ABSTRACT
Sap-sucking hemipterans host specialized, heritable microorganisms that supplement their diet with essential nutrients. These microbes show unusual features that provide a unique perspective on the coevolution of host-symbiont systems but are still poorly understood. Here, we combine microscopy with high-throughput sequencing to revisit 80-year-old reports on the diversity of symbiont transmission modes in a broadly distributed planthopper family, Dictyopharidae. We show that in seven species examined, the ancestral nutritional symbionts Sulcia and Vidania producing essential amino acids are complemented by co-primary symbionts, either Arsenophonus or Sodalis, acquired several times independently by different host lineages and contributing to the biosynthesis of B vitamins. These symbionts reside within separate bacteriomes within the abdominal cavity, although in females Vidania also occupies bacteriocytes in the rectal organ. Notably, the symbionts are transovarially transmitted from mothers to offspring in two alternative ways. In most examined species, all nutritional symbionts simultaneously infect the posterior end of the full-grown oocytes and next gather in their perivitelline space. In contrast, in other species, Sodalis colonizes the cytoplasm of the anterior pole of young oocytes, forming a cluster separate from the “symbiont ball” formed by late-invading Sulcia and Vidania. Our results show how newly arriving microbes may utilize different strategies to establish long-term heritable symbiosis.
KEYWORDS: planthoppers, nutritional symbiosis, transovarial transmission, genomes
INTRODUCTION
Mutualistic relationships with heritable bacterial and/or fungal microorganisms have played crucial roles in the biology of multiple groups of insects, contributing significantly to their evolutionary and ecological success (1–3). The growing awareness of the diversity and importance of insect symbioses, in addition to the rapid development in sequencing-based techniques, has led to an increased interest in these associations. However, outside a few model species and some reasonably well-sampled clades, our knowledge of the diversity, evolution, and biological characteristics of the microbial symbionts and the microbial roles in the evolution of insect diversity remains limited (4, 5). Among insects, sap-sucking hemipterans are obligately dependent on heritable nutritional microbes that supplement their unbalanced diet with essential amino acids, vitamins, and cofactors (4, 6–8). Multiple symbiont combinations have been described from Auchenorrhyncha, a suborder comprising infraorders Fulgoromorpha (planthoppers) and Cicadomorpha (cicadas, spittlebugs, treehoppers, and leafhoppers) (4, 9). Their common ancestor that lived about 300 million years ago (MYA) is thought to have been colonized by two microbes, a Bacteroidetes member currently known as “Candidatus Sulcia muelleri” (further referred to as Sulcia) and a betaproteobacterium, variably known as “Ca. Nasuia deltocephalinicola,” “Ca. Zinderia insecticola,” or “Ca. Vidania fulgoroideae” (further referred to as Nasuia, Zinderia, and Vidania, respectively, or as beta-symbionts collectively) (9–12). These maternally transmitting and nutrient-providing symbionts have become obligate components of host biology. However, in many host clades, one or both became complemented or replaced by other microbes. In these multipartite symbioses, microbes share the responsibility for essential nutrient biosynthesis. For example, in known Cicadomorpha, Sulcia encodes pathways for producing 7 or 8 essential amino acids, whereas the remaining 3 or 2 amino acids are provided by its symbiotic partner (8, 9, 12). The situation can get more complicated when one of the ancient microbes gets replaced by another or when the host is colonized by more than two symbionts, and the nutritional functions become subdivided among a greater number of partners. This has occurred repeatedly in different lineages of Auchenorrhyncha, where additional symbionts have either taken over the beta-symbiont’s role or contribute to vitamin biosynthesis (4).
These intimate and intricate metabolic interdependencies between insects and associated microorganisms indicate a vital role of nutritional symbionts in host biology and the need to ensure reliable transmission of these essential partners across generations. Microbes can be transmitted from mothers to offspring (vertically) or through the environment (horizontally), and within these transmission modes, a variety of transmission routes exists (13). In sap-feeding hemipteran insects, transovarial transmission through female germ cells, at a certain stage of their development, predominates (14–16). In some insects, symbionts infect undifferentiated germ cells, or young, previtellogenic oocytes (16). However, in most hemipteran taxa, including all Auchenorrhyncha studied to date, they invade ovarioles containing older (vitellogenic or choriogenic) oocytes. The complementation or replacement of ancient, coadapted heritable nutritional symbionts by newly arriving microbes, while likely beneficial to the hosts because of their greater metabolic capacity and efficiency (17, 18), creates apparent challenges for their transmission. Host lineages that acquired new symbionts have to adopt existing mechanisms or develop new traits and mechanisms for their effective vertical transmission, which is crucial for the fixation of new symbiosis (19), being also a matter of life or death to the newly established symbionts. The evolution of the symbiont transmission and the symbiont replacements are inseparably linked, and we need to study one to understand the other.
While summarizing decades of microscopy-based research on Auchenorrhyncha symbioses, Buchner (14) famously wrote about “the veritable fairyland of insect symbiosis,” apparently referring to the diversity of microbes in different host clades, as well as their transmission mechanisms. However, he lacked tools to fully characterize the evolution of symbioses across the auchenorrhynchan phylogeny. The popularization of DNA sequencing-based techniques enabled such investigation, but our knowledge is still restricted to a few Auchenorrhyncha clades, mainly within Cicadomorpha (12, 19, 20). While diagnostic screens revealed Sulcia, Vidania, and often other bacteria or fungi in most planthopper families (21), in only one species so far have nutritional symbionts been characterized using genomics. In the Hawaiian cixiid Oliarus filicicola, Vidania produces seven essential amino acids, and Sulcia synthesizes three, whereas a more recently acquired gammaproteobacterial symbiont, Purcelliella, contributes B vitamins (22). This rearrangement of Sulcia and Vidania nutritional responsibilities shed new light on the evolution of planthopper symbioses and how infections with additional microbes influence them. However, our understanding of symbioses in this diverse, widespread, and ecologically significant insect clade remains very limited.
This work aimed to provide an insight into the diversity and biology of symbioses in the large, diverse planthopper family Dictyopharidae. Previously, eight members of this family were shown to possess the bacteria Vidania and usually also Sulcia (21), but microscopic observations conducted by Müller (23, 24) and summarized by Buchner (14) indicated that they also host a third symbiont. They reported that these additional symbionts transmitted either together or separately from Sulcia and Vidania, suggesting their independent origins or other unusual phenomena, making this family a valuable system for the exploration of symbiont complementation and genome evolution.
Here, we report the results of microscopy and sequencing-based investigations of symbiotic bacteria associated with seven species belonging to the Dictyopharidae. We survey the diversity of the symbionts and explore their nutritional roles. In particular, we focus on how symbionts in this group are transmitted across generations. We describe and discuss how the auchenorrhynchan symbiont transmission can be separated in time and space, how this may have evolved, and what it may mean for the host.
RESULTS
Dictyopharidae planthoppers harbor (at least) three types of heritable symbionts.
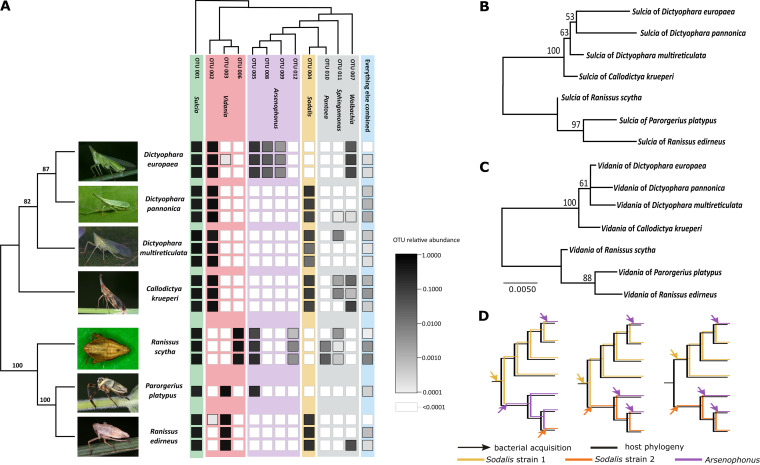
Amplicon sequencing-derived mitochondrial cytochrome oxidase I (COI) sequences for the representative specimens of seven experimental species confirmed their morphology-based identifications (Fig. 1A; see also Table S3A and B in the supplemental material). The phylogeny revealed two well-supported clades corresponding to subfamilies Dictyopharinae and Orgerinae; it also showed that Ranissus edirneus is more closely related to Parorgerius platypus than to Ranissus scytha, in agreement with the proposed taxonomic revision of the subfamily Orgerinae (25). We successfully amplified the bacterial 16S rRNA gene V4 region from the abdomens of all experimental individuals from these seven Dictyopharidae species. The total number of 16S rRNA gene reads passed through all analysis steps was 452,949, or 23,840 per sample on average (Table S3C). Clustering with 97% identity cutoff identified 67 operational taxonomic units (OTUs).
FIG 1.
The bacterial communities in Dictyopharidae planthoppers. (A) The diversity and relative abundance of bacteria in replicate specimens from seven experimental species. Insect phylogeny (Maximum Likelihood [ML]) is based on concatenated marker gene sequences (COI, CytB, 18S rRNA, and 28S rRNA); bootstrap values are shown above the nodes. The bacterial tree is based on representative sequences of the 16S rRNA V4 region for different OTUs. (B and C) ML phylogenies for Sulcia and Vidania symbionts from seven experimental species, based on full-length 16S rRNA sequences. (D) Three of many possible scenarios of Sodalis/Arsenophonus acquisition and replacement during Dictyopharidae diversification.
OTU tables. Download Table S3, XLSX file, 1.2 MB (1.2MB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All studied individuals hosted Sulcia and Vidania and, additionally, either Sodalis or Arsenophonus (Fig. 1A and Table S3C). Together, these symbionts comprised >97% of reads in each of the libraries. The single-nucleotide-resolution data (Table S3D) for these dominant symbionts revealed high similarity among strains of the slow-evolving symbiont Sulcia genotypes and a greater distance among strains of Vidania from different Dictyopharidae clades, which grouped into three distinct 97% OTUs. Callodictya krueperi, Dictyophara multireticulata, Dictyophara pannonica, and R. edirneus hosted different genotypes of Sodalis (clustering to one OTU). D. europaea, S. scytha, and P. platypus hosted Arsenophonus, typically different genotypes from more than one OTU. However, the consistent relative abundance of genotypes in replicate individuals suggested that these genotypes/OTUs correspond to different rRNA operons within the genome of a single Arsenophonus strain (Fig. 1A). Less-abundant microbial OTUs present in some species included Wolbachia, Pantoea, and Sphingomonas (Fig. 1A). All the remaining OTUs combined accounted for 0.2% of the total number of reads, and many of them represented contaminants or symbiont-derived sequences that accumulated large numbers of errors. We did not consider them further.
