Summary
One significant drawback of existing bioprinted tissues is their lack of shelf-availability caused by complications in both fabrication and storage. Here, we report a cryobioprinting strategy for simultaneously fabricating and storing cell-laden volumetric tissue constructs through seamlessly combining extrusion bioprinting and cryopreservation. The cryobioprinting performance was investigated by designing, fabricating, and storing cell-laden constructs made of our optimized cryoprotective gelatin-based bioinks using a freezing plate with precisely controllable temperature. The in situ freezing process further promoted the printability of cell-laden hydrogel bioinks to achieve freeform structures otherwise inconvenient with direct extrusion bioprinting. The effects of bioink composition on printability and cell viability were evaluated. The functionality of the method was finally investigated using cell differentiation and chick ex ovo assays. The results confirmed the feasibility and efficacy of cryobioprinting as a single-step method for concurrent tissue biofabrication and storage.
Keywords: biofabrication, 3D bioprinting, cryopreservation, cryobioprinting, tissue engineering, regenerative medicine
Graphical Abstract
eTOC blurb
Cryobioprinting potentially enpowers accelerating the transition of biofabricated artificial tissue constructs from research labs to clinics. The method helps bioengineers to fabricate cell-laden tissue constructs and store them for extended periods of time. This feature adds more flexibility in shelf-availability of biofabricated tissue constructs. The test samples were bioprinted using gelatin methacryloyl-based hydrogels, cryoprotective agents, and various human cells. Our findings showed that the cells remained viable and functional after 3 months of cryobioprinting and cryopreservation in liquid nitrogen.
Introduction
Three-dimensional (3D) bioprinting has paved a new avenue for fabricating intricate cell-laden tissue constructs1,2. So far, bioprinting has been adopted in myriad applications such as tissue engineering3,4, tissue model engineering5, and organoid production6, among others. There are, however, challenges regarding the fabrication and storage of shelf-ready 3D-bioprinted tissue constructs. Due to the intrinsic complexities involved in most bioprinting processes, including the broadly adopted extrusion bioprinting, using this method as an on-site fabrication technique can be cumbersome or sometimes impractical under a number of scenarios. Furthermore, the lack of a functional approach for long-term storage of cell-laden tissue constructs7 precludes the shelf-availability of pre-made bioprinted products.
Combining the fabrication and storage methods (i.e., bioprinting and cryopreservation) is a potential solution to address the obstacles mentioned above. As such, the bioprinted tissues can be fabricated in advance off-site, allowing direct storage in cryogenic conditions for later academic or clinical applications. The main challenge that should be addressed in this method is the ice crystal-formation and recrystallization, which may negatively affect cell viability in the fabricated products. The bioink components should be properly designed to efficiently preserve cells during bioprinting and storage/resuscitation periods. Various polymers, such as poly(vinyl alcohol) and poly(ethylene glycol), either in the form of bulk8 or self-assembled nanoparticles9, have shown promising capability in inhibiting ice-recrystallization. Cryoprotective agents (CPAs) are also believed to have a critical effect on eliminating or minimizing ice crystal-formation10. In the absence of CPAs, the ice crystals grow within the scaffold and may jeopardize cell viability by damaging the cell membranes over the freezing/thawing cycles11. Dimethylsulfoxide (DMSO) is a traditional CPA widely used in cryopreservation to minimize ice crystal-formation12–14. Besides, CPAs with larger molecular weights can protect the cells from osmotic damages caused by extracellular hypertonic and hypotonic conditions over the freezing and thawing periods15. Among these CPAs, various disaccharides such as sucrose16, trehalose17, lactose18, and maltose19 together with trisaccharides such as melezitose19 and raffinose20 have shown promising cryoprotective effects.
Here, we report an unconventional technology, termed cryobioprinting, that exploits 3D cell-laden bioprinting and cryopreservation methods to concomitantly fabricate and store bioprinted tissues for on-demand applications. By using this embodiment, the products are stored for a long(er)-term period and used after crosslinking, thawing, and resuscitating. The idea of cryogenic 3D printing of acellular inks has been recently proposed21–24. However, the incorporation of living cells within the bioink and bioprinting under cryogenic conditions, i.e., cryobioprinting as we define, remains very limited and requires many more considerations to ensure the efficacy of the method.
As schematically briefed in Fig. 1, a cell-laden hydrogel precursor (i.e., the bioink) is used for bioprinting tissue constructs on a customized freezing plate. The frozen bioprinted samples are immediately further cryopreserved for short- and long-term storage. When needed, the cryopreserved tissue constructs can be readily transported under cryogenic conditions, crosslinked, and revived on-site for subsequent usage. Gelatin methacryloyl (GelMA)25–27 and various CPAs are employed to design the optimum bioink. We investigate the effect of bioink composition (GelMA, DMSO, and one of the six types of saccharides) on the cryopreservation and resuscitation of various cells. The optimized bioink configuration is selected based on the cell viability results. A range of two-dimensional (2D) patterns and 3D structures are cryobioprinted with the optimized bioink to confirm the bioprinting performance. After resuscitating the samples, the functionality and stemness of the encapsulated human mesenchymal stem cells (hMSCs) are verified via osteogenic, adipogenic, and chondrogenic differentiations. Furthermore, the angiogenesis potential of the cryobioprinted constructs is assessed using the ex ovo chorioallantoic membrane (CAM) assay. Our results show that cryobioprinting provides a high-fidelity strategy to fabricate shelf-ready cell-laden constructs that can be broadly used in applications including but not limited to tissue engineering, regenerative medicine, tissue model engineering, and drug screening.
Fig. 1. Schematic illustration of cryobioprinting of tissue constructs for simultaneous cryopreservation.
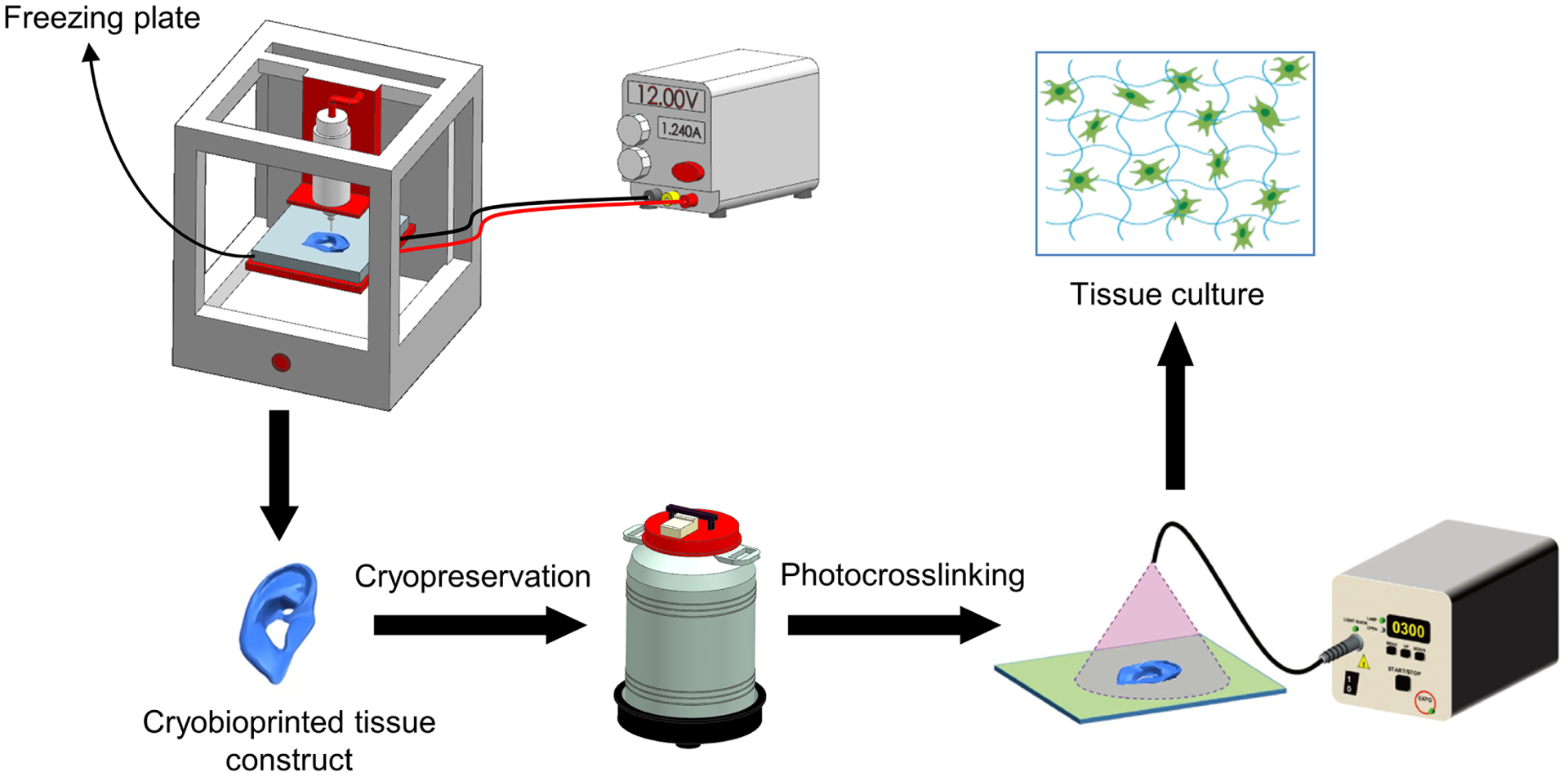
The cell-laden bioink is cryobioprinted on a freezing plate with tightly controlled temperature and kept in cryogenic conditions for long(er)-term preservation. The cryobioprinted structure can be then transferred, revived, and crosslinked immediately before subsequent culturing to achieve tissue-formation at the desired usage site. Figure created using BioRender.
Results
Freezing plate
A thermoelectric cooler, which is a solid-state heat pump and works based on the Peltier effect28,29, was designed and fabricated to maintain tightly controlled low temperatures on the substrate surface (fig. S1). This adjustable device transfers the heat from the printing stage, generating a precisely controllable temperature drop. The freezing plate temperature was adjusted via changing the input voltage or current, as shown in tables S1 and S2. The two tables also suggest that the effect of environmental temperature on the freezing plate’s temperature was negligible. Upon extrusion from the bioprinter nozzle and contact with the freezing plate, the bioink rapidly solidifies in situ and forms a stable structure, as shown in movie S1.
