Abstract
Quantifying the frequency of shifts to new host plants within diverse clades of specialist herbivorous insects is critically important to understand whether and how host shifts contribute to the origin of species. Oak gall wasps (Hymenoptera: Cynipidae: Cynipini) comprise a tribe of ∼1000 species of phytophagous insects that induce gall formation on various organs of trees in the family Fagacae—primarily the oaks (genus Quercus; ∼435 sp.). The association of oak gall wasps with oaks is ancient (∼50 my), and most oak species are galled by one or more gall wasp species. Despite the diversity of both gall wasp species and their plant associations, previous phylogenetic work has not identified the strong signal of host plant shifting among oak gall wasps that has been found in other phytophagous insect systems. However, most emphasis has been on the Western Palearctic and not the Nearctic where both oaks and oak gall wasps are considerably more species rich. We collected 86 species of Nearctic oak gall wasps from most of the major clades of Nearctic oaks and sequenced >1000 Ultraconserved Elements (UCEs) and flanking sequences to infer wasp phylogenies. We assessed the relationships of Nearctic gall wasps to one another and, by leveraging previously published UCE data, to the Palearctic fauna. We then used phylogenies to infer historical patterns of shifts among host tree species and tree organs. Our results indicate that oak gall wasps have moved between the Palearctic and Nearctic at least four times, that some Palearctic wasp clades have their proximate origin in the Nearctic, and that gall wasps have shifted within and between oak tree sections, subsections, and organs considerably more often than previous data have suggested. Given that host shifts have been demonstrated to drive reproductive isolation between host‐associated populations in other phytophagous insects, our analyses of Nearctic gall wasps suggest that host shifts are key drivers of speciation in this clade, especially in hotspots of oak diversity. Although formal assessment of this hypothesis requires further study, two putatively oligophagous gall wasp species in our dataset show signals of host‐associated genetic differentiation unconfounded by geographic distance, suggestive of barriers to gene flow associated with the use of alternative host plants.
Keywords: Cynipidae, galls, host‐associated differentiation, incipient speciation, Quercus
Estimates of the number of insect species range from 1.5 to 30 million, such that by most accounts they are the most species‐rich class of animal (Erwin 1982; May 1990; Stork 1993; although see Larsen et al. 2017). The vast majority of insects are herbivorous or otherwise parasitic and use plants, fungi, or other animals as hosts for some part of their life cycle (Price 1980; Strong 1988), a fundamental relationship that has inspired a long history of research into the underlying causes of insects’ evolutionary success (Mitter et al. 1988; Jaenike 1990; Nosil 2002; Forister et al. 2012). One particularly compelling question is how important are shifts to new hosts to the origin of diversity? On one hand, shifts to new hosts in some insect systems can drive the evolution of reproductive isolation between insect populations using the ancestral and derived hosts (Craig et al. 2001; Berlocher and Feder 2002; Drès and Mallet 2002; Matsubayashi et al. 2010; Forbes et al. 2017). On the other hand, if such host shifts occur only rarely, most parasitic insect diversity could be a result of co‐cladogenesis between insects and their hosts. Such co‐cladogenesis is a pattern seen in some highly specialized plant‐insect systems (Roderick 1997; Machado et al. 2001; Rønsted et al. 2005; McLeish et al. 2007), although even some of these show evidence for host shifts (Wang et al. 2016, 2021). Inferring phylogenetic relationships for large clades of parasitic insects in the context of their host use patterns can therefore provide important clues about the relative importance of these two alternative mechanisms.
The oak gall wasps (Hymenoptera: Cynipidae: Cynipini) are a species‐rich clade of ∼1000 phytophagous (plant‐feeding) insects that primarily use Quercus (oak) species as hosts (Stone et al. 2002; Melika and Abrahamson 2002; Buffington et al. 2020; Melika et al. 2021a, 2021b). Female gall wasps oviposit into meristematic tissue, inducing abnormal, but highly structured, outgrowths of tissue (galls), inside of which the larvae develop (Stone and Schönrogge 2003; Csóka et al. 2005; Martinson et al. 2022). Gall wasps can attack many parts (organs) of oaks, including leaves, stems, buds, flowers, petioles, roots, and acorns, and many gall wasp species alternate between sexual (gamic) and asexual (agamic) generations (Stone et al. 2002), with each generation inducing different gall morphologies often on different organs on the same or different tree species (Pujade‐Villar et al. 2001; Egan et al. 2018). These differences in gall morphology, location, and the morphology of the agamic and gamic adult gall wasps have in some instances led to different generations being classified as different species or even different genera (Lund et al. 1998; Pujade‐Villar et al. 2001; Melika and Abrahamson 2002). Conversely, similarities in gall and adult wasp morphology have led to many genera describing para‐ or polyphyletic groups (Cooke 2018; Melika et al. 2021a). Several gall wasp species are also known from only one generation, likely because the alternate generation galls are inconspicuous or hidden, for example, on roots or under bark (Weld 1959; Hood et al. 2018). The complexity of the life cycle and biology of gall wasps makes them fantastic systems for asking myriad ecological and evolutionary questions but has simultaneously complicated understanding of their evolution and phylogeny.
Oaks are also a species‐rich and taxonomically complex group, although their phylogeny is better resolved than that of the gall wasps. There are ∼435 oak species worldwide, currently organized into two subgenera (Cerris and Quercus; Hipp et al. 2020), each containing several sections further divided into subsections (Table 1). The split between subgenus Cerris and subgenus Quercus is estimated to be 40−50 Ma (McIver and Basinger 1999; Hipp et al. 2018), with subgenus Cerris confined entirely to the Palearctic and subgenus Quercus primarily in the Nearctic (Hipp et al. 2020). Only two relatively small clades of subgenus Quercus have dispersed back to Eurasia (the Palearctic roburoids and part of section Ponticae; Kremer and Hipp 2020; Manos and Hipp 2021). At the species level, oaks are considerably more diverse in the Nearctic (Kremer and Hipp 2020), where the two most species‐rich sections (sect. Quercus [white oaks] and sect. Lobatae [red oaks]) broadly overlap geographically with one another and the ranges of three other smaller sections (Virentes, Ponticae, and Protobalanus; Denk et al. 2017; Hipp et al. 2018; Manos and Hipp 2021). Oak gall wasps are also most species rich in the Nearctic, with an estimated 700 species (Krombein et al. 1979; Penzes et al. 2018; Melika et al. 2021b).
Table 1.
Classification of the subgenera, sections, and subsections of genus Quercus following Manos and Hipp (2021). Some names in the “subsections” column are not currently recognized as true subsections, but nevertheless represent apparently monophyletic clades of oaks (Hipp et al. 2018, 2020; Manos and Hipp 2021). Oak clades not represented by any gall wasps in this study are indicated by an asterisk
| Subgenus | Section | Subsection (Clades) | Biogeography |
|---|---|---|---|
| Cerris | Cerris | Palearctic | |
| Ilex* | Palearctic | ||
| Cyclobalanopsis* | Palearctic | ||
| Quercus | Lobatae | Agrifoliae | Nearctic—Pacific coast |
| Palustres | Nearctic—Central USA | ||
| Coccineae | Nearctic—Central USA | ||
| Phellos | Nearctic—Central USA | ||
| “Texas red oaks”* | Nearctic—Texas | ||
| Erythromexicana | Nearctic—Mexico | ||
| Protobalanus | Nearctic—Southwest USA and Northwest Mexico | ||
| Ponticae* | Western Palearctic and CA | ||
| Virentes | Nearctic—Southeast USA and Mexico | ||
| Quercus | Dumosae | Nearctic—Pacific coast | |
| Prinoideae | Nearctic | ||
| Albae | Nearctic—Central USA | ||
| Roburoids (incl. Mesobalanus) | Palearctic | ||
| Stellatae | Nearctic | ||
| Polymorphae* | Nearctic—Southwest USA to Guatemala | ||
| Leucomexicana | Nearctic—Southwest USA and Mexico |
If host shifting has contributed to the evolution of oak gall wasp diversity, then phylogenies of oak gall wasps should recover patterns of historical host shifts among oak subgenera, sections, subsections, and species. To date, the most ambitious phylogenetic study directly focused on the role of host shifting in oak gall wasp speciation sequenced two nuclear genes and a mitochondrial gene for 84 species of gall wasp in the Western Palearctic (Stone et al. 2009). Resultant trees showed that this assemblage of gall wasps sorted into clades organized by host tree associations with limited evidence of regular shifts among the oak subgenera and sections. However, a test for gall wasp host shifts limited to the Palearctic—where cynipid and oak diversity are well described but oak diversity is limited to three relatively species‐poor and phylogenetically divergent groups (Manos and Hipp 2021)—ultimately may not have adequate power to resolve the overall role of host shifting in oak gall wasp diversity at a global scale, especially if more shifting occurs at or below the oak section level. Further, new advances in global and continental oak phylogeny that have resolved relationships below the section level (Hipp et al. 2018, Hipp et al. 2020; Manos and Hipp 2021) now allow for assessing the role of host shifting at progressively finer scales.
Just as shifts among host trees may be correlated with diversification of oak gall wasps, the fact that different species induce galls on very different plant organs also raises the question of whether shifts to new organs—even within the same tree species—could lead to the evolution of reproductive isolation and contribute to speciation. Indeed, in one clade of Palearctic Andricus, shifts among tree organs do appear to be common, even as tree host shifts appear rarer (Cook et al. 2002). With a shift to new organs on the same plant, one emergent reproductive barrier could be habitat isolation (Carroll and Boyd 1992; Berlocher and Feder 2002; Feder and Forbes 2007; Hood et al. 2015), wherein discrimination among plant organs isolates insect populations from one another. Another possibility is allochronic isolation, with populations of gall wasps becoming isolated temporally due to their induction of galls on organs produced by trees at different times during the year. Temporal (allochronic) isolation has been shown to be a strong contributor to incipient speciation in other insects (Feder and Filchak 1999; Joy and Crespi 2007; Inskeep et al. 2021; Mattsson et al. 2022). New phylogenies of Nearctic gall wasps reared from a diversity of galled organs across a diversity of oak species should allow for evaluation of whether shifts among different host organs are correlated with speciation events in gall wasps.
Another question important to understanding patterns and drivers of diversification of oak gall wasps, although one entangled with host association, is what is the phylogenetic relationship of the most species‐rich gall wasp assemblage—the Nearctic gall wasps—to those in the Palearctic and in other biogeographic regions? The first serious attempts at understanding the phylogeny began with Kinsey (1920, 1936) who—prior to the advent of molecular phylogenetics—used wasp morphology, gall characters, geography, host ranges, and other nonmolecular characters to develop his hypothesis that the cradle of gall wasp diversity is present‐day Mexico. Most modern studies have employed one to three loci to infer relationships, and in general results have conflicted with Kinsey's view, putting the likely origin of oak galling in the Palearctic (Stone et al. 2009; Andersen et al. 2021). However, these studies have often used no (Stone et al. 2009) or a limited number of (Ronquist et al. 2015; Andersen et al. 2021) Nearctic gall wasp samples, or have used Nearctic wasps but no Palearctic wasps (Melika et al. 2021a). One recent three‐locus study that included both Nearctic and Palearctic samples maintains Palearctic wasps as basal to the Nearctic wasps, but with intercalated clades of Palearctic and Nearctic wasps (Nicholls et al. 2017), suggesting the possibility of multiple movements between continents throughout their evolutionary history. However, relationships among clades in this study were not sufficiently well resolved to assess this question directly.
Three recent studies have employed a multilocus genome‐level approach using Ultraconserved Elements (UCEs; Faircloth et al. 2012) to address oak gall wasp phylogenetic relationships. First, Cooke (2018) used 17 Palearctic and 52 Nearctic wasp species in an attempt to resolve questions about the monophyly of several oak gall wasp genera. Next, Blaimer et al. (2020) sequenced some Cynipini as part of a larger study of Cynipoidea and found a likely point of origin for oak galling approximately ∼50 MYA, roughly coincident with the split between subgenus Cerris and subgenus Quercus oaks (Hipp et al. 2018). Most recently, Brandão‐Dias et al. (2022) added additional data harvested from published Cynipini genomes to expand the Blaimer et al. (2020) UCE gall wasp dataset. Although not the main focus of any of these studies, in all cases the resultant phylogenies appear to show the Palearctic fauna as being paraphyletic. Cooke's (2018) study also showed a Nearctic oak gall wasp (Protobalandricus spectabilis) as being basal to all other oak gall wasps. An expanded Nearctic gall wasp phylogeny will aid in resolving these relationships.
