SUMMARY
Adaptation to changing environments and immune evasion is pivotal for fitness of pathogens. Yet, the underlying mechanisms remain largely unknown. Adaptation is governed by dynamic transcriptional re-programming, which is tightly connected to chromatin architecture. Here, we report a pivotal role for the HIR histone chaperone complex in modulating virulence of the human fungal pathogen Candida albicans. Genetic ablation of HIR function alters chromatin accessibility linked to aberrant transcriptional responses to protein as nitrogen source. This accelerates metabolic adaptation and increases the release of extracellular proteases, which enables scavenging of alternative nitrogen sources. Furthermore, HIR controls fungal virulence, as HIR1 deletion leads to differential recognition by immune cells and hypervirulence in a mouse model of systemic infection. This work provides mechanistic insights into chromatin-coupled regulatory mechanisms that fine-tune pathogen gene expression and virulence. Furthermore, the data point toward the requirement of refined screening approaches to exploit chromatin modifications as antifungal strategies.
In brief
Jenull et al. show that the HIR histone chaperone controls chromatin accessibility and transcription of genes mediating nitrogen assimilation of the human fungal pathogen Candida albicans. They further report that HIR1 ablation alters host interaction and promotes virulence, demonstrating that perturbed chromatin homeostasis fine-tunes pathogen fitness.
Graphical Abstract
INTRODUCTION
Infectious diseases pose a major threat to plant, animal, and human life (Fisher et al., 2020; Wu et al., 2020). The opportunistic human fungal pathogen Candida albicans colonizes mucosal surfaces, including the gastro-intestinal (GI) tract, as part of the normal microbiota. However, C. albicans can cause superficial infections in healthy individuals as well as life-threatening systemic diseases in immunocompromised people (Romo and Kumamoto, 2020). Importantly, fungal infections pose serious challenges in clinical settings, owing to poor diagnostic tools and emerging antifungal drug resistance (Berman and Krysan, 2020; Pappas et al., 2018).
Niches colonized or infected by C. albicans often differ strikingly in their nutrient availability, co-habiting microorganisms, as well as immune surveillance (Alves et al., 2020; Drummond and Lionakis, 2019; Miramón and Lorenz, 2017). A constant supply with nutrients is required to ensure viability and to repair macromolecules damaged upon host-imposed stress (Dühring et al., 2015; Valentine, 2007). C. albicans has adopted different strategies to survive as a commensal colonizer. The marked morphogenetic changes and the ability to sense and assimilate various nutrients makes C. albicans a successful colonizer and opportunistic pathogen capable of efficient immune escape (Ene et al., 2014; Höfs et al., 2016). For instance, C. albicans assimilates numerous nitrogen sources, including non-preferred sources such as protein when preferred sources such as ammonium or glutamine become limiting (Dunkel et al., 2014; Martínez and Ljungdahl, 2005). Nutrient assimilation from protein relies on secreted aspartic proteases (SAPs) for degradation and oligopeptide transporters (OPTs) for uptake (Morschhäuser, 2011).
Eukaryotic cells trigger dynamic transcriptional reprogramming upon facing environmental stimuli, including short-term adaptations to external triggers such as adverse stress conditions (Gasch et al., 2000; Reik, 2007; Vihervaara et al., 2018). Full-flexed transcriptomic reprogramming requires transcriptional regulators, as well as changes in the chromatin structure that establish permissive or repressive chromatin states. Chromatin architecture is thus a highly dynamic platform, which maintains cellular states but also mediates the complex modulation of developmental processes and cellular adaptations upon environmental signals (Klemm et al., 2019; Lai and Pugh, 2017; Zaret and Mango, 2016). Among other factors, histone chaperones play fundamental roles in maintaining and fine-tuning chromatin homeostasis during developmental processes and cellular adaptations (Banaszynski et al., 2013; Cheloufi and Hochedlinger, 2017). Histone chaperones can facilitate chromatin assembly coupled to replication and transcription (Hammond et al., 2017). The HIR histone chaperone complex, or HIRA in higher eukaryotes, mediates the incorporation of histone H3-H4 dimers into nucleosomes independent from DNA replication. HIR acts as transcriptional co-regulator affecting developmental processes in human, animal, or plant cells (Amin et al., 2013; Banaszynski et al., 2013; Duc et al., 2015; Sadasivam and Huang, 2016; Tagami et al., 2004) but also regulates gene expression in unicellular yeasts, including S. cerevisiae (Fillingham et al., 2009; Spector et al., 1997). As in S. cerevisiae (Prochasson et al., 2005), the C. albicans HIR complex consists of the Hir1, Hir2, Hir3, and Hpc2 subunits (Jenull et al., 2017; Stevenson and Liu, 2013). Besides transcriptional repression of histone genes (Stevenson and Liu, 2013), HIR affects sensitivity to antifungal azoles (Tscherner et al., 2015) and hyphal initiation in C. albicans (Jenull et al., 2017). Given the crucial role of chromatin regulators in gene-expression control, it is not surprising that an imbalance of histones or a lack of various chromatin modifiers alter fungal morphogenesis and virulence (Lee et al., 2015; Lopes da Rosa et al., 2010; Tscherner et al., 2015; Wurtele et al., 2010; Zacchi et al., 2010). Thus, targeting chromatin function appeared as new antifungal strategy (Kuchler et al., 2016). However, mechanistic studies of how an inhibition of histone modifiers affects fungal chromatin states are scarce.
Here, we present extensive mechanistic insights on how ablation of the HIR subunit Hir1 in C. albicans alters chromatin landscapes and transcriptional control of genes required for nutrient assimilation from proteins. Strikingly, C. albicans lacking HIR1 is hypervirulent in systemic infections in vivo. Moreover, less neutrophils, the main innate antifungal effector cells (Romani et al., 1997), are recruited in infections by hir1Δ/Δ cells. This work dissects chromatin-related aspects of fungal pathophysiology with relevance to therapeutic approaches. Thus, promising antifungal strategies targeting chromatin-modifying factors will have to assess the impact of drug modulators on HIR-related chromatin states to avoid potential adverse effects.
RESULTS
Deletion of HIR1 drives proteolytic activities
We recently demonstrated that hir1Δ/Δ C. albicans cells have impaired hyphal formation (Jenull et al., 2017). In follow-up experiments, we also analyzed colony morphology in response to fetal calf serum (FCS) on synthetic complete (SC) medium. Interestingly, hir1Δ/Δ cells produced on turbid zones around the colonies on SC-FCS medium, which was not observed in the absence of FCS (Figure 1A). C. albicans releases extracellular proteases when proteins serve as the major nitrogen source (Banerjee et al., 1991; Hube et al., 1994), creating a turbid halo around colonies (Bernardo et al., 2008). We reasoned that these turbid zones around hir1Δ/Δ colonies were due to increased proteolytic activity degrading bovine serum albumin (BSA) present in FCS. Indeed, hir1Δ/Δ colonies displayed large halos and grew better on YCB-BSA with BSA as the major nitrogen source, which was fully reverted to the wild-type (WT) phenotype upon re-integration of HIR1 into its native locus (Figure 1B). Similarly, hir1Δ/Δ cells showed a striking growth advantage in liquid YCB-BSA when compared to the WT, but only in the presence of BSA (Figure 1C). Since YCB medium contains histidine, methionine, and tryptophan in the micromolar range, we also assessed growth of WT and hir1Δ/Δ cells in yeast nitrogen base (YNB)-BSA without additional amino acids, thus making BSA the only nitrogen source. Like in YCB-BSA medium, growth in YNB-BSA showed the same phenotype, independent of the supplemented BSA concentration (Figure S1A; Figure 1D). As noted in previous studies (Dabas and Morschhäuser, 2008; Martínez and Ljungdahl, 2005), C. albicans can efficiently utilize BSA as a nitrogen source as indicated by the exponential growth rate once fungal cells have adapted (Figure 1C; Figure S1A). WT cells showed a long lag phase of about 48 h, while hir1Δ/Δ cells resumed exponential growth after 24 h. Notably, cells were washed with dH2O in our study, while an aliquot from YPD-grown cells was directly transferred into YCB-BSA medium in other reports (Dabas and Morschhäuser, 2008). This enhances fungal growth on protein due to the presence of additional extracellular amino acids, which stimulate SAP expression (Hube et al., 1994). Besides, growth on alternative carbon sources in otherwise rich conditions was not affected by the loss of HIR1 (Figures S1B–S1D), but hir1Δ/Δ cells growing on glucose entered the stationary phase earlier than the WT (Figure S1B).
Figure 1. Deletion of HIR1 drives proteolytic activities.
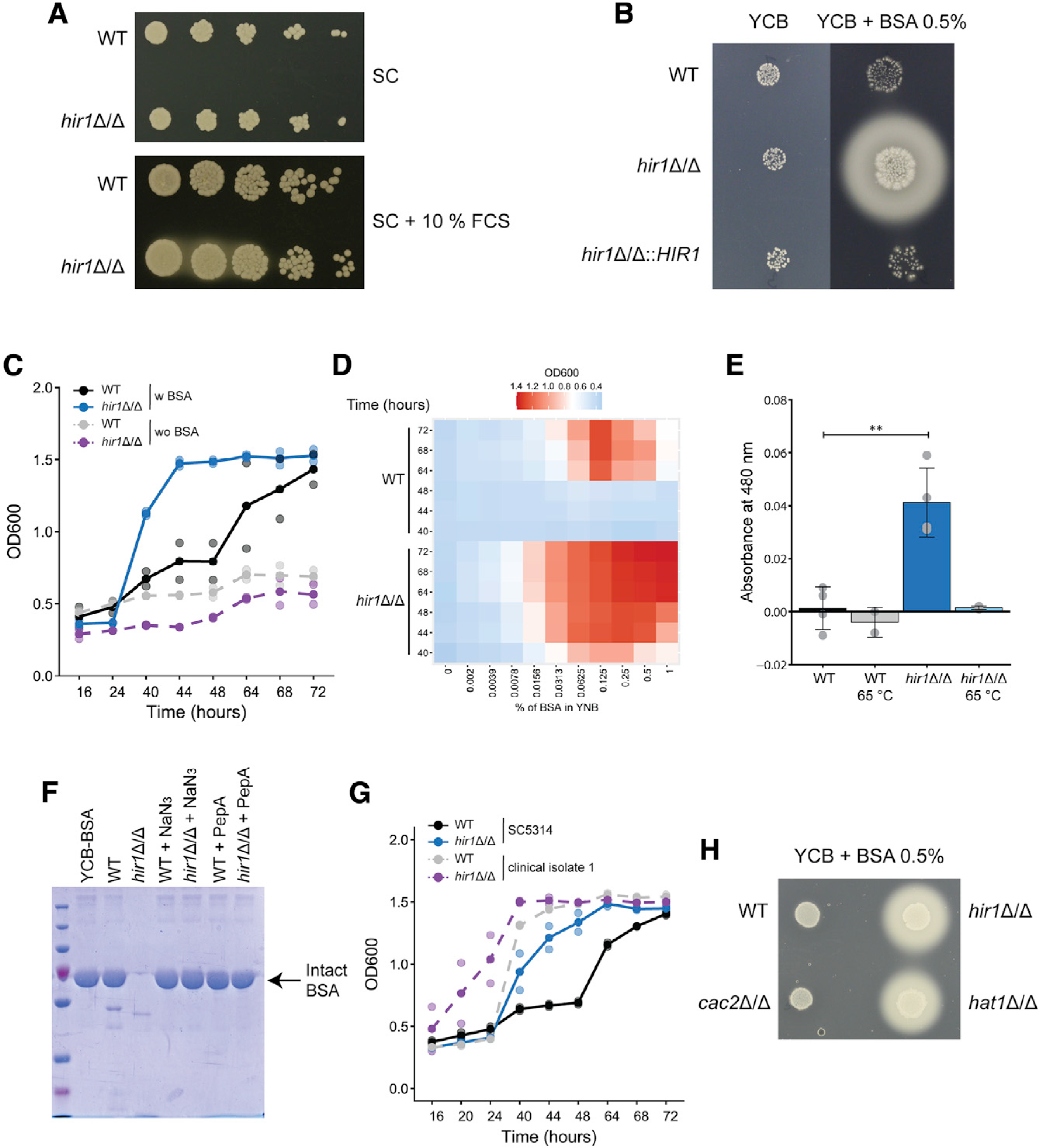
(A) Spot dilution assay on SC agar medium ±10% FCS. Images were taken after 1 day at 30°C and are representative of 3 biological replicates. Brightness (−50) and contrast (+20) were adjusted.
(B) Images of colony spots after 3 days at 30°C and are representative of 3 biological replicates.
(C) Growth of the indicated C. albicans strains in liquid YCB-BSA at 30°C. See also Figure S1E. Graphs show the mean (solid dots) and single measurements (opaque dots) from 2 biological replicates.
(D) Growth of C. albicans in YNB supplemented with the indicated percentage of BSA at 30°C. Colors of the heatmap indicate mean OD600 values from 3–4 biological replicates.
(E) Azocasein assay with YCB-BSA 24 h culture supernatants heat-treated (65°C, 10 min) or not treated prior the assay. Graphs show the mean ± SD from 2–4 biological replicates. ***p < 0.001 with Student’s t test after equal variance testing (F test).
(F) Coomassie staining of supernatants from 16 h YCB-BSA cultures, non-treated or treated with 0.01% sodium azide (NaN3) or 1 μM PepstatinA (PepA). The arrow indicates full-length BSA. The gel image is representative of 2 biological replicates.
(G) Growth of the indicated C. albicans strains and clinical isolates in liquid YCB-BSA at 30°C. See also Figure S1F. Graphs show the mean (solid dots) and single measurement values (opaque dots) from 2 biological replicates.
(H) Images of colony spots as in (B) after 4 days at 30°C, which are representative of 2 biological replicates.
w, with; wo, without. See also Figure S1.
Nitrogen acquisition from protein by C. albicans requires extracellular SAPs (Hube et al., 1997; Staib et al., 2008). Indeed, supernatants from 24 h hir1Δ/Δ YCB-BSA cultures had strong and heat-sensitive proteolytic activities as shown by azocasein assays (Figure 1E). Furthermore, the majority of BSA was degraded by hir1Δ/Δ cells after 16 h in YCB-BSA (Figure 1F), but not by the WT. The BSA decay required both active metabolism and SAP activity, since sodium azide (NaN3) and the SAP inhibitor Pepstatin A (PepA) abrogated BSA cleavage (Figure 1F) and growth in YCB-BSA (Figure S1E). Because of intra-species diversity regarding morphology, virulence, and host responses (Braunsdorf and LeibundGut-Landmann, 2018; Huang et al., 2019), we further tested two unrelated C. albicans clinical isolates. Both clinical isolates grew better on protein than the laboratory strain SC5314, and clinical isolate 10 displayed growth kinetics comparable to its HIR1-deficient isogenic variant (Figure S1F). Deleting HIR1 further enhanced growth on protein in both clinical isolates (Figure 1G; Figure S1F), suggesting that Hir1 regulates proteolytic growth independent of strain backgrounds.
Finally, we also tested a mutant lacking the CAC2 subunit of the CAF-1 histone chaperone complex, which couples chromatin assembly to DNA replication (Hammond et al., 2017). Unlike hir1Δ/Δ cells, BSA utilization as the main nitrogen source was unaffected upon CAC2 deletion (Figure 1H). Interestingly enough, loss of the histone acetyl transferase (HAT) Hat1, which functions in de novo nucleosome assembly upstream of both CAF-1 and HIR (Grover et al., 2018), phenocopied the lack of HIR1 (Figure 1H). These data demonstrate that HIR, but not CAF-1, controls the release of extracellular proteolytic activities through the replication-independent chromatin assembly pathway.
Hir1 is required for transcriptional adaptation to protein as the major nitrogen source
We next analyzed the global transcriptional response of WT and hir1Δ/Δ cells during the metabolic switch from standard laboratory growth in YPD to protein-rich conditions, as Hir1 and its homologs control gene-expression patterns (Nashun et al., 2015; Spector et al., 1997). To do this, YPD cultures (t0) from both genotypes were subjected to growth in YCB-BSA for 4 and 8 h (t4 and t8, respectively), followed by RNA sequencing (RNA-seq) analysis. We detected 412 genes at least 1.5-fold up- or downregulated after 4 h of growth in YCB-BSA, and 597 genes after 8 h in hir1Δ/Δ cells when compared to the WT control (Figures S2A and S2B). To better understand the differences between the transcriptional reprogramming of WT and hir1Δ/Δ cells, we directly compared both genotypes during the adaptation to growth on protein (Figures 2A and 2B). Strikingly, the general transcriptomic profiles of WT and hir1Δ/Δ cells showed a strong correlation (R = 0.75, Pearson’s correlation) during the switch from YPD growth (t0) to protein as major nitrogen source (Figure 2A; YCB-BSA 8 h, t8). Even genes with significant differential expression (FDR <0.05) in hir1Δ/Δ cells after 8 h growth in YCB-BSA, showed a similar transcriptional induction or repression during growth in YCB-BSA in both genotypes. For instance, the pH-regulated RBR1 cell-wall protein gene (Lotz et al., 2004), was among the most highly upregulated genes in both genotypes after 8 h in YCB-BSA, but transcript levels were approximately 6-fold higher in the hir1Δ/Δ mutant than in the WT strain (Figure 2A; Figure S2B). In addition, gene-expression changes mounted between 4 and 8 h of growth in YCB-BSA (t4 versus t8) correlated less well (R = 0.59, Pearson’s correlation; Figure 2B) between WT and hir1Δ/Δ cells, implying distinct adaptive mechanisms among strains when protein is the main nitrogen source. This was evident by the striking upregulation of the SAP2 protease and the OPT1 oligopeptide transporter in the hir1Δ/Δ strain (Figure 2B). In contrast, minor transcriptional induction of those genes was observed in WT cells grown in YPD growth (t0) when compared to 4 or 8 h of growth in YCB-BSA (t8 versus t0 or t8 versus t4, respectively; Figures 2A and 2B). These data suggest that loss of HIR1 affects transcriptional adaptation of specific gene sets during growth on protein, rather than general transcriptional dysregulation.
Figure 2. Hir1 is required for transcriptional adaptation to protein as the major nitrogen source.
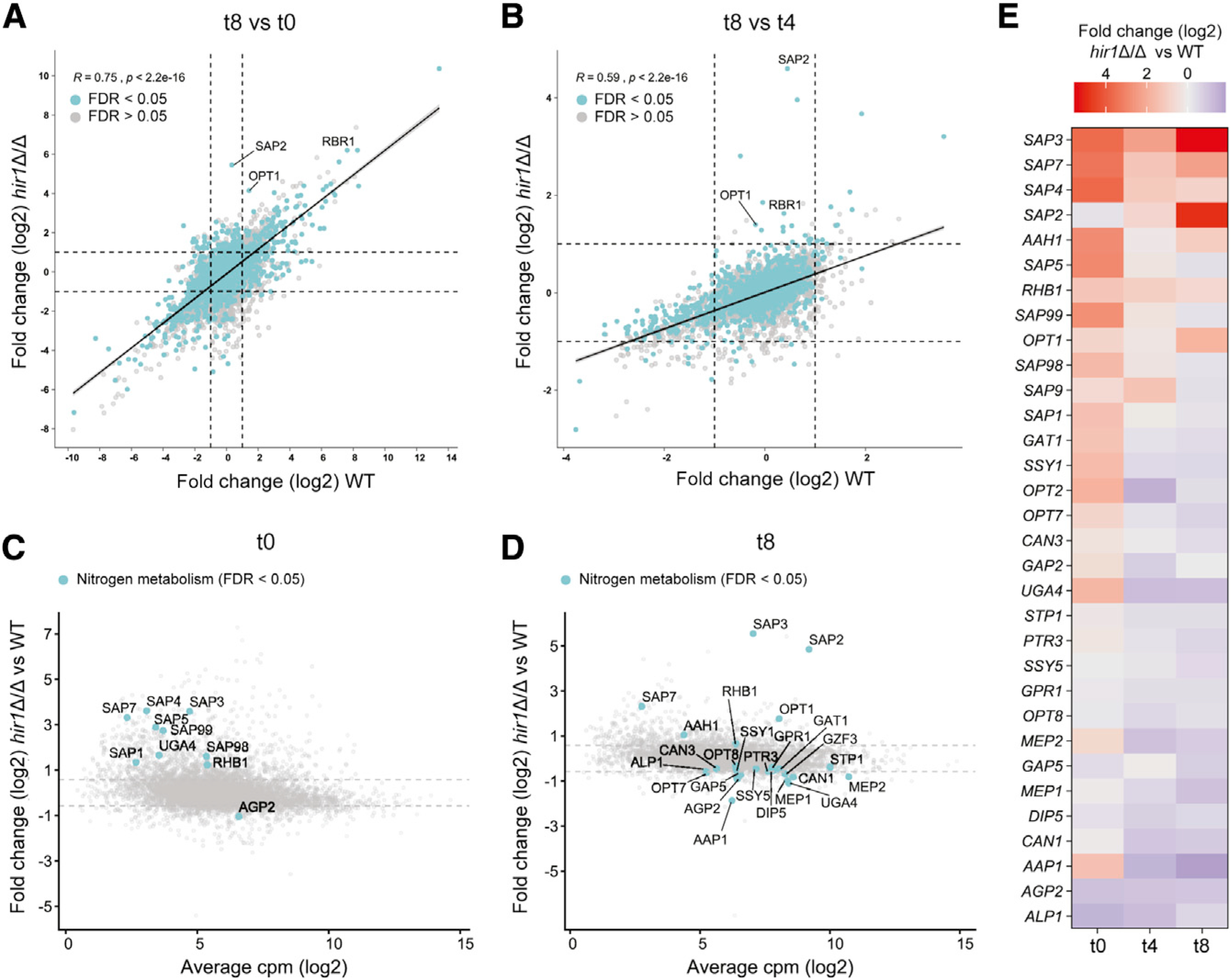
(A and B) The log2-fold change in mRNA abundance in the WT (x axis) is plotted against the log2-fold change in mRNA levels in hir1Δ/Δ cells (y axis) after 8 h of growth in YCB-BSA (t8) relative to YPD growth (t8 versus t0; A) or after 8 h growth in YCB-BSA relative to 4 h in YCB-BSA (t8 versus t4; B). Turquoise dots represent DEGs (FDR <0.05) in hir1Δ/Δ versus WT at t8. Dashed gray lines indicate log2-fold changes of 0.58 and −0.58. Linear regression lines are indicated. The Pearson’s correlation coefficient (R) and the corresponding p value are shown.
