Abstract
In this study, we examined endophytic fungi in leaves of Mandevilla catimbauensis, an endemic plant species found in the Brazilian dry forest (Caatinga), and endophytic fungi’s potential to produce L-asparaginase (L-ASNase). In total, 66 endophytes were isolated, and the leaf-fragment colonisation rate was 11.78%. Based on morphology, internal transcribed spacer (ITS), and partial large subunit (LSU) of ribosomal DNA sequencing, the endophytic fungi isolated belonged to six Ascomycota orders (Botryosphaeriales, Capnodiales, Diaporthales, Eurotiales, Marthamycetales, and Pleosporales). Phyllosticta species were the most frequent endophytes isolated (23 isolates [45.1%] from two species). The Shannon–Wiener and Fisher alpha index average values were 0.56 and 3.26, respectively. Twenty endophytes were randomly selected for the L-ASNase production test, of which fourteen isolates showed potential to produce the enzyme (0.48–2.22 U g−1), especially Phyllosticta catimbauensis URM 7672 (2.22 U g−1) and Cladosporium sp. G45 (2.11 U g−1). Phyllosticta catimbauensis URM 7672 was selected for the partial optimisation of L-ASNase production because of its ability to generate considerable amounts of enzyme. We obtained the highest L-ASNase activity (3.47 U g−1), representing an increase of 36.02% in enzymatic production, under the following experimental conditions: a pH of 4.2, 1.0% inoculum concentration, and 2.5% L-asparagine concentration. Our study showed that M. catimbauensis harbours an important diversity of endophytic fungi with biotechnological potential for L-ASNase production.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-021-00505-3.
Keywords: Caatinga forest, Endophytes, Fungal enzyme, Phyllosticta, L-asparaginase optimisation
Introduction
Endophytic fungi inhabit intra- and inter-cellular plant tissues without causing harmful effects on plants. These microorganisms may alter gene expression, modulate biosynthetic pathways, mitigate stressful conditions in plants, and play an important role in establishing the plant defence system against potential pathogens. Endophytic fungi are a potential resource for biosynthesis, biotransformation, and biodegradation because they produce many secondary metabolites [1, 2]. Therefore, investigating the fungi inhabiting plant species with medicinal properties can lead to the discovery of new metabolites with potential bioactivity [2].
Many studies on endophytic fungi have been conducted in humid and temperate tropical forests [3, 4]. However, arid, semiarid, and desert areas, such as the Brazilian tropical dry forest (Caatinga), are also promising environments in terms of fungal species diversity [5–7]. Studies conducted in Caatinga sites have contributed to the description of new fungal endophytes with biotechnological potential [6–8]. These studies have described new fungal endophytes, suggesting that the real mycobiome richness has been underestimated [2, 5, 9].
The Caatinga is a tropical dry forest covering a large part of northeastern Brazil and harbours multiple numbers of plants, animals, and microorganisms [10–13]. Considering the large-scale destruction because of anthropogenic interference and to guarantee the maintenance of Caatinga diversity, some areas of this ecosystem have been selected to be conservation units. One important conservation unit is the Catimbau National Park (Parque Nacional do Catimbau), the second largest archaeological park in Brazil, which presents a lush landscape and rich diversity [14]. Catimbau National Park harbours several endemic and exclusive plant species, including Mandevilla catimbauensis Souza-Silva, Rapini & JF Morales (Apocynaceae), a rare and endemic species that is vulnerable to extinction [15, 16]. Mandevilla catimbauensis is an herbaceous and climbing plant mainly recognised by its twining habit and it is only found in the Catimbau National Park [15, 17]. Mandevilla is considered the largest neotropical genus of the family Apocynaceae, and it is represented by about 40 species in Brazil [15]. Mandevilla species have medicinal importance in secondary chemical compound production with anti-inflammatory, antinociceptive, and antipyretic properties [18].
Several fungi isolated from dry environments have shown a potential for production of biomolecules of industrial interest, such as L-asparaginase (L-ASNase) [6, 7, 9]. L-ASNase has been used as a drug in anti-leukaemia chemotherapy, but there are several adverse effects when this enzyme is produced by bacteria, which has led the scientific community to search for alternative sources [19]. In addition to its clinical use, L-ASNase has been used by the food industry to reduce the formation of acrylamide, a harmful toxin formed during high-temperature food processing [20]. The L-ASNase used by the food industry has a fungal origin [21], and the increased use of the enzyme has motivated the search for fungal resources capable of meeting the high-L-ASNase demand of pharmaceutical and food industries [19, 22].
Endophytic fungi are a limitless source of novel metabolites, and endophytes from plants growing in particular ecological niches may have the ability to produce a variety of secondary metabolites [1]. In this study, we investigated, for the first time, the diversity and biotechnological potential of endophytic fungi from the plant M. catimbauensis, which grows in the Caatinga dry forest. We investigated the hypothesis that leaves of M. catimbauensis harbour a rich and diverse endophytic fungal community with the potential to produce L-ASNase. In order to test this hypothesis, our study aimed to analyse the diversity of endophytic fungi living in healthy M. catimbauensis leaves and verify the capacity of the endophytes to produce L-ASNase.
Materials and methods
Study area
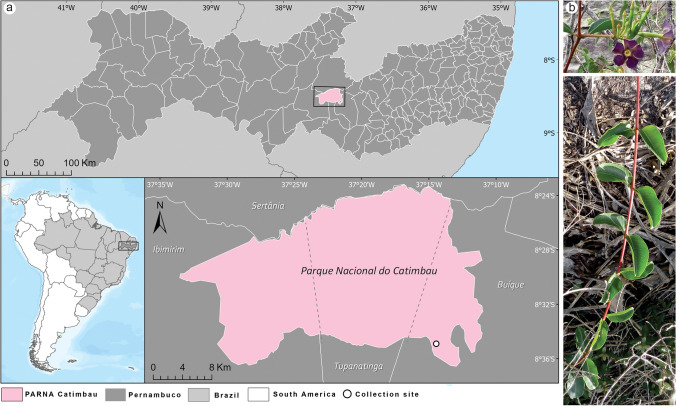
This study was carried out at the Parque Nacional do Catimbau (8° 24′ 00″ and 8° 36′ 35″ S; 37° 09′ 30″ and 37° 14′ 40″ W), a federal conservation unit in the Caatinga dry forest, Pernambuco State, Brazil (Fig. 1a). The climate is semiarid and is classified as BSh according to Köppen’s scale [23]. It has an average annual temperature and rainfall of 23 °C and 650–1100 mm, respectively, with March and October being the wettest and driest months, respectively. The park contains heterogeneous vegetation, with four distinct vegetation types: rupestrian grasslands, semi-deciduous and evergreen vegetation, and small deciduous trees and shrubs [12].
