Abstract
Accumulation of misfolded proteins in ER activates the unfolded protein response (UPR), a multifunctional signaling pathway that is important for cell survival. The UPR is regulated by three ER transmembrane sensors, one of which is inositol-requiring protein 1 (IRE1). IRE1 activates a transcription factor, X-box-binding protein 1 (XBP1), by removing a 26-base intron from XBP1 mRNA that generates spliced XBP1 mRNA (XBP1s). To search for XBP1 transcriptional targets, we utilized an XBP1s-inducible human cell line to limit XBP1 expression in a controlled manner. We also verified the identified XBP1-dependent genes with specific silencing of this transcription factor during pharmacological ER stress induction with both an N-linked glycosylation inhibitor (tunicamycin) and a non-competitive inhibitor of the sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) (thapsigargin). We then compared those results to the XBP1s-induced cell line without pharmacological ER stress induction. Using next‐generation sequencing followed by bioinformatic analysis of XBP1-binding motifs, we defined an XBP1 regulatory network and identified XBP1 as a repressor of PUMA (a proapoptotic gene) and IRE1 mRNA expression during the UPR. Our results indicate impairing IRE1 activity during ER stress conditions accelerates cell death in ER-stressed cells, whereas elevating XBP1 expression during ER stress using an inducible cell line correlated with a clear prosurvival effect and reduced PUMA protein expression. Although further studies will be required to test the underlying molecular mechanisms involved in the relationship between these genes with XBP1, these studies identify a novel repressive role of XBP1 during the UPR.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-03952-1.
Keywords: XBP1s, UPR, ER stress, BBC3, XBP1u, ERN1
Introduction
Endoplasmic reticulum (ER) stress can disrupt the folding and maturation of the secretory and membrane proteins and lead to the accumulation of unfolded proteins in the ER lumen, interruption of lipid synthesis, and deregulation of cellular calcium levels [1, 2]. The buildup of misfolded proteins in ER leads to the activation of the unfolded protein response (UPR), a multifunctional signaling pathway that either promotes cell recovery [3], or initiates cell death if the ER stress remains unmitigated [4]. The UPR signaling pathways are initiated by three ER transmembrane sensors: inositol-requiring protein 1 (IRE1), protein kinase RNA-like ER kinase (PERK) and activating transcription factor 6 (ATF6). IRE1 removes a 26-base intron from X-box-binding protein 1 (XBP1) mRNA in an unconventional splicing reaction that results in a translational frameshift that leads to the production of a functional and highly active spliced XBP1 (XBP1) transcription factor [5–9]. XBP1 enhances the expression of ER-resident chaperones and genes involved in ER-associated protein degradation (ERAD) [10] and promotes ER expansion [9]. The ER protein load is reduced by PERK-mediated phosphorylation of eIF2α which inhibits most protein synthesis, by ER-associated degradation of misfolded proteins, and by IRE1-mediated mRNA cleavage and degradation [3]. However, if ER stress remains unmitigated, the UPR utilizes the same pathways to promote cell death by activating the intrinsic apoptotic pathways [11–16].
The interruption of the UPR transitions from prosurvival to apoptosis and the alteration of cell fate decisions contribute to the pathomechanisms of a number of human diseases including diabetes mellitus, cancer, and neurodegenerative and respiratory disorders [17]. To facilitate novel interventions for treating these disorders, it is important to understand the mechanisms governing the UPR pathways. Although we know the many of the details of the UPR pathways that contribute to cellular survival or apoptosis [18–20], it remains unclear how these signals determine the cell fate transitions in vivo. Understanding molecular mechanisms underlying UPR-related cell fate decisions is very challenging given that the experimental models rely on pharmacological ER stressors that are used at different concentrations and utilize divergent mechanisms for disturbing ER homeostasis [21–24]. Furthermore, previous studies have shown that UPR signaling has distinct consequences that ultimately depend on the nature and intensity of the stimulus as well as the specific cell type involved [18].
Although previous studies suggested that the transcriptional activity XBP1 is important in deciding cell fate in the UPR [25–30], the information regarding XBP1’s direct role in modulating the transition between survival to apoptosis is limited. Therefore, the studies presented here were designed to select for XBP1-specific transcriptional targets and their roles in cell fate decisions. In our approach, we used inducible human cell lines that allowed for comparable and controlled expression of spliced XBP1 and unspliced XBP1 proteins. We also verified the identified XBP1-dependent genes with specific silencing of this transcription factor during mild pharmacological ER stress induction with both an N-linked glycosylation inhibitor (tunicamycin, Tm) as well as a non-competitive inhibitor of the sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) (thapsigargin, Tg). Using next‐generation sequencing (NGS) followed by bioinformatic analysis of XBP1-binding motifs, and validation using XBP1-specific silencing and quantitative real‐time PCR (qRT‐PCR), we defined an XBP1-dependent regulatory network and identified XBP1 as a repressor of both PUMA and IRE1 expression during the UPR. This approach not only confirmed previously known XBP1 roles during UPR, but also resulted in the identification of novel targets of this transcription factor that could fine-tune cell fate decisions. Furthermore, we show that XBP1 can modulate the PERK pathway activity via modulation of both CHOP and growth arrest and DNA damage-inducible protein (GADD34) mRNA expression. Although further studies will be required to test the underlying molecular mechanisms involved in the relationship between these genes with XBP1, the studies presented here identify a novel regulatory role of XBP1 during the UPR.
Materials and methods
Cell lines and culture conditions
HeLa S3 cells were obtained from the American Type Culture Collection (CCL‐2.2; Manassas, VA, USA). Cells were cultured in Minimum Essential Modified Eagle’s Medium (Thermo Fisher Scientific, Waltham, MA, USA) with 2 mM l‐glutamine (MilliporeSigma, Burlington, MA, USA), antibiotics (100 U/ml of penicillin and 100 µg/ml of streptomycin (MilliporeSigma), and 10% fetal bovine serum in a humidified incubator at 37 °C in 5% CO2 in 6‐well plates. Cells were allowed to grow to 70–80% confluence before the start of the experiments. The culture conditions for the human Schwann cells (SNF96.2), human epidermal keratinocytes (HaCaT), 10 donor-pooled human umbilical vein endothelial cells (HUVEC), and human bronchial epithelial cells (16HBE14o-) were the same as previously described in [21, 31].
The inducible HeLa S3 XBP1s and XBP1u cell lines were constructed beginning with vectors containing the cDNA sequences of XBP1s (NM_001079539.1) and XBP1u (NM_005080.3) that were obtained from GeneCopoeia (Rockville, MD, US; XBP1s cat. no. EX-Z4299 and XBP1u cat. no. EX-F0758). The ORFs sequences were verified with Sanger sequencing, and XBP1s and XBP1u cDNAs were restricted with EcoRI/MluI and EcoRI/BamHI, respectively, and cloned into the pCW57-MCS1-P2A-MCS2 (Hygro) vector that permits doxycycline-controlled inducible lentiviral expression [32]. pCW57-MCS1-P2A-MCS2 (Hygro) was a gift from Adam Karpf (Addgene plasmid # 80922; http://n2t.net/addgene:80922). The correct pCW57-XBP1s and pCW57-XBP1u insert sequences were verified with Sanger sequencing. These vectors along with VSV-G envelope expressing plasmid (pMD2.G) and lentiviral packaging plasmid (psPAX2) were used to transfect HEK-293 cells (ATCC CRL-1573) to generate lentiviruses carrying the XBP1s or XBP1u transgenes. The pMD2.G and psPAX2 plasmids were a gift from Didier Trono (Addgene plasmid # 12259; http://n2t.net/addgene:12259 and Addgene plasmid # 12260; http://n2t.net/addgene:12260, respectively). The lentiviruses were also used to transduce Hela S3 cells [33, 34]. Finally, following hygromycin b selection (300 µg/ml, Sigma) and qPCR verification of 24 h doxycycline induction (400 µg/ml, D3072 MilliporeSigma) of XBP1s and XBP1u mRNAs in HeLa S3 cells, two stable clonal cell lines capable of stable inducible expression of XBP1s (Hela-XBP1s) and XBP1u (HeLa-XBP1u) were obtained. These cell lines were cultured in Minimum Essential Modified Eagle’s Medium with 2 mM l‐glutamine, hygromycin B (300 µg/ml) and 10% tetracycline free fetal bovine serum (Takara Bio, USA) in a humidified incubator at 37 °C in 5% CO2 in 6‐well plates. Cells were allowed to grow to 70–80% confluence before the start of the experiments.
Induction of ER stress and activation of the UPR
Pharmacological induction of ER stress and activation of the UPR were performed as we previously described [21]. Briefly, cells were treated with the compounds for the time periods specified: tunicamycin (Tm 2.5 or 0.5 μg/ml; Sigma, T7765), thapsigargin (Tg 50 or 2.5 nM; Sigma, T9033). CTRL cells were treated with vehicle CTRL, DMSO (< 0.5% v/v; Sigma, D2650). Furthermore, to verify IRE1 activity, cells treated were 20 µM 4µ8C (an IRE1 inhibitor, Sigma-Aldrich, SML0949) dissolved in DMSO (Sigma-Aldrich, St. Louis, MI, USA) [35].
Real-time cell viability assay
For real‐time monitoring of cell viability, we applied the xCELLigence system as we described previously [36]. Briefly, HeLa cells (12,000 cells per well) were seeded in the 16‐well PC plates (00300600890, ACEA Biosciences Inc., San Diego, CA, USA) 24 h prior to the experiment. CTRL cells were cultured in the presence of DMSO vehicle. Treated cells were incubated with ER stressors for the next 24 h, and every 15 min, the cell conductances (cell index) were recorded. All experiments were performed in triplicate with three independent repeats. RTCA software v. 1.2.1 (ACEA Biosciences, Inc, San Diego, CA, USA) was used to calculate the normalized cell index and the cell growth curve slopes.
Monitoring caspase 3 and caspase 7 activity
The caspase 7 is considered to be redundant with caspase 3 because these enzymes share an optimal peptide recognition sequence and have several endogenous protein substrates in common [37]. While our main goal was to assess caspase 3 activity, the commercially available assays do not distinguish between these two cysteine proteases. Hence, we applied the caspase‐Glo 3/7 assay (Promega, Madison, WI, USA) to measure relative caspase activity as described previously [21, 36]. Briefly, cells the day after transfection with the specified siRNA were seeded onto 96-well luminescence assay white plates with clear bottoms (Corning Inc., 3903). The next day, the cells were treated with ER stressors or vehicle (0.1% DMSO) for indicated time points. Following treatment, cells were washed with PBS and the Caspase-Glo 3/7 assays (Promega) were performed in accordance with the manufacturer’s instructions using the GloMax-Multi + Detection System (Promega). The results were normalized to the values obtained from the vehicle control treatments.
siRNA transfections
siRNAs against XBP1 (Ambion assay id s14915) and BBC3 (Ambion assay id s25840) were purchased from Ambion. HeLa cells were transfected using the Lipofectamine RNAiMax (Invitrogen 13778030) according to manufacturer’s protocol. The siRNAs were used at final concentrations of 30 nM. The transfected cells were cultured for 2 days prior to further analysis. Ambion siRNA Negative Control 1 (Ambion assay id MC22484) was used as a control.
