138].
Mediator-TF condensates.
Diverse TF activation domains have been reported to form molecular condensates with the Mediator coactivator. For example, OCT4 and GCN4 activation domains form phased-separated droplets with Med1 and Med15 through their intrinsically disordered regions [31]. This condensate formation is proposed to activate transcription by increasing the local concentration of the transcription machinery. Related to this, in budding yeast, high levels of transcription coincide with “fuzzy” (weak multivalent) interactions between the disordered low complexity domains of Gcn5 and Med15. The interface between Gcn5 and Med15 is dynamic, with multiple conformations and orientations that form a condensate-like state [139, 140]. Activator induced conformational changes in mammalian Mediator have also been detected by EM analysis [141]. An open question is whether the conformational flexibility of Mediator as the consequence of TF interactions contributes in any way to condensates at promoters.
Concluding remarks and perspectives
Numerous in vivo and in vitro methods have defined direct cofactor targets of TFs as being chromatin remodelers like SWI-SNF, PIC assembly factors like TFIID, and Pol II recruitment factors like Mediator. Together these factors work to ensure access to promoter regions and efficient loading of the transcription machinery. There are many more proteins whose full functionality remain uncharacterized that are likely necessary for this type of promoter regulation. The open questions that remain and will serve as the foundation for future work include: do all promoters involve enhancers? Are enhancers organized in the same way and interact with similar sets of cofactors? Are there other regulatory elements that regulate transcription via different sets of interactions with the core transcription machinery? Identifying the function of the many hundreds of components that reside within promoter/enhancer chromatin and sets of TF-cofactor-PIC trifectas that coalesce into hubs will continue to be a fertile ground for fundamental scientific discoveries.
Highlights.
Transcription factors (TFs) engage in three main levels of regulation
TFs target chromatin remodelers and modifiers to regulate DNA access
TFs target initiation factors like TFIID, although not widespread in yeast
TFs target Mediator to augment Pol II loading at promoters
Acknowledgements
This work was supported by NIH grants GM059055 and ES013768 to BFP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
BFP has a financial interest in Peconic, LLC, which uses the ChIP-exo technology implemented in this study and could potentially benefit from the outcomes of this research.
References
- [1].Stadhouders R, Vidal E, Serra F, Di Stefano B, Le Dily F, Quilez J, et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet. 2018;50:238–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vihervaara A, Mahat DB, Guertin MJ, Chu T, Danko CG, Lis JT, et al. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun. 2017;8:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Reja R, Vinayachandran V, Ghosh S, Pugh BF. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev. 2015;29:1942–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Weingarten-Gabbay S, Nir R, Lubliner S, Sharon E, Kalma Y, Weinberger A, et al. Systematic interrogation of human promoters. Genome Res. 2019;29:171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Plank JL, Dean A. Enhancer function: mechanistic and genome-wide insights come together. Mol Cell. 2014;55:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vo Ngoc L, Huang CY, Cassidy CJ, Medrano C, Kadonaga JT. Identification of the human DPR core promoter element using machine learning. Nature. 2020;585:459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–35. [PubMed] [Google Scholar]
- [10].Lifton RP, Goldberg ML, Karp RW, Hogness DS. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42 Pt 2:1047–51. [DOI] [PubMed] [Google Scholar]
- [11].Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–8. [DOI] [PubMed] [Google Scholar]
- [12].Buratowski S, Hahn S, Sharp PA, Guarente L. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature. 1988;334:37–42. [DOI] [PubMed] [Google Scholar]
- [13].Vo Ngoc L, Kassavetis GA, Kadonaga JT. The RNA Polymerase II Core Promoter in Drosophila. Genetics. 2019;212:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tippens ND, Liang J, Leung AK, Wierbowski SD, Ozer A, Booth JG, et al. Transcription imparts architecture, function and logic to enhancer units. Nat Genet. 2020;52:1067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Andersson R, Sandelin A. Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet. 2020;21:71–87. [DOI] [PubMed] [Google Scholar]
