Abstract
The origin of the eukaryotic cell is a major open question in biology. Asgard archaea are the closest known prokaryotic relatives of eukaryotes, and their genomes encode various eukaryotic signature proteins, indicating some elements of cellular complexity prior to the emergence of the first eukaryotic cell. Yet, microscopic evidence to demonstrate the cellular structure of uncultivated Asgard archaea in the environment is thus far lacking. We used primer-free sequencing to retrieve 715 almost full-length Loki- and Heimdallarchaeota 16S rRNA sequences and designed novel oligonucleotide probes to visualize their cells in marine sediments (Aarhus Bay, Denmark) using catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH). Super-resolution microscopy revealed 1–2 µm large, coccoid cells, sometimes occurring as aggregates. Remarkably, the DNA staining was spatially separated from ribosome-originated FISH signals by 50–280 nm. This suggests that the genomic material is condensed and spatially distinct in a particular location and could indicate compartmentalization or membrane invagination in Asgard archaeal cells.
Subject terms: Soil microbiology, Microbial ecology, Archaeal physiology
Introduction
The origin of the eukaryotic cell is a major unresolved puzzle in the history of life. Several lines of evidence suggest that a merger between an archaeal host [1] and an Alphaproteobacteria-related symbiont [2] constituted a key event in the evolution of the eukaryotic cell. The archaeal host was likely an ancestral Asgard archaeon, as recent phylogenomic analyses showed that the Asgard archaea superphylum (e.g., Loki-, Thor-, Odin-, Heimdall-, and Helarchaeota) comprises the closest known extant prokaryotic relatives of eukaryotes [1, 3, 4]. Genomes of Asgard archaea are also enriched in eukaryotic signature proteins (ESPs) that are homologous to eukaryotic proteins involved in membrane trafficking, vesicle formation and/or transportation, protein ubiquitinylation, and cytoskeleton formation [1, 3, 4]. The presence of these ESPs suggests that the archaeal host already possessed some building blocks of cellular complexity before the first eukaryotic cell emerged. Microscopic investigations of the first cultured Lokiarchaeon “Candidatus Prometheoarchaeum syntrophicum” strain MK-D1 revealed thin, sometimes branched, membrane protrusions with cytosolic connection but no visible intracellular membrane structures [5]. This provided the first glimpse into the cell biology of Lokiarchaeota. However, Asgard archaea are highly diverse [6] and the exact branching point of eukaryotes within the superphylum is still uncertain [1, 7]. Therefore, visualization of Asgard archaeal cells in the environment is essential for a comprehensive understanding of their cellular structure and morphological diversity. Before their metagenomic identification, when Lokiarchaeota were known as Marine Benthic Group B, Knittel and colleagues visualized their cells in marine sediments [8] yet methodical limitations did not allow to discern single cells and to draw any in-depth conclusion on their morphology. A recent study suggested that Loki- and Heimdallarchaeota cells from brackish lake sediment had highly diverse morphologies and cell sizes up to 12 µm, allegedly with condensed DNA [9].
Here, we visualize Loki- and Heimdallarchaeota cells from marine sediments (Aarhus Bay, Denmark) by catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) and super-resolution microscopy. We captured the 16S rRNA sequence diversity of these two phyla using a recently established primer-free rRNA sequencing method [10]. Asgard archaeal 16S rRNA sequence diversity is poorly covered by common 16S rRNA gene sequence primer sets [10]. The primer-free approach therefore enabled us to obtain almost full-length 16S rRNA sequences of a diverse range of Loki- and Heimdallarchaeota populating the sediments and to design two specific oligonucleotide probes with high coverage for each of the two phyla. This allowed unambiguous visualization of Loki- and Heimdallarchaeota cells using dual-probe hybridizations. Our results invariably show 1–2 µm large, coccoid cells with spatially separated DNA and riboplasm, suggesting a potential for compartmentalization or membrane invagination in Asgard archaeal cells.
