SUMMARY
Schizophrenia has a heritability of 60–80%, much of which is attributable to common risk alleles. Here, in a 2-stage genome-wide association study of up to 76,755 people with schizophrenia and 243,649 controls, we report common variant associations at 287 distinct genomic loci. Associations were concentrated in genes expressed in CNS neurons, excitatory and inhibitory, but not other tissues or cell types. Using fine-mapping and functional genomic data, we identify 120 genes (106 protein-coding) as likely to underpin associations at some of these loci, including 16 genes with credible causal non-synonymous or UTR variation. We also implicate fundamental processes related to neuronal function, including synaptic organisation, differentiation, and transmission. Fine-mapped candidates were enriched for genes associated with rare disruptive coding variants in people with schizophrenia, including the glutamate receptor subunit GRIN2A and transcription factor SP4, and were also enriched for genes implicated by such variants in neurodevelopmental disorders. We identify biological processes relevant to schizophrenia pathophysiology, show convergence of common and rare variant associations in schizophrenia and neurodevelopmental disorders, and provide a rich resource of prioritised genes and variants to advance mechanistic studies.
INTRODUCTION
Schizophrenia typically manifests in late adolescence or early adulthood1 and is associated with reduced life expectancy, elevated risk of suicide2, serious physical illnesses3, and substantial health and social costs. Treatments are at least partially effective in most people, but many have chronic symptoms, and adverse treatment effects are common4. There is a need for novel therapeutic target discovery, a process impeded by our limited understanding of pathophysiology.
Much of the between-individual variation in risk is genetic, involving large numbers of common alleles,5 rare copy number variants (CNVs)6, and rare coding variants (RCVs)7,8. A recent genome-wide association study (GWAS) reported 176 genomic loci containing common alleles associated with schizophrenia9 but the causal variants driving these associations and the biological consequences of these variants are largely unknown. To increase our understanding of the common variant contribution to schizophrenia, we performed the largest GWAS of the disorder to date and analysed the findings to prioritise variants, genes and biological processes that contribute to pathogenesis.
RESULTS
Association Meta-Analysis
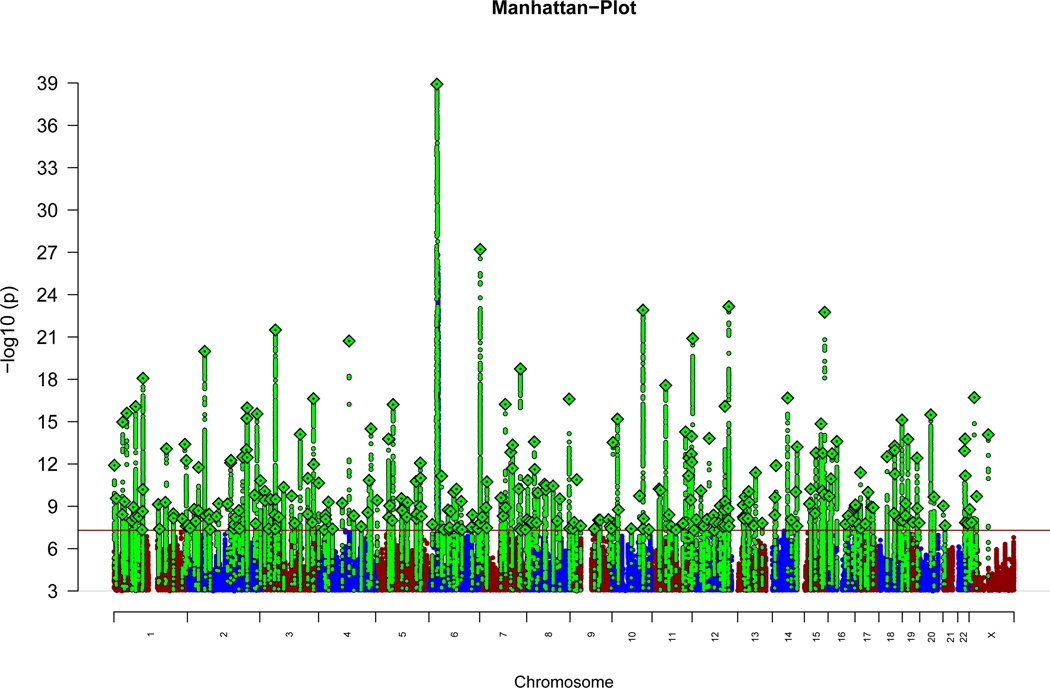
We carried out a primary GWAS in 74,776 cases and 101,023 controls followed by an Extended GWAS which included additional data for the most significant SNPs (Methods). In the primary GWAS, we combined by meta-analysis i) individual genotypes from a core PGC dataset of 90 cohorts of European (EUR) and East Asian (ASN) ancestry from the Psychiatric Genomics Consortium (PGC) totalling 67,390 cases and 94,015 controls. ii) summary-level data from 7,386 cases and 7,008 controls from 9 cohorts of African-American (AA) and Latino (LAT) ancestry10. We analysed up to 7,585,078 SNPs with MAF ≥ 1% in 175,799 individuals of whom 74.3% were EUR, 17.5% ASN, 5.7% AA, and 2.5% LAT (Supplementary Cohort Descriptions). This primary GWAS identified 313 independent SNPs (linkage disequilibrium (LD) r2 < 0.1) that exceeded genome-wide significance (p<5×10−8) (Extended Data Figure 1; Supplementary Table 1), spanning 263 distinct loci.
In the Extended GWAS, we meta-analysed the primary GWAS results with summary statistics from deCODE Genetics (1,979 cases, 142,626 controls) for index SNPs with P<10−5 and identified 342 LD-independent significant SNPS (Supplementary Table 2) located in 287 loci (Supplementary Table 3; Supplementary Figures 1–2). Comparisons with the 128 associations (108 loci) we reported in 2014 are provided (Supplementary Note); one association (rs3768644; chr2:72.3Mb) is no longer supported11.
Separate GWAS for males and females had a genetic correlation statistically indistinguishable from 1 (rg=0.992, SE 0.024). These and other analyses (Supplementary Note) show that common variant genetic liability to schizophrenia is essentially identical in males and females despite reported sex differences in age at onset, symptom profile, course, and outcome12.
SNP-based heritability and Polygenic Prediction
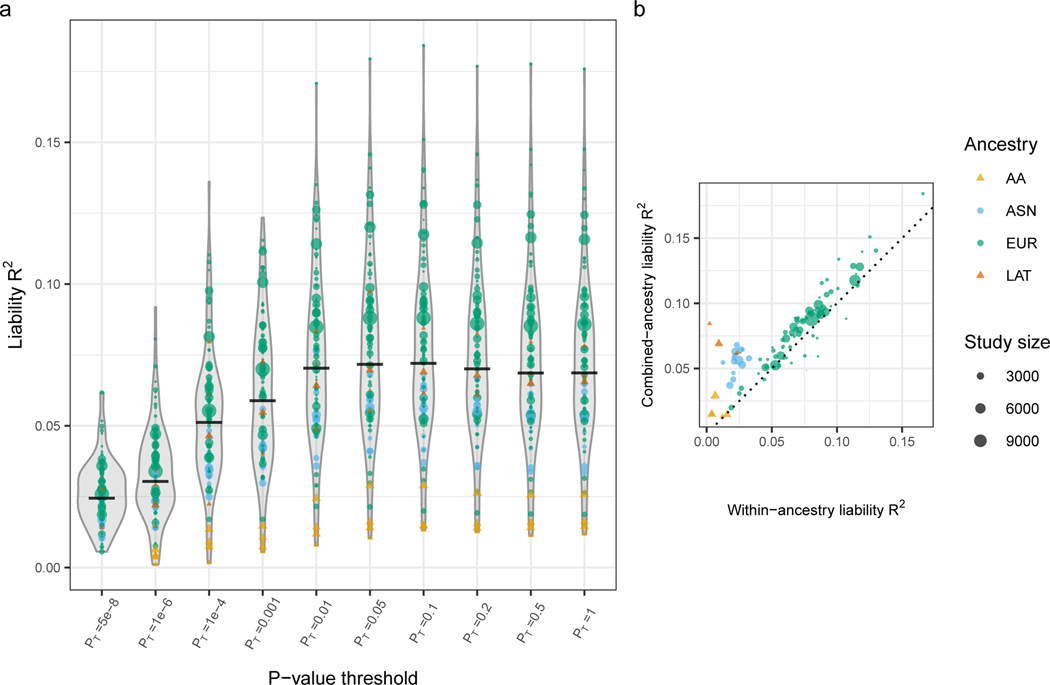
In the EUR sample, the SNP-based heritability (h2SNP) (i.e. proportion of variance in liability attributable to all measured SNPs) was estimated13 to be 0.24 (SE 0.007). Using the all ancestry primary GWAS as the discovery sample, polygenic risk score (PRS) analysis explained a median of 0.073 of variance in liability (SNPs with GWAS p<0.05), and 0.024 when restricted to genome-wide significant SNPs. For almost all cohorts, PRS had more explanatory power based on risk alleles derived from the larger combined ancestry GWAS than from the matched ancestry GWAS; given the ancestry specific sample sizes, unsurprisingly9, this effect was strongest for the non-EUR samples (Extended Data Figure 2 Supplementary Table 5).
PRS explained most variance in liability in cohorts of European ancestry (again a result of the ancestry composition of the GWAS9) and in samples which by ascertainment likely include the most severe cases (hospitalised patients or those treated with clozapine) (Supplementary Note). However, even in EUR cohorts, the median Area Under the Receiver Operating Characteristic Curve (AUC) is only 0.72, meaning the liability explained is insufficient for predicting diagnosis in the general population. Nevertheless, as a quantitative estimate of liability to schizophrenia, PRS has applications in research, and in those contexts, PRS can index substantial differences in liability between individuals in the primary GWAS. Compared to the lowest centile of PRS, the highest centile of PRS has an OR for schizophrenia of 39 (95% CI=29–53), and 5.6 (CI 4.9–6.5) when the top centile is compared with the remaining 99% of individuals (Supplementary Table 6). An extended discussion of heritability and polygenic prediction is provided in the Supplementary Note.
Post-GWAS processing
We next performed a number of secondary analyses in the core PGC dataset in which individual genotypes were available based on fully aligned QC and imputation procedures, and where the data in the HRC reference dataset allowed us to account for LD.
Gene Set Enrichments
Tissue and cell types
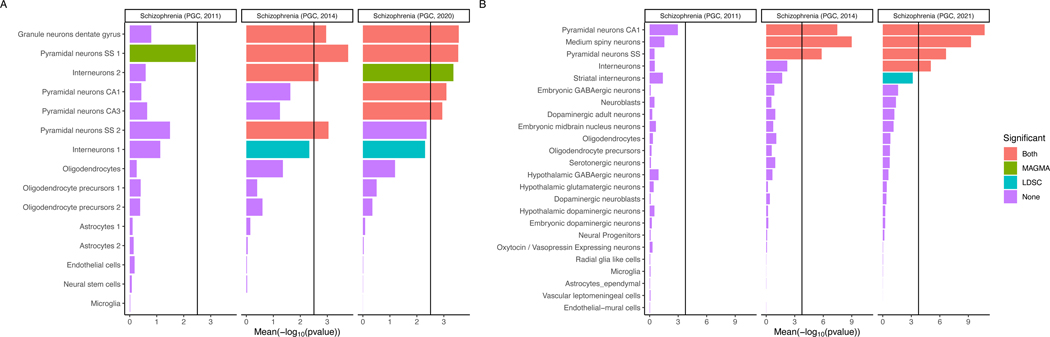
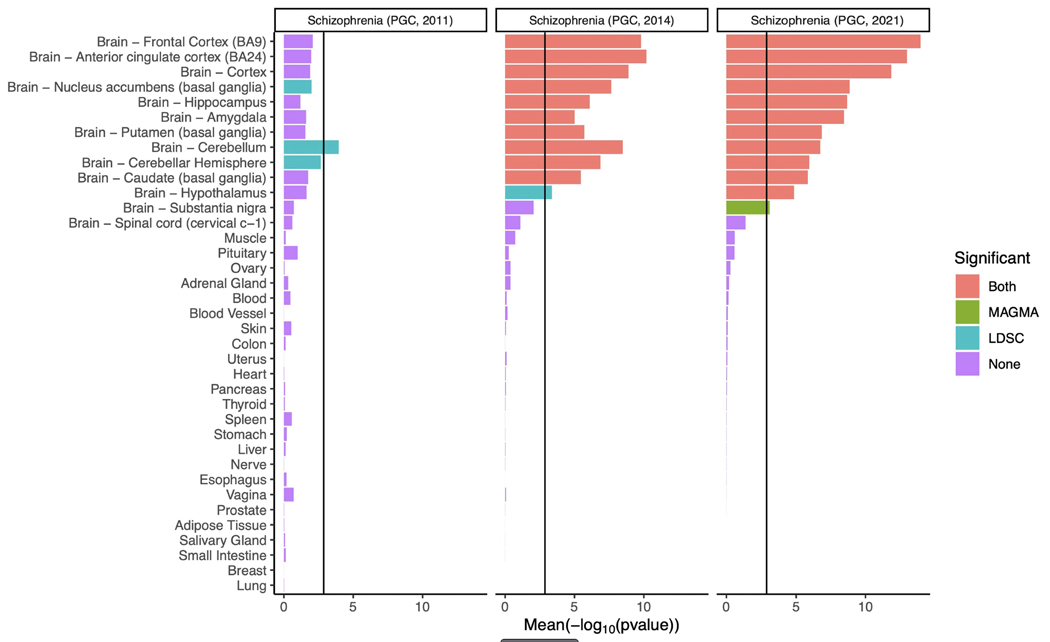
Genes with relatively high specificity for bulk expression in every tested region of human brain14 were significantly enriched for associations (Extended Data Figure 3. Comparison with our earlier studies11,15 shows increasingly clear contrast between the enrichments in brain and non-brain tissues. More strongly than in prior studies16, from human single cell expression data17, we found associations were enriched in genes with high expression in excitatory glutamatergic neurons from cerebral cortex and hippocampus (pyramidal CA1 and CA3 cells, and granule cells of dentate gyrus) and also human cortical inhibitory interneurons (Figure 4a). In mouse single-cell RNA-seq data16, we found similar patterns of enrichments in genes with high expression in excitatory glutamatergic pyramidal neurons from the cortex and hippocampus (Figure 4b), and inhibitory cortical interneurons. We also found associations were enriched in inhibitory medium spiny neurons, the predominant cells of the striatum.
Figure 4: Associations between schizophrenia and cell types from multiple brain regions in human and mouse.
a, b, The mean of the evidence (−log10 P value) obtained from two methods (MAGMA and LDSC) for testing GWAS data for enrichment of associations in genes with high expression in cell types. 15 human cell types (derived from single nuclei) from the cortex and hippocampus (a) and 24 cell types (derived from single-cell RNA-seq) from 5 different brain regions in mouse (cortex, hippocampus, striatum, midbrain and hypothalamus) and from specific enrichments of oligodendrocytes, serotonergic neurons, dopaminergic neurons and cortical parvalbuminergic interneurons (b). Bar colour indicates whether the cell type is significantly associated with both methods, one method or none. The black vertical line represents the significance threshold corrected for the total number of cell types tested in each analysis. Results obtained for previous iterations of schizophrenia GWAS12,18 are shown for comparison. Pyramidal SS, pyramidal neurons from the somatosensory cortex; pyramidal CA1/CA3, pyramidal neurons from the CA1/CA3 region of the hippocampus. Where types of cell (such as interneurons) formed sub-clusters in the source data, these are designated by the suffix 1 or 2.
Supportive results were also obtained using a different dataset of 265 cell types in the mouse central and peripheral nervous system18. Very strong enrichments were again seen for genes expressed in excitatory glutamatergic neurons of the cortex (especially the deep layers) and hippocampus but also the amygdala (Supplementary Figure 3). Highly significant enrichments were also seen for other neuronal populations, including as above, inhibitory medium spiny neurones in striatum, but also both excitatory and inhibitory neurons from the midbrain, thalamus and hindbrain, and inhibitory cells from the hippocampus. There was little evidence for enrichment of genes with highly specific expression in glia or microglia. Overall, the findings across all the datasets are consistent with the hypothesis that schizophrenia is primarily a disorder of neuronal function, but do not suggest that pathology is restricted to a circumscribed brain region.
Associations enriched in Neuronal Ontologies
Of 7,315 gene ontology (GO) classifications 24 were associated with schizophrenia (Supplementary Table 7). All were relevant to neuronal function including development, differentiation, and synaptic transmission, and involved multiple cellular components including ion channels, synapses, and both axon and dendritic annotations. Using the expert-curated ontology of the SynGO consortium19, we further examined the synaptic signal and found that conditionally significant annotations were mainly within postsynaptic terms (Supplementary Tables 8, 9), although enrichment was also found for genes involved in synaptic organisation and signalling.
Gene Prioritisation
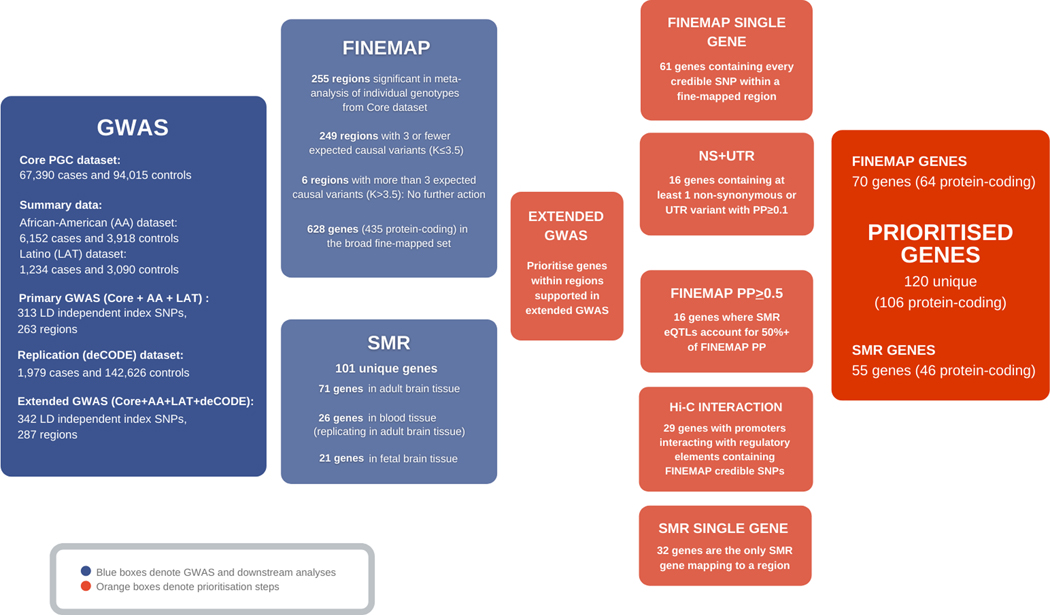
To facilitate biological interpretation and laboratory follow up, we sought to prioritise specific variants and genes most likely to explain associations using a combination of fine-mapping, transcriptomic analysis, and functional genomic annotations. The initial steps in these procedures were necessarily based on 293 index SNPs (255 loci) that attained significance in the core PGC dataset (Methods, Supplementary Table 10), we then focussed on the loci that remained significant in the full Extended GWAS to maximise robustness (Figure 1).
Figure 1: Overview of GWAS and gene prioritisation.
Flow diagram summarising GWAS, fine-mapping and SMR analyses and how these informed gene prioritisation.
Fine-mapping
We performed stepwise analyses (Supplementary Note), conditioning associations in loci on their index SNP (and any subsequent conditionally independent associations) to identify regions that contained independent signals (conditional p<10−6). This analysis supported the existence of independent associations in ~10% of loci (Supplementary Table 10b).
We also employed the Bayesian fine-mapping method implemented in FINEMAP20 to infer the most likely number of distinct causal variants driving our GWAS results. FINEMAP was based on 255 regions determined by the LD clumping procedure (Supplementary Table 11e), after merging clumps if their boundaries physically overlapped and excluding the extended MHC region (Methods). For regions predicted to contain 3 or fewer causal variants (N=249; Figure 1; Supplementary Tables 11a, 11b), we extracted from FINEMAP the posterior probabilities (PP) of being causal for every SNP across the region, and constructed credible sets of SNPs that cumulatively capture 95% of the regional PP (Supplementary Note).
For 33 regions, the 95% credible set contained 5 or fewer SNPs (Supplementary Table 11c) and for 9, only a single SNP. We highlight rs4766428 (PP>0.99) which is the only credible SNP in a locus that contains 25 genes and is located within ATP2A2. Mutations in ATP2A2 cause Darier Disease21, which co-segregates with bipolar disorder in several multiplex pedigrees and is associated with bipolar disorder and schizophrenia at a population level22. ATP2A2 encodes a sarcoplasmic/endoplasmic reticulum calcium pump, suggesting that its role in schizophrenia pathogenesis may be through regulating neuronal cytoplasmic calcium levels. The likely relevance of calcium metabolism is also suggested by enrichment for associations in and around voltage-gated calcium channels (Supplementary Tables 3 and 7).
We denote as our broad fine-map set 628 genes (435 protein coding) that contained at least one credible SNP (Figure 1). To identify the most credible causal genes, we prioritised those mapping to the 287 loci that were genome-wide significant in our Extended GWAS that also contained a) at least one nonsynonymous (NS) or untranslated region (UTR) variant with a PP> 0.1 b) the entire credible set (Supplementary Tables 13, 14). These protein-coding genes had a greater than 3-fold enrichment for loss of function intolerance compared with other protein-coding genes within the loci that were not tagged by credible SNPs (Supplementary Table 15; Supplementary Note), supporting our strategy to delimit credible causal genes.
Among the 70 FINEMAP prioritised genes (64 protein-coding) were 16 genes (protein-coding by definition) based on NS or UTR variants (Supplementary Table 13). These include SLC39A8 in which rs13107325, previously a moderately high credible SNP23, is now strongly supported as causal (PP > 0.99). Other non-synonymous variants with high PP were found in genes with minimal functional characterization including THAP8, WSCD2, and in two E3 ubiquitin ligases PJA1 and CUL9. Missense and UTR variants prioritised interferon regulatory factor 3 (IRF3 while KLF6, a transcription factor, was highlighted by three variants in the 3’ UTR. Finally, we identified 61 genes (55 protein-coding) in which the 95% credible set is restricted to a single gene (Supplementary Table 14).
Prioritisation by Gene Expression
To detect GWAS associations that are credibly explained by eQTLs, that is, variants that influence gene expression, we used summary-based Mendelian randomisation (SMR)24 to find evidence that GWAS signals co-localise with eQTLs (from adult brain25, fetal brain26 or whole blood27) and the HEIDI test24 to then reject co-localisations due to LD between distinct schizophrenia-associated and eQTL variants (Supplementary Table 16). To retain brain relevance, we considered only findings from blood that replicated in brain. After removing duplicates identified in multiple tissues (Supplementary Tables 17a–c), we identified 101 SMR-implicated genes (Supplementary Table 17d); the use of alternative methodologies supported the robustness of the SMR findings (Supplementary Note and Supplementary Table 17e).