Here, we report the largest UCE‐based phylogenetic study of Nearctic oak gall wasps to date. We sequence UCEs for 86 species reared from galls collected from 24 species of oak across the continental United States. We paired these with existing UCE data from Palearctic and other Nearctic oak gall wasps (Blaimer et al. 2020; Brandão‐Dias et al. 2022), such that our set spanned six of the eight oak sections and 11 of the 13 subsections of oak sections Lobatae and Quercus. Combining our new collections with existing Palearctic data allows us to re‐examine relationships among the Nearctic and Palearctic Cynipini. With this accomplished, our careful records of the host oak species and organ from which each gall was collected allow us to ask how often gall wasps have shifted among oak tree sections, subsections, and organs. After considering oak gall wasp biogeography and host shifting at a Holarctic scale, we synthesize our findings and explore, in detail, two gall wasp clades (Andricus quercuspetiolicola and a clade of Disholcaspis wasps) that show evidence of host‐associated genetic differentiation indicative of ongoing or recent divergence related to host plant shifts.
Material and Methods
COLLECTIONS
We collected oak galls from August 2015 to September 2019. Most collection sites were located in the eastern half of the United States, supplemented by smaller collections from the Southwest and the Pacific Coast (Fig. S1). Our strategy was not to collect all of the ∼700 estimated Nearctic gall wasp species, but to generate a subsample reflecting a wide geographic distribution spanning several different ecological dimensions (on different tree species, host organs, etc.). We reared insects directly from galls by placing gall collections of the same type and same collection date for each locality into deli cups with the base removed and replaced with a fine gauze to allow air flow. Most collections were stored in an incubator that rotated through four 3‐month artificial “seasons” of light:dark (LD), temperature, and relative humidity (RH) settings, as follows: Spring: 13:12 LD, 18°C day, 5°C night, 75% RH; Summer: 15:9 LD, 24°C day, 17°C night, 85% RH; Fall: 14:10 LD, 20°C day, 14°C night, 75% RH; Winter: 10:14 LD, 10°C day, 5°C night, 75% RH. Some galls from warmer climates were kept at room temperature on a lab bench to avoid dramatic changes in temperature, including simulated winter conditions. All cups were checked daily for up to 3 years for emergent insects, which were placed into 95% ethanol in microcentrifuge tubes labeled with collection metadata and the emergence date. Gall wasps were initially identified to species based on the morphological characteristics of the galls from which they emerged, and by their tree host, following a variety of resources (Weld and von Dalla Torre 1952; Weld 1957, 1959, 1960; Melika and Abrahamson 2002; Gallformers.org 2021; Russo 2021). Although gall morphology was our primary indicator of gall wasp species identity, the assumed wasp genus was verified morphologically from reared wasps against keys (Weld and von Dalla Torre 1952; Zimmerman 2018; Melika et al. 2021a) and all wasps selected for sequencing were photographed (see below) such that their identity could be confirmed as needed. We refer to some gall wasps reared from as‐yet‐undescribed galls by their description on gallformers.org as of December 2021, which includes the best‐attempt genus name, followed by the host tree from which that gall was first recorded, and a description of its morphology (e.g., “Callirhytis_q_stellata_pentagonal_cluster”).
DNA EXTRACTIONS
We extracted DNA from the whole adult bodies of 209 individuals representing 81 named species and five undescribed oak gall wasp species, and two Synergus individuals to use as outgroups (Table S1). In cases where we had reared >1 gall wasp individuals of the same species from different tree hosts and/or different geographic regions, we included these replicates. We photographed one fore wing and the lateral side of each wasp body prior to destructive extractions (File S1) and for many species deposited voucher specimens representing the same or similar collections into the Frost Entomological Museum at Pennsylvania State University or the National Museum of Natural History (Table S1; USNMENT_01788005‐064, PSUC_FEM_248712‐35). For 38 individuals sampled before 2018, we extracted DNA using a Qiagen DNeasy kit (Redwood City, CA). For the remaining 173 samples, we used a CTAB/PCI approach modified from Chen et al. (2010) because we found this method increases both the quality and quantity of the DNA yield.
UCE SEQUENCING
We prepared libraries for each individual using the Kapa Hyper Prep library preparation kit (Kapa Biosystems Inc., Wilmington, MA) and enzymatic fragmentation of DNA. To ensure the correct distribution of fragment lengths for UCE sequencing (300−800 bp), we double size‐selected fragments from each individual with AMpure Beads and verified fragment distributions using a Bioanalyzer. Libraries were then pooled and hybridized following the MyBaits protocol (ArborBiosciences, Ann Arbor, MI) using the Hym v2P bait set (Branstetter et al. 2017). We confirmed UCE loci enrichment for pooled samples using relative qPCR. We then sequenced the entire pooled library on one lane of a NovaSeq6000 (Illumina, Inc. San Diego, CA) at the Iowa Institute of Human Genetics at the University of Iowa. We followed the Phyluce version 1.7 pipeline (Faircloth 2016) to process our UCE loci. We first trimmed remaining adapters and primers using Trimmomatic (Bolger et al. 2014). We then assembled de novo contigs using SPAdes (Bankevich et al. 2012). Next, the UCE loci were aligned and internally trimmed using PHYLUCE (Faircloth 2016) and MAFFT version 7 (Katoh and Standley 2013).
We generated 2719−2,347,711 raw sequencing reads per taxon (median: 692,910) that we assembled with Spades to generate 1645−120,122 contigs per taxon (median: 9604). One Disholcaspis quercusglobulus sample was removed from the dataset due to low capture of UCE loci (2719 raw reads, 175 contigs, 91 UCE loci), probably the result of too little DNA being included in the final pooled library sample. Realized capture of the 2590 target UCE loci ranged from a minimum of 472 to a maximum of 1607 loci. Further details on UCE capture and assembly can be found in Table S2.
To investigate evolutionary relationships between Palearctic and Nearctic gall wasps, we added UCE loci from 16 additional taxa previously published by Branstetter et al. (2017), Blaimer et al. (2020), and Brandão‐Dias et al. (2022). We did not use UCE data from Cooke (2018) because these were not publicly available. We also bioinformatically extracted UCE loci from eight existing genomes following the protocol in Phyluce 1.7 (Faircloth 2016) again using the Hym v2P bait set. We intentionally selected gall wasp species from other geographical regions and/or not previously represented in our dataset. We also only used samples with >300 UCE loci recovered, a minimum we set arbitrarily but with the goal of maximizing data completeness. Samples added from other sources can be found in Table S3.
PHYLOGENETIC INFERENCE
We constructed phylogenetic trees for the entire combined dataset (235 individuals, representing 106 named species, five unknown species) using both a concatenated matrix and a multiple‐species coalescence approach, comparing resultant tree topologies across methods. For the concatenated matrix approach, we assembled three different data matrices composed only of the UCE loci found in at least 50%, 75%, and 90% of all individuals. These three matrices resulted in 1325, 1077, and 464 loci, respectively. Total alignment lengths of sequences from the three matrices were 442,593, 381,400, and 184,665 bp. We used AMAS (Borowiec 2016) to determine alignment length, percent missing data, AT and GC content, number and proportion of variable sites, and the number and proportion of parsimony informative sites (Table S4). We partitioned the matrix with SWSC‐EN (Tagliacollo and Lanfear 2018) and PartitionFinder2 (Lanfear et al. 2017) using the “rclusterf” algorithm (Lanfear et al. 2017) to test three models: GTR, GTR+G, and GTR+I+G. We then generated Maximum Likelihood (ML) trees with IQ‐TREE2 (Minh et al. 2020) for each of the concatenated matrices with 1000 Ultrafast bootstraps (Hoang et al. 2018) and 1000 SH‐likelihood ratio tests (Guindon et al. 2010). For the multiple‐species coalescence approach, we generated gene trees for each UCE locus with IQ‐TREE with ModelFinder (Kalyaanamoorthy et al. 2017) to determine the best model of substitution for each locus. We then used ASTRAL‐III (Zhang et al. 2018) to estimate a species tree from the combined pool of gene trees.
In addition to a tree containing both Palearctic and Nearctic gall wasps, we constructed a phylogeny consisting of only our new North American samples. Because we had used fresh specimens where other studies had included primarily museum samples, this “North American only” dataset has the advantage of having many more UCE loci recovered per individual, increasing the amount of phylogenetically informative sites. We again generated phylogenies using concatenated matrices and a multiple species coalescence approach as previously described.
BIOGEOGRAPHY AND HOST ASSOCIATIONS
To investigate how tree host and biogeographic history may have influenced gall wasp evolution, we mapped various levels of oak tree taxonomic organization onto our gall wasp phylogenies. The genus Quercus is composed of two subgenera, Cerris and Quercus, each of which is further divided into sections (Hipp et al. 2018), and further still into several subsections (Manos and Hipp 2021; Table 1). Oaks in subgenus Cerris are only found in the Palearctic, whereas members of three sections of the subgenus Quercus—Lobatae (red oaks), Virentes (live oaks), and Protobalanus (golden oaks)—are almost exclusively restricted to the Nearctic. Section Quercus (white oaks) straddles the Palearctic and Nearctic, and section Ponticae (intermediate oaks) is found only on the Pacific coast of North America and on the eastern coast of the Black Sea (Manos and Hipp 2021). Within the North American oaks in subgenus Quercus, sections and subsections tend to be restricted to one of three biogeographic regions: Eastern North America, the Southwest (representing the northern tip of a Mexican and Central American oak flora), or the Pacific coast (California, north to British Columbia) (Hipp et al. 2018; Manos and Hipp 2021).
We estimated the ancestral state for host associations of gall wasps using R package Phytools (Revell 2012). We coded the gall wasp‐associated traits based on host plant taxonomy (oak sections Quercus, Lobatae, Virentes, and Cerris; non‐oak genera Chrysolepis and Castanea), and for the three Western Palearctic gall wasp species that alternated between Quercus and Cerris we coded the alternate host plants as separate states. We pruned the 75% complete matrix tree down to a single specimen per taxon and performed marginal reconstructions and selected the equal rates (ER) model based on AIC score. The resulting phylogenetic tree was built using the package “ape” in R version 4.1.1 (Paradis et al. 2004; R Core Team 2021).
We also investigated evolutionary switches in the location of the gall on different host tree organs (e.g., bud, leaf, stem). Due to the cyclically parthenogenetic life cycle of most gall wasps, the two generations often induce galls on different plant organs, such that shifts to new organs could occur in one or both generations. We combined our own observations from collections with accounts from the literature (relying especially on gallformers.org) to map gall locations for both sexual (gamic) and sexual (agamic) gall generations for each species in our North American phylogeny, which we then used to tally a minimum number of host organ shifts for both the sexual and asexual generations.
Results
PHYLOGENY AND BIOGEOGRAPHY OF CYNIPINI
Both the multi‐species coalescence approach and concatenated matrix approaches (Figs. S2–S5) generated essentially the same topology (see below). We chose the concatenated matrix approach resulting from the 75% complete data matrix (1077 loci) as the basis for figures in this manuscript because it provided a middle ground between optimizing the amount of shared data among specimens and maximizing the total number of loci. Figure 1 shows the topology of the combined Palearctic and Nearctic tree (collapsed at the species level; see Fig. S3 for the full tree). Neither the Palearctic nor Nearctic gall wasps were monophyletic in any of the inferred trees (Figs. 1, S2−S5). The Palearctic gall wasps formed four independent clades in all tree topologies, with the Cerris‐associated Pseudoneuroterus saliens and Plagiotrochus suberi and the chestnut‐galling Dryocosmus kuriphilus being basal to all other gall wasps (Fig. 1). The remaining Palearctic gall wasps split into three clades within the larger North American fauna: a clade of Andricus wasps galling oaks of subgenus Quercus or alternating between oaks of subgenera Cerris and Quercus, a single Andricus species (A. inflator) on subgenus Quercus, and a clade that pairs Neuroterus quercusbaccarum with Cynips divisa, both on subgenus Quercus (Fig. 1). This topology implies a minimum of four movements of gall wasps between the Palearctic and Nearctic: one presumed original colonization of the Nearctic from the Palearctic, and then three clades subsequently “returning” to the Palearctic, either at the same or at different times. We say “minimum of four” because our sampling strategy did not capture all ∼700 species of Nearctic gall wasps, and evidence for additional cross‐continental exchanges might emerge with more sampling.
