Abstract
The circadian clock regulates tissue homeostasis through temporal control of tissue-specific clock-controlled genes. In articular cartilage, disruptions in the circadian clock are linked to a procatabolic state. In the presence of inflammation, the cartilage circadian clock is disrupted, which further contributes to the pathogenesis of diseases such as osteoarthritis. Using synthetic biology and tissue engineering, we developed and tested genetically engineered cartilage from murine induced pluripotent stem cells (miPSCs) capable of preserving the circadian clock in the presence of inflammation. We found that circadian rhythms arise following chondrogenic differentiation of miPSCs. Exposure of tissue-engineered cartilage to the inflammatory cytokine interleukin-1 (IL-1) disrupted circadian rhythms and degraded the cartilage matrix. All three inflammation-resistant approaches showed protection against IL-1–induced degradation and loss of circadian rhythms. These synthetic gene circuits reveal a unique approach to support daily rhythms in cartilage and provide a strategy for creating cell-based therapies to preserve the circadian clock.
Stem cells were engineered to protect against loss of their circadian rhythm caused by the inflammatory cytokine interleukin-1.
INTRODUCTION
The circadian clock is an internal genetic timing mechanism that exists in the brain and nearly all cells in peripheral tissues, operating on a roughly 24-hour period (1–3). The circadian clock coordinates tissue-specific physiology with different cycles such as light and darkness, body temperature, and rest and activity (1). The core clock mechanism, composed of transcriptional activators, Bmal1 and Clock, and transcriptional repressors, Per1/2 and Cry1/2, form an autoregulatory negative feedback loop that drives rhythmic gene expression. This transcription-translation feedback loop regulates output genes, such as Rev-erbs, Dbp, and Nfil3 (2, 3). The circadian clock drives expression of many other genes, referred to as clock-controlled genes, that are tissue specific and play important roles in maintaining tissue homeostasis through their temporal nature (1, 4–6). Disruptions in the circadian clock and subsequent disruptions in expression of these clock-controlled genes have been linked to several diseases such as obesity, diabetes, cardiovascular disease, and osteoarthritis (OA) (2, 4, 7, 8).
Clock genes play a critical role in maintaining tissue homeostasis in many different musculoskeletal tissues including muscle, tendon, bone, and articular cartilage (4, 9). Articular cartilage consists of an abundant extracellular matrix (ECM) synthetized by the only resident cell type, chondrocytes (10–12). Articular cartilage is avascular and aneural and is maintained through a balance between anabolic and catabolic activities. Disruption to this balance, for example, through increased inflammation, drives the degradation of articular cartilage, which is a key characteristic of OA, the leading cause of pain and disability worldwide (13–15). It has been shown that many anabolic and catabolic pathways in articular cartilage have different daily dynamics, driven by a functional circadian clock in chondrocytes, that help maintain cartilage homeostasis (4, 9, 16–23). Several cellular functions exhibit time of day–dependent activity, including metabolic, ECM remodeling, and catabolic pathways (4, 20), and disruptions in the clock cause loss in rhythmic expression and altered expression of these pathways (20). For example, the cartilage-specific knockout (KO) of one of the core clock genes, Bmal1, results in a catabolic response and cartilage degradation in mice (18). These findings suggest that the disruption of the circadian clock in cartilage could be an important contributor to the pathogenesis of OA (24).
Inflammation is a key driver in the pathogenesis of OA, characterized by increased levels of proinflammatory cytokines such as interleukin-1 (IL-1) and tumor necrosis factor–α (TNFα), which, in turn, induce ECM degradation through the enhanced production of degradative enzymes and other proinflammatory mediators that shift chondrocyte activity to a procatabolic state (25–29). Inflammation has been shown to disrupt circadian rhythms in cartilage (30). The addition of IL-1 to cartilage explants caused a loss in circadian rhythms that was only rescued with the administration of dexamethasone (dex), an anti-inflammatory agent (30). Anticytokine therapies such as IL-1 receptor antagonist (IL-1Ra; anakinra) have shown promise in alleviating symptoms of posttraumatic OA (31–33); however, systemically delivered anticytokine therapies are administered at high doses, which may have off-target effects, including an increased susceptibility to infection and certain autoimmune diseases (34) as well as limited tissue regeneration and repair (35–37).
Because of the drawbacks with anticytokine therapies and lack of effective disease-modifying treatments to address both the symptoms and structural changes of OA (38–40), we used genetic engineering to develop various self-regulating cellular systems to address these limitations (41). Using CRISPR-Cas9 gene editing, we created murine induced pluripotent stem cells (miPSCs) lacking a functional IL-1 receptor 1 (IL1R1) that can successfully be differentiated into tissue-engineered cartilage that is protected from IL-1–induced matrix degradation (IL1R1 KO) (42). We also used CRISPR-Cas9 gene editing in miPSCs to incorporate IL-1Ra, a competitive antagonist of IL-1, downstream of the Ccl2 locus (Ccl2-IL1Ra) to drive controlled production of IL-1Ra in response to inflammation and subsequent activation of Ccl2 (43). These cells were also able to protect cartilage from inflammation-mediated degradation (43). Furthermore, we have also developed a synthetic promoter that is activated by nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) to drive production of our desired transgene, IL-1Ra (NFκBr-IL1Ra) (44). The NF-κB signaling pathway is a key inflammatory pathway in articular cartilage that is activated by various cytokines such as IL-1, leading to the up-regulation of other proinflammatory mediators. This lentiviral gene therapy has been shown to inhibit IL-1 in response to inflammation or mechanical loading in miPSC-derived tissue-engineered cartilage and also in primary porcine chondrocytes (44, 45).
Because of the critical role of the circadian clock in maintaining cartilage tissue homeostasis, as evidenced by altered gene expression (20) and cartilage degradation (18) caused by disruptions in circadian rhythms, and the recent finding that inflammation can drive disruption in the chondrocyte circadian clock, the goal of this study was to develop tissue-engineered cartilage with genetically engineered stem cells to preserve the circadian clock in response to inflammation (Fig. 1). Therefore, we sought to establish a platform for testing the role of circadian rhythms in cartilage maintenance and accomplished this by first testing when circadian rhythms arise while differentiating our miPSCs into chondrocytes and then the consequences of inflammatory signals on chondrocyte circadian rhythms and cartilage integrity, before testing the ability of our genetically engineered circuits to preserve circadian rhythms. However, note that undifferentiated stem or progenitor cells generally do not exhibit circadian rhythms (46). Therefore, we first tested the hypothesis that circadian rhythms and cartilage matrix develop together as a part of chondrogenic differentiation in miPSCs. We next determined the effect of inflammatory cytokines on circadian cycling in our tissue-engineered pellets and determined that IL-1α and IL-1β, but not TNFα, drive disruptions in circadian rhythm. We then tested the ability of our genetically engineered cell-based therapies targeting IL-1 to protect the clock from IL-1–induced disruption. We show the ability of our self-regulating synthetic gene circuits to preserve daily rhythmicity and protect against inflammation-mediated disruption and cartilage degradation. These findings further our understanding of the circadian rhythm coordination in cartilage and its disruption in response to inflammatory cytokines and introduce the concept of a “clock-preserving” mechanism through the combination of synthetic biology and tissue engineering of stem cell gene circuits.
