Summary
Hyperimmune immunoglobulin (hCoV-2IG) generated from SARS-CoV-2 convalescent plasma (CP) are under evaluation in clinical trials. Here we explored the antibody epitope repertoire, and virus neutralizing capacity of six hCoV-2IG batches as well as nine CP against SARS-CoV-2 and emerging variants of concern (VOCs). Epitope-mapping by gene-fragment phage display library spanning the SARS-CoV-2 spike demonstrated broad recognition of multiple antigenic sites spanning the entire spike that was higher for hCoV-2IG than CP, with predominant binding to the fusion peptide. In the pseudovirus neutralization assay and in the wild-type SARS-CoV-2 PRNT assay, hCoV-2IG lots showed higher titers against the WA-1 strain compared with CP. Neutralization of VOCs were reduced to different extent by hCoV-2IG lots but were higher than CP. Significant reduction of hCoV-2IG binding was observed to RBD-E484K followed by RBD-N501Y (but not RBD-K417N). This study suggests that post-exposure treatment with hCoV-2IG could be preferable to CP.
Subject areas: biological sciences, immunology, immune response
Graphical abstract
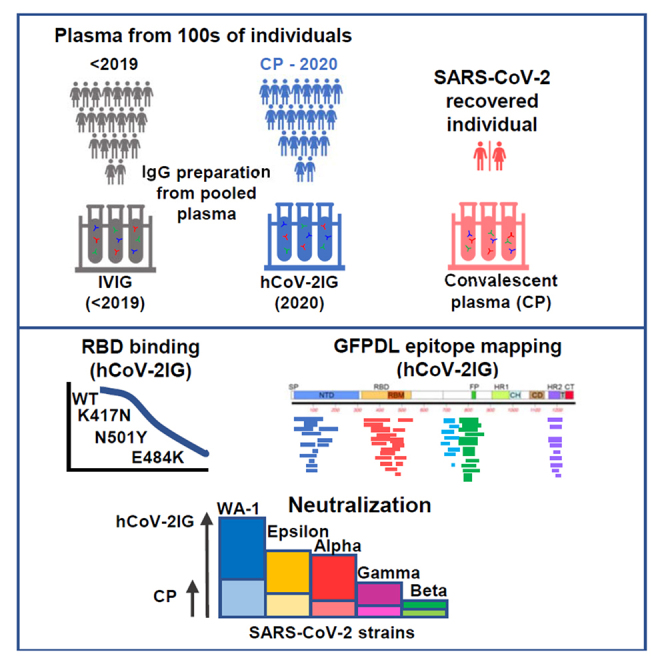
Highlights
-
•
SARS-CoV-2 hCoV-2IG demonstrate highly diverse antibody epitope profile
-
•
SARS-CoV-2 hCoV-2IG lots neutralized SARS-CoV-2 variants better than CP
-
•
Significant reduction of hCoV-2IG binding to RBD-E484K compared with unmutated RBD
-
•
Higher hCoV-2IG dose would be required for SARS-CoV-2 variant infected patients
Biological sciences; Immunology; Immune response
Introduction
An expedited access to treatment of patients with COVID-19 with convalescent plasma was issued by FDA via Emergency Use Authorization on August 23, 2020. Additional studies, including randomized, controlled trials, have provided data to further inform the safety and efficacy of COVID-19 convalescent plasma. Based on assessment of these data, potential clinical benefit of transfusion of COVID-19 convalescent plasma in hospitalized patients with COVID-19 is associated with high neutralizing titer units administered early in the course of disease (Casadevall et al., 2020; Joyner et al., 2021).
Intravenous immunoglobulins (IVIGs) are a more concentrated form of IgG preparations fractionated from large number of plasma units that are prescreened for the presence of high titer anti-spike antibodies and predetermined SARS-CoV-2 neutralization titers (Vandeberg et al., 2021). Several hyperimmune immunoglobulin (hCoV-2IG) lots are currently being evaluated in clinical trials. The effectiveness of hCoV-2 IG products may be hampered by evolving SARS-CoV-2 and the emergence of new variants with high transmissibility rates and mutations in the Receptor Binding Domain (RBD) which are less susceptible to antibodies from recovered COVID-19 patients. The main variants of concern (VOCs) are the B.1.1.7 spreading from the UK, the B1.351 spreading in South Africa (SA), and the P.1 that appeared in northeast Brazil and found in Japan (JP). In the US, several variants were identified recently including California (CA) variant B.1.429 (Kupferschmidt, 2021a, 2021b, 2021c; McCormick et al., 2021; Wibmer et al., 2021). While the impact of VOCs on post-infection sera and post-vaccination antibodies have been evaluated in several studies, the impact of such VOCs on therapeutic hCoV-2IG for treatment of COVID-19 is unknown (Zhou et al., 2021a) (Casadevall et al., 2021a; Ho et al., 2021; Wang et al., 2021b; Wibmer et al., 2021; Wu et al., 2021b).
The phage display technique is suitable to display properly folded and conformationally dependent proteins, as it has been widely used for display of large functionally active antibodies, enzymes, hormones, and viral and mammalian proteins. We have adapted this gene-fragment phage display library (GFPDL) technology for unbiased, comprehensive analyses of post-infection and post-vaccination antibody epitope repertoires for multiple viral pathogens including SARS-CoV-2, Ebola virus, highly pathogenic avian influenza virus, respiratory syncytial virus and Zika virus (Fuentes et al., 2016; Khurana et al., 2009, 2018; Ravichandran et al., 2020).
In the current study we probed the antibody quality of 6 hCoV-2IG products. In addition to using SARS-CoV-2-spike-GFPDL, Surface Plasmon Resonance (SPR) was used to measure antibody binding to SARS-CoV-2 spike protein receptor binding domain (RBD) representing WA-1 as well RBD mutants engineered to express key amino acid mutations of the VOCs. Neutralization capacity of the hCoV-2IG lots against the SARS-CoV-2 WA-1 strain and several VOC (CA, UK, JP, SA) was measured in a pseudovirion neutralization assay (PsVNA) as well as a classical PRNT assay. For comparison with hCoV-2IG, we evaluated nine convalescent plasma samples from recovered COVID-19 patients and 16 IVIG preparations that were manufactured with pre-pandemic plasma units prior to August 2019.
Results
SARS-CoV-2 spike antibody epitope repertoires of six hCoV-2IG batches
The spike protein is the antigen of choice for development of vaccines and therapeutics against SARS-CoV-2. To decipher the epitope-specificity of the SARS-CoV-2 spike-specific antibodies in an unbiased manner, we subjected the six hCoV-2IG lots to antibody epitope profiling with a highly diverse SARS-CoV-2 spike GFPDL with >107.1 unique phage clones displaying epitopes of 18–500 amino acid residues across the SARS-CoV-2 spike. In preliminary studies to characterize the SARS-CoV-2 spike GFPDL, epitope-mapping of monoclonal antibodies (MAbs) targeting SARS-CoV-2 spike or RBD identified the expected linear or conformation-dependent epitopes recognized by these MAbs. Recently, we showed that SARS-CoV-2 spike GFPDL can recognize both linear, conformational and neutralizing epitopes in the post-vaccination sera of rabbits and post-SARS-CoV-2 infection sera in adults and the elderly. Adsorption of immune sera with the SARS-CoV-2-S-GFPDL removed >90% of the anti-spike binding antibody in post-infection human sera (Ravichandran et al., 2020, 2021; Tang et al., 2021).
