Abstract
Purpose:
To investigate the therapeutic role of a novel telomere-directed inhibitor, 6-thio-2’-deoxyguanosine (THIO) in gliomas both in vitro and in vivo.
Experimental Design:
A panel of human and mouse glioma cell lines were used to test therapeutic efficacy of THIO using cell viability assays, flow cytometric analyses and immunofluorescence. Integrated analyses of RNA sequencing and reverse phase protein array data revealed the potential anti-tumor mechanisms of THIO. Four patient-derived xenografts (PDX), two patient-derived organoid (PDOs) and two xenografts of human glioma cell lines were used to further investigate the therapeutic efficacy of THIO.
Results:
THIO was effective in the majority of human and mouse glioma cell lines with no obvious toxicity against normal astrocytes. THIO as a monotherapy demonstrated efficacy in three glioma cell lines that had acquired resistance to TMZ. In addition, THIO showed efficacy in 4 human glioma cell lines grown as neurospheres by inducing apoptotic cell death. Mechanistically, THIO induced telomeric DNA damage not only in glioma cell lines but also in PDX tumor specimens. Integrated computational analyses of transcriptomic and proteomic data indicated that THIO significantly inhibited cell invasion, stem cell and proliferation pathways while triggering DNA damage and apoptosis. Importantly, THIO significantly decreased tumor proliferation in two PDO models and reduced the tumor size of a GBM xenograft and a PDX model.
Conclusions:
The current study established the therapeutic role of THIO in primary and recurrent gliomas and revealed the acute induction of telomeric DNA damage as a primary anti-tumor mechanism of THIO in gliomas.
Introduction
Gliomas are the most common primary tumor of the central nervous system (CNS), which account for approximately 80% of malignant primary tumors (1). CNS malignancies are classified into WHO grade I to IV based on level of tumor differentiation (2). Grade IV gliomas are often referred to as glioblastoma (GBM), and account for one of the most aggressive CNS tumors. Despite standard therapies for GBM, including surgery followed by postoperative irradiation with concomitant and adjuvant temozolomide (TMZ), most patients will die within 2 years from diagnosis with a median survival of 15 to 17 months along with a 5-year overall survival rate of 5.5% (3).
In many human cancers, telomere maintenance is critical for immortality (4). Cancer cells maintain full-length telomeres through telomerase-dependent mechanisms (85-90%). Telomerase consists of the telomerase reverse transcriptase (TERT) protein component, a functional telomerase RNA component (TERC) and other specialized proteins (5). Telomerase synthesizes telomeres by the catalytic protein TERT and RNA template TERC to maintain telomere length in almost all cancer cells (5). The expression of TERT is the rate-limiting step in telomerase activity. TERT promoter mutations create a binding site for the E26 transformation specific (ETS) transcription factor GA-binding protein (GABP), which enhances the expression of TERT (6,7). C250T and C228T, the most common TERT promoter mutations, are detected in 83% of primary glioblastomas (8). The analysis of 18,430 samples across 31 cancer types identified TERT promoter mutations in 89% of GBM and 45% of low grade glioma (LGG) (9). Therefore, telomerase is an attractive therapeutic target in gliomas.
6-thio-2’-deoxyguanosine (6-thio-dG; abbreviated as THIO) is a telomerase substrate precursor nucleoside analogue that is readily incorporated into newly synthesized telomeres by telomerase (10). As a telomerase-dependent telomere uncapping agent, the efficacy of THIO does not depend on the progressive shortening of telomere length. Compared to other direct telomerase inhibitors, THIO leads to DNA damage preferentially localized in telomeres which rapidly results in cell death in telomerase-positive tumor cells but not telomerase-inactive normal cells (10). So far, THIO has been studied in different types of cancer including non-small cell lung cancer (NSCLC), melanoma, colon cancer and pediatric brain tumors (11). Specifically, THIO induced telomere dysfunction in telomerase-positive colon cancer cells and human fibroblasts where hTERT was over-expressed, but did not in telomerase-silent normal cells (12). THIO also significantly suppressed tumor growth of A549 lung cancer cells in vivo with minimal toxicity observed in normal organs of mice (12). THIO significantly inhibited cell viability, proliferation, and tumor growth of melanoma cells that were resistant to inhibitors targeting the MAPK pathway (MAPKi) and/or immune checkpoint inhibitors, both in vitro and in vivo (13). THIO downregulated AXL, which is a well-known marker of therapy resistance for malignant melanoma (13). Similarly, THIO resulted in tumor reduction of both targeted therapy- and chemotherapy-resistant NSCLC human cells in vivo (14). THIO not only induced telomeric and genomic DNA damage but also stimulated a ROS-mediated adaptive response. Telomere-associated DNA damage caused by THIO can activate the host cytosolic DNA sensing STING/interferon I pathway and enhance the efficacy of immunotherapy in advanced cancers (15). THIO also strongly suppressed cell growth of telomerase-positive pediatric brain tumors, including diffuse intrinsic pontine glioma (DIPG), high-grade glioma (HGG), and medulloblastoma (11). THIO has also been demonstrated to successfully penetrat the blood-brain barrier (BBB) and specifically target tumors in an orthotopic patient-derived xenograft model of DIPG (11). Taken together, these studies demonstrate that THIO not only promotes cell death in telomerase-positive tumors but also inhibits therapy-resistant tumors in both in vitro and in vivo models. Furthermore, the ability of THIO to penetrate the BBB is important for treating CNS tumors. These previous studies prompted us to investigate THIO in adult gliomas based on the following rationale: (1) gliomas harbor a high frequency of TERT promoter mutations and (2) THIO not only induces telomeric DNA damage and subsequent cell death in vitro but also crosses the BBB allowing direct targeting of tumors in vivo as a telomerase-mediated telomere inhibitor.
In the current study, we investigated the therapeutic efficacy of THIO in a broad panel of 17 primary glioma cell lines, 3 mouse cell lines, 6 TMZ-resistant glioma cell lines, 4 neurospheres, 2 patient-derived organoids (PDOs) and 2 xenografts of human glioma cell lines and demonstrated that THIO suppressed the proliferation of both TMZ-sensitive and -resistant glioma cells in 2D and neurospheres in 3D derived glioma cell lines by inducing telomeric DNA damage. In addition, tumor proliferation was inhibited in two PDO models, and tumor regression was observed in the xenograft model of a human glioma cell line and a GBM PDX. Integrated analyses of transcriptomic and proteomic data indicated that the molecular action of THIO involved the regulation of various signaling pathways, including cell proliferation, invasion and cell stemness pathways in glioma cells. These preclinical findings highlight the therapeutic role of THIO in gliomas and provide a strong scientific rationale for propelling this telomerase inhibitor into clinical trials to provide a novel and alternative therapeutic strategy for patients with primary and recurrent glioma.
Materials and Methods
Cell lines
The glioma cell line, U251MG was kindly provided to us by Dr. Yiping He (Duke University). Patient-derived glioma cell lines (D2363PXA, D645PXA, D2159MG and D2224MG), patient-derived neurospheres (13.0302), and mouse cell lines (GL261, SMA560 and CT2A) were kindly provided to us by Dr. Darell Bigner (Duke University). TS-603 was obtained from Memorial Sloan Kettering Cancer Center through the material transfer agreement (MTA). The other glioma cell lines (A172, M059K, H4, Hs683, M059J, LN18, LN229, U87MG, U118MG, U138MG, DBTRG-05MG and T98G) used in this study were obtained from ATCC. The normal human astrocytes (NHA) were ordered from Duke Cell Culture Facility. All cell lines used in this study were subject to cell line authentication by STR fingerprinting and Mycoplasma detection by the MycoAlert™ PLUS Mycoplasma Detection Kit (Lonza, LT07-703). The number of passages of all cell lines from thawing to the completion of described experiments did not exceed five. U87MG, T98G, TS-603 and 13.0302 neurospheres grow in stem cell media (Stemcell technologies, 5751) supplemented with proliferation supplement (Stemcell technologies, 5751), 20ng/mL H-EGF (Stemcell technologies, 78006), 10ng/mL H-bFGF (Stemcell technologies, 78003) and 2μg/mL Heparin (Stemcell technologies, 7980). The rest of glioma cell lines were cultured in in Dulbecco's Modified Eagle Medium (DMEM) (ThermoFisher, 11995073) supplemented with 10% fetal bovine serum (FBS) (Hyclone, SH30071.03). Astrocytes were cultured in Astrocyte Growth Medium (Sigma, 821-500). Cells were incubated in a humidified atmosphere with 5% CO2 at 37°C.
Drug preparation
THIO (12) was dissolved in DMSO:water (1:1) to prepare a stock solution of 50 mM, aliquoted, and stored −20 °C. Temozolomize (Sigma-Aldrich, 14163) was dissolved in DMSO to prepare a stock solution of 100 mM, and stored −20 °C. BIBR1532 (Cayman Chemical, 16608) was dissolved in DMSO to prepare a stock solution of 100mM, and stored −20 °C.
FACS analysis of apoptosis and cell death
Adherent cells were harvested with 0.25% Trypsin-EDTA (Gibco, 25200072), and washed once with 1× PBS (Gibco, 10010-023). Cells were then pelleted and stained with PSVue 643 (MTTI, P-1006) at 5μM and propidium iodide (PI) (Alfa Aesar, 25535-16-4) at 10 μg/mL diluted in 1× PBS for 5 minutes in the dark. Cells were then immediately analyzed by FACS using a flow cytometer and at least 5,000 cells per sample were acquired.
