Abstract
Aging is impacted by interventions across species, often converging on metabolic pathways. Transcription factors regulate longevity yet approaches for their pharmacological modulation to exert geroprotection remain sparse. We show that increased expression of the transcription factor Grainyhead 1 (GRH-1) promotes lifespan and pathogen resistance in Caenorhabditis elegans. A compound screen identifies FDA-approved drugs able to activate human GRHL1 and promote nematodal GRH-1-dependent longevity. GRHL1 activity is regulated by post-translational lysine methylation and the phosphoinositide (PI) 3-kinase C2A. Consistently, nematodal longevity following impairment of the PI 3-kinase or insulin/IGF-1 receptor requires grh-1. In BXD mice, Grhl1 expression is positively correlated with lifespan and insulin sensitivity. In humans, GRHL1 expression positively correlates with insulin receptor signaling and also with lifespan. Fasting blood glucose levels, including in individuals with type 2 diabetes, are negatively correlated with GRHL1 expression. Thereby, GRH-1/GRHL1 is identified as a pharmacologically malleable transcription factor impacting insulin signaling and lifespan.
Subject terms: Ageing, Type 2 diabetes
Life- and healthspan of organisms can be modulated by dietary, genetic, or pharmacological interventions, which often affect metabolic pathways. Here the authors report that Grainyhead 1 is an evolutionarily conserved, drug-inducible transcription factor that promotes longevity in C. elegans, and thus a potential target for the development of geroprotective drugs.
Introduction
Research of the last decades provides compelling evidence to support the notion that aging, including that of humans, can to some extent be modulated. Life- and healthspan of different organisms, such as Caenorhabditis elegans, Drosophila melanogaster, and mice, are extendable by dietary1, genetic2, and pharmacological interventions3, often converging on metabolic pathways4. To develop pharmacological treatments to counteract aging-associated diseases5,6 and the general functional decline during in particular human aging, one promising recent approach is the identification of evolutionarily conserved and pharmacologically modulable targets controlling the aging process across species7. This strategy allows the use of experimentally accessible cell culture models and comparatively simple animal model organisms for the rational selection of novel lead compounds and repurposed drugs to potentially serve as geroprotectors, before transitioning toward more complex, costly, and time-consuming preclinical studies and first-in-human trials.
Aberrations in transcriptional networks have been identified as early drivers of aging and one of its potential root causes8. Reflecting this, transcription factors (TFs), many of which have been shown repeatedly and with remarkable evolutionary consistency to control development9, metabolism10, and stress responses11, are also increasingly linked to regulation of longevity12. Some TFs are demonstrated to impact lifespan of model organisms in a conserved manner and, at least by correlation, already implied to contribute to human longevity, as perhaps currently best represented by Forkhead box subfamily O (FOXO) TFs13, as well as the nuclear factor erythroid 2-related factor 2 (NFE2L2 aka NRF2)14.
In a previous study from our laboratory, impairment of Grainyhead 1 (GRH-1) in C. elegans, the sole nematodal orthologue of mammalian Grainyhead-like (GRHL) TFs15,16, was identified to shorten lifespan17. We now show that limited overexpression of grh-1 promotes nematodal lifespan without impairing motility or fertility, while also improving pathogen resistance of C. elegans. Small-molecule compounds, including the FDA-approved phytochemical papaverine, are identified as human GRHL1 activators, capable of extending nematodal lifespan through GRH-1. Human GRHL1 activity is here shown as controlled by lysine dimethylation and regulated by a methylase (KMT2D; SET-16 in C. elegans), demethylase (KDM1A; LSD-1 and SPR-5 in C. elegans), and PI3 kinase (PIK3C2A; PIKI-1 in C. elegans) in response to papaverine treatment. Consistent with PI3Ks being involved in insulin/IGF-1 signaling (IIS)18, we observe that nematodal grh-1 is necessary for longevity upon IIS impairment.
Supporting an evolutionarily conserved link of Grainyhead transcription factors to aging and IIS, we find Grhl1 expression in the BXD mouse genetic reference population19 to positively correlate with increased lifespan and improved insulin sensitivity. Furthermore, we find that GRHL1 expression in humans correlates with insulin receptor signaling and, similar to what is observed in BXD mice, that its increased expression predicts prolonged adult lifespan. In addition, human GRHL1 expression negatively correlates with fasting blood glucose levels, notably also including in individuals with type 2 diabetes.
Together, these results identify GRH-1/GRHL1 as a drug-inducible aging-related TF linked to regulation of insulin signaling and longevity and that substances activating GRHL1, such as papaverine, could potentially serve as lead compounds for the development of geroprotective drugs.
Results
Grainyhead 1 transcription factor regulates aging of nematodes
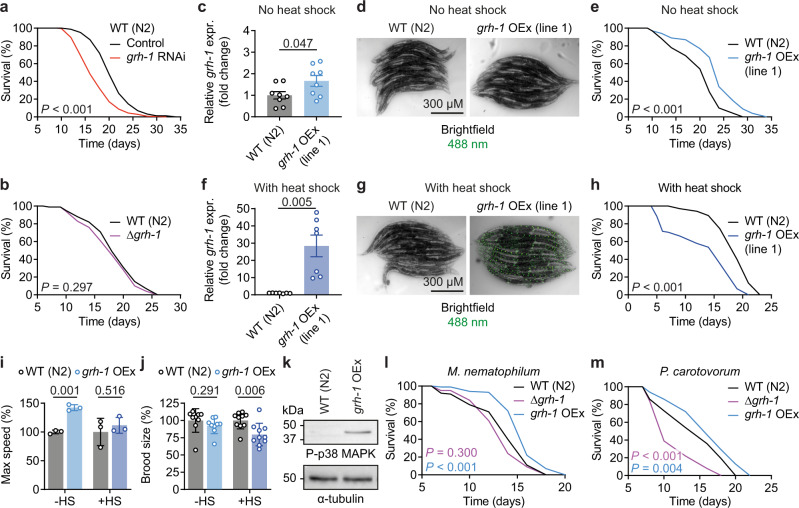
GRHL TFs are conserved from fungi to mammals, with mice and humans possessing three functionally distinct orthologues (Grhl/GRHL1-3) and nematodal GRH-1 most closely related to Grhl1/GRHL115,16. Post-developmental impairment of C. elegans grh-1 in nematodes by RNA interference (RNAi) significantly shortened lifespan (mean −17.0%, P < 0.001; Fig. 1a), as previously described17. On average, in three experiments, mean lifespan with grh-1 RNAi was reduced by 17.3 ± 10.9% compared to feeding with the L4440 control vector. Notably, lifespan of the grh-1 deletion strain VC2072 (Δgrh-1), carrying the grh-1 frameshift deletion allele gk96020, was not significantly different compared to wild-type N2 nematodes (WT) (Fig. 1b). See Supplementary Data 1 for detailed statistics of all lifespan assays and repeats thereof throughout this study.
Fig. 1. GRH-1 is a longevity-promoting transcription factor.
a Lifespan assay of wild-type (WT) N2 nematodes on post-developmental grh-1 RNAi compared to control vector L4440. b Lifespan assay on OP50 bacteria using a grh-1 deletion strain (null mutant VC2072) vs. WT (N2). c–e WT (N2) vs. grh-1 OEx line 1 (without heat shock) RT-qPCR of grh-1 with n = 8 for both (c), microscopy images to detect GRH-1::GFP (d), and lifespan on OP50 bacteria (e). f–h Same experiments as in (c–e) but in each case using heat shocked grh-1 OEx line 1 vs. respective WT (N2) control (RT-qPCR with n = 7 for both). i, j Motility (i) and fertility (j) assays of WT (N2) vs. grh-1 OEx sans (−HS) or with (+HS) heat shock (motility assay with n = 3 for all; fertility assay with n = 9 for −HS and n = 10 for +HS). k Western blot of phospho-p38 MAPK in WT (N2) and grh-1 OEx with α-tubulin as loading control. l, m Lifespan assay of WT (N2) vs. Δgrh-1 and grh-1 OEx on pathogenic M. nematophilum (l) or P. carotovorum bacteria (m). Data in bar graphs are mean ± SEM, with sample sizes as stated and individual data points representing biological replicates. P-values were determined with two-tailed unequal variances t-tests of the indicated comparisons of unpaired control vs. treatment groups. P-values of C. elegans lifespan assays were determined by log-rank test. See Supplementary Data 1 for detailed lifespan assay statistics, Supplementary Fig. 1a, d for microscopy images corresponding to (d and g), and Supplementary Fig. 2f for full western blot images corresponding to (k). Source data are provided as a Source data file.
Given these divergent results following grh-1 impairment vs. deletion and to further test if GRH-1 is indeed involved in the regulation of nematodal lifespan, two independent strains (named line 1 and 2) overexpressing grh-1::gfp under control of the heat shock-inducible hsp-16.2-promoter (grh-1 OEx) were generated. Without heat shock induction, grh-1 overexpression of grh-1 OEx line 1 was ~2-fold (P = 0.047; Fig. 1c), with no detectable nuclear GFP signal (Fig. 1d and Supplementary Fig. 1a), and lifespan of this strain significantly extended (mean + 16.3%, P < 0.001; Fig. 1e). This longevity phenotype was confirmed with the independently generated hsp-16.2p::grh-1::gfp OEx line 2, which also displayed limited grh-1 overexpression (~3-fold) and a significantly extended lifespan in absence of heat shock induction (Supplementary Fig. 1b, c). Transient heat shock of grh-1 OEx line 1 resulted in a stronger ~30-fold grh-1 overexpression (P = 0.005; Fig. 1f), visible nuclear accumulation of GRH-1::GFP (Fig. 1g and Supplementary Fig. 1d), and reduction of lifespan compared to WT nematodes subjected to the same heat shock treatment in parallel (mean −29.6%, P < 0.001; Fig. 1h). Similar results were again obtained with the independently generated grh-1 OEx line 2, which also displayed strongly increased grh-1 overexpression together with shortened lifespan after heat shock (Supplementary Fig. 1e, f). Considering the overall identical results using these two independent grh-1 OEx lines in initial lifespan assays, all following experiments were conducted using line 1, hereafter simply referred to as grh-1 OEx. Treatment of grh-1 OEx with grh-1 RNAi abolished longevity, thus indicating that increased grh-1 is indeed responsible for extending the lifespan of this strain (Supplementary Fig. 1g). In addition, overexpression of grh-1 under control of its endogenous promoter resulted in an intermediate ~10-fold increase of grh-1 mRNA and also shortened lifespan (Supplementary Fig. 1h, i), with the effect on lifespan tendentially less pronounced than in the heat shocked grh-1 OEx lines. Together, this indicates a dose-dependent effect of grh-1 expression on lifespan, with both its reduction by RNAi and stronger overexpression shortening lifespan, while limited overexpression promotes longevity.
To assess effects of grh-1 overexpression on overall fitness, the motility and fertility of non-heat shocked grh-1 OEx nematodes were assayed. Limited grh-1 overexpression significantly increased maximum speed (Fig. 1i) and did not affect brood size (Fig. 1j). In contrast, inducing grh-1 overexpression by heat shock treatment abolished positive effects on motility and led to reduced fertility (Fig. 1i, j), underscoring a dose-dependent effect of grh-1 not only on lifespan but also on health parameters. Given the protective role of limited grh-1 overexpression, all subsequent experiments were performed with the grh-1 OEx strain in absence of a heat shock.
GRH-1 promotes MAPK signaling and pathogen resistance
RNA-Seq of WT vs. grh-1 OEx (Supplementary Data 2) revealed MAP kinase activity (GO:0004707) as the most significantly enriched pathway among transcripts increased by grh-1 overexpression (Supplementary Fig. 2a) and pmk-1, encoding a human p38 MAP kinase orthologue, as the most significantly upregulated gene thereof (log2 ratio = 1.056, P = 4.3 × 10−10). Notably, both pmk-2 and pmk-3, which also belong to the MAP kinase activity pathway, were likewise significantly overexpressed in grh-1 OEx (Supplementary Data 2). Furthermore, as determined by two independent bioinformatic prediction tools used to scan all genes whose expression was increased in grh-1 OEx, pmk-1 was determined to contain GRH-1 binding motifs within its predicted promoter (Supplementary Fig. 2b and Supplementary Data 3), altogether indicating it as a possible direct target gene of GRH-1. Gene set enrichment analysis (GSEA) of human genes positively correlating with human GRHL1 expression in non-gender specific tissues identified the MAP kinase activity pathway (GO:0004707) as also significantly enriched among them (see Supplementary Fig. 2c for pathways correlating with nematodal grh-1 and with human GRHL1 by GSEA). In particular, three out of four human pmk-1 orthologues (MAPK11, MAPK13, and MAPK14) were found positively correlated with GRHL1 expression in muscle (Supplementary Fig. 2d), and similar correlations were observed with GRHL1 in liver (Supplementary Fig. 2e). This indicates an evolutionarily conserved link between Grainyhead TFs and MAPK signaling.
Consistent with the MAPK pathway being regulated by GRH-1, increased levels of phosphorylated p38 MAPK were detected in grh-1 OEx compared to WT (Fig. 1k and Supplementary Fig. 2f). In addition, lifespan-extension of grh-1 OEx was abolished by pmk-1 RNAi (Supplementary Fig. 2g), indicating this gene as required for mediating longevity of grh-1 OEx. Consistent with pmk-1 regulating the C. elegans immune response21,22, grh-1 OEx conferred resistance against two pathogenic bacterial strains, i.e., Microbacterium nematophilum (Fig. 1l) and Pectobacterium carotovorum (Fig. 1m). Since a combined lifespan assay of grh-1 RNAi bacteria together with these pathogenic bacteria was not feasible, the effect of M. nematophilum and P. carotovorum in absence of grh-1 was checked instead. When fed with P. carotovorum, lifespan of Δgrh-1 was reduced, both in comparison to WT and grh-1 OEx (mean −18.1% vs. WT and −27.5% vs. grh-1 OEx, P < 0.001; Fig. 1m).
