Abstract
Objective
To summarize the application of non-targeted metabolomics in epidemiological studies that assessed metabolite and metabolic pathway alterations associated with per- and polyfluoroalkyl substances (PFAS) exposure.
Recent Findings
Eleven human studies published before April 1st, 2021 were identified through database searches (PubMed, Dimensions, Web of Science Core Collection, Embase, Scopus), and citation chaining (Citationchaser). The sample sizes of these studies ranged from 40-965, involving children and adolescents (n=3), non-pregnant adults (n=5), or pregnant women (n=3). High-resolution liquid chromatography–mass spectrometry was the primary analytical platform to measure both PFAS and metabolome. PFAS were measured in either plasma (n=6) or serum (n=5), while metabolomic profiles were assessed using plasma (n=6), serum (n=4), or urine (n=1). Four types of PFAS (perfluorooctane sulfonate (n=11), perfluorooctanoic acid (n=10), perfluorohexan sulfonate (n=9), perfluorononanoic acid (n=5)) and PFAS mixtures (n=7) were the most studied. We found that alterations to tryptophan metabolism and the urea cycle were most reported PFAS-associated metabolomic signatures. Numerous lipid metabolites were also suggested to be associated with PFAS exposure, especially key metabolites in glycerophospholipid metabolism which is critical for biological membrane functions, and fatty acids and carnitines which are relevant to the energy supply pathway of fatty acid oxidation. Other important metabolome changes reported included the tricarboxylic acid (TCA) cycle regarding energy generation and purine and pyrimidine metabolism in cellular energy systems.
Conclusions
There is growing interest in using non-targeted metabolomics to study the human physiological changes associated with PFAS exposure. Multiple PFAS were reported to be associated with alterations in amino acid and lipid metabolism, but these results are driven by one predominant type of pathway analysis thus require further confirmation. Standardizing research methods and reporting are recommended to facilitate result comparison. Future studies should consider potential differences in study methodology, use of prospective design, and confounding bias and measurement errors.
Keywords: Exposome, Metabolomics, Persistent Organic Pollutants, Perfluorinated Compounds, Polyfluoroalkyl Substances
1. Introduction
Recent developments in mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy have allowed for a comprehensive and quantitative high-resolution phenotyping of non-targeted metabolic alterations at the molecular level (Holmes et al. 2008; Hu et al. 2020; Jones et al. 2012). This advanced non-targeted workflow can comprehensively and simultaneously assess hundreds or thousands of exogenous chemicals, their metabolites, and associated endogenous metabolic perturbations in small volumes of biological samples (Jin et al. 2021; Tzoulaki et al. 2014). Pathway-mapping and network modeling can further place identified metabolite features into interconnected biological pathways and into context with upstream genes and proteins (Johnson et al. 2016). Furthermore, the characterization of metabolic fingerprints and the deciphering of pathways can be linked to disease risk factors in the general population. This approach is known as the metabolome-wide association study (MWAS), a conceptual and technological tool to reveal the complex cellular mechanisms underlying environmentally mediated diseases. Briefly, the metabolome markers could be important intermediates to investigate the intricate triple relationship between exposure, molecular effect, and clinical outcomes (Bictash et al. 2010; Cai et al. 2020; Chadeau-Hyam et al. 2011; Lu et al. 2019; Nicholson et al. 2008; Rattray et al. 2018).
There has been increasing concern with respect to the potential health effects of per- and polyfluoroalkyl substances (PFAS) (Giesy and Kannan 2001; Sunderland et al. 2019). PFAS are a group of fluorinated chemicals widely used in industrial and commercial applications, including kitchenware, food packaging, clothing, carpeting and coating (Houde et al. 2006; Lau 2015; Wang et al. 2017b). PFAS are persistent and ubiquitous in the environment, and frequently detected in populations globally (Bjerregaard-Olesen et al. 2016; Calafat et al. 2019; Lau et al. 2007; Seo et al. 2018). The half-life of the most commonly identified PFAS in humans is about 4 to 8 years (Olsen et al. 2007). Contaminated water and food intake, indoor air inhalation and skin contact with the household environment are likely major exposure routes in human populations (De Silva et al. 2021; Lau 2015; Sunderland et al. 2019). Epidemiological studies have reported that PFAS exposures were associated with various human health risks, including child and adult adiposity and other cardiometabolic functions (Kahn et al. 2020; Rappazzo et al. 2017), cancers (Bartell and Vieira 2021; Steenland and Winquist 2021), fetal growth and childhood neurodevelopmental outcomes (Liew et al. 2018), and immunological health conditions (Chang et al. 2016). However, the underlying biological mechanisms how the exposures lead to these reported adverse health outcomes remain inconclusive.
Toxicological studies in laboratory animals or human cell lines have examined potential mechanisms of action for PFAS exposure, which include elevated oxidative stress (Chen et al. 2017b; Liu et al. 2016), shift from carbohydrate metabolism to fatty acid oxidation (Bjork et al. 2011), nuclear lipid hyperaccumulation (Li et al. 2017), suppression of glutamate-related neurological pathway (Wang et al. 2019), and loss of gap junction intercellular communication (Upham et al. 2009). Some metabolomics studies in in vivo or in vitro models have associated PFAS exposure with the metabolism of lipids, amino acids and purines (Gong et al. 2019; Ortiz-Villanueva et al. 2018; Zhang et al. 2021), but the dosage in experimental studies might not be comparable to exposure of the general population. Human studies of biological responses associated with PFAS exposures have predominantly focused on selected and targeted lipid or hormone markers or biomarkers of liver function (Donat-Vargas et al. 2019; Geiger et al. 2014; Gleason et al. 2015). In addition, increasing numbers of epidemiological studies have employed non-targeted metabolomics to identify early effect predictors for health risks due to PFAS exposure, but no studies to date have evaluated the methodology and robustness of findings in these environmental metabolomics studies.
The purpose of this scoping review is to provide an assessment of the current evidence regarding non-targeted metabolomics and associations with PFAS exposure in humans. We evaluated the study characteristics, research methods, metabolites identified, and metabolic pathways reported to be associated with specific PFAS chemicals in these studies. We also identified knowledge gaps and made recommendations for future research that examines alterations of the human metabolome associated with PFAS and related environmental chemical exposures.
2. Material and methods
We conducted a scoping review in compliance with the PRISMA methodology for Scoping Reviews (Tricco et al. 2018). This review summarized the literature on epidemiological studies through database searching (PubMed, Dimensions, Web of Science Core Collection as licensed at Yale, Embase via Ovid, Scopus), and citation chaining (via Citationchaser (Haddaway et al. 2021), which uses Lens as its data source). The searched terms used in PubMed were: (PFAS[tw] OR PFASs[tw] OR perfluoro*[tw] OR polyfluoro*[tw] OR PFOS[tw] OR PFOA[tw] OR PFNA[tw] OR PFHxS[tw] OR PFSA[tw] OR PFCA[tw] OR PFOSA[tw] OR PFDA[tw] OR PFUnDA[tw] OR PFDeA[tw] OR PFDoA[tw] OR PFHpA[tw] OR PFUdA[tw] OR EtFOSAA[tw] OR MeFOSAA[tw]) AND (metabolomics[MeSH Terms] OR metabolome[MeSH Terms] OR metabolomics[tw] OR metabolic[tw] OR metabolome[tw] OR metabonomics[tw]) AND (non-targeted[tw] OR non-target[tw] OR untargeted[tw] OR untarget[tw] OR mwas[tw] OR metabolome-wide[tw]) NOT (animal[mh] NOT human[mh]). Similar terms and logic were applied in other databases (Appendix A). We reviewed the titles of all study items found by the search and reviewed abstracts and full articles when necessary to identify studies meeting our inclusion criteria. The inclusion criteria were: 1) published epidemiological original articles, 2) publication date range from the year 2000 to 1st April 2021, 3) no language limit, 4) with at least one type of PFAS measured in participants’ biofluid samples, 5) with non-targeted metabolomics applied in participants’ biofluid samples.
Information was abstracted based on the publication in print and any further appendices provided by the authors using a tabular format. We extracted study characteristics, including study design, country/location (with project names if applicable), study population, sample size, measured PFAS types, sample collection, analytical platform of both PFAS exposure and metabolome, statistical analysis, and main findings on metabolomic features and pathways. In summarizing study findings related to exposure and metabolomic associations, we focused on the four most studied PFAS compounds which were reported in at least three studies (PFOS (n=11), PFOA (n=10), PFHxS (n=9), and PFNA (n=5)) and well-defined PFAS mixtures (n=7). The recent scientific evaluation from European Food Safety Authority (EFSA) has also focused on these four PFAS because they are more comparable in terms of observed levels in human blood, as well as several toxicokinetic effects and health effects in animals (Chain et al. 2020). We first presented the significant metabolomic features and pathways (along with defined statistical significance levels) reported in each study. Whenever available, we recorded metabolomic features with identification confidence levels in the Metabolomics Standards Initiative (MSI) reporting criteria (Schymanski et al. 2014; Sumner et al. 2007). Briefly, the confidence of identification is often communicated with the following five identification levels: fully identified compounds (level 1); putatively annotated compounds (level 2); putatively characterized compound classes (level 3); unassigned compounds (level 4-5) (Schymanski et al. 2014; Sumner et al. 2007). Level 1 identifications were confirmed via an accurate matching on a reference standard with MS, MS/MS and retention time. Level 2 features were identified based on accurate mass and fragmentation patterns using mass spectra in literature or external laboratory data without retention time information. Level 3 features were assigned to a chemical class rather than the exact structure, using a combination of accurate mass, mass spectra and fragmentation pattern knowledge, and retention time window. Level 4 and 5 features were those unable to be assigned with a possible structure, but only molecular formula or exact mass could be affirmed. In our main table (Table 1), we listed confirmed level 1 features, or all assigned features (level 1-3) when specific confidence levels were unavailable, while unassigned compounds (level 4-5) were excluded in this study. Next, we counted the overlap of reported significant associations between aforementioned four individual PFAS or PFAS mixtures and metabolites/pathways across studies. We used stacked barplots in the ggplot2 R package to present the findings (R Development Core Team 2021; Wickham H 2016). A significant association included was defined as a reported false discovery rate (FDR) value <.05 if not otherwise specified in Table 1 or its footnotes. All assigned metabolomic features (level 1 to 3) were also included in the quantitative description, and the features/pathways associated with PFAS were summed by the study numbers. We presented the results for individual PFAS or PFAS mixtures separately. Definitions of PFAS mixtures in each applicable study could be found in Supplementary Table 1. For visualization, metabolites and metabolic pathways were grouped based on chemical taxonomy in human metabolome database (HMDB) (https://hmdb.ca/) and metabolism category in Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/), and ranked by the total number of studies regardless of associated PFAS types.
Table 1.