(C and D) The average log2 cpm value (x axis) is plotted against the log2-fold change in mRNA abundance between hir1Δ/Δ and WT cells at t0 (C) or t8 (D). Selected genes involved in nitrogen metabolism with differential expression (FDR <0.05) are highlighted. Gray dashed lines indicate log2-fold changes of 0.58 and −0.58.
(E) Heatmap showing genes related to nitrogen metabolism differentially expressed at least at one of the tested growth conditions.
DEG, differentially expressed gene; cpm, counts per million reads; FDR, false discovery rate; t0 (YPD), t4 (YCB-BSA 4 h), t8 (YCB-BSA 8 h). See also Figure S2.
Next, we assessed whether HIR1 ablation affects genes implicated in nitrogen-sensing, starvation signaling, or nitrogen acquisition during YPD growth (Martínez and Ljungdahl, 2005; Morschhäuser, 2011; Ramachandra et al., 2014; Rutherford et al., 2019). Indeed, several SAPs, the amino acid permease gene AGP2 (Lan et al., 2002), the γ-aminobutyric acid permease UGA4 (Limjindaporn et al., 2003) and RHB1, encoding a small G-protein involved in TOR signaling (Chen et al., 2012), were differentially expressed in hir1Δ/Δ cells when compared to the WT grown in YPD (t0; Figure 2C). The shift to YCB-BSA additionally rewired transcription of nitrogen metabolism genes, with SAP2, SAP3, SAP7, and OPT1 among the highest upregulated genes in hir1Δ/Δ cells (Figure 2D). Assessing the dynamics of differentially expressed genes (DEGs) related to nitrogen metabolism revealed constitutive elevated expression of SAP3, SAP7, and RHB1 throughout the time course, while SAP2 and OPT1 became highly upregulated in hir1Δ/Δ cells only during growth on protein (Figure 2E). Additionally, several amino acid and ammonium permeases and sensor systems, including GAP2, GAP5 (Kraidlova et al., 2016), SSY1 (Martínez and Ljungdahl, 2005), MEP1, and MEP2 (Dunkel et al., 2014) were differentially expressed. Gene ontology (GO) term analysis indicated aromatic amino acid biosynthesis (adjusted p value 1.19E-08) and transmembrane transport (adjusted p value 0.0118) to be enriched for upregulated genes in hir1Δ/Δ cells during YPD growth (Figure S2C; Table S3). In addition, genes with increased expression in the hir1Δ/Δ mutant after 8 h YCB-BSA (t8) were linked to cell-cycle functions (adjusted p value 2.40E-05), suggesting re-entrance of hir1Δ/Δ cells into the cell cycle upon sufficient nitrogen assimilation (Figure S2C; Table S3). We further confirmed the differential expression of SAP2–3 and SAP7–8 in YNB-BSA, with SAP3 being markedly upregulated in hir1Δ/Δ cells independent of the growth conditions (Figure S2D). Taken together, these data suggest that hir1Δ/Δ cells more efficiently adapt to utilize proteins as nitrogen sources.
Hir1 controls chromatin accessibility of loci related to nitrogen metabolism
Since Hir1 facilitates histone deposition at gene regulatory regions (Pchelintsev et al., 2013), we employed an assay for transposase-accessible chromatin using sequencing (ATAC-seq) (Buenrostro et al., 2013) to link chromatin accessibility with transcriptional regulation. Thus, WT and hir1Δ/Δ strains were cultured in the same conditions as used for RNA-seq analysis and subjected to ATAC-seq. When assessing differential nucleosome-free ATAC-seq peak abundance between WT and hir1Δ/Δ cells in YPD medium (t0) or upon growth on protein (YCB-BSA 4 h, t4, or 8 h, t8), we identified several hundred genomic regions with increased chromatin accessibility upon loss of HIR1 (Figure S3B; Figure 3A; Table S4). In line with the RNA-seq data, deletion of HIR1 affected chromatin accessibilities at loci upstream of nitrogen metabolism genes during growth on protein (Figure 3A) but also in YPD culture (Figure S3B). For instance, upstream regions of SAP2, OPT1, SAP3, and RHB1 showed an increased chromatin accessibility in hir1Δ/Δ cells during YPD growth (t0) or in response to protein as major nitrogen source (t4 and t8; Figure 3B; Figure S3C). Moreover, upstream regions of nitrogen sensor and permease genes, including MEP1, MEP2, and GAP2 or the nitrogen-regulated transcription factors GAT1 and STP1 (Limjindaporn et al., 2003; Martínez and Ljungdahl, 2005) showed predominantly increased chromatin accessibility in hir1Δ/Δ cells during growth on protein (Figure 3B). Consequently, GO term analysis of genes with increased chromatin accessibility in hir1Δ/Δ cells after 4 or 8 h in YCB-BSA showed enrichment for responses to nutrient levels (adjusted p value 0.0263) and nitrogen utilization (adjusted p value 0.0357; Figure S3D; Table S5). These results demonstrate that HIR1 ablation alters the chromatin landscapes upstream of genes related to nitrogen metabolism not only in the presence of the alternative nitrogen source protein but also when preferred nitrogen sources in YPD medium are available. This was further substantiated when we assessed the chromatin accessibility upstream of DEGs in the hir1Δ/Δ mutant after 8 h of growth in YCB-BSA (RNA-seq t8). We found that those DEGs showed elevated nucleosome-free ATAC-seq read signals, stretching across at least a 1 kb region upstream the transcription start site (TSS), already during growth in YPD in the hir1Δ/Δ mutant (t0; Figure 3C, top graph), while this was less apparent for non-DEGs (Figure 3C, bottom graph). Accordingly, about 35% of differentially abundant ATAC-seq peaks in hir1Δ/Δ cells were upstream of genes with deregulated transcription in response to 4 or 8 h of growth in YCB-BSA (Figure S3E).
Figure 3. Hir1 controls chromatin accessibility of loci related to nitrogen metabolism.
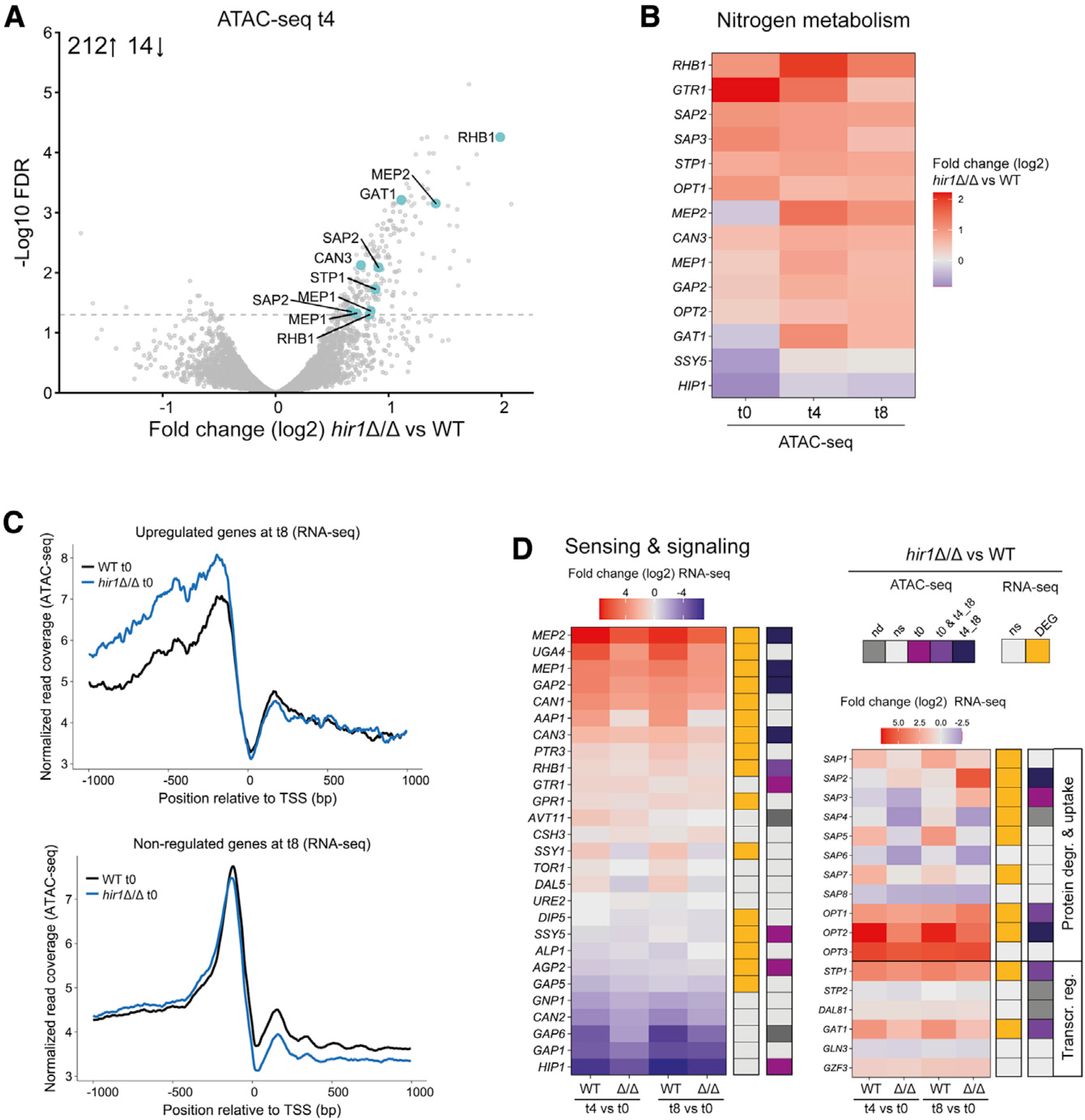
(A) Volcano plot depicting the log2-fold change in ATAC-seq peak signals after 4 h in YCB-BSA (t4; x axis) between the WT and hir1Δ/Δ cells. Each dot represents one ATAC-seq peak, which was annotated to the next adjacent gene. Turquoise color indicates selected genes involved in nitrogen metabolism. The gray dashed line represents a FDR of 0.05. The number insert illustrates the number of significantly up- or downregulated peaks (FDR <0.05).
(B) Heatmap of genes involved in nitrogen metabolism with differential ATAC-seq peak signals (peaks located max. 2 kb upstream the TSS; FDR <0.05) during at least one growth condition.
(C) Normalized ATAC-seq reads from cells grown in YPD (t0) plotted as coverage tracks around the TSS of all genes that are transcriptionally upregulated (FDR <0.05 and log2-fold change >0.58; top graph) or not differentially expressed (FDR >0.05; bottom graph) in hir1Δ/Δ versus WT after 8 h growth in YCB-BSA medium (t8).
(D) Heatmaps of selected genes related to nitrogen sensing and assimilation. The colored code indicates whether a gene is differentially regulated (FDR <0.05) in the RNA-seq and ATAC-seq dataset at any (RNA-seq) or the indicated time point (ATAC-seq).
FDR, false discovery rate; TSS, transcription start site; DEG, differentially expressed gene; ns, not significant (FDR >0.05); nd, not detected; degr., degradation; Transcr. reg, transcriptional regulation. See also Figure S3.
The integration of ATAC-seq and RNA-seq data indicate a divergent control of nitrogen metabolism in hir1Δ/Δ cells in different growth phases, which may also impact the adaptation kinetics to proteolytic growth on protein. Thus, we directly compared how WT and hir1Δ/Δ cells regulate selected genes mediating nitrogen-sensing, signal transduction, and extracellular protein catabolism during the switch from YPD to YCB-BSA. The majority genes showed a decreased amplitude of transcriptional induction and repression in hir1Δ/Δ cells upon media switch. Out of 27 genes, 16 were transcriptionally deregulated, and 9 showed an altered chromatin accessibility, out of which 5 were affected already in YPD (t0) in the hir1Δ/Δ cells (Figure 3D, left graph). Moreover, SAP3 was exclusively upregulated in hir1Δ/Δ cells growing in YCB-BSA (Figure 3D, right graph). In summary, these data strongly suggest that Hir1 controls permissive chromatin states upstream of nitrogen metabolism genes, which affects transcriptional adaptation to proteolytic growth on protein.
Hir1-mediated proteolytic activity is linked to Sap2 and SPS-sensor control
Mass spectrometry confirmed elevated Sap2, Sap3, and Sap8 levels in YCB-BSA supernatants in hir1Δ/Δ cells when compared to the WT control (Table S6). Likewise, prolonged growth for 3 days in SC medium containing ammonium sulfate and an amino acid mix as nitrogen source increased Sap2, Sap3, and Sap8 abundance in supernatants from hir1Δ/Δ cells when compared to the WT and the HIR1 complemented strain (Figure S3F; Table S6).
As Sap2 is the major and essential protease for nitrogen assimilation from protein (Hube et al., 1997; Staib et al., 2008), we tested the requirement of Sap2 for the growth advantage of hir1Δ/Δ cells on protein. The accelerated secretion of Sap2 by the hir1Δ/Δ mutant during growth in YCB-BSA (Figure S4A) coin-cided with BSA degradation in cultures from hir1Δ/Δ cells (Figure S4B). Loss of SAP2 impaired BSA degradation of hir1Δ/Δ cells on solid medium (Figure 4A), albeit residual proteolytic activities were still detectable in the hir1Δ/Δ sap2Δ/Δ double mutant. Unlike the sap2Δ/Δ single mutant, hir1Δ/Δ sap2Δ/Δ cells grew in YCB-BSA medium with almost WT kinetics (Figure 4B). Hence, we reasoned that Sap3 may compensate for the loss of SAP2. Indeed, growth of hir1Δ/Δ cells in YCB-BSA medium was completely abrogated by genetic removal of both SAP2 and SAP3 (Figure 4C), which also impaired BSA degradation (Figure S4C). Of note, ablation of solely SAP3 attenuated growth in YCB-BSA, BSA degradation, and Sap2 secretion kinetics in the WT and in the hir1Δ/Δ background (Figure 4C; Figures S4D and S4E). In summary, these data demonstrate that most extracellular proteolytic activities of hir1Δ/Δ cells are due to the deregulation of SAP2 and SAP3.
Figure 4. Hir1-mediated proteolytic activity is linked to Sap2 and SPS-sensor control.
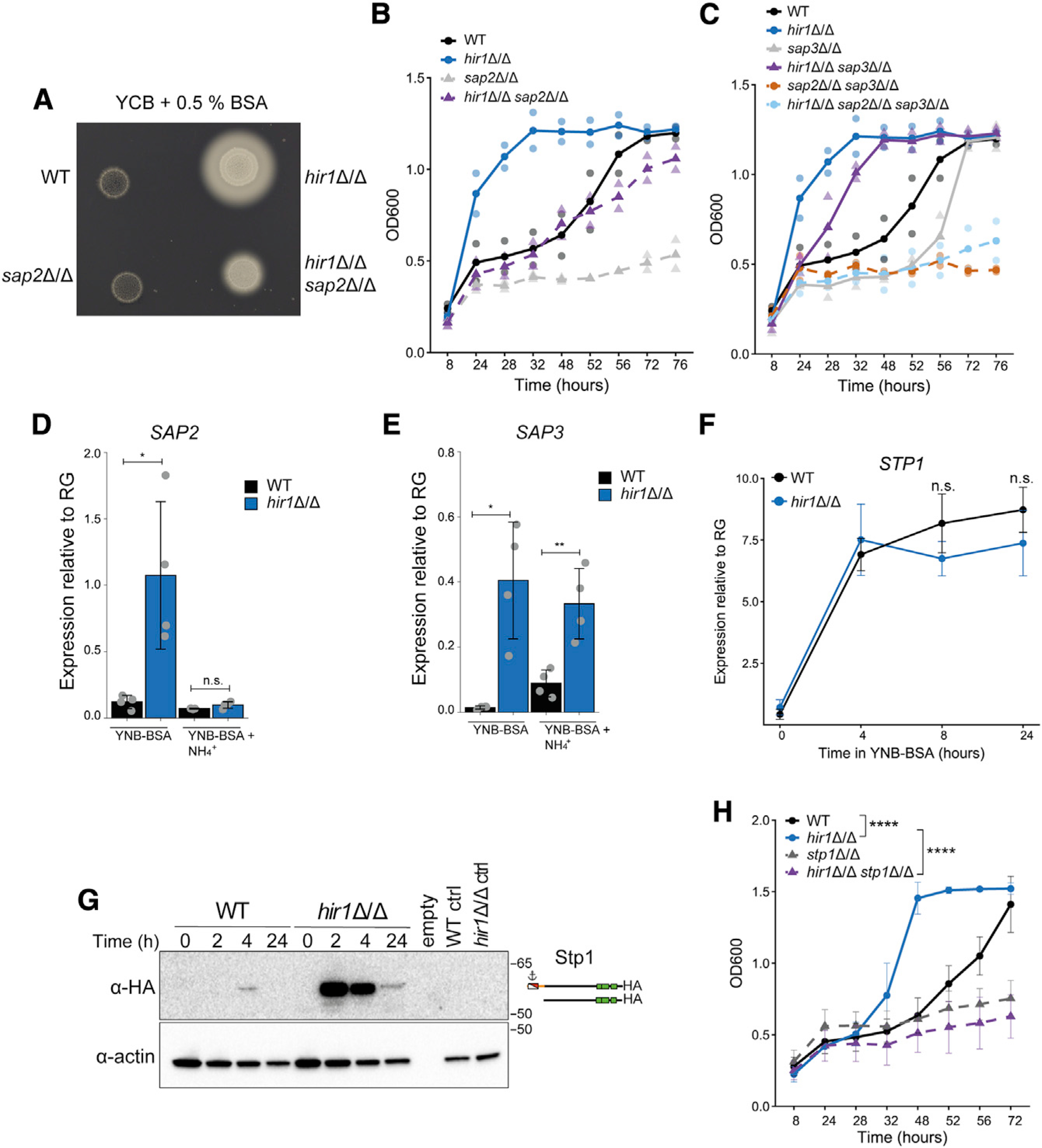
(A) Colony spot assay on YCB-BSA agar plates after 4 days at 30°C. Images are representative of 3 biological replicates.
(B and C) C. albicans growth in liquid YCB-BSA at 30°C. The indicated genotypes were analyzed always together in one experiment but were split into (B) and (C) for clarity. Graphs show the mean (solid dots) and single measurements (opaque dots) from 2 biological replicates.
(D and E) Quantitative real-time PCR analysis of SAP2 (D) and SAP3 (E) relative to the reference gene (RG) PAT1 after 8 h in YNB-BSA with or without 20 mM ammonium sulfate (+ NH4+) at 30°C. Graphs show the mean ± SD from 4 biological replicates. ns = p > 0.05, *p < 0.05, **p < 0.01 with one-way ANOVA followed by Tukey’s multiple comparison.
(F) Quantitative real-time PCR analysis of STP1 expression relative to PAT1 in YNB-BSA. Graphs show the mean ± SD from 3 biological replicates. Indicated p values were calculated with Student’s t test after equal variance testing (F test).