FACS analysis of cell division
Cells were stained with CellTrace Violet (Invitrogen, C34557) at 5μM and plated into 12-well plates with 50,000 cells/well. Adherent cells were treated with THIO at 5μM or control. Adherent cells were collected and detected the fluorescent intensity of CellTrace Violet by FACS at day 0-day 4.
Cell viability assay
Cells were plated into 96-well plates with 3,000-5,000 cells/well for 24 hours prior to treatment with THIO at titrated doses of 100, 25, 10, 5, 2.5, 1, 0.1, 0.01 μM for 96 hours. 10 μL Cell Count Kit-8 (CCK-8) (APEBIO, K1018) reagent was added into each well and incubated with cells for 3 hours at 37 °C. And then absorbance was measured at 450 nm by Tecan plate reader. Dose response curves were generated and IC50 were calculated by Graphpad Prism. All samples were analyzed in quadruplicate and standard deviations were from 2-3 independent experiments.
Immunofluorescence assay
M059K cells were plated into a 24-well plate. 24 hours later, cells were treated with THIO at different concentrations, including 0, 1, 2.5, 5 μM. 96 hours after treatment, cells were fixed with 4% paraformaldehyde (ThermoFisher, 28908) for 15 minutes at room temperature. Cells were rinsed three times with 1× PBS for 5 minutes each, permeabilized with PBS containing 0.3% Triton X-100 (ThermoFisher, 9002-93-1) for 10 minutes and blocked with 5% normal goat serum (Cell Signaling Technologies, 5425) for 1 hour. Then, cells were incubated with anti-Ki-67 fluorescent antibody (Cell Signaling Technologies, 11882) overnight at 4 °C in the dark. Cells were rinsed three times with 1× PBS for 5 minutes each, stained with DAPI (Cell Signaling Technologies, 8961) for 5 minutes and then washed four times with 1× PBS. Images were obtained by a fluorescence microscope.
Western blot
Protein was extracted from cells cultured in 6-well plates using RIPA lysis buffer (ThermoFisher, 89900) with protease inhibitors (ThermoFisher, 78440). A total of 30 μg protein lysates was separated by 4-12% NuPage Bis-Tris gels (ThermoFisher, NP0336) and then transferred to polyvinylidene fluoride (PVDF) membranes (BioRad, 1620264). The membranes were blocked with 5% Bovine Serum Albumin (BSA) (Sigma-Aldrich, A7906-100G), probed with primary antibodies overnight at 4 °C, and then incubated with the relevant secondary antibody for 1 hour at room temperature. Bands were visualized using ECL (ThermoFisher, 34080) with BioRad imaging system. The antibodies included anti-cleaved-PARP (Cell Signaling Technologies, 5625), anti-p21 (Cell Signaling Technologies, 2947), anti- p27kip1 (Cell Signaling Technologies, 3686), anti-Rb (Cell Signaling Technologies, 9309), anti-p-Rb (Cell Signaling Technologies, 8516), anti-AXL (Proteintech, 13196-1-AP), anti-ATRX (Novus Biologicals, NBP1-32851), anti-PDGFRβ (Proteintech, 13449-1-AP), anti-HSP70 (Proteintech, 10995-1-AP), anti-γH2AX (Cell Signaling Technologies, 9718T), anti-β-actin (Cell Signaling Technologies, 12620S), anti-rabbit-IgG-HRP (Cell Signaling Technologies, 7074S), and anti-mouse-IgG-HRP (Cell Signaling Technologies, 7076S) per instruction from the data sheet.
RT-qPCR
Total RNA was extracted from cultured cells by miRNeasy Mini Kit (Qiagen, 217004) according to the manufacturer’s instructions. 1 μg of total RNA was reverse transcribed into complementary DNA (cDNA) using cDNA synthesis kit (ThermoFisher, K1651). Amplification of cDNA product was performed using specific primers with the Fast SYBR™ Green Master Mix (ThermoFisher, 4385610) on a Real-Time PCR detection system (BioRad). Samples were analyzed in quadruplicates, and β-tubulin levels were used for normalization. Primers sequences for RT-qPCR were as follows: hTERT: forward primer, 5′-ATGCGACAGTTCGTGGCTCA-3′, and reverse primer, 5′-ATCCCCTGGCACTGGACGTA-3′; and β-tubulin: forward primer, 5′-TGGACTCTGTTCGCTCAGGT-3, and reverse primer, 5′-TGCCTCCTTCCGTACCACAT-3′.
Relative telomere length detection
Genomic DNA (gDNA) was extracted from cultured cells using DNA Mini Kit (Qiagen, 51304) following the manufacturer’s instructions. 20 ng gDNA was used to conduct quantitative PCR (qPCR). Telomere (T) and single copy gene (S) 36B4 were amplified using specific primers with the Fast SYBR™ Green Master Mix (ThermoFisher, 4385610). Primers sequence for qPCR were as follows: Telg: 5’-ACACTAAGGTTTGGGTTTGGGTTTGGGTTTGGGTTAGTGT-3’; Telc: 5’-TGTTAGGTATCCCTATCCCTATCCCTATCCCTATCCCTAACA-3’; and 36B4u: 5’-CAGCAAGTGGGAAGGTGTAATCC-3’; 36B4d: 5’-CCCATTCTATCATCAACGGGTACAA-3’. Relative T/S ratios will reflect relative telomere length as introduced in (16).
Crystal violet staining
Cells were plated into 12-well tissue culture plates at a density of 2,500 cells/well as biological duplicates in drug-free medium. Once cells were attached, cells were treated with drug including TMZ, BIBR1532 and THIO. The medium supplemented with drugs was refreshed every 3 or 4 days for 8-14 days. Cells in plates were then stained with methanol containing 0.05% crystal violet for 30 minutes at room temperature. After extensive washing with distilled water, cells were air-dried and subjected to image acquisition using BioRad Imager. Acetic acid (20%) was used to dissociate crystal violet for quantification. Acetic acid was added into wells (150 μL/well) and incubated for 10 minutes on a shaker. The optical density of acetic acid diluted in water at the ratio of 1:10 was measured at 590 nm by the Tecan plate reader.
Neurosphere formation and treatment
T98G, U87MG, TS603 and 13.0302 cells were cultured in neurobasal medium (Stemcell technologies, 5751) containing 10% proliferation supplement (Stemcell technologies, 5751), 20ng/mL H-EGF (Stemcell technologies, 78006), 10ng/mL H-bFGF (Stemcell technologies, 78003) and 2μg/mL Heparin (Stemcell technologies, 7980). Once neurospheres were formed, they were dissociated by Accutase (VWR, 490007-741) into single cells. Single cells were seeded into 12-well plates (5,000 cells/well) and treated with control medium or THIO at 5 μM for 12 days. Phase contrast images of neurospheres were acquired by ECHO Revolve Microscope at day 3, 6, 9 and 12. At day 12, neurospheres were stained using Live Dead Cell Viability Assay Kit (Sigma-Aldrich, CBA415) and imaged by ECHO Revolve Microscope.
Senescence assay
Senescence β-Galactosidase Staining Kit (Cell Signaling Technologies, 9860) was used to detect senescent cells after treatment with THIO. Briefly, cells were fixed by 1× fixative solution for 15 minutes at room temperature and then incubated with 1× staining solution containing 1mg/mL X-gal in 37°C incubator (no CO2) overnight following the manufacturer’s instruction. Bright field images were acquired by ECHO Revolve Microscope. The numbers of positively and negatively stained cells were counted to quantify the percentage of Senescence β-Galactosidase positive cells.
DNA damage detection
Protein lysates were obtained from H4, D2363PXA, M059K, A172, T98G, Hs638, LN229 and U87MG cells which were treated with or without THIO at 5uM for 72 hours. Western blot was performed to detect the expression of γH2AX. In addition, H4 cells were plated into 24-well plate at a density of 10,000 cells/well. 24 hours later, cells were treated with THIO at different concentrations of 0, 1, 2.5, 5 μM for 48 hours. And then, cells were fixed and used for immunofluorescence staining of γH2AX. Immunofluorescence staining of γH2AX was conducted for tumor specimens derived from D09-0500MG, D09-0394MG, and D10-0021MG PDX models treated with THIO at 2.5 mg/kg. Specifically, 5 μM tissue sections are deparaffinized with xylene (2 × 5 minutes), 100% ethanol (2 × 2 minutes), 95% ethanol (1 × 2 minutes), 75% ethanol (1 × 2 minutes), and 50% ethanol (1 × 2 minutes) and then washed with tap water (2 × 3 minutes). Deparaffinized tissue sections are incubated in sodium citrate buffer (10 mM Na-citrate, 0.05 % Tween 20, pH=6.0) at microwave for 20 minutes to retrieve antigens. After tissue sections cool down, they are rinsed with 1 × PBS for 5 minutes and incubated with blocking buffer (4 % BSA in PBST) for 30 minutes. Sections are incubated with phospho-histone H2AX antibody (1:500) (Cell Signaling, 9718S) in blocking buffer at 4C for O/N. Following 2 × 5 minutes washes with PBST, tissue sections are incubated with Alexaflour 568 conjugated goat anti-Rabbit in blocking buffer at RT for 1 hr. Sections are washed sequentially with PBST (3 × 5 minutes) and PBS (1 × 5 minutes). The slides are mounted with Vectashield mounting medium with DAPI. Images were captured at 63× magnification with an Axio ImagerZ2 equipped with an automatic metaphase capture system (Coolcube1 camera) and analyzed with ImageJ software.
