Abstract
Ess2, also known as Dgcr14, is a transcriptional co-regulator of CD4+ T cells. Ess2 is located in a chromosomal region, the loss of which has been associated with 22q11.2 deletion syndrome (22q11DS), which causes heart defects, skeletal abnormalities, and immunodeficiency. However, the specific association of Ess2 with 22q11DS remains unclear. To elucidate the role of Ess2 in T-cell development, we generated Ess2 floxed (Ess2fl/fl) and CD4+ T cell–specific Ess2 KO (Ess2ΔCD4/ΔCD4) mice using the Cre/loxP system. Interestingly, Ess2ΔCD4/ΔCD4 mice exhibited reduced naïve T-cell numbers in the spleen, while the number of thymocytes (CD4−CD8−, CD4+CD8+, CD4+CD8−, and CD4−CD8+) in the thymus remained unchanged. Furthermore, Ess2ΔCD4/ΔCD4 mice had decreased NKT cells and increased γδT cells in the thymus and spleen. A genome-wide expression analysis using RNA-seq revealed that Ess2 deletion alters the expression of many genes in CD4 single-positive thymocytes, including genes related to the immune system and Myc target genes. In addition, Ess2 enhanced the transcriptional activity of c-Myc. Some genes identified as Ess2 targets in mice show expressional correlation with ESS2 in human immune cells. Moreover, Ess2ΔCD4/ΔCD4 naïve CD4+ T cells did not maintain survival in response to IL-7. Our results suggest that Ess2 plays a critical role in post-thymic T-cell survival through the Myc and IL-7 signaling pathways.
Keywords: CD4+, T-cell transcription, Myc
Abbreviations: ChIP, chromatin immunoprecipitation assay; DAVID, Database for Annotation, Visualization, and Integrated Discovery; DN, double-negative; GSEA, gene set enrichment analysis; KO, knockout; NK, natural killer; qPCR, quantitative polymerase chain reaction
Transcriptional co-regulators contribute to gene expression by interacting with DNA-bound transcription factors (1). The functions of transcription co-regulators are diverse, including histone modifications, chromatin structure conversion, and chromosome structure changes. Because they regulate multiple transcription factors, transcriptional co-regulator functions in vivo are complex and still under investigation. We recently identified Ess2 (also known as Dgcr14 or Es2) as a transcriptional co-regulator of retinoid-related orphan nuclear receptor gamma/gamma-t (RORγ/γt) through biochemical purification and matrix-assisted laser desorption ionization–time of flight mass spectrometry analysis (2).
In humans, the ESS2 gene was first cloned as an expression sequence tag located in the 22q11.2 locus, which is related to the 22q11.2 deletion syndrome (22q11DS; also known as DiGeorge syndrome or CATCH 22 syndrome) (3, 4). 22q11DS is a disorder caused by a small deletion, located near the middle of chromosome 22, specifically designated q11.2 (5, 6, 7). Common signs and symptoms of this syndrome include heart abnormalities, cleft palate, distinctive facial features, and schizophrenia. In addition, 22q11DS patients are often immunodeficient and experience recurrent infections, and some patients develop autoimmune disorders, including rheumatoid arthritis and Graves’ disease (8). While genetically modified mouse models expressing 22q11DS phenotypes have been reported, immunodeficiency-related genes within 22q11.2 have not yet been identified.
RORγ/γt are members of the nuclear receptor superfamily, and they regulate TH17 cell development (9, 10). Ess2 regulates Il17a mRNA levels, which are induced during TH17 cell development through RORγ/γt (2). Additionally, the genes ribosomal S6 kinase 2 (Rsk2; Rps6ka3) and bromodomain adjacent to zinc finger domain 1B (Baz1b) associate with Ess2 to regulate the transcriptional activities of RORγ/γt in TH17 cells (2). Ess2 mRNA expression levels are higher in TH17 cells than in naïve, TH1, TH2, or induced regulatory T cells (iTregs) (2). The Ess2 protein localizes in the nucleus where it interacts with both transcription factors and spliceosomes (11, 12, 13). The function of Ess2 during T-cell development in vivo is poorly understood because homozygous Ess2 mutant mice are embryonically lethal (14).
Our previous studies (2) suggested the possibility that Ess2 regulates T-cell development and differentiation in vivo. To elucidate the function of the Ess2 gene during T-cell development in vivo, we generated Ess2 gene floxed mice (Ess2fl/fl) and CD4-specific Ess2 knockout (KO) mice (Ess2ΔCD4/ΔCD4) by crossing CD4-Cre transgenic mice (15) and Ess2fl/fl mice. Interestingly, these mice survive but have reduced naïve T-cell numbers. Our studies on these mice have elucidated the role of the Ess2 gene during T-cell development.
Results
Generation of Ess2ΔCD4/ΔCD4 mice
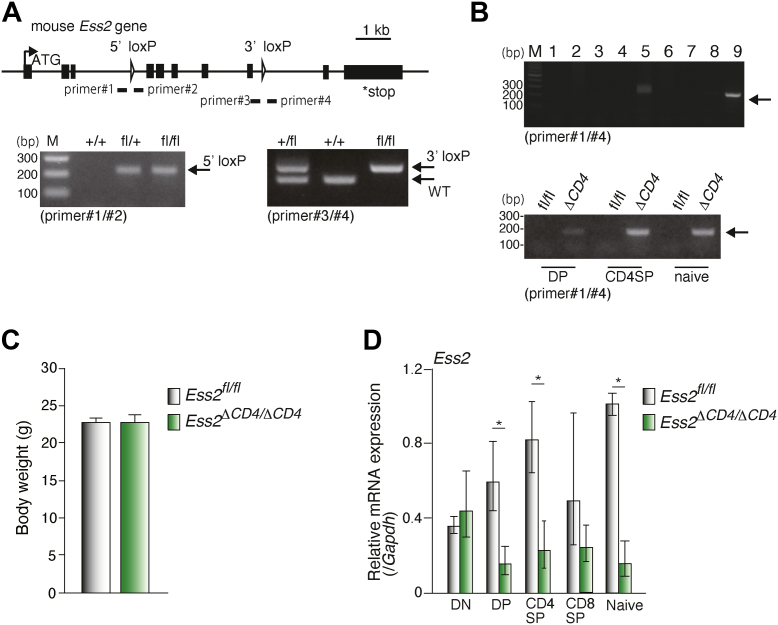
To elucidate Ess2 function in vivo, we generated Ess2ΔCD4/ΔCD4 mice using the Cre-loxP system (Fig. S1A). We first generated Ess2 KO (Ess2−/−) and Ess2+/− mice by inserting a splicing acceptor into the exon 2, FRP, and loxP sequences (Fig. S1A). Consistent with previous reports, Ess2−/− mice were embryonically lethal (Fig. S1, B and C) (14). However, Ess2+/− mice survived and did not have any abnormal appearances or bodyweight loss (data not shown). We then crossed Ess2+/− mice with FLP-expressing transgenic mice (RIKEN No. RBRC01834, see Experimental procedures) to generate Ess2fl/fl mice (Figs. S1A and 1A).
Figure 1.
