Abstract
In the yeast Saccharomyces cerevisiae, chromosomes terminate with a repetitive sequence [poly(TG1–3)] 350 to 500 bp in length. Strains with a mutation of TEL1, a homolog of the human gene (ATM) mutated in patients with ataxia telangiectasia, have short but stable telomeric repeats. Mutations of TLC1 (encoding the RNA subunit of telomerase) result in strains that have continually shortening telomeres and a gradual loss of cell viability; survivors of senescence arise as a consequence of a Rad52p-dependent recombination events that amplify telomeric and subtelomeric repeats. We show that a mutation in MEC1 (a gene related in sequence to TEL1 and ATM) reduces telomere length and that tel1 mec1 double mutant strains have a senescent phenotype similar to that found in tlc1 strains. As observed in tlc1 strains, survivors of senescence in the tel1 mec1 strains occur by a Rad52p-dependent amplification of telomeric and subtelomeric repeats. In addition, we find that strains with both tel1 and tlc1 mutations have a delayed loss of cell viability compared to strains with the single tlc1 mutation. This result argues that the role of Tel1p in telomere maintenance is not solely a direct activation of telomerase.
Most eukaryotic chromosomes end with simple repetitive DNA sequences (7). In the yeast Saccharomyces cerevisiae, wild-type strains have poly(TG1–3) tracts 350 to 500 bp in length (31, 42, 43). Within a yeast cell population, telomeric tracts, even for a single chromosome, range in size by about ± 50 bp (43). This variability in telomere length, as well as the identification of mutants with telomeric tracts that are longer or shorter than those in wild-type strains, suggests that telomere length is likely to reflect a balance between mechanisms that extend or contract these terminal repeats.
In many eukaryotes, including yeast, the most important mechanism for extending telomeric repeats is telomerase (7, 45). This RNA-protein enzyme complex extends the G-rich strand of the telomere 5′ to 3′, using telomeric repeats encoded within the RNA component of the enzyme as a template. The extended G-rich strand is thought to be copied 5′ to 3′ by conventional DNA polymerases to yield the complementary C-rich strand (7). In yeast, the RNA component of telomerase is encoded by TLC1 (33), and the protein component with reverse transcriptase activity is encoded by EST2 (16). In strains with mutations in either of these genes, telomeric tracts shorten with each cell division, resulting in gradual loss of cell viability (13, 33). This same senescent phenotype is observed in strains with mutations in several other EST (ever shorter telomeres) genes, including EST1, EST3, and CDC13/EST4 (13, 19). Since mutations in all EST genes have the same phenotype and since double-mutant est strains have the same phenotype as the single mutants, it has been suggested that all Est proteins function in the same pathway of telomere maintenance (13, 23, 40), although not necessarily as components of telomerase (15).
Although yeast strains with a mutation in EST or TLC1 genes undergo dramatic loss of cell viability associated with loss of telomeric repeats, fast-growing survivors arise within the mutant cultures (18, 19). In these survivors, amplification of telomeric and subtelomeric repeats occurs by a RAD52-dependent recombinational process (18). These extra repeats may act as buffers to prevent loss of essential DNA sequences located near the chromosome ends, a process that is analogous to the effects of the telomere-specific transposable elements found in Drosophila (25).
Mutations in several genes result in short telomeric tracts without leading to cell death. For example, the telomeric tracts in tel1 strains are about 50 bp in length, about sevenfold shorter than in wild-type strains (20). In addition, tel1 strains have slightly higher rates of chromosome loss and mitotic recombination (6). The TEL1 gene encodes a very large (322-kDa) protein with homology to the human tumor suppressor gene ATM (29). Tel1p also shares homology with a number of lipid and/or protein kinases required for the function of DNA damage-sensitive checkpoints, including MEC1 (11, 44), and in certain genetic backgrounds, Tel1p affects the sensitivity of the cell to DNA-damaging agents (21). For example, tel1 mec1 strains are considerably more sensitive to X rays than mec1 strains, whereas tel1 single-mutant strains are not X-ray sensitive (6, 21). Mec1p and Tel1p are involved, directly or indirectly, in phosphorylation of the checkpoint protein Rad53 (28). In addition, other proteins involved in DNA replication or checkpoint function, including replication protein A (2), Rad9p (5, 39), and Ddc1p (24), show Mec1p-dependent phosphorylation.
In contrast to Mec1p (44), Tel1p has a relatively minor role in the cellular response to DNA damage, and this role is evident only in strains lacking Mec1p. One interpretation of this result is that the target protein (or proteins) in the checkpoint pathway involved in the repair of DNA damage is a poor substrate for Tel1p but a good substrate for Mec1p. Although there is no direct biochemical evidence that Tel1p is a protein kinase, point mutations in the kinase domain of Tel1p result in short telomeres (6). The role of the Tel1p in telomere replication is not understood. One possibility is that Tel1p-mediated phosphorylation of a protein subunit of telomerase is required for the optimal activity of telomerase. As described below, our comparison of the phenotypes of tel1, tlc1, and double-mutant tel1 tlc1 strains suggests that this model is unlikely.
Since Tel1p shows homology with a number of other yeast proteins, including Mec1p, Tor1p, and Tor2p, Greenwell et al. (6) examined telomere lengths in strains mutated for mec1, tor1, or tor2. Since MEC1 and TOR2 are essential genes, only nonnull alleles were examined. No effect on telomere length was observed for any of the three mutations. We decided to reinvestigate the effect of the mec1 mutation on telomere length for two reasons. First, the mec1 strain previously examined was subsequently found to contain two mutations, mec1-1 (an allele eliminating the essential function of Mec1p) and sml1 (a mutation that suppressed the lethal effects of mec1-1 [26]); Sml1p is a negative regulator of deoxynucleoside triphosphate pools (46). Thus, an effect of the mec1-1 mutation on telomere length could have been hidden by the coexisting sml1 mutation. Second, mutations in the Schizosaccharomyces pombe rad3 gene, a homologue of MEC1 of S. cerevisiae, result in shortened telomeres (3), and strains with mutations in both rad3 and tel1, an S. pombe homologue of TEL1, lose all telomeric sequences (22).
As described below, we found that the mec1-21 allele (28) results in short telomeres and that strains with the tel1 mec1-21 genotype have telomeres shorter than those of either single mutant and exhibit an associated senescent phenotype. We suggest that both Tel1p and Mec1p have two different types of target proteins, one specific for cellular responses to DNA damage and one specific for telomere maintenance. In addition, an epistasis analysis of tel1, mec1, and tlc1 indicates that the essential role of Tel1p and Mec1p in telomere maintenance is not exerted by directly activating telomerase.
MATERIALS AND METHODS
Yeast strains and plasmids.
Yeast strains and plasmids used in this study are described in Table 1. All strains were isogenic (except for alterations introduced by transformation) with W303a (a leu2-3,112 his3-11,15 ura3-1 ade2-1 trp1-1 can1-100 [40]) or AMY125 (α ade5-1 his7-2 leu2-3,112 trp1-289 ura3-52 [39]).
TABLE 1.
