Abstract
Many physiologic effects of l-glutamate, the major excitatory neurotransmitter in the mammalian central nervous system, are mediated via signaling by ionotropic glutamate receptors (iGluRs). These ligand-gated ion channels are critical to brain function and are centrally implicated in numerous psychiatric and neurologic disorders. There are different classes of iGluRs with a variety of receptor subtypes in each class that play distinct roles in neuronal functions. The diversity in iGluR subtypes, with their unique functional properties and physiologic roles, has motivated a large number of studies. Our understanding of receptor subtypes has advanced considerably since the first iGluR subunit gene was cloned in 1989, and the research focus has expanded to encompass facets of biology that have been recently discovered and to exploit experimental paradigms made possible by technological advances. Here, we review insights from more than 3 decades of iGluR studies with an emphasis on the progress that has occurred in the past decade. We cover structure, function, pharmacology, roles in neurophysiology, and therapeutic implications for all classes of receptors assembled from the subunits encoded by the 18 ionotropic glutamate receptor genes.
Significance Statement
Glutamate receptors play important roles in virtually all aspects of brain function and are either involved in mediating some clinical features of neurological disease or represent a therapeutic target for treatment. Therefore, understanding the structure, function, and pharmacology of this class of receptors will advance our understanding of many aspects of brain function at molecular, cellular, and system levels and provide new opportunities to treat patients.
I. Introduction
Glutamate mediates most excitatory neurotransmission in the mammalian central nervous system (CNS) via binding to metabotropic glutamate receptors (mGluRs), which are G protein–coupled receptors, and ionotropic glutamate receptors (iGluRs), which are cation-permeable ligand-gated ion channels. The activation of mGluRs and iGluRs produces distinct cellular responses on vastly different time scales. The iGluRs are divided into different functional classes, namely α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors, kainate receptors, N-methyl-d-aspartate (NMDA) receptors, and GluD receptors (also known as delta or δ receptors) (Fig. 1). Functional classes were initially identified by pharmacological properties, such as a selective activating agonist (Watkins and Jane, 2006). However, the distinction between the classes was precisely defined when cloning of the different subunits revealed strong correlation between sequence identity and pharmacological properties of receptor subtypes (Seeburg, 1993; Hollmann and Heinemann, 1994). The GluD receptors are unusual in this respect, since these receptors were cloned and categorized as iGluRs based on sequence identity, yet it remains uncertain whether they form ion channels that function by passing current in the CNS (Kakegawa et al., 2007b; Kakegawa et al., 2009; Schmid et al., 2009; Matsuda et al., 2010; Elegheert et al., 2016; Dadak et al., 2017). There are also examples in which specific iGluR subtypes that typically mediate current responses (i.e., ionotropic signaling) also appear capable of promoting intracellular signaling (i.e., metabotropic signaling) through a variety of mechanisms [reviewed in Dore et al. (2016), Perez-Otano et al. (2016), Valbuena and Lerma (2016, 2021), Yuzaki and Aricescu (2017), Reiner and Levitz (2018)].
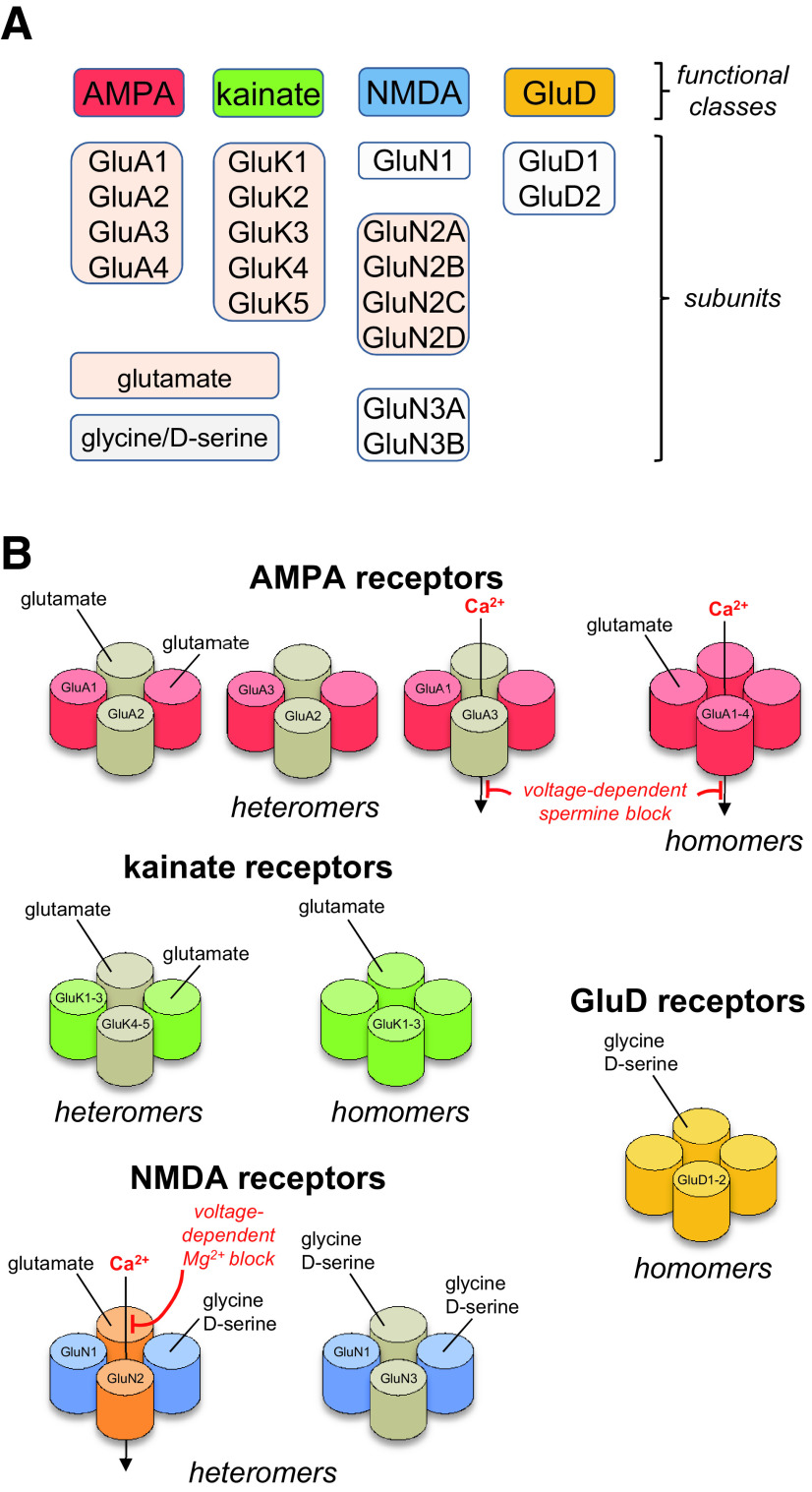
Fig. 1.
Structural and functional diversity of iGluRs. (A) Ionotropic glutamate receptors are divided into four functional classes: AMPA, kainate, NMDA, and GluD receptors. Multiple subunits have been cloned in each of these classes that bind either glutamate or glycine/D-serine. (B) Subunits from separate functional classes are unable to assemble as functional ion channels, but subunits within each functional class can assemble guided by specific sets of rules. AMPA receptor subunits can form functional homomers and heteromers, but GluA2-containing heteromers prevail throughout the CNS. Kainate receptor subunits GluK1-3 can assemble as functional homomers and heteromers, but GluK4-5 must coexpress with GluK1-3 to form functional receptors. NMDA receptors are strictly heteromeric receptors with two GluN1 subunits and two GluN2 subunits (GluN1/2) or two GluN3 subunits (GluN1/3). Triheteromeric NMDA receptors (not shown) contain two GluN1 subunits and two different GluN2 subunits (i.e., three types of subunits). GluD subunits form homomers that appear incapable of mediating ionotropic signaling.
Remarkable variation exists in the physiologic roles and functional properties among AMPA, kainate, and NMDA receptors. Vesicular presynaptic release of glutamate at central synapses activates excitatory postsynaptic currents (EPSCs) that give rise to excitatory postsynaptic potentials (EPSPs), which typically comprise a rapidly rising and decaying component mediated by AMPA receptors and a component mediated by NMDA receptors with a markedly slower time course (Hestrin et al., 1990; Sah et al., 1990; Trussell et al., 1993; Geiger et al., 1997) (Section IV. Receptor Activation, Deactivation, and Desensitization). In some synapses, activation of postsynaptic kainate receptors produces EPSCs with a decay time course that is intermediate to those of AMPA and NMDA receptors (Contractor et al., 2011). Unlike AMPA and kainate receptors, NMDA receptors exhibit voltage-dependent Mg2+ block and high permeability to Ca2+, and their activation requires simultaneous binding of two different agonists, glycine (or d-serine) and glutamate (Fig. 1). Vesicular glutamate release mediates the phasic activation of NMDA receptors, as tonic levels of extracellular glycine (or d-serine) can bind to synaptic and extrasynaptic NMDA receptors (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers). NMDA receptor ion channels are blocked by physiologic levels of extracellular Mg2+ at resting membrane potentials, but presynaptic glutamate release and subsequent rapid activation of AMPA/kainate receptors or other mechanisms, such as back propagation of action potentials (Tazerart et al., 2020), can depolarize the membrane potential, thereby relieving the voltage-dependent Mg2+ block of NMDA receptors and allowing the flow of inward current (i.e., positive ions moving into the cell) (Mayer et al., 1984; Nowak et al., 1984) (Section IV. Receptor Activation, Deactivation, and Desensitization). Thus, NMDA receptors serve as coincidence detectors that require simultaneous presynaptic glutamate release and postsynaptic depolarization to produce a slow Ca2+-permeable component of the EPSC (Bourne and Nicoll, 1993; Seeburg et al., 1995). The resulting increase in intracellular Ca2+ can trigger signaling events in the postsynaptic neuron that are accompanied by changes in synaptic efficacy and neuronal morphology (i.e., synaptic plasticity), a cellular correlate of memory and learning (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity).
Given the many roles of iGluRs in normal brain function, it is not surprising that their dysregulation is involved in numerous pathophysiological conditions (Lau and Zukin, 2007; Traynelis et al., 2010; Paoletti et al., 2013; Parsons and Raymond, 2014). Historically, there has been limited success in the development of iGluR ligands as therapeutic agents, but drug discovery efforts have been reinvigorated by the approval of the AMPA receptor antagonist perampanel as an antiepileptic drug and the NMDA receptor antagonist ketamine as an antidepressant. Furthermore, our improved understanding of disease mechanisms and genetics as well as the physiologic roles (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity) and localization (Section VI. Developmental and Regional Expression in the Central Nervous System) of iGluRs has increased interest in selective targeting of these receptors (Sections VII. Pharmacology of Orthosteric Ligands and Channel Blockers and IX. Exogenous Positive and Negative Allosteric Modulators) for potential treatment of CNS disorders (Section X. Glutamate Receptors in Disease).
In the recent decade, advances in X-ray crystallography and cryo-EM have increased available structural data for iGluR subtypes, and this information has catalyzed studies exploring the relationship between structure, function, and allosteric modulation of iGluRs (Sections II. Receptor Structure and VIII. Endogenous Allosteric Regulation). Similarly, our understanding of auxiliary subunits that associate with native iGluRs has increased profoundly (Section III. Auxiliary Subunits). These auxiliary subunits play key roles in the regulation of receptor biogenesis and trafficking as well as receptor function and allosteric regulation and account for much of the diversity in neuronal receptor function reported at distinct excitatory synapses (Section IV. Receptor Activation, Deactivation, and Desensitization). Technical advances have made it possible to determine the expression profiles of iGluR subtypes with increased precision at cellular and subcellular levels (Sections V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity and VI. Developmental and Regional Expression in the Central Nervous System), which has highlighted the diverse roles for iGluR subtypes within individual cells and in neural circuits. In this review, we summarize our understanding of the functional and physiologic roles of iGluRs, with an emphasis on mammalian receptors and aspects of their roles that have seen notable progress in this decade. We focus here on recognizing foundational studies that elucidated new principles and precedents. Although we have made efforts to recognize the full range of studies that have contributed to progress in the field, we regret that space has not allowed us to recognize in a truly comprehensive fashion all of the work that underpins each conceptual advance, and many excellent papers could not be included.
II. Receptor Structure
All iGluRs are integral membrane proteins assembled from four large (>850 residues) multidomain subunits to form an ion channel, which is a central ion-permeable pore that spans the membrane. The four iGluR classes are composed of different subunits: GluA1-4 for AMPA receptors (encoded by the GRIA1-4 genes); GluK1-5 for kainate receptors (encoded by GRIK1-5); GluN1, GluN2A-D, and GluN3A-B for NMDA receptors (encoded by GRIN1, GRIN2A-D, GRIN3A-B); and GluD1-2 for GluD receptors (encoded by GRID1-2) (Fig. 1). Most iGluR subunits bind glutamate as the activating endogenous agonist, with the exception of GluN1, GluN3A-B, and GluD1-2, which primarily bind glycine and d-serine. This diversity in subunit composition and endogenous agonists gives rise to variation in structural and functional properties between AMPA, kainate, NMDA, and GluD receptors and between receptor subtypes within each functional class.
A. Subunit Stoichiometry and Domain Organization
Functional iGluRs are assembled only by subunits from within the same class; for example, AMPA and kainate receptor subunits do not assemble to form a functional receptor (Fig. 1). AMPA receptor subunits form functional homotetramers and heterotetramers [reviewed in Hollmann and Heinemann (1994), Bowie (2012), Herguedas et al. (2013)], but GluA2-containing heterotetramers are most common in the CNS (Wenthold et al., 1996; Lu et al., 2009; Zhao et al., 2019). Kainate receptor subunits GluK1-3 can also express as functional homotetramers and heterotetramers, but GluK4-5 must coassemble with at least one GluK1-3 subunit to form functional heteromeric kainate receptors (Herb et al., 1992; Cui and Mayer, 1999; Jaskolski et al., 2005a; Reiner et al., 2012; Meyerson et al., 2014, 2016). Homomeric AMPA and kainate receptors have been extensively studied in heterologous expression systems (e.g., HEK293 cells or Xenopus oocytes), but the extent to which these homomeric receptors are found in native tissue is not understood. NMDA receptors are obligate heterotetramers composed of two glycine-binding GluN1 subunits and either two identical glutamate-binding GluN2 subunits (i.e., diheteromeric receptors containing two types of subunits), or two different GluN2 subunits (i.e., triheteromeric receptors containing three types of subunits) (Monyer et al., 1992; Ulbrich and Isacoff, 2007, 2008; Hansen et al., 2014; Karakas and Furukawa, 2014; Lee et al., 2014; Lu et al., 2017). NMDA receptors composed of two GluN1 and two GluN3 subunits (GluN1/3), which do not bind glutamate, are expressed in neurons (Grand et al., 2018; Otsu et al., 2019; Zhu et al., 2020), and triheteromeric receptors composed of GluN1, GluN2, and GluN3 subunits (GluN1/GluN2/GluN3) might also exist [reviewed in Perez-Otano et al. (2016), but see Ulbrich and Isacoff (2008)]. GluD1 and GluD2 subunits form homotetramers that serve as trans-synaptic adhesion complexes (i.e., synapse organizers) or require as yet unidentified factors to form functional ion channels (Mayat et al., 1995; Zuo et al., 1997; Kohda et al., 2000; Naur et al., 2007; Elegheert et al., 2016; Burada et al., 2020b). Whether GluD1 and GluD2 can assemble as heteromeric receptors is unresolved.
The iGluR subunits share a common modular design and membrane topology despite having modest primary sequence identity (Wo and Oswald, 1995; Paas, 1998) (Fig. 2). Each subunit includes a large extracellular region comprising the N-terminal domain (NTD), also referred to as amino-terminal domain (ATD), which mediates receptor assembly, trafficking and functional regulation, and the agonist binding domain (ABD), also referred to as ligand binding domain (LBD), which harbors binding sites for agonists, competitive antagonists, and some allosteric modulators. Each ABD is composed of two segments of the polypeptide chain, S1 and S2, separated by two transmembrane helices (M1 and M3) and a membrane reentrant loop (M2). The M1-M2-M3 regions together with a fourth transmembrane helix (M4) compose the transmembrane domain (TMD) (Fig. 2). The cytoplasmic C-terminal domain (CTD) directs receptor localization and regulation, is a locus of post-translational modifications (e.g., phosphorylation and palmitoylation), and varies in length and sequence among subunits (Sections II.E. The Intracellular C-Terminal Domain and II.F. Alternative Splicing, RNA Editing, and Post-Translational Modifications).
Fig. 2.
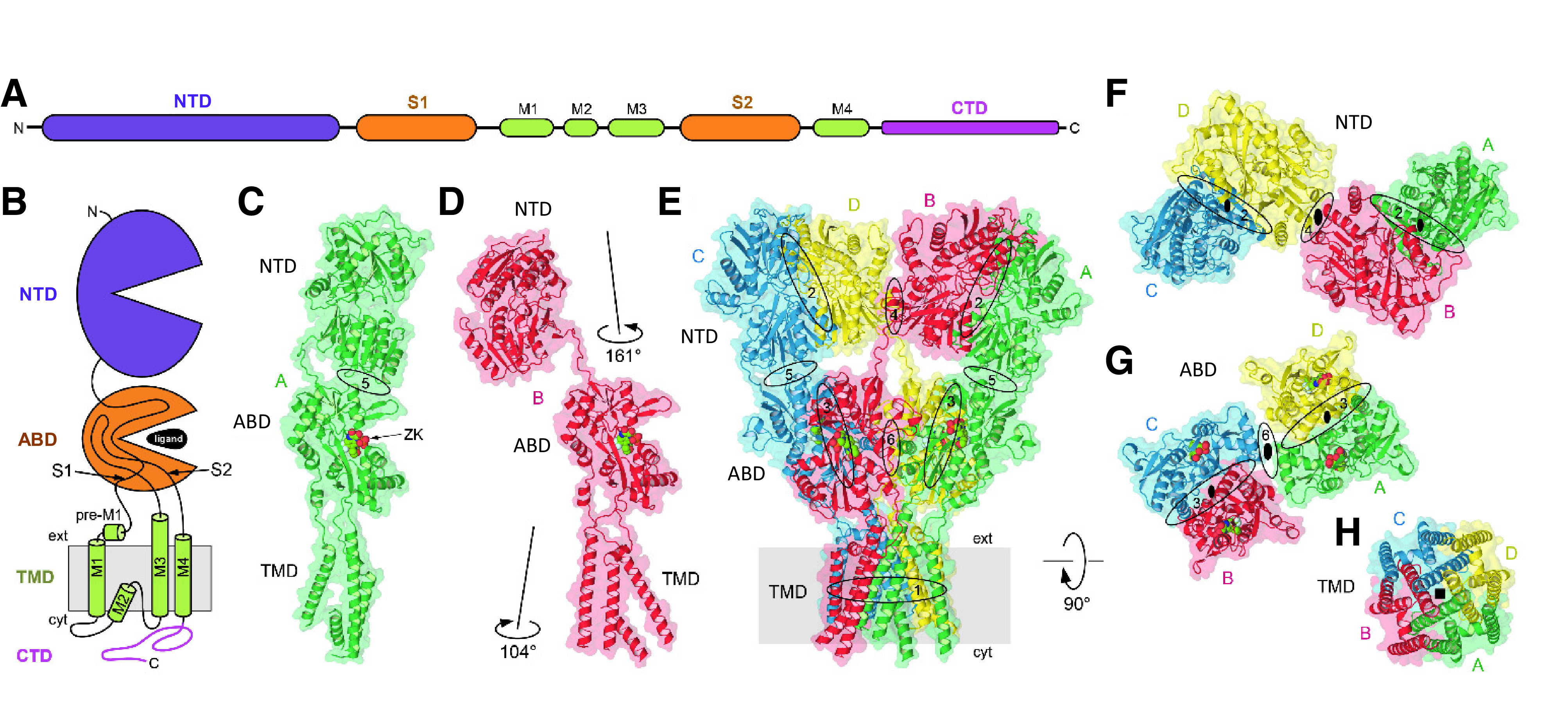
Common architecture of iGluRs. (A and B) Linear representation of the iGluR subunit polypeptide chain and cartoon of the iGluR subunit topology, illustrating the NTD, the S1 and S2 segments that together form the ABD, the TMD formed by M1-M2-M3 and M4, and the CTD. (C and D) Structures of distinct subunit conformations adopted in homomeric GluA2 receptors with similarly oriented ABDs. Oval in (C) indicates the NTD-ABD interface that is present in subunit positions A and C but not in positions B and D. Different orientations of NTD and TMD in subunit B compared with subunit A are indicated in (D). (E) Structure of GluA2 homomer (PDB: 3KG2) viewed parallel to the membrane, with interfaces indicated by ovals and labeled with numbers that increase as the interface surface area decreases. (F–H) The NTD (F), ABD (G), and TMD (H) layers viewed extracellularly perpendicular to the membrane, with the black ovals indicating the receptor overall (larger symbols at dimer-dimer interface) and NTD and ABD dimers local (smaller symbols within dimers) 2-fold rotational symmetry and black square indicating the 4-fold symmetry of the TMD layer. The subunit domains arrange as dimer-of-dimers, but switch dimer partner between the ABD and NTD layers (i.e., subunit crossover).
The iGluR subunits assemble into a tetrameric receptor with a layered architecture comprising the most extracellular NTD layer at the top followed by the ABD layer sandwiched between the NTD and TMD layers and the structurally unresolved intracellular CTD layer (Nakagawa et al., 2005; Nakagawa et al., 2006; Sobolevsky et al., 2009; Karakas and Furukawa, 2014; Meyerson et al., 2016; Burada et al., 2020b) (Fig. 2). In the tetramer, the four TMDs come together to form the cation-selective ion channel. The four subunits reside in positions within the tetramer (denoted A, B, C, and D) that can be divided into two diagonal pairs, A/C and B/D, based on a 2-fold rotational symmetry of the extracellular region (Fig. 2). As a result of this 2-fold symmetry, the conformations are similar for subunits within these pairs (i.e., A and C are similar) and different between the two subunit pairs (i.e., A and B are different) (Fig. 2). Thus, the same subunit must adopt two distinct conformations in homomeric iGluRs. In homomeric GluA2 receptors, for example, these differences are emphasized by the presence of an intrasubunit NTD-ABD interface in subunits A and C and its absence in subunits B and D (Fig. 2). The conformational differences between the subunit pairs typically reveal themselves as variation in the relative domain positioning and conformations of the peptide linkers that connect the NTD, ABD, and TMD layers, whereas the individual domains maintain similar conformations.
Pairs of subunit dimers assemble together as a tetramer by forming a number of intersubunit interfaces, including NTD dimer, ABD dimer, and TMD interfaces and weak interfaces between the two NTD dimers and between the two ABD dimers (Fig. 2) (Sun et al., 2002; Kumar et al., 2009; Karakas et al., 2011; Rossmann et al., 2011). The network of interfaces maintains the tetrameric structure of receptors composed of complex multidomain subunits while also allowing the conformational freedom that makes iGluRs highly dynamic structures. With the exception of homomeric GluD1 and GluD2 receptors, there is an intriguing swapping of domains between the NTD and ABD layers in iGluR structures (Fig. 3). Domain swapping results in NTD dimers formed by A/B and C/D subunits, whereas ABD dimers are formed by subunits A/D and B/C (Fig. 3). The domain swapping in iGluRs consequently creates a symmetry mismatch between the NTD and ABD layers, and the overall 2-fold symmetry of the extracellular region mismatches the pseudo 4-fold symmetry of the TMD layer forming the ion channel (Fig. 2). The only known exceptions to this canonical architecture are the homotetrameric GluD1 and GluD2 receptors, which do not exhibit domain swapping of subunits between the NTD and ABD layers (Burada et al., 2020a,b). Otherwise, the overall domain arrangement and symmetrical organization are preserved in AMPA, kainate, and NMDA receptors (Fig. 3).
Fig. 3.
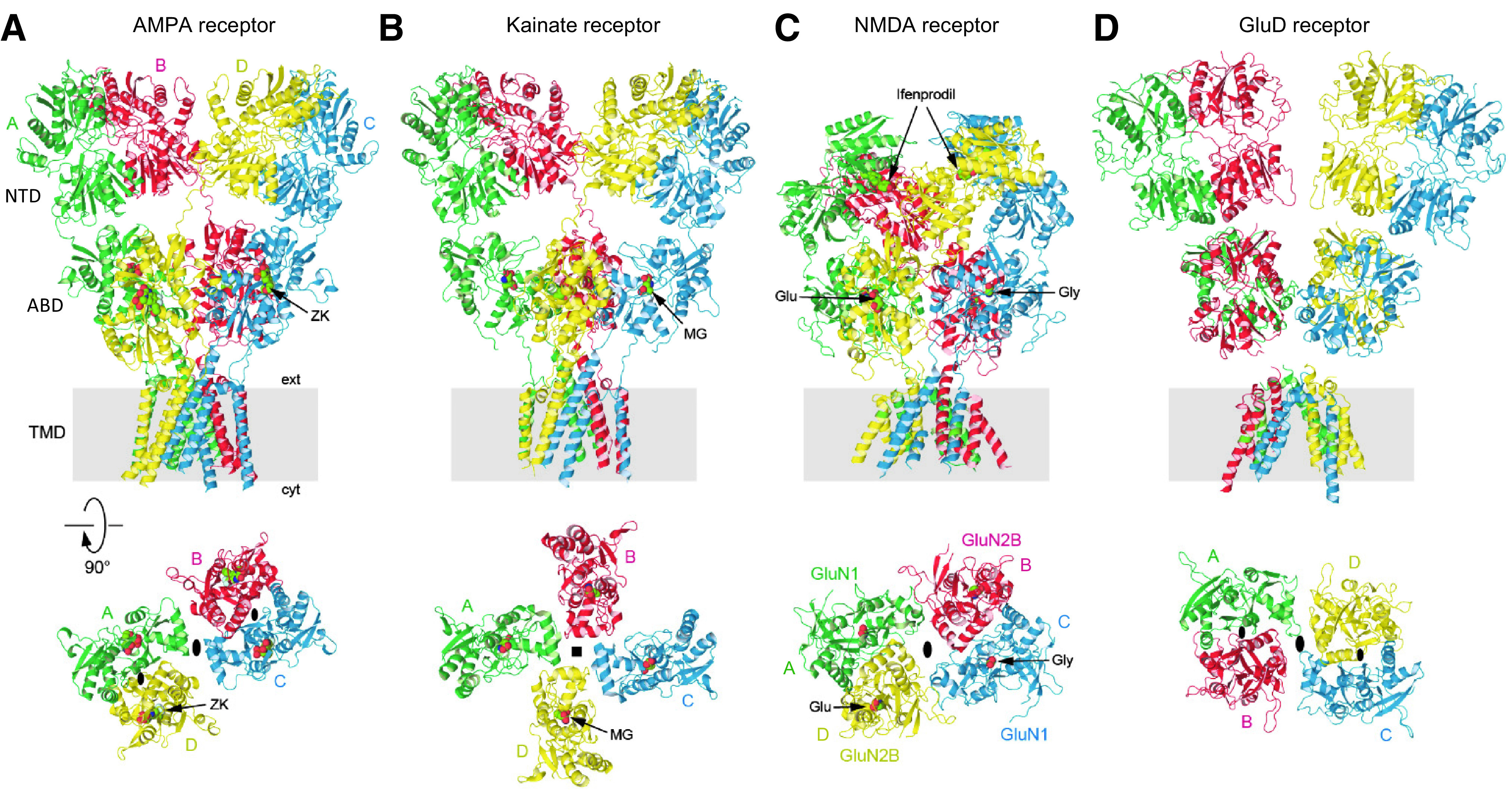
Representative structures of iGluR subtypes. (A) Homomeric GluA2 AMPA receptor in complex with competitive antagonist ZK 200775 (ZK) (PDB: 3KG2). (B) Homomeric GluK2 kainate receptor in complex with agonist SYM2081 (MG) (PDB: 5KUF). (C) GluN1/2B NMDA receptor in complex with Glu and Gly and the allosteric inhibitor ifenprodil (PDB: 4PE5). (D) Homomeric GluD1 receptor in complex with Ca2+ and the competitive antagonist 7-CKA (ligands not resolved) (PDB: 6KSS). The upper row shows iGluRs viewed parallel to the membrane, and the lower row shows ABD layers viewed extracellularly (GluK2 is presumably in the desensitized state). Black ovals indicate the receptor overall (larger symbols at dimer-dimer interface) and NTD and ABD dimers local (smaller symbols within dimers) 2-fold rotational symmetry, and black square indicates the 4-fold symmetry. GluD1 is unique by not having domain swapping between ABD and NTD layers.
Numerous iGluRs have been identified in nonmammals (Mayer and Jegla, 2018), such as insects (Benton et al., 2009; Li et al., 2016b; Prieto-Godino et al., 2016), worms (Maricq et al., 1995), plants (Lam et al., 1998), and single-cell organisms (Chen et al., 1999a; Lomash et al., 2013; Alberstein et al., 2015). The subunits in those organisms share their modular domain organization and architecture with the mammalian receptors, and the structures of isolated ABDs are similar (Mayer et al., 2001; Lomash et al., 2013; Alberstein et al., 2015; Han et al., 2015b; Li et al., 2016b; Alfieri et al., 2020; Stroebel and Paoletti, 2021; Gangwar et al., 2021). However, there are substantial differences in pharmacology, gating kinetics (Lomash et al., 2013; Alberstein et al., 2015; Prieto-Godino et al., 2017), and the role of auxiliary subunits (Zheng et al., 2004; Walker et al., 2006; Wang et al., 2012; Han et al., 2015b).
1. Structures of Tetrameric Ionotropic Glutamate Receptor Subtypes
The initial glimpses of tetrameric AMPA receptor structures at low resolution (∼20–30 Å) (Nakagawa et al., 2005; Nakagawa et al., 2006; Midgett and Madden, 2008) were followed by the first X-ray structure of the tetrameric GluA2 receptor at a resolution of 3.6 Å (Sobolevsky et al., 2009). Tremendous progress has been made in the past decade using X-ray crystallography and cryo-EM to obtain insights into the different functional states (e.g., closed, open, and desensitized conformations) (Section IV. Receptor Activation, Deactivation, and Desensitization), the structural basis for allosteric modulation (Section IX. Exogenous Positive and Negative Allosteric Modulators), and regulation by auxiliary subunits (Section III. Auxiliary Subunits).
The majority of tetrameric iGluR structures were preceded by more than a decade of studies with high-resolution structures of individual subunit ABDs and NTDs that in some cases formed physiologically relevant homodimer or heterodimer structures. Resolution of isolated domains provided the first views of subunit interfaces and resulted in structural models for activation, antagonism, and desensitization that are central to our appreciation of iGluR function (Sections II.B. The Extracellular Agonist Binding Domain and II.C. The Extracellular N-Terminal Domain). Importantly, these isolated ABD and NTD structures have been recapitulated in recent structures of tetrameric iGluRs. Before discussing the structures of individual ABDs and NTDs (Sections II.B. The Extracellular Agonist Binding Domain and II.C. The Extracellular N-Terminal Domain), we will summarize the major discoveries provided by X-ray and cryo-EM structures of tetrameric iGluRs. These structures were all obtained with subunits lacking their CTDs (Section II.E. The Intracellular C-Terminal Domain).
a. Structures of tetrameric AMPA receptors
After the first crystal structure of the homotetrameric GluA2 receptor in the closed, competitive antagonist-bound state (Sobolevsky et al., 2009), structural efforts focused on solving tetrameric AMPA receptor structures in additional conformational states. It took 5 years before new crystal structures of GluA2 in complex with agonists were reported (Chen et al., 2014b; Durr et al., 2014; Yelshanskaya et al., 2014). The channels in these structures were all in the closed conformation despite having been resolved with full or partial agonists as well as with the positive allosteric modulators (R,R)-2b or con-ikot-ikot, a peptide toxin from the Conus striatus cone snail. The ABDs in these complexes adopted incompletely closed conformations that could be interpreted as transitional states visited en route to channel opening; they were therefore referred to as preactive states (Twomey and Sobolevsky, 2018). X-ray crystallography yielded apo state structures of GluA2 in the absence of agonist binding (Durr et al., 2014; Yelshanskaya et al., 2016b). These apo structures adopted the same conformation as the competitive antagonist-bound structure (Sobolevsky et al., 2009), confirming that the antagonist-bound structures are faithful representations of the AMPA receptor closed state.
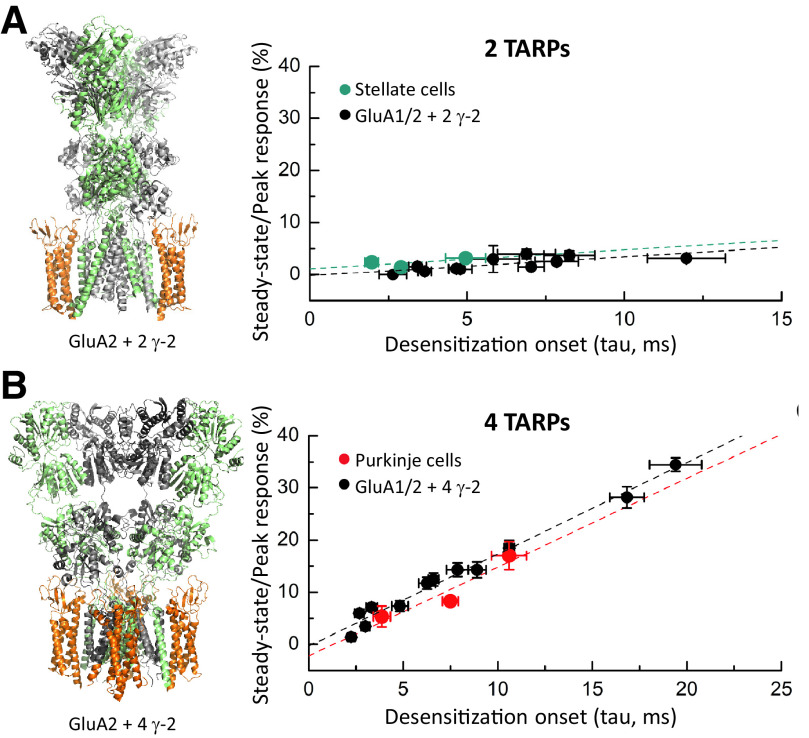
The “resolution revolution” in cryo-EM (Kuhlbrandt, 2014) made visualization of both open and desensitized states possible once the AMPA receptor interactome was elucidated, and multiple auxiliary subunits were isolated (Jackson and Nicoll, 2011a; Bettler and Fakler, 2017) (Section III. Auxiliary Subunits). To solve the open-state structures (Chen et al., 2017a; Twomey et al., 2017a), GluA2 was fused or coexpressed with the auxiliary subunit stargazin [transmembrane AMPA receptor regulatory protein (TARP) γ-2], which promotes channel opening, and GluA2 was bound to the agonists glutamate or quisqualate and the positive allosteric modulators cyclothiazide or (R,R)-2b. The desensitized state was resolved in structures of GluA2 either fused to the desensitization-promoting auxiliary subunit germ cell–specific gene 1-like (GSG1L) or coexpressed with stargazin (TARP γ-2) (both with quisqualate) (Chen et al., 2017a; Twomey et al., 2017b) (Section III. Auxiliary Subunits).
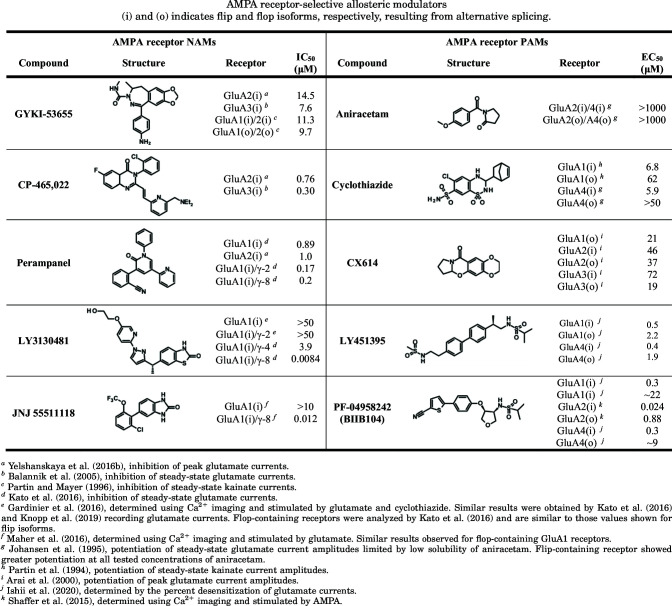
Noncompetitive inhibition (i.e., negative allosteric modulation, NAM) of AMPA receptors was studied by cocrystallizing GluA2 with the antiepileptic drug perampanel and mechanistically related GYKI-53655 and CP-465,022 (Yelshanskaya et al., 2016b). These noncompetitive inhibitors bound at the extracellular collar of the channel in a cavity formed by the pre-M1 linker and top portions of M3 and M4, where the NAMs were proposed to act as wedges to prevent conformational changes in the ion channel associated with pore opening (Twomey et al., 2017a; Twomey and Sobolevsky, 2018) (Section IX. Exogenous Positive and Negative Allosteric Modulators). More recently, X-ray crystallography was used to determine the site of action for noncompetitive inhibition by trans-4-butylcyclohexane carboxylic acid (4-BCCA), which binds in the side portals leading from membrane to the ion channel pore (Yelshanskaya et al., 2020), similar to local anesthetics in voltage-gated Na+ channels.
A variety of positively charged small molecules and peptide toxins occlude ion permeation of iGluRs through pore blockade (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers). The structural basis for channel block of AMPA receptors was elucidated using cryo-EM and open-state structures of the GluA2-stargazin complex (Twomey et al., 2018). Toxins and toxin-like molecules act as high-affinity pore blockers of calcium-permeable AMPA receptors by placing their bulky hydrophobic head groups in a central ion channel cavity and their positively charged polyamine tails into the narrow, negatively charged selectivity filter [reviewed in Bowie (2018)]. The negatively charged selectivity filter was also proposed to bind the endogenous polyamines spermine and spermidine, which lack the bulky hydrophobic head groups found in toxins (Twomey et al., 2018). As a result, intracellular polyamines act as low-affinity blockers of calcium-permeable AMPA receptors and yield the characteristic inward rectification of AMPA receptor-mediated currents by occluding outward current flow at depolarized potentials (Section VIII. Endogenous Allosteric Regulation). Comparison of the blocker-bound open-state structures with the closed-state structure allowed visualization of the molecular mechanism of blocker trapping (Twomey et al., 2018). Cryo-EM also made it possible to study AMPA receptor complexes that included auxiliary subunits as well as heteromeric assemblies of AMPA receptor subunits. In heteromeric AMPA receptors, the subunits appeared to arrange with the GluA2 subunit preferentially occupying the B/D positions (Herguedas et al., 2019; Zhao et al., 2019), and this placement may be influenced by features of the signal peptide preceding the NTD (He et al., 2016), but see also Duan et al. (2018).
b. Structures of tetrameric kainate receptors
Efforts toward structural resolution of tetrameric kainate receptors have not been as successful as those for AMPA receptors. The initial low-resolution structure at ∼21 Å (Schauder et al., 2013), which was followed by cryo-EM structure at 7.6 Å of the homotetrameric GluK2 receptor obtained in the presence of the high-affinity agonist 2S,4R-4-methylglutamate (SYM2081), revealed a surprising 4-fold symmetry of the ABD layer (Meyerson et al., 2014) (Fig. 3). The resolution of this structure, which was proposed to represent a desensitized receptor state, was further improved to 3.8 Å and was accompanied by a low-resolution structure at 11.6 Å of GluK2 in the presence of the competitive antagonist LY466195 that was reminiscent of the closed-state structure of GluA2 (Meyerson et al., 2016). More recently, low-resolution structures of GluK3 were obtained in the presence of kainate or SYM2081 as well as competitive antagonists UBP301 and UBP310, with the latter showing significant asymmetric distortions at the ABD layer (Kumari et al., 2019, 2020). The gating mechanism of kainate receptors is unknown because neither the apo nor the open-state structures of kainate receptors have been resolved. When compared with the closed-state AMPA receptor structure, the putative desensitized state structures of kainate receptors predict unusually large conformational changes associated with desensitization (Meyerson et al., 2014, 2016; Kumari et al., 2019, 2020). Recently, the cryo-EM structure of GluK2/5 heteromer was solved in the apo, antagonist-bound, and desensitized states (Khanra et al., 2021). The receptor assembles with two copies of each subunit with the GluK5 subunits positioned proximal to the channel, and GluK2 subunits but not GluK5 subunits undergo major structural rearrangements during desensitization, which is mediated by the peptide linkers connecting the pore helices. These data suggest that GluK2 and GluK5 subunits of the heteromer fulfill distinct roles to bring about channel desensitization and activation, respectively. The desensitized structures of kainate receptor homomers and heteromers reveal that kainate receptors undergo unusually large conformational changes to reach the desensitized state(s) compared with AMPA receptors (Meyerson et al., 2014, 2016; Kumari et al., 2019, 2020; Khanra et al., 2021).
c. Structures of tetrameric NMDA receptors
Structural studies of NMDA receptors lagged behind those of AMPA receptors because of the technical difficulties associated with assembling two different subunits GluN1 and GluN2, which are essential for formation of functional channels. The first two X-ray structures of GluN1/2B NMDA receptors represented the allosterically inhibited state in which the ion channel is closed, the GluN1 and GluN2B ABDs are bound to agonists, and the GluN1/2B NTD dimer interfaces are occupied by the GluN2B-selective NAMs, ifenprodil or Ro25-6981 (Karakas and Furukawa, 2014; Lee et al., 2014) (Fig. 3). The GluN1 and GluN2 subunits are arranged in the 1-2-1-2 order in NTD, ABD, and TMD domain layers similar to that of AMPA and kainate receptors. That is, the NTDs and ABDs are arranged as a dimer of GluN1-GluN2 heterodimers, and the heterodimeric pairs swap between the NTD and ABD layers. GluN1 has a conformation similar to subunits in the A/C position in AMPA and kainate receptors, whereas GluN2 has a conformation similar to subunits in the B/D position [e.g., see Salussolia et al. (2011b)] (Fig. 3).
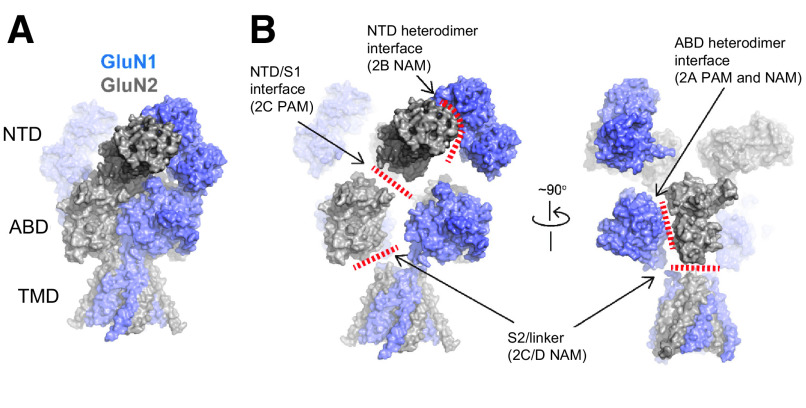
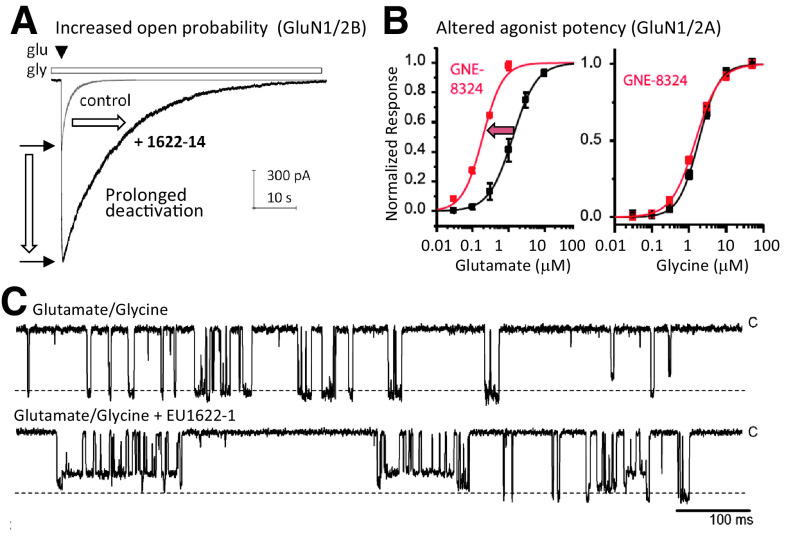
One key distinctive feature of NMDA receptors is the more extensive packing of NTDs and ABDs compared with non-NMDA receptors, which enables tight interdomain coupling. The existence of the NTD-ABD intradimer interface in each of the four subunits as well as additional interfaces between the NTD of one subunit and the ABD of the neighboring subunit create a more compact balloon shape of the NMDA receptor compared with AMPA and kainate receptors (Karakas and Furukawa, 2014; Lee et al., 2014; Tajima et al., 2016; Zhu et al., 2016; Lu et al., 2017; Regan et al., 2018) (Figs. 2 and 3). In this arrangement, changes in domain conformations and subunit orientation in the NTD layer would affect those of the ABD layer and vice versa. This structural feature is consistent with involvement of NMDA receptor NTDs in functional regulation, including deactivation kinetics, open probability, and agonist potency (Gielen et al., 2009; Yuan et al., 2009). Furthermore, small compounds and antibodies that bind NMDA receptor NTDs (Dalmau et al., 2008; Karakas et al., 2011; Chan et al., 2020) or between the NTD and ABD (Khatri et al., 2014) allosterically modulate receptor function. By contrast, the NTDs in AMPA and kainate receptors minimally regulate their function, consistent with the limited NTD-ABD interaction (Fig. 3). Previous studies on structures of the isolated NTDs and ABDs revealed that GluN2A or GluN2B NTD clamshells are closed in the presence of allosteric inhibitors, such as Zn2+, ifenprodil, and protons (i.e., low pH), but open in the absence of allosteric inhibitors (e.g., at higher pH 7.4–8; Section VIII. Endogenous Allosteric Regulation). ABD clamshells adopt a closed conformation in the presence of agonists and a more open conformation in the apo state or in presence of competitive antagonists (Fig. 4 and Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers). At the level of a tetramer, the patterns of conformational changes are more complex as NTDs and ABDs from GluN1 and GluN2 move in a concerted manner (Chou et al., 2020). In the presence of agonists at physiologic pH, the receptors reside in three major conformations, two nonactive states, and one active state, which are characterized by different orientations between the two GluN1-GluN2 ABD heterodimers and changes in the GluN1-GluN2 NTD heterodimer interfaces (Chou et al., 2020). There is tight conformational coupling of NTDs and ABDs that provide a mechanism by which the function of NMDA receptors can be regulated by targeting either of the domains (Sections VIII. Endogenous Allosteric Regulation and IX. Exogenous Positive and Negative Allosteric Modulators).
Fig. 4.
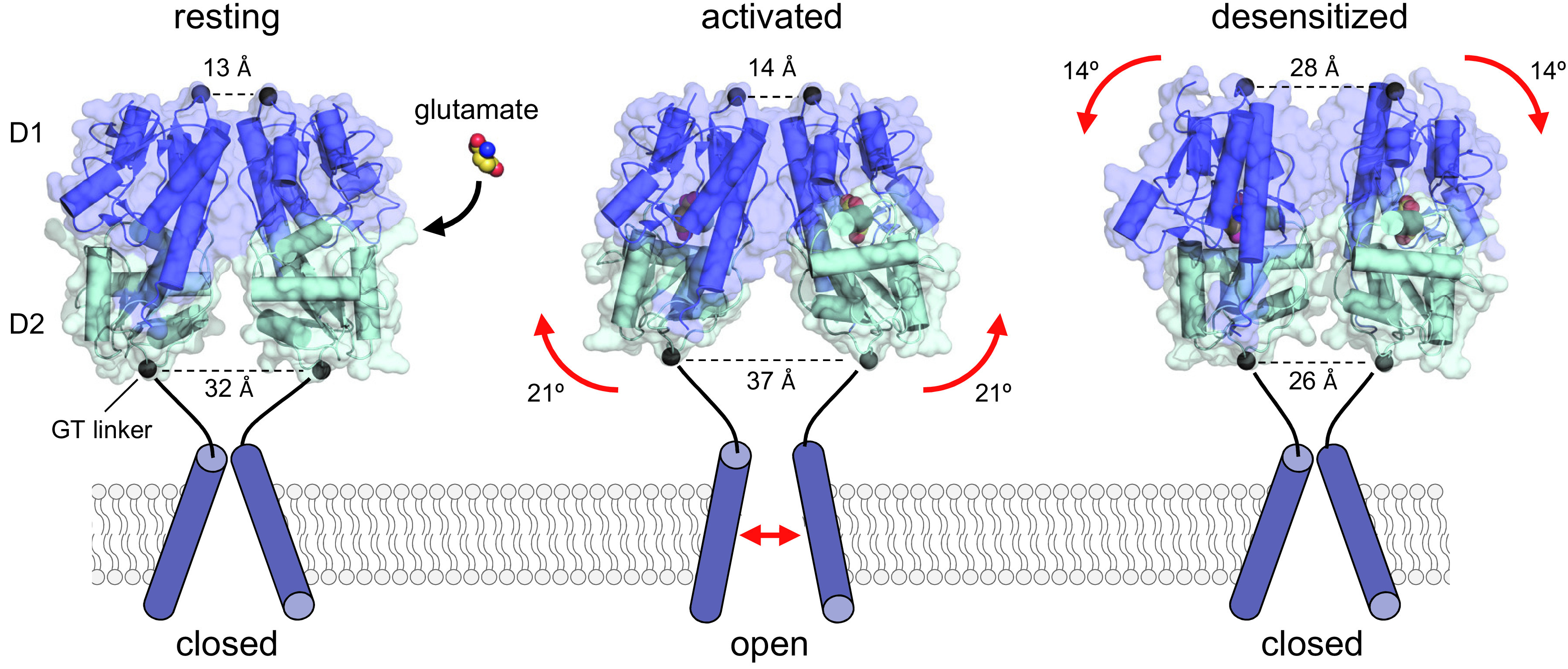
Conformational changes in the AMPA receptor ABDs during activation and desensitization. Crystal structures of the GluA2 ABD homodimer in conformations representing the resting apo state (PDB: 1FT0), the activated glutamate-bound state (PDB: 1FTJ), and the desensitized state (PDB: 2I3V). The ABDs are shaped as bilobed structures, with the agonist binding site located within the cleft between the two lobes referred to as D1 and D2. The Gly-Thr (GT) peptide linker positioned at the bottom of D2 replaces the TMDs in the full-length subunits. Agonist binding induces cleft closure in the ABD, thereby separating the linkers that replace the TMD. By contrast, the Cα-Cα distance between Gly residues at the top of the dimer remains relatively unchanged between apo and glutamate-bound ABD structures. After glutamate binding and ABD cleft closure, separation of the linkers can trigger reorientation of the transmembrane helices and channel opening. The activated receptor conformation is unstable, resulting in either reopening of the ABD or rearrangement at the ABD dimer interface, which results in desensitization by permitting repositioning the transmembrane helices to close the channel while glutamate remains bound.
Channel blockers of NMDA receptors, such as memantine and ketamine, have therapeutic potential in a range of CNS disorders (Section X. Glutamate Receptors in Disease). Crystal structures described the binding within the pore of MK-801, a high-affinity channel blocker in the same class as memantine and ketamine (Lu et al., 2017; Regan et al., 2018; Song et al., 2018). Truncation of the NTDs yielded crystals with an improved diffraction quality (3.2 Å) that showed that loss of the NTD resulted in two types of subunit arrangements in ABDs (Song et al., 2018). One conformation was consistent with the tetrameric receptor containing the NTDs, and the other conformation displayed swapped ABD dimer pairs, underscoring the importance of NTDs in promoting appropriate subunit assembly and receptor function. These studies also revealed the role of the Asn residues at and adjacent to the Q/R/N site at the apex of the M2 reentrant loop (Sections II.D. The Transmembrane Domain and VIII. Endogenous Allosteric Regulation) in binding of channel blockers (Song et al., 2018). Moreover, vestibules extended from the pore to the cell membrane, which could provide a potential path through which small blocker molecules might exit the closed channel (Song et al., 2018).
The majority of NMDA receptors in glutamatergic synapses on excitatory neurons are presumably represented by the triheteromeric GluN1/2A/2B subtype (Al-Hallaq et al., 2007; Luo et al., 1997; Sheng et al., 1994). One of the key functional features of the triheteromeric GluN1/2A/2B receptor is that its functional properties resemble those of the diheteromeric GluN1/2A NMDA receptor compared with the diheteromeric GluN1/2B NMDA receptor (Hatton and Paoletti, 2005; Hansen et al., 2014; Stroebel et al., 2014; Cheriyan et al., 2016; Sun et al., 2017b; Yi et al., 2019) (Section VIII. Endogenous Allosteric Regulation). Cryo-EM revealed that triheteromeric receptors were formed from a GluN1-GluN2A-GluN1-GluN2B subunit arrangement (Lu et al., 2017) in a similar pattern to the diheteromeric GluN1/2A (Jalali-Yazdi et al., 2018; Zhang et al., 2018b; Wang et al., 2021) and GluN1/2B receptors (Karakas and Furukawa, 2014; Lee et al., 2014). An important structural feature that likely shapes GluN1/2A/2B receptor function is the extensive interaction between the GluN2A NTD and the GluN1 ABD compared with that between the GluN2B NTD and the GluN1 ABD. The GluN1/2A/2B receptor structure was determined for protein samples prepared at low pH (6.5), suggesting the functional state represents a proton-inhibited state (Lu et al., 2017).
NMDA receptors containing the GluN1-1b splice isoform harbor a 21-amino-acid loop motif in the NTD, which is encoded by exon 5 (hereafter the N1 cassette, Section II.F. Alternative Splicing, RNA Editing, and Post-Translational Modifications), and exhibit lower sensitivity to proton inhibition (Traynelis et al., 1995) and faster deactivation kinetics compared with receptors lacking the N1 cassette (e.g., in the GluN1-1a isoform) (Rumbaugh et al., 2000; Vance et al., 2012; Regan et al., 2018; Yi et al., 2018, 2019). Cryo-EM analysis with GluN1-1b/2B receptors revealed that residues of the N1 cassette are located at the NTD-ABD domain interface on “top” of the heterodimer interface between GluN1 and GluN2B ABDs and in vicinity of the GluN2B NTD-ABD linker, indicating that the N1 cassette can control pH sensitivity by influencing the strength of the GluN1 and GluN2 ABD dimer interface (Regan et al., 2018) (Section VIII. Endogenous Allosteric Regulation). GluN1/2A receptors are inhibited by protons and Zn2+ in a synergistic manner, and cryo-EM structures have been obtained in various pH values (pH 6.1 – 8) and Zn2+ concentrations (1 µM and 1 mM) (Section VIII. Endogenous Allosteric Regulation).
d. Structures of tetrameric GluD receptors
GluD receptors bind d-serine (and glycine) but appear incapable of producing current in response to agonist binding (Section IV. Receptor Activation, Deactivation, and Desensitization). Isolated GluD2 ABDs in the apo form crystallize as homodimers in the presence of Ca2+ ions, which bind to and stabilize the ABD dimer interface (Naur et al., 2007; Chaudhry et al., 2009a; Hansen et al., 2009). GluD2 receptors may “predesensitize” in response to agonist binding, thereby precluding a current response, in part due to weak ABD dimer interactions (Chaudhry et al., 2009a; Hansen et al., 2009), but see Tapken et al. (2017) and Chin et al. (2020). Binding of Ca2+ to GluD2 cannot stabilize the ABD dimer interface enough to prevent this “predesensitization,” but extracellular Ca2+ ions can potentiate spontaneously activated currents mediated by GluD2 receptors carrying the lurcher mutation GluD2-A654T (Wollmuth et al., 2000; Hansen et al., 2009). The residues that coordinate Ca2+ ions in GluD2 are conserved in GluD1, and spontaneously active GluD1 receptors (GluD1-F655A) are also potentiated by Ca2+ (Yadav et al., 2011). d-Serine binding reduces spontaneously active currents from both GluD2-A654T and GluD1-F655A, presumably by triggering desensitization (Naur et al., 2007; Hansen et al., 2009; Yadav et al., 2011) (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity).
Cryo-EM structures of the tetrameric GluD1 receptor in two conformations (compact and splayed) and the tetrameric GluD2 receptor in a single conformation have been solved at low resolution (Burada et al., 2020a,b). These structures were determined in the presence of Ca2+ and the competitive antagonist 7-chlorokynurenic acid (7-CKA), which binds to the GluD2 ABD (Kristensen et al., 2016a). Thus, these structures are presumed to reflect “nonactive” states in the absence of agonist binding and with the ABD dimer interface stabilized by Ca2+ binding. These structures revealed that both GluD1 and GluD2 receptors assemble with a unique architecture in which domain swapping is not observed between the NTD and ABD layers (Fig. 3) and are the only known exception to the canonical architecture of AMPA, kainate, and NMDA receptors. The GluD structures maintained the distinct three-layered arrangement of the NTD, ABD, and TMD regions, and both the NTDs and ABDs are arranged as 2-fold symmetric dimers. The GluD1 receptor adopted an overall Y-shaped structure with the NTDs and ABDs in 2-fold symmetric dimer-of-dimers arrangement (Burada et al., 2020b) similar to other iGluRs. However, the NTD and ABD layers of A/B and C/D subunit positions pack directly on top of each other perpendicular to the membrane because of the nonswapped architecture of GluD1, and the two linear NTD-ABD arms of A/B and C/D subunit pairs adopted splayed conformations relative to each other (Fig. 3). The GluD2 structure was different from GluD1 by showing an asymmetric organization of the NTD and ABD layers, in which the NTD-ABD domain arm of subunits in the C/D position adopts a bent conformation that enables the upper regions of the C/D NTDs to interact with the lower regions of the A/B NTDs (Burada et al., 2020a). It is unresolved whether the apparent higher conformational freedom in the GluD NTD layers compared with other tetrameric iGluR structures is facilitated by the unique nonswapped architecture or is caused by the experimental conditions (Burada et al., 2020a,b). This conformational freedom might be important for the unique roles of postsynaptic GluD receptors as synaptic organizer proteins through interactions between their NTD and secreted cerebellin (Cbln) 1-4 proteins, which at the same time interact with presynaptic neurexins (Yuzaki and Aricescu, 2017; Yuzaki, 2018) (Sections IV. Receptor Activation, Deactivation, and Desensitization and V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity). Thus, the conformational freedom during cryo-EM imaging of GluD may reflect the absence of these stabilizing trans-synaptic interactions that would otherwise limit domain movements.
B. The Extracellular Agonist Binding Domain
The extracellular ABD comprises the S1 and S2 amino acid segments (Stern-Bach et al., 1994; Kuusinen et al., 1995; Arvola and Keinanen, 1996), which are separated by the M1 and M3 transmembrane helices and the M2 pore loop (Hollmann et al., 1994; Wo and Oswald, 1994) (Fig. 2). The ABD is joined to the NTD through the NTD-S1 polypeptide linker and to the TMD through the S1-M1, M3-S2, and S2-M4 linkers. The ABD-TMD linkers transmit conformational changes resulting from agonist binding to channel gating, whereas the NTD-ABD linkers alter ABD dynamics after binding of allosteric modulators to the NTD (Sections VIII. Endogenous Allosteric Regulation and IX. Exogenous Positive and Negative Allosteric Modulators). The S1 and S2 segments fold into bilobed ABD structures with an upper lobe (D1) and a lower, membrane-proximal lobe (D2). The binding site for agonists and competitive antagonists resides in the cleft between the D1 and D2 lobes (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers), and binding sites for both negative and positive allosteric modulators exist at the interface formed by adjacent ABDs (Section IX. Exogenous Positive and Negative Allosteric Modulators). The conservation in tertiary structure is remarkable among vertebrates and invertebrates, presumably due to evolution from a common bacterial ancestor, but subtle variations at key amino acid positions in the ABD result in different agonist binding properties across species (Mayer, 2021; Stroebel and Paoletti, 2021).
1. AMPA Receptor Agonist Binding Domains
The S1 and S2 segments were identified as two discontinuous segments, each of ∼150 amino acid residues, that when exchanged between the GluA3 AMPA receptor subunit and the GluK2 kainate receptor subunit also converted agonist pharmacology (Stern-Bach et al., 1994). Soluble AMPA receptor ABDs could be generated by linking the S1 and S2 segments with an artificial peptide linker (Kuusinen et al., 1995; Arvola and Keinanen, 1996) and exhibited ligand binding profiles comparable to those in full-length receptors, suggesting that structural integrity and characteristics of the agonist binding pocket are retained in isolated ABDs. These insights led to the first crystal structure of the GluA2 ABD with kainate bound in the cleft formed by the D1 and D2 lobes (Armstrong et al., 1998). The structure of the GluA2 ABD without bound ligand (apo form) or in complex with the competitive antagonist DNQX revealed that competitive antagonists stabilize the open apo conformation of the ABD, and agonists stabilize a closed conformation (Armstrong and Gouaux, 2000). Thus, agonist-induced closure of the ABD was proposed to be the conformational change initiating receptor activation (Fig. 4). This idea was supported by ABD structures in complex with various partial and full agonists, demonstrating that the extent of ligand-induced ABD closure around the agonist correlates with the extent of AMPA receptor activation and desensitization (Armstrong and Gouaux, 2000; Hogner et al., 2002; Jin et al., 2003). In the majority of these studies, the GluA2 ABDs crystallize as “back-to-back” dimers in which the dimer interface is formed exclusively by contacts in D1, whereas D2 is free to move after agonist binding (Armstrong and Gouaux, 2000; Sun et al., 2002; Jin et al., 2003, 2005; Horning and Mayer, 2004). Thus, when ABDs adopt the closed conformation after agonist binding, the pair of linkers at the bottom of D2 that connect to the TMD swing apart in the dimeric ABD structures, and this movement can transmit agonist binding to channel opening in full-length receptors by triggering reorientation of transmembrane helices (Fig. 4). Evaluation of GluA2 ABD structures established the mechanism by which mutations at the dimer interface (e.g., GluA2-L483Y) or positive allosteric modulators (e.g., cyclothiazide) can stabilize the dimer interface and thereby reduce desensitization (Sun et al., 2002; Horning and Mayer, 2004; Jin et al., 2005; Armstrong et al., 2006; Weston et al., 2006b; Yonkunas et al., 2017). These studies suggested a structural mechanism for iGluR desensitization in which agonist-induced ABD closure and movements of D1 and D2 relative to each other produce instability at the TMD and at the ABD dimer interface. This instability can subsequently be mitigated by either ABD reopening or by rearrangement at the ABD dimer interface (Fig. 4). ABD reopening is the first step in the process of agonist dissociation and receptor deactivation, whereas rearrangement at the ABD dimer interface would result in desensitization by enabling repositioning of transmembrane helices to a more relaxed conformation, thereby closing the ion channel. These initial insights from AMPA receptor ABD structures provided the iGluR field with the first structural framework for receptor activation, deactivation, and desensitization (Fig. 4), and hundreds of AMPA receptor ABD structures [e.g., see Pohlsgaard et al., (2011)] have revealed key ligand-receptor interactions that mediate binding and subunit selectivity of agonists and competitive antagonists (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers).
In the past decade, structural studies on AMPA receptor ABDs have looked beyond the initial models to more dynamic mechanisms that can explain partial agonism (i.e., agonist efficacy) and account for the behavior of ABDs in full-length receptors. In general, strong correlation is observed between agonist efficacy on full-length AMPA receptors and the degree of ligand-induced ABD cleft closure in crystal structures of GluA2 ABDs (Hogner et al., 2002; Jin et al., 2002, 2003). However, these static crystal structures only capture a single conformation of the isolated ABD that may be favored by contacts in the crystal lattice and could be further influenced by the lack of NTD and TMD interactions. These caveats to structures of the isolated ABDs have become more salient as additional structural data emerged [e.g., see Holm et al. (2005) and Zhang et al. (2008b)]. Furthermore, the cleft closure/agonist efficacy relationship observed in most isolated GluA2 ABDs is less evident in structures of GluA3 and GluA4 ABDs (Gill et al., 2008; Ahmed et al., 2009b; Birdsey-Benson et al., 2010; Poon et al., 2011) as well as kainate receptor ABDs [reviewed in Mollerud et al. (2017a)], and this correlation is not found in NMDA receptor ABDs (Furukawa and Gouaux, 2003; Inanobe et al., 2005; Vance et al., 2011; Hansen et al., 2013; Wang et al., 2020).
To elucidate the mechanisms governing agonist efficacy, the dynamic behavior of AMPA receptor ABDs has been investigated using molecular dynamics simulations, fluorescence resonance energy transfer (FRET), small-angle X-ray scattering, and NMR spectroscopy (Madden et al., 2005; Ahmed et al., 2007; Lau and Roux, 2007, 2011; Gonzalez et al., 2008; Maltsev et al., 2008; Fenwick and Oswald, 2010; Landes et al., 2011; Ramaswamy et al., 2012). These studies show that the ABDs sample a range of conformations in response to ligand binding in addition to the conformations observed in crystal structures. Thus, the evolving consensus is that agonist efficacy, presumably in all iGluR subunits, reflects the frequency at which the ABD samples a closed-cleft conformation permissible for channel opening. Support for this conformational selection mechanism of agonist efficacy came from molecular dynamics simulations using umbrella sampling to estimate the free energy landscapes that govern conformational changes in the GluA2 ABD in response to agonist and antagonist binding (Lau and Roux, 2007, 2011). These studies suggested that isolated ABDs are dynamic even when bound to agonists or antagonists and populate a range of conformations to varying degrees, with agonists adopting closed conformations more frequently than antagonists. The dynamic behaviors in these simulations are consistent with SAXS and single-molecule FRET measurements, which also showed that isolated GluA2 ABDs access a wide range of conformations in solution (Madden et al., 2005; Landes et al., 2011; Ramaswamy et al., 2012). Additional support was provided using NMR spectroscopy to show that the average GluA2 ABD conformations in solution are not strongly correlated with agonist efficacy (Maltsev et al., 2008; Ahmed et al., 2009a; Fenwick and Oswald, 2010). Evaluation of hydrogen bonding showed that different interlobe hydrogen bonds are formed between ABD residues and full or partial agonists, confirming these agents stabilize different receptor conformations (Ahmed et al., 2013). Crystal structures of the GluA2 ABD locked in the closed cleft conformation using disulfide bonds showed that the fully closed ABD conformation is capable of accommodating binding of kainate, iodowillardiine, and even the antagonist CNQX (Ahmed et al., 2011a). Thus, it is possible for partial agonists and even some antagonists to sample a fully closed ABD conformation by rearranging side chains in the agonist binding pocket. However, the fully closed ABD conformation sampled by partial agonists and antagonists is energetically unfavorable and would therefore occur with low probability (Lau and Roux, 2007, 2011; Landes et al., 2011; Postila et al., 2011; Ramaswamy et al., 2012). In summary, these studies on the isolated GluA2 ABDs suggest that channel gating is triggered by a closed cleft conformation of the ABD, which could be accessed by all agonists, but that it is the stability of this closed cleft conformation of the ABD as well as stability of the ABD dimer interface that primarily determines agonist efficacy. Mechanisms of receptor activation will be discussed in more detail in Section IV. Receptor Activation, Deactivation, and Desensitization.
An important limitation to isolated ABDs is the lack of interacting domains (e.g., NTDs and TMDs) in the context of the full tetrameric receptor. Consequently, the dynamic behavior of ABDs will likely be more complex in tetrameric receptors. Recent studies that investigated crystal structures of tetrameric GluA2 ABD assemblies rather than the abundant dimer ABD structures revealed that the ABD layer can adopt multiple distinct conformations in response to agonist binding and that full agonists appear more effective at inducing active ABD conformations than do partial agonists (Lau et al., 2013; Baranovic et al., 2016; Salazar et al., 2017).
AMPA receptor positive allosteric modulators (PAMs) exert their effects by increasing the stability of the ABD dimer interface to slow entry into the desensitized state or by slowing receptor deactivation (Jin et al., 2005; Mitchell and Fleck, 2007). Structures revealed that different PAMs can have distinct binding modes at different subsites within the ABD dimer interface (Sun et al., 2002; Jin et al., 2005; Kaae et al., 2007; Hald et al., 2009; Ptak et al., 2009; Sobolevsky et al., 2009; Ahmed et al., 2010; Timm et al., 2011; Krintel et al., 2013; Norholm et al., 2013; Goffin et al., 2018; Laulumaa et al., 2018). This discovery has stimulated development of newer classes of modulators with improved potency (Kaae et al., 2007; Ahmed and Oswald, 2010; Pohlsgaard et al., 2011; Ptak et al., 2014; Laulumaa et al., 2018) (Section IX. Exogenous Positive and Negative Allosteric Modulators).
2. Kainate Receptor Agonist Binding Domains
Since the first X-ray crystal structures of isolated GluK1 and GluK2 ABDs in complex with agonists (Mayer, 2005; Mayer et al., 2006; Hald et al., 2007), there has been a rapid expansion in available kainate subunit ABD structures [reviewed in Mollerud et al. (2017a)], including GluK3 and GluK4 ABDs (Venskutonytė et al., 2011b, 2012, 2014; Veran et al., 2012; Assaf et al., 2013; Kristensen et al., 2016b; Mollerud et al., 2017b; Brogi et al., 2018; Kumari et al., 2019). These structures provided insights into the mechanisms of desensitization, the distinct features of their agonist binding pockets that enabled design of agonists and competitive antagonists (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers), and the structural basis for kainate receptor modulation by cations and anions (Section VIII. Endogenous Allosteric Regulation).
Kainate receptor ABDs generally crystallize as “back-to-back” dimers similar to the GluA2 AMPA receptor ABDs [reviewed in Mollerud et al. (2017a)]. Like AMPA receptors, kainate receptors require rearrangement at the ABD dimer interface to enter the desensitized state. For example, the ABD structure of the nondesensitizing GluA2-L483Y mutant (Sun et al., 2002) was used as a guide to introduce cysteine mutations that stabilize the ABD dimer interface and produce nondesensitizing versions of GluK1, GluK2, and GluK3 receptors (Weston et al., 2006b). Furthermore, structures have revealed the binding sites for Na+ and Cl- ions at the ABD dimer interface and showed that binding of these ions affects desensitization by limiting dimer interface rearrangement (Plested and Mayer, 2007, 2009; Plested et al., 2008; Chaudhry et al., 2009a; Dawe et al., 2013) (Section VIII. Endogenous Allosteric Regulation). The introduction of a Lys residue at the dimer interface (e.g., GluK2-D776K) designed to substitute for the positive charge provided by Na+ enhances kainate receptor activation and prevents desensitization, whereas mutations that disrupt Na+ binding impair receptor activation (Nayeem et al., 2009, 2011; Dawe et al., 2013). Extracellular Zn2+ slows desensitization and enhances glutamate potency selectively for GluK3-containing kainate receptors, and this effect is mediated by Asp759, which is unique to the GluK3 ABD among the kainate receptor subunits (Veran et al., 2012). Structures of the isolated GluK3 ABD suggested that Zn2+ reduces desensitization by stabilizing the ABD dimer assembly (Veran et al., 2012).
Kainate receptors recover markedly slower from desensitization compared with AMPA receptors, which has been attributed in part to interactions between the D1 and D2 lobes that are conserved in kainate receptor subunits but absent in AMPA receptors and that stabilize the closed-cleft agonist-bound ABD conformation (Weston et al., 2006a; Chaudhry et al., 2009b; Wied et al., 2019). Disruptions of these interlobe D1-D2 contacts reduced agonist potency, accelerated receptor deactivation, and enabled faster recovery from desensitization.
The development of allosteric modulators of kainate receptor function has lagged behind AMPA and NMDA receptors (Section IX. Exogenous Positive and Negative Allosteric Modulators). However, one study reported structures of the isolated GluK1 ABD dimer with positive allosteric modulators bound at two equivalent sites in the dimer interface (Larsen et al., 2017). These modulators, which are nonselective and more potent at AMPA receptors, enhanced kainate receptor responses by slowing the rate of desensitization.
3. NMDA Receptor Agonist Binding Domains
Crystal structures of isolated ABDs from GluN1, GluN2A, GluN2D, GluN3A, and GluN3B subunits have been solved in complex with agonists and competitive antagonists (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers), as well as GluN2A-selective allosteric modulators (Section IX. Exogenous Positive and Negative Allosteric Modulators). Structures for the isolated GluN1 ABD in complex with full and partial agonists as well as competitive antagonists (Furukawa and Gouaux, 2003; Inanobe et al., 2005) were followed by a structure of the GluN2A ABD in complex with the GluN1 ABD, which provided the first structural insights into the GluN1-GluN2 subunit interface (Furukawa et al., 2005). The stabilization of GluN1/2 ABDs in the closed conformation by agonist binding and in the open conformation by binding of competitive antagonists is similar to the structural changes found for AMPA and kainate receptor ABDs (Furukawa and Gouaux, 2003; Kvist et al., 2013b; Jespersen et al., 2014; Yi et al., 2016; Lind et al., 2017; Romero-Hernandez and Furukawa, 2017; Chou et al., 2020; Wang et al., 2020; Wang et al., 2021). The agonist-mediated ABD closure in GluN1 and GluN2 subunits is enforced by the formation of interlobe interactions between residues in the D1 and D2 lobes that stabilize the agonist-bound ABD structure (Kalbaugh et al., 2004; Paganelli et al., 2013; Yu and Lau, 2018). Unlike structures of GluA2 AMPA receptor ABDs in complex with partial agonists, the GluN1 and GluN2 ABD structures with bound partial agonists (e.g., d-cycloserine, ACPC, and ACBC in GluN1 or NMDA, homoquinolinate, and Pr-NHP5G in GluN2) show almost identical domain closure compared with structures with full agonists (Inanobe et al., 2005; Vance et al., 2011; Hansen et al., 2013; Chou et al., 2020; Wang et al., 2021). Studies of the dynamic behavior of the isolated GluN1 and GluN2 ABDs suggest that they fluctuate between open and closed cleft conformations even in the absence of agonist, but binding of full agonists favors the fully closed ABD conformations more than binding of partial agonists does (Rambhadran et al., 2011; Yao et al., 2013; Dai et al., 2015; Dai and Zhou, 2015; Dolino et al., 2015, 2016). This conformational selection mechanism likely accounts for partial agonist efficacy in NMDA receptors and is similar to the mechanism of partial agonism in AMPA receptor ABDs (Section II.B.1. AMPA Receptor Agonist Binding Domains).
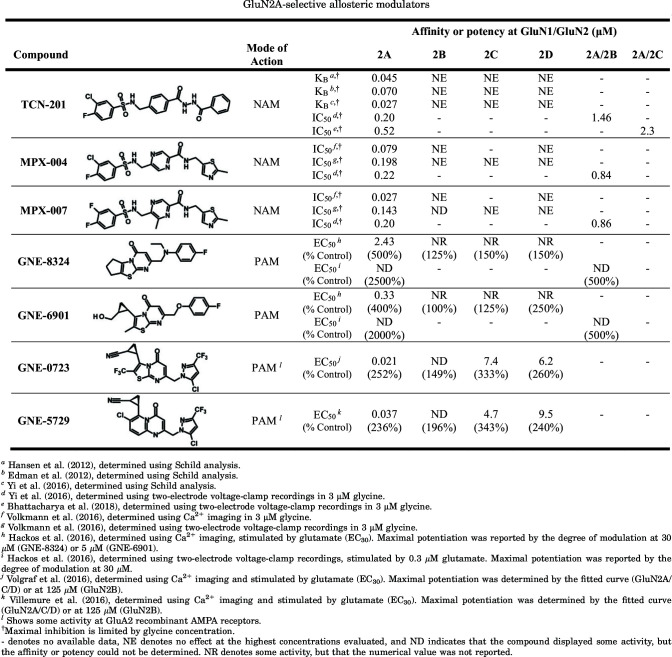
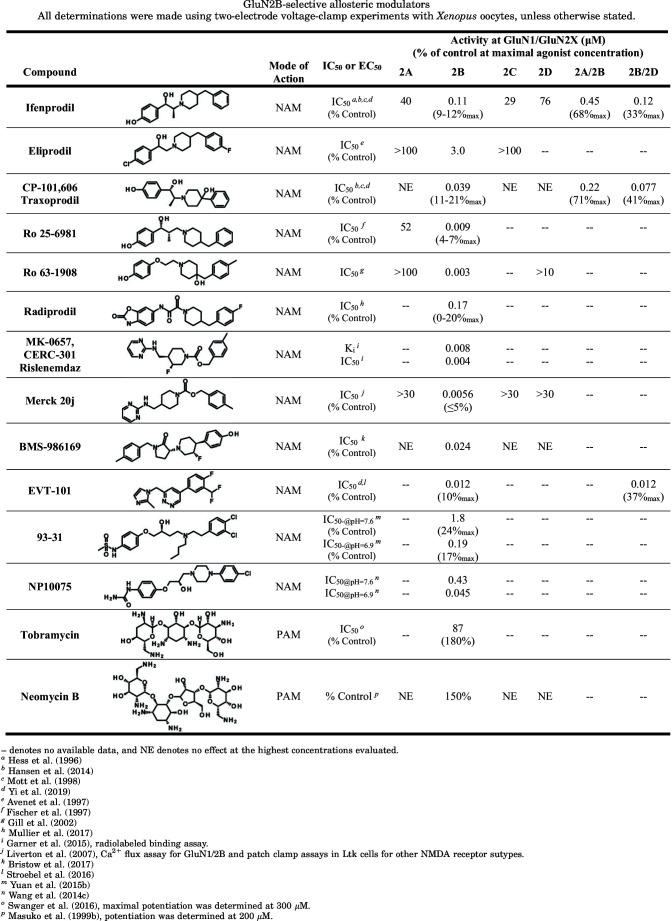
The ABD dimer interface in GluN1/2 receptors is largely unchanged after agonist binding (Borschel et al., 2011; Chou et al., 2020), and NMDA receptors display less desensitization compared with AMPA and kainate receptors (Section IV. Receptor Activation, Deactivation, and Desensitization). Nonpolar interactions between hydrophobic residues stabilize the ABD interface in NMDA receptors and mediate heterodimerization of GluN1 and GluN2 ABDs (Furukawa et al., 2005). Despite these stabilizing interactions, dynamics within the ABD heterodimer interface can influence factors controlling deactivation, such as agonist dissociation or channel open time (Furukawa et al., 2005; Borschel et al., 2015). Interfaces within the ABD heterodimer and between ABD heterodimers are important for mediating allosteric modulation of NMDA receptor function by NTD-binding modulators, such as ifenprodil, Zn2+, and protons (Gielen et al., 2008; Esmenjaud et al., 2019) (Section VIII. Endogenous Allosteric Regulation). The interface between ABD heterodimers also mediates a glutamate-dependent increase in glycine dissociation rate and vice versa (i.e., negative cooperativity between binding of the two agonists) (Durham et al., 2020) (Section IV. Receptor Activation, Deactivation, and Desensitization). Furthermore, the GluN1-GluN2A ABD heterodimer interface contains binding sites for both positive and negative allosteric modulators with selectivity for GluN2A-containing NMDA receptors (Hackos et al., 2016; Villemure et al., 2016; Volgraf et al., 2016; Yi et al., 2016) (Section IX. Exogenous Positive and Negative Allosteric Modulators). Mutations at Tyr535 in GluN1, a residue located centrally in the ABD dimer interface and whose aromatic ring is in a similar position to the positive allosteric modulator aniracetam in AMPA receptors, altered the deactivation time course and could enable potentiation of NMDA receptor responses by aniracetam (Furukawa et al., 2005; Borschel et al., 2015). Structures have revealed variation between agonist binding pockets of GluN1 and GluN3 subunits, although both bind glycine and d-serine (Furukawa and Gouaux, 2003; Inanobe et al., 2005; Yao et al., 2008, 2013), and these structural differences have been exploited in the development of GluN3-preferring orthosteric ligands (Kvist et al., 2013a,b) (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers).
4. GluD Receptor Agonist Binding Domains
The GluD subunits were initially designated as orphan subunits, since their activating endogenous agonist was unknown, and ion channel currents could only be measured from spontaneously active GluD2 receptors containing the lurcher mutation GluD2-A654T [e.g., see Yuzaki and Aricescu (2017)]. However, the orphan subunit designation changed when crystal structures of the isolated GluD2 ABD identified d-serine as a ligand capable of inducing ABD cleft closure that results in reduced spontaneously active currents from GluD2-A654T receptors (Naur et al., 2007). Later, d-serine was established as an endogenous GluD2 receptor agonist that mediates metabotropic signaling; that is, wild-type GluD receptors appear incapable of producing current responses to agonist binding (Kakegawa et al., 2011) (Section IV. Receptor Activation, Deactivation, and Desensitization). The ABD dimer interface in GluD2 receptors contains a binding site for Ca2+ ions, as described in Section II.B.4. GluD Receptor Agonist Binding Domains.
C. The Extracellular N-Terminal Domain
The ∼400 N-terminal residues of iGluRs fold into the semi-autonomous NTD that forms a bilobate structure, and like the ABDs, the NTDs assemble into local dimers. Modulation of receptor function via the NTD has so far only been described for NMDA receptors [reviewed in Zhu and Paoletti (2015) and Hansen et al. (2018)]. Deletion of non-NMDA receptor NTDs leads to minor changes in gating kinetics upon expression in heterologous systems (Horning and Mayer, 2004; Plested and Mayer, 2007; Cais et al., 2014; Moykkynen et al., 2014). By contrast, removal of the NTD produces marked changes in NMDA receptor properties (Gielen et al., 2009; Yuan et al., 2009). The NTDs also play key roles in receptor biogenesis [reviewed in Hansen et al. (2010a) and Herguedas et al. (2013)] and emerging roles in receptor organization at synapses (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity).
1. N-Terminal Domain Structure and Organization
Crystal structures of NTDs have been determined for members from all four iGluR families as homodimers or heterodimers (Jin et al., 2009; Karakas et al., 2009, 2011; Kumar et al., 2009, 2011; Farina et al., 2011; Sukumaran et al., 2011; Yao et al., 2011; Dutta et al., 2012; Elegheert et al., 2016; Herguedas et al., 2016, 2019). The two lobes are connected by three hinges, and the upper (R1) lobe encompasses a highly sequence-conserved region that mediates dimer formation in all iGluRs (Herguedas et al., 2013). Each NTD in the dimer contributes two α-helices and a loop [also termed “flap” (Jin et al., 2009)], which wedges into the dimer interface and differs in length among subunits. The lower (R2) lobes are less conserved and form interface contacts in all non-NMDA receptors contributing to dimer stability in a subtype-specific manner. As a result, AMPA, kainate, and GluD receptor NTDs form rigid dimeric assemblies that are glued together with low nM affinity, which likely plays a role during subunit assembly (Rossmann et al., 2011; Zhao et al., 2017). An exception is the AMPA receptor GluA3 NTD, which exhibits a spectrum of conformations due to repulsion between the R2 lobes caused by charged residues (Sukumaran et al., 2011; Dutta et al., 2012). The GluA3 R2 interface has the capacity to bind ligands and coordinates a phosphate ion, potentially modulating interface strength (Lee et al., 2019). Dynamic movement is unique to GluA3 NTD homodimers and is not apparent in GluA2/3 heteromers (Herguedas et al., 2016), which form tight contacts, as seen for all other AMPA receptor NTDs. The functional consequence of these GluA3-specific structural features is unknown. Although interlayer contacts between the NTD and ABD layers are loose in non-NMDA receptors, contact points have been described in AMPA receptor structures (Sobolevsky et al., 2009; Herguedas et al., 2019; Nakagawa, 2019) and are also predicted in the GluD receptor structures that harbor an additional α-helix that projects toward the ABD (Elegheert et al., 2016). The crystal structure of GluA2 with the Conus snail toxin (con-ikot-ikot) wedged between NTD and ABD also confirms the flexibility that exists between the two layers in AMPA receptors (Chen et al., 2014b).
2. Allosteric Function of NMDA Receptor N-Terminal Domains
The lower (R2) lobes in NMDA receptors are signal transducers triggered by binding of Zn2+, polyamines, and synthetic modulators. NMDA receptors also lack a loop element in the upper NTD lobe that wedges into the NTD dimer interface in non-NMDA receptors. These two structural features confer flexibility and allosteric activity in NMDA receptor NTDs: Lack of the loop element facilitates twisting motions of the NTD monomer (Karakas et al., 2009; Zhu et al., 2013), whereas the “loose” R2 lobes permit intradimer and interdimer motions as well as interlayer communication with the ABD (Tajima et al., 2016) (Section IX. Exogenous Positive and Negative Allosteric Modulators). The R2 interface is proposed to be a structural determinant of polyamine modulation, which can stabilize contacts in GluN1/2B NTD heterodimers, exerting positive allosteric action (Mony et al., 2011) (Section VIII. Endogenous Allosteric Regulation). This region is also regulated by alternative splicing of exon 5 in the GluN1 NTD, which encodes a positively charged loop segment that, like polyamines, affects deactivation kinetics and pH sensitivity (Section II.A.1.c. Structures of tetrameric N-Methyl-d-aspartate receptors).
D. The Transmembrane Domain
Within an individual subunit, the TMD consists of three transmembrane helices, M1, M3, and M4, and a reentrant M2 pore loop (Fig. 5). The transmembrane helices are connected to the ABD by peptide linkers S1-M1, M3-S2, and S2-M4 (Fig. 5). The ion channel is formed by assembly of the four subunit TMDs. When viewed down an axis perpendicular to the membrane, the individual TMDs are positioned in a largely symmetrical fashion around the central axis of the pore or permeation pathway (Sobolevsky et al., 2009; Karakas and Furukawa, 2014; Lee et al., 2014; Meyerson et al., 2014; Burada et al., 2020b).
Fig. 5.

Transmembrane domain topology of iGluRs. (A) Organization within a single subunit monomer illustrating the general arrangement of the upper and lower lobes of the ABD (D1 and D2), the TMD, and the ABD-TMD (ABD-TMD linkers) (PDB: 5WEK). (B) Side view of TMD composed of M1, M3, and M4 transmembrane helices and the membrane re-entrant loop M2. Only the two subunits at positions B and D of the receptor are shown. (C) Cytoplasmic view of the TMD through the central ion channel pore. (D) Key functional features of the TMD pore. The M3 segments contain SYTANLAAF, the most highly conserved motif in iGluRs. At their apex, the M3 segments form an activation gate that prevents the flux of ions in the closed state. The M2 pore loop forms the narrow constriction. (E) M4 wrapping, in which the M4 segment of one subunit is associated with the M1-M2-M3 segment of an adjacent subunit. In AMPA receptors, the M4 is required for receptor assembly, which is presumably stabilized by interactions between M4 and M1-M2-M3 of an adjacent subunit. Positions in M4 where single Trp substitutions block tetramerization are located along a specific face of the helix (red), referred to as “VLGAVE” (V, valine; L, leucine; G, glycine; A, alanine; V, valine; E, glutamate).
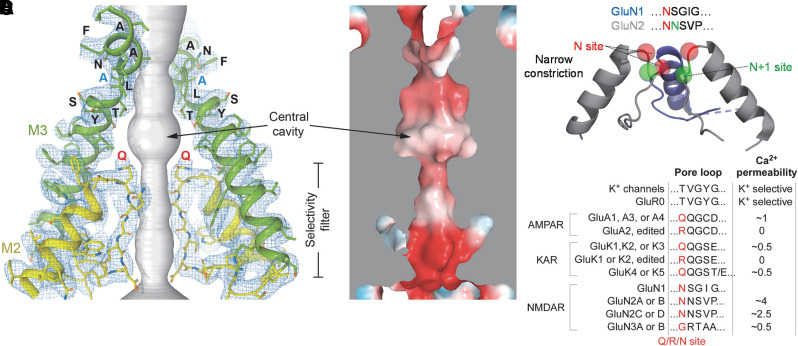
The iGluRs are members of the pore loop superfamily of ion channels that includes different K+ channels, voltage-gated Na+ and Ca2+ channels, cyclic-nucleotide–gated channels, and transient receptor potential channels. This superfamily is defined by a pore domain consisting of two membrane-spanning segments joined by a non–membrane-spanning pore loop that enters and exits on the same side of the membrane (i.e., re-entrant loop). In iGluRs, the pore domain consists of the M1 and M3 transmembrane helices and an M2 pore loop re-entering the membrane from the intracellular side (Wollmuth and Sobolevsky, 2004; Traynelis et al., 2010; Huettner, 2015; Tikhonov and Zhorov, 2020). This M1-M2-M3 pore domain is present in all iGluRs across phyla, but the M4 segment is a eukaryote-specific transmembrane helix that is located to the periphery of the pore domain (Stroebel and Paoletti, 2021).
The iGluR pore domain shares some sequence similarity to the pore domain of K+ channels but is inverted in the membrane by 180° (Wo and Oswald, 1995; Wood et al., 1995; Tikhonov and Zhorov, 2020). This relationship to K+ channels was supported by the identification of a prokaryotic iGluR0 that is missing the NTD and the M4 helix but has an iGluR-like ABD attached to an inverted K+-selective ion channel (Chen et al., 1999a) with the “TXVGYG” sequence of the selectivity filter found in K+ channels (Janovjak et al., 2011). Glutamate-driven ion channel gating can be recreated when a minimal viral K+ channel replaces M1-M3 in the background of a eukaryotic iGluR (Schonrock et al., 2019). Structures of iGluR TMDs have confirmed the structural homology of the iGluR pore domain with that for K+ channels (Sobolevsky et al., 2009; Karakas and Furukawa, 2014; Lee et al., 2014; Meyerson et al., 2014).
1. The Pore Domain or Ion Channel Core
The M3 transmembrane helices line the extracellular half of the permeation pathway (Beck et al., 1999; Sobolevsky et al., 2002a), whereas the M2 pore loops line the intracellular half of the pore (Kuner et al., 1996, 2001) (Fig. 5). The iGluR structures revealed a crossing of the M3 helices at their extracellular apex (Sobolevsky et al., 2009; Karakas and Furukawa, 2014; Lee et al., 2014; Meyerson et al., 2014). This helical bundle crossing of the M3 segments forms an activation gate, which is the structural element that precludes the flux of ions in the closed state (Chang and Kuo, 2008; Sobolevsky et al., 2009; Traynelis et al., 2010) (Section IV. Receptor Activation, Deactivation, and Desensitization). For ion flux to occur across the membrane, the M3 segments must be splayed away from the central axis of the pore (Chen et al., 2017a; Twomey et al., 2017a; Twomey and Sobolevsky, 2018). This general mechanism of pore opening appears conserved across AMPA, kainate, and NMDA receptors (Wilding and Huettner, 2019, 2020) and possibly also in GluD receptors (Hansen et al., 2009; Schmid et al., 2009).
The extracellular part of the M3 segment around the helical bundle crossing contains the SYTANLAAF motif, the most highly conserved motif in iGluR subunits. Residues in SYTANLAAF are elements of the activation gate (Sobolevsky et al., 2009) as well as the site of the lurcher mutation (SYTANLAAF) that initially identified the M3 segment as a gating element (Zuo et al., 1997). Cryo-EM structures of AMPA receptors in the open state revealed an alanine hinge in SYTANLAAF that allows the activation gate to open (Twomey et al., 2017a). Although open-state structures do not presently exist for other iGluRs, mutations at this site in NMDA receptors (Sobolevsky et al., 2007) and GluD1 receptors (Yadav et al., 2011) strongly influence gating, suggesting that this site carries out a common role across iGluRs. In addition, patient-derived variants in these regions can perturb iGluR function (Section X. Glutamate Receptors in Disease).
2. The Membrane M2 Re-Entrant Pore Loop
The M2 loop forms a narrow constriction in the pore that controls ion selectivity, including Ca2+ permeation, single-channel conductance, and channel block (Wollmuth and Sobolevsky, 2004; Traynelis et al., 2010; Glasgow et al., 2015; Huettner, 2015; Hansen et al., 2018). The narrow constriction resides about halfway across the membrane near the apex of the re-entrant M2 pore loop (Fig. 5; Section IV. Receptor Activation, Deactivation, and Desensitization for more detail).
Structures of tetrameric AMPA receptors have indicated that in the closed state, the M2 loops from the different subunits are positioned in close apposition, whereas in the open state they are spread apart (Twomey et al., 2017a). Although the tip of M2 appears to function structurally as a lower gate (Twomey et al., 2017a), the functional significance of the gating-associated M2 loop rearrangement is unknown [but see Smith and Howe (2000), Smith et al. (2000), and Prieto and Wollmuth (2010)]. In heteromeric GluA1/2 receptors, the unedited GluA1(Q) subunits occupy positions A and C in the tetramer, whereas the edited GluA2(R) subunits occupy positions B and D (see Section II.F. Alternative Splicing, RNA Editing, and Post-Translational Modifications for description of RNA editing). This subunit order places the M2 loop tip Q/R/N residues in alternating manner, with Arg residues projecting their side chains along the pore axis toward the central cavity (Herguedas et al., 2019). The M2 loop in NMDA receptors is a locus for gain-of-function human variants that can reduce Mg2+ block, alter Ca2+ permeability, and in some cases alter channel function and surface expression (Fedele et al., 2018; Vyklicky et al., 2018; Li et al., 2019a; Marwick et al., 2019) (Section X. Glutamate Receptors in Disease).
3. The Transmembrane M4 Helix and Receptor Assembly
The M4 helix is a eukaryote-specific transmembrane segment located at the periphery of the pore domain. Notably, the M4 segments of one subunit are associated with the pore domain (M1-M2-M3) of an adjacent subunit (Sobolevsky et al., 2009; Karakas and Furukawa, 2014; Lee et al., 2014; Meyerson et al., 2014). The significance of this trans-arrangement of the M4 segments is unknown, but it may help stabilize the tetramer (Gan et al., 2015). The M4 segment is a critical component for tetramerization of AMPA receptor subunits (Salussolia et al., 2011a, 2013). Without the M4 segment, GluA1 as well as GluA2 subunits fail to form tetrameric assemblies (Salussolia et al., 2013). In AMPA receptor structures [e.g., Sobolevsky et al. (2009)], the M4 from each subunit is fit into a groove formed by the M1 and M3 helices of an adjacent subunit (Fig. 5). Trp substitutions in this “M4 interaction face” (e.g., V795W and E813W) but not on the backside of the helix (e.g., L811W) disrupt tetramerization (Salussolia et al., 2013; Gan et al., 2016). The extracellular third of the AMPA receptor M4 segments may also play more prominent roles in receptor gating (Schmid et al., 2007; Yuan et al., 2014; Yelshanskaya et al., 2017; Shi et al., 2019). The role of M4 segments in NMDA receptor assembly is more complex (Meddows et al., 2001; Cao et al., 2011). Indeed, in contrast to AMPA receptors, a tryptophan screen of the M4 segments in GluN1 and GluN2A subunits revealed no dramatic role in receptor assembly (Amin et al., 2017). On the other hand, the M4 segments are required for functional NMDA receptors (Schorge and Colquhoun, 2003), which may reflect a critical role in gating (Ren et al., 2003a, 2008; Honse et al., 2004; Terhag et al., 2010; Perszyk et al., 2020a).
Upon channel opening in AMPA receptors, the S2-M4 linker that connects the ABD to the extracellular side of M4 shows complete unwinding of the short pre-M4 helices and stretching of the S2-M4 linkers toward the central pore axis to contribute to the ion permeation pathway (Twomey et al., 2017a; Twomey and Sobolevsky, 2018). This finding helps to explain why M4 is not only critical for the receptor tetrameric assembly but has effects on iGluR gating and is implicated in human pathologies (Section X. Glutamate Receptors in Disease). The intracellular part of M4 is directly attached to the highly modifiable intracellular CTD, which can also affect receptor gating (Maki et al., 2012; Murphy et al., 2014) and Ca2+ permeability (Aman et al., 2014).
E. The Intracellular C-Terminal Domain
The CTD of all glutamate receptor subunits resides within the intracellular compartment and is highly diverse even within each subtype class (Traynelis et al., 2010). The diversification of the NMDA receptor CTD among the four GluN2 subunits is proposed to allow specialized and complex roles in neuronal function (Ryan et al., 2008, 2013; Warnet et al., 2020). The CTD varies in length between the different receptor subtypes, being shortest for GluN1, AMPA, and kainate receptor subunits and longest for GluN2 subunits, with multiple splice isoforms described within the AMPA, kainate, and NMDA receptor CTDs (Traynelis et al., 2010) (Section II.F. Alternative Splicing, RNA Editing, and Post-Translational Modifications). The CTD shares no sequence homology to any known proteins, and no structural information exists other than for small portions of the CTD for various subunits (Ataman et al., 2007; Jenkins et al., 2014). The prevailing hypothesis is that the CTDs are flexible and lack overall tertiary structure (i.e., they are intrinsically disordered) (Choi et al., 2013; Chatterjee et al., 2019), which allows them to bind a host of intracellular binding partners with important functional and structural consequences as well as respond to phosphorylation, sometimes with structural changes (Choi et al., 2013; Jenkins et al., 2014; Chatterjee et al., 2019) [reviewed in Purkey and Dell'Acqua (2020) and Warnet et al. (2020)]. Similar to many transmembrane proteins, the intracellular CTDs of iGluR subunits participate in trafficking to the membrane, localization, and degradation [reviewed in Anggono and Huganir (2012), Evans et al. (2019), and Warnet et al. (2020)]. Multiple reports suggest the CTD can move during receptor activation and can impact receptor function (Sager et al., 2009; Maki et al., 2013; Aman et al., 2014; Aow et al., 2015; Dore et al., 2015; Zachariassen et al., 2016; Amin et al., 2018; Sapkota et al., 2019). Yet, removal of the CTD does not abolish function for AMPA, kainate, or NMDA receptors but does alter some functional properties (Yan et al., 2004; Puddifoot et al., 2009; Maki et al., 2012; Punnakkal et al., 2012; Jenkins et al., 2014).
Several endoplasmic reticulum (ER) retention signals reside in alternatively spliced exons of the GluN1 CTD (Scott et al., 2001, 2003; Horak and Wenthold, 2009) and have also been suggested in the CTD of GluN2B (Hawkins et al., 2004) and the CTD of GluN2C (Lichnerova et al., 2014) [also reviewed in Horak et al. (2014)]. Some isoforms of kainate receptor subunits (Section II.F. Alternative Splicing, RNA Editing, and Post-Translational Modifications) also contain discrete regions in their CTDs that promote forward trafficking in GluK2a and GluK3a (Yan et al., 2004; Jaskolski et al., 2005b) or ER retention of GluK1-2b and GluK5 subunits (Ren et al., 2003b, 2003c; Nasu-Nishimura et al., 2006; Hong et al., 2019). There is also a short span of conserved sequence immediately C-terminal to M4 in GluN2 subunits that participates in trafficking (Vissel et al., 2001a; Hawkins et al., 2004). Endocytic motifs that facilitate surface delivery of subunits have been identified within CTDs of GluN1, GluN2A, GluN2B, and GluN3A, and these motifs could mediate changes in NMDA receptor subunit composition during synaptic plasticity (Lavezzari et al., 2004; Scott et al., 2004; Chowdhury et al., 2013). Moreover, there is a nuclear localization signal within the intracellular CTD of GluN1 that could participate in activity-dependent nuclear signaling (Zhou and Duan, 2018).
All iGluR subunits can bind to a number of intracellular scaffold and signaling proteins [reviewed by Coussen (2009), Perez-Otano et al. (2016), Yuzaki and Aricescu (2017), Jacobi and von Engelhardt (2018), Wild and Dell'Acqua (2018), Buonarati et al. (2019), Evans et al. (2019), Matt et al. (2019), Vieira et al. (2020), and Warnet et al. (2020)]. Figure 6 provides a depiction of many of the proteins that can bind to glutamate receptor CTDs, including the PDZ domain ligands that the different iGluRs harbor at their C termini [comprehensive tables are also found in Traynelis et al. (2010)]. Classes of iGluR binding partners include scaffolding, anchoring, cytoskeletal, adaptor, ATPase, and signaling proteins. In addition to indirect linkage of iGluRs to signaling proteins mediated via scaffold proteins, several iGluR subunits bind directly to components of signaling complexes, including GluA1 to cGMP-dependent protein kinase II (PKG) (Serulle et al., 2007), GluA4 to protein kinase C (PKC) (Correia et al., 2003), and NMDA receptor and kainate receptor subunits to Ca2+/calmodulin-dependent protein kinase II (CaMKII) [(Gardoni et al., 1998; Strack and Colbran, 1998; Leonard et al., 1999, 2002; Strack et al., 2000; Carta et al., 2013); reviewed in Bayer and Schulman (2019)], and death-associated protein kinase 1 (DAPK1) (Tu et al., 2010) as well as several tyrosine kinases and phosphatases [reviewed in Groveman et al. (2012) and Won and Roche (2021)]. These interactions can influence intracellular signaling networks and may provide spatial and temporal specificity to receptor function. For example, recent studies have indicated that competition between DAPK1 versus CaMKII binding to the GluN2B CTD helps determine whether NMDA receptor activation of CaMKII signaling will promote long-term depression (LTD) versus long-term potentiation (LTP), respectively (Coultrap et al., 2014; Goodell et al., 2017) (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity). Synaptic iGluRs are embedded within a specialization of the postsynaptic membrane known as the postsynaptic density (PSD), which is formed by an elaborate protein-protein interaction network that is prominently organized by the iGluR-associated scaffolding, cytoskeletal, and signaling proteins [reviewed in Sheng and Kim (2011)] (Fig. 6). Emerging evidence indicates that liquid-liquid phase separation mediated by the multivalent protein-protein interactions within the PSD is important for organizing the synapse to control synaptic transmission and signaling that mediates plasticity [reviewed in Chen et al. (2020b)].
Fig. 6.
Interactions with scaffolding, anchoring, adaptor, and signaling protein complexes in the postsynaptic density (PSD). Depictions of representative CTD binding partners for NMDA receptors (top, left), AMPA receptors (top, right), GluD receptors (bottom, left), and kainate receptors (bottom, right). Note PSD interactions are only shown for select iGluR subunits as indicated and PDZ domain scaffold proteins (medium blue), other scaffolding proteins (peach), kinase/phosphatase anchoring proteins (aqua), protein kinases (green), protein phosphatases (red), second messenger-generating enzymes (dark gray), regulators of small GTPases (light yellow), adaptor proteins (olive), ATPase trafficking regulator (light purple), and F-actin (orange). GKAP, guanylate kinase-associated protein; NSF, N-ethylmaleimide sensitive factor; PICK1, protein interacting with PRKCA 1; Pyk, proline-rich tyrosine kinase; SAP, synapse-associated protein; SHANK, SH3 and multiple ankyrin repeat domains protein; SPAR, spine-associated RapGAP; STEP, striatal-enriched protein tyrosine phosphatase; SynGAP, synaptic Ras GTPase-activating protein; nNOS, neuronal nitric oxide synthase.
The CTD regulates multiple important roles in neuronal function, which is a conclusion supported by genetic deletion studies that remove the CTD or parts thereof. For example, removal of a large portion of the CTD from GluN2B produces a lethal phenotype (Mori et al., 1998; Sprengel et al., 1998). Deletion of the GluN2A CTD reduces GluN2A-containing synaptic receptors, with consequent deficits in cellular models of learning (Sprengel et al., 1998; Steigerwald et al., 2000). Similar effects are observed in mice lacking the GluN2C CTD (Rossi et al., 2002). Studies in which the GluN2A and GluN2B CTDs are swapped or the binding site for CaMKII in GluN2B is disrupted support a critical role of the CTD in synapse development, plasticity, and brain function [(Barria and Malinow, 2005; Gambrill and Barria, 2011; Halt et al., 2012); but see McKay et al. (2018)] and suggest differential sensitivity to excitotoxicity (Martel et al., 2012). Mice in which the GluA1 CTD was removed were still able to undergo forms of synaptic plasticity, arguing that post-translational modification of the GluA1 CTD is not required for LTP expression (Diaz-Alonso et al., 2020). However, other studies have found impacts on both LTP and LTD in mice expressing chimeras swapping the CTDs of GluA1 and GluA2 or point mutations eliminating phosphorylation sites in the GluA1 CTD (Lee et al., 2003; Zhou et al., 2018) (see Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity for additional details).
F. Alternative Splicing, RNA Editing, and Post-Translational Modifications
In addition to differences in subunit composition, diversity in function and pharmacology of iGluR subtypes can also be influenced by RNA editing, alternative splicing, and post-translational modifications of iGluR subunits.
1. Regulation of Receptor Function by RNA Editing
RNA editing is a post-transcriptional modification in which adenosine deaminases acting on RNA (ADARs) alter sequences by converting a specific adenosine nucleotide to inosine (A-to-I), which is read as guanosine during translation of the mRNA (Gott and Emeson, 2000; Bass, 2002). Depending on the modified codon, RNA editing can change the amino acid sequence of the encoded gene product. AMPA receptor GluA2, GluA3, and GluA4 and kainate receptor GluK1 and GluK2 subunits undergo RNA editing to shape their functional properties and physiologic roles [reviewed in Hood and Emeson (2012) and Filippini et al. (2017)] (Fig. 7).
Fig. 7.
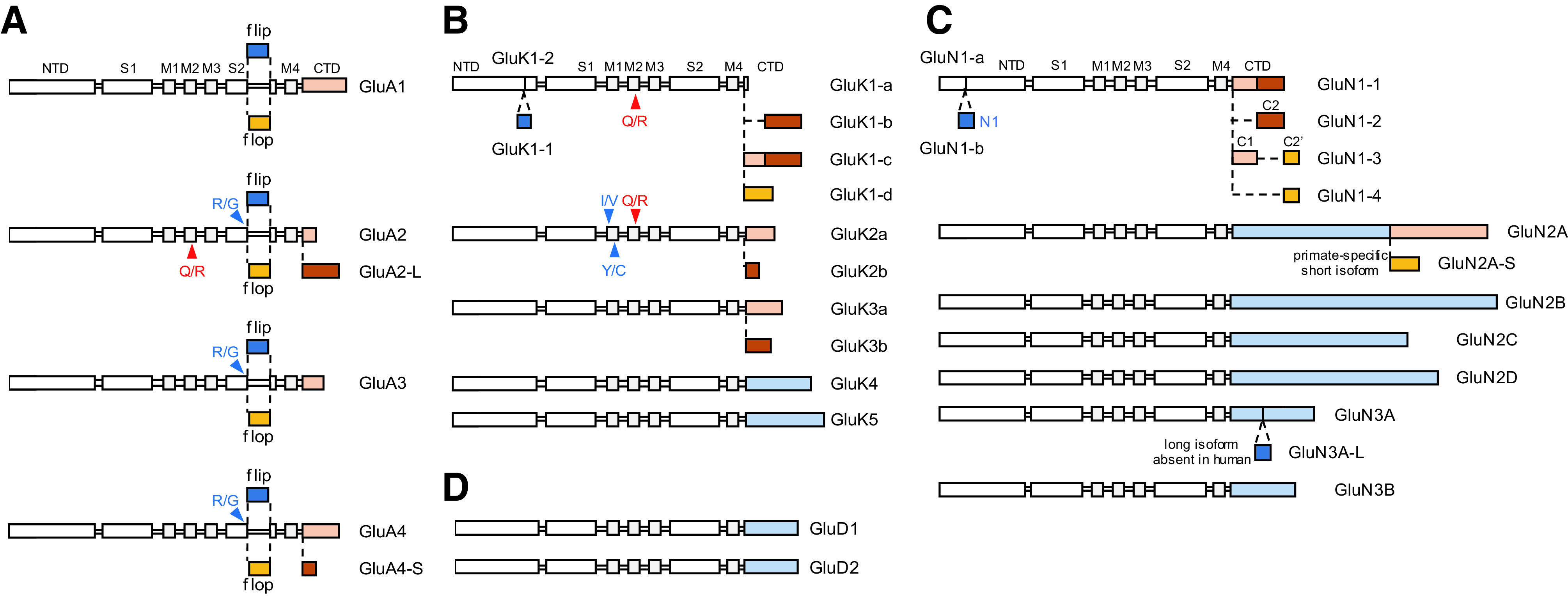
RNA editing and alternative splicing of iGluR subunits. Linear depictions of the polypeptide chains illustrate sites of amino acid changes resulting from RNA editing (indicated by arrows) and isoforms resulting from alternative RNA splicing (indicated by dashed lines). The subunits are not scaled accurately but indicate the relative sizes. (A) AMPA receptor subunits exist as either flip or flop isoforms that differ in the S2 region of the ABD. Alternative splicing also produces two isoforms of GluA2 and GluA4 with different lengths of their CTDs. RNA editing at the Gln/Arg (Q/R; also denoted Q/R/N) site in the M2 region of GluA2 regulates Ca2+ permeability and channel rectification of GluA2-containing AMPA receptors. GluA3 and GluA4 also undergo RNA editing at the Arg/Gly (R/G) site in the S2 region of the ABD. (B) Alternative splicing results in GluK1 isoforms with (GluK1-1) or without (GluK1-2) an insertion in the NTD. GluK1-3 also have variable CTDs as a result of alternative splicing. GluK1 and GluK2 undergo RNA editing at Q/R site in the M2 region, similar to GluA2. GluK2 undergoes additional RNA editing at Ile/Val (I/V) and Tyr/Cys (Y/C) sites in the M1 region. (C) GluN1 exists in 8 different isoforms because of alternative splicing of exon 5, exon 21, and exon 22; N1 (21 aa) in the NTD is encoded by exon 5, and C1 (37 aa) in the CTD is encoded by exon 21, whereas C2 (38 aa) in the CTD is encoded by exon 22. Deletion of exon 22 creates a shift in the open reading frame, resulting in the alternate exon 22′ that encodes C2′ (22 aa). GluN2A exists in two isoforms with short and long CTDs, although the short form appears to be primate-specific (i.e., not found in rodents). Alternative splicing creates the GluN3A-L isoform with a 20-aa insertion in the CTD, but this long isoform is not found in human. (D) GluD subunits do not appear to exist in multiple isoforms.
The permeation and channel block properties of GluA2-containing AMPA receptors and GluK1- and GluK2-containing kainate receptors are profoundly modified by RNA editing at the Q/R site (i.e., Gln is converted to Arg; also denoted the Q/R/N site) located at the narrow constriction formed by the M2 pore loop (Sommer et al., 1991) (Section IV. Receptor Activation, Deactivation, and Desensitization). AMPA or kainate receptors containing the edited forms (e.g., GluA2(R)), which place a charged residue (Arg) within the pore, are virtually impermeable to Ca2+, permeable to Cl-, and insensitive to polyamine channel block (Verdoorn et al., 1991; Egebjerg and Heinemann, 1993; Burnashev et al., 1995, 1996) (Section VIII. Endogenous Allosteric Regulation). These receptors possess a nearly linear current/voltage (I/V) relationship and show substantially reduced single-channel conductance (Section IV. Receptor Activation, Deactivation, and Desensitization, Supplemental Tables 1 and 2). By contrast, AMPA or kainate receptors that lack edited subunits or contain the unedited forms (e.g., GluA2(Q)) are permeable to Ca2+ and sensitive to polyamine channel block and display inwardly rectifying I/V relationships [reviewed in Bowie (2018)]. In the CNS, virtually all mRNAs for GluA2 are edited at the Q/R site throughout development (Sommer et al., 1991; Burnashev et al., 1992a), and native AMPA receptor subtypes are therefore typically categorized as Ca2+-permeable (GluA2-lacking) or Ca2+-impermeable (GluA2-containing) receptors (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity). RNA editing at the Q/R site in GluK1 and GluK2 subunits occurs with low efficiency early in development but increases to ∼40% and ∼80%, respectively, within the first postnatal days (Belcher and Howe, 1997; Paschen et al., 1997; Bernard et al., 1999).
AMPA receptor subunits GluA2, GluA3, and GluA4 can also undergo RNA editing at the R/G site located in the S2 segment of the ABD, preceding the flip/flop region that undergoes alternative splicing (Fig. 7). Editing at this R/G site increases with neuronal maturation and is influenced by neuronal activity (Lomeli et al., 1994; Orlandi et al., 2011; Sanjana et al., 2012; Balik et al., 2013). AMPA receptors with GluA2-flop subunits that are edited at the R/G site display faster rates of desensitization and faster recovery from desensitization (Lomeli et al., 1994; Krampfl et al., 2002). However, the vicinity to the flip/flop region makes these effects of R/G editing effective in flop but not in flip isoforms of GluA2 (Wen et al., 2017). Furthermore, both RNA editing at the Q/R and R/G sites in AMPA receptors influences subunit biogenesis and receptor assembly (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity).
In addition to Q/R editing, the GluK2 kainate receptor subunit can undergo RNA editing at I/V and Y/C sites located in the M1 transmembrane helix (Fig. 7). Editing at I/V and Y/C sites in GluK2 affects permeation properties in a manner dependent on editing at the Q/R site, with the effects of Q/R editing on Ca2+ permeability being enhanced by editing at the I/V and Y/C sites (Kohler et al., 1993; Burnashev et al., 1995).
2. Regulation of Receptor Function by Alternative Splicing
The mutually exclusive alternative splicing of exons 14 and 15 in all four AMPA receptor subunits produces two isoforms termed flip and flop that are different in the S2 segment of the ABD [(Sommer et al., 1990); reviewed in Sukumaran et al. (2012)] (Fig. 7). The flip/flop region influences the rates of desensitization and deactivation (Section IV. Receptor Activation, Deactivation, and Desensitization) and the effects of auxiliary subunits (Section III. Auxiliary Subunits) as well as sensitivity to allosteric modulators that bind at the ABD dimer interface (Sections VIII. Endogenous Allosteric Regulation and IX. Exogenous Positive and Negative Allosteric Modulators). Alternative splicing to produce flip/flop isoforms is developmentally regulated, with flip isoforms prominent from birth and throughout development, and flop isoforms occurring after birth and reaching adult levels at P14 in rodents (Monyer et al., 1991). Flop isoforms generally display faster desensitization compared with flip isoforms (Mosbacher et al., 1994), and flop isoforms are generally less sensitive to AMPA receptor PAMs, such as cyclothiazide (Section IX. Exogenous Positive and Negative Allosteric Modulators). The complexity among AMPA receptor subunits is enhanced by additional alternative splicing to create variation in the CTD of GluA2 and GluA4 subunits [reviewed in Shepherd and Huganir (2007) and Diering and Huganir (2018)] (Fig. 7). GluA1 and GluA4 and the GluA2-L isoform have long CTDs, whereas the predominant splice form of GluA2 has a short CTD similar to GluA3 and the GluA4-S isoform. The splice isoforms of AMPA receptor CTDs influence interactions with specific intracellular binding partners and affect regulation by post-translational modifications (e.g., protein phosphorylation) [reviewed in Shepherd and Huganir (2007) and Diering and Huganir (2018)].
Alternative splicing can produce two isoforms of the NTD in the GluK1 kainate receptor subunit, which are denoted GluK1-1 (15-amino-acid insertion) and GluK1-2 (Bettler et al., 1990) (Fig. 7). The CTD of GluK1 also exists in several isoforms, GluK1a, GluK1b, and GluK1c (Sommer et al., 1992) as well as an additional isoform, GluK1d, which is only found in human (Gregor et al., 1993; Barbon and Barlati, 2000). Splice isoforms of the CTDs in GluK2 and GluK3 subunits have also been reported (Gregor et al., 1993; Schiffer et al., 1997; Barbon et al., 2001) (Fig. 7). The variation in the CTDs of kainate receptor subunit isoforms influences cell surface trafficking, subcellular localization, and interactions with intracellular proteins involved in tuning of neuronal functions [reviewed in Contractor et al. (2011), Lerma and Marques (2013), Pahl et al. (2014), and Valbuena and Lerma (2021)].
The GluN1 NMDA receptor subunit occurs in eight different isoforms that arise from alternative splicing of three exons (Durand et al., 1992; Nakanishi et al., 1992; Sugihara et al., 1992; Hollmann et al., 1993) (Fig. 7). Exon 5 encodes 21 amino acids in the NTD (i.e., the N1 region), exon 21 encodes 37 amino acids (C1) in the CTD, and exon 22 encodes 38 amino acids (C2) in the CTD. Deletion of exon 22 removes the stop codon, resulting in the inclusion of 22 amino acids (C2’). The different GluN1 isoforms have distinct regional and developmental expression patterns (Section VI. Developmental and Regional Expression in the Central Nervous System), and alternative splicing of exon 5 produces marked differences in NMDA receptor function (Sections IV. Receptor Activation, Deactivation, and Desensitization and V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity) and pharmacology (Sections VIII. Endogenous Allosteric Regulation and IX. Exogenous Positive and Negative Allosteric Modulators). Alternative splicing produces a long and short form of the CTD in the GluN2A subunit, but the short form, GluN2A-S, which lacks 343 amino acids, appears to be specific to primates and has not been found in rodents (Warming et al., 2019). By contrast, a long isoform of the GluN3A CTD, GluN3A-L, which has an insertion of 20 amino acids, has been found in rodents (Sun et al., 1998) but is lacking from human (Andersson et al., 2001).
3. Post-Translational Modifications of Glutamate Receptor Subunits
All iGluR subunits are subject to various forms of post-translational modifications, such as glycosylation (Standley and Baudry, 2000) and disulfide bond formation in extracellular domains, S-nitrosylation (Nakamura and Lipton, 2016; Morris et al., 2018a), ubiquitylation (Goo et al., 2015), palmitoylation (Thomas and Huganir, 2013; Han et al., 2015a; Sohn and Park, 2019; Hayashi, 2021), and phosphorylation of intracellular residues (Sanz-Clemente et al., 2013; Wang et al., 2014d; Ghafari et al., 2015; Purkey and Dell'Acqua, 2020) as well as proteolytic cleavage (Wiera and Mozrzymas, 2015; Curcio et al., 2016). In many cases, the post-translational modifications are dynamically regulated to control receptor trafficking to the membrane, subcellular localization, and degradation (Anggono and Huganir, 2012; Pahl et al., 2014; Evans et al., 2019; Purkey and Dell'Acqua, 2020; Warnet et al., 2020). All iGluR subunits are glycosylated to facilitate proper subunit folding during biogenesis and shape the functional properties of the receptor (Standley and Baudry, 2000). Numerous specific sites for phosphorylation, S-nitrosylation, ubiquitylation, and palmitoylation have been reported in iGluR subunits that are critical to their physiologic roles and regulation in the CNS. Many excellent reviews summarize the physiologic effects mediated by these post-translational modifications (Traynelis et al., 2010; Gladding and Raymond, 2011; Mao et al., 2011; Lussier et al., 2015; Diering and Huganir, 2018).
III. Auxiliary Subunits
Auxiliary subunits are integral but not obligatory members of the glutamate receptor complex and directly interact with the receptor and modulate its function and localization but do not assemble alone to create receptors that bind agonist and undergo activation or gating. The past decade has seen a large number of studies that validate the essential role of auxiliary subunits in modifying all aspects of receptor function. The interfaces formed by auxiliary subunits and glutamate receptor subunits are also drug targets with the potential to tune receptor function selectively (Section IX. Exogenous Positive and Negative Allosteric Modulators).
A. AMPA Receptor Auxiliary Subunits
A substantial proportion of native AMPA receptors contain auxiliary subunits that include a surprisingly diverse range of structurally unrelated proteins (Jackson and Nicoll, 2011a; Straub and Tomita, 2012; Greger et al., 2017; Chen and Gouaux, 2019; Twomey et al., 2019; Kamalova and Nakagawa, 2021). AMPA receptor auxiliary subunits include TARPs (Chen et al., 2000; Tomita et al., 2003, 2005a; Kato et al., 2008), cornichons (CNIHs) (Schwenk et al., 2009), cysteine‐knot AMPA receptor modulating proteins (CKAMPS) (termed shisa proteins in the genome database) (von Engelhardt et al., 2010; Farrow et al., 2015), GSG1L (Schwenk et al., 2012; Shanks et al., 2012), and synapse differentiation-induced gene 4 (SynDIG4) [also known as proline rich transmembrane protein 1 (Prrt1)] (Matt et al., 2018) (Fig. 8). Auxiliary subunits can influence receptor localization, trafficking, pharmacology, response time course, ion permeation, and block by intracellular polyamines (Figs. 8 and 9). The expression patterns of auxiliary subunits vary substantially in the brain because each has different upstream promotors that respond to distinct signaling pathways (Tomita et al., 2003; Kato et al., 2007; Schwenk et al., 2014; Yamasaki et al., 2016).
Fig. 8.

Functional effects of different classes of AMPA receptor auxiliary subunits. The top panel illustrates membrane topologies of AMPA receptor auxiliary subunits (CKAMPs and SynDIG4 are based on prediction). The lower panel summarizes functional characteristics of AMPA receptors associated with the respective auxiliary subunits. Adapted with permission from Kamalova and Nakagawa (2021).
Fig. 9.

Examples of the action of AMPA receptor auxiliary subunits. (A) Recordings from cerebellar granule cells from wild type and stargazer (stg) mice that lack TARP γ-2. Left panel: whole cell synaptic currents evoked by mossy fiber stimulation. A near-complete loss of AMPA receptor-mediated synaptic transmission was observed in stg mice (slower NMDA receptor component remains). Right panel: Spontaneous synaptic currents were significantly reduced in homozygous stg/stg mice compared with heterozygous +/stg mice. (B) Outside-out patch-clamp recording of recombinant GluA4 and GluA4 with TARP γ-2 (stg) expressed in HEK cells. Receptors were activated by fast glutamate application and single-channel unitary currents were resolved in the tails of peak currents. Longer bursts or clusters of openings were observed in in the presence of the TARP γ-2. (C) Right panel: whole cell current recordings from hippocampal CA1 pyramidal cells evoked by stimulation of Schaffer collaterals before and after LTP induction in wild type (+/+) and TARP γ-8 KO mice. Left panel: average amplitudes of evoked synaptic responses before and after the induction of LTP. (D) Whole-cell current recordings from mossy hilar cells in the hippocampus. Spontaneous synaptic currents were compared between cells treated with short hairpin RNA (shRNA) of CNIH2 (red and blue are different cells) and untreated wild-type cells. Reproduced with permission from Hashimoto et al. (1999) (Copyright 1999 Society for Neuroscience) (A), Chen et al. (2000) (A), Tomita et al. (2005a) (B), Rouach et al. (2005) (C), and Boudkkazi et al. (2014) (D).
1. Transmembrane AMPA Receptor Regulatory Proteins (TARPs)
Stargazin (TARP γ-2) was the first AMPA receptor auxiliary subunit discovered. It is homologous to the γ-1 subunit of voltage-gated calcium channels and is the protein encoded by the causative gene for the stargazer mutant mouse, whose phenotypes are cerebellar ataxia, abnormal head tossing, and absence seizures (Letts et al., 1998). Recordings from cerebellar granule cells in stargazer mice revealed that AMPA receptors were absent from the plasma membrane (Hashimoto et al., 1999; Chen et al., 2000). Stargazin regulates both trafficking and gating of AMPA receptors (Chen et al., 2000; Tomita et al., 2005a) and thus is the founding member of a family of proteins referred to as TARPs, which consists of three subfamilies based on phylogeny: type Ia (γ-2 and γ-3), type Ib (γ-4, and γ-8), and type II (γ-5 and γ-7) (Tomita et al., 2003; Kato et al., 2008; Jackson and Nicoll, 2011a).
a. TARP structure
The architecture of AMPA receptor and type I TARP complexes in open and closed conformations has been studied using cryo-EM (Twomey et al., 2016, 2017b; Zhao et al., 2016b; Chen et al., 2017a; Herguedas et al., 2019) (Fig. 10). The stoichiometries observed are one, two, or four γ-2 per homomeric GluA2 (Twomey et al., 2016, 2017b, 2018; Zhao et al., 2016b; Chen et al., 2017a), whereas two copies of γ-8 were found in heteromeric GluA1/2 receptors (Gill et al., 2011; Herguedas et al., 2019). The current view is that multiple types of TARPs and also cornichons (see below) can coassemble into the same synaptic AMPA receptor complex (Kato et al., 2010; Studniarczyk et al., 2013; Khodosevich et al., 2014; Zhao et al., 2019). TARPs are homologous to claudins and share a similar overall architecture (Suzuki et al., 2014; Saitoh et al., 2015; Nakamura et al., 2019). The topology of TARPs comprises four transmembrane helices (TM1-4) with a cytoplasmic N terminus immediately preceding TM1 (Figs. 8 and 10). The extracellular domain is shaped like a hand and consists of five β-strands, an extracellular helix, and four flexible loops. The TM3 and TM4 of TARPs interface with the molecular surface formed by M1 and M4 of adjacent AMPA receptor subunits (Twomey et al., 2016), indicating that AMPA receptor subunit assembly is required for TARP binding (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity). As a consequence of the overall 2-fold symmetry of AMPA receptors, two distinct TARP binding sites are present and referred to as the A′/C′ and B′/D′ sites (Zhao et al., 2016b), which bear no relation to the A/C and B/D subunit positions defined within the AMPA receptor tetramer (Section II. Receptor Structure). Each of the two modes of association induces different interaction interfaces between the TARPs and AMPA receptor subunits, creating potentially unique drug binding sites (see Section IX. Exogenous Positive and Negative Allosteric Modulators). The extracellular domain arrangement of TARPs relative to the ABD and the ABD-TMD linkers in AMPA receptors differ substantially in the two types of TARP binding sites. The A′/C′ sites beneath ABD dimers are sterically more restricted than the B′/D′ sites, which are located beneath the ABD dimer of dimers. Hence, TARPs may shape function of AMPA receptors differently depending on which binding site they occupy within the receptor complex. Lipids are preferentially recruited to the surface adjacent to TM4 of γ-8 in the GluA1/GluA2/γ-8 complex (Fig. 10) and may contribute to TARP-dependent modulation of function (Herguedas et al., 2019).
Fig. 10.
Structures of AMPA receptor in complex with various auxiliary subunits. Ribbon diagrams are derived from cryo-EM maps of AMPA receptors in complex with TARP γ-2 (PDB: 5WEO), TARP γ-8 (PDB: 6QKC), GSG1L (PDB:5WEL), and CNIH3 (PDB: 6PEQ). Only the ABD and TMD of the AMPA receptor tetramers (GluA2: light orange, GluA1: light yellow) are shown. Side (left) and bottom (right) views are displayed. γ-2, γ-8, GSG1L, and CNIH3 bind to identical surfaces of AMPA receptor tetramer. The binding sites are indicated as A′, B′, C′, and D′ site. The overall 2-fold symmetric architecture of AMPA receptor tetramers produces two types of binding interfaces: the equivalent B′/D′ sites and equivalent A′/C′ sites.
A cytoplasmic C-terminal tail extends from TM4 and terminates with a PDZ domain binding motif in type I TARPs, whereas type II TARPs lack this motif. The interaction between the intracellular C terminus and PSD-95 and possibly other synaptic scaffolding proteins can contribute to diffusional trapping of the AMPA receptors into the postsynaptic density (Bats et al., 2007; Opazo et al., 2010; Constals et al., 2015). Such spatial reorganization of postsynaptic AMPA receptors at the millisecond time scale can contribute to synaptic plasticity (Heine et al., 2008; Penn et al., 2017). Both the PDZ domain binding motif and the membrane proximal elements in the C terminus of type I TARPs (γ-2 and γ-8) interact with PSD-95, and the formation of the complex induces a macromolecular condensation that results in small organelle-like subcellular compartments devoid of surrounding membrane, a phenomenon referred to as liquid-phase condensation (Zeng et al., 2019) (Section II.F. Alternative Splicing, RNA Editing, and Post-Translational Modifications). Phosphorylation at the C terminus of type I TARPs regulates the interaction with PSD-95 (Tomita et al., 2005b; Sumioka et al., 2010, 2011; Sheng et al., 2018; Zeng et al., 2019). Furthermore, the C-terminal tail of γ-2 interacts with adaptor proteins that regulate receptor endocytosis (Matsuda et al., 2008). The regulation of this interaction by phosphorylation is suggested as a mechanism for LTD (Matsuda et al., 2013). The membrane proximal region in the C terminus of γ-2 also binds to the plasticity-regulated protein Arc (Zhang et al., 2015) but with unknown functional consequence.
b. Functional effects of TARPs
Stargazin γ-2 increases the duration and frequency of AMPA receptor channel openings to higher conductance levels (Tomita et al., 2005a; Zhang et al., 2014a). TARPs also alter the pharmacological properties of AMPA receptors; in the presence of γ-2, the partial agonist kainate induces current with substantially greater amplitude (Tomita et al., 2006), and competitive antagonist CNQX but not NBQX is converted to a partial agonist (Menuz et al., 2007). Measurements of NBQX binding and unbinding rates as well as luminescence resonance energy transfer measurements indicated that γ-2 facilitated a higher degree of cleft closure of the ABD (MacLean et al., 2014), which might underlie the higher agonist efficacy observed in the presence of γ-2 (MacLean et al., 2014). Thus, the small degree of ABD closure induced by CNQX is sufficient for receptor activation in the presence of γ-2. TARPs also alter the effects of positive allosteric modulators (e.g., LY-450295 and CX-614) at AMPA receptors (Schober et al., 2011; Radin et al., 2018a,b).
In the presence of TARPs, desensitization is less prominent (Coombs et al., 2017), and the deactivation time course after removal of glutamate is slowed, prolonging the response time course and increasing charge transfer (Figs. 8 and 9, see Table 2). The rate of recovery from desensitization changes in a manner dependent on the combination of TARPs and AMPA receptor subunits. γ-2 and γ-8 accelerate recovery from desensitization for GluA1-containing receptors (Priel et al., 2005; Gill et al., 2012), whereas γ-8 slows recovery from desensitization for GluA2- and GluA3-containing receptors (Gill et al., 2012; Cais et al., 2014) (Fig. 11). As many as four TARPs may associate concurrently with a single AMPA receptor, and the characteristics of gating modulation by TARPs depend on the stoichiometry (Miguez-Cabello et al., 2020). Figure 11 and Table 2 summarize the effect of TARPs on AMPA receptor function. The association of AMPA receptors with γ-4, γ-7, and γ-8 induces resensitization upon prolonged (several seconds) exposure to glutamate (Kato et al., 2010). Resensitization is defined as a slowly increasing steady-state current after desensitization in the continued presence of glutamate (Supplemental Fig. 1). This phenomenon involves an extracellular linker domain in TARP γ-8 (Riva et al., 2017) and involves transitions to higher conductance levels while the receptor is in the desensitized state, which is similar to effects observed at the single-channel level for compounds that block desensitization (e.g., cyclothiazide) (Carrillo et al., 2020b) (Supplemental Fig. 1). Compounds that bind at the interface between γ-8 and the AMPA receptor TMD block resensitization (Kato et al., 2016; Maher et al., 2016; Dohrke et al., 2020) (Section IX. Exogenous Positive and Negative Allosteric Modulators). AMPA receptors with γ-2 and γ-8 have been shown to undergo superactivation upon repetitive high-frequency activation (i.e., response amplitudes increase with each subsequent activation), a phenomena that has also been termed resensitization (Carbone and Plested, 2016). Such resensitization properties mediated by TARPs may be relevant in synapses with slow glutamate clearance, such as, for example, mossy fiber terminals in the hippocampal CA3 region (Savtchenko and Rusakov, 2004).
TABLE 2.
Modulation of AMPA receptor properties by auxiliary subunits
Values at 2–3 significant figures are from the text or tables; when values are not reported, measurements were made from figures.
| Receptor a | Subunit b | Mean γ c | EC50,SS d (EC50,PEAK) e | τ-Deactivate f | τ-Desensitize f | τ-Recovery g |
Steady State/Peak
Current Ratio |
|---|---|---|---|---|---|---|---|
| pS | μM (mM) | ms | ms | ms | |||
| GluA1 | – | 16–20 | 15-37 (0.9–1.6) | 0.5–2.9 | 2.3–4.1 | 83–160 | 0.003–0.019 |
| GluA1 | γ−2 | ↑ 27 4,6 | ↓ 5–6 (0.27–0.46) 2,6,8,10,11 | ↑ 1.4–3.3 2,8,10,15,18,28,31 | ↑ 3.9–7.4 2,4,8,10,28,31 | ↓ 55–170 6,15 | ↑ 0.02–0.09 2,8,9,28 |
| GluA1 | γ−3 | ↑ 274 | ↓ 5.3 11 | ↑ 2.2–3.0 15,28,31 | ↑ 5.0–5.5 2,28,31 | – | ↑ 0.02 9,28 |
| GluA1 | γ−4 | 26–32 3 | ↓ 20 11 | ↑ 2.4–12 3,15,28,31 | ↑ 5.2–9.6 3,28,31 | – | ↑ 0.052 28 |
| GluA1 | γ−5 | ↑ 44 3 | ↑ 100 21 | 1.3 10 | 2.3–4 3,10 | – | – |
| GluA1 | γ−7 | – | 20 10 | ↑ 1.8 10 | ↑ 6 10 | – | – |
| GluA1 | γ−8 | – | ↓ 17 11 | ↑ 2.5–7.7 15,19,20,28,31 | ↑ 5.9–12 19,20,23,28,31 | ↓ 65–150 15,19 | ↑ 0.047–0.17 19,23,28 |
| GluA1 | GSG1L | ↓ 11 1 | – | – | – | – | – |
| GluA1 | CNIH2 | ↑ 30 2 | ↓ 9.4 (0.92) 2 | ↑ 4.0–9 2,15,20 | ↑ 5.5–13 2,20 | ↓ 135 15 | 0.067 2 |
| GluA1 | CNIH3 | ↑ 28 2 | – | ↑ 3.3 2 | ↑ 5.0 2 | – | 0.041 2 |
| GluA1 | CKAMP39 | – | ↓ 8.0 7 | 1.9 7 | 3.0 7 | ↑ 950 7 | 0.008 7 |
| GluA1 | CKAMP44 | – | ↓ 3.6 7 | – | 3.3 23 | – | ↓ 0.008 23 |
| GluA1 | CKAMP52 | – | ↓ 4.8 7 | 1.9 7 | 3.8 7 | 96 7 | ↑ 0.016 7 |
| GluA1 | CKAMP59 | – | – | 1.9 7 | 3.7–4.5 7,12 | 95–120 7,12 | 0.007 7 |
| GluA1 | SynDIG4 | – | – | ↑ 4.8 19 | ↑ 5.9 19 | 206 19 | 0.021 19 |
| GluA2 | – | 5.1 | 50 | 1.0 | 10 | – | 0.01–0.12 |
| GluA2 | γ−2 | 3.8 27 | – | ↑ 2.5–3.7 21,27 | 10–13 21,27 | – | ↑ 0.25–0.53 9,21,27 |
| GluA2 | γ−3 | – | – | – | – | – | ↑ 0.03 9 |
| GluA2 | γ−5 | 6.5 3 | ↑ 300 21 | 1.8 21 | ↓ 7 21 | – | ↓ 0.005–0.02 9,21 |
| GluA2 | γ−8 | – | – | – | ↑ 17 27 | – | ↑ 0.30 27 |
| GluA2 | GSG1L | – | – | – | 12 27 | – | 0.01427 |
| GluA2Q | – | 19–22 | 28–170 | 0.55–1.8 | 4.8–9.2 | 15–22 | 0.018–0.05 |
| GluA2Q | γ−2 | ↑ 30–31 2,3,27 | – | ↑ 0.67–5.0 22,24,27,30 | ↑ 10–45 2,22,24–26,30 | 13–22 22,30 | ↑ 0.07–0.27 2,8,22,24-26,30 |
| GluA2Q | γ−3 | – | – | – | ↑ 12 30 | 22 30 | ↑ 0.08 30 |
| GluA2Q | γ−4 | – | – | – | ↑ 13 30 | ↑ 67 30 | ↑ 0.05 30 |
| GluA2Q | γ−5 | ↑ 26 3 | 160 21 | – | 7.6 3 | 19 3 | – |
| GluA2Q | γ−7 | – | – | – | ↑ 16 24 | – | – |
| GluA2Q | γ−8 | ↑ 35 27 | – | – | ↑ 11–25 26,30 | ↑ 78 30 | ↑ 0.075–0.25 26,30 |
| GluA2Q | GSG1L | ↓ 12–13 1,27 | – | ↑ 5.9 13 | 9–11 5,13,27 | ↑ 160–200 5,13 | 0.032 13 |
| GluA2Q | CNIH2 | ↑ 29 2 | – | – | ↑ 12 2 | – | ↑0.089 2 |
| GluA2Q | CNIH3 | ↑ 33 2 | – | ↑ 12 2 | ↑ 26–36 2,25 | – | ↑0.11–0.13 2,25 |
| GluA2Q | CKAMP39 | – | ↓ 11 7 | 1.6 7 | ↑ 4.4 7 | ↑ 78 7 | ↑0.005 7 |
| GluA2Q | CKAMP44 | – | ↓ 11 7 | – | – | ↑ 130 23 | – |
| GluA2Q | CKAMP52 | – | ↓ 1.7 7 | ↑ 2.1 7 | 7.8 7 | 16 7 | ↑ 0.050 7 |
| GluA2Q | CKAMP59 | – | – | 1.7 7 | 7.7 7 | 25 7 | 0.020 7 |
| GluA4 | – | 20 | (0.81) | 0.6 | 3.5 | – | 0.014 |
| GluA4 | γ−2 | ↑ 31 3 | ↓ (0.39) 33 | 1.7 29 | ↑ 5.7 3 | – | ↑ 0.078 9 |
| GluA4 | γ−3 | ↑ 33 3 | – | – | ↑5.4 3 | – | – |
| GluA4 | γ−4 | ↑ 34 3 | ↓ (0.65) 33 | 9 3 | ↑12 3 | – | – |
| GluA4 | γ−5 | ↑ 36 3 | – | 2 3 | 3.0 3 | – | 0.014 21 |
| GluA4 | γ−7 | ↑ 33 3 | – | – | 3.3 3 | – | 0.056 21 |
| GluA4 | γ−8 | ↑ 37 3 | – | 1.6 29 | ↑ 7.8 3 | – | – |
| GluA1/2 | – | 3.0–4.4 | 40 | 0.8–4.5 | 4.8–6.3 | 58–87 | 0.020–0.046 |
| GluA1/2 | γ−2 | ↑ 6.2–7.1 2,17 | – | ↑ 3.4 14 | ↑ 6–7.7 2,14 | – | ↑ 0.16–0.20 2,9 |
| GluA1/2 | γ−5 | – | 80 21 | – | – | – | 0.018 9 |
| GluA1/2 | γ−8 | ↑ 6.4 17 | – | ↑ 1.7–9.5 19,29 | ↑ 11–33 19,32,34 | ↑ 110–160 19,32,34 | ↑ 0.085–0.18 19,32 |
| GluA1/2 | GSG1L | – | – | ↑ 2.5 34 | 8.1 34 | ↑ 580 34 | – |
| GluA1/2 | CNIH2 | 5.6 2 | – | ↑ 8.3 14 | ↑ 14 2,14,34 | 69–75 14,34 | ↑ 0.15–0.25 2,14,34 |
| GluA1/2 | CNIH3 | 5.1 2 | – | ↑ 1.3–7 2,14 | ↑ 11–22 2,14 | 45 14 | ↑ 0.13–0.20 2,14 |
| GluA1/2 | CKAMP52 | – | – | ↑ 5.8 16 | ↑ 6.0 16 | ↑ 110 16 | ↑ 0.12 16 |
| GluA1/2 | SynDIG4 | – | – | ↑ 5.3 19 | 5.8 19 | 62 19 | 0.021 19 |
| GluA2/4 | – | – | – | 0.71 | 0.92 | 39 | 0.01 |
| GluA2/4 | CNIH2 | – | – | ↑ 2.6 14 | ↑ 3.4 14 | 48 14 | ↑ 0.10 14 |
| GluA2/4 | CNIH3 | – | – | ↑ 2.6 14 | ↑ 3.5 14 | 45 14 | ↑ 0.10 14 |
a All receptors are rat flip splice isoforms, except GluA2/4 that was GluA2-flop/GluA4-flop. See Turetsky et al. (2005), Kato et al. (2007), Soto et al. (2009), Tomita et al. (2006), Dawe et al. (2016) and Ishii et al. (2020) for data on the effects of auxiliary subunits on different AMPA receptor splice isoforms. GluA2Q indicates cDNA for the unedited GluA2 with Gln at the Q/R/N site. Some authors report the median [e.g., Farrow et al. (2015)]. There is minimal information available for GluA3 with γ−5 increasing the conductance and the steady state to peak current ratio (Soto et al., 2009; Turetsky et al., 2005).
b Data for receptors in the absence of auxiliary subunits are only from the references providing data about auxiliary subunit for that receptor combination. Time constants that were considerably slower than the range of others were omitted under the assumption that solution exchange was insufficient to resolve the response time course accurately. For TARPs, some variability in reported values reflects different stoichiometry (Miguez-Cabello et al., 2020). Values are mean range; some papers reported median values.
c Weighted mean chord conductance was determined from variance analysis.
d EC50 was determined from the steady-state (SS) response from oocytes or transfected HEK cells; Farrow et al. (2015), Kato et al. (2008), and Priel et al. (2005) determined EC50 in the presence of cyclothiazide; GluA1-L497Y abolished effects of γ−2 on EC50h.
e EC50 was determined from the peak response to rapid glutamate application in HEK cells or macropatches from Xenopus oocytes.
f When more than one exponential described the deactivation or desensitization time course, the weighted mean time constants are given.
g The time course of recovery from desensitization was estimated from a single exponential, although it can be more complex (Bowie and Lange, 2002; Robert and Howe, 2003). When multiple exponential components were fitted to the time course, the weighted mean time constant is given. See Lomeli et al. (1994) for R/G RNA editing control of the rate of recovery from desensitization.
↑ and ↓ indicate that the measured parameter was reported to be significantly greater than or less than that for the receptor lacking the auxiliary subunit and was used when either statistical tests were reported or reported values did not have overlapping confidence intervals. ND indicates data were not compared with control. When conflicting results were reported, no direction is shown.
(Table 2 legend, cont.) 1 McGee et al. (2015), 2 Coombs et al. (2012), 3 Soto et al. (2009), 4 Soto et al. (2014), 5 Shanks et al. (2012), 6 Coombs et al. (2017), 7 Farrow et al. (2015), 8 Priel et al. (2005), 9 Turetsky et al. (2005), 10 Kato et al. (2007), 11 Kott et al. (2007), 12 Schmitz et al. (2017), 13 Twomey et al. (2017b), 14 (Schwenk et al., 2009), 15 Gill et al. (2012), 16 Klaassen et al. (2016), 17 Kristensen et al. (2011), GluA1 contains mutations S831A and S845A, 18 Tomita et al. (2006), 19 Matt et al. (2018), 20 Kato et al. (2010), 21 Kato et al. (2008), 22 Twomey et al. (2016), some measures from tandem fusion protein of GluA2Q and γ−2, 23 Khodosevich et al. (2014), 24 Dawe et al. (2016), 25 Hawken et al. (2017), 26 Riva et al. (2017), 27 Coombs et al. (2019), 28 Cho et al. (2007), 29 Ishii et al. (2020), 30 Cais et al. (2014), 31 Milstein et al. (2007), 32 Herguedas et al. (2019), 33 Pierce and Niu (2019), 34 Schwenk et al. (2012).
Fig. 11.
Gating modulation of GluA1 and GluA1/GluA2 AMPA receptors by auxiliary subunits. (A) Channel conductance (γMEAN), (B) deactivation time constant (TauDEACTIVATE), (C) desensitization time constant (TauDESENS), and (D) ratio of amplitudes of steady state (ISS) over peak currents (IPEAK) are depicted by the location of the colored balloon that represents various auxiliary subunits in complex with the indicated receptor. The upper number line represents effects on the homomeric GluA1 (A1), whereas the lower number line represents effects on the heteromeric GluA1/2 (A1/A2). The white balloons indicate values for AMPA receptors without any auxiliary subunits. Values are from Table 2 and associated references. GL, GSG1L; CN, CNIH; CK, CKAMP; SD, SynDIG4. TARP subtypes are indicated as γ-2, γ-3, γ-4, γ-5 and γ-8.
The efficacy for activation of steady-state currents by agonists can also be altered by TARPs and shows a bell-shaped curve with a decrease in amplitude at high glutamate concentration (>100 µM) (Morimoto-Tomita et al., 2009). Whether the inactivation at high glutamate concentration is mediated by intrinsic gating properties or physical decoupling of TARPs from AMPA receptor has not been fully resolved (Morimoto-Tomita et al., 2009; Semenov et al., 2012; Straub and Tomita, 2012; Coombs et al., 2017). CaMKII can potentiate homomeric GluA1 channels lacking TARPs, but association of γ-2 or γ-8 with GluA1/2 receptors is necessary for the increase in channel conductance after phosphorylation of Ser831 in GluA1 (Kristensen et al., 2011). TARPs can also reduce the affinity of polyamine block of Ca2+-permeable AMPA receptors (Brown et al., 2018).
c. Structural determinants of TARP action
The regions of the extracellular and cytoplasmic domains of γ-2 and γ-4 that impact receptor function have been identified (Tomita et al., 2005a; Turetsky et al., 2005; Cho et al., 2007). The residues in the lower lobe of the AMPA receptor ABD are critical for γ-2–mediated gating modulation (Dawe et al., 2016) and are hypothesized to interact with the extracellular domain of TARPs (Twomey et al., 2019). The action of γ-2 in slowing desensitization of GluA2 AMPA receptors is substantially attenuated in flop isoforms compared with flip isoforms (Dawe et al., 2019). Within the flip/flop cassette, residue 775 (Ser in flip, Asn in flop) is a key determinant that governs γ-2–mediated slowing of desensitization. In addition, γ-2 overcomes the effect of a mutation in the ABD that destabilizes the agonist-bound closed clamshell conformation (MacLean et al., 2014). For type I TARPs, residues in the β1-β2 loop (Fig. 8) of the extracellular domain control the slowing of desensitization (Hawken et al., 2017; Riva et al., 2017). When the cytoplasmic C terminus of γ-2 is deleted, TARP-mediated modulation of receptor function is attenuated (Priel et al., 2005; Tomita et al., 2005a; Turetsky et al., 2005). Furthermore, membrane-embedded elements that form the interaction interface between AMPA receptor subunits and TARPs were also mapped as critical sites for gating modulation (Ben-Yaacov et al., 2017; Hawken et al., 2017).
FRET investigations showed a TARP-induced shift to a more compact conformation of the ABD layer in AMPA receptors when bound to agonists (MacLean et al., 2014; Shaikh et al., 2016; Carrillo et al., 2020b). Treatment of receptors with Cys substituted into the ABD dimer interface with bifunctional crosslinkers of varying lengths can also produce a shift toward a more compact arrangement in the extracellular domains of AMPA receptors associated with γ-2 and γ-8, which correlates with higher steady-state currents for γ-2 and resensitization for γ-8 (Baranovic and Plested, 2018).
d. Different modulatory actions of type-I and type-II TARPs
Functional modulation of AMPA receptors by type II TARPs (γ-5 and γ-7) differs from type I TARPs in their selective actions on different AMPA receptor subunits and in their mixture of positive and negative effects on gating and trafficking (Kato et al., 2007, 2008; Soto et al., 2009; Bats et al., 2012; Studniarczyk et al., 2013). Similar to type I TARPs, type II TARPs increase channel conductance and attenuate polyamine block with γ-5 having less of an effect than γ-7 (Soto et al., 2009). γ-5 is unique among all TARPs in its ability to reduce peak open probability of GluA2-lacking, Ca2+-permeable AMPA receptors. As observed in Bergmann glial cells, in which γ-5 and Ca2+-permeable AMPA receptors are abundantly expressed, γ-5 increases conductance and reduces peak open probability but does not change the rate of desensitization of GluA1 and GluA4 subunit-containing AMPA receptors (Soto et al., 2009). The long cytoplasmic tails of GluA1 and GluA4 are molecular determinants for the selective action of γ-5 on these subunits (Kato et al., 2008; Soto et al., 2009). The expression of γ-5 in the CNS is spatially restricted and the lowest among the TARPs, albeit γ-5 is enriched in the hippocampal CA2 region and in Bergmann glia (Fukaya et al., 2005b; Schwenk et al., 2012, 2014; Shanks et al., 2012).
TARP γ-7 increases the conductance of AMPA receptors and suppresses synaptic trafficking of GluA2-containing, Ca2+-impermeable AMPA receptors (Studniarczyk et al., 2013). γ-7 is expressed broadly in the cerebellum and is found in granule cells, Purkinje cells, stellate cells, and Bergmann glial cells (Yamazaki et al., 2010). The function of γ-7 is best understood in granule cells and stellate cells (Bats et al., 2012; Studniarczyk et al., 2013), where it mediates resensitization of GluA1-containing AMPA receptor currents, similar to γ-4 and γ-8 (Kato et al., 2008, 2010). Notably, although CNQX acts as an agonist at AMPA receptors associated with γ-2, CNQX remains an antagonist at AMPA receptors associated with γ-7 (Bats et al., 2012).
Initial studies concluded that cerebellar granule cells in stargazer mice, which are deficient in γ-2 (Letts et al., 1998), lack surface expression of AMPA receptors (Chen et al., 1999b, 2000; Hashimoto et al., 1999). However, mice lacking both γ-2 and γ-7 had AMPA receptor-mediated synaptic currents in cerebellar granule cells, albeit reduced relative to wild-type mice and similar to the phenotype of heterozygous γ-2 KO (+/−) mice, suggesting a more complex interaction between the auxiliary subunits (Yamazaki et al., 2015). Surprisingly, synaptic AMPA receptor-mediated currents were observed upon knockdown of γ-7 in granule cells from γ-2 KO mice, reversing the absence of AMPA receptor-mediated synaptic events in these γ-2 KO mice (Studniarczyk et al., 2013). Furthermore, overexpression of γ-7 but not γ-5 rescued AMPA receptor surface expression in cerebellar granule cells of γ-2 KO mice (Kato et al., 2007). These disparate observations can be explained by a mechanism in which knockdown of γ-7 in granule cells removes its negative effect on trafficking of GluA2-containing AMPA receptors, thereby allowing synaptic expression in the absence of γ-2, whereas overexpression reduces the proportion of GluA2-containing AMPA receptors relative to GluA2-lacking AMPA receptors. In cerebellar Purkinje cells of γ-2 KO mice, AMPA receptor-mediated EPSCs are reduced but not eliminated because of a functional compensation by γ-7 (Yamazaki et al., 2010, 2015). γ-2 is necessary in Purkinje cells for maintaining basal EPSC amplitudes at climbing fiber synapses (Kawata et al., 2014). AMPA receptor-mediated EPSCs in neurons of the reticular thalamic nucleus are reduced in stargazer mice (Menuz and Nicoll, 2008; Chetkovich, 2009). In the absence of γ-2, the refinement at the retinogeniculate synapse is disrupted during experience-dependent synaptic plasticity (Louros et al., 2014). Furthermore, γ-2–containing AMPA receptors exhibit a slow component of current decay that underlies EPSCs with unusual kinetics in unipolar brush cells in the cerebellar granule cell layer (Balmer and Trussell, 2019).
Rare de novo mutations in TARPs and other auxiliary subunits might contribute to neurologic deficits. For example, the p.V143L variant in TM3 of human γ-2 has been found in a patient with intellectual disability (Hamdan et al., 2011). Double and triple TARP knockout mice express varying degrees of deficits in AMPA receptor-mediated synaptic transmission and in some cases lethality (Jackson and Nicoll, 2011a). In addition, TM3 and TM4, which form the interaction sites with AMPA receptors, are intolerant to variation in the healthy population (Supplemental Figs. 2 and 3).
2. Germ Cell–Specific Gene 1-Like (GSG1L) Protein
GSG1L, a distant homolog of TARPs (Fig. 8), was identified by mass spectrometry among the proteins that copurify with native AMPA receptors (Schwenk et al., 2012; Shanks et al., 2012). GSG1L stands out from other auxiliary subunits for having a strong overall negative modulatory function (McGee et al., 2015; Gu et al., 2016a; Mao et al., 2017). Although GSG1L can slow deactivation and desensitization, it stabilizes the desensitized state (Twomey et al., 2017b) and slows recovery from desensitization more than other auxiliary subunits (Schwenk et al., 2012; Shanks et al., 2012) (see Table 2 in Section IV. Receptor Activation, Deactivation, and Desensitization). Additionally, GSG1L reduces single-channel conductance and calcium permeability of Ca2+-permeable AMPA receptors (McGee et al., 2015) (Fig. 11). Overexpression of GSG1L decreases the amplitude of evoked EPSCs (McGee et al., 2015; Gu et al., 2016a; Mao et al., 2017), whereas GSG1L KO rats exhibit increased hippocampal LTP (Gu et al., 2016a). GSG1L is enriched in the anterior thalamic nuclei, where it reduces short-term facilitation of EPSCs by slowing the recovery from desensitization (Kamalova et al., 2020). Most of the GSG1L gene appears to be relatively tolerant to genetic variation (Supplemental Fig. 4).
Like TARPs, GSG1L binds to the surface formed by M1 and M4 of adjacent subunits in AMPA receptors (Twomey et al., 2017b). Cryo-EM structures contain only two GSG1L subunits per AMPA receptor tetramer, preferring the B′/D′ locations (Twomey et al., 2017a,b), which may be a hallmark of bulkier auxiliary subunits, including γ-8 (Fig. 10). GSG1L lacks the extracellular helix present in TARPs and instead has a longer second transmembrane domain extending to the extracellular space (Fig. 8). In addition, the β1-β2 loop is longer than that of the TARPs, and transferring the β1-β2 loop of TARP γ-2 into the corresponding location in GSG1L also transfers γ-2–like characteristics to GSG1L (Riva et al., 2017; Twomey et al., 2017b).
3. Cornichon 2 and 3 Proteins (CNIHs)
The cornichon family consists of CNIH1-4, evolutionarily conserved membrane proteins of ∼18 kDa (Roth et al., 1995; Powers and Barlowe, 1998; Hwang et al., 1999; Bokel et al., 2006), but only CNIH2 and CNIH3 are AMPA receptor auxiliary subunits (Schwenk et al., 2009) (Fig. 8). Cryo-EM structures of CNIH3 in complex with homomeric GluA2 receptors demonstrate that CNIH3 is composed of four transmembrane helices (TM1-4) with N and C termini on the extracellular side (Nakagawa, 2019) (Figs. 8 and 10); other cornichons likely have the same topology (Hessa et al., 2007). The lack of substantial extracellular domains is in agreement with the absence of N-glycans on CNIH2 (Zheng et al., 2015). CNIH2/3 subunits promote forward trafficking of AMPA receptors, possibly by regulating ER export (Harmel et al., 2012; Herring et al., 2013). CNIHs also potentiate AMPA receptor gating by increasing glutamate potency and slowing both deactivation and desensitization (Schwenk et al., 2009; Coombs et al., 2012) (Fig. 11, see Table 2 in Section IV. Receptor Activation, Deactivation, and Desensitization). Although recovery from desensitization is not affected by CNIH2/3 (Schwenk et al., 2009), polyamine block is attenuated, and channel conductance is increased (Coombs et al., 2012; Brown et al., 2018).
CNIH2 is as abundantly expressed as various TARPs in the hippocampus, cortex, and striatum (Schwenk et al., 2014), and CNIH2 expression is higher than CNIH3 in the hippocampus (Mauric et al., 2013; Schwenk et al., 2014), with a more severe synaptic phenotype of CNIH2 KO compared with CNIH3 KO mice (Herring et al., 2013). AMPA receptor-mediated synaptic currents in neurons from CNIH2/3 double-KO mice exhibit reduced amplitudes and faster time courses, which are thought to be the consequence of a decrease in the number of GluA1-containing heteromeric AMPA receptors (Herring et al., 2013). The effects of CNIH2 overexpression in the background of different TARP KO mice vary substantially. In the γ-8 KO background, overexpression of CNIH2 slows AMPA receptor-mediated evoked EPSCs in hippocampal CA1 neurons, consistent with a slowing of AMPA receptor deactivation by CNIH2 (Herring et al., 2013). However, overexpression of CNIH2 does not rescue the loss of surface AMPA receptor expression in cerebellar granule cells of γ-2 KO mice (Shi et al., 2010). CNIH2 shapes AMPA receptor-mediated EPSCs at mossy fiber synapses in the mossy cells of the dentate gyrus, as demonstrated by faster time course after knockdown of CNIH2 (Boudkkazi et al., 2014) (Fig. 9).
CNIH2 and CNIH3 slow AMPA receptor desensitization substantially compared with TARPs (Schwenk et al., 2009; Coombs et al., 2012) (Figs. 8 and 11). Unlike TARPs and GSG1L, whose extracellular domains are in contact with the AMPA receptor ABD (Twomey et al., 2019), CNIH2/3 have minimal extension in the extracellular space (Nakagawa, 2019), and thus gating modulation may not require direct interaction between the ABD and CNIH2/3. CNIH3 binds to the surface generated by M1 and M4 of adjacent subunits in homomeric GluA2 receptors, the same location that TARPs and GSG1L bind, which agrees with the functional competition observed between TARPs and CNIHs (Gill et al., 2011) (Fig. 10). The stoichiometry of the recombinant GluA2/CNIH3 complex revealed by the cryo-EM structure is 4:4 (Nakagawa, 2019) (Fig. 10), but competition with TARPs likely reduces the number of CNIHs associated with neuronal AMPA receptors (Kato et al., 2010; Schwenk et al., 2012). Assembly with CNIH2/3 precludes resensitization of AMPA receptors complexed with γ-8 or γ-4, which could explain why hippocampal CA1 pyramidal cell AMPA receptors do not exhibit resensitization despite containing γ-8 (Kato et al., 2010). The CNIH2/3 segment of the second transmembrane helix, which is absent in CNIH1/4, extends into the cytoplasm and partially contributes to gating modulation (Shanks et al., 2014). TM1 and TM2 of CNIH3 interface with the AMPA receptor, with the critical structural determinants of CNIH3-mediated gating modulation formed by Phe3, Phe5, and Phe8 of CNIH3 and several amino acids in the M1 and M4 of GluA2 close to the extracellular space (Hawken et al., 2017; Nakagawa, 2019). The amino acids of GluA2 that contact CNIH3 are conserved in other AMPA receptor subunits but are different in kainate receptor subunits that cannot bind CNIH3.
4. Cysteine‐Knot AMPA Receptor Modulating Proteins (CKAMPs)
The CKAMP family consists of CKAMP39 (shisa8), CKAMP44 (shisa9), CKAMP52 (shisa6), and CKAMP59 (shisa7) (von Engelhardt, 2019). CKAMP genes are referred to by their alternate nomenclature, SHISA6-9. CKAMP44, 52, and 59 are AMPA receptor auxiliary subunits (Farrow et al., 2015; Klaassen et al., 2016). CKAMP39 modulates AMPA receptor gating in non-neuronal cells, but its neuronal relevance is not yet established. Although CKAMP44 and CKAMP52 only colocalize with glutamatergic synapses, CKAMP59 might also serve as a GABAA receptor auxiliary subunit (Han et al., 2019).
CKAMP44, a 44-kDa brain-specific protein, was first identified as a protein that copurifies with brain AMPA receptors by mass spectrometry (von Engelhardt et al., 2010) and was detected in various AMPA receptor interactome studies (Schwenk et al., 2012, 2014; Shanks et al., 2012). CKAMP44 includes a cysteine knot motif in the N-terminal extracellular domain, a single transmembrane domain, and a cytoplasmic domain that terminates with a type II PDZ domain binding motif (Fig. 8). The PDZ domain binding motif interacts with several scaffold proteins, including PICK1 (Karataeva et al., 2014). A subpopulation of AMPA receptor complexes in the brain contains both CKAMP44 and TARPs (γ-2 or γ-8) (von Engelhardt et al., 2010; Khodosevich et al., 2014).
CKAMP44 increases glutamate potency, slows deactivation, speeds desensitization, and slows recovery from desensitization (von Engelhardt et al., 2010; Khodosevich et al., 2014; Farrow et al., 2015) (Fig. 11; see Table 2 in Section IV. Receptor Activation, Deactivation, and Desensitization), possibly by stabilizing the closed-cleft conformation of the ABD and promoting desensitization (von Engelhardt et al., 2010). In the hippocampal dentate gyrus and the lateral geniculate nucleus of the thalamus, CKAMP44 modulates short-term plasticity as well as the number of functional synaptic AMPA receptors (von Engelhardt et al., 2010; Chen et al., 2018). The abnormal synaptic and behavioral phenotypes of CKAMP52 KO and CKAMP59 KO mice support their roles in regulating AMPA receptor function in cerebellar motor learning and hippocampal contextual learning (Klaassen et al., 2016; Schmitz et al., 2017; Peter et al., 2020).
5. Synapse Differentiation-Induced Gene 4 (SynDIG4)
SynDIG4, also known as Prrt1, was identified as a synapse differentiation-induced gene (Diaz et al., 2002; Kalashnikova et al., 2010) and is also found among the AMPA receptor binding proteins in proteomics studies (von Engelhardt et al., 2010; Schwenk et al., 2012; Shanks et al., 2012). Prrt2, a homolog of Prrt1, also copurifies with AMPA receptors in proteomic studies and is reported to have a presynaptic function related to vesicle release (Valente et al., 2016; Coleman et al., 2018; Tan et al., 2018). SynDIG4 is predicted to have its N terminus in the cytoplasm, one transmembrane segment, and an additional membrane-associated domain in the C terminus (Fig. 8).
SynDIG4 slows deactivation and desensitization of AMPA receptors in a subunit-dependent manner, producing stronger modulation of homomeric GluA1 receptors compared with heteromeric GluA1/2 receptors (Fig. 11; see Table 2 in Section IV. Receptor Activation, Deactivation, and Desensitization). SynDIG4 colocalizes with GluA1 subunits at nonsynaptic sites and SynDIG4 KO mice have reduced AMPA receptor-mediated mEPSCs as well as deficits in LTP (Matt et al., 2018). Based on these results, it is proposed that SynDIG4 regulates the extrasynaptic pool of AMPA receptors. Because AMPA receptor/SynDIG4 complexes are localized outside of dendritic spines, its role as a bona fide auxiliary subunit remains to be established.