Generation of Ess2fl/fland Ess2ΔCD4/ΔCD4mice.A, targeting strategy for the generation of CD4+ T-cell-specific Ess2 knockout (Ess2ΔCD4/ΔCD4) mice (upper panel). Examples of the PCR genotyping for Ess2fl/fl (fl/fl), Ess2fl/+ (fl/+), and WT (+/+) mice (bottom panels). B, upper panel, PCR genotyping of tissues prepared from Ess2ΔCD4/ΔCD4 mice. 1; Brain, 2; Lung, 3; Heart, 4; Liver, 5; Kidney, 6; Adipose, 7; Muscle, 8; Testis, and 9; CD4+ T cells; Bottom panel, PCR genotyping for Ess2fl/fl (fl/fl) and Ess2ΔCD4/ΔCD4(ΔCD4) mice in CD4+CD8+ thymocytes (DP), CD4+CD8− thymocytes (CD4SP), and naive T cells (naive). C, the body weights of Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice used in Figures 2 and 3. D, RT-qPCR of Ess2 mRNA in thymocytes and naïve T cells normalized to the level of Gapdh mRNA expression. Three to four mice were used in each experiment. p values were calculated using Student’s t test. ∗p < 0.05. qPCR, quantitative polymerase chain reaction.
To determine the role of Ess2 during T-cell development, we generated Ess2ΔCD4/ΔCD4 by crossing Ess2fl/fl with CD4-Cre transgenic mice [Fig. 1B, (15)]. Ess2fl/fl littermates were used as controls. We confirmed the recombination of the Ess2 allele in CD4 single-positive (CD4SP) and double-positive (DP) thymocytes from Ess2ΔCD4/ΔCD4 mice by the detection of a 152-bp fragment (Fig. 1B). Ess2ΔCD4/ΔCD4 mice appear normal and have a similar body weight compared to Ess2fl/fl mice (Fig. 1C). Ess2 mRNA expression levels were significantly reduced in DP thymocytes isolated from Ess2ΔCD4/ΔCD4 mice but not in double-negative (DN) thymocytes (Fig. 1D). These results demonstrated that the Ess2 gene in Ess2ΔCD4/ΔCD4 mice was partially deleted in DP thymocytes and completely deleted in CD4SP thymocytes.
Ess2ΔCD4/ΔCD4 mice had reduced numbers of peripheral naïve T cells
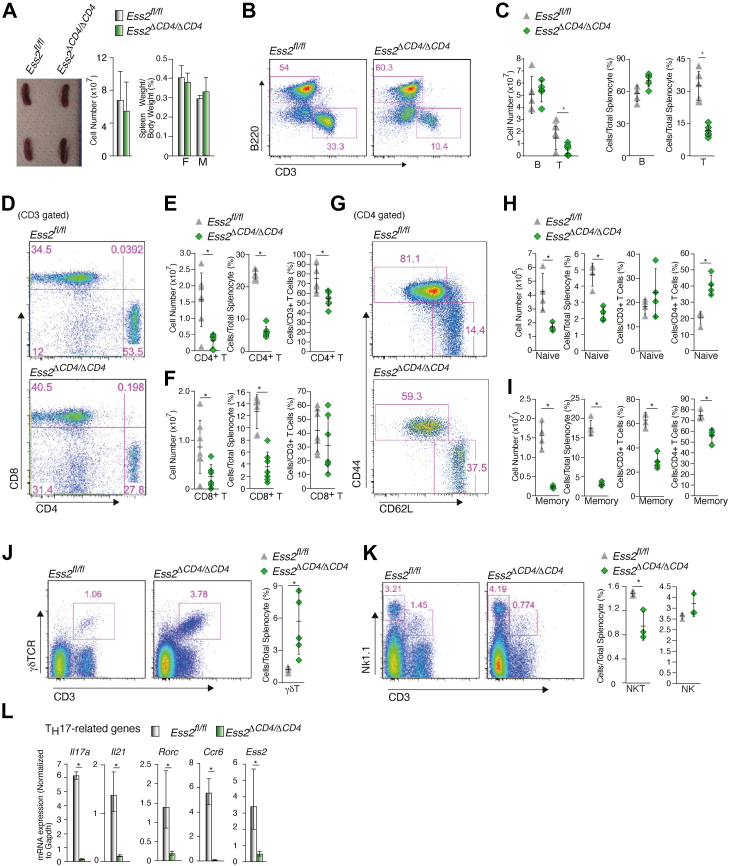
We next analyzed the number of peripheral T cells in the spleens of Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice. There were no abnormalities in the spleen, and the number of splenocytes was similar in both Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice (Fig. 2A). However, the numbers of CD3+ (Fig. 2, B and C), CD4+, and CD8+ T cells (Fig. 2, D–F) were all lower in the spleens of Ess2ΔCD4/ΔCD4 mice compared to Ess2fl/fl mice. The ratios of CD4+ T cells to splenocytes and CD8+ T cells to splenocytes were both significantly decreased in Ess2ΔCD4/ΔCD4 mice (Fig. 2, E and F). The ratio of CD4+ T cells to CD3+ T cells was reduced in Ess2ΔCD4/ΔCD4 mice, whereas the ratio of CD8+ T cells to CD3+ T cells was not significantly different (Fig. 2, E and F). In addition, the number of naïve CD4+ T cells was only 30% of those in Ess2fl/fl mice (Fig. 2, G and H). While the ratio of naïve CD4+ T cells to total splenocytes was also lower in Ess2ΔCD4/ΔCD4 mice, the ratio of naïve CD4+ T cells to total CD4+ T cells was higher, when compared to the Ess2fl/fl mice (Fig. 2H). However, unlike the naïve cells, the ratio of Ess2ΔCD4/ΔCD4 memory CD4+ T cells to total CD3+ T cells or CD4+ T cells was significantly lower than that of the Ess2fl/fl mice (Fig. 2I).
Figure 2.
Ess2ΔCD4/ΔCD4mice show aberrant T-cell development in splenocytes.A, representative spleens (left panel), quantification of splenocytes (middle panel), and spleen weights (right panel) in Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice. Scale bar = 1 mm. For flow cytometry, the following experiments were carried out under the same conditions: panels B and C; panels D–F; and panels G–I. B, representative population of B and T cells among the splenocytes of Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice as measured by flow cytometry. C, quantification of B cells and T cells in Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice. D, a representative population of CD4+ or CD8+ T cells as assessed by flow cytometry. E, quantification of CD4+ T cells in splenocytes and total CD3+ T cells. F, quantification of CD8+ T cells in splenocytes and total CD3+ T cells from. G, a representative population of naïve and memory CD4+ T cells as assessed by flow cytometry. H, quantification of naïve CD4+ T cells in total splenocytes, total CD3+ T cells, and total CD4+ T cells from splenocytes. I, quantification of memory CD4+ T cells in total splenocytes, total CD3+ T cells, and total CD4+ T cells from splenocytes. J, a representative population of γδT cells as assessed by flow cytometry (left and middle panels) and the quantification of γδT cells in splenocytes (right panel). K, a representative population of NKT cells as assessed by flow cytometry (left and middle panels) and the quantification of NKT cells in splenocytes (right panel). L, RT-qPCR analysis of primary CD4+ T cells cultured under TH17 conditions. The mRNA levels of all genes were normalized to the level of Gapdh mRNA expression. Each experiment was performed at least three times and the results are presented as the mean ± SD. Each experiment used 3 to 7 mice. p values were calculated using Student’s t test, ∗p < 0.05. qPCR, quantitative polymerase chain reaction.
Interestingly, Ess2ΔCD4/ΔCD4 mice had significantly more γδT cells and fewer natural killer (NK) T cells than Ess2fl/fl mice (Fig. 2, J and K). There was no significant difference in regulatory T cells (Fig. S2A). Next, naïve CD4+ T cells were cultured under TH17 conditions (Fig. 2L). As expected, the mRNA expression levels of TH17-related genes (Il17a, Il21, Rorc, and Ccr6) were lower in the Ess2ΔCD4/ΔCD4 mice. These data suggest that the loss of the Ess2 gene alters T-cell development in the spleen of Ess2ΔCD4/ΔCD4 mice.