Telomerase activity (TRAP) assay
The TRAP assay was performed with TRAPeze RT Telomerase Detection Kit (S7710) according to the manufacturer's instruction. Specifically, 1×106 cells were washed once in 1× PBS and homogenised in 200 μL of CHAPS lysis buffer. After 30 minutes of incubation on ice, the lysates were centrifuged at 12,000×g for 15 minutes at 4°C, and the supernatant was rapidly frozen and stored at −80°C. Samples were thawed and protein concentration was determined using the DC Protein Assay Kit (#5000111, BioRad). Samples were then mixed at equal protein amounts with 5× TRAPEZE RT Reaction mix and Taq polymerase for a final volume of 25 μL and were run in triplicates on an optical clear PCR plate in a BioRad CFX96 Real Time Thermocycler. Heat inactivated controls for each sample were run on the same plate and Relative Telomerase Activity quantification was performed using a standard curve of TSR8 control templates according to manufacturer's instructions.
Telomere dysfunction induced foci (TIF) assay
D2363PXA cells were seeded at the density of 100.000 on slides and let them attached overnight. Cells were then treated with 1.5 μM THIO for 48 hours. Slides were then rinsed twice with 1× PBS and fixed in 4% formaldehyde (Thermo Fisher) in PBS for 10 minutes. Then, cells were washed twice in 1× PBS shaking and permeabilized in 0.2% PBST for 10 minutes, on ice. Following permeabilization, slides were washed in 1× PBS for 5 minutes and blocked with 1% BSA/PBS for 30 minutes at RT. Primary mouse γH2AX antibody (Millipore) was diluted 1:200 in blocking solution and incubated on cells ON at 4°C. Then, slides were washed in 1× PBS shaking for 10 minutes, four times. Cells were incubated with Alexafluor 488 conjugated goat anti-mouse secondary antibody (Invitrogen) for 1 hour at RT, then washed four times with 1× PBS, for 10 minutes. Cells were fixed in 4% formaldehyde in PBS for 20 minutes at RT and slides washed twice in 1× PBS shaking, ten minutes each. Slides were then sequentially dehydrated with 70%, 90%, 100% ethanol and let them air dried. The slides were denatured for 3 minutes at 80°C with 20 μL of hybridization mixture contained 70% deionized formamide, 1M Tris pH 7.2, 8.56% buffer MgCl2, 5% maleic blocking reagent, 12 μg/mL Cy5-conjugated PNA Tel-C (CCCTAA) probe (Panagene, Korea), and incubated overnight at 4°C in a humid chamber. Slides were washed two times for 15 minutes in wash solution containing 70% formamide, 10mM Tris pH 7.2, 0.1% BSA, and washed three times for 5 minutes in a solution containing 0.1 M Tris pH 7.5, 0.15 M NaCl, and 0.08% Tween-20. The slides were dehydrated by ethanol series, air-dried, and counterstained with Vectashield/DAPI (Vector Laboratories, Burlingame, CA). Images were captured at 63× magnification with an Axio Imager Z2 (Carl Zeiss, Oberkochen, Germany). equipped with an automatic capture system (Metafer, Metasystems) and analyzed with ISIS software (Metasystems).
In vivo study
Male and female athymic mice (nu/nu genotype, Balb/c background, 6–8 weeks old) were used for this antitumor study. The animals were maintained in Thoren ventilated cage and rack system (Hazleton, PA). All animal procedures conformed to the Institutional Animal Care and Use Committee’s and the National Institute of Health’s guidelines. The glioma cell lines U87MG (a representative sensitive cell line) and U251MG (a representative resistant cell line) were used in in vivo experiments. The cell suspension was injected s.c. into the right flank of the athymic mouse at an inoculation volume of 100 μL. For the s.c. tumor studies, groups of 7-10 mice were stratified by tumor volume and were treated when the median tumor volumes were an average of 100 mm3. Groups of tumor-bearing animals received either vehicle control or THIO at a dose of 3mg/kg intraperitoneally (i.p.) every other day for a total of 10 treatments. Subcutaneous tumors were measured twice weekly with hand-held vernier calipers (Scientific Products, McGraw, IL). Tumor volumes, V, were calculated with the following formula: V (mm3) = [(width)2 × (length)]/2. The response of the s.c. xenografts to treatment was assessed by a delay in tumor growth and differences in median values. Growth delay, expressed as T-C, was defined as the difference in days between the median time required for the tumors in the treated (T) and control (C) animals to reach a volume five times greater than that measured at the start of the treatment and/or reach a minimum volume of > 1000 mm3. Statistical analysis was performed using a SAS statistical analysis program. The Wilcoxon rank order test and Student t test were used (17-23). Athymic Ncr-nu/nu mice (6-8 weeks old) were subcutaneously injected with PDX cell lines respectively including D09-0500MG, D09-0394MG and D10-0021MG. After tumor establishment with an average volume of 200 to 300 mm3, mice were injected intraperitoneally with DMSO-PBS (vehicle) or 3mg/kg THIO three times before euthanization. Tumors were resected from mice and made into Formalin-fixed paraffin-embedded (FFPE) sections for DNA damage detection. In the PDX models, female NCG mice (6-8 weeks old) were subcutaneously implanted with GBM PDX tissue. After tumor establishment with an average volume of 100 to 200 mm3, mice were randomly assigned to be injected intraperitoneally with DMSO-PBS (vehicle) or THIO at 3 mg/kg every other day (5 mice in each group). Subcutaneous tumors were measured every other day with hand-held vernier calipers. After 10-days treatment of THIO, mice were sacrificed. Tumor volumes were calculated with the following formula: volume = [(width)2 × length]/2.
Genomic DNA Extraction and whole exome sequencing for patient-derived cell lines
Cell line genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega). Peripheral blood was obtained with informed consent and genomic DNA extracted using the Gentra® Puregene® kit (Qiagen, Germantown, MD), following the manufacturer’s protocol. Germline and cell line genomic DNA were subjected to exome capture with the Agilent Sure Select Human All Exon V6 kit (Agilent, Santa Clara, CA) and sequenced on the HiSeq PE150 platform (Illumina), all performed by Novogene Corporation Inc. (Sacramento, CA). The raw fastq sequence files were aligned to B37 human reference genome from broad Institute by BWA (version 0.7.12). Then the Sam files were transformed to Bam format and sorted by Samtools (version 1.10). Mark-Duplicates and BQSR were performed by Picard (version 2.20.1) and GATK (version 4.1.1.0) on the Bam files. Database SNV and golden indels from broad Institute were marked in the Bams to be removed in the results. PON (Panel of Normal) was built by the unmatched blood samples from the patients in the same cohort, which was used to filter out the germline mutations in the results. The SNVs and Indels were called by GATK Mutect2. After calling, the variants were filtered by GATK to control the false positives. Then the variants were further filtered in R with the criterion: depth of tumor > 10 and alternative allele frequency > 0.1. The final variants were transformed to TSV format and submitted to CoMutplotter platform (http://tardis.cgu.edu.tw/comutplotter). In the CoMutplotter, the variants were annotated by Oncotator (version 1.9.9.0) and the results of different samples were integrated to find the unique and common variants.
RNA sequencing (RNAseq)
Total RNA was purified from M059K, H4, D2224MG and DBTRG-05MG cells by miRNeasy Mini Kit (Qiagen, 217004) according to the manufacturer’s instructions. The complemental DNA (cDNA) libraries were constructed and sequenced by Genewiz. Sequencing was carried out using a 150 paired-end configuration on the Illumina sequencing platform (HiSeq™ 2500). RNA-seq data were deposited into Gene Expression Omnibus (M059K and H4-accession number GSE160619, D2224MG and DBTRG-05MG-accession number GSE180874).
Reverse phase protein array (RPPA)
The reverse-phase protein array (RPPA) assay was performed in the RPPA Core Facility at M.D. Anderson Cancer Center using 80 μg protein per sample. Experimental details are awailable at https://www.mdanderson.org/research/research-resources/core-facilities/functional-proteomics-rppa-core/rppa-process.html. The RPPA data can be accessed from Supplemental Table 4.
Statistical Analysis
Two-sided Welch t-test was conducted to compare control and treatment samples. Heatmap was used to visualize data. Hierarchical clustering used ‘Canberra’ as distance measurement method and ‘Complete’ as the agglomeration method. Survival analysis was conducted with the Kaplan-Meier method. The log-rank test was used to determine statistical significance associate with survival analysis. The quality of Fastq sequence data were checked by Fastqc(v0.11.9) (24). Then the reads were aligned to human genome GRCH38 by STAR(v2.7.3a) (25). The quality of BAM files were checked by Picard CollectRnaSeqMetrics (v2.20.8) (26). Then BAM files were sorted by name using Samtools(v1.10) (27). Counting was performed by HTSeq(v0.11.2) (28). DESeq2(v1.28.1) (29) in R (v4.0.2) was used for differential expression analysis. Then the pre-ranked GSEA (gene score enrichment analysis) was performed by using fgsea(v1.14.0) (30) in R.
Data Availability Statement
RNA sequencing data generated in this study were deposited at Gene Expression Omnibus (GEO). Accession numbers are GSE160619 and GSE180874.
Results
Short-term treatment with THIO impaired cell viability and division of glioma cells
To evaluate the therapeutic efficacy of THIO in gliomas, we first assembled a panel of 3 mouse glioma cell lines and 17 human glioma cell lines including GBM cell lines (n=13), low grade glioma (LGG) cell lines (n=2), oligodendroglioma lines (n=1), and pleomorphic xanthoastrocytomas (PXA) (n=2). Among 18 cell lines sequenced, 78% harbored C228T or C250T mutations in TERT promoter (TERTp). In addition to TERTp mutations, we found that 22%, 76%, and 16% of the cell lines harbored ATRX, TP53 or IDH1 mutations, respectively (Supplementary Table 1).
