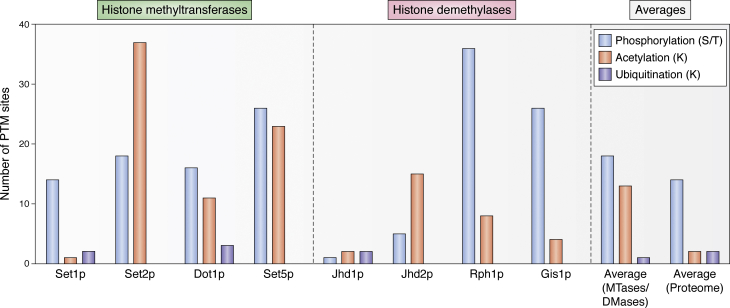
Abstract
Histone lysine methylation is a key epigenetic modification that regulates eukaryotic transcription. Here, we comprehensively review the function and regulation of the histone methylation network in the budding yeast and model eukaryote, Saccharomyces cerevisiae. First, we outline the lysine methylation sites that are found on histone proteins in yeast (H3K4me1/2/3, H3K36me1/2/3, H3K79me1/2/3, and H4K5/8/12me1) and discuss their biological and cellular roles. Next, we detail the reduced but evolutionarily conserved suite of methyltransferase (Set1p, Set2p, Dot1p, and Set5p) and demethylase (Jhd1p, Jhd2p, Rph1p, and Gis1p) enzymes that are known to control histone lysine methylation in budding yeast cells. Specifically, we illustrate the domain architecture of the methylation enzymes and highlight the structural features that are required for their respective functions and molecular interactions. Finally, we discuss the prevalence of post-translational modifications on yeast histone methylation enzymes and how phosphorylation, acetylation, and ubiquitination in particular are emerging as key regulators of enzyme function. We note that it will be possible to completely connect the histone methylation network to the cell’s signaling system, given that all methylation sites and cognate enzymes are known, most phosphosites on the enzymes are known, and the mapping of kinases to phosphosites is tractable owing to the modest set of protein kinases in yeast. Moving forward, we expect that the rich variety of post-translational modifications that decorates the histone methylation machinery will explain many of the unresolved questions surrounding the function and dynamics of this intricate epigenetic network.
Keywords: chromatin, histone methylation, Saccharomyces cerevisiae, epigenetics, kinase, phosphorylation, methyltransferase, demethylase, post-translational modification, post-translational regulation
The abbreviations used are: AdoMet, S-adenosyl-L-methionine; AID, autoinhibitory domain; AWS, associated with SET; BAH, bromo-adjacent homology; COMPASS, complex of proteins associated with Set1; C2H2, Cys2-His2; CTD, C-terminal domain; DSB, double strand break; Dot1p, disruptor of telomeric silencing 1; Gis1p, Gig1-2 suppressor 1; HAT, histone acetyltransferase; HDAC, histone deacetylase; Jhd1p, JmjC domain–containing histone demethylase 1; Jhd2p, JmjC domain–containing histone demethylase 2; JmjC, Jumonji C; JmjN, Jumonji N; LSD, lysine-specific demethylase; MYND, myeloid translocation protein, Nervy, Deaf; NHEJ, nonhomologous end joining; PHD, plant homeodomain; PRMT, protein arginine methyltransferase; PTM, post-translational modification; RNAPII, RNA polymerase II; Rph1, regulator of Phr1; RRM, RNA recognition motif; SET, Su(var)3-9, Enhancer of Zeste, Trithorax; Sir, silent information regulator; SRI, Set2 Rbp1 interacting; ZF, zinc finger; 7βS, seven-β-strand
Within the eukaryotic cell, genetic material is packaged into chromatin. The basic repeating unit of chromatin is the nucleosome, which comprises 146 base pairs of linear DNA wrapped approximately 1.6 times around an octamer of core histone proteins (two copies each of H2A, H2B, H3, and H4) (1). Each of these four histone families possesses a highly conserved and structured histone fold domain toward the center of the nucleosome, as well as a disordered N-terminal tail that protrudes from the nucleosomal core (2). The spatial accessibility of histone tails makes them available for post-translational modification (PTM), and indeed a number of different modification types have been identified in these unstructured regions, including methylation (3), acetylation (4), phosphorylation (5, 6), and ubiquitination (7). Such modifications are known to regulate gene expression by either affecting chromatin compaction or by serving as binding platforms for transcriptional coregulators that harbor domains to specifically recognize modified histone residues. Given their central role in transcription, it is unsurprising that aberrant modification of histones has been linked to the pathogenesis of human cancers (8, 9), neurodevelopmental defects (10), and autoimmune diseases (11). In the budding yeast Saccharomyces cerevisiae, dysregulated histone PTMs are associated with deleterious growth phenotypes (12, 13, 14) as well as altered apoptotic cell death and lifespan-resetting pathways (15).
Histone methylation is a key epigenetic modification that regulates many nuclear processes, including transcription (16), DNA replication (17), and DNA repair (18). It refers to the covalent attachment of methyl (CH3) group(s) to the amino acid side chains of lysine or arginine residues on histone proteins. Lysine residues can be mono-, di-, or trimethylated on their ε-amino group (3), whereas arginine residues can be mono-, asymmetrically di-, or symmetrically dimethylated on their terminal guanidinium group (19). Unlike other modifications that affect chromatin folding through an electrostatic mechanism (e.g., acetylation, phosphorylation), methylation does not alter the charge of lysine or arginine side chains. Instead, methylated residues constitute recognition sites for a range of transcription factors and associated regulatory proteins, which in turn elicit downstream changes in gene expression (20). These effector proteins carry “reader” interaction interfaces, such as chromodomains (21), PHD (22), and Tudor domains (23), which specifically bind methyl-lysine and methyl-arginine residues. Although some histone modifications simply denote either an open or closed chromatin conformation, methylation has a more nuanced role. Accordingly, specific methyl-lysine sites on histones can have either activating or repressive effects on transcription depending on their position and methylation state (16). In budding yeast, lysine methylation sites at H3K4, H3K36, and H3K79 are enriched within transcriptionally active euchromatin and are predominantly associated with gene expression. Strikingly, these methyl marks can also promote a repressed chromatin landscape depending on their location within transcriptional units, thus highlighting their functional diversity. All eukaryotic histone methylation sites display unique chromosomal signatures, both throughout gene bodies and within noncoding and regulatory elements (e.g., promoters, enhancers) (24, 25). Crucially, the abundance and distribution of specific methyl marks changes markedly during cellular growth (12), differentiation (26), and in response to exogenous perturbation (27), to bring about widespread transcriptional reprogramming.
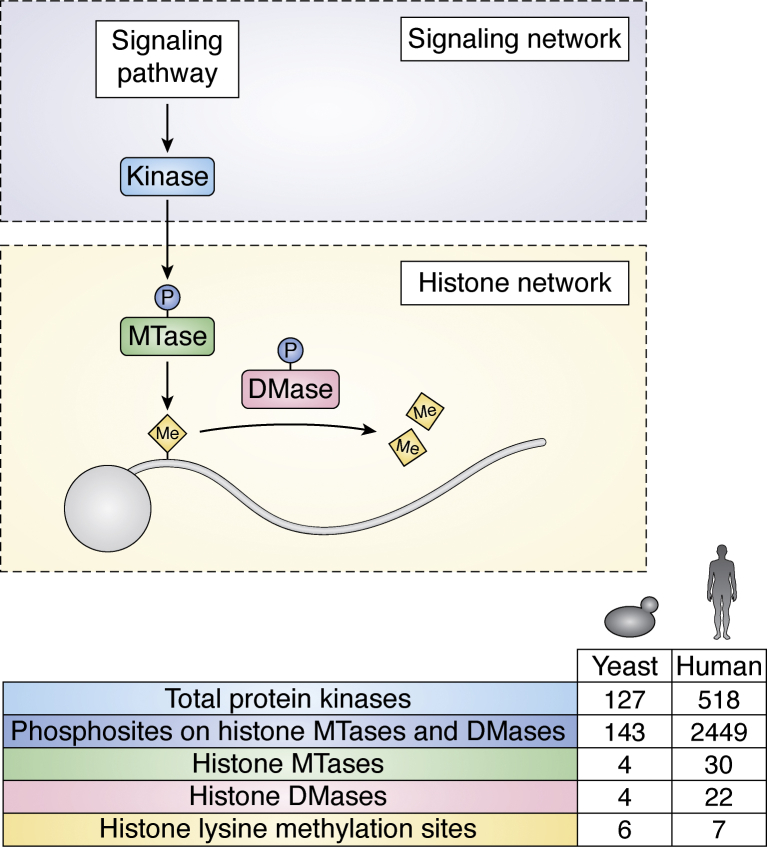
The histone methylation network is exquisitely conserved across eukaryotes, from the methylation sites themselves to the enzymatic machinery responsible for their regulation (28, 29, 30). In the human cell, there are seven histone lysine methylation sites that are controlled by the counteracting activities of 30 methyltransferases and 22 demethylases (31, 32, 33). This system is highly complex and challenging to interrogate experimentally given the number, redundancy, and overlapping site specificity of its constituent members. By contrast, S. cerevisiae is a eukaryotic model organism in which many of the foundational discoveries of histones and chromatin biology have been made (34). In yeast, the histone methylation system is substantially simplified, comprising six histone lysine methyl marks and only four methyltransferases and four demethylases (Fig. 1). Crucially, almost all yeast histone methyl marks and enzymes have a mammalian counterpart, thus underscoring the high degree of evolutionary conservation of this system (Table 1). Deciphering the function and regulation of the histone methylation network in yeast will therefore be of relevance to all eukaryotes and will provide new insights into epigenetic processes in higher organisms. This is of pharmacological importance given the association of human histone methylation enzymes with disease etiology (35, 36, 37) and their emergence as promising therapeutic targets for anticancer drug development (38, 39, 40, 41). Histone methylation sites and enzymes are also conserved, albeit to varying extents, in lower eukaryotes that impact on human health, such as protozoans (42, 43, 44). Budding yeast also serves as an excellent model for epigenetic gene regulation in these species and may therefore provide insights into the design of antiparasitic drugs.
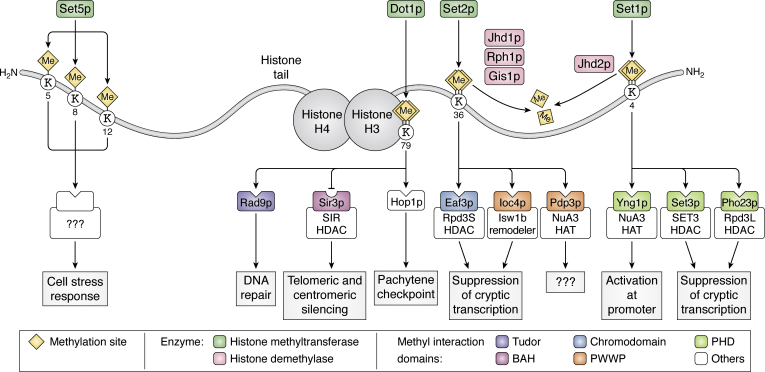
Figure 1.
Histone lysine methylation network in budding yeast. All histone lysine methylation sites in yeast are depicted along histone proteins as yellow diamonds. The upstream methyltransferase and demethylase enzymes that control these sites are shown in green and pink, respectively. Histone methyl marks are recognized by downstream effector proteins that harbor methyl-reader domains and are colored according to the key (bottom). Methylation can also inhibit binding of proteins to chromatin, notably the bromo-adjacent homology (BAH) domain of Sir3p (magenta), which is blocked by H3K79 methylation. The functional outcomes of histone methylation sites and the recruitment of specific effector proteins and complexes are shown in gray boxes. For ease of visualization, only a single copy of histones H3 and H4 has been illustrated, whereas both copies of histones H2A and H2B have been omitted.
Table 1.
Histone lysine methylation enzymes in budding yeast
| Typea | Enzyme | UniProt ID | SGD ID | Alias | Specificity | ECb | Chromosome | Coordinates | Copies/cellc | Human orthologd |
|---|---|---|---|---|---|---|---|---|---|---|
| MTase | Set1p | P38827 | YHR119W | KMT2 | H3K4me1/2/3 | 2.1.1.354 | VIII | 346043–349285 | 172 | SETD1A |
| Set2p | P46995 | YJL168C | KMT3 | H3K36me1/2/3 | 2.1.1.359 | X | 102227–104428 | 217 | SETD2 | |
| Dot1p | Q04089 | YDR440W | KMT4 | H3K79me1/2/3 | 2.1.1.360 | IV | 1342493–1344241 | 2160 | DOT1L | |
| Set5p | P38890 | YHR207C | - | H4K5/8/12me1 | - | VIII | 514905–516485 | 5000 | SMYD3 | |
| DMase | Jhd1p | P40034 | YER051W | KDM2 | H3K36me1/2 | 1.14.11.27 | V | 254656–256134 | 784 | FBXL11 |
| Jhd2p | P47156 | YJR119C | KDM5 | H3K4me1/2/3 | 1.14.11.67 | X | 644304–646490 | 290 | JARID1C | |
| Rph1p | P39956 | YER169W | KDM4 | H3K36me2/3 | 1.14.11.27 | V | 523369–525759 | 2229 | JMJD2A | |
| Gis1p | Q03833 | YDR096W | KDM4 | H3K36me1/2 | 1.14.11.27 | IV | 637139–639823 | 432 | JMJD2A |
Despite the importance that S. cerevisiae has played in our understanding of histone methylation, and the high conservation of these processes in higher eukaryotes, there has been no systematic analysis of the literature to date for this key epigenetic modification in yeast. To this end, here we present a comprehensive review of the histone lysine methylation system in yeast. We first outline the histone methylation sites and their contribution to both molecular and cellular processes. Next, we provide a detailed examination of the histone methyltransferases and demethylases that control these marks, with a particular focus on their structure, function, and regulation. Finally, we discuss the PTMs that are known to exist on histone methylation enzymes and how they are emerging as key regulators of enzyme function. We expect the latter will ultimately explain many of the intricacies of the histone methylation network in due course.
Histone methylation sites
Many of the histone methylation sites were first discovered through Edman sequencing of bulk histones after metabolic labeling (45), following which their roles in transcriptional regulation and other cellular processes have been established (46, 47). Strikingly, many of the major activating methylation sites are conserved among eukaryotes, whereas the repressive marks are more variable in their evolutionary conservation (30). This is exemplified in S. cerevisiae which carries the transcriptionally activating histone lysine methylations at H3K4, H3K36, and H3K79 but not the repressive H3K9, H3K27, and H4K20 methylation sites (Fig. 1) (48). More recently, monomethylation of H4K5, H4K8, and H4K12 has been identified in yeast (49). Here, we summarize the histone lysine methyl marks in S. cerevisiae, their genomic distribution, biological function, and how they cross talk with other histone PTMs.
H3K4me1/2/3
Histone H3 lysine 4 (H3K4) exhibits three methylation states, each of which have distinct functions and positional signatures across the yeast genome. Functionally, all three states are associated with transcriptional activation in a variety of eukaryotic species (50). Trimethylation of H3K4 (H3K4me3) is concentrated within promoter regions and toward the 5′-ends of actively transcribed genes (Fig. 2A) (51). H3K4me3 is highly correlated with transcription rates, active RNA polymerase II (RNAPII) occupancy, and histone acetylation (52), and its enrichment at transcription start sites is highly conserved across eukaryotes (53, 54). By contrast, the distribution and function of dimethylated H3K4 (H3K4me2) is variable between yeast and vertebrates (50). In S. cerevisiae, H3K4me2 is most abundant toward the middle of coding regions and is associated with both transcriptionally poised and active regions (Fig. 2A). The majority of H3K4me2 in vertebrates, however, colocalizes with H3K4me3 in discrete zones (~5–20 nucleosomes in length) proximal to highly transcribed genes (53). Monomethylated H3K4 (H3K4me1) peaks toward the 3′-ends of transcriptional units (Fig. 2A) and is considered a hallmark of active enhancers in metazoans (25, 55). Of importance, patterning of all H3K4 methylation states along active genes is not static or universal and changes dynamically depending on the frequency and rate of transcription elongation (56) and in response to transcriptional stress (57) in a gene-specific manner.
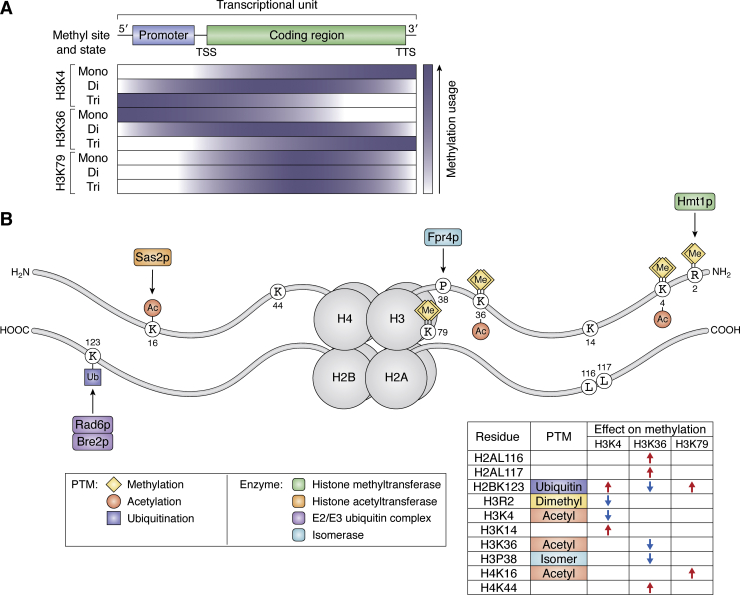
Figure 2.
Genomic distribution of histone lysine methylation sites in budding yeast and their regulation by other histone residues and modifications.A, the abundance of the mono-, di-, and trimethylated forms of H3K4 (top), H3K36 (middle), and H3K79 (bottom) along an active transcriptional unit is depicted by a color intensity gradient (indigo). B, regulation of budding yeast histone methylation sites by other histone residues and PTMs including acetylation (Ac; orange), ubiquitination (Ub; purple), and methylation (Me; yellow). The upstream modifying enzymes responsible for these PTMs are colored according to the key (bottom). The effects of these histone residues and PTMs are tabulated for each major lysine methylation site, where red arrows denote a stimulatory effect and blue arrows indicate an inhibitory effect. For both panels, histone H4 monomethylation sites at K5, K8, and K12 have been omitted given that little is known about the distribution of these modifications along genes and their cross talk with other features of the chromatin landscape. PTM, post-translational modification; TSS, transcription start site; TTS, transcription termination site.
H3K4 methylation is a nuanced epigenetic modification that can participate in both gene activation and repression (58). In its canonical role, methylation of H3K4 is most commonly associated with transcriptional activation and is found abundantly within genes being actively transcribed by RNAPII (59). It is required for the normal induction of transcription in yeast (60, 61, 62), which is achieved by the recruitment of specific chromatin modifiers (e.g., Chd1p, Isw1p, Yng1p, Pho23p, Set3p (63)) via their methyl “reader” domains (Fig. 1). These effectors have varying degrees of preference for distinct methylation states of H3K4 (64). For example, Yng1p binds H3K4 trimethylated chromatin using its plant homeodomain (PHD) finger (65) and subsequently associates with the yeast NuA3 histone acetyltransferase (HAT) complex to catalyze histone acetylation at the promoter and thus activate transcription. As an additional layer of regulation, H3K4me2 predominantly recruits histone deacetylases (HDACs, e.g., Set3p) throughout the body of a gene to prevent cryptic transcriptional initiation sites (66). Little is known about the molecular functions of H3K4me1 in transcriptional activation in yeast; however, it has been speculated that its presence is simply a transitional state between unmodified and dimethylated H3K4 (67). With respect to repression, H3K4 methylation has been found to be involved in silencing several heterochromatic genomic regions (e.g., telomeres (68), ribosomal DNA clusters (12), HML mating-type locus (69)). Accordingly, several ribosomal biosynthesis genes are downregulated by H3K4 methylation during multiple stresses (70). Simultaneous loss of H3K4me3 and H3K4me2 in yeast results in increased steady-state mRNA levels and delayed repression kinetics for certain gene groups in vivo (71), whereas H3K4me1 specifically inhibits gene expression induced by osmotic stress (72). These repressive effects are also mediated by chromatin modifiers and their cognate reader domains; Pho23p recognizes H3K4me3 and recruits the transcriptionally repressive Rpd3L HDAC complex (Fig. 1) (73).
At the cellular level, H3K4 methylation is involved in several key cellular processes and its dysregulation contributes to deleterious phenotypes. This modification has been shown to mediate yeast cell cycle progression and assembly of the mitotic spindle (74), DNA damage response and genomic stability (75), and mRNA splicing (76), and serves as an important molecular trigger for cell death (77). Of interest, increased levels of overall H3K4 methylation have been reported to act as a memory of recent transcriptional activity that allows genes to be rapidly switched on or off in response to stimuli (78). Yeast cells with defective H3K4 methylation have decreased viability owing to improper repair of double-strand DNA breaks (DSBs) by nonhomologous end-joining (NHEJ) (75). They also display increased cell death during chronological aging (77) and are sensitive to certain antifungal drugs (e.g., Brefeldin A) owing to abnormal expression of ergosterol biosynthesis enzymes (e.g., HMGCR) (79).