Fig. 1.
a The geographical location of the Parque Nacional do Catimbau (Catimbau National Park), Brazilian tropical dry forest (Caatinga). b Mandevilla catimbauensis in the Catimbau National Park
Plant collection and isolation of endophytic fungi
Mandevilla catimbauensis leaves (Fig. 1b) were sampled in May 2015 during one of the longest drought periods in a sedimentary area of the Caatinga dry forest from Serra de Jerusalém (at an altitude of 910 m), Catimbau National Park, Buíque municipality (Fig. 1a). Leaves from 20 individuals were randomly collected, packed in paper bags, and processed in the laboratory within 24 h. The endophytic fungi were isolated by processing the leaves according to the method proposed by Bezerra et al. [24]. After disinfection, the leaf fragments were placed in Petri dishes containing potato dextrose agar (PDA) medium supplemented with chloramphenicol (100 mg L−1) and tetracycline (50 mg L−1) to restrict bacterial growth. The Petri dishes were incubated at 28 ± 2 °C for up to 30 days under a natural light cycle. Fungal endophyte growth was observed every day, and any colony found was isolated, purified, and maintained on PDA for later identification. The efficacy of the surface sterilisation process was verified by inoculating 1 mL of water used during the last rinses onto Petri dishes containing the same medium, followed by incubation under the same conditions.
A specimen of M. catimbauensis has been deposited in the Herbário UFP – Geraldo Mariz under number 81.210, and representative cultures of the endophytes are deposited in the URM culture collection (Micoteca URM Profa. Maria Auxiliadora Cavalcanti). Both collections are located at the Universidade Federal de Pernambuco (Recife, Brazil). The plant collections were authorised by MMA/ICMBio (SISBIO number 48492–1).
DNA extraction, PCR, sequencing, and phylogenetic analyses
All the isolated endophytic fungi were cultured on PDA, and the DNA was extracted using the Wizard® Genomic DNA Purification kit (Promega, USA) according to the manufacturer’s instructions. The internal transcribed spacer (ITS) and part of the nuclear ribosomal small subunit (LSU) regions of the rDNA were used to identify endophytic fungi. PCR, sequencing, and sequence analyses were then performed as described by Bezerra et al. [25].
Searches using the BLASTn tool in the GenBank database at NCBI were used to verify the phylogenetic relationships among all the ITS and LSU rDNA sequences. Subsequently, selected sequences deposited in GenBank were aligned with the ITS sequences obtained in this study to verify their relationships based on maximum likelihood (ML) analysis. Sequence editing, alignments, and phylogenetic analysis were conducted according to the procedures described earlier [25], and the ML analysis used 1000 bootstrap replicates and GTR + I + G as the best nucleotide model.
The sequences obtained in this study were deposited in the GenBank database (ITS = MT569893–MT569930 and LSU = MT569931–MT569974) (Online Resource Table 1). The sequence alignment used to perform the phylogenetic analysis was deposited in TreeBASE (study S26389).
Colonisation rate and absolute and relative frequencies
The endophytic colonisation rate (CR) was calculated by considering the ratio between the number of colonised fragments (Nf) and the total number of fragments (Nt) taken from the plant tissue (CR = Nf / Nt × 100). The absolute frequency represents the total number of endophytes isolated, and the relative frequency is the number of isolates of each genus divided by the total number of isolates and multiplied by 100.
Ecological analyses
We calculated the richness (S), Shannon–Wiener diversity index (H′), and Fisher’s alpha value for all samples. Richness was defined as the number of species in each sample, and the Shannon–Wiener diversity index was calculated using the following equation: H′ = − Σ(Pi ln(Pi)), where Pi = ni / N and ni = number of endophytic isolates of taxon i and N = total number of endophytic isolates of all taxa [26]. For statistical purposes, the H′ values were converted to Exp (H′). Fisher’s alpha value was calculated using the equation: S = α × ln (l + n / α), where S is the number of taxa, n is the number of isolates, and α is Fisher’s alpha value [27]. Species accumulation curves were determined, and observed richness was compared to estimated richness using the first-order Jackknife (Jackknife 1). All analyses were performed using R software v.3.5.0 (R Development Core Team 2018) and the “agricolae” [28], “vegan” [29], and “iNEXT” packages [30].
L-ASNase production in liquid medium
The L-ASNase production process was based on the studies by Loureiro et al. [31] and Silva et al. [6]. During the pre-fermentative stage, biomass production was induced using Czapek Dox’s Medium (CDM) [32], as modified by Pádua et al. [7] [glucose (14.0 g L−1), L-asparagine (10.0 g L−1), KH2PO4 (1.52 g L−1), KCl (0.52 g L−1), MgSO4.7H2O (0.52 g L−1), CuNO3.3H2O (0.01 g L−1), ZnSO4.7H2O (0.01 g L−1), FeSO4.7H2O (0.01 g L−1), and (NH4)2SO4 (2.0 g L−1) at pH 6.2]. Erlenmeyer flasks (250 mL) containing 100 mL of CDM were inoculated with five discs (5 mm) of fungal mycelium that had been allowed to grow for 7 days on malt extract agar. These flasks were incubated at 30 °C for 96 h at 120 rpm. The cultures were filtered using Whatman no. 1 filter paper, and the biomass obtained was used to determine enzyme production. During the fermentative stage, the biomass obtained in the previous stage was inoculated into the modified CDM medium (as described above), and the modifications were that the glucose concentration was adjusted to 2.0 g L−1 and no (NH4)2SO4 was added. The inoculated media were incubated at 30 °C for 96 h at 120 rpm. Finally, the cultures were filtered using Whatman no. 1 filter paper. The enzymatic activity of the obtained biomass was quantified as described below.
L-ASNase activity
The L-ASNase activity was determined according to Drainas et al. [33], as modified by Silva et al. [6] and Pádua et al. [7]. A total of 1.5 mL Tris–HCl buffer (20 mM, pH 8.6) and 0.1 g of fungal biomass from each culture obtained after the fermentation step were vortexed. Then, 0.2 mL of L-asparagine solution (100 mM) and 0.2 mL of stock hydroxylamine solution (1 M, pH 7.0) were added, and the samples were incubated for 30 min at 37 °C and 150 rpm. The reaction was stopped by adding 0.5 mL of FeCl3 reagent [10% (w/v) FeCl3 plus 5% (w/v) trichloroacetic acid in 0.66 mol L−1 HCl] to all the samples, including the blank samples (Tris–HCl and mycelium). The reaction mixtures were then centrifuged for 15 min at 6000 rpm and 4 °C to remove the precipitates. Absorbance was measured at 500 nm against the blank samples that received L-asparagine and hydroxylamine solutions after 30 min of incubation. One unit of L-ASNase was defined as the amount of enzyme that releases 1 μmol of β-hydroxamic aspartic acid per minute.
