Abstract
The development of physiologically relevant in vitro colorectal cancer (CRC) models is vital for advancing understanding of tumor biology. Although CRC patient-derived xenografts (PDX) recapitulate key patient tumor characteristics and demonstrate high concordance with clinical outcomes, the use of this in vivo model is costly and low-throughput. Here we report the establishment and in-depth characterization of an in vitro tissue-engineered CRC model using PDX cells. To form the 3D engineered CRC-PDX (3D-eCRC-PDX) tissues, CRC PDX tumors were expanded in vivo, dissociated, and the isolated cells encapsulated within PEG-fibrinogen hydrogels. Following PEG-fibrinogen encapsulation, cells remain viable and proliferate within 3D-eCRC-PDX tissues. Tumor cell subpopulations, including human cancer and mouse stromal cells, are maintained in long-term culture (29 days); cellular subpopulations increase ratiometrically over time. 3D-eCRC-PDX tissues mimic the mechanical stiffness of originating tumors. ECM protein production by cells in the 3D-eCRC-PDX tissues resulted in approximately 57% of proteins observed in the CRC-PDX tumors also being present in the 3D-eCRC-PDX tissues on Day 22. Furthermore, we show congruence in enriched gene ontology molecular functions and Hallmark gene sets in 3D-eCRC-PDX tissues and CRC-PDX tumors compared to normal colon tissue, while prognostic Kaplan-Meier plots for overall and relapse free survival did not reveal significant differences between CRC-PDX tumors and 3D-eCRC-PDX tissues. Our results demonstrate high batch-to-batch consistency and strong correlation between our in vitro tissue-engineered PDX-CRC model and the originating in vivo PDX tumors, providing a foundation for future studies of disease progression and tumorigenic mechanisms.
Keywords: engineered tumor model, cancer tissue engineering, PDX, hydrogel, colon cancer
1. Introduction
Colorectal cancer (CRC) remains the third most prevalent cancer and the third highest cause of cancer-related deaths among both men and women in the United States with approximately 145,600 new cases and 51,020 deaths in 2019 [1–6]. Although there has been an overall decreasing trend in CRC rates over the past several decades, the incidence in individuals under the age of 55 has increased [1–6]. Additionally, despite advances in early CRC detection and targeted treatment strategies, the 5-year relative survival rate of patients with distant-stage CRC (malignant CRC metastasized to distal regions of the body) is only 14% [1–6].
To investigate tumor biology, in vivo and in vitro models are utilized to mimic the patient tumor milieu. However, deficiencies in existing models have impeded the success of clinical trials. In part, this deficiency can be attributed to the inability of pre-clinical models to accurately represent disease pathophysiology and the complex tumor microenvironment [7]. Tumors are heterogeneous neoplastic tissues composed of multiple cell types such tumor cells, cancer stem cells, endothelial cells, immune cells, cancer-associated fibroblasts, and other stromal cells, within and surrounded by extracellular matrix (ECM) [8–10]. Intratumoral cell-cell and cell-matrix interactions play crucial roles in tumor progression, cancer cell invasion and migration, and tumor metastasis, as well as in anti-cancer therapeutic response [11–18]. Therefore, the presence of cancer-associated cells together with cancer cells within a matrix is essential to accurately modeling the tumor microenvironment.
Improved in vitro and in vivo model recapitulation of the patient tumor microenvironment can be achieved in part through consideration of the model cellular components. Standard cell lines lack the extensive heterogeneity observed in patient tumors. Furthermore, the phenotype, genetics, and molecular characteristics of standard cell lines have been changed due to extended culture in vitro, thus resulting in poor correspondence to patient tumors [19, 20]. Patient-derived xenograft (PDX) models more accurately mimic patient tumors both genotypically and phenotypically [19, 21]. In most cases, it has been shown that PDX line propagation in immunocompromised mice maintains patient tumor molecular characteristics, gene expression, and mutations [22–24]. These PDX models are a useful tool to examine tumor biology with a higher degree of physiological relevance compared to cell lines [19]. However, employing PDX models is costly and time-consuming due to the low throughput nature of expansion and testing in mice [25–27]. Furthermore, the number of experimental conditions that can be tested using in vivo PDX models is limited, and the complexity of these in vivo models limits the ability to probe real-time cell-cell, cell-matrix, and cell-drug interactions.
The most common in vitro cancer cell culture model relies on two-dimensional (2D) culture of cells. However, the culture of PDX cells in 2D monolayers results in substantial changes in the ratios of cell types present in the overall population, thereby resulting in a cancer cell in vitro model that no longer mimics the in vivo PDX cell subpopulations [28, 29]. Furthermore, PDX tumors often contain cell subpopulations that do not adhere to the surface of the culture flask, preventing direct contact between non-anchorage and anchorage-dependent cell types, thereby hindering cell-cell interactions important for preserving in vivo cellular phenotypes. Moreover, the absence of a supporting matrix in 2D models precludes multi-dimensional cell-matrix interactions which modulate tumor pathophysiology. To overcome the inherent limitations of 2D models, scaffold-free three-dimensional (3D) platforms for the culture of PDX cells have been developed, including those for CRC PDX cells. For instance, Kondo et al. [30] generated cancer tissue-originated spheroids, formed from isolated aggregates of human CRC PDX cells. Other studies have developed organoids from either primary CRC obtained directly from patients or CRC PDX as novel models for high-throughput drug screening and investigation of disease mechanisms [31, 32]. In all of these cancer models, however, aggregates or spheroids selectively incorporated only some of the cell types from the primary or PDX tumor. Inclusion of all cellular subpopulations may be advantageous in recapitulating the parental tumor microenvironment more fully.
Formation of 3D engineered tissues using biomimetic scaffolds has emerged as an important tool for mimicking the native tumor tissue microenvironment and supporting the inclusion of all cell subpopulations. In comparison to 2D and aggregate cell culture, engineered cancer tissue models can more closely recapitulate in vivo tumor dimensionality and cell-cell and cell-matrix interactions [33–35]. Most of the research in cancer tissue engineering currently employs standard cancer cell lines [36–43]. Recent studies have begun to employ patient-derived cells to form engineered cancer tissues, including colorectal cancer [44, 45], prostate cancer [46], and glioblastoma tissues [47–49]. In these studies, 3D scaffolds have been utilized to mimic tumor microenvironmental cues with high relevance; however, there is a need to examine the potential of 3D scaffold-based in vitro models to support maintenance and expansion of mixed CRC-PDX tumor cell subpopulations, including both human cancer and mouse stromal cells, and investigate the extent to which such 3D in vitro models mimic the parental CRC-PDX tumor.
Here we examine the ability of CRC-PDX tumor cells to be encapsulated and maintained within poly(ethylene glycol)-fibrinogen (PEG-Fb) hydrogels and investigate the extent to which this 3D engineered CRC tissue model recapitulates the originating in vivo PDX tumors, including comparison of gene expression profiles through transcriptomic analysis. Prior work has shown that PEG-Fb supports the formation of engineered breast cancer tissues [36, 50, 51]. Capable of being remodeled by encapsulated cells, PEG-Fb provides cell adhesion sites and facilitates the incorporation of the heterogeneous cell types, including fibroblasts co-cultured with cancer cell lines [51]. To construct the 3D engineered CRC PDX (3D-eCRC-PDX) tissue model, PDX cells propagated in vivo from a patient diagnosed with stage II CRC adenocarcinoma were dissociated and encapsulated within the PEG-Fb matrix. To assess the ability of this 3D-eCRC-PDX tissue model to maintain parental tumor cell subpopulations throughout long-term culture, we monitored temporal variations in cell subpopulations in comparison to 2D CRC PDX (2D-CRCPDX) cell culture. Due to recent findings that tumor tissue mechanical stiffness plays an important role in cell phenotype and genotype, as well as cell-cell, cell-matrix, and cell-drug interactions [50, 52–57], we quantified the mechanical stiffness of the 3D-eCRC-PDX tissues and compared it to that of CRC-PDX tumors. ECM proteins from CRC-PDX tumors and 3D-eCRCPDX tissues were identified using proteomics; protein production by cells in the 3D-eCRC-PDX tissues resulted in approximately 57% of proteins observed in the CRC-PDX tumors also being present in the 3D-eCRC-PDX tissues on Day 22. Most notably, we found that 3D-eCRC-PDX tissues not only support the growth of CRC PDX cells from this patient line in long-term culture, but also mimic the originating CRC-PDX tumors in terms of cell subpopulation proportions, gene expression, and mechanical stiffness, providing evidence that these engineered tissues may be capable of supporting the culture of CRC PDX cells in vitro.
2. Materials and Methods
Fabrication of PDMS Molds
Polydimethylsiloxane (PDMS) molds were fabricated using the SYLGARD® 184 Silicone Elastomer Kit (Dow Corning). SYLGARD® 184 Silicone Elastomer base and SYLGARD® 184 Silicone Elastomer curing agent were mixed thoroughly at a ratio of 10:1 (w/w). The mixture was then degassed using a desiccator equipped with a vacuum pump for 15 minutes to remove any air bubbles. The liquid PDMS was poured onto a glass, sandwiched by another glass, and cured in an oven at 70 °C for 1 hour. The PDMS layer was then separated from the glasses, and 1-inch PDMS disks, each with ten 4 mm diameter wells were made using a hole-punch kit.
Synthesis of PEGDA and PEG-Fb
Poly(ethylene glycol) diacrylate (PEGDA) was synthesized following previously established methods [36]. The conjugation of fibrinogen to PEG was also carried out according to previously described methods [58]. The characterization of PEG-Fb was performed by evaluating PEGylation efficiency and the ability of PEG-Fb to photocrosslink and form 3D hydrogels [59].
Propagation of CRC PDX Cells in Mice
All experimental procedures were approved by the Animal Care and Use Committee. A frozen stage II CRC adenocarcinoma PDX line from a white female patient pre-treatment was kindly provided by BioConversant, LLC. To establish xenografts from frozen PDX cells, approximately 1.4 × 106 tumor cells were mixed at a 1:1 ratio with Matrix High Concentration, Phenol-Red free Matrigel (Corning) and injected into the flank of a NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NOD-SCID) mouse (The Jackson Laboratory). To propagate the PDX line, the freshly excised tumor (~0.8 g) was washed in sterile PBS, minced in approximately 2.5 mL DMEM with 100 U/mL penicillin and 100 μg/mL streptomycin, and approximately 200 μL of the minced tumor suspension was injected into the flank of a NOD-SCID mouse. Experiments discussed in this study were performed using CRC-PDX tumors that were propagated in the absence of Matrigel and were passaged 3–4 times in mice.
CRC-PDX Tumor Dissociation and Cell Isolation
Excised CRC-PDX tumors were dissociated according to the methods described in the Supplementary Methods, and all PDX cells, including both human cancer cells and mouse stromal cells [28, 60], were isolated for subsequent in vitro culture as 2D-CRC-PDX cells and 3D-eCRC-PDX tissues.
Formation and Culture of 3D-eCRC-PDX Tissues
To create the 3D-eCRC-PDX tissues, all isolated CRC PDX cells (i.e. both the adherent and nonadherent cell populations later observed in 2D culture) were mixed with a polymer precursor solution containing PEG-Fb with a final concentration of 1.5% (v/v) triethanolamine (TEOA) (Acros Organics), 37 mM 1-vinyl-2-pyrrolidinone (NVP) (Sigma-Aldrich), and 0.1 mM Eosin Y in PBS at a density of 20×106 cells/mL. The cell-laden precursor solution was gently pipetted into the prefabricated PDMS mold wells (10 μl/well) and photocrosslinked with visible light for 2 minutes. Molds were then removed, and cell culture media added. The 3D-eCRC-PDX tissues were maintained in long-term culture (29 days) in a standard incubator with a temperature of 37 °C and 5% CO2 with media refreshes every 2–3 days.