Results and discussion
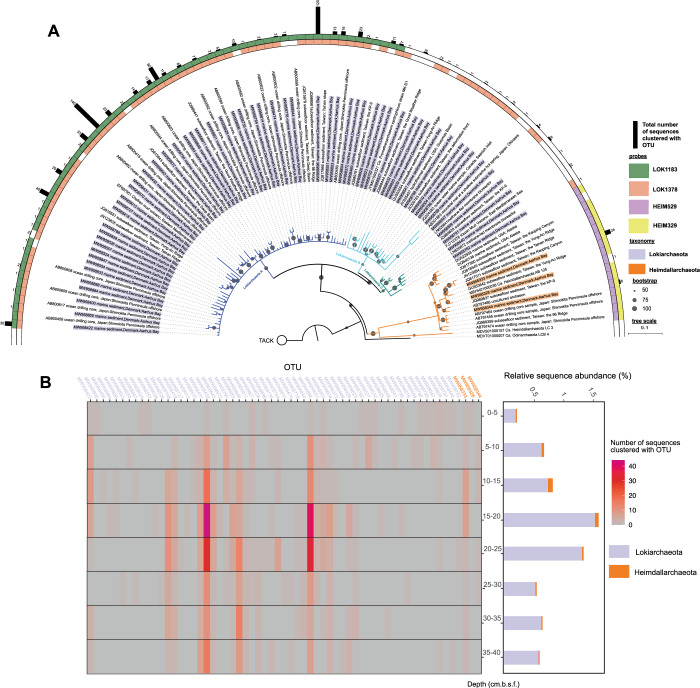
We retrieved 684 Lokiarchaeota and 31 Heimdallarchaeota near-full-length 16S rRNA sequences from sequence libraries generated from sediment sampled at 27 m water depth in 5 cm intervals between 0 and 40 cm below seafloor (cm.b.s.f) in Aarhus Bay (Supplementary Information). The maximum relative read abundance of Lokiarchaeota was 1.6% at 15–20 cm.b.s.f. and 0.1% for Heimdallarchaeota at 10–15 cm.b.s.f. (Fig. 1). The sequences were grouped into 58 Loki- and 3 Heimdallarchaeota operational taxonomic units (OTUs) using a 98% sequence identity threshold and formed three distinct Lokiarchaeota clades and one monophyletic Heimdallarchaeota cluster (Fig. 1). The primer-free sequencing of RNA extracts enabled us to broadly sample the Asgard archaeal diversity in Aarhus Bay sediments and provided a solid database to design oligonucleotide probes for their visualization.
Fig. 1. Phylogenetic analysis and depth distribution of Loki- and Heimdallarchaeota 16S rRNA sequences from Aarhus Bay sediments.
A Maximum likelihood phylogeny of Loki- and Heimdallarchaeota operational taxonomic units (OTUs) and related sequences selected from the SILVA database (v. 132). Specificities of FISH probes and the number of sequences constituting each OTU are also depicted. TACK archaea were selected as outgroup. Bar: 0.1 substitutions per nucleotide position. B Heatmap and relative abundances of Loki- and Heimdallarchaeota sequences at different sediment depths.
Based on the newly retrieved full-length sequences, we designed four novel oligonucleotide probes specifically targeting Loki- and Heimdallarchaeota 16S rRNA with high coverage (Fig. 1, Supplementary Table 1). Probe LOK1183 targets almost all sequences in Lokiarchaeota Clade A, which contains 92% of the retrieved Lokiarchaeota sequences from Aarhus Bay sediments, while probe LOK1378 targets 85% of the sequences in all three Lokiarchaeota clades. Probe HEIM329 and HEIM529 each target >97% of the retrieved Heimdallarchaeota sequences. All designed probes cover >89% sequences in their target groups in the SILVA database (v. 132). The two Lokiarchaeota probes match 5 and 10 different non-target sequences in the SILVA database (v. 132), respectively, while the Heimdallarchaeota probes have no match outside their target group. The broad coverage and high specificity suggest that our probes can also be used to detect Loki- and Heimdallarchaeota in other habitats. Furthermore, designing two probes for each phylum enabled us to identify Lokiarchaeota clade A and Heimdallarchaeota cells via double hybridizations with two distinct dyes and thus confidently distinguish true- and false-positive signals (Supplementary Fig. 1). The general archaeal probe ARC915 also targets Lokiarchaeota and thereby provided yet another control for specific hybridization of the two Lokiarchaeota-specific probes, while the non-sense probe NON338 served as the negative control. We also designed competitor probes to minimize the theoretical false-positive hybridizations with the most frequent one and two mismatches [11] in the SILVA database (v. 132) and helper probes to facilitate probe binding [12]. This comprehensive experimental design with appropriate controls enabled reliable detection of low-abundant Loki- and Heimdallarchaeota cells in Aarhus Bay sediments.