Phylogenetic trees of Sulcia and Vidania based on full-length 16S rRNA gene sequences (Fig. 1B and C) are congruent with the phylogeny of the seven experimental Dictyopharidae species (Fig. 1A), or a broader range of planthoppers (Fig. S1A and B), as expected for symbionts codiversifying with hosts. In contrast, 16S rRNA gene phylogenies for Sodalis and Arsenophonus from diverse hosts, while poorly resolved and supported, were suggestive of independent origins of at least some of the cosymbionts of Dictyopharidae (Fig. S1C and D). This hypothesis was further supported using phylogenomics: phylogenies reconstructed based on 129 conserved single-copy protein-coding genes of symbiotic enterobacteria and relatives, constructed using reference alignments and following the methods of McCutcheon and colleagues (18) and including C. krueperi, D. multireticulata, and R. scytha symbionts, strongly suggest that these three symbionts originated from independent infections (Fig. S1E). These data, combined with information on the distribution of the two symbiont clades across the host phylogeny and differences in transmission patterns and genomics characteristics (described below), indicate several independent infections by Arsenophonus/Sodalis or their repeated replacements by other strains of these symbionts (Fig. 1B to D and Fig. 2 to 5). Unfortunately, the currently available data do not allow for a reconstruction of the order of these infections. Three of many possible scenarios, all assuming relatively few replacement events and shared ancestry of some strains, are presented in Fig. 1D. However, the number of independent infections or replacements along the tree branches could have been much greater. In fact, patterns such as the 16S rRNA amplicon genotype diversity across D. pannonica individuals (Table S3D) and some morphological differences (Fig. S3A) among Sodalis cell populations hint at the possibility of recent or perhaps ongoing replacements.
FIG 2.
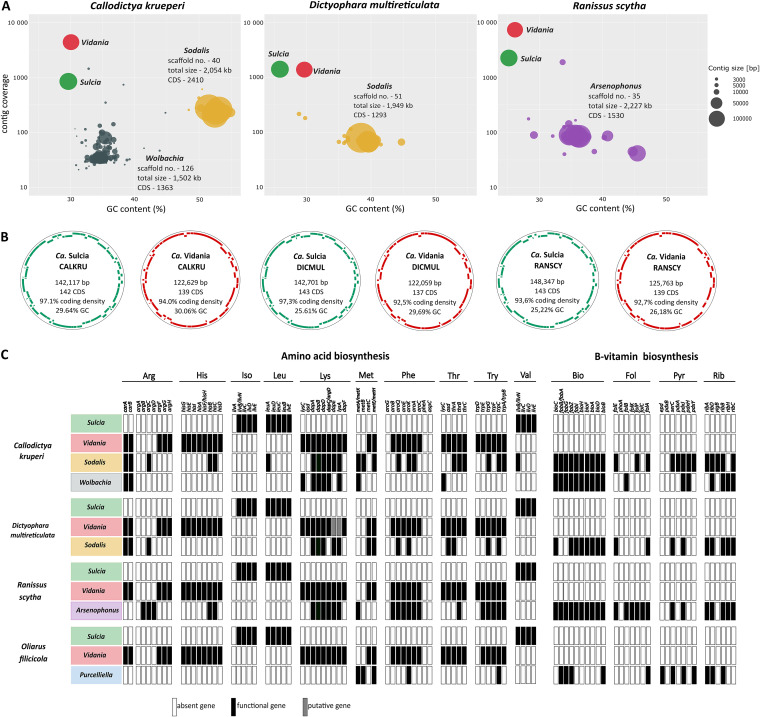
The summary of metagenomic data for three Dictyopharidae species. (A) Taxon-annotated GC content-coverage plots for metagenomic assemblies. Colored blobs represent scaffolds corresponding to the identified symbiont genome fragments. (B) Circular diagrams of Sulcia and Vidania genomes from three species, showing gene positions on forward and reverse strands and basic genome characteristics. (C) Contribution of symbionts to amino acid and B vitamin biosynthesis pathways. Standard abbreviations for amino acid, B vitamin, and gene names are used. Bio, biotin; Fol, folate; Pyr, pyridoxine; Rib, riboflavin.
FIG 3.
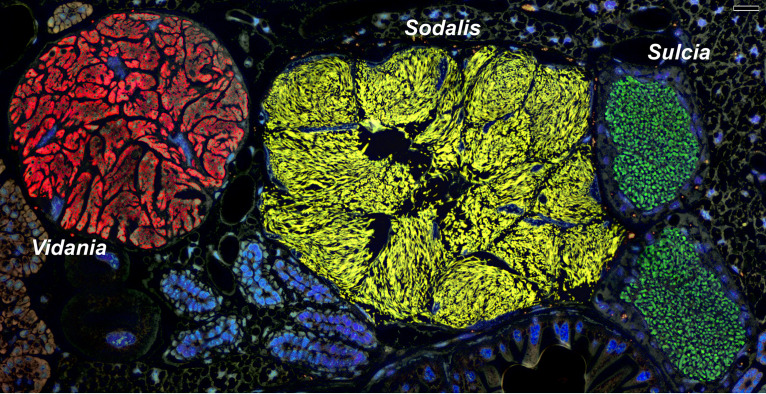
Fluorescence in situ hybridization (FISH) demonstrates how in C. krueperi, each of its three co-primary symbionts inhabits a distinct bacteriome. Transverse section through the insect’s abdomens. The image is oriented in dorsal-ventral position. Specific probes for Vidania (red), Sulcia (green), and Sodalis (yellow) were used. Blue represents cell nuclei stained with DAPI (4′,6-diamidino-2-phenylindole). Confocal laser microscope (CLM), bar = 20 μm.
FIG 4.
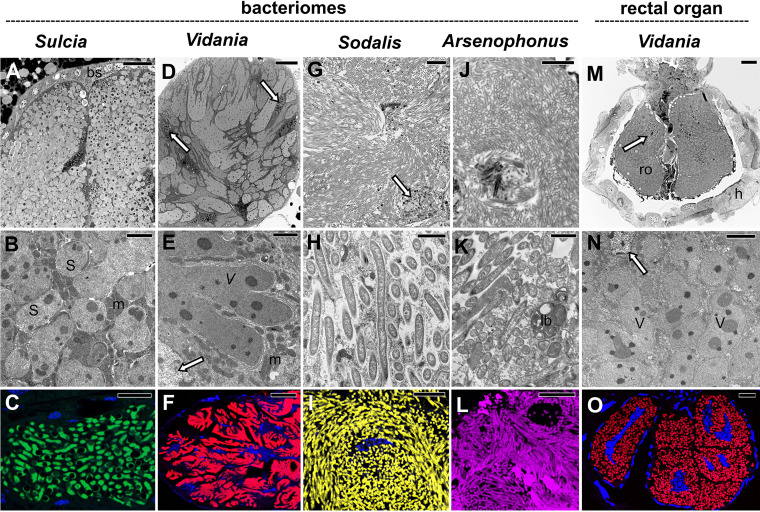
Tissue localization and morphology of symbionts in the Dictyopharidae species examined. Top row (A, D, G, J, and M): the organization of symbionts within bacteriomes or the rectal organ. Light microscopy (LM), bar = 20 μm. Middle row (B, E, H, K, and N): the ultrastructure of symbiont cells. Transmission electron microscopy (TEM), bar = 2 μm. Bottom row (C, F, I, L, and O): fluorescence in situ hybridization (FISH) microphotographs of symbiont cells within the bacteriome or rectal organ. Probes specific to each of the symbionts were used. Blue represents cell nuclei stained with DAPI. Confocal laser microscope (CLM), bar = 20 μm. Insect species: (A) D. multireticulata, (B and L) R. scytha, (C, E to I, M, and N) C. krueperi, (D and O) D. pannonica, (J and K) D. europaea. Abbreviations and symbol: bs, bacteriome sheath; h, hindgut; lb, lamellar body; m, mitochondrion; ro, rectal organ; S, Sulcia; V, Vidania; white arrow, bacteriocyte nucleus.
FIG 5.
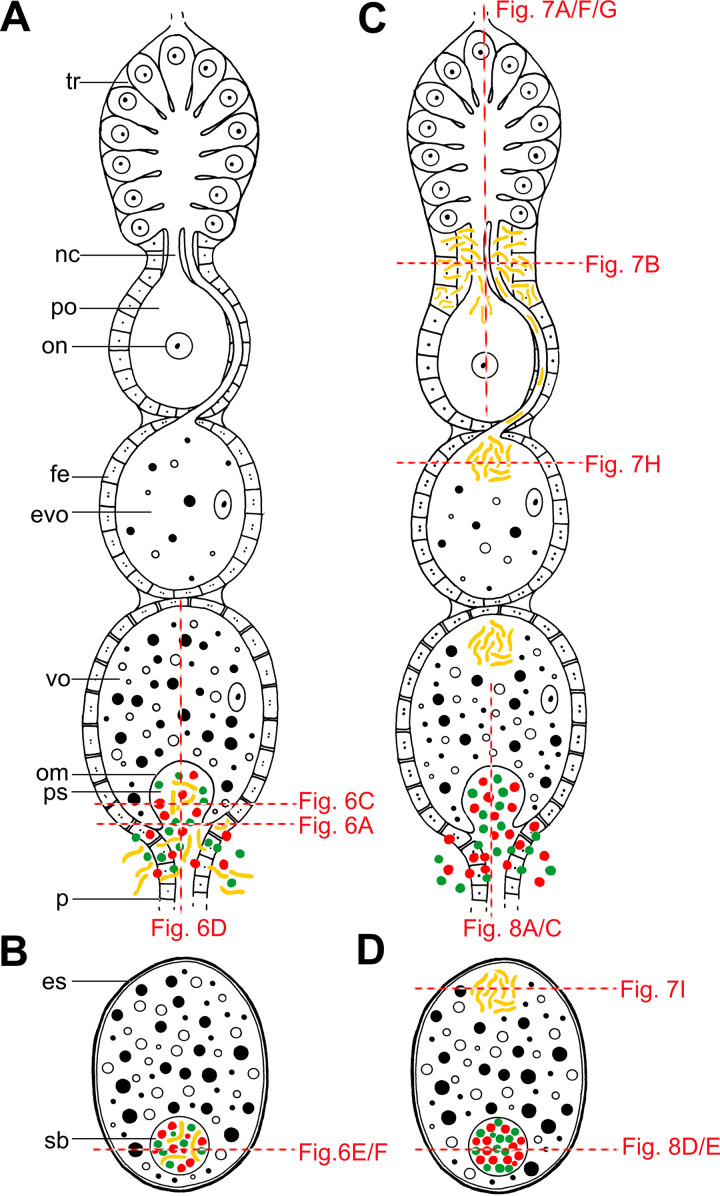
Schematic representation of the alternative modes of the symbiont transmission in Dictyopharidae planthoppers. (A) Simultaneous transmission of all types of nutritional symbionts through the follicular epithelium surrounding the posterior pole of terminal oocyte. (B) Full-grown oocyte with a “symbiont ball” containing three types of symbionts. The transmission mode shown in panels A and B is used by the large majority of Auchenorrhyncha, including subfamily Orgerinae and Arsenophonus-infected D. europaea from subfamily Dictyopharinae. (C) Spatially and temporally separated transmission of different symbionts (Sodalis versus Sulcia and Vidania). Sodalis infects previtellogenic and early-vitellogenic oocytes, whereas the remaining symbionts invade terminal vitellogenic oocytes. (D) Full-grown oocyte with the accumulation of Sodalis bacteria at the anterior pole and “symbiont ball” with Sulcia and Vidania cells at the posterior pole. This mode of transmission is unique to Sodalis-infected members of the subfamily Dictyopharinae. Red dashed lines indicate regions shown in panels of Fig. 6 to 8. Abbreviations: tr, tropharium; nc, nutritive cord; po, previtellogenic oocyte; on, oocyte nucleus; fe, follicular epithelium; evo, early-vitellogenic oocyte; vo, vitellogenic oocyte; om, oocyte membrane; p, pedicel; ps, perivitelline space; es, eggshells; sb, “symbiont ball.”