2D CRC PDX Cell Culture
CRC cells isolated from the CRC-PDX tumors were cultured in T-75 standard cell culture flasks using the same cell culture media as was employed for the 3D-eCRC-PDX tissues. These 2D cultured CRC PDX (2D-CRC-PDX) cells were passaged after 8 days of culture and subsequently passaged every 7 days up to a total of 29 days. Importantly, since non-adherent cell types (mostly cell aggregates) were present, the spent media containing the 2D-CRC2D-CRC-PDX cells was also collected and centrifuged at 300 ×g. The non-adherent cell pellets were then resuspended in fresh media and recombined with the seeded adherent cells. All of the cultured cells were maintained in a standard incubator with a temperature of 37 °C and 5% CO2, with media changes every 2–3 days.
To investigate the cell growth curve or the increase in the total number of 2D-CRC-PDX cells over time, 2×105 cells/cm2 were seeded into ten T-25 standard cell culture flasks, one of which was used for cell counting every 24 hours (Days 1–11) using 0.25% Trypsin-EDTA (Corning) to detach the cells. The spent media containing the 2D-CRC-PDX non-adherent cells was also collected and included in cell counting. Following staining with trypan blue, the live cells were then counted using a hemocytometer. Additionally, 2×105 cells/cm2 were seeded onto a 24-well plate to investigate the viability of cells in 2D-CRC-PDX culture on Day 1 by Live/Dead staining.
Cell Viability Assay and Image Acquisition and Analysis
A cell viability assay was performed using the LIVE/DEAD® Viability/Cytotoxicity Kit (ThermoFisher Scientific) according to the manufacturer’s protocol. To quantify the percentage of live cells, Hoechst 33342 (Calbiochem) (20 μg/mL) was used to counterstain the cell nuclei. The fluorescently labeled cells were observed, and images and Z-stacks were obtained using a Nikon Eclipse TE2000 Inverted Microscopes equipped with a Nikon A1 plus Confocal Microscope System. The confocal microscopic images of the 2D-CRC-PDX cells and Z-stacks of the 3D-eCRC-PDX tissues were analyzed using ImageJ software (version: 2.0.0-rc-68/1.52e). The percentage of viable cells was calculated by dividing the manually quantified number of viable cells (total cell nuclei count minus dead cells) by the total number of cell nuclei (total number of cells). Further details on the staining and imaging procedures are provided in the Supplementary Methods.
Image Acquisition and Morphological Analysis of 3D-eCRC-PDX Tissues and 2D-CRC Cells
The 3D-eCRC-PDX tissues were cultured in vitro over a 29-day period and imaged every 7 days. Phase contrast images and Z-stacks of 3D-eCRC-PDX tissues were obtained using an inverted Nikon Eclipse Ti microscope equipped with an Andor Luca S camera and analyzed using ImageJ software (version: 2.0.0-rc-68/1.52e) to quantify the area, major axis, minor axis, circularity, and aspect ratio of the tumor cell colonies within 3D-eCRC-PDX tissues. For 2D-CRC-PDX cells, phase contrast images were obtained prior to cell passaging.
By selecting random regions from phase contrast images or Z-stacks, three 3D-eCRC-PDX tissue batches with at least 50 tumor cell colonies from each batch, were analyzed and the data exported to MS Excel. The diameter represented in the Results section is the mean geometric diameter, which was calculated using Equation 1.
| (Equation 1) |
Cell Subpopulation Investigation by Flow Cytometry
To investigate the cell subpopulations using flow cytometry, the CRC-PDX tumors, 3D-eCRC-PDX tissues, and 2D-CRC-PDX cells were dissociated to obtain single cells, and the numbers of viable cells were manually counted post-dissociation as described above. Detailed cell staining and flow cytometry protocols are provided in the Supplementary Methods. Briefly, to identify human-derived cells, mouse-derived cells, CRC cells, and proliferating cells the following antibodies were used, respectively: β−2-microglobulin (B2M) mouse IgG2a mAb (OriGene Technologies) conjugated to Zenon™ R-phycoerythrin (PE) mouse IgG2a (ThermoFisher Scientific), MHC Class I H-2 Db antibody (H2Db) mouse IgG2a mAb (OriGene Technologies) conjugated to the Zenon™ PE mouse IgG2a, cytokeratin 20 (CK20) rabbit IgG mAb (Cell Signaling Technology) conjugated to Zenon™ Alexa Fluor™ 647 rabbit IgG (ThermoFisher Scientific), and Ki-67 rabbit IgG pAb (abcam) conjugated to the Zenon™ Alexa Fluor™ 647 rabbit IgG.
To exclude dead cells and debris from the analysis, cells were labeled with Zombie Green™ dye (Zombie Green™ Fixable Viability Kit, BioLegend). To block nonspecific binding on the cell surface, cold blocking buffer (0.5 % (w/v) Bovine Serum Albumin (BSA) (Sigma-Aldrich) and 10 % (v/v) FBS in 1X PBS) containing Human BD Fc Block™ (BD Biosciences) (12 μg/mL) and Mouse BD Fc Block™ (BD Biosciences) (5 μg/mL) was used. To fix and permeabilize the cells, 0.5 mL of cold Foxp3 Fixation/Permeabilization (eBioscience™) working solution (prepared according to manufacturer protocol) was used for each sample tube. To block nonspecific binding for intracellular staining, FACS buffer (0.5 % (w/v) BSA and 10 % (v/v) FBS in 1X Permeabilization Buffer) was used. Compensation beads (ThermoFisher Scientific) were used according to the manufacturer protocol to accurately set the compensation in the event of fluorochrome overlap. To monitor for cell autofluorescence, unstained cell controls were also used. To set an accurate compensation for Zombie Green™ dye, a dead control cell population was used. A BD Accuri C6 cytometer was used to quantify labeled cells and subsequently, FlowJo software (version 10.0.7) was employed for data analysis.
In addition to investigating the percentage of cell subpopulations in culture over time, we also assessed the change in the total number of cells and the number of cells in each subpopulation. To calculate the number of cells in each subpopulation, the population percentage quantified via flow cytometry was multiplied by the total number of viable cells counted as described above. To account for obligatory cell passaging required for maintenance in the 2D-CRC-PDX culture, total numbers of 2D-CRC-PDX cells were calculated based on cell counts at each time point and the number of cells seeded at each passage.
Morphological Study of Cells by Staining
To study the cell morphology within both 3D-eCRC-PDX tissues and 2D-CRC-PDX cultures, the cells were labeled with Hoechst 33342, B2M mouse IgG2a mAb with goat anti-mouse IgG Alexa Fluor™ 488 (ThermoFisher Scientific), Alexa Fluor™ 568 Phalloidin (ThermoFisher Scientific), and CK20 rabbit IgG mAb conjugated to Zenon™ Alexa Fluor™ 647 rabbit IgG to observe the cell nuclei, human-derived cells, f-Actin filaments, and CRC cells, respectively. The cells were imaged using a Nikon confocal microscope. Further detail is provided in the Supplementary Methods.
Histological Examination of 3D-eCRC-PDX Tissues and CRC-PDX Tumors
To visualize both cellular and tissue microanatomy, a portion of the CRC-PDX tumors and entire 3D-eCRC-PDX tissues were sequentially sectioned and embedded in paraffin. The paraffin-embedded sections of CRC-PDX tumors and 3D-eCRC-PDX tissues were stained using hematoxylin and eosin (H&E). The stained sections were scanned using an Aperio CS2 scanner (Leica Biosystems) at 40x magnification. The digital images were viewed using Aperio ImageScope (Ver. 12.3.2.8013). Further details on the H&E staining protocol are provided in the Supplementary Methods.
RT-qPCR Gene Expression
Total RNA was extracted from CRC-PDX tumors, 2D-CRC-PDX cells, and 3D-eCRC-PDX tissues using the RNeasy Plus Micro Kit (Qiagen). The analysis was carried out for 3–4 separately obtained tumors and the corresponding batches of 3D-eCRC-PDX tissues and 2D-CRC-PDX cultures. Total RNA (50 ng) was used for cDNA synthesis using SuperScript™ IV First-Strand Synthesis System (Invitrogen). Quantitative polymerase chain reaction (qPCR) amplifications were performed using RT2 SYBR Green qPCR Mastermix (Qiagen) with amplification conditions: 40 cycles of 15 seconds of denaturation at 95 °C followed by 1-minute annealing. Reactions were performed in triplicate, and data were calculated using the ΔΔCt method. Gene expression was normalized by using B2M and GAPDH as reference genes.
Morphological Analysis of 3D-eCRC-PDX Tissues
To study whole-tissue morphological changes of 3D-eCRC-PDX tissues over time, images were obtained from a CellScale MicroSquisher system (side view of the tissues) and through phase-contrast microscopy (top view of the tissues) and used to quantify the volume (top and side views of the tissues), mean geometric diameter (top view of the tissues using Equation 1), circularity (side view of the tissues using Equation 2), and aspect ratio (side view of the tissues using Equation 3) of the 3D-eCRC-PDX tissues using ImageJ software. A minimum of 3 samples per time point per batch were analyzed.
| (Equation 2) |
| (Equation 3) |
Mechanical Stiffness
The mechanical stiffness of CRC-PDX tumors and 3D-eCRC-PDX tissues was evaluated using a CellScale MicroSquisher system and the associated SquisherJoy software according to the previously published protocol [50]. Briefly, the geometric core, midpoint, and periphery of CRC-PDX tumors were cut into disk-shaped slices with a diameter of approximately 3 mm and a thickness of approximately 1 mm (thus resembling the 3D-eCRC-PDX tissue geometry) using a sterile scalpel (Bard-Parker®) and a surgical punch (Sklar) (Suppl. Figure 1). All CRC-PDX tumor slices and 3D-eCRC-PDX tissues were subjected to parallel compression testing under physiological temperatures in a PBS bath using the MicroSquisher apparatus. The compression system was equipped with a 558.8 μm gauge cantilever beam and a square microplate. All samples were preconditioned to 30% deformation for 3 compressions before data were collected. The force versus displacement data were then obtained via compression and relaxation cycles at a rate of 10 μm/s to 30% deformation. The data were then converted to stress-strain curves, linear least-squares fitting was performed on the linear portion of the curves, and the Young’s moduli of the tissue samples were calculated. Two separate cell culture batches of CRC-PDX tumors and 3D-eCRC-PDX tissues were examined. For 3D-eCRC-PDX tissues, the mechanical stiffness was quantified on Days 1, 8, 15, 22, and 29 post-encapsulation for a minimum of three samples. For CRC-PDX tumors, three pieces of each region were measured for each batch.
Proteomics Analysis
To isolate ECM enriched fractions of protein CRC-PDX tumors (obtained after 31–42 days of tumor growth) and 3D-eCRC-PDX tissues (obtained on the 22nd day of in vitro culture) were processed via Millipore Compartmental Protein Extraction Kit following the manufacturer’s instruction. Of note, samples were weighed prior to processing, and buffer volume was adjusted appropriately based on the sample weight. Following sequential extraction of cytoplasmic, nuclear, membrane, and cytoskeletal proteins via the kit [61, 62], the remaining pellet was washed once with 500 μL of PBS and reconstituted in 2x Novex NuPAGE LDS sample buffer (Invitrogen) supplemented with 2x NuPAGE sample reducing agent (Invitrogen). ECM-enriched fractions were then sonicated in an ultrasonic water bath for 20min. at ambient temperature. Debris was removed by centrifugation for 20 min. at 16,000 ×g, 4°C. Proteomics workup was carried out as previously reported [63]. Briefly, the protein fractions were quantified; ~20 μg of protein per sample was then reduced with DTT and denatured at 70°C for 10min. prior to loading onto 10% Bis-Tris Protein gels and separating as a short stack run. The gels were stained overnight with colloidal Coomassie for visualization purposes, each lane was cut into 6-MW fractions and equilibrated in 100 mM ammonium bicarbonate (AmBc), and each gel plug was then digested overnight with Trypsin Gold, Mass Spectrometry Grade (Promega) following manufacturer’s instruction. Peptide extracts were reconstituted in 0.1% Formic Acid/ ddH2O at 0.1 μg/μL. Mass spectrometry was carried out, and the data were processed, searched, filtered, grouped, and quantified using normalized spectral counting as previously reported in detail [63].
