Abstract
Purpose
To identify molecular features that distinguish individuals with shared clinical features of granulomatous uveitis.
Design
Cross-sectional observational study.
Participants
Four eyes from patients with active granulomatous uveitis.
Methods
We performed single-cell RNA sequencing with antigen-receptor sequence analysis to obtain an unbiased gene expression survey of ocular immune cells and to identify clonally expanded lymphocytes.
Main Outcomes Measures
For each inflamed eye, we measured the proportion of distinct immune cell types, the amount of B- or T-cell clonal expansion, and the transcriptional profile of T and B cells.
Results
Each individual showed robust clonal expansion arising from a single T- or B-cell lineage, suggesting distinct, antigen-driven pathogenic processes in each patient. This variability in clonal expansion was mirrored by individual variability in CD4 T-cell populations, whereas ocular CD8 T cells and B cells were more similar transcriptionally among patients. Finally, ocular B cells displayed evidence of class switching and plasmablast differentiation within the ocular microenvironment, providing additional support for antigen-driven immune responses in granulomatous uveitis.
Conclusions
Collectively, our study identified both conserved and individualized features of granulomatous uveitis, illuminating parallel pathophysiologic mechanisms and suggesting that future personalized therapeutic approaches may be warranted.
Keywords: B cells, single cell RNA sequencing, clonal expansion, T cells, Uveitis
Abbreviations and Acronyms: ABC, activated B cell; AC, anterior chamber; BCR, B-cell receptor; BSA, bovine serum albumin; FBS, fetal bovine serum; HLA, human leukocyte antigen; ILC, innate lymphoid cell; MHC, major histocompatibility complex; NK, natural killer; PBMC, peripheral blood mononuclear cell; PBS, phosphate buffered saline; PCA, principle component analysis; RPMI, Roswell Park Memorial Institute Media; scRNAseq, single-cell RNA sequencing; TCR, T-cell receptor; Treg, regulatory T; TH1, T helper 1; TH17, T helper 17; UMI, unique molecular identifier
Uveitis, or ocular inflammation, encompasses multiple individual disease entities that can be classified according to anatomic involvement (anterior, intermediate, posterior, choroiditis, retinal vasculitis), by known infectious causes (syphilis, toxoplasmosis), or by systemic disease association (sarcoidosis, multiple sclerosis, Behçet’s disease). However, at least half of all patients have idiopathic disease, with no known infectious cause or systemic disease association. Although specific causes can be separated based on clinically defined diagnostic criteria, significant interpatient heterogeneity exists in disease manifestations, severity, and therapeutic response. Likewise, empiric drug selection for immune suppression leads to variable success rates of 50% to 80%,1, 2, 3 leaving some patients at significant risk for disease progression and vision loss. Unrecognized molecular factors leading to clinical heterogeneity may underlie the variability in therapeutic response, highlighting the need for improved disease characterization based on well-defined pathophysiologic mechanisms.
Granulomatous uveitis is a term applied to ocular inflammation that is associated with specific clinical features, including characteristic cellular deposits on the corneal endothelium; so-called mutton fat keratic precipitates4; nodules in the iris, trabecular meshwork, optic nerve, retina, and choroid; granulomatous-appearing vitreous opacities; and segmental or nodular periphlebitis. Similar to granulomatous inflammation elsewhere, histologic analysis of ocular granulomas has demonstrated a predominance of T cells, as well as the presence of macrophages, dendritic cells, and B cells.5,6 However, the pathophysiologic contributions of these individual cell types are not known.
In this study, we performed a clinical and molecular comparison of 4 patients with similar gender, race, and ethnicity with a shared diagnosis of granulomatous uveitis without an associated systemic disease, yet demonstrated notable differences in their clinical courses and therapeutic responses. To determine whether the cellular and molecular features present in each patient were similar or unique, we obtained an unbiased molecular analysis of the inflammatory cells present in the anterior chamber (AC) by using single-cell RNA sequencing (scRNAseq). This technique is well suited for deep molecular phenotyping of the relatively small number of cells that can be isolated from the eye. We paired this analysis with T-cell and B-cell receptor sequencing to identify clonally expanded lymphocytes indicative of antigen-driven responses in granulomatous uveitis.
Methods
Patients with Uveitis and Sample Collection
Granulomatous uveitis was identified by pathognomonic findings, that is, characteristic deposits of inflammatory cells on the inner surface of the cornea (i.e., mutton-fat keratic precipitates). Aqueous humor and blood samples were collected from 4 patients (Table 1) with active anterior granulomatous uveitis defined by at least 1+ AC cell (>6 cells/high-powered field)7 and evidence of granulomatous uveitis based on the treating ophthalmologist’s examination findings, such as granulomatous keratoprecipitates. Because all patients had bilateral disease, the eye with more AC cells was chosen to be sampled to increase yield. Ocular and blood samples were collected concurrently in 3 patients (patients UV150, UV170, and UV174). In 1 individual, patient UV031, the blood was drawn on a subsequent appointment because of challenges with phlebotomy; however, the patient still had active disease during the follow-up of 2+ AC cell. Relevant clinical and laboratory evaluations were performed to exclude infectious causes and systemic rheumatologic conditions (Table 2).
Table 1.
Patient Demographics
| Patient No. | Age (yrs) | Gender | Race | Ethnicity |
|---|---|---|---|---|
| UV031 | 67 | F | Black | Not Hispanic |
| UV150 | 28 | F | Black | Not Hispanic |
| UV170 | 85 | F | Black | Not Hispanic |
| UV174 | 60 | F | Black | Not Hispanic |
F = female.
Table 2.
Clinical Data
| Patient | Eye Sampled | Inflammation Grade (SUN) | Duration of Current Flare (Symptoms) | Topical Steroids∗ | Anatomic Involvement | Disease Duration | Disease Course | Eyes Involved | Systemic Therapies∗ | Ocular Surgical History before Sampling | Prior Therapies | Angiotensin Converting Enzyme | CXR | Rheumatology Evaluation | Follow-up Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UV031 | Left | 2+ cell, 2+ flare | Chronic, uncontrolled for months | PF thrice daily for months | Anterior/intermediate | 8 yrs | Chronic | Both | MTX, ETN | Bilateral cataract surgeries | Oral prednisone, STK, PF eye drops | 20 | Minimal right lung base scarring, no LAD or granulomas | No evidence of systemic rheumatologic disease | ETN stopped, ADA started, MTX continued with questionable adherence and persistent inflammation |
| UV150 | Right | 1+ cell, 1+ flare | 90 days | PF 6×/day for 3 wks | Anterior/intermediate | 2 yrs | Chronic | Both | None | None | Oral prednisone, PF eye drops | 22 | Focal opacification of left lingual, no LAD or granulomas | ANA 1:80, possible history of malar rash; however, no systemic disease found | Uncontrolled with MTX and IFX; serial Ozurdex, Durezol; bilateral cataract and glaucoma surgeries (AGVI) |
| UV170 | Left | 2+ cell, 1+ flare | 8 days | PF 4 times daily for 2 days | Anterior, with inactive appearing MFC scars | 2 yrs | Recurrent | Both | None | Bilateral cataract surgeries | PF eye drops | 23 | No LAD or granulomas | No evidence of systemic disease | One subsequent anterior uveitis flare treated with PF |
| UV174 | Left | 3+ cell, 2+ flare | 30 days | No drops | Anterior/intermediate | 8 yrs | Recurrent | Both | None | Bilateral cataract surgeries | Multiple STK, PF eye drops | 24 | No LAD or granulomas | Not performed, no symptoms consistent with systemic rheumatologic disease | Controlled with BL vitrectomy and Retiserts |
ADA = adalimumab; AGVI = Ahmed glaucoma valve implantation; ANA = antinuclear antibodies; Durezol = difluprednate eye drops; ETN = etanercept; IFX = infliximab; LAD = lymphadenopathy; MFC = multifocal choroiditis; MTX = methotrexate; Ozurdex = injectable intravitreal dexamethasone implant that releases steroid for 3 mos; PF = prednisolone forte or prednisolone acetate eye drops; Retisert = surgically placed fluocinolone implant; STK = periocular (subtenon) Kenalog injection; SUN = Standardization in Uveitis Nomenclature criteria for quantifying anterior chamber cell and flare.
