Abstract
Three-dimensional (3D) organization of the human genome plays an essential role in all DNA-templated processes, including gene transcription, gene regulation, and DNA replication. Computational modeling can be an effective way of building high-resolution genome structures and improving our understanding of these molecular processes. However, it faces significant challenges as the human genome consists of over 6 × 109 base pairs, a system size that exceeds the capacity of traditional modeling approaches. In this perspective, we review the progress that has been made in modeling the human genome. Coarse-grained models parameterized to reproduce experimental data via the maximum entropy optimization algorithm serve as effective means to study genome organization at various length scales. They have provided insight into the principles of whole-genome organization and enabled de novo predictions of chromosome structures from epigenetic modifications. Applications of these models at a near-atomistic resolution further revealed physicochemical interactions that drive the phase separation of disordered proteins and dictate chromatin stability in situ. We conclude with an outlook on the opportunities and challenges in studying chromosome dynamics.
I. INTRODUCTION
The genome is often hailed as the blueprint of life. It instructs the development and operation of an entire organism. Advancement in sequencing techniques has made possible the determination of nucleic acid sequences that make up the genome for many species, including the completion of the human genome project.1,2 These sequences provide valuable information about the genome’s function and a glimpse into the meaning of life at the atomic level.3 They allowed discovering and cataloging coding regions or genes that produce protein molecules and define the distinctive features of individual cells. Notably, genes only make up a small fraction of the genome. While the rest of the genome is not transcribed into proteins, it helps encode cellular diversity found in multicellular organisms by regulating the amount of proteins genes produced.4
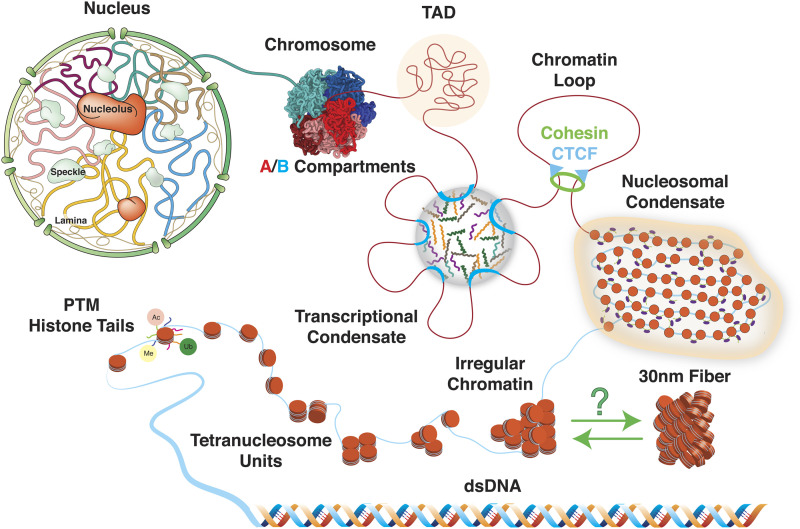
One of the means the non-coding regions regulate gene expression is through three-dimensional (3D) genome organization. The genome, much like enzymes, must fold in 3D to form active sites that catalyze the progression of chemical reactions.5–7 As shown in Fig. 1, the first layer of eukaryotic genome organization is the formation of nucleosomes by wrapping DNA around histone proteins. Access to nucleosomal DNA is restricted due to its tight binding with histones.8 Therefore, regulating nucleosome conformation serves as an effective strategy for controlling genome accessibility and modulating gene expression levels.9,10 Similar arguments can be made for a string of nucleosomes, i.e., chromatin, as compacting chromatin by stacking nucleosomes close to each other will again limit DNA accessibility and exclude the binding of the transcriptional machinery.11,12 At even larger scales, genome folding could bring regulatory elements (enhancers and promoters) that are far apart in the linear sequence (10kb to 1Mb) to spatial proximity.13 Such contacts could achieve targeted control of gene expression by selectively localizing the transcriptional machinery to cell-type-specific genes. A detailed characterization of the genome organization and its dependence on the non-coding regions could improve our understanding of gene regulation in eukaryotes.
FIG. 1.
Illustration of the many layers of three-dimensional genome organization. In eukaryotes, the double-stranded DNA first wraps around histone proteins (orange) to form nucleosomes. The N-terminal tails of histone proteins are subject to a wide range of post-translational modifications (PTM), including acetylation (Ac), methylation (Me), and ubiquitination (Ub). A string of nucleosomes, or chromatin, may compact into irregular structures and nucleosomal condensates, although regular structures have also been seen in vitro. At larger scales, genomic segments that are far apart in sequence can come in contact due to the formation of chromatin loops, transcriptional condensates, and topologically associating domains (TAD). TADs of similar properties may phase separate, resulting in the compartmentalization of chromosomes into regions enriched with heterochromatin (B compartment, blue) or euchromatin (A compartment, red). These two chromatin types differ in their compactness, gene density, and nuclear localization. Inside the nucleus, individual chromosomes often occupy non-overlapping regions to form territories.
Many questions regarding what the genome structure is and how the genome folds remain to be addressed.14 Globally, the genome organization appears to be poised on the border between order and disorder. While many studies have uncovered non-random structural features, the genome is largely amorphous and does not adopt a single unique conformation.15 How specific interactions emerge from the large pool of non-specific contacts is mostly unknown.16 It is tempting to assume that the genome adopts a similar sequence to structure relationship as that has been widely accepted for proteins. If so, what exactly is the sequence that dictates the genome organization? The answer to this question is not apparent and cannot be simply the sequence of nucleotides. Epigenetic modifications17–19 may be the key to forming distinct genome organizations to encode unique gene expression profiles in cells that share identical DNA sequences. Equally important questions arise at smaller scales on the level of single genes (∼10kb). While a seemingly more straightforward problem, the structure for a string of nucleosomes remains controversial.20–22 There is an ongoing debate regarding whether chromatin in situ adopts the same set of conformations as those seen in vitro. Its interaction with protein molecules inside the nucleus may drive chromatin into phase-separated condensates,23,24 which renders a high-resolution structural characterization challenging due to its dynamic nature. Addressing these questions will improve our understanding of gene regulatory mechanisms and is promising for uncovering novel genome engineering approaches to alter the genome organization for more targeted and reversible therapeutic approaches. We hope this perspective may introduce the chemical physics community to the fascinating genome folding problem and inspire more theoretical and physical chemistry research efforts.
Much like its success in studying protein folding, computational modeling could be powerful tools for interpreting experimental data, exploring mechanistic hypotheses, and predicting genome organization de novo.25–27 Many simulation tools introduced for protein folding can indeed be transferred to study genome organization. Computational modeling will face unique challenges not seen in protein folding as well, presenting opportunities for the development of new methods and theories. First, the human genome consists of over 6 × 109 base pairs, a system size that exceeds the capacity of all-atom explicit solvent simulations. Even a small chromatin segment, when fully solvated, can lead to systems of several millions of atoms in size. For whole-genome modeling, coarse-graining will be necessary. Second, while the physicochemical interactions that stabilize protein structures are relatively well understood, the same cannot be said for the genome. In particular, many long-range contacts between genomic segments are mediated by protein molecules, the identity of which has yet to be revealed. Since the system’s exact composition is lacking, multi-scale approaches28,29 that aim at deriving coarse-grained force fields from atomistic simulations are inapplicable for parameterizing whole-genome models. Further development of the coarse-grained modeling strategy is needed. Finally, the genome is inherently a non-equilibrium system, and its conformational ensemble, in principle, cannot be described with equilibrium statistical mechanics.30 Efficient algorithms are needed to simulate the impact of ATP-driven enzymes that consume energy to break the detailed balance, so are theoretical tools for interpreting simulation results.
In this perspective, we will review the progress in modeling the human genome and outline the challenges that lie ahead. It is organized as follows: We first discuss modeling efforts at the entire chromosome and the whole-genome level. Particular focus will be devoted to the data-driven mechanistic modeling approach that is effective at unraveling the principles of genome organization and enabling de novo structural prediction. In Sec. III, we transition to studies on a much smaller scale for a string of nucleosomes. Arguments will be made to near-atomistic models that can potentially bridge the gap between mesoscopic models and atomistic simulations to characterize chromatin organization in situ. While the modeling strategies in Secs. II and III differ significantly, maximum entropy optimization serves as an effective strategy for parameterizing coarse-grained models in both cases. We acknowledge that chromatin organization between the two scales considered here is of critical importance as well, and see the excellent reviews on such topics.31–33 Finally, we provide an outlook on the opportunities and challenges in modeling chromosome dynamics.
II. STRUCTURE FOR THE ENTIRE GENOME
The human genome consists of over 6 × 109 base pairs, and each one of the 46 chromosomes accounts for tens or hundreds of million bases (MB) in length. When fully extended, the genome accounts for ∼2 m. Formation of nucleosomes that are 10 nm in size and 200 bp in sequence length reduces the genome length by almost an order of magnitude to 0.3 m, a number that still vastly exceeds the size of a typical cell nucleus (∼10 μm in diameter). Therefore, additional folding and compaction must occur beyond nucleosomes to fit the genome inside the nucleus. We note that most probable polymer configurations are not straight lines but resemble random coils with size Rg ∝ N1/2a,34 where N is the number of nucleosomes and a is the nucleosome diameter. For a naive estimation with all chromosomes connected together, we have N ≈ 3 × 107, a = 10 nm, and Rg ≈ 55 µm. Further collapsing coiled configurations into globules that scale as N1/3a ≈ 3 µm does allow the genome to fit inside the nucleus. Great progress has been made toward understanding how the folding proceeds and what the final folded configuration is. Many research groups have contributed to addressing this inherently multi-scale problem using a wide range of tools.
A. Experimental characterization of genome organization
The genome is large and amorphous, and the nucleus environment is heterogeneous and crowded. These features render a high-resolution structural characterization of genome organization challenging. Many well-established techniques that succeed at producing atomic structures for proteins, including x-ray crystallography and Nuclear Magnetic Resonance (NMR), are not directly applicable to the genome. Instead, structural information of the genome was often derived from electron microscopy (EM) or fluorescence in situ hybridization (FISH). Applications of EM imaging have led to the discovery of different chromatin types, including heterochromatin and euchromatin that differ in their degree of compactness, gene density, and nuclear localization.35,36 Electron micrographs of metaphase chromosomes swollen by divalent ions further revealed the presence of radial loops.37 Meanwhile, FISH, which uses a probe DNA sequence attached with a fluorophore to hybridize with a corresponding chromosome region via base pairing, allows direct visualization of specific genomic loci with fluorescence microscopy. Experiments that employ multiple probes to measure the spatial distance between genomic loci support the presence of looped structures in interphase as well.38–40 Labeling whole chromosomes further revealed that they tend to demix and occupy non-overlapping spatial regions termed chromosome territories.41–43 While imaging-based techniques have successfully characterized chromosome morphology and large scale organization, traditionally, they have not been effective at studying fine-scale structures due to the limited resolution.
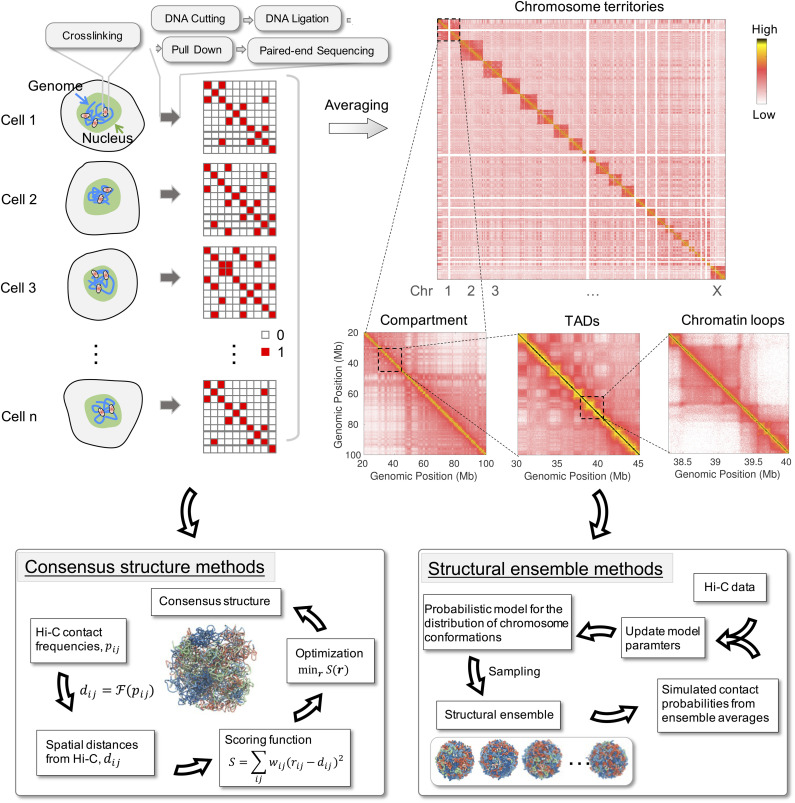
High-throughput sequencing-based techniques, including genome-wide chromosome conformation capture (Hi-C) and related methods,44–46 have served as effective alternatives for studying genome organization. As shown in Fig. 2, Hi-C experiments start by cross-linking DNA segments in spatial proximity inside the nucleus using formaldehyde. The genome is then fragmented with restriction enzymes to result in pairs of contacting segments. Sequencing these contacting pairs reveals their identity, which can be mapped to the reference genome to determine the genomic position of individual loci. For a single structure, this protocol produces a binary contact matrix, each entry of which being either 0 or 1 to indicate whether the corresponding genomic pair is in contact or not. When averaged over a population of millions of cells, as often done in experiments, the protocol produces a contact frequency, or probability, map. The contact map provides rich information on the arrangement of the genome inside the nucleus. Its resolution is bound by the frequency at which restriction enzymes cut the DNA. Since the average spacing between sequences recognized by restriction enzymes is ∼1kb, Hi-C experiments can, in principle, characterize genome organization at high resolution.
FIG. 2.
Computational modeling of genome organization with Hi-C data. Top: illustration of the experimental protocol used in population Hi-C experiments (see the text for details). An example contact probability map for the genome from GM12878 cells is shown on the right, with the probability decreasing from yellow to red and to white. Bottom: illustration of the two popular methods used in building genome structures from Hi-C data. In consensus structure methods (left), pairwise Hi-C contact frequencies (pij) are first transformed to distances (dij) via a mapping function . These distances can be used as constraints to refine computer models and derive consensus structures for the genome. An ensemble of structures can also be used (right) to reproduce Hi-C contact frequencies without converting them to distances. These structural ensemble methods often describe the structures with a probabilistic model and use iterative algorithms to update model parameters.
Hi-C experiments have confirmed findings from imaging studies and revealed many previously unknown features of genome organization.47–49 A typical contact probability map from GM12878 cells is shown in Fig. 2, where the probability decreases from yellow to red and to white. Consistent with the formation of chromosome territories, individual chromosomes appear as squares of high contact frequency along the diagonal. Zooming into individual chromosomes revealed block-wise checkerboard patterns that indicate the presence of two chromatin types, often termed A/B compartments45 and correlated with euchromatin and heterochromatin. The yellow squares appearing along the diagonal line support the formation of topologically associating domains (TADs) with enriched contacts inside the domain than contacts across domains.50–52 A subset of the TADs exhibits strong contact signals at the corner, supporting the formation of looped structures with pronounced interactions localizing at the ends.51
We note that Hi-C experiments are not without limitations. In particular, averaging over a population of cells is often necessary to achieve improved statistics but also casts doubt on the relevance of observed structural motifs in individual cells.53–55 Hi-C experiments also do not directly measure 3D distances. Sometimes, a higher contact frequency does not translate into a shorter distance,56,57 rendering a structural interpretation of the contact map less straightforward. The recent development of super-resolution imaging58–61 and in situ genome sequencing62 techniques can potentially overcome the shortcomings of traditional imaging and Hi-C to provide high-resolution single-cell structural models for the genome.43,63
B. Computational modeling of genome organization
Computational modeling has been valuable for studying genome organization. In addition to providing intuitive 3D structural views of the genome, computational approaches are effective at falsifying hypotheses and identifying the ones that are consistent with experimental data.26,27,64,65 Due to the complexity of the genome organization and our limited knowledge of the folding mechanism, various modeling strategies have been employed in prior studies. We classify them as data-driven approaches that mainly rely on experimental data for structural modeling and hypothesis-driven approaches in which experimental data mostly serve for validation purposes.
1. Data-driven modeling approaches
a. Consensus structure refinement approaches.
Hi-C experiments measure the contact probabilities between genomic loci. Assuming a direct mapping between the probabilities and spatial distances exists, one can convert Hi-C data into a distance matrix. Consensus structures can be derived from this matrix by minimizing scoring functions that measure the difference between simulated and experimental distances (Fig. 2). We note that similar ideas have been widely used in predicting protein structures66 in which the distances between amino acids can be derived from NMR measurements.
The scoring functions are often complex and are of high dimension. Numerous algorithms have been introduced for their efficient optimization to study genome organization in different organisms.67–78 Duan et al. constructed a three-dimensional model of the Saccharomyces cerevisiae genome using the interior-point gradient-based methods.67 The model effectively captures intra- and inter-chromosomal contacts and reveals notable structural features, including individual chromosomal folding and centromere-anchored inter-chromosomal interactions. Molecular dynamics simulations,73 Monte Carlo sampling,76 simulated annealing,69,70,75 Bayesian inference,77 and manifold embedding78–80 have also been used for structure building. These techniques allow for the construction of multiple models consistent with experimental data and have provided insight into the connection between 3D structures and 1D genomic features. Notably, Yildirim and Feig76 derived structural models for Caulobacter crescentus at the single base-pair resolution, thanks to its small genome size. The Mozziconacci laboratory introduced a 3D shortest-path reconstruction method based on the multidimensional scaling-based algorithm.68 This method is computationally efficient and can produce robust results for sparse and noisy contact maps. Algorithms that separate out intra- and inter-chromosomal contacts for high-resolution structural reconstruction at low computational costs have also been developed.74 Besides the applications on population Hi-C data, distance-based methods have recently been generalized to single-cell Hi-C81–84 and ChIA-PET data.71,72
b. Structural ensemble refinement approaches.
The pursuit of a consensus structure, while desirable for simplified interpretations, can mask the intrinsic heterogeneity of genome organization within a cell population. Since population Hi-C experiments report the contact frequencies averaged over many cells, there will likely be an ensemble of structures that collectively reproduce the data.53,81
To account for large conformational fluctuations around the mean structure, Sasai and co-workers designed a modeling approach using statistical potentials centered at the distances converted from contact probabilities.85,86 When applying this approach to the yeast genome, the authors explained experimental chromosome distributions and uncovered a correlation between the transcriptional level of genes and their spatial distribution.
Numerous groups have developed computational techniques to directly fit Hi-C data with an ensemble of structures and avoid converting experimental contact frequencies to spatial distances, the functional form for which remains unknown.87–95 For example, Alber and co-workers introduced a probabilistic framework based on the maximum likelihood optimization algorithm to reproduce Hi-C contacts with averages from a population of thousands of genome structures.87–89 Applications of this algorithm revealed the presence of centromere clusters in GM12878 cells and the role of these clusters in positioning chromosomes in the nucleus. Liang and co-workers introduced CHROMATIX, a method that first identifies a minimal set of sufficient interactions and deconvolutes these interactions into a population of single-cell contact states that can then be used for structure reconstruction.90–93 This method succeeds in characterizing higher-order interactions between multiple genomic regions and structural changes during embryogenesis in Drosophila.
Zhang and Wolynes took a more statistical mechanical approach by transforming the problem of structural refinement into parameterization of pairwise interactions between genomic loci, i.e., force field optimization.94,96 In particular, assuming that the genome organization from a population of cells can be approximated with the Boltzmann distribution of an energy function, finding the set of structures that reproduce experimental data is equivalent to determining the corresponding energy function. Using experimental data to parameterize or improve force field is indeed a topic well studied in the protein folding community. Low-resolution data, including small-angle x-ray scattering (SAXS), Förster resonance energy transfer (FRET), and nuclear magnetic resonance (NMR), are frequently used to improve the determination of protein structures.97–99 While many functional forms can be defined for the energy function, the one that introduces the least bias, i.e., the optimal model, is derived by maximizing the excess entropy based on the information theory.100
The starting point of the Zhang and Wolynes approach is a beads-on-a-string model to mimic the continuity of the DNA molecule, where each bead represents a genomic segment of fixed length. A corresponding energy function, , which usually contains terms that account for bonding, bending, and excluding volume effect, can be defined to evaluate the stability of polymer configurations. Additional energy terms that mimic the confinement effect of neighboring chromosomes and the nuclear envelope can also be included. The equilibrium Boltzmann distribution is
| (1) |
where β = 1/kBT. Defining a threshold function f(rij; ro) that switches from 0 to 1 as the distance rij decreases below ro, one can compute the contact probability for a pair of genomic loci as . ro can be viewed as the minimal contact distance necessary for cross-linking. Since U(r) does not capture any biological interactions that chromosomes experience inside the nucleus, the simulated contact probabilities are not expected to reproduce Hi-C data.