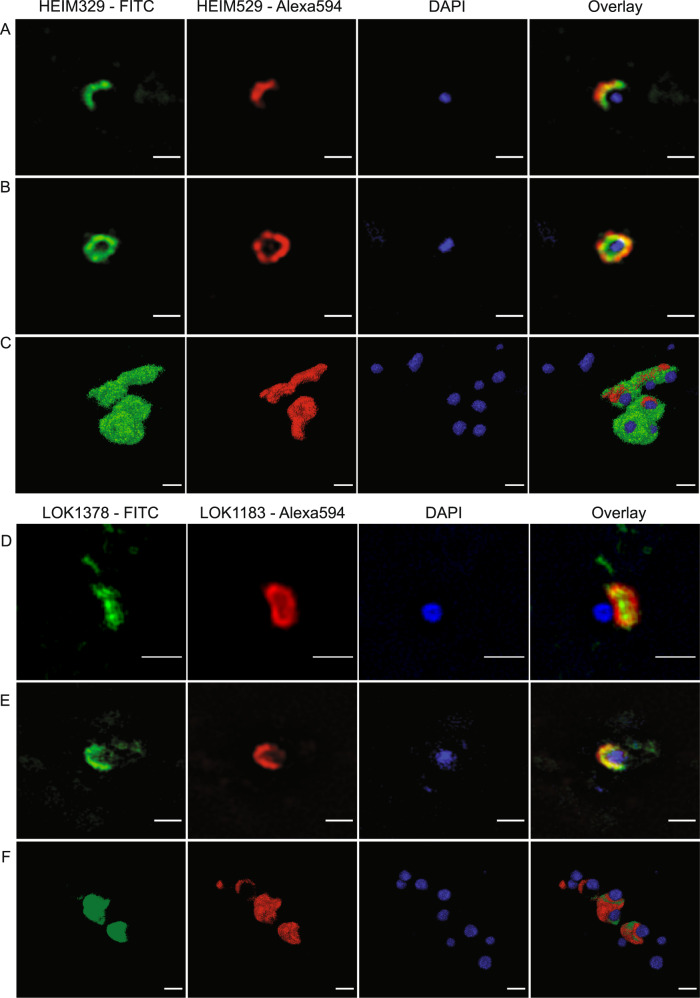
We used both confocal laser scanning microscopy (CLSM) and three-dimensional super-resolution structured illumination (SR-SIM) microscopy for detailed imaging of dual-labeled Loki- and Heimdallarchaeota signals. Loki- and Heimdallarchaeota cells featured coccoid shapes and often formed clusters (Fig. 2) (Supplementary Fig. 2). Based on SR-SIM imaging, Lokiarchaeota cells (n = 18) were 1.27 ± 0.24 µm in diameter and 1.43 ± 0.25 µm in length, while the width and the length of Heimdallarchaeota cells (n = 11) were 1.30 ± 0.20 µm and 1.37 ± 0.21 µm, respectively (Supplementary Table 2). In addition, we observed a few large (>3 µm) ovoid and filamentous cells, resembling some of the Lokiarchaeota morphotypes reported from lake sediment [9]; however, we never detected these cell types in double hybridizations with two probes (Supplementary Fig. 1P–R), and therefore consider them false-positives.
Fig. 2. Visualization of Loki- and Heimdallarchaeota cells in Aarhus Bay sediments by CARD-FISH.
Probe names and the dyes are indicated for each panel. Representative cell morphotypes were imaged in a super-resolution structured illumination microscope (SR-SIM; panels (A), (B), (D), (E)) and confocal laser scanning microscope (CLSM; panels (C) and (F)). For SR-SIM images, single slices from the center of the focal plane are shown. For CLSM images, three-dimensional (3D) surface reconstructions are depicted. All z-stack images taken in CLSM are included in Supplementary Fig. 2. 360° rotation of 3D reconstructed images are also provided in Supplementary Video. Negative and positive controls are shown in Supplementary Fig. 1 together with large ovoid and filamentous false-positive signals. Images are representative of dual labeled Lokiarchaeota (n = 72) and Heimdallarchaeota (n = 70) cells in five individual experiments using two different sediment cores taken from the same sampling site. The scale bar is 1 µm.