All identified proteins were converted from UniProtKB AC/ID to UniProtKB using the Retrieve/ID Mapping feature in UniProt to obtain primary gene names. The primary gene names were processed using MatrisomeAnnotator (http://matrisome.org/) [64] to identify ECM and ECM-associated proteins and subsequently categorized into ECM glycoproteins, collagens, proteoglycans, ECM regulators, ECM-affiliated proteins and secreted factors. The categorized proteins detected in only one out of six samples (CRC-PDX tumors and 3D-eCRC-PDX tissues) were removed from the analysis. The proteins were then employed to plot Euler diagrams using the eulerr package in RStudio (Version 1.2.1335) for each sample. Moreover, the categorized proteins from three samples (CRC-PDX tumors or 3D-eCRC-PDX tissues) were pooled and presented for the comparison between CRC-PDX tumors and 3D-eCRC-PDX tissues. For the pooled data, a protein was indicated as present when the protein was detected in at least two out three samples. Further details on the proteomics analysis protocol are provided in the Supplementary Methods.
Transcriptomic Analysis
RNeasy Plus Micro Kits (Qiagen) were used to isolate total RNA from frozen CRC-PDX tumors (obtained after 31–42 days of tumor growth) and 3D-eCRC-PDX tissues (obtained on the 15th day of in vitro culture) following the manufacturer’s protocol. Procedures for RNA-seq were performed at the Genomic Services Laboratory (GSL), HudsonAlpha Institute for Biotechnology. Extracted total RNA initial QC quantification was done using a Qubit Fluorometer (Invitrogen), and the quality of the extracted RNA was evaluated using an Agilent 2100 Bioanalyzer. RNA with a RIN score of 7.0 or higher was used for RNA-seq library preparation using an Illumina TruSeq Stranded RNA Kit with Ribo-Zero Gold and sequenced on NovaSeq sequencers at 100PE.
Sequencing reads were aligned to both the human GRCh38 and mouse GRCm38 reference genomes and counted with STAR [65] using aRNApipe, a RNA-seq analysis pipeline developed at the HusdonAlpha Institute for Biotechnology [66]. Approximately 62%−65% of the reads uniquely mapped to the human GRCh38 reference genome, while approximately 12%−21% of the reads uniquely mapped to the mouse GRCm38 reference genome (Suppl. Table 1). Human mapped raw counts obtained from STAR were used for differential expression analysis with the R package DESeq2 [67]. False discovery rates (FDR) p value less than 0.05 were considered significantly differentially expressed and genes with log2 fold change greater than 1.5 were used for downstream gene set analyses.
CRC Consensus Molecular Subtype Analysis
Human mapped raw counts were used for consensus molecular subtype analysis with the R package CMScaller [68], with default RNA-seq settings. CMScaller uses a nearest template prediction algorithm [69] to assign each sample to a consensus molecular subtype (CMS) [70]. In brief, CMScaller computes cosine distances for each sample and assigns it to a CMS based on the smallest distance to the CMS template. Prediction FDR p value greater than 0.05 resulted in no classification.
TCGA Analysis
The R package TCGAbiolinks [71] was used to obtain HT-Seq gene counts from the Genomic Data Commons for CRC tumor and tumor-adjacent tissue as a part of the TCGA-COAD [72] project. CRC-PDX tumor and 3D-eCRC-PDX tissue HT-Seq gene counts and TCGA counts were merged by Ensembl gene ID’s and processed as described above with DESeq2. The combined sequence data were variance stabilized using the DESeq method.
GSEA Analysis
GSEA was performed using the desktop GSEA program (version 4.0.3) [73] and the Molecular Signature Database (MSigDB) Hallmarks v7.0 gene sets. GSEA analysis was performed using 1000 permutations and the phenotype permutation type. Annotations of human ensemble gene ID to MSigDB.v7.0 was performed. Enrichment plots and a heatmap were presented to visualize the data. Normalized enrichment score (NES) and the false discovery rate (FDR) q-value which presents that the estimated probability that the NES represents a false positive finding were reported in the Enrichment plots.
Gene Ontology Analysis
The web-based application GOrilla [74] was used to identify enriched gene molecular function ontologies between CRC-PDX tumor and 3D-eCRC-PDX tissue samples and TCGA CRC tumor-adjacent tissue. The application REViGO [75] was used to eliminate redundant GO terms and visualize the enriched terms using the TreeMap function which shows a two-level hierarchy of GO terms. The following REViGO settings were used: allowed similarity, (0.5, small); and semantic similarity measure, Resnik (normalized).
Kaplan–Meier survival analysis
To examine expression of the top 16 significantly upregulated genes between CRC-PDX tumors and 3D-eCRC-PDX tissues in the precomputed GEO datasets GSE17536, GSE14333, GSE17537, and TCGA-COAD, the PROGgeneV2 tool was used [76, 77]. All cohorts were adjusted for age, stage, and gender covariates and bifurcated based on median expression. The hazard ratios, 95% confidence intervals, and p values were reported.
Statistical Analysis
The outliers were removed using the ROUT method with coefficient Q = 1%. The normality of distribution and equality of variance were then checked. T-test was performed when comparing the means of two groups. One-way ANOVA followed by Tukey post hoc test was performed when comparing the means of more than two groups. Two-way ANOVA followed by Tukey was performed for Morphological Analysis and Mechanical Stiffness. Two-way ANOVA followed by Bonferroni-Šídák post hoc test was performed when comparing groups to a control group (Flow Cytometry). Linear trend analysis was performed using one-way ANOVA with a post-test for linear trend. The rates of increase or decrease (slopes) were found using linear regression analysis. p value ≤ 0.05 was considered statistically significant unless otherwise stated. GraphPad Prism 7.0 (GraphPad Software, San Diego, California USA, www.graphpad.com) was used for the above analysis. Hierarchical cluster analysis was carried out using heatmap.plus package in RStudio (Version 1.2.1335).
3. Results
CRC-PDX Tumor Cell Viability Maintained Following 3D Encapsulation
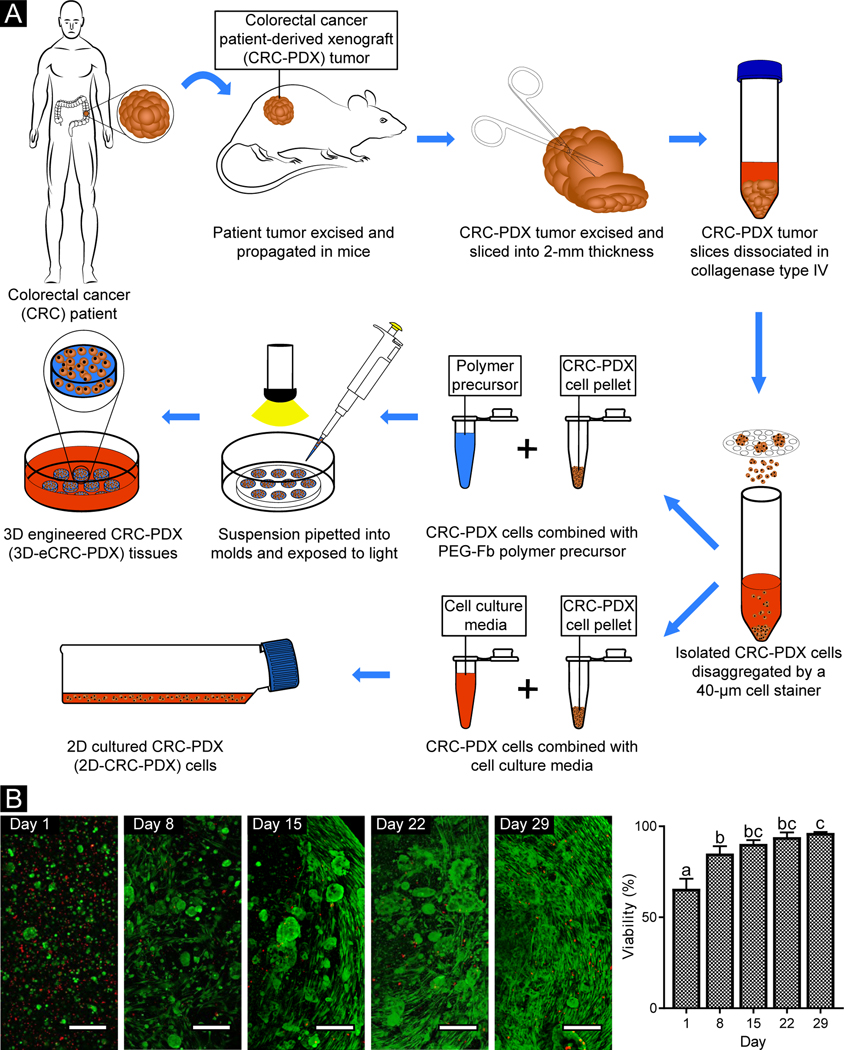
Using an optimized tumor dissociation method, we obtained 14.6 ± 0.6 million viable cells/gram of initial tissue (n = 5 CRC-PDX tumors). The isolated cells were then encapsulated in PEG-fibrinogen (PEGylation efficiency = 90 ± 4%) to create the 3D-eCRC-PDX tissues and separately cultured as 2D-CRC-PDX cells. A schematic of the process is shown in Figure 1.
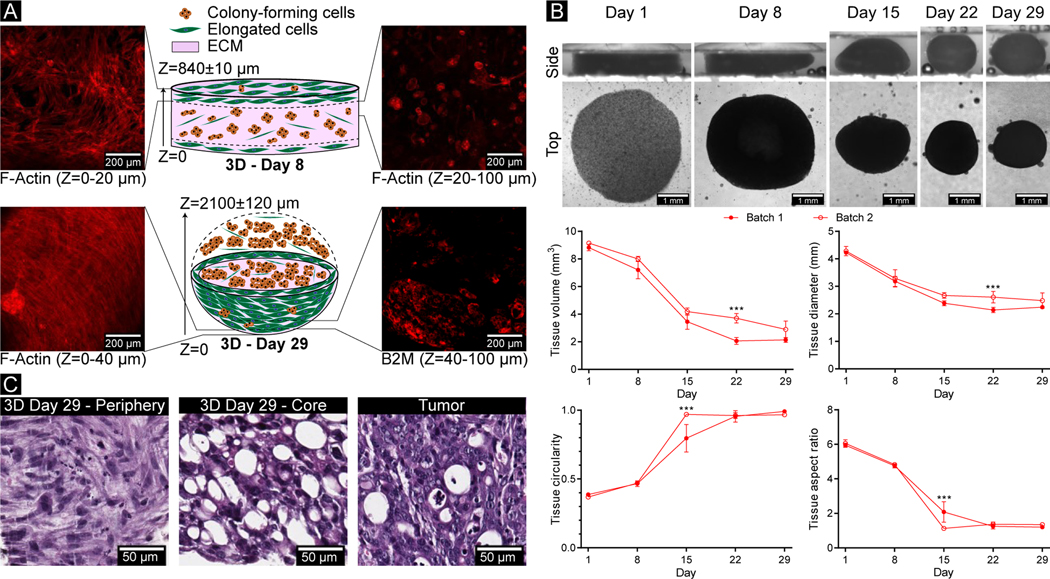
Figure 1. Following encapsulation in PEG-Fb, CRC-PDX tumor cells were viable during long-term culture.
(A) All PDX cells, including both human cancer cells and mouse stromal cells, were isolated from CRC-PDX tumors and used to create 3D-eCRC-PDX tissues and cultured as 2D-CRC-PDX cells. (B) Representative confocal images of live (green) and dead (red) cells within 3D-eCRC-PDX tissues (scale bar = 200 μm). Day 1 cell viability was equivalent to that of non-encapsulated cells (Suppl. Figure 2). Cell elongation and increasing cell colony size were observed from Day 1 to Day 29 post-encapsulation. As a result of cell growth, the percentage of viable cells within the 3D-eCRC-PDX tissues increased significantly during in vitro culture. Bar = mean ± SD. Means that do not share a letter are significantly different (p ≤ 0.05; n = 3 tissues).