We used three approaches to prioritise genes from these 101 candidates (Supplementary Note; Supplementary Tables 17f, 17g, 18). We identified (i) 32 genes as the single SMR-implicated gene at the locus or through conditional analysis of a locus containing multiple candidates: (ii) 16 genes where the putatively causal eQTLs captured 50% or more of the FINEMAP posterior probability (iii) 29 genes where chromatin conformation analysis (Hi-C analysis of adult and fetal brain) suggested that a promoter of that gene interacted with a putative regulatory element containing a FINEMAP credible SNP28.
After removing duplicates, there were 55 SMR/SMR-Hi-C prioritised genes (Supplementary Table 12) of which 46 were protein-coding. Genes where putatively causal eQTLs captured a particularly high FINEMAP PP (>95%) (Supplementary Table 17g) include ACE encoding angiotensin converting enzyme, the target of a major class of anti-hypertensive drugs (schizophrenia under-expression), DCLK3 encoding a neuroprotective kinase29(schizophrenia under-expression) and SNAP91 (discussed below; schizophrenia over-expression).
Combining all approaches, FINEMAP and SMR, we prioritised 120 genes of which 106 are protein-coding (Figure 1; Extended Data Table 1).
Synaptic Location and Function of Prioritised Genes
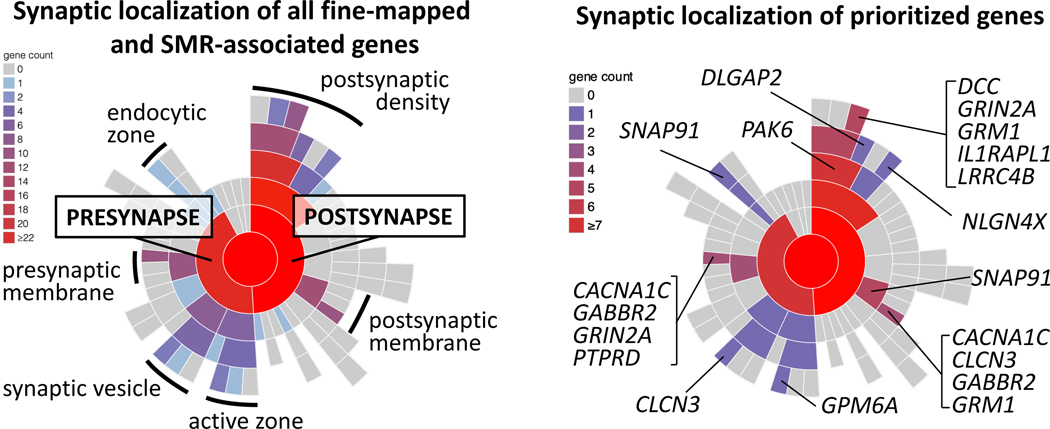
Following the findings from the genome-wide enrichment tests, we examined prioritised genes in the context of synaptic location and function in the SynGO database19 (Figure 3. Of the 106 proteins encoded, 15 have synaptic annotations (Supplementary Table 19); 7 postsynaptic, 5 both pre- and post- synaptic, 2 presynaptic, and 1 gene is not mapped to any specific compartment.
Figure 3: Mapping of all FINEMAP/SMR genes (A) and prioritised genes (B) to synaptic locations using SYNGO.
Sunburst plots depict synaptic locations with child terms in concentric rings, starting with the synapse (center), pre- and postsynaptic locations in the first ring and child terms in subsequent ring. The number of genes in each term is indicated by the colour scheme in the legend. A) FINEMAP/SMR genes are protein coding genes tagged by at least one credible SNP identified by FINEMAP and/or associated using SMR (N=470) of which N=58 are SynGO annotated, 51 to cellular components. B) Prioritised (Extended Data Table 1; N=106) of which 15 are SynGO annotated, 14 to cellular components.
The results are consistent with the genome-wide enrichment tests pointing to postsynaptic pathology. However, many prioritised genes had additional locations suggesting that presynaptic pathology may also be involved. The encoded proteins map to 16 unique biological terms in the hierarchy (Supplementary Table 19), but there are specific themes. Multiple genes encode receptors and ion channels, including voltage-gated calcium and chloride channels (CACNA1C, CLCN3), metabotropic receptors (glutamate (GRM1) and GABA (GABBR2)), and the ligand-gated NMDA receptor subunit (GRIN2A). Others involve proteins playing a role in endocytosis (SNAP91), synaptic organisation and differentiation (DLGAP2, LRRC4B, GPM6A, PAK6), including PTPRD a receptor protein tyrosine phosphatase presynaptic organizer that trans‐synaptically interacts with multiple postsynaptic cell adhesion molecules (e.g. IL1RAPL1), and modulation of chemical transmission (MAPK3, DCC, CLCN3, DLGAP2). The diversity of synaptic proteins identified in this study suggests multiple functional interactions of schizophrenia risk converging on synapses. It remains to be determined whether these interactions occur at a limited set of specific synapse types, or whether the diversity points to multiple types in different brain regions.
Convergence of Common and Rare Variant Associations
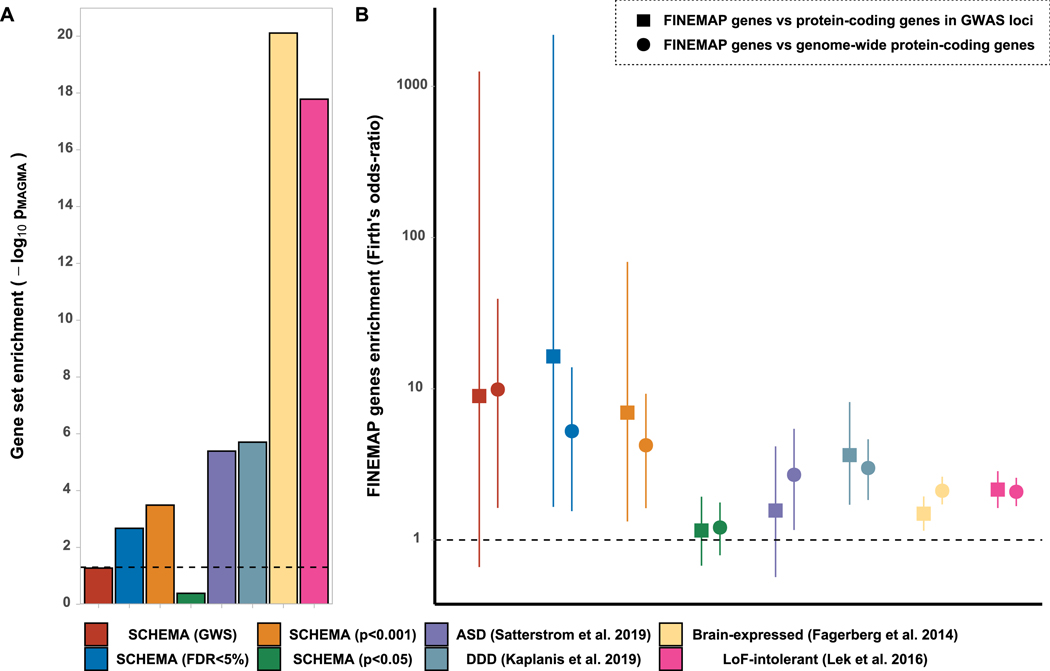
The Schizophrenia Exome Sequencing Meta-Analysis (SCHEMA) consortium (companion paper) identified 32 genes with damaging ultra-rare mutations associated with schizophrenia (FDR<0.05), including 10 at exome-wide significance. We found both sets of genes were enriched for common variant associations, as were more weakly associated SCHEMA genes down to uncorrected P<0.001 (Figure 2a, Supplementary Tables 20, 21). Moreover, within associated loci, protein coding genes containing one or more FINEMAP credible SNPs were enriched for SCHEMA genes relative to other protein-coding genes (Figure 2b; Supplementary Table 21). There are rare variant overlaps in liability to schizophrenia, autism spectrum disorder (ASD) and developmental disorder (DD)8,30,31. We tested for and found that genes in which rare variants increase risk of ASD and DD32,33 are also enriched for schizophrenia common variant associations. Moreover, they are also enriched among genes containing FINEMAP credible SNPs (Figure 2 Supplementary Tables 20, 21).
Figure 2: Gene set enrichment tests at genome-wide level and for protein coding genes containing FINEMAP credible SNPs.
Gene sets tested were retrieved from sequencing studies of schizophrenia (SCHEMA; companion paper), autism-spectrum disorder33 and developmental disorders32. Sets representing genes that are intolerant to loss-of function mutations40 (LoF-intolerant) and brain-expressed genes41are also shown. A) MAGMA gene set enrichment analysis, dotted line indicates nominal significance (p=0.05). B) Logistic regression (with Firth’s bias reduction method) showing the odds-ratio (and 95% CI) for association between protein-coding genes containing at least 1 credible FINEMAP SNP (N=418 after excluding genes with no LoF-intolerance data) and genes from the sets indicated. Odds-ratios are relative to protein-coding genes within GWAS K≤3.5 loci (1,283 genes, squares) or across the genome excluding the xMHC (19,547 genes; circles). Dotted line indicates no enrichment.
Convergences between rare variants and fine-mapped GWAS signals have been previously observed in other traits e.g.,34,35, suggesting that genes most strongly implicated by fine-mapping and which have additional support from rare variant data are compelling candidates. Of the 10 exome-wide significant genes identified by SCHEMA36, two were prioritised candidates from fine-mapping; GRIN2A encoding a glutamatergic NMDA receptor subunit, and SP4, a transcription factor highly expressed in brain and which is regulated by NMDA transmission, and also regulates NMDA receptor abundance37. Two other genes supported by SCHEMA at FDR<0.05 had strong support from fine-mapping: STAG1, which is involved in controlling chromosome segregation and regulating gene expression, and FAM120A, which encodes an RNA binding protein. SNPs mapping to these genes had cumulative FINEMAP PP of 0.88 and 0.72 respectively (Supplementary Table 11b). The prioritised fine-mapped set also contained 4 genes implicated in DD; a transcriptional regulator (BCL11B), the well-known CACNA1C38, and genes mentioned elsewhere in this paper (GRIN2A and SLC39A8). Genes encoding additional transcriptional regulators are also of note; RERE, FOXP1 and MYT1L. RERE was prioritised by SMR and is associated with DD. FOXP1 and MYT1L are associated with both DD and ASD and met our fine-mapping prioritisation criteria in the core PGC dataset (Supplementary Table 12).
DISCUSSION
We have performed the largest GWAS of schizophrenia to date and in doing so, identify a substantial increase in the number of associated loci. We show that genes we prioritise within associated loci by fine-mapping are enriched for those with an increased burden of rare deleterious mutations in schizophrenia, and identify GRIN2A, SP4, STAG1, and FAM120A as specific genes where the convergence of rare and common variant associations strongly supports their pathogenic role in the disorder. Importantly, this convergence also implies that the pathogenic relevance of altered function of these genes extends beyond the small proportion of cases carrying rare mutations. We also demonstrate that common variant schizophrenia associations are enriched at genes implicated in neurodevelopmental disorders, opening the door for using the increasing power of rare variant studies of those disorders to further prioritise genes from GWAS studies. Exploiting this, in addition to GRIN2A we identify BCL11B, CACNA1C, RERE, FOXP1, MYT1L and SLC39A8 as genes with strong support.
Enrichment of common variant associations was restricted to genes expressed in CNS neurons, both excitatory and inhibitory, and fundamental biological processes related to neuronal function. This points to neurons as the most important site of pathology in the disorder. We also show that genes with high relative specificity for expression in almost all tested brain regions are enriched for genetic association. This suggests that abnormal neuronal function in schizophrenia is not confined to a small number of brain structures, which in turn might explain its diverse psychopathology, association with a broad range of cognitive impairments, and lack of regional specificity in neuroimaging measures1.
Disrupted neuronal function in schizophrenia is unlikely to be restricted to the synapse, but the concentration of associations in genes with pre- and post-synaptic locations, and with functions related to synaptic organisation, differentiation and transmission, point to the pathophysiological importance of these neuronal compartments and their attendant functions. This is further supported by studies showing substantial effects on schizophrenia risk of CNVs39 and rare damaging coding variants in genes with similar functions, including some of the same genes (SCHEMA; companion paper). Genomic studies, therefore, converge in highlighting these areas of biology as targets for research aiming for a mechanistic understanding of the disorder; the large number of prioritised genes and variants identified here offer an unprecedented empirically-supported resource for that endeavour.
Ethics
The study protocols were approved by the institutional review board at each centre involved with recruitment. Informed consent and permission to share the data were obtained from all subjects, in compliance with the guidelines specified by the recruiting centres’ institutional review boards. Genotyping of samples recruited in mainland China were processed and analysed by Chinese groups on Chinese local servers, to comply with the Human Genetic Resources Administrative Regulations.
ONLINE METHODS
Overview of Samples
Details of each of the samples (including sample size, ancestry, and whether included in the previous publication by the PGC) are given in Supplementary Cohort Descriptions. The core PGC dataset included 90 cohorts for which we had individual level genotype data fully processed under a uniform pipeline. This core dataset contains genotypes on 161,405 unrelated subjects; 67,390 schizophrenia/schizoaffective disorder cases and 94,015 controls, equivalent in power to 73,189 of each. A parent-proband trio is considered to comprise one case and one control. Approximately half (31,914 cases and 47,176 controls) of the samples were not included in the previous GWAS of the PGC1. Around 80% of the probands (53,386 cases and 77,258 controls) were of European Ancestry, and the remainder (14,004 cases and 16757 controls) were of East Asian ancestry2. We additionally included in the Primary GWAS summary statistics from 9 cohorts comprising African-American (AA; 6152 cases 3918 controls) and Latino (1234 cases, 3090 controls) participants; the combined sample is equivalent in power to 6,551 each of cases and controls. 1249 LD – independent (r2 > 0.1) Variants showing evidence for association (P< 1×10−5) were further meta-analysed with an additional dataset of 1,979 cases and 142,626 controls of European ancestry obtained from deCODE genetics, thus the final analysis represents 320,404 diploid genomes.
Association Analysis
Technical Quality Control of the 90 cohorts comprising the primary PGC sample.
Technical Quality control was performed on the core PGC cohorts separately according to standards developed by the PGC3 including SNP missingness < 0.05 (before sample removal); subject missingness < 0.02; autosomal heterozygosity deviation (| Fhet | < 0.2); SNP missingness < 0.02 (after sample removal); difference in SNP missingness between cases and controls < 0.02; and SNP Hardy-Weinberg equilibrium (HWE: P > 10−6 in controls or P > 10−10 in cases). For family-based cohorts we excluded individuals with more than 10,000 Mendelian errors and SNPs with more than 4 Mendelian errors. For X-Chromosomal genotypes we applied an additional round of the above QC to the male and female subgroups separately.
Genomic Quality Control: Principal Component Analysis (PCA) and Relatedness Checking in the core PGC dataset
We performed PCA for all 90 cohorts separately using SNPs with high imputation quality (INFO >0.8), low missingness (<1%), MAF>0.05 and in relative linkage equilibrium (LD) after 2 iterations of LD pruning (r2 < 0.2, 200 SNP windows). We removed well known long-range-LD areas (MHC and chr8 inversion). Thus, we retained between 57K and 95K autosomal SNPs in each cohort. SNPs present in all 90 cohorts (N=7,561) were used for robust relatedness testing using PLINK v1.94; pairs of subjects with PIHAT > 0.2 were identified and one member of each pair removed at random, preferentially retaining cases and trio members over case-control members.
To control for false positive associations due to inflated test statistics we evaluated the effectiveness of the primary technical and genomic quality control parameters on the genome-wide inflation of test statistics using the lambda GC (median)5 and as necessary made the QC parameters more stringent until this value was between 1.0 and 1.4 (before inclusion of principal components as covariates) and/or between 1.0 and 1.15 after inclusion of PCA covariates. Additionally, we applied loose PCA filters for strongly stratified datasets even if we did not observe strong inflation of test statistics in order to retrieve reliable test statistics (see Supplementary Figure 4). Since the core PGC cohorts came from many distinct centres, countries, and continents, various measures (e.g., tightening of the technical QC parameters and/or genomic quality control) had to be taken in an iterative process to achieve this goal.
Supplementary Table 22 lists detailed per cohort exclusion numbers for individuals in the non-Asian samples. The Asian cohorts were sufficiently homogeneous as they did not show marked population structure in principal component analyses. The exclusion numbers for individuals during technical QC are in most cohorts low. For six cohorts (marked in yellow in Supplementary Table 22) it was necessary to exclude more than 100 cases during genomic QC so that Lambda GC fell within the window mentioned above. Supplementary Figure 4 gives details about this process and explains why the excluded cases could not be used with the presently available control cohorts for this manuscript.
Imputation of the core PGC dataset
Genotype imputation of case-control cohorts was performed using the pre-phasing/imputation stepwise approach implemented in EAGLE 26 / MINIMAC37 (with 132 genomic windows of variable size and default parameters). The imputation reference consisted of 54,330 phased haplotypes with 36,678,882 variants from the publicly available HRC reference, release 1.18 Chromosome X imputation was conducted using individuals passing quality control for the autosomal analysis. ChrX imputation and association analysis was performed separately for males and females. For trio-based cohorts, families with multiple (N) affected offspring were split into N parent-offspring trios, duplicating the parental genotype information. Trios were phased with SHAPEIT 39. We created pseudo-controls based on the non-transmitted alleles from the parents. Phased case-pseudo-control genotypes were then taken forward to the IMPUTE4 algorithm10 into the above HRC reference panel.
Association / Meta-analysis
In each individual cohort, association testing was based on an additive logistic regression model using PLINK11. As covariates we used a subset of the first 20 principal components (PCA), derived within each cohort. By default, we included the first 4 PCAs and thereafter every PCA that was nominally significantly associated (p<0.05) to case-control status. PCAs in trios were only used to remove extreme ancestry outliers. We conducted a meta-analysis of the results (including the 9 cohorts comprising African-American and Latino participants) using a standard error inverse-weighted fixed effects model. For chrX, gene dosages in males were scored 0 or 2, in females, 0/1/2. We summarised the associations as number of independently associated index SNPs. Index SNPs were LD independent and had r2 < 0.1 within 3 Mb windows. We recorded the left and rightmost variant with r2<0.1 to an index SNP to define an associated clump. To define loci, we added a 50kb window on each side of the LD clump and combined overlapping LD-clumps into a single locus.
Due to the strong signal and high linkage disequilibrium in the MHC, only one SNP was kept from the extended MHC region (chr6:25–35Mb).
We additionally examined the X chromosome for evidence of heterogeneity between the sexes and X chromosome dosage compensation using the methods described by Lee and colleagues12,13 (Supplementary Note). To minimise possible confounding effects of ancestry on effect sizes by sex, we restricted this analysis to those of European ancestry.
We obtained summary association results from deCODE genetics for 1,228 index SNPs (P < 1×10−5) based on 1,979 cases and 142,626 controls of European ancestry. Genotyping was carried out at deCODE Genetics. We used this sample to establish that SNP associations from the primary GWAS replicated en masse in an independent sample (see Supplementary Note) by showing the directions of effect of index SNPs differed from the null hypothesis of randomly oriented effects and also comparing the expected number of same direction effects with those if all associations were true, taking into account the discovery magnitude of effect, and the replication effect-estimate precision (Supplementary Note).
The summary statistics from deCODE were combined with those from our primary GWAS dataset using an inverse variance-weighted fixed effects model. Similarly to the discovery meta-analysis (see above) we merged overlapping LD-clumps to a total of 287 distinct genomic regions (5 on the X-chromosome) with at least one genome-wide significant signal.
Polygenic Prediction
We estimated the cumulative contribution of SNPs to polygenic risk of schizophrenia using a series of leave-one-out polygenic prediction analyses based on LD-clumping and P-value thresholding (P+T)14 (also known as C+T) using PLINK11. For calculating polygenic scores, we included the most significant SNP for any pair of SNPs within <500kb and with LD R2 >0.1. We included only those with minor allele frequency >1%. We considered a range of P-value thresholds; 5×10−8, 1×10−6, 1×10−4, 1×10−3, 1×10−2, 5×10−2, 1×10−1, 2×10−1, 5×10−1 and 1.0. We performed logistic regression analysis within each case-control sample, to assess the relationship between case status and PRS (P+T) quantiles. The same principal components used for each GWAS were used as covariates for this analysis. Whenever the number of controls at a quantile was fewer than 5 times the number of covariates15, or if the higher bound for the PRS Odds Ratio (OR) became infinity, Firth’s penalised likelihood method was used to compute regression statistics, as implemented in the R package “logistf”16. ORs from these calculations were then meta-analysed using a fixed-effects model in the R package “metafor”17. To ensure stability of the estimates, meta-analysis was conservatively restricted to case-control samples which contained more than 10 individuals in the top 1% PRS, with at least one of them being a control. Analogous analyses were conducted to assess the ORs between individuals at the top and bottom quantiles. To assess the performance of PRS as a predictor of schizophrenia case status, we calculated liability R2, Nagelkerke’s R2 following Lee et. al. 201218 and a combined area under the receiver operating characteristic curve (AUROC). Both liability R2 and Nagelkerke’s R2 included any principal components marginally associated with the outcome within each cohort, in the baseline model. AUROC was estimated using the non-parametric meta-analysis implemented in the R package “nsROC”19. Polygenic score analysis of the African-American and Latino cohorts were conducted by the authors of the study reporting those datasets20.
Secondary analyses in core PGC dataset
Some of the secondary analyses (Gene-set enrichments, conditional SNP association analyses, fine-mapping) necessitate access to individual level data, require identical QC and imputation procedures, and/or an accurate LD reference panel meaning these analyses could only be reliably performed in a subset of the dataset. The following analyses focussed on the core PGC dataset for which these conditions are met.
Gene Set Enrichments
Tissue and cell types
We collected bulk RNA-seq data across 53 human tissues (GTEx v8, median across samples)21; from a study of 19,550 nuclei from frozen adult human post-mortem hippocampus and prefrontal cortex representing 16 different cell types22; from a study of ~10,000 single cells from 5 mouse brain regions (cortex, hippocampus, hypothalamus, midbrain and striatum, in addition to specific enrichments for oligodendrocytes, dopaminergic neurons, serotonergic neurons and cortical parvalbuminergic interneurons) that identified 24 cell types23; from a study of~500,000 single cells from the mouse nervous system (19 regions) that identified 265 cell types24.
Datasets were processed uniformly25. First, we calculated the mean expression for each gene for each type of data if these statistics were not provided by the authors. We used the pre-computed median expression (transcript per million (TPM)) across individuals for the GTEx tissues (v8). For the GTEx dataset, we excluded tissues with less than 100 samples, merged tissues by organ (with the exception of brain tissues), excluded non-natural tissues (e.g. EBV-transformed lymphocytes) and testis (outlier in hierarchical clustering), resulting in 37 tissues. Genes without unique names and genes not expressed in any cell types were excluded. We scaled the expression data to 1M Unique Molecular Identifiers (UMIs) or TPM for each cell type/tissue. After scaling, we excluded non-protein coding genes, and, for mouse datasets, genes that had no expert curated 1:1 orthologs between mouse and human (Mouse Genome Informatics, The Jackson laboratory, version 11/22/2016). We then calculated a metric of gene expression specificity by dividing the expression of each gene in each cell type/tissue by the total expression of that gene in all cell types/tissue, leading to values ranging from 0 to 1 for each gene (0: meaning that the gene is not expressed in that cell type/tissue, 1 that 100% of the expression of that gene is performed in that cell type/tissue). We selected the 10% most specific genes per cell type (or tissue) with an expression level of at least 1TPM, or 1 UMI per million, for downstream analyses and used MAGMA v1.0826 to test whether they were enriched for genetic associations. We performed a one-sided test as we were only interested in enrichments for genetic associations (in contrast with depletions). We also applied partitioned LD score regression (LDSC) as described27 to the top 10% genes for each cell type for heritability enrichment. We selected the one-sided coefficient z-score p-value as a measure of the association of the cell type/tissue with schizophrenia.
