Abstract
The human gut microbiota are critical for preserving the health status because they are required for digestion and nutrient acquisition, the development of the immune system, and energy metabolism. The gut microbial composition is greatly influenced by the colonization of the recalcitrant pathogen Helicobacter pylori (H. pylori) and the conventional antibiotic regimens that follow. H. pylori is considered to be the main microorganism in gastric carcinogenesis, and it appears to be required for the early stages of the process. However, a non-H. pylori microbiota profile is also suggested, primarily in the later stages of tumorigenesis. On the other hand, specific groups of gut microbes may produce beneficial byproducts such as short-chain fatty acids (acetate, butyrate, and propionate) that can modulate inflammation and tumorigenesis pathways. In this review, we aim to present how H. pylori influences the population of the gut microbiota to modify the host immunity and trigger the development of gastric carcinogenesis. We will also highlight the effect of the gut microbiota on immunotherapeutic approaches such as immune checkpoint blockade in cancer treatment to present a perspective for further development of innovative therapeutic paradigms to prevent the progression of H. pylori-induced stomach cancer.
Keywords: Helicobacter pylori, gut microbiota, gastric carcinogenesis, immune system homeostasis, probiotics, immunotherapy
1 Introduction
Helicobacter pylori (H. pylori) is a Gram-negative spiral-shaped pathogen colonizing the host stomach and correlated with various gastric disorders such as chronic active gastritis, peptic ulcer disease (PUD), mucosa-associated lymphoid tissue (MALT) lymphoma and gastric carcinoma (Yamade et al., 2011; Sheh and Fox, 2013; Hooi et al., 2017). Gastric cancer is the third most prevalent cause of cancer-related death in both genders globally (Rawla and Barsouk, 2019; Choi et al., 2020). It has been suggested that more than 50% of the world’s population is infected with H. pylori, but it remains asymptomatic in the majority of the general population (Sheh and Fox, 2013; Hooi et al., 2017). The severity of H. pylori infection largely relies on the host genetics, immune response, and microbiome composition, as well as on bacterial virulence, and environmental conditions (Ding et al., 2010; Rawla and Barsouk, 2019). It has been long assumed that the human stomach is sterile owing to its harsh acidic environment; however, the discovery of H. pylori in 1982 revealed that the human gastric tissue holds a core microbiome (Warren and Marshall, 1983; Kusters et al., 2006).
It is now well documented that a normal gut microbiome is critically associated with the modulation of the host immune response, energy metabolism, and pathogen colonization resistance (Belkaid and Hand, 2014; Pascale et al., 2019). H. pylori can modify the α-diversity of the normal gastric microbiome that can result in dysbiosis and the development of gastric carcinoma (Hooi et al., 2017; Yang et al., 2019). The imbalance in the gastric microbiome community can result in continuous stimulation of the host immune system, which can in turn cause chronic inflammation of gastric mucosa or trigger tumor growth (Nasr et al., 2020; Zhang and Pan, 2020). Development of immunotherapy agents such as immune checkpoint inhibitors (ICIs) and also probiotic co-supplementation along with antibiotic regimens have been suggested as promising modalities for the treatment of H. pylori infection and gastric carcinoma (Qureshi et al., 2019; Rezasoltani et al., 2021). In recent years, cancer immunotherapy achieved brilliant breakthroughs in the field of clinical oncology by extending the lives of patients suffering from rapidly lethal tumors. The significance of immunotherapy in cancer research and treatment will develop expeditiously in the next decades as novel treatment strategies, and brand new druggable targets are expected to be discovered accordingly (Waldman et al., 2020). Here, we intend to highlight the latest and available literature regarding the role of H. pylori infection in perturbation of the gut microbiota and the probable role of dysbiosis in gastric carcinogenesis and tumorigenesis. We further discuss the reports for crosstalk between the gut microbiome and immunotherapeutic approaches in gastric carcinoma.
1.1 The human gut microbiota
It has been established that the human microbiota is the collection of a variety of microorganisms including bacteria, archaea, fungi, viruses, and parasites, which occupy various niches on or within human tissues and biofluids. The impact of these microbes on the human body may be beneficial or harmful (Stasiewicz and Karpinski, 2021). It is established for many years that the human body is hosting remarkable microbiota and includes 10-fold more microorganisms (1014) than human cells (Savage, 1977). These microorganisms are found inside and outside the human body exposed to the external environment, including the oral cavity, skin, respiratory, urogenital, and gut tracts. Among these potential niches, the gut has the highest density and variety of microbiota due to its ideal growth conditions (Ohland and Jobin, 2015). The complex ecosystem residing inside or passing through the digestive tract is termed the gut microbiome (Kastl et al., 2020). The oral cavity is enriched with over 700 bacterial species and thereby has the second largest level of α-diversity after that of the gut microbiome ( Deo and Deshmukh, 2019 ). A recent study has demonstrated that in a healthy oral cavity, the predominant bacterial genera are 12 Gram-positive microorganisms including Corynebacterium, Abiotrophia, Peptostreptococcus, Streptococcus, Pseudoramibacter, Stomatococcus, Actinomyces, Propionibacterium, Bifidobacterium, Eubacterium, Lactobacillus, and Rothia, as well as Gram-negative bacteria including Eikenella, Moraxella, Neisseria, Prevotella, Veillonella, Campylobacter, Capnocytophaga, Treponema, Desulfobacter, Desulfovibrio, Fusobacterium, Hemophilus, Leptotrichia, Selemonas, Simonsiella, and Wolinella (Deo and Deshmukh, 2019). It has been well understood that the oral microbiome are associated with multiple oral diseases. This association may arise from the ability of many oral bacteria to influence the oral inflammatory microenvironment (Lu et al., 2019).
Currently, the esophagus is suggested to contain a diverse microbiota, yet our understanding of the esophageal microbial composition and activity remains limited compared to that of the intestinal microbiota (Deshpande et al., 2018). However, several pieces of research have exhibited the correlation between the esophageal microorganisms and multiple esophageal illnesses following the advances in next-generation sequencing (NGS) techniques (Park and Lee, 2020). Moreover, many researchers have explored the structure of the esophageal microbiome in a normal esophagus and various esophageal disorders. Streptococcus is the dominant bacterial genus in the esophagus of a normal host followed by Haemophilus, Neisseria, Prevotella, and Veillonella ( Jung et al., 2022 ). Nevertheless, Gram-negative bacteria such as Prevotella, are more prevalent in an abnormal esophagus, such as in esophageal cancer, Barrett’s esophagus, gastroesophageal reflux disease (GERD), and eosinophilic esophagitis (EoE) (Park and Lee, 2020).
As mentioned above, the gastric microbiota is composed of several microorganisms, which have been presented in the human body since birth (Schmidt et al., 2018; Altveş et al., 2019). Owing to the acidic condition of the stomach, the gastric environment was previously considered as an unsuitable niche for the colonization of bacteria (Ansari and Yamaoka, 2017). The discovery of H. pylori by Marshall in 1982 revealed that various microbes could reside in the stomach (Warren and Marshall, 1983; Garcia-Castillo et al., 2018). The most abundant gastric microbiota phyla are Proteobacteria and Firmicutes, followed by Bacteroides, Actinobacteria, and Fusobacteria (Cao and Yu, 2015; Rinninella et al., 2019).
The gut microbial population can communicate with one another and their hosts by switching genes on and off via metabolically active chemicals (Gao et al., 2018b). The gut microbiota are involved in human metabolism by producing enzymes, which are not expressed by the human genome, such as those involved in the breakdown of polysaccharides and polyphenols (Rowland et al., 2018). Dietary fibers and other undigested materials are consumed by gut colonizing bacteria, which can create short-chain fatty acids (SCFA). In a mutualistic connection, the microbes benefit the host cell physiology, whereas the host supplies an appropriate habitat for the bacteria to reside and grow (Rinninella et al., 2019; Pascale et al., 2019). The presence of the gut microbiota is vital for preserving human health, absorption of nutrients, synthesis of some vitamins, drug metabolism, immunomodulation, and defense against pathogens colonization (Collado et al., 2009; Cao and Yu, 2015; Magne et al., 2020).
The gut microbiota are very dynamic and could be affected by different factors such as host lifestyle, antibiotic therapy, long-term proton pump inhibitors (PPIs) consumption, and H. pylori infection (Schmidt et al., 2018; Zhang et al., 2019). Perturbation in the gut homeostasis has been correlated with a wide spectrum of behavior to mental disorders (Moran-Ramos et al., 2020). H. pylori infection can increase the stomach pH and induce chronic inflammation of the gut epithelium; therefore, affect the microenvironment of the stomach, causing extensive changes in the diversity and structure of the gut microbiota (Mohammadi et al., 2020).
2 H. pylori and the gastric microbiota
2.1 The gastric microbiota in a healthy stomach
Prior to the discovery of H. pylori, the stomach was assumed to be free of microbes owing to its harsh and acidic environment (Yang et al., 2013). As the culture-based techniques are the keystone of microbiological research, H. pylori was presumed as the sole microorganism with the capacity of replication in the severe gastric environment for many decades (Stockbruegger, 1985). Based on culture-independent approaches, such as NGS, fluorescent in situ hybridization (FISH), and dot-blot hybridization analysis of the gastric microbiome, it is now revealed that the bacteria in the human stomach are more dense and diverse than previously assumed (Li et al., 2009).
In more than 20% of the NGS-based studies, Tenericutes, Bacteroidetes, Firmicutes, Actinobacteria, Spirochetes, Proteobacteria, Fusobacteria, and TM7 are among the bacterial phyla constituting the normal gastric microbiota. Moreover, Neisseria, Hemophilus, Fusobacterium, Prevotella, Veillonella, and Streptococcus were the six most commonly reported genera of the gastric microbiota in a healthy stomach (Rajilic‐Stojanovic et al., 2020). Another study performed in Sweden demonstrated that Firmicutes (42%), Bacteroidetes (24%), Proteobacteria (17%), Actinobacteria (7%), and Fusobacteria (6%) were allocated to the five most common phyla in the gastric mucosa of H. pylori-negative individuals. Additionally, Streptococcus (Firmicutes, 24%), Prevotella (Bacteroidetes, 23%), Veillonella (Firmicutes, 6%), Fusobacterium (Fusobacteria, 5%), Haemophilus (Proteobacteria, 4%), Neisseria (Proteobacteria, 4%), and Gemella (Firmicutes, 4%) were reported as the dominant genera among the inherent gastric microbiota (Ndegwa et al., 2020). These findings are consistent with earlier research that showed Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes as the most common taxa in the gastric mucus layer, yet their proportions vary between different studies and geographical regions. Although Prevotella, Veillonella, Streptococcus, Neisseria, Fusobacterium, and Haemophilus are the main genera among the gastric microbiota, data on the variety of the human microbiome are not uniform, indicating regional variances in the microbial abundance (Ozbey et al., 2020; Rajilic‐Stojanovic et al., 2020; Miftahussurur et al., 2020).
2.2 Impact of H. pylori on the gastric microbiota
It has been demonstrated that H. pylori possesses well-developed adaptation mechanisms that accelerate bacterial growth in the severe acidic niche of the stomach and promote the incidence of developing long-term infection (Ansari and Yamaoka, 2017). H. pylori is the most frequent microorganism detected in the gastric microbial ecology of an infected individual, accounting for 40% to 90% of the gastric microbiota (Chen et al., 2021). In general, H. pylori colonization results in a substantial downregulation in the overall α-diversity of the gastric microbiome. According to a recent study on the gastric microbial structure, the normal stomach demonstrated a significantly higher microbial α-diversity using the Shannon index analysis, compared to individuals with chronic H. pylori gastritis and atrophic gastritis (Ndegwa et al., 2020). It has been further indicated that H. pylori oncoproteins, especially cytotoxin-associated gene A (CagA), can disturb the gastric environment and trigger microbial dysbiosis (Ansari and Yamaoka, 2017).
Recently, it has been reported that the gastric microbiota of H. pylori-positive and H. pylori-negative subjects are enriched by the common phyla, albeit with different rates of relative abundance (Vasapolli et al., 2019). Proteobacteria (68.7%) are the most prevalent phyla in H. pylori-positive individuals, followed by Firmicutes (14.7%), Bacteroidetes (8.3%), and Actinobacteria (6%). However, in an H. pylori-negative population, the proportions of the gastric microbiota were reported as Proteobacteria (52.6%), Firmicutes (26.4%), Bacteroidetes (12%), and Actinobacteria (6.4%) (Llorca et al., 2017; Li et al., 2017). Furthermore, H. pylori infection will disturb the Firmicutes/Bacteroidetes (F/B) ratio and probably change the physiological state of the individual, especially in asymptomatic subjects (Sheh and Fox, 2013; Khosravi et al., 2016; Kienesberger et al., 2016). Firmicutes, Bacteroidetes, Actinobacteria, Cyanobacteria, and Fusobacteria were among the dominant commensal microbiota enriched after H. pylori eradication. Specific genera that increased among the gastric microbiota following H. pylori eradication included Prevotella, Gemella, Porphyromonas, Alloprevotella, Veillonella, Neisseria, Streptococcus, Rothia, and Haemophilus (Guo et al., 2020).
Due to the connection of the stomach to the esophagus, oral cavity, and duodenum, bacteria from the mouth, pharynx, nose, respiratory tract, esophagus, and small intestine can enter the stomach. Therefore, the occurrence of several gastrointestinal diseases, such as esophagitis, Barrett’s esophagus, esophageal carcinoma, gastritis, gastric cancer, small intestine bacterial overgrowth, irritable bowel syndrome (IBS), and celiac disease depends on the structure of the gut microbiota (Wang and Yang, 2013). Aside from the influence of H. pylori on the gastric microbial structure, several investigations have exhibited the essential impact of this bacterium on the gut microbial community. It has also been demonstrated that there are multiple host-microbe and microbe-microbe interactions between H. pylori and other bacterial species found in the digestive tract (Engstrand and Lindberg, 2013). It has been further revealed that H. pylori modifies the gut microbial community in three ways: first by modifying the level of stomach acidity (Goddard and Spiller, 1996; Navabi et al., 2013; Espinoza et al., 2018), second by supplying substrates favorable for the colonization of other bacterial species (Bauerfeind et al., 1997; Ziebarth et al., 1997), and third by changing the lifestyle and diet patterns of the host (Dong et al., 2017; Noto and Peek Jr, 2017). The correlation between the gut microbiota and H. pylori infection or gastric carcinoma in humans is yet to be fully elucidated (Gao et al., 2018a; Park et al., 2019).
2.3 H. pylori, the gut microbiota, and gastric cancer
Approximately 75% of all gastric cancer subjects are caused by H. pylori infection, which significantly contributes to the progression of low-grade MALT lymphoma, a subcategory of primary gastric lymphoma, representing about 30-40% of extranodal lymphomas (Fehlings et al., 2012; Noto and Peek Jr, 2017). H. pylori colonizes the gastric mucosa using different virulence factors such as cag pathogenicity island (cagPAI), CagA, vacuolating cytotoxin A (VacA), outer membrane proteins (OMPs), peptidoglycan, IceA as well as several adherence factors such as BabA, SabA, and OipA, which are involved in the carcinogenesis of gastric tissue (Cao and Yu, 2015; Espinoza et al., 2018; Ohno and Satoh-Takayama, 2020). During H. pylori infection, VacA stimulates CagA accumulation in the gastric epithelial cells. cagPAI encodes a syringe-like structure named type IV secretion system (T4SS) to facilitate the efficient delivery of CagA oncoprotein into the host cell. It is well documented that H. pylori strains expressing CagA are substantially correlated with high grades of inflammation in the gastric epithelium, resulting in chronic gastritis and gastric atrophy (Cao and Yu, 2015). While H. pylori bacteria are dominated in the stomach, their lipopolysaccharide (LPS) and surface proteins are released in the gastric lamina propria, after which they might provoke macrophages to increase the expression of pro-inflammatory cytokines such as interleukin-1β (IL-1β), IL-8, IL-17, tumor necrosis factors-α (TNF-α), and ultimately increases the risk of developing H. pylori-induced gastric carcinoma (Naito and Yoshikawa, 2002; Fehlings et al., 2012; Guo et al., 2013).
IL-8, a potent chemoattractant cytokine secreted by a plethora of tissue and blood cells, may promote the production of CD11b/CD18 in leukocytes and intercellular adhesion molecule-1 (ICAM-1) in endothelial cells, leading to the transmigration of leukocytes from the bloodstream to the gastric mucosa (Espinoza et al., 2018). Alteration in the gut microbial structure can elevate the secretion levels of systemic and local pro-inflammatory mediators as well as the expression of cancer-related genes, which ultimately contribute to the progression of gastric diseases (Zhang et al., 2019).
