Abstract
Background:
We investigated the mutational landscape of circulating tumor DNA (ctDNA) in predicting tumor response to first-line treatment in patients with metastatic colorectal cancer (mCRC).
Methods:
We included 41 patients with initially unresectable mCRC, treated with 5-fluorouracil/leucovorin/oxaliplatin (FOLFOX)/5-fluorouracil/leucovorin/irinotecan (FOLFIRI) with/without bevacizumab (Bev)/cetuximab (Cet). Blood samples were prospectively collected at two timepoints: at baseline and after four cycles of first-line treatment. Mutational status of 1086 genes were studied in ctDNA by targeted next-generation sequencing (NGS). Molecular mutational burden (MMB) was defined as mean mutation frequency among obtained mutations for each gene. To evaluate the association between molecular characteristics of cfDNA and therapeutic response better, we divided these patients into MMB-high and MMB-low group according to the median value of MMB (0.3).
Results:
Among the 41 enrolled patients, alterations of six genes (TRIM24, SPEN, RNF43, PRKAR1A, KRAS, and KDM5 C) were found at baseline. Baseline MMB of six genes was significantly lower in partial response (PR)/stable disease (SD) patients than progression disease (PD) patients (p = 0.0012). Further analysis demonstrated that genomic profiling of ctDNA from pretreatment blood samples was significantly different between PR/SD (non-PD) group and PD group. By comparing the baseline levels of KRAS MMB in the two subgroups, we found that PD cases were all MMB-high, whereas non-PD cases were mainly in MMB-low subgroup. Furthermore, patients with low-KRAS MMB had superior response rate, significantly longer progression-free survival (PFS) and longer overall survival (OS) than high-KRAS MMB group.
Conclusions:
This prospective and serial genomic profiling study revealed the utility of ctDNA in predicting clinical outcomes in mCRC patients under first-line treatment. Levels of KRAS MMB might aid in monitoring therapeutic efficacy in mCRC patients at pretreatment/after four cycles of first-line treatment.
Keywords: circulating tumor DNA, KRAS, metastatic colorectal cancer, molecular mutational burden, prognosis
Introduction
Colorectal cancer (CRC) is the third most common cause of cancer death in both men and women in the United States and ranks second when men and women are combined. 1 Combination chemotherapy regimens with a fluoropyrimidine plus oxaliplatin or irinotecan (FOLFOX or FOLFIRI) remain the current standard of care in the first-line treatment of metastatic colorectal cancer (mCRC). 2 Antiepidermal growth factor receptor (EGFR) antibodies combined with fluoropyrimidine-based chemotherapy may provide further clinical benefit in left-sided RAS wild-type cancers. Antivascular endothelial growth factor (VEGF) therapy is a reasonable choice for patients with RAS wild-type right-sided tumors and RAS-mutant tumors. Initially unresectable mCRC means those for whom intensive treatment is appropriate with the goal of cytoreduction (tumor shrinkage) and conversion to resectable disease; or those who need intensive treatment, although they will never make it to resection or local and ablative treatment (LAT), since they need a rapid reduction in tumor burden owing to impending clinical threat, organ dysfunction or severe symptoms, and those for whom intensive treatment is not necessary and where the goal is disease control. 3 CRC is a lethal disease with heterogeneous outcomes and drug responses. There is an urgent need to explore predictive biomarkers that can aid in diagnostic and therapeutic decision-making.
Cell-free DNA (cfDNA), which is released into the bloodstream by apoptotic or necrotic tumor cells, refers to fragmented DNA found in the noncellular component of the blood. 4 Analysis of cfDNA provides a more sensitive method of detecting malignancies than imaging or other conventional approaches. Clinical applications of cfDNA can be exploited in several ways, 5 such as early detection of colorectal cancer prior to the emergence of clinical symptoms or radiological manifestations and in the assessment of minimal residual disease (MRD). The application of cfDNA also seems to assist in tailoring therapy intensity in the neoadjuvant/adjuvant settings and in monitoring tumor response in palliative treatment.6–9 Furthermore, alterations of cfDNA might aid in tracking tumor heterogeneity and clonal evolution that lead to the emergency of resistance as well as in guiding the most appropriate therapies.10,11 Cell-free DNA from plasma is emerging as a minimally invasive adjunct to standard tumor biopsies and becoming a valuable approach for molecular testing, new insights into clonal evolution, tumor detection, and monitoring.
Previous studies have revealed that ctDNA levels in plasma might detect residual/recurrent disease, assess treatment response, monitor disease progression, and guide subsequent therapy. An analysis of 230 patients with resected stage II colon cancer showed that circulating tumor DNA (ctDNA) detection after stage II colon cancer resection identified patients at high risk of recurrence and provided direct evidence of residual disease. 12 A prospective multicenter study of 104 patients with locally advanced rectal cancer (LARC) demonstrated that serial ctDNA could predict tumor response to neoadjuvant chemoradiotherapy (nCRT). 13 A post-induction ctDNA analysis following first-line treatment in patients with metastatic colorectal cancer (mCRC) revealed that ctDNA quantification in post-induction plasma may serve as a prognostic biomarker for mCRC post-treatment outcomes. 14 The prognostic/predictive value of cfDNA has been indicated in other types of cancer as well. 15 For example, quantification of somatic variants in cfDNA provided dynamic insights into therapeutic efficacy and disease relapse in patients with small-cell-lung cancer (SCLC). 16 In a population-based prospective study, cfDNA had prognostic implications and contributed to estimating tumor burden, determining the mutational profile and genetic classification in patients with diffuse large B-cell lymphoma (DLBCL). 17
In this study, we aimed to explore the prognostic significance of molecular landscape of ctDNA for first-line therapy in patients with advanced CRC. KRAS molecular mutational burden (MMB) was used to address the correlation between molecular alterations of ctDNA and treatment response.
