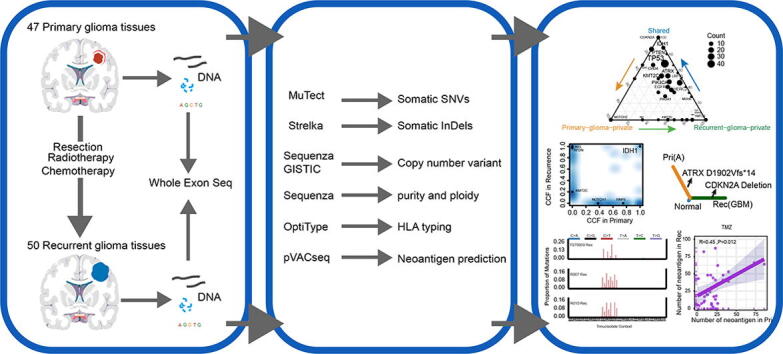
Graphical abstract
Keywords: Recurrent glioma, Whole-exome sequencing, Clonality, Immunogenicity, Evolution
Abbreviations: WES, Whole-exome sequencing; SNVs, Single-nucleotide variants; CNAs, Copy number alterations; CCF, Cancer cell fraction; VAF, Variant allele frequency; PI3K, phosphor-inositide 3-kinase; RTK, Receptor tyrosine kinase; MAPK, Mitogen-activated protein kinase; IDH, Isocitrate dehydrogenase; TERT, Telomerase reverse transcriptase; GBM, Glioblastoma multiforme; Indels, Insertions and deletions
Abstract
Diffuse glioma is a highly heterogeneous central nervous system tumor that is refractory to conventional therapy. Residual glioma cells escape from surgery and chemoradiotherapy, leading to lethal recurrence. Understanding the molecular mechanism of this recurrence process is critical to the development of successful therapies. Here, we analyzed whole-exome sequencing (WES) data of 97 paired primary and recurrent samples from 46 patients with glioma via a uniform pipeline. Clonality and phylogenetic analyses revealed that branching evolution was widespread in the recurrent process of gliomas. Recurrent tumors continued to evolve independently with chemoradiotherapy and harbored multiple recurrence-selected genetic alterations, such as amplification of PPFIBP1, PDE4DIP, and KRAS, deletion of TNFRSF14, DCC, CDKN2A, and MSH6, and mutations in ATRX, ARID1A, KEL, TP53, MSH6, and KMT2B. Meanwhile, truncal variants within partial driver genes were identified among primary and recurrent gliomas, suggesting that they might be ideal therapeutic targets. Intriguingly, the immunogenicity of recurrent gliomas did not increase significantly compared to the primary tumors. Genomic analysis of recurrent gliomas provided an opportunity to identify potentially clinically informative alterations not detected in clinically sampled primary tumors.
1. Introduction
Accumulating evidence suggests that cancer occurs via an accumulation of somatic genomic alterations in the process of clonal evolution. These genomic aberrations arise randomly but create an opportunity for the acquisition of selective advantage and generate intra-tumor heterogeneity (ITH). Such ITH allows cancer cells to resist treatment, eventually resulting in tumor metastasis and recurrence.
Compared to other tumors, diffuse gliomas are a heterogeneous collection of the most common malignant brain tumors and patients have a poor prognosis [1]. Despite significant improvements in systemic therapies, recurrence around the surgical cavity develops in most types of glioma [2], [3]. Nevertheless, no consensus on the standard treatment for recurrent or progressive glioma has yet been established. Paradoxically, comprehensive genomic sequencing studies over the past two decades have achieved molecular characterization of gliomagenesis, including activating or inactivating alterations in phosphoinositide 3-kinase (PI3K), receptor tyrosine kinase (RTK), and mitogen-activated protein kinase (MAPK) pathways as well as IDH1/2, TP53, TERT [4], ATRX, 1p/19-codeletion, deletion of chromosome 10 and amplification of chromosome 7 [5], [6]. However, previous studies have focused mainly on untreated tumors, and studies of recurrent gliomas remain limited. This is mainly due to the difficulty in achieving adequate tissue at recurrence or the time of death. Understanding the process of tumor recurrence is a prerequisite for the development of effective treatments for recurrent tumors. To date, genomic landscape analyses of longitudinal samples have demonstrated that recurrent tumors display variable degrees of genetic diversity [7], [8], [9]. The genetic alterations in the RB and AKT-mTOR pathways were confirmed to be involved in malignant progression. Distally recurred glioblastoma multiforme (GBM) displays high levels of genetic diversity [10]. The Glioma Longitudinal Analysis (GLASS) Consortium pooled datasets at institutions worldwide and reconstructed the evolutionary trajectories of 222 patients with glioma to help to understand treatment failures and tumor progression [9]. However, an understanding of the evolutionary history of tumor recurrence and the timing of mutational processes involved in tumor evolution remains limited. The relationship between the process of recurrence, immunogenicity, and chemoradiotherapy is also unclear.
In the present study, we analyzed the WES data of 97 paired primary and recurrent glioma samples from 46 patients using a uniform bioinformatics pipeline. We used these data to determine evolutionary trajectories of recurrence. We also revealed how mutational processes vary over time during the course of recurrence and provide a more comprehensive understanding of the clonality, mutation signatures, and immunogenicity of recurrent gliomas.
2. Materials and methods
2.1. WES of paired primary and recurrent gliomas
Twenty-seven of the 46 patients with primary and recurrent paired gliomas were recruited from the Sun Yat-sen University Cancer Center (SYSUCC) (Guangzhou, China) with written informed consent. The study was approved by the Ethics Committee of SYSUCC (B2020-314-01). All specimens were verified as gliomas by experienced pathologists and reviewed to determine the histological grade. Corresponding tumor tissues and blood were selected for DNA isolation. Each patient was matched with a corresponding normal blood sample and used as a control. Sequencing libraries were generated using the SureSelect Human All Exon V6 kit (Agilent Technologies, CA, USA) for sequencing on the Illumina Hiseq platform. The remaining 19 glioma patients from the INCB cohort originated from the Besta Brain Tumor Biobank and data were downloaded from European Nucleotide Archive (ENA, PRJNA320312) [7]. Complete clinical characteristics of all sequenced samples are provided in Table1 and Supplementary Table 1.
Table 1.
Demographics and clinicopathologic characteristics of patients in our study.