Printability and fidelity
Printability of potential cryoprotective bioinks with different DMSO concentrations was evaluated. Living cells were not used at this step, as the presence of the cells was assumed not to significantly affect the results of interest. The effect of parameters such as the pressure, the printhead moving speed, and the nozzle size was examined by cryobioprinting a grid structure using 5% (w/v) GelMA containing different DMSO concentrations. We used a 27G nozzle for cryobioprinting jobs since it generally yielded a better printing quality than a 30G nozzle (fig. S2). A lower printhead moving speed improved spatial resolution within the range of speed and pressure values investigated (quantitatively analyzed in fig. S3). As expected, increased pressure while keeping printhead speed constant at 10 mm s−1 caused a considerable enlargement in the filament diameter.
The cryobioprinted structures were either immediately crosslinked or cryopreserved in a liquid nitrogen tank for on-demand use. The ultraviolet (UV) exposure times needed for crosslinking 4-layer, 8×8-mm2 GelMA grid patterns with different DMSO concentrations were studied. The samples that were not exposed to UV fully dissolved in phosphate-buffered saline (PBS) after shaking overnight. As shown in fig. S4, 10 s of UV exposure resulted in a partially crosslinked sample, which did not maintain its integrity after shaking. Samples that underwent more prolonged UV exposure exhibited an acceptable fidelity, regardless of the DMSO concentration. The minimum essential UV exposure time, i.e., 20 s, was subsequently used for crosslinking the cell-laden grid structures without compromising cell viability. This minimal UV exposure is deemed to be safe for the living cells30. In addition, an identical crosslinking regime was used for all the samples, including the control, during the sample-preparation for biological tests. As a result, the effect of crosslinking method, even if minimal, was omitted from the analysis.
UV crosslinking was the first step after taking out the samples from the liquid nitrogen tank or other cryopreservation conditions. The samples were then rapidly thawed in pre-warmed PBS at 37 °C to avoid ice crystal-formation and simultaneously remove the residual CPAs. No noticeable damage, such as visible cracks, was observed after thawing. Representative cryobioprinted structures from simple 2D patterns to sophisticated 3D constructs were successfully created from GelMA- and gelatin-based bioinks (Fig. 2, A to R). Multi-material 2D and 3D constructs were subsequently fabricated from GelMA and gelatin, as shown in Fig. 2, M to R. Remarkably, we used 5% (w/v) GelMA or gelatin as the base bioink to create freeform constructs (Fig. 2, P to R), some of which could be complicated via conventional direct extrusion bioprinting methods.
Fig. 2. Sample cryobioprinted constructs and simulation results for fabricating a two-layer grid structure.
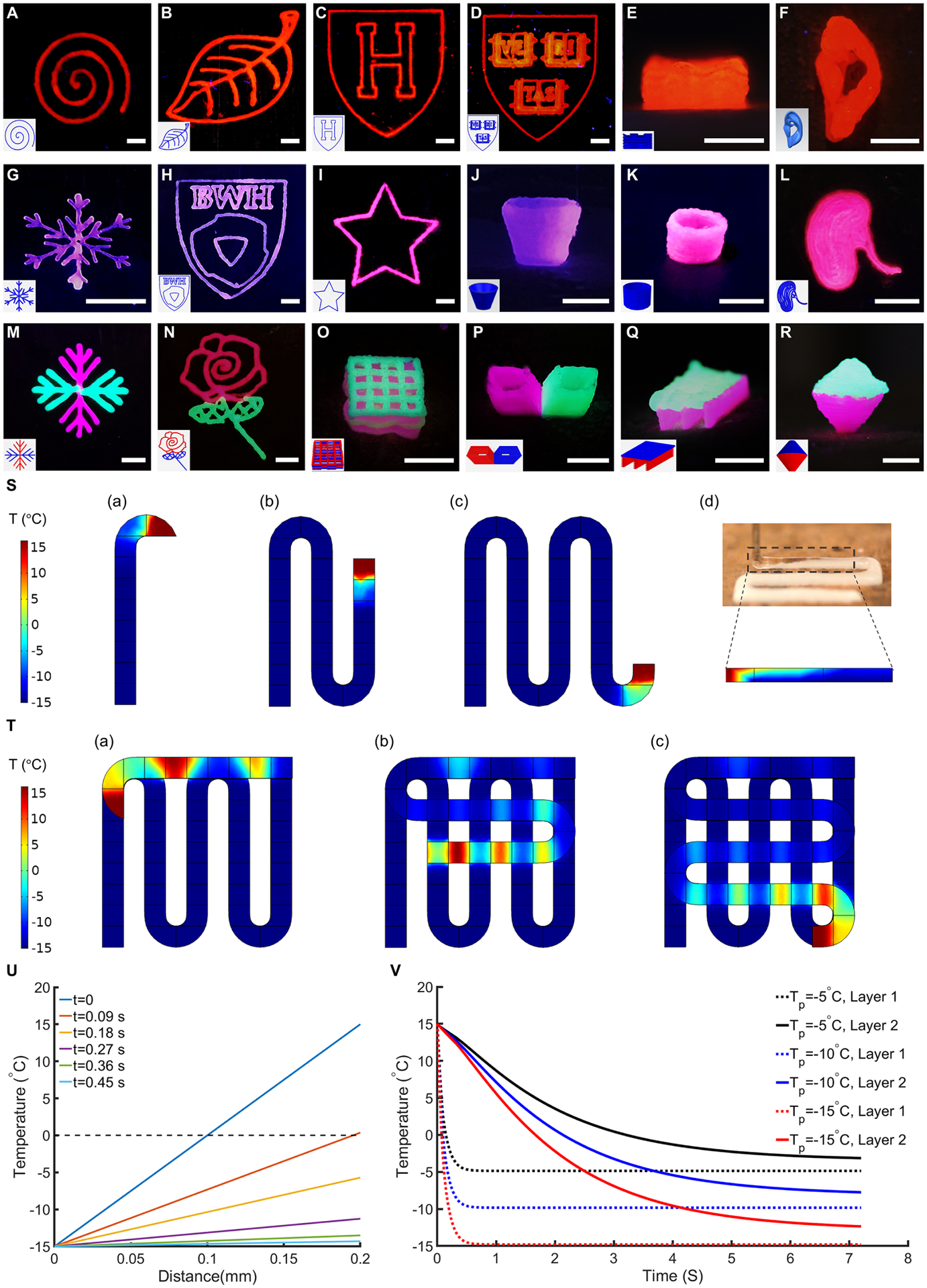
(A to R), Cryobioprinted 2D patterns, 3D structures, and freeform multi-material models made of (A to F) GelMA, (G to L) gelatin, and (M to R) GelMA+gelatin. Scale bars: 5 mm. (S), Temperature gradient in the first layer of a grid structure during cryobioprinting. The initial temperature of the bioink was set to 15 °C, and the freezing plate was simulated as a constant −15 °C surface. Panels (a) to (c) demonstrate the progress of the cryobioprinting, and panel (d) exhibits the freezing frontier lines in the experiment and the simulation. (T), Temperature gradient in the second layer of the grid structure. The regions that were in contact with the first layer froze faster. (U), Temperature changes through the thickness of the first layer during cryobioprinting. At 0.09 s after the bioink touched the freezing plate, the entire cross-section was frozen, and after 0.45 s, the bioink surface temperature reached approximately −14.5 °C. Since the dominant mechanism of heat-transfer through the thickness was conduction, the temperature change was linear. (V), Comparing the average temperature over the freezing period for the first and the second layers using different values of Tp.
Heat-transfer simulation
The layer-by-layer cryobioprinting of a grid structure was modeled in COMSOL Multiphysics® to understand the mechanism of heat-transfer during the process. Fig. 2S and movie S2 show the simulation of heat-transfer in the first layer while it is being cryobioprinted. Since the first layer directly contacts the freezing plate (−15 °C), the heat-transfer from the bioink to the freezing plate occurs instantaneously. Once the bioink contacts the freezing plate, the frozen region expands through the deposited material until the entire layer is frozen. Experimentally, the frontier of the frozen bioink perfectly matched that of the simulated model (Fig. 2S(d)), indicating a good agreement between the simulation and the experiment. The freezing regime in the second layer is slower since the entire layer is not touching the underlying frozen layer, as shown in Fig. 2T.
Temperature change through the first layer’s filament thickness was plotted for different time steps to analyze the cryobioprinting procedure quantitatively (Fig. 2U). At t=0, the first layer’s bottom surface is exposed to the freezing plate temperature, while the top of the layer remains at 15 °C. Since the heat conduction in the bioink obeys Fourier’s law, the temperature through the thickness changes linearly with the distance under a constant temperature gradient. At t=0.09 s, the whole bioink is frozen, and the top surface temperature reaches approximately 0 °C. This trend continues until the entire simulated section reaches an isothermal steady state at t=0.45 s. In Fig. 2V, the average surface temperatures of the layers are compared for three different freezing plate temperatures (Tp). For Tp = −15 °C, the top surface temperature of the second layer reaches 0 °C, after 1.7 s, which corresponds to the freezing point of the bioink. Increasing Tp to −10 °C and −5 °C results in a slower freezing regime in the second layer, while the rapid temperature change in the first layer is not remarkably affected. As expected, the time needed to completely freeze the second layer is longer than that of the first layer for all three Tp values.
Ice crystal-formation
One critical factor that affects cell viability during cryopreservation is the formation of ice crystals31. The spicular ice crystals can damage the cell membrane, resulting in lower cell viability. Movies S3 to S9 represent real-time recordings of the ice crystal-formation in GelMA hydrogel groups with different CPA concentrations, and fig. S5A shows the corresponding time-lapse images. The addition of CPAs noticeably affected the shape of ice crystals. In the control sample (without DMSO), the ice crystals were more spicular and formed faster than the other study groups. Increasing the DMSO concentration led to the formation of fine ice crystals; however, no remarkable improvement in ice-crystallization was observed when the saccharide concentration was increased, as the saccharide merely prevents osmotic damage to the cells. The fractal dimension (FD) theory, in conjunction with an image processing method, was employed to quantitatively compare the smoothness and the growth rate of the ice crystals. The FD value quantifies the geometry’s irregularities. In our analysis, ice crystals’ boundaries were extracted from the brightfield images and digitized, as depicted in fig. S5B. The FD value for the resulting quasi-circle geometry was then calculated using equation (1)32.
| (1) |
, where P and A stand for the perimeter and the area of the quasi-circle geometry, respectively. FD is an indicator of how the quasi-circle geometry deviates from a circle. When the geometry of interest is a perfect circle, the FD equals 1. As the irregularities and sharp edges appear in the geometry, the FD increases. For the freezing bioink, when ice crystals form, the geometry of the unfrozen region starts to diverge from a perfect circle, leading to an increased FD. The trends of changes in FD for freezing bioinks with different DMSO concentrations are shown in fig. S5C. In the control sample, i.e., DMSO-free, the highly spicular ice crystals formed rapidly, and therefore, the FD value was higher than those of the other groups. When 90% of the field of view was frozen, the FD values were recorded and compared (fig. S5D). It is evident that the addition of DMSO significantly decreased the FD value. In fig. S5E, the percentages of the frozen areas were plotted over time to compare the freezing rates. The control sample froze faster than the other samples, and as the DMSO concentration increased, the bioink started to freeze slower.
