Abstract
Purpose
Here, we aimed to elucidate the differences in microbiota composition between patients with gout and those with asymptomatic hyperuricemia (asHU) and determine the effect of uric acid-lowering therapy (ULT) on the gut microbiome.
Materials and Methods
Stool samples from patients with asHU (n=8) and three groups of gout patients, i.e., acute gout patients before ULT (0ULT, n=14), the same acute gout patients after 30-day ULT (30ULT, n=9), and chronic gout patients after ≥6-month ULT (cULT, n=18) were collected and analyzed using 16S rRNA gene-based pyrosequencing. The composition of microbial taxonomy and communities, species diversity, and relationships among microbial communities were elucidated by bioinformatic analysis.
Results
Gout patients showed less diverse gut microbiota than asHU patients. The microbiota of the asHU group exhibited a higher Firmicutes-to-Bacteroidetes (F/B) ratio and lower Prevotella-to-Bacteroides (P/B) ratio than the gout group; significantly, the F/B ratio increased in gout patients after ULT. Moreover, a balanced enterotype populated asHU patients compared to gout patients. Notably, the gut microbiota in asHU patients had a higher proportion of taxa with potentially anti-inflammatory effects compared to the gut microbiota in gout patients.
Conclusion
We found that microbial composition differs between asHU and gout patients. The differential gut microbiota in asHU patients may protect against gout development, whereas that in gout patients may have a role in gout provocation. ULT in gout patients altered the gut microbiota, and may help alleviate gout pathology and mitigate gout progression.
Keywords: Gastrointestinal microbiome, gout, hyperuricemia, uric acid
INTRODUCTION
Hyperuricemia is a strong risk factor for gout and exerts its pathological effects in a non-linear concentration-dependent manner.1 However, not all patients with hyperuricemia develop clinical gout, and only one-third of patients show progression to gout. In contrast, some patients with gout remain normouricemic.2
In patients with hyperuricemia, several factors can trigger a gout flare. The deposition of monosodium urate crystals in connective tissues is believed to be a trigger3; however, 76% of patients with asymptomatic hyperuricemia (asHU) do not have monosodium urate crystal deposition.2 In patients with gout, a flared inflammatory response is often triggered after a heavy meal or alcohol consumption. Studies have reported that some dietary lipids or alcohol consumption can directly trigger gout flares via the activation of the NALP3 inflammasome by binding to Toll-like receptors.4
The role of the intestines in uric acid regulation has been investigated. Although the kidneys are responsible for most uric acid excretion, the intestines contribute to 25% of uric acid excretion, which is further enhanced during renal dysfunction.5 ATP-binding cassette, subfamily G2 is one of the major urate secretion transporter involved in uric acid excretion from the intestines.6 In hyperuricemia, the extra-renal excretion of abundant serum uric acid creates an intestinal environment rich in uric acid content.7
These findings have raised the possibility that the intestinal microbiota are affected by high serum uric acid levels.8 Indeed, a remarkable study showed that the gut microbiota are altered in patients with gout compared to healthy controls, and that gout patients have a distinct gut microbial signature compared to healthy individuals.9 Another study reported that some metabolites present in the fecal samples of gout patients are involved in uric acid excretion, purine metabolism, and the inflammatory response.10 The positive association between gout-related metabolites and microbial taxa in patients with gout suggests that a high uric acid concentration and uric acid metabolites interact with the gut microbiota. Therefore, the gut microbiota in gout patients may play a role in gout pathophysiology and serve as a new target for the diagnosis and treatment of gout.8 However, the aforementioned studies were conducted on the assumption that patients with asHU and gout have comparable microbiota composition; thus, gout development in the hyperuricemia condition has not been explained.
Hence, in this study, we aimed to investigate whether patients with asHU and gout have differential gut microbiota signatures. Additionally, we investigated whether uric acid-lowering therapy (ULT) can change the microbiota composition.
MATERIALS AND METHODS
Survey of population ecological information
In total, 40 patients were enrolled in this study. The study cohort comprised four groups: one group of asHU patients (n=8) and three groups of gout patients, namely, acute gout patients before ULT (0ULT, n=14), the same acute gout patients after 30-day ULT (30ULT, n=9), and chronic gout patients after ≥6-month ULT (cULT, n=18). Patients with asHU included those who visited a health screening center or outpatient clinic and were incidentally diagnosed with hyperuricemia without a previous acute gout attack. A diagnosis of asHU was confirmed by elevated serum urate concentrations (>8.0 mg/dL). All patients with gout fulfilled the American College of Rheumatology/European League Against Rheumatism gout classification criteria,11 were more than 19 years of age, and had not taken antibiotic treatment within one month prior to study enrollment. Patients with active systemic infectious diseases were excluded from the study. Patient sex, age, duration of gout, medication dose, and comorbidities such as diabetes, hypertension, or chronic kidney disease were noted. The study protocol was approved by the ethics committee of Gangnam Severance Hospital (IRB protocol 2016-0124-001), and written informed consent was obtained from all patients.
Sample preparation and experiment
Stool and serum collection
Before stool sample collection, all patients were required to submit a diary with details of food intake during the previous 3 days, although no restriction was imposed on their diet except for antibiotic use. Stool samples (5–10 g) were collected in the morning before breakfast and stored at -20℃ until further processing. Serum samples were collected on the day of the clinic visit.
Fecal DNA isolation and pyrosequencing
The extraction of bacterial DNA was performed using FastDNA SPIN Kit for Soil (MP Biomedical, Santa Ana, CA, USA). The quality of the extracted DNA was evaluated by performing 0.8% agarose gel electrophoresis, and the DNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). All DNA samples were stored at -20℃ until further processing. Polymerase chain reaction (PCR) was performed using extracted DNA with primers targeting the V3–V4 regions of the 16S rRNA gene. For bacterial amplification, the primers 341F (5′-TCGTCGGCAGCGTC-AGATGT GTATAAGAGACAG-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GTCTCGTGGGCTCGG-AGATGTGTATAAGAGA CAG-GACTACHVGGGTATCTAATCC-3′) were used. The amplifications were carried out under the following conditions: initial denaturation at 95℃ for 3 min, followed by 25 cycles of denaturation at 95℃ for 30 sec, primer annealing at 55℃ for 30 sec, and extension at 72℃ for 30 sec, with a final elongation at 72℃ for 5 min. Then, secondary amplification for attaching the Illumina NexTera barcode was performed with i5 forward primer (5′-AATGATACGGCGACCACCGAGATCTACAC-XXXXXXXX-TCGTCGGCAGCGTC-3′; X indicates the barcode region) and i7 reverse primer (5′-CAAGCAGAAGACGGCA TACGAGAT-XXXXXXXX-AGTCTCGTGGGCTCGG-3′). Mixed amplicons were pooled, and sequencing was performed at ChunLab, Inc. (Seoul, Korea) on an Illumina MiSeq Sequencing System (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions.