To evaluate cell viability, the encapsulated cells within the 3D-eCRC-PDX tissues were labeled with Live/Dead dye and counterstained with Hoechst 33342. Based on confocal z-stack analysis, 67 ± 4% of encapsulated cells within the 3D-eCRC-PDX tissues were viable on Day 1 (Figure 1B). To examine whether cell viability was reduced due to the tumor dissociation process, the cell encapsulation process, or a combination of both procedures, we performed viability assays on 2D-CRC-PDX cells on Day 1 post-tumor dissociation. Image analysis revealed that approximately 66 ± 6% of the cells were viable (Suppl. Figure 2) with no significant difference between 2D-CRC-PDX cells and the cells encapsulated within the 3D-eCRC-PDX tissues. Therefore, the lower viability of tumor cells observed within the 3D-eCRC-PDX tissues on Day 1 was primarily a result of the tumor dissociation process, as expected. Prior investigation of CRC-PDX tumor dissociation by Petit et al. reported similar post-dissociation cell viability [78].
Long-term viability was also examined to determine the temporal impact of in vitro culture on cell survival in the 3D-eCRC-PDX tissues. The percentage of viable cells on Day 8, 15, 22, and 29 was 85 ± 6%, 90 ± 2%, 94 ± 3%, and 96 ± 1%, respectively (Figure 1B), which was significantly higher than Day 1 (p ≤ 0.0003). Furthermore, the cell viability was significantly higher on Day 29, as compared to Day 8 (p = 0.0149). The high viability at the later time points is a result of cell proliferation and a subsequent overall increase in the total number of viable cells throughout the long-term culture, as can be seen in the confocal images (Figure 1B). The cells encapsulated within the 3D-eCRC-PDX tissues proliferated (quantified via flow cytometry, as reported in detail below) and altered their morphological characteristics, with some cells forming colonies and others elongating within the biomaterial matrix (Figure 1B). Elongated cells were first observed on Day 8 and increased in abundance over time, as visualized on Day 29.
Growth and Morphological Analysis of Tumor Cell Colonies within 3D-eCRC-PDX Tissues
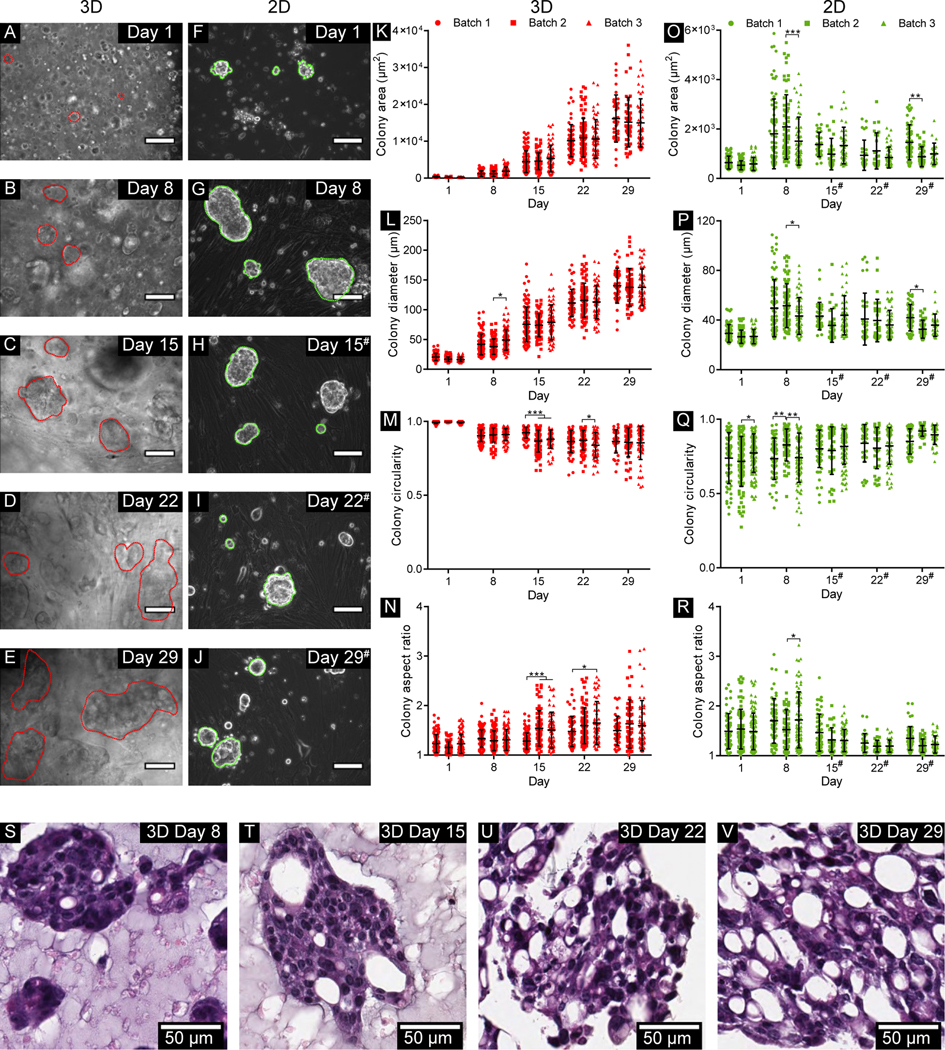
To assess the colony-forming ability of the tumor cells in the 3D-eCRC-PDX tissues and temporal changes in the colony size and morphology, cells within the 3D-eCRC-PDX tissues and in 2D-CRC-PDX culture were imaged over a period of 29 days. Three batches of 3D-eCRC-PDX tissues and 2D-CRC-PDX cultures were separately prepared, each from separate tumors, and analyzed. Phase-contrast z-stacks showed that the cell colony area and diameter within the 3D-eCRC-PDX tissues ranged from approximately 70 to 850 μm2 and 10 to 40 μm on Day 1, respectively (Figure 2A–E, K, L). However, for 2D-CRC-PDX cells, the cell colony area and diameter ranged from approximately 300 to 1,300 μm2 and 20 to 50 μm on Day 1, respectively (Figure 2F–J, O, P). Cell colony area and diameter within 3D-eCRC-PDX tissues increased from Day 1 to Day 29 for each batch based on one-way ANOVA with a post-test for linear trend (p < 0.0001); observed increases were consistent between batches with no significant difference between the slopes based on linear regression analysis. Within the 3D-eCRC-PDX tissues on Day 29, the cell colony area and diameter ranged from approximately 3,500 to 35,000 μm2 and from approximately 70 to 220 μm, respectively (Figure 2A–E, K, L). This increase in the size of CRC PDX cell colonies within the 3D-eCRC-PDX tissues over time was also observed using H&E staining (Figure 2S–V). For 2D-CRC-PDX cells, passaging was necessary after 8 days of in vitro culture to prevent cell death (Suppl. Figure 3). The 2D-CRC-PDX cell colonies increased in size from Day 1 to Day 8; however, following passaging the subsequent colonies did not grow as large as those quantified on Day 8 (Figure 2F–J, O, P).
Figure 2. Morphological analysis of cell colonies within 3D-eCRC-PDX tissues and in 2D-CRC-PDX cultures.
Representative phase contrast images of cell colonies (A-E) within the 3D-eCRC-PDX tissues (outlined by red dotted lines) and (F-J) in the 2D-CRC-PDX cultures (outlined by green dotted lines) over 29 days of culture (scale bar = 100 μm). Within the 3D-eCRC-PDX tissues, an increase in (K) cell colony area and (L) colony diameter, a (M) decrease in colony circularity, and an (N) increase in cell colony aspect ratio were observed from Day 1 to Day 29; these changes were generally consistent between batches. (O-P) 2D-CRC-PDX cell colony area and diameter increased from Day 1 to Day 8; however, following passaging on Day 8 the subsequent colonies did not grow as large as prior to passaging. For 2D-CRC-PDX cells, (Q) colony circularity increased and (R) colony aspect ratio decreased from Day 1 to Day 29. (S-V) Increasing CRC PDX colony size was also observed in histological sections of 3D-eCRCPDX tissues over time. Data are mean ± SD, and each dot represents one colony. At least 50 cell colonies within 3D-eCRC-PDX tissues or in 2D-CRC-PDX cultures were examined for each of 3 separately prepared batches. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. # indicates that the 2D-CRC-PDX cells were passaged at the previous time point.
Cell growth and morphology within the 3D-eCRC-PDX tissues were consistent between batches. No significant differences in 3D-eCRC-PDX tissue mean cell colony area were observed between batches at any time point; the only significant difference between batches in mean colony diameter occurred on Day 8 between batches 2 and 3 (p = 0.0017) (Figure 2K, L). Furthermore, the number of viable cells within each 3D-eCRC-PDX tissue was counted after dissociation for flow cytometry analysis. The total number of viable cells increased linearly from Day 1 to Day 29 based on one-way ANOVA with a post-test for linear trend (p < 0.0001); the data showed that this increase did not differ significantly between batches based on linear regression analysis (Suppl. Figure 4). In contrast, for 2D-CRC-PDX cells significant differences were observed between batches, including differences in both cell colony area and diameter on Day 8 and Day 29 (p = 0.0121 and p = 0.0187, respectively) (Figure 2O, P). In 2D-CRC-PDX cell culture, the total number of viable cells for all three batches was found to increase from Day 1 to Day 29 based on one-way ANOVA with a post-test for linear trend (p < 0.0001); however, in contrast with 3D-eCRC-PDX tissues, the slopes differed significantly for the 2D-CRC-PDX cells (p = 0.0274).
Circularity and aspect ratio of the cells and cell colonies, which are indicators of invasiveness [79–81], were also analyzed. Within the 3D-eCRC-PDX tissues, circularity of the cell colonies decreased and aspect ratio increased from Day 1 to Day 29 based on one-way ANOVA with a post-test for linear trend (p < 0.0001) (Figure 2M, N). Circularity ranged from 0.97 to 1.00 on Day 1 and from 0.55 to 0.97 on Day 29; the aspect ratio ranged from 1.00 to 1.80 on Day 1 and from 1.00 to 3.15 on Day 29. However, for 2D-CRC-PDX cells circularity increased and aspect ratio decreased from Day 1 to Day 29 (one-way ANOVA, post-test for linear trend, p < 0.0001) (Figure 2Q, R). Circularity ranged from 0.27 to 0.97 on Day 1 and from 0.72 to 0.98 on Day 29, while the aspect ratio ranged from 1.00 to 2.58 on Day 8 and between 1.00 to 2.00 on Day 29. Significant differences in cell colony circularity and aspect ratio were not consistently observed between batches (Figure 2M, N, Q, R).
