Abstract
The constitutive androstane receptor (CAR) is a nuclear receptor that plays a crucial role in regulating xenobiotic metabolism and detoxification, energy homeostasis, and cell proliferation by modulating the transcription of numerous target genes. CAR activation has been established as the mode of action by which phenobarbital-like nongenotoxic carcinogens promote liver tumor formation in rodents. This paradigm, however, appears to be unrelated to the function of human CAR (hCAR) in hepatocellular carcinoma (HCC), which remains poorly understood. Here, we show that hCAR expression is significantly lower in HCC than that in adjacent nontumor tissues and, importantly, reduced hCAR expression is associated with a worse HCC prognosis. We also show overexpression of hCAR in human hepatoma cells (HepG2 and Hep3B) profoundly suppressed cell proliferation, cell cycle progression, soft-agar colony formation, and the growth of xenografts in nude mice. RNA-Seq analysis revealed that the expression of erythropoietin (EPO), a pleiotropic growth factor, was markedly repressed by hCAR in hepatoma cells. Addition of recombinant EPO in HepG2 cells partially rescued hCAR-suppressed cell viability. Mechanistically, we showed that overexpressing hCAR repressed mitogenic EPO–EPO receptor signaling through dephosphorylation of signal transducer and activator of transcription 3, AKT, and extracellular signal–regulated kinase 1/2. Furthermore, we found that hCAR downregulates EPO expression by repressing the expression and activity of hepatocyte nuclear factor 4 alpha, a key transcription factor regulating EPO expression. Collectively, our results suggest that hCAR plays a tumor suppressive role in HCC development, which differs from that of rodent CAR and offers insight into the hCAR–hepatocyte nuclear factor 4 alpha–EPO axis in human liver tumorigenesis.
Keywords: human CAR, HCC, hepatoma cell proliferation, EPO, HNF4α
Abbreviations: CAR, constitutive androstane receptor; cDNA, complementary DNA; Dox, doxycycline; DR2, direct repeat 2; EPO, erythropoietin; EPOR, EPO receptor; ERK1/2, extracellular signal–regulated kinase 1/2; hCAR, human CAR; HCC, hepatocellular carcinoma; HEK293T, human embryonic kidney 293T cell line; HIF, hypoxia-inducible factor; HNF4α, hepatocyte nuclear factor 4 alpha; LBD, ligand-binding domain; PB, phenobarbital; STAT, signal transducer and activator of transcription; TCGA, The Cancer Genome Atlas; Tet, tetracycline
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related mortality worldwide and one of the fastest rising malignancies in the United States (1, 2). Despite improved screening and surveillance for early diagnosis and more treatment options, the 5-year survival rate of patients with HCC remains low (3). The dismal outcome has been attributed mainly to the highly metastatic nature and the angiogenic microenvironment of HCC. The pathogenesis of HCC is complex and multifactorial with risk factors, including hepatitis virus infection, chronic alcohol consumption, aflatoxin-B1 contamination, and more recently, the progression of fatty liver to nonalcoholic steatohepatitis, fibrosis, and cirrhosis (4). Mechanistically, dysregulation of hepatic transcription factors including nuclear receptors contributes significantly to the development and progression of HCC (5).
The constitutive androstane receptor (CAR, NR1I3), a member of the nuclear receptor superfamily, is a xenobiotic sensor primarily expressed in the liver that regulates the transcription of numerous genes associated with drug metabolism, detoxification, and clearance (6, 7). Upregulation of drug-metabolizing enzymes and transporters by CAR often accelerates biotransformation of drugs, leading to decreased therapeutic efficacy, altered drug toxicity, or increased bioactivation of prodrugs. In addition to these well-established roles in drug disposition, accumulating evidence extends the function of CAR to the regulation of various physiological and pathophysiological processes, including energy homeostasis, liver cell proliferation, and tumorigenesis. For instance, phenobarbital (PB) and 1,4-Bis [2-(3,5-dichloropyridyloxy)] benzene are prototypical CAR activators and known nongenotoxic carcinogens that promote the development of liver cancer in rodents (8, 9). Using CAR−/− mice, studies have established activation of CAR as a key event in rodent liver tumor formation stimulated by PB and related compounds, by which CAR activation results in altered hepatic gene expression and increased cell proliferation (10, 11, 12).
Notably, although human CAR (hCAR) exhibits several common features with its rodent counterparts, significant interspecies differences of CAR in energy homeostasis and cell proliferation have been observed (13, 14, 15). Structural comparison of classic nuclear receptors from different species revealed >90% sequence conservation in the ligand-binding domain (LBD) regions; nevertheless, the LBD sequences of hCAR and mouse CAR share only 71% amino acid homology (7, 16). Our understanding of the role hCAR plays in cancer development is particularly limited, and the underlying molecular mechanisms are largely unknown. Interestingly, PB-induced replicative DNA synthesis and cell proliferation in rats and mice were not observed in either cultured human hepatocytes in vitro or in chimeric mice with humanized liver in vivo (17, 18). Moreover, epidemiological studies suggest that PB does not increase liver tumor incidence in humans, even after long-term treatment at doses producing plasma concentrations that are carcinogenic in rodents (19). Our previous genome-wide transcriptome study indicated that numerous cell proliferation–associated genes were upregulated in the hCAR−/− HepaRG cell line (20). Collectively, these studies support a new paradigm wherein the role of hCAR in HCC development is in stark contrast to that of its rodent counterparts.
In the current study, we investigated the role of hCAR in HCC development and cell proliferation by using integrative approaches including high-resolution array-based gene profiling in HCC clinical samples coupled with patient prognosis; RNA-Seq transcriptome analysis; and cell-based gene regulation, proliferation, colony formation, cell signaling analyses, as well as in vivo xenograft experiments. We found that hCAR expression is significantly lower in HCC than that in adjacent nontumor tissues, and lower hCAR expression is associated with a poorer prognosis for HCC. Ectopic overexpression of hCAR attenuates the growth of hepatoma cells both in vitro and in vivo. In addition, we reveal an important role of erythropoietin (EPO) in CAR-mediated HCC suppression. The expression of EPO in HCC is inversely correlated with that of hCAR, and HCC patients with high EPO/low hCAR are associated with decreased overall survival. We also demonstrate that hCAR transcriptionally downregulates EPO by repressing hepatocyte nuclear factor 4 alpha (HNF4α). Thus, our findings support an innovative suppressive role of hCAR in HCC development and hepatoma cell proliferation, which may open the door to novel biomarkers and potential therapeutics.
Results
Downregulation of hCAR in HCC is associated with poor patient outcomes
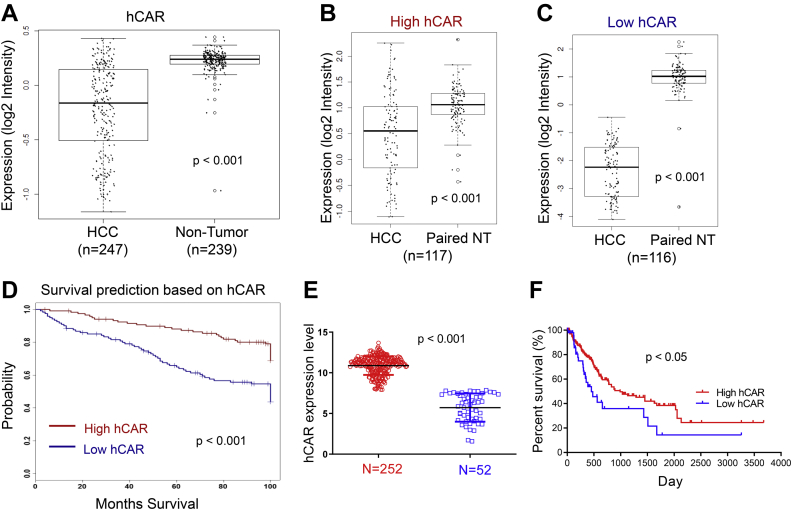
To investigate the potential roles of CAR in human liver cancer, the expression of hCAR in HCC versus nontumor liver tissues and its influence on the overall survival of patients with HCC were analyzed using the Liver Hepatocellular Carcinoma cohort as described elsewhere (21). As shown in Figure 1A, Affymetrix mRNA expression data indicated that mRNA expression of hCAR in HCC (n = 247) is significantly lower than that in nontumor liver tissues (n = 239, p < 0.001). To test whether hCAR expression is associated with clinical outcomes of patients with HCC, HCC samples in this cohort were subdivided into high (>average; n = 117) and low (<average; n = 116) hCAR groups based on the median values of mRNA expression. Notably, while hCAR expression in both groups is significantly lower than in their paired nontumors (Fig. 1, B and C), poorer overall survival rate is associated with lower levels of hCAR in patients with HCC (Fig. 1D; log-rank test, p < 0.001). A similar survival trend was observed by bioinformatic analysis of an independent HCC dataset from The Cancer Genome Atlas (TCGA) through SurvExpress (Catedra de Bioinformatica) (22), in which higher hCAR level is associated with longer survival, whereas lower hCAR expression is linked to poor prognosis in patients with HCC (Fig. 1, E and F). Taken together, these observations suggest that hCAR may exhibit tumor suppressive roles in human liver cancer.
Figure 1.
CAR downregulation correlates with poor survival in patients with liver cancer.A, the microarray analysis of hCAR expression in 247 HCC and 239 nontumor liver tissues reveals expression of hCAR is significantly lower in HCC. B and C, HCC samples were further divided into high and low hCAR clusters, and both have lower levels of hCAR in comparison to their paired controls. D, HCC patients with relatively low levels of hCAR are associated with poor survival (log-rank test, p < 0.05; n = 116). E and F, the analysis of hCAR expression and survival using TCGA datasets demonstrated that low expression of hCAR is associated with poor overall survival. CAR, constitutive androstane receptor; hCAR, human CAR; HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas.