Figure 1.
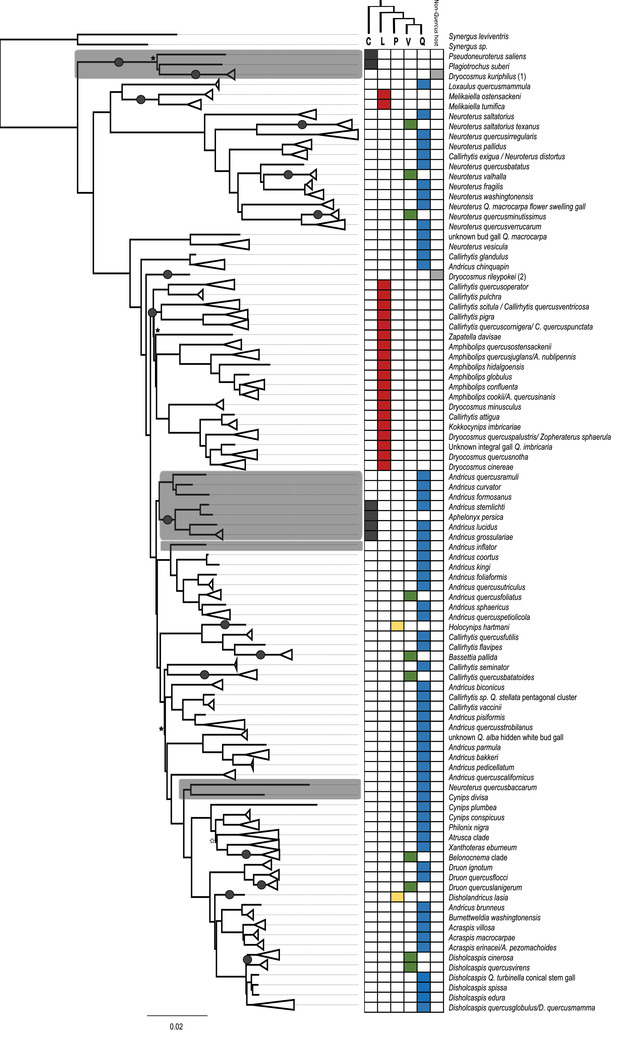
Phylogenetic tree of Nearctic and Palearctic oak gall wasps represented by the partitioned 75% complete data matrix. Clades have been collapsed to the species, or rarely genus, level (see Fig. S3 for the expanded tree). Tips with more than one species name indicate apparent polyphyly or the closing of life cycles (see File S2). All nodes have 100% bootstrap support, except for those indicated with black (<100%) or white (<80%) stars. Clades shaded in gray comprise Palearctic species. The grid to the right of the tree indicates the association of each gall with an oak section (or two sections, in the case of the alternating‐host Palearctic Andricus clade). The cladogram above the right‐hand grid illustrates the relationship among the oak sections: C = Cerris, L = Lobatae, P = Protobalanus, V = Virentes, Q = Quercus. (1) = from galls on chestnut (Castanea); (2) = from galls on Chrysolepis. Dark circles, superficially resembling galls, on some branches of the phylogeny indicate changes in host tree section, assuming an ancestral association with oaks in subgenus Quercus, section Quercus as suggested by Ancestral State Reconstruction (Fig. S6).
As in several previous phylogenetic treatments of the Cynipini (Rokas et al. 2003; Ács et al. 2010; Ronquist et al. 2015; Cooke 2018; Andersen et al. 2021; Brandão‐Dias et al. 2022), several gall wasp genera, including Andricus, Callirhytis, Cynipis, Neuroterus, and Dryocosmus, were found to be either para‐ or polyphyletic in our tree topologies (Figs. 1, 2, 3, 4). Of the genera represented in the tree by more than three species, only three genera maintain monophyly: Amphibolips, Acraspis, and Disholcaspis, the latter only because of a recent revision (Melika et al. 2021a) after Cooke (2018) had found Disholcaspis to be polyphyletic. Similarly, some species with different species names were sufficiently similar such that they likely represent the alternate generations of the same species (Fig. S3). These instances are coded in figures with a forward slash between the two different species names and further discussion regarding their status may be found in File S2.
Figure 2.
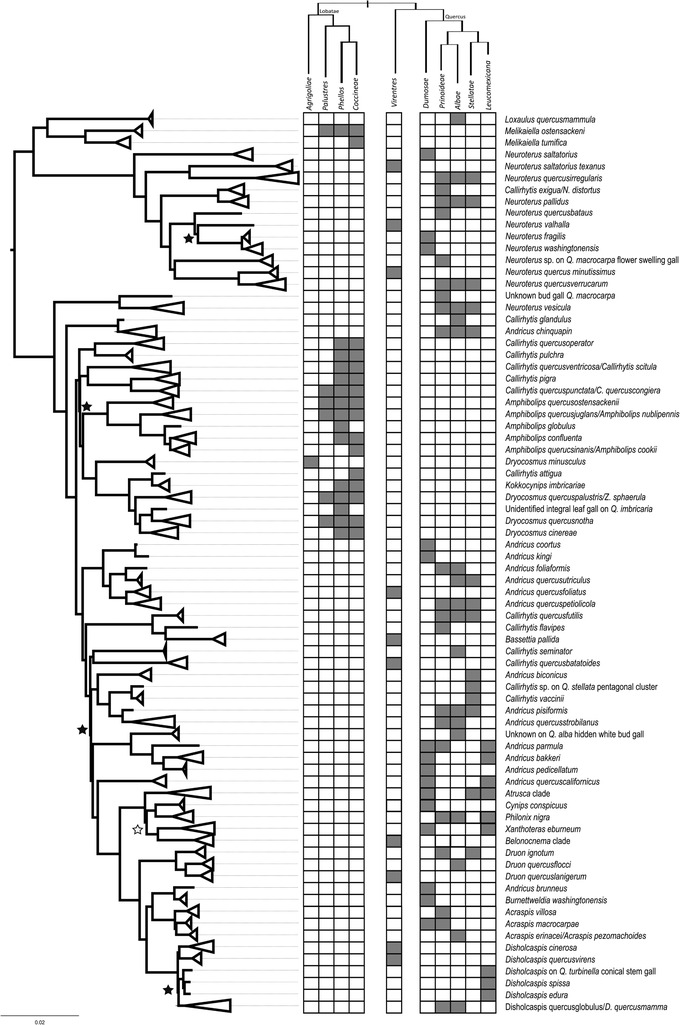
Phylogenetic representation of Nearctic gall wasps based on partitioned 75% completeness data matrix (see Fig. S8 for the expanded tree). Clades have been collapsed to the species or genus level as in Figure 1. Gray boxes in the table to the right of the tree represent known host associations—including those from which we collected insects and those from the literature—for each gall wasp species at the level of oak subsection. The cladogram above the table represents the evolutionary relationships among subsections within sections Lobatae, Virentes, and Quercus (section Virentes has no subsections). All nodes have 100% bootstrap support, except for those indicated with black (<100%) or white (<80%) stars. Although this tree implies that many oak gall wasp species induce galls on trees across >1 subsection, it may instead be the case that many taxa are species complexes, each species having a more restricted host range (see Discussion).
Figure 3.
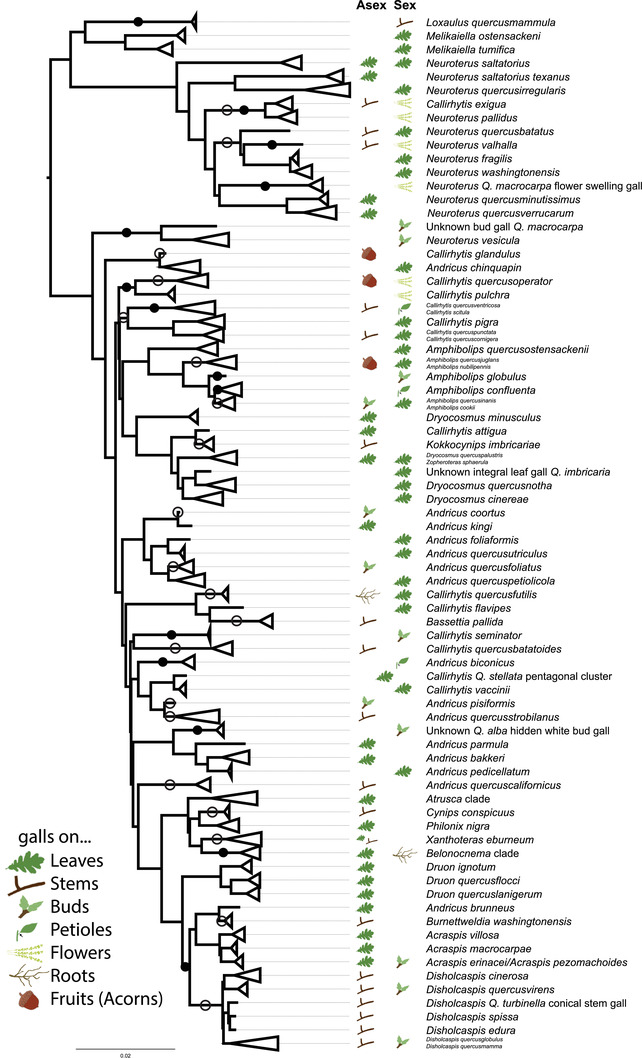
Known locations of galls on oaks for our Nearctic gall wasp set. Many cynipid gall wasps undergo cyclical parthenogenesis and have both asexual (“Asex”) and sexual (“Sex”) generations that gall different oak organs. Empty spaces indicate missing data and are in many cases due to the gall wasp having been described only from one generation. It is also possible that in some cases only one generation exists (e.g., Andricus quercuscalifornicus), although this is apparently rare in the Cynipini (Pujade‐Villar et al. 2001; Stone et al. 2008). Although taxon sampling and incomplete data prevent a formal reconstruction of location and number of host switches, we have mapped one possible minimum‐change scenario onto branches of the phylogeny: dark circles indicate changes in host organ by the sexual generation, assuming an ancestral association with leaves. Open circles indicate changes in host organ by the asexual generation, again assuming an ancestral association with leaves. Myriad other scenarios exist but in all cases would show that changes in host organ use have been common for both gall wasp generations.
Figure 4.
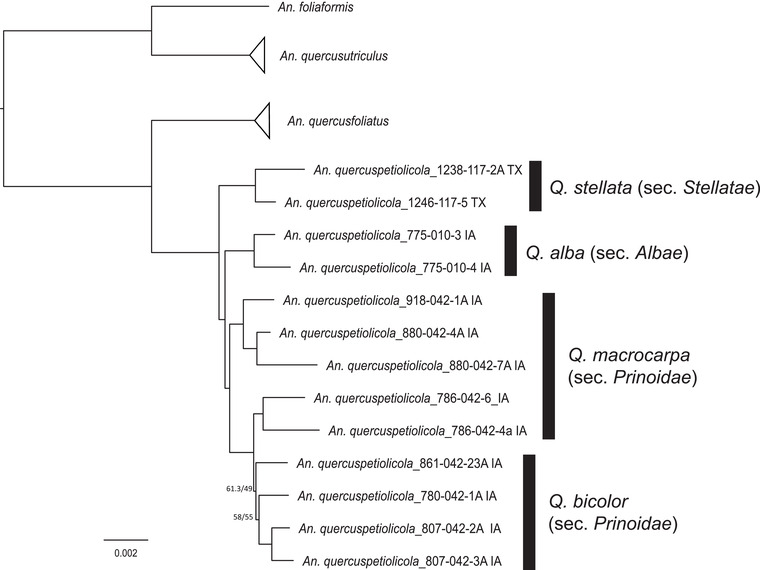
Host‐associated genetic differentiation in Andricus quercuspetiolicola. (a) Phylogeny of A. quercuspetiolicola based on a concatenated 75% data matrix of UCE loci. Branch support is bootstrap values/SH‐likelihood values. Black bars and annotations indicate host tree species and the Quercus section from which we collected wasps. Support values show bootstrap values followed by SH‐likelihood ratios (100/100 when none are shown).