Splat assay
To further confirm the effect of CPAs on inhibiting ice-recrystallization, a modified splat assay was employed. Cryomicrographs of annealed ice wafers (fig. S6A) clearly demonstrated that ice crystals were formed during the freezing of GelMA. The results showed that the average size of the ice crystals significantly decreased when DMSO concentration was increased (fig. S6B). Smaller crystal size is an indication of enhanced ice recrystallization-inhibition8,33.
Cryoprotective bioink design based on cell viability assays
A series of cell viability experiments were conducted using different concentrations of DMSO and saccharides to find the optimized configuration of CPAs in GelMA. Since monosaccharides generally show less protective effects regardless of concentration and freezing trend34, we focused on using disaccharides (lactose, maltose, sucrose, trehalose) and trisaccharides (raffinose, melezitose) (fig. S7) along with DMSO as the CPAs. The cell-laden bioink was used to produce samples containing different concentrations of DMSO and saccharides at room temperature (RT). The cast samples were then directly transferred to −80 °C for 24 h, conventionally cryopreserved in liquid nitrogen for 72 h, crosslinked via UV exposure, and underwent live/dead assays. Fig. 3A shows a representative set of live/dead images for immortalized mouse myoblast cells (C2C12) cryopreserved in 5% (w/v) GelMA with different DMSO and melezitose concentrations. By using the fluorescence images, the cell viability ratios of C2C12 cells were determined (fig. S8). Regardless of the DMSO concentration, the saccharide-free groups generally showed a lower cell viability rate since the presence of saccharides avoids osmotic shock during the thawing process15. It is, therefore, concluded that the saccharide is an essential part of a cryoprotective bioink.
Fig. 3. Cell viability in cryopreserved GelMA constructs with different concentrations and types of CPAs using seven different cell types.
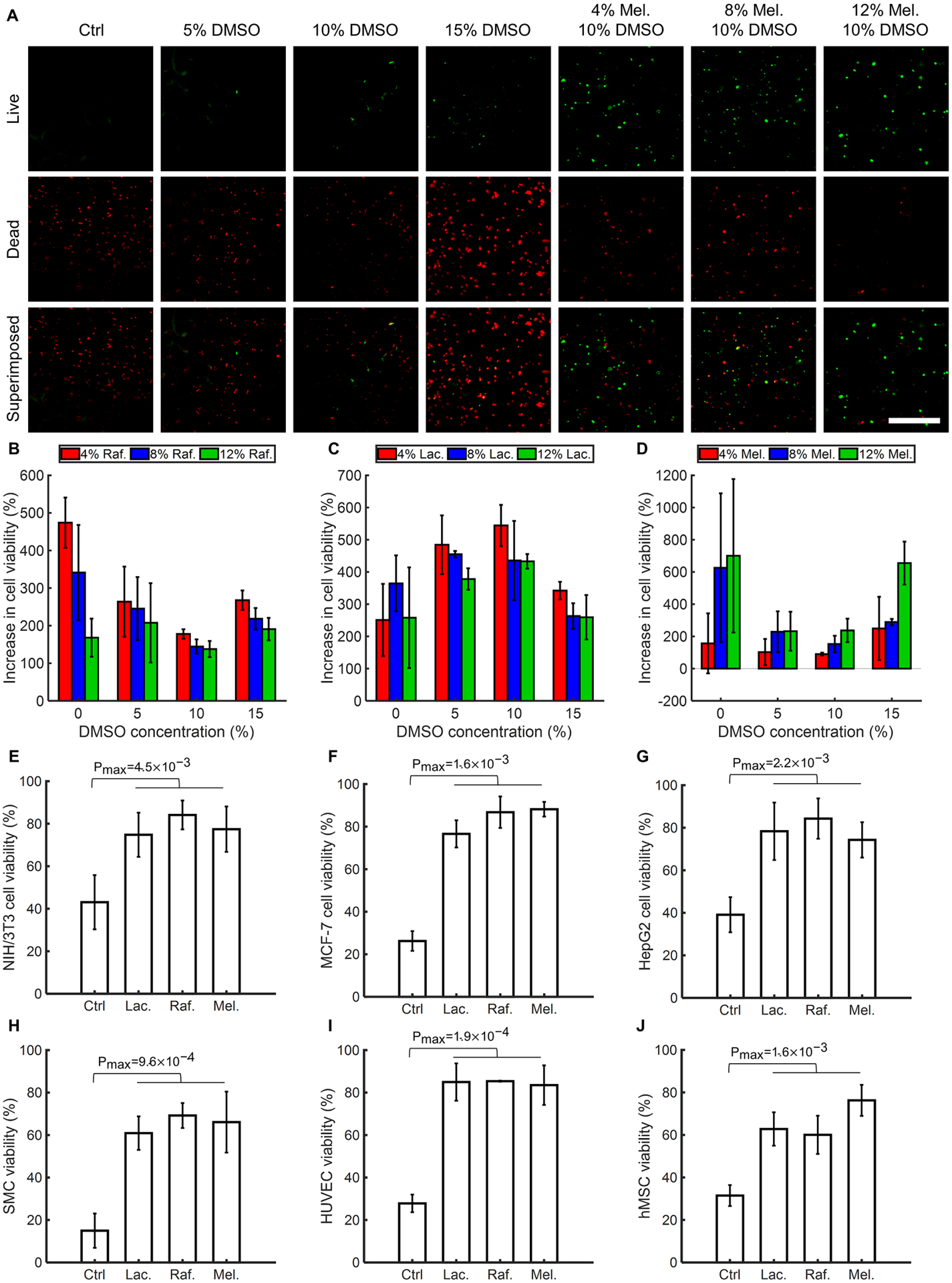
(A), Representative fluorescence images of the C2C12 cells encapsulated in bioinks, cryopreserved for 72 h, and resuscitated before staining/imaging. The cryopreserved bioinks were prepared using 5% (w/v) GelMA, different concentrations of DMSO, and different concentrations of saccharides. (B to D), Quantified increase in cell viability post-cryopreservation for 72 h due to supplementing the cryoprotective bioink with various saccharides: (B), Raffinose (Raf.). (C), Lactose (Lac.). (D), Melezitose (Mel.). (E to J), Significant increase in cell viability of various cell types when selected CPAs were employed. n = 3. (Ctrl: Control)
Cell viability of each group was normalized to the control group with the same DMSO concentration to choose the most functional saccharide and its optimum concentration (Fig. 3, B to D and fig. S9). The most effective saccharides that led to a higher cell viability increase were 12% (w/v) melezitose, 4% (w/v) raffinose, and 4% (w/v) lactose. The increase in cell viability of the groups with respect to the DMSO-free group with the same saccharide concentration was calculated to identify the most effective DMSO concentration. As shown in fig. S10, the optimum concentration of DMSO was 10% (v/v) for the selected sugar types. As a result, three combinations of CPAs were selected to be used with 5% (w/v) GelMA for the next set of experiments: 12% (w/v) melezitose+10% (v/v) DMSO, 4% (w/v) lactose+10% (v/v) DMSO, and 4% (w/v) raffinose+10% (v/v) DMSO. In the following sections, we refer to these selected groups using the names of their saccharides only, considering that they all contained 10% (v/v) DMSO and 5% (w/v) GelMA.
Using different cell types to assess the effectiveness of the selected CPAs
The efficacy of the CPAs varies with the cryopreserved cell type and cell size35. We studied the cell viability of six other cell types, including mouse embryonic fibroblast cell line (NIH/3T3), human liver cancer cell line (HepG2), human breast cancer cell line (MCF-7), primary human smooth muscle cells (SMCs), primary human umbilical vein endothelial cells (HUVECs), and primary hMSCs, using the three selected bioink groups to assess the feasibility of our method. As demonstrated in Fig. 3, E to J, all the cell types were significantly more viable after 72 h of cryopreservation in the selected groups comparing to the control group, i.e., the group without any CPAs. The high cell viability values for all the cell types confirmed the effectiveness of all three CPA combinations. Given the cell viability rates, the melezitose group was selected for subsequent cryobioprinting jobs, as it yielded a cell viability rate of ~80% for the investigated cell types. The fluorescence images of the studied cell types are presented in figs. S11 to S15.
Effect of the CPAs on the cells before cryobioprinting
In extrusion bioprinting, it is inevitable to keep the cells in contact with the bioink/CPAs for several minutes if not longer. Contact with the CPAs over an extended time period before cryobioprinting may jeopardize cell viability due to toxicity of the DMSO and the prolonged exposure to osmotic pressure caused by the presence of saccharide molecules. We quantified the cell viability of the NIH/3T3 fibroblasts encapsulated in GelMA/CPA after being kept at RT for different time periods. As shown in fig. S16, increased exposure time yields lower cell viability. We, therefore, minimized exposure time for our cryobioprinting jobs by using small aliquots of bioinks for each round. The viability drop within 60 min of exposure was still quite acceptable at >80%.
Effect of freezing rate on cell viability
The sample freezing rate during the cryobioprinting process has a critical impact on ice crystal-formation and cell viability. Different freezing rates were achieved by changing the Tp. Given the dynamic nature of cryobioprinting, a constant freezing rate cannot be achieved throughout the construct. Table S3 presents the average freezing rates in the first two layers of the cryobioprinted grid structure using different values of Tp. As expected, the freezing rate is higher when a lower temperature is used for the freezing plate. Since the second layer does not directly touch the freezing plate, the freezing rate is remarkably lower comparing to the first layer. As a result, the top layers generally have a moderate freezing rate, closer to what is used in conventional cryopreservation (i.e., ~1 °C min−1). Representative live/dead images of the NIH/3T3 fibroblasts cryobioprinted on the freezing plate with various temperatures, −5 °C, −10 °C, −15 °C, and −20 °C, are shown in fig. S17. The samples were stained and imaged immediately after cryobioprinting. For cases where Tp was −15 °C, the cell viability was not significantly different from the original unbioprinted bioink. However, decreasing the Tp to −20 °C caused a major decline in cell viability (only ~60% viable cells).