Methylation at H3K4 is known to cross talk with multiple other histone modifications. Perhaps the most well-studied example of eukaryotic PTM cross talk is the interplay between H3K4 methylation and H2B ubiquitination (Fig. 2B). Studies in S. cerevisiae first demonstrated that monoubiquitination at H2BK123 is required for subsequent H3K4 methylation (80, 81, 82), a trans-regulatory mechanism that has since been elucidated for several human H3K4 methyltransferase complexes (83, 84). Histone H2BK123 monoubiquitination, and thus H3K4 methylation, requires the yeast E2 ubiquitin-conjugating enzyme, Rad6p, and its cognate E3 ligase, Bre1p (85), and is involved in the transcriptional silencing of telomeric genes (81). The mechanism of cross talk will be discussed in greater detail later in the article (see “Histone methyltransferase” section). Curiously, despite conservation in mammalian cells, this histone methylation/ubiquitination interplay is absent in the fission yeast, Schizosaccharomyces pombe (86). Other examples of H3K4 methyl cross talk include its negative regulation by adjacent arginine asymmetric dimethylation at H3R2 (Fig. 2B), which abrogates H3K4me3 via spatial occlusion of the methylation machinery (87). A systematic histone mutagenesis screen revealed that H3K14, which is known to be acetylated by Gcn5p and Sas3p in vivo, is a critical residue for H3K4me3 levels, suggesting a potential cis-regulatory cross talk between histone methylation and acetylation through an unknown mechanism (88). Finally, antagonism between acetylation and methylation of H3K4 serves to fine-tune the deposition of these competing modifications, whereby H3K4 methylation limits H3K4 acetylation at promoters, and vice versa (89).
H3K36me1/2/3
Histone H3 lysine 36 (H3K36) can be co-transcriptionally modified by the addition of one (H3K36me1), two (H3K36me2), or three (H3K36me3) methyl groups in budding yeast and other eukaryotes (90, 91). All three methylation states of H3K36 accumulate in transcribed regions, making this modification a hallmark of active transcriptional elongation (13). Genome-wide localization studies in S. cerevisiae have shown that levels of H3K36 methylation increase in a 5′-to-3′ gradient along active transcriptional units, the direct converse of H3K4me patterning (Fig. 2A). Consequently, H3K36me1 is predominantly found at the 5′-end of gene bodies, and H3K36me2 and H3K36me3 are concentrated at their 3′-ends (52, 92, 93). Trimethylation at H3K36 in particular is highly correlated with active transcription and recruits distinct reader proteins to maintain a permissive transcriptional landscape (Fig. 1) (94). Indeed, H3K36 methylation does not spread to adjacent loci downstream of transcription termination sites and exhibits a relatively short epigenetic memory for recent transcriptional activity (95). Methylation at H3K36 is also conserved in lower eukaryotic microbes where it plays a critical role in the regulation of chromatin-templated processes. For example, in the pathogenic protozoan Plasmodium falciparum, H3K36 trimethylation by PfSETvs represses transcription of virtually all virulence genes in infected erythrocytes (44).
In budding yeast, H3K36 methylation regulates chromatin structure and transcriptional fidelity through the recruitment of specific chromatin modifiers involved in diverse cellular pathways (96). Within the context of transcription, distinct methylation states of H3K36 are known to differentially engage three major macromolecular complexes: (1) Rpd3S, (2) Isw1b, and (3) NuA3 (Fig. 1). With respect to the former, the Rpd3S HDAC complex specifically binds H3K36 methylated chromatin via the chromodomain of its constituent member, Eaf3p (97, 98, 99). This interaction is further stabilized by the PHD domain of another Rpd3S subunit, Rco1p (100). Rpd3S is thus preferentially targeted to H3K36me throughout coding regions where it promotes widespread histone deacetylation in the wake of transcription (101, 102, 103). This is required for the suppression of spurious transcriptional initiation from cryptic internal promoters (104). A similar regulatory mechanism is retained, although embellished, in human cells, where the mammalian ortholog of Eaf3p, MRG15, binds trimethylated H3K36 and can interact with a mammalian Rpd3S-like complex or the H3K4me2/3 demethylation machinery to coordinate transcription (105, 106). Second, the Isw1b chromatin remodeling complex is recruited to H3K36 methylation by the proline-tryptophan-tryptophan-proline (PWWP) domain–containing subunit, Ioc4p (107, 108), as evidenced by the co-localization of Isw1b with H3K36me at the mid- and 3′ regions of transcribed genes (Fig. 1). This complex works cooperatively with Rdp3S to re-establish a heterochromatic conformation following transcription in order to prevent production of intragenic transcripts. Third, in terms of the NuA3 HAT complex, the PWWP domain within its Pdp3p subunit selectively recognizes H3K36 trimethylation (Fig. 1) (109). In addition, the PHD finger in Nto1p of this complex has been shown to bind H3K36me3 in vitro (110), and H3K36 methylation is necessary for NuA3 chromatin binding (111). Taken together, these observations hint at a regulatory role for NuA3-mediated histone acetylation at actively transcribed gene bodies; however, this remains to be elucidated experimentally (94).
In normal yeast cells, H3K36 methylation is involved in the regulation of many genomic and transcriptomic processes, including DNA replication and repair (104, 112, 113), 5-methylcytosine deposition (114), and pre-mRNA splicing (76, 115, 116). Of interest, these distinct functions are linked to unique methylation states of H3K36. Appropriate temporal patterning of H3K36me3 and H3K36me2 around DSBs is required for maintenance and repair of chromatin structure at DNA damage sites (112), whereas H3K36me1 has been reported to regulate the formation of DNA replication origins (113) via an unknown reader protein. It has also been shown that H3K36 methylation is associated with yeast cellular aging (117), and deficits in sustaining this modification over time are related to increased cryptic transcription at certain loci in older cells (118). As with other histone methyl marks, disruption of H3K36me results in mild-to-severe growth and nutritional phenotypes in S. cerevisiae. H3K36me-null cells are sensitive to nutrient stress (14) and have a shortened life span due to aberrant initiation of cryptic transcription within gene bodies (13). Antisense transcripts produced from such spurious events impair gene expression from the sense strand (14, 119), and thus several yeast strains deficient in H3K36 methylation are synthetically sick or lethal with transcriptional elongation mutants (120).
Cross talk between H3K36 methylation and other epigenetic modifications occurs in budding yeast cells. Chromatin immunoprecipitation sequencing experiments have shown that genome-wide occupancy of H3K36ac and H3K36me are inversely related (121), raising the fascinating prospect that acetylation and methylation of a single histone lysine residue may display functional interplay to mediate chromatin-templated processes. Indeed, an acetyl/methyl switch at H3K36 has been found to control DSB repair pathway choice in fission yeast whereby trimethylation, in contrast to its role in transcriptional activation, compacts chromatin and promotes NHEJ, while counteracting Gcn5-dependent acetylation enhances chromatin accessibility and encourages repair by homologous recombination (122). Other examples of interplay include the trans-regulation of H3K36me deposition by H4K44 (123) and H2AL116/L117 (124) residues (Fig. 2B). In these cases, the unmodified H4 and H2A amino acids are required for the correct positioning of the H3K36 methylation machinery on nucleosomal substrates. Unlike methylation of H3K4 and H3K79, which require prior H2BK123ub, ubiquitination acts as a negative effector of H3K36 methylation (125). This occurs indirectly through the Ctk1p kinase, which needs H2BK123 to be deubiquitinated by SAGA-associated Ubp8p in order to phosphorylate the C-terminal domain (CTD) of RNAPII (126). Another negative regulator of H3K36 methylation is the histone proline isomerase, Fpr4p, which alters the adjacent H3P38 residue into a configuration that renders H3K36 unsuitable for trimethylation (Fig. 2B) (127).
H3K79me1/2/3
Histone H3 lysine 79 (H3K79) is a conserved eukaryotic methylation site that is uniquely located within the globular core of the nucleosomal architecture (128), as opposed to on a histone tail (Fig. 1). H3K79 exists in three methylation states; monomethyl (H3K79me1), dimethyl (H3K79me2), and trimethyl (H3K79me3), the latter of which is the most prevalent in vivo (~50%) (129). The distinct functions and genomic distributions of these methylation states, however, are poorly understood. Genome-wide maps have revealed that H3K79 methylation broadly occurs in a uniform fashion throughout the coding region of actively transcribed genes (Fig. 2A) (52, 130, 131) and is thus associated with transcriptional activation. Curiously, in S. cerevisiae, the vast majority (~90%) of H3K79 is methylated; however, in mammals, H3K79 is predominantly unmodified (129). Akin to other epigenetic marks, H3K79 methylation patterning across the yeast genome is dynamic. Levels of H3K79me3 remain unchanged throughout the cell cycle, whereas H3K79me2 levels increase gradually through the G1/S and G2/M phase transitions (132).
The precise mechanism by which H3K79 methylation regulates transcription and other biological processes is still an open question in the field of epigenetics (94). This is largely due to a paucity in understanding of reader domains and proteins specific to this methylated residue (23, 133). In budding yeast, H3K79 methylation is known to interact with two major effector proteins, which, in turn, coordinate two major cellular processes: (1) transcription and (2) DNA repair (Fig. 1). With respect to transcriptional regulation, H3K79 methylation is required for the proper formation of heterochromatin at telomeric and centromeric regions of the chromosome. This is mediated by the recruitment of Sir (silent information regulator) proteins and their cognate SIR HDAC complex (134). The presence of all three methylation states of H3K79 inhibits the binding of the bromo-adjacent homology (BAH) domain of Sir3p and prevents assembly of the repressive SIR complex and its resultant deacetylation within euchromatic regions (Fig. 1) (135, 136). Accordingly, defective H3K79 methylation causes aberrant redistribution of Sir proteins to actively transcribed genes and thereby impairs proper silencing of telomeres and cryptic mating-type (HML/HMR) loci (129, 137). The interplay between Sir proteins and histone methylation is bidirectional, as Sir3p competes with the H3K79 methylation machinery for binding to a basic patch of histone H4 (135, 138), as discussed in detail later. Of interest, the importance of fluctuations in nucleosomal occupancy of Sir proteins has been questioned (139), suggesting that there are additional layers of complexity to be uncovered.
In addition to its canonical role in transcription, H3K79 methylation has also been shown to mediate passage through several key checkpoints during yeast cellular growth and reproduction and in response to stress. First, this modification regulates the DNA damage checkpoints throughout the cell cycle by recruiting the checkpoint adaptor protein, Rad9p, via its methyl-binding Tudor domain (Fig. 1) (140). This interaction inhibits the production of single-stranded DNA at DSBs and at uncapped telomeres, suggesting that H3K79 plays a role in the resection of damaged DNA and its repair by homologous recombination (132, 141). Moreover, Rad9p is critical in the maintenance of single-stranded DNA during NHEJ in late G2 phase (141). The human ortholog of Rad9p, 53BP1, is similarly recruited to DSBs by H3K79 methylation (142). H3K79 methylation also plays a crucial role in progression through the pachytene checkpoint during meiosis (132). Although H3K79 methylation is virtually dispensable for unperturbed meiosis, it is essential in coordinating the checkpoint response to unrepaired DSBs and synapsis defects in certain yeast meiotic mutants (143, 144). This has been shown to occur through the chromosomal recruitment of Hop1p to H3K79me, which then activates Mek1p kinase in response to meiotic DNA damage (Fig. 1) (143). Unsurprisingly, yeast cells with deficient H3K79 methylation exhibit increased sensitivity to ionizing radiation (145) and are unable to initiate DNA damage repair (146) and meiotic recombination checkpoint functions (144).
Methylation at H3K79 can be positively and negatively regulated by several histone PTMs through cross talk. Analogous to H3K4me, monoubiquitination of H2BK123 by the E2-E3 complex Rad6p/Bre1p is a prerequisite for H3K79 trimethylation in S. cerevisiae (Fig. 2B) (131). These modifications lie in spatial proximity on the same exposed nucleosome surface, thus providing a structural basis for their interplay (147). Of importance, this cross talk is conserved in mammalian systems wherein dimethylation of H3K79 is stimulated by ubiquitination of H2BK120, which is equivalent to yeast H2BK123 (148). Although this was initially considered to be a unidirectional effect (82), it has since been shown that the H3K79 methyltransferase Dot1p promotes H2B ubiquitination via its N-terminal region, independent of its catalytic activity (149). Cross talk between ubiquitination and H3K79 methylation is further complicated by the input of the Rpd3L HDAC complex, which deacetylates its target genes in transcriptionally repressed regions devoid of H2BK123ub1 (150). As such, a subset of yeast genes have lower H3K79me3 and gene expression owing to antagonistic Rpd3L activity, a regulatory mechanism that is retained by human HDAC1 (150). Finally, acetylation at H4K16 in yeast by either Sas2p or Esa1p indirectly promotes H3K79 methylation through steric hindrance of Sir3p H4 binding (Fig. 2B) (135).
H4K5/8/12me1
It has recently been discovered that a cluster of lysine residues within the histone H4 N-terminal tail are subject to methylation in S. cerevisiae. In 2012, the laboratory of Or Gozani used systematic mutagenesis of the H4 tail, immunoblotting, and tandem mass spectrometry to identify monomethylation sites at H4K5, H4K8, and H4K12 in growing yeast cells (Fig. 1) (49). It is interesting that no evidence for di- or tri-methylation at these sites was found. H4K5 methylation is functionally conserved in mammalian cells where it is controlled by SMYD3 (151) and its dysregulation contributes to tumorigenesis (9, 151, 152, 153). Methylation sites at H4K8 and H4K12, however, are not retained in mammalian systems, suggesting they may serve specific functions in budding yeast. Although these methylation sites are newly discovered in comparison with other epigenetic modifications, their identification has added new layers of functionality to the H4 tail (154). Targeted studies have shown that, although loss of H4K5/8/12me1 resulted in only minor changes in global gene expression, these modifications play an important role in determining cellular fitness and responses to environmental stress (49). Unique from other histone methylation sites, these three lysine residues can functionally compensate for one another, indicating that they are unlikely to recruit different chromatin modifiers (Fig. 1) (155, 156, 157). With respect to PTM cross talk, it appears that H4 methylation sites cooperatively function with methylation at H3K4 as yeast strains deficient in H3K4 methylation and methylation at any of the H4 sites are sensitive to cellular stress. Moreover, the possibility of methylation/acetylation interplay has been suggested because H4K5, H4K8, and H4K12 are also known to be acetylated by Esa1p, a constituent of the NuA4 HAT complex in yeast (158, 159). Despite much progress, the exact molecular mechanisms underpinning H4K5/8/12me1 function and cross talk with H3K4 methylation and H4K5/8/12 acetylation are not well understood, and no reader proteins specific for this mark have been confirmed (Fig. 1). Considerable work is required to clarify the role of monomethylation sites at H4K5/8/12 in chromatin structure and function in yeast.
Histone methyltransferases
Despite the landmark discovery of histone methylation over 50 years ago (160), it was not until 2000 that the first histone methyltransferase was identified (161). This methyltransferase, SUV39H1, was shown to specifically trimethylate H3K9 in human cells where it controls the formation of repressive heterochromatin at pericentric and telomeric regions (162). Since then, numerous methyltransferases that target basic residues on histone proteins, particularly within their disordered N-terminal tails, have been identified in yeast and in other eukaryotes. Broadly, there are two evolutionarily conserved enzymatic families that catalyze the transfer of methyl group(s) from the metabolic donor S-adenosyl-L-methionine (AdoMet, also known as SAM) to the ε-amino group of lysine side chains on histone proteins (163). SET (Su(var)3-9, Enhancer of Zeste, Trithorax) domain methyltransferases harbor a SET catalytic domain, which forms a knot-like β-sheet structure that facilitates methyl transfer (164, 165). In S. cerevisiae, there are three SET domain–containing proteins that have bona fide histone methyltransferase activity: Set1p, Set2p, and Set5p, all of which methylate histones on their N-terminal tails (Fig. 1, Table 1) (164, 165). The seven-β-strand (7βS) family of methyltransferases is more diverse than the SET family, comprising both lysine and arginine protein methyltransferases, as well as DNA methyltransferases (32). Dot1p is the sole 7βS histone lysine methyltransferase in budding yeast and uniquely methylates histone H3 within its globular core (Fig. 1, Table 1) (166, 167). In this section, we discuss the structure, function, and conservation of the four histone lysine methyltransferases in S. cerevisiae and highlight recent efforts to understand their regulation.
Set1p (COMPASS)
SET domain-containing 1 (Set1p), also known as KMT2 or YTX1, was the first histone lysine methyltransferase to be discovered in S. cerevisiae. Early studies used sequence homology approaches to identify Set1p as a yeast member of the Trithorax gene family and revealed that, although not essential for viability, it plays key roles in the regulation of transcriptional silencing at mating-type loci and telomeres, in the maintenance of telomere length (69) and in DNA repair (168, 169). It was not until 2002 that the methyltransferase function of Set1p was investigated; Briggs et al. demonstrated that deletion of SET1 completely abolishes H3K4 methylation in vivo, manifesting in aberrant transcription at rDNA loci and a slow-growth phenotype (12). A number of groups have since confirmed, both in vivo and in vitro, that Set1p is the sole enzyme responsible for all three states of H3K4 methylation (Table 1) (68, 170, 171). Set1p is the largest histone methyltransferase in the yeast proteome, at 1080 amino acids in length, and comprises a SET catalytic domain (residues 938–1055) and a post-SET domain (residues 1064–1080) (Fig. 3). Set1p also harbors, in addition to its catalytic regions, several regulatory domains that control its enzymatic activity and interactions. An N-SET domain (residues 752–928) has been reported to enable the cross talk between Set1p-mediated H3K4 methylation and H2B ubiquitination (84), although these findings have been challenged (Fig. 3) (172). A highly conserved tandem RNA recognition motif, comprising RRM1 (residues 274–375, Protein Data Bank [PDB] ID: 2J8A (173)) and RRM2 (residues 376–579) toward the N-terminus of Set1p, is required for its capacity to trimethylate H3K4 (174) but is dispensable for dimethylation (Fig. 3) (175). This domain, as well as N-SET, allows Set1p to bind RNA in vitro and to interact with nascent transcripts in vivo (176). The positive regulatory effects of RRM are counterbalanced by a semiconserved and centrally located autoinhibitory domain (AID), which attenuates Set1p trimethyltransferase function (175). The precise residues that comprise Set1p AID are unknown and have thus been omitted from Figure 3; however, arginine 483 within this central region has been shown to be essential for autoinhibition.
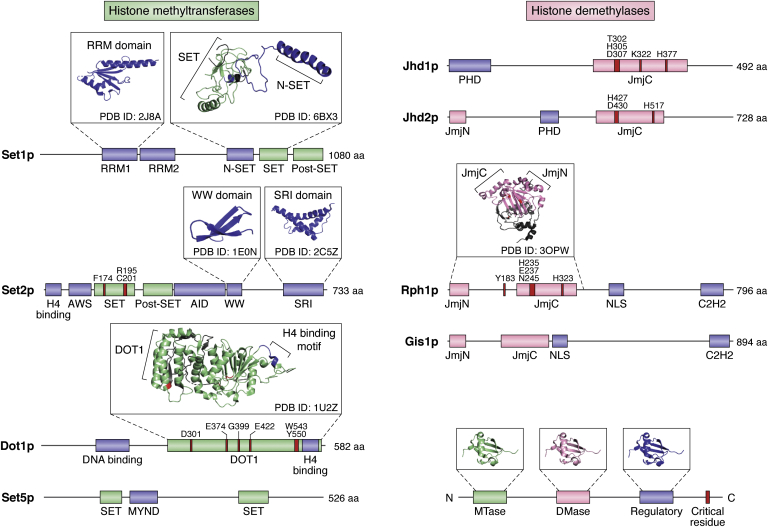
Figure 3.
Domain architecture and structural features of yeast histone methyltransferase and demethylase enzymes. Linear sequence maps of yeast histone methyltransferase (left panel) and demethylase (right panel) enzymes. Protein domains are displayed, to scale, for each enzyme. Methyltransferase and demethylase domains are shown in green and pink, respectively, whereas other regulatory and interaction domains are colored in blue. Amino acid (aa) residues that are critical for enzymatic activity are shown in crimson. To date, partial crystal structures have been resolved for the RNA recognition motif (RRM; Protein Data Bank [PDB] ID: 2J8A) and the SET methyltransferase domain (PDB ID: 6BX3) of Set1p, the tryptophan–tryptophan (WW; PDB ID: 1E0N) and Set2 Rbp1 interacting (SRI; PDB ID: 2C5Z) domains of Set2p, the DOT1 methyltransferase domain (PDB ID: 1U2Z) of Dot1p, and the JmjN and JmjC demethylase domains (PDB ID: 3OPW) of Rph1p. Structures are depicted as ribbon diagrams in inset boxes and colored according to the region of the linear sequence map to which they correspond. AID, autoinhibitory domain; AWS, associated with SET; C2H2, Cys2-His2; MYND, myeloid translocation protein, Nervy, Deaf; NLS, nuclear localization signal; PHD, plant homeodomain.