A small molecule screen identifies activators of human GRHL1
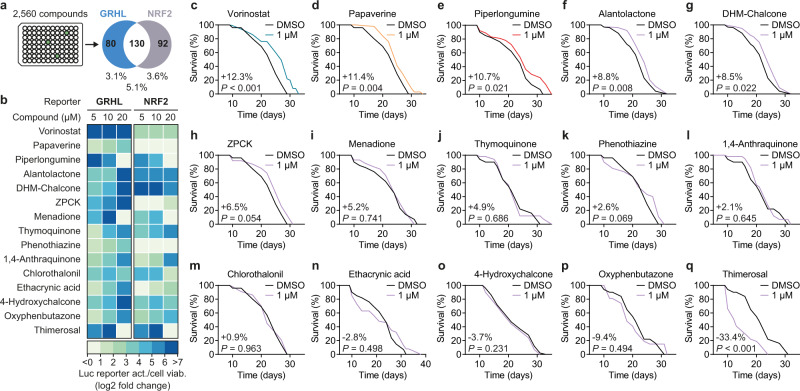
Having established Grainyhead 1 as an aging-related TF in nematodes, a screening to identify compounds modulating its activity was performed. For this purpose, a human HEK293 GRHL reporter was generated (GRHLWT-LUC), in which several repeats of the mammalian GRHL consensus binding sequence16 control expression of a luciferase reporter gene (Supplementary Fig. 3a, b). GRHLWT-LUC was validated to be activated by treatment with the histone deacetylase inhibitor (HDACi) Scriptaid (63.7 ± 2.9 fold, P = 8.7 × 10−13; Supplementary Fig. 3c), selected as a positive control compound based on GRHL1 expression being strongly induced by HDACis23. Similarly, an independent human HEK293 reporter for the antioxidant response element (ARE) (Supplementary Fig. 3a, b) targeted by the redox-sensitive TF NRF224, was generated and confirmed as responsive to the canonical NRF2-activator sulforaphane25 (7.5 ± 0.4-fold, P = 2.3 × 10−9; Supplementary Fig. 3c). These two independent reporter cell lines were used in parallel for primary screening of a library of 2560 compounds, comprising phytochemicals and approved drugs, at 10 µM final concentration. Thereby, 5.1% of the library (130 compounds) were identified as activators (≥2-fold activation) of both the GRHL and NRF2 reporter, whereas 3.1% (80 compounds) only activated the GRHL and 3.6% (92 compounds) only the NRF2 reporter (Fig. 2a, Supplementary Data 4).
Fig. 2. Human GRHL1-activating compounds and their impact on nematodal lifespan.
a Schematic Venn diagram of hits activating a human GRHL and/or NRF2 luciferase reporter with fold change ≥2 at 10 µM in the primary screening. Indicated are total number of hits and hits as percentage of full library (2560 compounds). See Supplementary Data 4. b Heat map depicting log2 fold change reporter activation in a three-point dose–response assay (5, 10, and 20 µM) of the top 15 GRHL hit compounds on the reporters as in (a), normalized to cell viability (n = 2–3 per condition). c–q Lifespan assays, all using WT (N2) nematodes treated with the respective individual compound at a concentration of 1 µM in plate and vs. DMSO control. Lifespan graphs contain percentage change of mean lifespan compound vs. DMSO control and P-values as determined by log-rank test. See Supplementary Data 1 for detailed lifespan assay statistics. Source data are provided as a Source data file.
The 15 compounds with the highest activation of GRHLWT-LUC that were readily available for resourcing were defined as primary hits and evaluated in more detail (see list of compounds in Fig. 2b). First, a three-point dose–response reporter assay (5, 10, and 20 µM final concentration), in independent repeat experiments and normalized to cell viability, was performed. All 15 primary hits were confirmed as activators of GRHLWT-LUC and 13 also activated the NRF2 reporter (heatmap depicting log2 fold change of either reporter vs. DMSO control in Fig. 2b), with papaverine and phenothiazine identified as the only exclusive activators of the GRHL reporter. The strongest activation of the GRHL reporter, considering all three tested concentrations for each compound, was observed with the compounds 2′,4′-Dihydroxy-4-methoxychalcone (DHM-chalcone, 286.2 ± 36.6 SEM at 20 µM, P = 0.008), alantolactone (131.5 ± 6.8 SEM at 20 µM, P = 0.017), and vorinostat (114.9 ± 6.0 SEM at 5 µM, P = 0.001). Notably, the HDACi vorinostat, while having the lowest efficacy of the top three GRHL reporter activators and in particular compared to DHM-chalcone, was most consistent in activating GRHLWT-LUC across the tested range of concentrations, i.e., was observed to have the highest potency.
GRHL activators extend nematodal lifespan dependent on GRH-1
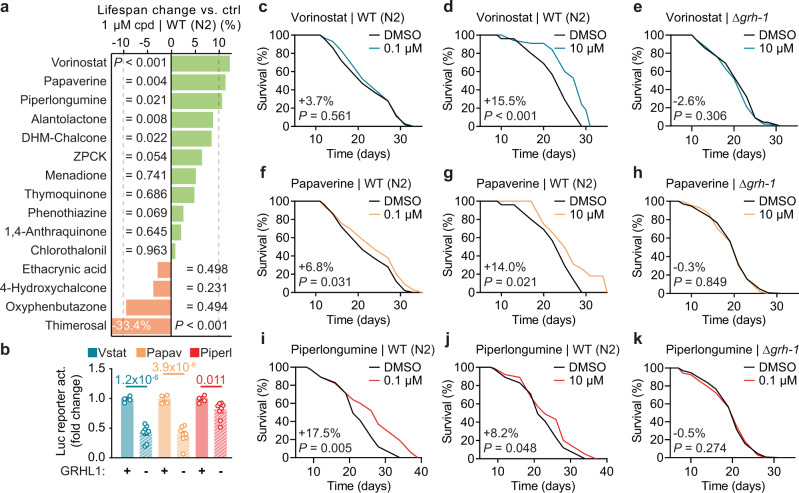
To explore whether GRHL reporter activation is indicative of compounds having longevity-promoting effects, all 15 confirmed primary hits were tested for their effect on lifespan of WT nematodes at a concentration of 1 µM (Fig. 2c–q). Overall, 9/15 compounds did not significantly alter lifespan at the tested concentration, and one compound (thimerosal, mean −33.4%, P < 0.001) shortened lifespan (Fig. 3a). The remaining compounds, including DHM-chalcone and alantolactone, significantly extended lifespan by +8.5–12.3% (Fig. 2c–g). The strongest effects (lifespan-extension >10%) were observed for the HDACi vorinostat (mean + 12.3%, P < 0.001) and two phytochemicals, papaverine (mean + 11.4%, P = 0.004) and piperlongumine (mean + 10.7%, P = 0.021). Both vorinostat and papaverine are compounds that are FDA-approved for pharmacological use. As the GRHL promoter element used here can be activated not only by GRHL1, but also by GRHL2 and GRHL316, specificity of vorinostat, papaverine, and piperlongumine in activating the reporter via GRHL1 was tested. To do so, using CRISPR/Cas9-mediated deletion, two independent GRHL reporter lines deficient for GRHL1 but nor 2 or 3 were generated (GRHLNULL-LUC; Supplementary Fig. 3d, e). Relative average activation of GRHLNULL-LUC by vorinostat, papaverine, and piperlongumine, compared to activation of GRHLWT-LUC without GRHL1 deletion (Fig. 3b), was significantly lower for all three compounds (vorinostat: 0.40 ± 0.05, P = 3.9 × 10−6; papaverine: 0.43 ± 0.05, P = 1.2 × 10−6; piperlongumine: 0.81 ± 0.05, P = 0.011), indicating that all partially act via GRHL1 to activate the human GRHL reporter.
Fig. 3. GRH-1 dependency of nematodal lifespan-extension elicited by selected GRHL1-activating compounds.
a Overview of impact of the respective compound (cpd) on lifespan of wild-type (WT) N2 nematodes at 1 µM in plate, as percentage change of mean lifespan vs. control treatment with DMSO (ctrl). Also see Fig. 2c–q. b Activation of GRHL reporter by the top 3 lifespan-extending compounds from (a) (Vstat = vorinostat, Papav = papaverine, Piperl = piperlongumine) at 5–10 µM in the presence (wild-type reporter with n = 4) or absence (two independent deletion reporters with n = 8 combined) of GRHL1. c, d Lifespan assays of vorinostat at 0.1 (c) or 10 µM (d) in plate, using WT (N2) nematodes and vs. DMSO control. e Lifespan assay of vorinostat at 10 µM in plate, using Δgrh-1 and vs. DMSO control. f, g Similar to (c), (d), using papaverine. h Similar to (e), using papaverine at 10 µM in plate. i, j Similar to (c), (d), using piperlongumine. k Similar to (e), using piperlongumine at 0.1 µM in plate. Data in bar graphs are mean ± SEM, with sample sizes as stated and individual data points representing biological replicates. P-values were determined with two-tailed unequal variances t-tests of the indicated comparisons of unpaired control vs. treatment groups. Lifespan graphs contain percentage change of mean lifespan compound vs. DMSO control and P-values as determined by log-rank test. See Supplementary Data 1 for detailed lifespan assay statistics. Source data are provided as a Source data file.
Vorinostat, papaverine, and piperlongumine were each tested at two additional concentrations (0.1 and 10 µM) for their effect on C. elegans lifespan (Fig. 3c, d, f, g, i, j). Vorinostat and papaverine had the most pronounced lifespan-extending effect at 10 µM (vorinostat: mean +15.5%, P < 0.001; papaverine: mean +14.0%, P = 0.021, Fig. 3d, g), whereas piperlongumine was identified to be most effective at 0.1 µM (mean + 17.5%, P = 0.005; Fig. 3i). When applying these concentrations of the respective compound to Δgrh-1, no changes in lifespan were observed anymore (Fig. 3e, h, k). This indicates that the tested compounds function through GRH-1 to exert their effects, and that this transcription factor is a mediator of pharmacologically induced longevity in nematodes. For subsequent experiments, we focused on the phytochemical papaverine, identified as the only lifespan-extending compound specific for activation of the GRHL reporter.
Analysis of the human GRHL1 protein interactome
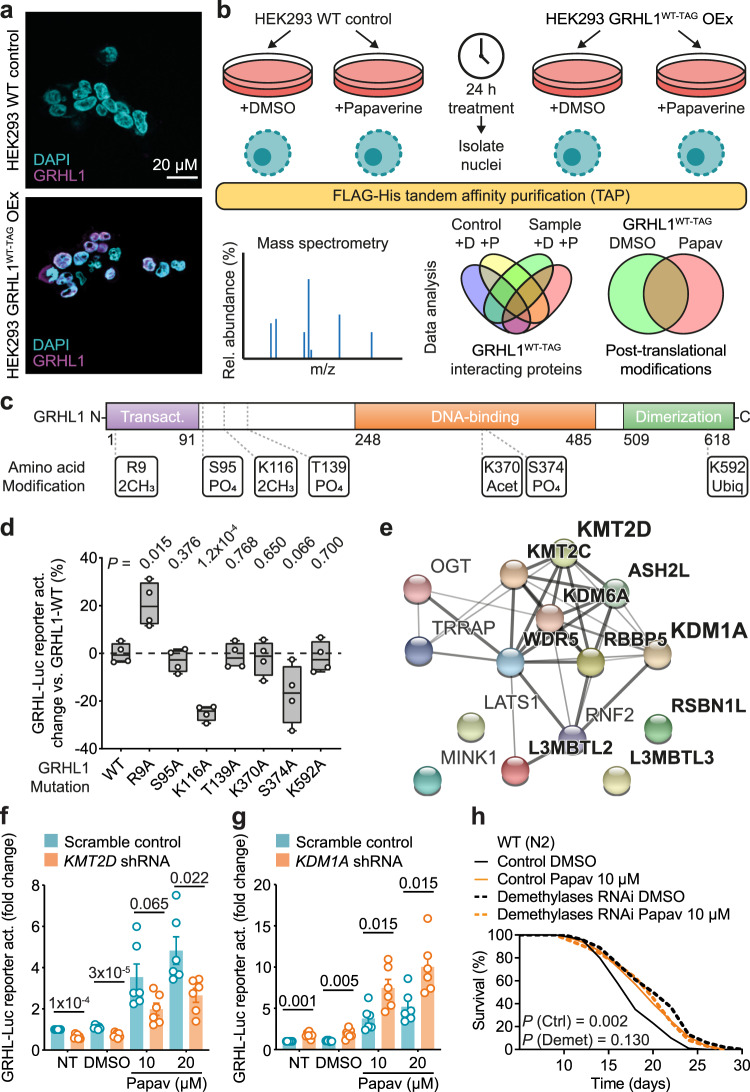
To investigate how papaverine might impact GRHL1 activity, a HEK293-derived cell line overexpressing C-terminally 3xFLAG-6xHis tagged wild-type human GRHL1 (GRHL1WT-TAG) was generated (Supplementary Fig. 3f, g). GRHL1WT-TAG was verified to be located in the nucleus (Fig. 4a and Supplementary Fig. 3h) and as suitable for tandem-affinity purification (Supplementary Fig. 3i). The GRHL1WT-TAG-overexpressing cell line, together with HEK293 WT cells as background control, was subjected to DMSO vs. papaverine treatment for 24 h, nuclei isolated, GRHL1WT-TAG purified, and GRHL1’s post-translational modifications (PTMs) and its protein interactome determined by liquid chromatography-mass spectrometry analysis (LC-MS; Fig. 4b, Supplementary Table 1 and Supplementary Data 5).
Fig. 4. GRH-1/GRHL1 are regulated by lysine methylation.
a Microscopy images of a HEK293 wild-type (WT) and the GRHL1WT-TAG-overexpressing cell line as DAPI staining and GRHL1 immunofluorescence overlay. See Supplementary Fig. 3h. b Experimental design of GRHL1WT-TAG tandem-affinity purification (TAP) from isolated nuclei and subsequent mass spectrometry. c Post-translational modifications (PTMs) of human GRHL1 identified only after treatment with papaverine. Position, amino acid, and type of PTM as indicated (2CH3 = dimethylation, PO4 = phosphorylation, Acet = acetylation, Ubiq = ubiquitylation). See Supplementary Table 1 and Supplementary Data 6. d Impact of mutating individual GRHL1 amino acids on GRHL reporter activation. Depicted is percentage change vs. activation with the GRHL1 WT control with n = 4 for all. Box plot center lines indicate the median, bounds of boxes indicate the 25th to 75th percentiles, and whiskers indicate minima and maxima. e Protein–protein interaction network of GRHL1 interactors classified as impacting PTMs. Edges represent known protein–protein interactions, with darker color indicating stronger data support. Proteins in bold are linked to K-methylation. Visualized with the help of STRING (https://string-db.org). See Supplementary Fig. 3j and Supplementary Data 5. f, g Activation of the GRHL reporter without (scramble control) or with concomitant knockdown of KMT2D (f) or KDM1A (g) by shRNA and indicated treatments (NT = no treatment, DMSO = vehicle control, Papav = papaverine at 10 or 20 µM). KMT2D and KDM1A scramble vs. shRNA with n = 7 for NT and DMSO and n = 6 for Papav. h Lifespan assay of WT (N2) nematodes on post-developmental lsd-1 + spr-5 demethylases RNAi compared to control vector L4440, and both conditions additionally treated with 10 µM papaverine or DMSO control. Data in bar graphs are mean ± SEM, with sample sizes as stated and individual data points representing biological replicates. P-values were determined with two-tailed unequal variances t-tests of the indicated comparisons of unpaired control vs. treatment groups. P-values of C. elegans lifespan assays were determined by log-rank test. See Supplementary Data 1 for detailed lifespan assay statistics. Source data are provided as a Source data file.
