Abstract
Limited knowledge of cellular and molecular mechanisms underlying hematopoietic stem cell and multipotent progenitor (HSC/MPP) expansion within their native niche has impeded the application of stem cell-based therapies for hematological malignancies. Here, we constructed a spatiotemporal transcriptome map of mouse fetal liver (FL) as a platform for hypothesis generation and subsequent experimental validation of novel regulatory mechanisms. Single-cell transcriptomics revealed three transcriptionally heterogeneous HSC/MPP subsets, among which a CD93-enriched subset exhibited enhanced stem cell properties. Moreover, by employing integrative analysis of single-cell and spatial transcriptomics, we identified novel HSC/MPP ‘pocket-like’ units (HSC PLUS), composed of niche cells (hepatoblasts, stromal cells, endothelial cells, and macrophages) and enriched with growth factors. Unexpectedly, macrophages showed an 11-fold enrichment in the HSC PLUS. Functionally, macrophage–HSC/MPP co-culture assay and candidate molecule testing, respectively, validated the supportive role of macrophages and growth factors (MDK, PTN, and IGFBP5) in HSC/MPP expansion. Finally, cross-species analysis and functional validation showed conserved cell–cell interactions and expansion mechanisms but divergent transcriptome signatures between mouse and human FL HSCs/MPPs. Taken together, these results provide an essential resource for understanding HSC/MPP development in FL, and novel insight into functional HSC/MPP expansion ex vivo.
Subject terms: Haematopoietic stem cells, Haematopoietic stem cells
Introduction
In mammals, hematopoietic stem cells and multipotent progenitors (HSCs/MPPs), occupying the top of hematopoietic hierarchy, display multilineage differentiation and self-renewal abilities.1 Studying HSC/MPP expansion holds great promise for regenerative medicine,2 and significant efforts have been devoted to fulfilling long-term ex vivo HSC/MPP expansion via genetic manipulation or optimization of culture conditions.3,4 Nevertheless, in current ex vivo HSC/MPP expansion culture, the key challenge is how to maintain the stem cell properties; therefore, a comprehensive understanding of HSC/MPP expansion within their native niche in vivo is required.
Highly vascularized fetal liver (FL) serves as the transitory developmental site for various sources of hematopoietic cells (HCs). In mice, the early wave of HCs colonizing the FL contains yolk sac erythro-myeloid progenitors (EMPs), which will generate erythroid-myeloid lineages, including macrophages.5,6 Subsequently, aorta-gonad-mesonephros (AGM)-derived HSCs migrate into the FL for maturation, expansion, and differentiation.7–9 Mounting evidence in mice demonstrates that FL structural niche cells, including endothelial cells (ECs), stromal cells, and hepatoblasts, interact with HSCs/MPPs via diverse growth factors, cytokines, and chemokines.10–13 These aforementioned findings preferentially focus on the effects of certain niche cell types or specific signaling molecules on HSC/MPP expansion. However, the mechanisms underlying the complex interplay among HSCs/MPPs and distinct niche cells, and the systematic regulatory network for HSC/MPP expansion remain elusive.
Single-cell RNA sequencing (scRNA-seq) represents a powerful tool to map cell atlas of single organ or multiple organs.14,15 However, the weakness of scRNA-seq is the loss of information about the cell-type spatial organization within intact organs. To expand our understanding of the spatial organization of diverse cell types in an unbiased manner, spatial transcriptomics (ST) has been developed.16 Here, to examine the organ-wide regulatory mechanisms of HSC/MPP expansion, we employed scRNA-seq and ST (including 10× Visium and Stereo-seq) to generate a spatiotemporal transcriptome map of mouse FL. With this map, we revealed the transcriptional heterogeneity of HSCs/MPPs, with a focus on a CD93+ subset exhibiting enhanced stem cell properties. We also decoded the architectural and molecular bases of HSC/MPP expansion. Furthermore, cross-species comparative analysis identified both conserved and divergent cellular and molecular events between mouse and human FL hematopoiesis. The map is accessible through an intuitive and interactive web portal at http://liulab.ioz.ac.cn/fetal_liver/. Together, our study reports a spatiotemporal transcriptome blueprint for understanding HSC/MPP development in FL and provides valuable insights into HSC/MPP expansion ex vivo.
Results
Single-cell transcriptomic atlas of the developing mouse FL
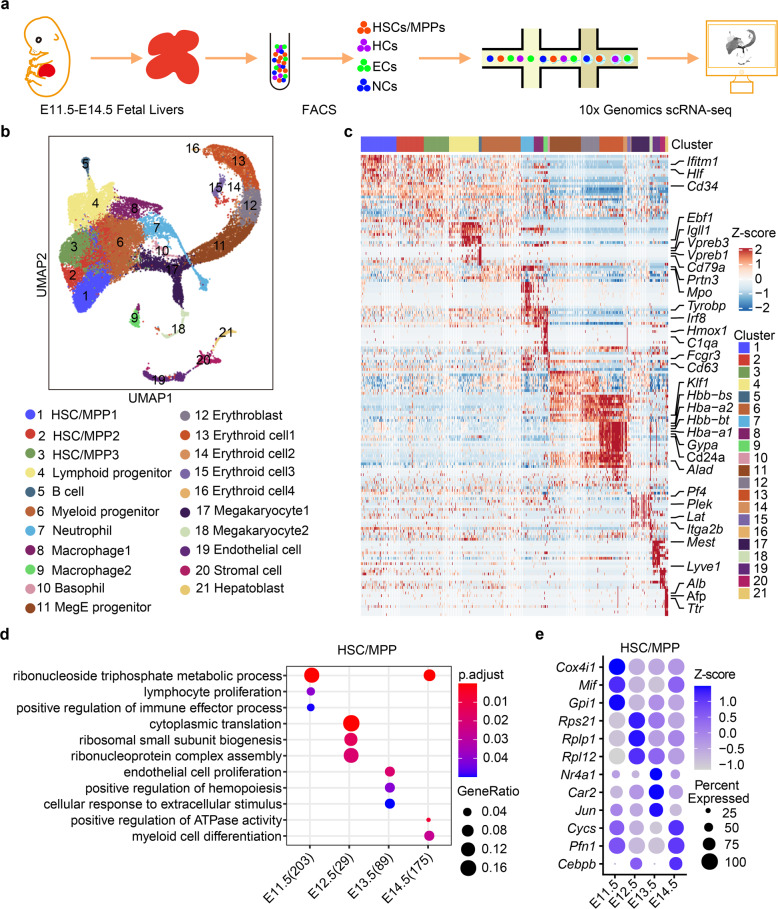
Previous studies have shown that FL-colonized HSCs/MPPs acquire definitive signatures from E11.5 to E12.5,17 followed by exhibiting expansion and differentiation characteristics from E12.5 to E14.5.7,18 To resolve the temporal dynamics of HSCs/MPPs and their niche at the transcriptomic level, we employed 10× Genomics to generate a single-cell resolution cell atlas of the developing FL at one-day intervals between E11.5 and E14.5 (Fig. 1a). To enrich HCs and ECs, we used fluorescence-activated cell sorting (FACS) at these time points to separately sort HCs, ECs, and non-hematopoietic/non-endothelial cells from the FL, and subsequently pooled them together in certain ratios for sequencing (see Materials and methods; Supplementary information, Fig. S1a, b). In total, 32,449 single cells were retained for Uniform Manifold Approximation and Projection (UMAP)19 analysis after quality control (Fig. 1b; Supplementary information, Fig. S1c, d). Based on the signature gene expression, we annotated 21 cell clusters (Fig. 1c; Supplementary information, Fig. S1e, f and Table S1), including 18 HC clusters and three structural niche cell clusters, and found that their proportions were dynamically correlated with the developmental stages (Supplementary information, Fig. S1g). Specifically, HSCs/MPPs (cluster (C) 1, C2, and C3) exhibited high expression levels of stem cell signature genes Mycn, Hlf, and Mecom.20,21 Lymphoid progenitors (C4) were marked by Ccr9, Il7r, and Flt3. Myeloid progenitors (C6) were characterized by the expression of Mpo, Spi1, and Ccl9.22 Three structural niche cell clusters consisted of C19 ECs (Lyve1, Kdr, and Pecam1), C20 stromal cells (Pdgfra, Alcam, and Ncam1), and C21 hepatoblasts (Afp, Alb, and Ttr) (Fig. 1c; Supplementary information, Fig. S1f).
Fig. 1. Single-cell transcriptomic atlas of the developing mouse FL.
a Schematic diagram of the procedures for tissue processing, cell isolation, and scRNA-seq profiling of FL. HSCs/MPPs, hematopoietic stem cells and multipotent progenitors; ECs, endothelial cells; HCs, hematopoietic cells; NCs, non-hematopoietic/non-endothelial cells. b UMAP visualization of all single cells colored by cell clusters in the FL. MegE progenitor, megakaryocytic-erythroid progenitor. c Heatmap showing row-scaled expression of the top 20 DEGs (Bonferroni-corrected P < 0.05, Wilcoxon Rank Sum test) per cluster. d Enriched GO terms in HSCs/MPPs during FL development. e Dot plots of the enriched DEGs in HSCs/MPPs during FL development.
The development of HSCs/MPPs and FL structural niche cells is tightly regulated by dynamic transcriptional programs.13,17 Accordingly, we focused on the transcriptional dynamics of HSCs/MPPs and structural niche cells. Analysis of differentially expressed genes (DEGs) was performed among four stages, and gene ontology (GO) analysis of DEGs in HSCs/MPPs identified that lymphoid program and immune response were enriched at E11.5 and E12.5, and hematopoietic development and mitotic cell cycle regulation were enriched at E13.5 and E14.5 (Fig. 1d, e). This suggests that the transcriptional dynamics of HSCs/MPPs in the developing FL underlies the maturation and expansion of HSCs/MPPs, which is consistent with previous reports.17,23 Moreover, GO analysis showed that mitotic cell cycle regulation and cell growth were also enriched in different types of structural niche cells, indicating a global increase in cell number and size to accommodate the FL expansion. Specifically, ECs exhibited vasculature sprouting at E11.5 and E12.5, and cell–cell adherent junctions at E13.5 and E14.5 (Supplementary information, Fig. S2a, b), representing a gradual formation of the FL vasculature. For the stromal cells, ribonucleoprotein biogenesis and RNA translation were enriched at E11.5 and E12.5, and extracellular structure organization and growth factor production were enriched at E13.5 and E14.5 (Supplementary information, Fig. S2c, d), suggesting that stromal cells provide structural support and signaling interaction for tissue development. GO analysis of hepatoblasts showed that ribosome biogenesis was enriched at E11.5 and E12.5, and metabolic regulation was enriched at E13.5 and E14.5 (Supplementary information, Fig. S2e, f). Taken together, we generated the first fully annotated single-cell resolution atlas of mouse FL, shown in a freely available website (Supplementary information, Video S1), to gain a comprehensive understanding of cellular composition dynamics, cell type-specific gene expression and stage-specific biological functions of both HSCs/MPPs and structural niche cells during FL development.