Materials and methods
Study design
This prospective cohort study was designed and implemented in Ruijin Hospital (Shanghai, China). A total of 41 patients with initially unresectable mCRC were enrolled from January 2019 to December 2020. Their blood samples were sequentially collected at baseline, and after four cycles of first-line therapy together with response evaluation. We analyzed eight initial variables at baseline, including ctDNA content fraction (CCF), tumor mutational burden (TMB), mean variant allele frequency (VAF), maximum VAF, copy number instability (CNI), copy number variant (CNV) burden, copy number gain (CNgain) burden, and copy number loss (CNloss) burden. Patients received four cycles of first-line chemotherapy with/without targeted therapy: FOLFOX or FOLFIRI ± bevacizumab or FOLFOX or FOLFIRI ± cetuximab (KRAS/NRAS/BRAF WT gene and left-sided tumors only). Clinical response and tumor burden were evaluated by the investigators based on Response Evaluation Criteria in Solid Tumors (RECIST) 1.1. All patients have signed the written informed consent for serial tumor genomic profiling. This study was approved by the Ethics Committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, and complied with the Declaration of Helsinki.
Plasma collection for biomarker analysis
Cell-free DNA (cfDNA) was extracted using the MagMAX Cell-Free DNA Isolation (thermo) following manufacturer instructions. Germline DNA was isolated from peripheral blood mononuclear cell (PBMC) using the TIANamp Blood DNA Kit (TIANGEN). DNA concentration was measured using Qubit dsDNA HS Assay kit or Qubit dsDNA BR Assay kit (Life Technologies). At least 10 ng cfDNA was required for DNA libraries preparation. Plasma samples were collected at two timepoints: pretreatment (named T1) and after four cycles of treatment (named T2).
Targeted capture sequencing and genomic data analysis
Genomic DNA was sheared into 150–200 base pairs (bp) fragments. Fragmented DNA libraries were constructed by KAPA HTP Library Preparation Kit (Illumina platforms; KAPA Biosystems) following producer’s instruction.
DNA was hybridized to two designed Genescope panels: (1) 543 genes (Genecast) that included tumor-related major genes, covering 1.7 Mb of the genome, and (2) 773 genes (Genecast) that included tumor-related major genes, covering 1.9 Mb of the genome. The final sequencing libraries were quantified using Qubit dsDNA HS Assay Kit (Thermo Fisher). The captured samples were subjected to Illumina Novaseq6000 for paired end sequencing.
Somatic mutation detection
Sequencing reads were processed using an in-house pipeline that contained Trimmomatic (v0.39) for reads adapter trimming and quality filtering, BWA (0.7.17) for mapping reads to the hg19 reference genome, Picard toolkit (version 2.1.0) for sorting and making duplicates, and Genome Analysis Tool Kit (version 3.7) for reads realignment. VarDict (version 1.5.1) was introduced for single-nucleotide variation (SNV) calling, while compound heterozygous mutations were merged with FreeBayes (version 1.2.0). The generated candidate mutations were annotated using ANNOVAR software tool and then filtered by ExAC, gnomAD, COSMIC, and dbSNP databases. Nonsynonymous mutations at the exonic and splicing regions were kept for the final mutation data set.
Estimation of ctDNA Content Fraction (CCF)
CCF of plasma samples were estimated by a maximum likelihood model based on SNVs and CNVs in the paired blood cell and plasma samples. Somatic and germline SNPs met the following criteria were used to build the model: (1) with a minimum depth of 50× in the paired samples; (2) not on genes with high polymorphism; (3) no InDels in the 50 bp upstream or downstream regions; (4) not in a copy number gain region; and (5) germline SNPs with significantly different variant allele frequencies (VAFs) in the paired samples, or somatic SNPs with VAFs significantly higher than background noise. These SNPs were defined as informative SNPs and clustered into multiple groups according to their VAFs, local copy numbers and hypothetic genotypes. The hypothetic genotypes in ctDNA were determined by the VAFs in the paired samples and the copy number in the plasma sample. Each cluster represents a unique ctDNA source. We then calculated the likelihood of observing SNPs under given CCFs in each cluster. By maximizing the likelihood, CCF of each cluster could therefore be estimated. Cluster with the highest CCF was considered to be from the main source of ctDNA, and its CCF was output as the final estimation.
TMB analysis
For the determination of samples’ tumor mutational burden (TMB) value, nonsynonymous somatic mutations at the exonic and splicing regions with variant frequency no less than 0.7% were quantified. Alterations likely or known to be bona fide oncogenic drivers were excluded. To calculate the TMB per megabase, the total number of mutations counted was divided by the size of the coding region of the targeted panel.
Copy number instability
After correction for GC content and length of target region using proprietary algorithms for each region, the read counts were transformed into log2 ratios and converted into Z-score based on Gaussian transformations versus a normal control group (n = 30). The target regions that satisfied the Z-score greater than the 95th percentile plus twice-times absolute standard deviation of the normal control group were retained, and these Z-score was summed as the CNI score.
Somatic copy number variant (CNV) and CNV burden
After correcting GC content, target region length, read count, the copy number, and gene specificity score (GCS) were calculated using 30 normal blood samples as control. GCS represents the degree of gene level difference between the case samples and control. Joint statistical significance test on GCS and absolute value of copy number together to determine CNV. CNV gain and CNV loss were defined as the CNV >2.5 and CNV <1.5, respectively.
Burden of CNV analysis includes measurements on burden of CNV, copy number gain (CNgain), and copy number loss (CNloss). Burden of CNgain/CNloss was defined as the total number of genes with CNgain/loss in one sample. The burden of CNV was calculated as the total number of genes with copy number gains or losses.
Definition of MMB
Mean variant allele frequencies (VAFs) for each gene was calculated and named as the molecular mutational burden (MMB). A total MMB of a gene set was calculated as the sum of the MMB of each gene.
Survival analysis
Univariate Cox regression analyses were carried out using survival and survminer packages in R. The hazard ratio (HR) and 95% confidence interval (CI) were calculated to identify features associated with progression-free survival and overall survival.
Statistical analysis
Wilcoxon rank sum test and Fisher’s exact test were applied for comparison of continuous variables and categorical variables between PD and non-PD groups, respectively. All diagrams and statistical analyses were done with R packages; p < 0.05 was considered statistically significant.
Results
Patient collection, baseline characteristics, and efficacy
Between January 2019 and December 2020, 41 patients with unresectable mCRC were enrolled. Their plasma samples were prospectively collected at baseline and after four cycles of first-line therapy (Figure 1). The clinical information of included patients at baseline are listed in Table 1. Briefly, 26 (63.41%) patients were male and 18 (43.90%) patients were 65 years or older. The vast majority of patients (35, 85.37%) received oxaliplatin-based first-line treatment.
