Significance
Despite recent therapeutic progress in cancer treatment, the metastatic establishment of cancers at distant organs remains the major cause of mortality in patients with solid tumors. The past decade has brought several advances in the understanding of metabolic phenotypes of tumors that are different from their adjacent nonmalignant tissues. Just recently, attention has been drawn to the fact that metastasizing tumor cells can display dynamic metabolic changes to survive in their changing microenvironment during the metastatic cascade. Here, we perform a comprehensive investigation of the extent of adaptation of metastatic triple negative breast cancer (TNBC) cells to their new microenvironment in the distant tissues. This study could reveal new therapeutic windows for developing more effective treatments of metastatic tumors.
Keywords: triple negative breast cancer, metastasis, gene expression, genome-scale metabolic models, systems biology
Abstract
Triple negative breast cancer (TNBC) metastases are assumed to exhibit similar functions in different organs as in the original primary tumor. However, studies of metastasis are often limited to a comparison of metastatic tumors with primary tumors of their origin, and little is known about the adaptation to the local environment of the metastatic sites. We therefore used transcriptomic data and metabolic network analyses to investigate whether metastatic tumors adapt their metabolism to the metastatic site and found that metastatic tumors adopt a metabolic signature with some similarity to primary tumors of their destinations. The extent of adaptation, however, varies across different organs, and metastatic tumors retain metabolic signatures associated with TNBC. Our findings suggest that a combination of anti-metastatic approaches and metabolic inhibitors selected specifically for different metastatic sites, rather than solely targeting TNBC primary tumors, may constitute a more effective treatment approach.
The most aggressive subtype of breast cancer is basal-like triple negative breast cancer (TNBC), which is associated with a poor prognosis, invasiveness, early relapse, and distant metastasis. Since TNBC tumors do not express progesterone receptor, estrogen receptor, or human EGF receptor-2 (HER2), they will not respond to hormonal therapy or medicines that target HER2 (1, 2). Therefore, available effective therapies for basal TNBC are still very limited. The main cause of death in patients with TNBC is metastasis, which allows tumor cells to migrate from the primary site into the circulatory system and invade and colonize other organs. During the metastatic process, cancer cells are faced with significant challenges in their new microenvironments, which they must overcome to survive (3). The ability of tumor cells to overcome these unique barriers and to meet biosynthetic and bioenergetic demands during the metastatic cascade is critical for successful colonization in other organs, yet remains poorly understood.
Metabolic reprogramming is known to be a general strategy of cancer cells to obtain nutrients from nutrient-deprived environments and to sustain their transformed state, promoting survival and uncontrolled proliferation (4). While the impact of metabolic reprogramming on the metastatic process is poorly understood, accumulating data suggest that metastatic cancer cells must prioritize different metabolic programs distinct from the primary tumor (TP) to survive during each step of the metastatic process and successfully colonize (5). Moreover, different metastatic sites can pose distinct metabolic challenges to the tumor cell, and meeting the metabolic demand of these host organs is crucial to maintain tumor cell proliferation (6). Understanding the mechanisms of molecular and metabolic plasticity in metastatic cancer cells may reveal clinical approaches to treat metastatic disease.
We therefore performed an analysis of RNA sequencing (RNA-seq) data from TNBC TPs, paired distant metastases in six different tissues, and TPs of these six tissues together with RNA-seq data from healthy tissue data obtained from Genotype-Tissue Expression (GTEx) to systematically investigate the tissue-specific characteristics of metastatic tumors (TMs). We conducted a comprehensive analysis of RNA-seq profiles from these TMs, their matched TPs, and healthy tissues, including batch correction, dimensionality reduction, clustering, deconvolution analyses, and gene set analysis (GSA), to study their characteristics. We then reconstructed their metabolic networks to investigate the metabolic features of TNBC metastatic tumors (TNBC-TMs) and compared them with those of TPs of the destination tissue. Our analysis showed that TNBC-TMs express a metabolic network that is in an intermediate state between TPs of the tissues of their origin and their destinations. Our metabolic network analysis also showed that each of the metastases had specific reactions that were not active in either their tissue of origin (TNBC TP) or metastatic site. Furthermore, our cross-tissue metabolic analysis allowed us to identify significantly changed metabolic pathways in cancers that may be considered as potential therapeutic targets.
Results
Integrative Analysis of TP, Metastatic, and Healthy Tissue Transcriptome Profiles.
We obtained RNA-seq count data from the Gene Expression Omnibus GSE110590 dataset for different cancer subtypes and their associated TMs (7). Samples with basal-like subtype of TNBC and their paired TMs in six different distant tissues (brain, lung, liver, lymph node, adrenal gland, and skin) were selected for this study (SI Appendix, Table S1). Throughout this paper, “breast-TP” refers to the basal-like subtype of TNBC. We also retrieved transcriptome profiles from The Cancer Genome Atlas (TCGA) for TPs and paired adjacent normal tissue (NT) samples corresponding to the tissues of origin and destination of the GSE110590 dataset. Healthy tissue profiles associated with the six TMs as well as breast were collected from the GTEx database (8). Our study therefore involved a comparison of metastatic TNBC transcriptomes with TPs, paired-normal, and healthy tissues corresponding to the TMs' tissue of origin (breast) and destinations (Fig. 1A). The collected count data from these three different sources were normalized and combined along their common genes. To reduce variation between datasets, we used log-transformed quantile-normalized transcript per million (TPM) in an empirical Bayes framework (9, 10). The resulting expression distributions and relative log expression analysis (11) of TPM and batch-corrected TPM showed that these three datasets from different resources can be analyzed jointly after normalization followed by batch effect removal and that the resulting batch-adjusted TPM of a gene was comparable across different samples (SI Appendix, Figs. S1 and S2).
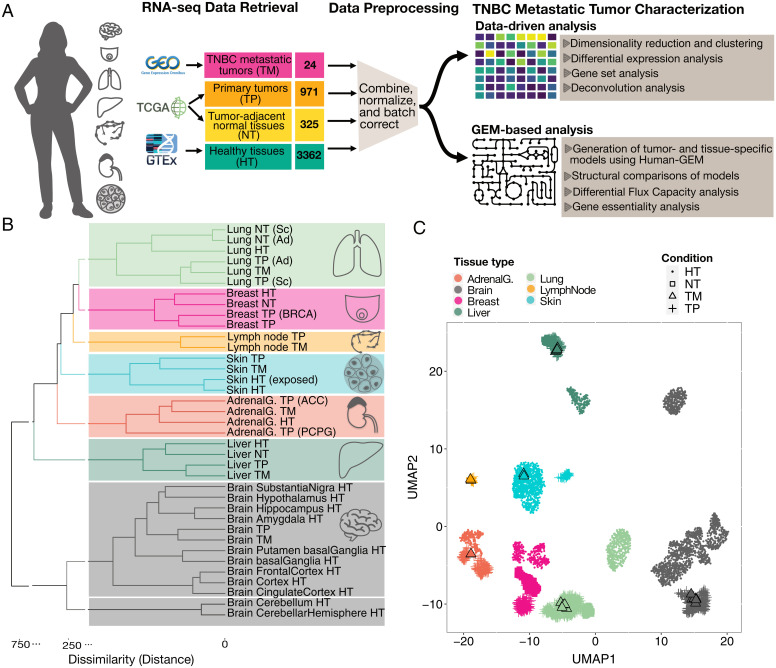
Fig. 1.