Ess2 does not contribute to DN/DP selection in the thymus
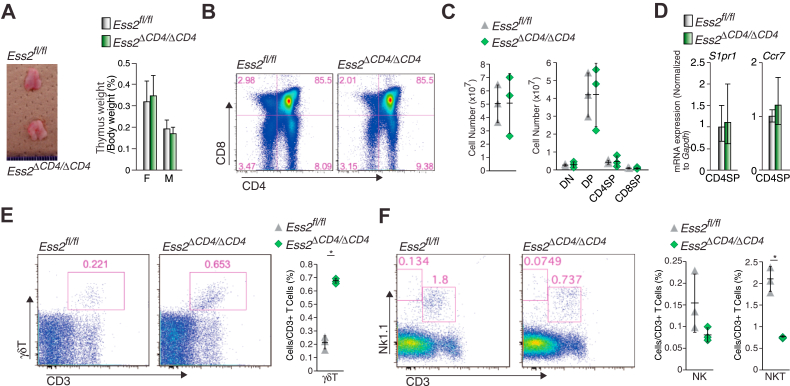
Our results indicated that Ess2 regulates naïve T-cell development and/or proliferation. To determine at what stage Ess2 is active in T-cell development, we determined the number of CD4SP and CD8SP thymocytes in the thymuses of Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice. Ess2 mRNA expression levels in DN and CD8SP thymocytes did not differ between Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice (Fig. 1D). These results suggest that the deletion of Ess2 in Ess2ΔCD4/ΔCD4 mice occurs during the DP stage. Thymus sizes and weights were similar in both mouse genotypes (Fig. 3A). Both positive and negative thymocyte selection occurred normally in Ess2ΔCD4/ΔCD4 mice (Figs. 3, B and C, and S2B). We next determined the expression of two thymocyte regulatory genes (Ccr7 and S1pr1) in CD4SP thymocytes by RT-quantitative PCR (qPCR) (Fig. 3D). The Ccr7 gene product regulates the export of positively selected thymocytes from the thymus (16, 17), and S1pr1 is highly upregulated in T-cell development to promote the exit of mature T cells from the thymus (18). Ccr7 and S1pr1 mRNA expression levels did not differ between Ess2ΔCD4/ΔCD4 and Ess2fl/fl mice (Fig. 3D). Splenic γδT cells (Fig. 3E) also increased, while NKT cells decreased (Fig. 3F) in the thymus of Ess2ΔCD4/ΔCD4 mice. These results indicate that Ess2 does not contribute to the positive/negative selection of T cells or their migration from the thymus. However, Ess2 does affect the development of γδT cells and NKT cells in the thymus.
Figure 3.
Ess2fl/fland Ess2ΔCD4/ΔCD4mice exhibit similar numbers of thymocytes.A, representative thymus (left panel) and thymus weights (right panel) of Ess2fl/fl and Ess2ΔCD4/ΔCD4. Scale bar = 1 mm. B, representative flow cytometry plots of CD4−CD8− (DN), CD4+CD8− (CD4P), CD4−CD8+ (CD8SP), and CD4+CD8+ (DP) thymocytes as assessed by flow cytometry. C, the whole cell number of thymocytes (left panel), DN, CD4SP, CD8SP, and DP cells in B (right panel). D, RT-qPCR of CD4SP-related genes (S1pr1 and Ccr7) in CD4SP cells collected from Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice. Three to five mice were used in each experiment. E, a representative population of γδT cells as assessed by flow cytometry (left and middle panels) and the quantification of γδT cells in thymocytes (right panel). F, a representative population of NKT cells as assessed by flow cytometry (left and middle panels) and the quantification of NKT cells in thymocytes (right panel). Each experiment used three mice. p values were calculated using Student’s t test, ∗p < 0.05. F, female; M, male; qPCR, quantitative polymerase chain reaction.
Ess2 regulates mRNA gene expression in CD4SP cells
Our results showed that an Ess2 deletion in CD4SP thymocytes reduces naïve T-cell numbers; however, the molecular mechanism is poorly understood. To investigate this mechanism, we compared the expression of several CD4+ T-cell-related genes in CD4SP thymocytes and naïve T cells from Ess2fl/fl and Ess2ΔCD4/ΔCD4mice; we did not observe any significant differences (Fig. S3).
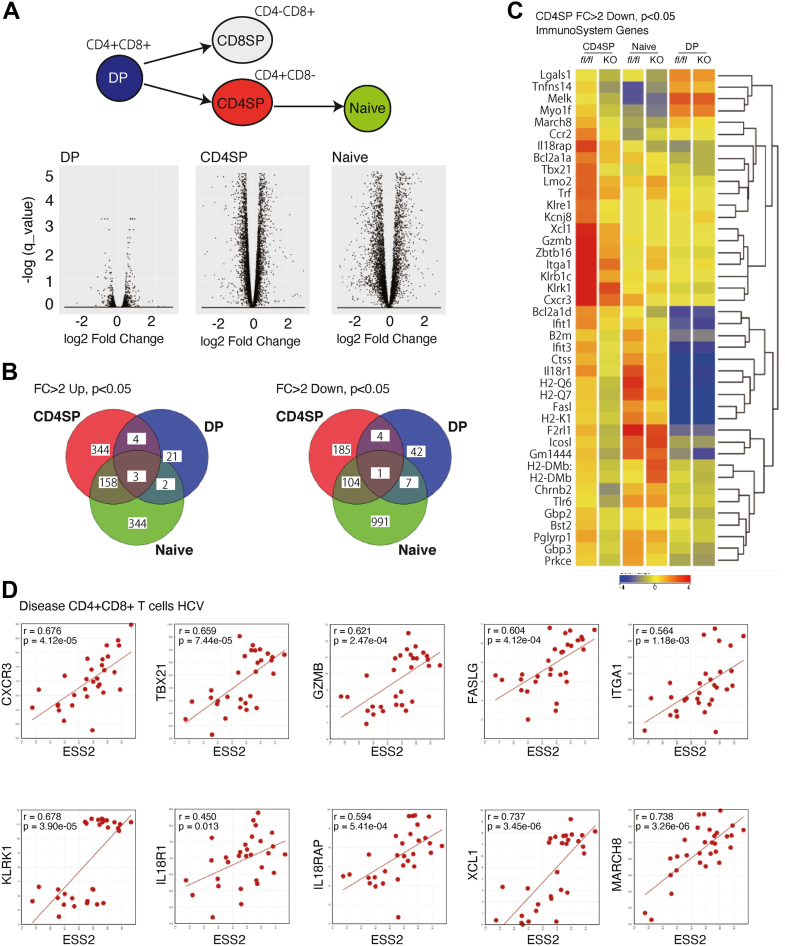
To elucidate the role of Ess2 in naïve T-cell maintenance, we analyzed the genome-wide RNA expression profiles of DP and CD4SP thymocytes as well as naïve CD4 T-cell splenocytes using RNA-seq (Figs. 4A and S4, A and B). Surprisingly, we observed many Ess2-dependent alterations in mRNA expression in CD4SP thymocytes but only a few in DP thymocytes (Fig. 4, A and B). Only a few splicing defects were seen in Ess2ΔCD4/ΔCD4 mice (data not shown). Although previous studies show that Ess2 interacts with both the spliceosome and transcription factors (13), our results indicate that Ess2 gene deficiency affects transcription starting at the CD4SP stage of development. However, the groups of genes differentially expressed between Ess2ΔCD4/ΔCD4 and Ess2fl/fl mice in CD4SP and naïve T cells overlap by only 10 to 30% (Fig. 4B). These results suggest that Ess2 regulates different gene sets in CD4SP cells than in naïve T cells.
Figure 4.