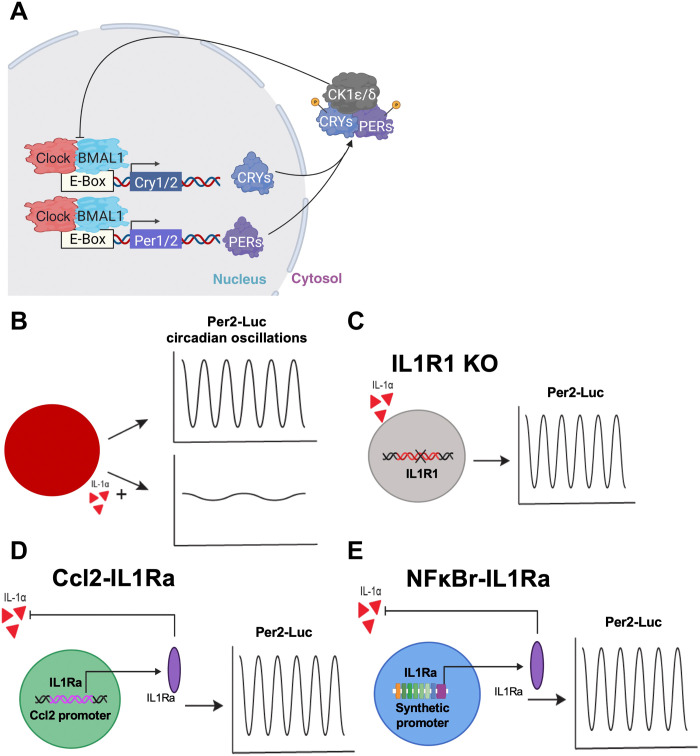
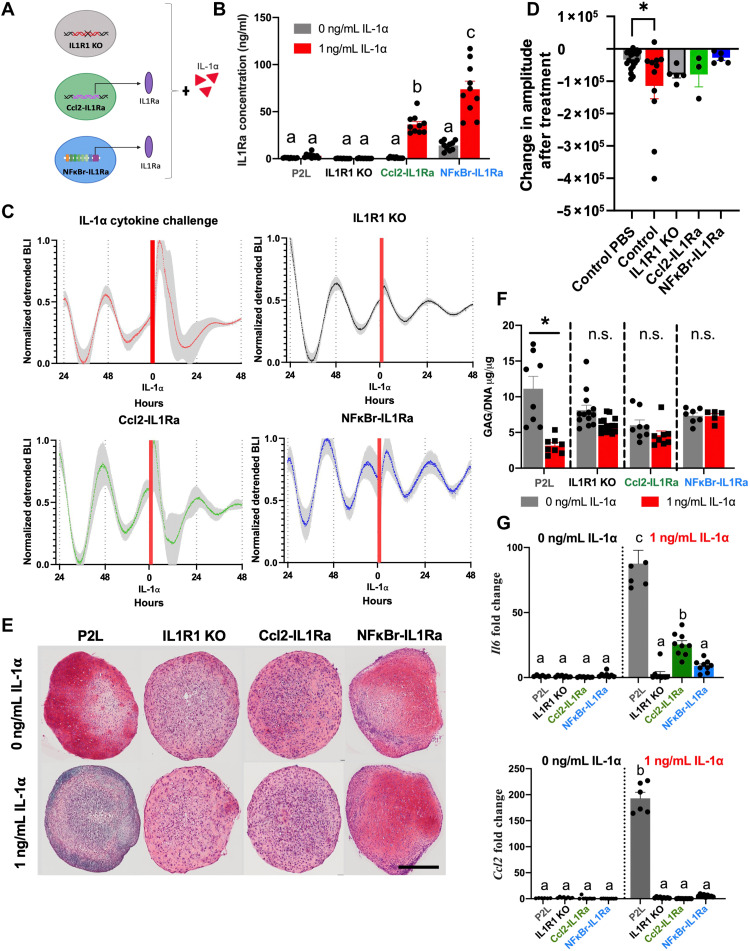
Fig. 1. Inflammation-resistant cell-based therapies and circadian measurements.
Tissue-engineered cartilage pellets with a circadian reporter (P2L) were tracked to measure their circadian clock. (A) Schematic of the molecular circadian transcription-translation feedback loop. (B) In the presence of IL-1α, the circadian clock was disrupted. Three different genetic engineering–based cell therapies were developed to be resistant to IL-1α: (C) IL1R1 KO, CRISPR-Cas9–edited cells lacking the IL1R1; (D) Ccl2-IL1Ra, CRISPR-Cas9–edited cells that produce an IL-1 antagonist, IL-1Ra, in response to inflammation and subsequent expression of Ccl2; and (E) NFκBr-IL1Ra, lentiviral gene therapy circuit with a synthetic promoter activated by NF-κB signaling driving production of IL-1Ra. The ability of these cell therapies to preserve the circadian clock in the presence of inflammation was tested.
RESULTS
The circadian clock develops in miPSCs with each stage of chondrogenesis
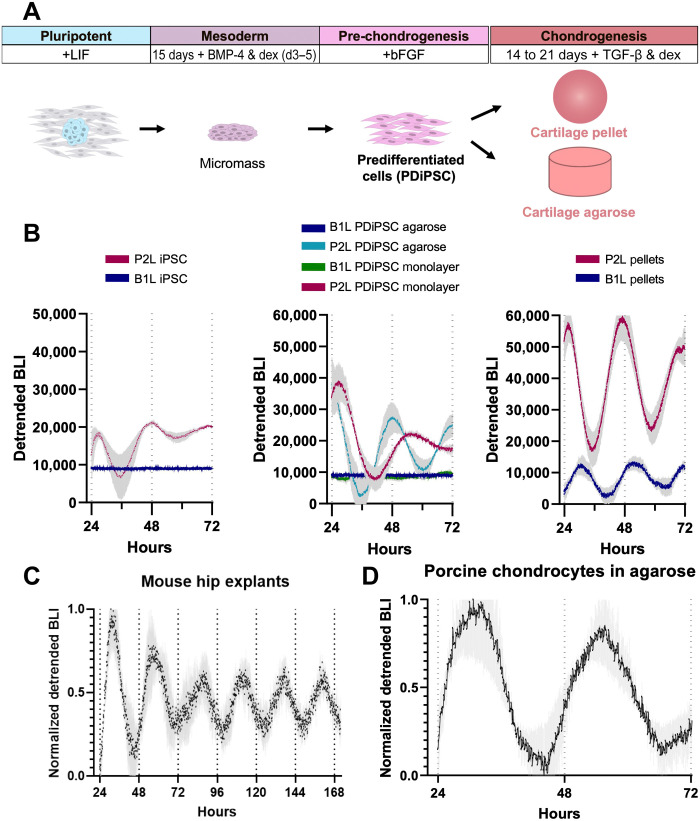
To assess the development of circadian rhythms during chondrogenesis, we transduced miPSCs and predifferentiated iPSCs (PDiPSCs) with Per2-Luc (P2L) or Bmal1-Luc (B1L) lentivirus and tracked the bioluminescence output of miPSCs, PDiPSCs in monolayer, PDiPSCs freshly cast in agarose, and chondrogenic cartilage pellets (Fig. 2A). We found that miPSCs expressed P2L and B1L, but with no evidence of circadian oscillation (e.g., periods >32 hours; Fig. 2B). In addition, at the PDiPSC differentiation stage, P2L-transduced PDiPSCs began to oscillate (average period = 28.1 ± 3.1 hours, means ± SEM), and B1L oscillation, which appears after Per2 expression in development (46), had not yet occurred (period length >32 hours; Fig. 2B). However, PDiPSCs cast in a three-dimensional (3D) agarose system showed enhanced rhythmicity in Per2 (average period = 26.3 ± 2.2 hours), while Bmal1 remained arrhythmic (period length >32 hours; Fig. 2B). Tissue-engineered cartilage pellets showed high amplitude oscillations of Per2 and Bmal1 in antiphase, indicating the circadian clock had fully developed in our tissue-engineered cartilage (P2L period = 23.2 ± 3.1 hours, B1L period = 21.5 ± 4.9 hours; Fig. 2B).
Fig. 2. Development of the circadian clock in miPSC chondrogenesis.
(A) Differentiation protocol of miPSCs to chondrocytes. miPSCs were subjected to high-density micromass culture to create PDiPSCs. PDiPSCs were then either cast in agarose or pelleted and cultured in chondrogenic media for 14 to 21 days to create tissue-engineered cartilage. (B) P2L bioluminescence intensity (BLI) of miPSCs (n = 10), PDiPSCs (n = 2), PDiPSCs in agarose (n = 4), and pellets (n = 10) (left) and B1L bioluminescence intensity (BLI) of miPSCs (n = 5), PDiPSCs (n = 4), PDiPSCs in agarose (n = 5) (middle), and pellets (n = 5) (right). (C) BLI of femoral head cartilage explants from PER2::LUC mice (n = 3, one hip per mouse). (D) P2L BLI in porcine chondrocytes cast in agarose (n = 5). Shaded region on graphs represents SEM.
Murine cartilage explants were taken from the femoral head of 10-week-old homozygous PER2::LUC mice (5). Bioluminescence recording of these cartilage explants demonstrated sustained circadian rhythms, as shown previously (Fig. 2C). Our tissue-engineered cartilage pellets had similar period length compared with PER2::LUC oscillations in the cartilage explants, further confirming the development and maturation of the circadian clock in our tissue-engineered cartilage (engineered cartilage period = 23.2 ± 3.1 hours, explant period = 23.9 ± 1.5 hours). In addition, we transduced primary porcine chondrocytes with our P2L lentivirus, and these porcine chondrocytes also showed sustained daily oscillations similar to our tissue-engineered pellets (period = 23.5 ± 3.3 hours; Fig. 2D). Together, these findings indicate that as murine iPSCs undergo chondrogenesis in tissue-engineered cartilage, they develop a circadian clock that has high amplitude and sustained oscillations, similar to that of primary chondrocytes or mature cartilage in another species.