2.3.1 The oral microbiota
The dynamic and polymicrobial oral microbiota directly affect the disease status of dental caries and periodontal diseases. There is solid evidence that the gastric microbiota of oral origin may play a role in gastric cancer development and progression. In spite of this, there is still much controversy regarding the relationship between the oral microbiota and gastric cancer progression as well as the role of H. pylori pathogenesis in this interaction. According to recent studies, patients with stomach cancer had more complicated oral bacteria than the normal control population. This might be due to the weak immunity of the patients with gastric cancer compared to healthy individuals (Sun et al., 2018; Bakhti and Latifi-Navid, 2021).
The presence of H. pylori in the gastric mucosa significantly influences gut colonization dynamics. It has been shown that H. pylori interacts with the oral microbiota and may even colonize in the oral cavity. Moreover, a significant increase in the α-diversity of the oral microbiota has been detected in H. pylori-positive individuals compared to H. pylori-negative individuals (Krzyzek and Gosciniak, 2018).
A recent study showed that the development of esophageal and gastric cancer may be associated with the oral pathobiont Fusobacterium spp., which isolated from dental plaques (F. nucleatum and F. periodontium) (Alon-Maimon et al., 2022). Dental plaque produces a key signaling substance called autoinducer-2 (AI-2), which is a chemorepellent agent, stimulating dispersion of H. pylori aggregates and biofilms and starting negative chemotaxis against the signal source (Anderson et al., 2015). Between the H. pylori-positive and H. pylori-negative groups, there were significant differences in the genera Actinomyces, Neisseria, Granulicatella, Helicobacter, Veillonella, Streptococcus, Fusobacterium, and Prevotella in the oral cavity. Additionally, in the bacterial profile of saliva or plaque of the patients infected with H. pylori, the higher relative abundance of Veillonella, Prevotella, Lactobacillus, Aggregatibacter, Streptococcus, and Megasphaera, and a lower proportion of Leptotrichia, Rothia, Capnocytophaga, Campylobacter, Tannerella, and Granulicatella was associated with stomach cancer (Dicksved et al., 2009). Furthermore, during the process from gastritis to stomach cancer, Lactobacillus bacteria may thus grow over other microorganisms in the oral cavity (Li et al., 2021). A study by Wu et al. showed that microbiota coating the tongue differ between healthy people and gastric cancer patients. Results from this study revealed that a higher relative abundance of Firmicutes and a lower relative abundance of Bacteroidetes were associated with the risk of developing gastric carcinoma (Wu et al., 2018). The prevalence of periodontal pathogens in the oral cavity specifically Treponema denticola, Tannerella forsythia, and Actinobacillus actinomycetemcomitans are currently considered significant risk factors for the development of precancerous stomach lesions (Sun et al., 2017). There are numerous fundamental mechanisms through which the oral bacteria can cause upper gastrointestinal cancer, such as releasing carcinogenic chemicals, stimulating chronic inflammation, and increasing the levels of certain bacterial metabolic pathways (Bakhti and Latifi-Navid, 2021).
2.3.2 The esophageal microbiota
The microbial composition of a normal esophagus is relatively conserved without distinct differences between the upper, middle, and lower esophagus. As with the oral microbiota, the esophageal microbiota comprises six major phyla, which include Firmicutes, Bacteroides, Actinobacteria, Proteobacteria, Fusobacteria, and TM7 (Gillespie et al., 2021). Alterations in the esophageal microbiome may be linked to the esophageal mucus injury and consequently disrupts the integrity of the epithelial barrier and results in the translocation of other microbes. There is a hypothesis that stands for the protective role of H. pylori against Barrett’s esophagus and esophageal adenocarcinoma (EAC), which might be contributed to the gastric acid output reduction (Anderson et al., 2008; Polyzos et al., 2018).
The dysbiotic status of the esophageal microbiota may play a crucial role in disturbing the epithelial barrier, causing chronic inflammation, inducing DNA damage, and promoting gastrointestinal carcinogenesis. Despite the lack of clarity around the direct contribution of the esophageal microbiota to gastric cancer, mounting data suggest that considerable changes in the esophageal microbial community may contribute to inflammation-induced gastric carcinogenesis (Sharma et al., 2022).
2.3.3 The gastric microbiota
It has been shown that Actinobacteria, Bacteroidetes, and Firmicutes are the main phyla in the stomach microbiota of H. pylori–negative subjects (Zhang et al., 2019). In contrast, the colonization of H. pylori in the host stomach will lead to the enrichment of Spirochetes and Proteobacteria phyla. Many carcinogenic metabolites such as N-nitroso compounds produced by Staphylococcus, Lactobacillus, and Escherichia coli are involved in various disadvantageous cellular mechanisms, which can trigger inflammation and tumor angiogenesis (Mowat et al., 2000; Wang et al., 2016; Pero et al., 2019). Furthermore, the balance of stomach pH may perturb during abnormal or disease states and leads to hypochlorhydria, which is correlated with higher levels of intragastric nitrite production and a greater risk of gastric cancer development. Consequently, a decreased gastric acidity elevates the risk of bacterial overgrowth and alters the indigenous community of the gastric microbiota. This will promote the colonization of pathobionts and those microbes with nitrosating capability, which are not frequently cultured from the microbial community of a healthy individual (Hunt et al., 2015). The promotion in the transformation of nitrogen compounds into carcinogenic N-nitroso compounds is promoted by chronic H2 receptor antagonists (H2RA) therapy that accelerate the colonization of nitrosating bacteria (Muscroft and Keighley, 1981; Stockbrugger et al., 1982; Fisher and Fisher, 2017; Piscione et al., 2021). As ascorbic acid is a powerful inhibitor of nitrosation reaction, these chemical reactions may take place when the stomach has the lowest gastric levels of ascorbic acid (Toh and Wilson, 2020). Furthermore, the rate of these chemical nitrosation reactions increases when the pH of the stomach is less than 4. In addition to H. pylori, some non-H. pylori bacteria were found to increase atrophy in patients following acid-suppressive medications (Li and Perez Perez, 2018). Based on a recent report, researchers discovered that patients taking acid-suppressing drugs such as H2 receptor blockers (H2RBs) or PPIs have a three-fold higher risk of developing infectious diarrhea compared to non-recipients (Martinsen et al., 2019).
Decreases in the ratio of beneficial Clostridium clusters XIVa and IV, including several butyrate-producing bacteria, have been also reported in multiple elderly individuals. As previously documented, the gastric cancer microbiota largely consists of several strains of the genera Streptococcus, Lactobacillus, Veillonella, and Prevotella (Aviles-Jimenez et al., 2014). Among the Streptococcus species, Streptococcus mitis and Streptococcus parasanguinis were found to be the predominant bacteria in the gastric mucosa of patients with gastric carcinoma (Dicksved et al., 2009). In another study, an overgrowth in Proteobacteria (including the genera Phyllobacterium, Achromobacter, and the families Xanthomonadaceae and Enterobacteriaceae) and the genera Lactobacillus, Clostridium, and Rhodococcus, was observed in gastric carcinoma (Ferreira et al., 2018). Moreover, Coker et al. indicated that the enrichment of Slackia exigua, Parvimonas micra, Fusobacterium nucleatum, Dialister pneumosintes, Prevotella intermedia, Prevotella oris, and Catonella morbi bacteria in the gastric microbiota elevates the risk of developing gastric cancer (Coker et al., 2018). Wang et al. in 2016 demonstrated that the microorganisms most frequently isolated from gastric cancer tissues were Lactobacillus, Shigella, Escherichia, Nitrospirae, fungerum, Burkholderia and Lachospiraceae (Stockbruegger, 1985). Rodríguez et al. concluded that Veilonella, Lactococcus, and Fusobacteriaceae (Fusobacterium and Leptotrichia) were among the bacterial taxa dominated in gastric cancer (Castaño-Rodríguez et al., 2017). Cavadas et al. demonstrated that a decrease in the proportion of Rhodococcus, Phyllobacterium, and Staphylococcus genera was correlated with a reduction in the microbial diversity of cancer cases, which was counterbalanced by enrichment in Bacillus, Enterobacter, Fusobacterium, and Sutterella genera (Cavadas et al., 2020). Lactobacillus, Paeniglutamicibacter, Glutamicibacter, Helicobacter, Enterococcus, and Carnobacterium genera were linked to gastric carcinoma in a case-control study in Mongolian patients (Gantuya et al., 2020). It has been further suggested that as precancerous lesions progressed to dysplasia and gastric cancer, the relative abundance of Capnocytophaga, Bacillus, and Prevotella bacteria increased, yet the relative abundance of Helicobacter genus decreased (Kadeerhan et al., 2021). Recently, Deng et al. (Deng et al., 2021) found an important but not statistically significant increase in Proteobacteria (mostly Pseudomonadales) in biopsies from antral gastric cancer compared to biopsies from antrum gastritis (from 38% to 60%) that was followed by a substantial reduction in Actinobacteria (from 15% to less than 1% of total sequencing reads). Also, they detected that Pseudomonadales and Erysipelotrichales have been enriched in individuals with H. pylori-negative antral gastric cancer, whereas Neisseriales were enriched in individuals with H. pylori-positive antral gastric carcinoma. Li et al. exhibited that the presence of Haemophilus, Serratia, Neisseria, and Stenotrophomonas was correlated with the development of gastric cancer (Li et al., 2017). Based on the recent findings that all types of gastric tumors are associated with an elevated enrichment of Enterobacteriaceae and a decreased proportion of Lactobacillaceae and Oscillibacter, it has been suggested that the correlation of higher Enterobacteriaceae abundance with gastric malignancies might be potentially introduced as a biomarker of gastric cancer (Sarhadi et al., 2021). These findings contradicted the results from Danish and Lithuanian studies, in which they found that the proportion of bacteria from the genera Gemella, Lactobacillus, Streptococcus, and Enterococcus (all Firmicutes) was higher in patients suffering from gastric cancer than in dyspeptic subjects, whereas the abundance of bacteria from the genera Staphylococcus, Actinomyces, and Corynebacterium was lower (Spiegelhauer et al., 2020). Another study revealed that H. pylori-infected individuals had significantly higher colonization of Proteobacteria bacteria and lower colonization of Actinobacteria, Streptococcus, Lactobacillus, Veillonella, and Prevotella bacteria, which may promote inflammation that could potentially accelerate cancer development (Aviles-Jimenez et al., 2014). In agreement with the aforementioned data, another investigation evaluated by Eun et al. reporting that Streptococcus, Lactobacillus, Veillonella, and Prevotella were the main bacterial genera in the gastric mucosa of individuals with stomach cancer (Eun et al., 2014). Wang et al. exhibited that Nitrospirae, Escherichia, Lactobacillus, Shigella, Burkholderia, and uncultured Lachnospiraceae were the main gastric microbiome found in individuals with gastric adenocarcinoma (Wang et al., 2016). A recent study speculated that the enrichment of Actinobacteria, Staphylococcus epidermidis and nitrosating/nitrate-reducing bacteria may lead to gastric cancer development. Furthermore, an enhancement in the population of Lactobacillus, Clostridium colicanis, and F. nucleatum bacteria represents diagnostic biomarkers for gastric cancer progression (Hsieh et al., 2018). Aggregatibacter, Neisseria, Streptococcus oralis, Alloprevotella, Streptococcus mitis, Streptococcus pneumoniae, and strain Porphyromonas endodontalis.t_GCF_000174815 are further suggested as the major stomach microbiome engaged in the development of gastric carcinoma (Hu et al., 2018). Additionally, several other studies have explored the effect of H. pylori infection on the diversity and structure of the gastric microbiome. In agreement with the aforementioned studies, the α-diversity of the gastric microbiota was reported higher in subjects with a healthy stomach mucosa, followed by the individuals with gastritis, intestinal metaplasia, precancerous lesions, and gastric cancer. This might suggest that disease progression reduces the mucosal microbial diversity and richness (Miftahussurur et al., 2020; Ndegwa et al., 2020; Rajilic‐Stojanovic et al., 2020).
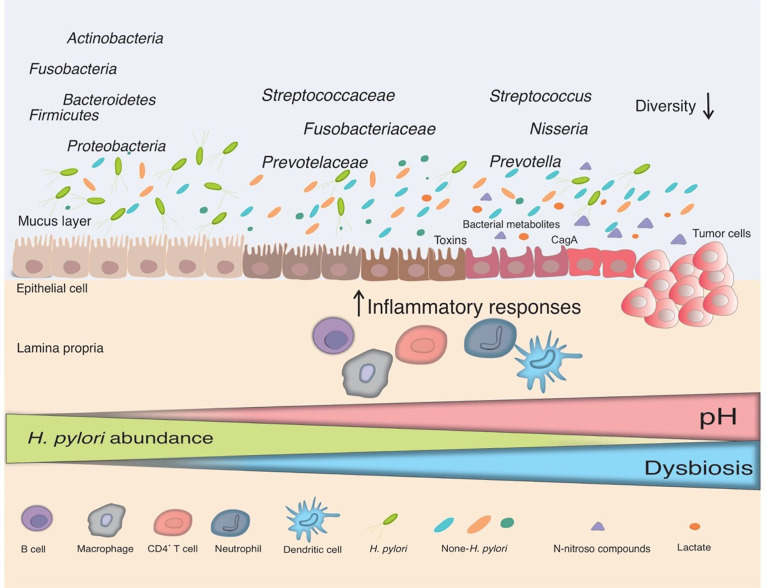
As depicted in Figure 1 , the overall evenness and diversity of the gastric microbiota in the lumen of patients with gastric carcinogenesis will decrease in comparison with healthy individuals. Some substantial alterations in the gastric microbiota composition of individuals suffering from gastric cancer are presented in Table 1 .
Figure 1.
The interplay between H. pylori and the gut microbiota. Infection with H. pylori causes and maintains an inflammatory response in the gastric mucosa, which leads to the loss of acid-secreting parietal cells and an elevation in gastric pH in certain infected people. H. pylori colonization declines in this changing environment and bacteria from other parts of the gut colonize the gastric niche, resulting in gut dysbiosis. Non-H. pylori bacteria promote gastric carcinogenesis through their own characteristics and microbial metabolites, such as N-nitroso compounds and lactate. The main putative mechanisms include induction of inflammatory response, modulation of the immune response, induction of DNA damage, and development of carcinogenesis.
Table 1.