Next, we performed cell viability assays for 17 human gliomas and 3 mouse glioma cell lines treated with THIO for 4 days and calculated the IC50 for each cell line. Normal human astrocytes (NHA) were included as the control (Fig. 1A). As expected, NHA were insensitive to THIO with IC50 over 100 μM. The median IC50 of human glioma cell lines treated with THIO for 4 days was 3.28 μM. Among all 17 human glioma cell lines we tested, U138MG, U251MG, DBTRG-05MG and D2224MG were identified as cell lines that had more intrinsic resistance to THIO (average IC50: 18.99 μM) when compared to the H4, D2159MG, A172, D2363PXA and M059K cell lines (average IC50: 1.42 μM).
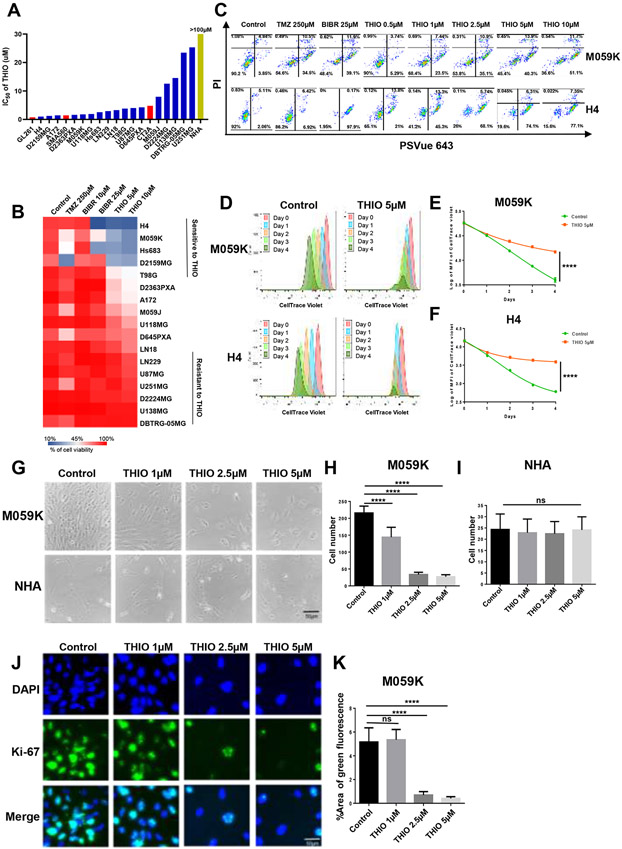
Figure 1.
Short-term treatment of glioma cell lines with THIO. (A) IC50 of THIO for 17 human glioma cell lines, 3 mouse glioma cell lines and normal human astrocyte (NHA). Cells were treated with THIO for 4 days at titrated doses ranging from 0.01 μM to 100 μM. And then CCK-8 assay was used to determine the cell viability. Graphpad Prism software was used to calculate IC50 of THIO. (B) Heatmap of cell viability for 17 glioma cell lines following treatment with the control, TMZ, BIBR1532 and THIO for 4 days at titrated doses. Cells were treated with TMZ, BIBR 1532 and THIO for 4 days and then stained with PI and PSVue 643 to distinguish live cells from apoptotic or dead cells by FACS (% of cell viability). (C) FACS data of M059K and H4. Cells were treated with THIO for 4 days at titrated doses and stained with PI and PSVue 643, and then analyzed by FACS. (D) M059K and H4 cells were labeled with CellTrace Violet and then treated with THIO at 5μM. The fluorescence intensity of CellTrace Violet was measured by FACS from Day 0 to Day 4. (E-F) The quantification of fluorescence intensity of CellTrace Violet in samples included in (D). (G) Phase contrast images of M059K and NHA cells treated with THIO at 0, 1, 2.5 and 5 μM for 4 days. (H-I) The quantification of cell number of M059K and NHA included in (G). Two experiments were ran and 9 fields for each group were measured. (J) Immunofluorescence staining images of M059K cells treated with or without THIO at 5μM for 4 days. Blue indicated the DAPI staining whereas green indicated the Ki-67 staining. (K) The quantification of the percentage of area with positive green fluorescence for samples shown in (J). Two experiments were ran and 9 fields for each group were measured.
Additionally, we performed a fluorescence-activated cell sorting (FACS)-based screening of 17 glioma cell lines that were treated with THIO at 5μM for 4 days and assessed the induction of apoptosis and cell death. TMZ, a standard-of-care therapy for treating patients with GBM (31), and BIBR1532 a known telomerase inhibitor (32), were included as a comparison against THIO. We defined sensitive cell lines as the percentage of viable cells less than 50%, whereas those with the percentage of viable cells greater than 90% were considered intrinsically resistant. We found that the short-term treatment with THIO for 4 days drastically inhibited cell viability in 5 out of 17 glioma cell lines (the mean viability is 25%), with a more profound effect observed in M059K, H4, Hs683, D2159MG and T98G cell lines. U87MG, LN229, U251MG, DBTRG-05MG, D2224MG and U138MG cell lines, however, were more resistant to THIO (the mean viability is 89.5%) (Fig. 1B). Furthermore, we noticed that glioma cell lines with higher IC50 of THIO (Fig. 1A) also demonstrated higher percentages of viable cells (Fig. 1B).
Next, we focused on two representative cell lines, M059K and H4 that were sensitive to THIO. We observed the efficacy of THIO in inhibiting cell viability and concluded that it was dose dependent (Fig. 1C). In order to determine whether THIO inhibited cell division of glioma cells, we labeled M059K and H4 cells with a cell membrane dye CellTrace Violet, which was first verified by the success of labeling with FACS (Supplementary Fig. 1A and B). Subsequently, we treated M059K and H4 cells with THIO at 5μM and monitored the fluorescence intensity of CellTrace Violet over a time course of 96 hours. THIO substantially and significantly inhibited cell division as compared to the control (p<0.0001) (Fig. 1D-F). The inhibition of cell division was accompanied by an increase in percentages of apoptotic and dead cells in both M059K and H4 cell lines (Supplementary Fig. 2C and D).
We observed that THIO led to up-regulation of the apoptosis-related protein cleaved-PARP, as well as an increase in the expression of cell cycle check point p21 and down-regulation of cell cycle-related proteins Rb and pRbSer807/811 in a time- and dose-dependent manner (Supplementary Fig. 1E and F).
To further evaluate if THIO is more selective towards tumor cells, we treated M059K (sensitive to THIO) and NHA with THIO at titrated concentrations for 4 days (Fig. 1G). As expected, THIO led to a significant reduction in cell number for M059K exclusively (Fig. 1H and I). Furthermore, we showed that THIO decreased the expression of the proliferation marker Ki-67 in M059K cells (Fig. 1J and K).
Taken together, we demonstrated that short-term treatment with THIO led to the induction of apoptosis and cell death as well as inhibited the cell viability and division in a subset of glioma cell lines in a time- and dose-dependent manner. This inhibitory effect by THIO was more selective towards tumor cells in a dose- and time-dependent way, which was not observed in normal cells.
Long-term treatment with THIO inhibited the cell proliferation and induced senescence and apoptosis
Having demonstrated that the short-term treatment with THIO impaired cell viability in a panel of glioma cell lines, we extended the analysis to the long-term treatment of a panel of 17 glioma cell lines with THIO for 8-13 days. We performed crystal violet staining and demonstrated that the long-term treatment with THIO impaired cell proliferation in 12 out of 17 glioma cell lines, including a subset that were intrinsically resistant to TMZ (Fig. 2A and B). Next, we investigated the efficacy of THIO in a panel of 6 glioma cell lines that acquired resistance to TMZ in culture over time, including H4-TR, LN229-TR, Hs683-TR, U87MG-TR, T98G-TR and M059K-TR. Long-term treatment with THIO for 8-10 days led to a decrease in cell viability of LN229-TR, Hs683-TR and H4-TR (Fig. 2C and D), whereas the efficacy of THIO for T98G-TR, M059K-TR and U87MG-TR was less pronounced (Supplementary Fig. 2A and B). In addition, we also performed the senescence-associated β-galactosidase (SA-β-gal) staining of glioma cells that were treated with THIO for 7 days. Treatment of M059K and H4 cells with THIO for 7 days led to the induction of about 50% and 30% SA-β-gal positive cells, respectively (Supplementary Fig. 2C-E). Additionally, treatment of DBTRG-05MG cells with THIO for 7 days also led to the induction of 12% SA-β-gal positive cells, although overall cell viability was minimally inhibited by THIO (Supplementary Fig. 2C and F).
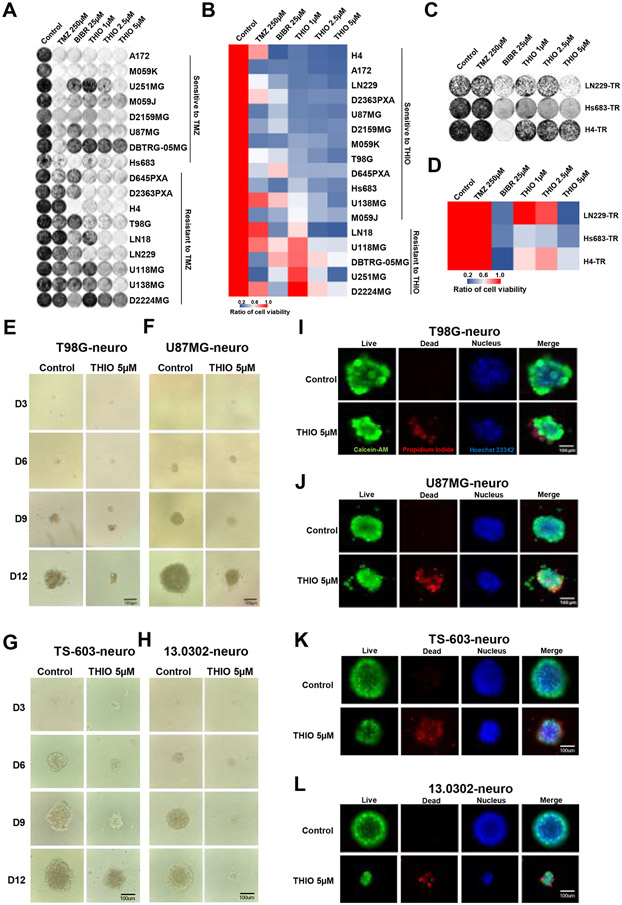
Figure 2.