Strain names, constructions, and relevant genotypesa
| Strain name | Construction or reference | Relevant genotype |
|---|---|---|
| W303a | 37 | Wild type, a mating type |
| W303α | 37 | Wild type, α mating type |
| SPY40 | 27; derived from W303a by transformation | a tel1::URA3 |
| SPY40FR | Isolation of 5FOAr SPY40 derivative | a tel1::ura3 |
| Y604 | Provided by Y. Sanchez and S. Elledge | a mec1-21 |
| PG28.8 | Transformation of W303a with XhoI-SalI fragment of pJL202b | a ura3Δ::HIS3 |
| PG30.1 | Transformation of PG28.8 with PCR fragmentc to replace TEL1 with URA3 | a ura3Δ::HIS3 tel1Δ::URA3 |
| PG30.1FR | Isolation of 5FOAr derivative of PG30.1 | a ura3Δ::HIS3 tel1Δ::ura3 |
| KRY20a | Transformation of PG30.1FR with HpaI-BamHI fragment of pDTK103d to generate ade3::hisG/URA3/hisG allele; isolation of 5FOAr derivative to generate ade3::hisG allele | a ura3Δ::HIS3 tel1Δ::ura3 ade3::hisG |
| KRY20α | Mating-type switch of KRY20a by using plasmid pGAL-HOe | α ura3Δ::HIS3 tel1Δ::ura3 ade3::hisG |
| KRY70 | Transformation of W303a with BamHI-treated pDTK102,f followed by isolation of 5FOAr derivative | a rad52::hisG |
| JMY73 | Transformation of W303a with PCR fragmentg to generate sml1::HIS3 | a sml1::HIS3 |
| KRY300 | Cross of W303a and W303α | Wild-type diploid, W303 background |
| KRY227 | Transformation of KRY300 with 6-kb SacI fragment of pPG47h | a/α tel1::URA3/TEL1 |
| KRY228 | Transformation of KRY300 with 2-kb XhoI fragment of pBLUE61::LEU2i | a/α tlc1::LEU2/TLC1 |
| KRY301 | Spore derived from KRY227 | a tel1::URA3 |
| KRY302 | Spore derived from KRY228 | α tlc1::LEU2 |
| KRY229 | Cross of KRY301 and KRY302 | a/α tel1::URA3/TEL1 tlc1::LEU2/TLC1 |
| KRY235 | Transformation of KRY228 with URA3 PCR fragmentj | a/α tlc1::LEU2/TLC1 ura3/URA3 |
| KRY303 | Spore derived from KRY235 | α tlc1::LEU2 URA3 |
| KRY236 | Cross of KRY303 and SPY40FR | a/α tel1::ura3/TEL1 tlc1::LEU2/TLC1 |
| ura3/URA3 | ||
| JMY300 | Cross of Y604 with KRY20α | a/α tel1::ura3/TEL1 mec1-21/MEC1 |
| ade3::hisG/ADE3 ura3::HIS3/ura3 | ||
| JMY300-1a | Spore derived from JMY300 | α tel1::ura3 mec1-21 |
| JMY300-2a | Spore derived from JMY300 | a tel1::ura3 mec1-21 |
| JMY300-3a | Spore derived from JMY300 | a tel1::ura3 mec1-21 ura3::HIS3 |
| JMY300-3aS | Fast-growing survivor derived from JMY300-3a | a tel1::ura3 mec1-21 ura3::HIS3 |
| KRY242 | Cross of KRY70 to JMY300-1a | a/α tel1::ura3/TEL1 mec1-21/MEC1 |
| rad52::hisG/RAD52 | ||
| KRY246 | Cross of W303α to JMY300-3aS | a/α tel1::ura3/TEL1 mec1-21/MEC1 |
| JMY301 | Cross of W303α to JMY300-2a | a/α tel1::ura3/TEL1 mec1-21/MEC1 |
| JMY302 | Transformation of JMY301 with PCR fragmentg to generate sml1::HIS3 | a/α tel1::ura3/TEL1 mec1-21/MEC1 |
| sml1::HIS3/SML1 | ||
| JMY303 | Cross of JMY73 to JMY300-1a | a/α tel1::ura3/TEL1 mec1-21/MEC1 |
| sml1::HIS3/SML1 | ||
| KRY306 | Spore derived from KRY229 | α tel1::URA3 tlc1::LEU2 |
| KRY238 | Cross of Y604 to KRY306 | a/α tel1::URA3/TEL1 mec1-21/MEC1 |
| tlc1::LEU2/TLC1 | ||
| AMY125 | Provided by A. Morrison | Wild type, α mating type |
| EAS16 | Provided by E. Sia, constructed by using plasmid pGAL-HOe | Wild type, a mating type |
| EAS20 | Cross of AMY125 to EAS16 | Wild-type diploid, AMY125 background |
| KRY230 | Transformation of EAS20 with 6-kb SacI fragment of pPG47h | a/α tel1::URA3/TEL1 |
| KRY231 | Transformation of EAS20 with 2-kb XhoI fragment of pBLUE61::LEU2i | a/α tlc1::LEU2/TLC1 |
| KRY304 | Spore derived from KRY230 | α tel1::URA3 |
| KRY305 | Spore derived from KRY231 | a tlc1::LEU2 |
| KRY232 | Cross of KRY304 and KRY305 | a/α tel1::URA3/TEL1 tlc1::LEU2/TLC1 |
Most of the strains used are isogenic (except for changes introduced by transformation) with W303a (a leu2-3,112 his3-11,15 ura3-1 ade2-1 trp1-1 can1-100). Strains listed below AMY125 are isogenic with AMY125 (α ade5-1 his7-2 leu2-3,112 trp1-289 ura3-52) except for changes introduced by transformation. All genes that differ from those of the progenitor genotype are listed. 5FOAr, 5-fluorooroate resistant.
Plasmid pJL202 (provided by J. Li) contains a yeast DNA fragment in which the coding sequence of URA3 is replaced with HIS3 (30).
The PCR fragment used to replace the coding sequence of TEL1 with URA3 was generated by using the primers 5′CCTTCAAAGAAAAGGGAAATCAGTGTAACATAGACGGATTGTACTGAGAGTGCACC and 5′CAAAAAAAAGAAGTATAAAGCATCTGCATAGCAATTACTGTGCGGTATTTCACACCG. The URA3 gene within pRS306 (32) was amplified with these primers, and the resulting DNA fragment used for transformation (41).
Plasmid pDTK103 (provided by D. Kirkpatrick) was derived from pDTK101 (BamHI-SalI fragment containing ADE3 inserted into BamHI/SalI-treated pNEB193) by insertion of a BamHI-BglII fragment containing hisG-URA3-hisG sequences derived from pNKY51 (1) into a BglII site within the ADE3 gene.
Use of plasmid pGAL-HO to do a mating-type switch is described by Herskowitz and Jensen (10).
Plasmid pDTK102 (provided by D. Kirkpatrick) was constructed by ligating a BamHI-BglII fragment containing hisG-URA3-hisG sequences derived from pNKY51 (1) to BglII-treated pSM20 (provided by D. Schild). The resulting plasmid (pDTK102) has a rad52::hisG-URA3-hisG gene.
The primers 5′CTTACGGTCTCACTAACCTCTCTTCAACTGCTCAATAATTTCCCGGGATCCGCTGCACGGTCCTG and 5′GTATGAAAGGAACTTTAGAAGTCCATTTCCTCGACCTTACCCTGGGCCTCGTTCAGAATGACACG (46) were used to amplify the HIS3 gene of yeast strain AS4 (35), generating a DNA fragment with the sml1::HIS3 allele.
Reference 6.
Reference 33.
The primers 5′GCTACATATAAGGAACGTGC and 5′TTTGCTGGCCGCATCTTCTC were used to amplify the URA3 gene in plasmid pRS306 (32).
Media and vegetative subculturing conditions.
Standard rich growth medium (YPD) and omission media (8) were used. Yeast strains were grown at 30°C, and diploids were sporulated at room temperature. Tetrad dissection procedures were standard (8). We examined the senescent phenotype by using two different, although similar, protocols. For spore cultures derived from the diploid strains JMY300, JMY302, JMY303, KRY238, or KRY242, spore colonies from the dissection plates were streaked onto rich growth medium (YPD) in quarter-plate sectors for single colonies (subcloning 1). After 2 days of growth at 30°C, a smear of cells derived from the first streak was restreaked on a second YPD plate (subcloning 2). This procedure was repeated, usually until 10 subclonings had been performed. Each subcloning involved about 20 cell divisions. For spore cultures derived from the diploid strains KRY229 and KRY232, similar procedures were used except that the incubation period between subclonings was 1 day instead of 2 days; each subcloning, therefore, involved about 10 cell divisions.
