Abstract
Environmental seasonality is a potent evolutionary force, capable of maintaining polymorphism, promoting phenotypic plasticity and causing bet-hedging. In Drosophila, environmental seasonality has been reported to affect life-history traits, tolerance to abiotic stressors and immunity. Oscillations in frequencies of alleles underlying fitness-related traits were also documented alongside SNPs across the genome. Here, we test for seasonal changes in two recombination characteristics, crossover rate and crossover interference, in a natural D. melanogaster population from India using morphological markers of the three major chromosomes. We show that winter flies, collected after the dry season, have significantly higher desiccation tolerance than their autumn counterparts. This difference proved to hold also for hybrids with three independent marker stocks, suggesting its genetic rather than plastic nature. Significant between-season changes are documented for crossover rate (in 9 of 13 studied intervals) and crossover interference (in four of eight studied pairs of intervals); both single and double crossovers were usually more frequent in the winter cohort. The winter flies also display weaker plasticity of both recombination characteristics to desiccation. We ascribe the observed differences to indirect selection on recombination caused by directional selection on desiccation tolerance. Our findings suggest that changes in recombination characteristics can arise even after a short period of seasonal adaptation (~8–10 generations).
Subject terms: Structural variation, Evolutionary biology, Evolutionary genetics
Introduction
Environmental seasonality plays an important role as an ecological factor, and its significance as a potent evolutionary force is becoming increasingly evident. The evolutionary consequences of within-year oscillations in selection directions and intensities considerably depend on the generation time. In perennials, exposure to environmental seasonality as lifespan-long regular background may select for pleiotropy and phenotypic plasticity. In annuals, whose developmental stages are distributed throughout a year, it may additionally select for fine-tuning of life-history traits and bet-hedging (Williams et al. 2017). Yet, seasonality effects in multivoltine species, having several generations per year, can be even more complex, leading to far-reaching population-level effects, including maintenance of balanced polymorphism (Haldane and Jayakar 1963; Korol et al. 1996; Wittmann et al. 2017), complex dynamics of allele frequencies (Kirzhner et al. 1995, 1996) and evolving dominance (Otto and Bourguet 1999; Connallon and Chenoweth 2019), in addition to those mentioned above. Moreover, multivoltine species seem to be the most appropriate models for addressing the intriguing interplay between different adaptations to seasonality, including the interaction between plastic and heritable responses to periodical environmental stressors.
Fruit flies are particularly informative models in seasonality studies. The population size of various Drosophila species has long been known to fluctuate during a year (Goldschmidt et al. 1955; Prakash and Reddy 1979). Later studies have also shown seasonal oscillation in several important fitness-related phenotypic traits, including desiccation tolerance (McKenzie and Parsons 1974; Parkash et al. 2011; Aggarwal et al. 2013), the activity of metabolic enzymes (Knibb 1986), life-history traits, resistance to heat, cold and starvation (Behrman et al. 2015) and innate immunity (Behrman et al. 2018). In a recent extensive genome-wide analysis, Bergland et al. (2014) identified hundreds of SNPs whose frequency oscillates among seasons; the authors related them to variation in adaptive phenotypic traits, first of all cold- and starvation tolerance.
In contrast to stress tolerance and other fitness-related traits considered in the above-mentioned studies, changes in recombination have never been studied in the context of seasonal adaptation, to the best of our knowledge. Typically, recombination does not directly affect the survival of the individual. However, it does affect the diversity of its progeny and, thereby, the genetic structure of the whole population in the next generation. This suggests that variation in recombination can be adaptive (Korol et al. 1994; Rice 2002; Stevison 2011; Ritz et al. 2017; Samuk et al. 2020)—similar to variation in fitness-related traits. Several studies have found that in natural populations recombination may vary in space—along certain environmental gradients (Saleem et al. 2001; Grishkan et al. 2003; Salomé et al. 2012; Dreissig et al. 2019; Neupane and Xu 2020). Variation of recombination in time is much less explored. To date, such changes were addressed only during the lifespan of an individual (King and Hayman 1978; Cobror et al. 1986; Saggoo et al. 2010) or phase-transition of a population (Nolte 1967). Here, we for the first time examine changes in recombination characteristics during seasonal adaptation of a population.
We hypothesised that if the above-mentioned fitness-related traits are oligo- or polygenic, and if seasonal changes in these traits at least partially result from genetic adaptation, then this will generate indirect selection for recombination. Such an assumption appeals to the first evolutionary explanation of recombination maintenance in nature—Weismann’s claim that recombination provides the raw material needed to adapt to a changing environment (Weismann 1889). The possibility of such indirect selection has been confirmed in several laboratory experiments that documented elevated recombination in certain genome regions upon selection for recombination-unrelated traits (Harinarayana and Murty 1971; Lobashev et al. 1973; Flexon and Rodell 1982; Zhuchenko et al. 1985; Gorodetsky et al. 1990; Gorlov et al. 1992; Korol and Iliadi 1994; Rodell et al. 2004; Aggarwal et al. 2015). In some of these experiments, changes in recombination were observed after quite a short selection: during 15 generations in fruit fly (Zhuchenko et al. 1985) and even 5 generations in cabbage (Harinarayana and Murty 1971). However, it remains unclear whether indirect selection for recombination can emerge in natural conditions during seasonal adaptation. To test this, we compared two recombination characteristics, crossover rate (CO rate) and crossover interference (CO interference), in two seasonal cohorts from the same natural population of Drosophila melanogaster. We expected that the winter flies, collected after a dry period, had evolved a higher desiccation tolerance compared to their autumn counterparts. The explicit test, conducted before the recombination-assessment experiments, has confirmed our assumption.
We also tested whether the two seasonal cohorts differ in the plasticity of both recombination characteristics (CO rate and CO interference) to desiccation stress, i.e., in desiccation-induced changes in these characteristics. Since the pioneering works by Plough (1917, 1921), both CO rate and CO interference have been recognised as traits exhibiting considerable plasticity to environmental stressors; see also Hayman and Parsons (1960), Grell (1978) and Parsons (1988). Empirical studies (although very limited) in general argue for a negative association between recombination plasticity of an organism and its fitness: genotypes with higher tolerance to a stressor tend to demonstrate a lower increase in CO rate and weaker relaxation of CO interference when they are exposed to this stressor (Kilias et al. 1979; Zhuchenko et al. 1986; Korol et al. 1994; Jackson et al. 2015; Aggarwal et al. 2019). In particular, in our recent study (Aggarwal et al. 2019), desiccation-tolerant D. melanogaster lines responded to desiccation stress less pronouncedly than their desiccation-sensitive counterparts, in terms of both CO rate and CO interference. Here, we examined seasonal variation in recombination plasticity in a natural Drosophila population. Given the higher desiccation tolerance of the winter flies, we expected them to experience milder stress upon desiccation hardening and, therefore, to exhibit a less pronounced recombination response to this factor.
Materials and methods
Flies and crosses
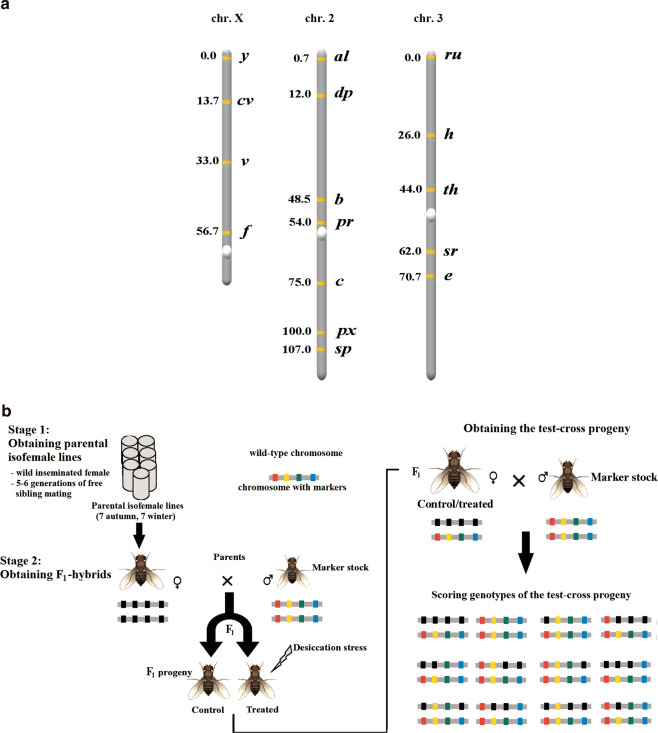
Recombination characteristics were assessed using visually distinguishable morphological markers. In total, we examined 16 markers in the large chromosomes: y, cv, v and f in chromosome X; al, dp, b, pr, c, px and sp in chromosome 2; and ru, h, th, sr and e in chromosome 3. The markers’ location is shown in Fig. 1A.