Long-term treatment of glioma cell lines with THIO. (A) Crystal violet staining images for 17 glioma cell lines. Cells were treated with control medium, TMZ, BIBR 1532 and THIO at indicated doses for 8-14 days. Then cells were fixed and stained with crystal violet followed by the acquisition of images. (B) The heatmap of quantification results of the crystal violet staining. (% relative to control) (C) Crystal violet staining images for TMZ-resistant (TR) cell lines treated with THIO. (D) The heatmap of quantification results of the crystal violet images of TR cells. (E-H) Phase contrast images of T98G, U87MG, TS-603 and 13.0302 in 3D. Cells were treated with THIO at 5 μM for 12 days and images were acquired at day 3, 6, 9 and 12. (I-L) Immunofluorescence images of T98G, U87MG, TS-603 and 13.0302 in 3D. Following treatment with THIO at 5 μM for 12 days, cells were stained with live/dead staining kit and images were acquired with ECHO Revolve Microscope. Green indicated live cells; red indicated dead cells; blue indicated nucleus.
In addition to adherent cell cultures, we also tested the inhibitory effect of THIO towards glioma neurospheres grown in three dimensions (3D). The T98G, U87MG, 13.0302 and TS-603 neurospheres were treated with THIO at 5 μM. Phase contrast images were taken at days 3, 6, 9 and 12 after the initial treatment with THIO (Fig. 2E-H). We determined the largest diameter of neuropsheres, which resulted in the suggestion that THIO inhibited the growth of neurospheres in a time-dependent manner (Supplementary Fig. 2G-J). At day 12, or upon the completion of the treatment, neurospheres were immediately stained with Calcein-AM and Propidium Iodide to label live and dead cells, respectively. Fluorescent images showed that THIO significantly induced more cell death compared to the control (Fig. 2I-L).
Taken together, long-term treatment with THIO not only inhibited cell viability of glioma cell lines but also induced cellular senescence. Importantly, THIO significantly inhibited the growth of neurospheres and induced cell death in the 3D culture system.
THIO Induced DNA damage
Having demonstrated the efficacy of THIO in a panel of glioma cell lines, we sought to identify a predictive biomarker of sensitivity to THIO. First, we assessed baseline levels of the relative telomere length of 17 glioma cell lines (Supplementary Fig. 3A; Supplementary Table 2) and conducted correlative analysis between IC50 of THIO and relative telomere length. We showed that there was no correlation between sensitivity and telomere length (Supplementary Fig. 3B), which is in line with findings reported in the NSCLC study (14). Furthermore, we measured the baseline level of expression of hTERT for 17 glioma cell lines via qRT-PCR (Supplementary Fig. 3C; Supplementary Table 2). Similarly, we obtained RNA sequencing (RNAseq) data of hTERT for 11 glioma cell lines that were profiled by the CCLE project (Supplementary Fig. 3D). In both cases, the analyses did not show correlation (Supplementary Fig. 3E and F). Finally, we calculated the enrichment scores of 5 telomere/telomerase-related gene signatures for 65 glioma cell lines included in the CCLE project (Supplementary Fig. 3G). Importantly, the analysis suggested that sensitivity of 11 glioma cell lines to THIO was positively and significantly correlated with enrichment scores of three telomere/telomerase-related gene sets including "Telomere Maintenance," "Telomere Pathway" and "TERT Targets" (Fig. 3A-C; Supplementary Table 3).
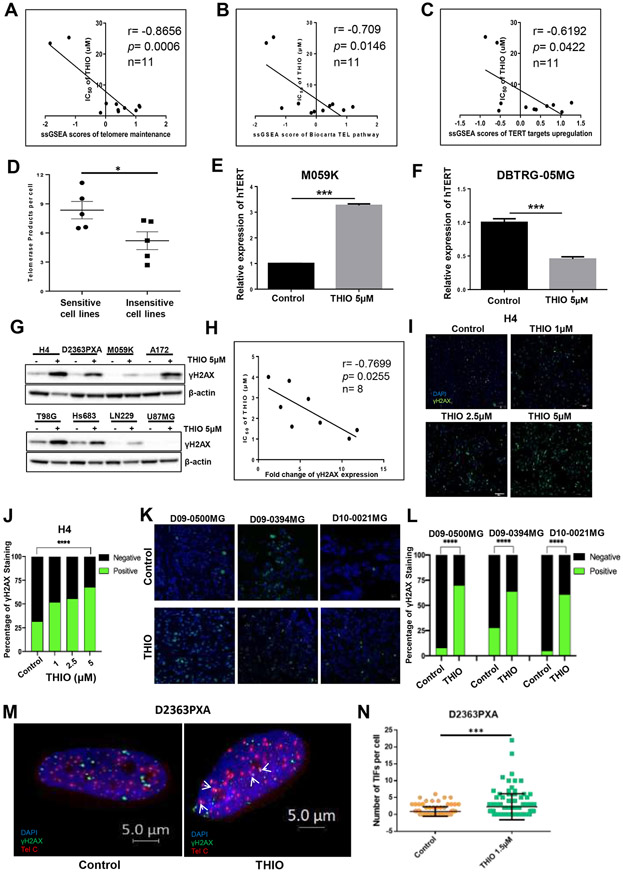
Figure 3.
THIO induced DNA damage in glioma cells. (A-C) The correlation of IC50 of THIO and the enrichment scores of telomere/telomerase-related gene signatures including "Telomere Maintenance," "Telomere Pathway" and "TERT Targets" in 11 glioma cell lines in CCLE. (D) Baseline telomerase activity of THIO sensitive cell lines and THIO resistant cell lines. (E-F) Relative expression of hTERT of M059K and DBTRG-05MG cells following treatment with THIO at 5 μM for 72 hours. (G) Western blot was performed to detect the protein expression of γH2AX in 8 glioma cell lines treated with THIO for 72 hours. (H) The correlation of IC50 of THIO and the fold change of γH2AX expression. (I) Immunofluorescence staining of γH2AX in M059K cells treated with THIO at 0, 1, 2.5 and 5 μM for 48 hours. Blue indicated DAPI and green indicated γH2AX. (J) Quantification of immunofluorescence staining of γH2AX for H4 cell lines treated with THIO at 0, 1, 2.5 and 5 μM for 72h. (K) Immunofluorescence staining of γH2AX for 3 PDX tumor models including D09-0500MG, D09-0394MG, and D10-0021MG. Tumor-bearing mice was treated with control or THIO at 2.5 mg/kg every other day. Blue indicated DAPI and green indicated γH2AX. (L) Quantification of immunofluorescence staining of γH2AX for 3 PDX tumor models.
(M) Telomere dysfunction induced foci (TIF) assay for the D2363PXA cell lines. D2363PXA cells were treated with THIO at 1.5 uM for 48h following by fixed and performed TIF analysis. (N) Quantification of number of TIF per cell.
Furthermore, we determined the relative telomere length of the sensitive cell line - M059K and that of the resistant cell line - DBTRG-05MG following treatment with THIO at 2.5 μM for 5 days by quantitative PCR. We observed that THIO did not have an impact on either cell line’s telomere length (Supplementary Fig. 3H and I). Similarly, and as expected, THIO did not have an impact on telomerase activity in either cell line as measured by the TRAP assay (Supplementary Fig. 3J and K). Moreover, we assessed the baseline telomerase activity of glioma cell lines including 5 cell lines more sensitive to THIO and 5 cell lines more resistant to THIO (Supplementary Fig. 3L). The telomerase activity of sensitive cell lines was significantly higher than that of insensitive cell lines (Fig. 3D). Moreover, the telomerase activity was negatively correlated with cell viability of these cell lines following THIO treatment (Supplementary Fig. 3M). Importantly, THIO significantly led to an increase in hTERT expression in M059K cells but a decrease in DBTRG-05MG cells (Fig. 3E and F).
To further investigate the impact of THIO on DNA damage, we treated 8 cell lines with THIO for 72 hours and observed an increase in the protein expression of γH2AX after afterwards, indicating that THIO induced DNA damage of glioma cells (Fig. 3G). We quantified the protein bands and determined the fold change of γH2AX expression in each cell line following THIO treatment (Supplementary Fig. 3N). We found γH2AX expression level to be significantly negatively correlated with IC50 of THIO (Fig. 3H). Furthermore, we performed immunofluorescence staining of H4 cells that were treated with THIO at titrated doses for 72 hours, and found that THIO significantly increased the percentage of γH2AX positive cells (Fig. 3I and J). To further corroborate in vitro data, we treated mice bearing three PDXs with THIO at 3 mg/kg for 3 times prior to harvesting tumors. Indeed, THIO induced DNA damage in all 3 PDX tumor models including D09-0500MG, D09-0394MG, and D10-0021MG as we observed a significant increase in γH2AX positive cells (Fig. 3K and L). To explore the telomeric DNA damage alterations, we performed telomere dysfunction induced foci (TIF) assay for the D2363PXA cell line following THIO treatment of 1.5 μM for 48 hours. We observed that THIO significantly induced DNA damage localized in the telomere regions (Fig. 3M and N).