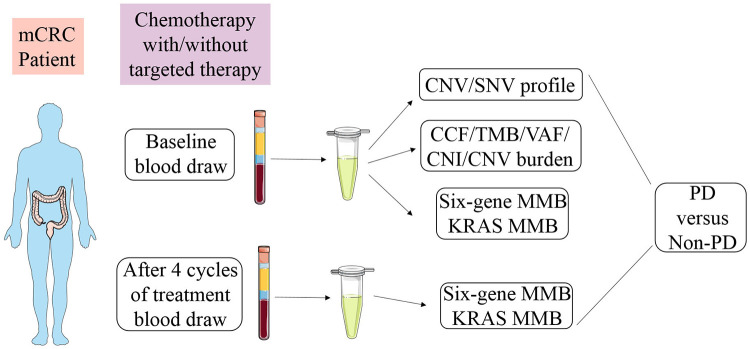
Figure 1.
Study schematic. Pre-treatment blood from mCRC patients receiving oxaliplatin/ irinotecan-based first-line therapy was collected and fractionated for ctDNA analysis. Early on-treatment blood was collected after four cycles of treatment within 2 months for ctDNA monitoring. Mutational parameters were tested for their association with clinical outcomes. Treatment response in advanced CRC patients receiving first-line therapy achieving objective response, partial response (PR), stable disease (SD), or progressive disease (PD) at the first scan by RECIST v1.1 criteria.
Table 1.
Summary of patient characteristics (N = 41).
| Patient characteristic | Total | % | |
|---|---|---|---|
| Age, years | <65 | 23 | 56.10 |
| ⩾65 | 18 | 43.90 | |
| Gender | Male | 26 | 63.41 |
| Female | 15 | 36.59 | |
| Primary tumor site | Right | 12 | 29.27 |
| Left | 14 | 34.15 | |
| Rectum | 14 | 34.15 | |
| Transverse colon | 1 | 2.44 | |
| Extent of metastatic disease | non-LLD | 27 | 65.85 |
| LLD | 14 | 34.15 | |
| First-line regimen | XELOX | 3 | 7.32 |
| FOLFOX | 19 | 46.34 | |
| FOLFOX-Bev | 8 | 19.51 | |
| FOLFOX-Cet | 5 | 12.20 | |
| FOLFIRI | 2 | 4.88 | |
| FOLFIRI-Bev | 3 | 7.32 | |
| FOLFIRI-Cet | 1 | 2.44 | |
| Response rate | PR | 22 | 53.66 |
| SD | 11 | 26.83 | |
| PD | 8 | 19.51 | |
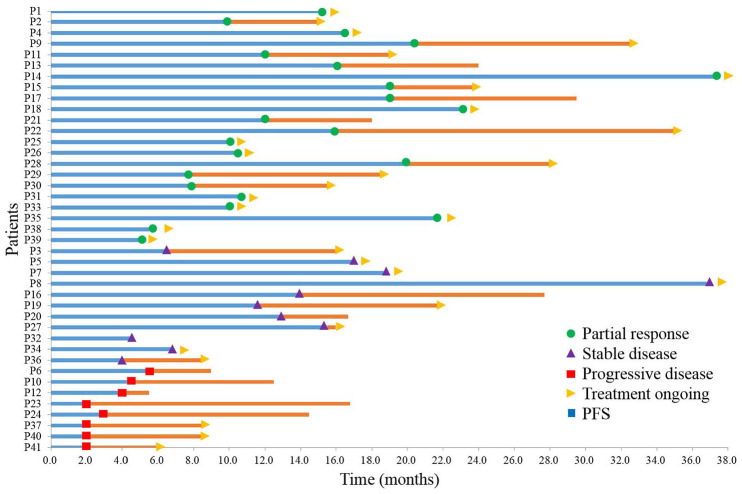
Of the 41 patients, 22 (53.66%) had confirmed partial response (PR), 11 (26.83%) had confirmed stable disease (SD), and 8 (19.51%) had progressive disease (PD) after four cycles of first-line therapy (Figure 2). By the time of data cutoff (March 31, 2021), 30 of 41 patients (73.17%) were still alive and their treatments were still ongoing (Figure 2).
Figure 2.
Clinical course for patients received first-line chemotherapy with or without targeted therapy. Swimming plot depicts duration of treatment, and RECIST v1.1 status at the first scan (green circle = partial response, purple triangles = stable disease, red squares = progressive disease). Time of censoring is shown (yellow triangles = treatment ongoing). Patient 13, 17, 21, 16, 20, 32, 6, 10, 12, 23, and 24 died at the time of the data cutoff. Survival time is depicted as horizontal lines (blue = PFS1, blue plus orange = OS).
Mutational profile in ctDNA from patients with metastatic colorectal cancer
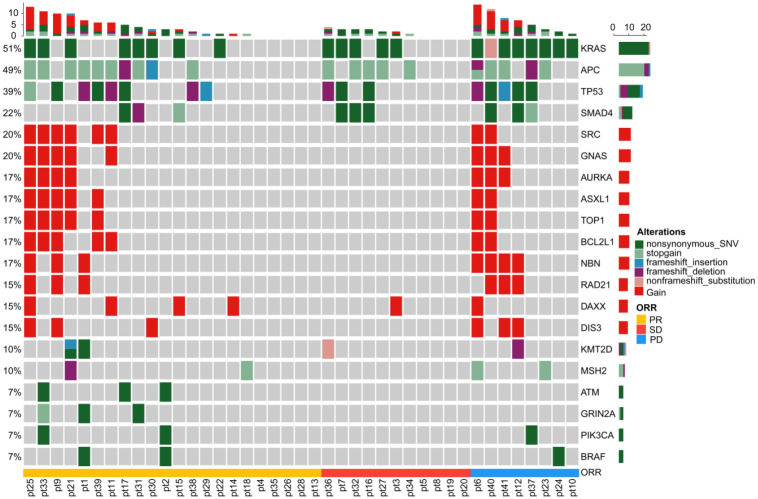
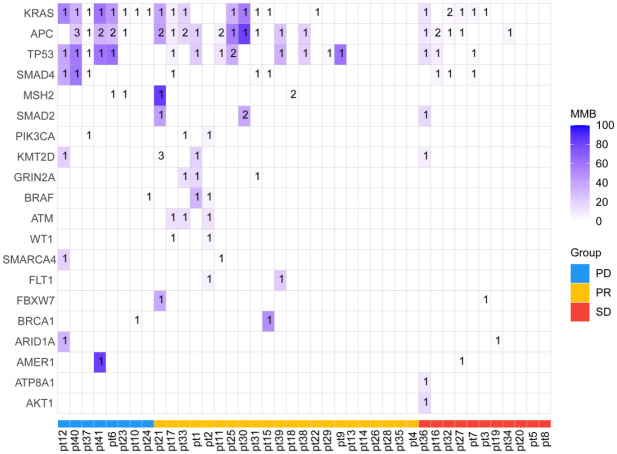
We sequenced genes from a cohort of 41 patients with mCRC based on a panel comprising 1086 genes. The single nucleotide variant (SNV), copy number variant (CNV), mutations, and structural variations of DNA isolated from 41 blood samples were analyzed. The landscape of the detected high-frequency (>5%) molecular alterations in the plasma at baseline was shown in Figure 3. The most frequently altered genes were KRAS, APC, TP53, SMAD4, SRC, GNASH, AURKA, ASXL1, TOP1, BCL2 L1, NBN, RAD21, DAXX, DIS3, KMT2D, and MSH2 (Figure 3).