Fig. 1. The location of the studied markers on the genetic map (A) and the overall scheme of experiment (B).
A The markers’ positions are stated in map units (cM). B Only crosses with the marker stock for one autosome are shown as an example.
The experimental scheme included three main stages: (1) establishing parental lines representing the two seasons, autumn and winter; (2) obtaining the F1-hybrids heterozygous for the examined markers, and rearing them under two regimes, either normal conditions or desiccation hardening; and (3) obtaining the test-cross progeny to assess the recombination characteristics in question (Fig. 1B).
To establish the parental lines (Fig. 1B, stage 1), we employed the isofemale-line approach, an effective tool to maintain the population heterogeneity (David et al. 2005). Each isofemale line was initiated by a single wild-collected inseminated adult female. These flies were collected using the net-sweeping method at the lowland locality Kalka in the Western Himalaya, India (latitude 30.85°N, longitude 76.54°E, altitude 656 m). The samples were collected at the end of climatic autumn (September 2017) and at the end of climatic winter (February 2018). The two seasons considerably differ in temperature and relative humidity: the autumn is warm and wet (average temperature 26.2 °C, relative humidity 72%) while the winter is cool and dry (average temperature 13.5 °C, relative humidity 51%) (IMD 2010). The progeny of each founder female underwent free sibling mating during five to six generations. Such families were maintained on the yeast-agar-sugar food medium under normal conditions (22 °C, relative humidity 65–70%) in standard Drosophila stock bottles (60 × 40 mm), with the progeny size >250 individuals per bottle. Bottles of the same volume were also used for further crosses.
For each season, we established >100 isofemale lines and then randomly chose seven of them for further assays. Thus, the implemented experimental scheme exploited the genetic variation released from 14 wild-type variants of each of the three major chromosomes during five to six rounds of both intra- and inter-chromosomal recombination. This variation represents a considerable part of the variation in the source population; see Dobzhansky (1946) and Dobzhansky et al. (1959) for related estimates.
Then, to obtain the F1-hybrids heterozygous for the above-mentioned markers (Fig. 1B, stage 2), we crossed the established parental isofemale lines with three standard laboratory marker stocks homozygous for these markers. The following marker stocks were used (according to the records of the Bloomington Drosophila Stock Centre): #1515 for chromosome X, #156 for chromosome 2 and #576 for chromosome 3. To this end, 20 virgin females (6-day-old post eclosion) were randomly chosen from each parental isofemale line and mated with 20 males from each marker stock. The obtained F1-hybrids are hereafter referred to as ‘the autumn hybrids’ and ‘the winter hybrids’, according to the origin of their parental isofemale lines. In each of 42 crosses (seven autumn and seven winter parental isofemale lines, each crossed with three marker stocks), virgin F1-females were divided into two groups: the treatment and the control ones. The treatment females were subjected to desiccation hardening—short-term sub-lethal desiccation stress (see the next subsection), while the control females were kept in normal conditions.
Finally, to obtain the test-cross progeny (Fig. 1B, stage 3), 20 treated and 20 non-treated F1-females were transferred to fresh food medium and mated with 20 males from the corresponding marker stock for 4 days. After mating, the F1-females laid eggs in fresh bottles for 48 h. The obtained test-cross progeny was scored for the morphological markers to assess the CO rate and CO interference.
Thus, the experimental scheme included 84 different test crosses: two seasons, seven parental isofemale lines per season, three marker stocks per parental isofemale line to obtain the heterozygous F1-hybrids, and two rearing regimes for each F1-hybrid (2 × 7 × 3 × 2 = 84). Supplementary Table S1 summarises the number of the scored test-cross progeny by seasons, lines, rearing regimes and chromosomes.
Desiccation-tolerance estimation and desiccation hardening
Desiccation tolerance was quantified as the complete lethal time (LT100)—the time till lethal dehydration of all flies in dry air. It was measured for all 14 parental isofemale lines (seven autumn and seven winter lines) and all 42 F1-hybrids (each parental isofemale line crossed with three different marker stocks). For each parental isofemale line and each hybrid, the assay consisted of ten replicates. In each replicate, ten virgin females (6 days after eclosion) were placed into a dry plastic vial containing 2 g of silica gel at the bottom. The vials were covered with a disk of foam and then placed into a desiccator chamber (Secador electronic desiccator cabinet; www.tarson.com) maintaining the relative humidity of 5–8%. The number of immobile flies was scored every 30 min during the first 10 h, and every 15 min thereafter.
Flies in the treatment group were subjected to desiccation hardening, a short-term exposure to dry air resulting in 5% mortality. The value of LT5 was calculated using the probit analysis. Importantly, the two seasonal cohorts appeared different in desiccation tolerance, which led also to different values of LT5: 4 h 15 min for the autumn hybrids and 4 h 30 min for their winter counterparts. Thus, the winter hybrids were subjected to 15 min longer desiccation hardening.
Statistical analysis
Desiccation tolerance was assessed based on ten replicates per each of 14 studied lines (seven autumn and seven winter ones). These data were analysed using the nested ANOVA, with line nested within season.
CO rates were assessed for each line and each of the two rearing regimes, normal conditions and desiccation hardening. The rates were first logit-transformed given their binomial nature. The effect of season was estimated using Student’s t-test for independent samples; the method’s assumptions were controlled by the Shapiro–Wilk test for normality and Levene’s test for the equality of variance. The effect of treatment was estimated using Student’s t-test for paired samples, separately for the autumn and the winter hybrids. Upon violation of the method’s assumption, the non-parametric signed-rank Wilcoxon test was applied instead. The modulating effect of desiccation tolerance was estimated using the repeated-measures ANCOVA, with treatment as the major factor and desiccation tolerance as the covariate. The analyses were performed using the Real Statistics Resource Pack and JASP 0.14.
CO interference was quantified using the coefficient of coincidence. These coefficients show the ratio of the actual frequency of double crossovers to the frequency expected under the assumption of independence. When recombination events in the two adjacent intervals suppress one another (positive CO interference), the coefficient is less than one. In the opposite situation, when the events stimulate one another (negative CO interference), the coefficient is more than one. Like CO rates, the coefficients of coincidence were assessed for each line and each rearing regime. The effects of the above-mentioned factors (season, line, treatment and their interaction) were estimated using the restricted maximum-likelihood analysis. Specifically, the likelihood function was maximised to estimate the coefficients of coincidence under the restriction that CO rates in both intervals are already estimated. A detailed description of the method is presented in Supplementary Text S2. We also analysed the chromosome-wide distribution of single and multiple crossover events using CODA software (Gauthier et al. 2011).
The significance values for the effects of different factors on CO rate and CO interference were FDR-corrected using the Benjamini–Hochberg method (Benjamini and Hochberg 1995).
Results
In the three following subsections, we analyse changes in desiccation tolerance, CO rate and its plasticity and CO interference and its plasticity.
Desiccation tolerance
We compared desiccation tolerance in the autumn and the winter isofemale lines. As expected, the winter lines showed on average higher resistance: 28.32 ± 0.44 h against 22.57 ± 0.36 h (LT100 ± SE). The effect of season appeared highly significant: F = 13.48, p = 3.2 · 10–3. Remarkably, the same pattern held in all three types of hybrids, with an even more pronounced effect: F from 24.0 to 38.5, p from 3.7 · 10–4 to 4.6 · 10–5 (Fig. 2). The nested ANOVA has also revealed a highly significant effect of line within season. The tolerance of all three types of hybrids strongly correlated with that of the parental isofemale lines: Pearson’s r from 0.91 to 0.95, p from 4.4 · 10–6 to 2.8 · 10–7. These correlations appeared stronger for the winter lines.
Fig. 2. The effect of season on desiccation tolerance in the autumn and the winter parental isofemale lines and their hybrids with three marker stocks.