Studies published on the gastric microbiome analysis in patients with gastric cancer.
| Authors/Year | Participants | Sample type | Microbiome analysis method | Taxon differences | Reference |
|---|---|---|---|---|---|
| Dicksved et al./2009 | 10 GC | GB | 16S rRNA gene sequencing (T-RFLP) | ↑ Streptococcus, Lactobacillus, Veillonella, Prevotella
↓ H. pylori |
(Dicksved et al., 2009) |
| Sjöstedt et al./1985 | NA | GJ | 16S rRNA gene sequencing | ↑ Proteobacteria, Firmicutes (genera Veilonella and Streptococcus, Bifidobacterium, Lactobacillus group | (Sjostedt et al., 1985) |
| Aviles-Jimenez et al./2014 | 5 intestinal-type GC | GB | Microarray G3 PhyloChip | ↑ Lactobacillus, Lachospiraceae
↓ Porphyromonas, Neisseria, S. sinensis |
(Aviles-Jimenez et al., 2014) |
| Eun et al./2014 | 11 GC | GM | 16S rRNA gene sequencing | ↑ Streptococcus, Lactobacillus, Veillonella, Prevotella | (Eun et al., 2014) |
| Maldonado-Contreras et al./2011 | NA | GB | 16S rRNA gene sequencing | ↑ Proteobacteria, Spirochetes, Acidobacteria ↓ Actinobacteria, Bacteroidetes, Firmicutes |
(Maldonado-Contreras et al., 2011) |
| Wang et al./2016 | 6 GC, 6 CG | GM | 16S rRNA gene sequencing | ↑ Lactobacillus, Escherichia, Shigella, Nitrospirae, Burkholderia, Fungerum, Lachospiraceae | (Wang et al., 2016) |
| Jo et al./2016 | 34 GC, 29 control | GM | 16S rRNA gene sequencing | ↑ Actinobacteria, S. epidermidis
↑ Nitrosating/nitrate reducing bacteria |
(Jo et al., 2016) |
| Castaño-Rodriguez et al./2017 | 12 GC, 20 dyspepsia | GB | Illumina MiSeq platform targeting the 16S rDNA | ↑ Lactococcus, Veilonella, Fusobacterium, Leptotrichia | (Castaño-Rodríguez et al., 2017) |
| Li et al./2017 | 9 GC | GB | Illumina MiSeq platform targeting the 16S rRNA | ↑ Flavobacterium, Klebsiella, Serratia marcescens, Stenotrophomonas, Achromobacter, Pseudomonas, Delftia, Ralstonia, Rhizobium, Elizabethkingia meningoseptica, Methyloversatiis, Cytophagacea | (Li et al., 2017) |
| Parsons et al./2017 | 95 GC | GB | 16S rRNA gene sequencing | ↑ Helicobacteraceae, Streptococcaceae, Fusobacteriaceae, Prevotellaceae
↓ Tannerella (Escherichia, Shigella, Salmonella, Treponema, Prevotella) |
(Parsons et al., 2017) |
| Yu et al./2017 | 160 GC | GB | 16S rRNA gene sequencing | ↑ Bacteriodetes, Firmicutes, Fusobacteria, Spirochetes ↓ Proteobacteria |
(Yu et al., 2017) |
| Ferreira et al./2018 | 54 GC, 81 CG | GM | 16S rRNA gene sequencing | ↑ Achromobacter, Lactobacillus, Clostridium, Rhodococcus, Citrobacter | (Ferreira et al., 2018) |
| Coker et al./2018 | 20 GC, 21 superficial gastritis, 23 atrophic gastritis, 17 IM | GM | 16S rRNA gene sequencing | ↑ Peptostreptococcus stomatis, S. anginosus, Parvimonas micra, S. exigua, D. pneumosintes
↓ Vogesella, Comamonadaceae, Acinetobacter |
(Coker et al., 2018) |
| Hu et al./2018 | 6 GC, 5 SG | GB | 16S rRNA gene sequencing | ↑ Neisseria, Alloprevotella, Aggregatibacter, S. mitis, Porphyromonoas endodontalis
↓ Sphingobium yanoikuyae |
(Hu et al., 2018) |
| Hsieh et al./2018 | 9 gastritis, 7 IM, 11 GC | GB | 16S rRNA gene sequencing | ↑ Lactobacillus, C. colicanis, F. nucleatum | (Hsieh et al., 2018) |
| Park et al./2019 | 55 GC, 19 IM, 62 CG | GB | 16S rRNA gene sequencing | ↑ Rhizobiales
↑ Cyanobacteria in H. pylori–negative |
(Park et al., 2019) |
| Gunathilake et al./2019 | 288 GC, 288 control | GB | 16S rRNA gene sequencing | ↑ P. copri, P. acnes | (Gunathilake et al., 2019) |
| Spiegelhauer et al./2020 | 22 dyspepsia, 12 GC | GB | 16S rRNA gene sequencing | ↑ Lactobacillus, Prevotella spp., Enterococcus spp., Streptococcus
↓ Actinomyces |
(Spiegelhauer et al., 2020) |
| Ndegwa et al./2020 | 316 GC | GB | 16S rRNA gene sequencing | ↑ Acinetobacter, Aeromonas, Asticcacaulis, Atopobium, DelftiaEwingella, Kocuria, Brevibacterium, Delftia, Halomonas, Kocuria, Lactococcus, Variovorax | (Ndegwa et al., 2020) |
| Cavadas et al./2020 | 164 healthy, 137 GC | GB | 16S rRNA gene sequencing | ↑ Bacillus, Enterobacter, Fusobacterium, Sutterella
↓ Rhodococcus, Phyllobacterium and Staphylococcus |
(Cavadas et al., 2020) |
| Gantuya et al./2020 | 48 GC, 20 gastritis, 40 atrophy, 40 IM. | GB | 16S rRNA gene sequencing | ↑ Lactobacilli, Enterococci, Carnobacterium, Glutamicibacter, Paeniglutamicibacter, Fusobacterium, Parvimonas | (Gantuya et al., 2020) |
| Deng et al./2021 | 25 CG, 34 GC | GB | 16S rRNA gene sequencing | ↑ Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, Fusobacteria | (Deng et al., 2021) |
| Kadeerhan et al./2021 | 193 GC | GB | 16S rRNA gene sequencing | ↑ Actinobacteria, Bacteroidetes, Firmicutes, Bacillus, Capnocytophaga, Prevotella | (Kadeerhan et al., 2021) |
CG, Chronic gastritis; GB, Gastric biopsies; GC, Gastric cancer; GJ, Gastric juice; GM; Gastric mucosa; IM, Intestinal metaplasia; NA, Not available.
2.3.4 The intestinal microbiota
The colon has the largest community of bacteria in the human digestive system, with a nearly 107-fold increase in population compared to the stomach (Sender et al., 2016). Crosstalk between H. pylori and other commensal bacteria along the gut has a great impact on the gastric and colonic microbiota and may influence the homeostasis of the entire system (Myllyluoma et al., 2007; Serrano et al., 2021). Due to the close interplay between H. pylori infection, the gut microbiome, and gastritis, the alterations in the gut microbiota could be associated with chronic gastrointestinal disorders (Ferreira et al., 2018; Yang et al., 2019). H. pylori infection leads to the enrichment of Firmicutes, Proteobacteria, Actinobacteria, and Acidobacteria, while reducing the proportion of Bacteroidetes phylum in the intestine of the patients suffering from gastric cancer (Gao et al., 2018a). Long-term infection with H. pylori further results in Akkermansia overgrowth in the colon (Heimesaat et al., 2014). A study by Wang et al. showed that gastric cancer patients have higher levels of Lactobacillus, Lachnospiraceae, Escherichia-Shigella, Nitrospirae, and Burkholderia compared to the control group, confirming earlier findings that Lactobacillus is widely colonized in gastric cancer (Wang et al., 2016). Although there is still no consensus on the role of the gut microbiome in gastric carcinogenesis, the dysbiotic microbial population may raise the risk for gastric cancer by increasing the inflammatory process in the stomach and inducing immunological responses (Ferreira et al., 2018). Additionally, microbial dysbiosis has a crucial role in tumor initiation and progression by inducing chronic inflammation, dysregulating the immune response, and producing toxins and metabolites (Lertpiriyapong et al., 2014). Several mechanism-oriented studies are required to identify the processes underlying microbial participation in the occurrence of gastric cancer and to prevent carcinogenesis.
3 The gut microbiota and the host immune system
It has been well documented that the microbiota in the stomach may have a role in cancer progression and responsiveness to anticancer treatment (Vivarelli et al., 2019). The interplay between the gut microbiota and gastric mucosal immunity is complicated and includes multifold interactions in homeostasis and disease (Zheng et al., 2020). The innate immune system provides the foremost barrier that acts immediately when an antigen enters the body, while the second line of defense against pathogens is the adaptive immune system (Thakur et al., 2019). Toll-like receptors (TLRs) are the main elements of the human innate immune response and perform fundamental roles in mediating inflammatory pathways by detecting pathogen-associated molecular patterns (PAMPs) such as LPS, flagellin, and peptidoglycan. Moreover, antigen-presenting cells (APCs), such as dendritic cells (DCs), develop in response to PAMPs via pattern recognition receptors (PRRs). DCs can directly stimulate CD8+ T cells and also can provoke naive T cells to generate CD4+ T cells, especially CD4+ T regulatory cells (Tregs), and Th17 cells. TLR-mediated signaling can trigger the transcription factor nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), interferon regulatory factor 3 (IRF3), and mitogen-activated protein (MAP) kinases p38 that initiate downstream signaling pathways (Kawasaki and Kawai, 2014). The main adaptor protein that mediates signals upon stimulation of TLRs is myeloid differentiation primary response gene 88 (MyD88) that enhances the systemic innate immune cell response (Wolz, 2016). MyD88 can recruit IL-1R-associated kinase family (IRAKs) and thereafter activates the MAP3 kinases, which regulates the activation of c-Jun N-terminal kinase (JNK), NF-κB, and MAP kinases p38 (Huang et al., 2004).
It has been elucidated that the gut microbiota oddly modulate the maturation and operation of innate and adaptive immunity. Adaptive immunity is made up of cellular and humoral immunity and is defined by the presence of lymphocytes including T and B cells. The development and function of the adaptive immunity are regulated by gut microbiome, meanwhile, the maturation of the gut microbial community is influenced by the host immune components (Lazar et al., 2018). The impact of the gut microbiota on the maturation of the host immune system were investigated in a study concluding that the microbiota can influence the activity of the gut and systemic immunity, including pathogen removal (Lee and Mazmanian, 2010; Wiertsema et al., 2021). Compared to normal mice models, germ-free (GF) mice, which have no microorganisms living inside or outside them, exhibit defective maturation of gut-associated lymphoid tissues (GALT), in addition to fewer and smaller Peyer’s patches, cellular mesenteric lymph nodes, and less cellular lamina propria of the small intestine. Consequently, the gut microbiota are essential for the development and function of multiple populations of gut immune cells.
The gut microbiota can boost epithelial cytokine expression that controls the activity of T and B lymphocytes, as well as macrophages (Cianci et al., 2019). The host inflammatory response is initiated by expressing particular cytokines such as IL-1β, TNF-α, IL-2, IL-6, IL-15, IL-21, and IL-23, while IL-10 and transforming growth factors-β (TGF-β) secretion can induce the host anti-inflammatory response (Kany et al., 2019). As a result, the inflammatory or homeostatic state of the gut is determined by the equilibrium between the ratios of pro- and anti-inflammatory cytokines.
The host immune system and cancer progression are intricately linked and thereby changes in the balance between the gut microbiota and the host immune response might trigger tumor development (Pagliari et al., 2017; Chen et al., 2021). The development of a single tumor cell into the primary tumor is termed tumor progression. The upregulation of cell proliferation and decreased cell apoptosis, which are enhanced by inflammation-driven mechanisms, have critical impacts on the initial tumor development. Several stimulating impacts of inflammation on carcinogenesis are applied at the level of tumor progression and most of the tumor promoters (Grivennikov et al., 2010).
3.1 Specific immune response during H. pylori infection and gastric carcinogenesis
H. pylori colonization leads to the aggressive stimulation of the innate and adaptive immunity and is a well-understood risk factor for the progression of stomach cancer. Normally, the host’s innate defense is activated when H. pylori resides in the stomach epithelium, leading to the secretion of pro-inflammatory and antibacterial agents by the gastric epithelium (Krzysiek‐Maczka et al., 2018). When the host’s prolonged inflammatory response fails to eliminate H. pylori, chronic gastritis develops, which may last through the lifespan and cause gastric atrophy, intestinal metaplasia, dysplasia, and intestinal-type gastric carcinoma (Sheh and Fox, 2013). It is suggested that H. pylori infection can initiate host inflammation by promoting the secretion of several inflammatory mediators from the gastric epithelium. These cytokines and chemokines attract innate immune cells including neutrophils, macrophages, DCs, natural killer (NK) cells, and lymphocytes (Robinson et al., 2007). IL-8, as a strong chemokine, promotes neutrophils and monocytes infiltration to the gastric mucosal surfaces. Thereafter, the DCs and monocytes induce the release of IL-6, TNF-α, and IL-1β (White et al., 2015). The stimulation of CD4+ T cells activity by IL-1β and IL-6 promotes the secretion of different cytokines such as IL-4, IL-5, IL-6, and interferon-γ (IFN-γ). On the other hand, H. pylori prevents the proliferation of CD4+ T cells by VacA (Beigier-Bompadre et al., 2011).
The reduction in the number of parietal cells developes a state of hypochlorhydria (pH >4) that can perturb the balance of the gastric microbiome. Multiple non-H. pylori species such as Streptococcus spp., Lactobacillus spp., Xanthomonas spp., Proteus spp., Klebsiella spp., Pseudomonas spp., Neisseria spp., E. coli, and Campylobacter jejuni have been detected in the stomach of hypochlorhydria subjects (Pereira et al., 2018). The precise processes through which the modified gut microbiome combine with H. pylori to cause gastric cancer are yet to be fully elucidated. Multiple studies have reported that the combination of H. pylori and other gut microorganisms may work together to raise the risk of stomach cancer. According to an investigation, there is a relationship between harmful shifts in the fecal microbial composition and the elevation in the production of pro-inflammatory cytokines (Wu et al., 2020). The symbiont Bacteroides fragilis producing polysaccharide A can downregulate pro-inflammatory cytokines production induced by H. pylori colonization (Troy and Kasper, 2010). The colonization of mice with enterotoxigenic B. fragilis (ETBF) resulted in the attraction of Th17-dependent myeloid cells to the tumor microenvironment (TME), which aided colon carcinogenesis (Orberg et al., 2017). Commensals in the gut, notably segmented filamentous bacteria (SFB), have been linked to gut immune development and the generation of IL-17 (Hedblom et al., 2018).
H. pylori infection has been suggested as the main cause of developing gastric adenocarcinoma so far. The Correa model of H. pylori-induced gastric pathogenesis and aging-related alterations in acid production supports the link between disease and age. The cumulative lifetime exposure to reactive oxygen and nitrogen species (RONS), pro-inflammatory cytokines, and tissue damages induced by H. pylori are coupled in elderly with a reducing ability to cope with antigens (immunosenescence) and decreased ability to regulate inflammatory responses (inflammaging) (Weiskopf et al., 2009). Immunosenescence, or the age-related reduction in immunity, is manifested in the elderly by diminished antigen presentation, diminished cytotoxic activity, the buildup of effector T cells, reduced naive T cell output and decreased B cell generation. Alterations in the structure of Firmicutes and an expansion in the proportion of Bacteroidetes, have been demonstrated to influence the gut microbiota of the elderly (Vemuri et al., 2018).
3.2 Effects of the gut microbiota on gastric cancer immunotherapy
Innate and adaptive immune cells, as well as pro-inflammatory mediators are involved in the initiation and expansion of inflammation (Luster et al., 2005). Moreover, chemokines (CCL2, CXCL12) are critical for recruiting the inflammatory cells to the site of infection. The gut microbiota have been demonstrated to influence the differentiation and function of M2-tumor-associated macrophages (M2-TAMs), tumor-associated neutrophils (TANs), DCs, and CD8+ T cells by their products, such as LPS, polysaccharide-dextran, deoxycholic acid (DCA), and SCFAs (Zhao et al., 2021).
The production of SCFAs by the gut microbiota such as butyrate, acetate, and propionate are demonstrated to have a major role in preventing carcinogenesis by stimulating apoptosis and growth arrest in malignant cells (Tang et al., 2011). SCFAs operate as extracellular and intracellular signal mediators and have a major part in the development and function of the host cell, particularly immune cells, attributing to epigenetic alteration and receptor-mediated signaling (Parada Venegas et al., 2019). These metabolites can block histone deacetylases (HDACs) in colonocytes and immune system activity to allow histone hyperacetylation. Similar to the use of HDAC inhibitors, the interaction of mononuclear cells and macrophages with SCFAs leads to reduced production of pro-inflammatory cytokines such as IL-6 and NF-κB. Additionally, by inhibiting HDAC, propionate and butyrate also prevent bone marrow stem cells from transforming into DCs, decreasing excessive inflammatory reactions. By inhibiting HDAC, SCFAs can impact the homeostasis of peripheral T cells and thereby promote the development of Tregs that critically control the intestinal inflammation and carcinogenesis (Furusawa et al., 2013; Arpaia et al., 2013). SCFAs are also involved in the upregulation of immunosuppressive IL-10 and TGF-β cytokines, as well as downregulation of pro-inflammatory cytokines in macrophages and neutrophils, which may lead to inhibition of Th17 cell development, suppression of inflammation, and carcinogenesis. Particularly, butyrate promotes the anticancer effectiveness of oxaliplatin and anti-PD-L1 therapy by directly increasing the cytotoxic CD8+ T cell response via ID2-dependent IL-12 signaling through its HDAC inhibitory action. SCFAs can operate either directly on cancer cells as HDAC inhibitors or indirectly by influencing the immune system through certain G-protein-coupled receptors (GPCRs) such as GPR41, GPR43, and GPR109A expressed in colonocytes and immune cells (Koh et al., 2016). Some studies revealed that acetate and propionate might stimulate Tregs in a GPR43-dependent manner, which results in reducing the severity of acute and chronic colitis (Furusawa et al., 2013). Other anti-tumor properties of SCFAs can be associated to their capability to promote mucin secretion and improve the integrity of gut epithelium by initiating the inflammasome and the peroxisome proliferator-activated receptor-γ (PPAR-γ) signaling pathway (Chen and Chen, 2021).