Figure 3.
The oncoprint diagrams of mutational profile at baseline of the 41 patients with mCRC. Upper panel: The frequency of listed driver genes. Middle panel: The matrix of mutations in a selection of frequently mutated genes. Each column represents one tumor sample, and each row represents one gene. Right panel: The total number of patients harboring mutations in each gene. Bottom annotation shows patient ID. Response to first-line treatment is also shown. PR, partial response; SD, stable disease; PD, progressive disease.
Impact of initial variables, including mutational profile and tumor burden, on response and outcome
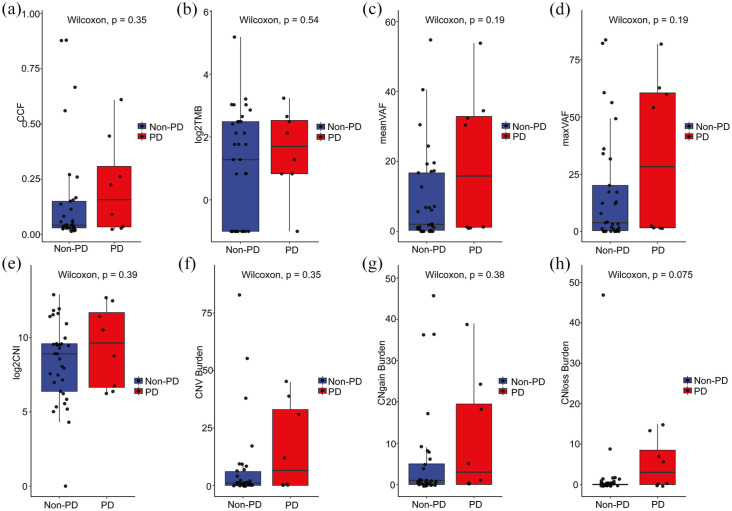
We compared the changes in mutational parameters, including CCF, TMB, mean VAF, maximum VAF, CNI, CNV burden, CNgain burden, and CNloss burden, between non-PD and PD group at baseline (Figure 4). The CCF was low level in non-PD group and high level in PD group (p = 0.35; Figure 4(a)). Differences were observed in the TMB levels between the two groups at baseline (p = 0.54; Figure 4(b)). Furthermore, pretreatment mean VAF levels were comparable between non-PD group and PD group (p = 0.19; Figure 4(c)). Use of maximum VAF (instead of mean) yielded similar associations (p = 0.19; Figure 4(d)). The pretreatment CNI levels were prone to be detected in PD group compared with non-PD group (p = 0.39; Figure 4(e)). These results suggested that changes in levels of mutational parameters at baseline might distinguish mCRC patients with different responses to first-line therapy (PD versus non-PD). However, there was no statistically significant difference between the two groups.
Figure 4.
Relationship between pretreatment mutational parameters and therapeutic response in 41 patients. (a) Box plot depicting the relationship between pretreatment CCF levels and clinical response. Each dot corresponds to one sample. (b) Correlation between TMB levels and therapeutic response. (c) Pretreatment VAF stratified by objective response (PR and SD versus PD) was not significantly associated with response in all patients. The horizontal bar represents the mean, the box represents the 25th and 75th percentiles, and whiskers ± 1.5 × interquartile range (IQR). (d) Pretreatment maximum VAF was not significantly associated with ORR in RECIST responders and nonresponders (p = 0.19). (e) There was no statistically significant difference between CNI levels and clinical benefit. (f), (g), and (h) The correlation between burden of tumor copy number changes (CNV, CNgain, and CNloss) and clinical benefit in all patients.
Influence of CNV burden on the response to first-line chemotherapy with/without targeted therapy in mCRC patients
To further explore the association between tumor somatic copy number changes and clinical response, we analyzed the predictive value of the CNV burden, including CNgain and CNloss burden, in mCRC patients who received first-line therapy (n = 41). We found that the level of CNV burden was prone to be detected in PD group compared with non-PD group (p = 0.35; Figure 4(f)). Accordingly, the burden of CNgain (p = 0.38; Figure 4(g)) or CNloss (p = 0.075; Figure 4(h)) was also comparable between PD group and non-PD group. Nevertheless, the changes in CNV, CNgain, and CNloss burden between PD and non-PD patients did not attain statistical significance.