At the time of sampling.
For uveitis patients, AC sampling was performed using a 25- to 30-gauge needle to extract approximately 100 to 200 μl of AC fluid and cells. The aqueous sample was centrifuged at 400g for 5 minutes and the aqueous fluid was removed and frozen at –80° C. Blood samples were obtained by venipuncture, collected into edetic acid tubes, and purified by Ficoll-Hypaque density gradient centrifugation. The AC cells and peripheral blood mononuclear cells (PBMCs) were cryopreserved in fetal bovine serum (FBS) with 10% dimethyl sulfoxide (DMSO) and stored at –140° C.
Patients Undergoing Cataract Surgery and Sample Collection
Patients without any clinical history of uveitis undergoing routine cataract surgery were sampled during surgery. At the beginning of surgery, a surgical corneal incision was made, through which the aqueous humor then was collected with a blunt cannula. Ten microliters of aqueous fluid were used for cell counts via a hemocytometer. The remaining aqueous fluid was centrifuged at 400g for 5 minutes, and the aqueous fluid was removed. Cells were resuspended in phosphate buffered saline (PBS) with 2% FBS for flow cytometry.
Flow Cytometry
Anterior chamber cells and PBMCs were stained with antibodies targeting CD45, CD3, CD4, CD8, and CD19 (Supplemental Table 1), washed in PBS with 2% FBS, fixed with 2% paraformaldehyde, and run on a FACSCanto (BD Biosciences).
Single-Cell RNA Sequencing
Frozen AC cells were thawed and washed once with FBS and once with PBS with 0.1% bovine serum albumin (BSA). Because of low cell numbers (5000–30 000 cells), minimal processing was performed to reduce cell loss. Peripheral blood mononuclear cells were thawed and washed twice with 10% Roswell Park Memorial Institute Media (RPMI) and then once with PBS with 0.1% BSA. Viability was more than 95% by trypan blue exclusion. Single-cell 5′ gene expression complementary DNA (cDNA) libraries were generated using the Chromium Controller (10x Genomics) platform for microdroplet-based, single-cell barcoding at the McDonnell Genome Institute (Washington University in St. Louis, St. Louis, MO) and were sequenced on the NovaSeq Sequencing System (Illumina).
Single-Cell RNA Expression Analysis
Sequencing reads were aligned to the human genome using Cell Ranger version 3.0.2 (10x Genomics). Quality control metrics are provided in Supplemental Table 2. Cells that had more than 11% of mitochondrial gene content were excluded from analysis. Given the cell-type variable gene number per unique molecular identifier (UMI) count, we did not use these methods to eliminate putative doublet cells. Instead, we identified doublets based on coexpression of lineage-defining markers. The publicly available Seurat R software package version 3.1 was used for normalization (SCTransform) and clustering of immune cells. Dimensionality reduction was carried out with the runPCA function, and t-distributed stochastic neighbor embedding was carried out with the runtSNE function.8 For lineage assignment, cells were assigned to a lineage or cell state based on canonical gene expression of the entire cluster to account for the effect of dropout.9 For lineage-specific analysis, immune cells within a cluster expressing either CD3D (T cells) or CD79A (B cells) were separated from the remaining clusters underwent subsequent principle component analysis (PCA) analysis, t-distributed stochastic neighbor embedding projection, and reclustering. Quantitative estimation of CD4 and CD8 T-cell proportions was based on the counts of ocular cells from CD4 or CD8 T-cell clusters (T1–T10). To generate expression values for heatmaps, mean expression of each gene for each cluster and patient was calculated. Heatmaps were generated using the publicly available web application Phantasus.10 Experimentally defined gene sets were obtained from previously published sources (Supplemental Tables 3 and 4).11, 12, 13, 14, 15, 16, 17, 18, 19 Heatmaps for individual genes represent the Z score of normalized average expression value of cells within the group of cells designated by the heatmap annotation bars (cluster, patient, and tissue). Heatmaps for gene sets represent average Z score of all genes in the set. Cells of a particular subtype were plotted only if that group represented at least 5% of the cells within that cluster.
Single-Cell T-Cell Receptor and B-Cell Receptor Processing
T-cell and B-cell enrichment libraries were generated with the Chromium Single Cell V(D)J Enrichment Kit (10x Genomics) at the McDonnell Genome Institute and were sequenced on the NovaSeq Sequencing System. Samples were aligned with Cell Ranger VDJ command. For T-cell receptor (TCR) downstream analysis, only clonotypes that (1) belonged to cells with transcriptional profile that passed quality control and (2) with 1 or 2 productive rearrangements for α and 1 productive rearrangement for β chain were used. Because of the intrinsic oligoclonality of unconventional T cells, mucosal associated T and natural killer T TCRs were filtered out based on V/J gene match.20 This allowed for a more direct comparison of conventional αβ T cells between the eye and blood as mucosal-associated T cells and natural killer T cells were only 0.00% to 0.25% of ocular T-cell clonotypes, but 0.0% to 2.0% of blood clonotypes. Clonotype correspondence with T-cell type was established based on expression of either CD4 or CD8A/CD8B genes. In cases when none of these genes was expressed because of gene dropouts, T-cell type was assigned based on cluster identity.
For B-cell receptor (BCR) analysis, only cells that (1) belonged to cells with transcriptional profile that passed quality control and (2) with 1 productive rearrangement for heavy chain and 1 productive rearrangement for light chain (either κ or λ) were included. Frequencies of clonotypes were counted based on number of cells that passed general expression quality control and share of each clonotype. To increase the sensitivity in identifying class-switched clonotypes for clonally expanded B cells, all B-cell clonotypes were interrogated for CDR3 homology, regardless of the number of productive heavy- or light-chain rearrangements.
HLA Genotyping
HLA genotyping was performed by Histogenetics LLC (Ossining, NY).
Data Availability
FASTQ files are available via dbGAP. Anonymized scRNA-seq is made available via the following links: (1) total CD45 eye plus PBMC, https://artyomovlab.wustl.edu/sce/?token=uveitis_eye_and_PBMC; (2) T-cells eye plus PBMCs, https://artyomovlab.wustl.edu/sce/?token=uveitis_eye_and_PBMC; (3) B-cells eye plus PMBCs, https://artyomovlab.wustl.edu/sce/?token=uveitis_eye_and_PBMC_B_cells; and (4) total CD45 eye only, https://artyomovlab.wustl.edu/sce/?token=uveitis_eye.
Study Approval
All human participants were enrolled after obtaining signed informed consent in accordance with the tenets of the Declaration of Helsinki and the institutional review board of Washington University in St. Louis (identifiers, 201704141 and 201912043). Written informed consent was received from participants before inclusion in the study.
Results
High-Resolution Profiling of Ocular Inflammatory Cells
To delineate the cellular and molecular components of granulomatous uveitis, we sampled the AC of 4 patients during an active disease flare. Although patient-to-patient variation was present in terms of age and duration of symptoms, most had inflammation localized to the anterior and vitreous chambers (i.e., anterior or intermediate uveitis), and 3 of 4 patients were not receiving systemic immunomodulatory therapy (Table 1, Table 2). We next identified clinical features unique to each patient. For example, 2 individuals had chronic inflammation (patients UV031 and UV150) that was recalcitrant to systemic immune suppression. One individual (patient UV150) was sampled before systemic immune suppression, and the other individual (patient UV031) was sampled after therapy failed for years. The other 2 patients (patients UV170 and UV174) had recurrent disease with flares separated by months to years, and 1 of these (patient UV170) also had evidence of prior chorioretinal inflammation.