Overexpression of hCAR inhibits the viability of hepatoma cells
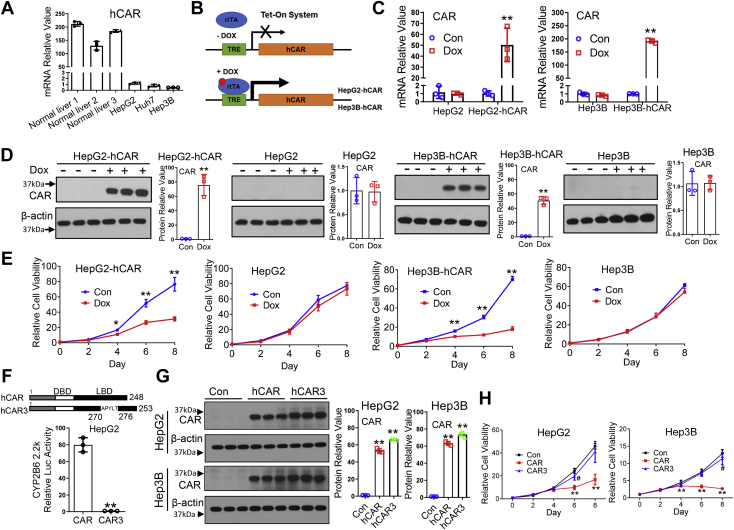
The role of hCAR in HCC cell viability was determined in experiments using HepG2 and Hep3B, two broadly used human hepatoma cell lines. We initially examined the basal mRNA levels of hCAR in normal human primary hepatocytes and several hepatoma cell lines. As shown in Figure 2A, compared with normal human hepatocytes, expression of hCAR in these tumor cells is negligible, which correlates well with observations between HCC and nontumor liver tissues described previously. This is also supported by data from analyzing a published MERAV database, where similar results were observed in seven human normal liver tissues and 45 liver cancer cell lines (Fig. S1). Ectopic overexpression of hCAR in HepG2 and Hep3B was achieved by using the Tet-On system as depicted in Figure 2B. Characterization of the two Tet-On-based–inducible HepG2-hCAR and Hep3B-hCAR stable lines revealed that treatment with doxycycline (Dox; 1 μg/ml) markedly increased expression of hCAR in Tet-On-hCAR–inducible but not normal HepG2 and Hep3B cell lines (Fig. 2C). Importantly, overexpression of hCAR significantly repressed the cell viability of HepG2-hCAR and Hep3B-hCAR cells in comparison with the vehicle control groups (Figs. 2, D, E, and S2), whereas Dox has no effects on the expression of hCAR or the growth of normal HepG2 and Hep3B cells. These results were further confirmed using a second cell viability experiment (Cell-Titer-Glo luminescent cell viability assay; Fig. S3).
Figure 2.
Overexpression of CAR suppresses the viability of hepatoma cells.A, endogenous mRNA expression of hCAR was measured using RT–PCR in human primary hepatocytes (three liver donors) and liver cancer cells (HepG2, Hep3B, and Huh7). B, schematic illustration of the generation of Tet-On-based hCAR-inducible hepatoma cell line. C, mRNA expression of hCAR was analyzed in HepG2-hCAR and Hep3B-hCAR cell lines as well as normal HepG2 and Hep3B cells treated with vehicle control (Con) or Dox (1 μg/ml). D, protein expression of hCAR in these cell lines is consistent with the mRNA changes 72 h after Dox or vehicle control treatment. E, overexpression of hCAR in HepG2-hCAR and Hep3B-hCAR cells results in suppression of cell growth in a time-dependent manner through 8 days of Dox treatment, whereas the growth of normal HepG2 and Hep3B was not significantly affected. F, HepG2 cells were transfected with the CYP2B6 luciferase reporter construct in the presence of hCAR or hCAR3 expression vector. Relative luciferase activity was measured using the Promega dual-luc reagent. In a separate experiment, (G) overexpression of hCAR or hCAR3 in HepG2 and Hep3B cells was achieved by infection of lentivirus-expressing hCAR or hCAR3 as detailed in the “Experimental procedures” section. H, relative cell viability in HepG2 and Hep3B cells infected with lentivirus-expressing empty vector (Con), hCAR, or hCAR3 was monitored for 8 days using CCK-8 reagents. Relative blot densitometry was quantified using ImageJ from three separately prepared cell experiments and normalized to the density of the loading control. Results are expressed as mean ± SD from at least three independent experiments. ∗p < 0.05 and ∗∗p < 0.01. CAR, constitutive androstane receptor; CCK-8, Cell Counting Kit-8; Dox, doxycycline; hCAR, human CAR.
It is worth noting that although our data indicated that overexpression of hCAR suppresses hepatoma cell proliferation, ectopic overexpression of a protein may lead to nonspecific cellular stress and cell growth arrest. To address this concern, we infected HepG2 and Hep3B cells with lentivirus expressing the WT-hCAR that is constitutively activated in hepatoma cell lines or a splice variant of hCAR (termed hCAR3), which exhibits low basal transcription activity as shown in a luciferase reporter experiment (Fig. 2F) and previous publications (23, 24). At comparable protein expression levels (Fig. 2G), the growth of HepG2 and Hep3B cells was markedly suppressed by hCAR but not by hCAR3 (Fig. 2H). This finding was further confirmed using HepG2-hCAR3 and Hep3B-hCAR3 stable cell lines as shown in Fig. S4. Overall, these results are of particular significance and suggest that hCAR exerts a specific tumor-suppressive function instead of a nonspecific cellular stress response.
Overexpression of hCAR represses cell cycle progression in HepG2 and Hep3B cells
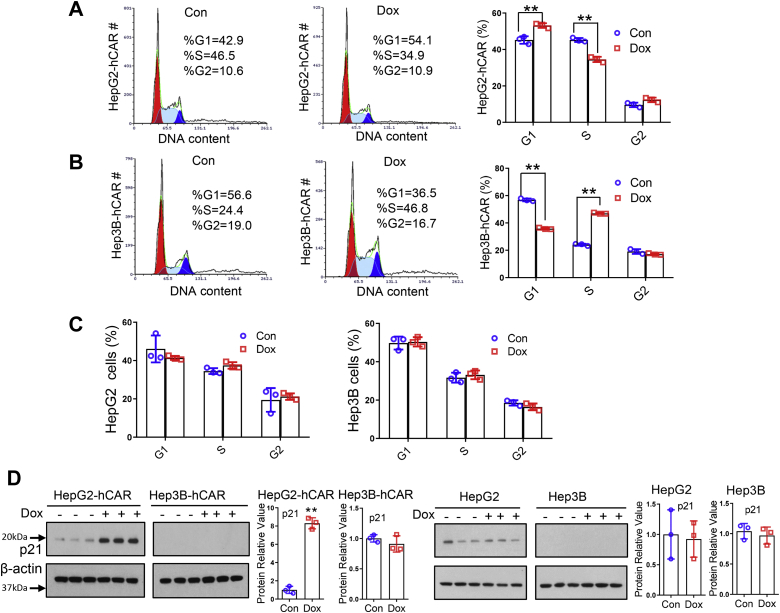
Next, we tested whether hCAR alters cell proliferation and cell cycle progression of hepatoma cells. Flow cytometry analysis revealed that overexpression of hCAR by Dox treatment in HepG2-hCAR cells resulted in significant G1 arrest with 54.1% cells in the G0/G1 phase versus 42.9% in the vehicle control group (p < 0.05), whereas the cell population in the S phase was decreased to 34.9% in comparison to 46.5% in control (p < 0.05) (Fig. 3A). Interestingly, unlike the observations in HepG2-hCAR, induction of hCAR in Hep3B-hCAR cells led to significant S phase detention, with 46.8% of hCAR-expressing cells in the S phase versus 24.4% of control cells (p < 0.05) (Fig. 3B). On the other hand, Dox treatment did not significantly alter the cell cycle progression of normal HepG2 and Hep3B cells (Fig. 3C). Together, these results indicate that overexpression of hCAR suppresses hepatoma cell proliferation by causing cell cycle arrest in the G1 phase for HepG2 cells and in the S phase for Hep3B cells, respectively. Given the differential status of the tumor-suppressor gene p53 in HepG2 (WT-p53) and Hep3B (p53-null) cells (25), we further analyzed the effects of hCAR on the expression of p21, a prototypical target gene of p53 and a known cyclin-dependent kinase inhibitor, in HepG2-hCAR and Hep3B-hCAR cells. As shown in Figure 3D, overexpression of hCAR induced the protein expression of p21 in HepG2-hCAR cells, whereas such a change was not observed in Hep3B-hCAR cells because of the undetectable expression of p21, suggesting that p21 contributes to the cell cycle arrest in HepG2-hCAR cells, whereas not in Hep3B-hCAR cells.
Figure 3.