Bacteroides thetaiotaomicron, an acetate-producing bacterium, enhances mucus secretion by promoting goblet cell development and mucus-related gene expression (Wrzosek et al., 2013). Moreover, butyrate-producing bacteria might restore vitamin D receptor (VDR) expression in the intestine and reduce the expansion of gut dysbiosis (Nabavi-Rad et al., 2022). Intestinal dysbiosis can enhance the release of LPS, which results in the production of cathepsin K (CTSK) in colorectal cancer (CRC) cells. Recent studies revealed a connection between the overexpression of CTSK and malignancies in colon cancer (Li et al., 2019; Calio et al., 2021). Additionally, CTSK activates the mammalian target of rapamycin (mTOR) pathway by interacting with TLR4 on the macrophage, causing M2 polarization and the generation of cytokines including IL-4 and IL-10. The conversion of M2 to M1 macrophage, a prospective target in cancer immunotherapy, is likely to be aided by the indigenous gut microbiota such as B. fragilis (Deng et al., 2016). B. fragilis has been reported to enhance the phagocytic functions of macrophages of the innate immune system and polarize them to an M1 phenotype (Frankenberg et al., 2008; Deng et al., 2016). Additionally, Bifidobacterium bifidum cell surface polysaccharides can activate the TLR2/MyD88 pathway and also suppress experimental colitis through Treg induction (Verma et al., 2018). As a result, it is plausible that PAMPs generated from the gut microbiota affect PRR, which controls the immune response to gut tumors. Consequently, the activation of NF-κB increases the expression of pro-inflammatory cytokines including TNF-α and IL-6, which might explain why immunosuppressive myeloid cells accumulated in TME (Zhang et al., 2021).
NK cells are effector lymphocytes of the innate immunity that modulate numerous forms of tumors by preventing their progression and development (Levy et al., 2011). NK cells can regulate tumor metastasis owing to their capacity to exert direct cellular cytotoxicity without prior sensitization and to release immunostimulatory cytokines such as IFN-γ. Nevertheless, NK cells show an impaired capability to infiltrate tumors in individuals suffering from cancer. Furthermore, in NK cell-based cancer immunotherapy, correcting NK cell dysfunction is a key requirement for immunotherapies against tumors (Borrego et al., 2016). Multiple medicines including immune checkpoint inhibitors (ICIs) and monoclonal antibodies have been developed to reverse the impaired NK cell function (Wang et al., 2021). A favorable gut microbiome can induce the accumulation of NK cells, which target tumor cells and enhance tumor surveillance. The development of precision medicine requires deep knowledge of the interaction between the gut microbiome and the immune response in cancer immunotherapy (Furusawa et al., 2013). Consequently, intestinal microbiota are attractive targets to manage inflammation-related cancers, as fecal microbiota transplantation (FMT) is becoming a popular way to enhance the gut microbiota and metabolome (Zhao et al., 2021).
The identification of proteins that suppress T cell responses such as Cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), programmed death-ligand 1 (PD-1) and its ligand PD-L1 opens up new opportunities for treating certain cancers having elevated production of such proteins. CTLA-4 and PD-1 are commonly found on activated T cells, while PD-1 ligands (PD-L1 and PD-L2) are found on a variety of cell types including antigen-presenting cells, innate immune cells, epithelial cells, and endothelial cells (Seidel et al., 2018). Activation of CTLA-4 and PD-1 can influence the proliferation of tumor-specific T cells and thereby protecting tumor cells in the TME. CTLA-4 controls the immune response just after T cells activation and PD-1 proceeds later to promote T cell death and subsequently terminate the immune response ( Figure 2 ). Anti-CTLA-4, PD-1, and PD-L1 inhibitors are the main types of checkpoint inhibitor immunotherapy drugs that have been developed and reported to be an efficient treatment for various malignant tumor types. The anti-PD-L1 inhibitor has been suggested for patients with advanced gastric cancer and positive PD-L1 expression. Ipilimumab is the single Food and Drug Administration (FDA)-approved ICI and the most prominent member of CTLA-4 inhibitor for cancer treatment (Naimi et al., 2022). Furthermore, PD-1 inhibitors nivolumab, pembrolizumab, and cemiplimab, as well as PD-L1 inhibitors atezolizumab, avelumab, and durvalumab are on the current list of authorized medicines (Vaddepally et al., 2020; Twomey and Zhang, 2021). Combining therapy of CTLA-4 and PD-1 blockers with checkpoint inhibitors can boost the immune response by stimulating the signaling pathways that lead to anticancer impact of T cells (Rotte, 2019).
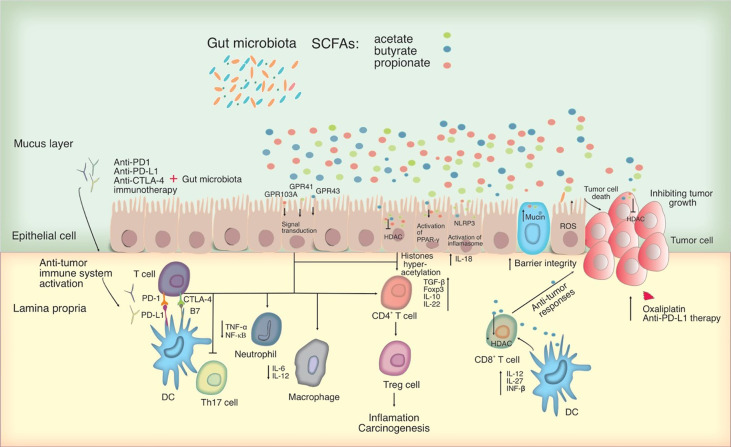
Figure 2.
Potential mechanisms for microbiota-mediated immunomodulation in tumor cells. SCFAs, which primarily consist of acetate, butyrate, and propionate can act as an HDAC inhibitor and influencing directly on cancer cells. By interacting with particular GPCRs including GPR41, GPR43 and GPR109A, SCFAs can have an effect on the immune system, leading to upregulation of immunosuppressive IL-10 and transforming growth factor-beta (TGF-β), downregulation of pro-inflammatory cytokines in macrophages and neutrophils, and inhibition of differentiation towards T helper type 17 (Th17) cells, thereby suppressing inflammation and carcinogenesis. SCFAs activate the inflammasome and the PPAR-γ pathway, promoting mucin production and improving epithelial integrity. SCFAs were also shown to activate the NLR family pyrin domain containing 3 (NLRP3) inflammasome, modulating the production of IL-18, which protects epithelial integrity. Significantly, SCFAs, in particular butyrate, may alter CD8+ T cell antitumor responses by influencing DC signaling pathways involving IL-12, IL-27, and IFN- β, all of which have an impact on tumor combination therapy. In this figure, the role of PD-1 and CTLA-4 in the priming and effector phases of anti-tumor immune responses is shown. Anti-CTLA-4 blocking antibodies may thereby restore T cell priming in lymph nodes, whereas PD-1 signaling inhibition may allow T cells to operate as tumor effectors. Other cell types in the tumor microenvironment, such as DCs, may also express PD-1 and hence be impacted by PD-1 inhibition. Blocking PD-1 and CTLA-4 may influence T helper cell profiles directly or indirectly by changing the microbiota. HDAC, Histone deacetylase; PD-L, Programmed death-ligand; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; PPAR-γ, peroxisome proliferator-activated receptor-γ; GPCRs, G-protein-coupled receptors.
According to new findings, the gut microbiota appears to have a substantial part in regulating responses to immunotherapies (Shui et al., 2020). The link between the gut microbiota and immunotherapy effectiveness was demonstrated by mice experiments, in which particular bacterial taxa were dominated in individuals, before checkpoint blockade therapy (Jain et al., 2021). According to an in vivo study, mice colonized with stool isolates from responders had a better outcome than mice colonized with stool specimens from non-responders with checkpoint blockade treatment. Intestinal bacteria appear to be necessary for the best response to cancer chemotherapy and immunotherapy via modulating myeloid cell activity (Hayase and Jenq, 2021). Two elements of the gut microbiota contribution to therapy effectiveness have been suggested, which include an adjuvant impact of translocated bacteria after the therapy-induced disruption of intestinal epithelial barrier and a priming impact given by the gut-inherent microbial community prior to the therapy, which explore the emerging role of microbiota in cancer immunotherapy (Gorjifard and Goldszmid, 2016).
In melanoma mice treated with anti-PD-1/PD-L1 drugs, the genus Bifidobacterium was discovered to be the main inducer for enhanced tumor-specific T cell responses and intratumoral CD8+ T cells (Shui et al., 2020; Liu et al., 2021). In a clinical trial, Akkermansia muciniphila supplementation was demonstrated to promote the efficacy of PD-1 blockade through stimulating the infiltration of CCR9+ CXCR3+ CD4+ T lymphocytes into TME in an IL-12-dependent manner (Routy et al., 2018). The higher proportion of Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium among the gut microbiota was presented with an increased response to PD-1 inhibitors (Matson et al., 2018). Furthermore, longer progression-free survival (PFS) in ipilimumab treatment of melanoma was positively associated with the proportion of Faecalibacterium and other Firmicutes bacteria, while negatively associated with the presence of Bacteroidetes genus (Chaput et al., 2017). Bacteroides caccae has been demonstrated as the main bacteria that present ICIs responders in various types of treatment. Regarding responders treated with anti-CTLA4 and anti-PD-1 immune checkpoint blockade, Faecalibacterium Prausnitzii, Bacteroides thetaiotamicron, and Holdemania filiformis were exhibited as the predominant bacteria (Frankel et al., 2017).
In an animal model of adoptive T cell transfer treatment, the gut microbiota was found to have an adjuvant impact during cancer immunotherapy (Vetizou et al., 2015). The translocation of bacteria into the mesenteric lymph nodes resulted in higher levels of circulating bacterial products, which activated DCs and enhanced the activity of transplanted T lymphocytes (Lathrop et al., 2011). Firmicutes phylum and Ruminococcaceae family are exhibited to be linked to both responses and toxicity, whereas the Bacteroidales order of the Bacteroidetes phylum was linked to a loss of response to immune checkpoint blockade. Taken together, despite the presence of major challenges in addressing the inherent contribution of microbiota in the regulation of cancer immunity, a great deal of evidence held the gut microbiota responsible for modifying the host antitumor immune response and immune checkpoint blockade response (Gopalakrishnan et al., 2018).
4 Microbiome-based therapeutic approaches in gastric cancer
It has become apparent that one of the major factors influencing the effectiveness of cancer treatments is the composition of the commensal microbiota (Soto Chervin and Gajewski, 2020). There is growing evidence to show that the interaction of the immune system and the gut microbiota within the host affects how the body reacts to cancer immunotherapy (Ma et al., 2019). Designing therapeutic treatments to improve the patient’s microbial structure is highly recommended, which necessitates an understanding of the mechanisms underpinning the efficacy of microbiome-based therapeutic interventions. Probiotic and/or prebiotic supplementation as well as FMT are considered the pioneer microbiome-modulating approaches, which are further discussed below.
4.1 Effects of probiotics on H. pylori, the gut microbiota, and gastric cancer
Probiotics have been proven to beneficially influence the gut microbiota, by temporarily or permanently shaping the composition of the gut microbiota (Hemarajata and Versalovic, 2013). Moreover, gut bacteria can interact directly with the host immune cells, triggering both innate and adaptive immune responses by their immune-modulating properties. Consequently, interactions between the gut microbiota and immune cells lead to the maturation of the host immune system throughout the lifespan (Pickard et al., 2017; Zhao and Elson, 2018; Chen et al., 2021). Additionally, probiotics can modify the effect of the gut microbiota on local immunological and inflammatory responses, through the release of particular cytokines such as IFNs, ILs, TNFs, and TGFs (Sang et al., 2010; Ebrahimpour-Koujan et al., 2020). However, probiotics can also interact with DCs, and bridge the innate and adaptive immune system. Intestinal epithelial cells and DCs may respond to the gut microbiota by their PRRs (Raheem et al., 2021). Following probiotic supplementation, the activated DCs induce a favorable immune response, inhibiting Th1-, Th2-, and Th17-mediated inflammatory response. By reducing the production of TLRs by secreting TNF-α inhibitory metabolites and preventing of NF-κB signaling in enterocytes, probiotics can further reduce intestinal inflammation (Raheem et al., 2021). Moreover, lactic acid produced by probiotics has been demonstrated in animal studies to modulate inflammation by regulating pro-inflammatory cytokines (Murosaki et al., 2000). In recent years, adjuvant therapy with probiotics has received significant attention to boost the success of H. pylori eradication (Goderska et al., 2018; Keikha and Karbalaei, 2021).
The progression of gastric malignancy is strongly correlated with the expression of CagA oncoprotein, as CagA-positive H. pylori This interaction is assumed to be mainly influenced by CagA-mediated overexpression of IL-8 in the gastric mucosa. Some probiotic strains such as Lactobacillus acidophilus, Lactobacillus bulgaricus, and Lactobacillus salivarius have been proved to reduce H. pylori-induced IL-8 upregulation (Kabir et al., 1997; Zhou et al., 2008; Yang et al., 2012). Based on recent studies, the administration of probiotics as a co-supplementary material with standard antibiotic regiments substantially increases H. pylori eradication rate (Eslami et al., 2019).
L. salivarius, Lactobacillus gasseri, Lactobacillus casei Shirota, Lactobacillus johnsonii La1, Lactobacillus rhamnosus GG, and Saccharomyces boulardii were recognized to have either anti-H. pylori properties or anti-inflammatory activity (Hsieh et al., 2012). Recent investigations suggested that probiotics may improve the host immune system and protect against cancer progression (Lu et al., 2021). Furthermore, probiotic co-supplementation with conventional antibiotic regimens can substantially increase the rate of H. pylori eradication (Penumetcha et al., 2021). Some probiotic strains can inhibit vacA and flaA virulence genes in H. pylori by producing substances such as reuterin, which is a potent antimicrobial agent expressed by Lactobacillus reuteri (Urrutia-Baca et al., 2018; Song et al., 2019). The metabolites produced by the probiotics including PG, surface proteins, secreted proteins, bacteriocins, cell wall polysaccharides, organic acids, hydrogen peroxide, indole, and extracellular vesicles have several favorable effects on the host by triggering various signaling pathways such as mucus secretion in goblet cells (Teame et al., 2020; Sankarapandian et al., 2022). Furthermore, many probiotic strains are expected to enrich the human gut, and particular species, such as Lactobacillus spp., can replicate in the human gastric environment and have the ability to prevent H. pylori colonization (Ji and Yang, 2020). Probiotics antagonize H. pylori through various mechanisms, including (a) secreting antibacterial compounds like organic acids, bacteriocins, and biosurfactants (b) inhibiting the colonization of H. pylori by occupying receptor sites or co-aggregation mechanism, (c) supportive role in the intestinal tissues by stimulating mucin synthesis, and (d) modulating the host immune response (Ji and Yang, 2020). Furthermore, these microbiota can express extensive amounts of lactic acid in the gastric lumen following successful colonization. Probiotics also reduce H. pylori growth by competing for binding sites or affecting the adhesion mechanism (Urrutia-Baca et al., 2018).
As previously discussed, probiotics have attracted a significant attention in medicine owing to their beneficial effects on different types of gastrointestinal cancers. However, a vast majority of clinical studies lack an appropriate long-term follow-up regarding the effect of probiotic supplementation as a biotherapeutic approach against cancer progression. Therefore, well-designed, randomized, double-blind, placebo-controlled clinical trial investigations are required to elucidate potential probiotic strains and their effective dosages, as an alternative therapy for cancer control (Javanmard et al., 2018). In addition, there are conflicting clinical outcomes concerning the effect of various probiotic strains on H. pylori eradication rate. Several studies have indicated no statistically significant difference between the probiotic supplemented group and the control group when it comes to the eradication rate of H. pylori (Armuzzi et al., 2001; Cremonini et al., 2002; Nista et al., 2004; Myllyluoma et al., 2005; Cindoruk et al., 2007; Manfredi et al., 2012; Navarro-Rodriguez et al., 2013; Shavakhi et al., 2013). Likewise, a recent meta-analysis study confirmed that the administration of probiotics to standard therapy regiments does not improve the eradication rates of H. pylori (Lu et al., 2016a). However, another meta-analysis study reported that probiotic supplements can improve H. pylori eradication rate, reduce antibiotic-related side effects, and decrease individual incidences of multiple symptoms (Tong et al., 2007; Zou et al., 2009; Wang et al., 2013; Lu et al., 2016b). It has been further suggested that probiotics may not directly eliminate H. pylori, but rather maintain low levels of the pathogen in the stomach; thereby, in combination with antibiotics, probiotics may increase the eradication rate and decrease the adverse effect of antibiotics (Gotteland et al., 2006). Consequently, the main advantage of probiotic supplementation is currently considered the reduction of antibiotic-related adverse effects (Felley et al., 2001; Cremonini et al., 2002; Tursi et al., 2004).