A summary of research methods and major findings reported in the 11 PFAS and non-targeted metabolomic studies.
| Publication | Location | Study design and sample size | PFAS and non-targeted metabolome measuresa | Analytical platform for PFAS and metabolome | Significant metabolomic features associated with PFAS (FDR <0.05b, confidence levels 1 to 3) | Significant pathways (p for pathway enrichment test <0.05) |
|---|---|---|---|---|---|---|
| (Schillemans et al. 2020) | Northern Sweden | 124 cases of type 2 diabetes (T2D) and 124 controls (40-60 years) enrolled during 1990-2003 and followed at 2000-2013 | PFHxS, PFOS, PFOA, PFNA, PFDA, PFUdA and metabolome were measured in fasting plasma samples at baseline and follow-up. | PFAS: LC-MS/MS; Metabolome: LC-QToF-MS (Agilent Technologies, USA) | PFAS levels were correlated with 171 features (0.16 ≤ |r| ≤ 0.37). Thirty-five features related to both PFAS and T2D and they predominantly represented glycerophospholipids and diacylglycerols. | n/a |
| (Chen et al. 2020) | California, US | 102 non-diabetic young adults (17-22 years) with a history of overweight/ obese enrolled during 2014-17 | PFOA, PFOS, PFHxS and metabolome were measured in fasting and 30-min glucose challenge plasma samples. | PFAS: LC-HRMS Metabolome: Orbitrap LC-HRMS (Dionex Ultimate 3000, Q-Exactive HF, Thermo Scientific) | Features of confidence level 1: proline, glutamine, methionine, citrulline, mannose, lysoPC (18:0), sphingosine, lactate, oxovalerate, glucose, multiple fatty acids. A targeted metabolomics analysis verified that higher PFOA exposure was associated with higher levels of glycerol (p=0.006) and higher fasting levels of short chain non-OH/DC acylcarnitines (p=0.046). | PFAS were associated with 16 pathways, especially the lipid pathways (e.g., fatty acid activation, glycosphingolipid metabolism) and the amino acid-related pathways (e.g., tryptophan metabolism, arginine and proline metabolism). |
| (Jin et al. 2020) | Atlanta, Georgia, US | 74 youths (7-19 years) with non-alcoholic fatty liver disease enrolled during 2007-15 | PFOA, PFOS, PFHxS and metabolome were measured in plasma samples. | PFAS: LC-HRMS Metabolome: Orbitrap LC–HRMS (Dionex Ultimate 3000, Q-Exactive HF, Thermo Scientific) | Six features (confidence levels 1 to 3), including phosphoethanolamine, tyrosine, phenylalanine, aspartate, creatine, betaine. | PFAS were associated with 21 pathways. The most affected pathways included tyrosine metabolism, aspartate and asparagine metabolism, glycine, serine, alanine and threonine metabolism. |
| (Li et al. 2020) | California, US | 404 pregnant women (an average age 25 years) enrolled during 1959-67. Fifty of these women developed breast cancer. | PFOS, PFOA, PFHxS and metabolome were measured in serum. | PFAS: online SPE-HPLC-MS/MS; Metabolome: Orbitrap LC–HRMS (Thermo Q-Exactive (Thermo Fisher, San Diego, CA)) | c Metabolite communities identified by a hierarchical community network model associated with the exposures were non-specific and shared among exposures. The most significant connections are shared with the lipid community. | b Metabolite communities associated with PFAS community (PFAS mixture) were enriched for fatty acid and mitochondria related pathways. |
| (Kingsley et al. 2019) | Ohio and Kentucky, US | 115 liveborn singletons at age 8 years enrolled during 2011-14 | PFOA, PFOS, PFNA, PFHxS and metabolome were measured in serum samples. | PFAS: online SPE-HPLC-MS/MS; Metabolome: Orbitrap LC–HRMS (Dionex Ultimate 3000, Q-Exactive, Thermo Scientific) | In the RPLC C18-negative mode, 17, 63, 47, and 29 features (confidence levels 1 to 4) were associated with serum PFOA, PFOS, PFNA, and PFHxS concentrations; In the HILIC-positive mode, 18, 253, 76, and 39 features (confidence levels 1 to 4) were associated with serum PFOA, PFOS, PFNA, and PFHxS concentrations, respectively, at FDR <20%. | Pathways associated with all four types of PFAS included arginine, proline, aspartate, asparagine, and butanoate metabolism. |
| (Hu et al. 2019) | California, US | 397 pregnant women (an average age 25 years) enrolled during 1959-67. Fifty of these women developed breast cancer. | PFOS, EtFOSAA and metabolome were measured in serum samples. | PFAS: online SPE-HPLC-MS/MS; Metabolome: Orbitrap LC–HRMS (Q-Exactive HF (Thermo Fisher)). | PFOS was associated with 34 features, and EtFOSAA was associated with 49 features. Features with confidence level 1 associated with PFOS included betaine, carnitine, creatine, adrenochrome, aminoisobutyric acid, arginine, asparagine, clupanodonyl carnitine, glutamate, hexadecenoyl carnitine, histidine, linoelaidyl carnitine, lysine, N1-Methyl-4-pyridone-5-carboxamide, N6,N6-dimethyl-L-lysine, peptide 2-(3-carboxy-3-aminopropyl)-L-histidine, phosphoserine, tetracosapentaenoyl carnitine. | Among the 12 associated pathways, the two common pathways that EtFOSAA and PFOS shared were glycine, threonine, alanine and serine metabolism, and urea cycle/amino group metabolism. |
| (Alderete et al. 2019) | Urban Los Angeles, California, US | Hispanic overweight youths (8-14 years) with a family history of T2D enrolled during 2001-12 and were followed at 1-to-3-yr. | PFOA, PFOS, PFHxS were measured in plasma at baseline and metabolome were measured in plasma in both baseline and follow-up. | PFAS: LC-HRMS Metabolome: Orbitrap LC–HRMS (Dionex Ultimate 3000, Q-Exactive HF, Thermo Scientific) | Plasma PFOA, PFOS and PFHxS were associated with 149, 298, and 17 metabolite features (confidence levels 1 to 4), respectively, at FDR <20%. | 24 metabolic pathways were associated with PFAS exposure, including significant alterations of lipids (e.g., glycosphingolipids, linoleic acid), and amino acids (e.g., aspartate and asparagine, tyrosine, arginine and proline). |
| (Maitre et al. 2018) | Catalonia and Basque Country, Spain | 750 pregnant women from Sabadell (2004-08) and Gipuzkoa (2006-08) with an average age 30 years. | PFHxS, PFNA, PFOA, PFOS in first-trimester serum and metabolome were measured in the first and the third trimester serum samples. | PFAS: HPLC-MS/MS Metabolome: NMR spectroscopy (Bruker Biospin, Rheinstetten, Germany) | In Gipuzkoa cohort, pregnanolone-3G, acetone, succinate were prospectively associated with PFHxS, while alanine, glycine, 3-hydroxybutyrate/3-aminoisobutyrate were prospectively associated with PFOA. However, these associations were not replicated in the Sabadell cohort. | n/a |
| (Salihovic et al. 2019) | Uppsala, Sweden | 965 older adults (aged 70 years) with equal gender distribution enrolled during 2001-04 | PFHxS, PFOS PFHpA, PFOA, PFNA, PFUdA and metabolome were measured in overnight fasting plasma samples | PFAS: UPLC-MS/MS; Metabolome: UPLC-Q-ToF-MS (Waters Corporation, Milford, USA) | Lipid related metabolites (e.g., glycerolipids, glycerophospholipids, and fatty acids), and amino acid and purine related metabolites were associated with PFAS. Features of confidence level 1 included L-proline, monoacylglycerol (16:1), phosphatidylcholine, uric acid. | Eight PFAS-associated metabolites mapped to human metabolic pathways were enriched in glycerophospholipid metabolism (p =0.004), linoleic acid metabolism (p = 0.04), and α-linoleic acid metabolism (p = 0.07). |
| (Wang et al. 2017a) | Shandong, China | 181 male adults (median age 34) without metabolic diseases | PFOA, PFOS and metabolome were measured in serum samples. | PFAS: LC-MS; Metabolome: UPLC/Orbitrap-MS (Thermo, USA) | 10 identified features (confidence levels 1 to 3) associated with PFAS included α-CEHC, arachidonic acid, D-glucurono-6,3-lactone, hypoxanthine, oxoglutaric acid, pyroglutamic acid, BH4, xanthine, DAHA, hydroxybutyric acid, at p<0.05. | d The lipid metabolism, xenobiotic detoxifying, antioxidation and the NO signal pathways may be affected by PFAS exposure. |
| (Lu et al. 2019) | Hubei, China | 40 workers (mean age 45) and 52 controls (mean age 50) from the general population enrolled in 2017 | PFBA, PFOA, PFBS, PFHxS, PFOS, 6:2 Cl-PFESA and metabolome were measured in overnight fasting plasma samples. | PFAS: HPLC-MS/MS; Metabolome: UPLC-Q Orbitrap MS system (Thermo, USA) GC-MS (Agilent 7890B/5977A Series GC/Mass Selective Detector system) | Features of confidence level 1 associated with PFAS included pyroglutamic acid, ornithine, hypoxanthine, DL-2-aminooctanoic acid, γ-CEHC, 3-hydroxyoctanoic acid, C18:2-CN, C18:1-CN, sebacic acid, methionine sulfoxide, GPC, azelaic acid, myo-inositol, piperine at p<0.05. | d PFAS were associated with lipid metabolism, amino acids metabolism, purine metabolism, inositol metabolism, retinol metabolism, and metabolism of alkaloids and their derivatives. |
Four studies used fasting samples (Schillemans, 2020; Chen, 2020; Salihovic, 2019; Lu, 2019). Six studies (Jin, 2020; Li, 2020; Kingsley, 2019; Hu, 2019; Maitre, 2018; Wang, 2017) used non-fasting samples. Alderete, 2019: Fasting and post-challenge plasma samples were collected at baseline but unclear which was for PFAS or metabolomics assays.
The significance level for metabolomic features in these studies was false discovery rate (FDR)<0.05 if not specified in the table.
Li, 2020: The significance of associations from PLS regression was assessed by permutation on both community member and sample labels. Instead of feature-level associations, the individual features are organized as members of communities, and the associations are tested at community level across exposome and metabolome.
Lu, 2019 conducted pathway analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, while Wang, 2017 did not specify the tools used for pathway analysis.
Abbreviations: α-CEHC: alpha-carboxyethyl hydroxychromanol; BH4, Tetrahydrobiopterin; 6:2 Cl-PFESA, 6:2 chlorinated polyfluorinated ether sulfonate; CN, acylcarnitine; DAHA: deoxyarabinohexonic acid; EtFOSAA, 2-(N-Ethyl-perfluorooctane sulfonamido) acetic acid; FDR, false discovery rate; γ-CEHC: gamma-carboxyethyl hydroxychroman; GC, gas chromatography; HRMS: high-resolution mass spectrometry; GPC, glycerophosphocholine; HPLC, high performance liquid chromatography; LC, liquid chromatography; LysoPC: lysoophosphatidylcholine; MS, mass spectrometry; NMR, nuclear magnetic resonance; NO, nitric oxide; PFAS, per- and polyfluoroalkyl substances; PFBA, perfluorobutanoic acid; PFBS, perfluorobutanesulfonic acid; PFDA, perfluorodecanoic acid; PFHpA, perfluoroheptanoic acid; PFHxS, perfluorohexane sulfonic acid; PFNA, perfluorononanoic acid; PFOA, perfluorooctanoic acid; PFOS, perfluorooctane sulfonate; QToF: quadrupole time-of-flight; RPLC, reversed phase liquid chromatography; SPE, solid-phase extraction; T2D, type 2 diabetes; UPLC: ultra-pressure liquid chromatography.
We utilized a scoring tool to evaluate the methodology and report quality of included studies, based on existing guidelines (Barnes et al. 2016a; Barnes et al. 2016b; Jin et al. 2021; Spicer et al. 2017; Sud et al. 2016; van der Werf et al. 2007). Five metrics were used including a total score of six (Supplementary Figure 1). The scores for each study were assessed independently by two authors (PG and QY) and subsequently confirmed by a third author (ZL).
3. Results
Figure 1 shows the process of study inclusion and exclusion using the PRISMA 2009 Flow Diagram. We identified a total of 11 eligible publications (2017-2021) written in English. Nine of the included publications were identified through the database searches, and two additional included publications were extracted through citation chaining. Most of the studies reviewed were scored (>3) for having a moderate to high methodological and reporting quality (Supplementary Table 2). Briefly, all studies have provided full description or relevant references on the study populations, the sample collection, and the analytical methods for PFAS exposure measurement. Three studies did not report complete information for data processing workflow including imputation of missing values (Alderete et al. 2019; Hu et al. 2019; Jin et al. 2020), but software used (e.g., xMSanalyzer) was reported. Five studies (Alderete et al. 2019; Jin et al. 2020; Li et al. 2020; Lu et al. 2019; Wang et al. 2017a) did not clearly state certainty of metabolite identification or did not capture high confidence metabolites (level 1-2).
Figure 1. PRISMA Flow diagram depicting process of article selection.
a WOS: Web of Science Core Collection as licensed at Yale Institution. Abbreviation: PFAS, per- and polyfluoroalkyl substances.