The most abundantly purified protein in the GRHL1WT-TAG samples reassuringly was GRHL1 itself. In addition, 130 additional proteins were identified as potential GRHL1WT-TAG interaction partners, i.e., fully absent or enriched ≥3-fold over the respective HEK293 WT control samples. Of these 130 proteins, 66 (50.8%) co-purified with GRHL1WT-TAG both after papaverine and DMSO control treatment, whereas 29 proteins (22.3%) were identified as potential GRHL1 interaction partners only following papaverine and 35 (26.9%) only following DMSO control treatment (Supplementary Data 5). All proteins were curated, and each assigned a category and classification reflecting their most prominent cellular function (Supplementary Fig. 3j and Supplementary Data 5).
The largest set of potential GRHL1 interaction partners as here identified is involved in transcriptional regulation (57 proteins), either as TFs themselves or as transcriptional coregulators. 19 of the potential GRHL1-interacting proteins are involved in chromatin organization and histone modification, consistent with a recently proposed role of human GRHL1-3 in regulating chromatin accessibility26, notably by mechanisms and interactors not yet elucidated in detail. The only two experimentally identified human GRHL1 interactors described so far, in a large-scale study of the human protein–protein interactome27, are GRHL2 and ZNF76. Our analysis confirmed GRHL2 as a potential interaction partner of GRHL1. Noteworthy, the GRHL1-GRHL2 interaction was observed only after papaverine treatment, suggesting that this heterodimer might be relevant in mediating transcriptional activation of target genes. While our analysis did not identify ZNF76 as an interaction partner, 14 potential interactors are other zinc finger proteins (including ZNF24, ZNF148, ZNF281, ZNF384, ZNF516, and ZNF638), suggesting a physiologically relevant link of GRHL1 to this highly abundant group of proteins that impacts a variety of processes in health and disease28.
GRHL1 activity is regulated by lysine methylation
In parallel to the above, a total of 24 amino acids of GRHL1WT-TAG were identified as post-translationally modified using stringent protein identification filters, with 7 thereby designated as modified only after papaverine treatment (Fig. 4c, Supplementary Table 1, and Supplementary Data 6). This suggests involvement of these PTMs in regulating GRHL1 activity, especially since no modifications were identified exclusively in DMSO control samples by this approach. Interestingly, monitoring the LC-MS traces of the identified peptides across all samples, an overall higher abundance of GRHL1 was observed in papaverine-treated samples. The modifications initially defined as papaverine-specific based on the peptide identification results could also be observed in DMSO control samples to some extent (Supplementary Data 7). These 7 GRHL1 PTMs, including lysine acetylations, arginine and lysine dimethylations, threonine and serine phosphorylations, and lysine ubiquitylation, were selected for follow-up experiments. Reassuringly, our approach also identified the only GRHL1 PTM already described in a large-scale study of the human phosphoproteome, i.e., phosphorylation of threonine 20829, in both the papaverine-treated and DMSO control samples.
To test whether the 7 GRHL1 PTMs only stringently identified in papaverine-treated samples are indeed involved in regulating GRHL1 activity, corresponding amino acid codons in the GRHL1WT-TAG overexpression construct were individually mutated to alanine. Each resulting vector (overexpressing tagged GRHL1 with mutation R9A, S95A, K116A, T139A, K370A, S374A, or K592A, respectively) was transiently transfected into the GRHLWT-LUC reporter and its relative effect on reporter activation, compared to transient transfection with the GRHL1WT-TAG vector, determined. This revealed that mutations R9A and K116A significantly impacted ability of GRHL1 to activate GRHLWT-LUC (Fig. 4d), with both R9 and K116 identified as dimethylated (me2) after papaverine treatment (Fig. 4c). The GRHL1R9A-TAG mutant displayed increased activation of the reporter (+20.5 ± 4.7%, P = 0.015), whereas activation by GRHL1K116A-TAG was reduced (−25.0 ± 1.6%, P = 1.1 × 10−4). This indicates that blocking R9 dimethylation promotes GRHL1 activity, thereby implying that R9me2 in WT GRHL1 is a repressing methylation that counteracts GRHL1 activity. Opposingly, when blocking dimethylation of K116, GRHL1 activated the transcriptional reporter less, from which follows that K116me2 in WT GRHL1 is likely an activating methylation.
To investigate this regulation of GRHL1 in more detail, we next considered all those GRHL1-interacting proteins which were categorized to be involved in impacting PTMs (Fig. 4e, Supplementary Fig. 3j, and Supplementary Data 5). Of the 15 proteins in this category, 10 are linked to K-methylation, among them K-methylases (KMT2C and KMT2D) and K-demethylases (KDM1A and KDM6A), and notably none to R-methylation or demethylation. Relevantly, transcriptional activity of p53 is repressed by KDM1A, likely by removal of an activating dimethylation mark of a single lysine residue30, similar to our findings that mutation of K116 in GRHL1 reduces transcriptional activity. Furthermore, KMT2C and KMT2D were recently also described as GRHL2-interacting proteins31.
Impact of K-methylases and -demethylases on GRHL1 activity was tested by stably transfecting the GRHLWT-LUC reporter with shRNAs against KMT2D or KDM1A. Downregulation of KMT2D reduced reporter activity, both in the basal state and after papaverine treatment (Fig. 4f). This result is congruent with the interpretation that dimethylation of GRHL1 K116 is an activating mark, which is impaired when KMT2D is knocked down. Conversely, and further supporting a role for K-methylation, impairment of KDM1A led to increased GRHL reporter activity, again both under basal and induced conditions (Fig. 4g). This suggests that KDM1A could be responsible for removing the activating dimethylation mark of GRHL1, hence GRHL1 activity being increased when KDM1A is downregulated. In C. elegans, simultaneous RNAi impairment of the demethylases lsd-1 and spr-5, which together encode the two highest confidence nematodal orthologues of human KDM1A, increased lifespan per se. Importantly, this longevity was not further extendable by additional treatment with papaverine (Fig. 4h), indicating a drug-gene interaction of papaverine with lysine demethylases also in nematodes.
Grainyhead TFs are linked to regulation of insulin signaling
To further elucidate the mechanism of GRHL1 activation by papaverine, we performed pull-down experiments using de novo synthesized biotinylated papaverine (Supplementary Fig. 4a) added to HEK293 cell extracts (Supplementary Fig. 4b). Thereby, 25 human proteins were identified as potential papaverine interactors, co-purifying exclusively or enriched ≥3-fold with biotinylated papaverine over control. Notably, the majority of these proteins are linked to insulin/IGF-1 and/or the related PI3K-AKT-mTOR signaling axis (Supplementary Fig. 4c and Supplementary Table 2). Among the proteins co-purifying with papaverine, we decided to focus in detail on the phosphoinositide 3-kinase (PI3K) PIK3C2A. PI3Ks have been previously reported to oppose KMT2D32 and to promote KDM1A33, both here identified as GRHL1 interactors influencing activation mediated by papaverine. In addition, papaverine has been independently linked to downregulation of PI3K signaling34. We hence hypothesized that papaverine might affect PIK3C2A, which in turn could lead to activation of KMT2D and inhibition of KDM1A, thus altering activity of GRHL1.
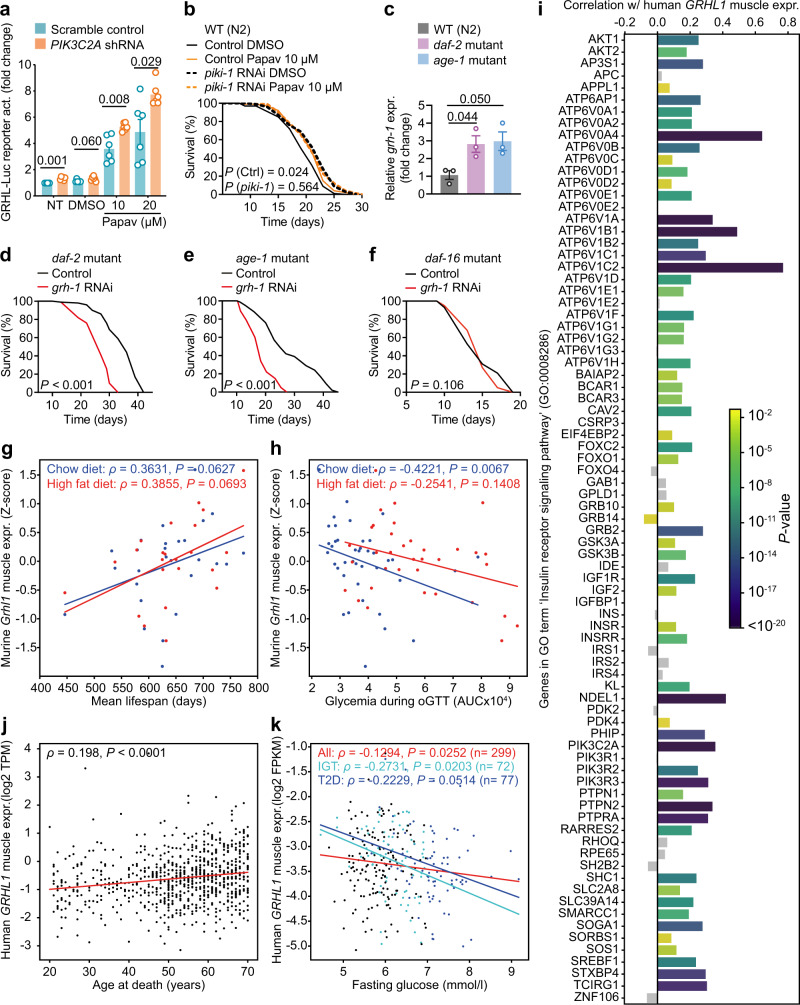
Impairing PIK3C2A by shRNA in the GRHLWT-LUC reporter (Fig. 5a), equivalent to the experiments performed for KDM1A and KMT2D, led to increased GRHL reporter activation, overall resembling the pattern observed for KDM1A impairment (Fig. 4g). The closest orthologue of human PIK3C2A in nematodes is a PI3K named PIKI-1. Treatment of nematodes with RNAi against piki-1 increased lifespan per se (Fig. 5b), similar to impairment of the nematodal KDM1A orthologues lsd-1 and spr-5 (Fig. 4h). Additional application of papaverine failed to further extend lifespan, again indicating epistatic interaction of this compound with PIKI-1/PIK3C2A (Fig. 5b). Together, these results are consistent with GRHL1 activity after papaverine treatment being promoted by K116 dimethylation and modulated by KMT2D, KDM1A, and PIK3C2A.
Fig. 5. Nematodal and mammalian Grainyhead TFs are linked to aging and insulin signaling.
a Activation of the GRHL reporter without (scramble control) or with concomitant knockdown of PIK3C2A by shRNA and indicated treatments (NT = no treatment, DMSO = vehicle control, Papav = papaverine at 10 or 20 µM; n = 6 for all). b Lifespan assay of wild-type (WT) N2 nematodes on post-developmental piki-1 RNAi compared to control vector L4440. Both conditions additionally treated with 10 µM papaverine or DMSO control. c RT-qPCR of grh-1, in WT (N2) vs. a daf-2 or age-1 mutant strain (n = 3 for all). d–f Lifespan assays with a daf-2 (d), age-1 (e), or daf-16 (f) mutant strain on post-developmental grh-1 RNAi compared to control vector L4440. g Spearman’s rho correlation between Grhl1 expression in muscle (Z-score normalized) and mean lifespan of chow and high-fat diet-fed mice from the BXD mouse genetic reference population. h Similar as in (g), depicting the correlation of Grhl1 expression in muscle with area under the curve (AUC) glycemia during oral glucose tolerance test (oGTT). i Pearson’s r correlation coefficients between GRHL1 and genes in GO term ‘Insulin receptor signaling pathway’ (GO: 0008286) in human muscle samples, derived from the GTEx database. Gray bars represent non-significant correlations (P-value > 0.05). j Spearman’s rank rho correlation between GRHL1 expression in human skeletal muscle and age at natural death from the GTEx v8 dataset (n = 621). k Spearman’s rank rho correlation between GRHL1 expression in human skeletal muscle and fasting blood glucose in the entire FUSION cohort (All) or in individuals with impaired glucose tolerance (IGT) or type 2 diabetes (T2D). Color of dots indicates IGT individuals (light blue), T2D individuals (blue), and individuals belonging to neither group (black). Data in bar graphs are mean ± SEM, with sample sizes as stated and individual data points representing biological replicates. P-values were determined with two-tailed unequal variances t-tests of the indicated comparisons of unpaired control vs. treatment groups. P-values of C. elegans lifespan assays were determined by log-rank test. See Supplementary Data 1 for detailed lifespan assay statistics. Source data are provided as a Source data file.
Given that PI3Ks are arguably best known as mediators of insulin/IGF-1 signaling35, we tested whether GRH-1 might be involved in this pathway. C. elegans loss-of-function mutations of daf-2 (encoding the insulin/IGF-1 receptor orthologue) and age-1 (encoding the canonical PI3K orthologue) are well described to extend lifespan in a manner requiring DAF-16/FOXO36–38, a downstream TF of IIS conserved from C. elegans to mammals13. In mRNA samples from both daf-2 and age-1 mutants, grh-1 expression was upregulated compared to WT (Fig. 5c), and application of grh-1 RNAi to either long-lived mutant markedly reduced their lifespan (Fig. 5d, e) to WT levels. Conversely, in the short-lived daf-16 mutant strain, application of grh-1 RNAi did not further reduce lifespan (Fig. 5f). This indicates a possible functional interaction between DAF-16/FOXO and GRH-1 as downstream transcriptional mediators of IIS. Consistently, extended lifespan of grh-1 OEx was fully abolished following impairment of aak-2 (AMPK), daf-16 (FOXO), or skn-1 (NRF2) by RNAi (Supplementary Fig. 4d–g), i.e., components established as required for IIS-mediated longevity38,39. Moreover, grh-1 overexpression was unable to further extend the longevity phenotype elicited by daf-2 RNAi treatment (Supplementary Fig. 4h).
Having uncovered a role for nematodal GRH-1, aside from affecting life- and healthspan, in impacting IIS, we next investigated whether these roles might be evolutionarily conserved in mammals. Strikingly, in the BXD mouse genetic reference population19 Grhl1 expression in skeletal muscle of mice on both a chow and high-fat diet predicts mean lifespan by trend (Fig. 5g). In addition, Grhl1 expression in skeletal muscle of animals on a chow diet negatively correlates with glycemia during oral glucose tolerance test (oGTT) (Fig. 5h), indicating increased insulin sensitivity. When considering murine Grhl1 expression in liver, a positive correlation with mean lifespan was again observed for animals fed with a chow diet, while such an effect was not detected for animals on a high-fat diet (Supplementary Fig. 5a). On neither diet a correlation of Grhl1 liver expression with glycemia during oGTT was found (Supplementary Fig. 5b).