Set1p is the only yeast histone methyltransferase that forms a catalytically active multimeric complex in vivo. It associates with seven other protein subunits, Bre2p (Cps60), Sdc1p (Cps25), Shg1p (Cps15), Spp1p (Cps40), Swd1p (Cps50), Swd2p (Cps35), and Swd3p (Cps30), to form a H3K4 methyltransferase complex known as COMPASS (complex of proteins associated with Set1) (177). Strikingly, of these subunits, only Swd2p is essential for yeast cell viability; however, this is likely due to its additional function within the RNA 3′-end processing and termination complex, APT (178). Although Set1p is the catalytic constituent of COMPASS, the other subunits, with the exception of Shg1p (179), each influence the stability and activity of the methyltransferase complex in distinct ways (180, 181, 182). For instance, the WD40 domain–containing subunits Swd1p and Swd3p are both required for COMPASS to catalyze all three states of H3K4 methylation, whereas Spp1p and Sdc1p are only needed for trimethylation (182, 183). Recently, structural studies have provided insights into the molecular stoichiometry and topology of COMPASS (184, 185). The complex is scaffolded by a core subcomplex involving two heteromeric interactions between Swd1p/Swd3p and Bre2p/Sdc1p (186). High-resolution cryogenic electron microscopy defined the three-dimensional structure of the COMPASS core, revealing a Y-shaped configuration wherein Swd1p/Swd3p localize at the top of adjacent lobes, whereas Bre2p/Sdc1p reside at the base (187). The SET domain of Set1p is located at the juncture of these subunits, thus creating a central canal that may regulate catalysis and product specificity of COMPASS. This macromolecular structure has been shown to be stabilized by an electrostatic interaction between a small basic patch within the N-SET domain of Set1p and an acidic patch toward the C-terminus of Swd1p (179). Crucially, the COMPASS complex forms a dimeric macromolecule in vivo, via the Sdc1p dimer interface, allowing COMPASS to efficiently deposit methylation at both copies of histone H3 within a single nucleosome (188). This symmetric H3K4 methylation by Set1p is the only known example of such a phenomenon in budding yeast.
Through its methylation of H3K4, Set1p is involved in the regulation of transcriptional initiation and early elongation. Set1p plays a key role in transcriptional activation; approximately 80% of S. cerevisiae genes are downregulated upon Set1p deletion (189). In certain contexts, Set1p can also function as a transcriptional repressor through its recruitment of HDAC complexes to chromatin where they antagonize nucleosome acetylation and remodeling of downstream promoters (Fig. 1) (66, 190). Strikingly, recent studies have proposed a model whereby the combined activities of Set1p and its cognate H3K4 demethylase, Jhd2p, cooperatively regulate genome-wide chromatin structure and thus gene expression, rather than opposing one another as logic may suggest (67, 188). The mechanistic details of this coregulation are discussed later (see Jhd2p section). Although the different degrees of H3K4 methylation are known to play distinct roles in transcriptional regulation (Fig. 2A) (66, 174), the precise molecular cues controlling the differential production of H3K4me1, me2, and me3 by Set1p are still largely unknown. Set1p contains two conserved RRMs that have been shown to bind RNA in vitro in a methyltransferase-independent manner and may affect COMPASS distribution along nascent mRNA transcripts and subsequent H3K4 methylation (Fig. 3) (176, 178). Set1p has also been shown to methylate the kinetochore component, Dam1p, making it the only histone methyltransferase in budding yeast to modify a nonhistone substrate (191). Here, Dam1p methylation negatively regulates its subsequent phosphorylation by the Aurora kinase, Ipl1p, in order to control chromosome segregation and cell viability. It remains unclear how Set1p selects its desired substrate (histone H3 or Dam1p) for methylation, and it is not known whether the activities of Set1p in transcriptional regulation and mitosis functionally interact.
There are two primary means by which the function of Set1p is regulated in the context of transcription. First, H3K4 methylation is controlled by H2B ubiquitination. Although H2BK123 ubiquitination is dispensable for H3K4 monomethylation, it is a requirement for COMPASS to catalyze both dimethylation and trimethylation (Fig. 2B) (131), suggesting that it may modulate Set1p processivity. The mechanisms underpinning this PTM cross talk have been intensely studied and rigorously debated in the literature. Initial studies proposed that Swd2p, the only essential COMPASS subunit, recognizes H2BK123-ubiquitinated chromatin and is thus required for the assembly of trimethylation-competent COMPASS (85, 192). It was later clarified that the N-SET domain of Set1p serves as a novel sensor of H2BK123ub and that this cross talk conditionally involves Spp1p but not Swd2p (84). Indeed, in vitro analysis of reconstituted COMPASS and H2Bub chromatin showed that the Spp1p PHDL domain, in conjunction with N-SET, interacts with Swd1p/Swd3p to facilitate H2Bub-dependent H3K4 methylation (193). However, in light of the lack of evidence showing a direct physical interaction between Spp1p or N-SET and H2BK123-ubiquitinated chromatin, the precise mechanisms that govern this interplay remain elusive. Second, Set1p is recruited to sites of transcriptional elongation through its phosphorylation-dependent interaction with the CTD of Rbp1p, a component of the RNAPII holoenzyme. In S. cerevisiae, the CTD of Rbp1p consists of 26 repeats of an evolutionarily conserved heptapeptide, of consensus sequence YSPTSPS (194). Both serine 2 and serine 5 within this repeat can be phosphorylated, and these modification isoforms show unique spatiotemporal profiles; serine 5–phosphorylated RNAPII is localized to promoter regions at initiation/early elongation stages, whereas its serine 2–phosphorylated counterpart is found throughout coding regions during elongation (195, 196). Set1p co-transcriptionally associates with serine 5–phosphorylated RNAPII (78) to establish a gradient of H3K4 methylation that peaks near the promoter and decreases throughout a gene’s body (Fig. 2A) (61). This 5′ concentration of H3K4 methylation is mediated by Kin28p, a TFIIH-associated kinase, which phosphorylates serine 5 of Rbp1p and thus triggers the transition between transcriptional initiation and elongation in response to cellular cues via COMPASS (78).
In addition to regulation by H2B ubiquitination and RNAPII phosphorylation, Set1p function is fine-tuned through a number of mechanisms. In one of the few examples of transcriptional control of genes encoding histone methylation proteins in S. cerevisiae, Gcn5p promotes the expression of SET1 and thus indirectly increases H3K4 trimethylation levels but not H3K4me2 and H3K4me1 (197). At the protein level, Hmt1p-mediated asymmetric dimethylation at H3R2 negatively regulates adjacent trimethylation of H3K4 by Set1p, thus highlighting functional cross talk between arginine and lysine methylation on histone proteins (Fig. 2B). H3R2me2a spatially occludes the COMPASS subunit, Spp1p, which is essential for H3K4 trimethylation (87). Other histone residues are known to regulate Set1p activity, namely, H3K14, which may positively cross talk with H2B ubiquitination and/or directly interact with COMPASS through electrostatic attraction to promote H3K4 methylation (88).
Set2p
SET domain-containing 2 (Set2p), also known as KMT3 or EZL1, is the sole H3K36-specific methyltransferase in the budding yeast proteome and is central to the regulation of transcriptional initiation and elongation. In 2002, Strahl et al. first purified and biochemically characterized Set2p from S. cerevisiae and demonstrated that it catalyzes the processive mono-, di-, and trimethylation of H3K36 through its catalytic SET domain in vivo (Table 1) (91, 198). This domain is comprised of AWS (associated with SET; residues 63–118), SET (residues 120–237), and post-SET (residues 244–260) motifs (Fig. 3) (199). Curiously, the isolated SET domain is capable of methylating free histones, whereas full-length Set2p preferentially acts upon nucleosomal substrates (200), suggesting that sequences distal to the methyltransferase domain regulate substrate specificity. Indeed, an N-terminal acidic patch (residues 31–39) and a C-terminal Set2-Rbp1 interacting (SRI; residues 619–718, PDB ID: 2C5Z (201)) domain control the association of Set2p with histone H4 (123, 124) and RNAPII (202), respectively. The tryptophan-tryptophan (WW) domain (residues 475–507, PDB ID: 1E0N (203)) is currently of no known function, although its deletion does not modulate H3K36 methylation levels or Set2p RNAPII binding (204). It has been speculated that it may control nonhistone methylation given that the WW domain of SETD2, the mammalian ortholog of Set2p, mediates its interaction with the Huntingtin protein (199, 205, 206). Yeast Set2p, however, has no known nonhistone substrates identified to date. Finally, an AID (residues 262–476) attenuates Set2p-mediated H3K36 trimethylation by antagonizing its catalytic activity and fine-tuning several functions of SRI (Fig. 3) (200).
In the context of transcription, Set2p recruits several key complexes that cooperate to re-establish a compact chromatin landscape in the wake of elongating RNAPII (207). Set2p co-transcriptionally modifies H3K36, which is in turn recognized by the chromodomain of Eaf3p, a subunit of the Rpd3S HDAC complex (Fig. 1) (120). Rpd3S functions to keep gene bodies deacetylated and thus restores chromatin structure between multiple rounds of transcription (120, 208, 209). This epigenetic resetting protects genes from inappropriate and bidirectional transcription, from cryptic initiation sites within open reading frames (97, 209, 210). Prevention of intragenic transcription by H3K36 methylation is conserved in human as deletion of mammalian SETD2 causes upregulation of spurious mRNA transcripts (211, 212), a phenotype shared by set2Δ yeast cells (208, 209). This repressive transcriptional environment is reinforced by the recruitment of Isw1b, a chromatin remodeling complex that binds H3K36me and reorganizes nucleosomes to allow Rpd3S-mediated deacetylation of neighboring nucleosomes (Fig. 1) (119, 213). More recently, it has been shown that Set2p suppresses the interaction of histone H3 with chaperones and thus impairs the incorporation of new histones, a phenomenon that typically occurs co-transcriptionally by histone exchange over open reading frames (91, 119). This incorporation of new histones dilutes existing histone PTMs in chromatin and increases levels of acetylation, a mark that is associated with soluble histone proteins (91). Although Set2p null yeast cells do not exhibit discernible growth phenotypes (99), many cellular pathways can be controlled by H3K36 methylation and Set2p, as discussed in the previous section. Set2p is involved in DNA damage response and repair (112, 122) and polyadenylation site selection (214) and has been shown to promote cellular aging through its regulation of telomeric silencing (117).
In terms of regulation, Set2p is directed to transcriptionally active regions of the genome through its association with phosphorylated RNAPII (99). This interaction was initially identified through chromatin immunoprecipitation assays and affinity purification–based approaches (215, 216, 217), and since then the precise mechanisms underlying such regulation have been illuminated. Although Set1p binds to the CTD heptapeptide repeat of the RNAPII subunit, Rbp1p, that is phosphorylated at serine 5 (discussed above), Set2p recognizes Rbp1p CTD heptapeptides that are phosphorylated at serine 2 (204, 215, 217). This recruitment of Set2p to elongating RNAPII throughout gene bodies occurs via its SRI domain (Fig. 3) (202). Serine 2 phosphorylation of Rbp1p CTD repeats is catalyzed predominantly by the CTD kinase I complex, of which Ctk1p is the catalytic subunit (120, 215), and to a lesser extent, the Bur1/Bur2 complex (218, 219). Perhaps unexpectedly, in vitro docking analyses clarified that the Set2p SRI domain preferentially binds serine 2/serine 5–diphosphorylated CTD repeats (201, 202), thus raising the possibility that its activity may be coordinately regulated by multiple upstream kinases (Ctk1p and Kin28p). Strikingly, H3K36me2 does not require Ctk1p-mediated RNAPII phosphorylation or the SRI domain of Set2p (212). By contrast, the deposition of H3K36me3 necessitates a range of regulatory factors (e.g., Spt6p, H3P38, CTD, Ctk1p, SRI) indicating that H3K36 trimethylation serves a specific function in transcriptional elongation that is distinct from H3K36me2. These observations provide fascinating insights into the methylation specificity of Set2p and may give us clues into the cellular cues that control the switch between its dimethyltransferase and trimethyltransferase functions.
With respect to substrate recognition, Set2p is controlled by several sequence and structural features. Recently, Liu et al. (220) solved a cryogenic electron microscopy structure of yeast Set2p complexed with a nucleosome substrate carrying an oncogenic histone H3 variant (H3K36M). This revealed a specific interaction between the α-N helix of histone H3 and the AWS domain of Set2p, an evolutionarily conserved binding mechanism that also mediates the nucleosomal association of human SETD2 (220). Substrate recognition is further controlled by the SRI domain as deletion of this region severely impairs Set2p chromatin binding. These observations are consistent with previous reports that the SRI domain can recognize nucleosomal linker DNA and thus fine-tune Set2p substrate specificity (200). Here, Set2p was shown to preferentially methylate nucleosomal substrates with longer linker DNA, and in vitro binding assays revealed that this preference is likely due to its SRI domain. It is interesting that SRI is not resolved in the electron density map of the yeast Set2p/nucleosome complex, suggesting that it may contact the nucleosome via nonspecific interactions (220). Taken together, these findings highlight a more diverse role for the SRI domain in the regulation of Set2p function than was previously appreciated. In addition to its canonical role in the initial recruitment of Set2p to RNAPII within transcribed regions, the SRI domain also (1) recognizes the linker DNA of nucleosomes and thus allows Set2p to travel alongside elongating RNAPII, and (2) suppresses the capacity of Set2p to methylate free histone proteins (200).
Several additional layers of regulation exist to intricately control Set2p activity in yeast cells (reviewed in (14)). First, Fpr4p-mediated isomerization of H3P38 negatively regulates Set2p through occluding its adjacent target lysine (Fig. 2B) (127). There is also emerging evidence that Set2p may be controlled by H2B ubiquitination (221). Although early reports showed that H3K36 methylation is unaffected by H2BK123 monoubiquitination, a recent cryogenic electron microscopy structure of Set2 from Chaetomium thermophilum revealed that its AWS domain interacts with ubiquitin on H2B, suggesting that ubiquitin assists in positioning Set2 on the nucleosome and thus stimulates activity (221). It will be necessary to confirm whether such regulation is conserved in Set2 homologs in yeast and higher eukaryotes.
Dot1p
Disruptor of telomeric silencing 1 (Dot1p), also known as KMT4 or PCH1, is an exquisitely conserved histone methyltransferase and is the sole enzyme that catalyzes H3K79 methylation in S. cerevisiae. In 1998, yeast Dot1p was originally identified as a high-copy disruptor of transcriptional silencing at telomeres, mating-type loci, and rDNA (144, 222). Using a range of in vitro and in vivo approaches, several groups independently confirmed that Dot1p, and its human ortholog DOT1L, are responsible for catalyzing mono-, di-, and trimethylation of lysine 79 within the globular core of histone H3 (Table 1, Fig. 1) (128, 129, 166, 167). Indeed, knockout of Dot1p leads to a complete loss of H3K79 methylation in yeast, and the same occurs on knockout of relevant homologs in other eukaryotes (129, 223, 224). There are two enzymatic properties of Dot1p that distinguish its activity from other histone methyltransferases. First, Dot1p can methylate chromatin but not free histones, indicating that its catalysis requires certain structural and/or PTM features of native chromatin (135). Second, H3K79 methylation occurs via a distributive kinetic mechanism whereby Dot1p establishes H3K79me1, H3K79me2, and H3K79me3 through repetitive rounds of binding and dissociation from its substrate (225, 226). This mode of action contrasts with the SET domain–containing methyltransferases that add multiple methyl groups in a processive manner (227, 228, 229). These observations are striking, especially considering the paucity of understanding regarding the distinct functions of the mono-, di-, and trimethylated forms of H3K79.
Crystallographic studies have resolved the elongated structure of the conserved core of Dot1p (PDB ID: 1M0R, 1U2Z (230)) and provided structural insights into its unique biology (Fig. 3). Unlike the three SET domain–containing enzymes, Dot1p is the only histone lysine methyltransferase in budding yeast that contains a 7βS catalytic domain (residues 254–568), a feature that is typically associated with class I arginine methyltransferases (PRMTs) (147). Within its 7βS domain, Dot1p harbors an active site that is surrounded by conserved hydrophobic residues and an AdoMet-binding motif that shows a high degree of sequence conservation with PRMTs (Fig. 3) (231). These structural differences may explain the distinct enzymatic mechanisms underpinning histone lysine methylation mediated by Dot1p in comparison with its SET domain–containing counterparts.
In S. cerevisiae, Dot1p regulates gene expression profiles involved in several key cellular processes. As is the case with much of the yeast histone methylation machinery, Dot1p can function as a transcriptional activator or repressor depending on cellular conditions. In concordance with its initial characterization, Dot1p is indispensable for transcriptional silencing at telomeres, and this suppression is dependent on its catalytic activity (225). Dot1p-mediated H3K79 methylation within euchromatin inhibits binding of the repressive Sir proteins, which specifically recognize unmethylated H3K79 via a BAH domain (Fig. 1) (232). Sir complexes are thus directed elsewhere in the genome to exert their silencing function, particularly within hypomethylated H3K79 chromatin, where they reciprocally block Dot1p methylation (129). This positive feedback loop between histone methylation and deacetylation enzymes provides an explanation for the gross silencing defects associated with aberrant localization of Sir proteins from heterochromatin caused by Dot1p overexpression or deletion (128, 129, 144). As discussed above, Dot1p and H3K79 methylation are crucial players in the pachytene meiotic checkpoint (143, 233), DNA damage response (140, 141, 234), and cell cycle progression (235, 236) through both direct and indirect mechanisms (reviewed in (147, 236)). More recently, Dot1p has been shown to have inherent histone chaperone and chromatin-remodeling activity through its nucleosome-binding domain and is involved in the regulation of nucleosomal dynamics and histone exchange in a methyltransferase-independent manner (237).
Dot1p activity is intricately regulated by various aspects of the epigenetic landscape, predominantly through short sequence features that direct its association with chromatin. Given its proximity to H3K79 within the three-dimensional nucleosomal architecture, monoubiquitination of H2BK123 by the Rad6p/Bre1p E2–E3 complex promotes Dot1p-mediated trimethylation (Fig. 2B) (62, 137, 238). Pulldown-based approaches have demonstrated that Dot1p directly binds ubiquitin through a lysine-rich region (residues 101–140) in the first half of its DNA/nucleosome binding domain (residues 105–172) and that deletion of this motif causes defects in H3K79me3 accumulation and subtelomeric gene silencing in vivo (239). This trans-regulatory mechanism is conserved in human, where DOT1L requires prior ubiquitination of H2BK120 by RNF20/40 for its methyltransferase activity (240). More recently, it has become apparent that Dot1p activity is also affected by histone acetylation (Fig. 2B). Mutational studies have shown that an acidic patch toward the C-terminus of yeast Dot1p (residues 557–561) contacts a short, basic sequence (residues 17–19) on histone H4 and that this charge-based interaction is essential for H3K79 di- and tri-methylation and proper telomeric silencing (138). The histone H4 N-terminal tail is also bound by Sir3p, a constituent of the SIR HDAC complex, where it negatively regulates Dot1p chromatin association through steric hindrance (135). Of importance, Sas2p-mediated acetylation at H4K16, a residue immediately adjacent to this shared H4 interaction interface, displaces Sir3p, but not Dot1p, and thus allosterically stimulates Dot1p H3K79 methyltransferase activity (241, 242). Taken together, the elaborate cross talk that exists between H3K79me, H4K16ac, and H2BK123ub1 highlights the convergence of distinct PTM signaling cascades that cooperate to demarcate telomeric boundaries and ensure optimal propagation of an epigenetic state throughout generations (243).
Set5p
SET domain–containing 5 (Set5p) is a histone H4 monomethyltransferase that has recently emerged as an important regulator of chromatin in budding yeast. In 2012, Green et al. used a biochemical approach to identify novel methylation sites at H4K5, H4K8, and H4K12 in S. cerevisiae (Fig. 1) and demonstrated that these marks are catalyzed by a previously uncharacterized histone methyltransferase, Set5p, both in vitro and in vivo (49). Set5p is a SMYD subfamily methyltransferase that characteristically harbors a split SET catalytic domain (residues 106–140 and 364–409) as well as an intervening zinc finger (ZF) domain known as MYND (myeloid translocation protein, Nervy, Deaf (244, 245)) that mediates chromatin association (Fig. 3) (246). The MYND, post-SET, and C-terminal regions of Set5p are required for its repression of subtelomeric genes (246). With respect to conservation, the functional ortholog of Set5p in metazoans is SMYD3, a fellow MYND-containing methyltransferase that methylates H4K5 both in vitro and in human cells (Table 1) (151). Significantly, dysregulation of SMYD3 activity has been implicated in tumorigenesis (9, 151, 152, 153), thus giving impetus to the functional investigation of yeast Set5p as it may provide insights into SMYD3-dependent oncogenesis in humans (154).