Correlation between molecular mutational burden levels and first-line treatment response
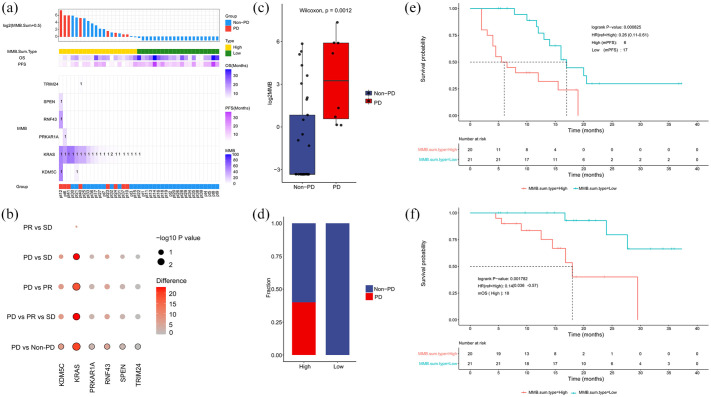
To investigate the association between molecular alterations of ctDNA and response to first-line therapy, we calculated baseline molecular mutational burden (MMB) in 41 patients (Figure 5). We found that alterations of six genes (named six-gene MMB) including TRIM24, SPEN, RNF43, PRKAR1A, KRAS, and KDM5 C were frequently mutated in 41 plasma samples at baseline (Figure 6(a)). In Figure 6(b), to screen significant genes related to therapeutic response, Wilcoxon rank sum test was used to compare the distribution of MMB between pairwise comparisons (PR versus SD, PD versus SD, PD versus PR, PD versus non-PD) and Kruskal–Wallis test for a comparison among three groups (PD versus PR versus SD). The difference of different group was also calculated, that is the mean MMB of the former minus the latter for pairwise comparisons and the maximum of mean MMB minus the minimum for a comparison among three groups. The deeper color from gray to red and the greater circle size represented significant difference. KRAS was the predominant one among the six hypermutated genes (Figure 6(b)). Compared with non-PD group, the gene count of significantly difference MMB was the most in PD group (Figure 6(b)). By summing up the total MMB of the six genes, we found that MMB was significantly lower in non-PD patients than PD patients (p = 0.0012; Figure 6(c)). To evaluate the prognostic value of MMB, we divided the patients into two groups (MMB-high versus MMB-low) based on the median value of MMB at baseline. By comparing the levels of MMB in the two subgroups, we found that PD group was all MMB-high, while non-PD cases were mainly in MMB-low subgroup (Figure 6(d)). Moreover, the median PFS in patients with low or high MMB was 17.0 months and 6.0 months, respectively (HR = 0.26, 95% CI = 0.11–0.61, p = 0.0008; Figure 6(e)). Patients with low MMB had markedly longer OS than those with high MMB (median OS: 18.0 months in MMB-high group, HR = 0.14, 95% CI = 0.036–0.57, p = 0.0018; Figure 6(f)).
Figure 5.
The molecular mutational burden at baseline of the 41 patients with mCRC. The matrix of mutations represents mutated genes assessed in liquid biopsy of mCRC patients; bottom annotation shows patient ID. Response to first-line treatment is also depicted.
PD, progressive disease; PR, partial response; SD, stable disease.
Figure 6.
Low molecular mutational burden was associated with superior response rate and longer survival time. (a) Alterations of six genes, including TRIM24, SPEN, RNF43, PRKAR1A, KRAS, and KDM5 C, were frequently observed in 41 plasma samples at baseline. (b) The significantly different genes in each group. All the significant gene entries have been illustrated in red, whereas other entries are in different degrees of red colors. (c) Molecular mutational burden of six genes was lower in non-PD group compared with PD group. (d) The distribution of patients with PD/non-PD in MMB-high and MMB-low subgroups. (e) PFS was longer in patients with low molecular mutational burden than patients with high molecular mutational burden. (f) OS was longer in low-MMB group than high-MMB group.
Relationship between levels of KRAS MMB and therapeutic response
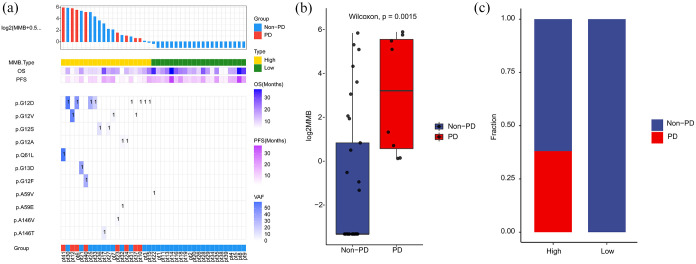
KRAS MMB was defined as the sum of alterations in KRAS including SNV and InDels. We found that alteration of KRAS G12 was the most frequently mutated point in the 41 blood samples at baseline (Figure 7(a)). Compared with non-PD group, patients with PD group had significantly higher values of KRAS MMB (p = 0.0015; Figure 7(b)). By comparing the levels of KRAS MMB in the two subgroups, we found that PD group was all MMB-high, while non-PD cases were mainly in MMB-low subgroup (Figure 7(c)). Moreover, patients with high levels of KRAS MMB had predominantly shorter PFS than those with low-KRAS MMB (p = 0.0008; Supplementary Figure S1A). Patients with high-KRAS MMB had markedly shorter OS than those with low-KRAS MMB (p = 0.0018; Supplementary Figure S1B).
Figure 7.
High-KRAS molecular mutational burden was related to inferior response rate and shorter survival time. (a) Summary of mutation sites of KRAS identified by individual patient at baseline. The variant allele frequencies for each mutation site are plotted on the right panel. (b) Molecular mutational burden of KRAS was higher in PD group than non-PD group. (c) The distribution of patients with PD/non-PD in high-KRAS MMB and low-KRAS MMB subgroups.
To further investigate the predictive value of KRAS MMB, we divided 41 patients into two groups (KRAS MUT versus KRAS WT) and explored the relationship between KRAS MUT/WT and survival. The median PFS in patients with KRAS MUT or WT was 6.5 months and 17.0 months, respectively (HR = 3.8, 95% CI = 1.6–9.1, p = 0.0010; Supplementary Figure S2A). Patients with KRAS MUT had markedly shorter OS than those with KRAS WT (median OS: 18.0 months in KRAS MUT group, HR = 4.6, 95% CI = 1.2–17, p = 0.0160; Supplementary Figure S2B).
To interpret the prognostic implications of MMB (KRAS, six-gene), we supplement multivariable Cox regression analyses including other prognostic markers in mCRC. Multivariable Cox analysis showed the prognostic implications of six-gene MMB at baseline (Supplementary Figure S3), the prognostic values of KRAS mutation status at baseline (Supplementary Figures S4 and S5). Overall, these data support the potential prognostic value of MMB (KRAS, six-gene) in mCRC patients.