Focusing lastly on human GRHL1, gene set enrichment analysis revealed its expression to consistently positively correlate with genes present in pathways related to insulin signaling across several tissues (Supplementary Fig. 5c). In particular, the majority of genes contained within the GO term ‘insulin receptor signaling pathway’ (GO:0008286), including PIK3C2A, are positively correlated with human GRHL1 expression in skeletal muscle (Fig. 5i), and similar correlations were observed with GRHL1 expression in liver (Supplementary Fig. 5d).
To further investigate whether human GRHL1 expression, following the observations in BXD mice, also correlates with lifespan and glucose tolerance, we analyzed the GTEx human cohort40. Thereby, we discovered that GRHL1 expression in skeletal muscle positively correlates with age at death (Fig. 5j and Supplementary Fig. 5e). Next, we analyzed the FUSION tissue biopsy study dataset41,42, which includes tissue biopsy gene expression data and the glucose tolerance status of participants (Supplementary Fig. 5f). Human skeletal muscle GRHL1 expression negatively correlates with fasting blood glucose not only when considering every participant of the cohort, but even more evidently so in individuals with impaired glucose tolerance (IGT), or type 2 diabetes (T2D) (Fig. 5k). These data suggest that GRHL1 is a critical regulator of glucose homeostasis also in humans. Taken together, these observations underscore a link of Grainyhead 1 both to regulation of lifespan and insulin signaling that is conserved from nematodes to mammals.
Discussion
Aging is arguably the most pervasive and simultaneously least understood biological phenomenon. Nevertheless, research over the last decades showed that aging (i) is influenced by environmental and genetic factors1,2; (ii) is a unifying risk factor for human diseases5; and (iii) can be impacted by various interventions across species3,6,7. The development of translational medicine applications to address human aging-associated diseases, and potentially to slow aging itself, requires identification of cellular targets relevant for controlling aging and externally modulable, e.g., by pharmacological treatment3,6,7. Here, we identified and characterized an evolutionarily conserved longevity-promoting transcription factor as a target for geroprotective interventions, further supporting the role transcriptional regulation is recognized to play in the aging process8,10–12.
Based on mammalian TF taxonomy, the specific protein belongs to the immunoglobulin fold superclass, and therein to the class of Grainyhead domain factors, and is termed Grhl1/GRHL1 in humans and mice, respectively, and GRH-1 in C. elegans. To date, Grainyhead TFs are mainly investigated regarding their role during murine development43,44, and for their complex influence on development and progression of various cancers45. More recently, a functional role for Grainyhead TFs in regulating the chromatin landscape and mediating altered epigenetic states has been described26. We now find that GRH-1 is also acting as an anti-aging TF in a dose-dependent manner. While the underlying mechanisms for this dose-dependency are subject to clarification, we believe this to be an important observation that highlights necessary considerations when aiming to manipulate TFs to counteract aging and in the context of addressing aging-associated diseases. As exemplified by NRF2’s dual role as both a potential tumor suppressor and oncogenic protein in various cancers46, the degree to which a particular aging- or disease-related TF is activated or inhibited, in addition to often observed tissue- and context-dependency of effects, needs to be considered when designing appropriate interventions. We further identified GRH-1 as involved in regulating MAPK signaling, a role likely conserved for human GRHL1, and to improve nematodal survival upon exposure to bacterial pathogens. Given the known role of Grainyhead TFs in regulating epithelial barrier formation across species, including in mammals44, it appears likely that mammalian Grhl1/GRHL1 might have comparable functions in opposing bacterial infections, which presents a relevant focus for future investigation. Our findings thus add to the already manifold functional roles of Grainyhead TFs, thereby highlighting the versatility of key transcriptional regulators in impacting diverse molecular, cellular, and physiological processes.
As proof-of-principle for developing pharmacological interventions targeting Grainyhead TFs with geroprotective drugs, we here established a human reporter cell line to identify small-molecule GRHL1 activators that also promote nematodal longevity via GRH-1. Among the strongest GRH-1-dependent lifespan-extending compounds here discovered was vorinostat, functionally classified as an HDAC-inhibitor. Interestingly, several studies have independently identified HDACis as candidate geroprotective compounds in nematodes, flies, mice, and human in vitro models47. With the underlying mechanisms still incompletely understood and actively investigated, we here provide evidence that individual TFs, such as GRH-1/Grhl1/GRHL1, might be responsible for mediating beneficial effects associated with HDACi-treatment. Piperlongumine, a plant alkaloid also exhibiting HDAC-inhibiting functions48, was likewise found here to extend nematodal lifespan dependent on GRH-1. Of note, piperlongumine has been described to act as a senolytic agent by specifically promoting cell death of senescent cultured fibroblasts49. The evidence we provide support it as a lead compound for developing pharmacological anti-aging treatments and overall indicate that utilization of in vitro models and comparatively simple non-vertebrate animal model organisms is a valid experimental strategy to identify candidate geroprotectors.
Among the human GRHL1-activating compounds extending nematodal lifespan, we specifically focused on papaverine as a representative activator, the only compound in our screen that failed to also activate NRF2. Papaverine is mainly known for its longstanding medical use as a vasodilator and its function as a non-selective phosphodiesterase inhibitor50, and to our knowledge, unlike vorinostat and piperlongumine, is hitherto not linked to regulation of lifespan or aging-related processes. We found treatment with papaverine to impact the human GRHL1 protein–protein interactome and several treatment-dependent potential interactors to be linked to aging and age-related pathologies. Examples are the TF MYC, a proto-oncogenic protein51 and core member of the MYC-MAX-MAD network, heterozygous deletion of which increases murine lifespan52; the RecQ helicase WRN, whose loss-of-function mutation causes the progeroid Werner syndrome in humans53,54; the two AAA + ATPases RUVBL1/PONTIN and RUVBL2/REPTIN, telomerase components essential for holoenzyme assembly55, with improper telomere maintenance being widely implicated in aging and disease56. Notably, this is the first comprehensive description of the human GRHL1 protein–protein interactome, which overall appears to support a role for Grainyhead TFs in the aging process. It should be noted, however, that further analyses of individual potential GRHL1-interacting proteins here identified, e.g., by performing reverse affinity purification experiments of candidates, are required to validate their physical interaction with GRHL1. Probing in detail the observed potential GRHL1 interaction partners in future studies thus might lead to confirmation of as yet unrecognized interconnections between different aging-related pathways.
Focusing for now on the regulation of GRHL1 itself, we have found its activity to be likely impacted by post-translational dimethylation of a specific lysine residue (K116) and to be opposingly modulated by a K-methylase (KMT2D) and K-demethylase (KDM1A aka LSD1), both here identified as potential GRHL1-interacting proteins. We furthermore observed nematodal lifespan-extension by papaverine to be impacted by RNAi of K-demethylase orthologues (lsd-1 and spr-5), thus indicating a potential mechanistic conservation of this signaling axis. Nevertheless, it currently remains unclear whether nematodal GRH-1 is indeed subject to similar post-translational modifications as GRHL1, in particular since K116 and also other amino acids found modified are not conserved between the C. elegans and human protein. Notably, our findings for human GRHL1 are consistent with a similar lysine-dependent regulation of p53 activity, also involving interaction with KDM1A/LSD130, with p53 structurally related to Grainyhead TFs and part of the same immunoglobulin fold superclass16. In addition, K-methylases were recently also described as interaction partners of GRHL231. This property is possibly related to the role of Grainyhead TFs in regulating the chromatin landscape by recruiting epigenetic modifiers26, for which our description of the GRHL1 protein–protein interactome provides a first comprehensive resource for studies in this regard.
By performing a protein pull-down using biotinylated papaverine, to further clarify the pathway leading to GRHL1 activation, we identified the PI3 kinase PIK3C2A as a potential papaverine interaction partner. It is important to note that in chemical proteomics, immobilization might alter activity and/or change the interaction profile of a lead compound depending on the attachment site. Therefore, we here used a differential proteomics approach to compare the relative abundance of proteins purified together with the probe (i.e., biotinylated papaverine) vs. those purified together with the biotinylated linker control, assessing only proteins that are enriched ≥3-fold as potential binding partners. As the control consists of the full structure up to the pharmacophore (i.e., papaverine), all differential binding partners are likely due to protein-compound interactions through the pharmacophore. Whereas it cannot be ruled out that certain binding partners might be missed due to the nature of the attachment site, the method here used enables identification of specific targets and avoids nonspecific binders. Notably, others independently found papaverine as linked to regulation of PI3K signaling34 and PI3Ks are previously reported to modulate the activity of both KMT2D32 and KDM1A33. Together with our findings, this suggests that papaverine might first act on PIK3C2A, which then in turn regulates KMT2D and KDM1A to alter the K-methylation state and thereby activity of GRHL1. Interestingly, GRHL1 and biotinylated papaverine share the probable serine carboxypeptidase CPVL (Swiss-Prot ID: Q9H3G5), an as yet only superficially characterized protein, as their only common potential interaction partner. While we have not followed up further on this finding in the context of the present study, it is a quite exciting prospect for future research.
Presently, we were particularly curious of the connection of PI3Ks to insulin signaling18, modulation of which is prominently linked to regulation of longevity across species36,37,57, and whether Grainyhead TFs might also be involved in this fundamental metabolic pathway. Indeed, we found grh-1 expression in nematodes to be increased in two long-lived loss-of-function mutants of the central IIS pathway components daf-2 (encoding the nematodal insulin/IGF-1 receptor orthologue) and age-1 (encoding the canonical IIS PI3K orthologue). Furthermore, GRH-1 is required to maintain the lifespan-extension of daf-2 and age-1 mutant strains, reminiscent of the transcription factor DAF-16/FOXO37,38.
Supporting an evolutionarily conserved role of Grainyhead TFs in aging and IIS, we found that in the BXD mouse genetic reference population19 Grhl1 expression especially in muscle of mice, and consistently so for animals undergoing different dietary interventions, not only positively correlates with an increased mean lifespan, but at the same time also improves insulin sensitivity, as evidenced by a negative correlation with blood glucose levels in an oral glucose tolerance test. Moreover, we found human GRHL1 expression to be consistently positively correlated with insulin signaling across tissues, and in particular in both muscle and liver to be strongly genetically correlated with the majority of insulin receptor signaling pathway components. Finally, highly reminiscent of the observations in BXD mice and thereby suggesting functional conservation of Grhl1/GRHL1 in mammals, we found that human skeletal muscle GRHL1 expression, in the GTEx cohort40, positively correlates with increased age at death and, in the FUSION tissue biopsy study dataset41,42, negatively with fasting blood glucose levels, including in individuals with impaired glucose tolerance or type 2 diabetes. Notably and in general, when attempting to interpret and potentially translate findings regarding the impact of IIS modulation in different species, similarities and discrepancies between, e.g., C. elegans and mammals need to be carefully considered. While daf-2 impairment by RNAi and daf-2 incomplete loss-of-function mutations both extend nematodal lifespan, partial impairment of the insulin receptor in mice does not37,58, and impaired insulin signaling in humans is linked to the development of metabolic syndrome59. By contrast, indicating a certain degree of evolutionary conservation, the glucometabolic effect of daf-2 impairment is very similar to that of impaired insulin signaling in mammalian cells, in both cases leading to impaired glucose uptake and transport60. Thus, while our findings clearly indicate a functional connection of Grainyhead TFs to insulin signaling in different species, including in humans, delineating their relevance for this central metabolic pathway in mammals will require detailed investigations and the use of more sophisticated models, e.g., to selectively impair or overexpress murine Grhl1 in liver or other organs and tissues important for IIS.
In summary, our results are consistent with Grainyhead TFs, in particular GRH-1/Grhl1/GRHL1, having an evolutionarily conserved role in regulating lifespan and insulin signaling in nematodes and mammals. They further identify these transcription factors as potential targets for the development of geroprotective drugs, for which small-molecule compounds such as papaverine, piperlongumine, and vorinostat might already serve as leads. Applying similar screening strategies, as here utilized for proof-of-principle and basic research purposes, to more extensive compound libraries can be expected to facilitate discovery of additional novel drug candidates in the quest to ameliorate metabolic diseases and slow human aging.
Methods
C. elegans strains
The following C. elegans strains used for this publication were provided by the Caenorhabditis Genetics Center (CGC, University of Minnesota, USA): Wild-type N2 (Bristol), RB754 aak-2 (ok524), TJ1052 age-1 (hx546), CB1370 daf-2 (e1370), CF1038 daf-16 (mu86), VC2072 grh-1 (gk960), RB1813 piki-1 (ok2346), EU31 skn-1 (zu135). Strains overexpressing grh-1 were generated as detailed below and are termed MIR260 (risIs31) (with hsp-16.2-promoter and referred to as grh-1 OEx line 1 in the main text), MIR261 (risIs31) (with hsp-16.2-promoter and referred to as grh-1 OEx line 2 in the main text), and MIR262 (risIs32) (with endogenous grh-1 promoter). For maintenance, nematodes were grown on Nematode Growth Medium (NGM) agar plates in 90 mm petri dishes at 20 °C using E. coli OP50 bacteria as a food source61. For experiments involving transient heat shock, nematodes on first day of adulthood were transferred to 33 °C for 1 h and then returned to 20 °C, with the respective wild-type control nematodes subjected to the same procedure. NGM agar plates, after pouring, were dried at room temperature for 1–2 days and then stored at 4 °C until further use.
Bacterial strains
E. coli OP50 bacteria (CGC) were streaked out on DYT agar plates and single colonies picked from such plates were cultured overnight at 37 °C and constant shaking in Erlenmeyer flasks containing liquid DYT medium. Bacterial overnight cultures were concentrated by centrifugation for 30 min at 3200 × g and 4 °C. The prepared bacteria were spotted on NGM agar plates and allowed to grow for 16–24 h prior to use.
E. coli HT115(DE3) RNAi bacteria (CGC) were streaked out on LB agar plates with 100 µg/ml Ampicillin and 12.5 µg/ml Tetracycline and single colonies picked from such plates were cultured overnight at 37 °C and constant shaking in Erlenmeyer flasks containing liquid LB medium with 100 µg/ml Ampicillin. Bacterial overnight cultures were concentrated by centrifugation for 30 min at 3200 × g and 4 °C. The prepared bacteria were spotted on NGM agar plates additionally containing 100 µg/ml Ampicillin and 1 mM Isopropyl-beta-D-thiogalactopyranoside (IPTG) and allowed to grow for 16–24 h prior to use (reagents from AppliChem).
For preparation of heat-inactivated (HIT) OP50, the overnight culture was pelleted by centrifugation as above, all DYT media removed, and bacteria resuspended in S-buffer supplemented with 1 M MgSO4 and 5 mg/ml Cholesterol to a 10-fold concentrated culture. Afterward, the bacterial suspension was placed in a 65 °C water bath for 45 min. HIT OP50 were spotted on NGM agar plates on the day of use and dried for 30 min before adding the worms.
Pathogenic bacterial strains used in this study were Microbacterium nematophilum (CGC, #CBX102) and Pectobacterium carotovorum (BCCM, #LMG2404). M. nematophilum was grown for 48 h in liquid LB medium at 37 °C and diluted 10-fold with OP50 or HT115 bacteria before being spotted on NGM agar plates. P. carotovorum was grown overnight at 37 °C in DYT medium at 37 °C and spotted on NGM agar plates pure.