Inflammation disrupts the circadian clock in native and tissue-engineered cartilage
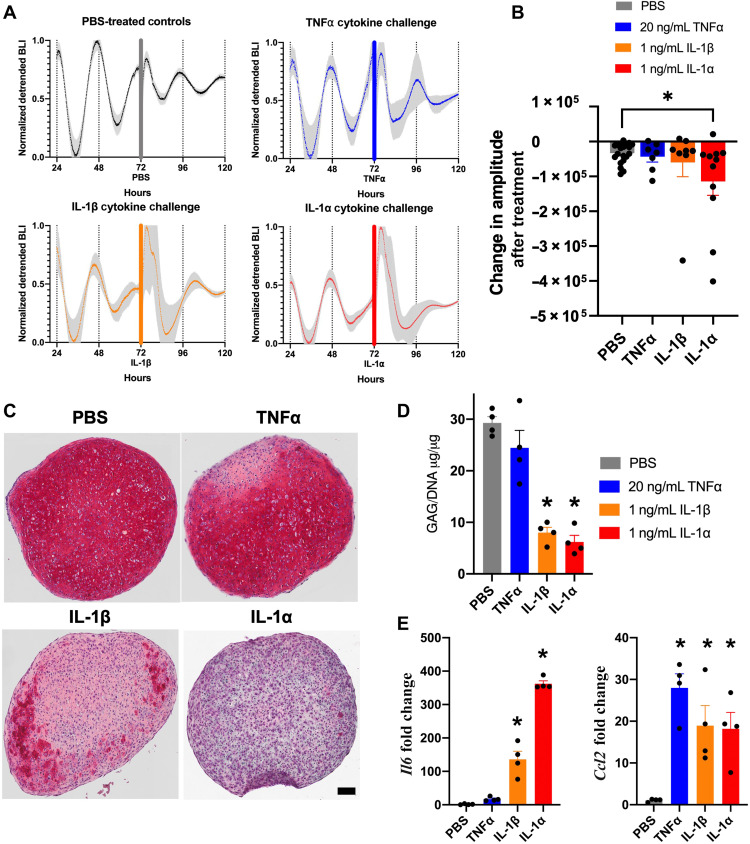
We next examined the effects of inflammatory cytokines IL-1α, IL-1β, and TNFα on circadian rhythms in tissue-engineered cartilage pellets. It has previously been shown that IL-1β causes disruption in the circadian rhythms of murine cartilage explants through loss in bioluminescence oscillations and changes in clock gene expression; however, TNFα had no effect (30). For these studies, we treated mature tissue-engineered cartilage transduced with the P2L lentiviral reporter with pathophysiologic levels of cytokines, IL-1α (1 ng/ml), IL-1β (1 ng/ml), TNFα (20 ng/ml), or phosphate-buffered saline (PBS) as a control. Bioluminescence tracking of P2L was recorded for 3 days before addition of cytokines. Cytokines were added in 2-μl volumes, and pellet bioluminescence was tracked for an additional 3 days after cytokine addition. To determine the effect of these cytokines on circadian rhythms, we analyzed both the period and decrease in amplitude, as these are considered the primary markers of circadian disruption. Decreased amplitude represents decreased oscillation in clock gene expression and weakening of circadian rhythms. Pellets subjected to inflammatory challenge were also collected for histological and biochemical analysis of a main component of articular cartilage, sulfated glycosaminoglycans (sGAGs), and analyzed for up-regulation of inflammatory genes Il6 and Ccl2. Histological and biochemical analysis of sGAG was performed to determine the effect of cytokines on cartilage matrix after addition of cytokine, with sGAG staining and concentration providing a sensitive measure of cytokine-induced cartilage matrix degradation. Before cytokine addition, all pellets showed circadian oscillations of P2L (n = 46; Fig. 3A). Pellets treated with PBS maintained circadian rhythms to the end of the recording (n = 20, pre-PBS period = 25.4 ± 3.1 hours, post-PBS period = 25.6 ± 7.2 hours; Fig. 3A) and exhibited a daily decrease in amplitude, seen in in vitro cultures, similar to undisturbed control pellets (fig. S1). Pellets treated with TNFα also continued to maintain circadian oscillations (n = 7, five of the seven were rhythmic after treatment, post-TNFα period = 27.6 ± 3.8 hours; Fig. 3A). Pellets treated with IL-1β showed a lengthening of period in all pellets (n = 8, six of the eight were rhythmic after treatment, post-IL1β period = 29.5 ± 4.6 hours; Fig. 3A). In contrast, pellets treated with IL-1α showed a rapid loss in circadian rhythms (n = 11, 6 of the 11 were rhythmic after treatment, post-IL1α period = 31.7 ± 6.6 hours; Fig. 3A) and decreased bioluminescence amplitude (P = 0.037; Fig. 3B). Nonnormalized, representative traces from each treatment group exhibited individual variability in mean bioluminescence, which justifies the display of normalized group-level data (fig. S2). However, all amplitude and period measurements were made from nonnormalized traces. Cartilage explants from PER2::LUC mice were also subjected to inflammatory challenge, and the bioluminescence output was recorded (Fig. 4). Similar to our tissue-engineered cartilage pellets, TNFα had no effect on the circadian rhythm of the cartilage explants; however, both IL-1α and IL-1β given at 1 ng/ml showed a rapid loss in circadian rhythms compared with controls and significant decrease in amplitude compared with PBS controls (P = 0.0021 and 0.0005, respectively; Fig. 4). The induction of PER2 bioluminescence that can be observed in all groups, including PBS treated, is a commonly observed phenomenon resulting from the role of PER2 in cellular stress response and regulation of mammalian target of rapamycin (mTOR), a cellular growth pathway (47, 48).The similar expression of circadian rhythms and responses to inflammatory cytokines of our tissue-engineered cartilage and ex vivo cartilage explants supports their utility as an in vitro model system.
Fig. 3. Tissue-engineered cartilage response to inflammatory cytokines.
(A) P2L bioluminescence traces of pellets pre and post cytokine [TNFα (n = 7), IL-1β (n = 8), IL-1α (n = 11), or PBS (n = 20)]. Tissue-engineered cartilage pellets were treated with TNFα, IL-1β, IL-1α, or PBS control for 72 hours. Bars show addition of cytokine or PBS control. Shaded region on graphs represents SEM. (B) P2L pellets treated with IL-1α show rapid decrease in bioluminescence followed by lower-amplitude circadian rhythm (P = 0.037). The amplitude due to treatment was quantified by measuring the difference in peak-trough amplitude 24 hours before and 48 hours following treatment. Asterisks represent significance compared with PBS control. (C) Safranin O staining for sGAGs (n = 4 per treatment; scale bar, 100 μM). Pellets treated with IL-1β and IL-1α show loss in stain. (D) Quantification of sGAGs normalized to DNA content within each pellet (n = 4 per treatment). Pellets treated with IL-1β and IL-1α show significant loss in sGAG compared with PBS controls (P < 0.0001). Asterisks represent significant differences as compared with PBS controls. (E) Gene expression analysis of Il6 and Ccl2 (n = 4 per treatment, fold change relative to PBS control). Only IL-1β and IL-1α treatment caused up-regulation of Il6 (P = 0.0002). All cytokine treatments caused up-regulation in Ccl2 (P < 0.05). Asterisks represent significant differences as compared with PBS controls.
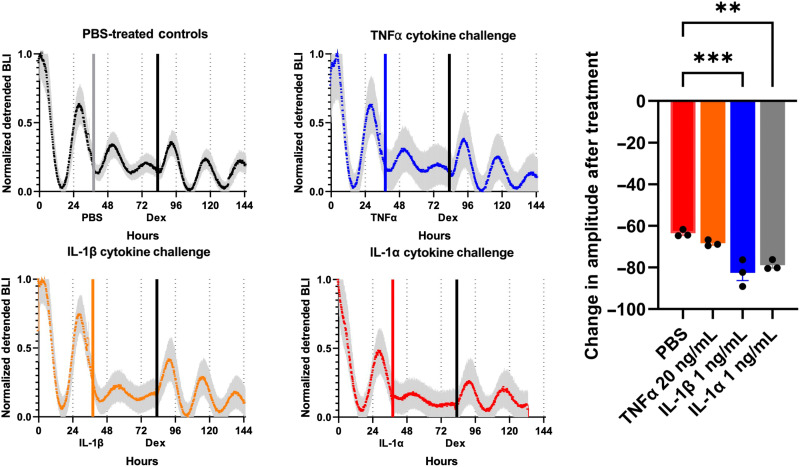
Fig. 4. Cartilage response to inflammatory cytokines.
Per2 expression in cartilage explants from the femoral head of PER2::LUC mice was recorded as bioluminescence intensity. Explants were treated with IL-1β (1 ng/ml), IL-1⍺ (1 ng/ml), TNF⍺ (20 ng/ml), or vehicle controls (n = 3 per each condition). Loss of circadian rhythm was seen in explants, given IL-1 and circadian rhythm was rescued with the administration of dex. Gray, blue, orange, and red bars indicate administration of PBS or cytokine; black bars indicate administration of dex. Shaded region on graph represents SEM. Asterisks represent significance compared to controls (P < 0.05). **P < 0.01; ***P < 0.05.
To better understand the effect of each cytokine on circadian rhythm and begin to elucidate the functional outputs of the cartilage clock, we examined the effect of cytokines on the degradation of cartilage ECM, since the circadian clock is known to maintain cartilage homeostasis (4, 9, 16–20). Pellets were stained with safranin O to look at sGAG content (n = 4 per group; Fig. 3C). In addition, sGAG content within pellets was quantified by biochemical analysis and normalized to DNA content within each pellet (n = 4 per group; Fig. 3D). The staining for sGAG showed rich red stain in the PBS control pellets and the pellets treated with TNFα, while pellets treated with IL-1β showed reduced sGAG staining, and pellets treated with IL-1α showed a loss in sGAG (Fig. 3D). The quantification of sGAG content matched the histological images, where the TNFα-treated pellets had similar sGAG content compared with PBS controls, whereas the IL-1β– and IL-1α–treated pellets showed significant sGAG loss compared with PBS controls (P < 0.0001; Fig. 3D).