Measurements of comparative growth rates and viability of tlc1 and tel1 tlc1 strains.
To compare the growth rates of tlc1 and tel1 tlc1 strains, we mixed spore colonies containing approximately equal numbers of cells of tlc1 and tel1 tlc1 strains of the same mating type (derived by sporulating either the diploid KRY229 or KRY236) and inoculated this mixture into 5 ml of rich growth medium. To maintain exponential growth in the culture, we diluted the cultures 1:100 every 24 h. The ratio between the two strains was determined by removing samples at 6- to 12-h intervals and plating a dilution of each sample on rich growth medium. After 2 days of growth at 30°C, the resulting colonies were replica plated to medium lacking leucine or uracil. Spores derived from KRY229 of the tel1 tlc1 genotype were Ura+ Leu+, and those of the tlc1 genotype were Ura− Leu+; spores derived from KRY236 of the tel1 tlc1 genotype were Ura− Leu+, and those of the tlc1 genotype were Ura+ Leu+ (Table 1).
We also performed two different assays of cell viability for tel1 tlc1 and tlc1 strains. One assay was to compare the number of cells as counted in the hemocytometer with the number of cells capable of colony formation; the second was to measure the fraction of cells in the culture capable of taking up the dye phloxine B (Sigma), which stains dead cells red (12). Cells were harvested from growth medium by centrifugation, incubated for 6 h at 30°C in YPD medium containing 15 μg of phloxine B per ml, and examined microscopically.
Southern analysis of telomere length.
Yeast DNA was isolated from vegetative cultures by standard methods (8). The DNA was treated with either XhoI or PstI, and the resulting fragments were separated by gel electrophoresis in 1% agarose gel. The fragments were transferred to a Hybond N+ nylon membrane and hybridized to a probe derived from a region of the Y′ element centromere-distal to the XhoI site. This probe was prepared by PCR amplification of pYT14 (31) by using the primers 5′ACACACTCTCTCACATCTACC and 5′TTGCGTTCCATGACGAGCGC. We used a Y′ probe rather than a poly(GT) probe (43) for two reasons. First, the poly(GT) probe hybridizes to both telomeric sequences and nontelomeric poly(GT) tracts (42); second, the poly(GT) probe hybridizes weakly to short telomeres. To quantitate the level of Y′ amplification in samples derived from senescence survivors, we rehybridized blots of PstI-treated DNA (previously hybridized to the Y′-specific probe) to a single-copy yeast DNA probe prepared by PCR amplification of genomic DNA with the primers 5′GTGGCGGTAGTTTTGGCGATTTTCTTTTGG and 5′TCACGGGATTTTATGCTCTGTAGTCCAATG. Blots were scanned with a PhosphorImager and analyzed by using ImageQuaNt software (Molecular Dynamics).
RESULTS
Rationale.
Mutations in many different yeast genes result in telomeres that are either shorter or longer than those in wild-type strains. With the exception of TLC1 and EST2, which encode the RNA and protein subunits, respectively, of telomerase, the roles of most of these genes in telomere maintenance are unknown. Below, we investigate genetic interactions between three mutations that result in short telomeres: mec1, tel1, and tlc1. By comparing the phenotypes of strains containing single- and double-mutant combinations of these genes (epistasis analysis), we conclude that Tel1p and Mec1p are required for telomere elongation in roles that are at least partially independent of telomerase.
Telomere length in tel1, mec1, and tel1 mec1 strains.
There are a number of yeast genes that encode proteins with C-terminal regions homologous to lipid/protein kinases (6, 21), including TEL1, MEC1, TOR1, and TOR2. Since mutations in the putative kinase domain of Tel1p result in short telomeres, we previously examined telomere length in strains with mutations in MEC1, TOR1, or TOR2 (6); no reduction in telomere length was observed in these strains. A subsequent analysis of the mec1 strain used in our study (DLY285; provided by T. Weinert) demonstrated that this strain contained two mutations (26, 46), the mec1-1 mutation (a recessive lethal) and a mutation in SML1 (suppressor of mec1 lethality). Consequently, we decided to reexamine telomere length by Southern analysis in a strain containing a different allele of mec1 in the absence of the sml1 suppressor.
In addition to the terminal poly(TG1–3) sequences, yeast chromosomes have subtelomeric repeats, X and Y′ (17). All telomeres have X repeats, and about half have one or more Y′ elements. The arrangement of these sequences (telomere to centromere) is poly(TG1–3)-Y′0–3-X. The terminal Y′ elements contain a PstI site located about 0.9 kb from the end of the chromosome. When genomic DNA is treated with PstI and hybridized to a Y′-specific probe derived from the region centromere-distal to the site (Fig. 1), the broad region of hybridization at 0.9 kb represents a composite of telomeric fragments from all Y′-containing telomeres. The DNA fragments of 3.5 and 4.8 kb represent tandemly arranged Y′ elements of two size classes (17).
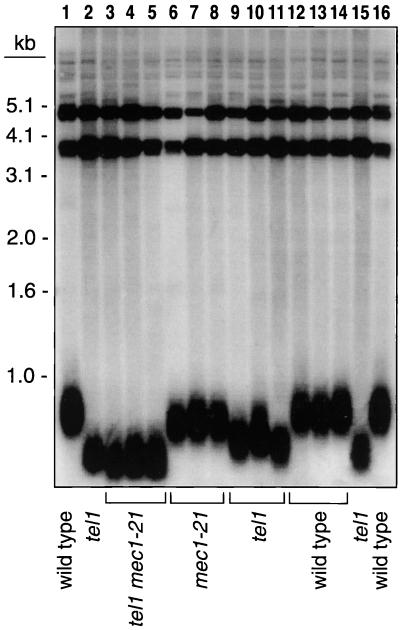
FIG. 1.
Telomere lengths in wild-type, tel1, mec1-21, and tel1 mec1-21 strains. A diploid strain heterozygous for tel1 and mec1-21 mutations was sporulated, and tetrads were dissected. In three tetrads in which all four genotypes were represented, DNA was isolated from spore cultures without subculturing; the strains had undergone about 35 cell divisions at the time of DNA extraction. The DNA was treated with PstI, and Southern analysis was performed. Strains analyzed: W303a (lanes 1 and 16); KRY20a (lanes 2 and 15); JMY300-1a, -2a, -3a (lanes 3 to 5, respectively); JMY300-1c, -2d, and -3d (lanes 6 to 8, respectively); JMY300-1d, -2b, and -3b (lanes 9 to 11, respectively); JMY300-1b, -2c, and -3c (lanes 12 to 14, respectively). Since the tel1 mutation exhibits a long phenotypic lag (20), the telomeres in the tel1 control strain KRY20a (lane 2), which had been subcultured for more than 100 doublings, were slightly shorter than those in the tel1 strains derived from the spores, which had not been subcultured.
We generated a series of haploids with wild-type, tel1, mec1-21, and tel1 mec1-21 genotypes by sporulating a diploid strain (JMY300) that was doubly heterozygous for the tel1 and mec1-21 mutations. DNA was isolated from spore cultures and examined by Southern analysis (Fig. 1). This analysis showed that mec1-21 strains had slightly (about 50 bp) shorter telomeres than wild-type strains. In addition, in strains of the tel1 mec1 genotype, telomeric repeats were slightly shorter than those in the tel1 single-mutant strains.