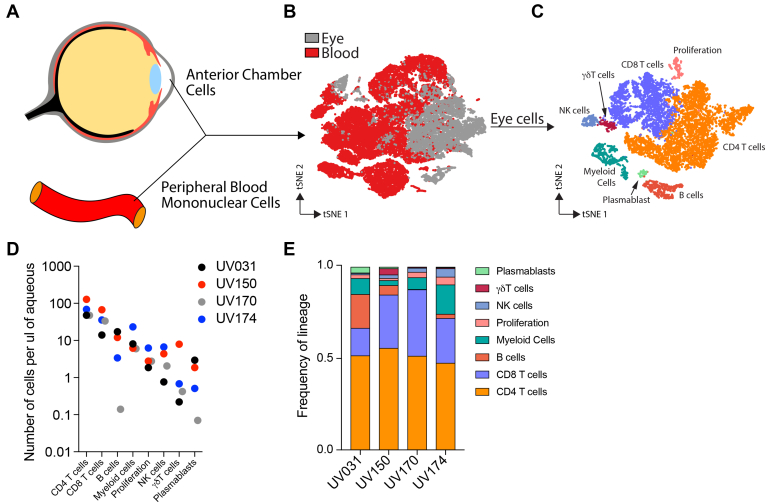
We used scRNAseq to examine the immune cells from the inflamed eyes of all 4 patients. Although it is broadly accepted that the AC is acellular in the healthy eye, previous reports have suggested that ocular lymphocytes can be extracted during routine cataract surgery,21,22 which could serve as a control for immune cells in granulomatous uveitis. However, when we analyzed aqueous fluid from 26 healthy eyes undergoing cataract surgery, no detectable hematopoietic cells were found in the anterior chamber (Supplemental Fig 1). In the absence of immune cells in healthy ocular samples for comparison, we used peripheral PBMCs from each patient as a control to identify transcriptional signatures unique to the inflamed eye during granulomatous uveitis (Fig 1A). We visualized the data in 2 dimensions via t-distributed stochastic neighbor embedding, an unsupervised nonlinear dimensionality reduction algorithm (Fig 1B) that plots cells with similar gene expression together, separating cells with distinct transcriptional programs.23,24
Figure 1.
High-resolution transcriptional profiling of ocular inflammatory cells. CD4 T cells are the most frequent immune cell across patients with variable contribution of other cell types. A, Diagram showing how cells were collected from the anterior chamber and the peripheral blood for single-cell RNA sequencing. B, Diagram showing t-distributed stochastic neighbor embedding (tSNE) visualization of blood and ocular immune cells colored by tissue source. C, Diagram showing ocular immune cells on a tSNE visualization, colored by cell type. D, Graph showing concentration of ocular immune cells for each patient and cell type. E, Bar graph showing frequency of ocular immune cell types for each patient. NK = natural killer.
Most ocular cells clustered separately from blood cells (Fig 1B), indicating that the inflamed eye imparts a distinct transcriptional signature on immune cells. Within the eye, we identified 8 major cell types based on canonical gene expression (Fig 1C, Supplemental Fig 2). CD4 T cells consistently were the most abundant cell type, representing approximately 50% of ocular cells in each patient; however, patient-to-patient variation was present in the proportions of the other cell types, that is, CD8 T cells, B cells, myeloid cells, natural killer (NK) cells, γδ T cells, and plasmablasts (Fig 1D, E). Because the transcriptional program associated with cellular proliferation exerted an effect on cell clustering that dominated over lineage identity, a separate cluster associated with cell division (proliferation) was composed of a mix of lineages, particularly T and B cells. Thus, scRNAseq analysis revealed commonalities in the intraocular immune cell milieu across all patients, while highlighting patient-specific differences.
We next defined the transcriptional program of immune cells within the eye. To accomplish this, we directly compared each ocular cell type with its peripheral blood counterpart by grouping our initial dataset into 4 cell types: myeloid cells (LYZ+), NK cells and innate lymphoid cells (ILCs) (NCR1+), T cells (CD3D+), and B cells (CD79A+; Supplemental Fig 3). This division allowed for a separate subanalysis of each cell type.
Innate Ocular Immune Cells Show Evidence of Antigen Presentation and Reduced Cytotoxicity
To study ocular myeloid cells, we compared LYZ+ myeloid cells from the eye and blood in a separate subanalysis of our larger dataset (Supplemental Fig 4). Ocular myeloid cell clusters expressed higher levels of major histocompatibility complex (MHC) II (HLA-DQA1) along with conventional dendritic cell genes (FLT3 and NDRG2), indicating they contained activated, antigen-presenting dendritic cells. However, robust differentiation of the clusters was limited. For example, macrophage genes (e.g., CD163) also were expressed by cells in the same clusters, suggesting that these 2 cell types were distinguished imperfectly, likely because of the overlapping gene expression between myeloid cell types or limited cell numbers (Supplemental Fig 4). We anticipate that recruiting additional patients and subsequent analysis of an expanded dataset will facilitate more robust analysis, and potentially better resolution, of numerically underrepresented myeloid cell subtypes.
To study NK cells, we similarly compared NCR1+ ocular and peripheral blood cells in a separate subanalysis (Supplemental Fig 5). Ocular NK cells expressed lower FCGR3A, GZMB, and CXCR3 compared with peripheral blood NK cells. Thus, most peripheral blood NK cells had transcriptional profiles similar to previously defined CD56dim NK cells, whereas uveitis NK cells were transcriptionally similar to less cytotoxic CD56bright NK cells (Supplemental Fig 5).
T-Cell or B-Cell Clones Undergo Extensive Expansion in Granulomatous Uveitis
To determine if differences in lymphocyte frequency could be the result of preferential expansion of certain T- or B-cell clones, we performed single-cell TCR and BCR sequencing. We first used the Gini coefficient to measure the diversity and clonality of ocular and blood T and B cells, with 0 representing no clonal expansion and 1 representing a monoclonal population. CD4 T-cell clonality was increased in the eye versus the blood in all 4 patients, whereas CD8 T-cell clonality was increased in 2 patients (patients UV150 and UV170; Fig 2A). B-cell clonality also was increased in the eye in 2 patients (patients UV031 and UV150; Fig 2A); however, fewer B cells were sampled in patients UV170 and UV174 (Fig 1D, E), which limited our ability to identify B-cell clonality in these patients conclusively.
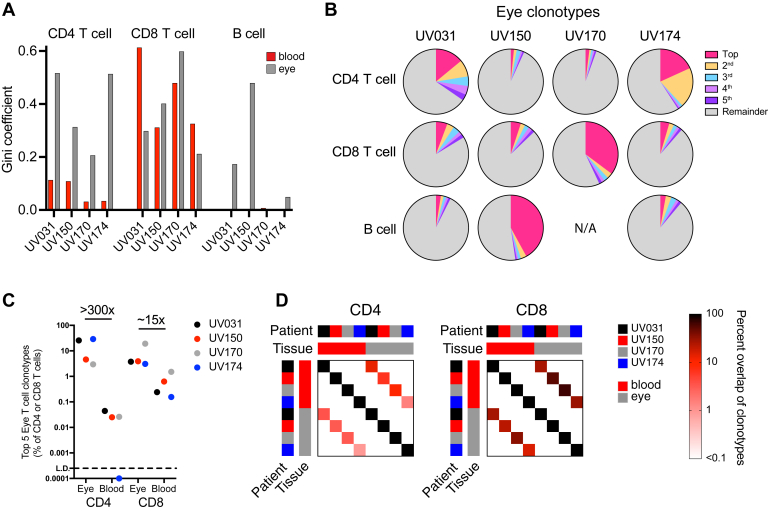
Figure 2.