Last, inflammatory genes Il6 and Ccl2 were examined in pellets treated with cytokines. IL-6 is a cytokine that up-regulates inflammatory pathways within the joint, and CCL-2 is a chemokine that attracts immune cells to the joint. While treatment of pellets with TNFα significantly up-regulated Ccl2 gene expression compared with PBS controls (n = 4, P = 0.0002), it did not significantly affect Il6 expression (n = 4; Fig. 3E). Addition of IL-1β and IL-1α significantly increased Ccl2 expression compared with PBS controls (n = 4, P = 0.0086 and 0.0122, respectively), as well as Il6, especially in the pellets treated with IL-1α (n = 4, P < 0.0001 for both IL-1β and IL-1α; Fig. 3E). These data also suggest that, although up-regulation of chemokines such as Ccl2 for immune cell recruitment is important, cytokines such as IL-1 and IL-6 are more likely to be involved in the disruption of circadian timing. Together, this suggests that inflammatory cytokines IL-1β and IL-1α can disrupt the cartilage circadian clock in cartilage explants and our tissue-engineered cartilage, and this effect might be through the up-regulation of inflammatory cytokines, such as IL-6, that lead to matrix degradation.
IL-1–resistant genetically engineered tissues are resistant to circadian disruption
Cartilage explants treated with IL-1α and IL-1β recovered the loss in circadian rhythms with the addition of dex, an anti-inflammatory agent (Fig. 4). Previously, it has been shown that forskolin, a circadian rhythm synchronizer, could not recover circadian rhythms in cartilage explants treated with cytokines and that only the administration of an anti-inflammatory agent could recover circadian disruptions due to inflammation (30). We therefore sought to test whether our anti-inflammatory cell therapies could preserve circadian cycling in the presence of IL-1α. For these studies, we used three cell lines designed to blunt the response to inflammation through different mechanisms (Fig. 5A). To test whether IL-1 acts through IL1R1 to disrupt circadian rhythms, we used miPSC CRISPR-Cas9 edited without IL1R1 (IL1R1 KO) (42). miPSCs edited with CRISPR-Cas9 to incorporate IL-1Ra downstream of the Ccl2 locus (Ccl2-IL1Ra) (43) allowed us to study the impact of endogenous circuit activation with rapid activation kinetics capable of inhibiting IL-1–mediated inflammation. A lentiviral circuit incorporating a synthetic promoter activated by the NF-κB signaling pathway that drives production of IL-1Ra (NFκBr-IL1Ra) (44) allowed us to study the impact of a synthetic activation system with increased IL-1Ra production compared with our Ccl2-IL1Ra system. All the miPSC lines or miPSCs were transduced with the lentiviral vector containing the P2L reporter and differentiated into tissue-engineered cartilage pellets. We then treated with IL-1α and assessed the ability of these pellets to maintain cartilage integrity and circadian expression.
Fig. 5. Engineered cell therapy response to IL-1α.
(A) Three different anti-inflammatory cell therapies expressing a P2L reporter were used to test the ability to protect the circadian rhythm of tissue-engineered cartilage: CRISPR-Cas9–edited miPSCs lacking the IL-1R (IL1R1 KO), CRISPR-Cas9–edited miPSCs that produce IL-1Ra downstream of the Ccl2 promoter (Ccl2-IL1Ra), and a lentiviral gene therapy composed of an NF-κB–responsive synthetic promoter that drives production of IL-1Ra (NFκBr-IL1Ra). (B) IL-1Ra concentration in response to IL-1α stimulation (n = 10 per group). The Ccl2-IL1Ra and NFκBr-IL1Ra groups showed increased IL-1Ra production in response to IL-1α. Different letters denote groups that are significantly different. b compared to a (P < 0.002), c compared to a or b (P < 0.0001). (C) P2L bioluminescence traces of pellets before and after cytokine (n = 13). All three cell therapy groups showed protection of the clock from IL-1–induced detrimental effects. Red bar represents addition of IL-1⍺. Shaded region on graph represents SEM. (D) Change in amplitude due to treatment was quantified by measuring the difference in peak-trough amplitude 24 hours before and 48 hours following treatment and was similar to normal damping of rhythms observed in PBS-treated samples. (E) Safranin O staining for sGAGs (n = 4 to 6 per group; scale bar, 100 μM). (F) Quantification of sGAGs normalized to DNA content within each pellet (n = 8 to 10 per treatment). Cell therapy groups showed no significant loss in sGAG in response to IL-1α. Asterisks denote the significance compared to PBS control within group. n.s., not significant. (G) Gene expression analysis of Il6 and Ccl2 (n = 6 to 8 per group fold change relative to P2L-untreated control). All cell therapy groups showed significantly less inflammatory gene expression compared with IL-1α–treated controls. Different letters denote groups that are significantly different. For Il6, b compared to a (P < 0.0045), and c compared to a or b (P < 0.0001). For Ccl2, b compared to a (P < 0.0001).
We found that all groups (P2L controls, IL1R1 KO, Ccl2-IL1Ra, and NFκBr-IL1Ra) had no baseline difference in IL-1Ra production (n = 10 per group; Fig. 5B). P2L and IL1R1 KO pellets also showed no difference in IL-1Ra production when treated with IL-1α (1 ng/ml). Both Ccl2-IL1Ra and NFκBr-IL1Ra had significantly increased IL-1Ra production in response to IL-1α (1 ng/ml), with the NFκBr-IL1Ra producing the highest amount of IL-1Ra (n = 10 per group, Ccl2-IL1Ra ~36 ng/ml IL-1Ra, NFκBr-IL1Ra ~74 ng/ml IL-1Ra, P < 0.0001).
After confirming that our self-regulating cells were producing sufficient IL-1Ra to inhibit IL-1 activity, we next tracked P2L through bioluminescence before the addition of cytokine and for 3 days after addition of IL-1α (1 ng/ml). All four groups before the addition of cytokine were rhythmic, showing that the process of gene editing did not disrupt the clock (n = 13, pre–IL-1α period = 24.6 ± 2.9 hours; Fig. 5C). All three of our cell therapy groups showed maintained circadian rhythms in response to IL-1α (n = 13, post–IL-1α period = 24.4 ± 2.0 hours; Fig. 5C), whereas the nonengineered controls became arrhythmic (n = 12; Fig. 5C). Cytokine-treated resistant pellets showed a decrease in amplitude comparable to what we observed in PBS-treated control pellets, which occurs naturally in culture in the absence of entraining cues (Fig. 5D). The three engineered systems all protected the cells from the cytokine-induced decreases in circadian rhythmicity, such that there were no significant changes in the fraction of circadian cultures, their period, or their amplitude compared with control cultures.
In addition, when examining staining and quantification of sGAG content of the cartilage ECM, the IL1R1 KO, Ccl2-IL1ra, and NFκBr-IL1Ra groups exhibited rich safranin O staining and no difference in sGAG content between control pellets (no cytokine treatment) and pellets treated with IL-1α (1 ng/ml) compared within each cell therapy group (n = 4 to 6 per group for histology, n = 8 to 10 per group for biochemical analysis; Fig. 5, E and F). Last, we found that the induction of Il6 by IL-1α (1 ng/ml) was markedly reduced in all engineered cell groups (n = 6 to 8, P < 0.0001; Fig. 5G), and induction of Ccl2 was blocked (n = 6 to 8, P < 0.0001; Fig. 5G). The partial blockade of IL-1α effects on Il6 72 hours after treatment in the Ccl2-IL1Ra group compared with other treatments (n = 6 to 8, P < 0.05; Fig. 5G) was consistent with the self-regulating response that rapidly inhibits IL-1 signaling (43) and the lower production of IL-1Ra. Together, these data demonstrate that our cell therapies edited to either be nonresponsive to IL-1α (IL1R1 KO) or be self-regulating and produce an anti-inflammatory therapeutic, IL-1Ra, can effectively protect their cartilage ECM from degradation and preserve the circadian clock in the presence of IL-1α.
DISCUSSION
By combining tissue engineering, synthetic biology, and circadian biology, we developed cell-based tissue constructs that are capable of preserving the cartilage circadian clock in response to disruptive stimuli such as IL-1–induced inflammation. While significant advances have been made in understanding the physiology of the circadian clock in recent years, the use of genetic approaches has not been used previously to create cell-based clock-preserving therapeutic systems. Here, we used chondrocytes and tissue-engineered cartilage as our model system, which provides a homogeneous cell population that avoids the complexities of interpreting data from multiple cell types. Using this system, we have been able to uncover characteristics of cartilage circadian rhythms and established a robust system for peripheral clock studies. These types of tools can be used to mitigate disease where circadian disruption plays a role (4), can be used to create better engineered tissues by ensuring tissue homeostasis through clock-controlled genes (4, 9), and may be used to uncover important characteristics of the circadian clock not yet known. In combination with tissue engineering, the use of genetically modified cells may allow the formation of functional bioartificial tissues with intrinsic clock-preserving characteristics.