- [17].O’Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31:76–82. [DOI] [PubMed] [Google Scholar]
- [18].Collas P, Dahl JA. Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front Biosci. 2008;13:929–43. [DOI] [PubMed] [Google Scholar]
- [19].Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pilon AM, Ajay SS, Kumar SA, Steiner LA, Cherukuri PF, Wincovitch S, et al. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood. 2011;118:e139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–26. [DOI] [PubMed] [Google Scholar]
- [23].Chang LH, Ghosh S, Noordermeer D. TADs and Their Borders: Free Movement or Building a Wall? J Mol Biol. 2020;432:643–52. [DOI] [PubMed] [Google Scholar]
- [24].Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Arzate-Mejia RG, Recillas-Targa F, Corces VG. Developing in 3D: the role of CTCF in cell differentiation. Development. 2018;145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Szabo Q, Bantignies F, Cavalli G. Principles of genome folding into topologically associating domains. Sci Adv. 2019;5:eaaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li Y, Haarhuis JHI, Sedeno Cacciatore A, Oldenkamp R, van Ruiten MS, Willems L, et al. The structural basis for cohesin-CTCF-anchored loops. Nature. 2020;578:472–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Barrington C, Georgopoulou D, Pezic D, Varsally W, Herrero J, Hadjur S. Enhancer accessibility and CTCF occupancy underlie asymmetric TAD architecture and cell type specific genome topology. Nat Commun. 2019;10:2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol. 2018;25:833–40. [DOI] [PubMed] [Google Scholar]
- [31].Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell. 2018;175:1842–55 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hahn S Phase Separation, Protein Disorder, and Enhancer Function. Cell. 2018;175:1723–5. [DOI] [PubMed] [Google Scholar]
- [37].McSwiggen DT, Mir M, Darzacq X, Tjian R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 2019;33:1619–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Peng Y, Zhang Y. Enhancer and super-enhancer: Positive regulators in gene transcription. Animal Model Exp Med. 2018;1:169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–60. [DOI] [PubMed] [Google Scholar]
- [40].Zamudio AV, Dall’Agnese A, Henninger JE, Manteiga JC, Afeyan LK, Hannett NM, et al. Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol Cell. 2019;76:753–66 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wilflingseder J, Willi M, Lee HK, Olauson H, Jankowski J, Ichimura T, et al. Enhancer and super-enhancer dynamics in repair after ischemic acute kidney injury. Nat Commun. 2020;11:3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen T, Dent SY. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 2014;15:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wieczorek E, Brand M, Jacq X, Tora L. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–91. [DOI] [PubMed] [Google Scholar]
- [44].Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, et al. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. [DOI] [PubMed] [Google Scholar]
- [45].Martinez E, Kundu TK, Fu J, Roeder RG. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–5. [DOI] [PubMed] [Google Scholar]
- [46].Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–77. [DOI] [PubMed] [Google Scholar]
- [48].Green MR. Eukaryotic transcription activation: right on target. Mol Cell. 2005;18:399–402. [DOI] [PubMed] [Google Scholar]
- [49].Rossi MJ, Lai WKM, Pugh BF. Simplified ChIP-exo assays. Nat Commun. 2018;9:2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rhee HS, Bataille AR, Zhang L, Pugh BF. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell. 2014;159:1377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vinayachandran V, Reja R, Rossi MJ, Park B, Rieber L, Mittal C, et al. Widespread and precise reprogramming of yeast protein-genome interactions in response to heat shock. Genome Res. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Badjatia N, Rossi MJ, Bataille AR, Mittal C, Lai WKM, Pugh BF. Acute stress drives global repression through two independent RNA polymerase II stalling events in Saccharomyces. Cell Rep. 2021;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, et al. Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell. 2016;167:709–21 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J Cell Biochem. 2002;87:117–25. [DOI] [PubMed] [Google Scholar]