Sequence processing and bioinformatic analysis
To improve data quality, low quality (<Q25) reads were filtered using the Trimmomatic (V0.32) read trimming tool (Usadel Lab, Aachen, Germany) and paired-end sequence data were merged using PANDAseq.12 Primers were then trimmed with ChunLab’s in-house program at a similarity cut-off of 0.8. Non-specific amplicons encoding non-16S rRNA genes were detected using the HMMER hmmsearch program (EMBL-EBI, Hinxton, Cambridge, UK) and were not considered for further sequence analysis. Sequences were denoised using DUDE-Seq, and non-redundant reads were extracted using the UCLUST clustering method.13 Bacterial taxonomic assignments were performed based on the EzBioCloud database using USEARCH (8.1.1861_i86linux32) followed by more precise pairwise alignment.13 UCHIME14 and the non-chimeric 16S rRNA database from EzBioCloud were used to detect chimera on reads with <97% similarity. Reads that were not identified at the species level (with <97% similarity) in the EzBioCloud database were compiled, and UCLUST5 was used to perform de novo clustering to generate operational taxonomic units (OTUs). Finally, OTUs with single reads (singletons) were omitted from further analysis. The bacterial taxonomic composition was evaluated from the phylum to the species level.
Quantitative PCR
Bacterial biomarkers discriminating patients before and after ULT were identified from sequencing analysis and further confirmed using quantitative PCR (qPCR). Quantification of specific taxa before and after ULT was performed using qPCR [a LightCycle 480 system (Roche Diagnostics, Basel, Switzerland)] with LightCycler FastStart DNA Master SYBR Green I (Roche Diagnostics). The primers used in the study were newly designed to target homologous sequences in the 16S rDNA of each bacteria to quantify the amount of bacteria in each sample (Supplementary Table 1, only online).
Statistical analyses
All statistical analyses were performed using R software (version 4.1.0; R Foundation for Statistical Computing, Vienna, Austria). Baseline characteristics of four groups of patients were compared using one-way analysis of variance. Categorical variables were compared using χ2 statistics. Alpha diversity was assessed using the Shannon index (evenness and richness), valid reads, and the number of observed species (OTUs). Beta diversity was assessed using a principal component (PCoA) score plot based on Bray-Curtis dissimilarity metrics, canonical correspondence, or redundancy analyses. The Wilcoxon rank-sum test was used to compare the taxa abundance ratio in the asHU group to that in the total gout group. To discover biomarkers with statistical differences among groups, the Kruskal-Wallis rank test with a p value of 0.05 was first used to compare differential microbiota abundance between groups. Then, a linear discriminant analysis (LDA) was performed to evaluate the influence of biomarkers on significantly different microbial groups based on the LDA scores. Linear discriminant analysis Effect Size (LEfSe) was used to determine potential markers among different groups. Pattern search function of the MicrobiomeAnalyst, a web-based tool, was used to compare abundance patterns in the dataset.15 Nonparametric Spearman’s test was used to test the correlation between microbial taxa and serum uric acid level. Moreover, Tax4Fun2, an R package,16 was used for the prediction of functional profiles from 16S rRNA gene sequences. Pathway analyses were conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
RESULTS
Characteristics of study subjects
The characteristics of enrolled patients are described in Table 1. All patients were male, and the mean age was comparable between groups. Gout patients had a statistically higher incidence of hypertension and chronic kidney disease compared to asHU patients. Serum creatinine concentration was significantly higher, and there was a non-significant trend for elevated C-reactive protein in patients with gout than for asHU patients, indicating the presence of comorbid conditions with an inflammatory burden in the gout patients compared to asHU patients. However, there were no statistical differences in age, alcohol or protein consumption, family history, or body mass index between asHU and gout patients. Disease duration of gout were significantly shorter in the acute gout patients compared to chronic gout patients (1.0 months vs. 41.4 months, p<0.001) reflecting that patients with both early and advanced phases of gout were enrolled. There were no statistical differences in the body mass index and comorbidities, except for a higher prevalence of chronic kidney disease (7.1% vs. 72.2%, p=0.001), between acute and chronic gout patients. Serum uric acid levels were elevated in patients with acute gout before ULT and in patients with asHU compared to patients with chronic gout who received ULT.
Table 1. Characteristics of Patients with asHU and Gout.
| asHU (n=8) | Acute gout (0ULT*) (n=14) | Chronic gout (cULT) (n=18) | p value | Post hoc p value (acute vs. chronic gout) | Post hoc p value (asHU vs. total gout) | ||