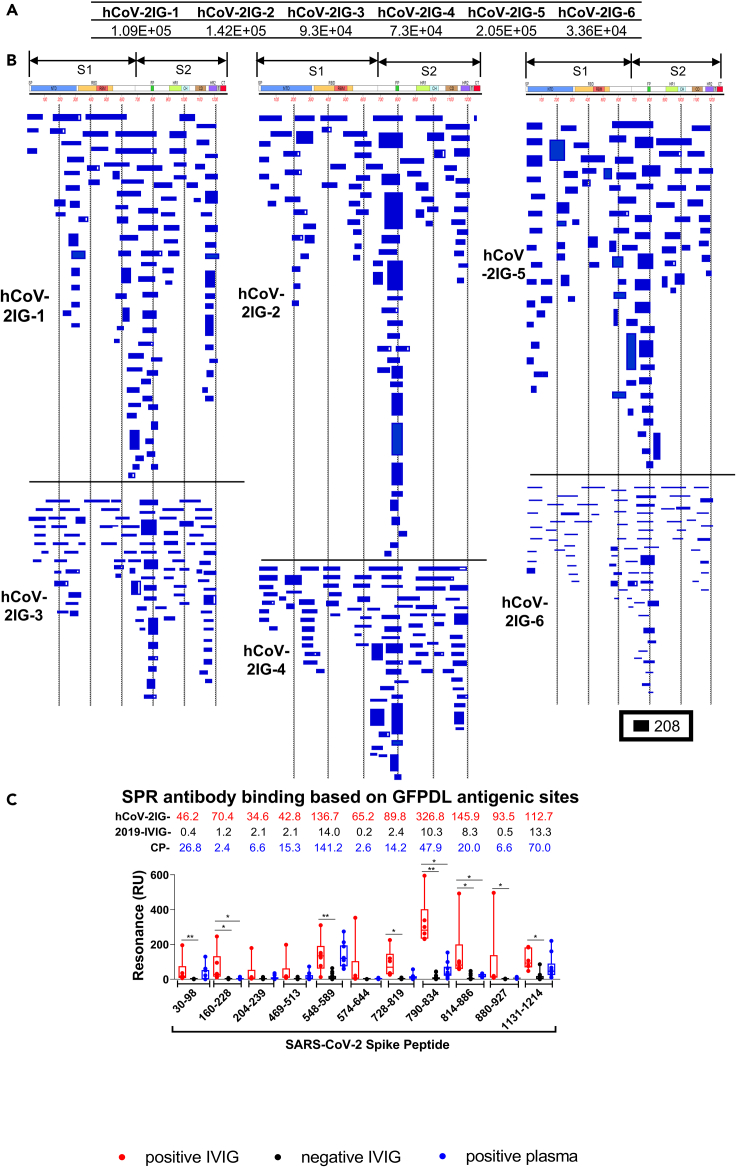
Six hCoV-2IG lots were used for SARS-CoV-2 GFPDL based epitope mapping as previously described (Fuentes et al., 2016, 2020; Khurana et al., 2009, 2018; Ravichandran et al., 2020). Similar numbers of phages were bound by IgG of each of these individual hCoV-2IG batches (3.4 × 104–2.1 × 105) (Figure 1A). The bound phages demonstrated a diverse epitope repertoire spanning the entire SARS-CoV-2 spike protein including N-terminal domain (NTD) and RBD in S1, and the fusion peptide (FP), β-rich connector domain (CD), heptad repeat 1 (HR1) and 2 (HR2) in S2 (Figures 1B and S1). Peptides representing the key immunodominant antigenic sites identified by GFPDL analysis were chemically synthesized and used to evaluate binding of each of the six hCoV-2IG batches, 16 pre-pandemic 2019-IVIG lots and 9 COVID-19 convalescent plasma (Figure 1C). The 2019-IVIG demonstrated minimal to no binding to the SARS-CoV-2 spike peptides. In aggregate, the convalescent plasma showed lower binding to epitopes spanning the entire spike in comparison with the hCoV-2IG (Figure 1C). In agreement with GFPDL analysis, the hCoV-2IG demonstrated highest antibody binding to the spike peptide 790–834 that contains the fusion peptide sequence (residues 788–806). Most of the GFPDL-identified antigenic site sequences recognized by hCoV-2IGs are conserved in the spike protein of various SARS-CoV-2 VOC (Figure S2 and Table S1). Together, the GFPDL analysis demonstrated that hCoV-2IGs contained very broad anti-spike antibody responses, targeting sites in NTD, RBD, C-terminal of S1, FP, that mapped to both the S1 and S2 spike domains.
Figure 1.
SARS-CoV-2 spike antibody epitope repertoires recognized by hCoV-2IG
SARS-CoV-2 spike GFPDL analyses of IgG antibodies in six batches of hCoV-2IG.
(A) Number of IgG bound phage clones selected using SARS-CoV-2 spike GFPDL for each of the six individual lots of hCoV-2IG (hCoV-2IG-1 to hCoV-2IG-6).
(B) Epitope repertoires of IgG antibody in hCoV-2IG batches and their alignment to the spike protein of SARS-CoV-2. Graphical distribution of representative clones with a frequency of >2, obtained after affinity selection, are shown. The horizontal position and the length of the bars indicate the alignment of peptide sequence displayed on the selected phage clone to its homologous sequence in the SARS-CoV-2 spike. The thickness of each bar represents the frequency of repetitively isolated phage. Scale value is shown enclosed in a black box beneath the alignments. The GFPDL affinity selection data was performed in duplicate (two independent experiments by researcher in the lab, who was blinded to sample identity), and a similar number of phage clones and epitope repertoire was observed in both phage display analysis.
(C) SPR binding of hCOV-2IG (n = 6; in red), control pre-pandemic 2019-IVIG (n = 16; in black) and convalescent plasma (n = 9; in blue) with SARS-CoV-2 spike antigenic site peptides identified using GFPDL analysis in Figure 1B. The amino acid designation is based on the SARS-CoV-2 spike protein sequence (Figure S1). Total antibody binding is represented in maximum resonance units (RU) in this figure for 10-fold serum dilution of CP, and 1mg/mL of 2019-IVIG or hCoV-2IG. The numbers above the peptides show the mean value for each respective group antibody binding to the peptide and is color-coded (6 hCOV-2IG in red, 16 2019-IVIG in black, and 9 CPs in blue). All SPR experiments were performed twice and the researchers performing the assay were blinded to sample identity. The variations for duplicate runs of SPR was <4%. The data shown are average values of two experimental runs. The statistical significances between the hCoV-2IG vs 2019-IVIG vs CP for antibody binding to each peptide were performed using multiple group comparisons by non-parametric (Kruskal-Wallis) statistical test using Dunn's post-hoc analysis in GraphPad prism. The differences were considered statistically significant with a 95% confidence interval when the p value was less than 0.05. (∗, p values of ≤0.05, ∗∗, p values of ≤0.01).