6. Auxiliary Subunits in Nonmammalian Systems
Homologs of vertebrate AMPA receptor auxiliary subunits are present in C. elegans, Drosophila, and Arabidopsis. In C. elegans, suppressor of lurcher protein 1 (SOL-1), a single-transmembrane domain protein with four extracellular complement Clr/Cls, sea urchin epidermal growth factor, bone morphogenetic protein 1 (CUB) domains, associates with the AMPA receptor homolog GLR-1 (Zheng et al., 2004, 2006; Wang et al., 2012). Homologs of mammalian TARPs, STG-1 and STG-2, are also conserved in C .elegans, but GLR-1 requires both types of auxiliary subunits (SOL-1 and STG-1, or SOL-1 and STG-2) for ion channel gating (Walker et al., 2006). SOL-2 is a homolog of mammalian Neto1 and Neto2, which are kainate receptor auxiliary subunits (see below). The soluble extracellular domain of SOL-1 requires SOL-2 to rescue the phenotype of SOL-1 mutants (Wang et al., 2012). Thus, SOL-2 interacts with SOL-1 and modulates gating of GLR-1, hence SOL-2 is yet another component of the native GLR complex in C. elegans. Unlike the mammalian CNIH2/3 that facilitate the ER export of AMPA receptors (Schwenk et al., 2019), the cornichon homolog CNI-1 in C. elegans retains GLR-1 in the ER (Brockie et al., 2013). In Arabidopsis, the cornichon homolog CORNICHON (AtCNIH) associates with iGluR homolog GLR and has a similar function as the C. elegans CNI-1 (Wudick et al., 2018). The results from nonvertebrate systems provide insight into the functional mechanisms of iGluR modulation by auxiliary subunits.
B. Kainate Receptor Auxiliary Subunits
Kainate receptors associate with the Neuropilin- and tolloid-like (Neto) proteins, which modify nearly all aspects of their function, enhance their biogenesis and trafficking, and promote their targeting to plasma membrane compartments in neurons. The two gene products in this family, Neto1 and Neto2, represent the only two kainate receptor-associated proteins shown to modify functional properties and neuronal localization both in vitro and in vivo (Copits and Swanson, 2012; Yan and Tomita, 2012). Neto1 and Neto2 mRNAs were cloned independently and initially referred to as Bctl1 and Bctl2 (Michishita et al., 2003; 2004). A proteomic screen of GluK2/3 subunit-associated proteins yielded Neto2, and coexpression of GluK2 with Neto2 greatly increased current amplitudes and altered agonist efficacy (Zhang et al., 2009b). Neto1 coexpression had an analogous effect on GluK2 receptor amplitudes (Zhang et al., 2009b) and was similarly proposed to be an auxiliary subunit for kainate receptors (Tang et al., 2011). Studies of kainate receptor function revealed that Neto2 coexpression slowed deactivation and desensitization and increased open probability of GluK2-containing kainate receptors, thereby shaping kainate receptor signaling in a fashion analogous to actions of auxiliary subunits on AMPA receptors (Zhang et al., 2009b) (Fig. 12).
Fig. 12.
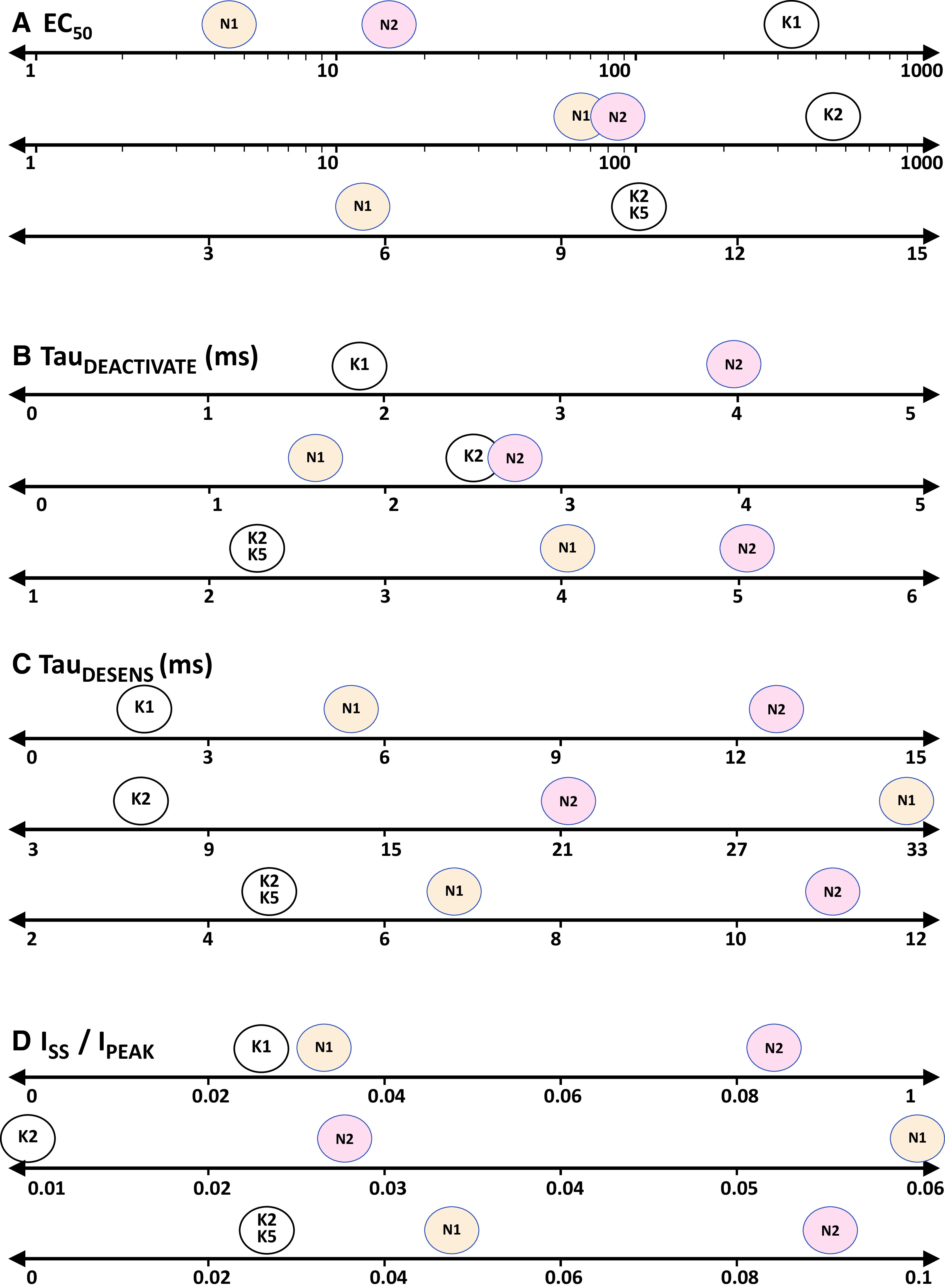
Gating modulation of GluK1, GluK2, and GluK2/GluK5 kainate receptors by Neto auxiliary subunits. (A) Glutamate EC50 (µM), (B) deactivation time constant (TauDEACTIVATE), (C) desensitization time constant (TauDESENS), and (D) ratio of amplitudes of steady state (ISS) over peak currents (IPEAK) are depicted by the location of the colored balloon that represents Neto1 (N1, orange) and Neto2 (N2, red) auxiliary subunits. White balloons indicate values for kainate receptors without auxiliary subunits. Values are from Table 3 and associated references.
Neto1 and Neto2 are modular proteins that each contain an extracellular segment comprising two CUB domains and a juxtamembrane low-density lipoprotein class A (LDLa) domain, a single-transmembrane helix, and an unstructured C-terminal cytoplasmic domain with a PDZ binding motif (Fig. 13). CUB domains are a conserved structural motif consisting of ∼110 amino acids organized into a β-sandwich of two sets of five β-sheets found in a large number of secreted and integral membrane proteins with diverse functions. The CUB domains in Neto proteins share modest sequence identity (∼35%) with analogous structures in neuropilin proteins, adhesion molecules that serve as receptors for secreted semaphorin axon guidance cues and vascular endothelial growth factor (Nakamura and Goshima, 2002; Pasterkamp, 2012). The LDLa domain is a 38-amino-acid cysteine-rich segment initially defined in the low-density lipoprotein receptor that precedes the transmembrane domain in Neto proteins. The single-transmembrane helices of Neto1 and Neto2 share as much sequence identity with each other (∼56%) as they do with disintegrin and ADAM22 (∼59%). The C-terminal tail of both Neto proteins contain PDZ ligands: type I in Neto1 (Ng et al., 2009) and type II in Neto2 (Tang et al., 2012).
Fig. 13.

Neto proteins modulate kainate receptor function. (A) Architecture of the Neto proteins is shown, adapted with permission from Copits and Swanson (2012). (B) The response time course of heteromeric GluK2/5 receptors to brief 1–2 milliseconds (arrow) or prolonged application of glutamate in the absence (black) and presence (red) of Neto1. (C) The response time course of GluK2/5 to brief 1–2 milliseconds (arrow) or prolonged application of glutamate in the absence (black) and presence (blue) of Neto2. Adapted with permission from Straub et al. (2011a) and Straub et al. (2011b). See Table 3 for time constants describing deactivation and desensitization.
1. Functional Effects of Neto Proteins
Modulation of recombinant kainate receptor gating and pharmacological properties by Neto proteins depends on both receptor subunit composition and the Neto protein assembled into the receptor complex (Table 3 in Section IV. Receptor Activation, Deactivation, and Desensitization). Functional kainate receptors can be homomeric assemblies of “low-affinity” GluK1, GluK2, or GluK3 subunits; addition of the “high-affinity” GluK4 or GluK5 subunits in heteromeric receptors alters many fundamental properties of receptor gating, in which “low-affinity” and “high-affinity” refer to the relative affinity and agonist potency for glutamate (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity). Neto2 assembly with homomeric GluK2 receptors slows deactivation and desensitization and increases the rate of recovery from desensitization (Zhang et al., 2009b; Copits et al., 2011; Fisher, 2015; Griffith and Swanson, 2015; Li et al., 2019b). Neto2 also increased peak open probability of homomeric GluK2 receptors, which in part occurs because of a predominance of high open-probability receptor states over slow time scales (Zhang et al., 2014a). The effect of Neto1 on homomeric GluK2 receptor gating is less clear and ranges from no apparent change in the time course of deactivation or desensitization of glutamate-evoked currents (Li et al., 2019b) to a 3-fold or greater slowing of desensitization (Fisher and Mott, 2013; Palacios-Filardo et al., 2016). The interaction between Neto1 and Neto2 with homomeric GluK1 receptors is more complex in that modulation of desensitization appears to be bidirectional. That is, assembly with Neto1 increased the rate of desensitization for homomeric GluK1 receptors (Copits et al., 2011; Fisher, 2015; Sheng et al., 2015), whereas Neto2 slowed desensitization (Copits et al., 2011; Straub et al., 2011b; Fisher, 2015; Sheng et al., 2015). In addition to these effects on receptor gating, assembly with either Neto1 or Neto2 decreased the EC50 of both homomeric GluK1 and GluK2 receptors, making the receptors more responsive to lower concentrations of glutamate (Fisher and Mott, 2013; Fisher, 2015) (Fig. 12). Neto proteins also reduce intracellular polyamine block of unedited kainate receptors (Fisher and Mott, 2012; Brown et al., 2016).
TABLE 3.
Modulation of kainate receptor properties by auxiliary subunits
Values at 2–3 significant figures are from the text or tables; when values are not reported, measurements were made from figures.
| Receptor | Subunit | EC50,PEAK c | EC50,SS d | τ-Deactivate e | τ-Desensitize e,g | τ-Recovery f,g |
Steady State/Peak
Current Ratio |
|---|---|---|---|---|---|---|---|
| μM | μM | ms | ms | ms | |||
| GluK1 a,b | – | 125–630 1,2 | 59 3 | 1.7 29 | 1.6–42 1,2,5-12,29 | 18–5100 7,10,29 | 0.01–0.04 10 |
| GluK1 | Neto1 | ↓ 4.4 2 | – | – | ↓ 2.6–5.2 2,7 | 301–1700 7 | 0.031 2 |
| GluK1 | Neto2 | ↓ 14 2 | – | 3.9 29 | ↑ 11–200 2,7,8,29 | ↓ 590 2,29 | ↑ 0.086 2 |
| GluK2a a | – | 234–1040 2,13-18 | 32–35 3,19 | 1.6–3.5 8,9,14,15,17-22 | 3.4–7 9,13,15,16,18-21,23-26 | 700–3020 8,13,15,16,20,21,23,25,27 | 0.001–0.008 2,13,19,20,23,24 |
| GluK2a | Neto1 | ↓ 60 25 | – | ∼1.6 22 | ↑ 32 25 | ↓ 240 25 | ↑ 0.06 25 |
| GluK2a | Neto2 | ↓ 77 2 | ↓ 15 19 | 2.6–3.3 8,19,22 | ↑ 15–24 8,9,19,26 | ↓ 800 8 | ↑ 0.01–0.027 2,19 |
| GluK1/5 b | – | 7.8 25 | 19 3 | – | 0.7–2.1 6,28,29 | 3800 29 | – |
| GluK1/5 | Neto1 | 5.2 25 | – | – | – | – | – |
| GluK1/5 | Neto2 | – | – | – | ↑ 3.3 29 | ↓ 450, ∼3800 29 | – |
| GluK2/4 | – | 43 25 | – | – | 4.4 27 | 2020 27 | – |
| GluK2/4 | Neto1 | ↓ 9.1 25 | – | – | – | – | – |
| GluK2/5 | – | 6.0–15 16,25 | 31 3 | 2.3–46 4,16, 29 | 1.8–6.0 4,6,8,9,16,25,26,29,30 | 1740–2700 16,25,29 | 0.02 4 |
| GluK2/5 | Neto1 | 5.0 25 | – | 3.9 4 | ↑ 6.2–8.2 4,25 | ↓ 250 25 | ↑ 0.064–0.082 4,25 |
| GluK2/5 | Neto2 | – | – | 5.0 29 | ↑ 11 26,29 | ↓ 61, ∼2500 29 | – |
| GluK3/5 | – | 6.0 25 | – | – | – | – | – |
| GluK3/5 | Neto1 | 4.8 25 | – | – | – | – | – |
a Unedited receptors or mutant kainate receptors with a glutamine at the Q/R/N site. All receptors are from rat except the human receptors reported in Alt et al. (2004).
b Values for GluK1-2a and GluK1-2b splice isoforms are pooled.
c EC50 was determined from the steady-state (SS) response from oocytes or transfected HEK cells.
d EC50 was determined from the peak response to rapid glutamate application in HEK cells or macropatches from Xenopus oocytes.
e If several exponentials described the deactivation or desensitization time course, the weighted mean time constant is given here.
f The time course of the recovery from desensitization was estimated from a single exponential.
g Onset and recovery from desensitization for GluK1-containing receptors are variable; the latter often shows two separable kinetic phases.
1 Sommer et al. (1992), 2 Fisher (2015), 3 Alt et al. (2004), 4 Straub et al. (2011a), 5 Swanson et al. (1997a), 6 Swanson et al. (1998), 7 Copits et al. (2011), 8 Vernon et al. (2017), 9 Copits et al. (2014), 10 Swanson and Heinemann (1998), 11 Larsen et al. (2017), 12 Ren et al. (2003c), 13 Traynelis and Wahl (1997), 14 Bowie (2002), 15 Kistler and Fleck (2007), 16 Barberis et al. (2008), 17 Perrais et al. (2009a), 18 Dawe et al. (2013), 19 Zhang et al. (2009b), 20 Heckmann et al. (1996), 21 Weston et al. (2006a), 22 Li et al. (2019b), 23 Bowie and Lange (2002), 24 Zhang et al. (2008e), 25 Fisher and Mott (2013), 26 Griffith and Swanson (2015), 27 Mott et al. (2010), 28 Swanson et al. (2002), 29 Straub et al. (2011b), 30 Fisher and Mott (2011).
Neto1 and Neto2 slow entry into and speed recovery from desensitization for heteromeric GluK5-containing kainate receptors (Fig. 13). Desensitization of glutamate-evoked currents from GluK1/5 and GluK2/5 receptors is rapid (Table 3 in Section IV. Receptor Activation, Deactivation, and Desensitization), but Neto1 assembly with heteromeric GluK2/5 receptors slowed both deactivation and desensitization and accelerated recovery from desensitization (Straub et al., 2011a). GluK1/5/Neto2 receptors desensitize slower than Neto2-less receptors by only about 2-fold (Straub et al., 2011b), which is in striking contrast to the profound slowing of Neto2 on homomeric GluK1 desensitization (Copits et al., 2011; Fisher, 2015; Vernon and Swanson, 2017). Desensitization of GluK2/5 receptors also is modestly slower when assembled with Neto2 (Straub et al., 2011b; Griffith and Swanson, 2015), and recovery from desensitization of GluK5-containing heteromeric receptors is faster with Neto2 (Straub et al., 2011b). The GluK5 subunit confers a higher affinity for glutamate on heteromeric kainate receptors (Barberis et al., 2008; Fisher and Mott, 2013); the addition of Neto1 only slightly increases glutamate potency at heteromeric GluK2/5 receptors, which is in contrast to its effect on homomeric kainate receptors (Fisher and Mott, 2013) (Fig. 12).
2. Structural Determinants of Neto Proteins
The LDLa domain was first identified as a determinant of Neto2 modulatory function (Fig. 13). Mutation of two Cys residues in this domain attenuated the increase in GluK2 receptor current amplitudes (Zhang et al., 2009b). A more extensive biochemical study with recombinant Neto1 and Neto2 reported that deletion of the second CUB domain (from the N terminus; CUB2) but not the CUB1 domain reduced association with GluK2 (Tang et al., 2011). Removal of both CUB domains eliminated association with GluK2 in the same study. In contrast, neither the extracellular LDLa domain nor the intracellular cytoplasmic tail were required for biochemical association with GluK2 (Tang et al., 2011).
The CUB1 domain in Neto1 and Neto2 coprecipitates with the NTD of GluK2, and this interaction occurs with a high affinity (Li et al., 2019b). Moreover, interactions between CUB1 and the GluK2 NTD were implicated as the domains that give rise to differential modulation of homomeric GluK2 receptor deactivation and desensitization by Neto1 and Neto2 (Li et al., 2019b). Modulation of deactivation, desensitization, and recovery from desensitization also required other downstream regions, referred to as the “core” of Neto proteins, with the CUB2 and LDLa likely playing key roles. A putative linker domain near the C terminus of the Neto CUB1 domain might interact with a cluster of residues on the most distal region of the GluK2 NTD (Li et al., 2019b). Functional studies with GluK2 mutants also identified the dimer interface of the ABD and the M3-S2 linker as being critical determinants of the modulatory effects of Neto2 on receptor gating (Griffith and Swanson, 2015). These data suggest that the modulatory effects of Neto proteins share a subset of sites of action with those observed in structural studies with AMPA receptors and their auxiliary subunits (Chen et al., 2017a; Herguedas et al., 2019; Nakagawa, 2019).
3. Neuronal Studies of Neto Proteins
The relevance of Neto auxiliary proteins to neuronal kainate receptor signaling has been explored in comparative studies using gene-targeted mice in which expression of one or both proteins is eliminated. The initial examination of Neto1 and Neto2 KO mice yielded evidence that Neto1 but not Neto2 assembles with neuronal kainate receptors and in part shapes the unusually slow decay of kainate receptor-mediated EPSCs at hippocampal mossy fiber to CA3 pyramidal cell synapses (Straub et al., 2011a; Tang et al., 2011). Genetic ablation of Neto1 accelerated the EPSC decay time constant (Straub et al., 2011a; Tang et al., 2011; Copits and Swanson, 2012; Yan and Tomita, 2012), suggesting that Neto1 selectively assembles with kainate receptors in mossy fiber synapses of CA3 pyramidal cells.
Neto2 does not participate as an auxiliary subunit at hippocampal mossy fiber synapses but has been implicated in kainate receptor signaling in other CNS regions. Initial biochemical evidence implicated Neto2 in synaptic delivery of GluK2-containing kainate receptors in cerebellar granule cells through a PDZ-dependent interaction with glutamate receptor-interacting protein (GRIP) (Tang et al., 2012). A comparative study of CA1 interneuron kainate receptors in Neto1 and Neto2 KO mice observed that presynaptic kainate receptors, which tonically suppress GABA release from CA1 cholecystokinin (CCK) and CB1-expressing interneurons, require Neto2 (and Neto1), whereas enhancement of excitability by somatodendritic kainate receptors depends on incorporation of Neto1 but not Neto2 (Wyeth et al., 2017). The roles of Neto1 and Neto2 were also differentiated for presynaptic kainate receptors, with Neto1 characterized as obligatory for kainate receptor signaling (possibly by regulating axonal targeting), whereas Neto2 increased glutamate potency for heterosynaptic receptor activation (Wyeth et al., 2017).
In the periphery, small- to medium-diameter nociceptive neurons in dorsal root ganglia (DRG) express kainate receptors (Vernon and Swanson, 2017) that modulate presynaptic release of glutamate at synapses in the spinal cord (Li et al., 1999). Neto2 is therefore likely most relevant to axonal or presynaptic receptors, similar to its role in the hippocampus. Expression of Neto2 was upregulated in response to axotomy and nerve injury, suggestive of a role in the response to axonal injury, which was further supported by the observation that axon outgrowth was attenuated in DRG neurons from Neto2 KO mice (Vernon and Swanson, 2017).
Knockout and overexpression studies have underscored the importance of Neto proteins to synaptic targeting of functional hippocampal kainate receptors (Copits et al., 2011; Sheng et al., 2015; Palacios-Filardo et al., 2016; Orav et al., 2017, 2019; Wyeth et al., 2017), although biochemical studies have been less definitive. Ablation of Neto1 either reduced (Tang et al., 2011; Wyeth et al., 2014) or had no effect (Straub et al., 2011a) on GluK2 and GluK5 immunoreactivity in hippocampal postsynaptic densities. Overexpression of kainate receptor subunits and Neto proteins in organotypic slice preparations showed that both Neto1 and Neto2 effectively promoted functional synaptic kainate receptors in CA1 pyramidal neurons when coexpressed with GluK1 subunits, which was attributed to selective targeting of the kainate receptor/Neto complexes to silent (AMPA receptor-less) synapses in these cells (Sheng et al., 2015). Moreover, both plasma membrane expression and synaptic targeting required distinct sets of residues in the C-terminal tails of Neto1 and Neto2 (Sheng et al., 2015), and serine phosphorylation of Neto2 occluded GluK1 synaptic targeting (Lomash et al., 2017). The promotion of synaptic targeting by Neto proteins was specific to coexpressed GluK1; in analogous experiments with GluK2, Neto1, or Neto2 coexpression did not alter the delivery of GluK2 to excitatory synapses (Sheng et al., 2017). These studies suggest that Neto proteins might differentially promote surface membrane expression and synaptic targeting of distinct kainate receptors, perhaps as a consequence of the presence (or absence) of forward-trafficking determinants in the receptor subunit CTDs (Ren et al., 2003b,c; Jaskolski et al., 2004; Yan et al., 2004).
Neto proteins in nonmammalian organisms play somewhat distinct roles that do not center on modulation of kainate receptor signaling. The C. elegans ortholog of Neto proteins, SOL-2, stabilizes synaptic localization and modified functional properties of the GLR-1 AMPA receptor (Wang et al., 2012). The Drosophila Neto proteins, Neto-α and Neto-β, are obligatory for synaptic accumulation of ionotropic glutamate receptors at neuromuscular junctions (Kim et al., 2012; Ramos et al., 2015) and, in the case of presynaptic Neto-α, for appropriate development of basal transmission and a form of homeostatic plasticity (Han et al., 2020).
Neto proteins also have been proposed to function in other capacities beyond those related to kainate receptor signaling. Neto1 was first suggested to be an NMDA receptor auxiliary protein, and synaptic NMDA receptors were altered in Neto1 KO mice (Ng et al., 2009; Wyeth et al., 2014). In contrast to kainate receptors, Neto1 coassembly has not been shown to modify NMDA receptor function or pharmacology, and it remains unclear whether Neto1 associates directly with NMDA receptors or indirectly as a component of a macromolecular protein complex (Cousins et al., 2013). Neto2 also was identified as an interacting protein for the K+-Cl- cotransporter KCC2, a regulator of the chloride equilibrium potential in neurons. GluK2 kainate receptor subunit was found subsequently to associate with KCC2 and influence surface expression of the transporter (Mahadevan et al., 2014; Pressey et al., 2017). Genetic targeting of either Neto2 or GluK2 disrupted chloride homeostasis in hippocampal neurons and, at least in the case of Neto2, enhanced seizure susceptibility (Mahadevan et al., 2015).
IV. Receptor Activation, Deactivation, and Desensitization
A. Receptor Activation and Deactivation
1. Glutamate Receptor Response Time Course and Synaptic Transmission
Glutamate receptors are best understood for their role in synaptic transmission, in which they are activated by rapidly released glutamate that reaches high concentration and endures in the synaptic cleft for a few milliseconds before diffusion and uptake reduce the concentration (Clements et al., 1992; Diamond, 2005; Budisantoso et al., 2013). The mechanisms that control iGluR channel opening and closing therefore dictate the postsynaptic iGluR response to brief synaptic pulses of glutamate (Lester et al., 1990; Silver et al., 1996a; Jonas, 2000; Traynelis et al., 2010; Paoletti et al., 2013), with the rate at which glutamate dissociates being the strongest determinant of the time course of synaptic currents (Fig. 14). For extrasynaptic or perisynaptic receptors, their affinity for glutamate will control their response to extrasynaptic glutamate after synaptic or glial release (Kessler, 2013) as well as the response to steady-state levels of glutamate in the extracellular space at concentrations near 80 nM (Moldavski et al., 2020). These properties vary for different subunit combinations for both AMPA and kainate receptors (Table 1). For NMDA receptors, the steady-state concentration of extracellular glycine [∼6 μM in cerebrospinal fluid (CSF); (D'Souza et al., 2000)] and d-serine [(∼2 μM in CSF; (Madeira et al., 2015)] will also impact receptor function. The glycine site is usually not saturated (Berger et al., 1998; Bergeron et al., 1998), and thus phasic changes in d-serine release from astrocytes or glycine release from nearby glycinergic terminals (Kalbaugh et al., 2009) and regional differences in glycine transporter expression (Zafra et al., 1995) could impact NMDA receptor function. When synaptic glutamate concentration remains elevated due to restricted diffusion at specialized synaptic structures or high-frequency release of vesicles, the rates into and out of the desensitized state shape the synaptic response time course [e.g., Kinney et al. (1997), Zampini et al. (2016)] (Fig. 14). The factors that impact receptor response characteristics create a vast palette of properties that synapses can choose from, allowing precise circuit refinement.
Fig. 14.
Time course of glutamate receptor–mediated EPSCs. (A) The average waveform for synaptic mEPSCs recorded from a hippocampal CA1 pyramidal cell in tetrodotoxin and 0.1 mM Mg2+; shaded area is the S.E.M. The AMPA receptor–mediated mEPSCs were isolated by inclusion of the NMDA receptor antagonist AP5, and NMDA receptor-mediated mEPSCs were the difference current in the absence and presence of AP5. Reproduced with permission from Perszyk et al. (2016). (B) Evoked mossy fiber synaptic currents recorded in hippocampal CA3 pyramidal neurons were mediated by AMPA receptors (black, VHOLD −70 mV) and kainate receptors (red) revealed by 50 µM GYKI 53655. The NMDA receptor-mediated component (blue) was recorded at +40 mV in CNQX. Modified from Straub et al. (2016) and unpublished data from Toshihiro Nomura and Anis Contractor. (C) The left panel shows an evoked AMPA receptor–mediated EPSC from a cerebellar unipolar brush cell. The right panel illustrates multiple release sites from a cerebellar mossy fiber onto a unipolar brush cell with synaptic geometry that restricts diffusion and confines glutamate to the synaptic space, thereby prolonging the postsynaptic response, especially after a train of EPSCs. Reproduced with permission from Kinney et al. (1997).
TABLE 1.
Kinetic parameters describing activation of iGluRs
Values at 2–3 significant figures are from the text or tables; when values are not reported, measurements were made from figures. Adapted with permission from Traynelis et al. (2010).
| Receptor a |
Glutamate EC50
b
(Glycine EC50) |
τ-Deactivate c | τ-Desensitize c,d | τ-Recovery d | Steady State/Peak Current Ratio |
|---|---|---|---|---|---|
| μM | ms | ms | ms | ||
| GluA1FLIP | 500–1600 1,11,50 | 0.7–1.5 2,3,36,50 | 2.5–4.1 2,3,32,36,50 | 111–147 2,11 | 0.002–0.032 2,4,5,50 |
| GluA1FLOP | 450 30 | 0.86–1.3 2,3,6,36 | 3.2–4.2 2,3,6,32,36 | 147–155 2,6 | 0.023–0.080 2,6,7 |
| GluA2FLIP | – | 1.0 47 | 10 47,48 | – | 0.01–0.12 48,54 |
| GluA2QFLIP e | 1390 8 | 0.62–1.1 8,36,50 | 5.0–9.9 8,32,36,47,50 | 11.7 8 | 0.068 8 |
| GluA2QFLOP e | 1140–1380 8,30 | 0.54–0.9 8,36 | 1.2–1.9 8,32,36 | 31.3 8 | 0.011 8 |
| GluA3FLIP | 1000–1970 7,31,f | 0.56 7 | 3.0–5.1 3,7,10,32 | 15–70 7,9,h | 0.024–0.054 7,9 |
| GluA3FLOP | 1100–1780 7,31,f | 0.63–1.05 6,7 | 1.1–2.8 3,6,7,10,32,36 | 55–142 6,7,35 | 0.01 7 |
| GluA4FLIP | 1810 11,f | 0.6 3 | 3.6–5.1 3,32 | 6–21 9,11,h | 0.006–0.04 9 |
| GluA4FLOP | 56 42,k,l | 0.6 3 | 0.9 3,32 | 31–43 9,h | 0.003 9 |
| GluA1FLIP/2FLIP | – | 0.8–4.5 50-53 | 4.8–6.3 33,50-53 | 28–87 33,51-53 | 0.009–0.041 33,50,52-54 |
| GluA1 FLOP/2FLOP | – | – | 2.8 33 | – | 0.007 54 |
| GluA1FLIP/2 FLOP | – | – | 3.4–4.4 33,54 | – | 0.006 54 |
| GluA3FLIP/2FLIP | – | – | 4.9 33 | 15–26 33 | 0.015–0.022 33 |
| GluA2FLIP/4 FLIP | – | 0.7 55 | 4.8–6.1 33,79 | 51 79 | – |
| GluA2FLOP/4 FLOP | – | 0.71 51 | 0.8–0.92 33,51 | 39 51 | 0.01 51 |
| GluK1-2a(Q) e,n | 631 12 | – | 1.6–13.4 12-14,56-58,67 | 18–50,5100 14,56,g | 0.01–0.04 14 |
| GluK1-2b(Q) | – | 1.7 59 | 1.6–42 56,59,60,76 | 2900 59 | – |
| GluK2a(Q) e | 427–1040 15,17,39,40,62,63 | 1.6–3.5 16,17, 37,39,57,58,61-63 | 3.4–7.0 15-17,37-40,58,61,63,64 | 700–3020 15-17,37,39,40,57,64 | 0.003–0.008 15-17,38 |
| GluK3a i | 5900–12300 18,57,62 65,66 | 0.7–1.5 57,62,65 | 3.8–8.4 18,57,65-67 | 900–1400 57,65,66 | 0.04 18 |
| GluK1-2a(Q)/5 m | 19 41,o | – | 1.4–2.1 44,67 | – | – |
| GluK1-2b(Q)/5 | – | – | 0.7 59 | 3800 59 | – |
| GluK2a(Q)/5 | 15–31 40,41,o | 2.3–46 40,59 | 1.8–5.9 40,57-59,64,67 | 1740–2700 40,59,64 | – |
| GluK3a/4 | – | – | 7.6 18 | – | 0.027 18 |
| GluK3a/5 | – | – | 5.3–6.6 18,67 | – | 0.035 18 |
| GluN1/2A p | 1.8–7.7 (0.86) 19,20 | 22–230 21,25,28,70 | 182 77 | 618 21 | 0.28–42 22,23 |
| GluN1-1b/2A | 3.42 (1.33) 70,k | 32–56 69,70 | – | – | – |
| GluN1/2B | 0.9–4 (0.34) 19,20,26,69 | 184–603 21,26,69-71 | 253–444 26,77 | 1010–2100 21,26 | 0.027–0.53 26,f,j |
| GluN1-1b/2B | 2.93 (0.29) 70,k | 144–155 69-71 | – | – | – |
| GluN1/2C | 1.0 (0.14) 27,34,k | 260–382 21,25,28 | 59–719 43 | – | 1.0 28 |
| GluN1/2D | 0.4 (0.12) 29,34,69,k | 1700–4400 21,24 | NA | NA | 1.0 24 |
| GluN1-1b/2D | 0.93 (0.15) 72,k | 700–992 69,72 | – | – | – |
| GluN1/2A/2B | 2.5–2.6 (0.63) 46,54,k | 57–143 46 | – | – | – |
| GluN1/2A/2C | 0.8–1.3 (0.48) 68,78,k | 301 68 | – | – | – |
| GluN1/2B/2D | 0.49 (0.30) 69,k | 1770 69 | – | – | – |
| GluN1/3A | – (48–81) 73,74,k | 205–442 74,75,q | 52 73,75 | 8 73 | 0.1–0.3 74,75 |
| GluN1/3B | –– | – | – | – | 0.2 75 |
a All receptors were from rat. For all experiments, the agonist was glutamate.
b Determined from the peak response to rapid glutamate application; see reference for glycine concentration used at NMDA receptors.
c Weighted time constant; two time constants can be detected for many receptors.
d See Lomeli et al. (1994) for RNA editing control of τ recovery.
e Unedited receptors had a glutamine at the Q/R/N site.
f Values predicted from simulations using rate constants.
g Onset and recovery from desensitization is variable from cell to cell.
h Recovery from desensitization produced by a 1-ms pulse of glutamate depended on the editing of an R/G site9.
i Splice isoforms 3a and 3b have similar rates.
j The ratio of steady state to peak current is typically higher in whole-cell recordings.
k Determined in Xenopus oocytes.
l Recorded in the presence of cyclothiazide.
m Determined by rapid application of glutamate; kainate-evoked currents desensitize with a dual exponential time course, with the fastest time constant being 15 ms (Herb et al., 1992).
n Glutamate desensitization of GluK1-2a receptors exhibits a time-dependent slowing (Copits et al., 2011; Swanson and Heinemann, 1998).
o The EC50 measurements from Alt et al. (2004) were made at equilibrium after treatment with con A and are not directly comparable to the peak EC50.
p Data are from GluN1-1a unless indicated otherwise.
q GluN1/GluN3 deactivation was recorded in the presence of CGP-78608 for two different redox states of GluN1.
NA, not applicable; GluN1/2D receptors show no desensitization in the continued presence of agonist.
1 Wahl et al. (1998), 2 Partin et al. (1995); Partin et al. (1996), 3 Mosbacher et al. (1994), 4 Banke et al. (2000), 5 Robert et al. (2001), 6 Banke et al. (2001), 7 Sekiguchi et al. (2002), 8 Koike et al. (2000), 9 Lomeli et al. (1994), 10 Grosskreutz et al. (2003), 11 Robert and Howe (2003), 12 Sommer et al. (1992), 13 Swanson et al. (1997a), 14 Swanson and Heinemann (1998), 15 Traynelis and Wahl (1997), 16 Heckmann et al. (1996), 17 Bowie (2002), Bowie and Lange (2002), 18 Schiffer et al. (1997), 19 Varney et al. (1996), 20 Chen et al. (2008), 21 Vicini et al. (1998), 22 Wyllie et al. (1996), 23 Krupp et al. (1998), 24 Wyllie et al. (1998), 25 Monyer et al. (1992), 26 Banke and Traynelis (2003), 27 Ishii et al. (1993), 28 Villarroel et al. (1998), 29 Ikeda et al. (1992), 30 Pei et al. (2009), 31 Pei et al. (2007), 32 Quirk et al. (2004), 33 Mosbacher et al. (1994), 34 Yuan et al. (2009), 35 Schlesinger et al. (2005), 36 Krampfl et al. (2001), 37 Weston et al. (2006b), 38 Zhang et al. (2008e), 39 Kistler and Fleck (2007), 40 Barberis et al. (2008), 41 Alt et al. (2004), 42 Plested et al. (2004), 43 Dravid et al. (2008), 44 Swanson et al. (2002), 45 Swanson et al. (1996), 46 Hansen et al. (2014), 47 Coombs et al. (2019), 48 Kato et al. (2008), 49 Turetsky et al. (2005), 50 Coombs et al. (2012), 51 Schwenk et al. (2009), 52 Klaassen et al. (2016), 53 Matt et al. (2018), 54Stroebel et al. (2014), 55 Ishii et al. (2020), 56 Copits et al. (2011), 57 Vernon et al. (2017), 58 Copits et al. (2014), 59 Straub et al. (2011b), 60 Larsen et al. (2017), 61 Zhang et al. (2009b), 62 Perrais et al. (2009a), 63 Dawe et al. (2013), 64 Fisher and Mott (2013), 65 Veran et al. (2012), 66 Kumari et al. (2019), 67 Swanson et al. (1998), 68 Bhattacharya et al. (2018), 69 Yi et al. (2019), 70 Yi et al. (2018), 71 Rumbaugh et al. (2000), 72 Vance et al. (2012), 73 Cummings and Popescu (2016), 74 Grand et al. (2018), 75 Hemelikova et al. (2019), 76 Ren et al. (2003c), 77 Erreger et al. (2005a), 78 Wafford et al. (1993), 79 Miguez-Cabello et al. (2020).
Synaptic AMPA receptors show a rapid activation and short response time course when glutamate concentration is elevated briefly in the cleft [e.g., Silver et al. (1992)], or a prolonged response time course when glutamate persists for longer periods at specialized synapses that entrap glutamate in the cleft (Kinney et al., 1997; Zampini et al., 2016) (Fig. 14). The high peak concentration of glutamate (estimated at 1.1 mM) in the cleft (Clements et al., 1992) ensures rapid binding, and the brief duration of elevated synaptic glutamate during low-frequency synaptic events minimizes receptor desensitization (Hestrin, 1992), which is profound for AMPA receptors and limits signaling to the millisecond range (Salazar et al., 2020). In a sense, AMPA receptor subunit/auxiliary subunit combinations that produce a brief response time course can emulate digital signaling in that synaptic glutamate release produces brief injections of current into the postsynaptic compartment (Raman and Trussell, 1995), which enables the postsynaptic neuron to respond to high-frequency signals. By contrast, AMPA receptor complexes (Section III. Auxiliary Subunits, see also below) or specialized synaptic geometry that slow the AMPA receptor response time course or reduce desensitization (Fig. 14) promote temporal summation and signal integration.
Expression of recombinant AMPA receptors in heterologous systems allows measurement of the deactivation rate after glutamate removal, which for AMPA receptors lacking auxiliary subunits follows an exponential time course with a time constant as brief as 1–5 milliseconds (Mosbacher et al., 1994). The onset of desensitization can be measured from the response to prolonged glutamate application (Fig. 15), and auxiliary subunits modify this response time course in synaptically relevant ways (Fig. 9) (Milstein and Nicoll, 2009). TARPs, CNIH proteins, GSG1L, CKAMP, and SynDIG4 can all prolong the deactivation time course up to several fold (Table 2), prolonging the duration of synaptic currents [e.g., Cho et al. (2007), McGee et al. (2015), Klaassen et al. (2016), Schmitz et al. (2017)]. AMPA receptor auxiliary subunits have variable effects on desensitization, which becomes relevant to signaling at high frequencies of firing and at specialized synapses with a prolonged synaptic glutamate time course (Table 2). Some auxiliary subunits alter the response properties to low concentrations of glutamate that might arise during glutamate spillover from neighboring synapses (Coombs et al., 2017). In addition, alternative splicing of the flip/flop cassette in AMPA receptors controls the rates of entry into and recovery from desensitization (Mosbacher et al., 1994; Koike et al., 2000; Quirk et al., 2004), the mobility of the resting or apo state of receptor ABDs (Dawe et al., 2019), and the efficiency of biogenesis in the ER (Coleman et al., 2006; Penn et al., 2008). The flip/flop cassette also comprises part of the binding pocket for positive allosteric modulators (Section IX. Exogenous Positive and Negative Allosteric Modulators), and RNA editing at the R/G site immediately preceding the flip/flop domain is an additional regulator of receptor kinetics (Lomeli et al., 1994) (Section II.F. Alternative Splicing, RNA Editing, and Post-Translational Modifications). All of these responses are filtered by membrane properties, which add an additional layer of complexity to the response time course (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity).
Fig. 15.
Glutamate receptor response time course. (A1) Responses to glutamate applications (gray bars) from GluA4 + γ-5 in an outside-out patch (VHOLD −60 mV). Reproduced with permission from Soto et al. (2009). (A2) Ten individual responses from a single GluA4-γ-2 tandem receptor in an outside-out patch (VHOLD, −100 mV) illustrate random channel activation and occasional lack of openings despite saturating glutamate that will produce full receptor occupancy. Reproduced with permission from Zhang et al. (2014a). (B1) Response from GluK2 with and without Neto2 in an outside-out patch (VHOLD −100 mV) illustrates rapid and intermediate time course of desensitization. (B2) Ten individual responses from a single GluK2+Neto2 receptor in an outside-out patch (−100 mV) illustrate an occasional lack of channel openings despite saturating glutamate. Reproduced with permission from Zhang et al. (2009b). (C1) Normalized whole-cell current responses from GluN1/2A, GluN1/2B, or GluN1/2A/2B receptors (VHOLD −60 mV) to 5-millisecond glutamate application. Reproduced with permission from Hansen et al. (2014). (C2) Ten individual responses from a single GluN1/2B receptor in an outside-out patch (VHOLD −80 mV) illustrate occasional lack of channel openings despite full agonist occupancy. Reproduced with permission from Erreger et al. (2005a). Glycine was present in all solutions.
Recombinant homomeric GluK1, GluK2, and GluK3 kainate receptors show a rapid activation profile with strong desensitization in the continued presence of agonist and recover from desensitization far more slowly than AMPA receptors (Table 1). GluK4 and GluK5 do not form homomeric channels on their own but coassemble with GluK1 and GluK2 to form heteromeric receptors with higher agonist affinity (Fig. 14). Postsynaptic kainate receptors are detected at only a subset of excitatory synapses in the brain and generally produce a slower time course for the synaptic current (Castillo et al., 1997; Kidd and Isaac, 1999, 2001; Contractor et al., 2011), which in part reflects the actions of GluK5 subunits incorporated into the receptor (Contractor et al., 2003; Barberis et al., 2008) (Table 1) as well as the association with Neto1 or Neto2 auxiliary subunits (Straub et al., 2011a; Vernon and Swanson, 2017). Agonist occupancy of GluK2 subunits evokes rapid desensitization (Barberis et al., 2008) and major rearrangment of GluK2 subunits within the heteromer complex (Khanra et al., 2021), suggesting that the slow response time course of native kainate receptors primarily reflects gating by GluK5 subunits. The association with Neto1 or Neto2 increases glutamate potency and slows desensitization for homomeric GluK1 and GluK2 receptors (Zhang et al., 2009b; Copits et al., 2011; Straub et al., 2011a,b) (Fig. 15; Table 3) (Section III. Auxiliary Subunits).
Synaptic NMDA receptors respond to rapid, brief synaptic pulse of glutamate with a slower time course than AMPA receptors, activating in several milliseconds (Table 1) and deactivating with a time course that can be orders of magnitude slower than that for AMPA receptors and slower than kainate receptors (Monyer et al., 1992; Vicini et al., 1998; Wyllie et al., 1998) Figs. 14 and 15). The deactivation time course varies with different GluN2 subunits, being fastest for GluN1/2A (time constant ∼35 milliseconds), which has a high open probability (∼0.5). The time constant describing deactivation is ∼200 milliseconds for GluN1/2B (open probability ∼0.1) and GluN1/2C (open probability 0.01) and takes several seconds for GluN1/2D receptors (open probability 0.01; Fig. 15; Table 1). Alternative splicing of exon 5 to produce isoforms that include 21 residues in the GluN1 NTD (e.g., GluN1-1b) also controls synaptic plasticity, alters the NMDA receptor pharmacology, increases open probability, and accelerates the deactivation time course (Rumbaugh et al., 2000; Kostakis et al., 2011; Das et al., 2012; Vance et al., 2012; Yi et al., 2018; Sengar et al., 2019) (Table 1).
The time course for glycine-activated GluN1/3 receptor responses is relatively rapid and followed by pH-dependent desensitization that may involve instability of the ABD heterodimer interface (Cummings and Popescu, 2016). Multiple lines of investigation suggest that the desensitization reflects autoinhibition (or predesensitization) after glycine binding to the GluN1 subunit (Awobuluyi et al., 2007; Madry et al., 2007; Kvist et al., 2013a; Grand et al., 2018). Thus, in contrast to GluN1/2 NMDA receptors (which require agonist occupancy of both GluN1 and GluN2 subunits), recombinant GluN1/3 receptors expressed in heterologous systems can open with agonist occupation of only the GluN3 subunit, whereas agonist binding to GluN1 produces strong desensitization (Awobuluyi et al., 2007; Madry et al., 2007; Kvist et al., 2013a; Grand et al., 2018). How this property influences GluN1/3 receptor function in the CNS, where ambient glycine should “predesensitize” the receptors (Grand et al., 2018; Otsu et al., 2019; Zhu et al., 2020) remains to be determined. Despite a lack of understanding of their activation mechanism, the description of glycine-activated currents in wild-type but not GluN3A KO neurons after block of glycine binding to GluN1 confirms neuronal expression of functional GluN1/3A receptors (Grand et al., 2018; Otsu et al., 2019; Zhu et al., 2020) (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity).
Finally, the GluD1 and GluD2 receptors encoded by GRID1 and GRID2 do not mediate a conventional ligand-gated current response (Araki et al., 1993; Lomeli et al., 1993). Although GluD2 harbors a d-serine binding site, and the occupancy of this site can close the bilobed ABD clamshell (Naur et al., 2007; Tapken et al., 2017; Chin et al., 2020), d-serine does not open the ion channel in GluD1 or GluD2 receptors expressed in heterologous systems. However, transplantation of the ABD of kainate and AMPA receptors onto GluD2 (Schmid et al., 2009) or removal of a disulfide bond in the GluD2 ABD (Hansen et al., 2009) enables opening the ion channel in response to agonist binding, demonstrating that the gating machinery and pore within GluD2 is intact (Yuzaki and Aricescu, 2017; Gantz et al., 2020). Introduction of GluD2 with a mutation in the d-serine binding site can diminish cerebellar LTD, suggesting d-serine exerts physiologically relevant effects through GluD2 receptors (Kakegawa et al., 2011). Studies have demonstrated an alternative mechanism for signaling by the GluD receptor family that involves trans-synaptic interactions and metabotropic signaling [reviewed in Yuzaki and Aricescu (2017)] (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity).
2. Single-Channel Properties of Glutamate Receptors
The single-channel properties of many AMPA, kainate, and NMDA receptor subunit combinations have been studied extensively in heterologous expression systems (Supplemental Table 1). NMDA receptors open to relatively large (20–50 pS) cation-selective conductance levels in neurons (Nowak et al., 1984; Cull-Candy and Usowicz, 1987; Jahr and Stevens, 1987) and in heterologous expression systems (Stern et al., 1992) after simultaneous binding of glutamate to both GluN2 subunits and glycine to both GluN1 subunits (Benveniste and Mayer, 1991; Laube et al., 1998). The requirement that four agonists must bind for channel opening has been demonstrated by the lack of a response from receptors with a single copy of a subunit harboring mutations in either the glycine or glutamate binding site in GluN1 and GluN2 subunits, respectively (Hansen et al., 2014; Yi et al., 2018). However, not every binding event produces channel opening (Fig. 15), raising the possibility that receptors can exist in a low-activity state (Erreger et al., 2005a; Amin et al., 2021a), which may vary with the GluN2 subunit (Popescu and Auerbach, 2003; Zhang et al., 2008c; Vance et al., 2013) and has also been suggested for AMPA and kainate receptors (Zhang et al., 2009b; Zhang et al., 2014a) (Fig. 15). This low-activity state could emerge as a central feature of receptor control, being a point of modulation by phosphorylation [e.g., Banke et al. (2000)] and a means for shifting postsynaptic receptor response amplitudes without changing the response time course. Examples of this low-activity state are evident for AMPA, kainate, and NMDA receptor single-channel responses to glutamate (Fig. 15), in which some receptors fail to open in response to brief agonist exposure. Transitions into and out of this low-activity state are hypothesized to occur on a slow time course (Banke et al., 2000; Zhang et al., 2008c; Vance et al., 2012; Amin et al., 2021a).
AMPA receptors open to four conductance levels, the amplitude of which is controlled by RNA editing, subunit composition, and auxiliary subunits (Fig. 16; Supplemental Table 1). In contrast to NMDA receptors that require occupancy of all agonist binding sites, AMPA receptor channels can open with a contribution from each subunit (Rosenmund et al., 1998; Coombs et al., 2017) (Fig. 16). Four conductance levels were therefore observed from a single AMPA receptor in an experiment in which four antagonist molecules were initially bound to the receptor, and then the receptor was allowed to bind agonist sequentially as each antagonist molecule was replaced by an agonist molecule (Rosenmund et al., 1998; Coombs et al., 2017). In this experiment, the four AMPA receptor subunits were detected as the unitary current amplitude increased in multiple steps, presumably because agonist binds immediately after the antagonist dissociates. That is, AMPA receptors opened to different conductance levels as the fraction of agonist-bound subunits was increased and the fraction of antagonist-bound channels decreased (Fig. 16). This experiment provided a rationale for the enigmatic subconductance levels that had been described for non-NMDA receptors [Swanson et al. (1996, 1997b); reviewed by Greger et al. (2017)] and suggested occupancy of different numbers of subunits was capable of opening the pore but with different properties.
Fig. 16.
Multiple conductance levels of AMPA receptors. Upper panel: Schematic of slow NBQX unbinding from an AMPA receptor, which allows the time course of channel activation by glutamate to be observed. Lower panel: Response from a single GluA1 + γ-2 (tandem) receptor in an outside-out patch that has NBQX bound to all subunits after rapid exchange into a solution lacking NBQX and containing 10 mM glutamate (VHOLD −60 mV). Each of four antagonist unbinding events is evident as glutamate occupies the available site following NBQX dissociation, which enables the opening to an individual sublevel, with chord conductance given (in pS). Reproduced with permission from Coombs et al. (2017).
Similar functional conclusions about the origin of multiple conductance levels were drawn from single-channel recordings from GluA1 receptors with phosphomimic substitution at Ser831 (i.e., S831E) (Kristensen et al., 2011), which increased the proportion of larger sublevels (Derkach et al., 1999; Derkach, 2003). The sublevel proportions could be quantitatively accounted for by a phosphorylation-induced increase in efficiency by which each subunit contributed to an active state, leading to an increase in the frequency of larger conductance levels as more subunits became active (Kristensen et al., 2011). In addition, reduction of desensitization via mutation at the ABD dimer interface also increased the proportion of large conductance openings (Zhang et al., 2017), whereas the AMPA receptor negative allosteric modulator perampanel reduced the proportion of large conductance levels, consistent with reduction in the contribution of each subunit to receptor activation (Yuan et al., 2018). Single-channel GluA2 openings induced by partial agonists, which occupy all four AMPA receptor agonist binding sites, also showed a correlation between the diminishing efficacy and a diminished ability to activate the largest conductance level (Jin et al., 2003; Poon et al., 2010, 2011; Kristensen et al., 2011). Many of the initial single-channel AMPA receptor studies (Supplemental Table 1) were performed in the absence of accessory subunits; however, more recently there has been considerable effort to understand how these accessory subunits control AMPA receptor gating (Table 2). Kainate receptors also open to at least three conductance levels (Supplemental Table 1).
GluN1/2A and GluN1/2B NMDA receptors, which contain two GluN1 and two of the same GluN2 subunit, primarily open to a single 50-pS conductance level in 1 mM Ca2+, with <10% of the openings to a ∼40-pS subconductance level (Stern et al., 1992). Agonist-bound GluN1/2A has a higher open probability than GluN1/2B. GluN1/2C and GluN1/2D channels show two lower conductance levels, briefer open time, and a lower open probability (Stern et al., 1992; Wyllie et al., 1996; Dravid et al., 2008; Vance et al., 2012) (Supplemental Table 1). Under certain recording conditions (such as the absence of divalent ions), GluN1/2A show prominent instability in both the mean open time and open probability, which produces periods of time with strikingly different single-channel characteristics, termed modal gating (Popescu and Auerbach, 2003). Other NMDA receptors have also been reported to show modal gating (Popescu and Auerbach, 2003; Amico-Ruvio and Popescu, 2010; Vance et al., 2013). Single-channel behaviors consistent with modal gating have been observed for neuronal NMDA receptors (Jahr and Stevens, 1987; Kleckner and Pallotta, 1995; Borschel et al., 2012), but how model gating relates to the physiologic roles of native receptors remains unclear. Triheteromeric NMDA receptors, which contain two GluN1 and two different GluN2 subunits, are found in adult neurons and show complex single-channel properties that are a unique and unpredictable blend of individual subunit properties rather than an average of properties of the two GluN2 subunits (Cheffings and Colquhoun, 2000; Brickley et al., 2003; Jones and Gibb, 2005; Bhattacharya et al., 2018) (Table 1). Some features of receptor function, such as deactivation time course, can be dominated by one subunit (Hansen et al., 2014; Sun et al., 2017b) (Fig. 15), whereas other functional and pharmacological features of the receptor draw on unique features of each subunit (Hansen et al., 2014; Stroebel et al., 2014; Cheriyan et al., 2016; Bhattacharya et al., 2018; Yi et al., 2018, 2019). This is evident in the pattern of single-channel activity for GluN1/2A/2C, which shows the burst behavior of GluN2A-containing receptors but the low open probability of GluN2C-containing receptors (Bhattacharya et al., 2018) (Fig. 17; Supplemental Table 1). Triheteromeric NMDA receptors show a range of subconductance levels that appear to be a combination of those observed for each diheteromeric receptor. For example, GluN1/2A/2C receptors show both high conductance levels (like GluN1/2A) and lower conductance levels (like GluN1/2C), with direct transitions between them, suggesting that the channel pore retains characteristics of diheteromeric receptors for each constituent GluN2 subunit. Similar results have been observed for recombinant GluN1/2A/2D (Cheffings and Colquhoun, 2000) and native GluN1/2B/2D receptors (Brickley et al., 2003; Jones and Gibb, 2005).
Fig. 17.
Single-channel properties of triheteromeric NMDA receptors. (A) Unitary currents for GluN1/2A, GluN1/2C, and GluN1/2A/2C NMDA receptors (VHOLD −80 mV). Openings of diheteromeric GluN1/2A and triheteromeric GluN1/2A/2C receptors are clustered into bursts (gray bars) separated by inactive periods. In contrast, openings of diheteromeric GluN1/2C receptors show no apparent burst structure. (B) Unitary currents in (A) are expanded to illustrate multiple conductance levels with direct sublevel transitions (asterisks). (C) Fitted amplitude histograms for GluN1/2A (left axis), GluN1/2A/2C (right axis), or GluN1/2C receptors (right axis) were fitted by the sum of 2–3 Gaussian distributions (smooth lines). (D) Representative open duration histograms for GluN1/2A, GluN1/2C, and GluN1/2A/2C receptors were fitted by multiple exponential components. Reproduced with permission from Bhattacharya et al. (2018).
Only a modest amount of data is available describing the single-channel properties of glycine-activated GluN1/3 channels. Glycine-activated single-channel currents arising from recombinant GluN1/GluN3B receptors expressed in Xenopus oocytes show two conductance levels (37 and 12 pS) (Chatterton et al., 2002) (Supplemental Table 1). An additional conductance level of 29–40 pS has been reported in patches excised from Xenopus oocytes or HEK cells expressing recombinant GluN1, GluN2A, and GluN3A (Das et al., 1998; Perez-Otano et al., 2001; Sasaki et al., 2002; Tong et al., 2008), although it is unclear whether this level arises from putative GluN1/2A/3A receptors or GluN1/3A receptors. Nonstationary variance analysis of glycine-activated native GluN1/3A receptors recorded in the presence of the competitive GluN1 antagonist CGP-78608 (to prevent predesensitization) suggested a weighted mean unitary conductance of 6 pS (assuming a reversal potential of 0 mV) (Zhu et al., 2020). There are no studies of the potential single-channel properties of wild-type GluD1 or GluD2 receptors, which do not show detectable channel opening in response to agonist binding.
B. Mechanisms Linking Agonist Binding to Channel Gating
Glutamate receptors are tetrameric assemblies of subunits that each contain a number of semi-autonomous domains, which interact through protein-protein interfaces as well as polypeptide linkers that couple the domains within the subunits (Figs. 2 and 18). To open the ion channel pore, the ABD and the TMD energetically communicate through a set of short polypeptide linkers that connect the ABD to TMD (Fig. 18).
Fig. 18.

The mechanism linking agonist binding to channel opening at the M3 gate. (A) Structure of GluA2 AMPA receptor illustrates the different subunit positions within the tetramer, termed A/C (occupied by GluN1 in NMDA receptors, blue) and B/D (occupied by GluN2, gray). The green arrows illustrate the glutamate binding site within the bilobed, clamshell-like ABD (PDB: 5WEK). (B1) Clamshell arrangement of an ABD dimer (left) and the two A/C (middle) and B/D (right) ABDs bound to glutamate, which produces clamshell closure. (B2) The left panel shows resting arrangement of the M3-S2 linkers and the M3/M2 membrane regions, which form the core of the ion permeation pathway. M1 and M4 transmembrane helices are omitted for clarity. The middle panel illustrates how agonist binding repositions the M3 helices of the A/C subunits, with a movement vertically displaced (red) distal to the extracellular surface of the membrane. The right panel illustrates how glutamate binding to the B/D subunits laterally displaces the M3 helices (red), causing a splaying at the Ala within the SYTANLAAF conserved motif. (B3) View of the extracellular side of the ion channel down the axis of the permeation path for closed (PDB: 5WEK) and open receptors (PDB: 5WEO). The side chains of Thr625, which form part of M3 activation gate, are shown.
1. The Core Gating Machinery
The basic structural changes that give rise to opening of the glutamate receptor pore are understood [reviewed in Plested (2016), Greger et al. (2017), Zhou (2017), Zhou and Wollmuth (2017), Hansen et al. (2018), Greger and Mayer (2019), Wang and Furukawa (2019)], with the best structural examples from AMPA receptors (Chen et al., 2017a; Twomey et al., 2017a; Twomey and Sobolevsky, 2018) (Supplemental Movie 1) and NMDA receptors (Tajima et al., 2016; Zhu et al., 2016; Chou et al., 2020; Wang et al., 2021) (Fig. 18). The bilobed ABD structures are comprised of an upper lobe (D1), a lower, membrane-proximal lobe (D2), and a binding site for agonists that resides in the cleft between the D1 and D2 lobes (Section II. Receptor Structure). Two ABDs form a “back-to-back” dimer with the dimer interface formed by the upper D1 lobes, and the ABD layer in the tetrameric receptor is formed by ABDs in a dimer-of-dimers arrangement (Figs. 3 and 18). In the absence of agonist, the bilobed ABD clamshell resides in an open conformation, and the activation gate (i.e., the physical structure that precludes the flux of ions in the closed state) located in the helical bundle crossing formed by the four M3 transmembrane helices is closed (Fig. 18). Agonist binding induces ABD clamshell closure, which translates into the lower D2 lobes moving away from the membrane (Fig. 18). The ABDs explore a wide range of conformations and side-chain orientations with agonist binding stabilizing a subset of closed clamshell states. Thus, ABD closure in response to agonist binding likely occurs by a conformational selection mechanism, whereby agonists alter the frequency and dwell times of fully closed conformations (Maltsev et al., 2008; Ramaswamy et al., 2012; Salazar et al., 2017) (Section II. Receptor Structure). The lower membrane-proximal D2 lobe is attached to M3-S2 linkers, which pull on the M3 helices as the agonist induces ABD clamshell closure. The tension created by ABD closure initiates conformational changes within the linkers that ultimately lead to rapid all-or-none opening of the ion channel pore (Armstrong and Gouaux, 2000; McFeeters and Oswald, 2002; Kazi et al., 2014; Ladislav et al., 2018; Černý et al., 2019). The transition of the M3 transmembrane helices from the closed to the open conformation permits the flux of ions across the membrane (Fig. 18). Although the apex of the M2 pore loops for all four subunits appear to adopt a position that could reflect its role as a second gate that controls ion flux, there is limited information on how it may participate in the process of channel gating (Premkumar et al., 1997; Buck et al., 2000; Chen et al., 2004b; Li et al., 2019a; Poulsen et al., 2019). Single-channel recordings of the relatively slow NMDA receptors have demonstrated at least two kinetically distinct conformational steps must occur after agonist binding but before the pore can open, and these may reflect any of a number of linker conformations, including side-chain rotation for key residues or repositioning of the polypeptide chain (Banke and Traynelis, 2003; Popescu and Auerbach, 2003; Auerbach and Zhou, 2005; Schorge et al., 2005; Gibb et al., 2018).
2. The Role of the M3-S2 Linker in Gating
Structures of AMPA receptors in the closed and open states have facilitated our understanding of how agonist binding to the ABD induces ion channel opening [Twomey et al. (2017a); see also Chen et al. (2017a)]. In the closed ion channel, the permeation pathway in homomeric GluA2 AMPA receptors is tightly sealed at the bundle crossing of the M3 helices by the side chains of Thr617, Ala621, Thr625, and Met629 (Sobolevsky et al., 2009). The first two of these residues, Thr617 and Ala621, are part of the most highly conserved sequence of iGluR subunits, which is referred to on the basis of its sequence as the SYTANLAAF motif. The linkers between the M3 helix and the S2 segment, which fold into part of the lower D2 lobe, take on distinct orientations dependent on the subunit position in the receptor (Sobolevsky et al., 2009; Karakas and Furukawa, 2014; Lee et al., 2014), with the M3-S2 linkers in the A/C position being perpendicular to the membrane and those in the B/D position nearly parallel (Fig. 18). Thus, the subunits in the A/C and B/D positions must undergo distinct displacements to move the M3 helices during gating. Helix αE is the first secondary structure in the D2 lobe after the M3-S2 linker. For A/C subunits, the predominant displacement of helix αE is perpendicular and away from the membrane (Fig. 18). By contrast, helix αE is displaced laterally and parallel to the plane of the membrane for B/D subunits. These movements are associated with alterations in the position of the M3 segments, which are distinct depending on the subunit position in the tetramer. For A/C subunits, one helical turn in the upper portion of the M3 helix unwinds, whereas for B/D subunits, the upper M3 unwinds, and a kink is generated at the conserved Ala618 (SYTANLAAF). These differences between subunits in the A/C and B/D positions are evident in the GluA1/2 heteromeric receptor, where the GluA1 M3 linkers position vertically, whereas those of the GluA2 subunit extend horizontally (Herguedas et al., 2019).
For NMDA receptors, less is known about the displacements of the M3 helices because of the absence of an open-state structure [but see Chou et al. (2020) and Wang et al. (2021)], but these movements are presumably comparable to those in AMPA receptors. Indeed, there are differences between the GluN1 subunits, which are in the A/C positions, and GluN2 subunits that are in the B/D positions (Fig. 3), and substitution at the conserved alanine in SYTANLAAF in the GluN2 subunit has strong effects on gating (Sobolevsky et al., 2007). In addition, the different conformational changes observed for A/C and B/D subunits may underlie the two kinetically distinct pregating steps that precede pore opening (Banke and Traynelis, 2003; Popescu and Auerbach, 2003; Auerbach and Zhou, 2005; Schorge et al., 2005) (Figs. 18 and 19).
Fig. 19.

Agonist-induced displacements of the outer structures prime the channel for pore opening. (A) Top-down view of a cartoon representation of the gating triad comprising the pre-M1 helix, the M3 helix, and the pre-M4 linker of the adjacent subunit. The cartoon illustrates two distinct steps required for channel opening that include movement of the pre-M1 helix and repositioning of the M4 linker (green, Pre-active state) that is hypothesized to occur prior to a secondary repositioning of the M3 helix and splaying at the Ala hinge (red) to reach the Active state and open the channel pore. (B) The GluN2 (B/D position in the tetramer) pre-M1 helix that lies parallel to the membrane, the GluN2 M3 transmembrane helix, and the GluN1 M4 transmembrane helix from one gating triad are shown as ribbon structures. In the transition from the closed (left, PDB: 5WEK) to the open (right, PDB: 5WEO) states, all three elements—the pre-M1 helix, and the tops of M4 and M3—undergo coordinated movements. The splaying of the M3 segments highlighted in red (right) is the final step in ion channel opening. It seems likely that the outer structures undergo displacements (pre-M1, and M4, green) prior to the final M3 displacement.
3. Transmembrane Helices M1 and M4 Surround the Pore-Forming Components
The inner M3/M3-S2 linker gating core is surrounded by the outer M1 and M4 transmembrane helices and the linkers connecting them to the ABD (i.e., the S1-M1 and S2-M4 linkers) (Sobolevsky et al., 2009; Karakas and Furukawa, 2014; Lee et al., 2014) (Fig. 19). These outer structures influence gating (Schmid et al., 2007; Talukder et al., 2010; Ren et al., 2012; Yelshanskaya et al., 2017; McDaniel et al., 2020), contain sites for allosteric modulators (Section IX. Exogenous Positive and Negative Allosteric Modulators), and must undergo agonist-induced movements for efficient pore opening to occur (Kazi et al., 2013) (Fig. 19). FRET measurements also show outward motions of the pre-M1 helix associated with agonist binding (Dolino et al., 2016; Durham et al., 2020). Although these outer structures each undergo agonist-dependent conformational changes (Dolino et al., 2017; Twomey et al., 2017a; Chou et al., 2020), the manner by which changes in the individual amino acid side chains and the polypeptide chain contribute to the process of opening the pore is unclear. In addition, it is unclear what structural features control the fixed size of the pore (0.55–0.8 nM) and resulting unitary conductance level (Supplemental Table 2). There could be changes within the networks of side-chain interactions after subtle rearrangements in the pre-M1 linker that alter the energetics of the closed state, bring about rapid pore dilation (McDaniel et al., 2020; Amin et al., 2021a; Iacobucci et al., 2021) in response to larger forces exerted along the polypeptide chain after agonist binding, cleft closure, and rotation of the ABD heterodimers, which produces the tension on the ABD-TMD linkers leading to vertical and horizontal displacement of the M3 helix (Twomey et al., 2017a; Chou et al., 2020).
Structures highlight the close link between the M3 helices, including the SYTANLAAF motif, the S1-M1 linker (most notably the pre-M1 helix), and the M4 transmembrane helix [Sobolevsky et al. (2009), Twomey et al. (2017a), Herguedas et al. (2019), Nakagawa (2019), Chou et al. (2020); reviewed in Perszyk et al. (2020a)]. These interacting motifs have been referred to as a “gating collar” in AMPA receptors (Yelshanskaya et al., 2017) and a “gating triad” in NMDA receptors (Gibb et al., 2018; Perszyk et al., 2020a; Amin et al., 2021a). These regions lack variation in the healthy population and are therefore under selective pressure, as departures from the wild-type residue will result in CNS diseases (Section X. Glutamate Receptors in Disease). These unbiased genetic data provide a strong argument that the interaction of these three elements is central to both the stability of the closed state and the process of channel opening.
4. The Pre-M1 Helix and S1-M1 Linker Control Pregating Steps
The roles of the pre-M1 helix and S1-M1 linker in the control of pregating steps have primarily been studied in NMDA receptors, as their limited desensitization and larger conductance states make them more amendable to single-channel recordings. Evaluation of single-channel shut-time durations of individual NMDA receptors activated by maximally effective concentrations of agonist suggests that they undergo multiple conformational changes between agonist binding and ion channel opening (i.e., pregating steps) (Gibb and Colquhoun, 1991; Banke and Traynelis, 2003; Popescu and Auerbach, 2003; Auerbach and Zhou, 2005; Schorge et al., 2005). Supplemental Figure 5 shows an example of how such shut times relate to hypothetical models of transitions among conformations in which shut times reflect closure of an agonist-bound channel followed by reversal of one (or more) pregating steps and then reactivation of those steps and reopening [reviewed in Hansen et al. (2018)]. Thus, the brief shut times contain information about the forward rates from specific conformations to the open state. One approach to a deeper understanding of receptor function is to connect the conformations with kinetic steps in the single-channel recordings. For example, the structural determinants of these pregating movements have been suggested to involve some change for the pre-M1 helix within the S1-M1 linker (Sobolevsky et al., 2009; Ogden et al., 2017; McDaniel et al., 2020; Amin et al., 2021a) (Fig. 19). The significance of the displacement of the GluN2 pre-M1 helix to channel gating is highlighted by the use of glutamate applications to outside-out patches containing single NMDA receptors. Here, decoupling the agonist-induced displacements of the GluN2A pre-M1 helix by insertion of glycine residues in the linker prevented NMDA receptors from contributing to rapid synaptic-like channel opening (Amin et al., 2021a).
The S1-M1 linker contains Pro557 in GluN1, Pro552 in GluN2A, and Pro553 in GluN2B, which are all highly conserved residues across a variety of species (Alsaloum et al., 2016). This conserved Pro residue is located in the short pre-M1 helix that lies parallel to the membrane, is in van der Waals contact with the M3 helix comprising the gate, and precedes the M1 transmembrane helix. Human variants and mutations that convert this conserved pre-M1 Pro in GluN2 into positively charged residues (e.g., GluN2A-P552R or GluN2A-P552K) dramatically slow the channel activation rate, which is governed by the rates of pregating steps (Ogden et al., 2017; Gibb et al., 2018). Thus, NMDA receptors lose their ability to respond rapidly to transient glutamate, suggesting that the conserved Pro controls agonist-induced movements that occur before opening of the ion channel. Alternatively, the positive substitutions make additional interactions that stabilize the closed conformation. Surprisingly, only a single intact Pro residue in one of two GluN2 subunits is sufficient to maintain fast NMDA receptor activation (Ogden et al., 2017). Hence, the normal displacement or reorientation of a single GluN2 pre-M1 helix is sufficient to allow fast pore opening to occur, suggesting that the S1-M1 linkers act independently and that the core gating machinery (M3-S2 linker and M3 helix) is energetically prepared for ion channel opening. Molecular dynamics simulations suggest that pre-M1 movements are independent of the actions of neighboring subunits, suggesting that this element could mediate subunit-specific features of NMDA receptor activity (Gibb et al., 2018; Černý et al., 2019; McDaniel et al., 2020). The same mutation in GluN1 does not alter NMDA receptor activation rate, emphasizing the distinct contribution to gating of GluN1 and GluN2 (Ogden et al., 2017). Other residues in pre-M1 play a role in gating, including GluN2A Leu550 (Iacobucci et al., 2021) and GluN2A Phe553, which is suggested to be embedded in an aromatic network that could stabilize the closed state and thus be a substrate for gating-induced reorganization prior to opening (McDaniel et al., 2020).
5. The Role of the S2-M4 Linker and the M4 Helix in Gating
The M4 helix does not contribute directly to the channel permeation pathway but rather serves a critical structural role and forms the binding sites for auxiliary subunits (Talukder et al., 2010). The NMDA receptor M4 helix contains sites that are important for pore opening. Deletion of the NMDA receptor M4 transmembrane helix results in no measurable current response, which could be rescued by coexpression of a cDNA encoding just the M4 transmembrane helix (Schorge and Colquhoun, 2003). Multiple function-changing de novo variants that are present in patients with neurologic disease but absent in the general population reside in the linker leading to the GluN1 and GluN2 M4 transmembrane helix (Perszyk et al., 2020a; Amin et al., 2021b) (Section X. Glutamate Receptors in Disease). For example, missense variants with strong effects on receptor function (Amin et al., 2018; Vyklicky et al., 2018) have been identified at a conserved Gly residue located at the juncture of the upper third and lower two-thirds of the M4 segment (GluN1-G815 and GluN2B-G820) (Partridge et al., 2004; Hamdan et al., 2014; Ohba et al., 2015; Lemke et al., 2016; Molnar et al., 2016; Platzer et al., 2017). These variants speed up the deactivation time course by restricting the ability of the pore-lining M3 helices to stay open (Amin et al., 2018). The conserved Gly in GluN1 have been suggested to act as a hinge, enabling the lower two-thirds of the GluN1 M4 to exist in either a “constrained” or “expanded” conformation (Amin et al., 2018) (Supplemental Fig. 6). Molecular dynamics simulations predict the GluN1 Gly815 variants will impact the M2 reentrant pore loop, consistent with a changed Ca2+ permeability in receptors with the GluN1-G815R mutation compared with wild type. If the ion channel pore is opened by the M3 gate (with pre-M1 movements before this), but the GluN1 M4 is “constrained,” the channel only shows brief openings and low Ca2+ permeability. By contrast, when the M3 gate is open, and the GluN1 M4 helices are “expanded” (Supplemental Fig. 6), the receptor enters into long-lived open states with higher Ca2+ permeability (Amin et al., 2018). In addition to these glycine residues, de novo missense variants in the pre-M4 region (e.g., GluN2A-M817V and GluN2A-L812M) produce functional changes (Section X. Glutamate Receptors in Disease). Evaluation of macroscopic current time course, single-channel properties, and molecular dynamics simulations suggests that GluN2A-M817V weakens the interaction between the GluN2A M4 helix and M1 and M3 helices in GluN1, potentially increasing movements of the pre-M1 helix (Chen et al., 2017b; Amin et al., 2021b).
C. Mechanisms of Partial Agonism
Partial agonists bind to the ABD but are not fully efficacious at saturating concentrations. A variety of partial agonists have been identified for AMPA, kainate, and NMDA receptors (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers), and a great deal of information exists regarding the functional and structural distinctions between partial agonists at AMPA receptors, in contrast to the minimal mechanistic insight for kainate receptors. Partial agonism involves the total conformational landscape that the ABD explores in the functioning tetrameric receptor rather than a single structural feature, such as the degree of cleft closure (Section II. Receptor Structure). Partial agonists are less efficacious than full agonists because their molecular shape produces ABD conformations that allow the receptor to spend more time in “unproductive” conformations that do not permit channel gating and less time in “productive” conformations that do permit channel gating (Salazar et al., 2017) (Section II. Receptor Structure). How “productive” ABD cleft closures translate to qualitatively different channel activation for partial and full agonists is complex.
The activation mechanism of AMPA receptors, whereby the single-channel conductance is dependent on the fraction of the four subunits that have bound agonist, has revealed interesting actions for partial agonists. Single-channel currents recorded in saturating concentrations of partial agonist, ensuring occupancy of all four AMPA receptor agonist binding sites, show a correlation between the diminishing efficacy and a diminished ability to activate the largest conductance levels (Jin et al., 2003; Poon et al., 2010, 2011; Kristensen et al., 2011). The changing relative proportion of the same four conductance levels allows a quantitative assessment of subunit-dependent efficacy, and the coupling efficiency between occupancy of a single subunit within a tetrameric receptor and channel opening can be estimated. A purely allosteric model in which all conductance levels can be accessed by receptors with any number of agonists bound with increasing affinity of larger conductances has been proposed (Dutta-Roy et al., 2015). However, it is not clear how this can be reconciled with the observation of sequential opening of increasing conductance levels as individual antagonist molecules dissociate from a single receptor and are replaced by agonist (Rosenmund et al., 1998; Coombs et al., 2017) (Fig. 16).
The mechanism of partial agonism at NMDA receptors differs from AMPA receptors at the single-channel level. Some single-channel studies suggest that glycine and glutamate partial agonists can impact the subunit-specific conformational changes in GluN1 and GluN2 (Fig. 18), respectively, that lead to gating (Banke and Traynelis, 2003; Erreger et al., 2005b). For one glutamate partial agonist (homoquinolinate), this may reflect distinct changes in the binding pocket, which shows increased mobility when bound to homoquinolinate compared with glutamate (Erreger et al., 2005b). However, other single-channel studies show distributed effects across multiple gating steps (Kussius and Popescu, 2009) and propose a mechanism of glycine and glutamate partial agonists that is subunit-independent, suggesting partial agonists stabilize slightly different families of conformers regardless of which subunit they bind to (Kussius et al., 2010).
D. Molecular Determinants of Desensitization
Desensitization is defined as the decrement of a response in the continued presence of a stimulus, with recovery occurring after withdrawal of the stimulus (Katz and Thesleff, 1957). Thus, desensitization represents a process that is usually slower than activation and results in closing, as opposed to opening, of the ion-channel pore in response to a persistent activating stimulus. Desensitization usually proceeds as an exponential decline of the current response in the continuous presence of the activating stimulus and could serve as a safety mechanism to prevent prolonged currents through the channel or repetitive activation (Fig. 15). Among iGluRs, the current decline in the continuous presence of glutamate is faster (<20 milliseconds) and more complete (>90% current amplitude reduction) for AMPA and kainate receptors than for NMDA receptors.
1. Desensitization and the Agonist Binding Domain Dimer Interface
Molecular determinants of iGluR desensitization have been explored by combining structural biology and molecular dynamics simulations of isolated ABDs with single-molecule approaches, mutagenesis, and functional studies of full-length receptors. Desensitization is best understood for AMPA and kainate receptors, which undergo similar rapid onset of desensitization but recover at vastly different rates, with kainate being as much as an order of magnitude slower [Table 1; Supplemental Movie 2; reviewed in Traynelis et al. (2010)]. Rearrangement of the ABD dimer D1-D1 interface after agonist binding plays a crucial role in the onset of desensitization for AMPA and kainate receptors. Binding of positive allosteric modulators, such as cyclothiazide, aniracetam, and CX614 (Section IX. Exogenous Positive and Negative Allosteric Modulators) at the ABD interface of AMPA receptors stabilize the interface and result in the attenuation or block of desensitization (Partin et al., 1995; Sun et al., 2002; Jin et al., 2005). Similarly, single-point mutations or covalent crosslinks that strengthen the D1-D1 interface in AMPA (Stern-Bach et al., 1998; Sun et al., 2002) or kainate (Weston et al., 2006b; Nayeem et al., 2009) receptors reduce desensitization. Kainate receptor desensitization is dependent on ions that bind at and influence the strength of the D1-D1 interface (Section VIII.B.1. Modulation of AMPA, Kainate, and GluD Receptors by Extracellular Ions).
Conformational changes at the ABD interface have been examined by crosslinking the AMPA and kainate receptor ABD dimers (Armstrong et al., 2006; Daniels et al., 2013; Yelshanskaya et al., 2016a; Baranovic and Plested, 2018; Salazar et al., 2020). The individual ABD clamshells maintained their maximally closed conformation similar to the open state, whereas the D1 lobes underwent significant separation. It was therefore hypothesized that desensitization involves rupture of the D1-D1 interface, which allows the D2 lobes and the linkers to the ion channel to adopt a closed state–like conformation (Armstrong et al., 2006). Consistent with luminescence resonance energy transfer measurements in AMPA and NMDA receptors (Gonzalez et al., 2010; Rambhadran et al., 2010), such D1-D1 rupture was proposed to dissociate the ABD dimers into individual ABD monomers in kainate receptors (Schauder et al., 2013). Crosslinking of the D1 lobes in NMDA receptors influenced receptor activation but had no effect on desensitization (Borschel et al., 2011).
Decoupling of conformational changes in ABDs from resulting changes in TMDs (i.e., gating) and rupture of the D1-D1 interface were confirmed in the desensitized state structures of intact AMPA receptors (Chen et al., 2017a; Twomey et al., 2017b). These structures show that individual ABDs in the desensitized state adopt closed clamshell conformation nearly identical to the open state (Supplemental Fig. 7). In contrast, the ABD dimers undergo rearrangements accompanied by rupture of the D1-D1 interface, which allows D2 lobes to come closer together by ∼15 Å and permit channel closure (Chen et al., 2017a; Twomey et al., 2017a,b) (Fig. 4). As a result of the D1-D1 interface rupture, the ABD dimers lose their 2-fold rotational symmetry; the ABDs in subunits occupying the A/C position rotate 14° away from their B/D dimer partners resulting in a more 4-fold symmetrical ABD tetramer arrangement signified by appearance of a cleft between the protomers of the ABD dimers (Supplemental Fig. 7). In this new ABD tetramer arrangement, the D2 portions facing the membrane acquire positions similar to the closed state, in which the ABD gating ring is significantly compressed compared with the open state (Supplemental Fig. 7). As a result, the ABD-TMD linkers return to the positions of a closed, nonconducting ion channel. Although the ion channel pore adopts nearly identical conformations in the desensitized and closed states, the rest of the molecule undergoes significant rearrangement. Relative to the ion channel, this is expressed as an overall shortening of the receptor on an axis perpendicular to the plane of the membrane by 5 Å in the desensitized state compared with the closed state and an 18° rigid-body rotation of the entire NTD layer. Consistent with luminescence resonance energy transfer, single-molecule FRET, electrophysiological, and mutagenesis experiments (Shaikh et al., 2016; Yelshanskaya et al., 2016a), the NTD layer maintains its tetrameric arrangement (Leuschner and Hoch, 1999; Ayalon and Stern-Bach, 2001; Papadakis et al., 2004; Ayalon et al., 2005; Jin et al., 2009).
Desensitization accompanied by dissociation of ABD dimers into individual ABD monomers is consistent with structures of tetrameric GluK2 and GluK3 kainate receptors solved under conditions favoring the desensitized state (Meyerson et al., 2014, 2016; Kumari et al., 2019). The extent of decoupling in these structures is much greater than that in AMPA receptors, which is consistent with conformational heterogeneity observed for AMPA and kainate receptors using single-molecule FRET (Litwin et al., 2019, 2020). The lack of open and closed state structures for kainate receptors prevents a comparison of gating-associated structural rearrangements between kainate and AMPA receptors.
2. Types of NMDA Receptor Desensitization
Many different types of NMDA receptor desensitization have been characterized functionally, including glycine-dependent desensitization (Mayer et al., 1989; Benveniste et al., 1990; Lester et al., 1993; Nahum-Levy et al., 2001; Regalado et al., 2001; Durham et al., 2020), calcium-dependent inactivation (Clark et al., 1990; Legendre et al., 1993; Rosenmund and Westbrook, 1993; Vyklicky, 1993; Medina et al., 1995; Vissel et al., 2002; Iacobucci and Popescu, 2017b, 2019, 2020), Zn2+-dependent desensitization (Section VIII. Endogenous Allosteric Regulation), and glycine- and Ca2+-independent desensitization (Chen et al., 2004b; Hu and Zheng, 2005; Sessoms-Sikes et al., 2005), but the molecular basis underlying such a diversity in types of desensitization remains largely unknown.
Glycine-dependent NMDA receptor desensitization only occurs in subsaturating glycine concentrations (Mayer et al., 1989) and is the result of a negative allosteric interaction between subunits such that binding of glutamate decreases glycine affinity and vice versa (Benveniste et al., 1990; Lester et al., 1993; Durham et al., 2020). When glutamate binds to the GluN2 ABD in the absence of near-saturating concentrations of glycine, the current will relax to a new level as glycine unbinds from the GluN1 subunit because of the allosteric reduction in glycine affinity. The glycine unbinding rate determines the time course for this form of desensitization, which could impact synaptic signaling. Recent studies suggest that the interactions between the heterodimer of the glycine binding GluN1 ABD and glutamate binding GluN2 ABD are involved in this negative cooperativity (Durham et al., 2020).
The mechanism of Zn2+-dependent desensitization involves a positive allosteric coupling between glutamate binding to the GluN2 ABD and Zn2+ binding to the GluN2A NTD, and this causes the receptor currents to decay to a new steady state with higher occupancy of the NTD by ambient Zn2+ after glutamate binding (i.e., since Zn2+ inhibits GluN2A-containing NMDA receptors) (Section VIII. Endogenous Allosteric Regulation).
NMDA receptors also desensitize during sustained increases in intracellular Ca2+ for several seconds (Clark et al., 1990; Legendre et al., 1993; Vyklicky, 1993; Rosenmund et al., 1995). This form of desensitization, also referred to as Ca2+-dependent inactivation, is most prominent for GluN2A-containing receptors and is more limited for GluN2B- and GluN2C-containing NMDA receptors (Medina et al., 1995; Krupp et al., 1996), apparently because of their lower open probabilities (Iacobucci and Popescu, 2020). An increase in the intracellular Ca2+ in the vicinity of the NMDA receptor is proposed to trigger uncoupling of the receptor from filamentous actin (Rosenmund and Westbrook, 1993), which reduces open probability (Rycroft and Gibb, 2004). In addition, calmodulin binding to the GluN1 CTD in some GluN1 isoforms can inhibit NMDA receptor function through a reduction in open time and open probability during receptor activations (Ehlers et al., 1996, 1998; Zhang et al., 1998; Krupp et al., 1999; Rycroft and Gibb, 2002). This can occur when calmodulin is close to the channel and senses Ca2+ arriving through the pore of the channel (Iacobucci and Popescu, 2017b). Moreover, Ca2+ entering one NMDA receptor can desensitize an adjacent receptor, providing a mechanism for negative coupling, and this effect is enhanced by PSD-95 clustering (Iacobucci and Popescu, 2019).
Most ligand-gated channels can desensitize in the continued presence of agonist by a mechanism thought to involve a conformational change to a stable and long-lived agonist-bound closed state, as described above for AMPA and kainate receptors. NMDA receptors can also desensitize in the continued presence of glutamate and glycine in a manner that is independent of glycine, Zn2+, and Ca2+, and this desensitization is sensitive to intracellular dialysis and perturbed by mutations throughout the receptor (Sather et al., 1990, 1992; Chen et al., 2004b; Hu and Zheng, 2005; Alsaloum et al., 2016).
E. Molecular Determinants of Ion Permeation
The ion-conducting pore of glutamate receptors is cation-selective and equally permeable to both K+ and Na+ ions (Chang et al., 1994; Jatzke et al., 2002). Under certain conditions, the RNA-edited R-forms of homomeric GluA2 and GluK2 are also anion-permeable (Burnashev et al., 1996), although the physiologic significance of this is unknown. At the resting membrane potential, opening of iGluRs is excitatory because of the strong electrochemical driving force for Na+ influx, leading to membrane depolarization that promotes neuronal firing (i.e., membrane potential approaches 0 mV). Many forms of glutamate receptor subtypes are permeable not only to monovalent cations but also to Ca2+ ions. Although the charge associated with Ca2+ may contribute to membrane depolarization, extracellular Ca2+ concentration is in the low mM range, and thus Ca2+ ions predominantly mediate intracellular signaling. Indeed, Ca2+ permeation and signaling often dominate how we think of NMDA receptor function as it contributes to their role in synaptic dynamics, neuronal development, and cellular pathology (Hunt and Castillo, 2012; Paoletti et al., 2013; Henley and Wilkinson, 2016; Chakraborty et al., 2017).
1. Permeation Pathway
The permeation pathway is largely formed by the M3 transmembrane segment and the re-entrant M2 pore loop (Fig. 20) with a topology common across all iGluR subtypes (Section II. Receptor Structure), although there are many differences in detail between the subtypes that are important to their role in physiology. Within the M2 loop resides a key site for ion permeation, the Q/R/N site (the Q/R site for non-NMDA receptors and the N site for NMDA receptors) (Hollmann et al., 1991; Hume et al., 1991; Burnashev et al., 1992a,b), although the GluN3 subunits have a Gly residue at this position. The Q/R/N site is located near the tip of the M2 loop and contributes to a narrow constriction within the pore (Fig. 20). The open conducting pore has a widened middle portion, the central cavity, which is located right above the Q/R/N site narrow constriction (Fig. 20). The central cavity has a hydrophobic character, but the entire open pore has an overall negative surface charge (Fig. 20), which is probably critical for cation selectivity of iGluRs. In particular, the intracellular portion of the pore that is lined by regions of M2 is negatively charged and has therefore been characterized as a “selectivity filter.” The presence of the Q/R/N site Arg residues in the open pore of heteromeric AMPA or kainate receptors that contain edited GluA2, GluK1, or GluK2 subunits introduces a positive charge at the narrow constriction of the pore, which reduces Ca2+ permeability and polyamine block in these channels (Section VIII. Endogenous Allosteric Regulation).
Fig. 20.
Structure of the ion permeation pathway. (A) Open-state structure of a homomeric GluA2 AMPA receptor (PDB: 5WEO). The ion conduction pathway is shown in gray, the pore-forming re-entrant M2 loop is in yellow, and the M3 gating helix is in green. Only two (B and D) out of four subunits are shown; the other two subunits (A and C) are removed for clarity. The blue mesh illustrates cryo-EM density. Residues in the SYTANLAAF motif, including the gating hinge Ala618 (blue) and the Q/R/N site glutamine (red), are labeled. (B) Electrostatic surface potential of the open pore with blue indicating a positive charge, red negative charge, and white neutral. (C) The re-entrant M2 pore loop for a GluN1/2 NMDA receptor is shown as a ribbon structure with the Q/R/N site Asn red, and a downstream adjacent Asn in GluN2 is green (indicated as N+1). (D) Summary of the Q/R/N site identity for iGluR subunits and qualitative assessment of Ca2+ permeability. Supplemental Table 2 summarizes measured Ca2+ permeabilities. KAR, kainate receptor. Panels C and D adapted with permission from Wollmuth (2018).
2. Structure of the Narrow Constriction in NMDA Receptors
In NMDA receptors, the narrow constriction is formed asymmetrically by different Asn residues; the Asn at the GluN1 Q/R/N site, referred to as the N site; and the Asn residue adjacent to the GluN2 Q/R/N site, referred to as the N+1 site (Fig. 20) (Wollmuth et al., 1996; Sobolevsky et al., 2002b; Song et al., 2018). Key features of GluN1/2 NMDA receptors are high Ca2+ permeability and strong voltage-dependent block of the pore by extracellular Mg2+ (Section VIII. Endogenous Allosteric Regulation). It remains unclear how the pore of NMDA receptors can distinguish between Ca2+, which is permeable, and Mg2+, which is largely impermeable (Mayer and Westbrook, 1987a; Ascher and Nowak, 1988; Mesbahi-Vasey et al., 2017). Determinants of both processes, Ca2+ permeation and Mg2+ block, are present in the M2 loop, but the asymmetry between subunits forming the narrow constriction may underlie this difference in permeation between Ca2+ and Mg2+. Consistent with this idea is the observation that the N site Asn residues from different subunits have distinct roles in Ca2+ permeation and Mg2+ block. Replacement of the GluN1 N site Asn with Gln strongly alters Ca2+ permeability while having only weak effects on Mg2+ block, whereas the same substitution at the GluN2 N site strongly alters Mg2+ block and only has weak effects on Ca2+ permeation [Burnashev et al. (1992b); reviewed by Traynelis et al. (2010)]. On the other hand, this substitution at the N+1 site in GluN2 strongly reduces Mg2+ block, in part by permitting greater Mg2+ permeation (Wollmuth et al., 1998).
3. Structural Determinants of Ca2+ Permeability in NMDA Receptors
GluN1/2 receptors have ∼3–4-fold higher Ca2+ permeability measured relative to monovalent ions (PCa/Pmonovalent) than non-NMDA receptors (Fig. 20) (Wollmuth, 2018). Diheteromeric GluN1/2A and GluN1/2B receptors show a higher Ca2+ permeation (PCa/Pmonovalent ∼4) than GluN1/2C and GluN1/2D receptors (PCa/Pmonovalent ∼2.5) (Supplemental Table 2). This difference is largely due to a residue at the S/L-site located in the M3 transmembrane helix (Siegler Retchless et al., 2012; Glasgow et al., 2015). The residue at the S/L-site, which is Ser in GluN2A/B and Leu in GluN2C/D, does not line the ion channel pore and does not interact directly with permeating ions. Instead, this residue may affect the positioning of Trp residues in the M2 loop of GluN1 (Siegler Retchless et al., 2012). This interaction between GluN1 and the GluN2 S/L-site also appears to be a key determinant of GluN2-specific variation in channel conductance and Mg2+ block (Siegler Retchless et al., 2012). Unlike GluN1/2 receptors, the diheteromeric GluN1/3 receptors have relatively low Ca2+ permeability and are less sensitive to Mg2+ block [Chatterton et al. (2002), Smothers and Woodward (2007), Madry et al. (2008), Pina-Crespo et al. (2010), Otsu et al. (2019); see also Yi et al. (2018)]. The reduced Ca2+ permeation of GluN3-containing NMDA receptors presumably reflects the residues at the N site and N+1 sites in GluN3 subunits that have Gly-Arg versus Asn-Asn in GluN2 subunits (Yi et al., 2018) (Fig. 20).
The mechanism of ion permeation in NMDA receptors follows Goldman-Hodgkin-Katz (GHK) assumptions at physiologic Ca2+ (Schneggenburger, 1998; Jatzke et al., 2002), suggesting independence of ion movements but deviates at lower and higher concentrations (Jatzke et al., 2002) and shows ion flux coupling (Wollmuth and Sakmann, 1998), which is a hallmark of ion-ion interactions. NMDA receptors also exhibit block by external Ca2+ ions, which appears as a reduction in the single-channel conductance (Premkumar and Auerbach, 1996; Wyllie et al., 1996; Maki and Popescu, 2014). High Ca2+ permeability and concurrent block by Ca2+ are not incompatible properties but may reflect multiple Ca2+ binding sites in the ion channel pore of NMDA receptors (Premkumar and Auerbach, 1996; Sharma and Stevens, 1996). One Ca2+ binding site is most likely at the narrow constriction, which is contributed at least in part by the N site in GluN1, whereas another Ca2+ binding site is formed by a cluster of charged residues in GluN1, the DRPEER motif, located at the external entrance to the ion channel (Karakas and Furukawa, 2014; Watanabe et al., 2002). The mechanism of Ca2+ permeation in Ca2+-permeable AMPA receptors is distinct to that in NMDA receptors (Wollmuth and Sakmann, 1998; Jatzke et al., 2002), which reflects the absence of a DRPEER-like motif in AMPA receptors (Jatzke et al., 2003).
4. Ca2+ Permeability in AMPA and Kainate Receptors
In non-NMDA receptors, the Gln (Q) codon at the Q/R/N site in GluA2, GluK1, and GluK2 subunits is edited at the mRNA level by adenosine deaminase to encode the positively charged Arg (R), which renders the channel Ca2+ impermeable and insensitive to intracellular polyamines [reviewed in Bass (2002) and Filippini et al. (2017)] (Figs. 7 and 20). Because of the dominance of the Q/R/N site in terms of pore properties (single-channel current, channel block, and Ca2+ permeation), AMPA receptors are categorized into GluA2-containing (Ca2+-impermeable, insensitive to polyamines) and GluA2-lacking (Ca2+-permeable AMPA receptors, blocked by polyamines) AMPA receptors (Section VIII. Endogenous Allosteric Regulation). The Q/R/N site of the GluA2 subunit is >99% edited in vivo, and this modification is crucial for survival (Higuchi et al., 2000). All other AMPA receptor subunits have a Gln at the analogous position in the pore loop, as they lack the pre-mRNA intronic sequence elements required for editing (Sommer et al., 1991; Bass, 2002; Penn et al., 2013). This Q/R/N site editing is dynamically regulated in the disease state (Wang et al., 2014a; Cueva Vargas et al., 2015; Li et al., 2015; Schmidt et al., 2015; Yamashita and Kwak, 2019).
Although mRNAs for GluK1-K2 kainate receptor subunits are also edited at the codon encoding the Q/R/N site, this editing is less complete in vivo than for GluA2 (Herb et al., 1996; Contractor et al., 2011). The Q/R/N site is edited only 40% in GluK1 with estimates for GluK2 editing ranging between <5% and 75% in the developing and adult rodent brain (Sommer et al., 1991; Puchalski et al., 1994; Schmitt et al., 1996).
5. Access of Ions to the Central Permeation Pathway
It is unclear how the extracellular permeant ions Na+ and Ca2+ access the ion-permeation pathway formed by the M3 transmembrane helix. The ABD is positioned on the extracellular end of the ion channel and is connected to the ion channel–forming TMD by the ABD-TMD linkers. These linkers are highly dynamic during pore opening and contain numerous charged side chains (Schmid et al., 2007; Talukder et al., 2010; Yelshanskaya et al., 2017). In addition, with pore opening, the S2-M4 linkers unwind and are positioned in close proximity to the central axis of the permeation pathway (Twomey et al., 2017a). Hence, the space near the ABD-TMD linkers could act as pathways or portals for permeant ions to access the central permeation pathway. How permeant ions might cross these portals and their impact on selectivity and single-channel conductance levels are unknown. These linkers are sites for modulation of receptor function (Section IX. Exogenous Positive and Negative Allosteric Modulators), some of which bind to linkers and alter ion-permeation properties and conductance (Perszyk et al., 2020b; Yelshanskaya et al., 2020). In addition, auxiliary subunits modulate Ca2+ permeability in AMPA receptors (Supplemental Table 2).
V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity
A. Receptor Assembly and Trafficking
For NMDA receptors and for kainate receptors containing GluK4 and GluK5, heteromerization is obligatory for functional expression at synapses and thus an important aspect of assembly. In addition, AMPA receptors with differing subunit compositions are selectively recruited during development in several forms of synaptic plasticity and under certain pathologic conditions, including epilepsy, addiction, and after ischemia (Cull-Candy et al., 2006; Clem and Huganir, 2010; Mameli et al., 2011; Purkey and Dell'Acqua, 2020), emphasizing the importance of different assembly routes. As is the case for all transmembrane (and secreted) proteins, iGluR biogenesis takes place in the ER and is tightly regulated during folding, assembly, association with biogenesis machinery, recruitment into coat protein complex II (COPII) transport vesicles, and ER export. Subunit folding likely occurs sequentially from the N terminus to the C terminus, aided by a set of general ER chaperones [reviewed in Greger et al. (2007)]. Each of four receptor domains (i.e., NTD, ABD, TMD, and CTD) contribute at different stages of biogenesis. Auxiliary subunits further expand this process for AMPA receptors [reviewed in Greger et al. (2007), Jackson and Nicoll (2011a), Haering et al. (2014)], which often coexist with various types of accessory proteins (Schwenk et al., 2012; Shanks et al., 2012; Jacobi and von Engelhardt, 2018) and for which dedicated ER assembly machinery has been identified (Schwenk et al., 2019). This scenario is also true for kainate receptors and their association with Neto1 and Neto2 auxiliary subunits (Copits and Swanson, 2012). At present, the most detailed information on receptor assembly and forward trafficking is available for AMPA receptors. In addition, although the different subtypes may share common features of assembly, they also exhibit critical differences.
1. Intrinsic Elements Regulating Receptor Assembly
The NTD has long been implicated in assembly (Ayalon and Stern-Bach, 2001) and is inserted into the ER membrane first, where it is subject to signal peptide cleavage and N-glycosylation and folds cotranslationally into a globule of ∼50 kDa. Once folded, the NTD forms dimers with either strong interface contacts (i.e., affinity in the nM range) for AMPA and kainate receptors (Rossmann et al., 2011; Zhao et al., 2017) or weaker (micromolar) contacts within NMDA receptors (Karakas et al., 2011). The current view for NMDA receptors is that the GluN1 NTD initially forms homodimers, which subsequently dissociate and reassemble as NTD heterodimers of GluN1 and GluN2 subunits to form mature heterotetrameric receptors (Atlason et al., 2007; Farina et al., 2011).
The ABDs of all iGluRs assemble into dimers of low affinity (Sun et al., 2002; Furukawa et al., 2005; Chaudhry et al., 2009a,b; Weston et al., 2006b). Although the NTDs drive subunit dimerization, the ABDs are involved in the dimerization of dimers step (i.e., tetramerization) (Fig. 21). That is, the ABDs dimerize during tetramer formation and thereby contribute to the domain swap that occurs between the NTD and ABD layers. In a GluA1/2 receptor, for example, the ABD of one GluA1 subunit dimerizes with the ABD of one GluA2 subunit, but its NTD dimerizes with the NTD of the other GluA2 subunit. The role of ABD dimers in tetramerization is based on a study comparing wild-type GluA2 dimers to those of the nondesensitizing GluA2-L483Y mutant that stabilizes ABD dimers (Sun et al., 2002; Shanks et al., 2010). In wild-type dimers, the NTDs and TMD are closely apposed, but the ABDs are separated, whereas the nondesensitizing mutation resulted in close apposition of the ABDs and an inability to tetramerize and leave the ER (Shanks et al., 2010). In AMPA receptors, the ABD is modulated by alternative RNA splicing to produce the flip/flop isoforms and by RNA editing (Sprengel and Seeburg, 1993), which shape biogenesis and the rate of ER export (Coleman et al., 2006; Greger et al., 2006; Penn et al., 2012). As part of a quality-control mechanism in the ER, it has been suggested that glutamate binds to AMPA and kainate receptor ABDs (Mah et al., 2005; Valluru et al., 2005; Penn et al., 2008), GluN2 ABDs (She et al., 2012), or that glycine binds the GluN1 and GluN3 ABDs (Kenny et al., 2009; Skrenkova et al., 2019, 2020) to promote forward trafficking.
Fig. 21.
AMPA receptor biogenesis in the ER. (A) The intrinsic assembly of AMPA receptor dimers occurs largely through interactions between their NTDs, whereas tetramerization (i.e., the dimerization of dimers) involves mostly TMDs and ABDs. These steps are aided by chaperones and assembly factors. (B) Extrinsic regulators based on the proposed “assembly line” model (Schwenk et al., 2019) are shown: (1) AMPA receptor monomers are associated with ABDH6, (2) dimerization is induced by coassociation with preassembled FRRS1L/CPT1c complexes, and (3) tetramerization is followed by (4) dissociation of ABDH6 and association of auxiliary subunits (TARP and/or CNIH). Dissociation of FRRS1L/CPT1c renders the AMPA receptor/auxiliary subunit complex competent for ER export via COPII vesicular carriers.
At the level of the TMD, the Q/R/N site at the apex of the M2 pore loop impedes homotetramer formation by RNA-edited GluA2 subunits (Greger et al., 2002, 2003). As a result, edited GluA2 has a greater ER dwell time than other AMPA receptor subunits, which may encourage incorporation of GluA2 into the heteromeric AMPA receptors that prevail in the brain. Similar observations have been made for kainate receptors, in which the GluK1 and GluK2 subunits undergo RNA editing, although to a lesser extent than does GluA2 (Ball et al., 2010; Evans et al., 2017). The M4 transmembrane helix also contributes to assembly (Section II. Receptor Structure). Deleting or mutating M4 in AMPA receptors impairs the transition from dimers to tetramers but does not impact dimerization (Salussolia et al., 2011a, 2013; Gan et al., 2015). The wrapping of M4 helices, in which the M4 helices from each subunit associate with the ion channel core (M1-M3) of the adjacent subunit (Fig. 5), may help overcome energy barriers and facilitate tetramer formation (Gan et al., 2016). The M4 helices in NMDA receptors appear to play a less prominent role in receptor assembly (Amin et al., 2017).
2. Regulators of AMPA Receptor Assembly
A number of accessory proteins associate with AMPA receptors in ER membranes and are not enriched at synapses, including ferric chelate reductase 1-like (Frrs1l), Cpt1c (carnitine palmitoyl transferase 1C), α/β-hydrolase domain–containing protein 6 (ABHD6), and PORCN (porcupine) (Brechet et al., 2017; Schwenk et al., 2019). Cpt1c knockdown reduces surface expression of GluA1, presumably because of the palmitoyl esterase activity of Cpt1c (Gratacos-Batlle et al., 2018). Palmitoylation by the palmitoyl transferase GODZ regulates forward trafficking of AMPA receptors, but its relation to the role of Cpt1c remains unclear (Hayashi et al., 2005). Mass spectrometry of AMPA receptors isolated from brain combined with native gel electrophoresis revealed complexes of different sizes corresponding to AMPA receptor assembly intermediates associated with these ER-based proteins. When reconstituted in heterologous cells, ABDH6 was predominantly associated with AMPA receptor subunit monomers in a complex that is also partly assembled with PORCN (Schwenk et al., 2019) (Fig. 21). In a subsequent step, ABDH6 is replaced by a complex consisting of Frrs1l and Cpt1c, facilitating AMPA receptor dimerization. Lastly, ER-export competence is achieved through TARPs and CNIH proteins (CNIH2 or CNIH3) joining the complex. Addition of these AMPA receptor auxiliary proteins (Section III. Auxiliary Subunits) results in dissociation of Frrs1l and Cpt1c, rendering the receptor ER export competent.
CNIH proteins are closely related to ER cargo export proteins originally identified in yeast and flies (Section III. Auxiliary Subunits). Similarly, CNIH proteins engage the COPII machinery that facilitates budding from the ER (Powers and Barlowe, 2002). This engagement involves the intracellular loops of CNIH proteins (Nakagawa, 2019). Related to these findings, knockdown of CNIH2, CNIH3, TARP γ-8, and Frrs1l reduces surface AMPA receptor protein and EPSC amplitudes (Rouach et al., 2005; Herring et al., 2013; Schwenk et al., 2019). Similarly, overexpression of ABHD6 reduced surface expression of AMPA receptors (Wei et al., 2017; Schwenk et al., 2019). Understanding the mechanics of this biogenesis pathway will require structural studies to reveal: i) the interplay between ABDH6, which stabilizes monomers, and the NTD, which drives dimerization via a stable dimer interface that forms once folding is complete (Rossmann et al., 2011; Schwenk et al., 2019); ii) the stoichiometry and architecture of the GluA-Frrs1l-Cptc1 complex; and iii) its interplay with TARPs and CNIH proteins prior to ER exit. Recent studies indicate that GluA1-containing receptors exiting the somatic ER follow the canonical secretory pathway passing through the somatic Golgi apparatus en route to the plasma membrane (Bowen et al., 2017). By contrast, GluA1-containing AMPA receptors exiting the ER in dendrites, which mostly lack the Golgi apparatus (Hanus and Ehlers, 2008; Hanus et al., 2016), instead enter a noncanonical secretory pathway passing first through the ER-Golgi intermediate compartment and then recycling endosomes on their way to the cell surface (Bowen et al., 2017).
3. Subunit Arrangement in the Tetrameric Assembly
The factors that determine subunit placement within a tetramer at the two conformationally different subunit pairs (A/C and B/D positions, Section II. Receptor Structure) are unknown. In NMDA receptors, GluN1 subunits occupy the A/C positions and GluN2 subunits reside in the B/D positions (Salussolia et al., 2011b; Karakas and Furukawa, 2014; Lee et al., 2014) but reorganize around the central axis of the channel if GluN1 and GluN2 NTDs are removed (Song et al., 2018), suggesting that the NTD is somehow involved in subunit placement. Structures of tetrameric AMPA receptors documented that the functionally critical GluA2 subunit occupies the B/D positions (Herguedas et al., 2019; Zhao et al., 2019), a finding that is consistent with functional data (He et al., 2016; Herguedas et al., 2019), although GluA2 at the A/C positions also forms functional receptors (Herguedas et al., 2016). Structures of isolated NTD heterodimers from heteromeric kainate receptors suggest that the GluK2 subunits occupy B/D positions and GluK5 the A/C positions (Kumar et al., 2011).
B. Regulation by Secreted Proteins
The NTDs of iGluRs project into the crowded environment of the synaptic cleft to face presynaptic glutamate release sites (Garcia-Nafria et al., 2016; Biederer et al., 2017). NTD associations with cleft components can cluster and organize iGluRs in synapses, and the formation of receptor clusters that increase the local receptor concentration has been observed by super-resolution microscopy for both AMPA and NMDA receptors (Savtchenko and Rusakov, 2013) (Section V.F. Mechanisms of Synaptic Plasticity). AMPA receptor nanoclusters locate directly beneath presynaptic glutamate release sites in synaptic “nanocolumns” (Tang et al., 2016), which enables full activation by high local agonist concentration. Hence, reorganization of existing synaptic AMPA receptors may be involved in plasticity (Biederer et al., 2017; Buonarati et al., 2019) (Section V.F. Mechanisms of Synaptic Plasticity). At CA1 synapses, deletion of the GluA2 NTD reduces EPSCs and increases receptor mobility in spines (Watson et al., 2017), whereas GluA1 NTD deletion reduces receptor expression at the synapse and inhibits the expression of LTP (Diaz-Alonso et al., 2017; Watson et al., 2017).
Clustering molecules targeting the NTD have been described for all four iGluR classes, with a prominent example being the cerebellins, a family of four secreted proteins (Cbln1-4) that bridge between presynaptic neurexin (Nrxn) isoforms and postsynaptic GluD receptors (Section V.E. Neuronal Functions of GluD Receptors). Structural data for this synaptic organizer complex illustrate how the globular (C-terminal) domain of Cbln1 interacts with the GluD2 NTD, whereas the N-terminal Cbln1 cysteine-rich domain associates specifically with Nrxn proteins that contain the splice site 4 insert (+SS4) (Elegheert et al., 2016). Based on these data, it has been proposed that two Cbln1 hexamers associate with a GluD2 receptor tetramer with nM binding affinity (Elegheert et al., 2016). Moreover, this interaction is not restricted to the cerebellum, since a related complex consisting of Nrxn(+SS4)-Cbln2-GluD1 is important for synapse formation and maintenance in the hippocampus (Tao et al., 2018), whereas Nrxn-Cbln4-GluD1 organizes inhibitory synapses in cortical pyramidal neurons (Fossati et al., 2019) (Section V.E. Neuronal Functions of GluD Receptors).
The best-described AMPA receptor clustering proteins are the neuronal pentraxin family of calcium-dependent lectins, of which two are secreted (Nptx1 and Nptx2), and one is transmembrane (Nptx receptor; NptxR) (Dodds et al., 1997; O'Brien et al., 1999, 2002; Cho et al., 2008; Lee et al., 2017b). Neuronal pentraxins localize to excitatory synapses, where their conserved C-terminal pentraxin domains directly interact with the NTD of AMPA receptors (Sia et al., 2007). Nptxs can assemble as homo- or hetero-hexameric complexes that interact through their globular pentraxin domain with the NTD of all four AMPA receptor subunits (Xu et al., 2003; Sia et al., 2007). Of these, Nptx2, which is also known as neuronal activity–regulated pentraxin (Narp), is regulated as an immediate early gene (IEG) (Xu et al., 2003) that is upregulated by neuronal activity (Tsui et al., 1996; O'Brien et al., 1999). Nptx2 is enriched at excitatory synapses in parvalbumin-positive interneurons (Chang et al., 2010) to promote activity-dependent accumulation of GluA4-containing AMPA receptors (Pelkey et al., 2015), which dictates interneuron activity to maintain circuit inhibitory/excitatory balance (Pelkey et al., 2017). During early postnatal development, increased Nptx2 expression coincides with GluA4 upregulation in parvalbumin-containing fast-spiking interneurons (Pelkey et al., 2017). Nptx1 and NptxR in axonal compartments are also important for recruitment of AMPA receptors to synapses (Sia et al., 2007). Although presynaptic interaction partners of Nptxs are not known, a recent study showed that synthetic fusion proteins of the Nptx1 pentraxin domain with the Nrxn-interacting region of Cbln1, termed CPTX, are capable of clustering AMPA receptors (Suzuki et al., 2020). The synthetic CPTX protein restored synaptic function in mutant mouse models for cerebellar ataxia, spinal cord injury, and Alzheimer’s disease, emphasizing the importance of receptor clustering at the synapse (Suzuki et al., 2020). Proteomic screens have unveiled other secreted proteins interacting with AMPA receptors, including noelin, brorin, and neuritin (Schwenk et al., 2012; Shanks et al., 2012), but their roles are currently unclear.
The secreted C1q-like proteins 2 and 3 (C1ql2 and C1ql3) bind to the GluK2 and GluK4 NTDs in vitro and are implicated in kainate receptor targeting at the hippocampal mossy fiber to CA3 region (Matsuda et al., 2016). The globular domain in C1ql binds to the GluK2 and GluK4 NTDs, whereas the C1ql N-terminal region interacts with a presynaptic Nrx3 isoform (Nrx3 +SS525b) and thereby recruits postsynaptic kainate receptors (Matsuda et al., 2016). NMDA receptor NTDs appear to interact with receptor tyrosine phosphatase EphB2 (Dalva et al., 2000; Washburn et al., 2020), although this is not a secreted protein. In C. elegans, however, a presynaptically secreted protein, NRAP-1, clusters NMDA receptors and shapes their function (Lei et al., 2017).
C. Roles of AMPA and Kainate Receptors in Neuronal Functions
The iGluRs play central roles in cellular and circuit development, synaptic transmission, plasticity, and signaling (see Fig. 22 for overview). For some features of iGluRs in neuronal functions, there are relatively few documented examples, and our knowledge is lacking, whereas other topics (e.g., synaptic plasticity) represent an enormous body of work with exhaustive data. Virtually all of the reported functional data are from mouse or rat neurons, and thus it is important to bear in mind there may be differences across species.
Fig. 22.
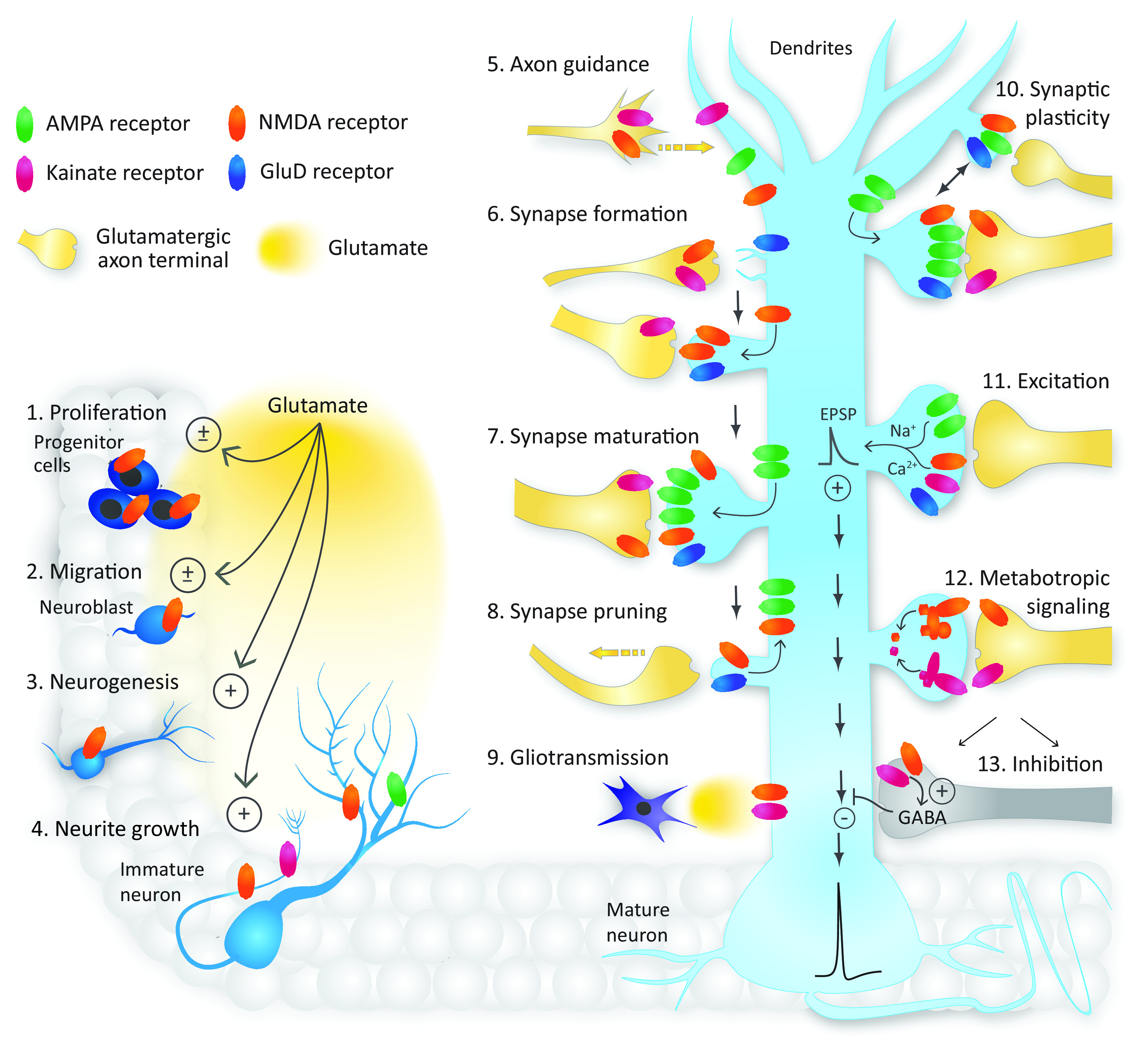
Roles of glutamate receptors in the CNS. The different iGluRs control diverse neuronal function beginning in embryonic development (left) and continuing in the mature neuron (right). NMDA receptors contribute to neuron development by regulating (1) neural progenitor cell proliferation, (2) migration, and (3) neurogenesis. NMDA receptor activation can either inhibit or enhance proliferation and migration. (4) NMDA and AMPA receptors play key roles in dendrite development, whereas NMDA and kainate receptors regulate axon growth and (5) axon guidance toward appropriate synaptic targets. (6) Synapse formation is regulated by postsynaptic GluD and NMDA receptors as well as presynaptic NMDA and kainate receptors, with kainate receptors playing a role in axon terminal maturation. (7) Synapse maturation involves the recruitment of AMPA receptors as well as continued activity of NMDA, kainate, and/or GluD receptors, whereas (8) synapse pruning involves NMDA and GluD receptors and the removal of AMPA receptors. (9) Extrasynaptic NMDA and kainate receptors regulate neuronal excitability in response to glutamate release from astrocytes. (10) Postsynaptic AMPA, NMDA, and GluD receptors have roles in short- and long-term synaptic plasticity, and NMDA and kainate receptors are implicated in presynaptic mechanisms underlying plasticity. (11) All iGluR subtypes (except GluD receptors) mediate depolarizing EPSPs that increase neuronal excitability and promote action-potential generation. (12) NMDA and kainate receptors have metabotropic signaling capabilities. (13) Presynaptic NMDA and kainate receptors are expressed at GABAergic synapses where they enhance inhibitory synaptic transmission.
1. AMPA Receptors and Development
The involvement of a particular gene in development is typically inferred by its early expression in cells that give rise to the organ of interest, often at an undifferentiated stage. In the case of neurons, that might also include a stage at which synaptic connectivity is absent or just beginning to be established. AMPA receptors are expressed in a number of systems during embryogenesis particularly during proliferative migratory stages (Mi et al., 2018). Thus, these receptors are assumed to play important roles in nervous system development (Metin et al., 2000), such as guiding or accelerating cell migration and controlling neurite extension (e.g., (Poluch et al., 2001; Poluch and Konig, 2002; Harlow et al., 2015). Indeed, immature and migrating cells express AMPA receptors as well as kainate and NMDA receptors prior to the expression of any functional synapses (Soria and Valdeolmillos, 2002; Lujan et al., 2005; Manent et al., 2006). The timing of their expression coincides with critical developmental windows in the CNS (Ambrogini et al., 2006; Huang et al., 2012; Xu et al., 2020) (Fig. 23), which also contains higher ambient glutamate levels than the adult brain (Hanson et al., 2019). Collectively, higher ambient glutamate levels, the developmental expression of iGluRs, and the recruitment of glutamatergic signaling regulate neuronal development (Kelsch et al., 2014; Akgul and McBain, 2016, 2020; Akgul et al., 2019), including the morphologic and electrical maturation of neurons (Manent et al., 2006; Yozu et al., 2008; Bortone and Polleux, 2009; De Marco Garcia et al., 2011; Le Magueresse and Monyer, 2013; Garcia et al., 2015; Chittajallu et al., 2017; De Marco Hanson et al., 2019).
Fig. 23.
Developmental roles of iGluRs in the CNS. The timeline shows receptors that play a developmental role in multiple early processes (left), as well as roles in adult neuronal function (right). The developmental expression patterns as well as subcellular localization of iGluR subunits result in subunit-specific functional roles through neuronal development. Only GluN1 and GluN2B are known to play roles in each developmental stage and across all subcellular compartments from neural progenitor cell proliferation through mature neuron function. Specific kainate receptors regulate axon and dendrite growth along with GluN1 and GluN2B, whereas GluN2D and specific AMPA receptor subunits regulate dendrite growth. Most subunits have established roles in synapse formation and/or maturation except for GluN2C and GluN2D. All receptor subtypes are expressed in the postsynaptic compartment and most contribute to synaptic plasticity. NMDA and kainate receptors are prominent in presynaptic and extrasynaptic compartments and can signal through noncanonical metabotropic pathways.
Studies with AMPA receptor KO mice have implicated early embryonic or postnatal expression of AMPA receptor subunits in key neurodevelopmental processes, including activity-dependent maturation of spinal cord motoneurons for GluA1 (Zhang et al., 2008a), dendritic spine complexity for GluA2 (Medvedev et al., 2008), sleep architecture for GluA3 (Steenland et al., 2008), and network excitability in cortical-thalamic circuits underlying models of absence epilepsy for GluA4 (Paz et al., 2011). Ablation or alteration of iGluRs in cortical or hippocampal pyramidal neurons during embryogenesis or in early development has consequences at both anatomic and physiologic levels that impact emerging nascent cortical microcircuits (Tashiro et al., 2006; Adesnik et al., 2008; Gray et al., 2011; Lu et al., 2011; Zhang et al., 2013). In hippocampal inhibitory interneurons, elimination of GluA1, GluA2, or GluA3 AMPA receptor subunits either during embryogenesis or postnatally has complex effects on morphologic, synaptic, and circuit features that can manifest as neural circuit disorders later in life (Belforte et al., 2010; Chittajallu et al., 2017; Akgul et al., 2019; Akgul and McBain, 2020). Embryonic elimination of NMDA receptor subunits can also impact AMPA receptor expression and synapse development and hinder the circuit integration of both hippocampal and neocortical neurons (Akgul and McBain, 2016; Chittajallu et al., 2017). As the brain develops, AMPA receptors continue to play key roles in establishing the correct connectivity. For example, GluA3 promotes the insertion of postsynaptic AMPA receptors at synapses between auditory nerve fibers and spherical bushy cells of the cochlear nucleus, which contain fast desensitizing flop isoforms of AMPA receptor subunits (Antunes et al., 2020). In addition, data from GluA3 KO mice suggest that GluA3 is required for the normal development of the presynaptic terminal (Antunes et al., 2020). GluA3- and GluA4-containing AMPA receptors also play a central role in maturation of synapses, such as the whisker-relay synapse in the thalamus (Wang et al., 2011b).
2. AMPA Receptor Function in Presynaptic Signaling
AMPA receptors are present in the presynaptic compartments of neurons in hippocampus, striatum, and spinal cord (Lu et al., 2002; Schenk et al., 2003; Fujiyama et al., 2004; Fiszman et al., 2007; Haglerod et al., 2017) and can potentially influence synaptic function by multiple means, including as autoreceptors that respond to glutamate in the synaptic cleft or as receptors on nearby terminals to impact vesicular release properties in response to glutamate that has diffused from a different synapse. Whereas the autoreceptor paradigm represents a conventional form of presynaptic regulation, the second form allows AMPA receptors to participate in intersynaptic communication. Presynaptic Ca2+-permeable AMPA receptors are increased in a model of inflammatory pain at spinal cord dorsal horn peptidergic axon terminals (Woodhams et al., 2019) and are also suggested to modulate synaptic transmission from nociceptive DRG neurons to dorsal horn neurons (Schenk and Matteoli, 2004; Shypshyna and Veselovsky, 2015).
3. AMPA Receptor Function in Postsynaptic Signaling
AMPA receptors are components of virtually all excitatory PSDs in mature synapses (Petralia and Wenthold, 1992; Polgar et al., 2008). Postsynaptic AMPA receptors are often the primary mediators of the depolarizing actions of synaptically released glutamate. This is in contrast to the role typically played by NMDA receptors (discussed below), which provide a more conditional or nuanced signaling capacity to neurons. Early modeling studies of neuronal circuits noted, however, that AMPA receptors need to have different signaling properties to effectively act as integrators of synaptic events and to account for ensemble firing behavior of the cortex and hippocampus (Shadlen and Newsome, 1994; Konig et al., 1996; Reyes et al., 1996; Alkondon et al., 2003). This is accomplished in a multitude of ways. For example, the time course of AMPA receptor deactivation, and hence the time synapses remain active, can be tuned by receptor subunits and alternate splicing of the flip/flop cassette as well as the type and stoichiometry of auxiliary subunits. The membrane time constant is a key determinant of EPSP time course, such that small changes in receptor deactivation rate may for some neurons and synapses exert only minimal effects on the EPSP decay time course. Furthermore, the voltage signal reaching the soma is typically smaller and slower than the original EPSP, which is filtered by the cable properties of the intervening membrane, particularly in neurons with large, elaborate dendritic arbors. In addition to the deactivation time course, desensitization time course will set the properties of frequency response regardless of membrane filtering of the EPSP, making these two parameters important underlying components of synaptic AMPA receptor function (Chen et al., 2002). These nuances to AMPA receptor signaling kinetics are fit-to-purpose through the differential expression of AMPA receptor subunits and auxiliary signaling proteins throughout the CNS (Greger et al., 2017; Jacobi and von Engelhardt, 2021) (Section III. Auxiliary Subunits).
Principal neurons express AMPA receptors with linear current-voltage (I/V) relationships and low Ca2+ permeability, indicating that these receptors are primarily GluA2-containing with slow deactivation and desensitization kinetics (Jonas et al., 1993, 1994; Geiger et al., 1995), which are well suited to act as temporal integrators of synaptic activity. CA1 pyramidal cells use voltage-gated (Ih) conductances to shape AMPA receptor EPSPs so that distally and proximally originating EPSPs end up having similar time courses (and temporal summation characteristics) at the soma (Magee and Cook, 2000). However, principal excitatory neurons and inhibitory interneurons in both the hippocampus and cortex express AMPA receptors with different gating and permeation properties (Jonas et al., 1994; Geiger et al., 1995). Moreover, interneuron expression profiles can be generally subdivided according to their embryonic origins, which arise either from the medial or caudal ganglionic eminence (MGE or CGE) of the developing telencephalon (Matta et al., 2013; Pelkey et al., 2017). The specific complements of AMPA receptors within interneurons may also determine their afferent inputs for feedforward and feedback inhibitory drive, which are critical for normal network function (Akgul and McBain, 2016). In addition, MGE-derived interneurons express AMPA receptors with higher Ca2+ permeability because of lower levels of the GluA2 subunit (Matta et al., 2013) and faster gating kinetics (Jonas et al., 1994; Geiger et al., 1995; Geiger et al., 1997). The hippocampal CA1 MGE-derived interneuron synapses are dominated by Ca2+-permeable, GluA2-lacking AMPA receptors with a small contribution from GluN2A-containing NMDA receptors, whereas CGE-derived interneurons synapses include GluA2-containing AMPA receptors with a larger contribution from GluN2B-containing NMDA receptors (Matta et al., 2013).
Differences in the recovery of paired-pulse depression (Jonas et al., 1994; Geiger et al., 1995) and the response time course (Jonas et al., 1994; Geiger et al., 1995; Geiger et al., 1997) suggested that principal neurons expressed AMPA receptors that are dominated by the GluA2-flip isoform, whereas AMPA receptors expressed by interneurons are dominated by the GluA4-flop isoform (Jonas et al., 1994). A similar distinction in the AMPA receptor gating kinetics was observed between principal cells and interneurons of the basal ganglia (Gotz et al., 1997) and cerebellum (Dawe et al., 2019), suggesting that neuronal circuits throughout the brain may share similar signaling capabilities due, in part, to the expression of different AMPA receptor subtypes (Fig. 24).
Fig. 24.
AMPA receptor expression in different neurons. (A) Upper panel: Current recordings illustrate how neuronal firing properties match AMPA receptor signaling. In the hippocampus, the CA1 pyramidal cells, hilar mossy cells, and hilar interneurons can be distinguished by their distinct firing properties. Lower panel: Current responses in excised outside-out patches to 1 millisecond (red) or prolonged (black) glutamate reveal that the gating kinetics of AMPA receptors are slower in pyramidal cells and mossy cells than in interneurons. Unpublished current and voltage-clamp records are from Ryan Alexander, Yuhao Yan, and Derek Bowie. (B) Upper panel: In the cerebellum, stellate and Purkinje cells show distinct firing properties. Lower panel: AMPA receptors in patches pulled from these neurons showed different responses to brief (green) and prolonged (black) application of glutamate. Reproduced with permission from Alexander et al. (2019) and Dawe et al. (2019).
Most individual neurons express several types of AMPA receptor subtypes with distinct subunit composition. For example, stratum lucidum hippocampal interneurons express both GluA2-containing or GluA2-lacking AMPA receptors when innervated by Schaffer collaterals from CA3 pyramidal neurons or mossy-fiber axons of dentate gyrus granule cells, respectively (Toth and McBain, 1998). Fusiform cells of the dorsal cochlear nucleus receive excitatory input onto their apical dendrites from granule cell parallel fibers and cochlear auditory nerve input onto their basal dendrites. Although several types of iGluR subunits are found at both synapses, GluA4 is selectively targeted to the basal dendrites and associated only with synapses formed by the auditory nerve (Hunter et al., 1993; Rubio and Wenthold, 1997; Wang et al., 1998). Since AMPA receptors containing the GluA4-flop subunit display a rapid response time course (Mosbacher et al., 1994), this may be a property essential for encoding timing information in sound processing (Raman et al., 1994). Miniature EPSCs (mEPSCs) that arise from auditory nerve inputs possess rapid kinetics and are blocked by PhTx (philanthotoxin), a selective inhibitor of GluA2-lacking AMPA receptors (Gardner et al., 1999). In contrast, mEPSCs associated with parallel fibers are resistant to PhTx and possess slower kinetics. These data suggest that in addition to GluR4 segregation, GluA2 is also differentially targeted across the cochlear neuron somato‐dendritic axis (Rubio and Wenthold, 1997).
Studies of both developmental and plasticity mechanisms, particularly in the hippocampus and cortex, suggest that there are more than one population of AMPA receptors at any given synapse. AMPA receptors at individual synapses can assemble as heteromeric assemblies of GluA1/2 or GluA2/3 subunits (Wenthold et al., 1996; Shi et al., 2001; Lu et al., 2009), which play different roles during synapse strengthening. GluA1/2 receptors traffic into synapses during periods of patterned activity after the activation of CaMKII, whereas GluA2/3 receptors constitutively cycle into and out of synapses (Shi et al., 2001). In addition, homomeric GluA1 receptors can be trafficked to synapses to mediate potentiation at some developmental stages and in response to different patterns of synaptic stimulation, neuromodulatory signaling, and drug exposure (Plant et al., 2006; Lu et al., 2007b; Yang et al., 2010; Clem and Huganir, 2010; Qian et al., 2012; Wolf and Tseng, 2012; Sanderson et al., 2016; Purkey et al., 2018).
In the cerebellum, migrating and mature cerebellar granule cells express GluA2/4 heteromeric receptors (Swanson et al., 1997b; Smith et al., 2000) as well as ultra-low conductance channels that arise from RNA-edited homomeric GluA2(R) receptors (Cull-Candy et al., 1988; Smith et al., 2000). In cerebellar stellate cells, high-frequency stimulation of parallel fiber input promotes a switch from GluA2-lacking to GluA2-containing AMPA receptors (Liu and Cull-Candy, 2000; Liu and Cull-Candy, 2005), which causes a change in the strength and Ca2+ permeability of the postsynaptic response. Light-evoked synaptic activation of NMDA receptors also induces a switch from GluA2-containing to GluA2-lacking AMPA receptors in certain retinal ganglion cells (Jones et al., 2012). In hippocampal parvalbumin-containing inhibitory neurons, excitatory synapses are populated by homomeric GluA1 receptors early in development, which then acquire GluA4 in a manner dependent on the secreted protein Nptx2 (Pelkey et al., 2015) (Section V.B. Regulation by Secreted Proteins). The expression of heteromeric GluA1/4 receptors endows rapid synaptic signaling to the parvalbumin-inhibitory interneuron. These findings establish that the composition and functionality of AMPA receptors in individual neurons and at individual synapses are regulated during development and during periods of sustained neuronal activity.
Much of this functional heterogeneity previously observed can in part be explained by AMPA receptor auxiliary proteins (Jackson and Nicoll, 2011a; Greger et al., 2017). Indeed, most native AMPA receptors assemble as a complex with a variety of auxiliary subunits, which can promote the forward trafficking of AMPA receptors to the plasma membrane (Section III. Auxiliary Subunits). Knockout/knockdown studies have demonstrated that TARPs and CNIH proteins shorten the time course of AMPA receptor–mediated synaptic events (Herring et al., 2013; Boudkkazi et al., 2014) consistent with a role in lengthening EPSCs (Table 2). Moreover, the slowing of recovery from desensitization when AMPA receptors coexpress with CKAMP44 (von Engelhardt et al., 2010) or GSG1L (Shanks et al., 2012) controls short-term plasticity and synaptic depression in dorsal lateral geniculate nucleus relay neurons (Chen et al., 2018) or at corticothalamic synapses (Kamalova et al., 2020).
Variations in the stoichiometry of the AMPA receptor/auxiliary subunit complex may regulate the time course of postsynaptic AMPA receptors. In the cerebellum, for example, stellate cells express AMPA receptors that coassemble with two TARP auxiliary subunits, whereas AMPA receptors expressed by cerebellar Purkinje cells possess four TARPs per receptor (Dawe et al., 2019) (Fig. 25). Earlier biochemical analysis of AMPA receptors in the cerebellum suggested that TARP stoichiometry was fixed (Kim et al., 2010), but it was not resolved whether the number of TARPs per AMPA receptor corresponded to partial (i.e., one, two, or three TARPs) or full occupancy (four TARPs) (Section III. Auxiliary Subunits). Structural studies have argued that the composition of AMPA receptor–TARP complexes can be variable, as indicated by analyses of AMPA receptors expressed by cerebellar stellate, granule, and Purkinje cells (Dawe et al., 2019; Miguez-Cabello et al., 2020). In the hippocampus, granule and CA1 pyramidal cells have an AMPA receptor–TARP stoichiometry of 1:2 or 1:4, respectively (Shi et al., 2009). Proteomic analysis suggests that a given AMPA receptor can interact with multiple different auxiliary subunits (Schwenk et al., 2012), and cryo-EM studies reveal four binding sites for TARPs, GSG1L, and CNIH3, creating opportunities for dual regulation (Section III. Auxiliary Subunits).
Fig. 25.
AMPA receptor/TARP stoichiometry influences function in a cell-specific manner in cerebellar neurons. (A) Left panel: Cryo-EM structures of GluA2-tandem TARP γ-2 complex with a stoichiometry of 2 auxiliary subunits (PDB: 5KBU). Right panel: Native and recombinant AMPA GluA1/2 receptor responses exhibit a linear relationship between the rate of the onset of desensitization (τ) and the degree of desensitization (steady-state/peak current ratio as %). The relationship between rate and extent of desensitization for stellate cells best matched that for recombinant AMPA receptors that had a constrained stoichiometry of two TARP γ-2 subunits per AMPA receptor. (B) Left panel: Cryo-EM structures of GluA2-tandem TARP γ-2 complex with a stoichiometry of 4 auxiliary subunits (PDB: 6DLZ). Right panel: Purkinje cell relationship between rate and extent of desensitization best matched with recombinant AMPA receptors that had a constrained stoichiometry of four TARP γ-2 subunits per AMPA receptor. Adapted with permission from Dawe et al. (2019).
4. AMPA Receptor Function in Glial Cells
Virtually all iGluRs have been identified in astrocytes, oligodendrocytes, and microglia (Ceprian and Fulton, 2019). For example, Ca2+-permeable AMPA receptor–mediated EPSCs have been recorded in oligodendrocyte precursor cells (Bergles et al., 2000), which detect glutamate released from axons in white matter (Kukley et al., 2007). AMPA receptor activation promotes oligodendrocyte precursor cell survival and, consequently, myelin production (Kougioumtzidou et al., 2017). In the cerebellum, Bergmann glial cells express AMPA receptors that are activated by glutamate released at parallel fiber and climbing fiber to Purkinje cell synapses (Bergles et al., 1997; Matsui and Jahr, 2004). These AMPA receptor signals have been suggested to guide Bergmann glia to envelop climbing fiber synapses onto Purkinje cells (Matsui and Jahr, 2004). Several reports suggest microglia also can express AMPA receptors, although the role in normal brain function and injury has not been fully explored (Noda et al., 2000; Christensen et al., 2006; Sivakumar et al., 2010; Wong et al., 2011).
5. Kainate Receptor Function in Development
Like AMPA receptors, kainate receptors are expressed early in life, suggesting a role in developmental processes (Fig. 23) (Section VI. Developmental and Regional Expression in the Central Nervous System). Kainate receptor activity promotes axon motility in DRG neurons (Tashiro et al., 2003), dendrite growth in cortical neurons (Monnerie and Le Roux, 2006), and maturation of the hippocampal mossy fiber-CA3 and CA3-CA1 synapses (Marchal and Mulle, 2004; Lauri et al., 2006). Mechanistic studies have uncovered roles for GluK1 and Neto2 in promoting DRG axon growth (Joseph et al., 2011; Vernon and Swanson, 2017). Furthermore, kainate application regulates DRG axon growth in a concentration-dependent manner via GluK5-mediated activation of collapsin-response-mediator proteins (CRMPs) and voltage-gated calcium channels (Marques et al., 2013; Quach et al., 2015). In the cortex, GluK2 promotes dendrite growth in pyramidal neurons, whereas GluK1 and Neto1 promote dendrite maturation in interneurons (Jack et al., 2019). GluK1 and GluK2 regulation of dendritogenesis requires downstream activation of NMDA receptors and voltage-gated calcium channels (Jack et al., 2019).
Overexpression of either GluK1, GluK2, or GluK3 increases presynaptic puncta and broadens the active zone in cultured neurons, whereas GluK2 or GluK5 knockdown reduces the density of presynaptic specializations (Sakha et al., 2016). At the Schaffer collateral to CA1 synapse in vivo, Neto1-mediated trafficking of GluK1 to axon terminals promotes synaptogenesis and synapse maturation (Vesikansa et al., 2012; Orav et al., 2017). Moreover, loss of GluK2 impairs structural and functional maturation of the complex presynaptic and postsynaptic specializations of mossy fiber to CA3 synapses (Lanore et al., 2012).
The kainate receptor auxiliary subunits Neto1 and Neto2 are also expressed early in development and thus may influence kainate receptor expression and function (Michishita et al., 2003, 2004). Axonal kainate receptors at immature Schaffer collateral synapses in the hippocampus tonically regulate glutamate release during synaptogenesis (Lauri et al., 2005; Lauri et al., 2006; Sakha et al., 2016). This functional role is absent in circuits from Neto1 KO mice, in which the targeting of kainate receptors to axonal domains is reduced (Orav et al., 2017). Neto1 is also required for postsynaptic targeting of kainate receptors in CA3 interneurons during development; genetic ablation does not alter basal levels of excitability but attenuates circuit activity in response to kainate receptor activation (Orav et al., 2019).
6. Kainate Receptor Function in Presynaptic Signaling
Presynaptic kainate receptors have well established roles in the bidirectional regulation of glutamate and GABA release, as discussed in several excellent reviews (Lerma and Marques, 2013; Sihra and Rodriguez-Moreno, 2013; Pressey and Woodin, 2021) (Fig. 26). The majority of this work has been in the hippocampus, but presynaptic kainate receptor function is also apparent in the amygdala (Braga et al., 2003, 2009; Aroniadou-Anderjaska et al., 2012; Negrete-Diaz et al., 2012), cortex (Jouhanneau et al., 2011; Andrade-Talavera et al., 2013; Gelsomino et al., 2013; Rodriguez-Moreno and Sihra, 2013), cingulate cortex (Wu et al., 2007b), cerebellum (Delaney and Jahr, 2002; Cervetto et al., 2010; Falcon-Moya et al., 2018), periaqueductal gray (Nakamura et al., 2010), striatum (Marshall et al., 2018), and thalamus (Miyata and Imoto, 2009). Presynaptic DRG kainate receptors regulate release of glutamate from synapses in the superficial dorsal horn of the spinal cord (Kerchner et al., 2001). A number of underlying mechanisms have been identified, including canonical ionotropic calcium-dependent mechanisms, G protein–mediated protein kinase A (PKA) or PKC signaling, and G protein –independent PKA signaling (Negrete-Diaz et al., 2018; Pressey and Woodin, 2021) (Fig. 26). More recently, several studies suggest presynaptic kainate receptors work cooperatively with cannabinoid receptors to control release of glutamate (Marshall et al., 2018) and GABA (Daw et al., 2010; Lourenco et al., 2010, 2011; Wyeth et al., 2017). In addition, kainate receptors modulate release of glutamate through presynaptic mechanisms at mossy fiber synapses (Contractor et al., 2000; Kamiya and Ozawa, 2000; Schmitz et al., 2000).
Fig. 26.

Neuronal functions of kainate receptors. (A1) Kainate receptors (KARs) regulate presynaptic GABA and glutamate release through second messenger-mediated signaling that involves G protein or by current flow. GABAR, GABA receptor. (A2) Upper panel: The kainate receptor agonist domoate reduced fEPSPs in CA1 stratum pyramidale, which was blocked by injection of pertussis toxin (PTX) into the hippocampus prior to slicing. Lower panel: Kainate reduced the frequency of GABAergic miniature inhibitory postsynaptic currents (mIPSCs) in CA1 pyramidal cells. (B1) Activation of kainate receptors on CA3 pyramidal cells alters intrinsic excitability by (B2, Upper panel) decreasing the afterhyperpolarization. Lower panel: The kainate receptor agonist ATPA decreased the AHP in CA3 interneurons. (C1) Kainate receptors mediate postsynaptic currents in response to presynaptic release of glutamate. (C2 upper panel) Evoked EPSCs recorded from a CA3 pyramidal cell in response to 20-Hz stimulation of mossy-fiber input or (lower panel) thalamocortical EPSCs in presence or absence of selective AMPA receptor inhibitor. Adapted with permission from Frerking et al. (2001), Rodriguez-Moreno and Lerma (1998) (A), Fisahn et al. (2005) (B), Segerstrale et al. (2010) (B), Rebola et al. (2007) and Kidd and Isaac (1999) (C) (Copyright 2001, 2007, 2010 Society for Neuroscience).
7. Kainate Receptor Function in Postsynaptic Signaling
By contrast to AMPA receptors, the roles played by postsynaptic kainate receptors in glutamatergic transmission have been difficult to define because of myriad modulatory effects ascribed to these receptors as well as their ability to signal via ionotropic and metabotropic pathways (Frerking and Nicoll, 2000; McBain and Fisahn, 2001; Lerma, 2003; Bowie, 2010; Perrais et al., 2010; Contractor et al., 2011; Akgul and McBain, 2016; Valbuena and Lerma, 2016). The first recordings of kainate receptors in acutely isolated sensory neurons from the rat DRG, which express GluK1 (Sato et al., 1993), exhibited rapid desensitization in response to the agonist kainate, which is in contrast to the nondesensitizing AMPA receptor response to kainate (Huettner, 1990; Patneau and Mayer, 1990). The agonists domoate, kainate, and glutamate all have a higher potency at kainate receptors compared with AMPA receptors, and kainate receptors recover from desensitization nearly ten times more slowly than AMPA receptors (Huettner, 1990; Patneau and Mayer, 1990; Jonas et al., 1994). In the CNS, kainate receptors were identified in cultured embryonic/early postnatal hippocampal neurons (Lerma et al., 1993; Ruano et al., 1995) and glial progenitor cells (Patneau et al., 1994; Puchalski et al., 1994), and were divided into low-affinity (GluK1, GluK2, and GluK3) and high-affinity (GluK4 and GluK5) subunits (Bettler et al., 1990; Egebjerg et al., 1991; Werner et al., 1991; Bettler et al., 1992; Herb et al., 1992; Sakimura et al., 1992). Kainate receptors are expressed throughout the CNS, with GluK2 and GluK5 subunits being the most abundant (Wisden and Seeburg, 1993), which is consistent with de novo GluK2 variants identified in patients with intellectual disability (Section X. Glutamate Receptors in Disease) and GluK2-mediated neurotoxicity in Huntington’s disease (Rubinsztein et al., 1997; MacDonald et al., 1999).
The noncompetitive NAMs of AMPA receptors GYKI-52466 and GYKI-53655 (Section IX. Exogenous Positive and Negative Allosteric Modulators) can isolate neuronal kainate receptor responses by blocking the almost 10-fold larger response mediated by AMPA receptors (Paternain et al., 1995) and have been used to demonstrate functional synaptic kainate receptors (Roche and Huganir, 1995; Castillo et al., 1997; Vignes and Collingridge, 1997) for both inhibitory interneurons and principal neurons (Chittajallu et al., 1996; Clarke et al., 1997; Rodriguez-Moreno et al., 1997; Cossart et al., 1998; Frerking et al., 1998; Bureau et al., 1999). Contrary to the rapid time course of recombinant kainate receptors studied in heterologous expression systems, most synaptic kainate receptors mediate a slow excitatory response that increases in amplitude during repetitive activation (Castillo et al., 1997; Vignes and Collingridge, 1997; Cossart et al., 1998; Frerking et al., 1998) (Fig. 26). However, not all synaptic kainate receptors exhibit slow gating behavior, as relatively rapid synaptic kainate receptor responses have been observed in the retina (DeVries and Schwartz, 1999), hippocampal interneurons (Cossart et al., 2002), and hippocampal granule cells in a model of temporal lobe epilepsy (Epsztein et al., 2005). In addition, in the ground squirrel retina, different OFF bipolar cell subtypes express a distinct complement of kainate (or AMPA) receptors to achieve different time courses of recovery from desensitization and, consequently, distinct responses to light-evoked reductions in glutamate release from photoreceptors (DeVries, 2000). To complicate matters further, kainate receptors expressed by hippocampal interneurons and DRG neurons can signal via a G protein–coupled metabotropic pathway that is proposed to diminish inhibitory neurotransmitter release by inhibiting voltage-gated Ca2+ channels (Rodriguez-Moreno and Lerma, 1998; Rozas et al., 2003; Rutkowska-Wlodarczyk et al., 2015) (Fig. 26). Another metabotropic action of kainate receptors on cellular excitability is the long-lasting inhibition of slow Ca2+-activated K+ channels expressed by CA1 hippocampal pyramidal cells (Melyan et al., 2002, 2004).
The response time course of recombinant GluK2/5 receptors closely matches the response time course of the slow synaptic kainate receptors (Barberis et al., 2008). Kinetic analysis of agonist response time course suggests that only partial occupancy of the receptor is necessary to bring about channel activation and slow deactivation (Barberis et al., 2008). Given that GluK2/5 have a higher apparent affinity for glutamate than GluK2 receptors alone (Barberis et al., 2008), it was proposed that occupancy of GluK5 brings about activation, whereas occupancy of GluK2 triggers desensitization (Mott et al., 2010; Fisher and Mott, 2011). Kainate receptor–mediated EPSCs are faster in GluK5 KO mice (Contractor et al., 2003) but not as fast as would be expected if GluK5 solely accounted for slow synaptic time course.
In addition to the role of GluK5 in slowing synaptic currents, Neto2 (Zhang et al., 2009b) and Neto1 (Straub et al., 2011a; Tang et al., 2011) further slow the kainate response time course [reviewed in Copits and Swanson (2012)] (Sections III. Auxiliary Subunits and IV. Receptor Activation, Deactivation, and Desensitization), fully accounting for the temporal characteristics of kainate receptor–mediated EPSCs (Tomita and Castillo, 2012). In the hippocamus, knockout of Neto1 but not Neto2 accelerates the time course of kainate-mediated synaptic events between mossy fibers and CA3 principal neurons (Straub et al., 2011a; Tang et al., 2011). Neto1 association with postsynaptic mossy fiber kainate receptors may lengthen the temporal window for summation of excitatory input and thereby increase the probability of spike firing (Straub et al., 2011a). Neto2 slows kainate responses in cultured hippocampal neurons (Zhang et al., 2009b; Copits et al., 2011; Straub et al., 2011a,b). Neto1 regulates somatodendritic kainate receptors in different interneuron types, whereas both Neto1 and Neto2 influence kainate receptors at presynaptic terminals (Wyeth et al., 2017).
Kainate receptor function has been examined at cortical synapses [e.g., Wu et al. (2005)], but the relevance of their signaling to higher-order cognitive processing is unexplored. In the entorhinal cortex, kainate receptors contribute to up-states (Digby et al., 2017), possibly through layer 3 pyramidal neurons (Cunningham et al., 2006; West et al., 2007). The pharmacological properties of these entorhinal currents and the expression patterns suggest that GluK2- and GluK5-containing receptors underlie this function (West et al., 2007; Beed et al., 2009). Kainate receptors also play a role in central pain and itch perception (Descalzi et al., 2013).
D. Roles of NMDA Receptors in Neuronal Functions
NMDA receptors are multifaceted signaling complexes with wide-ranging, context-dependent influence on neuronal function. NMDA receptors are localized to presynaptic, postsynaptic, and extrasynaptic compartments where they regulate neurogenesis, neurite development, synapse formation, synaptic transmission, synaptic plasticity, and neuronal excitability (Figs. 21 and 22).
1. Neurogenesis and Neuronal Migration
NMDA receptors are expressed early in CNS development to impact the generation of neurons as well as their migration. In Xenopus laevis, neural plate cells express NMDA receptor-mediated currents, and disrupting NMDA receptor activity impairs neural tube formation in chicken and X. laevis embryos (Andaloro et al., 1998; Rosenquist et al., 1999; Brauer and Rosenquist, 2002; Sequerra et al., 2018). In mammals, NMDA receptor expression does not begin until after neural tube closure (Bennett et al., 2006), and concordantly, complete GluN1 or GluN2B KO does not impact neurulation in mice (Forrest et al., 1994; Li et al., 1994). Subsequently, NMDA receptor activity supports mammalian neurogenesis in the fetal brain by promoting neural progenitor cell proliferation (Sadikot et al., 1998; Luk et al., 2003; Suzuki et al., 2006; Mochizuki et al., 2007; Li et al., 2011a). In addition, there is evidence to indicate NMDA receptor activity also has an impact of neural migration. NMDA receptors promote migration of cerebellar granule cells and glutamatergic cortical neurons in vitro (Komuro and Rakic, 1993; Behar et al., 1999; Hirai et al., 1999; Hirasawa et al., 2003; Mancini and Atchison, 2007; Tarnok et al., 2008). In vivo, NMDA receptor agonist and antagonist administration causes abnormal cortical architecture, which is indicative of altered radial neuronal migration (Marret et al., 1996; Reiprich et al., 2005). However, GluN1 deletion does not have significant effects on cortical architecture (Messersmith et al., 1997; Iwasato et al., 2000), although GRIN1 de novo variants have been associated with cortical abnormalities (Fry et al., 2018). Interestingly, AMPA but not NMDA receptors regulate hippocampal interneuron migration (Manent et al., 2006; Bortone and Polleux, 2009).
In contrast to early development, NMDA receptor activity can restrict neurogenesis as the brain matures. In the postnatal and adult dentate gyrus, NMDA receptor blockade increases neural progenitor cell proliferation (Gould et al., 1994; Cameron et al., 1995; Bernabeu and Sharp, 2000; Nacher et al., 2001; Kitayama et al., 2003; Nacher et al., 2003; Okuyama et al., 2004; Bursztajn et al., 2007; Bunk et al., 2014). This blockade can cause a long-lasting increase in adult-born neurons while also depleting the precursor cell population, potentially restricting behaviorally relevant neurogenesis that may be required at a later time (Joo et al., 2007; Petrus et al., 2009). This finding is important given the clinical use of memantine and ketamine, which accelerate progenitor cell proliferation and differentiation in the dentate gyrus (Maekawa et al., 2009; Namba et al., 2009; Zhao et al., 2014; Choi et al., 2016; Soumier et al., 2016; Ma et al., 2017; Michaelsson et al., 2019; Yamada and Jinno, 2019). Cell-specific GluN1 or GluN2B knockdown in postnatal subventricular and subgranular zone progenitor cells reduces survival of adult-born neurons in the olfactory bulb and dentate gyrus, respectively (Tashiro et al., 2006; Platel et al., 2010; Kelsch et al., 2012). These conflicting outcomes on adult-born neuronal survival between genetic and pharmacological manipulations suggest that there are unknown cell-type–specific roles for NMDA receptors in neurogenesis. Furthermore, the timing and mechanisms of NMDA receptors in transitioning from promoting proliferation in embryonic development to restricting proliferation in the adult are unknown.
2. Neurite Development
Subtype-specific NMDA receptor signaling has a complex role in the initial stages of neuronal differentiation both promoting and inhibiting growth of axons and dendrites. Investigations of topographic sensory map formation revealed that NMDA receptors regulate axonal arborization, and disrupting NMDA receptor function causes axons to overlap erroneously in the visual and somatosensory systems (Cline et al., 1987; Cline and Constantine-Paton, 1989; Simon et al., 1992; Iwasato et al., 1997; Rajan et al., 1999; Ramoa et al., 2001; Ruthazer et al., 2003; Lee et al., 2005; Yamasaki et al., 2014; Van Horn et al., 2017). Initially, dendritic NMDA receptors were thought to orchestrate axonal arborization through retrograde signaling (Schmidt, 2004), but more recent studies indicate axonal NMDA receptors regulate axon growth and pathfinding by controlling axon dynamics (Gao et al., 2018b). NMDA receptors also regulate presynaptic terminal maturation by promoting assembly of synaptic vesicle and active zone proteins (Corlew et al., 2007, 2008; Sceniak et al., 2012; Gill et al., 2015).
During embryonic and early postnatal development, NMDA receptors promote dendrite dynamics, growth, and branching in Xenopus tectal neurons and rat spinal motor neurons (Kalb, 1994; Rajan and Cline, 1998; Rajan et al., 1999). NMDA receptor activation is triggered by spontaneous glutamate release from developing axons to promote dendrite growth prior to axodendritic contact (Andreae and Burrone, 2015). However, in the P14 ferret brain, NMDA receptor blockade increases dendrite length in cortical neurons and dendrite branching in the lateral geniculate nucleus (Rocha and Sur, 1995; McAllister et al., 1996). These opposing results may be due to subunit-specific and context-dependent recruitment of NMDA receptor signaling via the GTPase RhoA, which promotes dendrite growth (Li et al., 2000, 2002; Sin et al., 2002) versus CaMKII signaling, which promotes dendrite stability (Wu and Cline, 1998; Gaudilliere et al., 2004; Ghiretti et al., 2013).
GluN2B signaling promotes dendrite growth, and GluN2A signaling facilitates dendrite refinement and stability in glutamatergic neurons (Espinosa et al., 2009; Sepulveda et al., 2010; Bustos et al., 2014; Sceniak et al., 2019). However, such effects are context-specific, as indicated by the finding that GluN2B-CaMKII signaling limited dendrite growth in immature cultured neurons (5 days in vitro) but enhanced growth in older neurons (12 days in vitro). Furthermore, artificially enhancing an interaction between GluN2A and CaMKII reactivated dendritogenesis in vivo (Bustos et al., 2017). NMDA receptors also regulate dendrite development in GABAergic interneurons (Chittajallu et al., 2017; Pla et al., 2018; Hanson et al., 2019), with GluN2D activation being required during a critical developmental window for proper cortical interneuron dendrite arborization (Hanson et al., 2019).
3. Synapse Formation
Many studies suggest that synaptogenesis triggers NMDA receptor clustering at nascent glutamatergic synapses (Cottrell et al., 2000; Washbourne et al., 2002), allowing NMDA receptors to regulate dendritic spine formation, maturation, and stabilization in developing and mature neurons (Engert and Bonhoeffer, 1999; Maletic-Savatic et al., 1999; McKinney, 2010; Kwon and Sabatini, 2011; Yasuda et al., 2011). Indeed, NMDA receptor activity has a well established role in synaptic refinement by controlling the pruning and strengthening of immature synapses during development (Rabacchi et al., 1992; Zhang et al., 2000; Luthi et al., 2001; Colonnese and Constantine-Paton, 2006; Zhang et al., 2013; Personius et al., 2016). However, depolarization can induce NMDA receptor clustering in the absence of synaptic contacts (Tao-Cheng et al., 2015), and several studies challenge the idea that glutamate release or NMDA receptors are required for dendritic spine formation in the developing nervous system (Verhage et al., 2000; Varoqueaux et al., 2002; Lu et al., 2013; Sigler et al., 2017). Thus, an alternative theory proposes that NMDA receptors control activity-dependent synapse refinement and elimination after circuit formation but are not required for neurite growth or synapse formation in developing neurons.
Several lines of evidence suggest GluN2B-mediated signaling supports dendritic spine dynamics and synapse refinement to exert a brake on synapse maturation, whereas GluN2A-mediated signaling hinders spine dynamics and promotes synapse maturation and stabilization (Ohno et al., 2010; Gray et al., 2011; Wang et al., 2011a; Henle et al., 2012; Kelsch et al., 2014; Wang et al., 2021). Subunit-specific regulation of synapse maturation depends upon distinct CTD interactions, with GluN2B-CaMKII signaling playing an important role [Alvarez et al. (2007), Gambrill and Barria (2011), El Gaamouch et al. (2012), Dupuis et al. (2014); but see McKay et al. (2018)]. GluN2B-containing NMDA receptors are constitutively trafficked to most forebrain glutamatergic synapses, whereas GluN2A trafficking is activity-dependent (Barria and Malinow, 2002; Aizenman and Cline, 2007; Storey et al., 2011).
NMDA receptors are also critical in inhibitory synapse development. Chronic NMDA receptor blockade alters GABAergic synapse number (Aamodt et al., 2000; Rosato-Siri et al., 2002; Henneberger et al., 2005). GluN1 deletion from hippocampal pyramidal neurons impaired inhibitory synapse formation via a mechanism requiring a calmodulin binding motif in the GluN1 CTD (Lu et al., 2013; Gu et al., 2016b). GluN2D-mediated signaling may be particularly involved, since GluN2D-selective inhibition reduces GABAergic synapse density in the adult cortex (Hanson et al., 2019). Interestingly, GluN2D is highly expressed in early postnatal development, but the role of GluN2D in the effects of GluN1 deletion or nonselective NMDA receptor blockers is unknown. In addition, NMDA receptors are embedded postsynaptically with GABA receptors at some developing GABAergic synapses, suggesting a unique developmental mechanism in formation of these synapses (Gundersen et al., 2004; Szabadits et al., 2011; Cserep et al., 2012).
4. Presynaptic Signaling
Presynaptic NMDA receptors control synaptic transmission by influencing neurotransmitter release (Aoki et al., 1994; Charton et al., 1999; Corlew et al., 2008). NMDA receptors enhance both spontaneous and evoked glutamate release at synapses in the cortex (Berretta and Jones, 1996; Woodhall et al., 2001; Sjostrom et al., 2003; Corlew et al., 2007; Larsen et al., 2011b), hippocampus (McGuinness et al., 2010; Prius-Mengual et al., 2019), cerebellum (Glitsch and Marty, 1999; Bidoret et al., 2009), and spinal cord (Robert et al., 1998; Bardoni et al., 2004). Presynaptic NMDA receptors also regulate neurotransmitter release at GABAergic synapses in the reticular nucleus of the thalamus (Crabtree et al., 2013), cortex (Pafundo et al., 2018), and cerebellum (Duguid and Smart, 2004; Rossi et al., 2012). Ambient glutamate likely activates NMDA receptors regulating spontaneous release, whereas action potential-driven release is likely mediated by autoreceptors or by glutamate spillover from adjacent synapses (Duguid and Smart, 2004). Distinct downstream signaling may also be involved, as a Mg2+-insensitive and Ca2+-independent pathway involving PKC and/or c-Jun N-terminal kinase is linked to regulation of spontaneous release, whereas a canonical Mg2+-sensitive mechanism is linked to evoked glutamate release (Kunz et al., 2013; Nistico et al., 2015; Abrahamsson et al., 2017; Marcelli et al., 2019).
NMDA receptor regulation of neurotransmitter release can be target-specific, as layer 5 cortical neurons express presynaptic NMDA receptors at synapses onto other principal cells and somatostatin-positive interneurons but not parvalbumin interneurons (Sjostrom et al., 2003; Buchanan et al., 2012). In addition, presynaptic NMDA receptors are present at only a subset of synaptic inputs onto a single neuron type (Brasier and Feldman, 2008; Corlew et al., 2008; Christie and Jahr, 2009; Banerjee et al., 2014; Larsen et al., 2014). GluN2B-containing receptors make up the majority of presynaptic NMDA receptors in the cortex, but GluN2D may also regulate presynaptic release in the hippocampus and cerebellum (Mameli et al., 2005; Berg et al., 2013; Dubois et al., 2016; Prius-Mengual et al., 2019). In addition, several studies implicate GluN2A in presynaptic regulation of neurotransmitter release in the spinal cord (Xie et al., 2016) and the parallel fiber–Purkinje cell synapse in the cerebellum (Bidoret et al., 2009).
5. Postsynaptic Signaling
The primary function of postsynaptic NMDA receptors is to transduce synaptic signals by mediating excitatory synaptic currents and initiating Ca2+-dependent intracellular signaling. Because depolarization is required to relieve channel block by extracellular Mg2+, NMDA receptor activity can detect coincident activity of presynaptic and postsynaptic neurons—a mechanism critical for some types of synaptic plasticity. A classic role of postsynaptic NMDA receptors is to modify the function, structure, and molecular composition of synapses via Ca2+-dependent activation of local intracellular signaling pathways, cytoskeleton dynamics, and synapse-to-nucleus signaling plasticity (Section V.F. Mechanisms of Synaptic Plasticity).