It may be possible to revisit some fundamental concepts about probiotics and concentrate on biologically relevant issues to ease the transition from empirical to patient-oriented therapeutics through high-throughput sequencing and experimental methods. Finally, the development of evidence-based policy should be constructed on large-scale randomized and blinded clinical trials, preferentially without commercial interests (Suez et al., 2019).
4.2 Effects of prebiotics on H. pylori, the gut microbiota, and gastric cancer
Prebiotics are indigestible food components that beneficially influence the host’s health by promoting the colonization and/or function of one or a small proportion of the gut commensals (Gibson and Roberfroid, 1995; Ziemer and Gibson, 1998; Serban, 2014). Inulin, oligofructose, lactulose, resistant starch, and wheat bran have all been employed in CRC research as prebiotic products. Combinations of probiotics and prebiotics, termed synbiotics, may result in additive or synergistic effects on gut function (Serban, 2014). Isomalto-oligosaccharides (IMO), fructo-oligosaccharides (FOS), galacto-oligosaccharides (GOS), lactulose, and resistant starch (the fraction of starch that resists digestion in the small intestine) are all commercially available prebiotics (Tymczyszyn et al., 2014).
Numerous investigations have looked into the health advantages of fiber ingestion, both with or without the prebiotic effect (Slavin, 2013). Epidemiologic investigations propose that sufficient fiber intake reliably reduces the risk of cardiovascular disease CVD and coronary heart disease (CHD), primarily within a decrease in low-density lipoprotein (LDL) levels (Threapleton et al., 2013). One of the prebiotic mechanisms of action is the growth-promoting impact on members of the Bifidobacterium and Lactobacillus genera, resulting in greater butyrate, acetate and lactate production (Lordan et al., 2020). SCFA production has several health benefits such as improving the gut barrier function, as well as mineral absorption (Ballan et al., 2020). Moreover, prebiotics similar to probiotics can modify the gut microbiota structure and function in particular on immune responses, pathogenic defense mechanisms, bowel function, and stool consistency (Gibson et al., 2017). The capacity of prebiotics to regulate the host immune responses is mainly by modulating the Th2 and Th1 response (Reid et al., 2003).
In a recent study, patients were supplemented with several probiotic strains including L. rhamnosus, L. casei, Streptococcus thermophilus, Bifidobacterium breve, L. acidophilus, Bifidobacterium longum, and L. bulgaricus, and co-supplemented with magnesium as well as fructooligosaccharide as prebiotic. According to this investigation, adding synbiotics to the conventional anti-H. pylori regime can help improve drug tolerance, while probiotics plus prebiotics could increase the rate of eradication significantly (Shafaghi et al., 2016).
Prebiotics are presented with an osmotic effect in the gut lumen, which can lead to diarrhea, abdominal pain, and gastroesophageal reflux following a large dose consumption. Moreover, prebiotic fermentation in the colon may cause gaseousness and bloating (Marteau and Seksik, 2004). Therefore, to develop recommendations for cancer prevention and therapy by prebiotic administration, well-designed, randomized, double-blind, placebo-controlled human trials utilizing probiotics and/or prebiotics are required (Serban, 2014).
4.3 Beneficial effects of FMT on H. pylori, the gut microbiota, and gastric cancer
The emergence of FMT application in various ill states is owing to its significant efficacy in treating recurrent Clostridioides difficile infection (rCDI) (Monaghan et al., 2021; Azimirad et al., 2022). The engraftment of microbial composition from a healthy donor to an appropriate patient thoroughly modifies the patient’s gut microbiota shifting it toward the donor’s microbial profile (Wang et al., 2019). SCFA-producing bacteria as well as mucin-producing bacteria such as A. muciniphila are reported in several studies to be enriched among the patient’s microbial community following a successful FMT (Ser et al., 2021).
Washed preparation of FMT was applied in a recent pilot study to eradicate H. pylori infection. The result of this study indicated the potential capacity of FMT in attenuating H. pylori infection to some extent (Ye et al., 2020). Considering the substantial influence of FMT on the intestinal microbiota compared to the gastric microbiota, clinical trials have investigated the impact of FMT on lower gastrointestinal disorders including CRC (Chen et al., 2019). Transplanting stool samples of patients suffering from CRC to GF mice was reported to increase the risk of developing intestinal inflammation and dysplasia in the animal models (Wong et al., 2017). However, the possible influence of intestinal microbiota on the progression of gastric cancer should be noted and animal studies and clinical trials should consider FMT as a stopgap treatment for gastric adenocarcinoma and precancerous lesions of gastric cancer.
5 Conclusion and future perspective
Recent advances in culture-independent techniques, mainly high-throughput sequencing and NGS technology, have expanded our understanding of the correlation between the gut microbiota and gastric carcinoma. Current studies have provided solid evidence regarding the effect of H. pylori infection on the gut microbial composition, microbial dysbiosis, and cancer progression. It is undoubted that substantial differences exist between the gut microbial composition of patients with gastric pathology, atrophic gastritis, IM, and gastric adenocarcinoma. Consequently, dysbiosis is suggested as a dynamic process that triggers and develops H. pylori-associated gastric carcinoma. Furthermore, the major role of the host immune system during H. pylori colonization, microbiota alteration, and cancer progression has encouraged researchers to develop innovative therapeutic paradigms in cancer immunotherapy. Pioneer investigations have proposed this therapeutic intervention as an applicable approach, which can be promoted by restoring the inherited microbial composition through probiotic supplementation or FMT. However, some challenges and bottlenecks remain to be addressed about the effective biomarkers to assess the immune condition of patients, the root of therapy failure, and the possibilities for predicting treatment outcomes. Exploring these challenges may shed light on novel treatment directions to reduce the risk of developing therapeutic side effects.
Author contributions
FF, BA, and AY contributed to the literature review and wrote the draft of the manuscript. AY and FF designed the figures for the manuscript. FF illustrated the figures for the manuscript. AY worked on concept and design of the study, interpreted the collected information and supervised the project. AS and MZ provided clinical advice and guidance for improving of the manuscript. AY, AN-R, and NS critically revised the final version of the manuscript. All authors approved the final version of the manuscript and the authorship list.
Funding
This study was financially supported by a research grant (no. RIGLD 1128, IR.SBMU.RIGLD.REC.1399.046) from the Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Acknowledgments
The authors would like to thank the members of the Foodborne and Waterborne Diseases Research Center at the Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Alon-Maimon T., Mandelboim O., Bachrach G. (2022). Fusobacterium nucleatum and cancer. Periodontology 89, 166–180. doi: 10.1111/prd.12426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altveş S., Yildiz H. K., Vural H. C. (2019). Interaction of the microbiota with the human body in health and diseases. Biosci. Microbiota Food Health 39, 19–023. doi: 10.12938/bmfh.19-023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. K., Huang J. Y., Wreden C., Sweeney E. G., Goers J., Remington S. J., et al. (2015). Chemorepulsion from the quorum signal autoinducer-2 promotes helicobacter pylori biofilm dispersal. mBio 6, e00379. doi: 10.1128/mBio.00379-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. A., Murphy S. J., Johnston B. T., Watson R. G. P., Ferguson H. R., Bamford K. B., et al. (2008). Relationship between <em<Helicobacter pylori</em< infection and gastric atrophy and the stages of the oesophageal inflammation, metaplasia, adenocarcinoma sequence: results from the FINBAR case–control study. Gut 57, 734. doi: 10.1136/gut.2007.132662 [DOI] [PubMed] [Google Scholar]
- Ansari S., Yamaoka Y. (2017). Survival of helicobacter pylori in gastric acidic territory. Helicobacter 22, e12386. doi: 10.1111/hel.12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armuzzi A., Cremonini F., Bartolozzi F., Canducci F., Candelli M., Ojetti V., et al. (2001). The effect of oral administration of lactobacillus GG on antibiotic-associated gastrointestinal side-effects during helicobacter pylori eradication therapy. Aliment Pharmacol. Ther. 15, 163–169. doi: 10.1046/j.1365-2036.2001.00923.x [DOI] [PubMed] [Google Scholar]
- Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., Deroos P., et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. doi: 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviles-Jimenez F., Vazquez-Jimenez F., Medrano-Guzman R., Mantilla A., Torres J. (2014). Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 4, 1–11. doi: 10.1038/srep04202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimirad M., Jo Y., Kim M. S., Jeong M., Shahrokh S., Asadzadeh Aghdaei H., et al. (2022). Alterations and prediction of functional profiles of gut microbiota after fecal microbiota transplantation for Iranian recurrent clostridioides difficile infection with underlying inflammatory bowel disease: A pilot study. J. Inflammation Res. 15, 105–116. doi: 10.2147/JIR.S338212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhti S. Z., Latifi-Navid S. (2021). Oral microbiota and helicobacter pylori in gastric carcinogenesis: what do we know and where next? BMC Microbiol. 21, 71. doi: 10.1186/s12866-021-02130-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballan R., Battistini C., Xavier-Santos D., Saad S. M. I. (2020). Interactions of probiotics and prebiotics with the gut microbiota. Prog. Mol. Biol. Trans. Sci. 171, 265–300. doi: 10.1016/bs.pmbts.2020.03.008 [DOI] [PubMed] [Google Scholar]
- Bauerfeind P., Garner R., Dunn B., Mobley H. (1997). Synthesis and activity of helicobacter pylori urease and catalase at low pH. Gut 40, 25–30. doi: 10.1136/gut.40.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigier-Bompadre M., Moos V., Belogolova E., Allers K., Schneider T., Churin Y., et al. (2011). Modulation of the CD4+ T-cell response by helicobacter pylori depends on known virulence factors and bacterial cholesterol and cholesterol α-glucoside content. J. Infect. Dis. 204, 1339–1348. doi: 10.1093/infdis/jir547 [DOI] [PubMed] [Google Scholar]
- Belkaid Y., Hand T. W. (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. doi: 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego F., Larrucea S., Solana R., Tarazona R. (2016). Editorial: NK cell-based cancer immunotherapy. Front. Immunol. 7. doi: 10.3389/fimmu.2016.00249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calio A., Brunelli M., Gobbo S., Argani P., Munari E., Netto G., et al. (2021). Cathepsin K: A novel diagnostic and predictive biomarker for renal tumors. Cancers (Basel) 13, 1–4. doi: 10.3390/cancers13102441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Yu J. (2015). Effect of helicobacter pylori infection on the composition of gastric microbiota in the development of gastric cancer. Gastrointestinal Tumors 2, 14–25. doi: 10.1159/000380893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Rodríguez N., Goh K.-L., Fock K. M., Mitchell H. M., Kaakoush N. O. (2017). Dysbiosis of the microbiome in gastric carcinogenesis. Sci. Rep. 7, 1–9. doi: 10.1038/s41598-017-16289-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavadas B., Camacho R., Ferreira J. C., Ferreira R. M., Figueiredo C., Brazma A., et al. (2020). Gastric microbiome diversities in gastric cancer patients from Europe and Asia mimic the human population structure and are partly driven by microbiome quantitative trait loci. Microorganisms 8, 1196. doi: 10.3390/microorganisms8081196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput N., Lepage P., Coutzac C., Soularue E., Le Roux K., Monot C., et al. (2017). Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 28, 1368–1379. doi: 10.1093/annonc/mdx108 [DOI] [PubMed] [Google Scholar]
- Chen Y., Chen Y.-X. (2021). Microbiota-associated metabolites and related immunoregulation in colorectal cancer. Cancers 13, 4054. doi: 10.3390/cancers13164054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-C., Liou J.-M., Lee Y.-C., Hong T.-C., El-Omar E. M., Wu M.-S. (2021). The interplay between helicobacter pylori and gastrointestinal microbiota. Gut Microbes 13, 1–22. doi: 10.1080/19490976.2021.1909459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Wu J., Jin D., Wang B., Cao H. (2019). Fecal microbiota transplantation in cancer management: Current status and perspectives. Int. J. Cancer 145, 2021–2031. doi: 10.1002/ijc.32003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I. J., Kim C. G., Lee J. Y., Kim Y.-I., Kook M.-C., Park B., et al. (2020). Family history of gastric cancer and helicobacter pylori treatment. New Engl. J. Med. 382, 427–436. doi: 10.1056/NEJMoa1909666 [DOI] [PubMed] [Google Scholar]
- Cianci R., Franza L., Schinzari G., Rossi E., Ianiro G., Tortora G., et al. (2019). The interplay between immunity and microbiota at intestinal immunological niche: the case of cancer. Int. J. Mol. Sci. 20, 501. doi: 10.3390/ijms20030501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cindoruk M., Erkan G., Karakan T., Dursun A., Unal S. (2007). Efficacy and safety of saccharomyces boulardii in the 14-day triple anti-helicobacter pylori therapy: a prospective randomized placebo-controlled double-blind study. Helicobacter 12, 309–316. doi: 10.1111/j.1523-5378.2007.00516.x [DOI] [PubMed] [Google Scholar]
- Coker O. O., Dai Z., Nie Y., Zhao G., Cao L., Nakatsu G., et al. (2018). Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 67, 1024–1032. doi: 10.1136/gutjnl-2017-314281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M. C., Isolauri E., Salminen S., Sanz Y. (2009). The impact of probiotic on gut health. Curr. Drug Metab. 10, 68–78. doi: 10.2174/138920009787048437 [DOI] [PubMed] [Google Scholar]
- Cremonini F., Di Caro S., Covino M., Armuzzi A., Gabrielli M., Santarelli L., et al. (2002). Effect of different probiotic preparations on anti-helicobacter pylori therapy-related side effects: a parallel group, triple blind, placebo-controlled study. Am. J. Gastroenterol. 97, 2744–2749. doi: 10.1111/j.1572-0241.2002.07063.x [DOI] [PubMed] [Google Scholar]
- Deng Y., Ding X., Song Q., Zhao G., Han L., Ding B., et al. (2021). Alterations in mucosa-associated microbiota in the stomach of patients with gastric cancer. Cell. Oncol., 44 1–14. doi: 10.1007/s13402-021-00596-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Li Z., Tan Y., Guo Z., Liu Y., Wang Y., et al. (2016). A novel strain of bacteroides fragilis enhances phagocytosis and polarises M1 macrophages. Sci. Rep. 6, 1–11. doi: 10.1038/srep29401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo P. N., Deshmukh R. (2019). Oral microbiome: Unveiling the fundamentals. J. Oral. Maxillofac. Pathol. JOMFP 23, 122–128. doi: 10.4103/jomfp.JOMFP_304_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande N. P., Riordan S. M., Castaño-Rodríguez N., Wilkins M. R., Kaakoush N. O. (2018). Signatures within the esophageal microbiome are associated with host genetics, age, and disease. Microbiome 6, 227. doi: 10.1186/s40168-018-0611-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicksved J., Lindberg M., Rosenquist M., Enroth H., Jansson J. K., Engstrand L. (2009). Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J. Med. Microbiol. 58, 509–516. doi: 10.1099/jmm.0.007302-0 [DOI] [PubMed] [Google Scholar]
- Ding S.-Z., Goldberg J. B., Hatakeyama M. (2010). Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol. 6, 851–862. doi: 10.2217/fon.10.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q., Xin Y., Wang L., Meng X., Yu X., Lu L., et al. (2017). Characterization of gastric microbiota in twins. Curr. Microbiol. 74, 224–229. doi: 10.1007/s00284-016-1176-8 [DOI] [PubMed] [Google Scholar]