|---|---|---|---|---|---|---|---|
| Male | 8 (100.0) | 14 (100.0) | 18 (100.0) | ||||
| Age (yr) | 56.5±12.9 | 49.7±15.8 | 50.0±17.4 | 0.413 | 0.962 | 0.298 | |
| Height (cm) | 171.5±4.9 | 172.8±4.8 | 174.6±7.9 | 0.230 | 0.456 | 0.364 | |
| Body weight (kg) | 76.0±13.8 | 74.2±8.5 | 80.9±14.9 | 0.263 | 0.158 | 0.697 | |
| BMI (kg/m2) | 25.7±3.5 | 24.8±2.4 | 26.6±5.2 | 0.473 | 0.230 | 0.939 | |
| 18.5–23 | 1 (12.5) | 3 (23.1) | 4 (23.5) | 0.443 | 0.233 | 0.638 | |
| ≥23 | 4 (50.0) | 5 (38.5) | 3 (17.6) | ||||
| ≥25 | 2 (25.0) | 5 (38.5) | 6 (35.3) | ||||
| ≥30 | 1 (12.5) | 0 (0.0) | 4 (23.5) | ||||
| Comorbidities | |||||||
| Hypertension | 0 (0.0) | 4 (28.6) | 7 (38.9) | 0.122 | 0.815 | 0.080 | |
| Dyslipidemia | 0 (0.0) | 3 (21.4) | 7 (38.9) | 0.100 | 0.501 | 0.165 | |
| Diabetes | 0 (0.0) | 0 (0.0) | 1 (5.6) | 0.534 | 0.999 | 0.999 | |
| CKD | 0 (0.0) | 1 (7.1) | 13 (72.2) | 0.001 | 0.001 | 0.034 | |
| Risk factors for gout | |||||||
| Protein/alcohol intake | 2 (25.0) | 10 (71.4) | 8 (44.4) | 0.091 | 0.243 | 0.236 | |
| Familyhistory of gout | 0 (0.0) | 2 (14.3) | 1 (5.6) | 0.433 | 0.819 | 0.881 | |
| Disease duration of gout | NA | 1.0±1.4 | 41.4±36.8 | NA | 0.001 | NA | |
| Laboratory findings | |||||||
| Total bilirubin (mg/dL) | 0.6±0.2 | 0.8±0.3 | 0.8±0.2 | 0.110 | 0.878 | 0.036 | |
| AST (IU/L) | 33.6±14.3 | 26.9±8.5 | 29.3±10.9 | 0.523 | 0.490 | 0.216 | |
| ALT (IU/L) | 26.9±5.2 | 28.6±17.3 | 31.8±20.9 | 0.477 | 0.649 | 0.363 | |
| ALP (IU/L) | 79.1±21.9 | 74.9±18.5 | 80.1±31.9 | 0.827 | 0.592 | 0.896 | |
| TG (mg/dL) | 285.5±57.7 | 266.4±127.5 | 220.0±134.9 | 0.303 | 0.443 | 0.515 | |
| Glucose (mg/dL) | 100.0±6.8 | 97.5±14.4 | 107.7±23.2 | 0.232 | 0.206 | 0.403 | |
| LDL-cholesterol (mg/dL) | 135.1±17.6 | 134.0±37.7 | 132.1±32.4 | 0.866 | 0.907 | 0.909 | |
| HbA1c (%) | 5.5±0.3 | 5.5±0.5 | 5.6±0.1 | 0.694 | 0.673 | 0.886 | |
| ESR (mm/h) | 12.7±13.5 | 24.7±26.8 | 16.6±12.7 | 0.660 | 0.538 | 0.335 | |
| CRP (mg/L) | 1.5±1.3 | 9.1±14.6 | 4.5±5.4 | 0.471 | 0.518 | 0.078 | |
| BUN (mg/dL) | 13.8±3.5 | 17.3±7.3 | 17.5±6.0 | 0.202 | 0.925 | 0.134 | |
| Uric acid (mg/dL) | 8.2±1.3 | 8.8±1.6 | 6.0±2.7 | 0.006 | 0.002 | 0.156 | |
| Creatinine(mg/dL) | 0.9±0.1 | 1.1±0.3 | 1.1±0.2 | 0.068 | 0.784 | 0.001 | |
| Creatinine clearance (mL/min/1.73 m2) | 92.8±12.9 | 83.5±26.2 | 80.3±22.9 | 0.220 | 0.718 | 0.220 | |
| ULT | 0.001 | 0.099 | 0.001 | ||||
| Allopurinol, n (%) | 0 (0.0) | 5 (45.5) | 2 (11.1) | ||||
| Febuxostat, n (%) | 0 (0.0) | 6 (54.5) | 16 (88.9) | ||||
Data are shown in mean±standard deviation or n (%).
*Only baseline characteristics were obtained from acute gout patients before ULT.
BMI, body mass index; CKD, chronic kidney disease; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALP, alkaline phosphatase; TG, triglyceride; LDL, low-density lipoprotein; BUN, blood urea nitrogen; ESR, erythrocyte sedimentation rate; CRP, C-reactive protein; ULT, uric acid-lowering therapy; NA, not applicable; asHU, asymptomatic hyperuricemia; 0ULT, acute gout patients before ULT; cULT, chronic gout patients after ≥6-month ULT.
Comparison of gut microbiota between asHU and gout patients
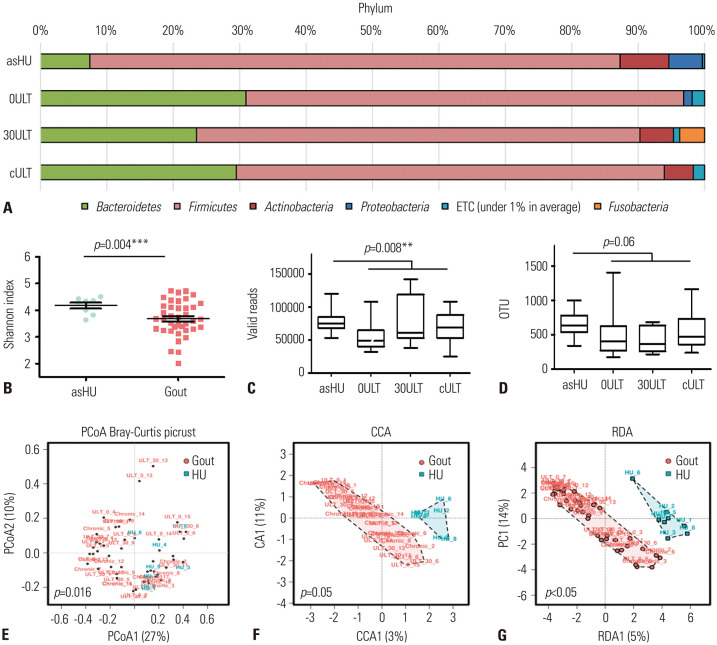
Variations in the bacterial phyla and family profiles were apparent between the asHU and gout groups (Fig. 1A and Supplementary Fig. 1, only online). Species richness, measured as the number of valid reads and OTUs, were statistically different between the asHU and whole gout groups (Fig. 1B–D). OTUs and valid reads showed a trend toward notably reduced microbiota diversity in acute gout patients compared to chronic gout patients, and gout patients showed more reduced microbiota diversity compared to asHU patients, indicating that the gut microbiome is different between asHU and gout patients. In gout patients, ULT may partially restore the microbiota composition, although the differences were not statistically significant. A PCoA score plot showed that gut microbiota of asHU and gout patients were separated (permutational multivariate analysis of variance F-value: 3.536; R2: 0.014; p=0.016) (Fig. 1E), indicating statistically significant community differences between asHU and gout patients. Beta diversity analysis, including canonical correspondence analysis and redundancy analysis, also showed that the microbiota composition was different between the asHU and gout groups (Fig. 1F and G).
Fig. 1. Distinct gut microbiota between patients with asHU and those with gout. (A) Average taxonomic compositions in the bacterial phyla. Variations were apparent between patients with asHU and gout patients. (B–D) The alpha diversity assessed using valid reads, the number of observed species (OTU), and Shannon diversity indices of different groups. (E–G) Principal component (PCoA) score plot based on Bray-Curtis dissimilarity for all participants. Each point represents the composition of the intestinal microbiota of each individual. ULT, uric acid-lowering therapy; asHU, asymptomatic hyperuricemia; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT; cULT, chronic gout patients after ≥6-month ULT; OTU, operational taxonomic unit.