Minor disagreements among trees using different approaches and data completeness matrices included (1) the clade including Callirhytis quercusscitula, C. quercusventricosa, C. pigra, C. quercuscornigera, and C. quercuspunctata was sometimes placed as sister to the clade including C. quercusoperator and C. pulchra instead of as sister to the rest of non‐Callirhytis gall wasps associated with section Lobatae oaks; (2) the clade including Andricus biconicus, an undescribed Callirhytis sp., C. vaccinii, An. pisiformis, and An. quercusstrobilanus was variably placed as sister to C. seminator and C. quercusbataoides or to a clade of Andricus species; and (3) the Cynips conspicuus and Philonix nigra clade was sometimes sister to and sometimes outgroup to the clade consisting of the Atrusca gall wasps, Xanthoteras eburneum, and the Belonocnema gall wasps.
HOST ASSOCIATION AND HOST SHIFTING
The ancestral state reconstruction recovered the oak subgenus Quercus as the ancestral host (Fig. S6). Based on this reconstruction, a minimum of 16 transitions among oaks of different sections are required to explain patterns in our tree (Fig. 1). As with inferences about geographic histories, these are minimum estimates because additional sampling of gall wasp species would only be expected to increase the number of inferred changes in host. Transitions between section Lobatae and section Quercus are estimated to have occurred at least twice, whereas those among the more closely related oak sections Quercus and Virentes have occurred at least nine times. The two gall wasp species, Holocynips hartmani and Disholandricus lasia, that were associated with oak section Protobalanus were embedded in two different clades of wasps, each associated with sections Quercus and Virentes (Fig. 1), implying at least two additional host shifts. Finally, a clade of Quercus‐associated Andricus wasps has apparently shifted back to subgenus Cerris, section Cerris, but only for one of their two generations. Two other gall wasp lineages have moved from oak hosts onto more distantly related hosts (Dryocosmus kuriphilus onto Castanea and Dryocosmus rileyipokei onto Chrysolepis).
Host shifts between subsections of oaks are also apparent in the phylogeny of North American cynipids, although quantifying the incidence of shifts between subsections is complicated by some individual gall wasp species that apparently use host trees across several subsections (Figs. 2, S7−S9). Although no gall wasp species in the North American assemblage apparently galls trees across multiple oak sections (i.e., only one host tree column is occupied for each Nearctic gall in Fig. 1), both historical records and our own collections show that many gall wasp species induce galls on trees from >1 subsection (i.e., multiple species occupy several subspecies columns in Fig. 2). Still, the presence of gall wasps on subsections Agrifoliae and Dumosae in multiple clades cannot be explained without invoking inter‐subsection host shifts.
The location at which galls are initiated and develop on host trees has also changed frequently across the Nearctic gall wasps (Fig. 3). In spite of a considerable amount of missing data regarding gall location for both generations of many species, a strong signal emerges: gall wasps in this phylogeny show evidence of at least 34 changes in location of their galls on oak organs (14 in the asexual generation, 20 in the sexual generation for one minimum evolution scenario; see Fig. 3). Within some clades of relatively recently diverged gall wasps, gall location appears to be more conserved. For instance, galls of asexual Disholcaspis (where known) are always on stems, whereas the galls of the corresponding sexual generation (again, where known) for all species are on buds. Similarly, species in the Acraspis, Atrusca, Belonocnema, and Druon clades show no apparent change in organ use within either generation (Melika and Abrahamson 2002; Zhang et al. 2021c) (Fig. 3). Whether this apparent conservation of host organ use within these clades is real or a function of our limited taxon sampling is unknown.
Discussion
Our oak gall wasp phylogenies lead to several new conclusions regarding the biogeographic history and evolution of this charismatic and speciose tribe of insects. Although we find additional support for a Palearctic origin of oak galling, a primary finding of our study is that some Palearctic clades appear to have their proximate origin in the Nearctic. We also uncovered evidence that host shifting has occurred frequently, including between tree species in different sections and subsections and among different tree organs. We discuss the implications of these results for the study of gall wasps as well as deeper implications for the role of variation in host plant use in the diversification of gall‐inducing insects.
OAK GALL WASP BIOGEOGRAPHY
Our phylogenies further crystallize a biogeographic history of oak gall wasps having their ultimate origin in the Palearctic, although some questions still remain. Oak subgenus Cerris (Palearctic only) split from subgenus Quercus (primarily Nearctic, with one clade in the Palearctic) in the early to mid‐Eocene, an estimated 50 Ma (Hipp et al. 2018). Likewise, the diversification of Cynipini into many of its extant lineages is estimated at ∼50 Ma (Blaimer et al. 2020). Together, these lines of evidence support the hypothesis of an ancestral split between Cerris‐ and Quercus‐associated gall wasps, with the origin of these gall wasps occurring in the Palearctic (as suggested by Stone et al. 2009). In our dataset, two of the three most basal oak gall wasps are the Palearctic endemic species Plagiotrochus suberi and Pseudoneuroterus saliens, which are associated with cork oak and Turkey oak (both subgenus Cerris; section Cerris) (the other wasp in the clade induces galls on chestnut trees). As the Palearctic is the only biogeographic realm where subsection Cerris oaks are endemic, a Palearctic origin for these oak gall wasps remains the most parsimonious hypothesis. One caveat to this inference drawn from our analysis is that our dataset did not include Protobalandricus spectabilis, a Nearctic oak gall wasp species associated with oaks in section Protobalanus. Protobalandricus spectabilis was inferred to be basal to all others by the phylogeny presented in Cooke (2018). However, the support for this relationship in Cooke (2018) was lower than for other inferred relationships. Moreover, two three‐gene phylogenies (Nicholls et al. 2017; Andersen et al. 2021) have instead grouped P. spectabilis as being sister to the other Nearctic species and not basal to all oak gall wasps. Future work that includes this taxon is required to resolve its relationships with the other oak gall wasps.
In contrast to our phylogenies’ apparently confirmatory nature with respect to the ultimate origins of oak gall wasps, our evolutionary trees rewrite the proximate origins of many extant oak gall wasp lineages. Although the present study and other previous genetically based phylogenies (Stone et al. 2009; Andersen et al. 2021) refute Kinsey's (1936) hypothesis of the Nearctic region as the point‐of‐origin for the oak‐gall wasp association, our phylogeny also shows three Palearctic clades embedded within the Nearctic gall wasps, with high support, which strongly suggests movements of oak gall wasps from the Nearctic back to the Palearctic. Hints of this biogeographic exchange can be seen in the UCE phylogenies of Cooke (2018), Blaimer et al. (2020), and Brandão‐Dias et al. (2022), with Cooke's (2018) phylogeny suggesting as many as five separate movements to the Palearctic. Thus, although gall wasps may not have originated in the Nearctic, this region must now be reconsidered as a cradle of most or all gall wasp diversity associated with Palearctic roburoid oaks (the only Palearctic clade of oaks in section Quercus). Importantly, this scenario matches current biogeographic hypotheses for the origins of the Palearctic roburoids themselves, which clearly originated in the Americas before moving into Asia and Europe (likely bringing Nearctic gall wasps with them) (Manos and Hipp 2021). One caveat is that the Eastern Palearctic remains undersampled, with just one species in our dataset (Andricus formosanus, from Taiwan; Blaimer et al. 2020). Although in our new combined Nearctic‐Palearctic phylogeny this wasp was embedded in one of the eastern Palearctic clades, more sampling should be made from this and other geographic areas.
A second caveat with respect to the combined Nearctic‐Palearctic dataset is that there was considerable difference in locus recovery between our new data from recently collected material and previously published UCEs. Because most representatives from the Palearctic region had 600 or fewer UCE loci represented (Tables S2 and S3), one might be mildly cautious about the phylogenetic placement of these wasps in our trees. We note, however, that the Nearctic representatives from Blaimer et al. (2020) (e.g., Amphibolips hidalgoensis) resolve within clades with their presumed close relatives and conspecifics (Fig. 1). Further, the topologies of our trees are consistent with those of Blaimer et al. (2020), and the general phylogenetic relationships among named taxa are similar to those recovered for the same species using a small number of Sanger‐sequenced loci (Stone et al. 2009; Ács et al. 2010; Ronquist et al. 2015; Andersen et al. 2021). For these reasons, the lower numbers of UCE loci for some samples are not likely to have caused major systematic (pun intended) errors in phylogenetic placement of the Palearctic species.
GALL WASP HOST SHIFTING ACROSS OAK SECTIONS AND ORGANS
Our data provide the clearest “big picture” assessment yet of the tribe‐wide relationships between host shifts and diversity in oak gall wasps. Our results demonstrate that gall wasps have moved between clades of oak tree hosts including at least 16 shifts among oak sections and some nonzero number of shifts among subsections within the same section (see below). These numbers do not include the previously published shift from section Ilex back to section Cerris by Plagiotrochus suberi (as the Ilex‐associated Plagiotrochus species are not present in our tree; see Stone et al. 2009). Some movements between oak sections could reasonably be interpreted as co‐cladogenesis; for instance, the deepest split between the three basal Palearctic gall wasps and the primarily Nearctic clade could have been coincident with subgenus Cerris splitting from subgenus Quercus. However other major changes in tree association (e.g., two independent transitions from section Quercus to section Lobatae, nine transitions from section Quercus to section Virentes, and two transitions onto non‐oak hosts) cannot all be explained by cospeciation of gall wasps with oaks and require at least some host shifting to have occurred.
Our conclusions differ from those of previous work that focused exclusively on Palearctic gall wasps (Stone et al. 2009; Ács et al. 2010). These studies found that gall wasps sorted largely into clades by their association with oak subgenera (Cerris, Quercus, or alternating between the two), and showed only one apparent complete shift among sections within Cerris (Stone et al. 2009). Based on this apparent minimal historical movement among hosts, these previous studies concluded that host use is strongly conserved in oak gall wasps, at least at the level of oak section. Indeed, in these previous studies, near monophyly among gall wasps associated with different tree sections has been the most reasonable emergent hypothesis based on gall wasp sampling and resultant phylogenies. In fact, if our UCE tree had been restricted to only the taxa that overlapped with those represented in the phylogenies of Andersen et al. (2021) and Stone et al. (2009), they would have shown essentially the same pattern.
Our differing conclusions from studies of primarily Palearctic species about the prevalence of host tree shifts in gall wasps are a result of our increased sampling of both gall wasps and oak sections from the Nearctic. Approximately 65% of oak species are found in the Nearctic (Nixon and Muller 1997; Manos et al. 1999; Hipp et al. 2018), including most of the subsections of oaks in subgenus Quercus, section Quercus (oaks from only one of these subsections occur in the western Palearctic; Manos and Hipp 2021), the entirety of sections Virentes and Protobalanus, and all four subsections of section Lobatae (no Virentes or Lobatae are endemic to the Palearctic) (Table 1). Conversely, the Western Palearctic, where most phylogenetic studies of cynipids have been conducted (Cook et al. 2002; Bailey et al. 2009; Stone et al. 2009), has just 29 endemic oak species (Govaerts and Frodin 1998). Oak gall wasp richness is also correspondingly higher in the Nearctic, amounting to 70%−80% of an estimated >1000 species globally (Melika and Abrahamson 2002; Buffington et al. 2020; Melika et al. 2021b). Our samples spanned most of the Nearctic oak sections and subsections, with the exception of some oak clades in Texas, Mexico, and Central America. These particular clades of oaks appear to have diversified recently and rapidly (Manos et al. 1999; Manos and Hipp 2021), such that their associated gall wasps will be an exciting future addition to these phylogenies. In general, as future work incorporates more gall wasp species, we expect estimates of the number of historical changes in host use to increase.