Comparison of basal TNBC metastatic and primary tumors, healthy tissues, NTs, and other TPs. (A) Study design. From GSE110590, we extracted only samples for basal-TNBC subtype including TPs (breast-TPs) and metastases in brain, lung, liver, lymph node, adrenal gland, and skin (TNBC-TMs) and matched them with the TPs of their metastatic organs and their associated NT adjacent to the tumor from TCGA and healthy tissues from GTEx. We performed identical processing of all samples to obtain TPM values. We then utilized several techniques to characterize the differences between TNBC-TMs, breast-TPs, and the TPs of the metastatic sites across tissue types. (B) General overview of integrated datasets using HHK clustering on median TPM values of each gene among all samples for each condition. The dendrogram is colored based on the optimal number of clusters as determined from the silhouette plot. BRCA represents breast invasive carcinoma; ACC, adrenocortical carcinoma; Ad, adenocarcinoma; Adrenal G., adrenal gland; PCPG, pheochromocytoma and paraganglioma; Sc, squamous cell carcinoma. (C) UMAP plot of all samples in the combined dataset. Points are colored by the associated tissue, and the shape of each point represents the condition: healthy tissue (HT), TM, TP, or NT.
To obtain a general overview of the integrated data, we performed Hybrid Hierarchical K-means (HHK) clustering (12, 13) on the gene expression profiles of all the conditions. The estimated optimal number of clusters by the silhouette coefficient (14) was used in the HHK algorithm for cutting (coloring) their associated dendrograms (Fig. 1B and SI Appendix, Fig. S3). The clustering dendrogram of median TPM across groups and also the uniform manifold approximation and projection (UMAP) (15) plot of all samples in the integrated dataset resulted in clustering by tissue and cancer type, showing that both metastatic and primary tumors are more similar to their surrounding tissue than to other tumors (Fig. 1C).
TNBC-TMs Exhibit Characteristics of Both Their Tissue of Origin and Their Destination.
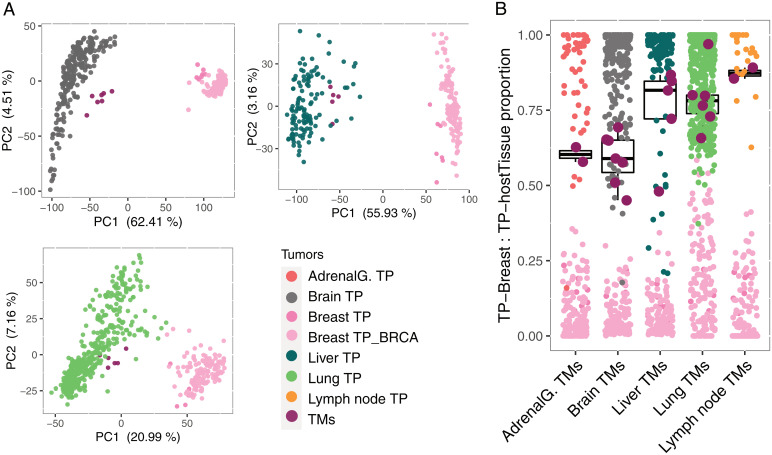
We next performed dimensionality reduction analysis for each metastatic tumor and TPs of the origin and the destination tissues. Principal component analysis (PCA) showed that TMs formed a distinct cluster much closer to TPs of the tissues of destination, while the two TP groups were clearly separated (Fig. 2A). This trend was observed for all the TM samples in brain, lung, and liver that had more than three metastatic samples. Thus, this analysis suggests that the expression profiles of TMs represent an intermediate state between TPs from the tissue of origin and the tissue of destination, with a profile closer to that of the destination tissue TP. To further investigate this phenomenon and quantify the similarities, we applied a deconvolution analysis pipeline (16) using median expression levels of TPs from the TMs’ tissue of origin and destination as references to quantify the “TP_origin:TP_destination” fraction for all metastatic samples. The result revealed substantial similarities among TMs and TPs of their destinations, although the trends of divergence from their tissue of origin varied across different metastatic sites (Fig. 2B). Reducing the number of signature genes used in the deconvolution analysis led to an increase in similarity between each of the TM profiles and TP profile of their destination tissue (SI Appendix, Fig. S4A). To assess whether infiltrating cells were responsible for the trend of divergence, we performed a Pearson correlation analysis between TM purity scores and their distances to breast TP. The analysis did not show any significant correlation between purity and the distance to the median expression level of breast TP as reference (SI Appendix, Fig. S4B).
Fig. 2.
Intermediate state of TNBC-TMs between breast-TPs and TPs of the metastatic organs. (A) PCA plot for breast-TP and the associated TM in distinct tissue types. In all three groups, TM samples (maroon) lie between breast-TP samples (pink) and the TP samples of their metastatic organs and are closer to TPs of their metastatic organs. (B) Deconvolution analysis of the TNBC-TM samples using median expression levels of breast-TPs and TPs of the metastatic destination tissues as references. The result of the analysis is the fraction of similarity of each TM sample (maroon) to the TPs of the destination tissue. A value of 1 on the y axis indicates the maximum proportion of “destination tissue_TP contribution”, meaning maximum similarity to median expression levels of “destination tissue_TP” as reference, and 0 indicates the minimum proportion of “destination tissue_TP contribution.” Boxplot represents the distribution of the proportion of “destination tissue_TP contribution” for TMs. The points, breast-TPs (pink), and TPs of the tissue of destination (colored by cancer type) deconvolution fractions are shown as references. AdrenalG., adrenal gland; BRCA represents breast invasive carcinoma.
Although the comparability of genes across the final integrated dataset was validated, the small number of metastatic samples in each destination tissue and the use of different resources for metastatic and primary tumors are potential limitations of the analysis. Therefore, the skin cutaneous melanoma (SKCM) cohort, as the only cancer in the TCGA project that is focused on metastatic cases, was used to validate our findings. We retrieved samples of distant metastases to the adrenal gland, brain, lymph nodes of the axilla, and lymph nodes of the head, face, and neck because these tissues were associated with larger sample sizes than the other distant metastatic regions and their sites of biopsy were similar to TP samples of the same tissue in TCGA. The aggregated samples were then normalized to allow comparison across datasets (SI Appendix, Fig. S5 A and B). A similar deconvolution analysis as described above was used to evaluate the similarity of SKCM TM expression profiles to those of TPs from their origin and destination tissues (SI Appendix, Fig. S5C). As was observed in the previous deconvolution analysis, reducing the number of signature genes used in the deconvolution increased the similarity between each of the TM profiles and the TP profile of their destination tissue (SI Appendix, Fig. S6A). These analyses also showed that melanoma distant metastases to lymph nodes maintained more similarities to their tissue of origin, suggesting a tumor-specific behavior for adaptation to metastatic sites. We repeated the Pearson correlation analysis between purity of TMs and their similarities to the median expression level of TPs of the host tissue. Lymph node–axilla TMs were selected due to a larger sample size, and the results did not show any significant correlation between immunohistchemistry (IHC) purity and similarity to the median expression level of TPs of the host tissue (SI Appendix, Fig. S6B). The SKCM cohort analysis further supports the divergence of TMs from their tumors of origin.
Gene Signatures and Biological Processes Differentiate TMs from Their Tissue of Origin.