Transcriptomic alterations induced by THIO treatment
To further elucidate the anti-tumor mechanisms of THIO at the molecular level, we treated two sensitive cell lines - M059K and H4 - and two resistant cell lines - D2224MG and DBTRG-05MG - at 5 μM for 72 hours. We interrogated the transcriptome by sequencing RNA (RNAseq) and investigated the alterations of gene expression and signaling pathways induced by THIO. First, we performed differential gene expression analysis in all four cell lines (Fig. 4A and B; Supplementary Fig. 4A-D). In M059K cells, a total of 376 genes were significantly altered following THIO treatment (p<0.05; ∣Log2 fold change (FC)∣>2.5), including 99 up-regulated and 277 down-regulated genes. In H4 cells, a total of 525 genes were significantly altered following THIO treatment (p<0.05; ∣Log2 FC∣>2.5), including 249 up-regulated and 276 down-regulated genes. In D2224MG cells, a total of 83 genes were significantly altered following treatment with THIO (p<0.05; ∣Log2 FC∣>2.5), including 61 up-regulated and 22 down-regulated genes. In DBTRG-05MG cells, a total of 296 genes were significantly altered following treatment with THIO (p<0.05; ∣Log2 FC∣>2.5), including 64 up-regulated and 232 down-regulated genes.
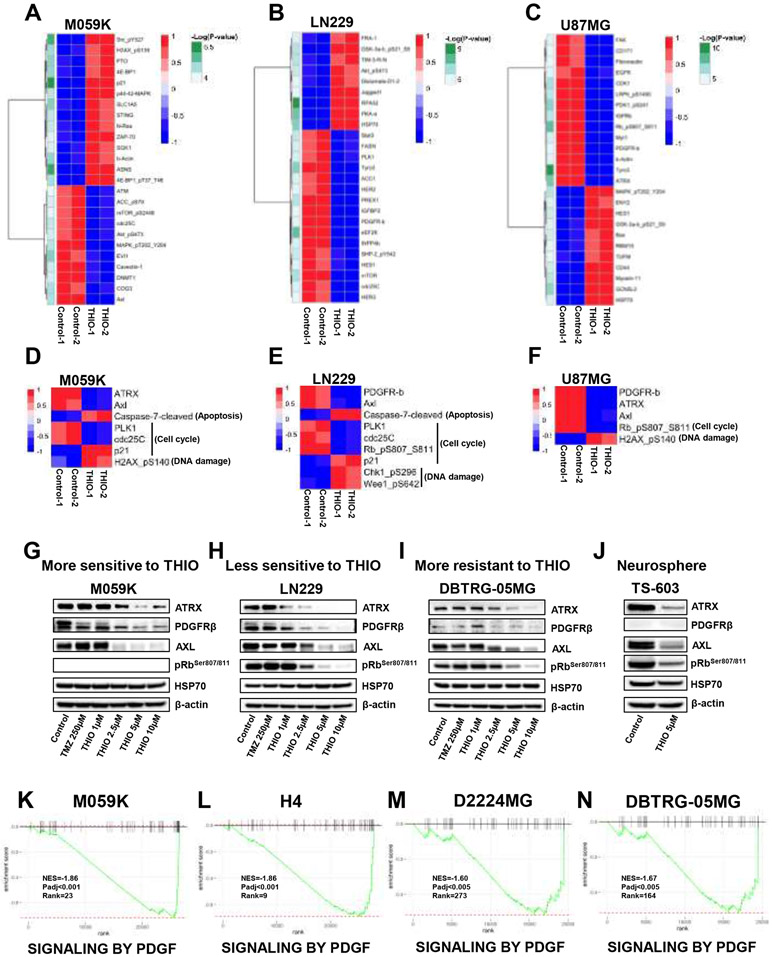
Figure 4.
Transcriptomics alterations induced by THIO. (A-B) The volcano plot of differentially expressed genes in M059K, H4 cells (more sensitive to THIO) and D2224MG, DBTRG-05MG (more resistant to THIO) treated with or without THIO for 72 hours. Log2 fold change and –Log10P were used as the X and Y axis, respectively. (C-F) fGSEA table of top 20 ranked down-regulated pathways in M059K, H4, D2224MG and DBTRG-05MG cells treated with or without THIO for 72 hours. (G-H) Single sample gene set enrichment analysis plots of down-regulated pathways in M059K and H4 cells treated with or without THIO for 72 hours, including ANASTASSIOU MULTICANCER INVASIVENESS SIGNATURE, BOQUEST STEM CELL UP, GLESIAS E2F TARGETS UP and RUIZ TNC TARGETS UP. (I) .The heatmap of single sample gene set enrichment analysis in M059K, H4, D2224MG and DBTRG-05MG.
Next, we performed gene set enrichment analysis (GSEA) of 3,368 chemical and genetic perturbation (CGP) gene sets and identified those gene sets that were enriched in significantly expressed genes. Specifically, we ranked the filtered pathways by normalized enrichment score (NES) and focused on the top 20 ranked pathways with the smallest NES (Fig. 4C-F; Supplementary Fig. 4E-H). Among the top 20 ranked pathways that were enriched in significantly down-regulated genes, 15 signaling pathways were shared between M059K and H4 cells treated with THIO (Supplementary Table 4A). Of note, among the 15 common pathways down-regulated by THIO, four pathways were involved in cell invasiveness, including ANASTASSIOU MULTICANCER INVASIVENESS, SCHUETZ BREAST CANCER DUCTAL INVASIVE, TURASHVILI BREAST LOBULAR VS DUCTAL NORMAL and ONDER CDH1 TARGETS. Four pathways were implicated in cell stemness, including BOQUEST STEM CELL, LEE NEURAL CREST STEM, MEBARKI HCC PROGENITOR WNT and WANG SMARCE1 TARGETS. Four pathways were related to cell proliferation, including IGLESIAS E2F TARGETS, KIM GLIS2 TARGETS, PLASARI TGFB1 and SENESE HDAC1 TARGETS. Moreover, THIO also inhibited the angiogenesis pathway, RUIZ TNC TARGETS, in M059K and H4. Importantly, those down-regulated pathways were not inhibited in D2224MG and DBTRG-05MG cell lines, which were more resistant to THIO (Fig. 4I).
Taken together, these findings suggested that THIO demonstrated anti-tumor activities via downregulating key tumor signaling pathways related to cell proliferation, invasion and stemness in gliomas.
Proteomics alterations induced by THIO
In order to further gain insights into molecular mechanisms of THIO at the protein level, we treated three representative glioma cell lines with THIO at 5μM for 72 hours, including M059K, LN229 and U87MG. We then conducted the reverse phase protein array (RPPA) experiment, in which 303 proteins (486 proteins for M059K) related to signaling transduction were included. The analysis of RPPA data demonstrated that 71, 78 and 70 proteins were differentially expressed in M059K, LN229 and U87MG cell lines, respectively (Fig. 5A-C, Supplementary Fig. 5A-C; Supplementary Table 5A). Specifically, we noticed that THIO significantly down-regulated protein expression of ATRX, PDGFRβ, AXL and cell cycle related proteins, and adaptively induced an increase in protein expression of heat shock protein 70 (HSP70), apoptosis-related proteins and DNA damage proteins (Fig. 5D-F).
Figure 5.
Proteomics alterations induced by THIO. (A-C) The heatmap of the top 25 significantly changed proteins in M059K, LN229 and U87MG cells. Cells were treated with or without THIO at 5 μM for 72 hours and then profiled by the RPPA assay. (D-F) The heatmap of common proteins regulated by THIO in M059K (D), LN229 (E) and U87MG (F) cells. (G-J) Western blot was performed to confirm the change of these proteins in M059K (more sensitive to THIO) (G), LN229 (less sensitive to THIO) (H), and DBTRG-05MG (more resistant to THIO) (I), and a neurosphere TS-603 (J) after treatment with THIO at 5 μM. Actin serves as a loading control. (K-N) fGSEA plots of SIGNALING BY PDGF pathway in M059K, H4, D2224MG and DBTRG-05MG cells.
In gliomas, ATRX is not only correlated with the alternative lengthening of telomeres (ALT), an alternative telomere maintenance mechanism, but also involved in many other functions, such as DNA damage, senescence, and genomic instability (33). Elevated AXL predicts poor overall survival and is related to the regulation of the immune microenvironment in glioblastoma (34). PDGFRβ belongs to tyrosine kinase receptors, which participates in multiple regulatory pathways by binding with its ligand in glioma (35). Inhibition of pRb promoted anti-glioma effects due to cell cycle arrest at G1 phase (36). Stress-induced HSP70 is an ATP-dependent molecular chaperone that associates with therapy-resistance and plays a key role in promoting cell survival (37).
To validate the RPPA results using representative cell lines, we treated M059K and H4 (more sensitive to THIO; average IC50 = 1.40 μM), LN229 and U87MG (less sensitive to THIO; average IC50 = 3.48 μM), and U251MG and DBTRG-05MG (more resistant to THIO; average IC50 = 24.41 μM) cells with THIO at 5 μM for 72 hours and performed the western blotting experiment (Fig. 5G-I; Supplementary Fig. 5D-F). Indeed, we were able to confirm that THIO led to the decreases in expression levels of ATRX, PDGFRβ, AXL and pRbSer807/811 and an increase in the expression level of HSP70 in H4, M059K, LN229 and U87MG cell lines, however, the effects were less pronounced in DBTRG-05MG and U251MG cell lines. We also treated TS-603 neurospheres grown in 3D with THIO at 5 μM for 7 days and observed that ATRX, PDGFRβ, AXL and pRbSer807/811 were substantially down-regulated (Fig. 5J). Consistent with RPPA results, GSEA results based on 1,604 Reactome gene sets revealed that the PDGF signaling pathway was significantly enriched in genes that were down-regulated in both in M059K and H4 cell lines treated with THIO, which was not the case for both D2224MG and DBTRG-05MG cell lines that are more resistant to THIO (Fig. 5K-N). The pathway analysis of RPPA data of M059K, LN229 and U87MG cells suggested that THIO inhibited cell cycle-related pathways and promoted DNA damage and cell apoptosis (Supplementary Fig. 5G and Supplementary Table 5B).