Partial optimisation of L‑ASNase production
A statistical experimental design (23 factorial design with four central points) [34, 35] was used to partially optimise L-ASNase production. We used a factorial design comprising 12 trials, three variables (concentration of L-asparagine, pH, and inoculum concentration), and four replications at the central point. The pre-fermentation conditions were the same as described above (see the “L-ASNase production in liquid medium” section). The modified CDM was also used for fermentation, where the variables used were adjusted according to the results of the experimental design, and the flasks were incubated at 120 rpm for 120 h at 30 °C. After the incubation period, the fungal biomass was filtered and used to determine enzyme activity [33 modified by 6 and 7]. Based on the factorial design results, additional experiments were carried out to increase the enzymatic production.
Statistical analysis of L-ASNase activity
The non-parametric Kruskal–Wallis test was used to determine if there was a statistically significant difference (p < 0.05) or a marginally significant difference (p < 0.1) between the results for L-ASNase production by endophytic fungi from M. catimbauensis. The results obtained from a 23 factorial design and the non-parametric Kruskal–Wallis test were processed using R software. This test was carried out to verify and evaluate the relationship between the selected independent variables and enzyme production. All calculations were randomly performed.
Results
A total of 560 leaf fragments from M. catimbauensis were randomly collected from 20 individuals. The colonisation rate for the leaf fragments was 11.78%, and a total of 66 endophytic fungi were isolated; however, 15 isolates did not grow after isolation. We identified 13 genera based on their morphological characteristics (macro and micromorphology) and analysis of the ITS or LSU rDNA sequences. The isolated endophytes belonged to six orders in Ascomycota (Botryosphaeriales, Capnodiales, Diaporthales, Eurotiales, Marthamycetales, and Pleosporales) (Online Resource Fig. 1). Phyllosticta isolates were the most frequent (45.10%) in M. catimbauensis, followed by Diaporthe (11.76%). Cercospora, Cladosporium, Marthamyces, Paracercosporidium, Paraconiothyrium, Parapyrenochaeta, Phaeophleospora, and Preussia, which were only present at low frequencies (one or two isolates) and were considered to be rare endophytes (Table 1).
Table 1.
Absolute (F) and relative (fr) (%) frequency, number of isolates, richness (S), and Shannon–Wiener (H′) and Fisher’s alpha (F-α) indexes diversity of endophytic fungi isolated from leaves of the medicinal plant Mandevilla catimbauensis, an endemic plant of the Caatinga dry forest, Brazil
| Endophytic fungi | Mandevilla catimbauensis individuals | F | fr | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |||
| Alternaria sp. (A. tomato species complex) | 1 | 1 | 1 | 3 | 5.88 | |||||||||||||||||
| Cercospora sp. | 1 | 1 | 1.96 | |||||||||||||||||||
| Cladosporium sp. (C. cladosporioides species complex) | 1 | 1 | 2 | 3.92 | ||||||||||||||||||
| Diaporthe caatingaensis | 1 | 1 | 1.96 | |||||||||||||||||||
| Diaporthe cf. acaciarum | 1 | 2 | 3 | 5.88 | ||||||||||||||||||
| Diaporthe heveae | 1 | 1 | 1.96 | |||||||||||||||||||
| Diaporthe sp. 1 | 1 | 1 | 1.96 | |||||||||||||||||||
| Marthamyces renga | 1 | 1 | 1.96 | |||||||||||||||||||
| Phaeophleospora eucalypticola | 1 | 1 | 1.96 | |||||||||||||||||||
| P. hymenocallidicola | 1 | 1 | 1.96 | |||||||||||||||||||
| Paracercosporidium sp. | 1 | 1 | 2 | 5.88 | ||||||||||||||||||
| Paraconiothyrium sp. | 1 | 1 | 1.96 | |||||||||||||||||||
| Parapyrenochaeta sp. | 1 | 1 | 1.96 | |||||||||||||||||||
| Phyllosticta capitalensis | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 4 | 2 | 15 | 29.40 | |||||||||||
| P. catimbauensis | 1 | 1 | 1 | 5 | 8 | 15.68 | ||||||||||||||||
| Preussia sp. | 1 | 1 | 1.96 | |||||||||||||||||||
| Pseudofusicoccum stromaticum | 1 | 2 | 3 | 5.88 | ||||||||||||||||||
| Talaromyces sp. (section Helici) | 1 | 2 | 2 | 5 | 9.8 | |||||||||||||||||
| Isolates | 3 | 2 | 2 | 2 | 1 | 1 | 3 | 3 | 3 | 9 | 4 | 2 | 8 | 6 | 2 | 51 | ||||||
| Richness (S) | 3 | 2 | 2 | 2 | 1 | 1 | 3 | 3 | 2 | 7 | 3 | 1 | 3 | 3 | 1 | |||||||
| Shannon–Wiener (H′) | 1.09 | 0.69 | 0.69 | 0.69 | 1.09 | 1.09 | 1.09 | 1.90 | 1.04 | 0.90 | 0.86 | |||||||||||
| Fisher’s alpha (F-α) | 3 | 26.78 | 5 | 0.79 | 2 | 2 | 0.79 | |||||||||||||||
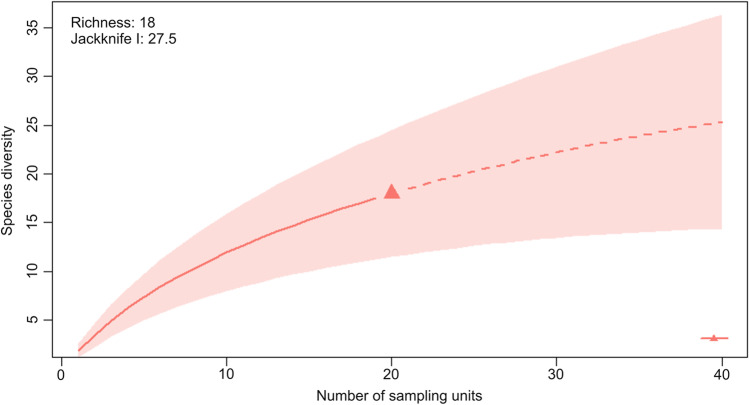
The total richness of the endophytic fungi was 18 taxa, and the estimated richness per individual M. catimbauensis varied from one to seven species, with an average of 1.9. The total diversity of the endophytic fungi, according to the Shannon index, ranged from 0.69 to 1.90, with an average of 0.56. The Fisher index values varied from 0.79 to 26.78, and the average was 3.26. The species accumulation curve did not reach a plateau, and the sampling effort was not sufficient to recover all the estimated richness using the first-order Chao Jackknife index (Chao Jackknife 1) (Fig. 2).
Fig. 2.
Species accumulation curve for endophytic fungi recovered from leaves of Mandevilla catimbauensis, an endemic plant of the Caatinga dry forest, Brazil
Of the 20 endophytic fungi tested in the liquid medium, 14 isolates showed a capacity to produce the enzyme L-ASNase, with the enzymatic activity varying between 0.48 and 2.22 U g−1 (Table 2). Phyllosticta catimbauensis URM 7672 and Cladosporium sp. G45 had the best results for the intracellular enzyme production (2.22 and 2.11 U g−1, respectively) (Table 2). Based on these results, the endophyte P. catimbauensis URM 7672 was selected for partial optimisation of L-ASNase production.