- [57].Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Raisner R, Kharbanda S, Jin L, Jeng E, Chan E, Merchant M, et al. Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell Rep. 2018;24:1722–9. [DOI] [PubMed] [Google Scholar]
- [59].Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell JE, Jr. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci U S A. 1996;93:15092–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Qin BY, Liu C, Srinath H, Lam SS, Correia JJ, Derynck R, et al. Crystal structure of IRF-3 in complex with CBP. Structure. 2005;13:1269–77. [DOI] [PubMed] [Google Scholar]
- [63].Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 2009;28:948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ortega E, Rengachari S, Ibrahim Z, Hoghoughi N, Gaucher J, Holehouse AS, et al. Transcription factor dimerization activates the p300 acetyltransferase. Nature. 2018;562:538–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].West JA, Cook A, Alver BH, Stadtfeld M, Deaton AM, Hochedlinger K, et al. Nucleosomal occupancy changes locally over key regulatory regions during cell differentiation and reprogramming. Nat Commun. 2014;5:4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].You JS, Kelly TK, De Carvalho DD, Taberlay PC, Liang G, Jones PA. OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc Natl Acad Sci U S A. 2011;108:14497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ding J, Xu H, Faiola F, Ma’ayan A, Wang J. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 2012;22:155–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].King HW, Klose RJ. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wu T, Kamikawa YF, Donohoe ME. Brd4’s Bromodomains Mediate Histone H3 Acetylation and Chromatin Remodeling in Pluripotent Cells through P300 and Brg1. Cell Rep. 2018;25:1756–71. [DOI] [PubMed] [Google Scholar]
- [71].Wu T, Pinto HB, Kamikawa YF, Donohoe ME. The BET family member BRD4 interacts with OCT4 and regulates pluripotency gene expression. Stem Cell Reports. 2015;4:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wang YL, Faiola F, Martinez E. Purification of multiprotein histone acetyltransferase complexes. Methods Mol Biol. 2012;809:427–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–78. [DOI] [PubMed] [Google Scholar]
- [74].Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–50. [DOI] [PubMed] [Google Scholar]
- [75].Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sermwittayawong D, Tan S. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. Embo J. 2006;25:3791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–85. [DOI] [PubMed] [Google Scholar]
- [78].Chen XF, Lehmann L, Lin JJ, Vashisht A, Schmidt R, Ferrari R, et al. Mediator and SAGA have distinct roles in Pol II preinitiation complex assembly and function. Cell Rep. 2012;2:1061–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. [DOI] [PubMed] [Google Scholar]
- [80].Donczew R, Warfield L, Pacheco D, Erijman A, Hahn S. Two roles for the yeast transcription coactivator SAGA and a set of genes redundantly regulated by TFIID and SAGA. Elife. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Reeves WM, Hahn S. Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol. 2005;25:9092–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lang SE, Hearing P. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene. 2003;22:2836–41. [DOI] [PubMed] [Google Scholar]
- [84].McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–74. [DOI] [PubMed] [Google Scholar]
- [85].Ard PG, Chatterjee C, Kunjibettu S, Adside LR, Gralinski LE, McMahon SB. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol Cell Biol. 2002;22:5650–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, et al. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. Embo J. 1999;18:5108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Liu X, Tesfai J, Evrard YA, Dent SY, Martinez E. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J Biol Chem. 2003;278:20405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zhang N, Ichikawa W, Faiola F, Lo SY, Liu X, Martinez E. MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits. Biochim Biophys Acta. 2014;1839:395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Gamper AM, Roeder RG. Multivalent binding of p53 to the STAGA complex mediates coactivator recruitment after UV damage. Mol Cell Biol. 2008;28:2517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Papadopoulos P, Gutierrez L, Demmers J, Scheer E, Pourfarzad F, Papageorgiou DN, et al. TAF10 Interacts with the GATA1 Transcription Factor and Controls Mouse Erythropoiesis. Mol Cell Biol. 2015;35:2103–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Sengupta T, Cohet N, Morle F, Bieker JJ. Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc Natl Acad Sci U S A. 2009;106:4213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Uesugi M, Verdine GL. The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc Natl Acad Sci U S A. 1999;96:14801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–3. [DOI] [PubMed] [Google Scholar]