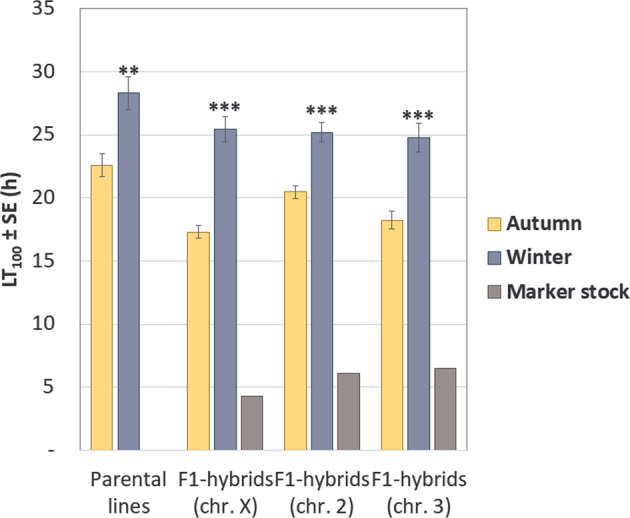
The effect is assessed using nested ANOVA, N = 70 per season (seven lines per season, ten replicates per line). The asterisks show the significance of the F-statistics: **p < 0.01; ***p < 0.001.
Crossover rate and its plasticity
First, we compared CO rates in the autumn and the winter hybrids reared in normal conditions. The two seasonal cohorts showed significantly different CO rates in nine intervals: two in chromosome X (cv–v and v–f), five in chromosome 2 (al–dp, dp–b, pr–c, c–px and px–sp) and two in chromosome 3 (h–th and sr–e). CO rates were usually higher in the winter hybrids, except for two intervals in chromosome 2 with the opposite pattern (dp–b and px–sp). The effect was the strongest for the pericentromeric interval pr–c (Fig. 3 and Supplementary Table S3).
Fig. 3. The effect of season on crossover rates in hybrids reared in normal conditions.
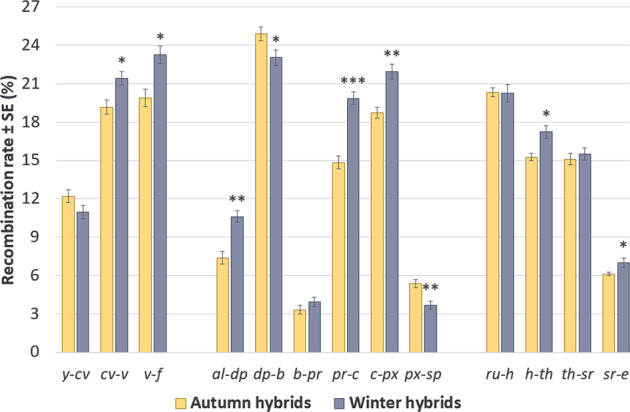
The effect is assessed using the t-test for independent samples, N = 7 per season. The asterisks show the FDR-corrected significance of the t-statistics: *pFDR < 0.1; **pFDR < 0.01; ***pFDR < 0.001.
Second, we examined the plasticity of CO rate to desiccation stress in the autumn and the winter hybrids. To this end, we compared, separately for each of the two seasonal cohorts, CO rates in flies reared under normal conditions with those in flies subjected to desiccation hardening. In the autumn hybrids, the treatment significantly increased CO rates in six intervals: two in chromosome X (cv–v and v–f), two in chromosome 2 (pr–c and c–px) and two in chromosome 3 (h–th and th–sr) (Fig. 4A and Supplementary Table S4). In contrast, the winter hybrids demonstrated a significant increase only in one interval, cv–v (much less significant than their autumn counterparts), and a significant decrease in another interval, al–dp (Fig. 4B and Supplementary Table S4).
Fig. 4. The effect of desiccation stress on crossover rates in the autumn (A) and the winter (B) hybrids.
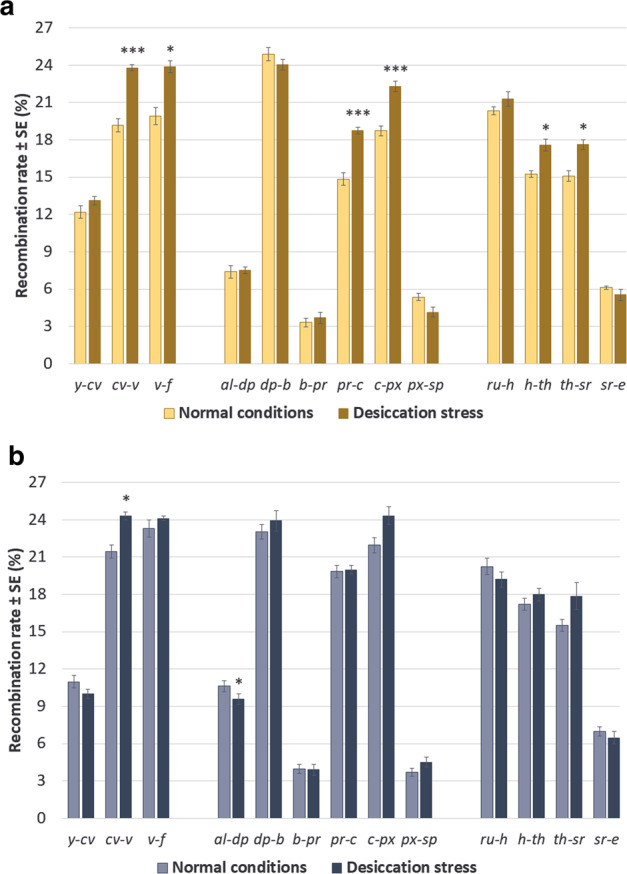
The effect is assessed using the t-test for paired samples, N = 7 per season. The asterisks show the FDR-corrected significance of the t-statistics: *pFDR < 0.1; **pFDR < 0.01; *** pFDR < 0.001.
Given the significant effect of line on desiccation tolerance (see the first subsection of the ‘Results’), we examined to which extent line modulates the effect of desiccation stress on CO rates. To this end, the line’s desiccation tolerance was introduced as a covariate into the repeated-measures ANCOVA with treatment as a major factor. No effect was found, neither for the autumn nor for the winter hybrids. However, when both seasonal cohorts were handled together, the tolerance did exert a significant modulating effect in four intervals: two in chromosome X (y–cv and cv–v), two in chromosome 2 (pr–c and px–sp) and one in chromosome 3 (ru–h) (Supplementary Table S5).
For all recombination-reactive intervals (i.e., those in which the effect of either season or treatment appeared significant), the segregation of both markers was close to 1:1, and the frequencies of both reciprocal recombinants changed rather concordantly (Supplementary Table S6). This suggests that differential transmission is an unlikely explanation for the observed changes in CO rates.
Crossover interference and its plasticity
First, we compared the coefficients of coincidence in the autumn and the winter hybrids reared in normal conditions. Bearing in mind rather moderate sample sizes of the test-cross progenies (~250 per line per rearing regime, see Supplementary Table S1), we analysed only those pairs of intervals where the distance between the markers exceeded 20 cM in each interval. The effect of season appeared significant for four pairs of intervals: one in chromosome X (cv–v–f), two in chromosome 2 (b–c–sp and pr–c–sp) and one in chromosome 3 (ru–h–th). Except for the pair ru–h–th, the winter hybrids demonstrated higher double CO rates, manifested as relaxation of positive CO interference or even emergence of negative CO interference. Notably, dp–b–c segment of chromosome 2 demonstrated significant negative interference in both seasonal cohorts (Table 1). The chromosome-level analysis (Gauthier et al. 2011), performed to complement the interval-level comparisons, has supported the revealed pattern: the frequency of double and multiple crossovers appeared higher in the winter hybrids, especially for chromosomes X and 2 (absolute increase by ~5%). This analysis has also confirmed a significant excess of double crossovers in chromosome 2 in both seasonal cohorts. In all mentioned cases, double and multiple crossovers increased at the expense of single ones (Supplementary Fig. S7A).
Table 1.
The effect of season on crossover interference in hybrids reared in normal conditions (based on the restricted maximum-likelihood analysis).
| Pair of intervals | Coefficient of coincidence ± SE | Effect of season | |||
|---|---|---|---|---|---|
| Autumn hybrids | Winter hybrids | χ2 | p | p (FDR)a | |
| Chromosome X | |||||
| y–cv–v | 0.872 ± 0.123 | 1.059 ± 0.130 | 1.083 | 0.298 | 0.477 |
| cv–v–f | 0.587 ± 0.080 | 1.115 ± 0.084 | 19.307 | 1.1 · 10−5 | 8.8 · 10−5 |
| Chromosome 2 | |||||
| dp–b–c | 1.497 ± 0.097 | 1.596 ± 0.086 | 0.589 | 0.443 | 0.506 |
| b–c–sp | 0.969 ± 0.093 | 1.271 ± 0.080 | 5.851 | 0.016 | 0.043 |
| pr–c–sp | 1.074 ± 0.106 | 1.353 ± 0.088 | 3.995 | 0.046 | 0.092 |
| Chromosome 3 | |||||
| ru–h–th | 1.128 ± 0.118 | 0.726 ± 0.095 | 7.110 | 7.7 · 10−3 | 0.031 |
| h–th–sr | 1.105 ± 0.140 | 1.041 ± 0.126 | 0.117 | 0.732 | 0.732 |
| h–th–e | 0.973 ± 0.111 | 0.855 ± 0.097 | 0.653 | 0.419 | 0.506 |
aFDR-corrected significances p (FDR) < 0.1 are bold.