Association between levels of KRAS MMB and clinical benefit after four cycles of first-line treatment
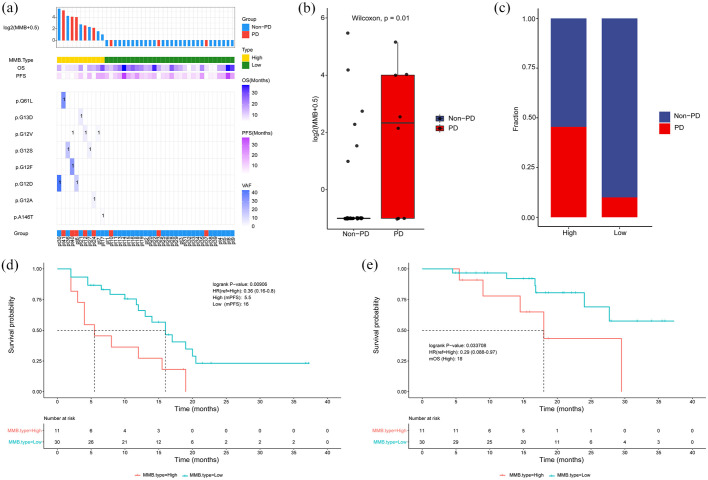
Then we explored the correlation between KRAS MMB levels and therapeutic efficacy after four cycles of treatment. The mutation sites of KRAS gene in the 41 blood samples after four cycles of treatment were illustrated in Figure 8(a). KRAS MMB was lower in non-PD patients than PD patients (p = 0.01; Figure 8(b)). PD cases were mainly in KRAS MMB-high subgroup, while non-PD patients were mainly in KRAS MMB-low subgroup (Figure 8(c)). The median PFS was 16.0 months in patients with low-KRAS MMB, which was longer than 5.5 months in patients with high-KRAS MMB (p = 0.0091; Figure 8(d)). The median OS was not reached in patients with low-KRAS MMB, which was longer than 18.0 months in patients with high-KRAS MMB (p = 0.0337; Figure 8(e)).
Figure 8.
Association between KRAS MMB and therapeutic response after four cycles of first-line treatment. (a) Summary of mutation sites of KRAS identified by individual patient after four cycles of first-line treatment. (b) KRAS MMB was lower in non-PD patients than PD patients. (c) The distribution of patients with PD/non-PD in MMB-high and MMB-low subgroups. (d) PFS was longer in patients with low-KRAS MMB than patients with high-KRAS MMB. (e) OS was longer in patients with low-KRAS MMB than patients with high-KRAS MMB.
Discussion
This study describes the predictive value of ctDNA in mCRC patients, detected by NGS before and after four cycles of first-line treatment. Our results indicated that six-gene MMB of frequently altered genes was markedly associated with therapeutic efficacy. There was a significantly difference in values of KRAS MMB between patients with partial response (PR)/stable disease (SD) versus progressive disease (PD) from pretreatment, and after four cycles of treatment blood samples, respectively. Furthermore, patients with low-KRAS MMB had significantly longer progression-free survival (PFS) and overall survival (OS) than those with high-KRAS MMB.
Colorectal cancer is a genomically heterogeneous disease, including both intra-tumor and inter-tumor heterogeneity. Due to the biological complexity and emergence of primary/acquired resistance, biopsy from a single site can hardly fully represent the clonal dynamics of metastatic disease. Therefore, ctDNA is a useful tool to detect genomic alterations and monitor clonal evolution in primary and metastatic sites. To the best of our knowledge, this study is the first to demonstrate that KRAS MMB is an efficient approach to predict first-line therapeutic response in mCRC patients. By comparing the baseline levels of KRAS MMB in the two subgroups, we observed that PD cases were mostly MMB-high, whereas non-PD cases were mainly in MMB-low subgroup. Furthermore, patients with low-KRAS MMB had superior treatment efficacy and significantly longer PFS/OS than patients with high-KRAS MMB, indicating that levels of KRAS MMB might predict the therapeutic response.
Cell-free DNA is derived from dying cells, detectable in plasma and is typically found as double-stranded fragments (average length of 150–200 base pairs). In patients without tumor tissue obtainable, ctDNA-based profiling might provide a reliable and noninvasive solution to meet the need for baseline molecular diagnosis. The clinical application of ctDNA, such as early detection of disease, prediction of treatment response, monitoring disease relapse, and identification of resistance mechanisms, has been extensively explored nowadays. In previous studies, ctDNA was used as a tool for predicting the therapeutic response in mCRC patients. However, ctDNA-based analysis is not currently ready to complement, or even ultimately replace radiological assessments in guiding systemic therapy. Consistently, our results also found that baseline mutational parameters, such as CCF, TMB, mean VAF, maximum VAF, cannot distinguish patients with PR/SD from those with PD.
In addition, KRAS MMB was used to explore the correlation between mutational landscape of ctDNA and therapeutic efficacy. Molecular mutational burden was different from tumor mutational burden. TMB measured the number of somatic mutations per megabase of interrogated genomic sequence in the tumor specimen. 18 CheckMate 227 trial was the first study that evaluated TMB as a biomarker in non-small-cell lung cancer (NSCLC) patients who received immunotherapy. CheckMate 227 showed that TMB was correlated with clinical response to combination therapy with nivolumab plus ipilimumab as a first-line regimen for advanced NSCLC. 19 In contrast, MMB was defined as the sum of mean mutation frequency value for each somatic mutation. 20 Therefore, the continuous and real-time changes of MMB could be used to monitoring treatment response dynamically. In our study, alterations of six genes were frequently mutated and identified at baseline. There was a robust correlation between six-gene MMB in ctDNA and therapeutic efficacy. Furthermore, PFS/OS was longer in MMB-low group than MMB-high group, indicating that six-gene MMB might be valuable to predict therapeutic efficacy and PFS/OS of first-line treatment in patients with metastatic CRC.