3.1. Study designs and populations
Table 1 presents the study characteristics of the included studies. Study samples were collected from nested case-control studies (n=3), prospective cohorts (n=4), cross-sectional studies among occupational workers (n=2) or in high-risk group (i.e., obesity history without diabetes, n=1) or patients of a specific disease (i.e., non-alcoholic fatty liver disease, n=1). The number of study participants in these studies ranged from 40 to 965. The age groups covered were broad and included children and adolescents (Alderete et al. 2019; Jin et al. 2020; Kingsley et al. 2019), young adults (Chen et al. 2020), middle-aged adults (Lu et al. 2019; Schillemans et al. 2020; Wang et al. 2017a), and elderly (Salihovic et al. 2019). There were three studies focused on pregnant women (Hu et al. 2019; Li et al. 2020; Maitre et al. 2018). The eleven studies were conducted in the United States (n=6), in Europe (n=3), and in China (n=2).
Nine studies assessed cross-sectional PFAS and metabolome associations using samples collected at the same study date. One study evaluated baseline PFAS exposure and metabolite changes over an average of 1.3 years of follow-up (Alderete et al. 2019). A pregnancy study assessed PFAS at baseline and metabolomic profiles twice in the first and the third trimesters (Maitre et al. 2018). Details of statistical models and covariate information are presented in Supplementary Table 3.
Four studies were designed to investigate metabolites or metabolic markers associated with PFAS exposures and also their relationship with metabolic health outcomes. Findings of these four studies are presented in Table 2. The investigated health outcomes included non-alcoholic fatty liver disease (NAFLD) (Jin et al. 2020), alterations in glucose homeostasis (Alderete et al. 2019), type 2 diabetes (T2D) (Schillemans et al. 2020), and cardiometabolic outcomes such as oral glucose tolerance test (OGTT) measures, body fat and lipid profiles (Chen et al. 2020).
Table 2.
A summary of studies reporting the metabolomic associations related to both PFAS exposures and health outcomes.
| Publication | Health outcomes | Main findings |
|---|---|---|
| (Schillemans et al. 2020) | Type 2 diabetes (T2D) | PFAS were associated with two groups of lipid species with opposite relations to T2D risk: glycerophospholipids were correlated positively with PFAS and were inversely associated with risk for T2D, while diacylglycerols were correlated positively with both PFAS and risk for T2D. |
| (Chen et al. 2020) | Cardiometabolic outcomes (OGTT measures, body fat and lipid profiles) | Increased lipolysis and fatty acid oxidation were contributing to the biological mechanisms linking PFAS exposure and impaired glucose metabolism among young adults. |
| (Jin et al. 2020) | Non-alcoholic fatty liver disease (NAFLD) | Each interquartile range increase of PFHxS was associated with increased odds for liver fibrosis, lobular inflammation, and higher NAFLD activity score. A cluster of children with nonalcoholic steatohepatitis was characterized by increased PFAS levels and altered metabolite patterns including higher plasma levels of phosphoethanolamine, tyrosine, phenylalanine, aspartate and creatine, and decreased plasma levels of betaine. |
| (Alderete et al. 2019) | Changes in glucose homeostasis from a baseline visit to a 1-to-3-yr visit among adolescents at risk of T2D | Higher PFAS exposure was associated with dysregulation of several lipid and amino acid pathways and longitudinal alterations in glucose homeostasis in Hispanic youth. |
Abbreviations: OGTT, oral glucose tolerance test; PFAS, per- and polyfluoroalkyl substances; PFHxS, perfluorohexane sulfonic acid; PFOA, perfluorooctanoic acid; PFOS, perfluorooctane sulfonate.
3.2. PFAS exposure assessment
Table 1 shows that 2 to 11 types of PFAS were investigated in these 11 studies. The most studied PFAS compounds (in at least 3 studies) were PFOS (n=11), PFOA (n=10), PFHxS (n=9), and PFNA (n=5). PFAS levels were measured in human plasma (n=6) or serum (n=5) samples. Among three studies in pregnant women, one study measured PFAS in the first-trimester serum samples (Maitre et al. 2018) while two studies used serum samples throughout pregnancy and/or from early postpartum period (1-3 days after delivery) (Hu et al. 2019; Li et al. 2020). In addition to studying individual PFAS compounds, seven studies evaluated PFAS mixtures (Supplementary Table 1). Three of these studies computed principal components as a composite variable representing PFAS burden (Alderete et al. 2019; Jin et al. 2020; Schillemans et al. 2020), two studies calculated “total PFAS” by a molarity sum of all the detected PFAS (Lu et al. 2019; Wang et al. 2017a), and one study constructed total PFAS using multivariable multiple linear regression (MMLR) (Salihovic et al. 2019).
All studies employed MS coupled with liquid chromatography (LC) to measure PFAS compounds (Table 1). Additional analytical details regarding PFAS exposure assessments can be found in Supplementary Table 4. All studies reported the overall frequencies of detection for each PFAS and only two studies did not report the limits of detection (Hu et al. 2019; Maitre et al. 2018). Eight studies have provided references for quality assurance and quality control procedures. In general, PFOS was detected at the highest concentrations in most studies, followed by PFOA. The highest concentrations of PFOS appeared in the China-based occupational cohort in 2017 (median ~909 ng/mL) (Wang et al. 2017a), followed by pregnant women in the U.S. CHDS cohort with samples collected in the 1960s (median ~42 ng/mL) (Hu et al. 2019; Li et al. 2020; Wang et al. 2011), and in the middle-aged adults from North Sweden in 1990s (median ~20 ng/mL) (Schillemans et al. 2020). The lowest concentrations of PFOS (median ~4 ng/mL) were reported in a study of U.S. children and adolescents between 2007 and 2015 (Jin et al. 2020). These findings are consistent with the temporal changes in perfluorinated compounds reported previously (Kato et al. 2011; Liu et al. 2021; Nyberg et al. 2018).
3.3. Non-targeted metabolomics analysis
Among all eleven studies, ten studies conducted non-targeted metabolomics analysis in plasma (n=6) or serum (n=4). Only one study (in pregnancy) examined urinary metabolomic profiles (Maitre et al. 2018).
For analytical platforms, ten studies used LC-MS for metabolomics in plasma or sera and one of them additionally used gas chromatography–mass spectrometry (GC-MS) (Lu et al. 2019). One urinary metabolomics (Maitre et al. 2018) analysis was conducted using 1H NMR spectroscopy (Beckonert et al. 2007). LC-MS appears to be the most common approach for metabolomic analyses related to PFAS exposures (Soltow et al. 2013), but there were various characteristics regarding types of ionization (i.e., electrospray ionization, electron ionization), modes of ionization (i.e., positive, negative), mass analyzer (i.e., orbitrap, quadrupole time-of-flight (QToF)), and types of columns. For mass analyzer, an orbitrap was used in eight studies and two used the QToF. Following LC–MS, six studies extracted raw data files and aligned them using apLCMS (Yu et al. 2009; Yu et al. 2013) with modifications by xMSanalyzer R package to improve feature detection, quality assessment, and annotation (Uppal et al. 2013). Three studies (Lu et al. 2019; Salihovic et al. 2019; Schillemans et al. 2020) took raw data into XCMS data processing workflow implemented in R (Ganna et al. 2015; Shi et al. 2017). One (Wang et al. 2017a) used SIEVE software to process the raw data (Zhang et al. 2014).
All studies have described the quality control and quality assurance approaches for the metabolomics analysis undertaken. Four studies reported that all individual samples were analyzed in triplicate (Chen et al. 2020; Hu et al. 2019; Jin et al. 2020; Kingsley et al. 2019), while this information was unclear in the other studies included. Only one study (Hu et al. 2019) included internal standards for non-targeted metabolomics analysis. Among the studies reviewed, three studies (Alderete et al. 2019; Hu et al. 2019; Jin et al. 2020) did not report the methods used for missing value data imputation, data normalization, or transformation. Different tools were used to address batch effects, including a cluster-based approach that calculated measurement drift per batch with batchCorr R package (Brunius et al. 2016), or using the ComBat method in xMSanalyzer software to correct the m/z features by batches (Johnson et al. 2007; Uppal et al. 2013). One study (Schillemans et al. 2020) used the former tool, three studies (Alderete et al. 2019; Jin et al. 2020; Kingsley et al. 2019) applied the latter method. It is unclear whether the three other studies that used xMSanalyzer software (Chen et al. 2020; Hu et al. 2019; Li et al. 2020) also conducted the ComBat method as this was not reported. Details regarding batch correction were not summarized in the remaining studies (Lu et al. 2019; Salihovic et al. 2019; Schillemans et al. 2020; Wang et al. 2017a). The heterogeneity in reporting for metabolomics studies has been previously noted, and suggestions have been made for improving reporting practices (Peter et al. 2021; Rattray et al. 2018).
3.4. Metabolite alterations associated with PFAS exposure
Eight studies reported significant associations between four dominant types of PFAS or PFAS mixtures and metabolites (identified to MSI levels 1-3), while the other three studies did not show feature-level associations (Alderete et al. 2019; Kingsley et al. 2019; Li et al. 2020). The full list of metabolites associated with PFAS is presented in Supplementary Table 5. For metabolite compound classes reported to be associated with PFAS exposures are in at least three studies, the numbers of associations organized by the compound class, the category of metabolite within the class, and the types of PFAS are shown in Figure 2.
Figure 2. A summary of the commonly detected metabolite compound classes (in at least three studies) and categories associated with four individual PFAS compounds and PFAS mixtures.
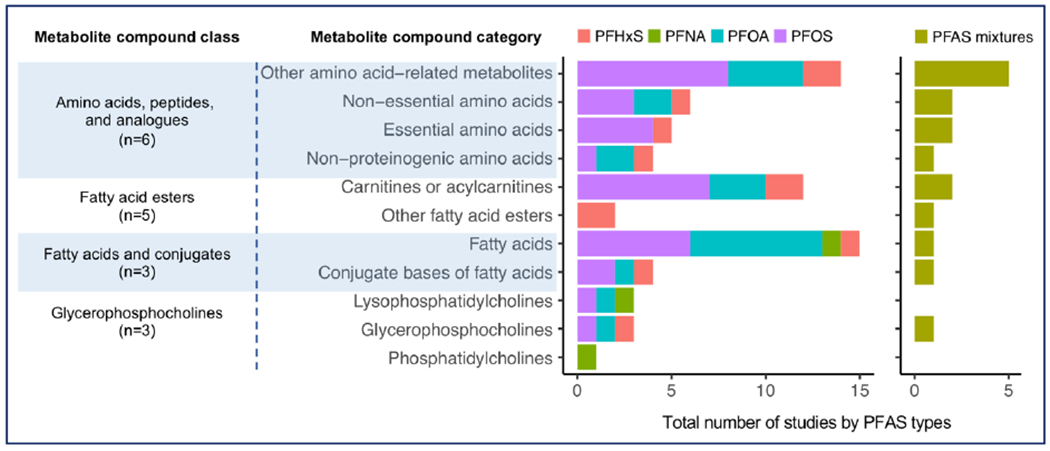
Individual PFAS compounds and the mixtures were differentiated in color, and the total number of significant associations summing across PFAS types reported in the eleven studies were organized according to the metabolite compound class and categories in human metabolome database (HMDB). Abbreviations: PFAS, per- and polyfluoroalkyl substances; PFHxS, perfluorohexane sulfonic acid; PFOS, perfluorooctane sulfonate; PFOA, perfluorooctanoic acid; PFNA, perfluorononanoic acid.
A total of 6 studies reported associations between PFAS and amino acid-related metabolites (Figure 2). Two specific amino acid-related metabolites were suggested in multiple studies which included pyroglutamic acid (Lu et al. 2019; Wang et al. 2017a) and either glutamate/glutamine (Chen et al. 2020; Hu et al. 2019), but the reported associations varied in both positive or negative directions (Supplementary Table 5). In addition, PFAS were also associated with lipids and lipid-like metabolites in the fatty acid esters or fatty acids and related conjugate classes. The specific lipid-related metabolites most commonly reported are carnitines/acylcarnitines (Chen et al. 2020; Hu et al. 2019; Lu et al. 2019) and glycerophosphocholines (Chen et al. 2020; Lu et al. 2019; Salihovic et al. 2019) . Most of these associations reported were related to PFOS and PFOA exposures while a few metabolites were detected for PFHxS or the mixture of PFAS compounds.
