Abstract
The genotypes of 78 strains of Helicobacter pylori from Calcutta, India (55 from ulcer patients and 23 from more-benign infections), were studied, with a focus on putative virulence genes and neutral DNA markers that were likely to be phylogenetically informative. PCR tests indicated that 80 to 90% of Calcutta strains carried the cag pathogenicity island (PAI) and potentially toxigenic vacAs1 alleles of the vacuolating cytotoxin gene (vacA), independent of disease status. This was higher than in the West (where cag PAI+ vacAs1 genotypes are disease associated) but lower than in east Asia. The iceA2 gene was weakly disease associated in Calcutta, whereas in the West the alternative but unrelated iceA1 gene at the same locus is weakly disease associated. DNA sequence motifs of vacAm1 (middle region) alleles formed a cluster that was distinct from those of east Asia and the West, whereas the cagA sequences of Calcutta and Western strains were closely related. An internal deletion found in 20% of Calcutta iceA1 genes was not seen in any of ∼200 strains studied from other geographic regions and thus seemed to be unique to this H. pylori population. Two mobile DNAs that were rare in east Asian strains were also common in Calcutta. About 90% of Calcutta strains were metronidazole resistant. These findings support the idea that H. pylori gene pools differ regionally and emphasize the potential importance of studies of Indian and other non-Western H. pylori populations in developing a global understanding of this gastric pathogen and associated disease.
Helicobacter pylori is a gastric pathogen that chronically infects more than half of all people worldwide (for reviews see references 48 and 64) and constitutes a major cause of peptic ulcer disease and an early risk factor for gastric cancer. It may also contribute to childhood malnutrition and increase the risk or severity of infection by other gastrointestinal pathogens such as Vibrio cholerae, especially in developing countries (18, 19). It appears to be one of the most genetically diverse of bacterial species, because DNA fingerprinting can distinguish any given isolate from most others (5, 56) and because of the ∼3 to 5% DNA sequence divergence typically found in essential genes from unrelated strains (3, 29). This mutational diversity is enhanced by a rich history of interstrain recombination (29, 40, 54). In contrast, most other well-studied bacterial species characterized to date are much more strongly clonal (see, e.g., references 28, 31, and 53).
The observed genetic diversity implies a lack of population-wide selection for just one or a few universally most fit H. pylori genotypes. Some of this may reflect preferential transmission within families and among people in close contact, not in large epidemics (11, 24, 50). Such a pattern means that no individual strain would compete simultaneously against many others (12, 38, 55). H. pylori diversity would also be enhanced if humans differ in traits that are important to individual strains (e.g., specificity or strength of immune and inflammatory responses or availability of receptors used for H. pylori adherence [26, 33]).
There are also indications of significant geographic differences among strains. For example, only one-half to two-thirds of U.S. and European strains carry the cag pathogenicity island (PAI), a 40-kb DNA segment many of whose genes seem to help induce interleukin 8 and thereby a strong and potentially damaging inflammatory response; such strains are recovered preferentially from persons with overt disease (4, 8, 9, 17). In contrast, nearly all east Asian strains carry the cag PAI independent of disease status (34, 47). Similarly, somewhat more than half of U.S. and European strains carry toxigenic (vacAs1) alleles of the vacuolating cytoxin gene, with other strains carrying nontoxigenic (vacAs2) alleles (in general, vacAs1 strains carry the cag PAI (65); nearly all east Asian strains carry vacAs1 alleles. Potentially more significant in terms of host interaction and evolution were findings that east Asian and Western strains differ markedly in DNA sequence motifs in the vacA and cagA genes (3, 34, 35, 46, 58, 62), since the proteins these genes encode probably each interact directly with host factors (CagA protein is translocated to host cells and is tyrosine phosphorylated in them but is not needed for interleukin 8 induction) (7, 45, 51). Even though less geographic partitioning was found in a sampling of housekeeping genes (3, 58), it is not clear whether the regional differences in cagA and vacA alleles reflect natural selection, random genetic drift including founder effects, or both during H. pylori evolution. It is noteworthy in this context that the two strains whose genome sequences have been determined (6, 57) and compared to better understand H. pylori genetic diversity (23) are from ethnic European patients (26695 from the United Kingdom [6]; J99 from a Caucasian in Pulaski, Tenn. [57; T. L. Cover, personal communication]) and thus may not be fully representative of H. pylori worldwide.
We began studies of genotypes of H. pylori strains of India, motivated by the differences between strains of east Asia and the West found to date and a sense that the peoples of the vast Indian subcontinent (some one-fifth of all humanity) may have been sufficiently isolated during much of human history to have allowed the emergence of a distinct H. pylori gene pool (37, 42, 63). It is well established that H. pylori infection and peptic (especially duodenal) ulcer disease are very common in India (1, 36, 44, 52) and that a large fraction of strains may be resistant to metronidazole (Mtzr) (2), but to our knowledge there has been very little analysis to date of the genotypes of the underlying H. pylori strains (20). Here we identify several markers that help distinguish H. pylori strains from Calcutta from those of east Asia and the West.
MATERIALS AND METHODS
Patient samples.
Adult ethnic Bengali patients of both sexes, age 21 to 65 years, who presented with gastric complaints were endoscoped at the Hospital of the Institute of Post Graduate Medical Education and Research in Calcutta, India, using well-washed and sterilized fiber optic endoscopes. Biopsies and gastric juice samples used in the present study were obtained during endoscopy with informed consent, under protocols approved by the institutional review boards of the Institute of Post Graduate Medical Education and Research and the National Institute of Cholera and Enteric Disease (Calcutta, India). Two biopsies were taken for culture, one from the gastric antrum and one from the corpus, and were stored at −70°C in 0.5 ml of brucella broth (Difco) with 15% glycerol until culture. Two milliliters of gastric juice was also collected during endoscopy in some cases and stored frozen at −70°C until use. Diagnoses of peptic ulcer disease were based on visual examination of the stomach and duodenum during endoscopy and also on any patient history of earlier peptic ulcer. If no evidence of peptic ulcer disease was found, the patient was considered to have a more-benign infection (nonulcer dyspepsia, gastritis only). Fifty-five of the patients had peptic ulcer disease, and 23 had gastritis only.
H. pylori culture.
Cultures were prepared by smearing single biopsy specimens on petri plates containing brain heart infusion (BHI) agar (Difco) supplemented with 7% horse blood, 0.4% IsoVitaleX, amphotericin B (8 μg/ml), trimethoprim (5 μg/ml), and vancomycin (6 μg/ml) and were incubated at 37°C in an atmosphere of 5% O2–10% CO2–85% N2 for 3 to 6 days. H. pylori colonies were identified based on their typical morphology, characteristic appearance on Gram staining, a positive urease test, and subsequent gene-specific PCR tests. The H. pylori cells that grew out from one biopsy on the primary culture plate were collected as a pooled population, and preserved in sterile BHI broth with 15% glycerol at −70°C. In general, only one such culture was analyzed per patient.
Resistance and susceptibility to metronidazole (MTZ) were scored by spotting aliquots of ∼107 exponentially growing H. pylori cells on BHI agar containing MTZ, typically at 8 μg/ml, and on MTZ-free control plates in parallel and monitoring the amount of growth after incubation. For more-sensitive scoring, 10-μl aliquots of a serially diluted suspension, ranging from 107 to 102 viable cells per aliquot, were spotted on BHI agar containing various fixed concentrations of MTZ, and survival (efficiency of colony formation) was scored after incubation as a function of MTZ dose.