Host shifts are also common between tree organs. For example, for the three species clade consisting of Amphibolips globulus, Am. confluenta, and Am. quercusinanis/Am. cookii, host tree association appears to have been somewhat conserved. Amphibolips globulus only induces galls on subsection Phellos oaks and Am. quercusinanis only on subsection Coccineae oaks, but Am. confluenta overlaps with both host ranges. However, these three species have shifted in their use of tree organs: Am. globulus induces galls on buds, Am. confluenta on petioles and young leaves, and Am. quercusinanis on leaves and sometimes flowers (Fig. 3). We find at least 34 such shifts between organs in our phylogeny (Fig. 3), with many more likely present but obscured because the location of development for some galls is unknown. These missing data regarding alternative galling sites, compounded with our incomplete sampling of the North American gall wasps, prevent a more robust analysis of whether host organ shifts occur more often with or without shifts in tree host. However, at a coarse level, many host shifts appear to occur without changes in tree section (Figs. 1, 3). This pattern echoes results from the Palearctic (Cook et al. 2002), suggesting that organ shifts may be relevant to lineage divergence even in the absence of host tree shifts. Shifts across different organs on the same host plant species on the surface may not seem sufficient to lead to reproductive isolation. However, temporal differences in when oak tree organs are suitable for gall induction may separate populations allochronically and create a powerful barrier to gene flow. Temporal isolation of this kind is known from many non‐galling insect systems (Malausa et al. 2005; Forbes et al. 2009; Powell et al. 2014; Boumans et al. 2017; Inskeep et al. 2021), some gall systems (Craig et al. 1993; Hood et al. 2019; Zhang et al. 2019), and even some instances where insects use the same host plants (Joy and Crespi 2007; Hippee et al. 2016, Hippee et al. 2021).
Although the opportunities for host shifts among oaks are much higher in the Nearctic due to the higher oak diversity, our finding of frequent host shifting was by no means a foregone conclusion; differences in oak chemistry and other life history and defensive traits might still have restricted colonization of highly divergent oaks. Indeed, it is clearly rarer for gall wasps to move across subgenera than it is for them to shift sections (Fig. 1). So, although the larger diversity of oaks and gall wasps in the Nearctic may simply mean that there are more opportunities for host shifts, it may also or instead be that some sections in the Nearctic are more closely related to one another than are the oak sections in the Palearctic, thereby making host shifting easier (e.g., Quercus and Virentes are sister sections and have the largest number of gall wasp shifts between them). On the other hand, similarities in oak defensive chemistry and other plant traits appear to be more related to biogeography and climate than phylogeny (Pearse and Hipp 2012; Moreira et al. 2018), so higher rates of gall wasp host shifting may really be a function of having a greater diversity of tree species in the same location.
HOST SHIFTS ACROSS OAK SUBSECTIONS AND SPECIES
We do not directly quantify the number of host tree shifts at the levels of oak subsection or species as describing host plant ranges for gall wasps is not straightforward. For example, although no single gall wasp species induces galls of the same generation on oaks from different sections (some Palearctic Andricus alternate between subgenera Cerris and Quercus but show fidelity to oak subgenus and section within each generation), many species induce galls on trees from different subsections within the same section (Fig. 2). One interpretation of this pattern is that gall wasps may often be to some extent oligophagous and able to move freely between closely related tree species without strong barriers to gene flow evolving. This could be because trees in the same section are more often chemically similar (although see Pearse and Hipp 2012, Moreira et al. 2018) or have more similar developmental pathways, perhaps making it easier for the gall wasp species to shift to some novel hosts. If this is the case, the evolution of reproductive isolation between gall wasps may require host trees to be more different in one or more important dimensions (e.g., chemistry, phenology, development) than is commonly observed at the subsection level. A second possible explanation for the apparently broad host ranges of many gall wasp species is that the picture may be clouded by how we define gall wasp “species.” With most species names relying largely on the morphology of the adult wasp, it may be that historically the names of some gall wasps have “lumped” together closely related but reproductively isolated lineages as revealed by Zhang et al. (2021c) for species of Belonocnema.
Although we did not design this study for a detailed assessment of species boundaries needed to disentangle the two above possibilities, our data provide two case studies that hint at cryptic host‐associated lineages within some gall wasp taxa. First, our samples included 13 individual Andricus quercuspetiolicola reared from five different oak tree species across both sympatric and allopatric sites. If we focus on this clade, we can test the hypothesis that An. quercuspetiolicola developing on the same hosts are more closely related to one another than sympatric wasps from different host plant species. Support for this hypothesis is shown when we enlarge and annotate the An. quercuspetiolicola clade from the 75% concatenated data matrix (Fig. 4): An. quercuspetiolicola gall wasps reared from Quercus stellata, Q. alba, and Q. bicolor sort into separate clades. Wasps associated with Q. macrocarpa were paraphyletic, but along with wasps reared from Q. bicolor form one clade, such that subsection (here, Prinoidae) could instead be the relevant taxonomic level of specialization for these wasps. Although the Q. stellata wasps were all from a site in Texas more than 1300 km from the others, wasps in the Q. bicolor clade were reared at sites within 10 miles of wasps galling Q. alba and Q. macrocarpa. The signal of closer genetic identity between allopatric populations developing on the same tree species than between sympatric populations developing on different tree species may be a preliminary signal that host association can act as a reproductive isolating barrier.
Further support for the hypothesis that some apparently oligophagous gall wasps are actually complexes of more specialized lineages comes from the clade containing Disholcaspis quercusmamma and D. quercusglobulus (Fig. 5a). Here again, wasps sort based on host tree species, independent of shared geography. Disholcaspis quercusglobulus reared from Q. stellata and Q. alba in Kentucky were less closely related to each other than to wasps reared from the same hosts in Missouri and Iowa, respectively. Similarly, D. quercusmamma collected from several sites in Iowa City, IA (3−7 km apart) on Quercus macrocarpa and Q. bicolor were less closely related to each other than to wasps from the same hosts collected in Spirit Lake, IA (352 km from Iowa City) and Reynoldsburg, OH (758 km from Iowa City). There has been some previous debate as to whether D. quercusglobulus and D. quercusmamma represent different species or simply induce galls of slightly different morphology on different oak species (McEwen et al. 2014). Our data now suggest that they are a complex of several putative species, each affiliated with different oak tree species.
Figure 5.
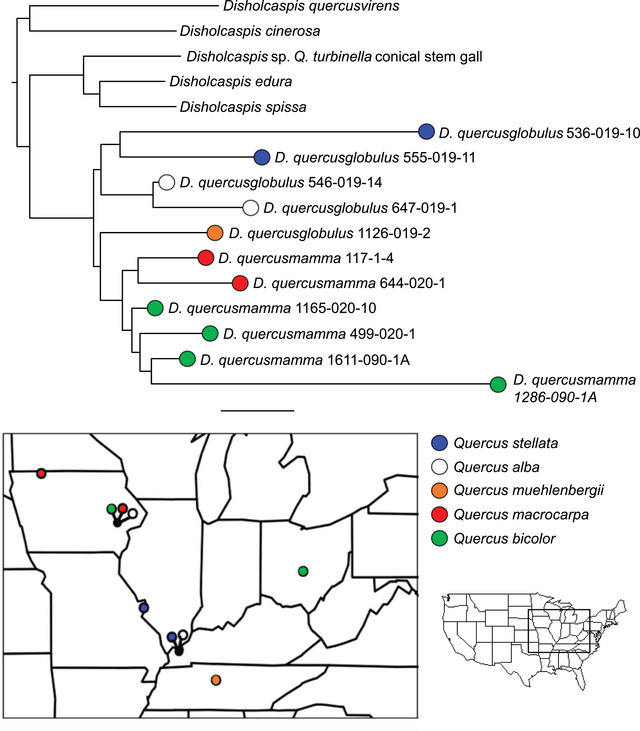
Host‐associated genetic differentiation in Disholcaspis quercusglobulus and Disholcaspis quercusmamma. Above: phylogeny of D. quercusglobulus and D. quercusmamma based on a concatenated 75% data matrix of UCE loci; branch support is bootstrap/SH‐likelihood values. Colored circles correspond to the gall wasp species ID and oak tree species from which each wasp was collected. Below: map of the upper Midwestern United States, showing location of collections. Collections from different oak tree species from the same site are indicated on the map by differently colored circles linked to a black circle, with the black circle indicating the location of the common collection site.
If both An. quercuspetiolicola and the Disholcaspis gall wasps (Figs. 4, 5) are complexes of several different host‐associated lineages, this further heightens the potential importance of host plant associations in understanding gall wasp diversity. Rather than many gall wasps being generalists with relatively broad host ranges (several tree species across >1 subsections as Fig. 2 implies), there may instead be more cryptic species than have been previously recognized, each quite highly specialized and inducing galls on only one or a few closely related tree species. In this scenario, host shifts among closely related tree species may be as frequent as they are between different organs on the same tree (Cook et al. 2002; this article), with shifts becoming less common—but still possible—as hosts become more taxonomically distant (subsections, sections, subgenera). We note as a caveat that in the ASTRAL tree (Fig. S5), An. quercuspetiolicola and Disholcaspis wasps did not sort as perfectly based on host tree species as in our concatenatedf matrix approach. This could be because these are recently diverged lineages, and resultant incomplete lineage sorting might confound resolution in multispecies coalescence approaches. Future work should use much denser sampling strategies from across larger geographic areas and multiple tree hosts to directly address host‐associated differentiation in these and other Nearctic gall wasps.
Although we provide strong evidence for shifts of gall wasps among oak subgenera, sections, subsections, species, and different tree organs, our analyses do not directly address the question of whether these host shifts actually drive speciation in gall wasps. Directly confronting whether host shifts result in new insect diversity requires identifying populations that are only recently diverged and/or actively diverging (i.e., documented ongoing gene flow) or for which a host shift has been documented historically (Forbes et al. 2017). Lineages meeting these conditions can then provide the framework for the comparative and or experimental analysis of the relationships between variation in components of fitness in relation to host plant use and assessment of levels of both pre‐ and postzygotic isolating mechanisms. Over a decade of work in one group of Nearctic gall wasps provides a template for addressing the role of host‐plant association in gall wasp speciation. The genus Belonocnema comprises three species that specialize on American live oaks (genus Quercus; subsection Virentes; Cavender‐Bares et al. 2015). Two species of Belonocnema, B. treatae and B. fossoria, co‐occur over a portion of their range across the coastal southeastern United States on the southern live oak, Q virginiana, and the sand live oak, Q. geminata, respectively (Zhang et al. 2021c), and represent two genetically diverged host‐associated genetic clusters (Driscoe et al. 2019). Several other studies provide examples of how the host plant influences the evolution of reproductive isolation and promotes speciation in this genus. These two sympatric gall wasps have evolved multiple barriers to gene flow, including temporal isolation (Hood et al. 2019), sexual isolation (Egan et al. 2012a), habitat isolation (Egan et al. 2012b), immigrant inviability and reduced fecundity (Zhang et al. 2021a), and context‐dependent reduced hybrid fitness (Zhang et al. 2021b). In total, these barriers combine to reduce gene flow by 95%–99% between host‐associated lineages (Hood et al. 2019; Zhang et al. 2021b). It would be fruitful to conduct similar studies in other Nearctic gall wasp genera to ascertain whether Belonocnema is unique or one of many similar systems.
If host shifts do prove to be a frequent driver of speciation in oak gall wasps, it will be exciting to then consider whether and how they affect the evolutionary histories of the many other insects associated with galls. Many parasitoids, inquilines, and hyperparasitoids have been reared from oak galls, with associates of some galls numbering more than 25 species (Joseph et al. 2011; Askew et al. 2013; Bird et al. 2013; Prior and Hellmann 2013; Forbes et al. 2016; Weinersmith et al. 2020). Recent work shows that many insect genera commonly associated with oak galls harbor species that specialize on one or a small subset of gall wasp hosts (Ward et al. 2019, 2020; Sheikh et al. 2022; Zhang et al. 2022). If gall wasps shift hosts, and are followed by concomitant shifts by natural enemies, the ecological dimensions relevant to reproductive isolation in the gall wasp might “cascade” during a host shift and promote reproductive isolation among insect natural enemies (Blair et al. 2005; Stireman et al. 2006; Abrahamson and Blair 2008; Forbes et al. 2009; Hood et al. 2015). Indeed, Zhang et al. (2019) found that the Synergus inquilines associated with different gall formers on live oaks were staggered in their emergence times, with differences tracking development of galls on different tree hosts. Host shifting in gall wasps may therefore be relevant not only to their own diversity, but for biodiversity in their larger communities of parasites, predators, and other gall associates. This promises to be an exciting area of future study.