Overexpression of CAR inhibits cell cycle progression in HepG2 and Hep3B cells. Cell cycle progression was analyzed in HepG2-hCAR and Hep3B-hCAR cells 3 days after treatment with vehicle control or Dox (1 μg/ml). The percentage of cells in different cell cycle phases was compared between vehicle control and Dox-treated HepG2-hCAR (A), Hep3B-hCAR (B), as well as normal HepG2 and Hep3B (C) cells. D, Western blotting was carried out to measure the protein levels of p21 in HepG2-hCAR, Hep3B-hCAR, HepG2, and Hep3B cells treated with vehicle control or Dox as detailed in the “Experimental procedures” section. Relative blot densitometry was quantified using ImageJ from three separately prepared cell experiments and normalized to the density of the loading control. Data were expressed as mean ± SD (n = 3). ∗∗p < 0.01. CAR, constitutive androstane receptor; Dox, doxycycline; hCAR, human CAR.
hCAR inhibits colony formation of hepatoma cells in vitro and the growth of hepatoma xenograft in vivo
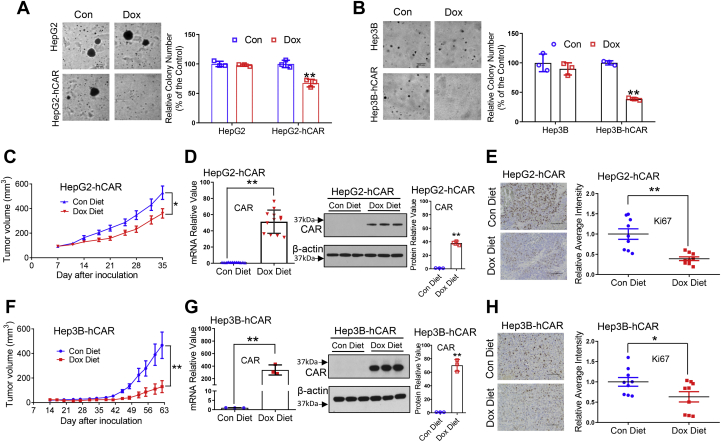
To determine whether overexpression of hCAR also attenuates tumorigenesis of hepatoma cells in vitro, we conducted soft-agar colony formation assays. Our results showed that Dox-triggered overexpression of hCAR in HepG2-hCAR and Hep3B-hCAR cell lines led to significantly fewer oncospheroids, whereas Dox does not affect colony formation in HepG2 and Hep3B cells (Fig. 4, A and B). We further investigated the role of hCAR in the growth of hepatoma cells in a xenograft model in vivo. HepG2-hCAR and Hep3B-hCAR cells were inoculated subcutaneously into nude mice as detailed under the “Experimental procedures” section. We showed that with Dox-diet feeding, the average volumes of HepG2-hCAR (Fig. 4C) and Hep3B-hCAR (Fig. 4F) xenografts were significantly lower than tumors in their respective control groups fed normal chow (p < 0.05). As expected, tumor expression of hCAR (mRNA and protein) was markedly increased in HepG2-hCAR (Fig. 4D) and Hep3B-hCAR (Fig. 4G) xenograft groups treated with Dox in comparison to their control groups. Immunohistochemical staining showed that the proliferation marker, Ki67, was lower in response to hCAR overexpression (Fig. 4, E and H). Collectively, these data indicate that hCAR plays a repressive role in human hepatoma cell proliferation and malignancy both in vivo and in vitro.
Figure 4.
Overexpression of hCAR inhibits colony formation of hepatoma cells in vitro and the growth of hepatoma xenograft in vivo. Normal and hCAR-inducible HepG2 and Hep3B cells were treated with Dox (1 μg/ml) or vehicle control for 72 h followed by a colony formation in soft-agar assay as described in the “Experimental procedures” section. Representative colony images and quantification colony formation are shown in A and B. The histograms represent mean ± SD of the cloning efficiency (%). Data presented are representative images from at least three independent experiments in A and B. For the xenograft experiment, 3 to 5 × 106 HepG2-hCAR or Hep3B-hCAR cells were inoculated into the nude mice for xenograft formation as detailed under the “Experimental procedures” section. The tumor growth curves of HepG2-hCAR (C) and Hep3B-hCAR xenografts (F) were measured in the control-diet and Dox-diet feeding groups during the experimental periods. RT–PCR and Western blotting were used to measure the mRNA and protein expressions of hCAR in the xenograft tumors (D and G). Relative blot densitometry was quantified using ImageJ from three separately prepared cell experiments and normalized to the density of the loading control. Expression of Ki67 was examined by immunohistochemistry (E and H); the scale bar represents 100 μm. Ki67 intensity was quantified using ImageJ and was expressed as mean ± SD (n = 9). ∗p < 0.05 and ∗∗p < 0.01. Dox, doxycycline; hCAR, human constitutive androstane receptor.
Genome-wide transcriptome analysis identifies EPO as a novel hCAR target gene contributing to hCAR-mediated suppression of hepatoma cell growth
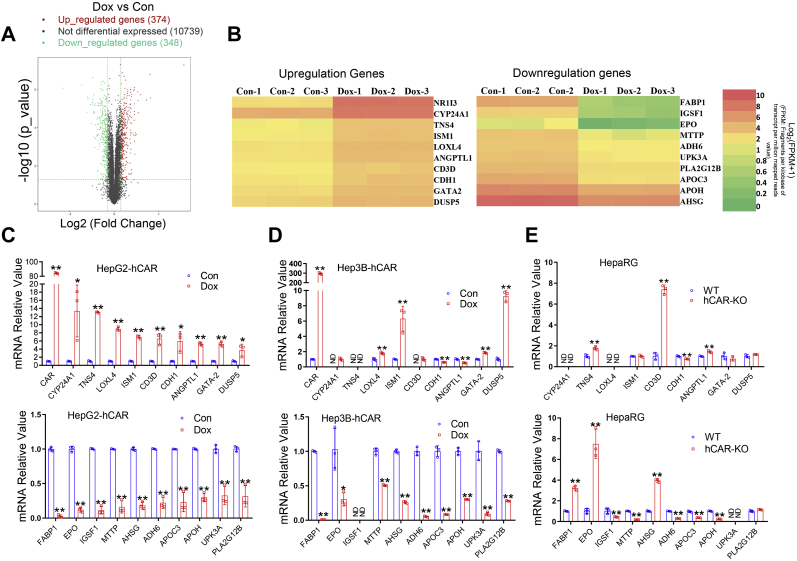
To obtain the global gene expression profile and identify novel determinants that impact hCAR regulation of hepatoma cell proliferation, we carried out an RNA-Seq assay on HepG2-hCAR cells challenged with Dox or vehicle control. Our results indicated that induced hCAR expression significantly upregulates 374 genes and downregulates 384 genes (Fig. 5A). A cluster of selected upregulated and downregulated genes with high fold changes was displayed in the heatmaps (Fig. 5B). A heatmap containing all significantly altered genes and gene ontology analyses of the RNA-Seq data are presented in supporting information (Figs. S5–S7). Key RNA-Seq findings were subsequently validated using RT–PCR in both HepG2-hCAR and Hep3B-hCAR cells. As expected, validation of 10 downregulated and 10 upregulated genes exhibited a complete match in HepG2-hCAR cells with which RNA-Seq was conducted (Fig. 5C). Notably, while the majority of hCAR-downregulated genes (9 of 10) showed consistent changes in Hep3B-hCAR cells, only 3 of 10 upregulated genes displayed the same trend of alteration in these cells (Fig. 5D). Given that all known HCC cell lines express extremely low endogenous hCAR, we next evaluated our RNA-Seq findings in fully differentiated HepaRG cells, a surrogate of human primary hepatocytes expressing functional hCAR, and an hCAR−/− HepaRG cell line (26). Interestingly, we found that expression of EPO, fatty acid–binding protein-1 (FABP1), and alpha-2-HS-glycoprotein (AHSG), three hCAR-repressed genes, was significantly enhanced in hCAR−/− HepaRG cells (Fig. 5E), whereas none of the selected hCAR-upregulated genes was remarkably repressed in the hCAR−/− HepaRG cells (Fig. 5E). Of the three genes (EPO, FABP1, and AHSG) that exhibited consistent changes across HepG2-hCAR, Hep3B-hCAR, and HepaRG cells, we chose EPO as a major focus of further investigation because previous studies indicated that EPO, a pleiotropic growth factor, is frequently overexpressed in HCC and is associated with increased tumor cell proliferation and poor prognosis (27).
Figure 5.
RNA-Seq analysis identifies EPO as a novel hCAR target gene. The total RNA from HepG2-hCAR was prepared 72 h after Dox (1 μg/ml) or vehicle control treatment for RNA-Seq analysis as detailed under the “Experimental procedures” section. A, the volcano plot of RNA-Seq data showed 374 upregulated and 348 downregulated genes in HepG2-hCAR cells after hCAR induction. B, heatmaps illustrate 10 selected upregulated and downregulated genes with high fold changes in HepG2-hCAR cells treated with Dox or vehicle control. RT–PCR validation of RNA-Seq analysis results for the 10 upregulated and 10 downregulated genes in HepG2-hCAR (C), Hep3B-hCAR (D), HepaRG WT and hCAR-KO cells (E), respectively. Data were collected from three independent experiments and expressed as mean ± SD in C–E. ∗p < 0.05 and ∗∗p < 0.01. Dox, doxycycline; EPO, erythropoietin; hCAR, human constitutive androstane receptor; ND, not detected.
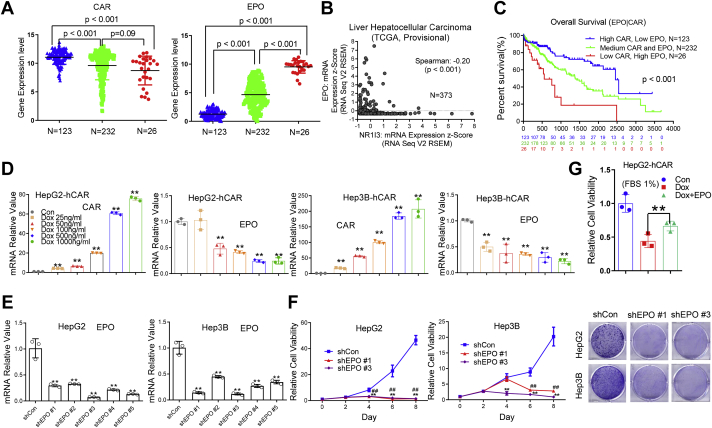
EPO is inversely correlated to hCAR expression in HCC and promotes proliferation of HepG2 and Hep3B cells
To examine the correlation between hCAR/EPO gene expression and HCC patient prognosis, we performed bioinformatic analysis of mRNA expression levels and patient survival in 381 HCC samples from a TCGA-liver-cancer dataset (SurvExpress) as detailed by Aguirre-Gamboa et al. (22). We found 123 HCC samples with elevated hCAR and low EPO expression; 232 HCC samples with medium levels of both hCAR and EPO; and 26 samples with low hCAR and high EPO levels, with a Spearman’s correlation coefficient of −0.2 (p < 0.001) (Fig. 6, A and B). Notably, HCC patients with low hCAR and high EPO levels exhibit poorer overall survival rates in comparison to the medium or low EPO/high hCAR groups (Fig. 6C), suggesting an inverse relationship between hCAR and EPO in HCC, and the pairing of low hCAR with high EPO may represent combined risks for HCC. The effects of hCAR on EPO expression were further analyzed in HepG2-hCAR and Hep3B-hCAR cell lines treated with vehicle control or Dox at multiple concentrations for 72 h. We demonstrated that Dox treatment led to a concentration-dependent increase of hCAR and decrease of EPO mRNA expression in both hCAR-inducible cell lines (Fig. 6D), whereas the same treatments did not significantly alter the expression of either hCAR or EPO in normal HepG2 and Hep3B cells (Fig. S8).