Table 2.
L-asparaginase activity produced by endophytic fungi isolated from leaves of the medicinal plant Mandevilla catimbauensis, an endemic plant of the Caatinga dry forest, Brazil
| Endophytic fungi | URM or isolate numbers | L-asparaginase activity (U g−1) |
|---|---|---|
| Alternaria sp. | G22 | 0.00 h |
| Alternaria sp. | G7 | 0.00 h |
| Cladosporium sp. | G45 | 2.11a |
| Diaporthe cf. acaciarum | G25 | 0.56 fg |
| D. heveae | URM 8162 | 0.80b |
| Marthamyces renga | G48 | 0.00 h |
| Phyllosticta capitalensis | URM 8159 | 0.00 h |
| P. capitalensis | G21 | 0.76 cd |
| P. catimbauensis | URM 7672 | 2.22a |
| P. catimbauensis | G12 | 0.48 g |
| P. catimbauensis | URM 7673 | 0.99bc |
| P. catimbauensis | G20 | 0.75de |
| P. catimbauensis | URM 7674 | 0.57ef |
| Pseudofusicoccum stromaticum | G6 | 0.61ef |
| P. stromaticum | G41 | 0.00 h |
| Paracercosporidium sp. | G46 | 0.48 g |
| Parapyrenochaeta sp. | URM 8160 | 0.79bc |
| Preussia sp. | URM 8161 | 0.95b |
| Talaromyces sp. | URM 8163 | 1.11b |
| Talaromyces sp. | URM 8164 | 0.00 h |
Values followed by the same letter do not differ significantly (p > 0.05) using the non-parametric test of Kruskal–Wallis
L-ASNase production using the experimental 23 factorial design varied between 0.61 and 2.25 U g−1 (Table 3). The highest L-ASNase production (2.25 U g−1) was obtained at pH 5.0, 1.5% L-asparagine, and 1.5% inoculum (Assay 6, Table 3). The analysis of variance results showed that the increase in L-asparagine production was statistically significant, whereas the pH and the interaction among the three explanatory variables were marginally significant, and the inoculum concentration was not significant (Online Resource Table 2). The lack of adjustment was not statistically significant, and the determination coefficients (R2) and adjusted R2 were 0.8418 and 0.7514, respectively. Based on the independent variables’ effects (Online Resource Fig. 2), an experimental sequence was performed with the adjusted variables to obtain a greater enzymatic production (Table 4). In this experimental design, 3.5 U g−1 of L-ASNase was obtained using 3.5% L-asparagine, pH 4.2, and 1.0% inoculum (Assay 5, Table 4). Table 4 shows the Kruskal–Wallis analysis, which resulted in a p-value of 0.0155. The best values for enzymatic production were produced by assays 4 (3.47 U g−1) and 5 (3.50 U g−1). According to the test results, there were no significant differences between these values. Assays 4 and 5 had the same pH (4.2) and inoculum concentration (1.0 g), but had different L-asparagine concentrations (assay 4 = 2.5 g and assay 5 = 3.5 g). The increase in L-ASNase enzymatic activity was 36.02% and 36.57% in assays 4 and 5, respectively.
Table 3.
Production of L-asparaginase by Phyllosticta catimbauensis URM 7672 in the 23 factorial experimental design
| Assay | L-asparagine (%) | pH | Inoculum concentration (biomass %) | L-Asparaginase activity (U g−1) |
|---|---|---|---|---|
| 1 | 0.5 | 5.0 | 0.5 | 0.61 |
| 2 | 1.5 | 5.0 | 0.5 | 2.03 |
| 3 | 0.5 | 7.0 | 0.5 | 1.07 |
| 4 | 1.5 | 7.0 | 0.5 | 1.38 |
| 5 | 0.5 | 5.0 | 1.5 | 1.46 |
| 6 | 1.5 | 5.0 | 1.5 | 2.25 |
| 7 | 0.5 | 7.0 | 1.5 | 0.67 |
| 8 | 1.5 | 7.0 | 1.5 | 1.81 |
| 9 | 1.0 | 6.0 | 1.0 | 1.70 |
| 10 | 1.0 | 6.0 | 1.0 | 1.85 |
| 11 | 1.0 | 6.0 | 1.0 | 1.46 |
| 12 | 1.0 | 6.0 | 1.0 | 1.54 |
Table 4.
Production of L-asparaginase by Phyllosticta catimbauensis URM 7672 obtained through an experimental sequence. Inoculum concentration was fixed in 1.0% fungal biomass
| Assay | L-asparagine (%) | pH | L-asparaginase activity |
|---|---|---|---|
| 1 | 1.5 | 5.0 | 2.33bc |
| 2 | 1.5 | 4.6 | 2.49b |
| 3 | 2.5 | 4.6 | 1.13c |
| 4 | 2.5 | 4.2 | 3.47a |
| 5 | 3.5 | 4.2 | 3.50a |
| 6 | 4.5 | 4.2 | 2.91ab |
Non-parametric test of Kruskal–Wallis (p = 0.0155). Values followed by the same letter do not differ statistically from each other
Discussion
In this study, we isolated 66 endophytic fungi from the leaves of M. catimbauensis, a plant species belonging to the Apocynaceae family. Studies on the endophytic fungi associated with this plant family have recorded between 11 and 22 endophytes [36]. The low recorded fungal colonisation rate (11.78%) in the leaves of M. catimbauensis may be because of the presence of latex in the plant composition, as latex has antibacterial and antifungal properties [37]. Another study on endophytic fungi isolated from the stems and roots of Nerium indicum (Apocynaceae) reported a total of 11 endophytic fungi isolated from healthy tissues of the plant (five from stems and six from roots) [38].
Different factors may have influenced the low diversity of fungal endophytes recovered from M. catimbauensis leaves (H′ = 0.69 to 1.90 and F-α = 0.79 to 26.78), for example, plant tissue type and age, micro-habitats, the relationship between fungal hosts, climate, vegetation changes, and human impact [39–41]. Variations in solar intensity and exposure are relevant factors associated with the composition of the endophytic fungal community [42]. Furthermore, ultraviolet radiation in semiarid regions, such as the Caatinga dry forest in Brazil, can negatively influence the diversity of these fungi [7].
Most of the endophytic fungi isolated in this study were pigmented, and Phyllosticta was the most frequently found genus. Pigmented fungi have been isolated in studies on various plant species from arid environments, and they may confer host plant tolerance to extreme environmental conditions, including ultraviolet radiation [43, 44]. Suryanarayanan et al. [43] studied the occurrence and distribution of endophytic fungi in tropical forests and observed that the most frequently isolated genera were Colletotrichum, Phyllosticta, and Diaporthe [Phomopsis]. Phyllosticta has been frequently isolated as endophytes from the leaves of various tropical plants [44], and endophytes belonging to Diaporthe are also frequently isolated [5, 7]. For example, Pádua et al. [7] reported that Diaporthe and Phyllosticta were the most common endophytes isolated from M. urundeuva occurring in the Caatinga forest of Brazil.