Neutralization capacity of CP and hCoV-2IG against the SARS-CoV-2 WA-1 and B.1.429, B1.1.7, P.1, B.1.351 VOCs
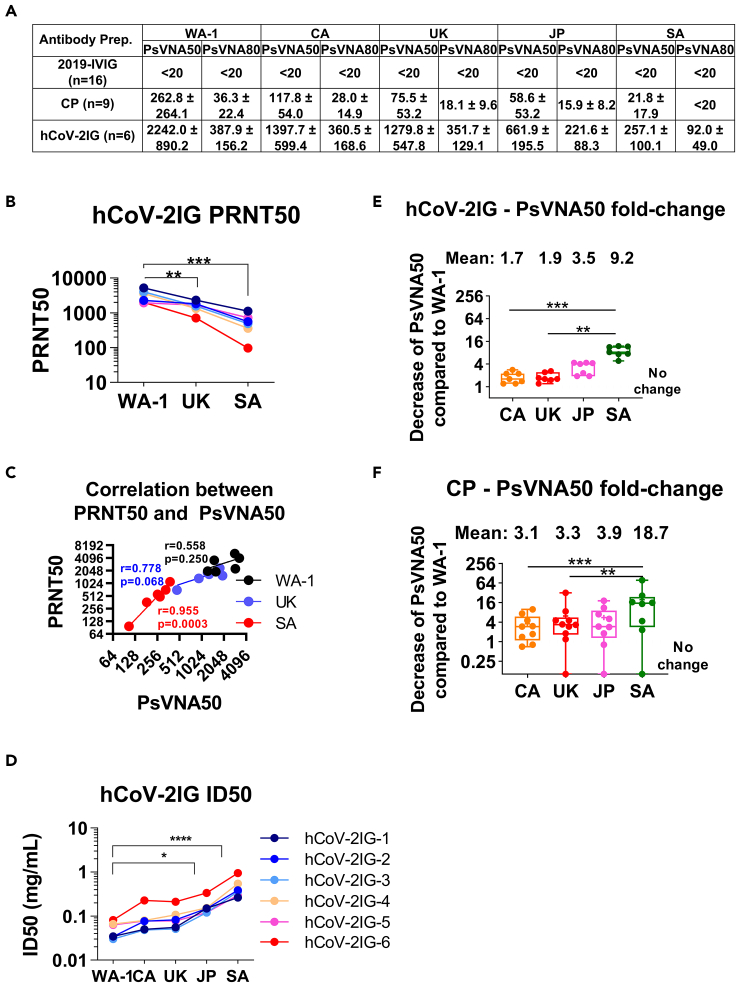
PsVNA was used to measure the neutralization activity for each of the six hCoV-2 IG, nine convalescent plasma (CP), and 16 pre-pandemic 2019-IVIG lots against the predominant SARS-CoV-2 WA-1 strain and the VOCs currently spreading around the globe; U.S./CA (B.1.429), UK (B.1.1.7), JP (P.1), and SA (B.1.351). Both 50% (PsVNA50) and 80% (PsVNA80) neutralization titers were calculated. The spike proteins mutations in the VOCs used for production of the pseudovirions are shown in Table S2.
All sixteen pre-pandemic 2019-IVIG preparations demonstrated titers of <20 PsVNA50 against SARS-CoV-2 strains (Figure 2A and Table S3). Among the nine CP lots tested against WA-1, variable PsVNA50 titers were observed, including one negative, one low (<1:80), six medium (≥1:160 < 1:640) and two high (>1:640). In contrast, all six hCoV-2IG lots exhibited high PsVNA50 titers against WA-1 ranging between 1:1,239-1:3,309. PsVNA80 titers for hCoV-2IG ranged between 1:168–1:593, but none of the CP lots showed PsVNA80 titers above 1:80 (range <20 to 1:74) against WA-1 (Table S3). Neutralization of the VOCs showed variable loss of titers as determined by either PsVNA50 or PsVNA80 for the hCoV-2IG and the CPs with the greatest reduction in titers measured against the SA VOC (Figures 2A, and Table S3).
Figure 2.
Neutralizing antibody titers and RBD binding antibodies of convalescent plasma and hCoV-2IG against various SARS-CoV-2 strains
(A) SARS-CoV-2 neutralizing antibody titers in each of the CP, 2019-IVIG and hCoV-2IG preparations as determined by pseudovirus neutralization assay in 293-ACE2-TMPRSS2 cells with SARS-CoV-2 WA-1 strain, CA variant (B.1.429), UK variant (B.1.1.7), JP variant (P.1) or SA variant (B.1.351). PsVNA50 (50% neutralization titer) and PsVNA80 (80% neutralization titer) titers for each of the control pre-pandemic 2019-IVIG (n = 16), convalescent plasma (n = 9) and hCoV-2IG (n = 6) were calculated with GraphPad prism version 8. Data show mean values ±SEM for PsVNA50 and PsVNA80 titers for each of the 3 antibody groups against the SARS-CoV-2 WA-1, CA, UK, JP and SA variants. If all CP lots or 2019-IVIG lots have titer <20, then they are shown as titer of <20. If any of the CP or 2019-IVIG showed PsVNA50 or PsVNA80 titers >20, then the average values are shown for the group against specific SARS-CoV-2 strain.
(B) End-point virus neutralization titers for six hCoV-2IG lots (different colors) using wild type authentic SARS-CoV-2 WA-1, UK and SA virus strains in a classical BSL3 neutralization assay based on a plaque assay was performed as described in Materials and Methods.
(C) Pearson two-tailed correlations are reported for the calculation of correlation of PRNT50 titers against wild-type SARS-CoV-2 strains (WA-1, UK or SA) and PsVNA50 titers against corresponding pseudovirions expressing either WA-1, UK or SA spike in pseudovirion neutralization assays for the six hCOV-2IG lots.
(D) Antibody concentration (in mg/mL) required for each of the six hCoV-2IG batches to achieve 50% neutralization of SARS-CoV-2 WA-1, CA, UK, JP or SA variants in PsVNA. Only the WA-1 strain had a significant difference compared to the JP and the SA strain.
(E and F) The fold-decrease in PsVNA50 neutralization titers against emerging variant strain CA (B.1.429), UK (B.1.1.7), JP (P.1) and SA (B.1.351) for six hCoV-2IG lots (E) and nine CP lots (F) in comparison with the SARS-CoV-2 WA-1 strain is shown. The numbers above the group indicates the mean fold-change for each variant.
For confirmation of the PsVNA neutralization titers, the six hCoV-2IG lots were also evaluated in a classical PRNT assay using VERO-E6 cells against authentic SARS-CoV-2 viruses representing WA-1 (USA-WA1/2020), UK-B1.1.7 (hCoV-19/England/204820464/2020), and SA-B1.351 (South Africa/KRISP-K005325/2020) strains (Figures 2B, and Table S4). Wild-type JP and CA strains were not available for the PRNT assay. Correlation of hCoV-2IG neutralization titers between PRNT50 and PsVNA50 were observed, as well as a similar decline in neutralization titers against the UK and SA VOC compared with WA-1 strain (Tables S3 and S4 and Figures 2B and 2C). Since all hCoV-2IG lots contain 100 mg/mL of IgG, it allowed calculation of ID50 values for the six hCoV-2IG lots against different SARS-CoV-2 strains (Table S5 and Figure 2D).