Figure 6.
EPO is inversely correlated to hCAR expression in HCC and promotes proliferation of HepG2 and Hep3B cells.A, hCAR and EPO gene expression was inversely correlated in patients with HCC. A total of 381 HCC samples were analyzed from a publicly accessible TCGA database through SurvExpress, and the overall survival was compared between low-hCAR–expressing (123), medium-hCAR–expressing (232), and high-hCAR expressing (26) groups. B, correlation analysis between hCAR and EPO reveal a Spearman’s correlation coefficient of −0.20 (p < 0.01). C, the pair of low hCAR with high EPO is associated with poor survival. D, in HepG2-hCAR and Hep3B-hCAR cells, treatment with Dox resulted in increased expression of hCAR and decreased expression of EPO in a concentration-dependent manner. E, expression of EPO in HepG2 cells was efficiently abolished after EPO knockdown via lentivirus shRNA targeting different regions of EPO (shEPO 1 to shEPO 5). F, depletion of EPO in HepG2 and Hep3B cells results in suppression of cell growth in a time-dependent manner. HepG2 and Hep3B cells were also visualized by Coomassie blue staining 8 days after infection with lentivirus shEPO 1, shEPO 3, or shCon. G, relative cell growth rate of HepG2-hCAR cells treated with Dox was partially rescued by addition of the recombinant human EPO treatment. Results are expressed as mean ± SD from three independent experiments in D–G. ##p < 0.01; ∗p < 0.05; and ∗∗p < 0.01. Dox, doxycycline; EPO, erythropoietin; hCAR, human constitutive androstane receptor; HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas.
We then evaluated the effects of EPO loss of function or gain of function on hepatoma cell proliferation using lentivirus-mediated EPO shRNAs or a commercially available recombinant EPO protein to alter intracellular EPO levels. All five EPO-specific shRNAs induced significant knockdown effects in HepG2 and Hep3B cells (Fig. 6E). shEPO 1 and shEPO 3 were randomly selected for subsequent cell viability studies. It is evident that both EPO shRNAs markedly repressed the growth of HepG2 and Hep3B cells in comparison with the shCon-infection control groups (Fig. 6F). In a separate experiment, reintroducing recombinant EPO (10 IU/ml; BioLegend, Inc) to Dox-treated HepG2-hCAR cells resulted in partial but statistically significant rescue of the hCAR-retarded cell proliferation (Fig. 6G). Together, these loss-of-function and gain-of-function studies reveal that EPO promotes hepatoma cell growth and may play an important role in hCAR-mediated HCC suppression.
Overexpression of hCAR represses EPO downstream signaling
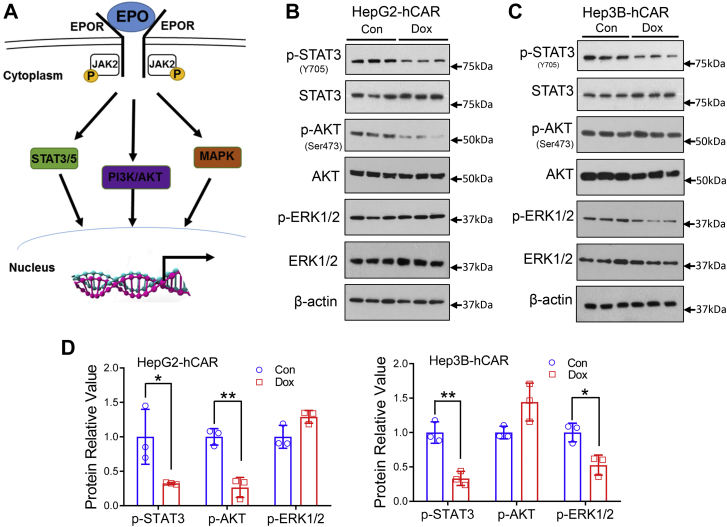
Mechanistically, EPO stimulates mitogenic and antiapoptotic reactions through the EPO–EPO receptor (EPOR) pathway by activating the Janus kinase 2–signal transducer and activator of transcription 3/5 (STAT3/5), PI3K–AKT, and the RAS–mitogen-activated protein kinase signaling cascades as illustrated in Figure 7A (28). Using HepG2-hCAR and Hep3B-hCAR cells treated with Dox or vehicle control, we showed that overexpressing hCAR reduces phosphorylation of STAT3 (Y705) and AKT (S473) but not extracellular signal–regulated kinase 1/2 (ERK1/2) (T183/Y185) in HepG2-hCAR (Fig. 7B), while dephosphorylating STAT3 (Y705) and ERK (T183/Y185) not AKT (S473) in Hep3B-hCAR cells (Fig. 7C). Densitometry analysis is shown in Figure 7D. These results highlight that hCAR-mediated downregulation of the EPO–EPOR signaling pathway may contribute to its repression of hepatoma cell proliferation.
Figure 7.
Overexpression of hCAR represses EPO downstream signaling in HepG2 and Hep3B cells.A, schematic illustration of the EPO–EPOR signaling pathway. B and C, HepG2-hCAR and Hep3B-hCAR cells were treated with vehicle control or Dox (1 μg/ml) for 72 h before Western blotting analysis of total and phosphorylation of STAT3 (p-STAT3-Y705), AKT (p-AKT-S473), and ERK1/2 (p-ERK1/2-T183/Y185). D, relative blot densitometry was quantified using ImageJ from three separately prepared cell experiments and normalized to the density of the loading control. Data are expressed as mean ± SD (n = 3). ∗p < 0.05 and ∗∗p < 0.01. Dox, doxycycline; EPO, erythropoietin; EPOR, EPO receptor; ERK1/2, extracellular signal–regulated kinase 1/2; hCAR, human constitutive androstane receptor; STAT3, signal transducer and activator of transcription 3.
hCAR downregulates EPO expression through suppression of HNF4α
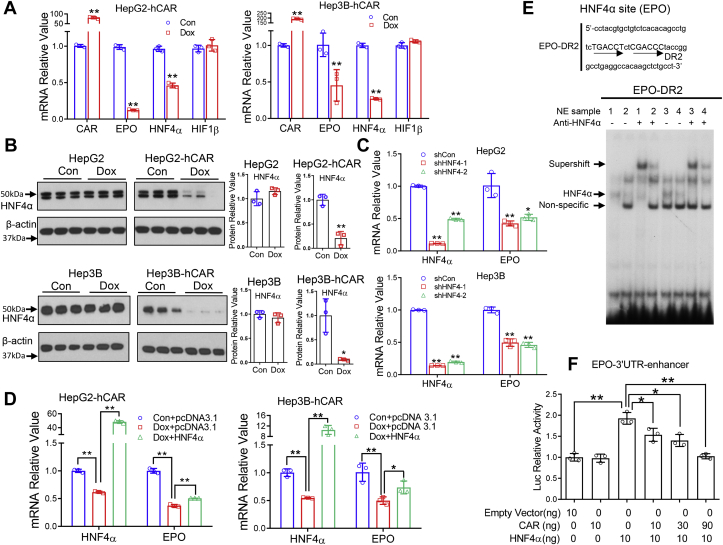
Next, we explored the mechanism whereby hCAR downregulates EPO. Hypoxia-inducible factors (HIFs) and HNF4α are known transcription factors that play key roles in stimulating EPO expression (29). Our results indicated that hCAR downregulates EPO in both HepG2 and Hep3B cells under normoxia cell culture conditions, and overexpression of hCAR resulted in repression of HNF4α but not HIF1 mRNA expression in HepG2-hCAR and Hep3B-hCAR cells (Fig. 8A). Dox treatment suppresses the protein levels of HNF4α in HepG2-hCAR and Hep3B-hCAR cells but not in normal HepG2 and Hep3B cells (Fig. 8B). We found that HNF4α knockdown significantly reduced the expression of EPO mRNA in HepG2 and Hep3B cells (Fig. 8C), whereas overexpression of HNF4α in HepG2-hCAR and Hep3B-hCAR cells partially rescued hCAR-repressed EPO expression (Fig. 8D). These findings suggest that hCAR-mediated downregulation of EPO is most likely a hypoxia-independent process, and HNF4α plays a key role in this response.