Mutations in five yeast genes (est1 to -4 and tlc1), including the RNA and protein subunits of telomerase, result in continually shortening telomeres and a senescent phenotype, a gradual loss of cell viability during vegetative subculturing (13, 19, 33). In the senescent cultures, fast-growing survivors are generated as a consequence of RAD52-dependent amplification of the subtelomeric Y′ repeats (18). As shown in Fig. 2a, although no senescence was observed for wild-type, mec1-21, or te11 strains, the tel1 mec1-21 strains had a senescent phenotype. As observed for the est and tlc1 strains, prolonged subculturing of the tel1 mec1-21 strains produced fast-growing survivors. Survivors of senescence, either in tel1 mec1-21 strains or in strains of other genotypes with the senescent phenotype described below, invariably contained amplified tandem Y′ elements (Fig. 3) and/or novel DNA fragments that hybridized to a telomeric probe; both classes of survivors have been observed previously in est1 mutants (18). To determine if the appearance of fast-growing survivors in the tel1 mec1-21 strains was dependent on Rad52p, we constructed and sporulated a diploid strain (KRY242) heterozygous for tel1, mec1-21, and rad52. All of the nine spores examined with the triple-mutant genotype were senescent but did not produce survivors (Fig. 2b). Thus, as observed previously for strains with est mutations, the ability to produce survivors in the tel1 mec1-21 strains is dependent on Rad52p.
FIG. 2.
Senescent phenotypes of tel1 mec1-21 and tel1 mec1-21 rad52 strains. Spores derived from JMY300 (JMY300-2a [tel1 mec1-21], JMY300-2b [tel1], JMY300-2c [wild type], and JMY300-2d [mec1-21]) or KRY242 (KRY242-8a [wild type], KRY242-8b [mec1-21 rad52], KRY242-8c [tel1 mec1-21 rad52], and KRY242-8d [tel1]) were vegetatively subcultured by streaking on plates containing rich growth medium (subcloning 1 [sc 1]). After 2 days at 30°C, each strain was restreaked onto a new plate (sc 2), and this protocol was continued for 10 subclonings; we calculate that there are about 20 cell divisions per subcloning.
FIG. 3.

Amplification of Y′ elements associated with survivors in the tel1 mec1-21 genotype. Southern analysis was done on PstI-treated DNA derived from the tel1 mec1-21 strain JMY300-3a after 1, 4, 7, and 10 subclonings (sc1, sc4, sc7, and sc10). Following hybridization to a Y′-specific probe, the blot was boiled to remove the probe and rehybridized to a single-copy probe (as described in Materials and Methods). By quantitating the hybridization to these two probes, we concluded that one class of tandem Y′ elements was amplified more than 10-fold in the 10th subcloning relative to the wild-type strain. By the 10th subcloning, most of the JMY300-3a cells were fast-growing survivors.
There was considerable heterogeneity in the rate of growth of different tel1 mec1-21 spores at different subculturings. About two-thirds of the tel1 mec1-21 spores grew more slowly than the isogenic wild-type strains even after one subcloning (Fig. 2a), although the growth rate after additional subclonings was even lower in most of these cultures. About one-third grew at approximately the same rate as the wild type during early subculturings, although growth slowed in the later subculturings. Several arguments support the conclusion that the tel1 mec1-21 strains have a senescent phenotype similar to that of tlc1, rather than a nonsenescent slow-growth phenotype. First, as discussed below, we observed similar growth variation in spores of the tel1 mec1-21 and tlc1 genotypes (otherwise isogenic); in previous studies of tlc1 strains (33), stochastic variation in growth rates was also observed. In a comparison of 40 pairs of spores derived from sporulating the diploid KRY238, we found that tel1 mec1-21 spores grew more slowly than tlc1 spores in 12 pairs, more rapidly than tlc1 spores in 15 pairs, and at about the same rate as tlc1 spores in 13 pairs. In addition, pairs of tlc1 spores derived from the same tetrad of KRY228 senesced at different rates in eight pairs and at approximately the same rate in four pairs. Second, the observed eventual death of all cells in the tel1 mec1-21 rad52 spore cultures is not consistent with a nonsenescent slow-growth pattern.
In S. pombe, tel1 rad3 strains (equivalent to S. cerevisiae tel1 mec1 strains) grow slowly and lose telomeric sequences (22). In fast-growing survivors of the tel1 rad3 strains, the chromosomes circularize and diploid strains with these circular chromosomes have greatly reduced spore viability. In S. cerevisiae, strains heterozygous for a single circular chromosome have reduced spore viability (61% viable spores) compared to a wild-type strain (90%), since recombination between a circular and a linear chromosome results in dicentric chromosomes (9). Thus, strains heterozygous for 16 circular chromosomes would be expected to have very poor spore viability. We crossed a tel1 mec1-21 survivor (JMY300-3aS) to W303α. The resulting diploid (KRY246) was sporulated, tetrads were dissected, and spore viability was determined. We also examined spore viability in an isogenic diploid strain (KRY300) resulting from a cross of two wild-type haploid parents. The percentages of spore viability were 80 for KRY246 and 92 for KRY300. In this survivor, therefore, it is unlikely that there is even a single circular chromosome.
Effect of sml1 on telomere length and cellular senescence.
The lethal effect of mec1-1 is suppressed by a mutation in the sml1 gene (26, 46). Since mutations of sml1 lead to elevations in deoxynucleoside triphosphate pools, Zhao et al. (46) suggested that Sml1p is a negative regulator of nucleotide pools and Mec1p and Rad53p are required to relieve this inhibition. To examine interactions between the tel1, mec1, and sml1 mutations, we analyzed spores derived from diploids (JMY302 and JMY303) heterozygous for all three mutations, resulting in strains of eight different genotypes.
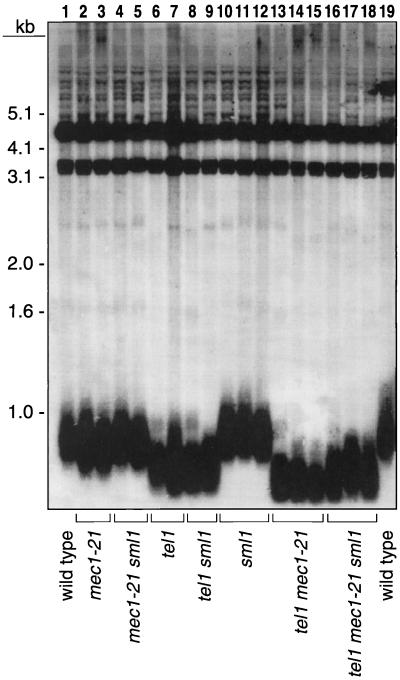
As shown in Fig. 4, of the eight genotypes, only tel1 mec1-21 or tel1 mec1-21 sml1 strains had a senescent phenotype. In eight tetrads that were analyzed with these genotypes, the triple mutant senesced more slowly than the double mutant in five tetrads (as in Fig. 4b) and at approximately the same rate in three tetrads. Thus, the sml1 mutation often delayed but did not suppress senescence. Telomere lengths in strains of eight genotypes are shown in Fig. 5. In general, sml1 had little effect on telomere length, although the telomeres of the mec1-21 sml1 strains were slightly longer (about 50 bp), resulting in telomeres of wild-type length. This effect is consistent with our earlier observation that telomeres in the mec1-1 sml1 strain were approximately the same length as those in the wild-type strain (6). In addition, the telomere lengths in tel1 mec1-21 sml1 strains had broader size distributions than those in the tel1 mec1-21 strains, possibly accounting for the delay in senescence described above.
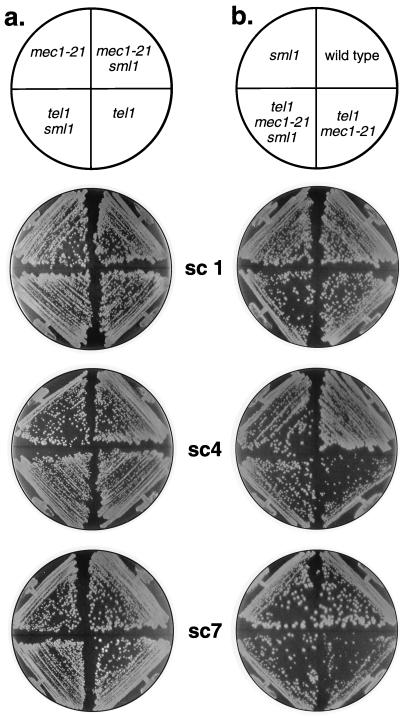
FIG. 4.