| INCB(N = 38) | SYSUCC(N = 59) | ||
|---|---|---|---|
| Sex (%) | Female | 12 (31.6) | 21 (35.6) |
| Male | 26 (68.4) | 38 (64.4) | |
| Age at diagnosis (years) | Mean (SD) | 55.0 (10.2) | 43.6 (14.9) |
| Pathological diagnosis (%) | |||
| Glioblastoma,IDH-wildtype | 36 (94.7) | 39 (66.1) | |
| Astrocytoma,IDH-mutant | 2 (5.3) | 14 (23.7) | |
| Oligodendroglioma,IDH-mutant, 1p/19q-codeleted |
0 | 4 (6.8) | |
| Unknown | 0 | 2 (3.4) | |
| Radiotherapy (%) | Yes | 19 (100.0) | 24 (77.4) |
| No | 0 | 7 (22.6) | |
| Chemotherapy (%) | Yes | 19 (100.0) | 20 (64.5) |
| No | 0 | 11 (35.5) | |
| Grades (%) | 4 | 38 (100.0) | 38 (66.7) |
| 2 | 0 | 8 (14.0) | |
| 3 | 0 | 11 (19.3) | |
| Normal control (%) | Yes | 38 (100.0) | 49 (83.1) |
| No | 0 | 10 (16.9) |
2.2. Somatic variant identification
After quality control using fastp (v.0.20.1) [11], paired reads were mapped to the reference human genome (UCSC hg19) by BWA-MEM (v.0.7.17) with default parameters [12]. Aligned reads were further processed with GATK (v.4.1.4) following the guidelines for best practice [13]. SNVs were called from paired tumor and blood control sequencing data by MuTect (v.1.1.5) [14]. Variant sites with fewer than 10 total reads or three variant reads in the tumor sample were removed. Further filtering was performed using Varscan (v2.4.2) with default parameters to remove SNVs with low average base quality, low average mapping quality, high average mismatch quality, and strand bias of variant reads [15]. Somatic insertions and deletions (InDels) were called using Strelka (v.2.9.2) with default parameters [16]. For 5 patients with no matching normal tissue available, we called SNVs from tumor and normal reference generated from 5 pooled unrelated normal blood control. These unrelated normal blood samples were sequenced using the same protocol. Each raw data of blood sample was downsampled and then pooled. The detailed analysis process can be found in previous studies [17]. These samples only were used for mutation burden analysis. InDel calling was not performed for tumors without matching normal tissue. SNVs and InDels were annotated with ANNOVAR [18]. Tumor mutation burden (TMB) was determined according to the number of somatic, coding base substitutions, and short insertions and deletions per megabase of the examined tumor genome.
2.3. Identification of putative driver mutations
To identify putative driver mutations, all non-silent variants were compared with potential driver genes derived from intOGen (https://www.intogen.org) or the Cancer Gene Census (https://cancer.sanger.ac.uk/census). Next, non-silent variants located in potential driver genes were classified as putative driver mutations if they satisfied one of the following criteria: (1) The function of the mutation was evaluated as “deleterious” by least one tool including SIFT [19], LRT [20], MutationTaster [21], Polyphen v2 [22], FATHMM [23], or PROVEAN [24]; (2) Identified as nonsense mutation, splicing point mutations and InDels in driver genes.
2.4. DNA somatic copy number alteration calling
Sequenza (v2.1.2) was used to estimate tumor purity and ploidy from paired tumor-normal WES data and to calculate allele-specific copy number profiles [25]. Only autosomes were used in the copy number alteration analysis. Samples with excessive noise and low tumor purity (P137694F/S, P176723F/S, P197462F/S, P247302F/S, P318050F/S, P398678S, P243757T, R011F, and R017S) were excluded. GISTIC (v.2.0.23) was used to identify significantly amplified and deleted regions in our dataset. Peaks with q-value < 0.05 were selected as significant results [26].
2.5. Phylogenetic tree construction
First, a binary matrix was constructed to indicate the presence or absence of each SNV/InDel. The phylogenetic tree for each patient was then constructed by the R package phangorn (v2.5.5) using the maximum parsimony method [27].
2.6. Mutational signature analysis
The R package Palimpsest (v2.0.0) was used to determine whether de novo mutation signatures represent new mutational processes or the previously described COSMIC signatures and to assign which signature gave rise to each mutation [28].
2.7. Cancer cell fraction estimation and clonality analysis
Excluding the samples with low tumor purity (n = 4), samples without matched blood controls (n = 10), and necrosis samples (n = 2), the cancer cell fraction of each SNV was estimated by integrating the Sequenza-derived integer copy number and tumor purity estimates with the variant allele frequency (VAF) as described by McGranahan et al. [29]. To determine the clonality of the mutation in each sample – that is, whether the mutation occurs in all or a fraction of the tumor cells – SNVs were classified as either clonal or subclonal mutations based on the 95% confidence interval (CI) of the cancer cell fraction (CCF). SNVs were defined as clonal if the 95% CI overlapped with one, and otherwise as subclonal. If InDels and somatic copy number alterations (SCNAs) were present in all samples of a patient, we defined this tumor as clonal, otherwise as subclonal.
2.8. Identification of somatic mutation immunogenicity
OptiType (v1.3.3) was used to produce accurate HLA class I typing predictions [30]. The tumor specific neoantigens were predicted using pVACseq (v.4.0.10, https://github.com/griffithlab/pVAC-Seq) [31]. In brief, all non-silent somatic SNVs and InDels were annotated using the variant effect predictor (VEP) from Ensembl (https://asia.ensembl.org/info/docs/tools/vep/index.html) and generating a list of peptides ranging nine amino acids in length with the mutated residues represented in each position. With patient-specific germline HLA class I typing, MHC class I binding affinity of both wild-type and mutant peptides were predicted by NetMHCpan (v4.1) [32], [33]. Neoantigens were identified as those with a predicted mutant peptide binding score ≤ 500 nM. All the downstream analyses were based on the inferred neoantigens and their corresponding wild-type peptides. After filtering neoantigens with predicted mutant peptide binding score > 500 nM, peptides were then further analyzed for quality based on differential agretopicity index (DAI), TCR recognition potential, and clonality. TCR recognition potential for each neoantigen was calculated based on a previous study [33], [34]. Given a neoantigen, the TCR recognition potential was the probability that a neoantigen was recognized by the TCR repertoire and calculated by alignment with a set of epitopes from IEDB database. High TCR recognition was set to an TCR recognition potential > 0.9 [33]. The DAI was calculated based on MHC-I affinity of wild-type (WTA) and mutant (MA) peptides arising from the same mutation [35]. Based on neoantigens and CCF, we classified neoantigens as clonal (present in all tumor cells) and subclonal (present only in a subset) neoantigens. All nonsynonymous SNVs and InDels were annotated as either in a region of copy number loss or not. Then, copy number loss-related neoantigens were counted. Lastly, we determined the origin of the corresponding neoantigen based on the origin of the mutation.
2.9. RNA-seq and immune deconvolution analysis
Raw RNA-seq reads from 26 primary or recurrent gliomas in the INCB cohort were downloaded from the European Nucleotide Archive (ENA, PRJNA320312). RNA-seq data of R017 and R018 were excluded from the downstream analysis due to the absence of paired primary samples. For the RNA-seq data of the remaining 24 samples, Salmon (v1.2.0) was used to quantify the expression of transcripts [36]. The TPM-normalized data were used in the immune deconvolution analysis by TIMER2.0 [37].
2.10. Statistical analysis
Statistical analyses were performed using R v4.0.2. Categorical variables were evaluated using χ2 tests. Continuous variables were evaluated using two-sided paired Wilcoxon signed-rank tests, or Wilcoxon rank-sum testing. For box plots, the box indicated the interquartile range (IQR), the middle line indicated the median, whiskers indicated the highest and lowest values within 1.5 × IQR from the box. All tests were two-sided, and P-values < 0.05 were considered statistically significant.
3. Results
3.1. Genomic characterization of paired primary and recurrent gliomas
We performed WES of 97 glioma tissues and 41 blood samples (including 38 previously reported glioma tissues and 19 blood samples). A total of 41 patients had primary tumors, recurrent tumors, and matched blood samples, whereas tumor tissues alone were available for the other five patients (Supplementary Fig. 1A). On average, 64.14% of coding bases in the exome were covered by ≥100× vhigh-quality reads (Supplementary Table 2). All samples were processed within a uniform bioinformatics pipeline to identify SNVs, InDels, and CNAs. In total, we detected 10,884 missense mutations, 548 nonsense mutations, and 553 frameshift mutations in protein-coding sequences. According to the 2021 WHO classification criteria for CNS tumors and the results of whole exome sequencing, there were 11 IDH mutant gliomas in this study, including 9 astrocytomas and 2 oligodendrogliomas, and the remaining IDH wild-type samples were all GBM. For patients P258689 and P432157, the pathology of the recurrent tissue was diagnosed as necrosis and gliosis, and these two cases were not retained for downstream analysis.