In quantifying clock gene expression in miPSCs, we observed that undifferentiated cells were not rhythmic and developed their circadian timing over the course of chondrogenic differentiation. This is consistent with prior reports that mouse embryonic stem cells and other stem/progenitor cells cannot generate circadian rhythms until they differentiate (46), suggesting that formation of a mature circadian clock is an integral characteristic of cellular differentiation. We detected daily rhythms in Per2, but not Bmal1, in PDiPSCs. Consistent with a prior report on clock development in mouse embryonic cells (46), this finding may indicate the beginning stages of rhythmogenesis and may reflect some limitations in the ability to detect weak rhythms in Bmal1 with this system. Furthermore, we observed that iPSC-derived chondrocytes or primary porcine chondrocytes in tissue-engineered cartilage pellets showed a similar circadian response as primary murine cartilage and responded similarly to inflammatory cytokines, indicating that this model of iPSC chondrogenesis replicates many of the features of native cartilage. In this respect, such in vitro model systems can be used to uncover key characteristics of the circadian clock development throughout chondrogenesis and in response to inflammatory cytokines that drive disease progression. This approach can be expanded to other iPSC differentiation lineages and additional stimuli to further understand the circadian clock in development.
Although it is clear that the circadian clock plays a critical role in articular cartilage physiology and OA pathology, the mechanisms by which clock genes maintain chondrocyte homeostasis remain to be determined. Here, we showed different responses of the circadian clock to cytokines in cartilage. By examining the effect of pathophysiologic concentrations (49) of the cytokines IL-1 and TNFα on the circadian clock as well as the cartilage ECM- and inflammation-associated genes, we determine that cytokines, IL-1β and IL-1α, that lead to matrix degradation and up-regulation of other proinflammatory cytokines such as Il6 drive the disruption in the circadian clock instead of cytokines such as TNFα that had no effect on the circadian clock, matrix degradation, or up-regulation of Il6. The difference between the effect of TNFα and IL-1β on daily rhythms has previously been attributed to the activation of NF-κB signaling pathway by IL-1β and lack of activation by TNFα (30). In addition, the loss in ECM components in pellets treated with IL-1 but not TNFα suggests that pathways activated by IL-1 leading to matrix degradation could also be driving the loss of daily rhythms, which, in turn, exacerbates matrix loss. As observed in cartilage treated with cytokine (30), the rapid damping of PER2 rhythmicity could be due to a reduction in the fraction of circadian cells or synchrony among rhythmic cells. Single-cell imaging could test the hypothesis that the treatment decreases both the fraction of circadian cells and synchrony among the cells. This information and this model system to test cytokines will help elucidate what each cytokine is directly acting on and help us uncover the direct link between inflammatory cytokines and the circadian clock.
An interesting observation made throughout this study was the maintenance of the circadian rhythms in cartilage explants, porcine chondrocytes, and tissue-engineered cartilage pellets over many days, where cells or tissues lack exogenous entrainment cues (50). Multiday recordings of adult mouse cartilage explants showed that oscillations were maintained for over 10 days without any perturbations such as media changes or plate movement. This type of maintained circadian clock is not common in tissue explants and highlights a potential interesting area of study (51, 52). While several previous studies have characterized the chondrocyte circadian clock and its influence on joint homeostasis, it is not known how the chondrocytes maintain their circadian clock within articular cartilage. It is hypothesized that changes in body temperature and factors within the synovial fluid help with clock synchronization (1, 9), but the prolonged maintenance of the circadian clock ex vivo without any synchronizing cues highlights that there might be a factor secreted by the chondrocytes themselves that maintains this synchrony, or the abundant ECM maintains the biochemical and biomechanical niche for synchrony (53). Future work looking into potential synchronizing factors will be key in uncovering this mechanism.
The ability of our genetic engineering approaches to maintain the circadian clock also opens a different area of therapeutic development. There are many diseases that are associated with disruptions in the circadian clock (2, 4, 7, 8). Using gene therapy approaches to create cell therapies that focus on preserving or synchronizing the clock can be a useful arsenal to help ameliorate diseases beyond OA. For example, in the context of clock preservation in the presence of inflammation, these circuits can be used to protect other musculoskeletal tissues subject to disease through IL-1–induced inflammation such as the intervertebral disc, which shows similar circadian rhythms and cytokine sensitivity as articular cartilage (2, 54). Using tissue engineering and creating cells that are capable of maintaining the circadian rhythm, beyond inflammation-driven circuits, opens the possibility of developing additional engineered cellular therapeutics against other conditions, such as age-related clock dampening or cancer (55, 56), through maintenance of the circadian clock and associated genes involved in tissue homeostasis (4, 9).
Using an engineered tissue that can protect itself against inflammation and preserving the circadian clock in response to inflammation can overcome the current limitations in OA therapies and articular cartilage repair. This work can be expanded to create clock-preserving cell therapies across many tissues by differentiating already developed iPSC lines into other tissues or using lentiviral approaches. For example, the NFκBr-IL1Ra lentiviral system has been used in both miPSCs and primary porcine chondrocytes for inflammation-activated and mechanoresponsive drug delivery (44, 45). Future work in comparing rhythmic transcriptomes and understanding the beneficial effects of rhythm preservation will additionally help further insights into the physiology of the cartilage clock. This approach opens an innovative frontier for creating cell-based therapies to maintain the circadian clock. Together, this framework for developing clock-preserving cell systems provides a new self-regulating approach to establish therapeutics and enhance tissue repair.
MATERIALS AND METHODS
Cell culture and differentiation
Murine-induced pluripotent stem cells
miPSCs, derived from tail fibroblasts from adult C57BL/6 mice and validated for pluripotency as described in (57, 58), were cultured in Dulbecco’s modified Eagle’s medium high glucose (DMEM-HG; Gibco), 20% lot selected fetal bovine serum (FBS; Atlanta Biologicals), 100 nM minimum essential medium nonessential amino acids (NEAA; Gibco), 55 μM 2-mercaptoethanol (2-me; Gibco), gentamicin (24 ng/ml; Gibco), and mouse leukemia inhibitory factor (LIF; 1000 U/ml; Millipore), and maintained on mitomycin C–treated mouse embryonic fibroblasts (Millipore).
miPSCs were differentiated toward a mesenchymal state using a high-density micromass culture (58). Differentiation medium contained DMEM-HG; 1% culture medium supplement containing recombinant human insulin, human transferrin, and sodium selenite (ITS+, Corning); 100 nM NEAA; 55 μM 2-me; gentamicin (24 ng/ml); l-ascorbic acid (50 μg/ml); and l-proline (40 μg/ml). On days 3 to 5, this medium was supplemented with 100 nM dex and bone morphogenetic protein 4 (BMP-4; 50 ng/ml; R&D Systems). After 15 days of culture, the micromasses were dissociated with pronase (MilliporeSigma) and collagenase type II (Worthington Biochemical), and the PDiPSCs were plated on gelatin-coated dishes in expansion medium containing DMEM-HG, 10% lot-selected FBS, 1% ITS+, 100 nM NEAA, 55 μM 2-me, 1% penicillin/streptomycin (P/S; Gibco), l-ascorbic acid (50 μg/ml), l-proline (40 μg/ml), and basic fibroblast growth factor (bFGF; 4 ng/ml; R&D Systems). To create a 3D PDiPSC system, passage 2 PDiPSCs were cast in agarose (4% molten type VII agarose; Sigma-Aldrich) at a final density of 100 million cells/ml in 1% agarose (58).
To create tissue-engineered cartilage, passage 2 PDiPSCs were pelleted by centrifugation of 250,000 cells (58). Pellets were cultured for 14 to 21 days in chondrogenic medium consisting of DMEM-HG, 1% ITS+, 100 nM NEAA, 55 μM 2-me, 1% P/S, l-ascorbic acid (50 μg/ml), l-proline (40 μg/ml), 100 nM dex, and transforming growth factor–β3 (TGF-β3; 10 ng/ml; R&D Systems).
Porcine primary chondrocytes
Full-thickness porcine articular chondrocytes were enzymatically isolated using collagenase II (Worthington Biochemical) from the femurs of pigs obtained from a local abattoir (~30 kg, 12 to 16 weeks old) postmortem in accordance with an exemption protocol by the Institutional Animal Care and Use Committee. Filtered cells were mixed 1:1 with 4% molten type VII agarose (Sigma-Aldrich), and the cell-agarose mixture was injected into a gel apparatus and allowed to set at room temperature. Chondrocyte-laden disks were punched out yielding engineered cartilage at a final concentration of 2% agarose and 15 to 20 million cells/ml. All constructs were cultured in chondrogenic medium for 2 to 21 days containing DMEM-HG, 10% FBS (Atlas Biologicals), 100 nM NEAA, 15 mM Hepes (Gibco), l-ascorbic acid (50 μg/ml), l-proline (40 μg/ml), and 1% P/S (59).
Circadian reporter design
Two luciferase reporter lentiviral vectors, one driven by the murine Bmal1 promoter (Addgene no. 182761) and one driven by the murine Period 2 (Per2) promoter (Addgene no. 182762), were used (54). These cassettes result in luciferase expression when the circadian promoters Bmal1 (B1L) and Per2 (P2L) are activated (20). These plasmids may be obtained by contacting the corresponding author. Bioluminescent reporters of core clock gene transcription are commonly used in the field of circadian biology to measure phase, period, and amplitude of rhythms in clock gene expression in real time.