Similar to our study, most investigations of microbial diversity have not sufficiently recovered all estimated species richness [7, 45–49]. For example, investigations of endophytic fungi associated with different plant hosts in dry tropical forests (Caatinga) in Brazil were not able to represent the estimated species richness [7, 47, 49]. Similarly, Oliveira et al. [48] did not isolate all expected endophytic fungi from the leaves of Cocos nucifera occurring in coconut crops in a region of the Atlantic Forest in Brazil. In a large study from the Canadian Arctic to the lowland tropical forest of central Panama, Arnold and Lutzoni [46] also demonstrated an inability to isolate all estimated endophytic species from the leaves of 28 hosts, showing that the expected species remarkably exceeded the numbers observed. Based on 1403 endophytes isolated from representative arctic, boreal, temperate, and tropical plants, the authors observed approximately 250 species, whereas 350 species were expected.
Most (70%) of the tested endophytic fungal isolates were able to produce L-ASNase. Phyllosticta catimbauensis, a species described in 2017 [50], had the highest ability to produce L-ASNase (URM 7672, 2.22 U g−1). Some studies have demonstrated that endophytic fungi have a high production capacity for L-ASNase [6, 7]. Furthermore, previous research has shown that some Phyllosticta species can produce novel bioactive metabolites [51], including anticancer drugs [7]. Other endophytes, such as Cladosporium sp. G45 and Talaromyces sp. URM 8163, have also demonstrated the capacity for L-ASNase production (2.11 U g−1 and 1.11 U g−1, respectively). Kumar et al. [52] reported Cladosporium sp. isolated from soil samples as the best producer of L-ASNase (3.74 U g−1). Species of Talaromyces have a huge biotechnological capacity to produce several metabolites. Krishnapura and Belur [53] partially purified and characterised the L-ASNase of T. pinophilus, an endophyte isolated from the rhizomes of Curcuma amada (Zingiberaceae), and they considered it a potential candidate for industrial and clinical trials because of its biochemical properties and high efficiency (174 U mL−1).
Similar to the present study, Pádua et al. [7] used biomass to quantify enzymatic activity based on the formation of β-hydroxamic aspartic acid. The authors reported that Diaporthe sp. URM 7793 was one of the best producers (2.41 U g−1), followed by Diaporthe sp. URM 7779 (2.00 U g−1) and Talaromyces sp. URM 7785 (1.91 U g−1). In addition, Pádua et al. [7] demonstrated the potential of Phyllosticta sp. URM 7787 to produce L-ASNase with an average of 0.57 U g−1. Silva et al. [6] evaluated the L-ASNase activities of endophytes isolated from the bromeliad Tillandsia catimbauensis and identified Talaromyces cf. cecidicola URM 7826 as the best producer (2.30 U g−1). In another study, Da Rocha et al. [54] evaluated 32 filamentous fungi isolated from the Caatinga and obtained an activity of 1.58 U g−1 using Aspergillus terreus URM 7732. However, Costa-Silva et al. [55] evaluated the conditions of biomass production by A. terreus CCT 7693 and obtained a production of 13.50 U g−1 after optimisation (the best condition was CDM containing glucose 2 g L−1, L-proline 10 g L−1, and an inoculum concentration of 4.8 × 108 spores mL−1 adjusted to pH 9.49 at 34.6 °C) of the enzyme production process. The superior production found by Costa-Silva et al. [55] is a basis for further studies on the conditions of L-ASNase production by P. catimbauensis URM 7672.
In this study, 2.5% L-asparagine and 1.0% fungal inoculum at pH 4.2 were the optimal parameters for enzyme production. Furthermore, the experimental design increased enzyme activity by 36.02%. According to Thakur et al. [56], the initial pH of the culture medium can influence enzyme production because it can affect nutrient availability. Kumar et al. [52] discovered that pH 5.8 was optimal for L-ASNase production by Cladosporium sp. isolated from soil. In another study, Farag et al. [57] optimised the fermentation parameters and obtained optimum conditions for L-ASNase production by Aspergillus terreus at pH 6.0 using dextrose and L-asparagine as sources of carbon and nitrogen, respectively. Similarly, Vieira et al. [58] optimised L-ASNase production by Penicillium sp., and the optimum conditions were pH 5.1, 1.2 g L−1 L-asparagine, and 3.0 g L−1 glucose. However, Dias and Sato [59] reported that 2% proline, 0.5% glucose, 0.2% L-asparagine, and pH 8.0 were ideal conditions for enzymatic production by Aspergillus oryzae.
Here, we present the first study on endophytic fungi associated with M. catimbauensis, an endemic species in the Brazilian tropical dry forest. Our findings showed that M. catimbauensis leaves are host to endophytic species with a capacity to produce L-ASNase. Among the 14 endophytic isolates that exhibited enzymatic activity (0.48–2.22 U g−1), P. catimbauensis URM 7672 (2.22 U g−1) and Cladosporium sp. G45 (2.11 U g−1) were the most promising enzyme producers. During the partial optimisation stage for L-ASNase production by P. catimbauensis URM 7672, L-asparagine concentration was found to be significant, and pH was found to be marginally significant, whereas the inoculum concentration was not significant. Furthermore, L-ASNase production by P. catimbauensis URM 7672 varied between 1.13 and 3.50 U g−1 when using the optimal experimental design conditions. The best conditions for L-ASNase production by P. catimbauensis URM 7672 (3.47 U g−1) were 2.5% L-asparagine and 1.0% inoculum at pH 4.2, considering that lower concentrations of L-asparagine did not produce statistically significant differences for the highest enzyme activity (3.50 U g−1). The production of secondary metabolites by endophytic fungi greatly impacts modern pharmaceutical products, which led us to explore this valuable biological resource to make the process more advantageous for human use [60]. The industrial importance of L-ASNase means that a search for new enzyme production sources is needed. Any new sources can contribute to biotechnological applications in the food and drug industries. These results suggest that the endophyte P. catimbauensis URM 7672 should be subjected to further optimisation studies that will investigate ways of optimising the production of L-ASNase. In addition, genetic manipulation, mutations, and other recombinant DNA techniques can contribute to obtaining higher L-ASNase production, as has been done for other substances [60]. Our results highlight the importance of protecting plant species in their natural environments to guarantee diversity and to allow their potential contribution to future biotechnological studies for promotion of a future eco-friendly and sustainable bio-economy.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors sincerely thank the Programa de Pós-Graduação em Biociência Animal of the Universidade Federal Rural de Pernambuco (UFRPE) and all members of the Departamento de Micologia Prof. Chaves Batista and of the Laboratório de Micologia Ambiental (both at the Universidade Federal de Pernambuco). We especially thank Dr. Laura Paiva, Dr. Alexandre Machado, Dr. Karla Freire, Dr. Renan Barbosa, Ana Patrícia Pádua (MSc), Aline Barboza (MSc), Tamara Caldas (MSc), and Thays Oliveira (MSc).