To improve the model’s biological relevance, correction terms can be introduced to the energy function. Maximum entropy optimization has become widely popular in recent years99,101–103 as an efficient means to improve the agreement between modeling and experiment. Under the maximum entropy framework, one seeks a new model, UME(r), and a corresponding probability distribution function . Importantly, for the new model to reproduce experimental contact probabilities, i.e., , while maximizing the excess entropy, −∫PME(r)ln[PME(r)/P(r)], its energy function has a unique solution defined as
| (2) |
where αij are Lagrangian multipliers, the value of which ensures the agreement between simulation and experiment. They can be determined via an efficient iterative optimization procedure, as detailed in Ref. 94. We refer the interested readers to Refs. 102 and 104–106 for detailed reviews on maximum entropy optimization.
From the energy function, an ensemble of chromosome structures can be constructed via molecular dynamics simulations. Notably, the structural ensemble is guaranteed to reproduce experimental contact probabilities. By inspecting the simulated structures with knot invariants, Zhang and Wolynes found that interphase chromosomes are free of topological entanglements. Their result generalizes beyond using the scaling exponent of contact probability as a function of sequence separation to support knot-free genome organization.45,107 Since the maximum entropy model matches the complete set of experimental data with minimal assumptions of genome organization, it produces the least biased or “most likely” chromosome structures based on the given knowledge. Conclusions drawn from these structures are thus more reliable than those based on specific features of Hi-C data. When applied to mitotic chromosomes, the maximum entropy approach further revealed cylindrical conformations with a helical twisting along the central axis. This twisting was later confirmed in experiments with higher resolution Hi-C data.108 Chu and Wang further studied the conformational transition between interphase and metaphase chromosomes and revealed a two-stage pathway for chromosome compaction.109
Tiana and co-workers applied a similar maximum entropy optimization approach to derive pairwise contact energies from Hi-C. Monte Carlo sampling of the resulting energy function predicted chromatin conformation heterogeneity within a single TAD and provided insight into asymmetric expression of X-chromosome inactivation.110,111 Shi and Thirumalai further applied the maximum entropy approach to recreating the structural ensemble by reproducing the pairwise mean distances converted from Hi-C probability.112 They characterized the heterogeneity of chromosome structures within the same cell type and across different types.
2. Hypothesis-driven modeling approaches
The data-driven modeling approaches are useful for reconstructing chromosome structures. However, they face difficulty at addressing questions regarding why chromosomes adopt specific conformations. Hypothesis-driven approaches are more suited for exploring genome organization mechanisms and have also been widely used in parallel.
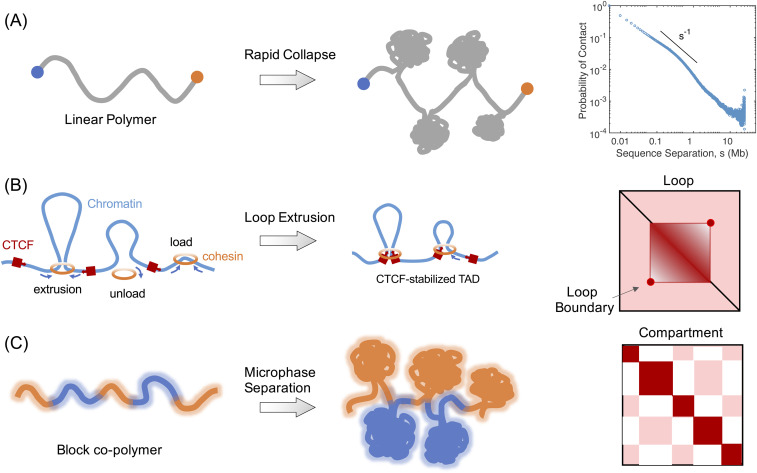
Given that chromosomes are long macromolecules, whose equilibrium conformations will become entangled under confinement, Grosberg et al. proposed the fractal globule as a model for chromosome organization113 [see Fig. 3(a)]. Conformations of the fractal globule are free of entanglements, the presence of which may hinder gene transcription and chromosome separation during mitosis. The fractal globule corresponds to a metastable state that forms when the polymer is rapidly collapsed from an extended conformation. It exhibits a different scaling law from equilibrium conformations regarding the decay of the contact probability as a function of sequence separation. Remarkably, early Hi-C experiments indeed appear to support the scaling exponent predicted by the fractal globule.45,107 Metastable conformations can persist over the lifetime of a cell, as shown by Rosa and Everaers,114 due to the slow relaxation time of polymer topology in a crowded environment.
FIG. 3.
Illustration of the various mechanisms proposed for genome folding. (a) The metastable fractal globule forms in a process that drives the rapid collapse of a long polymer from an expanded, knotless configuration. The two ends do not have enough time to take part in the collapse, and the polymer remains knotless. (b) In the extrusion model, chromatin loops form as a result of the processive movement of Cohesin molecules along the DNA. CTCF molecules act as blockers to stop Cohesin extrusion, explaining the accumulation of the two proteins at loop boundaries. (c) Microphase separation of block copolymers can lead to contact patterns similar to the compartmentalization seen in Hi-C maps.
It is worth mentioning that when interpreted strictly, as pointed out by Huang et al.,115 the fractal globule is probably too ideal of a model for genome organization. Its self-similarity property conflicts with the spatial heterogeneity of chromatin packing. Huang et al. further introduced a self-returning random walk (SRRW) model that achieves simultaneous high self-interacting frequency and high space-filling heterogeneity. This model makes several predictions consistent with experimental observations, including highly porous chromatin structures, flexible higher-order folding, and irregularly shaped chromosome territory.
In addition to their global topological organization, specific structural features of chromosomes have also been studied extensively. For example, loops have received significant attention in early models of interphase chromosomes to explain the scaling behavior of spatial distances as a function of genomic separation measured by FISH experiments.38,39,116,117 High-resolution Hi-C data provided direct evidence for the presence of loops in interphase chromosomes.51 Importantly, connecting the contact map with the underlying DNA sequence further allowed proposing and validating molecular mechanisms of loop formation. For example, the extrusion model118,119 assumes that these loops form by the processive movement of cohesin molecules that bring close genomic segments far apart in sequence until stopped by CTCF molecules [see Fig. 3(b)]. This model explains the enriched contact within a TAD and the flanking of TADs and loop boundaries with Cohesin and CTCF molecules. Several predictions of the extrusion model have been validated with perturbative Hi-C120–124 and in vitro experiments.125,126 Other groups have also highlighted the importance of protein binding in chromosome organization.127,128 In these models, the strong and specific interactions between protein and chromatin drive the contacts within a TAD and the separation between neighboring TADs.
At a larger scale, both imaging experiments and the checkerboard patterns in Hi-C contact maps support the compartmentalization of active and inactive chromatin. Compartmentalization can arise from the microphase separation seen in block copolymers.129–135 Block copolymers are reasonable models for chromosomes since different chromatin segments can exhibit different chemical properties due to their unique histone modification136 and protein association137,138 patterns. Additionally, non-equilibrium processes, such as transcription and chromatin remodeling, could contribute to the microphase separation as well. In a simplified model, these processes could give rise to higher effective temperature for active chromatin,139,140 and the hotter chromatin will separate from the cooler one as a result of their difference in mobility.141,142 Finally, Wang et al. treated the 3D chromosome arrangement as an ellipsoid packing problem to understand the correlation between chromosome positioning and cell geometry.143
As is evident from the above discussions, multiple models and mechanisms often explain chromosome organization at various scales and reproduce specific features of the experimental data equally well. It is plausible that numerous players co-exist to organize the genome. Quantitatively evaluating the relative significance of various mechanisms is challenging, however, due to the presence of free model parameters whose values are not known a priori.
C. Perspective: Data-driven mechanistic modeling
Coupling the data- and hypothesis-driven approaches together can potentially produce a powerful strategy for modeling genome organization. This strategy ensures the biological relevance of simulated genome structures since all model parameters will be derived from Hi-C data. In the meantime, it will be well suited for mechanistic investigation as the polymer model’s energy function will be designed explicitly from biological factors that are known to contribute to genome organization. Pioneering work by Di Pierro et al. has shown that a phase separation model among sub-compartments succeeds at reproducing Hi-C data and recapitulating many known aspects of chromosome organization.134
1. Exploring the principles of genome organization
We followed the data-driven mechanistic modeling strategy and introduced a polymer model to study the 3D organization of the human diploid genome at 1Mb resolution [Fig. 4(a)].144 One particular hypothesis that we aimed to evaluate is whether the whole-genome organization can be understood from phase separation of different chromatin types. Toward that end, we designed a block copolymer model that includes 46 polymers in spherical confinement. Each polymer represents a chromosome whose length is determined by the underlying DNA sequence. The spherical confinement mimics the effect of the nuclear envelope. A bead in the polymer was either labeled A or B to represent the two compartment types seen in Hi-C. The compartment profiles for individual chromosomes were determined from eigenanalysis on the correlation matrix of intra-chromosomal Hi-C contact maps.45 For this polymer model, the energy function is defined as follows:
| (3) |
where I represents indices over chromosomes and i and j represent individual genomic loci, respectively. U(rI) is the generic potential as that in Eq. (2) to account for polymer topology. The second term corresponds to the ideal chromosome potential that depends only on the sequence separation but not on compartment types. It accounts for the interactions within the same chromosome and could arise from protein-mediated contacts38,116–118,123,127,145,146 and spherical confinement.93,147 Compartment type (Ti ∈ {A, B}) specific interactions are included in the last term. The energy function, UGenome(r), can again be derived by maximizing the excess entropy while enforcing experimental constraints that measure the average contact probabilities at various sequence separations and between different compartment types.144 Parameters in the energy function, α(|i − j|) and α(Ti, Tj), can be uniquely determined with the iterative optimization algorithm mentioned in the Sec. II B 1 b.
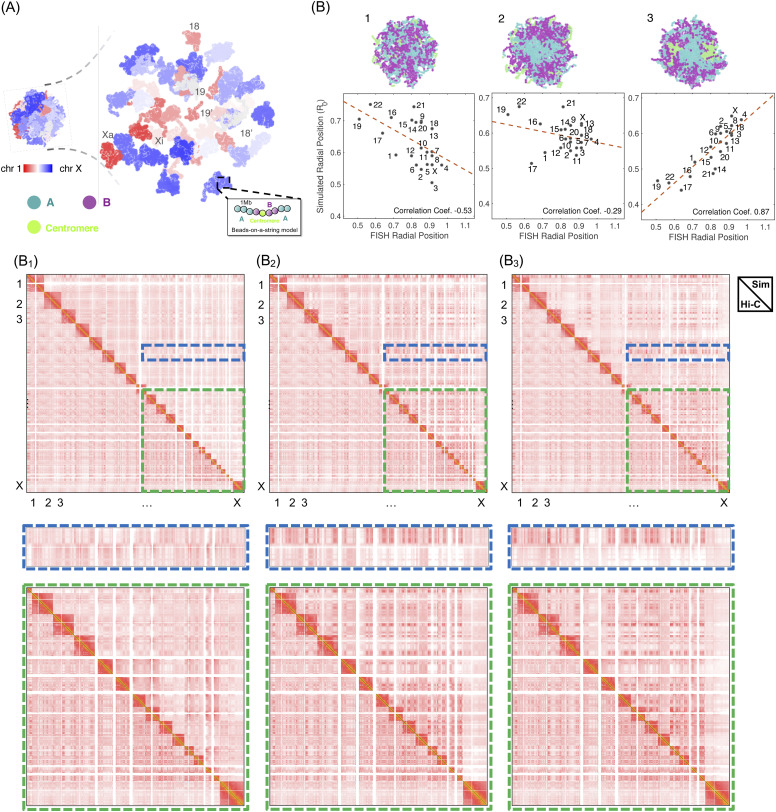
FIG. 4.
Data-driven mechanistic modeling of the whole-genome organization. (a) An example configuration of the diploid human genome colored from red to white and to blue with increasing chromosome ID. Each chromosome is modeled as a string of beads that can either be A (cyan) compartments, B (purple) compartments, or centromeres (green). (b) Example genome configurations colored by bead types (top), comparison between simulated and experimental chromosome radial positions (bottom), and comparison between simulated (upper triangle) and experimental (lower triangle) genome-wide contact maps for three genome models. In model 1, only one set of parameters was used to model intra- and inter-chromosomal interactions, while two sets of independent parameters were used in model 2. In model 3, in addition to the use of independent parameters for intra- and inter-chromosomal interactions, the centromeric regions were explicitly represented with a new type.
As shown in Fig. 4 B1, this model succeeds in recapitulating the microphase separation between the two compartments and the checkerboard pattern seen in Hi-C contact maps. However, a close examination of the simulated genome conformations suggests that heterochromatin (B compartments) is scattered over the entire nucleus rather than localizing to the nuclear periphery, as shown in EM images.36 Notably, the unique modeling strategy employed here ensures that the inconsistency between simulation and experiment cannot be resolved by fine-tuning the model parameters, the values of which were inferred from experimental data optimally. The hypotheses introduced in designing the model must be incorrect or incomplete. Therefore, cross-validating simulated structures with imaging experiments offers a feedback mechanism to falsify hypotheses of genome organization mechanisms.
After revisiting the model hypotheses, we found that decoupling the intra- and inter-chromosome interactions is necessary and sufficient for reproducing the peripheral localization of B compartments (Fig. 4 B2). This decoupling indicates the presence of different mechanisms that drive phase separation at various scales. Indeed, it is known that protein-mediated interactions dominate at intra-chromosome contacts.148 On the other hand, various nuclear landmarks, including lamina and speckles,149,150 could contribute to the aggregation of various chromosomes. These distinct mechanisms necessitate the use of two sets of independent parameters for intra- and inter-chromosome interactions.
We further found that a third compartment type that corresponds to the central region of chromosomes, or centromeres, is needed to reproduce the radial position of individual chromosomes (Fig. 4 B3). Including this type helps capture centromere clustering that has been seen in prior studies.151 We applied the modeling strategy to study the global rearrangement of genome organization upon tumorigenesis152 and observed a remarkable change in genome organization upon tumorigenesis. Unlike their interior localization in normal samples, A compartments in tumors are scattered across the entire nucleus. This observation was later confirmed by electron microscopy.
Most existing computer models do not explicitly include nuclear landmarks such as lamina, nucleoli, and speckles, although studies that account for them are emerging.85,86,153–156 Modeling the landmarks and the genome together could potentially circumvent the need for an artificial distinction between intra- and inter-chromosome interactions as in the current model. A careful investigation of the coupling between the genome and nuclear lamina may provide insight into the mechanical properties of the nucleus and the impact of mechanical forces on gene transcription.157,158 Such models could also reveal the role of chromatin network in both the kinetics and thermodynamics of phase separation that drives nucleolus and speckle formation.156,159,160
2. Predicting chromosome structures from epigenetic marks
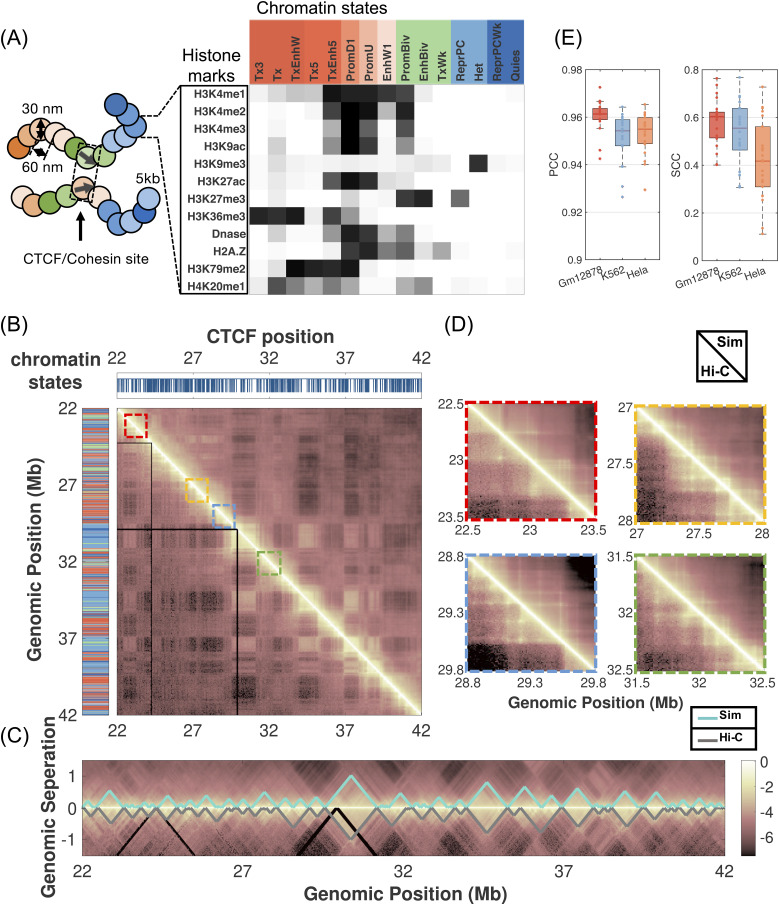
In addition to the whole-genome organization, we applied the data-driven mechanistic modeling strategy to study the structure of single chromosomes at 5kb resolution.161 The model’s higher resolution allows detailed characterization of fine-scale structures, including TADs and loops, which play essential roles in gene regulation. Chromosomes can again be viewed as block copolymers, but now the monomer types go beyond the A/B compartments used in whole-genome modeling. Instead, we represented a chromosome as a sequence of chromatin states that were defined as unique combinatorial patterns of histone modifications [see Fig. 5(a)]. Chromatin states and A/B compartments are related in many ways. In particular, the states marked with histone modifications H3K4me3 and H3K27ac often correspond to A compartments, while those marked with H3K9me3 or H3K27me3 overlap with B compartments. Chromatin states provide a more nuanced view of the variety of chromatin types and a molecular interpretation for the abstract compartments derived from Hi-C data. We note that the use of multiple representations for polymer modeling is not uncommon in the language of protein folding: while two types of amino acids, hydrophobic and hydrophilic, are sufficient to understand the collapse of protein molecules, the chemical specificity of individual amino acids becomes important for predicting high-resolution structures.
FIG. 5.
Predicting genome organization with a chromatin-state based polymer model. (a) Overview of the key elements of the computational model. The chromatin is modeled as a string of beads, each assigned with a chromatin state based on the corresponding combinatorial pattern of histone marks. Genomic regions bound by CTCF molecules are also identified to model CTCF mediated loop formation. The polymer model succeeds in quantitatively reproducing compartments (b), TADs (c), and chromatin loops (d) for chromosome 1 from GM12878 cells. (e) The polymer model is transferable across chromosomes and cell types as evidenced by the high correlation between simulated and experimental contact maps measured by Pearson correlation coefficients (PCC—left panel) and stratum-adjusted correlation coefficient (SCC—right panel).
We followed the same procedure as whole-genome modeling to define an energy function and derive parameters by enforcing simulated ensemble averages to match average experimental contact probabilities. Details of the model definition and parameterization can be found in Ref. 161. Computer simulations of this model provided a high-resolution structural characterization of chromatin loops, TADs, and compartments. They succeeded in quantitatively reproducing contact probabilities and power-law scaling of 3D contacts, as measured in Hi-C and super-resolution imaging experiments [Figs. 5(b)–5(d)]. In particular, we found that for chromosome 1 of GM12878 cells, the correlation between simulated and experimental contact map exceeds 0.96. The stratum-adjusted correlation coefficient (SCC),162 which considers the distance-dependence effect of contact maps by stratifying them according to the genomic distance, is 0.7. This value is comparable to that between the experimental replicates. We further examined the agreement between simulated and experimental contact maps using multiple feature-specific metrics. We found that our model reproduces over 74% of the CTCF-mediated chromatin loops, correctly identifies ∼80% of the TAD boundaries,163 and captures over 57% of the significant enhancer–promoter contacts.