DNA methods.
Chromosomal DNA was prepared by the CTAB (hexadecyltrimethyl ammonium bromide) extraction method (10) from confluent BHI agar plate cultures. DNA was also extracted from samples of gastric juice using a QIAamp DNA minikit (Qiagen Corporation, Chatsworth, Calif.).
Specific PCR was carried out in 20-μl volumes using 10 ng of DNA, 1 U of Taq polymerase (Promega, Madison, Wis.), 10 pmol of each primer per reaction, 0.25 mM (each) deoxynucleoside triphosphate, and 2 to 3 mM MgCl2 in standard PCR buffer for 30 cycles generally under the following conditions: 94°C for 40 s, 55°C for 40 s, and 72°C for a time chosen based on the size of the expected fragment (1 min/kb). Arbitrarily primed PCR (randomly amplified polymorphic DNA [RAPD]) DNA fingerprinting was carried out using buffer with 4 mM MgCl2 for 45 cycles of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min. PCR primers are listed in Table 1.
TABLE 1.
Primers used in this study
| Region | Primer | Nucleotide sequence |
|---|---|---|
| vacAs1 or vacAs2 | VA1-F | 5′-ATG GAA ATA CAA CAA ACA CAC |
| VA1-R | 5′-CTG CTT GAA TGC GCC AAA C | |
| vacAm1a | VA3-F | 5′-GGT CAA AAT GCG GTC ATG G |
| VA3-R | 5′-CCA TTG GTA CCT GTA GAA AC | |
| vacAm1b | VAm-F3 | 5′-GGC CCC AAT GCA GTC ATG GAT |
| VAm-R3 | 5′-GCT GTT AGT GCC TAA AGA AGC AT | |
| vacAm2 | VA4-F | 5′-GGA GCC CCA GGA AAC ATT G |
| VA4-R | 5′-CAT AAC TAG CGC CTT GCA C | |
| vacA, 0.7-kb middle | VAm-F | 5′-GCT CAT TAC GGC TTC CAC TAA TGT |
| VAm-R | 5′-GCG GTT ATT GTT GTT ATA AAG GGC TA | |
| cagA (5′ end) | cagA5 | 5′-GGC AAT GGT GGT CCT GGA GCT AGG C |
| cagA2 | 5′-GGA AAT CTT TAA TCT CAG TTC GG | |
| cagA (3′ end) | cagA-F40481 | 5′-AGG ATT TCA GCA AGG TAA CGC AAG C |
| CagA-R41660 | 5′-TAA GAT TTT TGG AAA CCA CCT TTT GTA T | |
| cag PAI empty site | Luni1 | 5′-ACA TTT TGG CTA AAT AAA CGC TG |
| R5280 | 5′-GGT TGC ACG CAT TTT CCC TTA ATC | |
| iceA1 | iceA1F | 5′-TAT TTC TGG AAC TTG CGC AAC CTG AT |
| M.Hpy1R | 5′-GGC CTA CAA CCG CAT GGA TAT | |
| iceA2 | cysSF | 5′-CGG CTG TAG GCA CTA AAG CTA |
| iceA2R | 5′-TCA ATC CTA TGT GAA ACA ATG ATC GTT | |
| iceA1 Δ94 bp | A1F673 | 5′-GGT GAG TCG TTG GGT AAG CGT TAC AGA ATT |
| A1R1174 | 5′-CAC AAC CAT CAT ATT CAG CCT CCC CCT CAT A | |
| IS605 orfA | ORF18F | 5′-CGC CTT GAT CGT TTC AGG ATT AGC |
| ORF18R | 5′-CAA CCA ACC GAA GCA AGC ATA ATC | |
| IS605 orfB | ORF19F | 5′-GGC TGT TCT AGG GTC GTG TAT AAC |
| ORF19R | 5′-CAA GCT AGA TGC AAT CTA GCT ACC | |
| IS606 | FB1 | 5′-GAA TGT AAT TCT ACC TAA TCC ATT C |
| RB8 | 5′-GAG AAA CCT TGA TTG TTC CAT G | |
| IS.Inv | X14 | 5′-GAA TTA TCG CAA GTT ACA GAG ATT G |
| X24 | 5′-GCA ATA TAA TCC GTT CTA AGA AAC |
PCR products were sequenced directly, after purification with the QIAquick gel extraction kit (Qiagen) using the BigDye Terminator cycle sequencing kit (Perkin-Elmer-Applied Biosystems, Foster City, Calif.) and an ABI automated sequencer. DNA sequence editing and analysis were performed with programs in the GCG package (Genetics Computer Group, Madison, Wis.), programs and data in the TIGR H. pylori genome database (57) (http://www.tigr.org/tdb/mdb /hpdb.html), and the Phylip program of J. Felsenstein (http://evolution.genetics.washington.edu/phylip.html).
Dot blot hybridization was performed using Hybond-N+ nylon membranes (Amersham Pharmacia Biotech, Piscataway, N.J.) containing ∼10- to 20-ng aliquots of genomic DNA per spot from each strain of interest and hybridization probes labeled using the enhanced chemiluminescence kit (ECL; Amersham Pharmacia Biotech) according to the manufacturer's instructions. Probes for the orfA and orfB segments of IS605 were generated by PCR from 26695 genomic DNA with primers ORF18F and ORF18R (370 bp) and ORF19F and ORF19R (661 bp) (39). Similarly, a probe for IS606 was generated from strain 84-183 genomic DNA with primers FB1 and FB8 (784 bp). A probe for the IS.Inv element region was generated from NCTC11637 genomic DNA using primers X14 and X24 (3.4 kb). PCR-amplified DNAs were purified using the Qiagen gel extraction kit prior to ECL labeling for use in hybridization.
Statistical analyses of iceA1 and iceA2 frequencies were kindly carried out by William Shannon, Division of Biostatistics in Medicine, Washington University Medical School, using the Cochran-Mantel-Haenszel test.
Nucleotide sequence accession numbers.
Sequences obtained during this study have been assigned the following GenBank accession numbers: AF217727 to AF217735 (vacAs region), AF220110 to AF220120 (vacAm region), AF202219 to AF202225 (cagA [5′ end]), AF222807 to AF222809 (cagA [3′ end]), and AF239991 to AF239994 (iceA1).
RESULTS
Genetic diversity and drug resistance of H. pylori strains in Calcutta.
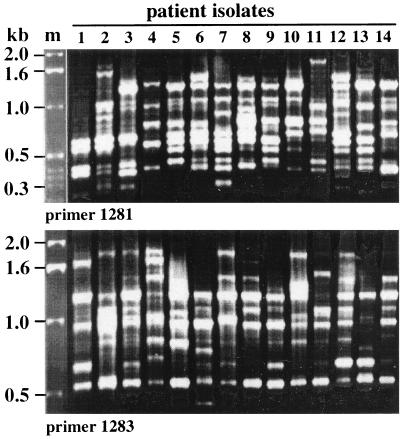
Arbitrarily primed PCR (RAPD) fingerprinting was carried out on DNAs from single-colony isolates of H. pylori from 14 patients with peptic ulcers to assess the overall diversity of strains in Calcutta. A different profile was obtained reproducibly from each isolate with each of several primers tested (Fig. 1), indicating that each isolate was unique in overall genotype. This genetic diversity was in accord with that seen with clinical isolates from other parts of the world.