Development of inflammation-resistant and self-regulating iPSCs
Three different cellular engineering approaches were used to test the ability to maintain the circadian clock in response to inflammatory cytokine IL-1α: A KO miPSC line lacking the IL1R1 (IL1R1 KO) (42), a CRISPR-Cas9–edited miPSC line that incorporates IL-1Ra downstream of the Ccl2 promoter (Ccl2-IL1Ra) (43), and cells transduced with a lentiviral gene therapy that includes a synthetic promoter activated by the NF-κB pathway that drives IL-1Ra production (NFκBr-IL1Ra) (44). IL1R1 KO–, Ccl2-IL1Ra–, and NFκBr-IL1Ra–transduced PDiPSCs were all transduced with P2L lentivirus and pelleted to grow into tissue-engineered cartilage for experimentation. These cell lines may be obtained by contacting the corresponding author.
IL1R1 KO miPSC line
The IL1R1 KO line was created using CRISPR-Cas9 technology to create a homozygous deletion of the IL1R1. Targeting sequences flanking exon 2 of Il1r1 and corresponding to 5′-gcttctgtgttgaagactca-3′ and 5′-gtagctgtgggcccacaacc-3′ were selected to generate the deletion of the IL1r1 signal peptide sequences. To produce single chimeric guide RNA (gRNA) expression vectors, complementary oligonucleotides containing each of the target sequences were cloned into an expression vector (Addgene plasmid no. 47108) (60). A plasmid encoding human codon optimized Streptococcus pyogenes Cas9 (hCas9) was also obtained from Addgene (plasmid no. 41815) (61). iPSCs were transfected using Lipofectamine 2000 (Life Technologies) following the manufacturer’s instructions to cotransfect gRNA plasmids and hCas9 plasmid. Posttransfection cells were subject to single-cell deposition and were screened for the appropriate deletion via genomic polymerase chain reaction (PCR). These cells do not respond to the presence of IL-1α and have been previously shown to be effective in mitigating inflammation-induced degradation of tissue-engineered cartilage (42).
Ccl2-IL1Ra miPSC line
The Ccl2-IL1Ra line was created using CRISPR-Cas9 technology to insert IL-1Ra downstream of the Ccl2 promoter, creating a heterozygous cell line. The hCas9 plasmid used in this cell line is the same as the plasmid used to create the IL1R1 KO line (Addgene no. 41815) (61). To target hCas9 to the Ccl2 locus, we used a gRNA targeting the start codon of the Ccl2 coding sequence using the complimentary oligonucleotides sgMcp1-4_S: 5′-caccGCTCTTCCTCCACCACCATGC-3′ and sgMcp1-4_AS: 5′-aaacGCATGGTGGTGGAGGAAGAGC-3′, where lowercase bases were used to clone into Bbs I–generated overhands in the expression vector (Addgene plasmid no. 47108) (60). Using the same protocol as for the IL1R1 KO line, iPSCs were transfected using Lipofectamine 2000 (Life Technologies), and cells were subjected to single-cell deposition. Clones were screened via genomic PCR. Ccl2 is a common gene up-regulated with inflammation in cartilage, and therefore, the Ccl2-IL1Ra cells effectively sense inflammation and produce an anti-inflammatory therapeutic in an autoregulated manner (43).
NFκBr-IL1Ra lentiviral vector
To create the NFκBr-IL1Ra lentiviral vector, a synthetic NF-κB–inducible promoter was designed to incorporate multiple NF-κB response elements as previously described (Addgene no. 183065) (44). A synthetic promoter was developed containing five consensus sequences approximating the NF-κB canonical recognition motif based on genes up-regulated through inflammatory challenge: InfB1, Il6, Mcp1, Adamts5, and Cxcl10. The specific sequence cloned was 5′-CGGGAATTCCGCTAGCACTAGTGGGACTTTCCCACTAGTGGGAAATTAGCCCGGGACTTTCCCGTCTCCTCGAGGGGACTTCCCA-3′. A TATA box derived from the minimal cytomegalovirus promoter was cloned between the synthetic promoter and downstream target gene, murine Il1rn, and an NF-κB–negative regulatory element (NRE-5′-AATTCCTCTGA-3′) was cloned upstream of the promoter to reduce background signal (44, 62, 63). This lentiviral vector was developed in our lab and has been shown to protect tissue-engineered cartilage from inflammation-mediated degradation and to be mechanoresponsive and used in porcine tissue–engineered cartilage (44, 45).
Lentivirus production and cell transduction
Human embryonic kidney 293T cells were cotransfected with second-generation packaging plasmid psPAX2 (no. 12260, Addgene), the envelope plasmid pMD2.G (no. 12259, Addgene), and the expression transfer vector (B1L, P2L, or NFκBr-IL1Ra plasmids) by calcium phosphate precipitation to make vesicular stomatitis virus glycoprotein pseudotyped lentivirus (64). The lentivirus was harvested at 24 and 48 hours after transfection and stored at −80°C until use. The functional titer of the virus was determined with quantitative real-time PCR to determine the number of lentiviral DNA copies integrated into the genome of transduced HeLa cells (64). For all cell transductions, virus was thawed on ice and diluted in medium to obtain the desired number of viral particles to achieve a multiplicity of infection of 3 (44). Polybrene was added to a concentration of 4 μg/ml to aid in transduction. The medium of the cells was aspirated and replaced with virus-containing medium, and cells were incubated for 24 hours before aspirating the viral medium.
Circadian clock characterization through bioluminescence recordings and imaging
miPSCs and PDiPSCs were transduced with either B1L or P2L virus. PDiPSCs were either cultured in monolayer, cast in agarose, or pelleted. miPSCs, PDiPSCs, PDiPSCs freshly cast in agarose (PDiPSC agarose), and mature pellets (tissue-engineered cartilage pellets) were plated in 35-mm petri dishes with 1 ml of recording medium containing d-luciferin (GoldBio), sealed with vacuum grease, and placed in a light-tight 36° incubator containing photomultiplier tubes (PMTs) (Hamamatsu Photonics). Each dish was placed under one PMT, and the bioluminescence was recorded as photons per 180 s. Bioluminescence data were detrended with a 24-hour moving average and analyzed in Chronostar 1.0 (65). Recoding medium contained DMEM powder (Sigma-Aldrich), B27 supplement (Invitrogen), P/S, l-glutamine (Invitrogen), Hepes (Sigma-Aldrich), and d-glucose (Invitrogen). Porcine chondrocytes were also transduced with P2L, cast in agarose, cultured, and then subjected to bioluminescence recordings in the dark incubator. As a control, the femoral head from 10-week-old PER2::LUC reporter mice (founders provided by J. Takahashi, University of Texas Southwestern) (5) was obtained, and the bioluminescent output from the cartilage explants was also recorded. In addition, dishes with pellets in recording media were recorded for 72 hours using an Andor iKon-M electron-multiplying charge-coupled device (Oxford Instruments) at a magnification of ×20 with 1-hour exposures and 2 × 2 binning. During multiday recordings, samples were maintained at 36° in darkness. Image sequences were concatenated into videos using ImageJ.
Inflammatory challenge and bioluminescence recordings
After 14 days of chondrogenic culture, pellets transduced with P2L underwent inflammatory challenge with TNFα (20 ng/ml), IL-1α (1 ng/ml), IL-1β (1 ng/ml), or a PBS control. Pellets were placed in recording medium, and bioluminescence was recorded for 72 hours. After 72 hours of recording, cytokine or PBS was added to the dish, and bioluminescence was recorded for an additional 72 hours. Pellets with a period of bioluminescence 18 to 32 hours, mean bioluminescence >30,000 photons, and precytokine cosine fit coefficient of determination (R2) value >0.8 were included in analysis. This led to the inclusion of 59 of the 60 pellets recorded for cytokine experiments. Damping is the normally occurring decrease in amplitude over time in the absence of entraining cues. The change in amplitude due to treatment was quantified by measuring the absolute difference in peak-trough amplitude 24 hours before and 48 hours following treatment. Additional pellets were treated with cytokines or PBS and stored at −80°C for quantitative reverse transcription PCR (qRT-PCR), −20°C for biochemical analysis, or fixed in 10% neutral buffered formalin (NBF). As a comparison, Per2::Luc mouse femoral head articular cartilage was dissected from 4- to 6-week-old mice. The cartilage explants were cultured in DMEM/F12 (without phenol red and FBS, with l-glutamine and NEAA). Explants were synchronized by 1 hour of treatment with 100 nM dex, and their bioluminescence was recorded in Lumicycle as described previously (18). Thirty-six hours after synchronization, explants underwent inflammatory challenge with TNFα (20 ng/ml), IL-1α (1 ng/ml), IL-1β (1 ng/ml), or a PBS control. After 48 hours of exposure to cytokines, 100 nM dex was added to the medium, and bioluminescence was recorded for another 48 hours. Damping of the circadian clock was assessed by measuring of the amplitude of peak before and after cytokine treatment, before the addition of dex. Explant bioluminescence was normalized by setting the minimum value of the trace to 0 and dividing all values by the maximum value of the trace, making the maximum value equal to 100%. IL1R1 KO, Ccl2-IL1Ra, NFκBr-IL1Ra, and P2L control pellets were all subjected to inflammatory challenge with IL-1α (1 ng/ml) or a PBS control. Bioluminescence was recorded 72 hours before addition of cytokine and for 72 hours after addition of cytokine. Additional pellets were treated with cytokines or PBS, media were collected, and pellets were stored at −80°C for qRT-PCR, −20°C for biochemical analysis, or fixed in 10% NBF.