ML phylogenies of symbionts based on 16S rRNA gene sequences. (A) ML phylogeny of Sulcia strains from diverse planthoppers, based on full-length sequences (1,373 shared nucleotide positions) of 16S rRNA gene. Only bootstrap values above 70% are shown. Sequences from the seven Dictyopharidae species studied here are highlighted using the colored font. (B) ML phylogeny of Vidania strains from diverse planthoppers based on full-length sequences (1,530 shared nucleotide positions) of 16S rRNA gene. Only bootstrap values above 70% are shown. Sequences from the seven Dictyopharidae species studied here are highlighted using the colored font. (C) ML phylogeny of Sodalis strains from different hosts based on 16S rRNA gene sequences. Only bootstrap values above 70% are shown. (D) ML phylogeny of Arsenophonus strains from different hosts based on 16S rRNA gene sequences. Only bootstrap values above 70% are shown. (E) ML phylogeny based on first and second codon positions of 129 conserved single-copy protein-coding genes of symbiotic enterobacteria and relatives. Only bootstrap values above 70% are shown. Download FIG S1, PDF file, 1.8 MB (1.8MB, pdf) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
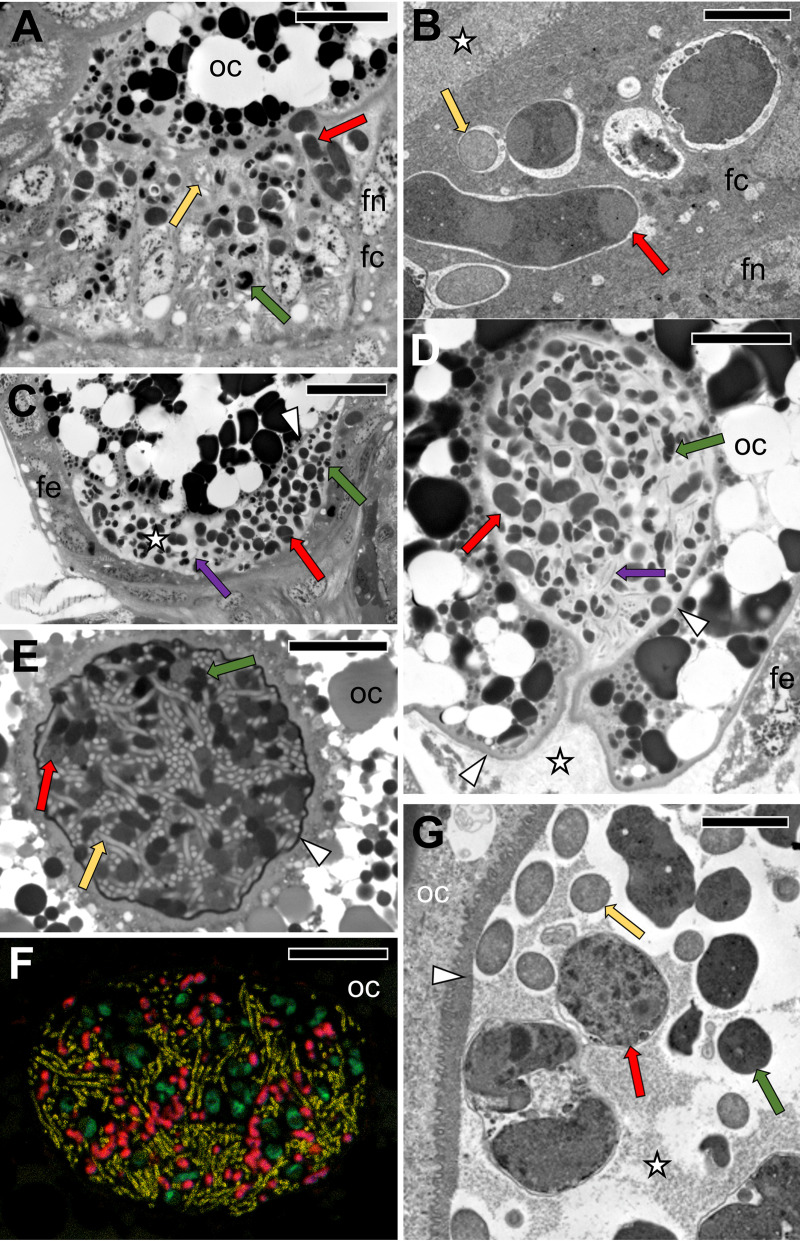
(A) Dictyophara pannonica. Two morphotypes of Sodalis symbiont (red and yellow arrows) within bacteriome of single individuals. TEM, bar = 2 μm. (B) Wolbachia symbiont cells within bacteriocytes of Dictyopharidae species (TEM). (1) Wolbachia cells (green arrows) in the cytoplasm of the Vidania (V) bacteriocyte from Callodictya krueperi. (2) Wolbachia cells (green arrows) in the cytoplasm of the Sulcia (S) bacteriocyte from Dictyophara europaea. TEM, bar = 2 μm. Download FIG S3, PDF file, 1.1 MB (1.1MB, pdf) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sulcia and Vidania provide essential amino acids to host insects, whereas Arsenophonus, Sodalis, and Wolbachia supplement their diet with B vitamins.
The symbiont genomes identified in the assemblies matched those identified by amplicon sequencing (Fig. 2A). The circular genomes of Sulcia and Vidania ranged in size between 142 and 148 kb and 122 to 125 kb, respectively, and were characterized by relatively low GC contents (25 to 30%) and high coverage (800 to 10,000×) (Fig. 2B). Sulcia and Vidania genomes were colinear relative to each other and the slightly larger reference genomes of O. filicicola (OLIH) symbionts (157 and 136 kb, respectively) (Fig. S2) (22). Our metagenomic analyses also confirmed the presence of Sodalis and Wolbachia in C. krueperi, Sodalis in D. multireticulata, and Arsenophonus in R. scytha. The genomic assemblies of these symbionts were fragmented; the number of scaffolds ranged between 23 and 126 and their total size between 1,500 kb and 2,220 kb. The scaffold read coverage was substantially lower than in the cases of Sulcia and Vidania but also relatively variable, particularly in the case of Arsenophonus (Table S4A).
The alignments of planthopper Sulcia and Vidania genomes. (A) PROMER alignments of final Sulcia genomes from three Dictyopharidae species, Callodictya krueperi (CALKRU), Dictyophara multireticulata (DICMUL), and Ranissus scytha (RANSCY), against the Sulcia genome from Oliarus filicicola (OLIH; family Cixiidae). (B) PROMER alignments of final Vidania genomes from three Dictyopharidae species, Callodictya krueperi (CALKRU), Dictyophara multireticulata (DICMUL), and Ranissus scytha (RANSCY), against the Vidania genome from Oliarus filicicola (OLIH; Cixiidae). Download FIG S2, PDF file, 0.1 MB (115KB, pdf) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome details. Download Table S4, XLSX file, 0.3 MB (282.3KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In terms of gene content, the newly characterized Sulcia and Vidania genomes are very similar to each other and the previously described OLIH symbiont genomes (22). Sulcia genomes include 142 to 143 predicted protein-coding genes, 27 to 29 identifiable tRNAs, and complete ribosomal operon, and their coding density ranges from 93.6% to 97.3%. Vidania genomes include 137 to 139 predicted protein-coding genes, 23 to 25 tRNAs, and complete ribosomal operon, with coding density ranging between 92.5% and 94% (Fig. 2B). In the two analyzed Sodalis genomes, Prokka identified 2,410 (CALKRU) and 1,132 (DICMUL) predicted protein-coding genes; in Arsenophonus, 1,530; and in Wolbachia, 1,363. However, because of the incompleteness of the assemblies of these cosymbiont genomes and challenges with pseudogene annotation, these numbers are approximate.
Sulcia and Vidania complement each other in provisioning host insects with 10 essential amino acids, in a way consistent among the three Dictyopharidae and OLIH (Fig. 2C). Sulcia participates in the biosynthesis of three essential amino acids including isoleucine, leucine, and valine, whereas Vidania is involved in synthesizing the remaining seven. Interestingly, some of the biosynthetic pathways (isoleucine, arginine, methionine, phenylalanine) were not complete, with genes missing from Sulcia and Vidania genomes (Fig. 2C). Some of these missing amino acid biosynthesis genes, and duplicate copies of some others, are present in Sodalis, Arsenophonus, and Wolbachia genomes. For example, in C. krueperi, Vidania lacks the first two genes essential for methionine biosynthesis from homoserine, but both are present in the Sodalis genome. Similarly, in D. multireticulata, Sodalis complements two pseudogenes (dapE, lysA) in the Vidania genome, part of the lysine biosynthesis pathway. Arsenophonus from R. scytha has the same set of phenylalanine and tryptophan biosynthesis genes as Vidania. In all Dictyopharidae, the gammaproteobacterial symbionts contain many genes in the lysine biosynthesis pathway and scattered genes from other pathways, and the list might get extended once assemblies are complete.
In addition to genes involved in amino acids’ biosynthesis, Sodalis, Arsenophonus, and Wolbachia contribute to the synthesis of B vitamins: biotin, folate, riboflavin, and pyridoxine (Fig. 2C). Sodalis and Wolbachia associated with C. krueperi as well as Arsenophonus of R. scytha contain full sets of biotin biosynthesis genes, whereas the genome of Sodalis of D. multireticulata carries 8 of 10 genes from that pathway. The Arsenophonus symbiont genome also carries the majority of genes involved in the biosynthesis of folate and riboflavin. Interestingly, in C. krueperi, both Sodalis and Wolbachia appear capable of riboflavin biosynthesis.
Nutritional symbionts of dictyopharids occupy distinct but adjacent bacteriocytes.
In all dictyopharids studied, nutritional symbionts (Sulcia, Vidania, and either Sodalis or Arsenophonus) reside within separate bacteriomes located close to each other within the insect abdomen (Fig. 3 and 4). Light microscopy observations revealed that bacteriomes harboring bacteria Sulcia, Sodalis, and Arsenophonus are made up of several bacteriocytes (Fig. 3 and 4A), whereas bacteriomes with Vidania are syncytial (Fig. 3 and 4D). Bacteriomes occupied by Sulcia and Vidania are surrounded by a thick or thin monolayered bacteriome sheath, respectively (Fig. 4A, C, D, and F). In contrast, bacteriomes with Arsenophonus and Sodalis are not covered by epithelial cells (Fig. 3).
Our histological, ultrastructural, and fluorescence in situ hybridization (FISH) analyses revealed that Sulcia cells are pleomorphic and possess large electron-dense inclusions in their cytoplasm (Fig. 3 and 4A to C). Vidania cells are giant and multilobed, with numerous electron-dense accumulations in the cytoplasm (Fig. 3 and 4D to F). The cells of gammaproteobacterial symbionts Sodalis and Arsenophonus are large, elongated, and morphologically similar in all host species (Fig. 3 and 4J to L). In particular, we did not observe distinct Arsenophonus morphotypes, further supporting our amplicon-based conclusions that different 16S OTUs correspond to different rRNA operons in the genome of a single strain (Fig. 4J to K).
In all species analyzed, bacteriocytes have large, polyploid nuclei, and their cytoplasm is tightly packed with symbionts, ribosomes, and mitochondria (Fig. 3 and 4). The density of mitochondria in bacteriocytes with Sulcia and Vidania seemed substantially higher than in bacteriocytes with Sodalis and Arsenophonus. Sulcia and Vidania tightly adhere to each other in the cytoplasm of their bacteriocytes, whereas Sodalis and Arsenophonus are somewhat less densely packed (Fig. 3 and 4G to L). Finally, ultrastructural observations in C. krueperi and D. europaea revealed small, rod-shaped bacteria in the cytoplasm of bacteriocytes housing Sulcia and Vidania (Fig. S3B). Their shape and size matched Wolbachia, detected in these species using amplicon sequencing.