The impact of NMDA receptor activation on neuronal function depends upon receptor composition, as the properties of the NMDA receptor will control the amount of Ca2+ entering the postsynaptic spine, with overall charge transfer through NMDA receptors impacted by open probability, deactivation time course, Mg2+ sensitivity, and Ca2+ permeability—all properties that vary with the GluN2 subunits in the NMDA receptor subtypes (Section IV. Receptor Activation, Deactivation, and Desensitization and Table 1). For example, GluN1/2A has a high open probability but fast kinetics, whereas GluN1/2B has lower open probability and deactivates more slowly. GluN1/2C has a very low open probability and deactivates slowly, whereas GluN1/2D has low open probability and deactivates very slowly. De novo missense mutations that have specific effects on response time course are clearly disease-causing (Swanger et al., 2016) (Section X. Glutamate Receptors in Disease), suggesting that the deactivation time course is an important determinant of circuit function as well as potential developmental and compensatory processes. Moreover, synapses expressing GluN2D, such as Schaffer collateral to hippocampal interneuron synapses, exhibit prolonged NMDA receptor–mediated currents compared with Schaffer collateral to pyramidal neuron synapses, which express GluN2A and GluN2B (von Engelhardt et al., 2015; Swanger et al., 2018; Yi et al., 2019, 2020). The recruitment of triheteromeric NMDA receptors, such as GluN1/2A/2B, GluN1/2A/2C, or GluN1/2B/2D, would allow additional diversity in synapse function given their unique properties relative to diheteromeric NMDA receptor subtypes (Hansen et al., 2014; Bhattacharya et al., 2018; Yi et al., 2019). In the cerebellum, GluN2C and GluN2D partner with GluN2A and GluN2B, respectively (Brickley et al., 2003; Bhattacharya et al., 2018), and may be recruited to synaptic or extrasynaptic sites after synapse formation. In cerebellar granule cells and cultured hippocampal neurons, GluN2C preferentially assembles with either GluN2A or GluN2B, with negligible surface expression of diheteromeric GluN1/2C, raising a question about whether GluN2C is predominantly present in triheteromeric receptors (Bhattacharya et al., 2018). In spinal and subthalamic neurons, synaptic currents appear to arise from GluN1/2B/2D receptors (Hildebrand et al., 2014; Swanger et al., 2015). The only evidence for diheteromeric GluN1/2D receptors is from Purkinje cells prior to establishing climbing fiber synapses (Misra et al., 2000).
The requirement for depolarization to relieve Mg2+ block and allow current flow in combination with the longer deactivation time of NMDA receptors allows their responses to summate during trains of synaptic input, which is necessary for synaptic activity to drive action-potential firing (Stuart and Spruston, 2015; Iacobucci and Popescu, 2017a). Thus, NMDA receptors are critical to the primary function of neurons, which is to integrate synaptic signals and transduce them into patterns of neuronal activity. Nonetheless, the interplay of NMDA and AMPA receptor activities in regard to membrane potential changes and current flow is complex, given the effects of depolarization on the driving forces for current flow [e.g., Poleg-Polsky and Diamond (2016)]. One form of dendritic integration is the nonlinear summation of NMDA receptor–mediated synaptic potentials that generate large, sometimes regenerative, dendritic potentials referred to as “NMDA spikes” (Schiller et al., 2000). In the cortex and thalamus, these depolarizing potentials enhance neuronal excitability and drive action potential generation directly (Losonczy and Magee, 2006; Major et al., 2008; Larkum et al., 2009; Manita et al., 2011; Lavzin et al., 2012; Smith et al., 2013; Farinella et al., 2014). NMDA receptors may generate depolarizing potentials indirectly via activating voltage-gated Ca2+ channels that facilitate dendritic calcium spikes capable of inducing action-potential bursts (Calton et al., 2000; Wei et al., 2001; Milojkovic et al., 2004; Polsky et al., 2009; Palmer et al., 2014). NMDA receptor–generated dendritic calcium spikes can also shape dendritic spine structural plasticity (Dittmer et al., 2019) and rapid signaling to the nucleus that can regulate gene transcription (Wild et al., 2019). One example is spike timing–dependent plasticity, in which NMDA receptor activation can produce synaptic plasticity depending on its relation to a back-propagating action potential in the dendrite, which can relieve Mg2+ block to enhance NMDA receptor activation if it precedes presynaptic release of glutamate or produce a different outcome if it follows glutamatergic transmission (Supplemental Fig. 8). In addition, NMDA receptors are predicted to contribute to burst firing in subthalamic neurons that may reflect integration of multiple synaptic inputs (Kubota and Rubin, 2011). Postsynaptic NMDA receptor activity also regulates synaptic transmission by activating small conductance Ca2+-activated potassium channels (SK channels) (Cai et al., 2004; Faber et al., 2005; Ngo-Anh et al., 2005; Faber, 2010; Seong et al., 2014; Bock and Stuart, 2016; Babiec et al., 2017) as well as large-conductance Ca2+- and voltage-gated K+ channels (BK channels) (Isaacson and Murphy, 2001; Zhang et al., 2018a), which shunt excitatory synaptic transmission (Kshatri et al., 2018).
6. Extrasynaptic NMDA Receptors
Extrasynaptic NMDA receptors include receptors expressed in somas, dendritic shafts, and perisynaptic regions. Morphologically, extrasynaptic receptors lie more than 100 nm away from the postsynaptic density, with perisynaptic receptors generally being 100–300 nm from the postsynaptic density (Kohr, 2006; Groc et al., 2009; Hardingham and Bading, 2010; Petralia et al., 2010). However, many physiologists define synaptic NMDA receptors as those activated by low-frequency synaptic activity, such as spontaneous synaptic release, and extrasynaptic receptors are not typically activated under these conditions (Tovar and Westbrook, 1999; Chen and Diamond, 2002; Harris and Pettit, 2007; Zhang and Diamond, 2009). Perisynaptic receptors are activated by spillover of synaptic glutamate during high-frequency activity (Asztely et al., 1997; Clark and Cull-Candy, 2002; Harris and Pettit, 2008; Herman et al., 2011; Wild et al., 2015), whereas NMDA receptors farther away from synapses on dendritic shafts and somas are likely activated by ambient glutamate (Sah et al., 1989; Cavelier and Attwell, 2005; Herman and Jahr, 2007) or glutamate released from astrocytes (Angulo et al., 2004; Bezzi et al., 2004; Fellin et al., 2004; Jourdain et al., 2007; Lee et al., 2007; Shigetomi et al., 2008; Bardoni et al., 2010; Hamilton and Attwell, 2010; Nie and Weng, 2010; Fleming et al., 2011; Oh et al., 2012; Park et al., 2015). It is also possible that synaptic NMDA receptors in quiescent synapses may be activated by glutamate spillover (“crosstalk”) from neighboring, active synapses (Arnth-Jensen et al., 2002; Scimemi et al., 2004; Henneberger et al., 2020), although the extent to which this occurs is debated (Barbour, 2001).
Extrasynaptic NMDA receptors were detected widely in the CNS more than 2 decades ago (Aoki et al., 1997; Clark et al., 1997; Rao and Craig, 1997; Kharazia and Weinberg, 1999; Valtschanoff et al., 1999; Momiyama, 2000), but we still lack a clear picture of how synaptic versus extrasynaptic receptors are organized, the degree to which receptors move between synaptic and extrasynaptic domains, and the physiologic and pathophysiological roles for these two receptor populations (Petralia, 2012; Papouin and Oliet, 2014; Parsons and Raymond, 2014; Zhou et al., 2015). Perisynaptic NMDA receptors may play a role in long-term synaptic plasticity but also can diffuse laterally into the synapse in an activity-dependent manner, which blurs the distinction between synaptic and extrasynaptic populations. Extrasynaptic receptors are also well positioned to detect glial release of glutamate, which can be triggered by G protein–coupled receptor activation as well as other signals (Mannaioni et al., 2001; Haroon et al., 2017; Kofuji and Araque, 2021). Furthermore, NMDA receptors in perisynaptic regions may have unique roles in dendritic excitability via activation of SK channels (Isaacson and Murphy, 2001) and “NMDA spikes” (Jourdain et al., 2007; Oikonomou et al., 2012) due to their localization near Ca2+- and voltage-activated channels.
Extrasynaptic NMDA receptors, particularly those expressed on dendritic shafts and somas, mediate tonic currents or slow inward currents that enhance neuronal excitability in response to ambient glutamate when the membrane potential is sufficiently depolarized (Sah et al., 1989; Herman and Jahr, 2007; Le Meur et al., 2007; Povysheva and Johnson, 2012; Wu et al., 2012). Tonic NMDA receptor currents may facilitate coupling between dendritic and somatic compartments, enhancing action potential firing in pyramidal neurons (Sah et al., 1989; Wu et al., 2012; Riebe et al., 2016). Furthermore, tonic NMDA receptor currents in interneurons regulate γ oscillatory activity (Mann and Mody, 2010), and slow inward currents activated by glial glutamate release contribute to the synchronization of neuronal firing (Angulo et al., 2004; Fellin et al., 2004, 2006; D'Ascenzo et al., 2007; Nie and Weng, 2010). The hypothesis that NMDA receptors containing GluN2C or GluN2D, which have reduced Mg2+ sensitivity and higher glutamate potency relative to GluN2A and GluN2B, mediate tonic currents is supported by studies in the substantia nigra (Morris et al., 2018b), thalamus (Zhang et al., 2012), and cortex (Hanson et al., 2019).
Extrasynaptic NMDA receptors were considered an important mediator of excitotoxicity in neurons in response to acute injury (Choi et al., 1987; Choi et al., 1988), as discussed in several excellent reviews (Lai et al., 2014; Li and Wang, 2016; Wang and Reddy, 2017; Olloquequi et al., 2018; Ge et al., 2020). Multiple studies suggest that extrasynaptic NMDA receptor signaling activates cell death pathways, whereas synaptic NMDA receptors can support cell survival (Sattler et al., 2000; Vanhoutte and Bading, 2003; Xu et al., 2009; Hardingham and Bading, 2010; Kaufman et al., 2012). Recent work contradicts these studies, however, by suggesting that synaptic receptors can contribute to excitotoxic cell death, and perhaps activation of both synaptic and extrasynaptic NMDA receptors is required to induce cell death (Papouin et al., 2012; Wroge et al., 2012; Zhou et al., 2013a,b). Thus, current evidence does not unequivocally support a rule for linking synaptic or extrasynaptic NMDA receptors to cell survival or death. The two populations likely make context-dependent contributions to excitotoxicity, and better methods for distinguishing and selectively manipulating signaling by synaptic and extrasynaptic receptors will be required to determine specific roles. Selective disruption of a novel noncanonical Ca2+-independent signaling pathway mediated by extrasynaptic NMDA receptors in complex with TRPM4 channels prevents neuronal death in models of stroke and retinal degeneration (Yan et al., 2020).
7. GluN3-Containing NMDA Receptors
Functional receptors that contain two GluN1 and two GluN3 subunits (GluN1/3) have been demonstrated in heterologous expression systems (e.g., Xenopus oocytes and HEK cells) (Chatterton et al., 2002; Smothers and Woodward, 2003, 2007, 2009; Wada et al., 2006; Awobuluyi et al., 2007; Madry et al., 2007, 2008, 2010; Ulbrich and Isacoff, 2007, 2008; Schuler et al., 2008; Cavara et al., 2009; Wee et al., 2010; McClymont et al., 2012; Kvist et al., 2013a,b; Balasuriya et al., 2014; Cummings and Popescu, 2016; Cummings et al., 2017; Grand et al., 2018; Kaniakova et al., 2018). NMDA receptors composed of GluN1, GluN2, and GluN3 subunits (GluN1/2/3) have also been suggested to exist [reviewed in Perez-Otano et al. (2016)], but the expression of GluN1/2/3 is ambiguous and the subunit stoichiometry is unresolved. The caveat that a mixed population of GluN1/3 and GluN1/2 receptors is expressed rather than GluN1/2/3 receptors cannot easily be excluded. This limited understanding of the subunit composition of neuronal GluN3-containing receptors has impeded studies of their physiologic roles. However, glycine-activated neuronal currents mediated by GluN3A-containing receptors have been recorded from wild-type mouse hippocampal CA1 neurons and are absent in GluN3A-deficient mice (Grand et al., 2018; Otsu et al., 2019; Zhu et al., 2020). These currents are insensitive to NMDA receptor antagonists, suggesting the receptor complex contains the glycine-binding GluN3A but not glutamate-binding GluN2 subunits. The neuronal glycine responses require the presence of the GluN1-selective competitive antagonist CGP-78,608, which prevents desensitization triggered by agonist binding to GluN1 (Section IV. Receptor Activation, Deactivation, and Desensitization), allowing glycine binding to GluN3A to activate the receptor. These data provide the first unambiguous evidence for native GluN1/3A receptors.
Despite this advance, GluN1/3 receptors have cryptic activation properties that obscure their physiologic roles. Although desensitization can be prevented by GluN1 binding site mutations or GluN1-selective inhibitors, GluN1/GluN3 receptors in the CNS will be tonically exposed to low glycine (or d-serine) concentrations (Bergeron et al., 1998; Billups and Attwell, 2003), resulting in occupancy at the high-affinity GluN1 glycine binding site that will presumably produce desensitization of the GluN1/3 receptors (Grand et al., 2018; Zhu et al., 2020). However, canonical ionotropic signaling mediated by GluN1/3 receptors may be dependent on an unknown biologic context that relieves this tonic desensitization (Grand et al., 2018).
GluN3A expression restricts synapse maturation (Perez-Otano et al., 2006; Roberts et al., 2009; Henson et al., 2012; Fiuza et al., 2013; Kehoe et al., 2014), and neurons in GluN3A KO mice have increased spine density compared with wild-type mice (Das et al., 1998). Studies of the role of GluN3A as a molecular brake on synapse maturation have focused on the intracellular CTD of GluN3A and its interactions with proteins involved in spine plasticity [reviewed in Perez-Otano et al. (2016)]. The postsynaptic scaffolding protein GIT1 forms an interaction with the GluN3A CTD that is critical to the prevention of synapse maturation (Fiuza et al., 2013; Kehoe et al., 2014) and facilitates activation of Rac1 and its effector PAK1 to promote synapse rearrangements and stabilization (Zhang et al., 2003, 2005). The GluN3A CTD has also been shown to interact with PP2A, microtubule associated protein 1S (MAP1S), MAP1B, plectin, CARP1, GPS2, and RHEB [Chan and Sucher (2001), Eriksson et al. (2007a,b), Eriksson et al. (2010); reviewed in Perez-Otano et al. (2016)]. Increased surface expression of GluN3A due to disrupted interaction with PACSIN1 has been suggested to contribute to behavioral impairments in Huntington’s disease (Marco et al., 2013, 2018; Mahfooz et al., 2016). In this disease, aggregation of huntingtin protein disrupts the subcellular localization of PACSIN1, thereby promoting cell surface expression of GluN3A-containing NMDA receptors (Marco et al., 2013). Genetic deletion of GluN3A slows motor and cognitive decline and reduces neuronal loss in a Huntington’s disease mouse model. GluN3A has been linked to cocaine-evoked synaptic changes, and cocaine exposure drives surface expression of GluN3A in the ventral tegmental area [Yuan et al. (2013), Creed et al. (2016); see also Huang et al. (2013), Huang et al. (2017)]. The physiologic roles of GluN3B-containing NMDA receptors are less understood. GluN3B KO mice show deficits in motor learning consistent with GluN3B expression in motoneurons (Section VI. Developmental and Regional Expression in the Central Nervous System) and show increased social interactions with familiar cage mates, decreased social interaction in a novel environment, and moderate anxiety-like behavior (Niemann et al., 2007).
E. Neuronal Functions of GluD Receptors
The GluD1 and GluD2 subunits show significant sequence similarity to glutamate receptor subunits, yet it remains uncertain whether they form functional ion channels (Kakegawa et al., 2007a; Schmid et al., 2009; Yuzaki and Aricescu, 2017). Rather, postsynaptic GluD receptors are shown to function as synaptic organizer proteins through interactions between their NTD and secreted Cbln1-4 proteins, which interact with presynaptic Nrxn (Yuzaki and Aricescu, 2017; Yuzaki, 2018; Dai et al., 2021). This Nrxn-Cbln-GluD trans-synaptic interaction is involved in the formation and maintenance of synapses in various brain regions. Furthermore, d-serine can bind to the ABD of GluD2 (Naur et al., 2007) and is an endogenous ligand of GluD2 receptors expressed in Purkinje cells, wherein d-serine binding triggers cerebellar LTD (Kakegawa et al., 2011). d-Serine can also bind the GluD1 ABD (Yadav et al., 2011).
1. GluD1 Receptors
GluD1 can bind to Cbln1 and Cbln2 (Yasumura et al., 2012), which in turn can interact with presynaptic Nrxn cell adhesion molecules (Ushkaryov et al., 1992; Ullrich et al., 1995; Reissner et al., 2013; Dai et al., 2021). The Nrxn-Cbln1-GluD complex has been studied in a coculture system in which artificial synapse can be formed between GluD expressing HEK293 cells and neurons expressing Nrxn when Cbln1 is provided into the growth medium (Matsuda and Yuzaki, 2011; Ryu et al., 2012; Elegheert et al., 2016). One Cbln1 hexamer can recruit one neurexin on the presynaptic terminal, and the Cbln1 hexamer can bind to one GluD dimer at the postsynaptic side through interaction with the GluD NTD (Lee et al., 2012; Cheng et al., 2016; Elegheert et al., 2016). GluD1 organizes excitatory synapses in the dorsal striatum through interactions with Cbln1 (Liu et al., 2020a). Ablating GluD1 reduces excitatory neurotransmission in medium spiny neurons, decreases vGlut2 protein levels in terminals from parafascicular thalamic neurons, and impairs behavioral flexibility (Liu et al., 2020a). Furthermore, GluD1 regulates the formation of synapses from parallel fibers to molecular layer interneurons in cerebellar cortex (Konno et al., 2014). GluD1 regulates the formation of excitatory synapses in hippocampal CA1 pyramidal cells through interactions with Cbln2; increased spine density follows overexpression of GluD1 in CA1 pyramidal cells, with a reciprocal reduction in spine density after knockdown of GluD1 (Tao et al., 2018, 2019). GluD1 receptors can also organize inhibitory synapses between cortical interneurons and pyramidal neurons via interactions with Cbln4, which is secreted by somatostatin-expressing interneurons (Yasumura et al., 2012; Favuzzi et al., 2019; Fossati et al., 2019).
d-Serine and glycine elicit postsynaptic signaling mediated by GluD1 that involves a guanine nucleotide exchange factor, ARHGEF12 and the regulatory subunit of protein phosphatase 1 (PPP1R12A) that may be involved in the formation of inhibitory synapses (Fossati et al., 2019). Studies with GluD1 KO mice have consistently found increased dendritic spine density in hippocampus and prefrontal cortex along with reduced expression of GluA1 and GluN2B (Yadav et al., 2012, 2013; Gupta et al., 2015). GluD1 KO mice show higher spontaneous activity, lower anxiety-like behavior, increased aggressiveness, reduced social interaction, deficits in learning, and altered cocaine-conditioned place preference (Yadav et al., 2012; Liu et al., 2018; Gantz et al., 2020; Nakamoto et al., 2020a). GluD1-dependent currents have been reported in raphe neurons in response to noradrenergic synaptic input, although the activating mechanism has not been elucidated (Gantz et al., 2020).
2. GluD2 Receptors
GluD2 receptors are expressed in cerebellar Purkinje cells at synapses in the distal dendrites that receive parallel fiber input but are excluded from synapses in proximal dendrites that receive climbing fiber and molecular layer interneuron input (Takayama et al., 1996; Landsend et al., 1997). GluD2 receptors in Purkinje cells bind to Cbln1, which is released from granule cells and also binds to presynaptic Nrxn located on the parallel fiber terminals (Matsuda et al., 2009, 2010; Uemura et al., 2010). The formation of the neurexin-Cbln1-GluD2 complex triggers accumulation of proteins associated with the presynaptic and postsynaptic sites (Hirai et al., 2005; Matsuda and Yuzaki, 2011; Ito-Ishida et al., 2012; Yamashita et al., 2013; Ichikawa et al., 2016). The number of parallel fiber to Purkinje cell synapses is reduced in GluD2 KO mice (Kashiwabuchi et al., 1995; Kurihara et al., 1997; Takeuchi et al., 2005), cerebellar LTD is impaired (Torashima et al., 2009), and the loss of synapses can be restored by introduction of GluD2 (Matsuda et al., 2010; Uemura et al., 2010). One Purkinje cell in adult wild-type mice is normally innervated by a single climbing fiber, but in GluD2 KO mice, multiple climbing fibers can synapse onto one Purkinje cell (Miyazaki et al., 2010; Hashizume et al., 2013). The density of molecular layer to Purkinje cell synapses is also increased in Cbln1 KO mice, which can be restored by recombinant Cbln1 protein in a manner dependent on the presence of GluD2 (Ito-Ishida et al., 2014).
GluD2 signaling is involved in cerebellar LTD in parallel fiber to Purkinje cell synapses (Yuzaki, 2013), which requires phosphorylation of AMPA receptors followed by their removal from synaptic sites (Kohda et al., 2013; Gallimore et al., 2016). The role of GluD2 in regulating AMPA receptor trafficking during cerebellar LTD requires the intracellular GluD2 CTD (Kohda et al., 2007, 2013; Torashima et al., 2009). In immature cerebellar slices, d-serine is released from Bergman glia in response to the burst stimulation of parallel fibers that trigger cerebellar LTD, which is dependent on d-serine binding to GluD2 (Kakegawa et al., 2011; Elegheert et al., 2016). Thus, d-serine could be an endogenous agonist of GluD2 receptors, although ion channel activity has not been detected in response to d-serine binding (Naur et al., 2007; Hansen et al., 2009; Kakegawa et al., 2011).
The importance of GluD2 in adaptive control of locomotion is illustrated by several mouse lines with mutations or deletions in the Grid2 gene (Hoxha et al., 2018). The lurcher mutation (GluD2-A654T) spontaneously opens the ion channel pore in the absence of agonist binding (Zuo et al., 1997; Kohda et al., 2000; Wollmuth et al., 2000). The constitutive inward currents in GluD2-expressing cells of lurcher mice result in ataxia and impaired motor skills as a result of Purkinje cell death (Hoxha et al., 2018). Multiple hotfoot mutations have been identified in mice with similar phenotypes as GluD2 KO mice, including deficits in cerebellar circuit function and impaired cerebellar LTD (Kashiwabuchi et al., 1995; Lalouette et al., 2001; Motohashi et al., 2007; Cendelin, 2014). Some hotfoot mutations as well as the more recently identified ts3 mutation (Miyoshi et al., 2014) are deletions in the open reading frame encoding GluD2, explaining the similar phenotype with GluD2 KO mice (Matsuda and Yuzaki, 2002; Wang et al., 2003; Motohashi et al., 2007; Takeuchi et al., 2012; Zanjani et al., 2016).
F. Mechanisms of Synaptic Plasticity
Synaptic plasticity, or the ability to bidirectionally modulate intercellular signaling strength, is an important feature of the CNS. Synaptic plasticity takes many different forms, producing changes in synaptic efficacy ranging from seconds to days that are thought to encode complex behaviors, such as learning, memory, sensory processing, emotional responses, addiction, and general cognition. The iGluRs play essential roles in several key forms of synaptic plasticity. This section will focus largely on work performed in the hippocampus and, in particular, the Schaffer collateral-CA1 synapse. Because of its laminar structure and well characterized connectivity, the hippocampal slice has been exhaustively used for studies of synaptic plasticity. However, synaptic plasticity is a key feature of neuronal signaling throughout the brain, including the neocortex (Sjostrom et al., 2001), basal ganglia (Kreitzer and Malenka, 2008), and cerebellum (Sjostrom et al., 2001). The field of synaptic plasticity is enormous, with many excellent reviews on specific models of synaptic plasticity (Abbott and Nelson, 2000; Citri and Malenka, 2008; Herring and Nicoll, 2016b; Nicoll, 2017; Diering and Huganir, 2018; Harris, 2020; Purkey and Dell'Acqua, 2020) as well as homeostatic plasticity (Turrigiano and Nelson, 2004; Turrigiano, 2012) and metaplasticity (Abraham, 2008; Lee et al., 2010b), which will not be discussed here.
1. Synaptic Organization and the Postsynaptic Density
The PSD at the Schaffer collateral-CA1 synapse, or the portion of the dendritic spine with a cluster of neurotransmitters receptors and signaling proteins, is directly apposed to presynaptic release sites and is organized into several nanoscale domains. This organization holds important implications for participation of glutamate receptors in synaptic plasticity. The center of the PSD contains several NMDA receptors, with AMPA receptor clusters adjacent to but not overlapping with NMDA receptor clusters (Shinohara, 2012; Fukata et al., 2013; MacGillavry et al., 2013; Nair et al., 2013; Tang et al., 2016; Kellermayer et al., 2018; Goncalves et al., 2020) (Fig. 27). AMPA receptor clusters may position directly underneath presynaptic neurotransmitter vesicular release sites, forming a nanoscale column of presynaptic release machinery and postsynaptic AMPA receptors, possibly aided by trans-synaptic neuroligins and Nrxns (Nair et al., 2013; Tang et al., 2016; Haas et al., 2018) (Fig. 27). However, nearly half of the postsynaptic AMPA receptors have been shown to be mobile within the PSD and are not associated with a particular nanodomain (Ashby et al., 2006; Heine et al., 2008; Makino and Malinow, 2009; Jacob and Weinberg, 2015; Goncalves et al., 2020) (Fig. 27). Each nanodomain is roughly 70 nm in diameter (MacGillavry et al., 2013; Nair et al., 2013), with adjacent nanodomains within the same synapse being approximately 100 nm apart (Haas et al., 2018; Choquet and Hosy, 2020). The glutamate concentration within the synapse rapidly diminishes to 500 µM or less about 100 nm outside of the activated nanodomain per given quanta, and this may be even faster in some regions because of the segregated architecture of the nanodomains (Nair et al., 2013; Haas et al., 2018; Choquet and Hosy, 2020) (Fig. 27). Recent work found that optogenetic recruitment of GluA1-containing AMPA receptors to the PSD in bulk, although sufficient to potentiate the strength of evoked transmission by increasing quantal event frequency, was not able to increase quantal amplitude. These findings support the model that only those AMPA receptors recruited to the “slots” in the PSD located near active sites of glutamate release are effectively engaged in synaptic transmission, whereas those that are recruited to the PSD distant from release sites are not (Sinnen et al., 2017). Since NMDA receptors are at the center of the PSD roughly 80–100 nm from each AMPA receptor nanodomain and require coagonists glutamate and glycine to bind as well as membrane depolarization to relieve Mg2+ block to generate current flow, their activation might require multiple presynaptic events at some synapses [Silver et al. (1996b), Helassa et al. (2018); but see Bekkers and Stevens (1989)]. This creates opportunities for plasticity to proceed through modulation of both postsynaptic receptors and presynaptic release.
Fig. 27.
Organization of the glutamatergic synapse. (A) The center of the PSD contains several NMDA receptors with adjacent AMPA receptor clusters. Presynaptic vesicular release machinery is positioned above each AMPA receptor cluster with transsynaptic anchoring proteins (e.g., neuroligins, neurexins) flanking the release sites. This alignment of presynaptic release machinery with postsynaptic AMPA receptors encased by neuroligins forms a 70–100-nm diameter domain. Intracellular postsynaptic scaffolding proteins have been omitted for clarity, as have mobile NMDA and AMPA receptors. (B) Simulations of glutamate concentration within the synaptic cleft after release of a single quanta suggest that neighboring AMPA receptor nanodomains might not see a sufficient concentration of glutamate for rapid activation. (C) Stimulated emission depletion (STED) imaging of cultured neurons demonstrates how AMPA receptors are clustered in dendrites (left), and single-particle tracking photoactivation localization microscopy (sptPALM) enables visualization of AMPA receptor nanodomains within dendritic spines (right, scale bar 200 nm). (D) sptPALM imaging revealed greater mobility of GluA2 than GluA1 within dendritic spines (left). Imaging of surface-expressed GluA2 subunits by universal point accumulation in nanoscale topography (uPAINT) imaging demonstrated its restricted mobility. (E) Stochastic optical reconstruction microscopy (STORM) imaging of immunostained AMPA and NMDA receptors in mouse brain revealed a diversity in the synaptic organization of receptor nanodomains, with some synapses displaying higher AMPA receptor content (top, left) and others higher NMDA receptor content (top, right). AMPA and NMDA receptor distribution within the postsynaptic area differed as some synapses displayed a high level of centrally located NMDA receptors (bottom, right), and other synapses had both AMPA and NMDA receptors located toward the periphery of the synapse. Adapted with permission from Choquet and Hosy (2020) (B), Nair et al. (2013) (C and D), and Dani et al. (2010) (E).
2. Short-Term Plasticity
Short-term plasticity refers to the ability of a synapse to increase or decrease postsynaptic signaling output and spine depolarization when pairs of presynaptic stimuli arrive within the same presynaptic bouton on a millisecond timescale. Short-term plasticity at excitatory synapses is important for several forms of memory (Ferguson et al., 2004; Jaaskelainen et al., 2011; Hansel and Mato, 2013), can serve to counterbalance frequency-dependent depression (Turecek et al., 2016), can exert frequency-dependent filtering (Jackman and Regehr, 2017), and can influence sensory processing (Fortune and Rose, 2002). Short-term plasticity only lasts for a matter of seconds to minutes, and both presynaptic and postsynaptic mechanisms are involved.
Short-term plasticity is classically divided into paired-pulse facilitation and paired-pulse depression, although other forms exist (Zucker and Regehr, 2002). Paired-pulse facilitation of AMPA receptor–mediated postsynaptic currents occurs when the second stimulus arrives in the presynaptic bouton before the first Ca2+ load is cleared. The further rise in terminal Ca2+ from the second action potential causes more neurotransmitter to be released by increasing the release probability, increasing the number of vesicles released, and engaging multiple release sites (Katz and Miledi, 1968). When the glutamate concentration in the synaptic cleft does not saturate AMPA receptors, increased glutamate release generates a larger postsynaptic current. The effect of Ca2+ during facilitation is also modulated by presynaptic Ca2+ buffering via calcium-binding proteins, such as calbindin, which compete for Ca2+ with release machinery (Blatow et al., 2003). Additionally, a postsynaptic mechanism for paired-pulse facilitation may involve activity-dependent relief of polyamine block acting on GluA2-lacking AMPA receptors, increasing postsynaptic depolarization (Bowie et al., 1998; Rozov and Burnashev, 1999; Toth et al., 2000).
Paired-pulse depression, the counterpoint to paired-pulse facilitation, is defined as a depressed postsy-naptic response to a second presynaptic stimulus compared with the preceding stimulus. Paired-pulse depression also occurs by both presynaptic and postsynaptic mechanisms. As even brief exposure to glutamate can desensitize AMPA receptors (Hestrin, 1992), the postsynaptic hypothesis of paired-pulse depression posits that the second pulse arrives within a time window during which many AMPA receptors are still desensitized and unable to respond to agonist (Koike-Tani et al., 2008). Indeed, desensitization shapes evoked postsynaptic responses at calyx synapses in the auditory pathway (Otis et al., 1996), although its impact at hippocampal CA3-CA1 synapses has been questioned (Hjelmstad et al., 1999). Alternatively, the second pulse may arrive while AMPA receptors are still substantially active, and the repeated exposure to glutamate may facilitate their desensitization, rendering them unresponsive to subsequent stimulation. Auxiliary subunits that alter AMPA receptor desensitization (Sections III. Auxiliary Subunits and IV. Receptor Activation, Deactivation, and Desensitization; Table 2) should impact this form of plasticity (Devi et al., 2020). AMPA receptors in the desensitized state can uncouple from PSD scaffolding proteins, potentially increasing their lateral mobility (Constals et al., 2015). Thus, an alternative mechanism for paired-pulse depression is a lack of new receptors to replace desensitized receptors that moved from the release site (Heine et al., 2008; Opazo et al., 2010; Constals et al., 2015), consistent with the observation that paired-pulse depression can be enhanced by reducing the mobile pool of extrasynaptic AMPA receptors either by crosslinking or by antibody tethering (Heine et al., 2008). Presynaptic models of paired-pulse depression center on vesicle depletion. Here, the readily releasable pool of neurotransmitter-loaded and docked presynaptic vesicles becomes depleted, particularly at synapses where a single action potential can generate the release of multiple vesicles (Betz, 1970; Wadiche and Jahr, 2001; Chen et al., 2004a). Such vesicular depletion combined with evidence showing presynaptic calcium spikes help prepare release machinery for subsequent fusion events (Hosoi et al., 2009) could generate a weakened second postsynaptic response.
It seems likely that different mechanisms for paired pulse facilitation and depression will be employed at different synapses. The form of short-term plasticity that each synapse may adopt is influenced by previous activity at that synapse. Synapses with a high presynaptic release probability, such as some cerebellar synapses, are prone to paired-pulse depression, whereas synapses with low vesicle release probability, such as some synapses in the hippocampus, are amenable to paired-pulse facilitation (Regehr, 2012). Presynaptic NMDA receptors (Chamberlain et al., 2008) and kainate receptors (Sun and Dobrunz, 2006) modestly influence short-term plasticity.
3. Long-Term Plasticity
LTP and LTD refer to forms of synaptic plasticity whereby specific stimulation of afferent glutamatergic fibers can induce long-lasting (sustained for hours and days) increases or decreases in synaptic signaling, respectively. Both LTP and LTD are thought to be the molecular correlates for encoding and storing long-term memories throughout the brain (Lisman, 1989; Nabavi et al., 2014). Although the exact molecular mechanisms involved in LTP and LTD are still under investigation, there is a consensus that iGluRs participate in all three phases (i.e., induction, expression, and maintenance) of long-term plasticity and that the endpoints ultimately result in changes in synaptic AMPA receptor content (Fig. 28). For an in-depth discussion of these topics, readers are referred to several excellent reviews on LTP (Nicoll, 2017; Diering and Huganir, 2018; Harris, 2020) and LTD (Collingridge et al., 2010).
Fig. 28.
Control of synaptic strength through modification of AMPA receptors. (A) Summary of four ways an excitatory synapse can change its properties through changes in either the number or subtype of AMPA receptors at the postsynaptic density (PSD). (B) Mechanisms underlying AMPA receptor trafficking during NMDA receptor-dependent LTP and LTD at hippocampal CA1 synapses. Under basal conditions (center), AMPA receptors undergo dynamic lateral exchange into and out of the PSD and recycling between the plasma membrane and endosomes. During induction of LTP (right), brief, high levels of Ca2+ influx through NMDA receptors activate a variety of protein kinase signaling pathways that promote PSD trapping of both Ca2+-permeable and Ca2+-impermeable AMPA receptors through lateral exchange and increased delivery to the extrasynaptic plasma membrane by exocytosis from recycling endosomes. During the induction LTD, prolonged, low, levels of NMDA receptors Ca2+ influx activate a variety of protein phosphatase and kinase pathways that promote AMPA receptor loss from the PSD and removal from the extrasynaptic plasma membrane by endocytosis. AMPA receptor trafficking during LTP and LTD is regulated by changes in both AMPA receptor CTD and TARP phosphorylation/dephosphorylation. AMPA receptor synaptic recruitment in LTP and removal in LTD are also coordinated with structural changes that increase and decrease, respectively, the number of PSD “slots” for AMPA receptors.
a. Long-term potentiation
A vast number of LTP studies have explored the Schaffer collateral-CA1 synapse, as this laminar pyramidal cell structure simplifies experimentation and allows recording of robust field excitatory postsynaptic potentials (fEPSPs). LTP induction at the Schaffer collateral-CA1 synapse requires sufficient postsynaptic depolarization to relieve Mg2+ block of NMDA receptors, coagonists glutamate and glycine binding to the NMDA receptor, and Ca2+ influx into the postsynaptic spine (Luscher and Malenka, 2012). In the classic paradigm, this is accomplished by a high-frequency burst or tetanic stimulation, which produces more persistent depolarization than a single synaptic event (Schwartzkroin and Wester, 1975). Pharmacological inhibition of NMDA receptors with competitive antagonists like amino-5-phosphonovaleric acid (AP5) (Collingridge et al., 1983), or channel blockers like MK-801 (Gilbert and Mack, 1990) and chelation of postsynaptic Ca2+ with EGTA (Lynch et al., 1983) prevent induction of LTP after tetanic stimulation. Multiple biochemical experiments have shown that a rise in postsynaptic Ca2+ via NMDA receptors activates intracellular signaling cascades, including PKA (Otmakhova et al., 2000), PKC (Malinow et al., 1989), PKG (Serulle et al., 2007), and CaMKII (Lisman et al., 2012), all of which help traffic, anchor, and sustain AMPA receptors within the PSD to produce a potentiated synaptic response (Fig. 28).
Several lines of early evidence suggested that the GluN2B subunit was the key driver for postsynaptic LTP induction, including the observation that RNA knockdown of GluN2B in hippocampus abolishes LTP at the Schaffer collateral-CA1 synapse (Kutsuwada et al., 1996; Clayton et al., 2002), and overexpression of GluN2B in hippocampus enhanced spatial learning (Tang et al., 2001). In addition, the GluN2B subunit harbors a CaMKII binding site on its intracellular CTD (Strack and Colbran, 1998; Leonard et al., 1999; Strack et al., 2000; Barria and Malinow, 2005) along with several phosphorylation sites critical for LTP (Rostas et al., 1996; Nakazawa et al., 2006). However, experiments using GluN2A KO mice indicate the involvement of the GluN2A in LTP induction, as noted by their lower level of potentiation compared with wild-type mice (Sakimura et al., 1995; Ito et al., 1997). GluN2A KO mice show deficits in spatial learning and memory, providing behavioral evidence to support the importance of GluN2A in LTP induction (Bannerman et al., 2008; Brigman et al., 2008).
Despite these observations, there is no clear evidence suggesting only one GluN2 subunit controls LTP induction and facilitates expression. Genetic deletion experiments carry the caveat of compensation, and pharmacological experiments also yield complex data. Furthermore, the majority of postsynaptic NMDA receptors at Schaffer collateral to CA1 synapses are triheteromeric assemblies containing two GluN1 subunits, one GluN2B subunit, and one GluN2A subunit (Sheng et al., 1994; Gray et al., 2011; Rauner and Kohr, 2011; Tovar et al., 2013; Yi et al., 2019). For example, the GluN2B-selective inhibitor ifenprodil can act on triheteromeric GluN1/2A/2B receptor complexes, and the competitive antagonist NVP-AAM007, which in error was described as GluN2A-selective (Frizelle et al., 2006; Lind et al., 2017), lacks sufficient selectivity from GluN2B-containing NMDA receptors to yield interpretable results. Thus, consistent with the triheteromeric view of NMDA receptors at hippocampal synapses, it is likely that both GluN2A and GluN2B subunits are important for LTP induction [e.g., Volianskis et al. (2013, 2015)].
A feature of GluN2 subunits is their distinct CTDs that harbor multiple binding sites for scaffolding proteins, kinases, and other binding partners (Section II. Receptor Structure). There may well be some GluN2-specific aspects to types and strength of LTP depending on, for example, CaMKII binding (Barria and Malinow, 2005; Zhou et al., 2007; Halt et al., 2012; Incontro et al., 2018). Notably, deletion of the CaMKII binding site on GluN2B reduces LTP, and transfer of this binding site to GluN2A increases LTP (Barria and Malinow, 2005; Halt et al., 2012). Whereas NMDA receptors and CaMKII are also required for induction of LTD, activation of a calcineurin-DAPK1 signaling pathway that prevents CaMKII from binding to GluN2B appears to be crucial for promoting LTD and preventing inappropriate synaptic potentiation (Coultrap et al., 2014; Goodell et al., 2017).
Since the initial description of LTP, many additional stimulus paradigms have been shown to elicit long-lasting potentiation, leading to the idea that multiple mechanisms exist by which synaptic strength can be enhanced. High-frequency stimulation applied in a series of bursts at a frequency within the θ range may be a more physiologic stimulus that mimics the endogenous θ rhythms of pyramidal cells. Although both tetanic and θ burst stimulus protocols can induce LTP, there may be distinctions between the underlying mechanisms. Application of several θ bursts produces a robust LTP signal, as these bursts can relieve Mg2+ block to allow increased Ca2+ flux through NMDA receptors (Larson and Munkacsy, 2015). Another relevant form of LTP induction involves temporal spike timing. In this Hebbian paradigm, presynaptic stimulation must precede postsynaptic action potential generation to potentiate the synapse [reviewed in Feldman (2012)]. This occurs presumably as a result of back-propagating action potentials that relieve Mg2+ block to increase the influx of Ca2+ through NMDA receptors (Dan and Poo, 2004). In addition, LTP at CA1 synapses and in cultured hippocampal neurons can also be induced chemically by a variety of means, including through the application of forskolin (Otmakhov et al., 2004) to activate adenylyl cyclase and rolipram to inhibit phosphodiesterases, which is sufficient to promote LTP induction, expression, and maintenance via activation of the PKA pathway along with spontaneous burst firing of CA3 neurons (Otmakhova et al., 2000). Likewise, some paradigms that use glycine application (to ensure saturation of GluN1) in low extracellular Mg2+ (to promote activation of synaptic NMDA receptors by glutamate released in response to spontaneous activity) also induce LTP [e.g., Musleh et al. (1997), Lu et al. (2001)). These forms of chemical LTP have allowed biochemical, imaging, and genetic approaches to better explore signaling systems in LTP, since chemical LTP can produce plastic changes at a much larger number of synapses than local electrical stimulation, although mechanisms may be somewhat different from LTP that is induced by electrical stimulation.
After induction, the expression of LTP is mediated by Ca2+-dependent signaling cascades that increase AMPA receptor numbers within the PSD, with activity-dependent trafficking of the AMPA receptor being a prevailing theory (Lledo et al., 1998; Lu et al., 2001) (Fig. 28). This process involves AMPA receptor trafficking to the plasma membrane, lateral diffusion into the synapse, and capture of the AMPA receptor by the PSD (Ahmed and Siegelbaum, 2009; Kennedy et al., 2010; Patterson et al., 2010; Opazo et al., 2012; Jurado et al., 2013; Penn et al., 2017; Wu et al., 2017) (Fig. 28). This idea was supported by studies of silent synapses in the developing CNS, which contain NMDA receptors but lack AMPA receptors (Isaac et al., 1995; Liao et al., 1995). The response of these synapses to various stimulation protocols highlighted a central role for AMPA receptor trafficking (Kerchner and Nicoll, 2008). One element of our understanding of extrasynaptic AMPA receptor recruitment into the PSD during the expression of LTP was the discovery that LTP induction generates extensive actin polymerization (Kim and Lisman, 1999; Matsuzaki et al., 2004; Harvey et al., 2008). By promoting actin polymerization, the activated dendritic spine can sustain an increased number of AMPA receptors within the PSD via an increase in spine-head diameter. Additionally, actin polymerization is concomitant with intracellular rearrangements with PDZ-containing scaffolding proteins such as PSD-95. The molecular shuffling in the dendritic spine actin cytoskeleton and the PSD induced by NMDA receptor signaling during LTP induction has been proposed to generate new PSD “slots” into which mobile surface AMPA receptors diffuse and become stabilized through interaction with AMPA receptor–associated TARPs (Opazo et al., 2012; Granger et al., 2013; Herring and Nicoll, 2016a) (Fig. 28). Roughly 70% of the AMPA receptor content of the PSD may enter by diffusion in the initial phase of LTP expression (Patterson et al., 2010). This finding was validated using antibody tethering to restrict the lateral mobility of surface AMPA receptors (Penn et al., 2017), which prevented synaptic enhancement after high-frequency stimulation of the Schaffer collateral pathway both in vitro and in vivo (Clarke and Johnson, 2006). However, antibody tethering was only able to inhibit initial LTP expression, whereas LTP at a lower amplitude persisted an hour after the high-frequency stimulus. Thus, it appears that the earliest LTP expression is mainly mediated by lateral diffusion of surface AMPA receptors followed by AMPA receptor trafficking by exocytosis to maintain increased AMPA receptor levels within the PSD. Early LTP could reflect phosphorylation of postsynaptic receptors and rapid capture of laterally diffusing receptors in the PSD followed by slower exocytic delivery of new AMPA receptors (Penn et al., 2017). Finally, late-phase LTP persisting longer than 3 hours requires new synthesis of plasticity-related proteins, including AMPA receptors (Sutton and Schuman, 2006; Buffington et al., 2014).
The AMPA receptor NTD appears to also play a central role in LTP maintenance, raising the possibility that NTD-interacting proteins in the synaptic cleft, such as Nptx1 and Nptx2, could also help define slots for AMPA receptors (Diaz-Alonso et al., 2017; Watson et al., 2017), perhaps anchoring the receptor proximal to presynaptic glutamate release sites (Biederer et al., 2017; Watson et al., 2017) (Section V.B. Regulation by Secreted Proteins).
LTP at its most fundamental level may only require structural rearrangements of the PSD to create additional “slots” that are then filled by AMPA receptors (independent of subunit composition) exchanging in and out of the synapse from a reserve pool of extrasynaptic receptors (Opazo et al., 2012; Granger et al., 2013; Herring and Nicoll, 2016b; Tang et al., 2016; Penn et al., 2017) (Fig. 28). Nonetheless, multiple studies have reported on the importance of GluA1 for the expression and maintenance of LTP (Vanderklish et al., 1992; Diering and Huganir, 2018). Data from multiple groups found that the majority of newly incorporated synaptic AMPA receptors in initial LTP expression are GluA2-lacking, Ca2+-permeable AMPA receptors [Plant et al. (2006), Guire et al. (2008), Jaafari et al. (2012), Purkey and Dell'Acqua (2020); but see Adesnik and Nicoll (2007)]. As the potentiated synapse transitions into the maintenance phase of LTP, these homomeric GluA1 are replaced with Ca2+-impermeable GluA1/2 receptors (Plant et al., 2006; Yang et al., 2010; Jaafari et al., 2012). In this regard, a number of studies have investigated the roles of GluA1 CTDs in AMPA receptor trafficking related to LTP expression. One approach has been to study the effects on LTP of CTD swapping in chimeric receptors. LTP was deficient in a transgenic animal in which native GluA1 was replaced with a chimeric GluA1 with the GluA2 CTD, but this deficiency was rescued by adding the GluA1 CTD to GluA2 (Zhou et al., 2018). Rescue experiments using triple-KO mouse lacking GluA1, GluA2, and GluA3 subunits (Lu et al., 2009), in which engineered receptors were reinserted into the neuron, found that LTP could still be recorded after GluA1 lacking the CTD was coexpressed with GluA2 (Granger et al., 2013). Similar results were found in a knock-in mouse in which the endogenous GluA1 was replaced with GluA1 lacking its CTD (Diaz-Alonso et al., 2020). Thus, the GluA1 CTD does not appear to be required for LTP expression. Nonetheless, a number of studies have demonstrated modulatory effects of GluA1 CTD phosphorylation. There are several key post-translational modifications on the GluA1 CTD, such as phosphorylation of Ser818 (Lin et al., 2009), Ser831 (Lee et al., 2000, 2003), and Ser845 (Lee et al., 2000, 2003), which have been shown to be important for LTP expression. These phosphorylation events activated by PKA, PKG, PKC, and CaMKII increase single-channel conductance and open probability (Derkach et al., 1999; Banke et al., 2000; Kristensen et al., 2011; Jenkins and Traynelis, 2012; Jenkins et al., 2014), help direct GluA1-containing AMPA receptors to the surface (Man et al., 2007; Serulle et al., 2007), and stabilize receptors in the PSD (Diering et al., 2016).
Although multiple studies suggest a particular role for GluA1, they do not rule out GluA1-independent mechanisms of LTP, consistent with findings from a number of studies suggesting that there is little involvement of homomeric GluA1 in LTP expression and no requirement for persistent Ca2+ influx for subsequent LTP maintenance (Adesnik and Nicoll, 2007; Gray et al., 2007; Granger et al., 2013). In considering these apparently conflicting results, differences in stimulation protocols and the potentially confounding effects of animal age must be considered (Guire et al., 2008; Park et al., 2016b; Purkey et al., 2018; Purkey and Dell'Acqua, 2020). In particular, the effects of genetic and pharmacologic manipulations can differ over the course of postnatal development, and genetic changes can elicit compensatory mechanisms (Zamanillo et al., 1999; Jensen et al., 2003; Kolleker et al., 2003; Lee et al., 2003; Lu et al., 2007b; Sanderson et al., 2016). For example, prior work found that postsynaptic PKA signaling and GluA1 Ser845 phosphorylation contribute to LTP in adult animals but are dispensable for LTP expression in juveniles (Lee et al., 2003; Lu et al., 2007b). These observations are consistent with findings mentioned above, such as those reported in Granger et al. (2013). It appears that the subunit composition of AMPA receptors already present in and acutely delivered to this reserve pool to be available for synaptic insertion may change during development and also be influenced by the induction stimulus employed as well as neuromodulatory input [reviewed in Purkey and Dell'Acqua (2020)]. Regardless, these differences among plasticity mechanisms proposed by different groups highlight both the diversity of mechanisms available to the synapse [e.g., Plant et al. (2006), Clem and Huganir (2010)] as well as a need for further exploration of the mechanisms of LTP expression and maintenance. Although there is enormous volume of work completed at the Schaffer collateral-CA1 synapse, there is ample precedent for both similar and different mechanisms at other central synapses. For example, the mossy fiber–CA3 synapse utilizes an NMDA receptor–independent LTP induction mechanism (Johnston et al., 1992; Nicoll and Schmitz, 2005).
b. Long-term depression
The induction of LTD has typically involved a low-frequency stimulus paradigm (Kemp et al., 2000) but can also be evoked by postsynaptic action potential generation before a presynaptic stimulation (i.e., spike-timing protocols) (Feldman, 2012) (Supplemental Fig. 8) or chemically by low concentrations of NMDA (Lee et al., 1998). These LTD induction paradigms suggest a need for low levels of intracellular Ca2+, as opposed to high levels needed to induce LTP. For NMDA receptor–dependent LTD, Ca2+ influx into the PSD activates several phosphatases, including calcineurin and protein phosphatase 1 (Mulkey et al., 1994) (Fig. 28). Low levels of Ca2+ promote phosphatase activity, which can promote dephosphorylation of GluA1 Ser831 and Ser845 (Lee et al., 2003) and TARPs (Tomita et al., 2005b), which can cause destabilization of AMPA receptor anchoring and removal from the PSD (Diering and Huganir, 2018). NMDA receptor–independent LTD is thought to rely on the metabotropic actions of mGluRs, mainly mGluR5, which activates PLC-PKC and tyrosine phosphatase signaling pathways (Oliet et al., 1997; Gladding et al., 2009). It remains a matter of investigation as to how NMDA receptor–dependent versus NMDA receptor–independent LTD induction mechanisms are coordinated to depress synaptic strength.
Although pharmacological studies initially indicated a primary role for GluN2B-containing receptors in LTD induction (Massey et al., 2004); but see (Morishita et al., 2007), LTD induction can proceed with either GluN2A or GluN2B alone, since genetic deletion of either subunit is insufficient to block LTD (Wong and Gray, 2018). Moreover, LTD expression does not rely on a specific AMPA receptor subtype being removed from the synapse, as confirmed via GluA1 and GluA2 KO experiments (Granger and Nicoll, 2014). There is no consensus on the identities of all of the intracellular proteins driving this phenomenon, but it is clear that local, postsynaptic scaffolding and highly regulated signaling crosstalk between protein phosphatase 1, calcineurin, PKA, DAPK1, p38-mitogen-activated protein kinase (MAPK), and CaMKII is crucial (Hu et al., 2006, 2007; Lu et al., 2007a; Jurado et al., 2010; Sanderson et al., 2012; Aow et al., 2015; Gao et al., 2018a; Goodell et al., 2017) (Fig. 28, see also Fig. 6). In particular, anchoring to the postsynaptic actin-associated scaffold protein neurabin is required for PP1 regulation of GluA1 phosphorylation, AMPA receptor trafficking, and synaptic depression during LTD (Hu et al., 2006, 2007; Gao et al., 2018a). In addition, PKA anchors to the scaffold protein A-kinase-anchoring protein 150 (AKAP150) along with the opposing phosphatase calcineurin, where it regulates GluA1 phosphorylation and plays a role controlling Ca2+-permeable AMPA receptor synaptic incorporation during NMDA receptor–dependent LTD, similar to its role during LTP [reviewed in Purkey and Dell'Acqua (2020)]. Using knock-in mice that are deficient in AKAP anchoring of either PKA or the opposing phosphatase calcineurin, Ca2+-permeable AMPA receptors were shown to be recruited to hippocampal synapses by anchored PKA during LTD induction but then rapidly removed by anchored calcineurin (Sanderson et al., 2016). Importantly, blocking Ca2+-permeable AMPA receptor recruitment, removal, or activity interferes with LTD. Thus, Ca2+-permeable AMPA receptor synaptic recruitment may be required to transiently augment NMDA receptor Ca2+ during LTD induction. Interestingly, NMDA receptor activation of the Ca2+-dependent kinases DAPK1 and CaMKII is also required for LTD, with calcineurin-dependent dephosphorylation and activation of DAPK1 maintaining its postsynaptic localization to prevent CaMKII from binding to GluN2B, which would instead result in LTP (Goodell et al., 2017). Whether Ca2+ influx through Ca2+-permeable AMPA receptors also contributes to DAPK1 and CaMKII signaling during LTD remains to be determined.
4. Nonionotropic Signaling and Plasticity
The CTD of some iGluRs responds to conformational changes within the ABD, initiating mechanisms for synaptic plasticity via intracellular signaling that are independent of current flow. NMDA receptor–dependent LTD can be induced by glutamate binding to the GluN2 subunit in the absence of ion flux [Nabavi et al. (2013), Stein et al. (2015, 2020); but see Babiec et al. (2014), Sanderson et al. (2016)]. Here, exposing synaptic receptors to glutamate but blocking the glycine site on GluN1 to block channel gating is sufficient for LTD induction and associated dendritic spine shrinkage. These data indicate that conformational changes within NMDA receptors enable metabotropic intracellular signaling that leads to LTD induction. Agonist binding to NMDA receptors independent of channel gating generates a conformational rearrangement of the CTD that promotes protein phosphatase 1 disengagement from the receptor complex (Aow et al., 2015; Dore et al., 2015). Sustained, nonionotropic signaling can lead to dephosphorylation of CaMKII potentially via newly released protein phosphatase 1, culminating in CaMKII removal from the NMDA receptor CTD (Dore et al., 2015). The diminished catalytic activity of CaMKII and its removal from NMDA receptors via dephosphorylation may be a priming factor in generating nonionotropic NMDA receptor–dependent LTD and associated dendritic spine shrinkage (Nabavi et al., 2013; Stein et al., 2015, 2020) and could also represent a mechanism for synaptic scaling in response to low presynaptic activity.
Glutamate binding to NMDA receptors can also activate p38 MAPK independent of glycine occupancy. Blockade of the GluN1 glycine binding site with antagonist when GluN2 is bound to glutamate reduces dendritic spine diameter in a manner dependent on p38 MAPK (Stein et al., 2015). This nonionotropic signaling is activated via neuronal nitric oxide synthase and cofilin, which promote cytoskeletal remodeling (Stein et al., 2020). Considering that some extracellular glycine is always present, even if phasically regulated, it is difficult to imagine a situation in which the GluN1 ABD would be unoccupied. However, given the higher potency of glutamate for NMDA receptors compared with AMPA receptors, glutamate spillover might preferentially bind to NMDA receptors without mediating ionotropic signaling due to Mg2+ block if the membrane is not adequately depolarized. Thus, nonionotropic spine shrinkage may influence synaptic strength via unconventional mechanisms [reviewed in Dore et al. (2017), Rajani et al. (2020)].
Although nonionotropic signaling has been described in AMPA receptors (Hayashi et al., 1999; Valbuena and Lerma, 2016), nonionotropic signaling via kainate receptors appears to be more robust and multifaceted. In addition to impacting membrane depolarization by catalyzing ion permeation, kainate receptors couple to Gαi/o proteins and to a lesser extent Gαq (Cunha et al., 1999; Ruiz et al., 2005). Acting presynaptically at the mossy fiber–CA3 and CA3-CA1 synapses, kainate receptor activation suppresses glutamate release onto pyramidal cells in a PKA-dependent manner (Lauri et al., 2005, 2006; Negrete-Diaz et al., 2006). Presynaptic kainate receptors also act through a PKC-dependent manner to depress GABA release from hippocampal interneurons and indirectly by stimulating the release of endocannabinoids that bind to CB1 receptors to attenuate GABA release (Rodriguez-Moreno and Lerma, 1998; Cunha et al., 2000; Lourenco et al., 2010). In addition, the metabotropic effects of GluK2-containing kainate receptors can control intrinsic excitability by dampening the magnitude of the slow Ca2+-activated K+ current, IAHP (Melyan et al., 2002; Fisahn et al., 2005; Segerstrale et al., 2010). Moreover, kainate receptors and their G protein signaling partners exhibit a bidirectional impact on neurite extension via interactions with cytoskeletal proteins, including cofilin, thus modulating overall synaptogenesis (Tashiro et al., 2003; Gelsomino et al., 2013; Marques et al., 2013).
5. Auxiliary Subunits and Plasticity
TARP auxiliary subunits play roles in plasticity by anchoring AMPA receptors within the PSD (Chen et al., 2000; Sumioka et al., 2010), promoting diffusional trapping in the PSD during LTP (Opazo et al., 2012), increasing channel conductance, slowing deactivation kinetics, and diminishing time spent in the desensitized state (Sections III. Auxiliary Subunits and IV. Receptor Activation, Deactivation, and Desensitization, Table 2). The best-studied TARP, γ-2 (Noebels et al., 1990), is required for long-term synaptic plasticity in cerebellar stellate cells (Jackson and Nicoll, 2011b) and CA1 pyramidal cells (Tomita et al., 2005b). Moreover, γ-8 is expressed along with γ-2 and is critical for the expression of hippocampal LTP, in part through promoting phosphorylation of AMPA receptor CTDs by CaMKII (Tomita et al., 2005b; Park et al., 2016a; Sheng et al., 2018) (Fig. 28). The kainate receptor auxiliary subunit Neto1 is critical for kainate receptor surface expression and stabilization within the PSD (Tang et al., 2011; Wyeth et al., 2014; Sheng et al., 2015). Neto1 is required for delivery of presynaptic kainate receptors, which regulate synaptogenesis at the Schaffer collateral-CA1 synapse (Orav et al., 2017), facilitate LTP induction (Ng et al., 2009), and modulate glutamate release onto GABAergic interneurons (Wyeth et al., 2017). Thus, given their involvement with multiple aspects of plasticity, auxiliary subunits could be therapeutic targets for diseases that involve maladaptive plasticity, such as epilepsy and addiction.
VI. Developmental and Regional Expression in the Central Nervous System
Cloning studies starting in 1989 enabled the localization of all iGluR subunit mRNAs in the mammalian central and peripheral nervous systems through in situ hybridization, revealing temporal and regional expression patterns. The production of subunit and splice isoform-specific antibodies together with advances in single-cell PCR yielded complementary maps of subunit protein distributions and finer-grained resolution of mRNA expression (Boulter et al., 1990; Sommer et al., 1990; Monyer et al., 1991, 1994; Bahn et al., 1994; Hollmann and Heinemann, 1994; Geiger et al., 1995; Dingledine et al., 1999). Many iGluR subunits exist as alternative splice isoforms (Fig. 7) whose expressions are regulated in a developmental and cell type–specific fashion (Monyer et al., 1991; Hadzic et al., 2017). An enormous volume of data has been generated in the intervening 30 years describing developmental, regional, cellular, and subcellular localization of all iGluR subunits. In addition, large-scale mapping and warehousing of gene transcript expression in publicly available databases provide a comprehensive (and growing) profile of subunit expression at the single-cell level (Schmidt et al., 2018; Armand et al., 2021). These resources include The Allen Brain Atlas (https://portal.brain-map.org/), DropViz (http://dropviz.org/), Mousebrain.org (http://mousebrain.org), the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell), and the Human Protein Atlas (http://www.proteinatlas.org/). We limit this review to a survey of iGluR expression that provides specific examples for each receptor class in several well studied brain regions [e.g., reviewed in Akgul and McBain (2016), Hadzic et al. (2017)].
A. AMPA Receptor Subunit Expression
AMPA receptor subunits are expressed in neurons and glia throughout the mammalian brain and spinal cord from an early stage of development (Bettler et al., 1990; Keinanen et al., 1990; Sommer et al., 1990; Monyer et al., 1991; Pellegrini-Giampietro et al., 1991; Tolle et al., 1993; Ritter et al., 2002; Hadzic et al., 2017). All mature excitatory synapses contain AMPA receptors, but mRNAs encoding all four AMPA receptor subunits are also present well before synaptogenesis, suggesting that the receptors play a role in the earliest stages of brain development.
1. Expression of GluA2 Controls AMPA Receptor Properties
The GluA2 subunit determines many functional properties of AMPA receptors (Sections IV. Receptor Activation, Deactivation, and Desensitization and V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity), and its expression pattern influences circuit function. RNA editing at the Q/R/N site of GluA2 changes the codon for Gln at the tip of the M2 pore loop to an Arg, which renders the pore impermeable to Ca2+ and insensitive intracellular polyamine. RNA editing of the GluA2 Q/R/N site occurs early in development with high fidelity (Sommer et al., 1991; Nutt and Kamboj, 1994; Kask et al., 1998). Thus, the Ca2+ permeability of AMPA receptor–mediated synaptic currents at an excitatory synapse depends on whether cells express GluA2 mRNA and in what quantity (Bochet et al., 1994; Geiger et al., 1995; Washburn et al., 1997).
GluA2 mRNA is expressed in all brain regions and is accompanied by transcripts for at least one other AMPA receptor subunit needed to form heteromeric receptors. As an example, GluA2 mRNA can be found in pyramidal neurons throughout the hippocampus as well as in dentate granule cells (Keinanen et al., 1990; Monyer et al., 1991). AMPA receptor–mediated EPSCs in principal neurons of the hippocampus and other parts of the CNS are typically Ca2+-impermeable and have approximately linear current-voltage relationships, a hallmark of GluA2-containing receptors (Hestrin et al., 1990; Perkel et al., 1990; Livsey et al., 1993). Some hippocampal interneurons, such as CGE-derived interneurons, express GluA2 and receive excitatory input to Ca2+-impermeable AMPA receptors (McBain and Dingledine, 1993; Matta et al., 2013; Akgul and McBain, 2016) (Fig. 29). Similarly, AMPA receptors in cerebellar Purkinje cells and striatal projection neurons contain GluA2 subunits despite being GABAergic (Perkel et al., 1990; Stefani et al., 1998). However, many GABAergic interneurons do not to express GluA2 and show AMPA receptor properties consistent with Ca2+ permeable receptors (Jonas et al., 1994; Geiger et al., 1995; Racca et al., 1996; Matta et al., 2013; Lalanne et al., 2016) (Fig. 29). Diversity occurs even at the level of single neurons. Hippocampal CA3 interneurons, for example, express AMPA receptors with distinct Ca2+ permeation properties (i.e., with or without GluA2 subunits) and differentially target those receptors to synapses from dentate granule cells (mossy fibers) or from CA3 pyramidal neurons in an afferent-specific manner (Toth and McBain, 1998). Similarly, a population of fast-spiking interneurons in the dentate gyrus localize Ca2+-permeable AMPA receptors to perforant path inputs but Ca2+-impermeable receptors to mossy-fiber synapses from granule cells (Sambandan et al., 2010).
Fig. 29.
AMPA receptor expression in hippocampal neurons. (A) Left panel: Hippocampal CA1 pyramidal cell filled with biotin, reproduced with permission from Yankova et al. (2001) (Copyright 2001 National Academy of Sciences, U.S.A.). Right panels: Evoked AMPA receptor-mediated EPSCs from CA1 pyramidal cells in hippocampal slices from floxed Gria1, Gria2, Gria3, or Gria1/2 mice injected with an rAAV1-Cre-GFP virus, which produced single-cell elimination of the desired gene product (green). Recordings from adjacent wild-type cells are in black; scale bars are 5 pA, 10 milliseconds. Adapted with permission from Lu et al. (2009). Reconstructions of MGE-derived (B) and CGE-derived (C) CA1 interneurons. MGE-derived interneurons are more sensitive to an inhibitor of GluA2-lacking AMPA receptors (PhTX-433), than CGE interneurons, suggesting that MGE-derived interneurons express more Ca2+-permeable AMPA receptors. Scale bars are 50 pA and 10 milliseconds. CRE, cAMP response element. Adapted with permission from Matta et al. (2013) and Lee et al. (2011).
Single-cell RNA-sequencing analyses in the hippocampus also found GluA2 transcripts expressed in all glutamatergic neurons: dentate granule cells, mossy cells, CA1-CA3 pyramidal neurons, and subicular neurons (Zeisel et al., 2015; Cembrowski et al., 2016b, 2018). Similarly, GluA2 mRNA was detected in nearly all principal excitatory neurons in the mouse cortex but only in a subset of interneurons (Tasic et al., 2016). The heterogeneity in expression of GluA2 and other AMPA receptor transcripts revealed by single-cell mapping studies underscores the limitations in considering any one cell type, even those as well characterized as hippocampal CA1 pyramidal neurons, as having a monolithic transcriptional identity (Cembrowski et al., 2016a).
2. Developmental Expression Patterns for AMPA Receptor Subunits
In contrast to GluA2, transcripts for GluA1, GluA3, and GluA4 are more heterogeneously distributed throughout in the CNS (Boulter et al., 1990; Keinanen et al., 1990; Monyer et al., 1991; Geiger et al., 1995). GluA1 and GluA3 are found in all hippocampal pyramidal neurons (Fig. 29) and dentate granule cells, where they assemble with GluA2 to form Ca2+-impermeable AMPA receptors (Hayashi et al., 2000; Passafaro et al., 2001; Shi et al., 2001; Meng et al., 2003; Lu et al., 2009; Granger et al., 2013). GluA4 transcripts, on the other hand, are expressed weakly in the mature principal cells of the hippocampus but are a key component of AMPA receptors on parvalbumin-containing interneurons (Keinanen et al., 1990; Monyer et al., 1991). GluA4 is highly expressed in cerebellar granule cells as part of heteromeric GluA2/4 receptors that underlie EPSCs at mossy-fiber synapses (Gallo et al., 1992; Lambolez et al., 1992). Elsewhere in the cerebellum, GluA1 is found in Purkinje cells, and GluA3 is expressed in molecular-layer interneurons and Purkinje cells (Keinanen et al., 1990; Lambolez et al., 1992). Bergmann glia express Ca2+-permeable AMPA receptors composed of GluA1 and GluA4 subunits (Keinanen et al., 1990; Gallo et al., 1992; Geiger et al., 1995). GluA1 and GluA4 are expressed by a subpopulation of striatal parvalbumin–positive interneurons but are absent from projection neurons and cholinergic interneurons. In contrast, AMPA receptors in striatal projection neurons are predominantly GluA2/3 receptors (Tallaksen-Greene and Albin, 1994; Bernard et al., 1997; Kwok et al., 1997).
AMPA receptor expression is developmentally regulated in brain region– and neuronal population–specific patterns. Although transcripts for AMPA receptor subunits can be detected as early as embryogenesis, it remains unknown whether functional receptors are produced at that stage. Transient postnatal expression of GluA4 in the hippocampus provides a striking example of developmental regulation. The restricted appearance of GluA4-containing receptors in pyramidal neurons plays a key role in the development of excitatory synapses and influences the mechanisms underlying synaptic plasticity, but the transcript is then downregulated and remains at nearly undetectable levels into adulthood (Zhu et al., 2000; Luchkina et al., 2014). GluA1 is expressed at high levels in early development of cerebellar granule cells and Purkinje neurons but is downregulated with maturation of this brain region (Martin et al., 1998), whereas the GluA4 expression increases in granule cells throughout development (Monyer et al., 1991). GluA2 shows only modest developmental peaks of mRNA expression in the hippocampus and other brain regions (Pellegrini-Giampietro et al., 1991).
3. Expression of AMPA Receptor Splice Isoforms
All four AMPA receptor subunit mRNAs undergo alternate splicing of the flip/flop cassette in the ABD (Fig. 7), which alters the response time course (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity). In general, flip isoforms are expressed early in development and maintained in the mature brain, whereas flop isoforms tend to arise at later phases of maturation after synaptogenesis (Monyer et al., 1994). In the rat hippocampus, cortex, and striatum, for example, the flip isoform of GluA1, GluA2, and GluA3 mRNAs is transcribed in excitatory neurons at postnatal day 1, whereas flop isoforms begin to express about a week later. There are a few exceptions to this general principle; GluA2-flop is expressed at much higher levels than GluA2-flip in the cerebellum during early postnatal development (Monyer et al., 1994). The regulation of flip versus flop expression is critical to the function of certain synapses, such as in the cochlear nucleus. Stimulation of auditory nerves elicits excitatory synaptic currents with fast decay kinetics in cochlear neurons compared with parallel fiber inputs to the same neurons. EPSCs with submillisecond kinetics are mediated by AMPA receptors containing flop-containing subunits (Gardner et al., 1999, 2001) consistent with the fast deactivation and desensitization observed in recombinant receptors (Mosbacher et al., 1994). Brainstem auditory neurons also express fast-decaying AMPA receptor currents indicative of flop-containing receptors (Raman et al., 1994; Ravindranathan et al., 2000). Thus, mammalian auditory systems rely on fast AMPA receptor kinetics to encode high-frequency sensory signals.
GluA2 and GluA4 subunits have alternative splice isoforms of the CTDs, which diversifies post-translational modifications and interactions with chaperones during biogenesis and synaptic targeting [e.g., Dev et al. (1999), Shi et al. (2001), Kolleker et al. (2003), Lee et al. (2004), Tigaret et al. (2006)] (Fig. 7). The GluA2 subunit CTD exists in two splice isoforms referred to as “short” and “long” isoforms (Kohler et al., 1994). Both long and short forms of GluA2 are detectable in brain lysates, and the long version plays a role in synaptic targeting of AMPA receptors after LTP in CA1 pyramidal neurons especially in juvenile animals (Kolleker et al., 2003). The “short” and “long” isoforms of GluA4 differ in their cerebellar expression; the long form is found in Bergmann glia, whereas the short form is expressed in granule neurons (Gallo et al., 1992).
In addition to central neurons, AMPA receptors are also found in peripheral neurons (Seifi and Swinny, 2016) and nearly all non-neuronal cells in the nervous system [reviewed in Ceprian and Fulton (2019)]. Indeed, transcripts for AMPA receptor subunits are detected in all types of glial cells (Conti et al., 1994; Noda et al., 2000; Talos et al., 2006; Christensen et al., 2016). AMPA receptor function has been confirmed in Bergmann glia (Geiger et al., 1995), oligodendrocyte precursor cells (Patneau et al., 1994; Bergles et al., 2000), astrocytes (Seifert and Steinhauser, 1995), and microglia (Noda et al., 2000).
B. Kainate Receptor Subunits
1. Regional Expression of Kainate Receptors Subunits
High-affinity binding sites for radiolabeled kainate reveal a high density of kainate receptors in the stratum lucidum of the CA3 region in the hippocampus, in the striatum, and in cerebellar granule cells, with less intense binding throughout other brain regions (Monaghan and Cotman, 1982). Subsequently, in situ hybridization shows a strong but not perfect correlation between high-affinity kainate binding sites in the hippocampus and the expression pattern of kainate receptor subunits GluK2 and GluK4 (Bettler et al., 1990; Egebjerg et al., 1991; Werner et al., 1991; Herb et al., 1992; Lomeli et al., 1992; Wisden and Seeburg, 1993; Bureau et al., 1999). The dense pattern of binding to a subregion of the CA3 arises from the targeting of GluK2/4-containing kainate receptors in CA3 pyramidal neurons to mossy-fiber synapses from dentate granule cells (Castillo et al., 1997; Vignes and Collingridge, 1997; Mulle et al., 1998). Deletion of GluK2 in mice eliminates high-affinity kainate binding sites in the CA3 stratum lucidum and in the dentate gyrus (Mulle et al., 1998).
To a first approximation, GluK1 is mostly expressed in interneurons, GluK2 is found in glutamatergic principal neurons, and GluK3 is found in a variety of neurons in several brain regions (Wisden and Seeburg, 1993). GluK4 is most restricted in its expression in the mature CNS and contrasts with the abundant and widespread distribution of GluK5. As with AMPA receptor subunits, however, these are generalizations, and expression varies within select neurons and brain regions. In the hippocampus, GluK1 transcripts are expressed predominantly by interneurons in the CA1 and CA3 regions (Bureau et al., 1999; Mulle et al., 2000; Paternain et al., 2000). GluK2-containing kainate receptors are found primarily in glutamatergic principal neurons, including pyramidal cells, hilar mossy cells, and dentate granule neurons as well as a subset of interneurons (Wisden and Seeburg, 1993; Bureau et al., 1999; Mulle et al., 2000; Paternain et al., 2000). GluK3 subunit mRNA is present in dentate granule cells and at very low levels elsewhere. GluK4 exhibits the most restricted expression pattern with dense expression in dentate granule cells and CA3 pyramidal cells but weak expression elsewhere (Kask et al., 2000) (Fig. 30). In fact, the GluK4 promoter was used to create a transgenic mouse with CA3-restricted expression of cAMP response element (CRE) recombinase (Nakazawa et al., 2002). Conversely, GluK5 is expressed in nearly all hippocampal regions and most neuronal subtypes (Herb et al., 1992; Wisden and Seeburg, 1993; Christensen et al., 2004) (Fig. 30). Kainate receptor signaling studies in most cases provide functional support for the distributions found in these studies (Fig. 30).
Fig. 30.
Kainate receptor expression in hippocampal CA3 pyramidal cells. (A) Electron micrograph showing mossy-fiber boutons (MFBs) and thorny excrescence spines (SPs) with gold particles for GluK2/3 labeling at the postsynaptic (blue) or the presynaptic membranes (red). (B) Left panel: GluK4 immunoperoxidase staining is evident in CA3 stratum lucidum (SL, arrowheads) but absent from the CA3 pyramidal cells in stratum pyramidale (SP, asterisk). Right panel: GluK5 staining is present in CA3 stratum lucidum and stratum pyramidale; SR indicates stratum radiatum. (C) Pairs of evoked mossy-fiber CA3 EPSCs recorded at several interstimulus intervals reveal paired-pulse facilitation in hippocampal slices from wild-type mice. Paired pulse facilitation is reduced in slices from GluK4 and GluK5 double-KO mice. (D) Mossy-fiber CA3 kainate receptor-mediated EPSCs isolated using the AMPA receptor inhibitor GYKI 53655 were evoked with short trains of stimulation (5 stimuli, 100 Hz) in slices from wild-type mice. Slices from GluK4 and GluK5 double KO mice show no detectable kainate receptor–mediated EPSCs. Reproduced with permission from Fernandes et al. (2009) (A, C, and D) and Darstein et al. (2003) (Copyright 2003 Society for Neuroscience) (B).
GluK3 and GluK5 mRNAs are expressed in most cortical layers in many regions, whereas GluK2 transcripts show dense expression in the cingulate cortex. GluK1 mRNA is distributed at a lower level throughout the cortex but with a notable density within the sensorimotor cortex. GluK4 signals are weaker than all other kainate receptor subunit transcripts (Wisden and Seeburg, 1993; Bahn et al., 1994). Subsequent RNA-sequencing analysis of broad classes of cortical neurons has identified GluK1 and GluK2 as differentially localized to cortical interneurons and GluK3 to pyramidal neurons in the S1 somatosensory cortex (Zeisel et al., 2015). Data from the Allen Brain Atlas (https://portal.brain-map.org/) confirm and extend the earlier in situ studies in the cortex: GluK1 is distributed throughout the lower layers of many cortical regions, likely in interneurons but also in a denser pattern in layer 4 in somatosensory cortices; GluK2 appears at highest levels in layer 5 neurons in a variety of neocortical regions; and GluK3 is found in all layers of the neocortex except layer 4.
In the cerebellum, mature granule cells and Golgi cells both express GluK2, and functional kainate receptors in these neurons are likely GluK2/5 (Wisden and Seeburg, 1993; Bahn et al., 1994; Ripellino et al., 1998; Bureau et al., 2000; Zhang et al., 2009b) in complex with the auxiliary subunit Neto2 (Tang et al., 2012). Kainate receptors in cerebellar granule cells are localized to presynaptic terminals at parallel fiber synapses and provide frequency-dependent homosynaptic regulation of glutamate release onto Purkinje neurons and Golgi cells (Delaney and Jahr, 2002). Purkinje cells express GluK1 and GluK4 mRNAs at low levels, and kainate receptors contribute a small component to climbing-fiber but not parallel-fiber synaptic currents (Huang et al., 2004). GluK3 transcripts are present in interneurons of the molecular layer, but their function in those cells remains unknown.
Kainate receptors are found in all other brain regions in overlapping expression patterns (Brandstatter et al., 1994). In the striatum, GluK2, GluK3, and GluK5 transcripts are most prominent (Wisden and Seeburg, 1993; Bischoff et al., 1997). GluK2-containing kainate receptors are present in striatal projection neurons, where they mediate a postsynaptic current at corticostriatal synapses (Fernandes et al., 2009) and modulate excitatory and inhibitory tone through indirect mechanisms involving A2A adenosine receptors (Chergui et al., 2000) or stimulation of endocannabinoid release (Marshall et al., 2018). GluK3 was localized to the substantia nigra pars compacta and ventral tegmental area, whereas GluK1 was expressed most prominently in the ventral pallidum and Islands of Calleja (Bischoff et al., 1997). Kainate receptor signaling can regulate GABAergic transmission in the basal ganglia (Jin and Smith, 2011).
Glutamate-activated currents in acutely dissociated DRG neurons almost entirely arise from GluK1/5 kainate receptors with Neto2 (Huettner, 1990; Sahara et al., 1997; Vernon and Swanson, 2017). Kainate receptor mRNAs are widely expressed in spinal cord neurons (Tolle et al., 1993). The localization of kainate receptors in nonpeptidergic nociceptors (Usoskin et al., 2015) and the analgesic activity of GluK1-selective antagonists [reviewed in Wu et al. (2007a), Bhangoo and Swanson (2013)], led to a drug discovery effort for pain relief in inflammatory hyperalgesia, postoperative, migraine-associated, and other forms of pain (Sang et al., 1998; Gilron et al., 2000; Sang et al., 2004). Central kainate receptors in the anterior cingulate cortex also play a role in pain and itch perception (Wu et al., 2007b).
2. Developmental Expression of Kainate Receptor Subunits
Kainate receptor expression begins early in embryonic development and changes in select regions before adulthood. For example, thalamocortical transmission in layer IV barrel cortex switches from kainate to AMPA receptor–mediated transmission (Bannister et al., 2005). In the hippocampus and other brain regions, GluK1 mRNA appears transiently in the early postnatal weeks before diminishing to undetectable levels as maturation progresses (Bahn et al., 1994). The early expression of GluK1- and Neto1-containing receptors in hippocampal pyramidal neurons regulates the development of nascent excitatory synapses (Lauri et al., 2005; Vesikansa et al., 2007, 2012; Clarke et al., 2014; Orav et al., 2017). Early studies of mossy-fiber kainate receptors using pharmacological tools and gene-targeted mice arrived at differing conclusions regarding contributions by GluK1 subunits. GluK1-selective antagonists block postsynaptic kainate receptor currents and NMDA receptor–independent LTP (Clarke et al., 1997; Bortolotto et al., 1999), but GluK1 KO mice show no apparent deficits in whole-cell kainate receptor currents or LTP (Contractor et al., 2000; Breustedt and Schmitz, 2004). This impasse may be explained by the transient postnatal expression of GluK1 subunits in CA3 pyramidal cells or compensatory gene expression in GluK1 KO mice. Developing synaptic connections between basolateral and central amygdalar neurons can be disrupted upon exogenous reduction or overexpression of GluK1 or GluK4 (Ryazantseva et al., 2020).
RNA editing of the Q/R/N site, first identified in GluA2 subunit transcripts, occurs in GluK1 and GluK2 mRNAs and progresses to different extents during an early neonatal time window (Sommer et al., 1991; Bernard and Khrestchatisky, 1994). Both GluK1 and GluK2 are substantially unedited during embryonic development in the rat and unlike GluA2 remain at least partially unedited into adulthood (Bernard and Khrestchatisky, 1994; Bernard et al., 1999). In the hippocampus, GluK1 is edited to ∼50%, whereas GluK2 mRNA is edited to a greater degree (70%–85%), and these proportions are generally consistent across many brain areas (Sommer et al., 1991; Bernard and Khrestchatisky, 1994; Bernard et al., 1999). RNA editing of the Q/R/N site in kainate receptors has similar functional consequences as for GluA2-containing AMPA receptors (i.e., elimination of Ca2+ permeability, reduced single-channel conductance, and insensitivity to intracellular polyamines), but the relevance of the changes in channel properties remains unclear. Mice expressing only unedited GluK2 are viable, exhibit no detectable behavioral or anatomic differences, and only a modest physiologic phenotype (Vissel et al., 2001b). In contrast, mice in which editing of GluA2 is genetically ablated develop lethal seizures within the first 3 postnatal weeks (Brusa et al., 1995), and mice lacking adenosine deaminase can be rescued from a similar phenotype by introducing alleles that encode edited GluA2 (Higuchi et al., 2000).
C. NMDA Receptor Subunits
1. Expression of GluN1/2 Receptors
Distinct expression patterns for GluN1 splice isoforms and GluN2 subtypes were established 3 decades ago, and recent advances in the sensitivity of single-cell sequencing and imaging methods have enriched our understanding of cell-type–specific gene expression. Furthermore, recently developed subtype-selective allosteric modulators (Section IX. Exogenous Positive and Negative Allosteric Modulators) have been used to assess functional contributions of NMDA receptor subtypes in specific cell types.
Consistent with its role as an obligate NMDA receptor subunit, GluN1 is expressed throughout the nervous system as early as E13 in rodents and gestational week 8 in humans and remains ubiquitous through adulthood (Moriyoshi et al., 1991; Watanabe et al., 1992; Ritter et al., 2001, 2002). In contrast, alternative splice isoforms of GluN1 are tightly regulated in a regional pattern that is established at birth and maintained through adulthood (Nakanishi et al., 1992; Laurie and Seeburg, 1994; Zhong et al., 1995; Paupard et al., 1997). The NTD splice isoforms of GluN1 influence NMDA receptor deactivation time course and pharmacology (Fig. 7 and Section IV. Receptor Activation, Deactivation, and Desensitization). The expression pattern of GluN1-b isoforms, which contain residues encoded by exon 5, is largely complementary to the pattern of those lacking exon 5 (GluN1-a isoforms). Exon 5–lacking transcripts are highly expressed in the telencephalon and weakly expressed in the thalamus, midbrain, and hindbrain, whereas transcripts containing exon 5 are abundant in the thalamus, midbrain, hindbrain, and spinal cord but restricted to only a few cortical areas, hippocampal CA3, and the dentate gyrus (Laurie and Seeburg, 1994; Luque et al., 1994; Paupard et al., 1997). Deletion of exon 5 thereby enables expression of only GluN1-a isoforms, disrupts developmental remodeling of NMDA receptors in the thalamus, and alters cortical excitatory connectivity (Liu et al., 2019a). However, hippocampal LTP is significantly enhanced in GluN1-a mice (i.e., mice with exon 5 deletion) compared with that in GluN1-b mice (i.e., mice with compulsory exon 5 expression), consistent with improved learning and spatial memory for GluN1-1a mice compared with GluN1-b mice (Sengar et al., 2019). In the cerebellum, 20% of GluN1 contains residues encoded by exon 5 early in development, compared with 80% of GluN1 in adult (Prybylowski et al., 2000), suggesting alternative splicing may be an important developmental factor (Paupard et al., 1997).
The expression of isoforms with alternative splicing in two regions of the intracellular CTD referred to as C1 and C2 (Fig. 7) is also regionally controlled with GluN1-2 isoforms expressed in nearly all brain regions, GluN1-1 isoforms restricted to the telencephalon and cerebellum, GluN1-3 isoforms displaying weak expression in the hippocampus, and GluN1-4 isoforms mirroring exon 5–containing transcripts with high expression in specific cortical areas, thalamus, midbrain, and hindbrain (Laurie and Seeburg, 1994; Paupard et al., 1997). Furthermore, cell-type–specific expression of splicing factors indicates that GluN1 splicing likely differs between excitatory and inhibitory neurons within specific brain regions (Furlanis et al., 2019). In addition, the C1 region contains a nuclear localization signal, raising the possibility of regulation by intracellular protease cleavage (Zhou and Duan, 2018).
The different spatial gene expression patterns of GluN2 subunits are major determinants of NMDA receptor functional diversity across the CNS. The GluN2B and GluN2D subunits are highly expressed during embryonic development, whereas GluN2A and GluN2C expression increases postnatally (Watanabe et al., 1992, 1993; Akazawa et al., 1994; Monyer et al., 1994; Wenzel et al., 1996, 1997). GluN2A mRNA expression in rodents begins a few days after birth and becomes highly expressed in nearly all CNS regions. GluN2B expression remains high in most brain regions through early postnatal development and then becomes restricted primarily to the forebrain. The increase in synaptic GluN2A expression and coordinated reduction in synaptic GluN2B expression is regulated by activity during development in the cortex, hippocampus, amygdala, thalamus, and cerebellum (Tovar and Westbrook, 1999; Philpot et al., 2001; Barria and Malinow, 2002; Lopez de Armentia and Sah, 2003; Gray et al., 2011). This developmental GluN2B-GluN2A switch occurs over timelines that differ across cortical regions and between GABAergic interneurons and principal glutamatergic cells within the forebrain (Dumas, 2005; Wang and Gao, 2009; Mierau et al., 2016). Furthermore, some cell types, such as neurons in the dorsal horn, do not undergo a developmental loss of synaptic GluN2B (Hildebrand et al., 2014; Mahmoud et al., 2020). The relative levels of synaptic GluN2A and GluN2B also differ between anatomically distinct inputs to a single neuron type in the cortex, hippocampus, striatum, thalamus, and retina (Fritschy et al., 1998; Kumar and Huguenard, 2003; Miyata and Imoto, 2006; Shinohara et al., 2008; Kalbaugh et al., 2009; Carta et al., 2018; Li and Pozzo-Miller, 2019).
GluN2C expression begins at approximately postnatal day 10 in the rodent, and it is abundantly expressed in cerebellar granule cells, lateral nuclei of the dorsal thalamus, and the glomerular layer and mitral cells of the olfactory bulb (Watanabe et al., 1993; Akazawa et al., 1994; Monyer et al., 1994). GluN2C is weakly expressed in the cortex, hippocampus, striatum, and amygdala, and recent evidence suggests that GluN2C is expressed primarily in astrocytes and not neurons in these regions (Karavanova et al., 2007; Ravikrishnan et al., 2018; Alsaad et al., 2019). Several reports suggest GluN2C is also expressed in oligodendrocytes in the white matter of the cerebellum, corpus callosum, and optic nerve (Karadottir et al., 2005; Salter and Fern, 2005; Micu et al., 2006; Burzomato et al., 2010; Doyle et al., 2018). In the spinal cord, GluN2C was weakly detected in the dorsal horn and non-neuronal cells in white and gray matter in the lumbar region (Tolle et al., 1993; Sundstrom et al., 1997; Shibata et al., 1999; Akesson et al., 2000) as well as the ventral horn early in postnatal development (Stegenga and Kalb, 2001), but was not detectable in the cervical spinal cord (Watanabe et al., 1994). Functional studies utilizing GluN2C-selective pharmacology support neuronal expression in the cerebellum, thalamus, and globus pallidus (Fernandez et al., 2017; Bhattacharya et al., 2018; Liu et al., 2019b, 2021).
GluN2D is widely expressed in the CNS during embryonic and early postnatal development, but after the first postnatal week GluN2D expression becomes restricted to GABAergic interneurons in the cortex and hippocampus (Fig. 31), cholinergic interneurons in the striatum, and select neurons within the thalamus, basal ganglia, bed nucleus of the stria terminalis, substantia gelatinosa of the spinal cord, and cerebellum (Tolle et al., 1993; Akazawa et al., 1994; Monyer et al., 1994; Standaert et al., 1994, 1996; Wenzel et al., 1996; Dubois et al., 2016; Salimando et al., 2020). GluN2D is not restricted to particular interneuron classes in the cortex and hippocampus and is commonly expressed in 2parvalbumin- and somatostatin-positive cells (von Engelhardt et al., 2015; Perszyk et al., 2016). Recent functional studies utilizing subtype-selective pharmacology support GluN2D expression in cortical and hippocampal interneurons (von Engelhardt et al., 2015; Perszyk et al., 2016; Swanger et al., 2018; Yi et al., 2019, 2020; Garst-Orozco et al., 2020), striatum (Feng et al., 2014; Zhang et al., 2014b,c; Nouhi et al., 2018), subthalamic nucleus (Swanger et al., 2015, 2018; Yi et al., 2020), substantia nigra (Pearlstein et al., 2015; Wu and Johnson, 2015; Morris et al., 2018b; Sitzia et al., 2020), and spinal cord (Hildebrand et al., 2014; Mahmoud et al., 2020).
Fig. 31.
NMDA receptor expression in hippocampal CA1 neurons. (A) Upper panel: fluorescence in situ hybridization for CB1 cannabinoid receptor, parvalbumnin, and Grin2d mRNA. Lower panel: the percentage of CA1 interneurons showing the GluN2 subunit expression based on interneuron classification as determined by single-cell reverse transcription PCR (RT-PCR). PV, parvalbumin; CCK, cholecystokinin; SOM, somatostatin; NPY, neuropeptide Y. Adapted with permission from Perszyk et al. (2016). (B) Paired recordings of outward NMDA receptor–mediated evoked EPSCs (VHOLD +40 mV) from wild-type CA1 pyramidal neurons (black) and pyramidal neurons lacking GluN2A, GluN2B, or both subunits (green). Scale bars as are 25 pA and 100 milliseconds. Adapted with permission from Gray et al. (2011). (C) Whole-cell current recording of evoked EPSCs from either CA1 pyramidal cells (C1) or interneurons (C2) in the absence (black) or presence (blue) of the GluN2A-selective negative allosteric modulator MPX-004, the GluN2B-selective negative allosteric modulator CP-101,606, or the GluN2C/D-selective negative allosteric modulator NAB-14. These data demonstrate pyramidal cell expression of GluN2A/B and interneuron expression of GluN2B/2D. Scale bars are 20 pA and 100 milliseconds. Adapted with permission from Yi et al. (2019) and Swanger et al. (2018) (Copyright 2018 American Chemical Society).
2. Expression of GluN3 Subunits
The spatial and temporal expression of GluN3A and GluN3B provides hints to the roles of these subunits [reviewed in Low and Wee (2010), Perez-Otano et al. (2016)]. Expression of the GluN3A protein is low in the brain during embryonic stages, peaking in the first two weeks of postnatal development and declining into adulthood (Chan and Sucher, 2001; Wong et al., 2002; Sucher et al., 2003; Mueller and Meador-Woodruff, 2005; Wee et al., 2016; Murillo et al., 2021). GluN3A mRNA is expressed in the thalamus, entorhinal cortex, subiculum, neocortex, hippocampus, spinal cord, medulla, pons, tegmentum, and hypothalamus (Ciabarra et al., 1995; Sucher et al., 1995; Das et al., 1998; Sun et al., 1998; Wong et al., 2002; Domingues et al., 2011; Murillo et al., 2021). Adult mice show distinct laminar GluN3A expression in the granular, mitral, and glomerular cell layers of the olfactory bulb (Lee et al., 2016b). GluN3A is functionally expressed in hippocampal CA1 principal cells (Grand et al., 2018; Zhu et al., 2020), neurons of the adult mouse medial habenula (Otsu et al., 2019), and possibly astrocytes [Lee et al. (2010a), Palygin et al. (2011), Zhang et al. (2014d); reviewed in Verkhratsky and Chvatal (2020)] and oligodendrocytes (Karadottir et al., 2005; Salter and Fern, 2005; Micu et al., 2006; Burzomato et al., 2010; Pina-Crespo et al., 2010). The expression of GluN3A is well timed to play a role in neuronal differentiation, migration, and synapse formation (Ciabarra et al., 1995; Sucher et al., 1995; Wong et al., 2002; Murillo et al., 2021). The spatial and temporal expression of GluN3A overlaps with the timing of spine and synapse development, and GluN3A KO mice show increased spine density (Section V.D.7. Kainate Receptor Function in Postsynaptic Signaling).
GluN3B has a different spatial and temporal expression than GluN3A (Chatterton et al., 2002; Matsuda et al., 2002; Wee et al., 2016), being either weakly expressed or absent in the embryonic and neonatal brain (Fukaya et al., 2005a; Ishihama et al., 2005; Ishihama and Turman, 2006; Liu and Wong-Riley, 2010; Wee et al., 2016). GluN3B expression starts to increase in the early postnatal period (P7) and maintain expression levels into adulthood (Wee et al., 2008, 2016). Spatially, GluN3B is highly expressed in pons, midbrain, medulla, and spinal cord but at low levels in forebrain and cerebellum (Matsuda et al., 2002; Wee et al., 2008). In the spinal cord, GluN3B shows a different temporal expression pattern compared with the brain. GluN3B is expressed in somatic motoneurons as early as embryonic day 16, peaking at postnatal day 28, and have been hypothesized to be neuroprotective for these neurons (Nishi et al., 2001; Matsuda et al., 2003; Fukaya et al., 2005a; Prithviraj and Inglis, 2008).
D. GluD Receptors
GluD1 expression is higher in younger animals, especially in the striatum and the anteroventral thalamic nucleus, whereas in the adult, GluD1 is expressed moderately in the cerebral cortex, hippocampus, dorsal raphe, striatum, central nucleus of the amygdala, and cerebellar cortex (Konno et al., 2014; Hepp et al., 2015; Nakamoto et al., 2020b). GluD1 expression is stronger than GluD2 in the hippocampus, dentate gyrus, piriform cortex, central nucleus of the amygdala, oval nucleus of the bed nucleus of the stria terminalis, and parafascicular thalamic nucleus (Nakamoto et al., 2020b). In the cerebellar cortex, GluD1 is expressed in molecular layer interneurons, where the receptors are localized at the parallel fiber–molecular layer interneuron synapses (Konno et al., 2014). In the auditory system, GluD1 mRNA is highly expressed in the inner hair cells, outer hair cells, spiral ganglia, and vestibular hair cells, and GluD1 KO mice showed hearing loss at high frequencies (>16 kHz) (Safieddine and Wenthold, 1997; Gao et al., 2007). The expression of GluD1 may depend on their role as trans-synaptic organizers mediated through the presynaptic terminal and thus may be related to expression of the Cbln family of proteins with which they interact (Section V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity).
GluD2 subunits expressed in cerebellar Purkinje cells play a role in motor coordination and motor learning [reviewed in Hirano (2006), Mandolesi et al. (2009), Yuzaki and Aricescu (2017)]. GluD2 is in distal dendrites of Purkinje cells that receive synaptic input from the parallel fibers, whereas proximal dendrites that receive input from climbing fibers (axons of inferior olivary nucleus) and molecular layer interneurons (stellate and basket cells) are devoid of GluD2 (Takayama et al., 1996; Landsend et al., 1997). GluD2 is also expressed in the olfactory glomerular layer, cingulate cortex, medial prefrontal cortex, retrosplenial granular cortex, olfactory tubercle, subiculum, striatum, anterodorsal thalamic nucleus, lateral septum, and arcuate hypothalamic nucleus (Nakamoto et al., 2020b). GluD2 mRNA is preferentially expressed in glutamatergic neurons in the cortical regions and by various neurons in subcortical regions (Nakamoto et al., 2020b).
E. Glutamate Receptor Subunits in Peripheral Tissues
Virtually all of the glutamate receptor subunits are expressed in peripheral tissues. This likely reflects the body’s ability to repurpose signaling molecules for the unique roles various cells play. There is ample precedent for this in biology, and although terminal differentiation can reduce active gene sets, every cell still has access to every gene. A comprehensive review of expression of glutamate receptor subunits in all peripheral tissues would be enormous, and thus we provide here only a summary of tissues and references for the most commonly discussed expression patterns (Supplemental Table 3).
VII. Pharmacology of Orthosteric Ligands and Channel Blockers
The functional iGluR classes were first identified based on the selective activation or inhibition by orthosteric ligands (i.e., agonists and competitive antagonists) or channel blockers [reviewed in Mayer and Westbrook (1987b), Collingridge and Lester (1989), Lodge (2009), Dawe et al. (2015), Lodge et al. (2019)]. After cloning of the different iGluR subunits, it became possible to quantitatively determine the activity of these ligands at recombinant iGluR subtypes expressed in heterologous expression systems. This approach facilitated the identification of new classes of orthosteric ligands and channel blockers with diverse chemical structures, some of which displayed subunit selectivity. More recently, X-ray crystallography and molecular dynamics have provided a more detailed understanding of binding mechanisms and the structural basis for subunit selectivity.
A. Agonists
The initial conformational change that triggers receptor activation is mediated by agonist binding and closure of the bilobed ABD, in which the upper and the lower (membrane-proximal) lobes are denoted D1 and D2, respectively. The agonist binding pocket is located in the cleft formed between D1 and D2, which are tethered by a highly flexible “hinge” region (Section II. Receptor Structure). Many structural features of agonist binding are conserved in all iGluR subunits, but some key differences exist that enable the design of subunit-selective ligands. Central to all iGluR agonists is the requirement that the chemical structure include a moiety equivalent to the backbone portion of glutamate and glycine/d-serine, namely the α-amino and α-carboxyl groups (i.e., the amino acid moiety). The region of the binding pocket that harbors the amino acid moiety primarily comprises residues from D1 and is similar in all iGluR subunits (Fig. 32). Agonists initially form interactions with the D1 lobe, primarily driven by the strong electrostatic interaction between the agonist α-carboxyl group and an arginine residue, which is conserved in all iGluR subunits. Once docked in the D1 region of the binding pocket, the agonist can recruit interactions with the D2 region, thereby closing the ABD (Abele et al., 2000; Armstrong and Gouaux, 2000; Cheng et al., 2005; Yu and Lau, 2018; Yu et al., 2018a). The iGluR subunits display greater structural variation in the D2 region of the pocket, consistent with the diversity in endogenous agonists (glutamate vs. glycine/d-serine). The agonist binding pockets are conserved among those iGluR subunits that bind glutamate (GluA1-4, GluK1-5, GluN2A-D) and are more variable in those subunits that bind glycine/d-serine (GluN1, GluN3A-B, GluD1-2) (Fig. 32). Because of this conservation, there are only few examples of agonists that display subunit selectivity.
Fig. 32.
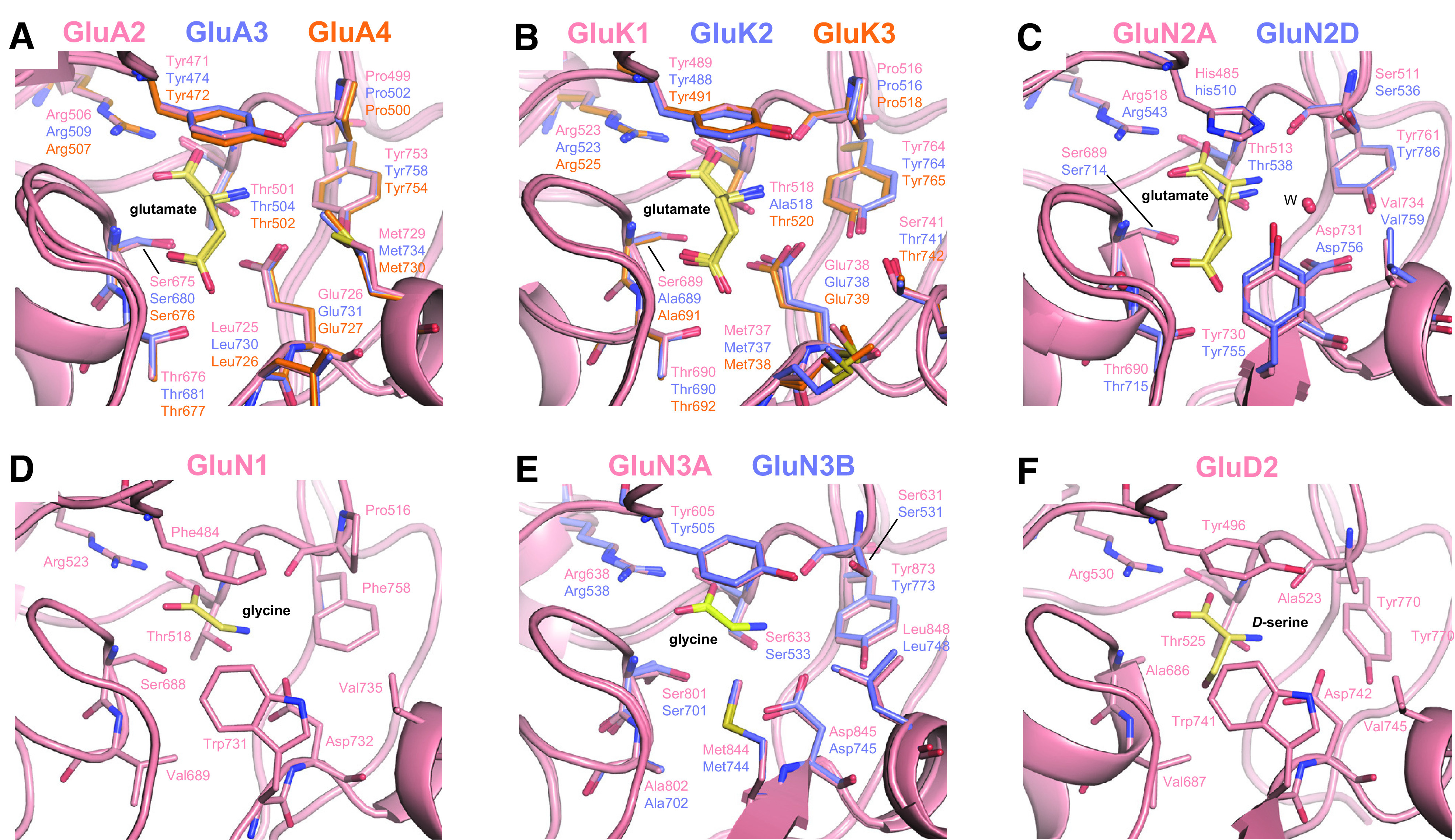
Agonist binding pockets in iGluR subunits. (A) Binding of glutamate (yellow) in the agonist binding pocket of GluA2 (PDB: 1FTJ), GluA3 (PDB: 3DLN), and GluA4 (PDB: 3EPE). Only side chains of key interacting residues are shown. (B) Binding of glutamate in GluK1 (PDB: 2F36), GluK2 (PDB: 1S7Y), and GluK3 (PDB: 4MH5). Compared with the agonist binding pocket of GluA2, there is a loss of a direct hydrogen bond to the α-amino group of glutamate at position Ala518 in GluK2, which is equivalent to Thr501 in GluA2. An additional water molecule forms a hydrogen bond to the α-amino group of glutamate in GluK2 (not shown). (C) Binding of glutamate in GluN2A (PDB: 5I57) and GluN2D (PDB: 3OEL). Compared with GluA2, the salt bridge between Asp731 and the α-amino group of glutamate is absent. Instead, the α-amino group of glutamate forms water-mediated hydrogen bonds to Asp731, Glu413, or Tyr761 (W indicates water). (D) Binding of glycine in GluN1 (PDB: 5I57). Specificity of GluN1 for glycine can be explained by the hydrophobic environment created by Val689 and the steric barrier formed by Trp731. (E) Binding of glycine in GluN3A (PDB: 2RC7) and GluN3B (PDB: 2RCA). Trp731 of GluN1 is replaced by GluN3A Met844. (F) Binding of D-serine in GluD2 (PDB: 2V3U).
1. AMPA and Kainate Receptor Agonists
A wide range of natural products, including kainate, ibotenic acid, domoate, quisqualate, willardiine, and dysiherbaine, and synthetic analogs thereof are agonists at AMPA and kainate receptors (Supplemental Tables 4 and 5). Considerable efforts have been devoted to the development of analogs of these natural products that are capable of discriminating between AMPA and kainate receptors but with limited success (Brauner-Osborne et al., 2000; Stensbol et al., 2002; Jane et al., 2009; Vogensen et al., 2011). For example, the agonist AMPA is a partial agonist at some kainate receptor subtypes (Herb et al., 1992; Swanson et al., 1996; Mott et al., 2010). Similarly, the agonist kainate is also a partial agonist at AMPA receptors (Patneau et al., 1993). Nonetheless, some agonists with selectivity for AMPA receptors over kainate receptors (except GluK1), and agonists with selectivity for kainate receptors have been developed (Supplemental Tables 4 and 5). By now, several hundred crystal structures of isolated AMPA and kainate receptor subunit ABDs provide detailed insights into ligand-receptor interactions, and these structures reveal the molecular basis for the selectivity of agonists between iGluR classes [reviewed in Pohlsgaard et al. (2011), Kumar and Mayer (2013), Mollerud et al. (2017a)] (Fig. 32).
AMPA receptor agonists show variation in the extent of desensitization they induce during prolonged activation. Glutamate and AMPA are full agonists at AMPA receptors (i.e., high agonist efficacy) that activate strongly desensitizing current responses, whereas kainate is a partial agonist (i.e., lower agonist efficacy) that induces rapid but incomplete desensitization (Patneau and Mayer, 1991; Patneau et al., 1993; Armstrong and Gouaux, 2000). Similarly, a series of 5-substituted willardiines act as partial agonists at AMPA receptors with varying levels of agonist efficacy and desensitization (Patneau et al., 1992; Jin et al., 2003). The correlation between agonist efficacy and desensitization for AMPA and kainate receptor agonists results in widely different values for agonist potencies and efficacies being reported for steady-state responses and peak responses, thereby complicating comparisons of agonist efficacy reported in studies using different functional assays. Similarly, it is difficult to compare agonist activities determined on native receptors with those determined on recombinant receptors, since neuronal receptors presumably express a mixed population of receptor subtypes with a varied complement of auxiliary subunits (Tables 2 and 3 in Section IV. Receptor Activation, Deactivation, and Desensitization). For example, auxiliary subunits can convert some competitive antagonists (e.g., DNQX and CNQX) into partial agonists for AMPA receptors (Section III.1.b. Functional effects of TARPs).
Efforts to develop agonists selective for a single AMPA receptor subunit have been unsuccessful. However, Br-HIBO, an analog of ibotenic acid (Coquelle et al., 2000), and CPW399, a willardiine analog (Campiani et al., 2001), can preferentially activate GluA1, GluA2, and GluA4 over GluA3 receptors (Supplemental Table 4). In part, this subunit preference is enabled by water-mediated hydrogen bonding between the agonists and a binding pocket Tyr in GluA1 and GluA2, which is Phe in GluA3 and GluA4 (Hogner et al., 2002; Frandsen et al., 2005). Molecular modeling predicted that the exchange of bromine for chlorine would improve selectivity, resulting in Cl-HIBO, which activates GluA1-2 subunits with 275- to 1600-fold selectivity over GluA3-4 subunits (Bjerrum et al., 2003; Ahmed et al., 2009b).
The AMPA receptor agonists AMPA, 2-amino-3-(3-hydroxy-5-tert-butylisoxazol-4-yl)propanoic acid (ATPA), 2-Me-Tet-AMPA, and 2-Bn-Tet-AMPA have increasingly bulky substituents in the 5-position of the isoxazole ring (Supplemental Table 4). Crystal structures revealed that these substituents point toward a side cavity in the agonist binding pocket that is formed by residues in both D1 and D2, including Met729 (Armstrong and Gouaux, 2000; Hogner et al., 2002; Lunn et al., 2003; Vogensen et al., 2007). The side chain of Met729 in GluA2 adopts widely different conformations to optimize the van der Waals interaction with the bulky substituents of the agonists (Fig. 32). Thus, the volume of the side cavity changes to accommodate agonists of varying sizes, resembling an induced fit mechanism of binding. The increase in bulk of the substituent in 2-Bn-Tet-AMPA (methyl to benzyl) resulted in >10-fold preference for GluA4 over GluA1-3 (Jensen et al., 2007), which might result from interactions with nonconserved residues in the cavity (Vogensen et al., 2007).
Kainoids are natural products composed of a 2-carboxypyrrolidine-3-acetic acid backbone, some of which are agonists at AMPA and kainate receptors, including domoate and kainate [reviewed in Jane et al. (2009), Tian et al. (2019)] (Supplemental Table 5). Heteromeric kainate receptor subtypes (e.g., GluK2/4 and GluK2/5) can have bell-shaped steady-state agonist concentration-response relationships due to initial binding of agonists to the high-affinity GluK4-5 subunits and, at higher agonist concentrations, binding to the low-affinity GluK1-3 subunits (Mott et al., 2010; Fisher and Mott, 2011, 2013; Fisher and Fisher, 2014). Agonist binding to GluK4-5 mediates activation with low desensitization (i.e., rising phase of the concentration-response curve), whereas agonist binding to GluK1-3 produces desensitization (i.e., falling phase). This dependence on subunit composition is at least in part due to the relationship between agonist binding (i.e., occupancy) and channel activation, in which partial occupancy of agonist binding sites in both homomeric or heteromeric receptors can be sufficient for activation but may not be sufficient for desensitization (Swanson et al., 2002; Fisher and Mott, 2011; Pinheiro et al., 2013; Fisher, 2014; Fisher and Fisher, 2014; Reiner and Isacoff, 2014; Pollok and Reiner, 2020).
The structural basis for the higher binding affinity of kainate at GluK4-5 over GluK1-3 has been investigated in a crystal structure of the isolated GluK4 ABD in complex with kainate (Kristensen et al., 2016b). In part, the higher binding affinity could be afforded by a more optimal shape complementarity with Ile670 in GluK4 (Ile669 in GluK5); the residue at this position is Leu in GluA1-4 and Val in GluK1-3. Two characteristics of the agonist binding pocket in GluA2 are conserved in GluK4-5. First, Met729 in GluA2, which undergoes major conformational changes to adopt the bound ligand (Pohlsgaard et al., 2011), is conserved in GluK4-5 and absent in GluK1-3. Second, an interdomain lock in GluA2 formed by Glu423 in D1 and Thr707 in D2 (Armstrong and Gouaux, 2000) is conserved in GluK4-5.
In contrast to kainate, the marine toxin dysiherbaine has high affinity for GluK1-2 and low affinity for GluK5 (Sakai et al., 2001; Swanson et al., 2002; Sanders et al., 2005, 2006). The high affinity of dysiherbaine for GluK1 promotes strong desensitization that persists minutes after agonist removal, enabling the use of dysiherbaine as a selective functional antagonist of GluK1 subunits in heteromeric GluK1/K5 receptors (Swanson et al., 2002). Neodysiherbaine is a naturally occurring analog that shows lower affinity for GluK1-2 and higher affinity for GluK5 compared with dysiherbaine (Sanders et al., 2005). However, the synthetic analogs 8-deoxy-neodysiherbaine (partial agonist), 9-deoxy-neodysiherbaine (full agonist), and MSVIII-19 show nanomolar affinity for GluK1 and more than 1000-fold selectivity for GluK1 over GluK2, GluK3, and GluK5 (Lash et al., 2008). MSVIII-19 was initially presumed to act as a GluK1 antagonist (Sanders et al., 2005), but a crystal structure of the isolated GluK1 ABD revealed full cleft closure, and functional studies show that MSVIII-19 is an agonist with very low efficacy (i.e., functional antagonist) (Frydenvang et al., 2009). Crystal structures of GluK1 and GluK2 ABDs bound to dysiherbaine analogs identified three nonconserved amino acids in the agonist binding pocket as the structural basis for the selectivity between GluK1 and GluK2 subunits (Unno et al., 2011).
Other agonists, including ATPA, 2-amino-3-(3-hydroxy-7,8-dihydro-6H-cyclohepta[d]isoxazol-4-yl)propionic acid (AHCP), 8-Me-AHCP, iodowillardiine, CBG-IV, (4R)-isopentyl glutamate, and LY339434, are potent agonists selective for GluK1, with almost no activity at GluK2 (Clarke et al., 1997; Jane et al., 1997; Small et al., 1998; Brehm et al., 2003; Bunch et al., 2009; Clausen et al., 2009; Juknaitė et al., 2012). This selectivity is, in some cases, mediated by steric occlusion in the agonist binding pocket that prevents the accommodation of bulky groups on agonists. For example, the larger binding cavity in GluK1 relieves steric occlusion of the bulky tert-butyl group of ATPA and the halogen atom of iodowillardiine (Mayer, 2005). The importance of cavity size has been further demonstrated by mutagenesis (Swanson et al., 1997a, 1998; Nielsen et al., 2003). Steric occlusion also mediates the selectivity for some agonists toward kainate receptors over AMPA receptors. The agonist binding cavities of kainate receptor subunits GluK1 and GluK2 are larger and can accommodate bulkier agonists compared with the AMPA receptor subunit GluA2 (Mayer, 2005; Naur et al., 2005; Venskutonytė et al., 2011b; Veran et al., 2012; Kristensen et al., 2016b). For example, GluA2 Leu671 is a smaller valine residue in GluK1-3 and Ile in GluK4-5, thereby enabling higher affinity binding by accommodating the 4-methyl group of SYM2081 or the 4-isoprenyl group of kainate (Armstrong et al., 1998, 2003; Mayer, 2005). Interestingly, the conformationally restricted glutamate/kainate analog CIP-AS has strong preference for GluK3 over GluK1, GluK2, and AMPA receptor subunits (Conti et al., 1999; Mollerud et al., 2017b).
The availability of crystallographic data for AMPA and kainate receptor ABDs has enabled molecular dynamics simulations to investigate mechanisms of agonist binding (Arinaminpathy et al., 2002, 2006; Lau and Roux, 2007, 2011; Postila et al., 2010, 2011; Sahai and Biggin, 2011; Yu and Lau, 2017; Yu et al., 2018a; Wied et al., 2019). Furthermore, ABD crystal structures have provided opportunities to design light-activated or light-inhibited AMPA and kainate receptor subunits [Volgraf et al. (2006), Gorostiza et al. (2007), Szobota et al. (2007), Numano et al. (2009), Janovjak et al. (2010), Reiner and Isacoff (2014), Carroll et al. (2015), Levitz et al. (2016); reviewed in Bregestovski et al. (2018)].
2. Agonists for Glutamate Binding NMDA Receptor Subunits (GluN2)
The GluN1/GluN2 NMDA receptors are unique among iGluRs by having a strict requirement of simultaneous binding of two distinct agonists for activation, namely glycine or d-serine to GluN1 and glutamate to GluN2 (Fig. 32 and Section II. Receptor Structure). The mechanism underlying selectivity of the agonist NMDA for GluN2 subunits was first suggested from a crystal structure of the glutamate/glycine-bound GluN1/2A ABD heterodimer (Furukawa et al., 2005) and later revealed in a GluN2D ABD structure in complex with NMDA (Vance et al., 2011). In the GluN2 binding pocket, a conserved Asp residue in D2 (Asp731 in GluN2A) is one methylene group shorter than the Glu residue found in AMPA and kainate receptor subunits (Fig. 32). Unlike the Glu residue in AMPA and kainate receptor subunits that directly interacts with the agonist, the Asp residue in GluN2 subunits interact with agonist α-amino groups via water-mediated hydrogen bonds (Furukawa et al., 2005; Vance et al., 2011; Hansen et al., 2013; Jespersen et al., 2014; Hackos et al., 2016; Volgraf et al., 2016; Yi et al., 2016; Chou et al., 2020). The reduced side chain of this Asp residue therefore creates space for the N-methyl group of NMDA in the GluN2 binding pocket, since this group can displace the water molecule that otherwise binds the α-amino group of glutamate (Vance et al., 2011). However, the position of the N-methyl group of NMDA in a hydrophilic subsite is less favorable, presumably resulting in the relative low potency of NMDA at the different NMDA receptor subtypes (Supplemental Table 6).
Several other endogenous agonists for GluN2 subunits have been identified, such as d- and l-aspartate, homocysteate, and cysteinesulfinate (Supplemental Table 6). Furthermore, a number of agonists that are selective for NMDA receptors over AMPA and kainate receptors are derived from the natural product ibotenic acid (or ibotenate) isolated from the toxic mushroom Amanita muscaria [reviewed in Risgaard et al. (2010)], such as the NMDA receptor-selective agonist AMAA (Madsen et al., 1996) (Supplemental Table 6). Glutamate adopts several conformations in the agonist binding pocket, but the introduction of ring systems in the agonist structure limits the number of possible conformations, resulting in improved binding affinity if the conformational restriction resembles a favorable conformation adopted by glutamate. Many GluN2 agonists are conformationally restricted glutamate analogs, including L-CCG-IV, trans-ACBD, cisACPD, and homoquinolinate (Supplemental Table 6). L-CCG-IV is the most potent conformationally restricted analog, and the potent agonist (RS)-(tetrazol-5-yl)glycine is conformationally restricted with a tetrazole as the carboxylate bioisostere (Schoepp et al., 1991) (Supplemental Table 6).
All the residues lining the agonist binding pocket are conserved among GluN2 subunits (Fig. 32), explaining why efforts to design GluN2-selective agonists have not been successful (Supplemental Table 6). However, agonist potencies display a graded variation among GluN2 subunits, with the lowest potency at GluN2A- and the highest potency at GluN2D-containing receptors (Erreger et al., 2007; Hansen et al., 2008) (Supplemental Table 6). This trend is profound for the agonist SYM2081, which also binds kainate receptors and displays a 46-fold lower EC50 value for GluN2D-containing receptors compared with GluN2A-containing NMDA receptors (Erreger et al., 2007). Most partial agonists also display variation in efficacy among GluN2 subunits (Supplemental Table 6). Consistent with the conserved agonist binding pocket, the structural determinants of the differences in agonist potency and efficacy between the GluN2 subunits reside in the NTD, suggesting that interdomain interactions between the ABD and the NTD influence the manner by which agonist binding leads to channel activation (Gielen et al., 2009; Yuan et al., 2009; Hansen et al., 2013). Crystal and cryo-EM structures have provided structural data that can facilitate studies of these interactions [Esmenjaud et al. (2019), Chou et al. (2020); reviewed in Wang and Furukawa (2019)].
Agonists with some GluN2 subunit selectivity in terms of agonist efficacy have been developed from the partial agonist NHP5G that relies on N-hydroxypyrazole as bioisostere for the distal carboxylate in the agonist aspartate (Clausen et al., 2004, 2008). Substituted NHP5G analogs showed GluN2 subunit selective agonist efficacy, with the strongest effects observed for ethyl- and propyl-NHP5G that have broad ranges of agonist efficacies among GluN2 subunits (Supplemental Table 6). The maximal responses to ethyl- and propyl-NHP5G relative to glutamate are 5% and ∼0%, 2D (/2A, but 72% and 45%, respectively, at GluN1/2D (Hansen et al., 2013). Single-channel recordings revealed that propyl-NHP5G activates brief channel openings of GluN1/2A with low probability, confirming that propyl-NHP5G is a partial agonist with very low efficacy (i.e., functional antagonist) at GluN1/2A (Hansen et al., 2013). The substituents of the NHP5G analogs protrude toward a Val residue in the GluN2 binding pocket, and the agonist efficacy of the NHP5G analogs correlated with the extent of predicted steric clash (i.e., Van der Waals interaction) with this Val residue (Hansen et al., 2005, 2013). Thus, larger substituents on NHP5G (e.g., propyl) reduce agonist efficacy. The 4-methyl substitution on the glutamate backbone in SYM2081 is predicted from molecular modeling to protrude toward the same Val residue in the GluN2 binding pocket (Erreger et al., 2007). The NHP5G analogs displayed promising discrimination between GluN2 subunits in terms of agonist efficacy, albeit the potencies of the analogs are relatively low (Supplemental Table 6). Efforts to increase potency while maintaining the variation in agonist efficacy among GluN2 subunits led to a series of 4-substituted CCG analogs (Risgaard et al., 2013) that combined the features of NHP5G that influence agonist efficacy with conformational restriction of the glutamate backbone to improve potency. The potencies of ethyl- and propyl-CCG considerably improved compared with ethyl- and propyl-NHP5G, but the CCG analogs did not show strong GluN2-specific efficacy (Supplemental Table 6).
Crystallographic data for GluN2 ABDs have facilitated molecular dynamics simulations to investigate the mechanisms of agonist binding to wild-type receptors (Erreger et al., 2007; Yao et al., 2013; Dai and Zhou, 2016; Yu and Lau, 2018) and NMDA receptors harboring human variants in the ABD (Swanger et al., 2016; Wells et al., 2018). This information has also enabled the design of light-activated or light-inhibited NMDA receptor subunits (Berlin et al., 2016). Extended molecular dynamics simulations suggested that positively charged residues on the surface of the GluN2A ABD facilitate glutamate binding by guiding (or “funneling”) the agonist to the binding pocket (Yu and Lau, 2018). Similarly, “guided-diffusion” mechanism for glutamate binding may also occur for the GluA2 ABD (Yu et al., 2018a). By contrast, the extended molecular dynamics simulations suggested that glycine binding to the GluN1 ABD occurs by “unguided diffusion” (Yu and Lau, 2018).
3. Agonists for Glycine/d-Serine Binding Subunits (GluN1, GluN3, GluD)
The agonist binding pockets of the glycine/d-serine–binding iGluR subunits lack D2 residues to stabilize a distal carboxylate group found in glutamate but form interactions between D1 residues and the agonist amino acid moiety similar to those in glutamate binding subunits (Fig. 32). The selectivity for either glycine or glutamate is primarily but not entirely mediated by two divergent residues that change the nature of the agonist binding pocket in GluN1 and GluN2 subunits (Furukawa and Gouaux, 2003; Furukawa et al., 2005; Inanobe et al., 2005). In GluN2A, the side chain of Tyr730 is in van der Waals contact with the γ-carboxylate of glutamate, which is further stabilized by hydrogen bonding to Thr690 (Fig. 32). The residue corresponding to GluN2A Tyr730 is the larger Trp731 in GluN1 that sterically occludes the γ-carboxylate of glutamate and provides a tighter fit with glycine or d-serine (Fig. 32). Furthermore, the hydrophobic Val689 in GluN1 replaces GluN2A Thr690, since this hydrogen bonding is not required for stabilization of glycine or d-serine. In addition to glycine, both d- and l- isomers of alanine and serine are GluN1 agonists (Supplemental Table 7).
d-Serine and glycine, endogenous coagonists of GluN1/2 NMDA receptors, are present in CSF at concentrations (2–6 μM) sufficient to result in partial or, in some cases, full saturation of GluN1 agonist binding sites [D'Souza et al. (2000), Madeira et al. (2015); reviewed in Mothet et al. (2015)]. Intraparenchymal concentrations of glycine and d-serine in vivo are not yet established, but concentrations in the 0.5–10 µM range have been reported (Bergeron et al., 1998; Billups and Attwell, 2003; Ishiwata et al., 2015; Bae et al., 2021). d-Serine in vivo is synthesized in neurons from l-serine by serine racemase (Wong et al., 2020), but d-serine can also be produced from astrocytes in primary cultures (i.e., in vitro) [reviewed in Coyle et al. (2020)]. Depletion of d-serine using enzymatic degradation or serine racemase KO mice reduces NMDA receptor signaling at synapses onto hippocampal CA3 and CA1 pyramidal neurons, prefrontal cortex, visual cortex, hypothalamic supraoptic nucleus, and nucleus accumbens (Panatier et al., 2006; Basu et al., 2009; Fossat et al., 2012; Papouin et al., 2012; Curcio et al., 2013; Rosenberg et al., 2013; Le Bail et al., 2015; Meunier et al., 2016). However, substantial NMDA receptor responses remain after depletion of d-serine, suggesting that glycine can also serve as coagonist at these synapses. The contributions of glycine and d-serine to the activation of NMDA receptors therefore appear to overlap, albeit there are differences between synapses in the preference for glycine or d-serine [reviewed in Mothet et al. (2015)]. In the mature hippocampus, d-serine is the main coagonist in Schaffer collateral to CA1 synapses, where there is a switch from glycine to d-serine as the coagonist during postnatal development (Le Bail et al., 2015) that parallels the timing of GluN2A expression in these neurons (Monyer et al., 1994; Gray et al., 2011; Rauner and Kohr, 2011; Rodenas-Ruano et al., 2012). By contrast, glycine is predominant in medial perforant path to dentate gyrus synapses (Le Bail et al., 2015).
The GluN2 subunits within the NMDA receptor assembly influence potencies of GluN1 agonists, with the lowest potency (i.e., highest EC50) typically observed for GluN2A and the highest potency (i.e., lowest EC50) observed for GluN2D (Supplemental Table 7). Several cyclic and halogenated analogs of glycine, including d-cycloserine, can act as GluN1 agonists (Supplemental Table 7). d-Cycloserine is a partial agonist of GluN2A-, GluN2B-, and GluN2D-containing NMDA receptors but is a superagonist at GluN2C-containing NMDA receptors (Sheinin et al., 2001; Dravid et al., 2010; Jessen et al., 2017). That is, d-cycloserine activates GluN2C-containing receptors with greater agonist efficacy than the endogenous agonist glycine (Fig. 33). d-Serine is also a superagonist at GluN2C-containing receptors compared with glycine, albeit with lower efficacy than d-cycloserine (Supplemental Table 7). A series of GluN1 agonists have been synthesized by replacing the hydroxy group of d-serine with an amido group, allowing the introduction of larger heterocyclic substitutions (Urwyler et al., 2009). These agonists display high binding affinity and a wide range of agonist efficacies. Evaluation of one of these agonists, (R)-2-amino-3-(4-(2-ethylphenyl)-1H-indole-2-carboxamido)propanoic acid (AICP) [R-26 in Urwyler et al. (2009)], at recombinant NMDA receptors found an unprecedented potency in the low nanomolar range (Jessen et al., 2017) (Fig. 33; Supplemental Table 7). AICP is full agonist at GluN1/2A, a partial agonist at GluN1/2B and GluN1/2D, and a highly efficacious superagonist at GluN1/2C receptors. Two additional agonists, 15a and 16a, displayed a similar GluN2-dependent activity at recombinant NMDA receptor subtypes (Urwyler et al., 2009; Maolanon et al., 2017) (Supplemental Table 7). Both 15a and 16a are superagonists at GluN1/2C receptors, but unlike AICP, they are partial agonists at GluN1/2A, suggesting that GluN1 agonists can be designed with a range of GluN2-dependent agonist efficacies (Maolanon et al., 2017). Structural determinants for the GluN2 subtype-dependent agonist efficacy of d-cycloserine, AICP, 15a, and 16a are located in both the GluN2 NTD and GluN2 ABD (Chen et al., 2008; Dravid et al., 2010; Jessen et al., 2017).
Fig. 33.
Glycine site NMDA receptor agonists with GluN2-specific activity. Concentration-response data for glycine site NMDA receptor agonists normalized to the maximal responses to glycine. All ligands are superagonists at GluN1/2C receptors, and D-cycloserine, 16a, and AICP display varying extents of partial agonism at GluN1/2A, GluN1/2B, and GluN1/2D receptor subtypes. Data are adapted with permission from Maolanon et al. (2017) (Copyright 2017 American Chemical Society) for D-serine and 16a and Jessen et al. (2017) for D-cycloserine and AICP.
GluN3A and GluN3B also bind glycine and d-serine, but several differences exist between the agonist binding pockets of GluN3 and GluN1 subunits, and the affinity of glycine for the isolated GluN3A ABD is over 600-fold higher than that for the GluN1 ABD (Yao and Mayer, 2006; Yao et al., 2008) (Fig. 32). The higher agonist affinity in the isolated GluN3A ABD is likely mediated by several unique interdomain interactions between D1 and D2 that are absent in the GluN1 ABD (Yao and Mayer, 2006; Yao et al., 2008, 2013). GluN1/3 receptors expressed in heterologous systems or neurons have cryptic activation properties in which agonist binding to GluN1 triggers strong desensitization, whereas agonist binding to GluN3 mediates activation (Sections IV. Receptor Activation, Deactivation, and Desensitization and V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity). GluN1/3 receptor desensitization is prevented by mutations in the GluN1 agonist binding site (Awobuluyi et al., 2007; Kvist et al., 2013a) or by the preapplication of highly GluN1-selective competitive antagonists, such as CGP-78608, that prevent glycine binding to GluN1 and enable activation of GluN1/3 receptors (Madry et al., 2007, 2008; Grand et al., 2018). Agonist potencies at functional GluN1/3 receptors behave differently from agonist affinities at isolated GluN1 and GluN3A ABDs, since glycine binds GluN1 in GluN1/2 receptors with high potency (EC50 ∼0.1–1 µM) and GluN3A in GluN1/3A receptors with low potency (EC50 ∼10–60 µM) (Kvist et al., 2013a; Grand et al., 2018). In GluN1/3 receptors, d-serine is a partial agonist compared with glycine (Chatterton et al., 2002; Smothers and Woodward, 2007), which may account for observations that d-serine can antagonize GluN1/GluN3 receptors (Awobuluyi et al., 2007).
Similar to GluN1 and GluN3 subunits, the ABD of GluD1 and GluD2 also binds d-serine and glycine (Naur et al., 2007; Hansen et al., 2009; Yadav et al., 2011; Tapken et al., 2017) (Fig. 32). There are no reports of current responses mediated by wild-type GluD receptors (Sections IV. Receptor Activation, Deactivation, and Desensitization and V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity), but agonists have been evaluated at GluD receptors containing mutations that cause spontaneously activity (Naur et al., 2007; Hansen et al., 2009; Yadav et al., 2011; Kristensen et al., 2016a; Tapken et al., 2017). The spontaneous activity of GluD2-A654T (i.e., the lurcher mutation) and GluD1-F655A is inhibited by agonist binding that mediates destabilization of the dimer interface and results in desensitization (Section IV. Receptor Activation, Deactivation, and Desensitization). Molecular dynamics simulations show that the free energy associated with d-serine binding to the GluD2 ABD is greater than that for other iGluR ABDs, suggesting that conformational rearrangements, such as desensitization, may occur in response to agonist binding to GluD2 and preclude channel gating (Chin et al., 2020), potentially explaining the lack of ion channel activity. Besides d-serine and glycine, β-fluoro-dl-alanine, l- and d-alanine, d-cysteine, and l-aspartate reduced responses at spontaneously active GluD2-A654T receptors by more than 25% (Naur et al., 2007; Kristensen et al., 2016a).
B. Competitive Antagonists
Crystal structures of isolated ABDs from iGluR subunits in complex with a range of agonists, partial agonists, and competitive antagonists have provided clues to the structural mechanisms governing agonist efficacy [i.e., the equilibrium constant for the transition from closed to open ion channels when the agonist is bound, e.g., see Colquhoun (1998)] (Sections II. Receptor Structure and IV. Receptor Activation, Deactivation, and Desensitization). Competitive antagonists are ligands that when bound to the receptor occlude the binding of agonists and prevent receptor activation. This is mediated by binding of the competitive antagonist into the agonist binding pocket but with no efficacy to trigger channel gating. In this regard, some agonists have been characterized as “functional antagonists,” such as dysiherbaine and MSVIII-19 at GluK1 (Swanson et al., 2002; Frydenvang et al., 2009) and propyl-NHP5G at GluN2A (Hansen et al., 2013), since they bind the agonist binding pocket with very low agonist efficacy and inhibit responses to full agonists without eliciting discernable current responses. The distinction between “competitive antagonist” and “functional antagonists” is blurred by the association of AMPA and kainate receptors with auxiliary subunits that influence agonist potency and efficacy (Section III. Auxiliary Subunits). The boost in agonist efficacy afforded by these auxiliary subunits can convert some competitive antagonists into AMPA receptor partial agonists (e.g., DNQX and CNQX at TARP-associated AMPA receptors) (Sections II. Receptor Structure and III. Auxiliary Subunits).
Competitive antagonists and agonists compete for receptor binding, and lower concentrations of a competitive antagonist can inhibit responses to low agonist concentrations, whereas higher antagonist concentrations are required to inhibit responses to high agonist concentrations. This means that competitive antagonist IC50 values determined from concentration-inhibition data are highly dependent on the concentration of agonist used to activate responses and the EC50 of the agonist. However, the binding affinity (Ki) of the competitive antagonist, which is independent of the agonist itself or the concentration of agonist used in the experiment, has often been estimated from IC50 values using the Cheng-Prusoff relationship (Cheng and Prusoff, 1973). Unfortunately, the Cheng-Prusoff relationship is not exact in the case of iGluRs, and accurate determinations of binding affinities require more elaborate pharmacological evaluation, such as Schild analysis [e.g., see Colquhoun (1998, 2007), Wyllie and Chen (2007)]. Despite the importance of the relationship between binding of competitive antagonists and agonists, failures to recognize it have resulted in misrepresentations of subunit selectivity for competitive iGluR antagonists, most notably for NVP-AAM077, which was incorrectly reported to be >100-fold selective for GluN1/2A over GluN1/2B receptors (Auberson et al., 2002) but is only 5- to 11-fold selective in terms of binding affinity (Frizelle et al., 2006; Lind et al., 2017).
The binding of competitive antagonists stabilizes open-cleft conformations of iGluR ABDs in crystal structures (Armstrong and Gouaux, 2000; Furukawa and Gouaux, 2003; Hogner et al., 2003; Mayer et al., 2006; Hald et al., 2007; Jespersen et al., 2014; Kristensen et al., 2016a). However, dynamic studies suggest that rather than stabilizing a single open conformation the binding of some competitive antagonists (e.g., CNQX and DNQX) permit a highly dynamic range of ABD conformations similar to the apo form (Ahmed et al., 2007, 2011a; Lau and Roux, 2007, 2011; Dai and Zhou, 2015). These antagonists interact with residues in the upper lobe D1 of the ABD and prevent engagement of the agonist amino acid moiety with the binding pocket (i.e., “shielding” mechanism) (Armstrong and Gouaux, 2000). By contrast, other competitive antagonists (e.g., UBP282 and ATPO) prevent the ABD from adopting the closed conformation by a “foot-in-the-door” mechanism (Hogner et al., 2003; Hald et al., 2007; Ahmed et al., 2009a). These competitive antagonists have extensive interactions with both D1 and D2 residues but impose sterical hindrance that prevents ABD closure to conformations capable of triggering channel gating. In some cases, as with the AMPA receptor antagonists NS1209 and UBP282, the ABD is stabilized in a hyperextended conformation compared with the apo form (Kasper et al., 2006; Ahmed et al., 2009a). Both “shielding” and “foot-in-the-door” antagonists crystallize with isolated iGluR ABDs in open or partially closed conformations, unlike agonists and “functional antagonists” that crystallize with ABDs in more closed conformations (Armstrong and Gouaux, 2000; Hogner et al., 2002; Inanobe et al., 2005; Mayer, 2005; Naur et al., 2007; Yao et al., 2008; Frydenvang et al., 2009; Unno et al., 2011; Hansen et al., 2013).
1. AMPA and Kainate Receptor Competitive Antagonists
The quinoxalinediones CNQX, DNQX, and NBQX are competitive AMPA and kainate receptor antagonists with selectivity over NMDA receptors (Honore et al., 1988; Sheardown et al., 1990). NBQX appears to be more selective for AMPA receptors over kainate receptors, whereas CNQX and DNQX appears to be less selective (Sheardown et al., 1990). However, CNQX and DNQX are also capable of binding NMDA receptors with low affinity (Lester et al., 1989; Sheardown et al., 1990). From these early quinoxalinediones, a range of analogs have been developed in the search for antagonists with drug-like properties as potential treatments of CNS disorders [e.g., see Mattes et al. (2010)] (Section X. Glutamate Receptors in Disease), several of which have been in clinical trials, including ZK 200775 (MPQX, fanapanel) (Turski et al., 1998), YM90K (Ohmori et al., 1994), YM872 (zonampanel) (Takahashi et al., 2002), and AMP397 (becampanel) (Auberson et al., 1999) (Supplemental Tables 8 and 9). Attempts to develop quinoxalinediones antagonists with subunit selectivity resulted in CNG-10310 with selectivity for AMPA over NMDA and kainate receptors and other analogs with preference for either GluK1 or GluK3 kainate receptor subunits (Demmer et al., 2015, 2017; Mollerud et al., 2019; Pallesen et al., 2019). The 2,4-quinazolinedione BGG492 (selurampanel) is an orally active competitive antagonist, which is structurally related to the quinoxalinediones but selective for AMPA receptors over kainate and NMDA receptors (Orain et al., 2017). The AMPA receptor competitive antagonist NS1209, which also binds to GluK1, was developed to replace the generally insoluble quinoxalinedione chemical scaffold (Nielsen et al., 1999; Kasper et al., 2006). NS-102, a competitive antagonist similar to NS1209, is selective for kainate over AMPA receptor subunits (Johansen et al., 1993; Verdoorn et al., 1994).
The antagonists ATPO and UBP282 bind AMPA receptor subunits as well as GluK1 (Moller et al., 1999; Dolman et al., 2005) (Supplemental Tables 8 and 9). A series based on the antagonist (2S,3R)-3-(3-carboxyphenyl)-pyrrolidine-2-carboxylic acid was designed to be nonselective among all glutamate-binding iGluR subunits (Larsen et al., 2011a) and included competitive antagonists with preference for GluK3 over GluK1 and AMPA receptor subunits (Krogsgaard-Larsen et al., 2015), GluK1 selectivity (Krogsgaard-Larsen et al., 2017), and selectivity for NMDA receptors over AMPA and kainate receptors (Kayser et al., 2020) (Supplemental Tables 8 and 9). A class of heterotricyclic glutamate analogs combined structural elements of kainate and neodysiherbaine A, resulting in IKM-159 with selectivity for AMPA receptors (Gill et al., 2010; Juknaitė et al., 2013).
GluK1-selective competitive antagonists include the decahydroisoquinolines LY382884 (O'Neill et al., 1998; Bortolotto et al., 1999) and LY466195 (Weiss et al., 2006) as well as the willardiine analogs UBP296, UBP302, and UBP304 (More et al., 2004; Dolman et al., 2005, 2006) (Supplemental Tables 8 and 9). The willardiine analogs UBP310 and UBP316 (ACET) are selective for both GluK1 and GluK3 subunits over GluK2 and AMPA receptors (More et al., 2004; Dolman et al., 2005, 2007; Dargan et al., 2009; Perrais et al., 2009b). Interestingly, GluK1-selective antagonists can be used as tools to reduce desensitization of heteromeric kainate receptors, such as GluK1/2 and GluK1/5, since partial occupancy of agonist binding sites in heteromeric kainate receptors can be sufficient for activation but may not be sufficient for strong desensitization (Pinheiro et al., 2013; Reiner and Isacoff, 2014; Pollok and Reiner, 2020). The mechanism accounting for the high GluK1 selectivity involves both differences in electrostatic and hydrogen bonding patterns as well as steric occlusion of binding to AMPA receptor subunits and GluK2-5 subunits (Mayer et al., 2006; Hald et al., 2007; Dargan et al., 2009; Alushin et al., 2011; Venskutonyte et al., 2011a).
2. Competitive Antagonists for Glutamate-Binding NMDA Receptor Subunits (GluN2)
Extending the glutamate backbone by just one carbon results in competitive antagonist activity at GluN2 subunits, as observed for the low-affinity antagonist (R)-aminoadipate (Biscoe et al., 1977). Greater affinity is obtained by replacing the distal carboxylate of (R)-aminoadipate with a phosphonate group, resulting in the prototypical GluN2 competitive antagonist (R)-AP5 (or D-APV) (Davies et al., 1981, 1982; Evans et al., 1982) that is widely used as a pharmacological tool to distinguish NMDA receptor–mediated responses from AMPA and kainate receptor responses. The addition of another carbon to the backbone of AP5 markedly reduces GluN2 binding affinity, but the addition of two carbons produces the highly potent antagonist (R)-AP7 (Perkins et al., 1981). Competitive antagonist affinity at GluN2 has been improved by constraining AP5 or AP7 with cyclic structures and by adding groups to this backbone to improve binding [reviewed in Monaghan and Jane (2009)]. Examples of such AP5 analogs include CGS 19755 (selfotel) and PMPA, whereas examples of AP7 analogs include D-CPP, D-CPPene, LY235959, NPC 17742, and SDZ 220-040 (Supplemental Table 10).
The residues lining the agonist binding pockets of GluN2 NMDA receptor subunits are fully conserved, and competitive antagonists with high selectivity for one GluN2 subunit have yet to be developed. Chemical scaffolds other than the AP5/AP7 structure have been explored, including NVP-AAM007 (or PEAQX) with high-affinity binding to GluN2 subunits (Auberson et al., 2002), but are shown to have insufficient selectivity to block only GluN2A receptors without also blocking GluN2B receptors (Frizelle et al., 2006; Lind et al., 2017) (Supplemental Table 10). The use of competitive GluN2 antagonists as pharmacological tools is further complicated by the nonequilibrium conditions of short exposure to high glutamate concentrations (∼1 mM for a few milliseconds) during excitatory synaptic transmission (Clements et al., 1992) as well as triheteromeric NMDA receptors containing two different GluN2 subunits [e.g., Lind et al. (2017)]. There are now better selective pharmacological tools with which to dissect the physiologic roles of GluN2 subunits (Section IX. Exogenous Positive and Negative Allosteric Modulators).
A series of 3-carboxypyrazoline amino acids is competitive antagonists with binding preferences similar to NVP-AAM007 among GluN2 subunits (Conti et al., 2010; Tamborini et al., 2016; Lind et al., 2017). ST3 in this series displays a 15-fold preference for GluN2A over GluN2B but intermediate affinity for GluN2C and GluN2D subunits (Supplemental Table 10). Crystal structures of the GluN1/2A ABD heterodimer in complex with ST3 or NVP-AAM007 have revealed the mechanism for the preference for GluN2A over GluN2B, which involves a cavity just outside the agonist binding pocket that the antagonists can exploit (Fig. 34). NVP-AAM077 and ST3 occupy this cavity, which extends toward the GluN1 ABD at the subunit interface (Lind et al., 2017; Romero-Hernandez and Furukawa, 2017).
Fig. 34.
Distinct binding modes of competitive NMDA receptor antagonists with GluN2 subunit preference. (A) Cartoon illustrating the GluN1/2A ABD heterodimer with glycine bound in GluN1 and antagonist bound in GluN2A. The upper D1 and lower D2 lobes are indicated. Below is an alignment of the amino-acid sequences of GluN2 subunits highlighting nonconserved residues that mediate the GluN2 preference of competitive antagonists. The GluN1 ABD is omitted for clarity in panels B–D. (B) Structure of the GluN2A ABD in complex with the GluN2C/D-preferring antagonist UBP791 (PDB: 6UZW). The phenanthrene rings of UBP791 are oriented along the cleft formed by the D1 and D2 lobes and contact nonconserved residues shown in blue in subsite II. (C and D) Structures of the GluN2A ABD in complex with the GluN2A-preferring antagonists ST3 (PDB: 5DDX) or NVP-AAM077 (PDB: 5U8C). These antagonists have substituents directed into a cavity of the GluN2A agonist binding site toward the subunit interface between GluN1 and GluN2A ABDs (subsite I).
Competitive GluN2 antagonists with a piperazine dicarboxylic acid scaffold and large, hydrophobic biphenyl or phenanthrene substituents, such as (2S*,3R*)-1-(phenanthrene-2-carbonyl)piperazine-2,3-dicarboxylic acid (PPDA), UBP141, and UBP145 (Feng et al., 2004, 2005; Morley et al., 2005; Costa et al., 2009; Irvine et al., 2012; Jespersen et al., 2014), have led to the high-affinity antagonists UBP791 and UBP1700, which show modest ∼15- to 40-fold preference for GluN2C/D over GluN2A/B subunits (Wang et al., 2020) (Supplemental Table 10). The structural basis for the GluN2C/D preference of these piperazine dicarboxylic acid antagonists is distinct from the mechanism that mediates GluN2A preference of ST3 and NVP-AAM077 (Fig. 34). The crystal structure of the GluN1/2A ABD heterodimer in complex with PPDA revealed that the large phenanthrene substituent is oriented in the opposite direction of the cavity that the substituents of ST3 and NVP-AAM077 exploit and toward another region of the GluN2 ABD with nonconserved residues (Jespersen et al., 2014). This insight was used in the design of UBP791 and UBP1700 to improve the modest GluN2C/D preference observed for PPDA (Wang et al., 2020). Similar rational design of other GluN2 competitive antagonists has been enabled by structural data (with or without the GluN1 ABD) (Furukawa et al., 2005; Vance et al., 2011; Hansen et al., 2013; Jespersen et al., 2014; Hackos et al., 2016; Volgraf et al., 2016; Yi et al., 2016; Lind et al., 2017; Romero-Hernandez and Furukawa, 2017; Villemure et al., 2016; Chou et al., 2020; Wang et al., 2020).
Conantokins are derived from the venom of predatory marine snails (Conus snails) and are peptides of 17–27 amino acids that are rich in the unusual γ-carboxyglutamate residues and lack disulfide bonds (Lewis et al., 2012). The conantokins produce both competitive and noncompetitive actions by binding to the GluN2 ABD (reviewed in (Prorok and Castellino, 2007; Twede et al., 2009a). Con-G, con-Pr3, and con-RIB inhibit GluN2B-containing NMDA receptors, with little inhibition of GluN2A-containing and GluN2C/D-containing receptors (Wittekindt et al., 2001; Sheng et al., 2007, 2009, 2010; Teichert et al., 2007; Gowd et al., 2012; Cheriyan et al., 2016). Con-Br and con-Bk-C are more potent for GluN2D-containing NMDA receptors than other conantokins (Twede et al., 2009b; Platt et al., 2014). Although con-G presumably binds to a site overlapping with the GluN2 agonist binding pocket, Met739 in GluN2B (Lys738 in GluN2A) located outside the agonist binding pocket has been implicated in the specificity of con-G (Sheng et al., 2009). Con-R and con-T are nonselective antagonists of NMDA receptors (Klein et al., 2001; Sheng et al., 2007; Teichert et al., 2007).
3. Competitive Antagonists for Glycine/d-Serine–Binding Subunits (GluN1, GluN3, GluD)
Most competitive GluN1 antagonists fall into two classes developed from the endogenous molecule kynurenic acid or quinoxalinediones, such as CNQX and DNQX [reviewed in Cai (2006), Monaghan and Jane (2009)]. The first includes high-affinity antagonists 7-CKA and 5,7-dichlorokynurenic acid (DCKA), L-683,344, L-689,560, L-701,324, and GV150526A (gavestinel), and the second class includes CGP 78608 and ACEA-1021 (Supplemental Table 11). Although ACEA-1021 suffers from poor solubility, the α-phosphoalanine–substituted CGP 78608 is highly selective for the GluN1 binding pocket and has improved solubility (Auberson et al., 1999). High-affinity GluN1 antagonists distinct from kynurenic acid have also been developed, including ZD 9379, MDL 29951, and MDL 105519 (Supplemental Table 11). Cycloleucine is also a competitive GluN1 antagonist, albeit with low affinity (Watson and Lanthorn, 1990). The identity of the GluN2 subunit in the NMDA receptor can change the binding affinities for competitive GluN1 antagonists by up to 10-fold (Ikeda et al., 1992; Kutsuwada et al., 1992; Priestley et al., 1995). The noble gas xenon exerts its anesthetic actions by a mechanism involving competitive inhibition of the GluN1, independent of potential effects on GABAergic transmission (Dickinson et al., 2007; Banks et al., 2010; Armstrong et al., 2012; Harris et al., 2013). Few competitive antagonists have been crystallized with the GluN1 ABD (with or without the GluN2A ABD) (Inanobe et al., 2005; Jespersen et al., 2014; Chou et al., 2020).
The complex activation properties of GluN1/3 receptors, in which agonist binding to GluN1 triggers strong desensitization and agonist binding to GluN3 mediates activation (Sections IV. Receptor Activation, Deactivation, and Desensitization and V. Glutamate Receptors in Neuronal Functions and Synaptic Plasticity) complicate studies of GluN3 pharmacology, and only a few studies have described competitive GluN3 antagonists (Yao and Mayer, 2006; Yao et al., 2008; Kvist et al., 2013a,b; Grand et al., 2018). Binding studies using isolated GluN1 and GluN3A ABDs demonstrated that competitive antagonists can bind both subunits but generally with higher affinity at the GluN1 ABD (Yao and Mayer, 2006). Functional studies with GluN1/3A revealed that the competitive antagonists TK40 and DCKA have ∼80- and ∼500-fold higher affinity, respectively, for the GluN1 subunit over GluN3 subunits (Kvist et al., 2013b). The binding affinities of TK40 estimated using the Cheng-Prusoff relationship were 15 µM at GluN3A and 4.9 µM at GluN3B (Kvist et al., 2013b). In silico screening of ligands targeting the agonist binding site of the GluN3A ABD in the open cleft conformations identified TK13, TK30, and TK80 as antagonists of GluN1/3 receptors, albeit with relatively low potency and modest selectivity for GluN1/3 over GluN1/2 receptors (Kvist et al., 2013a).
The competitive GluN1 antagonist CGP-78608 is a useful tool compound to “unmask” neuronal GluN1/3A receptors (Grand et al., 2018; Otsu et al., 2019; Zhu et al., 2020). Two features of CGP-78608 appear to be important for the effective “unmasking” of GluN1/3A receptors. First, CGP-78608 selectively binds with high affinity to the glycine binding pocket in GluN1 (Ki 6.4 nM), but not GluN3A (Ki 5.5 µM) (Yao and Mayer, 2006). Thus, CGP-78608 prevents GluN1/3 desensitization by acting as a competitive antagonist that selectively inhibits glycine binding to GluN1, thereby only allowing glycine binding to GluN3A. Second, CGP-78608 has a remarkably slow unbinding rate from GluN1. The time constant for CGP unbinding from GluN1 is ∼28 seconds (Grand et al., 2018), which enables activation of CGP-bound GluN1/3A by brief exposures to a high glycine concentration without desensitization due to glycine binding to GluN1 (Grand et al., 2018; Otsu et al., 2019; Zhu et al., 2020). The competitive GluN1 antagonist MDL-29951 similarly enhances responses from recombinant GluN1/3A and GluN1/3B receptors (Madry et al., 2007, 2008, 2010; Mesic et al., 2016), but is less effective at “unmasking” GluN1/3 receptors compared with CGP-78608 (Grand et al., 2018).
The pharmacology of GluD subunits is unexplored, largely because current responses mediated by wild-type GluD receptors have not been observed (Yuzaki and Aricescu, 2017). Pharmacological studies have been performed with GluD receptors containing mutations that cause spontaneously active receptors (e.g., GluD2-A654T) (Section IV. Receptor Activation, Deactivation, and Desensitization) and revealed that competitive GluN1 antagonists, including 7-CKA and L-689,560, can inhibit spontaneously active GluD2-A654T receptors by a competitive mechanism of action (Kristensen et al., 2016a).
C. Voltage-Dependent Channel Blockers
Channel blockers are a large and diverse class of molecules that bind within the channel pore-to-block ion flux. With few exceptions [e.g., Linsenbardt et al. (2013)], these compounds are positively charged at physiologic pH and require the channel to be open for binding. Consequently, inhibitory activity is both voltage- and channel activity–dependent, and these features of channel block are characterized as “voltage-dependent” and “use-dependent,” respectively. The inhibition by channel blockers is neither noncompetitive (i.e., negative allosteric modulation) nor competitive, and voltage-dependent channel block is therefore also denoted “un-competitive” antagonism. Although all compounds bind the channel to block ion flow, they are differentiated on the basis of mechanisms of block, which include 1) “sequential” or “foot-in-the-door” blockers that prevent channel closure (Benveniste and Mayer, 1995; Sobolevsky, 1999; Bolshakov et al., 2003; Barygin et al., 2009; Kaniakova et al., 2018), 2) blockers that become trapped inside the pore as the channel closes and require the channel to reopen to unbind (Sobolevsky and Yelshansky, 2000; Poulsen et al., 2015), and 3) partial trapping blockers that impede but do not prevent channel closure (Blanpied et al., 1997, 2005; Chen and Lipton, 1997; Mealing et al., 1999; Kotermanski et al., 2009; Johnson et al., 2015). Trapping blockers are retained in the pore after channel closure and agonist unbinding, remaining there until subsequent agonist exposure and channel reopening. Foot-in-door blockers prevent channel closure and must leave the pore before the receptor can deactivate, since the gate must close before the ABD bilobed clefts can reopen to allow agonist unbinding. Some compounds facilitate channel closure by allosteric mechanisms (Blanpied et al., 2005; Johnson et al., 2015). Endogenous Mg2+ and polyamines also block iGluR channels (Section VIII. Endogenous Allosteric Regulation).
1. Non–NMDA Receptor Channel Blockers
There is overlap in AMPA and kainate receptor channel blocker pharmacology, which is more extensively developed for AMPA receptors. Most AMPA and kainate receptor channel blockers incorporate a polyamine moiety, and a number are natural products from wasp, spider, snail, and snake venoms. These are highly potent AMPA and kainate receptor channel blockers, some of which also display potent channel block of NMDA receptors [reviewed in Andersen et al. (2006)]. In addition, synthetic polyamine derivatives, including IEM-1460 (Magazanik et al., 1997) and N1-naphthylacetylspermine (Koike et al., 1997), act primarily on GluA2-lacking, Ca2+-permeable AMPA receptors in a voltage-dependent manner. This selectivity derives from structural elements in the channel pore in proximity to the Q/R/N site at the apex of the M2 pore-lining loop (Sections II. Receptor Structure and VIII. Endogenous Allosteric Regulation). The amino groups of the polyamine-based blockers interact with residues that reside deeper in the pore than the Q/R/N site (Tikhonov et al., 2002; Twomey et al., 2018). These channel blockers have been useful pharmacological tools to probe the subunit composition of AMPA receptors [e.g., Plant et al. (2006)], although many also act on kainate receptors. There are variations on the blocking mechanism, exemplified by the phenylcyclohexyl derivative IEM-1925, which permeates and exits the channel on the intracellular side, allowing closed channels to escape from block (Tikhonova et al., 2008). The adamantane derivative IEM-1676 produces a voltage-dependent closed channel block from the intracellular compartment, in addition to open channel block from the extracellular compartment (Tikhonova et al., 2009). Some auxiliary subunits associated with the AMPA receptor reduce the inhibition of channel blockers (Kott et al., 2009; Jackson et al., 2011; Poulsen et al., 2014b; Soto et al., 2014) (Section III. Auxiliary Subunits). In addition, Joro spider toxin and philanthotoxin also block unedited GluK2 channels (Blaschke et al., 1993; Bahring and Mayer, 1998), and N1-naphthylacetylspermine blocks spontaneously active GluD1 and GluD2 receptor channels (Koike et al., 1997; Yadav et al., 2011).
Structure-activity studies of philanthotoxin derivatives highlight the importance of the polyamine moiety as well as the aromatic head group for blocker potency and AMPA receptor selectivity. Potent antagonists have at least two amino groups (Bolshakov et al., 2005). Shortening the polyamine chain of PhTX-343 decreased potency at AMPA receptors, and replacing the two secondary amines with either oxygen or methylene eliminated activity, whereas replacing only one with methylene improved potency 15-fold and increased selectivity for AMPA versus NMDA receptors (Mellor et al., 2003). PhTX-343 derivatives PhTX-56 and PhTX-74 differ in the number of amines and intervening methylenes in the polyamine tail (Kromann et al., 2002). PhTX-56 is 1000-fold selective for Ca2+-permeable over Ca2+-impermeable AMPA receptors, and 500-fold selective for AMPA over kainate receptors (Kromann et al., 2002). PhTX-74 was reported to display selectivity among the Ca2+-impermeable AMPA receptor by inhibiting GluA1/2 over GluA2/3 (Nilsen and England, 2007), but this selectivity has subsequently been challenged (Poulsen et al., 2014a). The role of the hydrophobic aromatic head group for potency and selectivity has been probed in PhTX-56 and PhTX-433 by replacement with a variety of structurally diverse moieties leading to derivatives with increased potency (Jensen et al., 2006; Frolund et al., 2010). Structure-activity studies for the more complex polyamine ArgTX-636 have demonstrated that modifications of both the polyamine chain and head group can increase potency or selectivity for AMPA receptors (Nelson et al., 2009; Poulsen et al., 2013).
2. NMDA Receptor Channel Blockers
Many synthetic NMDA receptor channel blockers have been developed in the search for new anesthetics and neuroprotective agents for use in acute brain injury (Section X. Glutamate Receptors in Disease). These include trapping blockers, such as ketamine, phencyclidine, and MK-801 as well as partial trapping blockers, amantadine, memantine, dextromethorphan and its metabolite dextrorphan, and lanicemine (AZD6765) (Supplemental Table 12). These two classes of compounds are also referred to as high- and low-affinity NMDA receptor channel blockers, respectively. The molecular determinants of the binding sites for NMDA channel blockers include residues within the M2 pore loop and residues in other pore-forming elements as well as the pre-M1 region (Johnson et al., 2015; Song et al., 2018). The structure-activity relationship underlying the trapping nature of blockers is unrelated to lipophilicity, and thus blockers are not capable of appreciably escaping through the membrane (Mealing et al., 1999, 2001; Bolshakov et al., 2005). The partial trapping blocker memantine can access multiple binding sites (Glasgow et al., 2018), one of which has been proposed to reside near the extracellular end of the pore and prevent full channel closure (Sobolevsky and Koshelev, 1998; Sobolevsky et al., 1998; Bolshakov et al., 2003).
Because the channel pore is relatively conserved among NMDA receptors, the channel blockers show only modest subunit selectivity for GluN2 subtypes (Yamakura et al., 1993; Dravid et al., 2007) (Supplemental Table 12). For example, MK-801 is ∼10-fold more potent for GluN2A- and GluN2B-containing receptors than GluN2C- and GluN2D-containing receptors (Dravid et al., 2007), whereas aryl-polyamine derivatives N1-dansyl-spermine and the tribenzyltriamine TB-3-4 are approximately 40-fold less potent at GluN2A- than GluN2D-containing NMDA receptors (Chao et al., 1997; Igarashi et al., 1997; Jin et al., 2007) (Supplemental Table 12). However, key residues in the permeation pathway alter the affinity of Mg2+, creating a small difference in affinity at GluN1/2A and GluN1/2B versus GluN1/2C and GluN1/2D (Siegler Retchless et al., 2012). This difference in Mg2+ affinity results in a modest enhancement of subunit selectivity for some channel blockers in the presence of physiologic Mg2+, such as memantine and ketamine, which show ∼10-fold selectivity for GluN1/2C and GluN1/2D over GluN1/2A and GluN1/2B (Kotermanski and Johnson, 2009; Yi et al., 2019). That is, memantine and ketamine compete with Mg2+ for access to their binding sites. Mg2+ has lower potency at GluN1/2C and GluN1/2D or dissociates more rapidly (Clarke and Johnson, 2006) (Section VIII. Endogenous Allosteric Regulation), which allows memantine and ketamine to gain access to their binding sites more readily at GluN1/2C and GluN1/2D compared with GluN1/2A and GluN1/2B. Ketamine blocks GluN2C-containing NMDA receptors at lower potency than other NMDA receptors in the presence of Mg2+, raising the possibility that this may contribute to its psychogenic actions (Khlestova et al., 2016).
As a consequence of the use dependence of inhibition, these agents may have significant selectivity for different populations of receptors in vivo dependent on their degree of activation. The trapping blockers, such as MK-801 and ketamine, are proposed to preferentially inhibit NMDA receptors on fast spiking interneurons over NMDA receptors on pyramidal neurons based on the higher level of activity of interneurons relative to pyramidal neurons (Widman and McMahon, 2018). These channel blockers could also show differences in their activity at extrasynaptic and synaptic NMDA receptors [reviewed by Johnson et al. (2015)]. This results in differential effects on γ oscillations, with low doses inducing γ oscillations, which are then suppressed at higher doses (Hiyoshi et al., 2014). Inhibitory interneurons but not pyramidal cells express GluN2D (Section VI. Developmental and Regional Expression in the Central Nervous System), which may result in higher potency of channel blockers at GluN2D-containing NMDA receptors in interneurons in physiologic Mg2+ (Kotermanski and Johnson, 2009; Yi et al., 2019). Furthermore, the degree of blocker trapping is also hypothesized to confer differential sensitivity at the circuit and functional level, evident in comparative studies of the trapping blockers lanicemine and ketamine; although both compounds induce γ oscillation in rodents, only ketamine induces locomotor hyperactivity (Sanacora et al., 2014). This dichotomy is evident in humans, in which both compounds induce γ oscillations, but only ketamine has psychotomimetic actions. Similarly, the lack of psychotomimetic side effects of memantine is attributed to the rapid unbinding rate that targets highly active NMDA receptors while preserving physiologic activity (Lipton, 2006, 2007).
Channel blockers continue to serve as important preclinical and clinical research tools. The approximate irreversibility of MK-801 channel block (Huettner and Bean, 1988) has been exploited to occlude synaptic NMDA receptors to allow the study of extrasynaptic receptors (Hardingham et al., 2002; Tovar and Westbrook, 2002; Harris and Pettit, 2007; Bordji et al., 2010; Liu et al., 2013). Moreover, the rate of MK-801 channel block can be used to assess open probability for NMDA receptors (Jahr, 1992; Chen et al., 1999c; Blanke and VanDongen, 2008; Gielen et al., 2009; Hansen et al., 2013).
VIII. Endogenous Allosteric Regulation
A. Voltage-Dependent Channel Block by Ions
The ion channel pores of iGluRs can be blocked by multiple endogenous modulators, including organic and inorganic cations in a voltage-dependent fashion. Hence, the extent of block depends on membrane potential, ensuring the channel block plays central roles in synaptic physiology. Below we review two of the most prominent endogenous channel blockers, intracellular polyamines at AMPA and kainate receptors and extracellular Mg2+ at NMDA receptors.