Second, we examined the plasticity of CO interference in the autumn and the winter hybrids. To this end, we compared, separately for each of the two seasonal cohorts, the coefficients of coincidence in flies reared under normal conditions with those in flies subjected to desiccation hardening. The treatment significantly increased double CO rate in one pair of intervals in the autumn hybrids (cv–v–f) and another pair in the winter hybrids (ru–h–th) (Table 2). The chromosome-level analysis showed that desiccation stress resulted in considerably higher double CO rates (absolute increase 5–6%) in chromosomes X and 2 of the autumn hybrids, at the expanse of single crossovers (Supplementary Fig. S7B). In their winter counterparts, the treatment caused an increase only in chromosome 2 (Supplementary Fig. S7C).
Table 2.
The effects of desiccation stress on crossover interference in the autumn and the winter hybrids (based on the restricted maximum-likelihood analysis).
| Pair of intervals | Season | Coefficient of coincidence ± SE | Effect of treatment | |||
|---|---|---|---|---|---|---|
| Normal conditions | Desiccation hardening | Za | p | p (FDR)b | ||
| Chromosome X | ||||||
| y–cv–v | Autumn | 0.872 ± 0.123 | 1.207 ± 0.116 | 1.928 | 0.054 | 0.216 |
| Winter | 1.059 ± 0.130 | 1.214 ± 0.133 | 0.793 | 0.427 | 0.663 | |
| cv–v–f | Autumn | 0.587 ± 0.080 | 1.047 ± 0.077 | 3.894 | 9.9 · 10−5 | 7.9 · 10−4 |
| Winter | 1.115 ± 0.084 | 1.008 ± 0.075 | −0.801 | 0.423 | 0.663 | |
| Chromosome 2 | ||||||
| dp–b–c | Autumn | 1.497 ± 0.097 | 1.640 ± 0.085 | 1.652 | 0.099 | 0.264 |
| Winter | 1.596 ± 0.086 | 1.698 ± 0.088 | 0.415 | 0.678 | 0.678 | |
| b–c–sp | Autumn | 0.969 ± 0.093 | 1.358 ± 0.077 | 0.810 | 0.418 | 0.557 |
| Winter | 1.271 ± 0.080 | 1.089 ±± 0.080 | 0.816 | 0.415 | 0.663 | |
| pr–c–sp | Autumn | 1.074 ± 0.106 | 1.431 ± 0.085 | 0.397 | 0.691 | 0.691 |
| Winter | 1.353 ± 0.088 | 1.157 ± 0.090 | 0.615 | 0.538 | 0.663 | |
| Chromosome 3 | ||||||
| ru–h–th | Autumn | 1.128 ± 0.118 | 1.183 ± 0.106 | 0.400 | 0.689 | 0.691 |
| Winter | 0.726 ± 0.095 | 1.190 ± 0.113 | 3.230 | 1.2 · 10−3 | 9.6 · 10−3 | |
| h–th–sr | Autumn | 1.105 ± 0.140 | 0.877 ± 0.109 | −1.277 | 0.202 | 0.404 |
| Winter | 1.041 ± 0.126 | 1.122 ± 0.117 | 0.553 | 0.580 | 0.663 | |
| th–sr–e | Autumn | 0.258 ± 0.125 | 0.426 ± 0.153 | 1.074 | 0.283 | 0.453 |
| Winter | 0.378 ± 0.136 | 0.374 ± 0.135 | 0.681 | 0.496 | 0.663 | |
aZ-statistics takes into account the direction of line effect (see Supplementary Text S2).
bFDR-corrected significances p (FDR) < 0.1 are bold.
Discussion
Seasonal changes in desiccation tolerance
The winter isofemale lines demonstrated significantly higher desiccation tolerance than their autumn counterparts. Moreover, an increased desiccation tolerance was observed not only in the winter isofemale lines themselves but also in their hybrids with three different marker stocks, and all four estimates of desiccation tolerance (measured in the parental lines and the three hybrids) significantly correlated (Fig. 1). These findings suggest that the observed increase in desiccation tolerance is considerably inheritable. One more argument for its genetic rather than plastic nature is the fact that the isofemale lines were kept over five to six generations in normal conditions before the desiccation-tolerance assay; probably, non-genetic (plastic) changes would have disappeared during this period. Thus, the population has evolved higher desiccation tolerance during just eight to ten generations separating the autumn and the winter samples (5 months, from September till February), i.e., much faster than in a similar artificial-selection experiment (Kang et al. 2016). Such a quick response to selection pressure is consistent with the conclusion of Hoffmann and Harshman (1999) that D. melanogaster has a very high heritability of desiccation tolerance, perhaps unusually high for the whole genus.
Flies’ migration does not seem to be an alternative explanation for the observed increase in desiccation tolerance. Migration to adjacent benign habitats may indeed be a potent adaptive strategy to cope with stressful abiotic conditions of the currently occupied locality. However, relative humidity and thermal regime in the neighbourhoods of Kalka locality remain considerably homogenous within each season (IMD 2010). Besides, in the previous field studies of natural Drosophila populations from Western Himalayas, Parkash’s group and we repeatedly reported a highly significant decline in the flies’ desiccation tolerance with relative humidity of the habitat (Parkash et al. 2009, 2011, 2014; Aggarwal et al. 2013). Such clines’ repeatability argues for the leading (compared to migration and drift) role of natural selection in shaping a trait’s variation in natural populations (Endler 1986).
Several empirical studies argue that desiccation tolerance in fruit flies may be associated with considerable metabolic costs. For example, resistant flies were shown to have several-fold more intensive metabolism of carbohydrates (Marron et al. 2003). Another implicit evidence may be a negative association between basal desiccation tolerance and its plasticity under desiccation hardening, reported for some Drosophila species (Kellermann et al. 2018). These findings imply that desiccation tolerance may decrease during the subsequent wet season, when its costs are no longer outbalanced by its adaptive value. And indeed, seasonal oscillations in desiccation tolerance have been documented in several field studies (McKenzie and Parsons 1974; Parkash et al. 2011; Aggarwal et al. 2013).
Seasonal changes in crossover rate
In nine intervals (cv–v, v–f, al–dp, dp–b, pr–c, c–px, px–sp, h–th and sr–e), CO rates significantly differed between the seasons. These genome regions remarkably overlap with those that had demonstrated an increase in CO rate after 48 generations of artificial directional selection for desiccation tolerance in our previous study (Aggarwal et al. 2015). We consider the observed changes in recombination in both situations to be genetic. Genetic variation for recombination, which serves as a precondition for such changes, is indeed present in natural D. melanogaster populations, as suggested by both explicit population-level surveys (Lawrence 1958; Broadhead et al. 1977; Brooks and Marks 1986; Comeron et al. 2012; Hunter et al. 2016) and successful artificial-selection experiments on decreased or increased recombination (Chinnici 1971; Kidwell 1972; Charlesworth and Charlesworth 1985). The question, however, is which forces drive the evolution of recombination under no direct selection on its rate, like in the current study or earlier experiments (Harinarayana and Murty 1971; Lobashev et al. 1973; Flexon and Rodell 1982; Zhuchenko et al. 1985; Gorodetsky et al. 1990; Gorlov et al. 1992; Korol and Iliadi 1994; Rodell et al. 2004; Aggarwal et al. 2015).
Theoretical models suggest that directional selection on a recombination-unrelated trait can generate indirect selection on increased recombination. Local episodes of such directional selection are considered relatively common in nature and are believed to be a powerful force in the evolution of recombination (Korol et al. 1994; Otto and Michalakis 1998; Barton 2010; Ortiz-Barrientos et al. 2016). Two non-excluding mechanisms are discussed within this indirect-selection explanation. According to the first one, if epistasis between the trait-affecting loci is negative, then negative linkage disequilibria (LD) will tend to emerge in the population and favour increased CO rates. This mechanism works in large populations, especially of species with a small number of chromosomes (Charlesworth 1993). However, the long-term advantage of increased recombination may be outbalanced by its short-term disadvantage. To avoid this, either negative epistasis must be not too strong or the recombination-modifying locus must be tightly linked to the trait-affecting loci (Barton 1995). According to the second mechanism, LDs (both positive and negative) between the trait-affecting loci permanently emerge due to drift. Yet, selection eliminates the positive LDs more easily, and the prevailing negative LDs again favour increased CO rate. This mechanism should work in small-to-moderate size populations, even when the fitness-related trait is controlled by purely multiplicative genes (Otto and Barton 2001; Barton and Otto 2005).