Human cell culture
HEK293 cells were obtained from a commercial source (CLS Cell Lines Service GmbH, #300192) and cultured in DMEM (Bioswisstec AG, #M 1434) with pH 7.4 and 4.5 g/l glucose, containing phenol red, 10% FBS, and 1% penicillin/streptomycin at 37 °C, 5% CO2, and 95% relative humidity. The different stable transgenic HEK293 cell lines used in this study are all derived from this initial cell line and were cultured under the same conditions, with the following antibiotics added: GRHL and ARE/NRF2 luciferase reporter and GRHL1 overexpressing cell lines +100 µg/ml hygromycin B; GRHL luciferase reporter with concomitant GRHL1 deletion +100 µg/ml hygromycin B and +0.25 µg/ml puromycin. For subculturing, HEK293 cells were commonly split at a 1:4–1:8 ratio every 2–3 days. Mouse embryonic fibroblasts (MEFs) derived from a conditional Grhl1 knockout mouse model were cultured using DMEM without phenol red but otherwise using the same general conditions as for HEK293 cells, with a split ratio of 1:2–1:4 every 3–4 days, and all MEF experiments were performed at a total passage number lower than five. Generation of the different cell lines and 4-hydroxytamoxifen induction of MEFs are detailed below.
Molecular cloning procedures
Vectors to overexpress grh-1 were generated with MultiSite Gateway cloning using the Gateway BP (#11789020) and LR (#11791020) Clonase II Enzyme mix according to the manufacturer’s instructions. The grh-1 full-length ORF without stop codons and with flanking attB1 and attB2 recombination sites was amplified from C. elegans cDNA using primers attB1_grh-1_fwd and attB2_grh-1_rev, with the amplified isoform identified as grh-1b by sequencing, and inserted into vector pDONR 221 (#12536017, all reagents from Thermo Fisher Scientific). A 1951 bp fragment of the endogenous grh-1 promoter with flanking attB4 and attB1R recombination sites was amplified from C. elegans gDNA, using primers attB4_grh-1p_f and attB1R_grh-1p_r. Similarly, the 348 bp hsp-16.2 promoter was amplified with primers attB4_hsp16.2p_fwd and attb1R_hsp16.2p_rev. Both promoters were individually inserted into pDONR P4-P1R. The different promoter-ORF combinations were inserted into the destination vector pdestMB14 (Addgene, #26415) in-frame with the sequence encoding a C-terminal GFP. The resulting vectors (pdestMB14_hsp-16.2p-grh-1-gfp, Addgene, #177762, and pdestMB14_grh-1p-grh-1-gfp, Addgene, #177761) were used to generate corresponding C. elegans grh-1 overexpressing strains: pdestMB14_hsp-16.2p-grh-1-gfp to generate MIR260 (risIs31) and MIR261 (risIs31), and pdestMB14_grh-1p-grh-1-gfp to generate MIR262 (risIs32).
Luciferase reporter vectors for GRHL and ARE/NRF2 are based on the vector pGL4.27 (Promega Corporation, #E8451). Synthetic promoter elements (SPE) with several repeats of the mammalian GRHL1-3 consensus binding sequence16 or the NRF2-bound antioxidant response element (ARE)24, flanked by 5′-KpnI and 3′-NheI restriction sites, were obtained by custom DNA synthesis and inserted in front of the pGL4.27 minimal promoter using standard restriction-ligation procedures. The resulting vectors (pGL4.27_GRHL-SPE, Addgene, #177774, and pGL4.27_ARE/NRF2-SPE, Addgene,#177775) were used to generate corresponding stable HEK293 luciferase reporter cell lines.
The vector to overexpress the full-length human GRHL1 ORF is based on vector pcDNA 3.1/Hygro(+) (Thermo Fisher Scientific, #V87020), which was first modified by inserting a sequence encoding a linker followed by a 3xFLAG-6xHis tag (L3F6H), using KpnI and BamHI restriction sites. The GRHL1 ORF without stop codon and with flanking NheI and KpnI restriction sites was amplified from HEK293 cDNA using primers GRHL1_NheI_fwd and GRHL1_KpnI_rev and inserted into pcDNA 3.1/Hygro(+)-L3F6H using standard restriction-ligation procedures. The resulting vector (pcDNA3.1_Hygro(+)_GRHL1-L3F6H, Addgene, #177765) was used to generate a HEK293 cell line stably overexpressing C-terminally 3xFLAG-6xHis tagged wild-type human GRHL1. For transient transfection of wild-type or mutated GRHL1 into the GRHL luciferase reporter cell line, individual amino acid codons in pcDNA3.1_Hygro(+)_GRHL1-L3F6H were mutated with the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, #E0554S) and primers selected with https://nebasechanger.neb.com/. The seven resulting vectors are also deposited at Addgene (#177767-177773).
Vectors for CRISPR/Cas9-mediated deletion of 2860 bp of human GRHL1 gDNA (partial exon 5, full exons 6 and 7, and partial exon 8) are based on vector pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene, #62988) and were cloned according to the detailed protocol provided by the Zhang lab62. Top and bottom strand oligonucleotides with 20 bp GRHL1-specific guide sequences were annealed and inserted into pSpCas9(BB)-2A-Puro (PX459) V2.0 using scarless cloning with the BbsI restriction enzyme. Oligonucleotides were designed to target sequences followed by NGG at their 3′-end in the genomic context, located in exon 5 (5′-TCCGCAGACTGAGATCCGAG-3′) and exon 8 (5′-CAACACTATCAGTAACATCG-3′), respectively. The resulting vectors (PX459_GRHL1-exon5, Addgene, #177763, and PX459_GRHL1-exon8, Addgene, #177764) were co-transfected into a HEK293 GRHL luciferase reporter cell line to obtain the respective reporter with a concomitant GRHL1 loss-of-function mutation. Sequences are listed in Supplementary Table 3.
Bombardment and generation of stable C. elegans strains
The pdestMB14 vectors for overexpression of grh-1 under control of the endogenous grh-1 promoter or the hsp-16.2 promoter were transformed into the unc-119-deficient C. elegans strain HT1593 (unc-119(ed3) III) by microparticle bombardment using the biolistic particle delivery system PDS-1000/He (Bio-Rad) according to the manufacturer’s instructions and previously described protocols63, with in total 7 µg of vector DNA per bombardment. Homozygous transgenic lines with stable integration of the respective vector (MIR260 (risIs31) and MIR261 (risIs31) with pdestMB14_hsp-16.2p-grh-1-gfp, and MIR262 (risIs32) with pdestMB14_grh-1p-grh-1-gfp) were selected based on GFP fluorescence (for hsp-16.2 promoter constructs, selection by GFP was performed after transient 1 h heat shock at 33 °C) and backcrossed at least four times against wild-type N2 nematodes before being used in any experiments.
Lipofection and generation of stable human cell lines
All transient and stable transfections of HEK293 cells were carried out using the Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, #L3000008) according to the manufacturer’s instructions. To obtain stable transgenic cell lines, lipofection was performed in 6-well plates and cells were transferred to 15 cm dishes after 24 h. Selection with the appropriate antibiotics was started after 48 h (hygromycin B at 200–300 µg/ml and puromycin at 0.5–1.0 µg/ml). After 7–12 days of selection, individual clones were picked using cloning discs and expanded for testing. Luciferase reporter cell lines were screened by performing a luciferase assay using appropriate positive control compounds, and GRHL1 overexpressing cell lines checked by western blot (see below). Homozygous partial deletion of GRHL1 in the GRHL luciferase reporter was confirmed by genotyping PCR using primers GRHL1_del_fwd, GRHL1_del_rev, and GRHL1_wt_rev. For generation of GRHL luciferase reporters with shRNA-mediated individual knockdown of KMT2D, KDM1A, or PIK3C2A, the reporter was transfected with a respective commercial shRNA or scramble control vector (all vectors from OriGene, KMT2D: #TL311461, KDM1A: #TL316984, PIK3C2A: #TL310432) carrying a puromycin resistance. Knockdown was confirmed by RT-qPCR using primers KMT2D_qPCR_fwd + KMT2D_qPCR_rev, KDM1A_qPCR_fwd + KDM1A_qPCR_rev, and PIK3C2A_qPCR_fwd + PIK3C2A_qPCR_rev, respectively. Monoclonal cell lines with the respective modifications were cultured for several passages and verified as stable before being used for the appropriate experiments. Sequences are listed in Supplementary Table 3.
Compound library and compound preparation
‘The Spectrum Collection’ compound library (MicroSource Discovery Systems), was obtained at 10 mM in DMSO in 32 × 96-well plates with 80 compounds per plate. Upon arrival, plates were aliquoted and stored at −80 °C until use. Individual compounds were sourced from different suppliers (CAS/Product #). abcr GmbH: 1,4-Anthraquinone (635-12-1/AB177277). Santa Cruz Biotechnology, Inc: Scriptaid (287383-59-9/sc-202807). Selleck Chemicals LLC: Alantolactone (546-43-0/S8318); piperlongumine (20069-09-4/S7551); vorinostat (149647-78-9/S1047). Sigma-Aldrich/Merck KGaA: 2′,4′-Dihydroxy-4-methoxychalcone (81674-91-1/IDF00081); 4-hydroxychalcone (20426-12-4/S350664); chlorothalonil (1897-45-6/36791); DL-sulforaphane (4478-93-7/S4441); ethacrynic acid (58-54-8/E1800000); menadione (58-27-5/M9429); oxyphenbutazone (129-20-4/SML0540); papaverine hydrochloride (61-25-6/P3510); phenothiazine (92-84-2/P14831); thimerosal (54-64-8/T5125); thymoquinone (490-91-5/274666); ZPCK (26049-94-5/860794). Compounds were stored at −20 °C and stock solutions for tissue culture and C. elegans experiments were freshly prepared at 10–20 mM in DMSO.
Synthesis of biotinylated papaverine
Papaverine hydrochloride (1) was mono-demethylated by treatment with sodium ethanethiolate in DMF resulting in a mixture of 6-desmethylpapaverine (2) and pacodine (3). (+)-Biotin N-hydroxysuccinimide ester (4) was coupled to 2-(methylamino)ethanol (5) resulting in biotinylated linker control (6). Pacodine (3) and biotinylated linker control (6) were condensed in a Mitsunobu reaction resulting in biotinylated papaverine (7). Numbers in parentheses correspond to compounds as depicted in Supplementary Fig. 4a. Detailed experimental procedures, references, and analytical data are contained in the Supplementary Methods.
Protein isolation and immunoblotting
For C. elegans total protein isolation, nematodes collected from three synchronized plates were washed three times with ice-cold S-Buffer and flash-frozen in liquid nitrogen. Frozen pellets were ground in a nitrogen-chilled mortar and suspended in 200 µl Sørensen buffer with 1x Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, #78440) and additionally 2 mM sodiumfluoride, 2 mM sodium orthovanadate, 1 mM PMSF, and 2 mM EDTA. Samples were centrifuged for 7 min at 12,000 × g and 4 °C to obtain cleared supernatants. Total protein from HEK293 cells was isolated using RIPA buffer with 1x Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, #78440). Trypsinized cells were counted, then pelleted by centrifugation for 5 min at 300 × g and washed once with ice-cold PBS, before adding 100 µl RIPA buffer per 2 × 106 cells. After 5 min incubation on ice, samples were sonicated at 50% amplitude for 3 × 10 s and centrifuged for 15 min at 14,000 × g and 4 °C. Supernatants were used for protein quantification with a standard BCA assay and then boiled for 10 min at 95 °C in 1x Laemmli sample buffer. Total protein extracts were used for standard SDS-PAGE and western blotting procedures using the primary and secondary antibodies listed below. The signal was visualized using ECL substrate (Bio-Rad, #1705061). Immunoblot images were acquired with the ChemiDoc MP Imaging System and Image Lab Touch Software V2.4 (Bio-Rad), using the following antibodies: Phospho-p38 MAPK (Thr180/Tyr182) antibody (Cell Signaling, #9211) at 1:1000 dilution with secondary anti-rabbit (Cell Signaling, #7074); α-Tubulin antibody (Sigma-Aldrich, #T6199) at 1:2000 dilution with secondary anti-mouse (Cell Signaling, #7076); FLAG M2 antibody (Sigma-Aldrich, #F3165) at 1:1000 dilution with secondary anti-mouse (Cell Signaling, #7076). All secondary antibodies were used at a dilution of 1:2000.
RNA extraction and RT-qPCR
For C. elegans total RNA isolation, nematodes collected from three synchronized plates were washed three times with ice-cold S-Buffer and flash-frozen in liquid nitrogen. Afterward, 500 µl ice-cold TRI Reagent (Molecular Research Center, #TR118) was added to the ground pellet. Concentration of the resulting RNA was measured with an LVis plate on a CLARIOstar microplate reader (BMG LABTECH) and stored at −80 °C until further use. Reverse transcription of RNA to cDNA was performed using the High-Capacity cDNA Reverse Transcription Kit together with Oligo(dT) primers following the manufacturer’s protocol. Quantitative real-time PCR was carried out on a ViiA 7 Real-Time PCR System using the SYBR Select Master Mix in 384 well plates according to the manufacturer’s instructions, and with the recommended cycling conditions. Samples were analyzed by the ddCT method using QuantStudio Real-Time PCR Software V1.3, with normalization to the reference gene Y45F10D.4 for C. elegans64. Instrument, reagents, and software from Thermo Fisher Scientific. Sequences are listed in Supplementary Table 3.
Next-generation sequencing (RNA-Seq)
To identify genes regulated by C. elegans GRH-1, RNA-Seq and data analysis was performed using three independent biological samples of total RNA extracted from either wild-type N2 nematodes or from the grh-1 overexpressing strain used throughout this study. Samples were obtained from synchronized populations, prepared as described below for lifespan assays (at 48 h post-development).
Library preparation
The quality of the isolated RNA was determined with a Qubit (1.0) Fluorometer (Life Technologies) and a Bioanalyzer 2100 (Agilent). Only those samples with a 260 nm/280 nm ratio between 1.8–2.1 and a 28S/18S ratio within 1.5–2 were further processed. The TruSeq RNA Sample Prep Kit v2 (Illumina, #RS-122-2001/2) was used in the succeeding steps. Briefly, total RNA samples (100–1000 ng) were poly A enriched and then reverse-transcribed into double-stranded cDNA. The cDNA samples were fragmented, end-repaired, and polyadenylated before ligation of TruSeq adapters containing the index for multiplexing. Fragments containing TruSeq adapters on both ends were selectively enriched with PCR. The quality and quantity of the enriched libraries were validated using Qubit (1.0) Fluorometer and the Caliper GX LabChip GX (Caliper Life Sciences). Products are a smear with an average fragment size of ~260 bp. The libraries were normalized to 10 nM in Tris-Cl 10 mM, pH 8.5 with 0.1% Tween-20.