In addition to producing high-resolution chromosome structures, the chromatin-state based model further enables de novo prediction. For example, it can be applied to any chromosome, as long as the corresponding sequence of chromatin state is provided as input. Unlike A/B compartments that can only be derived from Hi-C data, defining chromatin states only requires histone modifications, and no Hi-C data are needed for structure prediction. We performed additional simulations for chromosomes from GM12878, K562, and HeLa cells, which were not used in model parameterization. We found that the simulated Hi-C data are in good agreement with experimental results as well.
Several other groups have carried out predictive modeling of genome organization using histone modifications with great success as well.139,164,165 For example, Di Pierro et al. simulated chromosome structures at the 50kb resolution using chromatin types defined with histone modification profiles and trained to mimic genomic compartments.135 MacPherson et al. modeled chromosome 16 at the single nucleosome resolution.164 Using the H3K9me3 pattern derived from ChIP-seq signals, they reproduced the phase separation of euchromatin and heterochromatin seen in Hi-C contact maps. The accuracy of these epigenetic-mark-based models can be further improved. In particular, many protein molecules are known to mediate genomic contacts by directly engaging with the underlying nucleotides. A representation based purely on histone modifications is not sufficient to capture such DNA sequence-specific effects. Encouraging progress is being made at predicting these interactions from the DNA sequence using convolution neural networks.166–168 Incorporating these studies into the polymer model could provide a more accurate description of genome organization.
III. STRUCTURE FOR A STRING OF NUCLEOSOMES
The whole-genome modeling effort and many other studies169,170 firmly establish the importance of histone modifications in genome organization. Predicting chromosome structures from chromatin states provides further evidence for the sequence–structure relationship of the genome. This relationship suggests that tinkering the post-translational modifications could serve as effective means to alter genome organization and gene expression. Epigenome engineering that aims to modify such epigenetic marks rather than the genetic code has indeed been pursued as a complementary and less invasive approach to genome engineering.171,172
To effectively alter chromatin types for epigenome engineering and ensure robust and long-lasting changes in histone modifications, it is essential to study mechanisms that dictate their stability. Histone modifications are subject to constant perturbations from addition and removal enzymes inside the nucleus.173,174 The chromatin structure has been implicated in mediating the spread and maintenance of histone marks by these enzymes.136,175–180 Close contacts can facilitate the transfer of enzymes from modified nucleosomes to unmodified ones, introducing cooperativity among the modifications.181 Therefore, studying the stability of histone modifications necessitates a detailed characterization of chromatin organization at the resolution of tens of nucleosomes.
A. The 30 nm fiber
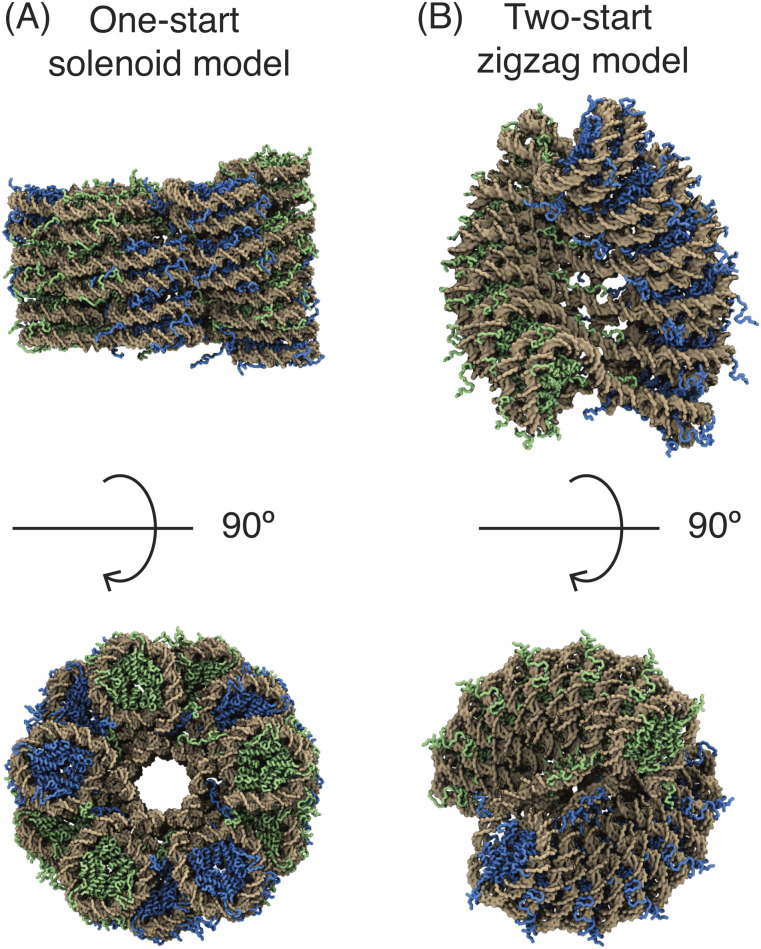
Packaging the 2-m long human genome inside a nucleus of 10 μm in diameter is a daunting task. Nucleosomes and TADs fold the genome at 200 bp and 1Mb, respectively. Additional compaction occurs to bridge the two rather different length scales. For example, Finch and Klug hypothesized that the 10 nm fiber as a string of nucleosomes could coil into a solenoid to form a 30 nm fiber.182 As shown in Fig. 6(a), nucleosomes are packed face-to-face around a central axis in the solenoid model, and each helical turn encloses six or seven nucleosomes. Additionally, Woodcock and co-workers introduced the two-start model based on the electron tomography data that exhibited two nucleosome-wide ribbons.183,184 The two-start model predicts a zigzagging of two nucleosomes that coil into a helical conformation of roughly 30 nm [Fig. 6(b)]. Other fibril models that differ in the nucleosome path have also been proposed.185 Support of these different structural models mostly come from EM images of the chromatin material extracted from the nucleus under harsh conditions. The use of in vitro reconstituted nucleosome arrays helped in removing sample heterogeneity in nucleosome spacing and made possible the determination of high-resolution structures with cryo-EM. These studies further support the presence of regular fibril conformations,186 and the more recent atomic structure favors the two-start zigzag model.187
FIG. 6.
Illustration of the two structural models proposed for chromatin fiber. A total of 24 nucleosomes are shown in panel (a) and 12 nucleosomes in panel (b). Histone proteins from the odd and even nucleosomes are shown in blue and green, respectively. The DNA molecule is indicated in gold.
However, the quest for 30 nm fibers inside the nucleus has often led to disappointing conclusions. Many research groups have failed to confirmed its presence employing a wide range of techniques, including cryo-EM,188 electron spectroscopic imaging,189 small-angle x-ray scattering,190,191 super-resolution imaging,192 Hi-C,122 and ChromEM tomography.193 It is becoming increasingly clear that chromatin organization at the kilobase scale is sensitive to a variety of factors,32,194 including salt concentration, nucleosome spacing, and interaction with non-histone proteins. A systematic characterization of these various factors will be essential for revealing key physicochemical interactions that drive chromatin folding and reconcile the seemingly contradictory experimental observations. However, it has proved challenging due to difficulties in precise chromatin engineering and assembly.
B. Mesoscopic modeling of chromatin fiber
Computational modeling offers an alternative and promising approach for studying chromatin organization at the kilobase scale. Compared to whole-genome modeling, the problem here is more straightforward, at least conceptually, since the system is well defined. All the components are known, and one can, in principle, carry out all-atom simulations for de novo structure prediction.195,196 Materese et al. have shown that such simulations could provide insight into the hydration and electrostatic environments near the nucleosome197 and the dependence of nucleosome elastic properties on histone variants.198 Shaytan et al. revealed a conformational coupling between histone tails and nucleosomal DNA, with histone tail binding promoting DNA bulging and twisting.199 The Collepardo-Guevara et al. characterized the role of histone modifications on chromatin compaction and the importance of linker histone H1 on chromatin hierarchical looping.200,201 Winogradoff and Aksimentiev further reported direct observation of spontaneous DNA unwrapping and characterized the role of CG content in such motions.202 Recently, the Wereszczynski laboratory applied atomistic simulations to study the impact of linker histones on the structure of an octa-nucleosome array.203 However, a minimum system for a string of nucleosomes with explicit solvation can easily exceed several millions of atoms,204 making it challenging to carry out such simulations over long timescales and quantify the stability of various chromatin conformations. Over the years, many coarse-grained models that differ in structural details and energetic terms have been introduced to study chromatin organization.
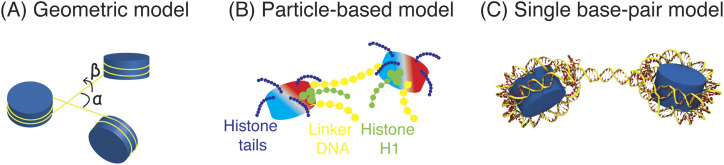
The two-angle model introduced by Woodcock et al. has been instrumental in providing a conceptual framework for studying the chromatin organization205 [see Fig. 7(a)]. The angles correspond to the DNA entry–exit angle of individual nucleosomes (α) and the relative rotational angle between two connecting nucleosomes (β). Varying the two angles can give rise to a wide variety of fibril conformations with different arrangements of nucleosomes,206,207 including the two popular models proposed for the 30-nm fiber: one-start solenoid and two-start zigzag. The dependence of chromatin conformation on histone modifications, histone H1 binding, and linker length can be understood from their impact on the angles.186,208
FIG. 7.
Illustration of the three different types of mesoscopic chromatin models that differ in the representation and energetic contributions. (a) In geometric models, the energetics of the chromatin fiber is fully specified by two angles that correspond to the DNA entry–exit angle of individual nucleosomes (α) and the relative rotational angle between two connecting nucleosomes (β). (b) Particle-based models allow for more accurate treatment of the flexibility of linker DNA and histone tails and histone H1. Inter-nucleosome interactions can be introduced to account for contributions from globular domains of histone proteins. (c) Models with DNA molecules at a single base-pair resolution have been introduced to characterize the bending and twisting of linker DNA with greater details.
The two angle model’s success encouraged the development of more refined models that go beyond geometric arguments. For example, accounting for the elasticity of linker DNAs and interactions between nucleosomes209 allowed Katritch et al. to reproduce the force-extension characteristics of chromatin fiber measured by single-molecule pulling experiments.210 Wedemann et al. further incorporated the electrostatic interactions between linker DNAs to quantify the stability and persistence length of the chromatin fiber and characterize the transition between different fiber structures induced by the binding of linker histone H1.211,212 Schlick and co-workers introduced a chromatin model that employs particle-based representations for histone tails, linker DNA, and linker histones and accounts for the charge distribution of the nucleosome core particle213,214 [Fig. 7(b)]. This model was shown to reproduce a variety of experimental observations, including the dependence of chromatin conformation on salt concentration213 and histone modifications,200,215 and the compaction of chromatin fiber upon the addition of divalent ions and linker histones.216 Lequieu et al. developed the 1CPN model that connects nucleosome core particles with flexible linker DNA.217 Notably, inter-nucleosome interactions were not computed from the mesoscopic representation but parameterized using free energy calculations carried out with a near-atomistic model. Such a multi-scale strategy could lead to a systematic improvement of model accuracy without sacrificing computational efficiency.
The strong dependence of chromatin organization on the linker DNA length further inspired the development of models with a single-base resolution for the DNA [Fig. 7(c)]. For example, Norouzi and Zhurkin used a knowledge-based potential to evaluate the elastic energy of the linker DNA deformations.218 Together with electrostatic interactions and specific internucleosomal interactions, the resulting model predicted the presence of different topoisomers that are favored by chromatin with linker DNA of 10n and 10n + 5 bp in length. The model was further applied to interpret experimental chromatin force-extension curves and dissect the contributions of nucleosome unwrapping and unstacking.219 de Jong et al.220 applied a rigid-base-pair DNA model to describe un- and re-wrapping of nucleosomal DNA and bending and twisting of linker DNA. Monte Carlo simulations with this model support the stability of different fiber models, two-start and one-start, for chromatin with linker lengths of 20 and 50 bp. A similar model that accounts for the bending and twisting of linker DNA was employed by Koslover et al.221 to study the dependence of the nucleosome path in chromatin fibers on the linker DNA length.186 The model predicts a number of energetically comparable configurations with different nucleosome–nucleosome interaction patterns, indicating a potential role for kinetic trapping in chromatin fiber formation.
C. Perspective: Near atomistic chromatin modeling
The prior studies have greatly enriched our understanding of the various conformations isolated chromatin can adopt. To further characterize chromatin organization in situ and reconcile the difficulty for detecting the 30 nm fiber inside the nucleus, one must study the complex interaction between chromatin and the nucleus environment.22,200,193 Increasing evidence suggests that intrinsically disordered proteins (IDPs) that interact strongly with chromatin can spontaneously form liquid droplets.222–228 Collective interactions with these proteins can drive the chromatin into highly dynamic conformations that differ significantly from rigid fibril structures.229,230 Therefore, accounting for protein chromatin interactions will be essential to model chromatin organization in situ but can be challenging with the existing mesoscopic models.
Implicit solvent near-atomistic models offer a promising approach for a detailed characterization of protein–protein and protein–DNA interactions at a single base and residue level. They allow a rigorous treatment of electrostatic, van der Waals, and other interactions between particles based on physical chemistry. Several models have been introduced for the DNA.231 For example, de Pablo and co-workers developed a DNA model that uses three sites per nucleotide (3SPN).232–234 Parameters of the model were chosen to reproduce the experimentally measured free energies of base stacking and hybridization. The model was shown to reproduce many properties of DNA molecules accurately, including the persistence length of both single and double-stranded DNA under physiological conditions, the dependence of persistence length on salt concentration and DNA sequence, DNA melting temperatures, and hybridization rate constants.
Integrating 3SPN DNA with similarly coarse-grained protein models has enabled both quantitative and qualitative investigations of chromatin organization. The de Pablo group modeled histone proteins at one bead per amino acid resolution with the atomistic-interaction based coarse-grained protein model (AICG).235 Interactions among amino acids were parameterized from energies and dynamics of all-atom models via a multiscale protocol. They applied the combined protein–DNA model to reveal that sequence-specific histone binding affinity of the DNA molecule is encoded in their shape.234 The group further uncovered the coupled role of the DNA-sequence, histone modifications, and chromatin remodelers in positioning nucleosomes236 and the dependence of nucleosome unwinding barrier on applied tension.237 Finally, they showed that the model succeeded in reproducing the binding strength between a pair of nucleosomes measured in DNA origami-based force spectrometer experiments.238,239 Using a closely related protein–DNA model, the Kenzaki and Takada studied the dynamics of DNA unwinding,240 transcription-factor binding to nucleosomes,241–243 structures of di- and tri-nucleosomes,244 and nucleosome sliding/remodeling and twist propagation.245,246 These studies highlight the usefulness of near-atomistic models in uncovering detailed mechanisms that are otherwise challenging to extract from either experimental studies or mesoscopic models.
To model a nucleosome at a high resolution, we combined 3SPN DNA with the associative memory, water-mediated, structure, and energy model (AWSEM) for protein introduced by Davtyan et al.247 Each amino acid was represented with three beads for the Cα, Cβ, and O sites. Interactions among amino acids were optimized following the energy landscape theory prescription to maximize the ratio of folding temperature over glass transition temperature and sculpt a funneled folding landscape for a set of training proteins.248–250 Similar to the studies of the de Pablo and the Takada groups, protein–DNA interactions were modeled with the Debye–Hückel theory to account for water’s dielectric properties and the screened electrostatic interaction. Because of the lack of base-specific hydrogen bonding between histone proteins and the nucleosomal DNA as seen in the crystal structure,251 electrostatic contributions are expected to dominate the interactions between them.
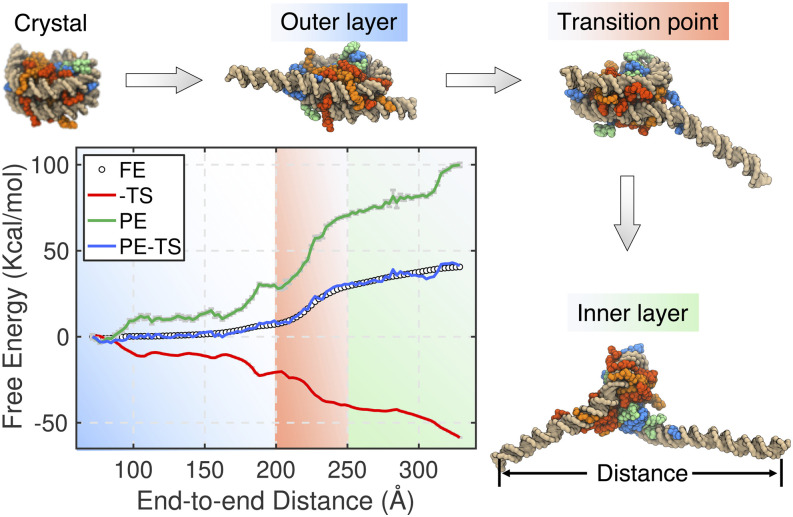
Using the near-atomistic model for protein and DNA molecules, we determined the free energy landscape for nucleosome unwinding.252,253 Our study revealed a sizable energetic barrier that decouples the unwinding of the 147 bp long DNA molecule into two separate processes (Fig. 8). This barrier height is in quantitative agreement with the value determined by Brower-Toland et al. using single-molecule force spectroscopy254 and by the Lequieu et al. using computer simulations of a closely related near-atomistic model.237 We note that the mechanistic origin of the energetic barrier has been the focus of several theoretical studies.255–257 Via a rigorous thermodynamic analysis, we found that the barrier mainly arises from a delayed loss of contacts between disordered histone tails and the DNA. This delay is caused by the dynamical relocation of disordered tails to preserve their DNA contacts. Surprisingly, the energetic penalty is largely offset by an entropic contribution from these newly freed tails. The enthalpy–entropy compensation mechanism provided a fresh perspective on nucleosome stability and highlighted the importance of studying chromatin organization at a high resolution.
FIG. 8.
Thermodynamics of nucleosome unwinding. The free energy (FE) profile (white dots) as a function of the DNA end-to-end distance supports a three-stage scenario for DNA unwinding. The first stage (blue) corresponds to the unwinding of the outer layer. In the second stage (orange), no significant DNA unwinding occurs, but free energy rises sharply. Finally, the inner layer begins to unwind at a modest free energy cost in the third stage (green). Example nucleosome configurations at different stages are provided on the side, with the DNA indicated in gold and histone proteins indicated in orange, red, blue, and green. The free energy barrier in the transition region is mostly dominated by energetic contributions (PE, green line), which are compensated by an increase in entropy (−TS, red line) from the freed histone tails.
We further studied the folding of the basic unit of the chromatin fiber, the tetra-nucleosome.258 The Cα protein model that represents each amino acid with one bead259,260 was used to reduce the computational cost. While lacking the sophisticated energy functions used in AWSEM, it suffices to capture the histone octamer’s conformational fluctuation around the crystal structure. Direct simulations of tetra-nucleosome folding from extended configurations to collapsed structures are not feasible. Folding will be hindered by slow dynamics that arise from the break and formation of strong, non-specific electrostatic contacts. An advanced sampling technique261 that combines metadynamics262 with temperature accelerated molecular dynamics simulations263,264 enabled efficient conformational sampling and produced various tetra-nucleosome configurations.
To further evaluate the stability of the simulated tetra-nucleosome configurations, we computed the free energy surface as a function of the six inter-nucleosomal distances (Fig. 9). The use of a large number of collective variables is needed to resolve the different conformations but poses challenges to traditional methods such as umbrella sampling. Instead, we determined the surface with a neural network approach by integrating mean forces collected at a series of preselected centers.265 The free energy surface supports the global stability of the stacked, zigzag configuration resolved in x-ray crystallography266 and cryo-EM187 as part of the 30 nm fiber. We determined the most probable folding pathways, or the minimum free energy paths, from open configurations to the zigzag structure using the finite-temperature string method.267 Notably, the pathways go through intermediate configurations that resemble chromatin configurations observed in situ.268,269 Our study suggests that chromatin configurations observed in situ are closely related to the in vitro fibril structures and may form as a result of local excitations or unfolding from the global minimum.