(G) Immunoblot analysis of 3xHA-tagged Stp1 during growth in YNB-BSA at 30°C. Immunoblot is representative of 3 biological replicates. The full-length and processed forms of Stp1–3xHA are schematically depicted. Untagged strains grown for 2 h in YNB-BSA served as control (ctrl).
(H) Growth in liquid YCB-BSA at 30°C. Graphs show the mean ± SD from 3 biological replicates. ****p < 0.0001 with one-way ANOVA followed by Tukey’s multiple comparison test at the 48 h time point after testing for equal variances (Bartlett’s test).
See also Figure S4.
The assimilation of less favorable nitrogen sources such as protein is usually repressed by favorable nitrogen sources such as ammonium, which is called nitrogen catabolite repression (NCR) (Dunkel et al., 2014). Therefore, we analyzed whether defects in transcriptional repression contribute to the hyper-induction of SAP2 and SAP3 in hir1Δ/Δ cells. The presence of 20 mM ammonium sulfate fully repressed enhanced SAP2 expression (Figure 4D), whereas upregulation of SAP3 was unaffected in hir1Δ/Δ cells (Figure 4E), suggesting distinct regulatory mechanisms controlling expression of SAP2 and SAP3.
SAP2 expression strictly depends on the transcription factor Stp1 (Martínez and Ljungdahl, 2005), whose expression is controlled by NCR-regulated GATA-type transcription factors Gat1 and Gln3 (Dabas and Morschhäuser, 2008). We next assessed a possible link between Hir1-mediated control of proteolytic activities and regulators of SAP2 expression. Notably, the kinetics of STP1 and GAT1 induction was unaffected by HIR1 deletion (Figure 4F; Figure S4F), which is consistent with the RNA-seq data (Figure 2E). However, Stp1 protein abundance was strikingly increased in hir1Δ/Δ cells switching from standard growth (time 0) conditions to protein-rich medium (Figure 4G). In addition to transcriptional regulation, Stp1 activity is regulated via proteolytic processing by the SPS-sensor component Ssy5 in response to amino acids (Andréasson et al., 2006). A minor band of processed and thus, activated, Stp1 was detectable after 2 h of growth on protein in the hir1Δ/Δ mutant (Figure 4G). Stp1 processing was confirmed by treating cultures with arginine and proline to trigger or not the SPS sensor, respectively (Silao et al., 2019) (Figure S4G). This increased protein levels of Stp1 provide a mechanistic link to the hyper-induction of SAP2 and enhanced growth on protein upon loss of HIR1. Likewise, the elevated induction of OPT1 (Figure 2E), another known Stp1 target in C. albicans (Martínez and Ljungdahl, 2005), was confirmed by quantitative real-time PCR (Figure S4H). These results provide further evidence that nitrogen sensing is altered in hir1Δ/Δ cells, which could be mediated by a hyper-excited SPS-sensor system linking enhanced Stp1 activity and proteolytic growth of hir1Δ/Δ cells. As expected, loss of STP1 or SSY5 abrogated the accelerated growth on protein of hir1Δ/Δ cells (Figure 4H; Figure S4I).
Hir1 affects fungal recognition by macrophages in vitro
Fungal metabolic states affect the fungal cell wall and thus impact the interaction with host immunity (Brown et al., 2014). Indeed, HIR1 deletion also affects the transcription of fungal cell-wall organization and biogenesis components (Figure S2C; adjusted p value 0.018). Therefore, we performed GO term analysis focusing on cellular components. Interestingly, loss of HIR1 affected more than 10% of genes associated with cell-wall function or the extracellular region, with the majority of genes having increased chromatin accessibility or upregulated transcription when compared to WT cells (Figures 5A and 5B; Table S7).
Figure 5. Hir1 affects fungal recognition by macrophages in vitro.
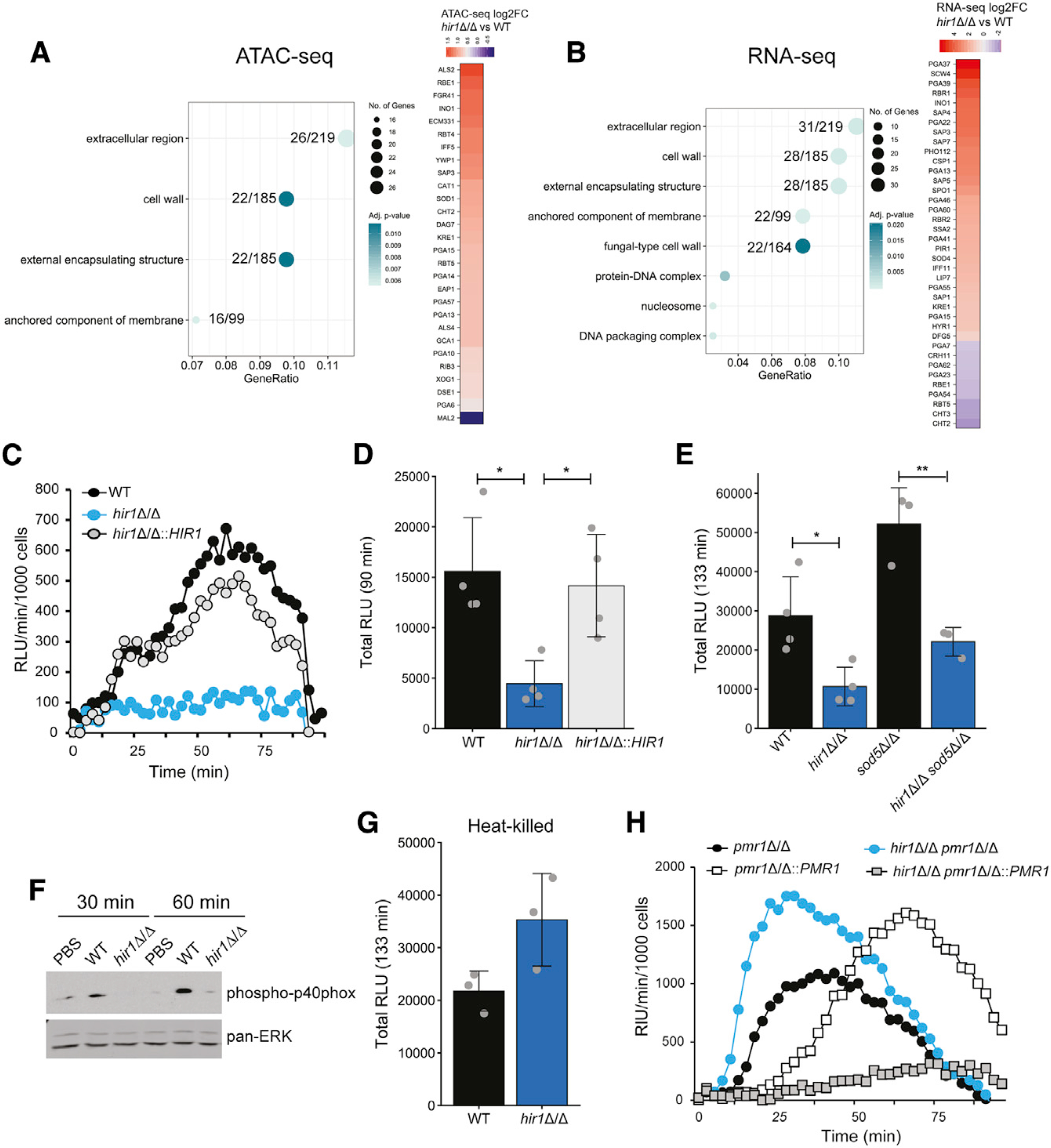
(A and B) GO enrichment analysis of ATAC-seq peaks (A) and RNA-seq signals (B) differentially regulated (FDR <0.05) in hir1Δ/Δ cells during growth in YPD. The GeneRatio denotes the number of genes enriched in the depicted GO term relative to the total number of input genes. The number of genes from the input dataset relative to the total number of genes associated with the plotted GO category is depicted.
(C–E) ROS assay of BMDMs challenged with the indicated strains, depicted as RLU per minute per 1,000 BMDMs over time (C) or as the total RLU after the indicated time (D and E). Graphs show the mean ± SD from 3–4 biological replicates. *p < 0.05, **p < 0.01 with one-way ANOVA followed by Tukey’s multiple comparison test after testing for equal variances (Bartlett’s test) for (D) and with Student’s t test after testing for equal variances (F-test) in (E).
(F) Immunoblot analysis of phosphorylated p40phox in BMDMs challenged with the indicated fungal strains or PBS. Blot is representative of 3 biological replicates.
(G) ROS assay of BMDMs infected with heat-killed C. albicans. Graphs show the mean ± SD from 3 biological replicates.
(H) ROS assay of BMDMs challenged with different fungal genotypes presented as RLU per minute per 1,000 BMDMs over time. Results are representative of 3 biological replicates.
FDR, false discovery rate; BMDMs, bone-marrow-derived macrophages; RLU, relative luciferase units. See also Figure S5.
The fungal cell wall is a major pathogen-associated molecular patterns (PAMPs) and, thus, the first line of contact with immune cells (Gow and Hube, 2012). Therefore, we analyzed interactions between hir1Δ/Δ cells and murine bone-marrow-derived macrophages (BMDMs), which initiate early antifungal responses (Netea et al., 2015). Remarkably, co-cultures of BMDMs and hir1Δ/Δ cells accumulated significantly less total ROS (Figures 5C and 5D; luminol) or extracellular ROS (Figure S5A; isoluminol) than BMDMs challenged with the WT or the revertant strain. Moreover, adding a second pro-inflammatory trigger such as zymosan or trehalose-6,6-dibehenate (TDB) (Gantner et al., 2003; Ishikawa et al., 2009) resulted in ROS accumulation in the presence of hir1Δ/Δ cells (Figures S5B and S5C), suggesting that hir1Δ/Δ cells do not actively impair ROS production in BMDMs facing secondary triggers. Further, deletion of SOD5, a key ROS detoxifier (Frohner et al., 2009), increased ROS in co-cultures with WT and hir1Δ/Δ cells, when compared to the corresponding single knockouts (Figure 5E). However, hir1Δ/Δ sod5Δ/Δ double mutants still triggered significantly less ROS when compared to BMDMs stimulated with the sod5Δ/Δ single mutant (Figure 5E). This implies that additional mechanisms account for impaired ROS responses of BMDMs challenged with hir1Δ/Δ cells. Therefore, we reasoned that an altered cell surface might dampen the oxidative burst mediated by the NADPH-oxidase complex (Nguyen et al., 2017). Indeed, BMDMs challenged by hir1Δ/Δ cells showed decreased activation of the NADPH-oxidase subunit p40phox (Figure 5F), suggesting that HIR1 ablation affects innate fungal recognition.
The outer fungal cell wall is composed of a network of mannans and mannosylated proteins, on top of the middle β-glucan layer that resides on the chitin skeleton. Especially β-glucans trigger pro-inflammatory responses via Dectin-1 signaling (Gow and Hube, 2012), while β-glucan masking by other cell-wall components may facilitate immune evasion (Ballou et al., 2016; Ene et al., 2013). Indeed, ectopic β-glucan exposure after heat-killing (Gantner et al., 2005) or deletion of the Golgi-resident Ca2+/Mn2+ ATPase PMR1 (Bates et al., 2005) fully restored ROS responses by BMDMs when challenged with hir1Δ/Δ cells (Figures 5G and 5H). Of note, exposure of β-glucans was not altered in the hir1Δ/Δ mutant during standard growth (Figures S5D and S5E) and alterations of the chitin content via deletion of the chitin synthase CHS3 (Lenardon et al., 2010) had no effect on ROS release during co-culture of BMDMs with hir1Δ/Δ cells (Figure S5F). Further, transcriptional induction of pro-inflammatory cytokines such as interleukin (IL)-1β and the chemokine CXCL1 in BMDMs stimulated by hir1Δ/Δ cells (Figures S5G and S5H) were similar to co-cultures of BMDMs with WT C. albicans. The same was true for fungal killing by BMDMs (Figure S5I). In summary, these results imply that loss of HIR1 distorts the normal cell-wall architecture otherwise triggering full ROS responses in BMDMs.
Loss of Hir1 alters fungal recognition in vivo and triggers hypervirulence in systemic infections
Deletion of HIR1 alters fungal recognition by BMDMs. Signals from macrophages are key to initiating leukocyte recruitment to infected tissues (Drummond and Lionakis, 2019; Lionakis et al., 2013). Hence, we next assessed the host response to hir1Δ/Δ cells in vivo using an intraperitoneal (i.p.) murine model of acute fungal infections suitable for testing leukocyte recruitment, with neutrophils being the predominant leukocyte population (Zwolanek et al., 2014; Figure S6). Interestingly, we found a decreased number of CD45+ cells in the peritoneum after 4 h of infection with hir1Δ/Δ cells when compared to the WT (Figure 6A). Moreover, we also observed a trend toward impaired neutrophil (Cd11b+ Ly6G+) recruitment, while infiltration of inflammatory monocytes (Cd11b+ Ly6G- Ly6Chi) was unaffected (Figures 6B and 6C). Tissue recruitment of neutrophils is elicited by cytokines and chemokines, such as CXCL1 and CXCL2 or IL-1β produced by tissue-resident immune cells (Drummond et al., 2019; Vonk et al., 2006). Peritoneal cells isolated after 4 h of i.p. infection with hir1Δ/Δ cells showed decreased expression of the CXC-chemokine Cxcl2 and Il1b (Figures 6D and 6E), indicating a differential recognition of hir1Δ/Δ cells by peritoneal immune cells, which may result in impaired leukocyte recruitment.
Figure 6. Deletion of hir1Δ/Δ alters in vivo host responses during acute infection.
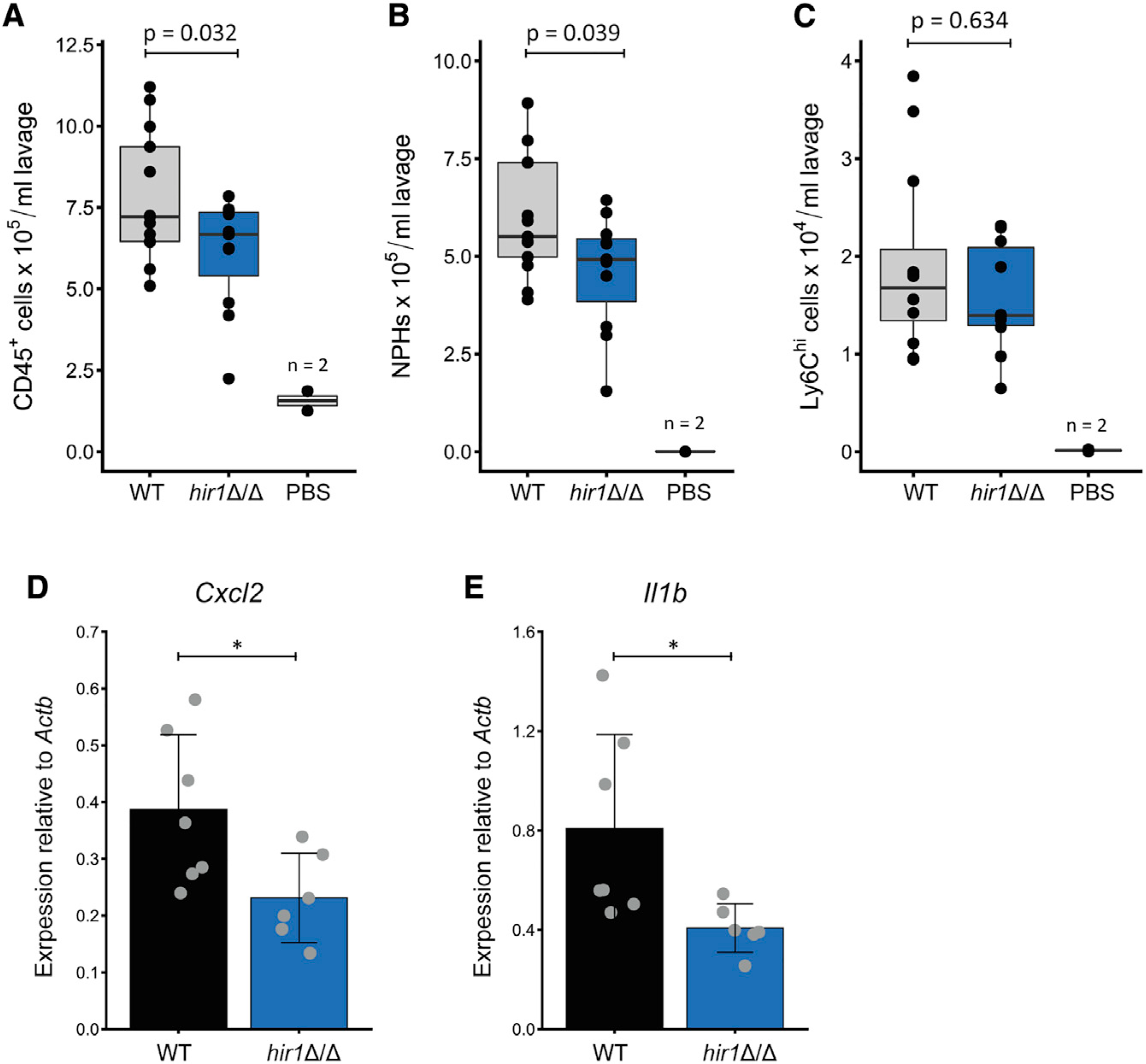
(A–C) Number of CD45+ cells (A), neutrophils (CD11b+ Ly6G+; B), and Cd11b+ Ly6Chi cells (C) in the peritoneum after 4 h of intraperitoneal (i.p.) mouse infection with the indicated fungal strain or PBS. Boxplots indicate median values (horizontal line), first and third quartiles (top and bottom hinges, respectively) with the whiskers extending 1.5-fold of the inter-quartile range. Data represent 13 (WT infected), 11 (hir1Δ/Δ infected), or 2 (PBS) animals pooled from 3 independent experiments. p values are derived from Student’s t test after equal variance testing (F test).
(D and E) Quantitative real-time PCR analysis for Cxcl2 (D) and Il1b (E) expression (relative to Actb [β-actin]) in peritoneal cells after 4 h of i.p. infection. Graphs show the mean value ± SD from 6–7 animals. Data are pooled from 2 independent experiments. *p < 0.05 with Student’s t test after equal variance testing (F test).
NPHs, neutrophils; MPHs, macrophages. See also Figure S6.
C. albicans is also frequently associated with chronic or acute superficial mucosal infections of vulvovaginal tracts or the oral cavity, with the latter manifested in oropharyngeal Candidiasis (OPC) (Pappas et al., 2018). Neutrophil recruitment is a major determinant for limiting fungal outgrowth (Trautwein-Weidner et al., 2015). Therefore, we tested the persistence of the hir1Δ/Δ mutant in an OPC infection model. Although deletion of HIR1 slightly increased fungal burdens in murine tongues after 1 day of infection (Figure S7A), histology indicated robust neutrophil infiltration upon hir1Δ/Δ or gene complemented strain infection (Figures S7B and S7C). Moreover, no differences in gene expression of Il1b, Csf3 (coding for granulocyte colony-stimulation factor [G-CSF]) and Cxcl2 (Figures S7D–S7F), which are implicated in neutrophil recruitment during mucosal C. albicans infections (Altmeier et al., 2016), were observed.
Because of the impaired neutrophil recruitment after intraperitoneal infections, we reasoned that the loss of HIR1 may affect fungal virulence during systemic infections, since early neutrophil recruitment is central to limiting C. albicans outgrowth (Lionakis et al., 2011; Romani et al., 1997). Strikingly, mice intravenously infected with the hir1Δ/Δ mutant showed a dramatic decrease in survival, as also reflected by accelerated loss of body weight when compared to the WT or the revertant strain (Figures 7A and 7B). Importantly, reintegration of HIR1 into its native locus fully abrogated the hypervirulence phenotype. In line with the increased virulence, fungal kidney burdens were approximately 10 times higher after 1 and 3 days of infection with hir1Δ/Δ cells (Figures 7C and 7D), while fungal burdens in spleen, liver, or lung were unaffected (Figures S7G–S7I). During systemic C. albicans infections, the kidney constitutes a main sink for fungal persistence due to a slower infiltration of neutrophils. Spleen and liver limit fungal growth more efficiently due to rapid recruitment of neutrophils and other inflammatory myeloid cells during the initial infection phase (Lionakis et al., 2011; Spellberg et al., 2005). Of note, differences in the transcriptional induction of Cxcl1, Cxcl2 and other pro-inflammatory cytokines (Il1b, Il6) or Icam-1, which enables endothelial leukocyte migration during tissue infiltration (Sadik et al., 2011), were not detected in kidney lysates after 1 day of infection with the WT or hir1Δ/Δ cells (Figures S7J–S7O). Likewise, expression of the kidney injury marker Kim-1 (Huo et al., 2010) was similar after 1 day of infection with WT and hir1Δ/Δ cells (Figure S7O). In addition, murine and human neutrophils mounted impaired ROS responses against hir1Δ/Δ cells (Figures 7E and 7F), without affecting fungal killing (Figure 7G). This suggests that the hypervirulence of hir1Δ/Δ cells is not a consequence of impaired fungal killing by neutrophils. It rather implies that loss of HIR1 alters fungal recognition by innate immune cells, which might compromise neutrophil recruitment. Consequently, fungal growth control upon the onset of acute or systemic infections is impaired.