AUTHOR CONTRIBUTIONS
AKGW and AAF designed the study. All authors made collections and/or reared animals. AKGW and SIS obtained the sequence data. AKGW, SIS, YMZ, and AAF conducted the analyses. AKGW and AAF wrote the manuscript. All authors contributed to revisions and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA ARCHIVING
Raw UCE sequences are available on the NCBI Short Read Archive (BioProject PRJNA827231). Alignment files, output trees, and files used in ASR analysis are available on the Dryad Digital Repository (https://doi.org/10.5061/dryad.37pvmcvn6) and at Zenodo (https://doi.org/10.5281/zenodo.6540755).
Associate Editor: J.‐P. Huang
Handling Editor: A. G. McAdam
Supporting information
Supplemental File 1. Lateral and (where available) wing pictures for representatives of each species or species group sequenced in this study.
Supplemental File 2 – Notes on Taxonomy
Supplemental Figure 1. Map of the 48 contiguous U.S. states showing collection locations (red dots) of oak gall wasps for this paper.
Supplemental Figure 2. Maximum likelihood phylogeny based on the Nearctic and Palearctic gall wasp 50% complete data matrix.
Supplemental Figure 3. Maximum likelihood phylogeny based on the Nearctic and Palearctic gall wasp 75% complete data matrix.
Supplemental Figure 4. Maximum likelihood phylogeny based on the Nearctic and Palearctic gall wasp 90% complete data matrix.
Supplemental Figure 5. ASTRAL‐III phylogeny based on the combined Nearctic and Palearctic gall wasp dataset.
Supplemental Figure 6. Ancestral State Reconstruction (ASR) for the Palearctic + Nearctic gall wasp dataset using the 75% complete data matrix phylogeny (Supplemental Figure 2) pruned to a single specimen per taxa.
Supplemental Figure 7. Maximum likelihood phylogeny based on the Nearctic gall wasp 50% complete data matrix.
Supplemental Figure 8. Maximum likelihood phylogeny based on the Nearctic gall wasp 75% complete data matrix.
Supplemental Figure 9. Maximum likelihood phylogeny based on the Nearctic gall wasp 90% complete data matrix.
Supplemental Table 1. Oak gall wasps (Hymenoptera: Cynipidae: Cynipini) used in this study.
Supplemental Table 2. Assembly statistics for UCE contigs. Raw read numbers are only given for samples sequenced for this study
Supplementary Table 3. Gall wasp UCE data from previous studies and published genomes that were used in this study. "n.d." indicates no data.
Supplemental Table 4. Alignment summaries for concatenated alignments
ACKNOWLEDGMENTS
The authors thank M. Blance, A. Driscoe, R. Busbee, C. Davis, B. Foley, E. Hu, R. Izen, S. Lee, K. McElroy, D. McGarry, S. Meadley‐Dunphy, K. Neely, M. Neiman, S. Pelini, C. Ruis, M. Shakally, S.‐A. Shzu, E. Tvedte, J. Verry, H. Widmayer, and C. Wilson for help with gall collection and/or insect rearing. A. Kranz helped identify some species based on their gall morphology. S. Hendrix and A. Hippee provided valuable comments on early manuscript drafts, and A. Hipp and G. Stone provided helpful feedback on a version of the manuscript that had been posted to a preprint archive. Funding for collections was provided to AAF by the University of Iowa, to AKGW by the Center for Global and Regional Environmental Research, and to KMP by the National Geographic Society and Binghamton University. Funding for library preparation and sequencing was provided to AKGW via an EECG award from the American Genetic Association. Mention of trade names or commercial products in this publication is solely to provide specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Contributor Information
Anna K. G. Ward, Email: anna-k-ward@uiowa.edu.
Andrew A. Forbes, Email: andrew-forbes@uiowa.edu.
LITERATURE CITED
- Abrahamson, W.G. & Blair, C.P. (2008) Sequential radiation through host‐race formation: herbivore diversity leads to diversity in natural enemies. Pp. 188–202 in Tilmon K. J., ed. Specialization, speciation and radiation: the evolutionary biology of herbivorous insects. Univ. of California Press, Berkeley, CA. [Google Scholar]
- Ács, Z. , Challis, R.J. , Bihari, P. , Blaxter, M. , Hayward, A. , Melika, G. , Csóka, G. , Pénzes, Z. , Pujade‐Villar, J. , Nieves‐Aldrey, J.‐L. , et al (2010) Phylogeny and DNA barcoding of inquiline oak gall wasps (Hymenoptera: Cynipidae) of the Western Palaearctic. Molecular Phylogenetics and Evolution, 55, 210–225. [DOI] [PubMed] [Google Scholar]
- Andersen, J.C. , Davis, M.J. , Schick, K.N. & Elkinton, J.S. (2021) Molecular placement of an outbreak‐causing gall wasp, Zapatella davisae (Hymenoptera: Cynipidae), with comments on phylogenetic arrangements in the tribe cynipini. J. Entomol. Sci., 56, 84–95. [Google Scholar]
- Askew, R.R. , Melika, G. , Pujade‐Villar, J. , Schönrogge, K. , Stone, G.N. & Nieves‐Aldrey, J.‐L. (2013) Catalogue of parasitoids and inquilines in cynipid oak galls in the West Palaearctic. Zootaxa, 3643, 1–133. [DOI] [PubMed] [Google Scholar]
- Bailey, R. , Schönrogge, K. , Cook, J.M. , Melika, G. , Csóka, G. , Thuróczy, C. & Stone, G.N. (2009) Host niches and defensive extended phenotypes structure parasitoid wasp communities. Plos Biology, 7, e1000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich, A. , Nurk, S. , Antipov, D. , Gurevich, A.A. , Dvorkin, M. , Kulikov, A.S. , Lesin, V.M. , Nikolenko, S.I. , Pham, S. , Prjibelski, A.D. , et al (2012) SPAdes: a new genome assembly algorithm and its applications to single‐cell sequencing. J. Comp. Biol., 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlocher, S.H. & Feder, J.L. (2002) Sympatric speciation in phytophagous insects: moving beyond controversy? Annual Review of Entomology, 47, 773–815. [DOI] [PubMed] [Google Scholar]
- Bird, J.P. , Melika, G. , Nicholls, J.A. , Stone, G.N. & Buss, E.A. (2013) Life history, natural enemies, and management of Disholcaspis quercusvirens (Hymenoptera: Cynipidae) on live oak trees. Journal of Economic Entomology, 106, 1747–1756. [DOI] [PubMed] [Google Scholar]
- Blaimer, B.B. , Gotzek, D. , Brady, S.G. & Buffington, M.L. (2020) Comprehensive phylogenomic analyses re‐write the evolution of parasitism within cynipoid wasps. Bmc Evolutionary Biology, 20, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair, C.P. , Abrahamson, W.G. , Jackman, J.A. & Tyrrell, L. (2005) Cryptic speciation and host‐race formation in a purportedly generalist tumbling flower beetle. Evolution; Internation Journal of Organic Evolution, 59, 304–316. [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. & Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec, M.L. (2016) AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ, 4, e1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumans, L. , Hogner, S. , Brittain, J. & Johnsen, A. (2017) Ecological speciation by temporal isolation in a population of the stonefly Leuctra hippopus (Plecoptera, Leuctridae). Ecol. Evol., 7, 1635–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão‐Dias, P.F.P. , Zhang, Y.M. , Pirro, S. , Vinson, C.C. , Weinersmith, K.L. , Ward, A.K. , Forbes, A.A. & Egan, S.P. (2022) Describing biodiversity in the genomics era: a new species of Nearctic Cynipidae gall wasp and its genome. Syst. Entomol., 47, 94–112. [Google Scholar]
- Branstetter, M.G. , Longino, J.T. , Ward, P.S. & Faircloth, B.C. (2017) Enriching the ant tree of life: enhanced UCE bait set for genome‐scale phylogenetics of ants and other Hymenoptera. Methods in Ecology and Evolution, 8, 768–776. [Google Scholar]
- Buffington, M.L. , Forshage, M. , Liljeblad, J. , Tang, C.‐T. & Van Noort, S. (2020) World Cynipoidea (Hymenoptera): a key to higher‐level groups. Insect. Syst. Divers., 4, 1–69. [Google Scholar]
- Carroll, S.P. & Boyd, C. (1992) Host race radiation in the soapberry bug: natural history with the history. Evolution; Internation Journal of Organic Evolution, 46, 1052–1069. [DOI] [PubMed] [Google Scholar]
- Cavender‐Bares, J. , González‐Rodríguez, A. , Eaton, D.A. , Hipp, A.A. , Beulke, A. & Manos, P.S. (2015) Phylogeny and biogeography of the American live oaks (Quercus subsection Virentes): a genomic and population genetics approach. Molecular Ecology, 24, 3668‐3687. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Rangasamy, M. , Tan, S.Y. , Wang, H. & Siegfried, B.D. (2010) Evaluation of five methods for total DNA extraction from western corn rootworm beetles. Plos One, 5, e11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, J.M. , Rokas, A. , Pagel, M. & Stone, G.N. (2002) Evolutionary shifts between host oak sections and host‐plant organs in Andricus gall wasps. Evolution; Internation Journal of Organic Evolution, 56, 1821–1830. [DOI] [PubMed] [Google Scholar]
- Cooke, C.L. (2018) Forest Micro‐Hymenoptera, including those attacking trees (Cynipidae oak gall wasps) and those potentially defending them (parasitic Pteromalidae). Ph.D. diss., University of Maryland, College Park, MD. [Google Scholar]
- Craig, T.P. , Itami, J.K. , Abrahamson, W.G. & Horner, J.D. , (1993) Behavioral evidence for host‐race formation in Eurosta solidaginis . Evolution; Internation Journal of Organic Evolution, 47, 1696–1710. [DOI] [PubMed] [Google Scholar]
- Craig, T.P. , Horner, J.D. & Itami, J.K. (2001) Genetics, experience, and host‐plant preference in Eurosta solidaginis: implications for host shifts and speciation. Evolution; Internation Journal of Organic Evolution, 55, 773–782. [DOI] [PubMed] [Google Scholar]
- Csóka, G. , Stone, G.N. & Melika, G. (2005) Biology, ecology and evolution of gall‐inducing Cynipidae. Pp. 573–642 in Raman A., Schaefer C. W., and Withers T. M., eds. Biology, ecology and evolution of gall‐inducing arthropods. Vol. 2. Science Publishers, Enfield, NH. [Google Scholar]
- Denk, T. , Velitzelos, D. , Güner, T.H. , Bouchal, J.M. , Grímsson, F. & Grimm, G.W. (2017) Taxonomy and palaeoecology of two widespread western Eurasian Neogene sclerophyllous oak species: Quercus drymeja Unger and Q. mediterranea Unger. Review of Palaeobotany and Palynology, 241, 98‐128. [Google Scholar]
- Drès, M. & Mallet, J. (2002) Host races in plant‐feeding insects and their importance in sympatric speciation. Philos. Trans. R. Soc. B Biol. Sci., 357, 471–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoe, A.L. , Nice, C.C. , Busbee, R.W. , Hood, G.R. , Egan, S.P. & Ott, J.R. (2019) Host plant associations and geography interact to shape diversification in a specialist insect herbivore. Molecular Ecology, 28, 4197–4211. [DOI] [PubMed] [Google Scholar]
- Egan, S.P. , Hood, G.R. , Feder, J.L. & Ott, J.R. (2012a) Divergent host plant use promotes reproductive isolation among cynipid gall wasp populations. Biology Letters, 8, 605–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, S.P. , Hood, G.R. & Ott, J.R. (2012b) Testing the role of habitat isolation among ecologically divergent gall wasp populations. Int. J. Ecol., 2012, 809897. [Google Scholar]
- Egan, S.P. , Hood, G.R. , Martinson, E.O. & Ott, J.R. (2018) Cynipid gall wasps. Current Biology, 28, R1370–R1374. [DOI] [PubMed] [Google Scholar]
- Erwin, T.L. (1982) Tropical forests: their richness in Coleoptera and other arthropod species. Coleopt. Bull., 36, 74–75. [Google Scholar]
- Faircloth, B.C. (2016) PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics, 32, 786–788. [DOI] [PubMed] [Google Scholar]
- Faircloth, B.C. , McCormack, J.E. , Crawford, N.G. , Harvey, M.G. , Brumfield, R.T. & Glenn, T.C. (2012) Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Systematic Biology, 61, 717–726. [DOI] [PubMed] [Google Scholar]
- Feder, J.L. & Forbes, A.A. (2007) Habitat avoidance and speciation for phytophagous insect specialists. Funct. Ecol., 21, 585–597. [Google Scholar]
- Feder, J.L. & Filchak, K.E. (1999) It's about time: the evidence for host plant‐mediated selection in the apple maggot fly, Rhagoletis pomonella, and its implications for fitness trade‐offs in phytophagous insects. Entomologia Experimentalis Et Applicata, 91, 211–225. [Google Scholar]
- Forbes, A.A. , Powell, T.H. , Stelinski, L.L. , Smith, J.J. & Feder, J.L. (2009) Sequential sympatric speciation across trophic levels. Science, 323, 776–779. [DOI] [PubMed] [Google Scholar]
- Forbes, A.A. , Hood, G.R. , Hall, M.C. , Lund, J. , Izen, R. , Egan, S.P. & Ott, J.R. (2016) Parasitoids, hyperparasitoids, and inquilines associated with the sexual and asexual generations of the gall former, Belonocnema treatae (Hymenoptera: Cynipidae). Annals of the Entomological Society of America, 109, 49–63. [Google Scholar]
- Forbes, A.A. , Devine, S.N. , Hippee, A.C. , Tvedte, E.S. , Ward, A.K. , Widmayer, H.A. & Wilson, C.J. (2017) Revisiting the particular role of host shifts in initiating insect speciation. Evolution; Internation Journal of Organic Evolution, 71, 1126–1137. [DOI] [PubMed] [Google Scholar]
- Forister, M.L. , Dyer, L.A. , Singer, M.S. , Stireman, J.O. III & Lill, J.T. (2012) Revisiting the evolution of ecological specialization, with emphasis on insect–plant interactions. Ecology, 93, 981–991. [DOI] [PubMed] [Google Scholar]
- Gallformers.org (2021) Gallformers.org. https://www.gallformers.org/.