Figure 8.
hCAR downregulates EPO expression through suppression of HNF4α.A, RT–PCR was used to analyze expression of hCAR, EPO, HNF4α, and HIF1β in HepG2-hCAR and Hep3B-hCAR cells 72 h after treatment with vehicle control or Dox (1 μg/ml) as detailed in the “Experimental procedures” section. B, the protein levels of HNF4α were analyzed by immunoblotting in HepG2-hCAR, Hep3B-hCAR, as well as normal HepG2 and Hep3B cells treated with vehicle control or Dox for 72 h. Relative blot densitometry was quantified using ImageJ from three separately prepared cell experiments and normalized to the density of the loading control. C, knockdown of HNF4α expression in HepG2 and Hep3B cells via lentivirus shRNA targeting different regions of HNF4α (shHNF4α-1 and shHNF4α-2) resulted in downregulation of EPO expression in these cells. D, overexpression of HNF4α partially rescues hCAR-mediated downregulation of EPO expression in HepG2-hCAR and Hep3B-hCAR stable cell lines. E, EMSA was performed to detect interaction between the EPO–DR2 motif and nuclear extract samples isolated from HepG2-hCAR and Hep3B-hCAR cells treated with vehicle control or Dox for 72 h. HNF4α antibody was used for measuring specific supershift of the HNF4α/DR2 band. 1: NE of HepG2-hCAR-control; 2: NE of HepG2-hCAR-Dox; 3: NE of Hep3B-hCAR-control; and 4: NE of Hep3B-hCAR-Dox. F, HEK293T cells were transfected with EPO-3′UTR-enhancer reporter construct in the presence of HNF4α and/or hCAR expression vectors as indicated, and luciferase activities were determined 48 h after transfection. Three independent measures from each group were analyzed and expressed as mean ± SD. ∗p < 0.05 and ∗∗p < 0.01. Dox, doxycycline; DR2, direct repeat 2; EPO, erythropoietin; hCAR, human constitutive androstane receptor; HEK293T, human embryonic kidney 293T cell line; HIF1β, hypoxia-inducible factor 1β; HNF4α, hepatocyte nuclear factor 4 alpha.
HNF4α is known to stimulate transcription of EPO through binding to a direct repeat 2 (DR2) motif located in the 3′UTR region of EPO (30). Subsequent EMSAs using nuclear extracts from HepG2-hCAR and Hep3B-hCAR cells with and without Dox treatment demonstrated that overexpression of hCAR reduced the interaction between HNF4α and the EPO–DR2 motif, and the specific HNF4α/DR2 band was further shifted (supershift) with the addition of an antibody against HNF4α (Fig. 8E). In human embryonic kidney 293T (HEK293T) cells transfected with the EPO-3′UTR-enhancer reporter plasmid, the EPO luciferase activity was significantly enhanced by cotransfection of an HNF4α expression vector (Fig. 8F). Notably, this HNF4α-mediated enhancement was concentration-dependently diminished by the inclusion of an hCAR expression plasmid. Together, these results indicate that hCAR represses EPO expression through inhibition of HNF4α expression and blocks its functional interaction with the regulatory region of EPO.
Discussion
Initially characterized as a xenobiotic sensor controlling the transcription of hepatic genes associated with drug metabolism and disposition, CAR has emerged as a signaling molecule that plays a crucial role in regulating energy homeostasis and cell proliferation (31). Accumulating evidence reveals that, by increasing hepatocyte replicative DNA synthesis and liver hypertrophy, CAR activation is a key event in liver tumor formation in rodents treated with PB-like compounds (9, 13). Studies using CAR−/− mice concluded that CAR is essential for PB-provoked murine liver tumor progression (11, 12). On the other hand, this mode of action for rodent liver tumor promotion appears to be unrelated to humans (32). The exact role of hCAR in HCC development was largely unknown.
In this report, we demonstrated that expression of hCAR in HCC is significantly lower than that in nontumor liver tissues, and low levels of hCAR expression correlate with poor overall survival of patients with HCC. In contrast, ectopic expression of hCAR in human hepatoma cells suppresses cell proliferation and colony formation in vitro and inhibits hepatoma xenograft growth in vivo. Mechanistically, we found that hCAR-mediated downregulation of the EPO–EPOR signaling through repression of HNF4α contributes significantly to its HCC-suppressive effects. Together, these findings reveal a novel anticancer function of hCAR in HCC that differs substantially from its rodent counterparts.
HCC is the fastest-rising cause of cancer-related death in the United States, even though the overall mortality rate for most other cancers has declined in recent years (33). The emerging importance of CAR in hepatocarcinogenesis is highlighted predominantly by studies in animals. While activation of CAR has been firmly established as a mode of action for rodent liver tumor formation by many nongenotoxic chemicals, such a mechanism cannot be extrapolated to humans largely because of the well-documented species differences between hCARs and rodent CARs (14, 20). Characterization of genetic alteration is generally considered the prototype for deciphering the association of genes with the development of human cancers. Analysis of mRNA expression in over 200 HCC cases and nontumor liver tissues in this study reveals that the expression level of hCAR varies over a wide range in HCC with most samples exhibiting significant suppression in comparison to that in nontumor tissues. While the exact mechanism of hCAR downregulation in HCC remains elusive, DNA methylation represents a common epigenetic modification that represses gene expression in cancers. Our findings are consistent with earlier observations indicating increased DNA methylation in the hCAR promoter in both human non–small cell lung cancer and HCC samples (34, 35). Importantly, in further stratification of HCC cases based on hCAR levels, we found that lower expression of hCAR is associated with a poorer prognosis of HCC, suggesting that loss of hCAR contributes to the progression of HCC.
Notably, all available immortalized liver tumor cell lines express negligible levels of endogenous hCAR, rendering experimental knockdown of this gene extremely challenging. Conversely, ectopic expression of hCAR using the Tet-On system provides regulated expression of hCAR in hepatoma cells by the addition or withdrawal of the highly stable tetracycline (Tet) derivative, Dox. We showed that overexpression of hCAR markedly represses the proliferation and tumorigenesis of HepG2 and Hep3B cells. Interestingly, although a known xenobiotic sensor, CAR is constitutively activated in immortalized hepatoma cells independent of chemical stimulation (36). Comparatively, an hCAR splice variant with an in-frame insertion of five amino acids in the highly conserved region of the LBD, termed hCAR3, exhibits minimal basal transcriptional activity in these hepatoma cells (24). We found that expression of hCAR3 at levels comparable to hCAR does not markedly repress hepatoma cell proliferation, implying that hCAR-mediated repression of cell proliferation is not a nonspecific stress-related event but rather is relevant to its intrinsic transcriptional activity.
While overexpression of hCAR suppressed the growth of both HepG2 and Hep3B cells, each of these cell lines exhibits common and unique genetic signatures that influence their cancer biology features. Indeed, our results revealed that hCAR arrests HepG2 cells in the G1 phase but allows Hep3B cells to skip G1 and be attenuated in the S phase of the cell cycle. It is well known that HepG2 cells express a normal form of p53, a key tumor suppressor, whereas Hep3B cells possess a homozygous deletion of p53 (25). We found that p53-mediated induction of p21 contributes significantly to hCAR-induced cell cycle arrest in HepG2 but not Hep3B cells.
Our genome-wide transcriptome analysis in HepG2-hCAR cells revealed that hCAR alters the expression of numerous genes associated with cell proliferation, apoptosis, angiogenesis, and oncogenic signaling. Particularly, several well-established genes with anticancer activity, such as epithelial cadherin (E-cadherin), isthmin-1 (ISM1), and the anti–angiogenic angioarrestin, are among the hCAR-upregulated genes, whereas genes having oncogenic function, such as EPO, alpha-fetoprotein (AFP), angiogenin, and insulin-like growth factor 2 (IGF2), were in the downregulated candidate list. Further validation of these findings by gain of function or loss of function of hCAR across HepG2-hCAR, Hep3B-hCAR, and HepaRG cells led to the discovery of EPO, a glycoprotein that exhibits angiogenesis and antiapoptosis functions, as a new candidate gene that correlates with hCAR-mediated suppression of HCC.
Erythrocytosis, a common paraneoplastic finding in patients with HCC, was deemed to be caused by increased production of EPO from HCC cells (37). Accumulated EPO in turn promotes HCC-associated angiogenesis, antiapoptosis, and cell proliferation (38). Our bioinformatic analysis of a TCGA cohort showed an inverse relationship between the expression of hCAR and EPO in HCC cases, and the low hCAR/high EPO expression cluster is associated with reduced overall survival rate in patients with HCC. The biological function of EPO in most cells is mediated through the canonical EPO–EPOR signaling pathway. Binding of EPO induces conformational changes and homodimerization of EPOR and triggers the activation of the downstream Janus kinase 2–STAT3/5, PI3K–AKT, and RAS–RAF–mitogen-activated protein kinase signaling cascades (39). We found that hCAR induces dephosphorylation of STAT3 in both HepG2 and Hep3B cells, whereas only dephosphorylating AKT in HepG2 cells and ERK1/2 in Hep3B cells, respectively. While this finding suggests that hCAR exhibits cell-specific shared and distinct effects on EPO–EPOR signaling, mechanisms underlying this observed discordance between HepaG2 and Hep3B are unknown. In this study, we also found that while depletion of EPO suppresses hepatoma cell proliferation, the addition of recombinant human EPO partially, but significantly, rescued hCAR-induced attenuation of cell growth. These findings are consistent with previous preclinical reports indicating that EPO promotes tumor cell proliferation and stem cell self-renewal (40) and further confirm EPO as a novel target of hCAR that contributes to hCAR-mediated HCC suppression.
Mechanistically, the expression of EPO is regulated at the transcriptional level in both hypoxia-dependent and hypoxia-independent manners with HIF1 and HNF4α as two key transcription factors. HIF1 and HNF4α upregulate EPO transcription by recognizing and binding to the hypoxia response element and an adjacent DR2 HNF4α element in the 3′ flanking region of the EPO gene (41). We found that overexpressing hCAR reduced the expression of HNF4α but not HIF1, and knockdown of HNF4α alone was sufficient to reduce EPO in both HepG2 and Hep3B cells. As liver enriched transcription factors, functional crosstalk between CAR and HNF4α has been documented previously. Miao et al. (42) reported that CAR can repress HNF4α transactivation by competing with HNF4α for binding to the DR1 motif in the promoter of CYP7A1 gene. Similar inhibitory crosstalk between CAR and HNF4α was also observed in the regulation of the gluconeogenic genes, phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) (43). In our gel shift and luciferase reporter experiments, we showed that overexpression of hCAR decreases the binding of HNF4α to the DR2 motif in EPO and reduces its transcriptional activity thereafter.