Highly expanded T- and B-cell clonotypes in the eye. Robust clonal expansion is seen in a single ocular T- or B-cell lineage, with ocular CD8 T-cell clonotypes more abundant in the blood compared with CD4 T-cell clonotypes. A, Bar graph showing T- and B-cell diversity and clonality in blood and ocular T or B cells for each indicated patient, as measured by the Gini coefficient, where on a scale from 0 to 1, 0 indicates that all sequences have the same frequency and 1 indicates that the repertoire is dominated by a single sequence. B, Pie charts showing the proportions of the 5 most frequent ocular CD4 T-cell, CD8 T-cell, or B-cell clonotypes for each individual. C, Graph showing composite frequency of the top 5 CD4 and CD8 T-cell clonotypes from (B) in both the eye and blood for each patient. D, Graphs showing percent overlap of all CD4 and CD8 T-cell clonotypes in the blood and eye from all patients. Percentage is calculated as the number of shared individual clonotypes in each row-column intersection divided by the total number of clonotypes in each column without regard to clonotype frequency. Overlap is detected only between blood and eye samples within a single patient, and not between patients.
We next determined whether the increased clonality was the result of expansion of a few select lymphocyte clones. Given the possibility of multiple cell-specific barcodes arising from the same cell,25 we set a cutoff of 8 or more identical BCR and TCR sequence reads as indicative of clonal expansion. We found that the increased clonality of ocular lymphocytes, associated with a Gini coefficient of more than 0.4, was the result of 2 to 5 expanded CD4 T-cell clonotypes (patients UV031 and UV174), a single expanded CD8 clonotype (patient UV170), or a single expanded B-cell clonotype (patient UV150; Fig 2B; Supplemental Tables 5–7). Thus, each individual had only 1 lymphocyte lineage with highly expanded clonotypes, revealing that despite the shared predominance of CD4 T cells in all 4 patients, the dominant clonally expanded lymphocyte was unique to each patient.
To explore whether the expanded ocular clonotypes also were found in the systemic circulation, we compared the relative frequency of the top 5 ocular clonotypes in each patient between eye and blood populations (Fig 2C). Each of the top 5 ocular CD4 or CD8 clonotypes were detected frequently in the eye, accounting for 3% to 30% of all CD4 or CD8 T cells. In the peripheral blood, the same top 5 ocular CD8 T-cell clonotypes were less common, constituting between 0.15% and 1.5% of blood CD8 T cells, whereas the top 5 ocular CD4 T-cell clonotypes were detected very rarely, at only 0.0% to 0.04% (Fig 2C). The 20-fold difference in tissue-specific enrichment between CD4 and CD8 T cells occurred despite similar CD4-to-CD8 T-cell ratios across all tissues and patients (1.0:3.3). Similarly, a higher percentage of all CD8 T-cell clonotypes were shared between the eye and blood compared with CD4 clonotypes within each patient (Fig 2D). Of note, no overlap of either eye or blood T-cell clonotypes was found between individuals (Fig 2D), which may be the result of differences in HLA genotype (Supplemental Table 8), antigenic stimulus, initial variable, diversity and joining (VDJ) recombination, or a combination thereof. Additionally, the greater CD8 T-cell clonality in the blood in 2 individuals (patients UV031 and UV170; Fig 2A) likely was the result of oligoclonal expansions against antigens unrelated to the eye. Ocular B-cell clones were not detected in the peripheral blood, potentially because of the lower frequency of B cells in the blood, and thus smaller number of B cells being sequenced. Collectively, these data suggest that clonally expanded CD4 T cells in granulomatous uveitis display greater ocular-specific enrichment than do CD8 T cells.
CD4 T Cells Show Individualized Combinations of Ocular T Helper 1, T Helper 1/T Helper 17, and Regulatory T Cells
A focused T-cell subanalysis was segregated into 12 unique clusters (Fig 3A). Minimal coclustering between eye and blood T cells again was observed, such that only 1 cluster (T8) contained cells from both tissues (Fig 3A), indicating that most ocular T cells were transcriptionally distinct from peripheral blood T cells. Although the proportion of blood T-cell clusters showed minimal variation across patients, ocular T-cell clustering was notably more variable (Supplemental Fig 6A), raising the possibility of individualized T-cell responses.
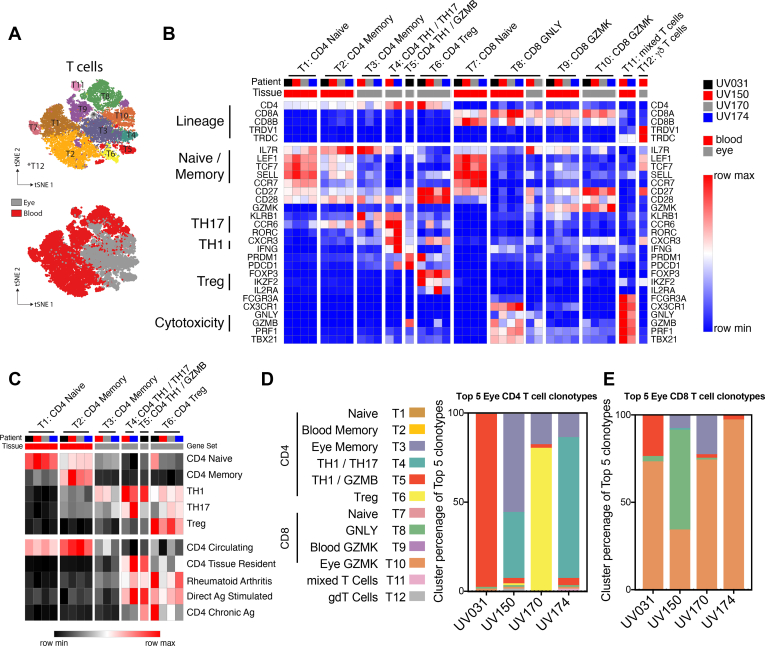
Figure 3.
CD4 T cells show individualized combinations of ocular TH1, TH1/TH17, and regulatory T (Treg) cells, whereas CD8 T cells share a common transcriptional program. Gene expression in effector CD4 T cells reflects both patient-to-patient variation and shared signatures of tissue residency, antigen exposure, and rheumatoid arthritis. In contrast, ocular CD8 T cells across patients share increased expression of intermediate differentiation markers and reduced expression of classic cytotoxic molecules. A, Diagram showing t-distributed stochastic neighbor embedding subanalysis of blood and ocular T cells colored by cluster (top panel) or tissue source (bottom panel). B, Heatmap representation of relative gene expression of cluster-defining genes (rows) for each patient (columns black, red, grey, blue) and tissue (blood, red; eye, grey). Clusters are annotated with cell lineages defined above. Samples with less than 5% contribution to each cluster were excluded. Clusters are annotated with lineage and functional subset. Genes defining specific T-cell states or functions are indicated. C, Heatmap representation of relative expression of cell lineage or state-defining gene set expression between T-cell clusters. D, E, Bar graphs showing percent of T-cell cluster occupancy for the top 5 ocular (D) CD4 and (E) CD8 T-cell clonotypes.