Figure 7. HIR1-deficient C. albicans is hypervirulent in vivo.
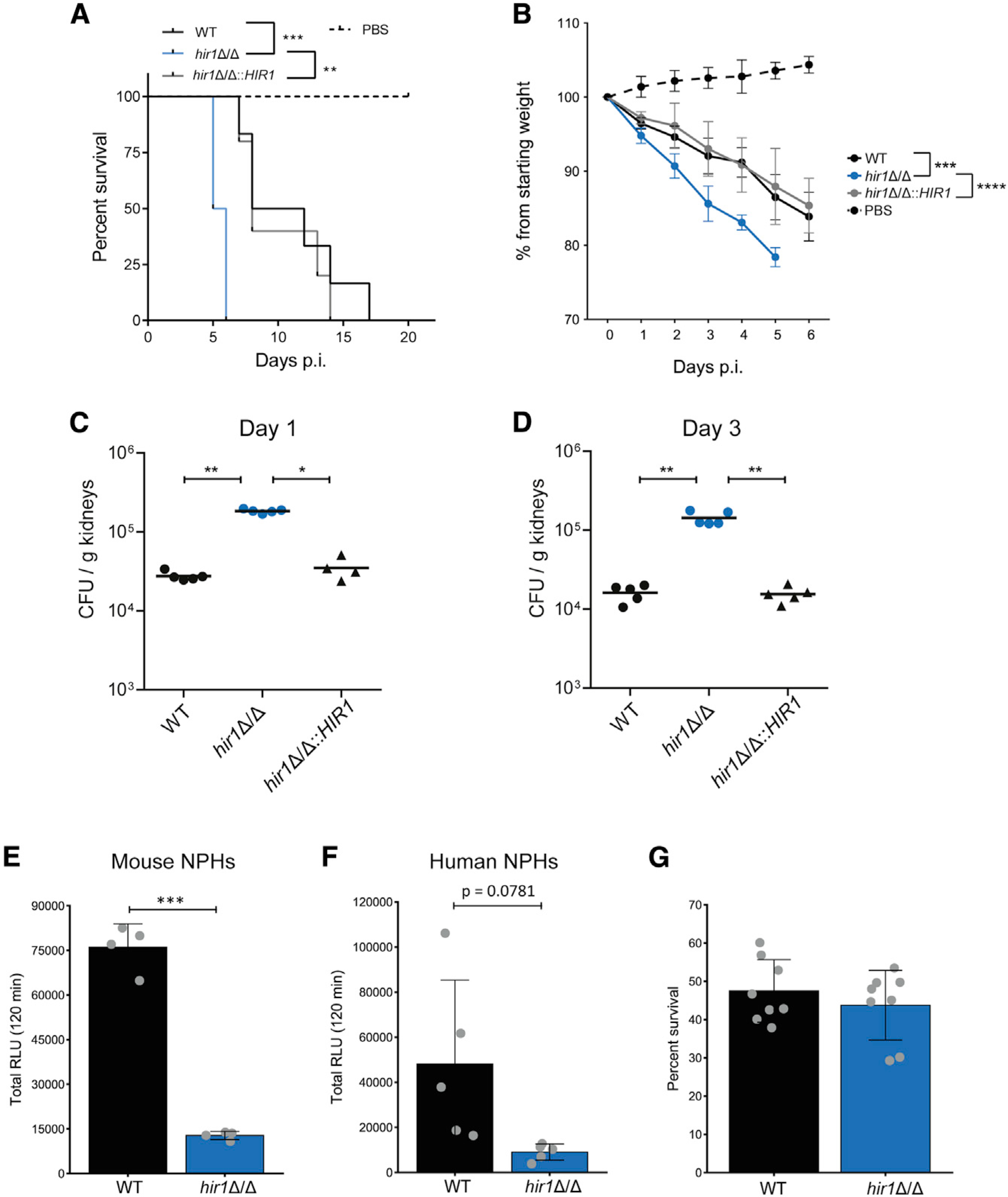
(A) Mouse survival after intravenous (i.v.) challenge with 5–6 mice per fungal infection group and with 2 mice for the PBS control. **p < 0.01, ***p < 0.001 with Mantel Cox log-rank test.
(B. Mouse weights corresponding to the experiment in A. Graph shows the mean ± SD. ***p < 0.001, ****p < 0.0001 with two-way repeated-measures ANOVA followed by Tukey’s multiple comparison test for weight loss within the first 4 days.
(C and D) Fungal burdens in kidneys at day 1 (C) and day 3 (D) post-i.v. infection. Each symbol corresponds to one animal. Horizontal lines indicate the mean from 4–5 animals. *p < 0.05, **p < 0.01 with Mann-Whitney U test).
(E and F) ROS assay of bone-marrow-derived murine neutrophils (E) or human neutrophils (F). Graphs show the mean ± SD from 4 replicates pooled from 2 independent experiments (E) or 5 blood donors (F). ***p < 0.001 with Student’s t test after equal variance testing (F-test).
(G) Fungal survival after 1 h of co-culture with bone marrow-derived murine neutrophils. Graphs show the mean ± SD from 8 replicates pooled from 4 independent experiments.
CFUs, colony forming units; NPHs, neutrophils. See also Figure S7.
DISCUSSION
Histone chaperones fulfill pivotal roles in cellular processes, owing to their essential requirement for chromatin homeostasis. By establishing permissive or repressive chromatin states, histone chaperones control gene expression in a broad sense but also guide context-specific processes such as cell-fate decisions or cellular adaptations (Cheloufi et al., 2015; Chen et al., 2020; Hammond et al., 2017; Li and Jiao, 2017). Here, we show that Hir1 determines chromatin landscapes accessible for transcription of genes that fine-tune responses to nitrogen sources. We propose that these mechanisms control metabolic flexibility in a context-dependent manner, such as fungal commensalism or pathogenic lifestyles. Moreover, HIR controls transcription of fungal cell-wall regulators, which might additionally determine fungal host interaction.
The function of HIR orthologs in transcription control is well established in higher eukaryotes (Hammond et al., 2017). Consequently, loss of Hira alters transcriptional dynamics and chromatin accessibility in different cellular contexts (Chen et al., 2020; Nashun et al., 2015). Of note, the C. albicans HIR complex exploits related mechanisms during fungal transitions from the unicellular yeast state to hyphal growth, whereby HIR1 deficiency alters the amplitudes of hyphal-specific gene expression (Jenull et al., 2017). The lack of fungal HIR might affect the histone supply chain during replication-independent responses, thereby decreasing nucleosome density in target genes and their stochastic activation potential (Flavahan et al., 2017). In line with this, we show that HIR ablation increases chromatin accessibility linked to altered transcriptional adaptations during the switch from growth on preferred nitrogen sources to the alternative source protein, which can be efficiently utilized by C. albicans once the initial adaptation is complete (Staib et al., 2008). Hence, we propose that HIR might act like a genetic valve affecting dynamic ranges of transcription in response to environmental or host immune stimuli by controlling chromatin states in C. albicans. Moreover, we observed that HIR1 loss leads to overly permissive chromatin states, thus offering an explanation for aberrant gene activity, since nucleosomes, and transcriptional regulators may compete for binding to regulatory regions (Klemm et al., 2019).
Despite qualitatively similar responses of WT and hir1Δ/Δ cells, the hyperinduction of SAP2 and related family members explain the increased proteolytic capacity of hir1Δ/Δ cells. Interestingly, STP1 also displayed elevated chromatin accessibility in hir1Δ/Δ cells, irrespective of growth conditions (Figure 3B). Although not coupled to enhanced transcription, this could still be a signature for differential regulation, not detectable at steady-state RNA levels. Indeed, Stp1 protein levels are strikingly increased in hir1Δ/Δ cells during early nitrogen assimilation from protein (Figure 4G), thus providing a mechanistic link for the hyperinduction of target genes, such as SAP2 and OPT1. We suggest that this reflects an altered metabolic state of hir1Δ/Δ cells, swiftly adapting to proteolytic growth on protein. Besides nitrogen-sensing and signaling components, SAP3 was most prominently affected upon loss of HIR1 independent of the available nitrogen sources (Figure 2B), which might cause an increased basal proteolytic state. Hence, peptide degradation products may further increase Stp1 protein stability and thus amplify SAP2 activation via the SPS-sensor system. Indeed, a link exists between proteolytic products and SAP2 regulation (Hube et al., 1994), probably involving the SPS sensor that responds to micromolar amounts of extracellular amino acids (Martínez and Ljungdahl, 2005). On a molecular and mechanistic level, our data demonstrate that these adaptive changes in nitrogen metabolism are tightly linked to altered chromatin landscapes, driving aberrant transcription of regulatory factors mediating metabolic adaptation.
Metabolic flexibility is key pathogen persistence in complex host microenvironments (Alves et al., 2020; Brown et al., 2014). Earlier reports about the requirement of individual SAPs for fungal virulence and tissue invasion have been contradictory (Correia et al., 2010; Lermann and Morschhäuser, 2008; Odds, 2008). The C. albicans SAP family member are regulated in a morphology- and growth phase-dependent manner (Hube et al., 1994). Hence, knockout studies might miss the fine-tuning of spatiotemporal SAP expression, as well as compensatory mechanisms activated in different host-niches. Indeed, SAP family members are expressed in a tissue-dependent manner and their in vivo expression depends on the infection stage and pathogen dosage (De Bernardis et al., 1995; Naglik et al., 1999; Schaller et al., 1998; Staib et al., 2000). Since host organs are generally rich in protein and amino acids (Miramón and Lorenz, 2017), ectopic SAP expression might therefore drive fungal fitness due to increased nutrient scavenging from peptides used as nitrogen and even carbon sources (Miramón et al., 2020; Vylkova et al., 2011). In addition, SAPs may degrade various host-defense molecules such as complement factors (Gropp et al., 2009), but also elicit pro-inflammatory responses, which established Sap2 as a candidate for therapeutic fungal vaccines (Pericolini et al., 2015). Thus, the hypervirulence of hir1Δ/Δ cells might be linked to deregulated SAP expression, which may promote fungal growth in vivo due to enhanced nutrient uptake and possibly drive immune evasion. Indeed, we find increased fungal kidney burdens and impaired neutrophil recruitment to the peritoneal cavity infected by hir1Δ/Δ cells (Figures 7C, 7D and Figures 6A and 6B). Assuming that neutrophil infiltration is potentially attenuated upon kidney invasion by hir1Δ/Δ cells, this offers a plausible explanation for the marked hypervirulence of hir1Δ/Δ cells, as neutrophil recruitment and activation is pivotal for the control of invasive C. albicans infections (Lionakis et al., 2011; Wirnsberger et al., 2014). While the precise contribution of SAP deregulation to Hir1-mediated virulence remains to be dissected, we provide compelling evidence that hir1Δ/Δ C. albicans is differentially recognized by innate phagocytes. This is manifested in a decreased activation of the phagocytic NADPH oxidase complex, an attenuated ROS release by innate phagocytes (Figure 5F and Figures 7E and 7F) upon stimulation with hir1Δ/Δ cells, as well as impaired peritoneal neutrophil recruitment and altered transcription of Il1b and Cxcl2 by peritoneal cells infected with hir1Δ/Δ cells (Figures 6B, 6D, and 6E). Under physiological conditions, neutrophils are usually not present in kidneys (Lionakis et al., 2011) or the peritoneal cavity (Zwolanek et al., 2014). Hence, tissue-resident immune cells release chemoattractant molecules, including CXCL2 receptor (CXCL2R) ligands or IL-1β, among many others, upon pathogen recognition (Drummond and Lionakis, 2019). Noteworthy, recognition of hir1Δ/Δ cells seems unaffected in a mucosal infection model (Figures S7D–S7F), highlighting well-known tissue-specific aspects of fungus-host interactions. Neutrophil recruitment during systemic and mucosal infections requires different effector cells and PRRs (Drummond and Lionakis, 2019; Netea et al., 2015; Swidergall et al., 2018). Interestingly enough, a recent study indicates that the C-type lectin-like receptor Dectin-1, recognizing fungal β-glucan (Brown and Gordon, 2001), is also involved in early neutrophil recruitment to the peritoneal cavity (Thompson et al., 2019). In addition to nitrogen assimilation, HIR controls chromatin accessibility upstream of genes associated with the fungal cell wall (Figures 5A and 5B). This might alter the fungal cell-wall architecture and thereby dampen immune recognition by PRRs (Gelis et al., 2012; Hall and Gow, 2013) as ectopic β-glucan exposure, such as after heat-killing or by inhibiting bulk protein mannosylation (Figures 5G and 5H), restores ROS release by macrophages facing hir1Δ/Δ cells. In line with this, major PRRs required for recognition of heat-killed and live C. albicans may differ (Gow et al., 2007), and the concerted action of distinct PRRs is required for tailored inflammatory responses during systemic fungal infections (Thompson et al., 2019).
In summary, we unequivocally establish a critical role for HIR-mediated chromatin assembly in fungal virulence. The combined action of aberrant transcription of genes shaping the fungal cell wall and those mediating nitrogen sensing or assimilation might act in concert to promote fungal fitness during systemic infections, and simultaneously enable immune evasion. The question arises why C. albicans employs a regulatory complex that restrains systemic fungal virulence and flexible nitrogen assimilation. As this fungus is a component of the microbiome in most healthy individuals (d’Enfert et al., 2020), it is tempting to propose that HIR-mediated chromatin homeostasis and transcriptional control are actually beneficial for the commensal state and prevent triggering strong host inflammatory responses otherwise initiating fungal clearance. This is reminiscent to the differential recognition of C. albicans yeast and hyphal stages at mucosal or epithelial barriers (Moyes et al., 2010). Notably, C. albicans mutants lacking the transcription factors Efg1 or Ume6 show enhanced competitive fitness in the GI tract (Witchley et al., 2019), while virulence is compromised in systemic Candidiasis (Banerjee et al., 2008; Park et al., 2005). Given that chromatin-targeting compounds are promising antifungal drug candidates (Kuchler et al., 2016; Pfaller et al., 2015), their impact on fungal virulence and adverse effects on host response must be carefully assessed. The HIR complex and orthologs cooperate and interact with other histone chaperones or chromatin modifying factors (Ferreira et al., 2011; Pchelintsev et al., 2013). Thus, antifungal strategies targeting chromatin must not interfere with HIR function, as this might lead to detrimental effects for the host. Moreover, in vivo hypervirulence phenotypes of C. albicans deletion mutants have been rarely observed until today. The Candida Genome Database (CGD) lists some 506 unique genes whose genetic ablation is associated with decreased/absent/delayed virulence in various model systems, while only 11 unique genes are linked to hypervirulence phenotypes (for recent reports, see Bahnan et al., 2012; Day et al., 2017; Kakade et al., 2019; Soloviev et al., 2011; Tams et al., 2019). Hence, our study serves as a basis to uncover yet-ill-defined aspects of chromatin biology in fungal pathophysiology, antifungal treatment strategies, and mechanisms of immune evasion.
STAR⋆METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Karl Kuchler (kar.kuchler@meduniwien.ac.at).
Materials availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Karl Kuchler (karl.kuchler@meduniwien.ac.at).