CD93-enriched HSCs/MPPs exhibit enhanced stem cell properties
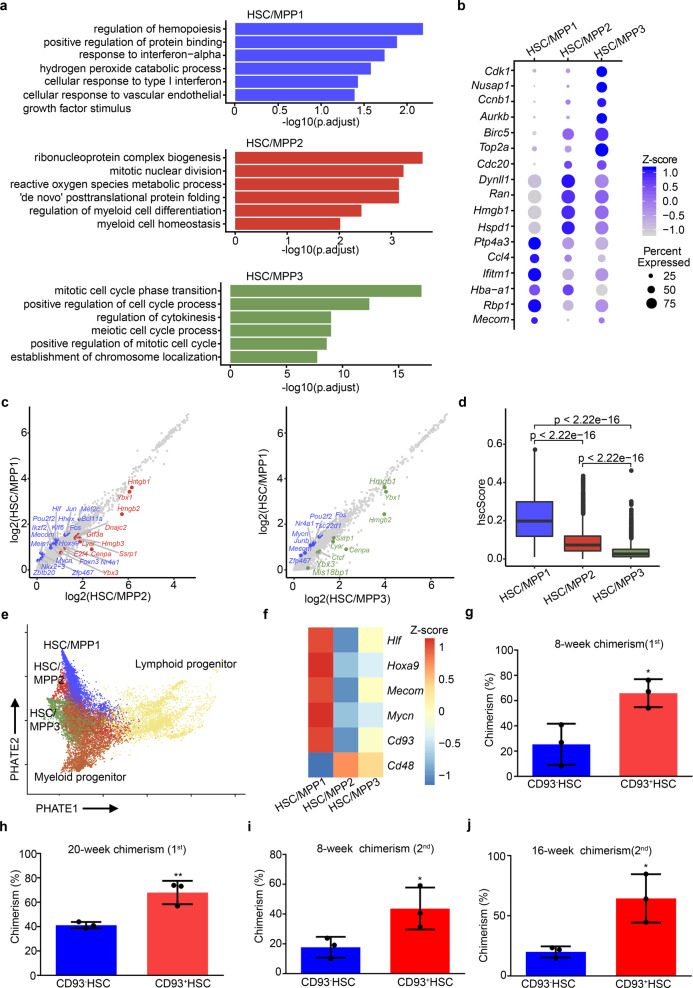
To further dissect the transcriptome features of HSCs/MPPs, we focused on HSC/MPP1–3 clusters and explored the underlying differences among them. GO and DEG analyses indicated enrichment of hematopoietic regulation and differentiation, active translation processes, and cell cycle regulation in HSC/MPP1–3, respectively (Fig. 2a, b). In HSC/MPP1, the expression of Mycn, Mecom, and Hlf was enriched; in HSC/MPP2, metabolic and lineage-specific genes were highly expressed; and in HSC/MPP3, cell cycle-related genes showed highly enriched expression (Fig. 2b, c). To further evaluate the stem cell properties of heterogeneous HSCs/MPPs, we calculated hscScore as previously reported,24 and found that the highest score was seen in HSC/MPP1 (Fig. 2d; Supplementary information, Table S2); meanwhile, trajectory analysis25 among HSC/MPP subsets and downstream myeloid and lymphoid progenitors revealed that HSC/MPP1 occupied the top of hematopoietic hierarchy (Fig. 2e). Overall, these results indicate that HSC/MPP1 exhibits the most robust stem cell transcriptome feature, compared to HSC/MPP2–3.
Fig. 2. CD93-enriched HSCs/MPPs exhibit enhanced stem cell properties.
a GO analysis among the three HSC/MPP subtypes. b Dot plots of the enriched DEGs in the three HSC/MPP subtypes. c Scatter plots of all gene expressions in HSC/MPP1 versus HSC/MPP2 (left) or versus HSC/MPP3 (right). Differentially expressed transcription factors are colored. d Boxplots showing the hscScore distribution in the three HSC/MPP subtypes. Student’s t-test. e Trajectory analysis by PHATE reconstructing the lineage differentiation from HSCs/MPPs to lymphoid and myeloid progenitors, respectively. f Heatmap showing the expression pattern of HSC/MPP signature genes and Cd93 in the three HSC/MPP subtypes. g Donor-derived chimerism in the peripheral blood (PB) of recipients at 8 weeks after primary transplantation using CD93+SLAM-LSK (CD93+ HSC) and CD93– HSC. n = 3 biological replicates. h Donor-derived chimerism in the PB of recipients at 20 weeks after primary transplantation using CD93+ HSC and CD93– HSC. n = 3 biological replicates. i Donor-derived chimerism in the PB of recipients at 8 weeks after secondary transplantation using CD93+ HSC and CD93– HSC. n = 3 biological replicates. j Donor-derived chimerism in the PB of recipients at 16 weeks after secondary transplantation using CD93+ HSC and CD93– HSC. n = 3 biological replicates. The statistical results are presented as means ± SEM. Student’s t-test; *P < 0.05; **P < 0.01.
To enrich HSC/MPP1 cells, we identified a cell-surface marker gene, Cd93, the expression of which was upregulated in HSC/MPP1 relative to HSC/MPP2–3 (Fig. 2f). Given that Cd93 (also known as AA4.1) has been shown to be expressed in hematopoietic progenitors,26–28 we asked whether the combination of CD93 and other HSC/MPP markers, such as Lineage–Sca-1+c-Kit+ (LSK), Flt3–LSK, and CD150+CD48–LSK (SLAM-LSK),29,30 can further purify HSCs/MPPs with robust stem cell properties. Flow cytometry analysis indicated that LSK (Supplementary information, Fig. S3a), SLAM-LSK (Supplementary information, Fig. S3c), and Flt3–LSK (Supplementary information, Fig. S3e) cells at E14.5 could be clearly divided into CD93+ and CD93– groups. Colony-forming unit (CFU) analysis showed that CD93+ cells (CD93+LSK, CD93+Flt3–LSK, and CD93+SLAM-LSK) exhibited a higher colony-forming ability than CD93– cells (Supplementary information, Fig. S3b, d, f). Among them, CD93+SLAM-LSK cells displayed the most robust ability to generate CFU-granulocyte, erythroid, macrophage, and megakaryocyte (CFU-GEMM) than other HSC/MPP groups (Supplementary information, Fig. S3d). To evaluate the engraftment ability of HSCs/MPPs purified by CD93+SLAM-LSK combination, we performed primary and secondary transplantation assays. The results showed that E14.5 CD93+SLAM-LSK cells exhibited higher reconstitution and self-renewal abilities than CD93–SLAM-LSK cells (Fig. 2g–j). Moreover, to determine whether CD93 can enrich HSCs/MPPs from heterogeneous LSK cells,31,32 we performed primary transplantation assay and the results showed that E14.5 CD93+LSK cells exhibited a higher short-term (8 weeks) and long-term (16 weeks) reconstitution abilities than LSK cells (Supplementary information, Fig. S3g); meanwhile, we also found that both E14.5 CD93High and CD93Mid LSK cells exhibited higher colony-forming and short-term (4 weeks) reconstitution abilities than the CD93Low LSK cells (Supplementary information, Fig. S3h, i). Furthermore, to reveal the functional role of CD93 in FL HSC/MPP development, we characterized the E14.5 FL HSC/MPP phenotype of CD93 genetic mutants (CD93 KO), whose knockout efficiency was validated at protein level (Supplementary information, Fig. S4a). Consequently, although the absolute number of whole FL cells and the proportion of undifferentiated Lineage– cells had no obvious change in the CD93 KO embryos (Supplementary information, Fig. S4b, c), the proportion of HSCs/MPPs (LSK cells and SLAM-LSK cells) was decreased in the CD93 KO embryos (Supplementary information, Fig. S4d–f). Taken together, CD93 is not only a surface marker for characterizing HSC/MPP heterogeneity, but also functionally required for FL HSC/MPP development.
Integration of scRNA-seq and ST decodes cell–cell interactions between HSCs/MPPs and niche cells
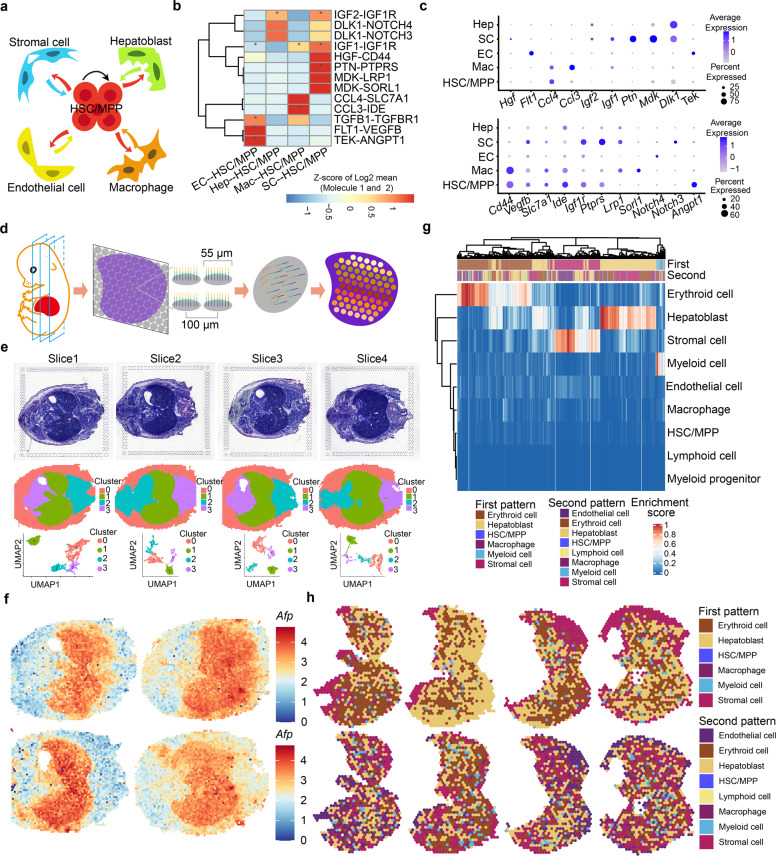
During embryogenesis, the FL provides a supportive niche for HSC/MPP development, however, the manner by which HSCs interact dynamically with their niche components remains elusive. To this end, we used CellPhoneDB33,34 to construct an unbiased HSC/MPP–niche cell interaction network, with a focus on structural niche cells and macrophages (Fig. 3a), the latter of which have been shown to act as a niche component for HSC/MPP development in the FL equivalent of zebrafish35 and mouse AGM region.36 In-depth analysis identified several well-known ligand–receptor interactions involved in HSC/MPP development, such as TGFΒ1–TGFΒR1, TEK–ANGPT1, and IGF2–IGF1R.37–39 Importantly, we identified several previously unrecognized ligand–receptor interactions (Fig. 3b, c), including those related to cell growth (FLT1–VEGFB, HGF–CD44, MDK–LPR1, IGF1–IGF1R, and PTN–PTPRS),40–43 cytokine recognition (CCL3–IDE and CCL4–SLC7A1),44 and Notch signaling (DLK1–NOTCH3 and DLK–NOTCH4).45
Fig. 3. Integration of scRNA-seq and ST decodes cell–cell interactions between HSCs/MPPs and niche cells.
a Schematic diagram indicating the cell–cell interactions between HSCs/MPPs and niche cells (endothelial cells, stromal cells, hepatoblasts and macrophages). b CellPhoneDB analysis showing the major HSC/MPP–niche cell interaction pairs. The asterisks mean P < 0.05 (permutation test) in corresponding cells. EC, endothelial cell; Hep, hepatoblast; SC, stromal cell; Mac, macrophage. c Dot plots of the ligand (top) and corresponding receptor (bottom) expression pattern of the major HSC/MPP–niche cell interaction pairs. d Schematic diagram of the procedures for 10× Visium ST experiments. e Hematoxylin and eosin (HE) staining of embryonic days (E)14.5 embryo tissue sections (top) included in ST analysis. Sections are shown in the dorsal (Slice 1) to the ventral direction (Slice 4); tissue sections (middle) and UMAP visualization (bottom) showing unbiased clustering of ST spots are presented. f Expression pattern of hepatoblast marker Afp in tissue sections. g Heatmap showing predicted enrichment scores of spots and two spot patterns of FL. h Spatial feature plots of the two spot patterns in FL sections.