Previous studies have reported that KRAS mutation is a prognostic biomarker in colorectal cancer. Mutations in KRAS, NRAS, and BRAF are associated with poor prognosis and resistance to regimens usually used for colorectal cancer. Patients with RAS mutation detected in plasma or in tumor tissue had significantly worse outcomes compared with wild-type counterparts. In consistent with the published literatures, we also found that patients with KRAS mutation had significantly shorter PFS/OS than those with KRAS wild type. Our study also explored the potential clinical meaningfulness of liquid biopsy to evaluate KRAS MMB in mCRC, considering the heterogeneity of KRAS mutation status in tumor tissue and the accessibility of tumor specimen. In our study, patients with low-KRAS MMB had superior response rate and markedly longer PFS and longer OS than patients with high-KRAS MMB. KRAS MMB in our study was comparable with KRAS mutation status. In addition, the use of NGS-based ctDNA profiling allowed us to concomitantly test both KRAS MMB and mutation status, as well as other genomic alterations with potential clinical implications.
There are several limitations in present research. First, owing to the accessibility of sequencing data and collection of clinical information, only 41 cases were enrolled in our study, which is relatively small and may lead to the potential selection bias. A large population-based prospective study is warranted in the future to validate the predictive efficacy of MMB. Second, we did not observe the significant differences between two groups (PD versus non-PD) when we compared the baseline mutational features, including CCF, TMB, mean VAF, maximum VAF, CNI, CNV burden, CNgain burden, and CNloss burden. Third, we only identified the baseline SNV/CNV profiles from ctDNA in plasma, whether it could well represent the SNV/CNV features in tumor specimen of mCRC remains future study. Last but not the least, exploring biomarkers to predict chemotherapy benefit seems less clinical meaning in the era of immunotherapy. As we all know, oxaliplatin-/irinotecan-based chemotherapy with/without targeted therapy still play an important role in the first-line treatment for advanced CRC patients. A substantial number of patients still need chemotherapy and might benefit from our explorative analysis.
In conclusion, our study revealed that the genomic profiling of ctDNA is related to clinical efficacy to first-line therapy in patients with metastatic CRC. Molecular mutational burden plays an important role in predicting treatment response, and KRAS MMB can monitor clinical efficacy and deserve further study. Last but not the least, our study shed light on the utility of ctDNA in predicting and monitoring the response of first-line treatment in patients with mCRC.
Supplemental Material
Supplemental material, sj-docx-1-tam-10.1177_17588359211070643 for Mutational landscape of circulating tumor DNA identifies distinct molecular features associated with therapeutic response in patients with metastatic colorectal cancer by Min Shi, Hong Yuan, Jun Ji, Shouwei Zhang, Qingyuan Li, Yawei Chen, Xiaoli Gong, Zhenggang Zhu and Jun Zhang in Therapeutic Advances in Medical Oncology
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xls-1-tam-10.1177_17588359211070643 for Mutational landscape of circulating tumor DNA identifies distinct molecular features associated with therapeutic response in patients with metastatic colorectal cancer by Min Shi, Hong Yuan, Jun Ji, Shouwei Zhang, Qingyuan Li, Yawei Chen, Xiaoli Gong, Zhenggang Zhu and Jun Zhang in Therapeutic Advances in Medical Oncology
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xls-2-tam-10.1177_17588359211070643 for Mutational landscape of circulating tumor DNA identifies distinct molecular features associated with therapeutic response in patients with metastatic colorectal cancer by Min Shi, Hong Yuan, Jun Ji, Shouwei Zhang, Qingyuan Li, Yawei Chen, Xiaoli Gong, Zhenggang Zhu and Jun Zhang in Therapeutic Advances in Medical Oncology
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xls-3-tam-10.1177_17588359211070643 for Mutational landscape of circulating tumor DNA identifies distinct molecular features associated with therapeutic response in patients with metastatic colorectal cancer by Min Shi, Hong Yuan, Jun Ji, Shouwei Zhang, Qingyuan Li, Yawei Chen, Xiaoli Gong, Zhenggang Zhu and Jun Zhang in Therapeutic Advances in Medical Oncology
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xls-4-tam-10.1177_17588359211070643 for Mutational landscape of circulating tumor DNA identifies distinct molecular features associated with therapeutic response in patients with metastatic colorectal cancer by Min Shi, Hong Yuan, Jun Ji, Shouwei Zhang, Qingyuan Li, Yawei Chen, Xiaoli Gong, Zhenggang Zhu and Jun Zhang in Therapeutic Advances in Medical Oncology
Footnotes
Author contributions: Min Shi: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Writing – original draft.
Hong Yuan: Data curation; Formal analysis; Funding acquisition; Methodology; Writing – original draft.
Jun Ji: Methodology; Project administration; Resources; Software; Validation; Visualization.
Shouwei Zhang: Formal analysis; Investigation; Methodology; Resources; Software; Validation; Visualization.
Qingyuan Li: Methodology; Resources; Software.
Yawei Chen: Methodology; Resources; Software.
Xiaoli Gong: Formal analysis; Investigation; Methodology; Resources; Software.
Zhenggang Zhu: Conceptualization; Investigation; Supervision; Validation; Visualization; Writing – review & editing.
Jun Zhang: Conceptualization; Funding acquisition; Investigation; Project administration; Supervision; Validation; Visualization; Writing – review & editing.
Conflict of interest statement: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: S. Zhang, Q. Li, Y. Chen, and X. Gong were employed by Genecast Biotechnology Co., Ltd. All the other authors declare no potential conflicts of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was supported by National Science Foundation of China (81972707 to J. Zhang, 81602411 to M. Shi, and 82102760 to H. Yuan) and collaborative innovation cluster project of Shanghai Municipal Health Commission (2020CXJQ03) to J. Zhang and Shanghai Rising-Star Program (20QA1406200) to M. Shi and Shanghai Municipal Commission of Health and Family Planning (20184Y0048) to M. Shi and Guangci Distinguished Young Scholars Training Program (GCQN-2018-A06) to M. Shi and Shanghai Sailing Program (21YF1440800) to H. Yuan and Shanghai Anticancer Association EYAS PROJECT (SACA-CY19C19) to H. Yuan.
ORCID iD: Jun Zhang  https://orcid.org/0000-0002-7973-8416
https://orcid.org/0000-0002-7973-8416
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Min Shi, Department of Oncology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Hong Yuan, Department of Oncology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Jun Ji, Department of Surgery, Shanghai Institute of Digestive Surgery, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Shouwei Zhang, Genecast Biotechnology Co., Ltd, Wuxi City, Jiangsu, China.