- [94].Yoon JW, Lamm M, Iannaccone S, Higashiyama N, Leong KF, Iannaccone P, et al. p53 modulates the activity of the GLI1 oncogene through interactions with the shared coactivator TAF9. DNA Repair (Amst). 2015;34:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol Cell Biol. 2007;27:297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Layer JH, Miller SG, Weil PA. Direct transactivator-transcription factor IID (TFIID) contacts drive yeast ribosomal protein gene transcription. J Biol Chem. 2010;285:15489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Papai G, Tripathi MK, Ruhlmann C, Layer JH, Weil PA, Schultz P. TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature. 2010;465:956–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rossi MJ, Kuntala PK, Lai WKM, Yamada N, Badjatia N, Mittal C, et al. High resolution protein architecture of the budding yeast genome. Nature. 2021;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Liu WL, Coleman RA, Ma E, Grob P, Yang JL, Zhang Y, et al. Structures of three distinct activator-TFIID complexes. Genes Dev. 2009;23:1510–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Saluja D, Vassallo MF, Tanese N. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol. 1998;18:5734–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Tanese N, Saluja D, Vassallo MF, Chen JL, Admon A. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci U S A. 1996;93:13611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hibino E, Inoue R, Sugiyama M, Kuwahara J, Matsuzaki K, Hoshino M. Interaction between intrinsically disordered regions in transcription factors Sp1 and TAF4. Protein Sci. 2016;25:2006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Gill G, Pascal E, Tseng ZH, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci U S A. 1994;91:192–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Wang X, Truckses DM, Takada S, Matsumura T, Tanese N, Jacobson RH. Conserved regionI of human coactivator TAF4 binds to a short hydrophobic motif present in transcriptional regulators. Proc Natl Acad Sci U S A. 2007;104:7839–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chen WY, Zhang J, Geng H, Du Z, Nakadai T, Roeder RG. A TAF4 coactivator function for E proteins that involves enhanced TFIID binding. Genes Dev. 2013;27:1596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Bhattacharya S, Lou X, Hwang P, Rajashankar KR, Wang X, Gustafsson JA, et al. Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D. Proc Natl Acad Sci U S A. 2014;111:9103–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wang H, Curran EC, Hinds TR, Wang EH, Zheng N. Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module. Cell Res. 2014;24:1433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Li HH, Li AG, Sheppard HM, Liu X. Phosphorylation on Thr-55 by TAF1 mediates degradation of p53: a role for TAF1 in cell G1 progression. Mol Cell. 2004;13:867–78. [DOI] [PubMed] [Google Scholar]
- [109].Li AG, Piluso LG, Cai X, Gadd BJ, Ladurner AG, Liu X. An acetylation switch in p53 mediates holo-TFIID recruitment. Mol Cell. 2007;28:408–21. [DOI] [PubMed] [Google Scholar]
- [110].Allende-Vega N, Saville MK, Meek DW. Transcription factor TAFII250 promotes Mdm2-dependent turnover of p53. Oncogene. 2007;26:4234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Coleman RA, Qiao Z, Singh SK, Peng CS, Cianfrocco M, Zhang Z, et al. p53 Dynamically Directs TFIID Assembly on Target Gene Promoters. Mol Cell Biol. 2017;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wei Y, Resetca D, Li Z, Johansson-Akhe I, Ahlner A, Helander S, et al. Multiple direct interactions of TBP with the MYC oncoprotein. Nat Struct Mol Biol. 2019;26:1035–43. [DOI] [PubMed] [Google Scholar]
- [113].Anandapadamanaban M, Andresen C, Helander S, Ohyama Y, Siponen MI, Lundstrom P, et al. High-resolution structure of TBP with TAF1 reveals anchoring patterns in transcriptional regulation. Nat Struct Mol Biol. 2013;20:1008–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Kotani T, Miyake T, Tsukihashi Y, Hinnebusch AG, Nakatani Y, Kawaichi M, et al. Identification of highly conserved amino-terminal segments of dTAFII230 and yTAFII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J Biol Chem. 1998;273:32254–64. [DOI] [PubMed] [Google Scholar]