3D-eCRC-PDX Tissues Maintained Cell Subpopulations
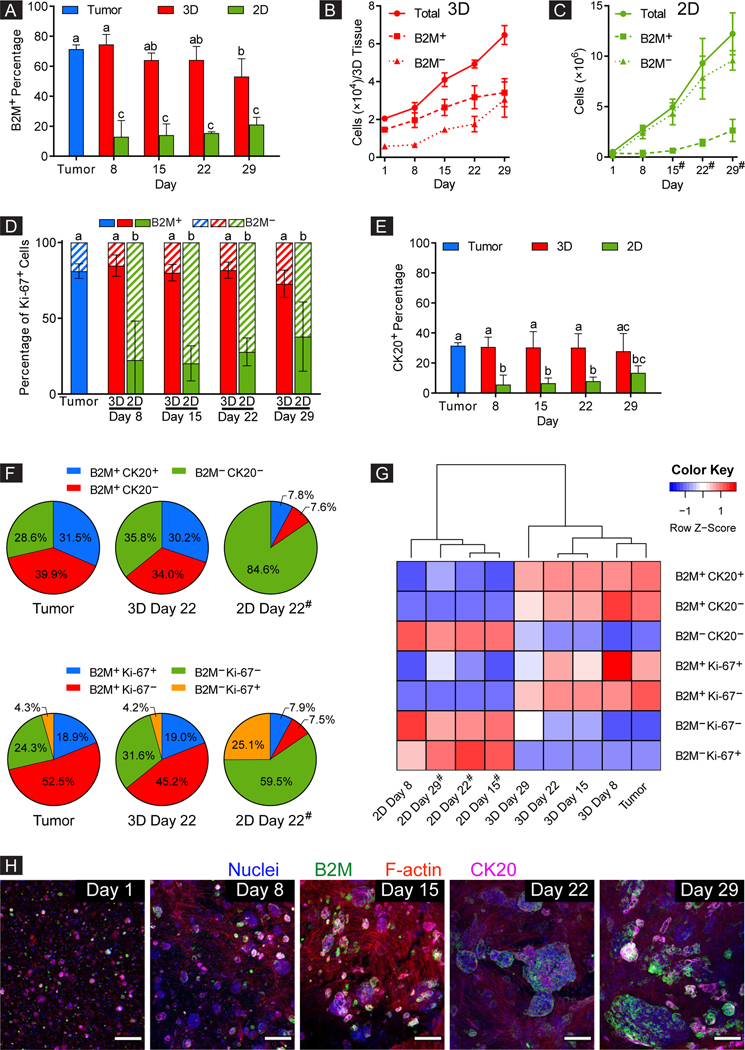
Using flow cytometry, we quantified both the human and mouse cell subpopulations from the CRC-PDX tumor, 3D-eCRC-PDX tissues, and 2D-CRC-PDX cultures; PDX models contain mouse stromal components in addition to the human malignant cells [28, 60] and maintaining both subpopulations is important for recapitulation of the tumor microenvironment. Human cells were identified using the cell marker β−2 microglobulin (B2M); the cells were also labeled with a mouse cell marker, H2Db, to confirm that those cells which were negative for B2M were the mouse cell subpopulation (Suppl. Figure 5). For dissociated CRC-PDX tumor cells, 71±3% of the cell population was positive for the B2M human marker (3 separately dissociated tumors, Figure 3A). Importantly, as the cells grew within the 3D-eCRC-PDX tissues, the human cell subpopulation (B2M+) percentage remained constant during long-term culture (through Day 22); there was an average percentage of 68±6% B2M+ cells (Days 8,15, 22) with no significant difference between the 3D-eCRC-PDX tissues and dissociated CRC-PDX tumors (n = 3 separately prepared batches). On Day 29, the percentage of B2M+ cells (53±12%) was significantly lower than that on Days 1 and 8 (71±3% and 74±7%, respectively, p < 0.003, Figure 3A). In sharp contrast to the 3D-eCRC-PDX tissues, the percentage of B2M+ cells in 2D-CRC-PDX cultures had already decreased by over 80 percent after 8 days of culture (from 71±3% to 13±11%, n = 3 separate batches, p < 0.0001)) and then remained constant over 29 days. It is important to note that whereas the 3D-eCRC-PDX tissues were maintained in continuous culture for 29 days, it was necessary to passage the 2D-CRC-PDX cells every 7 days starting on Day 8 to avoid cell confluence; data presented are prior to each passage for 2D-CRC-PDX cells.
Figure 3. 3D-eCRC-PDX tissues maintained the CRC-PDX tumor cell subpopulations during long-term culture, whereas the 2D-CRC-PDX cells did not.
(A) Percentage of human (B2M+) cells within the 3D-eCRC-PDX tissues remained constant during long-term culture (through Day 22), maintaining the originating CRC-PDX tumor composition. However, for 2D-CRC-PDX cells, the percentage of human cells decreased by over 80% after 8 days of culture. While the cell numbers increased over time, (B) the 3D-eCRC-PDX tissues maintained similar ratios of human cancer and mouse stromal cells to those of the originating CRC-PDX tumor, whereas in (C) 2D-CRC-PDX culture, mouse (B2M−) cells outgrew the human cells. The 3D-eCRC-PDX tissues maintained the percentages of (D) human proliferative (B2M+ Ki67+) and (E) CK20+ cells over 29 days of culture, whereas in 2D-CRC-PDX cultures these percentages decreased significantly after 8 days of culture and remained low thereafter. (F) In sharp contrast to the 2D-CRC-PDX cultures, cell subpopulations within the 3D-eCRC-PDX tissues were similar to the originating CRC-PDX tumors. (G) The 3D-eCRC-PDX tissue cell subpopulations clustered with the CRC-PDX tumors whereas the 2D-CRC-PDX cells clustered separately from both the 3D-eCRC-PDX tissues and CRC-PDX tumors. (H) Colony-forming cells within the 3DeCRC-PDX tissues were all B2M+ and partially CK20+, thereby being human cells. Conversely, the elongated cells were neither B2M+ nor CK20+, thereby being mouse stromal cells (scale bar = 200 μm). Data are mean ± SD. Means that do not share a letter are significantly different (p ≤ 0.05, n = 3 separate batches of cell culture). # indicates that the 2D-CRC-PDX cells were passaged at the previous time point.
In both the 3D-eCRC-PDX tissues and 2D-CRC-PDX cultures, the total number of viable cells, human cells, and mouse cells increased over time (Figure 3B, C). However, which cell subpopulations contributed to this increase differed drastically. In the 3D-eCRC-PDX tissues, both human and mouse cell subpopulations increased in number, maintaining similar ratios to the those of the original CRC-PDX tumor over time; no significant difference was observed between the rates of increase for the 3D-eCRC-PDX tissue B2M+ and B2M− subpopulations (based on linear regression analysis, p = 0.4695, Figure 3B). In sharp contrast, the mouse cells were the major contributor to the increase in total cell number in 2D-CRC-PDX cultures; although numbers of human cells increased over time, the rate was significantly lower (0.23±0.04 fold, p < 0.0001) than that for the mouse cell subpopulations.
To further investigate temporal cell subpopulation changes, Ki-67 was employed as a proliferative marker in tandem with B2M to identify human proliferative cells. Although only 23±4% of all cells harvested from the CRC-PDX tumor were Ki-67 positive (Suppl. Figure 6A), 81±5% of these Ki-67+ cells were human cells (B2M+) (Figure 3D). Remarkably, the 3D-eCRC-PDX tissues maintained this human proliferative cell subpopulation over 29 days of culture with no significant changes (85±7%, 80±5%, 82±5%, and 73±9% on Day 8, Day 15, Day 22, and Day 29, respectively), whereas in 2D-CRC-PDX cultures, a significant decrease (p = 0.0113) in the percentage of human proliferative cells was observed after 8 days of culture (from 81±5% to 22±26%) with the percentage remaining low thereafter.
For both 2D-CRC-PDX cultures and 3D-CRC-PDX tissues, approximately 20–30% of all cells (human and mouse) were Ki-67+ over the 29 days of culture (Suppl. Figure 6A). The lower human cell proliferation in 2D-CRC-PDX cultures was offset by higher mouse cell proliferation; therefore, less than 25% of 2D-CRC-PDX cells were B2M positive after a week of in vitro culture (Figure 3A). Moreover, total proliferative (Ki-67+) and non-proliferative (Ki-67−) cell numbers increased over time for both the 3D-eCRC-PDX tissues and 2D-CRC-PDX cultures; however, the major contributors to these increases differed. For 3D-eCRC-PDX tissues the rates of increase in human proliferative (B2M+ Ki67+) and mouse proliferative (B2M− Ki-67+) and human non-proliferative (B2M+ Ki-67−) and mouse non-proliferative (B2M− Ki-67−) cell numbers were similar (no significant difference), maintaining ratios equivalent to the original tumor. In sharp contrast, for 2D-CRC-PDX cultures, mouse cells were the major contributor to both the total proliferative and non-proliferative cell number increases, whereas human proliferative and non-proliferative cell numbers increased at significantly lower rates (0.53±0.07 and 0.12±0.01 fold, respectively, p ≤ 0.05) (Suppl. Figure 6B–E). Taken together, these results indicate that the maintenance of human CRC PDX cells and equivalent rates of increase in human and mouse cell subpopulations in the 3D-eCRC-PDX tissues throughout long-term culture was a result of maintenance of these proliferative cell subpopulations, whereas proliferative subpopulation changes in 2D-CRC-PDX cultures resulted in domination by the mouse cells over time.
To examine temporal changes in cancer cell subpopulations in the CRC-PDX tumor, 3D-eCRC-PDX tissues, and 2D-CRC-PDX cultures, cytokeratin 20 (CK20) was used to examine a CRC positive (CK20+) cell subpopulation. Even though CK20 is a CRC marker, not all CRC cells express the CK20 protein; the CK20 negative cell subpopulation may include other CRC cells [82–86]. As expected, the CK20+ cells were a subpopulation of the B2M+ cells while all mouse cells were negative for B2M as well as CK20 (Suppl. Figure 5). The number of cells in both B2M+ CK20+ and B2M+ CK20− subpopulations within the 3D-eCRC-PDX tissues and 2DCRC-PDX cultures increased over time (Suppl. Figure 6H, I). For the dissociated CRC-PDX tumors, 32±2% of cells were positive for CK20 (Figure 3E). Importantly, 3D-eCRC-PDX tissues were able to maintain a subpopulation of 32±2% CK20+ cells over 29 days of culture, whereas a significant decrease (p = 0.0001) in the percentage of CK20+ cells was observed by Day 8 in 2D-CRC-PDX cultures in relation to the CRC-PDX tumor (from 32±2% to 6±6%). This trend was also observed for the dual-labeled B2M+ and CK20− cell subpopulation (Suppl. Figure 6G).
Overall, we found that the percentage of key cell subpopulations within the 3D-eCRC-PDX tissues were maintained over time with no significant changes through Day 22 of culture as compared to the CRC-PDX tumors. Conversely, 2D-CRC-PDX cells exhibited significant changes within the initial days of culture (Figure 3F and Suppl. Figure 7). Moreover, cluster analysis of these cell subpopulations revealed that the 3D-eCRC-PDX tissues from all time points clustered in a group with the CRC-PDX tumors, with Day 8 of the 3D-eCRC-PDX tissues clustering closest (Figure 3G and Suppl. Figure 8). In contrast, the 2D-CRC-PDX cells clustered separately from both the 3D-eCRC-PDX tissues and CRC-PDX tumors.
Morphological study of the distinct cell types supported the results found using flow cytometry. To visualize the cells within the 3D-eCRC-PDX tissues and in 2D-CRC-PDX cultures, immunostaining for nuclei, B2M, F-actin, and CK20 was carried out using Hoechst 33342, anti-B2M (human marker), phalloidin, and anti-CK20, respectively. Two distinct subpopulations were noted—a subpopulation of colony-forming cells and another of elongated cells. All of the colony-forming cells were positive for B2M and a portion was positive for CK20, whereas none of the elongated cells were positive for either B2M or CK20. Therefore, we can infer that the elongated cell subpopulation was comprised of mouse stromal cells (Figure 3H and Suppl. Figure 9) which is consistent with observations that human stromal cells are replaced by mouse-derived cells in cancer xenografts [28, 60]. Consistent with these findings, labeled 2D-CRC-PDX cells also demonstrated that colony-forming cells were positive for CK20 and B2M, whereas the elongated cells were not positive for either maker (Suppl. Figure 10).