Ontology Gene sets
Gene set analyses were performed using MAGMA v1.0826. Gene boundaries were retrieved from Ensembl release 92 (GRCh37) using the “biomaRt” R package28 and expanded by 35 kb upstream and 10 kb downstream to include likely regulatory regions29. Gene-wide p-values were calculated from European and Asian summary statistics separately using the SNP-wise “mean” Imhof method, and meta-analysed within the software. LD reference data files were from the European and East Asian populations of the Haplotype Reference Consortium30. Within each gene set analysis, p-values were corrected for multiple testing using the Bonferroni procedure. Specifically, we tested the following gene sets:
Gene ontology: 7,315 sets extracted from the GO database (http://geneontology.org/, accession date: 09/11/2020) curated to include only annotations with experimental or phylogenetic supporting evidence.
SynGO ontology: Described elsewhere31, this collection was analysed as two subsets; “biological process” (135 gene sets) and “cellular component” (60 gene sets). We controlled for a set of 10,360 genes with detectable expression in brain tissue measured as Fragments Per Kilobase of transcript per Million mapped reads (FPKM)32 to detect synaptic signals above signals simply reflecting the property of brain expression. Exploiting the hierarchical structure of SynGO, gene sets were reconstructed using a “roll-up” method, in which parent categories contained all genes annotated to child categories. For stepwise conditional testing33, we prioritised the most specific child annotations34 (i.e. the lowest possible level) as regression covariates.
Conditional SNP Association Analyses
We performed stepwise conditional analyses of 248 loci that were genome wide significant in the core PGC dataset looking for independent associations. We performed association testing and meta-analysis across each locus, adding the allele dosages of the index SNP as a covariate. Where a second SNP had a conditional p-value of less than 1×10−6, we considered this as evidence for a second signal and repeated the process adding this as an additional covariate. We repeated this until no additional SNPs in the region achieved p<1×10-6. We also searched for long range dependencies. Here we tested the all pairs of independent signals for conditional independence (Supplementary Note).
Fine-mapping
We used FINEMAP35 to fine-map regions defined by LD clumps (r2>0.1), excluding the MHC locus due to its complex LD structure. Clumps which overlapped (without adding the additional 50kb used to define physically distinct loci) were combined. As fine-mapping requires data from all markers in the region36 we only performed fine-mapping on regions that attained genome-wide significance (GWS) in the core PGC GWAS. In total, we attempted to fine-map 255 non-overlapping regions (Supplementary Table 11e). Further details about the fine-mapping process are given in the Supplementary Note.
Summary-data-based Mendelian Randomization (SMR) analysis, FUSION and EpiXcan
We used SMR37 as our primary method to identify SNPs which might mediate association with schizophrenia through effects on gene expression. The significance for SMR is set at the Bonferroni corrected threshold of 0.05/M where M is the number of genes with significant eQTLs tested for a given tissue. Significant SMR associations imply colocalization of the schizophrenia associations with eQTL. We applied the HEIDI test37 to filter out SMR associations (PHEIDI < 0.01) due to linkage disequilibrium between SCZ-associated variants and eQTLs. cis-eQTL summary data were from three studies: fetal brain (N=120)38, adult brain (n = ~1,500)39 and blood (n = ~32,000)40. Linkage disequilibrium (LD) data required for the HEIDI test37 were estimated from the Health and Retirement Study (HRS)41 (n = 8,557). We included only genes with at least one cis-eQTL at PeQTL < 5×10−8, excluding those in MHC regions due to the complexity of this region. For blood, we included only genes with eQTLs in brain. This left 7,803 genes in blood, 10,890 genes in prefrontal cortex and 754 genes in fetal brain for analysis (see Supplementary Note for further details). SMR was performed using data from the primary GWAS. The results were then filtered to exclude significant SMR implicated genes where the eQTLs did not map within our definition of an associated locus in the Extended GWAS meta-analysis of our primary GWAS dataset and the dataset provided by deCODE genetics.
For genomic regions where there were multiple genes showing significant SMR associations, we attempted to resolve these with conditional analysis using GCTA-COJO42,43. We selected the top-associated cis-eQTL for one gene (or a set of genes sharing the same cis-eQTL) ran a COJO analysis in the schizophrenia GWAS data and the eQTL data for each of the other genes conditioning on the selected top cis-eQTL. We then re-ran the SMR and HEIDI analyses using these conditional GWAS and eQTL results.
We used FUSION44 and EpiXcan45 as tests of robustness of the SMR results. Details are supplied in the Supplementary Note as are our approaches to prioritising SMR associated genes.
DATA AVAILABILITY
Summary statistics for the “Extended”, “Core”, ancestry specific and sex-stratified analyses is available at “https://www.med.unc.edu/pgc/download-results/scz/”. Genotype data are available for a subset of cohorts, including dbGAP accession numbers and/or restrictions, as described in the Supplementary Information section “Cohort Descriptions”.
CODE AVAILABILITY
Core analysis code for RICOPILI can be found at “https://sites.google.com/a/broadinstitute.org/ricopili/”. This wraps PLINK (“https://www.cog-genomics.org/plink2/”), EIGENSOFT (“https://www.hsph.harvard.edu/alkes-price/software/”), EAGLE2 (“https://alkesgroup.broadinstitute.org/Eagle/”), MINIMAC3 (“https://genome.sph.umich.edu/wiki/Minimac3”), SHAPEIT3 (“https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html”), METAL (“https://genome.sph.umich.edu/wiki/METAL_Documentation”), LDSR (“https://github.com/bulik/ldsc”). For downstream analyses, FINEMAP can be found at “http://christianbenner.com/”, and our utility for meta-analysing cohort-specific LD matrices can be found at https://github.com/Pintaius/LDmergeFM. MAGMA can be found at ”https://ctg.cncr.nl/software/magma” and the GO gene sets and automated curation pipeline are provided in https://github.com/janetcharwood/pgc3-scz_wg-genesets. SMR is available at “https://cnsgenomics.com/software/smr/” and SbayesS at “https://cnsgenomics.com/software/gctb/”.
Extended Data
Extended Data Figure 1: Primary GWAS Manhattan plot.
The x-axis indicates chromosomal position and the y-axis is the significance of association (−log10(P)). The red line represents genome-wide significance level (5×10−8). SNPs in green are in linkage disequilibrium (LD; R2 >0.1) with index SNPs (diamonds) which represent LD independent genome-wide significant associations.
Extended Data Figure 2: Polygenic risk prediction.
A) Distributions of liability scale R2 across 98 left-out-cohorts for polygenic risk scores built from SNPs with different p-value thresholds. Distributions of liability R2 (assuming schizophrenia life-time risk of 1%) are shown for each p-value threshold, with point size representing size of the left-out cohort and colour representing ancestry. The median liability R2 is represented as a horizontal black line. B) Liability R2 of predicted and observed phenotypes in left-out cohorts using variants with p-value threshold p=0.05, from the fixed effect meta-analysis of variant effects, unadjusted for multiple comparisons. The polygenic risk scores are derived from two separate sets of leave-one-out GWAS meta-analyses: y-axis R2 based on the results of primary GWAS including all ancestries; x axis R2 based on cohorts of the same ancestry as the test samples. Circles denote core PGC samples. Triangles denote African American and Latino samples processed external to PGC by the providing author.
Extended Data Figure 3: Association between 37 human tissues and schizophrenia.
The mean of the evidence (-log10P) obtained from two methods (MAGMA, LDSC) for testing GWAS data for enrichment of association in genes with high expression in each tissue as determined from bulk RNA-seq20. The bar colour indicates whether gene expression in the tissue is significantly associated with both methods, one method or none. The black vertical line represents the significance threshold corrected for the total number of tissues tested in this experiment. We also analysed previous waves of PGC schizophrenia GWAS11,21 for comparison.
Extended Data Table1:
List of prioritized genes
List of genes meeting prioritisation criteria summarised in Figure 1. Index SNP: index associated SNP for the locus from the GWAS. Ensembl ID: Ensembl gene identifier. Symbol ID: HGNC gene symbol. Gene Biotype: as classified by Ensembl. FINEMAP and SMR priority genes: genes meeting the prioritisation criteria described in the text. Rare priority genes: genes implicated by rare coding variants in schizophrenia, autism spectrum disorders or developmental disorder. Full details regarding the prioritisation criteria for each gene are given inSupplementary Tables 11–18.
| Index SNP | Ensembl ID | Symbol ID | gene_biotype | FINEMAP priority gene | SMR priority gene | Rare priority gene |
|---|---|---|---|---|---|---|
|
| ||||||
| rs12712510 | ENSG00000231200 | AC068490.2 | lincRNA | • | ||
| rs6504163 | ENSG00000159640 | ACE | protein_coding | • | ||
| rs7575796 | ENSG00000115073 | ACTR1B | protein_coding | • | ||
| rs61833239 | ENSG00000117020 | AKT3 | protein_coding | • | ||
| rs6546857 | ENSG00000163016 | ALMS1P | pseudogene | • | ||
| rs9925915 | ENSG00000174939 | ASPHD1 | protein_coding | • | ||
| rs12285419 | ENSG00000175224 | ATG13 | protein_coding | • | ||
| rs4766428 | ENSG00000174437 | ATP2A2 | protein_coding | • | ||
| rs1540840 | ENSG00000127152 | BCL11B | protein_coding | • | • | |
| rs2304205 | ENSG00000126453 | BCL2L12 | protein_coding | • | ||
| rs3808581 | ENSG00000104765 | BNIP3L | protein_coding | • | ||
| rs2649999 | ENSG00000157895 | C12orf43 | protein_coding | • | ||
| rs10774034 | ENSG00000151067 | CACNA1C | protein_coding | • | • | |
| rs2944821 | ENSG00000183166 | CALN1 | protein_coding | • | ||
| rs6839635 | ENSG00000145354 | CISD2 | protein_coding | • | ||
| rs61405217 | ENSG00000109572 | CLCN3 | protein_coding | • | ||
| rs17194490 | ENSG00000144619 | CNTN4 | protein_coding | • | ||
| rs10127983 | ENSG00000143578 | CREB3L4 | protein_coding | • | ||
| rs2532240 | ENSG00000120088 | CRHR1 | protein_coding | • | ||
| 8:4180090_T_A | ENSG00000183117 | CSMD1 | protein_coding | • | ||
| rs715170 | ENSG00000206129 | CTD-2008L17.2 | lincRNA | • | ||
| rs113113059 | ENSG00000112659 | CUL9 | protein_coding | • | • | |
| rs10957321 | ENSG00000172817 | CYP7B1 | protein_coding | • | ||
| rs61828917 | ENSG00000117593 | DARS2 | protein_coding | • | ||
| rs4632195 | ENSG00000187323 | DCC | protein_coding | • | ||
| rs4678552 | ENSG00000163673 | DCLK3 | protein_coding | • | ||
| rs7816998 | ENSG00000085788 | DDHD2 | protein_coding | • | ||
| rs2600490 | ENSG00000198010 | DLGAP2 | protein_coding | • | ||
| rs8048039 | ENSG00000103423 | DNAJA3 | protein_coding | • | ||
| rs72728416 | ENSG00000188641 | DPYD | protein_coding | • | ||
| rs8175378 | ENSG00000170571 | EMB | protein_coding | • | ||
| rs999494 | ENSG00000135638 | EMX1 | protein_coding | • | ||
| rs11619756 | ENSG00000120658 | ENOX1 | protein_coding | • | ||
| rs959071 | ENSG00000262319 | ENSG00000262319 | antisense | • | ||
| rs4073003 | ENSG00000072134 | EPN2 | protein_coding | • | ||
| rs6925079 | ENSG00000188107 | EYS | protein_coding | • | ||
| rs815609 | ENSG00000055147 | FAM114A2 | protein_coding | • | ||
| rs4766428 | ENSG00000204856 | FAM216A | protein_coding | • | ||
| rs1006945 | ENSG00000101447 | FAM83D | protein_coding | • | ||
| rs58120505 | ENSG00000122687 | FTSJ2 | protein_coding | • | ||
| rs4702 | ENSG00000140564 | FURIN | protein_coding | • | • | |
| rs10985811 | ENSG00000136928 | GABBR2 | protein_coding | • | ||
| rs1858999 | ENSG00000167491 | GATAD2A | protein_coding | • | ||
| rs12498839 | ENSG00000150625 | GPM6A | protein_coding | • | ||
| rs12188094 | ENSG00000164199 | GPR98 | protein_coding | • | ||
| rs77502336 | ENSG00000023171 | GRAMD1B | protein_coding | • | ||
| rs9926049 | ENSG00000183454 | GRIN2A | protein_coding | • | • | |
| rs2206956 | ENSG00000152822 | GRM1 | protein_coding | • | ||
| rs11210892 | ENSG00000178922 | HYI | protein_coding | • | ||
| rs1378559 | ENSG00000169306 | IL1RAPL1 | protein_coding | • | ||
| rs38752 | ENSG00000184903 | IMMP2L | protein_coding | • | ||
| rs3814883 | ENSG00000169592 | INO80E | protein_coding | • | ||
| rs2304205 | ENSG00000126456 | IRF3 | protein_coding | • | ||
| rs2532240 | ENSG00000120071 | KANSL1 | protein_coding | • | • | |
| rs10243922 | ENSG00000122778 | KIAA1549 | protein_coding | • | ||
| rs17731 | ENSG00000067082 | KLF6 | protein_coding | • | ||
| rs459391 | ENSG00000224924 | LINC00320 | lincRNA | • | • | |
| rs9545047 | ENSG00000227676 | LINC01068 | lincRNA | • | ||
| rs28454198 | ENSG00000249307 | LINC01088 | antisense | • | ||
| rs2387414 | ENSG00000131409 | LRRC4B | protein_coding | • | ||
| rs59498392 | ENSG00000175324 | LSM1 | protein_coding | • | ||
| rs58120505 | ENSG00000002822 | MAD1L1 | protein_coding | • | ||
| rs35164357 | ENSG00000112893 | MAN2A1 | protein_coding | • | ||
| rs9925915 | ENSG00000102882 | MAPK3 | protein_coding | • | ||
| rs2532240 | ENSG00000186868 | MAPT | protein_coding | • | ||
| rs143116451 | ENSG00000175727 | MLXIP | protein_coding | • | ||
| rs2914983 | ENSG00000115540 | MOB4 | protein_coding | • | ||
| rs4793888 | ENSG00000153944 | MSI2 | protein_coding | • | ||
| rs11263770 | ENSG00000141140 | MYO19 | protein_coding | • | ||
| rs324017 | ENSG00000166886 | NAB2 | protein_coding | • | ||
| rs9545047 | ENSG00000102471 | NDFIP2 | protein_coding | • | ||
| rs2119242 | ENSG00000078114 | NEBL | protein_coding | • | ||
| rs1121296 | ENSG00000172260 | NEGR1 | protein_coding | • | ||
| rs5943629 | ENSG00000146938 | NLGN4X | protein_coding | • | ||
| rs9975024 | ENSG00000180530 | NRIP1 | protein_coding | • | ||
| rs11972718 | ENSG00000122584 | NXPH1 | protein_coding | • | ||
| rs1939514 | ENSG00000183715 | OPCML | protein_coding | • | ||
| rs56205728 | ENSG00000137843 | PAK6 | protein_coding | • | ||
| rs7432375 | ENSG00000114054 | PCCB | protein_coding | • | ||
| rs10069930 | ENSG00000204969 | PCDHA2 | protein_coding | • | ||
| rs246024 | ENSG00000204962 | PCDHA8 | protein_coding | • | ||
| rs35734242 | ENSG00000185619 | PCGF3 | protein_coding | • | ||
| rs58950470 | ENSG00000197136 | PCNXL3 | protein_coding | • | ||
| rs6588168 | ENSG00000184588 | PDE4B | protein_coding | • | ||
| rs2929278 | ENSG00000167004 | PDIA3 | protein_coding | • | ||
| rs34539323 | ENSG00000181191 | PJA1 | protein_coding | • | ||
| rs6673880 | ENSG00000149527 | PLCH2 | protein_coding | • | ||
| rs3813567 | ENSG00000041357 | PSMA4 | protein_coding | • | ||
| rs2890914 | ENSG00000153707 | PTPRD | protein_coding | • | ||
| rs61937595 | ENSG00000179912 | R3HDM2 | protein_coding | • | ||
| rs11121172 | ENSG00000142599 | RERE | protein_coding | • | • | |
| rs11227250 | ENSG00000172922 | RNASEH2C | protein_coding | • | ||
| rs13107325 | ENSG00000246560 | RP11–10L12.4 | antisense | • | ||
| rs6479487 | ENSG00000227603 | RP11–165J3.6 | antisense | • | ||
| rs505061 | ENSG00000234840 | RP11–399D6.2 | lincRNA | • | ||
| rs1198588 | ENSG00000259946 | RP11–490G2.2 | lincRNA | • | ||
| rs35351411 | ENSG00000259616 | RP11–507B12.2 | lincRNA | • | ||
| rs10035564 | ENSG00000272335 | RP11–53O19.3 | lincRNA | • | ||
| rs1915019 | ENSG00000253553 | RP11–586K2.1 | antisense | • | ||
| rs10873538 | ENSG00000256500 | RP11–73M18.2 | protein_coding | • | ||
| rs154433 | ENSG00000103037 | SETD6 | protein_coding | • | ||
| rs2914983 | ENSG00000115524 | SF3B1 | protein_coding | • | ||
| rs12652777 | ENSG00000170624 | SGCD | protein_coding | • | ||
| rs13107325 | ENSG00000138821 | SLC39A8 | protein_coding | • | • | |
| rs2909457 | ENSG00000144290 | SLC4A10 | protein_coding | • | ||
| rs6839635 | ENSG00000164037 | SLC9B1 | protein_coding | • | ||
| rs2022265 | ENSG00000065609 | SNAP91 | protein_coding | • | • | |
| rs7811417 | ENSG00000105866 | SP4 | protein_coding | • | • | |
| rs3810450 | ENSG00000161277 | THAP8 | protein_coding | • | ||
| rs704364 | ENSG00000163634 | THOC7 | protein_coding | • | ||
| rs7312697 | ENSG00000133687 | TMTC1 | protein_coding | • | ||
| rs1924377 | ENSG00000133107 | TRPC4 | protein_coding | • | ||
| rs13262595 | ENSG00000171045 | TSNARE1 | protein_coding | • | ||
| rs10861176 | ENSG00000198431 | TXNRD1 | protein_coding | • | ||
| rs10238960 | ENSG00000185274 | WBSCR17 | protein_coding | • | ||
| rs2929278 | ENSG00000092470 | WDR76 | protein_coding | • | ||
| rs3764002 | ENSG00000075035 | WSCD2 | protein_coding | • | ||
| rs11693094 | ENSG00000170396 | ZNF804A | protein_coding | • | ||
| rs72986630 | ENSG00000197933 | ZNF823 | protein_coding | • | • | |
| rs758749 | ENSG00000127903 | ZNF835 | protein_coding | • | ||
Supplementary Material
ACKNOWLEDGEMENTS
The National Institute of Mental Health (USA) provides core funding for the Psychiatric Genomics Consortium (PGC) under Award Number U01MH109514. The content is the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The work of the contributing groups was supported by numerous grants from governmental and charitable bodies as well as philanthropic donation (details in Supplementary Note). We acknowledge a substantial contribution from Pamela Sklar (deceased) as one of the PGC PIs, and Ed Scolnick, Chief Scientist Emeritus, Stanley Center of the Broad Institute, whose support for this study was vital. We acknowledge the Wellcome Trust Case Control Consortium for the provision of control genotype information. Membership of the Psychosis Endophenotype International Consortium, the SynGO consortium, the PsychENCODE Consortium, the eQTLGen consortium, the BIOS Consortium and the Indonesia Consortium are provided in the accompanying author and consortium XL file. We are grateful to Catrin Hopkins for illustrations.
The work at Cardiff University was additionally supported by Medical Research Council Centre Grant No. MR/L010305/1 and Program Grant No. G0800509. Dr. Shuhua Xu also gratefully acknowledges the support of the National Natural Science Foundation of China (NSFC) grant (31525014, 91731303, 31771388, 31961130380, and 32041008), the UK Royal Society-Newton Advanced Fellowship (NAF\R1\191094), Key Research Program of Frontier Sciences (QYZDJ-SSW-SYS009) and the Strategic Priority Research Program (XDB38000000) of the Chinese Academy of Sciences, and the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01). Dr. Ole Anreassen was supported by Research Council of Norway (283798, 262656, 248980, 273291, 248828, 248778, 223273); KG Jebsen Stiftelsen, South-East Norway Health Authority, EU H2020 # 847776. Béla Melegh was supported in part by the National Scientific Research Program (NKFIH) K 138669. Dr. Faraone is supported by the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement no 602805, the European Union’s Horizon 2020 research and innovation programme under grant agreements No 667302 & 728018 and NIMH grants 5R01MH101519 and U01 MH109536–01. Dr. Sintia Belangero was supported by FAPESP - Fundação de Amparo à Pesquisa do Estado de São Paulo (Brazil) - Grant numbers: 2010/08968–6 (S.I.B.); 2014/07280–1 (S.I.B.); 2007/58736–1 (M.AC.S.); 2011/50740–5 (R.A.B.); 2016/04983–7 (J.J.M.); 10/19176–3 (V.K.O. & S.I.B.); 12/12686–1 (M.L.S. & S.I.B.); CAPES - Coordenação de Aperfeiçoamento de Pessoal de Nível Superior Code 001. The Singapore team (Lee Jimmy, Liu Jianjun, Sim Kang, Chong Siow Chong, Mythily Subramanian) acknowledges the National Medical Research Council Translational and Clinical Research Flagship Programme (grant number: NMRC/TCR/003/2008). Milan Macek was supported by LM2018132, CZ.02.1.01/0.0/0.0/18_046/0015515 and IP6003 –VZFNM00064203 to MM Jr. Dr. Celso Arango has been funded by the Spanish Ministry of Science and Innovation. Instituto de Salud Carlos III (SAM16PE07CP1, PI16/02012, PI19/024), co-financed by ERDF Funds from the European Commission, “A way of making Europe”, CIBERSAM. Madrid Regional Government (B2017/BMD-3740 AGES-CM-2), European Union Structural Funds. European Union Seventh Framework Program; and European Union H2020 Program under the Innovative Medicines Initiative 2 Joint Undertaking (grant agreement No 115916, Project PRISM, and grant agreement No 777394, Project AIMS-2-TRIALS), Fundación Familia Alonso and Fundación Alicia Koplowitz. Dr. E. Bramon acknowledges support from: National Institute of Health Research UK (grant NIHR200756). Mental Health Research UK John Grace QC Scholarship 2018. An ESRC collaborative award 2020. BMA Margaret Temple Fellowship 2016. Medical Research Council New Investigator Award (G0901310) and MRC Centenary Award (G1100583), MRC project grant G1100583. National Institute of Health Research UK post-doctoral fellowship (PDA/02/06/016). NARSAD Young Investigator Awards 2005 and 2008. Wellcome Trust Research Training Fellowship, Wellcome Trust Case Control Consortium awards (085475/B/08/Z, 085475/Z/08/Z). European Commission Horizon 2020 (747429). NIHR Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust and King’s College London. NIHR Biomedical Research Centre at University College London Hospitals NHS Foundation Trust and University College London (UCLH BRC - Mental Health Theme). Dr. Dolores Moltó is funded by the European Regional Development Fund (ERDF)-Valencian Community 2014–2020, Spain. Dr. Elizabeth G. Atkinson was supported by the NIMH K01MH121659.