To determine whether the predicted signaling interactions indeed exist among the anatomically-organized cells, we performed 10× Visium ST using four slices from an E14.5 embryo along the dorsal–ventral axis, with a focus on the FL region (Fig. 3d–f). We also assessed the performance of ST using region-specific markers, such as lung and intestine regions (Supplementary information, Fig. S5a). The joint dataset of FL region consisted of 3791 individual spots after filtering (Supplementary information, Fig. S5b, c). To show the spatial organization of principal cell types in an unbiased manner, we performed deconvolution analysis46,47 by assigning an enrichment score for each spot with cell type signature genes derived from scRNA-seq. After deconvolution, two spot patterns (first pattern and second pattern) were shown based on the enrichment score of the top two cell types, and then mapped to the original FL region (Fig. 3g, h). As expected, erythroid cells and hepatoblasts showed the highest enrichment score in most spots, indicating that they are the predominated cell components of FL. To specifically determine the spatial organization of HSCs/MPPs and their interactive niche cells (ECs, stromal cells, hepatoblasts and macrophages), we regarded the spots as credible for one cell type, if the enrichment score of those cell types is higher than a threshold (80% to HSCs/MPPs and 70% to niche cells), and then mapped them to the original FL region (Supplementary information, Fig. S6a). To evaluate the performance of ST in resolving spatial organization, we examined the expression of cell type-specific genes in the candidate spots. As expected, Hlf and Mecom were selectively enriched in the HSC/MPP-localized spots (Supplementary information, Fig. S6b); Ly86 and Cd68 were specifically expressed in the macrophage-localized spots (Supplementary information, Fig. S6g); and the similar results were also observed in the structural niche cells-localized spots (Supplementary information, Fig. S6c–f). Taken together, ST profiles the spatial organization of FL for further decoding cell–cell interactions predicted by scRNA-seq (see below).
Identification of expansion units of HSCs/MPPs
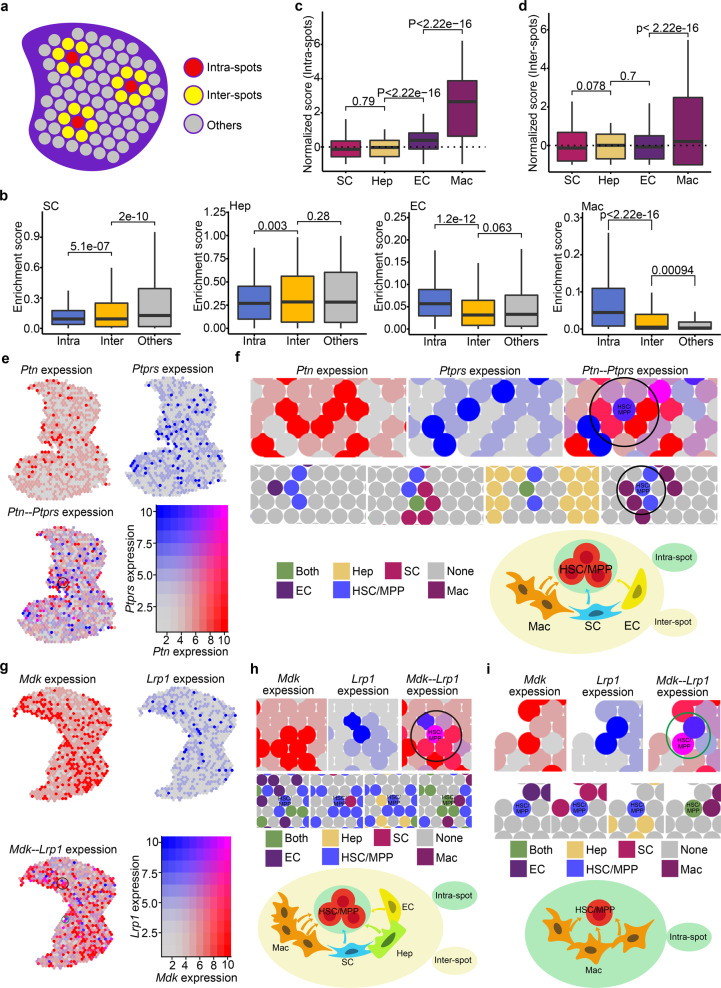
To determine the architectural basis of cell–cell interactions, we defined HSC/MPP-localized spots as intra-spots (indicating the closest relationship), HSC/MPP-surrounded spots as inter-spots (indicating the second closest relationship), and other distant spots (indicating nearly no interactive relationship) (Fig. 4a). For each niche cell type, analysis of enrichment score for different types of spots showed that EC with the highest score for intra-spots was close to HSCs/MPPs, which is consistent with a previous report13; hepatoblast and stromal cell with the highest scores respectively for inter-spots and other distant spots were less close to HSCs/MPPs; unexpectedly, macrophage with higher scores for intra- and inter-spots than that for other distant spots was considered as a novel niche component spatially close to HSCs/MPPs (Fig. 4b). To quantitatively compare the interaction between HSCs/MPPs and different niche cells, we defined an enrichment fold based on the ratio of the enrichment score median for each spot type to the enrichment score median for all spots. Consequently, we found that macrophage showed a 11.52-fold enrichment in the intra-spots and a 1.31-fold in the inter-spots, EC showed a 1.62-fold enrichment in the intra-spots, while hepatoblast and stromal cell showed less enrichment in the intra-spots (Fig. 4c, d; Supplementary information, Fig. S7a). Furthermore, to validate the spatial relationship at nearly single-cell resolution, we analyzed the mouse E13.5 FL ST data based on Stereo-seq, which is a sequencing-based spatially resolved transcriptomic technology with subcellular resolution.48 We defined 20 bins as a spot (10–15 μm in diameter), which may include 1–3 cell(s), and then annotated the spots (including intra-spotsStereo-seq, inter-spotsStereo-seq and other distant spotsStereo-seq) referring to the aforementioned pipeline. As a result, we found that macrophages showed a high enrichment both in intra-spotsStereo-seq and inter-spotsStereo-seq (Supplementary information, Fig. S7b). Finally, the consistent result was validated by CSOmap,49 an analytic method to reconstruct the cell spatial organization de novo based on scRNA-seq data (Supplementary information, Fig. S7c–f). Taken together, the results from three analytic methods of spatial information support that macrophage serve as an important niche cell with the closest relationship with HSCs/MPPs.
Fig. 4. Identification of expansion units of HSCs/MPPs.
a Schematic diagram indicating the intra-spots, inter-spots and others. Intra-spots, HSC/MPP-localized spots; Inter-spots, HSC/MPP-surrounded spots; Others, other distant spots. b Boxplots showing the enrichment score of niche cells in intra-spots, inter-spots, and others. c Boxplots showing the normalized (enrichment) score of niche cells in intra-spots. d Boxplots showing the normalized (enrichment) score of niche cells in inter-spots. e Co-expression pattern of Ptn and Ptprs in the FL section. f Spatial feature plots of Ptn−Ptprs-enriched HSC/MPP expansion units. Spatial feature plots showing the expression pattern of ligand gene Ptn in niche cells-localized spots (top, red spots), and receptor gene Ptprs in HSCs/MPPs-localized spots (top, blue spots). Spatial feature plots showing the cell types in HSC/MPP expansion units (middle). Schematic diagram indicating the Ptn−Ptprs-enriched HSC/MPP expansion units (bottom). g Co-expression pattern of Mdk and Lrp1 in the FL section. h Spatial feature plots of the Mdk−Lrp1-enriched HSC/MPP expansion units (inter-spots). i Spatial feature plots of the Mdk−Lrp1-enriched HSC/MPP expansion units (intra-spots).
At molecular level, we examined the spatial expression of interactive signals predicted by CellPhoneDB analysis, and found that genes encoding ligands, such as MDK and PTN, were highly expressed in niche cells of intra-spots and inter-spots (Fig. 4e–i), and genes related to receptors, such as LRP1 and PTPRS, were enriched in HSCs/MPPs-localized spots (Fig. 4e–i). These results indicate that spatial proximity facilitates the signaling interactions for functionally supporting HSC/MPP expansion. Given that the intra-spots and inter-spots were characterized by spatial proximity between cells and enriched interactive signals, we defined them as expansion units, in which HSCs/MPPs located at the core of spots and interacted with surrounding niche cell spots (Fig. 4f, h, i). Taken together, we demonstrate that FL HSCs/MPPs expand in a number of units, in which macrophages and several growth factors, including MDK and PTN, are highly enriched.