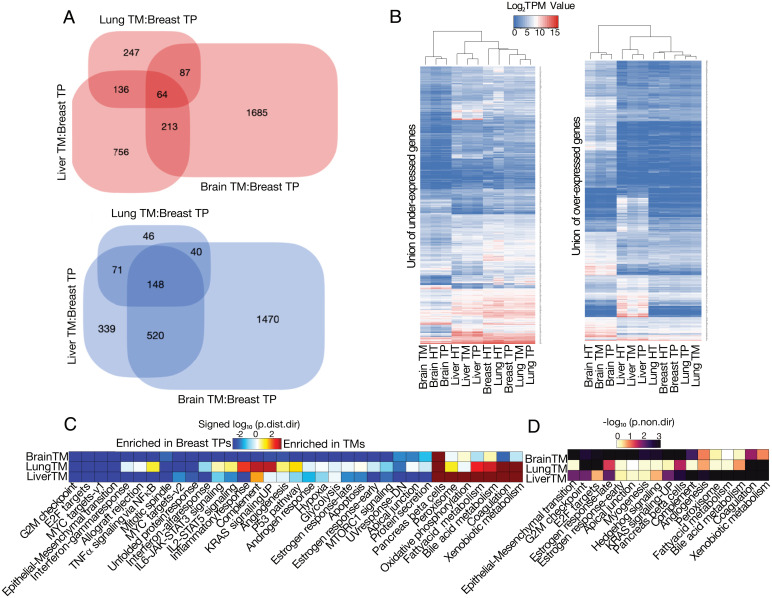
To further investigate the features that define the divergence between TNBC-TMs and their original TPs, we performed differential expression (DE) analysis (17) between the TM profiles (grouped by destination tissue) and the breast-TPs. DE analysis was performed for TNBC metastases to brain, liver, and lung, while metastases to adrenal gland and lymph node were excluded from this analysis because the number of samples was too low to achieve sufficient statistical power. The results showed widespread similarities in overexpressed and underexpressed genes across TNBC metastases to brain, liver, and lung. We identified 64 and 148 genes that were commonly overexpressed and underexpressed, respectively, in all TM profiles relative to breast-TPs (padj < 0.01 and |log2fold change (FC)| > 1) (Fig. 3A). However, each of the TM groups had specific over- and underexpressed genes that were not differentially expressed in either of the other groups relative to breast-TPs, suggesting an organ-specific adaptation of metastatic breast cancer cells when they colonize in their distant metastatic sites. The most changes in terms of differentially expressed genes (DEGs) relative to breast-TPs were seen in TNBC metastases to the brain. Patterns of gene expression among the specific over- or underexpressed genes for each of the TNBC-TM types showed that the expression pattern of TMs tended to be more similar to that of their tissues of destination than to their tissue of origin, particularly with TPs from the destination tissue (Fig. 3B).
Fig. 3.
Divergence of TNBC-TMs from breast-TPs based on DEGs and GSA. (A) Venn diagrams of the number of common and specific over-expressed genes (red) and under-expressed genes (blue) in TNBC-TMs compared with breast-TPs. (B) Heatmap of differentially over- and under-expressed genes of each metastasis. DEGs of each of the TMs distinct from the other metastases were combined, and the final subset of genes was extracted from breast-TP and TPs of the tissue of metastatic sites and their matched healthy tissues for brain, liver, and lung. The pattern of expression is similar to the tissue of destination in both under-expressed (Left) and over-expressed (Right) genes. (C) Results of the directional GSA of DE analysis results for TNBC metastases in lung, liver, and brain versus paired breast-TPs. Only the Hallmark gene set collection is shown here, and sets with <10 genes were excluded. The more significant (lower value) of the two directional P values for each gene set is shown in the heatmap as a log10-transformed value. The distinct directional gene set P values (padj.dist.dir) are calculated for coordinated increases (padj,dist-dir-up) and decreases (padj,dist-dir-down) in expression. The value is also “signed,” meaning that gene sets with a more significant decrease than increase (padj,dist-dir-down < padj,dist-dir-up) are negative (enriched in breast-TPs); otherwise, they are positive (enriched in TNBC-TMs). Only gene sets with a padj,dist-dir less than 0.01 in at least one TM are shown. (D) Nondirectional GSA results for three comparisons. The “p.non.directional” value for each gene set is filtered based on non.dir P values less than 0.01 and shown in the heatmap as a log10-transformed P value. HT, healthy tissue; IL, interleukin; NFκB, nuclear factor κB; UV, ultra violet; UVresponse-DN represents genes down-regulated in response to UV radiation.
To further understand the processes associated with the divergence between breast-TPs and their associated metastases to brain, lung, and liver, we performed directional and nondirectional GSAs (18) of the DE analysis results using the Kyoto Encyclopedia of Genes and Genomics (KEGG) and hallmark gene set collections (19). The directional GSA results revealed robust enrichment of metabolic pathways (xenobiotic metabolism, oxidative phosphorylation, fatty acid metabolism, bile acid metabolism, and retinol metabolism) and immune response functions (complement system and coagulation) in metastatic tumors compared to breast-TPs (Fig. 3C). In contrast, gene sets related to proliferation processes, signaling pathways, and the inflammatory response were significantly enriched in breast-TPs, such as G2M checkpoint, E2F targets, MYC targets, mitotic spindles, tumor necrosis factor (TNF)–α signaling, interferon responses, allograft rejection, and the unfolded protein response pathway, suggesting that genes associated with these pathways are down-regulated across metastatic tumors. When the direction of gene expression changes in the GSA was ignored (nondirectional), fewer gene sets were found to be significantly enriched in DEGs and were primarily associated with brain and liver metastases. Furthermore, metabolic pathways accounted for the most significant changes in TMs relative to TPs of their tissue of origin, although the extent and direction of these changes were adjusted specifically to the different metastatic sites (Fig. 3D and SI Appendix, Fig. S7). Gene sets associated with estrogen responses, which are signatures of breast cancers, were also underexpressed in TMs in comparison with breast-TPs, suggesting that TMs lose some of the defining characteristics of their parental tumors.
The other processes enriched in breast-TPs compared to metastases were associated with metastasis and the epithelial-mesenchymal transition (EMT), representing the early stages of the metastatic cascade. Other enriched signaling pathways and processes were MYC (20, 21), interleukins and their associated pathways (22, 23), interferon-α and -γ responses (24), the JAK-STAT3 pathway (25, 26), and TNFα and the signaling factors and transcription factors activated by TNFα (27). These pathways are considered key to the induction of invasion and metastasis in tumor cells. The protein secretion gene set and unfolded-protein response, which is associated with endoplasmic reticulum stress (28), were also enriched in breast-TPs versus TNBC-TMs. Enrichment of these signaling pathways and processes correlated with the enrichment of EMT and cell-cell junction and extracellular matrix–associated gene sets, reflecting the interplay of these pathways with feed-forward and feed-back loops to regulate plasticity of the EMT in tumor cells during metastasis (29).
In addition to the significant enrichment of EMT-associated processes in breast-TPs versus TNBC-TMs, expression of genes associated with metabolic pathways including glycolysis, bile acid metabolism, xenobiotic metabolism, and oxidative phosphorylation (OXPHOS) were also changed. The enrichment of EMT correlated with the enrichment of glycolysis in breast-TPs, whereas OXPHOS was enriched in TNBC-TMs. This observation supports the hypothesis that metabolic reprogramming and the metastatic cascade are linked to promote the stages of cancer progression and metastasis.