Gene Expression in CRC-PDX Tumors Was More Similar to 3D-eCRC-PDX Tissues than 2D-CRC-PDX Cells
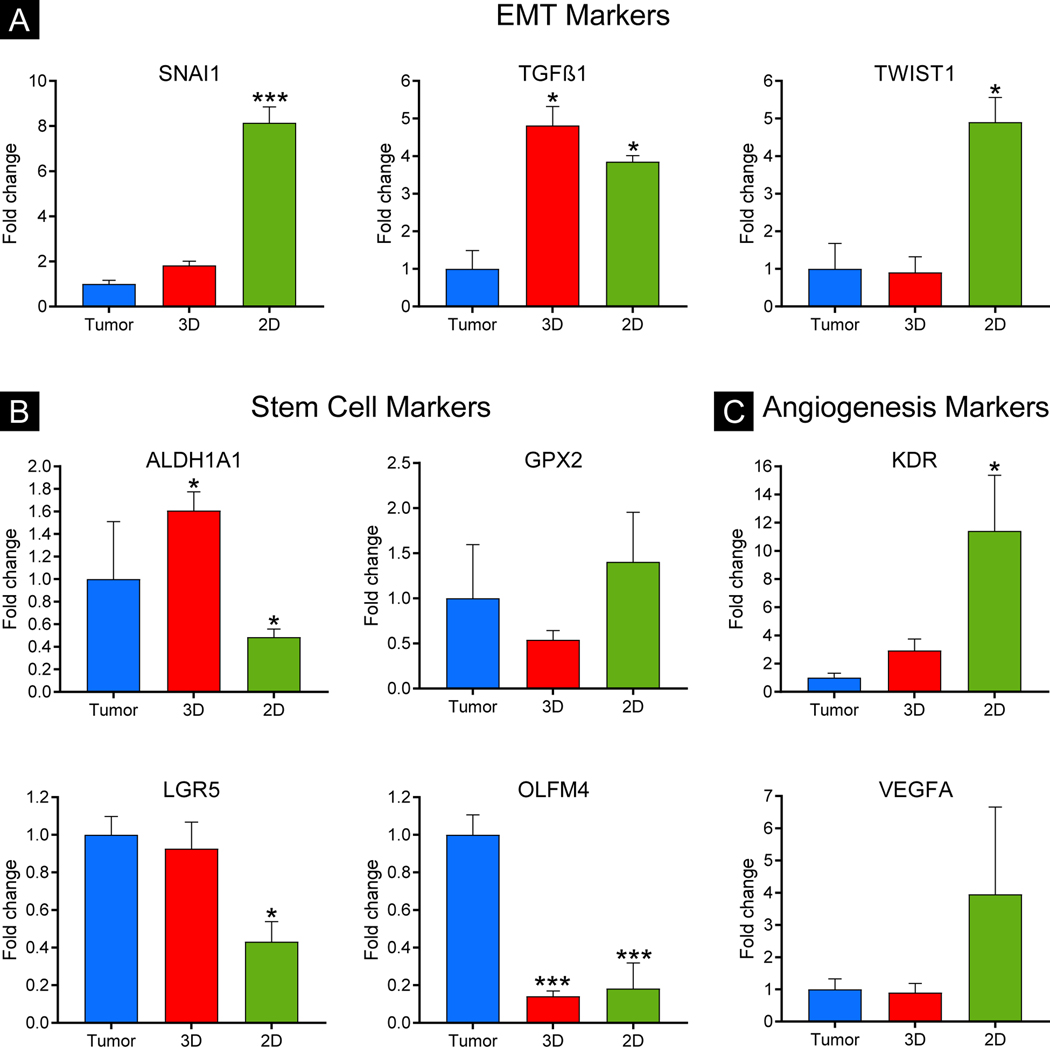
To examine gene expression of colon cancer markers of the epithelial to mesenchymal transition (EMT), stem cells, and angiogenesis, we performed RT-qPCR on total RNA isolated from CRC-PDX tumors, 3D-eCRC-PDX tissues, and 2D-CRC-PDX cells. We compared the expression of the colon cancer markers in the 3D-eCRC-PDX tissues and 2D-CRC-PDX cells to that in the CRC-PDX tumors. The expression of the EMT markers SNAI1 and TWIST1, which play a role in tumor metastasis, were 8.15 ± 0.71 and 4.91 ± 0.66 fold higher, respectively, in the 2D-CRC-PDX cells as compared to the CRC-PDX tumors (p = 0.001 and p = 0.005, respectively) (Figure 4A). However, no significant differences were found between the 3D-eCRC-PDX tissues and CRC-PDX tumors. The expression of the EMT marker TGFβ1 in both the 2D-CRC-PDX cells and 3D-eCRC-PDX tissues significantly increased by 3.85 ± 0.16 and 4.82 ± 0.51 fold, respectively, (p = 0.009 and p = 0.003, respectively). Next, we examined the expression of the stem cell marker genes ALDH1A1, GPX2, LGR5, and OLFM4. We observed a reduction in expression of ALDH1A1 (0.49 ± 0.7 fold), LGR5 (0.43 ± 0.11 fold) and OLFM4 (0.18 ± 0.14 fold) in the 2D-CRC-PDX cells (p = 0.016, p = 0.022, and p = 0.001, respectively) (Figure 4B). However, in the 3D-eCRC-PDX tissues, ALDH1A1 expression increased by 1.61 ± 0.17 fold (p = 0.006) and OLFM4 expression decreased by 0.14 ± 0.03 fold (p = 0.001). No significant difference in the expression of GPX2 was observed in the 2D-CRC-PDX cells and the 3D-eCRC-PDX tissues. Lastly, we examined the expression of the angiogenesis marker genes KDR and VEGFA (Figure 4C). We observed that KDR expression increased by 11.42 ± 3.95 fold in 2D-CRC-PDX cells (p = 0.028), while the VEGFA expression also increased but did not reach statistical significance (Figure 4C). In contrast, the expression of KDR and VEGFA was similar between the 3D-eCRC-PDX tissues and the CRC-PDX tumors. Taken together, our investigation into the expression of EMT, stem cell, and angiogenesis markers suggests that gene expression is more similar between the 3D-eCRC-PDX tissues and the CRC-PDX tumors, whereas significant differences were observed between 2D-CRC-PDX cells and the CRC-PDX tumors.
Figure 4. Gene expression of epithelial to mesenchymal transition (EMT), stem cell, and angiogenesis markers in CRC-PDX tumors was more similar to 3D-eCRC-PDX tissues than 2D-CRC-PDX cells.
Gene expression of (A) EMT markers, SNAI1 and TWIST1, (B) stem cell markers, GPX2 and LGR5, and (C) angiogenesis markers, KDR and VEGFA, in the 3De-CRC-PDX tissues was not significantly different compared to the CRC PDX tumors. In 2D-CRC-PDX cultures, however, gene expression of four out of these six markers (SNAI1, TWIST1, LGR5, and KDR) was significantly different compared to the CRC PDX tumors. Data are mean ± SE (*p ≤ 0.05 and ***p ≤ 0.001 compared to the CRC-PDX tumors. n = 3–4 separately obtained tumors and the corresponding batches of 3D-eCRC-PDX tissues and 2D-CRC-PDX cultures).
3D-eCRC-PDX Tissues Displayed Locational Heterogeneity and Cell-induced Contraction
Locational heterogeneity within the 3D-eCRC-PDX tissues was observed through immunostaining and confocal imaging. Elongated cells were primarily located on the outer layer (periphery) of the 3D-eCRC-PDX tissues, while the colony-forming cells were more prevalent within the 3D-eCRC-PDX tissues (Figure 5A). Similar locational heterogeneity of cell morphology was also observed in the H&E-stained 3D-eCRC-PDX tissues (Figure 5C). Furthermore, H&E staining illustrated that the colony size within the 3D-eCRC-PDX tissues increased over time and displayed similar cellular morphology to the CRC-PDX tumor on Days 22 and 29 (Figure 5C and Figure 2S–V). As the density of elongated cells increased on the periphery of the 3D-eCRC-PDX tissues, the 3D-eCRC-PDX tissues remodeled themselves from a disk shape on Days 1 and 8 to a near-spherical tissue by Day 29 (Figure 5A, B).
Figure 5. 3D-eCRC-PDX tissues showed locational heterogeneity and whole-tissue contraction over time.
(A) Locational heterogeneity was observed in the 3D-eCRC-PDX tissues through immunostaining and confocal imaging. A Z-stack projection of sequential images showed that elongated cells were predominantly located on the periphery of the 3D-eCRC-PDX tissues, whereas the colony-forming cells were present mostly inside the tissue. (B) Side views of the 3D-eCRC-PDX tissues demonstrated that the 3D-eCRC-PDX tissues were remodeled from a disk shape on Day 1 to a near-spherical tissue by Day 29 where the diameter of the disk decreased as the height increased. Top views of the 3D-eCRC-PDX tissues illustrate the cell-mediated contraction of 3D-eCRC-PDX tissues over 29 days of culture. The decrease in volume and diameter of the 3D-eCRC-PDX tissues, increase in circularity, and decrease in aspect ratio was associated with the morphological change from disk-like to a spherical shape. Data are mean ± SD (***p ≤ 0.001. n = at least 3 separate 3D-eCRC-PDX tissues). (C) Similar locational heterogeneity was observed in H&E stained 3D-eCRC-PDX tissues with elongated cells present primarily at the periphery and colony-forming cells present primarily at the core.
To examine the contraction of the 3D-eCRC-PDX tissues, which plays an important role in ECM remodeling, mechanical stiffness and cell phenotype, the size and dimensions of the 3D-eCRC-PDX tissues were investigated over 29 days in 2 separate batches. The 3D-eCRC-PDX tissue volume and diameter decreased significantly from Day 1 to Day 29 (Figure 5B). The volume of acellular control hydrogels increased (8.8±0.2%, p < 0.0001) from Day 1 to Day 8 and remained constant after Day 8 over 29 days of incubation at 37 °C and 5% CO2 (Suppl. Figure 11); this change in volume was a result of an increase in the acellular tissue thickness, whereas diameter remained constant. Thus, the 3D-eCRC-PDX tissue contraction and matrix remodeling were cell mediated. Furthermore, tissue circularity and tissue aspect ratio were examined over time to quantify changes in tissue morphology. As the volume and diameter decreased, the 3DeCRC-PDX tissue circularity increased and aspect ratio of the lateral surface (side view) decreased from Day 1 to Day 29, which was consistent with the change from the initial disk-like shape to a spherical shape; importantly, this result was consistent between batches (Figure 5B).
3D-eCRC-PDX Tissues Mimic the Mechanical Stiffness of CRC-PDX Tumors
To assess tissue mechanical stiffness, two CRC-PDX tumors and two paired batches of 3D-eCRC-PDX tissues were subjected to parallel plate compression testing. CRC-PDX tumor stiffness was found to vary significantly depending on the geometric region of the tumor from which the sample was taken. An increasing trend (p < 000.1) in stiffness was observed from the core to the periphery of the tumor for both tumors (Figure 6A). The core region of the CRC-PDX tumor was the least stiff (average of ~0.5 kPa), while the periphery region had the highest stiffness (average of ~2.8 kPa), with no significant difference between Tumor 1 and Tumor 2 for a given region.
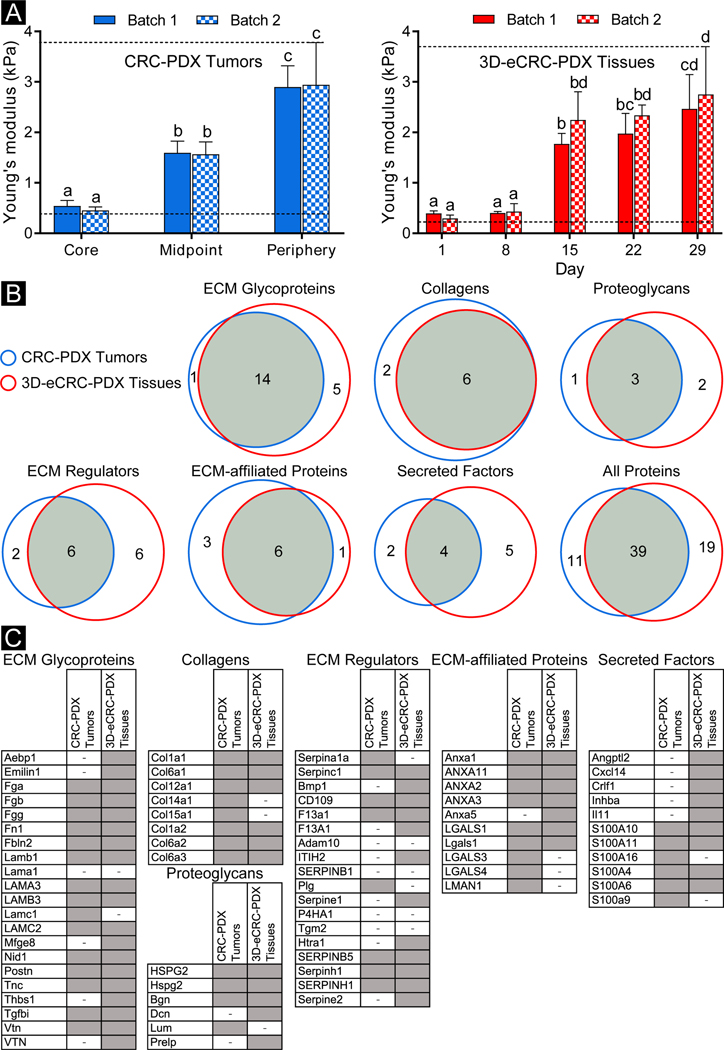
Figure 6. 3D-eCRC-PDX tissues and CRC-PDX tumors showed similar Young’s moduli and ECM proteins.