Besides bacteriomes, Vidania also occupies bacteriocytes in the rectal organ, localized in the invagination of hindgut epithelium (Fig. 4M to O). Vidania cells in the rectal organ differ in shape and size from Vidania localized in the bacteriomes.
Dictyopharids have developed different symbiont transmission strategies.
Histological observations of serial semithin sections revealed that nutritional symbionts of the studied Dictyopharidae are transmitted between generations transovarially. The details of Sulcia, Vidania, and Arsenophonus transmission agree with observations from other Auchenorrhyncha. Strikingly, we observed significant differences in the transmission of Sodalis (Fig. 5 to 8).
FIG 6.
The simultaneous transmission of three symbionts in selected Dictyopharidae, as shown in Fig. 5A and B. (A) The migration of Sulcia, Vidania, and Arsenophonus through follicular cells surrounding the posterior pole of the terminal oocyte. D. europaea, light microscopy (LM), bar = 20 μm. (B) Sodalis and Vidania in the cytoplasm of the follicular cell. R. edirneus, TEM, bar = 2 μm. (C and D) Symbiotic bacteria in the perivitelline space. D. europaea, LM, bar = 20 μm. (E) A “symbiont ball” containing bacteria Sulcia, Vidania, and Sodalis in the deep depression of the oolemma at the posterior pole of the terminal oocyte. R. edirneus, LM, bar = 20 μm. (F) In situ identification of symbionts in the “symbiont ball” within the terminal oocyte. R. edirneus, CLM, bar = 20 μm. (G) Fragment of the “symbiont ball” in the perivitelline space. R. edirneus, TEM, bar = 2 μm. Abbreviations and symbols: fc, follicular cell; fn, the nucleus of the follicular cell; oc, oocyte; asterisk, perivitelline space; arrowhead, oocyte membrane; red cells/arrows, Vidania; green cells/arrows, Sulcia; yellow cells/arrows, Sodalis; purple arrows, Arsenophonus.
FIG 7.
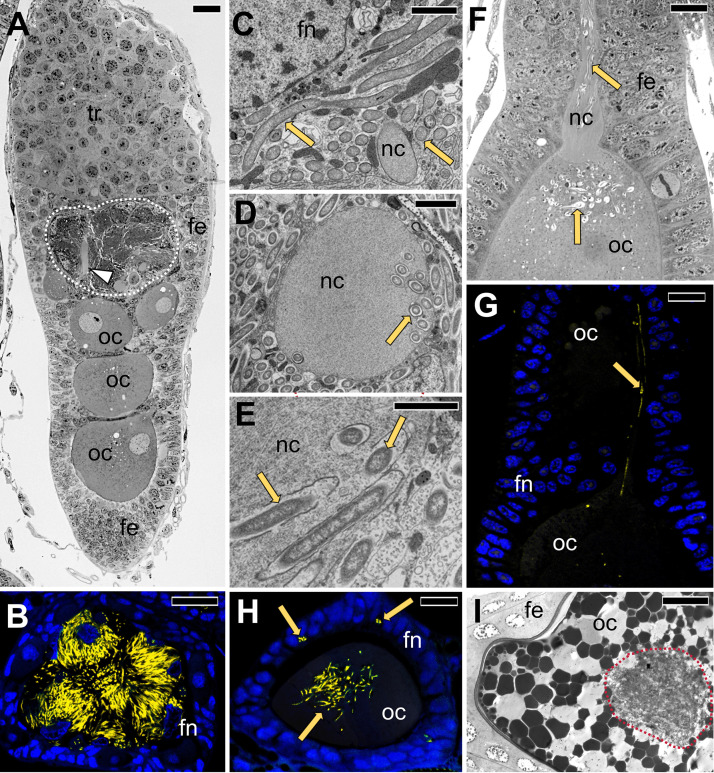
Transovarial transmission of Sodalis into the anterior pole of the developing oocyte in representatives of Dictyopharinae subfamily. (A) Longitudinal section through the ovariole in the previtellogenesis stage. Note numerous Sodalis cells in the region of the ovariole between tropharium and vitellarium (area surrounded with white dotted line). D. pannonica, LM, bar = 20 μm. (B) Cross-section through the neck region of the ovariole filled with prefollicular cells with bacteria Sodalis (marked in yellow); C. krueperi, confocal microscope, bar = 20 μm. (C) Fragment of the prefollicular cell with Sodalis occupying the neck region of the ovariole. D. multireticulata, TEM, bar = 2 μm. (D) Cross-section through the nutritive cord surrounded by Sodalis. Note Sodalis cells in the cytoplasm of the nutritive cord. C. krueperi, TEM, bar = 2 μm. (E) The higher magnification of Sodalis bacteria migrating via nutritive cord to the oocyte. C. krueperi. TEM, bar = 2 μm. (F and G) The transport of Sodalis via nutritive cord to the previtellogenic oocyte. C. krueperi. (F) LM, bar = 20 μm. (G) CLM, bar = 20 μm. (H) Accumulation of Sodalis cells in the cytoplasm of the anterior region of the previtellogenic oocyte. C. krueperi, CLM, bar = 20 μm. (I) Accumulation of Sodalis in the cytoplasm of the choriogenic oocyte. D. multireticulata, LM, bar = 20 μm. Abbreviations and symbol: fe, follicular epithelium; fn, the nucleus of a follicular cell; nc or arrowhead, nutritive cord; oc, oocyte; yellow cells/arrows, Sodalis.
FIG 8.
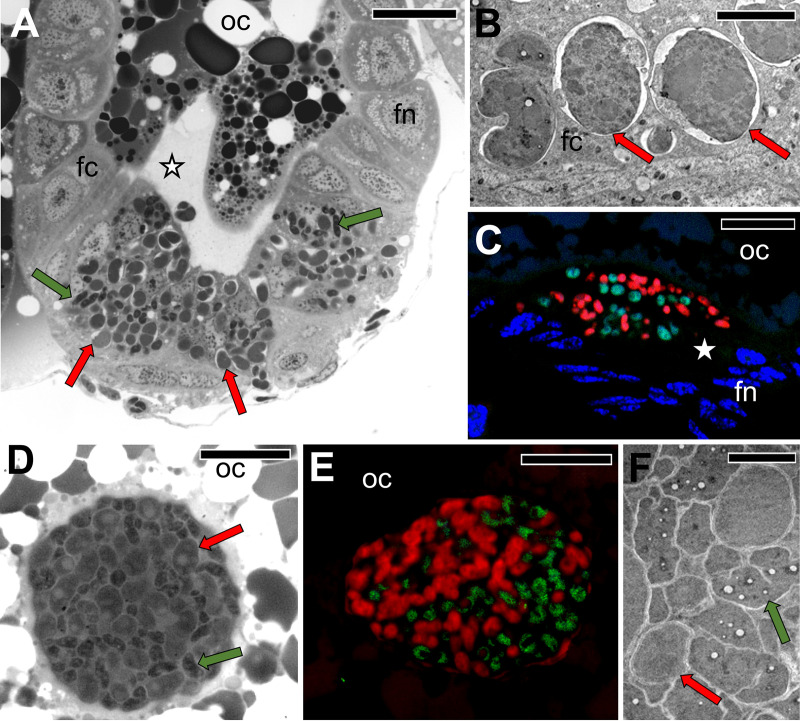
Transovarial transmission of Sulcia and Vidania into the posterior end of the ovariole in representatives of Dictyopharinae subfamily. (A) The migration of Sulcia and Vidania to the perivitelline space through the follicular epithelium surrounding the posterior pole of the terminal oocyte. D. multireticulata, LM, bar = 20 μm. (B) Vidania in the cytoplasm of the follicular cell. D. pannonica, TEM, bar = 2 μm. (C) Accumulation of Sulcia (green) and Vidania (red) in the perivitelline space. C. krueperi. Confocal microscope, bar = 20 μm. (D) A “symbiont ball” containing Sulcia and Vidania in the deep depression of the oolemma at the posterior pole of the terminal oocyte. D. multireticulata, LM, bar = 20 μm. (E) In situ identification of symbionts in the “symbiont ball” in the mature oocyte of C. krueperi. CLM, bar = 20 μm. (F) Fragment of “symbiont ball.” D. multireticulata, TEM, bar = 2 μm. Abbreviations and symbols: fc, follicular cell; fn, the nucleus of a follicular cell; oc, oocyte; asterisk, perivitelline space; red cells/arrows, Vidania; green cells/arrows, Sulcia.
In all species examined, Sulcia and Vidania simultaneously migrate and infect the posterior end of the ovariole during the choriogenesis stage of oocyte development (Fig. 5A and Fig. 6A). Symbionts migrate to the perivitelline space (=space between follicular epithelium and oolemma) via the cytoplasm of follicular cells surrounding the posterior pole of the terminal oocyte (Fig. 5, Fig. 6A and B, and Fig. 8A and B). These follicular cells are enlarged during symbiont migration, and their cytoplasm is full of bacterial cells (Fig. 6A and Fig. 8A). Arsenophonus (in D. europaea, R. scytha, and P. platypus) and Sodalis (in R. edirneus) are transmitted together with ancestral symbionts (Fig. 6A to D). After passing the follicular epithelium, symbionts gather in the perivitelline space. First, they create a cup-like accumulation (Fig. 6C) and then form a symbiont ball in the deep invagination of the oolemma (Fig. 6D to G).
In contrast, in C. krueperi, D. pannonica, and D. multireticulata, we observed the separation of symbiont transmission in time and space (Fig. 5C). Instead of entering the perivitelline space through follicular cells, Sodalis infects the region of the ovariole between the tropharium and vitellarium. We observed large accumulations of Sodalis cells in ovarioles containing oocytes in the previtellogenic stage of oogenesis. The follicular cells separating the tropharium from the vitellarium are tightly packed with bacteria (Fig. 7A to C). This specific region of the ovariole is penetrated by nutritive cords which connect oocytes developing in the vitellarium with trophocytes localized in the tropharium (Fig. 7A to C). Sodalis then infects previtellogenic oocytes using nutritive cords (Fig. 7D to H). They leave the follicular cells en masse, enter the nutritive cord area (Fig. 7D and E), and migrate toward the oocyte along microtubules (Fig. 7F to H). Then, Sodalis cells aggregate in the cytoplasm of the anterior pole of the oocyte (Fig. 7H) and stay in this form through the next stages of oogenesis (Fig. 7I). In these species, Sulcia and Vidania symbionts are transmitted to the perivitelline space of choriogenic oocytes (which contain clusters of Sodalis bacteria in the cytoplasm of the anterior pole) separately and, as summarized before, through follicular cells at their posterior end (Fig. 8A to C). They closely adhere to each other and form a characteristic “symbiont ball” (Fig. 8D to F).
In contrast to Sulcia, Sodalis, and Arsenophonus, which do not change their shape substantially during migration, Vidania undergoes significant morphological changes. In comparison to lobate Vidania cells within bacteriocytes, migrating Vidania cells (in the cytoplasm of follicular cells and in the “symbiont ball”) are smaller and more spherical (Fig. 6 and 8), resembling those occupying the rectal organ.
DISCUSSION
Repeated symbiont replacements shape the symbiosis in Dictyopharidae planthoppers.