Effect of the sml1 mutation on cellular senescence. The diploid strains JMY302 and JMY303 (heterozygous for tel1, mec1-21, and sml1) were sporulated, and tetrads were dissected. Spore cultures of the eight expected genotypes were vegetatively subcultured as described in Materials and Methods. In most tetrads, the rate of senescence in sml1 tel1 mec1-21 strains was delayed relative to the rate in tel1 mec1-21 strains. Strains analyzed: JMY302-11a (mec1 sml1), -11b (mec1), -11c (tel1), and -11d (tel1 sml1); JMY303-6a (wild-type), -6b (sml1), -6c (tel1 mec1), and -6d (tel1 mec1 sml1).
FIG. 5.
Effect of the sml1 mutation on telomere length. DNA was isolated from cultures of spores derived from the diploid JMY302 (heterozygous for tel1, mec1-21, and sml1). These samples were treated with PstI and examined by Southern analysis as described previously. Strains analyzed (from left to right): JMY302-6d, -11b, -25c, -11a, -22a, -11c, -22b, -11d, -25b, -6c, -9d, -12d, -6a, -9c, -12b, -6b, -9b, -12c, and -9a.
In conclusion, although tel1 and mec1-21 strains have short stable telomeres and do not undergo senescence, strains with both mutations undergo continual loss of telomeric repeats and an associated senescence. One interpretation of this result (discussed further below) is that Tel1p and Mec1p have functionally redundant roles in a pathway that is essential for telomere elongation. One model is that phosphorylation of a protein subunit of telomerase is essential for its function and that Tel1p or Mec1p is required for this phosphorylation. One prediction of this model is that the phenotype of a strain with a mutation in both TEL1 and a telomerase subunit should be identical to the phenotype of a strain with a single mutation eliminating telomerase activity such as tlc1 (33). If Tel1p and telomerase promote telomere elongation by independent pathways, one would expect the double mutant to have a different phenotype (for example, shorter telomeres and faster senescence) than either single mutant. Below, we describe evidence that tel1 tlc1 strains have an unexpected phenotype: the double-mutant strains have delayed senescence relative to tlc1 strains.
Genetic interactions between tel1 and tlc1.
A diploid strain (KRY229) heterozygous for null mutations of TEL1 and TLC1 was sporulated to generate isogenic strains with various combinations of these mutations. As shown in Fig. 6, cultures derived from spores of the tel1 tlc1 genotype underwent a delay in the onset of senescence compared to those of the tlc1 genotype. The difference in the growth behavior of the strains was usually evident at the first subculturing (about 30 cell divisions). We examined the patterns of senescence in 33 tetrads in which all four genotypes (wild type, tel1, tlc1, and tel1 tlc1) were represented. In 22 of these tetrads, the tel1 tlc1 strain had obviously delayed senescence. In 11 tetrads, the double mutant had only a slight delay or no obvious delay of senescence relative to the tlc1 strain. The double mutants were never observed to senesce faster than the tlc1 single mutants. In addition to the delay in onset of senescence, tel1 tlc1 strains had a corresponding delay in accumulation of telomerase-independent survivors. For example, in Fig. 6, survivors (large colonies) were much more common in subcultures 4 and 5 for the tlc1 strain than for the tel1 tlc1 strain.
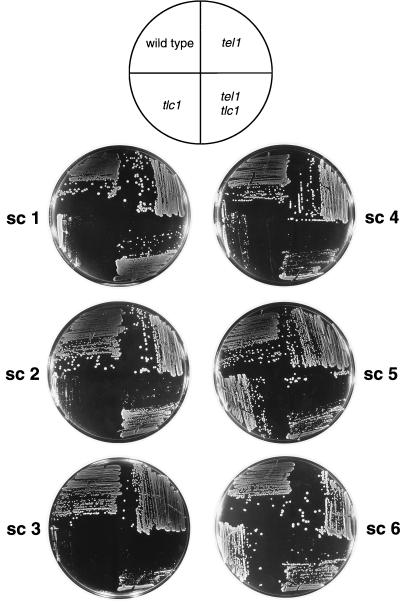
FIG. 6.
Comparison of senescence phenotypes of tlc1 and tel1 tlc1 strains. A diploid heterozygous for tlc1 and tel1 mutations was sporulated, and tetrads were dissected. Spore colonies (KRY229-6a [wild-type], KRY229-6b [tel1], KRY229-6c [tlc1], and KRY229-6d [tel1 tlc1]) were subcloned as described in the legend to Fig. 2 except that each subcloning was done at 24-h intervals. In general, strains with the tel1 tlc1 genotype had a delay in senescence compared to those of the tlc1 genotype. In addition, the fast-growing survivor strains appeared more quickly in the tlc1 strains than in the tel1 tlc1 strains.
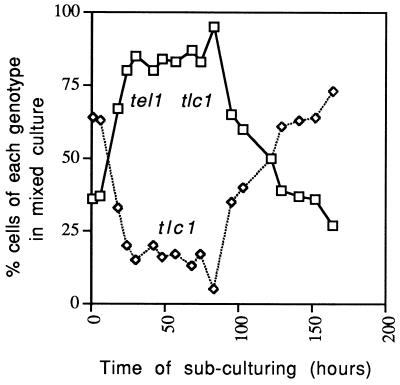
To confirm the difference in phenotypes between tlc1 and tel1 tlc1 strains, we also examined the relative growth rates of the two strains grown in competition in the same culture (Fig. 7). The genotypes of spore colonies derived from KRY229 could be rapidly tested on omission media, since the tlc1 was disrupted with a LEU2 insertion and tel1 was disrupted with URA3. Spore colonies with tlc1 and tel1 tlc1 genotypes derived from the same tetrad were resuspended in water, mixed, and inoculated into liquid rich growth medium. Since we found that the cultures had a doubling time of about 3 to 4 h, we diluted the cultures 1:100 every 24 h. The fraction of cells with each genotype was determined at 6- to 12-h intervals. During the first 80 h of growth, the tel1 tlc1 mutant grew more rapidly than the tlc1 strain. After 80 h of culturing, this trend was reversed. Our interpretation of this result, consistent with the observations described above, is that the tlc1 strain had an earlier onset of senescence and therefore a lower effective growth rate than the tel1 tlc1 strain during the early stages of subculturing. The switch in relative growth rates at later times (after 80 h) reflects the earlier production of survivors in the tlc1 strain relative to the tel1 tlc1 strain. In nine of nine competitive subculturing experiments, in the first 60 to 70 h, the tel1 tlc1 strain grew better than the tlc1 strain. In six of the nine experiments, after 80 h, there was a switch in relative growth rates in favor of the tlc1 strain, reflecting the earlier appearance of survivors in the tlc1 strain. In the other three experiments, the tlc1 strain grew more slowly than the tel1 tlc1 strain for the course of the experiment (usually 150 to 200 h of subculturing).
FIG. 7.
Comparative growth rates of tlc1 and tel1 tlc1 strains in mixed cultures. The diploia KRY229 was sporulated, and spores of the same mating type with the genotypes tlc1::LEU2 and tel1::URA3 tlc1::LEU2 were identified. Equal number of cells of pairs of such strains were mixed and inoculated into rich growth medium. The cultures were grown at 30°C. Exponential growth was maintained by dilution of the culture 1:100 into fresh medium every 24 h. Samples were taken every 6 to 12 h, and the ratio of the two strains was determined as described in Materials and Methods. Dotted lines and solid lines represent the data for the tlc1 (KRY229-13c) and tel1 tlc1 (KRY229-13d) strains, respectively.