3.5. Metabolic pathway alterations associated with PFAS exposure
Nine studies conducted pathway analysis to identify key altered metabolic pathways associated with PFAS exposure. Six of them used Mummichog pathway enrichment analysis (Li et al. 2013), which can directly map metabolites predicted in high-throughput metabolomics data to known metabolic networks and predicts functional activity (Li et al. 2013). One study (Salihovic et al. 2019) performed pathway enrichment and topology analysis using MetaboAnalyst 3.0 (Xia and Wishart 2016). One study (Lu et al. 2019) conducted pathway analysis using the KEGG database (http://www.genome.jp/kegg/). While one study (Wang et al. 2017a) did not provide information on the tools used for pathway analysis.
The full list of altered metabolic pathways significantly associated with four dominant PFAS subtypes or mixtures are shown in Supplementary Table 6. For metabolic pathways reported to be associated with PFAS in at least three studies, the numbers of associations organized by the category of metabolism, specific metabolism pathways, and the types of PFAS are shown in Figure 3.
Figure 3. A summary of the commonly detected metabolic pathways (in at least three studies) associated with four individual PFAS compounds and PFAS mixtures.
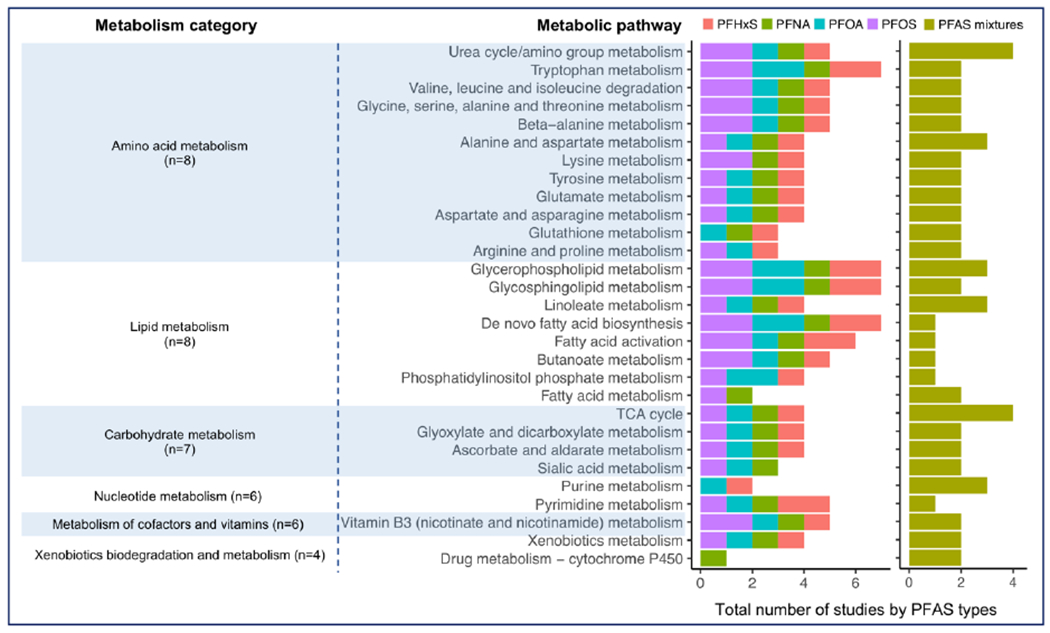
Individual PFAS compounds and the mixtures were differentiated in color and the total number of significant associations summing across PFAS types reported in the eleven studies were organized according to the metabolism category in Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Abbreviations: PFAS, per- and polyfluoroalkyl substances; PFHxS, perfluorohexane sulfonic acid; PFOS, perfluorooctane sulfonate; PFOA, perfluorooctanoic acid; PFNA, perfluorononanoic acid.
Similarly, amino acid metabolism and lipid metabolism were the most commonly suggested metabolic pathways associated with PFAS exposure (Figure 3). For amino acid metabolism, urea cycle/amino group metabolism (Alderete et al. 2019; Hu et al. 2019; Jin et al. 2020; Li et al. 2020; Lu et al. 2019), tryptophan metabolism (Chen et al. 2020; Kingsley et al. 2019; Li et al. 2020; Lu et al. 2019), valine, leucine and isoleucine degradation (Hu et al. 2019; Jin et al. 2020; Kingsley et al. 2019; Li et al. 2020), glycine, serine, alanine and threonine metabolism (Alderete et al. 2019; Hu et al. 2019; Jin et al. 2020; Kingsley et al. 2019), and beta-alanine metabolism (Alderete et al. 2019; Hu et al. 2019; Kingsley et al. 2019; Li et al. 2020) were associated with multiple PFAS exposure types and for PFOS was reported in at least two studies. For lipid metabolism, glycerophospholipid metabolism (Chen et al. 2020; Jin et al. 2020; Kingsley et al. 2019; Li et al. 2020; Salihovic et al. 2019), glycosphingolipid metabolism (Alderete et al. 2019; Chen et al. 2020; Kingsley et al. 2019; Li et al. 2020) , de novo fatty acid biosynthesis (Alderete et al. 2019; Chen et al. 2020; Kingsley et al. 2019), fatty acid activation (Chen et al. 2020; Kingsley et al. 2019; Li et al. 2020), they were associated with various PFAS exposure types and for both PFOS and PFOA were reported in at least two studies. Additional pathways were also suggested, such as carbohydrate metabolism (e.g., TCA cycle), nucleotide metabolism (i.e., purine and pyrimidine metabolism), and other pathways relevant to vitamins and xenobiotics metabolism. These metabolic pathways were linked with each of the individual PFAS compounds and their mixtures. These metabolic pathways were not specific to either plasma or serum samples (Supplementary Figure 2). Although there were only three studies in children and adolescents, this group covered metabolic pathways that were also reported for adults and pregnant women. The Mummichog algorithm contributed the most to the metabolic pathway summary, which also overlaps with pathways reported from using other statistical algorithms.
4. Discussion
The 11 studies we reviewed demonstrated the increasing utilization of the non-targeted metabolomics approach to screen for alterations of the human metabolome associated with specific and mixtures of PFAS exposure in wide demographic groups. We identified some overlapping PFAS exposure and metabolomic associations reported across studies, but these results were synthesized from a relatively low number of eligible studies thus require further scrutiny. Future studies are needed to investigate potential differences across demographic subgroups and research methodology and explore whether the biological pathways altered by PFAS exposures could lead to specific clinical outcomes using longitudinal designs. Standardization of research approach and reporting of metabolomic results (e.g., data processing workflow, confidence levels of metabolite identification, and pathway analyses procedures) are important to support cross-study comparisons. Potential influences from systematic errors, such as confounding bias and measurement errors, also need attention in future research.
Previous epidemiological research have suggested the effects of multiple types of PFAS on altering amino acid and lipid metabolism and cardiometabolic health risks (Chen et al. 2017a; Cheng et al. 2012; Chou et al. 2018; Farthing et al. 2015; Kovalik et al. 2021; Martinez et al. 2020; Qi et al. 2017; Senyavina et al. 2013). In non-targeted metabolomic associations we evaluated, a top-hit pathway related to PFAS exposure in the reviewed studies was urea cycle/amino group metabolism, which is an indispensable pathway to dispose of the excess nitrogen in the body (Alemany 2012; Ramos-Tovar and Muriel 2017). The disruption of urea cycle metabolism has implications on insulin resistance (Cao et al. 2019; Li et al. 2010) and diabetes-related heart failure in older adults (Kovalik et al. 2021; Razavi et al. 2020; Urpi-Sarda et al. 2019). Among metabolites involved in lipid metabolism associated with PFAS, carnitines/acylcarnitines are major molecules in the energy supply pathway of long chain fatty acid β-oxidation. Impaired fatty acid oxidation along with tissue lipid accumulation has been recognized as critical in the pathophysiology of obesity and insulin resistance (Bene et al. 2018; Mihalik et al. 2010), and cardiomyopathy (Kompare and Rizzo 2008; Wang et al. 2018). Another top-hit lipid pathway associated with PFAS exposure was glycerophospholipid metabolism, which has been previously linked to CVD progression (Chen et al. 2021), T2D and tuberculosis comorbidity (Lopez-Hernandez et al. 2019). Overall, these findings which associate PFAS exposure with changes to the endogenous metabolome corroborate results from epidemiological studies, suggesting that PFAS exposures might affect cardiometabolic health outcomes across life-span from childhood (Li et al. 2021a; Manzano-Salgado et al. 2017) to adulthood (Valvi et al. 2021; Zeeshan et al. 2021).
The metabolomic findings also implied that PFAS could have biological effects on several other human organs and physiological processes. For the kidney, one of the most significantly changed pathways for amino acid metabolism in the reviewed studies - tryptophan metabolism, is a sensitive and reliable indicator of renal injury, as the amount of L-tryptophan decreases with the development of chronic kidney disease (Chou et al. 2018; Gong et al. 2019). Abnormalities of purine and pyrimidine metabolism could also lead to hyperuricemia (Gong et al. 2019; Lv et al. 2013; Rathmann et al. 1998), a risk factor of chronic kidney disease (Gong et al. 2019). A growing body of toxicological and epidemiological evidence has indicated that PFAS compounds are emerging environmental threats to kidney health (Stanifer et al. 2018). In terms of the liver, some aberrant amino acids (e.g., glutamate/glutamine) and amino acid derivatives (e.g., pyroglutamic acid) have been recognized as prominent factors for the development of nonalcoholic steatohepatitis (Qi et al. 2017). Impaired fatty acid metabolic pathways included muscle damage (Lehmann et al. 2010) and hepatic dysfunction (Kompare and Rizzo 2008). The liver toxicity of PFAS has been widely documented in animal literature and recently also in clinical studies (Cave 2020; Gleason et al. 2015; Wu et al. 2018). Concerning the central nervous system, glutamate is a major excitatory neurotransmitter in the central nervous system which is critical for neuronal growth and maturation (Wang et al. 2019). The glycerophospholipid metabolism could affect fetal growth (Morillon et al. 2021), the brain neural membrane functions (Farooqui et al. 2000), and neuropsychiatric diseases (Healy-Stoffel and Levant 2018; Kalkman et al. 2021; Lin et al. 2010; Young and Conquer 2005). Finally, the TCA cycle and nucleotide metabolism (i.e., purine and pyrimidine metabolism) are critical for cellular homeostasis and energy generation related mechanisms (Martinez-Reyes and Chandel 2020; Nyhan 2005). A defective TCA cycle has been linked with a variety of clinical diseases in mechanistic studies, ranging from encephalopathies, neurodegenerative diseases, to cancers (Briere et al. 2006).
4.1. Quality of evidence and risk of bias
Approximately 40% of the metabolic pathways associated with PFAS exposure identified in this review were reported in at least three studies, and other sporadic and inconsistent reports could be affected by methodological issues, chance errors, or population differences. Although most studies reviewed received moderate to high methodological and reporting quality, an unstandardized data processing workflow for non-targeted metabolomics data, and the uncertainty of metabolite identifications bring challenges for findings synthesis across studies. First, the low quality of sample collected may introduce measurement errors, and information provided from each study generally did not allow the direct assessments of the influence of sample collection and storage. We may not rule out measurement errors of exposures for each individual study, which is a common issue for any meta-analyses. Quality control procedures should be described in all studies. Strengths and limitations of the two predominantly utilized analytical platforms for data acquisition and processing (i.e., MS and NMR spectroscopy) have been summarized elsewhere (Aderemi et al. 2021). Briefly, NMR spectroscopy has advantages in the reproducibility of result, which is also more quantitative, in addition the technique is non-destructive to the sample and is easier to prepare, whereas the MS approach provides much wider metabolite coverage and that comprehensive metabolite databases have been developed for metabolite identification. The raw data output from both MS and NMR spectroscopy requires pre-processing, including peak selection and alignment, baseline correction, and data preparations such as normalizing, centering, removing outliers and transforming data (Hivert et al. 2015). In addition, missing value imputation and correction for batch effects are also crucial to consider. Standardizing these data steps, and reporting, is important for cross-study comparison and synthesis.