Cell viability in different layers
Since the freezing regime is not homogenous throughout the thickness (Fig. 2, T and V), we investigated the cell viability of C2C12 myoblasts in different layers of the cryobioprinted constructs. Three representative layers with an average distance of 3 mm, termed upper, middle, and lower layers, were imaged on days 1, 3, and 7 after cryobioprinting. As shown in fig. S18, the cell viability was not noticeably affected by the distance from the freezing plate. The average cell viability within different layers of the scaffold was all about 85%, which is in an acceptable range.
Medium-term and long-term cryopreservation
Cryobioprinted hMSC-laden samples were cryopreserved for 14 and 28 days to study the effect of medium-term cryopreservation on cell viability. No significant difference between the cell viability rates of the cryopreserved samples in liquid nitrogen was observed after day 14 and day 28 (fig. S19). More excitingly, the viability of NIH/3T3 fibroblasts and HUVECs were further investigated after 3 months of cryopreservation and determined to be 83% and 86% after 7 days of culture, respectively (fig. S20), proving the efficacy of our cryobioprinting method for longer-term storage.
Cell differentiation
We used hMSCs to investigate their functionality through cell differentiation assays after cryobioprinting. The representative live/dead images for hMSCs after 72 h of cryopreservation are shown in Fig. 4A. Based on our medium-term results, we used cryobioprinted hMSC-laden samples after 14 days of cryopreservation in liquid nitrogen. On day 14, the samples were crosslinked, resuscitated, and cultured for 1 week before adding the differentiation media. The three types of cell differentiation, including osteogenic, chondrogenic, and adipogenic, were studied. In osteogenesis differentiation, runt-related transcription factor 2 (RUNX2, 1 week) and osteocalcin (2 and 3 weeks) immunostaining were conducted. Peroxisome proliferator-activated receptor gamma (PPARγ) and SRY-Box transcription factor-9 (SOX-9) immunostaining assays were used for assessing adipogenic and chondrogenic differentiation, respectively. In Fig. 4, B, E, and H, the fluorescence and immunostaining images of the encapsulated hMSCs after being cultured in differentiation media for 7 days, 14 days, and 21 days are presented. The quantitative analyses of osteogenesis (Fig. 4, C and D), chondrogenesis (Fig. 4, F and G), and adipogenesis (Fig. 4, I and J) revealed that the trends of differentiation in cryobioprinted samples were not notably different than those in control samples (i.e., the unfrozen samples). This finding proves that cryobioprinting did not negatively affect the functionality of hMSCs.
Fig. 4. Cell viability of hMSCs and cell differentiation assays in cryobioprinted GelMA/CPA constructs.
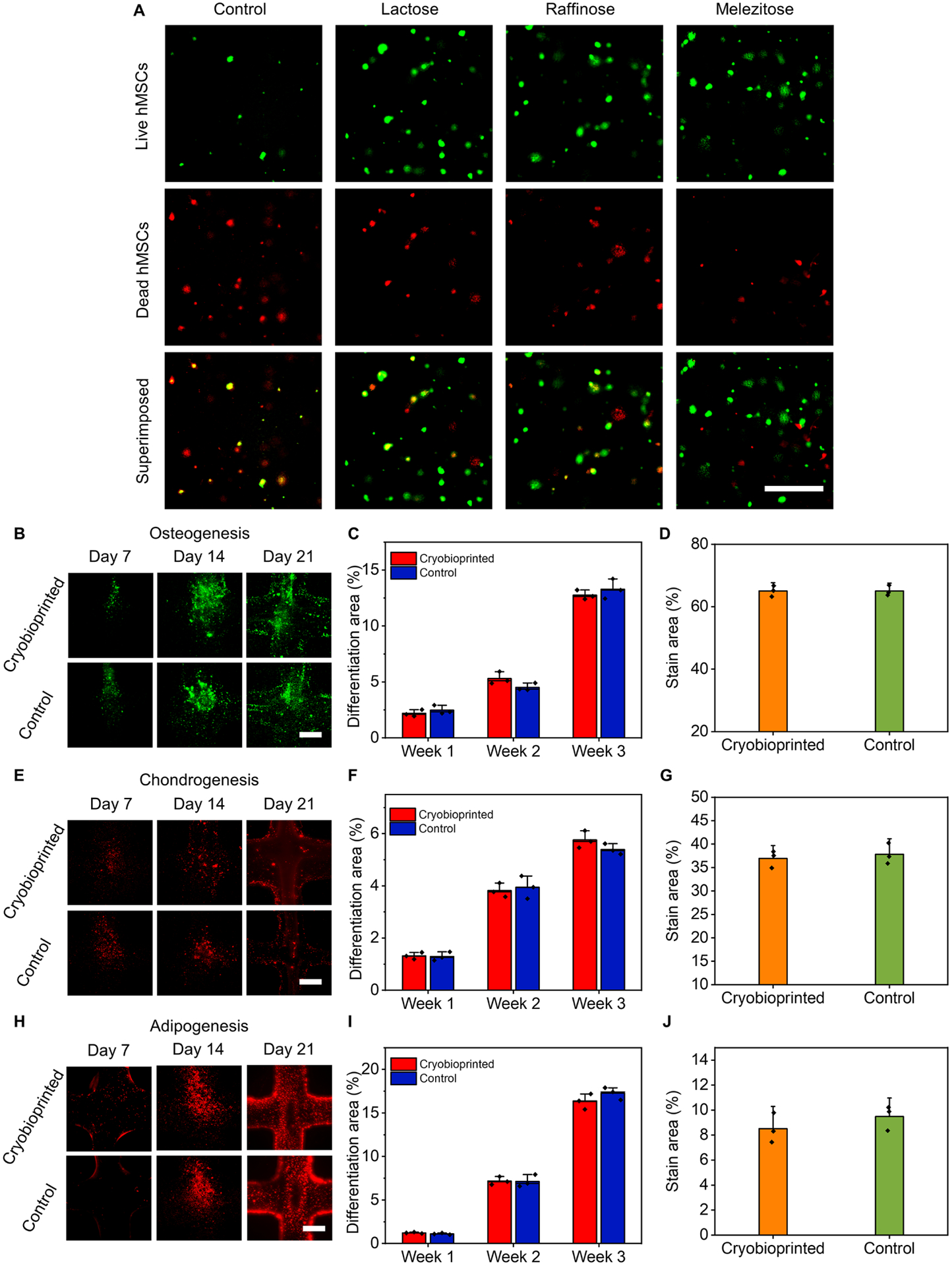
(A), Representative live (green)/dead (red) fluorescence images of hMSCs encapsulated in the three selected cryoprotective bioinks and cryopreserved for 72 h. (B to D), Osteogenic differentiation: (B), fluorescence micrographs, (C), semi-quantitative measurements of RUNX2 (1 week) and osteocalcin (2 and 3 weeks), and (D), quantification of Alizarin Red S-staining (3 weeks). (E to G), Chondrogenic differentiation: (E), fluorescence micrographs, (F), semi-quantitative measurements of SOX-9, and (G), quantification of Alcian blue staining (3 weeks). (H to J), Adipogenic differentiation: (H), fluorescence micrographs, (I), semi-quantitative measurements of PPARγ, and (J), quantification of Oil Red O staining (3 weeks). Scale bars in panels A, B, E, and H: 500 μm. n = 3.
CAM assay
CAM assay was used to evaluate angiogenesis potentials of the cryobioprinted scaffolds. The cryobioprinted constructs (previously stored in liquid nitrogen for 14 days) were implanted on day-7 ex ovo CAM and were collected after 7 days of additional incubation (Fig. 5A). The extents of angiogenesis in the cryobioprinted groups were determined using an image-processing method and histology (Fig. 5B). The average lengths of newly formed blood vessels (BVs) were measured to quantify the angiogenic responses surrounding the different cryobioprinted constructs. As presented in Fig. 5C, the smallest average length of BVs was associated with the control group (i.e., cryobioprinted GelMA+CPA) while adding either HUVECs or vascular endothelial growth factor (VEGF) resulted in a significant increase in the BV length. Cryobioprinted constructs with both HUVEC+VEGF (H&V) yielded the highest vascular growth. When the same bioink (H&V) was bioprinted at RT without freezing, similar vascular growth was achieved. These observations proved that the cryobioprinting procedure did not affect the functionality of HUVECs and VEGF.
Fig. 5. Chick ex ovo culture and CAM assay for investigating the vascularization in cryobioprinted scaffolds.