CONFLICTS OF INTEREST
Aarno Palotie is a member of Astra Zeneca’s Genomics Advisory Board. Veikko Salomaa has consulted for Novo Nordisk and Sanofi and has ongoing research collaboration with Bayer Ltd (both unrelated to the present study). Michael Green is a paid consultant for AiCure, Biogen, Lundbeck, and Roche, is a member of the Scientific Board of Cadent, and has received research funds from Forum.Gregory Light has consulted to Astellas, Forum, and Neuroverse. Keith Nuechterlein has research support from Janssen, Genentech, and Brain Plasticity Inc. Also has consulted to Astellas, MedinCell, Takeda, Teva, Genentech, Otsuka, Janssen, and Brain Plasticity Inc. David Cohen has reported past consultation for or the receipt of honoraria from Otsuka, Shire, Lundbeck, Roche and Janssen. Mark Daly is a founder of Maze Therapeutics. Anil K. Malhotra is a consultant to Genomind Inc, InformedDNA, and Concert Pharmaceuticals. Rodrigo Affonseca BressanOle has received research grants from Janssen; has been a forum consultant for Janssen and Sanof; Roche; speaker bureau for Ache, Janssen, Sanofi and Torrent. Cristiano Noto was on the speakers’ bureau and/or has acted as a consultant for Janssen and Daiichi-Sankyo in the last 12 months. Christos Pantelis has, for the last 3 years, served on an advisory board for Lundbeck and received honoraria for talks presented at educational meetings organized by Lundbeck. David A Collier is a full-time employee and stockholder of Eli Lilly and Company. Michael O’Donovan is supported by a collaborative research grant from Takeda Pharmaceuticals. Michael Owen is supported by a collaborative research grant from Takeda Pharmaceuticals. James Walters is supported by a collaborative research grant from Takeda Pharmaceuticals. Andrew Pocklington is supported by a collaborative research grant from Takeda Pharmaceuticals. Stephen R. Marder has consulted for the following companies: Roche, Sunovion, Lundbeck, Boeringer-Ingelheim, Acadia, and Merck. Srihari Gopal is a full time employee and shareholder Johnson & Johnson (AMEX: JNJ). Adam Savitz is an employee of Janssen Research & Development, LLC and own stock/stock options in the company. Qingqin Li is an employee of Janssen Research & Development, LLC and own stock/stock options in the company. Tony Kam-Thong is an employee of F.Hoffman-La Roche. Anna Rautanen is an employee of F.Hoffman-La Roche. Dheeraj Malhotra is an employee of F.Hoffman-La Roche. Sara Paciga is an employee of Pfizer Inc. Ole A. Andreassen is a consultant for HealthLytix, and received speaker’s honorarium from Lundbeck. Stephen Faraone has received income, potential income, travel expenses continuing education support and/or research support from, Akili Interactive Labs, Arbor, Genomind, Ironshore, Ondosis, Otsuka, Rhodes, Shire/Takeda, Sunovion, Supernus, Tris, and Vallon. With his institution, he has US patent US20130217707 A1 for the use of sodium-hydrogen exchange inhibitors in the treatment of ADHD. In previous years, he received support from: Alcobra, Aveksham, CogCubed, Eli Lilly, Enzymotec, Impact, Janssen, KemPharm, Lundbeck/Takeda, McNeil, Neurolifesciences, Neurovance, Novartis, Pfizer, and Vaya. He also receives royalties from books published by Guilford Press: Straight Talk about Your Child’s Mental Health; Oxford University Press: Schizophrenia: The Facts; and Elsevier: ADHD: Non-Pharmacologic Interventions. He is also Program Director of www.adhdinadults.com. Celso Arango has been a consultant to or has received honoraria or grants from Acadia, Angelini, Gedeon Richter, Janssen Cilag, Lundbeck, Minerva, Otsuka, Roche, Sage, Servier, Shire, Schering Plough, Sumitomo Dainippon Pharma, Sunovion and Takeda. Köksal Alptekin has received grants and honoraria for consulting work, lecturing and research from Abdi İbrahim, Abdi İbrahim Otsuka, Janssen, Ali Raif and TUBITAK.
Consortia
Indonesia Schizophrenia Consortium
• Nan Dai
• Qin Wenwen
• D. B. Wildenauer
• Feranindhya Agiananda
• Nurmiati Amir
• Ronald Antoni
• Tiana Arsianti
• Asmarahadi Asmarahadi
• H. Diatri
• Prianto Djatmiko
• Irmansyah Irmansyah
• Siti Khalimah
• Irmia Kusumadewi
• Profitasari Kusumaningrum
• Petrin R. Lukman
• Martina W. Nasrun
• N. S. Safyuni
• Prasetyawan Prasetyawan
• G. Semen
• Kristiana Siste
• Heriani Tobing
• Natalia Widiasih
• Tjhin Wiguna
• D. Wulandari
• None Evalina
• A. J. Hananto
• Joni H. Ismoyo
• T. M. Marini
• Supiyani Henuhili
• Muhammad Reza
• & Suzy Yusnadewi
PsychENCODE
• Alexej Abyzov
• Schahram Akbarian
• Allison Ashley-Koch
• Harm van Bakel
• Michael Breen
• Miguel Brown
• Julien Bryois
• Becky Carlyle
• Alex Charney
• Gerard Coetzee
• Gregory Crawford
• Stella Dracheva
• Prashant Emani
• Peggy Farnham
• Menachem Fromer
• Timur Galeev
• Mike Gandal
• Mark Gerstein
• Gina Giase
• Kiran Girdhar
• Fernando Goes
• Kay Grennan
• Mengting Gu
• Brittney Guerra
• Gamze Gursoy
• Gabriel Hoffman
• Thomas Hyde
• Andrew Jaffe
• Shan Jiang
• Yan Jiang
• Amira Kefi
• Yunjung Kim
• Robert Kitchen
• James A. Knowles
• Fides Lay
• Donghoon Lee
• Mingfeng Li
• Chunyu Liu
• Shuang Liu
• Eugenio Mattei
• Fabio Navarro
• Xinghua Pan
• Mette A. Peters
• Dalila Pinto
• Sirisha Pochareddy
• Damon Polioudakis
• Michael Purcaro
• Shaun Purcell
• Henry Pratt
• Tim Reddy
• Suhn Rhie
• Panagiotis Roussos
• Joel Rozowsky
• Stephan Sanders
• Nenad Sestan
• Anurag Sethi
• Xu Shi
• Annie Shieh
• Vivek Swarup
• Anna Szekely
• Daifeng Wang
• Jonathan Warrell
• Sherman Weissman
• Zhiping Weng
• Kevin White
• Jennifer Wiseman
• Heather Witt
• Hyejung Won
• Shannon Wood
• Feinan Wu
• Xuming Xu
• Lijing Yao
• & Peter Zandi
Psychosis Endophenotypes International Consortium
• Maria J. Arranz
• Steven Bakker
• Stephan Bender
• Elvira Bramon
• David A. Collier
• Benedicto Crepo-Facorro
• Jeremy Hall
• Conrad Iyegbe
• Assen V. Jablensky
• René Kahn
• Stephen Lawrie
• Cathryn Lewis
• Kuang Lin
• Don H. Linszen
• Ignacio Mata
• Andrew McIntosh
• Robin M. Murray
• Roel A. Ophoff
• Jim van Os
• John Powell
• Dan Rujescu
• Muriel Walshe
• & Matthias Weisbrod
The SynGO Consortium
• Tilmann Achsel
• Maria Andres-Alonso
• Claudia Bagni
• Àlex Bayés
• Thomas Biederer
• Nils Brose
• Tyler C. Brown
• John Jia En Chua
• Marcelo P. Coba
• L. Niels Cornelisse
• Arthur P. H. de Jong
• Jaime de Juan-Sanz
• Daniela C. Dieterich
• Guoping Feng
• Hana L. Goldschmidt
• Eckart D. Gundelfinger
• Casper Hoogenraad
• Richard L. Huganir
• Steven E. Hyman
• Cordelia Imig
• Reinhard Jahn
• Hwajin Jung
• Pascal S. Kaeser
• Eunjoon Kim
• Frank Koopmans
• Michael R. Kreutz
• Noa Lipstein
• Harold D. MacGillavry
• Robert Malenka
• Peter S. McPherson
• Vincent O’Connor
• Rainer Pielot
• Timothy A. Ryan
• Dnyanada Sahasrabudhe
• Carlo Sala
• Morgan Sheng
• Karl-Heinz Smalla
• August B. Smit
• Thomas C. Südhof
• Paul D. Thomas
• Ruud F. Toonen
• Jan R. T. van Weering
• Matthijs Verhage
• & Chiara Verpelli
Schizophrenia Working Group of the Psychiatric Genomics Consortium
• Vassily Trubetskoy
• Antonio F. Pardiñas
• Georgia Panagiotaropoulou
• Swapnil Awasthi
• Tim B. Bigdeli
• Charlotte A. Dennison
• Lynsey S. Hall
• Max Lam
• Oleksandr Frei
• Alexander L. Richards
• Jakob Grove
• Zhiqiang Li
• Mark Adams
• Ingrid Agartz
• Elizabeth G. Atkinson
• Esben Agerbo
• Mariam Al Eissa
• Margot Albus
• Madeline Alexander
• Behrooz Z. Alizadeh
• Köksal Alptekin
• Thomas D. Als
• Farooq Amin
• Volker Arolt
• Manuel Arrojo
• Lavinia Athanasiu
• Maria Helena Azevedo
• Silviu A. Bacanu
• Nicholas J. Bass
• Martin Begemann
• Richard A. Belliveau
• Judit Bene
• Beben Benyamin
• Sarah E. Bergen
• Giuseppe Blasi
• Julio Bobes
• Stefano Bonassi
• Alice Braun
• Rodrigo Affonseca Bressan
• Evelyn J. Bromet
• Richard Bruggeman
• Peter F. Buckley
• Randy L. Buckner
• Jonas Bybjerg-Grauholm
• Wiepke Cahn
• Murray J. Cairns
• Monica E. Calkins
• Vaughan J. Carr
• David Castle
• Stanley V. Catts
• Kimberley D. Chambert
• Raymond C. K. Chan
• Boris Chaumette
• Wei Cheng
• Eric F. C. Cheung
• Siow Ann Chong
• David Cohen
• Angèle Consoli
• Quirino Cordeiro
• Javier Costas
• Charles Curtis
• Michael Davidson
• Kenneth L. Davis
• Lieuwe de Haan
• Franziska Degenhardt
• Lynn E. DeLisi
• Ditte Demontis
• Faith Dickerson
• Dimitris Dikeos
• Timothy Dinan
• Srdjan Djurovic
• Jubao Duan
• Giuseppe Ducci
• Johan G. Eriksson
• Lourdes Fañanás
• Stephen V. Faraone
• Alessia Fiorentino
• Andreas Forstner
• Josef Frank
• Nelson B. Freimer
• Menachem Fromer
• Alessandra Frustaci
• Ary Gadelha
• Giulio Genovese
• Elliot S. Gershon
• Marianna Giannitelli
• Ina Giegling
• Paola Giusti-Rodríguez
• Stephanie Godard
• Jacqueline I. Goldstein
• Javier González Peñas
• Ana González-Pinto
• Srihari Gopal
• Jacob Gratten
• Michael F. Green
• Tiffany A. Greenwood
• Olivier Guillin
• Sinan Gülöksüz
• Raquel E. Gur
• Ruben C. Gur
• Blanca Gutiérrez
• Eric Hahn
• Hakon Hakonarson
• Vahram Haroutunian
• Annette M. Hartmann
• Carol Harvey
• Caroline Hayward
• Frans A. Henskens
• Stefan Herms
• Per Hoffmann
• Daniel P. Howrigan
• Masashi Ikeda
• Conrad Iyegbe
• Inge Joa
• Antonio Julià
• Anna K. Kähler
• Tony Kam-Thong
• Yoichiro Kamatani
• Sena Karachanak-Yankova
• Oussama Kebir
• Matthew C. Keller
• Brian J. Kelly
• Andrey Khrunin
• Sung-Wan Kim
• Janis Klovins
• Nikolay Kondratiev
• Bettina Konte
• Julia Kraft
• Michiaki Kubo
• Vaidutis Kučinskas
• Zita Ausrele Kučinskiene
• Agung Kusumawardhani
• Hana Kuzelova-Ptackova
• Stefano Landi
• Laura C. Lazzeroni
• Phil H. Lee
• Sophie E. Legge
• Douglas S. Lehrer
• Rebecca Lencer
• Bernard Lerer
• Miaoxin Li
• Jeffrey Lieberman
• Gregory A. Light
• Svetlana Limborska
• Chih-Min Liu
• Jouko Lönnqvist
• Carmel M. Loughland
• Jan Lubinski
• Jurjen J. Luykx
• Amy Lynham
• Milan Macek Jr
• Andrew Mackinnon
• Patrik K. E. Magnusson
• Brion S. Maher
• Wolfgang Maier
• Dolores Malaspina
• Jacques Mallet
• Stephen R. Marder
• Sara Marsal
• Alicia R. Martin
• Lourdes Martorell
• Manuel Mattheisen
• Robert W. McCarley
• Colm McDonald
• John J. McGrath
• Helena Medeiros
• Sandra Meier
• Bela Melegh
• Ingrid Melle
• Raquelle I. Mesholam-Gately
• Andres Metspalu
• Patricia T. Michie
• Lili Milani
• Vihra Milanova
• Marina Mitjans
• Espen Molden
• Esther Molina
• María Dolores Molto
• Valeria Mondelli
• Carmen Moreno
• Christopher P. Morley
• Gerard Muntané
• Kieran C. Murphy
• Inez Myin-Germeys
• Igor Nenadić
• Gerald Nestadt
• Liene Nikitina-Zake
• Cristiano Noto
• Keith H. Nuechterlein
• Niamh Louise O’Brien
• F. Anthony O’Neill
• Sang-Yun Oh
• Ann Olincy
• Vanessa Kiyomi Ota
• Christos Pantelis
• George N. Papadimitriou
• Mara Parellada
• Tiina Paunio
• Renata Pellegrino
• Sathish Periyasamy
• Diana O. Perkins
• Bruno Pfuhlmann
• Olli Pietiläinen
• Jonathan Pimm
• David Porteous
• John Powell
• Diego Quattrone
• Digby Quested
• Allen D. Radant
• Antonio Rampino
• Mark H. Rapaport
• Anna Rautanen
• Abraham Reichenberg
• Cheryl Roe
• Joshua L. Roffman
• Julian Roth
• Matthias Rothermundt
• Bart P. F. Rutten
• Safaa Saker-Delye
• Veikko Salomaa
• Julio Sanjuan
• Marcos Leite Santoro
• Adam Savitz
• Ulrich Schall
• Rodney J. Scott
• Larry J. Seidman
• Sally Isabel Sharp
• Jianxin Shi
• Larry J. Siever
• Kang Sim
• Nora Skarabis
• Petr Slominsky
• Hon-Cheong So
• Janet L. Sobell
• Erik Söderman
• Helen J. Stain
• Nils Eiel Steen
• Agnes A. Steixner-Kumar
• Elisabeth Stögmann
• William S. Stone
• Richard E. Straub
• Fabian Streit
• Eric Strengman
• T. Scott Stroup
• Mythily Subramaniam
• Catherine A. Sugar
• Jaana Suvisaari
• Dragan M. Svrakic
• Neal R. Swerdlow
• Jin P. Szatkiewicz
• Thi Minh Tam Ta
• Atsushi Takahashi
• Chikashi Terao
• Florence Thibaut
• Draga Toncheva
• Paul A. Tooney
• Silvia Torretta
• Sarah Tosato
• Gian Battista Tura
• Bruce I. Turetsky
• Alp Üçok
• Arne Vaaler
• Therese van Amelsvoort
• Ruud van Winkel
• Juha Veijola
• John Waddington
• Henrik Walter
• Anna Waterreus
• Bradley T. Webb
• Mark Weiser
• Nigel M. Williams
• Stephanie H. Witt
• Brandon K. Wormley
• Jing Qin Wu
• Zhida Xu
• Robert Yolken
• Clement C. Zai
• Wei Zhou
• Feng Zhu
• Fritz Zimprich
• Eşref Cem Atbaşoğlu
• Muhammad Ayub
• Alessandro Bertolino
• Donald W. Black
• Nicholas J. Bray
• Gerome Breen
• Nancy G. Buccola
• William F. Byerley
• Wei J. Chen
• C. Robert Cloninger
• Benedicto Crespo-Facorro
• Gary Donohoe
• Robert Freedman
• Cherrie Galletly
• Massimo Gennarelli
• David M. Hougaard
• Hai-Gwo Hwu
• Assen V. Jablensky
• Steven A. McCarroll
• Jennifer L. Moran
• Ole Mors
• Preben B. Mortensen
• Bertram Müller-Myhsok
• Amanda L. Neil
• Merete Nordentoft
• Michele T. Pato
• Tracey L. Petryshen
• Ann E. Pulver
• Thomas G. Schulze
• Jeremy M. Silverman
• Jordan W. Smoller
• Eli A. Stahl
• Debby W. Tsuang
• Elisabet Vilella
• Shi-Heng Wang
• Shuhua Xu
• Rolf Adolfsson
• Celso Arango
• Bernhard T. Baune
• Sintia Iole Belangero
• Anders D. Børglum
• David Braff
• Elvira Bramon
• Joseph D. Buxbaum
• Dominique Campion
• Jorge A. Cervilla
• Sven Cichon
• David A. Collier
• Aiden Corvin
• David Curtis
• Marta Di Forti
• Enrico Domenici
• Hannelore Ehrenreich
• Valentina Escott-Price
• Tõnu Esko
• Ayman H. Fanous
• Anna Gareeva
• Micha Gawlik
• Pablo V. Gejman
• Michael Gill
• Stephen J. Glatt
• Vera Golimbet
• Kyung Sue Hong
• Christina M. Hultman
• Steven E. Hyman
• Nakao Iwata
• Erik G. Jönsson
• René S. Kahn
• James L. Kennedy
• Elza Khusnutdinova
• George Kirov
• James A. Knowles
• Marie-Odile Krebs
• Claudine Laurent-Levinson
• Jimmy Lee
• Todd Lencz
• Douglas F. Levinson
• Qingqin S. Li
• Jianjun Liu
• Anil K. Malhotra
• Dheeraj Malhotra
• Andrew McIntosh
• Andrew McQuillin
• Paulo R. Menezes
• Vera A. Morgan
• Derek W. Morris
• Bryan J. Mowry
• Robin M. Murray
• Vishwajit Nimgaonkar
• Markus M. Nöthen
• Roel A. Ophoff
• Sara A. Paciga
• Aarno Palotie
• Carlos N. Pato
• Shengying Qin
• Marcella Rietschel
• Brien P. Riley
• Margarita Rivera
• Dan Rujescu
• Meram C. Saka
• Alan R. Sanders
• Sibylle G. Schwab
• Alessandro Serretti
• Pak C. Sham
• Yongyong Shi
• David St Clair
• Ming T. Tsuang
• Jim van Os
• Marquis P. Vawter
• Daniel R. Weinberger
• Thomas Werge
• Dieter B. Wildenauer
• Xin Yu
• Weihua Yue
• Peter A. Holmans
• Panos Roussos
• Evangelos Vassos
• Danielle Posthuma
• Ole A. Andreassen
• Kenneth S. Kendler
• Michael J. Owen
• Naomi R. Wray
• Mark J. Daly
• Hailiang Huang
• Benjamin M. Neale
• Patrick F. Sullivan
• Stephan Ripke
• James T. R. Walters
• Michael C. O’Donovan
Contributor Information
Vassily Trubetskoy, Department of Psychiatry and Psychotherapy, Charité - Universitätsmedizin, Berlin, Germany.
Georgia Panagiotaropoulou, Department of Psychiatry and Psychotherapy, Charité - Universitätsmedizin, Berlin, Germany.
Swapnil Awasthi, Department of Psychiatry and Psychotherapy, Charité - Universitätsmedizin, Berlin, Germany.
Alice Braun, Department of Psychiatry and Psychotherapy, Charité - Universitätsmedizin, Berlin, Germany.
Julia Kraft, Department of Psychiatry and Psychotherapy, Charité - Universitätsmedizin, Berlin, Germany.
Nora Skarabis, Department of Psychiatry and Psychotherapy, Charité - Universitätsmedizin, Berlin, Germany.
Henrik Walter, Department of Psychiatry and Psychotherapy, Charité - Universitätsmedizin, Berlin, Germany.
Stephan Ripke, Department of Psychiatry and Psychotherapy, Charité - Universitätsmedizin, Berlin, Germany.
Antonio F. Pardiñas, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Charlotte A. Dennison, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Lynsey S. Hall, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Janet C. Harwood, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Alexander L. Richards, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Sophie E. Legge, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Amy Lynham, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK.
Nigel M. Williams, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Nicholas J. Bray, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Valentina Escott-Price, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK.
George Kirov, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK.
Peter A. Holmans, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Andrew J. Pocklington, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Michael J. Owen, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
James T. R. Walters, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Michael C. O’Donovan, MRC Centre for Neuropsychiatric Genetics and Genomics, Division of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK
Ting Qi, Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland, Australia.
Julia Sidorenko, Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland, Australia.
Yang Wu, Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland, Australia.
Jian Zeng, Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland, Australia.
Jacob Gratten, Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland, Australia.
Peter M. Visscher, Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland, Australia
Jian Yang, Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland, Australia.
Naomi R. Wray, Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland, Australia
Ting Qi, School of Life Sciences, Westlake University, Hangzhou, China.
Jian Yang, School of Life Sciences, Westlake University, Hangzhou, China.
Tim B. Bigdeli, Department of Psychiatry and the Behavioral Sciences, SUNY Downstate Medical Center, New York, NY, USA Department of Psychiatry, Veterans Affairs New York Harbor Healthcare System, New York, NY, USA; Institute for Genomic Health, SUNY Downstate Medical Center, New York, NY, USA.
Ayman H. Fanous, Department of Psychiatry, Veterans Affairs New York Harbor Healthcare System, New York, NY, USA
Julien Bryois, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden.
Sarah E. Bergen, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden
Anna K. Kähler, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden
Patrik K. E. Magnusson, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden
Christina M. Hultman, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden
Patrick F. Sullivan, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden
Chia-Yen Chen, Biogen, Cambridge, MA, USA; Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA; Psychiatric and Neurodevelopmental Genetics Unit, Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA, USA.
Elizabeth G. Atkinson, Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Jacqueline I. Goldstein, Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Daniel P. Howrigan, Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Alicia R. Martin, Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Mark J. Daly, Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Hailiang Huang, Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA.
Benjamin M. Neale, Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA
Stephan Ripke, Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA.
Tian Ge, Psychiatric and Neurodevelopmental Genetics Unit, Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA, USA.