Isolation of RNA
Total RNA (containing both mRNA and miRNA) was isolated using miRNeasy kit (Qiagen). RNA concentrations were calculated based on the absorbance at 260 nm. RNA samples were stored at −70 °C until use.
Next‐generation RNA sequencing analyses
The RNA isolation and analyses were performed in HeLa-XBP1s and -XBP1u cells. Briefly, following XBP1 induction with doxycycline (24 h and 400 µg/ml final concentration) total RNA isolation, samples were validated with qRT‐PCR for ER stress activation prior to further analysis. Following rRNA depletion, the remaining RNA fraction was used for library construction and subjected to 100-bp single-end sequencing on an Illumina HiSeq 2000 instrument (San Diego, CA, USA). Sequencing reads were aligned to the Gencode human reference genome assembly (GRCh38 p7 Release 25) using STAR [38]. Transcript assembly and estimation of the relative abundance and tests for differential expression were carried out with Cufflinks and Cuffdiff [39]. The resulting data were validated with qRT‐PCR. The heat map generation and hierarchical clustering were performed with the Morpheus Web server (https://software.broadinstitute.org/morpheus). The Enrichr Web server (https://amp.pharm.mssm.edu/Enrichr/) [40] was applied to assign the NGS results into the ‘Gene Ontology Biological Process’ categories with the selection based on a False Discovery Rate Q‐value q < 0.05. Furthermore, the analyses were limited to experimentally verified interactions and no extended gene enrichment set analyses were performed.
Measurement of mRNA quantitative real-time PCR (qRT-PCR)
We used TaqMan One-Step RT-PCR Master Mix Reagents (Applied Biosystems) as described previously [41, 42] using the manufacturer’s protocol (retrotranscription: 15 min, 48 °C). For NGS data validation, 96 custom TaqMan expression array plates (id) were used according to the manufacturer’s instructions. The relative expressions were calculated using the 2−ΔΔCt method [43] with the glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and neutral ribosomal phosphoprotein P0 (RPLP0) genes as reference genes for the mRNA. TaqMan probes ids used are provided in Supplemental Table 1.
Western blots
The XBP1 protein detection was performed as described in [44]. Briefly, cells were lysed on ice for 15 min in RIPA buffer [150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris–HCl (pH 8.0)] supplemented with Protease Inhibitor Complete Mini (000000011836170001; Roche, Basel, Switzerland). The insoluble material was removed by centrifugation at 15,000g for 15 min. Protein concentrations were determined by Bio‐Rad Protein Assay [Bradford‐based method; Bio‐Rad, Hercules, CA, USA] using bovine serum albumin (BSA; MilliporeSigma) as the standard. Following the normalization of protein concentrations, lysates were mixed with an equal volume of 2 times Laemmli sample buffer (Bio-Rad) and incubated for 5 min at 95 °C before separation by SDS-PAGE on stain-free TGX gradient gels (Bio‐Rad). Following SDS-PAGE, the proteins were transferred to PVDF membranes (300 mA for 90 min at 4 °C). The membranes were then blocked with BSA dissolved in PBS and Tween-20 (3% BSA and 0.5% Tween-20) for 1–2 h followed by immunoblotting with the primary antibody for each experiment for spliced XBP1 (mAb 12782; diluted at 1:1000; Cell Signaling Technology, Danvers, MA, USA) or unspliced XBP1 (NBP1-77681; diluted at 1:700; Novus Biological USA). The unsliced XBP1 antibody has been independently validated with siRNA against XBP1 (Supplemental Figure S1). For PUMA, the monoclonal antibody MBS9131466 (MyBioSource Inc. San Diego, CA USA) was used for overnight incubations at 1:1500 dilution. For IRE1 (phosphor-S724), the monoclonal antibody ab243665 (Abcam, USA) was used at incubations at 1:1000 dilution. After the washing steps, the membranes were incubated with goat anti‐rabbit IgG (H + L) horseradish peroxidase-conjugated secondary antibodies (Bio‐Rad) and detected using ECL (Amresco, Solon, OH, USA). Densitometry was performed using Image Lab software v.4.1 (Bio-Rad).
XBP1 motif analysis
The promoters of the gene transcripts that were affected by XBP1 induction in the NGS experiments were analyzed for XBP1-binding sites. In each gene promoter sequence that was defined as a 20 kb window around the TSS, we examined only the open chromatin regions that were established in the HeLa S3 cell line by the ENCODE [45] project. We merged both DNase I-seq HeLa datasets found in Ensembl (v.79) [46]. We used the Nencki Genomics Database (v. 79_1) [47] to obtain genomic coordinates of these motif instances. For each gene, we calculated the number of instances found in the open chromatin regions.
Statistical analysis
Results were expressed as a mean ± standard deviation. Statistical significance was determined using the Student’s t-test and ANOVA on ranks with P values P ≤ 0.05 considered significant. The correlation was accessed via the Pearson product-moment correlation method.
Results
Since our working hypothesis was that the commonly used concentrations of ER stressors lead to non-physiological elevation of UPR signals and the potential for misassignment of UPR pathways targets or their role in cell fate decisions, we compared commonly used classical pharmacological stressors at high concentrations (high stress) and at low concentrations (low stress). We tested a glycosylation inhibitor tunicamycin (Tm) that is normally used at 2.5 µg/ml and a non-competitive inhibitor of the sarco/endoplasmic reticulum Ca2+ ATPase thapsigargin (Tg) that is normally used at 50 nM (high stress). For the low-stress model, we used Tm at 0.5 µg/ml and Tg at 2.5 nM. These concentrations were determined experimentally as the lowest concentrations that were able to induce XBP1, HSPA5 (BiP) and DITT3 (also known as CHOP) mRNAs by at least twofold after 6 h of treatment. We used HeLa cells since this is a common model system that has been employed in ER stress and UPR studies [48–51].
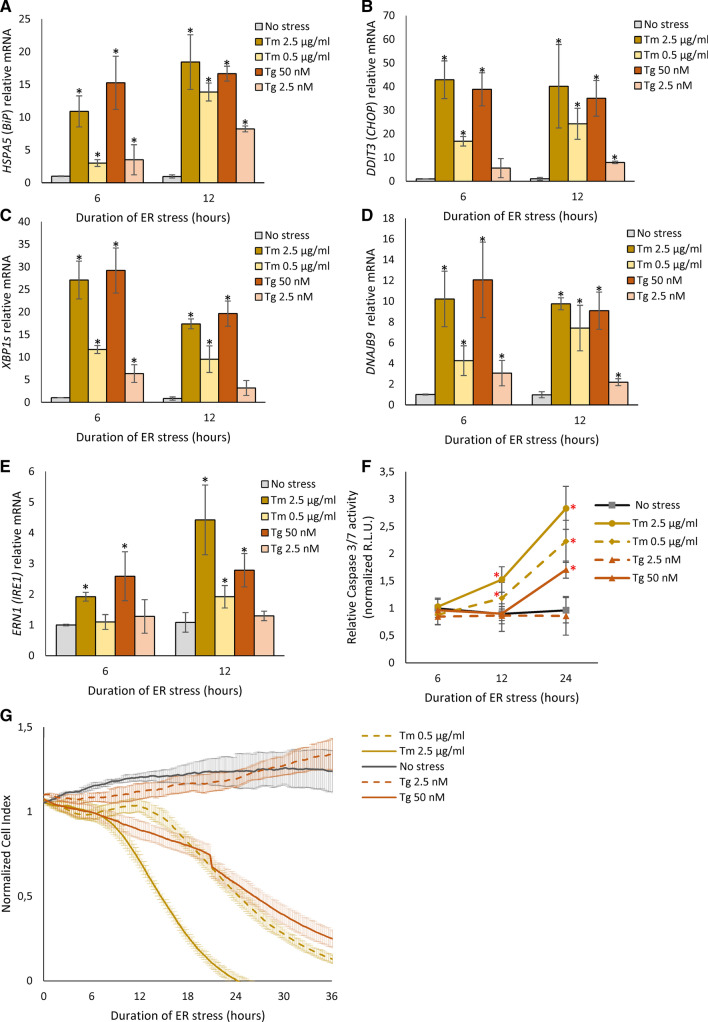
As shown in Fig. 1, both stress models were able to induce both prosurvival (HSPA5, XBP1s and DNAJB9) and apoptotic (DDIT3) reporters. The adaptive BiP chaperone mRNA levels (HSPA5) was continuously elevated in both the high and low-stress models (Fig. 1A). High ER stress was effectively induced with the commonly used Tm and Tg concentrations (2.5 µg/ml and 50 nM, respectively) [21, 44, 49]. High stress resulted in a 10- and 15-fold induction of HSPA5 mRNA with Tm and Tg, respectively, after 6 h and remained elevated after 12 h of treatment. In contrast, during mild ER stress conditions after 6 h, the HSPA5 mRNA was induced by ~ threefold by both Tm and Tg, and this dramatically increased after 12 h of treatment (Fig. 1A). The proapoptotic DDIT3 (CHOP) mRNA levels were also elevated 18- and 6-fold with Tm and Tg treatment, respectively, after 6 h of low-stress conditions and remained elevated after 12 h (Fig. 1B). The use of the higher concentrations of Tm and Tg resulted in ~ 40- and ~ 35-fold inductions of DDIT3 (CHOP) mRNA expression after both 6 and 12 h. A similar pattern of high- and low-stress treatments was seen with XPB1s (Fig. 1C) and DNAJB9 (Fig. 1D) mRNAs in that they were induced less during the low-stress conditions as might be expected. Interestingly, the XBP1s mRNA levels decreased after 12 h, whereas all the other mRNAs either increased or remained the same after high- and low-stress conditions. The DNAJB9 (DnaJ heat shock protein family (Hsp40) member B9) mRNA levels, a pro-adaptive chaperone and an XBP1 transcriptional target, were elevated as expected under both high- and low-stress conditions (Fig. 1D). Furthermore, ERN1 (IRE1) mRNA expression was induced after 6 h only by higher Tm and Tg concentrations, whereas in mild stress, ERN1 mRNA was only elevated by Tm treatment after 12 h (Fig. 1E).
Fig. 1.