Another important consideration is metabolite identification. Recent advances for metabolite identification have been summarized elsewhere (Nguyen et al. 2019a). To date, the most common method utilized for non-targeted studies is the traditional ion-centric approach, where the signals are compared to a standard reference library, such as the HMDB or METLIN (Montenegro-Burke et al. 2020) to identify metabolites (Hivert et al. 2015). However, given the large number of metabolites and the broad range of chemistries in reality, no reference library is complete to date (Nguyen et al. 2019a). Without stable isotope-labeled internal standards added to each sample for specific chemicals of interest, the non-targeted metabolomics analysis could have larger measurement errors in quantification but non-targeted approach has the advantage of being more feasible and cost-efficient compared to the targeted metabolomics approach (Chen et al. 2020). This is a general concern for current metabolomics studies in all fields which requires further development of metabolite identification techniques and the standardization of reporting.
In comparison, the exposure assessment methods and reporting of the common PFAS compounds in the reviewed studies were more consistent. All studies measured PFAS in blood plasma or serum, which has a 1:1 plasma to serum ratio in humans (Carpenter David et al. 2002; Ehresman et al. 2007). The studies did not utilize other methods to measure PFAS levels that might generate more measurement errors, such as geospatial modeling (Guelfo et al. 2018). The four main compounds of interest in this review generally had very high detection rates, but data steps to handle the levels below the limit of detection varied. A major issue in PFAS exposure assessment is the heterogeneity in the definition of PFAS mixture. Strategies to study environmental mixtures have evolved over time and the statistical modeling techniques are expected to advance (Carpenter David et al. 2002; Weisskopf et al. 2018). Only two of the eleven studies (Alderete et al. 2019; Maitre et al. 2018) collected repeated biological samples with a short time gap between the PFAS assessment and the measures of the metabolome.
Since metabolomics studies can identify thousands of features, multiple testing in statistical analyses needs to be considered. Findings for single/sporadic metabolites are subject to false positive results (type I errors) without multiple testing correction, but false negative results (type II errors) could occur when a strict statistical significance adjustment threshold was used. Pathway enrichment approaches that focus on multiple hits on the same metabolic pathways were suggested to be more robust against false positive findings and yield more consistent and replicable findings (Khatri et al. 2012; Nguyen et al. 2019b). This also underscores a key point to establishing the validity of pathway results –accurate identification of the upstream metabolites, as discussed above. Different methods for pathway enrichment analysis used could create discrepancies for cross-study comparisons. In our review, seven studies conducted pathway enrichment analysis based on an established algorithm within the software (e.g., the Mummichog algorithm), thereby the common findings are mostly driven by a single pathway analysis algorithm. The Mummichog program uses databases which encompass thousands of human metabolites such as KEGG, Recon1, and the Edinburgh human metabolic network, therefore the assignment of metabolites to pathways is based on canonical knowledge of metabolite pathways. Some pathways reported to be associated with PFAS exposure could also be affected by other environmental contaminants (Heindel et al. 2016; Li et al. 2021b), as many of the detoxification pathways are shared, and many of the contaminants are sex-steroid hormone receptor disruptors that share similar effects. Some might argue that this algorithm may systematically lead to ubiquitous endogenous pathways closely linked to oxidative stress and systematic inflammation (Li et al. 2021b; Samet and Wages 2018). However, three studies that did not use Mummichog algorithm also reported amino acid and lipid metabolism associated with PFAS exposure (Lu et al. 2019; Salihovic et al. 2019; Wang et al. 2017a), and these metabolic pathways were also detected in the studies using Mummichog algorithm (Supplementary Figure 2). Our exploratory approach is a timely initial assessment to look for overlaps in metabolomics signatures associated with PFAS exposure based on available evidence. Future mechanistic studies are needed to confirm these findings.
Systematic errors, particularly confounding bias, could influence the findings we reviewed. There is a general confusion in the epidemiological literature on whether the research performed is designed to conduct prediction modeling or to estimate the (causal) effect from defined exposures. If the study objected is to estimate exposure effect, confounding adjustment could follow rules and methods outlined in causal modeling, guided by knowledge in the literature (i.e., use of the directed acyclic graphs to select covariates which was only performed in one of the studies reviewed (Alderete et al. 2019). Generally, diet, lifestyle factors, and the underlying health status of the participants are key potential confounders when estimating PFAS exposure effects on health (Eick et al. 2021; Seshasayee et al. 2021). Demographic-specific variables, such as occupational factors or reproductive history in pregnancy cohorts, should be considered depending on the study setting. Confounding by other environmental pollutants can potentially influence the reported findings (Samet and Wages 2018), but a blind adjustment for multiple highly correlated exposure variables is not encouraged because that would lead to other problems such as a decrease in statistical efficiency and even the risk for bias amplification (Weisskopf et al. 2018). Other sources of bias should also be considered, including survival bias if PFAS exposures can influence mortality in the study setting (Liew et al. 2015; Mastrantonio et al. 2018), and measurements errors that influence both the exposure and the outcome variables. The literature in this field thus needs to pay more attention to these potential problems when correlating PFAS exposure data with markers generated from the human metabolome in order to derive valid inference.
4.2. Limitations of this review
This present review should be considered with several limitations. First, our review was limited by the number of eligible studies, heterogeneity in definitions of PFAS mixtures, and a wide confidence in the robustness of metabolite identification (using MSI levels 1 to 3) detected from the individual studies. In the description of study characteristics, there were some limits on the level of details provided by the individual reports. Moreover, our findings for the PFAS-associated metabolite and metabolic pathway changes across studies would be useful to point towards future investigation, but they should not be used to draw a definite conclusion with the current evidence available. As discussed above, there was considerable heterogeneity in the research methods in the current literature, including the definition for PFAS mixture, the data processing steps and identification of metabolites, and the approaches used for pathway enrichment analyses. Our approach prioritizes on summing and ranking the common and overlapping metabolite features or pathways associated with specific PFAS compounds from the articles we reviewed. This approach, however, would not capture true exposure-metabolomic associations that are demographic and health outcomes specific, e.g., true specific exposure-outcome associations that are not expected to show up across demographic groups from a wide range of studies.
4.3. Recommendations for future research
The findings of our review have identified a few research gaps and highlighted several important areas for future studies. First, longitudinal or repeated measurement of metabolic profiling are needed to distinguish timing-specific or cumulative exposure effects of PFAS on alterations of metabolome. Also, several steps could be taken to further strengthen the confidence and validity of findings derived using metabolomic data. Second, methods that improve the identifications of detected metabolites and the additional use of pathway analysis and replications of findings could rule out false positive results (Cai et al. 2020; Nguyen et al. 2019b). For example, to further investigate PFAS-related exposure-disease mechanisms, a “meet-in-the-middle” approach could be explored, e.g., studying PFAS, metabolome, and disease phenotypes relationships in the same study population (Chadeau-Hyam et al. 2011; Jin et al. 2020; Schillemans et al. 2020). Also, an incorporation of causal mediation analyses using metabolomic data might help to statistically quantify the biological mediating pathways between the association of an environmental exposure and the health outcome (Inoue et al. 2020). Third, biases that can threaten the validity of findings should also be addressed or acknowledged. For instance, all studies reviewed were in observational nature, but few considered potential biases due to confounding, selection bias and measurement errors. An incorporation of statistical methods that could be used to evaluate or adjust for these biases when analyzing high-dimensional biologic data in human cohorts are needed (Misra et al. 2019). More potential confounding factors should be evaluated in future studies, such as factors that can determine the level of PFAS exposures (e.g., diet, occupation, socio-economic profile, co-exposures to other chemicals, and chronic health conditions of participants) and influence the metabolomic signals. Studies of co-pollutant exposures are needed, but blindly co-adjusting multiple correlated exposure variables in one regression model should be avoided (Weisskopf et al. 2018). Forth, it is difficult to standardize the non-targeted metabolomic workflow between research labs due to the various analytical platforms and bioinformatics approaches used, and the inherent complexity of the metabolome for which there is not a one-size-fits-all approach to identify all metabolites within a sample. However, some standardization within the workflow would add robustness, improve the study quality of metabolomics research, and facilitate comparisons of findings across studies (Hernandez-Mesa et al. 2021). Fifth, research of the newer or less studied PFAS compounds with clear reports on quantification procedures (e.g., analytical platforms, LOD, frequencies of detection, missing data imputation) are needed. The studies we reviewed focused on the most common types of PFAS and their mixtures, while recent reports have suggested there are thousands of fluorinated chemicals have been manufactured and used (OECD 2018). The real PFAS body burden is underestimated when not counting for the emerging replacement of other major types of PFAS. Currently, no human studies could accurately quantify all possible PFAS chemicals, and this is awaiting advancement. Finally, literature review of metabolomics studies may benefit from using citation chaining tools and searching synonyms of “non-targeted metabolomics” thoroughly. For instance, the application of non-targeted metabolomics was described as “hypothesis-generating” or indicated by the analytical platform “orbitrap” in some studies (Lu et al. 2019; Wang et al. 2017a), which could be missed by using narrowly defined search terms.
5. Conclusions
In summary, high-resolution non-targeted metabolomics are increasingly being used to evaluate the biological impacts of PFAS exposures in epidemiological studies. Our review found that lipid- and amino acid-related pathways were most reported to be associated with four types of PFAS exposures, and that these PFAS-related metabolome alterations could have implications for cardiometabolic health and other chronic disease health risk. However, the overall body of literature is small, and heterogeneity of research methodology related to metabolomics feature identifications, data processing, and statistical output could influence the findings. The use of longitudinal measures, improvement in certainty of metabolite identification, adjustments for confounding and other sources of biases, and a more standardized procedure in reporting metabolomic findings are some key components identified for improvement.
Supplementary Material
Highlights.
Comprehensive review of research studies on metabolomics of PFAS exposures in humans
PFAS exposures are associated with disruption of amino acid, energy and lipid metabolism
Metabolic alterations associated with PFAS are implicated in cardiometabolic health
Research needs include prospective design, standardized reporting, and risk of bias
Acknowledgments
This project did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. PG is funded by the China Scholarship Council (201906380005). ZL is partly supported by the NIH/NIEHS Pathway to Independence Award (R00ES026729). The views expressed are those of the author(s) and not necessarily those of the funder(s).