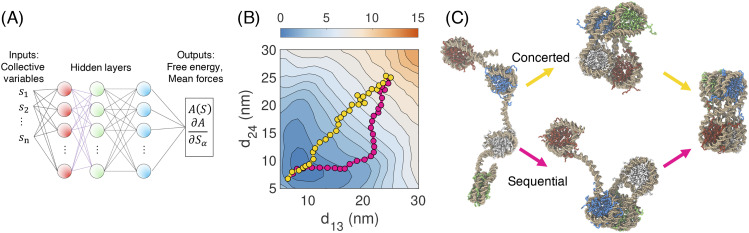
FIG. 9.
Stability and folding pathways of the tetra-nucleosome. (a) Illustration of the neural network approach for parameterizing high-dimensional free energy surfaces from mean forces. The neural network takes the six internucleosomal distances (S1, S2, …, Sn) as an input to compute the corresponding free energy [A(S)] and mean forces (). (b) Projection of the six-dimensional free energy profile to the distance between 1 and 3 (d13) and 2 and 4 (d24) nucleosomes. The sequential (pink) and concerted (yellow) pathway for tetra-nucleosome folding are shown on top of the free energy profile with energy unit kcal/mol. (c) Example tetra-nucleosome configurations along the two folding pathways. The DNA molecule is shown in gold, and the histone octamers are shown in green, white, blue, and red.
The folding intermediates bear comparable stability as the zigzag structure and can be further stabilized by configurational entropy, histone modifications, and variation in the secondary structure of the histone tail. Therefore, chromatin organization is sensitive to both thermal and chemical perturbations. Given the complexity of the nucleus environment, it is perhaps not too surprising to frequently observe the folding intermediates. Chromatin indeed favors more disordered configurations in which proteins mediate contact between non-neighboring nucleosomes,181 when the binding of the chromatin regulator, Polycomb Repressor Complex 2 (PRC2), was taken into account.
1. Study the role of phase separation in chromatin organization with MOFF
The techniques used for tetra-nucleosome folding can be generalized to longer chromatin to evaluate the stability of various fibril structures and the dependence of their stability on the linker DNA length. However, to study the impact of chromatin regulators, a more accurate protein force field is needed. Many chromatin regulators contain disordered regions, and recent evidence suggests that the phase separation of these proteins270–273 drives changes in the chromatin structure.222,223,234 We found that the existing coarse-grained force fields, which were often parameterized to fold globular proteins and predict protein structures,247,274,275 tend to produce overly compact conformations for IDPs. As such, there is widespread interest in developing force fields specifically suited for IDPs.276–278 Despite the advances, algorithms, which can drive systematic improvements in force field accuracy and ultimately reconcile differences between folded and unfolded models, remain scarce, although improvements have been made in the fully atomistic case.279–283
The maximum entropy optimization algorithm introduced in whole-genome modeling is well suited to improve the existing force fields for IDPs. For example, our group and others have incorporated low-resolution experimental data from small-angle x-ray scattering (SAXS) into computer models to refine structure prediction.284–286 However, an obvious drawback of this approach is that it cannot be applied to proteins for which no experimental data are available.
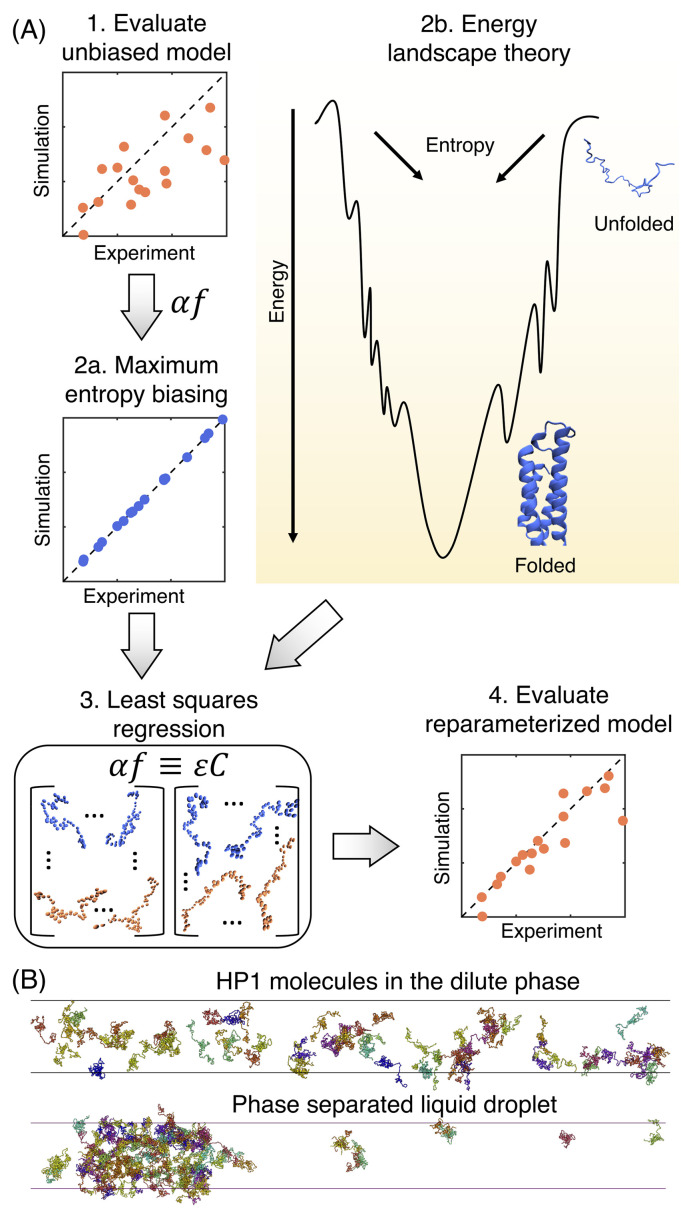
We developed another algorithm to parameterize a transferable force field for IDPs, which we term maximum entropy optimized force field for IDPs or MOFF-IDP.287 The algorithm consists of three steps, as outlined in Fig. 10(a). First, simulations are performed for a set of IDPs with an initial force field to evaluate the unbiased model. Then, maximum entropy biasing is used to determine the least biased correction energies (αf) that reproduce protein radii of gyration derived from SAXS experiments. Third, least squares regression parameterizes the protein-specific biasing energies with pairwise amino acid potentials (ɛC) to ensure transferability. These three steps can be repeated to improve the force field accuracy further. The resulting force field, MOFF-IDP, performed well for de novo prediction of IDP structural ensembles. For example, it captured the structural rearrangement in the epidermal growth factor receptor C-terminus domain upon phosphorylation.288
FIG. 10.
Coarse-grained protein force field, MOFF, enables large scale simulation of phase separation. (a) Illustration of the maximum entropy optimization algorithm for protein force field parameterization. The algorithm reparameterizes biasing energies (αf) determined from maximum entropy optimization with a weighted linear combination of contacts (ɛC). When solving the reparameterization algorithm with least squares regression, additional constraints can be included for globular proteins to ensure that the native conformations have the lowest energy (step 2b in yellow). (b) Example configurations of HP1 molecules in the dilute and condensed phase.
Just like how the force fields optimized for protein folding and structural prediction cannot be applied to IDPs, most IDP force fields, including MOFF-IDP, are not transferable to globular proteins either. Since both globular proteins and IDPs consist of the same 20 amino acids, it is plausible and desirable to have a consistent force field that describes both types of proteins equally. The maximum entropy optimization algorithm can be readily extended to include folded proteins to improve the force field transferability. Furthermore, it could be combined with other ideas introduced for force field optimization.
Recently, we extended the maximum entropy optimization algorithm with ideas from protein folding studies.289 For example, energy landscape theory, which succeeded in providing a conceptual framework for studying protein folding kinetics and thermodynamics, requires the protein force field to be funneled toward the native configuration for reliable and efficient folding of globular proteins.256 Therefore, we added globular proteins to the training set for the first two steps of the algorithm. We further enforced additional constraints for folded proteins when solving the reparameterization equations to require the energy of the native structures to be lower than that of the misfolded ones for these proteins [see step 2b of Fig. 10(a)]. The reparameterized force field (MOFF) succeeded in predicting the size of both globular and disordered proteins with consistent accuracy.
We have begun to utilize MOFF to study heterochromatin protein 1 (HP1), an essential chromatin regulator known to phase separate. Our results help explain the experimental observations of homolog specific phase separation [Fig. 10(b)].222,290 When combined with the DNA model mentioned in Sec. III C, MOFF could help address the role of phase-separating proteins in chromatin organization with near-atomistic details.
IV. PERSPECTIVE: CHROMOSOME DYNAMICS
Hi-C, imaging, and related methods have provided a comprehensive characterization of genome organization. To connect the structure with function, however, a detailed understanding of chromosome dynamics is required. A prominent example is the eukaryotic gene activation facilitated by the contact formation between enhancers and promoters. Whether these contacts are transient, as in the hit-and-run mechanism,291 or persistent292 remains controversial. Distinguishing the two mechanisms have significant implications on genome engineering to control gene expression. Chromosome movement is crucial for the sequence-based homology searches during double-strand break repair,293 gene rearrangements in antigen receptor repertoire establishment,294 and telomere length regulation295 as well. Therefore, a full understanding of the functional role of 3D genome organization can only be achieved by accounting for its dynamical component.
Live-cell imaging has provided great insight into chromatin dynamics.15,296 When the positions of individual loci were tracked over time, a subdiffusive, visco-elastic motion was observed.294,297,298 In contrast to regular diffusion, the mean squared displacement of genomic loci scales with time as tα with α < 1. Chromatin dynamics was further found to be highly heterogeneous and is sensitive to a variety of factors, including the nuclear localization,299,300 the length of the loci,301,302 and the concentration of lamin A/C protein.303 Mapping chromatin dynamics across the entire nucleus using Green fluorescent protein (GFP)-tagged histone H2B304 or fluorescent-labeled DNA305 uncovered surprisingly long-range correlation among genomic loci over the micrometer scale. This correlated motion is at least partially driven by ATP or transcription.
Experimental findings have inspired numerous theoretical studies.306,307 Much progress has been made to understand the origin of anomalous diffusion in a crowded environment. The elastic interaction between a locus and its neighboring consecutive segments because of the polymer nature of the genome alone could give rise to sub-diffusive behavior at intermediate timescales.308 The folded organization of chromosomes can lead to quantitative changes in the scaling exponent.309,310 Passive interactions between genomic loci and the nuclear compartments could further restrict their diffusivity via the so-called continuous-time random walk (CTRW).311 Finally, a particle moving through a viscoelastic environment will undergo subdiffusive motion over a range of time scales due to elastic stresses within the medium.294,312–314 This environment may, in fact, arise from the chromatin itself, which has been shown to exhibit glassy131 and gel-like property.315–317
While theoretical studies have been very successful at deriving general principles, they often have to introduce significant assumptions regarding the biological complexity of the underlying system to make the problem analytically solvable. Consequently, these studies face challenges in explaining the heterogeneity of single-locus dynamics. In addition, generalizing them to the whole genome is non-trivial. The correlated motion among loci is naturally a more complex problem. Hydrodynamics, motors, nuclear compartments, and phase separation all could potentially contribute to such a correlation. Computer simulations that treat the different factors on equal footing could quantify their contributions to chromosome dynamics. The detailed microscopic mechanism arising from numerical simulations could inspire more quantitative theories on chromatin dynamics.
The whole-genome model introduced in the Sec. II will be valuable for studying chromosome dynamics. For example, it will be straightforward to characterize the subdiffusive behavior of telomeres and understand the mechanistic origin that gives rise to the heterogeneity among different loci. In addition to the behavior of individual loci, the dynamical correlation among loci can also be characterized by computing the spatial–temporal correlation defined as
| (4) |
which quantifies the displacement correlations between loci separated by a distance r over the time interval Δt. The angular brackets represent averaging over time. Prior simulation studies have shown that correlated motions are present within individual chromosomes due to chain organization.318,319 The whole-genome model offers a unique opportunity to examine the correlation over scales beyond chromosome territories.304
A complete understanding of chromosome dynamics cannot be achieved without accounting for the role of ATP-driven remodeling enzymes. These enzymes can affect the dynamics of single loci320–322 and the large scale correlated motions. The whole-genome model can be modified straightforwardly to study non-equilibrium dynamics, at least approximately, by introducing colored noise to the equation of motion.323 In addition to numerical simulations, it is crucial to develop analytical approaches that help conceptualize the impact of non-equilibrium motors on the chromatin structure and dynamics. Wang and Wolynes introduced a perturbation theory to map the non-equilibrium steady state in terms of an effective temperature via a systematic expansion of the many-body master equation.139,324 This approach is appealing as it could open up the door of applying equilibrium statistical mechanical theories for non-equilibrium systems. We followed similar ideas140,325 to show that the effect of ATP-driven chromatin remodeling enzymes on nucleosome positioning could be well approximated by effective equilibrium models with rescaled temperatures and interactions. Numerical simulations support the theory’s accuracy in predicting both kinetic and steady-state quantities, including the effective temperature and the radial distribution function, in biologically relevant regimes. Generalizing these studies could complement numerical simulations to understand the role of molecular motors in genome organization.
It is worth noting that the force field derived from Hi-C data only provides an effective energy landscape, which is not guaranteed to reproduce dynamical measurements. The maximum caliber method,326,327 which is a generalization of the maximum entropy principle for dynamical trajectories, could be used to incorporate dynamical information328,329 and improve the dynamical properties of the whole-genome model.
AUTHORS’ CONTRIBUTIONS
X.L. and Y.Q. contributed equally to this work.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (Grant Nos. MCB-1715859 and MCB-2042362) and the National Institutes of Health (Grant No. R35GM133580). A.L. acknowledges support by the National Science Foundation Graduate Research Fellowship Program.
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.International Human Genome Sequencing, “Initial sequencing and analysis of the human genome,” Nature 409, 860–921 (2001). 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- 2.Liu X., Milshina N., Glasser K., Nelson K., Hannenhalli S., Chaturvedi K., Wolfe K., Gabor Miklos G. L., Carnes-Stine J., Turner R., Rodriguez R., Lewis M., Rowe W., Lu F., Caminha M., Kalush F., Brandon R., Zhang Q., Lei Y., Glodek A., Bafna V., Busam D., Thomas P. D., Vech C., Flanigan M., Peterson M., Wang A., Gluecksmann A., Sanders R., Kraft C., Wides R., Roberts R. J., Zhong W., Ye J., Gilbert D., Wang G., Mobarry C., Pratts E., Zhu X., Curry L., Fosler C., McIntosh T., Gire H., Neelam B., Spier G., Dahlke C., Zhang H., Sutton G. G., Venter J. C., Subramanian G., Stewart E., An H., Istrail S., Nguyen N., Ketchum K. A., Wu D., Sitter C., Kline L., Zhan M., Jordan C., Lippert R., Esparham S., Zhang J., Charlab R., Hart B., Abu-Threideh J., Gorokhov M., Evangelista C., Allen D., Xiao C., Scott R., Ma D., Muruganujan A., Kejariwal A., Zhong F., Tint N. N., Mural R. J., Hladun S., Garg N., Amanatides P., Ji R.-R., Ke Z., Kasha J., Adams M. D., Guan P., Pan S., Gu Z., Donnelly M., Lai Z., Beasley E., Suh E., Zheng X. H., Baldwin D., Heiman T. J., Wei M.-H., Peck J., Venter E., Yan C., Jordan J., Naik A. K., Hoover J., Nodell M., Guo N., Wetter J., Qureshi H., Awe A., Evans C. A., Sprague A., Simpson M., Howland T., Mays A. D., Nusskern D., Rusch D. B., Ge W., Francesco V. D., Levine A. J., Zhu S. C., Gocayne J. D., Yandell M., Basu A., McKusick V. A., Schwartz R., Remington K., Liang Y., Smith H. O., Rogers Y.-H., Wang X., Zinder N., Carter C., Sjolander K. V., Moy L., Majoros W., Moore H. M., Thomas R., Merkulov G. V., Baumhueter S., Salzberg S., Johnson J., Bonazzi V., Ballew R. M., Jennings D., Smith T., Wang Z. Y., Heil J., Delcher A., Myers E. W., Moy M., Narayan V. A., Dew I., Gan W., Higgins M. E., Wang J., Strong R., Baden H., Desilets R., Holt R. A., Hatton T., Stockwell T., Houck J., Gong F., Puri V., Kravitz S., Dodson K., Mann F., Karlak B., Koduru S., Shao W., Tse S., Lopez J., Chen L., Wen M., Clark A. G., Bolanos R., Biddick K., Gabrielian A. E., Nguyen T., Shue B., Eilbeck K., Yooseph S., Doup L., Pfannkoch C., Zhao Q., Beeson K., Zhao S., Halpern A., Fasulo D., Chandramouliswaran I., Davenport L., Cravchik A., Sato S., Heiner C., McCawley S., Danaher S., Deng Z., Windsor S., Ali F., May D., Zaveri K., Cheng M. L., Simon M., Carver A., Baxendale J., Broder S., Huson D. H., Hostin D., Lin X., Guigó R., Romblad D., Levy S., Hunkapiller M., Ibegwam C., Yao A., Haynes C., Ely D., Wang M., Nelson C., Chiang Y.-H., Nadeau J., Zheng L., Reardon M., Levitsky A., Harris M., Williams M., Ferriera S., Ruhfel B., Li P. W., Dunn P., Li J., Slayman C., Murphy B., Caulk P., Graham K., Wu M., Glanowski S., Florea L., Coyne M., Love A., Murphy S., Li Z., Lazareva B., Zaveri J., Xia A., Newman M., Wortman J. R., McDaniel J., Woodage T., McMullen I., Kagan L., Haynes J., Sun J., Center A., Campbell M. J., Smallwood M., Blick L., Diemer K., Henderson S., Kodira C. D., Winn-Deen E., Zandieh A., Zhang W., Walenz B., Gropman B., Barnstead M., Reinert K., Williams S., Mi H., Barrow I., Cargill M., Abril J. F., Narechania A., Dombroski M., Scott J., Dietz S., and Skupski M., “The sequence of the human genome,” Science 291, 1304–1351 (2002). 10.1126/science.1058040 [DOI] [Google Scholar]
- 3.Schrödinger E., What is Life?, Canto Classics (Cambridge University Press, 2014). [Google Scholar]
- 4.Roadmap Epigenomics Consortium, Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M. J., Amin V., Whitaker J. W., Schultz M. D., Ward L. D., Sarkar A., Quon G., Sandstrom R. S., Eaton M. L., Wu Y.-C., Pfenning A. R., Wang X., Claussnitzer M., Liu Y., Coarfa C., Harris R. A., Shoresh N., Epstein C. B., Gjoneska E., Leung D., Xie W., Hawkins R. D., Lister R., Hong C., Gascard P., Mungall A. J., Moore R., Chuah E., Tam A., Canfield T. K., Hansen R. S., Kaul R., Sabo P. J., Bansal M. S., Carles A., Dixon J. R., Farh K.-H., Feizi S., Karlic R., Kim A.-R., Kulkarni A., Li D., Lowdon R., Elliott G., Mercer T. R., Neph S. J., Onuchic V., Polak P., Rajagopal N., Ray P., Sallari R. C., Siebenthall K. T., Sinnott-Armstrong N. A., Stevens M., Thurman R. E., Wu J., Zhang B., Zhou X., Beaudet A. E., Boyer L. A., De Jager P. L., Farnham P. J., Fisher S. J., Haussler D., Jones S. J. M., Li W., Marra M. A., McManus M. T., Sunyaev S., Thomson J. A., Tlsty T. D., Tsai L.-H., Wang W., Waterland R. A., Zhang M. Q., Chadwick L. H., Bernstein B. E., Costello J. F., Ecker J. R., Hirst M., Meissner A., Milosavljevic A., Ren B., Stamatoyannopoulos J. A., Wang T., and Kellis M., “Integrative analysis of 111 reference human epigenomes,” Nature 518, 317–329 (2015). 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misteli T., “Beyond the sequence: Cellular organization of genome function,” Cell 128, 787–800 (2007). 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- 6.van Steensel B. and Furlong E. E. M., “The role of transcription in shaping the spatial organization of the genome,” Nat. Rev. Mol. Cell Biol. 20, 327 (2019). 10.1038/s41580-019-0114-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonev B. and Cavalli G., “Organization and function of the 3D genome,” Nat. Rev. Genet. 17, 661–678 (2016). 10.1038/nrg.2016.112 [DOI] [PubMed] [Google Scholar]
- 8.Luger K., Mäder A. W., Richmond D. K., Sargent D. F., and Richmond T. J., “Crystal structure of the nucleosome core particle at 2.8 Å resolution,” Nature 389, 251–260 (2002). 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 9.Lai W. K. M. and Pugh B. F., “Understanding nucleosome dynamics and their links to gene expression and DNA replication,” Nat. Rev. Mol. Cell Biol. 18, 548–562 (2017). 10.1038/nrm.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai L. and Morozov A. V., “Gene regulation by nucleosome positioning,” Trends Genet. 26, 476–483 (2010). 10.1016/j.tig.2010.08.003 [DOI] [PubMed] [Google Scholar]