(A) CRC-PDX tumor stiffness differed regionally with the core of the tumor having the lowest stiffness and the periphery having the highest stiffness, based on parallel plate compression testing. For two CRC-PDX tumors no statistical difference was found between samples taken from the same tumor region (p ≤ 0.05, n = 3 CRC-PDX tumor pieces per region per tumor). The stiffness of 3D-eCRC-PDX tissues increased over 29 days of culture with a significant increase from Day 8 to Day 15. No significant difference was found between two separate batches of 3D-eCRC-PDX tissues (p ≤ 0.05, n = 3 3D-eCRC-PDX tissues per timepoint per batch). Interestingly, the range of stiffness of the 3D-eCRC-PDX tissues was within that of the CRC-PDX tumors. Bar =mean ± SD. Means that do not share a letter are significantly different. The dashed lines show the ranges of stiffness. (B, C) ECM proteins from CRC-PDX tumors and 3D-eCRC-PDX tissues were identified using proteomics; 39 proteins (approximately 57%) were observed in both the CRC-PDX tumors and the 3D-eCRC-PDX tissues. ECM glycoproteins and collagens showed the highest overlap among all subtypes, whereas ECM regulators and secreted factors contained the least common proteins. A protein was indicated as present (gray cell) when detected in at least two out of three samples and as absent (cell with a minus sign) when detected in only one sample or not detected. Proteins presented in uppercase and lowercase letters are from human and mouse databases, respectively.
The average stiffness of the 3D-eCRC-PDX tissues increased temporally from Day 1 to Day 29, where a stiffness of ~0.4 kPa was quantified on Day 1 and ~2.8 kPa on Day 29, with no significant difference between Batch 1 and Batch 2 for a given time point (Figure 6A). Most notably, the range of stiffness of the 3D-eCRC-PDX tissues was similar to that of the CRC-PDX tumors, while the time-dependent stiffness of the 3D-eCRC-PDX tissues mimicked the region-dependent stiffness of the CRC-PDX tumor. To determine if the PEG-Fb biomaterial contributed to the increase in stiffness, acellular hydrogels were created and maintained in similar conditions to the cell-laden tissues. In contrast to the mechanical stiffness of 3D-eCRC-PDX tissues, we found that the stiffness of acellular hydrogels decreased from Day 1 to Day 8, primarily due to swelling, and then remained constant from Day 8 to Day 29 (Suppl. Figure 12). Therefore, the observed temporal increase in 3D-eCRC-PDX tissue stiffness is consistent with a cell-mediated mechanism.
Proteomic Analysis of CRC-PDX Tumors and 3D-eCRC-PDX Tissues
To compare the ECM compositions of CRC-PDX tumors (obtained after 31–42 days of tumor growth) and 3D-eCRC-PDX tissues (obtained on the 22nd day of in vitro culture), proteomic analysis using ECM protein enrichment and mass spectroscopy characterization was performed on three separate batches of paired CRC-PDX tumors and 3D-eCRC-PDX tissues. Between all samples, 872 unique proteins were identified, 74 of which were identified as matrisome [64]. Correlation in ECM proteins observed in CRC-PDX tumors and paired 3D-eCRC-PDX tissues was first examined on a batch-by-batch basis. Approximately 61% of identified ECM proteins (38 out of 62) were observed in both the batch 1 CRC-PDX tumor and its paired 3D-eCRC-PDX tissues, while approximately 10% were only identified in the CRC-PDX tumor and 29% only identified in the 3D-eCRC-PDX tissues (Suppl. Figure 13A). Similarly, in batch 2 approximately 54% of identified proteins (38 out of 70) were observed in both the CRC-PDX tumor and the 3D-eCRC-PDX tissues, whereas in batch 3, approximately 47% of proteins were found in both CRC-PDX tumors and 3D-eCRC-PDX tissues (Suppl. Figure 13B, C). Overall, when comparing the pooled ECM proteins identified, there were 39 proteins (approximately 57%) observed in both the CRC-PDX tumors and the 3D-eCRC-PDX tissues, whereas 11 proteins (approximately 16%) were only identified in the CRC-PDX tumors and 19 proteins (approximately 27%) were only identified in the 3D-eCRC-PDX tissues (Fig 6B). In addition, we evaluated the similarity between CRC-PDX tumors; approximately 60% of ECM proteins within CRC-PDX tumors were the same for all three batches, 28% of the ECM proteins were present in at least two batches, and 12% of the ECM proteins were specific to each batch (Suppl. Figure 14A). Similarly, within 3D-eCRC-PDX tissues, 60% of ECM proteins were found to be the same among all batches, 25% were present in at least two batches, and 15% were batch-specific (Suppl. Figure 14B). Therefore, batch-to-batch consistency of the ECM proteins from the 3D-eCRC-PDX tissues was similar to that of the CRC-PDX tumors.
We further categorized the ECM proteins into ECM glycoproteins, collagens, proteoglycans, ECM regulators, ECM-affiliated proteins, and secreted factors. The number of ECM proteins per each subtype (e.g. ECM Glycoproteins) found in the CRC-PDX tumors and 3D-eCRC-PDX tissues are presented in Figure 6B and Suppl. Figure 14A, B. Identified ECM proteins for all CRC-PDX tumors and 3D-eCRC-PDX tissues from all batches are also reported by subtype in a heatmap (Figure 6C and Suppl. Figure 15). We found that ECM glycoproteins, collagens, proteoglycans, and ECM-affiliated proteins exhibited the most ECM proteins shared between CRC-PDX tumors and 3D-eCRC-PDX tissues. ECM glycoproteins and Collagens showed the highest overlap among all subtypes, whereas ECM regulators and secreted factors contained the least common proteins. In general, a higher total number of ECM proteins were identified in the 3D-eCRC-PDX tissues, thereby resulting in a higher number of proteins that were specific to the 3D-eCRC-PDX tissues.
Molecular characterization of CRC-PDX Tumors and 3D-eCRC-PDX Tissues
Transcriptome analysis using RNA seq was performed to undertake a molecular characterization of the CRC-PDX tumors and the 3D-eCRC-PDX tissues. To assess the CRC molecular subtype [70] we classified the CRC-PDX tumors and the 3D-eCRC-PDX tissues together with 478 primary CRC tumors from the Cancer Genome Atlas Colon Adenocarcinoma (TCGA-COAD) cohort [87]. Our classification of TCGA-COAD cohort tumors resulted in the majority of tumors to be classified as consensus molecular subtype (CMS) 2 and 4 which is consistent with previous classifications of CRC tumors [70, 88] (Figure 7A). We observed that both the CRC-PDX tumors and the 3D-eCRC-PDX tissues classified as CMS1. Thus, the process of engineering our CRC PDX tissue did not alter the CRC molecular subtype.
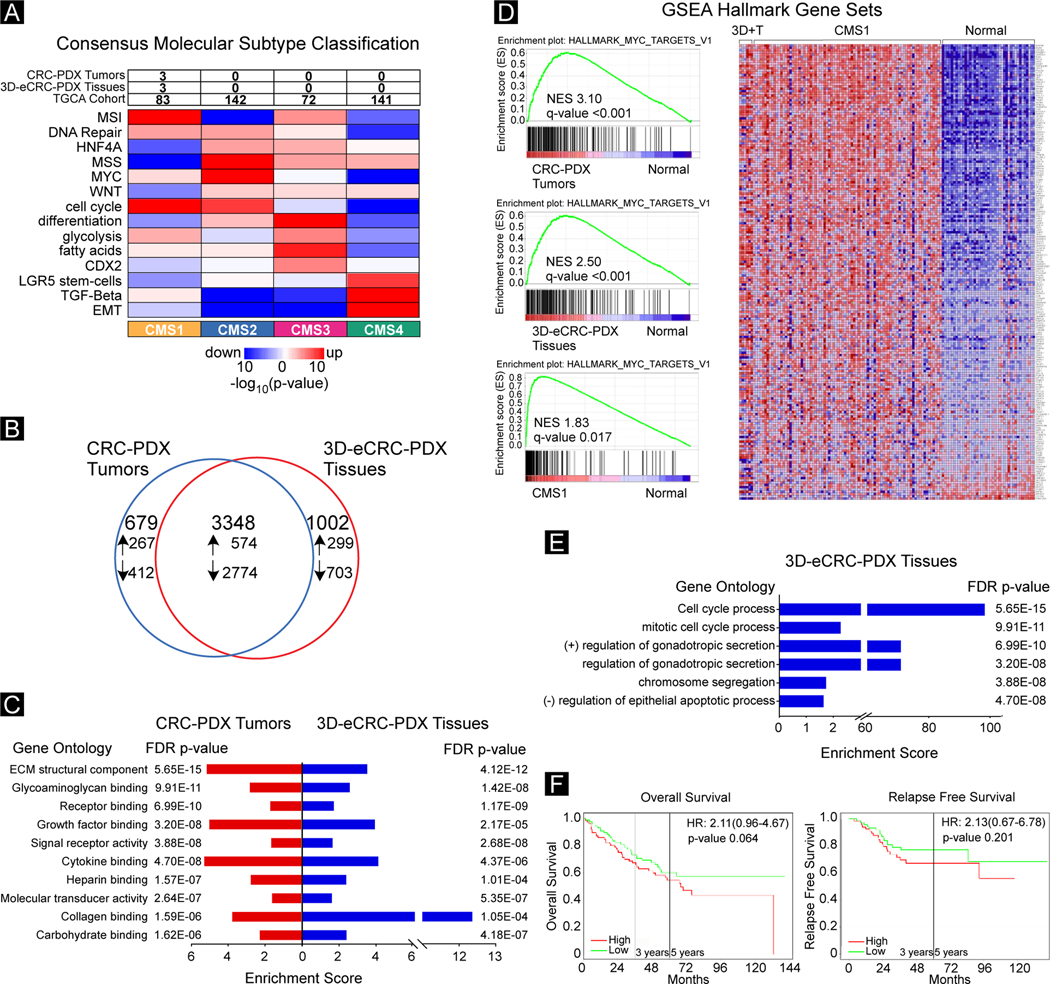
Figure 7. Transcriptomic characterization of 3D-eCRC-PDX tissues demonstrate that 3D-eCRC-PDX tissues recapitulate key molecular characteristics of CRC-PDX tumors.
(A) CRC Consensus Molecular Subtype Analysis. CRC-PDX tumor (n = 3) and 3D-eCRC-PDX tissue (n = 3) HT-Seq gene counts and TCGA-COAD counts were used to assess CRC consensus molecular subtype. (B) DEGs in CRC-PDX tumor and 3D-eCRC-PDX tissue compared to normal colon tissue (n = 41) from the TCGA-COAD. The number of DEGs that were downregulated (down arrow) or upregulated (up arrow) are shown. (C) Enriched gene molecular function ontologies between CRC-PDX tumors and 3D-eCRC-PDX tissues and normal colon tissue (n = 41) are shown. (D) GSEA Analysis. The top enriched Hallmarks gene sets in CRCPDX tumors (upper left panel), 3D-eCRC-PDX tissues (middle left panel) and patient CRC tumor TCGA-COAD samples (n = 84) (lower left panel) compared to normal colon tissue (n = 41). Normalized enrichment score (NES) and the FDR q-value are shown. Heatmap of CRCPDX tumors, 3D-eCRC-PDX tissues, and patient CRC tumors (n = 84) and normal colon tissue (right panel). (E) Gene ontology analysis of molecular functions enriched in DEGs between CRC-PDX tumors and 3D-eCRC-PDX tissues. (F) Prognostic Kaplan–Meier survival analysis using DEGs upregulated between CRC-PDX tumors and 3D-eCRC-PDX tissues in the precomputed GEO dataset GSE17536 (n = 87) adjusted for age, stage, and gender covariates and bifurcated based on median expression was examined for overall survival (left panel) and relapse free survival (right panel). The hazard ratios, 95% confidence intervals, and p values are shown.