Since its relatively recent discovery, several groups have sought to understand the function and regulation of Set5p in budding yeast cells. Initial genetic screens identified a functional link between Set5p and Set1p as cells lacking both histone methyltransferases show increased sensitivity to genotoxic and cellular stress (49). It was later found through RNA-sequencing analysis of set5Δset1Δ double-knockout cells that these enzymes play a synergistic role in transcriptional repression at repetitive regions, particularly near telomeres and at transposable elements (247). Although the precise mechanism underlying the cross talk between Set5p and Set1p in telomeric maintenance is unclear, it has been proposed that the transcriptional silencing that they mediate is dependent on H4K5 and H4K8 acetylation but not on H4K16ac and Sir proteins (248). Indeed, deletion of Yng2p, a subunit of the NuA4 HAT complex, in set5Δ yeast cells manifests in a slow growth phenotype, suggesting a functional cooperation between acetylation and methylation of H4K5/8/12 in cellular fitness (49, 154). Moreover, it has been reported that overexpression of Set5p confers tolerance to acetic acid, as well as to oxidative, osmotic, and heat stress, and also improves cellular growth and alcoholic fermentation (249, 250). Taken together, these findings suggest that Set5p, through its H4K5/8/12 methyltransferase activity, mediates specific gene expression programs to maintain genomic stability in response to yeast cellular stress. Nonetheless, the mechanism through which Set5p transduces signaling cascades at chromatin are entirely unknown (251) and is therefore an exciting avenue for future research.
Histone demethylases
Until relatively recently, histone methylation was believed to be a stable and heritable epigenetic modification (37). The identification of the first histone demethylase in 2004, mammalian lysine-specific demethylase 1 (LSD1), revealed that histone lysine methylation is enzymatically reversible and thus dynamic (252). LSD family demethylases are transcriptional repressors that specifically demethylate H3K4me1 and H3K4me2 in a flavin adenine dinucleotide (FAD)-dependent reaction (252, 253) but are unable to catalyze demethylation of trimethyl-lysine owing to the conformation of their active site pocket (254). This discovery was swiftly followed by the identification and characterization of another class of histone demethylases that contain a Jumonji C (JmjC) catalytic domain and are capable of removing all three lysine methylation states (255). JmjC enzymes demethylate substrate lysines through an oxidative reaction that utilizes Fe(II) and α-ketoglutarate as cofactors (256), and, unlike LSD demethylases, are highly conserved from yeast to human (254). Based on sequence homology predictions, five JmjC domain–containing proteins were identified in the budding yeast proteome: Jhd1p, Jhd2p, Rph1p, Gis1p, and Ecm5p (257). Of these, Jhd1p, Jhd2p, Rph1p, and Gis1p were subsequently shown to have bona fide histone demethylase activity through gene deletion screens and targeted biochemical assays (Fig. 1, Table 1) (258). In the following section, we examine the structure, function, conservation, and regulation of the four histone demethylase enzymes in budding yeast.
Jhd1p
JmjC domain–containing histone demethylase 1 (Jhd1p), also known as JHDM1 or KDM2, was the first histone demethylase to be identified in budding yeast. In 2006, Tsukada et al. (256) demonstrated that yeast Jhd1p and mammalian JHDM1 specifically demethylate mono- and dimethylated H3K36 in the presence of Fe(II) and α-ketoglutarate in vitro, generating formaldehyde and succinate as by-products (Table 1). This was subsequently verified in vivo where overexpression of Jhd1p reduces global H3K36 methylation levels (259), whereas deletion leads to accumulation of both monomethylated and dimethylated H3K36 states (258). Structurally, Jhd1p is a 492-amino-acid protein that comprises an N-terminal PHD domain (residues 6–70) and a JmjC catalytic domain (residues 254–409) and is the only histone demethylase in S. cerevisiae to lack a Jumonji N (JmjN) domain (Fig. 3) (260). Mutational studies have revealed that the JmjC domain and adjacent sequences are crucial for Jhd1p enzymatic activity, whereas the PHD domain is dispensable (259). Indeed, several point mutations within the cofactor-binding cleft of the JmjC domain (T302A, H305A, Y315A) significantly abrogate its H3K36 demethylase activity (Fig. 3) (256, 259). Although not required for catalysis, the PHD domain of Jhd1p specifically binds trimethylated H3K4 with high affinity in vitro (110), suggesting potential cis-regulatory cross talk between H3K36 demethylation and H3K4 methylation.
Jhd1p carries out an important, albeit subtle, role in the maintenance of transcriptional fidelity in S. cerevisiae. The H3K36me1/2 demethylase function of Jhd1p is required to remove repressive H3K36 methylation toward the 5′-ends of gene bodies and thus promote transcriptional elongation (254, 261). It has been proposed that Jhd1p may be recruited to transcription start sites, demarcated by H3K4 trimethylated chromatin, via its PHD domain where it can oppose repressive H3K36 dimethylation (262). Jhd1p has also been implicated in the regulation of pre-mRNA splicing through its genetic interactions with RNA splicing factors (116). Despite its apparent involvement in epigenetic processes, there are conflicting reports into the phenotypic effects of Jhd1p knockout in S. cerevisiae. One study observed no consequence for jhd1Δ in any of the functional assays performed (263), whereas another demonstrated only subtle changes in the chromosomal distribution of H3K36me2 upon removal of Jhd1p activity (259). Conversely, and in support of its biological significance, overexpression of Jhd1p was found to bypass the cellular requirement for the transcriptional elongation factor, Bur1p, and suppress the growth defect of bur1Δ yeast cells (264). Given these discrepancies, it is plausible that the function of Jhd1p may be masked by the redundancy of H3K36 demethylases in budding yeast under certain conditions (Fig. 1). In addition to Jhd1p, the paralogous enzymes Rph1p and Gis1p also possess H3K36 demethylase activity (see respective sections); however, they are each geared toward different H3K36 methylation states thus suggesting distinct molecular functions (Table 1) (265). Further work is required to clarify the role of Jhd1p, elucidate its regulation, and disentangle the overlapping activities of H3K36 demethylases in S. cerevisiae.
Jhd2p
JmjC domain–containing histone demethylase 2 (Jhd2p), also known as KDM5, is a member of the highly conserved JARID1 family and is the sole H3K4 demethylase in budding yeast. In 2007, Jhd2p was first purified from S. cerevisiae as a monomeric subunit with H3K4 demethylase activity in vitro (266). Through gene knockout and overexpression studies in yeast, several groups have since demonstrated the capacity of Jhd2p to demethylate all degrees of H3K4 methylation in vivo (260, 267), although with preferential affinity for trimethylated H3K4 (Table 1) (268). With respect to structure, Jhd2p is a 728-amino-acid protein that comprises JmjN (residues 4–47) and JmjC (residues 381–549) demethylase domains, as well as a PHD finger domain (residues 235–285), which associates with chromatin independently of H3K4 methylation and the H3 N-terminal tail (Fig. 3) (269). Mutation of the first histidine residue of the Fe(II)-binding motif (H427A) within the JmjC domain abolishes Jhd2p H3K4me3 demethylase activity (270).
Despite its seemingly central role in transcriptional regulation, deletion of Jhd2p manifests in only mild molecular and cellular phenotypes in S. cerevisiae. Under standard laboratory conditions, JHD2 null cells do not exhibit any significant differences in levels of H3K4me1/2/3 on bulk histones (257), nor do they show appreciable alterations in gene expression (67), thus confounding the study of this important regulatory protein using budding yeast as a model system (271). A systematic phenotypic analysis revealed a surprising lack of defects in the Jhd2p deletion yeast strain, with the exception of a subtle enhancement of telomeric silencing, an effect that was predictably reversed by overexpression (266). It has been proposed that jhd2Δ cells cannot properly initiate transcriptional silencing and are therefore seldom isolated from screens for loss-of-silencing phenotypes, which are typically performed during steady-state growth (272).
Notwithstanding a paucity of confirmed phenotypes, several targeted studies have interrogated the molecular function of Jhd2p and elucidated many aspects of its biology. Highlighting its diverse function, Jhd2p can either serve as a transcriptional activator or repressor depending on its genomic localization and cellular context. For example, Jhd2p recruitment positively regulates the transcription of genes encoding ribosomal proteins and Rap1-bound genes (273), whereas its activity inhibits expression of the Spt6p/Spn1p, FACT, and NNS transcriptional regulatory complexes (271). At the cellular level, Jhd2p was shown to globally repress spurious intergenic transcription during spore differentiation in postmeiotic yeast cells through its H3K4 demethylase activity and thereby promote gene transcription in the face of developmentally programmed transcriptional quiescence (271, 273). Modulation of Jhd2p during gametogenesis results in widespread and premature transcriptional downregulation and the aberrant production of stress-sensitive spores (273, 274). Although Jhd2p itself is upregulated upon entry into sporulation and is critical for the production of healthy meiotic progeny (273), its function is dispensable for gametogenesis-induced rejuvenation in budding yeast, suggesting functional redundancy in lifespan-resetting pathways (275). In addition to its canonical role in the regulation of sporulation, Jhd2p also mediates other important cellular processes, including rDNA repeat stability and silencing (261) and mRNA processing through 3′-UTR cleavage (276).
Recent work has sought to investigate the regulatory processes underpinning Jhd2p function and has duly uncovered how its activity is affected by specific modifications that can be present on histones. Jhd2p preferentially acts on nucleosomes that have only one histone H3 subunit trimethylated at K4 and thereby works cooperatively with dimeric COMPASS to focus symmetric H3K4me3 onto selected promoter nucleosomes (188). These findings suggest that Jhd2p primarily acts to reduce erroneous asymmetric methylation of nucleosomes under certain conditions and to provide correctional stability to gene expression programs in concert with COMPASS. Jhd2p-mediated H3K4me3 demethylation is also known to be negatively regulated by adjacent H3K14 acetylation by Gcn5p, thus highlighting opposing roles for HATs and Jhd2p in controlling deposition of H3K4 methylation (277). Finally, ubiquitination at H2BK123 sterically hinders Jhd2p from accessing its critical H2A-binding sites (H2AF26 and H2AQ57) thereby constituting an additional layer of trans-histone regulation of H3K4 methylation (269).
Rph1p
Regulator of PHR1 (Rph1p), also known as KDM4, is arguably the most functionally important of the three H3K36-specific demethylases in S. cerevisiae. Rph1p was originally identified as a major transcriptional repressor of the DNA repair gene, PHR1, which encodes a photolyase required for the light-dependent repair of pyrimidine dimers (278). Since then, Rph1p has been classified as a JMJD2 family enzyme that specifically demethylates H3K36me3 and H3K36me2 both in vitro and in vivo (Table 1) (254, 258, 279). Curiously, Rph1p is also capable of removing H3K9 methylation, a mammalian histone modification not found in budding yeast chromatin (279). In terms of its domain architecture, Rph1p is a 796-amino-acid protein that comprises JmjN (residues 14–55) and JmjC (residues 193–355) catalytic domains, as well as a C-terminal Cys2-His2 (C2H2) ZF domain that is responsible for DNA binding (Fig. 3) (279). The ZF domain of Rph1p shows complete sequence identity with its paralogous demethylase, Gis1p, which arose from the whole genome duplication in ancestral S. cerevisiae (280). Motif-based analysis of Rph1p identified a bipartite nuclear localization signal of sequence KRISSFQEQPLNKLLKR (residues 455–471) that may mediate its nuclear import, although this remains to be confirmed experimentally. The crystal structure of the catalytic core of Rph1p, both in apo form and in complex with its requisite cofactors, has been resolved (PDB ID: 3OPW and 3OPT (281)). This revealed that the substrate-binding cleft is formed by structural elements of the JmjC domain, a long β-hairpin, and a mixed structural motif (Fig. 3) (281). In vitro biochemical assays have demonstrated that mutations of several key residues within the Fe(II) (H235A, E237A, H323A) and α-ketoglutarate (Y183A, N245A, K253A)-binding pockets of the JmjC domain eliminate Rph1p demethylase activity.
Rph1p is the endogenous histone demethylase that controls H3K36me3 levels during active transcriptional elongation. Akin to other histone methylation enzymes in budding yeast, Rph1p can function as either a transcriptional activator or repressor in a context-dependent manner (282). Accordingly, the paralogous enzymes Rph1p and Gis1p can act on overlapping sets of genes as well as on distinct targets, and their respective activities and specificities vary throughout the cellular growth cycle. Although the enzymatic activity of Rph1p is not required for its role in growth phase–dependent gene regulation, the capacity for Rph1p to activate or repress transcription in vivo is enhanced by active demethylase function (283, 284). In terms of its repressive functionality, ~70% of mRNA transcripts are upregulated upon Rph1p knockout in S. cerevisiae, a large subset of which are involved in responses to DNA damage as well as to environmental and oxidative stress (283, 285, 286). Dysregulation of Rph1p-mediated PHR1 silencing contributes to deleterious growth defects; yeast cells overexpressing Rph1p are hypersensitive to UV irradiation (258). Conversely, several studies have shown that Rph1p promotes transcriptional elongation by removing H3K36 methylation within transcribed regions and thus preventing repressive histone deacetylation by the Rpd3S HDAC complex (254). This was confirmed through synthetic lethality experiments that showed that overexpression of H3K36 demethylases Rph1p and Jhd1p bypasses the cellular requirement for the transcriptional elongation factor Bur1p. It has been proposed that Rph1p, but not Jhd1p, directly opposes Rdp3S function through its specific demethylation of H3K36me3 (263), the methylation state recognized and bound by the Rdp3S subunit, Eaf3p (287). This may explain the more severe phenotypes exhibited by rph1Δ cells in comparison with their jhd1Δ counterparts, although this hypothesis will require further experimental investigation.
Recent work has suggested that Rph1p is a master transcriptional regulator of several key biological pathways (285). The contribution of the histone demethylase activity of Rph1p to these processes, however, is disputed. First, Rph1p is known to be a critical player in the environmental and oxidative stress response in S. cerevisiae (280, 288) as genes responding to such stimuli are overrepresented in differential expression analyses of Rph1 null cells (280). Under normal conditions, Rph1p represses transcription at these loci but is immediately dissociated from chromatin upon stress-induced signaling, thereby allowing expression of signal-responsive genes (289). This mechanism of post-translational regulation is discussed in greater detail later. The repression of stress response genes by Rph1p under physiological conditions is mediated, at least in part, by its demethylase activity (288). By contrast, Rph1p is a negative transcriptional regulator of autophagy in a manner that is independent of its catalytic activity (290). In a nutrient-replete environment, Rph1p represses expression of a subset of ATG genes, particularly ATG7, by binding their respective promoters and thus preventing autophagetic induction (291). Upon nitrogen starvation, signaling cascades trigger the phosphorylation-mediated dissociation of Rph1p and alleviate transcriptional repression (mechanism discussed later), facilitating autophagy (290). It will be interesting to determine the role, if any, of the histone demethylase function of Rph1p, and its mammalian JMJD2 family orthologs, in the regulation of autophagetic processes in yeast and in other eukaryotes.
Gis1p
Gig1-2 suppressor 1 (Gis1p) is a functional paralog of Rph1p/KDM4. However, its histone demethylase activity in vivo remains contentious. In 1999, Gis1p was first discovered and named as a multicopy suppressor of the Gal− phenotype of snf1/mig1/srb8 triple mutant yeast cells (292). Subsequent work revealed the largely redundant roles of Gis1p and Rph1p as transcriptional repressors of the photolyase, PHR1 (278), and it was not until 2007 that the putative demethylase function of Gis1p was investigated (Table 1) (258). Structurally, Gis1p is the largest histone demethylase in S. cerevisiae at 894 amino acids and is composed of JmjN (residues 12–53) and JmjC (residues 170–324) catalytic domains, in addition to a highly conserved C-terminal C2H2 ZF domain (residues 828–882) that is identical in sequence in Rph1p and mediates DNA binding (Fig. 3). Similar to Rph1p, motif-based analysis of Gis1p revealed a bipartite nuclear localization signal of sequence RKQPLKCGCGNKKEERK (residues 316–332); however, the functional role of this motif has not been verified. Although no crystal structure has been resolved for Gis1p, structural studies have revealed that the JmjN and JmjC domains physically interact in three dimensions, via two β sheets, to form a structural unit that stabilizes full-length Gis1p and regulates its selective proteolysis by the proteasome (293).
In its canonical role, Gis1p serves as a transcription factor that modulates the expression of genes involved in nutrient signaling, oxidative stress, and aging (294). It is capable of functioning as both a transcriptional repressor (278, 295) and a transcriptional activator (296) depending on its cellular context, an observation that may be explained by its numerous and diverse binding partners (297). Although transcriptional regulation by Rph1p predominantly occurs through STRE motifs (282), the repressive and activating roles of Gis1p are mediated by the post-diauxic shift element present in the promoters of its target genes (298). Appropriately, there are over 100 genes, including SSA3 and HSP12 (296), that are regulated by Gis1p during the diauxic transition in response to RAS/cAMP signaling and Rim15p activation (298, 299). The exact nature of this signaling cascade is discussed in greater detail later. Of interest, the biological function of Gis1p may be more diverse than was first appreciated, as two-hybrid approaches have shown that it interacts with at least 19 yeast proteins that are enriched for functions in transcription, SUMOylation, and DNA repair (297).
Although Gis1p contains a JmjC domain, it is still not clear whether this enzyme possesses bona fide H3K36 demethylase activity. Indeed, several groups have provided evidence either in support or in dispute of its active demethylase status. Mass spectrometric analysis of in vivo histone modifications revealed accumulation of H3K36me2 and H3K36me1 upon Gis1p deletion, suggesting that Gis1p specifically demethylates the lower H3K36 methylation states (Table 1) (258). These findings, however, could not be recapitulated in vitro, as enzymatic assays failed to demonstrate demethylase activity of Gis1p toward any of the three major histone lysine methylation sites in budding yeast (H3K4, H3K36, H3K79) (259, 260, 284). Crucially, Gis1p carries a single point mutation (H292Y) within the Fe(II)-binding cleft of the JmjC domain that is predicted bioinformatically to render its demethylase function inactive (255). An equivalent substitution (H305A) in the fission yeast demethylase, Epa1p, wholly ablates its enzymatic activity (256). There are also conflicting reports as to the contribution of the Gis1p demethylase domain to its role in transcriptional activation and repression. One study observed that mutation of Fe(II)- and α-ketoglutarate-binding residues (H204A, K222A), and even deletion of the entire JmjC domain, did not abolish Gis1p-mediated transcriptional activation during glucose starvation and sporulation, suggesting that its demethylase function is dispensable (300). Conversely, yeast cells expressing catalytically inactive Gis1p showed increased transcriptional activation of the hydrophilin, GRE1, but not SSA3 (293), thus providing backing for the histone demethylase activity of Gis1p and its role in transcriptional processes, albeit in a gene-dependent manner. Moving forward, significant work is needed to further investigate the putative demethylase function of Gis1p and illuminate its biochemical and functional features.
Post-translational regulation of histone methylation enzymes
Considerable progress has been made in the identification of histone lysine methyltransferases and demethylases in S. cerevisiae and mapping these enzymes to their respective histone targets. Recent studies have also shed light on how the activity of histone methylation enzymes can be controlled by their protein–protein interactions with various components of the transcriptional machinery, and through cross talk with other epigenetic modifications. However, these layers of regulation only explain a small proportion of the complexity that exists within the histone lysine methylation network in budding yeast. We reason that many of the unresolved questions surrounding the function and dynamics of this intricate epigenetic system may be clarified by a largely overlooked aspect of histone methyltransferase and demethylase biology, that being their post-translational modification and regulation.
There is substantial evidence that histone methylation enzymes in budding yeast are extensively post-translationally modified. The vast majority of these PTM sites have been identified through high-throughput proteomic screens that typically involve affinity-based enrichment for specific modification types and their subsequent analysis by high-resolution, bottom-up mass spectrometry of peptides. To canvass the PTM topology of histone methylation proteins, we mined several publicly available proteomic datasets (246, 301, 302, 303, 304, 305, 306, 307, 308, 309, 310, 311, 312, 313, 314, 315, 316, 317, 318, 319, 320, 321) and PTM databases (including UniProt (322), PhosphoGRID (323), and YAAM (324)), including one study that achieved near-complete characterization of the purified enzymes (315). Our collation of PTMs comprises a total of 251 modification sites, of which phosphorylation (143 sites), acetylation (101 sites), and ubiquitination (seven sites) were the most commonly found (Fig. 4). These PTMs represent those identified in a range of yeast cellular growth states and under different growth conditions. Despite efforts to identify other PTM types (e.g., protein methylation, acylation, crotonylation, ADP-ribosylation) on this family of enzymes (315), no such sites have been observed to date. This does not, however, definitively rule out the possibility of their occurrence in vivo. In this section, we will discuss the general biological roles of phosphorylation, acetylation, and ubiquitination and highlight their prevalence on histone methyltransferase and demethylase enzymes within the context of the budding yeast proteome. We will also review the relatively small number of targeted studies that have determined how specific PTM sites affect enzyme function and/or identified the upstream modifying enzymes that convey signaling information to the histone methylation network via PTM.
Figure 4.