Functional validation of the role of niche cells in promoting HSC/MPP expansion
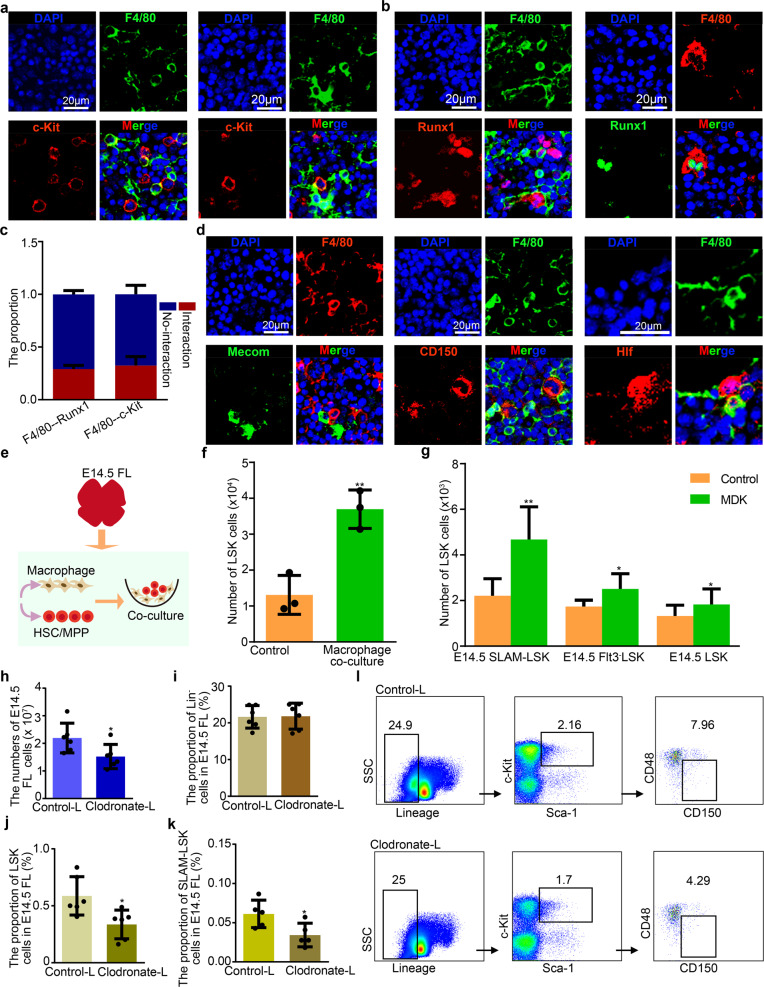
To determine how macrophages modulate HSC/MPP expansion in the FL, we firstly examined the spatial relationship between HSCs/MPPs and macrophages at E14.5. Immunofluorescence assay revealed two spatial patterns, one was that HSCs/MPPs attach proximal macrophages, the other was that HSCs/MPPs are surrounded by macrophages (Fig. 5a–d). Of note, the latter pattern is similar to that between hematopoietic stem and progenitor cells and ECs in the FL equivalent of zebrafish,50 suggesting that macrophages may promote HSC/MPP expansion via forming a ‘pocket-like’ architecture. Functionally, E14.5 HSCs/MPPs were co-cultured with macrophages or cultured in the defined medium supplemented with macrophage-derived MDK. As expected, we found that HSC/MPP expansion was enhanced in macrophage co-cultured or MDK-supplemented systems, suggesting that macrophages can facilitate HSC/MPP expansion via secreting MDK factor (Fig. 5e–g). Furthermore, to determine whether the depletion of macrophages within FL niche will affect HSC/MPP expansion, macrophages were first depleted by using apoptosis-inducing clodronate-liposomes (Supplementary information, Fig. S8a–d),36 and then we found that the absolute number of FL cells and the proportion of HSCs/MPPs (LSK cells and SLAM-LSK cells) were decreased at E14.5 (Fig. 5h–l).
Fig. 5. Macrophages promoting HSC/MPP expansion.
a Immunofluorescence analysis showing the expression of F4/80 (representing macrophages) and c-Kit (representing HSCs/MPPs) in E14.5 FL cryosections. b Immunofluorescence analysis showing the expression of F4/80 (representing macrophages), and Runx1 (representing HSCs/MPPs) in E14.5 FL cryosections. c The proportion of HSCs/MPPs with and without cellular interaction, respectively (n > 3 biological replicates). d Immunofluorescence analysis showing the expression of F4/80 (representing macrophages), CD150 (representing HSCs/MPPs), Mecom (representing HSCs/MPPs) and Hlf (representing HSCs/MPPs) in E14.5 FL cryosections. e Schematic diagram of the procedures for HSC/MPP co-culture with macrophages. f The number of HSCs/MPPs (LSK cells) derived from macrophage and HSC/MPP co-culture system. Macrophages were a combination of F4/80low and F4/80high macrophages at 1:1 ratio. n = 3 biological replicates. g The number of HSCs/MPPs (LSK cells) derived from the MDK (50 ng/mL)-supplemented culture of 100 HSCs/MPPs (LSK, Flt3–LSK and SLAM-LSK cells). n = 3 biological replicates. h Total cell number in E14.5 FL after Clodronate-liposome and Control-liposome treatment. Clodronate-liposome, Clodronate-L; Control-liposome, Control-L. n > 3 biological replicates. i The proportion of Lineage– (Lin–) cells in E14.5 FL after Clodronate-liposome and Control-liposome treatment. n > 3 biological replicates. j The proportion of LSK cells in E14.5 FL after Clodronate-liposome and Control-liposome treatment. n > 3 biological replicates. k The proportion of SLAM-LSK cells in E14.5 FL after Clodronate-liposome and Control-liposome treatment. n > 3 biological replicates. l Flow cytometry analysis showing the proportions of Lin–, LSK, SLAM-LSK cells in E14.5 FL after Clodronate-liposome and Control-liposome treatment. Scale bars, 20 μm. The statistical results are presented as means ± SEM. Student’s t-test; *P < 0.05; **P < 0.01.
Given that the FL is a transitory reservoir for both EMP- and HSC/MPP-derived macrophages,51 we thus performed bioinformatic analysis, immunofluorescence imaging and co-culture assay to determine which types of macrophages are in close contact to HSCs/MPPs. Firstly, by performing analyses of signature gene expression, developmental trajectory, and transcriptomic correlation, we found that macrophage1 (Mac1) and macrophage2 (Mac2) were derived from HSCs/MPPs and EMPs, respectively (Supplementary information, Fig. S9a–e). Then, we examined the spatial relationship between macrophages (Mac1 and Mac2) and HSCs/MPPs by 10× Visium ST. As a result, we found that both Mac1 and Mac2 exhibited a high enrichment in the intra- and inter-spots (Supplementary information, Fig. S10a), indicating that they are both in close contact to HSCs/MPPs. Moreover, immunofluorescence imaging showed that both Mac1 (marked by Ccr252,53) and Mac2 (marked by Iba1 and LXRα/β52,54) exhibited an adjacent relationship with HSCs/MPPs (marked by Runx1, c-Kit and CD15013,31,55) at E14.5 (Supplementary information, Fig. S10b–g). Finally, to examine the effect of Mac1 and Mac2 on the HSC/MPP expansion, we sorted F4/80low macrophages (CD45+CD11b+F4/80low,53,56 similar to Mac1), F4/80high macrophages (CD45+CD11blowF4/80high53,56, similar to Mac2), and performed co-culture assay with mouse HSCs/MPPs (LSK) at E14.5, respectively. As a result, we found that HSC/MPP expansion was enhanced both in Mac1- and Mac2-co-cultured systems (Supplementary information, Fig. S10h). Taken together, these results suggest that both EMP- and HSC/MPP-derived macrophages play important roles in supporting HSC/MPP expansion.
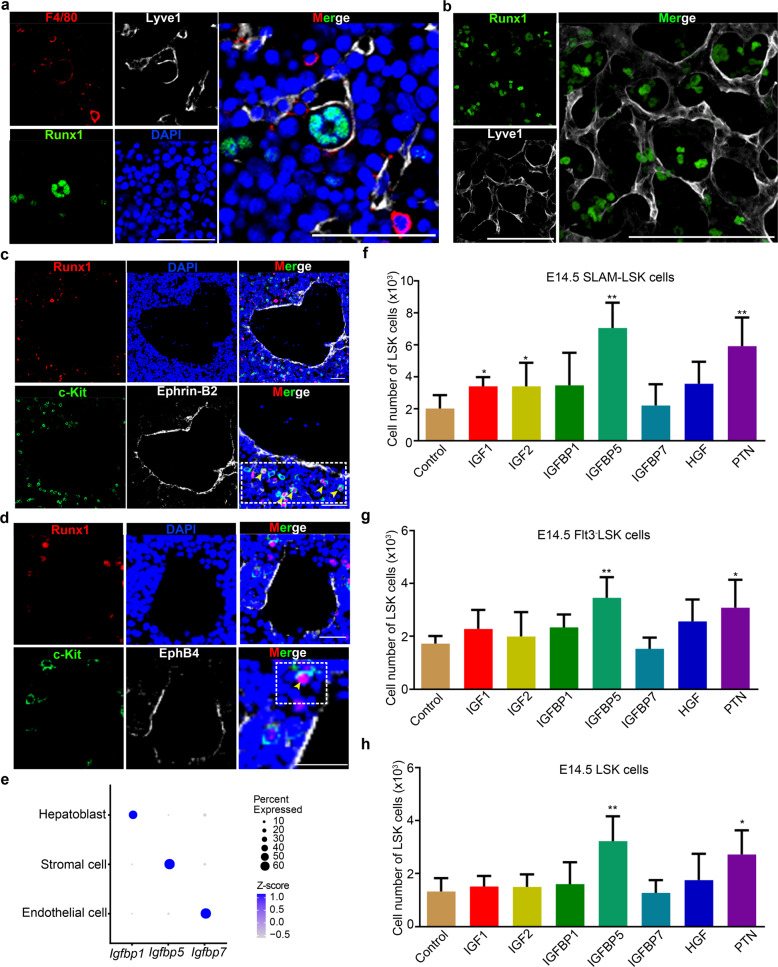
In addition to macrophages, a meaningful proportion of HSC/MPP expansion units were also composed of three structural niche cells (Supplementary information, Fig. S7g–i). Among them, a closer spatial relationship between HSCs/MPPs and ECs was uncovered by 10× Visium ST (Fig. 4b–d, Supplementary information, Fig. S7a). To characterize EC-forming niche of HSCs/MPPs in detail, we detected their spatial relationship. Immunofluorescence assay revealed that HSCs/MPPs were surrounded by ECs (Fig. 6a, b), and most of the HSCs/MPPs were close to arterial portal vessels labeled with EphrinB2,13 with a few HSCs/MPPs residing around hepatic veins labeled with EphB457 at E14.5 (Fig. 6c, d). These results indicate that HSCs/MPPs show preferential spatial association adjacent to a putative portal vessel niche in the FL. To functionally determine whether the predicted interactive signals derived from structural niche cells can regulate HSC/MPP expansion, we performed HSC/MPP expansion assay at E14.5 in the defined medium supplemented with IGF1, IGF2, HGF, and PTN, respectively. Given that the function of IGFs is regulated by their carrier proteins, IGFBPs58,59, we also examined the role of IGFBP1/5/7, whose gene expression was enriched in hepatoblasts, stromal cells, and ECs, respectively (Fig. 6e). Consequently, HSC/MPP expansion in IGFBP5- and PTN-supplemented culture systems was more dramatically enhanced than that in IGF1-, IGF2-, HGF-, IGFBP1- and IGFBP7-supplemented culture systems (Fig. 6f–h). Taken together, functional analysis validates the potential cell–cell interactions and underlying molecular mechanisms supporting HSC/MPP expansion.