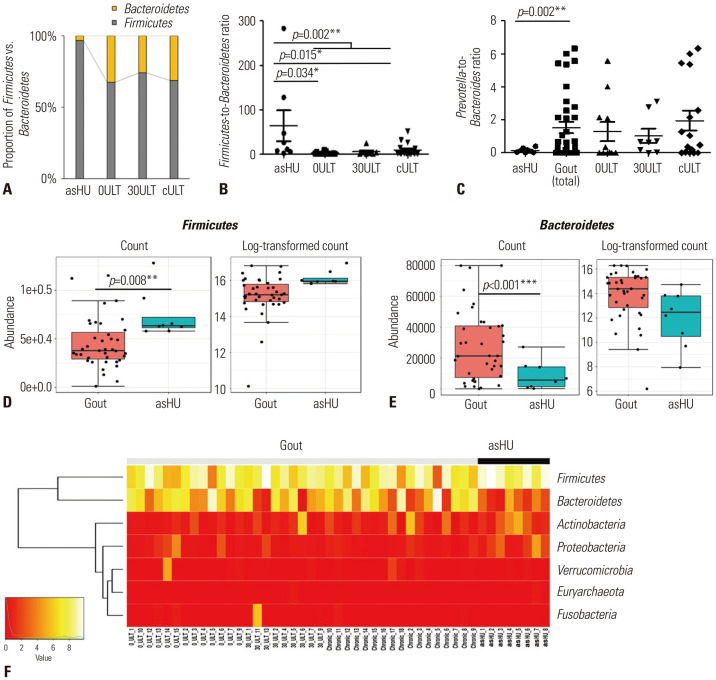
To detect the microbial groups with significantly different compositions in different patient groups, we first analyzed the ratio of Firmicutes (%)-to-Bacteroidetes (%) (F/B) at the phylum level and Prevotella (%)-to-Bacteroides (%) (P/B) ratio at the genus level. Our results showed that the relative abundance of Firmicutes was higher in the asHU group than in any gout group (Fig. 2A). The F/B ratio at the phylum level increased after ULT (Fig. 2B). The F/B ratio was significantly higher in asHU patients than in total gout patients (p=0.002, Wilcoxon rank-sum test). The taxonomic composition at the genus level revealed that the P/B ratio was significantly lower in asHU patients than in gout patients (p=0.002, Wilcoxon rank-sum test) (Fig. 2C). Moreover, asHU patients had a significantly low proportion of Bacteroidetes and a high proportion of Firmicutes (Fig. 2D–F) compared to gout patients.
Fig. 2. Bacterial family analysis. (A) Proportion of Firmicutes vs. Bacteroidetes. (B) Firmicutes-to-Bacteroidetes ratio (Firmicutes (%)/Bacteroidetes (%)) at the phylum level. (C) Prevotella-to-Bacteroides ratio. (D) Difference in the abundance of Firmicutes. (E) Difference in the abundance of Bacteroidetes. (F) Heatmap at the phylum level. ULT, uric acid-lowering therapy; asHU, asymptomatic hyperuricemia; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT; cULT, chronic gout patients after ≥6-month ULT.
At the family level, the proportion of Prevotellaceae and Bacteroidaceae was lower, whereas that of Lachnospiraceae was higher, in asHU patients than in gout patients (Supplementary Fig. 2A, only online). Next, the patients were grouped into harboring different enterotypes based on the dominant bacteria clusters: Prevotellaceae-, Lachnospiraceae-, Rumiococcaceae-, and Bacteroidaceae-dominant enterotypes and balanced enterotype (with no particular dominant microbiota) (Supplementary Fig. 2B, only online). Balanced enterotype populated asHU patients compared to 0ULT patients, whereas the proportion of the Bacteroidaceae-dominant enterotype was absent in asHU. After 30-day ULT, the number of patients with Bacteroidaceae-dominant enterotype was lower than that in the 0ULT group; however, these changes reverted in the cULT group, indicating that although ULT induces changes in the intestinal microbiome, it does not maintain these changes over a longer duration.
Differential taxa between asHU and gout patients
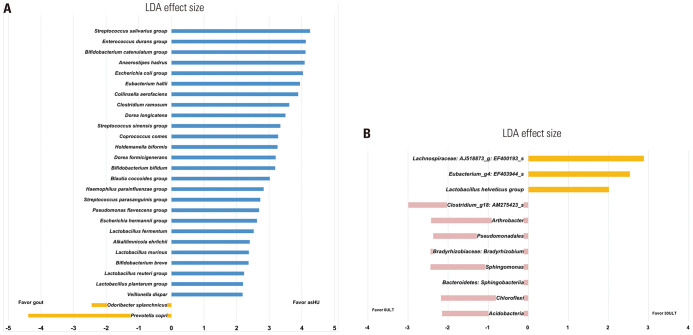
Based on the results of the Kruskal-Wallis test, we identified a significantly different taxa composition in the gut microbiome between asHU and gout patients (p<0.05) (Table 2). Next, bacterial taxa that were differentially represented between groups and had an LDA score of >2 were further analyzed (Fig. 3A). LEfSe analysis confirmed that Prevotella copri and Odoribacter splanchnicus were enriched in gout patients. In the asHU group, Streptococcus salivarius, S. parasanguinis, S. sinensis, Enterococcus durans, Anaerostipes hadrus, Bifidobacterium catenulatum, B. breve, B. bifidum, Lactobacillus plantarum, L. reuteri, L. murinus, and L. fermentum were enriched.
Table 2. Taxa That Were Significantly Different between Patients with Asymptomatic Hyperuricemia and Gout Patients Based on the Kruskal-Wallis Test (p<0.05).