Data and code availability
Data supporting the findings of this manuscript are available from the lead contact upon reasonable request. RNA-seq and ATAC-seq raw data have been deposited at the Gene Expression Omnibus (GEO), mass spectrometry data from SC media supernatants have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD018163. Data are available as of the date of publication.Accession numbers are listed in the key resources table. All original code has been deposited at github and is publicly available as of the date of publication. DOIs are listed in the key resources. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Purified anti-mouse CD16/32 antibody (93) | BioLegend | Cat#101302; RRID:AB_312800 |
| PE anti-mouse F4/80 antibody (BM8) | BioLegend | Cat#123110; RRID:AB_893498 |
| APC/Cy7 anti-mouse/human CD11b antibody (M1/70) | BioLegend | Cat#101226; RRID:AB_830642 |
| Pacific Blue anti-mouse Ly-6C antibody (HK1.4) | BioLegend | Cat#128014; RRID:AB_1732079 |
| PE anti-mouse CD45 antibody (30-F11) | BioLegend | Cat#103105; RRID:AB_312970 |
| FITC anti-mouse Ly-6G antibody (1A8) | BioLegend | Cat#127606; RRID:AB_1236494 |
| Fc(human):Dectin-1 antibody | AdiopenGen | Cat#AG-40B-0138-C050 |
| Alexa Fluor® 488 anti-human IgG Fc (HP6017) | BioLegend | Cat#409322; RRID:AB_2563437 |
| Rabbit anti-phospho-p40phox (Thr154) antibody | Cell Signaling | Cat#4311; RRID: AB_330690 |
| Mouse anti-panERK antibody | BD Biosciences | Cat#610123; RRID:AB_397529 |
| Rat anti-HA antibody, HRP | Roche | Cat#12013819001; RRID:AB_390917 |
| Goat anti-mouse IgG (H+L) secondary antibody, HRP | Thermo Fisher Scientific | Cat#31430; RRID:AB_228307 |
| Mouse anti-β-actin (D6A8) antibody | Abcam | Cat#8224; RRID:AB_449644 |
| Goat anti-rabbit IgG (H+L) secondary antibody, IRDye 800CW | LI-COR | Cat#926–32214; RRID:AB_621846 |
| Goat anti-mouse IgG (H+L) secondary antibody, IRDye 800CW | LI-COR | Cat#926–32210; RRID:AB_621842 |
| Goat anti-rabbit IgG (H+L) secondary antibody, HRP | Cell Signaling | Cat#7074; RRID:AB_2099233 |
| Rabbit anti-Sap2 polyclonal antibody | Kuchler Laboratory | N/A |
|
Chemicals, peptides, and recombinant proteins | ||
| Yeast extract | BD Biosciences | Cat#212720 |
| Peptone | BD Biosciences | Cat#211820 |
| Dulbecco’s Phosphate Buffered Saline | Sigma-Aldrich | Cat#D8537 |
| DMEM | Thermo Fisher Scientific | Cat#11584486 |
| RPMI-1640 | Thermo Fisher Scientific | Cat#21875 |
| Fetal calf serum | Sigma-Aldrich | Cat#F7524 |
| Bovine serum albumin | Sigma-Aldrich | Cat#A2153 |
| Yeast carbon base | Sigma-Aldrich | Cat#Y3627 |
| Yeast nitrogen base withoug ammonium sulfate and aa | BD Biosciences | Cat#233520 |
| Pepstatin A | Sigma-Aldrich | Cat#516481 |
| Zymosan A | Sigma-Aldrich | Cat#Z4250 |
| Trehalose-6,6-dibehenate (TDB) | InvivoGen | Cat#INV-tlrl-tdb |
| RNA/ater™ Stabilization Solution | Thermo Fisher Scientific | Cat#AM7022 |
| Nextera TDE1 | Illumina | Cat#15027865 |
| Complete, EDTA-free Protease Inhibitor Cocktail Tablets | Roche | Cat#04693132001 |
| Zombie Aqua Fixable Viability Dye | BioLegend | Cat#423101 |
| Fixation buffer | BioLegend | Cat#420801 |
|
Critical commercial assays | ||
| MinElute PCR purification kit | QIAGEN | Cat#28006 |
| RNeasy MinElute Cleanup kit | QIAGEN | Cat#74204 |
|
Deposited data | ||
| RNA-seq raw data | This study | GEO: GSE157411 |
| ATAC-seq raw data | This study | GEO: GSE157411 |
| Mass-spectrometry data from SC medium culture supernatants | This study | PRIDE: PXD018163 |
|
Experimental models: Primary cells | ||
| Murine bone marrow derived macrophages | This study | N/A |
| Murine bone marrow derived neutrophils | This study | N/A |
|
Experimental models: Organisms/strains | ||
| C57BL/6J mice | Jackson Laboratory | Cat#000664; RRID:IMSR_JAX:000664 |
| C. albicans: wild type (WT) | (Gillum et al., 1984) | N/A |
| C. albicans: Clinical isolate 1 | Birgit Willinger | AKH No. 2284 |
| C. albicans: Clinical isolate 10 | Birgit Willinger | AKH No. 2275 |
| C. albicans: hir1Δ/Δ | (Tscherner et al., 2015) | CASJ019 |
| C. albicans: hir1 Δ/Δ::HIR1 | (Jenull et al., 2017) | CASJ013 |
| C. albicans: Clinical isolate 1 hir1Δ/Δ | This study | CASJ255 |
| C. albicans: Clinical isolate 10 hir1 Δ/Δ | This study | CASJ252 |
| C. albicans: hat1 Δ/Δ | (Tscherner et al., 2015) | CA-MT014 |
| C. albicans: cac2Δ/Δ | (Tscherner et al., 2015) | CA-MT363 |
| C. albicans: sap2Δ/Δ | (Staib et al., 2008) | SAP2MS4A |
| C. albicans: sap2Δ/Δ hir1 Δ/Δ | This study | CASJ261 |
| C. albicans: sap3Δ/Δ | This study | CASJ301 |
| C. albicans: hir1 Δ/Δ sap3Δ/Δ | This study | CASJ304 |
| C. albicans: sap2Δ/Δ sap3Δ/Δ | This study | CASJ308 |
| C. albicans: sap2Δ/Δ sap3Δ/Δ hir1 Δ/Δ | This study | CASJ310 |
| C. albicans: stpl Δ/Δ | This study | CASJ403 |
| C. albicans: hir1 Δ/Δ stp1Δ/Δ | This study | CASJ405 |
| C. albicans: ssy5Δ/Δ | (Miramon and Lorenz, 2016) | CaPM23 |
| C. albicans: ssy5Δ/Δ hir1 Δ/Δ | This study | CASJ420 |
| C. albicans: pmr1 Δ/Δ | This study | CASJ224 |
| C. albicans: hir1 Δ/Δ pmr1 Δ/Δ | This study | CASJ226 |
| C. albicans: pmr1 Δ/Δ::PMR1 | This study | CASJ239 |
| C. albicans: hir1 Δ/Δ pmr1Δ/Δ::PMR1 | This study | CASJ241 |
| C. albicans: chs3Δ/Δ | This study | CA-MT611 |
| C. albicans: chs3Δ/Δ hir1Δ/Δ | This study | CASJ262 |
|
Oligonucleotides | ||
| See Table S1 for full oligonucleotide list | N/A | |
|
Recombinant DNA | ||
| Plasmid: pSFS3b | (Tscherner et al., 2012) | N/A |
| Plasmid: YEp352 | (Hill et al., 1986) | N/A |
| Plasmid: pFA6a-3xHA-SAT1-FLP | This study | pFA6a-3HA-SAT 1-FLP |
| Plasmid: YEp-SAP3urdr-FLP-NAT1 | This study | ECSJ071 |
| Plasmid: YEp-PMR1urdr-FLP-NAT1 | This study | ECSJ031 |
| Plasmid: YEp-PMR1 reint-FLP-NATI | This study | ECSJ082 |
| Plasmid: YEp-CHS3urdr-FLP-NAT1 | This study | YEp352-NAT1-CHS3urdr |
| Plasmid: Yep-STP1urdr-FLP-NAT1 | This study | ECSJ075 |
|
Software and algorithms | ||
| fastQC v v0.11.8 | (Andrews, 2010) | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| cutadapt v1.18 | https://cutadapt.readthedocs.io/en/stable/ | |
| NextGenMap v0.5.5 | (Sedlazeck et al., 2013) | https://cibiv.github.io/NextGenMap/ |
| Picard | Broad Institute | http://broadinstitute.github.io/picard/ |
| BEDTools | https://github.com/arq5x/bedtools2 | |
| HTseq | (Anders et al., 2015) | https://htseq.readthedocs.io/en/master/ |
| edgeR | (Robinson et al., 2010) | https://www.bioconductor.org/packages/release/bioc/html/edgeR.html |
| MACS2 v2.1.2 | (Zhang et al., 2008) | https://github.com/macs3-project/MACS |
| ChIPseeker | (Yu et al., 2015) | http://bioconductor.org/packages/release/bioc/html/ChIPseeker.html |
| deepTools2 | (Ramírez et al., 2016) | https://deeptools.readthedocs.io/en/develop/ |
| clusterProfiler | (Yu et al., 2012) | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| MaxQuant v1.6.2.6 | (Cox and Mann, 2008) | http://www.maxquant.org/ |
| ProteinPilot v5.0.1 | SCIEX | https://sciex.com/products/software/proteinpilot-software |
| Peakview v2.2 | SCIEX | https://sciex.com/products/software/peakview-software |
| MSStats | (Choi et al., 2014) | https://www.bioconductor.org/packages/release/bioc/html/MSstats.html |
| R v3.6.1 | R Core Team 2019 | https://www.R-project.org/ |
| RStudio v1.3.1093 | RStudio Team 2020 | https://www.rstudio.com/ |
| FlowJo software version 7.6.5 and 10.7.1 | FlowJo | N/A |
| Realplex software 2.0 | Eppendorf | N/A |
| GraphPad Prism version 6.0 | GraphPad | N/A |
| RNA-seq analysis script | This study | https://github.com/tschemic/RNAseq_analysis |
| ATAC-seq analysis script | (Jenull et al., 2020) | https://github.com/tschemic/ATACseq_analysis |
|
Deposited raw data | ||
| RNA-seq raw data | This study | GSE157411 (GSE157599 SuperSeries) |
| ATAC-seq raw data | This study | GSE157568 (GSE157599 SuperSeries) |
| Mass spectrometry raw data (SC culture supernatants) | This study | PXD018163 |
|
Other | ||
| RNA 6000 Nano Kit | Agilent | Cat#5067–1511 |
| High Sensitivity DNA Kit | Agilent | Cat# 5067–4626 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics statement
All animal experiments were evaluated by the ethics committee of the Medical University of Vienna and approved by the Federal Ministry of Science and Research, Vienna, Austria (bmwfw-68.205/0212-WF/V/3b/2016 and BMBWF-66.009/0436-V/3b/2019) or by the Veterinary office of the Canton Zurich, Switzerland (license number 166/2018) in accordance with the guidelines of the Swiss Animals Protection Law.
Intravenous mouse infection model
C57BL/6J mice were housed under specific pathogen-free conditions in the animal facility of the Max Perutz Labs Vienna. Mice breeding and maintenance was in accordance with ethical animal license protocols complying with the current Austrian law. For mice infections, C. albicans strains were grown overnight in YPD at 30°C shaking at 200 rpm to an OD600 between 1–2 and washed twice with PBS (Sigma-Aldrich). The final cell pellet was resuspended in phosphate buffered saline (PBS; Sigma-Aldrich) and fungal cells were quantified on a CASY cell counter (Roche). Fungal cells were then adjusted to the required infection concentration in PBS (Sigma-Aldrich). An aliquot was plated on YPD agar plates to assess the accuracy of the infection dosage. For the disseminated Candidiasis model, 1×105 fungal cells per 21.5 g mouse (male, 8–12 weeks old) in 100 μL were injected intravenously (i.v.) into the lateral tail vein of. Infected mice were monitored daily and sacrificed by cervical dislocation at the indicated time point. For survival experiments, murine body weight was monitored daily for the indicated time.
To assess the fungal burden, kidney homogenates were processed using an Ika T10 Basic Ultra-Turrax homogenizer (Ika, Staufen) as described previously (Riedelberger et al., 2020) and the remaining homogenate was stored at −80°C in aliquots. Spleen, liver, and lung fungal burdens were assessed at day 1 post i.v. infection as described earlier (Riedelberger et al., 2020). Gene expression analysis from whole kidneys was done from i.v. infected animals including a PBS-injected control. After 1 day post i.v. infection, both kidneys were collected and stored in RNAlater™ solution (ThermoFisher Scientific). Kidney RNA was isolated by homogenizing the organ in TRI reagent (LabConsulting) using an Ika T10 Basic Ultra-Turrax homogenizer. 1 mL of kidney lysate was used for RNA isolation, cDNA synthesis and qPCR analysis as described below in the method details section.
Intraperitoneal mouse infection model
For intraperitoneal infection, C. albicans cultures were prepared as described above and 1×107 fungal cells in 250 μL PBS (Sigma-Aldrich) were injected intraperitoneally (i.p.) in 8–12 weeks C57BL/6J mice. To determine leukocyte recruitment, female mice were used. After 4 hours post i.p. infection, the peritoneum was lavaged twice with 8 mL sterile PBS (Sigma-Aldrich) containing 10 mM EDTA. Peritoneal cells were collected (300 × g, 6 minutes, 4°C) and red blood cells (RBC) were lysed with RBC lysis buffer (0.01 M Tris-HCl buffer pH = 7.0 containing 8.3 g/l NH4Cl; all Sigma-Aldrich) for 2 minutes at room temperature. After the addition of 10 mL PBS, cells were harvested again and resuspended in 1 mL PBS. Cells (> 6 μm) were quantified using a CASY cell counter (Roche) and all cells were subjected to live/dead staining using the Zombie Aqua™ Fixable dye (BioLegend) with simultaneous blocking of CD32/CD16 (Biolegend) for 15 minutes at room temperature in PBS. Cells were then washed twice with PBS/2% BSA, fixed for 10 minutes at room temperature with fixation buffer (BioLegend), washed twice with PBS and stored overnight at 4°C in PBS. Leukocyte populations were then determined by staining 2 × 106 cells with the following panel in PBS/2% BSA: PE-labeled anti-CD45 (30-F11, BioLegend), APC-Cy7-labeled anti- CD11b (M1/70, BioLegend), FITC-labeled anti-Ly6G (1A8, BioLegend), Pacific blue-labeled anti-Ly6C (HK1.4, BioLegend) and APC-labeled anti-F4/80 antibody (BM8, BioLegend). Samples were measured on a LSRFortessa (BD Biosciences) cytometer including unstained and single stained controls. Flow cytometry data were analyzed using FlowJo (Flowjo LLC, version 10.7.1.). For gene expression analysis from peritoneal cells, male and female mice were used, which were distributed equally among the experimental groups. Peritoneal cells were harvested after 4 hours post i.p. infection by lavage as described above. Collected lavage fluid was centrifuged at 300 × g for 6 minutes at 4°C, cell pellets were resuspended in 1 mL TRI reagent (LabConsulting) and stored at −80°C until further processed. RNA extraction, cDNA synthesis and qPCR analysis were done as described for BMDMs in the Method Details section.
Oropharyngeal Candidiasis infection model
Female mice were infected sublingually with 2.5 × 106 C. albicans yeast cells as described (Solis and Filler, 2012), without immunosuppression. For determination of the fungal burden, the tongue of euthanized animals was removed, homogenized in sterile 0.05% NP40 in dH2O for 3 min at 25 Hz using a Tissue Lyzer (QIAGEN) and serial dilutions were plated on YPD agar containing 100 μg/ml Ampicillin. For histology, tissue was fixed in 4% PBS-buffered paraformaldehyde overnight and embedded in paraffin. Sagittal sections (9μm) were stained with Periodic-acidic Schiff (PAS) reagent, counterstained with Hematoxylin, and mounted with Pertex (Biosystem) according to standard protocols. Images were acquired with a digital slide scanner (NanoZoomer 2.0-HT, Hamamatsu) and analyzed with NDP.view2. Isolation of total RNA from bulk tongues was carried out according to standard protocols using TRI Reagent (Sigma Aldrich). cDNA was generated by RevertAid reverse transcriptase (ThermoFisher Scientific) and qPCR was performed using SYBR Green (Roche) on a QuantStudio 7 Flex (Life Technologies) instrument. All qRT-PCR assays were performed in duplicate and the relative expression (rel. expr.) of each gene was determined after normalization to β-actin transcript levels.
Human whole blood
Human whole blood samples were drawn from healthy adult female volunteers (24–30 years old) with informed consent and were generously provided by Ernst Müllner (Medical University of Vienna). Whole blood samples were collected into heparin tubes (Greiner-Bio) in accordance to the protocol approved by the Ethics Committee of the Medical University of Vienna (1043/2015) and registered at https://www.clinicaltrials.gov/ (NCT02639780).
Bone marrow-derived immune cells and human neutrophils experiments
Bone marrow isolation from C57BL/6J mice and differentiation of bone marrow-derived macrophages (BMDMs) was done essentially as described earlier (Riedelberger et al., 2020). Neutrophils were isolated from murine tibias and femurs using a Percoll-gradient (GE Healthcare) (Majer et al., 2012). Human neutrophils were isolated from peripheral whole blood of healthy volunteers by double density gradient centrifugation using Histopaque-1119 and -1077 (both Sigma-Aldrich). Blood was centrifuged at 1000 × g for 10 minutes at room temperature and plasma was carefully aspirated. The remaining sample was filled up with Hank’s balanced salt solution (HBSS; without Mg2+ and Ca2+, GIBCO) supplemented with 4% heat-inactivated (hi) FCS to the original volume (4 ml), slowly layered on the Histopaque double gradient and centrifuged at 800 × g for 35 minutes at room temperature. Neutrophils were collected from the Histopaque-1077 and -1119 interphase and diluted with HBSS supplemented with 4% hiFCS. After centrifugation at 300 × g for 10 minutes, red blood cells (RBC) were lysed with RBC lysis buffer (0.01 M Tris-HCl buffer pH 7.0 containing 8.3 g/l NH4Cl; all Sigma-Aldrich) for 1 minute at room temperature. After the addition of 10 mL HBSS/4% hiFCS, cells were harvested at 300 × g for 10 minutes, resuspended in HBSS/4% hiFCS and immediately used for ROS assays after cell counting on a CASY cell counter (Roche).
For co-culture experiments, C. albicans was grown in YPD at 30°C shaking at 200 rpm to the logarithmic growth phase and washed two times with phosphate buffered saline (PBS). Leukocytes were either challenged with live C. albicans (at the indicated multiplicity of infection [MOI]), heat-killed C. albicans, 75 μg Zymosan (Sigma-Aldrich) or 1 μg Trehalose-6,6-dibehenate (TDB; Invivogen). Heat-killing was done by incubating fungal cells at 70°C for 10 minutes. Loss of fungal viability was confirmed by plating an aliquot of heat-killed cells on YPD agar medium.
METHOD DETAILS
Media and fungal growth conditions
C. albicans was routinely grown on YPD medium (1% yeast extract, 2% peptone and 2% glucose [all BD Biosciences]) at 30°C with 200 rpm shaking. For solid medium, 2% Bacto agar (BD Biosciences) was added. Synthetic complete (SC; 1.7 g/L yeast nitrogen base without amino acids and ammonium sulfate [BD Biosciences], 5 g/L ammonium sulfate [Sigma-Aldrich], amino acid mix and 2% glucose [all BD Biosciences]) medium was prepared as previously described (Kaiser et al., 1994). YCB-BSA medium was composed of 23.4 g/L yeast carbon base (Sigma-Aldrich), adjusted with HCl to pH 4 and 1 g/L (liquid formulation) or 5 g/L (solid medium) BSA (Sigma-Aldrich). YNB-BSA medium was composed of 1.7 g/L yeast nitrogen base without amino acids and ammonium sulfate (BD Biosciences), 1% glucose (BD Biosciences) and, 1 g/L BSA (Sigma-Aldrich). For solid medium, 2% Bacto agar (BD Biosciences) was added.
To test growth on BSA as the major nitrogen source in liquid medium, strains were grown overnight in YPD, washed twice in distilled water (dH2O) and diluted in YCB-BSA medium or in YNB-BSA medium to a final OD600 of 0.5 and further grown at 30°C for the indicated time. To generate growth curves in YCB-BSA or YNB-BSA media the starting OD600 was 0.2. Growth of C. albicans on solid YCB-BSA medium or SC medium supplemented with 10% FCS (Sigma-Aldrich) was tested as described above, but cells were counted on a CASY cell counter (Roche) after washing with dH2O. Cells were diluted to 2×105 cells per ml and 5 μL were spotted on YCB-BSA medium supplemented with 2% Bacto agar (BD Biosciences). Images were taken after indicated time at 30°C. Where specified, YCB-BSA and YNB-BSA media were supplemented with 20 mM ammonium sulfate, 1 μM Pepstatin A or 0.01% sodium azide (all Sigma-Aldrich).
Plasmid and fungal strain constructions
Fungal strains and plasmids used in this study are listed in the Key resources table. All C. albicans strains were derived from the MTL a/α clinical isolate SC5314 (Gillum et al., 1984).
The construction of the HIR1 mutant (Tscherner et al., 2015), the HIR1 complemented strain (Jenull et al., 2017), the SOD5 (Frohner et al., 2009), SSY5 (Miramón and Lorenz, 2016) and SAP2 (Staib et al., 2008) deletion strains have been described earlier. The plasmids for SAP3, STP1, CHS3, and PMR1 deletion were created by the fusion of around 500 bp of up- and downstream sequence from the target ORF with a FRT-FLP-NAT1-FRT fragment from pSFS3b (Tscherner et al., 2012) and the YEp352 (Hill et al., 1986) backbone containing an ampicillin resistance marker and an E. coli origin of replication. These fragments were fused together using a Gibson assembly approach (Torella et al., 2014) for PMR1, SAP3, STP1 deletion plasmids and an in vivo cloning approach using the E. coli strain EL350 (Lee et al., 2001) for CHS3 gene targeting. The plasmid for the gene complementation of PMR1 was done via PCR amplifying the PMR1 ORF including 500 bp upstream region and fusing it with the FRT-FLP-NAT1-FRT fragment, the PMR1 downstream region and the YEp352 backbone like for the PMR1 deletion. For C-terminal HA tagging of Stp1, an approximately 4.8 kB PCR cassette was amplified from plasmid pFA6a-3xHA- FLP-SAT1 with primers containing 94 and 99 bp overhangs homologous to the STP1 up- and downstream region, respectively. The plasmid pFA6a-3xHA-FLP-SAT1 was derived from pFA6a-3HA-His3MX6 (Longtine et al., 1998) by exchanging the His3MX6 marker with the SAT1 marker and the Flp recombinase derived from pSFS2a (Reuss et al., 2004) after digestion with ApaI and SacI (FastDigest, ThermoFischer Scientific). Transformation of C. albicans was done by the lithium acetate method and electroporation (Reuss et al., 2004).
Correct genomic integration of the deletion constructs, gene complementation cassettes, tagging cassettes and the loss of the target gene were verified via colony PCR as described earlier (Tscherner et al., 2015). Briefly, colony material was resuspended in 40 μL dH2O and incubated 10 minutes at 95°C. Cell debris was spun down and 5 μL of the supernatant was used for the PCR reaction. The PCR was carried out with the DreamTaq Green DNA Polymerase (ThermoFisher Scientific) according to the manufacturer’s instructions. See Table S1 for all oligos used in this study.
Azocasein assay
Proteolytic activity from culture supernatants can be quantified using the azocasein assay, whereby azo dye-labeled casein serves as a proteolytic substrate. Azo-labeled peptides are then liberated via proteolytic activity, which remain soluble after protein precipitation with trichloracetic acid allowing for simple photometric quantification (Coêlho et al., 2016). Azocasein (Sigma-Aldrich) assays were performed form supernatants of YCB-BSA cultures grown for 24 hours at 30°C. Azocasein stocks (25 mg/ml in PBS) were centrifuged for 10 minutes at 21,000 × g at room temperature. The supernatant was removed and the pellet was resuspended in 1 mL PBS, which is designated as the “washed” azocasein stock. For the assay, washed azocasein was diluted to 5 mg/ml in 50 mM sodium acetate buffer (pH 4). A 100 μL aliquot of diluted azocasein was mixed with an equal volume of cell-free culture supernatants and incubated for 2 hours at 30°C. Intact azocasein was precipitated by adding 20 μL of 50% trichloroacetic acid (TCA) and incubation for 10 minutes on ice. The precipitate was harvested at 18,800 × g for 10 minutes at 4°C and 100 μL of the supernatant was transferred to a 96-well microtiter plate (Starlab) and mixed with 50 μL 2 M sodium hydroxide (Sigma-Aldrich). After 1 minute incubation, the absorbance was measured at 450 nm on a Victor Nivo plate reader (PerkinElmer).
RNA isolation, cDNA synthesis and qPCR
RNA extraction and cDNA synthesis from fungal cells was done using TRI reagent (LabConsulting) exactly as described previously (Tscherner et al., 2012). For RNA isolation from BMDMs challenged with C. albicans at a MOI of 5:1 (fungi to BMDMs), 1 mL of TRI reagent was added per 35 mm dish (containing 1×106 BMDMs), collected using a cell scraper (Starlab) and transferred into 1.5 mL tube on ice. After centrifugation at 14,000 × g for 15 min at 4°C, the supernatant was transferred into a fresh tube containing 200 μl chloroform (Sigma-Aldrich). Further steps and cDNA synthesis were done exactly as for fungal cells (Tscherner et al., 2012).