Compared to the WA-1 strain, the average PsVNA50 of the hCoV-2IG against CA, UK, JP and SA VOC were reduced by 1.7, 1.9, 3.5, and 9.2-fold respectively (Figure 2E). Since the amount of SARS-CoV-2 specific IgG in CP lots is more variable and 5–10 fold lower compared with the hyperimmune hCoV-2IG, the CPs exhibited greater loss of neutralizing activities against the variants in comparison with hCoV-2IG. The average PsVNA50 of the CP against the CA, UK, JP, and SA VOC were reduced by 3.1, 3.3, 3.9, and 18.7-fold, respectively (Table S3 and Figure 2F). PsVNA80 titers against the UK and JP VOC for all 9 CPs were lower (∼2-fold) and were minimal or negligible against the SA VOC (Figures 2A and Table S3). For hCoV-2IG, the PsVNA80 titers were similar for the WA-1, CA and UK strains, reduced by 1.75-fold against the JP variant, and decreased by 4.3-fold for SA VOC (Figure 2A).
Antibody binding of hCoV-2IG batches to RBD and RBD mutants: K417N, N501Y, and E484K
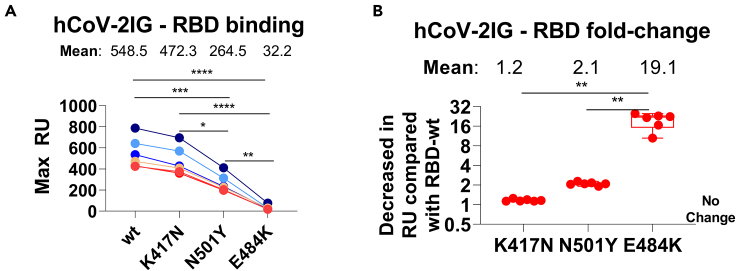
Many of the mutations in the spike protein of the different SARS-CoV-2 VOCs are unique (Figure S2), but a few key mutations among these strains are shared by VOCs as shown in Table S3. N501Y is shared among the UK, JP, and SA variants. E484K is shared between the JP and SA variants, and K417 is mutated to T in the JP variant, and to N in the SA variant. These key mutated residues have been shown to impact binding and neutralizing activity of antibodies in the post-infection and post-vaccination sera as well as multiple MAbs isolated from COVID-19 patients (Casadevall et al., 2021a; Ho et al., 2021; Wang et al., 2021b; Wibmer et al., 2021; Wu et al., 2021b; Zhou et al., 2021a). To further explore the possible contribution of the key mutations in binding of hCoV-2IG batches, purified RBD proteins with individual mutations (K417N, N501Y, and E484K) were analyzed in SPR based antibody binding assays (Figure 3A). The K417N had minimal to no impact on hCoV-2IG binding. The hCoV-2IG binding to RBD-N501Y was reduced by ∼2-fold compared with WA-1 RBD. However, binding to RBD-E484K resulted in an average 19-fold reduction in hCoV-2IG binding compared with the WA-1 RBD (Figure 3B).
Figure 3.
RBD binding antibodies of convalescent plasma and hCoV-2IG against various SARS-CoV-2 RBD mutant proteins
(A and B) Total antibody binding (Max RU) of 1mg/mL for the six batches of hCoV-2IG (hCoV-2IG-1 to hCoV-2IG-6) to purified WA-1 RBD (RBD-wt) and RBD mutants: RBD-K417N, RBD-N501Y and RBD-E484K by SPR (A). The numbers above the group show the mean antibody binding for each RBD. (B) The fold-decrease in antibody binding to mutants RBD-K417N, RBD-N501Y and RBD-E484K of hCoV-2IG in comparison with RBD-wt from WA-1 strain was calculated from the data in Panel A. The numbers above the group shows the mean fold-change for each mutant RBD. All SPR experiments were performed twice and the researchers performing the assay were blinded to sample identity. The variations for duplicate runs of SPR was <5%. The data shown are average values of two experimental runs. The statistical significances between the variants for hCoV-2IG were performed using One-way ANOVA using Tukey's pairwise multiple comparison test in GraphPad prism. The differences were considered statistically significant with a 95% confidence interval when the p value was less than 0.05. (∗, p values of ≤0.05, ∗∗, p values of ≤0.01, ∗∗∗, p values of ≤0.001, ∗∗∗∗, p ≤ 0.0001).
Discussion
In the current study we conducted analyses on six lots of hyperimmune globulin (hCoV-2IG) manufactured from plasma units collected from SARS-CoV-2 recovered individuals in 2020. Antibody epitope repertoire, neutralization of SARS-CoV-2 and VOCs, and binding to spike peptides as well as recombinant RBD expressing individual mutations observed in the VOC were evaluated in comparison with 9 CP and 16 pre-pandemic 2019-IVIG.
The antibody epitope repertoires using SARS-CoV-2 spike GFPDL identified a diverse epitope fingerprint of both short and large antigenic sites spanning the entire spike protein. The hCoV-2IG antibodies most frequently bound to sites in the NTD, S1/S2 cleavage site, fusion peptide and heptad repeat domains in S2. In recent studies with monoclonal antibodies isolated from SARS-CoV-2 memory cells from COVID-19 patients, multiple neutralizing antibodies were identified that targeted the RBD, S1-NTD, S2, and S protein trimer (Andreano and Rappuoli, 2021; Fagiani et al., 2020; Wrapp et al., 2020). The most potent neutralizing antibodies that target directly the RBM/ACE2 interface were isolated at low frequency (Andreano et al., 2021). Furthermore, Dejnirattisai et al reported recently that among 377 MAbs isolated from 77 COVID-19 recovered individuals 53% bound to epitopes in S2, 34% bound to S1, 21% bound RBD and 11% bound NTD (Zhou et al., 2021a). Our data is also in agreement with a study using VirScan expressing 56mer or 20mer peptides across the spike that demonstrated several antibody binding hotspots around amino acids 550–600, 800, and 1,200. These sites share a significant level of homology with several seasonal coronaviruses including hCoV-HKU1and hCoV-229E and hCoV-OC43 (Shrock et al., 2020). It is expected that long-lived memory B cells (IgG/IgA) specific for conserved sites on the coronavirus spike will be recalled shortly after SARS-CoV-2 infection and contribute to the antibody pool in CP. While not all the dominant antibody sites on the spike are targeted by neutralizing antibodies, they may be sites of virus evolution/escape due to other antibody functions. It will be of interest to determine whether vaccines based on 2-P stabilized spike induced antibodies with a similarly broad epitope profile or a more RBD-focused repertoire.
In light of the rapid spread of SARS-CoV-2 VOCs around the globe (McCormick et al., 2021), it is important to evaluate the therapeutic potential of both CP and hCoV-2IG against both early circulating SARS-CoV-2 strains and the emerging VOCs (Fontanet et al., 2021; Mascola et al., 2021; Wang et al., 2021b). In this study, we evaluated six lots of hyperimmune globulin (hCoV-2IG) manufactured from plasma units collected from SARS-CoV-2 recovered individuals in 2020. All six hCoV-2IG lots demonstrated a small decline in neutralization titers (and increase in ID50) against CA, UK (∼2-fold) followed by JP (∼3.5-fold) VOC, and a significant decrease in neutralization activity against SA VOC (∼9-fold). The nine CP evaluated demonstrated a range of neutralization titers compared with the hCoV-2IG against the WA-1 strain, but a more pronounced reduction in PsVNA50 titers against the VOCs compared with hCoV-2IG.