Abbreviations
- α-CEHC
alpha-carboxyethyl hydroxychromanol
- γ-CEHC
gamma-carboxyethyl hydroxychroman
- BH4
Tetrahydrobiopterin
- C18:2-CN
acylcarnitine 18:2
- C18:1-CN
acylcarnitine 18:1
- DAHA
deoxyarabinohexonic acid
- FDR
false discovery rate
- GPC
glycerophosphocholine
- HILIC
hydrophilic interaction liquid chromatography
- HMDB
Human Metabolome Database
- HPLC
high performance liquid chromatography
- HRMS
high-resolution mass spectrometry
- LysoPC
lysoophosphatidylcholine
- NFALD
non-alcoholic fatty liver
- NMR
nuclear magnetic resonance
- NO
nitric oxide
- NR
not reported
- OGTT
oral glucose tolerance test
- OH/DC
hydroxyl-/dicarboxyl- acylcarnitine
- POPs
persistent organic pollutants
- 6:2 Cl-PFESA
6:2 chlorinated polyfluorinated ether sulfonate
- EtFOSAA
2-(N-Ethyl-perfluorooctane sulfonamido) acetic acid
- MeFOSAA
2-(N-Methyl-perfluorooctane sulfonamido) acetic acid
- PFAS
per- and polyfluoroalkyl substances
- PFBA
perfluorobutanoic acid
- PFBS
perfluorobutanesulfonic acid
- PFHxS
perfluorohexane sulfonic acid
- PFOS
perfluorooctane sulfonate
- PFOA
perfluorooctanoic acid
- PFDA
perfluorodecanoic acid
- PFNA
perfluorononanoic acid
- PFDoA
perfluorododecanoic acid
- PFHpA
perfluoroheptanoic acid
- PFOSA
perfluorooctanesulfonamide
- PFUdA
perfluoroundecanoic acid
- QToF
quadrupole time-of-flight
- RPLC
reversed phase liquid chromatography
- Short-chain non-OH/DC
Short-chain non-hydroxyl-/dicarboxyl acylcarnitine
- SPE
solid-phase extraction
- TCA cycle
Tricarboxylic acid cycle
- T2D
type 2 diabetes
- UPLC
ultra-pressure liquid chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aderemi AV; Ayeleso AO; Oyedapo OO; Mukwevho E Metabolomics: A scoping review of its role as a tool for disease biomarker discovery in selected non-communicable diseases. Metabolites 2021;11:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderete TL; Jin R; Walker DI; Valvi D; Chen Z; Jones DP; Peng C; Gilliland FD; Berhane K; Conti DV; Goran MI; Chatzi L Perfluoroalkyl substances, metabolomic profiling, and alterations in glucose homeostasis among overweight and obese Hispanic children: A proof-of-concept analysis. Environ Int 2019;126:445–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemany M The problem of nitrogen disposal in the obese. Nutr Res Rev 2012;25:18–28 [DOI] [PubMed] [Google Scholar]
- Barnes S; Benton HP; Casazza K; Cooper SJ; Cui X; Du X; Engler JA; Kabarowski JH; Li S; Pathmasiri W; Prasain JK; Renfrow MB; Tiwari HK Training in metabolomics research. II. Processing and statistical analysis of metabolomics data, metabolite identification, pathway analysis, applications of metabolomics and its future. J Mass Spectrom 2016a;51:535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes S; Benton HP; Casazza K; Cooper SJ; Cui X; Du X; Engler JA; Kabarowski JH; Li S; Pathmasiri W; Prasain JK; Renfrow MB; Tiwari HK Training in metabolomics research. I. Designing the experiment, collecting and extracting samples and generating metabolomics data. J Mass Spectrom 2016b;51:ii–iii [DOI] [PubMed] [Google Scholar]
- Bartell SM; Vieira VM Critical review on PFOA, kidney cancer, and testicular cancer. J Air Waste Manag Assoc 2021;71:663–679 [DOI] [PubMed] [Google Scholar]
- Beckonert O; Keun HC; Ebbels TM; Bundy J; Holmes E; Lindon JC; Nicholson JK Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc 2007;2:2692–2703 [DOI] [PubMed] [Google Scholar]
- Bene J; Hadzsiev K; Melegh B Role of carnitine and its derivatives in the development and management of type 2 diabetes. Nutr Diabetes 2018;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bictash M; Ebbels TM; Chan Q; Loo RL; Yap IK; Brown IJ; de Iorio M; Daviglus ML; Holmes E; Stamler J; Nicholson JK; Elliott P Opening up the “Black Box”: metabolic phenotyping and metabolome-wide association studies in epidemiology. J Clin Epidemiol 2010;63:970–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerregaard-Olesen C; Bach CC; Long M; Ghisari M; Bossi R; Bech BH; Nohr EA; Henriksen TB; Olsen J; Bonefeld-Jorgensen EC Time trends of perfluorinated alkyl acids in serum from Danish pregnant women 2008-2013. Environ Int 2016;91:14–21 [DOI] [PubMed] [Google Scholar]
- Bjork JA; Butenhoff JL; Wallace KB Multiplicity of nuclear receptor activation by PFOA and PFOS in primary human and rodent hepatocytes. Toxicology 2011;288:8–17 [DOI] [PubMed] [Google Scholar]
- Briere JJ; Favier J; Gimenez-Roqueplo AP; Rustin P Tricarboxylic acid cycle dysfunction as a cause of human diseases and tumor formation. Am J Physiol Cell Physiol 2006;291:C1114–1120 [DOI] [PubMed] [Google Scholar]
- Brunius C; Shi L; Landberg R Large-scale untargeted LC-MS metabolomics data correction using between-batch feature alignment and cluster-based within-batch signal intensity drift correction. Metabolomics 2016;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y; Rosen Vollmar AK; Johnson CH Analyzing Metabolomics Data for Environmental Health and Exposome Research. Methods Mol Biol 2020;2104:447–467 [DOI] [PubMed] [Google Scholar]
- Calafat AM; Kato K; Hubbard K; Jia T; Botelho JC; Wong LY Legacy and alternative per- and polyfluoroalkyl substances in the U.S. general population: Paired serum-urine data from the 2013-2014 National Health and Nutrition Examination Survey. Environ Int 2019;131:105048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YF; Li J; Zhang Z; Liu J; Sun XY; Feng XF; Luo HH; Yang W; Li SN; Yang X; Fang ZZ Plasma Levels of Amino Acids Related to Urea Cycle and Risk of Type 2 Diabetes Mellitus in Chinese Adults. Front Endocrinol (Lausanne) 2019;10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter David O; Arcaro K; Spink David C Understanding the human health effects of chemical mixtures. Environ Health Perspect 2002;110:25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cave MC Environmental Pollution and the Developmental Origins of Childhood Liver Disease. Hepatology 2020;72:1518–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadeau-Hyam M; Athersuch TJ; Keun HC; De Iorio M; Ebbels TM; Jenab M; Sacerdote C ; Bruce SJ; Holmes E; Vineis P Meeting-in-the-middle using metabolic profiling - a strategy for the identification of intermediate biomarkers in cohort studies. Biomarkers 2011;16:83–88 [DOI] [PubMed] [Google Scholar]
- Chain, E.Panel O.C.i.t.F.; Schrenk D; Bignami M; Bodin L; Chipman JK; del Mazo J; Grasl-Kraupp B; Hogstrand C; Hoogenboom L; Leblanc J-C; Nebbia CS; Nielsen E; Ntzani E; Petersen A; Sand S; Vleminckx C; Wallace H; Barregård L; Ceccatelli S; Cravedi J-P; Halldorsson TI; Haug LS; Johansson N; Knutsen HK; Rose M; Roudot A-C; Van Loveren H; Vollmer G; Mackay K; Riolo F; Schwerdtle T. Risk to human health related to the presence of perfluoroalkyl substances in food. EFSA Journal 2020;18:e06223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang ET; Adami HO; Boffetta P; Wedner HJ; Mandel JS A critical review of perfluorooctanoate and perfluorooctanesulfonate exposure and immunological health conditions in humans. Crit Rev Toxicol 2016;46:279–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H; Wang Z; Qin M; Zhang B; Lin L; Ma Q; Liu C; Chen X; Li H; Lai W; Zhong S Comprehensive Metabolomics Identified the Prominent Role of Glycerophospholipid Metabolism in Coronary Artery Disease Progression. Front Mol Biosci 2021;8:632950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T; He P; Tan Y; Xu D Biomarker identification and pathway analysis of preeclampsia based on serum metabolomics. Biochem Biophys Res Commun 2017a;485:119–125 [DOI] [PubMed] [Google Scholar]
- Chen Y; Zhou L; Xu J; Zhang L; Li M; Xie X; Xie Y; Luo D; Zhang D; Yu X; Yang B; Kuang H Maternal exposure to perfluorooctanoic acid inhibits luteal function via oxidative stress and apoptosis in pregnant mice. Reprod Toxicol 2017b;69:159–166 [DOI] [PubMed] [Google Scholar]
- Chen Z; Yang T; Walker DI; Thomas DC; Qiu C; Chatzi L; Alderete TL; Kim JS; Conti DV; Breton CV; Liang D; Hauser ER; Jones DP; Gilliland FD Dysregulated lipid and fatty acid metabolism link perfluoroalkyl substances exposure and impaired glucose metabolism in young adults. Environ Int 2020;145:106091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S; Rhee EP; Larson MG; Lewis GD; McCabe EL; Shen D; Palma MJ; Roberts LD; Dejam A; Souza AL; Deik AA; Magnusson M; Fox CS; O’Donnell CJ; Vasan RS; Melander O; Clish CB; Gerszten RE; Wang TJ Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation 2012;125:2222–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J; Liu R; Yu J; Liu X; Zhao X; Li Y; Liu L; Sun C Fasting serum alphahydroxybutyrate and pyroglutamic acid as important metabolites for detecting isolated post-challenge diabetes based on organic acid profiles. J Chromatogr B Analyt Technol Biomed Life Sci 2018;1100–1101:6–16 [DOI] [PubMed] [Google Scholar]
- De Silva AO; Armitage JM; Bruton TA; Dassuncao C; Heiger-Bernays W; Hu XC; Karrman A; Kelly B; Ng C; Robuck A; Sun M; Webster TF; Sunderland EM PFAS Exposure Pathways for Humans and Wildlife: A Synthesis of Current Knowledge and Key Gaps in Understanding. Environ Toxicol Chem 2021;40:631–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donat-Vargas C; Bergdahl IA; Tornevi A; Wennberg M; Sommar J; Koponen J; Kiviranta H; Akesson A Associations between repeated measure of plasma perfluoroalkyl substances and cardiometabolic risk factors. Environ Int 2019;124:58–65 [DOI] [PubMed] [Google Scholar]
- Ehresman DJ; Froehlich JW; Olsen GW; Chang SC; Butenhoff JL Comparison of human whole blood, plasma, and serum matrices for the determination of perfluorooctanesulfonate (PFOS), perfluorooctanoate (PFOA), and other fluorochemicals. Environ Res 2007;103:176–184 [DOI] [PubMed] [Google Scholar]
- Eick SM; Goin DE; Trowbridge J; Cushing L; Smith SC; Park JS; DeMicco E; Padula AM; Woodruff TJ; Morello-Frosch R Dietary predictors of prenatal per- and polyfluoroalkyl substances exposure. J Expo Sci Environ Epidemiol 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui AA; Horrocks LA; Farooqui T Glycerophospholipids in brain: their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chem Phys Lipids 2000;106:1–29 [DOI] [PubMed] [Google Scholar]
- Farthing DE; Farthing CA; Xi L Inosine and hypoxanthine as novel biomarkers for cardiac ischemia: from bench to point-of-care. Exp Biol Med (Maywood) 2015;240:821–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganna A; Fall T; Salihovic S; Lee W; Broeckling CD; Kumar J; Hägg S; Stenemo M; Magnusson PKE; Prenni JE; Lind L; Pawitan Y; Ingelsson E Large-scale non-targeted metabolomic profiling in three human population-based studies. Metabolomics 2015;12:4 [Google Scholar]
- Geiger SD; Xiao J; Ducatman A; Frisbee S; Innes K; Shankar A The association between PFOA, PFOS and serum lipid levels in adolescents. Chemosphere 2014;98:78–83 [DOI] [PubMed] [Google Scholar]
- Giesy JP; Kannan K Global distribution of perfluorooctane sulfonate in wildlife. Environ Sci Technol 2001;35:1339–1342 [DOI] [PubMed] [Google Scholar]
- Gleason JA; Post GB; Fagliano JA Associations of perfluorinated chemical serum concentrations and biomarkers of liver function and uric acid in the US population (NHANES), 2007-2010. Environ Res 2015;136:8–14 [DOI] [PubMed] [Google Scholar]
- Gong X; Yang C; Hong Y; Chung ACK; Cai Z PFOA and PFOS promote diabetic renal injury in vitro by impairing the metabolisms of amino acids and purines. Sci Total Environ 2019;676:72–86 [DOI] [PubMed] [Google Scholar]
- Guelfo JL; Marlow T; Klein DM; Savitz DA; Frickel S; Crimi M; Suuberg EM Evaluation and Management Strategies for Per- and Polyfluoroalkyl Substances (PFASs) in Drinking Water Aquifers: Perspectives from Impacted U.S. Northeast Communities. Environ Health Perspect 2018;126:065001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddaway NR; Grainger MJ; Gray CT citationchaser: an R package for forward and backward citations chasing in academic searching. 2021 [DOI] [PubMed] [Google Scholar]