- 11.Li B., Carey M., and Workman J. L., “The role of chromatin during transcription,” Cell 128, 707–719 (2007). 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- 12.Narlikar G. J., Sundaramoorthy R., and Owen-Hughes T., “Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes,” Cell 154, 490–503 (2013). 10.1016/j.cell.2013.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorkin D. U., Leung D., and Ren B., “The 3D genome in transcriptional regulation and pluripotency,” Cell Stem Cell 14, 762–775 (2014). 10.1016/j.stem.2014.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B. and Wolynes P. G., “Genomic energy landscapes,” Biophys. J. 112, 427–433 (2017). 10.1016/j.bpj.2016.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cremer T., Cremer M., Hübner B., Strickfaden H., Smeets D., Popken J., Sterr M., Markaki Y., Rippe K., and Cremer C., “The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments,” FEBS Lett. 589, 2931–2943 (2015). 10.1016/j.febslet.2015.05.037 [DOI] [PubMed] [Google Scholar]
- 16.van Arensbergen J., van Steensel B., and Bussemaker H. J., “In search of the determinants of enhancer–promoter interaction specificity,” Trends Cell Biol. 24, 695–702 (2014). 10.1016/j.tcb.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strahl B. D. and Allis C. D., “The language of covalent histone modifications,” Nature 403, 41–45 (2000). 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 18.Rando O. J., “Combinatorial complexity in chromatin structure and function: Revisiting the histone code,” Curr. Opin. Genet. Dev. 22, 148–155 (2012). 10.1016/j.gde.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou V. W., Goren A., and Bernstein B. E., “Charting histone modifications and the functional organization of mammalian genomes,” Nat. Rev. Genet. 12, 7–18 (2011). 10.1038/nrg2905 [DOI] [PubMed] [Google Scholar]
- 20.Maeshima K., Hihara S., and Eltsov M., “Chromatin structure: Does the 30-nm fibre exist in vivo?,” Curr. Opin. Cell Biol. 22, 291–297 (2010). 10.1016/j.ceb.2010.03.001 [DOI] [PubMed] [Google Scholar]
- 21.Fussner E., Ching R. W., and Bazett-Jones D. P., “Living without 30 nm chromatin fibers,” Trends Biochem. Sci. 36, 1–6 (2011). 10.1016/j.tibs.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 22.Luger K., Dechassa M. L., and Tremethick D. J., “New insights into nucleosome and chromatin structure: An ordered state or a disordered affair?,” Nat. Rev. Mol. Cell Biol. 13, 436–447 (2012). 10.1038/nrm3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanulli S., Trnka M. J., Dharmarajan V., Tibble R. W., Pascal B. D., Burlingame A. L., Griffin P. R., Gross J. D., and Narlikar G. J., “HP1 reshapes nucleosome core to promote phase separation of heterochromatin,” Nature 575, 390–394 (2019). 10.1038/s41586-019-1669-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson B. A., Doolittle L. K., Schneider M. W. G., Jensen L. E., Gamarra N., Henry L., Gerlich D. W., Redding S., and Rosen M. K., “Organization of chromatin by intrinsic and regulated phase separation,” Cell 179, 470–484.e21 (2019). 10.1016/j.cell.2019.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marti-Renom M. A. and Mirny L. A., “Bridging the resolution gap in structural modeling of 3D genome organization,” PLoS Comput. Biol. 7, e1002125 (2011). 10.1371/journal.pcbi.1002125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dekker J., Marti-Renom M. A., and Mirny L. A., “Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data,” Nat. Rev. Genet. 14, 390–403 (2013). 10.1038/nrg3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brackey C. A., Marenduzzo D., and Gilbert N., “Mechanistic modeling of chromatin folding to understand function,” Nat. Methods 17, 767–775 (2020). 10.1038/s41592-020-0852-6 [DOI] [PubMed] [Google Scholar]
- 28.Saunders M. G. and Voth G. A., “Coarse-graining methods for computational biology,” Annu. Rev. Biophys. 42, 73–93 (2013). 10.1146/annurev-biophys-083012-130348 [DOI] [PubMed] [Google Scholar]
- 29.Noid W. G., “Perspective: Coarse-grained models for biomolecular systems,” J. Chem. Phys. 139, 090901 (2013). 10.1063/1.4818908 [DOI] [PubMed] [Google Scholar]
- 30.Zidovska A., “The self-stirred genome: Large-scale chromatin dynamics, its biophysical origins and implications,” Curr. Opin. Genet. Dev. 61, 83–90 (2020). 10.1016/j.gde.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moller J. and de Pablo J. J., “Bottom-up meets top-down: The crossroads of multiscale chromatin modeling,” Biophys. J. 118, 2057–2065 (2020). 10.1016/j.bpj.2020.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozer G., Luque A., and Schlick T., “The chromatin fiber: Multiscale problems and approaches,” Curr. Opin. Struct. Biol. 31, 124–139 (2015). 10.1016/j.sbi.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bendandi A., Dante S., Zia S. R., Diaspro A., and Rocchia W., “Chromatin compaction multiscale modeling: A complex synergy between theory, simulation, and experiment,” Front. Mol. Biosci. 7, 15 (2020). 10.3389/fmolb.2020.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Gennes P. G., Scaling Concepts in Polymer Physics (Cornell University Press, 1979). [Google Scholar]
- 35.Heitz E., Das Heterochromatin der Moose (Bornträger, 1928). [Google Scholar]
- 36.Fedorova E. and Zink D., “Nuclear architecture and gene regulation,” Biochim. Biophys. Acta, Mol. Cell Res. 1783, 2174–2184 (2008). 10.1016/j.bbamcr.2008.07.018 [DOI] [PubMed] [Google Scholar]
- 37.Marsden M. P. F. and Laemmli U. K., “Metaphase chromosome structure: Evidence for a radial loop model,” Cell 17, 849–858 (1979). 10.1016/0092-8674(79)90325-8 [DOI] [PubMed] [Google Scholar]
- 38.Sachs R. K., van den Engh G., Trask B., Yokota H., and Hearst J. E., “A random-walk/giant-loop model for interphase chromosomes,” Proc. Natl. Acad. Sci. U. S. A. 92, 2710–2714 (1995). 10.1073/pnas.92.7.2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mu C., Nkel È., Eils R., Dietzel S., Zink D., Mehring C., Wedemann G., Cremer T., and Rg Langowski J. È., “Compartmentalization of interphase chromosomes observed in simulation and experiment,” J. Mol. Biol. 285, 1053–1065 (1999). 10.1006/jmbi.1998.2361 [DOI] [PubMed] [Google Scholar]
- 40.Bickmore W. A. and Oghene K., “Visualizing the spatial relationships between defined DNA sequences and the axial region of extracted metaphase chromosomes,” Cell 84, 95–104 (1996). 10.1016/s0092-8674(00)80996-4 [DOI] [PubMed] [Google Scholar]
- 41.Cremer T. and Cremer C., “Chromosome territories, nuclear architecture and gene regulation in mammalian cells,” Nat. Rev. Genet. 2, 292–301 (2001). 10.1038/35066075 [DOI] [PubMed] [Google Scholar]
- 42.Bolzer A., Kreth G., Solovei I., Koehler D., Saracoglu K., Fauth C., Müller S., Eils R., Cremer C., Speicher M. R., and Cremer T., “Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes,” PLoS Biol. 3, e157 (2005). 10.1371/journal.pbio.0030157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su J.-H., Zheng P., Kinrot S. S., Bintu B., and Zhuang X., “Genome-scale imaging of the 3D organization and transcriptional activity of chromatin,” Cell 182, 1641–1659.e26 (2020). 10.1016/j.cell.2020.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dekker J., Rippe K., Dekker M., and Kleckner N., “Capturing chromosome conformation,” Science 295, 1306–1311 (2002). 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- 45.Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L. A., Lander E. S., and Dekker J., “Comprehensive mapping of long-range interactions reveals folding principles of the human genome,” Science 326, 289–293 (2009). 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wit E. D. and Laat W. D., “A decade of 3C technologies: Insights into nuclear organization,” Genes Dev. 26, 11–24 (2012). 10.1101/gad.179804.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibcus J. H. and Dekker J., “The hierarchy of the 3D genome,” Mol. Cell 49, 773–782 (2013). 10.1016/j.molcel.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraser J., Williamson I., Bickmore W. A., and Dostie J., “An overview of genome organization and how we got there: From FISH to Hi-C,” Microbiol. Mol. Biol. Rev. 79, 347–372 (2015). 10.1128/mmbr.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt A. D., Hu M., and Ren B., “Genome-wide mapping and analysis of chromosome architecture,” Nat. Rev. Mol. Cell Biol. 17, 743–755 (2016). 10.1038/nrm.2016.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pope B. D., Ryba T., Dileep V., Yue F., Wu W., Denas O., Vera D. L., Wang Y., Hansen R. S., Canfield T. K., Thurman R. E., Cheng Y., Gülsoy G., Dennis J. H., Snyder M. P., Stamatoyannopoulos J. A., Taylor J., Hardison R. C., Kahveci T., Ren B., and Gilbert D. M., “Topologically associating domains are stable units of replication-timing regulation,” Nature 515, 402–405 (2014). 10.1038/nature13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao S. S. P., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., Robinson J. T., Sanborn A. L., Machol I., Omer A. D., Lander E. S., and Aiden E. L., “A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping,” Cell 159, 1665–1680 (2014). 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dekker J. and Heard E., “Structural and functional diversity of topologically associating domains,” FEBS Lett. 589, 2877–2884 (2015). 10.1016/j.febslet.2015.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bintu B., Mateo L. J., Su J.-H., Sinnott-Armstrong N. A., Parker M., Kinrot S., Yamaya K., Boettiger A. N., and Zhuang X., “Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells,” Science 362, eaau1783 (2018). 10.1126/science.aau1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beagan J. A. and Phillips-Cremins J. E., “On the existence and functionality of topologically associating domains,” Nat. Genet. 52, 8–16 (2020). 10.1038/s41588-019-0561-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie W. J., Qi Y., and Zhang B., “Characterizing chromatin folding coordinate and landscape with deep learning,” PLoS Comput. Biol. 16, e1008262 (2020). 10.1371/journal.pcbi.1008262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fudenberg G. and Imakaev M., “FISH-ing for captured contacts: Towards reconciling FISH and 3C,” Nat. Methods 14, 673–678 (2017). 10.1038/nmeth.4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi G. and Thirumalai D., “Conformational heterogeneity in human interphase chromosome organization reconciles the FISH and Hi-C paradox,” Nat. Commun. 10, 3894 (2019). 10.1038/s41467-019-11897-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beliveau B. J., Boettiger A. N., Nir G., Bintu B., Yin P., Zhuang X., and Wu C.-t., “In situ super-resolution imaging of genomic DNA with OligoSTORM and OligoDNA-PAINT,” in Super-Resolution Microscopy: Methods and Protocols (Springer, New York, NY, 2017), pp. 231–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang S., Su J.-H., Beliveau B. J., Bintu B., Moffitt J. R., Wu C.-t., and Zhuang X., “Spatial organization of chromatin domains and compartments in single chromosomes,” Science 353, 598–602 (2016). 10.1126/science.aaf8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nir G., Farabella I., Pérez Estrada C., Ebeling C. G., Beliveau B. J., Sasaki H. M., Lee S. D., Nguyen S. C., McCole R. B., Chattoraj S., Erceg J., AlHaj Abed J., Martins N. M. C., Nguyen H. Q., Hannan M. A., Russell S., Durand N. C., Rao S. S. P., Kishi J. Y., Soler-Vila P., Di Pierro M., Onuchic J. N., Callahan S. P., Schreiner J. M., Stuckey J. A., Yin P., Aiden E. L., Marti-Renom M. A., and Wu C.-t., “Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling,” PLoS Genet. 14, e1007872 (2018). 10.1371/journal.pgen.1007872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie L., Dong P., Chen X., Hsieh T. S., Banala S., De Marzio M., English B. P., Qi Y., Jung S. K., Kieffer-Kwon K. R.et al. , “3D ATAC-PALM: Super-resolution imaging of the accessible genome,” Nat. Methods 17, 430–436 (2020). 10.1038/s41592-020-0775-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Payne A. C., Chiang Z. D., Reginato P. L., Mangiameli S. M., Murray E. M., Yao C.-C., Markoulaki S., Earl A. S., Labade A. S., Jaenisch R., Church G. M., Boyden E. S., Buenrostro J. D., and Chen F., “In situ genome sequencing resolves DNA sequence and structure in intact biological samples,” Science 371, eaay3446 (2020). 10.1126/science.aay3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takei Y., Yun J., Zheng S., Ollikainen N., Pierson N., White J., Shah S., Thomassie J., Suo S., Eng C.-H. L.et al. , “Integrated spatial genomics reveals global architecture of single nuclei,” Nature 590, 344 (2021). 10.1038/s41586-020-03126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meluzzi D. and Arya G., “Computational approaches for inferring 3D conformations of chromatin from chromosome conformation capture data,” Methods 181-182, 24 (2019). 10.1016/j.ymeth.2019.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou R. and Gao Y. Q., “Polymer models for the mechanisms of chromatin 3D folding: Review and perspective,” Phys. Chem. Chem. Phys. 22, 20189–20201 (2020). 10.1039/d0cp01877e [DOI] [PubMed] [Google Scholar]
- 66.Seffernick J. T. and Lindert S., “Hybrid methods for combined experimental and computational determination of protein structure,” J. Chem. Phys. 153, 240901 (2020). 10.1063/5.0026025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y. J., Lee C., Shendure J., Fields S., Blau C. A., and Noble W. S., “A three-dimensional model of the yeast genome,” Nature 465, 363 (2010). 10.1038/nature08973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lesne A., Riposo J., Roger P., Cournac A., and Mozziconacci J., “3D genome reconstruction from chromosomal contacts,” Nat. Methods 11, 1141–1143 (2014). 10.1038/nmeth.3104 [DOI] [PubMed] [Google Scholar]
- 69.Baù D., Sanyal A., Lajoie B. R., Capriotti E., Byron M., Lawrence J. B., Dekker J., and Marti-Renom M. A., “The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules,” Nat. Struct. Mol. Biol. 18, 107–115 (2011). 10.1038/nsmb.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baù D. and Marti-Renom M. A., “Genome structure determination via 3C-based data integration by the integrative modeling platform,” Methods 58, 300–306 (2012). 10.1016/j.ymeth.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 71.Szalaj P., Michalski P. J., Wróblewski P., Tang Z., Kadlof M., Mazzocco G., Ruan Y., and Plewczynski D., “3D-GNOME: An integrated web service for structural modeling of the 3D genome,” Nucleic Acids Res. 44, W288–W293 (2016). 10.1093/nar/gkw437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szalaj P., Tang Z., Michalski P., Pietal M. J., Luo O. J., Sadowski M., Li X., Radew K., Ruan Y., and Plewczynski D., “An integrated 3-dimensional genome modeling engine for data-driven simulation of spatial genome organization,” Genome Res. 26, 1697–1709 (2016). 10.1101/gr.205062.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie W. J., Meng L., Liu S., Zhang L., Cai X., and Gao Y. Q., “Structural modeling of chromatin integrates genome features and reveals chromosome folding principle,” Sci. Rep. 7, 2818 (2017). 10.1038/s41598-017-02923-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Segal M. R. and Bengtsson H. L., “Reconstruction of 3D genome architecture via a two-stage algorithm,” BMC Bioinf. 16, 373 (2015). 10.1186/s12859-015-0799-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adhikari B., Trieu T., and Cheng J., “Chromosome3D: Reconstructing three-dimensional chromosomal structures from Hi-C interaction frequency data using distance geometry simulated annealing,” BMC Genomics 17, 886 (2016). 10.1186/s12864-016-3210-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yildirim A. and Feig M., “High-resolution 3D models of Caulobacter crescentus chromosome reveal genome structural variability and organization,” Nucleic Acids Res. 46, 3937–3952 (2018). 10.1093/nar/gky141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu M., Deng K., Qin Z., Dixon J., Selvaraj S., Fang J., Ren B., and Liu J. S., “Bayesian inference of spatial organizations of chromosomes,” PLoS Comput. Biol. 9, e1002893 (2013). 10.1371/journal.pcbi.1002893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu G., Deng W., Hu H., Ma R., Zhang S., Yang J., Peng J., Kaplan T., and Zeng J., “Reconstructing spatial organizations of chromosomes through manifold learning,” Nucleic Acids Res. 46, e50 (2018). 10.1093/nar/gky065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paulsen J., Gramstad O., and Collas P., “Manifold based optimization for single-cell 3D genome reconstruction,” PLoS Comput. Biol. 11, e1004396 (2015). 10.1371/journal.pcbi.1004396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guarnera E., Tan Z. W., and Berezovsky I. N., “Three-dimensional chromatin ensemble reconstruction via stochastic embedding,” (to be published, 2021). 10.1016/j.str.2021.01.008 [DOI] [PubMed]
- 81.Nagano T., Lubling Y., Stevens T. J., Schoenfelder S., Yaffe E., Dean W., Laue E. D., Tanay A., and Fraser P., “Single-cell Hi-C reveals cell-to-cell variability in chromosome structure,” Nature 502, 59–64 (2013). 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stevens T. J., Lando D., Basu S., Atkinson L. P., Cao Y., Lee S. F., Leeb M., Wohlfahrt K. J., Boucher W., O’Shaughnessy-Kirwan A.et al. , “3D structures of individual mammalian genomes studied by single-cell Hi-C,” Nature 544, 59–64 (2017). 10.1038/nature21429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramani V., Deng X., Qiu R., Gunderson K. L., Steemers F. J., Disteche C. M., Noble W. S., Duan Z., and Shendure J., “Massively multiplex single-cell Hi-C,” Nat. Methods 14, 263–266 (2017). 10.1038/nmeth.4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan L., Xing D., Chang C.-H., Li H., and Xie X. S., “Three-dimensional genome structures of single diploid human cells,” Science 361, 924–928 (2018). 10.1126/science.aat5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tokuda N., Terada T. P., and Sasai M., “Dynamical modeling of three-dimensional genome organization in interphase budding yeast,” Biophys. J. 102, 296–304 (2012). 10.1016/j.bpj.2011.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tokuda N. and Sasai M., “Heterogeneous spatial distribution of transcriptional activity in budding yeast nuclei,” Biophys. J. 112, 491–504 (2017). 10.1016/j.bpj.2016.11.3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tjong H., Li W., Kalhor R., Dai C., Hao S., Gong K., Zhou Y., Li H., Zhou X. J., Le Gros M. A., Larabell C. A., Chen L., and Alber F., “Population-based 3D genome structure analysis reveals driving forces in spatial genome organization,” Proc. Natl. Acad. Sci. U. S. A. 113, E1663–E1672 (2016). 10.1073/pnas.1512577113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kalhor R., Tjong H., Jayathilaka N., Alber F., and Chen L., “Genome architectures revealed by tethered chromosome conformation capture and population-based modeling,” Nat. Biotechnol. 30, 90–98 (2012). 10.1038/nbt.2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tjong H., Gong K., Chen L., and Alber F., “Physical tethering and volume exclusion determine higher-order genome organization in budding yeast,” Genome Res. 22, 1295–1305 (2012). 10.1101/gr.129437.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perez-Rathke A., Sun Q., Wang B., Boeva V., Shao Z., and Liang J., “CHROMATIX: Computing the functional landscape of many-body chromatin interactions in transcriptionally active loci from deconvolved single cells,” Genome Biol. 21, 13 (2020). 10.1186/s13059-019-1904-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sun Q., Perez-Rathke A., Czajkowsky D. M., Shao Z., and Liang J., “High-resolution single-cell 3D-models of chromatin ensembles during Drosophila embryogenesis,” Nat. Comm. 12, 205 (2021). 10.1038/s41467-020-20490-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gürsoy G., Xu Y., and Liang J., “Spatial organization of the budding yeast genome in the cell nucleus and identification of specific chromatin interactions from multi-chromosome constrained chromatin model,” PLoS Comput. Biol. 13, e1005658 (2017). 10.1371/journal.pcbi.1005658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gürsoy G., Xu Y., Kenter A. L., and Liang J., “Spatial confinement is a major determinant of the folding landscape of human chromosomes,” Nucleic Acids Res. 42, 8223–8230 (2014). 10.1093/nar/gku462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang B. and Wolynes P. G., “Topology, structures, and energy landscapes of human chromosomes,” Proc. Natl. Acad. Sci. U. S. A. 112, 6062–6067 (2015). 10.1073/pnas.1506257112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meluzzi D. and Arya G., “Recovering ensembles of chromatin conformations from contact probabilities,” Nucleic Acids Res. 41, 63–75 (2013). 10.1093/nar/gks1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang B. and Wolynes P. P., “Shape transitions and chiral symmetry breaking in the energy landscape of the mitotic chromosome,” Phys. Rev. Lett. 116, 248101 (2016). 10.1103/physrevlett.116.248101 [DOI] [PubMed] [Google Scholar]