There are limitations to our study. Despite trying multiple antibodies, we were consistently unsuccessful in assaying EPO expression by immunoblotting. It is likely other laboratories encountered the same difficulty—in several publications, RT–PCR rather than Western blotting was used to assay EPO expression (27, 44). Hence, we were able to measure only EPO mRNA. Also, our liver cancer model is not entirely “human”; as a xenograft model in nude mice, the inoculated human hepatoma cells were grown in a murine physiological environment. Because of the extrahepatic nature of our xenograft model and the fact that hCAR expression in stable cell lines was driven by the Tet-On system, this model cannot be used to address the effects of PB-like compounds on hCAR activity and liver cancer in vivo. Additional unanswered questions, such as how hCAR downregulates HNF4α and whether HNF4α is the only transcription factor bridging hCAR and EPO, require studies beyond the scope of the current communication.
In summary, we find that hCAR exhibits potential tumor-suppressive roles in HCC development, which contrast with the documented tumor promotion effects of its rodent analogs. Low expression of hCAR in HCC is associated with a poor HCC prognosis. Ectopic overexpression of hCAR attenuates the growth of hepatoma cells both in vitro and in vivo. Mechanistically, we identified EPO as a novel hCAR-downregulated target through modulation of HNF4α. Repression of the EPO–EPOR signaling cascade contributes significantly to hCAR-mediated HCC suppression. While future studies to fully delineate the comprehensive mechanisms underlying hCAR suppression of HCC are warranted, our current findings support an important perception that hCAR plays a suppressive role in HCC that may offer novel therapeutic targets.
Experimental procedures
Chemicals and biological reagents
Dox and 5-bromodeoxyuridine were purchased from Sigma–Aldrich. Primers for real-time RT–PCR were synthesized by Integrated DNA Technologies, Inc. The Dual-Luciferase Reporter Assay System was purchased from Promega. Recombinant human EPO (carrier free) was purchased from BioLegend (catalog no.: 587102). All other cell culture reagents were purchased from Life Technologies or Sigma–Aldrich.
Transcriptome profiling and data analysis
We used gene expression data derived from a cohort consisting of 247 tumor and 239 nontumor specimens derived from 247 patients with HCC as described previously (45). Among 488 tumor specimens from 247 patients with HCC, we used tumor and matched nontumor pairs from 233 patients with HCC to stratify the high and low groups based on the hCAR expression. The microarray data are publicly available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) with accession number GSE14520. Transcriptome data were processed as described previously (45). Briefly, we normalized raw gene expression data using the robust multiarray average method and global median centering (46) and calculated average for the expression of genes with more than one probe set. Kaplan–Meier survival analysis was performed based on the survival R package and p value from log-rank test based on the Cox proportional hazards model. Permutation t test was calculated based on the perm R package by 1000 resamplings.
Plasmid constructs
Lentiviral expression plasmids of WT hCAR (CAR) and an alternative splicing variant of hCAR (CAR3) were generated by cloning the full-length complementary DNA (cDNA) into the EcoRI site of a modified pWPXLd lentiviral vector as described previously (47). The knockdown lentiviral plasmids of EPO (shEPO 1: TRCN0000372137; shEPO 2: TRCN0000058643; shEPO 3: TRCN0000058645; shEPO 4: TRCN0000058646; and shEPO 5: TRCN0000058647) and HNF4α (shHNF4α-1: TRCN0000019189; shHNF4α-2: TRCN0000019193) were purchased from Sigma–Aldrich. The pCR3-CAR, pCR3-CAR3, and CYP2B6-2.2kb reporter plasmids were described previously (48, 49). Luciferase reporter plasmids containing the EPO 3′UTR enhancer region were PCR amplified. The PCR primer sequences are listed in Table S1. The PCR products were subcloned into the KpnI and XhoI sites of pGL3-TK vector and termed EPO-3′UTR-enhancer.
Cell culture and lentiviral infection
HepG2 and Hep3B cells were obtained from the American Tissue Culture Collection, cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in humidified incubator at 37 °C and 5% CO2. HepaRG and HepaRG-hCAR-knockout cell lines and related culture medium used were obtained from Sigma–Aldrich. The authenticity of these cell lines was confirmed by short tandem repeat profiling analysis. Human hepatocytes were obtained from BioIVT and cultured as described previously (50). Lentivirus packaging was carried out following the protocol described previously (51). Seventy-two hours after infection, total RNA and proteins were prepared for RT–PCR and Western blot analysis.
Generation of Dox-inducible hCAR stable cell lines
hCAR expression lentivirus plasmid was generated by cloning the full-length cDNA into the EcoRI site of a pCW57-GFP-2A-MCS lentiviral vector (Tet-regulated expression system; Addgene [catalog no.: 71783]). For generation of Dox-inducible CAR overexpression stable cell lines, HepG2 and Hep3B cells were infected with Dox-inducible CAR lentivirus for 72 h and then refed with fresh medium including the selection drug. Cells were refed every 2 days until uninfected control cells were completely killed, which took 4 to 5 days for puromycin (1 μg/ml). The positive infection cells were plated 1 cell/well in 96-well plate. The cells grown up from a single cell were selected, and Dox induction was tested again. The cell lines with the best Dox induction for overexpression of hCAR, named HepG2-hCAR and Hep3B-hCAR, were selected for use in this study.
Cell viability analysis
Cell proliferation was assayed using Cell Counting Kit-8 kit (Enzo Life Sciences, Inc) according to the manufacturer's instructions. The HepG2, HepG2-hCAR, Hep3B, and Hep3B-hCAR cells were seeded at the density of 0.5 × 103 or 1 × 103 per well in 96-well plates, respectively. The HepG2-hCAR and Hep3B-hCAR cells were treated with vehicle control or Dox (1 μg/ml), and normal HepG2 and Hep3B cells were infected with CAR, CAR3, and control lentivirus on day 0. At the time points of days 0, 2, 4, 6, and 8, Cell Counting Kit-8 reagent was added into each well followed by incubation at 37 °C for 1.5 h. The absorbance was read at 450 nm on a 96-well plate reader (Spectramax M5; Molecular Devices). Relative cell growth rate was normalized against control on day 0.
Soft-agar assay
HepG2-hCAR and Hep3B-hCAR cells were trypsinized and suspended in culture medium containing 0.3% melting agar and 10% fetal bovine serum. Cell suspensions were then plated in 6-well cell culture plates precovered with 0.6% agar gel at a density of 5 × 103 or 1 × 104 per well. The semisolid agar gels containing cells were fed with Dulbecco's modified Eagle's medium including 1 μg/ml Dox every 2 days. The number of colonies from three randomly chosen fields per well were counted under a phase-contrast microscope about 2 or 3 weeks after HepG2-hCAR or Hep3B-hCAR cells plating, respectively. The relative colony number was calculated according to the following formula: relative colony number (%) = number of clones in Dox-treated group/number of colonies in control group × 100%. The experiment was carried out in triplicate.
Animal experiments
All procedures of the animal studies were approved by the Institutional Animal Care and Use Committee of the University of Maryland School of Medicine. Mice were housed in laminar flow cabinets under specific pathogen-free conditions at room temperature with a 24 h night/day cycle and fed with pellets and water ad libitum. HepG2-hCAR and Hep3B-hCAR cells (3–5 × 106) were inoculated subcutaneously in the left and right flanks of 6- to 7-week-old female Athymic Nude-Foxn1nu mice (Envigo). When xenografts reached around 100 mm3, tumor-bearing mice were randomized divided into control and treated groups (n = 8/group). The control diet (F4207; Bio-Serv) or Dox-containing diet (F4107; 600 mg/kg Dox; Bio-Serv) was administered for the duration of the study. Mice were checked every other day for toxicity and mortality. Tumors were measured twice a week with Vernier calipers. Tumor volumes were calculated by the formula: r12 × r22/3 (r1 is the shortest and r2 is the longest diameter). The tumors were dissected at the end of the experiments. RNA and protein samples of the tumors were harvested for further analysis. Immunohistochemical staining of mouse xenograft tumors was carried out as previously described (52). Quantification of immunohistochemical staining was analyzed using ImageJ/Fiji (version: 1.53c; the National Institutes of Health).
Transient transfection assays
HepG2 cells cultured in 24-well plates were transfected with CYP2B6-2.2 kb reporter construct and control plasmid (pRL-Tk) in the presence of pCR3-CAR or pCR3-CAR3 using X-tremeGENE 9 DNA Transfection Reagent (Roche Diagnostics Corporation). HEK293T cells cultured in 24-well plates were transfected with EPO-3′UTR-enhancer reporter plasmid and control plasmid (pRL-Tk) in the presence of pCR3-CAR or pcDNA3.1-HNF4α using X-tremeGENE 9 DNA Transfection Reagent (Roche Diagnostics Corporation). About 48 h after transfection, cell lysates were assayed for luciferase activities normalized against the activities of cotransfected renilla luciferase using Dual-Luciferase Kit. Data were represented as mean ± SD of three individual transfections.
Flow cytometry analysis
HepG2-hCAR and Hep3B-hCAR cells were plated 5 × 104/well on 6-well plates and treated with vehicle control or 1 μg/ml Dox 72 h to induce overexpression of CAR. After washing, the cells from each group were resuspended in 200 μl PBS and fixed by adding 800 μl of ice-cold 100% ethanol. The fixed cells were resuspended in 500 μl propidium iodide staining solution (50 μg/ml propidium iodide, 0.1% Triton X-100, and 0.2 mg/ml of RNase A), vortexed, and incubated for 20 min at 37 °C. The percentage of cells in different cell-cycle phases was determined by flow cytometry BD FACS Canto II (BD Biosciences).