The observed seasonal changes in desiccation tolerance seem to be mostly inheritable and likely reflect a short-term but powerful episode of natural selection, which makes the indirect-selection explanation quite plausible. Probably, this selection was much broader than just for desiccation tolerance: during winters, the population copes also with other stressors, like low temperature, low food availability, higher pathogen prevalence, etc. Seasonal changes in resistance to such stressors have been shown to include a considerable genetic component (Bergland et al. 2014; Behrman et al. 2015, 2018) (but see (Ayrinhac et al. 2004; Stone et al. 2020) for the opposite findings). Thus, such presumed episodes of natural selection should be more complex than those previously created in the lab. The multidimensional selection usually favours recombination better, as suggested by theoretical models with Hill-Robertson interference or rugged adaptive landscapes (Hadany and Beker 2003a; Roze and Barton 2006; Weissman 2014; Whitlock et al. 2016) (but see Kondrashov and Kondrashov 2001; Misevic et al. 2009 for even more complicated and less predictable scenarios).
We believe that the observed increased CO rates in the winter cohort represent longer, persistent oscillatory dynamics, analogously to variation documented for many SNP alleles across the D. melanogaster genome (Bergland et al. 2014). Since Dobzhansky et al. (1972), natural populations have been known to exhibit considerable genetic homeostasis. Thus, when selection for desiccation tolerance is over, the population will probably tend to return to its previous genetic structure. As a result, the occurring stabilising selection may decrease recombination to the initial level, as suggested by some experimental-evolution studies (Zhuchenko et al. 1985; Gorodetsky et al. 1990; Rodell et al. 2004). Besides, recombination-modifying genes may affect recombination simultaneously in different genome regions. Increased recombination in some of these regions may be maladaptive if it disrupts successful LD, which may give rise to selection on decreased recombination in the next season.
Several alternatives to the indirect-selection explanation exist. First, recombination-increasing alleles might initially be linked, by chance, to the favourable selected alleles or haplotypes. An implicit objection comes from studies where CO rate raised even when selection acted in two opposite directions: positive or negative geotaxis (Korol and Iliadi 1994), high or low sternopleural bristle number (Rodell et al. 2004), tolerance to hypoxia or hyperoxia (Aggarwal et al. 2015). The observed between-season differences in CO rates may also result from random changes in frequencies of recombination-modifying alleles. This scenario, however, seems less realistic given the remarkable concordance of the results within each season (insignificant line effect). A similar concordance was also observed in our previous artificial-selection experiments (Aggarwal et al. 2015). Finally, the differences may be associated with epigenetic changes in recombination that have succeeded to pass through generations. Indeed, several studies have reported changes in CO rate in untreated individuals whose parents and even grandparents were subjected to a stressor (Nolte 1967; Zhuchenko et al. 1983; Molinier et al. 2006; Boyko and Kovalchuk 2011). Yet, it seems unrealistic that such ‘epigenetic memory’ had lasted for five to six generations during which isofemale lines were kept in normal lab conditions before the recombination-assessment experiment.
It should be mentioned that our results may be to some extent affected by the rearing conditions. Indeed, since recombination in Drosophila is sensitive to desiccation and temperature, the autumn and winter hybrids may be differentially adapted to the employed laboratory regime. If so, the observed between-season changes in CO rate are caused simultaneously by two factors: the seasonally evolved differences in recombination control and recombination plasticity. To dissect these confound effects, one should uncover the whole reaction norm of recombination for each genotype, i.e., assess CO rate across the range of environmental conditions. However, this challenging task was beyond the scope of the current project. To the best of our knowledge, no empirical study has used such an exhausting scheme at the level of genome regions. The only study that compared reaction norms of different genotypes is one of Zhuchenko et al. (1986), where three cultivars of tomato plants, different in their temperature preferences, were examined across a range of temperature regimes. However, recombination there was assessed cytologically, based on chiasma frequencies. Here, we partially addressed this problem by comparing the desiccation-induced changes in recombination characteristics in the two seasonal cohorts. Importantly, the desiccation hardening used to test for plastic changes in recombination took into account differential desiccation tolerance of the two seasonal cohorts: more tolerant winter hybrids were subjected to a longer treatment than their autumn counterparts.
Desiccation-induced changes in crossover rate and their modulation by season and the genotype’s desiccation tolerance
In six intervals (cv–v, v–f, pr–c, c–px and th–sr), CO rates significantly increased in flies subjected to desiccation hardening, which contributes to extremely limited evidence for the recombinogenic effect of desiccation (Verde 2003; Aggarwal et al. 2015). Such an effect can be explained mechanistically, via desiccation-induced DNA damage and interaction between the DNA repair and recombination machinery (Dherin et al. 2000; Sekelsky 2017). In addition, higher CO rates in desiccation-stressed flies may be caused by intensified transcription, which may increase meiotic DNA accessibility to recombination machinery due to chromatin unfolding (Comeron et al. 2012; Adrian and Comeron 2013). Testing for this association in our material requires explicit assessment of gene expression and a much higher resolution of recombination analysis. Recombination response to desiccation hardening was higher in the autumn hybrids, even despite shorter treatment. Such a negative association between recombination plasticity and stress tolerance was previously observed in several empirical studies (Kilias et al. 1979; Zhuchenko et al. 1986; Korol et al. 1994; Jackson et al. 2015), including our recent experiment with desiccation-treated fruit flies (Aggarwal et al. 2019). Moreover, theoretical models have demonstrated that such association may be advantageous in changing environments (Hadany and Beker 2003b; Rybnikov et al. 2017).
It is worth mentioning that the season- and the treatment-associated changes in CO rate are of different nature. For the season comparisons, the changes likely result from indirect selection on recombination while in the latter they may reflect differential gene expression in the stressed and the unstressed flies. This explains why the genome regions that appeared reactive in the two comparisons (winter versus autumn and treatment versus control) do not necessarily coincide. Yet, in our analysis they do considerably overlap (compare Figs 3 and 4A). This fact additionally suggests that selection for desiccation tolerance is an important component of the presumed multivariate selection pressure.
Seasonal and desiccation-induced changes in crossover interference
The seasonal changes in CO interference appeared significant for four pairs of intervals (cv–v–f, b–c–sp, pr–c–sp and ru–h–th). Moreover, in two former pairs, double CO rates in the winter hybrids increased to such an extent that the initial positive interference had turned into a significant negative. Remarkably, the pair cv–v–f in chromosome X was highly reactive also in our previous study, where negative interference emerged upon long-term artificial selection for desiccation, as well as for resistance to hypoxia and hyperoxia (Aggarwal et al. 2015). At the same time, several pairs that were reactive in that evolutionary experiment (e.g., y–cv–v) did not show significant seasonal changes in the studied natural population. An interesting finding is the ru–h–th pair of intervals in chromosome 3, where double CO rate significantly decreased in the winter hybrids. Less pronounced changes in CO interference observed in the current study compared to the mentioned lab experiments can be explained by a much shorter period of selection: ~8–10 versus ~50–200 generations. The initially high double CO rate found in the autumn cohort may be another important factor. The negative interference observed in several regions, especially in chromosome 2, might have resulted from an unknown recent selection episode and probably hampered further evolution of this recombination feature. The presence of negative CO interference in natural populations or its appearance due to environmental or genomic stresses is debated (Denell and Keppy 1979; Peng et al. 2000; Campbell et al. 2015; Otto and Payseur 2019), and our finding contributes to the limited empirical evidence for this phenomenon.
Desiccation hardening caused a significant increase in double CO rates in two pairs of intervals, cv–v–f and ru–h–th. A higher frequency of double crossovers was also observed in the chromosome-level analysis. The similar interference-relaxing effect was earlier reported also for other environmental stressors, such as heat (Plough 1921; Graubard 1934; Hayman and Parsons 1960; Grell 1978), hypoxia and hyperoxia (Aggarwal et al. 2015) and even for intrinsic genome stress caused by meiosis-deregulating mutations (Baker and Hall 1976; Szauter 1984; Zetka and Rose 1995; Séguéla-Arnaud et al. 2015).