Histological and biochemical analysis of pellet cultures
After 72 hours of inflammatory challenge, pellets were washed with PBS and fixed in 10% NBF for 24 hours, paraffin embedded, and sectioned at 8-μm thickness. Slides were stained for safranin O/hematoxylin/fast green (66).
For biochemical analysis, pellets were digested overnight in papain (125 μg/ml) at 65°C. DNA content was measured with PicoGreen assay (Thermo Fisher Scientific), and total sGAG content was measured using a 1,9-dimethylmethylene blue assay at 525-nm wavelength (67).
Gene expression with qRT-PCR
Pellets were homogenized using a miniature beat beater, lysed in buffer (RL), and RNA was isolated following the manufacturer’s protocol (total RNA purification, Norgen Biotek). RT was performed using SuperScript (VILO) complementary DNA master mix (Invitrogen). qRT-PCR was performed using Fast SYBR Green Master Mix (Applied Biosystems). Primer pairs were synthesized by Integrated DNA Technologies Inc.: Ccl2 (forward, 5′-GGCTCAGCCAGATGCAGTTAA-3′; reverse, 5′-CCTACTCATTGGGATCATCTTGCT-3′), Il6 (forward, 5′-GAGGATACCACTCCCAACAGACC-3′; reverse, 5′-AAGTGCATCATCGTTGTTCATACA-3′), and r18s (forward, 5′-CGGCTACCACATCCAAGGAA-3′; reverse, 5′-GGGCCTCGAAAGAGTCCTGT-3′). Data are reported as fold changes and were calculated using the ΔΔCT method and are shown relative to the P2L PBS control group, and ribosomal 18S is used as the reference gene.
Enzyme-linked immunosorbent assays
After 72 hours of inflammatory challenge, culture media were collected from pellets and stored at −20°C. IL-1Ra concentration was measured with DuoSet enzyme-linked immunosorbent assay (ELISA) specific to mouse IL-1Ra/IL-1F3 (R&D Systems).
Statistical analysis
Statistical analysis was performed using Prism. A one-way analysis of variance (ANOVA) with Dunnett’s post hoc test was used to analyze all biochemistry data for P2L pellets given TNFα, IL-1α, IL-1β, or PBS with PBS as the control (α = 0.05). A one-way ANOVA was used to analyze change in amplitude for pellets given TNF-α, IL-1α, IL-1β, or PBS with PBS as the control (α = 0.05). In addition, a one-way ANOVA with Dunnett’s post hoc test was used to analyze biochemistry data for IL1R1 KO, Ccl2-IL1Ra, and NFκBr-IL1Ra pellet experiments within each group with 0 ng/ml dose as the control (α = 0.05). A two-way ANOVA with Tukey’s post hoc test was used to analyze all qRT-PCR data and the ELISA data for IL1R1 KO, Ccl2-IL1Ra, and NFκBr-IL1Ra pellet experiments (α = 0.05). Statistical analysis for cartilage explants and their response to inflammatory cytokines was performed using a one-way ANOVA comparing the values of the peak after control or cytokine treatment.
Acknowledgments
Funding: This work was supported by the Shriners Hospitals for Children, the National Institutes of Health (AR072870, AG46927, AG15768, AR072999, AR073752, and AR074992), the Philip and Sima Needleman Fellowship from the Washington University Center of Regenerative Medicine (to L.P.), a NRSA fellowship (F31CA250161 to A.R.D.), a Versus Arthritis Senior Research Fellowship Award (20875 to Q.-J.M.), and a Medical Research Council, UK (MRC) grant (MR/K019392/1 to Q.J.-M.).
Author contributions: L.P. and F.G. conceived the project. L.P., A.R.D., E.D.H., Q.-J.M., and F.G. designed the experiments. L.P. and A.R.D. conducted the circadian development, tissue engineered cartilage inflammatory challenge, and clock preservation experiments. M.D. conducted the cartilage explant inflammatory challenge experiments. L.P., A.R.D., and F.G. wrote the manuscript. All authors read, edited, and approved the final manuscript.
Competing interests: F.G. is an employee and shareholder in Cytex Therapeutics Inc. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 and S2
REFERENCES AND NOTES
- 1.Hastings M. H., Reddy A. B., Maywood E. S., A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 4, 649–661 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L., The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat. Rev. Genet. 9, 764–775 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roenneberg T., Merrow M., Circadian clocks—The fall and rise of physiology. Nat. Rev. Mol. Cell Biol. 6, 965–971 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Dudek M. M., Meng Q.-J., Running on time: The role of circadian clocks in the musculoskeletal system. Biochem. 463, 1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S., PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patke A., Young M. W., Axelrod S., Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 21, 67–84 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Shi S. Q., Ansari T. S., McGuinness O. P., Wasserman D. H., Johnson C. H., Circadian disruption leads to insulin resistance and obesity. Curr. Biol. 23, 372–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chellappa S. L., Vujovic N., Williams J. S., Scheer F., Impact of circadian disruption on cardiovascular function and disease. Trends Endocrinol. Metab. 30, 767–779 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang N., Meng Q. J., Circadian clocks in articular cartilage and bone: A compass in the sea of matrices. J. Biol. Rhythms 31, 415–427 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Mansour J. M., Biomechanics of cartilage. Kinesiol. Mech. Pathomechanics Hum. Mov. 66–79 (2009). [Google Scholar]
- 11.Lin Z., Willers C., Xu J., Zheng M.-H., The chondrocyte: Biology and clinical application. Tissue Eng. 12, 1971–1984 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Sophia Fox A. J., Bedi A., Rodeo S. A., The basic science of articular cartilage: Structure, composition, and function. Sports Health 1, 461–468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martel-Pelletier J., Boileau C., Pelletier J. P., Roughley P. J., Cartilage in normal and osteoarthritis conditions. Best Pract. Res. Clin. Rheumatol. 22, 351–384 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Goldring M. B., Goldring S. R., Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann. N. Y. Acad. Sci. 1192, 230–237 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Goldring M. B., Golding S. R., Osteoarthritis. J. Cell. Physiol. 213, 626–634 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Andersson M. L. E., Petersson I. F., Karlsson K. E., Jonsson E. N., Månsson B., Heinegård D., Saxne T., Diurnal variation in serum levels of cartilage oligomeric matrix protein in patients with knee osteoarthritis or rheumatoid arthritis. Ann. Rheum. Dis. 65, 1490–1494 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudek M., Angelucci C., Pathiranage D., Wang P., Mallikarjun V., Lawless C., Swift J., Kadler K. E., Boot-Handford R. P., Hoyland J. A., Lamande S. R., Bateman J. F., Meng Q. J., Circadian time series proteomics reveals daily dynamics in cartilage physiology. Osteoarthr. Cartil. 29, 739–749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudek M., Gossan N., Yang N., Im H. J., Ruckshanthi J. P., Yoshitane H., Li X., Jin D., Wang P., Boudiffa M., Bellantuono I., Fukada Y., Boot-Handford R. P., Meng Q. J., The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. J. Clin. Invest. 126, 365–376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong S. Y., Stabler T. V., Criscione L. G., Elliott A. L., Jordan J. M., Kraus V. B., Diurnal variation of serum and urine biomarkers in patients with radiographic knee osteoarthritis. Arthritis Rheum. 54, 2496–2504 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Gossan N., Zeef L., Hensman J., Hughes A., Bateman J. F., Rowley L., Little C. B., Piggins H. D., Rattray M., B.-Handford R. P., Meng Q. J., The circadian clock in murine chondrocytes regulates genes controlling key aspects of cartilage homeostasis. Arthritis Rheum. 65, 2334–2345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dernie F., Adeyoju D., A matter of time: Circadian clocks in osteoarthritis and the potential of chronotherapy. Exp. Gerontol. 143, 111163 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Chen G., Zhao H., Ma S., Chen L., Wu G., Zhu Y., Zhu J., Ma C., Zhao H., Circadian rhythm protein bmal1 modulates cartilage gene expression in temporomandibular joint osteoarthritis via the MAPK/ERK pathway. Front. Pharmacol. 11, 527744 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bekki H., Duffy T., Okubo N., Olmer M., A.-Garcia O., Lamia K., Kay S., Lotz M., Suppression of circadian clock protein cryptochrome 2 promotes osteoarthritis. Osteoarthr. Cartil. 28, 966–976 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gossan N., Boot-Handford R., Meng Q. J., Ageing and osteoarthritis: A circadian rhythm connection. Biogerontology 16, 209–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigoglou S., Papavassiliou A. G., The NF-κB signalling pathway in osteoarthritis. Int. J. Biochem. Cell Biol. 45, 2580–2584 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Wehling P., Reinecke J., Baltzer A. W. A., Granrath M., Schulitz K. P., Schultz C., Krauspe R., Whiteside T. W., Elder E., Ghivizzani S. C., Robbins P. D., Evans C. H., Clinical responses to gene therapy in joints of two subjects with rheuamtoid arthritis. Hum. Gene Ther. 20, 97–101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapoor M., M.-Pelletier J., Lajeunesse D., Pelletier J. P., Fahmi H., Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 7, 33–42 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Goldring M. B., Otero M., Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 23, 471–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastiaansen-Jenniskens Y., Saris D., Creemers L. B., Pro- and anti-inflammatory cytokine profiles in osteoarthritis. Cartilage 2, 81–97 (2017). [Google Scholar]