Gene transcription analysis from cDNA samples was done by quantitative real-time PCR (qPCR) using the Luna Universal qPCR Master Mix (New England Biolabs). Amplification curves were analyzed using the Realplex Software (Eppendorf) and relative quantification of qPCR products was done using the efficiency corrected ΔΔCt method (Pfaffl, 2001). RIP1 was used as a reference gene for fungal gene transcription analysis (Hnisz et al., 2010) and Actb for BMDMs, peritoneal cells and kidney samples. See Table S1 for all oligos used in this study.
RNA-seq sample preparation and bioinformatic analysis
WT and hir1Δ/Δ cells were grown for approximately 16 hours in YPD at 30°C shaking at 200 rpm and then prepared for YCB-BSA medium inoculation as described above. After 4 and 8 hours of culture at 30°C shaking at 200 rpm, cells were harvested at 1,500 × g for 3 minutes at 4°C. The cell pellet was washed 1x with ice-cold dH2O and then immediately snap-frozen and stored at −80°C until further processed. Additionally, an aliquot from the YPD culture was included in the sample preparation workflow as time point 0 control. Fungal RNA for RNA-seq analysis was isolated as described above, except that RNA was cleaned up after DNase treatment using the RNeasy MiniElute Cleanup kit (QIAGEN). Quality of DNase-treated RNA was assessed on a Bioanalyzer using a RNA 6000 Nano chip (Agilent). Removal of ribosomal RNA, library preparation and sequencing was done at the VBCF NGS Unit (https://www.viennabiocenter.org/facilities/next-generation-sequencing) in Vienna, Austria. Samples were sequenced in a 50 bp single-end read mode on a HiSeq4000 instrument. For all conditions, three biological replicates were sequenced.
Quality control of raw sequencing reads was done using fastQC v0.11.8 (Andrews, 2010). TrueSeq (Illumina) adapters were trimmed using cutadapt v1.18 (https://cutadapt.readthedocs.io/en/stable/; settings: -q 30 -O 1) and the reads were mapped onto the C. albicans SC5314 genome (assembly 22, version A22 s07-m01-r88; http://www.candidagenome.org/) using NextGenMap v0.5.5 (Sedlazeck et al., 2013) (settings: -b -Q 30). Optical read duplicates were removed using Picard tools (Broad Institute, https://broadinstitute.github.io/picard/, settings: MarkDuplicates REMOVE_DUPLICATES = true VALIDATION_STRINGENCY = LENIENT; Broad Institute) and ribosomal reads were removed using the ‘intersect’ tool from BEDTools with –v settings (https://github.com/arq5x/bedtools2). Read counting was done using HTseq (Anders et al., 2015) using the union mode and the genomic annotation from C. albicans assembly 22 (version A22 s07-m01-r88; settings: -f bam -t gene -i ID). Differential gene expression analysis was done using edgeR (Robinson et al., 2010). The false discovery rate (FDR) represents p values adjusted for multiple testing with the Benjamini-Hochberg method (Benjamini and Hochberg, 1995). Cut-off for significant differential expression was set to a maximum FDR of 0.05. GO term enrichment analysis was performed using the ‘enrichGO’ function from the clusterProfiler package (Yu et al., 2012). For visualization of RNA-seq data with the Integrative Genomics Viewer (IGV; (Robinson et al., 2011)), normalized read coverage files (bigwig) were created using the deepTools2 ‘bamCoverage’ function ((Ramírez et al., 2016); settings: –bs 5–normalizeUsing CPM). To display the mean normalized read coverage from three biological replicates, bigwig files from each single replicate were merged using a custom script, which additionally normalizes for library size differences. Table S2 presents the edgeR analysis result of the RNA-seq data.
ATAC-seq library preparation and bioinformatic analysis
Culture conditions for ATAC-seq analysis were essentially as for RNA-seq. ATAC-seq sample preparation was based on a previously published protocol (Schep et al., 2015) and was carried out as described earlier (Jenull et al., 2020). Briefly, spheroplasts were generated by treatment with 2 mg/ml Zymolase 100T (Sigma-Aldrich). To control for the sequence bias of the Tn5 transposase (TDE1) (Goryshin et al., 1998), genomic DNA (gDNA) was isolated from YPD-grown WT and hir1Δ/Δ cells as described previously (Jenull et al., 2020) and included in the sample preparation workflow. For tagmentation, 5×106 spheroplasts or 0.5 ng of naked gDNA were resuspended in the tagmentation reaction mix (12.50 μL Nextera 2xTD buffer, 2.00 μL Nextera TDE1 [all Illumina], 1 μL 25x protease inhibitor cocktail [Roche], 0.25 μL 1% Digitonin [New England Biolabs] and 9.75 μL nuclease-free dH2O [ThermoFisher Scientific]) and incubated at 37°C for 30 minutes. The tagmentation reaction was immediately purified using a QIAGEN MiniElute PCR purification kit and elution was done in 12 μL elution buffer provided by the kit. PCR amplification and size selection of ATAC-seq libraries was done as described earlier (Jenull et al., 2020). The quality of purified libraries was analyzed on a Bioanalyzer using a High Sensitivity DNA chip (Agilent) by following the manufacturer’s instructions. ATAC-seq libraries from WT and the hir1Δ/Δ mutant displayed a typical nucleosomal organization (Buenrostro et al., 2013), which was not observed in gDNA samples (Figure S3A). For sequencing, ATAC-seq libraries were prepared from three biological replicates for each condition and pooled in equimolar ratios. Sequencing was done in 75 bp paired-end read mode on a HiSeq 3000/4000 system at the Biomedical Sequencing Facility (BSF; https://cemm.at/research/facilities/biomedical-sequencing-facility-bsf/) at the Center of Molecular Medicine (CeMM) in Vienna, Austria.
Quality control of raw sequencing reads was done using fastQC v0.11.8 (Andrews, 2010). Adaptor trimming from sequencing reads was done with cutadapt v1.18 (https://cutadapt.readthedocs.io/en/stable/; settings:–interleaved -q 30 -O 1). Reads were aligned to the haploid C. albicans SC5314 genome (assembly 22, version A22 s07-m01-r88; http://www.candidagenome.org/) using NextGenMap v0.5.5 (Sedlazeck et al., 2013) (settings: -b -p -Q 30). Optical read duplicates were removed with Picard tools (Broad Institute, https://broadinstitute.github.io/picard/, settings: MarkDuplicates REMOVE_DUPLICATES = true VALIDATION_STRINGENCY = LENIENT; Broad Institute, https://broadinstitute.github.io/picard/) and mitochondrial reads were removed using the ‘intersect’ tool from BEDTools with –v settings (https://github.com/arq5x/bedtools2). Aligned bam files were further split according to the fragment lengths of the sequencing read pairs as done previously (Buenrostro et al., 2013). Read fragments below 100 bp were considered originating from nucleosome-free regions and were used for further analysis. Normalized read coverage files (bigwig) of nucleosome-free read fragments were created using the deepTools2 ‘bamCoverage’ function ((Ramírez et al., 2016); settings: -e -bs 5 –normalizeUsing CPM) and visualized using the IGV (Robinson et al., 2011). Coverage plots of nucleosome-free ATAC-seq reads across the indicated gene subsets were created by first computing a read coverage matrix using the deepTools2 ‘computeMatrix’ function ((Ramírez et al., 2016); settings: reference-point –a 1000 –b 1000 –bs 5) of merged bigwig files. The tabular output was used to plot the normalized read coverage with ggplot2 (Wickham, 2016) in RStudio (R version 3.6.1; (RStudio-Team, 2016)).
Peak-calling for each individual sample was done with MACS2 v2.1.2 using the ‘callpeak’ function ((Zhang et al., 2008), settings: -f BAMPE -g 14521502) using the gDNA sample as background control. Peaks from all samples and replicates were merged and converted to the gff format for read counting using HTseq ((Anders et al., 2015); settings: -f bam -s no -t peak). Peak annotation was done using the ChIPseeker Bioconductor package (Yu et al., 2015) with the’annotatePeak’ function (options: tssRegion = c(−2000, 0), genomicAnnotationPriority = c(“Promoter,” “5UTR,” “3UTR,” “Exon,” “Intron,” “Downstream,” “Intergenic”)). Differential accessible peak detection was done using the edgeR package (Robinson et al., 2010) with the generalized linear model (glm) approach. P values, adjusted with the Benjamini-Hochberg method (Benjamini and Hochberg, 1995), are presented as FDR. Table S4 contains the edgeR analysis result. GO term analysis of genes with significantly differential peak signal in upstream regions (max. 2 kb upstream of the TSS) in hir1Δ/Δ cells was done using the ‘enrichGO’ function from the clusterProfiler package (Yu et al., 2012).
Mass Spectrometry analysis of culture supernatants
Cell-free supernatants from 16 hours YNB-BSA (0.025% BSA) cultures grown at 30°C were used for Mass-Spec analysis. Collected supernatants were lyophilized and dissolved in 400 μl of water for Albuvoid treatment for albumin depletion (Zheng et al., 2015). Albumin-free enriched secretory proteome was eluted from beads in two sequential reactions using 200 μl of AlbuVoid elution buffer (AVEB), vortexing for 10 min, and centrifugating for 4 min at 10,000 rpm. For LC-MS sample preparation, an in-gel digestion protocol was applied. Briefly, 400 μl of eluted sample were mixed with 400 μl of 2x Laemmli buffer (LAEMMLI, 1970) and 40 μl of 1 M dithiothreitol (DTT), heated for 5 min at 95°C, followed by the addition of urea to a final concentration of 8 M. The sample was then filtered with a 10 kDa filter (Amicon Ultra-15 Centrifugal Filter Unit), 50 μl sample was attained, washed with 400 μl of 8 M urea and separated with sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel. Electrophoresis was conducted until prestained standards completely entered the gel. Gels were fixed, stained with Coomassie blue and excised gel slices were digested with sequencing-grade trypsin (Promega), according to the manufacture’s protocol. Peptides were extracted from the gel pieces using 60% acetonitrile (ACN) and 5% formic acid, and the supernatants were pooled prior to sample drying with a vacuum concentrator, and stored at −80°C until LC-MS/MS. Nano-LC-MSMS was performed as described previously (Zheng et al., 2015) using a Dionex rapid-separation liquid chromatography (RSLC) system interfaced with a LTQ Orbitrap Velos (ThermoFisher, San Jose, CA). The LC-MS/MS raw data was analyzed using the MaxQuant software version 1.6.2.6 (http://www.maxquant.org/) (Cox and Mann, 2008) with the Andromeda search engine against the C. albicans proteome (downloaded from UniProt [https://uniprot.org]) with carbamidomethylation on cysteine as fixed modification, oxidation of methionine, and deamidation on asparagine as variable modifications. Peptide-protein matched spectra allowed less than 1% FDR. The relative abundance of peptides in supernatants were reported along with differential protein abundance in the hir1Δ/Δ strain compared with the WT.
For the secretome analysis from SC medium cultures, strains were grown in 50 mL SC medium in an orbital shaker at 30°C and 150–200 rpm for 3 days, and harvested by centrifugation. The top 35 mL of supernatant was transferred to a new tube, 1 mM benzamidine and 1 mM EDTA were added as protease inhibitors, and the supernatants were re-centrifuged. The top 25 mL of supernatant were transferred to 3 kDa Amicon columns (Amicon Ultra-15 Centrifugal Filter Unit, UFC 900324) and concentrated following the manufacturer’s recommendations. 100 μL of the concentrated sample were precipitated overnight in 4 volumes of methanol:acetone in a LoBind Eppendorf tube, as previously described (Zacchi et al., 2020a, 2020b). The supernatant was eliminated after high speed centrifugation, and air-dried proteins were resuspended in 50 mM ammonium bicarbonate supplemented with 10 mM DTT and 1 μg of trypsin. After an overnight incubation at 37°C, peptides were desalted using C18 ZipTips (Millipore) and analyzed by LC-ESI-MS/MS at the Mass Spectrometry Facility of the School of Chemistry and Molecular Biosciences, University of Queensland. Desalted peptides were analyzed by LC-ESI-MS/MS using a Prominence nanoLC system (Shimadzu) and TripleTof 5600 mass spectrometer with a Nanospray III interface (SCIEX) as previously described (Xu et al., 2015; Zacchi and Schulz, 2016). Peptides were separated with buffer A (1% acetonitrile and 0.1% formic acid) and buffer B (80% acetonitrile with 0.1% formic acid) with a gradient of 10%–60% buffer B over 45 min. Gas and voltage settings were adjusted as required. For data-dependent acquisition (DDA), an MS TOF scan from m/z of 350–1800 was performed for 0.5 s followed by DDA of MS/MS with automated collision energy (CE) selection of the top 20 peptides from m/z of 40–1800 for 0.05 s per spectrum. Identical LC conditions were used for SWATH-MS, with an MS-TOF scan from m/z of 350–1800 for 0.05 s followed by high-sensitivity information-independent acquisition with 26 m/z isolation windows with 1 m/z window overlap each for 0.1 s across an m/z range of 400–1250. CE was automatically assigned by the Analyst software (SCIEX) based on m/z window ranges. Peptide identification was performed as previously described (Xu et al., 2015; Zacchi and Schulz, 2016) using ProteinPilot 5.0.1 (SCIEX), searching a FASTA file containing the C. albicans proteome (downloaded from UniProt (https://www.uniprot.org/) on 23 March 2020; Proteome ID UP000000559, with 6035 proteins) merged with a common contaminants database, using standard settings (sample type, identification; Cysteine alkylation, acrylamide; instrument, TripleTof 5600; species: none selected; ID focus, biological modifications; enzyme, trypsin; search effort, thorough ID). FDR analysis using ProteinPilot was performed on all searches with limits of 99% identification confidence and 1% local FDR. The ProteinPilot search results were used as ion libraries for SWATH analyses, using as input the number of proteins identified with 1% global FDR by ProteinPilot. Peptide abundance was measured with Peakview v2.2 (SCIEX) using the following standard settings: shared peptides not imported; 6 transitions/peptide; 99% peptide confidence threshold; excluding modified peptides, 1% FDR; 6 min XIC extraction window; and 75 ppm XIC width. All experiments were performed with three biological replicates. The output from Peakview was re-processed using an in-house developed script to eliminate the value of peptides measured with a FDR > 0.01, as previously described (Kerr et al., 2019). Statistical analyses for SWATH-MS proteomics were performed using MSStats in R (Choi et al., 2014; Xu et al., 2015; Zacchi and Schulz, 2016). Comparisons with p < 10−5 were considered significant. The final analysis result is presented in Table S6.
Fluorescent staining of β-glucans and flow cytometry analysis
C. albicans strains were grown to the logarithmic growth phase in YPD at 30°C, harvested at 1500 × g for 3 minutes and washed twice with PBS. To stain for β-glucans, 2×106 cells were washed three times with FACS buffer (1% FCS [GIBCO], 0.5 mM EDTA, 0.1% Tween-20 [both Sigma-Aldrich] in PBS) and incubated with 5 ng/m of Fc (human):Dectin-1 (AdipoGen) in FACS buffer on ice for 60 minutes. Cells were washed three times with FACS buffer and subsequently incubated with 2 μg/ml Alexa Fluor® 488 anti-human IgG Fc antibody (Biolegend) on ice for 45 minutes. Cells were then washed three times in FACS buffer, resuspended in 500 μL PBS and analyzed on a LSRFortessa (BD Biosciences) with the settings described earlier (Nogueira et al., 2017). In addition, FSC-W and FSC-H channels were monitored in order to discriminate cell doublets. Flow cytometry data were analyzed using FlowJo (Flowjo LLC, version 7.6.5.).
In vitro killing assay
In vitro C. albicans killing assays were performed with 1 × 105 BMDMs or neutrophils at an MOI of 1:10 (fungi to immune cell) in 96-well plates (Starlab). After 1 hour (neutrophils) or 3 hours (BMDMs) of co-culture, C. albicans survival was determined exactly as described previously (Majer et al., 2012).
Reactive oxygen species assay
Intra- and extracellular reactive oxygen species (ROS) accumulation was monitored at an MOI of 5:1 using a luminol-dependent chemiluminescence assay as described earlier (Frohner et al., 2009). Briefly, 4 × 104 BMDMs or neutrophils were suspended in DMEM without phenol red (Sigma-Aldrich) or Hank’s balanced salt solution (HBSS; with Mg2+ and Ca2+; GIBCO) in a well of a white 96-well luminometer plate (Nunc) and mixed with 50 μL HBSS containing 200 μM luminol (intra- and extracellular ROS) or 600 μM isoluminol (extracellular ROS) and 16 U horseradish peroxidase (HRP) type IV (all Sigma-Aldrich). Immediately afterward, 50 μL HBSS containing the indicated C. albicans strains, heat-killed C. albicans, 75 μg Zymosan or 1 μg TDB were added. Chemiluminescence was monitored on a Victor V3 or a Victor Nivo microplate reader (both PerkinElmer). As controls, BMDMs or neutrophils without stimulation and C. albicans alone were included. The detected relative luciferase units (RLU) are expressed as RLU per min per 1000 immune cells or as total RLU after the indicated time.
Immunoblotting
For time course analysis of Stp1 protein levels and proteolytic processing, cells of the indicated strains expressing Stp1-HA from overnight YPD cultures (30°C) were collected, washed 3 times with dH2O, and then inoculated into prewarmed YNB-BSA medium at an OD600 of 1. Cultures (60 ml) were incubated shaking at 30°C for the indicated period of time before aliquoting 10 mL of culture for lysate preparation. For Stp1 processing, a 10 mL aliquot of culture was taken after 2 hours and then immediately induced with 500 μM of either arginine or proline (Sigma-Aldrich). After 5 min, cells were immediately harvested for protein extraction or snap-frozen in liquid nitrogen for storage at −80°C until further processed. Collected cells were washed once with ice-cold dH2O, and then adjusted to an OD600 of 6. Whole cell lysates were prepared from 500 μL of adjusted cell suspension using NaOH/TCA method and then subjected to SDS-PAGE and electroblotting onto nitrocellulose (GE Healthcare Life sciences) (Silao et al., 2019). Membranes were blocked in 5% (Sap2 blots) or 10% (Stp1–3xHA blots) skimmed milk in TBST buffer (Tris-buffered saline containing 0.1% Tween-20 [Sigma-Aldrich]) at room temperature for 45 to 60 minutes. For Sap2 secretion analysis, YCB-BSA cultures were spun down and 5 μL of culture supernatant were mixed with 5 μL 2x Laemmli buffer (Laemmli, 1970) prior SDS-PAGE and immunoblot analysis. For Stp1-HA detection, HRP-conjugated anti-HA antibody (Pierce) was used at 1:2500 dilution in 5% skimmed milk/TBST. For loading control, a monoclonal β-actin primary antibody (Abcam) was used at 1:5000 dilution in 5% skimmed milk/TBST. Sap2 was detected with a self-made polyclonal rabbit anti-Sap2 antibody diluted 1:1000 in TBST buffer.
Co-culture of 1×106 BMDMs with C. albicans for protein extraction was performed at a MOI of 5:1 (fungi to BMDMs) for the indicated time in DMEM medium without FCS in 35 mm dishes (Starlab). Protein extracts were prepared by scraping the cells in hot 4 × SDS sample buffer (200 mM Tris-HCl pH 6.8, 36% Glycerol, 5% SDS, 0.2% Bromphenol Blue, 1% 2-mercaptoethanol [Sigma-Aldrich]). Lysates were collected in fresh 1.5 mL tubes on ice, sonicated in a water bath for 20 s and boiled for 5 minutes at 95°C. Aliquots of the protein extracts were analyzed via immunoblotting as described above. Blots were probed with primary antibodies against rabbit anti-phospho-p40phox at a dilution of 1:1000 (Thr154; all Cell Signaling Technologies) and mouse anti-panERK at a dilution of 1:5000 (BD Biosciences). Immunoreactive bands were visualized by an enhanced chemiluminescent detection system (Super-Signal Dura West Extended Duration Substrate; Pierce) using a ChemiDoc MP system (BioRad) for Stp1-HA and fungal β-actin levels, which were detected by a HRP-conjugated anti-HA antibody (Roche) and a HRP-conjugated goat anti-mouse secondary antibody (Pierce), respectively, diluted 1:10000 in 5% skimmed milk/TBST. Detection of phospho-p40phox was visualized using a HRP-conjugated goat-anti-rabbit antibody (Cell Signaling Technologies) diluted 1:3000 in 5% skimmed milk/TBST and CL-Xposure films (ThermoFisher Scientific). Sap2 and panERK detection was imaged using IRDye® 800 goat anti-rabbit (Sap2) or anti-mouse (panERK) antibodies (1:10000 dilution in TBST) and the Odyssey imaging system (LI-COR®).