ER stress-induced changes in A BIP, B CHOP, C XBP1s, D DNAJB9, and E IRE1 mRNA levels in HeLa cells. The results from three independent experiments (n = 9) are plotted normalized to GAPDH and RPLP0 mRNA levels and expressed as a fold change over the no-stress controls. Error bars represent standard deviations. Significant changes (P value P < 0.05) are marked with an asterisk. ER stressors used: Tm (2.5 µg/ml), Tm (0.5 µg/ml), Tg (50 nM)) and Tg (2.5 nM). F HeLa cells were treated with ER stressors ((2.5 µg/ml), Tm (0.5 µg/ml), Tg (50 nM) and Tg (2.5 nM)) for time points specified. The caspase 3/7 activity was monitored by luminescence and expressed in relative light units (RLU). Cells for each time point were seeded in triplicate, and the experiments repeated three times. Error bars represent standard deviations. G The cell conductances (expressed as normalized cell index) were accessed every 15 min following a 36-h treatment with Tm (2.5 µg/ml), Tm (0.5 µg/ml), Tg (50 nM) and Tg (2.5 nM). DMSO was used in the no-stress conditions (CTRL). The conductances were normalized to the last value prior to experiment start. Representative results from three independent experiments measurements (n = 9) are plotted
We next tested the effects of high and low stress on cell viability and proliferation. As shown in Fig. 1F, the Tm treatments at high- and low-stress conditions elevated caspase 3/7 activity at 12 and 24 h, whereas the Tg treatment elevated the caspase activity only at 24 h during high stress conditions (Fig. 1F). The lack of significant apoptotic signal for the lower Tg concentration was consistent with the lower induction of apoptotic mRNAs. These observations were also consistent with the results of real-time monitoring of HeLa cell proliferation (Fig. 1G). The mild ER Tg-induced stress had no significant effect on HeLa proliferation up to 36 h, while treatment with higher Tg concentration significantly limited cell growth throughout the entire time course. Whereas, both Tm concentrations affected HeLa growth and demonstrated a clear indication of apoptosis after 18 h (Fig. 1G). Taken together, the data illustrate that lower stress conditions still activate the UPR and may provide a more physiological model to delineate the differences between the adaptive and apoptotic UPR signaling pathways.
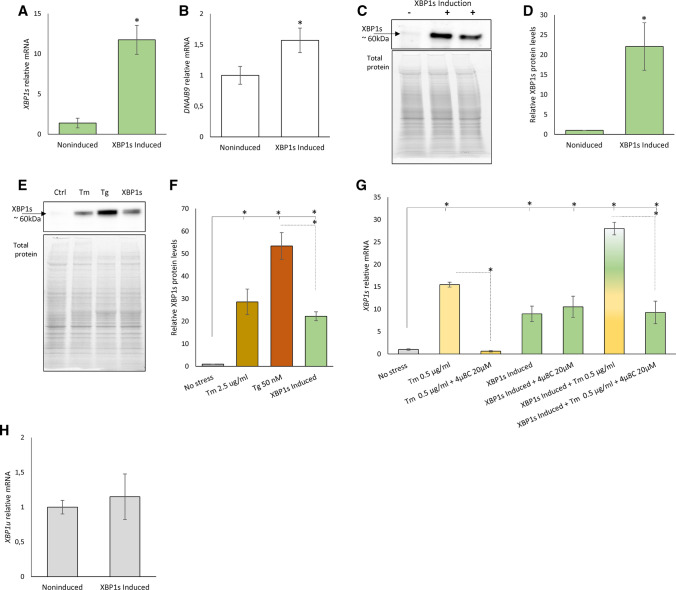
To follow XBP1’s role in the UPR, we created an inducible HeLa cell line model in which we could express low mRNA levels of this transcription factor that might mimic the levels observed during mild ER stress. Following the expansion and validation of individual clones expressing XBP1s under an inducible promoter, we selected a cell line that upon induction, stably expressed about an 11-fold increase XBP1s mRNA (HeLa-XBP1s) when compared to noninduced cells (Fig. 2A). The obtained XBP1 levels were sufficient to induce the expression of DNAJB9 mRNA, an XBP1 transcriptional target gene (Fig. 2B). In this case, the DNAJB9 mRNA was induced only about 1.5-fold, whereas in the corresponding ER stress model the DNAJB9 expression was higher (about 3–4-fold), suggesting that other transcriptional mechanisms may be responsible for this gene induction during UPR. The levels of XBP1s mRNA induction resulted in XBP1 protein levels (Fig. 2C) comparable to those observed after 6 h of ER stress-induced with 2.5 µg/ml Tm and about half of the protein levels observed with 50 nM Tg (Fig. 2 EF), despite the fact that both of these stressors induced XBP1s mRNA comparably (Fig. 1C). The exogenous XBP1 signals were independent of IRE1 activity since 4µ8C, a specific IRE1 activity inhibitor, had no effect on the induced XBP1s mRNA levels [52] (Fig. 2G). Furthermore, the XBP1 induction did not lead to accumulation of XBP1u mRNA (Fig. 2H). Notably, in the presence of doxycycline, the XBP1 protein was stably expressed up to a week (Supplemental Figure S2AB), showing a trend to accumulate after a prolonged time of induction. Finally, the doxycycline used for the induction of XBP1s did not induce ER stress (Supplemental Figure S2C) and siRNA knockdown of XBP1s mRNA induction with the inducible cell line dramatically reduced the XBP1s mRNA levels (Supplemental Figure S2D).
Fig. 2.
The HeLa-XBP1s cell line induced for 24 h accumulates A XBP1 (green) and B DNAJB9 (white) mRNA. The results from three independent experiments (n = 9) are plotted normalized to GAPDH and RPLP0 mRNA levels and expressed as a fold change over the noninduced cells. Error bars represent standard deviations. Significant changes (P value P < 0.05) are marked with an asterisk. The corresponding changes in XBP1 protein levels were evaluated by Western blot (C) normalized to total protein levels (D) and related noninduced control or evaluated by Western blot and to compared to ER stress induced by Tm (2.5 µg/ml, brown) Tg (50 nM, red) treatment for 6 h (E) and related to the no-stress control (F). *P < 0.05 was considered significant. G The exogenous XBP1s mRNA levels were independent IRE1 activity as shown with 4µ8C (20 µM), a specific IRE1 activity inhibitor. The yellow–green bars depict the conditions where XBP1s expression was induced in the presence of Tm and 4µ8C. Tm was used at 0.5 µg/ml concentration for 6 h (yellow). H The exogenous XBP1s expression does not lead to the accumulation of XBP1u mRNA (grey). The results from three independent experiments (n = 9) are plotted normalized to GAPDH and RPLP0 mRNA levels and expressed as a fold change over the noninduced cells. Error bars represent standard deviations. Significant changes (P value P < 0.05) are marked with an asterisk
In our analysis of XBP1‐affected factors, we obtained RNA samples from the HeLa-XBP1s cell line under control conditions (no induction) and after 24 h of induction, and subjected both to RNA‐seq analysis. Notably in these NGS analysis, inductions of all XBP1 isoform mRNAs were accessed as a one XBP1 gene change and in the range of fivefold and that is reflected by about 2 log2 fold change. The isoform-dedicated analysis, however, indicated that the XBP1s mRNA induction was about 20-fold. In this analysis, we focused only on genes that were specifically affected by induction by at least a log-fold (twofold change) and had a P value below 0.05.
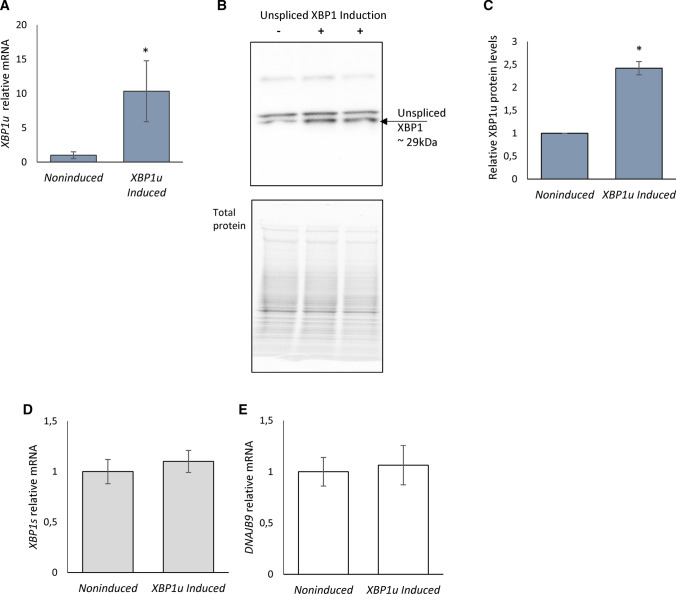
We were aware that by applying such “loose” selection parameters, however, would result in a large group of genes. Given this concern, we performed independent validations of the XBP1 predicted targets. Furthermore, given the large number of identified genes fulfilling this criterion could also result from the doxycycline treatment. As a control, therefore, the RNA-seq data obtained from a comparable inducible HeLa cell line expressing low levels of XBP1u (HeLa-XBP1u) were examined. These induced genes were omitted from further analysis. As previously mentioned, XBP1s mRNA results from ER stress-activated IRE1 splicing of a 26 nt unconventional intron in the coding region of unspliced XBP1 (XBP1u) that causes a frameshift. The XBP1 protein (~ 48 kDa) has the same N-terminus, but a longer and distinct C-terminus compared to the unspliced XBP1 protein (~ 29 kDa) [5, 53]. More importantly, the new C-terminus in XBP1 contains the transactivation domain [5, 53]. The induction levels of XBP1u mRNA in these cells were in the tenfold range that did result in unspliced XBP1 protein expression (Fig. 3A–C) but did not result in XBP1s mRNA accumulation (Fig. 3D) or any increase in XBP1 transcriptional activity (Fig. 3E) or protein (Supplemental Figure S1B). The unspliced XBP1 protein levels are in agreement with previous reports that this protein is rapidly degraded [54]. Furthermore, XBP1u induction was performed under no ER stress conditions to avoid the possibility of a negative impact of unspliced XBP1 protein on spliced XBP1 [55]. Given this, the RNA-seq data obtained from the induced HeLa-XBP1u cell line were an appropriate control for our experiments.
Fig. 3.