Amplicon sequencing, metagenomics, and microscopy data agreed that all studied species of Dictyopharidae host heritable nutritional symbionts Sulcia and Vidania. We know that Sulcia infected the common ancestor of all Auchenorrhyncha that lived about 300 MYA, and Vidania, together with spittlebug-associated Zinderia and leafhopper-associated Nasuia, appears to represent a similarly ancient lineage (2, 17, 26). We found surprisingly few differences in the organization or contents of genomes of these widely retained symbionts (14, 21, 23, 24) between Dictyopharidae and the only other planthopper studied using genomics, O. filicicola OLIH, even though they are separated by about 200 MY of evolution (2). Dictyopharidae symbiont genomes are somewhat smaller than those in OLIH (142 to 148 kb versus 156 kb for Sulcia, 122 to 126 kb versus 136 kb for Vidania), but they are colinear and carry essentially the same set of nutrient biosynthesis genes. Sulcia CALKRU has the most compact genome of many strains of this symbiont characterized so far. Vidania falls among bacteria with the smallest known genomes, alongside other hemipteran symbionts: Nasuia (≥110 kb), Tremblaya (≥138 kb), Carsonella (≥164 kb), and Hodgkinia (≥144 kb, but note that individual genomes in fragmented complexes can be much smaller) (7, 27–30).
The reduction of symbiont genomes in long-lasting symbiotic interactions is a natural consequence of the lack of DNA repair genes or recombination, allowing for the progressive accumulation of deleterious mutations (31). Combined with the increase in rates of evolution, this leads to rapid loss of genes and pathways engaged in metabolic processes redundant during intracellular life and is thought to negatively affect the function and efficiency of others, including those involved in essential cellular processes as well as biosynthesis of nutrients (32–34). This opens up a path to their complementation or replacement by newly arriving, more versatile microorganisms. From this evolutionary perspective, both the complementation and replacement of “old” symbionts by “new” ones should allow host insects to refresh and reset their biosynthetic capacity and likely positively influence fitness and perhaps extend their ecological niche (17, 18). The radiation of insect clades whose ancestors acquired new symbionts, including cicadas, sharpshooter leafhoppers, aphids, carpenter ants, and others, is suggestive of a competitive edge that the nutritional symbiont acquisition or replacement can provide (35, 36).
In addition to the two ancestral symbionts, all studied Dictyopharidae harbor heritable, bacteriocyte-associated enterobacteria, either Sodalis or Arsenophonus, apparently acquired several times independently. These two microbial clades have repeatedly infected diverse insects, assuming nutritional functions especially in sap- and blood-feeders (2, 37–40). Arsenophonus can also act as a facultative symbiont, reproductive manipulator, or insect-vectored plant pathogen (2, 41–43). In Auchenorrhyncha, Sodalis and Arsenophonus are usually associated with ancestral symbionts, presumably complementing them (40). For example, they have been reported alongside ancestral symbionts in leafhoppers Macrosteles laevis and Cicadella viridis and in the spittlebug Aphrophora quadrinotata (10, 12, 44). Other times, they appear to have replaced one of the ancient symbionts. For example, the loss of Zinderia and the acquisition of Sodalis in modern Philaenini spittlebugs seems to be linked (12). There is little information on how often such replacements happen, but similarly to what was suggested in mealybugs (28), serial replacements are a likely explanation for the diversity of Sodalis/Arsenophonus symbionts within the studied Dictyopharidae. We will get a much more complete picture of the incidence and significance of this symbiont complementation and replacement as we survey gamma-symbiont distributions more systematically and use larger numbers of whole genomes for phylogenetic reconstruction events (17, 18).
The remarkable conservation of tissue localization and functions in dynamic planthopper symbioses.
In all Dictyopharidae species analyzed, as in other planthoppers examined previously, the ancestral symbionts Sulcia and Vidania are localized in distinct, spatially separated bacteriomes within the insect body cavity (14, 45–47). This contrasts with the situation in Cicadomorpha, where the ancestral symbionts always inhabit the same bacteriome: Sulcia, as a rule, occupies bacteriocytes within the outer layer of the bacteriome, whereas the coresiding ancestral symbiont (Nasuia, Zinderia, or Hodgkinia) occupies the central portion (11, 14, 29, 48). The differences in the spatial organization of symbiont-housing organs may be due to chance but could also be related to their mutual relations and nutritional capabilities. In Cicadomorpha, co-primary symbionts are thought to share some cofactors and metabolites, mainly involved in the biosynthesis of energetically expensive amino acids such as methionine or histidine (4), and this could be facilitated by the proximity of bacteriocytes inhabited by different symbionts.
Gammaproteobacterial symbionts, including Arsenophonus and Sodalis, localize differently in different insect hosts. The gammaproteobacterial symbionts of Dictyopharidae, despite their varied origins, are always located within bacteriomes separated from, but adjacent to, those occupied by Sulcia and Vidania. In other Auchenorrhyncha, they can also occur in separate bacteriomes or, alternatively, colonize distinct bacteriocytes within existing bacteriomes, but they are also observed in the cytoplasm of the same bacteriocytes as ancestral symbionts, or even inside other symbionts’ cells (10, 11, 20). In other insects, they may be dispersed across host tissues other than bacteriocytes, including gut epithelium cells, fat body cells, and hemolymph (49, 50). Interestingly, in some systems, independently acquired microbes tend to inhabit the same tissue compartments, suggestive of preadaptations that make it a particularly hospitable place. A striking example is gammaproteobacterial symbionts of Pseudococcinae mealybugs, always residing inside the cells of the ancient Tremblaya symbiont (28).
In females of all Dictyopharidae, similarly to previously studied planthopper species, we observed two morphotypes of Vidania occupying distinct bacteriomes in the body cavity and rectal organ. Buchner (14) and Bressan and Mulligan (46) suggested that symbionts derived from the rectal organ are the infectious form of Vidania that are transmitted to the progeny. Future transcriptomic studies should clarify the roles of these morphotypes, but our microscopic observations agree with the view that Vidania cells which are transmitted to the ovary represent the rectal organ morphotype.
In Cicadomorpha, Sulcia carries genes involved in the biosynthesis of seven to eight amino acids, while the companion symbiont is responsible for the provisioning of the remaining two or three. In Fulgoromorpha, the relative contributions of the companion symbionts are reversed. The ability to synthesize arginine, lysine, phenylalanine, and threonine has been lost by Sulcia, and they are produced by Vidania instead. Notably, the nearly identical biosynthetic capabilities in Dictyopharidae and the divergent cixiid, O. filicicola, reveal how stable their ancestral nutritional symbionts can be over 200 MY of evolution alongside changing gammaproteobacterial partners. The consistent differences in Sulcia biosynthetic capabilities between Cicadomorpha and Fulgoromorpha suggest that the two clades have separated soon after the acquisition of the ancestral beta-symbiont, assuming that there was indeed one, as proposed but not unambiguously demonstrated (17). Later, stochastic or other factors must have caused differential gene and pathway loss among the partner symbionts in the ancestors of the two infraorders.
In planthoppers, at least those studied to date, B vitamin biosynthesis has been outsourced to additional gammaproteobacterial symbionts, for example, Purcelliella in O. filicicola or Arsenophonus in Nilaparvata lugens (22, 51). Indeed, all characterized planthopper-associated Sodalis and Arsenophonus strains possess largely complete sets of genes involved in riboflavin and biotin production. Additionally, Sodalis of C. krueperi and Arsenophonus of R. scytha may be able to synthesize pyridoxine and folate, respectively. A similar biosynthetic potential was found in Sodalis-related symbionts of mealybugs (28) and Arsenophonus living in symbiotic relations with other insects such as the wasp Nasonia vitripennis and many whiteflies and louse flies (39, 40, 52). Moreover, both Sodalis and Arsenophonus possess some genes from essential amino acid biosynthesis pathways. Most of them overlap those retained on the Vidania genome, but some complement Vidania in methionine and lysine production. Such biosynthetic pathway complementarity between the host and symbionts has been demonstrated experimentally in the mealybug system (18) but is likely to be a more universal phenomenon (28, 29). It would be interesting to explore more broadly whether there is a general trend toward complementarity, how it is affected by serial co-primary symbiont replacements, and whether it can facilitate the replacement of ancient symbionts.
Newly arriving symbionts utilize different strategies for transovarial transmission.
The crucial role of nutritional symbionts in insect biology is evidenced by the diversity of their vertical transmission routes (15, 16). Transovarial transmission is probably the most reliable means of providing the full symbiont complement to all offspring. In Auchenorrhyncha, ancient symbionts are transmitted in a consistent, conserved way, crossing the follicular cells surrounding the posterior pole of the vitellogenic oocytes and then forming the “symbiont ball” near its posterior end (16). Additional symbionts generally follow the same path (11, 48, 53), including Arsenophonus and Sodalis in some of the Dictyopharidae species presented here. In others, Sodalis has adopted a very different transmission strategy, invading the ”neck region” of the ovariole from where it is carried to the cytoplasm of previtellogenic oocytes via the nutritive cords. Müller (23, 24) and Buchner (14) emphasized that the diversity of strategies that newly arriving insect symbionts adopt help us understand the general patterns of symbiosis evolution and replacement. The fact that closely related microbes have adopted different strategies in related host species is particularly notable. Nonetheless, the spatiotemporal separation of transovarial symbiont transmission is not unique to this insect clade. For example, in the planthopper Cixius nervosus, one of its symbionts, unidentified so far, was observed to infect undifferentiated cystocytes, while others infect full-grown oocytes (16).
In other groups of hemipterans and more divergent insects, gammaproteobacterial nutritional symbionts have adopted an even wider range of transovarial transmission strategies. For instance, in some scale insects, bacteria invade larval ovaries and germ cells at a very early stage, before they differentiate into oocytes and trophocytes, and are later present in all germ cells in the ovariole (16). In some heteropteran bugs but also carpenter ants, symbiotic gammaproteobacteria infect early previtellogenic oocytes directly: infect follicular cells, gather in their cytoplasms, and then enter the oocyte’s cytoplasm via an endocytic pathway. In both cases, initially, symbionts are dispersed in the entire ooplasm, but during vitellogenesis they accumulate and form a “symbiont ball” (54, 55). In some host lineages, enterobacterial symbionts have adopted more exotic transmission strategies. In Cicadella viridis and Macrosteles laevis, gammaproteobacterial symbionts (Sodalis and Arsenophonus, respectively) live inside and are moved to the oocyte within Sulcia cells (10, 44). In the scale insect Puto superbus, whole, intact bacteriocytes containing Sodalis are transferred into ovarioles (56). In turn, in tsetse flies, Sodalis glossinidius is vertically carried into the developing larva via milk gland secretions (57).
Fitness differences between these alternative transmission strategies are not apparent. It is likely that they vary in efficiency, including in the proportion of cells departing from the bacteriomes that arrive within the oocytes, or the energy needed for each symbiont cell to complete the journey. It is also probable that there are differences in the maintenance costs of the mechanisms necessary to move the cells between bacteriomes and oocytes and, later, to the target regions of the developing embryo. Transporting gammaproteobacterial symbionts along the same path as Sulcia and Vidania could theoretically allow the host to reuse the existing cellular machinery in a relatively efficient manner but that would depend on the specificity of the mechanisms, something that we have virtually no knowledge of. Yet, another aspect relevant to fitness, especially in the long run, is the symbiont bottleneck size, which may affect the strength of genetic drift and the symbiont evolutionary rates (53). However, at the moment, we know little about the magnitude of any such differences or how important they might be. But ultimately, and most critically, all mechanisms appear to result in reliable transmission of all symbionts to every offspring.