Author contribution
Gianne R. Araújo-Magalhães, Cristina M. Souza-Motta, and Keila A. Moreira implemented the project; Gianne R. Araújo-Magalhães collected the plants; Gianne R. Araújo-Magalhães and Leticia F. da Silva performed the fungal endophytes isolation and L-asparaginase production parts of the study; Gualberto S. Agamez-Montalvo designed the experiment and statistically analysed the L-Asparaginase production results; Iolanda R. da Silva performed the ecological analysis; Jadson D.P. Bezerra performed the phylogenetic analysis; Gianne R. Araújo-Magalhães, Marília H.C. Maciel, and Jadson D.P. Bezerra wrote the paper. All the authors reviewed and approved the final version of the paper.
Funding
Fundação de Amparo à Ciência e Tecnologia de Pernambuco (FACEPE, financial support and scholarship IBPG-0970-5.05/14), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) in Brazil.
Data availability
Plant specimen is deposited in the Herbário UFP – Geraldo Mariz and endophytic fungi cultures in the URM culture collection, both collections at the Universidade Federal de Pernambuco (Recife, Brazil). DNA sequences are deposited in the GenBank database and sequence alignment in TreeBASE.
Code availability
Not applicable.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gianne R. Araújo-Magalhães and Marília H.C. Maciel contributed equally to this work.
Contributor Information
Jadson D. P. Bezerra, Email: jadsondpb@gmail.com, Email: jadsonbezerra@ufg.br
Keila A. Moreira, Email: moreirakeila@hotmail.com, Email: keila.moreira@ufape.edu.br
References
- 1.Strobel G, Castillo U, Harper J. Natural products from endophytic microorganisms. J Nat Prod. 2004;67:257–268. doi: 10.1021/np030397v. [DOI] [PubMed] [Google Scholar]
- 2.Dhayanithy G, Subban K, Chelliah J. Diversity and biological activities of endophytic fungi associated with Catharanthus roseus. BMC Microbiol. 2019;19:22. doi: 10.1186/s12866-019-1386-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold AE, Maynard Z, Gilbert GS. Fungal endophytes in dicotyledonous neotropical trees: patterns of abundance and diversity. Mycol Res. 2001;105:1502–1507. doi: 10.1017/S0953756201004956. [DOI] [Google Scholar]
- 4.Banerjee D (2011) Endophytic fungal diversity in tropical and subtropical plants. Res J Microbiol 6:54–62. 10.3923/jm.2011.54.62
- 5.Bezerra JDP, Machado AR, Firmino AL, Rosado AWC, Souza CAF, Souza-Motta CM, Freire KTLS, Paiva LM, Magalhães OMC, Pereira OL, Crous PW, Oliveira TGL, Abreu VP, Fan X. Mycological diversity description I. Acta Bot Bras. 2018;32:656–666. doi: 10.1590/0102-33062018abb0154. [DOI] [Google Scholar]
- 6.Silva LF, Freire KTLS, Araújo-Magalhães GR, Agamez-Montalvo GS, Sousa MA, Costa-Silva TA, Paiva LM, Pessoa-Junior A, Bezerra JDP, Souza-Motta CM. Penicillium and Talaromyces endophytes from Tillandsia catimbauensis, a bromeliad endemic in the Brazilian tropical dry forest, and their potential for L-asparaginase production. World J Microbiol Biotechnol. 2018;34:162. doi: 10.1007/s11274-018-2547-z. [DOI] [PubMed] [Google Scholar]
- 7.Pádua APSL, Freire KTLS, Oliveira TGL, Silva LF, Araújo-Magalhães GR, Agamez-Montalvo GS, Silva IR, Bezerra JDP, Souza-Motta CM. Fungal endophyte diversity in the leaves of the medicinal plant Myracrodruon urundeuva in a Brazilian dry tropical forest and their capacity to produce L-asparaginase. Acta Bot Bras. 2019;33:39–49. doi: 10.1590/0102-33062018abb0108. [DOI] [Google Scholar]
- 8.Ferreira MC, Cantrell CL, Wedge DE, Gonçalves VN, Jacob MR, Khan S, Rosa CA, Rosa LH. Diversity of the endophytic fungi associated with the ancient and narrowly endemic neotropical plant Vellozia gigantea from the endangered Brazilian rupestrian grasslands. Biochem Syst Ecol. 2017;71:163–169. doi: 10.1016/j.bse.2017.02.006. [DOI] [Google Scholar]
- 9.Santos MGS, Bezerra JDP, Svedese VM, Sousa MA, Silva DCV, Maciel MHC, Paiva LM, Porto ALF, Souza-Motta CM. Screening of endophytic fungi from cactus of the Brazilian tropical dry forest according to their L-asparaginase activity. Sydowia. 2015;67:147–156. doi: 10.12905/0380.sydowia67-2015-0147. [DOI] [Google Scholar]
- 10.Cruz R, Lima JS, Fonseca JC, Fernandes MJS, Lima DMM, Duda GP, Moreira KA, Souza-Motta CM. Diversity of filamentous fungi of area from Brazilian Caatinga and high-level tannase production using mango (Mangifera indica L.) and Surinam cherry (Eugenia uniflora L.) leaves under SSF. Adv in Microbiol. 2013;3:52–60. doi: 10.4236/aim.2013.38A009. [DOI] [Google Scholar]
- 11.Cruz R, Ramos SMS, Fonseca JC, Souza-Motta CM, Moreira KA. Anthropization effects on the filamentous fungal community of the Brazilian Catimbau National Park. Rev Bras Cienc Solo. 2017;41:1–13. doi: 10.1590/18069657rbcs20160373. [DOI] [Google Scholar]
- 12.Silva JMC, Leal IR, Tabarelli M. Caatinga: the largest tropical dry forest region in South America. Cham: Springer; 2018. [Google Scholar]
- 13.Barbosa RDN, Bezerra JDP, Santos ACS, Melo RFR, Houbraken J, Oliveira NT, Souza-Motta CM. Brazilian tropical dry forest (Caatinga) in the spotlight: an overview of species of Aspergillus, Penicillium and Talaromyces (Eurotiales) and the description of P. vascosobrinhous sp. nov. Acta Bot Bras. 2020;34:409–429. doi: 10.1590/0102-33062019abb0411. [DOI] [Google Scholar]
- 14.Rocha LGM, Drummond JA, Ganem RS. Parques nacionais brasileiros: problemas fundiários e alternativas para sua resolução. Rev Sociol Polít. 2010;18:205–226. doi: 10.1590/S0104-44782010000200013. [DOI] [Google Scholar]
- 15.Souza-Silva RF, Rapini A, Morales JF. Mandevilla catimbauensis (Apocynaceae), a new species from the semi-arid Region, Pernambuco, Brazil. Ed J of Botany. 2010;67:1–5. doi: 10.1017/S0960428609990230. [DOI] [Google Scholar]