1. Channel Block of AMPA and Kainate Receptors by Polyamines
Ca2+-permeable AMPA receptors, which lack the Q/R-edited GluA2 subunit (Fig. 7), are blocked by the cytoplasmic polyamines spermine, spermidine, and putrescine [Bowie and Mayer (1995), Kamboj et al. (1995), Koh et al. (1995); reviewed in Bowie (2018)], that are present in all cells (Pegg, 2016). Because of the voltage dependence of the channel block (Fig. 35), cytoplasmic polyamines can finely tune cellular excitability by regulating the number of channels available for activation (Bowie et al., 1998; Rozov et al., 1998; Rozov and Burnashev, 1999), much like inward rectifier K+ channels (Nichols and Lee, 2018). The unedited AMPA receptor subunits GluA1, GluA3, and GluA4 show similar polyamine sensitivity, whereas the kainate receptor GluK3 subunit has higher polyamine sensitivity (Perrais et al., 2009b) than GluK1 and GluK2 (Bowie and Mayer, 1995; Cui and Mayer, 1999). The AMPA receptor auxiliary subunits, TARP and CNIH, and kainate receptor auxiliary subunits, Neto1 and Neto2, attenuate channel block by polyamines (Soto et al., 2007; Coombs et al., 2012; Fisher and Mott, 2012; Brown et al., 2016, 2018) (Fig. 35; Table 4).
Fig. 35.
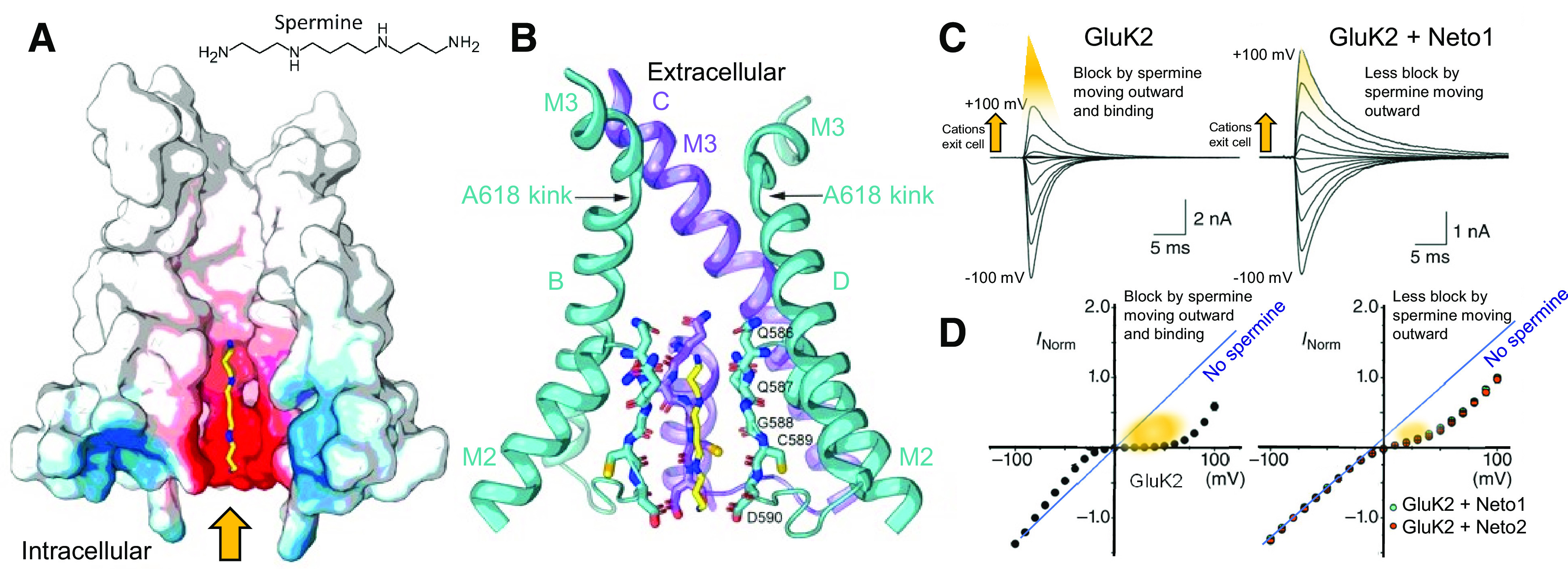
Polyamine binding within the AMPA receptor channel pore. (A) Structure of the endogenous polyamine spermine is shown within a cross-section of the GluA2(Q) AMPA receptor pore to illustrate the electroneutral cavity (white) above the Q/R/N site and electronegative cavity (red) of the inner pore where endogenous polyamines (yellow for carbon and blue for nitrogen) are proposed to bind. (B) Structure of the pore illustrates residues in the M2 reentrant loop that participate in polyamine binding (PDB: 6DM1). Adapted with permission from Twomey et al. (2018). (C) Electrophysiological traces of polyamine block of GluK2 (left) and GluK2 + Neto2 (right). (D) Current-voltage plots show the rectifying nature of polyamine block (indicated as gold), which is weaker when kainate receptors are coassembled with Neto1 or Neto2. Adapted with permission from Brown et al. (2016).
TABLE 4.
Effect of auxiliary subunits on polyamine block of AMPA and kainate receptors
Comparison of the permeation and block properties of homomeric and heteromeric AMPA and kainate receptors in the presence or absence of auxiliary subunits. The apparent affinity of spermine block (KD,0mV) was estimated from modified Boltzmann fits of conductance-voltage plots, which also provided the voltage-dependence of the onset and relief of block. The relative permeabilities of spermine to Na+ (PSpm/PNa+) or Ca2+ to Na+ (PCa2+/PNa+) were estimated by comparing the reversal potential in external solutions that contained spermine or Ca2+ to that obtained for Na+-based solutions. The rectification ratio corresponds to the ratio of current measured at +80 mV (or +60 mV for Ref. 2) and −80 mV holding potential.
|
Auxiliary
Subunit |
KD,0mV | Onset of Block | Relief of Block | PSpm/PNa+ | PCa2+/Na+ |
Rectification Index I +80 mV/I –80 mV |
|
|---|---|---|---|---|---|---|---|
| mM | mV | mV | mV | ||||
| GluA1 1,2 | – | 1.5 | – | – | – | 2.7 | 0.03–0.10 |
| GluA1 2,3 | γ-2 | – | – | – | – | ↑ 4.7 | ↑ 0.17 |
| GluA1 2,3 | CNIH3 | – | – | – | – | ↑ 5.1 | 0.07 |
| GluA2(Q) 3 | – | 1.8 | –14.1 | 14.5 | 0.28 | – | 0.49 |
| GluA2(Q) 2,3 | γ-2 | ↑ 17.9 | −13.4 | 15.7 | ↑ 1.1 | – | ↑ 0.87 |
| GluA2(Q) 2,3 | CNIH3 | ↑ 5.1 | −11.7 | 16.3 | ↑ 4.4 | – | ↑ 0.73 |
| GluK2 4,6,7,8 | 2.5–5.8 | –16 | 20.4 | – | 2.3 | 0.07–0.2 | |
| GluK2 4,8 | Neto1 | ↑ 71 | −24 | ↑ 43.3 | – | 2.3 | ↑ 0.66 |
| GluK2 4,8 | Neto2 | ↑ 39 | −18.5 | ↑ 48.6 | 0.042 | 2.3 | ↑ 0.48–0.64 |
| GluK3 5 | – | 0.042 | –21.6 | ND | – | – | <0.01 |
| GluK2/5 4 | – | 119 | –25.5 | 69.7 | 0.024 | 3.1 | 0.89 |
1 Bowie and Mayer (1995), 2 Coombs et al. (2012), 3 Brown et al. (2018), 4 Brown et al. (2016), 5 Perrais et al. (2009a), 6 Bahring et al. (1997), 7 Bowie et al. (1998), 8 Fisher and Mott (2012).
Both Ca2+ permeability and polyamine block depend on the Q/R/N site editing, with unedited AMPA and kainate receptors (Q) being Ca2+-permeable and blocked by polyamines, whereas edited AMPA and kainate receptors (R) are neither Ca2+-permeable nor blocked by polyamines (Burnashev et al., 1992a, 1996; Kohler et al., 1993; Bowie and Mayer, 1995; Cui and Mayer, 1999). This Ca2+ impermeability and polyamine insensitivity are thought to be due to the electrostatic repulsive effect of the positively charged arginine at the Q/R/N site at the narrowest region of the pore (Bowie, 2018). Outside of this common structural determinant, Ca2+ permeability and polyamine block arise by different mechanisms. Mutations in the M3 transmembrane helix disrupt Ca2+ permeability but leave polyamine block intact (Jatzke et al., 2003), whereas mutations in the M2 pore loop that are distinct from the Q/R/N site disrupt polyamine block but do not alter Ca2+ permeability (Dingledine et al., 1992). Similarly, substituting an Asn at the Q/R/N site, as in NMDA receptors, eliminates polyamine block while leaving divalent permeability intact (Burnashev et al., 1992a; Dingledine et al., 1992).
The structure of the ion-permeation pathway was recently resolved in cryo-EM structures of the GluA2(Q) AMPA receptor pore in complex with the polyamine blockers, including ArgTX-636, N1-naphthylacetylspermine, and IEM-1460, that block GluA2-lacking AMPA receptors from the external side in a use-dependent manner (Twomey et al., 2018) (Fig. 35). These blockers contain extended polyamine tails and a bulky, hydrophobic head group, which gets trapped in the narrow constriction of the permeation pathway. The intrapore electric field gives rise to the steep voltage dependence of polyamine block (Brown et al., 2016, 2018).
GluK2/5 heteromers attenuate polyamine block through a structural change in the pore helices that reduces the binding affinity and enhances permeation of the blocker (Brown et al., 2016). This is achieved through a conformationally rigid proline residue that is found in GluK4-5 but is absent from GluK1-3 subunits. Thus, heteromers containing either GluK4 or GluK5 should exhibit diminished polyamine block. In addition, the nanomolar affinity of polyamine block on GluK3 kainate receptors is due to residues in the M2 pore loop that are absent from GluK1 and GluK2 (Perrais et al., 2009b).
2. Block of NMDA Receptors by Extracellular Mg2+
NMDA receptors are blocked by extracellular Mg2+ in a voltage-dependent manner (Mayer et al., 1984; Nowak et al., 1984) (Fig. 36), which allows them to act as coincident detectors, requiring both postsynaptic depolarization as well as presynaptic release of glutamate to pass current. NMDA receptors are also blocked by intracellular Mg2+ (Johnson and Ascher, 1990; Li-Smerin et al., 2000; Li-Smerin et al., 2001), although the physiologic significance is not well understood. GluN1/2A and GluN1/2B receptors are more strongly blocked by extracellular Mg2+ than GluN1/2C and GluN1/2D (Monyer et al., 1994; Kuner and Schoepfer, 1996; Clarke and Johnson, 2006; Qian and Johnson, 2006; Siegler Retchless et al., 2012) (Fig. 36). At a −100 mV holding potential, the IC50 values for block by external Mg2+ are 2 µM, 2 µM, 14 µM, and 10 µM for GluN1/2A, GluN1/2B, GluN1/2C, and GluN1/2D, respectively (Kuner and Schoepfer, 1996). The millimolar concentrations of Mg2+ in CSF will produce strong block of native GluN1/2 NMDA receptors at resting membrane potentials of around −60 mV. Although the extent of this block would be attenuated for GluN2C- and GluN2D-containing NMDA receptors, block at 1 mM Mg2+ will still be sufficient to generate a region of negative slope conductance and thus allow GluN2C- and GluN2D-containing receptors to serve as coincidence detectors (Fig. 36). The dependence of Mg2+ block on the GluN2 subunit is influenced by multiple structural elements, with a main determinant being a residue in the M3 transmembrane helix (Siegler Retchless et al., 2012). This residue is a Ser in GluN2A/B and a Leu in GluN2C/D (i.e., the S/L site) and does not line the channel pore but may interact with a Trp residue in the M2 pore loop of GluN1 (Siegler Retchless et al., 2012). This interaction between GluN1 and the S/L site in GluN2 is also a key determinant of GluN2-specific variation in channel conductance and Ca2+-permeability (Siegler Retchless et al., 2012).
Fig. 36.
Voltage-dependent Mg2+ block of NMDA receptors. (A) Space fill of GluN1 (blue)/GluN2B (gray) NMDA receptor (PDB: 6WHS). The red box denotes the TMD. (B1) Cartoon illustrates direction of net current flow during periods when Mg2+ block occurs (red), when block Mg2+ is absent or modest (orange), and for outward current with no Mg2+ block (blue). (B2) Current-voltage relationship was recorded in the absence and presence of Mg2+; the smooth curve is a fit of the Woodhull equation to the data. Adapted with permission from Perszyk et al. (2020b). (C) Single channels recorded in outside-out patches obtained from cultured neurons in response to NMDA in the absence and presence of extracellular Mg2+. The red arrows indicate brief closures that increase in frequency with increasing Mg2+ concentration and likely reflect well-resolved individual blockages by Mg2+. Reproduced with permission from Ascher and Nowak (1988).
The rates for block and unblock by extracellular Mg2+ differ for NMDA receptors with different GluN2 subunits. GluN1/2C and GluN1/2D receptors show rapid Mg2+ unblock with a monoexponential time course, whereas GluN1/2A and GluN1/2B receptors show both a rapid and a slow component of unblock, the latter of which is weakly voltage-dependent (Clarke and Johnson, 2006). Variations in the unblocking rate might arise from interactions of Mg2+ with elements of the gate (Kampa et al., 2004; Vargas-Caballero and Robinson, 2004) as well as intrinsic voltage-dependent gating of GluN2A- and GluN2B-containing receptors (Clarke and Johnson, 2008; Clarke et al., 2013).
How does the pore of the NMDA receptor distinguish between the two divalent ions, permeable Ca2+ and largely impermeable Mg2+? Structural determinants of both Ca2+ permeation and Mg2+ block reside in the M2 pore loop (Burnashev et al., 1992b). However, mutations in the M2 pore loop of GluN1 and GluN2 subunits do not affect Ca2+ permeation and Mg2+ block equally (Burnashev et al., 1992b; Wollmuth et al., 1998), suggesting an asymmetry between subunits [e.g., Sobolevsky et al. (2002b)]. Part of the distinction between Ca2+ and Mg2+ involve differences in hydration energy; the smaller Mg2+ holds on to its water shell more tightly than the larger Ca2+. Furthermore, the preferred coordination numbers are different for Ca2+ and Mg2+ (7–8 and 6, respectively), resulting in specific interactions with elements in the M2 pore loop that could distinguish between Ca2+ and Mg2+ (Mesbahi-Vasey et al., 2017).
B. Modulation of Receptor Function by Ions
1. Modulation of AMPA, Kainate, and GluD Receptors by Extracellular Ions
Mechanistic insights into AMPA, kainate, and GluD receptors have resulted from evaluations of how external anions and cations regulate channel gating via binding to the interface between ABD dimers [reviewed in Bowie (2010), Plested (2011), Dawe et al. (2015)] (Fig. 37; Supplemental Fig. 9). Examination of anion modulation revealed that the intrinsic mobility of the resting or apo state of the AMPA receptors differs according to alternative splicing of the flip/flop cassette (Dawe et al., 2019). The study of cation regulation of AMPA receptors has suggested a structural explanation for the occurrence of modal gating of AMPA receptor–TARP signaling complexes. When associated with TARPs, AMPA receptors cycle through different gating modes of high and low open probability (Zhang et al., 2014a; Howe, 2015). Periods of low open probability are proposed to be governed by electrostatic interactions at the ABD dimer interfaces, whereas periods of high open-channel probability are proposed to be mediated by reversible AMPA receptor-TARP electrostatic interactions (Dawe et al., 2016; Twomey et al., 2016). These findings provide a potential explanation for single-channel data showing that the AMPA receptor–TARP subunit complexes exhibit modal gating (Zhang et al., 2014a).
Fig. 37.
Cation binding pockets of non-NMDA glutamate receptors in the ABD dimer interface. (A) Structures of the isolated ABD dimers of GluK2 (left, PDB: 3G3F) and GluA2 (right, PDB: 4IGT) highlight the cation binding sites (magenta) for Na+, Li+, and Ca2+ located at the apex of ABD dimer interface. The single-anion (Cl-, green) pocket of GluK2 is absent from AMPA receptors (left). (B) Expanded view highlights the residues that make up the cation binding pocket of GluK2 (left) and GluA2 (right) (C) Glutamate-activated currents from GluK2 kainate receptors (left) or GluA2 AMPA receptors (right) were recorded in different external cations. Adapted with permission from Wong et al. (2006) (Copyright 2006 Society for Neuroscience) and Dawe et al. (2016).
a. Cation and anion binding sites in AMPA receptors
The first indication of the regulatory effect of anions on AMPA receptors was the anomalous effect of the chaotropic anion, thiocyanate (SCN-), on channel gating (Bowie and Smart, 1993; Partin et al., 1996). Halide ions with a large atomic radius (e.g., I-) accelerate desensitization kinetics, and smaller anions (e.g., F-) slow AMPA receptor desensitization (Dawe et al., 2019). The structure of the GluA2 ABD homodimer revealed an anion binding site with Br- close to the base of the D1-D1 dimer interface (Dawe et al., 2019). Several water molecules surround each Br-, separating them from the Ser/Asn residue, which is the main difference between the flip/flop isoforms and alternative splicing therefore influences anion regulation (Dawe et al., 2019). Analysis of single AMPA receptors by atomic force microscopy offered a unifying explanation for the multiple effects of flip/flop splicing (Section IV. Receptor Activation, Deactivation, and Desensitization) by suggesting the cassette controls the NTD mobility of the apo or resting state of the receptors (Dawe et al., 2019). As a result, the apo state primes the AMPA receptor prior to activation and dictates its responsiveness to channel activators, allosteric modulators, and auxiliary subunits. The more mobile nature of the NTD for resting flop-dominant AMPA receptors gives rise to faster channel gating, weak allosteric regulation by anions, and modest effects of TARP auxiliary subunits (Dawe et al., 2019). By contrast, the moderate mobility of resting flip-dominant AMPA receptors gives rise to slower channel gating and robust allosteric regulation by anions and TARP auxiliary subunits (Dawe et al., 2019).
AMPA receptors also possess a cation binding pocket, which accommodates small cations such as Li+ (Assaf et al., 2013) (Fig. 37), and stabilizes the electrostatic interactions at the ABD dimer interface (Dawe et al., 2016). However, all AMPA receptor subunits possess a positively charged Lys residue near the cation binding pocket (Fig. 37), which curtails the residency time of cations in the pocket, and cation binding to AMPA receptors is not essential for channel gating (Dawe et al., 2016). By contrast, the equivalent position in kainate receptors possesses a nonpolar, hydrophobic residue (i.e., Ile, Met, Leu, or Val) that permits larger cations to bind (see below).
b. Cation and anion binding sites in kainate receptors
External anions and cations regulate the amplitude and time course of homomeric GluK1-3 kainate receptor responses (Bowie, 2002; Bowie and Lange, 2002; Paternain et al., 2003; Wong et al., 2006, 2007; Plested and Mayer, 2007; Plested et al., 2008). The effect of anions and cations on heteromeric kainate receptors containing either the GluK4 or GluK5 subunit is less clear (Paternain et al., 2003; Plested and Mayer, 2007). Three observations helped determine the structural basis for ion-dependent regulation of kainate receptors. First, external alkali metals and halide ions have effects distinct from both external pH and divalent cations (Wong et al., 2007). Second, ion substitution experiments demonstrated that kainate receptors require the binding of external Na+ for activation (Wong et al., 2006; Bowie, 2010; Maclean et al., 2011). Third, mutation of Met770 in the ABD dimer interface to a positively charged amino acid (Lys or Arg) disrupted anion and cation effects (Paternain et al., 2003; Wong et al., 2006, 2007; Nayeem et al., 2011).
The discrete anion and cation binding sites were resolved by X-ray crystallography at the kainate receptor ABD dimer interface (Plested and Mayer, 2007; Plested et al., 2008). Both GluK1 and GluK2 subunits possess a single anion binding pocket at the ABD dimer interface, whereas two cation pockets per dimer sit above at the apex flanking the anion site (Plested and Mayer, 2007; Plested et al., 2008; Nayeem et al., 2009, 2011) (Fig. 37). A similar arrangement has also been modeled from structural data of low-affinity GluK3 subunits (Venskutonytė et al., 2011b; Veran et al., 2012). However, the GluK3 subunit uniquely possesses Zn2+ binding sites that are located at the base of the ABD dimer interface below the anion binding sites in AMPA receptors (Veran et al., 2012). Occupancy of the Zn2+ binding sites in GluK3 and GluK2/3 attenuates receptor desensitization, accounting for the potentiating effect of Zn2+ on the agonist response (Veran et al., 2012) (Section VIII.B.2. Modulation of Glutamate Receptor Function by Extracellular Zn2+).
Single-channel studies and molecular dynamics suggested that occupancy of the cation binding pocket sustains channel activation and slows the onset of desensitization by stabilizing the ABD dimer interface (Dawe et al., 2013, 2015; Musgaard and Biggin, 2016). This idea was supported by a nondesensitizing mutant kainate receptor with an Arg in the cation pocket to stabilize the ABD dimer interface (Nayeem et al., 2009). Covalent cysteine crosslinking of the ABD dimer interface produces a similar outcome (Weston et al., 2006b); however, these engineered disulfide bonds may lock receptors out of the open state in addition to preventing desensitization (Daniels et al., 2013; Musgaard and Biggin, 2016). Electrophysiological and structural data suggest that the anion and cation binding sites are intact in GluK4 and GluK5 (Kristensen et al., 2016b; Plested et al., 2008). Furthermore, Neto2 attenuates cation sensitivity of homomeric and heteromeric kainate receptors (Griffith and Swanson, 2015).
c. Cation binding in GluD receptors
The GluD1 and GluD2 receptors do not form functional channels alone or when coexpressed with other glutamate receptor subunits but are spontaneously activated by mutations in the conserved M3 SYTANLAAF motif (Sections IV. Receptor Activation, Deactivation, and Desensitization and X. Glutamate Receptors in Disease). Glycine and d-serine can bind to the ABD of GluD1 and GluD2, and this binding reduces currents from spontaneously active mutant receptors (Naur et al., 2007; Yadav et al., 2011). GluD receptors contain binding sites at the ABD interface for Ca2+, the occupancy of which counteracts the inhibitory effects of d-serine on spontaneously active GluD receptors (Hansen et al., 2009; Yadav et al., 2011). Thus, d-serine binding induces rearrangements at the dimer interface of the GluD2 (Hansen et al., 2009) similar to those observed for desensitization of AMPA and kainate receptors. By contrast, Ca2+ stabilizes the ABD dimer interface in a manner reminiscent of monovalent cation binding to AMPA and kainate receptors (Hansen et al., 2009).
2. Modulation of Glutamate Receptor Function by Extracellular Zn2+
The transition metal ion Zn2+ is used by the CNS for a range of important actions, tightly regulated, concentrated in presynaptic vesicles by the Zn2+ transporter ZnT-3, and released from excitatory synaptic terminals in an activity-dependent manner (Smart et al., 2004; Marger et al., 2014). The zinc transporter ZnT-1 colocalizes with and binds to GluN2A in the postsynaptic terminal (Mellone et al., 2015; Krall et al., 2020). Extracellular Zn2+ binds with high affinity to the bilobed GluN2A NTD, with a nanomolar IC50 (Williams, 1996; Chen et al., 1997; Paoletti et al., 1997, 2000; Traynelis et al., 1998; Choi and Lipton, 1999; Fayyazuddin et al., 2000; Low et al., 2000; Rachline et al., 2005; Karakas et al., 2009) and inhibits receptors by decreasing open probability (Erreger and Traynelis, 2008; Amico-Ruvio et al., 2011) (Table 5). Physiologic saline solutions are contaminated by Zn2+ in the high nanomolar range that will inhibit GluN2A-containing NMDA receptors under most experimental conditions (Paoletti et al., 1997). To study Zn2+ modulation, it is necessary to use a buffer system to accurately control Zn2+ concentrations, and Zn2+-chelators, such as tricine or EDTA, in the extracellular solution are needed to eliminate contaminant Zn2+ (Paoletti et al., 1997). A shortcoming of both tricine and EDTA chelators is their slow rate of Zn2+ binding, which is insufficient to chelate synaptically released Zn2+ (Anderson et al., 2015). More recent studies have used the chelator, ZX1, which binds Zn2+ with high affinity (1 nM) and a rate that is 200-fold faster than tricine and EDTA (Anderson et al., 2015; Kalappa et al., 2015; Krall et al., 2020).
TABLE 5.
Allosteric modulation of glutamate receptors by extracellular Zn2+ and protons
Proton IC50 was determined using an activity coefficient of 0.8. Zn2+ IC50 was determined at pH 7.3. – denotes no available data, and ND indicates data are reported, but the EC50 or IC50 values were not determined. Data are for rat recombinant receptors expressed in oocytes or HEK cells. * indicates that protons or Zn2+ potentiated the response.
| Construct | Zn2+ IC50 | H+ IC50 |
|---|---|---|
| μM | μM (pH) | |
| GluN1/2A | 0.02 1 | 0.080–0.22 (pH 6.74–7.20) 2 |
| GluN1-1ac2/ GluN1-1ac1/2A | 0.091 3 | 0.20 (pH 6.80) 3 |
| GluN1-1ac2/ GluN1-1bc1/2A | 0.095 3 | 0.25 (pH 6.70) 3 |
| GluN1-1bc2/ GluN1-1bc1/2A | 0.17 3 | 0.33 (pH 6.58) 3 |
| GluN1/2A/2B | 0.058 4 | – |
| GluN1/2A/2C | 0.040 5 | – |
| GluN1/2B | 2.5 1 | 0.040–0.060 (pH 7.3–7.5) 2,3 |
| GluN1-1ac2/ GluN1-1ac1/2B | – | 0.036 (pH 7.54) 3 |
| GluN1-1ac2/ GluN1-1bc1/2B | – | 0.08 (pH 7.19) 3 |
| GluN1-1bc2/ GluN1-1bc1/2B | – | 0.18 (pH 6.83) 3 |
| GluN1/2C | 23 1 | 0.83 (pH 6.2) 6 |
| GluN1/2D | 14 1 | 0.065 (pH 7.3) 2 |
| GluN1/3A | ND * 7 | 0.099 * (pH 7.1) 7 |
| GluN1/3B | – | ND 8 |
| GluA1 | – | 0.63 (pH 6.3) 2 |
| GluA2(Q) | 430 9 | 0.50 (pH 6.4) 2 |
| GluA2/3 | 700 10 | – |
| GluA3 | 1000 10 | 0.79 (pH 6.2) 2 |
| GluA4c | – | 1.3 (pH 6.0) 2 |
| GluK1(Q) | 40 10 | 0.16 (pH 6.9) 11 |
| GluK2(Q) | 30 10 | 0.16–1.3 (pH 6.0–6.9) 11,12 |
| GluK2(R) | 67 10 | 0.13 (pH 7.0) 11 |
| GluK2(Q)/4 | – | 0.099 (pH 7.1) * 11 |
| GluK2(Q)/5 | 6.5 10 | 0.0016 (pH 8.9), 2.0 (pH 5.8) 11 |
| GluK2(R)/4 | 1.5 10 | – |
| GluK2(R)/5 | 3.0 10 | – |
| GluK3 | 46 * 13 | – |
| GluK2/3 | 480 * 13 | – |
| GluD2Lc | – | 0.042 (pH 7.5) 14 |
1 Traynelis et al. (1998), 2 Traynelis et al. (1995), 3 Yi et al. (2018), 4 Hansen et al. (2014), 5 Bhattacharya et al. (2018), 6 Low et al. (2003), 7 Cummings and Popescu (2016), 8 Cavara et al. (2009), GluN1/3B receptors were inhibited by protons, 9 Carrillo et al. (2020a), 10 Mott et al. (2008), 11 Mott et al. (2003), 12 Wong et al. (2007), 13 Veran et al. (2012), GluK3 was potentiated at lower Zn2+ and inhibited at higher Zn2+, 14 Williams et al. (2003), proton inhibition of GluD2 harboring the lurcher mutation (GluD2Lc) was incomplete.
The IC50 of Zn2+ inhibition at GluN1/2B and GluN1/2D receptors is in the µM range, with minimal activity at GluN2C-containing NMDA receptors (Paoletti et al., 1997; Traynelis et al., 1998). Zn2+ binding can also produce voltage-independent inhibition, although at concentrations above 10 μM, by blocking the channel pore in a manner similar to Mg2+ (Williams, 1996; Traynelis et al., 1998; Choi and Lipton, 1999; Low et al., 2000). Triheteromeric GluN1/2A/2B receptors show high sensitivity to Zn2+ with an IC50 value close to that of the GluN2A subunit, suggesting a dominant effect of GluN2A, but there is a modest reduction in inhibition at maximally effective concentrations of Zn2+ (Hatton and Paoletti, 2005; Hansen et al., 2014; Stroebel et al., 2014). Alternative splice isoforms of GluN1 reduce Zn2+ sensitivity, with receptors containing 21 residue in the NTD encoded by exon 5 being less sensitive to Zn2+ (Zheng et al., 1994; Traynelis et al., 1998; Yi et al., 2018). These residues establish additional contacts that stabilize the NTD-ABD and ABD-ABD interfaces (Regan et al., 2018).
An important aspect of Zn2+ inhibition is the positive allosteric intrasubunit interaction observed between Zn2+ binding to the NTD and glutamate binding to the ABD (Zheng et al., 2001; Erreger and Traynelis, 2005). Both ligands enhance the affinity of one another, such that glutamate binding to a receptor in subsaturating Zn2+ will cause the system to relax to a new equilibrium as Zn2+ re-equilibrates and more Zn2+ binds to the higher affinity site to inhibit the receptor. This will produce an apparent desensitization, as the response diminishes in the presence of agonist and extracellular Zn2+ (Chen et al., 1997) (Fig. 38). The time course for Zn2+-dependent desensitization is determined by the rate of Zn2+ binding (Erreger and Traynelis, 2005).
Fig. 38.
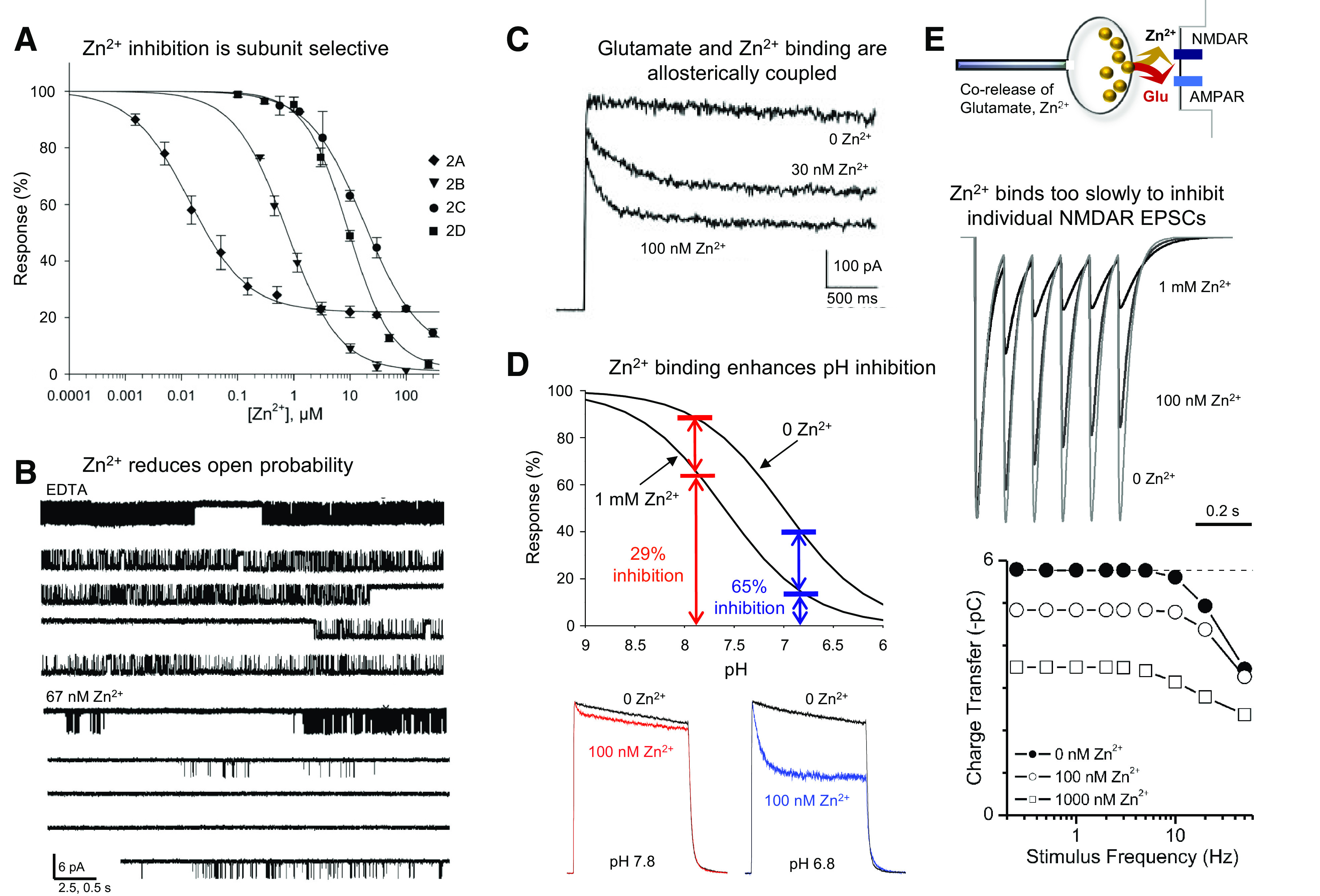
Negative allosteric modulation of NMDA receptors by extracellular Zn2+. (A) Concentration-response curve shows inhibition by Zn2+ of GluN1/2 receptors recorded at +50 mV at which voltage-dependent channel block does not contribute to inhibition. (B) GluN1-N615G/2A channels, which are not sensitive to channel block by Zn2+, recorded in cell-attached patches in the absence or presence of extracellular Zn2+. (C) GluN1/2A responses recorded at +40 mV in response to rapid application of glutamate with or without buffered Zn2+. The relaxation reflects a time-dependent Zn2+ association. (D) Proton inhibition curves recorded in the absence and presence of Zn2+ revealed a Zn2+-induced enhancement of proton inhibition. GluN1/2A responses recorded at +40 mV show greater Zn2+ inhibition at pH 6.8 than at pH 7.8, as predicted by the shift in the proton inhibition curve. (E) Cartoon illustrating corelease of Zn2+ and glutamate from presynaptic terminals. Simulation of NMDA-component EPSCs from a model that incorporates the positive allosteric regulation between Zn2+ and glutamate and corelease of Zn2+ (Erreger and Traynelis, 2005). The association rates determined for Zn2+ are too slow to allow synaptically released submicromolar Zn2+ to bind to NMDA receptors during the first EPSC, but inhibition is established with higher frequency release. Panels were reproduced or adapted with permission from Rachline et al. (2005) (Copyright 2005 Society for Neuroscience) (A), Amico-Ruvio et al. (2011) (B), Erreger et al. (2005a) (C and E), Low et al. (2000) (Copyright 2000 National Academy of Sciences, U.S.A.) (D), and Zheng et al. (2001) (D).
Zn2+ in synaptic vesicles is released in an activity-dependent manner (Tamano et al., 2016; McAllister and Dyck, 2017). The levels of Zn2+ in the synaptic cleft have been the subject of debate, with most estimates in the range of 1–10 μM, but no clear consensus because of technical and biologic complexities (Frederickson et al., 2006; Kay and Toth, 2008). Studies of Zn2+ action on NMDA receptors have provided rates for Zn2+ binding and unbinding in addition to defining the allosteric interaction between Zn2+ and glutamate (Erreger and Traynelis, 2005, 2008; Amico-Ruvio et al., 2011). These results allow the estimation of binding to NMDA receptors during simulated corelease of Zn2+ and glutamate, which suggest that the IC50 for inhibition of NMDA receptor-mediated EPSCs by synaptically released Zn2+ will be higher than that determined at steady state in the continuous presence of glutamate. The Zn2+ association rate is too slow at low concentrations to inhibit GluN2A-containing NMDA receptors before a single EPSC is terminated, suggesting that tonic Zn2+ but not necessarily synaptically released Zn2+ produces strong NMDA receptor inhibition during a single EPSC. Figure 38 shows that nanomolar concentrations of extracellular Zn2+ that saturate under steady-state conditions do not alter a single EPSC when coreleased, suggesting NMDA receptor inhibition requires higher concentrations of Zn2+ in the cleft or high-frequency firing to allow glutamate occupancy to increase the binding rate of Zn2+ (Erreger and Traynelis, 2005; Vergnano et al., 2014; Anderson et al., 2015).
Zn2+ binds within the cleft of the bilobed GluN2A NTD and interacts with four residues, His44, His128, Glu266, and Asp282, but in GluN2B, Zn2+ can only form two interactions with His127 and Glu284 (Low et al., 2000; Paoletti et al., 2000; Karakas et al., 2009; Romero-Hernandez et al., 2016) (Fig. 39). This binding site is supporting the idea that inhibition is due to NTD cleft closure induced by Zn2+ binding (Sirrieh et al., 2015a). Cryo-EM structures of the GluN1/2A receptor suggested a mechanism for Zn2+ inhibition (Jalali-Yazdi et al., 2018) in which the presence of Zn2+ facilitated closure of GluN2A NTD bilobes similar to that in crystal structure of the isolated GluN1/2A NTD heterodimer (Romero-Hernandez et al., 2016). The cryo-EM structures showed that Zn2+ binding also separated the GluN1/2A NTD heterodimer pairs, splaying the dimer of NTD dimers as well as GluN1/2A ABD dimer interfaces (Fig. 40). The rupture of intersubunit contacts was suggested to allow the membrane proximal domains of the ABD to move closer, which could relieve tension on the gate after agonist binding and ABD clamshell closure around agonist, favoring channel closure.
Fig. 39.

Coordination site for Zn2+ binding in GluN2A and GluN2B NTDs. (A) Crystal structure of GluN1 (slate)/GluN2B (gray) heterodimeric NTDs (PDB: 5TPW). Zn2+ binds within the cleft of the bilobed clamshell-like NTD. (B) Four residues within the bilobed GluN2A NTD coordinate Zn2+, resulting in high-affinity binding. (C) Crystal structure of Zn2+-bound GluN2B NTD (PDB: 3JPY). Zn2+ is coordinated by only two residue side chains, which accounts for its lower affinity compared with GluN2A.
Fig. 40.
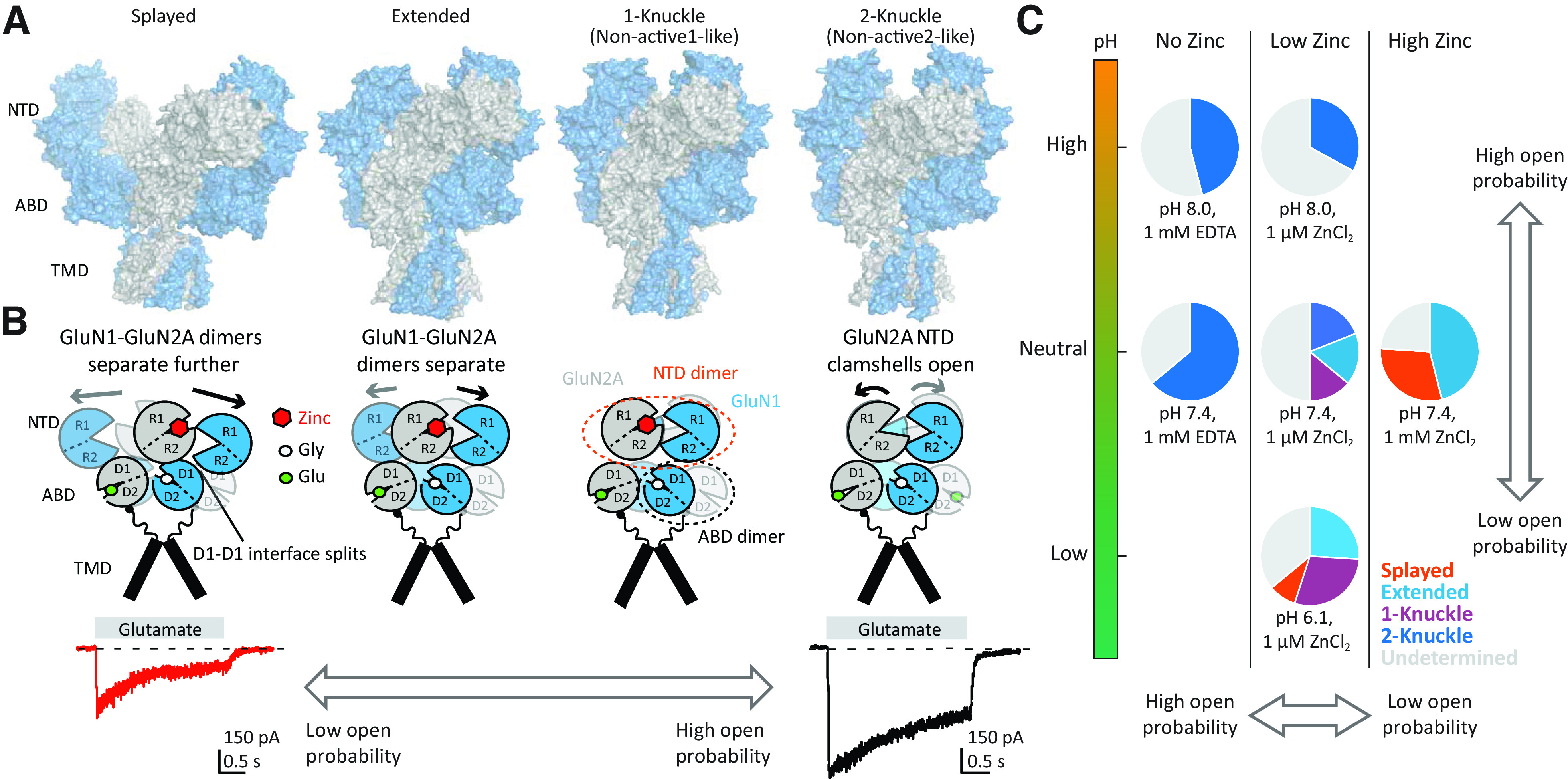
Structural basis of allosteric modulation of GluN1/2A NMDA receptors. (A) Structures of GluN1/2A NMDA receptors in the presence of glycine and glutamate (Gly and Glu) and in the presence and absence of Zn2+ at various pH values. Structures in surface presentation for Super-splayed (PDB: 6MMJ), Extended (PDB: 6MMM), 1-knuckle (PDB: 6MMK or 6IRF), and 2-knuckle (PDB: 6MMP or 6IRA). Intermediate conformational states are omitted for simplicity. GluN1 and GluN2A subunits are blue and gray, respectively. (B) Schematic presentations of domain and subunit arrangements of GluN1/2A NMDA receptors in various conformations in panel A. The 1-knuckle and 2-knuckle structures are similar to Non-active1 and Non-active2 structures, respectively, in which 2-knuckle contains open GluN2A NTD clamshells and 1-knuckle has closed GluN2A NTD clamshells. At neutral to high pH (7.4 to 8) with no Zn2+, the receptors are in 2-knuckle whereas at low pH (pH 6.3) with no Zn2+ (PDB: 6IRF) they are in 1-knuckle. Addition of Zn2+ at 1 µM at pH 6.1 splits GluN1-GluN2A NTD dimer pairs into Extended and Super-splayed conformations. At pH 7.4, the populations of Extended and Super-splayed increase as the Zn2+ concentration increases from low (1 µM) to high (1 mM). Currents at the bottom were recorded in the presence versus absence of EDTA and show inhibition by contaminant Zn2+ (unpublished data from Lonnie P. Wollmuth). (C) The relative occurrence of these structure classes in various conditions (no, low, high Zn2+; low, neutral, high pH). The pairs of structure classes, 2-Knuckle-Sym and 2-Knuckle-Asym, Extended and Extended-2, and Splayed and Super Splayed were combined for simplicity.
Multiple lines of evidence have suggested that the inhibitory actions of extracellular Zn2+ can be functionally understood through enhancement of proton inhibition (Choi and Lipton, 1999; Low et al., 2003; Gielen et al., 2008; Jalali-Yazdi et al., 2018). That is, Zn2+-bound receptors remain active but show altered pKa for proton inhibition, such that the degree of Zn2+-induced inhibition can be quantitatively predicted by the shift in tonic proton inhibition (Figs. 38 and 40). Structural studies have identified an ensemble of distinct Zn2+-bound conformations, with Zn2+ shifting the relative proportion of conformations that reduced interactions between the two heterodimer NTDs in a proton-dependent manner. These data showed that both protonation and Zn2+ binding induce a similar degree of closure of the GluN2A NTD (Jalali-Yazdi et al., 2018; Zhang et al., 2018b). In the absence of Zn2+ at pH 7.8, there are two interaction sites between the two NTD heterodimers (2-knuckle; Fig. 40), and the GluN2A NTD bilobes are open. At pH 6.3 with no zinc, however, NTD dimers rearrange to interact only at one site (1-knuckle; Fig. 40). Therefore, alkaline pH favors opening of GluN2A NTD bilobes and the 2-knuckle conformation with two interactions, whereas increasing proton concentrations favor the 1-knuckle conformations. In 1 µM Zn2+ at pH 7.4, the receptors are in 1-knuckle conformation along with more extended conformations that further separate the two heterodimeric NTDs (Extended; Fig. 40), whereas high (nonphysiologic) 1 mM Zn2+ at pH 7.4 even further separates the two heterodimeric NTDs (Super-splayed; Fig. 40). Finally, 1 µM unbuffered Zn2+ at pH 6.1 showed mostly 1-knuckle and Extended conformations plus the Super-splayed conformation, implying that proton and Zn2+ may synergistically separate the NTD dimers. These NTD conformational changes propagate through the receptor to the ABD layer to reduce the tension in the linkers connecting the ABDs to the channel pore (Gielen et al., 2008; Esmenjaud et al., 2019) (Fig. 40).
AMPA receptors show weak and biphasic sensitivity to extracellular Zn2+, with potentiation at 10s of µM and inhibition at higher concentrations (Rassendren et al., 1990; Kreitzer et al., 2009; Sun et al., 2010a,b; Carrillo et al., 2020a). Zn2+ regulation of AMPA receptors may be physiologically relevant in the dorsal cochlear nucleus where AMPA receptors are inhibited by synaptically released Zn2+ (Kalappa et al., 2015). Kainate receptors are inhibited by Zn2+ with strong subunit selectivity (Mott et al., 2003; Mott et al., 2008), with GluK1 receptors being sensitive to submicromolar levels of Zn2+ (Mott et al., 2003), whereas recombinant GluK2/5 are inhibited by higher concentrations of Zn2+ (Fukushima et al., 2003). GluK3 responses are potentiated by Zn2+, which reduces desensitization through stabilization of the ABD dimer interface (Veran et al., 2012). It has been proposed that LTP of synapses between hippocampal granule cell mossy fibers and CA3 pyramidal neurons is influenced by Zn2+ binding to presynaptic GluK2/3 kainate receptors (Contractor et al., 2001; Pinheiro et al., 2007; Pinheiro and Mulle, 2008). Data from mice lacking zinc transporters suggested that synaptically released Zn2+ inhibits postsynaptic kainate receptors at mossy fiber synapses (Mott et al., 2008). Similarly, exogenous application of Zn2+ can inhibit kainate receptors in the olfactory bulb (Blakemore and Trombley, 2020). Similar to NMDA receptors, there is a relationship between Zn2+ inhibition and extracellular protons, wherein Zn2+ inhibition is reduced in acidic pH, the opposite of NMDA receptors (Mott et al., 2008). Zn2+ can also facilitate glycine-activated GluN1/3 responses (Madry et al., 2008; Cummings and Popescu, 2016; Cummings et al., 2017) (Table 5).
C. Modulation by Protons and Polyamines
1. Modulation by Extracellular Protons
All iGluRs are sensitive to extracellular protons. AMPA receptors show modest proton sensitivity, and spontaneously active GluD2 receptors are pH-sensitive (Table 5). AMPA receptors show reduced open probability and enhanced rate of desensitization without a change in conductance at low pH (Lei et al., 2001). Block of desensitization by cyclothiazide eliminates pH inhibition, with rapidly desensitizing flop isoforms showing more sensitivity to pH than flip isoforms (Ihle and Patneau, 2000; Lei et al., 2001). Protons have inhibitory actions on most kainate receptors with IC50 values for homomeric GluK1 and GluK2 receptors corresponding to pH 6.9 (Table 5). Heteromeric kainate receptors that contain GluK5 are less sensitive to protons, and GluK2/4 receptors are potentiated by acidic pH (Mott et al., 2003).
NMDA receptors are fully inhibited by protons with an IC50 of 50–100 nM, corresponding to pH 7.4–7.0 (Giffard et al., 1990; Tang et al., 1990; Traynelis and Cull-Candy, 1990, 1991; Vyklicky et al., 1990). As a result, NMDA receptors will be tonically inhibited by protons under physiologic conditions because extracellular pH roughly matches the proton IC50. NMDA receptors are thus poised to respond to small changes in extracellular pH that occur under physiologic conditions due to release of protons from acidic synaptic vesicles or movement of protons or bicarbonate across the plasma membrane by pumps and channels (DeVries, 2001; Chesler, 2003; Chen and Chesler, 2015; Beckwith-Cohen et al., 2019; Stawarski et al., 2020). Such pH changes are challenging to measure given technical difficulties associated with ion-sensitive electrodes, rapid translocation of protons in solution, the active processes by which protons and bicarbonate move between compartments (Parker and Boron, 2013; Zhao et al., 2016a), and bicarbonate regulation via carbonic anhydrase (Ruusuvuori and Kaila, 2014; Stawarski et al., 2020). However, pH-sensitive fluorescent dyes, such as pH-sensitive GFP derivatives, provide new opportunities to monitor pH without perturbing the integrity of the tissue (Lee et al., 2016a). Pathologic conditions including ischemia (Kaplan et al., 1987; Katsura et al., 1992; Katsura and Siesjo, 1998) and seizures (Balestrino and Somjen, 1988; Raimondo et al., 2016) produce an extracellular acidification, decreasing pH to levels that can inhibit NMDA receptors (Chesler and Kaila, 1992; Chesler, 2003). Penumbral pH during ischemia can reach 6.8–6.9, with reduction in the infarct core reaching 6.5 (Mutch and Hansen, 1984; Smith et al., 1986; Nedergaard et al., 1991; Katsura and Siesjo, 1998), rendering proton inhibition of NMDA receptors important in the context of pathology.
GluN2 subunits control the sensitivity of NMDA receptors to extracellular protons (Traynelis et al., 1995), wherein GluN2A-, GluN2B-, and GluN2D-containing NMDA receptors all have proton IC50 values in the physiologic pH range (7.0–7.4) (Table 5). By contrast, GluN1/2C receptors have a proton IC50 value near pH 6 (Traynelis et al., 1995; Low et al., 2003). Furthermore, proton sensitivity is controlled by alternative splicing of exon 5 in GluN1, and this effect is dependent on the number of alternatively spliced GluN1 subunits in the receptor complex (Section IV.A.1. Glutamate Receptor Response Time Course and Synaptic Transmission and Table 5).
Proton inhibition is voltage-independent and does not affect glutamate binding, although reduction in pH modestly reduces glycine potency (Tang et al., 1990; Traynelis and Cull-Candy, 1990; Vyklicky et al., 1990; Banke et al., 2005). Protons reduce channel open probability without strong effects on response time course or single-channel conductance, and channel open time seems more sensitive to extracellular pH in GluN1/2A compared with GluN1/2B receptors (Banke et al., 2005; Dravid et al., 2007; Erreger and Traynelis, 2008). The complete structural determinants contributing to proton inhibition are unknown, although mutations at multiple residues in the NTD (Williams et al., 1995; Masuko et al., 1999a), ABD interface (Gielen et al., 2008; Cummings and Popescu, 2016), linkers and pore-forming elements (Kashiwagi et al., 1997; Low et al., 2003), and reentrant pore loop (Mott et al., 1998; Li et al., 2019a) can influence pH sensitivity consistent with cryo-EM structures that show conformational changes in the entire receptor structure at low pH (Zhang et al., 2018b). Some pore blockers acting at the M2 pore loop have pH-sensitive potencies, further emphasizing tight coupling between protons and gating (Dravid et al., 2007). In addition, removal of the GluN2 NTD does not eliminate the pH sensitivity, confirming that the residues underlying pH sensitivity do not alone reside in this region (Low et al., 2003; Gielen et al., 2008). Rather, multiple sites throughout the receptor synergize to sensitize NMDA receptors to the extracellular protons (Low et al., 2003; Gielen et al., 2008).
Cryo-EM studies of GluN1/2A receptors in alkaline (pH 7.8) and acidic conditions (pH 6.3) reveal NTD domain interactions and rearrangements that may reflect proton-inhibited states of the receptor (Lu et al., 2017; Zhang et al., 2018b). These studies suggest that under alkaline conditions, the GluN2A bilobed NTD adopts an open conformation that is twisted, whereas acidification (like Zn2+ binding) closes the bilobed domains, rearranging the tetrameric NTD and ABD layers (Zhang et al., 2018b). These data suggest that the NTD is a major substrate upon which reduced pH exerts its actions (Zhang et al., 2018b; Esmenjaud et al., 2019).
The mechanism of action of NMDA receptor NTD modulators appears to involve a change of the receptor’s proton sensitivity (e.g., pKa), which alters the degree of tonic inhibition at resting pH. Like Zn2+, the negative allosteric modulator ifenprodil enhances proton sensitivity and increases tonic inhibition at resting pH (Pahk and Williams, 1997; Traynelis et al., 1998; Mott et al., 1998; Choi and Lipton, 1999; Erreger and Traynelis, 2008; Bhatt et al., 2013). The shared effects of Zn2+ binding and protonation on the NTD heterodimer (Jalali-Yazdi et al., 2018; Zhang et al., 2018b) mirror functional studies showing quantitatively that Zn2+ inhibition can be accounted for by enhanced proton sensitivity (Erreger and Traynelis, 2005, 2008; Gielen et al., 2008). Alternatively, positive allosteric modulators, including extracellular polyamines, extracellular Mg2+, and some aminoglycosides, reduce the sensitivity of GluN2B-containing NMDA receptors to extracellular pH by reducing tonic proton inhibition (Paoletti et al., 1995; Traynelis et al., 1995; Kumamoto, 1996; Masuko et al., 1999a; Mony et al., 2011).
In contrast to inhibitory actions of protons on GluN1/2 NMDA receptors, acidification has been reported to potentiate glycine-activated current responses of GluN1/3A receptors with an EC50 value near physiologic pH but (like NMDA receptors) inhibits GluN1/3B receptors (Cummings and Popescu, 2016) (Table 5). Glycine-activated GluN1/3-mediated currents desensitize because of glycine binding to the GluN1 subunit (Awobuluyi et al., 2007; Madry et al., 2007; Kvist et al., 2013a), but increased proton concentration enhances glycine-activated currents in part through slower onset of desensitization and faster recovery from desensitization (Cummings and Popescu, 2016). The potentiation of GluN1/3A currents by extracellular pH is independent of alternative splicing of GluN1, independent of the actions of Zn2+, and dependent on residues at the GluN1/3A ABD dimer interface (Cummings and Popescu, 2016).
2. Modulation by Extracellular Polyamines
Intracellular polyamines have well established roles in the control of cell growth, interactions with nucleic acids (Fredriksson et al., 2019; Igarashi and Kashiwagi, 2019), and a high-affinity pore block of K+ channels, AMPA receptors, and kainate receptors (Section VIII.A.1. Channel Block of AMPA and Kainate Receptors by Polyamines). Extracellular polyamines, such as spermine and spermidine, are positive allosteric modulators of GluN2B-containing NMDA receptors function with EC50 values of ∼100 μM (Zhang et al., 1994). The actions of polyamines appear to reflect a relief of tonic proton inhibition, such that the pH sensitivity is reduced in the presence of polyamines. However, higher concentrations polyamines produce voltage-dependent channel block (Williams, 1997). Polyamine potentiation is absent in triheteromeric GluN1/2A/2B receptors (Hansen et al., 2014; Stroebel et al., 2014; Cheriyan et al., 2016; Yi et al., 2018), whereas polyamine potentiation is conserved but reduced in triheteromeric GluN1/2B/2D receptors (Yi et al., 2018).
Polyamines have been proposed to exert their potentiating actions through their interaction with clusters of negatively charged residues in the lower R2 lobes of bilobed GluN1 and GluN2B NTDs (Masuko et al., 1999a; Mony et al., 2011). Although the location of this binding site on the NTD remains to be identified, FRET studies suggest that spermine binding opens the GluN2B NTD clamshell (Sirrieh et al., 2015b). The actions of polyamines are attenuated by inclusion of residues encoded by GluN1 exon 5 (Durand et al., 1993; Zhang et al., 1994; Traynelis et al., 1995; Yi et al., 2018). Aminoglycosides are positively charged at physiologic pH and potentiate GluN2B-containing receptors, presumably by a similar mechanism as polyamines (Masuko et al., 1999b). The positively charged spermine has been proposed to shield negatively charged residues in the NTD, thereby eliminating electrostatic repulsion between the lower NTD lobes (Mony et al., 2011). Consistent with this model, extracellular Mg2+, in addition to its pore-blocking ability, can enhance GluN1/2B responses at millimolar concentrations (Paoletti et al., 1995), a phenomenon that is visible for outward currents at positive holding potentials, at which Mg2+ channel block is absent and the potentiating effects of Mg2+ prevail. Histamine also mediates GluN2B-selective potentiation that is suggested to involve the same site that responds to Mg2+ and polyamines, but structural determinants of this effect as well as its role in neurons remain elusive (Bekkers, 1993, 1996; Williams, 1994; Saybasili et al., 1995; Watanabe et al., 2008; Burban et al., 2010).
D. Neurosteroids
Several endogenous neurosteroids can influence circuit function through their ability to modulate NMDA receptor activity (Ratner et al., 2019); however, the mechanisms, subcellular localization (Chisari et al., 2019), and properties of these agents are complex. Pregnenolone sulfate (20-oxo-5-pregnen-3β-yl sulfate, PS) both inhibits and potentiates NMDA receptor activity over a range of potencies (Gibbs et al., 2006; Horak et al., 2006) (Table 6) and inhibits AMPA, kainate, and GABA receptors (Park-Chung et al., 1994). Strong potentiating actions of PS are evident when applied before receptor activation, whereas inhibitory actions arise when coapplied continuously with agonist (Horak et al., 2006). The dual actions of PS lead to divergent effects that depend on the GluN2 subunit; when applied during steady-state NMDA receptor responses, GluN1/2A and GluN1/2B are potentiated, and GluN1/2C and GluN1/2D are inhibited (Horak et al., 2006). PS produces a modest enhancement of glutamate potency at GluN1/2B (Horak et al., 2004) and slows desensitization and deactivation at GluN1/2A and GluN1/2B (Ceccon et al., 2001). PS prolongs channel open time in nondialyzed cells (Chopra et al., 2015) but has no effect on single-channel conductance (Wong and Moss, 1994). These actions of PS may require an intact intracellular environment (Petrovic et al., 2009) and could be accompanied by effects on NMDA receptor trafficking (Kostakis et al., 2013). Structural determinants underlying the actions of PS have been identified in the S2 segment of the ABD, the S2-M4 linker, and the TMD (Jang et al., 2004; Horak et al., 2006; Kostakis et al., 2011; Burnell et al., 2019), and the actions of PS may involve rearrangement of residues from M1 and M4 in GluN2 and M3 in GluN1 (Hrcka Krausova et al., 2020).
TABLE 6.
Other endogenous neurosteroids include the nonselective NAM pregnanolone sulfate (referred to as 3α5βS or PA-S) and the nonselective PAM 24(S)-hydroxycholesterol (24(S)-HC) (Park-Chung et al., 1994; Malayev et al., 2002; Borovska et al., 2012; Paul et al., 2013; Vyklicky et al., 2015), which acts at the TMD of NMDA receptors (Wilding et al., 2016). 24(S)-HC has been shown to have numerous effects on circuit function, including modulation of synaptic plasticity and effects of ethanol on learning (Izumi et al., 2021). The endogenous neurosteroid 25-hydroxycholesterol has been referred to as a silent or neutral modulator, with minimal effects on NMDA and AMPA receptors on its own but nearly complete block of the potentiating effects of 24(S)-HC, suggesting 25-hydroxycholesterol is a natural counterbalance to the potentiating actions of 24(S)-HC at NMDA receptors (Linsenbardt et al., 2014; Sun et al., 2017a). Some of the endogenous neurosteroid NAMs are agonist-dependent (Petrovic et al., 2005; Borovska et al., 2012; Vyklicky et al., 2015) and promote receptor desensitization without changing single-channel open time (Kussius et al., 2009). The neurosteroid pregnanolone sulfate can control NMDA receptor function in a manner dependent on palmitoylation (Hubalkova et al., 2021), and these steroid derivatives in general may partition into the membrane en route to their active site, which could alter their concentration-response relationship (Malayev et al., 2002; Borovska et al., 2012; Vyklicky et al., 2015). Cholesterol also modulates NMDA receptor function, and its removal inhibits receptor activity (Korinek et al., 2015), suggesting that the membrane environment influences NMDA receptor activity and could be an important determinant of neurosteroid action. 24(S)-HC can also compensate for effects on NMDA receptor function of cholesterol depletion (Hrcka Krausova et al., 2020). Unlike for GABA receptors (Laverty et al., 2017), a binding site for endogenous neurosteroids on NMDA receptors has not been clearly defined. A subset of neurosteroid inhibitors also have voltage-dependent actions, suggesting that they may inhibit NMDA receptors by blocking the channel pore (Vyklicky et al., 2015).
IX. Exogenous Positive and Negative Allosteric Modulators
A. AMPA and Kainate Receptor Modulators
AMPA and kainate receptor function can be positively and negatively tuned by allosteric modulators that interact with receptor subunits at sites that are distinct from the orthosteric agonist binding site. PAMs that act on AMPA receptors are extensively studied because of their therapeutic potential as cognitive enhancers and have become useful research tools to explore the structural changes that underlie AMPA receptor deactivation and desensitization. AMPA receptor–selective negative allosteric modulators (NAMs) are also useful tools, and AMPA receptor inhibitors have facilitated the characterization of neuronal kainate receptors. The AMPA receptor NAM perampanel has received FDA approval as an adjunctive therapy for partial onset seizures and for tonic-clonic seizures resistant to conventional therapeutics (Section X. Glutamate Receptors in Disease).
The development of kainate receptor allosteric modulators has lagged behind that for AMPA receptors. Agents that interact with oligosaccharide conjugates on the receptors and thereby constrain gating are PAMs of kainate receptors, but only a single study has reported PAMs that bind at the kainate receptor ABD dimer interface (Larsen et al., 2017) (see below). Likewise, kainate receptor NAMs consist only of compounds with weak crossreactivity described for compounds characterized as AMPA receptor NAMs and nonselective fatty acids. Nonetheless, new allosteric modulators for AMPA and kainate receptors remain ripe for exploration, as can be appreciated by the development of auxiliary subunit-dependent PAMs and NAMs for AMPA receptors (Section X. Glutamate Receptors in Disease).
1. AMPA Receptor Negative Modulators
The well characterized AMPA receptor NAMs reduce channel activity by disrupting the coupling between agonist binding to the ABD and conformational changes of the TMD that result in pore opening. These include the 2,3-benzodiazepines, the quinazoline inhibitors, and bipyridine compounds. However, none of these AMPA receptor NAMs show marked sensitivity to subunit composition (Bleakman et al., 1996; Lazzaro et al., 2002; Zwart et al., 2014). AMPA receptor NAMs that act on different allosteric sites (Chang et al., 2016) or that exhibit auxiliary subunit-dependent activity (Maher et al., 2017) provide new, fruitful directions for exploration of AMPA receptor modulation (Section X. Glutamate Receptors in Disease).
A family of 2,3-benzodiazepines first synthesized thirty years ago (Tarnawa et al., 1989; Bleakman et al., 1996; Lodge et al., 1996) is now used routinely to test the role of AMPA receptor signaling in physiologic processes. The mostly widely studied 2,3-benzodiazepine compounds include GYKI52466, GYKI-53405 (or LY-293606), and GYKI-53655 (or LY-300168) (Table 7). GYKI-53405 is the racemate of the early clinical candidate talampanel (Lodge et al., 1996). The negative modulation of AMPA receptors by 2,3-benzodiazepines was first described for agonist-evoked currents from AMPA receptors expressed by cultured hippocampal neurons (Donevan and Rogawski, 1993). Their anticonvulsant potential was noted soon after reports on their inhibitory activity on AMPA receptors (Smith et al., 1991; Loscher and Honack, 1994), but motor impairments proved to be a hallmark of strong inhibition of AMPA receptors in the CNS (Yamaguchi et al., 1993).
TABLE 7.
Soon after their development, GYKI-52466 and GYKI-53655 were employed to dissociate AMPA and kainate receptor currents in hippocampal and DRG neurons (Paternain et al., 1995; Wilding and Huettner, 1995). An important caveat to studies using GYKI-52466 and GYK-53655 to differentiate AMPA and kainate receptor signaling is that they are not absolutely AMPA receptor-selective and partially inhibit some kainate receptor subtypes at concentrations typically used for complete ablation of AMPA receptor signaling (30–50 μM) (Wilding and Huettner, 1995; Bleakman et al., 1996; Perrais et al., 2009b). Nonetheless, 2,3-benzodiazepine AMPA receptor NAMs define much of what we understand about neuronal kainate receptor function [reviewed in Contractor et al. (2011), Lerma and Marques (2013)].
A second chemotype of AMPA receptor NAM was derived from a quinazoline anticonvulsant, and synthetic efforts yielded CP-465,022 (Table 7). This compound inhibits AMPA receptor-mediated Ca2+ uptake into neurons with nanomolar potency, exhibits efficacy in animal seizure models (Welch et al., 2001), and shares an allosteric binding site with GYKI-52466 (Menniti et al., 2000; Balannik et al., 2005; Yelshanskaya et al., 2016b). Like the GYKI compounds, the anticonvulsant activity of CP-465,022 in animal seizure models occurred at doses only modestly lower than those that decreased locomotor activity (Menniti et al., 2003). Quinazoline scaffolds have been modified in the search for new anticonvulsants, some of which also act on NMDA receptors (Mosley et al., 2010; Ugale and Bari, 2014).
A high-throughput screen for structurally novel AMPA receptor NAMs identified a third chemical scaffold interacting with the GYKI binding site exemplified by perampanel (Hanada et al., 2011; Hibi et al., 2012). Perampanel inhibits neuronal AMPA receptors with nanomolar potency and is without apparent effect on NMDA or kainate receptor currents (Hanada et al., 2011; Ceolin et al., 2012; Chen et al., 2014a). Perampanel shares a binding site on AMPA receptors with CP-465,022 and GYKI-53655 (Hanada et al., 2011; Yelshanskaya et al., 2016b), and inhibits AMPA receptor currents with ∼10-fold higher potency compared with GYK-53655 (Yelshanskaya et al., 2016b) (Table 7).
The site of action of AMPA receptor NAMs discussed above resides in the linkers that transduce the energy of agonist binding at the bilobed ABD to open the pore via movement of the M3 transmembrane helix (Yelshanskaya et al., 2016b). The S1-M1, M3-S2, and S2-M4 linkers, referred to as a gating triad in NMDA receptors (Fig. 19), undergo rearrangements that are critical to receptor gating (Sobolevsky et al., 2009; Chen et al., 2017a; Twomey et al., 2017a) and confer a state dependence to the binding affinity of GYKI-53655 and CP-465,022. Both NAMs bind with a higher affinity to the resting state as compared with the agonist-bound activated conformation (Balannik et al., 2005). Chimeric studies first suggested that the S1-M1 and S2-M4 linkers on adjacent subunits were key domains for GYKI-53655 and CP-465,022 actions (Balannik et al., 2005). Determination of the site of action came with crystallographic structures of the homomeric GluA2 receptor in complex with GYKI-53655, CP-465,022, or perampanel, which bound largely within individual subunits (Yelshanskaya et al., 2016b). The structures also underscored the overlap in atomic contacts between each of the three NAMs and the binding pocket (Hanada et al., 2011) (Fig. 41). Residues that directly interact with the noncompetitive inhibitors are located within the S1-M1 and S2-M4 linkers and the extracellular segments of the M3 and M4 domains (Yelshanskaya et al., 2016b). Molecular dynamic simulations of NAM binding and further mutagenesis of putative interacting residues suggest that the NAM binding pocket in AMPA receptors is flexible enough to allow for interaction of diverse chemical structures that assume distinct poses within the pocket (Narangoda et al., 2019; Stenum-Berg et al., 2019). The mechanism of negative allosteric modulation was envisioned as constraining conformational changes required for channel opening by stabilizing the closed state of the receptor (Balannik et al., 2005; Yelshanskaya et al., 2016b). Single-channel studies support a nuanced view in which NAMs alter channel behavior when occupancy of binding sites is subsaturating (Shi et al., 2019; Yuan et al., 2018). An analogous site harbors the structural determinants for multiple NMDA receptor allosteric modulators (see below).
Fig. 41.

Sites of AMPA receptor modulation. (A) Structure of GluA2 AMPA receptor (PDB: 6DM1) with four subunits colored pink, gray, light green, and light blue. The positive allosteric modulator Conus striatus cone snail toxin con-ikot-ikot (PDB: 4U5B) is shown in red, and small-molecule regulators are shown in sticks (yellow). (B) An expanded view of the ABD dimer with glutamate and cyclothiazide (CTZ) bound (PDB: 1LBC). The inset shows an expanded view of the positive allosteric modulator binding site at the ABD dimer interface with CTZ (yellow; PDB: 1LBC), aniracetam (green; PDB: 2AL5), LY404187 (pink; PDB: 3KGC), CX614 (cyan; PDB: 2AL4), and a dimeric biarylpropylsulfonamide (BPSA, dark blue; PDB: 3BBR). (C) An expanded view of the TMD, with the front and back subunits (B and D) removed for clarity. The negative allosteric modulators perampanel (PDB: 5L1F) and 4-BCCA (PDB: 6XSR) are shown. The inset shows an expanded view of the binding site at the ion channel extracellular collar with CP-465,022 (blue; PDB: 5L1E), perampanel (yellow; PDB: 5L1F), and GYKI53655 (dark green; PDB: 5L1H).
A new class of AMPA receptor NAM was discovered through the isolation of bioactive constituents of the ketogenic diet used for treatment of drug-resistant epilepsies. The formulation of a medium-chain triglyceride ketogenic diet contains several fatty acids that include decanoic acid, which has anticonvulsant activity (Wlaz et al., 2012; Chang et al., 2013, 2016). The mechanism of antiseizure actions of decanoic acid includes direct inhibition of AMPA receptors at relatively high millimolar concentrations. Molecular modeling suggested that decanoic acid binds to residues within the M3 domain, potentially acting as a NAM with a site of action distinct from the pre-M1/M3 site discussed above (Chang et al., 2016). This hypothesis received experimental support from the demonstration of a synergistic effect of perampanel and decanoic acid for AMPA receptor inhibition and in in vitro seizure assays (Augustin et al., 2018). Concurrent screening of octanoic acid derivatives identified 4-BCCA as having anticonvulsant activity in in vitro and in vivo seizure models (Chang et al., 2015b). Crystallographic resolution of 4-BCCA in complex with GluA2 showed that the fatty acid resides at the interface between the M1 and M3 helices of adjacent subunits (Yelshanskaya et al., 2020) (Fig. 41). 4-BCCA may inhibit AMPA receptors by direct occlusion of permeation through the channel, imposing constraints on M3 dynamics underlying channel opening, or destabilization of the pore by displacing lipids in the membrane (Yelshanskaya et al., 2020).
NAMs with selectivity between AMPA receptors in different brain regions have potential to minimize adverse effects while maintaining efficacy as anticonvulsants or other therapeutically useful effects (Maher et al., 2017). A strategy to identify such compounds predicated on the differential regional distribution of AMPA receptor auxiliary subunits (Maher et al., 2017) yielded two NAMs that were selective for AMPA receptors complexed with TARP γ-8, LY-3130481 (or CERC-611) (Gardinier et al., 2016; Kato et al., 2016) and JNJ-55511118 (Maher et al., 2016). These NAMs preferentially bind AMPA receptors localized to the hippocampus based on the distribution of γ-8 (Tomita et al., 2003; Fukaya et al., 2006). For example, LY-3130481 inhibits AMPA receptors in hippocampal neurons but not Purkinje cells, which is consistent with a selective effect on AMPA receptors associated with γ-8 but not γ-2 (Kato et al., 2016; Maher et al., 2016). Similarly, γ-2- and CNIH3-selective compounds were recently described (Azumaya et al., 2017), as well as newer γ-8–selective AMPA receptor NAMs (Savall et al., 2018). Although the mechanism of action of auxiliary subunit-selective NAMs remains to be elucidated, studies using mutagenesis, domain-swapping, and computational modeling suggest a role for specific residues in the transmembrane segments of γ-8 (Kato et al., 2016; Maher et al., 2016; Lee et al., 2017a). The development of these and related modulators opens a new chapter in the search for drugs targeting AMPA receptors with regional specificity and reduced adverse effects.
2. AMPA Receptor Positive Modulators
Positive allosteric modulators selective for AMPA receptors have provided insights into excitatory synaptic signaling and AMPA receptor structure and function (Arai and Kessler, 2007; Partin, 2015). The point of initiation was the discovery that the nootropic drug aniracetam enhanced AMPA receptor current amplitudes (Ito et al., 1990). Aniracetam increases the peak amplitude of EPSCs (Ito et al., 1990; Tang et al., 1991) and enhances hippocampal LTP (Arai and Lynch, 1992). Aniracetam gave rise to a family of benzamide AMPA PAMs that include the early CX-series compounds termed “ampakines” (Hampson et al., 1998a,b). Subsequently, several distinct chemotypes of AMPA receptor PAMs have been characterized that bind to conserved sites located on the AMPA receptor ABD interface in at least three different modes (Fig. 41). For some PAMs, including cyclothiazide, two molecules bind at equivalent sites on each side of the dimer interface (Sun et al., 2002; Hald et al., 2009; Ptak et al., 2009; Norholm et al., 2013). For other PAMs, such as aniracetam and CX614, one molecule binds but with two overlapping conformations in a more central site within the dimer interface (Jin et al., 2005; Krintel et al., 2013; Goffin et al., 2018). Finally, the PAM N,N′-(1,4-Phenylenedi-2,1-ethanediyl)bis-2-propanesulfonamide (CMPDA) has a symmetrical chemical structure and binds to a central site (Kaae et al., 2007; Sobolevsky et al., 2009; Ahmed et al., 2010; Timm et al., 2011; Laulumaa et al., 2018). The AMPA receptor PAMs increase the stability of the ABD dimer interface to slow the transition into the desensitized state, and a subset of PAMs also slows receptor deactivation (Jin et al., 2005; Mitchell and Fleck, 2007; Partin, 2015; Shaffer et al., 2015). Alternative splicing to flip/flop isoforms of AMPA receptor subunits creates differences at the ABD dimer interface, and PAMs can be isoform-preferring (Partin et al., 1996). Most AMPA receptor PAMs are selective for the flip isoform, with notable exceptions being flop-selective 4-[2-(phenylsulfonylamino)ethylthio]-2,6-difluoro-phenoxyacetamide (PEPA) (Sekiguchi et al., 1997) and flop-preferring aniracetam (Johansen et al., 1995; Partin et al., 1996). The discovery of the distinct binding modes of AMPA receptor PAMs, with subsites spanning the dimer interface (Fig. 41), stimulated the development of dimeric PAMs that interact with multiple subsites with increased affinity (Kaae et al., 2007; Ahmed and Oswald, 2010; Ptak et al., 2014; Laulumaa et al., 2018). Characterization of early AMPA receptor PAMs has been described in several excellent reviews (Partin, 2015; Brogi et al., 2019).
AMPA receptor PAMs are functionally diverse with a range of different chemotypes (Partin, 2015; Brogi et al., 2019). Benzamide derivatives, such as aniracetam, CX-516, and other ampakines, primarily slow deactivation of AMPA receptors (Tang et al., 1991; Arai et al., 2002). In contrast, benzothiadiazines, such as the diuretic cyclothiazide, block entry into the desensitized state and have modest effects on deactivation (Partin et al., 1993; Dzubay and Jahr, 1999). Cyclothiazide exhibits greater efficacy on flip-containing isoforms by virtue of a specific interaction with a Ser in the flip splice cassette (Ser754 in GluA2) (Partin et al., 1994, 1995). A recent medicinal chemistry campaign yielded a high-affinity series of benzothiadiazines, including some with nanomolar EC50 values for potentiation of AMPA receptor signaling (Goffin et al., 2018). The biarylpropylsulfonamides include PEPA (flop-specific) and the high-affinity clinical candidate LY-451395 (or mibampator) (flip-specific) (Shepherd et al., 2002; Ishii et al., 2020). The tetrahydrofuran ether PF-04958242 (also known as BIIB104) (Table 7) completely occludes the desensitization state in flip-containing receptors, with less dependence on the flip/flop isoform for deactivation (Shaffer et al., 2015; Ishii et al., 2020). A comparative study with BIIB104 and LY-451395 revealed that TARP auxiliary subunits shape the effects on deactivation of AMPA receptors and eliminate most differential effects of flip/flop splicing (Ishii et al., 2020). These data predict that the potentiating activity of PAMs could show regiospecificity at synapses that follows the expression pattern of auxiliary subunits (Tomita et al., 2006) (Fig. 42). The functional actions of new agents on AMPA receptors so far have been similar to existing PAMs (Partin, 2015; Bretin et al., 2017; Brogi et al., 2019; Ward et al., 2020).
Fig. 42.
Modulation of AMPA receptors by PAMs is sensitive to RNA splicing and auxiliary subunits. Left panels: The whole cell current response of homomeric GluA4-flip (A) or GluA4-flop (B) splice isoforms recorded in response to brief application of 10 mM glutamate in the absence or presence of 10 µM of the AMPA receptor PAM BIIB104. Right panels: The current response of GluA4-flip and GluA4-flop receptors in response to prolonged application of 10 mM glutamate in the absence or presence of 10 µM BIIB104 to measure the rate and extent of desensitization. (C) The effect of 1 µM BIIB104 is shown on hippocampal Schaffer collateral to CA1 pyramidal cell EPSCs (upper panel) or on mossy fiber to cerebellar granule cell EPSCs. Reproduced with permission from Ishii et al. (2020). (D) Left panels: Outside-out patch recordings of glutamate-evoked currents from cells expressing AMPA receptors coexpressed with either γ-2 or γ-8 TARPs in the presence (red) and absence (black) of 1 µM JNJ-55511118. Right panels: Outside-out patch recordings of glutamate-evoked currents in patches from hippocampal or cerebellar neurons in the presence (red) and absence (black) of 1 µM JNJ-55511118. Glutamate (10 mM) was applied during the time depicted by the gray bar. Reproduced with permission from Maher et al. (2016).
AMPA receptor PAMs have been considered for a variety of therapeutic applications based on preclinical data in which animals treated with such compounds show improved performance in cognitive tasks (Section X. Glutamate Receptors in Disease). Procognitive effects have been demonstrated for AMPA receptor PAMs of many different chemotypes [e.g., Granger et al. (1993), Larson et al. (1995), Hampson et al. (1998a,b), O'Neill et al. (2004)]. Although the mechanisms by which AMPA receptor PAMs lead to cognitive improvement are not fully understood (Lynch and Gall, 2006), PAMs prolong the decay of AMPA receptor–mediated synaptic currents (e.g., Ito et al., 1990; Tang et al., 1991; Arai et al., 1994) and facilitate induction and magnitude of LTP (Arai and Lynch, 1992; Staubli et al., 1994). One line of inquiry focuses on the up-regulation of brain-derived neurotrophic factor (BDNF) expression, enhanced circuit excitability, and subsequent homeostatic normalization of activity (Lauterborn et al., 2000; Jourdi et al., 2009). AMPA receptor PAM-induced BDNF release has diverse activities on synaptic plasticity, spine stability, and other processes central to both memory formation and retention of cognitive efficacy [e.g., Simmons et al. (2011), Kramar et al. (2012), Lauterborn et al. (2016)].
Resolution of the binding sites for AMPA receptor PAMs has enabled the development of mechanistic models for the conformational changes that accompany deactivation and desensitization processes. As described in Section IV. Receptor Activation, Deactivation, and Desensitization, desensitization of AMPA receptors involves distinct reorganization of ABD dimer-of-dimers and transmission of those structural changes to the pore domain. Desensitization is the functional feature most profoundly altered by cyclothiazide and the finding that it stabilizes the D1 interface implicated decoupling of the D1 interface as a key constituent of desensitization (Sun et al., 2002). A hydrogen bond between cyclothiazide and Ser754 (in GluA2) in the flip/flop cassette explained the higher potency of the PAM on flip-containing AMPA receptors (Partin et al., 1995; Sun et al., 2002). Aniracetam and CX-614 (Table 7), which slow deactivation, interact at single sites in the interdomain hinge region with a small degree of overlap (Jin et al., 2005). The crystal structures led to the proposal that binding of the PAMs stabilized the closed, agonist-bound state of the receptor and thereby slowed agonist unbinding and deactivation (Jin et al., 2005). The binding mode of CX-516 was similar to that of aniracetam and CX-614 (Krintel et al., 2013). Structural resolution of the high-affinity biarylpropylsulfonamide (R,R)-2a in complex with the GluA2 ABD revealed a new symmetrical binding mode in which a single PAM molecule bridges the two cyclothiazide binding pockets (Kaae et al., 2007). A related PAM, (R,R)-2b, was used to explore activation mechanisms in GluA2 (Chen et al., 2014b). Insights into the flop-isoform selectivity of PEPA arose from structure of the PAM in complex with the GluA2 and GluA3 ABDs, which demonstrated that the amide group in the PAM forms a hydrogen bond with Asn754 (Ahmed et al., 2010). These data and an analysis of a series of thiazide derivatives led to the compartmentalization of the U-shaped binding cavity at the ABD interface into five subsites that are differentially occupied by different PAMs (Ptak et al., 2009, 2014).
The predatory Conus striatus cone snail toxin known as con-ikot-ikot acts as an AMPA receptor PAM and interacts at sites distinct from the ABD interface described above (Walker et al., 2009). The peptide toxin potentiates steady-state currents from AMPA receptors with somewhat greater efficacy on flop isoforms but has no detectable effect on NMDA or kainate receptors. X-ray crystallographic resolution of the toxin in complex with nearly full-length GluA2 receptors confirmed that dimeric con-ikot-ikot intercalates between the NTD and ABD of all four subunits in the GluA2 receptor (Chen et al., 2014b) (Fig. 41), reminiscent of the proposed site of action of modulatory lectins on kainate receptor gating (Fay and Bowie, 2006; Copits et al., 2014) as well as the GluN2C-selective NMDA receptor PAM, PYD-106 (Khatri et al., 2014). Negatively charged regions of the con-ikot-ikot toxin form ionic and polar contacts with positive residues on the outer surface of the ABD that are unique to AMPA receptors (Walker et al., 2009; Chen et al., 2014b). Binding of the toxin involved extensive intersubunits contacts by the dimeric con-ikot-ikot toxin and was state-dependent by requiring agonist binding and ABD cleft closure. Based on these insights, potentiation was proposed to result from a constraint imposed by the toxin on the conformational changes that underlie desensitization.
3. Kainate Receptor Negative and Positive Modulators
Few selective allosteric modulators of kainate receptors have been identified. Given the structural differences between AMPA and kainate receptors, the paucity of kainate receptor modulators likely reflects a relative lack of attention in drug development rather than intransigence as molecular targets. Nonetheless, kainate receptor inhibition reduces pain and seizures in animal models [reviewed in Bhangoo and Swanson (2013), Crepel and Mulle (2015)]. Thus, in principle, kainate receptor-selective NAMs represent a potentially fruitful avenue of exploration.
Kainate receptors are inhibited with low potency and efficacy by AMPA receptor NAMs, such as GYKI-52466, GYKI-53655, and CP-465,022, likely because binding site residues on the surface of the TMD are partially conserved between AMPA and kainate receptors (Wilding and Huettner, 1995; Bleakman et al., 1996; Lazzaro et al., 2002; Perrais et al., 2009b). A variety of cis-unsaturated fatty acids, including arachidonic acid and docosahexanoic acid, also reversibly inhibit kainate receptor signaling by acting within the TMD (Wilding et al., 1998), but AMPA and NMDA modulation by these molecules has also been reported (Miller et al., 1992; Kovalchuk et al., 1994). Curiously, inhibition requires RNA editing of the Q/R/N site to the positively charged Arg despite the voltage independence of inhibition by these NAMs (Wilding et al., 1998, 2005, 2008, 2010).
One study has reported PAMs for kainate receptors BPAM344 and BPAM521, with activity similar to the numerous AMPA receptors PAMs that bind the ABD interface (Larsen et al., 2017). Structures of the isolated GluK1 ABD dimer show that these PAMs bind at two equivalent sites in the ABD dimer interface (Larsen et al., 2017). These PAMs enhance kainate receptor responses with low potency by slowing the rate of desensitization but are nonselective and more potent at AMPA receptors (Larsen et al., 2017). Nonetheless, these PAMs demonstrate that the ABD dimer interface of kainate receptors can be targeted for modulation of receptor desensitization.
Irreversible potentiation of both AMPA and kainate receptor currents by the high-mannose binding lectin, Concanavalin A (ConA), (Mathers and Usherwood, 1976; Mayer and Vyklicky, 1989) is relatively selective for kainate receptors (Partin et al., 1993). The high mannose glycan chains that form binding sites for ConA and other lectins are distributed throughout extracellular regions of kainate receptor subunits (Everts et al., 1999), although sugars within the region between the NTD and the ABD appear to be important for potentiation (Fay and Bowie, 2006; Copits et al., 2014). Analysis of ConA activity with agonists of varying efficacy revealed potentiation was dependent upon the conformational state of the ABD; ConA was least efficacious when applied to receptors occupied by full agonists, and the degree of potentiation was inversely correlated with agonist efficacy (Fay and Bowie, 2006). ConA does not alter glutamate potency or deactivation kinetics (Bowie et al., 2003) and is without effect on synaptic kainate receptors (Ito et al., 2004), making it of limited utility as a pharmacological probe of native receptor function. The mechanism by which ConA potentiates kainate receptor currents was initially proposed to be through a reduction in desensitization (Partin et al., 1993; Wilding and Huettner, 1997); alternate ideas include promotion of channel opening from closed states (Paternain et al., 1998) and a lectin-dependent shift in relative contributions from distinct open states (Bowie et al., 2003).
A family of mammalian β-galactoside–selective lectins known as galectins might act as endogenous modulators of iGluR function (Copits et al., 2014). ConA and other lectins used for kainate receptor potentiation are derived from plants and have no obvious mammalian orthologs, and thus the physiologic relevance (if any) of their PAM activity was an open question. ConA binds specifically to immature high-mannose glycans that are conjugated to receptor subunits in the ER (Thalhammer et al., 2002). N-Glycan processing of integral membrane proteins proceeds through their biogenesis in the Golgi and trans-Golgi compartments, and remodeling results in the incorporation of complex oligosaccharide chains. Glycans conjugated to kainate and AMPA receptors can be processed to contain repeating polylactosamine disaccharides, which serve as the ligand for galectins. Human galectin-1 and eel congerin-1 galectins slow or occlude desensitization of recombinant AMPA and kainate receptors to differing degrees (Copits et al., 2014). Glycans located in the linker domain between the NTD and ABD were necessary components for galectin modulation. Galectin-1 is expressed in a variety of neurons in the CNS, suggesting they could act as endogenous PAMs, and indeed both galectin-1 and congerin-1 slowed desensitization of kainate receptor currents in DRG neurons (Copits et al., 2014).
B. NMDA Receptor Modulators
A number of competitive antagonists and channel blockers of NMDA receptors have served as effective pharmacological tools to distinguish the activity of NMDA receptors from kainate and AMPA receptors. The different mechanistic effects of these ligands have been used in clever ways to explore principles of NMDA receptor signaling [e.g., Clements et al. (1992), Hessler et al. (1993), Murthy et al. (1997), Tovar and Westbrook (2002)]. Several NMDA receptor channel blockers have been approved by the FDA for therapeutic use, including memantine for Alzheimer disease and esketamine for depression (Section X. Glutamate Receptors in Disease). However, the recent focus of preclinical research has been on the identification of NMDA receptor subunit-selective modulators. The competitive antagonists and channel blockers bind to the most conserved portions of the receptor subunits and thus have limited GluN2 subunit selectivity (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers). Like AMPA and kainate receptors, NMDA receptors contain multiple unique intraprotein and interprotein interfaces at which small molecules can bind to modulate domain movements during activation of the receptor (Fig. 43). A number of PAMs and NAMs with diverse mechanisms of action and subunit selectivity have been identified, providing new subunit-selective tool compounds to further advance our understanding of NMDA receptor function and provide substrates for new therapeutics.
Fig. 43.
Protein-protein domain and subunit interfaces in NMDA receptors. A) Surface presentation of GluN1/2B structure (PDB: 6WHS). B) The subunits and domains are translationally shifted to reveal and emphasize the inter– and intra–protein-protein interfaces within the multimeric protein complex. These sites indicated by red lines are locations for potential allosteric modulator binding.
1. GluN2A-Selective Negative Modulators
A series of GluN2A-selective NAMs that includes TCN-201 has been described (Bettini et al., 2010). These compounds are potent (i.e., KB for TCN-201 was 27–70 nM) (Table 8) and selective for GluN2A over other GluN2 subunits (Edman et al., 2012; Hansen et al., 2012; McKay et al., 2012; Yi et al., 2016) (Fig. 44). TCN-201 inhibition of GluN2A is insensitive to glutamate concentration but is diminished in high concentrations of glycine, which complicates its utility in neurophysiological studies. However, TCN-201 is a negative allosteric modulator of glycine affinity rather than directly competing with glycine binding (Hansen et al., 2012) (Table 9). Crystallographic data show that TCN-201 binds at the GluN1 and GluN2A ABD heterodimer interface with a pose containing π-stacking aromatic rings as a result of a turn produced by the sulfonamide group (Yi et al., 2016) (Fig. 44). MPX-004 is GluN2A-selective NAM related to TCN-201 that binds to the same modulatory site but with improved potency (Table 8), improved solubility, and more complete inhibition at saturating concentrations (Volkmann et al., 2016; Yi et al., 2016).
TABLE 8.
Fig. 44.
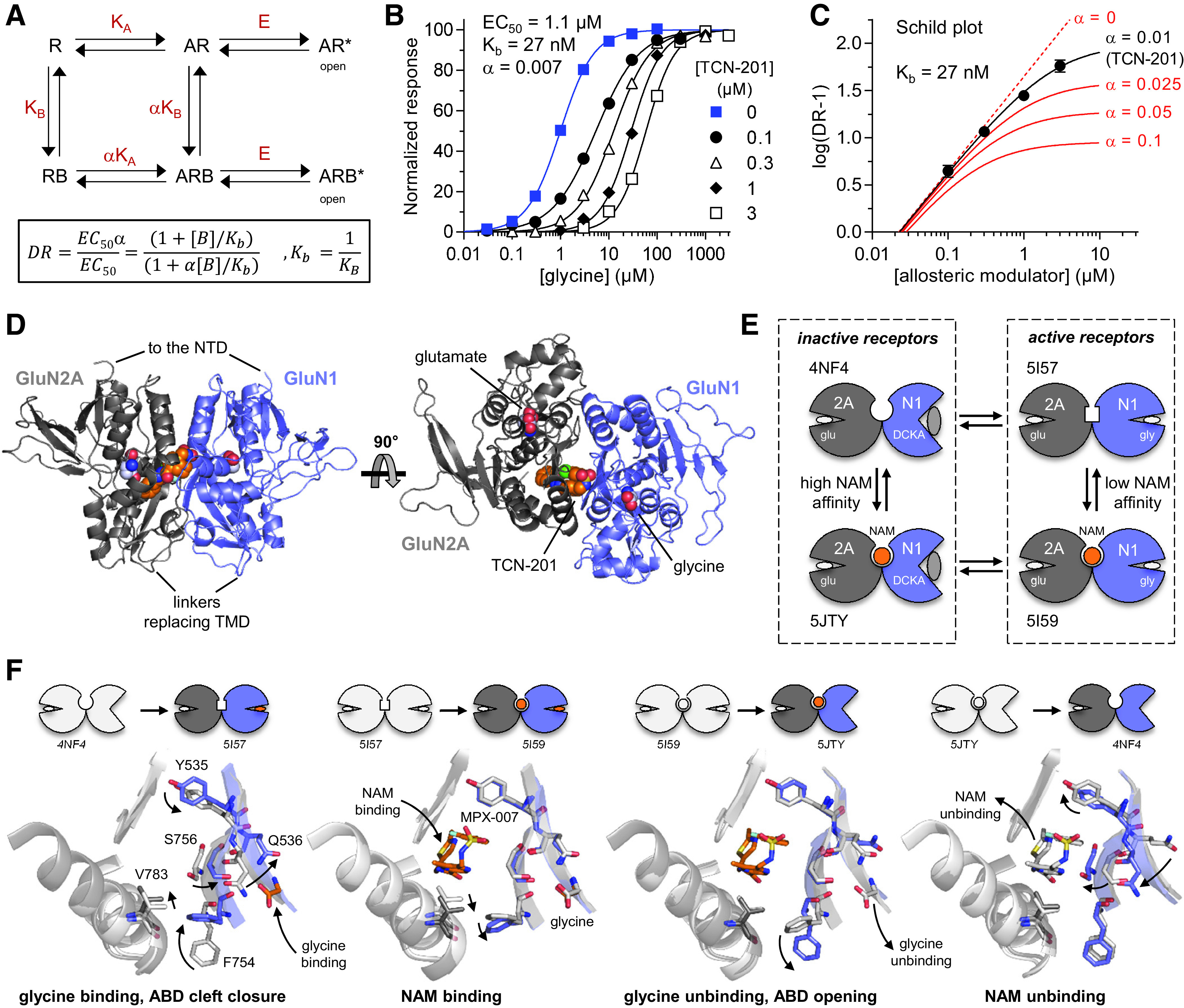
Mechanism of negative allosteric modulation by GluN2A-selective NMDA receptor antagonists. (A) Model for inhibition by negative modulation of glycine binding without changing agonist efficacy (E). A is the agonist, B is the NAM, and R is the receptor. KA and KB are equilibrium association constants for agonist and NAM, respectively. KA is changed by the allosteric constant α upon NAM binding and vice versa. DR is the dose ratio (i.e., the ratio of agonist EC50 values in presence and absence of modulator). Kb is the NAM dissociation constant. (B) Glycine concentration-response data for GluN1/2A in the absence or presence of the NAM, TCN-201, are analyzed by simultaneously fitting all data to the DR equation and the Hill equation using global nonlinear regression, yielding glycine EC50, TCN-201 binding constant (Kb), and allosteric constant α that describe all the experimental data. (C) DR values derived from fitting individual concentration-response data shown in (B) are plotted as a function of TCN-201 concentration, which also illustrates the effects of changing the allosteric constant α but with constant Kb. Competitive antagonists have α = 0 (dashed line). (D) Crystal structure of the agonist-bound GluN1/2A ABD heterodimer in complex with TCN-201 (PDB: 5I56). (E) Cartoon illustrating conformations of GluN1/2A ABD crystal structures that represent states in the NAM inhibition cycle [i.e., a Monod-Wyman-Changeux (MWC) model] (PDB codes are listed). Glutamate is continuously bound to the GluN2A ABD, and the competitive antagonist DCKA stabilizes the open (i.e., glycine-lacking) state of the GluN1 ABD. NAM binding stabilizes the inactive receptor with no glycine bound to GluN1 subunits. (F) Structural changes in modulatory binding site during the NAM inhibition cycle. Binding of NAM, in this case MPX-007, displaces GluN2A V783, and this “push” is accompanied by a steric effect on GluN1 F754, which undergoes movements during opening and closure of the GluN1 ABD. DCKA is omitted from PDB: 4NF4 and PDB: 5JTY. Modified with permission from Yi et al. (2016).
TABLE 9.
Distinct mechanisms of action of NMDA receptor modulators
| Selectivity | Mode of Action | Modulator | Open Probability † | Glutamate EC50 | Glycine EC50 |
Tau
Deact |
Unitary Conductance |
Ionic
Permeability |
|---|---|---|---|---|---|---|---|---|
| GluN2A | NAM | TCN-201 | – | NE | ↑ | ↑ | – | – |
| GluN2A | PAM | GNE-8324 | ↑ | ↓ | NE | ↑ | – | – |
| GluN2B | NAM | ifenprodil | ↓ | ↓ | – | ↑ | NE | – |
| GluN2C | PAM | PYD-106 | ↑ | ↓ | NE | ↑ | NE | – |
| GluN2C/D | NAM | QNZ-46 | ↓ | ↓ | NE | ↑ | NE | – |
| GluN2C/D | PAM | (+)-CIQ | ↑ | NE | NE* | NE* | NE | – |
| pan | NAM# | EU1794-4 | ↑ | ↓ | ↓ | ↑ | ↓ | ↓ pCa /pNa |
| pan | PAM | EU1622-14 | ↑ | ↓ | ↓ | ↑ | ↓ | ↓ pCa /pNa |
| pan | PAM | (S)-EU1180-55 | ↑ | ↓ | ↓ | ↑ | NE | NE |
| pan | PAM | GNE-9278 | ↑ | ↓ | ↓ | ↑ | – | – |
| pan | PAM | PTC-174 | ↑ (2A ↓) | ↓ | ↓ | ↑ | – | – |
| pan | PAM | EU-1794-27 | ↑ (2A ↓) | ↓ | ↓ | ↑ | – | – |
– denotes no available data, NE denotes no effect.
* (+)-CIQ had an effect only on GluN1/2C.
† Open probability was estimated from the overall effect on macroscopic peak responses and assumed no change in single-channel conductance or number of active receptors.
# EU1794-4 increased open probability but decreased single channel conductance, and the overall net effect was a reduction in macroscopic current, hence the designation of NAM (Perszyk et al., 2018; Perszyk et al., 2021).
↑ and ↓ indicate that the measured parameter was reported to be significantly greater than or less than the response in the absence of modulator and was used when either statistical tests were reported or reported values did not have overlapping confidence intervals.
Site-directed mutagenesis has identified a number of residues, such as GluN2A Val783, that are critical for NAM activity of TCN-201 and related analogs (Hansen et al., 2012). These GluN2A-selective NAMs stabilize the open conformation of the GluN1 ABD to facilitate glycine unbinding and reduce glycine potency (Yi et al., 2016). Crystallographic studies have captured structures for all four potential conformations predicted from the Monod-Wyman-Changeux model for allosteric action (Fig. 44). These structures show that NAM binding mediates a shift in the position of GluN2A Val783 that correlates with NAM actions. The subunit selectivity of the NAMs is mediated by GluN2A Val783, which is Phe in GluN2B and Leu in GluN2C/D (Hansen et al., 2012; Yi et al., 2016). Thus, key residues, including GluN2A Val783, form a molecular switch that distinguishes low- and high-affinity NAM binding states, establishes selectivity, and results in allosteric inhibition (Yi et al., 2016) (Fig. 44)
2. GluN2A-Selective Positive Modulators
GluN2A-selective PAMs possessing more than 10-fold selectivity for GluN2A over other GluN2 subunits, typified by GNE-0723, GNE-5729, GNE-6901, and GNE-8324, have been described (Hackos et al., 2016; Volgraf et al., 2016; Villemure et al., 2016) (Table 8). These compounds bind to the GluN1 and GluN2A ABD heterodimer interface at a site that partially overlaps the binding sites of TCN-201 and MPX-004 (Fig. 44), showing that occupancy of this pocket can produce either allosteric inhibition or potentiation. The GluN2A-selective PAMs interact with the same residue (GluN2A Val783) that controls subunit selectivity of GluN2A NAMs (Hansen et al., 2012; Hackos et al., 2016; Yi et al., 2016). However, binding of the GluN2A PAMs at this site has no effect on the position of GluN2A Val783 or on glycine potency for GluN1/2A, which is in contrast to the GluN2A NAMs (Hackos et al., 2016; Yi et al., 2016). Some analogs of the GluN2A PAMs also have activity at AMPA receptors (e.g., GNE-3419), with a similar potency as at NMDA receptors (Hackos et al., 2016).
The GluN2A-selective PAMs increase open probability, increase glutamate potency, and slow the rate of deactivation after removal of glutamate (Hackos and Hanson, 2017) (Fig. 45; Table 9). A complex structure-activity relationship exists among the GluN2A-selective modulators in terms of their efficacy and ability to slow the deactivation rate (Volgraf et al., 2016). The potentiation produced by some compounds in the GNE series of GluN2A-selective PAMs depends on the concentration of glutamate. GNE-8324 appears more potent when glutamate concentrations are higher, whereas GNE-6901 does not show a strong dependence on agonist concentration (Hackos et al., 2016). Although both compounds potentiate the response to maximal agonist concentrations, GNE-8324 (but not GNE-6901) enhances glutamate potency. These compounds also show distinct actions on NMDA receptor responses in hippocampal neurons (Hackos et al., 2016). GNE-6901 potentiates NMDA receptor-mediated EPSCs from hippocampal CA1 interneurons and pyramidal neurons, whereas GNE-8324 only potentiates NMDA receptor-mediated EPSCs from hippocampal CA1 interneurons (Hackos et al., 2016; Yao et al., 2018).
Fig. 45.
NMDA receptor modulators can have multiple effects on the receptor function. (A) Modulators can increase the open probability, estimated here from the current amplitude ratio in presence to absence of drug for responses to saturating concentrations of agonists. Modulators can also alter the deactivation time course in response to rapid removal of agonist. (B) Modulators can alter the agonist potency both through direct actions on agonist affinity or through efficacy-induced changes in EC50. Some modulators can modify unitary conductance, suggesting they alter the shape, configuration, and properties of the open channel. Modified with permission from Perszyk et al. (2020b) (A and C) and Hackos et al. (2016) (B).
The GluN2A PAMs have been evaluated in animal models of Alzheimer disease and Dravet syndrome, wherein GNE-0723 improved cognitive function and increased the power of low-frequency oscillations, which are perturbed in these diseases (Hanson et al., 2020). Another analog, GNE-5729, reversed anesthetic effects of ketamine and accelerated recovery from anesthetic and analgesic effects from a mixture of ketamine, fentanyl, and dexmedetomidine (Li et al., 2020).
3. GluN2B-Selective Negative Modulators
The GluN2B NAMs are the prototype class of subunit-selective NMDA receptor modulators, stemming from the discovery in 1993 that ifenprodil possessed this unique pharmacology (Williams, 1993). Ifenprodil inhibits GluN1/2B receptors with nanomolar potency and 200- to 400-fold selectivity over other GluN1/2 NMDA receptors (Table 10). The inhibition of GluN1/2A by high concentrations (>3–10 μM, depending on membrane potential) of ifenprodil reflects low-affinity nonselective channel block (Williams, 1993; Hansen et al., 2010b, 2014). The binding site for ifenprodil resides at the interface between the GluN1 and GluN2B NTD heterodimer (Masuko et al., 1999a; Karakas et al., 2011) (Fig. 46). Residues in GluN2B in contact with ifenprodil are conserved in GluN2A, but differences between GluN2A and GluN2B conformations within NTDs and in GluN1-GluN2 interactions limit accessibility of ifenprodil to the GluN1-GluN2A NTD interface, accounting for the subunit selectivity (Karakas et al., 2011; Romero-Hernandez et al., 2016).
TABLE 10.
Fig. 46.
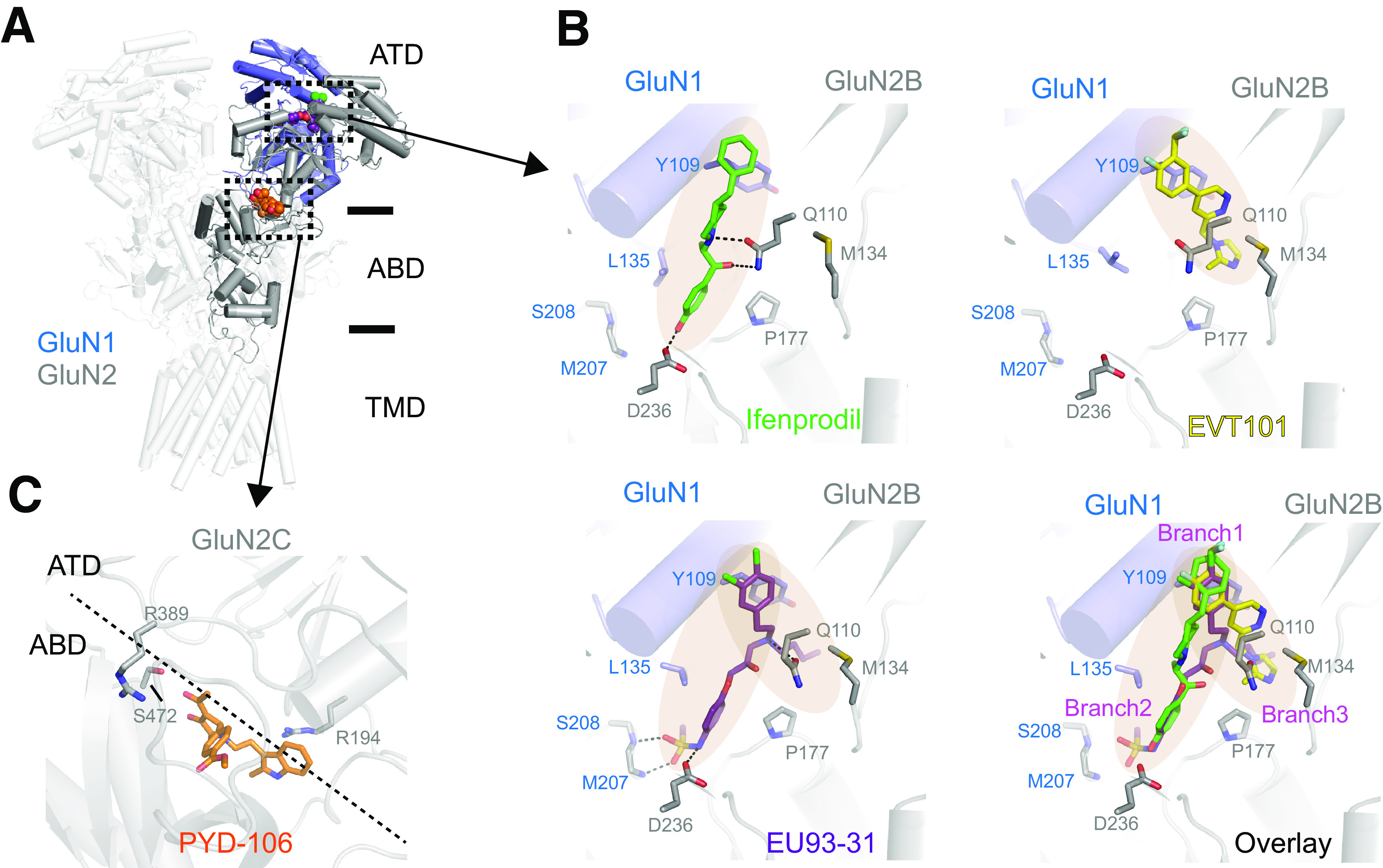
Multiple binding poses for GluN2B-selective negative allosteric modulators. (A) Binding site for GluN2B-selective NAMs at the GluN1/2B NTD heterodimer interface. The structure is the hybrid from GluN1/2B heterotetramer (PDB: 6WHR) and GluN1/2B NTD heterodimer (PDB: 6E7U). (B) Binding poses for different GluN2B-selective NAMs show occupancy of each arm or both arms of the binding pocket at the interface between the GluN1/2B NTD heterodimer (PDB: 3QEL, 5EWM, 6E7U for ifenprodil-, EVT-101-, and EU93-31–bound structures). (C) The proposed binding site for the GluN2C-selective PAM PYD-106 is shown at the interface between the GluN2C NTD and ABD in a model from Kaiser et al. (2018).
Ifenprodil inhibits currents activated using saturating agonist concentrations by 80%–90% (Williams, 1993; Kew et al., 1996; Mott et al., 1998; Masuko et al., 1999a). This submaximal inhibition is a consequence of prolonged channel shut times and a reduction in open durations, resulting in a decrease in open probability of approximately 75% (Amico-Ruvio et al., 2012; Bhatt et al., 2013). The magnitude of ifenprodil inhibition is reduced with increasing glycine concentrations (Williams, 1993). Ifenprodil modulation increases glutamate affinity, and ifenprodil can therefore enhance NMDA receptor responses activated by low levels of glutamate (Kew et al., 1996) (Table 9). The positive allosteric coupling between ifenprodil and glutamate binding is similar to what occurs with Zn2+ modulation (Sections IV.D.2. Types of NMDA Receptor Desensitization and VIII. Endogenous Allosteric Regulation).
Ifenprodil is widely used as a tool compound but has actions at multiple targets, including Ca2+ channels (Church et al., 1994; Bath et al., 1996; Delaney et al., 2012), σ receptors (Hashimoto and London, 1995), adrenergic receptors (Chenard et al., 1991), and serotonin receptors (McCool and Lovinger, 1995), which complicated utility in the clinic. However, GluN2B NAMs are better tolerated than most high-affinity and nonselective NMDA receptor channel blockers and competitive antagonists, and thus there is high therapeutic potential for this class of NAMs. For this reason, novel GluN2B-selective NAMs have been developed by both industry and academic groups, which resulted in many classes of GluN2B-selective NAMs with diverse scaffolds (Santangelo et al., 2012; Hashimoto et al., 2013; Lai et al., 2014; Shipton and Paulsen, 2013; Strong et al., 2014; Liu et al., 2020b). A number of structural analogs of ifenprodil with a shared piperidine core have been developed and exhibit improved potency and selectivity, including Ro 25-6981 (Fischer et al., 1997), CP-101,606 (Chenard et al., 1995), radiprodil (Mullier et al., 2017), Merck-20j, and MK-0657 (Liverton et al., 2007; Addy et al., 2009) (Table 10). Alternative scaffolds include propanolamines (Tahirovic et al., 2008), benzimidazoles (McCauley et al., 2004; Davies et al., 2012), cyclic benzamidines (Nguyen et al., 2007), amino cyclopentanes (Layton et al., 2011), piperidinyl pyrrolidinones (Marcin et al., 2018), and other compounds (Claiborne et al., 2003; McIntyre et al., 2009; Mosley et al., 2009; Brown et al., 2011; Beinat et al., 2014; Buemi et al., 2014; Bristow et al., 2017; Dey et al., 2018; Thum et al., 2018; Zscherp et al., 2018; Zampieri et al., 2019; Zhang et al., 2019). Molecules with two aromatic rings separated by a linker also cross over to act selectively on GluN2B, including the dopamine receptor antagonist haloperidol, the σ and dopamine receptor antagonist trifluperidol, the H3 histamine receptor antagonists clobenpropit and iodophenpropit, the β-adrenergic receptor agonist nylidrin, and the TRPV1 receptor antagonist capsazepine (Supplemental Table 13).
The binding site of GluN2B-selective NAMs at the NTD heterodimer interface of GluN1 and GluN2B has been shown in crystal structures for a number of GluN2B-selective NAMs, including ifenprodil, Ro 25-6981, and EVT-101 (Karakas et al., 2011; Stroebel et al., 2016) as well as EU93-31 and six analogs (Regan et al., 2019) (Fig. 46). Multiple studies suggest a mechanism of inhibition that involves NAM-induced closure of the clamshell-shaped NTD and repositioning of NTD heterodimers in relation to the ABD (Burger et al., 2012; Tajima et al., 2016; Chou et al., 2020), which controls receptor activation properties (Esmenjaud et al., 2019) (Supplemental Movie 3). In the presence of agonists, the NMDA receptors reside in three major conformations including Non-active1, Non-active2, and Active (Tajima et al., 2016; Chou et al., 2020) (Fig. 47), which are distinguished by the relative orientation of the two GluN1-GluN2 ABD heterodimers and conformational changes in the GluN1-GluN2 NTD heterodimer interfaces that harbor the ifenprodil binding site. In Non-active1 and Non-active2 conformations, the R2 lobes of the GluN1 and GluN2 NTDs are far apart (∼17 Å) at the GluN1-GluN2 NTD heterodimer interfaces, and the GluN1-GluN2B ABD heterodimer pairs are rolled toward the membrane, which extends the GluN2 ABD-TMD linker (Fig. 47). Non-active1 has a closed GluN2B NTD clamshell similar to the ifenprodil-bound form, whereas Non-active2 has an open NTD clamshell. However, neither Non-active1 nor Non-active2 reorient the GluN1-GluN2B NTD interface to promote changes in the arrangement of the GluN1-GluN2B ABD dimers, which are required for channel opening. In the Active conformation, the GluN2B NTD clamshell is open, and the R2 lobes of the GluN1 and GluN2 NTDs move closer (∼12 Å) to each other, which allows the GluN1-GluN2B ABD heterodimer pairs to roll up to pull on the GluN2 ABD-TMD linkers to open the pore. The GluN2B NAM ifenprodil binds the GluN1-GluN2B NTD interface and stabilizes the Non-active1–like conformation, thereby favoring channel closure (Tajima et al., 2016; Chou et al., 2020) (Fig. 47). The overall structural changes produced by GluN2B NAMs leading to reduced channel open probability are similar to those produced by inhibition of GluN2A-containing receptors by Zn2+ and H+ (Jalali-Yazdi et al., 2018).
Fig. 47.
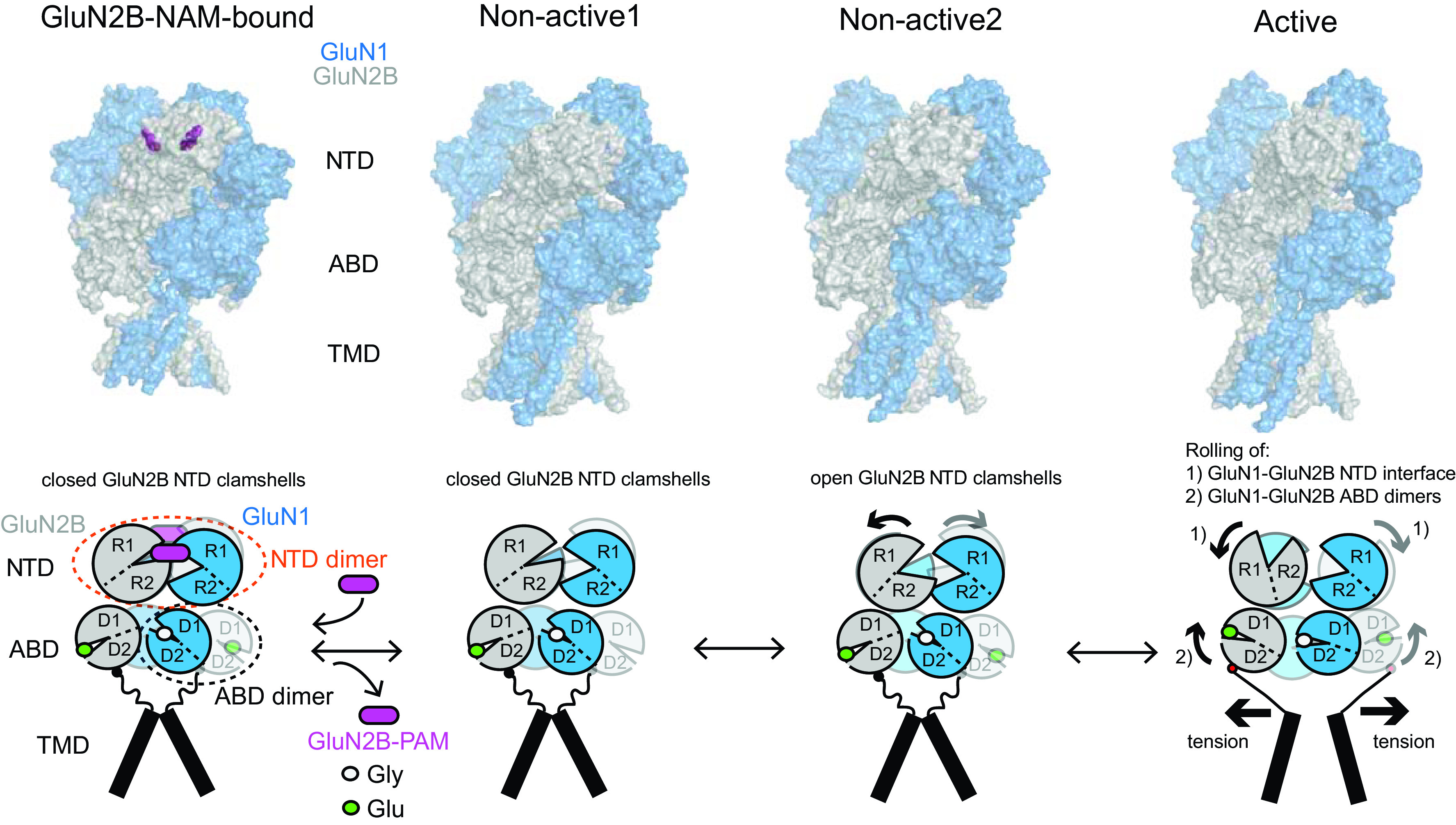
Structural basis of allosteric modulation of GluN1/2B NMDA receptors. Structures of GluN1/2B in the presence of agonists and the NAM ifenprodil. (A) Structures representing GluN2B-NAM–bound (PDB: 4PE5), Non-active1 (PDB: 6WHR), Non-active2 (PDB: 6WHS), and Active states (PDB: 6WI1). The GluN1 and GluN2B subunits are colored in slate and gray, respectively. The two ifenprodil molecules are represented by magenta spheres. In all of the structures, glycine (Gly) and glutamate (Glu) are present at ABDs but are omitted for clarity. (B) Schematic illustration of domain and subunit arrangement of GluN1/2B in conformations from (A). In Active state, the GluN2B NTD clamshell is open, the GluN1/2B NTD heterodimer interfaces roll (1), and the GluN1-GluN2B ABD heterodimers roll (2) to create tensions in the ABD-TMD linkers, that leads to channel opening. In Non-active1 and Non-active2, no rolling occurs in NTDs and ABDs, however, they differ by whether the GluN2B NTD clamshell is open (Non-active2) or closed (Non-active1). The population of Non-active1 increases with lower pH. GluN2B-NAMs targeting the subunit interfaces of GluN1/2B NTDs stabilize the closed GluN2B NTD clamshell, thereby stabilizing the conformation similar to Non-active1.
An unexpected result of structural analysis of GluN2B NAM binding was the identification of a different portion of the binding pocket between the GluN1 and GluN2B NTDs (Fig. 46). Binding of EVT-101 occurs within this space and adopts a different binding mode compared with ifenprodil (Stroebel et al., 2016). This space is also the site of action of EU93-31, which is unique in that it superimposes onto both ifenprodil and EVT-101 binding modes (Regan et al., 2019), thereby occupying all portions of the binding pocket. The appreciation of this new allosteric site could lead to structure-based design and new classes of GluN2B-selective ligands with different pharmacological and functional properties.
Two series of GluN2B antagonists, piperazines and propanolamines, have been developed through optimization of sensitivity to extracellular pH, which creates context dependence that should render them more effective inhibitors at reduced pH, which occurs during acute injury (Wang et al., 2014c; Yuan et al., 2015b; Myers et al., 2021). The pH sensitivity of GluN2B-selective NAM action (Pahk and Williams, 1997) arises from allosteric coupling between proton and GluN2B-selective NAM activities (Mott et al., 1998) and pH-dependent association and dissociation rates for NAM binding (Yuan et al., 2015b). Multiple residues in the NTD underlie pH sensitivity, including a pair of closely positioned glutamate residues in GluN2B as well as GluN1 His134, which forms hydrophobic interactions with nearby GluN1 Ile133 in the hydrophobic cage surrounding the pocket in which the n-butyl chain of 93-31 resides (Yuan et al., 2015b; Regan et al., 2019). Protonation of His134 is hypothesized to weaken the interaction with GluN1 Ile133, allowing the residue to strengthen its van der Waals contacts with 93-31.
Triheteromeric NMDA receptors that contain two different GluN2 subunits show altered responses to GluN2B-selective NAMs. Ifenprodil inhibits triheteromeric GluN1/2A/2B receptors with >3-fold reduced potency and a reduced degree of inhibition, which at saturating concentrations was ∼90% for GluN1/2B and ∼30% for GluN1/2A/2B (Hatton and Paoletti, 2005; Hansen et al., 2014; Stroebel et al., 2014). GluN1/2B/2D receptors show similar IC50 for ifenprodil as diheteromeric GluN1/2B receptors, but a reduced degree of inhibition (67%) at saturating concentrations compared with diheteromeric receptors (81%); other GluN2B-selective NAMs (EVT-101, CP-101,606) behave similarly (Yi et al., 2019).
4. GluN2B-Selective Positive Modulators
Interest in GluN2B-selective PAMs was catalyzed by the demonstration that overexpression of GluN2B in mice enhanced performance in cognitive tasks (Tang et al., 1999; Wang et al., 2014b). Cations, such as the polyamines spermine and spermidine, remain the best-characterized examples of GluN2B-selective PAMs (VIII. Endogenous Allosteric Regulation). Although the precise location of polyamine binding site(s) on the NTD remains to be identified, spermine binding may open the GluN2B NTD clamshell (Sirrieh et al., 2015a). Aminoglycoside antibiotics neomycin, kanamycin, and tobramycin also potentiate GluN2B-containing receptors by a similar mechanism of action (Masuko et al., 1999b; Swanger et al., 2016; Tang et al., 2020). Aminoglycosides appear to enhance glycine but not glutamate potency (Masuko et al., 1999b) and potentiate GluN1/2B responses without altering deactivation time course (Tang et al., 2020), consistent with a spermine-like mechanism involving relief of proton inhibition (Traynelis et al., 1995). Polyamines and presumably aminoglycosides are proposed to interact with clusters of negatively charged residues in the R2 lobes of bilobed GluN1 and GluN2B NTDs to shield negatively charged residues in the NTDs (Section VIII.C.2. Modulation by Extracellular Polyamines). One positive allosteric modulator has also been described with GluN2B preference [compound 97 in Strong et al. (2017)], raising the possibility that selectivity might be achieved at sites other than the NTD, but there remains a lack of drug-like potent and efficacious GluN2B-selective PAMs.
5. GluN2C-Selective Positive Modulators
A series of pyrollidinones stereo-selectively potentiate GluN1/2C receptor responses to saturating concentration of agonists by 2-fold with potencies in the low μM range (Khatri et al., 2014; Zimmerman et al., 2014) (Table 11). PYD-106 is selective for the GluN1/2C receptors but does not potentiate triheteromeric GluN1/2A/2C or GluN1/2B/2C receptors (Khatri et al., 2014; Bhattacharya et al., 2018; Kaiser et al., 2018). PYD-106 has a weak effect on glutamate potency and modestly prolongs the glutamate deactivation time course (Khatri et al., 2014) (Table 9). The structural determinants of action of PYD-106 reside at the interface of the GluN2C NTD and the upper lobe of the GluN2C ABD (Fig. 43), and modeling suggests that PYD-106 fits within a pocket at the GluN2C NTD-ABD interface, which is consistent with mutagenesis studies (Kaiser et al., 2018; Khatri et al., 2014). Expression of mutant GluN1/2C NMDA receptors with only a single PYD-106 binding site can still be potentiated, and this binding pocket can be transferred to other chimeric GluN2 subunits (Kaiser et al., 2018).
TABLE 11.
6. GluN2C- and GluN2D-Selective Negative Modulators
The first compounds with strong GluN2C and GluN2D selectivity (hereafter GluN2C/D selectivity) contained a quinazolin-4-one (QNZ) core; QNZ-46 is the prototypical compound in this class with ∼50-fold selectivity for GluN2C/D-containing NMDA receptors and potency in the low µM range (Mosley et al., 2010; Hansen and Traynelis, 2011) (Table 11). QNZ-46 has minimal actions on AMPA and kainate receptors, even though it shares a chemical scaffold with the AMPA-selective NAM CP-465,022 (Menniti et al., 2000). Inhibition by QNZ-46 is dependent on glutamate but not glycine in that glutamate binding increases QNZ-46 potency, and QNZ-46 must first dissociate before glutamate can unbind (Hansen and Traynelis, 2011) (Table 9). The structural determinants of QNZ-46 action reside in the membrane-proximal surface of the GluN2D ABD near the pre-M1 and M3 helices (Hansen and Traynelis, 2011). A recent GluA2 AMPA receptor crystal structure that shows CP-465,022 bound to the upper portion of the TMD raises the possibility that, given the similarity in the core structure of QNZ-46 and CP-465,022, both molecules bind to the same region (Hansen and Traynelis, 2011; Yelshanskaya et al., 2016b). QNZ-46 has been suggested to act as a neuroprotectant through inhibition of the response to axonal release of glutamate (Doyle et al., 2018).
A series of compounds with a dihydroquinolone-pyrazoline (DQP) core have properties similar to QNZ-46 (Acker et al., 2011). DQP-1105, a representative member of this class, is approximately 50-fold selective for GluN2C/D-containing NMDA receptors with low-µM IC50 values (Acker et al., 2011) (Table 11). Like QNZ-46, inhibition by DQP-1105 is dependent on glutamate binding and has structural determinants in the membrane-proximal surface of the GluN2 ABD (Acker et al., 2011). DQP-1105 inhibits NMDA receptor–mediated EPSCs onto subthalamic neurons, which express GluN2B and GluN2D (Swanger et al., 2015). Some DQP-1105 analogs have nanomolar IC50 values, making this series more potent and selective than QNZ-46 (Acker et al., 2013) (Table 11).
A class of negative allosteric modulators that are highly selective for GluN2C/D-containing NMDA receptors includes an N-aryl benzamide core with NAB-14 as the prototypical member showing >200-fold selectivity for GluN2C/D over GluN2A/B subunits (Swanger et al., 2018) (Table 11). The structural determinants of NAB-14 activity are distinct from QNZ-46 and DQP-1105 but overlap with those of the positive modulator CIQ (see below). NAB-14 is active at synaptic NMDA receptors in hippocampal interneurons, which express GluN2B and GluN2D (Section VI. Developmental and Regional Expression in the Central Nervous System), has minimal off-target actions, and is modestly brain-penetrant (Swanger et al., 2018; Yi et al., 2019).
7. GluN2C- and GluN2D-Selective Positive Modulators
CIQ was the first GluN2C/2D-selective positive allosteric modulator of NMDA receptors (Mullasseril et al., 2010) (Table 11). A structure-activity relationship has been developed for the class of modulators that includes CIQ, showing stereo-selective actions with robust selectivity for GluN2C/D-containing receptors and nanomolar EC50 values for some analogs (Santangelo Freel et al., 2013, 2014; Strong et al., 2017; Epplin et al., 2020). CIQ and related analogs potentiate the responses of triheteromeric NMDA receptors with only one copy of either GluN2C or GluN2D at a reduced efficacy (Mullasseril et al., 2010; Bhattacharya et al., 2018; Yi et al., 2019) (Table 11). The structural determinants of CIQ potentiation of GluN1/2D reside within the M1 transmembrane helix and the pre-M1 helix in the GluN2D subunit (Mullasseril et al., 2010; Ogden and Traynelis, 2013). However, it is unknown whether these regions directly contribute to the CIQ binding site. Whereas racemic CIQ and the active enantiomer (+)-CIQ have been used as tool compounds to explore the role of GluN2D in neuronal function (Yamamoto et al., 2013; Hildebrand et al., 2014; Ogden et al., 2014; Suryavanshi et al., 2014; Zhang et al., 2014b; Swanger et al., 2015; Perszyk et al., 2016), its utility is limited by poor physicochemical characteristics, including low solubility. Nevertheless, CIQ has revealed a role for GluN2D in excitatory synaptic input to subthalamic neurons (Swanger et al., 2015), excitatory drive onto prefrontal parvalbumin positive fast spiking interneurons (Garst-Orozco et al., 2020), prepulse inhibition and preclinical models of schizophrenic behaviors (Gawai et al., 2020), and the striatal response to degeneration of dopaminergic neurons (Nouhi et al., 2018). Within this compound series, (+)-EU1180-453 has improved physicochemical properties, is soluble, brain-penetrant, and shows minimal off-target actions, making it an improved tool compound for potentiation of GluN2C/2D-containing NMDA receptors (Epplin et al., 2020).
Development of the CIQ scaffold led to the discovery of (R)-(+)-EU1180-55, an analog selective for GluN2C/D-containing NMDA receptors with enhanced potency (Strong et al., 2017, 2021) (Table 11). Its enantiomer, (S)-(−)-EU1180-55, is a PAM at GluN1/2B, GluN1/2C, and GluN1/2D receptors activated by saturating concentrations of agonist and can also potentiate GluN1/2A at subsaturating agonist concentrations (Strong et al., 2021). These two enantiomers preferentially bind to distinct pockets on the tetrameric NMDA receptors, with the (+) enantiomer preferring a pocket that includes the GluN2 pre-M1 helix and the (-) enantiomer preferring the pocket including the GluN1 pre-M1 helix (Strong et al., 2021) (Fig. 48). The existence of two distinct pockets raises important questions about all classes of compounds that bind in this receptor region, particularly the allosteric modulators with complex pharmacology, which could bind to one, the other, or both pre-M1 sites (see below).
Fig. 48.
Multiple binding pockets at the TMD-ABD interface. (A) The GluN1/2D structure [model built from PDB: 6WHS, Strong et al. (2021)] with GluN1 shown in blue and GluN2D in gray. The pre-M1/M3 domain of NMDA receptors exhibits pseudo–4-fold symmetry due to the tetrameric arrangement GluN1-GluN2-GluN1-GluN2. (B) The pseudo–4-fold symmetry creates two distinct binding pockets for modulators that include either the GluN1 pre-M1/M3 helices shown with (S)-EU1180-55 docked or GluN2D pre-M1/M3 helices shown with (R)-EU1180-55 docked. Modified with permission from Strong et al. (2021) (Copyright 2021 American Chemical Society).
8. NMDA Receptor Modulators with Complex Pharmacology
Several modulators have different degrees and forms of modulation across different NMDA receptor subtypes. Although these compounds may appear nonselective, they show varied actions that suggest nuanced mechanisms or biased modulation of different modalities of the NMDA receptor. Furthermore, efficacy or potency can vary among NMDA receptor subtypes, creating opportunities to preferentially modulate certain circuits with specific GluN2 subunit combinations. In addition, the ability to act at multiple subunits raises possibilities for unique pharmacology at triheteromeric NMDA receptors.
Structure-activity studies of competitive antagonists with modest selectivity for GluN1/2C and GluN1/2D receptors, including PPDA and UBP-141 (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers), led to discovery of a class of allosteric modulators with complex GluN2 subunit selectivity, including UBP-710, UBP-512, and UBP-551 with naphthalene or phenanthrene cores (Costa et al., 2010). UBP-710 displays concentration-dependent effects that include potentiation of GluN1/2A and GluN1/2B at 100 μM but inhibition of GluN1/2C and GluN1/2D at a higher concentration range (Costa et al., 2010). UBP-551 may selectively enhance the response of GluN2D-containing NMDA receptors but with a biphasic concentration-effect relationship and a maximal potentiation of GluN1/2D observed at 30 μM; UBP-551 inhibits other NMDA receptor subtypes (Costa et al., 2010). Experiments with chimeric receptors suggest the action of these modulators is influenced by the S2 segment of the GluN2 ABD (Costa et al., 2010).
The EU1794 series of nonselective NAMs of NMDA receptors produce incomplete inhibition at saturating NAM concentrations (Katzman et al., 2015) (Table 12). This incomplete inhibition reflects a reduction in single-channel amplitude for the analog EU1794-4, which is accompanied by a change in relative ionic permeability (Perszyk et al., 2021). The property of incomplete inhibition did not remove the ability of EU1794-2 [compound 4 in Katzman et al. (2015)] to act as a neuroprotective agent in suppressing excitotoxicity. Several EU1794 analogs act as PAMs, in some cases with a single methyl group sufficient to convert NAM to PAM activity (Perszyk et al., 2018). Both PAMs and NAMs compete with one another and share overlapping structural determinants that involve the pre-M1 and M3 regions, and the binding of both PAMs and NAMs is dependent on glutamate and glycine binding. The PAM EU1794-27 robustly increased the potency for glutamate and glycine (Perszyk et al., 2018). The NAM EU1794-4 potentiates NMDA receptor responses to subsaturating agonist concentrations but inhibits responses to saturating agonist concentrations (Perszyk et al., 2018). This was due to allosteric enhancement of agonist potency (to a similar degree as the PAM analog EU1794-27), raising the possibility that EU1794-4 could enhance NMDA receptor responses to low agonist concentration, perhaps at extrasynaptic sites, but reduce the response to high levels of agonist within the synaptic cleft.
TABLE 12.
A number of nonselective and highly efficacious PAMs have been described, including GNE-9278 (Wang et al., 2017), (S)-EU1180-55 (and its potent analog (−)-EU1180-154 [compound 142 in Strong et al. (2017) and UBP-684 (Sapkota et al., 2017)]) (Table 12). GNE-9278 robustly potentiates all NMDA receptor subtypes and can enhance agonist potency for some subunit combinations; the actions of GNE-9278 appear to be dependent on glutamate binding (Wang et al., 2017). GNE-9278 is selective for NMDA receptors over AMPA receptors, and the structural determinants of action include residues on M1 and M3 helices of GluN1 (Wang et al., 2017). Similarly, (−)-EU1180-154 potentiates GluN1/2B, GluN1/2C, and GluN1/2D NMDA receptors with EC50 values of 380–580 nM, is selective for NMDA receptors over AMPA receptors, and increases glutamate potency and prolongs the deactivation time course, with structural determinants of action in the TMD (Strong et al., 2017; Strong et al., 2021). UBP-684 is a 2-napthoic acid derivative with PAM activity at all recombinant GluN1/2 NMDA receptors and EC50 values in the 30–40 μM range (Sapkota et al., 2017). UBP684 prolongs the deactivation time course after removal of glutamate but not glycine, and the mechanism is proposed to involve a stabilization of the ABD in the active conformation (Chopra et al., 2017; Sapkota et al., 2017).
The EU1622 series of PAMs possess the remarkable ability to alter ion permeation properties, namely channel conductance and relative calcium permeability along with other receptor properties (Perszyk et al., 2020b). EU1622-14 shares many properties with the other PAMs that lack GluN2 subunit selectivity in that they can potentiate the response to saturating agonist concentration, prolong deactivation, and enhance agonist potency (Fig. 45; Table 12). Single-channel analysis revealed that EU1622 series compounds reduced channel conductance (Fig. 45) but increased open probability (Perszyk et al., 2020b). Permeant ion replacement studies show that the NMDA receptor channel becomes less permeable to Ca2+ in the presence of EU1622-14, and the net effect of EU1622-14 modulation is to increase in Na+ influx with a minimal increase in Ca2+ influx (Perszyk et al., 2020b). This observed decrease in Ca2+ permeability has not yet been observed with other NMDA receptor PAMs and expands the ways of altering NMDA receptor function (Perszyk et al., 2020b) (Table 9).
PTC-174 potentiates GluN1/2C and GluN1/2D activity more than 10-fold and produces 2-fold potentiation of GluN1/2B activity but inhibits GluN1/2A activity by up to 50% (Yi et al., 2020) (Table 12). PTC-174 potentiates triheteromeric GluN1/2B/2D receptors to an intermediate level between GluN1/2B and GluN1/2D but is only weakly efficacious at triheteromeric GluN1/2A/2C receptors (Yi et al., 2020). PTC-174 potentiates NMDA receptor–mediated responses in subthalamic nucleus neurons and hippocampal CA1 interneurons, but not hippocampal CA1 pyramidal neurons (Yi et al., 2020). PTC-174 reduced amphetamine- and MK-801–induced hyperlocomotion in vivo and appears to reduce impulsivity in preclinical models (Callahan et al., 2020).
9. Neurosteroid NMDA Receptor Modulators
The actions of endogenous neurosteroids (Section VIII. Endogenous Allosteric Regulation) stimulated synthetic work on the steroid backbone as a scaffold from which to design potential therapeutic agents (Vales et al., 2012; Ishikawa et al., 2018; La et al., 2019). Substantial information exists about several classes of compounds, including analogs of 24(S)-HC, which are more potent than the endogenous PS and have different structural determinants that include the TMD and connecting linkers (Paul et al., 2013; Wilding et al., 2016). The 24(S)-HC analogs SGE-201 and SGE-301 (Table 12) enhance open probability and are more potent and efficacious PAMs than 24(S)-HC at all NMDA receptors, with nM potency for SGE-201 at native NMDA receptors (Paul et al., 2013; La et al., 2019). The endogenous 24(S)-HC modulates neuronal function but does not saturate its binding site on NMDA receptors (Sun et al., 2016).
A range of other steroid derivatives and steroid mimics with NAM activity at NMDA receptors have been described (Blanco et al., 2018; Smidkova et al., 2019; Chvojkova et al., 2020). For example, a class of amide analogs of pregnanolone inhibited all diheteromeric GluN1/2 subtype combinations more potently than the parent compound, but some had only modest selectivity for NMDA over GABA receptors (Rambousek et al., 2011; Adla et al., 2017). Perhydrophenanthrene sulfates also inhibit NMDA receptors (Slavikova et al., 2016). Replacing the sulfate on pregnanolone sulfate with carboxylic acid with a variable linker alters the apparent use dependence (Vyklicky et al., 2016), whereas D-ring modifications increase potency (Kudova et al., 2015). Neurosteroid analogs that can inhibit NMDA receptors and potentiate GABA receptors have been described, which may have unique therapeutically relevant attributes (Ziolkowski et al., 2021).
10. GluN3-Selective Modulators
EU1180-438 is a GluN3-selective NAM with low-µM IC50 and no detectable effects on GluN1/2A-D, GluA1-4, or GluK1-2 receptor responses (Zhu et al., 2020). This NAM inhibits recombinant and native glycine-activated GluN1/3 receptors when responses are prevented from desensitizing either by mutations within the glycine binding site in GluN1 or in the presence of the GluN1-selective antagonist CGP-78608 (Zhu et al., 2020). Inhibition does not involve a change in the EC50 for glycine, is not voltage-dependent, and cannot be overcome by increased glycine concentration. Compound EU1180-438 does not inhibit NMDA receptor-mediated responses activated by NMDA in hippocampal CA1 pyramidal cells but inhibits GluN1/3A receptor-mediated glycine responses recorded from the same neurons in the presence of CGP-78,608 (Zhu et al., 2020). Structural determinants reside in the GluN3 pre-M1 helix similar to both GluN2C/2D-selective NAMs and PAMs as well as nonselective NMDA receptor modulators.
X. Glutamate Receptors in Disease
A. Overview of Focus Areas in Drug Discovery
The first foray into drug development targeting iGluRs was the discovery of phencyclidine and analogs in the 1950s and 1960s (Supplemental Table 12). These agents were recognized for their potential as dissociative anesthetics and analgesics but also for their disturbing “schizophrenomimetic” effects (Luby et al., 1959). Among these agents, ketamine was approved as an anesthetic in 1970 at a time when our understanding of glutamatergic neurotransmission was just beginning to develop. The next milestone for iGluR drug development was research indicating that exposure of neurons to glutamate results in rapid neuronal death (Choi et al., 1988; Olney, 1994). These observations led to the hypothesis that elevated extracellular glutamate is a principal cause of neuronal death after brain injury. The additional observation that neurotoxicity could be limited by inhibiting NMDA receptors fostered a massive effort to develop NMDA receptor–targeted therapeutics as neuroprotective agents for stroke and traumatic brain injury [Muir and Lees (1995), Reinert and Bullock (1999); see Supplemental Tables S1–S3 in Yuan et al. (2015b)] and to explore the possibility that such agents may prevent brain damage during chronic neurodegenerative diseases (Doble, 1999). Unfortunately, despite extensive preclinical data demonstrating neuroprotection, multiple clinical studies in patients with acute brain injury did not find evidence of neuroprotection for the NMDA receptor–targeted agents tested (Ginsberg, 2008). The failure to translate the preclinical findings into clinical efficacy has been attributed to multiple factors, including the inability [see Saver (2013)] to administer neuroprotectants soon enough to prevent injury, dose-limiting side effects, patient and injury heterogeneity, and lack of quantifiable endpoints. Nevertheless, the NMDA receptor ligands that resulted from this work created an expanded pharmacological toolbox that continues to support research. Furthermore, the NMDA receptor channel blocker memantine was approved as a therapeutic for Alzheimer’s disease in 2003, and the channel blocker esketamine has been approved for treatment-resistant depression (Supplemental Table 12). Amantadine is commonly prescribed for l-DOPA–induced dyskinesias in Parkinson disease (AlShimemeri et al., 2020) and generally considered a low-affinity NMDA receptor channel blocker (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers) (Supplemental Table 12). Despite relatively modest therapeutic benefit, amantadine and memantine are currently the most prescribed NMDA receptor–targeted therapies. These approvals together with appreciation of NMDA receptor roles in genetic and autoimmune disorders are fueling a renaissance in the development of NMDA receptor–targeted therapeutics, with an emphasis of subunit-selective agents that can target one cell population or brain region over another.
The hypothesis that excessive glutamate signaling is neurotoxic also fostered an interest in AMPA receptor antagonists as neuroprotectants for stroke, traumatic brain injury, and neurodegenerative conditions (Buchan et al., 1993). Although these efforts failed because of some of the same reasons as those for NMDA receptor antagonists, this work also provided new pharmacological tools to explore the therapeutic potential of AMPA receptor inhibition. This culminated in the approval of a noncompetitive AMPA receptor antagonist perampanel as an anticonvulsant (Greenwood and Valdes, 2016). Furthermore, the notion that increases in synaptic AMPA receptors are associated with synaptic plasticity underlying learning and memory stimulated an interest in potentiation of AMPA receptors to enhance cognitive function (Lynch, 2006).
An important advance has been in identification and analyses of the impact of genetic variation in iGluRs on brain dysfunction as well as basic receptor function. Large-scale genetic sequencing efforts and the functional studies that followed are both redefining and subdividing some neurologic disorders, which has the potential to reorient drug discovery. Variation across the full spectrum of iGluR genes has been connected to functional alterations in these receptors that are, in turn, being associated with specific neurologic phenotypes (Swanger et al., 2016; Platzer et al., 2017; Strehlow et al., 2019). Efforts to evaluate how human genetic variation in both coding and noncoding regions impacts iGluR function and homeostasis in brain function can facilitate the development of diagnostic criteria for CNS diseases, improve preclinical testing in new animal models, and allow identification of target subpopulations who could benefit from new treatments.
The discussion below considers primarily three pharmacological parameters: 1) potency refers to the amount of drug required to obtain an effect (potent drugs act at low concentrations), 2) efficacy indicates the extent of maximal effect produced by a drug, and 3) selectivity considers whether a drug is more potent or effective at a particular receptor type compared with other receptors.
B. AMPA and Kainate Receptors as Therapeutic Targets
Multiple research programs in the 1980s were unable to develop antagonists with selectivity for AMPA over kainate receptors or vice versa, which led to grouping these two subtypes together as “non-NMDA receptors.” Although negative allosteric modulators were eventually developed that were selective for AMPA receptors, there has been only limited success in identifying drug-like compounds that are selective for kainate receptors. Consequently, there has been little progress on the clinical development of kainate receptor-selective agents. This contrasts with the considerable progress in the clinical development of AMPA and NMDA receptor-targeted agents, which will be reviewed below.
1. AMPA Receptor Antagonists
Development of AMPA receptor antagonists was initially driven by interest in these agents as neuroprotectant alternatives to NMDA receptor antagonists (Buchan et al., 1993). Two classes of compounds emerged (Mattes et al., 2010). The competitive antagonists, of which the prototype was the quinoxalinedione NBQX, inhibited both AMPA and kainate receptors and were found to be neuroprotective in a broad range of animal models of acute brain injury (Catarzi et al., 2007). However, the quinoxalinediones had significant pharmaceutical issues that hampered clinical development, including poor solubility and a propensity to precipitate in the kidney and other acidic compartments. The noncompetitive antagonists exemplified by the prototype GYKI-52466 are AMPA receptor–selective (Solyom and Tarnawa, 2002) and have better pharmaceutical properties compared with quinoxalinedione analogs but have less robust neuroprotective effects (Menniti et al., 2003). Only the AMPA receptor competitive antagonist ZK 200775 (fanapanel) was evaluated for neuroprotective efficacy in a clinical trial in patients with stroke but was associated with worsening of symptoms (Walters et al., 2005). Nonetheless, preclinical research suggested a number of potential therapeutic utilities for AMPA receptor antagonists, and some positive clinical data were obtained in migraine and neuropathic pain and against the symptoms of amyotrophic lateral sclerosis [reviewed in Mattes et al. (2010)]. Unfortunately, AMPA receptor inhibition also produced CNS depressant effects at doses in the therapeutic range.
Epilepsy is the recurrence of unprovoked seizures caused by abnormal, highly synchronous firing of neurons within a restricted brain region or brain hemisphere or generalized to the entire brain (Moshe et al., 2015). AMPA receptor antagonists inhibit seizures in a wide variety of preclinical models (Rogawski, 2013), and the past decade saw the first approval of an AMPA receptor antagonist, perampanel, for the treatment of different forms of epilepsy. Perampanel is a negative allosteric modulator that is highly selective for AMPA receptors over kainate and NMDA receptors but shows little selectivity for the different AMPA receptor subtypes (Hanada, 2014). Perampanel was approved in 2012 as an adjunctive treatment of partial-onset seizures and in 2015 for primary generalized tonic-clonic seizures in patients 12 years and older (Greenwood and Valdes, 2016). Investigations into the use of perampanel against other types of seizure disorders are ongoing (Potschka and Trinka, 2019). The antiseizure efficacy of perampanel is dose-dependent and, as with previous AMPA receptor antagonists, so too are side effects (Greenwood and Valdes, 2016). These include dizziness in 30%–40% of patients and somnolence in ∼15% of patients at efficacious doses. Nonetheless, perampanel is the first clinically tolerated AMPA receptor antagonist that can be administered chronically to reduce the incidence of recurrent seizures.
The competitive antagonist BGG492 (selurampanel) was also advanced into clinical testing (Faught, 2014) and as with quinoxalinediones, BGG492 inhibits both AMPA and kainate receptors. BGG492 dose-dependently reduced seizurogenic activity in patients with epilepsy that were sensitive to seizures induced by flashing light (photoparoxysmal response induced by intermittent photic stimulation) (Kasteleijn-Nolst Trenite et al., 2015). However, the compound failed to show efficacy when tested as an adjunct in patients with focal-onset seizures; the side effects appeared similar to those seen with perampanel (Elger et al., 2017).
AMPA receptors associate with multiple auxiliary subunits that have significant effects on their function and sensitivity to modulators (Sections III. Auxiliary Subunits and IV. Receptor Activation, Deactivation, and Desensitization). TARPs are differentially deployed in neurons throughout the CNS and may confer AMPA receptor subtype selectivity for modulators. The recent AMPA receptor NAMs JNJ-55511118 and LY3130481 show high selectivity for AMPA receptors associated with γ-8 over AMPA receptors associated with γ-2, γ-3, γ-4, or γ-7 (Gardinier et al., 2016; Kato et al., 2016; Maher et al., 2016).
2. AMPA Receptor Positive Allosteric Modulators
There is a strong rationale for the development of AMPA receptor PAMs given that increases in synaptic strength associated with synaptic plasticity can result from an increase in the number of synaptic AMPA receptors (Brogi et al., 2019). Hence, AMPA receptor PAMs may have therapeutic utility in disorders with a decline in cognitive function, including Alzheimer’s disease and schizophrenia. Although iGluR PAMs have been approached with caution because of potential for seizure induction and/or neurotoxicity, significant progress has been made in the development of AMPA receptor PAMs.
A class of AMPA receptor PAMs were derived from the nootropic aniracetam termed “AMPAkines” (Lynch, 2006). These compounds slow AMPA receptor deactivation and reduce desensitization with relatively low potency and efficacy (Section IX. Exogenous Positive and Negative Allosteric Modulators). A number of AMPAkines have been tested for procognitive effects in humans, but results were mixed, and none achieved approval for neuropsychiatric indications, although they were safe and well tolerated (Partin, 2015).
A second class of AMPA receptor PAMs, for which a prototype is LY404187 (Quirk and Nisenbaum, 2002; Pirotte et al., 2013; Ward et al., 2015), have high potency and efficacy and act within the ABD dimer interface at a similar site as the AMPAkines (Section IX. Exogenous Positive and Negative Allosteric Modulators). Compounds from this broad pharmacological class have procognitive effects in a variety of preclinical animal models (Quirk and Nisenbaum, 2002; Black, 2005). Unfortunately, these compounds also produce the toxicities predicted from high-efficacy AMPA receptor PAMs, including seizures and neurotoxicity (Shaffer et al., 2013). Another compound in this class, LY451395 (mibampator), was tested for procognitive (Chappell et al., 2007) and behavioral effects (Trzepacz et al., 2013) in Alzheimer’s disease but showed limited efficacy.
More encouraging are positive early findings for an AMPA receptor PAM, PF-04958242 (also known as BIIB104) (Shaffer et al., 2015). Preclinical studies indicate that this compound has an improved therapeutic index relative to LY451395, allowing for more flexible dosing in humans (Shaffer et al., 2015). PF-04958242 ameliorated working memory deficits in healthy volunteers when memory was impaired by treatment with ketamine (Ranganathan et al., 2017). PF-04958242 has lower efficacy at AMPA receptors associated with γ-2, the predominant TARP in cerebellum, compared with receptors associated with γ-8, expressed in hippocampus. LY451395 is equally efficacious at receptors associated with γ-8 and γ-2 (Ishii et al., 2020). If AMPA receptor PAM efficacy with regard to working memory is mediated by hippocampal receptors and toxicity from, for example, potentiation of cerebellar receptors (Shaffer et al., 2013), then the preference of AMPA receptor modulators for γ-8 may improve the therapeutic index (Kato et al., 2016; Maher et al., 2017).
C. NMDA Receptors as Therapeutic Targets
1. NMDA Receptor Antagonists
a. Ketamine as antidepressant
In 2000, Berman, Krystal, and colleagues observed that patients with depression experienced an alleviation of their symptoms in the hours after receiving a short intravenous infusion of the pan-NMDA receptor channel blocker ketamine (Berman et al., 2000) (Supplemental Table 12). After replication of this rapid onset antidepressant effect 6 years later (Zarate et al., 2006), interest increased and short intravenous infusions of racemic R,S-ketamine yielded a robust antidepressant response that 1) develops within hours, 2) may last for days to weeks, and 3) is sustainable with repeated doses (Aan Het Rot et al., 2010; Cornwell et al., 2012; Murrough et al., 2013; Diamond et al., 2014). Ketamine infusion also rapidly reduces suicidal ideation (Price et al., 2009; Murrough et al., 2015; Krystal et al., 2019) and has reported benefit in patients suffering bipolar depression (Zarate et al., 2012), obsessive-compulsive disorder (Rodriguez et al., 2013), and post-traumatic stress disorder (Feder et al., 2014). Ketamine is undergoing clinical testing for patients with Rett syndrome (Katz et al., 2016). Indeed, the antidepressant activity of ketamine has garnered considerable interest in psychiatry and invigorated drug discovery efforts to target NMDA receptors in depression (Duman and Aghajanian, 2012; Niciu et al., 2014).
Based on the clinical research findings, ketamine is being adopted in clinical practice for use in treatment of depression and other neuropsychiatric disorders. Recent studies are beginning to define optimal dosing regimens for acute and sustained use (Fava et al., 2020; Shiroma et al., 2020). However, there remain impediments to the widespread use of ketamine, including administration by intravenous infusion, mechanism-based psychotomimetic effects, cognitive disturbances that may occur during the infusion period, and substantial abuse potential that complicates access and distribution. These issues require that ketamine be administered in a clinical setting and that patients be monitored during and after drug administration. A follow-on approach to intravenous ketamine is esketamine, the S-isomer of ketamine formulated as a nasal spray. Esketamine was approved for depression in the United States and the European Union in 2019 (Kim et al., 2019; Mahase, 2019) as the first mechanistically novel antidepressant that provides symptomatic relief to depressed patients within hours or days as opposed to weeks. The key point of differentiation for esketamine is self-administration by patients as a nasal spray. Nonetheless, because the psychotomimetic and cognitive liabilities remain, the drug must still be taken under the supervision of a health care professional in a clinic, similarly to intravenous R,S-ketamine. Notably, the antidepressant efficacy reported in clinical trials appeared less robust than that for R,S-ketamine infusion (Swainson et al., 2019).
b. NMDA receptor antagonists in the treatment of depression
The successful repurposing of ketamine has stimulated interest in finding new agents that have an antidepressant efficacy similar to ketamine but with a more benign side-effect profile and improved ease of use. In addition, there is significant interest in agents that may prolong the antidepressant effects of a ketamine-like drug. Other NMDA receptor channel blockers have been under evaluation for efficacy against depression. Lanicemine (AZD6765) differs from ketamine in having a faster rate of dissociation from the channel pore hypothesized to result in less cognitive side effect liability (Mealing et al., 1999) (Supplemental Table 12). Although intravenous administration of lanicemine appeared better tolerated than ketamine, it failed to demonstrate consistent antidepressant activity in clinical trials (Sanacora et al., 2014, Sanacora et al., 2017). Dextromethorphan is another low-affinity channel blocker that is orally bioavailable but rapidly metabolized by CYP2D6, a polymorphic hepatic cytochrome P450 (Taylor et al., 2016) (Supplemental Table 12). To overcome this metabolic liability, dextromethorphan has been formulated with CYP2D6 inhibitors, such as bupropion [AXS-05; Wilkinson and Sanacora (2019)], which is a norepinephrine–dopamine reuptake inhibitor with slowly developing antidepressant properties. REL-1017 (dextromethadone) is also an NMDA receptor channel blocker in development as a rapid onset antidepressant in patients with treatment-resistant depression (Fogaca et al., 2019; Hanania et al., 2020) (Supplemental Table 12).
Another class of NMDA receptor antagonists, the GluN2B-selective NAMs (Section IX. Exogenous Positive and Negative Allosteric Modulators), are under evaluation for efficacy against depression, again with mixed results. CP-101,606, the clinical prototype for this class, was reported in 2008 to have rapid onset antidepressant activity after a single intravenous infusion in a small number of treatment-resistant patients (Preskorn et al., 2008). Several subjects in this study and other studies (Merchant et al., 1999; Nutt et al., 2008) experienced cognitive disruption and psychotomimetic effects similar to those produced by ketamine.
Rapastinel (GLYX-13) is an amidated tetrapeptide proposed to have the characteristics of a glycine site partial agonist and is reported to enhance NMDA receptor responses at low concentrations and suppress NMDA receptor responses at higher concentrations (Zhang et al., 2008d; Moskal et al., 2017; Donello et al., 2019). Rapastinel has biochemical and behavioral effects in a number of preclinical models hypothesized to reflect the antidepressant mechanisms of ketamine (Burgdorf et al., 2015; Pereira et al., 2019; Banerjee et al., 2020; Kato and Duman, 2020) and may disinhibit hippocampal neurons in acute brain slices (Widman and McMahon, 2018). A single intravenous infusion of rapastinel produced an antidepressant effect within 1 day that lasted for approximately 7 days, but this was not replicated in subsequent clinical studies (Kato and Duman, 2020).
An intriguing aspect of the antidepressant activity of ketamine is that the clinical response develops and is sustained after the drug and its metabolites have been cleared. In contrast, the psychotomimetic effects track closely with drug exposure (Shaffer et al., 2014). These results suggest that the antidepressant effects arise not from NMDA receptor inhibition per se but from the CNS response to that inhibition. This type of mechanism might be characterized as one related to metaplasticity, “the plasticity of synaptic plasticity” (Abraham and Bear, 1996). This form of plasticity could underlie the effects that an acute change in synaptic function (produced by rapid ketamine infusion) has on the ability of subsequent stimuli to effect new change (Turrigiano, 2008; Cooper and Bear, 2012). An intriguing hypothesis is that antidepressant effects of NMDA receptor inhibitors can be interpreted as a variation on this theme (Fig. 49). There is considerable evidence that the brief NMDA receptor inhibition produced by ketamine induces a “rebound” that ultimately results in an upregulation of AMPA receptor activity and a sustained increase in synaptic reactivity and plasticity (Krystal et al., 2019). The effect of ketamine to induce persistent changes in synaptic function engages intracellular signaling mechanisms that are suggested to include the mammalian target of rapamycin (mTOR) pathway as well as the potential regulation of BDNF (Li et al., 2010; Kavalali and Monteggia, 2012; Krystal et al., 2019). The mixed clinical results with other NMDA receptor antagonists might reflect a variable ability to induce such a rebound effect at the molecular level, although this has not been established. Thus, back-translating the molecular basis for such differences in clinical efficacy could afford insight into the underlying disease biology and provide clues to improving and sustaining efficacy while reducing side effects. However, the caveat to this line of research is the well known vagaries of clinical trials that yield both false positives and negatives, making distinctions among agents difficult. Additional clinical data will provide better clarity and perhaps enable improved understanding of mechanism. There are two research questions that bear investigation:
Fig. 49.
Mechanism for the antidepressant effect of NMDA receptor antagonists. Synaptic potentiation on the vertical axis is a hypothetical construct that captures 1) the fact that blocking NMDA receptors reduces activity at some synapses in the CNS, and 2) the wealth of preclinical and clinical data that indicate there is an increase in synaptic strength after NMDA receptor blockade, putatively resulting from the insertion of AMPA receptors into some population of synapses. Initially, acute NMDA receptor (NMDAr) inhibition reduces synaptic potentiation. This correlates closely in time with, and putatively causes, the psychotomimetic side effects (psychosis) that track the drug-exposure time course. NMDA receptor inhibition also triggers a synaptic potentiation (rebound) that outlasts the period of acute NMDA receptor inhibition. When synaptic potentiation exceeds a theoretical threshold (depression threshold), depressive symptoms are reduced (antidepressant response). In most cases, the NMDA receptor induced synaptic potentiation is transient and when this potentiation wanes to below the depression threshold, depressive symptoms return. The population of NMDA receptors that mediate the antidepressant response are not known at present. However, clinical data indicate that ketamine and the GluN2B-selective NAM CP-101,606 cause psychotomimetic effects and a subsequent antidepressant response.
The first question is the degree to which antidepressant efficacy is related to the kinetics of NMDA receptor inhibition. The standard infusion protocol for R,S-ketamine (0.5 mg/kg infused over 40 minutes) results in rapid inhibition of NMDA receptors that resolves quickly after the end of the infusion based on cognitive disruption as a functional biomarker. Given that the antidepressant response may be an adaptive process to NMDA receptor inhibition, the kinetics of inhibition may be critical to efficacy. This relatively simple research question has yet to be systematically explored in either preclinical or clinical studies. Resolving this question, ideally using R,S-ketamine, will facilitate interpretation of clinical differences in efficacy among NMDA receptor modulators.
The second question is the mechanistic implications of the similarities and differences in effects of GluN2B NAMs (e.g., CP-101,606) and channel-blocking NMDA antagonists (e.g., ketamine) in their antidepressant activity. Numerous preclinical studies indicate channel blockers and GluN2B NAMs have similar effects on measures hypothesized to underlie induction of an antidepressant response [e.g., Li et al. (2011b), Duman et al. (2012), Graef et al. (2015), Pochwat et al. (2017)]. Intravenous CP-101,606 has a clinical antidepressant and side-effect profile similar to intravenous ketamine (Preskorn et al., 2008). Consistent with the similarity in psychotomimetic effects observed in humans, some GluN2B NAMs have discriminative stimulus and reinforcing properties that overlap with channel blockers in rats and nonhuman primates (Chaperon et al., 2003; Nicholson et al., 2007). Based on these clinical and preclinical data, it is not unreasonable to hypothesize that the underlying molecular mechanisms for antidepressant efficacy and side-effect liabilities may be similar for GluN2B NAMs and channel blockers. This hypothesis has several implications. If ketamine and GluN2B NAMs in fact share the same mechanism, then it would argue against a role for ketamine metabolites as primary mediators of antidepressant responses (Zanos et al., 2016; Yang et al., 2019; Highland et al., 2021), since the GluN2B NAMs do not produce such metabolites. This argument is consistent with conclusions from a recent analysis that found no correlation between levels of ketamine metabolite and antidepressant response across several clinical studies of ketamine (Abdallah, 2020). Furthermore, a key difference between NMDA receptor channel blockers and GluN2B NAMs is that the latter do not enhance cortical γ oscillations, in contrast to the induction of γ oscillations by the channel blockers (Kocsis, 2012; Keavy et al., 2016; Nagy et al., 2016). These findings argue against a role for γ oscillations induced by cortical glutamate release as triggering the antidepressant response (Krystal et al., 2013). This lack of effect of the GluN2B NAMs on γ oscillations also indicates that γ oscillations per se do not underlie the psychotomimetic experiences. The mechanistic points of convergence (or divergence) between these two classes of NMDA receptor antagonists should be informative about the functional endpoints in preclinical models and clinical profiles. Thus, pinpointing mechanisms of ketamine and GluN2B NAMs may illuminate the molecular mechanisms that underlie the antidepressant response.
c. NMDA receptor antagonists in other CNS diseases
The first of the iGluR-targeted therapeutics, the phencyclidine-related NMDA receptor channel blockers, were developed as anesthetics and analgesics and are used as such as acute treatments. Considerable research indicates a role for NMDA receptors in mediating acute pain sensitivity as well as plasticity in circuits that results in the development of chronic pain (Costigan et al., 2009). Thus, there is has been a long-standing interest in NMDA receptor antagonists to treat chronic pain (Costigan et al., 2009; Aiyer et al., 2018) and as an adjunct to the use of opiates to augment analgesia and prevent tachyphylaxis (Trujillo and Akil, 1991). Only limited clinical data using ketamine as the probe (Niesters et al., 2014; Bell et al., 2017; Kreutzwiser and Tawfic, 2019) support these idea, and work in this vein is impeded by the hurdles imposed by ketamine’s psychotomimetic side effects. Hopefully, newer iGluR modulators will better define and target pain pathways with fewer side effects.