EMT-associated transcription factors orchestrate profound metabolic reprogramming that allows tumor cells to survive in an ever-changing microenvironment, though accumulating evidence suggests that the relationship between EMT and metabolism is mutual, and under some conditions such as hypoxia and nutrient deprivation, metabolic changes can promote EMT (30) and start the metastatic cascade. For example, increased expression of hexokinase 2 (HK2), which catalyzes the irreversible reaction of glucose to glucose-6-phosphate as the first rate-limiting step of glycolysis, can facilitate EMT by decreasing the pH of the tumor microenvironment, activating intracellular signaling including snail/ERK2, and up-regulating matrix metalloprotease activity (31). Increased expression of lactate dehydrogenases (LDHs), which convert pyruvate to lactate, induce lactate accumulation in the tumor microenvironment (32). Extracellular lactate would acidify the tumor microenvironment and facilitate matrix degradation (33) and immune evasion (34), resulting in migration and mobility of tumor cells. Both HK2 and LDHA/B were underexpressed in TNBC-TMs relative to breast-TPs, which supports metastatic colonization, including seeding, formation of tumor cell–matrix interactions, extracellular matrix remodeling, and finally, outgrowth (SI Appendix, Fig. S8A). Metastatic colonization in distant organs relies on increased adenosine 5′-triphosphate (ATP) production through the mitochondrial OXPHOS pathway to sustain high proliferative capacities of tumor cells (35, 36). For example, NDUFB8 and NDUFS7, which are subunits of the mitochondrial membrane respiratory chain reduced nicotinamide-adenine dinucleotide dehydrogenase (Complex I), and SLC25A4 as a mitochondrial adenosine 5′-diphosphate/ATP transporter were overexpressed in TMs compared with breast TPs. Furthermore, Aldolase B (ALDOB), which induces the incorporation of fructose into glycogen and lipids, was overexpressed in TMs compared with breast TPs, presumably to sustain highly proliferative capacity of tumor cells and promote metastatic colonization (36–38).
Significantly changed genes associated with EMT, glycolysis, and OXPHOS pathways showed a mixture of expression changes between TMs and TPs of their tissue of origin, while the direction and extent of these changes were different for each destination tissue, indicating a context-dependent behavior in different microenvironments. Furthermore, some genes associated with “Pancreas Beta Cell” in the hallmark gene set, which was highly enriched in TMs compared to breast-TPs, showed a strong tissue dependency. The overexpressed genes, including FOXA2, SLC2A2, PKLR, and HNF1A, are liver-enriched genes, and STXBP1, SST, ABCC8, CHGA, and NKX2-2 are brain-enriched genes (39), which were overexpressed in liver-TM and brain-TM compared to breast-TP (SI Appendix, Fig. S8B), respectively, further supporting the context dependency of reprogramming of TMs.
Altogether, these results emphasize the reprogramming of TMs to adapt to the new environment of their destination organs to survive and colonize, while these processes could be reversed again under some circumstances to promote EMT followed by metastasis and cellular migration to other tissues.
Metabolic Modeling of TNBC TPs and Paired Metastases in Different Tissues.
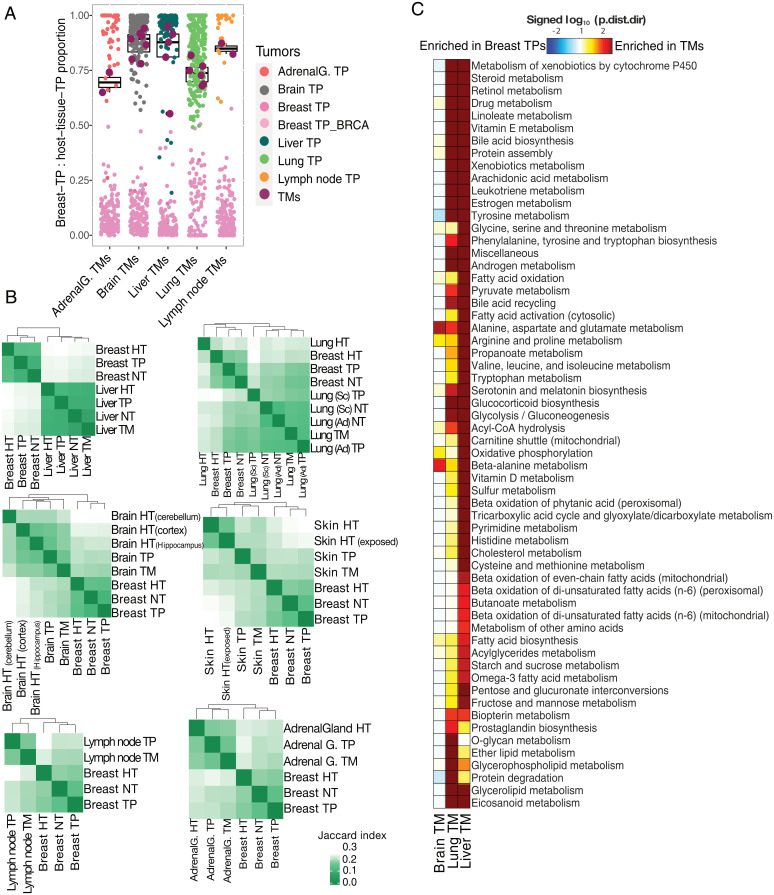
Consistent with the GSA results, deconvolution analysis using only genes associated with metabolism exhibited a more striking coordinated shift of metastatic samples to TPs of their destination sites than was observed when all genes were considered (Fig. 4A). Repeating the same analysis for SKCM TMs provided further support of the significant coordinated metabolic shift toward TPs of their destination tissues when using only metabolic genes (SI Appendix, Fig. S9). Demonstration of metabolic reprogramming requires not only gene expression data but also cellular context, including information of the associated reactions and metabolites.
Fig. 4.
Divergence of TNBC-TMs from breast-TPs based on metabolic signatures. (A) Deconvolution analysis of the TNBC-TM samples using median expression levels of metabolism-associated genes in breast-TPs and TPs of the tissue of destination as references. The result of the analysis is the fraction of similarity of each TM sample (maroon) to the TPs of the tissue of its destination based on only their metabolic genes. Boxplot represents the distribution of the proportion of “destination tissue_TP contribution” for TMs. (B) Heatmaps showing comparison of reaction content of GEMs specific to TNBC-TMs in distinct tissues with breast-TPs as their tumor of origin and GEMs specific to TPs of the tissue of destination and their matched healthy tissue (HT) and NTs, based on the Jaccard index. (C) Subsystem directional GSA results for the comparisons of breast-TPs in distinct tissues with breast-TPs. Shown are the log10-transformed “distinct directional” P values (p.dist.dir) for subsystems with P value < 0.01 in at least one comparison. The log10-transformed P values are signed, meaning that gene sets significantly enriched in expression increases are positive, while those enriched more in expression decreases are negative. Ad, adenocarcinoma; AdrenalG., adrenal gland; BRCA represents breast invasive carcinoma; Sc, squamous cell carcinoma.
To further investigate the metabolic adaptation of TNBC-TMs to their metastatic sites, we employed genome-scale metabolic modeling. Genome-scale metabolic models (GEMs) provide a scaffold to integrate gene expression data and interpret the information in a metabolic context. Using the Human1 GEM (40) as a reference of human metabolism, we generated 33 condition-specific models that included four groups of healthy tissue-, TM-, TP-, and paired NT–specific models based on RNA-seq data. Structural comparison of the TM models with models of their tissue of origin and metastatic site showed that metabolic signatures of metastases are more similar to that of their destination organs than that of their tissue of origin (Fig. 4B). An overview of relationships across reaction structures of all the models using UMAP revealed a strict and distant segregation of brain and liver clusters from the others, highlighting extensive metabolic reprogramming of TM cells based on their metastatic sites to ensure successful colonization (SI Appendix, Fig. S10). These findings based on the reaction content of each model again reaffirmed the extensive metabolic divergence of TMs from the TPs of their origin tissue.