To identify distinct CD4 T-cell states in the blood and the inflamed eye, we examined canonical gene expression as well as unique gene sets that characterized each cluster (Fig 3B, Supplemental Figs 6B and 7). Canonical genes were used to identify naïve CD4 T cells (T1) in the blood and memory CD4 T cells in the blood (T2) and eye (T3; Fig 3B). Additional genes upregulated in distinct ocular clusters revealed effector T helper (TH1)/TH17 cells (T4), effector TH1 cells expressing GZMB (granzyme B) (T5), and regulatory T (Treg) cells (T6; Fig 3B). Furthermore, we used empirically defined gene sets to validate the assignment of naïve, memory, TH1, TH17, and Treg cell states (Fig 3C). Thus, CD4 T cells in patients with granulomatous uveitis comprise individualized combinations of TH1, TH1/TH17, and Treg cells that were determined more clearly because of the large numbers of T cells, in contrast to innate cells.
Having delineated the types of T cells present in the eye, we determined which T-cell states were represented by the expanded clonotypes, because these clones may represent the dominantly pathogenic cells in each individual. To determine the transcriptional profile of expanded CD4 T-cell clones, we collated the top 5 clonotypes that were most abundant in the eye compared with the blood for each of the 4 patients. This analysis included CD4 T-cell clonotypes with both robust (patients UV031 and UV174) and mild (patients UV150 and UV170) clonal expansion. We found that the transcriptional program of these ocular-biased CD4 T-cell clonotypes varied by individual (Fig 3D). In 3 of the 4 patients (patients UV031, UV150, and UV174), the expanded CD4 T-cell clonotypes were TH1/TH17 cells (T4) or TH1 cells (T5). By contrast, for the fourth individual (patient UV170), the expanded CD4 T-cell clonotypes were mainly Tregs (T6; Fig 3D). In this individual, the expanded Treg population coexisted with clonally expanded CD8 T cells (Fig 2B), suggesting an immunoregulatory instead of a proinflammatory function for CD4 T cells in this patient. Thus, the patient-specific variability in clonal expansion is paralleled by CD4 T-cell variation.
CD4 T Cells Share Signatures of Tissue Residency, Autoimmunity, and Antigen Stimulation
Given the robust enrichment of clonally expanded CD4 T cells in the eye, we explored whether specific clusters would have evidence of tissue residency. We found that a gene signature of tissue-resident CD4 T cells26 was expressed preferentially in the TH1 and TH1/TH17 effector clusters (T4 and T5) in the eye, whereas a circulating gene signature was enriched in the blood (T1 and T2; Fig 3C). The effector CD4 T cells in the eye also shared similar transcriptional programming with peripheral helper T cells from rheumatoid arthritis joints11 (Fig 3C). Consistent with the presence of clonally expanded cells in clusters T4, T5, and T6, these cells also expressed genes upregulated in response to direct antigen stimulation in all patients (Fig 3C). In contrast, genes associated with chronic antigen exposure were expressed selectively by both effector and regulatory CD4 T cells from patient UV031, consistent with that individual’s history of prolonged, uncontrolled inflammation (Fig 3C). Thus, compared with memory CD4 T cells within the eye, ocular effector CD4 T cells expressed signatures of tissue residency, antigen-specific stimulation, and peripheral helper T cells.
Ocular CD8 T Cells Share a Common Transcriptional Program Characterized by Reduced Expression of Cytotoxic Molecules
In contrast to CD4 T cells, CD8 T cells demonstrated more transcriptional uniformity in the eye. Circulating naïve CD8 T cells (T7) were identified by SELL, CCR7, TCF7, and LEF1, whereas circulating memory CD8 T cells were divided by CX3CR1, GNLY, and GZMB expression (T8) versus CD27 and GZMK expression (T9), markers of late and intermediate CD8 T-cell differentiation, respectively (Fig 3B).27,28 However, ocular CD8 T cells were located principally in a separate cluster (T10) with features of intermediate differentiation (CD27, CD28, and GZMK) and reduced expression of canonical cytotoxic molecules (GNLY, GZMB, and PRF1; Fig 3B). In addition, the minor population of ocular CD8 T cells that clustered with late differentiated blood cells (T8) similarly showed lower expression of GNLY, GZMB, and PRF1 compared with blood cells within the same cluster (Fig 3B), suggesting either selective recruitment or transcriptional reprogramming of less cytotoxic CD8 T cells within the inflamed eye. Of note, a small number of ocular CD8 T cells clustered with ocular memory CD4 T cells (T3) and TH1 cells (T5; Fig 3B, Supplemental Figs 6B and 7).
Pairing of TCR sequencing with transcriptional analysis revealed that the clonally expanded CD8 T cells from patient UV170 were located within the CD27 and GZMK cluster (T10; Fig 3E). Additionally, the top CD8 T-cell clonotypes from the other patients also largely clustered in T10 (Fig 3E), or in the transcriptionally similar GNLY cluster in the case of patient UV150. Thus, clonally expanded CD8 T cells in the inflamed eye are enriched in transcripts associated with intermediate differentiation, polyfunctional cytokine expression, and reduced cytotoxicity.
Ocular B Cells in Granulomatous Uveitis Share a Transcriptional Profile Characteristic of Chronic Autoimmune B Cells
Targeted analysis of ocular and blood B cells (Supplemental Fig 3) revealed 9 clusters with ocular and blood cells occupying both shared and unique clusters (Fig 4A). B-cell clusters were distributed similarly among 3 of 4 patients in the peripheral blood; however, the frequency of ocular B cells varied by patient (Supplemental Fig 8A). Notably, patient UV170 had a relative paucity of ocular and peripheral blood B cells (Supplemental Fig 8B) and, although transcriptionally similar to the other ocular samples, was excluded from subsequent analysis. Notably, although her recurrent disease was long-standing, her uveitis flare was fairly acute at the time of sampling, whereas the other 3 patients had longer-standing ocular inflammation; thus, aqueous B cells may be a feature of chronic granulomatous uveitis.
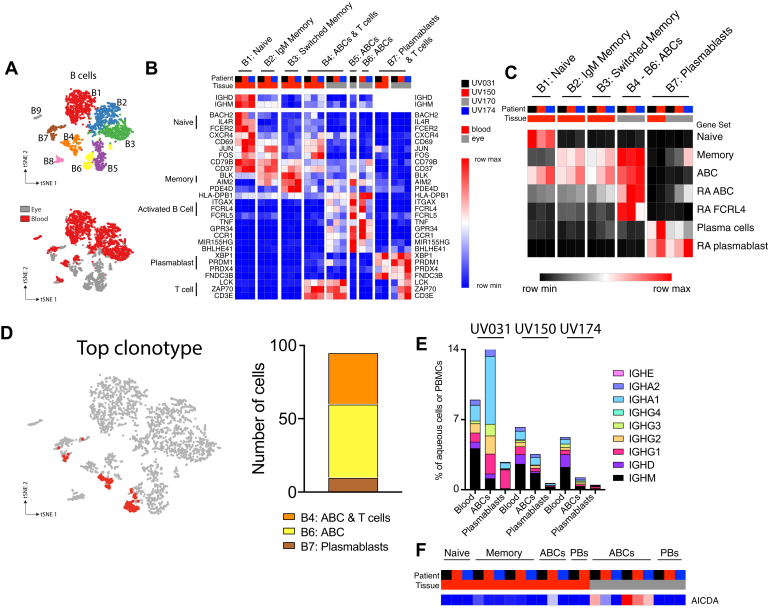
Figure 4.