FIG. 1.
Arbitrarily primed PCR (RAPD) fingerprint patterns from 14 independent H. pylori isolates, each from a different Calcutta resident with peptic ulcer disease.
Pools of H. pylori that had been cultured from individual biopsies from 55 patients with ulcers (the 14 surveyed in Fig. 1 plus another 41) and also from 18 patients with more-benign (gastritis-only) infections were tested for drug susceptibility (see Materials and Methods). None of these 73 cultures were resistant to clarithromycin (0.5 μg/ml), which is in accord with macrolides not being used very often in this Calcutta population. In contrast, 66 of them (90%) were resistant to at least an 8-μg/ml concentration of MTZ (Table 2). Four of the seven nominally Mtzs cultures grew on medium with 3 μg of MTZ/ml, indicating a leaky resistance phenotype, whereas the other three cultures were killed on this medium and were as sensitive as our standard Mtzs laboratory strains (J99, 26695). These data are in accord with an earlier report (2) that the frequency of Mtzr H. pylori is extremely high in India, although some truly Mtzs strains can still also be found.
TABLE 2.
Distribution of genetic or phenotypic characteristics of H. pylori from Calcutta in relation to disease status
| Trait or markerb | Patient disease statusa
|
|
|---|---|---|
| Peptic ulcer | Gastritis only | |
| Mtzrc | 49/55 | 17/18 |
| cag PAI+ onlyd | 46/55 | 18/23 |
| cag PAI+,− mixedd | 7/55 | 4/23 |
| cag PAI− onlyd | 2/55 | 1/23 |
| vacAs1 only | 49/55 | 18/23 |
| vacAs2 only | 2/55 | 1/23 |
| vacAs1, -s2 mixed | 4/55 | 4/23 |
| vacAm1c only | 31/55 | 12/23 |
| vacAm2 only | 19/55 | 8/23 |
| vacAm1c, -m2 mixed | 4/55 | 3/23 |
| iceA1e | 32/55 | 16/23 |
| iceA2 | 21/55 | 5/23 |
| iceA1, iceA2 mixed | 1/55 | 2/23 |
| IS605f | 16/55 | 3/18 |
| IS606 | 11/55 | 2/18 |
| IS.Inv | 10/55 | 4/18 |
Number with trait/number scored.
Distribution of DNA markers was determined by PCR or hybridization, as illustrated in the figures.
Resistant to 8 μg of MTZ/ml in agar dilution test. Four of the seven strains that were unable to grow on medium with 8 μg of MTZ/ml grew on medium with just 3 μg of MTZ/ml, indicating leaky resistance phenotypes (see text). The possibility that many of these Mtzr infections were mixed and contained only a small fraction of Mtzr cells and many Mtzs cells (as was common in a Peruvian population) (12) was tested by streaking representative Mtzr cultures on MTZ-free medium: Two single colonies from each of 10 such cultures were found to be Mtzr, whereas each of the two single colonies from an 11th Mtzr culture was Mtzs. This suggests that, despite occasional Mtzs Mtzr mixed infections, the majority of cells in most nominally Mtzr infections in Calcutta are indeed Mtzr.
cag PAI+ only, infection with strains carrying cag PAI only; cag PAI+,− mixed, mixed infection with strains carrying the cag PAI and strains lacking the cag PAI; cag PAI− only, infection with strains lacking the cag PAI only.
Just one culture failed to give iceA1 or iceA2 amplification. Of the 10 iceA1 genes (of 44 tested) that contained the 94-bp deletion (Fig. 6) not found in H. pylori from any other region studied to date, 8 were from ulcer patients and 2 were from gastritis-only patients.
Thirty-three of the 73 cultures contained just one type of mobile element, 5 cultures contained two of them, 1 culture contained all three elements, and 34 cultures lacked all three elements. Tests of 8 single colonies from each of 11 cultures carrying IS606 and each of 10 carrying IS.Inv (in total, 88 and 80 single colonies, respectively) identified these elements in every case, indicating that these estimates, based on hybridization to pools of H. pylori cultured from individual biopsies, are probably accurate.
cag PAI in H. pylori from Calcutta.
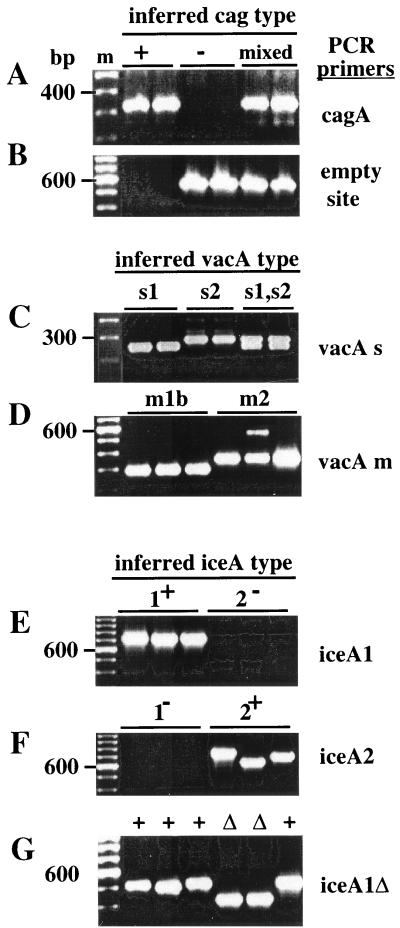
The presence or absence of the cag PAI was scored by PCR with specific primers (4) using DNAs extracted from cultured strains or gastric juice. Products indicative of the cag PAI (Fig. 2A) were obtained with primers specific for the cagA gene from the great majority of strains: 53 of 55 cultures from patients with overt disease and 22 of 23 patients with benign infections. The three cultures from which no cag PAI-specific PCR product was obtained yielded an empty-site product of the expected 550-bp size (Fig. 2B), indicating that they truly lacked the cag PAI (4). In addition, products corresponding to the empty site (Fig. 2A and B, lanes labeled mixed) were also obtained from 11 of the patients found to be infected with strains carrying the cag PAI, indicating that they had mixed infections, i.e., a mixture of strains with and without the cag PAI.
FIG. 2.
Representative sequence-specific PCR tests. (A) Test for the presence of cagA gene using primers cagA5 and cagA2. (B) Test for the presence of the cag PAI empty site using primers Luni1 and R5280. (C) Test for the presence of vacAs1 versus vacAs2 alleles using primers VA1-F and VA1-R (generating 259- and 286-bp products from vacAs1 and -s2 alleles, respectively). (D) Test for the presence of vacAm1 using primers VAm-F and VAm-R and vacAm2 alleles using primers VA4-F and VA4-R. (E) Test for iceA1 using primers iceA1F and M.Hpy1R. (F) Test for iceA2 using primers cysF and iceA2R. (G) Test for the presence or absence of the 94-bp deletion in iceA1 using primers A1F673 and A1R1174.