Taken together, our findings not only suggested that THIO treatment induced cell apoptosis via inhibiting key driving genes but also highlighted the important role of DNA damage that might underlie the therapeutic efficacy of THIO.
THIO delayed glioma growth in vivo
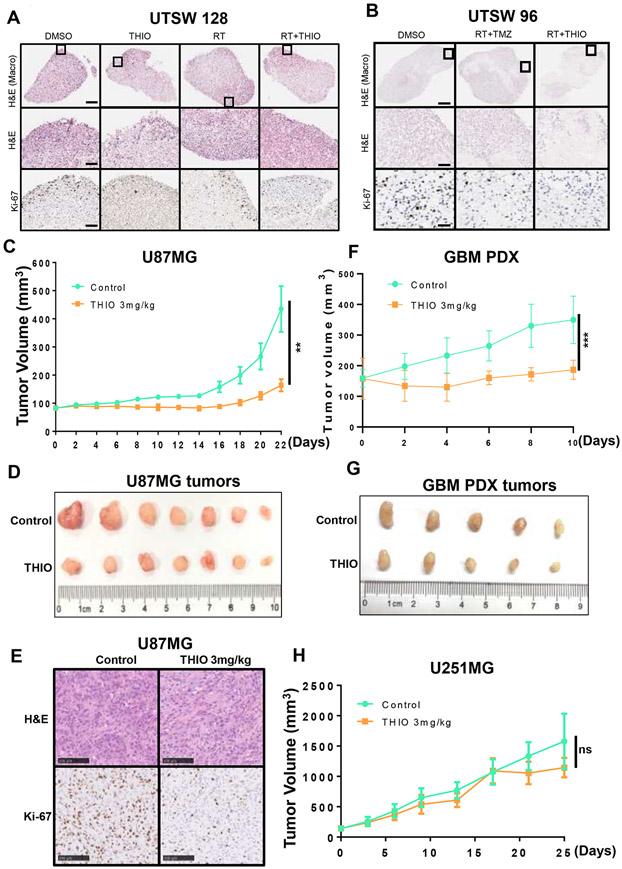
Next, we determined to explore the efficacy of THIO in patient-derived organoids (PDOs) models and xenograft models of human GBM cell line. First, we developed two PDOs models (Supplementary Fig. 6A) and treated UTSW128 PDO with DMSO as the control, THIO at 3 μM, radiotherapy (RT) at 10Gy, or the combination of RT and THIO. We also treated UTSW96 PDO with DMSO, the combination of RT with TMZ at 50 μM, or the combination of RT and THIO (Fig. 6A and B). Our data showed that THIO monotherapy reduced the expression of Ki-67 in the UTSW128 PDO and the combination of RT plus THIO further decreased its expression (Fig. 6A). Similarly, the combination of RT plus TMZ reduced the number of Ki-67+ cells in the UTSW96 PDO and no detectable Ki-67+ proliferating cells were observed for the combination of RT plus THIO (Fig. 6B). These results indicated that THIO in combination with standard-of-care treatments for GBM may be more effective in inhibiting tumor proliferation.
Figure 6.
Anti-tumor effect of THIO in vivo. (A-B) THIO inhibited cell proliferation in a primary GBM organoid model. (A) The GBM organoids (UTSW128) were treated with the DMSO control, THIO (3 μM), RT (10Gy), or the combination of RT and THIO. THIO was re-dosed 48 and 96 hours later and organoids were fixed on Day 6 for the H&E and Ki-67 IHC staining. (B) The GBM organoids (UTSW96) were treated with the DMSO control, the combination of RT (10Gy) and TMZ (50 μM), or the combination of RT and THIO (3 μM). TMZ or THIO was re-dosed 48 and 96 hours later and organoids were fixed on Day 6 for the H&E and Ki-67 IHC staining. (C) The tumor growth curve of U87MG tumor. Tumor-bearing mice were treated with control or THIO at 3 mg/kg every other day. (D) The picture of U87MG tumors in THIO treatment group and control group. (E) The H&E and Ki-67 IHC staining of U87MG tumors. (F) The tumor growth curve of GBM PDX tumor. Tumor-bearing mice were treated with control or THIO at 3 mg/kg every other day. (G) The picture of GBM PDX tumors in THIO treatment group and control group. (H) The tumor growth curve of U251MG tumor. Tumor-bearing mice were treated with control or THIO at 3 mg/kg every other day.
Furthermore, we xenografted U87MG cells subcutaneously that were relatively sensitive to THIO as exhibited in vitro. Mice were treated with the control or THIO at 3mg/kg. The results demonstrated that THIO significantly delayed U87MG tumor growth (Fig. 6C and D). The average tumor weight in the THIO group was lower than that in the control group (Supplementary Fig. 6B). Moreover, the IHC staining of tumor tissues suggested that THIO reduced the expression of Ki-67 (Fig. 6E). Treatment with THIO did not affect the body weight of mice (Supplementary Fig. 6C). We also established GBM PDX model to assess tumor suppression of THIO. The results showed that THIO significantly delayed the growth of PDX tumors (Fig. 6F and G). The average tumor weight in the THIO group was lower than that in the control group (Supplementary Fig. 6D). We also evaluated the anti-tumor effect of THIO for the U251MG subcutaneous model which was relatively resistant to THIO as exhibited in vitro. As expected, U251MG tumors did not respond to THIO (Fig. 6H). It is worth noting that THIO did not cause significant changes in mouse body weight in three xenograft experiments (Supplementary Fig. 6C, 6E and 6F).
Taken together, our data highlighted the efficacy of THIO in all GBM PDO, xenograft model of huamn glioma cell line and xenograft model of GBM PDX.
Discussion
Approximately 80% of gliomas harbor TERTp mutations resulting in the activation of the telomerase signaling pathway, whereas telomerase activity is not seen in most normal astrocytes. We have reasoned that telomerase inhibitors may be active in gliomas, for which novel therapies are urgently needed. In this study, we reported, for the first time, that THIO can be used as a novel telomerase-directed telomere inhibitor with promising and effective therapeutic efficacy for primary and recurrent gliomas.
We investigated the therapeutic efficacy of THIO by using a broad panel of 17 human glioma cell lines and 3 mouse cell lines and demonstrated that THIO is effective in the majority of glioma cell lines with minimal toxicity against normal astrocytes. Short-term treatment with THIO impaired cell viability and cell division of glioma cells, and long-term treatment with THIO inhibited cell proliferation and induced apoptosis with surviving cells undergoing senescence. THIO as a monotherapy demonstrated efficacy in three glioma cell lines with acquired resistance to TMZ (denoted as TMZ-R), indicating that THIO might be used as a potential salvage therapeutic strategy for patients with recurrent gliomas. Of note, in addition to primary and TMZ-R glioma cell lines grown as adherent cell cultures, THIO also showed potent efficacy in 4 glioma cell lines grown as neurospheres in a 3D cell culture system by inducing apoptosis and cell death.
We also aimed to identify predictive biomarkers for glioma cell lines treated with THIO. We did not establish a correlation between either telomere length or TERT expression and sensitivity to THIO, but did find that enrichment scores of three telomere/telomerase gene sets significantly correlated with sensitivities to THIO. This suggested that telomerase activity (not expression) might dictate the response of glioma cell lines to THIO. Indeed, our data are consistent with the conclusion based on a panel of metastatic melanoma from a previous study (13).
To further explore the anti-tumor mechanisms of THIO in gliomas at the molecular level, we performed integrated computational analyses of RNAseq and RPPA data derived from glioma cell lines treated with THIO. We discovered that cell invasion, cancer stem cell and proliferation pathways were all significantly inhibited, suggesting that THIO impedes cell proliferation via down-regulating key tumor signaling pathways related to cell invasion, stem cell character and dedifferentiation. Mechanistically, THIO induced DNA damage not only in glioma cell lines but also in PDX tumor specimens. Moreover, DNA damage was localized to the telomere region. Importantly, the analysis of RPPA data at the pathway level also suggested that THIO induced DNA damage in three glioma cell lines treated with THIO.
Further, we evaluated the anti-tumor effects of THIO in patient-derived organoid models, xenograft models of human glioma cell line and PDX model. Our data demonstrated that THIO, as a monotherapy or in combination with standard therapies (RT or TMZ), decreased the expression of Ki-67 in PDO models. We also established two glioma cell line xenograft models and indicated that THIO significantly delayed the tumor growth in vivo with little toxicity to mice. Moreover, the results of GBM PDX model further confirmed the anti-tumor effect of THIO.
While breakthroughs of immunotherapies have been achieved in cancer treatment, only a subgroup of patients benefit from these therapies and attain a durable clinical response. In gliomas, the effects of immunotherapies emerging from clinical trials are disappointing; however, combination therapies are still promising for these patients. Notably, a recent study has demonstrated that THIO treatment can activate the host cytosolic DNA sensing STING/interferon I pathway and overcome the resistance to anti-PD-L1 treatment in advanced cancers(15). This study also provided a rationale for combining THIO with immunotherapy in gliomas. Therefore, future studies may attempt to combine THIO with immunotherapy.