QUANTIFICATION AND STATISTICAL ANALYSIS
All data in this study are presented as mean ± standard deviation (SD), unless otherwise stated. The number of biological replicates is stated in each figure legend. Statistical analysis was done in R studio (RStudio Team, 2020) and GraphPad Prism 6. Two-group comparisons were done with two-tailed Student’s t test after equal variance testing (F-test). Multi-group comparison were conducted using the Bartlett’s test for equal variance testing and a one-way ANOVA followed by a Tukey’s multiple comparison test, unless otherwise stated. Non-parametric distributions were analyzed using a Mann-Whitney test. Mouse survival curves were analyzed using the log-rank test (Mantel-Cox) and visualized using GraphPad Prism 6. Pearson correlation and associated p values were calculated in R using the ‘stat_cor’ function from the ggpubr package (https://github.com/kassambara/ggpubr/). The number of biological replicates and type of applied statistical test is stated in each figure legend. P values are indicated in the relevant figure panels and in figure legends. The following p values were considered: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns = p > 0.05.
Supplementary Material
Highlights.
The HIR histone chaperone controls nutritional adaptation and fungal virulence
HIR1 modulates chromatin accessibility and transcription of metabolic genes
Innate immune cells show impaired recognition of fungal pathogens lacking HIR1
Genetic ablation of HIR drives hypervirulence in systemic fungal infections
ACKNOWLEDGMENTS
This work was funded by the Austrian Science Fund FWF (ChromFunVir; P-32582) and, in part, by the FWF-SFB-070-HIT subproject B08 from the Austrian Science Fund FWF to K.K. In addition, support came partially by a National Institutes of Health grant to N.C. and K.K. (R01AI124499). M.T. was supported by an Erwin Schroedinger Return Fellowship (J3835) of the Austrian Science Fund FWF. P.O.L.,F.G.S., and R.S. were supported by the EU grant MC-ITN-606786 (ImResFun) and grants from the Swedish Research Council VR-2015-04202 and 2019-01547 (P.O.L). Work in the LeibundGut-Landmann-lab is supported by the Sinergia program of the Swiss National Science Foundation (CRSII5_173863). L.F.Z. was funded by a Promoting Women Fellowship from the University of Queensland, Australia. We thankfully acknowledge Joachim Morschhäuser for sharing strains and plasmids, Birgit Willinger for providing clinical C. albicans isolates, Samir Jawhara for help with neutrophil assays, Michael Bonelli for advice on human neutrophil collection, and Art&Science (Dorotea Fracchiolla, www.my-art-science.at) for preparing the graphical abstract.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109406.
REFERENCES
- Altmeier S, Toska A, Sparber F, Teijeira A, Halin C, and LeibundGut-Landmann S (2016). IL-1 Coordinates the Neutrophil Response to C. albicans in the Oral Mucosa. PLoS Pathog. 12, e1005882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves R, Barata-Antunes C, Casal M, Brown AJP, Van Dijck P, and Paiva S (2020). Adapting to survive: How Candida overcomes host-imposed constraints during human colonization. PLoS Pathog. 16, e1008478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin AD, Vishnoi N, and Prochasson P (2013). A global requirement for the HIR complex in the assembly of chromatin. Biochim. Biophys. Acta 1819, 264–276. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Heessen S, and Ljungdahl PO (2006). Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 20, 1563–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010). A Quality Control Tool for High Throughput Sequence Data, Fast, QC.
- Bahnan W, Koussa J, Younes S, Abi Rizk M, Khalil B, El Sitt S, Hanna S, El-Sibai M, and Khalaf RA (2012). Deletion of the Candida albicans PIR32 results in increased virulence, stress response, and upregulation of cell wall chitin deposition. Mycopathologia 174, 107–119. [DOI] [PubMed] [Google Scholar]
- Ballou ER, Avelar GM, Childers DS, Mackie J, Bain JM, Wagener J, Kastora SL, Panea MD, Hardison SE, Walker LA, et al. (2016). Lactate signalling regulates fungal β-glucan masking and immune evasion. Nat. Microbiol. 2, 16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsässer SJ, Chapgier A, Goldberg AD, Canaani E, et al. (2013). Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Ganesan K, and Datta A (1991). Induction of secretory acid proteinase in Candida albicans. J. Gen. Microbiol. 137, 2455–2461. [DOI] [PubMed] [Google Scholar]
- Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, López-Ribot JL, and Kadosh D (2008). UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19, 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, MacCallum DM, Bertram G, Munro CA, Hughes HB, Buurman ET, Brown AJP, Odds FC, and Gow NAR (2005). Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 280, 23408–23415. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- Berman J, and Krysan DJ (2020). Drug resistance and tolerance in fungi. Nat. Rev. Microbiol. 18, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo SM, Khalique Z, Kot J, Jones JK, and Lee SA (2008). Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genet. Biol. 45, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunsdorf C, and LeibundGut-Landmann S (2018). Modulation of the fungal-host interaction by the intra-species diversity of C. albicans. Pathogens 7, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, and Gordon S (2001). Immune recognition. A new receptor for β-glucans. Nature 413, 36–37. [DOI] [PubMed] [Google Scholar]
- Brown AJP, Brown GD, Netea MG, and Gow NAR (2014). Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 22, 614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, and Hochedlinger K (2017). Emerging roles of the histone chaperone CAF-1 in cellular plasticity. Curr. Opin. Genet. Dev. 46, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Elling U, Hopfgartner B, Jung YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Euong Ang C, Tenen D, et al. (2015). The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Lin CY, Tsai PW, Yang CY, Hsieh WP, and Lan CY (2012). Rhb1 regulates the expression of secreted aspartic protease 2 through the TOR signaling pathway in Candida albicans. Eukaryot. Cell 11, 168–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Sun MA, Warzecha C, Bachu M, Dey A, Wu T, Adams PD, Macfarlan T, Love P, and Ozato K (2020). HIRA, a DiGeorge Syndrome Candidate Gene, Confers Proper Chromatin Accessibility on HSCs and Supports All Stages of Hematopoiesis. Cell Rep. 30, 2136–2149.e4. [DOI] [PubMed] [Google Scholar]
- Choi M, Chang C-Y, Clough T, Broudy D, Killeen T, MacLean B, and Vitek O (2014). MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526. [DOI] [PubMed] [Google Scholar]
- Coêlho DF, Saturnino TP, Fernandes FF, Mazzola PG, Silveira E, and Tambourgi EB (2016). Azocasein Substrate for Determination of Proteolytic Activity: Reexamining a Traditional Method Using Bromelain Samples. BioMed Res. Int. 2016, 8409183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia A, Lermann U, Teixeira L, Cerca F, Botelho S, da Costa RM, Sampaio P, Gärtner F, Morschhäuser J, Vilanova M, and Pais C (2010). Limited role of secreted aspartyl proteinases Sap1 to Sap6 in Candida albicans virulence and host immune response in murine hematogenously disseminated candidiasis. Infect. Immun. 78, 4839–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- d’Enfert C, Kaune A-K, Alaban L-R, Chakraborty S, Cole N, Delavy M, Kosmala D, Marsaux B, Fróis-Martins R, Morelli M, et al. (2020). The impact of the Fungus-Host-Microbiota interplay upon Candida albicans infections: current knowledge and new perspectives. FEMS Microbiol. Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabas N, and Morschhäuser J (2008). A transcription factor regulatory cascade controls secreted aspartic protease expression in Candida albicans. Mol. Microbiol. 69, 586–602. [DOI] [PubMed] [Google Scholar]
- Day AM, Smith DA, Ikeh MAC, Haider M, Herrero-de-Dios CM, Brown AJP, Morgan BA, Erwig LP, MacCallum DM, and Quinn J (2017). Blocking two-component signalling enhances Candida albicans virulence and reveals adaptive mechanisms that counteract sustained SAPK activation. PLoS Pathog. 13, e1006131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bernardis F, Cassone A, Sturtevant J, and Calderone R (1995). Expression of Candida albicans SAP1 and SAP2 in experimental vaginitis. Infect. Immun. 63, 1887–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RA, and Lionakis MS (2019). Organ-specific mechanisms linking innate and adaptive antifungal immunity. Semin. Cell Dev. Biol. 89, 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RA, Swamydas M, Oikonomou V, Zhai B, Dambuza IM, Schaefer BC, Bohrer AC, Mayer-Barber KD, Lira SA, Iwakura Y, et al. (2019). CARD9+ microglia promote antifungal immunity via IL-1β- and CXCL1-mediated neutrophil recruitment. Nat. Immunol. 20, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc C, Benoit M, Le Goff S, Simon L, Poulet A, Cotterell S, Tatout C, and Probst AV (2015). The histone chaperone complex HIR maintains nucleosome occupancy and counterbalances impaired histone deposition in CAF-1 complex mutants. Plant J. 81, 707–722. [DOI] [PubMed] [Google Scholar]
- Dühring S, Germerodt S, Skerka C, Zipfel PF, Dandekar T, and Schuster S (2015). Host-pathogen interactions between the human innate immune system and Candida albicans-understanding and modeling defense and evasion strategies. Front. Microbiol. 6, 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkel N, Biswas K, Hiller E, Fellenberg K, Satheesh SV, Rupp S, and Morschhäuser J (2014). Control of morphogenesis, protease secretion and gene expression in Candida albicans by the preferred nitrogen source ammonium. Microbiology (Reading) 160, 1599–1608. [DOI] [PubMed] [Google Scholar]
- Ene IV, Cheng S-C, Netea MG, and Brown AJP (2013). Growth of Candida albicans cells on the physiologically relevant carbon source lactate affects their recognition and phagocytosis by immune cells. Infect. Immun. 81, 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Brunke S, Brown AJP, and Hube B (2014). Metabolism in fungal pathogenesis. Cold Spring Harb. Perspect. Med. 4, a019695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira ME, Flaherty K, and Prochasson P (2011). The Saccharomyces cerevisiae histone chaperone Rtt106 mediates the cell cycle recruitment of SWI/SNF and RSC to the HIR-dependent histone genes. PLoS ONE 6, e21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J, Kainth P, Lambert JP, van Bakel H, Tsui K, Peña-Castillo L, Nislow C, Figeys D, Hughes TR, Greenblatt J, and Andrews BJ (2009). Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35, 340–351. [DOI] [PubMed] [Google Scholar]
- Fisher MC, Gurr SJ, Cuomo CA, Blehert DS, Jin H, Stukenbrock EH, Stajich JE, Kahmann R, Boone C, Denning DW, et al. (2020). Threats Posed by the Fungal Kingdom to Humans, Wildlife, and Agriculture. MBio 11, e00449, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, and Bernstein BE (2017). Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohner IE, Bourgeois C, Yatsyk K, Majer O, and Kuchler K (2009). Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 71, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner BN, Simmons RM, Canavera SJ, Akira S, and Underhill DM (2003). Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 197, 1107–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner BN, Simmons RM, and Underhill DM (2005). Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24, 1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, and Brown PO (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelis S, de Groot PWJ, Castillo L, Moragues MD, Sentandreu R, Gómez MM, and Valentín E (2012). Pga13 in Candida albicans is localized in the cell wall and influences cell surface properties, morphogenesis and virulence. Fungal Genet. Biol. 49, 322–331. [DOI] [PubMed] [Google Scholar]
- Gillum AM, Tsay EYH, and Kirsch DR (1984). Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198, 179–182. [DOI] [PubMed] [Google Scholar]
- Goryshin IY, Miller JA, Kil YV, Lanzov VA, and Reznikoff WS (1998). Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. USA 95, 10716–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NAR, and Hube B (2012). Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15, 406–412. [DOI] [PubMed] [Google Scholar]
- Gow NAR, Netea MG, Munro CA, Ferwerda G, Bates S, Mora-Montes HM, Walker L, Jansen T, Jacobs L, Tsoni V, et al. (2007). Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 196, 1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp K, Schild L, Schindler S, Hube B, Zipfel PF, and Skerka C (2009). The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol. Immunol. 47, 465–475. [DOI] [PubMed] [Google Scholar]
- Grover P, Asa JS, and Campos EI (2018). H3-H4 Histone Chaperone Pathways. Annu. Rev. Genet. 52, 109–130. [DOI] [PubMed] [Google Scholar]
- Hall RA, and Gow NAR (2013). Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol. Microbiol. 90, 1147–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond CM, Strømme CB, Huang H, Patel DJ, and Groth A (2017). Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, and Tzagoloff A (1986). Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2, 163–167. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Majer O, Frohner IE, Komnenovic V, and Kuchler K (2010). The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog. 6, e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfs S, Mogavero S, and Hube B (2016). Interaction of Candida albicans with host cells: virulence factors, host defense, escape strategies, and the microbiota. J. Microbiol. 54, 149–169. [DOI] [PubMed] [Google Scholar]
- Huang MY, Woolford CA, May G, McManus CJ, and Mitchell AP (2019). Circuit diversification in a biofilm regulatory network. PLoS Pathog. 15, e1007787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube B, Monod M, Schofield DA, Brown AJP, and Gow NAR (1994). Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14, 87–99. [DOI] [PubMed] [Google Scholar]
- Hube B, Sanglard D, Odds FC, Hess D, Monod M, Schäfer W, Brown AJ, and Gow NA (1997). Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect. Immun. 65, 3529–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo W, Zhang K, Nie Z, Li Q, and Jin F (2010). Kidney injury molecule-1 (KIM-1): a novel kidney-specific injury molecule playing potential double-edged functions in kidney injury. Transplant. Rev. (Orlando) 24, 143–146. [DOI] [PubMed] [Google Scholar]
- Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, and Yamasaki S (2009). Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J. Exp. Med. 206, 2879–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenull S, Tscherner M, Gulati M, Nobile CJ, Chauhan N, and Kuchler K (2017). The Candida albicans HIR histone chaperone regulates the yeast-to-hyphae transition by controlling the sensitivity to morphogenesis signals. Sci. Rep. 7, 8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenull S, Tscherner M, Mair T, and Kuchler K (2020). ATAC-Seq Identifies Chromatin Landscapes Linked to the Regulation of Oxidative Stress in the Human Fungal Pathogen Candida albicans. J. Fungi (Basel) 6, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, and Mitchell A (1994). Methods in Yeast Genetics. A Laboratory Course Manual (Cold Spring Harbor Laboratory Press). [Google Scholar]
- Kakade P, Mahadik K, Balaji KN, Sanyal K, and Nagaraja V (2019). Two negative regulators of biofilm development exhibit functional divergence in conferring virulence potential to Candida albicans. FEMS Yeast Res. 19, Published online March 1, 2019. 10.1093/femsyr/foy078. [DOI] [PubMed] [Google Scholar]
- Kerr ED, Phung TK, Caboche CH, Fox GP, Platz GJ, and Schulz BL (2019). The intrinsic and regulated proteomes of barley seeds in response to fungal infection. Anal. Biochem. 580, 30–35. [DOI] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, and Greenleaf WJ (2019). Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220. [DOI] [PubMed] [Google Scholar]
- Kraidlova L, Schrevens S, Tournu H, Van Zeebroeck G, Sychrova H, and Van Dijck P (2016). Characterization of the Candida albicans Amino Acid Permease Family: Gap2 Is the Only General Amino Acid Permease and Gap4 Is an S-Adenosylmethionine (SAM) Transporter Required for SAM-Induced Morphogenesis. MSphere 1, e00284, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K, Jenull S, Shivarathri R, and Chauhan N (2016). Fungal KATs/KDACs: A New Highway to Better Antifungal Drugs? PLoS Pathog. 12, e1005938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lai WKM, and Pugh BF (2017). Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 18, 548–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan C-Y, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, and Agabian N (2002). Metabolic specialization associated with phenotypic switching in Candidaalbicans. Proc. Natl. Acad. Sci. USA 99, 14907–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, and Copeland NG (2001). A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65. [DOI] [PubMed] [Google Scholar]
- Lee J-E, Oh J-H, Ku M, Kim J, Lee J-S, and Kang S-O (2015). Ssn6 has dual roles in Candida albicans filament development through the interaction with Rpd31. FEBS Lett. 589, 513–520. [DOI] [PubMed] [Google Scholar]
- Lenardon MD, Milne SA, Mora-Montes HM, Kaffarnik FAR, Peck SC, Brown AJP, Munro CA, and Gow NAR (2010). Phosphorylation regulates polarisation of chitin synthesis in Candida albicans. J. Cell Sci. 123, 2199–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermann U, and Morschhäuser J (2008). Secreted aspartic proteases are not required for invasion of reconstituted human epithelia by Candida albicans. Microbiology (Reading) 154, 3281–3295. [DOI] [PubMed] [Google Scholar]
- Li Y, and Jiao J (2017). Histone chaperone HIRA regulates neural progenitor cell proliferation and neurogenesis via β-catenin. J. Cell Biol. 216, 1975–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limjindaporn T, Khalaf RA, and Fonzi WA (2003). Nitrogen metabolism and virulence of Candida albicans require the GATA-type transcriptional activator encoded by GAT1. Mol. Microbiol. 50, 993–1004. [DOI] [PubMed] [Google Scholar]
- Lionakis MS, Lim JK, Lee CCR, and Murphy PM (2011). Organ-specific innate immune responses in a mouse model of invasive candidiasis. J. Innate Immun. 3, 180–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, Swamydas M, Fischer BG, Plantinga TS, Johnson MD, Jaeger M, Green NM, Masedunskas A, Weigert R, Mikelis C, et al. (2013). CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J. Clin. Invest. 123, 5035–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Lopes da Rosa J, Boyartchuk VL, Zhu LJ, and Kaufman PD (2010). Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc. Natl. Acad. Sci. USA 107, 1594–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz H, Sohn K, Brunner H, Muhlschlegel FA, and Rupp S (2004). RBR1, a novel pH-regulated cell wall gene of Candida albicans, is repressed by RIM101 and activated by NRG1. Eukaryot. Cell 3, 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer O, Bourgeois C, Zwolanek F, Lassnig C, Kerjaschki D, Mack M, Müller M, and Kuchler K (2012). Type I interferons promote fatal immunopathology by regulating inflammatory monocytes and neutrophils during Candida infections. PLoS Pathog. 8, e1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, and Ljungdahl PO (2005). Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol. Cell. Biol. 25, 9435–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramón P, and Lorenz MC (2016). The SPS amino acid sensor mediates nutrient acquisition and immune evasion in Candida albicans. Cell. Microbiol. 18, 1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramón P, and Lorenz MC (2017). A feast for Candida: Metabolic plasticity confers an edge for virulence. PLoS Pathog. 13, e1006144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramón P, Pountain AW, van Hoof A, and Lorenz MC (2020). The Paralogous Transcription Factors Stp1 and Stp2 of Candida albicans Have Distinct Functions in Nutrient Acquisition and Host Interaction. Infect. Immun. 88, e00763, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhäuser J (2011). Nitrogen regulation of morphogenesis and protease secretion in Candida albicans. Int. J. Med. Microbiol. 301, 390–394. [DOI] [PubMed] [Google Scholar]
- Moyes DL, Runglall M, Murciano C, Shen C, Nayar D, Thavaraj S, Kohli A, Islam A, Mora-Montes H, Challacombe SJ, and Naglik JR (2010). A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe 8, 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik JR, Newport G, White TC, Fernandes-Naglik LL, Greenspan JS, Greenspan D, Sweet SP, Challacombe SJ, and Agabian N (1999). In vivo analysis of secreted aspartyl proteinase expression in human oral candidiasis. Infect. Immun. 67, 2482–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashun B, Hill PWS, Smallwood SA, Dharmalingam G, Amouroux R, Clark SJ, Sharma V, Ndjetehe E, Pelczar P, Festenstein RJ, et al. (2015). Continuous Histone Replacement by Hira Is Essential for Normal Transcriptional Regulation and De Novo DNA Methylation during Mouse Oogenesis. Mol. Cell 60, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Joosten LAB, van der Meer JWM, Kullberg BJ, and van de Veerdonk FL (2015). Immune defence against Candida fungal infections. Nat. Rev. Immunol. 15, 630–642. [DOI] [PubMed] [Google Scholar]
- Nguyen GT, Green ER, and Mecsas J (2017). Neutrophils to the ROScue: Mechanisms of NADPH oxidase activation and bacterial resistance. Front. Cell. Infect. Microbiol. 7, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira MF, Istel F, Jenull S, Walker LA, Gow NA, and Lion T (2017). Quantitative Analysis of Candida Cell Wall Components by Flow Cytometry with Triple-Fluorescence Staining. J. Microbiol. Mod. Tech. Published online December 31, 2017. 10.15744/2575-5498.2.101. [DOI] [Google Scholar]
- Odds FC (2008). Secreted proteinases and Candida albicans virulence. Microbiology (Reading) 154, 3245–3246. [DOI] [PubMed] [Google Scholar]
- Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, and Kullberg BJ (2018). Invasive candidiasis. Nat. Rev. Dis. Primers 4, 18026. [DOI] [PubMed] [Google Scholar]
- Park H, Myers CL, Sheppard DC, Phan QT, Sanchez AAE, E Edwards J, and Filler SG (2005). Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell. Microbiol. 7, 499–510. [DOI] [PubMed] [Google Scholar]
- Pchelintsev NA, McBryan T, Rai TS, van Tuyn J, Ray-Gallet D, Almouzni G, and Adams PD (2013). Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 3, 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericolini E, Gabrielli E, Amacker M, Kasper L, Roselletti E, Luciano E, Sabbatini S, Kaeser M, Moser C, Hube B, et al. (2015). Secretory aspartyl proteinases cause vaginitis and can mediate vaginitis caused by Candida albicans in mice. MBio 6, e00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Rhomberg PR, Messer SA, and Castanheira M (2015). In vitro activity of a Hos2 deacetylase inhibitor, MGCD290, in combination with echinocandins against echinocandin-resistant Candida species. Diagn. Microbiol. Infect. Dis. 81, 259–263. [DOI] [PubMed] [Google Scholar]
- Prochasson P, Florens L, Swanson SK, Washburn MP, and Workman JL (2005). The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19, 2534–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandra S, Linde J, Brock M, Guthke R, Hube B, and Brunke S (2014). Regulatory networks controlling nitrogen sensing and uptake in Candida albicans. PLoS ONE 9, e92734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44 (W1), W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W (2007). Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432. [DOI] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, and Morschhäuser J (2004). The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341, 119–127. [DOI] [PubMed] [Google Scholar]
- Riedelberger M, Penninger P, Tscherner M, Seifert M, Jenull S, Brunnhofer C, Scheidl B, Tsymala I, Bourgeois C, Petryshyn A, et al. (2020). Type I Interferon Response Dysregulates Host Iron Homeostasis and Enhances Candida glabrata Infection. Cell Host Microbe 27, 454–466.e8. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani L, Mencacci A, Cenci E, Del Sero G, Bistoni F, and Puccetti P (1997). An immunoregulatory role for neutrophils in CD4+ T helper subset selection in mice with candidiasis. J. Immunol. 158, 2356–2362. [PubMed] [Google Scholar]
- Romo JA, and Kumamoto CA (2020). On Commensalism of Candida. J. Fungi (Basel) 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio-Team (2016). RStudio: Integrated Development for R.