Post-translational modifications on yeast histone methylation enzymes. PTM sites were from a recent systematic characterization of yeast histone methyltransferases and demethylases (315) and high-throughput phosphoproteomic and acetylomic studies (246, 301, 302, 303, 304, 305, 306, 307, 308, 309, 310, 311, 312, 313, 314, 315, 316, 317, 318, 319, 320, 321). For each modification type, the number of residues within each protein that has been shown to be post-translationally modified in at least one study has been graphed as a bar chart. For phosphorylation (blue), this refers to the number of phosphorylated serine (S) and threonine (T) residues, whereas for acetylation (orange) and ubiquitination (purple), it refers to the number of modified lysine (K) residues. For all eight histone methylation enzymes, the average (mean) number of PTM sites was calculated and shown in the right panel. Proteome values were calculated based on a total of 6275 protein-coding yeast genes (345) and all known protein phosphorylation (87,703), acetylation (10,035), and ubiquitination (14,880) sites from the YAAM database (324). PTM, post-translational modification.
Phosphorylation
Phosphorylation is the second most abundant PTM in the eukaryotic cell (325) and is arguably the most extensively studied. It predominantly refers to the transfer of the γ-phosphoryl group of ATP to the polar amino acid side chains of serine, threonine, or tyrosine residues by protein kinases (326). Phosphorylation is a hallmark of signaling cascades and is involved in the regulation of an array of key cellular processes, including growth, differentiation, and apoptosis (327). The altered physicochemical properties of phosphorylated residues allow their cognate proteins to recognize and bind specific interaction partners and thus carry out specific functions. Phosphorylation can also bring about allosteric changes in protein structure that can modulate enzymatic activity and interactions (328). Accordingly, many enzymes and receptors become activated upon phosphorylation and are deactivated through dephosphorylation by a phosphatase. There are also instances where a phosphorylation event inactivates an enzyme’s activity or blocks a specific interaction.
The histone lysine methylation network in budding yeast receives substantial input from upstream signaling pathways. A recent systematic characterization of all S. cerevisiae histone methyltransferases and demethylases defined a comprehensive set of phosphosites occurring on these enzymes in vivo for mid–log phase yeast cells grown under standard laboratory conditions (315). This study, in conjunction with previous high-throughput phosphoproteomic analyses, identified a total of 107 and 35 phosphosites on serine and threonine residues, respectively. This corresponds to an average of 18 phosphosites per histone methylation protein, which is slightly higher than the total S. cerevisiae proteome wherein proteins carry, on average, 14 phosphosites (Fig. 4) (324). Of interest, specific enzymes are phosphorylated to markedly different extents; the paralogous H3K36 demethylases Rph1p and Gis1p carry 36 and 26 sites, respectively, whereas the Jumonji domain–containing enzymes Jhd1p and Jhd2p are phosphorylated at just one and five sites, respectively (315). These observations are striking and suggest that these enzymes, and thus the histone methylation sites that they control, may be differentially regulated by cellular signaling. Enzymes with a large number of phosphosites have the potential to be intricately regulated by multiple independent signals, whereas those that have fewer sites may be predominantly controlled through other means, for example, at the level of transcription (33).
There is emerging evidence that phosphorylation can functionally modulate the activity of histone methyltransferase and demethylase enzymes. In the mammalian cell, several targeted studies have investigated the phosphoregulation of histone methylation proteins and discovered how specific phosphosites regulate diverse aspects of their biology, including their enzymatic activity, protein–protein interactions, genomic localization, and proteasomal degradation (33). Such studies have also begun to illuminate the wide range of human signaling pathways that transmit signaling data to the histone methylation network (33). By contrast, in S. cerevisiae, only a handful of low-throughput studies have interrogated phosphosite function using targeted experimental approaches. These studies have justifiably focused on the two most extensively phosphorylated enzymes, Rph1p and Gis1p. For Rph1p, phosphorylation at serine 652 potentially contributes to its dissociation from chromatin in response to DNA damage, and the checkpoint kinase Rad53p was implicated as its upstream regulator (Fig. 5) (283). Given that phosphorylation is highly abundant in this region of Rph1p (315), it is likely that other phosphosites may also control its chromatin association. Systematic mutagenesis revealed that phosphorylation at serine 412 of Rph1p is responsive to intracellular AdoMet levels and is required, but not sufficient, to activate its H3K36 demethylase function (329). Rph1p is also known to be controlled by kinase Rim15p, which phosphorylates and inhibits its activity upon nitrogen starvation to facilitate autophagic induction (290). The specific phosphosite, however, could not be determined. Rim15p is also known to indirectly control the function of Gis1p (Fig. 5), highlighting the multiple layers of information this kinase inputs to the methylation system. Here, Rim15p phosphorylates endosulfines to directly inhibit the Cdc55-protein phosphatase 2A, which thus preserves phosphorylation at serine 425 of Gis1p and facilitates its binding at promoters of nutrient-related genes (330). Finally, a high-throughput phosphoproteomic study established a candidate kinase for multiple phosphosites, showing that sites at serine residues 139, 561, and 575 of Rph1p and 425 and 696 of Gis1p are downregulated upon deletion of the key cell cycle kinase, Cdc28p (fungal homolog of Cdk1) (309). This, however, may be an indirect effect and will require validation through targeted approaches.
Figure 5.
Post-translational regulation of yeast histone methylation enzymes. The functional effects of phosphorylation (blue), acetylation (orange), and ubiquitination (purple) sites on histone methyltransferase (green) and demethylase (pink) biology are shown. Where known, the amino acid residues that carry functional PTM sites are displayed above their cognate enzyme. For Set5p, its catalytic activity and chromatin association are regulated by a cluster of ten phosphosites corresponding to phosphorylation at S458, S461, S462, S466, S475, S476, T511, S512, S517, and S520. The upstream modifying enzymes responsible for PTM sites are shown if known and colored according to the key. Instances where the upstream enzyme is not known are denoted by question marks. For some PTMs, the environmental signals that trigger their deposition are illustrated in gray boxes. The downstream functional effects of PTM sites are also shown. Although the number of targeted studies into the post-translational regulation of yeast histone methylation enzymes is modest, it is apparent that PTMs can affect their catalytic activity, chromatin binding, genomic localization, and degradation. PTM, post-translational modification.
An outstanding question regarding the role of phosphorylation is the degree to which phosphosites cross talk with one another or function cooperatively to fine-tune enzyme activity. This is particularly pertinent for extensively phosphorylated enzymes, such as Rph1p and Gis1p. Although the above-mentioned examples illustrate the capacity for individual phosphosites to modulate function, there are also instances where multiple sites are required to elicit functional effects. For example, the catalytic activity and chromatin association of Set5p are cooperatively controlled by a cluster of 10 phosphosites (S458, S461, S462, S466, S475, S476, T511, S512, S517, S520) within its C-terminal region, as opposed to the presence or absence of specific sites (Fig. 5) (246). More recently, the H3K36 methyltransferase activity of Set2p was shown to be regulated by a phosphorylation cluster (S6, S8, S10) within its acidic and intrinsically disordered N-terminal region (315). These phosphosites may work in conjunction with an adjacent histone H4 recognition motif to direct Set2p chromatin association through an electrostatic mechanism (Fig. 5). Moving forward, considerable work will be required to clarify the prevalence of PTM cross talk on S. cerevisiae histone methylation enzymes.
Acetylation
Acetylation is a major PTM that plays manifold functions in the regulation of metabolic processes, in particular. It is catalyzed by acetyltransferases that enzymatically transfer acetate (CH3CO) from the donor molecule, acetyl-CoA, to either the α-amine of the protein N-terminus or the ε-amino group of lysine side chains (331). Although protein acetylation was originally studied within the context of transcriptional regulation (4), it is becoming increasingly apparent that many nonhistone proteins are acetylated and that this modification affects diverse aspects of their function, including catalytic activity, stability, and subcellular localization (reviewed in (332)).
Histone methylation enzymes in S. cerevisiae are heavily acetylated, harboring a total of 101 acetyl sites across the family of eight proteins (302, 308, 315, 320). This averages out to ~13 acetyl sites per methylation protein, which represents a marked enrichment compared with the entire yeast proteome, in which proteins carry, on average, ~2 acetylation sites (Fig. 4). This overrepresentation of acetylation is likely to be of functional significance given that this PTM is dependent on the availability of acetyl-CoA, a key metabolite in many biochemical reactions. It is plausible that acetylation of the histone methylation machinery may provide a regulatory link between cellular metabolism and transcription, whereby specific gene expression programs are activated or repressed in response to metabolic signals (333). Akin to phosphosite distribution, histone methyltransferases and demethylases are also acetylated to markedly different extents, suggesting that some enzymes, and thus the histone methyl marks they control, are likely to be more responsive to the metabolic status of the yeast cell than others.
Although acetylation is widespread on histone methyltransferases and demethylases, very little is known about how specific acetyl sites regulate enzymatic function. To date, a single targeted study has investigated the regulatory effects of acetylation (285). Using a combination of in vitro and in vivo assays, Li et al. demonstrated that Gcn5p-mediated acetylation of the H3K36 demethylase Rph1p is required for its autophagic degradation in response to DNA damage stress; however, the specific site of acetylation could not be localized (Fig. 5) (285). The paucity of understanding regarding acetylation is likely due to the challenges in prioritizing acetyl sites for in-depth analysis, as discussed above for phosphosites. Given the complexities of protein acetylation, high-throughput mass spectrometry data are unable to distinguish acetyltransferase-mediated sites, of high regulatory importance, from nonenzymatic acetylation events, which are an artefact of the spontaneous reaction between acetyl-CoA and lysine (334). This spontaneous acetylation is more widespread throughout the eukaryotic cell than was previously appreciated; an array-based screen of 6000 human proteins revealed that ~1600 can be chemically acetylated in vitro (334). There is also limited concordance between acetylation sites reported on histone methylation enzymes in different acetylomic datasets (315), although the number of proteome-scale studies of acetylation in S. cerevisiae is modest. Considerable future work is required to characterize the full complement of acetylation sites occurring on these enzymes in vivo and to subsequently establish their regulatory capacity through targeted studies.
Ubiquitination
Ubiquitination refers to the post-translational conjugation of ubiquitin, a 76-residue polypeptide, to the ε-amino group of a substrate lysine residue. This reaction occurs through a three-step process involving an E1 activating enzyme, E2 conjugating enzyme, and an E3 ligase, the last of which is the central determinant of specificity in ubiquitin signaling (335). Substrate proteins modified with one or a few ubiquitin molecules are generally targeted for proteolysis via endocytosis in the yeast vacuole (336). Monoubiquitination can also regulate the intracellular endocytic trafficking of client proteins (337). By contrast, substrates marked with polymeric ubiquitin chains are degraded through the 26S proteasome (338).
Ubiquitination on histone methylation enzymes in S. cerevisiae is poorly understood. Only seven sites of ubiquitination have been reported across the family of eight enzymes to date (Fig. 4). This may reflect the relative lack of studies that have used ubiquitin enrichment techniques (e.g., anti-ubiquitin antibodies) prior to the identification of ubiquitination sites on yeast histone methyltransferases and demethylases (discussed in detail later). Although this modification is underrepresented on histone methyltransferases and demethylases, two studies have shown that ubiquitination regulates their degradation in vivo. First, Mersman et al. demonstrated that polyubiquitination of the H3K4 demethylase, Jhd2p, by the E3 ubiquitin ligase, Not4p, controls its turnover by the proteasome and thus H3K4me3 levels and gene expression (Fig. 5) (268). This regulation is conserved in mammals, where human NOT4 can also polyubiquitinate JARID1C, the ortholog of Jhd2p. In another example, Dronamraju et al. showed that the H3K36 methyltransferase, Set2p, is targeted for proteasomal degradation by the Anaphase Promoting Complex/Cyclosome (APC/C) complex in a cell cycle–dependent manner (Fig. 5) (339). Strikingly, human SETD2, the ortholog of Set2p, is also regulated by APC/C, thus highlighting an evolutionarily conserved mechanism of regulation for H3K36 methylation in cell cycle progression. In both these studies, the specific sites of ubiquitination could not be identified, meaning that further work is required to understand the molecular details of this ubiquitin-mediated degradation.
Does the lack of ubiquitination sites on the histone methylation machinery reflect their biological abundance or the technical challenges associated with their detection? We suspect that ubiquitination sites on these enzymes may be more prevalent than currently appreciated; however, the study of this modification is challenging using high-throughput mass spectrometric approaches without enrichment for ubiquitinated proteins or peptides. This is further complicated by the low abundance of most histone methyltransferases and demethylases in the yeast cell (Table 1), which makes them hard to detect in complex biological samples (e.g., cell lysates). Given the difficulties involved in confidently identifying ubiquitin sites through mass spectrometry (340), targeted approaches (e.g., immunoblotting, protein purification) remain necessary to characterize the ubiquitination landscape of specific proteins. Further studies of this nature will illuminate the full complement of ubiquitination sites on histone methylation enzymes in budding yeast and pave the way for their functional characterization.
Future challenges: a complete phosphoregulatory network
Phosphorylation is emerging as a key regulator of the yeast histone methylation network. Phosphoproteomic analyses have revealed that methyltransferase and demethylase enzymes are extensively phosphorylated (Fig. 4). Although still modest in number, targeted approaches have demonstrated that phosphorylation can control aspects of enzyme function and identified some of the upstream signaling kinases and pathways to which the histone methyltransferases and demethylases are connected (Fig. 5). These observations support an investigation into the phosphorylation of the S. cerevisiae histone methylation network in a holistic manner. This would involve the construction of a complete phosphoregulatory network wherein all phosphosites on methylation enzymes have a known upstream kinase and a known function (Fig. 6).
Figure 6.
Feasibility of constructing a complete phosphoregulatory network of histone lysine methylation in yeast. A comparison of the histone methylation networks in budding yeast (Saccharomyces cerevisiae) and human (Homo sapiens). Yeast possesses almost the same number of histone lysine methylation sites (yellow) as human; however, they have substantially fewer methyltransferase (green) and demethylase (pink) enzymes responsible for their regulation. As a result, there is a much smaller number of phosphosites (blue) present on the histone methylation machinery to investigate experimentally. Systematic kinase mapping is also conceivable in yeast, given the relatively modest number of protein kinases (baby blue) encoded in their genome.
The generation of a complete phosphoregulatory network of histone methylation in S. cerevisiae is conceptually feasible for a number of reasons. First, the histone lysine methyltransferases and demethylases are well characterized and their histone substrates are known and largely undisputed. This contrasts with the mammalian cell where there is still contention surrounding the substrate specificity of many human methylation enzymes (341, 342, 343, 344). It is therefore possible to experimentally monitor the downstream changes in histone methylation marks upon modulation of specific phosphosites on yeast histone methyltransferases and demethylases. Second, the regulatory network of histone lysine methylation in yeast is substantially simplified in comparison with its mammalian counterpart (Fig. 6), making this a tractable system for in-depth characterization. Compared with human cells, S. cerevisiae possesses a smaller number of histone lysine methyltransferases and demethylases (8 in yeast versus 52 in human), which therefore carry a smaller number of phosphosites (143 in yeast versus 2449 in human) for targeted analysis. There are also far fewer protein kinases encoded in the yeast genome (127 in yeast versus 518 in human), thus simplifying kinase discovery experiments (Fig. 6).
Considering the above mentioned advantages of S. cerevisiae as a model for the study of histone methylation, it is conceivable that a systems-level analysis of its phosphoregulation is a possible and indeed attractive prospect for future research. Through the use of targeted experimental approaches (e.g., site-directed mutagenesis, functional assays), studies should first seek to identify regulatory phosphosites on histone methyltransferases and demethylases in vivo and to then employ kinase mapping techniques (e.g., in vitro assays, gene knockouts, BioID) to establish the connections of enzymes with upstream signaling cascades. If completed, such a regulatory network would be the first of its kind for any epigenetic modification in any eukaryotic species. It would reveal the connections of the histone-based gene regulatory network with upstream signaling pathways and thus provide insights into the signal responsiveness of this epigenetic modification. Given the evolutionary conservation of histone methylation sites and enzymes, the generation of this network in yeast would likely provide clues into the regulatory mechanisms underpinning histone methylation and demethylation in human cells and in other higher eukaryotes.
Concluding remarks
The histone lysine methylation network in S. cerevisiae is well characterized. Many studies have investigated the biological and cellular functions of histone methylation sites and shown that the loss of specific sites manifests in deleterious growth phenotypes in budding yeast. Furthermore, all histone lysine methylation marks have a known methyltransferase responsible for their deposition, and many sites have a known corresponding demethylase to remove methylation as required. Given the lack of additional putative histone methylation enzymes in the S. cerevisiae proteome, it is likely that the four currently known methyltransferases (Set1p, Set2p, Dot1p, and Set5p) and four known demethylases (Jhd1p, Jhd2p, Rph1p, and Gis1p) represent the full complement of enzymes that control this epigenetic modification.
Despite extensive efforts to identify histone methylation sites and map enzymes to their respective histone substrates, there are still a number of outstanding questions surrounding the function and regulation of this network. In particular, there is a paucity of understanding of how histone methylation enzymes are recruited to specific locations within the yeast genome to mediate changes in gene expression in response to stimuli. This is a critical gap in our knowledge as we only poorly understand how the yeast cell controls when and where the methyl marks are made in chromatin.
There is emerging evidence that the histone methylation machinery is post-translationally modified and intricately regulated in the budding yeast cell. Recent targeted and high-throughput proteomic studies have demonstrated that yeast histone methyltransferases and demethylases are decorated with a large number of PTMs, particularly phosphorylation and acetylation, and to a lesser extent, ubiquitination. Although many PTM sites have been identified, only a handful of studies have sought to investigate the functional effects of single PTMs or to identify their upstream modifying enzymes. Crucially, these studies have shown that PTMs can regulate diverse aspects of methyltransferase and demethylase biology, including their enzymatic activity, chromatin association, genomic localization, and stability. These observations are consistent with those in the mammalian cell, where phosphorylation, in particular, is pervasive on human histone methyltransferases and demethylases and is known to control the function of some enzymes (33).
Moving forward, considerable work is required to holistically investigate the post-translational regulation of histone methyltransferases and demethylases in yeast and to connect this gene regulatory system to the cell’s signaling network. As discussed earlier, this is particularly pertinent for phosphorylation given that phosphosites have been systematically mapped and that phosphorylation is involved in many intracellular signaling pathways. With further work, it will be fascinating to learn how PTMs control when and where histone methylation enzymes act in yeast chromatin and which intracellular and extracellular cues trigger the transmission of signaling information to the histone methylation network to fine-tune its function.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Mandy Wong and Tyler Chapman for their preliminary research and Dr Joshua Hamey and Tara Bartolec for valuable input and suggestions.
Author contributions
R. J. S and M. R. W. conceptualization; R. J. S. visualization; R. J. S. writing-original draft; R. J. S. and M. R. W. writing-review and editing; M. R. W. supervision; M. R. W. funding acquisition.
Funding and additional information
R. J. S. was the recipient of an Australian Research Training Program Scholarship. This work was supported by Australian Research Council Grants DP170100108 and DP200100129 to M. R. W.