In conclusion, our study established a therapeutic role of THIO in primary and recurrent gliomas and revealed the induction of DNA damage as a primary anti-tumor mechanism of THIO in gliomas. Our findings, which are based on the analyses of glioma cell lines and patients’ tumor specimens, provide a proof-of-principle study for demonstrating the efficacy of THIO in the pre-clinical study setting. Further studies are warranted in order to bring this telomerase-mediated telomere inhibitor that penetrates the blood brain barrier into clinical trials in order to provide a novel and alternative therapeutic strategy for patients with primary and recurrent gliomas.
Supplementary Material
Translational Relevance.
About 80% of gliomas harbor TERT promoter mutations, which result in telomerase activation. Patients with glioma inevitably relapse on standard-of-care therapies providing an unmet need to identify novel therapies to improve patient survival outcomes. In the current study, we demonstrated the efficacy of 6-thio-dG (THIO), a telomerase targeting drug, in TMZ-sensitive and -resistant glioma cell lines, as well as neurospheres, patient-derived organoids and xenograft model of a human GBM cell line. THIO induces telomere associated DNA damage and inhibits key intrinsic signalling pathways used by tumors. Our results, based on preclinical models of glioma, not only highlight the potential therapeutic role of THIO but also provide a rationale for further testing THIO in the clinical setting for patients with primary and recurrent gliomas.
Acknowledgement
This research was supported by National Cancer Institute Grant 1U19CA264385-01 (D.M.A. and J.W.S.). U.H. is funded by National Institutes of Health (grant number R01CA136533). M.H., K.F. and G.M. are funded by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. S.Y. and S.W are funded by China Scholarship Council. Y.C. is funded by National Nature Science Foundation of China (grant number 31771549).
Footnotes
Conflict of interest statement
K. F. has/had served on the Board of Directors of Loxo Oncology, Clovis Oncology, Strata Oncology, Vivid Biosciences, Checkmate Pharmaceuticals, Kinnate Pharmaceuticals and Scorpion Therapeutics; Corporate Advisory Board of X4 Pharmaceuticals; Scientific Advisory Boards of PIC Therapeutics, Sanofi, Amgen, Asana, Adaptimmune, Aeglea, Shattuck Labs, Tolero, Apricity, Oncoceutics, Fog Pharma, Neon, Tvardi, xCures, Monopteros, Vibliome, and ALX Oncology; and as consultant to Lilly, Novartis, Genentech, BMS, Merck, Takeda, Verastem, Boston Biomedical, Pierre Fabre, Debiopharm; and received research funding from Novartis and Sanofi.
References
- 1.Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee Sh U. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pacific journal of cancer prevention : APJCP 2017;18:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rychly B, Sidlova H, Danis D. [The 2007 World Health Organisation classification of tumours of the central nervous system, comparison with 2000 classification]. Ceskoslovenska patologie 2008;44:35–6 [PubMed] [Google Scholar]
- 3.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009-2013. Neuro-oncology 2016;18:v1–v75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugarman ET, Zhang G, Shay JW. In perspective: An update on telomere targeting in cancer. Molecular carcinogenesis 2019;58:1581–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shay JW, Wright WE. Telomerase activity in human cancer. Current opinion in oncology 1996;8:66–71 [DOI] [PubMed] [Google Scholar]
- 6.Mancini A, Xavier-Magalhaes A, Woods WS, Nguyen KT, Amen AM, Hayes JL, et al. Disruption of the beta1L Isoform of GABP Reverses Glioblastoma Replicative Immortality in a TERT Promoter Mutation-Dependent Manner. Cancer cell 2018;34:513–28 e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell RJ, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 2015;348:1036–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA Jr., et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proceedings of the National Academy of Sciences of the United States of America 2013;110:6021–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barthel FP, Wei W, Tang M, Martinez-Ledesma E, Hu X, Amin SB, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nature genetics 2017;49:349–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mender I, Gryaznov S, Shay JW. A novel telomerase substrate precursor rapidly induces telomere dysfunction in telomerase positive cancer cells but not telomerase silent normal cells. Oncoscience 2015;2:693–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sengupta S, Sobo M, Lee K, Senthil Kumar S, White AR, Mender I, et al. Induced Telomere Damage to Treat Telomerase Expressing Therapy-Resistant Pediatric Brain Tumors. Molecular cancer therapeutics 2018;17:1504–14 [DOI] [PubMed] [Google Scholar]
- 12.Mender I, Gryaznov S, Dikmen ZG, Wright WE, Shay JW. Induction of telomere dysfunction mediated by the telomerase substrate precursor 6-thio-2'-deoxyguanosine. Cancer discovery 2015;5:82–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang G, Wu LW, Mender I, Barzily-Rokni M, Hammond MR, Ope O, et al. Induction of Telomere Dysfunction Prolongs Disease Control of Therapy-Resistant Melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research 2018;24:4771–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mender I, LaRanger R, Luitel K, Peyton M, Girard L, Lai TP, et al. Telomerase-Mediated Strategy for Overcoming Non-Small Cell Lung Cancer Targeted Therapy and Chemotherapy Resistance. Neoplasia 2018;20:826–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mender I, Zhang A, Ren Z, Han C, Deng Y, Siteni S, et al. Telomere Stress Potentiates STING-Dependent Anti-tumor Immunity. Cancer cell 2020;38:400–11 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic acids research 2002;30:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman HS, Colvin OM, Skapek SX, Ludeman SM, Elion GB, Schold SC Jr., et al. Experimental chemotherapy of human medulloblastoma cell lines and transplantable xenografts with bifunctional alkylating agents. Cancer research 1988;48:4189–95 [PubMed] [Google Scholar]
- 18.Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Current protocols in pharmacology 2011;Chapter 14:Unit 14 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandramohan V, Bao X, Kato Kaneko M, Kato Y, Keir ST, Szafranski SE, et al. Recombinant anti-podoplanin (NZ-1) immunotoxin for the treatment of malignant brain tumors. International journal of cancer 2013;132:2339–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman HS, Houghton PJ, Schold SC, Keir S, Bigner DD. Activity of 9-dimethylaminomethyl-10-hydroxycamptothecin against pediatric and adult central nervous system tumor xenografts. Cancer chemotherapy and pharmacology 1994;34:171–4 [DOI] [PubMed] [Google Scholar]
- 21.Keir ST, Dewhirst MW, Kirkpatrick JP, Bigner DD, Batinic-Haberle I. Cellular redox modulator, ortho Mn(III) meso-tetrakis(N-n-hexylpyridinium-2-yl)porphyrin, MnTnHex-2-PyP(5+) in the treatment of brain tumors. Anti-cancer agents in medicinal chemistry 2011;11:202–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keir ST, Friedman HS, Reardon DA, Bigner DD, Gray LA. Mibefradil, a novel therapy for glioblastoma multiforme: cell cycle synchronization and interlaced therapy in a murine model. Journal of neuro-oncology 2013;111:97–102 [DOI] [PubMed] [Google Scholar]
- 23.Keir ST, Hausheer F, Lawless AA, Bigner DD, Friedman HS. Therapeutic activity of 7-[(2-trimethylsilyl)ethyl)]-20 (S)-camptothecin against central nervous system tumor-derived xenografts in athymic mice. Cancer chemotherapy and pharmacology 2001;48:83–7 [DOI] [PubMed] [Google Scholar]
- 24.Andrews S FastQC: a quality control tool for high throughput sequence data. 2010 [Google Scholar]
- 25.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toolkit P. Broad Institute, GitHub Repository. http://broadinstitute.github.io/picard/. 2019
- 27.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reitman ZJ, Winkler F, Elia AEH. New Directions in the Treatment of Glioblastoma. Seminars in neurology 2018;38:50–61 [DOI] [PubMed] [Google Scholar]
- 32.Ding X, Cheng J, Pang Q, Wei X, Zhang X, Wang P, et al. BIBR1532, a Selective Telomerase Inhibitor, Enhances Radiosensitivity of Non-Small Cell Lung Cancer Through Increasing Telomere Dysfunction and ATM/CHK1 Inhibition. International journal of radiation oncology, biology, physics 2019;105:861–74 [DOI] [PubMed] [Google Scholar]
- 33.Haase S, Garcia-Fabiani MB, Carney S, Altshuler D, Nunez FJ, Mendez FM, et al. Mutant ATRX: uncovering a new therapeutic target for glioma. Expert opinion on therapeutic targets 2018;22:599–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadahiro H, Kang KD, Gibson JT, Minata M, Yu H, Shi J, et al. Activation of the Receptor Tyrosine Kinase AXL Regulates the Immune Microenvironment in Glioblastoma. Cancer research 2018;78:3002–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nazarenko I, Hede SM, He X, Hedren A, Thompson J, Lindstrom MS, et al. PDGF and PDGF receptors in glioma. Upsala journal of medical sciences 2012;117:99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S, Tang Y, Yuan X, Yuan D, Liu J, Li B, et al. Inhibition of Rb and mTOR signaling associates with synergistic anticancer effect of palbociclib and erlotinib in glioblastoma cells. Investigational new drugs 2018;36:961–9 [DOI] [PubMed] [Google Scholar]
- 37.Balaburski GM, Leu JI, Beeharry N, Hayik S, Andrake MD, Zhang G, et al. A modified HSP70 inhibitor shows broad activity as an anticancer agent. Molecular cancer research : MCR 2013;11:219–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data generated in this study were deposited at Gene Expression Omnibus (GEO). Accession numbers are GSE160619 and GSE180874.