There has been a longstanding interest to develop iGluR targeted drugs to more effectively treat l-DOPA–induced dyskinesias and to treat primary parkinsonian symptoms (Blandini et al., 1996; Ahmed et al., 2011b). The weak NMDA receptor channel blocker amantadine is used clinically in patients with Parkinson’s disease to reduce the side effects that develop with chronic l-DOPA therapy. There have been some clinical studies of GluN2B NAMs in this regard; however, results to date have been mixed, with no evidence for acute effects on primary symptoms and one positive and one negative result with regard to reducing dyskinesias (Nutt et al., 2008; Herring et al., 2017). It will be of interest to investigate the effects of new agents targeting GluN2D-containing NMDA receptors given the notable expression of this NMDA receptor subtype in different nodes of the basal ganglia circuitry (Standaert et al., 1994) and the effects of some of these agents on the activity of neurons in the subthalamic nucleus (Swanger et al., 2015). NMDA receptors could also be targets in Huntington’s disease to block excitotoxic damage to striatal neurons and thereby moderate the progression of this neurodegenerative condition (Young et al., 1988; Cepeda et al., 2001). This interest is supported by data indicating that genetic variation in GluN2A and GluN2B can impact disease progression [Arning et al. (2005); see also Mahfooz et al. (2016)]. Extrasynaptic GluN2B-containing receptors are thought to drive neurotoxicity in mouse models of Huntington’s disease (Dau et al., 2014), but this hypothesis has not been tested in clinical trials. Some clinical data indicate that the weak NMDA receptor antagonist amantadine reduces chorea in patients with Huntington’s disease (Verhagen Metman et al., 2002), suggesting there may be scope for developing iGluR-targeted symptomatic therapies.
Nuedexta is a combination of dextromethorphan and the CYP2D6 inhibitor quinidine (Taylor et al., 2016) that was approved in 2013 for reducing the symptoms of pseudobulbar affect, a disorder of uncontrollable crying and laughing that occurs in the absence of a congruent emotional state (Miller et al., 2011). Pseudobulbar affect frequently emerges secondary to brain injury or with neurodegenerative disease (Cruz, 2013). Nuedexta is used to treat pseudobulbar affect in a variety of patients with neurologic dysfunction, including amyotrophic lateral sclerosis, multiple sclerosis, stroke, traumatic brain injury, Alzheimer’s disease, and Parkinson’s disease (Yang and Deeks, 2015). The actions of Nuedexta on pseudobulbar affect result from a continuous inhibition of NMDA receptors, although both dextromethorphan and quinidine have other targets (Taylor et al., 2016). Pseudobulbar affect is both an affective and a neurologic disorder that results from disruption of neural networks controlling the motor output of emotions. Nuedexta appears to have limited efficacy against affective symptoms when tested as an antidepressant (Nofziger et al., 2019).
2. Enhancement of NMDA Receptor Function
NMDA receptor channel blockers can induce a state redolent of the symptoms of schizophrenia in healthy individuals, leading to the hypothesis that NMDA receptor hypofunction is the molecular basis for these symptoms (Javitt and Zukin, 1991; Olney et al., 1999; Krystal et al., 2003). This hypothesis has driven an interest in augmenting NMDA receptor signaling as a therapeutic approach to schizophrenia and, more broadly, to disorders of cognitive impairment (Brogi et al., 2019; Menniti et al., 2013). However, enthusiasm for discovery of such agents has been tempered by concerns about irreversible neurotoxicity. Synaptic NMDA receptors depend on synaptic release of glutamate for activation, but the strength of phasic activation by glutamate can be modulated by the tonic presence of agonists that bind the GluN1 agonist binding site (Panatier et al., 2006; Yang and Svensson, 2008; Wolosker and Balu, 2020). The idea to target the GluN1 agonist binding site in the development of new therapeutic strategies has received considerable attention and has led to clinical trials with various ligands that act at the GluN1 glycine binding site (Szakacs et al., 2012; Balu and Coyle, 2015; Goff, 2015; Schade and Paulus, 2016). Industry approached this idea through development of inhibitors of the GlyT1 transporter to increase glycine availability (Chue, 2013). The results of clinical studies of both approaches suggest that augmenting glycine-site agonist availability has a modest effect in schizophrenia, primarily reducing negative symptoms (Beck et al., 2016). Preclinical and clinical studies with the GlyT1 inhibitor biopterin (RG1678) revealed an inverted U-shaped dose response (D'Souza et al., 2018; Pinard et al., 2018), with reduced effect at higher drug concentrations, which could reflect off-target actions or tachyphylaxis induced by persistent glycine site occupancy (Quartermain et al., 1994).
d-Cycloserine, an NMDA receptor glycine site agonist with GluN2-dependent efficacy (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers) facilitates extinction of fear in rodents [reviewed in Davis et al. (2006), Hofmann et al. (2015), Singewald et al. (2015)]. This reduction in fear is observed only if d-cycloserine is given in combination with extinction training (Walker et al., 2002; Ledgerwood et al., 2003). Furthermore, it remains unresolved whether enhancement of GluN2C-containing NMDA receptors by d-cycloserine (Fig. 33) or partial agonism at other NMDA receptors is required for the behavioral effects (Sotres-Bayon et al., 2007, 2009; Hillman et al., 2011; Dalton et al., 2012; Gupta et al., 2013; Ogden et al., 2014). The effects of d-cycloserine on fear extinction may arise from its actions in basolateral amygdala (Walker et al., 2002; Ledgerwood et al., 2003; Mao et al., 2006), medial prefrontal cortex, or hippocampus (Chang and Maren, 2011; Inoue et al., 2013; Ren et al., 2013; Sierra et al., 2016). d-Cycloserine also facilitates extinction of conditioned drug response to alcohol and cocaine [reviewed Myers and Carlezon (2012)]. In clinical settings, it is unclear whether d-cycloserine facilitates cognitive behavioral therapy for anxiety disorders, with some studies showing positive effects (Ressler et al., 2004; Hofmann et al., 2006a,b; Kushner et al., 2007; Otto et al., 2010) and other studies indicating small or no effect (Burkner et al., 2017; Mataix-Cols et al., 2017). It is now acknowledged that d-cycloserine may be effective for cognitive behavioral therapy in anxiety disorders, albeit only under certain conditions and that variables such as number of sessions, dosing regimen, and success of behavioral therapy, may dictate effectiveness (Bowers and Ressler, 2015; Otto et al., 2016; Smits et al., 2020).
A series of spirocyclic-β-lactams similar to the tetrapeptide GLYX-13 (rapastinel) have been reported to act as NMDA receptor PAMs (Khan et al., 2018; Barth et al., 2020). These compounds are suggested to potentiate both metabotropic and ionotropic NMDA receptor signaling, and the metabotropic activity may underlie potential therapeutic by enhancing synaptic plasticity (Bowers et al., 2020).
D. Clinical Implications of Disease-Associated Glutamate Receptor Genetic Variants
Rapid advances in DNA-sequencing technologies have enabled genome-wide sequencing as a means of diagnosis and have yielded an unprecedented amount of genetic data from both patients and healthy individuals. Informatics tools, such as the residual variation intolerance score (RVIS) (Petrovski et al., 2013) can be used to analyze genetic variation in the healthy population. These scores indicate that most (but not all) of the genes encoding iGluR subunits show fewer single nucleotide polymorphisms than most genes, making them among the genes least tolerant to missense mutations (Petrovski et al., 2013) (Table 13). The intolerance to variation for most iGluR genes raises the likelihood that missense variants in these genes will result in disease. Comparison of the observed and expected frequency of genetic missense variations at specific amino acid positions in the healthy population, with the observed/expected ratio referred to as the missense tolerance ratio, can identify regions of a gene that are under purifying selection, also known as negative selection or the selective removal of alleles that are deleterious (Traynelis et al., 2017; Perszyk et al., 2021). Moreover, there are also distinct intolerant regions for each gene product, consistent with the fact that the different iGluR subunits play unique roles in brain function despite high homology and similar functional properties within a family.
TABLE 13.
Summary of genetic variants in iGluR genes
RVIS is the Residual Variation Intolerance Score given as a percentile rank among all genes (Petrovski et al., 2013) (http://genic-intolerance.org/). An RVIS score of 2.55 (e.g., GRIA1) means that 97.45% of all genes have more variation in the healthy population than GRIA1. All variants counted for this table were not present in healthy individuals (gnomAD database; evaluated on July 02, 2020). Many variants have multiple phenotypes, and this table is only a snapshot of the current literature, which is disproportionally weighted by different diagnostic procedures. “Others” includes large-scale chromosomal deletion, translocation, inversion, and duplication.
| Gene | Protein | RVIS | Variant types | Locations | Phenotypes | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Missense | Nonsense | Frame shift | Splice | Other | ATD | ABD | TMD-linker | CTD | EPI | ID | ASD | ADHD | MD | SCZ/BP | |||
| % | |||||||||||||||||
| GRIA1 | GluA1 | 2.55 | 12 | 1 | 2 | 0 | 0 | 6 | 4 | 5 | 0 | 0 | 11 | 1 | 0 | 0 | 0 |
| GRIA2 | GluA2 | 10.8 | 19 | 1 | 3 | 2 | 6 | 6 | 2 | 16 | 0 | 9 | 30 | 17 | 1 | 5 | 0 |
| GRIA3 | GluA3 | 5.49 | 32 | 2 | 3 | 0 | 9 | 7 | 10 | 20 | 0 | 5 | 29 | 6 | 0 | 3 | 0 |
| GRIA4 | GluA4 | 4.57 | 6 | 0 | 0 | 0 | 0 | 0 | 2 | 4 | 0 | 3 | 5 | 0 | 0 | 2 | 0 |
| GRIK1 | GluK1 | 14.3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| GRIK2 | GluK2 | 7.78 | 11 | 2 | 1 | 5 | 2 | 5 | 1 | 7 | 1 | 3 | 10 | 1 | 1 | 1 | 2 |
| GRIK3 | GluK3 | 4.91 | 2 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| GRIK4 | GluK4 | 13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| GRIK5 | GluK5 | 21.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GRIN1 | GluN1 | 6.92 | 86 | 4 | 1 | 0 | 0 | 15 | 24 | 45 | 7 | 27 | 38 | 4 | 0 | 21 | 3 |
| GRIN2A | GluN2A | 1.96 | 209 | 24 | 36 | 13 | 29 | 67 | 82 | 50 | 80 | 203 | 190 | 19 | 5 | 28 | 5 |
| GRIN2B | GluN2B | 1.28 | 194 | 22 | 22 | 6 | 14 | 39 | 65 | 56 | 78 | 99 | 187 | 35 | 2 | 10 | 5 |
| GRIN2C | GluN2C | 62 | 13 | 1 | 5 | 0 | 0 | 6 | 5 | 1 | 7 | 1 | 4 | 8 | 0 | 0 | 7 |
| GRIN2D | GluN2D | 11.7 | 27 | 0 | 0 | 1 | 0 | 3 | 8 | 9 | 7 | 13 | 13 | 5 | 0 | 0 | 9 |
| GRIN3A | GluN3A | 67.6 | 11 | 2 | 0 | 0 | 0 | 7 | 3 | 1 | 2 | 0 | 1 | 5 | 0 | 0 | 4 |
| GRIN3B | GluN3B | 98.8 | 6 | 0 | 2 | 0 | 0 | 1 | 3 | 1 | 3 | 0 | 0 | 2 | 0 | 4 | 2 |
| GRID1 | GluD1 | 4.21 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GRID2 | GluD2 | 8.77 | 5 | 2 | 0 | 0 | 3 | 3 | 0 | 4 | 0 | 0 | 1 | 2 | 0 | 7 | 1 |
TMD-linker, transmembrane domains (M1-4) and linker regions (S1-M1, M1-M2, M2-M3, M3-S2, and S2-M4); ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder; BP, bipolar disorder; Epi, epilepsy/seizures; ID, intellectual disability; MD, movement disorders; SCZ/BP, schizophrenia/bipolar.
1. NMDA Receptor De Novo Variants
Evaluation of missense tolerance ratio of observed/expected variation indicates that there are multiple subdomains in GRIN genes that are intolerant to variation in healthy individuals (e.g., SYTANLAAF in the M3 transmembrane helix) (Fig. 50). The pre-M1/M1 and pre-M4/M4 regions that couple agonist binding to channel opening showed intolerance to variation in GRIN genes (Chen et al., 2017b; Ogden et al., 2017; Perszyk et al., 2020a) (Fig. 50). In addition, the residues encoded by exon 5 in the GluN1 NTD are intolerant to variation. There appear to be more NMDA receptor variants in patients with neurologic disease than all of the other iGluR genes combined (Table 13). There are over 700 reported genetic variants in GRIN genes that are absent in the healthy population (Genome Aggregation Database, gnomAD). These genetic variants occurred in all NMDA receptor subunits and include missense, nonsense, frameshift, splice site, and large-scale variations, such as chromosomal translocation, deletion, or inversion. These variants have been found in all NMDA receptor subunits with 13% in GRIN1 encoding GluN1, 44% in GRIN2A encoding GluN2A, 37% in GRIN2B encoding GluN2B, 2.8% in GRIN2C encoding GluN2C, and 3.8% in GRIN2D encoding GluN2D (Table 13). The variants are scattered across all domains throughout the mature protein. A recent study of a cohort of over 200 patients hosting GRIN2A missense variants suggested that the variants located in NTD and ABD have less severe phenotypes, whereas variants in TMD and linker regions are associated with more severe phenotypes and a poor prognosis (Strehlow et al., 2019). However, this generalization may not hold for other GRIN genes, given different regional intolerance, roles, and expression patterns.
Fig. 50.

Regional genetic intolerance in NMDA receptor GRIN genes. (A) The ratio of observed/expected GRIN variants in the general population (OE-ratio) below the 10th percentile are red and indicate the regions under selection (Traynelis et al., 2017). The 21 amino acids in GluN1 encoded by exon 5 are highlighted in blue and show almost no variation in the general population. The regions highlighted with a dashed box have insufficient data available. Data shown are a sliding average of ratios for 31 residues. (B) NMDA receptor ABD/TMD domains are intolerant to genetic variation. Observed/expected variant ratios (OE-ratio) below the 10th percentile are colored magenta, indicating the regions under selection. Homology models for diheteromeric GluN1/2A receptor were generated from two template structures (PDB: 5FXH and 4PE5).
GRIN1 variants are present in patients with a set of broad neurologic and neuropsychiatric disorders, and many of the patients showed comorbidity. Among the patients with clinical information provided, the major phenotypes the patients presented are developmental delay/intellectual disability followed in order of prevalence by epilepsy/seizures, autistic features, movement disorders, symptoms of schizophrenia, and attention deficit hyperactivity disorder (Table 13, Supplemental Table 14).
Among GRIN2A variants, epilepsy/seizures were the most common phenotype and were followed by intellectual disability and movement disorders (Supplemental Table 14). Patients with GRIN2A-related epilepsy/seizures frequently presented with some aspects of the epilepsy-aphasia syndrome and displayed idiopathic focal epilepsy, atypical benign partial epilepsy, atypical childhood epilepsy with centrotemporal spikes, benign childhood epilepsy, benign epilepsy with centrotemporal spikes, continuous spike-and-wave during sleep syndrome, early-onset epileptic encephalopathy, and Landau-Kleffner syndrome (Strehlow et al., 2019).
In patients with GRIN2B variants, features of intellectual disability/developmental delay were present in almost all patients (94%) and followed in order of prevalence by epilepsy and autism spectrum disorder (Table 13, Supplemental Table 14), consistent with the roles of GluN2B in early development. In the cohort of patients studied, most GRIN2A variants were related to epilepsy/seizures, whereas most GRIN2B variants occurred in patients with neurodevelopmental disorders, such as intellectual disability and autism (Hu et al., 2016; XiangWei et al., 2018; Xu and Luo, 2018; Myers et al., 2019).
a. Functional evaluations of NMDA receptor variants
An understanding of the mechanisms by which the disease-associated variants produce a clinical phenotype requires functional evaluation of the variant receptors. This functional understanding is essential to classify variants of unknown significance and develop therapeutic options and offers a logical way to stratify patients, improve personalized therapy, and facilitate the clinical testing. Among the ∼700 known GRIN variants, functional evaluation has been published in peer-reviewed literature for well over 100 variants (Supplemental Table 14; see also Center for Functional Evaluation of Rare Variants, http://functionalvariants.emory.edu/). The published functional evaluations of disease-associated variants range from evaluation of a single parameter (i.e., current amplitudes or agonist potency) to comprehensive evaluation of multiple parameters in vitro and in vivo. Multiple parameters can be assessed in vitro using recombinant receptors and native cells/tissues from transgenic animals to determine current amplitude, agonist (glutamate and glycine) potency, voltage-dependent Mg2+ block, sensitivity to endogenous negative modulators (Zn2+ and protons), channel open probability, channel activation time course, glutamate deactivation time course (which can be predictive of synaptic response time course), desensitization, long-term potentiation/depression, receptor trafficking, and neuronal excitotoxicity (Swanger et al., 2016; Addis et al., 2017; Ogden et al., 2017; Amin et al., 2018; Fedele et al., 2018; Vyklicky et al., 2018; XiangWei et al., 2018, 2019; Li et al., 2019a; Sceniak et al., 2019; Strehlow et al., 2019; Amador et al., 2020; Shin et al., 2020; Skrenkova et al., 2020). These in vitro evaluations can be combined for many variants to provide a quantitative prediction of the overall effects that variants have on NMDA receptor pharmacology, function, and receptor localization. Although this approach has the capacity to approximate the contribution of the GRIN variants to NMDA receptor-mediated synaptic and nonsynaptic signaling [Swanger et al. (2016); see also XiangWei et al. (2018)], it will be critical to evaluate function, surface expression, and subcellular localization of variants in neurons. If the variants are not trafficked to the neuronal surface or incorporated into the synapse, then a functionally null phenotype would be seen in transgenic animals or transfected neurons.
The term gain of function is defined as any variant-related increase in NMDA receptor–mediated signaling and could arise from enhanced agonist potency (e.g., glutamate and glycine), reduced sensitivity to voltage-dependent Mg2+ block, reduced sensitivity to endogenous negative modulators, prolonged synaptic response time course, enhanced gating or open probability, reduced receptor desensitization, and enhanced receptor cell surface trafficking. Each of these effects could differentially impact synaptic and perisynaptic receptors, which are exposed to different concentrations of glutamate [e.g., Swanger et al. (2016), Moldavski et al. (2020)]. Ultimately, these variants will likely have different clinical manifestations, since they would be expected to alter circuit function in different ways. Similarly, loss-of-function variants, defined as producing a reduction in NMDA receptor–mediated signaling, may arise from reduced agonist potency, enhanced sensitivity to voltage-dependent Mg2+ block or other endogenous negative modulators, shortened deactivation time course, reduced response amplitude, and decreased receptor cell surface trafficking. Although these parameters can be easily measured in vitro, compensatory and developmental changes could produce equally (or more) important changes in the expression profile of other genes and connectivity of neural circuits. Furthermore, it is unclear if loss-of-function variants caused by nonsense mutations or deletions will behave the same as missense loss-of-function variants, which may assemble with other subunits and could exert a dominant negative effect. Although global designations of variant function are helpful, clinical phenotypes are likely driven by variant-induced changes in specific receptor properties rather than the overall net effect captured by the gain- and loss-of-function categorization.
The complexity of the effects of GRIN variants on clinical phenotypes is illustrated by the finding that both gain- and loss-of-function variants in the same gene can result in similar neurologic symptoms, such as seizure disorders. For instance, GRIN2A gain-of-function human variants L812M (Yuan et al., 2014) and P552R (Ogden et al., 2017) and loss-of-function GRIN2A variants D731N (Gao et al., 2017) and V685G (Swanger et al., 2016) are observed in patients with infantile onset epilepsy and cognitive impairment. This is intriguing, as one might hypothesize that the loss of excitatory synaptic GluN2A subunits would decrease excitability rather than promote an epileptic phenotype. This paradoxical observation is also seen for loss-of-function GRIN2A truncation variants, which display seizure phenotypes (Carvill et al., 2013; Lemke et al., 2013; Lesca et al., 2013), and Grin2A KO mice that show hyperexcitability (Salmi et al., 2018, 2019). These observations suggest that the loss of signaling from GluN2A-containing NMDA receptors must be placed into a developmental context, whereby a change in NMDA receptor function in particular cells (i.e., interneuron vs. principal cell) impacts a critical developmental window and likely changes the overall balance of excitation and inhibition. Moreover, functional evaluation of variants can result in conflicting results from multiple parameters. For example, the GRIN2A variant A643D showed enhanced glutamate potency but decreased current response and reduced cell surface expression (Fernandez-Marmiesse et al., 2018). Therefore, evaluation of only one or two aspects of the functional effects of variants is insufficient for reaching a conclusion on the physiologic consequence of a given variant and could lead to ineffective therapeutic strategies.
Another important aspect of functional studies of disease-associated variants is that they provide an unbiased indicator of regions of the receptor critical for function and human health. That is, the location of these disease-associated variants can point to regions of the receptor that mediate important functions. For example, disease-associated variants that were present in three closely positioned and intolerant regions (pre-M1, M3, and pre-M4/M4) showed strong functional changes, suggesting that they are well positioned to control channel gating [Chen et al. (2017b), Ogden et al. (2017), Amin et al. (2018), McDaniel et al. (2020); see Perszyk et al. (2020a), Amin et al. (2021b)] (Fig. 51). Deeper investigation of functional properties on all GRIN variants will reveal new features of NMDA receptors in regions that are important for normal brain function, including variants in the CTD that impact functions of the receptors distinct from those that are detected in studies of their electrophysiological properties in recombinant systems (Liu et al., 2017; Mota Vieira et al., 2020).
Fig. 51.

Variants in the pre-M1 helix, M3, and pre-M4/M4 regions identify a region controlling gating. (A) Three intolerant regions lie close to one another, potentially allowing side chains to be in van der Waals contact to form a gating control element. These three regions include the adjacent GluN1 pre-M1 helix (magenta), GluN1 M3/SYTANLAAF (gray), and GluN2 pre-M4 linker/M4 (green) of the NMDA receptor. A second gating triad (not shown) also exists for the GluN2 pre-M1 helix, GluN2 M3/SYTANLAAF, and GluN1 pre-M4 linker/M4. (B) M3 variant GRIN2D-p.V667I prolongs the deactivation time course and increases channel open probability. (C) Pre-M1 variant GRIN2A-p.P552R enhances agonist potency, slows activation and prolongs deactivation time course. (D) Pre-M4 linker variant GRIN2A-p.L812M enhances agonist potency, prolongs the deactivation time course, and increases channel open probability. WT, wildtype. C: closed, O: open. Adapted with permission from Yuan et al. (2015a) (A), Li et al. (2016a) (B), Ogden et al. (2017) (C), and Yuan et al. (2014) (D).
b. Studies in transgenic mice harboring disease-associated variants
Homozygous mice harboring a knock-in of a human de novo gain-of-function GRIN2A variant S644G (in the SYTANLAAF motif in M3) displayed lethal tonic-clonic seizures at 3 weeks of age (Amador et al., 2020). Heterozygous mice exhibited an altered seizure threshold, hyperactivity, increased repetitive behaviors, and decreased anxiety-like behaviors. Both homozygous and heterozygous mice showed decreased hippocampal thickness. The possibility that GRIN variants produce profound developmental changes emphasizes the need for anatomic information at different developmental time points. Mice harboring the GRIN2A variant N615S show reduced Mg2+ block, reduced seizure threshold, and altered network activity (Bertocchi et al., 2021). Heterozygous mice hosting knock-in human loss-of-function GRIN2B variant C456Y displayed hypoactivity, anxiolytic-like behavior, and repetitive self-grooming with normal social interaction, social communication, learning, and memory (Shin et al., 2020). These studies suggest that the phenotype determined in heterologous systems is predictive of that observed in neurons. Similarly, mice expressing reduced levels of GRIN1 show deficits similar to some patients (Intson et al., 2019; Mielnik et al., 2020), and mice lacking GRIN2A show hyperexcitability (Salmi et al., 2018, 2019), consistent with patients showing seizures with GRIN2A truncations (Lemke et al., 2013).
A number of studies also show variant NMDA receptors transfected into neurons produce similar effects as observed in heterologous expression systems. A gain-of-function variant GluN2A-P552R slowed the rise time for NMDA receptor–mediated EPSCs and prolonged their duration in vitro, as expected from data obtained from expression in heterologous systems (Ogden et al., 2017). Both GluN2A-P552R and GluN2D-V667I were neurotoxic when transfected into cultured neurons (Li et al., 2016a; Ogden et al., 2017). Another variant, GluN2A-S1459G, present at a conserved CaMKII phosphorylation site altered phosphorylation and NMDA receptor trafficking when transfected into cultured neurons and modified the frequency of mEPSCs (Mota Vieira et al., 2020). A loss of function variant that truncates the GluN2A subunit at residue 724 produced aberrations in neuronal morphology in vitro, with shorter dendrites, fewer branches, and other abnormal features (Sceniak et al., 2019). Thus, work in native systems largely validates the conclusions drawn from heterologous expression systems.
c. Pharmacological modulation of NMDA receptor variants
The determination of mechanisms by which GRIN variants alter NMDA receptor function raises the possibility of mitigating physiologic consequences of the functional effects with pharmacological modulation. This assumes that clinical phenotypes are driven at least in part by continued functional changes of variant proteins. Clinical phenotypes could also be dominated by the impact of functional changes on an abnormal developmental trajectory that is irreversible and includes compensatory changes in gene expression and circuitry alterations. A small set of NMDA receptor channel blockers and negative allosteric modulators has been evaluated for potency and efficacy at function-altering variants (Pierson et al., 2014; Li et al., 2016a; Swanger et al., 2016; Chen et al., 2017b; Mullier et al., 2017; Ogden et al., 2017; Platzer et al., 2017; XiangWei et al., 2019; Amador et al., 2020; Chen et al., 2020a; Xu et al., 2021). Some of these channel blockers (memantine, dextromethorphan) appear to be safe in pediatric patients (Chez et al., 2007). However, different gain-of-function GRIN variants showed differential sensitivity to the channel blockers (Chen et al., 2017b; Ogden et al., 2017; Pierson et al., 2014; Xu et al., 2021), demonstrating the necessity to determine the sensitivity of each variant to any therapeutic agent contemplated as a potential treatment. Several NMDA receptor PAMs (i.e., endogenous neurosteroid 24(S)-HC, pregnenolone sulfate, and FDA-approved aminoglycosides) and coagonists at the glycine-binding site (i.e., d-serine, l-serine, and d-cycloserine) potentiate responses from NMDA receptors with loss-of-function GRIN1, GRIN2A, and GRIN2B variants identified in patients with neurodevelopmental and neuropsychiatric disorders (Swanger et al., 2016; Addis et al., 2017; Vyklicky et al., 2018; Soto et al., 2019; Tang et al., 2020). Early treatment with d-cycloserine on young transgenic mice harboring a loss-of-function GRIN2B variant C456Y variant appear to rectify NMDA receptor–dependent synaptic long-term depression and improve anxiolytic-like behaviors in adult mice (Shin et al., 2020). Chronic treatment with either dextromethorphan, radiprodil (GluN2B-selective NAM), or Nuedexta (dextromethorphan + quinidine) of homozygous mice hosting a GRIN2A variant encoding S644G significantly delayed the onset of lethal seizures (Amador et al., 2020). Treatment of a loss-of-function Grin1 mouse with expression of wild-type gene improved cognitive function, suggesting it may be possible to mitigate some aspects of neurodevelopmental disorders arising from variants that reduce receptor expression (Mielnik et al., 2020). These preclinical studies provide evidence that treatment may be possible using clinically useful pharmacological and genetic approaches.
So-called “N of 1” trials with memantine, dextromethorphan, or subanesthetic ketamine have been performed in a small number of pediatric patients harboring gain-of-function GRIN2A, GRIN2B, and GRIN2D variants that were associated with severe clinical consequences. These patients showed drug-resistant epilepsy and early onset epileptic encephalopathy and had a divergent response to off-label use of these agents, with some patients showing reduced seizure burden and others showing no effect [Pierson et al. (2014), Li et al. (2016a), Platzer et al. (2017), XiangWei et al. (2019), Amador et al. (2020; Xu et al., 2021); see XiangWei et al. (2018), Camp and Yuan (2020)]. l-Serine, a nonessential amino acid, has been used as a dietary supplement in a patient with a GRIN2B variant P553T who presented with severe developmental encephalopathy (Soto et al., 2019). Addition of l-serine in this open-label trial appeared to improve motor, cognitive, and communication skills in this patient. These divergent responses reinforce the complexity of personalized therapy and emphasize the need for well designed double-blinded prospective clinical trials.
2. AMPA Receptors De Novo Variants
Since AMPA receptors play vital roles in fast excitatory synaptic transmission, it is not surprising that the GRIA genes are intolerant to genetic variation in regions that are involved in receptor function (Table 13). In addition, there are clear differences in intolerance between the GRIA genes that code for different regions within each subunit. For example, the pre-M1/M1 and M4 regions showed an opposite intolerance to genetic variation between GRIA1 and GRIA2 genes, with the residues encoding M3 and M4 being intolerant in GRIA1 compared with only a portion of M3 and pre-M1/M1 showing intolerance in GRIA2 (Fig. 52). This suggests the two subunits encoded by GRIA1 and GRIA2, often expressed together in cortical structures, have different underlying mechanisms that, when coassembled in heteromeric complexes and with different auxiliary subunits, contribute to a unique functional specificity for the heteromeric receptor.
Fig. 52.
Regional intolerance in AMPA glutamate receptor genes. (A) The ratio of observed/expected GRIA variants (OE-ratio) below the 10th percentile are shown in red and suggest regions/domains that are under selection (Traynelis et al., 2017). Regions highlighted with a dashed box have insufficient data available. M1 and M4 are sites for TARP, GSG1L, and CNIH binding, highlighted by blue squares. (B) Intolerance analysis of genetic variation across functional domains of GluA1 and GluA2. (A–D) Side views of the tetrameric receptor (upper panels) and a single subunit (chain A for GluA1, chain D for GluA2, lower panels) of a GluA/2 receptor based on the crystal structure (PDB: 6QKZ). GluA1 subunit is light yellow, and GluA2 subunit is light blue; regions with an OE-ratio below the 10th percentile are magenta and indicate the regions under selection.
After the first report of a disease-associated GRIA variant (Wu et al., 2007c), a large number of genetic variations (>100) scattered across all four GRIA genes have been identified (Chiyonobu et al., 2007; Wu et al., 2007c; Bonnet et al., 2009; Poot et al., 2010; Tzschach et al., 2010; Hamdan et al., 2011; Hackmann et al., 2013; Di Benedetto et al., 2014; Philips et al., 2014; Jin et al., 2015; Davies et al., 2017; Geisheker et al., 2017; Martin et al., 2017; Salpietro et al., 2019; Piard et al., 2020). Among these, 98 variants are not present in healthy individuals (gnomAD database) (Table 13). The most common site for the disease-associated variants is in the highly conserved TMD-linker regions, followed by the ABD and NTD (Table 13). Among 84 variants with known phenotypes, over 89% of the patients presented with some degree of intellectual disability, followed in order of prevalence by autistic spectrum disorder, seizure disorders, and movement disorders (Supplemental Table 14). Interestingly, 77% of patients hosting GRIA2 variants showed speech or language problems. GRIA3 resides on the X chromosome and is a candidate gene for X-linked intellectual disability (Chiyonobu et al., 2007). Taken together, these data indicate that rare genetic variation within the critical subdomains of GRIA genes are linked to subsets of patients with neurodevelopmental disorders.
Several studies have been performed to explore how GRIA variants impact receptor function and neuronal signaling (Wu et al., 2007c; Davies et al., 2017; Geisheker et al., 2017; Martin et al., 2017; Salpietro et al., 2019; Piard et al., 2020). These experiments range from in vitro studies of recombinant receptors expressed in heterologous systems to studies in transgenic animals harboring a disease-associated GRIA variant. One study of 28 unrelated patients with neurodevelopmental disorders harboring de novo GRIA2 variants indicated that most variants (Salpietro et al., 2019), when coexpressed with wild-type GluA1, reduced the current amplitude and decreased surface expression (Salpietro et al., 2019).
The SYTANLAAF motif in M3 that controls channel gating is highly conserved across all glutamate receptors and is a common site for disease-associated variants. Changes in the SYTANLAAF motif have been reported to influence channel activation in multiple studies (Sections II. Receptor Structure and IV. Receptor Activation, Deactivation, and Desensitization). A specific recurrent variant (A636T in GRIA1 gene; SYTANLAAF) in an identical position to the mouse Lurcher mutation in the GRID2 gene (Zuo et al., 1997) has been observed in multiple patients with intellectual disability, autism, and/or language problems (Geisheker et al., 2017). GluA1-A636T produced constitutive currents, which may reflect either spontaneous channel activity or increased sensitivity to contaminant levels of glutamate (Geisheker et al., 2017). Another missense variant GluA3-A653T in this motif (SYTANLAAF) was identified in siblings with intellectual disability and severe sleep-wake cycle dysregulation (Davies et al., 2017). Evaluation of recombinant receptors containing the GluA3-A653T variant revealed reduced glutamate potency and diminished desensitization without a change in GluA3 receptor surface expression. Transgenic mice hosting the same variant showed motor learning deficits with decreased current amplitude and frequency of spontaneous EPSCs onto cerebellar Purkinje cells (Davies et al., 2017).
3. Kainate Receptors De Novo Variants
Several disease-associated genetic variants in GRIK genes have been reported (Jamain et al., 2002; Strutz-Seebohm et al., 2006; Motazacker et al., 2007; Bonaglia et al., 2008; Pickard et al., 2008; Whalley et al., 2009; Han et al., 2010; Hamdan et al., 2011; Knight et al., 2012; Takenouchi et al., 2014; Cordoba et al., 2015; Guzman et al., 2017), although there seem to be fewer GRIK variants (28) than observed for the GRIA (98) and GRIN (706) gene families (Table 13). This is reflected in the reduced intolerance score, which places most GRIK genes as more tolerant to variation. Subdomains in the different GRIK genes showed a divergent intolerance to variation in healthy individuals, with a regional intolerance pattern that is different from AMPA and NMDA receptors (Supplemental Fig. 10). The kainate receptor NTD and TMD linker harbors the most variants (Table 13). The clinical phenotypes in patients with disease-associated GRIK variants include intellectual disability, movement disorders, autism, and epilepsy as well as schizophrenia and bipolar disorders. Single-nucleotide polymorphism–association studies have highlighted a potential role of the GRIK2 gene in autism (Jamain et al., 2002; Shuang et al., 2004; Dutta et al., 2007; Kim et al., 2007; Holt et al., 2010; Casey et al., 2012; Griswold et al., 2012). A GRIK2 missense variant, M867I, identified in a patient with autism is located in the CTD and showed modest ∼1.6-fold slowing of the desensitization time course (Han et al., 2010). GluK2-A657T in the lurcher position (SYTANLAAF in M3) identified in a pediatric patient with intellectual disability, ataxia, hypotonia, and motor and speech delay produces constitutive activity of heteromeric GluK2/5 receptors (Guzman et al., 2017).
4. De Novo Variants in GluN3 and GluD Subunits
Over two dozen missense variants in GRIN3A and GRIN3B genes that are absent in the healthy population have been identified in patients with neurologic and neuropsychiatric disorders (Niemann et al., 2008; Liu et al., 2009; Hamdan et al., 2011; Myers et al., 2011; Tarabeux et al., 2011; Yang et al., 2015; Gaynor et al., 2016; Yu et al., 2018b), but these two subunits appear more tolerant to variation than most other glutamate receptor subunits. These disease-associated variants are most common in the NTD, followed by ABD, CTD, and TMD-linker (Table 13), matching the locations of regional intolerance to variation in healthy individuals (Supplemental Fig. 11). The clinical phenotypes of patients include amyotrophic lateral sclerosis, autism, intellectual disability, bipolar disorders, and schizophrenia.
Variants in the GRID1 gene are associated with various neurologic conditions (Fallin et al., 2005; Guo et al., 2007; Glessner et al., 2009; Smith et al., 2009; Treutlein et al., 2009; Muglia et al., 2010; Greenwood et al., 2011; Nord et al., 2011; Edwards et al., 2012; Griswold et al., 2012; Zhang et al., 2018c). Genome-wide association studies found that GRID1 is a candidate gene for schizophrenia (Chen et al., 2011), with genetic variations in the promoter region of GRID1. Furthermore, several schizophrenia susceptibility genes are targeted by miR-346, which is located in a GRID1 intron (Zhu et al., 2009). Intronic deletions of GRID1 also have been found in patients with autism spectrum disorder (Glessner et al., 2009; Smith et al., 2009; Nord et al., 2011; Griswold et al., 2012). Deletions in the chromosome 10q22–q23 region where GRID1 is located are associated with neurodevelopmental abnormalities, cognitive impairment, features of autism spectrum disorder, and hyperactivity (Balciuniene et al., 2007).
Variations in the GRID2 gene are associated with schizophrenia-related symptoms (Greenwood et al., 2011, 2016; Tsuang et al., 2018), attention deficit hyperactivity disorder (Zhang et al., 2021), and autism spectrum disorder (Gazzellone et al., 2014; Pinto et al., 2014). De novo and inherited missense mutations and exon deletions in the GRID2 gene have been reported in patients with cerebellar ataxia, autism spectrum disorder, intellectual disability, and schizophrenia as well as some symptoms unrelated to the cerebellum, such as paraplegia and retinal dystrophy [Hills et al. (2013), Utine et al. (2013), Maier et al. (2014), Coutelier et al. (2015), Van Schil et al. (2015), Ali et al. (2017), Taghdiri et al. (2019), Hetzelt et al. (2020); see also Huang et al. (2014), Veerapandiyan et al. (2017), Taghdiri et al. (2019)]. GRID2, located on chromosome 4q22.1, is also a candidate gene in “4q deletion syndrome,” which is associated with autism spectrum disorder, intellectual disability, and attention deficit hyperactivity disorder (Strehle et al., 2012). Human phenotype associated with deletions in GRID2 includes cerebellar ataxia, tonic upgaze, nystagmus, and developmental delay (Hills et al., 2013; Utine et al., 2013). In one study of congenital ataxia, three missense variations in the GRID2 gene were reported (Ala654Thr, Ala654Asp, Leu656Val) (Coutelier et al., 2015). The first two variants affect the same alanine (SYTANLAAF in M3) as the lurcher mutation in mice (Zuo et al., 1997), and the patients show motor deficits that are consistent with behavior observed in mice expressing the GluD2 lurcher mutation, which include ataxia and jerky movement of the hind limbs (Kashiwabuchi et al., 1995; Zuo et al., 1997; Lalouette et al., 1998).
E. Glutamate Receptors in Autoimmune Diseases
A variety of nervous-system disorders are associated with antibodies that target self-antigens, including ion channels. The most well known and perhaps best-defined example is myasthenia gravis, in which autoantibodies target the nicotinic acetylcholine receptor as well as associated proteins at the neuromuscular junction to disrupt muscle strength (Gilhus et al., 2019). However, there are many examples of autoimmune channelopathies, including for GABAA receptors (Pruss and Kirmse, 2018), voltage-gated K+ channels (van Sonderen et al., 2017), and aquaporin (Soltys et al., 2019). There are enormous challenges in relating the presence of autoantibodies to any disease progression. Autoantibodies are often found in healthy patients and may be a natural immune response and a progression of aging (Pan et al., 2019). This is particularly acute for NMDA receptors that show a wide distribution not only in neuronal tissue but also non-neuronal tissue including B and T cells (Ehrenreich, 2018) (Supplemental Table 3). Furthermore, isolated antibodies are often polyclonal and polyspecific, targeting multiple epitopes on ion channels as well as associated proteins. Finally, although autoantibodies can show correlations to a disease phenotype, it is often unclear whether the autoantibodies cause the disease or whether they only shape some symptom or some behavioral phenotype.
Autoantibodies against iGluRs are detectable in several neurologic conditions, with strong evidence for contribution to disease pathology in some cases (Pleasure, 2008; Levite, 2014; Dalmau et al., 2017; Tay et al., 2017). Autoantibodies against iGluRs, including AMPA and NMDA receptors, have been discovered in patients with autoimmune encephalitis and paraneoplastic syndromes (Gardoni et al., 2021). The first identified anti-iGluR autoantibody was associated with Rasmussen’s encephalitis, in which the antibody epitope was on GluA3 (Rogers et al., 1994). Since then, numerous antibodies targeting iGluRs have been discovered along with putative mechanisms of disease (Table 14).
TABLE 14.
Anti-glutamate receptor autoantibodies with pathophysiological mechanisms
| Disease | Subunit Epitope | Pathophysiological Mechanisms |
|---|---|---|
| Limbic encephalitis (AMPA receptor encephalitis) | GluA1/A2 | • Decreases in synaptic density of AMPA receptors through internalization (Haselmann et al., 2018; Lai et al., 2009; Peng et al., 2015) • Acute decreases in AMPA receptor-mediated currents (Gleichman et al., 2014; Haselmann et al., 2018; Peng et al., 2015) • Decreases in LTP (Haselmann et al., 2018; Peng et al., 2015) • Behavioral and memory deficits (Haselmann et al., 2018) |
| Rasmussen’s encephalitis | GluA3 | • AMPA receptor–mediated excitotoxicity (He et al., 1998) • Potentiate AMPA receptors most likely as positive allosteric modulators but may also act as agonists (Carlson et al., 1997; Twyman et al., 1995) • Complement-mediated damage (Whitney and McNamara, 2000) |
| Epilepsy | GluA3 | • AMPA receptor-mediated excitotoxicity (Levite et al., 1999) • Potentiate AMPA receptors most likely as positive allosteric modulators but may also act as agonists (Cohen-Kashi Malina et al., 2006; Levite et al., 1999) • Behavioral deficits (Ganor et al., 2014) |
| Anti-NMDA receptor encephalitis | GluN1 | • Decreases in synaptic density of NMDA receptors through impaired surface diffusion and internalization (Castillo-Gomez et al., 2017; Hughes et al., 2010; Kreye et al., 2016; Ladepeche et al., 2018; Moscato et al., 2014; Planaguma et al., 2015) • Displacement from the EphrinB2 receptor that stabilizes synaptic NMDA receptors (Mikasova et al., 2012; Planaguma et al., 2015) • No acute changes in NMDA receptor–mediated currents (Castillo-Gomez et al., 2017), but see (Moscato et al., 2014) • Chronic decreases in NMDA receptor–mediated currents (Hughes et al., 2010; Kreye et al., 2016; Moscato et al., 2014) • Decreases in synaptic plasticity (Mikasova et al., 2012; Planaguma et al., 2015) • Behavioral and memory deficits (Planaguma et al., 2015) |
| Acute psychosis/ schizophrenia | GluN1 | • Disruption in EphrinB2 receptor interactions (see above) • Decrease in synaptic density of NMDA receptors through impaired surface diffusion (see above) • Decrease in synaptic plasticity (Jezequel et al., 2017) |
| Systemic lupus erythematosus | GluN2A/2B | • NMDA receptor–dependent excitotoxicity (DeGiorgio et al., 2001; Faust et al., 2010; Gono et al., 2011; Kapadia et al., 2017; Kowal et al., 2006; Kowal et al., 2004) • Acute changes in NMDA receptor–mediated currents (Faust et al., 2010; Gono et al., 2011; Kapadia et al., 2017) • Behavioral and memory deficits (Chang et al., 2015a; Kapadia et al., 2017; Nestor et al., 2018) • Recruitment of microglia (Nestor et al., 2018) • Primarily seems to act via GluN2A (Chan et al., 2020) |
1. Anti–AMPA Receptor Autoantibodies
Limbic encephalitis is characterized by inflammation of the limbic system and other parts of the brain and has an autoimmune origin. A subset of patients with limbic encephalitis may present with anti-GluA1, anti-GluA2, or anti-GluA1/GluA2 autoantibodies (also denoted “anti-AMPA receptor encephalitis,” but this term is not universally adopted). GluA1/2 autoantibodies bind to the NTD and ABD of GluA1 and GluA2 subunits, with no specific epitope (Gleichman et al., 2014). Anti–AMPA receptor autoantibodies have a diverse array of pathophysiological mechanisms at the synapse. Anti-GluA1/2 increases internalization of synaptic AMPA receptors leading to a chronic decrease in AMPA receptor function and decreased plasticity (Lai et al., 2009; Peng et al., 2015; Haselmann et al., 2018). The decrease in excitability from these antibodies is accompanied by homeostatic decreases in inhibition mediated by GABAA receptors (Peng et al., 2015). Impairment of synaptic plasticity can lead to the memory deficiencies seen in passive-transfer murine models with patient anti-GluA2 autoantibodies (Haselmann et al., 2018). Although these antibodies have epitopes in GluA1 and GluA2, they can also affect GluA3-containing heteromers (e.g., GluA2/3 receptors), decreasing GluA3-containing AMPA receptor content at the synapse (Lai et al., 2009; Peng et al., 2015). Interestingly, there is at least one reported case of anti-GluA2 autoantibodies found in a separate disease, nonfamilial olivopontocerebellar degeneration, in which they may function as positive allosteric modulators (Gahring et al., 1997).
Anti-GluA3 autoantibodies were discovered in a patient with Rasmussen’s encephalitis (Rogers et al., 1994), a debilitating disease that causes motor impairment, seizures, hemiparesis, and cognitive dysfunction primarily in children (Bien et al., 2005) as well as other seizure disorders (Levite, 2014). Several anti-GluA3 autoantibodies, including those identified in Rasmussen’s encephalitis, appear to bind to residues 372–395 in the GluA3 NTD (Levite et al., 1999; Mantegazza et al., 2002; Ganor et al., 2005a) and are thought to act as PAMs mediating the pathogenic effects through direct agonist activity and complement-mediated cell death (Twyman et al., 1995; Carlson et al., 1997; He et al., 1998; Levite et al., 1999; Whitney and McNamara, 2000; Cohen-Kashi Malina et al., 2006). Although anti-GluA3 autoantibodies have been found in patients with Rasmussen’s encephalitis, it remains unclear whether they are the cause of the disease syndrome (Watson et al., 2004; Bien et al., 2005; Schneider-Hohendorf et al., 2016). To date, immunosuppressants and hemispherectomy of the affected cortical hemisphere are the mainstay treatments for Rasmussen’s encephalitis. Animal models immunized with the anti-GluA3 epitope-containing peptide develop behavioral deficits and neuronal cell death (Ganor et al., 2005b, 2014; Goldberg-Stern et al., 2014), but it is unclear how these outcomes relate to the clinical phenotype. Anti-GluN2 antibodies have also been found in some patients with Rasmussen’s encephalitis, but it is unclear what their roles, if any, are in this disease (Takahashi et al., 2005).
2. Anti–NMDA Receptor Autoantibodies and Anti–NMDA Receptor Encephalitis
A variety of anti-GluN1 and anti-GluN2 autoantibodies have been reported in patients with diseases ranging from stroke to autism spectrum disorders, but with no known pathophysiological mechanisms (Bokesch et al., 2006; Hammer et al., 2014; Steiner et al., 2014; Zerche et al., 2015). Here, we will focus on those NMDA receptor–directed autoantibodies that have been well characterized. Perhaps the best-characterized anti-iGluR autoantibodies are those targeting GluN1 found in anti-NMDA receptor encephalitis (Dalmau et al., 2017). Anti-NMDA receptor encephalitis is characterized by a prodromal flu-like malaise followed by acute psychosis, paranoia, seizures, cognitive dysfunction, memory loss, and catatonia. The disease often affects women and may arise from ovarian teratomas that express NMDA receptors, exposing these receptors to the immune system in such a way as to induce formation of autoantibodies (Tuzun et al., 2009; Titulaer et al., 2013). Germline anti-GluN1 antibody-producing B cells and plasma cells that have escaped tolerance checkpoints may also be another cause (Irani et al., 2010; Kreye et al., 2016; Wenke et al., 2019). Anti-NMDA receptor encephalitis may also develop as a sequelae of herpes simplex virus encephalitis (Pruss et al., 2012; Hacohen et al., 2014).
Anti-GluN1 autoantibodies have at least one epitope near the hinge region of the bilobed NTD in GluN1, with critical residues at Asn368 and Gly369 (Gleichman et al., 2012; Kreye et al., 2016). The primary disease mechanism involves enhanced internalization of NMDA receptors. The literature is ambiguous about direct effects on NMDA receptor ion channel function, with some showing no effect, whereas others show a decrease or increase in channel function (Gleichman et al., 2012; Moscato et al., 2014; Castillo-Gomez et al., 2017). Nevertheless, anti-GluN1 antibodies have been shown to decrease synaptic content of NMDA receptors, leading to a chronic decrease in EPSCs and LTP in the hippocampus after long periods of exposure to the antibody (Mikasova et al., 2012; Moscato et al., 2014; Planaguma et al., 2016). This decrease in synaptic NMDA receptor content is mediated by the antibody disrupting the interaction between NMDA receptors and synaptic anchoring proteins, such as the EphrinB2 receptor, altering NMDA receptor surface diffusion dynamics and triggering NMDA receptor internalization (Hughes et al., 2010; Mikasova et al., 2012; Moscato et al., 2014; Planaguma et al., 2015; Kreye et al., 2016; Planaguma et al., 2016; Ladepeche et al., 2018). Passive transfer of anti-GluN1 autoantibodies onto murine models caused behavioral deficits and memory impairment (Planaguma et al., 2015), and in other instances epilepsy without memory deficits (Taraschenko et al., 2019). Anti-GluN1 antibodies do not appear to cause apoptosis, promote complement deposition, or increase brain lymphocytic infiltrates, suggesting that most of the pathophysiological effects observed stem from antibody-mediated NMDA receptor hypofunction (Planaguma et al., 2015).
Anti-GluN1 autoantibodies are also implicated in spontaneous acute psychosis/schizophrenia cases, which share some clinical features to those found in anti-NMDA receptor encephalitis (Jezequel et al., 2017; Lennox et al., 2017). The antibodies associated with the development of psychosis decrease synaptic NMDA receptor content, disrupt EphrinB2 receptor interactions, and lead to decreases in hippocampal LTP. However, the anti-GluN1 autoantibodies associated with psychosis/schizophrenia do not compete with anti-NMDA receptor encephalitis antibodies, and they do not appear to bind to the same Asn368/Gly369 motif in the NTD (Castillo-Gomez et al., 2017; Jezequel et al., 2017). Interestingly, a few healthy controls in the psychosis study also express anti-GluN1 autoantibodies that decrease synaptic NMDA receptor content, but not to the extent of psychosis patients (Jezequel et al., 2017). As all of the antibodies were isotype-controlled (i.e., all IgG), this would suggest that there may be intrinsic differences in the sample concentration, specific epitope, or avidity of the anti-GluN1 autoantibodies. Indeed, anti-GluN1 autoantibodies isolated from patients with anti-NMDA receptor encephalitis, healthy controls, and schizophrenia all demonstrated the capacity to internalize NMDA receptors and decrease NMDA receptor–mediated currents regardless of epitope-specificity and antibody isotype (Castillo-Gomez et al., 2017). Recent studies have suggested that clonal variations in anti-GluN1 autoantibodies could account for intrinsic differences in avidity for the NMDA receptor (Kreye et al., 2016; Ly et al., 2018). Thus, a similar titer of anti-GluN1 antibodies from one patient that elicits a clinical phenotype may not necessarily evoke a similar response in others, contributing to the variation in clinical presentation.
Anti-NMDA receptor encephalitis patients benefit from immunoglobulin-depleting treatments, including plasmapheresis and intravenous immunoglobulin (Titulaer et al., 2013). Second-line drugs that specifically target B cells also appear to eliminate symptoms for patients that are refractory to steroids and first-line immunosuppressants. Given that NMDA receptor hypofunction is implicated as a mechanistic feature of the disease, the use of positive allosteric modulators (PAMs) has been explored in experimental anti-NMDA receptor encephalitis models with some recovery of synaptic function (Warikoo et al., 2018; Mannara et al., 2020).
3. Anti–NMDA Receptor Autoantibodies in Systemic Lupus Erythematosus
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by multiorgan involvement. Anti-NMDA receptor autoantibodies were first described in CSF samples from a patient with SLE with declining cognitive function (DeGiorgio et al., 2001). This antibody bound to a pentapeptide consensus sequence in the GluN2 subunit called the DWEYS motif (amino acid sequence is DWDYS in GluN2A and EWDYG in GluN2B). This antibody also bound double-stranded DNA; anti–double-stranded DNA antibodies are a hallmark of SLE (Tsokos, 2011). Thus, these antibodies were later designated as “DNRAbs” (DNA and NMDA receptor–reactive antibodies) to distinguish them from other anti-GluN2 autoantibodies in SLE (Husebye et al., 2005; Gono et al., 2011; Chang et al., 2015a; Tay et al., 2017; Nestor et al., 2018). DNRAbs and anti-GluN2 autoantibodies are reactive to the same peptide sequence containing the GluN2 epitope and both promote cell death through enhancing NMDA receptor activity (DeGiorgio et al., 2001; Faust et al., 2010; Gono et al., 2011; Kapadia et al., 2017).
Based on the DWEYS epitope, approximately 30%–40% of patients with SLE are positive for anti-GluN2 autoantibodies (Tay et al., 2017), which may be associated with cognitive dysfunction and diffuse neuropsychiatric symptoms (Husebye et al., 2005; Omdal et al., 2005; Fragoso-Loyo et al., 2008; Yang et al., 2017; Schwarting et al., 2019). Whereas serum samples often show no significant correlation between anti-GluN2 antibodies and neuropsychiatric symptoms in SLE (Hanly et al., 2006; Harrison et al., 2006; Petri et al., 2010), antibody presence in CSF samples correlates with neuropsychiatric symptoms (Arinuma et al., 2008; Fragoso-Loyo et al., 2008; Hirohata et al., 2014; Lauvsnes et al., 2014). This suggests that the blood-brain barrier may be important for determining whether the pathogenic anti-GluN2 autoantibodies can affect the brain. Patients with SLE with neuropsychiatric symptoms benefit from immunoglobulin depletion and B cell–targeting drugs, but the mechanism is unclear since SLE is a multifactorial and complex disease with many other autoantibodies and systemic inflammatory processes occurring beyond anti-GluN2 autoantibodies (Milstone et al., 2005; Lim et al., 2010).
Because of the well defined epitope in the GluN2 subunits (DWEYS), a variety of approaches have been employed to study the mechanism of anti-GluN2 autoantibodies in causing symptoms. The first study of DNRAbs employed passive transfer of human SLE antibodies from CSF into mice and onto primary neuronal cultures. These DNRAbs were isolated from patient CSF using affinity chromatography with a DWEYS-peptide–conjugated column. DNRAbs caused neuronal apoptosis, but neurons were protected when antibodies were applied with the high-affinity NMDA receptor channel blocker MK-801 (DeGiorgio et al., 2001). DNRAbs eluted from postmortem brains of patients with SLE with cognitive impairment also caused neuronal apoptosis in the hippocampal CA1 region (Kowal et al., 2006).
Mouse models that endogenously generate DNRAbs were created by immunizing with the DWEYS peptide and then administering lipopolysaccharide (LPS) to induce systemic inflammation and permeabilize the blood-brain barrier (Kowal et al., 2004; Chang et al., 2015a). Without LPS, mice with circulating DNRAbs (DNRAb+) do not evidence hippocampal cell death. By contrast, after LPS treatment DNRAb+ mice display reduced neuronal numbers in the hippocampal CA1 region along with increased apoptotic cells (Kowal et al., 2006). If epinephrine is used in place of LPS to induce blood-brain barrier breakdown, the amygdala becomes the central target of DNRAbs; it is not clear why epinephrine and LPS differentially localize DNRAbs to different parts of the brain (Huerta et al., 2006). Still, patients with SLE show microstructural defects in the hippocampus with cognitive dysfunction and neuropsychiatric symptoms (Appenzeller et al., 2006; Lauvsnes et al., 2014; Mackay et al., 2019). Hence, LPS treatment after DWEYS immunization may model better the role of DNRAbs in cognitive dysfunction in SLE. The availability of monoclonal DNRAbs developed from patients with SLE has circumvented the limited patient antibody samples, enabling mechanistic studies (Zhang et al., 2009a).
The murine LPS/DNRAb+ models as well as DNRAb monoclonal antibodies have provided insights into the mechanisms and functional effects of DNRAbs. The CSF concentrations of DNRAbs in a cohort of SLE patients with neuropsychiatric dysfunction was approximately 30–180 μg/mL (median: ∼70 μg/mL) (Faust et al., 2010). G11 is a monoclonal antibody derived from a patient with lupus that specifically binds to GluN2A- or GluN2B-containing NMDA receptors. At clinically relevant concentrations, G11 acutely increased NMDA receptor EPSPs and caused NMDA receptor–dependent cell death in the hippocampus, which could be prevented by NMDA receptor antagonists (Faust et al., 2010). Using the LPS/DNRAb+ model, chronic changes in the brain were observed, including decreased hippocampal dendritic complexity, decreased object-place memory discrimination, and hippocampal place field expansion (Chang et al., 2015a). DNRAbs require complement immune response (C1q deposition) to mediate these chronic changes but not for inducing acute neuronal cell death (Nestor et al., 2018). Although G11 binds to both GluN2A and GluN2B subunits, the deficits associated with DNRAbs in the murine model were blocked in GluN2A KO but not GluN2B KO mice, indicating that most of the pathology is associated with the GluN2A subunit (Chan et al., 2020).
F. Perspectives and Directions for Glutamate Receptor–Targeted Drug Development
The past decade saw the FDA approval of three glutamate receptor targeted drugs: perampanel for epilepsy, dextromethorphan for pseudobulbar affect, and esketamine for depression. The success of ketamine and the approval of esketamine in 2019 is noteworthy, since standard-of-care therapeutics that target aminergic signaling are marginally effective and require weeks of treatment to gain benefit (Krystal et al., 2019). In contrast, ketamine and esketamine show efficacy within hours of a single administration (Niciu et al., 2014; Abdallah et al., 2015). For the pharmaceutical industry, the success of ketamine and the approval of esketamine shows that significant breakthroughs in CNS drug discovery remain a realistic goal. In addition, the clinical development of ketamine and esketamine frame several important considerations for successful development of new iGluR therapeutics.
Most notable is the mechanism underlying ketamine’s antidepressant activity that appears to involve a long-term response to a short drug exposure, which may lead to persistent changes in synaptic function that could contribute to the antidepressant response (Krystal et al., 2019). Given the inherent plasticity of glutamatergic signaling, other iGluR-targeted therapeutics might also induce some degree of a change in synaptic function. This may be true for the iGluR modulators that mimic physiologic synaptic plasticity. Thus, managing the brain’s response to an iGluR-targeted drug may be as important as managing the primary pharmacological response. For example, the disappointing results in schizophrenia trials with drugs aimed to enhance NMDA receptor responses by increasing glycine concentration may be due to both the modest effects of increasing glycine over existing levels that support NMDA receptor activation as well as tachyphylaxis (i.e., diminishing response to successive doses of a drug) (Quartermain et al., 1994), which may be caused by glycine-dependent priming of NMDA receptor internalization and subsequent downregulation (Rajani et al., 2020). The advantage of managing such regulatory mechanisms is illustrated by use of intermittent dosing with d-cycloserine to achieve a more robust therapeutic effect compared with chronically administered glycinergic agents (Goff et al., 2008), although the distinct actions of d-cycloserine, for example on GluN2C, cannot be ruled out as a distinguishing feature (Section VII. Pharmacology of Orthosteric Ligands and Channel Blockers). Nevertheless, exploring nontraditional pharmacokinetics, dose levels, and dose intervals may be a key element of successful development of new therapeutics.
Another challenge in the development of iGluR-targeted therapeutics will be in realizing efficacy while balancing side-effect liabilities. Although this is a tenet in all drug discovery, it is a particular challenge for iGluR drug development given that glutamate signaling is the principal functional activity within the CNS. Consequently, although regulating glutamate receptor signaling has strong therapeutic potential, such approaches also bear significant risk of adverse effects. This is well illustrated in the use of ketamine and esketamine, which produces psychotomimetic side effects and cognitive disruption during acute administration prior to the onset of therapeutic benefit as an antidepressant. One path to agents with favorable therapeutic indices may be selective targeting of receptor subtypes that are responsible for efficacy while avoiding more global effects that may result in unwanted side effects mediated by other glutamate receptor subtype. The discovery of negative allosteric modulators of GluN2B-containing NMDA receptors first highlighted the potential utility of subtype-selective drugs. Although the therapeutic benefits and liabilities of the GluN2B NAMs continue to be evaluated, an interesting aspect of the allosteric mechanism of these compounds is that there can be variation in the degree of receptor inhibition and other properties among the GluN2B NAMs [e.g., Wang et al. (2014c), Yuan et al. (2015b)]. This offers additional opportunity for fine tuning of efficacy in affected regions versus side-effect liabilities.
The variable actions of allosteric modulators that bind at a shared site but bring about different actions is an emerging theme in pharmacology. Another new approach is targeting iGluR auxiliary subunits for AMPA and potentially kainate receptors (Rosenbaum et al., 2020). Thus, the mechanistic classes of therapeutically relevant ligands define a broad pharmacology that offers opportunities to select agents for development that balance efficacy with side-effect liabilities. The challenge will be navigating this complex pharmacology in the context of the current drug-development paradigm, which is a race to achieve proof of concept in randomized, double-anonymized, placebo-controlled clinical trials before useful patent life runs out.
A related challenge for the development of new iGluR-targeted therapeutics is the definition of therapeutic indications based on disease classifications that continue to evolve. CNS drug development, approval, and commercialization proceed based on indications largely defined by DSM-V clinical characterization as opposed to the underlying neurobiological deficits or mechanism of drug action (Lilienfeld and Treadway, 2016; Krueger et al., 2018). The conundrum that this situation presents is illustrated by the expanding list of potential clinical utilities of ketamine, which include neuropsychiatric conditions, such as suicidality, addiction, and obsessive-compulsive disorder as well as the neurodevelopmental disorder Rett syndrome. Presumably, the potential efficacy of ketamine in these indications derives from the underlying molecular mechanism for these different conditions. Thus, it is not useful to label ketamine and esketamine as antidepressants, particularly in the case in which such drugs might be applied to nonpsychiatric conditions such as Rett syndrome. However, it is not currently possible to seek approval based on a descriptive label of mechanism that captures all of the therapeutic possibilities. The result is that the development of such agents may be narrowed based on market opportunities that differ across potential therapeutic uses. New paradigms will be needed to capture the full therapeutic utility of iGluR-targeted drugs and to increase the success rate in getting drugs approved without forcing each candidate into a narrowly defined or suboptimal development path.
Lastly, insight into the roles of genetic variation is redefining many neurologic disorders, with significant impact on drug discovery as well as the development of genetic therapies. A number of specific neurologic phenotypes have been connected to functional alterations in properties of iGluRs resulting from missense, nonsense, splice site variation, deletions, and duplications of their respective genes. Efforts to evaluate how human genetic variation in both coding and noncoding regions impact iGluR properties, homeostasis, and brain function have the potential to facilitate the development of diagnostic criteria for CNS disease, improve preclinical testing in animal models, and allow identification of target subpopulations who could benefit from pharmacological and genetic treatments.
XI. Conclusion
The past decade has witnessed an explosive increase in knowledge about the brain, neuronal function, and the molecular organization of the neuron. It is impossible to even begin to capture the volume of new information available that is relevant to glutamate receptors. Despite our best efforts, we have only scratched the surface of the knowledge base, and, because of space limitations, have side-stepped countless complex roles that the glutamate receptors are involved in. Yet, within this volume of new information, several key themes emerge that will drive future research efforts. We have summarized six areas that either did not exist 10 years ago or have been transformed by technical and conceptual advances over the past decade. First, whereas the molecular age in glutamate receptors was initiated over 30 years ago with the first single-channel recordings of glutamate receptors (Nowak et al., 1984; Cull-Candy and Usowicz, 1987; Jahr and Stevens, 1987) and subsequent cloning of a mammalian glutamate receptor gene (Hollmann et al., 1989), and the structural era dawned a decade later (Armstrong et al., 1998), we have now clearly entered the age of structural biology. The number of structures of glutamate receptor subunits and domains available has increased by orders of magnitude, and new approaches, such as cryo–electron microscopy (EM) are poised to enable even more complex structural analysis of proteins. This information has been on one hand enormously satisfying for those who have long labored building hypothetical structural models from indirect experimentation, yet on the other hand, has whet one’s appetite for even more detail. Second, super-resolution imaging techniques, RNA seq, proteomics, and molecular analyses have provided a view into subcellular events that would have been hard to imagine a decade ago. The enumeration of the proteins and molecules involved in neuronal processes has set the stage for connecting these players into a system of organized events. Third, there has been a renaissance in pharmacology and chemical biology. There are a multitude of new tools and probes for scientists to use, and a growing understanding of binding sites on proteins that will fuel structure-based design. The lion’s share of these tools are allosteric modulators, which lend themselves to biased modulation of multiple aspects of ion channel function and clever experimental design. This toolbox enables physiologic experiments and behavioral studies and is the fertile ground from which new therapeutic approaches spring to address unmet clinical needs. Fourth, technology-driven reduction in the cost of whole exome sequencing has produced a tidal wave of genetic data that impacts how we assess the roles of individual amino acids within protein subunits. This has stratified patients across a spectrum of diseases by providing diagnoses for patients, hope for families, and focus for the research communities. Moreover, identification of genes capable of causing neurologic disease can provide insight into these diseases, create new animal models, and catalyze development of both new drugs and genetic approaches. Advances in understanding autoimmune conditions involving these receptors similarly create new opportunities for advances. Fifth, an appreciation of the molecular composition of the receptor, from its auxiliary subunits to its intracellular and transcellular binding partners, has exploded. There are a dizzying array of ways to make a glutamate receptor, and it is clear that cells have a continuum of signaling properties to choose from as they build glutamatergic connections. Sixth, machine learning, artificial intelligence, graphical processing units, and the relentless increase in computational power have created new ways to analyze data and enabled new approaches as well as public databases that catalyze progress. These themes have already transformed how we think and what we do as a research community and will drive activity along many fronts. Moreover, taking stock of these advances, as can and has be done for previous decades (Dingledine et al., 1999; Traynelis et al., 2010), reminds us that the future is bright and that persistent, thoughtful, creative, and collaborative efforts stand an excellent chance of solving the most stubborn and intractable problems.
Acknowledgments
We are grateful to James Allen, Timothy A. Benke, Britton Barbee, Noele Certain, Kelvin Chan, Anis Contractor, Ian D. Coombs, Jan Egebjerg, Mark Farrant, Jesse E. Hanson, Anders S. Kristensen, Gabrielle Moody, Scott J. Myers, Roger Nicoll, Evan Spruston, Nelson Spruston, and Francis C. K. Tan for critical comments and helpful discussion as well as the reviewers for their work and willingness to help improve the manuscript. We also thank Anis Contractor, Toshihior Nomura, Lonnie P. Wollmuth, Derek Bowie, Ryan P.D. Alexander, and Yuhao Yan for sharing unpublished data.
Abbreviations
- ABD
agonist binding domain
- ABHD6
α/β-hydrolase domain–containing protein 6
- AHCP
2-amino-3-(3-hydroxy-7,8-dihydro-6H-cyclohepta[d]isoxazol-4-yl)propionic acid
- AICP
(R)-2-amino-3-(4-(2-ethylphenyl)-1H-indole-2-carboxamido)propanoic acid
- AKAP
A-kinase-anchoring protein
- AMPA
α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate
- AMPAR
AMPA receptor
- AP5
amino-5-phosphonovaleric acid
- ATPA
2-amino-3-(3-hydroxy-5-tert-butylisoxazol-4-yl)propanoic acid
- 4-BCCA
4-butylcyclohexane carboxylic acid
- BDNF
brain-derived neurotrophic factor
- CaMKII
Ca2+/calmodulin-dependent protein kinase II
- CGE
caudal ganglionic eminence
- Cbln
cerebellin
- 7-CKA
7-chlorokynurenic acid
- CKAMP
cysteine‐knot AMPA receptor modulating protein
- CNIH
protein cornichon homolog
- CNS
central nervous system
- ConA
Concanavalin A
- COPII
coat protein complex II
- CSF
cerebrospinal fluid
- CTD
C-terminal domain
- CUB
bone morphogenetic protein
- DAPK1
death-associated protein kinase 1
- DCKA
5,7-dichlorokynurenic acid
- DNRAb
DNA and NMDA receptor–reactive antibody
- DQP
dihydroquinolone-pyrazoline
- DRG
dorsal root ganglion
- EM
electron microscopy
- EPSC
excitatory postsynaptic current
- EPSP
excitatory postsynaptic potential
- ER
endoplasmic reticulum
- FDA
Food and Drug Administration
- fEPSP
field EPSP
- FRET
fluorescence resonance energy transfer
- Frrs1l
ferric chelate reductase 1-like
- GRIP
glutamate receptor-interacting protein
- GSG1L
germ cell–specific gene 1-like
- HEK
human embryonic kidney
- iGluR
ionotropic glutamate receptor
- KB
equilibrium dissociation constant from functional assays
- Ki
equilibrium dissociation constant from radioligand binding studies
- KO
knockout
- LDLa
low-density lipoprotein class A
- LPS
lipopolysaccharide
- LTD
long-term depression
- LTP
long-term potentiation
- MAP
microtubule associated protein
- mEPSC
miniature EPSC
- MGE
medial ganglionic eminence
- mGluR
metabotropic glutamate receptor
- NAM
negative allosteric modulation
- NMDA
N-methyl-d-aspartate
- MAPK
mitogen-activated protein kinase
- NMDAR
NMDA receptor
- Nrxn
neurexin
- NTD
N-terminal domain
- PAM
positive allosteric modulator
- PDB
Protein Data Bank
- PEPA
4-[2-(phenylsulfonylamino)ethylthio]-2,6-difluoro-phenoxyacetamide
- PhTx
philanthotoxin
- PKA
protein kinase A
- PKC
protein kinase C
- PKG
cGMP-dependent protein kinase II
- PP1
protein phosphatase 1
- PPDA
(2S*,3R*)-1-(phenanthrene-2-carbonyl)piperazine-2,3-dicarboxylic acid
- Prrt1
proline rich transmembrane protein 1
- PS
pregnenolone sulfate
- PSD
postsynaptic density
- QNZ
quinazolin-4-one
- RVIS
residual variation intolerance score
- 24(S)-hydroxycholesterol
24(S)-HC
- SLE
systemic lupus erythematosus
- SOL-1
suppressor of lurcher protein 1
- SynDIG4
synapse differentiation-induced gene 4
- TARP
transmembrane AMPA receptor regulatory protein
- TM
transmembrane
- TMD
transmembrane domain
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Hansen, Wollmuth, Bowie, Furukawa, Menniti, Sobolevsky, Swanson, Swanger, Greger, Nakagawa, McBain, Jayaraman, Low, Dell’Acqua, Diamond, Camp, Perszyk, Yuan, Traynelis.
Footnotes
K.B.H. was supported by National Institutes of Health National Institute of Neurological Disorders and Stroke [Grants NS097536 and NS116055] and National Institute of General Medical Sciences [Grant GM103546]; L.P.W. was supported by National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS088479]; A.I.S was supported by National Institutes of Health National Institute of Neurological Disorders and Stroke [Grants NS083660 and NS107253] and National Cancer Institute [Grant CA206573] and the National Science Foundation [1818086]; I.H.G. was supported by UK Research and Innovation [MC_U105174197] and the Biotechnology and Biological Sciences Research Council [BB/N002113/1]; H.F. was supported by the National Institutes of Health National Institutes of Health National Institute of Neurological Disorders and Stroke [Grants NS111745 and NS113632] and National Institute of Mental Health [Grant MH085926], Robertson funds at Cold Spring Harbor Laboratory, Doug Fox Alzheimer’s Fund, Austin’s Purpose, Heartfelt Wing Alzheimer’s Fund, and the Gertrude and Louis Feil Family Trust; C-M.L. was supported by the National University of Singapore [R184000261101]; T.N. was supported by National Institutes of Health National Institute of Mental Health [Grant MH123474]; C.J.M. was supported by the Intramural Research Program of the National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development; J.S.D. was supported by the Intramural Research Program of the National Institutes of Health National Institute of Neurologic Disorders and Stroke; D.B. was supported by operating grants from the Canadian Institutes of Health Research [FRN 136832, FRN 142431, FRN 162317]; M.L.D. was supported by the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS040701]; G.T.S. was supported by National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS105502]; V.J. was supported by National Institutes of Health National Institute of General Medical Sciences [Grant GM122528]; S.A.S. was supported by the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS105804]; H.Y. was supported by National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development [Grant HD082373]; C.C. was supported by National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS113530]; and S.F.T. was supported by the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS111619].
K.B.H is principal investigator on a research grant from Janssen Research and Development to the University of Montana. F.S.M is co-inventor on Pfizer- and Novartis-owned Intellectual Property that includes allosteric NMDA receptor modulators; is Chief Scientific Officer of MindImmune Therapeutics, Inc.; and is a consultant to Gilgamesh Therapeutics, Inc. H.F. was a principal investigator on a research grant from Allergan. H.Y. is principal investigator on a research grant from Sage Therapeutics to Emory University School of Medicine. S.F.T. is principal investigator on research grants from Biogen and Janssen to Emory; is a member of the Scientific Advisory Board for Eumentis Inc, Sage Therapeutics, the GRIN2B Foundation, the CureGRIN Foundation; is cofounder of NeurOp Inc and Agrithera Inc.; has received licensing fees and royalties from Emory; and is co-inventor on Emory-owned Intellectual Property that includes allosteric modulators of NMDA receptor function. All other authors have no conflicts of interest to declare.
 This article has supplemental material available at pharmrev.aspetjournals.org.
This article has supplemental material available at pharmrev.aspetjournals.org.
References
- Aamodt SM, Shi J, Colonnese MT, Veras W, Constantine-Paton M (2000) Chronic NMDA exposure accelerates development of GABAergic inhibition in the superior colliculus. J Neurophysiol 83:1580–1591. [DOI] [PubMed] [Google Scholar]
- aan het Rot M, Collins KA, Murrough JW, Perez AM, Reich DL, Charney DS, Mathew SJ (2010) Safety and efficacy of repeated-dose intravenous ketamine for treatment-resistant depression. Biol Psychiatry 67:139–145. [DOI] [PubMed] [Google Scholar]
- Abbott LF, Nelson SB (2000) Synaptic plasticity: taming the beast. Nat Neurosci 3 (Suppl):1178–1183. [DOI] [PubMed] [Google Scholar]
- Abdallah CG (2020) (2R,6R)-Hydroxynorketamine (HNK) plasma level predicts poor antidepressant response: is this the end of the HNK pipeline? Neuropsychopharmacology 45:1245–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah CG, Sanacora G, Duman RS, Krystal JH (2015) Ketamine and rapid-acting antidepressants: a window into a new neurobiology for mood disorder therapeutics. Annu Rev Med 66:509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abele R, Keinanen K, Madden DR (2000) Agonist-induced isomerization in a glutamate receptor ligand-binding domain. A kinetic and mutagenetic analysis. J Biol Chem 275:21355–21363. [DOI] [PubMed] [Google Scholar]
- Abraham WC (2008) Metaplasticity: tuning synapses and networks for plasticity. Nat Rev Neurosci 9:387. [DOI] [PubMed] [Google Scholar]
- Abraham WC, Bear MF (1996) Metaplasticity: the plasticity of synaptic plasticity. Trends Neurosci 19:126–130. [DOI] [PubMed] [Google Scholar]
- Abrahamsson TChou CYCLi SYMancino ACosta RPBrock JANuro EBuchanan KAElgar DBlackman AV, et al. (2017) Differential regulation of evoked and spontaneous release by presynaptic NMDA receptors. Neuron 96:839–855.e5. [DOI] [PubMed] [Google Scholar]
- Acker TM, Khatri A, Vance KM, Slabber C, Bacsa J, Snyder JP, Traynelis SF, Liotta DC (2013) Structure-activity relationships and pharmacophore model of a noncompetitive pyrazoline containing class of GluN2C/GluN2D selective antagonists. J Med Chem 56:6434–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acker TMYuan HHansen KBVance KMOgden KKJensen HSBurger PBMullasseril PSnyder JPLiotta DC, et al. (2011) Mechanism for noncompetitive inhibition by novel GluN2C/D N-methyl-D-aspartate receptor subunit-selective modulators. Mol Pharmacol 80:782–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addis L, Virdee JK, Vidler LR, Collier DA, Pal DK, Ursu D (2017) Epilepsy-associated GRIN2A mutations reduce NMDA receptor trafficking and agonist potency - molecular profiling and functional rescue. Sci Rep 7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addy CAssaid CHreniuk DStroh MXu YHerring WJEllenbogen AJinnah HAKirby LLeibowitz MT, et al. (2009) Single-dose administration of MK-0657, an NR2B-selective NMDA antagonist, does not result in clinically meaningful improvement in motor function in patients with moderate Parkinson’s disease. J Clin Pharmacol 49:856–864. [DOI] [PubMed] [Google Scholar]
- Adesnik H, Li G, During MJ, Pleasure SJ, Nicoll RA (2008) NMDA receptors inhibit synapse unsilencing during brain development. Proc Natl Acad Sci USA 105:5597–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adesnik H, Nicoll RA (2007) Conservation of glutamate receptor 2-containing AMPA receptors during long-term potentiation. J Neurosci 27:4598–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adla SKSlavikova BSmidkova MTloustova ESvoboda MVyklicky VKrausova BHubalkova PNekardova MHolubova K, et al. (2017) Physicochemical and biological properties of novel amide-based steroidal inhibitors of NMDA receptors. Steroids 117:52–61. [DOI] [PubMed] [Google Scholar]
- Ahmed AH, Loh AP, Jane DE, Oswald RE (2007) Dynamics of the S1S2 glutamate binding domain of GluR2 measured using 19F NMR spectroscopy. J Biol Chem 282:12773–12784. [DOI] [PubMed] [Google Scholar]
- Ahmed AH, Oswald RE (2010) Piracetam defines a new binding site for allosteric modulators of alpha-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA) receptors. J Med Chem 53:2197–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AH, Ptak CP, Fenwick MK, Hsieh CL, Weiland GA, Oswald RE (2013) Dynamics of cleft closure of the GluA2 ligand-binding domain in the presence of full and partial agonists revealed by hydrogen-deuterium exchange. J Biol Chem 288:27658–27666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AH, Ptak CP, Oswald RE (2010) Molecular mechanism of flop selectivity and subsite recognition for an AMPA receptor allosteric modulator: structures of GluA2 and GluA3 in complexes with PEPA. Biochemistry 49:2843–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AH, Thompson MD, Fenwick MK, Romero B, Loh AP, Jane DE, Sondermann H, Oswald RE (2009a) Mechanisms of antagonism of the GluR2 AMPA receptor: structure and dynamics of the complex of two willardiine antagonists with the glutamate binding domain. Biochemistry 48:3894–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AH, Wang S, Chuang HH, Oswald RE (2011a) Mechanism of AMPA receptor activation by partial agonists: disulfide trapping of closed lobe conformations. J Biol Chem 286:35257–35266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AH, Wang Q, Sondermann H, Oswald RE (2009b) Structure of the S1S2 glutamate binding domain of GLuR3. Proteins 75:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed I, Bose SK, Pavese N, Ramlackhansingh A, Turkheimer F, Hotton G, Hammers A, Brooks DJ (2011b) Glutamate NMDA receptor dysregulation in Parkinson’s disease with dyskinesias. Brain 134:979–986. [DOI] [PubMed] [Google Scholar]
- Ahmed MS, Siegelbaum SA (2009) Recruitment of N-Type Ca(2+) channels during LTP enhances low release efficacy of hippocampal CA1 perforant path synapses. Neuron 63:372–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyer R, Mehta N, Gungor S, Gulati A (2018) A systematic review of NMDA receptor antagonists for treatment of neuropathic pain in clinical practice. Clin J Pain 34:450–467. [DOI] [PubMed] [Google Scholar]
- Aizenman CD, Cline HT (2007) Enhanced visual activity in vivo forms nascent synapses in the developing retinotectal projection. J Neurophysiol 97:2949–2957. [DOI] [PubMed] [Google Scholar]
- Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N (1994) Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J Comp Neurol 347:150–160. [DOI] [PubMed] [Google Scholar]
- Akesson E, Kjaeldgaard A, Samuelsson EB, Seiger A, Sundström E (2000) Ionotropic glutamate receptor expression in human spinal cord during first trimester development. Brain Res Dev Brain Res 119:55–63. [DOI] [PubMed] [Google Scholar]
- Akgül G, Abebe D, Yuan XQ, Auville K, McBain CJ (2019) The role of AMPARs in the maturation and integration of caudal ganglionic eminence-derived interneurons into developing hippocampal microcircuits. Sci Rep 9:5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akgül G, McBain CJ (2016) Diverse roles for ionotropic glutamate receptors on inhibitory interneurons in developing and adult brain. J Physiol 594:5471–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akgül G, McBain CJ (2020) AMPA receptor deletion in developing MGE-derived hippocampal interneurons causes a redistribution of excitatory synapses and attenuates postnatal network oscillatory activity. Sci Rep 10:1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hallaq RA, Conrads TP, Veenstra TD, Wenthold RJ (2007) NMDA di-heteromeric receptor populations and associated proteins in rat hippocampus. J Neurosci 27:8334–8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberstein R, Grey R, Zimmet A, Simmons DK, Mayer ML (2015) Glycine activated ion channel subunits encoded by ctenophore glutamate receptor genes. Proc Natl Acad Sci USA 112:E6048–E6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RPD, Mitry J, Sareen V, Khadra A, Bowie D (2019) Cerebellar stellate cell excitability is coordinated by shifts in the gating behavior of voltage-gated Na+ and A-type K+ channels. eNeuro 6:ENEURO.0126-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfieri ADoccula FGPederzoli RGrenzi MBonza MCLuoni LCandeo ARomano Armada NBarbiroli AValentini G, et al. (2020) The structural bases for agonist diversity in an Arabidopsis thaliana glutamate receptor-like channel. Proc Natl Acad Sci USA 117:752–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Zulfiqar S, Klar J, Wikström J, Ullah F, Khan A, Abdullah U, Baig S, Dahl N (2017) Homozygous GRID2 missense mutation predicts a shift in the D-serine binding domain of GluD2 in a case with generalized brain atrophy and unusual clinical features. BMC Med Genet 18:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkondon M, Pereira EF, Albuquerque EX (2003) NMDA and AMPA receptors contribute to the nicotinic cholinergic excitation of CA1 interneurons in the rat hippocampus. J Neurophysiol 90:1613–1625. [DOI] [PubMed] [Google Scholar]
- Alsaad HA, DeKorver NW, Mao Z, Dravid SM, Arikkath J, Monaghan DT (2019) In the telencephalon, GluN2C NMDA receptor subunit mRNA is predominately expressed in glial cells and GluN2D mRNA in interneurons. Neurochem Res 44:61–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaloum M, Kazi R, Gan Q, Amin J, Wollmuth LP (2016) A molecular determinant of subtype-specific desensitization in ionotropic glutamate receptors. J Neurosci 36:2617–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlShimemeri S, Fox SH, Visanji NP (2020) Emerging drugs for the treatment of L-DOPA-induced dyskinesia: an update. Expert Opin Emerg Drugs 25:131–144. [DOI] [PubMed] [Google Scholar]
- Alt A, Weiss B, Ogden AM, Knauss JL, Oler J, Ho K, Large TH, Bleakman D (2004) Pharmacological characterization of glutamatergic agonists and antagonists at recombinant human homomeric and heteromeric kainate receptors in vitro. Neuropharmacology 46:793–806. [DOI] [PubMed] [Google Scholar]
- Alushin GM, Jane D, Mayer ML (2011) Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists. Neuropharmacology 60:126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez VA, Ridenour DA, Sabatini BL (2007) Distinct structural and ionotropic roles of NMDA receptors in controlling spine and synapse stability. J Neurosci 27:7365–7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador ABostick CDOlson HPeters JCamp CRKrizay DChen WHan WTang WKanber A, et al. (2020) Modelling and treating GRIN2A developmental and epileptic encephalopathy in mice. Brain 143:2039–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman TK, Maki BA, Ruffino TJ, Kasperek EM, Popescu GK (2014) Separate intramolecular targets for protein kinase A control N-methyl-D-aspartate receptor gating and Ca2+ permeability. J Biol Chem 289:18805–18817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrogini P, Minelli A, Lattanzi D, Ciuffoli S, Fanelli M, Cuppini R (2006) Synaptically-silent immature neurons show gaba and glutamate receptor-mediated currents in adult rat dentate gyrus. Arch Ital Biol 144:115–126. [PubMed] [Google Scholar]
- Amico-Ruvio SA, Murthy SE, Smith TP, Popescu GK (2011) Zinc effects on NMDA receptor gating kinetics. Biophys J 100:1910–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amico-Ruvio SA, Paganelli MA, Myers JM, Popescu GK (2012) Ifenprodil effects on GluN2B-containing glutamate receptors. Mol Pharmacol 82:1074–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amico-Ruvio SA, Popescu GK (2010) Stationary gating of GluN1/GluN2B receptors in intact membrane patches. Biophys J 98:1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin JB, Gochman A, He M, Certain N, Wollmuth LP (2021a) NMDA receptors require multiple pre-opening gating steps for efficient synaptic activity. Neuron 109:488–501.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin JB, Leng X, Gochman A, Zhou HX, Wollmuth LP (2018) A conserved glycine harboring disease-associated mutations permits NMDA receptor slow deactivation and high Ca2+ permeability. Nat Commun 9:3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin JB, Moody GR, Wollmuth LP (2021b) From bedside-to-bench: What disease-associated variants are teaching us about the NMDA receptor. J Physiol 599:397–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin JB, Salussolia CL, Chan K, Regan MC, Dai J, Zhou HX, Furukawa H, Bowen ME, Wollmuth LP (2017) Divergent roles of a peripheral transmembrane segment in AMPA and NMDA receptors. J Gen Physiol 149:661–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andaloro VJ, Monaghan DT, Rosenquist TH (1998) Dextromethorphan and other N-methyl-D-aspartate receptor antagonists are teratogenic in the avian embryo model. Pediatr Res 43:1–7. [DOI] [PubMed] [Google Scholar]
- Andersen TF, Tikhonov DB, Bølcho U, Bolshakov K, Nelson JK, Pluteanu F, Mellor IR, Egebjerg J, Strømgaard K (2006) Uncompetitive antagonism of AMPA receptors: Mechanistic insights from studies of polyamine toxin derivatives. J Med Chem 49:5414–5423. [DOI] [PubMed] [Google Scholar]
- Anderson CT, Radford RJ, Zastrow ML, Zhang DY, Apfel UP, Lippard SJ, Tzounopoulos T (2015) Modulation of extrasynaptic NMDA receptors by synaptic and tonic zinc. Proc Natl Acad Sci USA 112:E2705–E2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson O, Stenqvist A, Attersand A, von Euler G (2001) Nucleotide sequence, genomic organization, and chromosomal localization of genes encoding the human NMDA receptor subunits NR3A and NR3B. Genomics 78:178–184. [DOI] [PubMed] [Google Scholar]
- Andrade-Talavera Y, Duque-Feria P, Sihra TS, Rodríguez-Moreno A (2013) Pre-synaptic kainate receptor-mediated facilitation of glutamate release involves PKA and Ca(2+) -calmodulin at thalamocortical synapses. J Neurochem 126:565–578. [DOI] [PubMed] [Google Scholar]
- Andreae LC, Burrone J (2015) Spontaneous neurotransmitter release shapes dendritic arbors via long-range activation of NMDA receptors. Cell Rep 10:873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggono V, Huganir RL (2012) Regulation of AMPA receptor trafficking and synaptic plasticity. Curr Opin Neurobiol 22:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo MC, Kozlov AS, Charpak S, Audinat E (2004) Glutamate released from glial cells synchronizes neuronal activity in the hippocampus. J Neurosci 24:6920–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes FM, Rubio ME, Kandler K (2020) Role of GluA3 AMPA receptor subunits in the presynaptic and postsynaptic maturation of synaptic transmission and plasticity of endbulb-bushy cell synapses in the cochlear nucleus. J Neurosci 40:2471–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki C, Rhee J, Lubin M, Dawson TM (1997) NMDA-R1 subunit of the cerebral cortex co-localizes with neuronal nitric oxide synthase at pre- and postsynaptic sites and in spines. Brain Res 750:25–40. [DOI] [PubMed] [Google Scholar]
- Aoki C, Venkatesan C, Go CG, Mong JA, Dawson TM (1994) Cellular and subcellular localization of NMDA-R1 subunit immunoreactivity in the visual cortex of adult and neonatal rats. J Neurosci 14:5202–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aow J, Dore K, Malinow R (2015) Conformational signaling required for synaptic plasticity by the NMDA receptor complex. Proc Natl Acad Sci USA 112:14711–14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller S, Carnevalle AD, Li LM, Costallat LT, Cendes F (2006) Hippocampal atrophy in systemic lupus erythematosus. Ann Rheum Dis 65:1585–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai A, Kessler M, Xiao P, Ambros-Ingerson J, Rogers G, Lynch G (1994) A centrally active drug that modulates AMPA receptor gated currents. Brain Res 638:343–346. [DOI] [PubMed] [Google Scholar]
- Arai A, Lynch G (1992) Factors regulating the magnitude of long-term potentiation induced by theta pattern stimulation. Brain Res 598:173–184. [DOI] [PubMed] [Google Scholar]
- Arai AC, Kessler M (2007) Pharmacology of ampakine modulators: from AMPA receptors to synapses and behavior. Curr Drug Targets 8:583–602. [DOI] [PubMed] [Google Scholar]
- Arai AC, Kessler M, Rogers G, Lynch G (2000) Effects of the potent ampakine CX614 on hippocampal and recombinant AMPA receptors: interactions with cyclothiazide and GYKI 52466. Mol Pharmacol 58:802–813. [DOI] [PubMed] [Google Scholar]
- Arai AC, Xia YF, Rogers G, Lynch G, Kessler M (2002) Benzamide-type AMPA receptor modulators form two subfamilies with distinct modes of action. J Pharmacol Exp Ther 303:1075–1085. [DOI] [PubMed] [Google Scholar]
- Araki K, Meguro H, Kushiya E, Takayama C, Inoue Y, Mishina M (1993) Selective expression of the glutamate receptor channel delta 2 subunit in cerebellar Purkinje cells. Biochem Biophys Res Commun 197:1267–1276. [DOI] [PubMed] [Google Scholar]
- Arinaminpathy Y, Sansom MS, Biggin PC (2002) Molecular dynamics simulations of the ligand-binding domain of the ionotropic glutamate receptor GluR2. Biophys J 82:676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arinaminpathy Y, Sansom MS, Biggin PC (2006) Binding site flexibility: molecular simulation of partial and full agonists within a glutamate receptor. Mol Pharmacol 69:11–18. [DOI] [PubMed] [Google Scholar]
- Arinuma Y, Yanagida T, Hirohata S (2008) Association of cerebrospinal fluid anti-NR2 glutamate receptor antibodies with diffuse neuropsychiatric systemic lupus erythematosus. Arthritis Rheum 58:1130–1135. [DOI] [PubMed] [Google Scholar]
- Armand EJ, Li J, Xie F, Luo C, Mukamel EA (2021) Single-cell sequencing of brain cell transcriptomes and epigenomes. Neuron 109:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E (2000) Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28:165–181. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E (2006) Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell 127:85–97. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Mayer M, Gouaux E (2003) Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic adjustment of agonist-induced conformational changes. Proc Natl Acad Sci USA 100:5736–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Sun Y, Chen GQ, Gouaux E (1998) Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature 395:913–917. [DOI] [PubMed] [Google Scholar]
- Armstrong SP, Banks PJ, McKitrick TJ, Geldart CH, Edge CJ, Babla R, Simillis C, Franks NP, Dickinson R (2012) Identification of two mutations (F758W and F758Y) in the N-methyl-D-aspartate receptor glycine-binding site that selectively prevent competitive inhibition by xenon without affecting glycine binding. Anesthesiology 117:38–47. [DOI] [PubMed] [Google Scholar]
- Arning L, Kraus PH, Valentin S, Saft C, Andrich J, Epplen JT (2005) NR2A and NR2B receptor gene variations modify age at onset in Huntington disease. Neurogenetics 6:25–28. [DOI] [PubMed] [Google Scholar]
- Arnth-Jensen N, Jabaudon D, Scanziani M (2002) Cooperation between independent hippocampal synapses is controlled by glutamate uptake. Nat Neurosci 5:325–331. [DOI] [PubMed] [Google Scholar]
- Aroniadou-Anderjaska V, Pidoplichko VI, Figueiredo TH, Almeida-Suhett CP, Prager EM, Braga MF (2012) Presynaptic facilitation of glutamate release in the basolateral amygdala: a mechanism for the anxiogenic and seizurogenic function of GluK1 receptors. Neuroscience 221:157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvola M, Keinänen K (1996) Characterization of the ligand-binding domains of glutamate receptor (GluR)-B and GluR-D subunits expressed in Escherichia coli as periplasmic proteins. J Biol Chem 271:15527–15532. [DOI] [PubMed] [Google Scholar]
- Ascher P, Nowak L (1988) The role of divalent cations in the N-methyl-D-aspartate responses of mouse central neurones in culture. J Physiol 399:247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby MC, Maier SR, Nishimune A, Henley JM (2006) Lateral diffusion drives constitutive exchange of AMPA receptors at dendritic spines and is regulated by spine morphology. J Neurosci 26:7046–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf ZLarsen APVenskutonytė RHan LAbrahamsen BNielsen BGajhede MKastrup JSJensen AAPickering DS, et al. (2013) Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters. J Med Chem 56:1614–1628. [DOI] [PubMed] [Google Scholar]
- Asztely F, Erdemli G, Kullmann DM (1997) Extrasynaptic glutamate spillover in the hippocampus: dependence on temperature and the role of active glutamate uptake. Neuron 18:281–293. [DOI] [PubMed] [Google Scholar]
- Ataman ZA, Gakhar L, Sorensen BR, Hell JW, Shea MA (2007) The NMDA receptor NR1 C1 region bound to calmodulin: structural insights into functional differences between homologous domains. Structure 15:1603–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlason PT, Garside ML, Meddows E, Whiting P, McIlhinney RA (2007) N-Methyl-D-aspartate (NMDA) receptor subunit NR1 forms the substrate for oligomeric assembly of the NMDA receptor. J Biol Chem 282:25299–25307. [DOI] [PubMed] [Google Scholar]
- Auberson YP, Acklin P, Bischoff S, Moretti R, Ofner S, Schmutz M, Veenstra SJ (1999) N-phosphonoalkyl-5-aminomethylquinoxaline-2,3-diones: in vivo active AMPA and NMDA(glycine) antagonists. Bioorg Med Chem Lett 9:249–254. [DOI] [PubMed] [Google Scholar]
- Auberson YP, Allgeier H, Bischoff S, Lingenhoehl K, Moretti R, Schmutz M (2002) 5-Phosphonomethylquinoxalinediones as competitive NMDA receptor antagonists with a preference for the human 1A/2A, rather than 1A/2B receptor composition. Bioorg Med Chem Lett 12:1099–1102. [DOI] [PubMed] [Google Scholar]
- Auerbach A, Zhou Y (2005) Gating reaction mechanisms for NMDA receptor channels. J Neurosci 25:7914–7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin KWilliams SCunningham MDevlin AMFriedrich MJayasekera AHussain MAHolliman DMitchell PJenkins A, et al. (2018) Perampanel and decanoic acid show synergistic action against AMPA receptors and seizures. Epilepsia 59:e172–e178. [DOI] [PubMed] [Google Scholar]
- Avenet P, Léonardon J, Besnard F, Graham D, Depoortere H, Scatton B (1997) Antagonist properties of eliprodil and other NMDA receptor antagonists at rat NR1A/NR2A and NR1A/NR2B receptors expressed in Xenopus oocytes. Neurosci Lett 223:133–136. [DOI] [PubMed] [Google Scholar]
- Awobuluyi M, Yang J, Ye Y, Chatterton JE, Godzik A, Lipton SA, Zhang D (2007) Subunit-specific roles of glycine-binding domains in activation of NR1/NR3 N-methyl-D-aspartate receptors. Mol Pharmacol 71:112–122. [DOI] [PubMed] [Google Scholar]
- Ayalon G, Segev E, Elgavish S, Stern-Bach Y (2005) Two regions in the N-terminal domain of ionotropic glutamate receptor 3 form the subunit oligomerization interfaces that control subtype-specific receptor assembly. J Biol Chem 280:15053–15060. [DOI] [PubMed] [Google Scholar]
- Ayalon G, Stern-Bach Y (2001) Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron 31:103–113. [DOI] [PubMed] [Google Scholar]
- Azumaya CM, Days EL, Vinson PN, Stauffer S, Sulikowski G, Weaver CD, Nakagawa T (2017) Screening for AMPA receptor auxiliary subunit specific modulators. PLoS One 12:e0174742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiec WE, Guglietta R, Jami SA, Morishita W, Malenka RC, O’Dell TJ (2014) Ionotropic NMDA receptor signaling is required for the induction of long-term depression in the mouse hippocampal CA1 region. J Neurosci 34:5285–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiec WE, Jami SA, Guglietta R, Chen PB, O’Dell TJ (2017) Differential regulation of NMDA receptor-mediated transmission by SK channels underlies dorsal-ventral differences in dynamics of Schaffer collateral synaptic function. J Neurosci 37:1950–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae MRoh JDKim YKim SSHan HMYang EKang HLee SKim JYKang R, et al. (2021) SLC6A20 transporter: a novel regulator of brain glycine homeostasis and NMDAR function. EMBO Mol Med 13:e12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn S, Volk B, Wisden W (1994) Kainate receptor gene expression in the developing rat brain. J Neurosci 14:5525–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähring R, Bowie D, Benveniste M, Mayer ML (1997) Permeation and block of rat GluR6 glutamate receptor channels by internal and external polyamines. J Physiol 502:575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähring R, Mayer ML (1998) An analysis of philanthotoxin block for recombinant rat GluR6(Q) glutamate receptor channels. J Physiol 509:635–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balannik V, Menniti FS, Paternain AV, Lerma J, Stern-Bach Y (2005) Molecular mechanism of AMPA receptor noncompetitive antagonism. Neuron 48:279–288. [DOI] [PubMed] [Google Scholar]
- Balasuriya D, Takahashi H, Srivats S, Edwardson JM (2014) Activation-induced structural change in the GluN1/GluN3A excitatory glycine receptor. Biochem Biophys Res Commun 450:1452–1457. [DOI] [PubMed] [Google Scholar]
- Balciuniene JFeng NIyadurai KHirsch BCharnas LBill BREasterday MCStaaf JOseth LCzapansky-Beilman D, et al. (2007) Recurrent 10q22-q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am J Hum Genet 80:938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrino M, Somjen GG (1988) Concentration of carbon dioxide, interstitial pH and synaptic transmission in hippocampal formation of the rat. J Physiol 396:247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balik A, Penn AC, Nemoda Z, Greger IH (2013) Activity-regulated RNA editing in select neuronal subfields in hippocampus. Nucleic Acids Res 41:1124–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball SM, Atlason PT, Shittu-Balogun OO, Molnár E (2010) Assembly and intracellular distribution of kainate receptors is determined by RNA editing and subunit composition. J Neurochem 114:1805–1818. [DOI] [PubMed] [Google Scholar]
- Balmer TS, Trussell LO (2019) Selective targeting of unipolar brush cell subtypes by cerebellar mossy fibers. eLife 8:e44964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu DT, Coyle JT (2015) The NMDA receptor ‘glycine modulatory site’ in schizophrenia: D-serine, glycine, and beyond. Curr Opin Pharmacol 20:109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, González-Rueda A, Sampaio-Baptista C, Paulsen O, Rodríguez-Moreno A (2014) Distinct mechanisms of spike timing-dependent LTD at vertical and horizontal inputs onto L2/3 pyramidal neurons in mouse barrel cortex. Physiol Rep 2:e00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P, Donello JE, Hare B, Duman RS (2020) Rapastinel, an NMDAR positive modulator, produces distinct behavioral, sleep, and EEG profiles compared with ketamine. Behav Brain Res 391:112706. [DOI] [PubMed] [Google Scholar]
- Banke TG, Bowie D, Lee H, Huganir RL, Schousboe A, Traynelis SF (2000) Control of GluR1 AMPA receptor function by cAMP-dependent protein kinase. J Neurosci 20:89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banke TG, Dravid SM, Traynelis SF (2005) Protons trap NR1/NR2B NMDA receptors in a nonconducting state. J Neurosci 25:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banke TG, Greenwood JR, Christensen JK, Liljefors T, Traynelis SF, Schousboe A, Pickering DS (2001) Identification of amino acid residues in GluR1 responsible for ligand binding and desensitization. J Neurosci 21:3052–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banke TG, Traynelis SF (2003) Activation of NR1/NR2B NMDA receptors. Nat Neurosci 6:144–152. [DOI] [PubMed] [Google Scholar]
- Banks P, Franks NP, Dickinson R (2010) Competitive inhibition at the glycine site of the N-methyl-D-aspartate receptor mediates xenon neuroprotection against hypoxia-ischemia. Anesthesiology 112:614–622. [DOI] [PubMed] [Google Scholar]
- Bannerman DMNiewoehner BLyon LRomberg CSchmitt WBTaylor ASanderson DJCottam JSprengel RSeeburg PH, et al. (2008) NMDA receptor subunit NR2A is required for rapidly acquired spatial working memory but not incremental spatial reference memory. J Neurosci 28:3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister NJ, Benke TA, Mellor J, Scott H, Gürdal E, Crabtree JW, Isaac JT (2005) Developmental changes in AMPA and kainate receptor-mediated quantal transmission at thalamocortical synapses in the barrel cortex. J Neurosci 25:5259–5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovic J, Chebli M, Salazar H, Carbone AL, Faelber K, Lau AY, Daumke O, Plested AJ (2016) Dynamics of the ligand binding domain layer during AMPA receptor activation. Biophys J 110:896–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovic J, Plested AJ (2018) Auxiliary subunits keep AMPA receptors compact during activation and desensitization. eLife 7:e40548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis A, Sachidhanandam S, Mulle C (2008) GluR6/KA2 kainate receptors mediate slow-deactivating currents. J Neurosci 28:6402–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbon A, Barlati S (2000) Genomic organization, proposed alternative splicing mechanisms, and RNA editing structure of GRIK1. Cytogenet Cell Genet 88:236–239. [DOI] [PubMed] [Google Scholar]
- Barbon A, Vallini I, Barlati S (2001) Genomic organization of the human GRIK2 gene and evidence for multiple splicing variants. Gene 274:187–197. [DOI] [PubMed] [Google Scholar]
- Barbour B (2001) An evaluation of synapse independence. J Neurosci 21:7969–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardoni R, Ghirri A, Zonta M, Betelli C, Vitale G, Ruggieri V, Sandrini M, Carmignoto G (2010) Glutamate-mediated astrocyte-to-neuron signalling in the rat dorsal horn. J Physiol 588:831–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardoni R, Torsney C, Tong CK, Prandini M, MacDermott AB (2004) Presynaptic NMDA receptors modulate glutamate release from primary sensory neurons in rat spinal cord dorsal horn. J Neurosci 24:2774–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barria A, Malinow R (2002) Subunit-specific NMDA receptor trafficking to synapses. Neuron 35:345–353. [DOI] [PubMed] [Google Scholar]
- Barria A, Malinow R (2005) NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron 48:289–301. [DOI] [PubMed] [Google Scholar]
- Barth AL, Schneider JS, Johnston TH, Hill MP, Brotchie JM, Moskal JR, Cearley CN (2020) NYX-458 improves cognitive performance in a primate Parkinson’s disease model. Mov Disord 35:640–649. [DOI] [PubMed] [Google Scholar]
- Barygin OI, Gmiro VE, Kim KKh, Magazanik LG, Tikhonov DB (2009) Blockade of NMDA receptor channels by 9-aminoacridine and its derivatives. Neurosci Lett 451:29–33. [DOI] [PubMed] [Google Scholar]
- Bass BL (2002) RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu ACTsai GEMa CLEhmsen JTMustafa AKHan LJiang ZIBenneyworth MAFroimowitz MPLange N, et al. (2009) Targeted disruption of serine racemase affects glutamatergic neurotransmission and behavior. Mol Psychiatry 14:719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath CP, Farrell LN, Gilmore J, Ward MA, Hicks CA, O’Neill MJ, Bleakman D (1996) The effects of ifenprodil and eliprodil on voltage-dependent Ca2+ channels and in gerbil global cerebral ischaemia. Eur J Pharmacol 299:103–112. [DOI] [PubMed] [Google Scholar]
- Bats C, Groc L, Choquet D (2007) The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron 53:719–734. [DOI] [PubMed] [Google Scholar]
- Bats C, Soto D, Studniarczyk D, Farrant M, Cull-Candy SG (2012) Channel properties reveal differential expression of TARPed and TARPless AMPARs in stargazer neurons. Nat Neurosci 15:853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer KU, Schulman H (2019) CaM kinase: Still inspiring at 40. Neuron 103:380–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C, Wollmuth LP, Seeburg PH, Sakmann B, Kuner T (1999) NMDAR channel segments forming the extracellular vestibule inferred from the accessibility of substituted cysteines. Neuron 22:559–570. [DOI] [PubMed] [Google Scholar]
- Beck K, Javitt DC, Howes OD (2016) Targeting glutamate to treat schizophrenia: lessons from recent clinical studies. Psychopharmacology (Berl) 233:2425–2428. [DOI] [PubMed] [Google Scholar]
- Beckwith-Cohen B, Holzhausen LC, Wang TM, Rajappa R, Kramer RH (2019) Localizing proton-mediated inhibitory feedback at the retinal horizontal cell-cone synapse with genetically-encoded pH probes. J Neurosci 39:651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beed PS, Salmen B, Schmitz D (2009) GluK2-mediated excitability within the superficial layers of the entorhinal cortex. PLoS One 4:e5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar TN, Scott CA, Greene CL, Wen X, Smith SV, Maric D, Liu QY, Colton CA, Barker JL (1999) Glutamate acting at NMDA receptors stimulates embryonic cortical neuronal migration. J Neurosci 19:4449–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinat C, Banister SD, Hoban J, Tsanaktsidis J, Metaxas A, Windhorst AD, Kassiou M (2014) Structure-activity relationships of N-substituted 4-(trifluoromethoxy)benzamidines with affinity for GluN2B-containing NMDA receptors. Bioorg Med Chem Lett 24:828–830. [DOI] [PubMed] [Google Scholar]
- Bekkers JM (1993) Enhancement by histamine of NMDA-mediated synaptic transmission in the hippocampus. Science 261:104–106. [DOI] [PubMed] [Google Scholar]
- Bekkers JM, Stevens CF (1989) NMDA and non-NMDA receptors are co-localized at individual excitatory synapses in cultured rat hippocampus. Nature 341:230–233. [DOI] [PubMed] [Google Scholar]
- Bekkers JM, Vidovic M, Ymer S (1996) Differential effects of histamine on the N-methyl-D-aspartate channel in hippocampal slices and cultures. Neuroscience 72:669–677. [DOI] [PubMed] [Google Scholar]
- Belcher SM, Howe JR (1997) Characterization of RNA editing of the glutamate-receptor subunits GluR5 and GluR6 in granule cells during cerebellar development. Brain Res Mol Brain Res 52:130–138. [DOI] [PubMed] [Google Scholar]
- Belforte JE, Zsiros V, Sklar ER, Jiang Z, Yu G, Li Y, Quinlan EM, Nakazawa K (2010) Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nat Neurosci 13:76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RF, Eccleston C, Kalso EA (2017) Ketamine as an adjuvant to opioids for cancer pain. Cochrane Database Syst Rev 6:CD003351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yaacov A, Gillor M, Haham T, Parsai A, Qneibi M, Stern-Bach Y (2017) Molecular mechanism of AMPA receptor modulation by TARP/Stargazin. Neuron 93:1126–1137.e4. [DOI] [PubMed] [Google Scholar]
- Bennett GD, Moser K, Chaudoin T, Rosenquist TH (2006) The expression of the NR1-subunit of the NMDA receptor during mouse and early chicken development. Reprod Toxicol 22:536–541. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB (2009) Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136:149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste M, Clements J, Vyklický L Jr, Mayer ML (1990) A kinetic analysis of the modulation of N-methyl-D-aspartic acid receptors by glycine in mouse cultured hippocampal neurones. J Physiol 428:333–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste M, Mayer ML (1991) Structure-activity analysis of binding kinetics for NMDA receptor competitive antagonists: the influence of conformational restriction. Br J Pharmacol 104:207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste M, Mayer ML (1995) Trapping of glutamate and glycine during open channel block of rat hippocampal neuron NMDA receptors by 9-aminoacridine. J Physiol 483:367–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg LK, Larsson M, Morland C, Gundersen V (2013) Pre- and postsynaptic localization of NMDA receptor subunits at hippocampal mossy fibre synapses. Neuroscience 230:139–150. [DOI] [PubMed] [Google Scholar]
- Berger AJ, Dieudonné S, Ascher P (1998) Glycine uptake governs glycine site occupancy at NMDA receptors of excitatory synapses. J Neurophysiol 80:3336–3340. [DOI] [PubMed] [Google Scholar]
- Bergeron R, Meyer TM, Coyle JT, Greene RW (1998) Modulation of N-methyl-D-aspartate receptor function by glycine transport. Proc Natl Acad Sci USA 95:15730–15734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergles DE, Dzubay JA, Jahr CE (1997) Glutamate transporter currents in bergmann glial cells follow the time course of extrasynaptic glutamate. Proc Natl Acad Sci USA 94:14821–14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergles DE, Roberts JD, Somogyi P, Jahr CE (2000) Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature 405:187–191. [DOI] [PubMed] [Google Scholar]
- Berlin S, Szobota S, Reiner A, Carroll EC, Kienzler MA, Guyon A, Xiao T, Trauner D, Isacoff EY (2016) A family of photoswitchable NMDA receptors. eLife 5:e12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman RM, Cappiello A, Anand A, Oren DA, Heninger GR, Charney DS, Krystal JH (2000) Antidepressant effects of ketamine in depressed patients. Biol Psychiatry 47:351–354. [DOI] [PubMed] [Google Scholar]
- Bernabeu R, Sharp FR (2000) NMDA and AMPA/kainate glutamate receptors modulate dentate neurogenesis and CA3 synapsin-I in normal and ischemic hippocampus. J Cereb Blood Flow Metab 20:1669–1680. [DOI] [PubMed] [Google Scholar]
- Bernard A, Ferhat L, Dessi F, Charton G, Represa A, Ben-Ari Y, Khrestchatisky M (1999) Q/R editing of the rat GluR5 and GluR6 kainate receptors in vivo and in vitro: evidence for independent developmental, pathological and cellular regulation. Eur J Neurosci 11:604–616. [DOI] [PubMed] [Google Scholar]
- Bernard A, Khrestchatisky M (1994) Assessing the extent of RNA editing in the TMII regions of GluR5 and GluR6 kainate receptors during rat brain development. J Neurochem 62:2057–2060. [DOI] [PubMed] [Google Scholar]
- Bernard V, Somogyi P, Bolam JP (1997) Cellular, subcellular, and subsynaptic distribution of AMPA-type glutamate receptor subunits in the neostriatum of the rat. J Neurosci 17:819–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta N, Jones RS (1996) Tonic facilitation of glutamate release by presynaptic N-methyl-D-aspartate autoreceptors in the entorhinal cortex. Neuroscience 75:339–344. [DOI] [PubMed] [Google Scholar]
- Bertocchi IEltokhi ARozov AChi VNJensen VBus TPawlak VSerafino MSonntag HYang B, et al. (2021) Voltage-independent GluN2A-type NMDA receptor Ca2+ signaling promotes audiogenic seizures, attentional and cognitive deficits in mice. Commun Biol 4:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettini ESava AGriffante CCarignani CBuson ACapelli AMNegri MAndreetta FSenar-Sancho SAGuiral L, et al. (2010) Identification and characterization of novel NMDA receptor antagonists selective for NR2A- over NR2B-containing receptors. J Pharmacol Exp Ther 335:636–644. [DOI] [PubMed] [Google Scholar]
- Bettler B, Boulter J, Hermans-Borgmeyer I, O’Shea-Greenfield A, Deneris ES, Moll C, Borgmeyer U, Hollmann M, Heinemann S (1990) Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron 5:583–595. [DOI] [PubMed] [Google Scholar]
- Bettler B, Egebjerg J, Sharma G, Pecht G, Hermans-Borgmeyer I, Moll C, Stevens CF, Heinemann S (1992) Cloning of a putative glutamate receptor: a low affinity kainate-binding subunit. Neuron 8:257–265. [DOI] [PubMed] [Google Scholar]
- Bettler B, Fakler B (2017) Ionotropic AMPA-type glutamate and metabotropic GABAB receptors: determining cellular physiology by proteomes. Curr Opin Neurobiol 45:16–23. [DOI] [PubMed] [Google Scholar]
- Betz WJ (1970) Depression of transmitter release at the neuromuscular junction of the frog. J Physiol 206:629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzi P, Gundersen V, Galbete JL, Seifert G, Steinhäuser C, Pilati E, Volterra A (2004) Astrocytes contain a vesicular compartment that is competent for regulated exocytosis of glutamate. Nat Neurosci 7:613–620. [DOI] [PubMed] [Google Scholar]
- Bhangoo SK, Swanson GT (2013) Kainate receptor signaling in pain pathways. Mol Pharmacol 83:307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt JM, Prakash A, Suryavanshi PS, Dravid SM (2013) Effect of ifenprodil on GluN1/GluN2B N-methyl-D-aspartate receptor gating. Mol Pharmacol 83:9–21. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Khatri A, Swanger SA, DiRaddo JO, Yi F, Hansen KB, Yuan H, Traynelis SF (2018) Triheteromeric GluN1/GluN2A/GluN2C NMDARs with unique single-channel properties are the dominant receptor population in cerebellar granule cells. Neuron 99:315–328.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidoret C, Ayon A, Barbour B, Casado M (2009) Presynaptic NR2A-containing NMDA receptors implement a high-pass filter synaptic plasticity rule. Proc Natl Acad Sci USA 106:14126–14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Kaeser PS, Blanpied TA (2017) Transcellular nanoalignment of synaptic function. Neuron 96:680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien CGGranata TAntozzi CCross JHDulac OKurthen MLassmann HMantegazza RVillemure JGSpreafico R, et al. (2005) Pathogenesis, diagnosis and treatment of Rasmussen encephalitis: a European consensus statement. Brain 128:454–471. [DOI] [PubMed] [Google Scholar]
- Billups D, Attwell D (2003) Active release of glycine or D-serine saturates the glycine site of NMDA receptors at the cerebellar mossy fibre to granule cell synapse. Eur J Neurosci 18:2975–2980. [DOI] [PubMed] [Google Scholar]
- Birdsey-Benson A, Gill A, Henderson LP, Madden DR (2010) Enhanced efficacy without further cleft closure: reevaluating twist as a source of agonist efficacy in AMPA receptors. J Neurosci 30:1463–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff S, Barhanin J, Bettler B, Mulle C, Heinemann S (1997) Spatial distribution of kainate receptor subunit mRNA in the mouse basal ganglia and ventral mesencephalon. J Comp Neurol 379:541–562. [DOI] [PubMed] [Google Scholar]
- Biscoe TJ, Evans RH, Francis AA, Martin MR, Watkins JC, Davies J, Dray A (1977) D-alpha-Aminoadipate as a selective antagonist of amino acid-induced and synaptic excitation of mammalian spinal neurones. Nature 270:743–745. [DOI] [PubMed] [Google Scholar]
- Bjerrum EJ, Kristensen AS, Pickering DS, Greenwood JR, Nielsen B, Liljefors T, Schousboe A, Bräuner-Osborne H, Madsen U (2003) Design, synthesis, and pharmacology of a highly subtype-selective GluR1/2 agonist, (RS)-2-amino-3-(4-chloro-3-hydroxy-5-isoxazolyl)propionic acid (Cl-HIBO). J Med Chem 46:2246–2249. [DOI] [PubMed] [Google Scholar]
- Black MD (2005) Therapeutic potential of positive AMPA modulators and their relationship to AMPA receptor subunits. A review of preclinical data. Psychopharmacology (Berl) 179:154–163. [DOI] [PubMed] [Google Scholar]
- Blakemore LJ, Trombley PQ (2020) Zinc modulates olfactory bulb kainate receptors. Neuroscience 428:252–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco MJ, La D, Coughlin Q, Newman CA, Griffin AM, Harrison BL, Salituro FG (2018) Breakthroughs in neuroactive steroid drug discovery. Bioorg Med Chem Lett 28:61–70. [DOI] [PubMed] [Google Scholar]
- Blandini F, Porter RH, Greenamyre JT (1996) Glutamate and Parkinson’s disease. Mol Neurobiol 12:73–94. [DOI] [PubMed] [Google Scholar]
- Blanke ML, VanDongen AM (2008) Constitutive activation of the N-methyl-D-aspartate receptor via cleft-spanning disulfide bonds. J Biol Chem 283:21519–21529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Boeckman FA, Aizenman E, Johnson JW (1997) Trapping channel block of NMDA-activated responses by amantadine and memantine. J Neurophysiol 77:309–323. [DOI] [PubMed] [Google Scholar]
- Blanpied TA, Clarke RJ, Johnson JW (2005) Amantadine inhibits NMDA receptors by accelerating channel closure during channel block. J Neurosci 25:3312–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke M, Keller BU, Rivosecchi R, Hollmann M, Heinemann S, Konnerth A (1993) A single amino acid determines the subunit-specific spider toxin block of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionate/kainate receptor channels. Proc Natl Acad Sci USA 90:6528–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatow M, Caputi A, Burnashev N, Monyer H, Rozov A (2003) Ca2+ buffer saturation underlies paired pulse facilitation in calbindin-D28k-containing terminals. Neuron 38:79–88. [DOI] [PubMed] [Google Scholar]
- Bleakman DBallyk BASchoepp DDPalmer AJBath CPSharpe EFWoolley MLBufton HRKamboj RKTarnawa I, et al. (1996) Activity of 2,3-benzodiazepines at native rat and recombinant human glutamate receptors in vitro: stereospecificity and selectivity profiles. Neuropharmacology 35:1689–1702. [DOI] [PubMed] [Google Scholar]
- Bochet P, Audinat E, Lambolez B, Crépel F, Rossier J, Iino M, Tsuzuki K, Ozawa S (1994) Subunit composition at the single-cell level explains functional properties of a glutamate-gated channel. Neuron 12:383–388. [DOI] [PubMed] [Google Scholar]
- Bock T, Stuart GJ (2016) Impact of calcium-activated potassium channels on NMDA spikes in cortical layer 5 pyramidal neurons. J Neurophysiol 115:1740–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bökel C, Dass S, Wilsch-Bräuninger M, Roth S (2006) Drosophila Cornichon acts as cargo receptor for ER export of the TGFalpha-like growth factor Gurken. Development 133:459–470. [DOI] [PubMed] [Google Scholar]
- Bokesch PM, Izykenova GA, Justice JB, Easley KA, Dambinova SA (2006) NMDA receptor antibodies predict adverse neurological outcome after cardiac surgery in high-risk patients. Stroke 37:1432–1436. [DOI] [PubMed] [Google Scholar]
- Bolshakov KV, Gmiro VE, Tikhonov DB, Magazanik LG (2003) Determinants of trapping block of N-methyl-d-aspartate receptor channels. J Neurochem 87:56–65. [DOI] [PubMed] [Google Scholar]
- Bolshakov KV, Kim KH, Potapjeva NN, Gmiro VE, Tikhonov DB, Usherwood PN, Mellor IR, Magazanik LG (2005) Design of antagonists for NMDA and AMPA receptors. Neuropharmacology 49:144–155. [DOI] [PubMed] [Google Scholar]
- Bonaglia MCCiccone RGimelli GGimelli SMarelli SVerheij JGiorda RGrasso RBorgatti RPagone F, et al. (2008) Detailed phenotype-genotype study in five patients with chromosome 6q16 deletion: narrowing the critical region for Prader-Willi-like phenotype. Eur J Hum Genet 16:1443–1449. [DOI] [PubMed] [Google Scholar]
- Bonnet C, Leheup B, Béri M, Philippe C, Grégoire MJ, Jonveaux P (2009) Aberrant GRIA3 transcripts with multi-exon duplications in a family with X-linked mental retardation. Am J Med Genet A 149A:1280–1289. [DOI] [PubMed] [Google Scholar]
- Bordji K, Becerril-Ortega J, Nicole O, Buisson A (2010) Activation of extrasynaptic, but not synaptic, NMDA receptors modifies amyloid precursor protein expression pattern and increases amyloid-ß production. J Neurosci 30:15927–15942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovska J, Vyklicky V, Stastna E, Kapras V, Slavikova B, Horak M, Chodounska H, Vyklicky L Jr (2012) Access of inhibitory neurosteroids to the NMDA receptor. Br J Pharmacol 166:1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borschel WF, Cummings KA, Tindell LK, Popescu GK (2015) Kinetic contributions to gating by interactions unique to N-methyl-D-aspartate (NMDA) receptors. J Biol Chem 290:26846–26855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borschel WF, Murthy SE, Kasperek EM, Popescu GK (2011) NMDA receptor activation requires remodelling of intersubunit contacts within ligand-binding heterodimers. Nat Commun 2:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borschel WF, Myers JM, Kasperek EM, Smith TP, Graziane NM, Nowak LM, Popescu GK (2012) Gating reaction mechanism of neuronal NMDA receptors. J Neurophysiol 108:3105–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotto ZAClarke VRDelany CMParry MCSmolders IVignes MHo KHMiu PBrinton BTFantaske R, et al. (1999) Kainate receptors are involved in synaptic plasticity. Nature 402:297–301. [DOI] [PubMed] [Google Scholar]
- Bortone D, Polleux F (2009) KCC2 expression promotes the termination of cortical interneuron migration in a voltage-sensitive calcium-dependent manner. Neuron 62:53–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudkkazi S, Brechet A, Schwenk J, Fakler B (2014) Cornichon2 dictates the time course of excitatory transmission at individual hippocampal synapses. Neuron 82:848–858. [DOI] [PubMed] [Google Scholar]
- Boulter J, Hollmann M, O’Shea-Greenfield A, Hartley M, Deneris E, Maron C, Heinemann S (1990) Molecular cloning and functional expression of glutamate receptor subunit genes. Science 249:1033–1037. [DOI] [PubMed] [Google Scholar]
- Bourne HR, Nicoll R (1993) Molecular machines integrate coincident synaptic signals. Cell 72 (Suppl):65–75. [DOI] [PubMed] [Google Scholar]
- Bowen AB, Bourke AM, Hiester BG, Hanus C, Kennedy MJ (2017) Golgi-independent secretory trafficking through recycling endosomes in neuronal dendrites and spines. eLife 6:e27362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers ME, Ressler KJ (2015) An overview of translationally informed treatments for posttraumatic stress disorder: Animal models of Pavlovian fear conditioning to human clinical trials. Biol Psychiatry 78:E15–E27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers MS, Cacheaux LP, Sahu SU, Schmidt ME, Sennello JA, Leaderbrand K, Khan MA, Kroes RA, Moskal JR (2020) NYX-2925 induces metabotropic N-methyl-d-aspartate receptor (NMDAR) signaling that enhances synaptic NMDAR and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor. J Neurochem 152:523–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D (2002) External anions and cations distinguish between AMPA and kainate receptor gating mechanisms. J Physiol 539:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D (2010) Ion-dependent gating of kainate receptors. J Physiol 588:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D (2012) Redefining the classification of AMPA-selective ionotropic glutamate receptors. J Physiol 590:49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D (2018) Polyamine-mediated channel block of ionotropic glutamate receptors and its regulation by auxiliary proteins. J Biol Chem 293:18789–18802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D, Garcia EP, Marshall J, Traynelis SF, Lange GD (2003) Allosteric regulation and spatial distribution of kainate receptors bound to ancillary proteins. J Physiol 547:373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D, Lange GD (2002) Functional stoichiometry of glutamate receptor desensitization. J Neurosci 22:3392–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D, Lange GD, Mayer ML (1998) Activity-dependent modulation of glutamate receptors by polyamines. J Neurosci 18:8175–8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D, Mayer ML (1995) Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron 15:453–462. [DOI] [PubMed] [Google Scholar]
- Bowie D, Smart TG (1993) Thiocyanate ions selectively antagonize AMPA-evoked responses in Xenopus laevis oocytes microinjected with rat brain mRNA. Br J Pharmacol 109:779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga MF, Aroniadou-Anderjaska V, Li H, Rogawski MA (2009) Topiramate reduces excitability in the basolateral amygdala by selectively inhibiting GluK1 (GluR5) kainate receptors on interneurons and positively modulating GABAA receptors on principal neurons. J Pharmacol Exp Ther 330:558–566. [DOI] [PubMed] [Google Scholar]
- Braga MF, Aroniadou-Anderjaska V, Xie J, Li H (2003) Bidirectional modulation of GABA release by presynaptic glutamate receptor 5 kainate receptors in the basolateral amygdala. J Neurosci 23:442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstätter JH, Hartveit E, Sassoè-Pognetto M, Wässle H (1994) Expression of NMDA and high-affinity kainate receptor subunit mRNAs in the adult rat retina. Eur J Neurosci 6:1100–1112. [DOI] [PubMed] [Google Scholar]
- Brasier DJ, Feldman DE (2008) Synapse-specific expression of functional presynaptic NMDA receptors in rat somatosensory cortex. J Neurosci 28:2199–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer PR, Rosenquist TH (2002) Effect of elevated homocysteine on cardiac neural crest migration in vitro. Dev Dyn 224:222–230. [DOI] [PubMed] [Google Scholar]
- Bräuner-Osborne H, Egebjerg J, Nielsen EO, Madsen U, Krogsgaard-Larsen P (2000) Ligands for glutamate receptors: design and therapeutic prospects. J Med Chem 43:2609–2645. [DOI] [PubMed] [Google Scholar]
- Brechet ABuchert RSchwenk JBoudkkazi SZolles GSiquier-Pernet KSchaber IBildl WSaadi ABole-Feysot C, et al. (2017) AMPA-receptor specific biogenesis complexes control synaptic transmission and intellectual ability. Nat Commun 8:15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregestovski P, Maleeva G, Gorostiza P (2018) Light-induced regulation of ligand-gated channel activity. Br J Pharmacol 175:1892–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm L, Greenwood JR, Hansen KB, Nielsen B, Egebjerg J, Stensbøl TB, Bräuner-Osborne H, Sløk FA, Kronborg TT, Krogsgaard-Larsen P (2003) (S)-2-Amino-3-(3-hydroxy-7,8-dihydro-6H-cyclohepta[d]isoxazol-4-yl)propionic acid, a potent and selective agonist at the GluR5 subtype of ionotropic glutamate receptors. Synthesis, modeling, and molecular pharmacology. J Med Chem 46:1350–1358. [DOI] [PubMed] [Google Scholar]
- Bretin SLouis CSeguin LWagner SThomas JYChallal SRogez NAlbinet KIop FVillain N, et al. (2017) Pharmacological characterisation of S 47445, a novel positive allosteric modulator of AMPA receptors. PLoS One 12:e0184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breustedt J, Schmitz D (2004) Assessing the role of GLUK5 and GLUK6 at hippocampal mossy fiber synapses. J Neurosci 24:10093–10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickley SG, Misra C, Mok MH, Mishina M, Cull-Candy SG (2003) NR2B and NR2D subunits coassemble in cerebellar Golgi cells to form a distinct NMDA receptor subtype restricted to extrasynaptic sites. J Neurosci 23:4958–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigman JL, Feyder M, Saksida LM, Bussey TJ, Mishina M, Holmes A (2008) Impaired discrimination learning in mice lacking the NMDA receptor NR2A subunit. Learn Mem 15:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristow LJGulia JWeed MRSrikumar BNLi YWGraef JDNaidu PSSanmathi CAher JBastia T, et al. (2017) Preclinical characterization of (R)-3-((3S,4S)-3-fluoro-4-(4-hydroxyphenyl)piperidin-1-yl)-1-(4-methylbenzyl)pyrrolidin-2-one (BMS-986169), a novel, intravenous, glutamate N-methyl-d-aspartate 2B receptor negative allosteric modulator with potential in major depressive disorder. J Pharmacol Exp Ther 363:377–393. [DOI] [PubMed] [Google Scholar]
- Brockie PJJensen MMellem JEJensen EYamasaki TWang RMaxfield DThacker CHoerndli FDunn PJ, et al. (2013) Cornichons control ER export of AMPA receptors to regulate synaptic excitability. Neuron 80:129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogi SBrindisi MButini SKshirsagar GUMaramai SChemi GGemma SCampiani GNovellino EFiorenzani P, et al. (2018) ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic acid (AMPA) and kainate receptor ligands: Further exploration of bioisosteric replacements and structural and biological investigation. J Med Chem 61:2124–2130. [DOI] [PubMed] [Google Scholar]
- Brogi S, Campiani G, Brindisi M, Butini S (2019) Allosteric modulation of ionotropic glutamate receptors: An outlook on new therapeutic approaches to treat central nervous system disorders. ACS Med Chem Lett 10:228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DGMaier DLSylvester MAHoerter TNMenhaji-Klotz ELasota CCHirata LTWilkins DEScott CWTrivedi S, et al. (2011) 2,6-Disubstituted pyrazines and related analogs as NR2B site antagonists of the NMDA receptor with anti-depressant activity. Bioorg Med Chem Lett 21:3399–3403. [DOI] [PubMed] [Google Scholar]
- Brown PM, Aurousseau MR, Musgaard M, Biggin PC, Bowie D (2016) Kainate receptor pore-forming and auxiliary subunits regulate channel block by a novel mechanism. J Physiol 594:1821–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PMGE, McGuire H, Bowie D (2018) Stargazin and cornichon-3 relieve polyamine block of AMPA receptors by enhancing blocker permeation. J Gen Physiol 150:67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R (1995) Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 270:1677–1680. [DOI] [PubMed] [Google Scholar]
- Buchan AM, Lesiuk H, Barnes KA, Li H, Huang ZG, Smith KE, Xue D (1993) AMPA antagonists: do they hold more promise for clinical stroke trials than NMDA antagonists? Stroke 24(12, Suppl)I148–I152. [PubMed] [Google Scholar]
- Buchanan KA, Blackman AV, Moreau AW, Elgar D, Costa RP, Lalanne T, Tudor Jones AA, Oyrer J, Sjöström PJ (2012) Target-specific expression of presynaptic NMDA receptors in neocortical microcircuits. Neuron 75:451–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck DP, Howitt SM, Clements JD (2000) NMDA channel gating is influenced by a tryptophan residue in the M2 domain but calcium permeation is not altered. Biophys J 79:2454–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budisantoso T, Harada H, Kamasawa N, Fukazawa Y, Shigemoto R, Matsui K (2013) Evaluation of glutamate concentration transient in the synaptic cleft of the rat calyx of Held. J Physiol 591:219–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buemi MR, De Luca L, Ferro S, Gitto R (2014) Targeting GluN2B-containing N-methyl-D-aspartate receptors: design, synthesis, and binding affinity evaluation of novel 3-substituted indoles. Arch Pharm (Weinheim) 347:533–539. [DOI] [PubMed] [Google Scholar]
- Buffington SA, Huang W, Costa-Mattioli M (2014) Translational control in synaptic plasticity and cognitive dysfunction. Annu Rev Neurosci 37:17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch L, Gefflaut T, Alaux S, Sagot E, Nielsen B, Pickering DS (2009) Pharmacological characterization of (4R)-alkyl glutamate analogues at the ionotropic glutamate receptors--focus on subtypes iGlu(5-7). Eur J Pharmacol 609:1–4. [DOI] [PubMed] [Google Scholar]
- Bunk EC, König HG, Prehn JH, Kirby BP (2014) Effect of the N-methyl-D-aspartate NR2B subunit antagonist ifenprodil on precursor cell proliferation in the hippocampus. J Neurosci Res 92:679–691. [DOI] [PubMed] [Google Scholar]
- Buonarati OR, Hammes EA, Watson JF, Greger IH, Hell JW (2019) Mechanisms of postsynaptic localization of AMPA-type glutamate receptors and their regulation during long-term potentiation. Sci Signal 12:eaar6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burada AP, Vinnakota R, Kumar J (2020a) The architecture of GluD2 ionotropic delta glutamate receptor elucidated by cryo-EM. J Struct Biol 211:107546. [DOI] [PubMed] [Google Scholar]
- Burada AP, Vinnakota R, Kumar J (2020b) Cryo-EM structures of the ionotropic glutamate receptor GluD1 reveal a non-swapped architecture. Nat Struct Mol Biol 27:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burban A, Faucard R, Armand V, Bayard C, Vorobjev V, Arrang JM (2010) Histamine potentiates N-methyl-D-aspartate receptors by interacting with an allosteric site distinct from the polyamine binding site. J Pharmacol Exp Ther 332:912–921. [DOI] [PubMed] [Google Scholar]
- Bureau I, Bischoff S, Heinemann SF, Mulle C (1999) Kainate receptor-mediated responses in the CA1 field of wild-type and GluR6-deficient mice. J Neurosci 19:653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau I, Dieudonne S, Coussen F, Mulle C (2000) Kainate receptor-mediated synaptic currents in cerebellar Golgi cells are not shaped by diffusion of glutamate. Proc Natl Acad Sci USA 97:6838–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorf JZhang XLWeiss CGross ABoikess SRKroes RAKhan MABurch RMRex CSDisterhoft JF, et al. (2015) The long-lasting antidepressant effects of rapastinel (GLYX-13) are associated with a metaplasticity process in the medial prefrontal cortex and hippocampus. Neuroscience 308:202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger PB, Yuan H, Karakas E, Geballe M, Furukawa H, Liotta DC, Snyder JP, Traynelis SF (2012) Mapping the binding of GluN2B-selective N-methyl-D-aspartate receptor negative allosteric modulators. Mol Pharmacol 82:344–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkner PC, Bittner N, Holling H, Buhlmann U (2017) D-cycloserine augmentation of behavior therapy for anxiety and obsessive-compulsive disorders: A meta-analysis. PLoS One 12:e0173660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnashev N, Monyer H, Seeburg PH, Sakmann B (1992a) Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron 8:189–198. [DOI] [PubMed] [Google Scholar]
- Burnashev N, Schoepfer R, Monyer H, Ruppersberg JP, Günther W, Seeburg PH, Sakmann B (1992b) Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science 257:1415–1419. [DOI] [PubMed] [Google Scholar]
- Burnashev N, Villarroel A, Sakmann B (1996) Dimensions and ion selectivity of recombinant AMPA and kainate receptor channels and their dependence on Q/R site residues. J Physiol 496:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnashev N, Zhou Z, Neher E, Sakmann B (1995) Fractional calcium currents through recombinant GluR channels of the NMDA, AMPA and kainate receptor subtypes. J Physiol 485:403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell ES, Irvine M, Fang G, Sapkota K, Jane DE, Monaghan DT (2019) Positive and negative allosteric modulators of N-methyl-d-aspartate (NMDA) receptors: Structure-activity relationships and mechanisms of action. J Med Chem 62:3–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursztajn S, Falls WA, Berman SA, Friedman MJ (2007) Cell proliferation in the brains of NMDAR NR1 transgenic mice. Brain Res 1172:10–20. [DOI] [PubMed] [Google Scholar]
- Burzomato V, Frugier G, Pérez-Otaño I, Kittler JT, Attwell D (2010) The receptor subunits generating NMDA receptor mediated currents in oligodendrocytes. J Physiol 588:3403–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos FJJury NMartinez PAmpuero ECampos MAbarzúa SJaramillo KIbing SMardones MDHaensgen H, et al. (2017) NMDA receptor subunit composition controls dendritogenesis of hippocampal neurons through CAMKII, CREB-P, and H3K27ac. J Cell Physiol 232:3677–3692. [DOI] [PubMed] [Google Scholar]
- Bustos FJVarela-Nallar LCampos MHenriquez BPhillips MOpazo CAguayo LGMontecino MConstantine-Paton MInestrosa NC, et al. (2014) PSD95 suppresses dendritic arbor development in mature hippocampal neurons by occluding the clustering of NR2B-NMDA receptors. PLoS One 9:e94037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SX (2006) Glycine/NMDA receptor antagonists as potential CNS therapeutic agents: ACEA-1021 and related compounds. Curr Top Med Chem 6:651–662. [DOI] [PubMed] [Google Scholar]
- Cai X, Liang CW, Muralidharan S, Kao JP, Tang CM, Thompson SM (2004) Unique roles of SK and Kv4.2 potassium channels in dendritic integration. Neuron 44:351–364. [DOI] [PubMed] [Google Scholar]
- Cais O, Herguedas B, Krol K, Cull-Candy SG, Farrant M, Greger IH (2014) Mapping the interaction sites between AMPA receptors and TARPs reveals a role for the receptor N-terminal domain in channel gating. Cell Rep 9:728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan PM, Terry AV Jr, Nelson FR, Volkmann RA, Vinod AB, Zainuddin M, Menniti FS (2020) Modulating inhibitory response control through potentiation of GluN2D subunit-containing NMDA receptors. Neuropharmacology 173:107994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calton JL, Kang MH, Wilson WA, Moore SD (2000) NMDA-receptor-dependent synaptic activation of voltage-dependent calcium channels in basolateral amygdala. J Neurophysiol 83:685–692. [DOI] [PubMed] [Google Scholar]
- Cameron HA, McEwen BS, Gould E (1995) Regulation of adult neurogenesis by excitatory input and NMDA receptor activation in the dentate gyrus. J Neurosci 15:4687–4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp CR, Yuan H (2020) GRIN2D/GluN2D NMDA receptor: Unique features and its contribution to pediatric developmental and epileptic encephalopathy. Eur J Paediatr Neurol 24:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campiani GMorelli ENacci VFattorusso CRamunno ANovellino EGreenwood JLiljefors TGriffiths RSinclair C, et al. (2001) Characterization of the 1H-cyclopentapyrimidine-2,4(1H,3H)-dione derivative (S)-CPW399 as a novel, potent, and subtype-selective AMPA receptor full agonist with partial desensitization properties. J Med Chem 44:4501–4504. [DOI] [PubMed] [Google Scholar]
- Cao JY, Qiu S, Zhang J, Wang JJ, Zhang XM, Luo JH (2011) Transmembrane region of N-methyl-D-aspartate receptor (NMDAR) subunit is required for receptor subunit assembly. J Biol Chem 286:27698–27705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone AL, Plested AJ (2016) Superactivation of AMPA receptors by auxiliary proteins. Nat Commun 7:10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson NG, Gahring LC, Twyman RE, Rogers SW (1997) Identification of amino acids in the glutamate receptor, GluR3, important for antibody-binding and receptor-specific activation. J Biol Chem 272:11295–11301. [DOI] [PubMed] [Google Scholar]
- Carrillo E, Bhatia NK, Akimzhanov AM, Jayaraman V (2020a) Activity dependent inhibition of AMPA receptors by Zn2. J Neurosci 40:8629–8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo E, Shaikh SA, Berka V, Durham RJ, Litwin DB, Lee G, MacLean DM, Nowak LM, Jayaraman V (2020b) Mechanism of modulation of AMPA receptors by TARP-γ8. J Gen Physiol 152:e201912451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll EC, Berlin S, Levitz J, Kienzler MA, Yuan Z, Madsen D, Larsen DS, Isacoff EY (2015) Two-photon brightness of azobenzene photoswitches designed for glutamate receptor optogenetics. Proc Natl Acad Sci USA 112:E776–E785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta M, Opazo P, Veran J, Athané A, Choquet D, Coussen F, Mulle C (2013) CaMKII-dependent phosphorylation of GluK5 mediates plasticity of kainate receptors. EMBO J 32:496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta M, Srikumar BN, Gorlewicz A, Rebola N, Mulle C (2018) Activity-dependent control of NMDA receptor subunit composition at hippocampal mossy fibre synapses. J Physiol 596:703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvill GLRegan BMYendle SCO’Roak BJLozovaya NBruneau NBurnashev NKhan ACook JGeraghty E, et al. (2013) GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet 45:1073–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JPMagalhaes TConroy JMRegan RShah NAnney RShields DCAbrahams BSAlmeida JBacchelli E, et al. (2012) A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet 131:565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Gómez EOliveira BTapken DBertrand SKlein-Schmidt CPan HZafeiriou PSteiner JJurek BTrippe R, et al. (2017) All naturally occurring autoantibodies against the NMDA receptor subunit NR1 have pathogenic potential irrespective of epitope and immunoglobulin class. Mol Psychiatry 22:1776–1784. [DOI] [PubMed] [Google Scholar]
- Castillo PE, Malenka RC, Nicoll RA (1997) Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature 388:182–186. [DOI] [PubMed] [Google Scholar]
- Catarzi D, Colotta V, Varano F (2007) Competitive AMPA receptor antagonists. Med Res Rev 27:239–278. [DOI] [PubMed] [Google Scholar]
- Cavara NA, Orth A, Hollmann M (2009) Effects of NR1 splicing on NR1/NR3B-type excitatory glycine receptors. BMC Neurosci 10:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavelier P, Attwell D (2005) Tonic release of glutamate by a DIDS-sensitive mechanism in rat hippocampal slices. J Physiol 564:397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccon M, Rumbaugh G, Vicini S (2001) Distinct effect of pregnenolone sulfate on NMDA receptor subtypes. Neuropharmacology 40:491–500. [DOI] [PubMed] [Google Scholar]
- Cembrowski MS, Bachman JL, Wang L, Sugino K, Shields BC, Spruston N (2016a) Spatial gene-expression gradients underlie prominent heterogeneity of CA1 pyramidal neurons. Neuron 89:351–368. [DOI] [PubMed] [Google Scholar]
- Cembrowski MS, Wang L, Lemire AL, Copeland M, DiLisio SF, Clements J, Spruston N (2018) The subiculum is a patchwork of discrete subregions. eLife 7:e37701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cembrowski MS, Wang L, Sugino K, Shields BC, Spruston N (2016b) Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. eLife 5:e14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendelin J (2014) From mice to men: lessons from mutant ataxic mice. Cerebellum Ataxias 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceolin L, Bortolotto ZA, Bannister N, Collingridge GL, Lodge D, Volianskis A (2012) A novel anti-epileptic agent, perampanel, selectively inhibits AMPA receptor-mediated synaptic transmission in the hippocampus. Neurochem Int 61:517–522. [DOI] [PubMed] [Google Scholar]
- Cepeda C, Ariano MA, Calvert CR, Flores-Hernández J, Chandler SH, Leavitt BR, Hayden MR, Levine MS (2001) NMDA receptor function in mouse models of Huntington disease. J Neurosci Res 66:525–539. [DOI] [PubMed] [Google Scholar]
- Ceprian M, Fulton D (2019) Glial cell AMPA receptors in nervous system health, injury and disease. Int J Mol Sci 20:2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Černý J, Božíková P, Balík A, Marques SM, Vyklický L (2019) NMDA receptor opening and closing-transitions of a molecular machine revealed by molecular dynamics. Biomolecules 9:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervetto C, Maura G, Marcoli M (2010) Inhibition of presynaptic release-facilitatory kainate autoreceptors by extracellular cyclic GMP. J Pharmacol Exp Ther 332:210–219. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Murphy S, Coleman N (2017) The role of NMDA receptors in neural stem cell proliferation and differentiation. Stem Cells Dev 26:798–807. [DOI] [PubMed] [Google Scholar]
- Chamberlain SE, Yang J, Jones RS (2008) The role of NMDA receptor subtypes in short-term plasticity in the rat entorhinal cortex. Neural Plast 2008:872456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Nestor J, Huerta TS, Certain N, Moody G, Kowal C, Huerta PT, Volpe BT, Diamond B, Wollmuth LP (2020) Lupus autoantibodies act as positive allosteric modulators at GluN2A-containing NMDA receptors and impair spatial memory. Nat Commun 11:1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SF, Sucher NJ (2001) An NMDA receptor signaling complex with protein phosphatase 2A. J Neurosci 21:7985–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Maren S (2011) Medial prefrontal cortex activation facilitates re-extinction of fear in rats. Learn Mem 18:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EHVolpe BTMackay MAranow CWatson PKowal CStorbeck JMattis PBerlin RChen H, et al. (2015a) Selective impairment of spatial cognition caused by autoantibodies to the N-methyl-D-aspartate receptor. EBioMedicine 2:755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Ciani S, Kidokoro Y (1994) Ion permeation properties of the glutamate receptor channel in cultured embryonic Drosophila myotubes. J Physiol 476:1–16. [PMC free article] [PubMed] [Google Scholar]
- Chang HR, Kuo CC (2008) The activation gate and gating mechanism of the NMDA receptor. J Neurosci 28:1546–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MC, Park JM, Pelkey KA, Grabenstatter HL, Xu D, Linden DJ, Sutula TP, McBain CJ, Worley PF (2010) Narp regulates homeostatic scaling of excitatory synapses on parvalbumin-expressing interneurons. Nat Neurosci 13:1090–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Augustin K, Boddum K, Williams S, Sun M, Terschak JA, Hardege JD, Chen PE, Walker MC, Williams RS (2016) Seizure control by decanoic acid through direct AMPA receptor inhibition. Brain 139:431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Terbach N, Plant N, Chen PE, Walker MC, Williams RS (2013) Seizure control by ketogenic diet-associated medium chain fatty acids. Neuropharmacology 69:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Zuckermann AM, Williams S, Close AJ, Cano-Jaimez M, McEvoy JP, Spencer J, Walker MC, Williams RS (2015b) Seizure control by derivatives of medium chain fatty acids associated with the ketogenic diet show novel branching-point structure for enhanced potency. J Pharmacol Exp Ther 352:43–52. [DOI] [PubMed] [Google Scholar]
- Chao J, Seiler N, Renault J, Kashiwagi K, Masuko T, Igarashi K, Williams K (1997) N1-dansyl-spermine and N1-(n-octanesulfonyl)-spermine, novel glutamate receptor antagonists: block and permeation of N-methyl-D-aspartate receptors. Mol Pharmacol 51:861–871. [DOI] [PubMed] [Google Scholar]
- Chaperon F, Müller W, Auberson YP, Tricklebank MD, Neijt HC (2003) Substitution for PCP, disruption of prepulse inhibition and hyperactivity induced by N-methyl-D-aspartate receptor antagonists: preferential involvement of the NR2B rather than NR2A subunit. Behav Pharmacol 14:477–487. [DOI] [PubMed] [Google Scholar]
- Chappell AS, Gonzales C, Williams J, Witte MM, Mohs RC, Sperling R (2007) AMPA potentiator treatment of cognitive deficits in Alzheimer disease. Neurology 68:1008–1012. [DOI] [PubMed] [Google Scholar]
- Charton JP, Herkert M, Becker CM, Schröder H (1999) Cellular and subcellular localization of the 2B-subunit of the NMDA receptor in the adult rat telencephalon. Brain Res 816:609–617. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Ade C, Nurik CE, Carrejo NC, Dutta C, Jayaraman V, Landes CF (2019) Phosphorylation induces conformational rigidity at the C-terminal domain of AMPA receptors. J Phys Chem B 123:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton JEAwobuluyi MPremkumar LSTakahashi HTalantova MShin YCui JTu SSevarino KANakanishi N, et al. (2002) Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature 415:793–798. [DOI] [PubMed] [Google Scholar]
- Chaudhry C, Plested AJ, Schuck P, Mayer ML (2009a) Energetics of glutamate receptor ligand binding domain dimer assembly are modulated by allosteric ions. Proc Natl Acad Sci USA 106:12329–12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry C, Weston MC, Schuck P, Rosenmund C, Mayer ML (2009b) Stability of ligand-binding domain dimer assembly controls kainate receptor desensitization. EMBO J 28:1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheffings CM, Colquhoun D (2000) Single channel analysis of a novel NMDA channel from Xenopus oocytes expressing recombinant NR1a, NR2A and NR2D subunits. J Physiol 526:481–491. [PubMed] [Google Scholar]
- Chen C, Blitz DM, Regehr WG (2002) Contributions of receptor desensitization and saturation to plasticity at the retinogeniculate synapse. Neuron 33:779–788. [DOI] [PubMed] [Google Scholar]
- Chen CY, Matt L, Hell JW, Rogawski MA (2014a) Perampanel inhibition of AMPA receptor currents in cultured hippocampal neurons. PLoS One 9:e108021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Harata NC, Tsien RW (2004a) Paired-pulse depression of unitary quantal amplitude at single hippocampal synapses. Proc Natl Acad Sci USA 101:1063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, Cui C, Mayer ML, Gouaux E (1999a) Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature 402:817–821. [DOI] [PubMed] [Google Scholar]
- Chen HS, Lipton SA (1997) Mechanism of memantine block of NMDA-activated channels in rat retinal ganglion cells: uncompetitive antagonism. J Physiol 499:27–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Chesler M (2015) Autocrine boost of NMDAR current in hippocampal CA1 pyramidal neurons by a PMCA-dependent, perisynaptic, extracellular pH shift. J Neurosci 35:873–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Bao S, Qiao X, Thompson RF (1999b) Impaired cerebellar synapse maturation in waggler, a mutant mouse with a disrupted neuronal calcium channel gamma subunit. Proc Natl Acad Sci USA 96:12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA (2000) Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408:936–943. [DOI] [PubMed] [Google Scholar]
- Chen L, Dürr KL, Gouaux E (2014b) X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science 345:1021–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Li B, Murphy TH, Raymond LA (2004b) Site within N-methyl-D-aspartate receptor pore modulates channel gating. Mol Pharmacol 65:157–164. [DOI] [PubMed] [Google Scholar]
- Chen N, Luo T, Raymond LA (1999c) Subtype-dependence of NMDA receptor channel open probability. J Neurosci 19:6844–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Moshaver A, Raymond LA (1997) Differential sensitivity of recombinant N-methyl-D-aspartate receptor subtypes to zinc inhibition. Mol Pharmacol 51:1015–1023. [DOI] [PubMed] [Google Scholar]
- Chen PEGeballe MTKatz EErreger KLivesey MRO’Toole KKLe PLee CJSnyder JPTraynelis SF, et al. (2008) Modulation of glycine potency in rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J Physiol 586:227–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Diamond JS (2002) Synaptically released glutamate activates extrasynaptic NMDA receptors on cells in the ganglion cell layer of rat retina. J Neurosci 22:2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Gouaux E (2019) Structure and mechanism of AMPA receptor - auxiliary protein complexes. Curr Opin Struct Biol 54:104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhao Y, Wang Y, Shekhar M, Tajkhorshid E, Gouaux E (2017a) Activation and desensitization mechanism of AMPA receptor-TARP complex by cryo-EM. Cell 170:1234–1246.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Tankovic A, Burger PB, Kusumoto H, Traynelis SF, Yuan H (2017b) Functional evaluation of a de novo GRIN2A mutation identified in a patient with profound global developmental delay and refractory epilepsy. Mol Pharmacol 91:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Aslam M, Gollisch T, Allen K, von Engelhardt J (2018) CKAMP44 modulates integration of visual inputs in the lateral geniculate nucleus. Nat Commun 9:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Keramidas A, Harvey RJ, Lynch JW (2020a) Effects of GluN2A and GluN2B gain-of-function epilepsy mutations on synaptic currents mediated by diheteromeric and triheteromeric NMDA receptors. Neurobiol Dis 140:104850. [DOI] [PubMed] [Google Scholar]
- Chen XLee GMaher BSFanous AHChen JZhao ZGuo Avan den Oord ESullivan PFShi J, et al. ; GROUP investigators; International Schizophrenia Consortium (2011) GWA study data mining and independent replication identify cardiomyopathy-associated 5 (CMYA5) as a risk gene for schizophrenia. Mol Psychiatry 16:1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wu X, Wu H, Zhang M (2020b) Phase separation at the synapse. Nat Neurosci 23:301–310. [DOI] [PubMed] [Google Scholar]
- Chenard BLBordner JButler TWChambers LKCollins MADe Costa DLDucat MFDumont MLFox CBMena EE, et al. (1995) (1S,2S)-1-(4-hydroxyphenyl)-2-(4-hydroxy-4-phenylpiperidino)-1-propanol: a potent new neuroprotectant which blocks N-methyl-D-aspartate responses. J Med Chem 38:3138–3145. [DOI] [PubMed] [Google Scholar]
- Chenard BL, Shalaby IA, Koe BK, Ronau RT, Butler TW, Prochniak MA, Schmidt AW, Fox CB (1991) Separation of alpha 1 adrenergic and N-methyl-D-aspartate antagonist activity in a series of ifenprodil compounds. J Med Chem 34:3085–3090. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Du M, Ramanoudjame G, Jayaraman V (2005) Evolution of glutamate interactions during binding to a glutamate receptor. Nat Chem Biol 1:329–332. [DOI] [PubMed] [Google Scholar]
- Cheng S, Seven AB, Wang J, Skiniotis G, Özkan E (2016) Conformational plasticity in the transsynaptic neurexin-cerebellin-glutamate receptor adhesion complex. Structure 24:2163–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22:3099–3108. [DOI] [PubMed] [Google Scholar]
- Chergui K, Bouron A, Normand E, Mulle C (2000) Functional GluR6 kainate receptors in the striatum: indirect downregulation of synaptic transmission. J Neurosci 20:2175–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheriyan J, Balsara RD, Hansen KB, Castellino FJ (2016) Pharmacology of triheteromeric N-methyl-D-aspartate receptors. Neurosci Lett 617:240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler M (2003) Regulation and modulation of pH in the brain. Physiol Rev 83:1183–1221. [DOI] [PubMed] [Google Scholar]
- Chesler M, Kaila K (1992) Modulation of pH by neuronal activity. Trends Neurosci 15:396–402. [DOI] [PubMed] [Google Scholar]
- Chetkovich D (2009) Thalamic reticular neurons are unexcited by new stargazer seizure mechanism. Epilepsy Curr 9:59–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chez MG, Burton Q, Dowling T, Chang M, Khanna P, Kramer C (2007) Memantine as adjunctive therapy in children diagnosed with autistic spectrum disorders: an observation of initial clinical response and maintenance tolerability. J Child Neurol 22:574–579. [DOI] [PubMed] [Google Scholar]
- Chin AC, Yovanno RA, Wied TJ, Gershman A, Lau AY (2020) D-Serine potently drives ligand-binding domain closure in the ionotropic glutamate receptor GluD2. Structure 28:1168–1178.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari M, Wilding TJ, Brunwasser S, Krishnan K, Qian M, Benz A, Huettner JE, Zorumski CF, Covey DF, Mennerick S (2019) Visualizing pregnenolone sulfate-like modulators of NMDA receptor function reveals intracellular and plasma-membrane localization. Neuropharmacology 144:91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittajallu R, Vignes M, Dev KK, Barnes JM, Collingridge GL, Henley JM (1996) Regulation of glutamate release by presynaptic kainate receptors in the hippocampus. Nature 379:78–81. [DOI] [PubMed] [Google Scholar]
- Chittajallu RWester JCCraig MTBarksdale EYuan XQAkgül GFang CCollins DHunt SPelkey KA, et al. (2017) Afferent specific role of NMDA receptors for the circuit integration of hippocampal neurogliaform cells. Nat Commun 8:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyonobu THayashi SKobayashi KMorimoto MMiyanomae YNishimura ANishimoto AIto CImoto ISugimoto T, et al. (2007) Partial tandem duplication of GRIA3 in a male with mental retardation. Am J Med Genet A 143A:1448–1455. [DOI] [PubMed] [Google Scholar]
- Cho CH, St-Gelais F, Zhang W, Tomita S, Howe JR (2007) Two families of TARP isoforms that have distinct effects on the kinetic properties of AMPA receptors and synaptic currents. Neuron 55:890–904. [DOI] [PubMed] [Google Scholar]
- Cho RWPark JMWolff SBXu DHopf CKim JAReddy RCPetralia RSPerin MSLinden DJ, et al. (2008) mGluR1/5-dependent long-term depression requires the regulated ectodomain cleavage of neuronal pentraxin NPR by TACE. Neuron 57:858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW, Koh JY, Peters S (1988) Pharmacology of glutamate neurotoxicity in cortical cell culture: attenuation by NMDA antagonists. J Neurosci 8:185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW, Maulucci-Gedde M, Kriegstein AR (1987) Glutamate neurotoxicity in cortical cell culture. J Neurosci 7:357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Lee SH, Chang HL, Son H (2016) Hippocampal VEGF is necessary for antidepressant-like behaviors but not sufficient for antidepressant-like effects of ketamine in rats. Biochim Biophys Acta 1862:1247–1254. [DOI] [PubMed] [Google Scholar]
- Choi UB, Kazi R, Stenzoski N, Wollmuth LP, Uversky VN, Bowen ME (2013) Modulating the intrinsic disorder in the cytoplasmic domain alters the biological activity of the N-methyl-D-aspartate-sensitive glutamate receptor. J Biol Chem 288:22506–22515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YB, Lipton SA (1999) Identification and mechanism of action of two histidine residues underlying high-affinity Zn2+ inhibition of the NMDA receptor. Neuron 23:171–180. [DOI] [PubMed] [Google Scholar]