RNA-seq analysis of CD4SP, DP, and naïve T cells from Ess2ΔCD4/ΔCD4and Ess2fl/flmice.A, a volcano plot of CD4SP, DP, and naïve T cells from Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice. B, a Venn diagram of fold change (FC) in DP thymocytes, CD4SP thymocytes, and naive T cells from Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice. C, heatmap of immune system genes with a fold change (FC) <2 down in CD4SP thymocytes from Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice. The immune system gene set was defined by Strand NGS. D, correlation between human ESS2 and some immune system genes in humans using the R2 database (disease CD4+CD8+ T cells HCV).
We confirmed the changes in expression of immune-related genes in CD4SP cells; 42 genes had more than a two-fold difference in expression and some genes were significantly differently expressed in naïve T cells but not DP cells (Fig. 4C). To further investigate the clinical significance of ESS2 expression, we used the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). Interestingly, several Ess2-regulated genes were positively correlated with ESS2 expression in the human disease database of CD4+ and CD8+ T cells isolated from hepatitis C virus patients (19) (GEO source: GSE49954, Fig. 4D). These results suggest that they are involved in the ESS2-dependent regulation of the immune system in humans and that ESS2 expression may be a favorable prognostic factor in autoimmune diseases.
Ess2 regulates Myc target genes
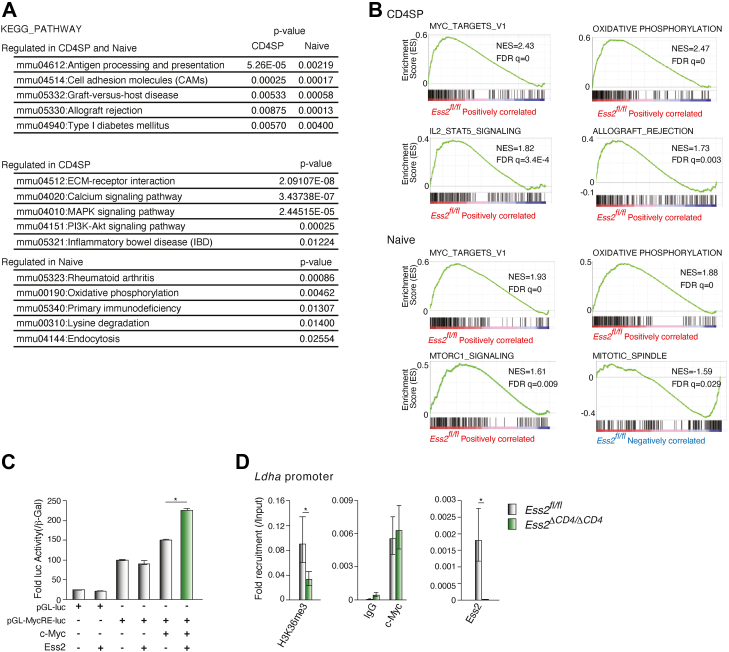
Next, we performed a Database for Annotation, Visualization, and Integrated Discovery (DAVID) pathway analysis and a gene set enrichment analysis (GSEA). The DAVID pathway analysis revealed that genes involved in antigen processing and presentation, cell adhesion, graft-versus-host disease, allograft rejection, and type I diabetes mellitus were significantly altered in both Ess2ΔCD4/ΔCD4 CD4SP thymocytes and naïve T cells compared to Ess2fl/fl cells (Fig. 5A). These results show that Ess2 transcriptionally regulates genes from multiple immune pathways.
Figure 5.
DAVID and GSEA analyses in Ess2fl/fland Ess2ΔCD4/ΔCD4mice.A, a DAVID analysis of highly scored gene sets in CD4SP thymocytes or naïve T cells. B, representative results of a GSEA analysis in CD4SP and naïve T cells. Among high-scoring gene sets, the immune system–related gene sets were selected (Table S2). C, luciferase reporter assay using the luciferase reporter (Myc3Elb-luc), which contains three Myc-binding elements. After transfection of each vector, the HEK293 cells were lysed and a luciferase reporter assay performed. Transcriptional activity was normalized to β-gal activity. D, ChIP-qPCR analysis of the Ldha promoter using antibodies to Ess2, c-Myc, and histone H3K36me3 in primary naïve CD4+ T cells. Isolated naïve CD4+ T cells were lysed, and the ChIP analyses were performed using the antibodies indicated. qPCR was performed with the primers described in Table S1. Each experiment was performed at least three times, and the results are presented as the mean ± SD. p values were calculated using a Student’s t test, ∗p < 0.05. ChIP, chromatin immunoprecipitation assay; DAVID, Database for Annotation, Visualization, and Integrated Discovery; GSEA, gene set enrichment analysis; qPCR, quantitative polymerase chain reaction.
We next analyzed the HALLMARK gene sets with a GSEA. Both the Myc and oxidative phosphorylation pathways were also altered in Ess2ΔCD4/ΔCD4 CD4SP thymocytes and naïve T cells (Fig. 5B). In Ess2ΔCD4/ΔCD4 CD4SP thymocytes, IL-2/STAT5-signaling- and allograft-rejection-related gene sets were altered. Other genetic pathways were also changed in Ess2ΔCD4/ΔCD4 mice (Fig. S6A and Table S2). Myc acts as an essential transcription factor in T cells, regulating metabolic reprogramming and proliferation (20). Interestingly, a recent report demonstrated an interaction between Myc and Ess2 through proteomic analysis (21). Based on these observations, we hypothesized that Ess2 acts as a transcriptional co-regulator of Myc. To elucidate the association between Ess2 and Myc, a luciferase reporter assay was performed by overexpressing Ess2 and c-Myc with a luciferase reporter vector (Myc3Elb-Luc) that contained three Myc-binding elements in the HEK293 cell line. As shown in Figure 5C, Ess2 enhanced the transcriptional activity of c-Myc in a dose-dependent manner. Additionally, chromatin immunoprecipitation assay (ChIP)-qPCR analysis demonstrated that naïve CD4+ T cells from Ess2ΔCD4/ΔCD4 mice decrease the recruitment of Ess2 to the Ldha promoter, which contains a Myc-binding element (22). Although naïve CD4+ T cells from Ess2ΔCD4/ΔCD4 mice did not alter the recruitment of c-Myc to the Ldha promoter (Fig. 5D), the levels of the transcriptionally active histone mark, histone H3K36me3, decreased in these cells. Moreover, immunofluorescent staining of overexpressed c-Myc and Ess2-GFP in U2OS cells revealed their co-localization in the nucleus (Fig. S5). These results establish that Ess2 has a direct impact on the transcriptional activity of c-Myc.
ESS2 expression correlates with T-cell-related genes in immune disease patients
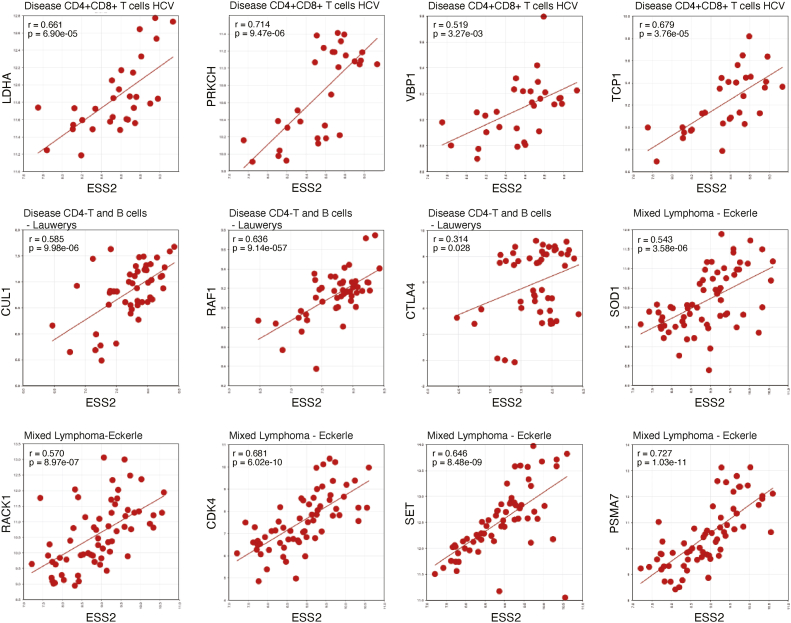
Next, we examined the R2 database and found a significant, positive correlation between ESS2 expression and the T-cell-related genes identified by the GSEA, specifically in lymphocytes of humans infected with hepatitis C virus, Mixed Lymphoma - Eckerle [(23), GEO source: GSE14879], Disease CD4-T and B cells - Lauwerys (GEO source: GSE4588), and mixed Crohn disease [(24), GEO source: GSE9686] (Figs. 6 and S6, A and B). As expected, ESS2 expression levels significantly correlated with some of the GSEA-identified genes in human patients. These results show that ESS2 may have similar functions in humans and mice, which is through Myc-targeted gene regulation.