| Patient category | Taxonomy | p value | FDR-adjusted p value |
|---|---|---|---|
| Favor gout | Prevotella copri | <0.001 | 0.009 |
| Odoribacter splanchnicus | 0.026 | 0.254 | |
| Enterococcus faecalis | 0.002 | 0.147 | |
| Ruminococcus faecis | 0.041 | 0.372 | |
| Megamonas funiformis | 0.048 | 0.419 | |
| Brevibacterium iodinum group | 0.019 | 0.233 | |
| Enterococcus casseliflavus group | 0.001 | 0.110 | |
| Favor asymptomatic hyperuricemia | Streptococcus salivarius group | <0.001 | 0.110 |
| Bifidobacterium breve | <0.001 | 0.110 | |
| Lactobacillus murinus | <0.001 | 0.110 | |
| Streptococcus sinensis group | 0.001 | 0.110 | |
| Alkalilimnicola ehrlichii | 0.001 | 0.110 | |
| Escherichia hermannii group | 0.001 | 0.110 | |
| Pseudomonas flavescens group | 0.002 | 0.131 | |
| Haemophilus parainfluenzae group | 0.002 | 0.168 | |
| Lactobacillus plantarum group | 0.002 | 0.176 | |
| Streptococcus parasanguinis group | 0.003 | 0.197 | |
| Dorea formicigenerans | 0.003 | 0.233 | |
| Coprococcus comes | 0.004 | 0.233 | |
| Enterococcus durans group | 0.004 | 0.233 | |
| Anaerostipes hadrus | 0.007 | 0.233 | |
| Blautia coccoides group | 0.009 | 0.233 | |
| Dorea longicatena | 0.009 | 0.233 | |
| Lactobacillus reuteri group | 0.012 | 0.233 | |
| Veillonella dispar | 0.014 | 0.233 | |
| Clostridium ramosum | 0.018 | 0.233 | |
| Collinsella aerofaciens | 0.018 | 0.233 | |
| Escherichia coli group | 0.023 | 0.233 | |
| Lactobacillus fermentum | 0.025 | 0.244 | |
| Holdemanella biformis | 0.028 | 0.265 | |
| Bifidobacterium catenulatum group | 0.028 | 0.269 | |
| Bifidobacterium bifidum | 0.048 | 0.419 |
FDR, false discovery rate.
Fig. 3. LDA Effect Size (LEfSe) plot of taxonomic biomarkers identified based on the fecal samples of patients. (A) Comparison of asHU and gout patients. Bacterial taxa that were differentially represented between asHU and gout patients (LDA score >2). Left bars indicate enrichment within the samples of gout patients, whereas right bars indicate enrichment within the samples of asHU patients. (B) Comparison of acute gout patients before and after ULT. Bacterial taxa that were differentially present between groups (LDA score >2). Left bars indicate enrichment within the samples of acute gout patients before ULT, whereas right bars indicate enrichment within the samples of acute gout patients after 30-day ULT. asHU, asymptomatic hyperuricemia; LDA, linear discriminant analysis; ULT, uric acid-lowering therapy; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT.
Co-occurrence pattern and uric acid correlation of specific taxon
We identified abundance patterns based on correlation analysis using specific taxon. The microbial clusters of Lactobacillus spp. and Bifidobacterium spp. are presented in Supplementary Fig. 3 (only online). Interestingly, Lactobacillus spp. showed a co-occurrence pattern with Bifidobacterium spp., suggesting symbiotic interactions. On the other hand, both Lactobacillus spp. and Bifidobacterium spp. had negative co-occurrence patterns with P. copri. Notably, nonparametric Spearman correlation showed a negative correlation between serum uric acid level and the proportion of P. copri (Supplementary Fig. 4, only online).
Comparison of gut microbiota before and after uric acid-lowering therapy in acute gout patients
Next, we assessed whether ULT can modulate microbiota composition in patients with acute gout. Minimal changes were observed in alpha diversity before and after the intervention in acute gout patients (Fig. 1C and D), indicating that the microbiota composition did not change. However, beta diversity analysis with canonical correspondence (p=0.05) and redundancy analyses (p=0.05) showed that the composition of the gut microbiome before and after 30-day ULT was significantly different (Supplementary Fig. 5, only online). Significant taxonomic changes were observed in the fecal samples of patients before ULT and after 30-day ULT (p<0.05) based on the Kruskal-Wallis test (Table 3). The proportion of L. helveticus and L. plantarum was significantly higher in the 30ULT group, whereas that of Bradyrhizobium (taxon id: Bradyrhizobium_uc) was higher in the 0ULT group. The results were consistent with qPCR analysis, which showed that the proportion of Bradyrhizobium was significantly higher in acute gout patients who experienced acute gout attacks and never received ULT, and this proportion decreased after ULT (Supplementary Table 2, only online). Furthermore, nonparametric Spearman correlation test showed a significant correlation between serum uric acid level and the proportion of Bradyrhizobium (Supplementary Fig. 4, only online). Using LEfSe, the unique taxa that were differentially present in acute gout patients before and after ULT were identified (Fig. 3B).
Table 3. Taxa That Were Significantly Different in Gout Patients before (0ULT) and after 30-day ULT (30ULT) Based on the Kruskal-Wallis Test (p<0.05).
| Patient category | Taxon name | p value | FDR-adjusted p value |
|---|---|---|---|
| Favor gout | Arthrobacter globiformis group | 0.029 | 0.029 |
| Bradyrhizobium_uc | 0.002 | 0.002 | |
| Sphingomonas_uc | 0.005 | 0.005 | |
| Clostridium_g18: AM275423_s | 0.012 | 0.012 | |
| Favor 30ULT | Lachnospiraceae: AJ518873_g: EF400193_s | 0.040 | 0.041 |
| Eubacterium_g: EF403944_s | 0.029 | 0.030 | |
| Lactobacillus helveticus group | 0.029 | 0.030 | |
| Lactobacillus plantarum group | 0.029 | 0.030 | |
| Oxalobacter: KI392030_s | 0.029 | 0.030 |
FDR, false discovery rate; ULT, uric acid-lowering therapy.
Functional prediction of bacteria using pathway analysis
Pathway analysis using the KEGG database revealed that genes involved in “apoptosis,” “lysosome,” “rheumatoid arthritis,” and “osteoclast differentiation” were over-represented among the significantly predicted genes in gout patients compared to asHU patients (p<0.05) (Supplementary Fig. 6A, only online). Furthermore, the pathway related to the “biosynthesis of unsaturated fatty acids” was more abundant in the after 30-day treatment group than in the before-treatment group (Supplementary Fig. 6B, only online).
DISCUSSION
Here, we examined microbial markers that were enriched or depleted in the asHU and gout groups at different taxonomy levels. Based on the abundance of microbiota, we identified a gout and asHU classifier that could prevent or provoke clinical gout. Subsequently, a microbiome transformation was identified that may be functionally affected by ULT. Overall, we found that gut microbiota composition differs significantly between asHU and gout patients, and proposed microbial markers that may influence the restoration of microbiota composition. The intestinal tract plays an important role in lowering the uric acid level and alters the gut microbiome in gout patients.7,9 We observed that the F/B ratio was significantly higher in asHU patients than in acute gout and chronic gout patients; this ratio increased in gout patients after ULT, indicating that the gut microbiota composition was restored during ULT. Additionally, the gut microbiota in asHU patients had a lower P/B ratio but was more diverse. The F/B ratio has been suggested as an index of gut microbiome health, and is associated with obesity, insulin resistance, dyslipidemia, and other related diseases; it is also used as a biomarker for obesity-associated phenotype,17 whereas the P/B ratio predicts body weight and fat-loss success.18 Therefore, higher F/B and lower P/B ratios in asHU patients than in gout patients indicated that the microbiome of asHU patients is associated with metabolic diseases, whereas that of gout patients is associated with an inflammatory or immune-related disease. Studies have shown that gout is more likely an autoinflammatory disease than a metabolic syndrome. For example, a Guangzhou study analyzing the gut microbiome function and metabolome suggested that gout is more similar to rheumatoid arthritis and ankylosing spondylitis than to obesity and type-2 diabetes.19 Another previous study reported that the number of Faecalibacterium prausnitzii, which exerts anti-inflammatory effects, is lower in gout patients than in healthy controls, whereas that of Bacteroides caccae, which induces inflammation, is higher in gout patients.9 However, previous studies were conducted on the assumption that patients with asHU and gout have comparable microbiota composition; therefore, gout development in the hyperuricemia condition has not been explained. Our findings revealed previously unknown differences in the gut microbiota composition between asHU and gout patients and will provide insights for future research in this field.