Why do some symbionts adopt different ways of transmission than others, then? We suspect that this is a combination of preadaptations, existing constraints, and chance events. The transmission strategy, but also final tissue localization, could be related to the microbe’s biology prior to the transition to the endosymbiotic lifestyle. The presence of specific genes or pathways in the microbe’s genome, and mechanisms that direct them toward, or facilitate entry into, certain cell types, is likely to drive the differences (58). At the same time, the reutilization of existing host-encoded mechanisms, including those for the transmission of older symbionts, is likely to play a significant role, probably increasing over time as symbiont genomes degenerate and erode away. For example, Spiroplasma and Wolbachia were shown to utilize the receptors involved in the transport of vitellogenin (protein precursors of yolk) into oocytes to enable entry into developing eggs (59, 60). We suspect that stochastic processes—random changes affecting the affinity between host and symbiont structural molecules at early stages of infection—play a significant role in determining the way the symbionts end up transmitting.
Conclusions.
The analyses of multipartner associations in Hemiptera provide unique insight into the factors shaping the evolution of nutritional symbiosis, highlighting the evolutionary significance and broad relevance of symbiont replacements which happened repeatedly in planthoppers and other hemipterans (12, 17, 19, 28, 61–64). While the list of nutritional symbionts that have reached a relatively stable stage and remained largely unchanged for millions or hundreds of millions of years extends beyond the familiar names such as Sulcia, Buchnera, Blochmannia, Baumannia, etc., it is also becoming clear that this is not a universal fate for newly acquired symbionts. Even related host species may harbor independently acquired symbiont strains with genomes at various stages of genomic degradation. It suggests that the “symbiont acquisition—genomic degradation—replacement” cycle is more frequent than often thought (17, 19, 63). This means that newly arriving microbes regularly face the challenge of establishing residence within host tissues and developing effective means of transmission across host generations. While there are differences among host clades in the organization of the symbiont-containing tissue, replacing symbionts often arrive at the same location within the host. Moreover, they generally undergo similar genome-reductive processes and converge on the same roles. Yet despite many common features, the detailed strategies utilized by the replacing symbionts may vary. Alternative strategies of intergenerational symbiont transmission in Dictyopharidae planthoppers are a key example. Our understanding of factors determining how the choice of strategies adopted by new symbionts, preadaptations, and random chance influence the outcomes of new infections is extremely limited. However, in the world of heritable nutritional symbioses that is proving unexpectedly dynamic, with many symbionts going through a rapid cycle of serial symbiont replacements, the balance between deterministic and stochastic factors is likely to determine the outcome of many symbioses.
MATERIALS AND METHODS
Study species.
We investigated symbioses in seven planthopper species from two subfamilies within the family Dictyopharidae. From subfamily Dictyopharinae, we characterized four species: Callodictya krueperi (Fieber, 1872), Dictyophara pannonica (Germar, 1830), Dictyophara multireticulata (Mulsant & Rey, 1855), and Dictyophara europaea (Linnaeus, 1767). From Orgerinae, we studied three species: Ranissus (Schizorgerius) scytha (Oshanin, 1913), Ranissus (Ranissus) edirneus (Dlabola, 1957), and Parorgerius platypus (Fieber, 1866). Adult specimens were sampled from a single population of each species in either Bulgaria or Poland between 2016 and 2018 (see Table S1 in the supplemental material), preserved in 80% ethanol or glutaraldehyde, and stored at 4°C until processing. Representative specimens from each species were identified based on morphological characteristics, and identities were confirmed using marker gene sequencing.
Collection details. Download Table S1, XLSX file, 0.01 MB (9.5KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Amplicon-based microbiome screen.
(i) Library preparation and sequencing. We sequenced amplicons for the V4 hypervariable region of the bacterial 16S rRNA gene and host mitochondrial cytochrome oxidase I (COI) genes simultaneously. DNA extracted using the Bio-Trace extraction kit (Eurx, Gdańsk, Poland) from dissected insect abdomens for up to three individual insects per species (plus negative controls) was used for amplicon library preparations following a modified two-step preparation protocol (65). In the first round of PCR, we amplified two marker regions of interest, using template-specific primers 515F/806R (66, 67) and COIBF3/COIBR2 (68) with Illumina adapter stubs. The bead-purified PCR products were used as the template for the second, indexing PCR. Pooled libraries were sequenced on an Illumina MiSeq v3 lane (2 × 300-bp reads) at the Institute of Environmental Sciences of Jagiellonian University. The primer sequences and detailed protocols are provided in Table S2.
Protocols. Download Table S2, XLSX file, 0.02 MB (23.6KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(ii) Analyses of amplicon sequencing data. We processed 16S rRNA and COI gene amplicon data using mothur v. 1.43.0 (69) following a pipeline detailed in Text S1. Initially, all amplicon data sets were split into bins corresponding to the two target genes. For both bins, we assembled reads into contigs. Quality-filtered, dereplicated contigs, with singletons removed, were aligned against the reference databases, screening 16S rRNA gene alignments for chimeras using UCHIME and then classifying sequences taxonomically. Finally, the sequences were clustered at 97% identity level using the nearest-neighbor algorithm and divided into OTUs.
Bioinformatic pipelines for the analysis of COI and 16S rRNA amplicon data. Download Text S1, DOCX file, 0.02 MB (16.5KB, docx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metagenomic library preparation and sequencing.
We sequenced bacteriome metagenomic libraries for three species: C. krueperi (specimen ID: CALKRU), D. multireticulata (DICMUL), and R. scytha (RANSCY). DNA from dissected bacteriomes of individual insects, extracted using the Sherlock AX kit (A&A Biotechnology, Gdynia, Poland), was fragmented using a Covaris E220 sonicator and used for metagenomic library preparation using the NEBNext Ultra II DNA Library Prep kit for Illumina (New England BioLabs), with the target insert length of 350 bp. The library pool, including three target species and other samples, was sequenced on an Illumina HiSeq X SBS lane by NGXBio (San Francisco, CA, USA).
Metagenome characterization and symbiont genome annotation.
Metagenomic reads were quality filtered using the ‘iu-filter-quality-minoche’ program included in illumina-utils software v1.4.4 (70) with default parameters (71). Contigs were assembled using Megahit v1.2.9 (k-mer 255, min contig size 1,000) (72). Because of the known issue of index swapping that occurs during cluster formation and sequencing on Illumina platforms (73, 74), which can lead to cross-contamination among samples in multiplexed lanes, we filtered the resulting assemblies for cross-contamination: we discarded all contigs that had >10× (and typically >200×)-greater coverage estimated based on strictly mapped reads from another library with one out of two indices shared than based on reads from the same library.
We identified symbiont contigs using BLASTN and tblastx searches against a custom database containing genomes of multiple known insect symbionts, verifying the identifications using coverage and GC content information (computed using BBTools v. 38.78). Then, for Sulcia and Vidania contigs, we confirmed their circularity and contiguity by read mapping and visualization on Tablet v. 1.20.12.24 (75) and the presence of overlapping ends. We rearranged the circularized genomes to ensure the same orientation and start position as in those published previously. Arsenophonus, Sodalis, and Wolbachia genomes were represented by multiple contigs, and we did not attempt to close them.
The genomic contigs of the three latter symbionts were annotated using Prokka v.1.14.6 (76), with the default parameters. For Sulcia and Vidania, Prokka but also InterProScan and GhostKOALA annotation attempts left multiple obvious gaps and unannotated or hypothetical proteins, and hence, we decided to use a custom workflow, modified from reference 29. Annotation was conducted by recursive searches for a manually curated set of alignments of protein-coding, rRNA, and noncoding RNA (ncRNA) genes from previously characterized genomes using HMMER 3.1b2 (77). Any open reading frames of at least 300 nucleotides that had not been annotated by the script were manually searched using hmmer and blastx/tblastx against UniProt and NCBI databases. All genes annotated as “hypothetical” or unannotated were carefully manually compared against the top hits in other microorganisms using blastp (https://blast.ncbi.nlm.nih.gov) and HMMER 3.3 (https://www.ebi.ac.uk/Tools/hmmer), resulting in the discovery of additional genes. Reference-based annotations of rRNA genes were supplemented by rRNA searches using RNAmmer v. 1.2 (78) and tRNA searches using tRNAscan-SE v. 1.4 (79). For all genes, we aligned all copies classified as functional using mafft v. 7.221 (80). In the case of protein-coding genes, alignments were conducted in protein space and reverse translated to nucleotide space.
The taxon-annotated GC-coverage plots for symbiont contigs were drawn using R v. 4.0.2 (R Development Core Team) with the ggplot2 package (81). Genomes were visualized using DNAPlotter GUI. In order to reconstruct amino acid and B vitamin biosynthetic pathways, we translated circular Sulcia and Vidania genomes as well as Sodalis, Arsenophonus, and Wolbachia contigs to amino acids and annotated them against KEGG with GhostKOALA (genus_prokaryotes) (82). After that, the presence of genes involved in biosynthetic pathways was checked manually.
Cloning, amplification, and phylogenetic and phylogenomic analyses.
For the four species for which metagenomes were not sequenced (D. pannonica, D. europaea, R. edirneus, and P. platypus), we obtained full-length 16S rRNA gene sequences of symbionts through molecular cloning in Escherichia coli cells as described previously (47). We also PCR amplified planthopper mitochondrial CytB and nuclear 18S and 28S rRNA genes (Table S2). Purified PCR products were Sanger sequenced by Genomed S.A. (Warsaw, Poland). Trimmed reads were merged into contigs and aligned, and alignments were inspected using CodonCode Aligner v. 8.0.2 (CodonCode Corp., Centerville, MA, USA).
We conducted phylogenetic analyses of concatenated insect marker genes and bacterial 16S rRNA genes, including those obtained from metagenomes using MEGA 7 software (83), using the Maximum Likelihood algorithm assuming the GTR (for insect genes) and GTR+GAMMA (for bacterial genes) models and with 1,000 bootstrap replicates.
Phylogenomic analyses of gammaproteobacterial symbiont genomes closely followed methods described in the supplement of a recent review that provided a genome-based phylogeny for 93 enterobacterial symbionts and their relatives (63). We used curated alignments of 129 conserved single-copy protein-coding genes provided by the authors and HMMER to identify the top homolog in the annotation for the three Dictyopharidae symbionts and mafft to align all sequences in amino acid space. We used RAxML v.8 (84) to reconstruct the phylogeny based on first and second codon positions in the concatenated alignment set, specifying GTR+GAMMA as the model of evolution, and with 100 bootstraps computed as support.
Microscopy.
(i) Light (LM) and electron (TEM) microscopy. Partially dissected abdomens of females of each of the seven species were fixed in the field in 2.5% glutaraldehyde solution in 0.1 M phosphate buffer (pH 7.4). In the laboratory, after washing with the same buffer with the addition of sucrose (5.8%), they were postfixed in a 1% solution of osmium tetroxide, dehydrated in ethanol and acetone series, and embedded in epoxy resin Epon 812 (Serva, Heidelberg, Germany). For histological analyses, the resin blocks were cut into serial, semithin sections, stained in 1% methylene blue in 1% borax, and observed under the Nikon Eclipse 80i light microscope. Ultrathin sections for ultrastructural analyses were contrasted with lead citrate and uranyl acetate and observed under the JEOL JEM 2100 electron transmission microscope.
(ii) Fluorescence microscopy. For fluorescence in situ hybridization (FISH), insects preserved in 80% ethanol were rehydrated and postfixed in 4% paraformaldehyde for 2 h. Then, the material was dehydrated again in the increasing concentration of ethanol and acetone and embedded in Technovit 8100 resin (Kulzer, Wehrheim, Germany). Resin blocks were cut into semithin sections and hybridized overnight at room temperature with symbiont-specific probes (Table S2). After hybridization, the slides were washed in phosphate-buffered saline (PBS), dried, covered with ProLong Gold antifade reagent (Life Technologies), and examined using a confocal laser scanning microscope, Zeiss Axio Observer LSM 710.