Figure 6.
Correlation between human ESS2 and Myc target genes. The correlation between ESS2 and identified Myc target genes in humans using the R2 database. The datasets used are shown in each graph.
Ess2 regulates IL-7-dependent CD4+ T-cell maintenance
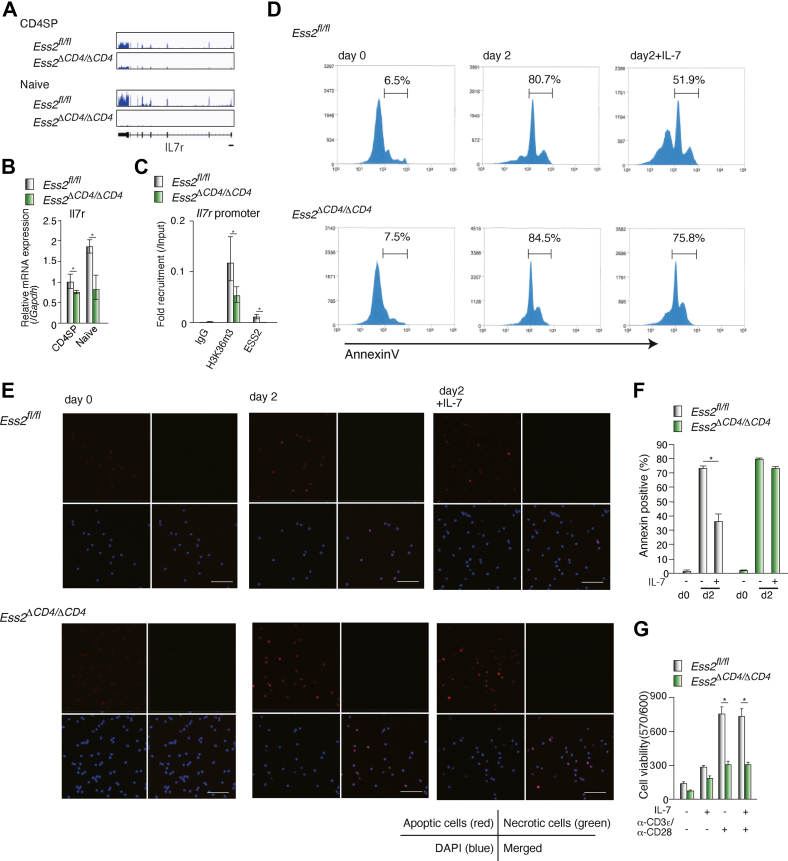
The IL-7 signaling pathway regulates naïve T-cell survival and proliferation (25, 26), and since IL-7R mRNA was reduced in CD4SP cells isolated from Ess2ΔCD4/ΔCD4 mice (Figs. 7, A and B and S4B; Table S3), we hypothesized that Ess2 may regulate IL-7-dependent CD4+ T-cell maintenance. ChIP-qPCR analysis showed that deletion of the Ess2 gene decreased the recruitment of the histone H3K36me3 marker to the Il7r promoter (Fig. 7C). We cultured naïve CD4+ T cells from Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice with or without IL-7 for 3 days. As expected, IL-7 maintained the survival of cells isolated from Ess2fl/fl but not from Ess2ΔCD4/ΔCD4 (Fig. 7D). We observed a similar number of apoptotic Ess2ΔCD4/ΔCD4 naïve CD4+ T cells regardless of the IL-7 treatment (Fig. 7, E and F). Although IL-2 synergizes with IL-7 to enhance T-cell proliferation, we did not observe alterations in the IL-2-related gene expression of Ess2ΔCD4/ΔCD4 naïve CD4+ T cells (Fig. S3). Next, we examined cell survival, stimulated by anti-CD3ε/CD28 antibodies, with and without IL-7. As shown in Figure 7G, Ess2ΔCD4/ΔCD4 naïve CD4+ T cells reduced TCR-activation-dependent proliferative effects. These results suggest that Ess2 regulates naïve T-cell survival through the IL-7 signaling pathway.
Figure 7.
Ess2 regulates IL-7-dependent cell survival in naïve CD4+T cells.Ess2fl/fl and Ess2ΔCD4/ΔCD4 naïve T cells were cultured with or without 100 ng ml−1 of IL-7. A, RNA-seq peaks in the IL7r promoter from CD4SP cells and naïve T cells by IGV. B, RT-qPCR of Il7r in CD4SP cells and naïve T cells normalized to the level of Gapdh mRNA expression. Three to five mice were used in each experiment, ∗p < 0.05. C, ChIP-qPCR analysis of the Il7r promoter with anti-Ess2 and anti-histoneH3K36me3 antibodies in primary naïve CD4+ T cells. D, representative apoptotic cells stained by Annexin V using flow cytometry (MoFlo XDP). Central and left peaks were Annexin-positive cells, and the percentages are described. E, representative confocal microscopy of apoptotic (red), necrotic (green), and DAPI (blue) staining; Scale bar = 50 μm. F, quantification of apoptotic cells. The number of cells counted is from 50 to 80. Each sample was counted in at least five areas. G, Presto blue staining of naïve CD4+ T cells stimulated with and without anti-CD3ε/CD28 antibodies and/or IL-7 for 2 days. Each experiment was performed at least three times, and the results are presented as the mean ± SD. p values were calculated using a Student’s t test, ∗p < 0.05. ChIP, chromatin immunoprecipitation assay; qPCR, quantitative polymerase chain reaction.
Discussion
In this study, we successfully generated Ess2fl/fl mice and CD4-specific Ess2 KO mice for the first time. As previously described (14), we found that Ess2−/− mice had an early embryonic lethality (Fig. S1C). These results indicate that Ess2 plays an essential role in early embryogenesis, although the precise mechanism is unclear.