Ocular B cells are activated B cells with transcriptional similarities to synovial B cells in rheumatoid arthritis (RA). Clonally expanded B cells demonstrate evidence of plasmablast differentiation and class switching. A, Diagram showing t-distributed stochastic neighbor embedding subanalysis of blood and ocular B cells colored by cluster (top panel) or by tissue source (bottom panel). B–E, Clusters B8 and B9 were excluded from further analysis because these clusters represented predominantly myeloid cells and erythrocyte precursors, respectively. B, Heatmap representation of relative gene expression of cluster-defining genes (rows) for each patient (columns black, red, grey, blue) and tissue (blood, red; eye, grey). Clusters are annotated with cell lineages defined above. Genes specific to B-cell subsets and T cells are indicated with labeled brackets. Samples with less than 5% contribution to each cluster were excluded. C, Heatmap representation of relative expression of cell lineage or state-defining gene set expression between B-cell clusters. Samples with less than 5% contribution to each cluster were excluded. D, Diagram showing distribution of the top expanded B-cell clonotype from patient UV150 between activated B cell (ABC; B4 and B6) and plasmablast (B7) clusters. E, Concentration of each immunoglobulin subclass in blood, ocular ABCs, and ocular plasmablasts clusters for each patient. Relative frequency of switched non-immunoglobulin M (IgM) subclass increases with differentiation. F, Relative expression of AICDA, found primarily in ABCs. PB = plasmablast.
The cellular identity of each B-cell cluster was defined based on expression of canonical genes (Fig 4B, Supplemental Figs 8C and 9). Most ocular B cells (B4–B6) were activated B cells (ABCs), characterized by expression of ITGAX (CD11c) and FCRL4 in addition to genes shared with peripheral blood memory B cells (Fig 4B). A smaller proportion of ocular B cells were identified as plasmablasts (B7) based on their expression of XBP1 and PRDM1 (Fig 4B).
The T-cell transcript CD3E and full-length TCR sequences were detected in ABCs and plasmablast clusters B4 and B7 (Fig 4B, Supplemental Figs 8C, 10A, B; Supplemental Table 9), likely because of doublet cells, or 2 cells that were simultaneously sequenced in the same droplet. Although these doublets may represent nonspecifically associated cells partitioned to the same droplet scRNAseq, it remains possible that they reflect biologically relevant immunologic synapses because this phenomenon occurred only in select clusters, yet was present in all patients, in both blood and ocular B cells. Alternatively, dual expressing cells with BCR and TCR expression recently were described in the peripheral blood of patients with autoimmune diabetes29; however, the gene expression profile of dual expressing cells was not shared by the doublets from our dataset (Supplemental Fig 10C). Nevertheless, additional image-based and functional analyses would need to be performed to determine whether autoimmune uveitis contains similar pathogenic dual expressing cells.
Ocular ABCs cells also expressed high levels of MHC class II molecules (Fig 4B), suggesting a central role in antigen-driven inflammation. Indeed, gene ontology pathway analysis confirmed that ocular ABCs were enriched for expression of genes associated with antigen presentation (Supplemental Table 10). This finding supports the possibility that the T cell–B cell doublets we identified represent immunologic synapses. Furthermore, ocular ABCs highly expressed TNF (Fig 4B), further supporting a central pathophysiologic role for these cells in granulomatous uveitis. Activated B cells, also referred to as aging-related or autoimmune B cells, are found in multiple chronic inflammatory diseases, including rheumatoid arthritis and lupus nephritis.11,30 Therefore, we used published gene sets to compare ocular B cells in granulomatous uveitis with B cells found in another autoimmune disease, rheumatoid arthritis (Fig 4C).11 We found that ocular ABCs and plasmablasts were transcriptionally similar to the ABCs and plasmablasts isolated from rheumatoid arthritis synovium in 2 separate studies (Fig 4C). Thus, B cells in granulomatous uveitis are characterized by features suggestive of antigen presentation and pathogenicity consistent with chronic autoimmunity.
Evidence for Antigen-Driven B-Cell Differentiation
We next asked whether the clonal expansion of B cells that we observed in 1 patient could shed additional light on the B-cell responses in granulomatous uveitis. We found that the dominant clone in patient UV150 was present in both the ABC and plasmablast clusters (Fig 4D), suggesting that cellular differentiation into antibody-secreting cells occurred within this clonally expanded population. Although most of the BCRs sequenced in the patient UV150 dominant clone were of the immunoglobulin M subclass, a single immunoglobulin A BCR was sequenced, which suggests that class switching occurred within this clonally expanded B-cell population (Supplemental Fig 11). Furthermore, the distribution of immunoglobulin classes within ocular ABCs and plasmablasts was skewed toward switched (non-immunoglobulin M classes) compared with peripheral blood B cells (Fig 4E), suggesting that class switching also may have occurred alongside plasmablast differentiation in all 3 patients. In support of this, AICDA (activation-induced cytidine deaminase), the molecule responsible for immunoglobulin class switching, was expressed by ABCs in all 3 individuals (Fig 4F). Taken together, the data suggest that antigen-driven B-cell clonal expansion and differentiation occur in granulomatous uveitis. Although only 1 patient demonstrated a very large clonally expanded B-cell population, smaller clonal populations were found in the other individuals that were not included because of the rigorous level of detection we set, but may have been analyzable readily had more B cells been sequenced. Thus, in addition to a shared transcriptional profile consistent with antigen presentation to T cells, ocular B cells undergo antidriven responses in granulomatous uveitis.
Discussion
In this study, we explored the transcriptional profile of immune cells in granulomatous uveitis. We coupled transcriptional profiling with antigen-receptor sequence analysis in each individual to identify the putatively antigen-expanded immune cells. This allowed us to draw conclusions about shared features in granulomatous uveitis, as well as to identify patient-specific features, the latter of which suggests that an individualized therapeutic approach may be warranted in the future.
Although the sample size was small, we found 5 key features that were conserved across patients. First, immune cells in the inflamed eye expressed a transcriptional profile that was distinct from peripheral blood immune cells. Second, a shared tissue-residency signature distinguished ocular effector CD4 T cells from peripheral blood T cells across most patients, suggesting that disease relapses could result from local reactivation of adaptive immune cells. Third, ocular B cells showed evidence of antigen-driven differentiation and antigen presentation to T cells, suggesting that they play a central role in the antigen-driven immune responses in some patients with granulomatous uveitis. Fourth, CD8 T cells shared a similar transcriptional program across individuals, revealing a shared inflammatory mechanism characterized by cytokine secretion, rather than cellular cytotoxicity. Finally, ocular CD4 T and B cells shared transcriptional signatures with synovial CD4 T and B cells from patients with rheumatoid arthritis, revealing potential overlapping features of chronic human autoimmunity in disparate organs. Thus, although a similar clinical phenotype may result from different antigenic stimuli in individual patients, as illustrated by the presence of patient-specific expanded clonotypes, these patient-unique triggers seem to result in shared immunologic processes, a finding with important implications for targeted therapies.
B cells have been described in chronic nongranulomatous31, 32, 33 and granulomatous34 uveitis and seem to participate in the formation of ectopic lymphoid structures during chronic uveitis.35 This report, to our knowledge, provides the first evidence of clonal expansion and class switching within the ocular microenvironment. Additionally, the expression of TNF and antigen-presentation genes suggests a pathogenic mechanism whereby antigen-expanded B cells promote antigen-driven T-cell activation. We surmise that B-cell–targeted therapies like rituximab may be particularly beneficial in chronic granulomatous uveitis, particularly where clonal expansion of B cells can be demonstrated.
Although TNF expression in ABCs was shared by our patients, the level of expression was lower in 1 patient (patient UV031) in whom, at the time of sampling, tumor necrosis factor α-inhibitor therapy was failing. It is tempting to speculate that the level of TNF expression therefore could be predictive of response to therapy. However, another patient (patient UV150) in whom tumor necrosis factor α-inhibitor therapy subsequently failed, showed robust TNF expression in ocular ABCs, suggesting that TNF expression level alone may be insufficient to predict treatment response.