Upon 24-h induction, the HeLa-XBP1u cell line accumulates XBP1u mRNA. A The results from three independent experiments (n = 9) are plotted normalized to GAPDH and RPLP0 mRNA levels and expressed as a fold change over the noninduced cells. Error bars represent standard deviations. Significant changes (P value P < 0.05) are marked with an asterisk. The corresponding changes in unspliced XBP1 protein levels were evaluated by Western blot (B) normalized to total protein levels (C) and related noninduced control. *P < 0.05 was considered significant. The exogenous XBP1u expression does not lead to the accumulation of XBP1s (D) and DNAJB9 (E) mRNA. The results from three independent experiments (n = 9) are plotted normalized to GAPDH and RPLP0 mRNA levels and expressed as a fold change over the noninduced cells. Error bars represent standard deviations. Significant changes (P value P < 0.05) are marked with an asterisk
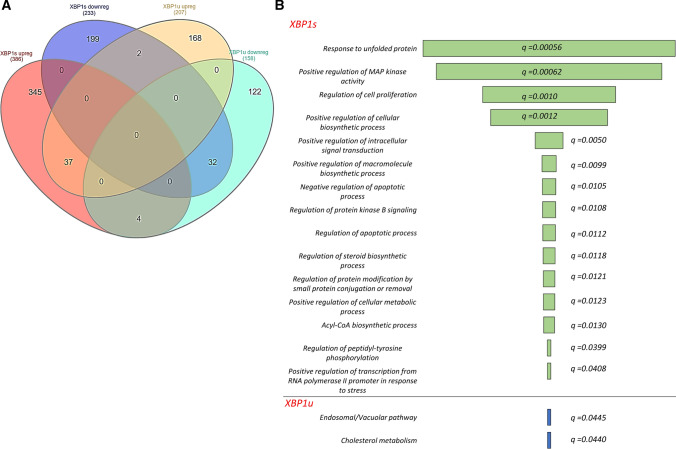
The overlap of the RNA-seq data obtained from induced spliced XBP1 and unspliced XBP1 expression systems is illustrated in a Venn diagram in Fig. 4A. The functional assignment of XBP1 modulated genes was performed with Enrichr Web server using a strict criterion (P value P < 0.05 and q value q < 0.05). As shown in Fig. 4B, this analysis of expression profiles specific for cells expressing XBP1u did not revealed any specific activation of ER stress, UPR signaling or apoptotic pathways. Nevertheless, some of the unspliced XBP1-related expression changes could be assigned to cholesterol metabolism and the endosomal pathway. Furthermore, analysis of genes that were common between spliced and unspliced XBP1 mRNA expression profiles did not result in any significant functional assignment. Notably, because of XBP1s induction, the mRNA of 345 and 199 genes were induced or reduced, respectively. Furthermore, this expression profile correlated well with changes related to the UPR that included protein folding, cellular proliferation, and negative regulation of apoptotic pathways (Fig. 4B).
Fig. 4.
The impact of the exogenic XBP1u and XBP1s induction on HeLa genome-wide cellular mRNA profiles and their potential functional consequences. A The Venn diagram [56] represents the general distribution of mRNAs that were significantly (P < 0.05) affected by XBP1s and XBP1u transgenes—Supplemental Table 2C. B Transcripts reduced and induced upon XBP1 induction are marked with dark blue and red, respectively, whereas mRNAs reduced and induced upon unspliced XBP1 induction are marked with light blue and yellow, respectively. The Gene Ontology assignment of the cellular functions of mRNAs potentially regulated by the spliced XBP1 (XBP1s) or unspliced XBP1 (XBP1u) as assigned by the Enrichr Web server—Supplemental Table 2D [40]. The green bar color depicts the q value less than 0.05. The longer bars have the lower q values
Given that XBP1’s impact on gene expression can result from both direct and indirect effects with transcription factors originating from the other UPR branches (PERK and ATF6), we decided to narrow our verification set to the genes that contained potential promoter regions for XBP1. To verify the potential direct role of XBP1 during the UPR, the identified gene locations were analyzed for the presence of XBP1-binding motifs (Fig. 5A). Our analysis was in HeLa cells, and we focused only on transcriptionally active chromatin regions.
Fig. 5.
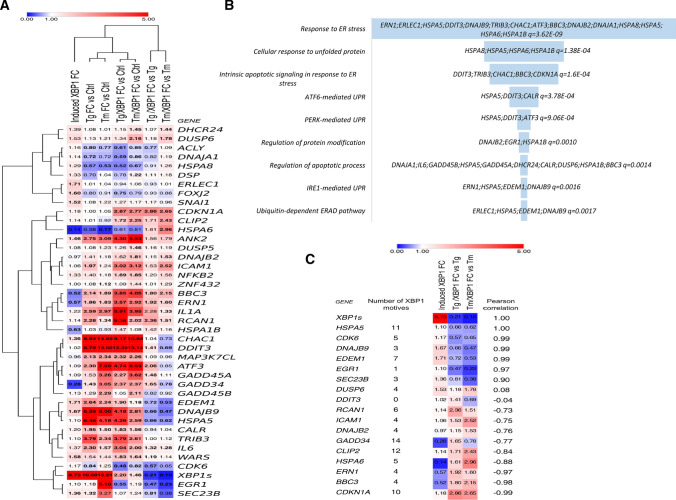
The heat map representing expression changes of all verified potentially XBP1-dependent genes identified in induced XBP1s expression experiments (Induced XBP1) and separately in silencing XBP1 during mild stress experiments in HeLa cells (Tg/XBP1s and Tm/XBP1s). A The logo of XBP1-binding motif M00402 (consensus: GACGTGkCmtww, where k = G or T; m = A or C; w = A or T) from the Hocomoco v. 9 motif library. B Heat maps were generated and organized according to hierarchical clustering of expression changes with the Morpheus software (Morpheus, https://software.broadinstitute.org/morpheus). C FC depicts “fold change”, Tm—tunicamycin 0.5 µg/ml induced ER stress for 6 h, Tg—thapsigargin 2.5 nM induced ER stress for 6 h, whereas Tm/XBP1 S and Tg/XBP1 S depict XBP1 silencing in each of these stress models. D The Enrichr Web server (https://amp.pharm.mssm.edu/Enrichr/) was applied to assign the qPCR results into the “Gene Ontology Biological Process” categories with a selection based on a q value q < 0.05. Heat map representing the mRNAs that display the most correlated changes with XBP1s mRNA levels as calculated by Pearson correlations
This resulted in the selection of 58 genes potentially directly regulated by spliced XBP1 that were then validated in 3 independent biological replicates using 96-well qPCR arrays (Supplemental Table 2AB). The expression changes were accessed following XBP1 overexpression as well as silencing with specific siRNA against XBP1 during 6 h of mild ER stress (induced with Tm or Tg). Furthermore, in our validation set, we included HSPA5 (BiP) and DDIT3 (CHOP) mRNAs that were previously also reported to be regulated by XBP1 [57]. Interestingly, the CHOP region did not contain any potential XBP1-binding motifs, whereas in BiP there were 11 sites. This approach identified significant changes in 40 of the transcripts (Table 1).
Table 1.
Summary of mRNA changes observed during XBP1s induction, ER stress-induced with Tg or Tm, and Tg or Tm ER stress together with XBP1 silencing (Tg and Tm/XBP1 S)
| Gene | Number of XBP1 motifs | Induced XBP1s FC | Tg FC vs Ctrl | Tg/XBP1 S FC vs Ctrl | Tg /XBP1 S FC vs Tg | Tm FC vs Ctrl | Tm/XBP1 S FC vs Ctrl | Tm/XBP1 S FC vs Tm |
|---|---|---|---|---|---|---|---|---|
| ACLY | 1 | 1.15 | 0.80 | 0.61 | 0.77 | 0.77 | 0.85 | 1.09 |
| ANK2 | 1 | 1.45 | 2.75 | 4.30 | 1.56 | 3.09 | 5.53 | 1.79 |
| ATF3 | 6 | 1.09 | 2.30 | 4.74 | 2.06 | 7.69 | 6.53 | 0.85 |
| BBC3 | 4 | 0.51 | 2.14 | 3.85 | 1.80 | 1.89 | 4.05 | 2.15 |
| CALR | 14 | 1.19 | 1.95 | 1.83 | 0.94 | 1.50 | 1.56 | 1.04 |
| CDK6 | 5 | 1.17 | 0.84 | 0.48 | 0.57 | 1.25 | 0.82 | 0.65 |
| CDKN1A | 10 | 1.18 | 1.00 | 2.87 | 2.86 | 1.05 | 2.77 | 2.65 |
| CHAC1 | 1 | 1.36 | 8.84 | 9.17 | 1.04 | 14.85 | 10.84 | 0.73 |
| DDIT3 | 0 | 1.02 | 8.78 | 12.38 | 1.41 | 19.00 | 13.14 | 0.69 |
| CLIP2 | 12 | 1.13 | 1.01 | 1.72 | 1.71 | 0.92 | 2.25 | 2.43 |
| DHCR24 | 2 | 1.39 | 1.08 | 1.15 | 1.07 | 1.01 | 1.45 | 1.44 |
| DNAJA1 | 4 | 1.13 | 0.72 | 0.59 | 0.82 | 0.72 | 0.86 | 1.19 |
| DNAJB2 | 4 | 0.96 | 1.41 | 1.62 | 1.15 | 1.18 | 1.81 | 1.53 |
| DNAJB9 | 3 | 1.67 | 6.36 | 4.18 | 0.66 | 6.00 | 2.81 | 0.47 |
| DSP | 2 | 1.33 | 0.70 | 0.78 | 1.11 | 1.04 | 1.22 | 1.18 |
| DUSP5 | 12 | 1.08 | 1.08 | 1.26 | 1.16 | 1.23 | 1.46 | 1.19 |
| DUSP6 | 4 | 1.52 | 1.13 | 1.34 | 1.18 | 1.21 | 2.16 | 1.78 |
| EDEM1 | 7 | 1.71 | 2.64 | 1.90 | 0.72 | 2.24 | 1.18 | 0.53 |
| EGR1 | 1 | 1.09 | 1.18 | 0.55 | 0.47 | 5.10 | 1.19 | 0.23 |
| ERLEC1 | 6 | 1.70 | 1.01 | 0.94 | 0.93 | 1.04 | 1.06 | 1.01 |
| ERN1 | 4 | 0.56 | 1.86 | 3.57 | 1.92 | 1.83 | 2.92 | 1.60 |
| FOXJ2 | 4 | 1.59 | 0.80 | 0.75 | 0.93 | 0.91 | 0.79 | 0.86 |
| GADD45A | 2 | 1.08 | 1.53 | 2.27 | 1.48 | 3.26 | 3.62 | 1.11 |
| GADD45B | 2 | 1.12 | 1.29 | 1.05 | 0.82 | 2.29 | 2.11 | 0.92 |
| HSPA1B | 13 | 0.63 | 1.03 | 1.47 | 1.42 | 0.93 | 1.08 | 1.16 |
| HSPA5 | 11 | 1.10 | 6.46 | 4.26 | 0.66 | 4.18 | 2.59 | 0.62 |
| HSPA6 | 5 | 0.13 | 0.38 | 0.61 | 1.61 | 0.17 | 0.51 | 2.96 |
| HSPA8 | 1 | 1.29 | 0.57 | 0.52 | 0.91 | 0.53 | 0.67 | 1.26 |
| ICAM1 | 4 | 1.06 | 1.97 | 3.02 | 1.53 | 1.24 | 3.12 | 2.52 |
| IL1A | 3 | 1.21 | 2.59 | 5.91 | 2.28 | 2.97 | 3.95 | 1.33 |
| IL6 | 4 | 1.37 | 2.30 | 3.04 | 1.32 | 1.57 | 2.00 | 1.28 |
| MAP3K7CL | 4 | 0.95 | 2.13 | 2.32 | 1.09 | 2.34 | 2.26 | 0.96 |
| NFKB2 | 11 | 1.32 | 1.40 | 1.69 | 1.20 | 1.18 | 1.85 | 1.56 |
| RCAN1 | 6 | 1.13 | 2.28 | 5.38 | 2.36 | 1.34 | 2.02 | 1.51 |
| SEC23B | 3 | 1.36 | 1.32 | 1.07 | 0.81 | 3.27 | 1.24 | 0.38 |
| SNAI1 | 5 | 1.51 | 1.08 | 1.27 | 1.17 | 1.22 | 1.17 | 0.96 |
| TRIB3 | 5 | 1.10 | 3.79 | 3.79 | 1.00 | 2.34 | 2.61 | 1.12 |
| WARS | 3 | 1.58 | 1.54 | 1.83 | 1.19 | 1.44 | 1.64 | 1.14 |
| GADD34 | 14 | 0.27 | 1.43 | 2.37 | 1.65 | 3.05 | 2.37 | 0.78 |
| ZNF432 | 3 | 1.01 | 1.08 | 1.09 | 1.01 | 1.12 | 1.44 | 1.29 |
| XBP1s | 8.73 | 10.60 | 2.20 | 0.21 | 15.21 | 1.46 | 0.10 |
As shown in Table 1 and Supplemental Table 2A, we observed that following XBP1s induction, 8 mRNAs were significantly induced that included ankyrin 2 (ANK2), glutathione-specific gamma-glutamylcyclotransferase 1 (CHAC1), ER-Resident Protein ERdj4, DNAJB9, ER degradation-enhancing alpha-mannosidase-like protein 1 (EDEM1), ER lectin 1 (ERLEC1), forkhead box J2 (FOXJ2), SEC23B, and interferon-induced protein 53 (WARS). Notably, ERLEC1, WARS and SEC23B have been previously identified as XBP1-dependent genes in MCF-7 and HEK-293 cells [58, 59], whereas CHAC1 has been identified recently as an important regulator of UPR-associated ferroptosis [60]. Furthermore, 5 mRNAs were significantly reduced upon XBP1s induction and included (1) IRE1 (ERN1), (2) growth arrest and DNA damage-inducible protein (GADD34) [61], a crucial PERK pathway regulator, (3) PUMA (BBC3), an important ER stress-related proapoptotic factor [62]), (4) heat shock protein family A (Hsp70) member 1B (HSPA1B), and (5) heat shock protein family A (Hsp70) member 6 (HSPA6).