Conclusions
Organisms in seasonal environments must integrate information from multiple cues to synchronise transitions between life-history stages, imposing direct and indirect selection on multiple fitness-related traits and functions. The herein presented study of a D. melanogaster population subject to humid autumns and dry winters in India suggests that this may also include recombination characteristics. We provide new evidence for between-season changes in both CO rate and CO interference in natural populations, probably indirectly driven by selection on seasonality-associated stress resistance. Seasonal adaptation to desiccation also affects the plasticity of recombination, making recombination response to desiccation stress less pronounced in more tolerant genotypes. Notably, these changes accumulate within a short period (eight to ten generations) of adaptation to a natural seasonal stressor.
Some assumptions raised in the current study require further elaboration, Still, it may be viewed as a first attempt to embed the question of recombination evolution into the framework of seasonal adaptation. It prompts further surveys to better understand the critical issues of this intriguing evolutionary interplay: the relation between genetic and plastic changes, between phenotypic traits and SNP alleles and between fitness loci and modifiers of recombination.
Supplementary information
Acknowledgements
We are grateful to Prof. SC Lakhotia (Banaras Hindu University, India) for kindly providing all the laboratory facilities to conduct the experiments. We thank Prof. II Dzeverin (Schmalhausen Institute of Zoology, National Academy of Sciences of Ukraine) and two anonymous reviewers for their helpful comments on the manuscript. SR is deeply thankful to Nataliya and Sandra Rybnikova for their assistance in surveying the literature and processing the data.
Funding
The study was supported by DS Kothari postdoctoral research project (grant F.4-2/2006(BSR)/BL/16-17/0330 University Grants Commission, India) sanctioned to DDA, the Israel Science Foundation (grant 1844/17), and the Israeli Ministry of Aliyah and Integration.
Data availability
The raw data (desiccation tolerance of all examined lines and genotypes of all test-cross progeny) are publicly available at Dryad: 10.5061/dryad.g1jwstqr8.
Competing interests
The authors declare no competing interests.
Footnotes
Associate editor: Louise Johnson.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Dau Dayal Aggarwal, Sviatoslav Rybnikov.
Contributor Information
Dau Dayal Aggarwal, Email: daudayal@south.du.ac.in.
Sviatoslav Rybnikov, Email: sviatoslav.rybnikov@gmail.com.
Abraham B. Korol, Email: korol@evo.haifa.ac.il
Supplementary information
The online version contains supplementary material available at 10.1038/s41437-021-00449-2.
References
- Adrian AB, Comeron JM. The Drosophila early ovarian transcriptome provides insight to the molecular causes of recombination rate variation across genomes. BMC Genomics. 2013;14:794. doi: 10.1186/1471-2164-14-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal DD, Ranga P, Kalra B, Parkash R, Rashkovetsky E, Bantis LE. Rapid effects of humidity acclimation on stress resistance in Drosophila melanogaster. Comp Biochem Physiol A Mol Integr Physiol. 2013;166:81–90. doi: 10.1016/j.cbpa.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Aggarwal DD, Rashkovetsky E, Michalak P, Cohen I, Ronin YI, Zhou D, et al. Experimental evolution of recombination and crossover interference in Drosophila caused by directional selection for stress-related traits. BMC Biol. 2015;13:101. doi: 10.1186/s12915-015-0206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal DD, Rybnikov S, Cohen I, Frenkel Z, Rashkovetsky E, Michalak P, et al. Desiccation-induced changes in recombination rate and interference in Drosophila melanogaster: evidence for fitness-dependent plasticity. Genetica. 2019;147:291–302. doi: 10.1007/s10709-019-00070-6. [DOI] [PubMed] [Google Scholar]
- Ayrinhac A, Debat V, Gibert P, Kister AG, Legout H, Moreteau B, et al. Cold adaptation in geographical populations of Drosophila melanogaster: phenotypic plasticity is more important than genetic variability. Funct Ecol. 2004;18:700–706. doi: 10.1111/j.0269-8463.2004.00904.x. [DOI] [Google Scholar]
- Baker BS, Hall JC. Meiotic mutants: genic control of meiotic recombination and chromosome segregation. In: Ashburner M, Novitski E, editors. The genetics and biology of Drosophila. New York, NY: Academic Press; 1976. pp. 351–434. [Google Scholar]
- Barton NH. A general model for the evolution of recombination. Genet Res. 1995;65:123–244. doi: 10.1017/S0016672300033140. [DOI] [PubMed] [Google Scholar]
- Barton NH. Genetic linkage and natural selection. Philos Trans R Soc B Biol Sci. 2010;365:2559–2569. doi: 10.1098/rstb.2010.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Otto SP. Evolution of recombination due to random drift. Genetics. 2005;169:2353–2370. doi: 10.1534/genetics.104.032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman EL, Howick VM, Kapun M, Staubach F, Bergland AO, Petrov DA, et al. Rapid seasonal evolution in innate immunity of wild Drosophila melanogaster. Proc R Soc B Biol Sci. 2018;285:20172599. doi: 10.1098/rspb.2017.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman EL, Watson SS, O’Brien KR, Heschel MS, Schmidt PS. Seasonal variation in life history traits in two Drosophila species. J Evol Biol. 2015;28:1691–1704. doi: 10.1111/jeb.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Bergland AO, Behrman EL, O’Brien KR, Schmidt PS, Petrov DA. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 2014;10:e1004775. doi: 10.1371/journal.pgen.1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A, Kovalchuk I. Genetic and epigenetic effects of plant-pathogen interactions: an evolutionary perspective. Mol Plant. 2011;4:1014–1023. doi: 10.1093/mp/ssr022. [DOI] [PubMed] [Google Scholar]
- Broadhead RS, Kidwell JF, Kidwell MG. Variation of the recombination fraction in Drosophila melanogaster females. J Hered. 1977;68:323–326. doi: 10.1093/oxfordjournals.jhered.a108846. [DOI] [PubMed] [Google Scholar]
- Brooks LD, Marks RW. The organization of genetic variation for recombination in Drosophila melanogaster. Genetics. 1986;114:525–547. doi: 10.1093/genetics/114.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Furlotte NA, Eriksson N, Hinds D, Auton A. Escape from crossover interference increases with maternal age. Nat Commun. 2015;6:6260. doi: 10.1038/ncomms7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Directional selection and the evolution of sex and recombination. Genet Res. 1993;61:205–224. doi: 10.1017/S0016672300031372. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. Genetic variation in recombination in Drosophila. I. responses to selection and preliminary genetic analysis. Heredity. 1985;54:71–83. doi: 10.1038/hdy.1985.10. [DOI] [Google Scholar]
- Chinnici JP. Modification of recombination frequency in Drosophila. I. Selection for increased and decreased crossing over. Genetics. 1971;69:71–83. doi: 10.1093/genetics/69.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobror O, Olmo E, Odierna G, Angelini F, Ciarcia G. Cyclic variation of chiasma frequency and distribution in Podarcis sicula (Reptilia: Lacertidae) Genetica. 1986;71:31–37. doi: 10.1007/BF00123230. [DOI] [Google Scholar]
- Comeron JM, Ratnappan R, Bailin S. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 2012;8:e1002905. doi: 10.1371/journal.pgen.1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Chenoweth SF. Dominance reversals and the maintenance of genetic variation for fitness. PLoS Biol. 2019;17:e3000118. doi: 10.1371/journal.pbio.3000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JR, Gibert P, Legout H, Pétavy G, Capy P, Moreteau B. Isofemale lines in Drosophila: an empirical approach to quantitative trait analysis in natural populations. Heredity. 2005;94:3–12. doi: 10.1038/sj.hdy.6800562. [DOI] [PubMed] [Google Scholar]