Twelve single-colony isolates from each of five such mixed infections were tested further, and at least one isolate lacking the cag PAI was obtained from each of 3 of them. Paired isolates with and without the cag PAI from these three cases were DNA fingerprinted, and a different RAPD profile was found in each case (data not shown). This indicated that these coexisting strains with and without the cag PAI were not related to one another and had in each case probably resulted from separate infections by strains with and without the cag PAI, not by a single infection with a strain carrying the cag PAI and then cag PAI excision or a reciprocal cag PAI acquisition by an infecting strain lacking the cag PAI.
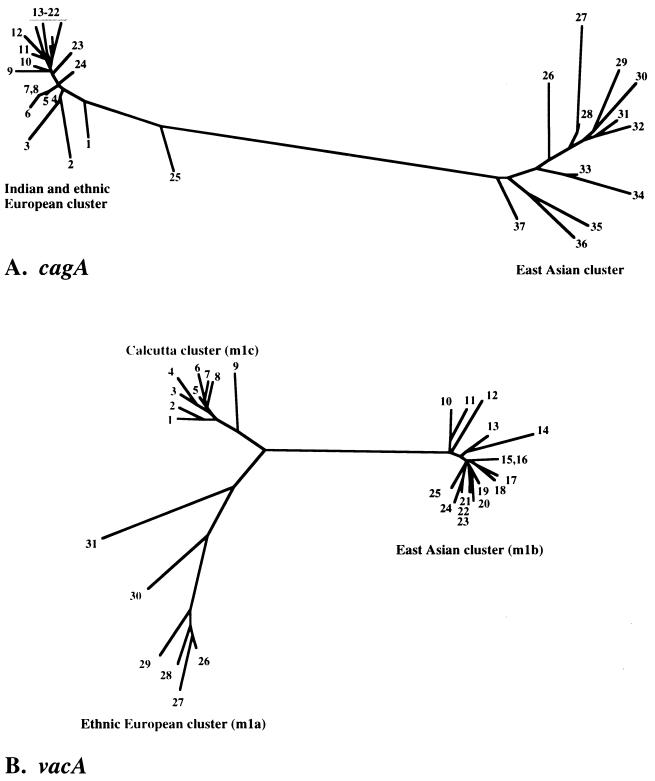
To assess the phylogenetic relationship between Indian cag PAIs and those from other regions, we focused first on a segment near the 5′ end of the cagA gene that had been used to distinguish east Asian and U.S. and European strains (58). This segment was amplified by PCR from seven Indian strains and sequenced directly. The sequences obtained were closely related to one another and also to those from Western strains, but not to those from Chinese and Japanese strains (Fig. 3A). A 1-kb segment near the 3′ end of cagA that was similarly useful for distinguishing east Asian from Western strains (66) was sequenced from three other strains (GenBank accession no. AF222807 to AF222809). These sequences also clustered with those from ethnic European strains (94 to 96% DNA sequence match), not with east Asian sequences (79 to 80%).
FIG. 3.
Phylogenetic trees of sequences within the cagA gene and vacAm1 alleles. Sequences from non-Indian strains that were used here were from public databases, as indicated. (A) Phylogenetic tree based on an informative 220-bp segment of cagA (62) of H. pylori strains determined in this study (Calcutta strains 5 to 7, 16 to 18, and 23) or reported by others. The tree was generated using PHYLIP (Phylogeny Inference Package), version 3.5c, of J. Felsenstein (see Materials and Methods). The strains used are as follows (GenBank accession numbers are in parentheses): 1, Dutch161A (AJ252965); 2, 26695 (AE000569); 3, Peru8C (AF198478); 4, Lith5-1 (AJ239734); 5, India10 (AF202222); 6, India19 (AF202225); 7, India3 (AF202219); 8, Peru24C (AF198473); 9, Peru2B (AF198474); 10, Dutch292 (AJ252971); 11, Peru35B (AF198476); 12, Dutch79 (AJ252970); 13, Gambia4659 (AF198468); 14, Peru4A (AF198477); 15, Gambia4797 (AF198469); 16, India18 (AF202224); 17, India9 (AF202221); 18, India7 (AF202220); 19, Dutch25 (AJ252968); 20, Dutch419 (AJ252974); 21, Guatemala88 (AF198472); 22, South Africa19 (AF198470); 23, India17 (AF202223); 24, Dutch107 (AJ252963); 25, Peru34B (AF198475); 26, ChinaR47 (AJ252985); 27, HongKong77 (AF198485); 28, Thailand88-28 (AJ239722); 29, ChinaR27 (AJ252979); 30, HongKong97-42 (AJ239733); 31, HongKong81 (AF198486); 32, ChinaR40 (AJ252982); 33, ChinaR59 (AJ252986); 34, ChinaR29 (AJ252980); 35, JapanF32 (AJ239726); 36, JapanGC4 (AF198484); 37, ChinaR48 (AJ252983). (B) Tree based on informative 650-bp segment of vacA gene containing vacAm1 alleles of Calcutta H. pylori strains determined in this study (strains 1 to 9 except strain 7) or reported by others. The tree was generated using PHYLIP (Phylogeny Inference Package), version 3.5c, of J. Felsenstein (see Materials and Methods). The sequences of east Asian and ethnic European vacAm1 alleles were taken from GenBank. Each number in this figure indicates the vacAm1 sequence from a given strain, as follows (GenBank accession numbers in parentheses): 1, India19 (AF220111); 2, India226 (AF220115); 3, India89 (AF220114); 4, India48 (AF220112); 5, India18 (AF220110); 6, India230 (AF220117); 7, GermanyMz19 (AJ006967); 8, India66 (AF220113); 9, India227 (AF220116); 10, JapanF52 (AF049631); 11, JapanF55 (AF049632); 12, ChinaR59 (AF035611); 13, JapanF63 (AF049635); 14, ChinaR13 (AF035610); 15, JapanF42 (AF049626); 16, Japan94 (AF049640); 17, JapanF72 (AF049651); 18, JapanF73 (AF049652); 19, JapanF47 (AF049629); 20, JapanF57 (AF049634); 21, JapanF35 (AF049625); 22, JapanF61 (AF049645); 23, JapanF36 (AF049462); 24, JapanF64 (AF049647); 25, JapanF45 (AF0496628); 26, Poland1492 (AF097570); 27, Poland278 (AF097571); 28, NCTC11637 (AF049653); 29, J99 (AE001511); 30, NCTC11638 (U07145); 31, 26695 (AE000598).
vacA (vacuolating cytotoxin gene).
The types of alleles at the 5′ end of the vacuolating cytotoxin gene (vacA) (vacAs1, generally toxigenic; vacAs2, generally nontoxigenic) were assessed, based on sizes of PCR products generated with appropriate vacA-specific primers (Fig. 2C). Of the 55 cultures from ulcer patients tested, 49 yielded a 259-bp fragment, indicating vacAs1 alleles, two yielded a 286-bp fragment, indicating vacAs2 alleles, and the remaining four yielded both the 259- and 286-bp fragments, indicating mixed infections (Table 2). Each infection that seemed to be due solely to a strain carrying vacAs2 in this test had also been scored as due solely to strain lacking the cag PAI in tests above. Similarly, the single-colony isolates lacking the cag PAI obtained from three mixed infections also carried the vacAs2 allele.