We also observed individual variation of effector CD4 T cells. Specifically, effector CD4 T cells in 1 individual (patient UV031) were transcriptionally distinct from effector CD4 T cells in the rest of the cohort (e.g., patients UV150 and UV174). We propose that uncontrolled disease over years in patient UV031 drove this difference, because persistent inflammation and antigen stimulation are dominant factors in CD4 T-cell differentiation.13,36 However, the influence of systemic immunotherapy with methotrexate and etanercept on this altered transcriptional program cannot be excluded.
Although CD4 T cells have been considered the main pathogenic lymphocyte in granulomatous inflammation based on their numeric predominance,37 our study highlights that the frequency of a particular lymphocyte lineage does not predict the degree of clonal expansion. In the most extreme case for 1 patient, B cells accounted for only 5% of ocular lymphocytes, but had the highest degree of clonal expansion. The individualized clonal expansion of T or B cells offers a new framework to understand better the heterogeneity of immune responses seen in human disease38,39 and ultimately may provide a new platform to classify granulomatous uveitis.
Within our dataset, 2 individuals (patients UV150 and UV170) showed a single expanded clonotype from either B or CD8 T cells, respectively, whereas the other 2 patients (patients UV031 and UV174) showed 2 or more codominant clonotypes from CD4 T cells. One possibility is that patients UV150 and UV170 had a shorter disease history of only 2 years compared with 8 years for patients UV031 and UV174, which may have been insufficient time for additional expanded clonotypes to develop. This interpretation may suggest an important role for early and aggressive control of ocular inflammation before additional clonotypes develop and make the inflammatory process more refractory to intervention.
The case of patient UV170, with a clonally expanded CD8 T-cell population and a paucity of effector CD4 T cells and B cells, may suggest that the clonally expanded cells are the sole drivers of inflammation and a potential patient-specific, therapeutic target. In this case, future therapies aimed at elimination of clonally expanded cells have the potential to cure the disease. However, it remains possible that the robust clonal expansion in B or T cells simply reflects the very first clone(s) to be activated and does not represent the singular mediator of ocular inflammation after clinical disease already has been established. In this scenario, therapies aimed at elimination of clonally expanded cells would provide only temporary relief until less frequent clones expand and restart the inflammatory process (Supplemental Fig 12). Thus, future studies likely will need to couple longitudinal ocular sampling with novel, targeted interventions against pathogenic cells40 to discriminate between these 2 possibilities.
Collectively, these data reveal unprecedented details about granulomatous ocular inflammation. Despite limiting our analysis to cells in the aqueous fluid, rather than iris tissue or keratoprecipitates, we identified highly expanded T- and B-cell clonotypes, consistent with antigen-driven responses that were specific to each individual’s disease. These data shed light on pathophysiologic differences between patients with similar clinical diagnoses and suggest that an individualized approach to therapeutic intervention may be warranted. During disease flares, evidence for clonally expanded effector cell populations may guide future therapies specifically to target those cells. Because our analysis is based on a single sampling of each patient, it remains to be determined if the same T- or B-cell clonotypes are expanded in subsequent disease flares. Likewise, future studies will address whether the same lymphocyte clonotypes drive multiorgan disease or whether distinct localized responses arise in parallel in different organs. Finally, TCR and BCR sequencing at the single cell level ultimately may allow for the discovery of antigenic triggers in noninfectious disorders.
Finally, this study provides a foundation for future studies of patients with inflammatory eye disease. Beyond oligoclonality of antigen receptors, differences were found in cellular composition and transcriptional profiles that may reflect disease activity, response to therapy, duration of illness, age, or other factors that could not be discerned in the small number of patients studied to date. Larger numbers of patients will need to be studied in detail to determine if a more refined molecular classification of granulomatous and other forms of uveitis can provide diagnostic and prognostic benefits beyond the current use of clinical criteria alone.
Acknowledgments
The authors thank the 4 uveitis patients and 26 cataract surgery patients in this study for their very generous donations of ocular fluid; Dr. Prabha Ranganathan for clinical evaluation of patients; Drs. Michael Stock, Alex Barsam, and Richard Weider for supervision of collection of ocular fluid during cataract surgery; and Dr. Paul Allen, Dr. Alfred Kim, Dr. Tarin Bigley, and members of the Yokoyama Lab for manuscript review and critical feedback.
Manuscript no. D-20-00029.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s): All authors have completed and submitted the ICMJE disclosures form.
The author(s) have made the following disclosure(s): L.M.H.: Nonfinancial support – Pfizer
Supported by the Barnes-Jewish Hospital Foundation; the Rheumatic Diseases Research Resource-Based Center (grant no.: P30 AR073752); Research to Prevent Blindness, Inc., New York, New York (unrestricted grant to the Department of Ophthalmology and Visual Sciences, Washington University); the National Institutes of Health, Bethesda, Maryland (grant no.: T32 AR007279 [M.A.P.]); the Rheumatology Research Foundation (Scientist Development Award [M.A.P.]); and the Shawn Hu and Angela Zeng Graduate Fellowship (E.E.). The funding organizations had no role in the design or conduct of this research. This manuscript was edited by the Scientific Editing Service supported by the Institute of Clinical and Translational Sciences at Washington University (National Institutes of Health grant no.: UL1 TR002345).
HUMAN SUBJECTS: Human subjects were included in this study. The human ethics committees at Washington University in St. Louis approved the study. All research adhered to the tenets of the Declaration of Helsinki. All participants provided informed consent.
No animal subjects were included in this study.
Author Contributions:
Conception and design: Hassman, M.A.Paley, G.L.Paley, Yokoyama
Analysis and interpretation: Hassman, M.A.Paley, Esaulova, Ruzycki, Artyomov, Yokoyama
Data collection: Hassman, M.A.Paley, G.L.Paley, Linskey, Laurent, Feigl-Lenzen, Springer, Montana, Hong, Enright, James
Obtained funding: Study was performed as part of regular employment duties at Washington University School of Medicine. No additional funding was provided.