- Denell RE, Keppy DO. The nature of genetic recombination near the third chromosome centromere of Drosophila melanogaster. Genetics. 1979;93:117–130. doi: 10.1093/genetics/93.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dherin C, Dizdaroglu M, Doerflinger H, Boiteux S, Radicella JP. Repair of oxidative DNA damage in Drosophila melanogaster: identification and characterization of dOgg1, a second DNA glycosylase activity for 8-hydroxyguanine and formamidopyrimidines. Nucleic Acids Res. 2000;28:4583–4592. doi: 10.1093/nar/28.23.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of natural populations. XIII. Recombination and variability in populations of Drosophila pseudoobscura. Genetics. 1946;31:269–290. doi: 10.1093/genetics/31.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, Levene H, Spassky B. Effects of selection and migration on geotactic and phototactic behaviour of Drosophila. 3. Proc R Soc B Biol Sci. 1972;180:21–41. doi: 10.1098/rspb.1972.0003. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T, Levene H, Spassky B, Spassky N. Release of genetic variability through recombination. III. Drosophila prosaltans. Genetics. 1959;44:75–92. doi: 10.1093/genetics/44.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreissig S, Mascher M, Heckmann S, Purugganan M. Variation in recombination rate is shaped by domestication and environmental conditions in barley. Mol Biol Evol. 2019;36:2029–2039. doi: 10.1093/molbev/msz141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler JA. Natural selection in the wild. Princeton, NJ: Princeton University Press; 1986. [Google Scholar]
- Flexon PB, Rodell CF. Genetic recombination and directional selection for DDT resistance in Drosophila melanogaster. Nature. 1982;298:6720674. doi: 10.1038/298672a0. [DOI] [PubMed] [Google Scholar]
- Gauthier F, Martin OC, Falque M. CODA (crossover distribution analyzer): quantitative characterization of crossover position patterns along chromosomes. BMC Bioinforma. 2011;12:27. doi: 10.1186/1471-2105-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt E, Wahrman J, Ledermann-Klein A, Weiss R. A two years’ survey of population dynamics in Drosophila melanogaster. Evolution. 1955;9:353–366. doi: 10.1111/j.1558-5646.1955.tb01547.x. [DOI] [Google Scholar]
- Gorlov I, Schuler L, Bunger L, Borodin P. Chiasma frequency in strains of mice selected for litter size and for high body weight. Theor Appl Genet. 1992;84:640–642. doi: 10.1007/BF00224163. [DOI] [PubMed] [Google Scholar]
- Gorodetsky BP, Zhuchenko AA, Korol AB. Efficiency of feedback selection for recombination in Drosophila. Genetika. 1990;26:1942–1952. [PubMed] [Google Scholar]
- Graubard MA. Temperature effect on interference and crossing over. Genetics. 1934;19:83–94. doi: 10.1093/genetics/19.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell RF. A comparison of heat and interchromosomal effects on recombination and interference in Drosophila melanogaster. Genetics. 1978;89:65–77. doi: 10.1093/genetics/89.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishkan I, Korol AB, Nevo E, Wasser SP. Ecological stress and sex evolution in soil microfungi. Proc R Soc B Biol Sci. 2003;270:13–18. doi: 10.1098/rspb.2002.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadany L, Beker T. Fitness-associated recombination on rugged adaptive landscapes. J Evol Biol. 2003;16:862–870. doi: 10.1046/j.1420-9101.2003.00586.x. [DOI] [PubMed] [Google Scholar]
- Hadany L, Beker T. On the evolutionary advantage of fitness-associated recombination. Genetics. 2003;165:2167–2179. doi: 10.1093/genetics/165.4.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS, Jayakar SD. Polymorphism due to selection of varying direction. J Genet. 1963;58:237–242. doi: 10.1007/BF02986143. [DOI] [Google Scholar]
- Harinarayana G, Murty BR. Cytological regulation of recombination in Pennisetum and Brassica. Cytologia. 1971;36:435–448. doi: 10.1508/cytologia.36.435. [DOI] [Google Scholar]
- Hayman DL, Parsons PA. The effect of temperature, age and an inversion on recombination values and interference in the X-chromosome of Drosophila melanogaster. Genetics. 1960;32:74–88. doi: 10.1007/BF01816087. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Harshman LG. Desiccation and starvation resistance in Drosophila: patterns of variation at the species, population and intrapopulation levels. Heredity. 1999;83:637–643. doi: 10.1046/j.1365-2540.1999.00649.x. [DOI] [PubMed] [Google Scholar]
- Hunter CM, Huang W, Mackay TFC, Singh ND. The genetic architecture of natural variation in recombination rate in Drosophila melanogaster. PLoS Genet. 2016;12:e1005951. doi: 10.1371/journal.pgen.1005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMD . Climatological normals 1981-2010: climatological tables of observatories in India 1981-2010. New Delhi: India Meteorological Department; 2010. [Google Scholar]
- Jackson S, Nielsen DM, Singh ND. Increased exposure to acute thermal stress is associated with a non-linear increase in recombination frequency and an independent linear decrease in fitness in Drosophila. BMC Evol Biol. 2015;15:175. doi: 10.1186/s12862-015-0452-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Aggarwal DD, Rashkovetsky E, Korol AB, Michalak P. Rapid genomic changes in Drosophila melanogaster adapting to desiccation stress in an experimental evolution system. BMC Genomics. 2016;17:233. doi: 10.1186/s12864-016-2556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann V, Hoffmann AA, Overgaard J, Loeschcke V, Sgrò CM. Plasticity for desiccation tolerance across Drosophila species is affected by phylogeny and climate in complex ways. Proc R Soc B Biol Sci. 2018;285:20180048. doi: 10.1098/rspb.2018.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. Genetic change of recombination value in Drosophila melanogaster. I. Artificial selection for high and low recombination and some properties of recombination-modifying genes. Genetics. 1972;70:419–432. doi: 10.1093/genetics/70.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilias G, Alahiotis SN, Onoufriou A. The alcohol dehydrogenase locus affects meiotic crossing-over in Drosophila melanogaster. Genetica. 1979;50:173–177. doi: 10.1007/BF00122042. [DOI] [Google Scholar]
- King M, Hayman D. Seasonal variation of chiasma frequency in Phyllodactylus marmoratus (Gray) (Gekkonidae – Reptilia) Chromosoma. 1978;69:131–154. doi: 10.1007/BF00329913. [DOI] [PubMed] [Google Scholar]
- Kirzhner VM, Korol AB, Nevo E. Complex dynamics of multilocus systems subjected to cyclical selection. Proc Natl Acad Sci USA. 1996;93:6532–6535. doi: 10.1073/pnas.93.13.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirzhner VM, Korol AB, Ronin YI, Nevo E. Genetic supercycles caused by cyclical selection. Proc Natl Acad Sci USA. 1995;92:7130–7133. doi: 10.1073/pnas.92.15.7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb WR. Temporal variation of Drosophila melanogaster ADH allele frequencies, inversion freqencies, and population sizes. Genetica. 1986;71:175–190. doi: 10.1007/BF00057691. [DOI] [Google Scholar]
- Kondrashov FA, Kondrashov AS. Multidimensional epistasis and the disadvantage of sex. Proc Natl Acad Sci USA. 2001;98:12089–12092. doi: 10.1073/pnas.211214298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korol AB, Iliadi KG. Increased recombination frequencies resulting from directional selection for geotaxis in Drosophila. Heredity. 1994;72:64–68. doi: 10.1038/hdy.1994.7. [DOI] [PubMed] [Google Scholar]
- Korol AB, Kirzhner VM, Ronin YI, Nevo E. Cyclical environmental changes as a factor maintaining genetic polymorphism. 2. Diploid selection for an additive trait. Evolution. 1996;50:1432–1441. doi: 10.1111/j.1558-5646.1996.tb03917.x. [DOI] [PubMed] [Google Scholar]
- Korol AB, Preygel IA, Preygel SI. Recombination variability and evolution. London: Chapman & Hall; 1994. [Google Scholar]
- Lawrence MJ. Genotypic control of crossing-over on the first chromosome of Drosophila melanogaster. Nature. 1958;182:889–890. doi: 10.1038/182889a0. [DOI] [PubMed] [Google Scholar]
- Lobashev ME, Ponomarenko VV, Polyanskaya GG, Tsapygina RI. The role of the nervous system in the regulation of various genetic and cytogenetic processes. J Evol Biochem Physiol. 1973;9:349–355. [PubMed] [Google Scholar]
- Marron MT, Markow TA, Kain KJ, Gibbs AG. Effects of starvation and desiccation on energy metabolism in desert and mesic Drosophila. J Insect Physiol. 2003;49:261–270. doi: 10.1016/S0022-1910(02)00287-1. [DOI] [PubMed] [Google Scholar]
- McKenzie A, Parsons PA. The genetic architecture of resistance to desiccation in populations of Drosophila melanogaster and D. simulans. Aust J Biol Sci. 1974;27:441–456. doi: 10.1071/BI9740441. [DOI] [PubMed] [Google Scholar]