To obtain a more comprehensive picture of the metabolic alterations that metastatic basal TNBC tumor cells undergo to overcome distinct metabolic challenges posed by microenvironments of their metastatic sites, we sought to identify metabolic pathways and metabolites associated with genes that are significantly differentially expressed (metastasis vs. breast-TP). We performed a GSA for which gene sets were metabolic pathways (subsystems) or metabolites of the metabolic network. The subsystem GSA revealed an enrichment of pathways associated with amino acid metabolism, lipid metabolism, xenobiotic metabolism, bile acid metabolism, glycolysis/gluconeogenesis, and OXPHOS in metastases (Fig. 4C). The GSA results show that TNBC-TMs employed distinct metabolic signatures dependent on their host tissues, meaning that the extent and direction of the enriched gene sets were different among liver, lung, and brain metastases.
Metastatic TNBC Cells Exhibit Divergent Metabolic Features with Some Similarities to TPs of Their Destination Tissues.
We next assessed whether metastatic TNBC cells retain any metabolic features from their parental tumors in their distinct metastatic sites. The models were grouped by metastatic tissue, including each of the TNBC-TMs, breast-TPs, TPs of metastatic site, and their matched healthy and NT models. By comparing the reactions included in the models of each group, we found that TMs had specific reactions that were not included in the other models of their group (Fig. 5A and Dataset S1). Furthermore, TM models contained some common reactions with their parental breast-TPs that were not present in other models in the group, indicating that metastatic TNBC tumors retained some unique signatures from their origin (Fig. 5A and Dataset S2). We next extracted the associated subsystems from the metabolic models to identify which metabolic pathways are potentially important during metastasis and colonization of a new microenvironment. Metastatic-specific reactions were mostly associated with transport (43.7%) and peptide metabolism (9.15%), though exchange reactions and bile acid biosynthesis were also identified as important (SI Appendix, Fig. S11A). The retained signature reactions from breast-TPs in their TMs also belonged to transport reactions, drug metabolism, exchange reactions, bile acid biosynthesis, and bile acid recycling subsystems (SI Appendix, Fig. S11B). Taken together, these results highlight that the metabolic phenotype of TNBC-TMs shows an intermediate state between their parental tumors and the TPs of their metastatic sites by retaining signatures from their parental tumors. Furthermore, the TM GEMs contain some specific reactions that may support the ability of TMs to overcome the challenges of colonization and proliferation in their new microenvironments.
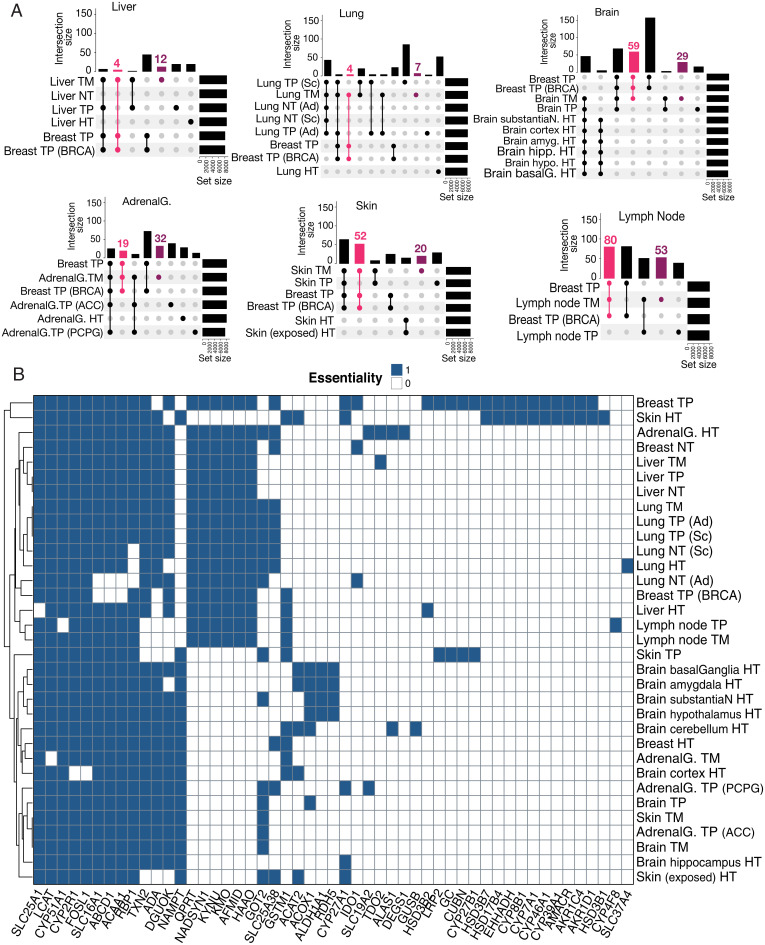
Fig. 5.
Metabolic signatures and essential genes as potential drug targets. (A) UpSet plot of comparison combinations in each group. Each row corresponds to a condition, and each bar shows a different combination. The filled-in cells show which condition is part of an intersection, and the lines connecting the filled-in cells show in which direction the plot should be read. TM-specific reactions are colored maroon, and common reactions between breast-TPs and TNBC-TMs are colored pink. (B) Genes predicted to be essential for biomass production are colored blue in each condition. Among the essential genes, those that are not essential in most healthy models can be considered potential drug targets for cancer therapy. ACC, adrenocortical carcinoma; Ad, adenocarcinoma; AdrenalG., adrenal gland; amyg., amygdala; basalG, basal ganglia; BRCA represents breast invasive carcinoma; hipp., hippocampus; HT, healthy tissue; hypo., hypothalamus; PCPG, pheochromocytoma and paraganglioma; Sc, squamous cell carcinoma; substantiaN, substantia nigra.
The results presented above highlight tissue-specific adaptation patterns by which TM cells survive in their secondary sites. We therefore sought to compare the metabolic functionality of these TMs with the TPs of their destination tissues to elucidate the metabolic shift that occurs in TNBC-TMs. To this end, we used the GECKO (41) framework, which integrates GEMs with enzyme kinetic data to generate enzyme-constrained GEMs (ecGEMs) to calculate flux distributions for functional comparison of metabolic models (40). We conducted flux variability analysis on each of the ecGEMs of TPs and TMs under the same nutrient conditions while maximizing biomass production (SI Appendix, Fig. S12). To quantify the flux differences, the fold changes of reaction capacities of both the TM and destination tissue TP ecGEMs versus the breast-TPs were calculated. Comparison of reaction flux capacities showed that metabolic reprogramming of TNBC-TMs in some parts is correlated with metabolic programs of TPs of their metastatic sites, which differ depending on the organ. However, TNBC-TMs and TPs of their metastatic sites could also have specific metabolic characteristics compared with breast-TPs.