Fig. 6. Structure niche cells promoting HSC/MPP expansion.
a Immunofluorescence analysis showing the expression of Lyve1 (representing sinusoid ECs) and Runx1 (representing HSCs/MPPs) in E14.5 FL cryosections. Scale bar, 50 μm. b Immunofluorescence analysis showing the expression of Lyve1 (representing sinusoid ECs) and Runx1 (representing HSCs/MPPs) in E14.5 FL cryosections. Scale bar, 50 μm. c Immunofluorescence analysis showing the expression of c-Kit (representing HSCs/MPPs), Runx1 (representing HSCs/MPPs), and EphrinB2 (representing arterial portal vessels) in E14.5 FL cryosections. Yellow arrowheads indicate the c-Kit and Runx1 double-positive HSCs/MPPs. Scale bar, 50 μm. d Immunofluorescence analysis showing the expression of c-Kit (representing HSCs/MPPs), Runx1 (representing HSCs/MPPs), and EphB4 (representing veins) in E14.5 FL cryosections. Yellow arrowheads indicate the c-Kit and Runx1 double-positive HSCs/MPPs. Scale bar, 25 μm. e Dot plots of the expression patterns of Igfbp1, Igfbp5, and Igfbp7 in three structural niche cells (endothelial cell, stromal cell, and hepatoblast). f The number of HSCs/MPPs (LSK cells) derived from the factors (50 ng/mL of each factor)-supplemented culture of 100 SLAM-LSK cells from E14.5 FL. n = 3 biological replicates. g The number of HSCs/MPPs (LSK cells) derived from the factors (50 ng/mL of each factor)-supplemented culture of 100 Flt3– LSK cells from E14.5 FL. n = 3 biological replicates. h The number of HSCs/MPPs (LSK cells) derived from the factors (50 ng/mL of each factor)-supplemented culture of 100 LSK cells from E14.5 FL. n = 3 biological replicates. The statistical results are presented as means ± SEM. Student’s t-test; *P < 0.05; **P < 0.01.
Cross-species analysis of FL hematopoiesis between mice and humans
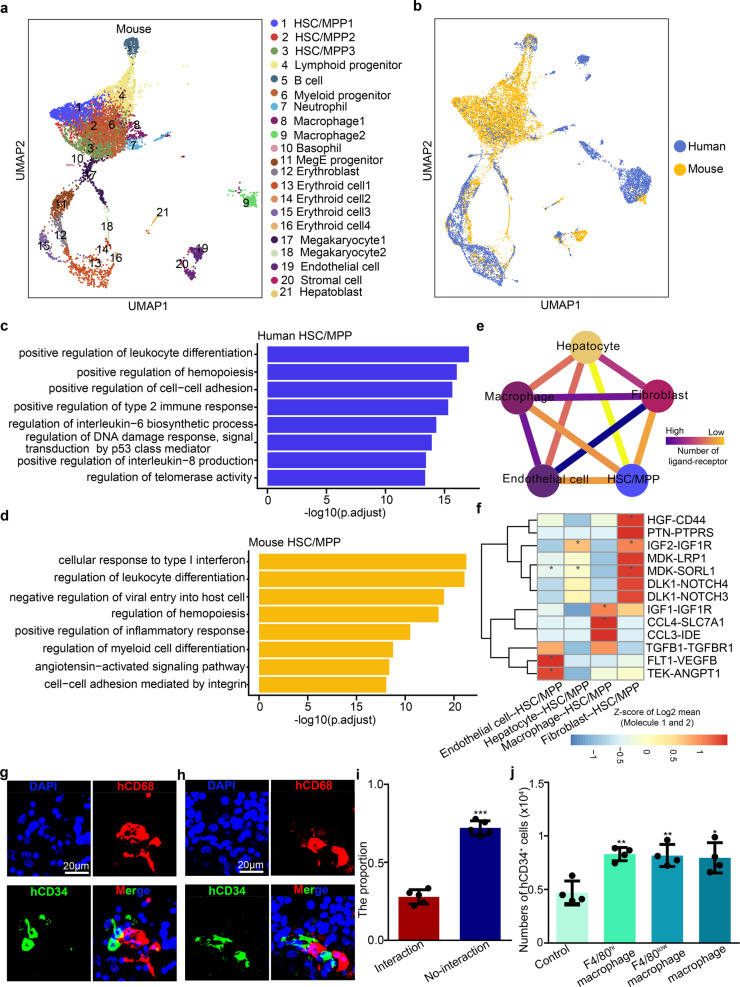
To analyze the conservation and divergence of FL hematopoiesis between mice and humans, we compared our scRNA-seq data with a recently published scRNA-seq atlas of the human FL.15 A total of 6325 single cells from the human FL at 11 post-conception weeks (PCW) and 8404 cells from the mouse FL at E14.5 were subjected to unsupervised UMAP analysis (Fig. 7a; Supplementary information, Fig. S11a). The integrative analysis showed that the FL cell types and HSC/MPP multilineage (erythrocyte/myelocyte/lymphocyte) differentiation routes were well mixed between these two species (Fig. 7b). Moreover, comparison of mouse and human FL cell types showed that most cell types were highly correlated (Supplementary information, Fig. S11b). Taken together, these data reveal conserved cell types and hematopoietic differentiation routes between mouse and human FL.
Fig. 7. Cross-species analysis of FL hematopoiesis between mice and humans.
a UMAP visualization of all cell types in the mouse E14.5 FL. Numbers indicate cell types; each dot represents one cell. b UMAP visualization of the FL cell populations from mice (E14.5) and humans (11 post-conception weeks). Yellow indicates the mouse cell population and blue indicates the human cell population. c GO analysis of the top DEGs related to biological processes in human HSCs/MPPs. d GO analysis of the top DEGs related to biological processes in mouse HSCs/MPPs. e Ligand–receptor interaction network showing the potential communications between human HSCs/MPPs and niche cells. f Heatmap showing the major human HSC/MPP−niche cell interaction pairs. The asterisks mean P < 0.05 (permutation test) in corresponding cells. g–h Immunofluorescence analysis showing the expression of CD68 (representing macrophages) and CD34 (representing HSCs/MPPs) in human 11 post-conception week FL section. Scale bars, 20 μm. i The proportion of HSCs/MPPs with and without cellular interaction, respectively (n > 3 biological replicates). j The number of human CD34+ HSCs/MPPs derived from mouse macrophage and HSC/MPP co-culture system. The statistical data are the cell number of CD34+ cells (total number of co-cultured cells × the proportion of CD34+ cells in total co-cultured cells) after co-culture. F4/80low macrophages were CD45+CD11b+F4/80low macrophages; F4/80high macrophages were CD45+CD11blowF4/80high macrophages; macrophages were a combination of F4/80low and F4/80high macrophages at 1:1 ratio. n > 3 biological replicates. The statistical results are presented as means ± SEM. Student’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001.
To compare the transcriptome features of FL HSCs/MPPs between mice and humans, we performed DEG analysis between HSCs/MPPs and other cell clusters for each species and identified 39 canonical HSC/MPP signature genes that are conserved in humans and mice (Supplementary information, Fig. S11c). Moreover, we performed GO analysis of the DEGs between mouse and human HSCs/MPPs. The results showed that the GO terms ‘regulation of DNA damage response’, ‘regulation of interleukin-6 biosynthetic process’, and ‘regulation of telomerase activity’ were enriched in human HSCs/MPPs (Fig. 7c), supporting the idea that human HSCs/MPPs are susceptible to DNA damage.60,61 By contrast, the GO terms ‘positive regulation of inflammatory response’, ‘cellular response to type I interferon’, and ‘negative regulation of viral entry into host cells’ were enriched in mouse HSCs/MPPs, supporting the notion that mouse HSCs/MPPs are responsive to inflammatory signals (Fig. 7d).62 Given the different living environments between mice and humans, mouse HSCs/MPPs may be more resistant to pathogen-derived inflammation, whereas human HSCs/MPPs are likely more susceptible to radiation-induced DNA damage. The presence of these divergent transcriptome features of HSCs/MPPs therefore indicates that despite overall similarity, there also exist species-specific mechanisms of the adaptive response to external stimuli.
To determine whether the mechanisms of HSC/MPP expansion in mice are conserved in humans, we also constructed an HSC/MPP–niche cell interaction network in the human FL using CellPhoneDB. We found that HSCs/MPPs displayed stronger interactions with macrophages and ECs than with the other two niche cells (Fig. 7e), which was also validated by CSOmap analysis (Supplementary information, Fig. S11d, e). In addition, we found that the interactions related to cell growth, cell adhesion, cytokine recognition, and Notch signaling identified in mice also existed in humans (Fig. 7f). Functionally, we performed immunofluorescence imaging and validated the cell–cell interaction between macrophages (marked by CD68) and HSCs/MPPs (marked by CD34) in human FL at 11 PCW (Fig. 7g–i); meanwhile, human cord blood (CB) CD34+ HSCs/MPPs were co-cultured with mouse macrophages. After four-day co-culture, we found that the number of co-cultured CB CD34+ HSCs/MPPs was more than (1.7 folds) that of CB HSCs/MPPs without macrophage co-culture, consistent with the notion that macrophages play a conserved role in supporting human HSC/MPP expansion (Fig. 7j). Taken together, these data are consistent with a model whereby many mechanisms of FL HSC expansion are conserved between humans and mice, but there are also species-specific transcriptome signatures in HSCs/MPPs.
Discussion
Using a high-throughput scRNA-seq and high-spatial-resolution ST framework, we generated an integrated spatiotemporal transcriptome atlas of FL in mice (Supplementary information, Fig. S12). scRNA-seq and functional analyses revealed that CD93 can be applied to purify a subset with enhanced stem cell properties from heterogeneous FL HSCs/MPPs. Given that CD93-marked type 1 HSC precursors in mouse AGM region do not show enhanced reconstitution ability than the counterparts after co-culture with stromal cells,63 we reason that hematopoietic cell heterogeneity characterized by CD93 may be cell-type- and developmental stage-dependent. Importantly, by integrating transcriptome, immunostaining, and functional analyses, we identified the ‘pocket-like’ units for HSC/MPP expansion, termed as HSC PLUS. Furthermore, although cross-species comparison of the FL cell atlas between mice and humans showed some divergent transcriptome signatures of HSCs/MPPs indicative of a species-specific mechanism of the adaptive response to external stimuli, the overall transcriptional similarity as well as conserved expansion mechanisms of HSCs/MPPs underscores the values of mouse models to provide guidance for the expansion of functional human HSCs/MPPs ex vivo.