- Ebrahimpour-Koujan S., Milajerdi A., Larijani B., Esmaillzadeh A. (2020). Effects of probiotics on salivary cytokines and immunoglobulines: a systematic review and meta-analysis on clinical trials. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-67037-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrand L., Lindberg M. (2013). Helicobacter pylori and the gastric microbiota. Best Pract. Res. Clin. Gastroenterol. 27, 39–45. doi: 10.1016/j.bpg.2013.03.016 [DOI] [PubMed] [Google Scholar]
- Eslami M., Yousefi B., Kokhaei P., Jazayeri Moghadas A., Sadighi Moghadam B., Arabkari V., et al. (2019). Are probiotics useful for therapy of helicobacter pylori diseases? Comp. Immunol. Microbiol. Infect. Dis. 64, 99–108. doi: 10.1016/j.cimid.2019.02.010 [DOI] [PubMed] [Google Scholar]
- Espinoza J. L., Matsumoto A., Tanaka H., Matsumura I. (2018). Gastric microbiota: An emerging player in helicobacter pylori-induced gastric malignancies. Cancer Lett. 414, 147–152. doi: 10.1016/j.canlet.2017.11.009 [DOI] [PubMed] [Google Scholar]
- Eun C. S., Kim B. K., Han D. S., Kim S. Y., Kim K. M., Choi B. Y., et al. (2014). Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 19, 407–416. doi: 10.1111/hel.12145 [DOI] [PubMed] [Google Scholar]
- Fehlings M., Drobbe L., Moos V., Renner Viveros P., Hagen J., Beigier-Bompadre M., et al. (2012). Comparative analysis of the interaction of helicobacter pylori with human dendritic cells, macrophages, and monocytes. Infect. Immun. 80, 2724–2734. doi: 10.1128/IAI.00381-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felley C. P., Corthésy-Theulaz I., Rivero J. L., Sipponen P., Kaufmann M., Bauerfeind P., et al. (2001). Favourable effect of an acidified milk (LC-1) on helicobacter pylori gastritis in man. Eur. J. Gastroenterol. Hepatol. 13, 25–29. doi: 10.1097/00042737-200101000-00005 [DOI] [PubMed] [Google Scholar]
- Ferreira R. M., Pereira-Marques J., Pinto-Ribeiro I., Costa J. L., Carneiro F., Machado J. C., et al. (2018). Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 67, 226–236. doi: 10.1136/gutjnl-2017-314205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher L., Fisher A. (2017). Acid-suppressive therapy and risk of infections: pros and cons. Clin. Drug Invest. 37, 587–624. doi: 10.1007/s40261-017-0519-y [DOI] [PubMed] [Google Scholar]
- Frankel A. E., Coughlin L. A., Kim J., Froehlich T. W., Xie Y., Frenkel E. P., et al. (2017). Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia 19, 848–855. doi: 10.1016/j.neo.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg T., Kirschnek S., Häcker H., Häcker G. (2008). Phagocytosis-induced apoptosis of macrophages is linked to uptake, killing and degradation of bacteria. Eur. J. Immunol. 38, 204–215. doi: 10.1002/eji.200737379 [DOI] [PubMed] [Google Scholar]
- Furusawa Y., Obata Y., Fukuda S., Endo T. A., Nakato G., Takahashi D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. doi: 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- Gantuya B., El Serag H. B., Matsumoto T., Ajami N. J., Uchida T., Oyuntsetseg K., et al. (2020). Gastric mucosal microbiota in a Mongolian population with gastric cancer and precursor conditions. Alimentary Pharmacol. Ther. 51, 770–780. doi: 10.1111/apt.15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Xu K., Liu H., Liu G., Bai M., Peng C., et al. (2018. b). Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J.-J., Zhang Y., Gerhard M., Mejias-Luque R., Zhang L., Vieth M., et al. (2018. a). Association between gut microbiota and helicobacter pylori-related gastric lesions in a high-risk population of gastric cancer. Front. Cell. Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Castillo V., Zelaya H., Ilabaca A., Espinoza-Monje M., Komatsu R., Albarracín L., et al. (2018). Lactobacillus fermentum UCO-979C beneficially modulates the innate immune response triggered by helicobacter pylori infection in vitro . Beneficial Microbes 9, 829–841. doi: 10.3920/BM2018.0019 [DOI] [PubMed] [Google Scholar]
- Gibson G. R., Hutkins R., Sanders M. E., Prescott S. L., Reimer R. A., Salminen S. J., et al. (2017). Expert consensus document: The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14, 491–502. doi: 10.1038/nrgastro.2017.75 [DOI] [PubMed] [Google Scholar]
- Gibson G. R., Roberfroid M. B. (1995). Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125, 1401–1412. doi: 10.1093/jn/125.6.1401 [DOI] [PubMed] [Google Scholar]
- Gillespie M. R., Rai V., Agrawal S., Nandipati K. C. (2021). The role of microbiota in the pathogenesis of esophageal adenocarcinoma. Biol. (Basel) 10, 1–23. doi: 10.3390/biology10080697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard A., Spiller R. (1996). The effect of omeprazole on gastric juice viscosity, pH and bacterial counts. Alimentary Pharmacol. Ther. 10, 105–109. doi: 10.1111/j.1365-2036.1996.tb00183.x [DOI] [PubMed] [Google Scholar]
- Goderska K., Agudo Pena S., Alarcon T. (2018). Helicobacter pylori treatment: antibiotics or probiotics. Appl. Microbiol. Biotechnol. 102, 1–7. doi: 10.1007/s00253-017-8535-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Helmink B. A., Spencer C. N., Reuben A., Wargo J. A. (2018). The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 33, 570–580. doi: 10.1016/j.ccell.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjifard S., Goldszmid R. S. (2016). Microbiota-myeloid cell crosstalk beyond the gut. J. Leukocyte Biol. 100, 865–879. doi: 10.1189/jlb.3RI0516-222R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotteland M., Brunser O., Cruchet S. (2006). Systematic review: are probiotics useful in controlling gastric colonization by helicobacter pylori? Aliment Pharmacol. Ther. 23, 1077–1086. doi: 10.1111/j.1365-2036.2006.02868.x [DOI] [PubMed] [Google Scholar]
- Grivennikov S. I., Greten F. R., Karin M. (2010). Immunity, inflammation, and cancer. Cell 140, 883–899. doi: 10.1016/j.cell.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunathilake M. N., Lee J., Choi I. J., Kim Y.-I., Ahn Y., Park C., et al. (2019). Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-50054-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Qian J.-M., Zhao Y.-Q., Li X.-B., Zhang J.-Z. (2013). Effects of IL-1β on the proliferation and apoptosis of gastric epithelial cells and acid secretion from isolated rabbit parietal cells. Mol. Med. Rep. 7, 299–305. doi: 10.3892/mmr.2012.1165 [DOI] [PubMed] [Google Scholar]
- Guo Y., Zhang Y., Gerhard M., Gao J.-J., Mejias-Luque R., Zhang L., et al. (2020). Effect of helicobacter pylori on gastrointestinal microbiota: a population-based study in linqu, a high-risk area of gastric cancer. Gut 69, 1598–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayase E., Jenq R. R. (2021). Role of the intestinal microbiome and microbial-derived metabolites in immune checkpoint blockade immunotherapy of cancer. Genome Med. 13, 1–11. doi: 10.1186/s13073-021-00923-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedblom G. A., Reiland H. A., Sylte M. J., Johnson T. J., Baumler D. J. (2018). Segmented filamentous bacteria - metabolism meets immunity. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimesaat M. M., Fischer A., Plickert R., Wiedemann T., Loddenkemper C., Göbel U. B., et al. (2014). Helicobacter pylori induced gastric immunopathology is associated with distinct microbiota changes in the large intestines of long-term infected Mongolian gerbils. PloS One 9, e100362. doi: 10.1371/journal.pone.0100362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemarajata P., Versalovic J. (2013). Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 6, 39–51. doi: 10.1177/1756283X12459294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooi J. K., Lai W. Y., Ng W. K., Suen M. M., Underwood F. E., Tanyingoh D., et al. (2017). Global prevalence of helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 153, 420–429. doi: 10.1053/j.gastro.2017.04.022 [DOI] [PubMed] [Google Scholar]
- Hsieh P.-S., Tsai Y.-C., Chen Y.-C., Teh S.-F., Ou C.-M., King V. A.-E. (2012). Eradication of helicobacter pylori infection by the probiotic strains lactobacillus johnsonii MH-68 and l. salivarius ssp. salicinius AP-32. Helicobacter 17, 466–477. doi: 10.1111/j.1523-5378.2012.00992.x [DOI] [PubMed] [Google Scholar]
- Hsieh Y.-Y., Tung S.-Y., Pan H.-Y., Yen C.-W., Xu H.-W., Lin Y.-J., et al. (2018). Increased abundance of clostridium and fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-017-18596-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Yang J., Lin Y., Walker C., Cheng J., Liu Z.-G., et al. (2004). Differential regulation of interleukin 1 receptor and toll-like receptor signaling by MEKK3. Nat. Immunol. 5, 98–103. doi: 10.1038/ni1014 [DOI] [PubMed] [Google Scholar]
- Hunt R., Camilleri M., Crowe S., El-Omar E., Fox J., Kuipers E., et al. (2015). The stomach in health and disease. Gut 64, 1650–1668. doi: 10.1136/gutjnl-2014-307595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.-L., Pang W., Huang Y., Zhang Y., Zhang C.-J. (2018). The gastric microbiome is perturbed in advanced gastric adenocarcinoma identified through shotgun metagenomics. Front. Cell. Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain T., Sharma P., Are A. C., Vickers S. M., Dudeja V. (2021). New insights into the cancer–Microbiome–Immune axis: Decrypting a decade of discoveries. Front. Immunol. 12. doi: 10.3389/fimmu.2021.622064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanmard A., Ashtari S., Sabet B., Davoodi S. H., Rostami-Nejad M., Esmaeil Akbari M., et al. (2018). Probiotics and their role in gastrointestinal cancers prevention and treatment; an overview. Gastroenterol. Hepatol. Bed Bench 11, 284–295. [PMC free article] [PubMed] [Google Scholar]
- Ji J., Yang H. (2020). Using probiotics as supplementation for helicobacter pylori antibiotic therapy. Int. J. Mol. Sci. 21, 1136. doi: 10.3390/ijms21031136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo H. J., Kim J., Kim N., Park J. H., Nam R. H., Seok Y. J., et al. (2016). Analysis of gastric microbiota by pyrosequencing: minor role of bacteria other than helicobacter pylori in the gastric carcinogenesis. Helicobacter 21, 364–374. doi: 10.1111/hel.12293 [DOI] [PubMed] [Google Scholar]
- Jung D. H., Youn Y. H., Kim D. H., Lim C. H., Lim H. S., Moon H. S., et al. (2022). Esophageal microbiota and nutritional intakes in patients with achalasia before and after peroral endoscopic myotomy. J. Neurogastroenterol. Motil. 28, 237–246. doi: 10.5056/jnm21057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir A., Aiba Y., Takagi A., Kamiya S., Miwa T., Koga Y. (1997). Prevention of helicobacter pylori infection by lactobacilli in a gnotobiotic murine model. Gut 41, 49–55. doi: 10.1136/gut.41.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadeerhan G., Gerhard M., Gao J.-J., Mejías-Luque R., Zhang L., Vieth M., et al. (2021). Microbiota alteration at different stages in gastric lesion progression: a population-based study in linqu, China. Am. J. Cancer Res. 11, 561. [PMC free article] [PubMed] [Google Scholar]
- Kany S., Vollrath J. T., Relja B. (2019). Cytokines in inflammatory disease. Int. J. Mol. Sci. 20, 6008. doi: 10.3390/ijms20236008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastl A. J., Jr., Terry N. A., WU G. D., ALBENBERG L. G. (2020). The structure and function of the human small intestinal microbiota: Current understanding and future directions. Cell Mol. Gastroenterol. Hepatol. 9, 33–45. doi: 10.1016/j.jcmgh.2019.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T., Kawai T. (2014). Toll-like receptor signaling pathways. Front. Immunol. 5. doi: 10.3389/fimmu.2014.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keikha M., Karbalaei M. (2021). Probiotics as the live microscopic fighters against helicobacter pylori gastric infections. BMC Gastroenterol. 21, 388–388. doi: 10.1186/s12876-021-01977-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi Y., Loke M. F., Goh K. L., Vadivelu J. (2016). Proteomics analysis revealed that crosstalk between helicobacter pylori and streptococcus mitis may enhance bacterial survival and reduces carcinogenesis. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger S., Cox L., Livanos A., Zhang X.-S., Chung J., Perez-Perez G., et al. (2016). Gastric helicobacter pylori infection affects local and distant microbial populations and host responses. Cell Rep. 14, 1395–1407. doi: 10.1016/j.celrep.2016.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. (2016). From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345. doi: 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- Krzysiek-Maczka G., Targosz A., Szczyrk U., Strzałka M., Sliwowski Z., Brzozowski T., et al. (2018). Role of helicobacter pylori infection in cancer-associated fibroblast-induced epithelial-mesenchymal transition in vitro . Helicobacter 23, e12538. doi: 10.1111/hel.12538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzek P., Gosciniak G. (2018). Oral helicobacter pylori: Interactions with host and microbial flora of the oral cavity. Dent. Med. Probl. 55, 75–82. doi: 10.17219/dmp/81259 [DOI] [PubMed] [Google Scholar]
- Kusters J. G., Van Vliet A. H., Kuipers E. J. (2006). Pathogenesis of helicobacter pylori infection. Clin. Microbiol. Rev. 19, 449–490. doi: 10.1128/CMR.00054-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop S. K., Bloom S. M., Rao S. M., Nutsch K., Lio C. W., Santacruz N., et al. (2011). Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254. doi: 10.1038/nature10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar V., Ditu L.-M., Pircalabioru G. G., Gheorghe I., Curutiu C., Holban A. M., et al. (2018). Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front. Immunol. 9. doi: 10.3389/fimmu.2018.01830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. K., Mazmanian S. K. (2010). Has the microbiota played a critical role in the evolution of the adaptive immune system? science 330, 1768–1773. doi: 10.1126/science.1195568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertpiriyapong K., Whary M. T., Muthupalani S., Lofgren J. L., Gamazon E. R., Feng Y., et al. (2014). Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 63, 54–63. doi: 10.1136/gutjnl-2013-305178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy E. M., Roberti M. P., Mordoh J. (2011). Natural killer cells in human cancer: from biological functions to clinical applications. J. BioMed. Biotechnol. 2011, 676198. doi: 10.1155/2011/676198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. P., Liu J. X., Lu L. L., Wang L. L., Xu L., Guo Z. H., et al. (2021). Overgrowth of lactobacillus in gastric cancer. World J. Gastrointest. Oncol. 13, 1099–1108. doi: 10.4251/wjgo.v13.i9.1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Perez Perez G. I. (2018). Is there a role for the non-helicobacter pylori bacteria in the risk of developing gastric cancer? Int. J. Mol. Sci. 19, 1353. doi: 10.3390/ijms19051353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. H., Qin Y., Sham P. C., Lau K., Chu K.-M., Leung W. K. (2017). Alterations in gastric microbiota after h. pylori eradication and in different histological stages of gastric carcinogenesis. Sci. Rep. 7, 1–8. doi: 10.1038/srep44935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen Y., Zhang S., Dong L. (2021). Gut microbiota-mediated immunomodulation in tumor. J. Exp. Clin. Cancer Res. 40, 1–20. doi: 10.1186/s13046-021-01983-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-X., Wong G. L.-H., To K.-F., Wong V. W.-S., Lai L. H., Chow D. K.-L., et al. (2009). Bacterial microbiota profiling in gastritis without helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PloS One 4, e7985. doi: 10.1371/journal.pone.0007985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zhou R., Wang H., Li W., Pan M., Yao X., et al. (2019). Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ 26, 2447–2463. doi: 10.1038/s41418-019-0312-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca L., Pérez-Pérez G., Urruzuno P., Martinez M. J., Iizumi T., Gao Z., et al. (2017). Characterization of the gastric microbiota in a pediatric population according to helicobacter pylori status. Pediatr. Infect. Dis. J. 36, 173–178. [DOI] [PubMed] [Google Scholar]