Max Lam, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Tian Ge, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Elizabeth G. Atkinson, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Richard A. Belliveau, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Kimberley D. Chambert, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Giulio Genovese, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Phil H. Lee, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Alicia R. Martin, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Olli Pietiläinen, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Steven A. McCarroll, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Jennifer L. Moran, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Jordan W. Smoller, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Tyler C. Brown, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Guoping Feng, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Steven E. Hyman, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Morgan Sheng, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Steven E. Hyman, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Hailiang Huang, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Benjamin M. Neale, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Max Lam, Research Division, Institute of Mental Health, Singapore, Republic of Singapore.
Siow Ann Chong, Research Division, Institute of Mental Health, Singapore, Republic of Singapore.
Mythily Subramaniam, Research Division, Institute of Mental Health, Singapore, Republic of Singapore.
Max Lam, Division of Psychiatry Research, Zucker Hillside Hospital, Glen Oaks, NY, USA.
Todd Lencz, Division of Psychiatry Research, Zucker Hillside Hospital, Glen Oaks, NY, USA.
Anil K. Malhotra, Division of Psychiatry Research, Zucker Hillside Hospital, Glen Oaks, NY, USA
Kyoko Watanabe, Department of Complex Trait Genetics, Center for Neurogenomics and Cognitive Research, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
Oleksandr Frei, NORMENT Centre, Division of Mental Health and Addiction, University of Oslo, Oslo, Norway.
Ingrid Agartz, NORMENT Centre, Division of Mental Health and Addiction, University of Oslo, Oslo, Norway.
Lavinia Athanasiu, NORMENT Centre, Division of Mental Health and Addiction, University of Oslo, Oslo, Norway.
Ingrid Melle, NORMENT Centre, Division of Mental Health and Addiction, University of Oslo, Oslo, Norway.
Ole A. Andreassen, NORMENT Centre, Division of Mental Health and Addiction, University of Oslo, Oslo, Norway
Oleksandr Frei, Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway.
Lavinia Athanasiu, Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway.
Ingrid Melle, Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway.
Nils Eiel Steen, Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway.
Ole A. Andreassen, Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway
Oleksandr Frei, Center for Bioinformatics, Department of Informatics, University of Oslo, Oslo, Norway.
Tian Ge, Department of Psychiatry, Harvard Medical School, Boston, MA, USA.
Lynn E. DeLisi, Department of Psychiatry, Harvard Medical School, Boston, MA, USA
Raquelle I. Mesholam-Gately, Department of Psychiatry, Harvard Medical School, Boston, MA, USA
Larry J. Seidman, Department of Psychiatry, Harvard Medical School, Boston, MA, USA
Frank Koopmans, Department of Molecular and Cellular Neurobiology, Center for Neurogenomics and Cognitive Research, Faculty of Science, Amsterdam Neuroscience, Vrije Universiteit, Amsterdam, The Netherlands.
Sigurdur Magnusson, deCODE Genetics, Amgen, Reykjavik, Iceland.
Hreinn Stefánsson, deCODE Genetics, Amgen, Reykjavik, Iceland.
Kari Stefansson, deCODE Genetics, Amgen, Reykjavik, Iceland.
Jakob Grove, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), Aarhus, Denmark.
Esben Agerbo, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), Aarhus, Denmark; National Centre for Register-based Research, Aarhus University, Aarhus, Denmark.
Thomas D. Als, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), Aarhus, Denmark
Jonas Bybjerg-Grauholm, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), Aarhus, Denmark.
Ditte Demontis, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), Aarhus, Denmark.
David M. Hougaard, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), Aarhus, Denmark
Ole Mors, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), Aarhus, Denmark.
Preben B. Mortensen, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), Aarhus, Denmark National Centre for Register-based Research, Aarhus University, Aarhus, Denmark.
Merete Nordentoft, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), Aarhus, Denmark.
Anders D. Børglum, The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), Aarhus, Denmark
Jakob Grove, Department of Biomedicine and Centre for Integrative Sequencing (iSEQ), Aarhus University, Aarhus, Denmark.
Thomas D. Als, Department of Biomedicine and Centre for Integrative Sequencing (iSEQ), Aarhus University, Aarhus, Denmark
Ditte Demontis, Department of Biomedicine and Centre for Integrative Sequencing (iSEQ), Aarhus University, Aarhus, Denmark.
Manuel Mattheisen, Department of Biomedicine and Centre for Integrative Sequencing (iSEQ), Aarhus University, Aarhus, Denmark.
Anders D. Børglum, Department of Biomedicine and Centre for Integrative Sequencing (iSEQ), Aarhus University, Aarhus, Denmark
Jakob Grove, Center for Genomics and Personalized Medicine, Aarhus, Denmark.
Thomas D. Als, Center for Genomics and Personalized Medicine, Aarhus, Denmark
Ditte Demontis, Center for Genomics and Personalized Medicine, Aarhus, Denmark.
Anders D. Børglum, Center for Genomics and Personalized Medicine, Aarhus, Denmark
Minsoo Kim, Department of Psychiatry, Semel Institute, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, USA.
Michael J. Gandal, Department of Psychiatry, Semel Institute, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, USA
Zhiqiang Li, Affiliated Hospital of Qingdao University and Biomedical Sciences Institute of Qingdao University (Qingdao Branch of SJTU Bio-X Institutes), Qingdao University, Qingdao, China.
Yongyong Shi, Affiliated Hospital of Qingdao University and Biomedical Sciences Institute of Qingdao University (Qingdao Branch of SJTU Bio-X Institutes), Qingdao University, Qingdao, China.
Yongyong Shi, Affiliated Hospital of Qingdao University and Biomedical Sciences Institute of Qingdao University (Qingdao Branch of SJTU Bio-X Institutes), Qingdao University, Qingdao, China.
Zhiqiang Li, Bio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders (Ministry of Education), Collaborative Innovation Center for Brain Science, Shanghai Jiao Tong University, Shanghai, China.
Wei Zhou, Bio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders (Ministry of Education), Collaborative Innovation Center for Brain Science, Shanghai Jiao Tong University, Shanghai, China.
Shengying Qin, Bio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders (Ministry of Education), Collaborative Innovation Center for Brain Science, Shanghai Jiao Tong University, Shanghai, China.
Yongyong Shi, Bio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders (Ministry of Education), Collaborative Innovation Center for Brain Science, Shanghai Jiao Tong University, Shanghai, China.
Yongyong Shi, Bio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders (Ministry of Education), Collaborative Innovation Center for Brain Science, Shanghai Jiao Tong University, Shanghai, China.
Georgios Voloudakis, Department of Psychiatry, Pamela Sklar Division of Psychiatric Genomics, Friedman Brain Institute, Department of Genetics and Genomic Science and Institute for Data Science and Genomic Technology, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Wen Zhang, Department of Psychiatry, Pamela Sklar Division of Psychiatric Genomics, Friedman Brain Institute, Department of Genetics and Genomic Science and Institute for Data Science and Genomic Technology, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Panos Roussos, Department of Psychiatry, Pamela Sklar Division of Psychiatric Genomics, Friedman Brain Institute, Department of Genetics and Genomic Science and Institute for Data Science and Genomic Technology, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Wen Zhang, Department of Genetics and Genomic Sciences and Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Mark Adams, Division of Psychiatry, Centre for Clinical Brain Sciences, University of Edinburgh, Royal Edinburgh Hospital, Edinburgh, UK.
Andrew McIntosh, Division of Psychiatry, Centre for Clinical Brain Sciences, University of Edinburgh, Royal Edinburgh Hospital, Edinburgh, UK.
Ingrid Agartz, Department of Psychiatric Research, Diakonhjemmet Hospital, Oslo, Norway.
Ingrid Agartz, Centre for Psychiatry Research, Department of Clinical Neuroscience, Karolinska Institutet and Stockholm Health Care Services, Stockholm Region, Stockholm, Sweden.
Erik Söderman, Centre for Psychiatry Research, Department of Clinical Neuroscience, Karolinska Institutet and Stockholm Health Care Services, Stockholm Region, Stockholm, Sweden.
Erik G. Jönsson, Centre for Psychiatry Research, Department of Clinical Neuroscience, Karolinska Institutet and Stockholm Health Care Services, Stockholm Region, Stockholm, Sweden
John J. McGrath, National Centre for Register-based Research, Aarhus University, Aarhus, Denmark
Mariam Al Eissa, Molecular Psychiatry Laboratory, Division of Psychiatry, University College London, London, UK.
Nicholas J. Bass, Molecular Psychiatry Laboratory, Division of Psychiatry, University College London, London, UK
Alessia Fiorentino, Molecular Psychiatry Laboratory, Division of Psychiatry, University College London, London, UK.
Niamh Louise O’Brien, Molecular Psychiatry Laboratory, Division of Psychiatry, University College London, London, UK.
Jonathan Pimm, Molecular Psychiatry Laboratory, Division of Psychiatry, University College London, London, UK.
Sally Isabel Sharp, Molecular Psychiatry Laboratory, Division of Psychiatry, University College London, London, UK.
Andrew McQuillin, Molecular Psychiatry Laboratory, Division of Psychiatry, University College London, London, UK.
Margot Albus, Comedicum Lindwurmhof, Munich, Germany.
Madeline Alexander, Center for Depression, Anxiety and Stress Research, McLean Hospital, Belmont, MA, USA.
Behrooz Z. Alizadeh, University Medical Center Groningen, University Center for Psychiatry, Rob Giel Research Center, University of Groningen, Groningen, The Netherlands
Richard Bruggeman, University Medical Center Groningen, University Center for Psychiatry, Rob Giel Research Center, University of Groningen, Groningen, The Netherlands.
Behrooz Z. Alizadeh, Department of Epidemiology, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands
Köksal Alptekin, Department of Psychiatry, Dokuz Eylül University School of Medicine, Izmir, Turkey.
Köksal Alptekin, Department of Neuroscience, Dokuz Eylül University Graduate School of Health Sciences, Izmir, Turkey.
Farooq Amin, Department of Psychiatry and Behavioral Sciences, Emory University, Atlanta, GA, USA.
Volker Arolt, Department of Psychiatry, University of Münster, Münster, Germany.
Rebecca Lencer, Department of Psychiatry, University of Münster, Münster, Germany.
Matthias Rothermundt, Department of Psychiatry, University of Münster, Münster, Germany.
Bernhard T. Baune, Department of Psychiatry, University of Münster, Münster, Germany
Manuel Arrojo, Servizo de Psiquiatría, Complexo Hospitalario Universitario de Santiago de Compostela, Servizo Galego de Saúde (SERGAS), Santiago de Compostela, Spain.
Maria Helena Azevedo, Institute of Medical Psychology, Faculty of Medicine, University of Coimbra, Coimbra, Portugal.
Silviu A. Bacanu, Virginia Institute for Psychiatric and Behavioral Genetics, Department of Psychiatry, Virginia Commonwealth University, Richmond, VA, USA
Bradley T. Webb, Virginia Institute for Psychiatric and Behavioral Genetics, Department of Psychiatry, Virginia Commonwealth University, Richmond, VA, USA
Brandon K. Wormley, Virginia Institute for Psychiatric and Behavioral Genetics, Department of Psychiatry, Virginia Commonwealth University, Richmond, VA, USA
Brien P. Riley, Virginia Institute for Psychiatric and Behavioral Genetics, Department of Psychiatry, Virginia Commonwealth University, Richmond, VA, USA
Kenneth S. Kendler, Virginia Institute for Psychiatric and Behavioral Genetics, Department of Psychiatry, Virginia Commonwealth University, Richmond, VA, USA
Martin Begemann, Clinical Neuroscience, Max Planck Institute of Experimental Medicine, Göttingen, Germany.
Marina Mitjans, Clinical Neuroscience, Max Planck Institute of Experimental Medicine, Göttingen, Germany.
Agnes A. Steixner-Kumar, Clinical Neuroscience, Max Planck Institute of Experimental Medicine, Göttingen, Germany
Hannelore Ehrenreich, Clinical Neuroscience, Max Planck Institute of Experimental Medicine, Göttingen, Germany.
Judit Bene, Department of Medical Genetics, Medical School, University of Pécs, Pécs, Hungary.
Beben Benyamin, Australian Centre for Precision Health, University of South Australia Cancer Research Institute, University of South Australia, Adelaide, South Australia, Australia.
Beben Benyamin, UniSA Allied Health and Human Performance, University of South Australia, Adelaide, South Australia, Australia.
Beben Benyamin, South Australian Health and Medical Research Institute, Adelaide, South Australia, Australia.
Giuseppe Blasi, Department of Basic Medical Science, Neuroscience and Sense Organs, University of Bari ‘Aldo Moro’, Bari, Italy.
Antonio Rampino, Department of Basic Medical Science, Neuroscience and Sense Organs, University of Bari ‘Aldo Moro’, Bari, Italy.
Silvia Torretta, Department of Basic Medical Science, Neuroscience and Sense Organs, University of Bari ‘Aldo Moro’, Bari, Italy.
Alessandro Bertolino, Department of Basic Medical Science, Neuroscience and Sense Organs, University of Bari ‘Aldo Moro’, Bari, Italy.
Julio Bobes, Área de Psiquiatría-Universidad de Oviedo, Hospital Universitario Central de Asturias (HUCA), Asturias, Spain.
Julio Bobes, Instituto de Investigación Sanitaria del Principado de Asturias (ISPA), Oviedo, Asturias, Spain.
Julio Bobes, Centro de Investigación Biomédica en Red de Salud Mental, Oviedo, Asturias, Spain.
Stefano Bonassi, Unit of Clinical and Molecular Epidemiology, IRCCS San Raffaele Roma and San Raffaele University, Rome, Italy.
Rodrigo Affonseca Bressan, Department of Psychiatry, Universidade Federal de São Paulo, São Paulo, Brazil.
Ary Gadelha, Department of Psychiatry, Universidade Federal de São Paulo, São Paulo, Brazil.
Cristiano Noto, Department of Psychiatry, Universidade Federal de São Paulo, São Paulo, Brazil.
Rodrigo Affonseca Bressan, Laboratory of Integrative Neuroscience, Universidade Federal de São Paulo, São Paulo, Brazil.
Ary Gadelha, Laboratory of Integrative Neuroscience, Universidade Federal de São Paulo, São Paulo, Brazil.
Cristiano Noto, Laboratory of Integrative Neuroscience, Universidade Federal de São Paulo, São Paulo, Brazil.
Vanessa Kiyomi Ota, Laboratory of Integrative Neuroscience, Universidade Federal de São Paulo, São Paulo, Brazil.
Marcos Leite Santoro, Laboratory of Integrative Neuroscience, Universidade Federal de São Paulo, São Paulo, Brazil.
Sintia Iole Belangero, Laboratory of Integrative Neuroscience, Universidade Federal de São Paulo, São Paulo, Brazil.
Evelyn J. Bromet, Department of Psychiatry and Behavioural Health, Stony Brook University, Stony Brook, NY, USA
Richard Bruggeman, Department of Clinical and Developmental Neuropsychology, University of Groningen, Groningen, The Netherlands.
Peter F. Buckley, Health Science Center, University of Tennessee, Memphis, TN, USA
Randy L. Buckner, Department of Psychology, Harvard University, Cambridge, MA, USA
Jonas Bybjerg-Grauholm, Center for Neonatal Screening, Department for Congenital Disorders, Statens Serum Institut, Copenhagen, Denmark.
David M. Hougaard, Center for Neonatal Screening, Department for Congenital Disorders, Statens Serum Institut, Copenhagen, Denmark
Wiepke Cahn, University Medical Center Utrecht, Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, Utrecht, The Netherlands.
René S. Kahn, University Medical Center Utrecht, Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, Utrecht, The Netherlands
Wiepke Cahn, Altrecht, General Menthal Health Care, Utrecht, The Netherlands.
Murray J. Cairns, School of Biomedical Sciences and Pharmacy, University of Newcastle, Newcastle, New South Wales, Australia
Rodney J. Scott, School of Biomedical Sciences and Pharmacy, University of Newcastle, Newcastle, New South Wales, Australia
Paul A. Tooney, School of Biomedical Sciences and Pharmacy, University of Newcastle, Newcastle, New South Wales, Australia
Murray J. Cairns, Hunter Medical Research Institute, Newcastle, New South Wales, Australia
Ulrich Schall, Hunter Medical Research Institute, Newcastle, New South Wales, Australia.
Rodney J. Scott, Hunter Medical Research Institute, Newcastle, New South Wales, Australia
Paul A. Tooney, Hunter Medical Research Institute, Newcastle, New South Wales, Australia
Murray J. Cairns, Centre for Brain and Mental Health Research, University of Newcastle, Newcastle, New South Wales, Australia
Paul A. Tooney, Centre for Brain and Mental Health Research, University of Newcastle, Newcastle, New South Wales, Australia
Monica E. Calkins, Department of Psychiatry, University of Pennsylvania, Philadelphia, PA, USA
Raquel E. Gur, Department of Psychiatry, University of Pennsylvania, Philadelphia, PA, USA
Ruben C. Gur, Department of Psychiatry, University of Pennsylvania, Philadelphia, PA, USA
Bruce I. Turetsky, Department of Psychiatry, University of Pennsylvania, Philadelphia, PA, USA
Vaughan J. Carr, School of Psychiatry, University of New South Wales, Sydney, New South Wales, Australia
Vaughan J. Carr, Department of Psychiatry, Monash University, Melbourne, Victoria, Australia
Vaughan J. Carr, Neuroscience Research Australia, Sydney, New South Wales, Australia
David Castle, Department of Psychiatry, University of Melbourne, Parkville, Victoria, Australia.
Carol Harvey, Department of Psychiatry, University of Melbourne, Parkville, Victoria, Australia.
David Castle, St Vincent’s Hospital, Melbourne, Victoria, Australia.
Stanley V. Catts, Brain and Mind Centre, The University of Sydney, Sydney, New South Wales, Australia
Stanley V. Catts, School of Medicine, University of Queensland, Herston, Queensland, Australia
Raymond C. K. Chan, Institute of Psychology, Chinese Academy of Science, Beijing, China
Raymond C. K. Chan, Department of Psychology, University of Chinese Academy of Sciences, Beijing, China
Boris Chaumette, INSERM U1266, Institute of Psychiatry and Neuroscience of Paris, Université de Paris, GHU Paris Psychiatrie & Neurosciences, Paris, France.
Oussama Kebir, INSERM U1266, Institute of Psychiatry and Neuroscience of Paris, Université de Paris, GHU Paris Psychiatrie & Neurosciences, Paris, France.
Marie-Odile Krebs, INSERM U1266, Institute of Psychiatry and Neuroscience of Paris, Université de Paris, GHU Paris Psychiatrie & Neurosciences, Paris, France.
Boris Chaumette, Department of Psychiatry, McGill University, Montreal, Québec, Canada.
Wei Cheng, Department of Computer Science, University of North Carolina, Chapel Hill, NC, USA.
Eric F. C. Cheung, Castle Peak Hospital, Hong Kong, China
Siow Ann Chong, Saw Swee Hock School of Public Health, National University of Singapore, Singapore, Republic of Singapore.
Mythily Subramaniam, Saw Swee Hock School of Public Health, National University of Singapore, Singapore, Republic of Singapore.
David Cohen, Faculté de Médecine Sorbonne Université, Groupe de Recherche Clinique no. 15 - Troubles Psychiatriques et Développement (PSYDEV), Department of Child and Adolescent Psychiatry, Hôpital Universitaire de la Pitié-Salpêtrière, Paris, France.
Angèle Consoli, Faculté de Médecine Sorbonne Université, Groupe de Recherche Clinique no. 15 - Troubles Psychiatriques et Développement (PSYDEV), Department of Child and Adolescent Psychiatry, Hôpital Universitaire de la Pitié-Salpêtrière, Paris, France.
Marianna Giannitelli, Faculté de Médecine Sorbonne Université, Groupe de Recherche Clinique no. 15 - Troubles Psychiatriques et Développement (PSYDEV), Department of Child and Adolescent Psychiatry, Hôpital Universitaire de la Pitié-Salpêtrière, Paris, France.
Claudine Laurent-Levinson, Faculté de Médecine Sorbonne Université, Groupe de Recherche Clinique no. 15 - Troubles Psychiatriques et Développement (PSYDEV), Department of Child and Adolescent Psychiatry, Hôpital Universitaire de la Pitié-Salpêtrière, Paris, France.
David Cohen, Centre de Référence des Maladies Rares à Expression Psychiatrique, Department of Child and Adolescent Psychiatry, AP-HP Sorbonne Université, Hôpital Universitaire de la Pitié-Salpêtrière, Paris, France.
Angèle Consoli, Centre de Référence des Maladies Rares à Expression Psychiatrique, Department of Child and Adolescent Psychiatry, AP-HP Sorbonne Université, Hôpital Universitaire de la Pitié-Salpêtrière, Paris, France.
Marianna Giannitelli, Centre de Référence des Maladies Rares à Expression Psychiatrique, Department of Child and Adolescent Psychiatry, AP-HP Sorbonne Université, Hôpital Universitaire de la Pitié-Salpêtrière, Paris, France.
Claudine Laurent-Levinson, Centre de Référence des Maladies Rares à Expression Psychiatrique, Department of Child and Adolescent Psychiatry, AP-HP Sorbonne Université, Hôpital Universitaire de la Pitié-Salpêtrière, Paris, France.
David Cohen, Institut des Systèmes Intelligents et de Robotique (ISIR), CNRS UMR7222, Faculté des Sciences et Ingénierie, Sorbonne Université, Paris, France.
Quirino Cordeiro, Department of Psychiatry, Irmandade da Santa Casa de Misericórdia de São Paulo, São Paulo, Brazil.
Javier Costas, Instituto de Investigación Sanitaria (IDIS) de Santiago de Compostela, Complexo Hospitalario Universitario de Santiago de Compostela (CHUS), Servizo Galego de Saúde (SERGAS), Santiago de Compostela, Spain.
Charles Curtis, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK.
Diego Quattrone, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK.
Gerome Breen, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK.
David A. Collier, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK
Marta Di Forti, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK.
Evangelos Vassos, Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK.
Charles Curtis, National Institute for Health Research (NIHR) Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, London, UK.
Valeria Mondelli, National Institute for Health Research (NIHR) Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, London, UK.
Diego Quattrone, National Institute for Health Research (NIHR) Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, London, UK.
Therese van Amelsvoort, National Institute for Health Research (NIHR) Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, London, UK.
Marta Di Forti, National Institute for Health Research (NIHR) Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, London, UK.
Robin M. Murray, National Institute for Health Research (NIHR) Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, London, UK
Evangelos Vassos, National Institute for Health Research (NIHR) Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, London, UK.
Therese van Amelsvoort, National Institute for Health Research (NIHR) Maudsley Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, London, UK.
Michael Davidson, University of Nicosia Medical School, Nicosia, Cyprus.
Kenneth L. Davis, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA
Vahram Haroutunian, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Dolores Malaspina, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Abraham Reichenberg, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Larry J. Siever, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA
Jeremy M. Silverman, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA
Joseph D. Buxbaum, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA
René S. Kahn, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA
Lieuwe de Haan, Department of Psychiatry, Academic Medical Centre, University of Amsterdam, Amsterdam, The Netherlands.
Lieuwe de Haan, Department of Psychiatry, Academic Medical Centre, University of Amsterdam, Amsterdam, The Netherlands.
Lieuwe de Haan, Arkin, Institute for Mental Health, Amsterdam, The Netherlands.