A profound understanding of cellular and molecular mechanisms underlying HSC/MPP expansion is essential for establishing protocols of HSC/MPP expansion ex vivo. Previous studies have suggested that FL niche cells can support HSC/MPP expansion via paracrine signaling. For instance, ECs can secrete SCF, ANGPTL2, and IGF2 for HSC expansion13; hepatoblasts support HSC/MPP expansion by secreting ANGPTL3, IGF2, SCF, and TPO12,64; and stromal cells produce ANGPTL2/3 and IGF2 for HSC/MPP expansion.39,65 However, how these different types of cells interact with each other to form distinct functional tissues/organs remains unclear. By integrative analysis of spatiotemporal transcriptome atlas and functional experiments, we, for the first time, defined the HSC PLUS as the smallest functional units of FL. Within the HSC PLUS, macrophages and expansion-favoring factors including MDK, PTN, and IGFBP5 are enriched. Although the close spatial relationships between macrophages/ECs and HSCs/MPPs were also revealed by CSOmap analysis in human FL, further functional validation of the conserved mechanisms is warranted in future investigations. Taken together, our study provides a rich resource which deepens the understanding of the crosstalk between HSCs/MPPs and niche cells, and gives novel insights into the generation of HSC PLUS-based culture system for HSCs/MPPs ex vivo.
Macrophages play an important role in immune defense, however, extensive studies have demonstrated their multiple functional roles beyond immune defense. For example, macrophages repair the ruptured brain vessels in zebrafish via adhesion and mechanical traction;66 in mouse aorta, macrophages secret matrix metalloproteinase 9 to control the collagen production and prevent arterial stiffness.67 Our immunofluorescence, integrative transcriptomics, and functional analyses identify an uncharacterized role of FL-resident macrophages in facilitating HSC/MPP expansion. Interestingly, macrophages form a ‘pocket-like’ architecture surrounding the inside HSCs/MPPs, which is architecturally similar to EC ‘pocket’ observed in E11.5 mouse FL.50 Of note, the existence of ‘pocket-like’ architecture formed by different niche cells may be partially explained by that a specialized microenvironment could effectively enhance cell–cell interaction by increasing the concentration of local signaling molecules,50 such as Ccl21 from ECs55 and MDK from macrophages (in this study). Taken together, our unbiased characterization of HSC/MPP niche components updates the paradigm that HSCs/MPPs preferentially reside in the perivascular niche and are regulated by ECs, hepatoblasts, and stromal cells.10–13,50
In summary, our findings characterize HSC/MPP heterogeneity, expansion-favoring factors and architectural basis, and conserved expansion mechanisms in both mice and humans, which provides a valuable resource for studying HSC/MPP expansion. Further analysis of this resource will help to elucidate the mechanism underlying hematological malignancies of fetal origin.
Materials and methods
Experimental model
Mouse lines
Wild-type C57BL/6 mice were purchased from SPF (Beijing) Biotechnology Co., Ltd. CD93 genetic mutant mice were generated as previously described.68 B6.SJL (CD45.1) mice were purchased from the Institute of Laboratory Animal Science (ILAS), Chinese Academy of Medical Sciences (CAMS). All mice were bred under specific pathogen-free conditions (animal license number: SYXK (Beijing) 2018-0021). For transplantation assays, male CD45.1 mice were crossed with female CD45.2 mice to obtain CD45.1/.2 embryos; the day that the vaginal plug was detected was defined as embryonic day (E) 0. The present study was approved by the Ethical Review Committee of the Institute of Zoology, Chinese Academy of Sciences, China.
Human samples
Human embryos were obtained following electively terminated pregnancies at Haidian Maternal & Child Health Hospital in Beijing. All experiments were performed in accordance with protocols approved by the Peking University Institutional Review Board (PUIRB) (certificate number: IRB00001052-18083)69 and the Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences. Written informed consent was obtained before sample collection.
Human cord blood (CB) samples were collected from healthy volunteers after informed consents obtained. All experiments were performed in accordance with protocols approved by the Research Ethics Committee of Beijing Institute of Transfusion Medicine and the Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences. CD34+ HSCs/MPPs from human CB were performed in accordance with protocols.70,71
Flow cytometry
Single-cell suspensions from the FL were prepared by mechanical dissociation, and the cells were stained for 30 min at 4 °C. Antibodies used in this study included those against: Lineage-FITC (Biolegend, 133302), 7-AAD (Biolegend, 420404), CD117-APC (eBioscience, 17-1171-82), CD117-PE-Cyanine5 (eBioscience, 15-1171-81), CD117-APC-eFluor 780 (eBioscience, 47-1171-82), Ly-6A/E-PE-Cyanine7 (eBioscience, 25-5981-81), CD135-PE-Cyanine5 (eBioscience, 15-1351-82), CD135-APC (Biolegend, 135309), CD45.1-FITC (eBioscience, 11-0451-81), CD45.2-PE-Cyanine7 (eBioscience, 25-0454-80), CD3e-APC (eBioscience, 17-0031-81), B220-PE (eBioscience, 12-0452-81), CD11b-APC (eBioscience, 17-0112-81), Ly6G/Ly-6C-PE (eBioscience, 12-5931-81), CD150-PE (eBioscience, 12-1502-82), CD48-APC-eFluor780 (eBioscience, 47-0481-82), CD93-APC (eBioscience, 17-5892-81), CD45-FITC (eBioscience, 11-0451-82), CD68-PE-Cyanine7 (eBioscience, 25-0681-80), CD34-APC (BD, 560940) and APC-Cyanine7-F4/80 (Biolegend, 123117). 7-AAD viability staining solution was used to exclude dead cells. Flow cytometry was performed using MoFlo XDP (Beckman Coulter) and the analysis was carried out with Flowjo (vX.7.0).
HSC/MPP cell culture
One hundred HSCs/MPPs from E14.5 FL, including Lin–Sca-1+c-Kit+ (LSK), Flit3–LSK, CD150+CD48–LSK (SLAM-LSK) cells, were alone cultured in 100 μL StemSpan medium in a well of 96 U-well plate for 15 days. The culture medium was supplemented with 50 ng/mL SCF (Peprotech), 30 ng/mL FLT3L (Peprotech), 20 ng/mL TPO (Peprotech), and 10 ng/mL IL-6 (Peprotech). The expansion medium was supplemented with 50 ng/mL IGF1, IGF2, IGFBP1, IGFBP5, IGFBP7, PTN, MDK, respectively.
E14.5 HSCs/MPPs (LSK cells) at a density of 2 × 103 cells were co-cultured with 2 × 104 F4/80low macrophages (CD45+CD11b+F4/80low), F4/80high macrophages (CD45+CD11blowF4/80high) or macrophages (containing half of F4/80low and half of F4/80high macrophages) in 500 μL MyeloCult M5300 medium in a well of 24-well plate for 4 days. The culture medium was supplemented with 50 ng/mL SCF (Peprotech), 30 ng/mL FLT3L (Peprotech), 20 ng/mL TPO (Peprotech), and 10 ng/mL IL-6 (Peprotech).
Human CB CD34+ HSCs/MPPs at a density of 2 × 103 cells were co-cultured with 2 × 104 mouse macrophages in 500 μL StemSpan medium in a well of 24-well plate for 4 days. The culture medium was supplemented with 50 ng/mL Recombinant Human SCF (Peprotech), 50 ng/mL FLT3L (Peprotech), 50 ng/mL TPO (Peprotech).
Clodronate-liposome treatment
For the clodronate-liposome treatment, pregnant females were injected with Clodronate-liposome and Control-liposome (CP-005-005, LIPOSOME) as dose of 1 mg per 20 g mouse body weight at E11.5, E12.5 and E13.5, respectively, and the macrophage depletion and FL HSC/MPP phenotype were measured at E14.5.
Immunofluorescence
Immunofluorescence assay for embryos was performed as previously described.72 The E14.5 embryos or human FL (11 PCW) were fixed in 4% paraformaldehyde/PBS overnight at 4 °C. After gradient dehydration in 15% and 30% sucrose/PBS at 4 °C, these embryos were embedded and frozen in OCT overnight at −80 °C. The following day, embryos were sectioned to 10-μm-thick slides and blocked for 2 h at room temperature with 5% BSA/PBS containing 0.3% Triton X-100. After washing three times in PBS, 15 min/time on an orbital shaker, the slides were incubated overnight at 4 °C with primary antibodies diluted in 1% BSA/PBS. The following day, the slices were washed three times in PBS containing 0.1% Tween-20, 15 min/time on an orbital shaker, and incubated for 2 h at room temperature with secondary antibodies diluted in 1% BSA/PBS. Nuclei were stained with Hoechst 33342 (Invitrogen) and immunofluorescence images were obtained using a laser scanning confocal microscope (A1; Nikon). The primary antibodies used in the assays included those against: Runx1 (1:800; abcam), CD150 (1:200; BD), CD150 (1:200; Biolegend), c-Kit (1:300; BD), c-Kit (1:200; Biolegend), Hlf (1:100; ABclonal), Mecom (1:200; ABclonal), EphrinB2 (1:300; R&D), Ephb4 (1:300; R&D), Lyve1 (1:800; R&D), F4/80 (1:800; Santa Cruz), LXRα/β (1:500; Santa Cruz), Iba1 (1:500; Santa Cruz), Ccr2 (1:250; abcam), human anti-CD34 antibody (1:250; abcam), human anti-CD68 antibody (1:200; Biolegend). The secondary antibodies used in the assays included goat anti-rabbit IgG Alexa Fluor 488 (1:800; Life Technologies), goat anti-rabbit IgG Alexa Fluor 594 (1:800; Life Technologies), donkey anti-rat IgG Alexa Fluor 488 (1:800; Life Technologies), goat anti-rat IgG Alexa Fluor 594 (1:800; Life Technologies), goat anti-mouse IgG Alexa Fluor 488 (1:800; Life Technologies), goat anti-mouse IgG Alexa Fluor 555 (1:800; Life Technologies) and donkey anti-goat IgG Alexa Fluor 647 (1:300; Life Technologies),
Colony-forming unit (CFU) culture assay
Single-cell suspensions of E14.5 HSCs/MPPs sorted by the combination of CD93 and other HSC/MPP markers were cultured in M3434 medium (Stem Cell Technologies, 03434) at 37 °C with 5% CO2 for 10 days. Subsequently, the number of each type of colony, including CFU-E, CFU-GM, and CFU-GEMM, was counted based on morphology. More than three groups of independent experiments were analyzed. CFU-E, CFU-erythroid; CFU-GEMM, CFU-granulocyte, erythroid, macrophage, megakaryocyte; CFU-GM, CFU-granulocyte, macrophage.
Transplantation assay
CD45.2 recipients received a split lethal dose (9 Gy) of X-ray irradiation (RS2000). For the primary transplantation assay, a total of 500 CD93+ HSCs/CD93– HSCs or 2000 LSK/CD93+LSK cells were sorted from E14.5 FL along with 4 × 104 CD45.2 nucleated bone marrow (BM) cells and intravenously injected into the tail vein of the recipients. For the secondary transplantation assay, 1 × 106 BM cells from donor mice were mixed with 4 × 104 CD45.2 nucleated BM cells and intravenously injected into the tail vein of the recipients. The chimerism and the multi-lineage reconstitution ability were analyzed using peripheral blood (PB) from the recipients.