- 16.Santos S, Delgado Jr G, Alves M (2012) Espécies raras e ameaçadas no PARNA Catimbau. https://23d61a9f-4a20-4a84-8b38-d9ded0cf6a76.filesusr.com/ugd/4c522e_206273cf8c7d42a99e2f23153f824126.pdf. Accessed 27 Aug 2020
- 17.Delgado-Junior GC, Alves M. Diversity of climbing plants in Parque Nacional do Catimbau, Pernambuco, Brazil. Rodriguésia. 2017;68:347–377. doi: 10.1590/2175-7860201768206. [DOI] [Google Scholar]
- 18.Ribeiro RV, Mariano DB, Arunachalam K, Soares IM, Aguiar RWS, Ascêncio SD, Ribeiro M, Colodel EM, Martins DTO. Chemical characterization and toxicological assessment of hydroethanolic extract of Mandevilla velame xylopodium. Rev Bras Farmacogn. 2019;29:605–612. doi: 10.1016/j.bjp.2019.05.002. [DOI] [Google Scholar]
- 19.Costa-Silva TA, Costa IM, Biasoto HP, Lima GM, Silva C, Pessoa A, Monteiro G. Critical overview of the main features and techniques used for the evaluation of the clinical applicability of L-asparaginase as a biopharmaceutical to treat blood cancer. Blood Rev. 2020;43:100651. doi: 10.1016/j.blre.2020.100651. [DOI] [PubMed] [Google Scholar]
- 20.Jiao L, Chi H, Lu Z, Zhang C, Chia SR, Show PL, Tao Y, Lu F. Characterization of a novel type I L-asparaginase from Acinetobacter soli and its ability to inhibit acrylamide formation in potato chips. J Biosci Bioeng. 2020;129:672–678. doi: 10.1016/j.jbiosc.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Krishnakumar T, Visvanathan R. Acrylamide in food products: a review. J Food Process Technol. 2014;5:7. doi: 10.4172/2157-7110.1000344. [DOI] [Google Scholar]
- 22.Dourado C, Pinto CA, Cunha S, Casal S, Saraiva JA. A novel strategy of acrylamide mitigation in fried potatoes using asparaginase and high pressure technology. Innov Food Sci Emerg Tecnol. 2020;60:102310. doi: 10.1016/j.ifset.2020.102310. [DOI] [Google Scholar]
- 23.Köppen W. Climatología: Con un estudio de los climas de la Tierra. Mexico: Fondo de Cultura Economica; 1948. [Google Scholar]
- 24.Bezerra JDP, Nascimento CCF, Barbosa RN, Silva DCV, Svedese VM, Silva-Nogueira EB, Gomes BS, Paiva LM, Souza-Motta CM. Endophytic fungi from medicinal plant Bauhinia forficata: diversity and biotechnological potential. Braz J Microbiol. 2015;46:49–57. doi: 10.1590/S1517-838246120130657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bezerra JDP, Oliveira RJV, Paiva LM, Silva GA, Groenewald JZ, Crous PW, Souza-Motta CM. Bezerromycetales and Wiesneriomycetales ord. nov. (class Dothideomycetes), with two novel genera to accommodate endophytic fungi from Brazilian cactus. Mycol Progress. 2017;16:297–309. doi: 10.1007/s11557-016-1254-0. [DOI] [Google Scholar]
- 26.Shannon C, Weaver W. The mathematical theory of communication. Urbana: University of Illinois Press; 1949. [Google Scholar]
- 27.Fisher RA, Corbet AS, Williams CB. The relation between the number of species and the number of individuals in a random sample of an animal population. J Anim Ecol. 1943;12:42–58. doi: 10.2307/1411. [DOI] [Google Scholar]
- 28.Mendiburu F (2017) Agricolae: statistical procedures for agricultural research. R package version 1.2-5. https://CRAN.R-project.org/package=agricolae. Accessed 25 Apr 2020
- 29.Oksanen J, Blanchet FG, Friendly M et al. (2017) Package ‘vegan’: community ecology package in R package, version 2.4-0. https://github.com/vegandevs/vegan. Accessed 25 Apr 2020
- 30.Hsieh TC, Ma KH, Chao A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers) Methods Ecol Evol. 2016;7:1451–1456. doi: 10.1111/2041-210X.12613. [DOI] [Google Scholar]
- 31.Loureiro CB, Borges KS, Andrade AF, Tone LG, Said S. Purification and biochemical characterization of native and pegylated form of L-asparaginase from Aspergillus terreus and evaluation of its antiproliferative activity. Adv Microbiol. 2012;2:138–145. doi: 10.4236/aim.2012.22019. [DOI] [Google Scholar]
- 32.Gulati R, Saxena RK, Gupta R. A rapid plate assay for screening L-asparaginase producing micro-organisms. Lett Appl Microbiol. 1997;24:23–26. doi: 10.1046/j.1472-765X.1997.00331.x. [DOI] [PubMed] [Google Scholar]
- 33.Drainas C, Kinghorn JR, Pateman JA. Aspartic hydroxamate resistance and asparaginase regulation in the fungus Aspergillus nidulans. J Gen Microbiol. 1977;98:493–501. doi: 10.1099/00221287-98-2-493. [DOI] [Google Scholar]
- 34.Myers RH, Montgomery DC. Response surface methodology: process and product optimization using designed experiments. Hoboken: Wiley; 1995. [Google Scholar]
- 35.Box GEP, Hunter JS, Hunter WG. Statistics for experimenters: design, innovation, and discovery. New Jersey: Wiley; 2005. [Google Scholar]
- 36.Venieraki A, Dimou M, Katinakis P. Endophytic fungi residing in medicinal plants have the ability to produce the same or similar pharmacologically active secondary metabolites as their hosts. Hell Plant Prot J. 2017;10:51–66. doi: 10.1515/hppj-2017-0006. [DOI] [Google Scholar]
- 37.Kareem SO, Akpan I, Ojo OP. Antimicrobial activities of Calotropis procera on selected pathogenic microorganisms. Afr J Biomed Res. 2008;11:105–110. [Google Scholar]
- 38.Na R, Jiajia L, Dongliang Y, Yingzi P, Juan H, Xiong L, Nana Z, Jing Z, Yitian L. Indentification of vincamine indole alkaloids producing endophytic fungi isolated from Nerium indicum, Apocynaceae. Microbiol Res. 2016;192:114–121. doi: 10.1016/j.micres.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Nascimento TL, Oki Y, Lima DMM, Almeida-Cortez JS, Fernandes GW, Souza-Motta CM. Biodiversity of endophytic fungi in different leaf ages of Calotropis procera and their antimicrobial activity. Fungal Ecol. 2015;14:79–86. doi: 10.1016/j.funeco.2014.10.004. [DOI] [Google Scholar]