- Lordan C., Thapa D., Ross R. P., Cotter P. D. (2020). Potential for enriching next-generation health-promoting gut bacteria through prebiotics and other dietary components. Gut Microbes 11, 1–20. doi: 10.1080/19490976.2019.1613124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Dong S., Wu X., Jin R., Chen H. (2021). Probiotics in cancer. Front. Oncol. 11. doi: 10.3389/fonc.2021.638148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Sang J., He H., Wan X., Lin Y., Li L., et al. (2016. a). Probiotic supplementation does not improve eradication rate of helicobacter pylori infection compared to placebo based on standard therapy: a meta-analysis. Sci. Rep. 6, 23522. doi: 10.1038/srep23522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luster A. D., Alon R., Von Andrian U. H. (2005). Immune cell migration in inflammation: present and future therapeutic targets. Nat. Immunol. 6, 1182–1190. doi: 10.1038/ni1275 [DOI] [PubMed] [Google Scholar]
- Lu M., Xuan S., Wang Z. (2019). Oral microbiota: A new view of body health. Food Sci. Hum. Wellness 8, 8–15. doi: 10.1016/j.fshw.2018.12.001 [DOI] [Google Scholar]
- Lu M., Yu S., Deng J., Yan Q., Yang C., Xia G., et al. (2016. b). Efficacy of probiotic supplementation therapy for helicobacter pylori eradication: A meta-analysis of randomized controlled trials. PloS One 11, e0163743. doi: 10.1371/journal.pone.0163743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magne F., Gotteland M., Gauthier L., Zazueta A., Pesoa S., Navarrete P., et al. (2020). The firmicutes/bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients 12, 1474. doi: 10.3390/nu12051474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Contreras A., Goldfarb K. C., Godoy-Vitorino F., Karaoz U., Contreras M., Blaser M. J., et al. (2011). Structure of the human gastric bacterial community in relation to helicobacter pylori status. ISME J. 5, 574–579. doi: 10.1038/ismej.2010.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Mao Q., Xia W., Dong G., Yu C., Jiang F. (2019). Gut microbiota shapes the efficiency of cancer therapy. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi M., Bizzarri B., Sacchero R. I., Maccari S., Calabrese L., Fabbian F., et al. (2012). Helicobacter pylori infection in clinical practice: probiotics and a combination of probiotics + lactoferrin improve compliance, but not eradication, in sequential therapy. Helicobacter 17, 254–263. doi: 10.1111/j.1523-5378.2012.00944.x [DOI] [PubMed] [Google Scholar]
- Marteau P., Seksik P. (2004). Tolerance of probiotics and prebiotics. J. Clin. Gastroenterol. 38, S67–S69. doi: 10.1097/01.mcg.0000128929.37156.a7 [DOI] [PubMed] [Google Scholar]
- Martinsen T. C., Fossmark R., Waldum H. L. (2019). The phylogeny and biological function of gastric juice–microbiological consequences of removing gastric acid. Int. J. Mol. Sci. 20, 6031. doi: 10.3390/ijms20236031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M. L., et al. (2018). The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108. doi: 10.1126/science.aao3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftahussurur M., Waskito L. A., El-Serag H. B., Ajami N. J., Nusi I. A., Syam A. F., et al. (2020). Gastric microbiota and helicobacter pylori in Indonesian population. Helicobacter 25, e12695. doi: 10.1111/hel.12695 [DOI] [PubMed] [Google Scholar]
- Mohammadi S. O., Yadegar A., Kargar M., Mirjalali H., Kafilzadeh F. (2020). The impact of helicobacter pylori infection on gut microbiota-endocrine system axis; modulation of metabolic hormone levels and energy homeostasis. J. Diabetes Metab. Disord. 19, 1–7. doi: 10.1007/s40200-020-00608-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan T. M., Duggal N. A., Rosati E., Griffin R., Hughes J., Roach B., et al. (2021). A multi-factorial observational study on sequential fecal microbiota transplant in patients with medically refractory clostridioides difficile infection. Cells. 10, 1–24. doi: 10.3390/cells10113234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Ramos S., Lopez-Contreras B. E., Villarruel-Vazquez R., Ocampo-Medina E., Macias-Kauffer L., Martinez-Medina J. N., et al. (2020). Environmental and intrinsic factors shaping gut microbiota composition and diversity and its relation to metabolic health in children and early adolescents: A population-based study. Gut Microbes 11, 900–917. doi: 10.1080/19490976.2020.1712985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat C., Williams C., Gillen D., Hossack M., Gilmour D., Carswell A., et al. (2000). Omeprazole, helicobacter pylori status, and alterations in the intragastric milieu facilitating bacterial n-nitrosation. Gastroenterology 119, 339–347. doi: 10.1053/gast.2000.9367 [DOI] [PubMed] [Google Scholar]
- Murosaki S., Muroyama K., Yamamoto Y., Yoshikai Y. (2000). Antitumor effect of heat-killed lactobacillus plantarum l-137 through restoration of impaired interleukin-12 production in tumor-bearing mice. Cancer Immunol. Immunother. 49, 157–164. doi: 10.1007/s002620050615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscroft T., Keighley M. (1981). Cimetidine, gastric pH, and nitrosation. Lancet 317, 1002. doi: 10.1016/S0140-6736(81)91768-2 [DOI] [PubMed] [Google Scholar]
- Myllyluoma E., Ahlroos T., Veijola L., Rautelin H., Tynkkynen S., Korpela R. (2007). Effects of anti-helicobacter pylori treatment and probiotic supplementation on intestinal microbiota. Int. J. Antimicrob. Agents 29, 66–72. doi: 10.1016/j.ijantimicag.2006.08.034 [DOI] [PubMed] [Google Scholar]
- Myllyluoma E., Veijola L., Ahlroos T., Tynkkynen S., Kankuri E., Vapaatalo H., et al. (2005). Probiotic supplementation improves tolerance to helicobacter pylori eradication therapy–a placebo-controlled, double-blind randomized pilot study. Aliment Pharmacol. Ther. 21, 1263–1272. doi: 10.1111/j.1365-2036.2005.02448.x [DOI] [PubMed] [Google Scholar]
- Nabavi-Rad A., Azizi M., Jamshidizadeh S., Sadeghi A., Aghdaei H. A., Yadegar A., et al. (2022). The effects of vitamins and micronutrients on helicobacter pylori pathogenicity, survival, and eradication: A crosstalk between micronutrients and immune system. J. Immunol. Res. 2022, 4713684. doi: 10.1155/2022/4713684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naimi A., Mohammed R. N., Raji A., Chupradit S., Yumashev A. V., Suksatan W., et al. (2022). Tumor immunotherapies by immune checkpoint inhibitors (ICIs); the pros and cons. Cell Commun. Signal 20, 44. doi: 10.1186/s12964-022-00854-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Yoshikawa T. (2002). Molecular and cellular mechanisms involved in helicobacter pylori-induced inflammation and oxidative stress. Free Radical Biol. Med. 33, 323–336. doi: 10.1016/s0891-5849(02)00868-7 [DOI] [PubMed] [Google Scholar]
- Nasr R., Shamseddine A., Mukherji D., Nassar F., Temraz S. (2020). The crosstalk between microbiome and immune response in gastric cancer. Int. J. Mol. Sci. 21, 6586. doi: 10.3390/ijms21186586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navabi N., Johansson M. E., Raghavan S., Lindén S. K. (2013). Helicobacter pylori infection impairs the mucin production rate and turnover in the murine gastric mucosa. Infect. Immun. 81, 829–837. doi: 10.1128/IAI.01000-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Rodriguez T., Silva F. M., Barbuti R. C., Mattar R., Moraes-Filho J. P., De Oliveira M. N., et al. (2013). Association of a probiotic to a helicobacter pylori eradication regimen does not increase efficacy or decreases the adverse effects of the treatment: a prospective, randomized, double-blind, placebo-controlled study. BMC Gastroenterol. 13, 56. doi: 10.1186/1471-230x-13-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndegwa N., Ploner A., Andersson A. F., Zagai U., Andreasson A., Vieth M., et al. (2020). Gastric microbiota in a low–helicobacter pylori prevalence general population and their associations with gastric lesions. Clin. Trans. Gastroenterol. 11. doi: 10.14309/ctg.0000000000000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nista E. C., Candelli M., Cremonini F., Cazzato I. A., Zocco M. A., Franceschi F., et al. (2004). Bacillus clausii therapy to reduce side-effects of anti-helicobacter pylori treatment: randomized, double-blind, placebo controlled trial. Aliment Pharmacol. Ther. 20, 1181–1188. doi: 10.1111/j.1365-2036.2004.02274.x [DOI] [PubMed] [Google Scholar]
- Noto J. M., Peek R. M., Jr (2017). The gastric microbiome, its interaction with helicobacter pylori, and its potential role in the progression to stomach cancer. PloS Pathog. 13, e1006573. doi: 10.1371/journal.ppat.1006573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohland C. L., Jobin C. (2015). Microbial activities and intestinal homeostasis: A delicate balance between health and disease. Cell. Mol. Gastroenterol. Hepatol. 1, 28–40. doi: 10.1016/j.jcmgh.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H., Satoh-Takayama N. (2020). Stomach microbiota, helicobacter pylori, and group 2 innate lymphoid cells. Exp. Mol. Med. 52, 1377–1382. doi: 10.1038/s12276-020-00485-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orberg E. T., Fan H., Tam A. J., Dejea C. M., Shields C. D., Wu S., et al. (2017). The myeloid immune signature of enterotoxigenic bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 10, 421–433. doi: 10.1038/mi.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbey G., Sproston E., Hanafiah A. (2020). Helicobacter pylori infection and gastric microbiota. Euroasian J. Hepato-Gastroenterol. 10, 36. doi: 10.5005/jp-journals-10018-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliari D., Gambassi G., Piccirillo C. A., Cianci R. (2017). The intricate link among gut “immunological niche,” microbiota, and xenobiotics in intestinal pathology. Mediators Inflammation 2017. doi: 10.1155/2017/8390595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada Venegas D., de la Fuente M. K., Landskron G., Gonzalez M. J., Quera R., Dijkstra G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. H., Lee S. K. (2020). Exploring esophageal microbiomes in esophageal diseases: A systematic review. J. Neurogastroenterol. Motil. 26, 171–179. doi: 10.5056/jnm19240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. H., Lee A. R., Lee Y. R., Eun C. S., Lee S. K., Han D. S. (2019). Evaluation of gastric microbiome and metagenomic function in patients with intestinal metaplasia using 16S rRNA gene sequencing. Helicobacter 24, e12547. doi: 10.1111/hel.12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons B. N., Ijaz U. Z., D’Amore R., Burkitt M. D., Eccles R., Lenzi L., et al. (2017). Comparison of the human gastric microbiota in hypochlorhydric states arising as a result of helicobacter pylori-induced atrophic gastritis, autoimmune atrophic gastritis and proton pump inhibitor use. PloS Pathog. 13, e1006653. doi: 10.1371/journal.ppat.1006653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale A., Marchesi N., Govoni S., Coppola A., Gazzaruso C. (2019). The role of gut microbiota in obesity, diabetes mellitus, and effect of metformin: new insights into old diseases. Curr. Opin. Pharmacol. 49, 1–5. doi: 10.1016/j.coph.2019.03.011 [DOI] [PubMed] [Google Scholar]
- Penumetcha S. S., Ahluwalia S., Irfan R., Khan S. A., Rohit Reddy S., Vasquez Lopez M. E., et al. (2021). The efficacy of probiotics in the management of helicobacter pylori: A systematic review. Cureus 13, e20483. doi: 10.7759/cureus.20483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira V., Abraham P., Nallapeta S., Shetty A. (2018). Gastric bacterial flora in patients harbouring helicobacter pylori with or without chronic dyspepsia: analysis with matrix-assisted laser desorption ionization time-of-flight mass spectroscopy. BMC Gastroenterol. 18, 1–8. doi: 10.1186/s12876-018-0744-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pero R., Brancaccio M., Laneri S., De Biasi M.-G., Lombardo B., Scudiero O. (2019). A novel view of human helicobacter pylori infections: Interplay between microbiota and beta-defensins. Biomolecules 9, 237. doi: 10.3390/biom9060237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard J. M., Zeng M. Y., Caruso R., Núñez G. (2017). Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 279, 70–89. doi: 10.1111/imr.12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscione M., Mazzone M., Di Marcantonio M. C., Muraro R., Mincione G. (2021). Eradication of helicobacter pylori and gastric cancer: a controversial relationship. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.630852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyzos S. A., Zeglinas C., Artemaki F., Doulberis M., Kazakos E., Katsinelos P., et al. (2018). Helicobacter pylori infection and esophageal adenocarcinoma: a review and a personal view. Ann. Gastroenterol. 31, 8–13. doi: 10.20524/aog.2017.0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi N., Li P., GU Q. (2019). Probiotic therapy in helicobacter pylori infection: a potential strategy against a serious pathogen? Appl. Microbiol. Biotechnol. 103, 1573–1588. doi: 10.1007/s00253-018-09580-3 [DOI] [PubMed] [Google Scholar]
- Raheem A., Liang L., Zhang G., Cui S. (2021). Modulatory effects of probiotics during pathogenic infections with emphasis on immune regulation. Front. Immunol. 12. doi: 10.3389/fimmu.2021.616713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M., Figueiredo C., Smet A., Hansen R., Kupcinskas J., Rokkas T., et al. (2020). Systematic review: gastric microbiota in health and disease. Alimentary Pharmacol. Ther. 51, 582–602. doi: 10.1111/apt.15650 [DOI] [PubMed] [Google Scholar]
- Rawla P., Barsouk A. (2019). Epidemiology of gastric cancer: global trends, risk factors and prevention. Przeglad Gastroenterologiczny 14, 26. doi: 10.5114/pg.2018.80001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G., Sanders M., Gaskins H. R., Gibson G. R., Mercenier A., Rastall R., et al. (2003). New scientific paradigms for probiotics and prebiotics. J. Clin. Gastroenterol. 37, 105–118. doi: 10.1097/00004836-200308000-00004 [DOI] [PubMed] [Google Scholar]
- Rezasoltani S., Yadegar A., Asadzadeh Aghdaei H., Reza Zali M. (2021). Modulatory effects of gut microbiome in cancer immunotherapy: A novel paradigm for blockade of immune checkpoint inhibitors. Cancer Med. 10, 1141–1154. doi: 10.1002/cam4.3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinninella E., Raoul P., Cintoni M., Franceschi F., Miggiano G. A. D., Gasbarrini A., et al. (2019). What is the healthy gut microbiota composition? a changing ecosystem across age, environment, diet, and diseases. Microorganisms 7, 14. doi: 10.3390/microorganisms7010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K., Argent R. H., Atherton J. C. (2007). The inflammatory and immune response to helicobacter pylori infection. Best Pract. Res. Clin. Gastroenterol. 21, 237–259. doi: 10.1016/j.bpg.2007.01.001 [DOI] [PubMed] [Google Scholar]
- Rotte A. (2019). Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J. Exp. Clin. Cancer Res. 38, 1–12. doi: 10.1186/s13046-019-1259-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B., Le Chatelier E., Derosa L., Duong C. P. M., Alou M. T., Daillere R., et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. doi: 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- Rowland I., Gibson G., Heinken A., Scott K., Swann J., Thiele I., et al. (2018). Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 57, 1–24. doi: 10.1007/s00394-017-1445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L.-X., Chang B., Zhang W.-L., Wu X.-M., Li X.-H., Jiang M. (2010). Remission induction and maintenance effect of probiotics on ulcerative colitis: a meta-analysis. World J. Gastroenterol.: WJG 16, 1908. doi: 10.3748/wjg.v16.i15.1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankarapandian V., Venmathi Maran B. A., Rajendran R. L., Jogalekar M. P., Gurunagarajan S., Krishnamoorthy R., et al. (2022). An update on the effectiveness of probiotics in the prevention and treatment of cancer. Life (Basel) 12, 1-19. doi: 10.3390/life12010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhadi V., Mathew B., Kokkola A., Karla T., Tikkanen M., Rautelin H., et al. (2021). Gut microbiota of patients with different subtypes of gastric cancer and gastrointestinal stromal tumors. Gut Pathog. 13, 1–9. doi: 10.1186/s13099-021-00403-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage D. C. (1977). Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31, 107–133. doi: 10.1146/annurev.mi.31.100177.000543 [DOI] [PubMed] [Google Scholar]
- Schmidt T. S., Raes J., Bork P. (2018). The human gut microbiome: from association to modulation. Cell 172, 1198–1215. doi: 10.1016/j.cell.2018.02.044 [DOI] [PubMed] [Google Scholar]
- Seidel J. A., Otsuka A., Kabashima K. (2018). Anti-PD-1 and anti-CTLA-4 therapies in cancer: mechanisms of action, efficacy, and limitations. Front. Oncol. 8. doi: 10.3389/fonc.2018.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body. PloS Biol. 14, e1002533. doi: 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serban D. E. (2014). Gastrointestinal cancers: influence of gut microbiota, probiotics and prebiotics. Cancer Lett. 345, 258–270. doi: 10.1016/j.canlet.2013.08.013 [DOI] [PubMed] [Google Scholar]