In the competition experiments described above, the tel1 tlc1 strain had the wild-type URA3 and LEU2 genes, whereas the tlc1 strain had only the wild-type LEU2 gene. To rule out the possibility that the URA3 gene resulted in a selective advantage for the tel1 tlc1 strain, we performed competitive growth experiments with strains (derived by sporulating the diploid strain KRY236) of the genotypes tel1::ura3 tlc1::LEU2 and tlc1::LEU2 URA3. In three of three such experiments, the tel1::ura3 tlc1::LEU2 strain grew better than the tlc1::LEU2 URA3 strain during the early stages of subculturing. This result demonstrates that the more rapid growth of the double mutant strains derived from KRY229 is not a consequence of the URA3 gene used to make the tel1 mutation.
The more rapid growth of the tel1 tlc1 strains than of the tlc1 strains (Fig. 6 and 7) could reflect less cell death, a more rapid cell cycle, or both factors. To examine this issue, we measured doubling times and cell viability of wild-type, tlc1, and tel1 tlc1 strains subcultured separately for 3 days in liquid medium. In Table 2, we show data for strains derived from two different tetrads; except for the relevant mutations, all strains are isogenic. Although, as in other experiments, there was considerable stochastic variation in growth rates of tlc1 and tel1 tlc1 strains, certain patterns were consistent. The doubling time of the wild-type strains (measured by cell counts with a hemocytometer) was about 90 min and constant for each day of subcloning. In addition, cell viability measured by CFU or by the ability of the cells to exclude the dye phloxine B (15) was 100%. In contrast, tlc1 strains had a low growth rate on day 1, an extremely low growth rate on day 2, and a higher growth rate (reflecting the appearance of survivors) on day 3. The very low growth rate observed for the tlc1 strains on day 2 was associated with a very low percentage of cells (about 1%) capable of forming colonies; many of the cells incapable of colony formation, however, were capable of dye exclusion. On day 3, the increased fraction of tlc1 cells capable of colony formation correlated with the increased growth rate. In the tel1 tlc1 strains, the growth rate and cell viability were higher than observed for the tlc1 strains on days 1 and 2. In comparison to the tlc1 strains, the tel1 tlc1 strains had a lower growth rate and a smaller fraction of cells capable of colony formation on day 3 than on day 2, reflecting the delayed appearance of survivors. These results strongly suggest that at least part of the growth rate difference observed between tlc1 and tel1 tlc1 strains reflects differences in the rate of cell death. We cannot rule out the possibility that the cell cycle times of the two strains are also different. One complicating factor is that in examining the rate of division of individual cells microscopically, we observed considerable intrastrain heterogeneity in both tlc1 and tel1 tlc1 strains (data not shown).
TABLE 2.
Cell division time and cell viability of wild-type, tlc1, and tel1 tlc1 strains during vegetative subculturing in liquid mediuma
| Days of subculturingb | Genotype | Doubling time (min)c | % Viable cells
|
|
|---|---|---|---|---|
| CFUd | Dye exclusione | |||
| 1 | Wild type | 108, 84 | 100, 98 | 100, 100 |
| tlc1 | 744, 172 | 6, 29 | 97, 97 | |
| tel1 tlc1 | 180, 117 | 75, 56 | 100, 100 | |
| 2 | Wild type | 90, 90 | 100, 100 | 100, 100 |
| tlc1 | 1140, 1100 | 2, 1 | 76, 43 | |
| tel1 tlc1 | 204, 168 | 71, 22 | 97, 98 | |
| 3 | Wild type | 96, 84 | 100, 100 | 100, 100 |
| tlc1 | 96, 342 | 96, 13 | 98, 92 | |
| tel1 tlc1 | 228, 295 | 15, 9 | 97, 94 | |
A diploid strain heterozygous for the tel1 and tlc1 mutations (KRY229) was sporulated, and tetrads were dissected. Two tetratype tetrads were identified and, in two independent experiments, liquid cultures of strains of the wild-type, tlc1, and tel1 tlc1 genotypes were established. Cell growth rates and cell viability measurements were done as described below. Numbers separated by commas in each column represent data derived from two isogenic spores of different tetrads.
Spore colonies were inoculated into liquid YPD and grown overnight at 30°C. The following morning, cultures were diluted 1:100 in YPD, and growth at 30°C was continued (day 1). At the end of day 1, growth rates were calculated, and the cultures were diluted to yield a starting cell concentration of about 106 cells/ml at the start of day 2. At the end of day 2, this procedure was repeated.
At 2-h intervals for 8 h each day, the cell concentration was measured in a hemocytometer. Doubling times were determined by standard methods.
Samples from each time point were diluted and plated on solid YPD (four plates with about 200 cells/plate). After 3 days of growth at 30°C, colonies were counted. The values shown represent the number of colonies divided by the number of cells (based on hemocytometer counts), expressed as a percentage.
Cells were stained with the dye phloxine B (as described in Materials and Methods). The values represent the number of red (dye-stained) cells divided by the total number of cells examined, expressed as a percentage.
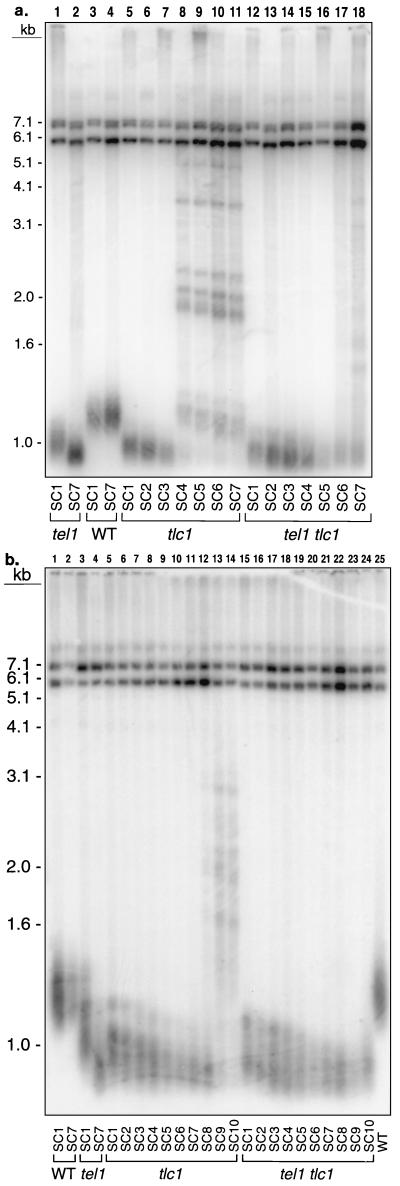
We also measured telomere lengths in tel1 tlc1 and tlc1 strains after various amounts of subculturing (Fig. 8a). Spore colonies derived from KRY229 were inoculated into liquid medium and grown continuously. Since cultures were diluted 1:100 every 24 h, each subcloning reflects about seven cell generations. Southern analysis, using a Y′-specific hybridization probe, was performed as described above. In both tel1 tlc1 and tlc1 strains, telomeres shortened during subcloning. Although telomere lengths in the two strains were similar, there were several differences in the details of the patterns. First, the amount of the terminal Y′-hybridizing fragment was greatly reduced in the tlc1 strain by the fourth subculturing; this fragment was retained through the seventh subculturing in the tel1 tlc1 strain. Second, amplification of Y′/poly(TG1–3)-hybridizing fragments greater than 1 kb in size was detected in the tlc1 strain by the fourth subcloning; this amplification was associated with fast-growing survivors. In the tel1 tlc1 strain, amplification was delayed until the seventh subcloning. Third, the distribution of telomere lengths of the Y′-hybridizing terminal fragment appeared broader in the tel1 tlc1 strain than in the tlc1 strain. These results suggest that the delay of senescence observed in tel1 tlc1 strains likely reflects a delay in the generation of cells that completely lack telomeric repeats.
FIG. 8.