- 97.Pitera J. W. and Chodera J. D., “On the use of experimental observations to bias simulated ensembles,” J. Chem. Theory Comput. 8, 3445–3451 (2012). 10.1021/ct300112v [DOI] [PubMed] [Google Scholar]
- 98.Boura E., Rozycki B., Herrick D. Z., Chung H. S., Vecer J., Eaton W. A., Cafiso D. S., Hummer G., and Hurley J. H., “Solution structure of the ESCRT-I complex by small-angle X-ray scattering, EPR, and FRET spectroscopy,” Proc. Natl. Acad. Sci. U. S. A. 108, 9437–9442 (2011). 10.1073/pnas.1101763108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roux B. and Weare J., “On the statistical equivalence of restrained-ensemble simulations with the maximum entropy method,” J. Chem. Phys. 138, 084107 (2013). 10.1063/1.4792208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jaynes E. T., “Information theory and statistical mechanics. II,” Phys. Rev. 108, 171–190 (1957). 10.1103/physrev.108.171 [DOI] [Google Scholar]
- 101.Crehuet R., Buigues P. J., Salvatella X., and Lindorff-Larsen K., “Bayesian-maximum-entropy reweighting of IDP ensembles based on NMR chemical shifts,” Entropy 21, 898 (2019). 10.3390/e21090898 [DOI] [Google Scholar]
- 102.Cesari A., Reißer S., and Bussi G., “Using the maximum entropy principle to combine simulations and solution experiments,” Computation 6, 15 (2018). 10.3390/computation6010015 [DOI] [Google Scholar]
- 103.Reppert M., Roy A. R., Tempkin J. O. B., Dinner A. R., and Tokmakoff A., “Refining disordered peptide ensembles with computational amide I spectroscopy: Application to elastin-like peptides,” J. Phys. Chem. B 120, 11395–11404 (2016). 10.1021/acs.jpcb.6b08678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Amirkulova D. B. and White A. D., “Recent advances in maximum entropy biasing techniques for molecular dynamics,” Mol. Simul. 45, 1285–1294 (2019). 10.1080/08927022.2019.1608988 [DOI] [Google Scholar]
- 105.Stein R. R., Marks D. S., and Sander C., “Inferring pairwise interactions from biological data using maximum-entropy probability models,” PLoS Comput. Biol. 11, e1004182 (2015). 10.1371/journal.pcbi.1004182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boomsma W., Ferkinghoff-Borg J., and Lindorff-Larsen K., “Combining experiments and simulations using the maximum entropy principle,” PLoS Comput. Biol. 10, e1003406 (2014). 10.1371/journal.pcbi.1003406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mirny L. A., “The fractal globule as a model of chromatin architecture in the cell,” Chromosome Res. 19, 37–51 (2011). 10.1007/s10577-010-9177-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gibcus J. H., Samejima K., Goloborodko A., Samejima I., Naumova N., Nuebler J., Kanemaki M. T., Xie L., Paulson J. R., Earnshaw W. C., Mirny L. A., and Dekker J., “A pathway for mitotic chromosome formation,” Science 359, eaao6135 (2018). 10.1126/science.aao6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chu X. and Wang J., “Conformational state switching and pathways of chromosome dynamics in cell cycle,” Appl. Phys. Rev. 7, 031403 (2020). 10.1063/5.0007316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Giorgetti L., Galupa R., Nora E. P., Piolot T., Lam F., Dekker J., Tiana G., and Heard E., “Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription,” Cell 157, 950–963 (2014). 10.1016/j.cell.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tiana G., Amitai A., Pollex T., Piolot T., Holcman D., Heard E., and Giorgetti L., “Structural fluctuations of the chromatin fiber within topologically associating domains,” Biophys. J. 110, 1234–1245 (2016). 10.1016/j.bpj.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shi G. and Thirumalai D., “From Hi-C contact map to three-dimensional organization of interphase human chromosomes,” Phys. Rev. X 11, 011051 (2021). 10.1103/physrevx.11.011051 [DOI] [Google Scholar]
- 113.Grosberg A. Y., Nechaev S. K., and Shakhnovich E. I., “The role of topological constraints in the kinetics of collapse of macromolecules,” J. Phys. 49, 2095–2100 (1988). 10.1051/jphys:0198800490120209500 [DOI] [Google Scholar]
- 114.Rosa A. and Everaers R., “Structure and dynamics of interphase chromosomes,” PLoS Comput. Biol. 4, e1000153 (2008). 10.1371/journal.pcbi.1000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huang K., Li Y., Shim A. R., Virk R. K. A., Agrawal V., Eshein A., Nap R. J., Almassalha L. M., Backman V., and Szleifer I., “Physical and data structure of 3D genome,” Sci. Adv. 6, eaay4055 (2020). 10.1126/sciadv.aay4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Münkel C. and Langowski J., “Chromosome structure described by a polymer model,” Phys. Rev. E 57, 5888–5896 (1998). 10.1103/physreve.57.5888 [DOI] [Google Scholar]
- 117.Bohn M. and Heermann D. W., “Diffusion-driven looping provides a consistent framework for chromatin organization,” PLoS One 5, e12218 (2010). 10.1371/journal.pone.0012218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sanborn A. L., Rao S. S. P., Huang S.-C., Durand N. C., Huntley M. H., Jewett A. I., Bochkov I. D., Chinnappan D., Cutkosky A., Li J., Geeting K. P., Gnirke A., Melnikov A., McKenna D., Stamenova E. K., Lander E. S., and Aiden E. L., “Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes,” Proc. Natl. Acad. Sci. U. S. A. 112, E6456–E6465 (2015). 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., and Mirny L. A., “Formation of chromosomal domains by loop extrusion,” Cell Rep. 15, 2038–2049 (2016). 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Haarhuis J. H. I., van der Weide R. H., Blomen V. A., Yáñez-Cuna J. O., Amendola M., van Ruiten M. S., Krijger P. H. L., Teunissen H., Medema R. H., van Steensel B., Brummelkamp T. R., de Wit E., and Rowland B. D., “The cohesin release factor WAPL restricts chromatin loop extension,” Cell 169, 693–707 (2017). 10.1016/j.cell.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fudenberg G., Abdennur N., Imakaev M., Goloborodko A., and Mirny L. A., “Emerging evidence of chromosome folding by loop extrusion,” Cold Spring Harbor Symp. Quant. Biol. 82, 45–55 (2017). 10.1101/sqb.2017.82.034710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., Fonseca N. A., Huber W., Haering C. H., Mirny L., and Spitz F., “Two independent modes of chromatin organization revealed by cohesin removal,” Nature 551, 51–56 (2017). 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nora E. P., Goloborodko A., Valton A.-L., Gibcus J. H., Uebersohn A., Abdennur N., Dekker J., Mirny L. A., and Bruneau B. G., “Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization,” Cell 169, 930–944.e22 (2017). 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rao S. S. P., Huang S.-C., Glenn St Hilaire B., Engreitz J. M., Perez E. M., Kieffer-Kwon K.-R., Sanborn A. L., Johnstone S. E., Bascom G. D., Bochkov I. D., Huang X., Shamim M. S., Shin J., Turner D., Ye Z., Omer A. D., Robinson J. T., Schlick T., Bernstein B. E., Casellas R., Lander E. S., and Aiden E. L., “Cohesin loss eliminates all loop domains,” Cell 171, 305–320.e324 (2017). 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim Y., Shi Z., Zhang H., Finkelstein I. J., and Yu H., “Human cohesin compacts DNA by loop extrusion,” Science 366, 1345–1349 (2019). 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Davidson I. F., Bauer B., Goetz D., Tang W., Wutz G., and Peters J.-M., “DNA loop extrusion by human cohesin,” Science 366, 1338–1345 (2019). 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- 127.Barbieri M., Chotalia M., Fraser J., Lavitas L.-M., Dostie J., Pombo A., and Nicodemi M., “Complexity of chromatin folding is captured by the strings and binders switch model,” Proc. Natl. Acad. Sci. U. S. A. 109, 16173–16178 (2012). 10.1073/pnas.1204799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brackley C. A., Johnson J., Kelly S., Cook P. R., and Marenduzzo D., “Simulated binding of transcription factors to active and inactive regions folds human chromosomes into loops, rosettes and topological domains,” Nucleic Acids Res. 44, 3503–3512 (2016). 10.1093/nar/gkw135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jost D., Carrivain P., Cavalli G., and Vaillant C., “Modeling epigenome folding: Formation and dynamics of topologically associated chromatin domains,” Nucleic Acids Res. 42, 9553–9561 (2014). 10.1093/nar/gku698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Di Pierro M., Zhang B., Aiden E. L., Wolynes P. G., and Onuchic J. N., “Transferable model for chromosome architecture,” Proc. Natl. Acad. Sci. U. S. A. 113, 12168–12173 (2016). 10.1073/pnas.1613607113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shi G., Liu L., Hyeon C., and Thirumalai D., “Interphase human chromosome exhibits out of equilibrium glassy dynamics,” Nat. Commun. 9, 3161 (2018). 10.1038/s41467-018-05606-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu S., Zhang L., Quan H., Tian H., Meng L., Yang L., Feng H., and Gao Y. Q., “From 1D sequence to 3D chromatin dynamics and cellular functions: A phase separation perspective,” Nucleic Acids Res. 46, 9367–9383 (2018). 10.1093/nar/gky633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Laghmach R., Di Pierro M., and Potoyan D. A., “Mesoscale liquid model of chromatin recapitulates nuclear order of eukaryotes,” Biophys. J. 118, 2130–2140 (2020). 10.1016/j.bpj.2019.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., and Sharp P. A., “A phase separation model for transcriptional control,” Cell 169, 13–23 (2017). 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Di Pierro M., Cheng R. R., Lieberman Aiden E., Wolynes P. G., and Onuchic J. N., “De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture,” Proc. Natl. Acad. Sci. U. S. A. 114, 12126–12131 (2017). 10.1073/pnas.1714980114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xie W. J. and Zhang B., “Learning the formation mechanism of domain-level chromatin states with epigenomics data,” Biophys. J. 116, 2047–2056 (2019). 10.1016/j.bpj.2019.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., Ward L. D., Brugman W., de Castro I. J., Kerkhoven R. M., Bussemaker H. J., and van Steensel B., “Systematic protein location mapping reveals five principal chromatin types in Drosophila cells,” Cell 143, 212–224 (2010). 10.1016/j.cell.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Erdel F. and Rippe K., “Formation of chromatin subcompartments by phase separation,” Biophys. J. 114, 2262–2270 (2018). 10.1016/j.bpj.2018.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang S. and Wolynes P. G., “Communication: Effective temperature and glassy dynamics of active matter,” J. Chem. Phys. 135, 000219–342 (2011). 10.1063/1.3624753 [DOI] [PubMed] [Google Scholar]
- 140.Jiang Z. and Zhang B., “Theory of active chromatin remodeling,” Phys. Rev. Lett. 123, 208102 (2019). 10.1103/physrevlett.123.208102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ganai N., Sengupta S., and Menon G. I., “Chromosome positioning from activity-based segregation,” Nucleic Acids Res. 42, 4145–4159 (2014). 10.1093/nar/gkt1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Grosberg A. Y. and Joanny J. F., “Nonequilibrium statistical mechanics of mixtures of particles in contact with different thermostats,” Phys. Rev. E 92, 032118 (2015). 10.1103/physreve.92.032118 [DOI] [PubMed] [Google Scholar]
- 143.Wang Y., Nagarajan M., Uhler C., and Shivashankar G. V., “Orientation and repositioning of chromosomes correlate with cell geometry-dependent gene expression,” Mol. Biol. Cell 28, 1997–2009 (2017). 10.1091/mbc.e16-12-0825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Qi Y., Reyes A., Johnstone S. E., Aryee M. J., Bernstein B. E., and Zhang B., “Data-driven polymer model for mechanistic exploration of diploid genome organization,” Biophys. J. 119, 1905–1916 (2020). 10.1016/j.bpj.2020.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Brackley C. A., Taylor S., Papantonis A., Cook P. R., and Marenduzzo D., “Nonspecific bridging-induced attraction drives clustering of DNA-binding proteins and genome organization,” Proc. Natl. Acad. Sci. U. S. A. 110, E3605–E3611 (2013). 10.1073/pnas.1302950110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Krepel D., Cheng R. R., Di Pierro M., and Onuchic J. N., “Deciphering the structure of the condensin protein complex,” Proc. Natl. Acad. Sci. U. S. A. 115, 11911–11916 (2018). 10.1073/pnas.1812770115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kang H., Yoon Y. G., Thirumalai D., and Hyeon C., “Confinement-induced glassy dynamics in a model for chromosome organization,” Phys. Rev. Lett. 115, 198102 (2015). 10.1103/physrevlett.115.198102 [DOI] [PubMed] [Google Scholar]
- 148.Strom A. R. and Brangwynne C. P., “The liquid nucleome—Phase transitions in the nucleus at a glance,” J. Cell Sci. 132, jcs235093 (2019). 10.1242/jcs.235093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen Y. and Belmont A. S., “Genome organization around nuclear speckles,” Curr. Opin. Genet. Dev. 55, 91–99 (2019). 10.1016/j.gde.2019.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.van Steensel B. and Belmont A. S., “Lamina-associated domains: Links with chromosome architecture, heterochromatin, and gene repression,” Cell 169, 780–791 (2017). 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wiblin A. E., Cui W., Clark A. J., and Bickmore W. A., “Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells,” J. Cell Sci. 118, 3861–3868 (2005). 10.1242/jcs.02500 [DOI] [PubMed] [Google Scholar]
- 152.Johnstone S. E., Reyes A., Qi Y., Adriaens C., Hegazi E., Pelka K., Chen J. H., Zou L. S., Drier Y., Hecht V.et al. , “Large-scale topological changes restrain malignant progression in colorectal cancer,” Cell 182, 1474–1489 (2020). 10.1016/j.cell.2020.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maji A., Ahmed J. A., Roy S., Chakrabarti B., and Mitra M. K., “A lamin-associated chromatin model for chromosome organization,” Biophys. J. 118, 3041 (2020). 10.1016/j.bpj.2020.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chiang M., Michieletto D., Brackley C. A., Rattanavirotkul N., Mohammed H., Marenduzzo D., and Chandra T., “Polymer modeling predicts chromosome reorganization in senescence,” Cell Rep. 28, 3212–3223.e6 (2019). 10.1016/j.celrep.2019.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li Q., Tjong H., Li X., Gong K., Zhou X. J., Chiolo I., and Alber F., “The three-dimensional genome organization of Drosophila melanogaster through data integration,” Genome Biol. 18, 145 (2017). 10.1186/s13059-017-1264-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Qi Y. and Zhang B., “Chromatin network retards droplet coalescence,” bioRxiv:433564 (2021). [DOI] [PMC free article] [PubMed]
- 157.Uhler C. and Shivashankar G. V., “Regulation of genome organization and gene expression by nuclear mechanotransduction,” Nat. Rev. Mol. Cell Biol. 18, 717–727 (2017). 10.1038/nrm.2017.101 [DOI] [PubMed] [Google Scholar]
- 158.Miroshnikova Y. A., Nava M. M., and Wickström S. A., “Emerging roles of mechanical forces in chromatin regulation,” J. Cell Sci. 130, 2243–2250 (2017). 10.1242/jcs.202192 [DOI] [PubMed] [Google Scholar]
- 159.Lee D. S., Wingreen N. S., and Brangwynne C. P., “Chromatin mechanics dictates subdiffusion and coarsening dynamics of embedded condensates,” Nat. Phys. 17, 531 (2021). 10.1038/s41567-020-01125-8 [DOI] [Google Scholar]
- 160.Zhang Y., Lee D. S., Meir Y., Brangwynne C. P., and Wingreen N. S., “Mechanical frustration of phase separation in the cell nucleus by chromatin,” Phys. Rev. Lett. (to be published, 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Qi Y. and Zhang B., “Predicting three-dimensional genome organization with chromatin states,” PLoS Comput. Biol. 15, e1007024 (2019). 10.1371/journal.pcbi.1007024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yang T., Zhang F., Yardımcı G. G., Song F., Hardison R. C., Noble W. S., Yue F., and Li Q., “HiCRep: Assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient,” Genome Res. 27, 1939–1949 (2017). 10.1101/gr.220640.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Serra F., Baù D., Goodstadt M., Castillo D., Filion G. J., and Marti-Renom M. A., “Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors,” PLoS Comput. Biol. 13, e1005665 (2017). 10.1371/journal.pcbi.1005665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.MacPherson Q., Beltran B., and Spakowitz A. J., “Bottom–up modeling of chromatin segregation due to epigenetic modifications,” Proc. Natl. Acad. Sci. U. S. A. 115, 12739–12744 (2018). 10.1073/pnas.1812268115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Esposito A., Bianco S., Chiariello A. M., Abraham A., Fiorillo L., Conte M., Campanile R., and Nicodemi M., “Polymer physics and machine learning reveal a combinatorial code linking chromatin 3D architecture to 1D epigenetics,” bioRxiv:433416 (2021). [DOI] [PubMed]
- 166.Schwessinger R., Gosden M., Downes D., Brown R. C., Oudelaar A. M., Telenius J., Teh Y. W., Lunter G., and Hughes J. R., “DeepC: Predicting 3D genome folding using megabase-scale transfer learning,” Nat. Methods 17, 1118–1124 (2020). 10.1038/s41592-020-0960-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fudenberg G., Kelley D. R., and Pollard K. S., “Predicting 3D genome folding from DNA sequence with Akita,” Nat. Methods 17, 1111–1117 (2020). 10.1038/s41592-020-0958-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li W., Wong W. H., and Jiang R., “DeepTACT: Predicting 3D chromatin contacts via bootstrapping deep learning,” Nucleic Acids Res. 47, e60 (2019). 10.1093/nar/gkz167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kouzarides T., “Chromatin modifications and their function,” Cell 128, 693–705 (2007). 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 170.Suganuma T. and Workman J. L., “Signals and combinatorial functions of histone modifications,” Annu. Rev. Biochem. 80, 473–499 (2011). 10.1146/annurev-biochem-061809-175347 [DOI] [PubMed] [Google Scholar]
- 171.Thakore P. I., Black J. B., Hilton I. B., and Gersbach C. A., “Editing the epigenome: Technologies for programmable transcription and epigenetic modulation,” Nat. Methods 13, 127–137 (2016). 10.1038/nmeth.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Park M., Keung A. J., and Khalil A. S., “The epigenome: The next substrate for engineering,” Genome Biol. 17, 183 (2016). 10.1186/s13059-016-1046-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ruthenburg A. J., Li H., Patel D. J., and David Allis C., “Multivalent engagement of chromatin modifications by linked binding modules,” Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007). 10.1038/nrm2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Imhof A., “Epigenetic regulators and histone modification,” Briefings Funct. Genomics Proteomics 5, 222–227 (2006). 10.1093/bfgp/ell030 [DOI] [PubMed] [Google Scholar]