- RStudio Team (2020). RStudio: Integrated Development Environment for R.
- Rutherford JC, Bahn YS, van den Berg B, Heitman J, and Xue C (2019). Nutrient and stress sensing in pathogenic yeasts. Front. Microbiol. 10, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam DA, and Huang DH (2016). Maintenance of Tissue Pluripotency by Epigenetic Factors Acting at Multiple Levels. PLoS Genet. 12, e1005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadik CD, Kim ND, and Luster AD (2011). Neutrophils cascading their way to inflammation. Trends Immunol. 32, 452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M, Schäfer W, Korting HC, and Hube B (1998). Differential expression of secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol. Microbiol. 29, 605–615. [DOI] [PubMed] [Google Scholar]
- Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, and Greenleaf WJ (2015). Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25, 1757–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, and von Haeseler A (2013). NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 29, 2790–2791. [DOI] [PubMed] [Google Scholar]
- Silao FGS, Ward M, Ryman K, Wallström A, Brindefalk B, Udekwu K, and Ljungdahl PO (2019). Mitochondrial proline catabolism activates Ras1/cAMP/PKA-induced filamentation in Candida albicans. PLoS Genet. 15, e1007976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis NV, and Filler SG (2012). Mouse model of oropharyngeal candidiasis. Nat. Protoc. 7, 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloviev DA, Jawhara S, and Fonzi WA (2011). Regulation of innate immune response to Candida albicans infections by αMβ2-Pra1p interaction. Infect. Immun. 79, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector MS, Raff A, DeSilva H, Lee K, and Osley MA (1997). Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 17, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg B, Ibrahim AS, Edwards JEJ Jr., and Filler SG (2005). Mice with disseminated candidiasis die of progressive sepsis. J. Infect. Dis. 192, 336–343. [DOI] [PubMed] [Google Scholar]
- Staib P, Kretschmar M, Nichterlein T, Hof H, and Morschhäuser J (2000). Differential activation of a Candida albicans virulence gene family during infection. Proc. Natl. Acad. Sci. USA 97, 6102–6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib P, Lermann U, Blass-Warmuth J, Degel B, Würzner R, Monod M, Schirmeister T, and Morschhäuser J (2008). Tetracycline-inducible expression of individual secreted aspartic proteases in Candida albicans allows isoenzyme-specific inhibitor screening. Antimicrob. Agents Chemother. 52, 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JS, and Liu H (2013). Nucleosome assembly factors CAF-1 and HIR modulate epigenetic switching frequencies in an H3K56 acetylation-associated manner in Candida albicans. Eukaryot. Cell 12, 591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidergall M, Solis NV, Lionakis MS, and Filler SG (2018). EphA2 is an epithelial cell pattern recognition receptor for fungal β-glucans. Nat. Microbiol. 3, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, and Nakatani Y (2004). Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61. [DOI] [PubMed] [Google Scholar]
- Tams RN, Cassilly CD, Anaokar S, Brewer WT, Dinsmore JT, Chen Y-L, Patton-Vogt J, and Reynolds TB (2019). Overproduction of Phospholipids by the Kennedy Pathway Leads to Hypervirulence in Candida albicans. Front. Microbiol. 10, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Davies LC, Liao CT, da Fonseca DM, Griffiths JS, Andrews R, Jones AV, Clement M, Brown GD, Humphreys IR, et al. (2019). The protective effect of inflammatory monocytes during systemic C. albicans infection is dependent on collaboration between C-type lectin-like receptors. PLoS Pathog. 15, e1007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torella JP, Lienert F, Boehm CR, Chen J-H, Way JC, and Silver PA (2014). Unique nucleotide sequence-guided assembly of repetitive DNA parts for synthetic biology applications. Nat. Protoc. 9, 2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein-Weidner K, Gladiator A, Nur S, Diethelm P, and LeibundGut-Landmann S (2015). IL-17-mediated antifungal defense in the oral mucosa is independent of neutrophils. Mucosal Immunol. 8, 221–231. [DOI] [PubMed] [Google Scholar]
- Tscherner M, Stappler E, Hnisz D, and Kuchler K (2012). The histone acetyltransferase Hat1 facilitates DNA damage repair and morphogenesis in Candida albicans. Mol. Microbiol. 86, 1197–1214. [DOI] [PubMed] [Google Scholar]
- Tscherner M, Zwolanek F, Jenull S, Sedlazeck FJ, Petryshyn A, Frohner IE, Mavrianos J, Chauhan N, von Haeseler A, and Kuchler K (2015). The Candida albicans Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways. PLoS Pathog. 11, e1005218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine DL (2007). Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 5, 316–323. [DOI] [PubMed] [Google Scholar]
- Vihervaara A, Duarte FM, and Lis JT (2018). Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 19, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk AG, Netea MG, van Krieken JH, Iwakura Y, van der Meer JWM, and Kullberg BJ (2006). Endogenous interleukin (IL)-1 α and IL-1 β are crucial for host defense against disseminated candidiasis. J. Infect. Dis. 193, 1419–1426. [DOI] [PubMed] [Google Scholar]
- Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, and Lorenz MC (2011). The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio 2, e00055, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag; ). [Google Scholar]
- Wirnsberger G, Zwolanek F, Stadlmann J, Tortola L, Liu SW, Perlot T, Järvinen P, Dürnberger G, Kozieradzki I, Sarao R, et al. (2014). Jagunal homolog 1 is a critical regulator of neutrophil function in fungal host defense. Nat. Genet. 46, 1028–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley JN, Penumetcha P, Abon NV, Woolford CA, Mitchell AP, and Noble SM (2019). Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection. Cell Host Microbe 25, 432–443.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele H, Tsao S, Lépine G, Mullick A, Tremblay J, Drogaris P, Lee E-H, Thibault P, Verreault A, and Raymond M (2010). Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat. Med. 16, 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Bailey U-M, and Schulz BL (2015). Automated measurement of site-specific N-glycosylation occupancy with SWATH-MS. Proteomics 15, 2177–2186. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, and He Q-Y (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, and He Q-Y (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383. [DOI] [PubMed] [Google Scholar]
- Zacchi LF, and Schulz BL (2016). SWATH-MS Glycoproteomics Reveals Consequences of Defects in the Glycosylation Machinery. Mol. Cell. Proteomics 15, 2435–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacchi LF, Schulz WL, and Davis DA (2010). HOS2 and HDA1 encode histone deacetylases with opposing roles in Candida albicans morphogenesis. PLoS ONE 5, e12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacchi LF, Recinos DR, Pegg CL, Phung TK, Napoli M, Aitken C, Sandford V, Mahler SM, Lee YY, Schulz BL, et al. (2020a). Analysis of coagulation factor IX in bioreactor cell culture medium predicts yield and quality of the purified product. BioRxiv, 2020.06.03.131177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacchi LF, Recinos DR, Otte E, Aitken C, Hunt T, Sandford V, Lee YY, Schulz BL, and Howard CB (2020b). S-Trap eliminates cell culture media polymeric surfactants for effective proteomic analysis of mammalian cell bioreactor supernatants. J. Proteome Res. 19, 2149–2158. [DOI] [PubMed] [Google Scholar]
- Zaret KS, and Mango SE (2016). Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 37, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhao C, Qian M, Roy S, Arpa A, Soherwardy A, and Kuruc M (2015). AlbuVoid™ Coupled to On-Bead Digestion - Tackling the Challenges of Serum Proteomics. J. Proteomics Bioinform. 8, 225–230. [Google Scholar]
- Zwolanek F, Riedelberger M, Stolz V, Jenull S, Istel F, Köprülü AD, Ellmeier W, and Kuchler K (2014). The non-receptor tyrosine kinase Tec controls assembly and activity of the noncanonical caspase-8 inflammasome. PLoS Pathog. 10, e1004525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this manuscript are available from the lead contact upon reasonable request. RNA-seq and ATAC-seq raw data have been deposited at the Gene Expression Omnibus (GEO), mass spectrometry data from SC media supernatants have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD018163. Data are available as of the date of publication.Accession numbers are listed in the key resources table. All original code has been deposited at github and is publicly available as of the date of publication. DOIs are listed in the key resources. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Purified anti-mouse CD16/32 antibody (93) | BioLegend | Cat#101302; RRID:AB_312800 |
| PE anti-mouse F4/80 antibody (BM8) | BioLegend | Cat#123110; RRID:AB_893498 |
| APC/Cy7 anti-mouse/human CD11b antibody (M1/70) | BioLegend | Cat#101226; RRID:AB_830642 |
| Pacific Blue anti-mouse Ly-6C antibody (HK1.4) | BioLegend | Cat#128014; RRID:AB_1732079 |
| PE anti-mouse CD45 antibody (30-F11) | BioLegend | Cat#103105; RRID:AB_312970 |
| FITC anti-mouse Ly-6G antibody (1A8) | BioLegend | Cat#127606; RRID:AB_1236494 |
| Fc(human):Dectin-1 antibody | AdiopenGen | Cat#AG-40B-0138-C050 |
| Alexa Fluor® 488 anti-human IgG Fc (HP6017) | BioLegend | Cat#409322; RRID:AB_2563437 |
| Rabbit anti-phospho-p40phox (Thr154) antibody | Cell Signaling | Cat#4311; RRID: AB_330690 |
| Mouse anti-panERK antibody | BD Biosciences | Cat#610123; RRID:AB_397529 |
| Rat anti-HA antibody, HRP | Roche | Cat#12013819001; RRID:AB_390917 |
| Goat anti-mouse IgG (H+L) secondary antibody, HRP | Thermo Fisher Scientific | Cat#31430; RRID:AB_228307 |
| Mouse anti-β-actin (D6A8) antibody | Abcam | Cat#8224; RRID:AB_449644 |
| Goat anti-rabbit IgG (H+L) secondary antibody, IRDye 800CW | LI-COR | Cat#926–32214; RRID:AB_621846 |
| Goat anti-mouse IgG (H+L) secondary antibody, IRDye 800CW | LI-COR | Cat#926–32210; RRID:AB_621842 |
| Goat anti-rabbit IgG (H+L) secondary antibody, HRP | Cell Signaling | Cat#7074; RRID:AB_2099233 |
| Rabbit anti-Sap2 polyclonal antibody | Kuchler Laboratory | N/A |
|
Chemicals, peptides, and recombinant proteins | ||
| Yeast extract | BD Biosciences | Cat#212720 |
| Peptone | BD Biosciences | Cat#211820 |
| Dulbecco’s Phosphate Buffered Saline | Sigma-Aldrich | Cat#D8537 |
| DMEM | Thermo Fisher Scientific | Cat#11584486 |
| RPMI-1640 | Thermo Fisher Scientific | Cat#21875 |
| Fetal calf serum | Sigma-Aldrich | Cat#F7524 |
| Bovine serum albumin | Sigma-Aldrich | Cat#A2153 |
| Yeast carbon base | Sigma-Aldrich | Cat#Y3627 |
| Yeast nitrogen base withoug ammonium sulfate and aa | BD Biosciences | Cat#233520 |
| Pepstatin A | Sigma-Aldrich | Cat#516481 |
| Zymosan A | Sigma-Aldrich | Cat#Z4250 |
| Trehalose-6,6-dibehenate (TDB) | InvivoGen | Cat#INV-tlrl-tdb |
| RNA/ater™ Stabilization Solution | Thermo Fisher Scientific | Cat#AM7022 |
| Nextera TDE1 | Illumina | Cat#15027865 |
| Complete, EDTA-free Protease Inhibitor Cocktail Tablets | Roche | Cat#04693132001 |
| Zombie Aqua Fixable Viability Dye | BioLegend | Cat#423101 |
| Fixation buffer | BioLegend | Cat#420801 |
|
Critical commercial assays | ||
| MinElute PCR purification kit | QIAGEN | Cat#28006 |
| RNeasy MinElute Cleanup kit | QIAGEN | Cat#74204 |
|
Deposited data | ||
| RNA-seq raw data | This study | GEO: GSE157411 |
| ATAC-seq raw data | This study | GEO: GSE157411 |
| Mass-spectrometry data from SC medium culture supernatants | This study | PRIDE: PXD018163 |
|
Experimental models: Primary cells | ||
| Murine bone marrow derived macrophages | This study | N/A |
| Murine bone marrow derived neutrophils | This study | N/A |
|
Experimental models: Organisms/strains | ||
| C57BL/6J mice | Jackson Laboratory | Cat#000664; RRID:IMSR_JAX:000664 |
| C. albicans: wild type (WT) | (Gillum et al., 1984) | N/A |
| C. albicans: Clinical isolate 1 | Birgit Willinger | AKH No. 2284 |
| C. albicans: Clinical isolate 10 | Birgit Willinger | AKH No. 2275 |
| C. albicans: hir1Δ/Δ | (Tscherner et al., 2015) | CASJ019 |
| C. albicans: hir1 Δ/Δ::HIR1 | (Jenull et al., 2017) | CASJ013 |
| C. albicans: Clinical isolate 1 hir1Δ/Δ | This study | CASJ255 |
| C. albicans: Clinical isolate 10 hir1 Δ/Δ | This study | CASJ252 |
| C. albicans: hat1 Δ/Δ | (Tscherner et al., 2015) | CA-MT014 |
| C. albicans: cac2Δ/Δ | (Tscherner et al., 2015) | CA-MT363 |
| C. albicans: sap2Δ/Δ | (Staib et al., 2008) | SAP2MS4A |
| C. albicans: sap2Δ/Δ hir1 Δ/Δ | This study | CASJ261 |
| C. albicans: sap3Δ/Δ | This study | CASJ301 |
| C. albicans: hir1 Δ/Δ sap3Δ/Δ | This study | CASJ304 |
| C. albicans: sap2Δ/Δ sap3Δ/Δ | This study | CASJ308 |
| C. albicans: sap2Δ/Δ sap3Δ/Δ hir1 Δ/Δ | This study | CASJ310 |
| C. albicans: stpl Δ/Δ | This study | CASJ403 |
| C. albicans: hir1 Δ/Δ stp1Δ/Δ | This study | CASJ405 |
| C. albicans: ssy5Δ/Δ | (Miramon and Lorenz, 2016) | CaPM23 |
| C. albicans: ssy5Δ/Δ hir1 Δ/Δ | This study | CASJ420 |
| C. albicans: pmr1 Δ/Δ | This study | CASJ224 |
| C. albicans: hir1 Δ/Δ pmr1 Δ/Δ | This study | CASJ226 |
| C. albicans: pmr1 Δ/Δ::PMR1 | This study | CASJ239 |
| C. albicans: hir1 Δ/Δ pmr1Δ/Δ::PMR1 | This study | CASJ241 |
| C. albicans: chs3Δ/Δ | This study | CA-MT611 |
| C. albicans: chs3Δ/Δ hir1Δ/Δ | This study | CASJ262 |
|
Oligonucleotides | ||
| See Table S1 for full oligonucleotide list | N/A | |
|
Recombinant DNA | ||
| Plasmid: pSFS3b | (Tscherner et al., 2012) | N/A |
| Plasmid: YEp352 | (Hill et al., 1986) | N/A |
| Plasmid: pFA6a-3xHA-SAT1-FLP | This study | pFA6a-3HA-SAT 1-FLP |
| Plasmid: YEp-SAP3urdr-FLP-NAT1 | This study | ECSJ071 |
| Plasmid: YEp-PMR1urdr-FLP-NAT1 | This study | ECSJ031 |
| Plasmid: YEp-PMR1 reint-FLP-NATI | This study | ECSJ082 |
| Plasmid: YEp-CHS3urdr-FLP-NAT1 | This study | YEp352-NAT1-CHS3urdr |
| Plasmid: Yep-STP1urdr-FLP-NAT1 | This study | ECSJ075 |
|
Software and algorithms | ||
| fastQC v v0.11.8 | (Andrews, 2010) | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| cutadapt v1.18 | https://cutadapt.readthedocs.io/en/stable/ | |
| NextGenMap v0.5.5 | (Sedlazeck et al., 2013) | https://cibiv.github.io/NextGenMap/ |
| Picard | Broad Institute | http://broadinstitute.github.io/picard/ |
| BEDTools | https://github.com/arq5x/bedtools2 | |
| HTseq | (Anders et al., 2015) | https://htseq.readthedocs.io/en/master/ |
| edgeR | (Robinson et al., 2010) | https://www.bioconductor.org/packages/release/bioc/html/edgeR.html |
| MACS2 v2.1.2 | (Zhang et al., 2008) | https://github.com/macs3-project/MACS |
| ChIPseeker | (Yu et al., 2015) | http://bioconductor.org/packages/release/bioc/html/ChIPseeker.html |
| deepTools2 | (Ramírez et al., 2016) | https://deeptools.readthedocs.io/en/develop/ |
| clusterProfiler | (Yu et al., 2012) | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| MaxQuant v1.6.2.6 | (Cox and Mann, 2008) | http://www.maxquant.org/ |
| ProteinPilot v5.0.1 | SCIEX | https://sciex.com/products/software/proteinpilot-software |
| Peakview v2.2 | SCIEX | https://sciex.com/products/software/peakview-software |
| MSStats | (Choi et al., 2014) | https://www.bioconductor.org/packages/release/bioc/html/MSstats.html |
| R v3.6.1 | R Core Team 2019 | https://www.R-project.org/ |
| RStudio v1.3.1093 | RStudio Team 2020 | https://www.rstudio.com/ |
| FlowJo software version 7.6.5 and 10.7.1 | FlowJo | N/A |
| Realplex software 2.0 | Eppendorf | N/A |
| GraphPad Prism version 6.0 | GraphPad | N/A |
| RNA-seq analysis script | This study | https://github.com/tschemic/RNAseq_analysis |
| ATAC-seq analysis script | (Jenull et al., 2020) | https://github.com/tschemic/ATACseq_analysis |
|
Deposited raw data | ||
| RNA-seq raw data | This study | GSE157411 (GSE157599 SuperSeries) |
| ATAC-seq raw data | This study | GSE157568 (GSE157599 SuperSeries) |
| Mass spectrometry raw data (SC culture supernatants) | This study | PXD018163 |
|
Other | ||
| RNA 6000 Nano Kit | Agilent | Cat#5067–1511 |
| High Sensitivity DNA Kit | Agilent | Cat# 5067–4626 |