The DNA stain (4′,6-diamidino-2-phenylindole; DAPI) in the FISH-identified Loki- and Heimdallarchaeota cells was consistently confined to a single spherical central or lateral position (Fig. 2), corroborating the signal pattern suggested for some of the Asgard archaeal cells in lake sediments [9]. Using SR-SIM, we could image a clear gap, which separated the DNA from the ribosome-originated FISH signals with an average width of 0.18 ± 0.07 µm in Heimdallarchaeota and 0.16 ± 0.13 µm in Lokiarchaeota cells (Supplementary Table 2). The spatial separation of DNA and ribosomes in Loki- and Heimdallarchaeota cells represents an unusual observation since DAPI and FISH signals generally overlap partially or completely in prokaryotic cells [13]. Accordingly, SR-SIM imaging of benthic bacteria in Aarhus Bay sediments demonstrated the prevalence of this overlapping signal pattern (Supplementary Fig. 3). Also, the separated DNA signal observed in Loki- and Heimdallarchaeota cells appeared different from the condensed DNA formation previously described, for example, in Escherichia coli cells [14] and the Thaumarcheota Cenarcheum symbiosum [15] and Nitrosopumilus maritimus [16]. To corroborate this, we performed SR-SIM imaging of CARD-FISH-labeled E.coli and N. maritimus cells. Although their DNA was condensed in particular cellular locations, their FISH and DAPI signals always overlapped, indicating that their DNA and ribosomes are partially co-localized and not fully separated (Supplementary Fig. 4).
To assess whether the gap between DAPI and FISH signals was indicative of an internal membrane, we tried various dyes to stain membranes of the CARD-FISH-labeled Asgard archaeal cells (Supplementary Information). However, none of these stainings was successful, not even for the outer cell membrane, most likely because cell membranes were disintegrated during the CARD-FISH protocol. We then used wheat germ agglutinin (WGA), a lectin primarily binding to N-acetyl-D-glucosamine but also other glycoconjugates and oligosaccharides [17] to at least be able to visualize the surfaces of Loki- and Heimdallarchaeota cells. WGA consistently decorated a cell surface that enclosed the proximal FISH and DAPI signals, suggesting that both signals originated from the same single cell (Supplementary Fig. 5). The WGA staining also demonstrated extracellular structures connected to some Heimdallarchaeota cells (Supplementary Fig. 5). These structures appear different than the membrane protrusions in the first cultured Lokiarchaeon “Ca. P. syntrophicum”, which has a considerably smaller cell size (550 nm in diameter) and does not possess the separated DNA and ribosome signals [5]. Our observations therefore indicate diverse cellular organizations and morphotypes within Asgard archaea superphylum.
Our combined results suggest that genomic material is condensed and spatially distinct from the riboplasm within the detected Loki- and Heimdallarchaeota cells. Considering the anticipated role of Asgard archaea in eukaryogenesis, in particular the presence of ESPs potentially involved in dynamic cytoskeleton formation [18] and membrane remodeling [4, 19], the separation of DNA- and ribosome-derived signals might be indicative of cellular compartmentalization. Alternatively, the observed pattern could be the result of a membrane invagination to form a nucleoid region, similar to membrane organizations for example in Planctomycetes cells [20] or Atribacter laminatus [21].
Our study demonstrates the first visualization of diverse Loki- and Heimdallarchaeota cells in the marine environment and provides a protocol for reliable in situ imaging of rare microorganisms in environmental samples. Future research should address the ultrastructure of Asgard archaeal cells using electron microscopy. This would help to elucidate the cell biology of Asgard archaea and provide insights into the emergence of subcellular complexity of the eukaryotic cell.
Supplementary information
Acknowledgements
We thank Ronny M. Baaske, Britta Poulsen, and Susanne Nielsen for excellent technical support; the captain and crew of RV Aurora for help during sediment sampling; Rikke Louise Meyer for providing access to the CLSM; Rosa Groth and Lærke U. Mortensen for their initial work on probe optimization. Martin Könneke and Evgenios Bouzetos are gratefully acknowledged for respectively providing N. maritimus and E.coli cultures. This work was supported by VILLUM Experiment grant (#17621) “FISHing for the ancestors of the eukaryotic cell” to KUK and AS, VILLUM Research grant (#15510) to MA, and by grants from European Research Council (ERC consolidator grant 817834), the Dutch Research Council (NWO-VICI grant VI.C.192.016), and the Moore–Simons Project on the Origin of the Eukaryotic Cell (Simons Foundation 735925LPI) to TJGE. BA dedicates this study to the memory of his beloved father Süreyya Avcı.