Effect of subculturing tlc1 and tel1 tlc1 strains on telomere length phenotypes. Strains of the tel1, tlc1, tel1 tlc1, and wild-type genotypes derived from the diploids KRY229 (W303a background) (a) and KRY232 (AMY125 background) (b) were inoculated into rich growth medium and grown at 30°C. Cultures were diluted 1:100 into fresh medium every 24 h, and samples for DNA isolation were harvested every 24 h. Each subculturing (SC) represents about 10 cell divisions. Southern analysis was performed on XhoI-treated DNA samples. In such samples, the terminal fragments from Y′-bearing telomeres are about 1.2 kb in size and the tandemly arranged Y′ elements yield DNA fragments of about 5.4 and 6.7 kb. Strains analyzed: (a) KRY229-6b (lanes 1 and 2), KRY229-6a (lanes 3 and 4), KRY229-6c (lanes 5 to 11), and KRY229-6d (lanes 12 to 18); (b) KRY232-7d (lanes 1, 2, and 25), KRY232-7b (lanes 3 and 4), KRY232-7c (lanes 5 to 14), and KRY232-7a (lanes 15 to 24).
In addition to examining the relative growth rates of tel1 tlc1 and tcl1 strains, we compared the growth rates of tel1 mec1 tlc1 and mec1 tlc1 strains to those of tlc1 strains. These strains were constructed by sporulating a diploid strain (KRY238) heterozygous for tel1, tlc1, and mec1 mutations. The growth rates of pairs of spores (each pair of spores derived from a different tetrad) with the relevant genotypes were compared in double-blind experiments. In a total of 37 pairwise comparisons, tel1 mec1 tlc1 strains grew much more slowly than tlc1 strains in 34 pairs and at about the same rate in 3 pairs. Strains with the mec1 tlc1 genotype usually senesced more rapidly than tlc1 strains, although the effect was not statistically significant (P = 0.1); strains with the mec1 tlc1 genotype grew more slowly than strains with the tlc1 genotype in 13 pairs, more rapidly in 5 pairs, and at approximately the same rate in 27 pairs. Thus, although both tel1 and mec1 mutations result in short telomeres as single mutants, these mutations result in different phenotypes in combination with tlc1.
Analysis of the phenotypic effects of tel1 tlc1 and tlc1 mutations in a different genetic background.
All of the strains described above were isogenic with W303a. To generalize our results, we constructed tel1 tlc1 and tlc1 strains in a different genetic background, that of the haploid strain AMY125 (36). We examined spores derived from a diploid (KRY232) of this genetic background that was heterozygous for tel1 and tlc1 mutations. The onset of senescence was delayed for both tel1 tlc1 and tlc1 strains in this background relative to the W303a background, but the tel1 tlc1 strains still senesced later than the tlc1 strain in six of nine spore pairs examined and at about the same time in three of nine pairs. In addition, survivors of senescence appeared more quickly in the tlc1 strains than in the tel1 tlc1 strains in nine of nine spore pairs.
We also examined telomere length in wild-type, tel1, tel1 tlc1, and tlc1 strains (Fig. 8b). Wild-type strains derived from AMY125 had telomeres that were about 150 bp longer than those in wild-type W303 strains; this difference is likely to account for the greater delay in senescence. In addition, as observed for the W303a-derived strains, we found that (i) the Y′-hybridizing telomeric fragment persisted through more subculturing for the tel1 tlc1 strain than for the tlc1 strain, (ii) the amplification of Y′/poly(TG1–3)-hybridizing fragments was observed later for the tel1 tlc1 strain than for the tlc1 strain, and (iii) the size distribution of the Y′-hybridizing telomeric fragment was greater for the tel1 tlc1 strain than for the tlc1 strain. In summary, the interactions between the tel1 and tlc1 mutations appear similar in the two different genetic backgrounds.
DISCUSSION
From the studies described above, we conclude that (i) both Tel1p and Mec1p are required to maintain wild-type telomere length, (ii) tel1 mec1 strains have very short telomeres and senesce at approximately the same rate as tlc1 strains, (iii) recombination is required to produce survivors in tel1 mec1 strains, (iv) at early subclonings, tlc1 strains have a lower growth rate than tel1 tlc1 strains, and (v) at early subclonings, tel1 mec1 tlc1 strains have a lower growth rate than tlc1 strains. These conclusions will be discussed below.
Tel1p and Mec1p are very large proteins (about 300 kDa) with significant regions of homology, particularly near the C terminus (9, 24). The conserved C-terminal domain is also found in other proteins (such as DNA-specific protein kinase) that have lipid and/or protein kinase activity. Strains with the mec1 mutation are sensitive to various DNA-damaging agents because of a defective checkpoint function (44). Strains with the tel1 mutation are not sensitive to DNA-damaging agents, but addition of a single extra copy of TEL1 can partially suppress the damage sensitivity of mec1 strains (24, 31). The phosphorylation of Rad53p, another checkpoint protein, is dependent on Mec1p function, although overexpression of Tel1p results in phosphorylation of Rad53p in the absence of Mec1p (31). These results suggest that Mec1p and Tel1p can both phosphorylate Rad53p in response to DNA damage but that Rad53p is a better substrate for Mec1p than for Tel1p.
Based on the phenotypes of the tel1, mec1, and tel1 mec1 strains, one interpretation of our results is that Mec1p and Tel1p both phosphorylate a protein required for telomere elongation. Telomeres in tel1 strains are much shorter than those in mec1 strains, suggesting that the Tel1p kinase operates on this substrate more efficiently than the Mec1p kinase. It should be emphasized, however, that there is no biochemical evidence that either Tel1p or Mec1p has kinase activity, although point mutations within the kinase domain of Tel1p result in short telomeres (9). One explanation of the observation that tel1 mec1 strains senesce with approximately the same kinetics as tlc1 strains is that the common substrate is a protein subunit of telomerase that cannot function unless phosphorylated. The simplest form of this model predicts that the rate of senescence will be the same for tlc1, tel1 tlc1, mec1 tlc1, and tel1 mec1 tlc1 strains, a prediction that is violated by our data. Although we cannot rule out a role of Tel1p and/or Mec1p in the telomerase pathway, the epistasis results are inconsistent with the possibility that the only function of Tel1p and Mec1p in regulating telomere length is to activate telomerase.
Although the hypothesis that Tel1p and Mec1p share a common substrate involved in regulating telomere length is appealing, some details of the epistasis results are difficult to explain by this model. For example, tel1 tlc1 strains senesce slower than tlc1 strains, whereas mec1 tlc1 strains senesce faster than tlc1 strains. For this reason, we prefer a model in which Tel1p and Mec1p have different target molecules. Although we do not understand the roles of Tel1p and Mec1p in regulating telomere length, we will discuss one possibility in detail and mention briefly a few other possibilities.
The model that will be discussed in detail is that Tel1p and Mec1p control accessibility of the telomeric sequences to telomerase (to lengthen the telomeric repeats) and cellular exonucleases (to shorten the telomeric tracts) by phosphorylation of target proteins (TelXp for Tel1p and TelYp for Mec1p) (Fig. 9); by this model, the Tel1p and Mec1p behave analogously to the protein complexes that control accessibility of promoters to the recombination machinery (41). We suggest that in tel1 strains (unphosphorylated TelXp, phosphorylated TelYp), access of the telomeres to telomerase is greatly reduced and access to exonucleases is slightly reduced, resulting in net telomere shortening. We suggest that in mec1 strains (phosphorylated TelXp, unphosphorylated TelYp), access of the telomeres to exonucleases is slightly increased with little effect on access to telomerase. Finally, in tel1 mec1 strains (unphosphorylated TelXp and TelYp), access of the telomeres to telomerase is severely reduced (although not eliminated) and access to exonucleases is substantially increased.
FIG. 9.
Telomere accessibility model for the functions of Tel1p and Mec1p. In this model, Tel1p and Mec1p affect the accessibility of the telomeres to telomerase and exonucleases by phosphorylation of target proteins (TelXp for Tel1p and TelYp for Mec1p) located at the telomeres. Phosphorylated forms of TelXp and TelYp are indicated by encircled “P’s.”