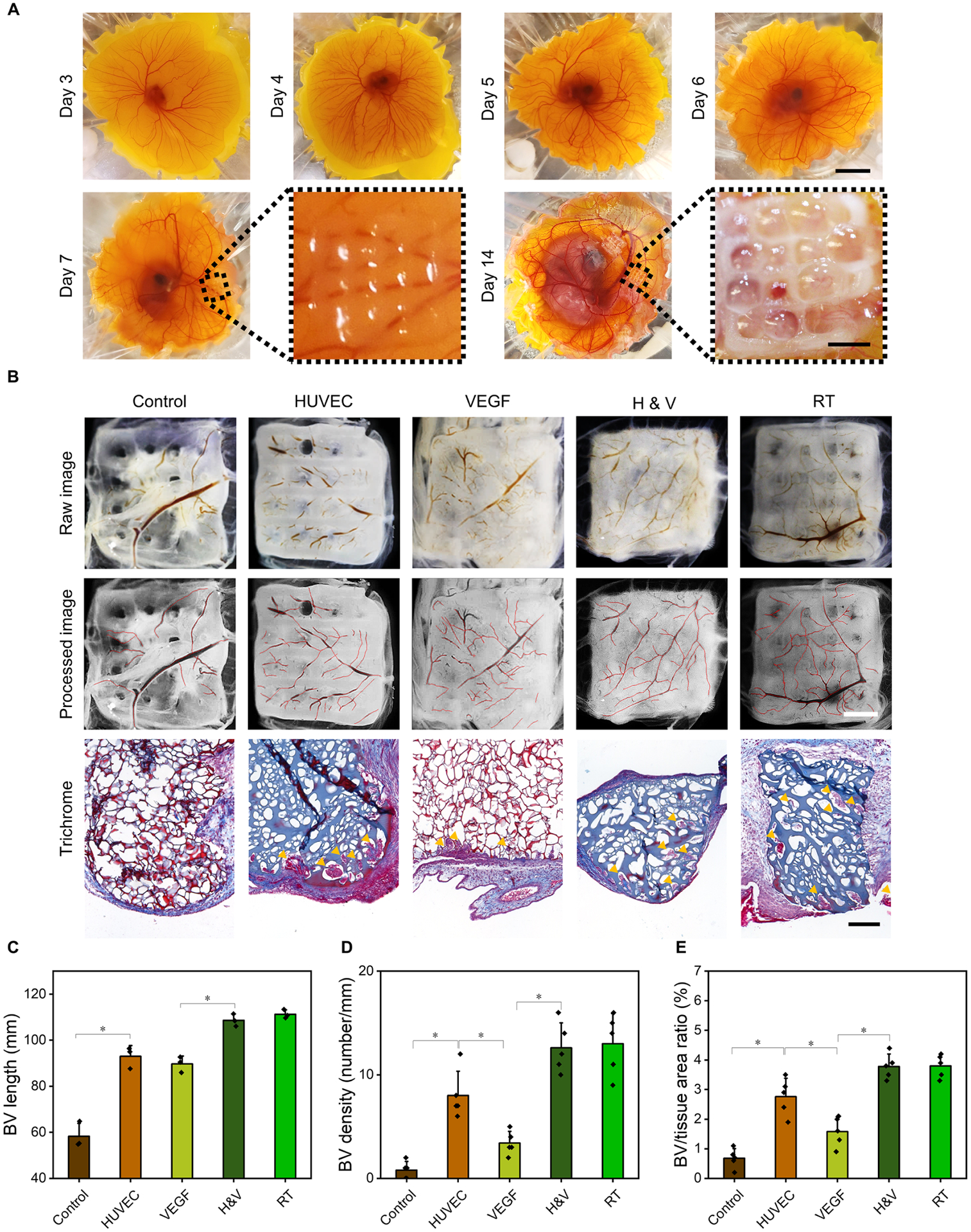
(A), Representative images of chick ex ovo culture with implanted cryobioprinted scaffolds for CAM assay in different timesteps. Scale bars: 2 cm for the low-magnification images and 2 mm for the high-magnification images. (B), Angiogenic abilities of the scaffolds and Goldner’s trichrome staining showing erythrocytes in red and collagen in green, where the blood vessels are indicated by yellow arrowheads (control: GelMA+CPA, HUVEC: GelMA+CPA+HUVECs, VEGF: GelMA+CPA+VEGF, H&V: GelMA+CPA+HUVECs+VEGF, and RT: GelMA+CPA+HUVECs+VEGF bioprinted at RT). Scale bars: 2 mm for optical images and 200 μm for histology images. (C to E), Quantification of the BV growth surrounding and within the cryobioprinted constructs by measuring different parameters: (C), lengths of the BVs surrounding the scaffolds, (D), BV densities, and (E), the ratio of BV-to-tissue areas within the scaffolds. n = 5; *P < 0.05.
Histological analyses further confirmed that the presence of HUVECs had a significant effect on CAM vessel formation within the 3D constructs (Fig. 5B). Almost no BVs grew into the control group, while the vascular infiltration in the HUVEC group was significantly higher. Similarly, vascular invasion in the H&V group was substantially higher than that of the VEGF group. Furthermore, neovascularization was observed deep in the H&V group, as shown in the histology images.
We also quantified the BV density (Fig. 5D) and the ratio of BV-to-tissue areas (Fig. 5E) within the constructs. BV density was calculated as the number of BVs within 1 mm of the interface along the scaffold/tissue border. The ratio of BV-to-tissue area was defined as the percentage of the BVs area over the area of the section36. The density of the BVs in the HUVEC group was significantly higher than that in the control group. Similarly, the H&V group showed a significantly higher BV density than the VEGF group. The ratio of BV-to-tissue areas also resulted in a similar trend.
Discussion
Considering the propensity to use shelf-ready products for clinical and research needs, the development of biofabrication methods capable of creating storable tissue constructs is of paramount importance. In this study, we invented and optimized cryobioprinting, a biofabrication process to simultaneously create and store cell-laden tissue constructs. Our results emphasized the unique synergy between bioprinting and cryopreservation, leading to faithful fabrication of tissue constructs with a decent cell viability rate. One of the key points in our approach was designing a freezing plate that allowed precisely controllable and stable temperature during the cryobioprinting procedure.
The limited printability window of soft materials, especially hydrogels, restricts a narrow boundary for choosing bioinks37. For example, 5% (w/v) GelMA bioink should be kept at approximately 15 °C or lower to achieve successfully bioprinted constructs38. In cryobioprinting, the deposited layers immediately freeze in situ. This rapid solidification enhances the printability of the bioink and fidelity of the 3D-bioprinted constructs, providing an expanded range of options to be used as bioinks. Free-standing and oblique constructs have been previously bioprinted using a cell-friendly supporting bath as the collecting substrate39–43. Our cryobioprinting approach offers another suitable method for bioprinting such constructs (Fig. 2, P to R), without any need for supporting baths, extra supports, or sacrificial materials. Another advantage of cryobioprinting is the capability of fabricating ready-to-use products. No washing steps are required to remove excessive supporting materials that exist in embedded bioprinting. This feature can transform cryobioprinting into a promising method for leveraging the scalability of the 3D bioprinting technology.
Recently, cylindrical hollow struts have been bioprinted in cryogenic conditions by implementing a core-shell nozzle44. While elegant, the presented method is relegated to fabricating fairly simple structures and is not applicable to bioprinted scaffolds with homogeneously dispersed cells. There are also intrinsic limitations associated with cryobioprinting of thick constructs using a freezing plate in our strategy. Although we were able to successfully fabricate relatively thick models, such as the ear model shown in Fig. 2F, printing beyond a certain thickness may slow down the heat-transfer from the top layers. Slower heat-transfer can negatively affect the printing fidelity of bioinks during the cryobioprinting procedure. Lowering the freezing plate temperature can potentially compensate for the decreased heat-transfer rate but may, on the other hand, compromise cell viability, as demonstrated in fig. S17. In addition, the long-term exposure of living cells to CPAs can be detrimental, as mentioned in the Results section. This issue can be addressed by using microfluidic systems45 for delivering the CPAs to bioink immediately before cryobioprinting. Additional optimizations are needed to investigate possible solutions for addressing these limitations and improving the technology.
In conventional cell cryopreservation, fetal bovine serum (FBS) is normally used as the CPA in place of saccharides. However, since the deviation of FBS contents might affect the cryopreservation efficiency in an unknown manner, we used well-defined saccharides for this purpose. As non-permeating CPAs, saccharides can preserve the cells during the thawing process by minimizing the osmotic shock46. It has been previously reported that raffinose in concentrations of 10–18% (w/v) is an effective CPA47. Other researchers have shown that melezitose and maltose offer acceptable cryoprotective effects19. Trehalose and sucrose may also reduce the intramembrane lateral stress, which leads to a practical cryopreservation approach48. DMSO is a permeating CPA that inhibits the formation of intracellular spicular ice crystals. Our findings showed that cell viability is a function of the type and the concentration of CPAs. Furthermore, the freezing regime had a critical effect on cell viability. For many types of cells, there is a so-called “temperature danger zone,” which lies between −15 °C to −60 °C49. During the freezing/resuscitation processes, the cells pass this range of temperatures twice. Staying longer periods in this zone may exacerbate cell damages. In addition, slow freezing causes a gradual increase in the solute concentration of the intra- and extracellular solutions, leading to a change in osmotic pressure50. On the other hand, the chance of intracellular ice crystallization and cell damage increases when the freezing procedure is too rapid51. These opposite effects contribute to reducing cell viability, thereby narrowing the range of optimum cryopreservation.
As thoroughly reviewed before52, the formation of sharp ice crystals at the ice nucleation point may jeopardize the viability of encapsulated cells. We found that the addition of CPAs decreased the sharpness and sizes of the ice crystals in the frozen hydrogel constructs. It was previously reported that the addition of CPAs lowers the freezing point of water and changes the ice crystals’ microstructure10. We observed a similar phenomenon for our GelMA-based cryoprotective bioinks. Avoiding the formation of spicular ice crystals is critical during freezing/thawing and plays a vital role in keeping the cells intact53. In complement to the benefits of CPAs, the hydrogels’ 3D network also reduces ice-crystal formation, thereby contributing to an intrinsic physical cryoprotective microenvironment for the cells31,54. This fact is in great accordance with previous observations reporting that cell-laden hydrogel scaffolds containing CPAs offer a decent cryopreservative environment leading to high cell viability and cell functionality after revival55.
Intriguingly, we stored cryobioprinted cell-laden constructs for up to 3 months without any significant reduction in cell viability, proving the capability of our method to fabricate shelf-ready products that could sustain extended storage prior to usage. Apart from viability, the functionality of the cryobioprinted cells is also essential to determine whether this fabrication and storage process has imposed any permanent damage to the cells. We used two methods to demonstrate how different types of cells could remain functional after being cryobioprinted. First, the capability of hMSCs in differentiation was investigated. It was shown that similar to normal hMSCs, cryobioprinted hMSCs were able to successfully differentiate to osteoblasts, chondrocytes, or adipocytes when exposed to proper differentiation media. Furthermore, we demonstrated that cryobioprinted HUVECs and VEGF could maintain their ability to induce angiogenesis when investigated in the CAM assay. These in vitro cell differentiation studies and ex ovo CAM assays shed light on the efficacy of the cryobioprinting method as a functional approach for creating cell-laden implants or tissue models.
Conclusion
In conclusion, cryobioprinting as a synergistic biofabrication method that combines the advantages of bioprinting and cryopreservation was developed. Our findings showed that cryobioprinting is a feasible method for fabricating storable cell-laden constructs to be used as the next generation of shelf-ready implants and tissue products. As the field of tissue engineering grows fast, these storable implants are envisioned to facilitate the clinical translation of the bioprinting method. Expanded applications towards laboratory and other usages, such as storable tissue models that can be readily shipped to desired sites, are also envisaged.