- 40.Koide RT, Ricks KD, Davis ER. Climate and dispersal influence the structure of leaf fungal endophyte communities of Quercus gambelii in the eastern Great Basin, USA. Fungal Ecol. 2017;30:19–28. doi: 10.1016/J.FUNECO.2017.08.002. [DOI] [Google Scholar]
- 41.Harrison JG, Griffin EA. The diversity and distribution of endophytes across biomes, plant phylogeny, and host tissues—how far have we come and where do we go from here? Environ Microbiol. 2020;22:2107–2123. doi: 10.1111/1462-2920.14968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez RJ, White JF, Jr, Arnold AE, Redman RS. Fungal endophytes: diversity and functional roles. New Phytol. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 43.Suryanarayanan TS, Murali TS, Venkatesan G. Occurrence and distribution of fungal endophytes in tropical forests across a rainfall gradient. Can J Bot. 2002;80:818–826. doi: 10.1139/b02-069. [DOI] [Google Scholar]
- 44.Pandey AK, Reddy MS, Suryanarayanan TS. ITS-RFLP and ITS sequence analysis of a foliar endophytic Phyllosticta from different tropical trees. Mycol Res. 2003;107:439–444. doi: 10.1017/S0953756203007494. [DOI] [PubMed] [Google Scholar]
- 45.Hilarino MPA, Silveira FAO, Oki Y, Rodrigues L, Santos JC, Corrêa Junior A, Fernandes GW, Rosa CA. Distribution of the endophytic fungi community in leaves of Bauhinia brevipes (Fabaceae) Acta Bot Bras. 2011;25:815–821. doi: 10.1590/S0102-33062011000400008. [DOI] [Google Scholar]
- 46.Arnold AE, Lutzoni F. Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology. 2007;88:541–549. doi: 10.1890/05-1459. [DOI] [PubMed] [Google Scholar]
- 47.Bezerra JDP, Santos MGS, Barbosa RN, Svedese VM, Lima DMM, Fernandes MJS, Gomes BS, Paiva LM, Almeida-Cortez JS, Souza-Motta CM. Fungal endophytes from cactus Cereus jamacaru in Brazilian tropical dry forest: a first study. Symbiosis. 2013;60:53–63. doi: 10.1007/s13199-013-0243-1. [DOI] [Google Scholar]
- 48.Oliveira RJV, Sousa NMF, Pinto Neto WP, Bezerra JL, Silva GA, Cavalcanti MAQ. Seasonality affects the community of endophytic fungi in coconut (Cocos nucifera) crop leaves. Acta Bot Bras. 2020;34:704–711. doi: 10.1590/0102-33062020abb0106. [DOI] [Google Scholar]
- 49.Oliveira TGL, Bezerra JDP, Silva IR, Souza-Motta CM, Magalhães OMC. Diversity of endophytic fungi in the leaflets and branches of Poincianella pyramidalis, an endemic species of Brazilian tropical dry forest. Acta Bot Bras. 2020;34:755–764. doi: 10.1590/0102-33062020abb0253. [DOI] [Google Scholar]
- 50.Crous PW, Wingfield MJ, Burgess TI, et al. Fungal planet description sheets: 625–715. Persoonia. 2017;39:270–467. doi: 10.3767/persoonia.2017.39.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wikee S, Lombard L, Crous PW, Nakashima C, Motohashi K, Chukeatirote E, Alias SA, McKenzie EHC, Hyde KD. Phyllosticta capitalensis, a widespread endophyte of plants. Fungal Divers. 2013;60:91–105. doi: 10.1007/s13225-013-0235-8. [DOI] [Google Scholar]
- 52.Kumar NSM, Ramasamy R, Manonmani HK. Production and optimization of L-asparaginase from Cladosporium sp. using agricultural residues in solid state fermentation. Ind Crops Prod. 2013;43:150–158. doi: 10.1016/j.indcrop.2012.07.023. [DOI] [Google Scholar]
- 53.Krishnapura PR, Belur PD. Partial purification and characterization of L-asparaginase from an endophytic Talaromyces pinophilus isolated from the rhizomes of Curcuma amada. J Mol Catal B Enzym. 2016;124:83–91. doi: 10.1016/j.molcatb.2015.12.007. [DOI] [Google Scholar]
- 54.Da Rocha WRV, Costa-Silva TA, Montalvo GSA, Feitosa VA, Machado SEF, de Souza Lima GM, Pessoa-Jr A, Alves HS. Screening and optimizing fermentation production of L-asparaginase by Aspergillus terreus strain S-18 isolated from the Brazilian Caatinga Biome. J Appl Microb. 2019;126:1426–1437. doi: 10.1111/jam.14221. [DOI] [PubMed] [Google Scholar]
- 55.Costa-Silva TA, Camacho-Córdova DI, Agamez-Montalvo GS, Parizotto LA, Sánchez-Moguel I, Pessoa-Jr A. Optimization of culture conditions and bench-scale production of anticancer enzyme L-asparaginase by submerged fermentation from Aspergillus terreus CCT 7693. Prep Biochem Biotechnol. 2019;49:95–104. doi: 10.1080/10826068.2018.1536990. [DOI] [PubMed] [Google Scholar]
- 56.Thakur M, Lincoln L, Niyonzima FN, More SS. Isolation, purification and characterization of fungal extracellular L-asparaginase from Mucor hiemalis. J Biocatal Biotransformation. 2013;2:2. doi: 10.4172/2324-9099.1000108. [DOI] [Google Scholar]
- 57.Farag AM, Hassan SW, Beltagy EA, El-Shenawy MA. Optimization of production of anti-tumor L-asparaginase by free and immobilized marine Aspergillus terreus. Egypt J Aquat Res. 2015;41:295–302. doi: 10.1016/j.ejar.2015.10.002. [DOI] [Google Scholar]
- 58.Vieira WF, Correa HT, Campos ES, Sette LD, Pessoa A, Jr, Cardoso VL, Filho UC. A novel multiple reactor system for the long-term production of L-asparaginase by Penicillium sp. LAMAI 505. Process Biochem. 2020;90:23–31. doi: 10.1016/j.procbio.2019.11.012. [DOI] [Google Scholar]
- 59.Dias FFG, Sato HH. Sequential optimization strategy for maximum L-asparaginase production from Aspergillus oryzae CCT 3940. Biocatal Agric Biotechnol. 2016;6:33–39. doi: 10.1016/j.bcab.2016.02.006. [DOI] [Google Scholar]
- 60.Banyal A, Thakur V, Thakur R, Kumar P. Endophytic microbial diversity: a new hope for the production of novel anti-tumor and anti-HIV agents as future therapeutics. Curr Microbiol. 2021 doi: 10.1007/s00284-021-02359-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plant specimen is deposited in the Herbário UFP – Geraldo Mariz and endophytic fungi cultures in the URM culture collection, both collections at the Universidade Federal de Pernambuco (Recife, Brazil). DNA sequences are deposited in the GenBank database and sequence alignment in TreeBASE.
Not applicable.