Most of the SARS-CoV-2 VOCs that have been spreading in different parts of the world have multiple mutations both in the spike and other genes. However, several VOCs share one or more mutations in the RBD (McCormick et al., 2021). Decrease in antibody binding to the RBD interface with ACE2 receptor is probably the key reason for loss of SARS-CoV-2 neutralizing activity against the VOCs (Casadevall et al., 2021a) (Ho et al., 2021; Wang et al., 2021b; Wibmer et al., 2021; Wu et al., 2021b; Zhou et al., 2021a). Interestingly, while most of the interactions between the 25 residues of the Receptor Binding Motif (RBM) and the predominant IGHV3-53/IGHV3-66 used by multiple neutralizing antibodies are mediated by hydrogen bonds, only K417 and E484 have been described to form a salt bridge resulting in a stronger interaction and higher immune pressure (Wu et al., 2020). We found that the E484K mutation, which is shared between Brazil/JP P.1 VOC and SA B.1.351 VOC, significantly reduced binding of hCoV-2IG to the RBD (19-fold reduction) compared with RBD-wt. In contrast, the K417N had only minimal effect on RBD binding and the N501Y reduced binding of the hCoV-2IG by 2-fold. These amino acids mutations were predicted to increase ACE2 binding, but not necessarily to escape neutralization. Therefore, virus neutralization by different hCoV-2IG lots may be impacted both by specific amino acid mutations in the RBD/RBM and by the specificity of the polyclonal antibodies that bind to other sites on the SARS-CoV-2 spike, as well as antibody affinity.
The correlate of protection in terms of antibody neutralizing titers is being investigated in ongoing vaccine efficacy trials and test-negative surveillance studies (Letizia et al., 2021). Preclinical studies in rhesus macaques showed that passive transfer of 250 mg/kg SARS-CoV-2 IgG one day after challenge, reduced the peak lung viral loads and cleared the virus by day 3 (McMahan et al., 2020). Current animal studies are exploring heterologous protection against VOC following infection or vaccination with the Wuhan-1 strain.
CP demonstrated significant loss of neutralizing activities against the emerging VOC, especially the SA B.1.351 (Casadevall et al., 2021a; Wang et al., 2021b; Wibmer et al., 2021). Additionally, reduction in post-vaccination titers was observed against the new variants, especially the B.1.351 VOC. But SARS-CoV-2 vaccines that elicit high and durable neutralization titers may still be effective against severe disease (Wu et al., 2021a; Zhou et al., 2021a).
Our study underscores the advantage of using hCoV-2IG products compared with CP for treatment of SARS-CoV-2 infected patients. The neutralizing titers were found to decline significantly after 3 or 4 months in recovered patients with COVID-19 (Girardin et al., 2021). Therefore, screening of multiple CP units using qualified virus neutralization assays before pooling or fractionation can ensure high neutralizing titer hCoV-2 IG products and lot to lot consistency. In the near future, post vaccination plasma may be included in the manufacturing of new hCoV-2IG products following recent modification to the FDA guidance (https://www.fda.gov/media/136798/download).
The added values of hCoV-2IG over CP is even more critical in the face of emerging more transmissible VOCs that are spreading in several countries around the globe. Casadevall et al. (Casadevall et al., 2021b) emphasized that antibody preparations should contain sufficiently high concentrations of specific immunoglobulin to mediate biological effect against SARS-CoV-2 and its variants and should be administered early post-exposure.
However, several concerns should be addressed during the selection of CP and release of new batches of hCoV-2IG. Around 10% of severe COVID patients were found to have autoantibodies against type I IFNs and other immunomodulatory proteins including cytokines, chemokines, complement components, and cell-surface proteins (Bastard et al., 2020; Wadman, 2020; Wang et al., 2021a). This risk may be higher for individual CP derived from severe patients than for the hCoV-2IG which are manufactured from large numbers of pooled COVID-19 CP, likely to result in overall dilution of such autoantibodies (Cagdas, 2021). Nevertheless, it may be beneficial to establish assays for detection of anti-IFNs antibodies during CP collection and hCoV-2IG manufacturing and release. Other side effects of transfusion-related injury such as hemolytic reactions and immunomodulation including antibody-dependent enhancement (ADE) (Lee et al., 2020; Zhou et al., 2021b) need to be followed after CP or hCoV-2IG administration. It is likely that a high ratio of neutralizing to non-neutralizing antibodies will reduce the risk of ADE. In the case of hCoV-2IG, high virus neutralization titers have been demonstrated in validated assays, while in the case of individual COVID-19 CP, virus neutralization titers were often measured in research lab assays or not at all. One potential advantage of CP over hCoV-2IG is the presence of IgM and IgA in addition to IgG, which could contribute to virus neutralization. However, early studies demonstrated that both IgM and IgA decline within 4 weeks post symptoms and may be limited during convalescence. Furthermore, IgG are likely to have higher avidity (Pichler et al., 2021; Sokal et al., 2021) that may impact therapeutic outcome (Davey et al., 2019).
In summary, both CP and hCoV-2IG demonstrated reduced neutralization titers ranging from ∼2–4 fold against UK & JP VOCs and ∼10–20 fold against SA VOC, with lower drop in titers for the hCoV-2IG. Thus, early treatment of infected patients with hCoV-2IG, could benefit from knowledge of the prevalent circulating SARS-CoV-2 strains. Our findings indicate that treatment of COVID-19 patients with hCoV-2IG/CP may still be feasible but would likely require administration of higher volumes, or repeated infusions, in patients infected with the emerging SARS-CoV-2 variants. A treatment approach could be to treat with a standard dose immediately, and if genotyping reveals a critical mutation, like the E484K, to adjust the hCoV-2IG dosing subsequently. This will require rapid development of RT-PCR based diagnostics or other diagnostic assays that are designed to differentiate the prior predominant strains (WA-1 like) from new emerging SARS-CoV-2 variants reported in California, New York, UK, India, SA, Brazil, Chile, and Japan. Furthermore, in countries where a new VOC becomes dominant, the manufacturing of new hCoV-2IG should incorporate screening of the plasma and of the hCoV-2IG lots for neutralization activities against VOC. This study suggests that in countries with multiple co-circulating SARS-CoV-2 variants, the identification of the infecting SARS-CoV-2 strain will be critical in determining the effectiveness of antibody therapy in early SARS-CoV-2 infected individuals with high risk for developing severe disease.
Limitations of the study
In the current study, we evaluated six lots of hCoV-2IG that were manufactured from CP collected early in the pandemic in US (first half of 2020). It is likely that the composition of newer lots of hCoV-2IG would demonstrate different capacity to neutralize VOC including the B.1.617.2 (Delta), which is spreading rapidly around the globe. When available we will attempt to expand our analyses of hCoV-2IG lots.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 16 intravenous immunoglobulin (IVIG) batches produced from plasma collected prior to August 2019 | Five companies | Deidentified and Blinded to identity |
| Six hCoV-2IG batches prepared from 250-400 COVID-19 convalescent plasma donors per lot | Three companies | Deidentified and Blinded to identity |
| Nine random CP lots were obtained from recovered COVID-19 patients between May-July 2020 | U.S. Plasma donors | Deidentified and Blinded to identity |
| Bacterial and virus strains | ||
| E coli TG1 | Khurana et al., 2011 | In house |
| SARS-CoV-2 pseudovirus particles | Ravichandran et al., 2020 | In house |
| SARS-CoV-2 GFPDL | Ravichandran et al., 2020 | In house |
| SARS-CoV-2 WA-1 (USA-WA1/2020), UK-B.1.1.7 (hCoV-19/England/204820464/2020), and SA-B.1.351 (South Africa/KRISP-K005325/2020) strains | CDC | In house |
| M13K07 | NEB | N0315S |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 spike receptor binding domain (RBD) | Sino Biologicals | 40592-V08H82 |
| SARS-CoV-2 RBD-K417N | Sino Biologicals | 40592-V08H59 |
| SARS-CoV-2 RBD-N501Y | Sino Biologicals | 40592-V08H82 |
| SARS-CoV-2 RBD-E484K | Sino Biologicals | 40592-V08H84 |
| Biotinylated human ACE2 | Acros Bio | AC2-H82E6 |
| Biotinylated SARS-CoV-2 peptides | This manuscript | In house |
| Experimental models: cell lines | ||
| Vero E6 cells | ATCC | CCL-81 |
| 293-ACE2-TMPRSS2 cells | Neerukonda et al., 2021 | In house |
Resource availability
Lead contact
Surender Khurana, Ph.D (Surender.Khurana@fda.hhs.gov).