Experimental Procedures
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yu Shrike Zhang (yszhang@research.bwh.harvard.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional datasets are available from the lead contact upon reasonable requests. All requests for raw and analyzed data and materials will be promptly reviewed by the Brigham and Women’s Hospital to verify whether the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a Material Transfer Agreement.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
GelMA synthesis
Unless otherwise mentioned, all materials were purchased from Sigma-Aldrich. GelMA was prepared based on a previously developed protocol26. Briefly, porcine skin type-A gelatin was dissolved in PBS with a concentration of 10% (w/v) at 50 °C. Methacrylic anhydride was then added drop-by-drop using a syringe pump until a concentration of 5% (v/v) was reached. The emulsion was mixed on a magnetic hot plate for 2 h at 50 °C to ensure homogeneity. GelMA was then diluted twice and dialyzed using distilled water at 40 °C for 5 days. The distilled water was changed every 12 h. The solution was filtered at 40 °C using a 0.22-μm Stericup-GP Sterile Vacuum Filtration System (Millipore). The filtered solution was then aliquoted into 25-mL lots and stored at −80 °C for 24 h. The frozen GelMA was lyophilized for 5 days at 0.2 mbar and 24 °C in a FreeZone Labconco™ freeze-dryer. The desired amount of lyophilized GelMA was then dissolved in a pre-mixed solution of PBS and 0.6% (v/v) 2-hydroxy-4-(2-hydroxyethoxy)-2-methylpropiophenone (Irgacure 2959, Sigma-Aldrich) as the photoinitiator. The resulting solution was used as the base bioink for cryobioprinting.
Bioink design
GelMA at a final concentration of 5% (w/v) was used as the main constituent of the bioink. DMSO, along with different saccharides listed in Table 1, were added as the CPAs. D-(+)-trehalose dihydrate, D-lactose, sucrose ultrapure, D-(+) raffinose pentahydrate, and D-(+)-melezitose hydrate were purchased from Alfa Aesar. The saccharides’ concentration (0%, 4%, 8%, 12% (w/v)) and the DMSO concentration (0%, 5%, 10%, 15% (v/v)) were systematically studied to identify the optimal values based on cell viability results.
Table 1.
The properties of the saccharides used in this study.
| Sugar | Molecular formula | Solubility at 20 °C (g mL−1) | Density (g cm−3) | Type |
|---|---|---|---|---|
| Trehalose | C12H22O11 | 0.689 | 1.58 | Disaccharide |
| Lactose | C12H22O11 | 0.216 | 1.52 | Disaccharide |
| Sucrose | C12H22O11 | 2.100 | 1.59 | Disaccharide |
| Raffinose | C18H32O16 | 0.203 | 1.80 | Trisaccharide |
| Melezitose | C18H32O16 | 0.781 | 1.80 | Trisaccharide |
| Maltose | C12H22O11 | 1.080 | 1.54 | Disaccharide |
Cryobioprinting
A continuous extrusion bioprinter, Allevi 2 (3D Systems), was used for cryobioprinting. The Repetier-Host (Hot-World GmbH & Co. KG) and Cura (Ultimaker) software packages were employed to process the models and generate the corresponding G-codes. A self-designed and manually assembled freezing plate (fig. S1) was used as the bioprinting substrate. The freezing plate temperature was set to −15 °C. The needle size (27G), pressure (30 psi), and printhead speed (5 mm s−1) were selected based on the printability results of fig. S2. The required time for printing each layer of an 8×8-mm2 grid structure was approximately 8 s. To enhance the viscosity of the bioink, the cell-encapsulated GelMA/CPA was kept at 4 °C for 20 min before bioprinting. The samples were bioprinted on the freezing plate and stored in −80 °C or liquid nitrogen.
Ice crystal imaging
The hydrogel groups were cast on the freezing plate to study the effect of DMSO concentration on ice crystal-formation. A volume of 25 μL of the precursor was deposited on the freezing plate. The process was recorded using a brightfield microscope eyepiece camera (GXCAM-D800 - GT Vision Ltd.) mounted on a Leica EZ4 stereomicroscope (Leica Camera AG). Different concentrations of DMSO and melezitose were used in the hydrogels. The ice crystal-formation was quantified by processing the video frames using an open-source code56 and a customized script in MATLAB (Mathworks).
Splat ice recrystallization-inhibition assay
The splat cooling assay was performed following the previously developed protocols8,9,33. Briefly, 10 μL of samples with different DMSO concentrations was added between two glass coverslips. The coverslips were placed on the freezing plate (Tp=−20 °C) for 30 min. Images were then obtained using an optical microscope (Nikon) and processed by ImageJ (National Institutes of Health) to determine the ice crystal size in each group.
Heat-transfer simulations
COMSOL Multiphysics® was employed to simulate heat-transfer in cryobioprinting a two-layer 8×8-mm2 grid structure. The layers were generated by sweeping a 2-mm-thick rectangle over a defined bioprinting path. The simulation parameters are listed in table S4. Heat-transfer in the porous hydrogel was modeled by defining proper boundary conditions and using time-marching schemes. The layers were divided into 45 sections. The computational domain was discretized using an unstructured grid of free tetrahedral meshes with a size between 0.28 mm and 0.12 mm.
The top and the side boundaries were defined so that the convection heat-transfer with the ambient medium was taken into account. The cryobioprinting procedure was simulated by imposing a step-function temperature decrease on the bottom surface of the bioprinted section, which contacts the freezing plate. The bottom surface of successive sections was exposed to the low temperature, i.e., the Tp, to simulate cryobioprinting deposition kinematics. By considering the printing speed, the sections were exposed to the low temperature every 0.16 s, one at a time. After the first layer was entirely deposited, the second layer was assembled section by section on top of the first layer. For the second layer, the continuity condition was applied to equate the temperature of the second layer’s bottom surface with that of the first layer’s top surface. As a simplifying assumption, the phase-change from liquid to solid was neglected to decrease the computation cost.
Cell culture
Seven types of cells, namely NIH/3T3 fibroblasts, C2C12 skeletal myoblasts, HepG2 hepatocellular carcinoma cells, MCF-7 breast cancer cells, SMCs, HUVECs, and hMSCs, were separately cultured. Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific) enriched with 10% (v/v) FBS (Thermo Fisher Scientific) was used for culturing NIH/3T3, C2C12, HepG2, and MCF-7 cells. SMC growth medium, which was supplemented with 5% (v/v) FBS, 0.1% (v/v) insulin, 0.2% (v/v) human fibroblast growth factor-basic (hFGF-b), 0.1% (v/v) gentamicin sulfate-amphotericin (GA-1000), and 0.1% (v/v) human epidermal growth factor (hEGF) (Lonza Biologics), was used for culturing SMCs. HUVECs were cultured in endothelial cell growth medium supplemented with 2% (v/v) FBS, 0.04% (v/v) hydrocortisone, 0.4% (v/v) hFGF-b, 0.1% (v/v) VEGF, 0.1% insulin-like growth factor-1 (IGF-1), 0.1% (v/v) ascorbic acid, 0.1% (v/v) hEGF, 0.1% (v/v) GA-1000, and 0.1% (v/v) heparin (Lonza Biologics). Mesenchymal stem cell growth medium supplemented with 0.1% (v/v) GA-1000, 2% (v/v) L-glutamine, and 10% (v/v) mesenchymal cell growth supplement (Lonza Biologics) was used to culture hMSCs. The cells were cultured in T75 flasks at 37 °C in a 5% CO2 humidified incubator. The media were changed every 3 days until confluency of approximately 80% was achieved. After removing the media and washing with Dulbecco’s PBS (DPBS) 1× (Alfa Aesar), the cells were dissociated using 0.05% (v/v) trypsin-ethylenediaminetetraacetic acid (trypsin-EDTA) (Thermo Fisher Scientific).
Cryopreservation
Ninety-six groups of GelMA samples (four DMSO concentrations, four saccharide concentrations, and six types of saccharides) were studied to identify the optimal CPA configuration. C2C12 cells were encapsulated in the GelMA/CPAs precursor. A volume of 50 μL of the cell-laden precursor was directly cast on a freezing plate in aseptic conditions. The frozen samples were immediately stored at −80 °C or in liquid nitrogen (−196 °C) for cryopreservation. On day 3, the samples were removed from cryogenic storage and directly crosslinked via UV exposure. The samples were washed in PBS to remove any excess CPAs in preparation for subsequent use.
Cell viability
Cell viability was quantified within unthawed cell-encapsulated samples to identify the optimal combination of CPAs. Following the crosslinking and washing steps, the hydrogel samples were cultured for a definite period and then stained using calcein-AM and ethidium homodimer-1 (Thermo Fisher Scientific) per the provider’s protocol. An inverted Eclipse-Ti fluorescence microscope (Nikon) was employed to image the live and dead cells in green and red channels, respectively. The ratio of live cells to the total captured cells in random regions of the samples was determined through processing the fluorescent images using ImageJ.
Cell differentiation
The hMSCs were encapsulated in the selected group of GelMA/CPA to be used as the bioink. The cryobioprinted samples were stored at −80 °C overnight and were then transferred to liquid nitrogen for cryopreservation. On day 14, the samples were UV-crosslinked and revived. After 3 days of culture, the three types of differentiation media were separately added. The constructs were cultured in DMEM (4.5 g L−1 of D-glucose) supplemented with 1-μM dexamethasone, 5-μg mL−1 insulin, and 50-μM indomethacin38 to induce adipogenesis. Osteogenesis was conducted by culturing the constructs in DMEM (1 g L−1 of D-glucose) supplemented with 10-mmol L−1 β-glycerophosphate, 300-μmol L−1 L-ascorbic acid, and 100-nmol L−1 dexamethasone38. For chondrogenic differentiation, the constructs were cultured in DMEM (4.5 g L−1 of D-glucose) supplemented with 50-μg L−1 ITS+ Premix Tissue Culture Supplement, 100-nmol L−1 dexamethasone, 1-μM ascorbate-2-phosphate, and 10-ng mL−1 transforming growth factor-β157. At certain time points, i.e., day 7, day14, and day 21, Oil Red O, Alizarin Red S, and Alcian blue (all from ScienCell Research Laboratories) were used for staining the samples and characterizing adipogenesis, osteogenesis, or chondrogenesis.
Immunostaining
For immunofluorescence staining, the anti-PPARγ, anti-RUNX2, anti-osteocalcin, or anti-SOX-9 antibodies (all from Abcam) were diluted in PBS to achieve a final concentration of 200 μg L−1 and incubated overnight at 4 °C for adipogenesis, osteogenesis, or chondrogenesis. The samples were washed with PBS and then stained with the corresponding secondary antibodies. The samples were washed again with PBS, and fluorescence micrographs were captured using an inverted fluorescence microscope.