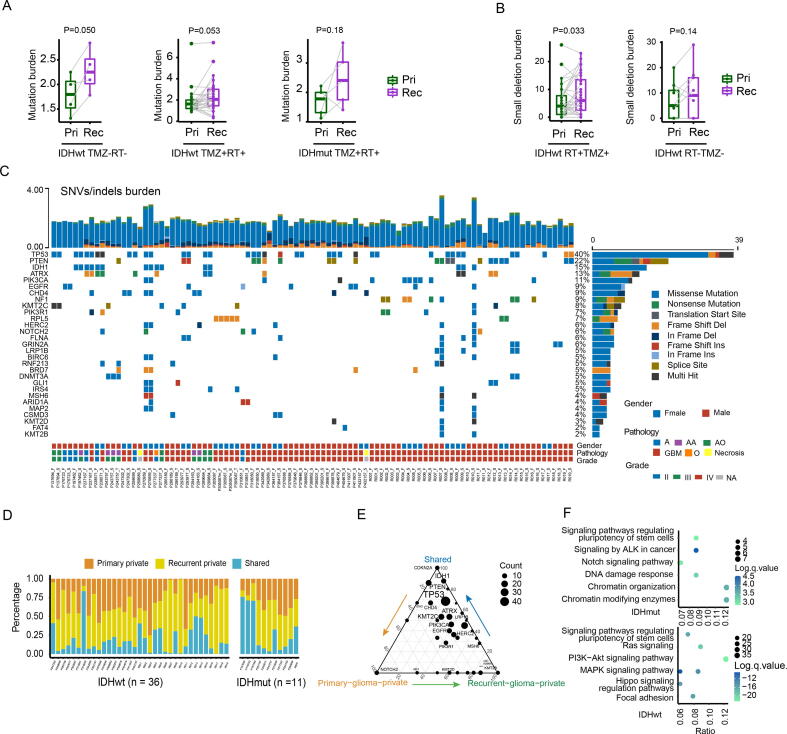
Excluding the three tumor samples with hypermutation (R007, R010, P270959) and five tumor samples without matching normal control available, the mutation burden of the recurrent GBM and IDH mutant glioma was not significantly higher than that of the primary counterpart (Fig. 1A, P = 0.050/0.053/0.18). According to previous studies [38], primary and recurrent hypermutation samples of P270959 were cases of de novo hypermutation (MSH6/F958Lfs*5), while R007 and R010 were post-treatment hypermutation (R007: MSH6/G841E/R792X/S198N; R010: MSH6/G300E/W917X). Consistent with the previous study [39], the number of small deletions acquired in recurrent radiotherapy-treated GBM was significantly higher than that of paired primary samples (Fig. 1B, P = 0.033). In total, 764 putative drivers SNVs/InDels were identified. Among them, some putative driver genes showed high concordance in the paired glioma samples (Fig. 1C). For instance, somatic mutations in TP53, IDH1, ATRX, PIK3CA, and EGFR showed concordance in the paired gliomas of most patients. The percentages of primary glioma-private or recurrent-private SNVs/InDels were highly variable across 46 patients (Fig. 1D), indicating varying degrees of genetic similarity between the primary and recurrent tumors. Among 764 putative driver mutations, those in TP53, IDH1, PTEN, ATRX, KMT2C, CDKN2A, and CDKN2C were shared by primary and recurrent gliomas (Fig. 1D). NOTCH2, KMT2B, CIC, and KMD5A tended to be private to the primary or recurrent glioma and thus, likely occurred following the acquisition of glioma initiation events (Fig. 1E). Pathway enrichment analysis showed that recurrent-private driver genes of IDH mutant gliomas were enriched in Notch signaling pathway, stem cell regulation-related pathway and chromatin modification, while GBM was enriched in PI3K-Akt, Ras, MAPK, Hippo signaling pathway, stem cell regulation-related pathway (Fig. 1F).
Fig. 1.
The landscape of driver mutations in paired primary and recurrent gliomas. (A) The boxplots depicted the mutation burden between primary and TMZ-untreated/TMZ-treated/ glioma samples. A two-sided paired t-test was applied for statistical testing. (B) The boxplots depicted the small-deletion burden between primary and TMZ-untreated/TMZ-treated/radiated GBM samples. A two-sided paired t-test was applied for statistical testing. (C) Oncoprint of functional driver mutations in 97 glioma samples. The upper stacked bar plots illustrate the total number of SNVs and indels in each sample. (D) The proportion of SNVs/indels that are shared, primary-private, or recurrent-private. (E) Ternary plot of mutation counts of driver genes in shared, primary-private, and recurrent-private mutation. (F) Recurrent-private driver mutations were used for pathway enrichment analysis. The Y-axis label represents the pathway, and X-axis label represents the gene term ratio (gene term ratio = gene numbers annotated in this pathway term/all gene numbers annotated in this pathway term). The bubble size represented the number of genes enriched in the pathway, and color showed the −log(q-value) of the enriched pathway. RT: Radiotherapy; TMZ: Temozolomide; Pri: Primary gliomas; Rec: Recurrent gliomas.
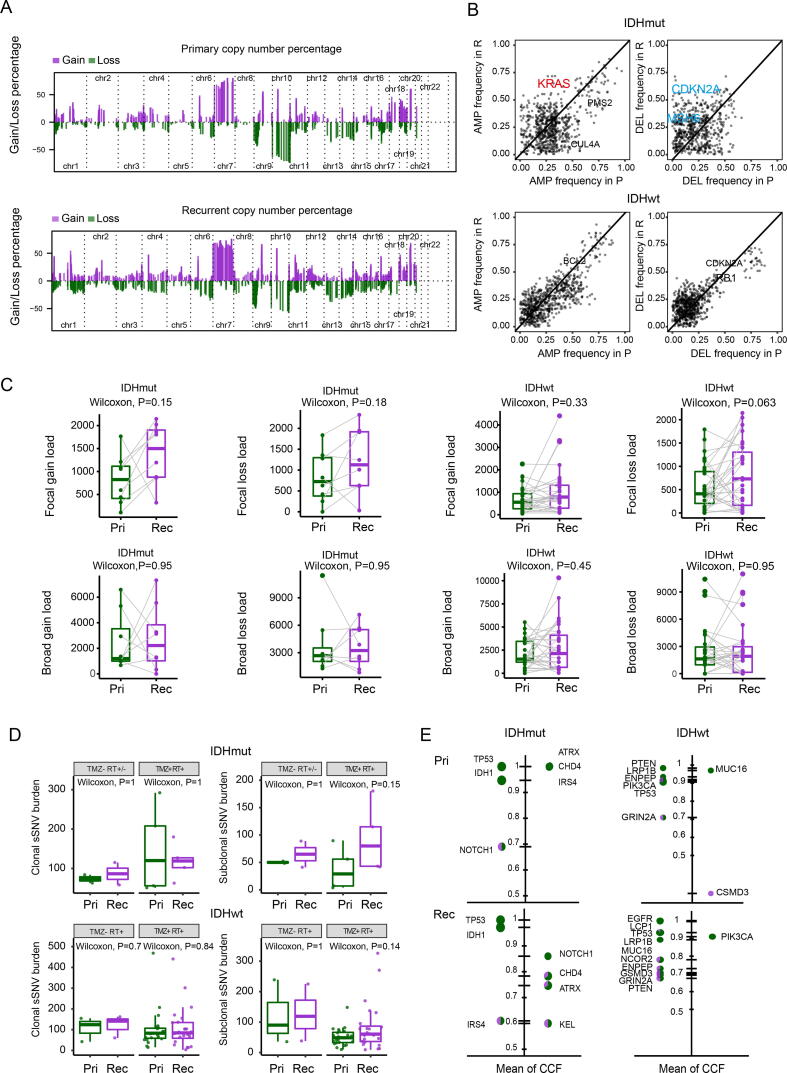
In addition, the overall copy number landscape was highly concordant between the primary and recurrent gliomas. Copy gain of chromosome 7 and loss of chromosome 10 were the most frequent events due to the inclusion of a high proportion of glioblastomas in our cohort (Fig. 2A and Supplementary Fig. 1B). However, several previously confirmed oncogenes and tumor suppressor genes were more frequently amplified or deleted in recurrent tumors compared to paired primary tumors, including CDKN2A, KRAS, and MSH6 (Fig. 2B). The Focal-level and arm-level copy number burden were also similar between primary and recurrent gliomas, regardless of IDH mutation (Fig. 2C). In addition, ploidy and tumor purity were comparable between the primary glioma and recurrent pairs (Supplementary Fig. 1C).