Unexpectedly, Ess2ΔCD4/ΔCD4 mice produced reduced CD3+ T-cell and NKT-cell numbers but had increased numbers of γδT cells. In Ess2ΔCD4/ΔCD4 mice, the ratio of CD4+ T cells to splenocytes or total CD3+ T cells decreased. Additionally, the ratio of naïve CD4+ T cells to total CD4+ T cells increased, while the ratio of memory CD4+ T cells to CD3+ T cells or total CD4+ T cells decreased. Such alterations in the ratios of naïve and memory CD4+ T cells in Ess2ΔCD4/ΔCD4 mice may be due to the different impact that ESS2 has on cell survival in naïve or memory CD4+ T cells. Further studies will be required in order to characterize this mechanism. Genome-wide RNA expression analyses revealed that Ess2 transcriptionally regulates several genes in thymic CD4SP cells. In particular, the expression of Myc target genes and oxidative phosphorylation pathway genes was decreased significantly in Ess2ΔCD4/ΔCD4 CD4SP thymocytes and naïve T cells, which may be the cause of reduced naïve T-cell counts in these mice. Interestingly, our findings demonstrated that Ess2 acts as a transcriptional co-regulator for the transcriptional activity of c-Myc and that Ess2 is co-localized with c-Myc in the nucleus. In addition, the expression levels of several Myc target genes, such as LDHA, RACK1, and CDK4, are correlated with ESS2 expression in immunodeficient patients (Fig. 6). These results suggest that Ess2 controls CD4+ T-cell survival and homeostasis via transcriptional regulation through c-Myc and RORγ/γt (Fig. S7). Additional studies may elucidate the detailed mechanism(s) behind Ess2-dependent transcriptional activation. Importantly, our findings show that deletion of Ess2 caused transcriptional dysfunction, rather than splicing defects. These findings suggest a significant role for Ess2 as a transcriptional co-regulator in the immune system.
Additionally, our findings show that Ess2ΔCD4/ΔCD4 mice exhibit dysregulation in both γδT and NKT cells. γδT cells develop from DN thymocytes in the thymus, and some transcription factors that regulate γδT-cell development have been identified (27). One such regulator is IL7R; CD4-Cre IL7R deficient mice similarly increased the number of thymic DN and γδT cells (28). Because Ess2ΔCD4/ΔCD4 naïve T cells exhibited reduced Il7r mRNA levels (Fig. 7, A and B), as well as disrupted IL-7 signal-dependent cell survival (Fig. 7, D–G), we conclude that Ess2-dependent regulation of the IL-7 signaling pathway may be required to maintain γδT-cell survival. It is unclear, however, whether Ess2 regulates the function of STAT5 and other downstream transcription factors in the IL-7 signaling pathway. Furthermore, there are many transcription factors involved in γδT-cell differentiation (27), and it is unclear which of these are regulated by Ess2. Future studies will focus on elucidating the function of Ess2 in γδT cells.
NKT cells are a heterogeneous subset of T lymphocytes that are developmentally and functionally distinct from conventional CD4+ and CD8+ T cells (29). While T cells and NKT cells both originate in the thymus, their selection requirements are divergent. NKT-cell development occurs in the thymus from a common precursor pool of DP thymocytes and requires several transcription factors, such as NF-κB, GATA3, and RORγt. Because Ess2 localizes to the nucleus and regulates RORγ/γt activity, NKT cells in Ess2ΔCD4/ΔCD4 mice may reduce RORγ/γt activity in DP thymocytes. Additional studies are required to elucidate the role of Ess2 on the transcriptional regulation of NKT cells.
Interestingly, human ESS2 expression correlated with the expression of several immune- and oxidative-phosphorylation-related genes in autoimmune disease patients (Figs. 4D and 6). These results suggest that Ess2 may also contribute to immunity and metabolism in humans. However, a more detailed analysis is necessary, and a functional analysis of Ess2 in a conditional KO mouse model may help elucidate the molecular mechanism of Ess2-related immune abnormalities. Our previous studies showed that Ess2 induces TH17 differentiation by enhancing the transcriptional activities of RORγ/γt (2). In Ess2ΔCD4/ΔCD4 mice, TH17-related gene mRNA levels were also suppressed in primary CD4+ T cells cultured under TH17 conditions (Fig. 2L). However, in this study, we found that Ess2 affects earlier stages of T-cell maturation which are independent of RORγ/γt. For future studies, additional Cre transgenic mice will be necessary to investigate the function of all TH cells in vivo.
RORγ KO mice regulate naïve T-cell proliferation through Bcl2 expression (30), which did not occur in Ess2ΔCD4/ΔCD4 mice (data not shown). These results show that the Ess2 protein associates with several transcription factors to regulate their activity, which may also contribute to Ess2-specific immune regulation.
Several mouse models of 22q11DS exist (31), including 22q11.2 deletion heterozygous mice and 22q11DS-associated transgenic mice (32, 33, 34). A recent study has reported the generation of a CRISPR-Cas-mediated deletion of 22q11DS (35). Several 22q11DS phenotypes can be analyzed using these mouse models, such as cardiac development, abnormal cranial base morphology, and schizophrenia. However, the causal genes within the 22q11.2 deletion region that are related to immunodeficiency have not yet been reported. We measured the concentration of several cytokines from the blood of Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice under normal conditions; however, no significant differences were observed (data not shown). Our results suggest that a more detailed analysis of Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice may provide additional insights into 22q11DS as it regards immunodeficiency.
In summary, we generated both Ess2fl/fl and Ess2ΔCD4/ΔCD4 mice and have demonstrated a role for Ess2 in post thymic T-cell survival. These findings will enable future work to elucidate the role of Ess2 in the regulation of the immune system.
Experimental procedures
Generation of heterozygous Ess2 (Ess2+/−), Ess2fl/f, and Ess2ΔCD4/ΔCD4 mice
To generate Ess2+/− mice, we purchased the Ess2 targeting vector (PG00129_Y_4_A10, Wellcome Trust Sanger Institute) from BACPAC Resources Center CHORI (Fig. S1A). The targeting vector was linearized and electroporated into RENKA embryonic stem (ES) cell lines derived from the C57BL/6 mouse strain (TransGenic Inc). After screening by genomic southern blot and PCR, Ess2 gene-targeted ES cells were isolated. Positive ES cell clones were then injected into blastocysts derived from C57BL/6 mice. Successfully injected embryos were transferred into the uteri of pseudopregnant foster mothers and the resulting chimeric mice crossed with C57BL/6 mice. Ess2+/− mice were identified in the F1 generation, demonstrating the germ-line transmission of the Ess2 mutation. Ess2+/− mice were then backcrossed for five generations with C57BL/6 background mice to obtain Ess2+/− mice. We generated the Ess2−/− mice by crossing male and female Ess2+/− mice. The heterozygous deletion of Ess2 was confirmed by genomic southern blotting (Fig. S1B).
To generate Ess2fl/fl mice (Fig. S1A), Ess2+/− mice were crossed with FLPe transgenic mice [RIKEN, BRC No. 01834_B6-Tg(CAG-FLPe)36 (36)]. Ess2fl/fl mice were then crossed with CD4-Cre transgenic mice (15), and finally male and female Ess2ΔCD4/+ mice were crossed to generate the Ess2ΔCD4/ΔCD4 mice. Ess2ΔCD4/ΔCD4 mice were confirmed with genotyping PCR using primer sets #1/#2, #3/#4 (Fig. 1A and Table S1), and #1/#4 (Fig. 1B).
All mice were raised in specific pathogen-free conditions. Animal experiments were performed according to national and institutional animal care and ethical guidelines and were approved by Nihon University.
Antibodies
For flow cytometry analysis, antibodies specific for CD3e (2C11), CD19 (6D5), CD11b (M1/70), CD11c (N418), CD4 (RM4-5), CD8a (GK1.5 or 53.67), DX5 (DX5), Gr-1 (RBC-8C5), Ter-119 (Ter-119), and CD62L (MEL14) were purchased from BD Biosciences, BioLegend, and eBioscience.
For ChIP-qPCR analysis, an Ess2 specific antibody was generated in our laboratory (13), and an antibody, specific for c-Myc (9402S), was purchased from Cell Signaling Technology.
Flow cytometry
Splenocytes and thymocytes were separated from 8- to 16-week-old mice and subjected to hypotonic red blood cell lysis (150 mM NH4Cl, 15 mM NaHCO3, and 0.1 mM EDTA) to generate a single-cell suspension. Single-cell suspensions were stained in FACS buffer (0.5% BSA, 2 mM EDTA in PBS) with Fc block containing Zombie red (Biolegend). All data were acquired on a BD LSR FortessaTM (BD Bioscience) and analyzed using FlowJo software (TreeStar).