In equivalent PCR tests of DNAs from cultured strains or gastric-juice samples from the 23 patients with benign infections, 18 yielded vacAs1 products, one yielded a vacAs2 product, and the other four yielded both vacAs1 and vacAs2 products, again indicating mixed infections. Thus, the associations of the cag PAI with vacAs1 and the absence of the cag PAI with vacAs2 observed here match those typically seen in the West. In contrast, the lack of association of the genotype in which vacAs2 is present and the cag PAI is absent with more-benign infection is distinct from what is typically seen in the West.
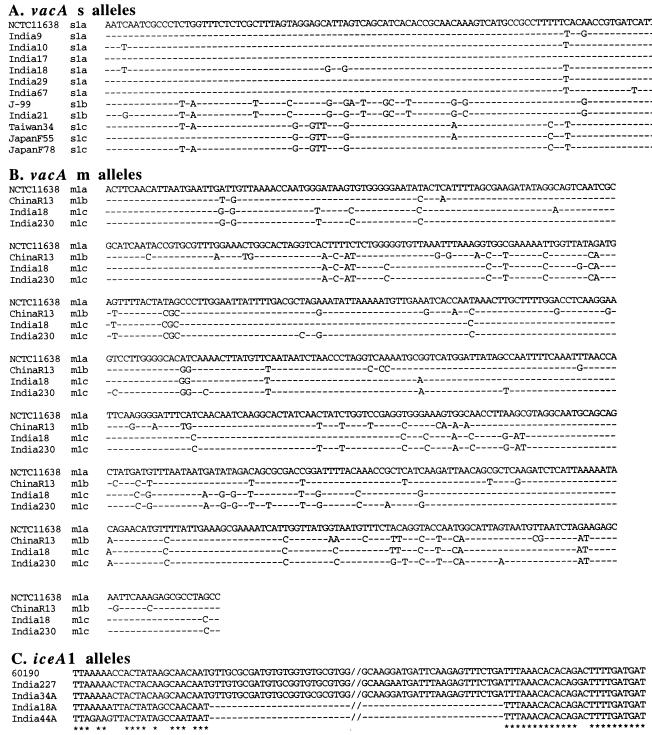
PCR products containing vacAs1 alleles of seven representative strains were sequenced directly. Six of them were of the vacAs1a allele type, and one was of the vacAs1b type (Fig. 4A), which are each common in European and U.S. populations. None of them were of the vacAs1c type, which was found in more than three-fourths of east Asian strains (35, 60; Y. Ito and T. Azuma, unpublished data). Two vacAs2 products were also sequenced and were found to be closely related to those of European or U.S. H. pylori strains (GenBank accession no. AF217727 and AF217733).
FIG. 4.
DNA sequence alignments, illustrating motifs found in H. pylori strains from Calcutta. Sequences from non-Indian strains that were used here were from public databases, as indicated. (A) vacAs region sequences, showing differences between vacAs1a, -s1b, and -s1c type alleles. vacAs1a alleles are common in the Calcutta H. pylori strains, whereas vacAs1c alleles are most common elsewhere in Asia. Each of the 97-bp sequences presented here starts at position 27 of a reference vacA open reading frame (GenBank accession no. U07145). Identical nucleotides are indicated by hyphens. GenBank accession numbers for the sequences depicted here are U07145 (NCTC11638), AF217728 (India9), AF217729 (India10), AF217730 (India17), AF217731 (India18), AF217734 (India29), AF217735 (India67), AE001511 (J99), AF217732 (India21), AF091830 (Taiwan34), AF049632 (Japan55), and AF049638 (Japan78). (B) vacAm1 middle region sequences, showing differences among vacAm1a, vacAm1b, and two of the representative vacAm1c allele types that we characterized. The vacAm1c alleles, which were common in Calcutta strains, were rare in populations from outside India that have been studied to date. Each of the 650-bp sequences presented here corresponds to nucleotides (nt) 2060 to 2709 of the vacA gene of reference strain NCTC11638 (Genbank accession no. U07145). GenBank accession numbers for the sequences depicted here are U07145 (NCTC11638), AF035610 (ChinaR13), AF220110 (India18; strain 5 in Fig. 3) and AF220117 (India230; strain 6 in Fig. 3). (C) Alignment of four iceA1 genes of Calcutta isolates, along with reference strain 60190 (GenBank accession no. U43917). The sequences presented here correspond to nt 426 to 558 in India227 (accession no. AF239991), nt 446 to 558 in India34A (accession no. AF239992), nt 430 to 478 in India18A (accession no. AF239993), and nt 423 to 471 in India44A (accession no. AF239994).
The alleles of the middle region of vacA, a part that seems to affect host cell specificity (45), were also scored by PCR (Fig. 2D) and by sequencing several representatives. Products were obtained with only vacAm1b primers from culture or gastric-juice samples from 43 of the 78 patients; conversely, products were obtained only with vacAm2 primers from 27 of the 78 patient samples; and products were obtained using both primer sets from another 7 of the patient samples, indicating mixed infections, (4 of these 7 were from patients with infections that were mixed in terms of vacAs1 and -s2 alleles, whereas the other 3 were not). Just one sample was exceptional in yielding an amplification product with the vacAm1b forward primer and the vacAm2 reverse primer, suggesting that it was a vacAm1-vacAm2 hybrid.
DNA sequencing of a 0.7-kb PCR fragment containing the vacA middle region from eight strains that had yielded products with vacAm1b-specific primers (five ulcer and three gastritis) showed each to be closely related to one another but distinct from both canonical vacAm1b alleles of east Asian strains and vacAm1a alleles of ethnic European strains (Fig. 3B and 4B). These Calcutta alleles, termed vacAm1c, were also closely related to an allele from an unusual isolate from Germany (Fig. 3B, no. 7) (32), but for which patient ethnicity, community contacts, and travel history are not known. The middle regions of two strains that had yielded products with vacAm2-specific primers were each closely related to those of canonical vacAm2 strains from other societies (GenBank accession no. AF220118 and AF220119), and the middle region of the strain that had yielded a PCR product only with vacAm1b forward and vacAm2 reverse primers was found indeed to contain a hybrid vacAm1c-vacAm2 allele (96 to 98% match with Indian vacAm1c type sequences proximally, 92 to 94% match with vacAm2 sequences distally; GenBank accession no. AF220120), reminiscent of the vacAm1b-vacAm2 recombinant allele found among H. pylori strains from Shanghai (where vacAm1b and -m2 alleles are common) (46).
Polymorphism for iceA1 and iceA2 genes.
PCR was used to distinguish between iceA1, the restriction endonuclease NlaIII homolog that is associated with virulence in the West and whose expression is induced by gastric epithelial cell contact (49, 61), and the iceA2 gene that, although unrelated in sequence, occupies the same chromosomal locus in strains lacking iceA1 (Fig. 2E and F). The iceA1 gene was found alone in 32 of the 55 (58%) peptic ulcer patients and in 16 of 23 (70%) gastritis-only patients, whereas the iceA2 gene was found alone in cultures from 21 of 55 (38%) peptic ulcer patients and 5 of 23 (22%) gastritis-only patients. A few patients had mixture of iceA1 and iceA2 strains (Table 2). This distribution of iceA1 and iceA2 genes in relation to disease status in the Calcutta population differed significantly from that found in Tennessee (49) and The Netherlands (61) (Cochran-Mantel-Haenszel chi square test result, P = 0.001), with iceA2 being weakly disease associated in Calcutta, rather than iceA1, as in Europe and the United States.