Lieuwe de Haan, Arkin, Institute for Mental Health, Amsterdam, The Netherlands.
Franziska Degenhardt, Institute of Human Genetics, University of Bonn, Bonn, Germany.
Andreas Forstner, Institute of Human Genetics, University of Bonn, Bonn, Germany.
Markus M. Nöthen, Institute of Human Genetics, University of Bonn, Bonn, Germany
Lynn E. DeLisi, Cambridge Health Alliance, Cambridge, MA, USA
Faith Dickerson, Sheppard Pratt Health System, Baltimore, MD, USA.
Dimitris Dikeos, First Department of Psychiatry, Medical School, National and Kapodistrian University of Athens, Eginition Hospital, Athens, Greece.
George N. Papadimitriou, First Department of Psychiatry, Medical School, National and Kapodistrian University of Athens, Eginition Hospital, Athens, Greece
Timothy Dinan, Department of Psychiatry and Neurobehavioural Sciences, University College Cork, Cork, Ireland.
Timothy Dinan, APC Microbiome Ireland, University College Cork, Cork, Ireland.
Srdjan Djurovic, NORMENT Centre, Department of Clinical Science, University of Bergen, Bergen, Norway.
Srdjan Djurovic, Department of Medical Genetics, Oslo University Hospital, Oslo, Norway.
Jubao Duan, Center for Psychiatric Genetics, NorthShore University HealthSystem, Evanston, IL, USA.
Pablo V. Gejman, Center for Psychiatric Genetics, NorthShore University HealthSystem, Evanston, IL, USA
Alan R. Sanders, Center for Psychiatric Genetics, NorthShore University HealthSystem, Evanston, IL, USA
Jubao Duan, Department of Psychiatry and Behavioral Neurosciences, The University of Chicago, Chicago, IL, USA.
Pablo V. Gejman, Department of Psychiatry and Behavioral Neurosciences, The University of Chicago, Chicago, IL, USA
Alan R. Sanders, Department of Psychiatry and Behavioral Neurosciences, The University of Chicago, Chicago, IL, USA
Giuseppe Ducci, Department of Mental Health, ASL Rome 1, Rome, Italy.
Frank Dudbridge, Department of Health Sciences, University of Leicester, Leicester, UK.
Johan G. Eriksson, Department of General Practice and Primary Health Care, University of Helsinki and Helsinki University Hospital, Helsinki, Finland
Johan G. Eriksson, Folkhälsan Research Center, Helsinki, Finland
Johan G. Eriksson, Department of Obstetrics and Gynecology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Republic of Singapore
Lourdes Fañanás, Department of Evolutionary Biology, Ecology and Environmental Sciences, Faculty of Biology, University of Barcelona, Barcelona, Spain.
Lourdes Fañanás, Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Madrid, Spain.
Javier González Peñas, Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Madrid, Spain.
Ana González-Pinto, Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Madrid, Spain.
María Dolores Molto, Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Madrid, Spain.
Carmen Moreno, Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Madrid, Spain.
Mara Parellada, Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Madrid, Spain.
Julio Sanjuan, Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Madrid, Spain.
Benedicto Crepo-Facorro, Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Madrid, Spain.
Ignacio Mata, Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Madrid, Spain.
Celso Arango, Centro de Investigación Biomédica en Red en Salud Mental (CIBERSAM), Madrid, Spain.
Stephen V. Faraone, Departments of Psychiatry and Neuroscience and Physiology, SUNY Upstate Medical University, Syracuse, NY, USA
Andreas Forstner, Centre for Human Genetics, University of Marburg, Marburg, Germany.
Josef Frank, Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, University of Heidelberg, Mannheim, Germany.
Fabian Streit, Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, University of Heidelberg, Mannheim, Germany.
Stephanie H. Witt, Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, University of Heidelberg, Mannheim, Germany
Marcella Rietschel, Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, University of Heidelberg, Mannheim, Germany.
Nelson B. Freimer, Department of Human Genetics, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, USA
Roel A. Ophoff, Department of Human Genetics, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, USA
Nelson B. Freimer, Department of Psychiatry and Biobehavioral Sciences, University of California Los Angeles, Los Angeles, CA, USA
Menachem Fromer, Division of Psychiatric Genomics, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Eli A. Stahl, Division of Psychiatric Genomics, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA
Alessandra Frustaci, Barnet, Enfield and Haringey Mental Health NHS Trust, St Ann’s Hospital, London, UK.
Elliot S. Gershon, Departments of Psychiatry and Human Genetics, University of Chicago, Chicago, IL, USA
Ina Giegling, Department of Psychiatry and Psychotherapy, Medical University of Vienna, Vienna, Austria.
Annette M. Hartmann, Department of Psychiatry and Psychotherapy, Medical University of Vienna, Vienna, Austria
Bettina Konte, Department of Psychiatry and Psychotherapy, Medical University of Vienna, Vienna, Austria.
Dan Rujescu, Department of Psychiatry and Psychotherapy, Medical University of Vienna, Vienna, Austria.
Paola Giusti-Rodríguez, Department of Genetics, University of North Carolina, Chapel Hill, NC, USA.
Jin P. Szatkiewicz, Department of Genetics, University of North Carolina, Chapel Hill, NC, USA
Patrick F. Sullivan, Department of Genetics, University of North Carolina, Chapel Hill, NC, USA
Stephanie Godard, Departments of Psychiatry and Human and Molecular Genetics, INSERM, Institut de Myologie, Hôpital de la Pitiè-Salpêtrière, Paris, France.
Javier González Peñas, Department of Child and Adolescent Psychiatry, Institute of Psychiatry and Mental Health, General Universitario Gregorio Marañón, School of Medicine, Universidad Complutense, IiSGM, Madrid, Spain.
Carmen Moreno, Department of Child and Adolescent Psychiatry, Institute of Psychiatry and Mental Health, General Universitario Gregorio Marañón, School of Medicine, Universidad Complutense, IiSGM, Madrid, Spain.
Mara Parellada, Department of Child and Adolescent Psychiatry, Institute of Psychiatry and Mental Health, General Universitario Gregorio Marañón, School of Medicine, Universidad Complutense, IiSGM, Madrid, Spain.
Celso Arango, Department of Child and Adolescent Psychiatry, Institute of Psychiatry and Mental Health, General Universitario Gregorio Marañón, School of Medicine, Universidad Complutense, IiSGM, Madrid, Spain.
Ana González-Pinto, BIOARABA Health Research Institute, OSI Araba, University Hospital, University of the Basque Country, Vitoria, Spain.
Srihari Gopal, Neuroscience Therapeutic Area, Janssen Research and Development, Titusville, NJ, USA.
Adam Savitz, Neuroscience Therapeutic Area, Janssen Research and Development, Titusville, NJ, USA.
Qingqin S. Li, Neuroscience Therapeutic Area, Janssen Research and Development, Titusville, NJ, USA
Jacob Gratten, Mater Research Institute, University of Queensland, Brisbane, Queensland, Australia.
Michael F. Green, Department of Psychiatry and Biobehavioral Sciences, Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, USA
Keith H. Nuechterlein, Department of Psychiatry and Biobehavioral Sciences, Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, USA
Catherine A. Sugar, Department of Psychiatry and Biobehavioral Sciences, Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, USA
Michael F. Green, VA Greater Los Angeles Healthcare System, Los Angeles, CA, USA
Tiffany A. Greenwood, Department of Psychiatry, University of California San Diego, La Jolla, CA, USA
Gregory A. Light, Department of Psychiatry, University of California San Diego, La Jolla, CA, USA
Neal R. Swerdlow, Department of Psychiatry, University of California San Diego, La Jolla, CA, USA
David Braff, Department of Psychiatry, University of California San Diego, La Jolla, CA, USA.
Olivier Guillin, INSERM, Rouen, France.
Dominique Campion, INSERM, Rouen, France.
Olivier Guillin, Centre Hospitalier du Rouvray, Rouen, France.
Dominique Campion, Centre Hospitalier du Rouvray, Rouen, France.
Olivier Guillin, UFR Santé, Université de Rouen Normandie, Rouen, France.
Sinan Gülöksüz, Department of Psychiatry and Neuropsychology, School for Mental Health and Neuroscience, Maastricht University Medical Centre, Maastricht, The Netherlands.
Jurjen J. Luykx, Department of Psychiatry and Neuropsychology, School for Mental Health and Neuroscience, Maastricht University Medical Centre, Maastricht, The Netherlands
Bart P. F. Rutten, Department of Psychiatry and Neuropsychology, School for Mental Health and Neuroscience, Maastricht University Medical Centre, Maastricht, The Netherlands
Therese van Amelsvoort, Department of Psychiatry and Neuropsychology, School for Mental Health and Neuroscience, Maastricht University Medical Centre, Maastricht, The Netherlands.
Ruud van Winkel, Department of Psychiatry and Neuropsychology, School for Mental Health and Neuroscience, Maastricht University Medical Centre, Maastricht, The Netherlands.
Therese van Amelsvoort, Department of Psychiatry and Neuropsychology, School for Mental Health and Neuroscience, Maastricht University Medical Centre, Maastricht, The Netherlands.
Ruud van Winkel, Department of Psychiatry and Neuropsychology, School for Mental Health and Neuroscience, Maastricht University Medical Centre, Maastricht, The Netherlands.
Sinan Gülöksüz, Department of Psychiatry, Yale School of Medicine, New Haven, CT, USA.
Blanca Gutiérrez, Department of Psychiatry, Faculty of Medicine and Biomedical Research Centre (CIBM), University of Granada, Granada, Spain.
Eric Hahn, Department of Psychiatry, Charité - Universitätsmedizin, Berlin, Germany.
Hakon Hakonarson, Children’s Hospital of Philadelphia, Leonard Madlyn Abramson Research Center, Philadelphia, PA, USA.
Renata Pellegrino, Children’s Hospital of Philadelphia, Leonard Madlyn Abramson Research Center, Philadelphia, PA, USA.
Vahram Haroutunian, Department of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Vahram Haroutunian, Mental Illness Research Clinical and Education Center (MIRECC), JJ Peters VA Medical Center, New York, NY, USA.
Carol Harvey, NorthWestern Mental Health, Melbourne, Victoria, Australia.
Christos Pantelis, NorthWestern Mental Health, Melbourne, Victoria, Australia.
Caroline Hayward, MRC Human Genetics Unit, University of Edinburgh, Institute of Genetics and Cancer, Western General Hospital, Edinburgh, UK.
Frans A. Henskens, School of Medicine and Public Health, University of Newcastle, Newcastle, New South Wales, Australia
Brian J. Kelly, School of Medicine and Public Health, University of Newcastle, Newcastle, New South Wales, Australia
Stefan Herms, Division of Medical Genetics, Department of Biomedicine, University of Basel, Basel, Switzerland.
Per Hoffmann, Division of Medical Genetics, Department of Biomedicine, University of Basel, Basel, Switzerland.
Daniel P. Howrigan, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Menachem Fromer, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Mark J. Daly, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Masashi Ikeda, Department of Psychiatry, Fujita Health University School of Medicine, Toyoake Aichi, Japan.
Nakao Iwata, Department of Psychiatry, Fujita Health University School of Medicine, Toyoake Aichi, Japan.
Conrad Iyegbe, Department of Psychosis Studies, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK.
Jim van Os, Department of Psychosis Studies, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK.
Inge Joa, Regional Centre for Clinical Research in Psychosis, Department of Psychiatry, Stavanger University Hospital, Stavanger, Norway.
Antonio Julià, Rheumatology Research Group, Vall d’Hebron Research Institute, Barcelona, Spain.
Sara Marsal, Rheumatology Research Group, Vall d’Hebron Research Institute, Barcelona, Spain.
Tony Kam-Thong, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, F. Hoffman-La Roche, Basel, Switzerland.
Anna Rautanen, Roche Pharma Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, F. Hoffman-La Roche, Basel, Switzerland.
Yoichiro Kamatani, Laboratory of Complex Trait Genomics, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Tokyo, Japan.
Yoichiro Kamatani, Laboratory for Statistical Analysis, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan.
Sena Karachanak-Yankova, Department of Medical Genetics, Medical University, Sofia, Bulgaria.
Draga Toncheva, Department of Medical Genetics, Medical University, Sofia, Bulgaria.
Sena Karachanak-Yankova, Department of Genetics, Faculty of Biology, Sofia University “St. Kliment Ohridski”, Sofia, Bulgaria.
Matthew C. Keller, Institute for Behavioural Genetics, University of Colorado Boulder, Boulder, CO, USA
Andrey Khrunin, Institute of Molecular Genetics of National Research Centre “Kurchatov Institute”, Moscow, Russia.
Svetlana Limborska, Institute of Molecular Genetics of National Research Centre “Kurchatov Institute”, Moscow, Russia.
Petr Slominsky, Institute of Molecular Genetics of National Research Centre “Kurchatov Institute”, Moscow, Russia.
Sung-Wan Kim, Department of Psychiatry, Chonnam National University Medical School, Gwangju, Korea.
Janis Klovins, Latvian Biomedical Research and Study Centre, Riga, Latvia.
Liene Nikitina-Zake, Latvian Biomedical Research and Study Centre, Riga, Latvia.
Nikolay Kondratiev, Mental Health Research Center, Moscow, Russian Federation.
Vera Golimbet, Mental Health Research Center, Moscow, Russian Federation.
Julia Kraft, Berlin School of Mind and Brain, Humboldt-Universität zu Berlin, Berlin, Germany.
Michiaki Kubo, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan.
Vaidutis Kučinskas, Faculty of Medicine, Vilnius University, Vilnius, Lithuania.
Zita Ausrele Kučinskiene, Faculty of Medicine, Vilnius University, Vilnius, Lithuania.
Agung Kusumawardhani, Psychiatry Department, University of Indonesia - Cipto Mangunkusumo National General Hospital, Jakarta, Indonesia.
Hana Kuzelova-Ptackova, Department of Psychiatry, 1st Faculty of Medicine and General University Hospital, Prague, Czech Republic.
Stefano Landi, Dipartimento di Biologia, Universita’ di Pisa, Pisa, Italy.
Laura C. Lazzeroni, Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, CA, USA
Douglas F. Levinson, Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, CA, USA
Laura C. Lazzeroni, Department of Biomedical Data Science, Stanford University, Stanford, CA, USA
Phil H. Lee, Psychiatric and Neurodevelopmental Genetics Unit, Department of Psychiatry and Center for Genomic Medicine, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA
Tracey L. Petryshen, Psychiatric and Neurodevelopmental Genetics Unit, Department of Psychiatry and Center for Genomic Medicine, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA
Jordan W. Smoller, Psychiatric and Neurodevelopmental Genetics Unit, Department of Psychiatry and Center for Genomic Medicine, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA
Douglas S. Lehrer, Department of Psychiatry, Wright State University, Dayton, OH, USA
Bernard Lerer, Department of Psychiatry, Hadassah-Hebrew University Medical Center, Jerusalem, Israel.
Miaoxin Li, Zhongshan School of Medicine and Key Laboratory of Tropical Diseases Control (SYSU), Sun Yat-sen University, Guangzhou, China.
Jeffrey Lieberman, Department of Psychiatry, Columbia University, New York, NY, USA.
T. Scott Stroup, Department of Psychiatry, Columbia University, New York, NY, USA.
Gregory A. Light, VISN 22, Mental Illness Research, Education and Clinical Center (MIRECC), VA San Diego Healthcare System, San Diego, CA, USA
David Braff, VISN 22, Mental Illness Research, Education and Clinical Center (MIRECC), VA San Diego Healthcare System, San Diego, CA, USA.
Chih-Min Liu, Department of Psychiatry, National Taiwan University Hospital, Taipei, Taiwan.
Chih-Min Liu, Neurobiology and Cognitive Science Center, National Taiwan University, Taipei, Taiwan.
Hai-Gwo Hwu, Neurobiology and Cognitive Science Center, National Taiwan University, Taipei, Taiwan.
Jouko Lönnqvist, Mental Health Unit, Department of Public Health Solutions, National Institute for Health and Welfare, Helsinki, Finland.
Jouko Lönnqvist, Department of Psychiatry, University of Helsinki, Helsinki, Finland.
Carmel M. Loughland, Hunter New England Health and University of Newcastle, Newcastle, New South Wales, Australia
Jan Lubinski, Department of Genetics and Pathology, International Hereditary Cancer Center, Pomeranian Medical University in Szczecin, Szczecin, Poland.
Jurjen J. Luykx, Department of Psychiatry, UMC Utrecht Brain Center, University Medical Centre Utrecht, Utrecht University, Utrecht, The Netherlands
Steven Bakker, Department of Psychiatry, UMC Utrecht Brain Center, University Medical Centre Utrecht, Utrecht University, Utrecht, The Netherlands.
René Kahn, Department of Psychiatry, UMC Utrecht Brain Center, University Medical Centre Utrecht, Utrecht University, Utrecht, The Netherlands.
Jurjen J. Luykx, Department of Translational Neuroscience, UMC Utrecht Brain Center, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands
Jurjen J. Luykx, Second Opinion Outpatient Clinic, GGNet Mental Health, Warnsveld, The Netherlands
Milan Macek, Jr, Department of Biology and Medical Genetics, 2nd Faculty of Medicine and University Hospital Motol, Prague, Czech Republic.
Andrew Mackinnon, Black Dog Institute, University of New South Wales, Randwick, New South Wales, Australia.
Andrew Mackinnon, Melbourne School of Population and Global Health, University of Melbourne, Melbourne, Victoria, Australia.
Brion S. Maher, Department of Mental Health, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, USA
Wolfgang Maier, Department for Neurodegenerative Diseases and Geriatric Psychiatry, University Hospital Bonn, Bonn, Germany.
Dolores Malaspina, Department of Genetics and Genomics, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Eşref Cem Atbaşoğlu, Department of Genetics and Genomics, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Jacques Mallet, Asfalia Biologics, iPEPS-ICM, Hôpital Universitaire de la Pitié-Salpêtrière, Paris, France.
Stephen R. Marder, Semel Institute for Neurosciene, University of California Los Angeles, Los Angeles, CA, USA
Alicia R. Martin, Department of Medicine, Harvard Medical School, Boston, MA, USA
Hailiang Huang, Department of Medicine, Harvard Medical School, Boston, MA, USA.
Lourdes Martorell, Hospital Universitari Institut Pere Mata, IISPV, Universitat Rovira i Virgili, CIBERSAM, Reus, Spain.
Gerard Muntané, Hospital Universitari Institut Pere Mata, IISPV, Universitat Rovira i Virgili, CIBERSAM, Reus, Spain.
Elisabet Vilella, Hospital Universitari Institut Pere Mata, IISPV, Universitat Rovira i Virgili, CIBERSAM, Reus, Spain.
Manuel Mattheisen, Department of Psychiatry, Dalhousie University, Halifax, Nova Scotia, Canada.
Sandra Meier, Department of Psychiatry, Dalhousie University, Halifax, Nova Scotia, Canada.
Manuel Mattheisen, Department of Community Health and Epidemiology, Dalhousie University, Halifax, Nova Scotia, Canada.
Manuel Mattheisen, Institute of Psychiatric Phenomics and Genomics (IPPG), University Hospital, LMU Munich, Munich, Germany.
Thomas G. Schulze, Institute of Psychiatric Phenomics and Genomics (IPPG), University Hospital, LMU Munich, Munich, Germany
Robert W. McCarley, VA Boston Health Care System, Brockton, MA, USA
Colm McDonald, Centre for Neuroimaging, Cognition and Genomics (NICOG), National University of Ireland Galway, Galway, Ireland.
Gary Donohoe, Centre for Neuroimaging, Cognition and Genomics (NICOG), National University of Ireland Galway, Galway, Ireland.
Derek W. Morris, Centre for Neuroimaging, Cognition and Genomics (NICOG), National University of Ireland Galway, Galway, Ireland
John J. McGrath, Queensland Brain Institute, University of Queensland, Brisbane, Queensland, Australia
Sathish Periyasamy, Queensland Brain Institute, University of Queensland, Brisbane, Queensland, Australia.
Bryan J. Mowry, Queensland Brain Institute, University of Queensland, Brisbane, Queensland, Australia
Naomi R. Wray, Queensland Brain Institute, University of Queensland, Brisbane, Queensland, Australia
John J. McGrath, Queensland Centre for Mental Health Research, The Park Centre for Mental Health, Brisbane, Queensland, Australia
Helena Medeiros, Department of Psychiatry and the Behavioral Sciences, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Janet L. Sobell, Department of Psychiatry and the Behavioral Sciences, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA
Helena Medeiros, College of Medicine, SUNY Downstate Health Sciences University, New York, NY, USA.
Sandra Meier, Department of Biomedicine, Aarhus University, Aarhus, Denmark.
Bela Melegh, Department of Medical Genetics, University of Pécs, School of Medicine, Pécs, Hungary.
Raquelle I. Mesholam-Gately, Massachusetts Mental Health Center Public Psychiatry Division of the Beth Israel Deaconess Medical Center, Boston, MA, USA
Larry J. Seidman, Massachusetts Mental Health Center Public Psychiatry Division of the Beth Israel Deaconess Medical Center, Boston, MA, USA
Andres Metspalu, Estonian Genome Center, Institute of Genomics, University of Tartu, Tartu, Estonia.
Lili Milani, Estonian Genome Center, Institute of Genomics, University of Tartu, Tartu, Estonia.
Tõnu Esko, Estonian Genome Center, Institute of Genomics, University of Tartu, Tartu, Estonia.
Patricia T. Michie, School of Psychology, University of Newcastle, Newcastle, New South Wales, Australia
Vihra Milanova, Psychiatric Clinic, Alexandrovska University Hospital, Sofia, Bulgaria.
Espen Molden, Department of Pharmacy, University of Oslo, Oslo, Norway.
Espen Molden, Center for Psychopharmacology, Diakonhjemmet Hospital, Oslo, Norway.
Esther Molina, Department of Nursing, Faculty of Health Sciences and Biomedical Research Centre (CIBM), University of Granada, Granada, Spain.
María Dolores Molto, Department of Genetics, Faculty of Biological Sciences, Universidad de Valencia, Valencia, Spain.
María Dolores Molto, Biomedical Research Institute INCLIVA, Valencia, Spain.
Julio Sanjuan, Biomedical Research Institute INCLIVA, Valencia, Spain.
Valeria Mondelli, Department of Psychological Medicine, Institute of Psychiatry, Psychology, and Neuroscience, King’s College London, London, UK.
Christopher P. Morley, Departments of Public Health and Preventive Medicine, Family Medicine, and Psychiatry and Behavioral Sciences, State University of New York, Upstate Medical University, Syracuse, NY, USA
Gerard Muntané, Institut de Biologia Evolutiva (UPF-CSIC), Departament de Ciències Experimentals i de la Salut, Universitat Pompeu Fabra, PRBB, Barcelona, Spain.
Kieran C. Murphy, Department of Psychiatry, Royal College of Surgeons in Ireland, Dublin, Ireland
Inez Myin-Germeys, Department for Neurosciences, Center for Contextual Psychiatry, KU Leuven, Leuven, Belgium.
Igor Nenadić, Cognitive Neuropsychiatry Laboratory, Department of Psychiatry and Psychotherapy, Philipps Universität Marburg, Marburg, Germany.
Igor Nenadić, Department of Psychiatry and Psychotherapy, Jena University Hospital, Jena, Germany.