RNA isolation and quantitative RT-PCR
Total RNA was extracted using TRIzol (Invitrogen). First-strand cDNA was synthesized from total RNA using PrimeScript Reverse Transcriptase (Takara Bio Inc). For qPCR, an ABI PRISM7000 (Thermo Fisher Scientific) or StepOne (Applied Biosystems) system was used with the Light Cycler SYBR Green I Master Mix (Takara). The relative quantitation value is expressed as 2−ΔCt, where ΔCt is the difference between the mean cycle threshold (Ct) values of triplicates for each sample and that of the Gapdh control. Primer sequences were previously described (2) and are listed (Table S1).
RNA-seq and bioinformatics
CD4 single-positive (CD4SP, CD4+CD8−), double-positive (DP, CD4+CD8+), and naïve T cells (CD4+CD62L+CD44−CD25−) were sorted with a FACSAria II cell sorter (BD Biosciences), and RNA was extracted with TRIzol (Invitrogen). Single-end RNA-seq was performed using the Illumina HiSeq 2500 (Illumina, Inc). We obtained 101-bp single reads from cDNA fragments. For data analysis, tRNA and rRNA reads were first filtered out using the UCSC Genome Browser. RNA-seq reads were aligned to the mouse genome (mm10 downloaded from Ensembl Genome Browser) using the software TopHat (version 2.0.8 (37)). The raw read counts for each gene were analyzed using the edgeR package.
Gene expression of annotated transcripts was calculated from mapped RNA-seq reads using Cuffdiff to obtain FPKM (fragments per kilobase of exon per million fragments mapped)-normalized gene expression values (38). Some data were normalized and visualized using Strand NGS software (Strand Life Sciences).
To visualize RNA-Seq signals, Bam files were generated and loaded into IGV [Integrative Genomics Viewer, Broad Institute, https://www.broadinstitute.org/igv/ (39)]. p-values of gene-list enrichment were calculated using binomial tests using the R software package. All RNA-seq analyses were performed using three biological replicates.
DAVID analysis and GSEA
RNA-seq data were normalized, and gene ontology term and pathway analyses were performed using the DAVID v6.8 (https://david.ncifcrf.gov/). For gene annotation enrichment analysis, gene names were input into the functional annotation-clustering tool in DAVID (40, 41). Functional annotation clustering was performed with the default criteria, and the enrichment score for each annotation cluster was determined. GSEA was performed using the GSEA software package (GSEA v2.2.3), and all gene set files were obtained from the GSEA website (www.broadinstitute.org/gsea/).
Primary naïve T-cell culture
Naïve CD4+ T cells (CD3+, CD4+, CD62Lhigh, CD44Nega) from spleens were isolated using the Naive CD4+ T-Cell Isolation Kit (Miltenyi Biotec) and cultured in 10% FBS RPMI1640 with or without 100 ng ml−1IL-7. Apoptotic and necrotic cells were identified using Apoptotic and Necrotic Detection Kits (AAT Bioquest Inc) by fluorescence confocal microscopy (LSM710, Carl Zeiss), and apoptotic cells were counted using ImageJ software (NIH). For measuring cell viability, primary cultured naïve T cells were stained with a PrestoBlue Cell Viability Reagent (A13261; Thermo Fisher Scientific) and fluorescence excision was measured at 570 nm and emission 600 nm using the Flex Station 3G (Molecular Devices).
For TH17 cell differentiation, naïve T cells were cultured under TH 17 conditions (1 μg ml−1 anti-CD3ε, 1 μg ml−1 anti-CD28, 1 μg ml−1 anti-IL-4, 1 μg ml−1 anti-IFN-γ, 10 ng ml−1 IL-6 [Peprotech], and 2 ng ml−1 TGF-β) for 3 days, and then the RNA was extracted.
Luciferase reporter assays
For the luciferase reporter assays, HEK293 cells were transfected using the Lipofectamine(R) 2000 (Thermo Fisher Scientific) reagent according to the manufacturer's instructions (42). Twenty-four hours after transfection, the cells were harvested and assayed for luciferase and β-galactosidase activity using a luminometer and a microplate reader (PHERAStar FS, BMG LABTECH). Co-transfection experiments used 150 ng of the reporter plasmid, 10 ng of the pCMX-β-galactosidase expression plasmid, 25 ng of the c-Myc expression plasmid, and 25 ng of the Ess2 expression plasmid for a total 210 ng of the plasmids. Plasmid concentrations were adjusted to equimolar concentrations with the addition of an empty pcDNA3 plasmid for each well in a 96-well plate. Luciferase data were normalized to the internal β-galactosidase control, and all results are presented as the mean ± standard deviation (SD). All transfections were performed in triplicate and repeated at least twice in independent experiments.
ChIP-qPCR analysis
A ChIP was performed according to the manufacturer’s instructions (MAGnify ChIP; Thermo Fisher Scientific). Briefly, cells were fixed with 1% formaldehyde (Sigma) and chromatin was sheared by sonication to average lengths between 300 and 500 bp. Chromatin was immunoprecipitated with control IgG or specific antibodies overnight at 4 °C and then incubated with protein A magnetic beads for an additional 2 h. After washing and elution, protein-DNA cross-links were disrupted by heating at 55 °C. Purified DNA was analyzed by qPCR using the StepOne system with SYBR green (Takara Bio). The relative quantitation value was expressed as 2−ΔCT, where ΔCT is the difference between the mean CT value, for triplicates of the sample, and that of the input control. The primer sequences used are shown in Table S1.
Statistical analyses
Data are presented as mean ± the SD. Equality of variances was assessed using an F-test. For all statistical analyses, except the RNA-seq analyses, comparisons between two groups were made using a two-tailed Student’s t test or two-tailed Welch’s t test, when the variances were equal or unequal, respectively. p values less than 0.05 were considered statistically significant. For GSEA, an FDR q-value less than 0.05 was considered statistically significant.
Data availability
The raw data of RNA-seq are publicly available on the GEO repository (Accession No. PRJNA575280). Ess2+/− mice (BRC09772) and Ess2fl/fl mice (RBRC09771) were registered with the RIKEN BRC. All remaining data are contained within this article and the supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of the article.
Acknowledgments
We thank the members of Prof Makishima's laboratory for helpful discussions and M. Mito (RIKEN) for technical assistance with RNA-seq and southern blotting. We appreciate the Collaborative Research Resources, School of Medicine, Keio University, for technical assistance with our experiments.
Author contributions
I. T. and S. T. conceptualization; I. T., S. H., K. Y., H. O., M. T., and A. Y. methodology; I. T., S. H., S. T., and S. N. data curation; I. T. writing-original draft; S. S. and H. O. software; S. S. and H. O. validation; T. N., T. K., S. N., and M. M. supervision; M. M. writing-review and editing.
Funding and additional information
This study was supported by the Mitsui Life Social Welfare Foundation, Bristol-Myers Squibb, and a Grant-in-Aid for Scientific Research (C; 25462382 and 18K09162).
Edited by Peter Cresswell
Contributor Information
Ichiro Takada, Email: takada.ichiro@nihon-u.ac.jp.
Makoto Makishima, Email: makishima.makoto@nihon-u.ac.jp.