Fig. 2.
Copy number alteration (CNA) analysis. (A) The percentage of somatic CNAs for primary gliomas and recurrent across 22 chromosomes, with gains in purple and losses in green. (B) The frequency of somatic CNAs in putative driver genes in paired primary gliomas and recurrent. (C) Box plots and paired lines depicting focal and broad CNA load in primary and matched recurrent samples across two subtypes. Wilcoxon signed-rank test P values were indicated. (D) Comparison of clonal and subclonal SNVs burden in primary and recurrent gliomas across two subtypes. A two-sided Wilcoxon rank-sum test was applied for statistical testing. (E) The proportion of the clonal (green) and subclonal (violet) SNVs of each selected driver gene in primary, and recurrent gliomas. The size of each pie indicated the mutation frequency of driver genes ordered by their mean CCF of SNVs. RT: Radiotherapy; TMZ: Temozolomide; Pri: Primary gliomas; Rec: Recurrent gliomas. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We next evaluated the clonality of mutations in paired primary tumors and recurrences. In total, 21,237 clonal mutations (median = 87) were detected in 81 samples. These mutations might have been acquired before or during the most recent complete selective sweep, which describes the process by which a new mutation emerges with high fitness and outcompetes all other subclones in the tumor. Therefore, clonal mutations included passenger mutations that preceded tumor initiation and the driver mutations present in each selective sweep. In addition, 12,861 subclonal SNVs were detected in the samples mentioned above (median = 56). These subclonal SNVs exist in only a small fraction of glioma cells, indicating these mutations might be a later event in gliomagenesis. In addition, the number of subclonal SNVs in TMZ-treated samples did not show a significant increase compared to the primary samples (Fig. 2D). In IDH mutant gliomas, the high incidence of clonal mutation of TP53, IDH1, ATRX, CHD4, NOTCH1, and IRS4 in both primary and recurrent samples (Fig. 2E). In GBM, a large proportion of PTEN, LRP1B, ENPEP, PIK3CA, TP53, and GRIN2A mutations were clonal mutations in both primary and recurrent samples. These results implied that these mutations were early events in gliomagenesis.
These observations suggested that recurrent gliomas harbor variable degrees of genetic relatedness to the primary samples. Shared clonal variants (truncal) mutations within driver genes might be ideal targets for drug development and precision medicine strategies.
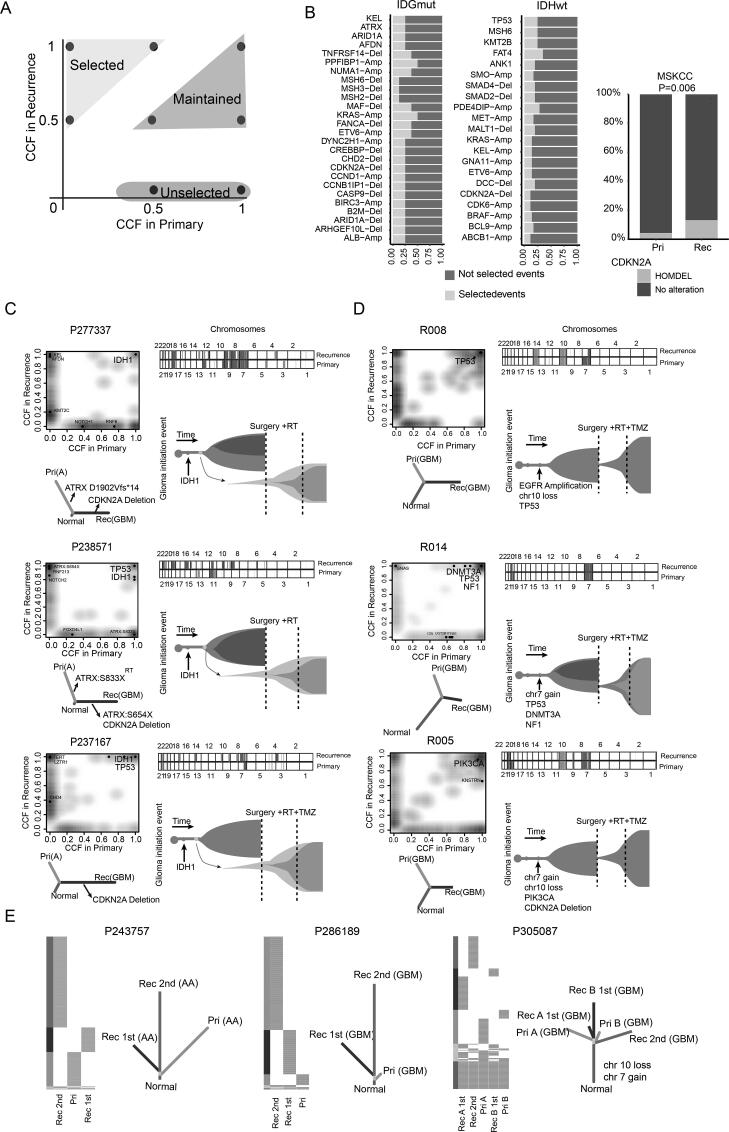
3.2. Selected events driving glioma recurrence
Taking advantage of the longitudinal samples in our cohort, we searched for recurrence-selected genetic features as defined previously [40] (Fig. 3A). Our results suggested that tumor cells select for a number of specific genomic alterations during recurrence. In IDH mutant gliomas, this included amplification of PPFIBP1 and KRAS, deletion of TNFRSF14, CDKN2A, and MSH6, and mutations in ATRX, ARID1A, and KEL. In GBM, mutations in TP53, MSH6, and KMT2B, amplification of PDE4DIP, and deletion of DCC were selected in recurrent samples. These results implied that these genomic alterations were potentially important in driving glioma recurrence (Fig. 3B). Especially in our cohort, all cases of IDH-mut astrocytoma that underwent malignant progression to GBM harbored an acquired deletion of CDKN2A in recurrences (Fig. 3C). This result was consistent with 2021 WHO classification of CNS tumors and previous studies [9], [41], which have defined CDKN2A deletion as a marker for high-grade astrocytoma. Furthermore, we collected two additional longitudinal datasets, including GLASS and MSKCC dataset. Among GBMs in the GLASS dataset, the proportion of mutation in MSH6, KMT2B, ANK1, TP53, and FAT4 in recurrent tumors was significantly higher than that in primary samples (Supplementary Fig. 2A). Among IDH-mut gliomas in the MSKCC and GLASS dataset, the proportion of deletions of CDKN2A in recurrent tumors was significantly higher than that in primary glioma samples (Fig. 3B and Supplementary Fig. 2B). In addition, the proportion of deletions of MSH6, amplification of KRAS, and mutation in ARID1A and AFDN in recurrent tumors was also significantly higher than that in primary glioma samples in the GLASS dataset. (Supplementary Fig. 2B). However, the remaining genomic alterations, such as amplification of ABCB1 and deletion of DCC, were found only in our cohort. These results implied the potential importance of these molecular features to drive recurrence.