- 175.Sood A. and Zhang B., “Quantifying epigenetic stability with minimum action paths,” Phys. Rev. E 101, 062409 (2020). 10.1103/physreve.101.062409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dodd I. B., Micheelsen M. A., Sneppen K., and Thon G., “Theoretical analysis of epigenetic cell memory by nucleosome modification,” Cell 129, 813–822 (2007). 10.1016/j.cell.2007.02.053 [DOI] [PubMed] [Google Scholar]
- 177.Erdel F. and Greene E. C., “Generalized nucleation and looping model for epigenetic memory of histone modifications,” Proc. Natl. Acad. Sci. U. S. A. 113, E4180–E4189 (2016). 10.1073/pnas.1605862113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Michieletto D., Orlandini E., and Marenduzzo D., “Polymer model with epigenetic recoloring reveals a pathway for the de novo establishment and 3D organization of chromatin domains,” Phys. Rev. X 6, 041047 (2016). 10.1103/physrevx.6.041047 [DOI] [Google Scholar]
- 179.Jost D. and Vaillant C., “Epigenomics in 3D: Importance of long-range spreading and specific interactions in epigenomic maintenance,” Nucleic Acids Res. 46, 2252–2264 (2018). 10.1093/nar/gky009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Katava M., Shi G., and Thirumalai D., “Chromatin dynamics controls epigenetic domain formation,” bioRxiv:427115 (2021). [DOI] [PMC free article] [PubMed]
- 181.Leicher R., Ge E. J., Lin X., Reynolds M. J., Xie W., Walz T., Zhang B., Muir T. W., and Liu S., “Single-molecule and in silico dissection of the interaction between Polycomb repressive complex 2 and chromatin,” Proc. Natl. Acad. Sci. U. S. A. 117, 30465–30475 (2020). 10.1073/pnas.2003395117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Finch J. T. and Klug A., “Solenoidal model for superstructure in chromatin,” Proc. Natl. Acad. Sci. U. S. A. 73, 1897–1901 (2006). 10.1073/pnas.73.6.1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Horowitz R. A., Agard D. A., Sedat J. W., and Woodcock C. L., “The three-dimensional architecture of chromatin in situ: Electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon,” J. Cell Biol. 125, 1–10 (1994). 10.1083/jcb.125.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Woodcock C. L., Frado L. L., and Rattner J. B., “The higher-order structure of chromatin: Evidence for a helical ribbon arrangement,” J. Cell Biol. 99, 42–52 (1984). 10.1083/jcb.99.1.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Williams S. P. and Langmore J. P., “Small angle x-ray scattering of chromatin. Radius and mass per unit length depend on linker length,” Biophys. J. 59, 606–618 (1991). 10.1016/s0006-3495(91)82276-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Robinson P. J. J., Fairall L., Huynh V. A. T., and Rhodes D., “EM measurements define the dimensions of the ‘30-nm’ chromatin fiber: Evidence for a compact, interdigitated structure,” Proc. Natl. Acad. Sci. U. S. A. 103, 6506–6511 (2006). 10.1073/pnas.0601212103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Song F., Chen P., Sun D., Wang M., Dong L., Liang D., Xu R.-M., Zhu P., and Li G., “Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units,” Science 344, 376–380 (2014). 10.1126/science.1251413 [DOI] [PubMed] [Google Scholar]
- 188.Eltsov M., MacLellan K. M., Maeshima K., Frangakis A. S., and Dubochet J., “Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ,” Proc. Natl. Acad. Sci. U. S. A. 105, 19732–19737 (2008). 10.1073/pnas.0810057105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ahmed K., Dehghani H., Rugg-Gunn P., Fussner E., Rossant J., and Bazett-Jones D. P., “Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo,” PLoS One 5, e10531 (2010). 10.1371/journal.pone.0010531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nishino Y., Eltsov M., Joti Y., Ito K., Takata H., Takahashi Y., Hihara S., Frangakis A. S., Imamoto N., Ishikawa T., and Maeshima K., “Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure,” EMBO J. 31, 1644–1653 (2012). 10.1038/emboj.2012.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Maeshima K., Rogge R., Tamura S., Joti Y., Hikima T., Szerlong H., Krause C., Herman J., Seidel E., DeLuca J., Ishikawa T., and Hansen J. C., “Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers,” EMBO J. 35, 1115–1132 (2016). 10.15252/embj.201592660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ricci M. A., Manzo C., García-Parajo M. F., Lakadamyali M., and Cosma M. P., “Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo,” Cell 160, 1145–1158 (2015). 10.1016/j.cell.2015.01.054 [DOI] [PubMed] [Google Scholar]
- 193.Ou H. D., Phan S., Deerinck T. J., Thor A., Ellisman M. H., and O’Shea C. C., “ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells,” Science 357, eaag0025 (2017). 10.1126/science.aag0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Maeshima K., Ide S., and Babokhov M., “Dynamic chromatin organization without the 30-nm fiber,” Curr. Opin. Cell Biol. 58, 95–104 (2019). 10.1016/j.ceb.2019.02.003 [DOI] [PubMed] [Google Scholar]
- 195.Bishop T. C., “Molecular dynamics simulations of a nucleosome and free DNA,” J. Biomol. Struct. Dyn. 22, 673–685 (2005). 10.1080/07391102.2005.10507034 [DOI] [PubMed] [Google Scholar]
- 196.Li Z. and Kono H., “Distinct roles of histone H3 and H2A tails in nucleosome stability,” Sci. Rep. 6, 31437 (2016). 10.1038/srep31437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Materese C. K., Savelyev A., and Papoian G. A., “Counterion atmosphere and hydration patterns near a nucleosome core particle,” J. Am. Chem. Soc. 131, 15005–15013 (2009). 10.1021/ja905376q [DOI] [PubMed] [Google Scholar]
- 198.Melters D. P., Pitman M., Rakshit T., Dimitriadis E. K., Bui M., Papoian G. A., and Dalal Y., “Intrinsic elasticity of nucleosomes is encoded by histone variants and calibrated by their binding partners,” Proc. Natl. Acad. Sci. U. S. A. 116, 24066–24074 (2019). 10.1073/pnas.1911880116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Shaytan A. K., Armeev G. A., Goncearenco A., Zhurkin V. B., Landsman D., and Panchenko A. R., “Coupling between histone conformations and DNA geometry in nucleosomes on a microsecond timescale: Atomistic insights into nucleosome functions,” J. Mol. Biol. 428, 221–237 (2016). 10.1016/j.jmb.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Collepardo-Guevara R., Portella G., Vendruscolo M., Frenkel D., Schlick T., and Orozco M., “Chromatin unfolding by epigenetic modifications explained by dramatic impairment of internucleosome interactions: A multiscale computational study,” J. Am. Chem. Soc. 137, 10205–10215 (2015). 10.1021/jacs.5b04086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sridhar A., Farr S. E., Portella G., Schlick T., Orozco M., and Collepardo-Guevara R., “Emergence of chromatin hierarchical loops from protein disorder and nucleosome asymmetry,” Proc. Natl. Acad. Sci. U. S. A. 117, 7216–7224 (2020). 10.1073/pnas.1910044117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Winogradoff D. and Aksimentiev A., “Molecular mechanism of spontaneous nucleosome unraveling,” J. Mol. Biol. 431, 323–335 (2019). 10.1016/j.jmb.2018.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Woods D. C., Rodríguez-Ropero F., and Wereszczynski J., “The dynamic influence of linker histone saturation within the poly-nucleosome array,” J. Mol. Biol. 433, 166902 (2021). 10.1016/j.jmb.2021.166902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jung J., Nishima W., Daniels M., Bascom G., Kobayashi C., Adedoyin A., Wall M., Lappala A., Phillips D., Fischer W., Tung C. S., Schlick T., Sugita Y., and Sanbonmatsu K. Y., “Scaling molecular dynamics beyond 100,000 processor cores for large-scale biophysical simulations,” J. Comput. Chem. 40, 1919–1930 (2019). 10.1002/jcc.25840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Woodcock C. L., Grigoryev S. A., Horowitz R. A., and Whitaker N., “A chromatin folding model that incorporates linker variability generates fibers resembling the native structures,” Proc. Natl. Acad. Sci. U. S. A. 90, 9021–9025 (1993). 10.1073/pnas.90.19.9021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schiessel H., Gelbart W. M., and Bruinsma R., “DNA folding: Structural and mechanical properties of the two-angle model for chromatin,” Biophys. J. 80, 1940–1956 (2001). 10.1016/s0006-3495(01)76164-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Diesinger P. M. and Heermann D. W., “Two-angle model and phase diagram for chromatin,” Phys. Rev. E 74, 031904 (2006). 10.1103/physreve.74.031904 [DOI] [PubMed] [Google Scholar]
- 208.Wong H., Victor J. M., and Mozziconacci J., “An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length,” PLoS One 2, e877 (2007). 10.1371/journal.pone.0000877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Katritch V., Bustamante C., and Olson W. K., “Pulling chromatin fibers: Computer simulations of direct physical micromanipulations,” J. Mol. Biol. 295, 29–40 (2000). 10.1006/jmbi.1999.3021 [DOI] [PubMed] [Google Scholar]
- 210.Cui Y. and Bustamante C., “Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure,” Proc. Natl. Acad. Sci. U. S. A. 97, 127–132 (2000). 10.1073/pnas.97.1.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wedemann G. and Langowski J., “Computer simulation of the 30-nanometer chromatin fiber,” Biophys. J. 82, 2847–2859 (2002). 10.1016/s0006-3495(02)75627-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kepper N., Foethke D., Stehr R., Wedemann G., and Rippe K., “Nucleosome geometry and internucleosomal interactions control the chromatin fiber conformation,” Biophys. J. 95, 3692–3705 (2008). 10.1529/biophysj.107.121079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sun J., Zhang Q., and Schlick T., “Electrostatic mechanism of nucleosomal array folding revealed by computer simulation,” Proc. Natl. Acad. Sci. U. S. A. 102, 8180–8185 (2005). 10.1073/pnas.0408867102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bascom G. D. and Schlick T., “Mesoscale modeling of chromatin fibers,” in Nuclear Architecture and Dynamics (Elsevier, 2018), pp. 123–147. [Google Scholar]
- 215.Arya G., Zhang Q., and Schlick T., “Flexible histone tails in a new mesoscopic oligonucleosome model,” Biophys. J. 91, 133–150 (2006). 10.1529/biophysj.106.083006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Grigoryev S. A., Arya G., Correll S., Woodcock C. L., and Schlick T., “Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions,” Proc. Natl. Acad. Sci. U. S. A. 106, 13317–13322 (2009). 10.1073/pnas.0903280106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lequieu J., Córdoba A., Moller J., and De Pablo J. J., “1CPN: A coarse-grained multi-scale model of chromatin,” J. Chem. Phys. 150, 215102 (2019). 10.1063/1.5092976 [DOI] [PubMed] [Google Scholar]
- 218.Norouzi D. and Zhurkin V. B., “Topological polymorphism of the two-start chromatin fiber,” Biophys. J. 108, 2591–2600 (2015). 10.1016/j.bpj.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Norouzi D. and Zhurkin V. B., “Dynamics of chromatin fibers: Comparison of Monte Carlo simulations with force spectroscopy,” Biophys. J. 115, 1644–1655 (2018). 10.1016/j.bpj.2018.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.de Jong B. E., Brouwer T. B., Kaczmarczyk A., Visscher B., and van Noort J., “Rigid basepair Monte Carlo simulations of one-start and two-start chromatin fiber unfolding by force,” Biophys. J. 115, 1848–1859 (2018). 10.1016/j.bpj.2018.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Koslover E. F., Fuller C. J., Straight A. F., and Spakowitz A. J., “Local geometry and elasticity in compact chromatin structure,” Biophys. J. 99, 3941–3950 (2010). 10.1016/j.bpj.2010.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Larson A. G., Elnatan D., Keenen M. M., Trnka M. J., Johnston J. B., Burlingame A. L., Agard D. A., Redding S., and Narlikar G. J., “Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin,” Nature 547, 236–240 (2017). 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Strom A. R., Emelyanov A. V., Mir M., Fyodorov D. V., Darzacq X., and Karpen G. H., “Phase separation drives heterochromatin domain formation,” Nature 547, 241–245 (2017). 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Plys A. J., Davis C. P., Kim J., Rizki G., Keenen M. M., Marr S. K., and Kingston R. E., “Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2,” Genes Dev. 33, 799–813 (2019). 10.1101/gad.326488.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tatavosian R., Kent S., Brown K., Yao T., Duc H. N., Huynh T. N., Zhen C. Y., Ma B., Wang H., and Ren X., “Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation,” J. Biol. Chem. 294, 1451–1463 (2019). 10.1074/jbc.ra118.006620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sabari B. R., Dall’Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., Malik S., Hnisz D., Lee T. I., Cisse I. I., Roeder R. G., Sharp P. A., Chakraborty A. K., and Young R. A., “Coactivator condensation at super-enhancers links phase separation and gene control,” Science 361, eaar3958 (2018). 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Boija A., Klein I. A., Sabari B. R., Dall’Agnese A., Coffey E. L., Zamudio A. V., Li C. H., Shrinivas K., Manteiga J. C., Hannett N. M., Abraham B. J., Afeyan L. K., Guo Y. E., Rimel J. K., Fant C. B., Schuijers J., Lee T. I., Taatjes D. J., and Young R. A., “Transcription factors activate genes through the phase-separation capacity of their activation domains,” Cell 175, 1842 (2018). 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G. M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., and Tjian R., “Imaging dynamic and selective low-complexity domain interactions that control gene transcription,” Science 361, eaar2555 (2018). 10.1126/science.aar2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Cheutin T., McNairn A. J., Jenuwein T., Gilbert D. M., Singh P. B., and Misteli T., “Maintenance of stable heterochromatin domains by dynamic HP1 binding,” Science 299, 721–725 (2003). 10.1126/science.1078572 [DOI] [PubMed] [Google Scholar]
- 230.Klosin A. and Hyman A. A., “Molecular biology: A liquid reservoir for silent chromatin,” Nature 547, 168–170 (2017). 10.1038/nature23089 [DOI] [PubMed] [Google Scholar]
- 231.Potoyan D. A., Savelyev A., and Papoian G. A., “Recent successes in coarse-grained modeling of DNA,” Wiley Interdiscip. Rev.: Comput. Mol. Sci. 3, 69–83 (2013). 10.1002/wcms.1114 [DOI] [Google Scholar]
- 232.Freeman G. S., Hinckley D. M., and De Pablo J. J., “A coarse-grain three-site-per-nucleotide model for DNA with explicit ions,” J. Chem. Phys. 135, 165104 (2011). 10.1063/1.3652956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hinckley D. M., Freeman G. S., Whitmer J. K., and De Pablo J. J., “An experimentally-informed coarse-grained 3-site-per-nucleotide model of DNA: Structure, thermodynamics, and dynamics of hybridization,” J. Chem. Phys. 139, 144903 (2013). 10.1063/1.4822042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Freeman G. S., Lequieu J. P., Hinckley D. M., Whitmer J. K., and De Pablo J. J., “DNA shape dominates sequence affinity in nucleosome formation,” Phys. Rev. Lett. 113, 168101 (2014). 10.1103/physrevlett.113.168101 [DOI] [PubMed] [Google Scholar]
- 235.Li W., Wolynes P. G., and Takada S., “Frustration, specific sequence dependence, and nonlinearity in large-amplitude fluctuations of allosteric proteins,” Proc. Natl. Acad. Sci. U. S. A. 108, 3504–3509 (2011). 10.1073/pnas.1018983108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lequieu J., Schwartz D. C., and de Pablo J. J., “In silico evidence for sequence-dependent nucleosome sliding,” Proc. Natl. Acad. Sci. U. S. A. 114, E9197–E9205 (2017). 10.1073/pnas.1705685114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lequieu J., Córdoba A., Schwartz D. C., and De Pablo J. J., “Tension-dependent free energies of nucleosome unwrapping,” ACS Cent. Sci. 2, 660–666 (2016). 10.1021/acscentsci.6b00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Moller J., Lequieu J., and De Pablo J. J., “The free energy landscape of internucleosome interactions and its relation to chromatin fiber structure,” ACS Cent. Sci. 5, 341–348 (2019). 10.1021/acscentsci.8b00836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Funke J. J., Ketterer P., Lieleg C., Schunter S., Korber P., Dietz H., Lieleg C., Funke J. J., Ketterer P., Dietz H., and Schunter S., “Uncovering the forces between nucleosomes using DNA origami,” Sci. Adv. 2, e1600974 (2016). 10.1126/sciadv.1600974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Kenzaki H. and Takada S., “Partial unwrapping and histone tail dynamics in nucleosome revealed by coarse-grained molecular simulations,” PLoS Comput. Biol. 11, e1004443 (2015). 10.1371/journal.pcbi.1004443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Watanabe S., Mishima Y., Shimizu M., Suetake I., and Takada S., “Interactions of HP1 bound to H3K9me3 dinucleosome by molecular simulations and biochemical assays,” Biophys. J. 114, 2336–2351 (2018). 10.1016/j.bpj.2018.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kanada R., Terakawa T., Kenzaki H., and Takada S., “Nucleosome crowding in chromatin slows the diffusion but can promote target search of proteins,” Biophys. J. 116, 2285–2295 (2019). 10.1016/j.bpj.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Tan C. and Takada S., “Nucleosome allostery in pioneer transcription factor binding,” Proc. Natl. Acad. Sci. U. S. A. 117, 20586–20596 (2020). 10.1073/pnas.2005500117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chang L. and Takada S., “Histone acetylation dependent energy landscapes in tri-nucleosome revealed by residue-resolved molecular simulations,” Sci. Rep. 6, 34441 (2016). 10.1038/srep34441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Brandani G. B., Niina T., Tan C., and Takada S., “DNA sliding in nucleosomes via twist defect propagation revealed by molecular simulations,” Nucleic Acids Res. 46, 2788–2801 (2018). 10.1093/nar/gky158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Brandani G. B. and Takada S., “Chromatin remodelers couple inchworm motion with twist-defect formation to slide nucleosomal DNA,” PLoS Comput. Biol. 14, e1006512 (2018). 10.1371/journal.pcbi.1006512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Davtyan A., Schafer N. P., Zheng W., Clementi C., Wolynes P. G., and Papoian G. A., “AWSEM-MD: Protein structure prediction using coarse-grained physical potentials and bioinformatically based local structure biasing,” J. Phys. Chem. B 116, 8494–8503 (2012). 10.1021/jp212541y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Onuchic J. N., Luthey-Schulten Z., and Wolynes P. G., “Theory of protein folding: The energy landscape perspective,” Annu. Rev. Phys. Chem. 48, 545–600 (1997). 10.1146/annurev.physchem.48.1.545 [DOI] [PubMed] [Google Scholar]
- 249.Goldstein R. A., Luthey-Schulten Z. A., and Wolynes P. G., “Optimal protein-folding codes from spin-glass theory,” Proc. Natl. Acad. Sci. U. S. A. 89, 4918–4922 (1992). 10.1073/pnas.89.11.4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Eastwood M. P., Hardin C., Luthey-Schulten Z., and Wolynes P. G., “Statistical mechanical refinement of protein structure prediction schemes: Cumulant expansion approach,” J. Chem. Phys. 117, 4602–4615 (2002). 10.1063/1.1494417 [DOI] [Google Scholar]