- Healy-Stoffel M; Levant B N-3 (Omega-3) Fatty Acids: Effects on Brain Dopamine Systems and Potential Role in the Etiology and Treatment of Neuropsychiatric Disorders. CNS Neurol Disord Drug Targets 2018;17:216–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindel JJ; Blumberg B; Cave M; Machtinger R; Mantovani A; Mendez MA; Nadal A; Palanza P; Panzica G; Sargis RM; Vandenberg LN; Saal F.S.v. Metabolism disrupting chemicals and metabolic disorders. 2016;68:3–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Mesa M; Le Bizec B; Dervilly G Metabolomics in chemical risk analysis - A review. Anal Chim Acta 2021;1154:338298. [DOI] [PubMed] [Google Scholar]
- Hivert MF; Perng W; Watkins SM; Newgard CS; Kenny LC; Kristal BS; Patti ME; Isganaitis E; DeMeo DL; Oken E; Gillman MW Metabolomics in the developmental origins of obesity and its cardiometabolic consequences. J Dev Orig Health Dis 2015;6:65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E; Wilson ID; Nicholson JK Metabolic phenotyping in health and disease. Cell 2008;134:714–717 [DOI] [PubMed] [Google Scholar]
- Houde M; Martin JW; Letcher RJ; Solomon KR; Muir DC Biological monitoring of polyfluoroalkyl substances: A review. Environ Sci Technol 2006;40:3463–3473 [DOI] [PubMed] [Google Scholar]
- Hu X; Li S; Cirillo P; Krigbaum N; Tran V; Ishikawa T; La Merrill MA; Jones DP; Cohn B Metabolome Wide Association Study of serum DDT and DDE in Pregnancy and Early Postpartum. Reprod Toxicol 2020;92:129–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X; Li S; Cirillo PM; Krigbaum NY; Tran V; Jones DP; Cohn BA Metabolome Wide Association Study of Serum Poly and Perfluoroalkyl Substances (PFASs) in Pregnancy and Early Postpartum. Reprod Toxicol 2019;87:70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K; Yan Q; Arah OA; Paul K; Walker DI; Jones DP; Ritz B Air Pollution and Adverse Pregnancy and Birth Outcomes: Mediation Analysis Using Metabolomic Profiles. Curr Environ Health Rep 2020;7:231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L; Pollitt KJG; Liew Z; Vollmar AKR; Vasiliou V; Johnson CH; Zhang Y Use of Untargeted Metabolomics to Explore the Air Pollution-Related Disease Continuum. Current Environmental Health Reports 2021:1–16 [DOI] [PubMed] [Google Scholar]
- Jin R; McConnell R; Catherine C; Xu S; Walker DI; Stratakis N; Jones DP; Miller GW; Peng C; Conti DV; Vos MB; Chatzi L Perfluoroalkyl substances and severity of nonalcoholic fatty liver in Children: An untargeted metabolomics approach. Environ Int 2020;134:105220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH; Ivanisevic J; Siuzdak G Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 2016;17:451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE; Li C; Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–127 [DOI] [PubMed] [Google Scholar]
- Jones DP; Park Y; Ziegler TR Nutritional metabolomics: progress in addressing complexity in diet and health. Annu Rev Nutr 2012;32:183–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn LG; Philippat C; Nakayama SF; Slama R; Trasande L Endocrine-disrupting chemicals: implications for human health. Lancet Diabetes Endocrinol 2020;8:703–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkman HO; Hersberger M; Walitza S; Berger GE Disentangling the Molecular Mechanisms of the Antidepressant Activity of Omega-3 Polyunsaturated Fatty Acid: A Comprehensive Review of the Literature. Int J Mol Sci 2021;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K; Wong LY; Jia LT; Kuklenyik Z; Calafat AM Trends in exposure to polyfluoroalkyl chemicals in the U.S. Population: 1999-2008. Environ Sci Technol 2011;45:8037–8045 [DOI] [PubMed] [Google Scholar]
- Khatri P; Sirota M; Butte AJ Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol 2012;8:e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley SL; Walker DI; Calafat AM; Chen A; Papandonatos GD; Xu Y; Jones DP; Lanphear BP; Pennell KD; Braun JM Metabolomics of childhood exposure to perfluoroalkyl substances: a cross-sectional study. Metabolomics 2019;15:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kompare M; Rizzo WB Mitochondrial fatty-acid oxidation disorders. Semin Pediatr Neurol 2008;15:140–149 [DOI] [PubMed] [Google Scholar]
- Kovalik JP; Zhao X; Gao F; Leng S; Chow V; Chew H; Teo LLY; Tan RS; Ewe SH; Tan HC; Wee HN; Lee LS; Ching J; Keng BMH; Koh WP; Zhong L; Koh AS Amino acid differences between diabetic older adults and non-diabetic older adults and their associations with cardiovascular function. J Mol Cell Cardiol 2021;158:63–71 [DOI] [PubMed] [Google Scholar]
- Lau C Perfluorinated Compounds: An Overview. in: DeWitt JC, ed. Toxicological Effects of Perfluoroalkyl and Polyfluoroalkyl Substances. Cham: Springer International Publishing; 2015 [Google Scholar]
- Lau C; Anitole K; Hodes C; Lai D; Pfahles-Hutchens A; Seed J Perfluoroalkyl acids: a review of monitoring and toxicological findings. Toxicol Sci 2007;99:366–394 [DOI] [PubMed] [Google Scholar]
- Lehmann R; Zhao X; Weigert C; Simon P; Fehrenbach E; Fritsche J; Machann J; Schick F; Wang J; Hoene M; Schleicher ED; Haring HU; Xu G; Niess AM Medium chain acylcarnitines dominate the metabolite pattern in humans under moderate intensity exercise and support lipid oxidation. PLoS One 2010;5:e11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K; Gao P; Xiang P; Zhang X; Cui X; Ma LQ Molecular mechanisms of PFOA-induced toxicity in animals and humans: Implications for health risks. Environ Int 2017;99:43–54 [DOI] [PubMed] [Google Scholar]
- Li LO; Hu YF; Wang L; Mitchell M; Berger A; Coleman RA Early hepatic insulin resistance in mice: a metabolomics analysis. Mol Endocrinol 2010;24:657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N; Liu Y; Papandonatos GD; Calafat AM; Eaton CB; Kelsey KT; Cecil KM; Kalkwarf HJ; Yolton K; Lanphear BP; Chen A; Braun JM Gestational and childhood exposure to per- and polyfluoroalkyl substances and cardiometabolic risk at age 12 years. Environ Int 2021a;147:106344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S; Cirillo P; Hu X; Tran V; Krigbaum N; Yu S; Jones DP; Cohn B Understanding mixed environmental exposures using metabolomics via a hierarchical community network model in a cohort of California women in 1960’s. Reprod Toxicol 2020;92:57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S; Park Y; Duraisingham S; Strobel FH; Khan N; Soltow QA; Jones DP; Pulendran B Predicting network activity from high throughput metabolomics. PLoS Comput Biol 2013;9:e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z; Liang D; Ye D; Chang HH; Ziegler TR; Jones DP; Ebelt ST Application of high-resolution metabolomics to identify biological pathways perturbed by traffic-related air pollution. Environ Res 2021b;193:110506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew Z; Goudarzi H; Oulhote Y Developmental Exposures to Perfluoroalkyl Substances (PFASs):An Update of Associated Health Outcomes. Curr Environ Health Rep 2018;5:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew Z; Olsen J; Cui X; Ritz B; Arah OA Bias from conditioning on live birth in pregnancy cohorts: an illustration based on neurodevelopment in children after prenatal exposure to organic pollutants. Int J Epidemiol 2015;44:345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PY; Huang SY; Su KP A meta-analytic review of polyunsaturated fatty acid compositions in patients with depression. Biol Psychiatry 2010;68:140–147 [DOI] [PubMed] [Google Scholar]
- Liu G; Zhang S; Yang K; Zhu L; Lin D Toxicity of perfluorooctane sulfonate and perfluorooctanoic acid to Escherichia coli: Membrane disruption, oxidative stress, and DNA damage induced cell inactivation and/or death. Environ Pollut 2016;214:806–815 [DOI] [PubMed] [Google Scholar]
- Liu L; Qu Y; Huang J; Weber R Per- and polyfluoroalkyl substances (PFASs) in Chinese drinking water: risk assessment and geographical distribution. Environmental Sciences Europe 2021;33:6 [Google Scholar]
- Lopez-Hernandez Y; Lara-Ramirez EE; Salgado-Bustamante M; Lopez JA; Oropeza-Valdez JJ; Jaime-Sanchez E; Castaneda-Delgado JE; Magana-Aquino M; Murgu M; Enciso-Moreno JA Glycerophospholipid Metabolism Alterations in Patients with Type 2 Diabetes Mellitus and Tuberculosis Comorbidity. Arch Med Res 2019;50:71–78 [DOI] [PubMed] [Google Scholar]
- Lu Y; Gao K; Li X; Tang Z; Xiang L; Zhao H; Fu J; Wang L; Zhu N; Cai Z; Liang Y; Wang Y; Jiang G Mass Spectrometry-Based Metabolomics Reveals Occupational Exposure to Per- and Polyfluoroalkyl Substances Relates to Oxidative Stress, Fatty Acid beta-Oxidation Disorder, and Kidney Injury in a Manufactory in China. Environ Sci Technol 2019;53:9800–9809 [DOI] [PubMed] [Google Scholar]
- Lv Q; Meng XF; He FF; Chen S; Su H; Xiong J; Gao P; Tian XJ; Liu JS; Zhu ZH; Huang K; Zhang C High serum uric acid and increased risk of type 2 diabetes: a systemic review and meta-analysis of prospective cohort studies. PLoS One 2013;8:e56864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre L; Robinson O; Martinez D; Toledano MB; Ibarluzea J; Marina LS; Sunyer J; Villanueva CM; Keun HC; Vrijheid M; Coen M Urine Metabolic Signatures of Multiple Environmental Pollutants in Pregnant Women: An Exposome Approach. Environ Sci Technol 2018;52:13469–13480 [DOI] [PubMed] [Google Scholar]
- Manzano-Salgado CB; Casas M; Lopez-Espinosa MJ; Ballester F; Iniguez C; Martinez D; Romaguera D; Fernandez-Barres S; Santa-Marina L; Basterretxea M; Schettgen T; Valvi D; Vioque J; Sunyer J; Vrijheid M Prenatal Exposure to Perfluoroalkyl Substances and Cardiometabolic Risk in Children from the Spanish INMA Birth Cohort Study. Environ Health Perspect 2017;125:097018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez PJ; Agudiez M; Molero D; Martin-Lorenzo M; Baldan-Martin M; Santiago-Hernandez A; Garcia-Segura JM; Madruga F; Cabrera M; Calvo E; Ruiz-Hurtado G;Barderas MG; Vivanco F; Ruilope LM; Alvarez-Llamas G Urinary metabolic signatures reflect cardiovascular risk in the young, middle-aged, and elderly populations. J Mol Med (Berl) 2020;98:1603–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Reyes I; Chandel NS Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 2020;11:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrantonio M; Bai E; Uccelli R; Cordiano V; Screpanti A; Crosignani P Drinking water contamination from perfluoroalkyl substances (PFAS): an ecological mortality study in the Veneto Region, Italy. Eur J Public Health 2018;28:180–185 [DOI] [PubMed] [Google Scholar]
- Mihalik SJ; Goodpaster BH; Kelley DE; Chace DH; Vockley J; Toledo FG; DeLany JP Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity (Silver Spring) 2010;18:1695–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra BB; Langefeld C; Olivier M; Cox LA Integrated omics: tools, advances and future approaches. J Mol Endocrinol 2019;62:R21–R45 [DOI] [PubMed] [Google Scholar]
- Montenegro-Burke JR; Guijas C; Siuzdak G METLIN: A Tandem Mass Spectral Library of Standards. in: Li S., ed. Computational Methods and Data Analysis for Metabolomics. New York, NY: Springer US; 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon AC; Leite DFB; Yakkundi S; Gethings LA; Thomas G; Baker PN; Kenny LC; English JA; McCarthy FP Glycerophospholipid and detoxification pathways associated with small for gestation age pathophysiology: discovery metabolomics analysis in the SCOPE cohort. Metabolomics 2021;17:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH; Nguyen CH; Mamitsuka H Recent advances and prospects of computational methods for metabolite identification: a review with emphasis on machine learning approaches. Brief Bioinform 2019a;20:2028–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TM; Shafi A; Nguyen T; Draghici S Identifying significantly impacted pathways: a comprehensive review and assessment. Genome Biol 2019b;20:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK; Holmes E; Elliott P The metabolome-wide association study: a new look at human disease risk factors. J Proteome Res 2008;7:3637–3638 [DOI] [PubMed] [Google Scholar]