Qingyuan Li, Genecast Biotechnology Co., Ltd, Wuxi City, Jiangsu, China.
Yawei Chen, Genecast Biotechnology Co., Ltd, Wuxi City, Jiangsu, China.
Xiaoli Gong, Genecast Biotechnology Co., Ltd, Wuxi City, Jiangsu, China.
Zhenggang Zhu, Department of Oncology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, ChinaDepartment of Surgery, Shanghai Institute of Digestive Surgery, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Jun Zhang, Department of Oncology, Rui Jin Hospital, Shanghai Jiao Tong University School of Medicine, No. 197 Ruijin er Road, Shanghai 200025, China; State Key Laboratory of Oncogenes and Related Genes, Shanghai Jiao Tong University, Shanghai, China.
References
- 1. Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA 2021; 71: 7–33. [DOI] [PubMed] [Google Scholar]
- 2. Burge M, Price T, Karapetis CS. First-line therapy for metastatic colorectal cancer: current perspectives and future directions. Asia Pac J Clin Oncol 2019; 15(Suppl. 1): 3–14. [DOI] [PubMed] [Google Scholar]
- 3. Van Cutsem E, Cervantes A, Adam R, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol 2016; 27: 1386–1422. [DOI] [PubMed] [Google Scholar]
- 4. Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med 2018; 379: 1754–1765. [DOI] [PubMed] [Google Scholar]
- 5. Dasari A, Morris VK, Allegra CJ, et al. CtDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper. Nat Rev Clin Oncol 2020; 17: 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015; 26: 1715–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jia N, Sun Z, Gao X, et al. Serial monitoring of circulating tumor DNA in patients with metastatic colorectal cancer to predict the therapeutic response. Front Genet 2019; 10: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hamfjord J, Guren TK, Dajani O, et al. Total circulating cell-free DNA as a prognostic biomarker in metastatic colorectal cancer before first-line oxaliplatin-based chemotherapy. Ann Oncol 2019; 30: 1088–1095. [DOI] [PubMed] [Google Scholar]
- 9. Garlan F, Laurent-Puig P, Sefrioui D, et al. Early evaluation of circulating tumor DNA as marker of therapeutic efficacy in metastatic colorectal cancer patients (PLACOL study). Clin Cancer Res 2017; 23: 5416–5425. [DOI] [PubMed] [Google Scholar]
- 10. Wang DS, Liu ZX, Lu YX, et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut 2019; 68: 1152–1161. [DOI] [PubMed] [Google Scholar]
- 11. Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017; 545: 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016; 8: 346ra392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou J, Wang C, Lin G, et al. Serial circulating tumor DNA in predicting and monitoring the effect of neoadjuvant chemoradiotherapy in patients with rectal cancer: a prospective multicenter study. Clin Cancer Res 2021; 27: 301–310. [DOI] [PubMed] [Google Scholar]
- 14. Max Ma X, Bendell JC, Hurwitz HI, et al. Disease monitoring using post-induction circulating tumor DNA analysis following first-line therapy in patients with metastatic colorectal cancer. Clin Cancer Res 2020; 26: 4010–4017. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Q, Luo J, Wu S, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov 2020; 10: 1842–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Almodovar K, Iams WT, Meador CB, et al. Longitudinal cell-free DNA analysis in patients with small cell lung cancer reveals dynamic insights into treatment efficacy and disease relapse. J Thorac Oncol 2018; 13: 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rivas-Delgado A, Nadeu F, Enjuanes A, et al. Mutational landscape and tumor burden assessed by cell-free DNA in diffuse large B-cell lymphoma in a population-based study. Clin Cancer Res 2021; 27: 513–521. [DOI] [PubMed] [Google Scholar]
- 18. Sha D, Jin Z, Budczies J, et al. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov 2020; 10: 1808–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018; 378: 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang T, Li X, Wang J, et al. Mutational landscape of cfDNA identifies distinct molecular features associated with therapeutic response to first-line platinum-based doublet chemotherapy in patients with advanced NSCLC. Theranostics 2017; 7: 4753–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tam-10.1177_17588359211070643 for Mutational landscape of circulating tumor DNA identifies distinct molecular features associated with therapeutic response in patients with metastatic colorectal cancer by Min Shi, Hong Yuan, Jun Ji, Shouwei Zhang, Qingyuan Li, Yawei Chen, Xiaoli Gong, Zhenggang Zhu and Jun Zhang in Therapeutic Advances in Medical Oncology
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xls-1-tam-10.1177_17588359211070643 for Mutational landscape of circulating tumor DNA identifies distinct molecular features associated with therapeutic response in patients with metastatic colorectal cancer by Min Shi, Hong Yuan, Jun Ji, Shouwei Zhang, Qingyuan Li, Yawei Chen, Xiaoli Gong, Zhenggang Zhu and Jun Zhang in Therapeutic Advances in Medical Oncology
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xls-2-tam-10.1177_17588359211070643 for Mutational landscape of circulating tumor DNA identifies distinct molecular features associated with therapeutic response in patients with metastatic colorectal cancer by Min Shi, Hong Yuan, Jun Ji, Shouwei Zhang, Qingyuan Li, Yawei Chen, Xiaoli Gong, Zhenggang Zhu and Jun Zhang in Therapeutic Advances in Medical Oncology
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xls-3-tam-10.1177_17588359211070643 for Mutational landscape of circulating tumor DNA identifies distinct molecular features associated with therapeutic response in patients with metastatic colorectal cancer by Min Shi, Hong Yuan, Jun Ji, Shouwei Zhang, Qingyuan Li, Yawei Chen, Xiaoli Gong, Zhenggang Zhu and Jun Zhang in Therapeutic Advances in Medical Oncology
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (https://creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xls-4-tam-10.1177_17588359211070643 for Mutational landscape of circulating tumor DNA identifies distinct molecular features associated with therapeutic response in patients with metastatic colorectal cancer by Min Shi, Hong Yuan, Jun Ji, Shouwei Zhang, Qingyuan Li, Yawei Chen, Xiaoli Gong, Zhenggang Zhu and Jun Zhang in Therapeutic Advances in Medical Oncology