- Chopra DA, Monaghan DT, Dravid SM (2015) Bidirectional effect of pregnenolone sulfate on GluN1/GluN2A N-methyl-D-aspartate receptor gating depending on extracellular calcium and intracellular milieu. Mol Pharmacol 88:650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra DA, Sapkota K, Irvine MW, Fang G, Jane DE, Monaghan DT, Dravid SM (2017) A single-channel mechanism for pharmacological potentiation of GluN1/GluN2A NMDA receptors. Sci Rep 7:6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet D, Hosy E (2020) AMPA receptor nanoscale dynamic organization and synaptic plasticities. Curr Opin Neurobiol 63:137–145. [DOI] [PubMed] [Google Scholar]
- Chou TH, Tajima N, Romero-Hernandez A, Furukawa H (2020) Structural basis of functional transitions in mammalian NMDA receptors. Cell 182:357–371.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Marco S, Brooks IM, Zandueta A, Rao Y, Haucke V, Wesseling JF, Tavalin SJ, Pérez-Otaño I (2013) Tyrosine phosphorylation regulates the endocytosis and surface expression of GluN3A-containing NMDA receptors. J Neurosci 33:4151–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JK, Paternain AV, Selak S, Ahring PK, Lerma J (2004) A mosaic of functional kainate receptors in hippocampal interneurons. J Neurosci 24:8986–8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen PC, Samadi-Bahrami Z, Pavlov V, Stys PK, Moore GRW (2016) Ionotropic glutamate receptor expression in human white matter. Neurosci Lett 630:1–8. [DOI] [PubMed] [Google Scholar]
- Christensen RN, Ha BK, Sun F, Bresnahan JC, Beattie MS (2006) Kainate induces rapid redistribution of the actin cytoskeleton in ameboid microglia. J Neurosci Res 84:170–181. [DOI] [PubMed] [Google Scholar]
- Christie JM, Jahr CE (2009) Selective expression of ligand-gated ion channels in L5 pyramidal cell axons. J Neurosci 29:11441–11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chue P (2013) Glycine reuptake inhibition as a new therapeutic approach in schizophrenia: focus on the glycine transporter 1 (GlyT1). Curr Pharm Des 19:1311–1320. [DOI] [PubMed] [Google Scholar]
- Church J, Fletcher EJ, Baxter K, MacDonald JF (1994) Blockade by ifenprodil of high voltage-activated Ca2+ channels in rat and mouse cultured hippocampal pyramidal neurones: comparison with N-methyl-D-aspartate receptor antagonist actions. Br J Pharmacol 113:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chvojkova M, Rambousek L, Chodounska H, Kudova E, Vales K (2020) Synthetic structural modifications of neurosteroid pregnanolone sulfate: Assessment of neuroprotective effects in vivo. Eur J Pharmacol 881:173187. [DOI] [PubMed] [Google Scholar]
- Ciabarra AM, Sullivan JM, Gahn LG, Pecht G, Heinemann S, Sevarino KA (1995) Cloning and characterization of chi-1: a developmentally regulated member of a novel class of the ionotropic glutamate receptor family. J Neurosci 15:6498–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Malenka RC (2008) Synaptic plasticity: multiple forms, functions, and mechanisms. Neuropsychopharmacology 33:18–41. [DOI] [PubMed] [Google Scholar]
- Claiborne CFMcCauley JALibby BECurtis NRDiggle HJKulagowski JJMichelson SRAnderson KDClaremon DAFreidinger RM, et al. (2003) Orally efficacious NR2B-selective NMDA receptor antagonists. Bioorg Med Chem Lett 13:697–700. [DOI] [PubMed] [Google Scholar]
- Clark BA, Cull-Candy SG (2002) Activity-dependent recruitment of extrasynaptic NMDA receptor activation at an AMPA receptor-only synapse. J Neurosci 22:4428–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BA, Farrant M, Cull-Candy SG (1997) A direct comparison of the single-channel properties of synaptic and extrasynaptic NMDA receptors. J Neurosci 17:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GD, Clifford DB, Zorumski CF (1990) The effect of agonist concentration, membrane voltage and calcium on N-methyl-D-aspartate receptor desensitization. Neuroscience 39:787–797. [DOI] [PubMed] [Google Scholar]
- Clarke RJ, Glasgow NG, Johnson JW (2013) Mechanistic and structural determinants of NMDA receptor voltage-dependent gating and slow Mg2+ unblock. J Neurosci 33:4140–4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RJ, Johnson JW (2006) NMDA receptor NR2 subunit dependence of the slow component of magnesium unblock. J Neurosci 26:5825–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RJ, Johnson JW (2008) Voltage-dependent gating of NR1/2B NMDA receptors. J Physiol 586:5727–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke VRBallyk BAHoo KHMandelzys APellizzari ABath CPThomas JSharpe EFDavies CHOrnstein PL, et al. (1997) A hippocampal GluR5 kainate receptor regulating inhibitory synaptic transmission. Nature 389:599–603. [DOI] [PubMed] [Google Scholar]
- Clarke VR, Molchanova SM, Hirvonen T, Taira T, Lauri SE (2014) Activity-dependent upregulation of presynaptic kainate receptors at immature CA3-CA1 synapses. J Neurosci 34:16902–16916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen RPChristensen CHansen KBGreenwood JRJørgensen LMicale NMadsen JCNielsen BEgebjerg JBräuner-Osborne H, et al. (2008) N-Hydroxypyrazolyl glycine derivatives as selective N-methyl-D-aspartic acid receptor ligands. J Med Chem 51:4179–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen RP, Hansen KB, Calí P, Nielsen B, Greenwood JR, Begtrup M, Egebjerg J, Bräuner-Osborne H (2004) The respective N-hydroxypyrazole analogues of the classical glutamate receptor ligands ibotenic acid and (RS)-2-amino-2-(3-hydroxy-5-methyl-4-isoxazolyl)acetic acid. Eur J Pharmacol 499:35–44. [DOI] [PubMed] [Google Scholar]
- Clausen RPNaur PKristensen ASGreenwood JRStrange MBräuner-Osborne HJensen AANielsen ASGeneser URinggaard LM, et al. (2009) The glutamate receptor GluR5 agonist (S)-2-amino-3-(3-hydroxy-7,8-dihydro-6H-cyclohepta[d]isoxazol-4-yl)propionic acid and the 8-methyl analogue: synthesis, molecular pharmacology, and biostructural characterization. J Med Chem 52:4911–4922. [DOI] [PubMed] [Google Scholar]
- Clayton DA, Mesches MH, Alvarez E, Bickford PC, Browning MD (2002) A hippocampal NR2B deficit can mimic age-related changes in long-term potentiation and spatial learning in the Fischer 344 rat. J Neurosci 22:3628–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RL, Huganir RL (2010) Calcium-permeable AMPA receptor dynamics mediate fear memory erasure. Science 330:1108–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements JD, Lester RA, Tong G, Jahr CE, Westbrook GL (1992) The time course of glutamate in the synaptic cleft. Science 258:1498–1501. [DOI] [PubMed] [Google Scholar]
- Cline HT, Constantine-Paton M (1989) NMDA receptor antagonists disrupt the retinotectal topographic map. Neuron 3:413–426. [DOI] [PubMed] [Google Scholar]
- Cline HT, Debski EA, Constantine-Paton M (1987) N-methyl-D-aspartate receptor antagonist desegregates eye-specific stripes. Proc Natl Acad Sci USA 84:4342–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Kashi Malina K, Ganor Y, Levite M, Teichberg VI (2006) Autoantibodies against an extracellular peptide of the GluR3 subtype of AMPA receptors activate both homomeric and heteromeric AMPA receptor channels. Neurochem Res 31:1181–1190. [DOI] [PubMed] [Google Scholar]
- Coleman J, Jouannot O, Ramakrishnan SK, Zanetti MN, Wang J, Salpietro V, Houlden H, Rothman JE, Krishnakumar SS (2018) PRRT2 regulates synaptic fusion by directly modulating SNARE complex assembly. Cell Rep 22:820–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SK, Möykkynen T, Cai C, von Ossowski L, Kuismanen E, Korpi ER, Keinänen K (2006) Isoform-specific early trafficking of AMPA receptor flip and flop variants. J Neurosci 26:11220–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge GL, Kehl SJ, McLennan H (1983) Excitatory amino acids in synaptic transmission in the Schaffer collateral-commissural pathway of the rat hippocampus. J Physiol 334:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge GL, Lester RA (1989) Excitatory amino acid receptors in the vertebrate central nervous system. Pharmacol Rev 41:143–210. [PubMed] [Google Scholar]
- Collingridge GL, Peineau S, Howland JG, Wang YT (2010) Long-term depression in the CNS. Nat Rev Neurosci 11:459–473. [DOI] [PubMed] [Google Scholar]
- Colonnese MT, Constantine-Paton M (2006) Developmental period for N-methyl-D-aspartate (NMDA) receptor-dependent synapse elimination correlated with visuotopic map refinement. J Comp Neurol 494:738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D (1998) Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol 125:924–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D (2007) Why the Schild method is better than Schild realised. Trends Pharmacol Sci 28:608–614. [DOI] [PubMed] [Google Scholar]
- Constals APenn ACCompans BToulmé EPhillipat AMarais SRetailleau NHafner ASCoussen FHosy E, et al. (2015) Glutamate-induced AMPA receptor desensitization increases their mobility and modulates short-term plasticity through unbinding from Stargazin. Neuron 85:787–803. [DOI] [PubMed] [Google Scholar]
- Conti F, Minelli A, Brecha NC (1994) Cellular localization and laminar distribution of AMPA glutamate receptor subunits mRNAs and proteins in the rat cerebral cortex. J Comp Neurol 350:241–259. [DOI] [PubMed] [Google Scholar]
- Conti P, De Amici M, De Sarro G, Rizzo M, Stensbøl TB, Bräuner-Osborne H, Madsen U, Toma L, De Micheli C (1999) Synthesis and enantiopharmacology of new AMPA-kainate receptor agonists. J Med Chem 42:4099–4107. [DOI] [PubMed] [Google Scholar]
- Conti PPinto ATamborini LMadsen UNielsen BBräuner-Osborne HHansen KBLanducci EPellegrini-Giampietro DEDe Sarro G, et al. (2010) Novel 3-carboxy- and 3-phosphonopyrazoline amino acids as potent and selective NMDA receptor antagonists: design, synthesis, and pharmacological characterization. ChemMedChem 5:1465–1475. [DOI] [PubMed] [Google Scholar]
- Contractor A, Mulle C, Swanson GT (2011) Kainate receptors coming of age: milestones of two decades of research. Trends Neurosci 34:154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Sailer AW, Darstein M, Maron C, Xu J, Swanson GT, Heinemann SF (2003) Loss of kainate receptor-mediated heterosynaptic facilitation of mossy-fiber synapses in KA2-/- mice. J Neurosci 23:422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Swanson G, Heinemann SF (2001) Kainate receptors are involved in short- and long-term plasticity at mossy fiber synapses in the hippocampus. Neuron 29:209–216. [DOI] [PubMed] [Google Scholar]
- Contractor A, Swanson GT, Sailer A, O’Gorman S, Heinemann SF (2000) Identification of the kainate receptor subunits underlying modulation of excitatory synaptic transmission in the CA3 region of the hippocampus. J Neurosci 20:8269–8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs ID, MacLean DM, Jayaraman V, Farrant M, Cull-Candy SG (2017) Dual effects of TARP γ-2 on glutamate efficacy can account for AMPA receptor autoinactivation. Cell Rep 20:1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs ID, Soto D, McGee TP, Gold MG, Farrant M, Cull-Candy SG (2019) Homomeric GluA2(R) AMPA receptors can conduct when desensitized. Nat Commun 10:4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs ID, Soto D, Zonouzi M, Renzi M, Shelley C, Farrant M, Cull-Candy SG (2012) Cornichons modify channel properties of recombinant and glial AMPA receptors. J Neurosci 32:9796–9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LN, Bear MF (2012) The BCM theory of synapse modification at 30: interaction of theory with experiment. Nat Rev Neurosci 13:798–810. [DOI] [PubMed] [Google Scholar]
- Copits BA, Robbins JS, Frausto S, Swanson GT (2011) Synaptic targeting and functional modulation of GluK1 kainate receptors by the auxiliary neuropilin and tolloid-like (NETO) proteins. J Neurosci 31:7334–7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copits BA, Swanson GT (2012) Dancing partners at the synapse: auxiliary subunits that shape kainate receptor function. Nat Rev Neurosci 13:675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copits BA, Vernon CG, Sakai R, Swanson GT (2014) Modulation of ionotropic glutamate receptor function by vertebrate galectins. J Physiol 592:2079–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquelle T, Christensen JK, Banke TG, Madsen U, Schousboe A, Pickering DS (2000) Agonist discrimination between AMPA receptor subtypes. Neuroreport 11:2643–2648. [DOI] [PubMed] [Google Scholar]
- Córdoba M, Rodriguez S, González Morón D, Medina N, Kauffman MA (2015) Expanding the spectrum of Grik2 mutations: intellectual disability, behavioural disorder, epilepsy and dystonia. Clin Genet 87:293–295. [DOI] [PubMed] [Google Scholar]
- Corlew R, Brasier DJ, Feldman DE, Philpot BD (2008) Presynaptic NMDA receptors: newly appreciated roles in cortical synaptic function and plasticity. Neuroscientist 14:609–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corlew R, Wang Y, Ghermazien H, Erisir A, Philpot BD (2007) Developmental switch in the contribution of presynaptic and postsynaptic NMDA receptors to long-term depression. J Neurosci 27:9835–9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornwell BR, Salvadore G, Furey M, Marquardt CA, Brutsche NE, Grillon C, Zarate CA Jr (2012) Synaptic potentiation is critical for rapid antidepressant response to ketamine in treatment-resistant major depression. Biol Psychiatry 72:555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia SS, Duarte CB, Faro CJ, Pires EV, Carvalho AL (2003) Protein kinase C gamma associates directly with the GluR4 alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate receptor subunit. Effect on receptor phosphorylation. J Biol Chem 278:6307–6313. [DOI] [PubMed] [Google Scholar]
- Cossart R, Epsztein J, Tyzio R, Becq H, Hirsch J, Ben-Ari Y, Crépel V (2002) Quantal release of glutamate generates pure kainate and mixed AMPA/kainate EPSCs in hippocampal neurons. Neuron 35:147–159. [DOI] [PubMed] [Google Scholar]
- Cossart R, Esclapez M, Hirsch JC, Bernard C, Ben-Ari Y (1998) GluR5 kainate receptor activation in interneurons increases tonic inhibition of pyramidal cells. Nat Neurosci 1:470–478. [DOI] [PubMed] [Google Scholar]
- Costa BM, Feng B, Tsintsadze TS, Morley RM, Irvine MW, Tsintsadze V, Lozovaya NA, Jane DE, Monaghan DT (2009) N-methyl-D-aspartate (NMDA) receptor NR2 subunit selectivity of a series of novel piperazine-2,3-dicarboxylate derivatives: preferential blockade of extrasynaptic NMDA receptors in the rat hippocampal CA3-CA1 synapse. J Pharmacol Exp Ther 331:618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa BM, Irvine MW, Fang G, Eaves RJ, Mayo-Martin MB, Skifter DA, Jane DE, Monaghan DT (2010) A novel family of negative and positive allosteric modulators of NMDA receptors. J Pharmacol Exp Ther 335:614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Scholz J, Woolf CJ (2009) Neuropathic pain: a maladaptive response of the nervous system to damage. Annu Rev Neurosci 32:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell JR, Dubé GR, Egles C, Liu G (2000) Distribution, density, and clustering of functional glutamate receptors before and after synaptogenesis in hippocampal neurons. J Neurophysiol 84:1573–1587. [DOI] [PubMed] [Google Scholar]
- Coultrap SJ, Freund RK, O’Leary H, Sanderson JL, Roche KW, Dell’Acqua ML, Bayer KU (2014) Autonomous CaMKII mediates both LTP and LTD using a mechanism for differential substrate site selection. Cell Rep 6:431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins SL, Innocent N, Stephenson FA (2013) Neto1 associates with the NMDA receptor/amyloid precursor protein complex. J Neurochem 126:554–564. [DOI] [PubMed] [Google Scholar]
- Coussen F (2009) Molecular determinants of kainate receptor trafficking. Neuroscience 158:25–35. [DOI] [PubMed] [Google Scholar]
- Coutelier MBurglen LMundwiller EAbada-Bendib MRodriguez DChantot-Bastaraud SRougeot CCournelle MAMilh MToutain A, et al. (2015) GRID2 mutations span from congenital to mild adult-onset cerebellar ataxia. Neurology 84:1751–1759. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Balu D, Wolosker H (2020) D-Serine, the shape-shifting NMDA receptor co-agonist. Neurochem Res 45:1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree JW, Lodge D, Bashir ZI, Isaac JT (2013) GABAA, NMDA and mGlu2 receptors tonically regulate inhibition and excitation in the thalamic reticular nucleus. Eur J Neurosci 37:850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creed M, Kaufling J, Fois GR, Jalabert M, Yuan T, Lüscher C, Georges F, Bellone C (2016) Cocaine exposure enhances the activity of ventral tegmental area dopamine neurons via calcium-impermeable NMDARs. J Neurosci 36:10759–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crépel V, Mulle C (2015) Physiopathology of kainate receptors in epilepsy. Curr Opin Pharmacol 20:83–88. [DOI] [PubMed] [Google Scholar]
- Cruz MP (2013) Nuedexta for the treatment of pseudobulbar affect: a condition of involuntary crying or laughing. P&T 38:325–328. [PMC free article] [PubMed] [Google Scholar]
- Cserép C, Szabadits E, Szőnyi A, Watanabe M, Freund TF, Nyiri G (2012) NMDA receptors in GABAergic synapses during postnatal development. PLoS One 7:e37753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cueva Vargas JL, Osswald IK, Unsain N, Aurousseau MR, Barker PA, Bowie D, Di Polo A (2015) Soluble tumor necrosis factor alpha promotes retinal ganglion cell death in glaucoma via calcium-permeable AMPA receptor activation. J Neurosci 35:12088–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Mayer ML (1999) Heteromeric kainate receptors formed by the coassembly of GluR5, GluR6, and GluR7. J Neurosci 19:8281–8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy S, Kelly L, Farrant M (2006) Regulation of Ca2+-permeable AMPA receptors: synaptic plasticity and beyond. Curr Opin Neurobiol 16:288–297. [DOI] [PubMed] [Google Scholar]
- Cull-Candy SG, Howe JR, Ogden DC (1988) Noise and single channels activated by excitatory amino acids in rat cerebellar granule neurones. J Physiol 400:189–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy SG, Usowicz MM (1987) Multiple-conductance channels activated by excitatory amino acids in cerebellar neurons. Nature 325:525–528. [DOI] [PubMed] [Google Scholar]
- Cummings KA, Belin S, Popescu GK (2017) Residues in the GluN1 C-terminal domain control kinetics and pharmacology of GluN1/GluN3A N-methyl-d-aspartate receptors. Neuropharmacology 119:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings KA, Popescu GK (2016) Protons potentiate GluN1/GluN3A currents by attenuating their desensitisation. Sci Rep 6:23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha RA, Malva JO, Ribeiro JA (1999) Kainate receptors coupled to G(i)/G(o) proteins in the rat hippocampus. Mol Pharmacol 56:429–433. [DOI] [PubMed] [Google Scholar]
- Cunha RA, Malva JO, Ribeiro JA (2000) Pertussis toxin prevents presynaptic inhibition by kainate receptors of rat hippocampal [(3)H]GABA release. FEBS Lett 469:159–162. [DOI] [PubMed] [Google Scholar]
- Cunningham MO, Pervouchine DD, Racca C, Kopell NJ, Davies CH, Jones RS, Traub RD, Whittington MA (2006) Neuronal metabolism governs cortical network response state. Proc Natl Acad Sci USA 103:5597–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio L, Podda MV, Leone L, Piacentini R, Mastrodonato A, Cappelletti P, Sacchi S, Pollegioni L, Grassi C, D’Ascenzo M (2013) Reduced D-serine levels in the nucleus accumbens of cocaine-treated rats hinder the induction of NMDA receptor-dependent synaptic plasticity. Brain 136:1216–1230. [DOI] [PubMed] [Google Scholar]
- Curcio M, Salazar IL, Mele M, Canzoniero LM, Duarte CB (2016) Calpains and neuronal damage in the ischemic brain: The swiss knife in synaptic injury. Prog Neurobiol 143:1–35. [DOI] [PubMed] [Google Scholar]
- D’Ascenzo M, Fellin T, Terunuma M, Revilla-Sanchez R, Meaney DF, Auberson YP, Moss SJ, Haydon PG (2007) mGluR5 stimulates gliotransmission in the nucleus accumbens. Proc Natl Acad Sci USA 104:1995–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza DC, Carson RE, Driesen N, Johannesen J, Ranganathan M, Krystal JH; Yale GlyT1 Study Group (2018) Dose-related target occupancy and effects on circuitry, behavior, and neuroplasticity of the glycine transporter-1 inhibitor PF-03463275 in healthy and schizophrenia subjects. Biol Psychiatry 84:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza DCGil RCassello KMorrissey KAbi-Saab DWhite JSturwold RBennett AKarper LPZuzarte E, et al. (2000) IV glycine and oral D-cycloserine effects on plasma and CSF amino acids in healthy humans. Biol Psychiatry 47:450–462. [DOI] [PubMed] [Google Scholar]
- Dadak S, Bouquier N, Goyet E, Fagni L, Levenes C, Perroy J (2017) mGlu1 receptor canonical signaling pathway contributes to the opening of the orphan GluD2 receptor. Neuropharmacology 115:92–99. [DOI] [PubMed] [Google Scholar]
- Dai J, Patzke C, Liakath-Ali K, Seigneur E, Südhof TC (2021) GluD1 is a signal transduction device disguised as an ionotropic receptor. Nature 595:261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Wollmuth LP, Zhou HX (2015) Mechanism-based mathematical model for gating of ionotropic glutamate receptors. J Phys Chem B 119:10934–10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Zhou HX (2015) Reduced curvature of ligand-binding domain free-energy surface underlies partial agonism at NMDA receptors. Structure 23:228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Zhou HX (2016) Semiclosed conformations of the ligand-binding domains of NMDA receptors during stationary gating. Biophys J 111:1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmau J, Geis C, Graus F (2017) Autoantibodies to synaptic receptors and neuronal cell surface proteins in autoimmune diseases of the central nervous system. Physiol Rev 97:839–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmau J, Gleichman AJ, Hughes EG, Rossi JE, Peng X, Lai M, Dessain SK, Rosenfeld MR, Balice-Gordon R, Lynch DR (2008) Anti-NMDA-receptor encephalitis: case series and analysis of the effects of antibodies. Lancet Neurol 7:1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton GL, Wu DC, Wang YT, Floresco SB, Phillips AG (2012) NMDA GluN2A and GluN2B receptors play separate roles in the induction of LTP and LTD in the amygdala and in the acquisition and extinction of conditioned fear. Neuropharmacology 62:797–806. [DOI] [PubMed] [Google Scholar]
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME (2000) EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell 103:945–956. [DOI] [PubMed] [Google Scholar]
- Dan Y, Poo MM (2004) Spike timing-dependent plasticity of neural circuits. Neuron 44:23–30. [DOI] [PubMed] [Google Scholar]
- Dani A, Huang B, Bergan J, Dulac C, Zhuang X (2010) Superresolution imaging of chemical synapses in the brain. Neuron 68:843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels BA, Andrews ED, Aurousseau MR, Accardi MV, Bowie D (2013) Crosslinking the ligand-binding domain dimer interface locks kainate receptors out of the main open state. J Physiol 591:3873–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargan SLClarke VRAlushin GMSherwood JLNisticò RBortolotto ZAOgden AMBleakman DDoherty AJLodge D, et al. (2009) ACET is a highly potent and specific kainate receptor antagonist: characterisation and effects on hippocampal mossy fibre function. Neuropharmacology 56:121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darstein M, Petralia RS, Swanson GT, Wenthold RJ, Heinemann SF (2003) Distribution of kainate receptor subunits at hippocampal mossy fiber synapses. J Neurosci 23:8013–8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SSasaki YFRothe TPremkumar LSTakasu MCrandall JEDikkes PConner DARayudu PVCheung W, et al. (1998) Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature 393:377–381. [DOI] [PubMed] [Google Scholar]
- Das SR, Jensen R, Kelsay R, Shumaker M, Bochart R, Brim B, Zamzow D, Magnusson KR (2012) Reducing expression of GluN1(0XX) subunit splice variants of the NMDA receptor interferes with spatial reference memory. Behav Brain Res 230:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dau A, Gladding CM, Sepers MD, Raymond LA (2014) Chronic blockade of extrasynaptic NMDA receptors ameliorates synaptic dysfunction and pro-death signaling in Huntington disease transgenic mice. Neurobiol Dis 62:533–542. [DOI] [PubMed] [Google Scholar]
- Davies BBrown LACais OWatson JClayton AJChang VTBiggs DPreece CHernandez-Pliego PKrohn J, et al. ; WGS500 Consortium (2017) A point mutation in the ion conduction pore of AMPA receptor GRIA3 causes dramatically perturbed sleep patterns as well as intellectual disability. Hum Mol Genet 26:3869–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DJCrowe MLucas NQuinn JMiller DDPritchard SGrose DBettini ECalcinaghi NVirginio C, et al. (2012) A novel series of benzimidazole NR2B-selective NMDA receptor antagonists. Bioorg Med Chem Lett 22:2620–2623. [DOI] [PubMed] [Google Scholar]
- Davies J, Evans RH, Jones AW, Smith DA, Watkins JC (1982) Differential activation and blockade of excitatory amino acid receptors in the mammalian and amphibian central nervous systems. Comp Biochem Physiol C Comp Pharmacol 72:211–224. [DOI] [PubMed] [Google Scholar]
- Davies J, Francis AA, Jones AW, Watkins JC (1981) 2-Amino-5-phosphonovalerate (2APV), a potent and selective antagonist of amino acid-induced and synaptic excitation. Neurosci Lett 21:77–81. [DOI] [PubMed] [Google Scholar]
- Davis M, Ressler K, Rothbaum BO, Richardson R (2006) Effects of D-cycloserine on extinction: translation from preclinical to clinical work. Biol Psychiatry 60:369–375. [DOI] [PubMed] [Google Scholar]
- Daw MI, Pelkey KA, Chittajallu R, McBain CJ (2010) Presynaptic kainate receptor activation preserves asynchronous GABA release despite the reduction in synchronous release from hippocampal cholecystokinin interneurons. J Neurosci 30:11202–11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe GB, Aurousseau MR, Daniels BA, Bowie D (2015) Retour aux sources: defining the structural basis of glutamate receptor activation. J Physiol 593:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe GBKadir MFVenskutonytė RPerozzo AMYan YAlexander RPDNavarrete CSantander EAArsenault MFuentes C, et al. (2019) Nanoscale mobility of the apo state and TARP stoichiometry dictate the gating behavior of alternatively spliced AMPA receptors. Neuron 102:976–992.e5. [DOI] [PubMed] [Google Scholar]
- Dawe GB, Musgaard M, Andrews ED, Daniels BA, Aurousseau MR, Biggin PC, Bowie D (2013) Defining the structural relationship between kainate-receptor deactivation and desensitization. Nat Struct Mol Biol 20:1054–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe GB, Musgaard M, Aurousseau MRP, Nayeem N, Green T, Biggin PC, Bowie D (2016) Distinct structural pathways coordinate the activation of AMPA receptor-auxiliary subunit complexes. Neuron 89:1264–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco García NV, Karayannis T, Fishell G (2011) Neuronal activity is required for the development of specific cortical interneuron subtypes. Nature 472:351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco García NV, Priya R, Tuncdemir SN, Fishell G, Karayannis T (2015) Sensory inputs control the integration of neurogliaform interneurons into cortical circuits. Nat Neurosci 18:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGiorgio LA, Konstantinov KN, Lee SC, Hardin JA, Volpe BT, Diamond B (2001) A subset of lupus anti-DNA antibodies cross-reacts with the NR2 glutamate receptor in systemic lupus erythematosus. Nat Med 7:1189–1193. [DOI] [PubMed] [Google Scholar]
- Delaney AJ, Jahr CE (2002) Kainate receptors differentially regulate release at two parallel fiber synapses. Neuron 36:475–482. [DOI] [PubMed] [Google Scholar]
- Delaney AJ, Power JM, Sah P (2012) Ifenprodil reduces excitatory synaptic transmission by blocking presynaptic P/Q type calcium channels. J Neurophysiol 107:1571–1575. [DOI] [PubMed] [Google Scholar]
- Demmer CS, Møller C, Brown PM, Han L, Pickering DS, Nielsen B, Bowie D, Frydenvang K, Kastrup JS, Bunch L (2015) Binding mode of an α-amino acid-linked quinoxaline-2,3-dione analogue at glutamate receptor subtype GluK1. ACS Chem Neurosci 6:845–854. [DOI] [PubMed] [Google Scholar]
- Demmer CS, Rombach D, Liu N, Nielsen B, Pickering DS, Bunch L (2017) Revisiting the quinoxalinedione scaffold in the construction of new ligands for the ionotropic glutamate receptors. ACS Chem Neurosci 8:2477–2495. [DOI] [PubMed] [Google Scholar]
- Derkach V, Barria A, Soderling TR (1999) Ca2+/calmodulin-kinase II enhances channel conductance of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate type glutamate receptors. Proc Natl Acad Sci USA 96:3269–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkach VA (2003) Silence analysis of AMPA receptor mutated at the CaM-kinase II phosphorylation site. Biophys J 84:1701–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descalzi G, Chen T, Koga K, Li XY, Yamada K, Zhuo M (2013) Cortical GluK1 kainate receptors modulate scratching in adult mice. J Neurochem 126:636–650. [DOI] [PubMed] [Google Scholar]
- Dev KK, Nishimune A, Henley JM, Nakanishi S (1999) The protein kinase C alpha binding protein PICK1 interacts with short but not long form alternative splice variants of AMPA receptor subunits. Neuropharmacology 38:635–644. [DOI] [PubMed] [Google Scholar]
- Devi SPS, Cheng Y, Tomita S, Howe JR, Zhang W (2020) TARPs modulate receptor-mediated paired-pulse depression and recovery from desensitization. J Neurosci 40:8233–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries SH (2000) Bipolar cells use kainate and AMPA receptors to filter visual information into separate channels. Neuron 28:847–856. [DOI] [PubMed] [Google Scholar]
- DeVries SH (2001) Exocytosed protons feedback to suppress the Ca2+ current in mammalian cone photoreceptors. Neuron 32:1107–1117. [DOI] [PubMed] [Google Scholar]
- DeVries SH, Schwartz EA (1999) Kainate receptors mediate synaptic transmission between cones and ‘Off’ bipolar cells in a mammalian retina. Nature 397:157–160. [DOI] [PubMed] [Google Scholar]
- Dey S, Schepmann D, Wünsch B (2018) 2-Methyltetrahydro-3-benzazepin-1-ols - The missing link in SAR of GluN2B selective NMDA receptor antagonists. Bioorg Med Chem 26:501–508. [DOI] [PubMed] [Google Scholar]
- Di Benedetto DMusumeci SAAvola EAlberti ABuono SScuderi CGrillo LGalesi OSpalletta AGiudice ML, et al. (2014) Definition of minimal duplicated region encompassing the XIAP and STAG2 genes in the Xq25 microduplication syndrome. Am J Med Genet A 164A:1923–1930. [DOI] [PubMed] [Google Scholar]
- Diamond JS (2005) Deriving the glutamate clearance time course from transporter currents in CA1 hippocampal astrocytes: transmitter uptake gets faster during development. J Neurosci 25:2906–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond PR, Farmery AD, Atkinson S, Haldar J, Williams N, Cowen PJ, Geddes JR, McShane R (2014) Ketamine infusions for treatment resistant depression: a series of 28 patients treated weekly or twice weekly in an ECT clinic. J Psychopharmacol 28:536–544. [DOI] [PubMed] [Google Scholar]
- Díaz E, Ge Y, Yang YH, Loh KC, Serafini TA, Okazaki Y, Hayashizaki Y, Speed TP, Ngai J, Scheiffele P (2002) Molecular analysis of gene expression in the developing pontocerebellar projection system. Neuron 36:417–434. [DOI] [PubMed] [Google Scholar]
- Díaz-Alonso J, Morishita W, Incontro S, Simms J, Holtzman J, Gill M, Mucke L, Malenka RC, Nicoll RA (2020) Long-term potentiation is independent of the C-tail of the GluA1 AMPA receptor subunit. eLife 9:e58042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Alonso J, Sun YJ, Granger AJ, Levy JM, Blankenship SM, Nicoll RA (2017) Subunit-specific role for the amino-terminal domain of AMPA receptors in synaptic targeting. Proc Natl Acad Sci USA 114:7136–7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson R, Peterson BK, Banks P, Simillis C, Martin JC, Valenzuela CA, Maze M, Franks NP (2007) Competitive inhibition at the glycine site of the N-methyl-D-aspartate receptor by the anesthetics xenon and isoflurane: evidence from molecular modeling and electrophysiology. Anesthesiology 107:756–767. [DOI] [PubMed] [Google Scholar]
- Diering GH, Heo S, Hussain NK, Liu B, Huganir RL (2016) Extensive phosphorylation of AMPA receptors in neurons. Proc Natl Acad Sci USA 113:E4920–E4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diering GH, Huganir RL (2018) The AMPA receptor code of synaptic plasticity. Neuron 100:314–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digby RJ, Bravo DS, Paulsen O, Magloire V (2017) Distinct mechanisms of Up state maintenance in the medial entorhinal cortex and neocortex. Neuropharmacology 113 (Pt A):543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF (1999) The glutamate receptor ion channels. Pharmacol Rev 51:7–61. [PubMed] [Google Scholar]
- Dingledine R, Hume RI, Heinemann SF (1992) Structural determinants of barium permeation and rectification in non-NMDA glutamate receptor channels. J Neurosci 12:4080–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer PJ, Dell’Acqua ML, Sather WA (2019) Synaptic crosstalk conferred by a zone of differentially regulated Ca2+ signaling in the dendritic shaft adjoining a potentiated spine. Proc Natl Acad Sci USA 116:13611–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doble A (1999) The role of excitotoxicity in neurodegenerative disease: implications for therapy. Pharmacol Ther 81:163–221. [DOI] [PubMed] [Google Scholar]
- Dodds DC, Omeis IA, Cushman SJ, Helms JA, Perin MS (1997) Neuronal pentraxin receptor, a novel putative integral membrane pentraxin that interacts with neuronal pentraxin 1 and 2 and taipoxin-associated calcium-binding protein 49. J Biol Chem 272:21488–21494. [DOI] [PubMed] [Google Scholar]
- Dohrke JN, Watson JF, Birchall K, Greger IH (2020) Characterizing the binding and function of TARP γ8-selective AMPA receptor modulators. J Biol Chem 295:14565–14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolino DM, Chatterjee S, MacLean DM, Flatebo C, Bishop LDC, Shaikh SA, Landes CF, Jayaraman V (2017) The structure-energy landscape of NMDA receptor gating. Nat Chem Biol 13:1232–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolino DM, Cooper D, Ramaswamy S, Jaurich H, Landes CF, Jayaraman V (2015) Structural dynamics of the glycine-binding domain of the N-methyl-D-aspartate receptor. J Biol Chem 290:797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolino DM, Rezaei Adariani S, Shaikh SA, Jayaraman V, Sanabria H (2016) Conformational selection and submillisecond dynamics of the ligand-binding domain of the N-methyl-d-aspartate receptor. J Biol Chem 291:16175–16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolman NP, More JC, Alt A, Knauss JL, Pentikäinen OT, Glasser CR, Bleakman D, Mayer ML, Collingridge GL, Jane DE (2007) Synthesis and pharmacological characterization of N3-substituted willardiine derivatives: role of the substituent at the 5-position of the uracil ring in the development of highly potent and selective GLUK5 kainate receptor antagonists. J Med Chem 50:1558–1570. [DOI] [PubMed] [Google Scholar]
- Dolman NP, More JC, Alt A, Knauss JL, Troop HM, Bleakman D, Collingridge GL, Jane DE (2006) Structure-activity relationship studies on N3-substituted willardiine derivatives acting as AMPA or kainate receptor antagonists. J Med Chem 49:2579–2592. [DOI] [PubMed] [Google Scholar]
- Dolman NPTroop HMMore JCAlt AKnauss JLNistico RJack SMorley RMBortolotto ZARoberts PJ, et al. (2005) Synthesis and pharmacology of willardiine derivatives acting as antagonists of kainate receptors. J Med Chem 48:7867–7881. [DOI] [PubMed] [Google Scholar]
- Domingues AM, Neugebauer KM, Fern R (2011) Identification of four functional NR3B isoforms in developing white matter reveals unexpected diversity among glutamate receptors. J Neurochem 117:449–460. [DOI] [PubMed] [Google Scholar]
- Donello JEBanerjee PLi YXGuo YXYoshitake TZhang XLMiry OKehr JStanton PKGross AL, et al. (2019) Positive N-methyl-D-aspartate receptor modulation by rapastinel promotes rapid and sustained antidepressant-like effects. Int J Neuropsychopharmacol 22:247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donevan SD, Rogawski MA (1993) GYKI 52466, a 2,3-benzodiazepine, is a highly selective, noncompetitive antagonist of AMPA/kainate receptor responses. Neuron 10:51–59. [DOI] [PubMed] [Google Scholar]
- Dore K, Aow J, Malinow R (2015) Agonist binding to the NMDA receptor drives movement of its cytoplasmic domain without ion flow. Proc Natl Acad Sci USA 112:14705–14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore K, Aow J, Malinow R (2016) The emergence of NMDA receptor metabotropic function: Insights from imaging. Front Synaptic Neurosci 8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore K, Stein IS, Brock JA, Castillo PE, Zito K, Sjöström PJ (2017) Unconventional NMDA receptor signaling. J Neurosci 37:10800–10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S, Hansen DB, Vella J, Bond P, Harper G, Zammit C, Valentino M, Fern R (2018) Vesicular glutamate release from central axons contributes to myelin damage. Nat Commun 9:1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid SM, Burger PB, Prakash A, Geballe MT, Yadav R, Le P, Vellano K, Snyder JP, Traynelis SF (2010) Structural determinants of D-cycloserine efficacy at the NR1/NR2C NMDA receptors. J Neurosci 30:2741–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid SMErreger KYuan HNicholson KLe PLyuboslavsky PAlmonte AMurray EMosely CBarber J, et al. (2007) Subunit-specific mechanisms and proton sensitivity of NMDA receptor channel block. J Physiol 581:107–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid SM, Prakash A, Traynelis SF (2008) Activation of recombinant NR1/NR2C NMDA receptors. J Physiol 586:4425–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan GF, Ye Y, Xu S, Tao W, Zhao S, Jin T, Nicoll RA, Shi YS, Sheng N (2018) Signal peptide represses GluK1 surface and synaptic trafficking through binding to amino-terminal domain. Nat Commun 9:4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois CJ, Lachamp PM, Sun L, Mishina M, Liu SJ (2016) Presynaptic GluN2D receptors detect glutamate spillover and regulate cerebellar GABA release. J Neurophysiol 115:271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguid IC, Smart TG (2004) Retrograde activation of presynaptic NMDA receptors enhances GABA release at cerebellar interneuron-Purkinje cell synapses. Nat Neurosci 7:525–533. [DOI] [PubMed] [Google Scholar]
- Duman RS, Aghajanian GK (2012) Synaptic dysfunction in depression: potential therapeutic targets. Science 338:68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman RS, Li N, Liu RJ, Duric V, Aghajanian G (2012) Signaling pathways underlying the rapid antidepressant actions of ketamine. Neuropharmacology 62:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas TC (2005) Developmental regulation of cognitive abilities: modified composition of a molecular switch turns on associative learning. Prog Neurobiol 76:189–211. [DOI] [PubMed] [Google Scholar]
- Dupuis JPLadépêche LSeth HBard LVarela JMikasova LBouchet DRogemond VHonnorat JHanse E, et al. (2014) Surface dynamics of GluN2B-NMDA receptors controls plasticity of maturing glutamate synapses. EMBO J 33:842–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand GM, Bennett MV, Zukin RS (1993) Splice variants of the N-methyl-D-aspartate receptor NR1 identify domains involved in regulation by polyamines and protein kinase C. Proc Natl Acad Sci USA 90:6731–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand GM, Gregor P, Zheng X, Bennett MV, Uhl GR, Zukin RS (1992) Cloning of an apparent splice variant of the rat N-methyl-D-aspartate receptor NMDAR1 with altered sensitivity to polyamines and activators of protein kinase C. Proc Natl Acad Sci USA 89:9359–9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham RJ, Paudyal N, Carrillo E, Bhatia NK, Maclean DM, Berka V, Dolino DM, Gorfe AA, Jayaraman V (2020) Conformational spread and dynamics in allostery of NMDA receptors. Proc Natl Acad Sci USA 117:3839–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr KL, Chen L, Stein RA, De Zorzi R, Folea IM, Walz T, Mchaourab HS, Gouaux E (2014) Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell 158:778–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Shrivastava IH, Sukumaran M, Greger IH, Bahar I (2012) Comparative dynamics of NMDA- and AMPA-glutamate receptor N-terminal domains. Structure 20:1838–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Das S, Guhathakurta S, Sen B, Sinha S, Chatterjee A, Ghosh S, Ahmed S, Ghosh S, Usha R (2007) Glutamate receptor 6 gene (GluR6 or GRIK2) polymorphisms in the Indian population: a genetic association study on autism spectrum disorder. Cell Mol Neurobiol 27:1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta-Roy R, Rosenmund C, Edelstein SJ, Le Novère N (2015) Ligand-dependent opening of the multiple AMPA receptor conductance states: a concerted model. PLoS One 10:e0116616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzubay JA, Jahr CE (1999) The concentration of synaptically released glutamate outside of the climbing fiber-Purkinje cell synaptic cleft. J Neurosci 19:5265–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman S, McKay S, Macdonald LJ, Samadi M, Livesey MR, Hardingham GE, Wyllie DJ (2012) TCN 201 selectively blocks GluN2A-containing NMDARs in a GluN1 co-agonist dependent but non-competitive manner. Neuropharmacology 63:441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards ACAliev FBierut LJBucholz KKEdenberg HHesselbrock VKramer JKuperman SNurnberger JI JrSchuckit MA, et al. (2012) Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr Genet 22:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egebjerg J, Bettler B, Hermans-Borgmeyer I, Heinemann S (1991) Cloning of a cDNA for a glutamate receptor subunit activated by kainate but not AMPA. Nature 351:745–748. [DOI] [PubMed] [Google Scholar]
- Egebjerg J, Heinemann SF (1993) Ca2+ permeability of unedited and edited versions of the kainate selective glutamate receptor GluR6. Proc Natl Acad Sci USA 90:755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD, Fung ET, O’Brien RJ, Huganir RL (1998) Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. J Neurosci 18:720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD, Zhang S, Bernhadt JP, Huganir RL (1996) Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell 84:745–755. [DOI] [PubMed] [Google Scholar]
- Ehrenreich H (2018) Autoantibodies against N-methyl-d-aspartate receptor 1 in health and disease. Curr Opin Neurol 31:306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gaamouch F, Buisson A, Moustié O, Lemieux M, Labrecque S, Bontempi B, De Koninck P, Nicole O (2012) Interaction between αCaMKII and GluN2B controls ERK-dependent plasticity. J Neurosci 32:10767–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elegheert JKakegawa WClay JEShanks NFBehiels EMatsuda KKohda KMiura ERossmann MMitakidis N, et al. (2016) Structural basis for integration of GluD receptors within synaptic organizer complexes. Science 353:295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elger CE, Hong SB, Brandt C, Mancione L, Han J, Strohmaier C (2017) BGG492 as an adjunctive treatment in patients with partial-onset seizures: A 12-week, randomized, double-blind, placebo-controlled, phase II dose-titration study with an open-label extension. Epilepsia 58:1217–1226. [DOI] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T (1999) Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature 399:66–70. [DOI] [PubMed] [Google Scholar]
- Epplin MP, Mohan A, Harris LD, Zhu Z, Strong KL, Bacsa J, Le P, Menaldino DS, Traynelis SF, Liotta DC (2020) Discovery of dihydropyrrolo[1,2-a]pyrazin-3(4H)-one-based second-generation GluN2C- and GluN2D-selective positive allosteric modulators (PAMs) of the N-methyl-d-aspartate (NMDA) receptor. J Med Chem 63:7569–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztein J, Represa A, Jorquera I, Ben-Ari Y, Crépel V (2005) Recurrent mossy fibers establish aberrant kainate receptor-operated synapses on granule cells from epileptic rats. J Neurosci 25:8229–8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Nilsson A, Samuelsson H, Samuelsson EB, Mo L, Akesson E, Benedikz E, Sundström E (2007a) On the role of NR3A in human NMDA receptors. Physiol Behav 92:54–59. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Samuelsson H, Björklund S, Tortosa E, Avila J, Samuelsson EB, Benedikz E, Sundström E (2010) MAP1B binds to the NMDA receptor subunit NR3A and affects NR3A protein concentrations. Neurosci Lett 475:33–37. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Samuelsson H, Samuelsson EB, Liu L, McKeehan WL, Benedikz E, Sundström E (2007b) The NMDAR subunit NR3A interacts with microtubule-associated protein 1S in the brain. Biochem Biophys Res Commun 361:127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Dravid SM, Banke TG, Wyllie DJ, Traynelis SF (2005a) Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J Physiol 563:345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Geballe MT, Dravid SM, Snyder JP, Wyllie DJ, Traynelis SF (2005b) Mechanism of partial agonism at NMDA receptors for a conformationally restricted glutamate analog. J Neurosci 25:7858–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger KGeballe MTKristensen AChen PEHansen KBLee CJYuan HLe PLyuboslavsky PNMicale N, et al. (2007) Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-D-aspartate glutamate receptors. Mol Pharmacol 72:907–920. [DOI] [PubMed] [Google Scholar]
- Erreger K, Traynelis SF (2005) Allosteric interaction between zinc and glutamate binding domains on NR2A causes desensitization of NMDA receptors. J Physiol 569:381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Traynelis SF (2008) Zinc inhibition of rat NR1/NR2A N-methyl-D-aspartate receptors. J Physiol 586:763–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmenjaud JB, Stroebel D, Chan K, Grand T, David M, Wollmuth LP, Taly A, Paoletti P (2019) An inter-dimer allosteric switch controls NMDA receptor activity. EMBO J 38:e99894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JS, Wheeler DG, Tsien RW, Luo L (2009) Uncoupling dendrite growth and patterning: single-cell knockout analysis of NMDA receptor 2B. Neuron 62:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AJ, Gurung S, Henley JM, Nakamura Y, Wilkinson KA (2019) Exciting times: New advances towards understanding the regulation and roles of kainate receptors. Neurochem Res 44:572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AJ, Gurung S, Wilkinson KA, Stephens DJ, Henley JM (2017) Assembly, secretory pathway trafficking, and surface delivery of kainate receptors is regulated by neuronal activity. Cell Rep 19:2613–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RH, Francis AA, Jones AW, Smith DA, Watkins JC (1982) The effects of a series of omega-phosphonic alpha-carboxylic amino acids on electrically evoked and excitant amino acid-induced responses in isolated spinal cord preparations. Br J Pharmacol 75:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts I, Petroski R, Kizelsztein P, Teichberg VI, Heinemann SF, Hollmann M (1999) Lectin-induced inhibition of desensitization of the kainate receptor GluR6 depends on the activation state and can be mediated by a single native or ectopic N-linked carbohydrate side chain. J Neurosci 19:916–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber ES (2010) Functional interplay between NMDA receptors, SK channels and voltage-gated Ca2+ channels regulates synaptic excitability in the medial prefrontal cortex. J Physiol 588:1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber ES, Delaney AJ, Sah P (2005) SK channels regulate excitatory synaptic transmission and plasticity in the lateral amygdala. Nat Neurosci 8:635–641. [DOI] [PubMed] [Google Scholar]
- Falcón-Moya R, Losada-Ruiz P, Sihra TS, Rodríguez-Moreno A (2018) Cerebellar kainate receptor-mediated facilitation of glutamate release requires Ca2+-calmodulin and PKA. Front Mol Neurosci 11:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallin MDLasseter VKAvramopoulos DNicodemus KKWolyniec PSMcGrath JASteel GNestadt GLiang KYHuganir RL, et al. (2005) Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet 77:918–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina AN, Blain KY, Maruo T, Kwiatkowski W, Choe S, Nakagawa T (2011) Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. J Neurosci 31:3565–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinella M, Ruedt DT, Gleeson P, Lanore F, Silver RA (2014) Glutamate-bound NMDARs arising from in vivo-like network activity extend spatio-temporal integration in a L5 cortical pyramidal cell model. PLOS Comput Biol 10:e1003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow P, Khodosevich K, Sapir Y, Schulmann A, Aslam M, Stern-Bach Y, Monyer H, von Engelhardt J (2015) Auxiliary subunits of the CKAMP family differentially modulate AMPA receptor properties. eLife 4:e09693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faught E (2014) BGG492 (selurampanel), an AMPA/kainate receptor antagonist drug for epilepsy. Expert Opin Investig Drugs 23:107–113. [DOI] [PubMed] [Google Scholar]
- Faust TWChang EHKowal CBerlin RGazaryan IGBertini EZhang JSanchez-Guerrero JFragoso-Loyo HEVolpe BT, et al. (2010) Neurotoxic lupus autoantibodies alter brain function through two distinct mechanisms. Proc Natl Acad Sci USA 107:18569–18574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava VARSarin LMLucchese ACDel Sant LMagalhães EDelfino RSTuena MANakahira CJackowski APAbdo G, et al. (2020) The probability of response after each subcutaneous injection of esketamine in treatment-resistant depression. Rev Psiquiatr Salud Ment 14:212–214. [DOI] [PubMed] [Google Scholar]
- Favuzzi EDeogracias RMarques-Smith AMaeso PJezequel JExposito-Alonso DBalia MKroon THinojosa AJMaraver EF, et al. (2019) Distinct molecular programs regulate synapse specificity in cortical inhibitory circuits. Science 363:413–417. [DOI] [PubMed] [Google Scholar]
- Fay AM, Bowie D (2006) Concanavalin-A reports agonist-induced conformational changes in the intact GluR6 kainate receptor. J Physiol 572:201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayyazuddin A, Villarroel A, Le Goff A, Lerma J, Neyton J (2000) Four residues of the extracellular N-terminal domain of the NR2A subunit control high-affinity Zn2+ binding to NMDA receptors. Neuron 25:683–694. [DOI] [PubMed] [Google Scholar]
- Fedele L, Newcombe J, Topf M, Gibb A, Harvey RJ, Smart TG (2018) Disease-associated missense mutations in GluN2B subunit alter NMDA receptor ligand binding and ion channel properties. Nat Commun 9:957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder AParides MKMurrough JWPerez AMMorgan JESaxena SKirkwood KAan Het Rot MLapidus KAWan LB, et al. (2014) Efficacy of intravenous ketamine for treatment of chronic posttraumatic stress disorder: a randomized clinical trial. JAMA Psychiatry 71:681–688. [DOI] [PubMed] [Google Scholar]
- Feldman DE (2012) The spike-timing dependence of plasticity. Neuron 75:556–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellin T, Pascual O, Gobbo S, Pozzan T, Haydon PG, Carmignoto G (2004) Neuronal synchrony mediated by astrocytic glutamate through activation of extrasynaptic NMDA receptors. Neuron 43:729–743. [DOI] [PubMed] [Google Scholar]
- Fellin T, Pozzan T, Carmignoto G (2006) Purinergic receptors mediate two distinct glutamate release pathways in hippocampal astrocytes. J Biol Chem 281:4274–4284. [DOI] [PubMed] [Google Scholar]
- Feng B, Morley RM, Jane DE, Monaghan DT (2005) The effect of competitive antagonist chain length on NMDA receptor subunit selectivity. Neuropharmacology 48:354–359. [DOI] [PubMed] [Google Scholar]
- Feng B, Tse HW, Skifter DA, Morley R, Jane DE, Monaghan DT (2004) Structure-activity analysis of a novel NR2C/NR2D-preferring NMDA receptor antagonist: 1-(phenanthrene-2-carbonyl) piperazine-2,3-dicarboxylic acid. Br J Pharmacol 141:508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng ZJ, Zhang X, Chergui K (2014) Allosteric modulation of NMDA receptors alters neurotransmission in the striatum of a mouse model of Parkinson’s disease. Exp Neurol 255:154–160. [DOI] [PubMed] [Google Scholar]
- Fenwick MK, Oswald RE (2010) On the mechanisms of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptor binding to glutamate and kainate. J Biol Chem 285:12334–12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson GD, Wang H, Herschman HR, Storm DR (2004) Altered hippocampal short-term plasticity and associative memory in synaptotagmin IV (-/-) mice. Hippocampus 14:964–974. [DOI] [PubMed] [Google Scholar]
- Fernandes HB, Catches JS, Petralia RS, Copits BA, Xu J, Russell TA, Swanson GT, Contractor A (2009) High-affinity kainate receptor subunits are necessary for ionotropic but not metabotropic signaling. Neuron 63:818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marmiesse A, Kusumoto H, Rekarte S, Roca I, Zhang J, Myers SJ, Traynelis SF, Couce ML, Gutierrez-Solana L, Yuan H (2018) A novel missense mutation in GRIN2A causes a nonepileptic neurodevelopmental disorder. Mov Disord 33:992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez LMJ, Pellegrini C, Vantomme G, Béard E, Lüthi A, Astori S (2017) Cortical afferents onto the nucleus Reticularis thalami promote plasticity of low-threshold excitability through GluN2C-NMDARs. Sci Rep 7:12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini A, Bonini D, La Via L, Barbon A (2017) The good and the bad of glutamate receptor RNA editing. Mol Neurobiol 54:6795–6805. [DOI] [PubMed] [Google Scholar]
- Fisahn A, Heinemann SF, McBain CJ (2005) The kainate receptor subunit GluR6 mediates metabotropic regulation of the slow and medium AHP currents in mouse hippocampal neurones. J Physiol 562:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, Mutel V, Trube G, Malherbe P, Kew JN, Mohacsi E, Heitz MP, Kemp JA (1997) Ro 25-6981, a highly potent and selective blocker of N-methyl-D-aspartate receptors containing the NR2B subunit. Characterization in vitro. J Pharmacol Exp Ther 283:1285–1292. [PubMed] [Google Scholar]
- Fisher JL (2014) The neurotoxin domoate causes long-lasting inhibition of the kainate receptor GluK5 subunit. Neuropharmacology 85:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JL (2015) The auxiliary subunits Neto1 and Neto2 have distinct, subunit-dependent effects at recombinant GluK1- and GluK2-containing kainate receptors. Neuropharmacology 99:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JL, Mott DD (2011) Distinct functional roles of subunits within the heteromeric kainate receptor. J Neurosci 31:17113–17122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JL, Mott DD (2012) The auxiliary subunits Neto1 and Neto2 reduce voltage-dependent inhibition of recombinant kainate receptors. J Neurosci 32:12928–12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JL, Mott DD (2013) Modulation of homomeric and heteromeric kainate receptors by the auxiliary subunit Neto1. J Physiol 591:4711–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MT, Fisher JL (2014) Contributions of different kainate receptor subunits to the properties of recombinant homomeric and heteromeric receptors. Neuroscience 278:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiszman ML, Erdélyi F, Szabó G, Vicini S (2007) Presynaptic AMPA and kainate receptors increase the size of GABAergic terminals and enhance GABA release. Neuropharmacology 52:1631–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiuza M, González-González I, Pérez-Otaño I (2013) GluN3A expression restricts spine maturation via inhibition of GIT1/Rac1 signaling. Proc Natl Acad Sci USA 110:20807–20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TM, Scott V, Naskar K, Joe N, Brown CH, Stern JE (2011) State-dependent changes in astrocyte regulation of extrasynaptic NMDA receptor signalling in neurosecretory neurons. J Physiol 589:3929–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogaça MV, Fukumoto K, Franklin T, Liu RJ, Duman CH, Vitolo OV, Duman RS (2019) N-Methyl-D-aspartate receptor antagonist d-methadone produces rapid, mTORC1-dependent antidepressant effects. Neuropsychopharmacology 44:2230–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest D, Yuzaki M, Soares HD, Ng L, Luk DC, Sheng M, Stewart CL, Morgan JI, Connor JA, Curran T (1994) Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron 13:325–338. [DOI] [PubMed] [Google Scholar]
- Fortune ES, Rose GJ (2002) Roles for short-term synaptic plasticity in behavior. J Physiol Paris 96:539–545. [DOI] [PubMed] [Google Scholar]
- Fossat PTurpin FRSacchi SDulong JShi TRivet JMSweedler JVPollegioni LMillan MJOliet SH, et al. (2012) Glial D-serine gates NMDA receptors at excitatory synapses in prefrontal cortex. Cereb Cortex 22:595–606. [DOI] [PubMed] [Google Scholar]
- Fossati M, Assendorp N, Gemin O, Colasse S, Dingli F, Arras G, Loew D, Charrier C (2019) Trans-synaptic signaling through the glutamate receptor delta-1 mediates inhibitory synapse formation in cortical pyramidal neurons. Neuron 104:1081–1094.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso-Loyo H, Cabiedes J, Orozco-Narváez A, Dávila-Maldonado L, Atisha-Fregoso Y, Diamond B, Llorente L, Sánchez-Guerrero J (2008) Serum and cerebrospinal fluid autoantibodies in patients with neuropsychiatric lupus erythematosus. Implications for diagnosis and pathogenesis. PLoS One 3:e3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen APickering DSVestergaard BKasper CNielsen BBGreenwood JRCampiani GFattorusso CGajhede MSchousboe A, et al. (2005) Tyr702 is an important determinant of agonist binding and domain closure of the ligand-binding core of GluR2. Mol Pharmacol 67:703–713. [DOI] [PubMed] [Google Scholar]
- Frederickson CJGiblin LJ 3rdBalaji RVMasalha RFrederickson CJZeng YLopez EVKoh JYChorin UBesser L, et al. (2006) Synaptic release of zinc from brain slices: factors governing release, imaging, and accurate calculation of concentration. J Neurosci Methods 154:19–29. [DOI] [PubMed] [Google Scholar]
- Fredriksson RSreedharan SNordenankar KAlsiö JLindberg FAHutchinson AEriksson ARoshanbin SCiuculete DMKlockars A, et al. (2019) The polyamine transporter Slc18b1(VPAT) is important for both short and long time memory and for regulation of polyamine content in the brain. PLoS Genet 15:e1008455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerking M, Malenka RC, Nicoll RA (1998) Synaptic activation of kainate receptors on hippocampal interneurons. Nat Neurosci 1:479–486. [DOI] [PubMed] [Google Scholar]
- Frerking M, Nicoll RA (2000) Synaptic kainate receptors. Curr Opin Neurobiol 10:342–351. [DOI] [PubMed] [Google Scholar]
- Frerking M, Schmitz D, Zhou Q, Johansen J, Nicoll RA (2001) Kainate receptors depress excitatory synaptic transmission at CA3-->CA1 synapses in the hippocampus via a direct presynaptic action. J Neurosci 21:2958–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschy JM, Weinmann O, Wenzel A, Benke D (1998) Synapse-specific localization of NMDA and GABA(A) receptor subunits revealed by antigen-retrieval immunohistochemistry. J Comp Neurol 390:194–210. [PubMed] [Google Scholar]
- Frizelle PA, Chen PE, Wyllie DJ (2006) Equilibrium constants for (R)-[(S)-1-(4-bromo-phenyl)-ethylamino]-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-5-yl)-methyl]-phosphonic acid (NVP-AAM077) acting at recombinant NR1/NR2A and NR1/NR2B N-methyl-D-aspartate receptors: Implications for studies of synaptic transmission. Mol Pharmacol 70:1022–1032. [DOI] [PubMed] [Google Scholar]
- Frølund S, Bella A, Kristensen AS, Ziegler HL, Witt M, Olsen CA, Strømgaard K, Franzyk H, Jaroszewski JW (2010) Assessment of structurally diverse philanthotoxin analogues for inhibitory activity on ionotropic glutamate receptor subtypes: discovery of nanomolar, nonselective, and use-dependent antagonists. J Med Chem 53:7441–7451. [DOI] [PubMed] [Google Scholar]
- Fry AEFawcett KAZelnik NYuan HThompson BANShemer-Meiri LCushion TDMugalaasi HSims DStoodley N, et al. (2018) De novo mutations in GRIN1 cause extensive bilateral polymicrogyria. Brain 141:698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydenvang KLash LLNaur PPostila PAPickering DSSmith CMGajhede MSasaki MSakai RPentikaïnen OT, et al. (2009) Full domain closure of the ligand-binding core of the ionotropic glutamate receptor iGluR5 induced by the high affinity agonist dysiherbaine and the functional antagonist 8,9-dideoxyneodysiherbaine. J Biol Chem 284:14219–14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama F, Kuramoto E, Okamoto K, Hioki H, Furuta T, Zhou L, Nomura S, Kaneko T (2004) Presynaptic localization of an AMPA-type glutamate receptor in corticostriatal and thalamostriatal axon terminals. Eur J Neurosci 20:3322–3330. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Dimitrov A, Boncompain G, Vielemeyer O, Perez F, Fukata M (2013) Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J Cell Biol 202:145–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya M, Hayashi Y, Watanabe M (2005a) NR2 to NR3B subunit switchover of NMDA receptors in early postnatal motoneurons. Eur J Neurosci 21:1432–1436. [DOI] [PubMed] [Google Scholar]
- Fukaya MTsujita MYamazaki MKushiya EAbe MAkashi KNatsume RKano MKamiya HWatanabe M, et al. (2006) Abundant distribution of TARP gamma-8 in synaptic and extrasynaptic surface of hippocampal neurons and its major role in AMPA receptor expression on spines and dendrites. Eur J Neurosci 24:2177–2190. [DOI] [PubMed] [Google Scholar]
- Fukaya M, Yamazaki M, Sakimura K, Watanabe M (2005b) Spatial diversity in gene expression for VDCCgamma subunit family in developing and adult mouse brains. Neurosci Res 53:376–383. [DOI] [PubMed] [Google Scholar]
- Fukushima T, Shingai R, Ogurusu T, Ichinose M (2003) Inhibition of willardiine-induced currents through rat GluR6/KA-2 kainate receptor channels by Zinc and other divalent cations. Neurosci Lett 349:107–110. [DOI] [PubMed] [Google Scholar]
- Furlanis E, Traunmüller L, Fucile G, Scheiffele P (2019) Landscape of ribosome-engaged transcript isoforms reveals extensive neuronal-cell-class-specific alternative splicing programs. Nat Neurosci 22:1709–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Gouaux E (2003) Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J 22:2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E (2005) Subunit arrangement and function in NMDA receptors. Nature 438:185–192. [DOI] [PubMed] [Google Scholar]
- Gahring LC, Rogers SW, Twyman RE (1997) Autoantibodies to glutamate receptor subunit GluR2 in nonfamilial olivopontocerebellar degeneration. Neurology 48:494–500. [DOI] [PubMed] [Google Scholar]
- Gallimore AR, Aricescu AR, Yuzaki M, Calinescu R (2016) A computational model for the AMPA receptor phosphorylation master switch regulating cerebellar long-term depression. PLOS Comput Biol 12:e1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo V, Upson LM, Hayes WP, Vyklicky L Jr, Winters CA, Buonanno A (1992) Molecular cloning and development analysis of a new glutamate receptor subunit isoform in cerebellum. J Neurosci 12:1010–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambrill AC, Barria A (2011) NMDA receptor subunit composition controls synaptogenesis and synapse stabilization. Proc Natl Acad Sci USA 108:5855–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q, Dai J, Zhou HX, Wollmuth LP (2016) The transmembrane domain mediates tetramerization of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. J Biol Chem 291:6595–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q, Salussolia CL, Wollmuth LP (2015) Assembly of AMPA receptors: mechanisms and regulation. J Physiol 593:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwar SP, Green MN, Michard E, Simon AA, Feijó JA, Sobolevsky AI (2021) Structure of the arabidopsis glutamate receptor-like channel GLR3.2 ligand-binding domain. Structure 29:161–169.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganor Y, Goldberg-Stern H, Blank M, Shoenfeld Y, Dobrynina LA, Kalashnikova L, Levite M (2005a) Antibodies to glutamate receptor subtype 3 (GluR3) are found in some patients suffering from epilepsy as the main disease, but not in patients whose epilepsy accompanies antiphospholipid syndrome or Sneddon’s syndrome. Autoimmunity 38:417–424. [DOI] [PubMed] [Google Scholar]
- Ganor Y, Goldberg-Stern H, Cohen R, Teichberg V, Levite M (2014) Glutamate receptor antibodies directed against AMPA receptors subunit 3 peptide B (GluR3B) can be produced in DBA/2J mice, lower seizure threshold and induce abnormal behavior. Psychoneuroendocrinology 42:106–117. [DOI] [PubMed] [Google Scholar]
- Ganor Y, Gottlieb M, Eilam R, Otmy H, Teichberg VI, Levite M (2005b) Immunization with the glutamate receptor-derived peptide GluR3B induces neuronal death and reactive gliosis, but confers partial protection from pentylenetetrazole-induced seizures. Exp Neurol 195:92–102. [DOI] [PubMed] [Google Scholar]
- Gantz SC, Moussawi K, Hake HS (2020) Delta glutamate receptor conductance drives excitation of mouse dorsal raphe neurons. eLife 9:e56054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Hu XD, Yang H, Xia H (2018a) Distinct roles of protein phosphatase 1 bound on neurabin and spinophilin and its regulation in AMPA receptor trafficking and LTD induction. Mol Neurobiol 55:7179–7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao JMaison SFWu XHirose KJones SMBayazitov ITian YMittleman GMatthews DBZakharenko SS, et al. (2007) Orphan glutamate receptor delta1 subunit required for high-frequency hearing. Mol Cell Biol 27:4500–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Stevenson TJ, Douglass AD, Barrios JP, Bonkowsky JL (2018b) The midline axon crossing decision is regulated through an activity-dependent mechanism by the NMDA receptor. eNeuro 5:ENEURO.0389-0317.2018. [DOI] [PMC free article] [PubMed]
- Gao KTankovic AZhang YKusumoto HZhang JChen WXiangWei WShaulsky GHHu CTraynelis SF, et al. (2017) A de novo loss-of-function GRIN2A mutation associated with childhood focal epilepsy and acquired epileptic aphasia. PLoS One 12:e0170818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Nafría J, Herguedas B, Watson JF, Greger IH (2016) The dynamic AMPA receptor extracellular region: a platform for synaptic protein interactions. J Physiol 594:5449–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardinier KMGernert DLPorter WJReel JKOrnstein PLSpinazze PStevens FCHahn PHollinshead SPMayhugh D, et al. (2016) Discovery of the first α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor antagonist dependent upon transmembrane AMPA receptor regulatory protein (TARP) γ-8. J Med Chem 59:4753–4768. [DOI] [PubMed] [Google Scholar]
- Gardner SM, Trussell LO, Oertel D (1999) Time course and permeation of synaptic AMPA receptors in cochlear nuclear neurons correlate with input. J Neurosci 19:8721–8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SM, Trussell LO, Oertel D (2001) Correlation of AMPA receptor subunit composition with synaptic input in the mammalian cochlear nuclei. J Neurosci 21:7428–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardoni F, Caputi A, Cimino M, Pastorino L, Cattabeni F, Di Luca M (1998) Calcium/calmodulin-dependent protein kinase II is associated with NR2A/B subunits of NMDA receptor in postsynaptic densities. J Neurochem 71:1733–1741. [DOI] [PubMed] [Google Scholar]
- Gardoni F, Stanic J, Scheggia D, Benussi A, Borroni B, Di Luca M (2021) NMDA and AMPA receptor autoantibodies in brain disorders: From molecular mechanisms to clinical features. Cells 10:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner RGopalakrishnan SMcCauley JABednar RAGaul SLMosser SDKiss LLynch JJPatel SFandozzi C, et al. (2015) Preclinical pharmacology and pharmacokinetics of CERC-301, a GluN2B-selective N-methyl-D-aspartate receptor antagonist. Pharmacol Res Perspect 3:e00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst-Orozco JMalik RLanz TAWeber MLXi HArion DEnwright JF 3rdLewis DAO’Donnell PSohal VS, et al. (2020) GluN2D-mediated excitatory drive onto medial prefrontal cortical PV+ fast-spiking inhibitory interneurons. PLoS One 15:e0233895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudillière B, Konishi Y, de la Iglesia N, Yao Gl, Bonni A (2004) A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron 41:229–241. [DOI] [PubMed] [Google Scholar]
- Gawai P, Upadhyay R, Gakare SG, Sarode L, Dravid SM, Ugale RR (2020) Antipsychotic-like profile of CIQ isomers in animal models of schizophrenia. Behav Pharmacol 31:524–534. [DOI] [PubMed] [Google Scholar]
- Gaynor SC, Breen ME, Monson ET, de Klerk K, Parsons M, DeLuca AP, Scheetz TE, Zandi PP, Potash JB, Willour VL (2016) A targeted sequencing study of glutamatergic candidate genes in suicide attempters with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet 171:1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzellone MJZhou XLionel ACUddin MThiruvahindrapuram BLiang SSun CWang JZou MTammimies K, et al. (2014) Copy number variation in Han Chinese individuals with autism spectrum disorder. J Neurodev Disord 6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Chen W, Axerio-Cilies P, Wang YT (2020) NMDARs in cell survival and death: Implications in stroke pathogenesis and treatment. Trends Mol Med 26:533–551. [DOI] [PubMed] [Google Scholar]
- Geiger JR, Lübke J, Roth A, Frotscher M, Jonas P (1997) Submillisecond AMPA receptor-mediated signaling at a principal neuron-interneuron synapse. Neuron 18:1009–1023. [DOI] [PubMed] [Google Scholar]
- Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H (1995) Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron 15:193–204. [DOI] [PubMed] [Google Scholar]
- Geisheker MRHeymann GWang TCoe BPTurner TNStessman HAFHoekzema KKvarnung MShaw MFriend K, et al. (2017) Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat Neurosci 20:1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsomino G, Menna E, Antonucci F, Rodighiero S, Riganti L, Mulle C, Benfenati F, Valtorta F, Verderio C, Matteoli M (2013) Kainate induces mobilization of synaptic vesicles at the growth cone through the activation of protein kinase A. Cereb Cortex 23:531–541. [DOI] [PubMed] [Google Scholar]
- Ghafari M, Keihan Falsafi S, Höger H, Bennett KL, Lubec G (2015) Identification of new phosphorylation sites of AMPA receptors in the rat hippocampus--A resource for neuroscience research. Proteomics Clin Appl 9:808–816. [DOI] [PubMed] [Google Scholar]
- Ghiretti AE, Kenny K, Marr MT 2nd, Paradis S (2013) CaMKII-dependent phosphorylation of the GTPase Rem2 is required to restrict dendritic complexity. J Neurosci 33:6504–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb AJ, Colquhoun D (1991) Glutamate activation of a single NMDA receptor-channel produces a cluster of channel openings. Proc Biol Sci 243:39–45. [DOI] [PubMed] [Google Scholar]
- Gibb AJ, Ogden KK, McDaniel MJ, Vance KM, Kell SA, Butch C, Burger P, Liotta DC, Traynelis SF (2018) A structurally derived model of subunit-dependent NMDA receptor function. J Physiol 596:4057–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs TT, Russek SJ, Farb DH (2006) Sulfated steroids as endogenous neuromodulators. Pharmacol Biochem Behav 84:555–567. [DOI] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P (2008) Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron 57:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P (2009) Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature 459:703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffard RG, Monyer H, Christine CW, Choi DW (1990) Acidosis reduces NMDA receptor activation, glutamate neurotoxicity, and oxygen-glucose deprivation neuronal injury in cortical cultures. Brain Res 506:339–342. [DOI] [PubMed] [Google Scholar]
- Gilbert ME, Mack CM (1990) The NMDA antagonist, MK-801, suppresses long-term potentiation, kindling, and kindling-induced potentiation in the perforant path of the unanesthetized rat. Brain Res 519:89–96. [DOI] [PubMed] [Google Scholar]
- Gilhus NE, Tzartos S, Evoli A, Palace J, Burns TM, Verschuuren JJGM (2019) Myasthenia gravis. Nat Rev Dis Primers 5:30. [DOI] [PubMed] [Google Scholar]
- Gill A, Birdsey-Benson A, Jones BL, Henderson LP, Madden DR (2008) Correlating AMPA receptor activation and cleft closure across subunits: crystal structures of the GluR4 ligand-binding domain in complex with full and partial agonists. Biochemistry 47:13831–13841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill I, Droubi S, Giovedi S, Fedder KN, Bury LA, Bosco F, Sceniak MP, Benfenati F, Sabo SL (2015) Presynaptic NMDA receptors - dynamics and distribution in developing axons in vitro and in vivo. J Cell Sci 128:768–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MB, Frausto S, Ikoma M, Sasaki M, Oikawa M, Sakai R, Swanson GT (2010) A series of structurally novel heterotricyclic alpha-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate receptor-selective antagonists. Br J Pharmacol 160:1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MB, Kato AS, Roberts MF, Yu H, Wang H, Tomita S, Bredt DS (2011) Cornichon-2 modulates AMPA receptor-transmembrane AMPA receptor regulatory protein assembly to dictate gating and pharmacology. J Neurosci 31:6928–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MB, Kato AS, Wang H, Bredt DS (2012) AMPA receptor modulation by cornichon-2 dictated by transmembrane AMPA receptor regulatory protein isoform. Eur J Neurosci 35:182–194. [DOI] [PubMed] [Google Scholar]
- Gill RAlanine ABourson AButtelmann BFischer GHeitz MPKew JNLevet-Trafit BLorez HPMalherbe P, et al. (2002) Pharmacological characterization of Ro 63-1908 (1-[2-(4-hydroxy-phenoxy)-ethyl]-4-(4-methyl-benzyl)-piperidin-4-ol), a novel subtype-selective N-methyl-D-aspartate antagonist. J Pharmacol Exp Ther 302:940–948. [DOI] [PubMed] [Google Scholar]
- Gilron I, Max MB, Lee G, Booher SL, Sang CN, Chappell AS, Dionne RA (2000) Effects of the 2-amino-3-hydroxy-5-methyl-4-isoxazole-proprionic acid/kainate antagonist LY293558 on spontaneous and evoked postoperative pain. Clin Pharmacol Ther 68:320–327. [DOI] [PubMed] [Google Scholar]
- Ginsberg MD (2008) Neuroprotection for ischemic stroke: past, present and future. Neuropharmacology 55:363–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladding CM, Collett VJ, Jia Z, Bashir ZI, Collingridge GL, Molnár E (2009) Tyrosine dephosphorylation regulates AMPAR internalisation in mGluR-LTD. Mol Cell Neurosci 40:267–279. [DOI] [PubMed] [Google Scholar]
- Gladding CM, Raymond LA (2011) Mechanisms underlying NMDA receptor synaptic/extrasynaptic distribution and function. Mol Cell Neurosci 48:308–320. [DOI] [PubMed] [Google Scholar]
- Glasgow NG, Siegler Retchless B, Johnson JW (2015) Molecular bases of NMDA receptor subtype-dependent properties. J Physiol 593:83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow NG, Wilcox MR, Johnson JW (2018) Effects of Mg2+ on recovery of NMDA receptors from inhibition by memantine and ketamine reveal properties of a second site. Neuropharmacology 137:344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleichman AJ, Panzer JA, Baumann BH, Dalmau J, Lynch DR (2014) Antigenic and mechanistic characterization of anti-AMPA receptor encephalitis. Ann Clin Transl Neurol 1:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleichman AJ, Spruce LA, Dalmau J, Seeholzer SH, Lynch DR (2012) Anti-NMDA receptor encephalitis antibody binding is dependent on amino acid identity of a small region within the GluN1 amino terminal domain. J Neurosci 32:11082–11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JTWang KCai GKorvatska OKim CEWood SZhang HEstes ABrune CWBradfield JP, et al. (2009) Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glitsch M, Marty A (1999) Presynaptic effects of NMDA in cerebellar Purkinje cells and interneurons. J Neurosci 19:511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff DC (2015) Drug development in schizophrenia: are glutamatergic targets still worth aiming at? Curr Opin Psychiatry 28:207–215. [DOI] [PubMed] [Google Scholar]
- Goff DC, Cather C, Gottlieb JD, Evins AE, Walsh J, Raeke L, Otto MW, Schoenfeld D, Green MF (2008) Once-weekly D-cycloserine effects on negative symptoms and cognition in schizophrenia: an exploratory study. Schizophr Res 106:320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin EDrapier TLarsen APGeubelle PPtak CPLaulumaa SRovinskaja KGilissen JTullio POlsen L, et al. (2018) 7-Phenoxy-substituted 3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxides as positive allosteric modulators of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors with nanomolar potency. J Med Chem 61:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg-Stern H, Ganor Y, Cohen R, Pollak L, Teichberg V, Levite M (2014) Glutamate receptor antibodies directed against AMPA receptors subunit 3 peptide B (GluR3B) associate with some cognitive/psychiatric/behavioral abnormalities in epilepsy patients. Psychoneuroendocrinology 40:221–231. [DOI] [PubMed] [Google Scholar]
- Goncalves J, Bartol TM, Camus C, Levet F, Menegolla AP, Sejnowski TJ, Sibarita JB, Vivaudou M, Choquet D, Hosy E (2020) Nanoscale co-organization and coactivation of AMPAR, NMDAR, and mGluR at excitatory synapses. Proc Natl Acad Sci USA 117:14503–14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gono T, Takarada T, Fukumori R, Kawaguchi Y, Kaneko H, Hanaoka M, Katsumata Y, Yoneda Y, Yamanaka H (2011) NR2-reactive antibody decreases cell viability through augmentation of Ca(2+) influx in systemic lupus erythematosus. Arthritis Rheum 63:3952–3959. [DOI] [PubMed] [Google Scholar]
- Gonzalez J, Du M, Parameshwaran K, Suppiramaniam V, Jayaraman V (2010) Role of dimer interface in activation and desensitization in AMPA receptors. Proc Natl Acad Sci USA 107:9891–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J, Rambhadran A, Du M, Jayaraman V (2008) LRET investigations of conformational changes in the ligand binding domain of a functional AMPA receptor. Biochemistry 47:10027–10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo MS, Scudder SL, Patrick GN (2015) Ubiquitin-dependent trafficking and turnover of ionotropic glutamate receptors. Front Mol Neurosci 8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell DJ, Zaegel V, Coultrap SJ, Hell JW, Bayer KU (2017) DAPK1 mediates LTD by making CaMKII/GluN2B binding LTP specific. Cell Rep 19:2231–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorostiza P, Volgraf M, Numano R, Szobota S, Trauner D, Isacoff EY (2007) Mechanisms of photoswitch conjugation and light activation of an ionotropic glutamate receptor. Proc Natl Acad Sci USA 104:10865–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JM, Emeson RB (2000) Functions and mechanisms of RNA editing. Annu Rev Genet 34:499–531. [DOI] [PubMed] [Google Scholar]
- Götz T, Kraushaar U, Geiger J, Lübke J, Berger T, Jonas P (1997) Functional properties of AMPA and NMDA receptors expressed in identified types of basal ganglia neurons. J Neurosci 17:204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould E, Cameron HA, McEwen BS (1994) Blockade of NMDA receptors increases cell death and birth in the developing rat dentate gyrus. J Comp Neurol 340:551–565. [DOI] [PubMed] [Google Scholar]
- Gowd KHHan TSTwede VGajewiak JSmith MDWatkins MPlatt RJToledo GWhite HSOlivera BM, et al. (2012) Conantokins derived from the Asprella clade impart conRl-B, an N-methyl d-aspartate receptor antagonist with a unique selectivity profile for NR2B subunits. Biochemistry 51:4685–4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef JDNewberry KNewton APieschl RShields ELuan FNSimmermacher JLuchetti DSchaeffer ELi YW, et al. (2015) Effect of acute NR2B antagonist treatment on long-term potentiation in the rat hippocampus. Brain Res 1609:31–39. [DOI] [PubMed] [Google Scholar]
- Grand T, Abi Gerges S, David M, Diana MA, Paoletti P (2018) Unmasking GluN1/GluN3A excitatory glycine NMDA receptors. Nat Commun 9:4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger AJ, Nicoll RA (2014) LTD expression is independent of glutamate receptor subtype. Front Synaptic Neurosci 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger AJ, Shi Y, Lu W, Cerpas M, Nicoll RA (2013) LTP requires a reserve pool of glutamate receptors independent of subunit type. Nature 493:495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger R, Staubli U, Davis M, Perez Y, Nilsson L, Rogers GA, Lynch G (1993) A drug that facilitates glutamatergic transmission reduces exploratory activity and improves performance in a learning-dependent task. Synapse 15:326–329. [DOI] [PubMed] [Google Scholar]
- Gratacòs-Batlle E, Olivella M, Sánchez-Fernández N, Yefimenko N, Miguez-Cabello F, Fadó R, Casals N, Gasull X, Ambrosio S, Soto D (2018) Mechanisms of CPT1C-dependent AMPAR trafficking enhancement. Front Mol Neurosci 11:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Fink AE, Sariñana J, Vissel B, O’Dell TJ (2007) Long-term potentiation in the hippocampal CA1 region does not require insertion and activation of GluR2-lacking AMPA receptors. J Neurophysiol 98:2488–2492. [DOI] [PubMed] [Google Scholar]
- Gray JA, Shi Y, Usui H, During MJ, Sakimura K, Nicoll RA (2011) Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron 71:1085–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood J, Valdes J (2016) Perampanel (Fycompa): A review of clinical efficacy and safety in epilepsy. P&T 41:683–698. [PMC free article] [PubMed] [Google Scholar]
- Greenwood TALazzeroni LCCalkins MEFreedman RGreen MFGur REGur RCLight GANuechterlein KHOlincy A, et al. (2016) Genetic assessment of additional endophenotypes from the Consortium on the Genetics of Schizophrenia Family Study. Schizophr Res 170:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TALazzeroni LCMurray SSCadenhead KSCalkins MEDobie DJGreen MFGur REGur RCHardiman G, et al. (2011) Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry 168:930–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Akamine P, Khatri L, Ziff EB (2006) Developmentally regulated, combinatorial RNA processing modulates AMPA receptor biogenesis. Neuron 51:85–97. [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Kong X, Ziff EB (2003) AMPA receptor tetramerization is mediated by Q/R editing. Neuron 40:763–774. [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Ziff EB (2002) RNA editing at arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron 34:759–772. [DOI] [PubMed] [Google Scholar]
- Greger IH, Mayer ML (2019) Structural biology of glutamate receptor ion channels: towards an understanding of mechanism. Curr Opin Struct Biol 57:185–195. [DOI] [PubMed] [Google Scholar]
- Greger IH, Watson JF, Cull-Candy SG (2017) Structural and functional architecture of AMPA-type glutamate receptors and their auxiliary proteins. Neuron 94:713–730. [DOI] [PubMed] [Google Scholar]
- Greger IH, Ziff EB, Penn AC (2007) Molecular determinants of AMPA receptor subunit assembly. Trends Neurosci 30:407–416. [DOI] [PubMed] [Google Scholar]
- Gregor P, O’Hara BF, Yang X, Uhl GR (1993) Expression and novel subunit isoforms of glutamate receptor genes GluR5 and GluR6. Neuroreport 4:1343–1346. [DOI] [PubMed] [Google Scholar]
- Griffith TN, Swanson GT (2015) Identification of critical functional determinants of kainate receptor modulation by auxiliary protein Neto2. J Physiol 593:4815–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold AJMa DCukier HNNations LDSchmidt MAChung RHJaworski JMSalyakina DKonidari IWhitehead PL, et al. (2012) Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet 21:3513–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groc L, Bard L, Choquet D (2009) Surface trafficking of N-methyl-D-aspartate receptors: physiological and pathological perspectives. Neuroscience 158:4–18. [DOI] [PubMed] [Google Scholar]
- Grosskreutz J, Zoerner A, Schlesinger F, Krampfl K, Dengler R, Bufler J (2003) Kinetic properties of human AMPA-type glutamate receptors expressed in HEK293 cells. Eur J Neurosci 17:1173–1178. [DOI] [PubMed] [Google Scholar]
- Groveman BR, Feng S, Fang XQ, Pflueger M, Lin SX, Bienkiewicz EA, Yu X (2012) The regulation of N-methyl-D-aspartate receptors by Src kinase. FEBS J 279:20–28. [DOI] [PubMed] [Google Scholar]
- Gu X, Mao X, Lussier MP, Hutchison MA, Zhou L, Hamra FK, Roche KW, Lu W (2016a) GSG1L suppresses AMPA receptor-mediated synaptic transmission and uniquely modulates AMPA receptor kinetics in hippocampal neurons. Nat Commun 7:10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Zhou L, Lu W (2016b) An NMDA receptor-dependent mechanism underlies inhibitory synapse development. Cell Rep 14:471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guire ES, Oh MC, Soderling TR, Derkach VA (2008) Recruitment of calcium-permeable AMPA receptors during synaptic potentiation is regulated by CaM-kinase I. J Neurosci 28:6000–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen V, Holten AT, Storm-Mathisen J (2004) GABAergic synapses in hippocampus exocytose aspartate on to NMDA receptors: quantitative immunogold evidence for co-transmission. Mol Cell Neurosci 26:156–165. [DOI] [PubMed] [Google Scholar]
- Guo SZHuang KShi YYTang WZhou JFeng GYZhu SMLiu HJChen YSun XD, et al. (2007) A case-control association study between the GRID1 gene and schizophrenia in the Chinese Northern Han population. Schizophr Res 93:385–390. [DOI] [PubMed] [Google Scholar]
- Gupta SC, Hillman BG, Prakash A, Ugale RR, Stairs DJ, Dravid SM (2013) Effect of D-cycloserine in conjunction with fear extinction training on extracellular signal-regulated kinase activation in the medial prefrontal cortex and amygdala in rat. Eur J Neurosci 37:1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SC, Yadav R, Pavuluri R, Morley BJ, Stairs DJ, Dravid SM (2015) Essential role of GluD1 in dendritic spine development and GluN2B to GluN2A NMDAR subunit switch in the cortex and hippocampus reveals ability of GluN2B inhibition in correcting hyperconnectivity. Neuropharmacology 93:274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán YF, Ramsey K, Stolz JR, Craig DW, Huentelman MJ, Narayanan V, Swanson GT (2017) A gain-of-function mutation in the GRIK2 gene causes neurodevelopmental deficits. Neurol Genet 3:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas KT, Compans B, Letellier M, Bartol TM, Grillo-Bosch D, Sejnowski TJ, Sainlos M, Choquet D, Thoumine O, Hosy E (2018) Pre-post synaptic alignment through neuroligin-1 tunes synaptic transmission efficiency. eLife 7:e31755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackmann K, Matko S, Gerlach EM, von der Hagen M, Klink B, Schrock E, Rump A, Di Donato N (2013) Partial deletion of GLRB and GRIA2 in a patient with intellectual disability. Eur J Hum Genet 21:112–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackos DH, Hanson JE (2017) Diverse modes of NMDA receptor positive allosteric modulation: Mechanisms and consequences. Neuropharmacology 112 (Pt A):34–45. [DOI] [PubMed] [Google Scholar]
- Hackos DHLupardus PJGrand TChen YWang TMReynen PGustafson AWallweber HJVolgraf MSellers BD, et al. (2016) Positive allosteric modulators of GluN2A-containing NMDARs with distinct modes of action and impacts on circuit function. Neuron 89:983–999. [DOI] [PubMed] [Google Scholar]
- Hacohen YDeiva KPettingill PWaters PSiddiqui AChretien PMenson ELin JPTardieu MVincent A, et al. (2014) N-methyl-D-aspartate receptor antibodies in post-herpes simplex virus encephalitis neurological relapse. Mov Disord 29:90–96. [DOI] [PubMed] [Google Scholar]
- Hadzic M, Jack A, Wahle P (2017) Ionotropic glutamate receptors: Which ones, when, and where in the mammalian neocortex. J Comp Neurol 525:976–1033. [DOI] [PubMed] [Google Scholar]
- Haering SC, Tapken D, Pahl S, Hollmann M (2014) Auxiliary subunits: shepherding AMPA receptors to the plasma membrane. Membranes (Basel) 4:469–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglerød C, Hussain S, Nakamura Y, Xia J, Haug FS, Ottersen OP, Henley JM, Davanger S (2017) Presynaptic PICK1 facilitates trafficking of AMPA-receptors between active zone and synaptic vesicle pool. Neuroscience 344:102–112. [DOI] [PubMed] [Google Scholar]
- Hald H, Ahring PK, Timmermann DB, Liljefors T, Gajhede M, Kastrup JS (2009) Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2. J Mol Biol 391:906–917. [DOI] [PubMed] [Google Scholar]
- Hald HNaur PPickering DSSprogøe DMadsen UTimmermann DBAhring PKLiljefors TSchousboe AEgebjerg J, et al. (2007) Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid. J Biol Chem 282:25726–25736. [DOI] [PubMed] [Google Scholar]
- Halt AR, Dallapiazza RF, Zhou Y, Stein IS, Qian H, Juntti S, Wojcik S, Brose N, Silva AJ, Hell JW (2012) CaMKII binding to GluN2B is critical during memory consolidation. EMBO J 31:1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FFGauthier JAraki YLin DTYoshizawa YHigashi KPark ARSpiegelman DDobrzeniecka SPiton A, et al. ; S2D Group (2011) Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet 88:306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FFSrour MCapo-Chichi JMDaoud HNassif CPatry LMassicotte CAmbalavanan ASpiegelman DDiallo O, et al. (2014) De novo mutations in moderate or severe intellectual disability. PLoS Genet 10:e1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton NB, Attwell D (2010) Do astrocytes really exocytose neurotransmitters? Nat Rev Neurosci 11:227–238. [DOI] [PubMed] [Google Scholar]
- Hammer CStepniak BSchneider APapiol STantra MBegemann MSirén ALPardo LASperling SMohd Jofrry S, et al. (2014) Neuropsychiatric disease relevance of circulating anti-NMDA receptor autoantibodies depends on blood-brain barrier integrity. Mol Psychiatry 19:1143–1149. [DOI] [PubMed] [Google Scholar]
- Hampson RE, Rogers G, Lynch G, Deadwyler SA (1998a) Facilitative effects of the ampakine CX516 on short-term memory in rats: correlations with hippocampal neuronal activity. J Neurosci 18:2748–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson RE, Rogers G, Lynch G, Deadwyler SA (1998b) Facilitative effects of the ampakine CX516 on short-term memory in rats: enhancement of delayed-nonmatch-to-sample performance. J Neurosci 18:2740–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Wu P, Wang F, Chen J (2015a) S-palmitoylation regulates AMPA receptors trafficking and function: a novel insight into synaptic regulation and therapeutics. Acta Pharm Sin B 5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TH, Dharkar P, Mayer ML, Serpe M (2015b) Functional reconstitution of Drosophila melanogaster NMJ glutamate receptors. Proc Natl Acad Sci USA 112:6182–6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han THVicidomini RRamos CIWang QNguyen PJarnik MLee CHStawarski MHernandez RXMacleod GT, et al. (2020) Neto-α controls synapse organization and homeostasis at the drosophila neuromuscular junction. Cell Rep 32:107866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han WLi JPelkey KAPandey SChen XWang YXWu KGe LLi TCastellano D, et al. (2019) Shisa7 is a GABAA receptor auxiliary subunit controlling benzodiazepine actions. Science 366:246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wang C, Park JS, Niu L (2010) Channel-opening kinetic mechanism for human wild-type GluK2 and the M867I mutant kainate receptor. Biochemistry 49:9207–9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T (2014) The discovery and development of perampanel for the treatment of epilepsy. Expert Opin Drug Discov 9:449–458. [DOI] [PubMed] [Google Scholar]
- Hanada T, Hashizume Y, Tokuhara N, Takenaka O, Kohmura N, Ogasawara A, Hatakeyama S, Ohgoh M, Ueno M, Nishizawa Y (2011) Perampanel: a novel, orally active, noncompetitive AMPA-receptor antagonist that reduces seizure activity in rodent models of epilepsy. Epilepsia 52:1331–1340. [DOI] [PubMed] [Google Scholar]
- Hanania T, Manfredi P, Inturrisi C, Vitolo OV (2020) The N-methyl-D-aspartate receptor antagonist d-methadone acutely improves depressive-like behavior in the forced swim test performance of rats. Exp Clin Psychopharmacol 28:196–201. [DOI] [PubMed] [Google Scholar]
- Hanly JG, Robichaud J, Fisk JD (2006) Anti-NR2 glutamate receptor antibodies and cognitive function in systemic lupus erythematosus. J Rheumatol 33:1553–1558. [PubMed] [Google Scholar]
- Hansel D, Mato G (2013) Short-term plasticity explains irregular persistent activity in working memory tasks. J Neurosci 33:133–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Bräuner-Osborne H, Egebjerg J (2008) Pharmacological characterization of ligands at recombinant NMDA receptor subtypes by electrophysiological recordings and intracellular calcium measurements. Comb Chem High Throughput Screen 11:304–315. [DOI] [PubMed] [Google Scholar]
- Hansen KB, Clausen RP, Bjerrum EJ, Bechmann C, Greenwood JR, Christensen C, Kristensen JL, Egebjerg J, Bräuner-Osborne H (2005) Tweaking agonist efficacy at N-methyl-D-aspartate receptors by site-directed mutagenesis. Mol Pharmacol 68:1510–1523. [DOI] [PubMed] [Google Scholar]
- Hansen KB, Furukawa H, Traynelis SF (2010a) Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol 78:535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KBMullasseril PDawit SKurtkaya NLYuan HVance KMOrr AGKvist TOgden KKLe P, et al. (2010b) Implementation of a fluorescence-based screening assay identifies histamine H3 receptor antagonists clobenpropit and iodophenpropit as subunit-selective N-methyl-D-aspartate receptor antagonists. J Pharmacol Exp Ther 333:650–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Naur P, Kurtkaya NL, Kristensen AS, Gajhede M, Kastrup JS, Traynelis SF (2009) Modulation of the dimer interface at ionotropic glutamate-like receptor delta2 by D-serine and extracellular calcium. J Neurosci 29:907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Ogden KK, Traynelis SF (2012) Subunit-selective allosteric inhibition of glycine binding to NMDA receptors. J Neurosci 32:6197–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Ogden KK, Yuan H, Traynelis SF (2014) Distinct functional and pharmacological properties of triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron 81:1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jørgensen L, Vance KM, Ogden KK, Clausen RP, Furukawa H, Traynelis SF (2013) Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol Pharmacol 84:114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Traynelis SF (2011) Structural and mechanistic determinants of a novel site for noncompetitive inhibition of GluN2D-containing NMDA receptors. J Neurosci 31:3650–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Yi F, Perszyk RE, Furukawa H, Wollmuth LP, Gibb AJ, Traynelis SF (2018) Structure, function, and allosteric modulation of NMDA receptors. J Gen Physiol 150:1081–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson EArmbruster MLau LASommer MEKlaft ZJSwanger SATraynelis SFMoss SJNoubary FChadchankar J, et al. (2019) Tonic activation of GluN2C/GluN2D-containing NMDA receptors by ambient glutamate facilitates cortical interneuron maturation. J Neurosci 39:3611–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson JEMa KElstrott JWeber MSaillet SKhan ASSimms JLiu BKim TAYu GQ, et al. (2020) GluN2A NMDA receptor enhancement improves brain oscillations, synchrony, and cognitive functions in Dravet syndrome and Alzheimer’s disease models. Cell Rep 30:381–396.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanus C, Ehlers MD (2008) Secretory outposts for the local processing of membrane cargo in neuronal dendrites. Traffic 9:1437–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanus C, Geptin H, Tushev G, Garg S, Alvarez-Castelao B, Sambandan S, Kochen L, Hafner AS, Langer JD, Schuman EM (2016) Unconventional secretory processing diversifies neuronal ion channel properties. eLife 5:e20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Bading H (2010) Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci 11:682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Fukunaga Y, Bading H (2002) Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci 5:405–414. [DOI] [PubMed] [Google Scholar]
- Harlow DE, Saul KE, Komuro H, Macklin WB (2015) Myelin proteolipid protein complexes with αv integrin and AMPA receptors in vivo and regulates AMPA-dependent oligodendrocyte progenitor cell migration through the modulation of cell-surface GluR2 expression. J Neurosci 35:12018–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmel N, Cokic B, Zolles G, Berkefeld H, Mauric V, Fakler B, Stein V, Klöcker N (2012) AMPA receptors commandeer an ancient cargo exporter for use as an auxiliary subunit for signaling. PLoS One 7:e30681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon E, Miller AH, Sanacora G (2017) Inflammation, glutamate, and glia: A trio of trouble in mood disorders. Neuropsychopharmacology 42:193–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AZ, Pettit DL (2007) Extrasynaptic and synaptic NMDA receptors form stable and uniform pools in rat hippocampal slices. J Physiol 584:509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AZ, Pettit DL (2008) Recruiting extrasynaptic NMDA receptors augments synaptic signaling. J Neurophysiol 99:524–533. [DOI] [PubMed] [Google Scholar]
- Harris K, Armstrong SP, Campos-Pires R, Kiru L, Franks NP, Dickinson R (2013) Neuroprotection against traumatic brain injury by xenon, but not argon, is mediated by inhibition at the N-methyl-D-aspartate receptor glycine site. Anesthesiology 119:1137–1148. [DOI] [PubMed] [Google Scholar]
- Harris KM (2020) Structural LTP: from synaptogenesis to regulated synapse enlargement and clustering. Curr Opin Neurobiol 63:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ, Ravdin LD, Lockshin MD (2006) Relationship between serum NR2a antibodies and cognitive dysfunction in systemic lupus erythematosus. Arthritis Rheum 54:2515–2522. [DOI] [PubMed] [Google Scholar]
- Harty TP, Rogawski MA (2000) Felbamate block of recombinant N-methyl-D-aspartate receptors: selectivity for the NR2B subunit. Epilepsy Res 39:47–55. [DOI] [PubMed] [Google Scholar]
- Harvey CD, Yasuda R, Zhong H, Svoboda K (2008) The spread of Ras activity triggered by activation of a single dendritic spine. Science 321:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselmann HMannara FWerner CPlanagumà JMiguez-Cabello FSchmidl LGrünewald BPetit-Pedrol MKirmse KClassen J, et al. (2018) Human autoantibodies against the AMPA receptor subunit GluA2 induce receptor reorganization and memory dysfunction. Neuron 100:91–105.e9. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Fukaya M, Qiao X, Sakimura K, Watanabe M, Kano M (1999) Impairment of AMPA receptor function in cerebellar granule cells of ataxic mutant mouse stargazer. J Neurosci 19:6027–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, London ED (1995) Interactions of erythro-ifenprodil, threo-ifenprodil, erythro-iodoifenprodil, and eliprodil with subtypes of sigma receptors. Eur J Pharmacol 273:307–310. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Malchow B, Falkai P, Schmitt A (2013) Glutamate modulators as potential therapeutic drugs in schizophrenia and affective disorders. Eur Arch Psychiatry Clin Neurosci 263:367–377. [DOI] [PubMed] [Google Scholar]
- Hashizume M, Miyazaki T, Sakimura K, Watanabe M, Kitamura K, Kano M (2013) Disruption of cerebellar microzonal organization in GluD2 (GluRδ2) knockout mouse. Front Neural Circuits 7:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton CJ, Paoletti P (2005) Modulation of triheteromeric NMDA receptors by N-terminal domain ligands. Neuron 46:261–274. [DOI] [PubMed] [Google Scholar]
- Hawken NM, Zaika EI, Nakagawa T (2017) Engineering defined membrane-embedded elements of AMPA receptor induces opposing gating modulation by cornichon 3 and stargazin. J Physiol 595:6517–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins LM, Prybylowski K, Chang K, Moussan C, Stephenson FA, Wenthold RJ (2004) Export from the endoplasmic reticulum of assembled N-methyl-d-aspartic acid receptors is controlled by a motif in the c terminus of the NR2 subunit. J Biol Chem 279:28903–28910. [DOI] [PubMed] [Google Scholar]
- Hayashi T (2021) Post-translational palmitoylation of ionotropic glutamate receptors in excitatory synaptic functions. Br J Pharmacol 178:784–797. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Rumbaugh G, Huganir RL (2005) Differential regulation of AMPA receptor subunit trafficking by palmitoylation of two distinct sites. Neuron 47:709–723. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Umemori H, Mishina M, Yamamoto T (1999) The AMPA receptor interacts with and signals through the protein tyrosine kinase Lyn. Nature 397:72–76. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Shi SH, Esteban JA, Piccini A, Poncer JC, Malinow R (2000) Driving AMPA receptors into synapses by LTP and CaMKII: requirement for GluR1 and PDZ domain interaction. Science 287:2262–2267. [DOI] [PubMed] [Google Scholar]
- He XP, Patel M, Whitney KD, Janumpalli S, Tenner A, McNamara JO (1998) Glutamate receptor GluR3 antibodies and death of cortical cells. Neuron 20:153–163. [DOI] [PubMed] [Google Scholar]
- He XYLi YJKalyanaraman CQiu LLChen CXiao QLiu WXZhang WYang JJChen G, et al. (2016) GluA1 signal peptide determines the spatial assembly of heteromeric AMPA receptors. Proc Natl Acad Sci USA 113:E5645–E5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann M, Bufler J, Franke C, Dudel J (1996) Kinetics of homomeric GluR6 glutamate receptor channels. Biophys J 71:1743–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine M, Groc L, Frischknecht R, Béïque JC, Lounis B, Rumbaugh G, Huganir RL, Cognet L, Choquet D (2008) Surface mobility of postsynaptic AMPARs tunes synaptic transmission. Science 320:201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helassa N, Dürst CD, Coates C, Kerruth S, Arif U, Schulze C, Wiegert JS, Geeves M, Oertner TG, Török K (2018) Ultrafast glutamate sensors resolve high-frequency release at Schaffer collateral synapses. Proc Natl Acad Sci USA 115:5594–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelikova K, Kolcheva M, Skrenkova K, Kaniakova M, Horak M (2019) Lectins modulate the functional properties of GluN1/GluN3-containing NMDA receptors. Neuropharmacology 157:107671. [DOI] [PubMed] [Google Scholar]
- Henle F, Dehmel M, Leemhuis J, Fischer C, Meyer DK (2012) Role of GluN2A and GluN2B subunits in the formation of filopodia and secondary dendrites in cultured hippocampal neurons. Naunyn Schmiedebergs Arch Pharmacol 385:171–180. [DOI] [PubMed] [Google Scholar]
- Henley JM, Wilkinson KA (2016) Synaptic AMPA receptor composition in development, plasticity and disease. Nat Rev Neurosci 17:337–350. [DOI] [PubMed] [Google Scholar]
- Henneberger CBard LPanatier AReynolds JPKopach OMedvedev NIMinge DHerde MKAnders SKraev I, et al. (2020) LTP induction boosts glutamate spillover by driving withdrawal of perisynaptic astroglia. Neuron 108:919–936.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberger C, Jüttner R, Schmidt SA, Walter J, Meier JC, Rothe T, Grantyn R (2005) GluR- and TrkB-mediated maturation of GABA receptor function during the period of eye opening. Eur J Neurosci 21:431–440. [DOI] [PubMed] [Google Scholar]
- Henson MA, Larsen RS, Lawson SN, Pérez-Otaño I, Nakanishi N, Lipton SA, Philpot BD (2012) Genetic deletion of NR3A accelerates glutamatergic synapse maturation. PLoS One 7:e42327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepp RHay YAAguado CLujan RDauphinot LPotier MCNomura SPoirel OEl Mestikawy SLambolez B, et al. (2015) Glutamate receptors of the delta family are widely expressed in the adult brain. Brain Struct Funct 220:2797–2815. [DOI] [PubMed] [Google Scholar]
- Herb A, Burnashev N, Werner P, Sakmann B, Wisden W, Seeburg PH (1992) The KA-2 subunit of excitatory amino acid receptors shows widespread expression in brain and forms ion channels with distantly related subunits. Neuron 8:775–785. [DOI] [PubMed] [Google Scholar]
- Herb A, Higuchi M, Sprengel R, Seeburg PH (1996) Q/R site editing in kainate receptor GluR5 and GluR6 pre-mRNAs requires distant intronic sequences. Proc Natl Acad Sci USA 93:1875–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herguedas B, García-Nafría J, Cais O, Fernández-Leiro R, Krieger J, Ho H, Greger IH (2016) Structure and organization of heteromeric AMPA-type glutamate receptors. Science 352:aad3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herguedas B, Krieger J, Greger IH (2013) Receptor heteromeric assembly-how it works and why it matters: the case of ionotropic glutamate receptors. Prog Mol Biol Transl Sci 117:361–386. [DOI] [PubMed] [Google Scholar]
- Herguedas B, Watson JF, Ho H, Cais O, García-Nafría J, Greger IH (2019) Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP γ8. Science 364:eaav9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Jahr CE (2007) Extracellular glutamate concentration in hippocampal slice. J Neurosci 27:9736–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Nahir B, Jahr CE (2011) Distribution of extracellular glutamate in the neuropil of hippocampus. PLoS One 6:e26501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Nicoll RA (2016a) Kalirin and Trio proteins serve critical roles in excitatory synaptic transmission and LTP. Proc Natl Acad Sci USA 113:2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Nicoll RA (2016b) Long-term potentiation: From CaMKII to AMPA receptor trafficking. Annu Rev Physiol 78:351–365. [DOI] [PubMed] [Google Scholar]
- Herring BE, Shi Y, Suh YH, Zheng CY, Blankenship SM, Roche KW, Nicoll RA (2013) Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron 77:1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring WJ, Assaid C, Budd K, Vargo R, Mazenko RS, Lines C, Ellenbogen A, Verhagen Metman L (2017) A phase Ib randomized controlled study to evaluate the effectiveness of a single-dose of the NR2B selective N-methyl-D-aspartate antagonist MK-0657 on Levodopa-induced dyskinesias and motor symptoms in patients with Parkinson disease. Clin Neuropharmacol 40:255–260. [DOI] [PubMed] [Google Scholar]
- Hess SD, Daggett LP, Crona J, Deal C, Lu CC, Urrutia A, Chavez-Noriega L, Ellis SB, Johnson EC, Veliçelebi G (1996) Cloning and functional characterization of human heteromeric N-methyl-D-aspartate receptors. J Pharmacol Exp Ther 278:808–816. [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G (2007) Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450:1026–1030. [DOI] [PubMed] [Google Scholar]
- Hessler NA, Shirke AM, Malinow R (1993) The probability of transmitter release at a mammalian central synapse. Nature 366:569–572. [DOI] [PubMed] [Google Scholar]
- Hestrin S (1992) Activation and desensitization of glutamate-activated channels mediating fast excitatory synaptic currents in the visual cortex. Neuron 9:991–999. [DOI] [PubMed] [Google Scholar]
- Hestrin S, Nicoll RA, Perkel DJ, Sah P (1990) Analysis of excitatory synaptic action in pyramidal cells using whole-cell recording from rat hippocampal slices. J Physiol 422:203–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzelt KLML, Kraus C, Kusnik S, Thiel CT, Uebe S, Ekici AB, Trollmann R, Reis A, Zweier C (2020) A case of severe autosomal recessive spinocerebellar ataxia type 18 with a novel nonsense variant in GRID2. Eur J Med Genet 63:103998. [DOI] [PubMed] [Google Scholar]
- Hibi S, Ueno K, Nagato S, Kawano K, Ito K, Norimine Y, Takenaka O, Hanada T, Yonaga M (2012) Discovery of 2-(2-oxo-1-phenyl-5-pyridin-2-yl-1,2-dihydropyridin-3-yl)benzonitrile (perampanel): a novel, noncompetitive α-amino-3-hydroxy-5-methyl-4-isoxazolepropanoic acid (AMPA) receptor antagonist. J Med Chem 55:10584–10600. [DOI] [PubMed] [Google Scholar]
- Highland JNZanos PRiggs LMGeorgiou PClark SMMorris PJMoaddel RThomas CJZarate CA JrPereira EFR, et al. (2021) Hydroxynorketamines: Pharmacology and potential therapeutic applications. Pharmacol Rev 73:763–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406:78–81. [DOI] [PubMed] [Google Scholar]
- Hildebrand ME, Pitcher GM, Harding EK, Li H, Beggs S, Salter MW (2014) GluN2B and GluN2D NMDARs dominate synaptic responses in the adult spinal cord. Sci Rep 4:4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman BG, Gupta SC, Stairs DJ, Buonanno A, Dravid SM (2011) Behavioral analysis of NR2C knockout mouse reveals deficit in acquisition of conditioned fear and working memory. Neurobiol Learn Mem 95:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills LBMasri AKonno KKakegawa WLam ATLim-Melia EChandy NHill RSPartlow JNAl-Saffar M, et al. (2013) Deletions in GRID2 lead to a recessive syndrome of cerebellar ataxia and tonic upgaze in humans. Neurology 81:1378–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai HPang ZBao DMiyazaki TLi LMiura EParris JRong YWatanabe MYuzaki M, et al. (2005) Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat Neurosci 8:1534–1541. [DOI] [PubMed] [Google Scholar]
- Hirai K, Yoshioka H, Kihara M, Hasegawa K, Sakamoto T, Sawada T, Fushiki S (1999) Inhibiting neuronal migration by blocking NMDA receptors in the embryonic rat cerebral cortex: a tissue culture study. Brain Res Dev Brain Res 114:63–67. [DOI] [PubMed] [Google Scholar]
- Hirano T (2006) Cerebellar regulation mechanisms learned from studies on GluRdelta2. Mol Neurobiol 33:1–16. [DOI] [PubMed] [Google Scholar]
- Hirasawa T, Wada H, Kohsaka S, Uchino S (2003) Inhibition of NMDA receptors induces delayed neuronal maturation and sustained proliferation of progenitor cells during neocortical development. J Neurosci Res 74:676–687. [DOI] [PubMed] [Google Scholar]
- Hirohata S, Sakuma Y, Yanagida T, Yoshio T (2014) Association of cerebrospinal fluid anti-Sm antibodies with acute confusional state in systemic lupus erythematosus. Arthritis Res Ther 16:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyoshi T, Kambe D, Karasawa J, Chaki S (2014) Differential effects of NMDA receptor antagonists at lower and higher doses on basal gamma band oscillation power in rat cortical electroencephalograms. Neuropharmacology 85:384–396. [DOI] [PubMed] [Google Scholar]
- Hjelmstad GO, Isaac JT, Nicoll RA, Malenka RC (1999) Lack of AMPA receptor desensitization during basal synaptic transmission in the hippocampal slice. J Neurophysiol 81:3096–3099. [DOI] [PubMed] [Google Scholar]
- Hofmann SG, Meuret AE, Smits JA, Simon NM, Pollack MH, Eisenmenger K, Shiekh M, Otto MW (2006a) Augmentation of exposure therapy with D-cycloserine for social anxiety disorder. Arch Gen Psychiatry 63:298–304. [DOI] [PubMed] [Google Scholar]
- Hofmann SG, Otto MW, Pollack MH, Smits JA (2015) D-cycloserine augmentation of cognitive behavioral therapy for anxiety disorders: an update. Curr Psychiatry Rep 17:532. [DOI] [PubMed] [Google Scholar]
- Hofmann SG, Pollack MH, Otto MW (2006b) Augmentation treatment of psychotherapy for anxiety disorders with D-cycloserine. CNS Drug Rev 12:208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogner A, Greenwood JR, Liljefors T, Lunn ML, Egebjerg J, Larsen IK, Gouaux E, Kastrup JS (2003) Competitive antagonism of AMPA receptors by ligands of different classes: crystal structure of ATPO bound to the GluR2 ligand-binding core, in comparison with DNQX. J Med Chem 46:214–221. [DOI] [PubMed] [Google Scholar]
- Hogner A, Kastrup JS, Jin R, Liljefors T, Mayer ML, Egebjerg J, Larsen IK, Gouaux E (2002) Structural basis for AMPA receptor activation and ligand selectivity: crystal structures of five agonist complexes with the GluR2 ligand-binding core. J Mol Biol 322:93–109. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Boulter J, Maron C, Beasley L, Sullivan J, Pecht G, Heinemann S (1993) Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron 10:943–954. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Hartley M, Heinemann S (1991) Ca2+ permeability of KA-AMPA--gated glutamate receptor channels depends on subunit composition. Science 252:851–853. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S (1994) Cloned glutamate receptors. Annu Rev Neurosci 17:31–108. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Maron C, Heinemann S (1994) N-glycosylation site tagging suggests a three transmembrane domain topology for the glutamate receptor GluR1. Neuron 13:1331–1343. [DOI] [PubMed] [Google Scholar]
- Hollmann M, O’Shea-Greenfield A, Rogers SW, Heinemann S (1989) Cloning by functional expression of a member of the glutamate receptor family. Nature 342:643–648. [DOI] [PubMed] [Google Scholar]
- Holm MM, Lunn ML, Traynelis SF, Kastrup JS, Egebjerg J (2005) Structural determinants of agonist-specific kinetics at the ionotropic glutamate receptor 2. Proc Natl Acad Sci USA 102:12053–12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RBarnby GMaestrini EBacchelli EBrocklebank DSousa IMulder EJKantojärvi KJärvelä IKlauck SM, et al. ; EU Autism MOLGEN Consortium (2010) Linkage and candidate gene studies of autism spectrum disorders in European populations. Eur J Hum Genet 18:1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Jeyifous O, Ronilo M, Marshall J, Green WN, Standley S (2019) A novel function for the ER retention signals in the C-terminus of kainate receptor subunit, GluK5. Biochim Biophys Acta Mol Cell Res 1866:459–473. [DOI] [PubMed] [Google Scholar]
- Honoré T, Davies SN, Drejer J, Fletcher EJ, Jacobsen P, Lodge D, Nielsen FE (1988) Quinoxalinediones: potent competitive non-NMDA glutamate receptor antagonists. Science 241:701–703. [DOI] [PubMed] [Google Scholar]
- Honse Y, Ren H, Lipsky RH, Peoples RW (2004) Sites in the fourth membrane-associated domain regulate alcohol sensitivity of the NMDA receptor. Neuropharmacology 46:647–654. [DOI] [PubMed] [Google Scholar]
- Hood JL, Emeson RB (2012) Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr Top Microbiol Immunol 353:61–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak M, Petralia RS, Kaniakova M, Sans N (2014) ER to synapse trafficking of NMDA receptors. Front Cell Neurosci 8:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak M, Vlcek K, Chodounska H, Vyklicky L Jr (2006) Subtype-dependence of N-methyl-D-aspartate receptor modulation by pregnenolone sulfate. Neuroscience 137:93–102. [DOI] [PubMed] [Google Scholar]
- Horak M, Vlcek K, Petrovic M, Chodounska H, Vyklicky L Jr (2004) Molecular mechanism of pregnenolone sulfate action at NR1/NR2B receptors. J Neurosci 24:10318–10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak M, Wenthold RJ (2009) Different roles of C-terminal cassettes in the trafficking of full-length NR1 subunits to the cell surface. J Biol Chem 284:9683–9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horning MS, Mayer ML (2004) Regulation of AMPA receptor gating by ligand binding core dimers. Neuron 41:379–388. [DOI] [PubMed] [Google Scholar]
- Hosoi N, Holt M, Sakaba T (2009) Calcium dependence of exo- and endocytotic coupling at a glutamatergic synapse. Neuron 63:216–229. [DOI] [PubMed] [Google Scholar]
- Howe JR (2015) Modulation of non-NMDA receptor gating by auxiliary subunits. J Physiol 593:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoxha E, Balbo I, Miniaci MC, Tempia F (2018) Purkinje cell signaling deficits in animal models of ataxia. Front Synaptic Neurosci 10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrcka Krausova BKysilov BCerny JVyklicky VSmejkalova TLadislav MBalik AKorinek MChodounska HKudova E, et al. (2020) Site of action of brain neurosteroid pregnenolone sulfate at the N-methyl-D-aspartate receptor. J Neurosci 40:5922–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Zheng F (2005) Molecular determinants of glycine-independent desensitization of NR1/NR2A receptors. J Pharmacol Exp Ther 313:563–569. [DOI] [PubMed] [Google Scholar]
- Hu C, Chen W, Myers SJ, Yuan H, Traynelis SF (2016) Human GRIN2B variants in neurodevelopmental disorders. J Pharmacol Sci 132:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XD, Huang Q, Roadcap DW, Shenolikar SS, Xia H (2006) Actin-associated neurabin-protein phosphatase-1 complex regulates hippocampal plasticity. J Neurochem 98:1841–1851. [DOI] [PubMed] [Google Scholar]
- Hu XD, Huang Q, Yang X, Xia H (2007) Differential regulation of AMPA receptor trafficking by neurabin-targeted synaptic protein phosphatase-1 in synaptic transmission and long-term depression in hippocampus. J Neurosci 27:4674–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Lin A, Dong H, Wang C (2014) The human δ2 glutamate receptor gene is not mutated in patients with spinocerebellar ataxia. Neural Regen Res 9:1068–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LC, Barclay M, Lee K, Peter S, Housley GD, Thorne PR, Montgomery JM (2012) Synaptic profiles during neurite extension, refinement and retraction in the developing cochlea. Neural Dev 7:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ni W, Zhang C (2017) Calcium-impermeable NMDA receptor: A novel target for addiction. Neurosci Bull 33:357–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Dykes-Hoberg M, Tanaka K, Rothstein JD, Bergles DE (2004) Climbing fiber activation of EAAT4 transporters and kainate receptors in cerebellar Purkinje cells. J Neurosci 24:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Schlüter OM, Dong Y (2013) An unusual suspect in cocaine addiction. Neuron 80:835–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubalkova PLadislav MVyklicky VSmejkalova THrcka Krausova BKysilov BKrusek JNaimová ZKorinek MChodounska H, et al. (2021) Palmitoylation controls NMDA receptor function and steroid sensitivity. J Neurosci 41:2119–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta PT, Kowal C, DeGiorgio LA, Volpe BT, Diamond B (2006) Immunity and behavior: antibodies alter emotion. Proc Natl Acad Sci USA 103:678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner JE (1990) Glutamate receptor channels in rat DRG neurons: activation by kainate and quisqualate and blockade of desensitization by Con A. Neuron 5:255–266. [DOI] [PubMed] [Google Scholar]
- Huettner JE (2015) Glutamate receptor pores. J Physiol 593:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner JE, Bean BP (1988) Block of N-methyl-D-aspartate-activated current by the anticonvulsant MK-801: selective binding to open channels. Proc Natl Acad Sci USA 85:1307–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes EG, Peng X, Gleichman AJ, Lai M, Zhou L, Tsou R, Parsons TD, Lynch DR, Dalmau J, Balice-Gordon RJ (2010) Cellular and synaptic mechanisms of anti-NMDA receptor encephalitis. J Neurosci 30:5866–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume RI, Dingledine R, Heinemann SF (1991) Identification of a site in glutamate receptor subunits that controls calcium permeability. Science 253:1028–1031. [DOI] [PubMed] [Google Scholar]
- Hunt DL, Castillo PE (2012) Synaptic plasticity of NMDA receptors: mechanisms and functional implications. Curr Opin Neurobiol 22:496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Petralia RS, Vu T, Wenthold RJ (1993) Expression of AMPA-selective glutamate receptor subunits in morphologically defined neurons of the mammalian cochlear nucleus. J Neurosci 13:1932–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye ES, Sthoeger ZM, Dayan M, Zinger H, Elbirt D, Levite M, Mozes E (2005) Autoantibodies to a NR2A peptide of the glutamate/NMDA receptor in sera of patients with systemic lupus erythematosus. Ann Rheum Dis 64:1210–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SY, Oh B, Zhang Z, Miller W, Solter D, Knowles BB (1999) The mouse cornichon gene family. Dev Genes Evol 209:120–125. [DOI] [PubMed] [Google Scholar]
- Iacobucci GJ, Popescu GK (2017a) NMDA receptors: linking physiological output to biophysical operation. Nat Rev Neurosci 18:236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci GJ, Popescu GK (2017b) Resident calmodulin primes NMDA receptors for Ca2+-dependent inactivation. Biophys J 113:2236–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci GJ, Popescu GK (2019) Spatial coupling tunes NMDA receptor responses via Ca2+ diffusion. J Neurosci 39:8831–8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci GJ, Popescu GK (2020) Ca2+-dependent inactivation of GluN2A and GluN2B NMDA receptors occurs by a common kinetic mechanism. Biophys J 118:798–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci GJ, Wen H, Helou M, Liu B, Zheng W, Popescu GK (2021) Cross-subunit interactions that stabilize open states mediate gating in NMDA receptors. Proc Natl Acad Sci USA 118:e2007511118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa R, Sakimura K, Watanabe M (2016) GluD2 endows parallel Fiber-Purkinje Cell Synapses with a High Regenerative Capacity. J Neurosci 36:4846–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K (2019) The functional role of polyamines in eukaryotic cells. Int J Biochem Cell Biol 107:104–115. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Shirahata A, Pahk AJ, Kashiwagi K, Williams K (1997) Benzyl-polyamines: novel, potent N-methyl-D-aspartate receptor antagonists. J Pharmacol Exp Ther 283:533–540. [PubMed] [Google Scholar]
- Ihle EC, Patneau DK (2000) Modulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor desensitization by extracellular protons. Mol Pharmacol 58:1204–1212. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa M, Mori H, Araki K, Sakimura K, Watanabe M, Inoue Y, Mishina M (1992) Cloning and expression of the epsilon 4 subunit of the NMDA receptor channel. FEBS Lett 313:34–38. [DOI] [PubMed] [Google Scholar]
- Inanobe A, Furukawa H, Gouaux E (2005) Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron 47:71–84. [DOI] [PubMed] [Google Scholar]
- Incontro S, Díaz-Alonso J, Iafrati J, Vieira M, Asensio CS, Sohal VS, Roche KW, Bender KJ, Nicoll RA (2018) The CaMKII/NMDA receptor complex controls hippocampal synaptic transmission by kinase-dependent and independent mechanisms. Nat Commun 9:2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Kamiyama H, Matsumoto M, Yanagawa Y, Hiraide S, Saito Y, Shimamura K, Togashi H (2013) Synaptic modulation via basolateral amygdala on the rat hippocampus-medial prefrontal cortex pathway in fear extinction. J Pharmacol Sci 123:267–278. [DOI] [PubMed] [Google Scholar]
- Intson K, van Eede MC, Islam R, Milenkovic M, Yan Y, Salahpour A, Henkelman RM, Ramsey AJ (2019) Progressive neuroanatomical changes caused by Grin1 loss-of-function mutation. Neurobiol Dis 132:104527. [DOI] [PubMed] [Google Scholar]
- Irani SRBera KWaters PZuliani LMaxwell SZandi MSFriese MAGalea IKullmann DMBeeson D, et al. (2010) N-methyl-D-aspartate antibody encephalitis: temporal progression of clinical and paraclinical observations in a predominantly non-paraneoplastic disorder of both sexes. Brain 133:1655–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine MWCosta BMDlaboga DCulley GRHulse RScholefield CLAtlason PFang GEaves RMorley R, et al. (2012) Piperazine-2,3-dicarboxylic acid derivatives as dual antagonists of NMDA and GluK1-containing kainate receptors. J Med Chem 55:327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine MW, Fang G, Sapkota K, Burnell ES, Volianskis A, Costa BM, Culley G, Collingridge GL, Monaghan DT, Jane DE (2019) Investigation of the structural requirements for N-methyl-D-aspartate receptor positive and negative allosteric modulators based on 2-naphthoic acid. Eur J Med Chem 164:471–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac JT, Nicoll RA, Malenka RC (1995) Evidence for silent synapses: implications for the expression of LTP. Neuron 15:427–434. [DOI] [PubMed] [Google Scholar]
- Isaacson JS, Murphy GJ (2001) Glutamate-mediated extrasynaptic inhibition: direct coupling of NMDA receptors to Ca(2+)-activated K+ channels. Neuron 31:1027–1034. [DOI] [PubMed] [Google Scholar]
- Ishihama K, Kogo M, Wakisaka S, Turman JE Jr (2005) Prenatal development of NMDA receptor composition and function in trigeminal neurons. Arch Histol Cytol 68:321–335. [DOI] [PubMed] [Google Scholar]
- Ishihama K, Turman JE Jr (2006) NR3 protein expression in trigeminal neurons during postnatal development. Brain Res 1095:12–16. [DOI] [PubMed] [Google Scholar]
- Ishii TMoriyoshi KSugihara HSakurada KKadotani HYokoi MAkazawa CShigemoto RMizuno NMasu M, et al. (1993) Molecular characterization of the family of the N-methyl-D-aspartate receptor subunits. J Biol Chem 268:2836–2843. [PubMed] [Google Scholar]
- Ishii T, Stolz JR, Swanson GT (2020) Auxiliary proteins are the predominant determinants of differential efficacy of clinical candidates acting as AMPA receptor positive allosteric modulators. Mol Pharmacol 97:336–350. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Yoshitomi T, Covey DF, Zorumski CF, Izumi Y (2018) Neurosteroids and oxysterols as potential therapeutic agents for glaucoma and Alzheimer’s disease. Neuropsychiatry (London) 8:344–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwata S, Umino A, Balu DT, Coyle JT, Nishikawa T (2015) Neuronal serine racemase regulates extracellular D-serine levels in the adult mouse hippocampus. J Neural Transm (Vienna) 122:1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito I, Futai K, Katagiri H, Watanabe M, Sakimura K, Mishina M, Sugiyama H (1997) Synapse-selective impairment of NMDA receptor functions in mice lacking NMDA receptor epsilon 1 or epsilon 2 subunit. J Physiol 500:401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito I, Tanabe S, Kohda A, Sugiyama H (1990) Allosteric potentiation of quisqualate receptors by a nootropic drug aniracetam. J Physiol 424:533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Contractor A, Swanson GT (2004) Attenuated plasticity of postsynaptic kainate receptors in hippocampal CA3 pyramidal neurons. J Neurosci 24:6228–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Ishida A, Kakegawa W, Kohda K, Miura E, Okabe S, Yuzaki M (2014) Cbln1 downregulates the formation and function of inhibitory synapses in mouse cerebellar Purkinje cells. Eur J Neurosci 39:1268–1280. [DOI] [PubMed] [Google Scholar]
- Ito-Ishida A, Miyazaki T, Miura E, Matsuda K, Watanabe M, Yuzaki M, Okabe S (2012) Presynaptically released Cbln1 induces dynamic axonal structural changes by interacting with GluD2 during cerebellar synapse formation. Neuron 76:549–564. [DOI] [PubMed] [Google Scholar]
- Iwasato T, Datwani A, Wolf AM, Nishiyama H, Taguchi Y, Tonegawa S, Knöpfel T, Erzurumlu RS, Itohara S (2000) Cortex-restricted disruption of NMDAR1 impairs neuronal patterns in the barrel cortex. Nature 406:726–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasato T, Erzurumlu RS, Huerta PT, Chen DF, Sasaoka T, Ulupinar E, Tonegawa S (1997) NMDA receptor-dependent refinement of somatotopic maps. Neuron 19:1201–1210. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Mennerick SJ, Doherty JJ, Zorumski CF (2021) Oxysterols modulate the acute effects of ethanol on hippocampal N-methyl-d-aspartate receptors, long-term potentiation, and learning. J Pharmacol Exp Ther 377:181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafari N, Henley JM, Hanley JG (2012) PICK1 mediates transient synaptic expression of GluA2-lacking AMPA receptors during glycine-induced AMPA receptor trafficking. J Neurosci 32:11618–11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jääskeläinen IP, Ahveninen J, Andermann ML, Belliveau JW, Raij T, Sams M (2011) Short-term plasticity as a neural mechanism supporting memory and attentional functions. Brain Res 1422:66–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A, Hamad MIK, Gonda S, Gralla S, Pahl S, Hollmann M, Wahle P (2019) Development of cortical pyramidal cell and interneuronal dendrites: a role for kainate receptor subunits and NETO1. Mol Neurobiol 56:4960–4979. [DOI] [PubMed] [Google Scholar]
- Jackman SL, Regehr WG (2017) The mechanisms and functions of synaptic facilitation. Neuron 94:447–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC, Milstein AD, Soto D, Farrant M, Cull-Candy SG, Nicoll RA (2011) Probing TARP modulation of AMPA receptor conductance with polyamine toxins. J Neurosci 31:7511–7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC, Nicoll RA (2011a) The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron 70:178–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC, Nicoll RA (2011b) Stargazin (TARP gamma-2) is required for compartment-specific AMPA receptor trafficking and synaptic plasticity in cerebellar stellate cells. J Neurosci 31:3939–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AL, Weinberg RJ (2015) The organization of AMPA receptor subunits at the postsynaptic membrane. Hippocampus 25:798–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi E, von Engelhardt J (2018) AMPA receptor complex constituents: Control of receptor assembly, membrane trafficking and subcellular localization. Mol Cell Neurosci 91:67–75. [DOI] [PubMed] [Google Scholar]
- Jacobi E, von Engelhardt J (2021) Modulation of information processing by AMPA receptor auxiliary subunits. J Physiol 599:471–483. [DOI] [PubMed] [Google Scholar]
- Jahr CE (1992) High probability opening of NMDA receptor channels by L-glutamate. Science 255:470–472. [DOI] [PubMed] [Google Scholar]
- Jahr CE, Stevens CF (1987) Glutamate activates multiple single channel conductances in hippocampal neurons. Nature 325:522–525. [DOI] [PubMed] [Google Scholar]
- Jalali-Yazdi F, Chowdhury S, Yoshioka C, Gouaux E (2018) Mechanisms for zinc and proton inhibition of the GluN1/GluN2A NMDA receptor. Cell 175:1520-1532 e1515. [DOI] [PMC free article] [PubMed]
- Jamain S, Betancur C, Quach H, Philippe A, Fellous M, Giros B, Gillberg C, Leboyer M, Bourgeron T; Paris Autism Research International Sibpair (PARIS) Study (2002) Linkage and association of the glutamate receptor 6 gene with autism. Mol Psychiatry 7:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jane DE, Hoo K, Kamboj R, Deverill M, Bleakman D, Mandelzys A (1997) Synthesis of willardiine and 6-azawillardiine analogs: pharmacological characterization on cloned homomeric human AMPA and kainate receptor subtypes. J Med Chem 40:3645–3650. [DOI] [PubMed] [Google Scholar]
- Jane DE, Lodge D, Collingridge GL (2009) Kainate receptors: pharmacology, function and therapeutic potential. Neuropharmacology 56:90–113. [DOI] [PubMed] [Google Scholar]
- Jang MK, Mierke DF, Russek SJ, Farb DH (2004) A steroid modulatory domain on NR2B controls N-methyl-D-aspartate receptor proton sensitivity. Proc Natl Acad Sci USA 101:8198–8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janovjak H, Sandoz G, Isacoff EY (2011) A modern ionotropic glutamate receptor with a K(+) selectivity signature sequence. Nat Commun 2:232. [DOI] [PubMed] [Google Scholar]
- Janovjak H, Szobota S, Wyart C, Trauner D, Isacoff EY (2010) A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nat Neurosci 13:1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskolski F, Coussen F, Mulle C (2005a) Subcellular localization and trafficking of kainate receptors. Trends Pharmacol Sci 26:20–26. [DOI] [PubMed] [Google Scholar]
- Jaskolski F, Coussen F, Nagarajan N, Normand E, Rosenmund C, Mulle C (2004) Subunit composition and alternative splicing regulate membrane delivery of kainate receptors. J Neurosci 24:2506–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskolski F, Normand E, Mulle C, Coussen F (2005b) Differential trafficking of GluR7 kainate receptor subunit splice variants. J Biol Chem 280:22968–22976. [DOI] [PubMed] [Google Scholar]
- Jatzke C, Hernandez M, Wollmuth LP (2003) Extracellular vestibule determinants of Ca2+ influx in Ca2+-permeable AMPA receptor channels. J Physiol 549:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jatzke C, Watanabe J, Wollmuth LP (2002) Voltage and concentration dependence of Ca(2+) permeability in recombinant glutamate receptor subtypes. J Physiol 538:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javitt DC, Zukin SR (1991) Recent advances in the phencyclidine model of schizophrenia. Am J Psychiatry 148:1301–1308. [DOI] [PubMed] [Google Scholar]
- Jenkins MA, Traynelis SF (2012) PKC phosphorylates GluA1-Ser831 to enhance AMPA receptor conductance. Channels (Austin) 6:60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins MA, Wells G, Bachman J, Snyder JP, Jenkins A, Huganir RL, Oswald RE, Traynelis SF (2014) Regulation of GluA1 α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor function by protein kinase C at serine-818 and threonine-840. Mol Pharmacol 85:618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AA, Christesen T, Bølcho U, Greenwood JR, Postorino G, Vogensen SB, Johansen TN, Egebjerg J, Bräuner-Osborne H, Clausen RP (2007) Functional characterization of Tet-AMPA [tetrazolyl-2-amino-3-(3-hydroxy-5-methyl- 4-isoxazolyl)propionic acid] analogues at ionotropic glutamate receptors GluR1-GluR4. The molecular basis for the functional selectivity profile of 2-Bn-Tet-AMPA. J Med Chem 50:4177–4185. [DOI] [PubMed] [Google Scholar]
- Jensen LS, Bølcho U, Egebjerg J, Strømgaard K (2006) Design, synthesis, and pharmacological characterization of polyamine toxin derivatives: potent ligands for the pore-forming region of AMPA receptors. ChemMedChem 1:419–428. [DOI] [PubMed] [Google Scholar]
- Jensen VKaiser KMBorchardt TAdelmann GRozov ABurnashev NBrix CFrotscher MAndersen PHvalby Ø, et al. (2003) A juvenile form of postsynaptic hippocampal long-term potentiation in mice deficient for the AMPA receptor subunit GluR-A. J Physiol 553:843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen A, Tajima N, Fernandez-Cuervo G, Garnier-Amblard EC, Furukawa H (2014) Structural insights into competitive antagonism in NMDA receptors. Neuron 81:366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen M, Frederiksen K, Yi F, Clausen RP, Hansen KB, Bräuner-Osborne H, Kilburn P, Damholt A (2017) Identification of AICP as a GluN2C-selective N-methyl-d-aspartate receptor superagonist at the GluN1 glycine site. Mol Pharmacol 92:151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jézéquel JJohansson EMDupuis JPRogemond VGréa HKellermayer BHamdani NLe Guen ERabu CLepleux M, et al. (2017) Dynamic disorganization of synaptic NMDA receptors triggered by autoantibodies from psychotic patients. Nat Commun 8:1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin LSugiyama HTakigawa MKatagiri DTomitori HNishimura KKaur NPhanstiel O 4thKitajima MTakayama H, et al. (2007) Comparative studies of anthraquinone- and anthracene-tetraamines as blockers of N-methyl-D-aspartate receptors. J Pharmacol Exp Ther 320:47–55. [DOI] [PubMed] [Google Scholar]
- Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E (2003) Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci 6:803–810. [DOI] [PubMed] [Google Scholar]
- Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM (2005) Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci 25:9027–9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Horning M, Mayer ML, Gouaux E (2002) Mechanism of activation and selectivity in a ligand-gated ion channel: structural and functional studies of GluR2 and quisqualate. Biochemistry 41:15635–15643. [DOI] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E (2009) Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J 28:1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin XT, Smith Y (2011) Localization and functions of kainate receptors in the basal ganglia. Adv Exp Med Biol 717:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Yu L, Geng J, Wang J, Jin X, Huang H (2015) A novel 47.2 Mb duplication on chromosomal bands Xq21.1-25 associated with mental retardation. Gene 567:98–102. [DOI] [PubMed] [Google Scholar]
- Johansen TH, Chaudhary A, Verdoorn TA (1995) Interactions among GYKI-52466, cyclothiazide, and aniracetam at recombinant AMPA and kainate receptors. Mol Pharmacol 48:946–955. [PubMed] [Google Scholar]
- Johansen TH, Drejer J, Wätjen F, Nielsen EO (1993) A novel non-NMDA receptor antagonist shows selective displacement of low-affinity [3H]kainate binding. Eur J Pharmacol 246:195–204. [DOI] [PubMed] [Google Scholar]
- Johnson JW, Ascher P (1990) Voltage-dependent block by intracellular Mg2+ of N-methyl-D-aspartate-activated channels. Biophys J 57:1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JW, Glasgow NG, Povysheva NV (2015) Recent insights into the mode of action of memantine and ketamine. Curr Opin Pharmacol 20:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston D, Williams S, Jaffe D, Gray R (1992) NMDA-receptor-independent long-term potentiation. Annu Rev Physiol 54:489–505. [DOI] [PubMed] [Google Scholar]
- Jonas P (2000) The time course of signaling at central glutamatergic synapses. News Physiol Sci 15:83–89. [DOI] [PubMed] [Google Scholar]
- Jonas P, Major G, Sakmann B (1993) Quantal components of unitary EPSCs at the mossy fibre synapse on CA3 pyramidal cells of rat hippocampus. J Physiol 472:615–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas P, Racca C, Sakmann B, Seeburg PH, Monyer H (1994) Differences in Ca2+ permeability of AMPA-type glutamate receptor channels in neocortical neurons caused by differential GluR-B subunit expression. Neuron 12:1281–1289. [DOI] [PubMed] [Google Scholar]
- Jones RS, Carroll RC, Nawy S (2012) Light-induced plasticity of synaptic AMPA receptor composition in retinal ganglion cells. Neuron 75:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Gibb AJ (2005) Functional NR2B- and NR2D-containing NMDA receptor channels in rat substantia nigra dopaminergic neurones. J Physiol 569:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JY, Kim BW, Lee JS, Park JY, Kim S, Yun YJ, Lee SH, Lee SH, Rhim H, Son H (2007) Activation of NMDA receptors increases proliferation and differentiation of hippocampal neural progenitor cells. J Cell Sci 120:1358–1370. [DOI] [PubMed] [Google Scholar]
- Joseph DJ, Williams DJ, MacDermott AB (2011) Modulation of neurite outgrowth by activation of calcium-permeable kainate receptors expressed by rat nociceptive-like dorsal root ganglion neurons. Dev Neurobiol 71:818–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhanneau JS, Ball SM, Molnár E, Isaac JT (2011) Mechanisms of bi-directional modulation of thalamocortical transmission in barrel cortex by presynaptic kainate receptors. Neuropharmacology 60:832–841. [DOI] [PubMed] [Google Scholar]
- Jourdain P, Bergersen LH, Bhaukaurally K, Bezzi P, Santello M, Domercq M, Matute C, Tonello F, Gundersen V, Volterra A (2007) Glutamate exocytosis from astrocytes controls synaptic strength. Nat Neurosci 10:331–339. [DOI] [PubMed] [Google Scholar]
- Jourdi H, Hsu YT, Zhou M, Qin Q, Bi X, Baudry M (2009) Positive AMPA receptor modulation rapidly stimulates BDNF release and increases dendritic mRNA translation. J Neurosci 29:8688–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juknaitė LSugamata YTokiwa KIshikawa YTakamizawa SEng ASakai RPickering DSFrydenvang KSwanson GT, et al. (2013) Studies on an (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA) receptor antagonist IKM-159: asymmetric synthesis, neuroactivity, and structural characterization. J Med Chem 56:2283–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juknaitė LVenskutonytė RAssaf ZFaure SGefflaut TAitken DJNielsen BGajhede MKastrup JSBunch L, et al. (2012) Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors. J Struct Biol 180:39–46. [DOI] [PubMed] [Google Scholar]
- Jurado S, Biou V, Malenka RC (2010) A calcineurin/AKAP complex is required for NMDA receptor-dependent long-term depression. Nat Neurosci 13:1053–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S, Goswami D, Zhang Y, Molina AJ, Südhof TC, Malenka RC (2013) LTP requires a unique postsynaptic SNARE fusion machinery. Neuron 77:542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaae BHHarpsøe KKastrup JSSanz ACPickering DSMetzler BClausen RPGajhede MSauerberg PLiljefors T, et al. (2007) Structural proof of a dimeric positive modulator bridging two identical AMPA receptor-binding sites. Chem Biol 14:1294–1303. [DOI] [PubMed] [Google Scholar]
- Kaiser TMKell SAKusumoto HShaulsky GBhattacharya SEpplin MPStrong KLMiller EJCox BDMenaldino DS, et al. (2018) The bioactive protein-ligand conformation of GluN2C-selective positive allosteric modulators bound to the NMDA receptor. Mol Pharmacol 93:141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa W, Kohda K, Yuzaki M (2007a) The delta2 ‘ionotropic’ glutamate receptor functions as a non-ionotropic receptor to control cerebellar synaptic plasticity. J Physiol 584:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa W, Miyazaki T, Hirai H, Motohashi J, Mishina M, Watanabe M, Yuzaki M (2007b) Ca2+ permeability of the channel pore is not essential for the delta2 glutamate receptor to regulate synaptic plasticity and motor coordination. J Physiol 579:729–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa W, Miyazaki T, Kohda K, Matsuda K, Emi K, Motohashi J, Watanabe M, Yuzaki M (2009) The N-terminal domain of GluD2 (GluRdelta2) recruits presynaptic terminals and regulates synaptogenesis in the cerebellum in vivo. J Neurosci 29:5738–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa WMiyoshi YHamase KMatsuda SMatsuda KKohda KEmi KMotohashi JKonno RZaitsu K, et al. (2011) D-serine regulates cerebellar LTD and motor coordination through the δ2 glutamate receptor. Nat Neurosci 14:603–611. [DOI] [PubMed] [Google Scholar]
- Kalappa BI, Anderson CT, Goldberg JM, Lippard SJ, Tzounopoulos T (2015) AMPA receptor inhibition by synaptically released zinc. Proc Natl Acad Sci USA 112:15749–15754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalashnikova E, Lorca RA, Kaur I, Barisone GA, Li B, Ishimaru T, Trimmer JS, Mohapatra DP, Díaz E (2010) SynDIG1: an activity-regulated, AMPA- receptor-interacting transmembrane protein that regulates excitatory synapse development. Neuron 65:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb RG (1994) Regulation of motor neuron dendrite growth by NMDA receptor activation. Development 120:3063–3071. [DOI] [PubMed] [Google Scholar]
- Kalbaugh TL, VanDongen HM, VanDongen AM (2004) Ligand-binding residues integrate affinity and efficacy in the NMDA receptor. Mol Pharmacol 66:209–219. [DOI] [PubMed] [Google Scholar]
- Kalbaugh TL, Zhang J, Diamond JS (2009) Coagonist release modulates NMDA receptor subtype contributions at synaptic inputs to retinal ganglion cells. J Neurosci 29:1469–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalova A, Futai K, Delpire E, Nakagawa T (2020) AMPA receptor auxiliary subunit GSG1L suppresses short-term facilitation in corticothalamic synapses and determines seizure susceptibility. Cell Rep 32:107921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalova A, Nakagawa T (2021) AMPA receptor structure and auxiliary subunits. J Physiol 599:453–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj SK, Swanson GT, Cull-Candy SG (1995) Intracellular spermine confers rectification on rat calcium-permeable AMPA and kainate receptors. J Physiol 486:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya H, Ozawa S (2000) Kainate receptor-mediated presynaptic inhibition at the mouse hippocampal mossy fibre synapse. J Physiol 523:653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampa BM, Clements J, Jonas P, Stuart GJ (2004) Kinetics of Mg2+ unblock of NMDA receptors: implications for spike-timing dependent synaptic plasticity. J Physiol 556:337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniakova MKleteckova LLichnerova KHolubova KSkrenkova KKorinek MKrusek JSmejkalova TKorabecny JVales K, et al. (2018) 7-Methoxyderivative of tacrine is a ‘foot-in-the-door’ open-channel blocker of GluN1/GluN2 and GluN1/GluN3 NMDA receptors with neuroprotective activity in vivo. Neuropharmacology 140:217–232. [DOI] [PubMed] [Google Scholar]
- Kapadia M, Bijelić D, Zhao H, Ma D, Stojanovich L, Milošević M, Andjus P, Šakić B (2017) Effects of sustained i.c.v. infusion of lupus CSF and autoantibodies on behavioral phenotype and neuronal calcium signaling. Acta Neuropathol Commun 5:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J, Dimlich RV, Biros MH, Hedges J (1987) Mechanisms of ischemic cerebral injury. Resuscitation 15:149–169. [DOI] [PubMed] [Google Scholar]
- Káradóttir R, Cavelier P, Bergersen LH, Attwell D (2005) NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature 438:1162–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Furukawa H (2014) Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 344:992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H (2009) Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J 28:3910–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H (2011) Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 475:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karataeva AR, Klaassen RV, Ströder J, Ruiperez-Alonso M, Hjorth JJ, van Nierop P, Spijker S, Mansvelder HD, Smit AB (2014) C-terminal interactors of the AMPA receptor auxiliary subunit Shisa9. PLoS One 9:e87360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karavanova I, Vasudevan K, Cheng J, Buonanno A (2007) Novel regional and developmental NMDA receptor expression patterns uncovered in NR2C subunit-beta-galactosidase knock-in mice. Mol Cell Neurosci 34:468–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwabuchi NIkeda KAraki KHirano TShibuki KTakayama CInoue YKutsuwada TYagi TKang Y, et al. (1995) Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluR delta 2 mutant mice. Cell 81:245–252. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K, Pahk AJ, Masuko T, Igarashi K, Williams K (1997) Block and modulation of N-methyl-D-aspartate receptors by polyamines and protons: role of amino acid residues in the transmembrane and pore-forming regions of NR1 and NR2 subunits. Mol Pharmacol 52:701–713. [DOI] [PubMed] [Google Scholar]
- Kask K, Jerecic J, Zamanillo D, Wilbertz J, Sprengel R, Seeburg PH (2000) Developmental profile of kainate receptor subunit KA1 revealed by Cre expression in YAC transgenic mice. Brain Res 876:55–61. [DOI] [PubMed] [Google Scholar]
- Kask K, Zamanillo D, Rozov A, Burnashev N, Sprengel R, Seeburg PH (1998) The AMPA receptor subunit GluR-B in its Q/R site-unedited form is not essential for brain development and function. Proc Natl Acad Sci USA 95:13777–13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper CPickering DSMirza OOlsen LKristensen ASGreenwood JRLiljefors TSchousboe AWätjen FGajhede M, et al. (2006) The structure of a mixed GluR2 ligand-binding core dimer in complex with (S)-glutamate and the antagonist (S)-NS1209. J Mol Biol 357:1184–1201. [DOI] [PubMed] [Google Scholar]
- Kasteleijn-Nolst Trenité DBrandt CMayer TRosenow FSchmidt BSteinhoff BJGardin AImbert GJohns DSagkriotis A, et al. (2015) Dose-dependent suppression of human photoparoxysmal response with the competitive AMPA/kainate receptor antagonist BGG492: Clear PK/PD relationship. Epilepsia 56:924–932. [DOI] [PubMed] [Google Scholar]
- Kato ASBurris KDGardinier KMGernert DLPorter WJReel JDing CTu YSchober DALee MR, et al. (2016) Forebrain-selective AMPA-receptor antagonism guided by TARP γ-8 as an antiepileptic mechanism. Nat Med 22:1496–1501. [DOI] [PubMed] [Google Scholar]
- Kato ASGill MBHo MTYu HTu YSiuda ERWang HQian YWNisenbaum ESTomita S, et al. (2010) Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron 68:1082–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Siuda ER, Nisenbaum ES, Bredt DS (2008) AMPA receptor subunit-specific regulation by a distinct family of type II TARPs. Neuron 59:986–996. [DOI] [PubMed] [Google Scholar]
- Kato ASZhou WMilstein ADKnierman MDSiuda ERDotzlaf JEYu HHale JENisenbaum ESNicoll RA, et al. (2007) New transmembrane AMPA receptor regulatory protein isoform, gamma-7, differentially regulates AMPA receptors. J Neurosci 27:4969–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Duman RS (2020) Rapastinel, a novel glutamatergic agent with ketamine-like antidepressant actions: Convergent mechanisms. Pharmacol Biochem Behav 188:172827. [DOI] [PubMed] [Google Scholar]
- Katsura K, Asplund B, Ekholm A, Siesjö BK (1992) Extra- and intracellular pH in the brain during ischaemia, related to tissue lactate content in normo- and hypercapnic rats. Eur J Neurosci 4:166–176. [DOI] [PubMed] [Google Scholar]
- Katsura K and Siesjo B (1998) Acid-base metabolism in ischemia, in pH and Brain Function (Kaila K and Ransom BR eds) p 563, Wiley-Liss, New York. [Google Scholar]
- Katz B, Miledi R (1968) The role of calcium in neuromuscular facilitation. J Physiol 195:481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz B, Thesleff S (1957) A study of the desensitization produced by acetylcholine at the motor end-plate. J Physiol 138:63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz DM, Menniti FS, Mather RJ (2016) N-Methyl-D-aspartate receptors, ketamine, and Rett syndrome: Something special on the road to treatments? Biol Psychiatry 79:710–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman BM, Perszyk RE, Yuan H, Tahirovic YA, Sotimehin AE, Traynelis SF, Liotta DC (2015) A novel class of negative allosteric modulators of NMDA receptor function. Bioorg Med Chem Lett 25:5583–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman AM, Milnerwood AJ, Sepers MD, Coquinco A, She K, Wang L, Lee H, Craig AM, Cynader M, Raymond LA (2012) Opposing roles of synaptic and extrasynaptic NMDA receptor signaling in cocultured striatal and cortical neurons. J Neurosci 32:3992–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavalali ET, Monteggia LM (2012) Synaptic mechanisms underlying rapid antidepressant action of ketamine. Am J Psychiatry 169:1150–1156. [DOI] [PubMed] [Google Scholar]
- Kawata S, Miyazaki T, Yamazaki M, Mikuni T, Yamasaki M, Hashimoto K, Watanabe M, Sakimura K, Kano M (2014) Global scaling down of excitatory postsynaptic responses in cerebellar Purkinje cells impairs developmental synapse elimination. Cell Rep 8:1119–1129. [DOI] [PubMed] [Google Scholar]
- Kay AR, Tóth K (2008) Is zinc a neuromodulator? Sci Signal 1:re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser SHansen JCStaudt MMoroz ALarsen YTemperini PYi FSyrenne JTKrogsgaard-Larsen NIliadis S, et al. (2020) Stereoselective synthesis of new (2S,3R)-3-carboxyphenyl)pyrrolidine-2-carboxylic acid analogues utilizing a C(sp3)-H activation strategy and structure-activity relationship studies at the ionotropic glutamate receptors. ACS Chem Neurosci 11:674–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi R, Dai J, Sweeney C, Zhou HX, Wollmuth LP (2014) Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat Neurosci 17:914–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi R, Gan Q, Talukder I, Markowitz M, Salussolia CL, Wollmuth LP (2013) Asynchronous movements prior to pore opening in NMDA receptors. J Neurosci 33:12052–12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keavy D, Bristow LJ, Sivarao DV, Batchelder M, King D, Thangathirupathy S, Macor JE, Weed MR (2016) The qEEG signature of selective NMDA NR2B negative allosteric modulators; A potential translational biomarker for drug development. PLoS One 11:e0152729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe LA, Bellone C, De Roo M, Zandueta A, Dey PN, Pérez-Otaño I, Muller D (2014) GluN3A promotes dendritic spine pruning and destabilization during postnatal development. J Neurosci 34:9213–9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinänen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH (1990) A family of AMPA-selective glutamate receptors. Science 249:556–560. [DOI] [PubMed] [Google Scholar]
- Kellermayer BFerreira JSDupuis JLevet FGrillo-Bosch DBard LLinarès-Loyez JBouchet DChoquet DRusakov DA, et al. (2018) Differential nanoscale topography and functional role of GluN2-NMDA receptor subtypes at glutamatergic synapses. Neuron 100:106–119.e7. [DOI] [PubMed] [Google Scholar]
- Kelsch W, Li Z, Eliava M, Goengrich C, Monyer H (2012) GluN2B-containing NMDA receptors promote wiring of adult-born neurons into olfactory bulb circuits. J Neurosci 32:12603–12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsch W, Li Z, Wieland S, Senkov O, Herb A, Göngrich C, Monyer H (2014) GluN2B-containing NMDA receptors promote glutamate synapse development in hippocampal interneurons. J Neurosci 34:16022–16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp N, McQueen J, Faulkes S, Bashir ZI (2000) Different forms of LTD in the CA1 region of the hippocampus: role of age and stimulus protocol. Eur J Neurosci 12:360–366. [DOI] [PubMed] [Google Scholar]
- Kennedy MJ, Davison IG, Robinson CG, Ehlers MD (2010) Syntaxin-4 defines a domain for activity-dependent exocytosis in dendritic spines. Cell 141:524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny AV, Cousins SL, Pinho L, Stephenson FA (2009) The integrity of the glycine co-agonist binding site of N-methyl-D-aspartate receptors is a functional quality control checkpoint for cell surface delivery. J Biol Chem 284:324–333. [DOI] [PubMed] [Google Scholar]
- Kerchner GA, Nicoll RA (2008) Silent synapses and the emergence of a postsynaptic mechanism for LTP. Nat Rev Neurosci 9:813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchner GA, Wilding TJ, Li P, Zhuo M, Huettner JE (2001) Presynaptic kainate receptors regulate spinal sensory transmission. J Neurosci 21:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler JP (2013) Control of cleft glutamate concentration and glutamate spill-out by perisynaptic glia: uptake and diffusion barriers. PLoS One 8:e70791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew JN, Trube G, Kemp JA (1996) A novel mechanism of activity-dependent NMDA receptor antagonism describes the effect of ifenprodil in rat cultured cortical neurones. J Physiol 497:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Houck DR, Gross AL, Zhang XL, Cearley C, Madsen TM, Kroes RA, Stanton PK, Burgdorf J, Moskal JR (2018) NYX-2925 is a novel NMDA receptor-specific spirocyclic-β-lactam that modulates synaptic plasticity processes associated with learning and memory. Int J Neuropsychopharmacol 21:242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanra N, Brown PM, Perozzo AM, Bowie D, Meyerson JR (2021) Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor. eLife 10:e66097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharazia VN, Weinberg RJ (1999) Immunogold localization of AMPA and NMDA receptors in somatic sensory cortex of albino rat. J Comp Neurol 412:292–302. [DOI] [PubMed] [Google Scholar]
- Khatri A, Burger PB, Swanger SA, Hansen KB, Zimmerman S, Karakas E, Liotta DC, Furukawa H, Snyder JP, Traynelis SF (2014) Structural determinants and mechanism of action of a GluN2C-selective NMDA receptor positive allosteric modulator. Mol Pharmacol 86:548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khlestova E, Johnson JW, Krystal JH, Lisman J (2016) The role of GluN2C-containing NMDA receptors in ketamine’s psychotogenic action and in schizophrenia models. J Neurosci 36:11151–11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodosevich K, Jacobi E, Farrow P, Schulmann A, Rusu A, Zhang L, Sprengel R, Monyer H, von Engelhardt J (2014) Coexpressed auxiliary subunits exhibit distinct modulatory profiles on AMPA receptor function. Neuron 83:601–615. [DOI] [PubMed] [Google Scholar]
- Kidd FL, Isaac JT (1999) Developmental and activity-dependent regulation of kainate receptors at thalamocortical synapses. Nature 400:569–573. [DOI] [PubMed] [Google Scholar]
- Kidd FL, Isaac JT (2001) Kinetics and activation of postsynaptic kainate receptors at thalamocortical synapses: role of glutamate clearance. J Neurophysiol 86:1139–1148. [DOI] [PubMed] [Google Scholar]
- Kim CH, Lisman JE (1999) A role of actin filament in synaptic transmission and long-term potentiation. J Neurosci 19:4314–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Farchione T, Potter A, Chen Q, Temple R (2019) Esketamine for treatment-resistant depression - first FDA-approved antidepressant in a new class. N Engl J Med 381:1–4. [DOI] [PubMed] [Google Scholar]
- Kim KS, Yan D, Tomita S (2010) Assembly and stoichiometry of the AMPA receptor and transmembrane AMPA receptor regulatory protein complex. J Neurosci 30:1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Kim JH, Park M, Cho IH, Yoo HJ (2007) Family-based association study between GRIK2 polymorphisms and autism spectrum disorders in the Korean trios. Neurosci Res 58:332–335. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Bao H, Bonanno L, Zhang B, Serpe M (2012) Drosophila Neto is essential for clustering glutamate receptors at the neuromuscular junction. Genes Dev 26:974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney GA, Overstreet LS, Slater NT (1997) Prolonged physiological entrapment of glutamate in the synaptic cleft of cerebellar unipolar brush cells. J Neurophysiol 78:1320–1333. [DOI] [PubMed] [Google Scholar]
- Kistler T, Fleck MW (2007) Functional consequences of natural substitutions in the GluR6 kainate receptor subunit ligand-binding site. Channels (Austin) 1:417–428. [DOI] [PubMed] [Google Scholar]
- Kitayama T, Yoneyama M, Yoneda Y (2003) Possible regulation by N-methyl-d-aspartate receptors of proliferative progenitor cells expressed in adult mouse hippocampal dentate gyrus. J Neurochem 84:767–780. [DOI] [PubMed] [Google Scholar]
- Klaassen RVStroeder JCoussen FHafner ASPetersen JDRenancio CSchmitz LJNormand ELodder JCRotaru DC, et al. (2016) Shisa6 traps AMPA receptors at postsynaptic sites and prevents their desensitization during synaptic activity. Nat Commun 7:10682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner NW, Pallotta BS (1995) Burst kinetics of single NMDA receptor currents in cell-attached patches from rat brain cortical neurons in culture. J Physiol 486:411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RC, Warder SE, Galdzicki Z, Castellino FJ, Prorok M (2001) Kinetic and mechanistic characterization of NMDA receptor antagonism by replacement and truncation variants of the conantokin peptides. Neuropharmacology 41:801–810. [DOI] [PubMed] [Google Scholar]
- Knight HM, Walker R, James R, Porteous DJ, Muir WJ, Blackwood DH, Pickard BS (2012) GRIK4/KA1 protein expression in human brain and correlation with bipolar disorder risk variant status. Am J Med Genet B Neuropsychiatr Genet 159B:21–29. [DOI] [PubMed] [Google Scholar]
- Knopp KLSimmons RMAGuo WAdams BLGardinier KMGernert DLOrnstein PLPorter WReel JDing C, et al. (2019) Modulation of TARP γ8-containing AMPA receptors as a novel therapeutic approach for chronic pain. J Pharmacol Exp Ther 369:345–363. [DOI] [PubMed] [Google Scholar]
- Kocsis B (2012) Differential role of NR2A and NR2B subunits in N-methyl-D-aspartate receptor antagonist-induced aberrant cortical gamma oscillations. Biol Psychiatry 71:987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji P, Araque A (2021) G-Protein-coupled receptors in astrocyte-neuron communication. Neuroscience 456:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh DS, Burnashev N, Jonas P (1995) Block of native Ca(2+)-permeable AMPA receptors in rat brain by intracellular polyamines generates double rectification. J Physiol 486:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda K, Kakegawa W, Matsuda S, Nakagami R, Kakiya N, Yuzaki M (2007) The extreme C-terminus of GluRdelta2 is essential for induction of long-term depression in cerebellar slices. Eur J Neurosci 25:1357–1362. [DOI] [PubMed] [Google Scholar]
- Kohda K, Kakegawa W, Matsuda S, Yamamoto T, Hirano H, Yuzaki M (2013) The δ2 glutamate receptor gates long-term depression by coordinating interactions between two AMPA receptor phosphorylation sites. Proc Natl Acad Sci USA 110:E948–E957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda K, Wang Y, Yuzaki M (2000) Mutation of a glutamate receptor motif reveals its role in gating and delta2 receptor channel properties. Nat Neurosci 3:315–322. [DOI] [PubMed] [Google Scholar]
- Köhler M, Burnashev N, Sakmann B, Seeburg PH (1993) Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron 10:491–500. [DOI] [PubMed] [Google Scholar]
- Köhler M, Kornau HC, Seeburg PH (1994) The organization of the gene for the functionally dominant alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor subunit GluR-B. J Biol Chem 269:17367–17370. [PubMed] [Google Scholar]
- Köhr G (2006) NMDA receptor function: subunit composition versus spatial distribution. Cell Tissue Res 326:439–446. [DOI] [PubMed] [Google Scholar]
- Koike M, Iino M, Ozawa S (1997) Blocking effect of 1-naphthyl acetyl spermine on Ca(2+)-permeable AMPA receptors in cultured rat hippocampal neurons. Neurosci Res 29:27–36. [DOI] [PubMed] [Google Scholar]
- Koike M, Tsukada S, Tsuzuki K, Kijima H, Ozawa S (2000) Regulation of kinetic properties of GluR2 AMPA receptor channels by alternative splicing. J Neurosci 20:2166–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Tani M, Kanda T, Saitoh N, Yamashita T, Takahashi T (2008) Involvement of AMPA receptor desensitization in short-term synaptic depression at the calyx of Held in developing rats. J Physiol 586:2263–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolleker A, Zhu JJ, Schupp BJ, Qin Y, Mack V, Borchardt T, Köhr G, Malinow R, Seeburg PH, Osten P (2003) Glutamatergic plasticity by synaptic delivery of GluR-B(long)-containing AMPA receptors. Neuron 40:1199–1212. [DOI] [PubMed] [Google Scholar]
- Komuro H, Rakic P (1993) Modulation of neuronal migration by NMDA receptors. Science 260:95–97. [DOI] [PubMed] [Google Scholar]
- König P, Engel AK, Singer W (1996) Integrator or coincidence detector? The role of the cortical neuron revisited. Trends Neurosci 19:130–137. [DOI] [PubMed] [Google Scholar]
- Konno K, Matsuda K, Nakamoto C, Uchigashima M, Miyazaki T, Yamasaki M, Sakimura K, Yuzaki M, Watanabe M (2014) Enriched expression of GluD1 in higher brain regions and its involvement in parallel fiber-interneuron synapse formation in the cerebellum. J Neurosci 34:7412–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek MVyklicky VBorovska JLichnerova KKaniakova MKrausova BKrusek JBalik ASmejkalova THorak M, et al. (2015) Cholesterol modulates open probability and desensitization of NMDA receptors. J Physiol 593:2279–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakis E, Jang MK, Russek SJ, Gibbs TT, Farb DH (2011) A steroid modulatory domain in NR2A collaborates with NR1 exon-5 to control NMDAR modulation by pregnenolone sulfate and protons. J Neurochem 119:486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakis E, Smith C, Jang MK, Martin SC, Richards KG, Russek SJ, Gibbs TT, Farb DH (2013) The neuroactive steroid pregnenolone sulfate stimulates trafficking of functional N-methyl D-aspartate receptors to the cell surface via a noncanonical, G protein, and Ca2+-dependent mechanism. Mol Pharmacol 84:261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotermanski SE, Johnson JW (2009) Mg2+ imparts NMDA receptor subtype selectivity to the Alzheimer’s drug memantine. J Neurosci 29:2774–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotermanski SE, Wood JT, Johnson JW (2009) Memantine binding to a superficial site on NMDA receptors contributes to partial trapping. J Physiol 587:4589–4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kott S, Sager C, Tapken D, Werner M, Hollmann M (2009) Comparative analysis of the pharmacology of GluR1 in complex with transmembrane AMPA receptor regulatory proteins gamma2, gamma3, gamma4, and gamma8. Neuroscience 158:78–88. [DOI] [PubMed] [Google Scholar]