In further studies, a characteristic 94-bp deletion was found by PCR near the 3′ end of iceA1 (Fig. 2G) from 10 of the 48 Calcutta strains (8 of the 32 from peptic ulcer patients and 2 of the 16 from gastritis-only patients), with the same deletion end points in each case sequenced (Fig. 4C). This deletion was not found in iceA1 genes from any of 211 strains from other geographic regions (Japan, Hong Kong, South Africa, Spain, North Europe, Alaska, or Peru; Y. Ito, T. Azuma, and D. E. Berg, unpublished data), suggesting that it may be useful as an Indian ethnic group-specific marker.
Markers that differ geographically but are not implicated in virulence.
The prevalence of several mobile DNAs was also studied, because of their potential utility for detecting ancient phylogenetic lineages. Hybridization and PCR tests identified sequences from IS606, IS605, and the recently discovered IS.Inv element (N. Akopyants, A. Raudonikiene, and D. E. Berg, unpublished data) in some 17 to 25% of cultures (Table 2), with carriage of each element apparently being independent of that of each of the others (see legend). The IS606 and IS.Inv elements are unrelated in sequence, and it is striking that each is much less common in east Asian strains than in Calcutta strains (<2% in each case in east Asia [A. K. Mukhopadhyay, Z. J. Pan, D. E. Berg, et al., unpublished data] versus ∼17% in Calcutta).
In other studies, we found that DNA sequence motifs at the right end of the cag PAI in Calcutta strains also differed markedly from those in other geographic regions (see Calcutta H. pylori strain GenBank accession no. AF190663, AF191015, AF191016, AF200689, AF201074, and AF201075) (41).
DISCUSSION
H. pylori strains from Calcutta were studied to better understand the global population genetic structure and evolution of this gastric pathogen, in particular, to test if the H. pylori gene pool in at least this part of India is distinct from that found in east Asia, Europe, or both and also to test putative virulence genes of H. pylori for disease associations in an Indian setting. We found that strains carrying the cag PAI and the potentially toxigenic vacAs1 alleles of the vacuolating cytotoxin gene (vacA) were more abundant in Calcutta than in the West (8) but that about 10 to 20% of Calcutta strains lacked the cag PAI entirely and contained vacAs2 (nontoxigenic) rather than vacAs1 (potentially toxigenic) alleles. In contrast, essentially all Chinese and Japanese strains studied to date carried cag PAI genes and vacAs1 alleles (34, 35, 46, 47). Both iceA2 and iceA1 were present in Calcutta populations, but iceA2 seemed to be somewhat disease associated, not iceA1, as in the West. An iceA1 DNA deletion motif found in about one-fifth of Calcutta strains also seemed to be a region-specific marker, and two mobile DNAs that were very rare in east Asian strains were each quite common in Calcutta strains. It will be of great interest to learn just how these and other polymorphic traits might be distributed in other parts of the Indian subcontinent, in peoples separated from those of Calcutta by distance, language, culture, ethnicity, and ancestry (13, 63).
Given the existence of some truly vacAs2 strains with the cag PAI in Calcutta, it is noteworthy that such strains were recovered at similar frequency from patients with ulcers and more-benign infections. In contrast, in the West vacAs2 strains lacking the cag PAI are recovered disproportionately from persons with benign infections (8). One explanation for strains lacking the cag PAI being equally associated with overt disease and benign infection assumes that infections by such strains in India are often mixed with infections by strains with the cag PAI, the latter strains being responsible for most of the observed pathology. Additional explanations include (i) host or environmental factors and (ii) additional bacterial genetic virulence determinants that might be specific to Indian strains and/or that might be more important than cag PAI and vacAs1 status as determinants of disease in the Indian setting. The second of these alternative explanations is supported by our finding of several genetic differences between Calcutta and other (non-Indian) H. pylori populations.
The evolutionary forces of natural selection and random genetic drift (which includes founder effects) may have each helped shape the gene pool of H. pylori in Calcutta and made it distinct from those in other parts of the world. The effect of natural selection may be best illustrated by the Mtzr of some 90% of H. pylori strains in Calcutta, in contrast to the much lower Mtzr frequencies (∼10 to 30%) in Japan and in the West. We have found that in India, as in other societies, Mtzr results from mutation of the chromosomal rdxA nitroreductase gene (HP0954), not the acquisition of new “resistance genes” (e.g., in plasmids or transposons) (30; J.-Y. Jeong, A. K. Mukhopadhyay, and D. E. Berg, unpublished data) and that MTZ is mutagenic (G. Sisson, J.-Y. Jeong, D. E. Berg, and P. S. Hoffman, unpublished data). As noted above, MTZ is used frequently against a variety of illnesses in India. The present abundance of Mtzr in Indian H. pylori strains can be ascribed to this frequent use of MTZ, generally at doses that may induce and select for resistant mutants of resident H. pylori strains without eradicating them (59).
The unrelated IS606 and IS.Inv mobile DNA elements were found in about 17% of Calcutta strains (present results) but in <2% of strains from China and Japan analyzed in the same way (A. K. Mukhopadhyay, Z. J. Pan, W. W. Su, and D. E. Berg, unpublished data). We suggest that this reflects random genetic drift, not selection. This is based on an assumption that these elements do not affect the risk of H. pylori infection, persistence, or virulence in a manner specific to particular human ethnic groups or geographic regions. Also possibly indicating involvement of genetic drift is our finding, mentioned above, of different DNA sequence motifs at the right end of the cag PAI (41). A model invoking genetic drift in the divergence of H. pylori in different regions would be in accord with its transmission preferentially within family groups and local communities, rather than in worldwide epidemics (11, 24, 40), and the relative separation of peoples of different ethnicities (including Indians from east Asians and from Europeans) during thousands of years of human history (13, 16).
The high frequency of the cag PAI and vacAs1 types among Calcutta H. pylori strains would be in accord with the high overall risk of H. pylori infection in India and the evolutionary consideration that high rates of transmission favor the emergence of more-virulent strains of a pathogen (27). There is also speculation, however, that H. pylori might have jumped recently from various animal species to humans, perhaps in early agricultural communities (21, 22, 25, 41, 43) versus an alternative in which humans have always carried H. pylori (14, 15). In the first scenario, any distinctive adaptive features of Calcutta H. pylori (abundance of cag PAI vacAs1 genotypes and/or particular DNA sequence motifs that distinguish these strains from others) might be a legacy of divergent selection pressures in different putative ancestral animal hosts, rather than in humans. Many of these questions may be resolved with the development of new cell culture and animal models that give further insight into the biological activities of H. pylori virulence proteins and their contributions to bacterial fitness and through further population genetic analyses of H. pylori elsewhere in India and in expatriate Indians living abroad.
ACKNOWLEDGMENTS
We are grateful to Bill Shannon, Division of Biostatistics in Medicine, Washington University Medical School, for statistical analyses, and to N. K. Ganguly, Indian Council of Medical Research, for his encouragement.
This work was supported in part by NIH grants AI38166 and DK53727 to D.E.B. and P30 DK52574 to Washington University.