Cluster generation and sequencing
The TruSeq PE Cluster Kit HS4000 or TruSeq SR Cluster Kit HS4000 (Illumina) were used for cluster generation using 10 pM of pooled normalized libraries on the cBOT. Sequencing was performed on the Illumina HiSeq 2000 single-end at 100 bp using the TruSeq SBS Kit HS4000 (Software: HCS Software for HiSeq 2500, 2000, 1500, and 1000 Systems V2.2.68; Illumina).
RNA-Seq data analysis
Bioinformatic analysis was performed using the R package ezRun (github.com/uzh/ezRun) within the data analysis framework SUSHI65. Raw reads were quality checked using FastQC and FastQ Screen66. Quality controlled reads (adapter trimmed, first and last 4 bases hard trimmed, minimum average quality Q10, minimum tail quality Q10, minimum read length 20 nt) were aligned to the C. elegans reference genome (Ensembl WBcel235) using the STAR aligner67. Expression counts were computed using featureCounts in the Bioconductor package Subread68. Differential expression analysis was performed using the Bioconductor DESeq2 package, where raw read counts were normalized using the quantile method, and differential expression was computed using the quasi-likelihood (QL) F-test69. Gene ontology (GO) enrichment analysis was performed using Bioconductor packages GOseq70 and GOstats71. Quality checkpoints, such as quality control of the alignment and count results, were implemented in ezRun and applied throughout the analysis workflow to ensure correct data interpretation.
The datasets generated for this study can be found in the NCBI’s Gene Expression Omnibus, GEO Series accession number GSE159077.
In silico GRH-1 binding sites analysis
Presence of GRH-1 binding sites in the predicted promoters of differentially expressed genes (DEGs) were determined using the BiocManager::TFBSTools function72 in R and the FIMO algorithm73 from the MEME Suite74 in parallel, using a GRH-1 position weight matrix available from literature15,17. Both analyses were run on the predicted promoter regions of all DEGs previously determined by RNA-Seq data analysis. Predicted promoter regions were defined as the sequences spanning from 2.5 kb upstream to 1 kb downstream of each DEG’s transcriptional start site. Only GRH-1 binding sites identified by both methods were considered further.
Tissue-specific correlation of human genes with GRHL1
GTEx database (v8) data for tissue-specific gene TPMs (version 2017-06-05_v8_RNASeQCv1.1.9) was obtained from www.gtexportal.org/home/datasets40. Pearson’s correlations between expression of GRHL1 and all the other genes were calculated with the Python package scipy.stats. Data is visualized with Python packages matplotlib and seaborn.
GSEA of human genes correlating with GRHL1
In each tissue, Pearson’s r correlation values of genes with P-values <0.05, obtained as described above, were used to calculate gene set enrichment analysis (GSEA) with the Python package gseapy, using gene set files retrieved from download.baderlab.org/EM_Genesets/current_release/Human/symbol (gene set assembly: Human_GO_AllPathways_with_GO_iea_March_01_2021_symbol.gmt). Data is visualized with Python packages matplotlib and seaborn.
BXD mice data analysis
Murine Grhl1 expression data and phenotypic traits were obtained from GeneNetwork2 (www.genenetwork.org/). Gene expression datasets analyzed: EPFLMouseMuscleCDRMAEx1112, EPFLMouseMuscleCDRMA1211, EPFLMouseMuscleHFDRMAEx1112, EPFLMouseMuscleHFDRMA1211, EPFLMouseLiverCDEx0413, EPFLMouseLiverCDRMA0413, EPFLMouseLiverCDRMA0818, EPFLMouseLiverHFDRMA0818, EPFLMouseLiverHFDRMA0413.
Phenotypic traits analyzed: 21450 (Lifespan); 17661, 17662 (glycemia during oGTT - AUC); 17663, 176634 (insulin during oGTT - AUC); 17603, 17604 (body weight percentage gain 8–28 weeks).
In each expression dataset, Grhl1 expression RMA in each BXD strain was Z-score normalized, and the average value of Grhl1 Z-score expression across the same tissue/condition (chow diet muscle, high-fat diet muscle, chow diet liver, high-fat diet liver) in each BXD strains was used to calculate Pearson’s r correlations with the phenotypic traits of interest. Z-score normalization and Pearson correlation coefficients were computed with Python package scipy.stats. Data is visualized with Python packages matplotlib and seaborn.
GTEX and FUSION dataset analysis
Data from Common Fund (CF) Genotype-Tissue Expression Project (GTEx) (phs000424.v8.p2) and The Finland-United States Investigation of NIDDM Genetics (FUSION) Tissue Biopsy Study (phs001048.v2.p1) datasets were obtained from dbGaP (approved request #28650: “Population genetics of human aging”).
For the GTEx dataset, only individuals who died of natural causes (field “death manner” DTHMNNR = Natural), were included in the analysis of Spearman rank correlation coefficients for age at death and for GRHL1 expression in skeletal muscle (ENSG00000134317). Data is represented as the scatterplot of age versus GRHL1 log2 TPM in skeletal muscle, and the linear regression line is shown to demonstrate correlation.
For the FUSION dataset, Spearman rank correlation coefficients for fasting blood glucose (field GL0) and expression of GRHL1 (ENSG00000134317) in skeletal muscle were computed in the entire population (All) or in individuals with impaired glucose tolerance (ogtt_status = IGT) or type 2 diabetes (ogtt_status = T2D). Data is represented as the scatterplot of fasting blood glucose versus GRHL1 log2 FPKM in skeletal muscle, and the linear regression line is shown to demonstrate correlation.
All analyses were performed using the Python package scipy.stats.
Confocal imaging
Worms were mounted on freshly prepared agarose pads (3% agarose in S-buffer) and paralyzed using 20 mM tetramisole75. Confocal microscopy was performed using an Olympus FluoView 3000 (Olympus Corporation) microscope with inverted stand. Fluoview FV31S-SW software was used for image acquisition. Single plane images of groups of worms were acquired using an UPLFLN 10 × 2 objective (exc/em GFP: 488/510), while Z-stacks of cells were acquired using an UPLSAPO 60X objective (exc/em Alexa 488: 499/520, Mitotracker: 578/598, DAPI: 359/461). Single plane images of cells were overlayed using Fiji ImageJ.
Immunofluorescence
Cells were seeded in wells of a 6-well plate, on top of a glass coverslip (VWR, #631-1567) and left to attach overnight. Media was removed and cells were washed once with PBS. Mitotracker Red (ThermoFisher, #M7513) was applied to the cells in DMEM at a final concentration of 250 nM and cells were incubated for 30 min at 37 °C in the incubator. All subsequent steps were performed in the dark. Cells were washed twice with PBS and then fixed with 4% PFA at 37 °C for 15 min. After two washes with PBS, cells were incubated with 0.2% Triton in PBS for 5 min at RT. Cells were then washed 3 times for 5 min each with PBS on a shaker. Blocking solution (2% BSA and 0.2% Triton in PBS) was added and cells were incubated on a shaker for 30 min at RT. Primary anti-FLAG antibody (Sigma-Aldrich, #F3165) was added at a dilution of 1:200 in blocking solution and cells were incubated for 1 h at 37 °C. Subsequently, cells were washed with PBS three times for 5 min each and secondary Alexa Fluor® 488 goat anti-mouse IgG antibody (ThermoFisher, #A-21141) was added at a dilution of 1:1000 in blocking solution. Afterward, cells were incubated for 1 h at 37 °C. Cells were washed with PBS three times for 5 min each and then coverslips were mounted using DAPI mounting media ProLong Diamond Antifade Mountant with DAPI (Invitrogen, #P36966) and incubated overnight at RT prior to imaging.
C. elegans RNAi and compound treatment
For RNAi-mediated gene knockdown experiments, E. coli HT115 bacteria were applied to nematodes by feeding as previously described76. The clones for RNAi of aak-2/T01C8.1, daf-16/R13H8.1, piki-1/F39B1.1, pmk-1/B0218.3, and skn-1/T19E7.2 were obtained from the RNAi ORF library v1.1 (Thermo Fisher Scientific). The clones for lsd-1/T08D10.2 and spr-5/Y40B1B.6 were obtained from the Ahringer RNAi library (Source BioScience). The clone for grh-1/Y48G8AR.1 was constructed as previously described17. The clone for daf-2/Y55D5A.5 was thankfully provided by the Kenyon lab. Compounds were dissolved in DMSO to a stock concentration of 20 mM and added to NGM agar before pouring. Final DMSO concentration in NGM was 0.5% All compound treatments were performed on HIT OP50. To maintain synchronized populations during long-term experiments, nematodes were washed off the plates into 15 ml tubes every day of the reproductive period, allowed to settle, and then washed again repeatedly until the supernatant was free of progeny. The clean nematodes were then transferred to the freshly prepared treatment plates.
C. elegans lifespan assays
All C. elegans lifespan assays, aside from heat shock experiments, were performed at 20 °C according to standard protocols as previously described, explicitly omitting FUdR77. Briefly, adult nematodes were allowed to lay eggs for four to 9 h and the resulting eggs incubated for 64 h at 20 °C on NGM agar plates inoculated with OP50 to obtain a synchronized population of young adult nematodes. For a typical lifespan assay, 100 young adult nematodes per condition were manually transferred to NGM agar plates (30–35 nematodes per 55 mm petri dish) inoculated with the respective bacteria as indicated. For the first 10–12 days, nematodes were transferred daily and afterward every 2–3 days. Nematodes showing no reaction to gentle stimulation were scored as dead. Nematodes that crawled off the plates, displayed internal hatching, or a protruding vulva were censored. For heat shock lifespan assays, nematodes on first day of adulthood were transferred to 33 °C for 1 h and then returned to 20 °C. This treatment was repeated every seven days for the remainder of the experiment’s duration and wild-type control nematodes were subjected to the same procedure throughout.
C. elegans health assays
To determine fertility, nematodes were synchronized as described above. Single L4 larvae were transferred onto individual plates inoculated with OP50 bacteria (10 plates per condition) and subsequently onto fresh plates every day of the reproductive period. Progeny were allowed to hatch and counted manually.
To determine motility, nematodes were synchronized and single L4 larvae transferred onto individual plates inoculated with OP50 bacteria and subsequently onto fresh plates for two days. Afterward, 15–20 nematodes were transferred to fresh plates and 30 s video clips were recorded (Leica M165FC microscope with Leica camera DFC 3000G). Three independent videos per condition were analyzed regarding the speed of each population. Videos were analyzed using Fiji78 with the wrMTrck plugin as described79.
Luciferase assay compound screening
Unless stated otherwise, HEK293 luciferase reporter cell lines were seeded into white clear bottom 96-well plates (Greiner Bio-One, #655098) in 100 µl full DMEM and at 5 × 104 cells/well, 24 h prior to compound treatment. Final DMSO concentration for all treatment and control conditions was adjusted to 0.5%. Readout was performed 24 h after treatment (i.e., in total 48 h after seeding), using a CLARIOstar microplate reader (BMG LABTECH) and the ONE-Glo Luciferase Assay System (Promega Corporation, #E6130) according to the manufacturers’ instructions. Details for individual assay setups are as follows:
Primary compound screening
Compounds from “The Spectrum Collection” library were pre-diluted from 10 mM stocks and added to wells to a final concentration of 10 µM. Positive control wells were treated with either 5 µM sulforaphane (NRF2/ARE reporter) or 2 µM scriptaid (GRHL reporter), i.e., the same compounds used for initial screening and verification of the reporters. Luciferase activity after compound treatment was normalized to DMSO controls and a two-fold induction of the respective reporter was used as cut-off for primary hit identification.
Dose–response confirmation of hit compounds
Resourced individual compounds were used in a three-point dose-response assay at 5, 10, and 20 µM final concentration, with two to three replicate plates at individual timepoints. Overall setup was as for primary compound screening, additionally using the non-lytic CellTiter-Fluor Cell Viability Assay (Promega Corporation, #G6082) according to the manufacturer’s instructions and prior to measuring luciferase activity. Luciferase activity was normalized to cell viability and DMSO controls.
GRHL1-dependency of hit compounds
Relative GRHL1-dependency of selected compounds at 5 and 10 µM final concentration was measured with a wild-type GRHL reporter (GRHLWT-LUC) and a GRHL reporter with a CRISPR/Cas9-mediated GRHL1 deletion (GRHLNULL-LUC), using two individual replicates per cell line and concentration. Luciferase activity was normalized to the respective DMSO control and relative GRHL1-dependency determined as GRHLNULL-LUC/GRHLWT-LUC fold change across both concentrations and two independent GRHLNULL-LUC cell lines.
Impact of GRHL1 mutation on GRHL reporter activation
GRHL reporter cells were seeded at 1 × 104 cells/well and otherwise as above. After 24 h, cells were individually transfected with either a wild-type or mutated GRHL1 overexpression vector or an empty vector control, using the Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, #L3000008). Readout was performed 48 h after transfection (in total 72 h after seeding). Two independent experiments with in total 4 individual replicate wells per condition were performed and activity determined as percentage change compared to transfection with the vector encoding wild-type GRHL1, normalized to the respective empty vector control sample.
Tandem-affinity purification of human GRHL1 from nuclei
Nuclei isolation
Wild-type HEK293 cells or a HEK293 cell line overexpressing C-terminally 3xFLAG-6xHis tagged wild-type human GRHL1 (GRHL1WT-TAG) were seeded into 15 cm plates at a split ratio of 1:8 and treated with 20 µM papaverine or 0.1% DMSO as vehicle control 24 h prior to the isolation procedure. For nuclei isolation, cells were trypsinized, counted, and 20 × 106 cells per sample transferred to a 50 ml tube. Cells were pelleted by centrifugation for 3 min at 1000 × g, washed once with ice-cold PBS, and pelleted again. Cell pellets were resuspended in 1 ml freshly prepared, filter-sterilized ice-cold hypotonic lysis buffer (0.01 M Tris-HCl pH 7.4, 0.01 M NaCl, 10% Glycerol, 0.3% NP-40, 1x Halt Protease and Phosphatase Inhibitor Cocktail from Thermo Fisher Scientific, #78440) and incubated on ice for 10 min. Afterward, nuclei were pelleted by centrifugation for 8 min at 800 × g and 4 °C, the supernatant containing the cytoplasmic fraction discarded, and nuclear pellets flash-frozen in liquid nitrogen and immediately stored at −80 °C until tandem-affinity purification.