- Kott S, Werner M, Körber C, Hollmann M (2007) Electrophysiological properties of AMPA receptors are differentially modulated depending on the associated member of the TARP family. J Neurosci 27:3780–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kougioumtzidou E, Shimizu T, Hamilton NB, Tohyama K, Sprengel R, Monyer H, Attwell D, Richardson WD (2017) Signalling through AMPA receptors on oligodendrocyte precursors promotes myelination by enhancing oligodendrocyte survival. eLife 6:e28080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk Y, Miller B, Sarantis M, Attwell D (1994) Arachidonic acid depresses non-NMDA receptor currents. Brain Res 643:287–295. [DOI] [PubMed] [Google Scholar]
- Kowal C, Degiorgio LA, Lee JY, Edgar MA, Huerta PT, Volpe BT, Diamond B (2006) Human lupus autoantibodies against NMDA receptors mediate cognitive impairment. Proc Natl Acad Sci USA 103:19854–19859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal C, DeGiorgio LA, Nakaoka T, Hetherington H, Huerta PT, Diamond B, Volpe BT (2004) Cognition and immunity; antibody impairs memory. Immunity 21:179–188. [DOI] [PubMed] [Google Scholar]
- Krall RF, Moutal A, Phillips MB, Asraf H, Johnson JW, Khanna R, Hershfinkel M, Aizenman E, Tzounopoulos T (2020) Synaptic zinc inhibition of NMDA receptors depends on the association of GluN2A with the zinc transporter ZnT1. Sci Adv 6:eabb1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramár EA, Chen LY, Lauterborn JC, Simmons DA, Gall CM, Lynch G (2012) BDNF upregulation rescues synaptic plasticity in middle-aged ovariectomized rats. Neurobiol Aging 33:708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krampfl K, Schlesinger F, Wolfes H, Dengler R, Bufler J (2001) Functional diversity of recombinant human AMPA type glutamate receptors: possible implications for selective vulnerability of motor neurons. J Neurol Sci 191:19–23. [DOI] [PubMed] [Google Scholar]
- Krampfl K, Schlesinger F, Zörner A, Kappler M, Dengler R, Bufler J (2002) Control of kinetic properties of GluR2 flop AMPA-type channels: impact of R/G nuclear editing. Eur J Neurosci 15:51–62. [DOI] [PubMed] [Google Scholar]
- Kreitzer AC, Malenka RC (2008) Striatal plasticity and basal ganglia circuit function. Neuron 60:543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitzer MA, Birnbaum AD, Qian H, Malchow RP (2009) Pharmacological characterization, localization, and regulation of ionotropic glutamate receptors in skate horizontal cells. Vis Neurosci 26:375–387. [DOI] [PubMed] [Google Scholar]
- Kreutzwiser D, Tawfic QA (2019) Expanding role of NMDA receptor antagonists in the management of pain. CNS Drugs 33:347–374. [DOI] [PubMed] [Google Scholar]
- Kreye JWenke NKChayka MLeubner JMurugan RMaier NJurek BLy LTBrandl DRost BR, et al. (2016) Human cerebrospinal fluid monoclonal N-methyl-D-aspartate receptor autoantibodies are sufficient for encephalitis pathogenesis. Brain 139:2641–2652. [DOI] [PubMed] [Google Scholar]
- Krintel C, Harpsøe K, Zachariassen LG, Peters D, Frydenvang K, Pickering DS, Gajhede M, Kastrup JS (2013) Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain. Acta Crystallogr D Biol Crystallogr 69:1645–1652. [DOI] [PubMed] [Google Scholar]
- Kristensen ASHansen KBNaur POlsen LKurtkaya NLDravid SMKvist TYi FPøhlsgaard JClausen RP, et al. (2016a) Pharmacology and structural analysis of ligand binding to the orthosteric site of glutamate-like GluD2 receptors. Mol Pharmacol 89:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen AS, Jenkins MA, Banke TG, Schousboe A, Makino Y, Johnson RC, Huganir R, Traynelis SF (2011) Mechanism of Ca2+/calmodulin-dependent kinase II regulation of AMPA receptor gating. Nat Neurosci 14:727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen O, Kristensen LB, Møllerud S, Frydenvang K, Pickering DS, Kastrup JS (2016b) The structure of a high-affinity kainate receptor: GluK4 ligand-binding domain crystallized with kainate. Structure 24:1582–1589. [DOI] [PubMed] [Google Scholar]
- Krogsgaard-Larsen NDelgar CGKoch KBrown PMMøller CHan LHuynh THHansen SWNielsen BBowie D, et al. (2017) Design and synthesis of a series of l-trans-4-substituted prolines as selective antagonists for the ionotropic glutamate receptors including functional and X-ray crystallographic studies of new subtype selective kainic acid receptor subtype 1 (GluK1) antagonist (2S,4R)-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid. J Med Chem 60:441–457. [DOI] [PubMed] [Google Scholar]
- Krogsgaard-Larsen NStorgaard MMøller CDemmer CSHansen JHan LMonrad RNNielsen BTapken DPickering DS, et al. (2015) Structure-activity relationship study of ionotropic glutamate receptor antagonist (2S,3R)-3-(3-carboxyphenyl)pyrrolidine-2-carboxylic Acid. J Med Chem 58:6131–6150. [DOI] [PubMed] [Google Scholar]
- Kromann H, Krikstolaityte S, Andersen AJ, Andersen K, Krogsgaard-Larsen P, Jaroszewski JW, Egebjerg J, Strømgaard K (2002) Solid-phase synthesis of polyamine toxin analogues: potent and selective antagonists of Ca2+-permeable AMPA receptors. J Med Chem 45:5745–5754. [DOI] [PubMed] [Google Scholar]
- Krueger RF, Kotov R, Watson D, Forbes MK, Eaton NR, Ruggero CJ, Simms LJ, Widiger TA, Achenbach TM, Bach B, Bagby RM, Bornovalova MA, Carpenter WT, Chmielewski M, Cicero DC, Clark LA, Conway C, DeClercq B, DeYoung CG, Docherty AR, Drislane LE, First MB, Forbush KT, Hallquist M, Haltigan JD, Hopwood CJ, Ivanova MY, Jonas KG, Latzman RD, Markon KE, Miller JD, Morey LC, Mullins-Sweatt SN, Ormel J, Patalay P, Patrick CJ, Pincus AL, Regier DA, Reininghaus U, Rescorla LA, Samuel DB, Sellbom M, Shackman AJ, Skodol A, Slade T, South SC, Sunderland M, Tackett JL, Venables NC, Waldman ID, Waszczuk MA, Waugh MH, Wright AGC, Zald DH, Zimmermann J (2018) Progress in achieving quantitative classification of psychopathology. World psychiatry: official journal of the World Psychiatric Association 17:282-293. [DOI] [PMC free article] [PubMed]
- Krupp JJ, Vissel B, Heinemann SF, Westbrook GL (1996) Calcium-dependent inactivation of recombinant N-methyl-D-aspartate receptors is NR2 subunit specific. Mol Pharmacol 50:1680–1688. [PubMed] [Google Scholar]
- Krupp JJ, Vissel B, Heinemann SF, Westbrook GL (1998) N-terminal domains in the NR2 subunit control desensitization of NMDA receptors. Neuron 20:317–327. [DOI] [PubMed] [Google Scholar]
- Krupp JJ, Vissel B, Thomas CG, Heinemann SF, Westbrook GL (1999) Interactions of calmodulin and alpha-actinin with the NR1 subunit modulate Ca2+-dependent inactivation of NMDA receptors. J Neurosci 19:1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal JH, Abdallah CG, Sanacora G, Charney DS, Duman RS (2019) Ketamine: A paradigm shift for depression research and Treatment. Neuron 101:774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal JH, D’Souza DC, Mathalon D, Perry E, Belger A, Hoffman R (2003) NMDA receptor antagonist effects, cortical glutamatergic function, and schizophrenia: toward a paradigm shift in medication development. Psychopharmacology (Berl) 169:215–233. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Sanacora G, Duman RS (2013) Rapid-acting glutamatergic antidepressants: the path to ketamine and beyond. Biol Psychiatry 73:1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshatri AS, Gonzalez-Hernandez A, Giraldez T (2018) Physiological roles and therapeutic potential of Ca2+ activated potassium channels in the nervous system. Front Mol Neurosci 11:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota S, Rubin JE (2011) NMDA-induced burst firing in a model subthalamic nucleus neuron. J Neurophysiol 106:527–537. [DOI] [PubMed] [Google Scholar]
- Kudova E, Chodounska H, Slavikova B, Budesinsky M, Nekardova M, Vyklicky V, Krausova B, Svehla P, Vyklicky L (2015) A new class of potent N-methyl-D-aspartate receptor inhibitors: Sulfated neuroactive steroids with lipophilic D-ring modifications. J Med Chem 58:5950–5966. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W (2014) Biochemistry. The resolution revolution. Science 343:1443–1444. [DOI] [PubMed] [Google Scholar]
- Kukley M, Capetillo-Zarate E, Dietrich D (2007) Vesicular glutamate release from axons in white matter. Nat Neurosci 10:311–320. [DOI] [PubMed] [Google Scholar]
- Kumamoto E (1996) Neuromodulation by Mg2+ and polyamines of excitatory amino acid currents in rodent neurones in culture. Magnes Res 9:317–327. [PubMed] [Google Scholar]
- Kumar J, Mayer ML (2013) Functional insights from glutamate receptor ion channel structures. Annu Rev Physiol 75:313–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Jin R, Mayer ML (2009) The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol 16:631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Mayer ML (2011) Structure and assembly mechanism for heteromeric kainate receptors. Neuron 71:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SS, Huguenard JR (2003) Pathway-specific differences in subunit composition of synaptic NMDA receptors on pyramidal neurons in neocortex. J Neurosci 23:10074–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari J, Bendre AD, Bhosale S, Vinnakota R, Burada AP, Tria G, Ravelli RBG, Peters PJ, Joshi M, Kumar J (2020) Structural dynamics of the GluK3-kainate receptor neurotransmitter binding domains revealed by cryo-EM. Int J Biol Macromol 149:1051–1058. [DOI] [PubMed] [Google Scholar]
- Kumari J, Vinnakota R, Kumar J (2019) Structural and functional insights into GluK3-kainate receptor desensitization and recovery. Sci Rep 9:10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner T, Beck C, Sakmann B, Seeburg PH (2001) Channel-lining residues of the AMPA receptor M2 segment: structural environment of the Q/R site and identification of the selectivity filter. J Neurosci 21:4162–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner T, Schoepfer R (1996) Multiple structural elements determine subunit specificity of Mg2+ block in NMDA receptor channels. J Neurosci 16:3549–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner T, Wollmuth LP, Karlin A, Seeburg PH, Sakmann B (1996) Structure of the NMDA receptor channel M2 segment inferred from the accessibility of substituted cysteines. Neuron 17:343–352. [DOI] [PubMed] [Google Scholar]
- Kunz PA, Roberts AC, Philpot BD (2013) Presynaptic NMDA receptor mechanisms for enhancing spontaneous neurotransmitter release. J Neurosci 33:7762–7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara H, Hashimoto K, Kano M, Takayama C, Sakimura K, Mishina M, Inoue Y, Watanabe M (1997) Impaired parallel fiber-->Purkinje cell synapse stabilization during cerebellar development of mutant mice lacking the glutamate receptor delta2 subunit. J Neurosci 17:9613–9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner MG, Kim SW, Donahue C, Thuras P, Adson D, Kotlyar M, McCabe J, Peterson J, Foa EB (2007) D-cycloserine augmented exposure therapy for obsessive-compulsive disorder. Biol Psychiatry 62:835–838. [DOI] [PubMed] [Google Scholar]
- Kussius CL, Kaur N, Popescu GK (2009) Pregnanolone sulfate promotes desensitization of activated NMDA receptors. J Neurosci 29:6819–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius CL, Popescu AM, Popescu GK (2010) Agonist-specific gating of NMDA receptors. Channels (Austin) 4:78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius CL, Popescu GK (2009) Kinetic basis of partial agonism at NMDA receptors. Nat Neurosci 12:1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsuwada TKashiwabuchi NMori HSakimura KKushiya EAraki KMeguro HMasaki HKumanishi TArakawa M, et al. (1992) Molecular diversity of the NMDA receptor channel. Nature 358:36–41. [DOI] [PubMed] [Google Scholar]
- Kutsuwada TSakimura KManabe TTakayama CKatakura NKushiya ENatsume RWatanabe MInoue YYagi T, et al. (1996) Impairment of suckling response, trigeminal neuronal pattern formation, and hippocampal LTD in NMDA receptor epsilon 2 subunit mutant mice. Neuron 16:333–344. [DOI] [PubMed] [Google Scholar]
- Kuusinen A, Arvola M, Keinänen K (1995) Molecular dissection of the agonist binding site of an AMPA receptor. EMBO J 14:6327–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist T, Greenwood JR, Hansen KB, Traynelis SF, Bräuner-Osborne H (2013a) Structure-based discovery of antagonists for GluN3-containing N-methyl-D-aspartate receptors. Neuropharmacology 75:324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist T, Steffensen TB, Greenwood JR, Mehrzad Tabrizi F, Hansen KB, Gajhede M, Pickering DS, Traynelis SF, Kastrup JS, Bräuner-Osborne H (2013b) Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site. J Biol Chem 288:33124–33135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok KH, Tse YC, Wong RN, Yung KK (1997) Cellular localization of GluR1, GluR2/3 and GluR4 glutamate receptor subunits in neurons of the rat neostriatum. Brain Res 778:43–55. [DOI] [PubMed] [Google Scholar]
- Kwon HB, Sabatini BL (2011) Glutamate induces de novo growth of functional spines in developing cortex. Nature 474:100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La DSSalituro FGMartinez Botella GGriffin AMBai ZAckley MADai JDoherty JJHarrison BLHoffmann EC, et al. (2019) Neuroactive steroid N-methyl-d-aspartate receptor positive allosteric modulators: Synthesis, SAR, and pharmacological activity. J Med Chem 62:7526–7542. [DOI] [PubMed] [Google Scholar]
- Ladépêche L, Planagumà J, Thakur S, Suárez I, Hara M, Borbely JS, Sandoval A, Laparra-Cuervo L, Dalmau J, Lakadamyali M (2018) NMDA receptor autoantibodies in autoimmune encephalitis cause a subunit-specific nanoscale redistribution of NMDA receptors. Cell Rep 23:3759–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladislav M, Cerny J, Krusek J, Horak M, Balik A, Vyklicky L (2018) The LILI motif of M3-S2 linkers is a component of the NMDA receptor channel gate. Front Mol Neurosci 11:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MHughes EGPeng XZhou LGleichman AJShu HMatà SKremens DVitaliani RGeschwind MD, et al. (2009) AMPA receptor antibodies in limbic encephalitis alter synaptic receptor location. Ann Neurol 65:424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai TW, Zhang S, Wang YT (2014) Excitotoxicity and stroke: identifying novel targets for neuroprotection. Prog Neurobiol 115:157–188. [DOI] [PubMed] [Google Scholar]
- Lalanne T, Oyrer J, Mancino A, Gregor E, Chung A, Huynh L, Burwell S, Maheux J, Farrant M, Sjöström PJ (2016) Synapse-specific expression of calcium-permeable AMPA receptors in neocortical layer 5. J Physiol 594:837–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalouette A, Guénet JL, Vriz S (1998) Hotfoot mouse mutations affect the delta 2 glutamate receptor gene and are allelic to lurcher. Genomics 50:9–13. [DOI] [PubMed] [Google Scholar]
- Lalouette A, Lohof A, Sotelo C, Guénet J, Mariani J (2001) Neurobiological effects of a null mutation depend on genetic context: comparison between two hotfoot alleles of the delta-2 ionotropic glutamate receptor. Neuroscience 105:443–455. [DOI] [PubMed] [Google Scholar]
- Lam HM, Chiu J, Hsieh MH, Meisel L, Oliveira IC, Shin M, Coruzzi G (1998) Glutamate-receptor genes in plants. Nature 396:125–126. [DOI] [PubMed] [Google Scholar]
- Lambolez B, Audinat E, Bochet P, Crépel F, Rossier J (1992) AMPA receptor subunits expressed by single Purkinje cells. Neuron 9:247–258. [DOI] [PubMed] [Google Scholar]
- Landes CF, Rambhadran A, Taylor JN, Salatan F, Jayaraman V (2011) Structural landscape of isolated agonist-binding domains from single AMPA receptors. Nat Chem Biol 7:168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsend AS, Amiry-Moghaddam M, Matsubara A, Bergersen L, Usami S, Wenthold RJ, Ottersen OP (1997) Differential localization of delta glutamate receptors in the rat cerebellum: coexpression with AMPA receptors in parallel fiber-spine synapses and absence from climbing fiber-spine synapses. J Neurosci 17:834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanore F, Labrousse VF, Szabo Z, Normand E, Blanchet C, Mulle C (2012) Deficits in morphofunctional maturation of hippocampal mossy fiber synapses in a mouse model of intellectual disability. J Neurosci 32:17882–17893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum ME, Nevian T, Sandler M, Polsky A, Schiller J (2009) Synaptic integration in tuft dendrites of layer 5 pyramidal neurons: a new unifying principle. Science 325:756–760. [DOI] [PubMed] [Google Scholar]
- Larsen AM, Venskutonytė R, Valadés EA, Nielsen B, Pickering DS, Bunch L (2011a) Discovery of a new class of ionotropic glutamate receptor antagonists by the rational design of (2S,3R)-3-(3-carboxyphenyl)-pyrrolidine-2-carboxylic acid. ACS Chem Neurosci 2:107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen AP, Fièvre S, Frydenvang K, Francotte P, Pirotte B, Kastrup JS, Mulle C (2017) Identification and structure-function study of positive allosteric modulators of kainate receptors. Mol Pharmacol 91:576–585. [DOI] [PubMed] [Google Scholar]
- Larsen RSCorlew RJHenson MARoberts ACMishina MWatanabe MLipton SANakanishi NPérez-Otaño IWeinberg RJ, et al. (2011b) NR3A-containing NMDARs promote neurotransmitter release and spike timing-dependent plasticity. Nat Neurosci 14:338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RS, Smith IT, Miriyala J, Han JE, Corlew RJ, Smith SL, Philpot BD (2014) Synapse-specific control of experience-dependent plasticity by presynaptic NMDA receptors. Neuron 83:879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J, Lieu T, Petchpradub V, LeDuc B, Ngo H, Rogers GA, Lynch G (1995) Facilitation of olfactory learning by a modulator of AMPA receptors. J Neurosci 15:8023–8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J, Munkácsy E (2015) Theta-burst LTP. Brain Res 1621:38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash LL, Sanders JM, Akiyama N, Shoji M, Postila P, Pentikäinen OT, Sasaki M, Sakai R, Swanson GT (2008) Novel analogs and stereoisomers of the marine toxin neodysiherbaine with specificity for kainate receptors. J Pharmacol Exp Ther 324:484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AY, Roux B (2007) The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure 15:1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AY, Roux B (2011) The hidden energetics of ligand binding and activation in a glutamate receptor. Nat Struct Mol Biol 18:283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AY, Salazar H, Blachowicz L, Ghisi V, Plested AJ, Roux B (2013) A conformational intermediate in glutamate receptor activation. Neuron 79:492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CG, Zukin RS (2007) NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci 8:413–426. [DOI] [PubMed] [Google Scholar]
- Laube B, Kuhse J, Betz H (1998) Evidence for a tetrameric structure of recombinant NMDA receptors. J Neurosci 18:2954–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulumaa S, Hansen KV, Masternak M, Drapier T, Francotte P, Pirotte B, Frydenvang K, Kastrup JS (2018) Crystal structures of potent dimeric positive allosteric modulators at the ligand-binding domain of the GluA2 receptor. ACS Med Chem Lett 10:243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauri SE, Segerstråle M, Vesikansa A, Maingret F, Mulle C, Collingridge GL, Isaac JT, Taira T (2005) Endogenous activation of kainate receptors regulates glutamate release and network activity in the developing hippocampus. J Neurosci 25:4473–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauri SE, Vesikansa A, Segerstråle M, Collingridge GL, Isaac JT, Taira T (2006) Functional maturation of CA1 synapses involves activity-dependent loss of tonic kainate receptor-mediated inhibition of glutamate release. Neuron 50:415–429. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Seeburg PH (1994) Regional and developmental heterogeneity in splicing of the rat brain NMDAR1 mRNA. J Neurosci 14:3180–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterborn JC, Lynch G, Vanderklish P, Arai A, Gall CM (2000) Positive modulation of AMPA receptors increases neurotrophin expression by hippocampal and cortical neurons. J Neurosci 20:8–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterborn JCPalmer LCJia YPham DTHou BWang WTrieu BHCox CDKantorovich SGall CM, et al. (2016) Chronic ampakine treatments stimulate dendritic growth and promote learning in middle-aged rats. J Neurosci 36:1636–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvsnes MB, Beyer MK, Kvaløy JT, Greve OJ, Appenzeller S, Kvivik I, Harboe E, Tjensvoll AB, Gøransson LG, Omdal R (2014) Association of hippocampal atrophy with cerebrospinal fluid antibodies against the NR2 subtype of the N-methyl-D-aspartate receptor in patients with systemic lupus erythematosus and patients with primary Sjögren’s syndrome. Arthritis Rheumatol 66:3387–3394. [DOI] [PubMed] [Google Scholar]
- Laverty D, Thomas P, Field M, Andersen OJ, Gold MG, Biggin PC, Gielen M, Smart TG (2017) Crystal structures of a GABAA-receptor chimera reveal new endogenous neurosteroid-binding sites. Nat Struct Mol Biol 24:977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavezzari G, McCallum J, Dewey CM, Roche KW (2004) Subunit-specific regulation of NMDA receptor endocytosis. J Neurosci 24:6383–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavzin M, Rapoport S, Polsky A, Garion L, Schiller J (2012) Nonlinear dendritic processing determines angular tuning of barrel cortex neurons in vivo. Nature 490:397–401. [DOI] [PubMed] [Google Scholar]
- Layton MEKelly MJ 3rdRodzinak KJSanderson PEYoung SDBednar RADilella AGMcDonald TPWang HMosser SD, et al. (2011) Discovery of 3-substituted aminocyclopentanes as potent and orally bioavailable NR2B subtype-selective NMDA antagonists. ACS Chem Neurosci 2:352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro JT, Paternain AV, Lerma J, Chenard BL, Ewing FE, Huang J, Welch WM, Ganong AH, Menniti FS (2002) Functional characterization of CP-465,022, a selective, noncompetitive AMPA receptor antagonist. Neuropharmacology 42:143–153. [DOI] [PubMed] [Google Scholar]
- Le Bail MMartineau MSacchi SYatsenko NRadzishevsky IConrod SAit Ouares KWolosker HPollegioni LBillard JM, et al. (2015) Identity of the NMDA receptor coagonist is synapse specific and developmentally regulated in the hippocampus. Proc Natl Acad Sci USA 112:E204–E213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgerwood L, Richardson R, Cranney J (2003) Effects of D-cycloserine on extinction of conditioned freezing. Behav Neurosci 117:341–349. [DOI] [PubMed] [Google Scholar]
- Lee CH, Lü W, Michel JC, Goehring A, Du J, Song X, Gouaux E (2014) NMDA receptor structures reveal subunit arrangement and pore architecture. Nature 511:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CJ, Mannaioni G, Yuan H, Woo DH, Gingrich MB, Traynelis SF (2007) Astrocytic control of synaptic NMDA receptors. J Physiol 581:1057–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Oh WC, Seong J, Kim J (2016a) Advanced fluorescence protein-based synapse-detectors. Front Synaptic Neurosci 8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Barbarosie M, Kameyama K, Bear MF, Huganir RL (2000) Regulation of distinct AMPA receptor phosphorylation sites during bidirectional synaptic plasticity. Nature 405:955–959. [DOI] [PubMed] [Google Scholar]
- Lee HK, Kameyama K, Huganir RL, Bear MF (1998) NMDA induces long-term synaptic depression and dephosphorylation of the GluR1 subunit of AMPA receptors in hippocampus. Neuron 21:1151–1162. [DOI] [PubMed] [Google Scholar]
- Lee HK, Min SS, Gallagher M, Kirkwood A (2005) NMDA receptor-independent long-term depression correlates with successful aging in rats. Nat Neurosci 8:1657–1659. [DOI] [PubMed] [Google Scholar]
- Lee HKTakamiya KHan JSMan HKim CHRumbaugh GYu SDing LHe CPetralia RS, et al. (2003) Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell 112:631–643. [DOI] [PubMed] [Google Scholar]
- Lee JH, Wei L, Deveau TC, Gu X, Yu SP (2016b) Expression of the NMDA receptor subunit GluN3A (NR3A) in the olfactory system and its regulatory role on olfaction in the adult mouse. Brain Struct Funct 221:3259–3273. [DOI] [PubMed] [Google Scholar]
- Lee JY, Krieger J, Herguedas B, García-Nafría J, Dutta A, Shaikh SA, Greger IH, Bahar I (2019) Druggability simulations and X-ray crystallography reveal a ligand-binding site in the GluA3 AMPA receptor N-terminal domain. Structure 27:241–252.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Ting KK, Adams S, Brew BJ, Chung R, Guillemin GJ (2010a) Characterisation of the expression of NMDA receptors in human astrocytes. PLoS One 5:e14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Yasuda R, Ehlers MD (2010b) Metaplasticity at single glutamatergic synapses. Neuron 66:859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MR, Gardinier KM, Gernert DL, Schober DA, Wright RA, Wang H, Qian Y, Witkin JM, Nisenbaum ES, Kato AS (2017a) Structural determinants of the γ-8 TARP dependent AMPA receptor antagonist. ACS Chem Neurosci 8:2631–2647. [DOI] [PubMed] [Google Scholar]
- Lee SH, Simonetta A, Sheng M (2004) Subunit rules governing the sorting of internalized AMPA receptors in hippocampal neurons. Neuron 43:221–236. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Uemura T, Yoshida T, Mishina M (2012) GluRδ2 assembles four neurexins into trans-synaptic triad to trigger synapse formation. J Neurosci 32:4688–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Wei M, Zhang C, Maxeiner S, Pak C, Calado Botelho S, Trotter J, Sterky FH, Südhof TC (2017b) Presynaptic neuronal pentraxin receptor organizes excitatory and inhibitory synapses. J Neurosci 37:1062–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Földy C, Szabadics J, Soltesz I (2011) Cell-type-specific CCK2 receptor signaling underlies the cholecystokinin-mediated selective excitation of hippocampal parvalbumin-positive fast-spiking basket cells. J Neurosci 31:10993–11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Rosenmund C, Westbrook GL (1993) Inactivation of NMDA channels in cultured hippocampal neurons by intracellular calcium. J Neurosci 13:674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei N, Mellem JE, Brockie PJ, Madsen DM, Maricq AV (2017) NRAP-1 is a presynaptically released NMDA receptor auxiliary protein that modifies synaptic strength. Neuron 96:1303–1316.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei S, Orser BA, Thatcher GR, Reynolds JN, MacDonald JF (2001) Positive allosteric modulators of AMPA receptors reduce proton-induced receptor desensitization in rat hippocampal neurons. J Neurophysiol 85:2030–2038. [DOI] [PubMed] [Google Scholar]
- Le Magueresse C, Monyer H (2013) GABAergic interneurons shape the functional maturation of the cortex. Neuron 77:388–405. [DOI] [PubMed] [Google Scholar]
- Le Meur K, Galante M, Angulo MC, Audinat E (2007) Tonic activation of NMDA receptors by ambient glutamate of non-synaptic origin in the rat hippocampus. J Physiol 580:373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JRGeider KHelbig KLHeyne HOSchütz HHentschel JCourage CDepienne CNava CHeron D, et al. (2016) Delineating the GRIN1 phenotypic spectrum: A distinct genetic NMDA receptor encephalopathy. Neurology 86:2171–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JRLal DReinthaler EMSteiner INothnagel MAlber MGeider KLaube BSchwake MFinsterwalder K, et al. (2013) Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat Genet 45:1067–1072. [DOI] [PubMed] [Google Scholar]
- Lennox BRPalmer-Cooper ECPollak THainsworth JMarks JJacobson LLang BFox HFerry BScoriels L, et al. ; PPiP study team (2017) Prevalence and clinical characteristics of serum neuronal cell surface antibodies in first-episode psychosis: a case-control study. Lancet Psychiatry 4:42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AS, Bayer KU, Merrill MA, Lim IA, Shea MA, Schulman H, Hell JW (2002) Regulation of calcium/calmodulin-dependent protein kinase II docking to N-methyl-D-aspartate receptors by calcium/calmodulin and alpha-actinin. J Biol Chem 277:48441–48448. [DOI] [PubMed] [Google Scholar]
- Leonard AS, Lim IA, Hemsworth DE, Horne MC, Hell JW (1999) Calcium/calmodulin-dependent protein kinase II is associated with the N-methyl-D-aspartate receptor. Proc Natl Acad Sci USA 96:3239–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma J (2003) Roles and rules of kainate receptors in synaptic transmission. Nat Rev Neurosci 4:481–495. [DOI] [PubMed] [Google Scholar]
- Lerma J, Marques JM (2013) Kainate receptors in health and disease. Neuron 80:292–311. [DOI] [PubMed] [Google Scholar]
- Lerma J, Paternain AV, Naranjo JR, Mellström B (1993) Functional kainate-selective glutamate receptors in cultured hippocampal neurons. Proc Natl Acad Sci USA 90:11688–11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesca GRudolf GBruneau NLozovaya NLabalme ABoutry-Kryza NSalmi MTsintsadze TAddis LMotte J, et al. (2013) GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet 45:1061–1066. [DOI] [PubMed] [Google Scholar]
- Lester RA, Clements JD, Westbrook GL, Jahr CE (1990) Channel kinetics determine the time course of NMDA receptor-mediated synaptic currents. Nature 346:565–567. [DOI] [PubMed] [Google Scholar]
- Lester RA, Quarum ML, Parker JD, Weber E, Jahr CE (1989) Interaction of 6-cyano-7-nitroquinoxaline-2,3-dione with the N-methyl-D-aspartate receptor-associated glycine binding site. Mol Pharmacol 35:565–570. [PubMed] [Google Scholar]
- Lester RA, Tong G, Jahr CE (1993) Interactions between the glycine and glutamate binding sites of the NMDA receptor. J Neurosci 13:1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts VA, Felix R, Biddlecome GH, Arikkath J, Mahaffey CL, Valenzuela A, Bartlett FS 2nd, Mori Y, Campbell KP, Frankel WN (1998) The mouse stargazer gene encodes a neuronal Ca2+-channel gamma subunit. Nat Genet 19:340–347. [DOI] [PubMed] [Google Scholar]
- Leuschner WD, Hoch W (1999) Subtype-specific assembly of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits is mediated by their n-terminal domains. J Biol Chem 274:16907–16916. [DOI] [PubMed] [Google Scholar]
- Levite M (2014) Glutamate receptor antibodies in neurological diseases: anti-AMPA-GluR3 antibodies, anti-NMDA-NR1 antibodies, anti-NMDA-NR2A/B antibodies, anti-mGluR1 antibodies or anti-mGluR5 antibodies are present in subpopulations of patients with either: epilepsy, encephalitis, cerebellar ataxia, systemic lupus erythematosus (SLE) and neuropsychiatric SLE, Sjogren’s syndrome, schizophrenia, mania or stroke. These autoimmune anti-glutamate receptor antibodies can bind neurons in few brain regions, activate glutamate receptors, decrease glutamate receptor’s expression, impair glutamate-induced signaling and function, activate blood brain barrier endothelial cells, kill neurons, damage the brain, induce behavioral/psychiatric/cognitive abnormalities and ataxia in animal models, and can be removed or silenced in some patients by immunotherapy. J Neural Transm (Vienna) 121:1029–1075. [DOI] [PubMed] [Google Scholar]
- Levite M, Fleidervish IA, Schwarz A, Pelled D, Futerman AH (1999) Autoantibodies to the glutamate receptor kill neurons via activation of the receptor ion channel. J Autoimmun 13:61–72. [DOI] [PubMed] [Google Scholar]
- Levitz J, Popescu AT, Reiner A, Isacoff EY (2016) A toolkit for orthogonal and in vivo optical manipulation of ionotropic glutamate receptors. Front Mol Neurosci 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RJ, Dutertre S, Vetter I, Christie MJ (2012) Conus venom peptide pharmacology. Pharmacol Rev 64:259–298. [DOI] [PubMed] [Google Scholar]
- Li CYan JTang DZhu JHuang CSun YHu RWang HFu CChen Y, et al. (2020) GluN2A-selective positive allosteric modulator-nalmefene-flumazenil reverses ketamine-fentanyl-dexmedetomidine-induced anesthesia and analgesia in rats. Sci Rep 10:5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DYuan HOrtiz-Gonzalez XRMarsh EDTian LMcCormick EMKosobucki GJChen WSchulien AJChiavacci R, et al. (2016a) GRIN2D recurrent de novo dominant mutation causes a severe epileptic encephalopathy treatable with NMDA receptor channel blockers. Am J Hum Genet 99:802–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZhang JTang WMizu RKKusumoto HXiangWei WXu YChen WAmin JBHu C, et al. (2019a) De novo GRIN variants in NMDA receptor M2 channel pore-forming loop are associated with neurological diseases. Hum Mutat 40:2393–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhang DQ, Wang XZ, Xu TJ (2011a) NR2B-containing NMDA receptors promote neural progenitor cell proliferation through CaMKIV/CREB pathway. Biochem Biophys Res Commun 411:667–672. [DOI] [PubMed] [Google Scholar]
- Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, Li XY, Aghajanian G, Duman RS (2010) mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science 329:959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Liu RJ, Dwyer JM, Banasr M, Lee B, Son H, Li XY, Aghajanian G, Duman RS (2011b) Glutamate N-methyl-D-aspartate receptor antagonists rapidly reverse behavioral and synaptic deficits caused by chronic stress exposure. Biol Psychiatry 69:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Wilding TJ, Kim SJ, Calejesan AA, Huettner JE, Zhuo M (1999) Kainate-receptor-mediated sensory synaptic transmission in mammalian spinal cord. Nature 397:161–164. [DOI] [PubMed] [Google Scholar]
- Li V, Wang YT (2016) Molecular mechanisms of NMDA receptor-mediated excitotoxicity: implications for neuroprotective therapeutics for stroke. Neural Regen Res 11:1752–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Pozzo-Miller L (2019) Differences in GluN2B-containing NMDA receptors result in distinct long-term plasticity at ipsilateral versus contralateral cortico-striatal synapses. eNeuro 6:ENEURO.0118-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dharkar P, Han TH, Serpe M, Lee CH, Mayer ML (2016b) Novel functional properties of drosophila CNS glutamate receptors. Neuron 92:1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Erzurumlu RS, Chen C, Jhaveri S, Tonegawa S (1994) Whisker-related neuronal patterns fail to develop in the trigeminal brainstem nuclei of NMDAR1 knockout mice. Cell 76:427–437. [DOI] [PubMed] [Google Scholar]
- Li YJDuan GFSun JHWu DYe CZang YYChen GQShi YYWang JZhang W, et al. (2019b) Neto proteins regulate gating of the kainate-type glutamate receptor GluK2 through two binding sites. J Biol Chem 294:17889–17902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Aizenman CD, Cline HT (2002) Regulation of rho GTPases by crosstalk and neuronal activity in vivo. Neuron 33:741–750. [DOI] [PubMed] [Google Scholar]
- Li Z, Tian Y, Tian N, Zhao X, Du C, Han L, Zhang H (2015) Aberrant alternative splicing pattern of ADAR2 downregulates adenosine-to-inosine editing in glioma. Oncol Rep 33:2845–2852. [DOI] [PubMed] [Google Scholar]
- Li Z, Van Aelst L, Cline HT (2000) Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat Neurosci 3:217–225. [DOI] [PubMed] [Google Scholar]
- Liao D, Hessler NA, Malinow R (1995) Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature 375:400–404. [DOI] [PubMed] [Google Scholar]
- Lichnerova K, Kaniakova M, Skrenkova K, Vyklicky L, Horak M (2014) Distinct regions within the GluN2C subunit regulate the surface delivery of NMDA receptors. Front Cell Neurosci 8:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilienfeld SO, Treadway MT (2016) Clashing diagnostic approaches: DSM-ICD versus RDoC. Annu Rev Clin Psychol 12:435–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KS, Cheong KL, Tan CT (2010) Periodic lateralized epileptiform discharges in neuropsychiatric lupus: association with cerebritis in magnetic resonance imaging and resolution after intravenous immunoglobulin. Lupus 19:748–752. [DOI] [PubMed] [Google Scholar]
- Lin DT, Makino Y, Sharma K, Hayashi T, Neve R, Takamiya K, Huganir RL (2009) Regulation of AMPA receptor extrasynaptic insertion by 4.1N, phosphorylation and palmitoylation. Nat Neurosci 12:879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind GE, Mou TC, Tamborini L, Pomper MG, De Micheli C, Conti P, Pinto A, Hansen KB (2017) Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits. Proc Natl Acad Sci USA 114:E6942–E6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsenbardt AJ, Chisari M, Yu A, Shu HJ, Zorumski CF, Mennerick S (2013) Noncompetitive, voltage-dependent NMDA receptor antagonism by hydrophobic anions. Mol Pharmacol 83:354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsenbardt AJ, Taylor A, Emnett CM, Doherty JJ, Krishnan K, Covey DF, Paul SM, Zorumski CF, Mennerick S (2014) Different oxysterols have opposing actions at N-methyl-D-aspartate receptors. Neuropharmacology 85:232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton SA (2006) Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat Rev Drug Discov 5:160–170. [DOI] [PubMed] [Google Scholar]
- Lipton SA (2007) Pathologically activated therapeutics for neuroprotection. Nat Rev Neurosci 8:803–808. [DOI] [PubMed] [Google Scholar]
- Lisman J (1989) A mechanism for the Hebb and the anti-Hebb processes underlying learning and memory. Proc Natl Acad Sci USA 86:9574–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Yasuda R, Raghavachari S (2012) Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci 13:169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Smerin Y, Aizenman E, Johnson JW (2000) Inhibition by intracellular Mg(2+) of recombinant N-methyl-D-aspartate receptors expressed in Chinese hamster ovary cells. J Pharmacol Exp Ther 292:1104–1110. [PubMed] [Google Scholar]
- Li-Smerin Y, Levitan ES, Johnson JW (2001) Free intracellular Mg(2+) concentration and inhibition of NMDA responses in cultured rat neurons. J Physiol 533:729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin DB, Carrillo E, Shaikh SA, Berka V, Jayaraman V (2019) The structural arrangement at intersubunit interfaces in homomeric kainate receptors. Sci Rep 9:6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin DB, Paudyal N, Carrillo E, Berka V, Jayaraman V (2020) The structural arrangement and dynamics of the heteromeric GluK2/GluK5 kainate receptor as determined by smFRET. Biochim Biophys Acta Biomembr 1862:183001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DD, Yang Q, Li ST (2013) Activation of extrasynaptic NMDA receptors induces LTD in rat hippocampal CA1 neurons. Brain Res Bull 93:10–16. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang H, Peterson M, Zhang W, Hou G, Zhang ZW (2019a) N-terminal alternative splicing of GluN1 regulates the maturation of excitatory synapses and seizure susceptibility. Proc Natl Acad Sci USA 116:21207–21212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HP, Lin WY, Liu SH, Wang WF, Tsai CH, Wu BT, Wang CK, Tsai FJ (2009) Genetic variation in N-methyl-D-aspartate receptor subunit NR3A but not NR3B influences susceptibility to Alzheimer’s disease. Dement Geriatr Cogn Disord 28:521–527. [DOI] [PubMed] [Google Scholar]
- Liu J, Gandhi PJ, Pavuluri R, Shelkar GP, Dravid SM (2018) Glutamate delta-1 receptor regulates cocaine-induced plasticity in the nucleus accumbens. Transl Psychiatry 8:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Shelkar GP, Gandhi PJ, Gawande DY, Hoover A, Villalba RM, Pavuluri R, Smith Y, Dravid SM (2020a) Striatal glutamate delta-1 receptor regulates behavioral flexibility and thalamostriatal connectivity. Neurobiol Dis 137:104746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Shelkar GP, Sarode LP, Gawande DY, Zhao F, Clausen RP, Ugale RR, Dravid SM (2021) Facilitation of GluN2C-containing NMDA receptors in the external globus pallidus increases firing of fast spiking neurons and improves motor function in a hemiparkinsonian mouse model. Neurobiol Dis 150:105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Shelkar GP, Zhao F, Clausen RP, Dravid SM (2019b) Modulation of burst firing of neurons in nucleus reticularis of the thalamus by GluN2C-containing NMDA receptors. Mol Pharmacol. [DOI] [PMC free article] [PubMed]
- Liu Q, Wong-Riley MT (2010) Postnatal development of N-methyl-D-aspartate receptor subunits 2A, 2B, 2C, 2D, and 3B immunoreactivity in brain stem respiratory nuclei of the rat. Neuroscience 171:637–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhou L, Yuan H, Vieira M, Sanz-Clemente A, Badger JD 2nd, Lu W, Traynelis SF, Roche KW (2017) A rare variant identified within the GluN2B C-terminus in a patient with autism affects NMDA receptor surface expression and spine density. J Neurosci 37:4093–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Cull-Candy SG (2005) Subunit interaction with PICK and GRIP controls Ca2+ permeability of AMPARs at cerebellar synapses. Nat Neurosci 8:768–775. [DOI] [PubMed] [Google Scholar]
- Liu SQ, Cull-Candy SG (2000) Synaptic activity at calcium-permeable AMPA receptors induces a switch in receptor subtype. Nature 405:454–458. [DOI] [PubMed] [Google Scholar]
- Liu W, Jiang X, Zu Y, Yang Y, Liu Y, Sun X, Xu Z, Ding H, Zhao Q (2020b) A comprehensive description of GluN2B-selective N-methyl-D-aspartate (NMDA) receptor antagonists. Eur J Med Chem 200:112447. [DOI] [PubMed] [Google Scholar]
- Liverton NJBednar RABednar BButcher JWClaiborne CFClaremon DACunningham MDiLella AGGaul SLLibby BE, et al. (2007) Identification and characterization of 4-methylbenzyl 4-[(pyrimidin-2-ylamino)methyl]piperidine-1-carboxylate, an orally bioavailable, brain penetrant NR2B selective N-methyl-D-aspartate receptor antagonist. J Med Chem 50:807–819. [DOI] [PubMed] [Google Scholar]
- Livsey CT, Costa E, Vicini S (1993) Glutamate-activated currents in outside-out patches from spiny versus aspiny hilar neurons of rat hippocampal slices. J Neurosci 13:5324–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lledo PM, Zhang X, Südhof TC, Malenka RC, Nicoll RA (1998) Postsynaptic membrane fusion and long-term potentiation. Science 279:399–403. [DOI] [PubMed] [Google Scholar]
- Lodge D (2009) The history of the pharmacology and cloning of ionotropic glutamate receptors and the development of idiosyncratic nomenclature. Neuropharmacology 56:6–21. [DOI] [PubMed] [Google Scholar]
- Lodge D, Bond A, O’Neill MJ, Hicks CA, Jones MG (1996) Stereoselective effects-of 2,3-benzodiazepines in vivo: electrophysiology and neuroprotection studies. Neuropharmacology 35:1681–1688. [DOI] [PubMed] [Google Scholar]
- Lodge D, Watkins JC, Bortolotto ZA, Jane DE, Volianskis A (2019) The 1980s: D-AP5, LTP and a decade of NMDA receptor discoveries. Neurochem Res 44:516–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomash RM, Sheng N, Li Y, Nicoll RA, Roche KW (2017) Phosphorylation of the kainate receptor (KAR) auxiliary subunit Neto2 at serine 409 regulates synaptic targeting of the KAR subunit GluK1. J Biol Chem 292:15369–15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomash S, Chittori S, Brown P, Mayer ML (2013) Anions mediate ligand binding in Adineta vaga glutamate receptor ion channels. Structure 21:414–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli H, Mosbacher J, Melcher T, Höger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH (1994) Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266:1709–1713. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Sprengel R, Laurie DJ, Köhr G, Herb A, Seeburg PH, Wisden W (1993) The rat delta-1 and delta-2 subunits extend the excitatory amino acid receptor family. FEBS Lett 315:318–322. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Wisden W, Köhler M, Keinänen K, Sommer B, Seeburg PH (1992) High-affinity kainate and domoate receptors in rat brain. FEBS Lett 307:139–143. [DOI] [PubMed] [Google Scholar]
- Lopez de Armentia M, Sah P (2003) Development and subunit composition of synaptic NMDA receptors in the amygdala: NR2B synapses in the adult central amygdala. J Neurosci 23:6876–6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löscher W, Hönack D (1994) Effects of the non-NMDA antagonists NBQX and the 2,3-benzodiazepine GYKI 52466 on different seizure types in mice: comparison with diazepam and interactions with flumazenil. Br J Pharmacol 113:1349–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losonczy A, Magee JC (2006) Integrative properties of radial oblique dendrites in hippocampal CA1 pyramidal neurons. Neuron 50:291–307. [DOI] [PubMed] [Google Scholar]
- Lourenço J, Cannich A, Carta M, Coussen F, Mulle C, Marsicano G (2010) Synaptic activation of kainate receptors gates presynaptic CB(1) signaling at GABAergic synapses. Nat Neurosci 13:197–204. [DOI] [PubMed] [Google Scholar]
- Lourenço J, Matias I, Marsicano G, Mulle C (2011) Pharmacological activation of kainate receptors drives endocannabinoid mobilization. J Neurosci 31:3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louros SR, Hooks BM, Litvina L, Carvalho AL, Chen C (2014) A role for stargazin in experience-dependent plasticity. Cell Rep 7:1614–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low CMLyuboslavsky PFrench ALe PWyatte KThiel WHMarchan EMIgarashi KKashiwagi KGernert K, et al. (2003) Molecular determinants of proton-sensitive N-methyl-D-aspartate receptor gating. Mol Pharmacol 63:1212–1222. [DOI] [PubMed] [Google Scholar]
- Low CM, Wee KS (2010) New insights into the not-so-new NR3 subunits of N-methyl-D-aspartate receptor: localization, structure, and function. Mol Pharmacol 78:1–11. [DOI] [PubMed] [Google Scholar]
- Low CM, Zheng F, Lyuboslavsky P, Traynelis SF (2000) Molecular determinants of coordinated proton and zinc inhibition of N-methyl-D-aspartate NR1/NR2A receptors. Proc Natl Acad Sci USA 97:11062–11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CR, Hwang SJ, Phend KD, Rustioni A, Valtschanoff JG (2002) Primary afferent terminals in spinal cord express presynaptic AMPA receptors. J Neurosci 22:9522–9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Helton TD, Blanpied TA, Rácz B, Newpher TM, Weinberg RJ, Ehlers MD (2007a) Postsynaptic positioning of endocytic zones and AMPA receptor cycling by physical coupling of dynamin-3 to Homer. Neuron 55:874–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Bushong EA, Shih TP, Ellisman MH, Nicoll RA (2013) The cell-autonomous role of excitatory synaptic transmission in the regulation of neuronal structure and function. Neuron 78:433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü W, Du J, Goehring A, Gouaux E (2017) Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science 355:eaal3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Gray JA, Granger AJ, During MJ, Nicoll RA (2011) Potentiation of synaptic AMPA receptors induced by the deletion of NMDA receptors requires the GluA2 subunit. J Neurophysiol 105:923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Man H, Ju W, Trimble WS, MacDonald JF, Wang YT (2001) Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron 29:243–254. [DOI] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH, Nicoll RA (2009) Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron 62:254–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Allen M, Halt AR, Weisenhaus M, Dallapiazza RF, Hall DD, Usachev YM, McKnight GS, Hell JW (2007b) Age-dependent requirement of AKAP150-anchored PKA and GluR2-lacking AMPA receptors in LTP. EMBO J 26:4879–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby ED, Cohen BD, Rosenbaum G, Gottlieb JS, Kelley R (1959) Study of a new schizophrenomimetic drug; sernyl. AMA Arch Neurol Psychiatry 81:363–369. [DOI] [PubMed] [Google Scholar]
- Luchkina NV, Huupponen J, Clarke VR, Coleman SK, Keinänen K, Taira T, Lauri SE (2014) Developmental switch in the kinase dependency of long-term potentiation depends on expression of GluA4 subunit-containing AMPA receptors. Proc Natl Acad Sci USA 111:4321–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luján R, Shigemoto R, López-Bendito G (2005) Glutamate and GABA receptor signalling in the developing brain. Neuroscience 130:567–580. [DOI] [PubMed] [Google Scholar]
- Luk KC, Kennedy TE, Sadikot AF (2003) Glutamate promotes proliferation of striatal neuronal progenitors by an NMDA receptor-mediated mechanism. J Neurosci 23:2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn ML, Hogner A, Stensbøl TB, Gouaux E, Egebjerg J, Kastrup JS (2003) Three-dimensional structure of the ligand-binding core of GluR2 in complex with the agonist (S)-ATPA: implications for receptor subunit selectivity. J Med Chem 46:872–875. [DOI] [PubMed] [Google Scholar]
- Luo J, Wang Y, Yasuda RP, Dunah AW, Wolfe BB (1997) The majority of N-methyl-D-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B). Mol Pharmacol 51:79–86. [DOI] [PubMed] [Google Scholar]
- Luque JM, Bleuel Z, Malherbe P, Richards JG (1994) Alternatively spliced isoforms of the N-methyl-D-aspartate receptor subunit 1 are differentially distributed within the rat spinal cord. Neuroscience 63:629–635. [DOI] [PubMed] [Google Scholar]
- Lüscher C, Malenka RC (2012) NMDA receptor-dependent long-term potentiation and long-term depression (LTP/LTD). Cold Spring Harb Perspect Biol 4:a005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier MP, Sanz-Clemente A, Roche KW (2015) Dynamic regulation of N-methyl-d-aspartate (NMDA) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors by posttranslational modifications. J Biol Chem 290:28596–28603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthi A, Schwyzer L, Mateos JM, Gähwiler BH, McKinney RA (2001) NMDA receptor activation limits the number of synaptic connections during hippocampal development. Nat Neurosci 4:1102–1107. [DOI] [PubMed] [Google Scholar]
- Ly LT, Kreye J, Jurek B, Leubner J, Scheibe F, Lemcke J, Wenke NK, Reincke SM, Prüss H (2018) Affinities of human NMDA receptor autoantibodies: implications for disease mechanisms and clinical diagnostics. J Neurol 265:2625–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch G (2006) Glutamate-based therapeutic approaches: ampakines. Curr Opin Pharmacol 6:82–88. [DOI] [PubMed] [Google Scholar]
- Lynch G, Gall CM (2006) Ampakines and the threefold path to cognitive enhancement. Trends Neurosci 29:554–562. [DOI] [PubMed] [Google Scholar]
- Lynch G, Larson J, Kelso S, Barrionuevo G, Schottler F (1983) Intracellular injections of EGTA block induction of hippocampal long-term potentiation. Nature 305:719–721. [DOI] [PubMed] [Google Scholar]
- Ma Z, Zang T, Birnbaum SG, Wang Z, Johnson JE, Zhang CL, Parada LF (2017) TrkB dependent adult hippocampal progenitor differentiation mediates sustained ketamine antidepressant response. Nat Commun 8:1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ME, Vonsattel JP, Shrinidhi J, Couropmitree NN, Cupples LA, Bird ED, Gusella JF, Myers RH (1999) Evidence for the GluR6 gene associated with younger onset age of Huntington’s disease. Neurology 53:1330–1332. [DOI] [PubMed] [Google Scholar]
- MacGillavry HD, Song Y, Raghavachari S, Blanpied TA (2013) Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron 78:615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay MVo ATang CCSmall MAnderson EWPloran EJStorbeck JBascetta BKang SAranow C, et al. (2019) Metabolic and microstructural alterations in the SLE brain correlate with cognitive impairment. JCI Insight 4:e124002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean DM, Ramaswamy SS, Du M, Howe JR, Jayaraman V (2014) Stargazin promotes closure of the AMPA receptor ligand-binding domain. J Gen Physiol 144:503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean DM, Wong AY, Fay AM, Bowie D (2011) Cations but not anions regulate the responsiveness of kainate receptors. J Neurosci 31:2136–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden DR, Armstrong N, Svergun D, Pérez J, Vachette P (2005) Solution X-ray scattering evidence for agonist- and antagonist-induced modulation of cleft closure in a glutamate receptor ligand-binding domain. J Biol Chem 280:23637–23642. [DOI] [PubMed] [Google Scholar]
- Madeira CLourenco MVVargas-Lopes CSuemoto CKBrandão COReis TLeite RELaks JJacob-Filho WPasqualucci CA, et al. (2015) d-serine levels in Alzheimer’s disease: implications for novel biomarker development. Transl Psychiatry 5:e561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madry C, Betz H, Geiger JR, Laube B (2008) Supralinear potentiation of NR1/NR3A excitatory glycine receptors by Zn2+ and NR1 antagonist. Proc Natl Acad Sci USA 105:12563–12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madry C, Betz H, Geiger JR, Laube B (2010) Potentiation of glycine-gated NR1/NR3A NMDA receptors relieves Ca-dependent outward rectification. Front Mol Neurosci 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madry C, Mesic I, Bartholomäus I, Nicke A, Betz H, Laube B (2007) Principal role of NR3 subunits in NR1/NR3 excitatory glycine receptor function. Biochem Biophys Res Commun 354:102–108. [DOI] [PubMed] [Google Scholar]
- Madsen U, Frydenvang K, Ebert B, Johansen TN, Brehm L, Krogsgaard-Larsen P (1996) N-methyl-D-aspartic acid receptor agonists: resolution, absolute stereochemistry, and pharmacology of the enantiomers of 2-amino-2-(3-hydroxy-5-methyl-4-isoxazolyl)acetic acid. J Med Chem 39:183–190. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Namba T, Suzuki E, Yuasa S, Kohsaka S, Uchino S (2009) NMDA receptor antagonist memantine promotes cell proliferation and production of mature granule neurons in the adult hippocampus. Neurosci Res 63:259–266. [DOI] [PubMed] [Google Scholar]
- Magazanik LG, Buldakova SL, Samoilova MV, Gmiro VE, Mellor IR, Usherwood PN (1997) Block of open channels of recombinant AMPA receptors and native AMPA/kainate receptors by adamantane derivatives. J Physiol 505:655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JC, Cook EP (2000) Somatic EPSP amplitude is independent of synapse location in hippocampal pyramidal neurons. Nat Neurosci 3:895–903. [DOI] [PubMed] [Google Scholar]
- Mah SJ, Cornell E, Mitchell NA, Fleck MW (2005) Glutamate receptor trafficking: endoplasmic reticulum quality control involves ligand binding and receptor function. J Neurosci 25:2215–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan V, Dargaei Z, Ivakine EA, Hartmann AM, Ng D, Chevrier J, Ormond J, Nothwang HG, McInnes RR, Woodin MA (2015) Neto2-null mice have impaired GABAergic inhibition and are susceptible to seizures. Front Cell Neurosci 9:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan VPressey JCActon BAUvarov PHuang MYChevrier JPuchalski ALi CMIvakine EAAiraksinen MS, et al. (2014) Kainate receptors coexist in a functional complex with KCC2 and regulate chloride homeostasis in hippocampal neurons. Cell Rep 7:1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E (2019) Esketamine is approved in Europe for treating resistant major depressive disorder. BMJ 367:l7069. [DOI] [PubMed] [Google Scholar]
- Maher MP, Matta JA, Gu S, Seierstad M, Bredt DS (2017) Getting a handle on neuropharmacology by targeting receptor-associated proteins. Neuron 96:989–1001. [DOI] [PubMed] [Google Scholar]
- Maher MPWu NRavula SAmeriks MKSavall BMLiu CLord BWyatt RMMatta JADugovic C, et al. (2016) Discovery and characterization of AMPA receptor modulators selective for TARP-γ8. J Pharmacol Exp Ther 357:394–414. [DOI] [PubMed] [Google Scholar]
- Mahfooz K, Marco S, Martínez-Turrillas R, Raja MK, Pérez-Otaño I, Wesseling JF (2016) GluN3A promotes NMDA spiking by enhancing synaptic transmission in Huntington’s disease models. Neurobiol Dis 93:47–56. [DOI] [PubMed] [Google Scholar]
- Mahmoud H, Martin N, Hildebrand ME (2020) Conserved contributions of NMDA receptor subtypes to synaptic responses in lamina II spinal neurons across early postnatal development. Mol Brain 13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A, Klopocki E, Horn D, Tzschach A, Holm T, Meyer R, Meyer T (2014) De novo partial deletion in GRID2 presenting with complicated spastic paraplegia. Muscle Nerve 49:289–292. [DOI] [PubMed] [Google Scholar]
- Major G, Polsky A, Denk W, Schiller J, Tank DW (2008) Spatiotemporally graded NMDA spike/plateau potentials in basal dendrites of neocortical pyramidal neurons. J Neurophysiol 99:2584–2601. [DOI] [PubMed] [Google Scholar]
- Maki BA, Aman TK, Amico-Ruvio SA, Kussius CL, Popescu GK (2012) C-terminal domains of N-methyl-D-aspartic acid receptor modulate unitary channel conductance and gating. J Biol Chem 287:36071–36080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki BA, Cole R, Popescu GK (2013) Two serine residues on GluN2A C-terminal tails control NMDA receptor current decay times. Channels (Austin) 7:126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki BA, Popescu GK (2014) Extracellular Ca(2+) ions reduce NMDA receptor conductance and gating. J Gen Physiol 144:379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H, Malinow R (2009) AMPA receptor incorporation into synapses during LTP: the role of lateral movement and exocytosis. Neuron 64:381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malayev A, Gibbs TT, Farb DH (2002) Inhibition of the NMDA response by pregnenolone sulphate reveals subtype selective modulation of NMDA receptors by sulphated steroids. Br J Pharmacol 135:901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K (1999) Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science 283:1923–1927. [DOI] [PubMed] [Google Scholar]
- Malinow R, Schulman H, Tsien RW (1989) Inhibition of postsynaptic PKC or CaMKII blocks induction but not expression of LTP. Science 245:862–866. [DOI] [PubMed] [Google Scholar]
- Maltsev AS, Ahmed AH, Fenwick MK, Jane DE, Oswald RE (2008) Mechanism of partial agonism at the GluR2 AMPA receptor: Measurements of lobe orientation in solution. Biochemistry 47:10600–10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mameli M, Bellone C, Brown MT, Lüscher C (2011) Cocaine inverts rules for synaptic plasticity of glutamate transmission in the ventral tegmental area. Nat Neurosci 14:414–416. [DOI] [PubMed] [Google Scholar]
- Mameli M, Carta M, Partridge LD, Valenzuela CF (2005) Neurosteroid-induced plasticity of immature synapses via retrograde modulation of presynaptic NMDA receptors. J Neurosci 25:2285–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man HY, Sekine-Aizawa Y, Huganir RL (2007) Regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor trafficking through PKA phosphorylation of the Glu receptor 1 subunit. Proc Natl Acad Sci USA 104:3579–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini JD, Atchison WD (2007) The NR2B subunit in NMDA receptors is functionally important during cerebellar granule cell migration. Neurosci Lett 429:87–90. [DOI] [PubMed] [Google Scholar]
- Mandolesi G, Cesa R, Autuori E, Strata P (2009) An orphan ionotropic glutamate receptor: the delta2 subunit. Neuroscience 158:67–77. [DOI] [PubMed] [Google Scholar]
- Manent JB, Jorquera I, Ben-Ari Y, Aniksztejn L, Represa A (2006) Glutamate acting on AMPA but not NMDA receptors modulates the migration of hippocampal interneurons. J Neurosci 26:5901–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manita S, Miyazaki K, Ross WN (2011) Synaptically activated Ca2+ waves and NMDA spikes locally suppress voltage-dependent Ca2+ signalling in rat pyramidal cell dendrites. J Physiol 589:4903–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EO, Mody I (2010) Control of hippocampal gamma oscillation frequency by tonic inhibition and excitation of interneurons. Nat Neurosci 13:205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannaioni G, Marino MJ, Valenti O, Traynelis SF, Conn PJ (2001) Metabotropic glutamate receptors 1 and 5 differentially regulate CA1 pyramidal cell function. J Neurosci 21:5925–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannara FRadosevic MPlanagumà JSoto DAguilar EGarcía-Serra AMaudes EPedreño MPaul SDoherty J, et al. (2020) Allosteric modulation of NMDA receptors prevents the antibody effects of patients with anti-NMDAR encephalitis. Brain 143:2709–2720. [DOI] [PubMed] [Google Scholar]
- Mantegazza R, Bernasconi P, Baggi F, Spreafico R, Ragona F, Antozzi C, Bernardi G, Granata T (2002) Antibodies against GluR3 peptides are not specific for Rasmussen’s encephalitis but are also present in epilepsy patients with severe, early onset disease and intractable seizures. J Neuroimmunol 131:179–185. [DOI] [PubMed] [Google Scholar]
- Mao LM, Guo ML, Jin DZ, Fibuch EE, Choe ES, Wang JQ (2011) Post-translational modification biology of glutamate receptors and drug addiction. Front Neuroanat 5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao SC, Hsiao YH, Gean PW (2006) Extinction training in conjunction with a partial agonist of the glycine site on the NMDA receptor erases memory trace. J Neurosci 26:8892–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Gu X, Lu W (2017) GSG1L regulates the strength of AMPA receptor-mediated synaptic transmission but not AMPA receptor kinetics in hippocampal dentate granule neurons. J Neurophysiol 117:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maolanon ARRisgaard RWang SYSnoep YPapangelis AYi FHolley DBarslund AFSvenstrup NHansen KB, et al. (2017) Subtype-specific agonists for NMDA receptor glycine binding sites. ACS Chem Neurosci 8:1681–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelli SIannuzzi FFiculle EMango DPieraccini SPellegrino SCorbo MSironi MPittaluga ANisticò R, et al. (2019) The selective disruption of presynaptic JNK2/STX1a interaction reduces NMDA receptor-dependent glutamate release. Sci Rep 9:7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C, Mulle C (2004) Postnatal maturation of mossy fibre excitatory transmission in mouse CA3 pyramidal cells: a potential role for kainate receptors. J Physiol 561:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcin LRWarrier JThangathirupathy SShi JKarageorge GNPearce BCNg APark HKempson JLi J, et al. (2018) BMS-986163, a negative allosteric modulator of GluN2B with potential utility in major depressive disorder. ACS Med Chem Lett 9:472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco SGiralt APetrovic MMPouladi MAMartínez-Turrillas RMartínez-Hernández JKaltenbach LSTorres-Peraza JGraham RKWatanabe M, et al. (2013) Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models. Nat Med 19:1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco S, Murillo A, Pérez-Otaño I (2018) RNAi-based GluN3A silencing prevents and reverses disease phenotypes induced by mutant huntingtin. Mol Ther 26:1965–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marger L, Schubert CR, Bertrand D (2014) Zinc: an underappreciated modulatory factor of brain function. Biochem Pharmacol 91:426–435. [DOI] [PubMed] [Google Scholar]
- Maricq AV, Peckol E, Driscoll M, Bargmann CI (1995) Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature 378:78–81. [DOI] [PubMed] [Google Scholar]
- Marques JM, Rodrigues RJ, Valbuena S, Rozas JL, Selak S, Marin P, Aller MI, Lerma J (2013) CRMP2 tethers kainate receptor activity to cytoskeleton dynamics during neuronal maturation. J Neurosci 33:18298–18310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marret S, Gressens P, Evrard P (1996) Arrest of neuronal migration by excitatory amino acids in hamster developing brain. Proc Natl Acad Sci USA 93:15463–15468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JJ, Xu J, Contractor A (2018) Kainate receptors inhibit glutamate release via mobilization of endocannabinoids in striatal direct pathway spiny projection neurons. J Neurosci 38:3901–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel MARyan TJBell KFFowler JHMcMahon AAl-Mubarak BKomiyama NHHorsburgh KKind PCGrant SG, et al. (2012) The subtype of GluN2 C-terminal domain determines the response to excitotoxic insults. Neuron 74:543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LJ, Furuta A, Blackstone CD (1998) AMPA receptor protein in developing rat brain: glutamate receptor-1 expression and localization change at regional, cellular, and subcellular levels with maturation. Neuroscience 83:917–928. [DOI] [PubMed] [Google Scholar]
- Martin SChamberlin AShinde DNHempel MStrom TMSchreiber AJohannsen JOusager LBLarsen MJHansen LK, et al. (2017) De novo variants in GRIA4 lead to intellectual disability with or without seizures and gait abnormalities. Am J Hum Genet 101:1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwick KFM, Hansen KB, Skehel PA, Hardingham GE, Wyllie DJA (2019) Functional assessment of triheteromeric NMDA receptors containing a human variant associated with epilepsy. J Physiol 597:1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey PV, Johnson BE, Moult PR, Auberson YP, Brown MW, Molnar E, Collingridge GL, Bashir ZI (2004) Differential roles of NR2A and NR2B-containing NMDA receptors in cortical long-term potentiation and long-term depression. J Neurosci 24:7821–7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuko T, Kashiwagi K, Kuno T, Nguyen ND, Pahk AJ, Fukuchi J, Igarashi K, Williams K (1999a) A regulatory domain (R1-R2) in the amino terminus of the N-methyl-D-aspartate receptor: effects of spermine, protons, and ifenprodil, and structural similarity to bacterial leucine/isoleucine/valine binding protein. Mol Pharmacol 55:957–969. [DOI] [PubMed] [Google Scholar]
- Masuko T, Kuno T, Kashiwagi K, Kusama T, Williams K, Igarashi K (1999b) Stimulatory and inhibitory properties of aminoglycoside antibiotics at N-methyl-D-aspartate receptors. J Pharmacol Exp Ther 290:1026–1033. [PubMed] [Google Scholar]
- Mataix-Cols DFernández de la Cruz LMonzani BRosenfield DAndersson EPérez-Vigil AFrumento Pde Kleine RADifede JDunlop BW, et al. ; the DCS Anxiety Consortium (2017) D-Cycloserine augmentation of exposure-based cognitive behavior therapy for anxiety, obsessive-compulsive, and posttraumatic stress disorders: A systematic review and meta-analysis of individual participant data. JAMA Psychiatry 74:501–510. [DOI] [PubMed] [Google Scholar]
- Mathers DA, Usherwood PN (1976) Concanavalin A blocks desensitisation of glutamate receptors on insect muscle fibres. Nature 259:409–411. [DOI] [PubMed] [Google Scholar]
- Matsuda KBudisantoso TMitakidis NSugaya YMiura EKakegawa WYamasaki MKonno KUchigashima MAbe M, et al. (2016) Transsynaptic modulation of kainate receptor functions by C1q-like proteins. Neuron 90:752–767. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Fletcher M, Kamiya Y, Yuzaki M (2003) Specific assembly with the NMDA receptor 3B subunit controls surface expression and calcium permeability of NMDA receptors. J Neurosci 23:10064–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Kamiya Y, Matsuda S, Yuzaki M (2002) Cloning and characterization of a novel NMDA receptor subunit NR3B: a dominant subunit that reduces calcium permeability. Brain Res Mol Brain Res 100:43–52. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Kondo T, Iijima T, Matsuda S, Watanabe M, Yuzaki M (2009) Cbln1 binds to specific postsynaptic sites at parallel fiber-Purkinje cell synapses in the cerebellum. Eur J Neurosci 29:707–717. [DOI] [PubMed] [Google Scholar]
- Matsuda KMiura EMiyazaki TKakegawa WEmi KNarumi SFukazawa YIto-Ishida AKondo TShigemoto R, et al. (2010) Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science 328:363–368. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Yuzaki M (2011) Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci 33:1447–1461. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Kakegawa W, Budisantoso T, Nomura T, Kohda K, Yuzaki M (2013) Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long-term depression. Nat Commun 4:2759. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Miura E, Matsuda K, Kakegawa W, Kohda K, Watanabe M, Yuzaki M (2008) Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron 57:730–745. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Yuzaki M (2002) Mutation in hotfoot-4J mice results in retention of delta2 glutamate receptors in ER. Eur J Neurosci 16:1507–1516. [DOI] [PubMed] [Google Scholar]
- Matsui K, Jahr CE (2004) Differential control of synaptic and ectopic vesicular release of glutamate. J Neurosci 24:8932–8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H (2004) Structural basis of long-term potentiation in single dendritic spines. Nature 429:761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt L, Kim K, Chowdhury D, Hell JW (2019) Role of palmitoylation of postsynaptic proteins in promoting synaptic plasticity. Front Mol Neurosci 12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt LKirk LMChenaux GSpeca DJPuhger KRPride MCQneibi MHaham TPlambeck KEStern-Bach Y, et al. (2018) SynDIG4/Prrt1 is required for excitatory synapse development and plasticity underlying cognitive function. Cell Rep 22:2246–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta JA, Pelkey KA, Craig MT, Chittajallu R, Jeffries BW, McBain CJ (2013) Developmental origin dictates interneuron AMPA and NMDA receptor subunit composition and plasticity. Nat Neurosci 16:1032–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes H, Carcache D, Kalkman HO, Koller M (2010) alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) antagonists: from bench to bedside. J Med Chem 53:5367–5382. [DOI] [PubMed] [Google Scholar]
- Mauric V, Mölders A, Harmel N, Heimrich B, Sergeeva OA, Klöcker N (2013) Ontogeny repeats the phylogenetic recruitment of the cargo exporter cornichon into AMPA receptor signaling complexes. Mol Cell Neurosci 56:10–17. [DOI] [PubMed] [Google Scholar]
- Mayat E, Petralia RS, Wang YX, Wenthold RJ (1995) Immunoprecipitation, immunoblotting, and immunocytochemistry studies suggest that glutamate receptor delta subunits form novel postsynaptic receptor complexes. J Neurosci 15:2533–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML (2005) Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron 45:539–552. [DOI] [PubMed] [Google Scholar]
- Mayer ML (2021) Glutamate receptors from diverse animal species exhibit unexpected structural and functional diversity. J Physiol 599:2605–2613. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Ghosal A, Dolman NP, Jane DE (2006) Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci 26:2852–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Jegla T (2018) Family matters. eLife 7:e43815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Olson R, Gouaux E (2001) Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state. J Mol Biol 311:815–836. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Vyklicky L Jr (1989) Concanavalin A selectively reduces desensitization of mammalian neuronal quisqualate receptors. Proc Natl Acad Sci USA 86:1411–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Vyklicky L Jr, Clements J (1989) Regulation of NMDA receptor desensitization in mouse hippocampal neurons by glycine. Nature 338:425–427. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL (1987a) Permeation and block of N-methyl-D-aspartic acid receptor channels by divalent cations in mouse cultured central neurones. J Physiol 394:501–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL (1987b) The physiology of excitatory amino acids in the vertebrate central nervous system. Prog Neurobiol 28:197–276. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL, Guthrie PB (1984) Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature 309:261–263. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC (1996) Neurotrophin regulation of cortical dendritic growth requires activity. Neuron 17:1057–1064. [DOI] [PubMed] [Google Scholar]
- McAllister BB, Dyck RH (2017) Zinc transporter 3 (ZnT3) and vesicular zinc in central nervous system function. Neurosci Biobehav Rev 80:329–350. [DOI] [PubMed] [Google Scholar]
- McBain CJ, Dingledine R (1993) Heterogeneity of synaptic glutamate receptors on CA3 stratum radiatum interneurones of rat hippocampus. J Physiol 462:373–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain CJ, Fisahn A (2001) Interneurons unbound. Nat Rev Neurosci 2:11–23. [DOI] [PubMed] [Google Scholar]
- McCauley JATheberge CRRomano JJBillings SBAnderson KDClaremon DAFreidinger RMBednar RAMosser SDGaul SL, et al. (2004) NR2B-selective N-methyl-D-aspartate antagonists: synthesis and evaluation of 5-substituted benzimidazoles. J Med Chem 47:2089–2096. [DOI] [PubMed] [Google Scholar]
- McClymont DW, Harris J, Mellor IR (2012) Open-channel blockade is less effective on GluN3B than GluN3A subunit-containing NMDA receptors. Eur J Pharmacol 686:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool BA, Lovinger DM (1995) Ifenprodil inhibition of the 5-hydroxytryptamine3 receptor. Neuropharmacology 34:621–629. [DOI] [PubMed] [Google Scholar]
- McDaniel MJ, Ogden KK, Kell SA, Burger PB, Liotta DC, Traynelis SF (2020) NMDA receptor channel gating control by the pre-M1 helix. J Gen Physiol 152:e201912362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFeeters RL, Oswald RE (2002) Structural mobility of the extracellular ligand-binding core of an ionotropic glutamate receptor. Analysis of NMR relaxation dynamics. Biochemistry 41:10472–10481. [DOI] [PubMed] [Google Scholar]
- McGee TP, Bats C, Farrant M, Cull-Candy SG (2015) Auxiliary subunit GSG1L acts to suppress calcium-permeable AMPA receptor function. J Neurosci 35:16171–16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness L, Taylor C, Taylor RD, Yau C, Langenhan T, Hart ML, Christian H, Tynan PW, Donnelly P, Emptage NJ (2010) Presynaptic NMDARs in the hippocampus facilitate transmitter release at theta frequency. Neuron 68:1109–1127. [DOI] [PubMed] [Google Scholar]
- McIntyre CJMcCauley JABednar BBednar RAButcher JWClaremon DACunningham MEFreidinger RMGaul SLHomnick CF, et al. (2009) Synthesis and evaluation of novel tricyclic benzo[4.5]cyclohepta[1.2]pyridine derivatives as NMDA/NR2B antagonists. Bioorg Med Chem Lett 19:5132–5135. [DOI] [PubMed] [Google Scholar]
- McKay S, Griffiths NH, Butters PA, Thubron EB, Hardingham GE, Wyllie DJ (2012) Direct pharmacological monitoring of the developmental switch in NMDA receptor subunit composition using TCN 213, a GluN2A-selective, glycine-dependent antagonist. Br J Pharmacol 166:924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SRyan TJMcQueen JIndersmitten TMarwick KFMHasel PKopanitsa MVBaxter PSMartel MAKind PC, et al. (2018) The developmental shift of NMDA receptor composition proceeds independently of GluN2 subunit-specific GluN2 C-terminal sequences. Cell Rep 25:841–851.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney RA (2010) Excitatory amino acid involvement in dendritic spine formation, maintenance and remodelling. J Physiol 588:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealing GA, Lanthorn TH, Murray CL, Small DL, Morley P (1999) Differences in degree of trapping of low-affinity uncompetitive N-methyl-D-aspartic acid receptor antagonists with similar kinetics of block. J Pharmacol Exp Ther 288:204–210. [PubMed] [Google Scholar]
- Mealing GA, Lanthorn TH, Small DL, Murray RJ, Mattes KC, Comas TM, Morley P (2001) Structural modifications to an N-methyl-D-aspartate receptor antagonist result in large differences in trapping block. J Pharmacol Exp Ther 297:906–914. [PubMed] [Google Scholar]
- Meddows E, Le Bourdelles B, Grimwood S, Wafford K, Sandhu S, Whiting P, McIlhinney RA (2001) Identification of molecular determinants that are important in the assembly of N-methyl-D-aspartate receptors. J Biol Chem 276:18795–18803. [DOI] [PubMed] [Google Scholar]
- Medina I, Filippova N, Charton G, Rougeole S, Ben-Ari Y, Khrestchatisky M, Bregestovski P (1995) Calcium-dependent inactivation of heteromeric NMDA receptor-channels expressed in human embryonic kidney cells. J Physiol 482:567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev NI, Rodríguez-Arellano JJ, Popov VI, Davies HA, Tigaret CM, Schoepfer R, Stewart MG (2008) The glutamate receptor 2 subunit controls post-synaptic density complexity and spine shape in the dentate gyrus. Eur J Neurosci 27:315–325. [DOI] [PubMed] [Google Scholar]
- Mellone M, Pelucchi S, Alberti L, Genazzani AA, Di Luca M, Gardoni F (2015) Zinc transporter-1: a novel NMDA receptor-binding protein at the postsynaptic density. J Neurochem 132:159–168. [DOI] [PubMed] [Google Scholar]
- Mellor IRBrier TJPluteanu FStrømgaard KSaghyan AEldursi NBrierley MJAndersen KJaroszewski JWKrogsgaard-Larsen P, et al. (2003) Modification of the philanthotoxin-343 polyamine moiety results in different structure-activity profiles at muscle nicotinic ACh, NMDA and AMPA receptors. Neuropharmacology 44:70–80. [DOI] [PubMed] [Google Scholar]
- Melyan Z, Lancaster B, Wheal HV (2004) Metabotropic regulation of intrinsic excitability by synaptic activation of kainate receptors. J Neurosci 24:4530–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melyan Z, Wheal HV, Lancaster B (2002) Metabotropic-mediated kainate receptor regulation of IsAHP and excitability in pyramidal cells. Neuron 34:107–114. [DOI] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Jia Z (2003) Synaptic transmission and plasticity in the absence of AMPA glutamate receptor GluR2 and GluR3. Neuron 39:163–176. [DOI] [PubMed] [Google Scholar]
- Menniti FS, Buchan AM, Chenard BL, Critchett DJ, Ganong AH, Guanowsky V, Seymour PA, Welch WM (2003) CP-465,022, a selective noncompetitive AMPA receptor antagonist, blocks AMPA receptors but is not neuroprotective in vivo. Stroke 34:171–176. [DOI] [PubMed] [Google Scholar]
- Menniti FSChenard BLCollins MBDucat MFElliott MLEwing FEHuang JIKelly KALazzaro JTPagnozzi MJ, et al. (2000) Characterization of the binding site for a novel class of noncompetitive alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor antagonists. Mol Pharmacol 58:1310–1317. [DOI] [PubMed] [Google Scholar]
- Menniti FS, Lindsley CW, Conn PJ, Pandit J, Zagouras P, Volkmann RA (2013) Allosteric modulators for the treatment of schizophrenia: targeting glutamatergic networks. Curr Top Med Chem 13:26–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz K, Nicoll RA (2008) Loss of inhibitory neuron AMPA receptors contributes to ataxia and epilepsy in stargazer mice. J Neurosci 28:10599–10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz K, Stroud RM, Nicoll RA, Hays FA (2007) TARP auxiliary subunits switch AMPA receptor antagonists into partial agonists. Science 318:815–817. [DOI] [PubMed] [Google Scholar]
- Merchant RE, Bullock MR, Carmack CA, Shah AK, Wilner KD, Ko G, Williams SA (1999) A double-blind, placebo-controlled study of the safety, tolerability and pharmacokinetics of CP-101,606 in patients with a mild or moderate traumatic brain injury. Ann N Y Acad Sci 890:42–50. [DOI] [PubMed] [Google Scholar]
- Mesbahi-Vasey S, Veras L, Yonkunas M, Johnson JW, Kurnikova MG (2017) All atom NMDA receptor transmembrane domain model development and simulations in lipid bilayers and water. PLoS One 12:e0177686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesic I, Madry C, Geider K, Bernhard M, Betz H, Laube B (2016) The N-terminal domain of the GluN3A subunit determines the efficacy of glycine-activated NMDA receptors. Neuropharmacology 105:133–141. [DOI] [PubMed] [Google Scholar]
- Messersmith EK, Feller MB, Zhang H, Shatz CJ (1997) Migration of neocortical neurons in the absence of functional NMDA receptors. Mol Cell Neurosci 9:347–357. [DOI] [PubMed] [Google Scholar]
- Métin C, Denizot JP, Ropert N (2000) Intermediate zone cells express calcium-permeable AMPA receptors and establish close contact with growing axons. J Neurosci 20:696–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier CN, Dallérac G, Le Roux N, Sacchi S, Levasseur G, Amar M, Pollegioni L, Mothet JP, Fossier P (2016) D-Serine and glycine differentially control neurotransmission during visual cortex critical period. PLoS One 11:e0151233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson JR, Chittori S, Merk A, Rao P, Han TH, Serpe M, Mayer ML, Subramaniam S (2016) Structural basis of kainate subtype glutamate receptor desensitization. Nature 537:567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S (2014) Structural mechanism of glutamate receptor activation and desensitization. Nature 514:328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi DLi ZLim LLi MMoissidis MYang YGao THu TXPratt TPrice DJ, et al. (2018) Early emergence of cortical interneuron diversity in the mouse embryo. Science 360:81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaëlsson HAndersson MSvensson JKarlsson LEhn JCulley GEngström ABergström NSavvidi PKuhn HG, et al. (2019) The novel antidepressant ketamine enhances dentate gyrus proliferation with no effects on synaptic plasticity or hippocampal function in depressive-like rats. Acta Physiol (Oxf) 225:e13211. [DOI] [PubMed] [Google Scholar]
- Michishita M, Ikeda T, Nakashiba T, Ogawa M, Tashiro K, Honjo T, Doi K, Itohara S, Endo S (2003) A novel gene, Btcl1, encoding CUB and LDLa domains is expressed in restricted areas of mouse brain. Biochem Biophys Res Commun 306:680–686. [DOI] [PubMed] [Google Scholar]
- Michishita M, Ikeda T, Nakashiba T, Ogawa M, Tashiro K, Honjo T, Doi K, Itohara S, Endo S (2004) Expression of Btcl2, a novel member of Btcl gene family, during development of the central nervous system. Brain Res Dev Brain Res 153:135–142. [DOI] [PubMed] [Google Scholar]
- Micu IJiang QCoderre ERidsdale AZhang LWoulfe JYin XTrapp BDMcRory JERehak R, et al. (2006) NMDA receptors mediate calcium accumulation in myelin during chemical ischaemia. Nature 439:988–992. [DOI] [PubMed] [Google Scholar]
- Midgett CR, Madden DR (2008) The quaternary structure of a calcium-permeable AMPA receptor: conservation of shape and symmetry across functionally distinct subunit assemblies. J Mol Biol 382:578–584. [DOI] [PubMed] [Google Scholar]
- Mielnik CABinko MAChen YFunk AJJohansson EMIntson KSivananthan NIslam RMilenkovic MHorsfall W, et al. (2020) Consequences of NMDA receptor deficiency can be rescued in the adult brain. Mol Psychiatry 26:2929–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierau SB, Patrizi A, Hensch TK, Fagiolini M (2016) Cell-specific regulation of N-methyl-D-aspartate receptor maturation by Mecp2 in cortical circuits. Biol Psychiatry 79:746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguez-Cabello F, Sánchez-Fernández N, Yefimenko N, Gasull X, Gratacòs-Batlle E, Soto D (2020) AMPAR/TARP stoichiometry differentially modulates channel properties. eLife 9:e53946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikasova L, De Rossi P, Bouchet D, Georges F, Rogemond V, Didelot A, Meissirel C, Honnorat J, Groc L (2012) Disrupted surface cross-talk between NMDA and Ephrin-B2 receptors in anti-NMDA encephalitis. Brain 135:1606–1621. [DOI] [PubMed] [Google Scholar]
- Miller A, Pratt H, Schiffer RB (2011) Pseudobulbar affect: the spectrum of clinical presentations, etiologies and treatments. Expert Rev Neurother 11:1077–1088. [DOI] [PubMed] [Google Scholar]
- Miller B, Sarantis M, Traynelis SF, Attwell D (1992) Potentiation of NMDA receptor currents by arachidonic acid. Nature 355:722–725. [DOI] [PubMed] [Google Scholar]
- Milojkovic BA, Radojicic MS, Goldman-Rakic PS, Antic SD (2004) Burst generation in rat pyramidal neurones by regenerative potentials elicited in a restricted part of the basilar dendritic tree. J Physiol 558:193–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein AD, Nicoll RA (2009) TARP modulation of synaptic AMPA receptor trafficking and gating depends on multiple intracellular domains. Proc Natl Acad Sci USA 106:11348–11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein AD, Zhou W, Karimzadegan S, Bredt DS, Nicoll RA (2007) TARP subtypes differentially and dose-dependently control synaptic AMPA receptor gating. Neuron 55:905–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstone AM, Meyers K, Elia J (2005) Treatment of acute neuropsychiatric lupus with intravenous immunoglobulin (IVIG): a case report and review of the literature. Clin Rheumatol 24:394–397. [DOI] [PubMed] [Google Scholar]
- Misra C, Brickley SG, Wyllie DJ, Cull-Candy SG (2000) Slow deactivation kinetics of NMDA receptors containing NR1 and NR2D subunits in rat cerebellar Purkinje cells. J Physiol 525:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell NA, Fleck MW (2007) Targeting AMPA receptor gating processes with allosteric modulators and mutations. Biophys J 92:2392–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M, Imoto K (2006) Different composition of glutamate receptors in corticothalamic and lemniscal synaptic responses and their roles in the firing responses of ventrobasal thalamic neurons in juvenile mice. J Physiol 575:161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M, Imoto K (2009) Contrary roles of kainate receptors in transmitter release at corticothalamic synapses onto thalamic relay and reticular neurons. J Physiol 587:999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Yamasaki M, Takeuchi T, Sakimura K, Mishina M, Watanabe M (2010) Ablation of glutamate receptor GluRδ2 in adult Purkinje cells causes multiple innervation of climbing fibers by inducing aberrant invasion to parallel fiber innervation territory. J Neurosci 30:15196–15209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi YYoshioka YSuzuki KMiyazaki TKoura MSaigoh KKajimura NMonobe YKusunoki SMatsuda J, et al. (2014) A new mouse allele of glutamate receptor delta 2 with cerebellar atrophy and progressive ataxia. PLoS One 9:e107867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Takagi N, Kurokawa K, Kawai T, Besshoh S, Tanonaka K, Takeo S (2007) Effect of NMDA receptor antagonist on proliferation of neurospheres from embryonic brain. Neurosci Lett 417:143–148. [DOI] [PubMed] [Google Scholar]
- Moldavski A, Behr J, Bading H, Bengtson CP (2020) A novel method using ambient glutamate for the electrophysiological quantification of extrasynaptic NMDA receptor function in acute brain slices. J Physiol 598:633–650. [DOI] [PubMed] [Google Scholar]
- Møller EH, Egebjerg J, Brehm L, Stensbøl TB, Johansen TN, Madsen U, Krogsgaard-Larsen P (1999) Resolution, absolute stereochemistry, and enantiopharmacology of the GluR1-4 and GluR5 antagonist 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid. Chirality 11:752–759. [DOI] [PubMed] [Google Scholar]
- Møllerud S, Frydenvang K, Pickering DS, Kastrup JS (2017a) Lessons from crystal structures of kainate receptors. Neuropharmacology 112 (Pt A):16–28. [DOI] [PubMed] [Google Scholar]
- Møllerud SHansen RBPallesen JTemperini PPasini DBornholt JNielsen BMamedova EChalupnik PPaternain AV, et al. (2019) N-(7-(1H-Imidazol-1-yl)-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl)benzamide, a new kainate receptor selective antagonist and analgesic: Synthesis, X-ray crystallography, structure-affinity relationships, and in vitro and in vivo pharmacology. ACS Chem Neurosci 10:4685–4695. [DOI] [PubMed] [Google Scholar]
- Møllerud SPinto AMarconi LFrydenvang KThorsen TSLaulumaa SVenskutonytė RWinther SMoral AMCTamborini L, et al. (2017b) Structure and affinity of two bicyclic glutamate analogues at AMPA and kainate receptors. ACS Chem Neurosci 8:2056–2064. [DOI] [PubMed] [Google Scholar]
- Molnár J, Szakács G, Tusnády GE (2016) Characterization of disease-associated mutations in human transmembrane proteins. PLoS One 11:e0151760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momiyama A (2000) Distinct synaptic and extrasynaptic NMDA receptors identified in dorsal horn neurones of the adult rat spinal cord. J Physiol 523:621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan DT, Cotman CW (1982) The distribution of [3H]kainic acid binding sites in rat CNS as determined by autoradiography. Brain Res 252:91–100. [DOI] [PubMed] [Google Scholar]
- Monaghan DT and Jane DE (2009) Pharmacology of NMDA Receptors, in Biology of the NMDA Receptor (Van Dongen AM, ed) pp 257–282, CRC Press/Taylor & Francis, Boca Raton, FL. [PubMed] [Google Scholar]
- Monnerie H, Le Roux PD (2006) Glutamate receptor agonist kainate enhances primary dendrite number and length from immature mouse cortical neurons in vitro. J Neurosci Res 83:944–956. [DOI] [PubMed] [Google Scholar]
- Mony L, Zhu S, Carvalho S, Paoletti P (2011) Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. EMBO J 30:3134–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH (1994) Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 12:529–540. [DOI] [PubMed] [Google Scholar]
- Monyer H, Seeburg PH, Wisden W (1991) Glutamate-operated channels: developmentally early and mature forms arise by alternative splicing. Neuron 6:799–810. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH (1992) Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 256:1217–1221. [DOI] [PubMed] [Google Scholar]
- More JCNistico RDolman NPClarke VRAlt AJOgden AMBuelens FPTroop HMKelland EEPilato F, et al. (2004) Characterisation of UBP296: a novel, potent and selective kainate receptor antagonist. Neuropharmacology 47:46–64. [DOI] [PubMed] [Google Scholar]
- Mori HManabe TWatanabe MSatoh YSuzuki NToki SNakamura KYagi TKushiya ETakahashi T, et al. (1998) Role of the carboxy-terminal region of the GluR epsilon2 subunit in synaptic localization of the NMDA receptor channel. Neuron 21:571–580. [DOI] [PubMed] [Google Scholar]
- Morimoto-Tomita M, Zhang W, Straub C, Cho CH, Kim KS, Howe JR, Tomita S (2009) Autoinactivation of neuronal AMPA receptors via glutamate-regulated TARP interaction. Neuron 61:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita W, Lu W, Smith GB, Nicoll RA, Bear MF, Malenka RC (2007) Activation of NR2B-containing NMDA receptors is not required for NMDA receptor-dependent long-term depression. Neuropharmacology 52:71–76. [DOI] [PubMed] [Google Scholar]
- Moriyoshi K, Masu M, Ishii T, Shigemoto R, Mizuno N, Nakanishi S (1991) Molecular cloning and characterization of the rat NMDA receptor. Nature 354:31–37. [DOI] [PubMed] [Google Scholar]
- Morley RM, Tse HW, Feng B, Miller JC, Monaghan DT, Jane DE (2005) Synthesis and pharmacology of N1-substituted piperazine-2,3-dicarboxylic acid derivatives acting as NMDA receptor antagonists. J Med Chem 48:2627–2637. [DOI] [PubMed] [Google Scholar]
- Morris G, Walder K, Carvalho AF, Tye SJ, Lucas K, Berk M, Maes M (2018a) The role of hypernitrosylation in the pathogenesis and pathophysiology of neuroprogressive diseases. Neurosci Biobehav Rev 84:453–469. [DOI] [PubMed] [Google Scholar]
- Morris PG, Mishina M, Jones S (2018b) Altered synaptic and extrasynaptic NMDA receptor properties in substantia nigra dopaminergic neurons from mice lacking the GluN2D subunit. Front Cell Neurosci 12:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbacher J, Schoepfer R, Monyer H, Burnashev N, Seeburg PH, Ruppersberg JP (1994) A molecular determinant for submillisecond desensitization in glutamate receptors. Science 266:1059–1062. [DOI] [PubMed] [Google Scholar]
- Moscato EH, Peng X, Jain A, Parsons TD, Dalmau J, Balice-Gordon RJ (2014) Acute mechanisms underlying antibody effects in anti-N-methyl-D-aspartate receptor encephalitis. Ann Neurol 76:108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshé SL, Perucca E, Ryvlin P, Tomson T (2015) Epilepsy: new advances. Lancet 385:884–898. [DOI] [PubMed] [Google Scholar]
- Moskal JR, Burgdorf JS, Stanton PK, Kroes RA, Disterhoft JF, Burch RM, Khan MA (2017) The development of rapastinel (formerly GLYX-13); A rapid acting and long lasting antidepressant. Curr Neuropharmacol 15:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley CA, Acker TM, Hansen KB, Mullasseril P, Andersen KT, Le P, Vellano KM, Bräuner-Osborne H, Liotta DC, Traynelis SF (2010) Quinazolin-4-one derivatives: A novel class of noncompetitive NR2C/D subunit-selective N-methyl-D-aspartate receptor antagonists. J Med Chem 53:5476–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley CAMyers SJMurray EESantangelo RTahirovic YAKurtkaya NMullasseril PYuan HLyuboslavsky PLe P, et al. (2009) Synthesis, structural activity-relationships, and biological evaluation of novel amide-based allosteric binding site antagonists in NR1A/NR2B N-methyl-D-aspartate receptors. Bioorg Med Chem 17:6463–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota Vieira M, Nguyen TA, Wu K, Badger JD 2nd, Collins BM, Anggono V, Lu W, Roche KW (2020) An epilepsy-associated GRIN2A rare variant disrupts CaMKIIα phosphorylation of GluN2A and NMDA receptor trafficking. Cell Rep 32:108104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motazacker MMRost BRHucho TGarshasbi MKahrizi KUllmann RAbedini SSNieh SEAmini SHGoswami C, et al. (2007) A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am J Hum Genet 81:792–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothet JP, Le Bail M, Billard JM (2015) Time and space profiling of NMDA receptor co-agonist functions. J Neurochem 135:210–225. [DOI] [PubMed] [Google Scholar]
- Motohashi J, Kakegawa W, Yuzaki M (2007) Ho15J: a new hotfoot allele in a hot spot in the gene encoding the delta2 glutamate receptor. Brain Res 1140:153–160. [DOI] [PubMed] [Google Scholar]
- Mott DD, Benveniste M, Dingledine RJ (2008) pH-dependent inhibition of kainate receptors by zinc. J Neurosci 28:1659–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott DD, Doherty JJ, Zhang S, Washburn MS, Fendley MJ, Lyuboslavsky P, Traynelis SF, Dingledine R (1998) Phenylethanolamines inhibit NMDA receptors by enhancing proton inhibition. Nat Neurosci 1:659–667. [DOI] [PubMed] [Google Scholar]
- Mott DD, Rojas A, Fisher JL, Dingledine RJ, Benveniste M (2010) Subunit-specific desensitization of heteromeric kainate receptors. J Physiol 588:683–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott DD, Washburn MS, Zhang S, Dingledine RJ (2003) Subunit-dependent modulation of kainate receptors by extracellular protons and polyamines. J Neurosci 23:1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möykkynen T, Coleman SK, Semenov A, Keinänen K (2014) The N-terminal domain modulates α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor desensitization. J Biol Chem 289:13197–13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller HT, Meador-Woodruff JH (2005) Distribution of the NMDA receptor NR3A subunit in the adult pig-tail macaque brain. J Chem Neuroanat 29:157–172. [DOI] [PubMed] [Google Scholar]
- Muglia PTozzi FGalwey NWFrancks CUpmanyu RKong XQAntoniades ADomenici EPerry JRothen S, et al. (2010) Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry 15:589–601. [DOI] [PubMed] [Google Scholar]
- Muir KW, Lees KR (1995) Clinical experience with excitatory amino acid antagonist drugs. Stroke 26:503–513. [DOI] [PubMed] [Google Scholar]
- Mulkey RM, Endo S, Shenolikar S, Malenka RC (1994) Involvement of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature 369:486–488. [DOI] [PubMed] [Google Scholar]
- Mullasseril PHansen KBVance KMOgden KKYuan HKurtkaya NLSantangelo ROrr AGLe PVellano KM, et al. (2010) A subunit-selective potentiator of NR2C- and NR2D-containing NMDA receptors. Nat Commun 1:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle CSailer APérez-Otaño IDickinson-Anson HCastillo PEBureau IMaron CGage FHMann JRBettler B, et al. (1998) Altered synaptic physiology and reduced susceptibility to kainate-induced seizures in GluR6-deficient mice. Nature 392:601–605. [DOI] [PubMed] [Google Scholar]
- Mulle C, Sailer A, Swanson GT, Brana C, O’Gorman S, Bettler B, Heinemann SF (2000) Subunit composition of kainate receptors in hippocampal interneurons. Neuron 28:475–484. [DOI] [PubMed] [Google Scholar]
- Mullier B, Wolff C, Sands ZA, Ghisdal P, Muglia P, Kaminski RM, André VM (2017) GRIN2B gain of function mutations are sensitive to radiprodil, a negative allosteric modulator of GluN2B-containing NMDA receptors. Neuropharmacology 123:322–331. [DOI] [PubMed] [Google Scholar]
- Murillo A, Navarro AI, Puelles E, Zhang Y, Petros TJ, Pérez-Otaño I (2021) Temporal dynamics and neuronal specificity of Grin3a expression in the mouse forebrain. Cereb Cortex 31:1914–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JAStein ISLau CGPeixoto RTAman TKKaneko NAromolaran KSaulnier JLPopescu GKSabatini BL, et al. (2014) Phosphorylation of Ser1166 on GluN2B by PKA is critical to synaptic NMDA receptor function and Ca2+ signaling in spines. J Neurosci 34:869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrough JW, Perez AM, Pillemer S, Stern J, Parides MK, aan het Rot M, Collins KA, Mathew SJ, Charney DS, Iosifescu DV (2013) Rapid and longer-term antidepressant effects of repeated ketamine infusions in treatment-resistant major depression. Biol Psychiatry 74:250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrough JWSoleimani LDeWilde KECollins KALapidus KAIacoviello BMLener MKautz MKim JStern JB, et al. (2015) Ketamine for rapid reduction of suicidal ideation: a randomized controlled trial. Psychol Med 45:3571–3580. [DOI] [PubMed] [Google Scholar]
- Murthy VN, Sejnowski TJ, Stevens CF (1997) Heterogeneous release properties of visualized individual hippocampal synapses. Neuron 18:599–612. [DOI] [PubMed] [Google Scholar]
- Musgaard M, Biggin PC (2016) Steered molecular dynamics simulations predict conformational stability of glutamate receptors. J Chem Inf Model 56:1787–1797. [DOI] [PubMed] [Google Scholar]
- Musleh W, Bi X, Tocco G, Yaghoubi S, Baudry M (1997) Glycine-induced long-term potentiation is associated with structural and functional modifications of alpha-amino-3-hydroxyl-5-methyl-4-isoxazolepropionic acid receptors. Proc Natl Acad Sci USA 94:9451–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutch WA, Hansen AJ (1984) Extracellular pH changes during spreading depression and cerebral ischemia: mechanisms of brain pH regulation. J Cereb Blood Flow Metab 4:17–27. [DOI] [PubMed] [Google Scholar]
- Myers KM, Carlezon WA Jr (2012) D-cycloserine effects on extinction of conditioned responses to drug-related cues. Biol Psychiatry 71:947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RACasals FGauthier JHamdan FFKeebler JBoyko ARBustamante CDPiton AMSpiegelman DHenrion E, et al. (2011) A population genetic approach to mapping neurological disorder genes using deep resequencing. PLoS Genet 7:e1001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SJRuppa KPWilson LJTahirovic YALyuboslavsky PMenaldino DSDentmon ZWKoszalka GWZaczek RDingledine RJ, et al. (2021) A GluN2B-selective inhibitor of NMDA receptor function with enhanced potency at acidic pH and oral bioavailability for clinical use. J Pharmacol Exp Ther 379:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SJ, Yuan H, Kang JQ, Tan FCK, Traynelis SF, Low CM (2019) Distinct roles of GRIN2A and GRIN2B variants in neurological conditions. F1000 Res 8:1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi S, Fox R, Proulx CD, Lin JY, Tsien RY, Malinow R (2014) Engineering a memory with LTD and LTP. Nature 511:348–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi S, Kessels HW, Alfonso S, Aow J, Fox R, Malinow R (2013) Metabotropic NMDA receptor function is required for NMDA receptor-dependent long-term depression. Proc Natl Acad Sci USA 110:4027–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacher J, Alonso-Llosa G, Rosell DR, McEwen BS (2003) NMDA receptor antagonist treatment increases the production of new neurons in the aged rat hippocampus. Neurobiol Aging 24:273–284. [DOI] [PubMed] [Google Scholar]
- Nacher J, Rosell DR, Alonso-Llosa G, McEwen BS (2001) NMDA receptor antagonist treatment induces a long-lasting increase in the number of proliferating cells, PSA-NCAM-immunoreactive granule neurons and radial glia in the adult rat dentate gyrus. Eur J Neurosci 13:512–520. [DOI] [PubMed] [Google Scholar]
- Nagy D, Stoiljkovic M, Menniti FS, Hajós M (2016) Differential effects of an NR2B NAM and ketamine on synaptic potentiation and gamma synchrony: Relevance to rapid-onset antidepressant efficacy. Neuropsychopharmacology 41:1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahum-Levy R, Lipinski D, Shavit S, Benveniste M (2001) Desensitization of NMDA receptor channels is modulated by glutamate agonists. Biophys J 80:2152–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D, Hosy E, Petersen JD, Constals A, Giannone G, Choquet D, Sibarita JB (2013) Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J Neurosci 33:13204–13224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T (2019) Structures of the AMPA receptor in complex with its auxiliary subunit cornichon. Science 366:1259–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T (2005) Structure and different conformational states of native AMPA receptor complexes. Nature 433:545–549. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Sheng M, Walz T (2006) Three-dimensional structure of an AMPA receptor without associated stargazin/TARP proteins. Biol Chem 387:179–187. [DOI] [PubMed] [Google Scholar]
- Nakamoto C, Kawamura M, Nakatsukasa E, Natsume R, Takao K, Watanabe M, Abe M, Takeuchi T, Sakimura K (2020a) GluD1 knockout mice with a pure C57BL/6N background show impaired fear memory, social interaction, and enhanced depressive-like behavior. PLoS One 15:e0229288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto CKonno KMiyazaki TNakatsukasa ENatsume RAbe MKawamura MFukazawa YShigemoto RYamasaki M, et al. (2020b) Expression mapping, quantification, and complex formation of GluD1 and GluD2 glutamate receptors in adult mouse brain. J Comp Neurol 528:1003–1027. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Goshima Y (2002) Structural and functional relation of neuropilins. Adv Exp Med Biol 515:55–69. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Choi KH, Choi SK, Do CS, Jun JH, Kwon HK, Lee SM, Moon RJ, Yi KJ, Jang IS (2010) Presynaptic kainate receptors increase GABAergic neurotransmission in rat periaqueductal gray neurons. Eur J Pharmacol 635:72–78. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Irie K, Tanaka H, Nishikawa K, Suzuki H, Saitoh Y, Tamura A, Tsukita S, Fujiyoshi Y (2019) Morphologic determinant of tight junctions revealed by claudin-3 structures. Nat Commun 10:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Lipton SA (2016) Protein S-nitrosylation as a therapeutic target for neurodegenerative diseases. Trends Pharmacol Sci 37:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Axel R, Shneider NA (1992) Alternative splicing generates functionally distinct N-methyl-D-aspartate receptors. Proc Natl Acad Sci USA 89:8552–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa KQuirk MCChitwood RAWatanabe MYeckel MFSun LDKato ACarr CAJohnston DWilson MA, et al. (2002) Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science 297:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa TKomai SWatabe AMKiyama YFukaya MArima-Yoshida FHorai RSudo KEbine KDelawary M, et al. (2006) NR2B tyrosine phosphorylation modulates fear learning as well as amygdaloid synaptic plasticity. EMBO J 25:2867–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T, Maekawa M, Yuasa S, Kohsaka S, Uchino S (2009) The Alzheimer’s disease drug memantine increases the number of radial glia-like progenitor cells in adult hippocampus. Glia 57:1082–1090. [DOI] [PubMed] [Google Scholar]
- Narangoda C, Sakipov SN, Kurnikova MG (2019) AMPA receptor noncompetitive inhibitors occupy a promiscuous binding site. ACS Chem Neurosci 10:4511–4521. [DOI] [PubMed] [Google Scholar]
- Nasu-Nishimura Y, Hurtado D, Braud S, Tang TT, Isaac JT, Roche KW (2006) Identification of an endoplasmic reticulum-retention motif in an intracellular loop of the kainate receptor subunit KA2. J Neurosci 26:7014–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur PHansen KBKristensen ASDravid SMPickering DSOlsen LVestergaard BEgebjerg JGajhede MTraynelis SF, et al. (2007) Ionotropic glutamate-like receptor delta2 binds D-serine and glycine. Proc Natl Acad Sci USA 104:14116–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P, Vestergaard B, Skov LK, Egebjerg J, Gajhede M, Kastrup JS (2005) Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate. FEBS Lett 579:1154–1160. [DOI] [PubMed] [Google Scholar]
- Nayeem N, Mayans O, Green T (2011) Conformational flexibility of the ligand-binding domain dimer in kainate receptor gating and desensitization. J Neurosci 31:2916–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayeem N, Zhang Y, Schweppe DK, Madden DR, Green T (2009) A nondesensitizing kainate receptor point mutant. Mol Pharmacol 76:534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard M, Kraig RP, Tanabe J, Pulsinelli WA (1991) Dynamics of interstitial and intracellular pH in evolving brain infarct. Am J Physiol 260:R581–R588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete-Díaz JV, Duque-Feria P, Andrade-Talavera Y, Carrión M, Flores G, Rodríguez-Moreno A (2012) Kainate receptor-mediated depression of glutamatergic transmission involving protein kinase A in the lateral amygdala. J Neurochem 121:36–43. [DOI] [PubMed] [Google Scholar]
- Negrete-Díaz JV, Sihra TS, Delgado-García JM, Rodríguez-Moreno A (2006) Kainate receptor-mediated inhibition of glutamate release involves protein kinase A in the mouse hippocampus. J Neurophysiol 96:1829–1837. [DOI] [PubMed] [Google Scholar]
- Negrete-Díaz JV, Sihra TS, Flores G, Rodríguez-Moreno A (2018) Non-canonical mechanisms of presynaptic kainate receptors controlling glutamate release. Front Mol Neurosci 11:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JK, Frølund SU, Tikhonov DB, Kristensen AS, Strømgaard K (2009) Synthesis and biological activity of argiotoxin 636 and analogues: selective antagonists for ionotropic glutamate receptors. Angew Chem Int Ed Engl 48:3087–3091. [DOI] [PubMed] [Google Scholar]
- Nestor JArinuma YHuerta TSKowal CNasiri EKello NFujieda YBialas AHammond TSriram U, et al. (2018) Lupus antibodies induce behavioral changes mediated by microglia and blocked by ACE inhibitors. J Exp Med 215:2554–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DPitcher GMSzilard RKSertié AKanisek MClapcote SJLipina TKalia LVJoo DMcKerlie C, et al. (2009) Neto1 is a novel CUB-domain NMDA receptor-interacting protein required for synaptic plasticity and learning. PLoS Biol 7:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo-Anh TJ, Bloodgood BL, Lin M, Sabatini BL, Maylie J, Adelman JP (2005) SK channels and NMDA receptors form a Ca2+-mediated feedback loop in dendritic spines. Nat Neurosci 8:642–649. [DOI] [PubMed] [Google Scholar]
- Nguyen KTClaiborne CFMcCauley JALibby BEClaremon DABednar RAMosser SDGaul SLConnolly TMCondra CL, et al. (2007) Cyclic benzamidines as orally efficacious NR2B-selective NMDA receptor antagonists. Bioorg Med Chem Lett 17:3997–4000. [DOI] [PubMed] [Google Scholar]
- Nichols CG, Lee SJ (2018) Polyamines and potassium channels: A 25-year romance. J Biol Chem 293:18779–18788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson KL, Mansbach RS, Menniti FS, Balster RL (2007) The phencyclidine-like discriminative stimulus effects and reinforcing properties of the NR2B-selective N-methyl-D-aspartate antagonist CP-101 606 in rats and rhesus monkeys. Behav Pharmacol 18:731–743. [DOI] [PubMed] [Google Scholar]
- Niciu MJ, Henter ID, Luckenbaugh DA, Zarate CA Jr, Charney DS (2014) Glutamate receptor antagonists as fast-acting therapeutic alternatives for the treatment of depression: ketamine and other compounds. Annu Rev Pharmacol Toxicol 54:119–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll RA (2017) A brief history of long-term potentiation. Neuron 93:281–290. [DOI] [PubMed] [Google Scholar]
- Nicoll RA, Schmitz D (2005) Synaptic plasticity at hippocampal mossy fibre synapses. Nat Rev Neurosci 6:863–876. [DOI] [PubMed] [Google Scholar]
- Nie H, Weng HR (2010) Impaired glial glutamate uptake induces extrasynaptic glutamate spillover in the spinal sensory synapses of neuropathic rats. J Neurophysiol 103:2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen EO, Varming T, Mathiesen C, Jensen LH, Moller A, Gouliaev AH, Wätjen F, Drejer J (1999) SPD 502: a water-soluble and in vivo long-lasting AMPA antagonist with neuroprotective activity. J Pharmacol Exp Ther 289:1492–1501. [PubMed] [Google Scholar]
- Nielsen MM, Liljefors T, Krogsgaard-Larsen P, Egebjerg J (2003) The selective activation of the glutamate receptor GluR5 by ATPA is controlled by serine 741. Mol Pharmacol 63:19–25. [DOI] [PubMed] [Google Scholar]
- Niemann SKanki HFukui YTakao KFukaya MHynynen MNChurchill MJShefner JMBronson RTBrown RH Jr, et al. (2007) Genetic ablation of NMDA receptor subunit NR3B in mouse reveals motoneuronal and nonmotoneuronal phenotypes. Eur J Neurosci 26:1407–1420. [DOI] [PubMed] [Google Scholar]
- Niemann S, Landers JE, Churchill MJ, Hosler B, Sapp P, Speed WC, Lahn BT, Kidd KK, Brown RH Jr, Hayashi Y (2008) Motoneuron-specific NR3B gene: no association with ALS and evidence for a common null allele. Neurology 70:666–676. [DOI] [PubMed] [Google Scholar]
- Niesters M, Martini C, Dahan A (2014) Ketamine for chronic pain: risks and benefits. Br J Clin Pharmacol 77:357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen A, England PM (2007) A subtype-selective, use-dependent inhibitor of native AMPA receptors. J Am Chem Soc 129:4902–4903. [DOI] [PubMed] [Google Scholar]
- Nishi M, Hinds H, Lu HP, Kawata M, Hayashi Y (2001) Motoneuron-specific expression of NR3B, a novel NMDA-type glutamate receptor subunit that works in a dominant-negative manner. J Neurosci 21:RC185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisticò RFlorenzano FMango DFerraina CGrilli MDi Prisco SNobili ASaccucci SD’Amelio MMorbin M, et al. (2015) Presynaptic c-Jun N-terminal Kinase 2 regulates NMDA receptor-dependent glutamate release. Sci Rep 5:9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M, Nakanishi H, Nabekura J, Akaike N (2000) AMPA-kainate subtypes of glutamate receptor in rat cerebral microglia. J Neurosci 20:251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noebels JL, Qiao X, Bronson RT, Spencer C, Davisson MT (1990) Stargazer: a new neurological mutant on chromosome 15 in the mouse with prolonged cortical seizures. Epilepsy Res 7:129–135. [DOI] [PubMed] [Google Scholar]
- Nofziger JL, Paxos C, Emshoff J, Mullen C (2019) Evaluation of dextromethorphan with select antidepressant therapy for the treatment of depression in the acute care psychiatric setting. Ment Health Clin 9:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord ASRoeb WDickel DEWalsh TKusenda MO’Connor KLMalhotra DMcCarthy SEStray SMTaylor SM, et al. ; STAART Psychopharmacology Network (2011) Reduced transcript expression of genes affected by inherited and de novo CNVs in autism. Eur J Hum Genet 19:727–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nørholm ABFrancotte POlsen LKrintel CFrydenvang KGoffin EChallal SDanober LBotez-Pop ILestage P, et al. (2013) Synthesis, pharmacological and structural characterization, and thermodynamic aspects of GluA2-positive allosteric modulators with a 3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide scaffold. J Med Chem 56:8736–8745. [DOI] [PubMed] [Google Scholar]
- Nouhi M, Zhang X, Yao N, Chergui K (2018) CIQ, a positive allosteric modulator of GluN2C/D-containing N-methyl-d-aspartate receptors, rescues striatal synaptic plasticity deficit in a mouse model of Parkinson’s disease. CNS Neurosci Ther 24:144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A (1984) Magnesium gates glutamate-activated channels in mouse central neurones. Nature 307:462–465. [DOI] [PubMed] [Google Scholar]
- Numano R, Szobota S, Lau AY, Gorostiza P, Volgraf M, Roux B, Trauner D, Isacoff EY (2009) Nanosculpting reversed wavelength sensitivity into a photoswitchable iGluR. Proc Natl Acad Sci USA 106:6814–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt JG, Gunzler SA, Kirchhoff T, Hogarth P, Weaver JL, Krams M, Jamerson B, Menniti FS, Landen JW (2008) Effects of a NR2B selective NMDA glutamate antagonist, CP-101,606, on dyskinesia and Parkinsonism. Mov Disord 23:1860–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Kamboj RK (1994) Differential RNA editing efficiency of AMPA receptor subunit GluR-2 in human brain. Neuroreport 5:1679–1683. [DOI] [PubMed] [Google Scholar]
- O’Brien R, Xu D, Mi R, Tang X, Hopf C, Worley P (2002) Synaptically targeted narp plays an essential role in the aggregation of AMPA receptors at excitatory synapses in cultured spinal neurons. J Neurosci 22:4487–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien RJ, Xu D, Petralia RS, Steward O, Huganir RL, Worley P (1999) Synaptic clustering of AMPA receptors by the extracellular immediate-early gene product Narp. Neuron 23:309–323. [DOI] [PubMed] [Google Scholar]
- O’Neill MJ, Bleakman D, Zimmerman DM, Nisenbaum ES (2004) AMPA receptor potentiators for the treatment of CNS disorders. Curr Drug Targets CNS Neurol Disord 3:181–194. [DOI] [PubMed] [Google Scholar]
- O’Neill MJ, Bond A, Ornstein PL, Ward MA, Hicks CA, Hoo K, Bleakman D, Lodge D (1998) Decahydroisoquinolines: novel competitive AMPA/kainate antagonists with neuroprotective effects in global cerebral ischaemia. Neuropharmacology 37:1211–1222. [DOI] [PubMed] [Google Scholar]
- Ogden KKChen WSwanger SAMcDaniel MJFan LZHu CTankovic AKusumoto HKosobucki GJSchulien AJ, et al. (2017) Molecular mechanism of disease-associated mutations in the pre-M1 helix of NMDA receptors and potential rescue pharmacology. PLoS Genet 13:e1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden KK, Khatri A, Traynelis SF, Heldt SA (2014) Potentiation of GluN2C/D NMDA receptor subtypes in the amygdala facilitates the retention of fear and extinction learning in mice. Neuropsychopharmacology 39:625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden KK, Traynelis SF (2013) Contribution of the M1 transmembrane helix and pre-M1 region to positive allosteric modulation and gating of N-methyl-D-aspartate receptors. Mol Pharmacol 83:1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Han KS, Park H, Woo DH, Kim HY, Traynelis SF, Lee CJ (2012) Protease activated receptor 1-induced glutamate release in cultured astrocytes is mediated by Bestrophin-1 channel but not by vesicular exocytosis. Mol Brain 5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba CShiina MTohyama JHaginoya KLerman-Sagie TOkamoto NBlumkin LLev DMukaida SNozaki F, et al. (2015) GRIN1 mutations cause encephalopathy with infantile-onset epilepsy, and hyperkinetic and stereotyped movement disorders. Epilepsia 56:841–848. [DOI] [PubMed] [Google Scholar]
- Ohmori J, Sakamoto S, Kubota H, Shimizu-Sasamata M, Okada M, Kawasaki S, Hidaka K, Togami J, Furuya T, Murase K (1994) 6-(1H-imidazol-1-yl)-7-nitro-2,3(1H,4H)-quinoxalinedione hydrochloride (YM90K) and related compounds: structure-activity relationships for the AMPA-type non-NMDA receptor. J Med Chem 37:467–475. [DOI] [PubMed] [Google Scholar]
- Ohno T, Maeda H, Murabe N, Kamiyama T, Yoshioka N, Mishina M, Sakurai M (2010) Specific involvement of postsynaptic GluN2B-containing NMDA receptors in the developmental elimination of corticospinal synapses. Proc Natl Acad Sci USA 107:15252–15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou KD, Short SM, Rich MT, Antic SD (2012) Extrasynaptic glutamate receptor activation as cellular bases for dynamic range compression in pyramidal neurons. Front Physiol 3:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama N, Takagi N, Kawai T, Miyake-Takagi K, Takeo S (2004) Phosphorylation of extracellular-regulating kinase in NMDA receptor antagonist-induced newly generated neurons in the adult rat dentate gyrus. J Neurochem 88:717–725. [DOI] [PubMed] [Google Scholar]
- Oliet SH, Malenka RC, Nicoll RA (1997) Two distinct forms of long-term depression coexist in CA1 hippocampal pyramidal cells. Neuron 18:969–982. [DOI] [PubMed] [Google Scholar]
- Olloquequi J, Cornejo-Córdova E, Verdaguer E, Soriano FX, Binvignat O, Auladell C, Camins A (2018) Excitotoxicity in the pathogenesis of neurological and psychiatric disorders: Therapeutic implications. J Psychopharmacol 32:265–275. [DOI] [PubMed] [Google Scholar]
- Olney JW (1994) New mechanisms of excitatory transmitter neurotoxicity. J Neural Transm Suppl 43:47–51. [PubMed] [Google Scholar]
- Olney JW, Newcomer JW, Farber NB (1999) NMDA receptor hypofunction model of schizophrenia. J Psychiatr Res 33:523–533. [DOI] [PubMed] [Google Scholar]
- Omdal R, Brokstad K, Waterloo K, Koldingsnes W, Jonsson R, Mellgren SI (2005) Neuropsychiatric disturbances in SLE are associated with antibodies against NMDA receptors. Eur J Neurol 12:392–398. [DOI] [PubMed] [Google Scholar]
- Opazo P, Labrecque S, Tigaret CM, Frouin A, Wiseman PW, De Koninck P, Choquet D (2010) CaMKII triggers the diffusional trapping of surface AMPARs through phosphorylation of stargazin. Neuron 67:239–252. [DOI] [PubMed] [Google Scholar]
- Opazo P, Sainlos M, Choquet D (2012) Regulation of AMPA receptor surface diffusion by PSD-95 slots. Curr Opin Neurobiol 22:453–460. [DOI] [PubMed] [Google Scholar]
- Orain DTasdelen EHaessig SKoller MPicard ADubois CLingenhoehl KDesrayaud SFloersheim PCarcache D, et al. (2017) Design and synthesis of selurampanel, a novel orally active and competitive AMPA receptor antagonist. ChemMedChem 12:197–201. [DOI] [PubMed] [Google Scholar]
- Orav E, Atanasova T, Shintyapina A, Kesaf S, Kokko M, Partanen J, Taira T, Lauri SE (2017) NETO1 guides development of glutamatergic connectivity in the hippocampus by regulating axonal kainate receptors. eNeuro 4:ENEURO.0048-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orav E, Dowavic I, Huupponen J, Taira T, Lauri SE (2019) NETO1 regulates postsynaptic kainate receptors in CA3 interneurons during circuit maturation. Mol Neurobiol 56:7473–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi C, La Via L, Bonini D, Mora C, Russo I, Barbon A, Barlati S (2011) AMPA receptor regulation at the mRNA and protein level in rat primary cortical cultures. PLoS One 6:e25350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otis T, Zhang S, Trussell LO (1996) Direct measurement of AMPA receptor desensitization induced by glutamatergic synaptic transmission. J Neurosci 16:7496–7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otmakhov N, Khibnik L, Otmakhova N, Carpenter S, Riahi S, Asrican B, Lisman J (2004) Forskolin-induced LTP in the CA1 hippocampal region is NMDA receptor dependent. J Neurophysiol 91:1955–1962. [DOI] [PubMed] [Google Scholar]
- Otmakhova NA, Otmakhov N, Mortenson LH, Lisman JE (2000) Inhibition of the cAMP pathway decreases early long-term potentiation at CA1 hippocampal synapses. J Neurosci 20:4446–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu YDarcq EPietrajtis KMátyás FSchwartz EBessaih TAbi Gerges SRousseau CVGrand TDieudonné S, et al. (2019) Control of aversion by glycine-gated GluN1/GluN3A NMDA receptors in the adult medial habenula. Science 366:250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto MW, Kredlow MA, Smits JAJ, Hofmann SG, Tolin DF, de Kleine RA, van Minnen A, Evins AE, Pollack MH (2016) Enhancement of psychosocial treatment with D-cycloserine: Models, moderators, and future directions. Biol Psychiatry 80:274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto MW, Tolin DF, Simon NM, Pearlson GD, Basden S, Meunier SA, Hofmann SG, Eisenmenger K, Krystal JH, Pollack MH (2010) Efficacy of d-cycloserine for enhancing response to cognitive-behavior therapy for panic disorder. Biol Psychiatry 67:365–370. [DOI] [PubMed] [Google Scholar]
- Paas Y (1998) The macro- and microarchitectures of the ligand-binding domain of glutamate receptors. Trends Neurosci 21:117–125. [DOI] [PubMed] [Google Scholar]
- Pafundo DE, Miyamae T, Lewis DA, Gonzalez-Burgos G (2018) Presynaptic effects of N-methyl-D-aspartate receptors enhance parvalbumin cell-mediated inhibition of pyramidal cells in mouse prefrontal cortex. Biol Psychiatry 84:460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganelli MA, Kussius CL, Popescu GK (2013) Role of cross-cleft contacts in NMDA receptor gating. PLoS One 8:e80953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahk AJ, Williams K (1997) Influence of extracellular pH on inhibition by ifenprodil at N-methyl-D-aspartate receptors in Xenopus oocytes. Neurosci Lett 225:29–32. [DOI] [PubMed] [Google Scholar]
- Pahl S, Tapken D, Haering SC, Hollmann M (2014) Trafficking of kainate receptors. Membranes (Basel) 4:565–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios-Filardo J, Aller MI, Lerma J (2016) Synaptic targeting of kainate receptors. Cereb Cortex 26:1464–1472. [DOI] [PubMed] [Google Scholar]
- Pallesen JMøllerud SFrydenvang KPickering DSBornholdt JNielsen BPasini DHan LMarconi LKastrup JS, et al. (2019) N1-Substituted quinoxaline-2,3-diones as kainate receptor antagonists: X-ray crystallography, structure-affinity relationships, and in vitro pharmacology. ACS Chem Neurosci 10:1841–1853. [DOI] [PubMed] [Google Scholar]
- Palmer LM, Shai AS, Reeve JE, Anderson HL, Paulsen O, Larkum ME (2014) NMDA spikes enhance action potential generation during sensory input. Nat Neurosci 17:383–390. [DOI] [PubMed] [Google Scholar]
- Palygin O, Lalo U, Pankratov Y (2011) Distinct pharmacological and functional properties of NMDA receptors in mouse cortical astrocytes. Br J Pharmacol 163:1755–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan HOliveira BSaher GDere ETapken DMitjans MSeidel JWesolowski JWakhloo DKlein-Schmidt C, et al. (2019) Uncoupling the widespread occurrence of anti-NMDAR1 autoantibodies from neuropsychiatric disease in a novel autoimmune model. Mol Psychiatry 24:1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panatier A, Theodosis DT, Mothet JP, Touquet B, Pollegioni L, Poulain DA, Oliet SH (2006) Glia-derived D-serine controls NMDA receptor activity and synaptic memory. Cell 125:775–784. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Ascher P, Neyton J (1997) High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J Neurosci 17:5711–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q (2013) NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci 14:383–400. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Neyton J, Ascher P (1995) Glycine-independent and subunit-specific potentiation of NMDA responses by extracellular Mg2+. Neuron 15:1109–1120. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Perin-Dureau F, Fayyazuddin A, Le Goff A, Callebaut I, Neyton J (2000) Molecular organization of a zinc binding n-terminal modulatory domain in a NMDA receptor subunit. Neuron 28:911–925. [DOI] [PubMed] [Google Scholar]
- Papadakis M, Hawkins LM, Stephenson FA (2004) Appropriate NR1-NR1 disulfide-linked homodimer formation is requisite for efficient expression of functional, cell surface N-methyl-D-aspartate NR1/NR2 receptors. J Biol Chem 279:14703–14712. [DOI] [PubMed] [Google Scholar]
- Papouin T, Ladépêche L, Ruel J, Sacchi S, Labasque M, Hanini M, Groc L, Pollegioni L, Mothet JP, Oliet SH (2012) Synaptic and extrasynaptic NMDA receptors are gated by different endogenous coagonists. Cell 150:633–646. [DOI] [PubMed] [Google Scholar]
- Papouin T, Oliet SH (2014) Organization, control and function of extrasynaptic NMDA receptors. Philos Trans R Soc Lond B Biol Sci 369:20130601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HHan KSSeo JLee JDravid SMWoo JChun HCho SBae JYAn H, et al. (2015) Channel-mediated astrocytic glutamate modulates hippocampal synaptic plasticity by activating postsynaptic NMDA receptors. Mol Brain 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chávez AE, Mineur YS, Morimoto-Tomita M, Lutzu S, Kim KS, Picciotto MR, Castillo PE, Tomita S (2016a) CaMKII phosphorylation of TARPγ-8 is a mediator of LTP and learning and memory. Neuron 92:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P, Sanderson TM, Amici M, Choi SL, Bortolotto ZA, Zhuo M, Kaang BK, Collingridge GL (2016b) Calcium-permeable AMPA receptors mediate the induction of the protein kinase A-dependent component of long-term potentiation in the hippocampus. J Neurosci 36:622–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park-Chung M, Wu FS, Farb DH (1994) 3 alpha-Hydroxy-5 beta-pregnan-20-one sulfate: a negative modulator of the NMDA-induced current in cultured neurons. Mol Pharmacol 46:146–150. [PubMed] [Google Scholar]
- Parker MD, Boron WF (2013) The divergence, actions, roles, and relatives of sodium-coupled bicarbonate transporters. Physiol Rev 93:803–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MP, Raymond LA (2014) Extrasynaptic NMDA receptor involvement in central nervous system disorders. Neuron 82:279–293. [DOI] [PubMed] [Google Scholar]
- Partin KM (2015) AMPA receptor potentiators: from drug design to cognitive enhancement. Curr Opin Pharmacol 20:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partin KM, Bowie D, Mayer ML (1995) Structural determinants of allosteric regulation in alternatively spliced AMPA receptors. Neuron 14:833–843. [DOI] [PubMed] [Google Scholar]
- Partin KM, Fleck MW, Mayer ML (1996) AMPA receptor flip/flop mutants affecting deactivation, desensitization, and modulation by cyclothiazide, aniracetam, and thiocyanate. J Neurosci 16:6634–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partin KM, Mayer ML (1996) Negative allosteric modulation of wild-type and mutant AMPA receptors by GYKI 53655. Mol Pharmacol 49:142–148. [PubMed] [Google Scholar]
- Partin KM, Patneau DK, Mayer ML (1994) Cyclothiazide differentially modulates desensitization of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor splice variants. Mol Pharmacol 46:129–138. [PubMed] [Google Scholar]
- Partin KM, Patneau DK, Winters CA, Mayer ML, Buonanno A (1993) Selective modulation of desensitization at AMPA versus kainate receptors by cyclothiazide and concanavalin A. Neuron 11:1069–1082. [DOI] [PubMed] [Google Scholar]
- Partridge AW, Therien AG, Deber CM (2004) Missense mutations in transmembrane domains of proteins: phenotypic propensity of polar residues for human disease. Proteins 54:648–656. [DOI] [PubMed] [Google Scholar]
- Paschen W, Schmitt J, Gissel C, Dux E (1997) Developmental changes of RNA editing of glutamate receptor subunits GluR5 and GluR6: in vivo versus in vitro. Brain Res Dev Brain Res 98:271–280. [DOI] [PubMed] [Google Scholar]
- Passafaro M, Piëch V, Sheng M (2001) Subunit-specific temporal and spatial patterns of AMPA receptor exocytosis in hippocampal neurons. Nat Neurosci 4:917–926. [DOI] [PubMed] [Google Scholar]
- Pasterkamp RJ (2012) Getting neural circuits into shape with semaphorins. Nat Rev Neurosci 13:605–618. [DOI] [PubMed] [Google Scholar]
- Paternain AV, Cohen A, Stern-Bach Y, Lerma J (2003) A role for extracellular Na+ in the channel gating of native and recombinant kainate receptors. J Neurosci 23:8641–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternain AV, Herrera MT, Nieto MA, Lerma J (2000) GluR5 and GluR6 kainate receptor subunits coexist in hippocampal neurons and coassemble to form functional receptors. J Neurosci 20:196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternain AV, Morales M, Lerma J (1995) Selective antagonism of AMPA receptors unmasks kainate receptor-mediated responses in hippocampal neurons. Neuron 14:185–189. [DOI] [PubMed] [Google Scholar]
- Paternain AV, Rodríguez-Moreno A, Villarroel A, Lerma J (1998) Activation and desensitization properties of native and recombinant kainate receptors. Neuropharmacology 37:1249–1259. [DOI] [PubMed] [Google Scholar]
- Patneau DK, Mayer ML (1990) Structure-activity relationships for amino acid transmitter candidates acting at N-methyl-D-aspartate and quisqualate receptors. J Neurosci 10:2385–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patneau DK, Mayer ML (1991) Kinetic analysis of interactions between kainate and AMPA: evidence for activation of a single receptor in mouse hippocampal neurons. Neuron 6:785–798. [DOI] [PubMed] [Google Scholar]
- Patneau DK, Mayer ML, Jane DE, Watkins JC (1992) Activation and desensitization of AMPA/kainate receptors by novel derivatives of willardiine. J Neurosci 12:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patneau DK, Vyklicky L Jr, Mayer ML (1993) Hippocampal neurons exhibit cyclothiazide-sensitive rapidly desensitizing responses to kainate. J Neurosci 13:3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patneau DK, Wright PW, Winters C, Mayer ML, Gallo V (1994) Glial cells of the oligodendrocyte lineage express both kainate- and AMPA-preferring subtypes of glutamate receptor. Neuron 12:357–371. [DOI] [PubMed] [Google Scholar]
- Patterson MA, Szatmari EM, Yasuda R (2010) AMPA receptors are exocytosed in stimulated spines and adjacent dendrites in a Ras-ERK-dependent manner during long-term potentiation. Proc Natl Acad Sci USA 107:15951–15956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SMDoherty JJRobichaud AJBelfort GMChow BYHammond RSCrawford DCLinsenbardt AJShu HJIzumi Y, et al. (2013) The major brain cholesterol metabolite 24(S)-hydroxycholesterol is a potent allosteric modulator of N-methyl-D-aspartate receptors. J Neurosci 33:17290–17300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupard MC, Friedman LK, Zukin RS (1997) Developmental regulation and cell-specific expression of N-methyl-D-aspartate receptor splice variants in rat hippocampus. Neuroscience 79:399–409. [DOI] [PubMed] [Google Scholar]
- Paz JT, Bryant AS, Peng K, Fenno L, Yizhar O, Frankel WN, Deisseroth K, Huguenard JR (2011) A new mode of corticothalamic transmission revealed in the Gria4(-/-) model of absence epilepsy. Nat Neurosci 14:1167–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlstein E, Gouty-Colomer LA, Michel FJ, Cloarec R, Hammond C (2015) Glutamatergic synaptic currents of nigral dopaminergic neurons follow a postnatal developmental sequence. Front Cell Neurosci 9:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE (2016) Functions of polyamines in mammals. J Biol Chem 291:14904–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei W, Huang Z, Niu L (2007) GluR3 flip and flop: differences in channel opening kinetics. Biochemistry 46:2027–2036. [DOI] [PubMed] [Google Scholar]
- Pei W, Huang Z, Wang C, Han Y, Park JS, Niu L (2009) Flip and flop: a molecular determinant for AMPA receptor channel opening. Biochemistry 48:3767–3777. [DOI] [PubMed] [Google Scholar]
- Pelkey KABarksdale ECraig MTYuan XSukumaran MVargish GAMitchell RMWyeth MSPetralia RSChittajallu R, et al. (2015) Pentraxins coordinate excitatory synapse maturation and circuit integration of parvalbumin interneurons. Neuron 85:1257–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkey KA, Chittajallu R, Craig MT, Tricoire L, Wester JC, McBain CJ (2017) Hippocampal GABAergic inhibitory interneurons. Physiol Rev 97:1619–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini-Giampietro DE, Bennett MV, Zukin RS (1991) Differential expression of three glutamate receptor genes in developing rat brain: an in situ hybridization study. Proc Natl Acad Sci USA 88:4157–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Hughes EG, Moscato EH, Parsons TD, Dalmau J, Balice-Gordon RJ (2015) Cellular plasticity induced by anti-α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor encephalitis antibodies. Ann Neurol 77:381–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn AC, Balik A, Greger IH (2013) Steric antisense inhibition of AMPA receptor Q/R editing reveals tight coupling to intronic editing sites and splicing. Nucleic Acids Res 41:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn AC, Balik A, Wozny C, Cais O, Greger IH (2012) Activity-mediated AMPA receptor remodeling, driven by alternative splicing in the ligand-binding domain. Neuron 76:503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn AC, Williams SR, Greger IH (2008) Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum. EMBO J 27:3056–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn AC, Zhang CL, Georges F, Royer L, Breillat C, Hosy E, Petersen JD, Humeau Y, Choquet D (2017) Hippocampal LTP and contextual learning require surface diffusion of AMPA receptors. Nature 549:384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira VS, Joca SRL, Harvey BH, Elfving B, Wegener G (2019) Esketamine and rapastinel, but not imipramine, have antidepressant-like effect in a treatment-resistant animal model of depression. Acta Neuropsychiatr 31:258–265. [DOI] [PubMed] [Google Scholar]
- Pérez-Otaño I, Larsen RS, Wesseling JF (2016) Emerging roles of GluN3-containing NMDA receptors in the CNS. Nat Rev Neurosci 17:623–635. [DOI] [PubMed] [Google Scholar]
- Pérez-Otaño I, Luján R, Tavalin SJ, Plomann M, Modregger J, Liu XB, Jones EG, Heinemann SF, Lo DC, Ehlers MD (2006) Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat Neurosci 9:611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Otano I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, Heinemann SF (2001) Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J Neurosci 21:1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkel DJ, Hestrin S, Sah P, Nicoll RA (1990) Excitatory synaptic currents in Purkinje cells. Proc Biol Sci 241:116–121. [DOI] [PubMed] [Google Scholar]
- Perkins MN, Stone TW, Collins JF, Curry K (1981) Phosphonate analogues of carboxylic acids as aminoacid antagonists on rat cortical neurones. Neurosci Lett 23:333–336. [DOI] [PubMed] [Google Scholar]
- Perrais D, Coussen F, Mulle C (2009a) Atypical functional properties of GluK3-containing kainate receptors. J Neurosci 29:15499–15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrais D, Pinheiro PS, Jane DE, Mulle C (2009b) Antagonism of recombinant and native GluK3-containing kainate receptors. Neuropharmacology 56:131–140. [DOI] [PubMed] [Google Scholar]
- Perrais D, Veran J, Mulle C (2010) Gating and permeation of kainate receptors: differences unveiled. Trends Pharmacol Sci 31:516–522. [DOI] [PubMed] [Google Scholar]
- Personius KE, Slusher BS, Udin SB (2016) Neuromuscular NMDA receptors modulate developmental synapse elimination. J Neurosci 36:8783–8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perszyk RKatzman BMKusumoto HKell SAEpplin MPTahirovic YAMoore RLMenaldino DBurger PLiotta DC, et al. (2018) An NMDAR positive and negative allosteric modulator series share a binding site and are interconverted by methyl groups. eLife 7:e34711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perszyk REDiRaddo JOStrong KLLow CMOgden KKKhatri AVargish GAPelkey KATricoire LLiotta DC, et al. (2016) GluN2D-Containing N-methyl-d-Aspartate Receptors Mediate Synaptic Transmission in Hippocampal Interneurons and Regulate Interneuron Activity. Mol Pharmacol 90:689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perszyk RE, Kristensen AS, Lyuboslavsky P, Traynelis SF (2021) 3-Dimensional Missense Tolerance Ratio Analysis. Genome Res 31:1447–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perszyk RE, Myers SJ, Yuan H, Gibb AJ, Furukawa H, Sobolevsky AI, Traynelis SF (2020a) Hodgkin-Huxley-Katz Prize Lecture: Genetic and pharmacological control of glutamate receptor channel through a highly conserved gating motif. J Physiol 598:3071–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perszyk RESwanger SAShelley CKhatri AFernandez-Cuervo GEpplin MPZhang JLe PBülow PGarnier-Amblard E, et al. (2020b) Biased modulators of NMDA receptors control channel opening and ion selectivity. Nat Chem Biol 16:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perszyk REZheng ZBanke TGZhang JXie LMcDaniel MJKatzman BMPelly SCYuan HLiotta DC, et al. (2021) The negative allosteric modulator EU1794-4 reduces single channel conductance and Ca2+ permeability of GluN1/GluN2A NMDA receptors. Mol Pharmacol 99:399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter SUrbanus BHAKlaassen RVWu BBoele HJAzizi SSlotman JAHoutsmuller ABSchonewille MHoebeek FE, et al. (2020) AMPAR auxiliary protein SHISA6 facilitates Purkinje cell synaptic excitability and procedural memory formation. Cell Rep 31:107515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS (2012) Distribution of extrasynaptic NMDA receptors on neurons. ScientificWorldJournal 2012:267120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Hua F, Yi Z, Zhou A, Ge L, Stephenson FA, Wenthold RJ (2010) Organization of NMDA receptors at extrasynaptic locations. Neuroscience 167:68–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wenthold RJ (1992) Light and electron immunocytochemical localization of AMPA-selective glutamate receptors in the rat brain. J Comp Neurol 318:329–354. [DOI] [PubMed] [Google Scholar]
- Petri M, Naqibuddin M, Carson KA, Wallace DJ, Weisman MH, Holliday SL, Sampedro M, Padilla PA, Brey RL (2010) Depression and cognitive impairment in newly diagnosed systemic lupus erythematosus. J Rheumatol 37:2032–2038. [DOI] [PubMed] [Google Scholar]
- Petrovic M, Sedlacek M, Cais O, Horak M, Chodounska H, Vyklicky L Jr (2009) Pregnenolone sulfate modulation of N-methyl-D-aspartate receptors is phosphorylation dependent. Neuroscience 160:616–628. [DOI] [PubMed] [Google Scholar]
- Petrovic M, Sedlacek M, Horak M, Chodounska H, Vyklický L Jr (2005) 20-oxo-5beta-pregnan-3alpha-yl sulfate is a use-dependent NMDA receptor inhibitor. J Neurosci 25:8439–8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB (2013) Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 9:e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrus DS, Fabel K, Kronenberg G, Winter C, Steiner B, Kempermann G (2009) NMDA and benzodiazepine receptors have synergistic and antagonistic effects on precursor cells in adult hippocampal neurogenesis. Eur J Neurosci 29:244–252. [DOI] [PubMed] [Google Scholar]
- Philips AKSirén AAvela KSomer MPeippo MAhvenainen MDoagu FArvio MKääriäinen HVan Esch H, et al. (2014) X-exome sequencing in Finnish families with intellectual disability--four novel mutations and two novel syndromic phenotypes. Orphanet J Rare Dis 9:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpot BD, Weisberg MP, Ramos MS, Sawtell NB, Tang YP, Tsien JZ, Bear MF (2001) Effect of transgenic overexpression of NR2B on NMDA receptor function and synaptic plasticity in visual cortex. Neuropharmacology 41:762–770. [DOI] [PubMed] [Google Scholar]
- Piard JBéreau MXiangWei WWirth TAmsallem DBuisson LRichard PLiu NXu YMyers SJ, et al. (2020) The GRIA3 c.2477G > A variant causes an exaggerated startle reflex, chorea, and multifocal myoclonus. Mov Disord 35:1224–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard BSKnight HMHamilton RSSoares DCWalker RBoyd JKMachell JMaclean AMcGhee KACondie A, et al. (2008) A common variant in the 3'UTR of the GRIK4 glutamate receptor gene affects transcript abundance and protects against bipolar disorder. Proc Natl Acad Sci USA 105:14940–14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce VD, Niu L (2019) Stargazin and γ4 slow the channel opening and closing rates of GluA4 AMPA receptors. Sci Rep 9:9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TMYuan HMarsh EDFuentes-Fajardo KAdams DRMarkello TGolas GSimeonov DRHolloman CTankovic A, et al. ; PhD for the NISC Comparative Sequencing Program (2014) GRIN2A mutation and early-onset epileptic encephalopathy: personalized therapy with memantine. Ann Clin Transl Neurol 1:190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña-Crespo JCTalantova MMicu IStates BChen HSTu SNakanishi NTong GZhang DHeinemann SF, et al. (2010) Excitatory glycine responses of CNS myelin mediated by NR1/NR3 “NMDA” receptor subunits. J Neurosci 30:11501–11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinard E, Borroni E, Koerner A, Umbricht D, Alberati D (2018) Glycine transporter type I (GlyT1) inhibitor, bitopertin: A journey from lab to patient. Chimia (Aarau) 72:477–484. [DOI] [PubMed] [Google Scholar]
- Pinheiro PS, Lanore F, Veran J, Artinian J, Blanchet C, Crépel V, Perrais D, Mulle C (2013) Selective block of postsynaptic kainate receptors reveals their function at hippocampal mossy fiber synapses. Cereb Cortex 23:323–331. [DOI] [PubMed] [Google Scholar]
- Pinheiro PS, Mulle C (2008) Presynaptic glutamate receptors: physiological functions and mechanisms of action. Nat Rev Neurosci 9:423–436. [DOI] [PubMed] [Google Scholar]
- Pinheiro PS, Perrais D, Coussen F, Barhanin J, Bettler B, Mann JR, Malva JO, Heinemann SF, Mulle C (2007) GluR7 is an essential subunit of presynaptic kainate autoreceptors at hippocampal mossy fiber synapses. Proc Natl Acad Sci USA 104:12181–12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto DDelaby EMerico DBarbosa MMerikangas AKlei LThiruvahindrapuram BXu XZiman RWang Z, et al. (2014) Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 94:677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirotte B, Francotte P, Goffin E, de Tullio P (2013) AMPA receptor positive allosteric modulators: a patent review. Expert Opin Ther Pat 23:615–628. [DOI] [PubMed] [Google Scholar]
- Pla RStanco AHoward MARubin ANVogt DMortimer NCobos IPotter GBLindtner SPrice JD, et al. (2018) Dlx1 and Dlx2 promote interneuron GABA synthesis, synaptogenesis, and dendritogenesis. Cereb Cortex 28:3797–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planagumà JHaselmann HMannara FPetit-Pedrol MGrünewald BAguilar ERöpke LMartín-García ETitulaer MJJercog P, et al. (2016) Ephrin-B2 prevents N-methyl-D-aspartate receptor antibody effects on memory and neuroplasticity. Ann Neurol 80:388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planagumà JLeypoldt FMannara FGutiérrez-Cuesta JMartín-García EAguilar ETitulaer MJPetit-Pedrol MJain ABalice-Gordon R, et al. (2015) Human N-methyl D-aspartate receptor antibodies alter memory and behaviour in mice. Brain 138:94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant K, Pelkey KA, Bortolotto ZA, Morita D, Terashima A, McBain CJ, Collingridge GL, Isaac JT (2006) Transient incorporation of native GluR2-lacking AMPA receptors during hippocampal long-term potentiation. Nat Neurosci 9:602–604. [DOI] [PubMed] [Google Scholar]
- Platel JC, Dave KA, Gordon V, Lacar B, Rubio ME, Bordey A (2010) NMDA receptors activated by subventricular zone astrocytic glutamate are critical for neuroblast survival prior to entering a synaptic network. Neuron 65:859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Curtice KJ, Twede VD, Watkins M, Gruszczyński P, Bulaj G, Horvath MP, Olivera BM (2014) From molecular phylogeny towards differentiating pharmacology for NMDA receptor subtypes. Toxicon 81:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzer KYuan HSchütz HWinschel AChen WHu CKusumoto HHeyne HOHelbig KLTang S, et al. (2017) GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects. J Med Genet 54:460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasure D (2008) Diagnostic and pathogenic significance of glutamate receptor autoantibodies. Arch Neurol 65:589–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plested AJ (2011) Kainate receptor modulation by sodium and chloride. Adv Exp Med Biol 717:93–113. [DOI] [PubMed] [Google Scholar]
- Plested AJ (2016) Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels. Nat Struct Mol Biol 23:494–502. [DOI] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML (2007) Structure and mechanism of kainate receptor modulation by anions. Neuron 53:829–841. [DOI] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML (2009) Engineering a high-affinity allosteric binding site for divalent cations in kainate receptors. Neuropharmacology 56:114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plested AJ, Vijayan R, Biggin PC, Mayer ML (2008) Molecular basis of kainate receptor modulation by sodium. Neuron 58:720–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plested AJ, Wildman SS, Lieb WR, Franks NP (2004) Determinants of the sensitivity of AMPA receptors to xenon. Anesthesiology 100:347–358. [DOI] [PubMed] [Google Scholar]
- Pochwat B, Rafało-Ulińska A, Domin H, Misztak P, Nowak G, Szewczyk B (2017) Involvement of extracellular signal-regulated kinase (ERK) in the short and long-lasting antidepressant-like activity of NMDA receptor antagonists (zinc and Ro 25-6981) in the forced swim test in rats. Neuropharmacology 125:333–342. [DOI] [PubMed] [Google Scholar]
- Pøhlsgaard J, Frydenvang K, Madsen U, Kastrup JS (2011) Lessons from more than 80 structures of the GluA2 ligand-binding domain in complex with agonists, antagonists and allosteric modulators. Neuropharmacology 60:135–150. [DOI] [PubMed] [Google Scholar]
- Poleg-Polsky A, Diamond JS (2016) Retinal circuitry balances contrast tuning of excitation and inhibition to enable reliable computation of direction selectivity. J Neurosci 36:5861–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgár E, Watanabe M, Hartmann B, Grant SG, Todd AJ (2008) Expression of AMPA receptor subunits at synapses in laminae I-III of the rodent spinal dorsal horn. Mol Pain 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollok S, Reiner A (2020) Subunit-selective iGluR antagonists can potentiate heteromeric receptor responses by blocking desensitization. Proc Natl Acad Sci USA 117:25851–25858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polsky A, Mel B, Schiller J (2009) Encoding and decoding bursts by NMDA spikes in basal dendrites of layer 5 pyramidal neurons. J Neurosci 29:11891–11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poluch S, Drian MJ, Durand M, Astier C, Benyamin Y, König N (2001) AMPA receptor activation leads to neurite retraction in tangentially migrating neurons in the intermediate zone of the embryonic rat neocortex. J Neurosci Res 63:35–44. [DOI] [PubMed] [Google Scholar]
- Poluch S, König N (2002) AMPA receptor activation induces GABA release from neurons migrating tangentially in the intermediate zone of embryonic rat neocortex. Eur J Neurosci 16:350–354. [DOI] [PubMed] [Google Scholar]
- Poon K, Ahmed AH, Nowak LM, Oswald RE (2011) Mechanisms of modal activation of GluA3 receptors. Mol Pharmacol 80:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon K, Nowak LM, Oswald RE (2010) Characterizing single-channel behavior of GluA3 receptors. Biophys J 99:1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Eleveld MJ, van ’t Slot R, Ploos van Amstel HK, Hochstenbach R (2010) Recurrent copy number changes in mentally retarded children harbour genes involved in cellular localization and the glutamate receptor complex. Eur J Hum Genet 18:39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu G, Auerbach A (2003) Modal gating of NMDA receptors and the shape of their synaptic response. Nat Neurosci 6:476–483. [DOI] [PubMed] [Google Scholar]
- Postila PA, Swanson GT, Pentikäinen OT (2010) Exploring kainate receptor pharmacology using molecular dynamics simulations. Neuropharmacology 58:515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postila PA, Ylilauri M, Pentikäinen OT (2011) Full and partial agonism of ionotropic glutamate receptors indicated by molecular dynamics simulations. J Chem Inf Model 51:1037–1047. [DOI] [PubMed] [Google Scholar]
- Potschka H, Trinka E (2019) Perampanel: Does it have broad-spectrum potential? Epilepsia 60 (Suppl 1):22–36. [DOI] [PubMed] [Google Scholar]
- Poulsen MH, Andersen J, Christensen R, Hansen KB, Traynelis SF, Strømgaard K, Kristensen AS (2015) Binding of ArgTX-636 in the NMDA receptor ion channel. J Mol Biol 427:176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen MH, Lucas S, Bach TB, Barslund AF, Wenzler C, Jensen CB, Kristensen AS, Strømgaard K (2013) Structure-activity relationship studies of argiotoxins: selective and potent inhibitors of ionotropic glutamate receptors. J Med Chem 56:1171–1181. [DOI] [PubMed] [Google Scholar]
- Poulsen MH, Lucas S, Strømgaard K, Kristensen AS (2014a) Evaluation of PhTX-74 as subtype-selective inhibitor of GluA2-containing AMPA receptors. Mol Pharmacol 85:261–268. [DOI] [PubMed] [Google Scholar]
- Poulsen MH, Lucas S, Strømgaard K, Kristensen AS (2014b) Inhibition of AMPA receptors by polyamine toxins is regulated by agonist efficacy and stargazin. Neurochem Res 39:1906–1913. [DOI] [PubMed] [Google Scholar]
- Poulsen MH, Poshtiban A, Klippenstein V, Ghisi V, Plested AJR (2019) Gating modules of the AMPA receptor pore domain revealed by unnatural amino acid mutagenesis. Proc Natl Acad Sci USA 116:13358–13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povysheva NV, Johnson JW (2012) Tonic NMDA receptor-mediated current in prefrontal cortical pyramidal cells and fast-spiking interneurons. J Neurophysiol 107:2232–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Barlowe C (1998) Transport of axl2p depends on erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J Cell Biol 142:1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J, Barlowe C (2002) Erv14p directs a transmembrane secretory protein into COPII-coated transport vesicles. Mol Biol Cell 13:880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar LS, Auerbach A (1996) Identification of a high affinity divalent cation binding site near the entrance of the NMDA receptor channel. Neuron 16:869–880. [DOI] [PubMed] [Google Scholar]
- Premkumar LS, Qin F, Auerbach A (1997) Subconductance states of a mutant NMDA receptor channel kinetics, calcium, and voltage dependence. J Gen Physiol 109:181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preskorn SH, Baker B, Kolluri S, Menniti FS, Krams M, Landen JW (2008) An innovative design to establish proof of concept of the antidepressant effects of the NR2B subunit selective N-methyl-D-aspartate antagonist, CP-101,606, in patients with treatment-refractory major depressive disorder. J Clin Psychopharmacol 28:631–637. [DOI] [PubMed] [Google Scholar]
- Pressey JCMahadevan VKhademullah CSDargaei ZChevrier JYe WHuang MChauhan AKMeas SJUvarov P, et al. (2017) A kainate receptor subunit promotes the recycling of the neuron-specific K+-Cl- co-transporter KCC2 in hippocampal neurons. J Biol Chem 292:6190–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressey JC, Woodin MA (2021) Kainate receptor regulation of synaptic inhibition in the hippocampus. J Physiol 599:485–492. [DOI] [PubMed] [Google Scholar]
- Price RB, Nock MK, Charney DS, Mathew SJ (2009) Effects of intravenous ketamine on explicit and implicit measures of suicidality in treatment-resistant depression. Biol Psychiatry 66:522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A, Kolleker A, Ayalon G, Gillor M, Osten P, Stern-Bach Y (2005) Stargazin reduces desensitization and slows deactivation of the AMPA-type glutamate receptors. J Neurosci 25:2682–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestley T, Laughton P, Myers J, Le Bourdellés B, Kerby J, Whiting PJ (1995) Pharmacological properties of recombinant human N-methyl-D-aspartate receptors comprising NR1a/NR2A and NR1a/NR2B subunit assemblies expressed in permanently transfected mouse fibroblast cells. Mol Pharmacol 48:841–848. [PubMed] [Google Scholar]
- Prieto ML, Wollmuth LP (2010) Gating modes in AMPA receptors. J Neurosci 30:4449–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Godino LL, Rytz R, Bargeton B, Abuin L, Arguello JR, Peraro MD, Benton R (2016) Olfactory receptor pseudo-pseudogenes. Nature 539:93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Godino LL, Rytz R, Cruchet S, Bargeton B, Abuin L, Silbering AF, Ruta V, Dal Peraro M, Benton R (2017) Evolution of acid-sensing olfactory circuits in drosophilids. Neuron 93:661–676.e6. [DOI] [PubMed] [Google Scholar]
- Prithviraj R, Inglis FM (2008) Expression of the N-methyl-D-aspartate receptor subunit NR3B regulates dendrite morphogenesis in spinal motor neurons. Neuroscience 155:145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prius-Mengual J, Pérez-Rodríguez M, Andrade-Talavera Y, Rodríguez-Moreno A (2019) NMDA receptors containing GluN2B/2C/2D subunits mediate an increase in glutamate release at hippocampal CA3-CA1 synapses. Mol Neurobiol 56:1694–1706. [DOI] [PubMed] [Google Scholar]
- Prorok M, Castellino FJ (2007) The molecular basis of conantokin antagonism of NMDA receptor function. Curr Drug Targets 8:633–642. [DOI] [PubMed] [Google Scholar]
- Prüss HFinke CHöltje MHofmann JKlingbeil CProbst CBorowski KAhnert-Hilger GHarms LSchwab JM, et al. (2012) N-methyl-D-aspartate receptor antibodies in herpes simplex encephalitis. Ann Neurol 72:902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüss H, Kirmse K (2018) Pathogenic role of autoantibodies against inhibitory synapses. Brain Res 1701:146–152. [DOI] [PubMed] [Google Scholar]
- Prybylowski K, Rumbaugh G, Wolfe BB, Vicini S (2000) Increased exon 5 expression alters extrasynaptic NMDA receptors in cerebellar neurons. J Neurochem 75:1140–1146. [DOI] [PubMed] [Google Scholar]
- Ptak CP, Ahmed AH, Oswald RE (2009) Probing the allosteric modulator binding site of GluR2 with thiazide derivatives. Biochemistry 48:8594–8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak CP, Hsieh CL, Weiland GA, Oswald RE (2014) Role of stoichiometry in the dimer-stabilizing effect of AMPA receptor allosteric modulators. ACS Chem Biol 9:128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchalski RBLouis JCBrose NTraynelis SFEgebjerg JKukekov VWenthold RJRogers SWLin FMoran T, et al. (1994) Selective RNA editing and subunit assembly of native glutamate receptors. Neuron 13:131–147. [DOI] [PubMed] [Google Scholar]
- Puddifoot CA, Chen PE, Schoepfer R, Wyllie DJ (2009) Pharmacological characterization of recombinant NR1/NR2A NMDA receptors with truncated and deleted carboxy termini expressed in Xenopus laevis oocytes. Br J Pharmacol 156:509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnakkal P, Jendritza P, Köhr G (2012) Influence of the intracellular GluN2 C-terminal domain on NMDA receptor function. Neuropharmacology 62:1985–1992. [DOI] [PubMed] [Google Scholar]
- Purkey AM, Dell’Acqua ML (2020) Phosphorylation-dependent regulation of Ca2+-permeable AMPA receptors during hippocampal synaptic plasticity. Front Synaptic Neurosci 12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkey AM, Woolfrey KM, Crosby KC, Stich DG, Chick WS, Aoto J, Dell’Acqua ML (2018) AKAP150 palmitoylation regulates synaptic incorporation of Ca2+-permeable AMPA receptors to control LTP. Cell Rep 25:974–987.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian A, Johnson JW (2006) Permeant ion effects on external Mg2+ block of NR1/2D NMDA receptors. J Neurosci 26:10899–10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Matt L, Zhang M, Nguyen M, Patriarchi T, Koval OM, Anderson ME, He K, Lee HK, Hell JW (2012) β2-Adrenergic receptor supports prolonged theta tetanus-induced LTP. J Neurophysiol 107:2703–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach TT, Honnorat J, Kolattukudy PE, Khanna R, Duchemin AM (2015) CRMPs: critical molecules for neurite morphogenesis and neuropsychiatric diseases. Mol Psychiatry 20:1037–1045. [DOI] [PubMed] [Google Scholar]
- Quartermain D, Mower J, Rafferty MF, Herting RL, Lanthorn TH (1994) Acute but not chronic activation of the NMDA-coupled glycine receptor with D-cycloserine facilitates learning and retention. Eur J Pharmacol 257:7–12. [DOI] [PubMed] [Google Scholar]
- Quirk JC, Nisenbaum ES (2002) LY404187: a novel positive allosteric modulator of AMPA receptors. CNS Drug Rev 8:255–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirk JC, Siuda ER, Nisenbaum ES (2004) Molecular determinants responsible for differences in desensitization kinetics of AMPA receptor splice variants. J Neurosci 24:11416–11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabacchi S, Bailly Y, Delhaye-Bouchaud N, Mariani J (1992) Involvement of the N-methyl D-aspartate (NMDA) receptor in synapse elimination during cerebellar development. Science 256:1823–1825. [DOI] [PubMed] [Google Scholar]
- Racca C, Catania MV, Monyer H, Sakmann B (1996) Expression of AMPA-glutamate receptor B subunit in rat hippocampal GABAergic neurons. Eur J Neurosci 8:1580–1590. [DOI] [PubMed] [Google Scholar]
- Rachline J, Perin-Dureau F, Le Goff A, Neyton J, Paoletti P (2005) The micromolar zinc-binding domain on the NMDA receptor subunit NR2B. J Neurosci 25:308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radin DP, Li YX, Rogers G, Purcell R, Lippa A (2018a) Stargazin differentially modulates ampakine gating kinetics and pharmacology. Biochem Pharmacol 148:308–314. [DOI] [PubMed] [Google Scholar]
- Radin DP, Li YX, Rogers G, Purcell R, Lippa A (2018b) Tarps differentially affect the pharmacology of ampakines. Biochem Pharmacol 154:446–451. [DOI] [PubMed] [Google Scholar]
- Raimondo JV, Tomes H, Irkle A, Kay L, Kellaway L, Markram H, Millar RP, Akerman CJ (2016) Tight coupling of astrocyte pH dynamics to epileptiform activity revealed by genetically encoded pH sensors. J Neurosci 36:7002–7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan I, Cline HT (1998) Glutamate receptor activity is required for normal development of tectal cell dendrites in vivo. J Neurosci 18:7836–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan I, Witte S, Cline HT (1999) NMDA receptor activity stabilizes presynaptic retinotectal axons and postsynaptic optic tectal cell dendrites in vivo. J Neurobiol 38:357–368. [DOI] [PubMed] [Google Scholar]
- Rajani V, Sengar AS, Salter MW (2020) Tripartite signalling by NMDA receptors. Mol Brain 13:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman IM, Trussell LO (1995) The mechanism of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptor desensitization after removal of glutamate. Biophys J 68:137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman IM, Zhang S, Trussell LO (1994) Pathway-specific variants of AMPA receptors and their contribution to neuronal signaling. J Neurosci 14:4998–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Cooper D, Poddar N, MacLean DM, Rambhadran A, Taylor JN, Uhm H, Landes CF, Jayaraman V (2012) Role of conformational dynamics in α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptor partial agonism. J Biol Chem 287:43557–43564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambhadran A, Gonzalez J, Jayaraman V (2010) Subunit arrangement in N-methyl-D-aspartate (NMDA) receptors. J Biol Chem 285:15296–15301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambhadran A, Gonzalez J, Jayaraman V (2011) Conformational changes at the agonist binding domain of the N-methyl-D-aspartic acid receptor. J Biol Chem 286:16953–16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambousek LBubenikova-Valesova VKacer PSyslova KKenney JHolubova KNajmanova VZach PSvoboda JStuchlik A, et al. (2011) Cellular and behavioural effects of a new steroidal inhibitor of the N-methyl-d-aspartate receptor 3α5β-pregnanolone glutamate. Neuropharmacology 61:61–68. [DOI] [PubMed] [Google Scholar]
- Ramoa AS, Mower AF, Liao D, Jafri SI (2001) Suppression of cortical NMDA receptor function prevents development of orientation selectivity in the primary visual cortex. J Neurosci 21:4299–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos CI, Igiesuorobo O, Wang Q, Serpe M (2015) Neto-mediated intracellular interactions shape postsynaptic composition at the Drosophila neuromuscular junction. PLoS Genet 11:e1005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan MDeMartinis NHuguenel BGaudreault FBednar MMShaffer CLGupta SCahill JSherif MAMancuso J, et al. (2017) Attenuation of ketamine-induced impairment in verbal learning and memory in healthy volunteers by the AMPA receptor potentiator PF-04958242. Mol Psychiatry 22:1633–1640. [DOI] [PubMed] [Google Scholar]
- Rao A, Craig AM (1997) Activity regulates the synaptic localization of the NMDA receptor in hippocampal neurons. Neuron 19:801–812. [DOI] [PubMed] [Google Scholar]
- Rassendren FA, Lory P, Pin JP, Nargeot J (1990) Zinc has opposite effects on NMDA and non-NMDA receptors expressed in Xenopus oocytes. Neuron 4:733–740. [DOI] [PubMed] [Google Scholar]
- Ratner MH, Kumaresan V, Farb DH (2019) Neurosteroid Actions in Memory and Neurologic/Neuropsychiatric Disorders. Front Endocrinol (Lausanne) 10:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauner C, Köhr G (2011) Triheteromeric NR1/NR2A/NR2B receptors constitute the major N-methyl-D-aspartate receptor population in adult hippocampal synapses. J Biol Chem 286:7558–7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikrishnan A, Gandhi PJ, Shelkar GP, Liu J, Pavuluri R, Dravid SM (2018) Region-specific expression of NMDA receptor GluN2C subunit in parvalbumin-positive neurons and astrocytes: Analysis of GluN2C expression using a novel reporter model. Neuroscience 380:49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranathan A, Donevan SD, Sugden SG, Greig A, Rao MS, Parks TN (2000) Contrasting molecular composition and channel properties of AMPA receptors on chick auditory and brainstem motor neurons. J Physiol 523:667–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebola N, Sachidhanandam S, Perrais D, Cunha RA, Mulle C (2007) Short-term plasticity of kainate receptor-mediated EPSCs induced by NMDA receptors at hippocampal mossy fiber synapses. J Neurosci 27:3987–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regalado MP, Villarroel A, Lerma J (2001) Intersubunit cooperativity in the NMDA receptor. Neuron 32:1085–1096. [DOI] [PubMed] [Google Scholar]
- Regan MC, Grant T, McDaniel MJ, Karakas E, Zhang J, Traynelis SF, Grigorieff N, Furukawa H (2018) Structural mechanism of functional modulation by gene splicing in NMDA receptors. Neuron 98:521–529.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan MC, Zhu Z, Yuan H, Myers SJ, Menaldino DS, Tahirovic YA, Liotta DC, Traynelis SF, Furukawa H (2019) Structural elements of a pH-sensitive inhibitor binding site in NMDA receptors. Nat Commun 10:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regehr WG (2012) Short-term presynaptic plasticity. Cold Spring Harb Perspect Biol 4:a005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Arant RJ, Isacoff EY (2012) Assembly stoichiometry of the GluK2/GluK5 kainate receptor complex. Cell Rep 1:234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Isacoff EY (2014) Tethered ligands reveal glutamate receptor desensitization depends on subunit occupancy. Nat Chem Biol 10:273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Levitz J (2018) Glutamatergic signaling in the central nervous system: Ionotropic and metabotropic receptors in concert. Neuron 98:1080–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert MM, Bullock R (1999) Clinical trials in head injury. Neurol Res 21:330–338. [DOI] [PubMed] [Google Scholar]
- Reiprich P, Kilb W, Luhmann HJ (2005) Neonatal NMDA receptor blockade disturbs neuronal migration in rat somatosensory cortex in vivo. Cereb Cortex 15:349–358. [DOI] [PubMed] [Google Scholar]
- Reissner C, Runkel F, Missler M (2013) Neurexins. Genome Biol 14:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Honse Y, Karp BJ, Lipsky RH, Peoples RW (2003a) A site in the fourth membrane-associated domain of the N-methyl-D-aspartate receptor regulates desensitization and ion channel gating. J Biol Chem 278:276–283. [DOI] [PubMed] [Google Scholar]