- Misevic D, Kouyos RD, Bonhoeffer S. Predicting the evolution of sex on complex fitness landscapes. PLoS Comput Biol. 2009;5:e1000510. doi: 10.1371/journal.pcbi.1000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- Neupane S, Xu S. Adaptive divergence of meiotic recombination rate in ecological speciation. Genome Biol Evol. 2020;12:1869–1881. doi: 10.1093/gbe/evaa182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte DJ. Phase transformation and chiasma formation in locusts. Chromosoma. 1967;21:123–139. doi: 10.1007/BF00343640. [DOI] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Engelstädter J, Rieseberg LH. Recombination rate evolution and the origin of species. Trends Ecol Evol. 2016;31:226–236. doi: 10.1016/j.tree.2015.12.016. [DOI] [PubMed] [Google Scholar]
- Otto SP, Barton NH. Selection for recombination in small populations. Evolution. 2001;55:1921–1931. doi: 10.1111/j.0014-3820.2001.tb01310.x. [DOI] [PubMed] [Google Scholar]
- Otto SP, Bourguet D. Balanced polymorphisms and the evolution of dominance. Am Nat. 1999;153:561–574. doi: 10.1086/303204. [DOI] [PubMed] [Google Scholar]
- Otto SP, Michalakis Y. The evolution of recombination in changing environments. Trends Ecol Evol. 1998;13:145–151. doi: 10.1016/S0169-5347(97)01260-3. [DOI] [PubMed] [Google Scholar]
- Otto SP, Payseur BA. Crossover interference: shedding light on the evolution of recombination. Annu Rev Genet. 2019;53:19–44. doi: 10.1146/annurev-genet-040119-093957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkash R, Aggarwal DD, Kalra B, Ranga P. Divergence of water balance mechanisms in two melanic Drosophila species from the western Himalayas. Comp Biochem Physiol Part A Mol Integr Physiol. 2011;158:531–541. doi: 10.1016/j.cbpa.2010.12.018. [DOI] [PubMed] [Google Scholar]
- Parkash R, Ranga P, Aggarwal DD. Developmental acclimation to low or high humidity conditions affect starvation and heat resistance of Drosophila melanogaster. Comp Biochem Physiol Part A Mol Integr Physiol. 2014;175:46–56. doi: 10.1016/j.cbpa.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Parkash R, Sharma V, Kalra B. Impact of body melanisation on desiccation resistance in montane populations of D. melanogaster: analysis of seasonal variation. J Insect Physiol. 2009;55:898–908. doi: 10.1016/j.jinsphys.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Parsons PA. Evolutionary rates: effects of stress upon recombination. Biol J Linn Soc. 1988;35:49–68. doi: 10.1111/j.1095-8312.1988.tb00458.x. [DOI] [Google Scholar]
- Peng J, Korol AB, Fahima T, Ro MS, Ronin YI, Li YC, et al. Molecular genetic maps in wild emmer wheat, Triticum dicoccoides: genome-wide coverage, massive negative interference, and putative quasi-linkage. Genome Res. 2000;10:1509–1531. doi: 10.1101/gr.150300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plough HH. The effect of temperature on crossingover in Drosophila. J Exp Zool. 1917;24:147–209. doi: 10.1002/jez.1400240202. [DOI] [Google Scholar]
- Plough HH. Further studies on the effect of temperature on crossing over. J Exp Zool. 1921;32:187–202. doi: 10.1002/jez.1400320202. [DOI] [Google Scholar]
- Prakash HS, Reddy GS. Seasonality and population fluctuations in the Drosophila of Western Ghats. Proc Anim Sci. 1979;88:193–204. doi: 10.1007/BF03179094. [DOI] [Google Scholar]
- Rice WR. Experimental tests of the adaptive significance of sexual recombination. Nat Rev Genet. 2002;3:241–251. doi: 10.1038/nrg760. [DOI] [PubMed] [Google Scholar]
- Ritz KR, Noor MAF, Singh ND. Variation in recombination rate: adaptive or not? Trends Genet. 2017;33:364–374. doi: 10.1016/j.tig.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Rodell CF, Schipper MR, Keenan DK. Modes of selection and recombination response in Drosophila melanogaster. J Hered. 2004;95:70–75. doi: 10.1093/jhered/esh016. [DOI] [PubMed] [Google Scholar]
- Roze D, Barton NH. The Hill-Robertson effect and the evolution of recombination. Genetics. 2006;173:1793–1811. doi: 10.1534/genetics.106.058586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybnikov SR, Frenkel ZM, Korol AB. What drives the evolution of condition-dependent recombination in diploids? Some insights from simulation modelling. Philos Trans R Soc B Biol Sci. 2017;372:20160460. doi: 10.1098/rstb.2016.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggoo MIS, Gupta RC, Kaur R. Seasonal variation in chiasma frequency among three morphotypes of Eclipta alba. Chromosom Bot. 2010;5:33–36. doi: 10.3199/iscb.5.33. [DOI] [Google Scholar]
- Saleem M, Lamb BC, Nevo E. Inherited differences in crossing over and gene conversion frequencies between wild strains of Sordaria fimicola from ‘Evolution Canyon’. Genetics. 2001;159:1573–1593. doi: 10.1093/genetics/159.4.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Bomblies K, Fitz J, Laitinen RAE, Warthmann N, Yant L, et al. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity. 2012;108:447–455. doi: 10.1038/hdy.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuk K, Manzano-Winkler B, Ritz KR, Noor MAF. Natural selection shapes variation in genome-wide recombination rate in Drosophila pseudoobscura. Curr Biol. 2020;30:1517–1528. doi: 10.1016/j.cub.2020.03.053. [DOI] [PubMed] [Google Scholar]
- Séguéla-Arnaud M, Crismani W, Larchevêque C, Mazel J, Froger N, Choinard S, et al. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc Natl Acad Sci USA. 2015;112:4713–4718. doi: 10.1073/pnas.1423107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. DNA repair in Drosophila: mutagens, models, and missing genes. Genetics. 2017;205:471–490. doi: 10.1534/genetics.116.186759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevison LS. Causes and consequences of recombination rate variation in drosophila [Thesis] Durham, NC: Duke University; 2011. [Google Scholar]
- Stone HM, Erickson PA, Bergland AO. Phenotypic plasticity, but not adaptive tracking, underlies seasonal variation in post-cold hardening freeze tolerance of Drosophila melanogaster. Ecol Evol. 2020;10:217–231. doi: 10.1002/ece3.5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szauter P. An analysis of regional constraints on exchange in Drosophila melanogaster using recombination-defective meiotic mutants. Genetics. 1984;106:45–71. doi: 10.1093/genetics/106.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde LA. The effect of stress on meiotic recombination in maize (Zea mays L.) [Thesis] Ames, IA: Iowa State University; 2003. [Google Scholar]
- Weismann A. The significance of sexual reproduction in the theory of natural selection. In: Poulton EB, Schonland S, Shipley AE, editors. Essays upon heredity and kindred biological problems. Oxford: Clarendon Press; 1889. pp. 251–332. [Google Scholar]
- Weissman DB (2014). Stress-induced variation can cause average mutation and recombination rates to be positively correlated with fitness. In: Artificial life 14: proceedings of the 14th international conference on the synthesis and simulation of living systems, pp 43–44
- Whitlock AOB, Peck KM, Azevedo RBR, Burch CL. An evolving genetic architecture interacts with Hill–Robertson interference to determine the benefit of sex. Genetics. 2016;203:923–936. doi: 10.1534/genetics.116.186916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CM, Ragland GJ, Betini G, Buckley LB, Cheviron ZA, Donohue K, et al. Understanding evolutionary impacts of seasonality: an introduction to the symposium. Integr Comp Biol. 2017;57:921–933. doi: 10.1093/icb/icx122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann MJ, Bergland AO, Feldman MW, Schmidt PS, Petrov DA. Seasonally fluctuating selection can maintain polymorphism at many loci via segregation lift. Proc Natl Acad Sci USA. 2017;114:E9932–E9941. doi: 10.1073/pnas.1702994114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Rose AM. Mutant rec-1 eliminates the meiotic pattern of crossing over in Caenorhabditis elegans. Genetics. 1995;141:1339–1349. doi: 10.1093/genetics/141.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuchenko AA, Korol AB, Gavrilenko TA, Kibenko TY. The relation between genotype adaptivity and reactivity of its recombination characteristics under temperature effect. Genetika. 1986;22:966–974. [Google Scholar]
- Zhuchenko AA, Korol AB, Kovtyukh PL. Change of the crossing-over frequency in Drosophila during selection for resistance to temperature fluctuations. Genetica. 1985;67:73–78. doi: 10.1007/BF02424463. [DOI] [Google Scholar]
- Zhuchenko AA, Kovtyukh LP, Korol AB. Testing the ‘ontogenetic memory’ hypothesis on Drosophila. Proc USSR Acad Sci. 1983;269:1493–1495. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data (desiccation tolerance of all examined lines and genotypes of all test-cross progeny) are publicly available at Dryad: 10.5061/dryad.g1jwstqr8.