CAM assay
The ex ovo chick CAM culture was undertaken following the guidelines used in the previous studies58,59. For preparing the VEGF group, the cryoprotective bioink was supplemented with 100 ng mL−1 of VEGF (PeproTech)60. The eggshells were carefully cracked to start the ex ovo culture 3 days after incubation. The cryobioprinted scaffolds were implanted in chick embryo CAM on day 7. The samples were then incubated for another 7 days until they were collected for imaging. We used 10% (v/v) formalin to fix the samples for inspecting the angiogenesis. An optical camera (EOS 60D, Canon) was used for photographing. The discernible BV lengths on the cryobioprinted constructs were measured using the NeuronJ tracing toolbox of ImageJ to quantify the extent of vascularization58.
Histology
The samples collected from the CAM assays were fixed in 10% (v/v) formalin for 24 h. The fixed samples were then dehydrated, embedded in paraffin wax, and then serially sectioned (5 μm in thickness). Goldner’s trichrome staining involved the use of Weigert’s hematoxylin, ponceau-fuchsin-azophloxin (erythrocytes), phosphomolybdic acid (cytoplasm), and light green (collagen)61. The BV density and the ratio of BV-to-tissue areas in the different groups were calculated36.
Statistical Analyses
All data are expressed as means ± standard deviations. Minimum triplicates were used for all the experiments. One-way analysis of variance (ANOVA) was used to compare the effect of different CPAs on cell viability. The two-tailed paired or unpaired student’s t-test was used for determining the significantly different values. A P-value of 0.05 was considered as the threshold.
Supplementary Material
Movie S1. Cryobioprinting procedure.
Movie S2. Heat-transfer simulation for cryobioprinting.
Movie S3. Ice crystal growth in 5% (w/v) GelMA control sample (CPA-free).
Movie S4. Ice crystal growth in 5% (w/v) GelMA + 5% (v/v) DMSO sample.
Movie S5. Ice crystal growth in 5% (w/v) GelMA + 10% (v/v) DMSO sample.
Movie S6. Ice crystal growth in 5% (w/v) GelMA + 15% (v/v) DMSO sample.
Movie S7. Ice crystal growth in 5% (w/v) GelMA + 4% (w/v) Melezitose sample.
Movie S8. Ice crystal growth in 5% (w/v) GelMA + 8% (w/v) Melezitose sample.
Movie S9. Ice crystal growth in 5% (w/v) GelMA + 12% (w/v) Melezitose sample.
Progress and Potential.
The lack of shelf-availability of cell-laden bioprinted products has been a hurdle in developing artificial tissue constructs. This work highlights the concept of cryobioprinting as an enabling method for concurrently biofabricating and cryopreserving cell-laden tissue constructs. The samples were fabricated using an extrusion bioprinter on a freezing plate as the build platform with accurately controllable low temperatures. The GelMA-based bioink composition was engineered using different types and concentrations of cryoprotective agents. Intriguingly, cell differentiation and angiogenesis data revealed that the cryobioprinted cells remained viable and functional after revival. Furthermore, the intrinsic nature of cryobioprinting enabled the user to fabricate freeform structures, which can be hardly possible to achieve using conventional extrusion bioprinting techniques. These findings shed light on the development of cryobioprinting as a functional biofabrication method.
Highlights.
Cryobioprinting is optimized for fabricating freeform storable tissue constructs
Shelf availability of cryobioprinted products is achieved
Cryoprotective bioinks help in maintaining the functionality of the cells
Acknowledgments
H.R. was supported by the FRQNT’s International Internship Award (279390), MITACS Globalink Research Award (IT14553), McGill’s Graduate Mobility Award, McGill’s Doctoral Internship Award, and the FRQNT’s Postdoctoral Fellowship (296447). X.Z. acknowledges the Program of China Scholarship Council (No.201807045057) and the High-Level Talent Internationalization Training Program of Henan Province (No.2019004). L.M. acknowledges the funding from the National Institute on Deafness and other Communication Disorders (NIDCD) of the National Institutes of Health (NIH) grant numbers R01DC005788 and R01DC014461. Y.S.Z. acknowledges the support from the Brigham Research Institute. The authors thank Jing Ge of the University of Arkansas for the insightful suggestions on chick ex ovo CAM assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Ravanbakhsh H, Karamzadeh V, Bao G, Mongeau L, Juncker D, Zhang YS (2021). Emerging Technologies in Multi-Material Bioprinting. Adv. Mater, 2104730. 10.1002/adma.202104730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinrich MA, Liu W, Jimenez A, Yang J, Akpek A, Liu X, et al. (2019). 3D Bioprinting: from Benches to Translational Applications. Small 15, 1805510. 10.1002/smll.201805510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang HW, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A (2016). A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat Biotechnol. 34, 312–319. 10.1038/nbt.3413. [DOI] [PubMed] [Google Scholar]
- 4.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA (2014). 3D Bioprinting of Vascularized , Heterogeneous Cell-Laden Tissue Constructs. Adv. Mater 26, 3124–3130. 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 5.Ma X, Liu J, Zhu W, Tang M, Lawrence N, Yu C, et al. (2018). 3D bioprinting of functional tissue models for personalized drug screening and in vitro disease modeling. Adv. Drug Deliv. Rev 132, 235–251. 10.1016/j.addr.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawlor KT, Vanslambrouck JM, Higgins JW, Chambon A, Bishard K, Arndt D, et al. (2021). Cellular extrusion bioprinting improves kidney organoid reproducibility and conformation. Nat. Mater 20, 260–271. 10.1038/s41563-020-00853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeon O, Lee YB, Hinton TJ, Feinberg AW, Alsberg E (2019). Cryopreserved cell-laden alginate microgel bioink for 3D bioprinting of living tissues. Mater. Today Chem 12, 61–70. 10.1016/j.mtchem.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell DE, Fayter AER, Deller RC, Hasan M, Gutierrez-Marcos J, Gibson MI (2019). Ice-recrystallization inhibiting polymers protect proteins against freeze-stress and enable glycerol-free cryostorage. Mater. Horizons 6, 364–368. 10.1039/C8MH00727F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgiou PG, Marton HL, Baker AN, Congdon TR, Whale TF, Gibson MI (2021). Polymer Self-Assembly Induced Enhancement of Ice Recrystallization Inhibition. J. Am. Chem. Soc 143, 7449–7461. 10.1021/jacs.1c01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang S, Lyu C, Zhao P, Li W, Kong W, Huang C, et al. (2019). Cryoprotectant enables structural control of porous scaffolds for exploration of cellular mechano-responsiveness in 3D. Nat. Commun 10, 1–14. 10.1038/s41467-019-11397-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H, Zhao G, Zhang Y, Xu J, Toth TL, He X (2017). Predehydration and Ice Seeding in the Presence of Trehalose Enable Cell Cryopreservation. ACS Biomater. Sci. Eng 3, 1758–1768. 10.1021/acsbiomaterials.7b00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahari L, Bein A, Yashunsky V, Braslavsky I (2018). Directional freezing for the cryopreservation of adherent mammalian cells on a substrate. PLoS One. 13, e0192265. 10.1371/journal.pone.0192265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stock W, Pinseel E, De Decker S, Sefbom J, Blommaert L, Chepurnova O, et al. (2018). Expanding the toolbox for cryopreservation of marine and freshwater diatoms. Sci. Rep 8, 1–9. 10.1038/s41598-018-22460-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Von Bomhard A, Elsässer A, Ritschl LM, Schwarz S, Rotter N (2016). Cryopreservation of endothelial cells in various cryoprotective agents and media - Vitrification versus slow freezing methods. PLoS One. 11, e0149660. 10.1371/journal.pone.0149660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solocinski J, Osgood Q, Wang M, Connolly A, Menze MA, Chakraborty N (2017). Effect of trehalose as an additive to dimethyl sulfoxide solutions on ice formation, cellular viability, and metabolism. Cryobiology. 75, 134–143. 10.1016/j.cryobiol.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Tsai S, Chong G, Meng PJ, Lin C (2018). Sugars as supplemental cryoprotectants for marine organisms. Rev Aquac. 10, 703–715. 10.1111/raq.12195. [DOI] [Google Scholar]
- 17.Malo C, Gil L, Gonzalez N, Cano R, de Blas I, Espinosa E (2010). Comparing sugar type supplementation for cryopreservation of boar semen in egg yolk based extender. Cryobiology. 61, 17–21. 10.1016/j.cryobiol.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Silva CG, Cunha ER, Blume GR, Malaquias JV, Báo SN, Martins CF (2015). Cryopreservation of boar sperm comparing different cryoprotectants associated in media based on powdered coconut water, lactose and trehalose. Cryobiology. 70, 90–94. 10.1016/j.cryobiol.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Hino T, Takabe M, Suzuki-migishima R, Yokoyama M (2007). Cryoprotective effects of various saccharides on cryopreserved mouse sperm from various strains. Reprod. Med. Biol 6, 229–233. 10.1111/j.1447-0578.2007.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eroglu A (2010). Cryopreservation of mammalian oocytes by using sugars: Intra- and extracellular raffinose with small amounts of dimethylsulfoxide yields high cryosurvival, fertilization, and development rates. Cryobiology. 60, S54–S59. 10.1016/j.cryobiol.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Z, Parisi C, Di Silvio L, Dini D, Forte AE (2017). Cryogenic 3D Printing of Super Soft Hydrogels. Sci. Rep 7, 1–11. 10.1038/s41598-017-16668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi L, Hu Y, Ullah MW, Ullah I, Ou H, Zhang W, et al. (2019). Cryogenic freeform extrusion bioprinting of decellularized small intestinal submucosa for potential applications in skin tissue engineering. Biofabrication. 11, 035023. 10.1088/1758-5090/ab15a9. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Zhao Q, Wang M (2017). Cryogenic 3D printing for producing hierarchical porous and rhBMP-2-loaded Ca-P/PLLA nanocomposite scaffolds for bone tissue engineering. Biofabrication. 9, 025031. 10.1088/1758-5090/aa71c9. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Florczyk SJ (2020). Freeze-FRESH: A 3D printing technique to produce biomaterial scaffolds with hierarchical porosity. Materials. 13, 354. 10.3390/ma13020354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loessner D, Meinert C, Kaemmerer E, Martine LC, Yue K, Levett PA, et al. (2016). Functionalization, preparation and use of cell-laden gelatin methacryloyl-based hydrogels as modular tissue culture platforms. Nat. Protoc 11, 727–746. 10.1038/nprot.2016.037. [DOI] [PubMed] [Google Scholar]