RNA-Seq
HepG2-hCAR cells were treated with vehicle control or Dox (1 μg/ml) for 72 h. RNA samples were isolated using the miRNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. Total RNA was quantified using NanoDrop ND-1000 instrument. About 1 to 2 μg of total RNA was used to prepare the sequencing library. In brief, total RNA is enriched by oligo (dT) magnetic beads (rRNA removed); RNA-Seq library was prepared by using KAPA Stranded RNA-Seq Library Prep Kit (Illumina), which incorporates dUTP into the second cDNA strand and renders the RNA-Seq library strand specific. The completed libraries were qualified with Agilent 2100 Bioanalyzer and quantified by absolute quantification quantitative PCR method. Sequencing was carried out using the Illumina HiSeq 4000 according to the manufacturer’s instructions. Sequencing was carried out by running 150 cycles.
Image analysis and base calling were performed using Solexa pipeline, version 1.8 (Off-Line Base Caller software). Sequence quality was examined using the FastQC software (Babraham Bioinformatics). The trimmed reads were aligned to reference genome using Hisat2 software (Johns Hopkins University). The transcript abundances for each sample were estimated with StringTie (Johns Hopkins University), and the fragments per kilobase of transcript per million mapped reads values for the gene and transcript levels were calculated with R package Ballgown. The differentially expressed genes and transcripts were filtered using R package Ballgown. The Volcano plot is constructed by plotting −log10(p value) on the y-axis, and log2(Fold_Change) in gene expression between the vehicle control and Dox groups on the x-axis.
EMSAs
HepG2-hCAR and Hep3B-hCAR cells were treated with vehicle control or Dox (1 μg/ml) for 72 h. Nuclear proteins were prepared using Nuclear Extraction Kit (active motif) according to the manufacturer’s instructions. EMSA was carried out as described previously (50). The EMSA oligonucleotide sequences are shown in Figure 8E.
RT–PCR analysis
Total RNA was isolated from cells using TRIzol reagent (Thermo Fisher Scientific) and reverse transcribed to cDNA using a High-Capacity cDNA Archive Kit (Applied Biosystems) following the manufacturer's instructions. RT–PCR assay was performed on an ABI StepOnePlus Real-Time PCR System (Applied Biosystems) using SYBR Green PCR Mastermix (Qiagen). Induction values were calculated according to the following equation: fold over control = 2ΔΔCt, where ΔCt represents the differences in cycle threshold numbers between the target gene and GAPDH, and ΔΔCt represents the relative change in these differences between control and treatment groups. All primer sequences are listed in Table S1.
Western blot analysis
Cell homogenate proteins were resolved on SDS–polyacrylamide gels (4–12% or 12%) and electrophoretically transferred onto polyvinylidene fluoride membranes. Subsequently, membranes were blocked with 5% milk or 5% bovine serum albumin and incubated with antibodies against CAR (1:1000 dilution), STAT3 (1:1000 dilution), phospho-STAT3 (1:1000 dilution), AKT (1:1000 dilution), phospho-AKT (1:1000 dilution), ERK1/2 (1:1000 dilution), phospho-ERK1/2 (1:1000 dilution), HNF4α (1:1000 dilution), p21 (1:1000 dilution), or β-actin (1:5000 dilution) at 4 °C overnight. The antibodies used in this study are listed in Table S2. Blots were washed and incubated with horseradish peroxidase secondary antibodies and developed with West Pico chemiluminescent substrates (Thermo Fisher Scientific). ImageJ (National Institutes of Health) was used for quantitation from three separately prepared cell experiments and normalized to the density of the loading control.
Statistical analysis
All data represent at least three independent experiments and were expressed as the mean ± SD. Statistical comparisons were made using one-way ANOVA followed by a post hoc Dunnett's test or Student's t test where appropriate. Statistical significance in tumor growth rates was tested by two-way repeated-measures ANOVA. Statistical significance was set at p < 0.05 and p < 0.01.
Data availability
All representative data are contained within this published article and its supporting information files.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Xi Yang (University of Maryland School of Medicine) for assistance in immunostaining experiments. We are also grateful to members of the Wang laboratory for constructive comments on the article.
Author contributions
Z. L., and H. W. conceptualization; Z. L., S. M. K., D. L., L. L., X. P., J. Z., T. S., J.-P. R., X. W. W., and H. W. formal analysis; Z. L., S. M. K., D. L., L. L., X. P., J. Z., T. S., J.-P. R., X. W. W., and H. W. investigation; Z. L. and H. W. writing–original draft; Z. L., S. M. K., D. L., L. L., X. P., J. Z., T. S., J.-P. R., M .N., X. W. W., and H. W. writing–review & editing.
Funding and additional information
This work was partly supported by the National Institutes of Health, United States, grants R01 CA262084 and GM121550 (to H. W.). S. M. K. and X. W. W. were supported by the grants ZIA-BC010313, ZIA-BC010876, ZIA BC 010877, and ZIA BC 011870, and T. S. and M. N. by Z01ES1005-01 from the Intramural Research Program of the National Institutes of Health, United States. J.-P. R. was supported by grant BX004890 from the Department of Veterans Affairs Biomedical Laboratory Research and Development Program, United States. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by F. Peter Guengerich
Supporting information
References
- 1.Roessler S., Long E.L., Budhu A., Chen Y., Zhao X., Ji J., Walker R., Jia H.L., Ye Q.H., Qin L.X., Tang Z.Y., He P., Hunter K.W., Thorgeirsson S.S., Meltzer P.S., et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–966.e12. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Goding Sauer A., Fedewa S.A., Butterly L.F., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Gentile D., Donadon M., Lleo A., Aghemo A., Roncalli M., di Tommaso L., Torzilli G. Surgical treatment of hepatocholangiocarcinoma: A systematic review. Liver Cancer. 2020;9:15–27. doi: 10.1159/000503719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farazi P.A., DePinho R.A. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat. Rev. Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 5.Vacca M., Degirolamo C., Massafra V., Polimeno L., Mariani-Costantini R., Palasciano G., Moschetta A. Nuclear receptors in regenerating liver and hepatocellular carcinoma. Mol. Cell Endocrinol. 2013;368:108–119. doi: 10.1016/j.mce.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 6.Zelko I., Negishi M. Phenobarbital-elicited activation of nuclear receptor CAR in induction of cytochrome P450 genes. Biochem. Biophys. Res. Commun. 2000;277:1–6. doi: 10.1006/bbrc.2000.3557. [DOI] [PubMed] [Google Scholar]
- 7.Qatanani M., Moore D.D. CAR, the continuously advancing receptor, in drug metabolism and disease. Curr. Drug Metab. 2005;6:329–339. doi: 10.2174/1389200054633899. [DOI] [PubMed] [Google Scholar]
- 8.Ward J.M., Lynch P., Riggs C. Rapid development of hepatocellular neoplasms in aging male C3H/HeNCr mice given phenobarbital. Cancer Lett. 1988;39:9–18. doi: 10.1016/0304-3835(88)90035-3. [DOI] [PubMed] [Google Scholar]
- 9.Diwan B.A., Lubet R.A., Ward J.M., Hrabie J.A., Rice J.M. Tumor-promoting and hepatocarcinogenic effects of 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) in DBA/2NCr and C57BL/6NCr mice and an apparent promoting effect on nasal cavity tumors but not on hepatocellular tumors in F344/NCr rats initiated with N-nitrosodiethylamine. Carcinogenesis. 1992;13:1893–1901. doi: 10.1093/carcin/13.10.1893. [DOI] [PubMed] [Google Scholar]
- 10.Yueh M.F., Taniguchi K., Chen S., Evans R.M., Hammock B.D., Karin M., Tukey R.H. The commonly used antimicrobial additive triclosan is a liver tumor promoter. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17200–17205. doi: 10.1073/pnas.1419119111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto Y., Moore R., Goldsworthy T.L., Negishi M., Maronpot R.R. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64:7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- 12.Huang W., Zhang J., Washington M., Liu J., Parant J.M., Lozano G., Moore D.D. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol. Endocrinol. 2005;19:1646–1653. doi: 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- 13.Lake B.G., Price R.J., Osimitz T.G. Mode of action analysis for pesticide-induced rodent liver tumours involving activation of the constitutive androstane receptor: Relevance to human cancer risk. Pest Manag. Sci. 2015;71:829–834. doi: 10.1002/ps.3854. [DOI] [PubMed] [Google Scholar]
- 14.Mackowiak B., Hodge J., Stern S., Wang H. The roles of xenobiotic receptors: Beyond chemical disposition. Drug Metab. Dispos. 2018;46:1361–1371. doi: 10.1124/dmd.118.081042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H., LeCluyse E.L. Role of orphan nuclear receptors in the regulation of drug-metabolising enzymes. Clin. Pharmacokinet. 2003;42:1331–1357. doi: 10.2165/00003088-200342150-00003. [DOI] [PubMed] [Google Scholar]
- 16.Moore L.B., Maglich J.M., McKee D.D., Wisely B., Willson T.M., Kliewer S.A., Lambert M.H., Moore J.T. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol. Endocrinol. 2002;16:977–986. doi: 10.1210/mend.16.5.0828. [DOI] [PubMed] [Google Scholar]
- 17.Parzefall W., Erber E., Sedivy R., Schulte-Hermann R. Testing for induction of DNA synthesis in human hepatocyte primary cultures by rat liver tumor promoters. Cancer Res. 1991;51:1143–1147. [PubMed] [Google Scholar]