- Govaerts, R. & Frodin, D.G. (1998) World checklist and bibliography of Fagales. Royal Botanic Gardens, Kew, Richmond, U.K. [Google Scholar]
- Guindon, S. , Dufayard, J.‐F. , Lefort, V. , Anisimova, M. , Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Hipp, A.L. , Manos, P.S. , González‐Rodríguez, A. , Hahn, M. , Kaproth, M. , McVay, J.D. , Avalos, S.V. & Cavender‐Bares, J. (2018) Sympatric parallel diversification of major oak clades in the Americas and the origins of Mexican species diversity. New Phytol., 217, 439–452. [DOI] [PubMed] [Google Scholar]
- Hipp, A.L. , Manos, P.S. , Hahn, M. , Avishai, M. , Bodénès, C. , Cavender‐Bares, J. , Crowl, A.A. , Deng, M. , Denk, T. , Fitz‐Gibbon, S. , et al (2020) Genomic landscape of the global oak phylogeny. New Phytol., 226, 1198–1212. [DOI] [PubMed] [Google Scholar]
- Hippee, A.C. , Elnes, M.E. , Armenta, J.S. , Condon, M.A. & Forbes, A.A. (2016) Divergence before the host shift? Prezygotic reproductive isolation among three varieties of a specialist fly on a single host plant. Ecological Entomology, 41, 389–399. [Google Scholar]
- Hippee, A.C. , Beer, M.A. , Bagley, R.K. , Condon, M.A. , Kitchen, A. , Lisowski, E.A. , Norrbom, A.L. & Forbes, A.A. (2021) Host shifting and host sharing in a genus of specialist flies diversifying alongside their sunflower hosts. Journal of Evolutionary Biology, 34, 364–379. [DOI] [PubMed] [Google Scholar]
- Hoang, D.T. , Chernomor, O. , von Haeseler, A. , Minh, B.Q. & Le, S.V. (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular biology and evolution, 35, 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, G.R. , Forbes, A.A. , Powell, T.H. , Egan, S.P. , Hamerlinck, G. , Smith, J.J. & Feder, J.L. (2015) Sequential divergence and the multiplicative origin of community diversity. Proc. Natl. Acad. Sci. USA, 112, E5980–E5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, G.R. , Zhang, L. , Topper, L. , Brandão‐Dias, P.F.P. , Del Pino, G.A. , Comerford, M.S. & Egan, S.P. (2018) Closing the life cycle of Andricus quercuslanigera (Hymenoptera: Cynipidae). Annals of the Entomological Society of America, 111, 103–113. [Google Scholar]
- Hood, G.R. , Zhang, L. , Hu, E.G. , Ott, J.R. & Egan, S.P. (2019) Cascading reproductive isolation: plant phenology drives temporal isolation among populations of a host‐specific herbivore. Evolution; Internation Journal of Organic Evolution, 73, 554–568. [DOI] [PubMed] [Google Scholar]
- Inskeep, K.A. , Doellman, M.M. , Powell, T.H.Q. , Berlocher, S.H. , Seifert, N.R. , Hood, G.R. , Ragland, G.J. , Meyers, P.J. & Feder, J.L. (2021) Divergent diapause life history timing drives both allochronic speciation and reticulate hybridization in an adaptive radiation of Rhagoletis flies. Molecular Ecology, 10.1111/mec.15908. [DOI] [PubMed] [Google Scholar]
- Jaenike, J. (1990) Host specialization in phytophagous insects. Annu. Rev. Ecol. Syst., 21, 243–273. [Google Scholar]
- Joseph, M.B. , Gentles, M. & Pearse, I.S. (2011) The parasitoid community of Andricus quercuscalifornicus and its association with gall size, phenology, and location. Biodiversity and Conservation, 20, 203–216. [Google Scholar]
- Joy, J.B. & Crespi, B.J. (2007) Adaptive radiation of gall‐inducing insects within a single host‐plant species. Evol. Int. J. Org. Evol., 61, 784–795. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy, S. , Minh, B.Q. , Wong, T.K.F. , Von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey, A.C. (1920) Phylogeny of cynipid genera and biological characteristics. Bull. Am. Mus. Nat. Hist., 42, 357–402. [Google Scholar]
- ——— (1936) The origin of the higher categories in Cynips . Indiana Univ. Publ. Sci. Ser., 4, 1–334. [Google Scholar]
- Kremer, A. & Hipp, A.L. (2020) Oaks: an evolutionary success story. New Phytol., 226, 987–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krombein, K.V. , Hurd, P.D. , Smith, D.R. & Burks, B.D. (1979) Catalog of Hymenoptera in America north of Mexico. Smithsonian Institution Press, Washington, D.C. [Google Scholar]
- Lanfear, R. , Frandsen, P.B. , Wright, A.M. , Senfeld, T. & Calcott, B. (2017) Partitionfinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular biology and evolution, 34, 772–773. [DOI] [PubMed] [Google Scholar]
- Larsen, B.B. , Miller, E.C. , Rhodes, M.K. & Wiens, J.J. (2017) Inordinate fondness multiplied and redistributed: the number of species on earth and the new pie of life. Quarterly Review of Biology, 92, 229–265. [Google Scholar]
- Lund, J.N. , Ott, J.R. & Lyon, R.J. (1998) Heterogony in Belonocnema treatae Mayr (Hymenoptera: Cynipidae). Proc. Entomol. Soc. Wash., 100, 755–763. [Google Scholar]
- Machado, C.A. , Jousselin, E. , Kjellberg, F. , Compton, S.G. & Herre, E.A. (2001) Phylogenetic relationships, historical biogeography and character evolution of fig‐pollinating wasps. Proc. R. Soc. B Biol. Sci., 268, 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malausa, T. , Bethenod, M.‐T. , Bontemps, A. , Bourguet, D. , Cornuet, J.‐M. & Ponsard, S. (2005) Assortative mating in sympatric host races of the European corn borer. Science, 308, 258–260. [DOI] [PubMed] [Google Scholar]
- Manos, P.S. & Hipp, A.L. (2021) An updated infrageneric classification of the North American oaks (Quercus subgenus Quercus): review of the contribution of phylogenomic data to biogeography and species diversity. Forests, 12, 786. [Google Scholar]
- Manos, P.S. , Doyle, J.J. & Nixon, K.C. (1999) Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae). Molecular Phylogenetics and Evolution, 12, 333–349. [DOI] [PubMed] [Google Scholar]
- Martinson, E.O. , Werren, J.H. & Egan, S.P. (2022) Organ‐specific gene expression shows a cynipid wasp repurposes oak host gene networks to create a complex and novel parasite‐specific organ. Molecular Ecology, 31, 3228–3240. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, K.W. , Ohshima, I. & Nosil, P. (2010) Ecological speciation in phytophagous insects. Entomologia Experimentalis Et Applicata, 134, 1–27. [Google Scholar]
- Mattsson, M. , Hood, G.R. , Yee, W.L. , Doellman, M.M. , Bruzzese, D.J. , Goughnour, R.B. , Driscoe, A.L. , Van Dexter, S. , Tait, C. , Glover, M.M. , et al (2022) Recursive adaptation in action: allochronic isolation and divergence of host‐associated populations of the apple maggot fly, Rhagoletis pomonella, following its recent introduction to the western USA. Entomologia Experimentalis Et Applicata, 170, 48–63. [Google Scholar]
- May, R.M. (1990) How many species? Philos. Trans. R. Soc. Lond. B. Biol. Sci., 330, 293–304. [Google Scholar]
- McEwen, C. , Digweed, S. , Nicholls, J.A. & Cranshaw, W. (2014) Description and biology of the sexual generation of Disholcaspis quercusmamma (Walsh and Riley) (Hymenoptera: Cynipidae), with notes on associated parasitoids. Proc. Entomol. Soc. Wash., 116, 294–310. [Google Scholar]
- McIver, E.E. & Basinger, J.F. (1999) Early Tertiary floral evolution in the Canadian High Arctic. Ann. Mo. Bot. Gard., 86, 523–545. [Google Scholar]
- McLeish, M.J. , Crespi, B.J. , Chapman, T.W. & Schwarz, M.P. (2007) Parallel diversification of Australian gall‐thrips on Acacia . Molecular Phylogenetics and Evolution, 43, 714–725. [DOI] [PubMed] [Google Scholar]
- Melika, G. & Abrahamson, W.G. (2002) Review of the world genera of oak cynipid wasps (Hymenoptera: Cynipidae, Cynipini). Pp. 150–190 in Melika G. and Thuróczy C., eds. Parasitic wasps: evolution, systematics, biodiversity and biological control. Agroinform, Budapest, Hungary. [Google Scholar]
- Melika, G. , Pujade‐Villar, J. , Nicholls, J.A. , Cuesta‐Porta, V. , Cooke‐McEwen, C. & Stone, G.N. (2021a) Three new Nearctic genera of oak cynipid gall wasps (Hymenoptera: Cynipidae: Cynipini): Burnettweldia Pujade‐Villar, Melika & Nicholls, Nichollsiella Melika, Pujade‐Villar & Stone, Disholandricus Melika, Pujade‐Villar & Nicholls; and re‐establishment of the genus Paracraspis Weld. Zootaxa, 4993, 1–81. [DOI] [PubMed] [Google Scholar]
- Melika, G. , Nicholls, J.A. , Abrahamson, W.G. , Buss, E.A. & Stone, G.N. (2021b) New species of Nearctic oak gall wasps (Hymenoptera: Cynipidae, Cynipini). Zootaxa, 5084, 1–131. [DOI] [PubMed] [Google Scholar]
- Minh, B.Q. , Schmidt, H.A. , Chernomor, O. , Schrempf, D. , Woodhams, M.D. , Von Haeseler, A. & Lanfear, R. , (2020) IQ‐TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular biology and evolution, 37, 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter, C. , Farrell, B. & Wiegmann, B. (1988) The phylogenetic study of adaptive zones: has phytophagy promoted insect diversification? American Naturalist, 132, 107–128. [Google Scholar]
- Moreira, X. , Abdala‐Roberts, L. , Galmán, A. , Francisco, M. , de la Fuente, M. , Butrón, A. & Rasmann, S. (2018) Assessing the influence of biogeographical region and phylogenetic history on chemical defences and herbivory in Quercus species. Phytochemistry, 153, 64–73. [DOI] [PubMed] [Google Scholar]
- Nicholls, J.A. , Melika, G. & Stone, G.N. , (2017) Sweet tetra‐trophic interactions: multiple evolution of nectar secretion, a defensive extended phenotype in Cynipid gall wasps. American Naturalist, 189, 67–77. [DOI] [PubMed] [Google Scholar]
- Nixon, K.C. & Muller, C.H. (1997) Quercus Linnaeus sect. Quercus . Pp. 471–506 in Flora of North America Editorial Committee , ed. Flora of North America. Vol. 3. Oxford Univ. Press, New York. [Google Scholar]
- Nosil, P. (2002) Transition rates between specialization and generalization in phytophagous insects. Evolution; Internation Journal of Organic Evolution, 56, 1701–1706. [DOI] [PubMed] [Google Scholar]
- Paradis, E. , Claude, J. & Strimmer, K. (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 20, 289–290. [DOI] [PubMed] [Google Scholar]
- Pearse, I.S. & Hipp, A.L. (2012) Global patterns of leaf defenses in oak species. Evolution; Internation Journal of Organic Evolution, 66, 2272–2286. [DOI] [PubMed] [Google Scholar]
- Penzes, Z. , Tang, C.‐T. , Stone, G.N. , Nicholls, J.A. , Schweger, S. , Bozso, M. & Melika, G. (2018) Current status of the oak gall wasp (Hymenoptera: Cynipidae: Cynipini) fauna of the Eastern Palaearctic and Oriental Regions. Zootaxa, 4433, 245–289. [DOI] [PubMed] [Google Scholar]