- 26.Gong J, Schuurmans CCL, van Genderen AM, Cao X, Li W, Cheng F, et al. (2020). Complexation-induced resolution enhancement of 3D-printed hydrogel constructs. Nat. Commun 11, 1–14. 10.1038/s41467-020-14997-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maharjan S, Alva J, Cámara C, Rubio AG, Hernández D, Delavaux C, et al. (2020). Symbiotic Photosynthetic Oxygenation within 3D-Bioprinted Vascularized Tissues. Matter. 4, 217–240. 10.1016/j.matt.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savage N (2009). Thermoelectric coolers. Nat. Photonics 3, 541–542. 10.1038/nphoton.2009.158 [DOI] [Google Scholar]
- 29.Kishore RA, Nozariasbmarz A, Poudel B, Sanghadasa M, Priya S (2019). Ultra-high performance wearable thermoelectric coolers with less materials. Nat. Commun 10, 1–13. 10.1038/s41467-019-09707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mironi-Harpaz I, Wang DY, Venkatraman S, Seliktar D (2012). Photopolymerization of cell-encapsulating hydrogels: Crosslinking efficiency versus cytotoxicity. Acta Biomater. 8, 1838–1848. 10.1016/j.actbio.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 31.Cheng Y, Yu Y, Zhang Y, Zhao G, Zhao Y (2019). Cold-Responsive Nanocapsules Enable the Sole-Cryoprotectant-Trehalose Cryopreservation of β Cell–Laden Hydrogels for Diabetes Treatment. Small. 15, 1904290. 10.1002/smll.201904290. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y (2020). Two Sets of Simple Formulae to Estimating Fractal Dimension of Irregular Boundaries. Math. Probl. Eng 2020, 7528703. 10.1155/2020/7528703. [DOI] [Google Scholar]
- 33.Tomczak MM, Marshall CB, Gilbert JA, Davies PL (2003). A facile method for determining ice recrystallization inhibition by antifreeze proteins. Biochem. Biophys. Res. Commun 311, 1041–1046. 10.1016/j.bbrc.2003.10.106. [DOI] [PubMed] [Google Scholar]
- 34.An TZ, Iwakiri M, Edashige K, Sakurai T, Kasai M (2000). Factors affecting the survival of frozen-thawed mouse spermatozoa. Cryobiology. 40, 237–249. 10.1006/cryo.2000.2245. [DOI] [PubMed] [Google Scholar]
- 35.Smith GD, Takayama S (2019). Cryopreservation and microfluidics: A focus on the oocyte. Reprod. Fertil. Dev 31, 93–104. 10.1071/RD18326. [DOI] [PubMed] [Google Scholar]
- 36.Choi SW, Zhang Y, Macewan MR, Xia Y (2013). Neovascularization in Biodegradable Inverse Opal Scaffolds with Uniform and Precisely Controlled Pore Sizes. Adv. Healthc. Mater 2, 145–154. 10.1002/adhm.201200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Wu C, Chu PK, Gelinsky M (2020). 3D printing of hydrogels: Rational design strategies and emerging biomedical applications. Mater. Sci. Eng. R Rep 140, 100543. 10.1016/j.mser.2020.100543. [DOI] [Google Scholar]
- 38.Ying G, Jiang N, Parra-Cantu C, Tang G, Zhang J, Wang H, et al. (2020). Bioprinted Injectable Hierarchically Porous Gelatin Methacryloyl Hydrogel Constructs with Shape-Memory Properties. Adv. Funct. Mater 30, 2003740. 10.1002/adfm.202003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu W, Deconinck A, Lewis JA (2011). Omnidirectional printing of 3D microvascular networks. Adv. Mater 23, 178–183. 10.1002/adma.201004625. [DOI] [PubMed] [Google Scholar]
- 40.Highley CB, Rodell CB, Burdick JA (2015). Direct 3D Printing of Shear-Thinning Hydrogels into Self-Healing Hydrogels. Adv. Mater 27, 5075–5079. 10.1002/adma.201501234. [DOI] [PubMed] [Google Scholar]
- 41.Bhattacharjee T, Zehnder SM, Rowe KG, Jain S, Nixon RM, Sawyer WG, et al. (2015). Writing in the granular gel medium. Sci. Adv 1, e1500655. 10.1126/sciadv.1500655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue HJ, et al. (2015). Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv 1, e1500758. 10.1126/sciadv.1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bao G, Jiang T, Ravanbakhsh H, Reyes A, Ma Z, Strong M, et al. (2020). Triggered micropore-forming bioprinting of porous viscoelastic hydrogels. Mater. Horizons 7, 2336–2347. 10.1039/D0MH00813C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JY, Kim GH (2020). A cryopreservable cell-laden GelMa-based scaffold fabricated using a 3D printing process supplemented with an in situ photo-crosslinking. J Ind Eng Chem. 85, 249–257. 10.1016/j.jiec.2020.02.007. [DOI] [Google Scholar]
- 45.du Chatinier DN, Figler KP, Agrawal P, Liu W, Zhang YS (2021). The potential of microfluidics-enhanced extrusion bioprinting. Biomicrofluidics. 15, 041304. 10.1063/5.0033280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright DL, Eroglu A, Toner M, Toth TL (2004). Use of sugars in cryopreserving human oocytes. Reprod. Biomed. Online 9, 179–186. 10.1016/s1472-6483(10)62127-x. [DOI] [PubMed] [Google Scholar]
- 47.Tada N, Sato M, Yamanoi J, Mizorogi T, Kasai K, Ogawa S (1990). Cryopreservation of mouse spermatozoa in the presence of raffinose and glycerol. J. Reprod. Fertil 89, 511–516. 10.1530/jrf.0.0890511. [DOI] [PubMed] [Google Scholar]
- 48.Yoon YH, Pope JM, Wolfe J (1998). The effects of solutes on the freezing/properties of and hydration forces in lipid lamellar phases. Biophys. J 74, 1949–1965. 10.1016/S0006-3495(98)77903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao G, Fu J (2017). Microfluidics for cryopreservation. Biotechnol. Adv 35, 323–336. 10.1016/j.biotechadv.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jang TH, Park SC, Yang JH, Kim JY, Seok JH, Park US, et al. (2017). Cryopreservation and its clinical application. Integr. Med. Res 6, 12–18. 10.1016/j.imr.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baust JG, Gao D, Baust JM (2009). Cryopreservation: An emerging paradigm change. Organogenesis. 5, 90–96. 10.4161/org.5.3.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.John Morris G, Acton E (2013). Controlled ice nucleation in cryopreservation - A review. Cryobiology. 66, 85–92. 10.1016/j.cryobiol.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 53.Deller RC, Vatish M, Mitchell DA, Gibson MI (2014). Synthetic polymers enable non-vitreous cellular cryopreservation by reducing ice crystal growth during thawing. Nat. Commun 5, 1–7. 10.1038/ncomms4244. [DOI] [PubMed] [Google Scholar]
- 54.Kanmani P, Satish Kumar R, Yuvaraj N, Paari KA, Pattukumar V, Arul V (2011). Cryopreservation and microencapsulation of a probiotic in alginate-chitosan capsules improves survival in simulated gastrointestinal conditions. Biotechnol. Bioprocess Eng 16, 1106–1114. 10.1007/s12257-011-0068-9. [DOI] [Google Scholar]
- 55.Cagol N, Bonani W, Maniglio D, Migliaresi C, Motta A (2018). Effect of cryopreservation on cell-laden hydrogels: Comparison of different cryoprotectants. Tissue Eng. Part C Methods 24, 20–31. 10.1089/ten.tec.2017.0258. [DOI] [PubMed] [Google Scholar]
- 56.Jiro. (2021). Grabit: Extract (pick out) data points off image files. MATLAB Central File Exchange, https://www.mathworks.com/matlabcentral/fileexchange/7173-grabit.
- 57.Solchaga LA, Penick KJ, Welter JF (2011). Chondrogenic differentiation of bone marrow-derived mesenchymal stem cells: tips and tricks. Methods Mol Biol. 698, 253–278. 10.1007/978-1-60761-999-4_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mangir N, Dikici S, Claeyssens F, Macneil S (2019). Using ex Ovo Chick Chorioallantoic Membrane (CAM) Assay to Evaluate the Biocompatibility and Angiogenic Response to Biomaterials. ACS Biomater. Sci. Eng 5, 3190–3200. 10.1021/acsbiomaterials.9b00172. [DOI] [PubMed] [Google Scholar]
- 59.Mousseau Y, Mollard S, Qiu H, Richard L, Cazal R, Nizou A, et al. (2014). In vitro 3D angiogenesis assay in egg white matrix: Comparison to Matrigel, compatibility to various species, and suitability for drug testing. Lab. Invest 94, 340–349. 10.1038/labinvest.2013.150. [DOI] [PubMed] [Google Scholar]
- 60.Wu Y, Al-Ameen MA, Ghosh G (2014). Integrated Effects of Matrix Mechanics and Vascular Endothelial Growth Factor (VEGF) on Capillary Sprouting. Ann Biomed Eng. 42, 1024–1036. 10.1007/s10439-014-0987-7. [DOI] [PubMed] [Google Scholar]
- 61.Cidonio G, Alcala-Orozco CR, Lim KS, Glinka M, Mutreja I, Kim YH, et al. (2019). Osteogenic and angiogenic tissue formation in high fidelity nanocomposite Laponite-gelatin bioinks. Biofabrication. 11, 35027. 10.1088/1758-5090/ab19fd. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Cryobioprinting procedure.
Movie S2. Heat-transfer simulation for cryobioprinting.
Movie S3. Ice crystal growth in 5% (w/v) GelMA control sample (CPA-free).
Movie S4. Ice crystal growth in 5% (w/v) GelMA + 5% (v/v) DMSO sample.
Movie S5. Ice crystal growth in 5% (w/v) GelMA + 10% (v/v) DMSO sample.
Movie S6. Ice crystal growth in 5% (w/v) GelMA + 15% (v/v) DMSO sample.
Movie S7. Ice crystal growth in 5% (w/v) GelMA + 4% (w/v) Melezitose sample.
Movie S8. Ice crystal growth in 5% (w/v) GelMA + 8% (w/v) Melezitose sample.
Movie S9. Ice crystal growth in 5% (w/v) GelMA + 12% (w/v) Melezitose sample.
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional datasets are available from the lead contact upon reasonable requests. All requests for raw and analyzed data and materials will be promptly reviewed by the Brigham and Women’s Hospital to verify whether the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a Material Transfer Agreement.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