Edited by Patrick Sung
References
- 1.Kornberg R.D. Chromatin structure: A repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Young N.L., DiMaggio P.A., Garcia B.A. The significance, development and progress of high-throughput combinatorial histone code analysis. Cell Mol. Life Sci. 2010;67:3983–4000. doi: 10.1007/s00018-010-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray K. The occurrence of iε-N-methyl lysine in histones. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 4.Allfrey V., Faulkner R., Mirsky A. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutierrez R.M., Hnilica L.S. Tissue specificity of histone phosphorylation. Science. 1967;157:1324–1325. doi: 10.1126/science.157.3794.1324. [DOI] [PubMed] [Google Scholar]
- 6.Kleinsmith L.J., Allfrey V.G., Mirsky A.E. Phosphoprotein metabolism in isolated lymphocyte nuclei. Proc. Natl. Acad. Sci. U. S. A. 1966;55:1182–1189. doi: 10.1073/pnas.55.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldknopf I., Taylor C.W., Baum R.M., Yeoman L.C., Olson M., Prestayko A., Busch H. Isolation and characterization of protein A24, a" histone-like" non-histone chromosomal protein. J. Biol. Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- 8.Fraga M.F., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 9.Hamamoto R., Furukawa Y., Morita M., Iimura Y., Silva F.P., Li M., Yagyu R., Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 10.Alarcón J.M., Malleret G., Touzani K., Vronskaya S., Ishii S., Kandel E.R., Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Huber L.C., Stanczyk J., Jüngel A., Gay S. Epigenetics in inflammatory rheumatic diseases. Arthritis Rheum. U. S. A. 2007;56:3523–3531. doi: 10.1002/art.22948. [DOI] [PubMed] [Google Scholar]
- 12.Briggs S.D., Bryk M., Strahl B.D., Cheung W.L., Davie J.K., Dent S.Y., Winston F., Allis C.D. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Gene Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J., Ahn J.H., Wang G.G. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol. Life Sci. 2019;76:2899–2916. doi: 10.1007/s00018-019-03144-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDaniel S.L., Hepperla A.J., Huang J., Dronamraju R., Adams A.T., Kulkarni V.G., Davis I.J., Strahl B.D. H3K36 methylation regulates nutrient stress response in Saccharomyces cerevisiae by enforcing transcriptional fidelity. Cell Rep. 2017;19:2371–2382. doi: 10.1016/j.celrep.2017.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahrenkrog B. Histone modifications as regulators of life and death in Saccharomyces cerevisiae. Microb. Cell. 2016;3:1–13. doi: 10.15698/mic2016.01.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black J.C., Van Rechem C., Whetstine J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu H., Maunakea A.K., Martin M.M., Huang L., Zhang Y., Ryan M., Kim R., Lin C.M., Zhao K., Aladjem M.I. Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle. Plos Genet. 2013;9 doi: 10.1371/journal.pgen.1003542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong F., Miller K.M. Histone methylation and the DNA damage response. Mutat. Res. Rev. Mutat. 2019;780:37–47. doi: 10.1016/j.mrrev.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bedford M.T., Richard S. Arginine methylation: An emerging regulatorof protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Martin C., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell. Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen P.R., Nietlispach D., Mott H.R., Callaghan J., Bannister A., Kouzarides T., Murzin A.G., Murzina N.V., Laue E.D. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 22.Shi X., Hong T., Walter K.L., Ewalt M., Michishita E., Hung T., Carney D., Pena P., Lan F., Kaadige M.R. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M.T. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 26.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 27.Weiner A., Hsieh T.-H.S., Appleboim A., Chen H.V., Rahat A., Amit I., Rando O.J., Friedman N. High-resolution chromatin dynamics during a yeast stress response. Mol. Cell. 2015;58:371–386. doi: 10.1016/j.molcel.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo Y.H., Li W.-H. Evolutionary conservation of histone modifications in mammals. Mol. Biol. Evol. 2012;29:1757–1767. doi: 10.1093/molbev/mss022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roguev A., Schaft D., Shevchenko A., Aasland R., Shevchenko A., Stewart A.F. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J. Biol. Chem. 2003;278:8487–8493. doi: 10.1074/jbc.M209562200. [DOI] [PubMed] [Google Scholar]
- 30.Fuchs J., Demidov D., Houben A., Schubert I. Chromosomal histone modification patterns–from conservation to diversity. Trends Plant Sci. 2006;11:199–208. doi: 10.1016/j.tplants.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Højfeldt J.W., Agger K., Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat. Rev. Drug Discov. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 32.Husmann D., Gozani O. Histone lysine methyltransferases in biology and disease. Nat. Struct. Mol. Biol. 2019;26:880–889. doi: 10.1038/s41594-019-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Separovich R.J., Pang C.N.I., Wilkins M.R. Controlling the controllers: Regulation of histone methylation by phosphosignalling. Trends Biochem. Sci. 2020;45:1035–1048. doi: 10.1016/j.tibs.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Rando O.J., Winston F. Chromatin and transcription in yeast. Genetics. 2012;190:351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donaldson-Collier M.C., Sungalee S., Zufferey M., Tavernari D., Katanayeva N., Battistello E., Mina M., Douglass K.M., Rey T., Raynaud F. EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat. Genet. 2019;51:517–528. doi: 10.1038/s41588-018-0338-y. [DOI] [PubMed] [Google Scholar]
- 36.Greer E.L., Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyun K., Jeon J., Park K., Kim J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017;49 doi: 10.1038/emm.2017.11. e324-e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Liu K., Qin S., Xu C., Min J. Epigenetic targets and drug discovery: Part 1: Histone methylation. Pharmacol. Therapeut. 2014;143:275–294. doi: 10.1016/j.pharmthera.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 39.McGrath J., Trojer P. Targeting histone lysine methylation in cancer. Pharmacol. Therapeut. 2015;150:1–22. doi: 10.1016/j.pharmthera.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Morera L., Lübbert M., Jung M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin. Epigenetics. 2016;8:1–16. doi: 10.1186/s13148-016-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song Y., Wu F., Wu J. Targeting histone methylation for cancer therapy: Enzymes, inhibitors, biological activity and perspectives. J. Hematol. Oncol. 2016;9:1–21. doi: 10.1186/s13045-016-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croken M.M., Nardelli S.C., Kim K. Chromatin modifications, epigenetics, and how protozoan parasites regulate their lives. Trends Parasitol. 2012;28:202–213. doi: 10.1016/j.pt.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emery-Corbin S.J., Hamey J.J., Balan B., Rojas-López L., Svärd S.G., Jex A.R. Eukaryote-conserved histone post-translational modification landscape in Giardia duodenalis revealed by mass spectrometry. Int. J. Parasitol. 2021;51:225–239. doi: 10.1016/j.ijpara.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Jiang L., Mu J., Zhang Q., Ni T., Srinivasan P., Rayavara K., Yang W., Turner L., Lavstsen T., Theander T.G. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013;499:223–227. doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Gene Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 46.Margueron R., Trojer P., Reinberg D. The key to development: Interpreting the histone code? Curr. Opin. Genet. Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Shi Y., Whetstine J.R. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y., Garcia B.A. Comprehensive catalog of currently documented histone modifications. C.S.H. Perspect. Biol. 2015;7:a025064. doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green E.M., Mas G., Young N.L., Garcia B.A., Gozani O. Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nat. Struct. Mol. Biol. 2012;19:361–363. doi: 10.1038/nsmb.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruthenburg A.J., Allis C.D., Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 51.Rando O.J. Global patterns of histone modifications. Curr. Opin. Genet. Dev. 2007;17:94–99. doi: 10.1016/j.gde.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 53.Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Iii, Gingeras T.R. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 54.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 55.Heintzman N.D., Ren B. Finding distal regulatory elements in the human genome. Curr. Opin. Genet. Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soares L.M., He P.C., Chun Y., Suh H., Kim T., Buratowski S. Determinants of histone H3K4 methylation patterns. Mol. Cell. 2017;68:773–785. doi: 10.1016/j.molcel.2017.10.013. e776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L., Schroeder S., Fong N., Bentley D.L. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress’. EMBO J. 2005;24:2379–2390. doi: 10.1038/sj.emboj.7600711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malik S., Bhaumik S.R. Mixed lineage leukemia: Histone H3 lysine 4 methyltransferases from yeast to human. FEBS J. 2010;277:1805–1821. doi: 10.1111/j.1742-4658.2010.07607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soares L.M., Radman-Livaja M., Lin S.G., Rando O.J., Buratowski S. Feedback control of Set1 protein levels is important for proper H3K4 methylation patterns. Cell Rep. 2014;6:961–972. doi: 10.1016/j.celrep.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhaumik S.R., Smith E., Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 61.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 63.Chong S.Y., Cutler S., Lin J.-J., Tsai C.-H., Tsai H.-K., Biggins S., Tsukiyama T., Lo Y.-C., Kao C.-F. H3K4 methylation at active genes mitigates transcription-replication conflicts during replication stress. Nat. Commun. 2020;11:1–16. doi: 10.1038/s41467-020-14595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sims R.J., Reinberg D. Histone H3 Lys 4 methylation: Caught in a bind? Gene Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 65.Martin B.J., McBurney K.L., Maltby V.E., Jensen K.N., Brind’Amour J., Howe L.J. Histone H3K4 and H3K36 methylation independently recruit the NuA3 histone acetyltransferase in Saccharomyces cerevisiae. Genetics. 2017;205:1113–1123. doi: 10.1534/genetics.116.199422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim T., Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramakrishnan S., Pokhrel S., Palani S., Pflueger C., Parnell T.J., Cairns B.R., Bhaskara S., Chandrasekharan M.B. Counteracting H3K4 methylation modulators Set1 and Jhd2 co-regulate chromatin dynamics and gene transcription. Nat. Commun. 2016;7:1–16. doi: 10.1038/ncomms11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krogan N.J., Dover J., Khorrami S., Greenblatt J.F., Schneider J., Johnston M., Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 69.Nislow C., Ray E., Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiner A., Chen H.V., Liu C.L., Rahat A., Klien A., Soares L., Gudipati M., Pfeffner J., Regev A., Buratowski S. Systematic dissection of roles for chromatin regulators in a yeast stress response. Plos Biol. 2012;10 doi: 10.1371/journal.pbio.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Margaritis T., Oreal V., Brabers N., Maestroni L., Vitaliano-Prunier A., Benschop J.J., van Hooff S., van Leenen D., Dargemont C., Geli V. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. Plos Genet. 2012;8 doi: 10.1371/journal.pgen.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nadal-Ribelles M., Mas G., Millán-Zambrano G., Solé C., Ammerer G., Chávez S., Posas F., de Nadal E. H3K4 monomethylation dictates nucleosome dynamics and chromatin remodeling at stress-responsive genes. Nucleic Acids Res. 2015;43:4937–4949. doi: 10.1093/nar/gkv220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang S.-S., Zhou B.O., Zhou J.-Q. Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol. Cell. Biol. 2011;31:3171–3181. doi: 10.1128/MCB.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beilharz T.H., Harrison P.F., Miles D.M., See M.M., Le U.M.M., Kalanon M., Curtis M.J., Hasan Q., Saksouk J., Margaritis T. Coordination of cell cycle progression and mitotic spindle assembly involves histone H3 lysine 4 methylation by Set1/COMPASS. Genetics. 2017;205:185–199. doi: 10.1534/genetics.116.194852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faucher D., Wellinger R.J. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. Plos Genet. 2010;6 doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hérissant L., Moehle E.A., Bertaccini D., Van Dorsselaer A., Schaeffer-Reiss C., Guthrie C., Dargemont C. H2B ubiquitylation modulates spliceosome assembly and function in budding yeast. Biol. Cell. 2014;106:126–138. doi: 10.1111/boc.201400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walter D., Matter A., Fahrenkrog B. Loss of histone H3 methylation at lysine 4 triggers apoptosis in Saccharomyces cerevisiae. Plos Genet. 2014;10 doi: 10.1371/journal.pgen.1004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ng H.H., Robert F., Young R.A., Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 79.South P.F., Harmeyer K.M., Serratore N.D., Briggs S.D. H3K4 methyltransferase Set1 is involved in maintenance of ergosterol homeostasis and resistance to Brefeldin A. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1016–E1025. doi: 10.1073/pnas.1215768110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Briggs S.D., Xiao T., Sun Z.-W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D. Trans-histone regulatory pathway in chromatin. Nature. 2002;418 doi: 10.1038/nature00970. 498-498. [DOI] [PubMed] [Google Scholar]
- 81.Dover J., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 82.Sun Z.-W., Allis C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 83.Kim J., Guermah M., McGinty R.K., Lee J.-S., Tang Z., Milne T.A., Shilatifard A., Muir T.W., Roeder R.G. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim J., Kim J.-A., McGinty R.K., Nguyen U.T., Muir T.W., Allis C.D., Roeder R.G. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol. Cell. 2013;49:1121–1133. doi: 10.1016/j.molcel.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee J.-S., Shukla A., Schneider J., Swanson S.K., Washburn M.P., Florens L., Bhaumik S.R., Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 86.Mikheyeva I.V., Grady P.J., Tamburini F.B., Lorenz D.R., Cam H.P. Multifaceted genome control by Set1 dependent and independent of H3K4 methylation and the Set1C/COMPASS complex. Plos Genet. 2014;10 doi: 10.1371/journal.pgen.1004740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kirmizis A., Santos-Rosa H., Penkett C.J., Singer M.A., Vermeulen M., Mann M., Bähler J., Green R.D., Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakanishi S., Sanderson B.W., Delventhal K.M., Bradford W.D., Staehling-Hampton K., Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guillemette B., Drogaris P., Lin H.-H.S., Armstrong H., Hiragami-Hamada K., Imhof A., Bonneil E., Thibault P., Verreault A., Festenstein R.J. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. Plos Genet. 2011;7 doi: 10.1371/journal.pgen.1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmähling S., Meiler A., Lee Y., Mohammed A., Finkl K., Tauscher K., Israel L., Wirth M., Philippou-Massier J., Blum H. Regulation and function of H3K36 di-methylation by the trithorax-group protein complex AMC. Development. 2018;145 doi: 10.1242/dev.163808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Venkatesh S., Smolle M., Li H., Gogol M.M., Saint M., Kumar S., Natarajan K., Workman J.L. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 92.Bell O., Wirbelauer C., Hild M., Scharf A.N., Schwaiger M., MacAlpine D.M., Zilbermann F., Van Leeuwen F., Bell S.P., Imhof A. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 2007;26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rao B., Shibata Y., Strahl B.D., Lieb J.D. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wozniak G.G., Strahl B.D. Hitting the ‘mark’: Interpreting lysine methylation in the context of active transcription. B.B.A. Gene Regul. Mech. 2014;1839:1353–1361. doi: 10.1016/j.bbagrm.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 95.Sein H., Värv S., Kristjuhan A. Distribution and maintenance of histone H3 lysine 36 trimethylation in transcribed locus. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lerner A.M., Hepperla A.J., Keele G.R., Meriesh H.A., Yumerefendi H., Restrepo D., Zimmerman S., Bear J.E., Kuhlman B., Davis I.J. An optogenetic switch for the Set2 methyltransferase provides evidence for transcription-dependent and-independent dynamics of H3K36 methylation. Genome Res. 2020;30:1605–1617. doi: 10.1101/gr.264283.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.-J., Anderson S., Yates J., Washburn M.P. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 98.Joshi A.A., Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 99.Keogh M.-C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 100.Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J.L. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 101.Govind C.K., Qiu H., Ginsburg D.S., Ruan C., Hofmeyer K., Hu C., Swaminathan V., Workman J.L., Li B., Hinnebusch A.G. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C (S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li B., Jackson J., Simon M.D., Fleharty B., Gogol M., Seidel C., Workman J.L., Shilatifard A. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J. Biol. Chem. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Youdell M.L., Kizer K.O., Kisseleva-Romanova E., Fuchs S.M., Duro E., Strahl B.D., Mellor J. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 2008;28:4915–4926. doi: 10.1128/MCB.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wagner E.J., Carpenter P.B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell. Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xie L., Pelz C., Wang W., Bashar A., Varlamova O., Shadle S., Impey S. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang P., Du J., Sun B., Dong X., Xu G., Zhou J., Huang Q., Liu Q., Hao Q., Ding J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maltby V.E., Martin B.J., Schulze J.M., Johnson I., Hentrich T., Sharma A., Kobor M.S., Howe L. Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Mol. Cell. Biol. 2012;32:3479–3485. doi: 10.1128/MCB.00389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smolle M., Venkatesh S., Gogol M.M., Li H., Zhang Y., Florens L., Washburn M.P., Workman J.L. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012;19:884. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gilbert T.M., McDaniel S.L., Byrum S.D., Cades J.A., Dancy B.C., Wade H., Tackett A.J., Strahl B.D., Taverna S.D. A PWWP domain-containing protein targets the NuA3 acetyltransferase complex via histone H3 lysine 36 trimethylation to coordinate transcriptional elongation at coding regions. Mol. Cell. Proteomics. 2014;13:2883–2895. doi: 10.1074/mcp.M114.038224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shi X., Kachirskaia I., Walter K.L., Kuo J.-H.A., Lake A., Davrazou F., Chan S.M., Martin D.G., Fingerman I.M., Briggs S.D. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martin D.G., Grimes D.E., Baetz K., Howe L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol. Cell. Biol. 2006;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jha D.K., Strahl B.D. An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat. Commun. 2014;5:1–13. doi: 10.1038/ncomms4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pryde F., Jain D., Kerr A., Curley R., Mariotti F.R., Vogelauer M. H3 k36 methylation helps determine the timing of cdc45 association with replication origins. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morselli M., Pastor W.A., Montanini B., Nee K., Ferrari R., Fu K., Bonora G., Rubbi L., Clark A.T., Ottonello S. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. eLife. 2015;4 doi: 10.7554/eLife.06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Leung C.S., Douglass S.M., Morselli M., Obusan M.B., Pavlyukov M.S., Pellegrini M., Johnson T.L. H3K36 methylation and the chromodomain protein Eaf3 are required for proper cotranscriptional spliceosome assembly. Cell Rep. 2019;27:3760–3769. doi: 10.1016/j.celrep.2019.05.100. e3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sorenson M.R., Jha D.K., Ucles S.A., Flood D.M., Strahl B.D., Stevens S.W., Kress T.L. Histone H3K36 methylation regulates pre-mRNA splicing in Saccharomyces cerevisiae. RNA Biol. 2016;13:412–426. doi: 10.1080/15476286.2016.1144009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ryu H.-Y., Rhie B.-H., Ahn S.H. Loss of the Set2 histone methyltransferase increases cellular lifespan in yeast cells. Biochem. Biophys. Res. Co. 2014;446:113–118. doi: 10.1016/j.bbrc.2014.02.061. [DOI] [PubMed] [Google Scholar]