Western blotting
Western blotting was performed as previously described.72 The proteins were extracted from E14.5 FL cells with cell lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, and 0.5% NP-40) containing protease inhibitor (No. 04693116001; Roche). The protein level of CD93 was detected using the following antibodies: anti-β-actin (1:2000; Cell Signaling Technology), anti-CD93 (1:1000; abcam).
Cell preparation for scRNA-seq
FACS was performed at four time points to separately sort hematopoietic cells (HCs), endothelial cells (ECs), and non-hematopoietic/non-endothelial cells (NCs) from the FL, and the cells were subsequently pooled together for sequencing. At E11.5, CD45+ HCs9,73 and CD45– non-HCs were pooled together for sequencing. At E12.5, E13.5, and E14.5, LSK cells74,75, CD45+ HCs, CD45–CD31+ ECs,76 and CD45–CD31– NCs were pooled together for sequencing.
Processing of scRNA-seq data
To capture individual cells, we utilized the Chromium Single Cell 2’ Reagent Version 1 Kit (10× Genomics). The quality of the sequencing reads was evaluated by FastQC and MultiQC. Cell Ranger v2.2.0 was used to align the sequencing reads to the mm10 mouse reference genome and quantitate the expression of transcripts in each cell. R v3.6.0 and the package Seurat v3.1.2 were used for the downstream analyses. Cells with fewer than 250 detected genes, less than 500 unique molecular identifiers (UMIs), or more than 10% mitochondrial UMIs were removed. Finally, we detected an average of 3503 genes per cell.
Batch correction and dimension reduction
To minimize technical variability due to sample collection time points while preserving biological variation, two functions in Seurat, FindIntegrationAnchors and IntegrateData, were used for batch correction among cells at the four embryonic stages. Specifically, only principal component analysis (PCA) and PCA-dependent analyses were performed using batch-corrected data.
Dimension reduction includes three stages of analysis: selection of variable genes (FindVariableGene), PCA (RunPCA, from variable genes), and uniform manifold approximation and projection (UMAP). After quantity control, we obtained 32,449 high-quality single cells for further analysis. Subsequently, we performed PCA using the 2000 most variable genes selected by the FindIntegrationAnchors function. The top 30 principal components (PCs) were selected for UMAP to obtain bidimensional coordinates for each cell.
To dissect the potential relationship among HSCs/MPPs, progenitors, and macrophages, we also performed PHATE analysis using the same 2000 genes.
Unsupervised clustering and annotation
We performed the FindNeighbors and FindClusters functions (resolution = 0.5) to cluster cells using the Louvain algorithm based on the top 30 PCs. The final number of cell clusters was found to be 21 by selecting a resolution of 0.5. The known markers used for cell cluster annotation are listed in Supplementary information, Table S1.
Identification of DEGs
We used the FindAllMarkers function (logfc. threshold = log(1.2)) based on normalized data to identify DEGs. P value adjustment was performed using Bonferroni correction based on the total number of genes in the dataset. DEGs with adjusted P > 0.05 were filtered out. GO analyses were performed using the R package cluster profiler.77
Cell–cell interaction analysis
CellPhoneDB33,34 was used to infer enriched ligand–receptor interactions based on the gene expression of a ligand in one cell type and its receptor in another cell type. Selected ligand–receptor pairs enriched between HSCs and niche cells or macrophages were visualized. We only considered the ligands expressed in more than 20%, and receptors expressed in more than 4%, of cells in one cluster.
10× Visium ST
Staining and imaging
E14.5 embryos were embedded in tissue-Tek (OCT) and snap-frozen using dry ice slurry. Cryosections were cut at 10-μm thickness, mounted onto the GEX arrays. Sections were placed on Thermocycler Adaptor with the active surface facing up, incubated for 1 min at 37 °C, and fixed for 30 min with methyl alcohol in –20 °C, followed by staining with H&E (Eosin, Dako CS701, Hematoxylin Dako S3309, bluing buffer CS702). The brightfield images were taken on a Leica DMI8 whole-slide scanner at 10× resolution.
Permeabilization and reverse transcription
Visium spatial gene expression was processed using Visium spatial gene expression slide and Reagent Kit (10× Genomics, PN-1000184). For each well, Slide Cassette was used to create leakproof wells for adding reagents. 70 μL Permeabilization enzyme was added and incubated at 37 °C for 20 min. Each well was washed with 100 μL SSC, and 75 μL reverse transcription Master Mix was added for cDNA Synthesis.
cDNA library preparation for sequencing
At the end of first-strand synthesis, remove reverse transcription Master Mix from the wells. Add 75 μL 0.08 M KOH and incubate 5 min at room temperature, then remove the KOH from wells and wash with 100 μL EB buffer. Add 75 μL Second Strand Mix to each well for second-strand synthesis. cDNA amplification was performed on a S1000TM Touch Thermal Cycler (Bio Rad). According to the manufacture’s introduction, Visium spatial libraries were constructed using Visium spatial Library construction kit (10× Genomics, PN-1000184). The libraries were finally sequenced using an Illumina Novaseq6000 sequencer with a sequencing depth of at least 100,000 reads per spot with pair-end 150 bp (PE150) reading strategy (performed by CapitalBio Technology, Beijing).
Processing of the ST data
To annotate spots, we applied the integration workflow introduced in Seurat v3, which enables the probabilistic transfer of cell types from the scRNA-seq data to the ST data. Specifically, we first identified pairwise correspondences between single cells and single spots to quantify the batch effect. Each spot was then annotated based on the transcriptomic similarity between spots and cell types in the scRNA-seq dataset. This probabilistic transfer procedure was implemented using the FindTransferAnchors (dims = 1:30) and TransferData (dims = 1:30) functions in Seurat with the combination of top 100 DEGs of each cell type.
Cross-species analysis of FL between mouse and human
We obtained single-cell transcriptome data of human FL from ArrayExpress under the accession code E-MTAB-7407. Firstly, we analyzed gene expression matrices of 6325 human FL cells at 11 post-conception weeks and 8404 mouse FL cells at E14.5 after filtering low-quality cells. We then filtered mouse–human ortholog genes based on the Mouse Genome Informatics (MGI) database (http://www.informatics.jax.org/downloads/reports/HMD_HumanPhenotype.rpt), and obtained 13,819 homologous genes with a one-to-one translation between humans and mice for further analysis. Next, the merge function was used to combine human and mouse FL cells, and further analysis was performed including UMAP, differential expression, and Pearson correlation using the same methods described above.
Quantitation and statistical analysis
GraphPad Prism 6 was used to analyze the data. Values are presented as the means ± SEM. Student’s unpaired two-tailed t-test was applied for comparisons unless otherwise indicated.
Supplementary information
Acknowledgements
We thank Yiwei Lai (BGI-ShenZhen, Shenzhen, China) for data transmission of E13.5 Stereo-seq dataset. This work was supported by grants from the National Key R&D Program of China (2018YFA0800200, 2018YFA0801000, 2016YFA0100500 to F.L., and 2016YFA0100103 to C.L.), the Strategic Priority Research Program of the Chinese Academy of Sciences, China (XDA16010207 to F.L.), the National Natural Science Foundation of China (31830061, 81530004, 31425016 to F.L., and 31871266 to C.L.), and NSFC Key Research Grant 71532001 (to C.L.). Part of the data analysis was performed on the High-Performance Computing Platform of the Center for Life Sciences, Peking University.
Author contributions
S.G. and F.L. conceived the project, analyzed the data, and wrote the paper; S.G. performed most of the experiments with help from Y.Z., G.L., B.H., D.M., and L.W.; Q.S. and Z.K. performed the bioinformatics analysis under the guidance of C.L.; J.J. provided CD93 genetic mutant mice; X.F. provided human CB sample; C.X. provided human FL sample; X.X. and L.L. provided E13.5 FL Stereo-seq dataset; B.G. interpreted the data and assisted with manuscript preparation. All authors read and approved the final manuscript.
Data availability
The scRNA-seq and 10× Visium ST datasets generated in the present study are available at https://bigd.big.ac.cn/gsub/ in BIG Submission Portal, under the accession numbers CRA002489 and CRA003651, respectively. The E13.5 Stereo-seq dataset used in the present study is kindly provided by Prof. Xun Xu and Prof. Longqi Liu.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Suwei Gao, Qiang Shi.
Contributor Information
Cheng Li, Email: cheng_li@pku.edu.cn.
Feng Liu, Email: liuf@ioz.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-021-00540-7.