- Ren H, Salous AK, Paul JM, Lamb KA, Dwyer DS, Peoples RW (2008) Functional interactions of alcohol-sensitive sites in the N-methyl-D-aspartate receptor M3 and M4 domains. J Biol Chem 283:8250–8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Zhao Y, Dwyer DS, Peoples RW (2012) Interactions among positions in the third and fourth membrane-associated domains at the intersubunit interface of the N-methyl-D-aspartate receptor forming sites of alcohol action. J Biol Chem 287:27302–27312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Li X, Zhang X, Li M, Wang Y, Ma Y (2013) The effects of intra-hippocampal microinfusion of D-cycloserine on fear extinction, and the expression of NMDA receptor subunit NR2B and neurogenesis in the hippocampus in rats. Prog Neuropsychopharmacol Biol Psychiatry 44:257–264. [DOI] [PubMed] [Google Scholar]
- Ren Z, Riley NJ, Garcia EP, Sanders JM, Swanson GT, Marshall J (2003b) Multiple trafficking signals regulate kainate receptor KA2 subunit surface expression. J Neurosci 23:6608–6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Riley NJ, Needleman LA, Sanders JM, Swanson GT, Marshall J (2003c) Cell surface expression of GluR5 kainate receptors is regulated by an endoplasmic reticulum retention signal. J Biol Chem 278:52700–52709. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Rothbaum BO, Tannenbaum L, Anderson P, Graap K, Zimand E, Hodges L, Davis M (2004) Cognitive enhancers as adjuncts to psychotherapy: use of D-cycloserine in phobic individuals to facilitate extinction of fear. Arch Gen Psychiatry 61:1136–1144. [DOI] [PubMed] [Google Scholar]
- Reyes AD, Rubel EW, Spain WJ (1996) In vitro analysis of optimal stimuli for phase-locking and time-delayed modulation of firing in avian nucleus laminaris neurons. J Neurosci 16:993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riebe I, Seth H, Culley G, Dósa Z, Radi S, Strand K, Fröjd V, Hanse E (2016) Tonically active NMDA receptors--a signalling mechanism critical for interneuronal excitability in the CA1 stratum radiatum. Eur J Neurosci 43:169–178. [DOI] [PubMed] [Google Scholar]
- Ripellino JA, Neve RL, Howe JR (1998) Expression and heteromeric interactions of non-N-methyl-D-aspartate glutamate receptor subunits in the developing and adult cerebellum. Neuroscience 82:485–497. [DOI] [PubMed] [Google Scholar]
- Risgaard R, Hansen KB, Clausen RP (2010) Partial agonists and subunit selectivity at NMDA receptors. Chemistry 16:13910–13918. [DOI] [PubMed] [Google Scholar]
- Risgaard R, Nielsen SD, Hansen KB, Jensen CM, Nielsen B, Traynelis SF, Clausen RP (2013) Development of 2′-substituted (2S,1‘R,2’S)-2-(carboxycyclopropyl)glycine analogues as potent N-methyl-d-aspartic acid receptor agonists. J Med Chem 56:4071–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter LM, Unis AS, Meador-Woodruff JH (2001) Ontogeny of ionotropic glutamate receptor expression in human fetal brain. Brain Res Dev Brain Res 127:123–133. [DOI] [PubMed] [Google Scholar]
- Ritter LM, Vazquez DM, Meador-Woodruff JH (2002) Ontogeny of ionotropic glutamate receptor subunit expression in the rat hippocampus. Brain Res Dev Brain Res 139:227–236. [DOI] [PubMed] [Google Scholar]
- Riva I, Eibl C, Volkmer R, Carbone AL, Plested AJ (2017) Control of AMPA receptor activity by the extracellular loops of auxiliary proteins. eLife 6:e28680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Black JA, Waxman SG (1998) Endogenous NMDA-receptor activation regulates glutamate release in cultured spinal neurons. J Neurophysiol 80:196–208. [DOI] [PubMed] [Google Scholar]
- Robert A, Howe JR (2003) How AMPA receptor desensitization depends on receptor occupancy. J Neurosci 23:847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Irizarry SN, Hughes TE, Howe JR (2001) Subunit interactions and AMPA receptor desensitization. J Neurosci 21:5574–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts ACDíez-García JRodriguiz RMLópez IPLuján RMartínez-Turrillas RPicó EHenson MABernardo DRJarrett TM, et al. (2009) Downregulation of NR3A-containing NMDARs is required for synapse maturation and memory consolidation. Neuron 63:342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha M, Sur M (1995) Rapid acquisition of dendritic spines by visual thalamic neurons after blockade of N-methyl-D-aspartate receptors. Proc Natl Acad Sci USA 92:8026–8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche KW, Huganir RL (1995) Synaptic expression of the high-affinity kainate receptor subunit KA2 in hippocampal cultures. Neuroscience 69:383–393. [DOI] [PubMed] [Google Scholar]
- Rodenas-Ruano A, Chávez AE, Cossio MJ, Castillo PE, Zukin RS (2012) REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci 15:1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Moreno A, Herreras O, Lerma J (1997) Kainate receptors presynaptically downregulate GABAergic inhibition in the rat hippocampus. Neuron 19:893–901. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Moreno A, Lerma J (1998) Kainate receptor modulation of GABA release involves a metabotropic function. Neuron 20:1211–1218. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Moreno A, Sihra TS (2013) Presynaptic kainate receptor-mediated facilitation of glutamate release involves Ca2+-calmodulin and PKA in cerebrocortical synaptosomes. FEBS Lett 587:788–792. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Kegeles LS, Levinson A, Feng T, Marcus SM, Vermes D, Flood P, Simpson HB (2013) Randomized controlled crossover trial of ketamine in obsessive-compulsive disorder: proof-of-concept. Neuropsychopharmacology 38:2475–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogawski MA (2013) AMPA receptors as a molecular target in epilepsy therapy. Acta Neurol Scand Suppl 197:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SW, Andrews PI, Gahring LC, Whisenand T, Cauley K, Crain B, Hughes TE, Heinemann SF, McNamara JO (1994) Autoantibodies to glutamate receptor GluR3 in Rasmussen’s encephalitis. Science 265:648–651. [DOI] [PubMed] [Google Scholar]
- Romero-Hernandez A, Furukawa H (2017) Novel mode of antagonist binding in NMDA receptors revealed by the crystal structure of the GluN1-GluN2A ligand-binding domain complexed to NVP-AAM077. Mol Pharmacol 92:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Hernandez A, Simorowski N, Karakas E, Furukawa H (2016) Molecular basis for subtype specificity and high-affinity zinc inhibition in the GluN1-GluN2A NMDA receptor amino-terminal domain. Neuron 92:1324–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato-Siri M, Grandolfo M, Ballerini L (2002) Activity-dependent modulation of GABAergic synapses in developing rat spinal networks in vitro. Eur J Neurosci 16:2123–2135. [DOI] [PubMed] [Google Scholar]
- Rosenbaum MI, Clemmensen LS, Bredt DS, Bettler B, Strømgaard K (2020) Targeting receptor complexes: a new dimension in drug discovery. Nat Rev Drug Discov 19:884–901. [DOI] [PubMed] [Google Scholar]
- Rosenberg DArtoul SSegal ACKolodney GRadzishevsky IDikopoltsev EFoltyn VNInoue RMori HBillard JM, et al. (2013) Neuronal D-serine and glycine release via the Asc-1 transporter regulates NMDA receptor-dependent synaptic activity. J Neurosci 33:3533–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Feltz A, Westbrook GL (1995) Calcium-dependent inactivation of synaptic NMDA receptors in hippocampal neurons. J Neurophysiol 73:427–430. [DOI] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens CF (1998) The tetrameric structure of a glutamate receptor channel. Science 280:1596–1599. [DOI] [PubMed] [Google Scholar]
- Rosenmund C, Westbrook GL (1993) Calcium-induced actin depolymerization reduces NMDA channel activity. Neuron 10:805–814. [DOI] [PubMed] [Google Scholar]
- Rosenquist TH, Schneider AM, Monogham DT (1999) N-methyl-D-aspartate receptor agonists modulate homocysteine-induced developmental abnormalities. FASEB J 13:1523–1531. [DOI] [PubMed] [Google Scholar]
- Rossi B, Ogden D, Llano I, Tan YP, Marty A, Collin T (2012) Current and calcium responses to local activation of axonal NMDA receptors in developing cerebellar molecular layer interneurons. PLoS One 7:e39983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi P, Sola E, Taglietti V, Borchardt T, Steigerwald F, Utvik JK, Ottersen OP, Köhr G, D’Angelo E (2002) NMDA receptor 2 (NR2) C-terminal control of NR open probability regulates synaptic transmission and plasticity at a cerebellar synapse. J Neurosci 22:9687–9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M, Sukumaran M, Penn AC, Veprintsev DB, Babu MM, Greger IH (2011) Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers. EMBO J 30:959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostas JA, Brent VA, Voss K, Errington ML, Bliss TV, Gurd JW (1996) Enhanced tyrosine phosphorylation of the 2B subunit of the N-methyl-D-aspartate receptor in long-term potentiation. Proc Natl Acad Sci USA 93:10452–10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg FS, Barcelo G, Schüpbach T (1995) Cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell 81:967–978. [DOI] [PubMed] [Google Scholar]
- Rouach N, Byrd K, Petralia RS, Elias GM, Adesnik H, Tomita S, Karimzadegan S, Kealey C, Bredt DS, Nicoll RA (2005) TARP gamma-8 controls hippocampal AMPA receptor number, distribution and synaptic plasticity. Nat Neurosci 8:1525–1533. [DOI] [PubMed] [Google Scholar]
- Rozas JL, Paternain AV, Lerma J (2003) Noncanonical signaling by ionotropic kainate receptors. Neuron 39:543–553. [DOI] [PubMed] [Google Scholar]
- Rozov A, Burnashev N (1999) Polyamine-dependent facilitation of postsynaptic AMPA receptors counteracts paired-pulse depression. Nature 401:594–598. [DOI] [PubMed] [Google Scholar]
- Rozov A, Zilberter Y, Wollmuth LP, Burnashev N (1998) Facilitation of currents through rat Ca2+-permeable AMPA receptor channels by activity-dependent relief from polyamine block. J Physiol 511:361–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano D, Lambolez B, Rossier J, Paternain AV, Lerma J (1995) Kainate receptor subunits expressed in single cultured hippocampal neurons: molecular and functional variants by RNA editing. Neuron 14:1009–1017. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Leggo J, Chiano M, Dodge A, Norbury G, Rosser E, Craufurd D (1997) Genotypes at the GluR6 kainate receptor locus are associated with variation in the age of onset of Huntington disease. Proc Natl Acad Sci USA 94:3872–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ME, Wenthold RJ (1997) Glutamate receptors are selectively targeted to postsynaptic sites in neurons. Neuron 18:939–950. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Sachidhanandam S, Utvik JK, Coussen F, Mulle C (2005) Distinct subunits in heteromeric kainate receptors mediate ionotropic and metabotropic function at hippocampal mossy fiber synapses. J Neurosci 25:11710–11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Prybylowski K, Wang JF, Vicini S (2000) Exon 5 and spermine regulate deactivation of NMDA receptor subtypes. J Neurophysiol 83:1300–1306. [DOI] [PubMed] [Google Scholar]
- Ruthazer ES, Akerman CJ, Cline HT (2003) Control of axon branch dynamics by correlated activity in vivo. Science 301:66–70. [DOI] [PubMed] [Google Scholar]
- Rutkowska-Wlodarczyk I, Aller MI, Valbuena S, Bologna JC, Prézeau L, Lerma J (2015) A proteomic analysis reveals the interaction of GluK1 ionotropic kainate receptor subunits with Go proteins. J Neurosci 35:5171–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruusuvuori E, Kaila K (2014) Carbonic anhydrases and brain pH in the control of neuronal excitability. Subcell Biochem 75:271–290. [DOI] [PubMed] [Google Scholar]
- Ryan TJ, Emes RD, Grant SG, Komiyama NH (2008) Evolution of NMDA receptor cytoplasmic interaction domains: implications for organisation of synaptic signalling complexes. BMC Neurosci 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan TJKopanitsa MVIndersmitten TNithianantharajah JAfinowi NOPettit CStanford LESprengel RSaksida LMBussey TJ, et al. (2013) Evolution of GluN2A/B cytoplasmic domains diversified vertebrate synaptic plasticity and behavior. Nat Neurosci 16:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazantseva M, Englund J, Shintyapina A, Huupponen J, Shteinikov V, Pitkänen A, Partanen JM, Lauri SE (2020) Kainate receptors regulate development of glutamatergic synaptic circuitry in the rodent amygdala. eLife 9:e52798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycroft BK, Gibb AJ (2002) Direct effects of calmodulin on NMDA receptor single-channel gating in rat hippocampal granule cells. J Neurosci 22:8860–8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycroft BK, Gibb AJ (2004) Regulation of single NMDA receptor channel activity by alpha-actinin and calmodulin in rat hippocampal granule cells. J Physiol 557:795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu K, Yokoyama M, Yamashita M, Hirano T (2012) Induction of excitatory and inhibitory presynaptic differentiation by GluD1. Biochem Biophys Res Commun 417:157–161. [DOI] [PubMed] [Google Scholar]
- Sadikot AF, Burhan AM, Belanger MC, and Sasseville R (1998) NMDA receptor antagonists influence early development of GABAergic interneurons in the mammalian striatum. Brain Res Dev Brain Res 105:35–42. [PubMed] [Google Scholar]
- Safieddine S, Wenthold RJ (1997) The glutamate receptor subunit delta1 is highly expressed in hair cells of the auditory and vestibular systems. J Neurosci 17:7523–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager C, Terhag J, Kott S, Hollmann M (2009) C-terminal domains of transmembrane alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA) receptor regulatory proteins not only facilitate trafficking but are major modulators of AMPA receptor function. J Biol Chem 284:32413–32424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah P, Hestrin S, Nicoll RA (1989) Tonic activation of NMDA receptors by ambient glutamate enhances excitability of neurons. Science 246:815–818. [DOI] [PubMed] [Google Scholar]
- Sah P, Hestrin S, Nicoll RA (1990) Properties of excitatory postsynaptic currents recorded in vitro from rat hippocampal interneurones. J Physiol 430:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai MA, Biggin PC (2011) Quantifying water-mediated protein-ligand interactions in a glutamate receptor: a DFT study. J Phys Chem B 115:7085–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara Y, Noro N, Iida Y, Soma K, Nakamura Y (1997) Glutamate receptor subunits GluR5 and KA-2 are coexpressed in rat trigeminal ganglion neurons. J Neurosci 17:6611–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y, Suzuki H, Tani K, Nishikawa K, Irie K, Ogura Y, Tamura A, Tsukita S, Fujiyoshi Y (2015) Tight junctions. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science 347:775–778. [DOI] [PubMed] [Google Scholar]
- Sakai R, Swanson GT, Shimamoto K, Green T, Contractor A, Ghetti A, Tamura-Horikawa Y, Oiwa C, Kamiya H (2001) Pharmacological properties of the potent epileptogenic amino acid dysiherbaine, a novel glutamate receptor agonist isolated from the marine sponge Dysidea herbacea. J Pharmacol Exp Ther 296:650–658. [PubMed] [Google Scholar]
- Sakha P, Vesikansa A, Orav E, Heikkinen J, Kukko-Lukjanov TK, Shintyapina A, Franssila S, Jokinen V, Huttunen HJ, Lauri SE (2016) Axonal kainate receptors modulate the strength of efferent connectivity by regulating presynaptic differentiation. Front Cell Neurosci 10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakimura KKutsuwada TIto IManabe TTakayama CKushiya EYagi TAizawa SInoue YSugiyama H, et al. (1995) Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor epsilon 1 subunit. Nature 373:151–155. [DOI] [PubMed] [Google Scholar]
- Sakimura K, Morita T, Kushiya E, Mishina M (1992) Primary structure and expression of the gamma 2 subunit of the glutamate receptor channel selective for kainate. Neuron 8:267–274. [DOI] [PubMed] [Google Scholar]
- Salazar H, Eibl C, Chebli M, Plested A (2017) Mechanism of partial agonism in AMPA-type glutamate receptors. Nat Commun 8:14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar H, Mischke S, Plested AJR (2020) Measurements of the timescale and conformational space of AMPA receptor desensitization. Biophys J 119:206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimando GJ, Hyun M, Boyt KM, Winder DG (2020) BNST GluN2D-containing NMDA receptors influence anxiety- and depressive-like behaviors and ModulateCell-specific excitatory/inhibitory synaptic balance. J Neurosci 40:3949–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmi M, Bolbos R, Bauer S, Minlebaev M, Burnashev N, Szepetowski P (2018) Transient microstructural brain anomalies and epileptiform discharges in mice defective for epilepsy and language-related NMDA receptor subunit gene Grin2a. Epilepsia 59:1919–1930. [DOI] [PubMed] [Google Scholar]
- Salmi MDel Gallo FMinlebaev MZakharov APauly VPerron PPons-Bennaceur ACorby-Pellegrino SAniksztejn LLenck-Santini PP, et al. (2019) Impaired vocal communication, sleep-related discharges, and transient alteration of slow-wave sleep in developing mice lacking the GluN2A subunit of N-methyl-d-aspartate receptors. Epilepsia 60:1424–1437. [DOI] [PubMed] [Google Scholar]
- Salpietro VDixon CLGuo HBello ODVandrovcova JEfthymiou SMaroofian RHeimer GBurglen LValence S, et al. ; SYNAPS Study Group (2019) AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat Commun 10:3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MG, Fern R (2005) NMDA receptors are expressed in developing oligodendrocyte processes and mediate injury. Nature 438:1167–1171. [DOI] [PubMed] [Google Scholar]
- Salussolia CL, Corrales A, Talukder I, Kazi R, Akgul G, Bowen M, Wollmuth LP (2011a) Interaction of the M4 segment with other transmembrane segments is required for surface expression of mammalian α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. J Biol Chem 286:40205–40218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia CL, Gan Q, Kazi R, Singh P, Allopenna J, Furukawa H, Wollmuth LP (2013) A eukaryotic specific transmembrane segment is required for tetramerization in AMPA receptors. J Neurosci 33:9840–9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia CL, Prodromou ML, Borker P, Wollmuth LP (2011b) Arrangement of subunits in functional NMDA receptors. J Neurosci 31:11295–11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandan S, Sauer JF, Vida I, Bartos M (2010) Associative plasticity at excitatory synapses facilitates recruitment of fast-spiking interneurons in the dentate gyrus. J Neurosci 30:11826–11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanacora GJohnson MRKhan AAtkinson SDRiesenberg RRSchronen JPBurke MAZajecka JMBarra LSu HL, et al. (2017) Adjunctive lanicemine (AZD6765) in patients with major depressive disorder and history of inadequate response to antidepressants: A randomized, placebo-controlled study. Neuropsychopharmacology 42:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanacora G, Smith MA, Pathak S, Su HL, Boeijinga PH, McCarthy DJ, Quirk MC (2014) Lanicemine: a low-trapping NMDA channel blocker produces sustained antidepressant efficacy with minimal psychotomimetic adverse effects. Mol Psychiatry 19:978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JM, Ito K, Settimo L, Pentikäinen OT, Shoji M, Sasaki M, Johnson MS, Sakai R, Swanson GT (2005) Divergent pharmacological activity of novel marine-derived excitatory amino acids on glutamate receptors. J Pharmacol Exp Ther 314:1068–1078. [DOI] [PubMed] [Google Scholar]
- Sanders JM, Pentikäinen OT, Settimo L, Pentikäinen U, Shoji M, Sasaki M, Sakai R, Johnson MS, Swanson GT (2006) Determination of binding site residues responsible for the subunit selectivity of novel marine-derived compounds on kainate receptors. Mol Pharmacol 69:1849–1860. [DOI] [PubMed] [Google Scholar]
- Sanderson JL, Gorski JA, Dell’Acqua ML (2016) NMDA receptor-dependent LTD requires transient synaptic incorporation of Ca2+-permeable AMPARs mediated by AKAP150-anchored PKA and calcineurin. Neuron 89:1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson JL, Gorski JA, Gibson ES, Lam P, Freund RK, Chick WS, Dell’Acqua ML (2012) AKAP150-anchored calcineurin regulates synaptic plasticity by limiting synaptic incorporation of Ca2+-permeable AMPA receptors. J Neurosci 32:15036–15052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang CN, Hostetter MP, Gracely RH, Chappell AS, Schoepp DD, Lee G, Whitcup S, Caruso R, Max MB (1998) AMPA/kainate antagonist LY293558 reduces capsaicin-evoked hyperalgesia but not pain in normal skin in humans. Anesthesiology 89:1060–1067. [DOI] [PubMed] [Google Scholar]
- Sang CNRamadan NMWallihan RGChappell ASFreitag FGSmith TRSilberstein SDJohnson KWPhebus LABleakman D, et al. (2004) LY293558, a novel AMPA/GluR5 antagonist, is efficacious and well-tolerated in acute migraine. Cephalalgia 24:596–602. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Levanon EY, Hueske EA, Ambrose JM, Li JB (2012) Activity-dependent A-to-I RNA editing in rat cortical neurons. Genetics 192:281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo Freel RM, Ogden KK, Strong KL, Khatri A, Chepiga KM, Jensen HS, Traynelis SF, Liotta DC (2013) Synthesis and structure activity relationship of tetrahydroisoquinoline-based potentiators of GluN2C and GluN2D containing N-methyl-D-aspartate receptors. J Med Chem 56:5351–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo Freel RM, Ogden KK, Strong KL, Khatri A, Chepiga KM, Jensen HS, Traynelis SF, Liotta DC (2014) Correction to synthesis and structure activity relationship of tetrahydroisoquinoline-based potentiators of GluN2C and GluN2D containing N-methyl-d-aspartate receptors. J Med Chem 57:4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo RM, Acker TM, Zimmerman SS, Katzman BM, Strong KL, Traynelis SF, Liotta DC (2012) Novel NMDA receptor modulators: an update. Expert Opin Ther Pat 22:1337–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Clemente A, Nicoll RA, Roche KW (2013) Diversity in NMDA receptor composition: many regulators, many consequences. Neuroscientist 19:62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota K, Dore K, Tang K, Irvine M, Fang G, Burnell ES, Malinow R, Jane DE, Monaghan DT (2019) The NMDA receptor intracellular C-terminal domains reciprocally interact with allosteric modulators. Biochem Pharmacol 159:140–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota KIrvine MWFang GBurnell ESBannister NVolianskis ACulley GRDravid SMCollingridge GLJane DE, et al. (2017) Mechanism and properties of positive allosteric modulation of N-methyl-d-aspartate receptors by 6-alkyl 2-naphthoic acid derivatives. Neuropharmacology 125:64–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YFRothe TPremkumar LSDas SCui JTalantova MVWong HKGong XChan SFZhang D, et al. (2002) Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons. J Neurophysiol 87:2052–2063. [DOI] [PubMed] [Google Scholar]
- Sather W, Dieudonné S, MacDonald JF, Ascher P (1992) Activation and desensitization of N-methyl-D-aspartate receptors in nucleated outside-out patches from mouse neurones. J Physiol 450:643–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather W, Johnson JW, Henderson G, Ascher P (1990) Glycine-insensitive desensitization of NMDA responses in cultured mouse embryonic neurons. Neuron 4:725–731. [DOI] [PubMed] [Google Scholar]
- Sato K, Kiyama H, Park HT, Tohyama M (1993) AMPA, KA and NMDA receptors are expressed in the rat DRG neurones. Neuroreport 4:1263–1265. [DOI] [PubMed] [Google Scholar]
- Sattler R, Xiong Z, Lu WY, MacDonald JF, Tymianski M (2000) Distinct roles of synaptic and extrasynaptic NMDA receptors in excitotoxicity. J Neurosci 20:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savall BMWu DSwanson DMSeierstad MWu NVives Martinez JGarcía Olmos BLord BCoe KKoudriakova T, et al. (2018) Discovery of imidazo[1,2-a]pyrazines and pyrazolo[1,5-c]pyrimidines as TARP γ-8 selective AMPAR negative modulators. ACS Med Chem Lett 10:267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saver JL (2013) The evolution of technology. Stroke 44(6, Suppl 1)S13–S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savtchenko LP, Rusakov DA (2004) Glutamate escape from a tortuous synaptic cleft of the hippocampal mossy fibre synapse. Neurochem Int 45:479–484. [DOI] [PubMed] [Google Scholar]
- Savtchenko LP, Rusakov DA (2013) Moderate AMPA receptor clustering on the nanoscale can efficiently potentiate synaptic current. Philos Trans R Soc Lond B Biol Sci 369:20130167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saybasili H, Stevens DR, Haas HL (1995) pH-dependent modulation of N-methyl-D-aspartate receptor-mediated synaptic currents by histamine in rat hippocampus in vitro. Neurosci Lett 199:225–227. [DOI] [PubMed] [Google Scholar]
- Sceniak MP, Berry CT, Sabo SL (2012) Facilitation of neocortical presynaptic terminal development by NMDA receptor activation. Neural Dev 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sceniak MP, Fedder KN, Wang Q, Droubi S, Babcock K, Patwardhan S, Wright-Zornes J, Pham L, Sabo SL (2019) An autism-associated mutation in GluN2B prevents NMDA receptor trafficking and interferes with dendrite growth. J Cell Sci 132:jcs232892. [DOI] [PubMed] [Google Scholar]
- Schade S, Paulus W (2016) D-Cycloserine in neuropsychiatric diseases: A systematic review. Int J Neuropsychopharmacol 19:pyv102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder DM, Kuybeda O, Zhang J, Klymko K, Bartesaghi A, Borgnia MJ, Mayer ML, Subramaniam S (2013) Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain. Proc Natl Acad Sci USA 110:5921–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk U, Matteoli M (2004) Presynaptic AMPA receptors: more than just ion channels? Biol Cell 96:257–260. [DOI] [PubMed] [Google Scholar]
- Schenk U, Verderio C, Benfenati F, Matteoli M (2003) Regulated delivery of AMPA receptor subunits to the presynaptic membrane. EMBO J 22:558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer HH, Swanson GT, Heinemann SF (1997) Rat GluR7 and a carboxy-terminal splice variant, GluR7b, are functional kainate receptor subunits with a low sensitivity to glutamate. Neuron 19:1141–1146. [DOI] [PubMed] [Google Scholar]
- Schiller J, Major G, Koester HJ, Schiller Y (2000) NMDA spikes in basal dendrites of cortical pyramidal neurons. Nature 404:285–289. [DOI] [PubMed] [Google Scholar]
- Schlesinger F, Tammena D, Krampfl K, Bufler J (2005) Desensitization and resensitization are independently regulated in human recombinant GluR subunit coassemblies. Synapse 55:176–182. [DOI] [PubMed] [Google Scholar]
- Schmid SM, Körber C, Herrmann S, Werner M, Hollmann M (2007) A domain linking the AMPA receptor agonist binding site to the ion pore controls gating and causes lurcher properties when mutated. J Neurosci 27:12230–12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SM, Kott S, Sager C, Huelsken T, Hollmann M (2009) The glutamate receptor subunit delta2 is capable of gating its intrinsic ion channel as revealed by ligand binding domain transplantation. Proc Natl Acad Sci USA 106:10320–10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HD, McFarland KN, Darnell SB, Huizenga MN, Sangrey GR, Cha JH, Pierce RC, Sadri-Vakili G (2015) ADAR2-dependent GluA2 editing regulates cocaine seeking. Mol Psychiatry 20:1460–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JT (2004) Activity-driven sharpening of the retinotectal projection: the search for retrograde synaptic signaling pathways. J Neurobiol 59:114–133. [DOI] [PubMed] [Google Scholar]
- Schmidt TSamaras PFrejno MGessulat SBarnert MKienegger HKrcmar HSchlegl JEhrlich HCAiche S, et al. (2018) ProteomicsDB. Nucleic Acids Res 46 (D1):D1271–D1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J, Dux E, Gissel C, Paschen W (1996) Regional analysis of developmental changes in the extent of GluR6 mRNA editing in rat brain. Brain Res Dev Brain Res 91:153–157. [DOI] [PubMed] [Google Scholar]
- Schmitz D, Frerking M, Nicoll RA (2000) Synaptic activation of presynaptic kainate receptors on hippocampal mossy fiber synapses. Neuron 27:327–338. [DOI] [PubMed] [Google Scholar]
- Schmitz LJMKlaassen RVRuiperez-Alonso MZamri AEStroeder JRao-Ruiz PLodder JCvan der Loo RJMansvelder HDSmit AB, et al. (2017) The AMPA receptor-associated protein Shisa7 regulates hippocampal synaptic function and contextual memory. eLife 6:e24192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneggenburger R (1998) Altered voltage dependence of fractional Ca2+ current in N-methyl-D-aspartate channel pore mutants with a decreased Ca2+ permeability. Biophys J 74:1790–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Hohendorf TMohan HBien CGBreuer JBecker AGörlich DKuhlmann TWidman GHerich SElpers C, et al. (2016) CD8(+) T-cell pathogenicity in Rasmussen encephalitis elucidated by large-scale T-cell receptor sequencing. Nat Commun 7:11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober DA, Gill MB, Yu H, Gernert DL, Jeffries MW, Ornstein PL, Kato AS, Felder CC, Bredt DS (2011) Transmembrane AMPA receptor regulatory proteins and cornichon-2 allosterically regulate AMPA receptor antagonists and potentiators. J Biol Chem 286:13134–13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepp DD, Smith CL, Lodge D, Millar JD, Leander JD, Sacaan AI, Lunn WH (1991) D,L-(tetrazol-5-yl) glycine: a novel and highly potent NMDA receptor agonist. Eur J Pharmacol 203:237–243. [DOI] [PubMed] [Google Scholar]
- Schönrock M, Thiel G, Laube B (2019) Coupling of a viral K+-channel with a glutamate-binding-domain highlights the modular design of ionotropic glutamate-receptors. Commun Biol 2:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorge S, Colquhoun D (2003) Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J Neurosci 23:1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorge S, Elenes S, Colquhoun D (2005) Maximum likelihood fitting of single channel NMDA activity with a mechanism composed of independent dimers of subunits. J Physiol 569:395–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüler T, Mesic I, Madry C, Bartholomäus I, Laube B (2008) Formation of NR1/NR2 and NR1/NR3 heterodimers constitutes the initial step in N-methyl-D-aspartate receptor assembly. J Biol Chem 283:37–46. [DOI] [PubMed] [Google Scholar]
- Schwarting AMöckel TLütgendorf FTriantafyllias KGrella SBoedecker SWeinmann AMeineck MSommer CSchermuly I, et al. (2019) Fatigue in SLE: diagnostic and pathogenic impact of anti-N-methyl-D-aspartate receptor (NMDAR) autoantibodies. Ann Rheum Dis 78:1226–1234. [DOI] [PubMed] [Google Scholar]
- Schwartzkroin PA, Wester K (1975) Long-lasting facilitation of a synaptic potential following tetanization in the in vitro hippocampal slice. Brain Res 89:107–119. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Baehrens D, Haupt A, Bildl W, Boudkkazi S, Roeper J, Fakler B, Schulte U (2014) Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron 84:41–54. [DOI] [PubMed] [Google Scholar]
- Schwenk JBoudkkazi SKocylowski MKBrechet AZolles GBus TCosta KKollewe AJordan JBank J, et al. (2019) An ER assembly line of AMPA-receptors controls excitatory neurotransmission and its plasticity. Neuron 104:680–692.e9. [DOI] [PubMed] [Google Scholar]
- Schwenk JHarmel NBrechet AZolles GBerkefeld HMüller CSBildl WBaehrens DHüber BKulik A, et al. (2012) High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron 74:621–633. [DOI] [PubMed] [Google Scholar]
- Schwenk JHarmel NZolles GBildl WKulik AHeimrich BChisaka OJonas PSchulte UFakler B, et al. (2009) Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323:1313–1319. [DOI] [PubMed] [Google Scholar]
- Scimemi A, Fine A, Kullmann DM, Rusakov DA (2004) NR2B-containing receptors mediate cross talk among hippocampal synapses. J Neurosci 24:4767–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DB, Blanpied TA, Ehlers MD (2003) Coordinated PKA and PKC phosphorylation suppresses RXR-mediated ER retention and regulates the surface delivery of NMDA receptors. Neuropharmacology 45:755–767. [DOI] [PubMed] [Google Scholar]
- Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD (2001) An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci 21:3063–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DB, Michailidis I, Mu Y, Logothetis D, Ehlers MD (2004) Endocytosis and degradative sorting of NMDA receptors by conserved membrane-proximal signals. J Neurosci 24:7096–7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg PH (1993) The TiPS/TINS lecture: the molecular biology of mammalian glutamate receptor channels. Trends Pharmacol Sci 14:297–303. [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Burnashev N, Köhr G, Kuner T, Sprengel R, Monyer H (1995) The NMDA receptor channel: molecular design of a coincidence detector. Recent Prog Horm Res 50:19–34. [DOI] [PubMed] [Google Scholar]
- Segerstråle M, Juuri J, Lanore F, Piepponen P, Lauri SE, Mulle C, Taira T (2010) High firing rate of neonatal hippocampal interneurons is caused by attenuation of afterhyperpolarizing potassium currents by tonically active kainate receptors. J Neurosci 30:6507–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G, Steinhäuser C (1995) Glial cells in the mouse hippocampus express AMPA receptors with an intermediate Ca2+ permeability. Eur J Neurosci 7:1872–1881. [DOI] [PubMed] [Google Scholar]
- Seifi M, Swinny JD (2016) Immunolocalization of AMPA receptor subunits within the enteric nervous system of the mouse colon and the effect of their activation on spontaneous colonic contractions. Neurogastroenterol Motil 28:705–720. [DOI] [PubMed] [Google Scholar]
- Sekiguchi M, Fleck MW, Mayer ML, Takeo J, Chiba Y, Yamashita S, Wada K (1997) A novel allosteric potentiator of AMPA receptors: 4--2-(phenylsulfonylamino)ethylthio--2,6-difluoro-phenoxyaceta mide. J Neurosci 17:5760–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi M, Nishikawa K, Aoki S, Wada K (2002) A desensitization-selective potentiator of AMPA-type glutamate receptors. Br J Pharmacol 136:1033–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenov A, Möykkynen T, Coleman SK, Korpi ER, Keinänen K (2012) Autoinactivation of the stargazin-AMPA receptor complex: subunit-dependency and independence from physical dissociation. PLoS One 7:e49282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengar ASLi HZhang WLeung CRamani AKSaw NMWang YTu YRoss PJScherer SW, et al. (2019) Control of long-term synaptic potentiation and learning by alternative splicing of the NMDA receptor subunit GluN1. Cell Rep 29:4285–4294.e5. [DOI] [PubMed] [Google Scholar]
- Seong HJ, Behnia R, Carter AG (2014) Impact of subthreshold membrane potential on synaptic responses at dendritic spines of layer 5 pyramidal neurons in the prefrontal cortex. J Neurophysiol 111:1960–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda FJ, Bustos FJ, Inostroza E, Zúñiga FA, Neve RL, Montecino M, van Zundert B (2010) Differential roles of NMDA receptor subtypes NR2A and NR2B in dendritic branch development and requirement of RasGRF1. J Neurophysiol 103:1758–1770. [DOI] [PubMed] [Google Scholar]
- Sequerra EB, Goyal R, Castro PA, Levin JB, Borodinsky LN (2018) NMDA receptor signaling is important for neural tube formation and for preventing antiepileptic drug-induced neural tube defects. J Neurosci 38:4762–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serulle Y, Zhang S, Ninan I, Puzzo D, McCarthy M, Khatri L, Arancio O, Ziff EB (2007) A GluR1-cGKII interaction regulates AMPA receptor trafficking. Neuron 56:670–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessoms-Sikes S, Honse Y, Lovinger DM, Colbran RJ (2005) CaMKIIalpha enhances the desensitization of NR2B-containing NMDA receptors by an autophosphorylation-dependent mechanism. Mol Cell Neurosci 29:139–147. [DOI] [PubMed] [Google Scholar]
- Shadlen MN, Newsome WT (1994) Noise, neural codes and cortical organization. Curr Opin Neurobiol 4:569–579. [DOI] [PubMed] [Google Scholar]
- Shaffer CLHurst RSScialis RJOsgood SMBryce DKHoffmann WELazzaro JTHanks ANLotarski SWeber ML, et al. (2013) Positive allosteric modulation of AMPA receptors from efficacy to toxicity: the interspecies exposure-response continuum of the novel potentiator PF-4778574. J Pharmacol Exp Ther 347:212–224. [DOI] [PubMed] [Google Scholar]
- Shaffer CL, Osgood SM, Smith DL, Liu J, Trapa PE (2014) Enhancing ketamine translational pharmacology via receptor occupancy normalization. Neuropharmacology 86:174–180. [DOI] [PubMed] [Google Scholar]
- Shaffer CLPatel NCSchwarz JScialis RJWei YHou XJXie LKarki KBryce DKOsgood SM, et al. (2015) The discovery and characterization of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor potentiator N-(3S,4S)-4-[4-(5-cyano-2-thienyl)phenoxy]tetrahydrofuran-3-ylpropane-2-sulfonamide (PF-04958242). J Med Chem 58:4291–4308. [DOI] [PubMed] [Google Scholar]
- Shaikh SA, Dolino DM, Lee G, Chatterjee S, MacLean DM, Flatebo C, Landes CF, Jayaraman V (2016) Stargazin modulation of AMPA receptors. Cell Rep 17:328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks NF, Cais O, Maruo T, Savas JN, Zaika EI, Azumaya CM, Yates JR 3rd, Greger I, Nakagawa T (2014) Molecular dissection of the interaction between the AMPA receptor and cornichon homolog-3. J Neurosci 34:12104–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks NF, Maruo T, Farina AN, Ellisman MH, Nakagawa T (2010) Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J Neurosci 30:2728–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks NFSavas JNMaruo TCais OHirao AOe SGhosh ANoda YGreger IHYates JR 3rd, et al. (2012) Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Rep 1:590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G, Stevens CF (1996) Interactions between two divalent ion binding sites in N-methyl-D-aspartate receptor channels. Proc Natl Acad Sci USA 93:14170–14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She K, Ferreira JS, Carvalho AL, Craig AM (2012) Glutamate binding to the GluN2B subunit controls surface trafficking of N-methyl-D-aspartate (NMDA) receptors. J Biol Chem 287:27432–27445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheardown MJ, Nielsen EO, Hansen AJ, Jacobsen P, Honoré T (1990) 2,3-Dihydroxy-6-nitro-7-sulfamoyl-benzo(F)quinoxaline: a neuroprotectant for cerebral ischemia. Science 247:571–574. [DOI] [PubMed] [Google Scholar]
- Sheinin A, Shavit S, Benveniste M (2001) Subunit specificity and mechanism of action of NMDA partial agonist D-cycloserine. Neuropharmacology 41:151–158. [DOI] [PubMed] [Google Scholar]
- Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY (1994) Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature 368:144–147. [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim E (2011) The postsynaptic organization of synapses. Cold Spring Harb Perspect Biol 3:a005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng N, Bemben MA, Díaz-Alonso J, Tao W, Shi YS, Nicoll RA (2018) LTP requires postsynaptic PDZ-domain interactions with glutamate receptor/auxiliary protein complexes. Proc Natl Acad Sci USA 115:3948–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng N, Shi YS, Lomash RM, Roche KW, Nicoll RA (2015) Neto auxiliary proteins control both the trafficking and biophysical properties of the kainate receptor GluK1. eLife 4:e11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng N, Shi YS, Nicoll RA (2017) Amino-terminal domains of kainate receptors determine the differential dependence on Neto auxiliary subunits for trafficking. Proc Natl Acad Sci USA 114:1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z, Dai Q, Prorok M, Castellino FJ (2007) Subtype-selective antagonism of N-methyl-D-aspartate receptor ion channels by synthetic conantokin peptides. Neuropharmacology 53:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z, Liang Z, Geiger JH, Prorok M, Castellino FJ (2009) The selectivity of conantokin-G for ion channel inhibition of NR2B subunit-containing NMDA receptors is regulated by amino acid residues in the S2 region of NR2B. Neuropharmacology 57:127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z, Prorok M, Castellino FJ (2010) Specific determinants of conantokins that dictate their selectivity for the NR2B subunit of N-methyl-D-aspartate receptors. Neuroscience 170:703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL (2007) The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol 23:613–643. [DOI] [PubMed] [Google Scholar]
- Shepherd TAAikins JABleakman DCantrell BERearick JPSimon RLSmith ECStephenson GAZimmerman DMMandelzys A, et al. (2002) Design and synthesis of a novel series of 1,2-disubstituted cyclopentanes as small, potent potentiators of 2-amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propanoic acid (AMPA) receptors. J Med Chem 45:2101–2111. [DOI] [PubMed] [Google Scholar]
- Shi EY, Yuan CL, Sipple MT, Srinivasan J, Ptak CP, Oswald RE, Nowak LM (2019) Noncompetitive antagonists induce cooperative AMPA receptor channel gating. J Gen Physiol 151:156–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Hayashi Y, Esteban JA, Malinow R (2001) Subunit-specific rules governing AMPA receptor trafficking to synapses in hippocampal pyramidal neurons. Cell 105:331–343. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lu W, Milstein AD, Nicoll RA (2009) The stoichiometry of AMPA receptors and TARPs varies by neuronal cell type. Neuron 62:633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Suh YH, Milstein AD, Isozaki K, Schmid SM, Roche KW, Nicoll RA (2010) Functional comparison of the effects of TARPs and cornichons on AMPA receptor trafficking and gating. Proc Natl Acad Sci USA 107:16315–16319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Watanabe M, Ichikawa R, Inoue Y, Koyanagi T (1999) Different expressions of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid and N-methyl-D-aspartate receptor subunit mRNAs between visceromotor and somatomotor neurons of the rat lumbosacral spinal cord. J Comp Neurol 404:172–182. [DOI] [PubMed] [Google Scholar]
- Shigetomi E, Bowser DN, Sofroniew MV, Khakh BS (2008) Two forms of astrocyte calcium excitability have distinct effects on NMDA receptor-mediated slow inward currents in pyramidal neurons. J Neurosci 28:6659–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin WKim KSerraz BCho YSKim DKang MLee EJLee HBae YCPaoletti P, et al. (2020) Early correction of synaptic long-term depression improves abnormal anxiety-like behavior in adult GluN2B-C456Y-mutant mice. PLoS Biol 18:e3000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y (2012) Quantification of postsynaptic density proteins: glutamate receptor subunits and scaffolding proteins. Hippocampus 22:942–953. [DOI] [PubMed] [Google Scholar]
- Shinohara Y, Hirase H, Watanabe M, Itakura M, Takahashi M, Shigemoto R (2008) Left-right asymmetry of the hippocampal synapses with differential subunit allocation of glutamate receptors. Proc Natl Acad Sci USA 105:19498–19503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipton OA, Paulsen O (2013) GluN2A and GluN2B subunit-containing NMDA receptors in hippocampal plasticity. Philos Trans R Soc Lond B Biol Sci 369:20130163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroma PR, Thuras P, Wels J, Albott CS, Erbes C, Tye S, Lim KO (2020) A randomized, double-blind, active placebo-controlled study of efficacy, safety, and durability of repeated vs single subanesthetic ketamine for treatment-resistant depression. Transl Psychiatry 10:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuang M, Liu J, Jia MX, Yang JZ, Wu SP, Gong XH, Ling YS, Ruan Y, Yang XL, Zhang D (2004) Family-based association study between autism and glutamate receptor 6 gene in Chinese Han trios. Am J Med Genet B Neuropsychiatr Genet 131B:48–50. [DOI] [PubMed] [Google Scholar]
- Shypshyna MS, Veselovsky NS (2015) Presynaptic Ca2+-permeable AMPA-receptors modulate paired-pulse depression in nociceptive sensory synapses. Neurosci Lett 585:1–5. [DOI] [PubMed] [Google Scholar]
- Sia GM, Béïque JC, Rumbaugh G, Cho R, Worley PF, Huganir RL (2007) Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron 55:87–102. [DOI] [PubMed] [Google Scholar]
- Siegler Retchless B, Gao W, Johnson JW (2012) A single GluN2 subunit residue controls NMDA receptor channel properties via intersubunit interaction. Nat Neurosci 15:406-413, S401-402. [DOI] [PMC free article] [PubMed]
- Sierra RO, Nítola LP, Duran JM, Prieto DR, León LA, Cardenas FP (2016) Medial orbitofrontal cortex lesion prevents facilitatory effects of d-cycloserine during fear extinction. Behav Brain Res 296:379–383. [DOI] [PubMed] [Google Scholar]
- Sigler A, Oh WC, Imig C, Altas B, Kawabe H, Cooper BH, Kwon HB, Rhee JS, Brose N (2017) Formation and Maintenance of Functional Spines in the Absence of Presynaptic Glutamate Release. Neuron 94:304–311.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sihra TS, Rodríguez-Moreno A (2013) Presynaptic kainate receptor-mediated bidirectional modulatory actions: mechanisms. Neurochem Int 62:982–987. [DOI] [PubMed] [Google Scholar]
- Silver RA, Colquhoun D, Cull-Candy SG, Edmonds B (1996a) Deactivation and desensitization of non-NMDA receptors in patches and the time course of EPSCs in rat cerebellar granule cells. J Physiol 493:167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver RA, Cull-Candy SG, Takahashi T (1996b) Non-NMDA glutamate receptor occupancy and open probability at a rat cerebellar synapse with single and multiple release sites. J Physiol 494:231–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver RA, Traynelis SF, Cull-Candy SG (1992) Rapid-time-course miniature and evoked excitatory currents at cerebellar synapses in situ. Nature 355:163–166. [DOI] [PubMed] [Google Scholar]
- Simmons DA, Mehta RA, Lauterborn JC, Gall CM, Lynch G (2011) Brief ampakine treatments slow the progression of Huntington’s disease phenotypes in R6/2 mice. Neurobiol Dis 41:436–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DK, Prusky GT, O’Leary DD, Constantine-Paton M (1992) N-methyl-D-aspartate receptor antagonists disrupt the formation of a mammalian neural map. Proc Natl Acad Sci USA 89:10593–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin WC, Haas K, Ruthazer ES, Cline HT (2002) Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature 419:475–480. [DOI] [PubMed] [Google Scholar]
- Singewald N, Schmuckermair C, Whittle N, Holmes A, Ressler KJ (2015) Pharmacology of cognitive enhancers for exposure-based therapy of fear, anxiety and trauma-related disorders. Pharmacol Ther 149:150–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnen BL, Bowen AB, Forte JS, Hiester BG, Crosby KC, Gibson ES, Dell’Acqua ML, Kennedy MJ (2017) Optogenetic control of synaptic composition and function. Neuron 93:646–660.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrieh RE, MacLean DM, Jayaraman V (2015a) A conserved structural mechanism of NMDA receptor inhibition: A comparison of ifenprodil and zinc. J Gen Physiol 146:173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrieh RE, MacLean DM, Jayaraman V (2015b) Subtype-dependent N-methyl-D-aspartate receptor amino-terminal domain conformations and modulation by spermine. J Biol Chem 290:12812–12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitzia G, Mantas I, Zhang X, Svenningsson P, Chergui K (2020) NMDA receptors are altered in the substantia nigra pars reticulata and their blockade ameliorates motor deficits in experimental parkinsonism. Neuropharmacology 174:108136. [DOI] [PubMed] [Google Scholar]
- Sivakumar V, Ling EA, Lu J, Kaur C (2010) Role of glutamate and its receptors and insulin-like growth factors in hypoxia induced periventricular white matter injury. Glia 58:507–523. [DOI] [PubMed] [Google Scholar]
- Sjöström PJ, Turrigiano GG, Nelson SB (2001) Rate, timing, and cooperativity jointly determine cortical synaptic plasticity. Neuron 32:1149–1164. [DOI] [PubMed] [Google Scholar]
- Sjöström PJ, Turrigiano GG, Nelson SB (2003) Neocortical LTD via coincident activation of presynaptic NMDA and cannabinoid receptors. Neuron 39:641–654. [DOI] [PubMed] [Google Scholar]
- Skrenkova K, Hemelikova K, Kolcheva M, Kortus S, Kaniakova M, Krausova B, Horak M (2019) Structural features in the glycine-binding sites of the GluN1 and GluN3A subunits regulate the surface delivery of NMDA receptors. Sci Rep 9:12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrenkova KSong JMKortus SKolcheva MNetolicky JHemelikova KKaniakova MKrausova BHKucera TKorabecny J, et al. (2020) The pathogenic S688Y mutation in the ligand-binding domain of the GluN1 subunit regulates the properties of NMDA receptors. Sci Rep 10:18576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavikova B, Chodounska H, Nekardova M, Vyklicky V, Ladislav M, Hubalkova P, Krausova B, Vyklicky L, Kudova E (2016) Neurosteroid-like inhibitors of N-methyl-d-aspartate receptor: Substituted 2-sulfates and 2-hemisuccinates of perhydrophenanthrene. J Med Chem 59:4724–4739. [DOI] [PubMed] [Google Scholar]
- Small B, Thomas J, Kemp M, Hoo K, Ballyk B, Deverill M, Ogden AM, Rubio A, Pedregal C, Bleakman D (1998) LY339434, a GluR5 kainate receptor agonist. Neuropharmacology 37:1261–1267. [DOI] [PubMed] [Google Scholar]
- Smart TG, Hosie AM, Miller PS (2004) Zn2+ ions: modulators of excitatory and inhibitory synaptic activity. Neuroscientist 10:432–442. [DOI] [PubMed] [Google Scholar]
- Smidkova M, Hajek M, Adla SK, Slavikova B, Chodounska H, Matousova M, Mertlikova-Kaiserova H, Kudova E (2019) Screening of novel 3α5β-neurosteroids for neuroprotective activity against glutamate- or NMDA-induced excitotoxicity. J Steroid Biochem Mol Biol 189:195–203. [DOI] [PubMed] [Google Scholar]
- Smith M, Spence MA, Flodman P (2009) Nuclear and mitochondrial genome defects in autisms. Ann N Y Acad Sci 1151:102–132. [DOI] [PubMed] [Google Scholar]
- Smith ML, von Hanwehr R, Siesjö BK (1986) Changes in extra- and intracellular pH in the brain during and following ischemia in hyperglycemic and in moderately hypoglycemic rats. J Cereb Blood Flow Metab 6:574–583. [DOI] [PubMed] [Google Scholar]
- Smith SE, Dürmüller N, Meldrum BS (1991) The non-N-methyl-D-aspartate receptor antagonists, GYKI 52466 and NBQX are anticonvulsant in two animal models of reflex epilepsy. Eur J Pharmacol 201:179–183. [DOI] [PubMed] [Google Scholar]
- Smith SL, Smith IT, Branco T, Häusser M (2013) Dendritic spikes enhance stimulus selectivity in cortical neurons in vivo. Nature 503:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TC, Howe JR (2000) Concentration-dependent substate behavior of native AMPA receptors. Nat Neurosci 3:992–997. [DOI] [PubMed] [Google Scholar]
- Smith TC, Wang LY, Howe JR (2000) Heterogeneous conductance levels of native AMPA receptors. J Neurosci 20:2073–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits JAJPollack MHRosenfield DOtto MWDowd SCarpenter JDutcher CDLewis EMWitcraft SMPapini S, et al. (2020) Dose timing of D-cycloserine to augment exposure therapy for social anxiety disorder: A randomized clinical trial. JAMA Netw Open 3:e206777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers CT, Woodward JJ (2003) Effect of the NR3 subunit on ethanol inhibition of recombinant NMDA receptors. Brain Res 987:117–121. [DOI] [PubMed] [Google Scholar]
- Smothers CT, Woodward JJ (2007) Pharmacological characterization of glycine-activated currents in HEK 293 cells expressing N-methyl-D-aspartate NR1 and NR3 subunits. J Pharmacol Exp Ther 322:739–748. [DOI] [PubMed] [Google Scholar]
- Smothers CT, Woodward JJ (2009) Expression of glycine-activated diheteromeric NR1/NR3 receptors in human embryonic kidney 293 cells Is NR1 splice variant-dependent. J Pharmacol Exp Ther 331:975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky A, Koshelev S (1998) Two blocking sites of amino-adamantane derivatives in open N-methyl-D-aspartate channels. Biophys J 74:1305–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI (1999) Two-component blocking kinetics of open NMDA channels by organic cations. Biochim Biophys Acta 1416:69–91. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Beck C, Wollmuth LP (2002a) Molecular rearrangements of the extracellular vestibule in NMDAR channels during gating. Neuron 33:75–85. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Koshelev SG, Khodorov BI (1998) Interaction of memantine and amantadine with agonist-unbound NMDA-receptor channels in acutely isolated rat hippocampal neurons. J Physiol 512:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Prodromou ML, Yelshansky MV, Wollmuth LP (2007) Subunit-specific contribution of pore-forming domains to NMDA receptor channel structure and gating. J Gen Physiol 129:509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rooney L, Wollmuth LP (2002b) Staggering of subunits in NMDAR channels. Biophys J 83:3304–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E (2009) X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462:745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Yelshansky MV (2000) The trapping block of NMDA receptor channels in acutely isolated rat hippocampal neurones. J Physiol 526:493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn H, Park M (2019) Palmitoylation-mediated synaptic regulation of AMPA receptor trafficking and function. Arch Pharm Res 42:426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltys J, Liu Y, Ritchie A, Wemlinger S, Schaller K, Schumann H, Owens GP, Bennett JL (2019) Membrane assembly of aquaporin-4 autoantibodies regulates classical complement activation in neuromyelitis optica. J Clin Invest 129:2000–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sólyom S, Tarnawa I (2002) Non-competitive AMPA antagonists of 2,3-benzodiazepine type. Curr Pharm Des 8:913–939. [DOI] [PubMed] [Google Scholar]
- Sommer B, Burnashev N, Verdoorn TA, Keinänen K, Sakmann B, Seeburg PH (1992) A glutamate receptor channel with high affinity for domoate and kainate. EMBO J 11:1651–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Keinänen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Köhler M, Takagi T, Sakmann B, Seeburg PH (1990) Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science 249:1580–1585. [DOI] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, Seeburg PH (1991) RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67:11–19. [DOI] [PubMed] [Google Scholar]
- Song X, Jensen MO, Jogini V, Stein RA, Lee CH, Mchaourab HS, Shaw DE, Gouaux E (2018) Mechanism of NMDA receptor channel block by MK-801 and memantine. Nature 556:515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria JM, Valdeolmillos M (2002) Receptor-activated calcium signals in tangentially migrating cortical cells. Cereb Cortex 12:831–839. [DOI] [PubMed] [Google Scholar]
- Soto D, Coombs ID, Gratacòs-Batlle E, Farrant M, Cull-Candy SG (2014) Molecular mechanisms contributing to TARP regulation of channel conductance and polyamine block of calcium-permeable AMPA receptors. J Neurosci 34:11673–11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto D, Coombs ID, Kelly L, Farrant M, Cull-Candy SG (2007) Stargazin attenuates intracellular polyamine block of calcium-permeable AMPA receptors. Nat Neurosci 10:1260–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto D, Coombs ID, Renzi M, Zonouzi M, Farrant M, Cull-Candy SG (2009) Selective regulation of long-form calcium-permeable AMPA receptors by an atypical TARP, gamma-5. Nat Neurosci 12:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto DOlivella MGrau CArmstrong JAlcon CGasull XSantos-Gómez ALocubiche SGómez de Salazar MGarcía-Díaz R, et al. (2019) L-Serine dietary supplementation is associated with clinical improvement of loss-of-function GRIN2B-related pediatric encephalopathy. Sci Signal 12:eaaw0936. [DOI] [PubMed] [Google Scholar]
- Sotres-Bayon F, Bush DE, LeDoux JE (2007) Acquisition of fear extinction requires activation of NR2B-containing NMDA receptors in the lateral amygdala. Neuropsychopharmacology 32:1929–1940. [DOI] [PubMed] [Google Scholar]
- Sotres-Bayon F, Diaz-Mataix L, Bush DE, LeDoux JE (2009) Dissociable roles for the ventromedial prefrontal cortex and amygdala in fear extinction: NR2B contribution. Cereb Cortex 19:474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumier A, Carter RM, Schoenfeld TJ, Cameron HA (2016) New hippocampal neurons mature rapidly in response to ketamine but are not required for its acute antidepressant effects on neophagia in rats. eNeuro 3:ENEURO.0116-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengel R, Seeburg PH (1993) The unique properties of glutamate receptor channels. FEBS Lett 325:90–94. [DOI] [PubMed] [Google Scholar]
- Sprengel RSuchanek BAmico CBrusa RBurnashev NRozov AHvalby OJensen VPaulsen OAndersen P, et al. (1998) Importance of the intracellular domain of NR2 subunits for NMDA receptor function in vivo. Cell 92:279–289. [DOI] [PubMed] [Google Scholar]
- Standaert DG, Landwehrmeyer GB, Kerner JA, Penney JB Jr, Young AB (1996) Expression of NMDAR2D glutamate receptor subunit mRNA in neurochemically identified interneurons in the rat neostriatum, neocortex and hippocampus. Brain Res Mol Brain Res 42:89–102. [DOI] [PubMed] [Google Scholar]
- Standaert DG, Testa CM, Young AB, Penney JB Jr (1994) Organization of N-methyl-D-aspartate glutamate receptor gene expression in the basal ganglia of the rat. J Comp Neurol 343:1–16. [DOI] [PubMed] [Google Scholar]
- Standley S, Baudry M (2000) The role of glycosylation in ionotropic glutamate receptor ligand binding, function, and trafficking. Cell Mol Life Sci 57:1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stäubli U, Perez Y, Xu FB, Rogers G, Ingvar M, Stone-Elander S, Lynch G (1994) Centrally active modulators of glutamate receptors facilitate the induction of long-term potentiation in vivo. Proc Natl Acad Sci USA 91:11158–11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawarski MHernandez RXFeghhi TBorycz JALu ZAgarwal ABReihl KDTavora RLau AWCMeinertzhagen IA, et al. (2020) Neuronal glutamatergic synaptic clefts alkalinize rather than acidify during neurotransmission. J Neurosci 40:1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenland HW, Kim SS, Zhuo M (2008) GluR3 subunit regulates sleep, breathing and seizure generation. Eur J Neurosci 27:1166–1173. [DOI] [PubMed] [Google Scholar]
- Stefani A, Chen Q, Flores-Hernandez J, Jiao Y, Reiner A, Surmeier DJ (1998) Physiological and molecular properties of AMPA/Kainate receptors expressed by striatal medium spiny neurons. Dev Neurosci 20:242–252. [DOI] [PubMed] [Google Scholar]
- Stegenga SL, Kalb RG (2001) Developmental regulation of N-methyl-D-aspartate- and kainate-type glutamate receptor expression in the rat spinal cord. Neuroscience 105:499–507. [DOI] [PubMed] [Google Scholar]
- Steigerwald F, Schulz TW, Schenker LT, Kennedy MB, Seeburg PH, Köhr G (2000) C-Terminal truncation of NR2A subunits impairs synaptic but not extrasynaptic localization of NMDA receptors. J Neurosci 20:4573–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein IS, Gray JA, Zito K (2015) Non-ionotropic NMDA receptor signaling drives activity-induced dendritic spine shrinkage. J Neurosci 35:12303–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein IS, Park DK, Flores JC, Jahncke JN, Zito K (2020) Molecular mechanisms of non-ionotropic NMDA receptor signaling in dendritic spine shrinkage. J Neurosci 40:3741–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner J, Teegen B, Schiltz K, Bernstein HG, Stoecker W, Bogerts B (2014) Prevalence of N-methyl-D-aspartate receptor autoantibodies in the peripheral blood: healthy control samples revisited. JAMA Psychiatry 71:838–839. [DOI] [PubMed] [Google Scholar]
- Stensbøl TB, Madsen U, Krogsgaard-Larsen P (2002) The AMPA receptor binding site: focus on agonists and competitive antagonists. Curr Pharm Des 8:857–872. [DOI] [PubMed] [Google Scholar]
- Stenum-Berg C, Musgaard M, Chavez-Abiega S, Thisted CL, Barrella L, Biggin PC, Kristensen AS (2019) Mutational analysis and modeling of negative allosteric modulator binding sites in AMPA receptors. Mol Pharmacol 96:835–850. [DOI] [PubMed] [Google Scholar]
- Stern-Bach Y, Bettler B, Hartley M, Sheppard PO, O’Hara PJ, Heinemann SF (1994) Agonist selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid-binding proteins. Neuron 13:1345–1357. [DOI] [PubMed] [Google Scholar]
- Stern-Bach Y, Russo S, Neuman M, Rosenmund C (1998) A point mutation in the glutamate binding site blocks desensitization of AMPA receptors. Neuron 21:907–918. [DOI] [PubMed] [Google Scholar]
- Stern P, Béhé P, Schoepfer R, Colquhoun D (1992) Single-channel conductances of NMDA receptors expressed from cloned cDNAs: comparison with native receptors. Proc Biol Sci 250:271–277. [DOI] [PubMed] [Google Scholar]
- Storey GP, Opitz-Araya X, Barria A (2011) Molecular determinants controlling NMDA receptor synaptic incorporation. J Neurosci 31:6311–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack S, Colbran RJ (1998) Autophosphorylation-dependent targeting of calcium/ calmodulin-dependent protein kinase II by the NR2B subunit of the N-methyl- D-aspartate receptor. J Biol Chem 273:20689–20692. [DOI] [PubMed] [Google Scholar]
- Strack S, McNeill RB, Colbran RJ (2000) Mechanism and regulation of calcium/calmodulin-dependent protein kinase II targeting to the NR2B subunit of the N-methyl-D-aspartate receptor. J Biol Chem 275:23798–23806. [DOI] [PubMed] [Google Scholar]
- Straub C, Hunt DL, Yamasaki M, Kim KS, Watanabe M, Castillo PE, Tomita S (2011a) Distinct functions of kainate receptors in the brain are determined by the auxiliary subunit Neto1. Nat Neurosci 14:866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub CNoam YNomura TYamasaki MYan DFernandes HBZhang PHowe JRWatanabe MContractor A, et al. (2016) Distinct subunit domains govern synaptic stability and specificity of the kainate receptor. Cell Rep 16:531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub C, Tomita S (2012) The regulation of glutamate receptor trafficking and function by TARPs and other transmembrane auxiliary subunits. Curr Opin Neurobiol 22:488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub C, Zhang W, Howe JR (2011b) Neto2 modulation of kainate receptors with different subunit compositions. J Neurosci 31:8078–8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehle EMYu LRosenfeld JADonkervoort SZhou YChen TJMartinez JEFan YSBarbouth DZhu H, et al. (2012) Genotype-phenotype analysis of 4q deletion syndrome: proposal of a critical region. Am J Med Genet A 158A:2139–2151. [DOI] [PubMed] [Google Scholar]
- Strehlow VHeyne HOVlaskamp DRMMarwick KFMRudolf Gde Bellescize JBiskup SBrilstra EHBrouwer OFCallenbach PMC, et al. ; GRIN2A study group (2019) GRIN2A-related disorders: genotype and functional consequence predict phenotype. Brain 142:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D, Buhl DL, Knafels JD, Chanda PK, Green M, Sciabola S, Mony L, Paoletti P, Pandit J (2016) A Novel Binding Mode Reveals Two Distinct Classes of NMDA Receptor GluN2B-selective Antagonists. Mol Pharmacol 89:541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D, Carvalho S, Grand T, Zhu S, Paoletti P (2014) Controlling NMDA receptor subunit composition using ectopic retention signals. J Neurosci 34:16630–16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D, Paoletti P (2021) Architecture and function of NMDA receptors: an evolutionary perspective. J Physiol 599:2615–2638. [DOI] [PubMed] [Google Scholar]
- Strong KL, Epplin MP, Bacsa J, Butch CJ, Burger PB, Menaldino DS, Traynelis SF, Liotta DC (2017) The structure-activity relationship of a tetrahydroisoquinoline class of N-methyl-d-aspartate receptor modulators that potentiates GluN2B-containing N-methyl-d-aspartate receptors. J Med Chem 60:5556–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong KLEpplin MPOgden KKBurger PBKaiser TMWilding TJKusumoto HCamp CRShaulsky GBhattacharya S, et al. (2021) Distinct GluN1 and GluN2 structural determinants for subunit-selective positive allosteric modulation of N-methyl-d-aspartate receptors. ACS Chem Neurosci 12:79–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong KL, Jing Y, Prosser AR, Traynelis SF, Liotta DC (2014) NMDA receptor modulators: an updated patent review (2013-2014). Expert Opin Ther Pat 24:1349–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutz-Seebohm N, Korniychuk G, Schwarz R, Baltaev R, Ureche ON, Mack AF, Ma ZL, Hollmann M, Lang F, Seebohm G (2006) Functional significance of the kainate receptor GluR6(M836I) mutation that is linked to autism. Cell Physiol Biochem 18:287–294. [DOI] [PubMed] [Google Scholar]
- Stuart GJ, Spruston N (2015) Dendritic integration: 60 years of progress. Nat Neurosci 18:1713–1721. [DOI] [PubMed] [Google Scholar]
- Studniarczyk D, Coombs I, Cull-Candy SG, Farrant M (2013) TARP γ-7 selectively enhances synaptic expression of calcium-permeable AMPARs. Nat Neurosci 16:1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucher NJ, Akbarian S, Chi CL, Leclerc CL, Awobuluyi M, Deitcher DL, Wu MK, Yuan JP, Jones EG, Lipton SA (1995) Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J Neurosci 15:6509–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucher NJ, Kohler K, Tenneti L, Wong HK, Gründer T, Fauser S, Wheeler-Schilling T, Nakanishi N, Lipton SA, Guenther E (2003) N-methyl-D-aspartate receptor subunit NR3A in the retina: developmental expression, cellular localization, and functional aspects. Invest Ophthalmol Vis Sci 44:4451–4456. [DOI] [PubMed] [Google Scholar]
- Sugihara H, Moriyoshi K, Ishii T, Masu M, Nakanishi S (1992) Structures and properties of seven isoforms of the NMDA receptor generated by alternative splicing. Biochem Biophys Res Commun 185:826–832. [DOI] [PubMed] [Google Scholar]
- Sukumaran M, Penn AC, Greger IH (2012) AMPA receptor assembly: atomic determinants and built-in modulators. Adv Exp Med Biol 970:241–264. [DOI] [PubMed] [Google Scholar]
- Sukumaran M, Rossmann M, Shrivastava I, Dutta A, Bahar I, Greger IH (2011) Dynamics and allosteric potential of the AMPA receptor N-terminal domain. EMBO J 30:972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumioka A, Brown TE, Kato AS, Bredt DS, Kauer JA, Tomita S (2011) PDZ binding of TARPγ-8 controls synaptic transmission but not synaptic plasticity. Nat Neurosci 14:1410–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumioka A, Yan D, Tomita S (2010) TARP phosphorylation regulates synaptic AMPA receptors through lipid bilayers. Neuron 66:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HY, Dobrunz LE (2006) Presynaptic kainate receptor activation is a novel mechanism for target cell-specific short-term facilitation at Schaffer collateral synapses. J Neurosci 26:10796–10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Margolis FL, Shipley MT, Lidow MS (1998) Identification of a long variant of mRNA encoding the NR3 subunit of the NMDA receptor: its regional distribution and developmental expression in the rat brain. FEBS Lett 441:392–396. [DOI] [PubMed] [Google Scholar]
- Sun MY, Izumi Y, Benz A, Zorumski CF, Mennerick S (2016) Endogenous 24S-hydroxycholesterol modulates NMDAR-mediated function in hippocampal slices. J Neurophysiol 115:1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun MY, Taylor A, Zorumski CF, Mennerick S (2017a) 24S-hydroxycholesterol and 25-hydroxycholesterol differentially impact hippocampal neuronal survival following oxygen-glucose deprivation. PLoS One 12:e0174416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Hansen KB, Jahr CE (2017b) Allosteric interactions between NMDA receptor subunits shape the developmental shift in channel properties. Neuron 94:58–64.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang XD, Liu X, Gong HQ, Liang PJ (2010a) Synaptic contribution of Ca2+-permeable and Ca2+-impermeable AMPA receptors on isolated carp retinal horizontal cells and their modulation by Zn2+. Brain Res 1317:60–68. [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E (2002) Mechanism of glutamate receptor desensitization. Nature 417:245–253. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zhang YT, Gong HQ, Liang PJ (2010b) The mechanisms of Zn2+ effects on Ca2+-permeable AMPA receptors on carp retinal horizontal cells. Brain Res 1345:103–109. [DOI] [PubMed] [Google Scholar]
- Sundström E, Whittemore S, Mo LL, Seiger A (1997) Analysis of NMDA receptors in the human spinal cord. Exp Neurol 148:407–413. [DOI] [PubMed] [Google Scholar]
- Suryavanshi PS, Ugale RR, Yilmazer-Hanke D, Stairs DJ, Dravid SM (2014) GluN2C/GluN2D subunit-selective NMDA receptor potentiator CIQ reverses MK-801-induced impairment in prepulse inhibition and working memory in Y-maze test in mice. Br J Pharmacol 171:799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM (2006) Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127:49–58. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y (2014) Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 344:304–307. [DOI] [PubMed] [Google Scholar]
- Suzuki KElegheert JSong ISasakura HSenkov OMatsuda KKakegawa WClayton AJChang VTFerrer-Ferrer M, et al. (2020) A synthetic synaptic organizer protein restores glutamatergic neuronal circuits. Science 369:eabb4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Nelson AD, Eickstaedt JB, Wallace K, Wright LS, Svendsen CN (2006) Glutamate enhances proliferation and neurogenesis in human neural progenitor cell cultures derived from the fetal cortex. Eur J Neurosci 24:645–653. [DOI] [PubMed] [Google Scholar]
- Swainson J, Thomas RK, Archer S, Chrenek C, MacKay MA, Baker G, Dursun S, Klassen LJ, Chokka P, Demas ML (2019) Esketamine for treatment resistant depression. Expert Rev Neurother 19:899–911. [DOI] [PubMed] [Google Scholar]
- Swanger SAChen WWells GBurger PBTankovic ABhattacharya SStrong KLHu CKusumoto HZhang J, et al. (2016) Mechanistic insight into NMDA receptor dysregulation by rare variants in the GluN2A and GluN2B agonist binding domains. Am J Hum Genet 99:1261–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanger SAVance KMAcker TMZimmerman SSDiRaddo JOMyers SJBundgaard CMosley CASummer SLMenaldino DS, et al. (2018) A novel negative allosteric modulator selective for GluN2C/2D-containing NMDA receptors inhibits synaptic transmission in hippocampal interneurons. ACS Chem Neurosci 9:306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanger SA, Vance KM, Pare JF, Sotty F, Fog K, Smith Y, Traynelis SF (2015) NMDA receptors containing the GluN2D subunit control neuronal function in the subthalamic nucleus. J Neurosci 35:15971–15983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson GT, Feldmeyer D, Kaneda M, Cull-Candy SG (1996) Effect of RNA editing and subunit co-assembly single-channel properties of recombinant kainate receptors. J Physiol 492:129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson GT, Gereau RW 4th, Green T, Heinemann SF (1997a) Identification of amino acid residues that control functional behavior in GluR5 and GluR6 kainate receptors. Neuron 19:913–926. [DOI] [PubMed] [Google Scholar]
- Swanson GT, Green T, Heinemann SF (1998) Kainate receptors exhibit differential sensitivities to (S)-5-iodowillardiine. Mol Pharmacol 53:942–949. [PubMed] [Google Scholar]
- Swanson GT, Green T, Sakai R, Contractor A, Che W, Kamiya H, Heinemann SF (2002) Differential activation of individual subunits in heteromeric kainate receptors. Neuron 34:589–598. [DOI] [PubMed] [Google Scholar]
- Swanson GT, Heinemann SF (1998) Heterogeneity of homomeric GluR5 kainate receptor desensitization expressed in HEK293 cells. J Physiol 513:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson GT, Kamboj SK, Cull-Candy SG (1997b) Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J Neurosci 17:58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadits E, Cserép C, Szonyi A, Fukazawa Y, Shigemoto R, Watanabe M, Itohara S, Freund TF, Nyiri G (2011) NMDA receptors in hippocampal GABAergic synapses and their role in nitric oxide signaling. J Neurosci 31:5893–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakacs R, Janka Z, Kalman J (2012) The “blue” side of glutamatergic neurotransmission: NMDA receptor antagonists as possible novel therapeutics for major depression. Neuropsychopharmacol Hung 14:29–40. [PubMed] [Google Scholar]
- Szobota SGorostiza PDel Bene FWyart CFortin DLKolstad KDTulyathan OVolgraf MNumano RAaron HL, et al. (2007) Remote control of neuronal activity with a light-gated glutamate receptor. Neuron 54:535–545. [DOI] [PubMed] [Google Scholar]
- Taghdiri M, Kashef A, Abbassi G, Moshtagh A, Sadatian N, Fardaei M, Najafi K, Kariminejad R (2019) Further delineation of the phenotype caused by a novel large homozygous deletion of GRID2 gene in an adult patient. Clin Case Rep 7:1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic YAGeballe MGruszecka-Kowalik EMyers SJLyuboslavsky PLe PFrench AIrier HChoi WBEasterling K, et al. (2008) Enantiomeric propanolamines as selective N-methyl-D-aspartate 2B receptor antagonists. J Med Chem 51:5506–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, Furukawa H (2016) Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature 534:63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Kohara A, Shishikura J, Kawasaki-Yatsugi S, Ni JW, Yatsugi S, Sakamoto S, Okada M, Shimizu-Sasamata M, Yamaguchi T (2002) YM872: a selective, potent and highly water-soluble alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor antagonist. CNS Drug Rev 8:337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi YMori HMishina MWatanabe MKondo NShimomura JKubota YMatsuda KFukushima KShiroma N, et al. (2005) Autoantibodies and cell-mediated autoimmunity to NMDA-type GluRepsilon2 in patients with Rasmussen’s encephalitis and chronic progressive epilepsia partialis continua. Epilepsia 46 (Suppl 5):152–158. [DOI] [PubMed] [Google Scholar]
- Takayama C, Nakagawa S, Watanabe M, Mishina M, Inoue Y (1996) Developmental changes in expression and distribution of the glutamate receptor channel delta 2 subunit according to the Purkinje cell maturation. Brain Res Dev Brain Res 92:147–155. [DOI] [PubMed] [Google Scholar]
- Takenouchi T, Hashida N, Torii C, Kosaki R, Takahashi T, Kosaki K (2014) 1p34.3 deletion involving GRIK3: Further clinical implication of GRIK family glutamate receptors in the pathogenesis of developmental delay. Am J Med Genet A 164A:456–460. [DOI] [PubMed] [Google Scholar]
- Takeuchi E, Sato Y, Miura E, Yamaura H, Yuzaki M, Yanagihara D (2012) Characteristics of gait ataxia in δ2 glutamate receptor mutant mice, ho15J. PLoS One 7:e47553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Miyazaki T, Watanabe M, Mori H, Sakimura K, Mishina M (2005) Control of synaptic connection by glutamate receptor delta2 in the adult cerebellum. J Neurosci 25:2146–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallaksen-Greene SJ, Albin RL (1994) Localization of AMPA-selective excitatory amino acid receptor subunits in identified populations of striatal neurons. Neuroscience 61:509–519. [DOI] [PubMed] [Google Scholar]
- Talos DM, Fishman RE, Park H, Folkerth RD, Follett PL, Volpe JJ, Jensen FE (2006) Developmental regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid receptor subunit expression in forebrain and relationship to regional susceptibility to hypoxic/ischemic injury. I. Rodent cerebral white matter and cortex. J Comp Neurol 497:42–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder I, Borker P, Wollmuth LP (2010) Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J Neurosci 30:11792–11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamano H, Koike Y, Nakada H, Shakushi Y, Takeda A (2016) Significance of synaptic Zn2+ signaling in zincergic and non-zincergic synapses in the hippocampus in cognition. J Trace Elem Med Biol 38:93–98. [DOI] [PubMed] [Google Scholar]
- Tamborini LChen YFoss CAPinto AHorti AGTraynelis SFDe Micheli CMease RCHansen KBConti P, et al. (2016) Development of radiolabeled ligands targeting the glutamate binding site of the N-methyl-d-aspartate receptor as potential imaging agents for brain. J Med Chem 59:11110–11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GHLiu YYWang LLi KZhang ZQLi HFYang ZFLi YLi DWu MY, et al. (2018) PRRT2 deficiency induces paroxysmal kinesigenic dyskinesia by regulating synaptic transmission in cerebellum. Cell Res 28:90–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AH, Chen H, Li TP, Metzbower SR, MacGillavry HD, Blanpied TA (2016) A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536:210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CM, Dichter M, Morad M (1990) Modulation of the N-methyl-D-aspartate channel by extracellular H+. Proc Natl Acad Sci USA 87:6445–6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CM, Shi QY, Katchman A, Lynch G (1991) Modulation of the time course of fast EPSCs and glutamate channel kinetics by aniracetam. Science 254:288–290. [DOI] [PubMed] [Google Scholar]
- Tang M, Ivakine E, Mahadevan V, Salter MW, McInnes RR (2012) Neto2 interacts with the scaffolding protein GRIP and regulates synaptic abundance of kainate receptors. PLoS One 7:e51433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Pelkey KA, Ng D, Ivakine E, McBain CJ, Salter MW, McInnes RR (2011) Neto1 is an auxiliary subunit of native synaptic kainate receptors. J Neurosci 31:10009–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Liu D, Traynelis SF, Yuan H (2020) Positive allosteric modulators that target NMDA receptors rectify loss-of-function GRIN variants associated with neurological and neuropsychiatric disorders. Neuropharmacology 177:108247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YP, Shimizu E, Dube GR, Rampon C, Kerchner GA, Zhuo M, Liu G, Tsien JZ (1999) Genetic enhancement of learning and memory in mice. Nature 401:63–69. [DOI] [PubMed] [Google Scholar]
- Tang YP, Wang H, Feng R, Kyin M, Tsien JZ (2001) Differential effects of enrichment on learning and memory function in NR2B transgenic mice. Neuropharmacology 41:779–790. [DOI] [PubMed] [Google Scholar]
- Tao W, Díaz-Alonso J, Sheng N, Nicoll RA (2018) Postsynaptic δ1 glutamate receptor assembles and maintains hippocampal synapses via Cbln2 and neurexin. Proc Natl Acad Sci USA 115:E5373–E5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Ma C, Bemben MA, Li KH, Burlingame AL, Zhang M, Nicoll RA (2019) Mechanisms underlying the synaptic trafficking of the glutamate delta receptor GluD1. Mol Psychiatry 24:1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao-Cheng JH, Azzam R, Crocker V, Winters CA, Reese T (2015) Depolarization of hippocampal neurons induces formation of nonsynaptic NMDA receptor islands resembling nascent postsynaptic densities. eNeuro 2:ENEURO.0066-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapken D, Steffensen TB, Leth R, Kristensen LB, Gerbola A, Gajhede M, Jørgensen FS, Olsen L, Kastrup JS (2017) The low binding affinity of D-serine at the ionotropic glutamate receptor GluD2 can be attributed to the hinge region. Sci Rep 7:46145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarabeux JKebir OGauthier JHamdan FFXiong LPiton ASpiegelman DHenrion ÉMillet BFathalli F, et al. ; S2D team (2011) Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl Psychiatry 1:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraschenko O, Fox HS, Pittock SJ, Zekeridou A, Gafurova M, Eldridge E, Liu J, Dravid SM, Dingledine R (2019) A mouse model of seizures in anti-N-methyl-d-aspartate receptor encephalitis. Epilepsia 60:452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnawa I, Farkas S, Berzsenyi P, Pataki A, Andrási F (1989) Electrophysiological studies with a 2,3-benzodiazepine muscle relaxant: GYKI 52466. Eur J Pharmacol 167:193–199. [DOI] [PubMed] [Google Scholar]
- Tárnok K, Czöndör K, Jelitai M, Czirók A, Schlett K (2008) NMDA receptor NR2B subunit over-expression increases cerebellar granule cell migratory activity. J Neurochem 104:818–829. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Dunaevsky A, Blazeski R, Mason CA, Yuste R (2003) Bidirectional regulation of hippocampal mossy fiber filopodial motility by kainate receptors: a two-step model of synaptogenesis. Neuron 38:773–784. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Sandler VM, Toni N, Zhao C, Gage FH (2006) NMDA-receptor-mediated, cell-specific integration of new neurons in adult dentate gyrus. Nature 442:929–933. [DOI] [PubMed] [Google Scholar]
- Tasic BMenon VNguyen TNKim TKJarsky TYao ZLevi BGray LTSorensen SADolbeare T, et al. (2016) Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay SH, Fairhurst AM, Mak A (2017) Clinical utility of circulating anti-N-methyl-d-aspartate receptor subunits NR2A/B antibody for the diagnosis of neuropsychiatric syndromes in systemic lupus erythematosus and Sjögren’s syndrome: An updated meta-analysis. Autoimmun Rev 16:114–122. [DOI] [PubMed] [Google Scholar]
- Taylor CP, Traynelis SF, Siffert J, Pope LE, Matsumoto RR (2016) Pharmacology of dextromethorphan: Relevance to dextromethorphan/quinidine (Nuedexta®) clinical use. Pharmacol Ther 164:170–182. [DOI] [PubMed] [Google Scholar]
- Tazerart S, Mitchell DE, Miranda-Rottmann S, Araya R (2020) A spike-timing-dependent plasticity rule for dendritic spines. Nat Commun 11:4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert RW, Jimenez EC, Twede V, Watkins M, Hollmann M, Bulaj G, Olivera BM (2007) Novel conantokins from Conus parius venom are specific antagonists of N-methyl-D-aspartate receptors. J Biol Chem 282:36905–36913. [DOI] [PubMed] [Google Scholar]
- Terhag J, Gottschling K, Hollmann M (2010) The transmembrane domain C of AMPA receptors is critically involved in receptor function and modulation. Front Mol Neurosci 3:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A, Everts I, Hollmann M (2002) Inhibition by lectins of glutamate receptor desensitization is determined by the lectin’s sugar specificity at kainate but not AMPA receptors. Mol Cell Neurosci 21:521–533. [DOI] [PubMed] [Google Scholar]
- Thomas GM, Huganir RL (2013) Palmitoylation-dependent regulation of glutamate receptors and their PDZ domain-containing partners. Biochem Soc Trans 41:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum S, Schepmann D, Kalinin DV, Ametamey SM, Wünsch B (2018) Replacement of the benzylpiperidine moiety with fluorinated phenylalkyl side chains for the development of GluN2B receptor ligands. ChemMedChem 13:2522–2529. [DOI] [PubMed] [Google Scholar]
- Tian Z, Clark BLM, Menard F (2019) Kainic acid-based agonists of glutamate receptors: SAR analysis and guidelines for analog design. ACS Chem Neurosci 10:4190–4198. [DOI] [PubMed] [Google Scholar]
- Tigaret CM, Thalhammer A, Rast GF, Specht CG, Auberson YP, Stewart MG, Schoepfer R (2006) Subunit dependencies of N-methyl-D-aspartate (NMDA) receptor-induced alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor internalization. Mol Pharmacol 69:1251–1259. [DOI] [PubMed] [Google Scholar]
- Tikhonov DB, Mellor JR, Usherwood PN, Magazanik LG (2002) Modeling of the pore domain of the GLUR1 channel: homology with K+ channel and binding of channel blockers. Biophys J 82:1884–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov DB, Zhorov BS (2020) The pore domain in glutamate-gated ion channels: Structure, drug binding and similarity with potassium channels. Biochim Biophys Acta Biomembr 1862:183401. [DOI] [PubMed] [Google Scholar]
- Tikhonova TB, Barygin OI, Gmiro VE, Tikhonov DB, Magazanik LG (2008) Organic blockers escape from trapping in the AMPA receptor channels by leaking into the cytoplasm. Neuropharmacology 54:653–664. [DOI] [PubMed] [Google Scholar]
- Tikhonova TB, Tikhonov DB, Magazanik LG (2009) Common binding site for externally and internally applied AMPA receptor channel blockers. J Mol Neurosci 39:169–174. [DOI] [PubMed] [Google Scholar]
- Timm DE, Benveniste M, Weeks AM, Nisenbaum ES, Partin KM (2011) Structural and functional analysis of two new positive allosteric modulators of GluA2 desensitization and deactivation. Mol Pharmacol 80:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titulaer MJMcCracken LGabilondo IIizuka TKawachi IBataller LTorrents ARosenfeld MRBalice-Gordon RGraus F, et al. (2013) Late-onset anti-NMDA receptor encephalitis. Neurology 81:1058–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tölle TR, Berthele A, Zieglgänsberger W, Seeburg PH, Wisden W (1993) The differential expression of 16 NMDA and non-NMDA receptor subunits in the rat spinal cord and in periaqueductal gray. J Neurosci 13:5009–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA, Bredt DS (2005a) Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature 435:1052–1058. [DOI] [PubMed] [Google Scholar]
- Tomita S, Castillo PE (2012) Neto1 and Neto2: auxiliary subunits that determine key properties of native kainate receptors. J Physiol 590:2217–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, Bredt DS (2003) Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J Cell Biol 161:805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Sekiguchi M, Wada K, Nicoll RA, Bredt DS (2006) Stargazin controls the pharmacology of AMPA receptor potentiators. Proc Natl Acad Sci USA 103:10064–10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Stein V, Stocker TJ, Nicoll RA, Bredt DS (2005b) Bidirectional synaptic plasticity regulated by phosphorylation of stargazin-like TARPs. Neuron 45:269–277. [DOI] [PubMed] [Google Scholar]
- Tong GTakahashi HTu SShin YTalantova MZago WXia PNie ZGoetz TZhang D, et al. (2008) Modulation of NMDA receptor properties and synaptic transmission by the NR3A subunit in mouse hippocampal and cerebrocortical neurons. J Neurophysiol 99:122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torashima T, Iizuka A, Horiuchi H, Mitsumura K, Yamasaki M, Koyama C, Takayama K, Iino M, Watanabe M, Hirai H (2009) Rescue of abnormal phenotypes in delta2 glutamate receptor-deficient mice by the extracellular N-terminal and intracellular C-terminal domains of the delta2 glutamate receptor. Eur J Neurosci 30:355–365. [DOI] [PubMed] [Google Scholar]
- Tóth K, McBain CJ (1998) Afferent-specific innervation of two distinct AMPA receptor subtypes on single hippocampal interneurons. Nat Neurosci 1:572–578. [DOI] [PubMed] [Google Scholar]
- Toth K, Suares G, Lawrence JJ, Philips-Tansey E, McBain CJ (2000) Differential mechanisms of transmission at three types of mossy fiber synapse. J Neurosci 20:8279–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar KR, McGinley MJ, Westbrook GL (2013) Triheteromeric NMDA receptors at hippocampal synapses. J Neurosci 33:9150–9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL (1999) The incorporation of NMDA receptors with a distinct subunit composition at nascent hippocampal synapses in vitro. J Neurosci 19:4180–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL (2002) Mobile NMDA receptors at hippocampal synapses. Neuron 34:255–264. [DOI] [PubMed] [Google Scholar]
- Traynelis J, Silk M, Wang Q, Berkovic SF, Liu L, Ascher DB, Balding DJ, Petrovski S (2017) Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res 27:1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Burgess MF, Zheng F, Lyuboslavsky P, Powers JL (1998) Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J Neurosci 18:6163–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Cull-Candy SG (1990) Proton inhibition of N-methyl-D-aspartate receptors in cerebellar neurons. Nature 345:347–350. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Cull-Candy SG (1991) Pharmacological properties and H+ sensitivity of excitatory amino acid receptor channels in rat cerebellar granule neurones. J Physiol 433:727–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Hartley M, Heinemann SF (1995) Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science 268:873–876. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wahl P (1997) Control of rat GluR6 glutamate receptor open probability by protein kinase A and calcineurin. J Physiol 503:513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62:405–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein JMühleisen TWFrank JMattheisen MHerms SLudwig KUTreutlein TSchmael CStrohmaier JBösshenz KV, et al. (2009) Dissection of phenotype reveals possible association between schizophrenia and Glutamate Receptor Delta 1 (GRID1) gene promoter. Schizophr Res 111:123–130. [DOI] [PubMed] [Google Scholar]
- Trujillo KA, Akil H (1991) Inhibition of morphine tolerance and dependence by the NMDA receptor antagonist MK-801. Science 251:85–87. [DOI] [PubMed] [Google Scholar]
- Trussell LO, Zhang S, Raman IM (1993) Desensitization of AMPA receptors upon multiquantal neurotransmitter release. Neuron 10:1185–1196. [DOI] [PubMed] [Google Scholar]
- Trzepacz PT, Cummings J, Konechnik T, Forrester TD, Chang C, Dennehy EB, Willis BA, Shuler C, Tabas LB, Lyketsos C (2013) Mibampator (LY451395) randomized clinical trial for agitation/aggression in Alzheimer’s disease. Int Psychogeriatr 25:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos GC (2011) Systemic lupus erythematosus. N Engl J Med 365:2110–2121. [DOI] [PubMed] [Google Scholar]
- Tsuang DW, Greenwood TA, Jayadev S, Davis M, Shutes-David A, Bird TD (2018) A genetic study of psychosis in Huntington’s disease: Evidence for the involvement of glutamate signaling pathways. J Huntingtons Dis 7:51–59. [DOI] [PubMed] [Google Scholar]
- Tsui CC, Copeland NG, Gilbert DJ, Jenkins NA, Barnes C, Worley PF (1996) Narp, a novel member of the pentraxin family, promotes neurite outgrowth and is dynamically regulated by neuronal activity. J Neurosci 16:2463–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu WXu XPeng LZhong XZhang WSoundarapandian MMBalel CWang MJia NZhang W, et al. (2010) DAPK1 interaction with NMDA receptor NR2B subunits mediates brain damage in stroke. Cell 140:222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecek J, Jackman SL, Regehr WG (2016) Synaptic specializations support frequency-independent Purkinje cell output from the cerebellar cortex. Cell Rep 17:3256–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turetsky D, Garringer E, Patneau DK (2005) Stargazin modulates native AMPA receptor functional properties by two distinct mechanisms. J Neurosci 25:7438–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano G (2012) Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harb Perspect Biol 4:a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG (2008) The self-tuning neuron: synaptic scaling of excitatory synapses. Cell 135:422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB (2004) Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci 5:97–107. [DOI] [PubMed] [Google Scholar]
- Turski L, Huth A, Sheardown M, McDonald F, Neuhaus R, Schneider HH, Dirnagl U, Wiegand F, Jacobsen P, Ottow E (1998) ZK200775: a phosphonate quinoxalinedione AMPA antagonist for neuroprotection in stroke and trauma. Proc Natl Acad Sci USA 95:10960–10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tüzün E, Zhou L, Baehring JM, Bannykh S, Rosenfeld MR, Dalmau J (2009) Evidence for antibody-mediated pathogenesis in anti-NMDAR encephalitis associated with ovarian teratoma. Acta Neuropathol 118:737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twede VD, Miljanich G, Olivera BM, Bulaj G (2009a) Neuroprotective and cardioprotective conopeptides: an emerging class of drug leads. Curr Opin Drug Discov Devel 12:231–239. [PMC free article] [PubMed] [Google Scholar]
- Twede VD, Teichert RW, Walker CS, Gruszczyński P, Kaźmierkiewicz R, Bulaj G, Olivera BM (2009b) Conantokin-Br from Conus brettinghami and selectivity determinants for the NR2D subunit of the NMDA receptor. Biochemistry 48:4063–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Sobolevsky AI (2018) Structural mechanisms of gating in ionotropic glutamate receptors. Biochemistry 57:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI (2016) Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science 353:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI (2017a) Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature 549:60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI (2017b) Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron 94:569–580.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Sobolevsky AI (2019) Structural and functional insights into transmembrane AMPA receptor regulatory protein complexes. J Gen Physiol 151:1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Vassilevski AA, Sobolevsky AI (2018) Mechanisms of channel block in calcium-permeable AMPA receptors. Neuron 99:956–968.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyman RE, Gahring LC, Spiess J, Rogers SW (1995) Glutamate receptor antibodies activate a subset of receptors and reveal an agonist binding site. Neuron 14:755–762. [DOI] [PubMed] [Google Scholar]
- Tzschach A, Menzel C, Erdogan F, Istifli ES, Rieger M, Ovens-Raeder A, Macke A, Ropers HH, Ullmann R, Kalscheuer V (2010) Characterization of an interstitial 4q32 deletion in a patient with mental retardation and a complex chromosome rearrangement. Am J Med Genet A 152A:1008–1012. [DOI] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M (2010) Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141:1068–1079. [DOI] [PubMed] [Google Scholar]
- Ugale VG, Bari SB (2014) Quinazolines: new horizons in anticonvulsant therapy. Eur J Med Chem 80:447–501. [DOI] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY (2007) Subunit counting in membrane-bound proteins. Nat Methods 4:319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY (2008) Rules of engagement for NMDA receptor subunits. Proc Natl Acad Sci USA 105:14163–14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich B, Ushkaryov YA, Südhof TC (1995) Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron 14:497–507. [DOI] [PubMed] [Google Scholar]
- Unno M, Shinohara M, Takayama K, Tanaka H, Teruya K, Doh-ura K, Sakai R, Sasaki M, Ikeda-Saito M (2011) Binding and selectivity of the marine toxin neodysiherbaine A and its synthetic analogues to GluK1 and GluK2 kainate receptors. J Mol Biol 413:667–683. [DOI] [PubMed] [Google Scholar]
- Urwyler S, Floersheim P, Roy BL, Koller M (2009) Drug design, in vitro pharmacology, and structure-activity relationships of 3-acylamino-2-aminopropionic acid derivatives, a novel class of partial agonists at the glycine site on the N-methyl-D-aspartate (NMDA) receptor complex. J Med Chem 52:5093–5107. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Südhof TC (1992) Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science 257:50–56. [DOI] [PubMed] [Google Scholar]
- Usoskin DFurlan AIslam SAbdo HLönnerberg PLou DHjerling-Leffler JHaeggström JKharchenko OKharchenko PV, et al. (2015) Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18:145–153. [DOI] [PubMed] [Google Scholar]
- Utine GE, Haliloğlu G, Salanci B, Çetinkaya A, Kiper PO, Alanay Y, Aktas D, Boduroğlu K, Alikaşifoğlu M (2013) A homozygous deletion in GRID2 causes a human phenotype with cerebellar ataxia and atrophy. J Child Neurol 28:926–932. [DOI] [PubMed] [Google Scholar]
- Valbuena S, Lerma J (2016) Non-canonical signaling, the hidden life of ligand-gated ion channels. Neuron 92:316–329. [DOI] [PubMed] [Google Scholar]
- Valbuena S, Lerma J (2021) Kainate receptors, homeostatic gatekeepers of synaptic plasticity. Neuroscience 456:17–26. [DOI] [PubMed] [Google Scholar]
- Valente PCastroflorio ERossi PFadda MSterlini BCervigni RIPrestigio CGiovedì SOnofri FMura E, et al. (2016) PRRT2 is a key component of the Ca(2+)-dependent neurotransmitter release machinery. Cell Rep 15:117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vales K, Rambousek L, Holubova K, Svoboda J, Bubenikova-Valesova V, Chodounska H, Vyklicky L, Stuchlik A (2012) 3α5β-Pregnanolone glutamate, a use-dependent NMDA antagonist, reversed spatial learning deficit in an animal model of schizophrenia. Behav Brain Res 235:82–88. [DOI] [PubMed] [Google Scholar]
- Valluru L, Xu J, Zhu Y, Yan S, Contractor A, Swanson GT (2005) Ligand binding is a critical requirement for plasma membrane expression of heteromeric kainate receptors. J Biol Chem 280:6085–6093. [DOI] [PubMed] [Google Scholar]
- Valtschanoff JG, Burette A, Wenthold RJ, Weinberg RJ (1999) Expression of NR2 receptor subunit in rat somatic sensory cortex: synaptic distribution and colocalization with NR1 and PSD-95. J Comp Neurol 410:599–611. [PubMed] [Google Scholar]
- Van Horn MR, Strasser A, Miraucourt LS, Pollegioni L, Ruthazer ES (2017) The gliotransmitter d-serine promotes synapse maturation and axonal stabilization in vivo. J Neurosci 37:6277–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schil KMeire FKarlstetter MBauwens MVerdin HCoppieters FScheiffert EVan Nechel CLangmann TDeconinck N, et al. (2015) Early-onset autosomal recessive cerebellar ataxia associated with retinal dystrophy: new human hotfoot phenotype caused by homozygous GRID2 deletion. Genet Med 17:291–299. [DOI] [PubMed] [Google Scholar]
- van Sonderen A, Petit-Pedrol M, Dalmau J, Titulaer MJ (2017) The value of LGI1, Caspr2 and voltage-gated potassium channel antibodies in encephalitis. Nat Rev Neurol 13:290–301. [DOI] [PubMed] [Google Scholar]
- Vance KM, Hansen KB, Traynelis SF (2012) GluN1 splice variant control of GluN1/GluN2D NMDA receptors. J Physiol 590:3857–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KM, Hansen KB, Traynelis SF (2013) Modal gating of GluN1/GluN2D NMDA receptors. Neuropharmacology 71:184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KM, Simorowski N, Traynelis SF, Furukawa H (2011) Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nat Commun 2:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderklish P, Neve R, Bahr BA, Arai A, Hennegriff M, Larson J, Lynch G (1992) Translational suppression of a glutamate receptor subunit impairs long-term potentiation. Synapse 12:333–337. [DOI] [PubMed] [Google Scholar]
- Vanhoutte P, Bading H (2003) Opposing roles of synaptic and extrasynaptic NMDA receptors in neuronal calcium signalling and BDNF gene regulation. Curr Opin Neurobiol 13:366–371. [DOI] [PubMed] [Google Scholar]
- Vargas-Caballero M, Robinson HP (2004) Fast and slow voltage-dependent dynamics of magnesium block in the NMDA receptor: the asymmetric trapping block model. J Neurosci 24:6171–6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varney MAJachec CDeal CHess SDDaggett LPSkvoretz RUrcan MMorrison JHMoran TJohnson EC, et al. (1996) Stable expression and characterization of recombinant human heteromeric N-methyl-D-aspartate receptor subtypes NMDAR1A/2A and NMDAR1A/2B in mammalian cells. J Pharmacol Exp Ther 279:367–378. [PubMed] [Google Scholar]
- Varoqueaux F, Sigler A, Rhee JS, Brose N, Enk C, Reim K, Rosenmund C (2002) Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc Natl Acad Sci USA 99:9037–9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerapandiyan A, Enner S, Thulasi V, Ming X (2017) A rare syndrome of GRID2 deletion in 2 siblings. Child Neurol Open 4:2329048X17726168. [DOI] [PMC free article] [PubMed]
- Venskutonyte RButini SCoccone SSGemma SBrindisi MKumar VGuarino EMaramai SValenti SAmir A, et al. (2011a) Selective kainate receptor (GluK1) ligands structurally based upon 1H-cyclopentapyrimidin-2,4(1H,3H)-dione: synthesis, molecular modeling, and pharmacological and biostructural characterization. J Med Chem 54:4793–4805. [DOI] [PubMed] [Google Scholar]
- Venskutonytė R, Frydenvang K, Gajhede M, Bunch L, Pickering DS, Kastrup JS (2011b) Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate. J Struct Biol 176:307–314. [DOI] [PubMed] [Google Scholar]
- Venskutonytė R, Frydenvang K, Hald H, Rabassa AC, Gajhede M, Ahring PK, Kastrup JS (2012) Kainate induces various domain closures in AMPA and kainate receptors. Neurochem Int 61:536–545. [DOI] [PubMed] [Google Scholar]
- Venskutonytė R, Larsen AP, Frydenvang K, Gajhede M, Sagot E, Assaf Z, Gefflaut T, Pickering DS, Bunch L, Kastrup JS (2014) Molecular recognition of two 2,4-syn-functionalized (S)-glutamate analogues by the kainate receptor GluK3 ligand binding domain. ChemMedChem 9:2254–2259. [DOI] [PubMed] [Google Scholar]
- Veran J, Kumar J, Pinheiro PS, Athané A, Mayer ML, Perrais D, Mulle C (2012) Zinc potentiates GluK3 glutamate receptor function by stabilizing the ligand binding domain dimer interface. Neuron 76:565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoorn TA, Burnashev N, Monyer H, Seeburg PH, Sakmann B (1991) Structural determinants of ion flow through recombinant glutamate receptor channels. Science 252:1715–1718. [DOI] [PubMed] [Google Scholar]
- Verdoorn TA, Johansen TH, Drejer J, Nielsen EO (1994) Selective block of recombinant glur6 receptors by NS-102, a novel non-NMDA receptor antagonist. Eur J Pharmacol 269:43–49. [DOI] [PubMed] [Google Scholar]
- Vergnano AM, Rebola N, Savtchenko LP, Pinheiro PS, Casado M, Kieffer BL, Rusakov DA, Mulle C, Paoletti P (2014) Zinc dynamics and action at excitatory synapses. Neuron 82:1101–1114. [DOI] [PubMed] [Google Scholar]
- Verhage MMaia ASPlomp JJBrussaard ABHeeroma JHVermeer HToonen RFHammer REvan den Berg TKMissler M, et al. (2000) Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science 287:864–869. [DOI] [PubMed] [Google Scholar]
- Verhagen Metman L, Morris MJ, Farmer C, Gillespie M, Mosby K, Wuu J, Chase TN (2002) Huntington’s disease: a randomized, controlled trial using the NMDA-antagonist amantadine. Neurology 59:694–699. [DOI] [PubMed] [Google Scholar]
- Verkhratsky A, Chvátal A (2020) NMDA receptors in astrocytes. Neurochem Res 45:122–133. [DOI] [PubMed] [Google Scholar]
- Vernon CG, Copits BA, Stolz JR, Guzmán YF, Swanson GT (2017) N-glycan content modulates kainate receptor functional properties. J Physiol 595:5913–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon CG, Swanson GT (2017) Neto2 assembles with kainate receptors in DRG neurons during development and modulates neurite outgrowth in adult sensory neurons. J Neurosci 37:3352–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesikansa A, Sakha P, Kuja-Panula J, Molchanova S, Rivera C, Huttunen HJ, Rauvala H, Taira T, Lauri SE (2012) Expression of GluK1c underlies the developmental switch in presynaptic kainate receptor function. Sci Rep 2:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesikansa A, Sallert M, Taira T, Lauri SE (2007) Activation of kainate receptors controls the number of functional glutamatergic synapses in the area CA1 of rat hippocampus. J Physiol 583:145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicini S, Wang JF, Li JH, Zhu WJ, Wang YH, Luo JH, Wolfe BB, Grayson DR (1998) Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J Neurophysiol 79:555–566. [DOI] [PubMed] [Google Scholar]
- Vieira M, Yong XLH, Roche KW, Anggono V (2020) Regulation of NMDA glutamate receptor functions by the GluN2 subunits. J Neurochem 154:121–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignes M, Collingridge GL (1997) The synaptic activation of kainate receptors. Nature 388:179–182. [DOI] [PubMed] [Google Scholar]
- Villarroel A, Regalado MP, Lerma J (1998) Glycine-independent NMDA receptor desensitization: localization of structural determinants. Neuron 20:329–339. [DOI] [PubMed] [Google Scholar]
- Villemure EVolgraf MJiang YWu GLy CQYuen PWLu ALuo XLiu MZhang S, et al. (2016) GluN2A-selective pyridopyrimidinone series of NMDAR positive allosteric modulators with an improved in vivo profile. ACS Med Chem Lett 8:84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissel B, Krupp JJ, Heinemann SF, Westbrook GL (2001a) A use-dependent tyrosine dephosphorylation of NMDA receptors is independent of ion flux. Nat Neurosci 4:587–596. [DOI] [PubMed] [Google Scholar]
- Vissel B, Krupp JJ, Heinemann SF, Westbrook GL (2002) Intracellular domains of NR2 alter calcium-dependent inactivation of N-methyl-D-aspartate receptors. Mol Pharmacol 61:595–605. [DOI] [PubMed] [Google Scholar]
- Vissel BRoyle GAChristie BRSchiffer HHGhetti ATritto TPerez-Otano IRadcliffe RASeamans JSejnowski T, et al. (2001b) The role of RNA editing of kainate receptors in synaptic plasticity and seizures. Neuron 29:217–227. [DOI] [PubMed] [Google Scholar]
- Vogensen SBFrydenvang KGreenwood JRPostorino GNielsen BPickering DSEbert BBølcho UEgebjerg JGajhede M, et al. (2007) A tetrazolyl-substituted subtype-selective AMPA receptor agonist. J Med Chem 50:2408–2414. [DOI] [PubMed] [Google Scholar]
- Vogensen SB, Greenwood JR, Bunch L, Clausen RP (2011) Glutamate receptor agonists: stereochemical aspects. Curr Top Med Chem 11:887–906. [DOI] [PubMed] [Google Scholar]
- Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D (2006) Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol 2:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volgraf MSellers BDJiang YWu GLy CQVillemure EPastor RMYuen PWLu ALuo X, et al. (2016) Discovery of GluN2A-selective NMDA receptor positive allosteric modulators (PAMs): Tuning deactivation kinetics via structure-based design. J Med Chem 59:2760–2779. [DOI] [PubMed] [Google Scholar]
- Volianskis A, Bannister N, Collett VJ, Irvine MW, Monaghan DT, Fitzjohn SM, Jensen MS, Jane DE, Collingridge GL (2013) Different NMDA receptor subtypes mediate induction of long-term potentiation and two forms of short-term potentiation at CA1 synapses in rat hippocampus in vitro. J Physiol 591:955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volianskis A, France G, Jensen MS, Bortolotto ZA, Jane DE, Collingridge GL (2015) Long-term potentiation and the role of N-methyl-D-aspartate receptors. Brain Res 1621:5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann RAFanger CMAnderson DRSirivolu VRPaschetto KGordon EVirginio CGleyzes MBuisson BSteidl E, et al. (2016) MPX-004 and MPX-007: New pharmacological tools to study the physiology of NMDA receptors containing the GluN2A subunit. PLoS One 11:e0148129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Engelhardt J (2019) AMPA receptor auxiliary proteins of the CKAMP family. Int J Mol Sci 20:1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Engelhardt J, Bocklisch C, Tönges L, Herb A, Mishina M, Monyer H (2015) GluN2D-containing NMDA receptors-mediate synaptic currents in hippocampal interneurons and pyramidal cells in juvenile mice. Front Cell Neurosci 9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Engelhardt J, Mack V, Sprengel R, Kavenstock N, Li KW, Stern-Bach Y, Smit AB, Seeburg PH, Monyer H (2010) CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science 327:1518–1522. [DOI] [PubMed] [Google Scholar]
- Vyklický L Jr (1993) Calcium-mediated modulation of N-methyl-D-aspartate (NMDA) responses in cultured rat hippocampal neurones. J Physiol 470:575–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyklický L Jr, Vlachová V, Krůsek J (1990) The effect of external pH changes on responses to excitatory amino acids in mouse hippocampal neurones. J Physiol 430:497–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyklicky VKrausova BCerny JBalik AZapotocky MNovotny MLichnerova KSmejkalova TKaniakova MKorinek M, et al. (2015) Block of NMDA receptor channels by endogenous neurosteroids: implications for the agonist induced conformational states of the channel vestibule. Sci Rep 5:10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyklicky VKrausova BCerny JLadislav MSmejkalova TKysilov BKorinek MDanacikova SHorak MChodounska H, et al. (2018) Surface expression, function, and pharmacology of disease-associated mutations in the membrane domain of the human GluN2B subunit. Front Mol Neurosci 11:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyklicky VSmejkalova TKrausova BBalik AKorinek MBorovska JHorak MChvojkova MKleteckova LVales K, et al. (2016) Preferential inhibition of tonically over phasically activated NMDA receptors by pregnane derivatives. J Neurosci 36:2161–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A, Takahashi H, Lipton SA, Chen HS (2006) NR3A modulates the outer vestibule of the “NMDA” receptor channel. J Neurosci 26:13156–13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadiche JI, Jahr CE (2001) Multivesicular release at climbing fiber-Purkinje cell synapses. Neuron 32:301–313. [DOI] [PubMed] [Google Scholar]
- Wafford KA, Bain CJ, Le Bourdelles B, Whiting PJ, Kemp JA (1993) Preferential co-assembly of recombinant NMDA receptors composed of three different subunits. Neuroreport 4:1347–1349. [DOI] [PubMed] [Google Scholar]
- Wahl P, Anker C, Traynelis SF, Egebjerg J, Rasmussen JS, Krogsgaard-Larsen P, Madsen U (1998) Antagonist properties of a phosphono isoxazole amino acid at glutamate R1-4 (R,S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid receptor subtypes. Mol Pharmacol 53:590–596. [DOI] [PubMed] [Google Scholar]
- Walker CS, Brockie PJ, Madsen DM, Francis MM, Zheng Y, Koduri S, Mellem JE, Strutz-Seebohm N, Maricq AV (2006) Reconstitution of invertebrate glutamate receptor function depends on stargazin-like proteins. Proc Natl Acad Sci USA 103:10781–10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CSJensen SEllison MMatta JALee WYImperial JSDuclos NBrockie PJMadsen DMIsaac JT, et al. (2009) A novel Conus snail polypeptide causes excitotoxicity by blocking desensitization of AMPA receptors. Curr Biol 19:900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DL, Ressler KJ, Lu KT, Davis M (2002) Facilitation of conditioned fear extinction by systemic administration or intra-amygdala infusions of D-cycloserine as assessed with fear-potentiated startle in rats. J Neurosci 22:2343–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MR, Kaste M, Lees KR, Diener HC, Hommel M, De Keyser J, Steiner H, Versavel M (2005) The AMPA antagonist ZK 200775 in patients with acute ischaemic stroke: a double-blind, multicentre, placebo-controlled safety and tolerability study. Cerebrovasc Dis 20:304–309. [DOI] [PubMed] [Google Scholar]
- Wang AL, Carroll RC, Nawy S (2014a) Down-regulation of the RNA editing enzyme ADAR2 contributes to RGC death in a mouse model of glaucoma. PLoS One 9:e91288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CC, Held RG, Chang SC, Yang L, Delpire E, Ghosh A, Hall BJ (2011a) A critical role for GluN2B-containing NMDA receptors in cortical development and function. Neuron 72:789–805. [DOI] [PubMed] [Google Scholar]
- Wang D, Jacobs SA, Tsien JZ (2014b) Targeting the NMDA receptor subunit NR2B for treating or preventing age-related memory decline. Expert Opin Ther Targets 18:1121–1130. [DOI] [PubMed] [Google Scholar]
- Wang H, James ML, Venkatraman TN, Wilson LJ, Lyuboslavsky P, Myers SJ, Lascola CD, Laskowitz DT (2014c) pH-sensitive NMDA inhibitors improve outcome in a murine model of SAH. Neurocrit Care 20:119–131. [DOI] [PubMed] [Google Scholar]
- Wang H, Liu H, Zhang ZW (2011b) Elimination of redundant synaptic inputs in the absence of synaptic strengthening. J Neurosci 31:16675–16684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HX, Gao WJ (2009) Cell type-specific development of NMDA receptors in the interneurons of rat prefrontal cortex. Neuropsychopharmacology 34:2028–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JQ, Guo ML, Jin DZ, Xue B, Fibuch EE, Mao LM (2014d) Roles of subunit phosphorylation in regulating glutamate receptor function. Eur J Pharmacol 728:183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Furukawa H (2019) Dissecting diverse functions of NMDA receptors by structural biology. Curr Opin Struct Biol 54:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JXIrvine MWBurnell ESSapkota KThatcher RJLi MSimorowski NVolianskis ACollingridge GLMonaghan DT, et al. (2020) Structural basis of subtype-selective competitive antagonism for GluN2C/2D-containing NMDA receptors. Nat Commun 11:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lv S, Stroebel D, Zhang J, Pan Y, Huang X, Zhang X, Paoletti P, Zhu S (2021) Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors. Neuron 109:2443–2456.e5. [DOI] [PubMed] [Google Scholar]
- Wang R, Mellem JE, Jensen M, Brockie PJ, Walker CS, Hoerndli FJ, Hauth L, Madsen DM, Maricq AV (2012) The SOL-2/Neto auxiliary protein modulates the function of AMPA-subtype ionotropic glutamate receptors. Neuron 75:838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Reddy PH (2017) Role of glutamate and NMDA receptors in Alzheimer’s disease. J Alzheimers Dis 57:1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TMBrown BMDeng LSellers BDLupardus PJWallweber HJAGustafson AWong EVolgraf MSchwarz JB, et al. (2017) A novel NMDA receptor positive allosteric modulator that acts via the transmembrane domain. Neuropharmacology 121:204–218. [DOI] [PubMed] [Google Scholar]
- Wang Y, Matsuda S, Drews V, Torashima T, Meisler MH, Yuzaki M (2003) A hot spot for hotfoot mutations in the gene encoding the delta2 glutamate receptor. Eur J Neurosci 17:1581–1590. [DOI] [PubMed] [Google Scholar]
- Wang YQ, Huang YH, Balakrishnan S, Liu L, Wang YT, Nestler EJ, Schlüter OM, Dong Y (2021) AMPA and NMDA receptor trafficking at cocaine-generated synapses. J Neurosci 41:1996–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Wenthold RJ, Ottersen OP, Petralia RS (1998) Endbulb synapses in the anteroventral cochlear nucleus express a specific subset of AMPA-type glutamate receptor subunits. J Neurosci 18:1148–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SEHarries MHAldegheri LBradford AMBallini EDawson LLacroix LPardoe JStarr KWeil A, et al. (2020) Pharmacological characterisation of MDI-222, a novel AMPA receptor positive allosteric modulator with an improved safety profile. J Psychopharmacol 34:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SE, Pennicott LE, Beswick P (2015) AMPA receptor-positive allosteric modulators for the treatment of schizophrenia: an overview of recent patent applications. Future Med Chem 7:473–491. [DOI] [PubMed] [Google Scholar]
- Warikoo NBrunwasser SJBenz AShu HJPaul SMLewis MDoherty JQuirk MPiccio LZorumski CF, et al. (2018) Positive allosteric modulation as a potential therapeutic strategy in anti-NMDA receptor encephalitis. J Neurosci 38:3218–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming HPegasiou CMPitera APKariis HHoughton SDKurbatskaya KAhmed AGrundy PVajramani GBulters D, et al. (2019) A primate-specific short GluN2A-NMDA receptor isoform is expressed in the human brain. Mol Brain 12:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnet XL, Bakke Krog H, Sevillano-Quispe OG, Poulsen H, Kjaergaard M (2020) The C-terminal domains of the NMDA receptor: How intrinsically disordered tails affect signalling, plasticity and disease. Eur J Neurosci. [DOI] [PubMed]
- Washbourne P, Bennett JE, McAllister AK (2002) Rapid recruitment of NMDA receptor transport packets to nascent synapses. Nat Neurosci 5:751–759. [DOI] [PubMed] [Google Scholar]
- Washburn HR, Xia NL, Zhou W, Mao YT, Dalva MB (2020) Positive surface charge of GluN1 N-terminus mediates the direct interaction with EphB2 and NMDAR mobility. Nat Commun 11:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MS, Numberger M, Zhang S, Dingledine R (1997) Differential dependence on GluR2 expression of three characteristic features of AMPA receptors. J Neurosci 17:9393–9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe C, Orito T, Watanabe H, Mizoguchi H, Yonezawa A, Yanai K, Mobarakeh JI, Onodera K, Sakurada T, Sakurada S (2008) Intrathecal high-dose histamine induces spinally-mediated nociceptive behavioral responses through a polyamine site of NMDA receptors. Eur J Pharmacol 581:54–63. [DOI] [PubMed] [Google Scholar]
- Watanabe J, Beck C, Kuner T, Premkumar LS, Wollmuth LP (2002) DRPEER: a motif in the extracellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. J Neurosci 22:10209–10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Inoue Y, Sakimura K, Mishina M (1992) Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport 3:1138–1140. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Inoue Y, Sakimura K, Mishina M (1993) Distinct spatio-temporal distributions of the NMDA receptor channel subunit mRNAs in the brain. Ann N Y Acad Sci 707:463–466. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Mishina M, Inoue Y (1994) Distinct spatiotemporal distributions of the N-methyl-D-aspartate receptor channel subunit mRNAs in the mouse cervical cord. J Comp Neurol 345:314–319. [DOI] [PubMed] [Google Scholar]
- Watkins JC, Jane DE (2006) The glutamate story. Br J Pharmacol 147 (Suppl 1):S100–S108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson GB, Lanthorn TH (1990) Pharmacological characteristics of cyclic homologues of glycine at the N-methyl-D-aspartate receptor-associated glycine site. Neuropharmacology 29:727–730. [DOI] [PubMed] [Google Scholar]
- Watson JF, Ho H, Greger IH (2017) Synaptic transmission and plasticity require AMPA receptor anchoring via its N-terminal domain. eLife 6:e23024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RJiang YBermudez IHoulihan LClover LMcKnight KCross JHHart IKRoubertie AValmier J, et al. (2004) Absence of antibodies to glutamate receptor type 3 (GluR3) in Rasmussen encephalitis. Neurology 63:43–50. [DOI] [PubMed] [Google Scholar]
- Wee KS, Tan FC, Cheong YP, Khanna S, Low CM (2016) Ontogenic profile and synaptic distribution of GluN3 proteins in the rat brain and hippocampal neurons. Neurochem Res 41:290–297. [DOI] [PubMed] [Google Scholar]
- Wee KS, Wee ZN, Chow NB, Low CM (2010) The distal carboxyl terminal of rat NR3B subunit regulates NR1-1a/NR3B and NR1-2a/NR3B surface trafficking. Neurochem Int 57:97–101. [DOI] [PubMed] [Google Scholar]
- Wee KS, Zhang Y, Khanna S, Low CM (2008) Immunolocalization of NMDA receptor subunit NR3B in selected structures in the rat forebrain, cerebellum, and lumbar spinal cord. J Comp Neurol 509:118–135. [DOI] [PubMed] [Google Scholar]
- Wei DS, Mei YA, Bagal A, Kao JP, Thompson SM, Tang CM (2001) Compartmentalized and binary behavior of terminal dendrites in hippocampal pyramidal neurons. Science 293:2272–2275. [DOI] [PubMed] [Google Scholar]
- Wei M, Jia M, Zhang J, Yu L, Zhao Y, Chen Y, Ma Y, Zhang W, Shi YS, Zhang C (2017) The inhibitory effect of α/β-hydrolase domain-containing 6 (ABHD6) on the surface targeting of GluA2- and GluA3-containing AMPA receptors. Front Mol Neurosci 10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss BAlt AOgden AMGates MDieckman DKClemens-Smith AHo KHJarvie KRizkalla GWright RA, et al. (2006) Pharmacological characterization of the competitive GLUK5 receptor antagonist decahydroisoquinoline LY466195 in vitro and in vivo. J Pharmacol Exp Ther 318:772–781. [DOI] [PubMed] [Google Scholar]
- Welch WMEwing FEHuang JMenniti FSPagnozzi MJKelly KSeymour PAGuanowsky VGuhan SGuinn MR, et al. (2001) Atropisomeric quinazolin-4-one derivatives are potent noncompetitive alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor antagonists. Bioorg Med Chem Lett 11:177–181. [DOI] [PubMed] [Google Scholar]
- Wells G, Yuan H, McDaniel MJ, Kusumoto H, Snyder JP, Liotta DC, Traynelis SF (2018) The GluN2B-Glu413Gly NMDA receptor variant arising from a de novo GRIN2B mutation promotes ligand-unbinding and domain opening. Proteins 86:1265–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Lin CY, Niu L (2017) R/G editing in GluA2Rflop modulates the functional difference between GluA1 flip and flop variants in GluA1/2R heteromeric channels. Sci Rep 7:13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenke NKKreye JAndrzejak Evan Casteren ALeubner JMurgueitio MSReincke SMSecker CSchmidl LGeis C, et al. (2019) N-methyl-D-aspartate receptor dysfunction by unmutated human antibodies against the NR1 subunit. Ann Neurol 85:771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenthold RJ, Petralia RS, Blahos J II, Niedzielski AS (1996) Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J Neurosci 16:1982–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A, Fritschy JM, Mohler H, Benke D (1997) NMDA receptor heterogeneity during postnatal development of the rat brain: differential expression of the NR2A, NR2B, and NR2C subunit proteins. J Neurochem 68:469–478. [DOI] [PubMed] [Google Scholar]
- Wenzel A, Villa M, Mohler H, Benke D (1996) Developmental and regional expression of NMDA receptor subtypes containing the NR2D subunit in rat brain. J Neurochem 66:1240–1248. [DOI] [PubMed] [Google Scholar]
- Werner P, Voigt M, Keinänen K, Wisden W, Seeburg PH (1991) Cloning of a putative high-affinity kainate receptor expressed predominantly in hippocampal CA3 cells. Nature 351:742–744. [DOI] [PubMed] [Google Scholar]
- West PJ, Dalpé-Charron A, Wilcox KS (2007) Differential contribution of kainate receptors to excitatory postsynaptic currents in superficial layer neurons of the rat medial entorhinal cortex. Neuroscience 146:1000–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MC, Gertler C, Mayer ML, Rosenmund C (2006a) Interdomain interactions in AMPA and kainate receptors regulate affinity for glutamate. J Neurosci 26:7650–7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML (2006b) Conformational restriction blocks glutamate receptor desensitization. Nat Struct Mol Biol 13:1120–1127. [DOI] [PubMed] [Google Scholar]
- Whalley HC, Pickard BS, McIntosh AM, Zuliani R, Johnstone EC, Blackwood DH, Lawrie SM, Muir WJ, Hall J (2009) A GRIK4 variant conferring protection against bipolar disorder modulates hippocampal function. Mol Psychiatry 14:467–468. [DOI] [PubMed] [Google Scholar]
- Whitney KD, McNamara JO (2000) GluR3 autoantibodies destroy neural cells in a complement-dependent manner modulated by complement regulatory proteins. J Neurosci 20:7307–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widman AJ, McMahon LL (2018) Disinhibition of CA1 pyramidal cells by low-dose ketamine and other antagonists with rapid antidepressant efficacy. Proc Natl Acad Sci USA 115:E3007–E3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wied TJ, Chin AC, Lau AY (2019) High conformational variability in the GluK2 kainate receptor ligand-binding domain. Structure 27:189–195.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiera G, Mozrzymas JW (2015) Extracellular proteolysis in structural and functional plasticity of mossy fiber synapses in hippocampus. Front Cell Neurosci 9:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild AR, Bollands M, Morris PG, Jones S (2015) Mechanisms regulating spill-over of synaptic glutamate to extrasynaptic NMDA receptors in mouse substantia nigra dopaminergic neurons. Eur J Neurosci 42:2633–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild AR, Dell’Acqua ML (2018) Potential for therapeutic targeting of AKAP signaling complexes in nervous system disorders. Pharmacol Ther 185:99–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild AR, Sinnen BL, Dittmer PJ, Kennedy MJ, Sather WA, Dell’Acqua ML (2019) Synapse-to-nucleus communication through NFAT is mediated by L-type Ca2+ channel Ca2+ spike propagation to the soma. Cell Rep 26:3537–3550.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Chai YH, Huettner JE (1998) Inhibition of rat neuronal kainate receptors by cis-unsaturated fatty acids. J Physiol 513:331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Chen K, Huettner JE (2010) Fatty acid modulation and polyamine block of GluK2 kainate receptors analyzed by scanning mutagenesis. J Gen Physiol 136:339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Fulling E, Zhou Y, Huettner JE (2008) Amino acid substitutions in the pore helix of GluR6 control inhibition by membrane fatty acids. J Gen Physiol 132:85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Huettner JE (1995) Differential antagonism of alpha-amino-3-hydroxy-5-methyl-4- isoxazolepropionic acid-preferring and kainate-preferring receptors by 2,3-benzodiazepines. Mol Pharmacol 47:582–587. [PubMed] [Google Scholar]
- Wilding TJ, Huettner JE (1997) Activation and desensitization of hippocampal kainate receptors. J Neurosci 17:2713–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Huettner JE (2019) Cadmium opens GluK2 kainate receptors with cysteine substitutions at the M3 helix bundle crossing. J Gen Physiol 151:435–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Huettner JE (2020) Cadmium activates AMPA and NMDA receptors with M3 helix cysteine substitutions. J Gen Physiol 152:e201912537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Lopez MN, Huettner JE (2016) Chimeric glutamate receptor subunits reveal the transmembrane domain is sufficient for NMDA receptor pore properties but some positive allosteric modulators require additional domains. J Neurosci 36:8815–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Zhou Y, Huettner JE (2005) Q/R site editing controls kainate receptor inhibition by membrane fatty acids. J Neurosci 25:9470–9478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson ST, Sanacora G (2019) A new generation of antidepressants: an update on the pharmaceutical pipeline for novel and rapid-acting therapeutics in mood disorders based on glutamate/GABA neurotransmitter systems. Drug Discov Today 24:606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K (1993) Ifenprodil discriminates subtypes of the N-methyl-D-aspartate receptor: selectivity and mechanisms at recombinant heteromeric receptors. Mol Pharmacol 44:851–859. [PubMed] [Google Scholar]
- Williams K (1994) Subunit-specific potentiation of recombinant N-methyl-D-aspartate receptors by histamine. Mol Pharmacol 46:531–541. [PubMed] [Google Scholar]
- Williams K (1996) Separating dual effects of zinc at recombinant N-methyl-D-aspartate receptors. Neurosci Lett 215:9–12. [DOI] [PubMed] [Google Scholar]
- Williams K (1997) Modulation and block of ion channels: a new biology of polyamines. Cell Signal 9:1–13. [DOI] [PubMed] [Google Scholar]
- Williams K, Dattilo M, Sabado TN, Kashiwagi K, Igarashi K (2003) Pharmacology of delta2 glutamate receptors: effects of pentamidine and protons. J Pharmacol Exp Ther 305:740–748. [DOI] [PubMed] [Google Scholar]
- Williams K, Kashiwagi K, Fukuchi J, Igarashi K (1995) An acidic amino acid in the N-methyl-D-aspartate receptor that is important for spermine stimulation. Mol Pharmacol 48:1087–1098. [PubMed] [Google Scholar]
- Wisden W, Seeburg PH (1993) A complex mosaic of high-affinity kainate receptors in rat brain. J Neurosci 13:3582–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekindt B, Malany S, Schemm R, Otvos L, Maccecchini ML, Laube B, Betz H (2001) Point mutations identify the glutamate binding pocket of the N-methyl-D-aspartate receptor as major site of conantokin-G inhibition. Neuropharmacology 41:753–761. [DOI] [PubMed] [Google Scholar]
- Wlaź P, Socała K, Nieoczym D, Łuszczki JJ, Zarnowska I, Zarnowski T, Czuczwar SJ, Gasior M (2012) Anticonvulsant profile of caprylic acid, a main constituent of the medium-chain triglyceride (MCT) ketogenic diet, in mice. Neuropharmacology 62:1882–1889. [DOI] [PubMed] [Google Scholar]
- Wo ZG, Oswald RE (1994) Transmembrane topology of two kainate receptor subunits revealed by N-glycosylation. Proc Natl Acad Sci USA 91:7154–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wo ZG, Oswald RE (1995) Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci 18:161–168. [DOI] [PubMed] [Google Scholar]
- Wolf ME, Tseng KY (2012) Calcium-permeable AMPA receptors in the VTA and nucleus accumbens after cocaine exposure: when, how, and why? Front Mol Neurosci 5:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP (2018) Ion permeation in ionotropic glutamate receptors: Still dynamic after all these years. Curr Opin Physiol 2:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP, Kuner T, Jatzke C, Seeburg PH, Heintz N, Zuo J (2000) The Lurcher mutation identifies delta 2 as an AMPA/kainate receptor-like channel that is potentiated by Ca(2+). J Neurosci 20:5973–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP, Kuner T, Sakmann B (1998) Adjacent asparagines in the NR2-subunit of the NMDA receptor channel control the voltage-dependent block by extracellular Mg2+. J Physiol 506:13–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP, Kuner T, Seeburg PH, Sakmann B (1996) Differential contribution of the NR1- and NR2A-subunits to the selectivity filter of recombinant NMDA receptor channels. J Physiol 491:779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP, Sakmann B (1998) Different mechanisms of Ca2+ transport in NMDA and Ca2+-permeable AMPA glutamate receptor channels. J Gen Physiol 112:623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP, Sobolevsky AI (2004) Structure and gating of the glutamate receptor ion channel. Trends Neurosci 27:321–328. [DOI] [PubMed] [Google Scholar]
- Wolosker H, Balu DT (2020) D-Serine as the gatekeeper of NMDA receptor activity: implications for the pharmacologic management of anxiety disorders. Transl Psychiatry 10:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S, Roche KW (2021) Regulation of glutamate receptors by striatal-enriched tyrosine phosphatase 61 (STEP61 ). J Physiol 599:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AY, Fay AM, Bowie D (2006) External ions are coactivators of kainate receptors. J Neurosci 26:5750–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AY, MacLean DM, Bowie D (2007) Na+/Cl- dipole couples agonist binding to kainate receptor activation. J Neurosci 27:6800–6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HKLiu XBMatos MFChan SFPérez-Otaño IBoysen MCui JNakanishi NTrimmer JSJones EG, et al. (2002) Temporal and regional expression of NMDA receptor subunit NR3A in the mammalian brain. J Comp Neurol 450:303–317. [DOI] [PubMed] [Google Scholar]
- Wong JM, Folorunso OO, Barragan EV, Berciu C, Harvey TL, Coyle JT, Balu DT, Gray JA (2020) Postsynaptic serine racemase regulates NMDA receptor function. J Neurosci 40:9564–9575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, Gray JA (2018) Long-term depression is independent of GluN2 subunit composition. J Neurosci 38:4462–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M, Moss RL (1994) Patch-clamp analysis of direct steroidal modulation of glutamate receptor-channels. J Neuroendocrinol 6:347–355. [DOI] [PubMed] [Google Scholar]
- Wong WT, Wang M, Li W (2011) Regulation of microglia by ionotropic glutamatergic and GABAergic neurotransmission. Neuron Glia Biol 7:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MW, VanDongen HM, VanDongen AM (1995) Structural conservation of ion conduction pathways in K channels and glutamate receptors. Proc Natl Acad Sci USA 92:4882–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhall G, Evans DI, Cunningham MO, Jones RS (2001) NR2B-containing NMDA autoreceptors at synapses on entorhinal cortical neurons. J Neurophysiol 86:1644–1651. [DOI] [PubMed] [Google Scholar]
- Woodhams SG, Markus R, Gowler PRW, Self TJ, Chapman V (2019) Cell type-specific super-resolution imaging reveals an increase in calcium-permeable AMPA receptors at spinal peptidergic terminals as an anatomical correlate of inflammatory pain. Pain 160:2641–2650. [DOI] [PubMed] [Google Scholar]
- Wroge CM, Hogins J, Eisenman L, Mennerick S (2012) Synaptic NMDA receptors mediate hypoxic excitotoxic death. J Neurosci 32:6732–6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Bacaj T, Morishita W, Goswami D, Arendt KL, Xu W, Chen L, Malenka RC, Südhof TC (2017) Postsynaptic synaptotagmins mediate AMPA receptor exocytosis during LTP. Nature 544:316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GY, Cline HT (1998) Stabilization of dendritic arbor structure in vivo by CaMKII. Science 279:222–226. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Ko SW, Zhuo M (2007a) Kainate receptors and pain: from dorsal root ganglion to the anterior cingulate cortex. Curr Pharm Des 13:1597–1605. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Xu H, Ren M, Zhuo M (2007b) Genetic and pharmacological studies of GluR5 modulation of inhibitory synaptic transmission in the anterior cingulate cortex of adult mice. Dev Neurobiol 67:146–157. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Zhao MG, Toyoda H, Ko SW, Zhuo M (2005) Kainate receptor-mediated synaptic transmission in the adult anterior cingulate cortex. J Neurophysiol 94:1805–1813. [DOI] [PubMed] [Google Scholar]
- Wu YArai ACRumbaugh GSrivastava AKTurner GHayashi TSuzuki EJiang YZhang LRodriguez J, et al. (2007c) Mutations in ionotropic AMPA receptor 3 alter channel properties and are associated with moderate cognitive impairment in humans. Proc Natl Acad Sci USA 104:18163–18168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YN, Johnson SW (2015) Memantine selectively blocks extrasynaptic NMDA receptors in rat substantia nigra dopamine neurons. Brain Res 1603:1–7. [DOI] [PubMed] [Google Scholar]
- Wu YW, Grebenyuk S, McHugh TJ, Rusakov DA, Semyanov A (2012) Backpropagating action potentials enable detection of extrasynaptic glutamate by NMDA receptors. Cell Rep 1:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudick MMPortes MTMichard ERosas-Santiago PLizzio MANunes COCampos CSanta Cruz Damineli DCarvalho JCLima PT, et al. (2018) CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca2+ homeostasis. Science 360:533–536. [DOI] [PubMed] [Google Scholar]
- Wyeth MS, Pelkey KA, Petralia RS, Salter MW, McInnes RR, McBain CJ (2014) Neto auxiliary protein interactions regulate kainate and NMDA receptor subunit localization at mossy fiber-CA3 pyramidal cell synapses. J Neurosci 34:622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyeth MSPelkey KAYuan XVargish GJohnston ADHunt SFang CAbebe DMahadevan VFisahn A, et al. (2017) Neto auxiliary subunits regulate interneuron somatodendritic and presynaptic kainate receptors to control network inhibition. Cell Rep 20:2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie DJ, Béhé P, Colquhoun D (1998) Single-channel activations and concentration jumps: comparison of recombinant NR1a/NR2A and NR1a/NR2D NMDA receptors. J Physiol 510:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie DJ, Béhé P, Nassar M, Schoepfer R, Colquhoun D (1996) Single-channel currents from recombinant NMDA NR1a/NR2D receptors expressed in Xenopus oocytes. Proc Biol Sci 263:1079–1086. [DOI] [PubMed] [Google Scholar]
- Wyllie DJ, Chen PE (2007) Taking the time to study competitive antagonism. Br J Pharmacol 150:541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XiangWei W, Jiang Y, Yuan H (2018) De novo mutations and rare variants occurring in NMDA receptors. Curr Opin Physiol 2:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XiangWei WKannan VXu YKosobucki GJSchulien AJKusumoto HMoufawad El Achkar CBhattacharya SLesca GNguyen S, et al. (2019) Heterogeneous clinical and functional features of GRIN2D-related developmental and epileptic encephalopathy. Brain 142:3009–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie JD, Chen SR, Chen H, Zeng WA, Pan HL (2016) Presynaptic N-methyl-d-aspartate (NMDA) receptor activity is increased through protein kinase C in paclitaxel-induced neuropathic pain. J Biol Chem 291:19364–19373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Hopf C, Reddy R, Cho RW, Guo L, Lanahan A, Petralia RS, Wenthold RJ, O’Brien RJ, Worley P (2003) Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron 39:513–528. [DOI] [PubMed] [Google Scholar]
- Xu J, Kurup P, Zhang Y, Goebel-Goody SM, Wu PH, Hawasli AH, Baum ML, Bibb JA, Lombroso PJ (2009) Extrasynaptic NMDA receptors couple preferentially to excitotoxicity via calpain-mediated cleavage of STEP. J Neurosci 29:9330–9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Löwel S, Schlüter OM (2020) Silent synapse-based mechanisms of critical period plasticity. Front Cell Neurosci 14:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XX, Luo JH (2018) Mutations of N-methyl-D-aspartate receptor subunits in epilepsy. Neurosci Bull 34:549–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Song R, Chen W, Strong K, Shrey D, Gedela S, Traynelis SF, Zhang G, Yuan H (2021) Recurrent seizure-related GRIN1 variant: Molecular mechanism and targeted therapy. Ann Clin Transl Neurol 8:1480–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Gupta SC, Hillman BG, Bhatt JM, Stairs DJ, Dravid SM (2012) Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS One 7:e32969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Hillman BG, Gupta SC, Suryavanshi P, Bhatt JM, Pavuluri R, Stairs DJ, Dravid SM (2013) Deletion of glutamate delta-1 receptor in mouse leads to enhanced working memory and deficit in fear conditioning. PLoS One 8:e60785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Rimerman R, Scofield MA, Dravid SM (2011) Mutations in the transmembrane domain M3 generate spontaneously open orphan glutamate δ1 receptor. Brain Res 1382:1–8. [DOI] [PubMed] [Google Scholar]
- Yamada J, Jinno S (2019) Potential link between antidepressant-like effects of ketamine and promotion of adult neurogenesis in the ventral hippocampus of mice. Neuropharmacology 158:107710. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Donevan SD, Rogawski MA (1993) Anticonvulsant activity of AMPA/kainate antagonists: comparison of GYKI 52466 and NBOX in maximal electroshock and chemoconvulsant seizure models. Epilepsy Res 15:179–184. [DOI] [PubMed] [Google Scholar]
- Yamakura T, Mori H, Masaki H, Shimoji K, Mishina M (1993) Different sensitivities of NMDA receptor channel subtypes to non-competitive antagonists. Neuroreport 4:687–690. [DOI] [PubMed] [Google Scholar]
- Yamamoto HKamegaya ESawada WHasegawa RYamamoto THagino YTakamatsu YImai KKoga HMishina M, et al. (2013) Involvement of the N-methyl-D-aspartate receptor GluN2D subunit in phencyclidine-induced motor impairment, gene expression, and increased Fos immunoreactivity. Mol Brain 6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki M, Fukaya M, Yamazaki M, Azechi H, Natsume R, Abe M, Sakimura K, Watanabe M (2016) TARP γ-2 and γ-8 differentially control AMPAR density across Schaffer collateral/commissural synapses in the hippocampal CA1 area. J Neurosci 36:4296–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki M, Okada R, Takasaki C, Toki S, Fukaya M, Natsume R, Sakimura K, Mishina M, Shirakawa T, Watanabe M (2014) Opposing role of NMDA receptor GluN2B and GluN2D in somatosensory development and maturation. J Neurosci 34:11534–11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Kawaguchi SY, Hirano T (2013) Contribution of postsynaptic GluD2 to presynaptic R-type Ca(2+) channel function, glutamate release and long-term potentiation at parallel fiber to Purkinje cell synapses. Cerebellum 12:657–666. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Kwak S (2019) Cell death cascade and molecular therapy in ADAR2-deficient motor neurons of ALS. Neurosci Res 144:4–13. [DOI] [PubMed] [Google Scholar]
- Yamazaki MFukaya MHashimoto KYamasaki MTsujita MItakura MAbe MNatsume RTakahashi MKano M, et al. (2010) TARPs gamma-2 and gamma-7 are essential for AMPA receptor expression in the cerebellum. Eur J Neurosci 31:2204–2220. [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Le Pichon CE, Jackson AC, Cerpas M, Sakimura K, Scearce-Levie K, Nicoll RA (2015) Relative contribution of TARPs γ-2 and γ-7 to cerebellar excitatory synaptic transmission and motor behavior. Proc Natl Acad Sci USA 112:E371–E379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Tomita S (2012) Defined criteria for auxiliary subunits of glutamate receptors. J Physiol 590:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Bengtson CP, Buchthal B, Hagenston AM, Bading H (2020) Coupling of NMDA receptors and TRPM4 guides discovery of unconventional neuroprotectants. Science 370:eaay3302. [DOI] [PubMed] [Google Scholar]
- Yan S, Sanders JM, Xu J, Zhu Y, Contractor A, Swanson GT (2004) A C-terminal determinant of GluR6 kainate receptor trafficking. J Neurosci 24:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Yang J, Luo A, Hashimoto K (2019) Molecular and cellular mechanisms underlying the antidepressant effects of ketamine enantiomers and its metabolites. Transl Psychiatry 9:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CR, Svensson KA (2008) Allosteric modulation of NMDA receptor via elevation of brain glycine and D-serine: the therapeutic potentials for schizophrenia. Pharmacol Ther 120:317–332. [DOI] [PubMed] [Google Scholar]
- Yang J, Wang S, Yang Z, Hodgkinson CA, Iarikova P, Ma JZ, Payne TJ, Goldman D, Li MD (2015) The contribution of rare and common variants in 30 genes to risk nicotine dependence. Mol Psychiatry 20:1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LP, Deeks ED (2015) Dextromethorphan/quinidine: a review of its use in adults with pseudobulbar affect. Drugs 75:83–90. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang XB, Zhou Q (2010) Perisynaptic GluR2-lacking AMPA receptors control the reversibility of synaptic and spines modifications. Proc Natl Acad Sci USA 107:11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yuan C, Shen SQ, Wang XE, Mei QH, Jiang WQ, Huang Q (2017) Autoantibodies to NR2A peptide of the glutamate/NMDA receptor in patients with seizure disorders in neuropsychiatric systemic lupus erythematosus. Mediators Inflamm 2017:5047898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankova M, Hart SA, Woolley CS (2001) Estrogen increases synaptic connectivity between single presynaptic inputs and multiple postsynaptic CA1 pyramidal cells: a serial electron-microscopic study. Proc Natl Acad Sci USA 98:3525–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Zong Y, Gu S, Zhou J, Xu H, Mathews II, Jin R (2011) Crystal structure of the glutamate receptor GluA1 N-terminal domain. Biochem J 438:255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Grand T, Hanson JE, Paoletti P, Zhou Q (2018) Higher ambient synaptic glutamate at inhibitory versus excitatory neurons differentially impacts NMDA receptor activity. Nat Commun 9:4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Belcher J, Berger AJ, Mayer ML, Lau AY (2013) Conformational analysis of NMDA receptor GluN1, GluN2, and GluN3 ligand-binding domains reveals subtype-specific characteristics. Structure 21:1788–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML (2008) Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. EMBO J 27:2158–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Mayer ML (2006) Characterization of a soluble ligand binding domain of the NMDA receptor regulatory subunit NR3A. J Neurosci 26:4559–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M, Johnson-Venkatesh EM, Zhang H, Parent JM, Sutton MA, Umemori H (2011) Multiple forms of activity-dependent competition refine hippocampal circuits in vivo. Neuron 70:1128–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura M, Yoshida T, Lee SJ, Uemura T, Joo JY, Mishina M (2012) Glutamate receptor δ1 induces preferentially inhibitory presynaptic differentiation of cortical neurons by interacting with neurexins through cerebellin precursor protein subtypes. J Neurochem 121:705–716. [DOI] [PubMed] [Google Scholar]
- Yelshanskaya MV, Li M, Sobolevsky AI (2014) Structure of an agonist-bound ionotropic glutamate receptor. Science 345:1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya MV, Mesbahi-Vasey S, Kurnikova MG, Sobolevsky AI (2017) Role of the ion channel extracellular collar in AMPA receptor gating. Sci Rep 7:1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya MV, Saotome K, Singh AK, Sobolevsky AI (2016a) Probing intersubunit interfaces in AMPA-subtype ionotropic glutamate receptors. Sci Rep 6:19082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya MV, Singh AK, Narangoda C, Williams RSB, Kurnikova MG, Sobolevsky AI (2020) Structural basis of AMPA receptor inhibition by trans-4-butylcyclohexane carboxylic acid. Br J Pharmacol. [DOI] [PMC free article] [PubMed]
- Yelshanskaya MV, Singh AK, Sampson JM, Narangoda C, Kurnikova M, Sobolevsky AI (2016b) Structural bases of noncompetitive inhibition of AMPA-subtype ionotropic glutamate receptors by antiepileptic drugs. Neuron 91:1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Bhattacharya S, Thompson CM, Traynelis SF, Hansen KB (2019) Functional and pharmacological properties of triheteromeric GluN1/2B/2D NMDA receptors. J Physiol 597:5495–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Mou TC, Dorsett KN, Volkmann RA, Menniti FS, Sprang SR, Hansen KB (2016) Structural basis for negative allosteric modulation of GluN2A-containing NMDA receptors. Neuron 91:1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Rouzbeh N, Hansen KB, Xu Y, Fanger CM, Gordon E, Paschetto K, Menniti FS, Volkmann RA (2020) PTC-174, a positive allosteric modulator of NMDA receptors containing GluN2C or GluN2D subunits. Neuropharmacology 173:107971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Zachariassen LG, Dorsett KN, Hansen KB (2018) Properties of triheteromeric N-methyl-d-aspartate receptors containing two distinct GluN1 isoforms. Mol Pharmacol 93:453–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonkunas M, Buddhadev M, Flores Canales JC, Kurnikova MG (2017) Configurational preference of the glutamate receptor ligand binding domain dimers. Biophys J 112:2291–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AB, Greenamyre JT, Hollingsworth Z, Albin R, D’Amato C, Shoulson I, Penney JB (1988) NMDA receptor losses in putamen from patients with Huntington’s disease. Science 241:981–983. [DOI] [PubMed] [Google Scholar]
- Yozu M, Tabata H, Konig N, Nakajima K (2008) Migratory behavior of presumptive interneurons is affected by AMPA receptor activation in slice cultures of embryonic mouse neocortex. Dev Neurosci 30:105–116. [DOI] [PubMed] [Google Scholar]
- Yu A, Lau AY (2017) Energetics of glutamate binding to an ionotropic glutamate receptor. J Phys Chem B 121:10436–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Lau AY (2018) Glutamate and glycine binding to the NMDA receptor. Structure 26:1035–1043.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Salazar H, Plested AJR, Lau AY (2018a) Neurotransmitter funneling optimizes glutamate receptor kinetics. Neuron 97:139–149.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YLin YTakasaki YWang CKimura HXing JIshizuka KToyama MKushima IMori D, et al. (2018b) Rare loss of function mutations in N-methyl-D-aspartate glutamate receptors and their contributions to schizophrenia susceptibility. Transl Psychiatry 8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CL, Shi EY, Srinivasan J, Ptak CP, Oswald RE, Nowak LM (2018) Modulation of AMPA receptor gating by the anticonvulsant drug, perampanel. ACS Med Chem Lett 10:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF (2009) Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci 29:12045–12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan HHansen KBZhang JPierson TMMarkello TCFajardo KVHolloman CMGolas GAdams DRBoerkoel CF, et al. (2014) Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat Commun 5:3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Low CM, Moody OA, Jenkins A, Traynelis SF (2015a) Ionotropic GABA and glutamate receptor mutations and human neurologic diseases. Mol Pharmacol 88:203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan HMyers SJWells GNicholson KLSwanger SALyuboslavsky PTahirovic YAMenaldino DSGanesh TWilson LJ, et al. (2015b) Context-dependent GluN2B-selective inhibitors of NMDA receptor function are neuroprotective with minimal side effects. Neuron 85:1305–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Mameli M, O’Connor EC, Dey PN, Verpelli C, Sala C, Perez-Otano I, Lüscher C, Bellone C (2013) Expression of cocaine-evoked synaptic plasticity by GluN3A-containing NMDA receptors. Neuron 80:1025–1038. [DOI] [PubMed] [Google Scholar]
- Yuzaki M (2013) Cerebellar LTD vs. motor learning-lessons learned from studying GluD2. Neural Netw 47:36–41. [DOI] [PubMed] [Google Scholar]
- Yuzaki M (2018) Two classes of secreted synaptic organizers in the central nervous system. Annu Rev Physiol 80:243–262. [DOI] [PubMed] [Google Scholar]
- Yuzaki M, Aricescu AR (2017) A GluD coming-of-age story. Trends Neurosci 40:138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariassen LG, Katchan L, Jensen AG, Pickering DS, Plested AJ, Kristensen AS (2016) Structural rearrangement of the intracellular domains during AMPA receptor activation. Proc Natl Acad Sci USA 113:E3950–E3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafra F, Aragón C, Olivares L, Danbolt NC, Giménez C, Storm-Mathisen J (1995) Glycine transporters are differentially expressed among CNS cells. J Neurosci 15:3952–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanillo DSprengel RHvalby OJensen VBurnashev NRozov AKaiser KMKöster HJBorchardt TWorley P, et al. (1999) Importance of AMPA receptors for hippocampal synaptic plasticity but not for spatial learning. Science 284:1805–1811. [DOI] [PubMed] [Google Scholar]
- Zampieri D, Fortuna S, Calabretti A, Romano M, Menegazzi R, Schepmann D, Wünsch B, Collina S, Zanon D, Mamolo MG (2019) Discovery of new potent dual sigma receptor/GluN2b ligands with antioxidant property as neuroprotective agents. Eur J Med Chem 180:268–282. [DOI] [PubMed] [Google Scholar]
- Zampini V, Liu JK, Diana MA, Maldonado PP, Brunel N, Dieudonné S (2016) Mechanisms and functional roles of glutamatergic synapse diversity in a cerebellar circuit. eLife 5:e15872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanjani HS, Vogel MW, Mariani J (2016) Deletion of the GluRδ2 receptor in the Hotfoot mouse mutant causes granule cell loss, delayed Purkinje cell death, and reductions in Purkinje cell dendritic tree area. Cerebellum 15:755–766. [DOI] [PubMed] [Google Scholar]
- Zanos PMoaddel RMorris PJGeorgiou PFischell JElmer GIAlkondon MYuan PPribut HJSingh NS, et al. (2016) NMDAR inhibition-independent antidepressant actions of ketamine metabolites. Nature 533:481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate CA Jr, Brutsche NE, Ibrahim L, Franco-Chaves J, Diazgranados N, Cravchik A, Selter J, Marquardt CA, Liberty V, Luckenbaugh DA (2012) Replication of ketamine’s antidepressant efficacy in bipolar depression: a randomized controlled add-on trial. Biol Psychiatry 71:939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate CA Jr, Singh JB, Carlson PJ, Brutsche NE, Ameli R, Luckenbaugh DA, Charney DS, Manji HK (2006) A randomized trial of an N-methyl-D-aspartate antagonist in treatment-resistant major depression. Arch Gen Psychiatry 63:856–864. [DOI] [PubMed] [Google Scholar]
- Zeisel AMuñoz-Manchado ABCodeluppi SLönnerberg PLa Manno GJuréus AMarques SMunguba HHe LBetsholtz C, et al. (2015) Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347:1138–1142. [DOI] [PubMed] [Google Scholar]
- Zeng M, Díaz-Alonso J, Ye F, Chen X, Xu J, Ji Z, Nicoll RA, Zhang M (2019) Phase separation-mediated TARP/MAGUK complex condensation and AMPA receptor synaptic transmission. Neuron 104:529–543.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerche MWeissenborn KOtt CDere EAsif ARWorthmann HHassouna IRentzsch KTryc ABDahm L, et al. (2015) Preexisting serum autoantibodies against the NMDAR subunit NR1 modulate evolution of lesion size in acute ischemic stroke. Stroke 46:1180–1186. [DOI] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Horwitz AF (2003) Synapse formation is regulated by the signaling adaptor GIT1. J Cell Biol 161:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF (2005) A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci 25:3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Diamond JS (2009) Subunit- and pathway-specific localization of NMDA receptors and scaffolding proteins at ganglion cell synapses in rat retina. J Neurosci 29:4274–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Guan X, Li Q, Meredith AL, Pan HL, Yan J (2018a) Glutamate-activated BK channel complexes formed with NMDA receptors. Proc Natl Acad Sci USA 115:E9006–E9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jacobi AM, Wang T, Berlin R, Volpe BT, Diamond B (2009a) Polyreactive autoantibodies in systemic lupus erythematosus have pathogenic potential. J Autoimmun 33:270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JBChang SXu PMiao MWu HZhang YZhang TWang HZhang JXie C, et al. (2018b) Structural basis of the proton sensitivity of human GluN1-GluN2A NMDA receptors. Cell Rep 25:3582–3590.e4. [DOI] [PubMed] [Google Scholar]
- Zhang L, Quan J, Zhao Y, Yang D, Zhao Q, Liu P, Cheng M, Ma C (2019) Design, synthesis and biological evaluation of 1-benzyl-5-oxopyrrolidine-2-carboximidamide derivatives as novel neuroprotective agents. Eur J Med Chem 182:111654. [DOI] [PubMed] [Google Scholar]
- Zhang LSchessl JWerner MBonnemann CXiong GMojsilovic-Petrovic JZhou WCohen ASeeburg PMisawa H, et al. (2008a) Role of GluR1 in activity-dependent motor system development. J Neurosci 28:9953–9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zheng X, Paupard MC, Wang AP, Santchi L, Friedman LK, Zukin RS, Bennett MV (1994) Spermine potentiation of recombinant N-methyl-D-aspartate receptors is affected by subunit composition. Proc Natl Acad Sci USA 91:10883–10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LI, Tao HW, Poo M (2000) Visual input induces long-term potentiation of developing retinotectal synapses. Nat Neurosci 3:708–715. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Huang X, Chen XZ, Li SY, Yao T, Wu J (2021) Association of gene variations in ionotropic glutamate receptor and attention-deficit/hyperactivity disorder in the Chinese population: A two-stage case-control study. J Atten Disord 25:1362–1373. [DOI] [PubMed] [Google Scholar]
- Zhang S, Ehlers MD, Bernhardt JP, Su CT, Huganir RL (1998) Calmodulin mediates calcium-dependent inactivation of N-methyl-D-aspartate receptors. Neuron 21:443–453. [DOI] [PubMed] [Google Scholar]
- Zhang T, Hou L, Chen DT, McMahon FJ, Wang JC, Rice JP (2018c) Exome sequencing of a large family identifies potential candidate genes contributing risk to bipolar disorder. Gene 645:119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Cho Y, Lolis E, Howe JR (2008b) Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating. J Neurosci 28:932–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Devi SP, Tomita S, Howe JR (2014a) Auxiliary proteins promote modal gating of AMPA- and kainate-type glutamate receptors. Eur J Neurosci 39:1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Eibl C, Weeks AM, Riva I, Li YJ, Plested AJR, Howe JR (2017) Unitary properties of AMPA receptors with reduced desensitization. Biophys J 113:2218–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Howe JR, Popescu GK (2008c) Distinct gating modes determine the biphasic relaxation of NMDA receptor currents. Nat Neurosci 11:1373–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WSt-Gelais FGrabner CPTrinidad JCSumioka AMorimoto-Tomita MKim KSStraub CBurlingame ALHowe JR, et al. (2009b) A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron 61:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu J, Ward MD, Yang S, Chuang YA, Xiao M, Li R, Leahy DJ, Worley PF (2015) Structural basis of arc binding to synaptic proteins: implications for cognitive disease. Neuron 86:490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Feng ZJ, Chergui K (2014b) Allosteric modulation of GluN2C/GluN2D-containing NMDA receptors bidirectionally modulates dopamine release: implication for Parkinson’s disease. Br J Pharmacol 171:3938–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Feng ZJ, Chergui K (2014c) GluN2D-containing NMDA receptors inhibit neurotransmission in the mouse striatum through a cholinergic mechanism: implication for Parkinson’s disease. J Neurochem 129:581–590. [DOI] [PubMed] [Google Scholar]
- Zhang XL, Sullivan JA, Moskal JR, Stanton PK (2008d) A NMDA receptor glycine site partial agonist, GLYX-13, simultaneously enhances LTP and reduces LTD at Schaffer collateral-CA1 synapses in hippocampus. Neuropharmacology 55:1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Buonanno A, Vertes RP, Hoover WB, Lisman JE (2012) NR2C in the thalamic reticular nucleus; effects of the NR2C knockout. PLoS One 7:e41908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YChen KSloan SABennett MLScholze ARO’Keeffe SPhatnani HPGuarnieri PCaneda CRuderisch N, et al. (2014d) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nayeem N, Green T (2008e) Mutations to the kainate receptor subunit GluR6 binding pocket that selectively affect domoate binding. Mol Pharmacol 74:1163–1169. [DOI] [PubMed] [Google Scholar]
- Zhang ZW, Peterson M, Liu H (2013) Essential role of postsynaptic NMDA receptors in developmental refinement of excitatory synapses. Proc Natl Acad Sci USA 110:1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Carney KE, Falgoust L, Pan JW, Sun D, Zhang Z (2016a) Emerging roles of Na+/H+ exchangers in epilepsy and developmental brain disorders. Prog Neurobiol 138-140:19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Lomash S, Chittori S, Glasser C, Mayer ML, Schuck P (2017) Preferential assembly of heteromeric kainate and AMPA receptor amino terminal domains. eLife 6:e32056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Li Y, Wei W, Savage S, Zhou L, Ma D (2014) Ketamine administered to pregnant rats in the second trimester causes long-lasting behavioral disorders in offspring. Neurobiol Dis 68:145–155. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chen S, Swensen AC, Qian WJ, Gouaux E (2019) Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science 364:355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen S, Yoshioka C, Baconguis I, Gouaux E (2016b) Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature 536:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CY, Chang K, Suh YH, Roche KW (2015) TARP γ-8 glycosylation regulates the surface expression of AMPA receptors. Biochem J 465:471–477. [DOI] [PubMed] [Google Scholar]
- Zheng F, Erreger K, Low CM, Banke T, Lee CJ, Conn PJ, Traynelis SF (2001) Allosteric interaction between the amino terminal domain and the ligand binding domain of NR2A. Nat Neurosci 4:894–901. [DOI] [PubMed] [Google Scholar]
- Zheng X, Zhang L, Durand GM, Bennett MV, Zukin RS (1994) Mutagenesis rescues spermine and Zn2+ potentiation of recombinant NMDA receptors. Neuron 12:811–818. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Brockie PJ, Mellem JE, Madsen DM, Walker CS, Francis MM, Maricq AV (2006) SOL-1 is an auxiliary subunit that modulates the gating of GLR-1 glutamate receptors in Caenorhabditis elegans. Proc Natl Acad Sci USA 103:1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Mellem JE, Brockie PJ, Madsen DM, Maricq AV (2004) SOL-1 is a CUB-domain protein required for GLR-1 glutamate receptor function in C. elegans. Nature 427:451–457. [DOI] [PubMed] [Google Scholar]
- Zhong J, Carrozza DP, Williams K, Pritchett DB, Molinoff PB (1995) Expression of mRNAs encoding subunits of the NMDA receptor in developing rat brain. J Neurochem 64:531–539. [DOI] [PubMed] [Google Scholar]
- Zhou HX (2017) Gating motions and stationary gating properties of ionotropic glutamate receptors: Computation meets electrophysiology. Acc Chem Res 50:814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Wollmuth LP (2017) Advancing NMDA receptor physiology by integrating multiple approaches. Trends Neurosci 40:129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Duan J (2018) The C-terminus of NMDAR GluN1-1a subunit translocates to nucleus and regulates synaptic function. Front Cell Neurosci 12:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Chen Z, Yun W, Ren J, Li C, Wang H (2015) Extrasynaptic NMDA receptor in excitotoxicity: Function revisited. Neuroscientist 21:337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Ding Q, Chen Z, Yun H, Wang H (2013a) Involvement of the GluN2A and GluN2B subunits in synaptic and extrasynaptic N-methyl-D-aspartate receptor function and neuronal excitotoxicity. J Biol Chem 288:24151–24159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Hollern D, Liao J, Andrechek E, Wang H (2013b) NMDA receptor-mediated excitotoxicity depends on the coactivation of synaptic and extrasynaptic receptors. Cell Death Dis 4:e560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Takahashi E, Li W, Halt A, Wiltgen B, Ehninger D, Li GD, Hell JW, Kennedy MB, Silva AJ (2007) Interactions between the NR2B receptor and CaMKII modulate synaptic plasticity and spatial learning. J Neurosci 27:13843–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Liu A, Xia S, Leung C, Qi J, Meng Y, Xie W, Park P, Collingridge GL, Jia Z (2018) The C-terminal tails of endogenous GluA1 and GluA2 differentially contribute to hippocampal synaptic plasticity and learning. Nat Neurosci 21:50–62. [DOI] [PubMed] [Google Scholar]
- Zhu JJ, Esteban JA, Hayashi Y, Malinow R (2000) Postnatal synaptic potentiation: delivery of GluR4-containing AMPA receptors by spontaneous activity. Nat Neurosci 3:1098–1106. [DOI] [PubMed] [Google Scholar]
- Zhu S, Paoletti P (2015) Allosteric modulators of NMDA receptors: multiple sites and mechanisms. Curr Opin Pharmacol 20:14–23. [DOI] [PubMed] [Google Scholar]
- Zhu S, Stein RA, Yoshioka C, Lee CH, Goehring A, Mchaourab HS, Gouaux E (2016) Mechanism of NMDA receptor inhibition and activation. Cell 165:704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Stroebel D, Yao CA, Taly A, Paoletti P (2013) Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nat Struct Mol Biol 20:477–485. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Kalbfleisch T, Brennan MD, Li Y (2009) A MicroRNA gene is hosted in an intron of a schizophrenia-susceptibility gene. Schizophr Res 109:86–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ZYi FEpplin MPLiu DSummer SLMizu RShaulsky GXiangWei WTang WBurger PB, et al. (2020) Negative allosteric modulation of GluN1/GluN3 NMDA receptors. Neuropharmacology 176:108117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman SS, Khatri A, Garnier-Amblard EC, Mullasseril P, Kurtkaya NL, Gyoneva S, Hansen KB, Traynelis SF, Liotta DC (2014) Design, synthesis, and structure-activity relationship of a novel series of GluN2C-selective potentiators. J Med Chem 57:2334–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski L, Mordukhovich I, Chen DM, Chisari M, Shu HJ, Lambert PM, Qian M, Zorumski CF, Covey DF, Mennerick S (2021) A neuroactive steroid with a therapeutically interesting constellation of actions at GABAA and NMDA receptors. Neuropharmacology 183:108358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zscherp R, Baumeister S, Schepmann D, Wünsch B (2018) Pyridine bioisosteres of potent GluN2B subunit containing NMDA receptor antagonists with benzo[7]annulene scaffold. Eur J Med Chem 157:397–404. [DOI] [PubMed] [Google Scholar]
- Zucker RS, Regehr WG (2002) Short-term synaptic plasticity. Annu Rev Physiol 64:355–405. [DOI] [PubMed] [Google Scholar]
- Zuo J, De Jager PL, Takahashi KA, Jiang W, Linden DJ, Heintz N (1997) Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature 388:769–773. [DOI] [PubMed] [Google Scholar]
- Zwart R, Sher E, Ping X, Jin X, Sims JR Jr, Chappell AS, Gleason SD, Hahn PJ, Gardinier K, Gernert DL, et al. (2014) Perampanel, an antagonist of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors, for the treatment of epilepsy: studies in human epileptic brain and nonepileptic brain and in rodent models. J Pharmacol Exp Ther 351:124–133. [DOI] [PubMed] [Google Scholar]