- 18.Yamada T., Okuda Y., Kushida M., Sumida K., Takeuchi H., Nagahori H., Fukuda T., Lake B.G., Cohen S.M., Kawamura S. Human hepatocytes support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogen sodium phenobarbital in an in vivo study using a chimeric mouse with humanized liver. Toxicol. Sci. 2014;142:137–157. doi: 10.1093/toxsci/kfu173. [DOI] [PubMed] [Google Scholar]
- 19.Olsen J.H., Schulgen G., Boice J.D., Jr., Whysner J., Travis L.B., Williams G.M., Johnson F.B., McGee J.O. Antiepileptic treatment and risk for hepatobiliary cancer and malignant lymphoma. Cancer Res. 1995;55:294–297. [PubMed] [Google Scholar]
- 20.Li D., Mackowiak B., Brayman T.G., Mitchell M., Zhang L., Huang S.M., Wang H. Genome-wide analysis of human constitutive androstane receptor (CAR) transcriptome in wild-type and CAR-knockout HepaRG cells. Biochem. Pharmacol. 2015;98:190–202. doi: 10.1016/j.bcp.2015.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parpart S., Roessler S., Dong F., Rao V., Takai A., Ji J., Qin L.X., Ye Q.H., Jia H.L., Tang Z.Y., Wang X.W. Modulation of miR-29 expression by alpha-fetoprotein is linked to the hepatocellular carcinoma epigenome. Hepatology. 2014;60:872–883. doi: 10.1002/hep.27200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguirre-Gamboa R., Gomez-Rueda H., Martinez-Ledesma E., Martinez-Torteya A., Chacolla-Huaringa R., Rodriguez-Barrientos A., Tamez-Pena J.G., Trevino V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Chen T., Cottrell J., Wang H. Nuclear translocation of adenoviral-enhanced yellow fluorescent protein-tagged-human constitutive androstane receptor (hCAR): A novel tool for screening hCAR activators in human primary hepatocytes. Drug Metab. Dispos. 2009;37:1098–1106. doi: 10.1124/dmd.108.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auerbach S.S., Stoner M.A., Su S., Omiecinski C.J. Retinoid X receptor-alpha-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3) Mol. Pharmacol. 2005;68:1239–1253. doi: 10.1124/mol.105.013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosaka M., Kang M.R., Yang G., Li L.C. Targeted p21WAF1/CIP1 activation by RNAa inhibits hepatocellular carcinoma cells. Nucleic Acid Ther. 2012;22:335–343. doi: 10.1089/nat.2012.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aninat C., Piton A., Glaise D., Le Charpentier T., Langouet S., Morel F., Guguen-Guillouzo C., Guillouzo A. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab. Dispos. 2006;34:75–83. doi: 10.1124/dmd.105.006759. [DOI] [PubMed] [Google Scholar]
- 27.Ke S., Chen S., Dong Z., Hong C.S., Zhang Q., Tang L., Yang P., Zhai J., Yan H., Shen F., Zhuang Z., Wen W., Wang H. Erythrocytosis in hepatocellular carcinoma portends poor prognosis by respiratory dysfunction secondary to mitochondrial DNA mutations. Hepatology. 2017;65:134–151. doi: 10.1002/hep.28889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debeljak N., Solar P., Sytkowski A.J. Erythropoietin and cancer: The unintended consequences of anemia correction. Front. Immunol. 2014;5:563. doi: 10.3389/fimmu.2014.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bunn H.F., Gu J., Huang L.E., Park J.W., Zhu H. Erythropoietin: A model system for studying oxygen-dependent gene regulation. J. Exp. Biol. 1998;201:1197–1201. doi: 10.1242/jeb.201.8.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galson D.L., Tsuchiya T., Tendler D.S., Huang L.E., Ren Y., Ogura T., Bunn H.F. The orphan receptor hepatic nuclear factor 4 functions as a transcriptional activator for tissue-specific and hypoxia-specific erythropoietin gene expression and is antagonized by EAR3/COUP-TF1. Mol. Cell Biol. 1995;15:2135–2144. doi: 10.1128/mcb.15.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackowiak B., Wang H. Mechanisms of xenobiotic receptor activation: Direct vs. indirect. Biochim. Biophys. Acta. 2016;1859:1130–1140. doi: 10.1016/j.bbagrm.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elcombe C.R., Peffer R.C., Wolf D.C., Bailey J., Bars R., Bell D., Cattley R.C., Ferguson S.S., Geter D., Goetz A., Goodman J.I., Hester S., Jacobs A., Omiecinski C.J., Schoeny R., et al. Mode of action and human relevance analysis for nuclear receptor-mediated liver toxicity: A case study with phenobarbital as a model constitutive androstane receptor (CAR) activator. Crit. Rev. Toxicol. 2014;44:64–82. doi: 10.3109/10408444.2013.835786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 34.Fukumasu H., Rochetti A.L., Pires P.R., Silva E.R., Mesquita L.G., Strefezzi R.F., De Carvalho D.D., Dagli M.L. Constitutive androstane receptor ligands modulate the anti-tumor efficacy of paclitaxel in non-small cell lung cancer cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0099484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang X., Ge L., Chen Z., Kong S., Liu W., Xu Y., Zeng S., Chen S. Methylation of the constitutive androstane receptor is involved in the suppression of CYP2C19 in hepatitis B virus-associated hepatocellular carcinoma. Drug Metab. Dispos. 2016;44:1643–1652. doi: 10.1124/dmd.116.070243. [DOI] [PubMed] [Google Scholar]
- 36.Honkakoski P., Zelko I., Sueyoshi T., Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol. Cell Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuyama M., Yamazaki O., Horii K., Higaki I., Kawai S., Mikami S., Higashino M., Oka H., Nakai T., Inoue T. Erythrocytosis caused by an erythropoietin-producing hepatocellular carcinoma. J. Surg. Oncol. 2000;75:197–202. doi: 10.1002/1096-9098(200011)75:3<197::aid-jso8>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 38.Wen Y., Zhou X., Lu M., He M., Tian Y., Liu L., Wang M., Tan W., Deng Y., Yang X., Mayer M.P., Zou F., Chen X. Bclaf1 promotes angiogenesis by regulating HIF-1alpha transcription in hepatocellular carcinoma. Oncogene. 2019;38:1845–1859. doi: 10.1038/s41388-018-0552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aapro M., Jelkmann W., Constantinescu S.N., Leyland-Jones B. Effects of erythropoietin receptors and erythropoiesis-stimulating agents on disease progression in cancer. Br. J. Cancer. 2012;106:1249–1258. doi: 10.1038/bjc.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright J.R., Ung Y.C., Julian J.A., Pritchard K.I., Whelan T.J., Smith C., Szechtman B., Roa W., Mulroy L., Rudinskas L., Gagnon B., Okawara G.S., Levine M.N. Randomized, double-blind, placebo-controlled trial of erythropoietin in non-small-cell lung cancer with disease-related anemia. J. Clin. Oncol. 2007;25:1027–1032. doi: 10.1200/JCO.2006.07.1514. [DOI] [PubMed] [Google Scholar]
- 41.Ebert B.L., Bunn H.F. Regulation of the erythropoietin gene. Blood. 1999;94:1864–1877. [PubMed] [Google Scholar]
- 42.Miao J., Fang S., Bae Y., Kemper J.K. Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J. Biol. Chem. 2006;281:14537–14546. doi: 10.1074/jbc.M510713200. [DOI] [PubMed] [Google Scholar]
- 43.Kachaylo E.M., Yarushkin A.A., Pustylnyak V.O. Constitutive androstane receptor activation by 2,4,6-triphenyldioxane-1,3 suppresses the expression of the gluconeogenic genes. Eur. J. Pharmacol. 2012;679:139–143. doi: 10.1016/j.ejphar.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Chen J., Connor K.M., Aderman C.M., Smith L.E. Erythropoietin deficiency decreases vascular stability in mice. J. Clin. Invest. 2008;118:526–533. doi: 10.1172/JCI33813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roessler S., Jia H.L., Budhu A., Forgues M., Ye Q.H., Lee J.S., Thorgeirsson S.S., Sun Z., Tang Z.Y., Qin L.X., Wang X.W. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 47.Li Z., Li D., Choi E.Y., Lapidus R., Zhang L., Huang S.M., Shapiro P., Wang H. Silencing of solute carrier family 13 member 5 disrupts energy homeostasis and inhibits proliferation of human hepatocarcinoma cells. J. Biol. Chem. 2017;292:13890–13901. doi: 10.1074/jbc.M117.783860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen T., Tompkins L.M., Li L., Li H., Kim G., Zheng Y., Wang H. A single amino acid controls the functional switch of human constitutive androstane receptor (CAR) 1 to the xenobiotic-sensitive splicing variant CAR3. J. Pharmacol. Exp. Ther. 2010;332:106–115. doi: 10.1124/jpet.109.159210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H., Faucette S., Sueyoshi T., Moore R., Ferguson S., Negishi M., LeCluyse E.L. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J. Biol. Chem. 2003;278:14146–14152. doi: 10.1074/jbc.M212482200. [DOI] [PubMed] [Google Scholar]
- 50.Li L., Li H., Garzel B., Yang H., Sueyoshi T., Li Q., Shu Y., Zhang J., Hu B., Heyward S., Moeller T., Xie W., Negishi M., Wang H. SLC13A5 is a novel transcriptional target of the pregnane X receptor and sensitizes drug-induced steatosis in human liver. Mol. Pharmacol. 2015;87:674–682. doi: 10.1124/mol.114.097287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubinson D.A., Dillon C.P., Kwiatkowski A.V., Sievers C., Yang L., Kopinja J., Rooney D.L., Zhang M., Ihrig M.M., McManus M.T., Gertler F.B., Scott M.L., Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 52.Guo Z., Dai B., Jiang T., Xu K., Xie Y., Kim O., Nesheiwat I., Kong X., Melamed J., Handratta V.D., Njar V.C., Brodie A.M., Yu L.R., Veenstra T.D., Chen H., et al. Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell. 2006;10:309–319. doi: 10.1016/j.ccr.2006.08.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All representative data are contained within this published article and its supporting information files.