Author contributions
BA, AS, and KUK designed the study. BA and KUK carried out sediment sampling and RNA extraction. JB and MA performed 16S rRNA sequencing. BA and TJGE did phylogenetic analysis. BA performed probe design and CARD-FISH experiments. BA and AS carried out CLSM analysis. NE and DN did SR-SIM measurements and WGA staining. BA, TJGE, AS, and KUK wrote the manuscript, and all authors edited and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Burak Avcı, Email: burak.avci@wur.nl.
Andreas Schramm, Email: andreas.schramm@bio.au.dk.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01098-3.
References
- 1.Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG. Archaea and the origin of eukaryotes. Nat Rev Microbiol. 2017;15:711–23. doi: 10.1038/nrmicro.2017.133. [DOI] [PubMed] [Google Scholar]
- 2.Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature. 2018;557:101–5. doi: 10.1038/s41586-018-0059-5. [DOI] [PubMed] [Google Scholar]
- 3.Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–9. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–8. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 5.Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, et al. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature. 2020;577:519–25. doi: 10.1038/s41586-019-1916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin X, Cai M, Liu Y, Zhou G, Richter-Heitmann T, Aromokeye DA, et al. Subgroup level differences of physiological activities in marine Lokiarchaeota. ISME J. 2021;15:848–61. doi: 10.1038/s41396-020-00818-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Makarova KS, Huang W-C, Wolf YI, Nikolskaya AN, Zhang X, et al. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature. 2021;593:553–7. doi: 10.1038/s41586-021-03494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knittel K, Lösekann T, Boetius A, Kort R, Amann R. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol. 2005;71:467–79. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salcher MM, Andrei AŞ, Bulzu PA, Keresztes ZG, Banciu HL, Ghai R. Visualization of Lokiarchaeia and Heimdallarchaeia (Asgardarchaeota) by fluorescence in situ hybridization and catalyzed reporter deposition (CARD-FISH) mSphere. 2020;5:e00686–20. doi: 10.1128/mSphere.00686-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karst SM, Dueholm MS, McIlroy SJ, Kirkegaard RH, Nielsen PH, Albertsen M. Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat Biotechnol. 2018;36:190–5. doi: 10.1038/nbt.4045. [DOI] [PubMed] [Google Scholar]
- 11.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. doi: 10.1016/S0723-2020(11)80121-9. [DOI] [Google Scholar]
- 12.Fuchs BM, Glöckner FO, Wulf J, Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 2000;66:3603–7. doi: 10.1128/AEM.66.8.3603-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLong EF, Taylor LT, Marsh TL, Preston CM. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–63. doi: 10.1128/AEM.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chai Q, Singh B, Peisker K, Metzendorf N, Ge X, Dasgupta S, et al. Organization of ribosomes and nucleoids in Escherichia coli cells during growth and in quiescence. J Biol Chem. 2014;289:11342–52. doi: 10.1074/jbc.M114.557348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preston CM, Wu KY, Molinski TF, DeLong EF. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–6. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 17.Chazotte B Labeling membrane glycoproteins or glycolipids with fluorescent wheat germ agglutinin. Cold Spring Harb Protoc. 2011;2011:pdb.prot5623. [DOI] [PubMed]
- 18.Akıl C, Robinson RC. Genomes of Asgard archaea encode profilins that regulate actin. Nature. 2018;562:439–43. doi: 10.1038/s41586-018-0548-6. [DOI] [PubMed] [Google Scholar]
- 19.Neveu E, Khalifeh D, Salamin N, Fasshauer D. Prototypic SNARE proteins are encoded in the genomes of Heimdallarchaeota, potentially bridging the gap between the prokaryotes and eukaryotes. Curr Biol. 2020;30:2468–80.e5. doi: 10.1016/j.cub.2020.04.060. [DOI] [PubMed] [Google Scholar]
- 20.Wiegand S, Jogler M, Jogler C. On the maverick Planctomycetes. FEMS Microbiol Rev. 2018;42:739–60. doi: 10.1093/femsre/fuy029. [DOI] [PubMed] [Google Scholar]
- 21.Katayama T, Nobu MK, Kusada H, Meng X-Y, Hosogi N, Uematsu K, et al. Isolation of a member of the candidate phylum ‘Atribacteria’ reveals a unique cell membrane structure. Nat Commun. 2020;11:6381. doi: 10.1038/s41467-020-20149-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.