- 30.Guo B., Yang N., Borysiewicz E., Dudek M., Williams J. L., Li J., Maywood E. S., Adamson A., Hastings M. H., Bateman J. F., White M. R. H., Boot-Handford R. P., Meng Q. J., Catabolic cytokines disrupt the circadian clock and the expression of clock-controlled genes in cartilage via an NFкB-dependent pathway. Osteoarthr. Cartil. 23, 1981–1988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choy E. H., Kavanaugh A. F., Jones S. A., The problem of choice: Current biologic agents and future prospects in RA. Nat. Rev. Rheumatol. 9, 154–163 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Furman B. D., Mangiapani D. S., Zeitler E., Bailey K. N., Horne P. H., Huebner J. L., Kraus V. B., Guilak F., Olson S. A., Targeting pro-inflammatory cytokines following joint injury: Acute intra-articular inhibition of interleukin-1 following knee injury prevents post-traumatic arthritis. Arthritis Res. Ther. 16, R134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chevalier X., Goupille P., Beaulieu A. D., Burch F. X., Bensen W. G., Conrozier T., Loeuille D., Kivitz A. J., Silver D., Appleton B. E., Intraarticular injection of anakinra in osteoarthritis of the knee: A multicenter, randomized, double-blind, placebo-controlled study. Arthritis Rheum. 61, 344–352 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Ramos-Casals M., Brito-Zeron P., Soto M. J., Cuadrado M. J., Khamashta M. A., Autoimmune diseases induced by TNF-targeted therapies. Best Pract. Res. Clin. Rheumatol. 22, 847–861 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Gopinath S. D., Rando T. A., Stem cell review series: Aging of the skeletal muscle stem cell niche. Aging Cell 7, 590–598 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Kimmerling K. A., Furman B. D., Mangiapani D. S., Moverman M. A., Sinclair S. M., Huebner J. L., Chilkoti A., Kraus V. B., Setton L. A., Guilak F., Olson S. A., Sustained intra-articular delivery of IL-1Ra from a thermally-responsive elastin-like polypeptide as a therapy for post-traumatic arthritis. Eur. Cell. Mater. 29, 124–140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mozzetta C., Minetti G., Puri P. L., Regenerative pharmacology in the treatment of genetic diseases: The paradigm of muscular dystrophy. Int. J. Biochem. Cell Biol. 41, 701–710 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans C. H., Ghivizzani S. C., Robbins P. D., Gene delivery to joints by intra-articular injection. Hum. Gene Ther. 29, 2–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews G. L., Hunter D. J., Emerging drugs for osteoarthritis. Expert Opin. Emerg. Drugs 16, 479–491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldring M. B., Berenbaum F., Emerging targets in osteoarthritis therapy. Curr. Opin. Pharmacol. 22, 51–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guilak F., Pferdehirt L., Ross A. K., Choi Y.-R., Collins K. H., Nims R. J., Katz D. B., Klimak M., Tabbaa S., Pham C. T. N., Designer stem cells: Genome engineering and the next generation of cell-based therapies. J. Orthop. Res. 37, 1287–1293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunger J. M., Zutshi A., Willard V. P., Gersbach C. A., Guilak F., CRISPR/cas9 editing of murine induced pluripotent stem cells for engineering inflammation-resistant tissues. Arthritis Rheumatol. 69, 1111–1121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunger J. M., Zutshi A., Willard V. P., Gersbach C. A., Guilak F., Genome engineering of stem cells for autonomously regulated, closed-loop delivery of biologic drugs. Stem Cell Reports 8, 1202–1213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pferdehirt L., Ross A. K., Brunger J. M., Guilak F., A synthetic gene circuit for self-regulating delivery of biologic drugs in engineered tissues. Tissue Eng. Part A 25, 809–820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nims R. J., Pferdehirt L., Ho N. B., Savadipour A., Lorentz J., Sohi S., Kassab J., Ross A. K., O’Conor C. J., Liedtke W. B., Zhang B., McNulty A. L., Guilak F., A synthetic mechanogenetic gene circuit for autonomous drug delivery in engineered tissues. Sci. Adv. 7, eabd9858 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yagita K., Horie K., Koinuma S., Nakamura W., Yamanaka I., Urasaki A., Shigeyoshi Y., Kawakami K., Shimada S., Takeda J., Uchiyama Y., Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc. Natl. Acad. Sci. U.S.A. 107, 3846–3851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magnone M. C., Langmesser S., Bezdek A. C., Tallone T., Rusconi S., Albrecht U., The mammalian circadian clock gene per2 modulates cell death in response to oxidative stress. Front. Neurol. 5, 289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu R., Dang F., Li P., Wang P., Xu Q., Liu Z., Li Y., Wu Y., Chen Y., Liu Y., The circadian protein period2 suppresses mTORC1 activity via recruiting Tsc1 to mTORC1 complex. Cell Metab. 29, 653–667.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 49.McNulty A. L., Rothfusz N. E., Leddy H. A., Guilak F., Synovial fluid concentrations and relative potency of interleukin-1 alpha and beta in cartilage and meniscus degradation. J. Orthop. Res. 31, 1039–1045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beaulé C., Granados-Fuentes D., Marpegan L., Herzog E. D., In vitrocircadian rhythms: Imaging and electrophysiology. Essays Biochem. 49, 103–117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abe M., Herzog E. D., Yamazaki S., Straume M., Tei H., Sakaki Y., Menaker M., Block G. D., Circadian rhythms in isolated brain regions. J. Neurosci. 22, 350–356 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamazaki S., Numano R., Abe M., Hida A., Takahashi R. I., Ueda M., Block G. D., Sakaki Y., Menaker M., Tei H., Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Williams J., Yang N., Wood A., Zindy E., Meng Q. J., Streuli C. H., Epithelial and stromal circadian clocks are inversely regulated by their mechano-matrix environment. J. Cell Sci. 131, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dudek M., Yang N., Ruckshanthi J. P. D., Williams J., Borysiewicz E., Wang P., Adamson A., Li J., Bateman J. F., White M. R., Boot-Handford R. P., Hoyland J. A., Meng Q. J., The intervertebral disc contains intrinsic circadian clocks that are regulated by age and cytokines and linked to degeneration. Ann. Rheum. Dis. 76, 576–584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altman B. J., Hsieh A. L., Sengupta A., Krishnanaiah S. Y., Stine Z. E., Walton Z. E., Gouw A. M., Venkataraman A., Li B., Goraksha-Hicks P., Diskin S. J., Bellovin D. I., Simon M. C., Rathmell J. C., Lazar M. A., Maris J. M., Felsher D. W., Hogenesch J. B., Weljie A. M., Dang C. V., MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab. 22, 1009–1019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hood S., Amir S., The aging clock: Circadian rhythms and later life. J. Clin. Invest. 127, 437–446 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carey B. W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M., Jaenisch R., Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. U.S.A. 106, 157–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diekman B. O., Christoforou N., Willard V. P., Sun H., Sanchez-Adams J., Leong K. W., Guilak F., Cartilage tissue engineering using differentiated and purified induced pluripotent stem cells. Proc. Natl. Acad. Sci. U.S.A. 109, 19172–19177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Conor C. J., Leddy H. A., Benefield H. C., Liedtke W. B., Guilak F., TRPV4-mediated mechanotransduction regulates the metabolic response of chondrocytes to dynamic loading. Pro. Natl. Acad. Sci. U.S.A. 111, 1316–1321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez-Pinera P., Ousterout D. G., Brunger J. M., Farin A. M., Glass K. A., Guilak F., Crawford G. E., Hartemink A. J., Gersbach C. A., Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat. Methods 10, 239–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., Church G. M., RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hou C. H., Huang J., Qian R. L., Identification of α NF-κB site in the negative regulatory element(ɛ-NRAII) of human ɛ-globin gene and its binding protein NF-κB p50 in the nuclei of K562 cells. Cell Res. 12, 79–82 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Nourbakhsh M., Hoffmann K., Hauser H., Interferon-beta promoters contain a DNA element that acts as a position-independent silencer on the NF-kappa B site. EMBO J. 12, 451–459 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salmon P., Trono D., Production and titration of lentiviral vectors. Curr. Protoc. Hum. Genet. 12, Unit 12.10 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Maier B., Lorenzen S., Finger A. M., Herzel H., Kramer A., Searching novel clock genes using RNAi-based screening. Methods Mol. Biol. 2130, 103–114 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Estes B. T., Diekman B. O., Gimble J. M., Guilak F., Isolation of adipose-derived stem cells and their induction to a chondrogenic phenotype. Nat. Protoc. 5, 1294–1311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Farndale R. W., Buttle D. J., Barrett A. J., Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim. Biophys. Acta 883, 173–177 (1986). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 and S2