Supporting information
References
- 1.Rosenfeld M.G., Lunyak V.V., Glass C.K. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 2.Takada I. DGCR14 induces Il17a gene expression through the RORgamma/BAZ1B/RSKS2 complex. Mol. Cell. Biol. 2015;35:344–355. doi: 10.1128/MCB.00926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindsay E.A., Rizzu P., Antonacci R., Jurecic V., Delmas-Mata J., Lee C.C., et al. A transcription map in the CATCH22 critical region: identification, mapping, and ordering of four novel transcripts expressed in heart. Genomics. 1996;32:104–112. doi: 10.1006/geno.1996.0082. [DOI] [PubMed] [Google Scholar]
- 4.Rizzu P., Lindsay E.A., Taylor C., O'Donnell H., Levy A., Scambler P., et al. Cloning and comparative mapping of a gene from the commonly deleted region of DiGeorge and Velocardiofacial syndromes conserved in C. elegans. Mamm. Genome. 1996;7:639–643. doi: 10.1007/s003359900197. [DOI] [PubMed] [Google Scholar]
- 5.Scambler P.J. The 22q11 deletion syndromes. Hum. Mol. Genet. 2000;9:2421–2426. doi: 10.1093/hmg/9.16.2421. [DOI] [PubMed] [Google Scholar]
- 6.Robin N.H., Shprintzen R.J. Defining the clinical spectrum of deletion 22q11.2. J. Pediatr. 2005;147:90–96. doi: 10.1016/j.jpeds.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 7.McDonald-McGinn D.M., Sullivan K.E. Chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/Velocardiofacial syndrome) Medicine (Baltimore) 2011;90:1–18. doi: 10.1097/MD.0b013e3182060469. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan K.E. Chromosome 22q11.2 deletion syndrome and DiGeorge syndrome. Immunol. Rev. 2019;287:186–201. doi: 10.1111/imr.12701. [DOI] [PubMed] [Google Scholar]
- 9.Mangelsdorf D.J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jetten A.M. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 2009;7 doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bessonov S., Anokhina M., Will C.L., Urlaub H., Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 12.Hegele A., Kamburov A., Grossmann A., Sourlis C., Wowro S., Weimann M., et al. Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 13.Takada I., Tsuchiya M., Yanaka K., Hidano S., Takahashi S., Kobayashi T., et al. Ess2 bridges transcriptional regulators and spliceosomal complexes via distinct interacting domains. Biochem. Biophys. Res. Commun. 2018;497:597–604. doi: 10.1016/j.bbrc.2018.02.110. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay E.A., Botta A., Jurecic V., Carattini-Rivera S., Cheah Y.C., Rosenblatt H.M., et al. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- 15.Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 16.Daley S.R., Hu D.Y., Goodnow C.C. Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-kappaB. J. Exp. Med. 2013;210:269–285. doi: 10.1084/jem.20121458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueno T., Saito F., Gray D.H., Kuse S., Hieshima K., Nakano H., et al. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J. Exp. Med. 2004;200:493–505. doi: 10.1084/jem.20040643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matloubian M., Lo C.G., Cinamon G., Lesneski M.J., Xu Y., Brinkmann V., et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 19.Zhao B.B., Zheng S.J., Gong L.L., Wang Y., Chen C.F., Jin W.J., et al. T lymphocytes from chronic HCV-infected patients are primed for activation-induced apoptosis and express unique pro-apoptotic gene signature. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalkat M., Resetca D., Lourenco C., Chan P.K., Wei Y., Shiah Y.J., et al. MYC protein interactome profiling Reveals functionally distinct regions that cooperate to drive tumorigenesis. Mol. Cell. 2018;72:836–848.e7. doi: 10.1016/j.molcel.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 22.Shim H., Dolde C., Lewis B.C., Wu C.S., Dang G., Jungmann R.A., et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckerle S., Brune V., Doring C., Tiacci E., Bohle V., Sundstrom C., et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia. 2009;23:2129–2138. doi: 10.1038/leu.2009.161. [DOI] [PubMed] [Google Scholar]
- 24.Carey R., Jurickova I., Ballard E., Bonkowski E., Han X., Xu H., et al. Activation of an IL-6:STAT3-dependent transcriptome in pediatric-onset inflammatory bowel disease. Inflamm. Bowel Dis. 2008;14:446–457. doi: 10.1002/ibd.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marrack P., Bender J., Hildeman D., Jordan M., Mitchell T., Murakami M., et al. Homeostasis of alpha beta TCR+ T cells. Nat. Immunol. 2000;1:107–111. doi: 10.1038/77778. [DOI] [PubMed] [Google Scholar]
- 26.Fry T.J., Mackall C.L. The many faces of IL-7: from lymphopoiesis to peripheral T cell maintenance. J. Immunol. 2005;174:6571–6576. doi: 10.4049/jimmunol.174.11.6571. [DOI] [PubMed] [Google Scholar]
- 27.Fiala G.J., Gomes A.Q., Silva-Santos B. From thymus to periphery: molecular basis of effector gammadelta-T cell differentiation. Immunol. Rev. 2020;298:47–60. doi: 10.1111/imr.12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tani-ichi S., Shimba A., Wagatsuma K., Miyachi H., Kitano S., Imai K., et al. Interleukin-7 receptor controls development and maturation of late stages of thymocyte subpopulations. Proc. Natl. Acad. Sci. U. S. A. 2013;110:612–617. doi: 10.1073/pnas.1219242110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das R., Sant'Angelo D.B., Nichols K.E. Transcriptional control of invariant NKT cell development. Immunol. Rev. 2010;238:195–215. doi: 10.1111/j.1600-065X.2010.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Z., Unutmaz D., Zou Y.R., Sunshine M.J., Pierani A., Brenner-Morton S., et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 31.Guna A., Butcher N.J., Bassett A.S. Comparative mapping of the 22q11.2 deletion region and the potential of simple model organisms. J. Neurodev. Disord. 2015;7:18. doi: 10.1186/s11689-015-9113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meechan D.W., Maynard T.M., Tucker E.S., Fernandez A., Karpinski B.A., Rothblat L.A., et al. Modeling a model: mouse genetics, 22q11.2 deletion syndrome, and disorders of cortical circuit development. Prog. Neurobiol. 2015;130:1–28. doi: 10.1016/j.pneurobio.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sumitomo A., Horike K., Hirai K., Butcher N., Boot E., Sakurai T., et al. A mouse model of 22q11.2 deletions: molecular and behavioral signatures of Parkinson's disease and schizophrenia. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aar6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motahari Z., Moody S.A., Maynard T.M., LaMantia A.S. In the line-up: deleted genes associated with DiGeorge/22q11.2 deletion syndrome: are they all suspects? J. Neurodev. Disord. 2019;11:7. doi: 10.1186/s11689-019-9267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito R., Koebis M., Nagai T., Shimizu K., Liao J., Wulaer B., et al. Comprehensive analysis of a novel mouse model of the 22q11.2 deletion syndrome: a model with the most common 3.0-Mb deletion at the human 22q11.2 locus. Transl. Psychiatry. 2020;10:35. doi: 10.1038/s41398-020-0723-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanki H., Suzuki H., Itohara S. High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp. Anim. 2006;55:137–141. doi: 10.1538/expanim.55.137. [DOI] [PubMed] [Google Scholar]
- 37.Kim D., Salzberg S.L. TopHat-fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12 doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 41.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parvin R., Noro E., Saito-Hakoda A., Shimada H., Suzuki S., Shimizu K., et al. Inhibitory effects of a novel PPAR-gamma agonist MEKT1 on pomc expression/ACTH secretion in AtT20 cells. PPAR Res. 2018;2018 doi: 10.1155/2018/5346272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of RNA-seq are publicly available on the GEO repository (Accession No. PRJNA575280). Ess2+/− mice (BRC09772) and Ess2fl/fl mice (RBRC09771) were registered with the RIKEN BRC. All remaining data are contained within this article and the supporting information.