Gerald Nestadt, Department of Psychiatry and Behavioral Sciences, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
Ann E. Pulver, Department of Psychiatry and Behavioral Sciences, Johns Hopkins University School of Medicine, Baltimore, MD, USA
F. Anthony O’Neill, Centre for Public Health, Institute of Clinical Sciences, Queen’s University Belfast, Belfast, UK.
Sang-Yun Oh, Department of Statistics and Applied Probability, University of California at Santa Barbara, Santa Barbara, CA, USA.
Sang-Yun Oh, Computational Research Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
Ann Olincy, Department of Psychiatry, University of Colorado Denver, Aurora, CO, USA.
Robert Freedman, Department of Psychiatry, University of Colorado Denver, Aurora, CO, USA.
Vanessa Kiyomi Ota, Department of Morphology and Genetics, Laboratorio de Genetica, Universidade Federal de São Paulo, São Paulo, Brazil.
Marcos Leite Santoro, Department of Morphology and Genetics, Laboratorio de Genetica, Universidade Federal de São Paulo, São Paulo, Brazil.
Sintia Iole Belangero, Department of Morphology and Genetics, Laboratorio de Genetica, Universidade Federal de São Paulo, São Paulo, Brazil.
Christos Pantelis, Melbourne Neuropsychiatry Centre, University of Melbourne and Melbourne Health, Melbourne, Victoria, Australia.
Christos Pantelis, The Florey Institute of Neuroscience and Mental Health, University of Melbourne, Parkville, Victoria, Australia.
Bernhard T. Baune, The Florey Institute of Neuroscience and Mental Health, University of Melbourne, Parkville, Victoria, Australia
Christos Pantelis, Department of Psychiatry, Melbourne Medical School, University of Melbourne, Parkville, Victoria, Australia.
Bernhard T. Baune, Department of Psychiatry, Melbourne Medical School, University of Melbourne, Parkville, Victoria, Australia
Tiina Paunio, Department of Public Health Solutions, Genomics and Biomarkers Unit, National Institute for Health and Welfare, Helsinki, Finland.
Tiina Paunio, Department of Psychiatry and SleepWell Research Program, Faculty of Medicine, University of Helsinki and Helsinki University Central Hospital, Helsinki, Finland.
Sathish Periyasamy, Queensland Centre for Mental Health Research, University of Queensland, Brisbane, Queensland, Australia.
Bryan J. Mowry, Queensland Centre for Mental Health Research, University of Queensland, Brisbane, Queensland, Australia
Diana O. Perkins, Department of Psychiatry, University of North Carolina, Chapel Hill, NC, USA
Patrick F. Sullivan, Department of Psychiatry, University of North Carolina, Chapel Hill, NC, USA
Bruno Pfuhlmann, Clinic of Psychiatry and Psychotherapy, Weißer Hirsch, Dresden, Germany.
Olli Pietiläinen, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA, USA.
Steven E. Hyman, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA, USA
Olli Pietiläinen, Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland.
Christian Benner, Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland.
Matti Pirinen, Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland.
Aarno Palotie, Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland.
Mark J. Daly, Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland
David Porteous, Centre for Genomic and Experimental Medicine, Institute of Genetics and Molecular Medicine, University of Edinburgh, Western General Hospital, Edinburgh, UK.
John Powell, Department of Basic and Clinical Neuroscience, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK.
Diego Quattrone, South London and Maudsley NHS Mental Health Foundation Trust, London, UK.
Marta Di Forti, South London and Maudsley NHS Mental Health Foundation Trust, London, UK.
Digby Quested, Oxford Health NHS Foundation Trust, Warneford Hospital, Oxford, UK.
Digby Quested, Department of Psychiatry, University of Oxford, Oxford, UK.
Allen D. Radant, Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA, USA
Debby W. Tsuang, Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA, USA
Allen D. Radant, VA Puget Sound Health Care System, Seattle, WA, USA
Debby W. Tsuang, VA Puget Sound Health Care System, Seattle, WA, USA
Mark H. Rapaport, Huntsman Mental Health Institute, Department of Psychiatry, University of Utah School of Medicine, Salt Lake City, UT, USA
Cheryl Roe, SUNY Upstate Medical University, Syracuse, NY, USA.
Chunyu Liu, SUNY Upstate Medical University, Syracuse, NY, USA.
Joshua L. Roffman, Department of Psychiatry, Massachusetts General Hospital, Boston, MA, USA
Jennifer L. Moran, Department of Psychiatry, Massachusetts General Hospital, Boston, MA, USA
Julian Roth, Department of Psychiatry, Psychosomatics and Psychotherapy, Julius-Maximilians-Universität Würzburg, Würzburg, Germany.
Micha Gawlik, Department of Psychiatry, Psychosomatics and Psychotherapy, Julius-Maximilians-Universität Würzburg, Würzburg, Germany.
Safaa Saker-Delye, Généthon, Evry, France.
Veikko Salomaa, THL–Finnish Institute for Health and Welfare, Helsinki, Finland.
Jaana Suvisaari, THL–Finnish Institute for Health and Welfare, Helsinki, Finland.
Julio Sanjuan, Department of Psychiatry, School of Medicine, University of Valencia, Hospital Clínico Universitario de Valencia, Valencia, Spain.
Ulrich Schall, Priority Centre for Brain and Mental Health Research, University of Newcastle, Mater Hospital, McAuley Centre, Newcastle, New South Wales, Australia.
Rodney J. Scott, Division of Molecular Medicine, NSW Health Pathology North, Newcastle, New South Wales, Australia
Jianxin Shi, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD, USA.
Larry J. Siever, James J. Peters VA Medical Center, Bronx, NY, USA
Jeremy M. Silverman, James J. Peters VA Medical Center, Bronx, NY, USA
Engilbert Sigurdsson, Faculty of Medicine, University of Iceland, Reykjavik, Iceland.
Engilbert Sigurdsson, Department of Psychiatry, Landspitali University Hospital, Reykjavik, Iceland.
Kang Sim, West Region, Institute of Mental Health, Singapore, Singapore.
Kang Sim, Yoo Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
Kang Sim, Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore.
Hon-Cheong So, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong, China.
Hon-Cheong So, Department of Psychiatry, The Chinese University of Hong Kong, Hong Kong, China.
Helen J. Stain, School of Social and Health Sciences, Leeds Trinity University, Leeds, UK
Helen J. Stain, TIPS - Network for Clinical Research in Psychosis, Stavanger University Hospital, Stavanger, Norway
Nils Eiel Steen, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Erik G. Jönsson, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, Oslo, Norway
Elisabeth Stögmann, Department of Neurology, Medical University of Vienna, Vienna, Austria.
Fritz Zimprich, NORMENT Centre, Institute of Clinical Medicine, University of Oslo, Oslo, Norway; Department of Neurology, Medical University of Vienna, Vienna, Austria.
William S. Stone, Harvard Medical School Department of Psychiatry at Beth Israel Deaconess Medical Center, Boston, MA, USA
William S. Stone, Massachusetts Mental Health Center, Boston, MA, USA
Richard E. Straub, Lieber Institute for Brain Development, Baltimore, MD, USA
Thomas Hyde, Lieber Institute for Brain Development, Baltimore, MD, USA.
Andrew Jaffe, Lieber Institute for Brain Development, Baltimore, MD, USA.
Daniel R. Weinberger, Lieber Institute for Brain Development, Baltimore, MD, USA
Eric Strengman, Department of Medical Genetics, University Medical Centre Utrecht, Utrecht, The Netherlands.
Catherine A. Sugar, Department of Biostatistics, Fielding School of Public Health, University of California Los Angeles, Los Angeles, CA, USA
Dragan M. Svrakic, Department of Psychiatry, Washington University, St Louis, MO, USA
C. Robert Cloninger, Department of Psychiatry, Washington University, St Louis, MO, USA.
Thi Minh Tam Ta, Department of Psychiatry, Charité – Universitätsmedizin Berlin, Berlin, Germany.
Thi Minh Tam Ta, Berlin Institute of Health (BIH), Berlin, Germany.
Atsushi Takahashi, Laboratory for Statistical and Translational Genetics, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan.
Chikashi Terao, Laboratory for Statistical and Translational Genetics, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan.
Florence Thibaut, Université de Paris, Faculté de Médecine, Hôpital Cochin-Tarnier, Paris, France.
Florence Thibaut, INSERM U1266, Institut de Psychiatrie et de Neurosciences, Paris, France.
Draga Toncheva, Bulgarian Academy of Science, Sofia, Bulgaria.
Sarah Tosato, Department of Neuroscience, Biomedicine and Movement Sciences, Section of Psychiatry, University of Verona, Verona, Italy.
Gian Battista Tura, Psychiatry Unit, IRCCS Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia, Italy.
Alp Üçok, Department of Psychiatry, Faculty of Medicine, Istanbul University, Istanbul, Turkey.
Arne Vaaler, Division of Mental Health, St. Olav’s Hospital, Trondheim University Hospital, Trondheim, Norway.
Arne Vaaler, Department of Mental Health, Norwegian University of Science and Technology, Trondheim, Norway.
Ruud van Winkel, Department of Neurosciences, Center for Clinical Psychiatry, KU Leuven, Leuven, Belgium.
Juha Veijola, Department of Psychiatry, Research Unit of Clinical Neuroscience, University of Oulu, Oulu, Finland.
Ruud van Winkel, Department of Psychiatry, Research Unit of Clinical Neuroscience, University of Oulu, Oulu, Finland.
Juha Veijola, Medical Research Center Oulu, Oulu University Hospital and University of Oulu, Oulu, Finland.
John Waddington, Molecular and Cellular Therapeutics, Royal College of Surgeons in Ireland, Dublin, Ireland.
Anna Waterreus, Neuropsychiatric Epidemiology Research Unit, School of Population and Global Health, University of Western Australia, Perth, Western Australia, Australia.
Vera A. Morgan, Neuropsychiatric Epidemiology Research Unit, School of Population and Global Health, University of Western Australia, Perth, Western Australia, Australia
Anna Waterreus, Centre for Clinical Research in Neuropsychiatry, University of Western Australia, Perth, Western Australia, Australia.
Assen V. Jablensky, Centre for Clinical Research in Neuropsychiatry, University of Western Australia, Perth, Western Australia, Australia
Vera A. Morgan, Centre for Clinical Research in Neuropsychiatry, University of Western Australia, Perth, Western Australia, Australia
Mark Weiser, Sheba Medical Center, Tel Hashomer, Israel.
Jing Qin Wu, Baker Heart and Diabetes Institute, Melbourne, Victoria, Australia.
Zhida Xu, Department of Psychiatry, GGz Centraal, Utrecht, The Netherlands.
Robert Yolken, Stanley Neurovirology Laboratory, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
Clement C. Zai, Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health, Toronto, Ontario, Canada
James L. Kennedy, Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health, Toronto, Ontario, Canada
Clement C. Zai, Department of Psychiatry, University of Toronto, Toronto, Ontario, Canada
James L. Kennedy, Department of Psychiatry, University of Toronto, Toronto, Ontario, Canada
Feng Zhu, Department of Psychiatry, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China.
Feng Zhu, Center for Translational Medicine, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China.
Eşref Cem Atbaşoğlu, Department of Psychiatry, School of Medicine, Ankara University, Ankara, Turkey.
Meram C. Saka, Department of Psychiatry, School of Medicine, Ankara University, Ankara, Turkey
Muhammad Ayub, Department of Psychiatry, Queens University Kingston, Kingston, Ontario, Canada.
Donald W. Black, Department of Psychiatry, University of Iowa Carver College of Medicine, Iowa City, IA, USA
Nancy G. Buccola, School of Nursing, Louisiana State University Health Sciences Center, New Orleans, LA, USA
William F. Byerley, Department of Psychiatry, University of California San Francisco, San Francisco, CA, USA
Wei J. Chen, Center for Neuropsychiatric Research, National Health Research Institutes, Zhunan Town, Taiwan
Wei J. Chen, Institute of Epidemiology and Preventive Medicine, College of Public Health, National Taiwan University, Taipei, Taiwan
Benedicto Crespo-Facorro, University of Sevilla, CIBERSAM IBiS, Seville, Spain.
Benedicto Crespo-Facorro, Hospital Universitario Virgen del Rocio, Department of Psychiatry, Universidad del Sevilla, Seville, Spain.
Cherrie Galletly, Discipline of Psychiatry, Adelaide Medical School, University of Adelaide, Adelaide, South Australia, Australia.
Cherrie Galletly, Ramsay Health Care (SA) Mental Health, Adelaide, South Australia, Australia.
Cherrie Galletly, Northern Adelaide Local Health Network, Adelaide, South Australia, Australia.
Massimo Gennarelli, Department of Molecular and Translational Medicine, University of Brescia, Brescia, Italy.
Massimo Gennarelli, Genetic Unit, IRCCS Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia, Italy.
Hai-Gwo Hwu, Department of Psychiatry, College of Medicine and National Taiwan University Hospital, National Taiwan University, Taipei, Taiwan.
Ole Mors, Psychosis Research Unit, Aarhus University Hospital, Aarhus, Denmark.
Bertram Müller-Myhsok, Max Planck Institute of Psychiatry, Munich, Germany.
Bertram Müller-Myhsok, Munich Cluster for Systems Neurology, Munich, Germany.
Bertram Müller-Myhsok, Department of Health Data Science, University of Liverpool, Liverpool, UK.
Amanda L. Neil, Menzies Institute for Medical Research, University of Tasmania, Hobart, Tasmania, Australia
Merete Nordentoft, Mental Health Services in the Capital Region of Denmark, Mental Health Center Copenhagen, University of Copenhagen, Copenhagen, Denmark.
Michele T. Pato, Rutgers University, Robert Wood Johnson Medical School, New Brunswick, NJ, USA
Carlos N. Pato, Rutgers University, Robert Wood Johnson Medical School, New Brunswick, NJ, USA
Michele T. Pato, Rutgers University, New Jersey Medical School, Newark, NJ, USA
Carlos N. Pato, Rutgers University, New Jersey Medical School, Newark, NJ, USA
Matti Pirinen, Department of Mathematics and Statistics, University of Helsinki, Helsinki, Finland.
Matti Pirinen, Department of Public Health, University of Helsinki, Helsinki, Finland.
Thomas G. Schulze, Department of Psychiatry and Behavioral Sciences, SUNY Upstate Medical University, Syracuse, NY, USA
Thomas G. Schulze, Department of Psychiatry and Psychotherapy, University Medical Center Göttingen, Göttingen, Germany
Thomas G. Schulze, Department of Psychiatry and Behavioral Sciences, The Johns Hopkins University, Baltimore, MD, USA
Eli A. Stahl, Program in Medical and Population Genetics, The Broad Institute of MIT and Harvard, Cambridge, MA, USA
Tõnu Esko, Program in Medical and Population Genetics, The Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Eli A. Stahl, Regeneron Genetics Center, Orange, CA, USA
Shi-Heng Wang, College of Public Health, China Medical University, Taichung, Taiwan.
Shuhua Xu, State Key Laboratory of Genetic Engineering and Ministry of Education (MOE) Key Laboratory of Contemporary Anthropology, Collaborative Innovation Center of Genetics and Development, Human Phenome Institute, School of Life Sciences, Fudan University, Shanghai, China.
Shuhua Xu, School of Life Science and Technology, ShanghaiTech University, Shanghai, China.
Shuhua Xu, Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, China.
Rolf Adolfsson, Department of Clinical Sciences, Psychiatry, Umeå University, Umeå, Sweden.
Elvira Bramon, Division of Psychiatry, Department of Mental Health Neuroscience, University College London, London, UK.
Jorge A. Cervilla, Department of Psychiatry, San Cecilio University Hospital, University of Granada, Granada, Spain
Sven Cichon, Institute of Medical Genetics and Pathology, University Hospital Basel, Basel, Switzerland.
Sven Cichon, Department of Biomedicine, University of Basel, Basel, Switzerland.
Sven Cichon, Institute of Neuroscience and Medicine (INM-1), Research Center Juelich, Juelich, Germany.
David A. Collier, Eli Lilly and Company, Windlesham, UK
David A. Collier, Eli Lilly and Company, Windlesham, UK
Aiden Corvin, Neuropsychiatric Genetics Research Group, Department of Psychiatry, Trinity College Dublin, Dublin, Ireland.
Michael Gill, Neuropsychiatric Genetics Research Group, Department of Psychiatry, Trinity College Dublin, Dublin, Ireland.
David Curtis, UCL Genetics Institute, University College London, London, UK.
David Curtis, Centre for Psychiatry, Queen Mary University London, London, UK.
Enrico Domenici, Department of Cellular, Computational and Integrative Biology, University of Trento, Trento, Italy.
Valentina Escott-Price, Dementia Research Institute, Cardiff University, Cardiff, UK.
Ayman H. Fanous, Department of Psychiatry, Phoenix VA Healthcare System, Phoenix, AZ, USA
Ayman H. Fanous, Banner-University Medical Center, Phoenix, AZ, USA
Anna Gareeva, Department of Human Molecular Genetics of the Institute of Biochemistry and Genetics of the Ufa Federal Research Center of the Russian Academy of Sciences (IBG UFRC RAS), Ufa, Russia.
Elza Khusnutdinova, Department of Human Molecular Genetics of the Institute of Biochemistry and Genetics of the Ufa Federal Research Center of the Russian Academy of Sciences IBG UFRC RAS), Ufa, Russia.
Anna Gareeva, Department of Human Molecular Genetics of the Institute of Biochemistry and Genetics of the Ufa Federal Research Center of the Russian Academy of Sciences (IBG UFRC RAS), Ufa, Russia.
Anna Gareeva, Federal State Educational Institution of Highest Education Bashkir State Medical University of Public Health Ministry of Russian Federation (BSMU), Ufa, Russia.
Elza Khusnutdinova, Federal State Educational Institution of Highest Education Bashkir State Medical University of Public Health Ministry of Russian Federation (BSMU), Ufa, Russia.
Anna Gareeva, Federal State Educational Institution of Highest Education Bashkir State Medical University of Public Health Ministry of Russian Federation (BSMU), Ufa, Russia.
Stephen J. Glatt, Psychiatric Genetic Epidemiology and Neurobiology Laboratory (PsychGENe lab), Department of Psychiatry and Behavioral Sciences, SUNY Upstate Medical University, Syracuse, NY, USA
Kyung Sue Hong, Department of Psychiatry, Sungkyunkwan University School of Medicine, Samsung Medical Center, Seoul, Korea.
James A. Knowles, Department of Psychiatry and Zilkha Neurogenetics Institute, Keck School of Medicine at University of Southern California, Los Angeles, CA, USA
James A. Knowles, Department of Cell Biology, State University of New York, Downstate Health Sciences University, New York, NY, USA
Jimmy Lee, Department of Psychosis, Institute of Mental Health, Singapore, Singapore.
Jimmy Lee, Neuroscience and Mental Health, Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore.
Todd Lencz, Institute of Behavioral Science, Feinstein Institutes for Medical Research, Manhasset, NY, USA.
Anil K. Malhotra, Institute of Behavioral Science, Feinstein Institutes for Medical Research, Manhasset, NY, USA
Todd Lencz, Department of Psychiatry, Zucker School of Medicine at Hofstra/Northwell, Hempstead, NY, USA.
Anil K. Malhotra, Department of Psychiatry, Zucker School of Medicine at Hofstra/Northwell, Hempstead, NY, USA
Jianjun Liu, Human Genetics, Genome Institute of Singapore, A*STAR, Singapore, Singapore.
Jianjun Liu, Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
Dheeraj Malhotra, Roche Pharma Research and Early Development, Roche Innovation Center Basel, F. Hoffman-La Roche, Basel, Switzerland.
Paulo R. Menezes, Department of Preventative Medicine, Faculdade de Medicina FMUSP, University of São Paulo, São Paulo, Brazil
Vishwajit Nimgaonkar, Department of Psychiatry, University of Pittsburgh, Pittsburgh, PA, USA.
Roel A. Ophoff, Center for Neurobehavioral Genetics, Semel Institute for Neuroscience and Human Behavior, University of California Los Angeles, Los Angeles, CA, USA
Roel A. Ophoff, Department of Psychiatry, Erasmus University Medical Center, Rotterdam, The Netherlands
Sara A. Paciga, Early Clinical Development, Pfizer Worldwide Research and Development, Groton, CT, USA
Aarno Palotie, Analytic and Translational Genetics Unit, Department of Medicine, Department of Neurology and Department of Psychiatry, Massachusetts General Hospital, Boston, MA, USA.
Aarno Palotie, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Stephan Ripke, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Shengying Qin, Shanghai Key Laboratory of Psychotic Disorders, Shanghai Mental Health Center, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Margarita Rivera, Department of Biochemistry and Molecular Biology II, Faculty of Pharmacy, University of Granada, Granada, Spain.
Margarita Rivera, Institute of Neurosciences, Biomedical Research Center (CIBM), University of Granada, Granada, Spain.
Sibylle G. Schwab, Faculty of Science, Medicine and Health, University of Wollongong, Wollongong, New South Wales, Australia
Sibylle G. Schwab, Illawarra Health and Medical Research Institute, Wollongong, New South Wales, Australia
Alessandro Serretti, Department of Biomedical and Neuromotor Sciences, University of Bologna, Bologna, Italy.
Pak C. Sham, Centre for PanorOmic Sciences, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China
Pak C. Sham, Centre for PanorOmic Sciences, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China
Pak C. Sham, State Key Laboratory of Brain and Cognitive Sciences, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China
Pak C. Sham, State Key Laboratory of Brain and Cognitive Sciences, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China
Pak C. Sham, Department of Psychiatry, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China
Pak C. Sham, Department of Psychiatry, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, China
David St Clair, Institute of Medical Sciences, University of Aberdeen, Aberdeen, UK.
Ming T. Tsuang, Center for Behavioral Genomics, Department of Psychiatry, University of California San Diego, La Jolla, CA, USA
Ming T. Tsuang, Institute of Genomic Medicine, University of California San Diego, La Jolla, CA, USA
Jim van Os, University Medical Center Utrecht, Department of Psychiatry, Utrecht, The Netherlands.
Marquis P. Vawter, Department of Psychiatry and Human Behavior, School of Medicine, University of California Irvine, Irvine, CA, USA
Thomas Werge, Institute of Biological Psychiatry, Mental Health Services, Copenhagen University Hospital, Copenhagen, Denmark.
Thomas Werge, Department of Clinical Medicine, University of Copenhagen, Copenhagen, Denmark.
David St Clair, Department of Clinical Medicine, University of Copenhagen, Copenhagen, Denmark.
Thomas Werge, Center for GeoGenetics, GLOBE Institute, University of Copenhagen, Copenhagen, Denmark.
Thomas Werge, The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH, Copenhagen, Denmark.
Jim van Os, The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH, Copenhagen, Denmark.
Dieter B. Wildenauer, School of Psychiatry and Clinical Neurosciences, University of Western Australia, Perth, Western Australia, Australia
Jim van Os, School of Psychiatry and Clinical Neurosciences, University of Western Australia, Perth, Western Australia, Australia.
Xin Yu, Peking University Sixth Hospital, Peking University Institute of Mental Health, Beijing, China.
Weihua Yue, Peking University Sixth Hospital, Peking University Institute of Mental Health, Beijing, China.
Xin Yu, National Clinical Research Center for Mental Disorders, NHC Key Laboratory of Mental Health (Peking University) and Chinese Academy of Medical Sciences Research Unit, Beijing, China.