References
- 1.Yamamoto R, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson AC, Igarashi KJ, Nakauchi H. Haematopoietic stem cell self-renewal in vivo and ex vivo. Nat. Rev. Genet. 2020;21:541–554. doi: 10.1038/s41576-020-0241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvanese V, et al. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature. 2019;576:281–286. doi: 10.1038/s41586-019-1790-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson AC, et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature. 2019;571:117–121. doi: 10.1038/s41586-019-1244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGrath KE, Frame JM, Palis J. Early hematopoiesis and macrophage development. Semin. Immunol. 2015;27:379–387. doi: 10.1016/j.smim.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoeffel G, Ginhoux F. Ontogeny of tissue-resident macrophages. Front. Immunol. 2015;6:486. doi: 10.3389/fimmu.2015.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ema H, Nakauchi H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood. 2000;95:2284–2288. [PubMed] [Google Scholar]
- 8.Mikkola HKA, Orkin SH. The journey of developing hematopoietic stem cells. Development. 2006;133:3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- 9.Rybtsov S, Ivanovs A, Zhao SL, Medvinsky A. Concealed expansion of immature precursors underpins acute burst of adult HSC activity in foetal liver. Development. 2016;143:1284–1289. doi: 10.1242/dev.131193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, Leslie J, Yang Y, Ding L. Hepatic stellate and endothelial cells maintain hematopoietic stem cells in the developing liver. J. Exp. Med. 2021;218:e20200882. doi: 10.1084/jem.20200882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swain A, Inoue T, Tan KS, Nakanishi Y, Sugiyama D. Intrinsic and extrinsic regulation of mammalian hematopoiesis in the fetal liver. Histol. Histopathol. 2014;29:1077–1082. doi: 10.14670/HH-29.1077. [DOI] [PubMed] [Google Scholar]
- 12.Schmelzer E. Hepatic progenitors of the fetal liver: interactions with hematopoietic stem cells. Differentiation. 2019;106:9–14. doi: 10.1016/j.diff.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Khan JA, et al. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science. 2016;351:176–180. doi: 10.1126/science.aad0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X, et al. Mapping the mouse cell Atlas by Microwell-Seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Popescu DM, et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ståhl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 17.McKinney-Freeman S, et al. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell. 2012;11:701–714. doi: 10.1016/j.stem.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao S, Liu F. Fetal liver: an ideal niche for hematopoietic stem cell expansion. Sci. China Life Sci. 2018;61:885–892. doi: 10.1007/s11427-018-9313-4. [DOI] [PubMed] [Google Scholar]
- 19.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. PR-domain-containing Mds1-Evi1 is critical for long-term hematopoietic stem cell function. Blood. 2011;118:3853–3861. doi: 10.1182/blood-2011-02-334680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokomizo T, et al. Hlf marks the developmental pathway for hematopoietic stem cells but not for erythro-myeloid progenitors. J. Exp. Med. 2019;216:1599–1614. doi: 10.1084/jem.20181399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang KL, et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 2017;20:1052–1061. doi: 10.1038/nn.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tober J, et al. Maturation of hematopoietic stem cells from prehematopoietic stem cells is accompanied by up-regulation of PD-L1. J. Exp. Med. 2018;215:645–659. doi: 10.1084/jem.20161594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamey FK, Gottgens B. Machine learning predicts putative hematopoietic stem cells within large single-cell transcriptomics data sets. Exp. Hematol. 2019;78:11–20. doi: 10.1016/j.exphem.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moon KR, et al. Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol. 2019;37:1482–1492. doi: 10.1038/s41587-019-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan CT, McKearn JP, Lemischka IR. Cellular and developmental properties of fetal hematopoietic stem cells. Cell. 1990;61:953–963. doi: 10.1016/0092-8674(90)90061-i. [DOI] [PubMed] [Google Scholar]
- 27.Rebel VI, et al. A comparison of long-term repopulating hematopoietic stem cells in fetal liver and adult bone marrow from the mouse. Exp. Hematol. 1996;24:638–648. [PubMed] [Google Scholar]
- 28.Harrison DE, Zhong RK, Jordan CT, Lemischka IR, Astle CM. Relative to adult marrow, fetal liver repopulates nearly five times more effectively long-term than short-term. Exp. Hematol. 1997;25:293–297. [PubMed] [Google Scholar]
- 29.Zeigler FC, Bennett B, Jordan CT, Bauer K, Matthews W. Cellular and molecular characterization of the role of the Flk-2 Flt-3 receptor tyrosine kinase in hematopoietic stem-cells. Blood. 1994;84:A415–A415. [PubMed] [Google Scholar]
- 30.Kim I, He SH, Yilmaz OH, Kiel MJ, Morrison SJ. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–744. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toyama H, Arai F, Hosokawa K, Ikushima YM, Suda T. N-cadherin+ HSCs in fetal liver exhibit higher long-term bone marrow reconstitution activity than N-cadherin- HSCs. Biochem. Biophys. Res. Commun. 2012;428:354–359. doi: 10.1016/j.bbrc.2012.10.058. [DOI] [PubMed] [Google Scholar]
- 32.Iwasaki H, Arai F, Kubota Y, Dahl M, Suda T. Endothelial protein C receptor-expressing hematopoietic stem cells reside in the perisinusoidal niche in fetal liver. Blood. 2010;116:544–553. doi: 10.1182/blood-2009-08-240903. [DOI] [PubMed] [Google Scholar]
- 33.Vento-Tormo R, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 35.Li D, et al. VCAM-1(+) macrophages guide the homing of HSPCs to a vascular niche. Nature. 2018;564:119–124. doi: 10.1038/s41586-018-0709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariani SA, et al. Pro-inflammatory aorta-associated macrophages are involved in embryonic development of hematopoietic stem cells. Immunity. 2019;50:1439–1452.e5. doi: 10.1016/j.immuni.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaidya A, Kale VP. TGF-beta signaling and its role in the regulation of hematopoietic stem cells. Syst. Synth. Biol. 2015;9:1–10. doi: 10.1007/s11693-015-9161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikushima YM, et al. Enhanced Angpt1/Tie2 signaling affects the differentiation and long-term repopulation ability of hematopoietic stem cells. Biochem. Biophys. Res. Commun. 2013;430:20–25. doi: 10.1016/j.bbrc.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Zhang CC, Lodish HF. Insulin-like growth factor 2 expressed in a novel fetal liver cell population is a growth factor for hematopoietic stem cells. Blood. 2004;103:2513–2521. doi: 10.1182/blood-2003-08-2955. [DOI] [PubMed] [Google Scholar]
- 40.Wong JC, et al. A glucocorticoid- and diet-responsive pathway toggles adipocyte precursor cell activity in vivo. Sci. Signal. 2016;9:ra103. doi: 10.1126/scisignal.aag0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caselli A, et al. IGF-1-mediated osteoblastic niche expansion enhances long-term hematopoietic stem cell engraftment after murine bone marrow transplantation. Stem Cells. 2013;31:2193–1204. doi: 10.1002/stem.1463. [DOI] [PubMed] [Google Scholar]
- 42.Gealekman O, et al. Enhanced angiogenesis in obesity and in response to PPARgamma activators through adipocyte VEGF and ANGPTL4 production. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1056–E1064. doi: 10.1152/ajpendo.90345.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imai A, et al. Adrenomedullin suppresses vascular endothelial growth factor-induced vascular hyperpermeability and inflammation in retinopathy. Am. J. Pathol. 2017;187:999–1015. doi: 10.1016/j.ajpath.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 44.Broxmeyer HE, Capitano M, Campbell TB, Hangoc G, Cooper S. Modulation of hematopoietic chemokine effects in vitro and in vivo by DPP-4/CD26. Stem Cells Dev. 2016;25:575–585. doi: 10.1089/scd.2016.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macedo DB, Kaiser UB. DLK1, notch signaling and the timing of puberty. Semin. Reprod. Med. 2019;37:174–181. doi: 10.1055/s-0039-3400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fawkner-Corbett D, et al. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell. 2021;184:810–26 e823. doi: 10.1016/j.cell.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asp M, et al. A spatiotemporal organ-wide gene expression and cell Atlas of the developing human heart, cell. Cell. 2019;179:1647–60 e1619. doi: 10.1016/j.cell.2019.11.025. [DOI] [PubMed] [Google Scholar]
- 48.Chen, A, et al. Large field of view-spatially resolved transcriptomics at nanoscale resolution. bioRxiv10.1101/2021.01.17.427004 (2021).
- 49.Ren X, et al. Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly. Cell Res. 2020;30:763–778. doi: 10.1038/s41422-020-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamplin OJ, et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell. 2015;160:241–252. doi: 10.1016/j.cell.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varol C, Mildner A, Jung S. Macrophages: development and tissue specialization. Annu. Rev. Immunol. 2015;33:643–675. doi: 10.1146/annurev-immunol-032414-112220. [DOI] [PubMed] [Google Scholar]
- 52.Yahara Y, et al. Erythromyeloid progenitors give rise to a population of osteoclasts that contribute to bone homeostasis and repair. Nat. Cell Biol. 2020;22:49–59. doi: 10.1038/s41556-019-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoeffel G, et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mass E, et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353:aaf4238. doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue Y, et al. The vascular niche regulates hematopoietic stem and progenitor cell lodgment and expansion via klf6a-ccl25b. Dev. Cell. 2017;42:349–362. doi: 10.1016/j.devcel.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 56.Gomez Perdiguero E, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ozgur E, et al. Distribution of EphB4 and EphrinB2 in normal and malignant urogenital tissue. Urol. Oncol. 2011;29:78–84. doi: 10.1016/j.urolonc.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 58.Allard JB, Duan C. IGF-binding proteins: why do they exist and why are there so many? Front. Endocrinol. (Lausanne) 2018;9:117. doi: 10.3389/fendo.2018.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bach LA. IGF-binding proteins. J. Mol. Endocrinol. 2018;61:T11–T28. doi: 10.1530/JME-17-0254. [DOI] [PubMed] [Google Scholar]
- 60.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 61.Milyavsky M, et al. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 2010;7:186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 62.Mohrin M, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou F, et al. Tracing haematopoietic stem cell formation at single-cell resolution. Nature. 2016;533:487–492. doi: 10.1038/nature17997. [DOI] [PubMed] [Google Scholar]
- 64.Chou S, Lodish HF. Fetal liver hepatic progenitors are supportive stromal cells for hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 2010;107:7799–7804. doi: 10.1073/pnas.1003586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang CC, et al. Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nat. Med. 2006;12:240–245. doi: 10.1038/nm1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu C, et al. Macrophages mediate the repair of brain vascular rupture through direct physical adhesion and mechanical traction. Immunity. 2016;44:1162–1176. doi: 10.1016/j.immuni.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Lim HY, et al. Hyaluronan receptor LYVE-1-expressing macrophages maintain arterial tone through hyaluronan-mediated regulation of smooth muscle cell collagen. Immunity. 2018;49:326–341 e327. doi: 10.1016/j.immuni.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 68.Liang Q, Su L, Zhang D, Jiao J. CD93 negatively regulates astrogenesis in response to MMRN2 through the transcriptional repressor ZFP503 in the developing brain. Proc. Natl. Acad. Sci. USA. 2020;117:9413–9422. doi: 10.1073/pnas.1922713117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, et al. Comparative analysis of cell lineage differentiation during hepatogenesis in humans and mice at the single-cell transcriptome level. Cell Res. 2020;30:1109–1126. doi: 10.1038/s41422-020-0378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ding N, et al. Global transcriptome analysis for identification of interactions between coding and noncoding RNAs during human erythroid differentiation. Front. Med. 2016;10:297–310. doi: 10.1007/s11684-016-0452-0. [DOI] [PubMed] [Google Scholar]
- 71.Ren, Y, et al. The dynamic interactive network of long non-coding RNAs and chromatin accessibility facilitates erythroid differentiation. bioRxiv10.1101/2021.03.02.433076 (2021).
- 72.Lv J, Wang L, Gao Y, Ding YQ, Liu F. 5-hydroxytryptamine synthesized in the aorta-gonad-mesonephros regulates hematopoietic stem and progenitor cell survival. J. Exp. Med. 2017;214:529–545. doi: 10.1084/jem.20150906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oberlin E, et al. VE-cadherin expression allows identification of a new class of hematopoietic stem cells within human embryonic liver. Blood. 2010;116:4444–4455. doi: 10.1182/blood-2010-03-272625. [DOI] [PubMed] [Google Scholar]
- 74.Chen Y, Haviernik P, Bunting KD, Yang YC. Cited2 is required for normal hematopoiesis in the murine fetal liver. Blood. 2007;110:2889–2898. doi: 10.1182/blood-2007-01-066316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lancini C, Gargiulo G, van den Berk PC, Citterio E. Quantitative analysis by next generation sequencing of hematopoietic stem and progenitor cells (LSK) and of splenic B cells transcriptomes from wild-type and Usp3-knockout mice. Data Brief. 2016;6:556–561. doi: 10.1016/j.dib.2015.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao Y, et al. ATF4 plays a pivotal role in the development of functional hematopoietic stem cells in mouse fetal liver. Blood. 2015;126:2383–2391. doi: 10.1182/blood-2015-03-633354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq and 10× Visium ST datasets generated in the present study are available at https://bigd.big.ac.cn/gsub/ in BIG Submission Portal, under the accession numbers CRA002489 and CRA003651, respectively. The E13.5 Stereo-seq dataset used in the present study is kindly provided by Prof. Xun Xu and Prof. Longqi Liu.