- Ser H.-L., Letchumanan V., Goh B.-H., Wong S. H., Lee L.-H. (2021). The use of fecal microbiome transplant in treating human diseases: Too early for poop? Front. Microbiol. 12. doi: 10.3389/fmicb.2021.519836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano C., Harris P. R., Smith P. D., Bimczok D. (2021). Interactions between h. pylori and the gastric microbiome: Impact on gastric homeostasis and disease. Curr. Opin. Physiol. 21, 57–64. doi: 10.1016/j.cophys.2021.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafaghi A., Pourkazemi A., Khosravani M., Asl S. F., Maafi A. A., Roshan Z. A., et al. (2016). The effect of probiotic plus prebiotic supplementation on the tolerance and efficacy of helicobacter pylori eradication quadruple therapy: a randomized prospective double blind controlled trial. Middle East J. Digestive Dis. 8, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma T., Gupta A., Chauhan R., Bhat A. A., Nisar S., Hashem S., et al. (2022). Cross-talk between the microbiome and chronic inflammation in esophageal cancer: potential driver of oncogenesis. Cancer Metastasis Rev. doi: 10.1007/s10555-022-10026-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavakhi A., Tabesh E., Yaghoutkar A., Hashemi H., Tabesh F., Khodadoostan M., et al. (2013). The effects of multistrain probiotic compound on bismuth-containing quadruple therapy for helicobacter pylori infection: a randomized placebo-controlled triple-blind study. Helicobacter 18, 280–284. doi: 10.1111/hel.12047 [DOI] [PubMed] [Google Scholar]
- Sheh A., Fox J. G. (2013). The role of the gastrointestinal microbiome in helicobacter pylori pathogenesis. Gut Microbes 4, 505–531. doi: 10.4161/gmic.26205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui L., Yang X., Li J., Yi C., Sun Q., Zhu H. (2020). Gut microbiome as a potential factor for modulating resistance to cancer immunotherapy. Front. Immunol. 10. doi: 10.3389/fimmu.2019.02989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostedt S., Heimdahl A., Kager L., Nord C. E. (1985). Microbial colonization of the oropharynx, esophagus and stomach in patients with gastric diseases. Eur. J. Clin. Microbiol. 4, 49–51. doi: 10.1007/BF02148660 [DOI] [PubMed] [Google Scholar]
- Slavin J. (2013). Fiber and prebiotics: mechanisms and health benefits. Nutrients 5, 1417–1435. doi: 10.3390/nu5041417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Zhou L., Liu D., Ge L., Li Y. (2019). Probiotic effect on helicobacter pylori attachment and inhibition of inflammation in human gastric epithelial cells. Exp. Ther. Med. 18, 1551–1562. doi: 10.3892/etm.2019.7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto Chervin C., Gajewski T. F. (2020). Microbiome-based interventions: therapeutic strategies in cancer immunotherapy. Immunooncol. Technol. 8, 12–20. doi: 10.1016/j.iotech.2020.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelhauer M. R., Kupcinskas J., Johannesen T. B., Urba M., Skieceviciene J., Jonaitis L., et al. (2020). Transient and persistent gastric microbiome: adherence of bacteria in gastric cancer and dyspeptic patient biopsies after washing. J. Clin. Med. 9, 1882. doi: 10.3390/jcm9061882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiewicz M., Karpinski T. M. (2021). The oral microbiota and its role in carcinogenesis. Semin. Cancer Biol. doi: 10.1016/j.semcancer.2021.11.002 [DOI] [PubMed] [Google Scholar]
- Stockbruegger R. (1985). Bacterial overgrowth as a consequence of reduced gastric acidity. Scandinavian J. Gastroenterol. 20, 7–15. doi: 10.3109/00365528509093749 [DOI] [PubMed] [Google Scholar]
- Stockbrugger R., Cotton P., Eugenides N., Bartholomew B., Hill M., Walters C. (1982). Intragastric nitrites, nitrosamines, and bacterial overgrowth during cimetidine treatment. Gut 23, 1048–1054. doi: 10.1136/gut.23.12.1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suez J., Zmora N., Segal E., Elinav E. (2019). The pros, cons, and many unknowns of probiotics. Nat. Med. 25, 716–729. doi: 10.1038/s41591-019-0439-x [DOI] [PubMed] [Google Scholar]
- Sun J. H., Li X. L., Yin J., Li Y. H., Hou B. X., Zhang Z. (2018). A screening method for gastric cancer by oral microbiome detection. Oncol. Rep. 39, 2217–2224. doi: 10.3892/or.2018.6286 [DOI] [PubMed] [Google Scholar]
- Sun J., Zhou M., Salazar C. R., Hays R., Bedi S., Chen Y., et al. (2017). Chronic periodontal disease, periodontal pathogen colonization, and increased risk of precancerous gastric lesions. J. Periodontol 88, 1124–1134. doi: 10.1902/jop.2017.160829 [DOI] [PubMed] [Google Scholar]
- Tang Y., Chen Y., Jiang H., Nie D. (2011). Short-chain fatty acids induced autophagy serves as an adaptive strategy for retarding mitochondria-mediated apoptotic cell death. Cell Death Differentiation 18, 602–618. doi: 10.1038/cdd.2010.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teame T., Wang A., Xie M., Zhang Z., Yang Y., Ding Q., et al. (2020). Paraprobiotics and postbiotics of probiotic lactobacilli, their positive effects on the host and action mechanisms: A review. Front. Nutr. 7. doi: 10.3389/fnut.2020.570344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A., Mikkelsen H., Jungersen G. (2019). Intracellular pathogens: host immunity and microbial persistence strategies. J. Immunol. Res. 2019 doi: 10.1155/2019/1356540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threapleton D. E., Greenwood D. C., Evans C. E., Cleghorn C. L., Nykjaer C., Woodhead C., et al. (2013). Dietary fibre intake and risk of cardiovascular disease: systematic review and meta-analysis. BMJ 347, f6879. doi: 10.1136/bmj.f6879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh J. W., Wilson R. B. (2020). Pathways of gastric carcinogenesis, helicobacter pylori virulence and interactions with antioxidant systems, vitamin c and phytochemicals. Int. J. Mol. Sci. 21, 6451. doi: 10.3390/ijms21176451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J. L., Ran Z. H., Shen J., Zhang C. X., Xiao S. D. (2007). Meta-analysis: the effect of supplementation with probiotics on eradication rates and adverse events during helicobacter pylori eradication therapy. Aliment Pharmacol. Ther. 25, 155–168. doi: 10.1111/j.1365-2036.2006.03179.x [DOI] [PubMed] [Google Scholar]
- Troy E. B., Kasper D. L. (2010). Beneficial effects of bacteroides fragilis polysaccharides on the immune system. Front. Biosci: J. Virtual Library 15, 25. doi: 10.2741/3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursi A., Brandimarte G., Giorgetti G. M., Modeo M. E. (2004). Effect of lactobacillus casei supplementation on the effectiveness and tolerability of a new second-line 10-day quadruple therapy after failure of a first attempt to cure helicobacter pylori infection. Med. Sci. Monit 10, CR662–CR666. [PubMed] [Google Scholar]
- Twomey J. D., Zhang B. (2021). Cancer immunotherapy update: FDA-approved checkpoint inhibitors and companion diagnostics. AAPS J. 23, 39. doi: 10.1208/s12248-021-00574-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymczyszyn E. E., Santos M. I., Costa M. D. C., Illanes A., Gomez-Zavaglia A. (2014). History, synthesis, properties, applications and regulatory issues of prebiotic oligosaccharides. Carbohydrates Appl. Med. 37/662 (2) [Google Scholar]
- Urrutia-Baca V. H., Escamilla-García E., de la Garza-Ramos M. A., Tamez-Guerra P., Gomez-Flores R., Urbina-Ríos C. S. (2018). In vitro antimicrobial activity and downregulation of virulence gene expression on helicobacter pylori by reuterin. Probiotics Antimicrobial Proteins 10, 168–175. doi: 10.1007/s12602-017-9342-2 [DOI] [PubMed] [Google Scholar]
- Vaddepally R. K., Kharel P., Pandey R., Garje R., Chandra A. B. (2020). Review of indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence. Cancers 12, 738. doi: 10.3390/cancers12030738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasapolli R., Schütte K., Schulz C., Vital M., Schomburg D., Pieper D. H., et al. (2019). Analysis of transcriptionally active bacteria throughout the gastrointestinal tract of healthy individuals. Gastroenterology 157, 1081–1092.e3. doi: 10.1053/j.gastro.2019.05.068 [DOI] [PubMed] [Google Scholar]
- Vemuri R., Gundamaraju R., Shastri M. D., Shukla S. D., Kalpurath K., Ball M., et al. (2018). Gut microbial changes, interactions, and their implications on human lifecycle: an ageing perspective. BioMed. Res. Int. 2018. doi: 10.1155/2018/4178607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Lee C., Jeun E.-J., Yi J., Kim K. S., Ghosh A., et al. (2018). Cell surface polysaccharides of bifidobacterium bifidum induce the generation of Foxp3+ regulatory T cells. Sci. Immunol. 3, 602–9. doi: 10.1126/sciimmunol.aat6975 [DOI] [PubMed] [Google Scholar]
- Vetizou M., Pitt J. M., Daillere R., Lepage P., Waldschmitt N., Flament C., et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. doi: 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivarelli S., Salemi R., Candido S., Falzone L., Santagati M., Stefani S., et al. (2019). Gut microbiota and cancer: from pathogenesis to therapy. Cancers 11, 38. doi: 10.3390/cancers11010038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. D., Fritz J. M., Lenardo M. J. (2020). A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 20, 651–668. doi: 10.1038/s41577-020-0306-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. H., Gao Q. Y., Fang J. Y. (2013). Meta-analysis of the efficacy and safety of lactobacillus-containing and bifidobacterium-containing probiotic compound preparation in helicobacter pylori eradication therapy. J. Clin. Gastroenterol. 47, 25–32. doi: 10.1097/MCG.0b013e318266f6cf [DOI] [PubMed] [Google Scholar]
- Wang J. W., Kuo C. H., Kuo F. C., Wang Y. K., Hsu W. H., Yu F. J., et al. (2019). Fecal microbiota transplantation: Review and update. J. Formos Med. Assoc. 118 Suppl 1, S23–S31. doi: 10.1016/j.jfma.2018.08.011 [DOI] [PubMed] [Google Scholar]
- Wang F., Lau J. K. C., Yu J. (2021). The role of natural killer cell in gastrointestinal cancer: killer or helper. Oncogene 40, 717–730. doi: 10.1038/s41388-020-01561-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. K., Yang Y. S. (2013). Upper gastrointestinal microbiota and digestive diseases. World J. Gastroenterol. 19, 1541–1550. doi: 10.3748/wjg.v19.i10.1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhou J., Xin Y., Geng C., Tian Z., Yu X., et al. (2016). Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur. J. Gastroenterol. Hepatol. 28, 261. doi: 10.1097/MEG.0000000000000542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren J. R., Marshall B. (1983). Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 321, 1273–1275. doi: 10.1016/S0140-6736(83)92719-8 [DOI] [PubMed] [Google Scholar]
- Weiskopf D., Weinberger B., Grubeck-Loebenstein B. (2009). The aging of the immune system. Transpl. Int. 22, 1041–1050. doi: 10.1111/j.1432-2277.2009.00927.x [DOI] [PubMed] [Google Scholar]
- White J. R., Winter J. A., Robinson K. (2015). Differential inflammatory response to helicobacter pylori infection: etiology and clinical outcomes. J. Inflammation Res. 8, 137. doi: 10.2147/JIR.S64888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertsema S. P., Van Bergenhenegouwen J., Garssen J., Knippels L. M. J. (2021). The interplay between the gut microbiome and the immune system in the context of infectious diseases throughout life and the role of nutrition in optimizing treatment strategies. Nutrients 13, 886. doi: 10.3390/nu13030886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz O.-O. (2016). MyD88 mutations in b cell tumors: constitutive signaling by TIR domain oligomerization (Universitätsbibliothek Tübingen; ). [Google Scholar]
- Wong S. H., Zhao L., Zhang X., Nakatsu G., Han J., Xu W., et al. (2017). Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology 153, 1621–1633 e6. doi: 10.1053/j.gastro.2017.08.022 [DOI] [PubMed] [Google Scholar]
- Wrzosek L., Miquel S., Noordine M.-L., Bouet S., Chevalier-Curt M. J., Robert V., et al. (2013). Bacteroides thetaiotaomicron and faecalibacterium prausnitziiinfluence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 11, 61. doi: 10.1186/1741-7007-11-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Xu S., Xiang C., Cao Q., Li Q., Huang J., et al. (2018). Tongue coating microbiota community and risk effect on gastric cancer. J. Cancer 9, 4039–4048. doi: 10.7150/jca.25280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zeng T., Deligios M., Milanesi L., Langille M. G., Zinellu A., et al. (2020). Age-related variation of bacterial and fungal communities in different body habitats across the young, elderly, and centenarians in Sardinia. Msphere 5, e00558–e00519. doi: 10.1128/mSphere.00558-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamade M., Sugimoto M., Uotani T., Nishino M., Kodaira C., Furuta T. (2011). Resistance of helicobacter pylori to quinolones and clarithromycin assessed by genetic testing in Japan. J. Gastroenterol. Hepatol. 26, 1457–1461. doi: 10.1111/j.1440-1746.2011.06815.x [DOI] [PubMed] [Google Scholar]
- Yang Y.-J., Chuang C.-C., Yang H.-B., Lu C.-C., Sheu B.-S. (2012). Lactobacillus acidophilus ameliorates h. pylori-induced gastric inflammation by inactivating the Smad7 and NFκB pathways. BMC Microbiol. 12, 1–8. doi: 10.1186/1471-2180-12-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I., Nell S., Suerbaum S. (2013). Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol. Rev. 37, 736–761. doi: 10.1111/1574-6976.12027 [DOI] [PubMed] [Google Scholar]
- Yang L., Zhang J., Xu J., Wei X., Yang J., Liu Y., et al. (2019). Helicobacter pylori infection aggravates dysbiosis of gut microbiome in children with gastritis. Front. Cell. Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z. N., Xia H. H., Zhang R., Li L., Wu L. H., Liu X. J., et al. (2020). The efficacy of washed microbiota transplantation on helicobacter pylori eradication: A pilot study. Gastroenterol. Res. Pract. 2020, 8825189. doi: 10.1155/2020/8825189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Torres J., Hu N., Medrano-Guzman R., Herrera-Goepfert R., Humphrys M. S., et al. (2017). Molecular characterization of the human stomach microbiota in gastric cancer patients. Front. Cell. Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Ma C., Zhang Z., Zhang H., Hu H. (2021). NF-kappaB signaling in inflammation and cancer. MedComm 2, 618–653. doi: 10.1002/mco2.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Pan Z. (2020). Influence of microbiota on immunity and immunotherapy for gastric and esophageal cancers. Gastroenterol. Rep. 8, 206–214. doi: 10.1093/gastro/goaa014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Shi D., Li M., Li Y., Wang X., Li W. (2019). The relationship between gastric microbiota and gastric disease. Scandinavian J. Gastroenterol. 54, 391–396. doi: 10.1080/00365521.2019.1591499 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Elson C. O. (2018). Adaptive immune education by gut microbiota antigens. Immunology 154, 28–37. doi: 10.1111/imm.12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Wu L., Yan G., Chen Y., Zhou M., Wu Y., et al. (2021). Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduction Targeted Ther. 6, 1–46. doi: 10.1038/s41392-021-00658-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Liwinski T., Elinav E. (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. doi: 10.1038/s41422-020-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Ma F.-Z., Deng X.-J., Yuan H., Ma H.-S. (2008). Lactobacilli inhibit interleukin-8 production induced by helicobacter pylori lipopolysaccharide-activated toll-like receptor 4. World J. Gastroenterol.: WJG 14, 5090. doi: 10.3748/wjg.14.5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebarth D., Spiegelhalder B., Bartsch H. (1997). N-nitrosation of medicinal drugs catalysed by bacteria from human saliva and gastro-intestinal tract, including helicobacter pylori. Carcinogenesis 18, 383–389. doi: 10.1093/carcin/18.2.383 [DOI] [PubMed] [Google Scholar]
- Ziemer C. J., Gibson G. R. (1998). An overview of probiotics, prebiotics and synbiotics in the functional food concept: perspectives and future strategies. Int. Dairy J. 8, 473–479. doi: 10.1016/S0958-6946(98)00071-5 [DOI] [Google Scholar]
- Zou J., Dong J., Yu X. (2009). Meta-analysis: Lactobacillus containing quadruple therapy versus standard triple first-line therapy for helicobacter pylori eradication. Helicobacter 14, 97–107. doi: 10.1111/j.1523-5378.2009.00716.x [DOI] [PubMed] [Google Scholar]