- 118.Sen P., Dang W., Donahue G., Dai J., Dorsey J., Cao X., Liu W., Cao K., Perry R., Lee J.Y. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Gene Dev. 2015;29:1362–1376. doi: 10.1101/gad.263707.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Venkatesh S., Li H., Gogol M.M., Workman J.L. Selective suppression of antisense transcription by Set2-mediated H3K36 methylation. Nat. Commun. 2016;7:1–14. doi: 10.1038/ncomms13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fuchs S.M., Kizer K.O., Braberg H., Krogan N.J., Strahl B.D. RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di-and trimethylation at lysine 36. J. Biol. Chem. 2012;287:3249–3256. doi: 10.1074/jbc.M111.273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Morris S.A., Rao B., Garcia B.A., Hake S.B., Diaz R.L., Shabanowitz J., Hunt D.F., Allis C.D., Lieb J.D., Strahl B.D. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 2007;282:7632–7640. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pai C.-C., Deegan R.S., Subramanian L., Gal C., Sarkar S., Blaikley E.J., Walker C., Hulme L., Bernhard E., Codlin S. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 2014;5:1–11. doi: 10.1038/ncomms5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Du H.-N., Fingerman I.M., Briggs S.D. Histone H3 K36 methylation is mediated by a trans-histone methylation pathway involving an interaction between Set2 and histone H4. Gene Dev. 2008;22:2786–2798. doi: 10.1101/gad.1700008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Du H.-N., Briggs S.D. A nucleosome surface formed by histone H4, H2A, and H3 residues is needed for proper histone H3 Lys36 methylation, histone acetylation, and repression of cryptic transcription. J. Biol. Chem. 2010;285:11704–11713. doi: 10.1074/jbc.M109.085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Batta K., Zhang Z., Yen K., Goffman D.B., Pugh B.F. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Gene Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wyce A., Xiao T., Whelan K.A., Kosman C., Walter W., Eick D., Hughes T.R., Krogan N.J., Strahl B.D., Berger S.L. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 127.Nelson C.J., Santos-Rosa H., Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 128.Ng H.H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Gene Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van Leeuwen F., Gafken P.R., Gottschling D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 130.Ng H.H., Ciccone D.N., Morshead K.B., Oettinger M.A., Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shahbazian M.D., Zhang K., Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 132.Farooq Z., Banday S., Pandita T.K., Altaf M. The many faces of histone H3K79 methylation. Mutat. Res. Rev. Mutat. 2016;768:46–52. doi: 10.1016/j.mrrev.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Botuyan M.V., Lee J., Ward I.M., Kim J.-E., Thompson J.R., Chen J., Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Norris A., Boeke J.D. Silent information regulator 3: The Goldilocks of the silencing complex. Gene Dev. 2010;24:115–122. doi: 10.1101/gad.1865510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Altaf M., Utley R.T., Lacoste N., Tan S., Briggs S.D., Côté J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Onishi M., Liou G.-G., Buchberger J.R., Walz T., Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 137.Ng H.H., Xu R.-M., Zhang Y., Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 138.Fingerman I.M., Li H.-C., Briggs S.D. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: Identification of a new trans-histone pathway. Gene Dev. 2007;21:2018–2029. doi: 10.1101/gad.1560607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kitada T., Kuryan B.G., Tran N.N.H., Song C., Xue Y., Carey M., Grunstein M. Mechanism for epigenetic variegation of gene expression at yeast telomeric heterochromatin. Gene Dev. 2012;26:2443–2455. doi: 10.1101/gad.201095.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wysocki R., Javaheri A., Allard S., Sha F., Côté J., Kron S.J. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lazzaro F., Sapountzi V., Granata M., Pellicioli A., Vaze M., Haber J.E., Plevani P., Lydall D., Muzi-Falconi M. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huyen Y., Zgheib O., DiTullio R.A., Jr., Gorgoulis V.G., Zacharatos P., Petty T.J., Sheston E.A., Mellert H.S., Stavridi E.S., Halazonetis T.D. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 143.Ontoso D., Acosta I., van Leeuwen F., Freire R., San-Segundo P.A. Dot1-dependent histone H3K79 methylation promotes activation of the Mek1 meiotic checkpoint effector kinase by regulating the Hop1 adaptor. Plos Genet. 2013;9 doi: 10.1371/journal.pgen.1003262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.San-Segundo P.A., Roeder G.S. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell. 2000;11:3601–3615. doi: 10.1091/mbc.11.10.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bostelman L.J., Keller A.M., Albrecht A.M., Arat A., Thompson J.S. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair. 2007;6:383–395. doi: 10.1016/j.dnarep.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 146.Giannattasio M., Lazzaro F., Plevani P., Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 147.Nguyen A.T., Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Gene Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhu B., Zheng Y., Pham A.-D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 149.van Welsem T., Korthout T., Ekkebus R., Morais D., Molenaar T.M., van Harten K., Poramba-Liyanage D.W., Sun S.M., Lenstra T.L., Srivas R. Dot1 promotes H2B ubiquitination by a methyltransferase-independent mechanism. Nucleic Acids Res. 2018;46:11251–11261. doi: 10.1093/nar/gky801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vlaming H., McLean C.M., Korthout T., Alemdehy M.F., Hendriks S., Lancini C., Palit S., Klarenbeek S., Kwesi-Maliepaard E.M., Molenaar T.M. Conserved crosstalk between histone deacetylation and H3K79 methylation generates DOT1L-dose dependency in HDAC1-deficient thymic lymphoma. EMBO J. 2019;38 doi: 10.15252/embj.2019101564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Van Aller G.S., Reynoird N., Barbash O., Huddleston M., Liu S., Zmoos A.-F., McDevitt P., Sinnamon R., Le B., Mas G. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics. 2012;7:340–343. doi: 10.4161/epi.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hamamoto R., Silva F.P., Tsuge M., Nishidate T., Katagiri T., Nakamura Y., Furukawa Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–118. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang S.-z., Luo X.-g., Shen J., Zou J.-n., Lu Y.-h., Xi T. Knockdown of SMYD3 by RNA interference inhibits cervical carcinoma cell growth and invasion in vitro. BMB Rep. 2008;41:294–299. doi: 10.5483/bmbrep.2008.41.4.294. [DOI] [PubMed] [Google Scholar]
- 154.Green E.M., Morrison A.J., Gozani O. New marks on the block: Set5 methylates H4 lysines 5, 8 and 12. Nucleus. 2012;3:335–339. doi: 10.4161/nucl.20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bird A.W., David Y.Y., Pray-Grant M.G., Qiu Q., Harmon K.E., Megee P.C., Grant P.A., Smith M.M., Christman M.F. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 156.Dion M.F., Altschuler S.J., Wu L.F., Rando O.J. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ma X.-J., Wu J., Altheim B.A., Schultz M.C., Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Allard S., Utley R.T., Savard J., Clarke A., Grant P., Brandl C.J., Pillus L., Workman J.L., Côté J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Clarke A.S., Lowell J.E., Jacobson S.J., Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Allfrey V.G., Mirsky A.E. Structural modifications of histones and their possible role in the regulation of RNA synthesis. Science. 1964;144 doi: 10.1126/science.144.3618.559. 559-559. [DOI] [PubMed] [Google Scholar]
- 161.Rea S., Eisenhaber F., O'carroll D., Strahl B.D., Sun Z.-W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 162.Peters A.H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 163.Guo H.-B., Guo H. Mechanism of histone methylation catalyzed by protein lysine methyltransferase SET7/9 and origin of product specificity. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8797–8802. doi: 10.1073/pnas.0702981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cheng X., Collins R.E., Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu. Rev. Biophys. Biomol. Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lanouette S., Mongeon V., Figeys D., Couture J.F. The functional diversity of protein lysine methylation. Mol. Syst. Biol. 2014;10:724. doi: 10.1002/msb.134974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Feng Q., Wang H., Ng H.H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 167.Lacoste N., Utley R.T., Hunter J.M., Poirier G.G., Côté J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 168.Corda Y., Schramke V., Longhese M.P., Smokvina T., Paciotti V., Brevet V., Gilson E., Géli V. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat. Genet. 1999;21:204–208. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- 169.Schramke V., Neecke H., Brevet V., Corda Y., Lucchini G., Longhese M.P., Gilson E., Géli V. The set1Δ mutation unveils a novel signaling pathway relayed by the Rad53-dependent hyperphosphorylation of replication protein A that leads to transcriptional activation of repair genes. Gene Dev. 2001;15:1845–1858. doi: 10.1101/gad.193901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Miller T., Krogan N.J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., Greenblatt J.F., Shilatifard A. Compass: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shilatifard A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Thornton J.L., Westfield G.H., Takahashi Y.-h., Cook M., Gao X., Woodfin A.R., Lee J.-S., Morgan M.A., Jackson J., Smith E.R. Context dependency of Set1/COMPASS-mediated histone H3 Lys4 trimethylation. Gene Dev. 2014;28:115–120. doi: 10.1101/gad.232215.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Trésaugues L., Dehé P.-M., Guérois R., Rodriguez-Gil A., Varlet I., Salah P., Pamblanco M., Luciano P., Quevillon-Cheruel S., Sollier J. Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J. Mol. Biol. 2006;359:1170–1181. doi: 10.1016/j.jmb.2006.04.050. [DOI] [PubMed] [Google Scholar]
- 174.Fingerman I.M., Wu C.-L., Wilson B.D., Briggs S.D. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:28761–28765. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schlichter A., Cairns B.R. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J. 2005;24:1222–1231. doi: 10.1038/sj.emboj.7600607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Luciano P., Jeon J., El-Kaoutari A., Challal D., Bonnet A., Barucco M., Candelli T., Jourquin F., Lesage P., Kim J. Binding to RNA regulates Set1 function. Cell Discov. 2017;3:1–19. doi: 10.1038/celldisc.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Soares L.M., Buratowski S. Yeast Swd2 is essential because of antagonism between Set1 histone methyltransferase complex and APT (associated with Pta1) termination factor. J. Biol. Chem. 2012;287:15219–15231. doi: 10.1074/jbc.M112.341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bae H.J., Dubarry M., Jeon J., Soares L.M., Dargemont C., Kim J., Geli V., Buratowski S. The Set1 N-terminal domain and Swd2 interact with RNA polymerase II CTD to recruit COMPASS. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-16082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mersman D.P., Du H.-N., Fingerman I.M., South P.F., Briggs S.D. Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. J. Biol. Chem. 2012;287:2652–2665. doi: 10.1074/jbc.M111.280867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Dehé P.-M., Dichtl B., Schaft D., Roguev A., Pamblanco M., Lebrun R., Rodríguez-Gil A., Mkandawire M., Landsberg K., Shevchenko A. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J. Biol. Chem. 2006;281:35404–35412. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- 181.Dehe P.-M., Geli V. The multiple faces of Set1. Biochem. Cell. Biol. 2006;84:536–548. doi: 10.1139/o06-081. [DOI] [PubMed] [Google Scholar]
- 182.Schneider J., Wood A., Lee J.-S., Schuster R., Dueker J., Maguire C., Swanson S.K., Florens L., Washburn M.P., Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 183.Takahashi Y.-H., Shilatifard A. Structural basis for H3K4 trimethylation by yeast Set1/COMPASS. Adv. Enzyme Regul. 2010;50:104–111. doi: 10.1016/j.advenzreg.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hsu P.L., Li H., Lau H.-T., Leonen C., Dhall A., Ong S.-E., Chatterjee C., Zheng N. Crystal structure of the COMPASS H3K4 methyltransferase catalytic module. Cell. 2018;174:1106–1116.e1109. doi: 10.1016/j.cell.2018.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Qu Q., Takahashi Y.-h., Yang Y., Hu H., Zhang Y., Brunzelle J.S., Couture J.-F., Shilatifard A., Skiniotis G. Structure and conformational dynamics of a COMPASS histone H3K4 methyltransferase complex. Cell. 2018;174:1117–1126.e1112. doi: 10.1016/j.cell.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Roguev A., Schaft D., Shevchenko A., Pijnappel W.P., Wilm M., Aasland R., Stewart A.F. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Takahashi Y.-h., Westfield G.H., Oleskie A.N., Trievel R.C., Shilatifard A., Skiniotis G. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20526–20531. doi: 10.1073/pnas.1109360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Choudhury R., Singh S., Arumugam S., Roguev A., Stewart A.F. The Set1 complex is dimeric and acts with Jhd2 demethylation to convey symmetrical H3K4 trimethylation. Gene Dev. 2019;33:550–564. doi: 10.1101/gad.322222.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Boa S., Coert C., Patterton H.G. Saccharomyces cerevisiae Set1p is a methyltransferase specific for lysine 4 of histone H3 and is required for efficient gene expression. Yeast. 2003;20:827–835. doi: 10.1002/yea.995. [DOI] [PubMed] [Google Scholar]
- 190.Pinskaya M., Gourvennec S., Morillon A. H3 lysine 4 di-and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang K., Lin W., Latham J.A., Riefler G.M., Schumacher J.M., Chan C., Tatchell K., Hawke D.H., Kobayashi R., Dent S.Y. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wu M., Wang P.F., Lee J.S., Martin-Brown S., Florens L., Washburn M., Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Jeon J., McGinty R.K., Muir T.W., Kim J.-A., Kim J. Crosstalk among Set1 complex subunits involved in H2B ubiquitylation-dependent H3K4 methylation. Nucleic Acids Res. 2018;46:11129–11143. doi: 10.1093/nar/gky920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Corden J.L. Tails of RNA polymerase II. Trends Biochem. Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 195.Komarnitsky P., Cho E.-J., Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Gene Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Licatalosi D.D., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 197.Gong X., Yu Q., Duan K., Tong Y., Zhang X., Mei Q., Lu L., Yu X., Li S. Histone acetyltransferase Gcn5 regulates gene expression by promoting the transcription of histone methyltransferase SET1. B.B.A. Gene Regul. Mech. 2020;1863:194603. doi: 10.1016/j.bbagrm.2020.194603. [DOI] [PubMed] [Google Scholar]
- 198.Strahl B.D., Grant P.A., Briggs S.D., Sun Z.-W., Bone J.R., Caldwell J.A., Mollah S., Cook R.G., Shabanowitz J., Hunt D.F. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.McDaniel S.L., Strahl B.D. Shaping the cellular landscape with Set2/SETD2 methylation. Cell. Mol. Life Sci. 2017;74:3317–3334. doi: 10.1007/s00018-017-2517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang Y., Niu Y., Li B. Balancing acts of SRI and an auto-inhibitory domain specify Set2 function at transcribed chromatin. Nucleic Acids Res. 2015;43:4881–4892. doi: 10.1093/nar/gkv393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Vojnic E., Simon B., Strahl B.D., Sattler M., Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J. Biol. Chem. 2006;281:13–15. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- 202.Kizer K.O., Phatnani H.P., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Macias M.J., Gervais V., Civera C., Oschkinat H. Structural analysis of WW domains and design of a WW prototype. Nat. Struct. Biol. 2000;7:375–379. doi: 10.1038/75144. [DOI] [PubMed] [Google Scholar]
- 204.Xiao T., Hall H., Kizer K.O., Shibata Y., Hall M.C., Borchers C.H., Strahl B.D. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Gene Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Faber P.W., Barnes G.T., Srinidhi J., Chen J., Gusella J.F., MacDonald M.E. Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 206.Gao Y.-G., Yang H., Zhao J., Jiang Y.-J., Hu H.-Y. Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin. Structure. 2014;22:378–386. doi: 10.1016/j.str.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 207.Jha D.K., Pfister S.X., Humphrey T.C., Strahl B.D. SET-ting the stage for DNA repair. Nat. Struct. Mol. Biol. 2014;21:655–657. doi: 10.1038/nsmb.2866. [DOI] [PubMed] [Google Scholar]
- 208.Li B., Gogol M., Carey M., Pattenden S.G., Seidel C., Workman J.L. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Gene Dev. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lickwar C.R., Rao B., Shabalin A.A., Nobel A.B., Strahl B.D., Lieb J.D. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hacker K.E., Fahey C.C., Shinsky S.A., Chiang Y.-C.J., DiFiore J.V., Jha D.K., Vo A.H., Shavit J.A., Davis I.J., Strahl B.D. Structure/function analysis of recurrent mutations in SETD2 protein reveals a critical and conserved role for a SET domain residue in maintaining protein stability and histone H3 Lys-36 trimethylation. J. Biol. Chem. 2016;291:21283–21295. doi: 10.1074/jbc.M116.739375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Carvalho S., Raposo A.C., Martins F.B., Grosso A.R., Sridhara S.C., Rino J., Carmo-Fonseca M., de Almeida S.F. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 2013;41:2881–2893. doi: 10.1093/nar/gks1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gopalakrishnan R., Marr S.K., Kingston R.E., Winston F. A conserved genetic interaction between Spt6 and Set2 regulates H3K36 methylation. Nucleic Acids Res. 2019;47:3888–3903. doi: 10.1093/nar/gkz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lee C.-H., Wu J., Li B. Chromatin remodelers fine-tune H3K36me-directed deacetylation of neighbor nucleosomes by Rpd3S. Mol. Cell. 2013;52:255–263. doi: 10.1016/j.molcel.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kaczmarek Michaels K., Mohd Mostafa S., Ruiz Capella J., Moore C.L. Regulation of alternative polyadenylation in the yeast Saccharomyces cerevisiae by histone H3K4 and H3K36 methyltransferases. Nucleic Acids Res. 2020;48:5407–5425. doi: 10.1093/nar/gkaa292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Krogan N.J., Kim M., Tong A., Golshani A., Cagney G., Richards D.P., Beattie B.K., Emili A., Boone C., Shilatifard A. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Li J., Moazed D., Gygi S.P. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 217.Schaft D., Roguev A., Kotovic K.M., Shevchenko A., Sarov M., Shevchenko A., Neugebauer K.M., Stewart A.F. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 2003;31:2475–2482. doi: 10.1093/nar/gkg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chu Y., Simic R., Warner M.H., Arndt K.M., Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 2007;26:4646–4656. doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chu Y., Sutton A., Sternglanz R., Prelich G. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 2006;26:3029–3038. doi: 10.1128/MCB.26.8.3029-3038.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Liu Y., Zhang Y., Xue H., Cao M., Bai G., Mu Z., Yao Y., Sun S., Fang D., Huang J. Cryo-EM structure of SETD2/Set2 methyltransferase bound to a nucleosome containing oncohistone mutations. Cell Discov. 2021;7:1–12. doi: 10.1038/s41421-021-00261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bilokapic S., Halic M. Nucleosome and ubiquitin position Set2 to methylate H3K36. Nat. Commun. 2019;10:1–9. doi: 10.1038/s41467-019-11726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Singer M.S., Kahana A., Wolf A.J., Meisinger L.L., Peterson S.E., Goggin C., Mahowald M., Gottschling D.E. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jones B., Su H., Bhat A., Lei H., Bajko J., Hevi S., Baltus G.A., Kadam S., Zhai H., Valdez R. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Shanower G.A., Muller M., Blanton J.L., Honti V., Gyurkovics H., Schedl P. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics. 2005;169:173–184. doi: 10.1534/genetics.104.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Frederiks F., Tzouros M., Oudgenoeg G., Van Welsem T., Fornerod M., Krijgsveld J., Van Leeuwen F. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat. Struct. Mol. Biol. 2008;15:550. doi: 10.1038/nsmb.1432. [DOI] [PubMed] [Google Scholar]
- 226.Stulemeijer I.J., De Vos D., Van Harten K., Joshi O.K., Blomberg O., Van Welsem T., Terweij M., Vlaming H., De Graaf E.L., Altelaar A.M. Dot1 histone methyltransferases share a distributive mechanism but have highly diverged catalytic properties. Sci. Rep. 2015;5:1–11. doi: 10.1038/srep09824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.De Vos D., Frederiks F., Terweij M., Van Welsem T., Verzijlbergen K.F., Iachina E., De Graaf E.L., Maarten Altelaar A., Oudgenoeg G., Heck A.J. Progressive methylation of ageing histones by Dot1 functions as a timer. EMBO Rep. 2011;12:956–962. doi: 10.1038/embor.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kwon T., Chang J.H., Kwak E., Lee C.W., Joachimiak A., Kim Y.C., Lee J.W., Cho Y. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9—AdoMet. EMBO J. 2003;22:292–303. doi: 10.1093/emboj/cdg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Patnaik D., Chin H.G., Esteve P.-O., Benner J., Jacobsen S.E., Pradhan S. Substrate specificity and kinetic mechanism of mammalian G9a histone H3 methyltransferase. J. Biol. Chem. 2004;279:53248–53258. doi: 10.1074/jbc.M409604200. [DOI] [PubMed] [Google Scholar]
- 230.Sawada K., Yang Z., Horton J.R., Collins R.E., Zhang X., Cheng X. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J. Biol. Chem. 2004;279:43296–43306. doi: 10.1074/jbc.M405902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Schubert H.L., Blumenthal R.M., Cheng X. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.van Welsem T., Frederiks F., Verzijlbergen K.F., Faber A.W., Nelson Z.W., Egan D.A., Gottschling D.E., van Leeuwen F. Synthetic lethal screens identify gene silencing processes in yeast and implicate the acetylated amino terminus of Sir3 in recognition of the nucleosome core. Mol. Cell. Biol. 2008;28:3861–3872. doi: 10.1128/MCB.02050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ismail M.B., Shinohara M., Shinohara A. Dot1-dependent histone H3K79 methylation promotes the formation of meiotic double-strand breaks in the absence of histone H3K4 methylation in budding yeast. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tatum D., Li S. Evidence that the histone methyltransferase Dot1 mediates global genomic repair by methylating histone H3 on lysine 79. J. Biol. Chem. 2011;286:17530–17535. doi: 10.1074/jbc.M111.241570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Janzen C.J., Hake S.B., Lowell J.E., Cross G.A. Selective di-or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol. Cell. 2006;23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 236.Kim W., Choi M., Kim J.-E. The histone methyltransferase Dot1/DOT1L as a critical regulator of the cell cycle. Cell Cycle. 2014;13:726–738. doi: 10.4161/cc.28104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lee S., Oh S., Jeong K., Jo H., Choi Y., Seo H.D., Kim M., Choe J., Kwon C.S., Lee D. Dot1 regulates nucleosome dynamics by its inherent histone chaperone activity in yeast. Nat. Commun. 2018;9:1–14. doi: 10.1038/s41467-017-02759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Schulze J.M., Jackson J., Nakanishi S., Gardner J.M., Hentrich T., Haug J., Johnston M., Jaspersen S.L., Kobor M.S., Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Oh S., Jeong K., Kim H., Kwon C.S., Lee D. A lysine-rich region in Dot1p is crucial for direct interaction with H2B ubiquitylation and high level methylation of H3K79. Biochem. Biophys. Res. Co. 2010;399:512–517. doi: 10.1016/j.bbrc.2010.07.100. [DOI] [PubMed] [Google Scholar]
- 240.Weake V.M., Workman J.L. Histone ubiquitination: Triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 241.Carmen A.A., Milne L., Grunstein M. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 2002;277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- 242.Millar C., Kurdistani S., Grunstein M. Acetylation of yeast histone H4 lysine 16: A switch for protein interactions in heterochromatin and euchromatin. Cold Spring Harb. Sym. 2004;69:193–200. doi: 10.1101/sqb.2004.69.193. [DOI] [PubMed] [Google Scholar]