Reduction in telomere length can be achieved either by reducing the rate of telomere elongation or by increasing the rate of telomere degradation. In the model described above, the short telomeres in tel1 strains reflect the first mechanism and the short telomeres in mec1 strains reflect the second. The senescent phenotype of tel1 mec1 reflects the superimposition of a decrease in telomere elongation and an increase in telomere degradation. Since the tel1 mutation reduces access of the telomeres to exonucleases, as well as to telomerase, in the absence of telomerase, tel1 tlc1 strains would be expected to growth faster than tlc1 strains during early subculturings (as observed). In contrast, since mec1 strains have an increased rate of telomere degradation, one would expect that the mec1 mutation would accelerate the senescence of tlc1 strains. To explain the lower growth rates of tel1 mec1 tlc1 strains than of tlc1 strains, we hypothesize that a small amount of telomerase-mediated telomere elongation occurs in tel1 mec1 strains; this level of elongation delays but does not prevent senescence. Elimination of telomerase in tel1 mec1 strains, therefore, leads to an accelerated rate of senescence.
Many variants of the model shown in Fig. 9 could be proposed. The restriction of telomere elongation or promotion of telomere degradation may involve mechanisms other than telomere accessibility. For example, Mec1p could directly inhibit exonucleases involved in telomere degradation. It is also possible that there are some overlaps in the functions of Tel1p and Mec1p, in addition to functional differences. Finally, it is possible that Tel1p and Mec1p or both proteins have more than one role in telomere elongation. For example, Tel1p could have an essential and direct function in activating a subunit of telomerase and an indirect effect on telomere length (possibly by affecting some parameter of the cell cycle).
Based on its homology to checkpoint genes and its overlapping function with Mec1p, an obvious alternative model is that Tel1p regulates a checkpoint that results in cell cycle arrest if telomeres are too short (6). If this checkpoint is similar to those involved in the response to DNA damage (44), several predictions can be made: (i) tlc1 strains with the checkpoint would have longer telomeres and would survive better than those lacking the checkpoint (tel1 mec1 tlc1 strains), (ii) tel1 tlc1 or mec1 tlc1 strains would have a phenotype intermediate between those of wild-type and tel1 mec1 tlc1 strains, and (iii) many of the cells in tlc1 strains would be arrested at a discrete part of the cell cycle; in cells lacking this checkpoint, cell cycle arrest would not occur. Although we observe that tlc1 strains senesce more slowly than the tel1 mec1 tlc1 strains (consistent with the checkpoint model), the observation that tlc1 strains have more dead cells during the early stages of subculturing than tel1 tlc1 strains is not explained by the simplest form of this model. The observation that tel1 tlc1 strains, although dividing more rapidly than tlc1 strains, have telomeres that are as long as or longer than those of tlc1 strains is also not expected if the tel1 mutation results in a partial checkpoint defect. Finally, we find cells arrested as doublets in cultures containing senescent cells in both tlc1 and tel1 mec1 tlc1 cultures (data not shown), suggesting the existence of an active checkpoint in both types of strains. Since tlc1 strains generate survivors more quickly than tel1 tlc1 strains, we cannot rule out versions of the checkpoint model in which the checkpoint response involves early activation of telomere elongation by recombination.
Regardless of the mechanistic details, our results demonstrate that Tel1p and Mec1p have important roles in telomere maintenance in S. cerevisiae. In a recent study, Naito et al. (22) identified a homolog of TEL1 in S. pombe. Strains with mutations in this gene and in rad3 (a homolog of MEC1) rapidly lose telomeric repeats and grow very slowly (22). The importance of the Tel1p and Mec1p in telomere elongation, therefore, is conserved between two distantly related fungi. In addition, since human cell lines expressing dominant-negative fragments of ATM (a homolog of Tel1p) have short telomeres and normal levels of telomerase (37), similar pathways may also exist in mammals. Activation of telomerase is found in many human cancers, and it has been suggested that telomerase may be a good target for anticancer drugs (6). Our results suggest that the pathways involving Tel1p and Mec1p may represent a good alternative target.
ACKNOWLEDGMENTS
We thank D. Gottschling for plasmid pBLUE61::LEU2, D. Kirkpatrick for plasmids pDTK102 and pDTK103, and Y. Sanchez and S. Elledge for S. cerevisiae Y604. We also thank R. Craven and V. Lundblad for helpful discussions and comments on the manuscript.
This research was supported by NIH grant GM24110.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brush G S, Morrow D M, Heiter P, Kelly T J. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahlen M, Olsson T, Kanter-Smoler G, Ramne A, Sunnerhagen P. Regulation of telomere length by checkpoint genes in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:611–621. doi: 10.1091/mbc.9.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Lange T. Telomeres and senescence: ending the debate. Science. 1998;279:334–335. doi: 10.1126/science.279.5349.334. [DOI] [PubMed] [Google Scholar]
- 5.Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 6.Greenwell P W, Kronmal S L, Porter S E, Gassenhuber J, Obermaier B, Petes T D. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 7.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 8.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 9.Haber J E, Thorburn P C, Rogers D. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics. 1984;106:185–205. doi: 10.1093/genetics/106.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herskowitz I, Jensen R E. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 1991;194:132–146. doi: 10.1016/0076-6879(91)94011-z. [DOI] [PubMed] [Google Scholar]
- 11.Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohli J, Hottinger H, Munz P, Strauss A, Thuriaux P. Genetic mapping in Schizosaccharomyces pombe by mitotic and meiotic analysis and induced haploidization. Genetics. 1977;87:471–489. doi: 10.1093/genetics/87.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J J, Zakian V A. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lingner J, Cech T R, Hughes T R, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro telomerase activity. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 17.Louis E J. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- 18.Lundblad V, Blackburn E H. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 19.Lundblad V, Szostak J W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 20.Lustig A J, Petes T D. Identification of yeast mutants with altered telomere structure. Proc Natl Acad Sci USA. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 22.Naito T, Matsuura A, Ishikawa F. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet. 1998;20:203–206. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- 23.Nugent C I, Hughes T R, Lue N F, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 24.Paciotti V, Lucchini G, Plevani P, Longhese M P. Mec1 is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pardue M L, Danilevskaya O N, Lowenhaupt K, Slot F, Traverse K L. Drosophila telomeres: new views on chromosome evolution. Trends Genet. 1996;12:48–52. doi: 10.1016/0168-9525(96)81399-0. [DOI] [PubMed] [Google Scholar]
- 26.Paulovich A G, Margulies R U, Garvik B M, Hartwell L H. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter S E, Greenwell P W, Ritchie K B, Petes T D. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez Y, Desany B A, Jones W L, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 29.Savitsky K, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 30.Schiestl R H, Petes T D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shampay J, Szostak J W, Blackburn E H. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 32.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 34.Smilenov L B, Morgan S E, Mellado W, Sawant S G, Kastan M B, Pandita T K. Influence of ATM function on telomere metabolism. Oncogene. 1997;15:2659–2665. doi: 10.1038/sj.onc.1201449. [DOI] [PubMed] [Google Scholar]
- 35.Stapleton A, Petes T D. The Tn3 β-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics. 1991;127:39–51. doi: 10.1093/genetics/127.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strand M, Prolla T A, Liskay R M, Petes T D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 37.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 38.Tsukiyama T, Wu C. Chromatin modeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 39.Vialard J E, Gilbert C S, Green C M, Lowndes N F. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virta-Pearlman V, Morris D K, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 41.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 42.Walmsley R M, Chan C S M, Tye B-K, Petes T D. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984;310:157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]
- 43.Walmsley R M, Petes T D. Genetic control of telomere length in yeast. Proc Natl Acad Sci USA. 1985;82:506–510. doi: 10.1073/pnas.82.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 45.Zakian V A. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu Rev Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]
- 46.Zhao X, Muller E G, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]