It is well known that prioritized metabolic strategies in TNBC include elevated glucose uptake, glutamine uptake, activity of LDHA/B, and decreased glutamine synthesis (42). The flux comparison analyses displayed either a coordinated decrease or no change in dependency of TMs on glutamine compared with breast-TPs, while the increased uptake of different resources, especially branched chain amino acids (BCAAs) as well as other nonessential amino acids, was observed. Some cancer cells are known to display glutamine addiction despite glutamine being a nonessential amino acid, where the high rate of glutamine uptake is required for uptake of additional amino acids and reduced nicotinamide-adenine dinucleotide phosphate (NADPH) production for redox control (43). Cancers become dependent on glutamine to provide nitrogen for nucleotide and protein synthesis, although the amount of this dependency varies across different cancer types (44). For example, brain TMs exhibited reduced dependency on glutamine metabolism and more dependency on the other amino acid pathways compared with breast-TPs, in correlation with the brain-TP metabolic program, but the capacity of glucose uptake remained unchanged (SI Appendix, Fig. S13). It is possible that the microenvironment of the brain plays a key role in this phenomenon. Although gene expression levels of glycolytic enzymes have been shown to increase in brain metastasis (45), the availability of glucose in the brain interstitial space is lower than that of blood (46, 47). Moreover, an increase in glucose uptake compared with breast tumors is not a known feature of breast cancer brain metastasis (48). Brain interstitial space contains high levels of glutamine and other nonessential amino acids and also BCAAs (49), which can serve as energy resources for metastasizing cancer cells to survive and thrive in the brain by mimicking brain cells (50). Our analysis showed a decrease in glutamine uptake capacity in both brain-TM and brain-TP compared with breast-TPs, demonstrating the extensive dependency of TNBC tumors on glutamine, which can be considered a significant signature of this cancer type. However, this may not be prioritized in their associated TMs to the same extent. Divergent metabolic patterns were also observed in the other TNBC-TMs (SI Appendix, Figs. S14 and S15). Overall, these results suggest that TNBC-TMs display different metabolic phenotypes distinct from the metabolic strategy of breast TPs in response to the TM microenvironment.
Identification of Metabolic Pathways and Genes as Potential Drug Targets for Cancer Treatment.
To determine important reactions and subsystems that could potentially constitute drug targets, we extracted the union of specific reactions from GEMs of TPs and TMs and excluded reactions that were involved in any of the healthy tissues or NT GEMs. The remaining reactions highlighted the importance and abundance of several metabolic pathways in metastatic and/or primary tumors (SI Appendix, Fig. S16 and Dataset S3). Abundant reactions belonged to transport reactions, peptide metabolism, protein degradation, drug metabolism, and cholesterol metabolic pathways. Among the extracted transport reactions, two reactions were associated with carnitine in fatty acid oxidation (FAO), which is important for cancer cells to overcome nutrient and energy deficiencies to survive and maintain uncontrolled proliferation (51, 52). The other interesting specific transport reactions in skin-TM and brain-TP involved xanthurenate, a catabolic derivative of tryptophan, suggesting the importance of tryptophan metabolism in cancer. Depletion of tryptophan from the local space has long been known to be a common feature of cancers to promote immune suppression, and several inhibitors of this pathway are currently undergoing clinical trials (44).
Finally, we conducted gene essentiality analysis on all the generated models to determine which genes, when deleted, reduce the production of biomass to near zero. Genes found to be essential across all models were excluded to ignore core metabolic reactions, which are required for basic metabolic processes in many tissues (Fig. 5B). Among the remaining essential genes, we considered those that were essential for metastatic and/or primary tumors but not essential for healthy and adjacent normal tissues to focus on potential drug targets with minimal side effects. Phenylalanine, tyrosine, and tryptophan metabolism-related genes, including tryptophan 2,3-dioxygenase (TDO2), indoleamine 2,3 dioxygenase (IDO1), kynureninase (KYNU), kynurenine monooxygenase (KMO), and arylformamidase (AFMID), were among the essential genes for breast-TP, lung-TM, liver-TM, lymph node–TM, and the TPs of destination tissues, with minimal effect on healthy tissue GEMs. A predicted essential gene for brain-TM and its matched TP (brain-TP), skin-TM and its matched TP (skin-TP), adrenal gland (PCPG)-TP, and adrenal gland (ACC)-TP was GOT2, which also belongs to tyrosine and tryptophan metabolism. These observations emphasize the key role of this metabolic subsystem, particularly tryptophan metabolism and the kynurenine pathway, in different cancers (44, 53, 54) and more importantly TNBC-TMs. The other predicted essential genes for metastatic and primary tumors were associated with nicotinate and nicotinamide metabolism (e.g., QPRT, NADSYN1, and HAAO) (55). Vitamin D metabolism-related genes including CUBN, LRP2, GC, and CYP27B1, which form an NADPH-dependent enzyme complex together, were essential for skin-TP and TNBC tumor growth. The essentiality of AKR1D1 and AKR1C4 for breast-TP growth highlighted the importance of α-keto reductases in bile acid biosynthesis and retinol metabolism pathways for breast cancers and specifically TNBC invasion (56–58). Some bile acid biosynthesis-related genes, including HSD3B7, AMACR, CYP39A1, CYP46A1, CYP7A1, and CYP8B1, and FAO-related genes such as EHHADH and HSD17B4 also showed essentiality for TNBC growth. An interesting observation is that most of the predicted essential genes for different cancers and TNBC distant metastases encode for NADPH-dependent enzymes. Available drugs that can target many of these genes and enzymes were collected from the DrugBank database (59) or from literature for those compounds that do not have any records in the database (SI Appendix, Table S2).
Collectively, these results highlight the importance of several metabolic pathways contributing to tumor viability, such as tryptophan metabolism, kynurenine pathway, nicotinate and nicotinamide metabolism, vitamin D metabolism, and bile acid biosynthesis and recycling, which may hold potential as drug targets in cancer and metastasis treatment.
Discussion
Metastasis is the primary contributor to death for patients with TNBC, which is associated with poor prognosis and an aggressive phenotype. The molecular mechanisms involved in TNBC metastasis continue to be investigated, and the metabolic reprogramming of TNBC tumors during metastasis, colonization, and growth in distant organs is an important component. It is clear that we must understand the metabolic characteristics of TNBC cells, which metastasize to different organs, in order to understand how these TMs overcome challenges posed by their ever-changing microenvironment during the metastatic cascade. This understanding is crucial for effective therapy and prevention.
Recently, several studies have explored how molecular mechanisms and metabolic strategies employed by cancer cells in their primary microenvironment change during the different stages of metastasis across different cancers (29, 42, 56, 60, 61). However, metabolic reprogramming of TMs in distant organs remains poorly understood. Moreover, given the critical importance of metabolic adaptation of tumor cells to their secondary tissues to support and enable colonization, survival, and uncontrolled proliferation in the new nutrient-deprived environment (5, 6), it has been suggested that TMs may exhibit phenotypes similar to those of the TPs of their metastatic sites. Here, we show the importance of targeting both metabolic reprogramming and the metastatic cascade using combination therapy. This approach is more adapted to the organ hosting the metastasis than to its original TP and could be beneficial for targeted treatment of TNBC-TMs.