Data availability.
Accession numbers for sequences of the planthopper symbionts discussed in this paper may be found in Table S5 in the supplemental material.
Accession numbers. Download Table S5, XLSX file, 0.07 MB (74.7KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Custom database. Download Table S6, XLSX file, 0.01 MB (11.9KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank John McCutcheon for permission to use the facilities at the University of Montana, advice, and valuable comments; Ada Jankowska, Monika Prus, and Mateusz Buczek for laboratory assistance; Marcin Walczak for providing D. europaea specimens; and Gernot Kunz and Ilia Gjonov for insect images.
This project was supported by the Polish National Science Centre grants 2017/26/D/NZ8/00799 (to A.M.) and 2018/30/E/NZ8/00880 (to P.Ł.) as well as Polish National Agency for Academic Exchange grant PPN/PPO/2018/1/00015 (P.Ł.). The open-access publication of this article was funded by the Priority Research Area BioS under the program “Excellence Initiative–Research University” at the Jagiellonian University in Krakow.
Author contributions: A.M. and P.Ł. designed research; A.S. conducted the sampling; A.M., D.C.F., M.K., T.S., and P.Ł. performed research and analyzed data; A.M., D.C.F., and P.Ł. wrote the paper. All authors reviewed the manuscript and approved the final version.
We declare no conflict of interests.
Footnotes
Citation Michalik A, Castillo Franco D, Kobiałka M, Szklarzewicz T, Stroiński A, Łukasik P. 2021. Alternative transmission patterns in independently acquired nutritional cosymbionts of Dictyopharidae planthoppers. mBio 12:e01228-21. https://doi.org/10.1128/mBio.01228-21.
Contributor Information
Anna Michalik, Email: a.michalik@uj.edu.pl.
Gordon M. Bennett, Univ. California Merced
Margaret J. McFall-Ngai, University of Hawaii at Manoa
REFERENCES
- 1.Feldhaar H. 2011. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol 36:533–543. doi: 10.1111/j.1365-2311.2011.01318.x. [DOI] [Google Scholar]
- 2.Johnson KP, Dietrich CH, Friedrich F, Beutel RG, Wipfler B, Peters RS, Allen JM, Petersen M, Donath A, Walden KKO, Kozlov AM, Podsiadlowski L, Mayer C, Meusemann K, Vasilikopoulos A, Waterhouse RM, Cameron SL, Weirauch C, Swanson DR, Percy DM, Hardy NB, Terry I, Liu S, Zhou X, Misof B, Robertson HM, Yoshizawa K. 2018. Phylogenomics and the evolution of hemipteroid insects. Proc Natl Acad Sci USA 115:12775–12780. doi: 10.1073/pnas.1815820115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 4.Douglas A. 2016. How multi-partner endosymbioses function. Nat Rev Microbiol 14:731–743. doi: 10.1038/nrmicro.2016.151. [DOI] [PubMed] [Google Scholar]
- 5.Frago E, Zytynska SE, Fatouros NE. 2020. Microbial symbionts of herbivorous species across the insect tree, p 111–159. In Mechanisms underlying microbial symbiosis. Academic Press Inc, San Diego, CA. [Google Scholar]
- 6.Douglas AE. 2009. The microbial dimension in insect nutritional ecology. Funct Ecol 23:38–47. doi: 10.1111/j.1365-2435.2008.01442.x. [DOI] [Google Scholar]
- 7.McCutcheon J, McDonald BR, Moran N. 2009. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA 106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCutcheon JP, Moran NA. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol 2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett GM, Moran NA. 2013. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol 5:1675–1688. doi: 10.1093/gbe/evt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobiałka M, Michalik A, Walczak M, Junkiert Ł, Szklarzewicz T. 2016. Sulcia symbiont of the leafhopper Macrosteles laevis (Ribaut, 1927) (Insecta, Hemiptera, Cicadellidae: Deltocephalinae) harbors Arsenophonus bacteria. Protoplasma 253:903–912. doi: 10.1007/s00709-015-0854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobiałka M, Michalik A, Szwedo J, Szklarzewicz T. 2018. Diversity of symbiotic microbiota in Deltocephalinae leafhoppers (Insecta, Hemiptera, Cicadellidae). Arthropod Struct Dev 47:268–278. doi: 10.1016/j.asd.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Koga R, Bennett GM, Cryan JR, Moran NA. 2013. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol 15:2073–2081. doi: 10.1111/1462-2920.12121. [DOI] [PubMed] [Google Scholar]
- 13.Bright M, Bulgheresi S. 2010. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol 8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York, NY. [Google Scholar]
- 15.Kikuchi Y. 2009. Endosymbiotic bacteria in insects: their diversity and culturability. Microbes Environ 24:195–204. doi: 10.1264/jsme2.me09140s. [DOI] [PubMed] [Google Scholar]
- 16.Szklarzewicz T, Michalik A. 2017. Transovarial transmission of symbionts in insects, p 43–67. In Kloc M (ed), Oocytes: maternal information and functions. Springer International Publishing, Cham, Switzerland. [DOI] [PubMed] [Google Scholar]
- 17.Bennett GM, Moran NA. 2015. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA 112:10169–10176. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bublitz DC, Chadwick GL, Magyar JS, Sandoz KM, Brooks DM, Mesnage S, Ladinsky MS, Garber AI, Bjorkman PJ, Orphan VJ, McCutcheon JP. 2019. Peptidoglycan production by an insect-bacterial mosaic. Cell 179:703–712.e7. doi: 10.1016/j.cell.2019.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao M, Bennett GM. 2020. Symbiont replacements reset the co-evolutionary relationship between insects and their heritable bacteria. ISME J 14:1384–1395. doi: 10.1038/s41396-020-0616-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobiałka M, Michalik A, Świerczewski D, Szklarzewicz T. 2020. Complex symbiotic systems of two treehopper species: Centrotus cornutus (Linnaeus, 1758) and Gargara genistae (Fabricius, 1775) (Hemiptera: Cicadomorpha: Membracoidea: Membracidae). Protoplasma 257:819–831. doi: 10.1007/s00709-019-01466-z. [DOI] [PubMed] [Google Scholar]
- 21.Urban JM, Cryan JR. 2012. Two ancient bacterial endosymbionts have coevolved with the planthoppers (Insecta: Hemiptera: Fulgoroidea). BMC Evol Biol 12:87. doi: 10.1186/1471-2148-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett GM, Mao M. 2018. Comparative genomics of a quadripartite symbiosis in a planthopper host reveals the origins and rearranged nutritional responsibilities of anciently diverged bacterial lineages. Environ Microbiol 20:4461–4472. doi: 10.1111/1462-2920.14367. [DOI] [PubMed] [Google Scholar]
- 23.Müller HJ. 1940. Die Symbiose der Fulgoroiden (Homoptera Cicadina). Zoologica 98:1–110. [Google Scholar]
- 24.Müller HJ. 1940. Die Symbiose der Fulgoroiden (Homoptera-Cicadina). Zoologica 98:111–220. [Google Scholar]
- 25.Emeljanov AF. 2003. New taxa and new data on distribution of the subfamily Orgeriinae in the Mediterranean (Homoptera, Dictyopharidae). Zoosyst Ross 11:311–319. [Google Scholar]
- 26.Moran NA, Tran P, Gerardo NM. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol 71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett GM, Abbà S, Kube M, Marzachì C. 2016. Complete genome sequences of the obligate symbionts “Candidatus Sulcia muelleri” and “Ca. Nasuia deltocephalinicola” from the pestiferous leafhopper Macrosteles quadripunctulatus (Hemiptera: Cicadellidae). Genome Announc 4:e01604-15. doi: 10.1128/genomeA.01604-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Husnik F, McCutcheon JP. 2016. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci USA 113:E5416–E5424. doi: 10.1073/pnas.1603910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Łukasik P, Nazario K, Van Leuven JT, Campbell MA, Meyer M, Michalik A, Pessacq P, Simon C, Veloso C, McCutcheon JP. 2018. Multiple origins of interdependent endosymbiotic complexes in a genus of cicadas. Proc Natl Acad Sci USA 115:E226–E235. doi: 10.1073/pnas.1712321115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakabachi A, Piel J, Malenovský I, Hirose Y. 2020. Comparative genomics underlines multiple roles of Profftella, an obligate symbiont of psyllids: providing toxins, vitamins, and carotenoids. Genome Biol Evol 12:1975–1987. doi: 10.1093/gbe/evaa175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourguignon T, Kinjo Y, Villa-Martín P, Coleman NV, Tang Q, Arab DA, Wang Z, Tokuda G, Hongoh Y, Ohkuma M, Ho SYW, Pigolotti S, Lo N. 2020. Increased mutation rate is linked to genome reduction in prokaryotes. Curr Biol 30:3848–3855.e4. doi: 10.1016/j.cub.2020.07.034. [DOI] [PubMed] [Google Scholar]
- 32.Boscaro V, Kolisko M, Felletti M, Vannini C, Lynn DH, Keeling PJ. 2017. Parallel genome reduction in symbionts descended from closely related free-living bacteria. Nat Ecol Evol 1:1160–1167. doi: 10.1038/s41559-017-0237-0. [DOI] [PubMed] [Google Scholar]
- 33.McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 34.Moran NA, Bennett GM. 2014. The tiniest tiny genomes. Annu Rev Microbiol 68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 35.Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE. 2009. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci USA 106:21236–21241. doi: 10.1073/pnas.0907926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sudakaran S, Kost C, Kaltenpoth M. 2017. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol 25:375–390. doi: 10.1016/j.tim.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 37.Boyd BM, Allen JM, Koga R, Fukatsu T, Sweet AD, Johnson KP, Reed DL. 2016. Two bacterial genera, Sodalis and Rickettsia, associated with the seal louse Proechinophthirus fluctus (Phthiraptera: Anoplura). Appl Environ Microbiol 82:3185–3197. doi: 10.1128/AEM.00282-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nováková E, Hypša V, Moran NA. 2009. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol 9:143. doi: 10.1186/1471-2180-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nováková E, Husník F, Šochová E, Hypša V. 2015. Arsenophonus and Sodalis symbionts in louse flies: an analogy to the Wigglesworthia and Sodalis system in tsetse flies. Appl Environ Microbiol 81:6189–6199. doi: 10.1128/AEM.01487-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos-Garcia D, Juravel K, Freilich S, Zchori-Fein E, Latorre A, Moya A, Morin S, Silva FJ. 2018. To B or not to B: comparative genomics suggests Arsenophonus as a source of B vitamins in whiteflies. Front Microbiol 9:2254. doi: 10.3389/fmicb.2018.02254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dittmer J, Lusseau T, Foissac X, Faoro F. 2021. Skipping the insect vector: plant stolon transmission of the phytopathogen Ca. Phlomobacter fragaria from the Arsenophonus clade of insect endosymbionts. Insects 12(2):93. doi: 10.3390/insects12020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Šochová E, Husník F, Nováková E, Halajian A, Hypša V. 2017. Arsenophonus and Sodalis replacements shape evolution of symbiosis in louse flies. PeerJ 5:e4099. doi: 10.7717/peerj.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor GP, Coghlin PC, Floate KD, Perlman SJ. 2011. The host range of the male-killing symbiont Arsenophonus nasoniae in filth fly parasitioids. J Invertebr Pathol 106:371–379. doi: 10.1016/j.jip.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Michalik A, Jankowska W, Kot M, Gołas A, Szklarzewicz T. 2014. Symbiosis in the green leafhopper, Cicadella viridis (Hemiptera, Cicadellidae). Association in statu nascendi? Arthropod Struct Dev 43:579–587. doi: 10.1016/j.asd.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Bressan A, Arneodo J, Simonato M, Haines WP, Boudon-Padieu E. 2009. Characterization and evolution of two bacteriome-inhabiting symbionts in cixiid planthoppers (Hemiptera: Fulgoromorpha: Pentastirini). Environ Microbiol 11:3265–3279. doi: 10.1111/j.1462-2920.2009.02055.x. [DOI] [PubMed] [Google Scholar]