- 243.Valencia-Sánchez M.I., De Ioannes P., Wang M., Truong D.M., Lee R., Armache J.-P., Boeke J.D., Armache K.-J. Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation. Science. 2021;371 doi: 10.1126/science.abc6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Calpena E., Palau F., Espinos C., Galindo M.I. Evolutionary history of the Smyd gene family in metazoans: A framework to identify the orthologs of human Smyd genes in Drosophila and other animal species. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Spellmon N., Holcomb J., Trescott L., Sirinupong N., Yang Z. Structure and function of SET and MYND domain-containing proteins. Int. J. Mol. Sci. 2015;16:1406–1428. doi: 10.3390/ijms16011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Jaiswal D., Turniansky R., Moresco J.J., Ikram S., Ramaprasad G., Akinwole A., Wolf J., Yates J.R., Green E.M. Function of the MYND domain and C-terminal region in regulating the subcellular localization and catalytic activity of the SMYD family lysine methyltransferase Set5. Mol. Cell. Biol. 2020;40 doi: 10.1128/MCB.00341-19. e00341-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Martín G.M., King D.A., Green E.M., Garcia-Nieto P.E., Alexander R., Collins S.R., Krogan N.J., Gozani O.P., Morrison A.J. Set5 and Set1 cooperate to repress gene expression at telomeres and retrotransposons. Epigenetics. 2014;9:513–522. doi: 10.4161/epi.27645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jezek M., Gast A., Choi G., Kulkarni R., Quijote J., Graham-Yooll A., Park D., Green E.M. The histone methyltransferases Set5 and Set1 have overlapping functions in gene silencing and telomere maintenance. Epigenetics. 2017;12:93–104. doi: 10.1080/15592294.2016.1265712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lamour J., Wan C., Zhang M., Zhao X., Den Haan R. Overexpression of endogenous stress-tolerance related genes in Saccharomyces cerevisiae improved strain robustness and production of heterologous cellobiohydrolase. F.E.M.S. Yeast Res. 2019;19 doi: 10.1093/femsyr/foz035. foz035. [DOI] [PubMed] [Google Scholar]
- 250.Zhang M.M., Zhao X.Q., Cheng C., Bai F.W. Improved growth and ethanol fermentation of Saccharomyces cerevisiae in the presence of acetic acid by overexpression of SET5 and PPR1. Biotechnol. J. 2015;10:1903–1911. doi: 10.1002/biot.201500508. [DOI] [PubMed] [Google Scholar]
- 251.Jaiswal D., Turniansky R., Green E.M. Choose your own adventure: The role of histone modifications in yeast cell fate. J. Mol. Biol. 2017;429:1946–1957. doi: 10.1016/j.jmb.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 253.Shi Y.-J., Matson C., Lan F., Iwase S., Baba T., Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 254.Kim T., Buratowski S. Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. J. Biol. Chem. 2007;282:20827–20835. doi: 10.1074/jbc.M703034200. [DOI] [PubMed] [Google Scholar]
- 255.Klose R.J., Kallin E.M., Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 256.Tsukada Y.-I., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 257.Kwon D.-W., Ahn S.H. Role of yeast JmjC-domain containing histone demethylases in actively transcribed regions. Biochem. Biophys. Res. Co. 2011;410:614–619. doi: 10.1016/j.bbrc.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 258.Tu S., Bulloch E.M., Yang L., Ren C., Huang W.-C., Hsu P.-H., Chen C.-H., Liao C.-L., Yu H.-M., Lo W.-S. Identification of histone demethylases in Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:14262–14271. doi: 10.1074/jbc.M609900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Fang J., Hogan G.J., Liang G., Lieb J.D., Zhang Y. The Saccharomyces cerevisiae histone demethylase Jhd1 fine-tunes the distribution of H3K36me2. Mol. Cell. Biol. 2007;27:5055–5065. doi: 10.1128/MCB.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Huang F., Chandrasekharan M.B., Chen Y.-C., Bhaskara S., Hiebert S.W., Sun Z.-W. The JmjN domain of Jhd2 is important for its protein stability, and the plant homeodomain (PHD) finger mediates its chromatin association independent of H3K4 methylation. J. Biol. Chem. 2010;285:24548–24561. doi: 10.1074/jbc.M110.117333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ryu H.-Y., Ahn S.H. Yeast histone H3 lysine 4 demethylase Jhd2 regulates mitotic ribosomal DNA condensation. B.M.C. Biol. 2014;12:1–16. doi: 10.1186/s12915-014-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Klose R.J., Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell. Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 263.Lee K.Y., Ranger M., Meneghini M.D. Combinatorial genetic control of Rpd3S through histone H3K4 and H3K36 methylation in budding Yeast. G3 Genes Genom. Genet. 2018;8:3411–3420. doi: 10.1534/g3.118.200589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ha S.D., Ham S., Kim M.Y., Kim J.H., Jang I., Lee B.B., Lee M.K., Hwang J.-T., Roh T.-Y., Kim T. Transcription-dependent targeting of Hda1C to hyperactive genes mediates H4-specific deacetylation in yeast. Nat. Commun. 2019;10:1–14. doi: 10.1038/s41467-019-12077-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Klose R.J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 266.Liang G., Klose R.J., Gardner K.E., Zhang Y. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat. Struct. Mol. Biol. 2007;14:243–245. doi: 10.1038/nsmb1204. [DOI] [PubMed] [Google Scholar]
- 267.Ingvarsdottir K., Edwards C., Lee M.G., Lee J.S., Schultz D.C., Shilatifard A., Shiekhattar R., Berger S.L. Histone H3 K4 demethylation during activation and attenuation of GAL1 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:7856–7864. doi: 10.1128/MCB.00801-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Mersman D.P., Du H.-N., Fingerman I.M., South P.F., Briggs S.D. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Gene Dev. 2009;23:951–962. doi: 10.1101/gad.1769209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Huang F., Ramakrishnan S., Pokhrel S., Pflueger C., Parnell T.J., Kasten M.M., Currie S.L., Bhachech N., Horikoshi M., Graves B.J. Interaction of the Jhd2 histone H3 Lys-4 demethylase with chromatin is controlled by histone H2A surfaces and restricted by H2B ubiquitination. J. Biol. Chem. 2015;290:28760–28777. doi: 10.1074/jbc.M115.693085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Seward D.J., Cubberley G., Kim S., Schonewald M., Zhang L., Tripet B., Bentley D.L. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat. Struct. Mol. Biol. 2007;14:240–242. doi: 10.1038/nsmb1200. [DOI] [PubMed] [Google Scholar]
- 271.Lee K.Y., Chen Z., Jiang R., Meneghini M.D. H3K4 methylation dependent and independent chromatin regulation by JHD2 and SET1 in budding yeast. G3 Genes Genom. Genet. 2018;8:1829–1839. doi: 10.1534/g3.118.200151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Osborne E.A., Dudoit S., Rine J. The establishment of gene silencing at single-cell resolution. Nat. Genet. 2009;41:800–806. doi: 10.1038/ng.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Xu M., Soloveychik M., Ranger M., Schertzberg M., Shah Z., Raisner R., Venkatasubrahmanyan S., Tsui K., Gebbia M., Hughes T. Timing of transcriptional quiescence during gametogenesis is controlled by global histone H3K4 demethylation. Dev. Cell. 2012;23:1059–1071. doi: 10.1016/j.devcel.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Verzijlbergen K.F., Marston A.L. A JARID family demethylase controls differentiation timing through global effects on transcription. Mol. Cell. 2012;48:489–490. doi: 10.1016/j.molcel.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 275.Ünal E., Amon A. Gamete formation resets the aging clock in yeast. Cold Spring Harb. Sym. 2011;76:73–80. doi: 10.1101/sqb.2011.76.011379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Blair L.P., Liu Z., Labitigan R.L.D., Wu L., Zheng D., Xia Z., Pearson E.L., Nazeer F.I., Cao J., Lang S.M. KDM5 lysine demethylases are involved in maintenance of 3′ UTR length. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1501662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Maltby V.E., Martin B.J., Brind’Amour J., Chruscicki A.T., McBurney K.L., Schulze J.M., Johnson I.J., Hills M., Hentrich T., Kobor M.S. Histone H3K4 demethylation is negatively regulated by histone H3 acetylation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2012;109:18505–18510. doi: 10.1073/pnas.1202070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Jang Y.K., Wang L., Sancar G.B. Are damage-responsive GIS1 and RPH1. Mol. Cell. Biol. 1999;19:7630. doi: 10.1128/mcb.19.11.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Klose R.J., Gardner K.E., Liang G., Erdjument-Bromage H., Tempst P., Zhang Y. Demethylation of histone H3K36 and H3K9 by Rph1: A vestige of an H3K9 methylation system in Saccharomyces cerevisiae? Mol. Cell. Biol. 2007;27:3951–3961. doi: 10.1128/MCB.02180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Liang C.-Y., Wang L.-C., Lo W.-S. Dissociation of the H3K36 demethylase Rph1 from chromatin mediates derepression of environmental stress-response genes under genotoxic stress in Saccharomyces cerevisiae. Mol. Biol. Cell. 2013;24:3251–3262. doi: 10.1091/mbc.E12-11-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Chang Y., Wu J., Tong X.-J., Zhou J.-Q., Ding J. Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: Insights into the substrate specificity and catalytic mechanism. Biochem. J. 2011;433:295–302. doi: 10.1042/BJ20101418. [DOI] [PubMed] [Google Scholar]
- 282.Westholm J.O., Tronnersjö S., Nordberg N., Olsson I., Komorowski J., Ronne H. Gis1 and Rph1 regulate glycerol and acetate metabolism in glucose depleted yeast cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Liang C.-Y., Hsu P.-H., Chou D.-F., Pan C.-Y., Wang L.-C., Huang W.-C., Tsai M.-D., Lo W.-S. The histone H3K36 demethylase Rph1/KDM4 regulates the expression of the photoreactivation gene PHR1. Nucleic Acids Res. 2011;39:4151–4165. doi: 10.1093/nar/gkr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Nordberg N., Olsson I., Carlsson M., Hu G.-Z., Westholm J.O., Ronne H. The histone demethylase activity of Rph1 is not essential for its role in the transcriptional response to nutrient signaling. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Li F., Zheng L.-D., Chen X., Zhao X., Briggs S.D., Du H.-N. Gcn5-mediated Rph1 acetylation regulates its autophagic degradation under DNA damage stress. Nucleic Acids Res. 2017;45:5183–5197. doi: 10.1093/nar/gkx129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Venters B.J., Wachi S., Mavrich T.N., Andersen B.E., Jena P., Sinnamon A.J., Jain P., Rolleri N.S., Jiang C., Hemeryck-Walsh C. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Steunou A.-L., Cramet M., Rossetto D., Aristizabal M.J., Lacoste N., Drouin S., Côté V., Paquet E., Utley R.T., Krogan N. Combined action of histone reader modules regulates NuA4 local acetyltransferase function but not its recruitment on the genome. Mol. Cell. Biol. 2016;36:2768–2781. doi: 10.1128/MCB.00112-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.McCauley B.S., Dang W. Histone methylation and aging: Lessons learned from model systems. B.B.A. Gene Regul. Mech. 2014;1839:1454–1462. doi: 10.1016/j.bbagrm.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Mohammad K., Dakik P., Medkour Y., McAuley M., Mitrofanova D., Titorenko V.I. Some metabolites act as second messengers in yeast chronological aging. Int. J. Mol. Sci. 2018;19:860. doi: 10.3390/ijms19030860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Bernard A., Jin M., González-Rodríguez P., Füllgrabe J., Delorme-Axford E., Backues S.K., Joseph B., Klionsky D.J. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr. Biol. 2015;25:546–555. doi: 10.1016/j.cub.2014.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Eapen V.V., Waterman D.P., Bernard A., Schiffmann N., Sayas E., Kamber R., Lemos B., Memisoglu G., Ang J., Mazella A. A pathway of targeted autophagy is induced by DNA damage in budding yeast. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1158–E1167. doi: 10.1073/pnas.1614364114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Balciunas D., Ronne H. Yeast genes GIS1–4: Multicopy suppressors of the Gal− phenotype of snf1 mig1 srb8/10/11 cells. Mol. Gen. Genet. 1999;262:589–599. doi: 10.1007/s004380051121. [DOI] [PubMed] [Google Scholar]
- 293.Quan Z., Oliver S.G., Zhang N. JmjN interacts with JmjC to ensure selective proteolysis of Gis1 by the proteasome. Microbiology. 2011;157:2694–2701. doi: 10.1099/mic.0.048199-0. [DOI] [PubMed] [Google Scholar]
- 294.Lal S., Comer J.M., Konduri P.C., Shah A., Wang T., Lewis A., Shoffner G., Guo F., Zhang L. Heme promotes transcriptional and demethylase activities of Gis1, a member of the histone demethylase JMJD2/KDM4 family. Nucleic Acids Res. 2018;46:215–228. doi: 10.1093/nar/gkx1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Oshiro J., Han G.-S., Iwanyshyn W.M., Conover K., Carman G.M. Regulation of the yeast DPP1-encoded diacylglycerol pyrophosphate phosphatase by transcription factor Gis1p. J. Biol. Chem. 2003;278:31495–31503. doi: 10.1074/jbc.M305452200. [DOI] [PubMed] [Google Scholar]
- 296.Pedruzzi I., Bürckert N., Egger P., De Virgilio C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 2000;19:2569–2579. doi: 10.1093/emboj/19.11.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Tronnersjö S., Hanefalk C., Balciunas D., Hu G.-Z., Nordberg N., Murén E., Ronne H. The jmjN and jmjC domains of the yeast zinc finger protein Gis1 interact with 19 proteins involved in transcription, sumoylation and DNA repair. Mol. Genet. Genomics. 2007;277:57–70. doi: 10.1007/s00438-006-0171-3. [DOI] [PubMed] [Google Scholar]
- 298.Zhang N., Wu J., Oliver S.G. Gis1 is required for transcriptional reprogramming of carbon metabolism and the stress response during transition into stationary phase in yeast. Microbiology. 2009;155:1690–1698. doi: 10.1099/mic.0.026377-0. [DOI] [PubMed] [Google Scholar]
- 299.Cameroni E., Hulo N., Roosen J., Winderickx J., Virgilio C.D. The novel yeast PAS kinase Rim15 orchestrates G0-associated antioxidant defense mechanisms. Cell Cycle. 2004;3:460–466. [PubMed] [Google Scholar]
- 300.Yu Y., Neiman A.M., Sternglanz R. The JmjC domain of Gis1 is dispensable for transcriptional activation. F.E.M.S. Yeast Res. 2010;10:793–801. doi: 10.1111/j.1567-1364.2010.00680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Beltrao P., Albanèse V., Kenner L.R., Swaney D.L., Burlingame A., Villén J., Lim W.A., Fraser J.S., Frydman J., Krogan N.J. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., Pedrioli P.G., Gerrits B., Picotti P., Lam H., Vitek O. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Chen S.-h., Albuquerque C.P., Liang J., Suhandynata R.T., Zhou H. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J. Biol. Chem. 2010;285:12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Chi A., Huttenhower C., Geer L.Y., Coon J.J., Syka J.E., Bai D.L., Shabanowitz J., Burke D.J., Troyanskaya O.G., Hunt D.F. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Gnad F., de Godoy L.M., Cox J., Neuhauser N., Ren S., Olsen J.V., Mann M. High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics. 2009;9:4642–4652. doi: 10.1002/pmic.200900144. [DOI] [PubMed] [Google Scholar]
- 307.Helbig A.O., Rosati S., Pijnappel P.W., van Breukelen B., Timmers M.H., Mohammed S., Slijper M., Heck A.J. Perturbation of the yeast N-acetyltransferase NatB induces elevation of protein phosphorylation levels. BMC Genomics. 2010;11:1–15. doi: 10.1186/1471-2164-11-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Henriksen P., Wagner S.A., Weinert B.T., Sharma S., Bačinskaja G., Rehman M., Juffer A.H., Walther T.C., Lisby M., Choudhary C. Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2012;11:1510–1522. doi: 10.1074/mcp.M112.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Holt L.J., Tuch B.B., Villén J., Johnson A.D., Gygi S.P., Morgan D.O. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., Gerrits B., Aebersold R., Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Gene Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Iesmantavicius V., Weinert B.T., Choudhary C. Convergence of ubiquitylation and phosphorylation signaling in rapamycin-treated yeast cells. Mol. Cell. Proteomics. 2014;13:1979–1992. doi: 10.1074/mcp.O113.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Lanz M.C., Yugandhar K., Gupta S., Sanford E.J., Faça V.M., Vega S., Joiner A.M., Fromme J.C., Yu H., Smolka M.B. In-depth and 3-dimensional exploration of the budding yeast phosphoproteome. EMBO Rep. 2021;22 doi: 10.15252/embr.202051121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Li J., Paulo J.A., Nusinow D.P., Huttlin E.L., Gygi S.P. Investigation of proteomic and phosphoproteomic responses to signaling network perturbations reveals functional pathway Organizations in yeast. Cell Rep. 2019;29:2092–2104. doi: 10.1016/j.celrep.2019.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Li X., Gerber S.A., Rudner A.D., Beausoleil S.A., Haas W., Villén J., Elias J.E., Gygi S.P. Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 315.Separovich R.J., Wong M.W., Chapman T.R., Slavich E., Hamey J.J., Wilkins M.R. Post-translational modification analysis of Saccharomyces cerevisiae histone methylation enzymes reveals phosphorylation sites of regulatory potential. J. Biol. Chem. 2020;296:100192. doi: 10.1074/jbc.RA120.015995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Smolka M.B., Albuquerque C.P., Chen S.-h., Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Soulard A., Cremonesi A., Moes S., Schütz F., Jenö P., Hall M.N. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell. 2010;21:3475–3486. doi: 10.1091/mbc.E10-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Swaney D.L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N.J., Villén J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tong Z., Kim M.-S., Pandey A., Espenshade P.J. Identification of candidate substrates for the Golgi Tul1 E3 ligase using quantitative diGly proteomics in yeast. Mol. Cell. Proteomics. 2014;13:2871–2882. doi: 10.1074/mcp.M114.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Weinert B.T., Iesmantavicius V., Moustafa T., Schölz C., Wagner S.A., Magnes C., Zechner R., Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 2014;10:833. doi: 10.15252/msb.156513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Winter D.L., Hart-Smith G., Wilkins M.R. Characterization of protein methyltransferases Rkm1, Rkm4, Efm4, Efm7, Set5 and Hmt1 reveals extensive post-translational modification. J. Mol. Biol. 2018;430:102–118. doi: 10.1016/j.jmb.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 322.Consortium U. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Sadowski I., Breitkreutz B.-J., Stark C., Su T.-C., Dahabieh M., Raithatha S., Bernhard W., Oughtred R., Dolinski K., Barreto K. The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database: Version 2.0 update. Database. 2013;2013 doi: 10.1093/database/bat026. bat026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Ledesma L., Sandoval E., Cruz-Martínez U., Escalante A.M., Mejía S., Moreno-Álvarez P., Ávila E., García E., Coello G., Torres-Quiroz F. Yaam: Yeast amino acid modifications database. Database. 2018;2018 doi: 10.1093/database/bax099. bax099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Khoury G.A., Baliban R.C., Floudas C.A. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci. Rep. 2011;1:1–5. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Ardito F., Giuliani M., Perrone D., Troiano G., Lo Muzio L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int. J. Mol. Med. 2017;40:271–280. doi: 10.3892/ijmm.2017.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Singh V., Ram M., Kumar R., Prasad R., Roy B.K., Singh K.K. Phosphorylation: Implications in cancer. Protein J. 2017;36:1–6. doi: 10.1007/s10930-017-9696-z. [DOI] [PubMed] [Google Scholar]
- 328.Tsai C.-J., Del Sol A., Nussinov R. Protein allostery, signal transmission and dynamics: A classification scheme of allosteric mechanisms. Mol. Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Ye C., Sutter B.M., Wang Y., Kuang Z., Zhao X., Yu Y., Tu B.P. Demethylation of the protein phosphatase PP2A promotes demethylation of histones to enable their function as a methyl group sink. Mol. Cell. 2019;73:1115–1126. doi: 10.1016/j.molcel.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Bontron S., Jaquenoud M., Vaga S., Talarek N., Bodenmiller B., Aebersold R., De Virgilio C. Yeast endosulfines control entry into quiescence and chronological life span by inhibiting protein phosphatase 2A. Cell Rep. 2013;3:16–22. doi: 10.1016/j.celrep.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 331.Drazic A., Myklebust L.M., Ree R., Arnesen T. The world of protein acetylation. B.B.A. Proteins Proteom. 2016;1864:1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 332.Narita T., Weinert B.T., Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell. Biol. 2019;20:156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 333.Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 334.Olia A.S., Barker K., McCullough C.E., Tang H.-Y., Speicher D.W., Qiu J., LaBaer J., Marmorstein R. Nonenzymatic protein acetylation detected by NAPPA protein arrays. A.C.S. Chem. Biol. 2015;10:2034–2047. doi: 10.1021/acschembio.5b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Pickart C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 336.Hicke L. Gettin’ down with ubiquitin: Turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- 337.Haglund K., Di Fiore P.P., Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 2003;28:598–604. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 338.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 339.Dronamraju R., Jha D.K., Eser U., Adams A.T., Dominguez D., Choudhury R., Chiang Y.-C., Rathmell W.K., Emanuele M.J., Churchman L.S. Set2 methyltransferase facilitates cell cycle progression by maintaining transcriptional fidelity. Nucleic Acids Res. 2018;46:1331–1344. doi: 10.1093/nar/gkx1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Nielsen M.L., Vermeulen M., Bonaldi T., Cox J., Moroder L., Mann M. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat. Methods. 2008;5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 341.Carlson S.M., Moore K.E., Sankaran S.M., Reynoird N., Elias J.E., Gozani O. A proteomic strategy identifies lysine methylation of splicing factor snRNP70 by the SETMAR enzyme. J. Biol. Chem. 2015;290:12040–12047. doi: 10.1074/jbc.M115.641530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Mas-y-Mas S., Barbon M., Teyssier C., Déméné H., Carvalho J.E., Bird L.E., Lebedev A., Fattori J., Schubert M., Dumas C. The human mixed lineage leukemia 5 (MLL5), a sequentially and structurally divergent SET domain-containing protein with no intrinsic catalytic activity. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Mazur P.K., Reynoird N., Khatri P., Jansen P.W., Wilkinson A.W., Liu S., Barbash O., Van Aller G.S., Huddleston M., Dhanak D. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510:283–287. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Wilkinson A.W., Diep J., Dai S., Liu S., Ooi Y.S., Song D., Li T.-M., Horton J.R., Zhang X., Liu C. SETD3 is an actin histidine methyltransferase that prevents primary dystocia. Nature. 2019;565:372–376. doi: 10.1038/s41586-018-0821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Liu W., Li L., Ye H., Chen H., Shen W., Zhong Y., Tian T., He H. From Saccharomyces cerevisiae to human: The important gene co-expression modules. Biomed. Rep. 2017;7:153–158. doi: 10.3892/br.2017.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Ghaemmaghami S., Huh W.-K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 347.The Alliance of Genome Resources Consortium Alliance of genome Resources Portal: Unified model organism research platform. Nucleic Acids Res. 2020;48:D650–D658. doi: 10.1093/nar/gkz813. [DOI] [PMC free article] [PubMed] [Google Scholar]