In the parallel XBP1-silencing experiments (Table 1 and Supplemental Table 2B), we observed that mRNA levels of well-known XBP1 targets such as DNAJB9 and EDEM1 [44, 57] as well as interleukin 6—IL6 [63] were significantly induced by both stressors and reduced upon XBP1 silencing. In addition, as previously reported, XBP1 silencing had a limited effect on HSPA5 (BiP) expression during ER stress [57]. Importantly, we also observed that ERN1, BBC3, and regulator of calcineurin 1 (RCAN1) were induced by both stressors and their levels were even higher after XBP1 silencing. Notably, we recently identified RCAN1 as an important prosurvival regulator of ER stress-induced cell fate decisions [21].
We also noted the induction of ER stress proapoptotic cyclin-dependent kinase inhibitor 1A (CDKN1A (p21)) and cytoplasmic linker 2 (CLIP2) expression upon XBP1 silencing. Interestingly, CLIP2 has been associated as a key gene for diabetes mellitus development [64]. The PERK-dependent cyclin-dependent kinase 6 (CDK6 [65]) expression was significantly reduced only by Tg and its levels became even lower upon XBP1 silencing during both Tm and Tg treatments. ANK2, ATF3 and GADD45A mRNAs were induced by both stressors and their levels were further elevated upon XBP1 silencing during Tg treatment. Notably, the UPR-induced proapoptotic GADD45A [21] has been proposed as an XBP1-dependent gene [57]. Furthermore, HSPA6 mRNA levels were significantly induced upon XBP1 silencing in Tm- and Tg-treated cells, but the expression of the HSPA6 gene was exceptionally low in HeLa cells, however, and therefore, this observation will require further verification.
We also observed that XBP1 silencing resulted in lower expression of the transcription factor involved in ER stress-related regulation of cell cycle progression early growth response 1—EGR1 [66] in both stress models. In addition, XBP1 silencing induced the anti-apoptotic dual-specificity phosphatase 6—DUSP6 gene [67] expression in Tm-treated cells. Notably, DDIT3 (CHOP) and GADD34 levels were significantly induced in both ER stress models, and further increased upon XBP1 silencing in Tg-treated cells whereas they were reduced in Tm-treated cells (Fig. 5A). Furthermore, we did not find any significant correlation between the potential number of XBP1-binding motifs and their connection to actual functional effects. Importantly, the functional analysis of all these verified XBP1-related genes indicated that they were almost exclusively connected to ER stress outcomes including all three UPR branches and ERAD, and the regulation of apoptotic processes (Fig. 5B).
Despite some discrepancies between the data obtained from the XBP1 induction and silencing in different ER stress models, we were able to define a group of genes whose expression was XBP1 dependent, and their expression was significantly affected in at least two out of three independent experimental approaches (XBP1 induction; silencing XBP1s during Tm treatment; or silencing XBP1s during Tg treatment) (Table 1). As shown in Fig. 5C, this resulted in a final selection of a group of 17 genes. In this gene set, the expression of six genes (HSPA5, CDK6, DNAJB9, EDEM1, EGR1, and SEC23B) was positively correlated with the XBP1 levels.
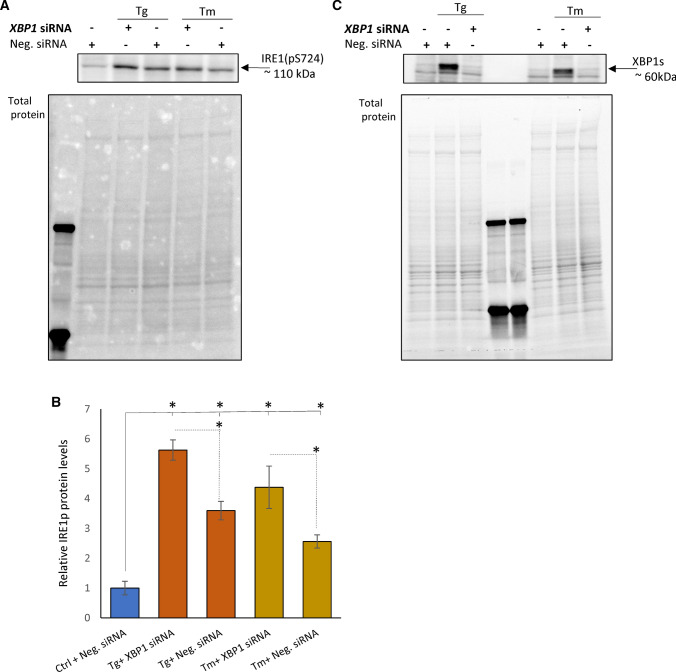
Notably, we also identified three genes, BBC3, ERN1 and HSPA6, whose expression was negatively correlated with the XBP1 expression in both ER stress models. Given that XBP1-mediated attenuation of ERN1 expression suggests a novel negative-feedback regulatory loop between XBP1 and IRE1, we also tested if XBP1-related reduction of ERN1 mRNA levels were reflected by IRE1 protein changes in Tm- and Tg-treated cells. In HeLa cells exposed to both Tm and Tg, the phosphorylated IRE1 protein levels were increased upon XBP1 silencing as shown in Fig. 6.
Fig. 6.
The XBP1s silencing is accompanied by reduced IRE1p protein levels in Tg- and Tm-treated HeLa cells. A Representative changes in IRE1p protein levels in HeLa cells treated with Tg (50 nM) and Tm (2.5 µg/ml) and Tg for 6 h in the presence or absence of a specific siRNA against XBP1 as evaluated by Western blot and B normalized to total protein levels and related to the no-stress control. The experiments were repeated three times. Error bars represent standard deviations and *P < 0.05 was considered significant. C The corresponding changes in XBP1 protein levels were evaluated by Western blot
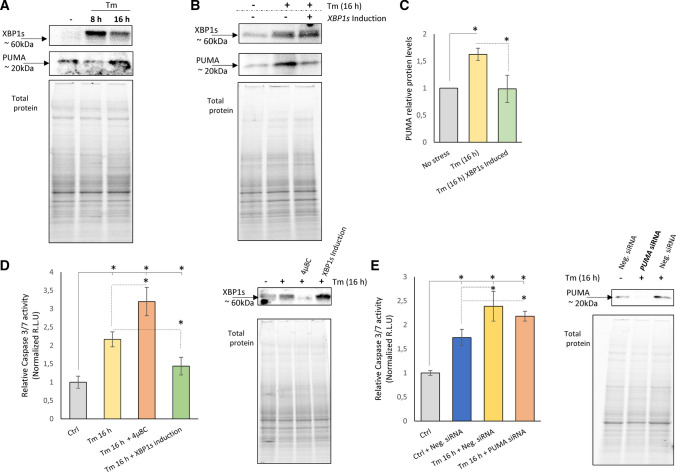
Given that BBC3-encoded PUMA protein was reported as an important apoptotic factor during UPR, we also tested if XBP1-related reduction of BBC3 mRNA levels were reflected by PUMA protein changes. As shown in Fig. 7A, in HeLa cells exposed to Tm (2.5 µg/ml), PUMA protein levels were negatively correlated with XBP1s expression and reduced after 8 h when the XBP1 expression is maximal. Furthermore, when XBP1 is reduced after 16 h of treatment, the PUMA levels rise. To support the idea of the negative correlation, we performed an experiment to test the idea of maintaining XBP1s levels by keeping transgenic XBP1s levels high at 16 h during Tm treatment to see the effect on PUMA expression (Fig. 7B, C). The results clearly show that higher XBP1s levels prevented PUMA accumulation during Tm ER stress at 16 h, confirming XPB1s repression on PUMA expression.
Fig. 7.