Overall responsibility: Hassman, M.A.Paley, Esaulova, Yokoyama
Supplementary Data
References
- 1.Jaffe G.J., Dick A.D., Brezin A.P., et al. Adalimumab in patients with active noninfectious uveitis. N Engl J Med. 2016;375(10):932–943. doi: 10.1056/NEJMoa1509852. [DOI] [PubMed] [Google Scholar]
- 2.Jabs D.A. Immunosuppression for the uveitides. Ophthalmology. 2018;125(2):193–202. doi: 10.1016/j.ophtha.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rathinam S.R., Gonzales J.A., Thundikandy R., et al. Effect of corticosteroid-sparing treatment with mycophenolate mofetil vs methotrexate on inflammation in patients with uveitis: a randomized clinical trial. JAMA. 2019;322(10):936–945. doi: 10.1001/jama.2019.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanski J.J., Bowling B., Nischal K.K., Pearson A. 7th ed. Elsevier/Saunders; Edinburgh; New York: 2011. Clinical Ophthalmology: A Systematic Approach; p. ix. [Google Scholar]
- 5.Chan C.C., BenEzra D., Hsu S.M., et al. Granulomas in sympathetic ophthalmia and sarcoidosis. Immunohistochemical study. Arch Ophthalmol. 1985;103(2):198–202. doi: 10.1001/archopht.1985.01050020050017. [DOI] [PubMed] [Google Scholar]
- 6.Chan C.C., Wetzig R.P., Palestine A.G., et al. Immunohistopathology of ocular sarcoidosis. Report of a case and discussion of immunopathogenesis. Arch Ophthalmol. 1987;105(10):1398–1402. doi: 10.1001/archopht.1987.01060100100036. [DOI] [PubMed] [Google Scholar]
- 7.Jabs D.A., Nussenblatt R.B., Rosenbaum J.T. Standardization of Uveitis Nomenclature Working Group. Standardization of uveitis nomenclature for reporting clinical data. Results of the First International Workshop. Am J Ophthalmol. 2005;140(3):509–516. doi: 10.1016/j.ajo.2005.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stuart T., Butler A., Hoffman P., et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902 e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kharchenko P.V., Silberstein L., Scadden D.T. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11(7):740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zenkova D.K.V., Sablina R., Artyomov M., Sergushichev A. Phantasus: visual and interactive gene expression analysis. https://genome.ifmo.ru/phantasus10.18129/B9.bioc.phantasus Available at: Accessed 12/01/2019. [DOI] [PMC free article] [PubMed]
- 11.Zhang F., Wei K., Slowikowski K., et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. 2019;20(7):928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arlehamn C.L., Seumois G., Gerasimova A., et al. Transcriptional profile of tuberculosis antigen-specific T cells reveals novel multifunctional features. J Immunol. 2014;193(6):2931–2940. doi: 10.4049/jimmunol.1401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford A., Angelosanto J.M., Kao C., et al. Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity. 2014;40(2):289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman A.M., Liu C.L., Green M.R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao D.A., Gurish M.F., Marshall J.L., et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 2017;542(7639):110–114. doi: 10.1038/nature20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bangs S.C., Baban D., Cattan H.J., et al. Human CD4+ memory T cells are preferential targets for bystander activation and apoptosis. J Immunol. 2009;182(4):1962–1971. doi: 10.4049/jimmunol.0802596. [DOI] [PubMed] [Google Scholar]
- 17.Ehrhardt G.R., Hijikata A., Kitamura H., et al. Discriminating gene expression profiles of memory B cell subpopulations. J Exp Med. 2008;205(8):1807–1817. doi: 10.1084/jem.20072682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amara K., Clay E., Yeo L., et al. B cells expressing the IgA receptor FcRL4 participate in the autoimmune response in patients with rheumatoid arthritis. J Autoimmun. 2017;81:34–43. doi: 10.1016/j.jaut.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sintes J., Gentile M., Zhang S., et al. mTOR intersects antibody-inducing signals from TACI in marginal zone B cells. Nat Commun. 2017;8(1):1462. doi: 10.1038/s41467-017-01602-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godfrey D.I., Uldrich A.P., McCluskey J., et al. The burgeoning family of unconventional T cells. Nat Immunol. 2015;16(11):1114–1123. doi: 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- 21.Avunduk A.M., Cetinkaya K., Kapicioglu Z. Comparison of T helper/T suppressor ratios in aqueous humor and blood samples by flow cytometric analysis. Ophthalmologica. 1997;211(2):84–86. doi: 10.1159/000310764. [DOI] [PubMed] [Google Scholar]
- 22.Avunduk A.M., Avunduk M.C., Tekelioglu Y., Kapicioglu Z. CD4+ T cell/CD8+ T cell ratio in the anterior chamber of the eye after penetrating injury and its comparison with normal aqueous samples. Jpn J Ophthalmol. 1998;42(3):204–207. doi: 10.1016/s0021-5155(97)00142-1. [DOI] [PubMed] [Google Scholar]
- 23.van der Maaten L., Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 24.Mair F., Hartmann F.J., Mrdjen D., et al. The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur J Immunol. 2016;46(1):34–43. doi: 10.1002/eji.201545774. [DOI] [PubMed] [Google Scholar]
- 25.Lareau C.A., Ma S., Duarte F.M., Buenrostro J.D. Inference and effects of barcode multiplets in droplet-based single-cell assays. Nature Communications from bioRxiv. 2020;11(1):866. doi: 10.1038/s41467-020-14667-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar B.V., Ma W., Miron M., et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017;20(12):2921–2934. doi: 10.1016/j.celrep.2017.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bengsch B., Ohtani T., Herati R.S., et al. Deep immune profiling by mass cytometry links human T and NK cell differentiation and cytotoxic molecule expression patterns. J Immunol Methods. 2018;453:3–10. doi: 10.1016/j.jim.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiniry B.E., Hunt P.W., Hecht F.M., et al. Differential expression of CD8(+) T cell cytotoxic effector molecules in blood and gastrointestinal mucosa in HIV-1 infection. J Immunol. 2018;200(5):1876–1888. doi: 10.4049/jimmunol.1701532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed R., Omidian Z., Giwa A., et al. A public BCR present in a unique dual-receptor-expressing lymphocyte from type 1 diabetes patients encodes a potent T cell autoantigen. Cell. 2019;177(6):1583–1599 e16. doi: 10.1016/j.cell.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arazi A., Rao D.A., Berthier C.C., et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol. 2019;20(7):902–914. doi: 10.1038/s41590-019-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.George R.K., Chan C.C., Whitcup S.M., Nussenblatt R.B. Ocular immunopathology of Behcet’s disease. Surv Ophthalmol. 1997;42(2):157–162. doi: 10.1016/s0039-6257(97)00026-x. [DOI] [PubMed] [Google Scholar]
- 32.Kalinina Ayuso V., van Dijk M.R., de Boer J.H. Infiltration of plasma cells in the iris of children with ANA-positive anterior uveitis. Invest Ophthalmol Vis Sci. 2015;56(11):6770–6778. doi: 10.1167/iovs.15-17351. [DOI] [PubMed] [Google Scholar]
- 33.Wildschutz L., Ackermann D., Witten A., et al. Transcriptomic and proteomic analysis of iris tissue and aqueous humor in juvenile idiopathic arthritis-associated uveitis. J Autoimmun. 2019;100:75–83. doi: 10.1016/j.jaut.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Lubin J.R., Albert D.M., Weinstein M. Sixty-five years of sympathetic ophthalmia. A clinicopathologic review of 105 cases (1913–1978) Ophthalmology. 1980;87(2):109–121. doi: 10.1016/s0161-6420(80)35270-6. [DOI] [PubMed] [Google Scholar]
- 35.Epps S.J., Coplin N., Luthert P.J., et al. Features of ectopic lymphoid-like structures in human uveitis. Exp Eye Res. 2020;191:107901. doi: 10.1016/j.exer.2019.107901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morou A., Palmer B.E., Kaufmann D.E. Distinctive features of CD4+ T cell dysfunction in chronic viral infections. Curr Opin HIV AIDS. 2014;9(5):446–451. doi: 10.1097/COH.0000000000000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iannuzzi M.C., Rybicki B.A., Teirstein A.S. Sarcoidosis. N Engl J Med. 2007;357(21):2153–2165. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell A.M., Kaiser Y., Falta M.T., et al. Shared alphabeta TCR usage in lungs of sarcoidosis patients with Lofgren’s syndrome. J Immunol. 2017;199(7):2279–2290. doi: 10.4049/jimmunol.1700570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forrester J.M., Wang Y., Ricalton N., et al. TCR expression of activated T cell clones in the lungs of patients with pulmonary sarcoidosis. J Immunol. 1994;153(9):4291–4302. [PubMed] [Google Scholar]
- 40.Ellebrecht C.T., Bhoj V.G., Nace A., et al. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science. 2016;353(6295):179–184. doi: 10.1126/science.aaf6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
FASTQ files are available via dbGAP. Anonymized scRNA-seq is made available via the following links: (1) total CD45 eye plus PBMC, https://artyomovlab.wustl.edu/sce/?token=uveitis_eye_and_PBMC; (2) T-cells eye plus PBMCs, https://artyomovlab.wustl.edu/sce/?token=uveitis_eye_and_PBMC; (3) B-cells eye plus PMBCs, https://artyomovlab.wustl.edu/sce/?token=uveitis_eye_and_PBMC_B_cells; and (4) total CD45 eye only, https://artyomovlab.wustl.edu/sce/?token=uveitis_eye.