Collectively, the results of previous studies and our present study indicate that the gut microbiota in asHU patients have anti-inflammatory properties and participate in uric acid processing. Some probiotic Lactobacillus strains can reduce serum uric acid levels and prevent renal changes and hypertension caused by hyperuricemia.20,21,22,23 L. paracasei suppresses NLRP3 inflammasome activation and inflammatory stress-induced caspase-1 activation by either promoting interleukin-10 production24 or inhibiting interleukin-1β secretion.25 Considering that NLRP inflammasome activation triggers acute gouty arthritis,4 the lactic acid bacteria may suppress gout development owing to their inhibitory effects on inflammasome activation. Therefore, the increased abundance of lactic acid bacteria after ULT may be attributed to the preventive and therapeutic effects observed against gout. Moreover, an increase in the proportion of Bifidobacterium in the gut microbiota of mice exerts beneficial effects in high-fat-diet-induced diabetes by improving glucose tolerance and glucose-induced insulin secretion and reducing inflammation development.26 Streptococcus salivarius,27 Anaerostipes hadrus,28 and Enterococcus durans29 have also been shown to have anti-inflammatory properties. Therefore, the abundance of these bacteria could play a protective role against gout development.
Gout-associated microbiota can trigger gout development when introduced into the gut of a healthy organism. For instance, healthy rats become hyperuricemic after receiving a fecal transplant from hyperuricemic rats.30 The changes in gut microbiome and metabolites, such as decreased level of short-chain fatty acids, may increase the possibility of gout development in susceptible individuals.31 Gut-dwelling P. copri is associated with the pathogenesis of rheumatoid arthritis, especially in pre-clinical stages.32 A putative role for P. copri in the pathogenesis of rheumatoid arthritis is via the P. copri 27-kD protein (Pc-p27) associated immune response.33 Given the distinct pathophysiologic features of rheumatoid arthritis and gout, it is not likely P. copri causes joint inflammation through the same mechanism in both diseases. Instead, the presence of abundant P. copri may reflect subclinical gut inflammation and gingival inflammation. Periodontal Prevotella intermedia, a different species, that has been implicated in periodontitis, was found to be significantly more abundant in gout patients than in healthy controls.34 Likewise, P. copri may lead to subclinical gastrointestinal inflammation, leading to increased permeability that allows pathogens to enter the circulatory system to induce immune responses such as inflammasome activation.35 Bradyrhizobium, a symbiotic abundant in the roots of many legumes, allows the plant to fix nitrous which is associated with purine metabolism and may increase serum uric acid when present in the host. Our findings showed an abundance of Bradyrhizobium in acute gout patients and a significant decrease in their proportion after 30-day ULT, suggestive of their potential to trigger an acute gout attack. However, whether this trigger is associated with metabolites or increased serum uric acid level remains unknown. Until now, the triggers for an acute gout attack in patients with hyperuricemia have been unknown; however, our study suggests that gout-specific microbiota are associated with gouty arthritis, and these results need to be validated in future studies.
In our study, microbiota diversity was high among asHU patients, low among acute gout patients, and restored among chronic gout patients. Moreover, acute gout patients harbored a low proportion of balanced enterotype and an increased proportion of specific strain-dominant type, suggesting that a balanced gut microbiota is important in addition to the presence of specific bacteria. Decreased microbiota diversity may be a trigger for an acute gout attack in hyperuricemic patients. The lack of microbial diversity, in terms of taxa diversity and microbial gene richness, is related to trigger, relapse, or treatment response in various diseases.36 The overall gut microbiota composition explains gut health better than changes in specific bacterial species.37 Hence, future studies should focus on restoring the gut microbiota in gout patients to those observed in healthy individuals, notably by improving the gut microbiota diversity.
Our results suggest that a disturbed microbiome by the dominant increase of “pathogens” with flora disequilibrium may provoke the risk of gout, whereas the abundance of “probionts” may protect patients with asHU from developing gout. The limitations of ULT include poor patient adherence to medication and systemic adverse effects, and it does not guarantee protection against gout attacks or end-organ damages.38 Our findings, which revealed the differences in microbiota composition between asHU and gout patients, have implications for gout prevention, diagnosis, and monitoring of gout pathology. The effort to modify gut microbiota using probionts may prevent gout development for at-risk patients in its occult stages and mitigate the activity of overt gout. Fecal transplantation or probiotics using beneficial microbiota may prevent gout in at-risk patients. Moreover, blood uric acid level, which is an unreliable serum marker as it does not differentiate gout and asHU, can be replaced by a commercialized personal microbiome analysis.
This study included Korean patients with gout and asHU, and there is a possibility of ethnic or geographical influences on the differential composition of the gut microbiome. Previous studies have investigated the lifestyle, dietary, and uncharacterized differences that collectively result in gut microbiota variation due to ethnicity.19,39 Therefore, the diet and lifestyle of Koreans could have affected the microbiome of gout patients and asHU patients in our results. However, our gout patients shared the characteristic microbial taxa of higher abundance of Prevotella and Bacteroides spp., which were previously reported as a microbial signature of gout in different countries.9,10,19,40
Our study had some limitations. We included limited samples only from men, and their diet was not strictly controlled. Future studies should include larger patient cohorts of both sexes. Moreover, both allopurinol and febuxostat were used as ULT, and the impact of medications on the microbiome was not investigated. This should be explored in future investigations.
In conclusion, gout and asHU patients harbor gut microbiota of different compositions, and specific taxa present in the gut microbiome of gout patients may play a role in provoking or preventing gout development. Further assessment to uncover the relationship between specific microbes and gout development and pathogenesis, as well as functional analysis related to uric acid processing and inflammation in the intestines, should be conducted.