- 251.Davey C. A., Sargent D. F., Luger K., Maeder A. W., and Richmond T. J., “Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution,” J. Mol. Biol. 319, 1097–1113 (2002). 10.1016/s0022-2836(02)00386-8 [DOI] [PubMed] [Google Scholar]
- 252.Zhang B., Zheng W., Papoian G. A., and Wolynes P. G., “Exploring the free energy landscape of nucleosomes,” J. Am. Chem. Soc. 138, 8126–8133 (2016). 10.1021/jacs.6b02893 [DOI] [PubMed] [Google Scholar]
- 253.Parsons T. and Zhang B., “Critical role of histone tail entropy in nucleosome unwinding,” J. Chem. Phys. 150, 185103 (2019). 10.1063/1.5085663 [DOI] [PubMed] [Google Scholar]
- 254.Brower-Toland B. D., Smith C. L., Yeh R. C., Lis J. T., Peterson C. L., and Wang M. D., “Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA,” Proc. Natl. Acad. Sci. U. S. A. 99, 1960–1965 (2002). 10.1073/pnas.022638399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Sudhanshu B., Mihardja S., Koslover E. F., Mehraeen S., Bustamante C., and Spakowitz A. J., “Tension-dependent structural deformation alters single-molecule transition kinetics,” Proc. Natl. Acad. Sci. U. S. A. 108, 1885–1890 (2011). 10.1073/pnas.1010047108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kulić I. M. and Schiessel H., “DNA spools under tension,” Phys. Rev. Lett. 92, 228101 (2004). 10.1103/PhysRevLett.92.228101 [DOI] [PubMed] [Google Scholar]
- 257.Dobrovolskaia I. V. and Arya G., “Dynamics of forced nucleosome unraveling and role of nonuniform histone–DNA interactions,” Biophys. J. 103, 989–998 (2012). 10.1016/j.bpj.2012.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ding X., Lin X., and Zhang B., “Stability and folding pathways of tetra-nucleosome from six-dimensional free energy surface,” Nat. Commun. 12, 1091 (2021). 10.1038/s41467-021-21377-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Whitford P. C., Noel J. K., Gosavi S., Schug A., Sanbonmatsu K. Y., and Onuchic J. N., “An all-atom structure-based potential for proteins: Bridging minimal models with all-atom empirical forcefields,” Proteins: Struct., Funct., Bioinf. 75, 430–441 (2009). 10.1002/prot.22253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Clementi C., Nymeyer H., and Onuchic J. N., “Topological and energetic factors: What determines the structural details of the transition state ensemble and ‘en-route’ intermediates for protein folding? An investigation for small globular proteins,” J. Mol. Biol. 298, 937–953 (2000). 10.1006/jmbi.2000.3693 [DOI] [PubMed] [Google Scholar]
- 261.Chen M., Cuendet M. A., and Tuckerman M. E., “Heating and flooding: A unified approach for rapid generation of free energy surfaces,” J. Chem. Phys. 137, 024102 (2012). 10.1063/1.4733389 [DOI] [PubMed] [Google Scholar]
- 262.Laio A. and Parrinello M., “Escaping free-energy minima,” Proc. Natl. Acad. Sci. U. S. A. 99, 12562–12566 (2002). 10.1073/pnas.202427399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Maragliano L. and Vanden-Eijnden E., “A temperature accelerated method for sampling free energy and determining reaction pathways in rare events simulations,” Chem. Phys. Lett. 426, 168–175 (2006). 10.1016/j.cplett.2006.05.062 [DOI] [Google Scholar]
- 264.Abrams J. B. and Tuckerman M. E., “Efficient and direct generation of multidimensional free energy surfaces via adiabatic dynamics without coordinate transformations,” J. Phys. Chem. B 112, 15742–15757 (2008). 10.1021/jp805039u [DOI] [PubMed] [Google Scholar]
- 265.Schneider E., Dai L., Topper R. Q., Drechsel-Grau C., and Tuckerman M. E., “Stochastic neural network approach for learning high-dimensional free energy surfaces,” Phys. Rev. Lett. 119, 150601 (2017). 10.1103/physrevlett.119.150601 [DOI] [PubMed] [Google Scholar]
- 266.Schalch T., Duda S., Sargent D. F., and Richmond T. J., “X-ray structure of a tetranucleosome and its implications for the chromatin fibre,” Nature 436, 138–141 (2005). 10.1038/nature03686 [DOI] [PubMed] [Google Scholar]
- 267.Vanden-Eijnden E. and Venturoli M., “Revisiting the finite temperature string method for the calculation of reaction tubes and free energies,” J. Chem. Phys. 130, 194103 (2009). 10.1063/1.3130083 [DOI] [PubMed] [Google Scholar]
- 268.Cai S., Böck D., Pilhofer M., and Gan L., “The in situ structures of mono-, di-, and trinucleosomes in human heterochromatin,” Mol. Biol. Cell 29, 2450–2457 (2018). 10.1091/mbc.e18-05-0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Ohno M., Ando T., Priest D. G., Kumar V., Yoshida Y., and Taniguchi Y., “Sub-nucleosomal genome structure reveals distinct nucleosome folding motifs,” Cell 176, 520–534.e25 (2019). 10.1016/j.cell.2018.12.014 [DOI] [PubMed] [Google Scholar]
- 270.Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Julicher F., and Hyman A. A., “Germline P granules are liquid droplets that localize by controlled dissolution/condensation,” Science 324, 1729–1732 (2009). 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- 271.van der Lee R., Buljan M., Lang B., Weatheritt R. J., Daughdrill G. W., Dunker A. K., Fuxreiter M., Gough J., Gsponer J., Jones D. T., Kim P. M., Kriwacki R. W., Oldfield C. J., Pappu R. V., Tompa P., Uversky V. N., Wright P. E., and Babu M. M., “Classification of intrinsically disordered regions and proteins,” Chem. Rev. 114, 6589–6631 (2014). 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Schneider R., Blackledge M., and Jensen M. R., “Elucidating binding mechanisms and dynamics of intrinsically disordered protein complexes using NMR spectroscopy,” Curr. Opin. Struct. Biol. 54, 10–18 (2019). 10.1016/j.sbi.2018.09.007 [DOI] [PubMed] [Google Scholar]
- 273.Hyman A. A., Weber C. A., and Jülicher F., “Liquid-liquid phase separation in biology,” Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- 274.De Jong D. H., Singh G., Bennett W. F. D., Arnarez C., Wassenaar T. A., Schäfer L. V., Periole X., Tieleman D. P., and Marrink S. J., “Improved parameters for the Martini coarse-grained protein force field,” J. Chem. Theory Comput. 9, 687–697 (2013). 10.1021/ct300646g [DOI] [PubMed] [Google Scholar]
- 275.Liwo A., Baranowski M., Czaplewski C., Gołaś E., He Y., Jagieła D., Krupa P., Maciejczyk M., Makowski M., Mozolewska M. A., Niadzvedtski A., Ołdziej S., Scheraga H. A., Sieradzan A. K., Ślusarz R., Wirecki T., Yin Y., and Zaborowski B., “A unified coarse-grained model of biological macromolecules based on mean-field multipole–multipole interactions,” J. Mol. Model. 20, 2306 (2014). 10.1007/s00894-014-2306-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Dignon G. L., Zheng W., Kim Y. C., Best R. B., and Mittal J., “Sequence determinants of protein phase behavior from a coarse-grained model,” PLoS Comput. Biol. 14, e1005941 (2018). 10.1371/journal.pcbi.1005941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Wu H., Wolynes P. G., and Papoian G. A., “AWSEM-IDP: A coarse-grained force field for intrinsically disordered proteins,” J. Phys. Chem. B 122, 11115–11125 (2018). 10.1021/acs.jpcb.8b05791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Baul U., Chakraborty D., Mugnai M. L., Straub J. E., and Thirumalai D., “Sequence effects on size, shape, and structural heterogeneity in intrinsically disordered proteins,” J. Phys. Chem. B 123, 3462–3474 (2019). 10.1021/acs.jpcb.9b02575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Best R. B., Zheng W., and Mittal J., “Balanced protein–water interactions improve properties of disorderd proteins and non-specific protein association,” J. Chem. Theory Comput. 10, 5113 (2014). 10.1021/ct500569b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Robustelli P., Piana S., and Shaw D. E., “Developing a molecular dynamics force field for both folded and disordered protein states,” Proc. Natl. Acad. Sci. U. S. A. 115, E4758–E4766 (2018). 10.1073/pnas.1800690115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Vitalis A. and Pappu R. V., “ABSINTH: A new continuum solvation model for simulations of polypeptides in aqueous solutions,” J. Comput. Chem. 30, 673–699 (2009). 10.1002/jcc.21005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Choi J.-M. and Pappu R. V., “Improvements to the ABSINTH force field for proteins based on experimentally derived amino acid specific backbone conformational statistics,” J. Chem. Theory Comput. 15, 1367–1382 (2019). 10.1021/acs.jctc.8b00573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B. L., Grubmüller H., and MacKerell A. D., “CHARMM36m: An improved force field for folded and intrinsically disordered proteins,” Nat. Methods 14, 71–73 (2016). 10.1038/nmeth.4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Latham A. P. and Zhang B., “Improving coarse-grained protein force fields with small-angle x-ray scattering data,” J. Phys. Chem. B 123, 1026–1034 (2019). 10.1021/acs.jpcb.8b10336 [DOI] [PubMed] [Google Scholar]
- 285.Yang S., Park S., Makowski L., and Roux B., “A rapid coarse residue-based computational method for x-ray solution scattering characterization of protein folds and multiple conformational states of large protein complexes,” Biophys. J. 96, 4449–4463 (2009). 10.1016/j.bpj.2009.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Rozycki B., Kim Y., and Hummer G., “SAXS Ensemble Refinement of ESCRT-III CHMP3 Conformational Transitions,” Structure 19, 109–116 (2011). 10.1016/j.str.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Latham A. P. and Zhang B., “Maximum entropy optimized force field for intrinsically disordered proteins,” J. Chem. Theory Comput. 16, 773–781 (2020). 10.1021/acs.jctc.9b00932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Regmi R., Srinivasan S., Latham A. P., Kukshal V., Cui W., Zhang B., Bose R., and Schlau-Cohen G. S., “Phosphorylation-dependent conformations of the disordered carboxyl-terminus domain in the epidermal growth factor receptor,” J. Phys. Chem. Lett. 11, 10037–10044 (2020). 10.1021/acs.jpclett.0c02327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Latham A. P. and Zhang B., “Consistent force field captures homolog resolved HP1 phase separation,” J. Chem. Theory Comput. 17(5), 3134–3144 (2021). 10.1021/acs.jctc.0c01220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Wang L., Gao Y., Zheng X., Liu C., Dong S., Li R., Zhang G., Wei Y., Qu H., Li Y., Allis C. D., Li G., Li H., and Li P., “Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism,” Mol. Cell 76, 646–659.e6 (2019). 10.1016/j.molcel.2019.08.019 [DOI] [PubMed] [Google Scholar]
- 291.Spivakov M., “Spurious transcription factor binding: Non-functional or genetically redundant?,” BioEssays 36, 798–806 (2014). 10.1002/bies.201400036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Chen H., Levo M., Barinov L., Fujioka M., Jaynes J. B., and Gregor T., “Dynamic interplay between enhancer–promoter topology and gene activity,” Nat. Genet. 50, 1296–1303 (2018). 10.1038/s41588-018-0175-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Seeber A., Hauer M. H., and Gasser S. M., “Chromosome dynamics in response to DNA damage,” Annu. Rev. Genet. 52, 295–319 (2018). 10.1146/annurev-genet-120417-031334 [DOI] [PubMed] [Google Scholar]
- 294.Lucas J. S., Zhang Y., Dudko O. K., and Murre C., “3D trajectories adopted by coding and regulatory DNA elements: First-passage times for genomic interactions,” Cell 158, 339–352 (2014). 10.1016/j.cell.2014.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Schmidt J. C., Zaug A. J., and Cech T. R., “Live cell imaging reveals the dynamics of telomerase recruitment to telomeres,” Cell 166, 1188–1197.e9 (2016). 10.1016/j.cell.2016.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Bystricky K., “Chromosome dynamics and folding in eukaryotes: Insights from live cell microscopy,” FEBS Lett. 589, 3014–3022 (2015). 10.1016/j.febslet.2015.07.012 [DOI] [PubMed] [Google Scholar]
- 297.Marshall W. F., Straight A., Marko J. F., Swedlow J., Dernburg A., Belmont A., Murray A. W., Agard D. A., and Sedat J. W., “Interphase chromosomes undergo constrained diffusional motion in living cells,” Curr. Biol. 7, 930–939 (1997). 10.1016/s0960-9822(06)00412-x [DOI] [PubMed] [Google Scholar]
- 298.Bronstein I., Israel Y., Kepten E., Mai S., Shav-Tal Y., Barkai E., and Garini Y., “Transient anomalous diffusion of telomeres in the nucleus of mammalian cells,” Phys. Rev. Lett. 103, 018102 (2009). 10.1103/physrevlett.103.018102 [DOI] [PubMed] [Google Scholar]
- 299.Hajjoul H., Mathon J., Ranchon H., Goiffon I., Mozziconacci J., Albert B., Carrivain P., Victor J.-M., Gadal O., Bystricky K., and Bancaud A., “High-throughput chromatin motion tracking in living yeast reveals the flexibility of the fiber throughout the genome,” Genome Res. 23, 1829–1838 (2013). 10.1101/gr.157008.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Shinkai S., Nozaki T., Maeshima K., and Togashi Y., “Dynamic nucleosome movement provides structural information of topological chromatin domains in living human cells,” PLoS Comput. Biol. 12, e1005136 (2016). 10.1371/journal.pcbi.1005136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Chen B., Gilbert L. A., Cimini B. A., Schnitzbauer J., Zhang W., Li G.-W., Park J., Blackburn E. H., Weissman J. S., Qi L. S., and Huang B., “Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system,” Cell 155, 1479–1491 (2013). 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Wang X., Kam Z., Carlton P. M., Xu L., Sedat J. W., and Blackburn E. H., “Rapid telomere motions in live human cells analyzed by highly time-resolved microscopy,” Epigenet. Chromatin 1, 4 (2008). 10.1186/1756-8935-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Bronshtein I., Kepten E., Kanter I., Berezin S., Lindner M., Redwood A. B., Mai S., Gonzalo S., Foisner R., Shav-Tal Y., and Garini Y., “Loss of lamin A function increases chromatin dynamics in the nuclear interior,” Nat. Commun. 6, 8044 (2015). 10.1038/ncomms9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Zidovska A., Weitz D. A., and Mitchison T. J., “Micron-scale coherence in interphase chromatin dynamics,” Proc. Natl. Acad. Sci. U. S. A. 110, 15555–15560 (2013). 10.1073/pnas.1220313110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Shaban H. A., Barth R., and Bystricky K., “Formation of correlated chromatin domains at nanoscale dynamic resolution during transcription,” Nucleic Acids Res. 46, e77 (2018). 10.1093/nar/gky269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Amitai A. and Holcman D., “Polymer physics of nuclear organization and function,” Phys. Rep. 678, 1–83 (2017). 10.1016/j.physrep.2017.02.002 [DOI] [Google Scholar]
- 307.Tortora M. M., Salari H., and Jost D., “Chromosome dynamics during interphase: A biophysical perspective,” Curr. Opin. Genet. Dev. 61, 37–43 (2020). 10.1016/j.gde.2020.03.001 [DOI] [PubMed] [Google Scholar]
- 308.Doi M. and Edwards S. F., The Theory of Polymer Dynamics (Oxford University Press, Oxford, 1986). [Google Scholar]
- 309.Tamm M. V., Nazarov L. I., Gavrilov A. A., and Chertovich A. V., “Anomalous diffusion in fractal globules,” Phys. Rev. Lett. 114, 178102 (2015). 10.1103/physrevlett.114.178102 [DOI] [PubMed] [Google Scholar]
- 310.Polovnikov K. E., Gherardi M., Cosentino-Lagomarsino M., and Tamm M. V., “Fractal folding and medium viscoelasticity contribute jointly to chromosome dynamics,” Phys. Rev. Lett. 120, 088101 (2018). 10.1103/physrevlett.120.088101 [DOI] [PubMed] [Google Scholar]
- 311.Montroll E. W. and Weiss G. H., “Random walks on lattices. II,” J. Math. Phys. 6, 167–181 (1965). 10.1063/1.1704269 [DOI] [Google Scholar]
- 312.Weber S. C., Theriot J. A., and Spakowitz A. J., “Subdiffusive motion of a polymer composed of subdiffusive monomers,” Phys. Rev. E 82, 011913 (2010). 10.1103/physreve.82.011913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Kepten E., Bronshtein I., and Garini Y., “Ergodicity convergence test suggests telomere motion obeys fractional dynamics,” Phys. Rev. E 83, 041919 (2011). 10.1103/physreve.83.041919 [DOI] [PubMed] [Google Scholar]
- 314.Burnecki K., Kepten E., Janczura J., Bronshtein I., Garini Y., and Weron A., “Universal algorithm for identification of fractional Brownian motion. A case of telomere subdiffusion,” Biophys. J. 103, 1839–1847 (2012). 10.1016/j.bpj.2012.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Khanna N., Zhang Y., Lucas J. S., Dudko O. K., and Murre C., “Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions,” Nat. Commun. 10, 2771 (2019). 10.1038/s41467-019-10628-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Strickfaden H., Tolsma T. O., Sharma A., Underhill D. A., Hansen J. C., and Hendzel M. J., “Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells,” Cell 183, 1772–1784 (2020). 10.1016/j.cell.2020.11.027 [DOI] [PubMed] [Google Scholar]
- 317.Biggs R. J., Liu N., Peng Y., Marko J. F., and Qiao H., “Micromanipulation of prophase I chromosomes from mouse spermatocytes reveals high stiffness and gel-like chromatin organization,” Commun. Biol. 3, 542 (2020). 10.1038/s42003-020-01265-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Liu L., Shi G., Thirumalai D., and Hyeon C., “Chain organization of human interphase chromosome determines the spatiotemporal dynamics of chromatin loci,” PLoS Comput. Biol. 14, e1006617 (2018). 10.1371/journal.pcbi.1006617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Pierro M. D., Potoyan D. A., Wolynes P. G., and Onuchic J. N., “Anomalous diffusion, spatial coherence, and viscoelasticity from the energy landscape of human chromosomes,” Proc. Natl. Acad. Sci. U. S. A. 115, 7753–7758 (2018). 10.1073/pnas.1806297115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Put S., Sakaue T., and Vanderzande C., “Active dynamics and spatially coherent motion in chromosomes subject to enzymatic force dipoles,” Phys. Rev. E 99, 032421 (2019). 10.1103/physreve.99.032421 [DOI] [PubMed] [Google Scholar]
- 321.Joo S., Durang X., Lee O.-c., and Jeon J.-H., “Anomalous diffusion for active Brownian particles cross-linked to a networked polymer: Langevin dynamics simulation and theory,” Soft Matter 16, 9188 (2020). 10.1039/d0sm01200a [DOI] [PubMed] [Google Scholar]
- 322.Osmanović D. and Rabin Y., “Dynamics of active Rouse chains,” Soft Matter 13, 963–968 (2017). 10.1039/c6sm02722a [DOI] [PubMed] [Google Scholar]
- 323.Farage T. F., Krinninger P., and Brader J. M., “Effective interactions in active Brownian suspensions,” Phys. Rev. E 91, 042310 (2015). 10.1103/physreve.91.042310 [DOI] [PubMed] [Google Scholar]
- 324.Wang S. and Wolynes P. G., “Tensegrity and motor-driven effective interactions in a model cytoskeleton,” J. Chem. Phys. 136, 145102 (2012). 10.1063/1.3702583 [DOI] [PubMed] [Google Scholar]
- 325.Jiang Z. and Zhang B., “On the role of transcription in positioning nucleosomes,” PLoS Comput. Biol. 17, e1008556 (2021). 10.1371/journal.pcbi.1008556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Pressé S., Ghosh K., Lee J., and Dill K. A., “Principles of maximum entropy and maximum caliber in statistical physics,” Rev. Mod. Phys. 85, 1115–1141 (2013). 10.1103/revmodphys.85.1115 [DOI] [Google Scholar]
- 327.Capelli R., Tiana G., and Camilloni C., “An implementation of the maximum-caliber principle by replica-averaged time-resolved restrained simulations,” J. Chem. Phys. 148, 184114 (2018). 10.1063/1.5030339 [DOI] [PubMed] [Google Scholar]
- 328.Guan J., Liu H., Shi X., Feng S., and Huang B., “Tracking multiple genomic elements using correlative CRISPR imaging and sequential DNA FISH,” Biophys. J. 112, 1077–1084 (2017). 10.1016/j.bpj.2017.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Takei Y., Shah S., Harvey S., Qi L. S., and Cai L., “Multiplexed dynamic imaging of genomic loci by combined CRISPR imaging and DNA sequential FISH,” Biophys. J. 112, 1773–1776 (2017). 10.1016/j.bpj.2017.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.