REFERENCES
- 1.Abraham P, Bhatia S J. Position paper on Helicobacter pylori in India. Indian J Gastroenterol. 1997;16(Suppl. 1):S29–S33. [PubMed] [Google Scholar]
- 2.Abraham P, Sandhu N, Naik S R. In vitro sensitivity of Helicobacter pylori in India. Indian J Gastroenterol. 1997;16(Suppl. 1):S20–S21. [PubMed] [Google Scholar]
- 3.Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z J, Suerbaum S, Thompson S A, van der Ende A, van Doorn L J. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 4.Akopyants N S, Clifton S W, Kersulyte D, Crabtree J E, Youree B E, Reece C A, Bukanov N O, Drazek E S, Roe B A, Berg D E. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–54. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 5.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 7.Asahi M, Azuma T, Ito S, Ito Y, Suto H, Nagai Y, Tsubokawa M, Tohyama Y, Maeda S, Omata M, Suzuki T, Sasakawa C. Helicobacter pylori CagA protein delivered to gastric epithelial cells can be tyrosine phosphorylated. J Exp Med. 2000;191:593–602. doi: 10.1084/jem.191.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atherton J C. H. pylori virulence factors. Br Med Bull. 1998;54:105–120. doi: 10.1093/oxfordjournals.bmb.a011662. [DOI] [PubMed] [Google Scholar]
- 9.Atherton J C, Cover T L, Twells R J, Morales M R, Hawkey C J, Blaser M J. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J Clin Microbiol. 1999;37:2979–2982. doi: 10.1128/jcm.37.9.2979-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1993. [Google Scholar]
- 11.Bamford K B, Bickley J, Collins J S, Johnston B T, Potts S, Boston V, Owen R J, Sloan J M. Helicobacter pylori: comparison of DNA fingerprints provides evidence for intrafamilial infection. Gut. 1993;34:1348–1350. doi: 10.1136/gut.34.10.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg D E, Gilman R H, Lelwala-Guruge J, Valdez Y, Watanabe J, Miyagi J, Akopyants N S, Srivastava K, Ramirez-Ramos A, Yoshiwara T H, Recavarren S, Leon-Barua R. Helicobacter pylori populations in Peruvian patients. Clin Infect Dis. 1997;25:996–1002. doi: 10.1086/516081. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharyya N P, Basu P, Das M, Pramanik S, Banerjee R, Roy B, Roychoudhury S, Majumder P P. Negligible male gene flow across ethnic boundaries in India, revealed by analysis of Y-chromosomal DNA polymorphisms. Genome Res. 1999;9:711–719. [PubMed] [Google Scholar]
- 14.Blaser M J. Helicobacters are indigenous to the human stomach: duodenal ulceration is due to changes in gastric microecology in the modern era. Gut. 1998;43:721–727. doi: 10.1136/gut.43.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaser M J. Hypothesis: the changing relationships of Helicobacter pylori and humans: implications for health and disease. J Infect Dis. 1999;179:1523–1530. doi: 10.1086/314785. [DOI] [PubMed] [Google Scholar]
- 16.Cavalli-Sforza L L, Menozzi P, Piazza A. The history and geography of human genes. Princeton, N.J: Princeton University Press; 1994. [Google Scholar]
- 17.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clemens J, Albert M J, Rao M, Qadri F, Huda S, Kay B, van Loon F P, Sack D, Pradhan B A, Sack R B. Impact of infection by Helicobacter pylori on the risk and severity of endemic cholera. J Infect Dis. 1995;171:1653–1656. doi: 10.1093/infdis/171.6.1653. [DOI] [PubMed] [Google Scholar]
- 19.Dale A, Thomas J E, Darboe M K, Coward W A, Harding M, Weaver L T. Helicobacter pylori infection, gastric acid secretion, and infant growth. J Pediatr Gastroenterol Nutr. 1998;26:393–397. doi: 10.1097/00005176-199804000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Dhar A, Sharma M P. Lacunae in data on Helicobacter pylori from India. Indian J Gastroenterol. 1997;16(Suppl. 1):S13–S15. [PubMed] [Google Scholar]
- 21.Diamond J. Guns, germs, and steel: the fates of human societies. W. W. New York, N.Y: Norton and Co.; 1997. [Google Scholar]
- 22.Dimola S, Caruso M L. Helicobacter pylori in animals affecting the human habitat through the food chain. Anticancer Res. 1999;19:3889–3894. [PubMed] [Google Scholar]
- 23.Doig P, de Jonge B L, Alm R A, Brown E D, Uria-Nickelsen M, Noonan B, Mills S D, Tummino P, Carmel G, Guild B C, Moir D T, Vovis G F, Trust T J. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol Mol Biol Rev. 1999;63:675–707. doi: 10.1128/mmbr.63.3.675-707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominici P, Bellentani S, Di Biase A R, Saccoccio G, Le Rose A, Masutti F, Viola L, Balli F, Tiribelli C, Grilli R, Fusillo M, Grossi E. Familial clustering of Helicobacter pylori infection: population based study. BMJ. 1999;319:537–540. doi: 10.1136/bmj.319.7209.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dore M P, Sepulveda A R, Osato M S, Realdi G, Graham D Y. Helicobacter pylori in sheep milk. Lancet. 1999;354:132. doi: 10.1016/S0140-6736(99)01724-9. [DOI] [PubMed] [Google Scholar]
- 26.Dubois A, Berg D E, Incecik E T, Fiala N, Heman-Ackah L M, Perez-Perez G I, Blaser M J. Transient and persistent infection of nonhuman primates with Helicobacter pylori: implications for human disease. Infect Immun. 1996;64:2885–2891. doi: 10.1128/iai.64.8.2885-2891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ewald P. Evolution of infectious disease. New York, N.Y: Oxford University Press; 1994. [Google Scholar]
- 28.Feil E J, Maiden M C, Achtman M, Spratt B G. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol. 1999;16:1496–1502. doi: 10.1093/oxfordjournals.molbev.a026061. [DOI] [PubMed] [Google Scholar]
- 29.Garner J A, Cover T L. Analysis of genetic diversity in cytotoxin-producing and non-cytotoxin-producing Helicobacter pylori strains. J Infect Dis. 1995;172:290–293. doi: 10.1093/infdis/172.1.290. [DOI] [PubMed] [Google Scholar]
- 30.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten S J O, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 31.Guttman D S, Dykhuizen D E. Detecting selective sweeps in naturally occurring Escherichia coli. Genetics. 1994;138:993–1003. doi: 10.1093/genetics/138.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S R, Schreiber H J, Bhakdi S, Loos M, Maeurer M J. vacA genotypes and genetic diversity in clinical isolates of Helicobacter pylori. Clin Diagn Lab Immunol. 1998;5:139–145. doi: 10.1128/cdli.5.2.139-145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ilver D, Arnqvist A, Ogren J, Frick I-M, Kersulyte D, Incecik E T, Berg D E, Covacci A, Engstrand L, Boren T. The Helicobacter pylori Lewis b blood group antigen binding adhesin revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 34.Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, Sato F, Kato T, Kohli Y, Kuriyama M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito Y, Azuma T, Ito S, Suto H, Miyaji H, Yamazaki Y, Kohli Y, Kuriyama M. Full-length sequence analysis of the vacA gene from cytotoxic and noncytotoxic Helicobacter pylori. J Infect Dis. 1998;178:1391–1398. doi: 10.1086/314435. [DOI] [PubMed] [Google Scholar]