Fig. 3.
Evolution patterns of recurrence in gliomas. (A) Schematic illustration for the definition of selected, maintained, or unselected alterations in recurrences. (B) The stacked bar plots showing the proportions of recurrence-selected alterations that occurred in more than two primary-recurrence pairs. Proportions of CDKN2A deletion between primary and recurrent IDH-mut gliomas in MSKCC. (C-D) Genetic divergence between primary and recurrent tumors. Two-dimensional density plot based on CCFs of all SNVs identified at the primary tumor (x-axis) and recurrence (y-axis). The dot represented the driver genes. Chromosomal copy-number plots for paired glioma samples; in each plot, light red = gain; red = Amplification; light blue = loss; blue = Deletion. Phylogenetic trees were generated by maximum parsimony based on mutational presence or absence, for which canonical glioma drivers genes are labeled. (E) Phylogenetic trees depicted the evolution patterns in samples with multiple recurrences. RT: Radiotherapy; TMZ: Temozolomide; Pri: Primary gliomas; Rec: Recurrent gliomas. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Corroborating previous studies [7], all primary tumor and recurrent samples were consistent with a branched evolution pattern, where both the primary tumor and the recurrence shared a common ancestor but continued to evolve separately. Among them, four primary/recurrence pairs with malignant progression to GBM shared an average of 17% of the mutations, suggesting a higher genetic divergence. Three of these shared a clonal driver mutation in IDH1, highlighting the critical role of IDH1 mutations in the initiation of gliomas (Fig. 3C and Supplementary Fig. 2C). In P238571, the primary tumor and recurrences derived from initial cells possessed IDH1 R132H, TP53 R141H, and ATRX mutations. Moreover, the recurrent tumor contained nonsense mutation sites in ATRX that were distinct from the primary tumor (Fig. 3C, middle panel), suggesting this convergent process occurred through strong selection for dysfunction of this gene. In addition, recurrence in the remaining cases without malignant progression also showed different levels of genetic divergence compared with primary tumor (Fig. 3D and Supplementary Fig. 3C). The phenomenon might be due to the fact that recurrent tumors could originate from one subpopulation that branched off early during tumorigenesis or much later. Furthermore, three multiple recurrent cases were used to provide a more comprehensive view of the recurrent process. For cases P243757 and P286189, the first recurrent tumors originated from one subclone that branched off early during gliomagenesis. The second recurrence also branched off from the first recurrent tumor at an earlier evolutionary stage. For P305087, the second recurrence branched off from the initial tumor at an earlier evolutionary stage than the cells seeding the first recurrence (Fig. 3E). These data indicated that the recurrence could arise from the dominant clone in the primary tumor, leading to genetic similarity or one subpopulation that branches off early in the clonal evolution of the primary tumor.
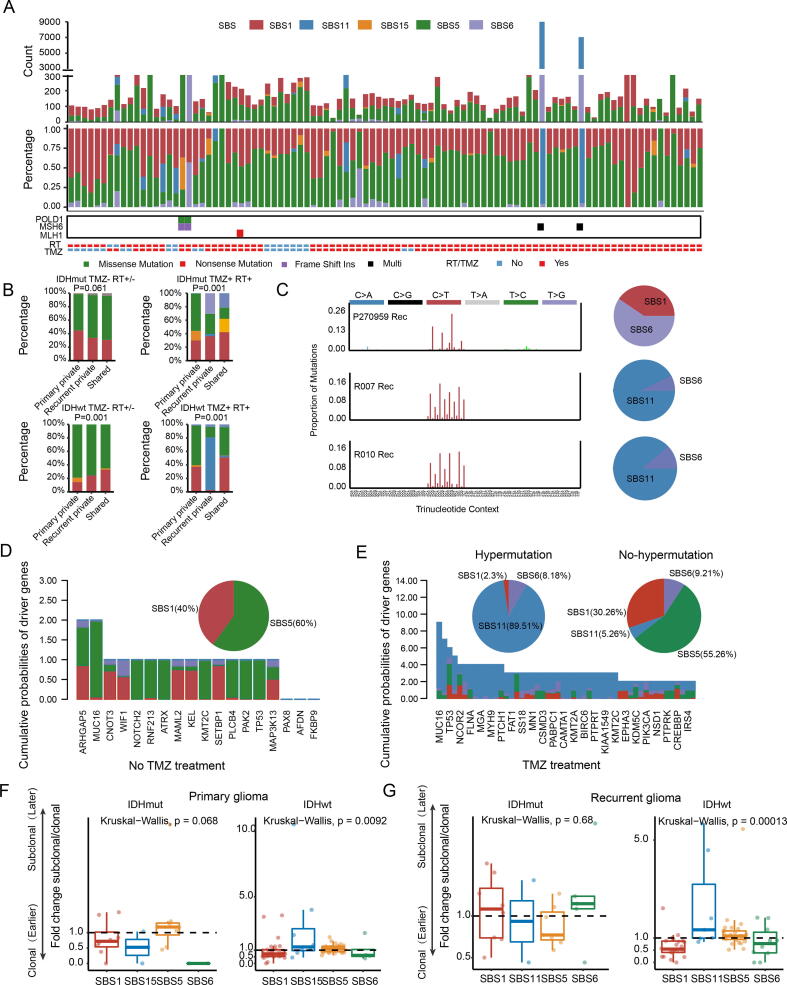
3.3. Temporal changes in mutational signatures
Mutational signatures are characteristic combinations of mutation types arising from both endogenous and external processes. Thus, the repertoire of somatic mutations could reflect the mutation processes or biological features associated with tumor evolution. To explore whether these mutational processes vary during tumor evolution, we identified five robust mutation signatures (1, 11, 15, 5, and 6) in all samples (Fig. 4A). Reflecting the age of the patient at diagnosis, signature 1/5 was ubiquitous in all glioma samples and dominated the shared mutations (Fig. 4B). Signature 11 was strongly enriched in recurrent-private mutations in TMZ-treated GBM samples, especially in samples with hypermutation (Fig. 4A-B). However, not all post-TMZ-treated samples harbored the activity of signature 11. Signatures 6 and 15, associated with defective DNA mismatch repair (MMR), were not always accompanied by deleterious mutations in the MMR pathway-related genes, suggesting the existence of other forms of deactivation, such as DNA methylation. In accordance with our previous results, cases with hypermutation harbored signatures 6, 15, and 11. Furthermore, the mutation profiles of these hypermutated samples showed that de novo hypermutated gliomas harbored mutation signatures 6 and 1, implicating MMR deficiency as the cause of hypermutation (Fig. 4C, upper panel). In contrast, mutation signature 11 was enriched in post-treatment hypermutated gliomas (Fig. 4C, middle and lower panel). To determine the origin of the driver mutations, we estimated the probability that SNVs were derived from each mutation process (Fig. 4D-E). In samples with hypermutation, the origin of driver mutations was focused mainly on signature 11 (Fig. 4E, left panel; SBS11: 89.51%; SBS1: 2.3%; SBS6: 8.18%). In contrast, the driver gene variants of samples without hypermutation consisted mainly of age-related signatures (Fig. 4D and E, right panel; SBS1: 40%/30.26%; SBS5: 60%/55.26%). Remarkably, our results also indicated that some MMR-related variants were caused by TMZ treatment (R007: MSH6 S198N, MSH6 G841E; R010: G300E, W917X).