The 24 h induction of XBP1s is accompanied by reduced PUMA protein levels and lower Caspase 3/7 activity in Tm-treated HeLa cells. A Representative changes in XBP1 and PUMA protein levels in HeLa cells treated with Tm (2.5 µg/ml) for 8 and 16 h as evaluated by Western blot. B The changes in XBP1 and PUMA protein levels in XBP1s-induced and -noninduced HeLa cells treated with Tm (2.5 µg/ml) 16 h were evaluated by Western blot (C) normalized to total protein levels and related to the noninduced, no-stress control, and *P < 0.05 was considered significant. XBP1s-induced (24 h) and -noninduced HeLa cells were treated with Tm (2.5 µg/ml) for 16 h in the presence or absence of 4µ8C (20 µM), a specific IRE1 activity inhibitor. D The caspase 3/7 activity was monitored by luminescence and expressed in relative light units (RLU). Cells for each time point were seeded in triplicate, and the experiments were repeated three times. Error bars represent standard deviations and *P < 0.05 was considered significant. The corresponding changes in XBP1 protein levels were evaluated by Western blot. E HeLa cells were treated with Tm (2.5 µg/ml) for 16 h in the presence or absence of a specific siRNA against BBC3 (PUMA). The caspase 3/7 activity was monitored by luminescence and expressed in RLU. Cells for each time point were seeded in triplicate, and the experiments were repeated three times. Error bars represent standard deviations, *P < 0.05 was considered significant. The corresponding changes in PUMA protein levels were evaluated by Western blot
To test XBP1’s impact on prosurvival UPR activity, we subjected HeLa cells to Tm-induced ER stress for 16 h and measured caspase 3/7 activity. We directly compared Tm-induced ER stress to Tm-induced stress in the presence of IRE1 inhibitor (4µ8C) which would prevent IRE1 RIDD activity during XBP1s mRNA formation. We found that impairing IRE1 activity accelerates cell death in ER-stressed cells, whereas XBP1s expression correlated with a clear prosurvival effect (Fig. 7D). It has been previously reported that upon XBP1 deficiency, IRE1 switches to perform RIDD [68]. Nevertheless, given that the PUMA mRNA sequence does not contain the IRE1-required consensus sequence (CUGCAG) [69], it seems unlikely that this transcript is degraded by RIDD. Furthermore, inhibiting this IRE1 activity can also prevent death receptor 5 (DR5) mRNA degradation and thus apoptosis engagement via caspase-8 [16]. Finally, since PUMA was reported to stimulate the intrinsic pathway of apoptosis [70, 71], we performed an analogous experiment and siRNA silenced PUMA (Fig. 7E). Interestingly, we did not observe any significant changes in caspase 3/7 activity related to PUMA depletion (Fig. 7E). PUMA or BH3 protein (Bid) can separately activate Bax [72, 73], and this could potentially lead to mitochondrial outer membrane permeability and apoptosis [74]. In the absence of PUMA, other BH3-only proteins (Bid and Bim) that are PERK dependent [75, 76] could still be efficiently activating caspase 3/7.
To generalize the findings to other cell types, we followed consequences of XBP1 silencing on both ERN1 and BBC3 mRNA levels in several other human cell lines exposed to ER stress that included Schwann Cells (SNF96.2), epidermal keratinocytes (HaCaT), umbilical vein endothelial cells (HUVEC) and bronchial epithelial cells (16HBE14o-). As shown in Supplemental Figure S3, BBC3 expression was significantly induced upon XBP1 silencing in all of these experimental models. Furthermore, ERN1 levels were modestly but significantly higher in the absence of XBP1s in all Tg-treated cell lines as well in HaCaTs and HUVECs treated with Tm (Supplemental Figure S3). These data support the general repressive role of XBP1 on ERN1 levels in a diverse group of human cell lines.
In summary, we identified eight genes whose expression was ER stress model-dependent and/or not affected by XBP1 induction alone: DUSP6, DDIT3, GADD34, RCAN1, ICAM1, DNAJB2, CLIP2 and CDKN1A. This set contains mainly genes that can be attributed to the PERK pathway activity (DDIT3, GADD34) and cell survival (DDIT3, GADD34, RCAN1 and CDKN1A), suggesting that their levels result from both XBP1 activity and other UPR pathways, including the PERK pathway.
Discussion
The IRE1-dependent pathway is the most ancient and conserved branch of UPR [25] and serves as the molecular timer and executor for ER stress-related cell death [77–79]. It is, therefore, not surprising that the role of its downstream transcription factor, XBP1, has been extensively studied [44, 49, 57–59, 80, 81]. The transcriptional targets of XBP1 are well identified and consist of ERAD components (EDEM1), chaperones (HSPA5, DNAJB9, and DNAJC3), and vesicle-trafficking components (SEC23B) [57–59, 80]. Furthermore, genes involved in the inflammatory responses (including IL6) [59], as well as genes not related to UPR pathways including adipocyte and myogenic differentiation (C/EBP and MIST1) have been proposed as tissue-dependent XBP1 transcriptional targets [82]. Consequently, the XBP1 has been widely accepted as an adaptive component of UPR that is responsible for facilitating protein folding and ER expansion.
Despite these advances, determining the global network of XBP1 transcriptional activity and its consequences on cell fate decisions remains less clear. Part of the problem in defining XBP1’s role involves analyzing its function in models that are often based on one type of ER stressor or are utilizing high levels of overexpression of XBP1. Furthermore, the studies often focus on a small subset of induced genes. In our approach, we exploited inducible cell lines capable of XBP1(s) and XBP1(u) lower expression levels and two models of pharmacological ER stress induction, glycosylation inhibition and disruption of ER calcium homeostasis. Using this system, we were able to demonstrate the induction of the main UPR mediators including HSPA5 (BIP), IRE1, XBP1s and CHOP. Notably, the levels of XBP1s mRNA obtained during cell line induction were on the low end of those observed in our ER stress models.
To follow the XBP1-related changes in transcriptome, we performed next-generation sequencing profiling in HeLa cells with the induced expression of XBP1s or XBP1u and focused on changes in gene expression related mainly to prosurvival and apoptotic UPR signaling pathways. Notably XBP1u expression did not result in any changes in which we could clearly assign to these activities. The unspliced XBP1 protein has been shown to be rapidly degraded and maintained at low levels and the XBP1u transgene transcript could not be processed to functional XBP1, and our results are in good agreement with previous reports [54].
Nevertheless, the XBP1s transgene induction resulted in wide changes of expression profiles of the genes involved in the UPR including DNAJB9 and EDEM1, stress responses, and regulation of cellular biosynthetic and apoptotic responses (Fig. 4). The results confirmed XBP1’s role as a crucial UPR mediator and potentially defined a large set of genes which resulted from XBP1 transcriptional activity. Importantly, following XBP1s induction, we did not observe some of the classical UPR activation genes since both BIP and CHOP mRNA levels remained relatively constant, and therefore, it is quite plausible that the observed transcriptomic changes often seen did not result specifically from XBP1 activity.
To further test this hypothesis, we selected a set of 58 genes (Supplemental Table 2) whose genomic locations were in the proximity of XBP1-binding motifs and DDIT3 and HSPA5 and validated them independently. The results revealed XBP1-related changes in 40 transcripts, most of which were related to the UPR stress responses and regulation of apoptosis (Fig. 5). The number of potential XBP1-binding motifs in the promoter regions of genes did not correlate well with their transcript expression levels, suggesting that other requirements such as the relative position from the transcriptional start site or the presence of other potential binding motifs (such as ATF6 for example [57, 59]) may be necessary to achieve efficient transcription. However, the effects of the number of transcription factor-binding motifs on expression are only observed for some transcription factors [83]. Nevertheless, homotypic clusters of motifs for some transcription factors are known to potentiate the effects of these factors on expression for some genes [84]. Therefore, we also tested our gene set for a correlation between XBP1 motif clusters and fold changes, but no significant effects were observed. Finally, taking into account extreme complexity and dynamic course of UPR signaling, it is important to note that XBP1 cooperates with the other arms of UPR to modulate transcriptomic profiles, rather than being a master regulator of gene expression. Nevertheless, most of the preselected genes displayed expression patterns that positively correlated with the elevated expression levels of XBP1.
Despite our best efforts to find the optimal mild ER stress conditions, Tm and Tg still have different effects on the course of the UPR signaling pathways. For example, the Tm-treated cells were more prone to apoptosis (Fig. 1). We suggest that these differences between Tm and Tg could result in differences in the expression profiles of the cell fate decisions and the PERK-induced genes that include DDIT3, GADD34, ATF3 and RCAN1. In general, silencing XBP1s during Tg treatment resulted in higher expression of DDIT3, GADD34, ATF3 and RCAN1, whereas a complete lack of XBP1 during Tm treatment resulted in reduced expression of these genes. Since all these transcripts are closely related to the PERK branch of UPR, there exists the intriguing possibility of XBP1-mediated crosstalk between this pathway and the IRE1 branch that determines cell fate decisions [85, 86]. This hypothesis is also supported by the observation that inhibiting the PERK arm of the UPR has a different impact on XBP1s mRNA levels in cells treated with Tm when compared to cells treated with Tg (Supplemental Figure S4). All these genes are also potentially regulated by the other UPR-related transcription factors including CHOP and ATF6 (Supplemental Table S3). Hence, our results suggest that the gene expression modulations by XBP1 can also be influenced by other UPR pathways. This hypothesis, however, and the related mechanisms controlling this will obviously require further study.
Despite the differences between the data obtained from the XBP1 induction and silencing in different ER stress models, we were able to define a group of genes whose expression was clearly XBP1 dependent (Fig. 5C). The expression of HSPA5, CDK6, DNAJB9, EDEM1, EGR1, and SEC23B was clearly positively correlated with the XBP1 levels. In all these genes, their expression was not only induced along with XBP1s induction, but it was also reduced when XBP1 was silenced in both stress models. However, HSPA5, EGR1 and CDK6 only correlate positively in our ER stress-induced conditions and not in an XBP1 overexpression model, suggesting that ATF6 may also be required for their expression induction [57, 59, 87].
Here, we have identified the 3 genes, BBC3 (PUMA), ERN1 (IRE1) and HSPA6 (BiP), whose expression was clearly negatively correlated with the XBP1, and their levels were reduced upon XBP1 induction and induced upon XBP1 silencing in both stress models. In the case of HSPA6, the expression levels in HeLa cells were extremely low, and therefore, may represent a cell-type-specific effect. The expression levels for the BBC3 and ERN1, however, were more robust and clearly indicated this reversal effect during siRNA silencing of XBP1s. Obviously, both BBC3 (PUMA) and ERN1 (IRE1) are crucial UPR regulators, and their XBP1-dependent repression reveals a novel regulatory mechanism in the UPR.
The ability of XBP1 to attenuate ERN1 expression and thus reduce IRE1 activity identifies novel negative-feedback regulatory loop between XBP1 and IRE1. Although this observation and potential consequences of such a regulation requires further verification, it is clear that other UPR branches have negative effects on IRE1 activity that include PERK [79] and ATF6 [88]. Nevertheless, the implications of this during ER stress are that XBP1 controls its own levels and cell fate by limiting IRE1 activity.
Furthermore, numerous reports proposed that PUMA as an important and PERK-related contributor to UPR-related cell death [89–91], since it can inhibit all prosurvival Bcl-2 family members and activate the intrinsic pathway of apoptosis [70, 71]. Hence, it is plausible that by preventing BBC3 (PUMA) accumulation, XBP1 modulates the extent of intrinsic apoptotic signaling and thus contributes to the adaptive UPR response. Although our data clearly show that XBP1s induction leads to a reduction of PUMA protein levels, and at the same time reduces the extent of apoptotic UPR signaling (Fig. 6), evaluating the exact contribution of XBP1’s role will require further study.
Taken together, our approach not only confirmed previously known XBP1 roles during the UPR [24], but also identified novel targets of this transcription factor that regulate the mechanisms of the UPR cell fate decisions. Having said that, our experimental approaches are limited by the complexity of UPR signaling pathways and all the other factors involved in this process, and therefore, further studies will be necessary to understand the complex relationships between XBP1 and all of its targets. One of the limitations here is that the time (at least 24 h) required to obtain sufficient XBP1 expression prevents the model from reproducing the acute stress response since IRE1 is transiently activated and rapidly downregulated by the PERK arm of the stress response [78].
In summary, the studies presented here have identified that XBP1 can repress expression of two key players involved in UPR, IRE1 and PUMA, and further studies are obviously required to decipher the molecular mechanisms underlying this observation.