- 36.Jain A K, Dayal V M. Helicobacter pylori recolonization and ulcer relapse after its eradication in India. Indian J Gastroenterol. 1997;16(Suppl. 1):S22–S24. [PubMed] [Google Scholar]
- 37.Jin L, Underhill P A, Doctor V, Davis R W, Shen P, Cavalli-Sforza L L, Oefner P J. Distribution of haplotypes from a chromosome 21 region distinguishes multiple prehistoric human migrations. Proc Natl Acad Sci USA. 1999;96:3796–3800. doi: 10.1073/pnas.96.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen M, Daskalopoulos G, Warburton V, Mitchell H M, Hazell S L. Multiple strain colonization and metronidazole resistance in Helicobacter pylori-infected patients: identification from sequential and multiple biopsy specimens. J Infect Dis. 1996;174:631–635. doi: 10.1093/infdis/174.3.631. [DOI] [PubMed] [Google Scholar]
- 39.Kersulyte D, Akopyants N S, Clifton S W, Roe B A, Berg D E. Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene. 1998;223:175–186. doi: 10.1016/s0378-1119(98)00164-4. [DOI] [PubMed] [Google Scholar]
- 40.Kersulyte D, Chalkauskas H, Berg D E. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31:31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- 41.Kersulyte D, Mukhopadhyay A K, Velapatiño B, Su W, Pan Z, Garcia C, Hernandez V, Valdez Y, Mistry R S, Gilman R H, Yuan Y, Gao H, Alarcón T, López-Brea M, Nair G B, Chowdhury A, Datta S, Shirai M, Nakazawa T, Ally R, Segal I, Wong B C Y, Lam S K, Olfat F, Borén T, Engstrand L, Torres O, Schneider R, Thomas J E, Czinn S, Berg D E. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182:3210–3218. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majumder P P, Roy B, Banerjee S, Chakraborty M, Dey B, Mukherjee N, Roy M, Thakurta P G, Sil S K. Human-specific insertion/deletion polymorphisms in Indian populations and their possible evolutionary implications. Eur J Hum Genet. 1999;7:435–446. doi: 10.1038/sj.ejhg.5200317. [DOI] [PubMed] [Google Scholar]
- 43.McNeil W H. Plagues and peoples. Garden City, N.Y: Anchor Press/Doubleday; 1976. [Google Scholar]
- 44.Misra S P, Misra V, Dwivedi M. Proceedings of the First National Workshop on Helicobacter pylori: the Indian scenario. Indian J Gastroenterol. 1998;17:114–115. [PubMed] [Google Scholar]
- 45.Pagliaccia C, de Bernard M, Lupetti P, Ji X, Burroni D, Cover T L, Papini E, Rappuoli R, Telford J L, Reyrat J M. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc Natl Acad Sci USA. 1998;95:10212–10217. doi: 10.1073/pnas.95.17.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan Z J, Berg D E, van der Hulst R W, Su W W, Raudonikiene A, Xiao S D, Dankert J, Tytgat G N, van der Ende A. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J Infect Dis. 1998;178:220–226. doi: 10.1086/515601. [DOI] [PubMed] [Google Scholar]
- 47.Pan Z J, van der Hulst R W, Feller M, Xiao S D, Tytgat G N, Dankert J, van der Ende A. Equally high prevalences of infection with cagA-positive Helicobacter pylori in Chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia. J Clin Microbiol. 1997;35:1344–1347. doi: 10.1128/jcm.35.6.1344-1347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsonnet J. Helicobacter and gastric adenocarcinoma. In: Parsonnet J, editor. Microbes and malignancy: infection as a cause of human cancers. New York, N.Y: Oxford University Press; 1999. pp. 372–408. [Google Scholar]
- 49.Peek R M, Jr, Thompson S A, Donahue J P, Tham K T, Atherton J C, Blaser M J, Miller G G. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physicians. 1998;110:531–544. [PubMed] [Google Scholar]
- 50.Sarker S A, Rahman M M, Mahalanabis D, Bardhan P K, Hildebrand P, Beglinger C, Gyr K. Prevalence of Helicobacter pylori infection in infants and family contacts in a poor Bangladesh community. Dig Dis Sci. 1995;40:2669–2672. doi: 10.1007/BF02220458. [DOI] [PubMed] [Google Scholar]
- 51.Segal E D, Cha J, Lo J, Falkow S, Tompkins L S. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci USA. 1999;96:14559–14564. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma M P. Duodenal ulcer and Helicobacter pylori: presidential oration, Indian Society of Gastroenterology. Indian J Gastroenterol. 1999;18:51–53. [PubMed] [Google Scholar]
- 53.Smith J M, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suerbaum S, Smith J M, Bapumia K, Morelli G, Smith N H, Kuntsmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor N S, Fox J G, Akopyants N S, Berg D E, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter F M, Correa P. Long term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tee W, Lambert J, Smallwood R, Schembri M, Ross B C, Dwyer B. Ribotyping of Helicobacter pylori from clinical specimens. J Clin Microbiol. 1992;30:1562–1567. doi: 10.1128/jcm.30.6.1562-1567.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 58.van der Ende A, Pan Z J, Bart A, van der Hulst R W, Feller M, Xiao S D, Tytgat G N, Dankert J. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect Immun. 1998;66:1822–1826. doi: 10.1128/iai.66.5.1822-1826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Wouden E J, Thijs J C, van Zwet A A, Kleibeuker J H. Review article: nitroimidazole resistance in Helicobacter pylori. Aliment Pharmacol Ther. 2000;14:7–14. doi: 10.1046/j.1365-2036.2000.00675.x. [DOI] [PubMed] [Google Scholar]
- 60.van Doorn L J, Figueiredo C, Megraud F, Pena S, Midolo P, Queiroz D M, Carneiro F, Vanderborght B, Pegado M D, Sanna R, De Boer W, Schneeberger P M, Correa P, Ng E K, Atherton J, Blaser M J, Quint W G. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/s0016-5085(99)70065-x. [DOI] [PubMed] [Google Scholar]
- 61.van Doorn L J, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, Quint W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/s0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 62.van Doorn L J, Figueiredo C, Sanna R, Blaser M J, Quint W G. Distinct variants of Helicobacter pylori cagA are associated with vacA subtypes. J Clin Microbiol. 1999;37:2306–2311. doi: 10.1128/jcm.37.7.2306-2311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watkins W S, Bamshad M, Dixon M E, Bhaskara Rao B, Naidu J M, Reddy P G, Prasad B V, Das P K, Reddy P C, Gai P B, Bhanu A, Kusuma Y S, Lum J K, Fischer P, Jorde L B. Multiple origins of the mtDNA 9-bp deletion in populations of South India. Am J Phys Anthropol. 1999;109:147–158. doi: 10.1002/(SICI)1096-8644(199906)109:2<147::AID-AJPA1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 64.Westblom T U, Czinn S J, Nedrud J G. Gastroduodenal disease and Helicobacter pylori: pathophysiology, diagnosis and treatment. Curr Top Microbiol Immunol. 1999;241:1–313. [Google Scholar]
- 65.Xiang Z, Censini S, Bayeli P F, Telford J L, Figura N, Rappuoli R, Covacci A. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun. 1995;63:94–98. doi: 10.1128/iai.63.1.94-98.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamaoka Y, Kodama T, Kashima K, Graham D Y, Sepulveda A R. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol. 1998;36:2258–2263. doi: 10.1128/jcm.36.8.2258-2263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]