Quantifying the distances of TMs from TPs of their origin and of their destinations revealed that TNBC-TMs in different tissues are more similar to the TPs of those metastatic sites. The extent of the divergence was dependent on metastatic tissue and reflected an intermediate state between the TPs of origin and destination. We identified genes that were significantly differentially expressed in TNBC-TMs in liver, lung, and brain compared with breast-TPs, and many were involved in EMT, metastasis-associated processes, and metabolic pathways. The extent and direction of significantly enriched gene sets varied across the TMs in different tissues. It has emerged that crosstalk between signaling pathways associated with metastasis (e.g., Snail/ERK2), and metabolic enzymes, including HK2 and LDHA/B, can facilitate EMT (30–34, 62). Furthermore, metabolic reprogramming to enhance fructose metabolism through up-regulation of ALDOB and ATP production through OXPHOS promotes the metastatic colonization at distant organs (35–38). Consistent with our results, Jin et al. (63) investigated breast cancer using single-cell RNA-seq data and showed that reprogramming of lipid metabolism facilitates colonization and adaptation to the brain microenvironment. The critical role of lipid metabolism and, more specifically, fatty acid biosynthesis in breast cancer brain metastasis was further supported by another study by Ferraro et al. (64).
Using only metabolic-associated genes, TNBC-TMs exhibited more similarity to TPs of the destination tissues. The results also confirmed the coordinated metabolic shift of TNBC-TMs from breast-TPs to TPs of their destination tissues. A similar pattern of metabolic shift was observed for SKCM TMs, suggesting that the coordinated metabolic shift of TMs from TPs of their origin toward TPs of their destination organs could be a common phenomenon in different types of TMs. It was recently demonstrated that highly metastatic breast cancer cells enhance their metastatic adaptation to their ever-changing microenvironments during stages of metastasis by engaging both glycolysis and OXPHOS as metabolic strategies (60). This phenomenon is required during the metastatic cascade to maximize metabolic flexibility, which allows TM cells to respond to rapidly changing metabolic demands and nutrient deprivation in their new microenvironment (42). Using gene sets extracted from the Human-GEM network, we observed significant enrichment in both glycolysis and OXPHOS pathways for lung, liver, and brain metastases compared to breast-TPs, although the extent of this increase was dependent on the metastatic site.
Since basal TNBC cells are known to prioritize specific metabolic programs, including excessive uptake of glutamine and glucose to ensure their survival and invasiveness (42), our results suggest a change in prioritized metabolic strategy of TNBC-TMs compared with breast-TPs. While the changed metabolic program in TNBC-TMs was consistent with TPs of their destination, the TMs also employed metabolic programs distinct from both TPs of their origin and destination. These functional changes were primarily associated with transport reactions and uptake capacity of different nutrients. In addition, specific reactions of TNBC-TMs as well as the retained metabolic signatures from the TPs of the origin were also associated with transport reactions, emphasizing the capability of TMs to obtain nutrients from their microenvironments to survive circulation and specific distal tissues.
We identified therapeutically relevant pathways by extracting reactions present only in cancer-specific models and by identifying genes uniquely essential to these models. Phenylalanine, tyrosine, and tryptophan metabolism, the kynurenine pathway, nicotinate and nicotinamide metabolism, vitamin D metabolism, and bile acid biosynthesis and recycling pathways were the most important subsystems among metabolism of cancer cells, which constitute potential drug targets for both primary and metastatic tumors. Enhanced expression of enzymes, including IDO1/IDO2/TDO2, the main rate-limiting enzymes in the kynurenine pathway, correlate with increased survival, EMT, drug resistance, and decreased anoikis in different cancers (54). Although inhibitors targeting IDO1 showed promising results in early-stage clinical trials for cancer, a phase 3 trial result was disappointing (65), suggesting that TDO2 might be able to substitute for IDO1 when it is inhibited. However, tryptophan metabolism remains an important target for cancer therapy. Therefore, some inhibitors for TDO2 and dual IDO1/TDO2 enzymes have been developed (53). Our results also suggest KYNU, KMO, AFMID, and GOT2 genes as potential drug targets from tryptophan metabolism, especially for TNBC, the paired TMs in lung and liver, and TPs of lung and liver. Moreover, the predicted importance of bile acid biosynthesis and recycling metabolic pathways in our analyses represent additional interesting approaches in therapeutic development for TNBC TP and TMs.
Since patients in the metastatic dataset were at the late stage of breast cancer and generally received chemotherapy compounds, the conclusions drawn might be affected by these treatments. Although none of the treatments had direct known effects on metabolism, it is a potential limitation of our study. Another potential limitation of our study could be small sample sizes. Future studies comparing matched, therapy-naive, postneoadjuvant therapy, and distant metastases with bigger sample sizes will be required to expand upon the evidence presented here on site-specific metabolic adaptation of TM cells.
In conclusion, TM cells differentially engage distinct metabolic strategies similar to those of TPs of the destination tissues to sustain their survival and proliferation depending on the local microenvironments in the metastatic sites. It is therefore important when developing therapeutic strategies involving metastasis to recognize that TNBC TMs are distinct cancer types that exhibit similarities to TPs of the metastatic tissues while retaining some key signatures from their parental TNBC TPs.
Materials and Methods
All the materials and methods associated with omics data collection and analysis are detailed in SI Appendix. This includes RNA-seq data retrieval, data processing, statistical methods, HHK clustering, dimensionality reduction, DE and GSA, network integrative analysis, and deconvolution analysis. All details on reconstruction, comparison, evaluation, and simulations of metabolic network models can also be found in SI Appendix.
Supplementary Material
Acknowledgments
The authors thank Marni B. Siegel (University of North Carolina) for providing the count data of metastatic breast cancer and Ivan Domenzain for valuable discussions. F.R. acknowledges a visiting scholarship from University of Tehran, Iran. This work was supported by funding from the Knut and Alice Wallenberg foundation (J.N.).
Footnotes
Reviewers: H.U.K., Korea Advanced Institute of Science and Technology; and E.R., NIH.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205456119/-/DCSupplemental.
Data, Materials, and Software Availability
All the datasets used in this manuscript are available in public repositories, and references are given in the text (see the “Data retrieval and processing” subsection in SI Appendix, SI Material and Methods). Correspondence and requests for materials should be addressed to nielsenj@chalmers.se. Reconstructed GEMs and ecGEMs for TPs, TMs, HTs and NTs are available at https://github.com/FaribaRoshanzamir/Metastatic-TNBC (66). All other methods and algorithms used in this study are publicly available, and refrences are given in the text (see SI Appendix, SI Material and Methods). All study data are included in the article and/or supporting information.
References
- 1.Dent R., et al. , Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 13, 4429–4434 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Gluz O., et al. , Triple-negative breast cancer--Current status and future directions. Ann. Oncol. 20, 1913–1927 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Massagué J., Obenauf A. C., Metastatic colonization by circulating tumour cells. Nature 529, 298–306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Elia I., Doglioni G., Fendt S. M., Metabolic hallmarks of metastasis formation. Trends Cell Biol. 28, 673–684 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Schild T., Low V., Blenis J., Gomes A. P., Unique metabolic adaptations dictate distal organ-specific metastatic colonization. Cancer Cell 33, 347–354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel M. B., et al. , Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J. Clin. Invest. 128, 1371–1383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keen J. C., Moore H. M., The genotype-tissue expression (GTEx) project: Linking clinical data with molecular analysis to advance personalized medicine. J. Pers. Med. 5, 22–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson W. E., Li C., Rabinovic A., Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Leek J., Johnson W. E., Jaffe A., Parker H., Storey J., The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandolfo L. C., Speed T. P., RLE plots : Visualizing unwanted variation in high dimensional data. PLoS One 13, e0191629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B., Tai P. C., Harrison R., Pan Y., “Novel hybrid hierarchical-K-means clustering method (H-K-means) for microarray analysis.” in 2005 IEEE Comput. Syst. Bioinforma. Conf. Work. Poster Abstr. (2005), pp. 105–108.