ACKNOWLEDGEMENTS
We would like to thank Hye Sun Lee, PhD, and Sinae Kim, MS (Biostatistics Collaboration Unit, Yonsei University College of Medicine) for statistical consultation.
This study was supported by a faculty research grant from Yonsei University College of Medicine (6-2017-0055).
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Hye Won Kim and Min-Chan Park.
- Data curation: Hye Won Kim and Eun-Jeong Yoon.
- Formal analysis: Hye Won Kim and Eun-Jeong Yoon.
- Funding acquisition: Min-Chan Park.
- Investigation: Hye Won Kim.
- Methodology: Hye Won Kim.
- Project administration: all authors.
- Resources: all authors.
- Software: Hye Won Kim.
- Supervision: Seok Hoon Jeong and Min-Chan Park.
- Validation: Seok Hoon Jeong and Min-Chan Park.
- Visualization: Hye Won Kim.
- Writing—original draft: Hye Won Kim and Eun-Jeong Yoon.
- Writing—review&editing: Hye Won Kim and Min-Chan Park.
- Approval of final manuscript: all authors.
SUPPLEMENTARY MATERIALS
List of Primers and Polymerase Chain Reaction Conditions Used in This Study
Changes in Gut Microbiome of Gout Patients before and after ULT Confirmed Using Quantitative PCR
Averaged taxonomic compositions in the bacterial (A) family and (B) species. Variations were apparent between asHU and gout patients. ULT, uric acid-lowering therapy; asHU, asymptomatic hyperuricemia; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT; cULT, chronic gout patients after ≥6-month ULT.
Enterotype analysis. (A) Composition of dominant bacterial families in patients with asHU and with gout. (B) Proportion of patients with dominant taxa group. ULT, uric acid-lowering therapy; asHU, asymptomatic hyperuricemia; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT; cULT, chronic gout patients after ≥6-month ULT.
Co-clustering patterns of Lactobacillus spp. and Bifidobacterium.spp.
Nonparametric Spearman correlation between serum uric acid level and abundance of microbiota.
Beta diversity analysis using canonical correspondence analysis and redundancy analysis for acute gout patients before and after ULT. Each point represents the composition of the intestinal microbiota of each individual. ULT, uric acid-lowering therapy; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT; CCA, canonical correspondence analysis; RDA, redundancy analysis.
Comparison of predicted KEGG pathway in (A) asHU and gout patients and in (B) 0ULT and 30ULT groups. An extended error bar plot indicating the differences in gut microbiota. Only statistically significant pathways with p<0.05 are shown. asHU, asymptomatic hyperuricemia; ULT, uric acid-lowering therapy; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT; KEGG, Kyoto Encyclopedia of Genes and Genomes.
References
- 1.Brucato A, Cianci F, Carnovale C. Management of hyperuricemia in asymptomatic patients: a critical appraisal. Eur J Intern Med. 2020;74:8–17. doi: 10.1016/j.ejim.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Dalbeth N, House ME, Aati O, Tan P, Franklin C, Horne A, et al. Urate crystal deposition in asymptomatic hyperuricaemia and symptomatic gout: a dual energy CT study. Ann Rheum Dis. 2015;74:908–911. doi: 10.1136/annrheumdis-2014-206397. [DOI] [PubMed] [Google Scholar]
- 3.Chhana A, Lee G, Dalbeth N. Factors influencing the crystallization of monosodium urate: a systematic literature review. BMC Musculoskelet Disord. 2015;16:296. doi: 10.1186/s12891-015-0762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joosten LA, Netea MG, Mylona E, Koenders MI, Malireddi RK, Oosting M, et al. Engagement of fatty acids with Toll-like receptor 2 drives interleukin-1β production via the ASC/caspase 1 pathway in monosodium urate monohydrate crystal-induced gouty arthritis. Arthritis Rheum. 2010;62:3237–3248. doi: 10.1002/art.27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosomi A, Nakanishi T, Fujita T, Tamai I. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. PLoS One. 2012;7:e30456. doi: 10.1371/journal.pone.0030456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takada T, Ichida K, Matsuo H, Nakayama A, Murakami K, Yamanashi Y, et al. ABCG2 dysfunction increases serum uric acid by decreased intestinal urate excretion. Nucleosides Nucleotides Nucleic Acids. 2014;33:275–281. doi: 10.1080/15257770.2013.854902. [DOI] [PubMed] [Google Scholar]
- 7.Yun Y, Yin H, Gao Z, Li Y, Gao T, Duan J, et al. Intestinal tract is an important organ for lowering serum uric acid in rats. PLoS One. 2017;12:e0190194. doi: 10.1371/journal.pone.0190194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Chen Y, Zhong H, Chen F, Regenstein J, Hu X, et al. The gut microbiota as a target to control hyperuricemia pathogenesis: Potential mechanisms and therapeutic strategies. Crit Rev Food Sci Nutr. 2021 Jan 22; doi: 10.1080/10408398.2021.1874287. [Epub]. Available at: [DOI] [PubMed] [Google Scholar]
- 9.Guo Z, Zhang J, Wang Z, Ang KY, Huang S, Hou Q, et al. Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep. 2016;6:20602. doi: 10.1038/srep20602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao T, Shao L, Li H, Xie Z, He Z, Wen C. Combined signature of the fecal microbiome and metabolome in patients with gout. Front Microbiol. 2017;8:268. doi: 10.3389/fmicb.2017.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neogi T, Jansen TL, Dalbeth N, Fransen J, Schumacher HR, Berendsen D, et al. 2015 gout classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheumatol. 2015;67:2557–2568. doi: 10.1002/art.39254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 14.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chong J, Liu P, Zhou G, Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. 2020;15:799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 16.Wemheuer F, Taylor JA, Daniel R, Johnston E, Meinicke P, Thomas T, et al. Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ Microbiome. 2020;15:11. doi: 10.1186/s40793-020-00358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 18.Hjorth MF, Blædel T, Bendtsen LQ, Lorenzen JK, Holm JB, Kiilerich P, et al. Prevotella-to-Bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: results from a post-hoc analysis. Int J Obes (Lond) 2019;43:149–157. doi: 10.1038/s41366-018-0093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu Y, Sun S, Huang Y, Gao Q, Xie X, Wang P, et al. Metagenomic analysis revealed the potential role of gut microbiome in gout. NPJ Biofilms Microbiomes. 2021;7:66. doi: 10.1038/s41522-021-00235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo YW, Hsieh SH, Chen JF, Liu CR, Chen CW, Huang YF, et al. Lactobacillus reuteri TSR332 and Lactobacillus fermentum TSF331 stabilize serum uric acid levels and prevent hyperuricemia in rats. PeerJ. 