Fig. 4.
Mutation signature analysis. (A) The distribution of each mutation signature in 97 glioma samples. The upper panel showed the mutation counts attributed to different signatures. The middle panel showed the percent. The lower panel showed the somatic mutation of the MMR encoding genes and clinical information. (B) Stacked bar plot showing the proportion of each mutation signature that was shared, primary-private or recurrent-private. The difference between the spectra for three groups across all cases was assessed using a χ2 test. C. 96-trinucleotide-motif plot of all single base substitutions (SBSs) in hypermutated cases. The pie plot showed the proportion of various mutational signatures. (D-E) Distribution of mutational signatures associated with driver gene mutations. The bar plots showed the cumulative probabilities of each driver gene mutation being due to each mutational process. The pie charts showed the percentage of mutations signatures across all driver gene mutations. (F-G) Comparison of subclonal(later)-to-clonal(earlier) mutation fold change caused by each mutation signature between GBMs and IDH mutant gliomas. Each sample was represented as a dot. RT: Radiotherapy; TMZ: Temozolomide; Pri: Primary gliomas; Rec: Recurrent gliomas.
In addition, the mutational processes were temporally dynamic within tumors. Previous studies demonstrated that clonal mutations represented relatively early events in tumor evolution, occurring before or at the time of tumor initiation, whereas subclonal mutations represent later events [29]. Thus, we classified SNVs in each sample as early (clonal) or late (subclonal). We then calculated the enrichment of subclonal mutations in SNVs contributed by each mutation signature. Signature 1 tended to appear earlier than the other mutation signatures, suggesting that a large proportion of early mutations represent non-cancer-specific mutational processes. In GBM samples, the SNVs induced by TMZ treatment were enriched in late variants (Fig. 4H). Together, these results suggested that TMZ-associated mutation tended to occur later and might play a key role in fostering subclonal expansions during the recurrence process.
3.4. Immunogenicity of paired primary and recurrent gliomas
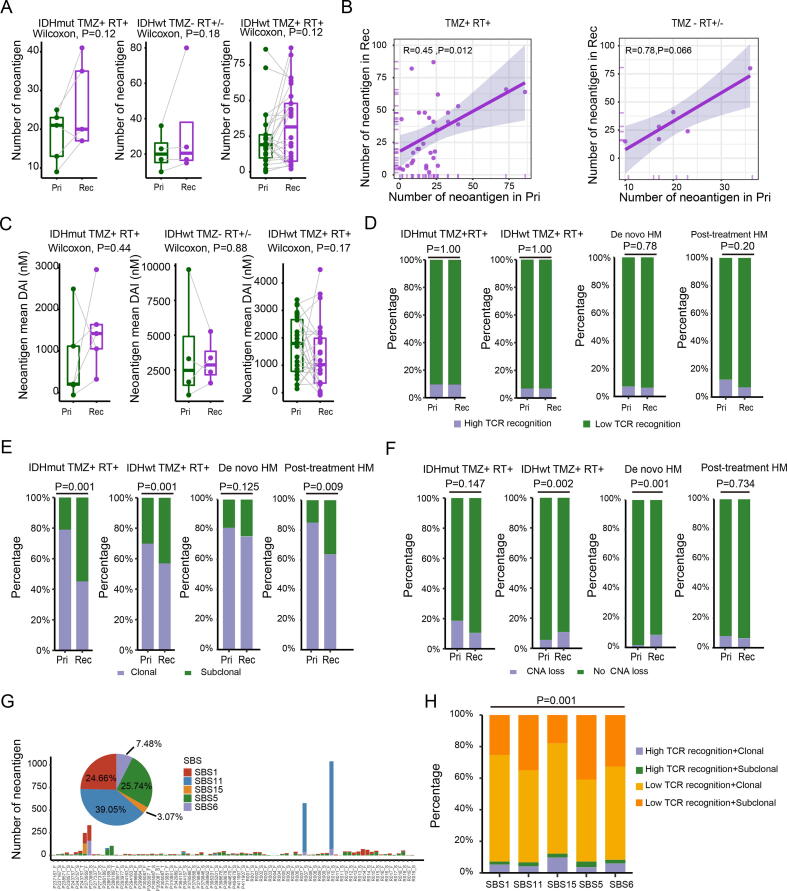
Mutations not only provide fitness through the activating crucial driver events or causing loss of tumor suppressor genes during evolution, but also generate neoantigens, which in turn produce an immune response against cancer cells. To compare the immunogenicity of primary and recurrent gliomas, we used the sequences of neopeptides caused by all SNVs/InDels to predict the quantity of the neoantigen. Excluding paired cases with hypermutation (n = 3), the quantity of neoantigen produced by recurrent tumors did not increase significantly compared to that produced by the primary tumors, regardless TMZ treatment or not (Fig. 5A; Wilcoxon test, P = 0.12/0.18/0.12). The number of neoantigens produced by post-treatment hypermutated recurrences increased more than in paired primary tumors (Supplementary Fig. 3A). In addition, the number of neoantigens in primary gliomas was positively correlated with the number of neoantigens of recurrent gliomas (Fig. 5B; R = 0.45/0.78, P = 0.012/0.066). To further identify the antitumor potential of these neoantigens and improve the true positive rate of neoantigen predictions, we used three previously described parameters, differential agretopicity index (DAI), TCR recognition, and clonality of the neoantigens, to evaluate the quality of neoantigens. The results showed no significant differences in the proportion of high TCR recognition (χ2 test, P = 1.00/1.00/0.78/0.20) and DAI (Wilcoxon test, P = 0.44/0.88/0.17) between the primary and recurrent gliomas (Fig. 5C-D and Supplementary Fig. 3B-C). Based on previous studies showing that clonal neoantigens induce stronger antitumor immunity, we compared the clonality of neoantigens in primary and recurrent gliomas using CCF. Compared to primary tumors, post-TMZ-treated recurrent GBMs and recurrent tumors of IDH mutant gliomas exhibited a significantly higher proportion of subclonal neoantigens (Fig. 5E; χ2 test, P = 0.001/0.001/ 0.009; Supplementary Fig. 3D). However, this trend was not significant in the de novo hypermutation samples and TMZ-untreated recurrent samples (Fig. 5E; χ2 test, P = 0.125; Supplementary Fig. 3D). Furthermore, the abundance of T cell infiltration was similar between primary and recurrent gliomas (Supplementary Fig. 3E; Wilcoxon test, P = 0.51/0.8). Taken together, these results indicated that the quantity and quality of neoantigens were similar among primary and recurrent gliomas. The extent of copy number loss-related neoantigens was then quantified. Although the CNV burden of primary and recurrent GBMs was similar (Fig. 2C), we found that the proportion of CNV loss-related neoantigens in post-TMZ-treated recurrent GBMs and de novo hypermutation samples were significantly higher than that in primary tumors (Fig. 5F; χ2 test, P = 0.002/0.001). Furthermore, we analyzed the quantity and immunogenicity of neoantigens caused by various mutation processes that were activated during tumor evolution. Our results showed that neoantigens were derived mainly from age-related processes (SBS1 and SBS5) or TMZ treatment (SBS11), especially in recurrent samples of R007 and R010 (Fig. 5G). Compared with other mutational processes, defective DNA mismatch repair (SBS15) was associated with a higher proportion of highly immunogenic neoantigens (Fig. 5H; χ2 test, P = 0.001). Conversely, TMZ-related signature 11 produced a lower proportion of high-quality neoantigens and a higher proportion of subclonal neoantigens (Supplementary Fig. 3G-H; χ2 test, P = 0.26). Overall, these results suggested that the immunogenicity of neoantigens produced by various mutational processes was not concordant with the process of glioma development. TMZ-induced mutations increased the total number of neoantigens, but did not necessarily activate an effective antitumor response, which might be due to the subclonal nature of neoantigens.