- [115].Patel AB, Louder RK, Greber BJ, Grunberg S, Luo J, Fang J, et al. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science. 2018;362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013;48:575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin Cell Dev Biol. 2011;22:759–68. [DOI] [PubMed] [Google Scholar]
- [118].Verger A, Monte D, Villeret V. Twenty years of Mediator complex structural studies. Biochem Soc Trans. 2019;47:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Plaschka C, Lariviere L, Wenzeck L, Seizl M, Hemann M, Tegunov D, et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518:376–80. [DOI] [PubMed] [Google Scholar]
- [120].Quevedo M, Meert L, Dekker MR, Dekkers DHW, Brandsma JH, van den Berg DLC, et al. Mediator complex interaction partners organize the transcriptional network that defines neural stem cells. Nat Commun. 2019;10:2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci U S A. 1996;93:8329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Sever R, Glass CK. Signaling by nuclear receptors. Cold Spring Harb Perspect Biol. 2013;5:a016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci U S A. 1998;95:7939–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Belorusova AY, Bourguet M, Hessmann S, Chalhoub S, Kieffer B, Cianferani S, et al. Molecular determinants of MED1 interaction with the DNA bound VDR-RXR heterodimer. Nucleic Acids Res. 2020;48:11199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Ren Y, Behre E, Ren Z, Zhang J, Wang Q, Fondell JD. Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol Cell Biol. 2000;20:5433–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Liu Z, Yao X, Yan G, Xu Y, Yan J, Zou W, et al. Mediator MED23 cooperates with RUNX2 to drive osteoblast differentiation and bone development. Nat Commun. 2016;7:11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–94. [DOI] [PubMed] [Google Scholar]
- [128].Stevens JL, Cantin GT, Wang G, Shevchenko A, Shevchenko A, Berk AJ. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science. 2002;296:755–8. [DOI] [PubMed] [Google Scholar]
- [129].Yin JW, Liang Y, Park JY, Chen D, Yao X, Xiao Q, et al. Mediator MED23 plays opposing roles in directing smooth muscle cell and adipocyte differentiation. Genes Dev. 2012;26:2192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Chu Y, Gomez Rosso L, Huang P, Wang Z, Xu Y, Yao X, et al. Liver Med23 ablation improves glucose and lipid metabolism through modulating FOXO1 activity. Cell Res. 2014;24:1250–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Yang X, Zhao M, Xia M, Liu Y, Yan J, Ji H, et al. Selective requirement for Mediator MED23 in Ras-active lung cancer. Proc Natl Acad Sci U S A. 2012;109:E2813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Sun Y, Zhu X, Chen X, Liu H, Xu Y, Chu Y, et al. The mediator subunit Med23 contributes to controlling T-cell activation and prevents autoimmunity. Nat Commun. 2014;5:5225. [DOI] [PubMed] [Google Scholar]
- [133].Fant CB, Taatjes DJ. Regulatory functions of the Mediator kinases CDK8 and CDK19. Transcription. 2019;10:76–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Poss ZC, Ebmeier CC, Odell AT, Tangpeerachaikul A, Lee T, Pelish HE, et al. Identification of Mediator Kinase Substrates in Human Cells using Cortistatin A and Quantitative Phosphoproteomics. Cell Rep. 2016;15:436–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526:273–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Steinparzer I, Sedlyarov V, Rubin JD, Eislmayr K, Galbraith MD, Levandowski CB, et al. Transcriptional Responses to IFN-gamma Require Mediator Kinase-Dependent Pause Release and Mechanistically Distinct CDK8 and CDK19 Functions. Mol Cell 2019;76:485–99 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A. 2014;111:7319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Tuttle LM, Pacheco D, Warfield L, Luo J, Ranish J, Hahn S, et al. Gcn4-Mediator Specificity Is Mediated by a Large and Dynamic Fuzzy Protein-Protein Complex. Cell Rep. 2018;22:3251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, et al. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell. 2011;44:942–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Taatjes DJ, Naar AM, Andel F 3rd, Nogales E, Tjian R Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–62. [DOI] [PubMed] [Google Scholar]