3xFLAG and 6xHis tandem-affinity purification
Nuclei isolated from GRHL1WT-TAG overexpressing cells and wild-type HEK293 cells were subjected to the same purification procedures, with the latter samples serving as background control for unspecific absorption of proteins to the utilized affinity matrices. For 3xFLAG purification, nuclear pellets were resuspended in 1 ml freshly prepared, filter-sterilized ice-cold FLAG lysis buffer (0.05 M Tris-HCl pH 7.4, 0.15 M NaCl, 10% Glycerol, 1% Triton X-100, 0.01% Tween-20, 1x Halt Protease and Phosphatase Inhibitor Cocktail from Thermo Fisher Scientific, #78440), briefly vortexed, and incubated on ice for 15 min. Afterward, samples were centrifuged for 10 min at 14,000 × g and 4 °C, and supernatants used as input for purification. Anti-FLAG M2 Magnetic Beads (Millipore/Merck KGaA, #M8823) were prepared in TBS according to the manufacturer’s instructions and 30 µl packed gel volume used per reaction. Supernatants were added to beads and incubated for 2 h at 4 °C with end-over-end rotation. After incubation, supernatants were discarded, and beads washed twice with 300 µl TBS + 10% Glycerol. Proteins were eluted with 150 µl His binding buffer (0.02 M HEPES-KOH pH 7.4, 0.3 M NaCl, 10% Glycerol, 0.02 M Imidazol-HCl pH 7.4, 0.01% Tween-20, 1x Halt Protease and Phosphatase Inhibitor Cocktail from Thermo Fisher Scientific, #78440) containing 150 ng/µl 3X FLAG peptide (Millipore/Merck KGaA, #F4799) by incubation for 30 min at 4 °C with end-over-end rotation. Supernatants with eluted proteins were adjusted to 700 µl with His binding buffer and added to 50 µl Dynabeads His-Tag Isolation and Pulldown (Thermo Fisher Scientific, #10104D) per reaction. After another incubation for 30 min at 4 °C with end-over-end rotation, Dynabeads were washed twice with 300 µl His binding buffer and once with 300 µl His binding buffer without Tween-20. During the last washing step, samples were transferred to new reaction tubes, any remaining supernatants discarded completely, and beads resuspended in 100 µl His binding buffer without Tween-20 per sample.
On-bead digestion
For each sample, the beads were washed once with 100 µl of digestion buffer (10 mM Tris + 2 mM CaCl2, pH 8.2). The washed beads were resuspended in 90 µl digestion buffer, and the proteins were on-bead digested using 10 µl of Sequencing Grade Trypsin (100 ng/µl in 10 mM HCl, Promega, #V5111). The digestion was carried out in a microwave instrument (Discover System, CEM) for 30 min at 5 W and 60 °C. The supernatants were collected, and the peptides were extracted from the beads with 100 µl of TFA-buffer (0.1% TFA in 10 mM Tris, 2 mM CaCl2, 50% acetonitrile). The supernatants were combined, and the samples were finally dried in a speed-vac, resolubilized in 20 µl of 0.1% formic acid and centrifuged at max speed (20,000 × g) for 10 min. Ten microliters of digested peptides were injected for shotgun liquid chromatography with tandem mass spectrometry (LC-MS/MS) as described below.
Protein pull-down using biotinylated papaverine
Cell lysis
Cells were collected from 15-cm dishes without trypsinization. Cells were centrifuged at 500 × g, 4 °C for 5 min and washed once with cold PBS. Cells were spun down again as above, and supernatant was discarded. Lysis buffer (20 mM HEPES pH 7.3, 50 mM KCl, 5 mM MgCl2, 0.01% NP40, 2 mM NaF, 2 mM Na3VO4) was added, cells were vortexed and incubated on ice for 15 min. Cells were sonicated 3 times for 2 s each at 50% amplitude and incubated on ice for 10 min. The lysate was centrifuged at 13,000 × g for 5 min and 4 °C. Supernatant was collected and protein concentration was measured using a standard BCA assay.
Bead preparation and pull-down
For each condition, 40 µl of streptavidin bead-suspension was used (Thermo Fisher Scientific, #11205D). Beads were equilibrated three times with lysis buffer and subsequently incubated with biotinylated papaverine (compound 7 in Supplementary Fig. 4a) or biotinylated linker control (compound 6 in Supplementary Fig. 4a) for 30 min at RT with rotation. Cell lysate was first incubated with biotinylated linker control for 3 h at 4 °C with rotation, then split into half and incubated with either biotinylated-linker-beads (as a control condition) or biotinylated-papaverine-beads for 4 h at 4 °C with rotation. After incubation, beads were washed five times with lysis buffer (w/o NP40, NaF, or Na3VO4) and stored at −80 °C prior to MS analysis. All experiments were performed in triplicate.
On-bead digestion
A variation of the on-bead digestion protocol from the Mann lab80 was used. Proteins were pre-digested at 30 °C for 15 min by adding 20 μl elution buffer (2 M urea in 50 mM Tris-HCl, pH 8.0) supplemented with 5 mM DTT and 100 ng Sequencing Grade Trypsin (Promega, #V5111). Next, iodoacetamide was added to a final concentration of 3 mM, and incubation was continued for another 15 min. Subsequently, proteins were eluted by adding two times (50 and 30 μl) elution buffer supplemented with 1 mM DTT and 5 ng/μL trypsin. Eluates were transferred to new microcentrifuge tubes, incubated at 32 °C overnight (or minimum 6 h). The digestion was stopped by adding 0.5% of TFA. All incubation steps were performed in a thermoshaker at 400 rpm gentle shaking. Digested peptides were desalted using self-packed C18 Stage-Tips81, vacuum dried, and resolubilized in 20 μl 3% acetonitrile, 0.1% formic acid for mass spectrometry analysis. Four microliters of digested peptides were injected for shotgun liquid chromatography with tandem mass spectrometry (LC-MS/MS) as described below.
Liquid chromatography-mass spectrometry analysis
Label-free shotgun LC-MS/MS was performed either on a Q Exactive mass spectrometer (Software: Exactive MS Series Instrument Control Software V2.9; Thermo Fisher Scientific) equipped with a nanoAcquity UPLC system (Waters Inc.) for the purified GRHL1WT-TAG and associated control samples, or on an Orbitrap Fusion Lumos mass spectrometer (Software: Orbitrap Tribrid MS Series Instrument Control Software V3.1; Thermo Fisher Scientific) equipped with an Acquity UPLC M-Class system (Waters Inc.) for the purified biotinylated papaverine and associated control samples. Peptide mixtures underwent reverse-phase chromatographic separation before nano-electrospray ionization (ESI) with a Digital PicoView nanospray source (New Objective). Peptides were eluted at a flow rate of 300 nl/min with a binary acetonitrile gradient system composed of (A) 0.1% formic acid in H2O and (B) 0.1% formic acid in acetonitrile. Gradient was set with the following conditions:
Q Exactive: BEH300 C18 column (1.7 µm, 75 µm × 150 mm, Waters Inc.); multistep gradient of 5–35% B in 90 min, 40% B in 5 min, and 80% B in 1 min.
Orbitrap Fusion Lumos: HSS C18 T3 Col 100A column (1.8 µm, 75 µm × 250 mm, Waters Inc.); multistep gradient of 5–35% B in 135 min, 40% B in 5 min, and 80% B in 1 min.
For the data-dependent analysis, mass spectrometers were set as follows:
Q Exactive: precursor scan range of 350–1500 m/z at a resolution of 70,000 at 200 m/z with AGC (automated gain control) target value of 3 × 106, followed by HCD (higher-energy collision dissociation) fragmentation on the 12 most intense signals per cycle with a normalized collision energy of 25.
Orbitrap Fusion Lumos: precursor scan range of 300–2000 m/z at a resolution of 120,000 at 200 m/z with AGC target value 4 × 106; followed by HCD fragmentation on the 15 most intense signals per cycle, with a collision energy of 35.
In both cases, only precursors with intensity above 25,000 were selected for MS/MS. Precursor masses previously selected for MS/MS measurement were excluded from further selection for 30 s (Orbitrap Fusion Lumos) or 40 s (Q Exactive), and the exclusion window was set at 10 ppm. All measurements were acquired using internal lock mass calibration on m/z 371.1010 and 445.1200.
Data analysis of GRHL1WT-TAG samples
For the identification of interactors, the raw data were converted into Mascot Generic Format files (.mgf) using ProteoWizard (http://proteowizard.sourceforge.net/), and the proteins were identified using the Mascot search engine (Matrix Science, version 2.5.1.3). Spectra were searched against a UniProtKB/Swiss-Prot Homo sapiens proteome database (taxonomy 9606, release 2018_09 [https://ftp.uniprot.org/pub/databases/uniprot/previous_major_releases/release-2018_09/knowledgebase]), concatenated to its reversed decoyed FASTA database. Methionine oxidation was set as variable modification, and enzyme specificity was set to trypsin allowing a maximum of two missed cleavages. A fragment ion mass tolerance of 0.030 Da and a parent ion tolerance of 10.0 PPM were set. Scaffold (Proteome Software Inc., version 4.10) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they achieved a false discovery rate (FDR) of <0.1% by the Scaffold Local FDR algorithm. Protein identifications were accepted if they achieved an FDR of <1.0% and contained at least 2 identified peptides.
The search engine PEAKS 8.5 (PEAKS X, Bioinformatic Solutions) was used for the characterization of post-translational modifications (PTMs) on protein GRHL1WT-TAG. Data were searched with a fragment ion mass tolerance of 0.03 Da and a parent ion tolerance of 15.0 PPM. Oxidation (M), Acetylation (K), Dimethylation (KR), Methylation (KR), Phosphorylation (STY), and Ubiquitination (K) were specified as a variable modification. A maximum of 3 missed cleavages were allowed. Peptide identifications were accepted if they achieved a peptide false discovery rate (FDR) of <0.1% and the proteins contained at least 2 identified peptides.
Detected GRHL1WT-TAG PTMs were considered as papaverine-specific if the MS/MS spectra could be identified in at least one of the papaverine-treated samples and not in any of the DMSO control samples. Proteins were considered as GRHL1 interactors if present in at least 2/3 samples GRHL1WT-TAG and either fully absent or enriched at least ≥3-fold over the respective control, based on sample average of unique peptide counts. In total 130 proteins were identified as potential GRHL1WT-TAG interaction partners. These proteins were curated manually, and each assigned a non-exclusive category and classification reflecting its most prominent cellular function, based on associated gene ontology terms and information available from UniProtKB/Swiss-Prot82 (Supplementary Data 5).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE83 partner repository with the dataset identifier PXD021768.
Data analysis of biotinylated papaverine pull-down samples
All raw data analyses were handled and annotated by the local laboratory information management system (LIMS) (10.1145/1739041.1739135), further analyzed with MaxQuant software suite version 1.6.3.3 (Max Planck Institute of Biochemistry, Munich) supported by the Andromeda search engine84. Spectra were searched against the UniProt Human proteome database encompassing 75,004 protein entries (proteome ID UP000005640, version from 2019-05-05), concatenated to its reversed decoyed FASTA database. Data were searched with carbamidomethylation as a fixed modification and protein N-terminal acetylation and methionine oxidation as variable modifications. A maximum of two missed cleavages were allowed while requiring strict trypsin specificity, and only peptides with a minimum sequence length of six were considered for further data analysis. Only peptides and proteins with a false discovery rate of <1% were accepted.
Proteins were considered as papaverine interactors if present in at least 2/3 replicates of the biotinylated papaverine samples and enriched ≥3-fold over control, based on sample averages of both label-free quantification intensities and unique peptide counts.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE83 partner repository with the dataset identifier PXD021808.
Statistical analyses
Statistical analyses for all data except those from lifespan assays were carried out using a t-test with appropriate parameters, i.e., a two-tailed unequal variances t-test used for comparison of the unpaired control vs. treatment groups. For comparing distributions between different groups in the lifespan assays, statistical calculations were performed using JMP software V9.0 (SAS Institute), applying the log-rank test. All other calculations were performed using Microsoft Excel 365 or GraphPad Prism V8.2.0 (GraphPad Software). P-values are reported in detail, without the use of arbitrary star ratings. However, in line with longstanding conventions used in the field, P-values <0.05 are generally referred to as indicating a significant difference, i.e., a rejection of the null hypothesis.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
C. elegans strains used in this work were provided by the Caenorhabditis Genetics Centre (Univ. of Minnesota, USA), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This study was funded by the Swiss National Science Foundation (#31003A_176127 to M.R.). The Ristow laboratory is further supported by the Horizon 2020 program of the European Union (Ageing with Elegans, #633589). The authors thank Cynthia Kenyon for providing the daf-2 RNAi clone, as well as Sarah Tailly and Uli Kodjadjiku for help with supporting experiments. The authors thank Laura Kunz and Paolo Nanni of the Functional Genomics Center Zurich (FGCZ) for their support in acquisition, analysis, and presentation of mass spectrometry data.
Source data
Author contributions
M.R. and F.F. conceived and supervised the project. G.G. and F.F. performed the majority of experiments, and J.Y.W., C.T., B.L., D.P., and K.Z. performed additional experiments. R.E. and E.C. were responsible for synthesis of biotinylated papaverine. E.A. performed all BXD, GTEx, and FUSION dataset analyses. G.G. and F.F. analyzed the majority of all other data and M.L.F., J.Y.W., and R.E. performed additional data analyses. F.F. and G.G. prepared all figures, with contributions from E.A. and R.E., and M.R. provided additional input. F.F., G.G., E.A., M.S., and M.R. co-wrote the manuscript with input from all authors.
Peer review information
Nature Communications thanks Ole Jensen and the other anonymous reviewer(s) for their contribution to the peer review this work. Peer reviewer reports are available.
Data availability
Publicly accessible databases and datasets used in this study are as follows and in the order they are referenced in the Methods section. RNA-Seq data analysis: C. elegans reference genome (Genome assembly WBcel235). Tissue-specific correlations of human genes with GRHL1: GTEx database (v8) data for tissue-specific gene TPMs (version 2017-06-05_v8_RNASeQCv1.1.9). GSEA of human genes correlating with GRHL1: Gene set assembly Human_GO_AllPathways_with_GO_iea_March_01_2021_symbol.gmt retrieved from [download.baderlab.org/EM_Genesets/current_release/Human/symbol]. BXD mice data analysis: Murine Grhl1 expression data and phenotypic traits were obtained from GeneNetwork2 [www.genenetwork.org]. Accession codes of gene expression datasets analyzed (accessed on 06.03.2021—detailed retrieval procedure can be found here: https://github.com/araldi/Grigolon-et-al_GRHL1_NatureCommunications/blob/main/BXD/1_Download_BXD_datasets.ipynb): EPFLMouseMuscleCDRMAEx1112 and EPFLMouseMuscleCDRMA1211 (muscle chow diet), EPFLMouseMuscleHFDRMAEx1112, EPFLMouseMuscleHFDRMA1211 (muscle high fat diet), EPFLMouseLiverCDEx0413, EPFLMouseLiverCDRMA0413, EPFLMouseLiverCDRMA0818 (liver chow diet); EPFLMouseLiverHFDRMA0818, EPFLMouseLiverHFDRMA0413, EPFLMouseLiverHFCEx0413 (liver high fat diet). Phenotypic traits analyzed (accessed on 06.03.2021—detailed retrieval procedure can be found here: https://github.com/araldi/Grigolon-et-al_GRHL1_NatureCommunications/blob/main/BXD/1_Download_BXD_datasets.ipynb): 21450 (Lifespan); 17661, 17662 (glycemia during oGTT - AUC); 17663, 176634 (insulin during oGTT - AUC); 17603, 17604 (body weight percentage gain 8–28 weeks). GTEX and FUSION dataset analysis: Common Fund (CF) Genotype-Tissue Expression Project (GTEx) (phs000424.v8.p2) and The Finland-United States Investigation of NIDDM Genetics (FUSION) Tissue Biopsy Study (phs001048.v2.p1) datasets were obtained from dbGaP (request #28650: “Population genetics of human aging”). Mass spectrometry data analysis: UniProtKB/Swiss-Prot Homo sapiens proteome database (taxonomy 9606, release 2018_09 [https://ftp.uniprot.org/pub/databases/uniprot/previous_major_releases/release-2018_09/knowledgebase/]). The RNA-Seq data generated in this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) database under GEO Series accession number GSE159077. The mass spectrometry proteomics data generated in this study have been deposited to the ProteomeXchange (PX) Consortium via the PRIDE database under dataset identifiers PXD021768 and PXD021808. All other data supporting the findings of this study are available within this paper, its Supplementary Information, and its Supplementary Files. Source data are provided with this paper.