- 46.Bressan A, Mulligan KL. 2013. Localization and morphological variation of three bacteriome-inhabiting symbionts within a planthopper of the genus Oliarus (Hemiptera: Cixiidae). Environ Microbiol Rep 5:499–505. doi: 10.1111/1758-2229.12051. [DOI] [PubMed] [Google Scholar]
- 47.Michalik A, Szwedo J, Stroiński A, Świerczewski D, Szklarzewicz T. 2018. Symbiotic cornucopia of the monophagous planthopper Ommatidiotus dissimilis (Fallén, 1806) (Hemiptera: Fulgoromorpha: Caliscelidae). Protoplasma 255:1317–1329. doi: 10.1007/s00709-018-1234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szklarzewicz T, Świerczewski D, Stroiński A, Michalik A. 2020. Conservatism and stability of the symbiotic system of the invasive alien treehopper Stictocephala bisonia (Hemiptera, Cicadomorpha, Membracidae). Ecol Entomol 45:876–885. doi: 10.1111/een.12861. [DOI] [Google Scholar]
- 49.Dale C, Maudlin I. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Syst Evol Bacteriol 49(Part 1):267–275. doi: 10.1099/00207713-49-1-267. [DOI] [PubMed] [Google Scholar]
- 50.Kuechler SM, Dettner K, Kehl S. 2011. Characterization of an obligate intracellular bacterium in the midgut epithelium of the bulrush bug Chilacis typhae (Heteroptera, Lygaeidae, Artheneinae). Appl Environ Microbiol 77:2869–2876. doi: 10.1128/AEM.02983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue J, Zhou X, Zhang C-X, Yu L-L, Fan H-W, Wang Z, Xu H-J, Xi Y, Zhu Z-R, Zhou W-W, Pan P-L, Li B-L, Colbourne JK, Noda H, Suetsugu Y, Kobayashi T, Zheng Y, Liu S, Zhang R, Liu Y, Luo Y-D, Fang D-M, Chen Y, Zhan D-L, Lv X-D, Cai Y, Wang Z-B, Huang H-J, Cheng R-L, Zhang X-C, Lou Y-H, Yu B, Zhuo J-C, Ye Y-X, Zhang W-Q, Shen Z-C, Yang H-M, Wang J, Wang J, Bao Y-Y, Cheng J-A. 2014. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol 15:521. doi: 10.1186/s13059-014-0521-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darby AC, Choi J-H, Wilkes T, Hughes MA, Werren JH, Hurst GDD, Colbourne JK. 2010. Characteristics of the genome of Arsenophonus nasoniae, son-killer bacterium of the wasp Nasonia. Insect Mol Biol 19(Suppl 1):75–89. doi: 10.1111/j.1365-2583.2009.00950.x. [DOI] [PubMed] [Google Scholar]
- 53.Campbell MA, Łukasik P, Meyer MC, Buckner M, Simon C, Veloso C, Michalik A, McCutcheon JP. 2018. Changes in endosymbiont complexity drive host-level compensatory adaptations in cicadas. mBio 9:e02104-18. doi: 10.1128/mBio.02104-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kupper M, Stigloher C, Feldhaar H, Gross R. 2016. Distribution of the obligate endosymbiont Blochmannia floridanus and expression analysis of putative immune genes in ovaries of the carpenter ant Camponotus floridanus. Arthropod Struct Dev 45:475–487. doi: 10.1016/j.asd.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Swiatoniowska M, Ogorzalek A, Golas A, Michalik A, Szklarzewicz T. 2013. Ultrastructure, distribution, and transovarial transmission of symbiotic microorganisms in Nysius ericae and Nithecus jacobaeae (Heteroptera: Lygaeidae: Orsillinae). Protoplasma 250:325–332. doi: 10.1007/s00709-012-0416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szklarzewicz T, Kalandyk-Kołodziejczyk M, Michalik K, Jankowska W, Michalik A. 2018. Symbiotic microorganisms in Puto superbus (Leonardi, 1907) (Insecta, Hemiptera, Coccomorpha: Putoidae). Protoplasma 255:129–138. doi: 10.1007/s00709-017-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aksoy S, Chen X, Hypsa V. 1997. Phylogeny and potential transmission routes of midgut-associated endosymbionts of tsetse (Diptera: Glossinidae). Insect Mol Biol 6:183–190. doi: 10.1111/j.1365-2583.1997.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 58.Dale C, Plague GR, Wang B, Ochman H, Moran NA. 2002. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Natl Acad Sci USA 99:12397–12402. doi: 10.1073/pnas.182213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo Y, Hoffmann AA, Xu X-Q, Mo P-W, Huang H-J, Gong J-T, Ju J-F, Hong X-Y. 2018. Vertical transmission of Wolbachia is associated with host vitellogenin in Laodelphax striatellus. Front Microbiol 9:2016. doi: 10.3389/fmicb.2018.02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herren JK, Paredes JC, Schüpfer F, Lemaitre B. 2013. Vertical transmission of a Drosophila endosymbiont via cooption of the yolk transport and internalization machinery. mBio 4:e00532-12. doi: 10.1128/mBio.00532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobiałka M, Michalik A, Walczak M, Szklarzewicz T. 2018. Dual “bacterial-fungal” symbiosis in Deltocephalinae leafhoppers (Insecta, Hemiptera, Cicadomorpha: Cicadellidae). Microb Ecol 75:771–782. doi: 10.1007/s00248-017-1075-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuura Y, Moriyama M, Łukasik P, Vanderpool D, Tanahashi M, Meng X-Y, McCutcheon JP, Fukatsu T. 2018. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc Natl Acad Sci USA 115:E5970–E5979. doi: 10.1073/pnas.1803245115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCutcheon JP, Boyd BM, Dale C. 2019. The life of an insect endosymbiont from the cradle to the grave. Curr Biol 29:R485–R495. doi: 10.1016/j.cub.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 64.Chong RA, Moran NA. 2018. Evolutionary loss and replacement of Buchnera, the obligate endosymbiont of aphids. ISME J 12:898–908. doi: 10.1038/s41396-017-0024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glenn TC. 2011. Field guide to next-generation DNA sequencers. Mol Ecol Resour 11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 66.Apprill A, McNally S, Parsons R, Weber L. 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 67.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 68.Elbrecht V, Braukmann TWA, Ivanova NV, Prosser SWJ, Hajibabaei M, Wright M, Zakharov EV, Hebert PDN, Steinke D. 2019. Validation of COI metabarcoding primers for terrestrial arthropods. PeerJ 7:e7745. doi: 10.7717/peerj.7745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eren AM, Vineis JH, Morrison HG, Sogin ML. 2013. A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS One 8:e66643. doi: 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li D, Luo R, Liu C-M, Leung C-M, Ting H-F, Sadakane K, Yamashita H, Lam T-W. 2016. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 73.Costello M, Fleharty M, Abreu J, Farjoun Y, Ferriera S, Holmes L, Granger B, Green L, Howd T, Mason T, Vicente G, Dasilva M, Brodeur W, DeSmet T, Dodge S, Lennon NJ, Gabriel S. 2018. Characterization and remediation of sample index swaps by non-redundant dual indexing on massively parallel sequencing platforms. BMC Genomics 19:332. doi: 10.1186/s12864-018-4703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Illumina. 2018. Effects of index misassignment on multiplexing and downstream analysis. Illumina, Inc, San Diego, CA. https://www.illumina.com/content/dam/illumina-marketing/documents/products/whitepapers/index-hopping-white-paper-770-2017-004.pdf.
- 75.Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 76.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 77.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wickham H. 2011. ggplot2. Wiley Interdiscip Rev Comput Stat 3:180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- 82.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 83.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
OTU tables. Download Table S3, XLSX file, 1.2 MB (1.2MB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ML phylogenies of symbionts based on 16S rRNA gene sequences. (A) ML phylogeny of Sulcia strains from diverse planthoppers, based on full-length sequences (1,373 shared nucleotide positions) of 16S rRNA gene. Only bootstrap values above 70% are shown. Sequences from the seven Dictyopharidae species studied here are highlighted using the colored font. (B) ML phylogeny of Vidania strains from diverse planthoppers based on full-length sequences (1,530 shared nucleotide positions) of 16S rRNA gene. Only bootstrap values above 70% are shown. Sequences from the seven Dictyopharidae species studied here are highlighted using the colored font. (C) ML phylogeny of Sodalis strains from different hosts based on 16S rRNA gene sequences. Only bootstrap values above 70% are shown. (D) ML phylogeny of Arsenophonus strains from different hosts based on 16S rRNA gene sequences. Only bootstrap values above 70% are shown. (E) ML phylogeny based on first and second codon positions of 129 conserved single-copy protein-coding genes of symbiotic enterobacteria and relatives. Only bootstrap values above 70% are shown. Download FIG S1, PDF file, 1.8 MB (1.8MB, pdf) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Dictyophara pannonica. Two morphotypes of Sodalis symbiont (red and yellow arrows) within bacteriome of single individuals. TEM, bar = 2 μm. (B) Wolbachia symbiont cells within bacteriocytes of Dictyopharidae species (TEM). (1) Wolbachia cells (green arrows) in the cytoplasm of the Vidania (V) bacteriocyte from Callodictya krueperi. (2) Wolbachia cells (green arrows) in the cytoplasm of the Sulcia (S) bacteriocyte from Dictyophara europaea. TEM, bar = 2 μm. Download FIG S3, PDF file, 1.1 MB (1.1MB, pdf) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The alignments of planthopper Sulcia and Vidania genomes. (A) PROMER alignments of final Sulcia genomes from three Dictyopharidae species, Callodictya krueperi (CALKRU), Dictyophara multireticulata (DICMUL), and Ranissus scytha (RANSCY), against the Sulcia genome from Oliarus filicicola (OLIH; family Cixiidae). (B) PROMER alignments of final Vidania genomes from three Dictyopharidae species, Callodictya krueperi (CALKRU), Dictyophara multireticulata (DICMUL), and Ranissus scytha (RANSCY), against the Vidania genome from Oliarus filicicola (OLIH; Cixiidae). Download FIG S2, PDF file, 0.1 MB (115KB, pdf) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome details. Download Table S4, XLSX file, 0.3 MB (282.3KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Collection details. Download Table S1, XLSX file, 0.01 MB (9.5KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Protocols. Download Table S2, XLSX file, 0.02 MB (23.6KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bioinformatic pipelines for the analysis of COI and 16S rRNA amplicon data. Download Text S1, DOCX file, 0.02 MB (16.5KB, docx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Accession numbers. Download Table S5, XLSX file, 0.07 MB (74.7KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Custom database. Download Table S6, XLSX file, 0.01 MB (11.9KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Accession numbers for sequences of the planthopper symbionts discussed in this paper may be found in Table S5 in the supplemental material.
Accession numbers. Download Table S5, XLSX file, 0.07 MB (74.7KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Custom database. Download Table S6, XLSX file, 0.01 MB (11.9KB, xlsx) .
Copyright © 2021 Michalik et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.