Materials availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Surender Khurana, Ph.D. (Surender.Khurana@fda.hhs.gov) under a material transfer agreement on reasonable request.
Experimental model and subject details
Study design
The objective of this study was to investigate various therapeutic polyclonal CP or purified hCoV-2-IG antibody preparations, which are being evaluated in clinical trials, for antibody binding and neutralizing capacity of important emerging SARS-CoV-2 variant of concern (VOC). Such variants of concern (VOC) are the United Kingdom (UK) variant B.1.1.7, California (CA) variant B.1.429, Japan (JP) variant P.1, and the South Africa (SA) variant B.1.351. There are multiple IVIG products approved by the FDA. These are polyclonal antibodies made from U.S. plasma donors. Each lot of product is derived from 10,000 or more donors. The manufacturing processes vary between manufacturers and usually include cold alcohol fractionation (Cohn-Oncley), anion-exchange and size-exclusion chromatography. Sixteen intravenous immunoglobulin (IVIG) batches were produced from plasma collected prior to August 2019 (each lot derived from >10,000 donors) from five manufacturers. The final product is sterile-filtered IgG (≥95%) and formulated at 100 mg/mL.
Six hCoV-2IG batches prepared from 250–400 COVID-19 CP donors per lot were obtained/purchased from three commercial companies under MTA to perform blinded (de-identified) antibody analysis. The plasma units used in the manufacturing of the hCoV-2IG batches were collected in 2020 prior to emergence of variants of concern in the U.S.
Nine random CP lots were obtained from recovered COVID-19 patients between May-July 2020, at least 30-days post-recovery from U.S. plasma donors. They were not selected based on any a priory criterion in order to represent a broad spectrum of antibody titers as reported in the literature. There was no attempt to isolate virus from the CP donors, so it is not known if any of them were infected with a variant. But based on the time of collection most of the infecting viruses were D614G in the US.
This study was approved by the Food and Drug Administration's Research Involving Human Subjects Committee (RIHSC #2020-04-02).
Ethics statement
This study was approved by the Food and Drug Administration's Research Involving Human Subjects Committee (RIHSC #2020-04-02). This study complied with all relevant ethical regulations for work with human participants, and informed consent was obtained. Samples were collected from adult subjects who provided informed consent to participate in the study. All assays performed fell within the permissible usages in the original informed consent.
Method details
Pseudovirus neutralization assay (PsVNA)
Antibody preparations were evaluated by SARS-CoV-2 pseudovirus neutralization assay (PsVNA) using WA-1 strain, UK variant (B.1.1.7 with spike mutations: H69-V70del, Y144del, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H), SA variant (B.1.351 strain with spike mutations L18F, D80A, D215G, L242-244del, R246I, K417N, E484K, N501Y, D614G, and A701V), CA variant (B.1.429 strain with spike mutations S13I, W152C, L452R, D614G) and JP variant (P.1 strain with spike mutations L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, T1027I, D614G, V1176F) (Figure S1 and Table S1).
Briefly, human codon-optimized cDNA encoding SARS-CoV-2 S glycoprotein of the WA-1, UK VOC, CA VOC, JP VOC and SA VOC were synthesized by GenScript and cloned into eukaryotic cell expression vector pcDNA 3.1 between the BamHI and XhoI sites. Pseudovirions were produced by co-transfection Lenti-X 293T cells with psPAX2(gag/pol), pTrip-luc lentiviral vector and pcDNA 3.1 SARS-CoV-2-spike-deltaC19, using Lipofectamine 3000. The supernatants were harvested at 48h post transfection and filtered through 0.45μm membranes and titrated using 293T-ACE2-TMPRSS2 cells (HEK 293T cells that express ACE2 and TMPRSS2 proteins).
For the neutralization assay, 50 μL of SARS-CoV-2 S pseudovirions were pre-incubated with an equal volume of medium containing serum at varying dilutions at room temperature for 1 h, then virus-antibody mixtures were added to 293T-ACE2-TMPRSS2 cells (Neerukonda et al., 2021) in a 96-well plate. The input virus with all three SARS-CoV-2 strains used in the current study were the same (2 × 105 Relative light units/50 μL/well). After a 3 h incubation, the inoculum was replaced with fresh medium. Cells were lysed 24 h later, and luciferase activity was measured using luciferin. Controls included cells only, virus without any antibody and positive sera. The cut-off value or the limit of detection for the neutralization assay is 1:10.
Classical wild-type SARS-CoV-2 virus neutralization assay
100 TCID50 of SARS-CoV-2 WA-1 (USA-WA1/2020), UK-B.1.1.7 (hCoV-19/England/204820464/2020), and SA-B.1.351 (South Africa/KRISP-K005325/2020) strains were incubated with 2-fold serial dilutions in a round bottom plate at 37°C for 1 hr. The virus-antibody mixture was then added to a 96-well plate with 5 × 104 Vero E6 cells. After 1 h the mixture was removed and replenished with fresh MEM containing 2% FBS. Cells were incubated at 37°C for an additional 48 hours, then fixed with 4% paraformaldehyde, followed by staining of cells with 0.1% crystal violet in 20% methanol. The PRNT50 and PRNT90 titers were calculated as the last serum dilution resulting in at least 50% and 90% SARS-CoV-2 neutralization, respectively. Wild-type SARS-CoV-2 CA and JP strains were not available to test in the BSL3 PRNT assay.
SARS-CoV-2 gene fragment phage display library (GFPDL) construction
DNA encoding the spike gene of SARS-CoV-2 isolate Wuhan-Hu-1 strain (GenBank: MN908947.3) was chemically synthesized and used for cloning. A gIII display-based phage vector, fSK-9-3, was used where the desired polypeptide can be displayed on the surface of the phage as a gIII-fusion protein. Purified DNA containing spike gene was digested with DNaseI to obtain gene fragments of 50–1,500 bp size range (18–500 amino acids) and used for GFPDL construction (Khurana et al., 2020; Ravichandran et al., 2020; Tang et al., 2021).
Affinity selection of SARS-CoV-2 GFPDL phages
Prior to panning of GFPDL with polyclonal hCoV-2IG antibodies, Ig components, which could non-specifically interact with phage proteins, were removed by incubation with UV-killed M13K07 phage-coated Petri dishes. Equal volumes of each of the six hCoV-2IG lots were used for GFPDL panning. GFPDL affinity selection was carried out in-solution with protein A/G resin. Briefly, the hCoV-2IG lot was incubated with the GFPDL and the protein A/G resin, the unbound phages were removed by PBST (PBS containing 0.1% Tween-20) wash followed by PBS. Bound phages were eluted by addition of 0.1 N Gly-HCl pH 2.2 and neutralized by adding 8 μL of 2 M Tris solution per 100 μL eluate. After panning, antibody-bound phage clones were amplified, the inserts were sequenced, and the sequences were aligned to the SARS-CoV-2 spike gene, to define the fine epitope specificity in these polyclonal hCoV-2IG lots.