- Powell, T.H.Q. , Forbes, A.A. , Hood, G.R. & Feder, J.L. (2014) Ecological adaptation and reproductive isolation in sympatry: genetic and phenotypic evidence for native host races of Rhagoletis pomonella . Molecular Ecology, 23, 688–704. [DOI] [PubMed] [Google Scholar]
- Price, P.W. (1980) Evolutionary biology of parasites. (MPB‐15), Volume 15. Princeton Univ. Press, Princeton, NJ. [PubMed] [Google Scholar]
- Prior, K.M. & Hellmann, J. (2013) Does enemy loss cause release? A biogeographical comparison of parasitoid effects on an introduced insect. Ecology, 94, 1015–1024. [DOI] [PubMed] [Google Scholar]
- Pujade‐Villar, J. , Bellido, D. , Segu, G. & Melika, G. (2001) Current state of knowledge of heterogony in Cynipidae (Hymenoptera, Cynipoidea). Sess. Conjunta Entomol., 11, 87–107. [Google Scholar]
- R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R‐project.org/. [Google Scholar]
- Revell, L.J. , (2012) phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3, 217–223. [Google Scholar]
- Roderick, G.K. (1997) Herbivorous insects and the Hawaiian silversword alliance: coevolution or cospeciation? Pac. Sci., 51, 440–449. [Google Scholar]
- Rokas, A. , Melika, G. , Abe, Y. , Nieves‐Aldrey, J.‐L. , Cook, J.M. & Stone, G.N. (2003) Lifecycle closure, lineage sorting, and hybridization revealed in a phylogenetic analysis of European oak gall wasps (Hymenoptera: Cynipidae: Cynipini) using mitochondrial sequence data. Molecular Phylogenetics and Evolution, 26, 36–45. [DOI] [PubMed] [Google Scholar]
- Ronquist, F. , Nieves‐Aldrey, J.‐L. , Buffington, M.L. , Liu, Z. , Liljeblad, J. & Nylander, J.A. (2015) Phylogeny, evolution and classification of gall wasps: the plot thickens. Plos One, 10, e0123301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rønsted, N. , Weiblen, G.D. , Cook, J.M. , Salamin, N. , Machado, C.A. & Savolainen, V. (2005) 60 million years of co‐divergence in the fig‐wasp symbiosis. Proc. R. Soc. B Biol. Sci., 272, 2593–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, R.A. (2021) Plant Galls of the Western United States. Princeton Univ. Press, Princeton, NJ. [Google Scholar]
- Sheikh, S.I. , Ward, A.K.G. , Zhang, Y.M. , Davis, C.K. , Zhang, L. , Egan, S.P. & Forbes, A.A. (2022) Ormyrus labotus (Hymenoptera: Ormyridae): another generalist that should not be a generalist is not a generalist. Insect Syst. Divers,. 6, 8. [Google Scholar]
- Stireman, J.O. III , Nason, J.D. , Heard, S.B. & Seehawer, J.M. (2006) Cascading host‐associated genetic differentiation in parasitoids of phytophagous insects. Proc. R. Soc. B Biol. Sci., 273, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, G.N. & Schönrogge, K. (2003) The adaptive significance of insect gall morphology. Trends in Ecology & Evolution, 18, 512–522. [Google Scholar]
- Stone, G.N. , Schönrogge, K. , Atkinson, R.J. , Bellido, D. & Pujade‐Villar, J. (2002) The population biology of oak gall wasps (Hymenoptera: Cynipidae). Ann. Rev. Entomol., 47, 633–668. [DOI] [PubMed] [Google Scholar]
- Stone, G.N. , Atkinson, R.J. , Rokas, A. , Nieves‐Aldrey, J.‐L. , Melika, G. , Ács, Z. , Csóka, G. , Hayward, A. , Bailey, R. , Buckee, C. , et al (2008) Evidence for widespread cryptic sexual generations in apparently purely asexual Andricus gall wasps. Molecular Ecology, 17, 652–665. [DOI] [PubMed] [Google Scholar]
- Stone, G.N. , Hernandez‐Lopez, A. , Nicholls, J.A. , di Pierro, E. , Pujade‐Villar, J. , Melika, G. & Cook, J.M. (2009) Extreme host plant conservatism during at least 20 million years of host plant pursuit by oak gall wasps. Evolution; Internation Journal of Organic Evolution, 63, 854–869. [DOI] [PubMed] [Google Scholar]
- Stork, N.E. (1993) How many species are there? Biodiversity and Conservation, 2, 215–232. [Google Scholar]
- Strong, D.R. (1988) Insect host range. Ecology, 69, 885–915. [Google Scholar]
- Tagliacollo, V.A. & Lanfear, R. (2018) Estimating improved partitioning schemes for ultraconserved elements. Molecular biology and evolution, 35, 1798–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Cannon, C.H. & Chen, J. , (2016) Pollinator sharing and gene flow among closely related sympatric dioecious fig taxa. Proc. R. Soc. B Biol. Sci., 283, 20152963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Zhang, X. , Herre, E.A. , McKey, D. , Machado, C.A. , Yu, W.B. , Cannon, C.H. , Arnold, M.L. , Pereira, R.A. , Ming, R. , et al (2021) Genomic evidence of prevalent hybridization throughout the evolutionary history of the fig‐wasp pollination mutualism. Nat. Comm., 12, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, A.K. , Khodor, O.S. , Egan, S.P. , Weinersmith, K.L. & Forbes, A.A. (2019) A keeper of many crypts: a behaviour‐manipulating parasite attacks a taxonomically diverse array of oak gall wasp species. Biology Letters, 15, 20190428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, A.K. , Sheikh, S.I. & Forbes, A.A. (2020) Diversity, host ranges, and potential drivers of speciation among the inquiline enemies of oak gall wasps (Hymenoptera: Cynipidae). Insect Syst. Divers., 4, 3. [Google Scholar]
- Weinersmith, K.L. , Forbes, A.A. , Ward, A.K.G. , Brandão‐Dias, P.F.P. , Zhang, Y.M. & Egan, S.P. (2020) Arthropod community associated with the asexual generation of Bassettia pallida Ashmead (Hymenoptera: Cynipidae). Annals of the Entomological Society of America, 113, 373–388. [Google Scholar]
- Weld, L.H. (1957) Cynipid galls of the Pacific Slope. Privately printed, Ann Arbor, MI. [Google Scholar]
- ——— (1959) Cynipid galls of the eastern United States. Privately printed, Ann Arbor, MI. [Google Scholar]
- ——— (1960) Cynipid galls of the southwest. Privately printed, Ann Arbor, MI. [Google Scholar]
- Weld, L.H. & von Dalla Torre, K.W. (1952) Cynipoidea (Hym.) 1905–1950, being a supplement to the Dalla Torre and Kieffer monograph. Privately printed, Ann Arbor, MI. [Google Scholar]
- Zhang, C. , Rabiee, M. , Sayyari, E. & Mirarab, S. , (2018) ASTRAL‐III: polynomial time species tree reconstruction from partially resolved gene trees. Bmc Bioinformatics [Electronic Resource], 19, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Hood, G.R. , Ott, J.R. & Egan, S.P. (2019) Temporal isolation between sympatric host plants cascades across multiple trophic levels of host‐associated insects. Biology Letters, 15, 20190572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Hood, G.R. , Roush, A.M. , Shzu, S. , Comerford, M.S. , Ott, J.R. & Egan, S.P. (2021a) Asymmetric, but opposing reductions in immigrant viability and fecundity promote reproductive isolation among host‐associated populations of an insect herbivore. Evolution; Internation Journal of Organic Evolution, 75, 476–489. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Hood, G.R. , Carroo, I. , Ott, J.R. & Egan, S.P. (2021b) Context‐dependent reproductive isolation: host plant variability drives fitness of hybrid herbivores. American Naturalist, 197, 732–739. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.M. , Egan, S.P. , Driscoe, A.L. & Ott, J.R. (2021c) One hundred and sixty years of taxonomic confusion resolved: Belonocnema (Hymenoptera: Cynipidae: Cynipini) gall wasps associated with live oaks in the USA. Zool. J. Linn. Soc., 193, 1234–1255, 10.1093/zoolinnean/zlab001 [DOI] [Google Scholar]
- Zhang, Y. M. , Sheikh, S. I. , Ward, A. K. G. , Forbes, A. A. , Prior, K. M. , Stone, G. N. , Gates, M. W. , Egan, S. P. , Zhang, L. , Davis, C. , Weinersmith, K. L. , Melika, G. , & Lucky, A. (2022). Delimiting the cryptic diversity and host preferences of Sycophila parasitoid wasps associated with oak galls using phylogenomic data. Mol. Ecol., 10.1111/mec.16582 [DOI] [PubMed] [Google Scholar]
- Zimmerman, J.R. (2018) A synopsis of oak gall wasps (Hymenoptera: Cynipidae) of the southwestern United States with a key and comments on each of the genera. J. Kans. Entomol. Soc., 91, 58–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File 1. Lateral and (where available) wing pictures for representatives of each species or species group sequenced in this study.
Supplemental File 2 – Notes on Taxonomy
Supplemental Figure 1. Map of the 48 contiguous U.S. states showing collection locations (red dots) of oak gall wasps for this paper.
Supplemental Figure 2. Maximum likelihood phylogeny based on the Nearctic and Palearctic gall wasp 50% complete data matrix.
Supplemental Figure 3. Maximum likelihood phylogeny based on the Nearctic and Palearctic gall wasp 75% complete data matrix.
Supplemental Figure 4. Maximum likelihood phylogeny based on the Nearctic and Palearctic gall wasp 90% complete data matrix.
Supplemental Figure 5. ASTRAL‐III phylogeny based on the combined Nearctic and Palearctic gall wasp dataset.
Supplemental Figure 6. Ancestral State Reconstruction (ASR) for the Palearctic + Nearctic gall wasp dataset using the 75% complete data matrix phylogeny (Supplemental Figure 2) pruned to a single specimen per taxa.
Supplemental Figure 7. Maximum likelihood phylogeny based on the Nearctic gall wasp 50% complete data matrix.
Supplemental Figure 8. Maximum likelihood phylogeny based on the Nearctic gall wasp 75% complete data matrix.
Supplemental Figure 9. Maximum likelihood phylogeny based on the Nearctic gall wasp 90% complete data matrix.
Supplemental Table 1. Oak gall wasps (Hymenoptera: Cynipidae: Cynipini) used in this study.
Supplemental Table 2. Assembly statistics for UCE contigs. Raw read numbers are only given for samples sequenced for this study
Supplementary Table 3. Gall wasp UCE data from previous studies and published genomes that were used in this study. "n.d." indicates no data.
Supplemental Table 4. Alignment summaries for concatenated alignments
Data Availability Statement
Raw UCE sequences are available on the NCBI Short Read Archive (BioProject PRJNA827231). Alignment files, output trees, and files used in ASR analysis are available on the Dryad Digital Repository (https://doi.org/10.5061/dryad.37pvmcvn6) and at Zenodo (https://doi.org/10.5281/zenodo.6540755).