The results from three independent experiments are normalized to GAPDH and RPLP0 mRNA levels and expressed as a fold change (FC) over the respective controls. For induction experiments during ER stress, normalization was performed against noninduced and non-ER stress exposed cells, whereas for silencing experiments, the noninduced negative control siRNA-transfected cells that were not exposed to ER stress were used as a control. Significant changes (P value P < 0.05) are marked in grey. The Supplemental Table 2AB contains all the individual values.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Anna Janszak-Jasiecka and Marcin Serocki for their kind assistance.
Abbreviations
- ACLY
ATP citrate lyase
- ANK2
Ankyrin 2
- ATF3
Activating transcription factor 3
- ATF6
Activating transcription factor 6
- BBC3
BCL2-Binding component 3
- BiP
Binding immunoglobulin protein (glucose-regulated protein 78, a.k.a. HSPA5)
- BSA
Bovine serum albumin
- CALR
Calreticulin
- CDK6
Cyclin-dependent kinase 6
- CDKN1A
Cyclin-dependent kinase inhibitor 1A (a.k.a p21)
- CEBPB
CCAAT-enhancer-binding protein beta (a.k.a C/EBP)
- CHAC1
ChaC glutathione-specific gamma-glutamylcyclotransferase 1
- CHOP
CCAAT-enhancer-binding protein homologous protein (a.k.a DDIT3)
- CLIP2
Cytoplasmic linker 2
- CTRL
Control
- DDIT3
DNA damage-inducible transcript 3 (a.k.a CHOP)
- DHCR24
24-Dehydrocholesterol reductase
- DMSO
Dimethyl sulfoxide
- DNAJA1
DnaJ heat shock protein family (Hsp40) member A1
- DNAJB2
DnaJ heat shock protein family (Hsp40) member B2
- DNAJB9
DnaJ heat shock protein family (Hsp40) member B9 (a.k.a ERdj4)
- DNAJC3
DnaJ heat shock protein family (Hsp40) member C3
- DSP
Desmoplakin
- DUSP6
Dual-specificity phosphatase 6
- ECL
Enhanced chemiluminescence
- EDEM1
ER degradation-enhancing alpha-mannosidase-like protein 1
- EGR1
Early growth response 1
- eIF2α
Eukaryotic translation initiation factor 2a
- ER
Endoplasmic reticulum
- ERAD
Endoplasmic reticulum-associated degradation
- ERLEC1
ER lectin 1
- ERN1
Endoplasmic reticulum to nucleus signaling 1 (a.k.a. IRE1)
- FOXJ2
Forkhead box J2
- GADD34
Growth arrest and DNA damage-inducible protein (a.k.a PPP1R15A)
- GADD45A
Growth arrest and DNA damage-inducible alpha
- GADD45B
Growth arrest and DNA damage-inducible beta
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- GEO
Gene expression omnibus
- HEK-293 cells
Human embryonic kidney cells
- HeLa
Human cervix adenocarcinoma cells
- HSPA1B
Heat shock protein family A (Hsp70) member 1B
- HSPA5
Heat shock protein family A (Hsp70) member 5, (a.k.a. BIP)
- HSPA6
Heat shock protein family (Hsp70) member 6
- HSPA8
Heat shock protein family A (Hsp70) member 8
- HUVECs
Human primary endothelial cells
- ICAM1
Intercellular adhesion molecule 1
- IL1A
Interleukin 1α
- IL6
Interleukin 6
- IRE1
Inositol-requiring protein 1
- MAP3K7
Mitogen-Activated Protein Kinase Kinase Kinase 7
- MCF-7
Human breast cancer cell line
- MIST1
Basic helix–loop–helix family member A15
- NFKB2
Nuclear factor kappa B subunit 2
- NGS
Next-generation sequencing
- PBS
Phosphate-buffered saline
- PERK
Protein kinase RNA-like ER kinase
- PUMA
BCL2-binding component 3 (a.k.a. BBC3)
- qPCR
Quantitative polymerase chain reaction
- qRT-PCR
Quantitative real-time PCR
- RLU
Relative light units
- RCAN1
Regulator of calcineurin 1
- RPLP0
Neutral ribosomal phosphoprotein P0
- RTCA
Real-time cell analysis
- SD
Standard deviations
- SEC23B
Coat complex II component
- SERCA
Sarco/endoplasmic reticulum Ca2 + ATPase
- siRNA
Small interfering RNA
- SNAI1
Snail family transcriptional repressor 1
- Tg
Thapsigargin
- Tm
Tunicamycin
- UPR
Unfolded protein response
- WARS
Tryptophanyl-tRNA synthetase
- XBP1
X-box-binding protein 1
- XBP1s
Spliced X-box-binding protein 1
- XBP1u
Unspliced form of XBP1
- ZNF432
Zinc finger protein 432
Author contributions
Conceived and designed the experiments: RB. Performed the experiments: MG, AS, and SB. Analyzed the data: RB, JFC, ACJK, DKC, PM, and MD. Contributed reagents/materials/resources: RB. Wrote the paper: RB and JFC. All the authors read the final version of the manuscript.
Funding
This research was funded by National Science Center “OPUS” Program under contract UMO-2020/37/B/NZ3/00861 to RB. J.F.C. was funded by an NIH P30 DK072482 and a Research Development Program Grant from the Cystic Fibrosis Foundation.
Data availability
Deep sequencing data were deposited in Gene Expression Omnibus (GEO) at accession number: GSE160416.
Declarations
Conflict of interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data or in the writing of the manuscript or in the decision to publish the results.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Berridge MJ. The endoplasmic reticulum: a multifunctional signaling organelle. Cell Calcium. 2002;32:235–249. doi: 10.1016/S0143416002001823. [DOI] [PubMed] [Google Scholar]
- 2.Urra H, Dufey E, Lisbona F, Rojas-Rivera D, Hetz C. When ER stress reaches a dead end. Biochem Biophys Acta. 2013;1833:3507–3517. doi: 10.1016/j.bbamcr.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Schroder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 4.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochem Biophys Acta. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 6.Back SH, Lee K, Vink E, Kaufman RJ. Cytoplasmic IRE1alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J Biol Chem. 2006;281:18691–18706. doi: 10.1074/jbc.M602030200. [DOI] [PubMed] [Google Scholar]
- 7.Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uemura A, Oku M, Mori K, Yoshida H. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J Cell Sci. 2009;122:2877–2886. doi: 10.1242/jcs.040584. [DOI] [PubMed] [Google Scholar]
- 9.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 10.Karagoz GE, Acosta-Alvear D, Walter P. The unfolded protein response: detecting and responding to fluctuations in the protein-folding capacity of the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2019 doi: 10.7554/eLife.43036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 12.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 13.Hitomi J, Katayama T, Eguchi Y, Kudo T, Taniguchi M, Koyama Y, Manabe T, Yamagishi S, Bando Y, Imaizumi K, Tsujimoto Y, Tohyama M. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J Cell Biol. 2004;165:347–356. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Lee B, Lee AS. Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J Biol Chem. 2006;281:7260–7270. doi: 10.1074/jbc.M509868200. [DOI] [PubMed] [Google Scholar]
- 15.Lam M, Marsters SA, Ashkenazi A, Walter P. Misfolded proteins bind and activate death receptor 5 to trigger apoptosis during unresolved endoplasmic reticulum stress. Elife. 2020 doi: 10.7554/eLife.522919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 19.Hetz C, Chevet E, Oakes SA. Proteostasis control by the unfolded protein response. Nat Cell Biol. 2015;17:829–838. doi: 10.1038/ncb3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shore GC, Papa FR, Oakes SA. Signaling cell death from the endoplasmic reticulum stress response. Curr Opin Cell Biol. 2011;23:143–149. doi: 10.1016/j.ceb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartoszewski R, Gebert M, Janaszak-Jasiecka A, Cabaj A, Kroliczewski J, Bartoszewska S, Sobolewska A, Crossman DK, Ochocka R, Kamysz W, Kalinowski L, Dabrowski M, Collawn JF. Genome-wide mRNA profiling identifies RCAN1 and GADD45A as regulators of the transitional switch from survival to apoptosis during ER stress. FEBS J. 2020;287:2923–2947. doi: 10.1111/febs.15195. [DOI] [PubMed] [Google Scholar]
- 22.Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. Plos Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karam R, Lou CH, Kroeger H, Huang L, Lin JH, Wilkinson MF. The unfolded protein response is shaped by the NMD pathway. Embo Rep. 2015;16:599–609. doi: 10.15252/embr.201439696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Zhang C, Wang A. Divergence and conservation of the major UPR Branch IRE1-bZIP signaling pathway across eukaryotes. Sci Rep. 2016;6:27362. doi: 10.1038/srep27362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Qi L. SUMO modification regulates the transcriptional activity of XBP1. Biochem J. 2010;429:95–102. doi: 10.1042/BJ20100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Sun C, Zhou Y, Lee J, Gokalp D, Herrema H, Park SW, Davis RJ, Ozcan U. p38 MAPK-mediated regulation of Xbp1s is crucial for glucose homeostasis. Nat Med. 2011;17:1251–1260. doi: 10.1038/nm.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang FM, Chen YJ, Ouyang HJ. Regulation of unfolded protein response modulator XBP1s by acetylation and deacetylation. Biochem J. 2011;433:245–252. doi: 10.1042/BJ20101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- 30.Majumder M, Huang C, Snider MD, Komar AA, Tanaka J, Kaufman RJ, Krokowski D, Hatzoglou M. A novel feedback loop regulates the response to endoplasmic reticulum stress via the cooperation of cytoplasmic splicing and mRNA translation. Mol Cell Biol. 2012;32:992–1003. doi: 10.1128/MCB.06665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartoszewski R, Serocki M, Janaszak-Jasiecka A, Bartoszewska S, Kochan-Jamrozy K, Piotrowski A, Kroliczewski J, Collawn JF. miR-200b downregulates Kruppel Like Factor 2 (KLF2) during acute hypoxia in human endothelial cells. Eur J Cell Biol. 2017;96:758–766. doi: 10.1016/j.ejcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barger CJ, Branick C, Chee L, Karpf AR. Pan-cancer analyses reveal genomic features of FOXM1 overexpression in cancer. Cancers (Basel). 2019;11:251. doi: 10.3390/cancers11020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malashicheva A, Kanzler B, Tolkunova E, Trono D, Tomilin A. Lentivirus as a tool for lineage-specific gene manipulations (vol 45, pg 456, 2007) Genesis. 2007;45:793–793. doi: 10.1002/dvg.20368. [DOI] [PubMed] [Google Scholar]