Code availability
The codes used for the analyses of BXD, FUSION, and GTEX data are available at https://github.com/araldi/Grigolon-et-al_GRHL1_NatureCommunications.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Giovanna Grigolon, Fabian Fischer.
These authors jointly supervised this work: Michael Ristow, Fabian Fischer.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-27732-4.
References
- 1.McCay CM. Retarded growth lifespan, ultimate body size, and age changes in the albino rat after feeding diets restricted in calories. J. Nutr. 1937;18:1–13. doi: 10.1111/j.1753-4887.1975.tb05227.x. [DOI] [PubMed] [Google Scholar]
- 2.Klass MR. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech. Ageing Dev. 1983;22:279–286. doi: 10.1016/0047-6374(83)90082-9. [DOI] [PubMed] [Google Scholar]
- 3.Barardo D, et al. The DrugAge database of aging-related drugs. Aging Cell. 2017;16:594–597. doi: 10.1111/acel.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finkel T. The metabolic regulation of aging. Nat. Med. 2015;21:1416–1423. doi: 10.1038/nm.3998. [DOI] [PubMed] [Google Scholar]
- 5.Kubben N, Misteli T. Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol. 2017;18:595–609. doi: 10.1038/nrm.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssens GE, Houtkooper RH. Identification of longevity compounds with minimized probabilities of side effects. Biogerontology. 2020;21:709–719. doi: 10.1007/s10522-020-09887-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaeberlein M, Rabinovitch PS, Martin GM. Healthy aging: the ultimate preventative medicine. Science. 2015;350:1191–1193. doi: 10.1126/science.aad3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Booth LN, Brunet A. The aging epigenome. Mol. Cell. 2016;62:728–744. doi: 10.1016/j.molcel.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol. Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 11.de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011;12:833–845. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- 12.Martinez Corrales G, Alic N. Evolutionary conservation of transcription factors affecting longevity. Trends Genet. 2020;36:373–382. doi: 10.1016/j.tig.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Martins R, Lithgow GJ, Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016;15:196–207. doi: 10.1111/acel.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis KN, Mele J, Hayes JD, Buffenstein R. Nrf2, a guardian of healthspan and gatekeeper of species longevity. Integr. Comp. Biol. 2010;50:829–843. doi: 10.1093/icb/icq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatesan K, McManus HR, Mello CC, Smith TF, Hansen U. Functional conservation between members of an ancient duplicated transcription factor family, LSF/Grainyhead. Nucleic Acids Res. 2003;31:4304–4316. doi: 10.1093/nar/gkg644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ming Q, et al. Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family. Nucleic Acids Res. 2018;46:2082–2095. doi: 10.1093/nar/gkx1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozanov L, et al. Redox-mediated regulation of aging and healthspan by an evolutionarily conserved transcription factor HLH-2/Tcf3/E2A. Redox Biol. 2020;32:101448. doi: 10.1016/j.redox.2020.101448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruderman NB, Kapeller R, White MF, Cantley LC. Activation of phosphatidylinositol 3-kinase by insulin. Proc. Natl Acad. Sci. USA. 1990;87:1411–1415. doi: 10.1073/pnas.87.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreux PA, et al. Systems Genetics of Metabolism: the Use of the BXD Murine Reference Panel for Multiscalar Integration of Traits. Cell. 2012;150:1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.C. elegans Deletion Mutant Consortium. Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G32, 1415–1425 (2012). [DOI] [PMC free article] [PubMed]
- 21.Shivers RP, et al. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 2010;6:e1000892. doi: 10.1371/journal.pgen.1000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolz DD, Tenor JL, Aballay A. A conserved PMK-1/p38 MAPK is required in C. elegans tissue-specific immune response to Y. pestis infection. J. Biol. Chem. 2010;285:10832–10840. doi: 10.1074/jbc.M109.091629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabian J, et al. GRHL1 acts as tumor suppressor in neuroblastoma and is negatively regulated by MYCN and HDAC3. Cancer Res. 2014;74:2604–2616. doi: 10.1158/0008-5472.CAN-13-1904. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharm. Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 25.Thimmulappa RK, et al. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 26.Jacobs J, et al. The transcription factor Grainy head primes epithelial enhancers for spatiotemporal activation by displacing nucleosomes. Nat. Genet. 2018;50:1011–1020. doi: 10.1038/s41588-018-0140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huttlin EL, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cassandri M, et al. Zinc-finger proteins in health and disease. Cell Death Disco. 2017;3:17071. doi: 10.1038/cddiscovery.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H, et al. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 2013;12:260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 30.Huang J, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 31.MacFawn I, et al. Grainyhead-like-2 confers NK-sensitivity through interactions with epigenetic modifiers. Mol. Immunol. 2019;105:137–149. doi: 10.1016/j.molimm.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toska E, et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science. 2017;355:1324–1330. doi: 10.1126/science.aah6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao G, et al. Lysine-specific demethylase 1 mediates epidermal growth factor signaling to promote cell migration in ovarian cancer cells. Sci. Rep. 2015;5:15344. doi: 10.1038/srep15344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H, Li LJ, Zhang HB, Wei AY. Papaverine selectively inhibits human prostate cancer cell (PC-3) growth by inducing mitochondrial mediated apoptosis, cell cycle arrest and downregulation of NF-kappaB/PI3K/Akt signalling pathway. J. BUON. 2017;22:112–118. [PubMed] [Google Scholar]
- 35.Hopkins BD, Goncalves MD, Cantley LC. Insulin-PI3K signalling: an evolutionarily insulated metabolic driver of cancer. Nat. Rev. Endocrinol. 2020;16:276–283. doi: 10.1038/s41574-020-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang RA. C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 38.Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zarse K, et al. Impaired insulin/IGF1-signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.TEx G. Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valle T, et al. Mapping genes for NIDDM. Design of the Finland-United States Investigation of NIDDM Genetics (FUSION) Study. Diabetes Care. 1998;21:949–958. doi: 10.2337/diacare.21.6.949. [DOI] [PubMed] [Google Scholar]
- 42.Scott LJ, et al. The genetic regulatory signature of type 2 diabetes in human skeletal muscle. Nat. Commun. 2016;7:11764. doi: 10.1038/ncomms11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Auden A, et al. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr. Patterns: GEP. 2006;6:964–970. doi: 10.1016/j.modgep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Wang S, Samakovlis C. Grainy head and its target genes in epithelial morphogenesis and wound healing. Curr. Top. Dev. Biol. 2012;98:35–63. doi: 10.1016/B978-0-12-386499-4.00002-1. [DOI] [PubMed] [Google Scholar]
- 45.Mlacki M, Kikulska A, Krzywinska E, Pawlak M, Wilanowski T. Recent discoveries concerning the involvement of transcription factors from the Grainyhead-like family in cancer. Exp. Biol. Med. 2015;240:1396–1401. doi: 10.1177/1535370215588924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Menegon S, Columbano A, Giordano S. The dual roles of NRF2 in cancer. Trends Mol. Med. 2016;22:578–593. doi: 10.1016/j.molmed.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 47.McIntyre RL, Daniels EG, Molenaars M, Houtkooper RH, Janssens GE. From molecular promise to preclinical results: HDAC inhibitors in the race for healthy aging drugs. EMBO Mol. Med. 2019;11:e9854. doi: 10.15252/emmm.201809854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thatikonda S, Pooladanda V, Sigalapalli DK, Godugu C. Piperlongumine regulates epigenetic modulation and alleviates psoriasis-like skin inflammation via inhibition of hyperproliferation and inflammation. Cell Death Dis. 2020;11:21. doi: 10.1038/s41419-019-2212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, et al. Discovery of piperlongumine as a potential novel lead for the development of senolytic agents. Aging. 2016;8:2915–2926. doi: 10.18632/aging.101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boswell-Smith V, Spina D, Page CP. Phosphodiesterase inhibitors. Br. J. Pharm. 2006;147(Suppl 1):S252–S257. doi: 10.1038/sj.bjp.0706495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaczmarek L, Hyland JK, Watt R, Rosenberg M, Baserga R. Microinjected c-myc as a competence factor. Science. 1985;228:1313–1315. doi: 10.1126/science.4001943. [DOI] [PubMed] [Google Scholar]
- 52.Hofmann JW, et al. Reduced expression of MYC increases longevity and enhances healthspan. Cell. 2015;160:477–488. doi: 10.1016/j.cell.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Werner CWO. Über Katarakt in Verbindung mit Sklerodermie (Schmidt & Klaunig, 1904).
- 54.Matsumoto T, Shimamoto A, Goto M, Furuichi Y. Impaired nuclear localization of defective DNA helicases in Werner’s syndrome. Nat. Genet. 1997;16:335–336. doi: 10.1038/ng0897-335. [DOI] [PubMed] [Google Scholar]
- 55.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 57.van Heemst D. Insulin, IGF-1 and longevity. Aging Dis. 2010;1:147–157. [PMC free article] [PubMed] [Google Scholar]
- 58.Merry TL, et al. Partial impairment of insulin receptor expression mimics fasting to prevent diet-induced fatty liver disease. Nat. Commun. 2020;11:2080. doi: 10.1038/s41467-020-15623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rask-Madsen C, Kahn CR. Tissue-specific insulin signaling, metabolic syndrome, and cardiovascular disease. Arterioscler Thromb. Vasc. Biol. 2012;32:2052–2059. doi: 10.1161/ATVBAHA.111.241919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zarse K, et al. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Segref A, Hoppe T. Analysis of ubiquitin-dependent proteolysis in Caenorhabditis elegans. Methods Mol. Biol. 2012;832:531–544. doi: 10.1007/978-1-61779-474-2_38. [DOI] [PubMed] [Google Scholar]
- 64.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hatakeyama M, et al. SUSHI: an exquisite recipe for fully documented, reproducible and reusable NGS data analysis. BMC Bioinforma. 2016;17:228. doi: 10.1186/s12859-016-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wingett SW, Andrews S. FastQ Screen: a tool for multi-genome mapping and quality control. F1000Res. 2018;7:1338. doi: 10.12688/f1000research.15931.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 72.Tan G, Lenhard B. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics. 2016;32:1555–1556. doi: 10.1093/bioinformatics/btw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ravichandran M, et al. Impairing L-threonine catabolism promotes healthspan through methylglyoxal-mediated proteohormesis. Cell Metab. 2018;27:914–925. doi: 10.1016/j.cmet.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 76.Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G. & Ahringer, J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002 (2001). [DOI] [PMC free article] [PubMed]
- 77.Schulz TJ, et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 78.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hahm JH, et al. C. elegans maximum velocity correlates with healthspan and is maintained in worms with an insulin receptor mutation. Nat. Commun. 2015;6:8919. doi: 10.1038/ncomms9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hubner NC, et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 82.UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Publicly accessible databases and datasets used in this study are as follows and in the order they are referenced in the Methods section. RNA-Seq data analysis: C. elegans reference genome (Genome assembly WBcel235). Tissue-specific correlations of human genes with GRHL1: GTEx database (v8) data for tissue-specific gene TPMs (version 2017-06-05_v8_RNASeQCv1.1.9). GSEA of human genes correlating with GRHL1: Gene set assembly Human_GO_AllPathways_with_GO_iea_March_01_2021_symbol.gmt retrieved from [download.baderlab.org/EM_Genesets/current_release/Human/symbol]. BXD mice data analysis: Murine Grhl1 expression data and phenotypic traits were obtained from GeneNetwork2 [www.genenetwork.org]. Accession codes of gene expression datasets analyzed (accessed on 06.03.2021—detailed retrieval procedure can be found here: https://github.com/araldi/Grigolon-et-al_GRHL1_NatureCommunications/blob/main/BXD/1_Download_BXD_datasets.ipynb): EPFLMouseMuscleCDRMAEx1112 and EPFLMouseMuscleCDRMA1211 (muscle chow diet), EPFLMouseMuscleHFDRMAEx1112, EPFLMouseMuscleHFDRMA1211 (muscle high fat diet), EPFLMouseLiverCDEx0413, EPFLMouseLiverCDRMA0413, EPFLMouseLiverCDRMA0818 (liver chow diet); EPFLMouseLiverHFDRMA0818, EPFLMouseLiverHFDRMA0413, EPFLMouseLiverHFCEx0413 (liver high fat diet). Phenotypic traits analyzed (accessed on 06.03.2021—detailed retrieval procedure can be found here: https://github.com/araldi/Grigolon-et-al_GRHL1_NatureCommunications/blob/main/BXD/1_Download_BXD_datasets.ipynb): 21450 (Lifespan); 17661, 17662 (glycemia during oGTT - AUC); 17663, 176634 (insulin during oGTT - AUC); 17603, 17604 (body weight percentage gain 8–28 weeks). GTEX and FUSION dataset analysis: Common Fund (CF) Genotype-Tissue Expression Project (GTEx) (phs000424.v8.p2) and The Finland-United States Investigation of NIDDM Genetics (FUSION) Tissue Biopsy Study (phs001048.v2.p1) datasets were obtained from dbGaP (request #28650: “Population genetics of human aging”). Mass spectrometry data analysis: UniProtKB/Swiss-Prot Homo sapiens proteome database (taxonomy 9606, release 2018_09 [https://ftp.uniprot.org/pub/databases/uniprot/previous_major_releases/release-2018_09/knowledgebase/]). The RNA-Seq data generated in this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) database under GEO Series accession number GSE159077. The mass spectrometry proteomics data generated in this study have been deposited to the ProteomeXchange (PX) Consortium via the PRIDE database under dataset identifiers PXD021768 and PXD021808. All other data supporting the findings of this study are available within this paper, its Supplementary Information, and its Supplementary Files. Source data are provided with this paper.
The codes used for the analyses of BXD, FUSION, and GTEX data are available at https://github.com/araldi/Grigolon-et-al_GRHL1_NatureCommunications.