Weihua Yue, National Clinical Research Center for Mental Disorders, NHC Key Laboratory of Mental Health (Peking University) and Chinese Academy of Medical Sciences Research Unit, Beijing, China.
Weihua Yue, PKU–IDG/McGovern Institute for Brain Research, Peking University, Beijing, China.
Panos Roussos, Mental Illness Research, Education, and Clinical Center (VISN 2 South), James J. Peters VA Medical Center, New York, NY, USA.
Evangelos Vassos, Oxford Health NHS Foundation Trust, Oxford, UK.
Matthijs Verhage, Department of Clinical Genetics, Center for Neurogenomics and Cognitive Research, University Medical Center Amsterdam, Amsterdam, The Netherlands.
Frank Koopmans, Department of Functional Genomics, Faculty of Exact Science, Center for Neurogenomics and Cognitive Research, VU University Amsterdam and VU Medical Center, Amsterdam, The Netherlands.
Dnyanada Sahasrabudhe, Department of Functional Genomics, Faculty of Exact Science, Center for Neurogenomics and Cognitive Research, VU University Amsterdam and VU Medical Center, Amsterdam, The Netherlands.
Ruud F. Toonen, Department of Functional Genomics, Faculty of Exact Science, Center for Neurogenomics and Cognitive Research, VU University Amsterdam and VU Medical Center, Amsterdam, The Netherlands
Matthijs Verhage, Department of Functional Genomics, Faculty of Exact Science, Center for Neurogenomics and Cognitive Research, VU University Amsterdam and VU Medical Center, Amsterdam, The Netherlands.
Matthijs Verhage, Department of Functional Genomics, Faculty of Exact Science, Center for Neurogenomics and Cognitive Research, VU University Amsterdam and VU Medical Center, Amsterdam, The Netherlands.
Danielle Posthuma, Department of Functional Genomics, Faculty of Exact Science, Center for Neurogenomics and Cognitive Research, VU University Amsterdam and VU Medical Center, Amsterdam, The Netherlands.
Jian Yang, Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, China.
Nan Dai, Western Australian Institute for Medical Research and Centre for Medical Research, University of Western Australia, Nedlands, Western Australia, Australia.
Qin Wenwen, Western Australian Institute for Medical Research and Centre for Medical Research, University of Western Australia, Nedlands, Western Australia, Australia.
D. B. Wildenauer, Western Australian Institute for Medical Research and Centre for Medical Research, University of Western Australia, Nedlands, Western Australia, Australia
Nan Dai, School of Psychiatry and Clinical Neurosciences, University of Western Australia, Crawley, Western Australia, Australia.
Qin Wenwen, School of Psychiatry and Clinical Neurosciences, University of Western Australia, Crawley, Western Australia, Australia.
D. B. Wildenauer, School of Psychiatry and Clinical Neurosciences, University of Western Australia, Crawley, Western Australia, Australia
Feranindhya Agiananda, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Nurmiati Amir, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Ronald Antoni, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Tiana Arsianti, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Asmarahadi Asmarahadi, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
H. Diatri, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia
Prianto Djatmiko, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Irmansyah Irmansyah, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Siti Khalimah, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Irmia Kusumadewi, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Profitasari Kusumaningrum, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Petrin R. Lukman, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia
Martina W. Nasrun, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia
N. S. Safyuni, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia
Prasetyawan Prasetyawan, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
G. Semen, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia
Kristiana Siste, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Heriani Tobing, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Natalia Widiasih, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Tjhin Wiguna, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
D. Wulandari, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia
None Evalina, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
A. J. Hananto, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia
Joni H. Ismoyo, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia
T. M. Marini, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia
Supiyani Henuhili, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Muhammad Reza, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Suzy Yusnadewi, Department of Psychiatry, University of Indonesia, Jakarta, Indonesia.
Alexej Abyzov, Mayo Clinic, Rochester, MN, USA.
Schahram Akbarian, Mount Sinai, New York, NY, USA.
Harm van Bakel, Mount Sinai, New York, NY, USA.
Michael Breen, Mount Sinai, New York, NY, USA.
Alex Charney, Mount Sinai, New York, NY, USA.
Stella Dracheva, Mount Sinai, New York, NY, USA.
Kiran Girdhar, Mount Sinai, New York, NY, USA.
Gabriel Hoffman, Mount Sinai, New York, NY, USA.
Yan Jiang, Mount Sinai, New York, NY, USA.
Dalila Pinto, Mount Sinai, New York, NY, USA.
Shaun Purcell, Mount Sinai, New York, NY, USA.
Panagiotis Roussos, Mount Sinai, New York, NY, USA.
Jennifer Wiseman, Mount Sinai, New York, NY, USA.
Allison Ashley-Koch, Duke University, Durham, NC, USA.
Gregory Crawford, Duke University, Durham, NC, USA.
Tim Reddy, Duke University, Durham, NC, USA.
Miguel Brown, University of Chicago, Chicago, IL, USA.
Kay Grennan, University of Chicago, Chicago, IL, USA.
Julien Bryois, Karolinska Institutet, Stockholm, Sweden.
Becky Carlyle, Yale University, New Haven, CT, USA.
Prashant Emani, Yale University, New Haven, CT, USA.
Timur Galeev, Yale University, New Haven, CT, USA.
Mark Gerstein, Yale University, New Haven, CT, USA.
Mengting Gu, Yale University, New Haven, CT, USA.
Brittney Guerra, Yale University, New Haven, CT, USA.
Gamze Gursoy, Yale University, New Haven, CT, USA.
Robert Kitchen, Yale University, New Haven, CT, USA.
Donghoon Lee, Yale University, New Haven, CT, USA.
Mingfeng Li, Yale University, New Haven, CT, USA.
Shuang Liu, Yale University, New Haven, CT, USA.
Fabio Navarro, Yale University, New Haven, CT, USA.
Xinghua Pan, Yale University, New Haven, CT, USA.
Sirisha Pochareddy, Yale University, New Haven, CT, USA.
Joel Rozowsky, Yale University, New Haven, CT, USA.
Nenad Sestan, Yale University, New Haven, CT, USA.
Anurag Sethi, Yale University, New Haven, CT, USA.
Xu Shi, Yale University, New Haven, CT, USA.
Anna Szekely, Yale University, New Haven, CT, USA.
Daifeng Wang, Yale University, New Haven, CT, USA.
Jonathan Warrell, Yale University, New Haven, CT, USA.
Sherman Weissman, Yale University, New Haven, CT, USA.
Feinan Wu, Yale University, New Haven, CT, USA.
Xuming Xu, Yale University, New Haven, CT, USA.
Gerard Coetzee, University of Southern California, Los Angeles, CA, USA.
Peggy Farnham, University of Southern California, Los Angeles, CA, USA.
Fides Lay, University of Southern California, Los Angeles, CA, USA.
Suhn Rhie, University of Southern California, Los Angeles, CA, USA.
Heather Witt, University of Southern California, Los Angeles, CA, USA.
Shannon Wood, University of Southern California, Los Angeles, CA, USA.
Lijing Yao, University of Southern California, Los Angeles, CA, USA.
Mike Gandal, University of California Los Angeles, Los Angeles, CA, USA.
Damon Polioudakis, University of California Los Angeles, Los Angeles, CA, USA.
Vivek Swarup, University of California Los Angeles, Los Angeles, CA, USA.
Hyejung Won, University of California Los Angeles, Los Angeles, CA, USA.
Gina Giase, University of Illinois at Chicago, Chicago, IL, USA.
Shan Jiang, University of Illinois at Chicago, Chicago, IL, USA.
Amira Kefi, University of Illinois at Chicago, Chicago, IL, USA.
Annie Shieh, University of Illinois at Chicago, Chicago, IL, USA.
Fernando Goes, Johns Hopkins University, Baltimore, MD, USA.
Peter Zandi, Johns Hopkins University, Baltimore, MD, USA.
Yunjung Kim, University of North Carolina - Chapel Hill, Chapel Hill, NC, USA.
James A. Knowles, SUNY Downstate Medical Center, New York, NY, USA
Eugenio Mattei, University of Massachusetts, Amherst, MA, USA.
Michael Purcaro, University of Massachusetts, Amherst, MA, USA.
Henry Pratt, University of Massachusetts, Amherst, MA, USA.
Mette A. Peters, Sage Bionetworks, Seattle, WA, USA
Stephan Sanders, University of California San Francisco, San Francisco, CA, USA.
Zhiping Weng, University of Massachusetts Medical School, Worcester, MA, USA.
Kevin White, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
Maria J. Arranz, King’s College London, London, UK
Elvira Bramon, King’s College London, London, UK.
Conrad Iyegbe, King’s College London, London, UK.
Cathryn Lewis, King’s College London, London, UK.
Kuang Lin, King’s College London, London, UK.
Robin M. Murray, King’s College London, London, UK
John Powell, King’s College London, London, UK.
Muriel Walshe, King’s College London, London, UK.
Maria J. Arranz, Fundació de Docència i Recerca Mútua de Terrassa, Universitat de Barcelona, Barcelona, Spain
Stephan Bender, Child and Adolescent Psychiatry, University of Technology Dresden, Dresden, Germany.
Stephan Bender, Section for Experimental Psychopathology, General Psychiatry, Heidelberg, Germany.
Matthias Weisbrod, Section for Experimental Psychopathology, General Psychiatry, Heidelberg, Germany.
Elvira Bramon, Institute of Cognitive Neuroscience, University College London, London, UK.
Benedicto Crepo-Facorro, University Hospital Marqués de Valdecilla, Instituto de Formación e Investigación Marqués de Valdecilla, University of Cantabria, Santander, Spain.
Ignacio Mata, University Hospital Marqués de Valdecilla, Instituto de Formación e Investigación Marqués de Valdecilla, University of Cantabria, Santander, Spain.
Jeremy Hall, Neuroscience and Mental Health Research Institute, Division of Psychiatry and Clinical Neuroscience, Cardiff University, Cardiff, UK.
Stephen Lawrie, Division of Psychiatry, University of Edinburgh, Edinburgh, UK.
Andrew McIntosh, Division of Psychiatry, University of Edinburgh, Edinburgh, UK.
Don H. Linszen, Department of Psychiatry, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands
Roel A. Ophoff, Department of Human Genetics, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, USA
Jim van Os, Maastricht University Medical Centre, South Limburg Mental Health Research and Teaching Network, EURON, Maastricht, The Netherlands.
Jim van Os, Institute of Psychiatry, King’s College London, London, UK.
Dan Rujescu, Department of Psychiatry, University of Halle, Halle, Germany.
Dan Rujescu, Department of Psychiatry, University of Munich, Munich, Germany.
Tilmann Achsel, Department of Fundamental Neurosciences, University of Lausanne, Lausanne, Switzerland.
Claudia Bagni, Department of Fundamental Neurosciences, University of Lausanne, Lausanne, Switzerland.
Maria Andres-Alonso, RG Neuroplasticity, Leibniz Institute for Neurobiology, Magdeburg, Germany.
Michael R. Kreutz, RG Neuroplasticity, Leibniz Institute for Neurobiology, Magdeburg, Germany
Àlex Bayés, Molecular Physiology of the Synapse Laboratory, Biomedical Research Institute Sant Pau, Barcelona, Spain.
Thomas Biederer, Department of Neurology, Yale School of Medicine, New Haven, CT, USA.
Nils Brose, Department of Molecular Neurobiology, Max Planck Institute of Experimental Medicine, Göttingen, Germany.
John Jia En Chua, LSI Neurobiology Programme, National University of, Singapore, Singapore.
Marcelo P. Coba, Zilkha Neurogenetic Institute and Department of Psychiatry and Behavioral Sciences, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA
L. Niels Cornelisse, Functional Genomics section, Department of Human Genetics, Center for Neurogenomics and Cognitive Research, Amsterdam University Medical Center, Amsterdam, The Netherlands.
Jan R. T. van Weering, Functional Genomics section, Department of Human Genetics, Center for Neurogenomics and Cognitive Research, Amsterdam University Medical Center, Amsterdam, The Netherlands
Arthur P. H. de Jong, Cell Biology, Neurobiology and Biophysics, Department of Biology, Faculty of Science, Utrecht University, Utrecht, The Netherlands
Harold D. MacGillavry, Cell Biology, Neurobiology and Biophysics, Department of Biology, Faculty of Science, Utrecht University, Utrecht, The Netherlands
Jaime de Juan-Sanz, Sorbonne Université, Institut du Cerveau ‐ Paris Brain Institute ‐ ICM, Inserm, CNRS, APHP, Hôpital de la Pitié Salpêtrière, Paris, France.
Daniela C. Dieterich, Institute for Pharmacology and Toxicology, Medical Faculty Otto-von-Guericke University Magdeburg, Magdeburg, Germany
Rainer Pielot, Institute for Pharmacology and Toxicology, Medical Faculty Otto-von-Guericke University Magdeburg, Magdeburg, Germany.
Karl-Heinz Smalla, Institute for Pharmacology and Toxicology, Medical Faculty Otto-von-Guericke University Magdeburg, Magdeburg, Germany.
Daniela C. Dieterich, Leibniz Institute for Neurobiology (LIN), Magdeburg, Germany
Eckart D. Gundelfinger, Leibniz Institute for Neurobiology (LIN), Magdeburg, Germany
Rainer Pielot, Leibniz Institute for Neurobiology (LIN), Magdeburg, Germany.
Karl-Heinz Smalla, Leibniz Institute for Neurobiology (LIN), Magdeburg, Germany.
Guoping Feng, McGovern Institute for Brain Research, Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology (MIT), Cambridge, MA, USA.
Hana L. Goldschmidt, Solomon H. Snyder Department of Neuroscience, Kavli Neuroscience Discovery Institute, The Johns Hopkins University School of Medicine, Baltimore, MD, USA
Richard L. Huganir, Solomon H. Snyder Department of Neuroscience, Kavli Neuroscience Discovery Institute, The Johns Hopkins University School of Medicine, Baltimore, MD, USA
Casper Hoogenraad, Department of Neuroscience, Genentech, South San Francisco, CA, USA.
Steven E. Hyman, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA, USA
Cordelia Imig, Department of Neuroscience, University of Copenhagen, Copenhagen, Denmark.
Reinhard Jahn, Laboratory of Neurobiology, Max-Planck Institute for Biophysical Chemistry, Göttingen, Germany.
Hwajin Jung, Center for Synaptic Brain Dysfunctions, Institute for Basic Science (IBS), Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, South Korea.
Eunjoon Kim, Center for Synaptic Brain Dysfunctions, Institute for Basic Science (IBS), Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, South Korea.
Pascal S. Kaeser, Department of Neurobiology, Harvard Medical School, Boston, MA, USA
Noa Lipstein, Department of Molecular Physiology and Cell Biology, Leibniz-Forschungsinstitut für Molekulare Pharmakologie, Berlin, Germany.
Robert Malenka, Nancy Pritzker Laboratory, Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, CA, USA.
Peter S. McPherson, Department of Neurology and Neurosurgery, Montreal Neurological Institute, McGill University, Montreal, Québec, Canada
Vincent O’Connor, Biological Sciences, University of Southampton, Southampton, UK.
Timothy A. Ryan, Department of Biochemistry, Weill Cornell Medicine, New York, NY, USA
Carlo Sala, CNR Neuroscience Institute, Milan, Italy.
Chiara Verpelli, CNR Neuroscience Institute, Milan, Italy.
August B. Smit, Department of Molecular and Cellular Neurobiology, Center for Neurogenomics and Cognitive Research, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands
Thomas C. Südhof, Department of Molecular and Cellular Physiology, Howard Hughes Medical Institute, Stanford University, Stanford, CA, USA
Paul D. Thomas, Division of Bioinformatics, Department of Preventive Medicine, Keck School of Medicine of USC, University of Southern California, Los Angeles, CA, USA
MAIN TEXT REFERENCES
- 1.Owen MJ Sawa A.& Mortensen PB Schizophrenia. The Lancet vol. 388 86–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plana-Ripoll O.et al. A comprehensive analysis of mortality-related health metrics associated with mental disorders: a nationwide, register-based cohort study. Lancet 394, 1827–1835 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Momen NC et al. Association between mental disorders and subsequent medical conditions. N. Engl. J. Med 382, 1721–1731 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jääskeläinen E.et al. A systematic review and meta-analysis of recovery in schizophrenia. Schizophr Bull. 39, 1296–1306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pocklington AJ et al. Novel Findings from CNVs Implicate Inhibitory and Excitatory Signaling Complexes in Schizophrenia. Neuron 86, 1203–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh T.et al. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat. Genet 49, 1167–1173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rees E.et al. De novo mutations identified by exome sequencing implicate rare missense variants in SLC6A1 in schizophrenia. Nat. Neurosci (2020) doi: 10.1038/s41593-019-0565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam M.et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet (2019) doi: 10.1038/s41588-019-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bigdeli TB et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol. Psychiatry 25, 2455–2467 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Räsänen S, Pakaslahti A, Syvälahti E, Jones PB & Isohanni M.Sex differences in schizophrenia: A review. Nord. J. Psychiatry 54, 37–45 (2000). [Google Scholar]
- 13.Zeng J.et al. Bayesian analysis of GWAS summary data reveals differential signatures of natural selection across human complex traits and functional genomic categories. bioRxiv 752527 (2019) doi: 10.1101/752527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguet F.et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genome-wide association study identifies five new schizophrenia loci. Nat. Genet 43, 969–978 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skene NG et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet 50, 825–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habib N.et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeisel A.et al. Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koopmans F.et al. SynGO: An Evidence-Based, Expert-Curated Knowledge Base for the Synapse. Neuron 103, 217–234.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benner C.et al. FINEMAP: Efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakuntabhai A.et al. Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nat. Genet 21, 271–277 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Cederlöf M.et al. The association between Darier disease, bipolar disorder, and schizophrenia revisited: A population-based family study. Bipolar Disord. 17, 340–344 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Pardiñas AF et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet 50, 381–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Z.et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet 48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Gandal MJ et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science (80-. ) 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien HE et al. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 19, 194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Võsa U.et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet 53, 1300–1310 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D.et al. Comprehensive functional genomic resource and integrative model for the human brain. Science (80-. ) 362, eaat8464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galvan L.et al. The Striatal Kinase DCLK3 Produces Neuroprotection Against Mutant Huntingtin - PubMed. Brain 141, 1434–1454 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rees E.et al. Analysis of intellectual disability copy number variants for association with schizophrenia. JAMA Psychiatry 73, 963–969 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fromer M.et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplanis J.et al. Integrating healthcare and research genetic data empowers the discovery of 49 novel developmental disorders. bioRxiv (2019) doi: 10.1101/797787. [DOI] [Google Scholar]
- 33.Satterstrom FK et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 180, 568–584.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y.et al. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat. Genet 49, 186–192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Y.et al. Rare genetic variants affecting urine metabolite levels link population variation to inborn errors of metabolism. Nat. Commun 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh T, Neale BM & Daly MJ Exome sequencing identifies rare coding variants in 10 genes which confer substantial risk for schizophrenia on behalf of the Schizophrenia Exome Meta-Analysis (SCHEMA) Consortium*. medRxiv 2020.09.18.20192815 (2020). [Google Scholar]
- 37.Priya A, Johar K.& Wong-Riley MTT Specificity protein 4 functionally regulates the transcription of NMDA receptor subunits GluN1, GluN2A, and GluN2B. Biochim. Biophys. Acta - Mol. Cell Res 1833, 2745–2756 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ripke S.et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet 45, 1150–1159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirov G.et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry 17, 142–153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lek M.et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fagerberg L.et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 1.Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam M.et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet (2019) doi: 10.1038/s41588-019-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam M.et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics (2019) doi: 10.1093/bioinformatics/btz633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purcell S, Neale B, Todd-Brown K, Thomas L.& Ferreira MA PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 81, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devlin B.& Roeder K.Genomic Control for Association Studies. Biometrics 55, 997–1004 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das S.et al. Next-generation genotype imputation service and methods. Nat. Genet 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haplotype Reference Consortium r1.1. https://ega-archive.org/datasets/EGAD00001002729.
- 9.O’Connell J.et al. Haplotype estimation for biobank-scale data sets. Nat. Genet 48, 817–820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bycroft C.et al. Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv 166298 (2017) doi: 10.1101/166298. [DOI] [Google Scholar]
- 11.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JJ et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet 50, 1112–1121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tukiainen T.et al. Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vittinghoff E.& McCulloch CE Relaxing the rule of ten events per variable in logistic and cox regression. Am. J. Epidemiol 165, 710–718 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Heinze G.& Ploner M.A SAS macro, S-PLUS library and R package to perform logistic regression without convergence problems. https://cemsiis.meduniwien.ac.at/fileadmin/user_upload/_imported/fileadmin/msi_akim/CeMSIIS/KB/programme/tr2_2004.pdf (2004).
- 17.Viechtbauer W.Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw 36, 1–48 (2010). [Google Scholar]
- 18.Lee SH, Goddard ME, Wray NR & Visscher PM A Better Coefficient of Determination for Genetic Profile Analysis. Genet. Epidemiol 36, 214–224 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Camblor P.Fully non-parametric receiver operating characteristic curve estimation for random-effects meta-analysis. Stat. Methods Med. Res 26, 5–20 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Bigdeli TB et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol. Psychiatry 25, 2455–2467 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguet F.et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habib N.et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skene NG et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet 50, 825–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeisel A.et al. Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryois J.et al. Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat. Genet 1–12 (2020) doi: 10.1038/s41588-020-0610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Leeuw CA, Mooij JM, Heskes T.& Posthuma D.MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLOS Comput. Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durinck S, Spellman PT, Birney E.& Huber W.Mapping identifiers for the integration of genomic datasets with the R/ Bioconductor package biomaRt. Nat. Protoc 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maston GA, Evans SK & Green MR Transcriptional Regulatory Elements in the Human Genome. Annu. Rev. Genomics Hum. Genet 7, 29–59 (2006). [DOI] [PubMed] [Google Scholar]
- 30.A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet 48, 1279–1283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koopmans F.et al. SynGO: An Evidence-Based, Expert-Curated Knowledge Base for the Synapse. Neuron 103, 217–234.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genovese G.et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat. Neurosci 19, 1433–1441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardiñas AF et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet 50, 381–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merico D, Isserlin R, Stueker O, Emili A.& Bader GD Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation. PLoS One 5, e13984 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benner C.et al. FINEMAP: Efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benner C.et al. Prospects of Fine-Mapping Trait-Associated Genomic Regions by Using Summary Statistics from Genome-wide Association Studies. Am. J. Hum. Genet 101, 539–551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Z.et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet 48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 38.O’Brien HE et al. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 19, 194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gandal MJ et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science (80-. ) 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Võsa U.et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv 447367 (2018) doi: 10.1101/447367. [DOI] [Google Scholar]
- 41.Sonnega A.et al. Cohort profile: The Health and Retirement Study (HRS). Int. J. Epidemiol 43, 576–585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Lee SH, Goddard ME & Visscher PM GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J.et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet 44, 369–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gusev A.et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet 48, 245–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W.et al. Integrative transcriptome imputation reveals tissue-specific and shared biological mechanisms mediating susceptibility to complex traits. Nat. Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics for the “Extended”, “Core”, ancestry specific and sex-stratified analyses is available at “https://www.med.unc.edu/pgc/download-results/scz/”. Genotype data are available for a subset of cohorts, including dbGAP accession numbers and/or restrictions, as described in the Supplementary Information section “Cohort Descriptions”.