- Nyberg E; Awad R; Bignert A; Ek C; Sallsten G; Benskin JP Inter-individual, inter-city, and temporal trends of per-and polyfluoroalkyl substances in human milk from Swedish mothers between 1972 and 2016. Environmental Science: Processes & Impacts 2018;20:1136–1147 [DOI] [PubMed] [Google Scholar]
- Nyhan WL Disorders of purine and pyrimidine metabolism. Mol Genet Metab 2005;86:25–33 [DOI] [PubMed] [Google Scholar]
- OECD. Toward a new comprehensive global database of per-and polyfluoroalkyl substances (PFASs): Summary report on updating the OECD 2007 list of per-and polyfluoroalkyl substances (PFASs). OECD Environment Directorate, Environment, Health and Safety Division Paris; …; 2018 [Google Scholar]
- Olsen GW; Burris JM; Ehresman DJ; Froehlich JW; Seacat AM; Butenhoff JL; Zobel LR Half-life of serum elimination of perfluorooctanesulfonate,perfluorohexanesulfonate, and perfluorooctanoate in retired fluorochemical production workers. Environ Health Perspect 2007;115:1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Villanueva E; Jaumot J; Martinez R; Navarro-Martin L; Pina B; Tauler R Assessment of endocrine disruptors effects on zebrafish (Danio rerio) embryos by untargeted LC-HRMS metabolomic analysis. Sci Total Environ 2018;635:156–166 [DOI] [PubMed] [Google Scholar]
- Peter KT; Phillips AL; Knolhoff AM; Gardinali PR; Manzano CA; Miller KE; Pristner M; Sabourin L; Sumarah MW; Warth B; Sobus JR Nontargeted Analysis Study Reporting Tool: A Framework to Improve Research Transparency and Reproducibility. Anal Chem 2021;93:13870–13879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S; Xu D; Li Q; Xie N; Xia J; Huo Q; Li P; Chen Q; Huang S Metabonomics screening of serum identifies pyroglutamate as a diagnostic biomarker for nonalcoholic steatohepatitis. Clin Chim Acta 2017;473:89–95 [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2021 [Google Scholar]
- Ramos-Tovar E; Muriel P Does Nutrition Matter in Liver Disease? in: Muriel P, ed. Liver Pathophysiology. Boston: Academic Press; 2017 [Google Scholar]
- Rappazzo KM; Coffman E; Hines EP Exposure to Perfluorinated Alkyl Substances and Health Outcomes in Children: A Systematic Review of the Epidemiologic Literature. Int J Environ Res Public Health 2017;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmann W; Funkhouser E; Dyer AR; Roseman JM Relations of hyperuricemia with the various components of the insulin resistance syndrome in young black and white adults: the CARDIA study. Ann Epidemiol 1998;8:250–261 [DOI] [PubMed] [Google Scholar]
- Rattray NJW; Deziel NC; Wallach JD; Khan SA; Vasiliou V; Ioannidis JPA; Johnson CH Beyond genomics: understanding exposotypes through metabolomics. Hum Genomics 2018;12:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi AC; Bazzano LA; He J; Fernandez C; Whelton SP; Krousel-Wood M; Li S; Nierenberg JL; Shi M; Li C; Mi X; Kinchen J; Kelly TN Novel Findings From a Metabolomics Study of Left Ventricular Diastolic Function: The Bogalusa Heart Study. J Am Heart Assoc 2020;9:e015118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salihovic S; Fall T; Ganna A; Broeckling CD; Prenni JE; Hyotylainen T; Karrman A; Lind PM; Ingelsson E; Lind L Identification of metabolic profiles associated with human exposure to perfluoroalkyl substances. J Expo Sci Environ Epidemiol 2019;29:196–205 [DOI] [PubMed] [Google Scholar]
- Samet JM; Wages PA Oxidative stress from environmental exposures. Current Opinion in Toxicology 2018;7:60–66 [PMC free article] [PubMed] [Google Scholar]
- Schillemans T; Shi L; Donat-Vargas C; Hanhineva K; Tornevi A; Johansson I; Koponen J; Kiviranta H; Rolandsson O; Bergdahl IA; Landberg R; Åkesson A; Brunius C Plasma metabolites associated with exposure to perfluoroalkyl substances and risk of type 2 diabetes - A nested case-control study. Environ Int 2020;146:106180. [DOI] [PubMed] [Google Scholar]
- Schymanski EL; Jeon J; Gulde R; Fenner K; Ruff M; Singer HP; Hollender J Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol 2014;48:2097–2098 [DOI] [PubMed] [Google Scholar]
- Senyavina NV; Khaustova SA; Grebennik TK; Pavlovich SV Analysis of purine metabolites in maternal serum for evaluating the risk of gestosis. Bull Exp Biol Med 2013;155:682–684 [DOI] [PubMed] [Google Scholar]
- Seo SH; Son MH; Choi SD; Lee DH; Chang YS Influence of exposure to perfluoroalkyl substances (PFASs) on the Korean general population: 10-year trend and health effects. Environ Int 2018;113:149–161 [DOI] [PubMed] [Google Scholar]
- Seshasayee SM; Rifas-Shiman SL; Chavarro JE; Carwile JL; Lin PD; Calafat AM; Sagiv SK; Oken E; Fleisch AF Dietary patterns and PFAS plasma concentrations in childhood: Project Viva, USA. Environ Int 2021;151:106415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L; Brunius C; Lindelof M; Shameh SA; Wu H; Lee I; Landberg R; Moazzami AA Targeted metabolomics reveals differences in the extended postprandial plasma metabolome of healthy subjects after intake of whole-grain rye porridges versus refined wheat bread. Mol Nutr Food Res 2017;61:1600924. [DOI] [PubMed] [Google Scholar]
- Soltow QA; Strobel FH; Mansfield KG; Wachtman L; Park Y; Jones DP High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DCFTMS) for study of the exposome. Metabolomics 2013;9:S132–S143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer RA; Salek R; Steinbeck C Compliance with minimum information guidelines in public metabolomics repositories. Sci Data 2017;4:170137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanifer JW; Stapleton HM; Souma T; Wittmer A; Zhao X; Boulware LE Perfluorinated Chemicals as Emerging Environmental Threats to Kidney Health: A Scoping Review. Clin J Am Soc Nephrol 2018;13:1479–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenland K; Winquist A PFAS and cancer, a scoping review of the epidemiologic evidence. Environ Res 2021;194:110690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud M; Fahy E; Cotter D; Azam K; Vadivelu I; Burant C; Edison A; Fiehn O; Higashi R; Nair KS; Sumner S; Subramaniam S Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res 2016;44:D463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW; Amberg A; Barrett D; Beale MH; Beger R; Daykin CA; Fan TW; Fiehn O; Goodacre R; Griffin JL; Hankemeier T; Hardy N; Harnly J; Higashi R; Kopka J; Lane AN; Lindon JC; Marriott P; Nicholls AW; Reily MD; Thaden JJ; Viant MR Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007;3:211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderland EM; Hu XC; Dassuncao C; Tokranov AK; Wagner CC; Allen JG A review of the pathways of human exposure to poly- and perfluoroalkyl substances (PFASs) and present understanding of health effects. J Expo Sci Environ Epidemiol 2019;29:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricco AC; Lillie E; Zarin W; O’Brien KK; Colquhoun H; Levac D; Moher D; Peters MDJ; Horsley T; Weeks L; Hempel S; Akl EA; Chang C; McGowan J; Stewart L; Hartling L; Aldcroft A; Wilson MG; Garritty C; Lewin S; Godfrey CM; Macdonald MT; Langlois EV; Soares-Weiser K; Moriarty J; Clifford T; Tuncalp O; Straus SE PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann Intern Med 2018;169:467–473 [DOI] [PubMed] [Google Scholar]
- Tzoulaki I; Ebbels TM; Valdes A; Elliott P; Ioannidis JP Design and analysis of metabolomics studies in epidemiologic research: a primer on-omic technologies. Am J Epidemiol 2014;180:129–139 [DOI] [PubMed] [Google Scholar]
- Upham BL; Park J-S; Babica P; Sovadinova I; Rummel AM; Trosko JE; Hirose A; Hasegawa R; Kanno J; Sai K Structure-activity-dependent regulation of cell communication by perfluorinated fatty acids using in vivo and in vitro model systems. National Institute of Environmental Health Sciences; 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K; Soltow QA; Strobel FH; Pittard WS; Gernert KM; Yu T; Jones DP xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urpi-Sarda M; Almanza-Aguilera E; Llorach R; Vazquez-Fresno R; Estruch R; Corella D; Sorli JV; Carmona F; Sanchez-Pla A; Salas-Salvado J; Andres-Lacueva C Non-targeted metabolomic biomarkers and metabotypes of type 2 diabetes: A cross-sectional study of PREDIMED trial participants. Diabetes Metab 2019;45:167–174 [DOI] [PubMed] [Google Scholar]
- Valvi D; Hojlund K; Coull BA; Nielsen F; Weihe P; Grandjean P Life-course Exposure to Perfluoroalkyl Substances in Relation to Markers of Glucose Homeostasis in Early Adulthood. J Clin Endocrinol Metab 2021;106:2495–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf MJ; Takors R; Smedsgaard J; Nielsen J; Ferenci T; Portais JC; Wittmann C; Hooks M; Tomassini A; Oldiges M; Fostel J; Sauer U Standard reporting requirements for biological samples in metabolomics experiments: microbial and in vitro biology experiments. Metabolomics 2007;3:189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M; Park JS; Petreas M Temporal changes in the levels of perfluorinated compounds in California women’s serum over the past 50 years. Environ Sci Technol 2011;45:7510–7516 [DOI] [PubMed] [Google Scholar]
- Wang X; Liu L; Zhang W; Zhang J; Du X; Huang Q; Tian M; Shen H Serum metabolome biomarkers associate low-level environmental perfluorinated compound exposure with oxidative /nitrosative stress in humans. Environ Pollut 2017a;229:168–176 [DOI] [PubMed] [Google Scholar]
- Wang Y; Wang L; Chang W; Zhang Y; Zhang Y; Liu W Neurotoxic effects of perfluoroalkyl acids: Neurobehavioral deficit and its molecular mechanism. Toxicol Lett 2019;305:65–72 [DOI] [PubMed] [Google Scholar]
- Wang Z; DeWitt JC; Higgins CP; Cousins IT A Never-Ending Story of Per- and Polyfluoroalkyl Substances (PFASs)? Environ Sci Technol 2017b;51:2508–2518 [DOI] [PubMed] [Google Scholar]
- Wang ZY; Liu YY; Liu GH; Lu HB; Mao CY l-Carnitine and heart disease. Life Sci 2018;194:88–97 [DOI] [PubMed] [Google Scholar]
- Weisskopf MG; Seals RM; Webster TF Bias Amplification in Epidemiologic Analysis of Exposure to Mixtures. Environ Health Perspect 2018;126:047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York; 2016 [Google Scholar]
- Wu X; Xie G; Xu X; Wu W; Yang B Adverse bioeffect of perfluorooctanoic acid on liver metabolic function in mice. Environ Sci Pollut Res Int 2018;25:4787–4793 [DOI] [PubMed] [Google Scholar]
- Xia J; Wishart DS Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinformatics 2016;55:14 10 11–14 10 91 [DOI] [PubMed] [Google Scholar]
- Young G; Conquer J Omega-3 fatty acids and neuropsychiatric disorders. Reprod Nutr Dev 2005;45:1–28 [DOI] [PubMed] [Google Scholar]
- Yu T; Park Y; Johnson JM; Jones DP apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics 2009;25:1930–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T; Park Y; Li S; Jones DP Hybrid feature detection and information accumulation using high-resolution LC-MS metabolomics data. J Proteome Res 2013;12:1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeshan M; Zhang YT; Yu S; Huang WZ; Zhou Y; Vinothkumar R; Chu C; Li QQ; Wu QZ; Ye WL; Zhou P; Dong P; Zeng XW; Hu LW; Yang BY; Shen X; Zhou Y; Dong GH Exposure to isomers of per- and polyfluoroalkyl substances increases the risk of diabetes and impairs glucose-homeostasis in Chinese adults: Isomers of C8 health project. Chemosphere 2021;278:130486. [DOI] [PubMed] [Google Scholar]
- Zhang J; Mu X; Xia Y; Martin FL; Hang W; Liu L; Tian M; Huang Q; Shen H Metabolomic analysis reveals a unique urinary pattern in normozoospermic infertile men. J Proteome Res 2014;13:3088–3099 [DOI] [PubMed] [Google Scholar]
- Zhang R; Yao Y; Tu L; Luan T; Chen B Non-targeted metabolomics of multiple human cells revealing differential toxic effects of perfluorooctanoic acid. J Hazard Mater 2021;409:125017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