Fig. 5.
Characteristics of neoantigens among primary and recurrent glioma. (A) Box plot and and paired lines showing comparison of the number of neoantigens between primary and recurrent samples. Wilcoxon signed-rank test P values were indicated. (B) Scatter plot showing the relationship between the number of neoantigens of primary and recurrent gliomas. Spearman's rank correlation coefficient and P values were indicated. (C) Box plot and and paired lines showing comparison of mean DAI of neoantigen between primary and recurrent gliomas across two subtypes. (D) Percent stacked bar chart showing the proportion of high the TCR recognition. (E) The proportion of neoantigens arising from clonal or subclonal mutations was shown in primary and recurrent gliomas. (F) Percent stacked bar chart showing the different proportions of copy number loss related neoantigen among primary and recurrent gliomas. (G) Number and distribution of neoantigens produced by different mutational processes. (H) Comparison of TCR recognition probability and clonality of neoantigens produced in different mutational processes. Statistical tests used can be found in materials and methods. HM: Hypermutation RT: Radiotherapy; TMZ: Temozolomide; Pri: Primary gliomas; Rec: Recurrent gliomas.
4. Discussion
In this study, we performed a systematic analysis of WES data of paired primary glioma and recurrence samples. We found that recurrent gliomas displayed variable degrees of genetic similarity to the initial gliomas and acquired unique characteristics. The evolutionary phylogenetic trees of longitudinal samples from SYSUCC cohort revealed that recurrence could arise from the dominant clone in the primary tumor leading to genetic similarity or clone branched off early in the clonal evolution process of the primary tumor, which brought a new perspective into therapy development. In accordance with a previous study [9], [43], primary and recurrent tumors shared gliomagenesis-related variants, including mutations in TP53, IDH1, PTEN, ATRX, PIK3CA, loss of chromosome 10, and gain of chromosome 7. In IDH mutant gliomas, clonal mutations of TP53, IDH1, ATRX, CHD4, NOTCH1, and IRS4 were shared in primary and recurrent samples. In GBM, PTEN, LRP1B, ENPEP, PIK3CA, TP53, and GRIN2A mutations were clonal mutations in both primary and recurrent samples. These results implied that these variants were early events in gliomagenesis. In addition to the previously reported RB and AKT-mTOR signaling pathways [10], recurrent-private variants were also involved in the Notch signaling pathway, stem cell regulation-related pathway, chromatin modification, Ras, MAPK, Hippo signaling pathway, and stem cell regulation-related pathway Consistent with this, accumulating evidence suggests that Notch signaling suppresses differentiation and maintains stem-like properties, contributing to glioblastoma tumorigenesis and treatment resistance [42]. We also identified several genetic alterations with the potential to drive IDH mutant gliomas or GBMs recurrences, including amplification of PPFIBP1, KRAS, and PDE4DIP, deletion of TNFRSF14, CDKN2A, DCC and MSH6, and mutations in ATRX, ARID1A, KEL, TP53, ANK1, FAT4, MSH6, and KMT2B. In the GLASS dataset, the proportion of mutation in MSH6, KMT2B, ANK1, TP53, FAT4, ARID1A, and AFDN, deletion of MSH6 and CDKN2A, amplification of KRAS in recurrent tumors was also significantly higher than that in primary samples. The other genomic alterations, such as amplification of ABCB1 and deletion of DCC, were only present in our cohort. These results showed that recurrent mechanisms of glioma were diverse.
In line with previous studies [9], [44], recurrences in our cohort were broadly classified as branched evolution. The branched recurrence pattern suggested that current treatment reduced the physical burden and clonal diversity of the tumor tissue, and recurrence-initiating glioma cells emerged from the residual subclones that survived therapy and originated from cells that branched off early during tumorigenesis or much later. Moreover, the de novo SNVs/InDels introduced by TMZ and radiation treatments complicated these recurrence processes, especially in the inactivation of the MMR pathway. Thus, the extent of genomic alterations in recurrent tumors is related not only to tumor heterogeneity but also to previous treatment. Treatments based on the analysis of the primary gliomas do not provide accurate guidance information for determining the most appropriate treatment for recurrent gliomas, especially earlier branch recurrences. Shared clonal mutations were ideal therapeutic targets, especially for personalized cancer vaccines. An IDH1(R132H)-specific peptide vaccine has been shown to be effective in a phase I trial against IDH1(R132H) gliomas [45]. In our study, we also observed that there was no significant increase in immunogenicity of IDH mutant gliomas or GBMs in terms of the quantity and quality of neoantigens. Although TMZ induced more de novo mutations, TMZ induced a smaller proportion of high-quality neoantigens than other mutagenic factors. This might partly explain the previously described phenomenon that hypermutated gliomas are characterized by a lack of prominent T cell infiltration. It can be speculated that these results explain the heterogeneity of immune checkpoint blockade responses of hypermutated gliomas, although this needs further verification. However, these results highlight the importance of considering the benefits of TMZ, including antitumor effects and the potential risk of inducing de novo mutations.
Some limitations of the present study should be noted. First, our study did not include multiregional sampling, and phylogenetic relationships were limited by the breadth of sampling and sequencing depth. Second, we used WES to characterize the genetic alterations of the primary and recurrent gliomas; however, we could not detect non-coding and structure variants. Third, the absence of molecular biology experimental validation was also one of the limitations of our study. Lastly, our study lacked other omics data as a supplement. Beyond genome evolution, cancers can also evolve through epigenetic modifications, which interact closely with the changing tumor microenvironment [46].
5. Conclusions
In summary, our study highlights the importance of studying the patterns of glioma recurrence and the impact of chemoradiotherapy on the intratumoral heterogeneity of recurrent gliomas. Future studies of paired primary and recurrent gliomas with multi-regional sampling and single-cell sequencing are required to further elucidate these processes.