- 13.Kassambara A. and Mundt F., Factoextra: Extract and visualize the results of multivariate data analyses. CRAN R Package version 1.0.7. (2020) http://www.sthda.com/english/rpkgs/factoextra
- 14.Rousseeuw P. J., Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987). [Google Scholar]
- 15.Mcinnes L., Healy J., Melville J., UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv [Preprint] (2018) 10.48550/arXiv.1802.03426. (Accessed 3 August 2020). [DOI]
- 16.Gong T., Szustakowski J. D., DeconRNASeq: A statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-seq data. Bioinformatics 29, 1083–1085 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Ritchie M. E., et al. , limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Väremo L., Nielsen J., Nookaew I., Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 41, 4378–4391 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberzon A., et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H., et al. , MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat. Cell Biol. 14, 567–574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H. Y., et al. , C-MYC drives breast cancer metastasis to the brain, but promotes synthetic lethality with TRAIL. Mol. Cancer Res. 17, 544–554 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Sullivan N. J., et al. , Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 28, 2940–2947 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson D. E., O’Keefe R. A., Grandis J. R., Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15, 139–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortiz A., Fuchs S. Y., Anti-metastatic functions of type 1 interferons: Foundation for the adjuvant therapy of cancer. Cytokine 89, 4–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng Y., Ross J. L., Cowell J. K., The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAK-STAT 3, e28086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lokau J., Schoeder V., Haybaeck J., Garbers C., Jak-stat signaling induced by interleukin-6 family cytokines in hepatocellular carcinoma. Cancers (Basel) 11, 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waters J. P., Pober J. S., Bradley J. R., Tumour necrosis factor and cancer. J. Pathol. 230, 241–248 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Liu J., Lin P. C., Zhou B. P., Inflammation fuels tumor progress and metastasis. Curr. Pharm. Des. 21, 3032–3040 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai J. H., Yang J., Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 27, 2192–2206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georgakopoulos-Soares I., Chartoumpekis D. V., Kyriazopoulou V., Zaravinos A., EMT factors and metabolic pathways in cancer. Front. Oncol. 10, 499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G., et al. , Deregulation of hexokinase II is associated with glycolysis, autophagy, and the epithelial-mesenchymal transition in tongue squamous cell carcinoma under hypoxia. BioMed Res. Int. 2018, 8480762 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie H., et al. , Targeting lactate dehydrogenase-A inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 19, 795–809 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothberg J. M., et al. , Acid-mediated tumor proteolysis: Contribution of cysteine cathepsins. Neoplasia 15, 1125–1137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goetze K., Walenta S., Ksiazkiewicz M., Kunz-Schughart L. A., Mueller-Klieser W., Lactate enhances motility of tumor cells and inhibits monocyte migration and cytokine release. Int. J. Oncol. 39, 453–463 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Molina J. R., et al. , An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 24, 1036–1046 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Davis R. T., et al. , Transcriptional diversity and bioenergetic shift in human breast cancer metastasis revealed by single-cell RNA sequencing. Nat. Cell Biol. 22, 310–320 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Bu P., et al. , Aldolase B-mediated fructose metabolism drives metabolic reprogramming of colon cancer liver metastasis. Cell Metab. 27, 1249–1262.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei Q., Qian Y., Yu J., Wong C. C., Metabolic rewiring in the promotion of cancer metastasis: Mechanisms and therapeutic implications. Oncogene 39, 6139–6156 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlén M., et al. , Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Robinson J. L., et al. , An atlas of human metabolism. Sci. Signal. 13, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sánchez B. J., et al. , Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 13, 935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehuédé C., Dupuy F., Rabinovitch R., Jones R. G., Siegel P. M., Metabolic plasticity as a determinant of tumor growth and metastasis. Cancer Res. 76, 5201–5208 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Wise D. R., Thompson C. B., Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurmi K., Haigis M. C., Nitrogen metabolism in cancer and immunity. Trends Cell Biol. 30, 408–424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen E. I., et al. , Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 67, 1472–1486 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Shestov A. A., et al. , Simultaneous measurement of glucose transport and utilization in the human brain. Am. J. Physiol. Endocrinol. Metab. 301, E1040–E1049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cengiz E., Tamborlane W. V., A tale of two compartments: Interstitial versus blood glucose monitoring. Diabetes Technol. Ther. 11 (suppl. 1), S11–S16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manohar K., Bhattacharya A., Mittal B. R., Low positive yield from routine inclusion of the brain in whole-body 18F-FDG PET/CT imaging for noncerebral malignancies: Results from a large population study. Nucl. Med. Commun. 34, 540–543 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Yudkoff M., Brain metabolism of branched-chain amino acids. Glia 21, 92–98 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Chen J., et al. , Gain of glucose-independent growth upon metastasis of breast cancer cells to the brain. Cancer Res. 75, 554–565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melone M. A. B., et al. , The carnitine system and cancer metabolic plasticity. Cell Death Dis. 9, 228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo X., et al. , Emerging roles of lipid metabolism in cancer metastasis. Mol. Cancer 16, 76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Opitz C. A., et al. , The therapeutic potential of targeting tryptophan catabolism in cancer. Br. J. Cancer 122, 30–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Q., et al. , Comprehensive analysis of the expression and prognosis for TDO2 in breast cancer. Mol. Ther. Oncolytics 17, 153–168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chowdhry S., et al. , NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 569, 570–575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Weverwijk A., et al. , Metabolic adaptability in metastatic breast cancer by AKR1B10-dependent balancing of glycolysis and fatty acid oxidation. Nat. Commun. 10, 2698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rižner T. L., Penning T. M., Role of aldo-keto reductase family 1 (AKR1) enzymes in human steroid metabolism. Steroids 79, 49–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X., et al. , AKR1B1 promotes basal-like breast cancer progression by a positive feedback loop that activates the EMT program. J. Exp. Med. 214, 1065–1079 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wishart D. S., et al. , DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 46 (D1), D1074–D1082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dupuy F., et al. , PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. 22, 577–589 (2015). [DOI] [PubMed] [Google Scholar]
- 61.LeBleu V. S., et al. , PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 16, 992–1003, 1–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sciacovelli M., Frezza C., Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. FEBS J. 284, 3132–3144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin X., et al. , A metastasis map of human cancer cell lines. Nature 588, 331–336 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferraro G. B., et al. , Fatty acid synthesis is required for breast cancer brain metastasis. Nat. Can. 2, 414–428 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garber K., A new cancer immunotherapy suffers a setback. Science 360, 588 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Roshanzamir F., et al., Metastatic triple negative breast cancer adapts its metabolism to destination tissues while retaining key metabolic signatures. GITHUB. https://github.com/FaribaRoshanzamir/Metastatic-TNBC. Deposited 10 August 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the datasets used in this manuscript are available in public repositories, and references are given in the text (see the “Data retrieval and processing” subsection in SI Appendix, SI Material and Methods). Correspondence and requests for materials should be addressed to nielsenj@chalmers.se. Reconstructed GEMs and ecGEMs for TPs, TMs, HTs and NTs are available at https://github.com/FaribaRoshanzamir/Metastatic-TNBC (66). All other methods and algorithms used in this study are publicly available, and refrences are given in the text (see SI Appendix, SI Material and Methods). All study data are included in the article and/or supporting information.