The GFPDL affinity selection was performed in duplicate (two independent experiments by a research fellow in the lab, who was blinded to sample identity). Similar numbers of bound phage clones and epitope repertoire were observed in the two GFPDL panning.
Proteins and peptides
Recombinant SARS-CoV-2 spike receptor binding domain (RBD) and its mutants were purchased from Sino Biologicals (RBD-wt; 40592-V08H82, RBD-K417N; 40592-V08H59, RBD-N501Y; 40592-V08H82 and RBD-E484K; 40592-V08H84). Recombinant purified RBD proteins used in the study were produced in HEK-293 mammalian cells. The native receptor-binding activity of the spike RBD proteins was determined by binding to the 5 μg/mL human ACE2 protein (Ravichandran et al., 2020, 2021; Tang et al., 2021). Biotinylated human ACE2 (AC2-H82E6) contains amino acid residues Gln 18 - Ser 740 expressed from HEK 293 cells was purchased from Acro Biosystems.
GFPDL identified antigenic sites representing peptides up to 70 amino acids long were chemically synthesized using F-moc chemistry, biotinylated at C-terminus, and purified using RP-HPLC.
Antibody binding kinetics to SARS-CoV-2 RBD mutants or SARS-CoV-2 peptides by surface plasmon resonance (SPR)
Steady-state equilibrium binding of hCoV-2IG lots was monitored at 25°C using a ProteOn surface plasmon resonance (BioRad). The purified recombinant SARS-CoV-2 RBD proteins were captured to a Ni-NTA sensor chip with 200 resonance units (RU) in the test flow channels. The protein density on the chip was optimized so as to measure monovalent interactions independent of the antibody isotype. The biotinylated SARS-CoV-2 peptides were captured on a NLC chip and used for peptide antibody profiling of hCoV-2IG, CP and 2019-IVIG lots.
Serial dilutions (1 mg/mL, 0.33 mg/mL and 0.11 mg/mL) of freshly prepared hCoV-2IG or 2019-IVIG or 10-fold dilution of CP in BSA-PBST buffer (PBS pH 7.4 buffer with Tween 20 and BSA) were injected at a flow rate of 50 μL/min (120 s contact duration) for association, and disassociation was performed over a 600-s interval. Responses from the protein surface were corrected for the response from a mock surface and for responses from a buffer-only injection. SPR was performed with serially diluted samples in this study. Total antibody binding was calculated with BioRad ProteOn manager software (version 3.1). All SPR experiments were performed twice, and the researchers performing the assay were blinded to sample identity. The maximum resonance units (Max RU) data shown in the figures are the calculated RU signals for the 1 mg/mL hCoV-2IG sample or 2019-IVIG or 10-fold dilution of CP.
Quantification and statistical analysis
All experimental data were analyzed using GraphPad Prism, version 9.0.1 (GraphPad software Inc, San Diego, CA) or R package. Differences between groups were analyzed using multiple group comparisons by non-parametric (Kruskal-Wallis) statistical test using Dunn's post-hoc analysis. The difference within each group was performed using one-way ANOVA using Tukey's pairwise multiple comparison test. The differences were considered statistically significant with a 95% confidence interval when the p value was less than 0.05. (∗, p values of ≤0.05, ∗∗, p values of ≤0.01, ∗∗∗, p values of ≤0.001, ∗∗∗∗, p ≤ 0.0001). Correlation analysis of PRNT and PsVNA titers were performed by computing Pearson's correlation coefficient in Graphpad.
Acknowledgments
We thank Keith Peden and Marina Zaitseva for their insightful review of the manuscript. We thank Carol Weiss for providing plasmid clones expressing SARS-CoV-2 variants and Dorothy Scott for providing convalescent plasma and hCoV-2IG.
The research work described in this manuscript was supported by FDA Medical Countermeasures Initiative (MCMi) grant # OCET 2021-1565 and NIH-NIAID IAA #AAI20040. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Author contributions
Designed research, S.K; clinical specimens and clinical data, H.G. and B.G; performed research, J.T., Y.L., S.R., G.G., C. H., S.C., T.W., and S.K; contributed to writing, S.K., H.G., and B.G.
Declaration of interests
The authors declare no competing interests.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103006.
Supplemental information
Data and code availability
This study did not generate any unique code or data sets. All data are shown in the manuscript figures and supplemental information. For questions regarding raw data and request for resources and reagents, please contact the Lead Contact, Surender Khurana, Ph.D. (Surender.Khurana@fda.hhs.gov). All software's used in this study are commercially available.
References
- Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021 doi: 10.1016/j.cell.2021.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Rappuoli R. Immunodominant antibody germlines in COVID-19. J. Exp. Med. 2021;218:e20210281. doi: 10.1084/jem.20210281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Beziat V. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagdas D. Convalescent plasma and hyperimmune globulin therapy in COVID-19. Expert Rev. Clin. Immunol. 2021;17:309–316. doi: 10.1080/1744666X.2021.1894927. [DOI] [PubMed] [Google Scholar]
- Casadevall A., Joyner M.J., Pirofski L.A. SARS-CoV-2 viral load and antibody responses: the case for convalescent plasma therapy. J. Clin. Invest. 2020;130:5112–5114. doi: 10.1172/JCI139760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Henderson J., Joyner M., Pirofski L.A. SARS-Cov2 variants and convalescent plasma: reality, fallacies, and opportunities. J. Clin. Invest. 2021;131:e148832. doi: 10.1172/JCI148832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L.A., Joyner M.J. The principles of antibody therapy for infectious diseases with relevance for COVID-19. mBio. 2021;12:e03372-20. doi: 10.1128/mBio.03372-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey R.T., Jr., Fernandez-Cruz E., Markowitz N., Pett S., Babiker A.G., Wentworth D., Khurana S., Engen N., Gordin F., Jain M.K. Anti-influenza hyperimmune intravenous immunoglobulin for adults with influenza A or B infection (FLU-IVIG): a double-blind, randomised, placebo-controlled trial. Lancet Respir. Med. 2019;7:951–963. doi: 10.1016/S2213-2600(19)30253-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagiani F., Catanzaro M., Lanni C. Molecular features of IGHV3-53-encoded antibodies elicited by SARS-CoV-2. Signal. Transduct. Target Ther. 2020;5:170. doi: 10.1038/s41392-020-00287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanet A., Autran B., Lina B., Kieny M.P., Karim S.S.A., Sridhar D. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet. 2021;397:952–954. doi: 10.1016/S0140-6736(21)00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes S., Coyle E.M., Beeler J., Golding H., Khurana S. Antigenic fingerprinting following primary RSV infection in young children identifies novel antigenic sites and reveals unlinked evolution of human antibody repertoires to fusion and attachment glycoproteins. PLoS Pathog. 2016;12:e1005554. doi: 10.1371/journal.ppat.1005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes S., Hahn M., Chilcote K., Chemaly R.F., Shah D.P., Ye X., Avadhanula V., Piedra P.A., Golding H., Khurana S. Antigenic fingerprinting of respiratory syncytial virus (RSV)-A-Infected hematopoietic cell transplant recipients reveals importance of mucosal anti-RSV G antibodies in control of RSV infection in humans. J. Infect. Dis. 2020;221:636–646. doi: 10.1093/infdis/jiz608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin R.C., Dupuis A.P., Payne A.F., Sullivan T.J., Strauss D., Parker M.M., McDonough K.A. Temporal analysis of serial donations reveals decrease in neutralizing capacity and justifies revised qualifying criteria for COVID-19 convalescent plasma. J. Infect. Dis. 2021;223:743–751. doi: 10.1093/infdis/jiaa803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D., Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. Res. Sq. 2021;Jan 29. doi: 10.21203/rs.3.rs-155394/v1. Submitted for publication. [DOI] [Google Scholar]