Funding
This work was supported by National Natural Science Foundation of China (No. 82072761, 81872059, and 81872299), National Basic Research Program of China (No. 2015CB755505), Guangzhou Science Technology and Innovation Project (201508020125), Guangdong Science and Technology Planning Project (No. 2016A020213004), Guangzhou Science Technology Project (No. 201508020125 and 201904010007), Shenzhen Science Technology Project (No. JCYJ20190807160011600), and the Fundamental Research Funds for the Central Universities, Sun Yat-sen University (2021qntd43). 5010 Medical Project of Sun Yat-sen University (No. 2012012).
Author contributions
ZPC and XL conceived the study and supervised the work. CL and FRC collected samples. SYX, ZQZ, and JW reviewed the cases. Sample processing was performed by CL. Bioinformatics analysis was performed by PFX. PFX, CL, and XL performed data analysis and data interpretation. PFX wrote the first draft of the manuscript. All authors edited and approved the final draft.
Data availability
Supplementary Table 3 included the mutation annotation format (MAF) file for all patients sequenced. The raw data of WES have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number HRA001707 that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human. MSKCC dataset used in this study can be downloaded from cBioPortal (https://www.cbioportal.org/). GLASS dataset used in this study can be downloaded from Synapse (https://www.synapse.org/#!Synapse:syn17038081/wiki/585622).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.04.034.
Contributor Information
Xin Li, Email: lixin253@mail.sysu.edu.cn.
Zhong-Ping Chen, Email: chenzhp@sysucc.org.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Weller M., van den Bent M., Hopkins K., Tonn J.C., Stupp R., Falini A., et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014;15(9):e395–e403. doi: 10.1016/S1470-2045(14)70011-7. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R., Taillibert S., Kanner A., Read W., Steinberg D., Lhermitte B., et al. Effect of Tumor-Treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA. 2017;318(23):2306–2316. doi: 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiba K., Lorbeer F.K., Shain A.H., Mcswiggen D.T., Schruf E., Oh A., et al. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science. 2017;357(6358):1416–1420. doi: 10.1126/science.aao0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceccarelli M., Barthel F.P., Malta T.M., Sabedot T.S., Salama S.R., Murray B.A., et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455(7216):1061-68. 10.1038/nature07385. [DOI] [PMC free article] [PubMed]
- 7.Wang J., Cazzato E., Ladewig E., Frattini V., Rosenbloom D.I.S., Zairis S., et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H., Zheng S., Amini S.S., Virk S.M., Mikkelsen T., Brat D.J., et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25(3):316–327. doi: 10.1101/gr.180612.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barthel F.P., Johnson K.C., Varn F.S., Moskalik A.D., Tanner G., Kocakavuk E., et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature. 2019;576(7785):112–120. doi: 10.1038/s41586-019-1775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson B.E., Mazor T., Hong C., Barnes M., Aihara K., Mclean C.Y., et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S., Zhou Y., Chen Y., Gu J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del A.G., Levy-Moonshine A., et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:10–11. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koboldt D.C., Zhang Q., Larson D.E., Shen D., Mclellan M.D., Lin L., et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S., Scheffler K., Halpern A.L., Bekritsky M.A., Noh E., Kallberg M., et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15(8):591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 17.Shi W., Ng C., Lim R.S., Jiang T., Kumar S., Li X., et al. Reliability of Whole-Exome sequencing for assessing intratumor genetic heterogeneity. Cell Rep. 2018;25(6):1446–1457. doi: 10.1016/j.celrep.2018.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucl Acids Res. 2010;38(16) doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19(9):1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 22.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shihab H.A., Gough J., Cooper D.N., Day I.N., Gaunt T.R. Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics. 2013;29(12):1504–1510. doi: 10.1093/bioinformatics/btt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7(10) doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Favero F., Joshi T., Marquard A.M., Birkbak N.J., Krzystanek M., Li Q., et al. Sequenza: Allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26(1):64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schliep K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics. 2011;27(4):592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letouzé E., Shinde J., Renault V., Couchy G., Blanc J., Tubacher E., et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun. 2017;8(1) doi: 10.1038/s41467-017-01358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mcgranahan N., Favero F., de Bruin E.C., Birkbak N.J., Szallasi Z., Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7(283):254r–283r. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szolek A., Schubert B., Mohr C., Sturm M., Feldhahn M., Kohlbacher O. OptiType: Precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30(23):3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hundal J., Carreno B.M., Petti A.A., Linette G.P., Griffith O.L., Mardis E.R., et al. PVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8(1):11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: Improved Peptide-MHC class i interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199(9):3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.łuksza M M., Riaz N., Makarov V., Balachandran V.P., Hellmann M.D., Solovyov A., et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551(7681):517–520. doi: 10.1038/nature24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balachandran V.P., Luksza M., Zhao J.N., Makarov V., Moral J.A., Remark R., et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richman L.P., Vonderheide R.H., Rech A.J. Neoantigen dissimilarity to the Self-Proteome predicts immunogenicity and response to immune checkpoint blockade. Cell Syst. 2019;9(4):375–382. doi: 10.1016/j.cels.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Körber V., Yang J., Barah P., Wu Y., Stichel D., Gu Z., et al. Evolutionary trajectories of IDHWT glioblastomas reveal a common path of early tumorigenesis instigated years ahead of initial diagnosis. Cancer Cell. 2019;35(4):692–704. doi: 10.1016/j.ccell.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Kocakavuk E., Anderson K.J., Varn F.S., Johnson K.C., Amin S.B., Sulman E.P., et al. Radiotherapy is associated with a deletion signature that contributes to poor outcomes in patients with cancer. Nat Genet. 2021;53(7):1088–1096. doi: 10.1038/s41588-021-00874-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turajlic S., Xu H., Litchfield K., Rowan A., Chambers T., Lopez J.I., et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell. 2018;173(3):581–594. doi: 10.1016/j.cell.2018.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louis D.N., Perry A., Wesseling P., Brat D.J., Cree I.A., Figarella-Branger D., et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021;23(8):1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bazzoni R., Bentivegna A. Role of notch signaling pathway in glioblastoma pathogenesis. Cancers (Basel) 2019;11(3) doi: 10.3390/cancers11030292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng H.K., Wong Q.H., Liu E.M., Li K.K. The new WHO molecular criteria for adult glioblastoma – Are we a step too far? Glioma. 2021;4:65–67. [Google Scholar]
- 44.Aldape K., Amin S.B., Ashley D.M., Barnholtz-Sloan J.S., Bates A.J., Beroukhim R., et al. Glioma through the looking GLASS: Molecular evolution of diffuse gliomas and the Glioma Longitudinal Analysis Consortium. Neuro-Oncology. 2018;20(7):873–884. doi: 10.1093/neuonc/noy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Platten M., Bunse L., Wick A., Bunse T., Le Cornet L., Harting I., et al. A vaccine targeting mutant IDH1 in newly diagnosed glioma. Nature. 2021;592(7854):463–468. doi: 10.1038/s41586-021-03363-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicholson J.G., Fine H.A. Diffuse glioma heterogeneity and its therapeutic implications. Cancer Discov. 2021;11(3):575–590. doi: 10.1158/2159-8290.CD-20-1474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary Table 3 included the mutation annotation format (MAF) file for all patients sequenced. The raw data of WES have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number HRA001707 that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human. MSKCC dataset used in this study can be downloaded from cBioPortal (https://www.cbioportal.org/). GLASS dataset used in this study can be downloaded from Synapse (https://www.synapse.org/#!Synapse:syn17038081/wiki/585622).