- 34.Gomez-Martinez M, Schmitz D, Hergovich A. Generation of stable human cell lines with Tetracycline-inducible (Tet-on) shRNA or cDNA expression. J Vis Exp. 2013 doi: 10.3791/50171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartoszewska S, Kroliczewski J, Crossman DK, Pogorzelska A, Baginski M, Collawn JF, Bartoszewski R. Triazoloacridone C-1305 impairs XBP1 splicing by acting as a potential IRE1alpha endoribonuclease inhibitor. Cell Mol Biol Lett. 2021;26:11. doi: 10.1186/s11658-021-00255-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gebert M, Bartoszewska S, Janaszak-Jasiecka A, Moszynska A, Cabaj A, Kroliczewski J, Madanecki P, Ochocka RJ, Crossman DK, Collawn JF, Bartoszewski R. PIWI proteins contribute to apoptosis during the UPR in human airway epithelial cells. Sci Rep. 2018;8:16431. doi: 10.1038/s41598-018-34861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamkanfi M, Kanneganti TD. Caspase-7: a protease involved in apoptosis and inflammation. Int J Biochem Cell Biol. 2010;42:21–24. doi: 10.1016/j.biocel.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartoszewska S, Kamysz W, Jakiela B, Sanak M, Kroliczewski J, Bebok Z, Bartoszewski R, Collawn JF. miR-200b downregulates CFTR during hypoxia in human lung epithelial cells. Cell Mol Biol Lett. 2017;22:23. doi: 10.1186/s11658-017-0054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moszynska A, Collawn JF, Bartoszewski R. IRE1 endoribonuclease activity modulates hypoxic HIF-1alpha signaling in human endothelial cells. Biomolecules. 2020 doi: 10.3390/biom10060895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Bartoszewska S, Cabaj A, Dabrowski M, Collawn JF, Bartoszewski R. miR-34c-5p modulates X-box-binding protein 1 (XBP1) expression during the adaptive phase of the unfolded protein response. FASEB J. 2019;33:11541–11554. doi: 10.1096/fj.201900600RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, Flicek P. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krystkowiak I, Lenart J, Debski K, Kuterba P, Petas M, Kaminska B, Dabrowski M. Nencki Genomics Database—Ensembl funcgen enhanced with intersections, user data and genome-wide TFBS motifs. Database. 2013 doi: 10.1093/database/bat069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008;15:1460–1471. doi: 10.1038/cdd.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartoszewski R, Rab A, Fu L, Bartoszewska S, Collawn J, Bebok Z. CFTR expression regulation by the unfolded protein response. Methods Enzymol. 2011;491:3–24. doi: 10.1016/B978-0-12-385928-0.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 51.Iurlaro R, Munoz-Pinedo C. Cell death induced by endoplasmic reticulum stress. FEBS J. 2016;283:2640–2652. doi: 10.1111/febs.13598. [DOI] [PubMed] [Google Scholar]
- 52.Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, Harding HP. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci USA. 2012;109:E869–878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J Biol Chem. 2006;281:5852–5860. doi: 10.1074/jbc.M509061200. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Misiewicz M, Dery MA, Foveau B, Jodoin J, Ruths D, LeBlanc AC. Identification of a novel endoplasmic reticulum stress response element regulated by XBP1. J Biol Chem. 2013;288:20378–20391. doi: 10.1074/jbc.M113.457242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 60.Gaudette BT, Jones DD, Bortnick A, Argon Y, Allman D. mTORC1 coordinates an immediate unfolded protein response-related transcriptome in activated B cells preceding antibody secretion. Nat Commun. 2020;11:723. doi: 10.1038/s41467-019-14032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta S, Giricz Z, Natoni A, Donnelly N, Deegan S, Szegezdi E, Samali A. NOXA contributes to the sensitivity of PERK-deficient cells to ER stress. Febs Lett. 2012;586:4023–4030. doi: 10.1016/j.febslet.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 63.Fang P, Xiang L, Huang S, Jin L, Zhou G, Zhuge L, Li J, Fan H, Zhou L, Pan C, Zheng Y. IRE1alpha-XBP1 signaling pathway regulates IL-6 expression and promotes progression of hepatocellular carcinoma. Oncol Lett. 2018;16:4729–4736. doi: 10.3892/ol.2018.9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu GM, Zeng HD, Zhang CY, Xu JW. Key genes associated with diabetes mellitus and hepatocellular carcinoma. Pathol Res Pract. 2019;215:152510. doi: 10.1016/j.prp.2019.152510. [DOI] [PubMed] [Google Scholar]
- 65.Brewer JW, Diehl JA. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc Natl Acad Sci USA. 2000;97:12625–12630. doi: 10.1073/pnas.220247197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shan J, Dudenhausen E, Kilberg MS. Induction of early growth response gene 1 (EGR1) by endoplasmic reticulum stress is mediated by the extracellular regulated kinase (ERK) arm of the MAPK pathways. Biochim Biophys Acta Mol Cell Res. 2019;1866:371–381. doi: 10.1016/j.bbamcr.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Piya S, Kim JY, Bae J, Seol DW, Moon AR, Kim TH. DUSP6 is a novel transcriptional target of p53 and regulates p53-mediated apoptosis by modulating expression levels of Bcl-2 family proteins. Febs Lett. 2012;586:4233–4240. doi: 10.1016/j.febslet.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 68.Tang C-HA, Chang S, Paton AW, Paton JC, Gabrilovich DI, Ploegh HL, Del Valle JR, Hu C-CA. Phosphorylation of IRE1 at S729 regulates RIDD in B cells and antibody production after immunization. J Cell Biol. 2018;217:1739–1755. doi: 10.1083/jcb.201709137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oikawa D, Tokuda M, Hosoda A, Iwawaki T. Identification of a consensus element recognized and cleaved by IRE1 alpha. Nucleic Acids Res. 2010;38:6265–6273. doi: 10.1093/nar/gkq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, Strasser A, Kluck RM, Adams JM, Huang DC. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 71.Gurzov EN, Germano CM, Cunha DA, Ortis F, Vanderwinden JM, Marchetti P, Zhang L, Eizirik DL. p53 up-regulated modulator of apoptosis (PUMA) activation contributes to pancreatic beta-cell apoptosis induced by proinflammatory cytokines and endoplasmic reticulum stress. J Biol Chem. 2010;285:19910–19920. doi: 10.1074/jbc.M110.122374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jabbour AM, Heraud JE, Daunt CP, Kaufmann T, Sandow J, O'Reilly LA, Callus BA, Lopez A, Strasser A, Vaux DL, Ekert PG. Puma indirectly activates Bax to cause apoptosis in the absence of Bid or Bim. Cell Death Differ. 2009;16:555–563. doi: 10.1038/cdd.2008.179. [DOI] [PubMed] [Google Scholar]
- 73.Garrison SP, Phillips DC, Jeffers JR, Chipuk JE, Parsons MJ, Rehg JE, Opferman JT, Green DR, Zambetti GP. Genetically defining the mechanism of Puma- and Bim-induced apoptosis. Cell Death Differ. 2012;19:642–649. doi: 10.1038/cdd.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pihán P, Carreras-Sureda A, Hetz C. BCL-2 family: integrating stress responses at the ER to control cell demise. Cell Death Differ. 2017;24:1478–1487. doi: 10.1038/cdd.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verfaillie T, Rubio N, Garg AD, Bultynck G, Rizzuto R, Decuypere JP, Piette J, Linehan C, Gupta S, Samali A, Agostinis P. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 2012;19:1880–1891. doi: 10.1038/cdd.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giam M, Huang DCS, Bouillet P. BH3-only proteins and their roles in programmed cell death. Oncogene. 2008;27:S128–S136. doi: 10.1038/onc.2009.50. [DOI] [PubMed] [Google Scholar]
- 77.Chen Y, Brandizzi F. IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 2013;23:547–555. doi: 10.1016/j.tcb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shemorry A, Harnoss JM, Guttman O, Marsters SA, Komuves LG, Lawrence DA, Ashkenazi A. Caspase-mediated cleavage of IRE1 controls apoptotic cell commitment during endoplasmic reticulum stress. Elife. 2019;8:47084. doi: 10.7554/eLife.47084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang TK, Lawrence DA, Lu M, Tan J, Harnoss JM, Marsters SA, Liu P, Sandoval W, Martin SE, Ashkenazi A. Coordination between two branches of the unfolded protein response determines apoptotic cell fate. Mol Cell. 2018;71:629–636. doi: 10.1016/j.molcel.2018.06.038. [DOI] [PubMed] [Google Scholar]
- 80.Byrd AE, Brewer JW. Intricately regulated: a cellular toolbox for fine-tuning XBP1 expression and activity. Cells. 2012;1:738–753. doi: 10.3390/cells1040738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bartoszewski R, Rab A, Twitty G, Stevenson L, Fortenberry J, Piotrowski A, Dumanski JP, Bebok Z. The mechanism of cystic fibrosis transmembrane conductance regulator transcriptional repression during the unfolded protein response. J Biol Chem. 2008;283:12154–12165. doi: 10.1074/jbc.M707610200. [DOI] [PubMed] [Google Scholar]
- 82.He Y, Sun S, Sha H, Liu Z, Yang L, Xue Z, Chen H, Qi L. Emerging roles for XBP1, a sUPeR transcription factor. Gene Expr. 2010;15:13–25. doi: 10.3727/105221610X12819686555051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weingarten-Gabbay S, Nir R, Lubliner S, Sharon E, Kalma Y, Weinberger A, Segal E. Systematic interrogation of human promoters. Genome Res. 2019;29:171–183. doi: 10.1101/gr.236075.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lavallee JF, Gray TA, Dumville J, Russell W, Cullum N. The effects of care bundles on patient outcomes: a systematic review and meta-analysis. Implement Sci. 2017;12:142. doi: 10.1186/s13012-017-0670-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gonen N, Sabath N, Burge CB, Shalgi R. Widespread PERK-dependent repression of ER targets in response to ER stress. Sci Rep. 2019;9:4330. doi: 10.1038/s41598-019-38705-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsuru A, Imai Y, Saito M, Kohno K. Novel mechanism of enhancing IRE1α-XBP1 signalling via the PERK-ATF4 pathway. Sci Rep. 2016;6:24217. doi: 10.1038/srep24217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 88.Walter F, O'Brien A, Concannon CG, Düssmann H, Prehn JHM. ER stress signaling has an activating transcription factor 6α (ATF6)-dependent “off-switch”. J Biol Chem. 2018;293:18270–18284. doi: 10.1074/jbc.RA118.002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wali JA, Rondas D, McKenzie MD, Zhao Y, Elkerbout L, Fynch S, Gurzov EN, Akira S, Mathieu C, Kay TWH, Overbergh L, Strasser A, Thomas HE. The proapoptotic BH3-only proteins Bim and Puma are downstream of endoplasmic reticulum and mitochondrial oxidative stress in pancreatic islets in response to glucotoxicity. Cell Death Dis. 2014;5:e1124–e1124. doi: 10.1038/cddis.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pagliarini V, Giglio P, Bernardoni P, De Zio D, Fimia GM, Piacentini M, Corazzari M. Downregulation of E2F1 during ER stress is required to induce apoptosis. J Cell Sci. 2015;128:1166–1179. doi: 10.1242/jcs.164103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep sequencing data were deposited in Gene Expression Omnibus (GEO) at accession number: GSE160416.