- Joyner M.J., Carter R.E., Senefeld J.W., Klassen S.A., Mills J.R., Johnson P.W., Theel E.S., Wiggins C.C., Bruno K.A., Klompas A.M. Convalescent plasma antibody levels and the risk of death from Covid-19. N. Engl. J. Med. 2021;384:1015–1027. doi: 10.1056/NEJMoa2031893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S., Coyle E.M., Manischewitz J., King L.R., Gao J., Germain R.N., Schwartzberg P.L., Tsang J.S., Golding H., the C.H.I.C. AS03-adjuvanted H5N1 vaccine promotes antibody diversity and affinity maturation, NAI titers, cross-clade H5N1 neutralization, but not H1N1 cross-subtype neutralization. NPJ Vaccin. 2018;3:40. doi: 10.1038/s41541-018-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S., Ravichandran S., Hahn M., Coyle E.M., Stonier S.W., Zak S.E., Kindrachuk J., Davey R.T., Jr., Dye J.M., Chertow D.S. Longitudinal human antibody repertoire against complete viral proteome from Ebola virus survivor reveals protective sites for vaccine design. Cell Host Microbe. 2020;27:262–276.e4. doi: 10.1016/j.chom.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S., Suguitan A.L., Jr., Rivera Y., Simmons C.P., Lanzavecchia A., Sallusto F., Manischewitz J., King L.R., Subbarao K., Golding H. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med. 2009;6:e1000049. doi: 10.1371/journal.pmed.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S., Verma N., Yewdell J.W., Hilbert A.K., Castellino F., Lattanzi M., Del Giudice G., Rappuoli R., Golding H. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med. 2011;3:85ra48. doi: 10.1126/scitranslmed.3002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K. Fast-spreading U.K. virus variant raises alarms. Science. 2021;371:9–10. doi: 10.1126/science.371.6524.9. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K. New mutations raise specter of 'immune escape'. Science. 2021;371:329–330. doi: 10.1126/science.371.6527.329. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K. Viral evolution may herald new pandemic phase. Science. 2021;371:108–109. doi: 10.1126/science.371.6525.108. [DOI] [PubMed] [Google Scholar]
- Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- Letizia A.G., Ge Y., Vangeti S., Goforth C., Weir D.L., Kuzmina N.A., Balinsky C.A., Chen H.W., Ewing D., Soares-Schanoski A. SARS-CoV-2 seropositivity and subsequent infection risk in healthy young adults: a prospective cohort study. Lancet Respir. Med. 2021;9:712–720. doi: 10.1016/S2213-2600(21)00158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., Graham B.S., Fauci A.S. SARS-CoV-2 viral variants-tackling a moving target. JAMA. 2021;325:1261–1262. doi: 10.1001/jama.2021.2088. [DOI] [PubMed] [Google Scholar]
- McCormick K.D., Jacobs J.L., Mellors J.W. The emerging plasticity of SARS-CoV-2. Science. 2021;371:1306–1308. doi: 10.1126/science.abg4493. [DOI] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2020;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neerukonda S.N., Vassell R., Herrup R., Liu S., Wang T., Takeda K., Yang Y., Lin T.L., Wang W., Weiss C.D. Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2. PLoS One. 2021;16:e0248348. doi: 10.1371/journal.pone.0248348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler D., Baumgartner M., Kimpel J., Rossler A., Riepler L., Bates K., Fleischer V., von Laer D., Borena W., Wurzner R. Marked increase in avidity of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) antibodies 7-8 months after infection is not diminished in old age. J. Infect Dis. 2021:jiab300. doi: 10.1093/infdis/jiab300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran S., Coyle E.M., Klenow L., Tang J., Grubbs G., Liu S., Wang T., Golding H., Khurana S. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl Med. 2020;12:eabc3539. doi: 10.1126/scitranslmed.abc3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran S., Lee Y., Grubbs G., Coyle E.M., Klenow L., Akasaka O., Koga M., Adachi E., Saito M., Nakachi I. Longitudinal antibody repertoire in "mild" versus "severe" COVID-19 patients reveals immune markers associated with disease severity and resolution. Sci. Adv. 2021;7:eabf2467. doi: 10.1126/sciadv.abf2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e4. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Ravichandran S., Lee Y., Grubbs G., Coyle E.M., Klenow L., Genser H., Golding H., Khurana S. Antibody affinity maturation and plasma IgA associate with clinical outcome in hospitalized COVID-19 patients. Nat. Commun. 2021;12:1221. doi: 10.1038/s41467-021-21463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeberg P., Cruz M., Diez J.M., Merritt W.K., Santos B., Trukawinski S., Wellhouse A., Jose M., Willis T. Production of anti-SARS-CoV-2 hyperimmune globulin from convalescent plasma. Transfusion. 2021;61:1705–1709. doi: 10.1111/trf.16378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman M. Flawed interferon response spurs severe illness. Science. 2020;369:1550–1551. doi: 10.1126/science.369.6511.1550. [DOI] [PubMed] [Google Scholar]
- Wang E.Y., Mao T., Klein J., Dai Y., Huck J.D., Jaycox J.R., Liu F., Zhou T., Israelow B., Wong P. Diverse functional autoantibodies in patients with COVID-19. Nature. 2021 doi: 10.1038/s41586-021-03631-y. [DOI] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Team V.-C.C.-R., Hoffmann M. Structural basis for potent neutralization of betacoronaviruses by single-domain Camelid antibodies. Cell. 2020;181:1004–1015.e5. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B.S. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.01.25.427948. [DOI] [Google Scholar]
- Wu N.C., Yuan M., Liu H., Lee C.D., Zhu X., Bangaru S., Torres J.L., Caniels T.G., Brouwer P.J.M., van Gils M.J. An alternative binding mode of IGHV3-53 antibodies to the SARS-CoV-2 receptor binding domain. Cell Rep. 2020;33:108274. doi: 10.1016/j.celrep.2020.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liu Z., Li S., Xu W., Zhang Q., Silva I.T., Li C., Wu Y., Jiang Q., Liu Z. Enhancement versus neutralization by SARS-CoV-2 antibodies from a convalescent donor associates with distinct epitopes on the RBD. Cell Rep. 2021;34:108699. doi: 10.1016/j.celrep.2021.108699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique code or data sets. All data are shown in the manuscript figures and supplemental information. For questions regarding raw data and request for resources and reagents, please contact the Lead Contact, Surender Khurana, Ph.D. (Surender.Khurana@fda.hhs.gov). All software's used in this study are commercially available.