2021;9:e11209. doi: 10.7717/peerj.11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Mei L, Deng Y, Liu Y, Wei X, Liu M, et al. Lactobacillus brevis DM9218 ameliorates fructose-induced hyperuricemia through inosine degradation and manipulation of intestinal dysbiosis. Nutrition. 2019;62:63–73. doi: 10.1016/j.nut.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 22.García-Arroyo FE, Gonzaga G, Muñoz-Jiménez I, Blas-Marron MG, Silverio O, Tapia E, et al. Probiotic supplements prevented oxonic acid-induced hyperuricemia and renal damage. PLoS One. 2018;13:e0202901. doi: 10.1371/journal.pone.0202901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu CL, Hou YH, Wang CS, Lin SW, Jhou BY, Chen CC, et al. Antiobesity and uric acid-lowering effect of Lactobacillus plantarum GKM3 in high-fat-diet-induced obese rats. J Am Coll Nutr. 2019;38:623–632. doi: 10.1080/07315724.2019.1571454. [DOI] [PubMed] [Google Scholar]
- 24.Yamazaki T, Ohshio K, Sugamata M, Morita Y. Lactic acid bacterium, Lactobacillus paracasei KW3110, suppresses inflammatory stress-induced caspase-1 activation by promoting interleukin-10 production in mouse and human immune cells. PLoS One. 2020;15:e0237754. doi: 10.1371/journal.pone.0237754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki H, Yamazaki T, Ohshio K, Sugamata M, Yoshikawa M, Kanauchi O, et al. A specific strain of lactic acid bacteria, lactobacillus paracasei, inhibits inflammasome activation in vitro and prevents inflammation-related disorders. J Immunol. 2020;205:811–821. doi: 10.4049/jimmunol.1900657. [DOI] [PubMed] [Google Scholar]
- 26.Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 27.Di Pierro F, Zanvit A, Nobili P, Risso P, Fornaini C. Cariogram outcome after 90 days of oral treatment with Streptococcus salivarius M18 in children at high risk for dental caries: results of a randomized, controlled study. Clin Cosmet Investig Dent. 2015;7:107–113. doi: 10.2147/CCIDE.S93066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kant R, Rasinkangas P, Satokari R, Pietilä TE, Palva A. Genome sequence of the butyrate-producing anaerobic bacterium anaerostipes hadrus PEL 85. Genome Announc. 2015;3:e00224-15. doi: 10.1128/genomeA.00224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carasi P, Racedo SM, Jacquot C, Elie AM, Serradell ML, Urdaci MC. Enterococcus durans EP1 a promising anti-inflammatory probiotic able to stimulate sIgA and to increase Faecalibacterium prausnitzii abundance. Front Immunol. 2017;8:88. doi: 10.3389/fimmu.2017.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Lv Q, Ren H, Gao L, Zhao P, Yang X, et al. The altered gut microbiota of high-purine-induced hyperuricemia rats and its correlation with hyperuricemia. PeerJ. 2020;8:e8664. doi: 10.7717/peerj.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vieira AT, Galvão I, Macia LM, Sernaglia ÉM, Vinolo MA, Garcia CC, et al. Dietary fiber and the short-chain fatty acid acetate promote resolution of neutrophilic inflammation in a model of gout in mice. J Leukoc Biol. 2017;101:275–284. doi: 10.1189/jlb.3A1015-453RRR. [DOI] [PubMed] [Google Scholar]
- 32.Bernard NJ. Rheumatoid arthritis: Prevotella copri associated with new-onset untreated RA. Nat Rev Rheumatol. 2014;10:2. doi: 10.1038/nrrheum.2013.187. [DOI] [PubMed] [Google Scholar]
- 33.Pianta A, Arvikar S, Strle K, Drouin EE, Wang Q, Costello CE, et al. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017;69:964–975. doi: 10.1002/art.40003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Cui L, Yan X, Zhao X, Cheng J, Zhou L, et al. Analysis of oral microbiota revealed high abundance of Prevotella intermedia in gout patients. Cell Physiol Biochem. 2018;49:1804–1812. doi: 10.1159/000493626. [DOI] [PubMed] [Google Scholar]
- 35.Maeda Y, Takeda K. Host-microbiota interactions in rheumatoid arthritis. Exp Mol Med. 2019;51:1–6. doi: 10.1038/s12276-019-0283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 37.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 38.Chalès G. How should we manage asymptomatic hyperuricemia? Joint Bone Spine. 2019;86:437–443. doi: 10.1016/j.jbspin.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Senghor B, Sokhna C, Ruimy R, Lagier JC. Gut microbiota diversity according to dietary habits and geographical provenance. Hum Microbiome J. 2018;7-8:1–9. [Google Scholar]
- 40.Méndez-Salazar EO, Vázquez-Mellado J, Casimiro-Soriguer CS, Dopazo J, Çubuk C, Zamudio-Cuevas Y, et al. Taxonomic variations in the gut microbiome of gout patients with and without tophi might have a functional impact on urate metabolism. Mol Med. 2021;27:50. doi: 10.1186/s10020-021-00311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of Primers and Polymerase Chain Reaction Conditions Used in This Study
Changes in Gut Microbiome of Gout Patients before and after ULT Confirmed Using Quantitative PCR
Averaged taxonomic compositions in the bacterial (A) family and (B) species. Variations were apparent between asHU and gout patients. ULT, uric acid-lowering therapy; asHU, asymptomatic hyperuricemia; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT; cULT, chronic gout patients after ≥6-month ULT.
Enterotype analysis. (A) Composition of dominant bacterial families in patients with asHU and with gout. (B) Proportion of patients with dominant taxa group. ULT, uric acid-lowering therapy; asHU, asymptomatic hyperuricemia; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT; cULT, chronic gout patients after ≥6-month ULT.
Co-clustering patterns of Lactobacillus spp. and Bifidobacterium.spp.
Nonparametric Spearman correlation between serum uric acid level and abundance of microbiota.
Beta diversity analysis using canonical correspondence analysis and redundancy analysis for acute gout patients before and after ULT. Each point represents the composition of the intestinal microbiota of each individual. ULT, uric acid-lowering therapy; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT; CCA, canonical correspondence analysis; RDA, redundancy analysis.
Comparison of predicted KEGG pathway in (A) asHU and gout patients and in (B) 0ULT and 30ULT groups. An extended error bar plot indicating the differences in gut microbiota. Only statistically significant pathways with p<0.05 are shown. asHU, asymptomatic hyperuricemia; ULT, uric acid-lowering therapy; 0ULT, acute gout patients before ULT; 30ULT, acute gout patients after 30-day ULT; KEGG, Kyoto Encyclopedia of Genes and Genomes.