To assess cancer-specific differentially expressed genes and gene ontology functions in the CRC-PDX tumors and the 3D-eCRC-PDX tissues, we determined differentially expressed genes (DEGs) in the CRC-PDX tumors and the 3D-eCRC-PDX tissues compared to tumor-adjacent normal colon tissue in the TGCA-COAD patient cohort. Approximately 66% of the DEGs were overlapping when the samples were compared (Figure 7B). ECM structural component (GO:000521) was the top significantly enriched gene ontology function term observed in both the CRC-PDX tumors (5.65E-15 FDR p value, 5.05 enrichment) and 3D-eCRC-PDX tissues (4.12E-12 FDR p value, 3.41 enrichment) (Figure 7C). Further, the top ten significantly (FDR p value < 0.05) enriched gene ontology function terms observed in the CRC-PDX tumors were also significantly enriched in the 3D-eCRC-PDX tissues. Because there is overlap between gene ontology terms, a REViGO analysis [75] was performed to remove redundant terms from the significantly enriched gene ontology function terms identified in the CRC-PDX tumors and the 3D-eCRC-PDX tissues. As shown in the TreeMaps in Suppl. Figure 16, enrichment in receptor binding, signaling receptor activity, extracellular matrix structural constituents were observed in both the CRC-PDX tumors and the 3D-eCRC-PDX tissues.
To gain cancer-related biological insight into gene expression in the CRC-PDX tumors and 3D-eCRC-PDX tissues, we performed gene set enrichment analysis (GSEA) using the Hallmark gene sets [89] from the Molecular Signatures Database (Broad Institute) [73]. The most enriched gene set (Myc targets v1, Normalized enrichment score 3.10, q-value < 0.001) in the CRC-PDX tumors (Figure 7D, upper left panel) was also the most enriched (Normalized enrichment score 2.50, q-value < 0.001) in the 3D-eCRC-PDX tissues (Figure 7D, middle left panel). We also observed that 8 of the top 10 Hallmark gene sets (Myc_Targets v1, E2F_Targets, Myc_Targets v2, DNA_Repair, Unfolded_Protein_Response, P53_Pathway, and MTORC1) significantly enriched in the CRC-PDX tumors were also significantly enriched in the 3D-eCRC-PDX tissues (Suppl. Figure 17, left and middle panels). To assess whether the Hallmark gene sets are also enriched in patient tumors, we next examined whether CMS1 classified primary tumors in the TCGA-COAD cohort were enriched for the 10 significantly enriched Hallmark gene sets in the CRC-PDX tumors and the 3D-eCRC-PDX tissues. The top enriched gene set in the CRC-PDX tumors and the 3D-eCRC-PDX tissues, Myc_Targets v1 gene set, was also significantly enriched (Normalized enrichment score 1.83, q-value < 0.017) in the CMS1 patient CRC tumors (Figure 7D, lower left panel). Indeed, 9 of the top 10 Hallmark gene sets significantly enriched in the CRC-PDX tumors were also significantly enriched in CMS1 patient CRC tumors (Suppl. Figure 17, right panel); only the P53_Pathway gene set was not significantly (q-value 0.174) enriched in the CMS1 patient CRC tumors (Suppl. Figure 17, right panel). To visualize the enrichment of the Myc targets v1 gene set in the CRC-PDX tumors, 3D-eCRC-PDX tissues, CMS1 patient CRC tumors, and tumor-adjacent normal colon tissue in the TGCA-COAD patient cohort, a heatmap was generated using the GSEA software. As shown in Figure 7D, right panel, tumor-adjacent normal colon tissue differentially expresses genes in the Myc targets v1 gene set compared to CRC-PDX tumors, 3D-eCRC-PDX tissues, and CMS1 patient CRC tumors.
To determine transcriptomic differences between the CRC-PDX tumors and the 3D-eCRC-PDX tissues, a differential expression analysis was performed. We observed 2193 DEGs (adj p value < 0.05, baseMean > 10, Log2-fold > 1.5). Gene ontology was performed to assess potential function differences between the CRC-PDX tumors and the 3D-eCRC-PDX tissues. The significantly (FDR p value < 0.05) regulated gene ontology function terms enriched in the 3D-eCRC-PDX tissues were cell cycle processes (Figure 7E). To examine the prognostic relevance of the top 16 upregulated DEGs in the 3D-eCRC-PDX tissues compared to the CRC-PDX tumors on overall survival and relapse free survival in CRC patients, we generated Kaplan-Meier plots [76]. In the CRC cohort GSE17536, there was not a significant difference in overall survival (p = 0.064) and relapse free survival (p = 0.201) in CRC patients with high expression of the 16 DEGs in the 3D-eCRC-PDX tissues (Figure 7F). We also did not observe significant differences in overall survival (p = 0.246) in the CRC cohort GSE17537 and the TGCA-COAD cohort (p = 0.697) and with relapse free survival (p = 0.112) in the CRC cohort GSE14333 (Suppl. Figure 18).
4. Discussion
In vitro 3D cancer tissues created using tissue engineering tools represent one of the new cancer disease models needed to advance our understanding of cancer biology. For CRC, 3D tissue-engineered platforms using biomaterials such as PEG-Fb [36], collagen I [38, 40, 41, 43], alginate [42], and decellularized matrices [39] have been used to study cell morphology, growth rate, and migration, cell-cell interactions, and drug testing. In each of these studies, however, standard cancer cell lines such as HCT 116, HT-29, LS 174T, and SW480 were used to create the in vitro disease model. While standard cancer cell lines provide valuable information in the field of cancer research, cell heterogeneity and molecular characteristics of standard cancer cell lines do not represent patient tumors [19, 20]. In contrast, PDX models are more advanced cancer models of the patient tumor which demonstrate high potential in clinical translatability due to the maintenance of key characteristics of the patient tumor [22] while still enabling assessment of reproducibility over time using the same PDX CRC line.
Here we developed a new cancer model for CRC combining the clinical translatability of PDX lines and the more high-throughput capacity of in vitro culture. We described the generation of the 3D-eCRC-PDX tissues as an in vitro tumor model and investigated the extent to which this in vitro model recapitulated the originating in vivo CRC-PDX tumors with a parallel comparison to 2D-CRC-PDX cultures. We found that, in contrast to 2D-CRC-PDX cultures, the 3D-eCRC-PDX tissues maintained the originating CRC-PDX tumor cell compositions during long-term culture, with the total cell numbers for each subpopulation increasing over time. Importantly, the 3D-eCRC-PDX tissues recapitulated key transcriptomic characteristics, ECM proteins, and mechanical stiffness of the CRC-PDX tumors.
Cell-cell and cell-matrix interactions can impact the cellular growth rate and thus, changes in distinct cell subpopulations may arise as the cells respond to their environment [28, 29, 90]. As PDXs are maintained and propagated in vivo, the resultant tumors are composed of both human and mouse cells. The inclusion of cancer and stromal PDX cell subpopulations is necessary for accurate recapitulation of the PDX tumor microenvironment. However, in the few studies that have reported the culture of PDX cells in vitro in a 3D microenvironment, the adherent cell subpopulation was selectively removed from the PDX cell population [46, 91]. None of these studies investigated whether the original PDX cell subpopulations could be maintained in a 3D engineered tissue model over time. Therefore, here we examined changes in distinct cell subpopulations using β−2 microglobulin (B2M) as a human cell marker, Ki-67 as a proliferative cell marker, and cytokeratin 20 (CK20) as a CRC cell marker. Importantly, we demonstrated that even as cell number increased during long-term culture, the 3D-eCRC-PDX tissues maintained the key cell subpopulations at ratios similar to the originating CRC-PDX tumor, whereas 2D-CRC-PDX cultures did not. Although there was an increasing trend in the human proliferative cell subpopulation in 2D-CRC-PDX cultures, the trend was not statistically significant. The changes in cell subpopulations of 2D-CRC-PDX cultures were consistent with the observations reported by Kodack et al. [29] as they attempted to establish cancer cell lines from multiple primary patient-derived tumors; in some cases, the stromal cells, primarily fibroblasts, outgrew the cancer cells, thereby hindering the tumor cell growth and resulting in the failure of cell line generation. Thus, the 3D-eCRC-PDX tissues has two advantages: 1) regulating the relative proliferation rates of human and mouse cells, and 2) regulating the relative percentage of mouse fibroblasts, in close comparison with CRC-PDX tumors.
Taken together, our results indicated that the 3D-eCRC-PDX tissues not only mimicked the cellular heterogeneity and cell population composition of the CRC-PDX tumors but were also able to maintain these cell populations in long-term in vitro culture. Notably, this maintenance of cell subpopulation ratios occurred simultaneously with temporal increases in cell numbers, an increase in the size of cell colonies, and an observed increasing presence of elongated cells over time within the 3D-eCRC-PDX tissues labeled with B2M+, CK20+, and phalloidin. Importantly, high reproducibility was observed between batches of the 3D-eCRCPDX tissues, which is important for downstream applications. Furthermore, consistent with flow cytometry results, gene expression analysis suggests that EMT, stem cell, and angiogenesis marker expression was more similar to the CRC-PDX tumors in the 3D-eCRC-PDX tissues than the 2D-CRC-PDX cells.
In addition, it must be noted that immunostaining revealed two distinct cell subpopulations in the in vitro cultures, colony-forming cells (B2M+) and elongated cells (B2M−). Since human stromal cells are replaced by mouse-derived cells in cancer xenografts [25, 78], our data suggest that all human cells (colony-forming cells) were CRC cells despite the fact that only a portion (~ 40%) of them positively expressed CK20. This result is consistent with immunohistochemical findings that 78% of stage II colon cancer patients had partial positivity for CK20 [92]. Importantly the CK20 protein is linked to metastasis and invasiveness, microsatellite instability, differentiation, and stage of CRC [82–85].
Transcriptomic analysis was performed to examine molecular similarities and differences between our CRC-PDX tumors and 3D-eCRC-PDX tissues. We first observed that the CRC-PDX tumors and 3D-eCRC-PDX tissues were classified as CMS1 tumors which is a molecular subtype associated with worse survival after relapse [70]. Even though we observed approximately 2000 DEGs between the CRC-PDX tumors and 3D-eCRC-PDX tissues, there was not a significant difference in prognostic Kaplan-Meier plots for overall survival and relapse free survival using the top 16 DEGs between the CRC-PDX tumors and 3D-eCRC-PDX tissues, suggesting that the top DEGs between the two models are not clinically relevant. Our observation of differential enrichment in the cell cycle molecular function between the CRCPDX tumors and 3D-eCRC-PDX tissues is consistent with differential growth between in vivo tumors and in vitro tissues [93].
To further assess transcriptomic similarities between our CRC-PDX tumors and 3D-eCRC-PDX tissues, we identified enriched gene ontology molecular functions and Hallmark gene sets in the CRC-PDX tumors and the 3D-eCRC-PDX tissues compared to tumor-adjacent normal colon tissue. We observed congruence in the enriched gene ontology molecular functions (top ten GO terms in CRC-PDX tumors were also significantly enriched in 3D-eCRC-PDX tissues) and Hallmark gene sets (eight of the top ten enriched gene sets in CRC-PDX tumors were also significantly enriched in 3D-eCRC-PDX tissues). Taken together, our molecular characterization results suggest that 3D-eCRC-PDX tissues transcriptionally resemble CRC-PDX tumors.
We next sought to identify and compare ECM proteins from both CRC-PDX tumors and 3D-eCRC-PDX tissues from three separate batches. ECM compositions of engineered CRC-PDX tissues have not been previously investigated. Patient CRC tumor ECM composition has been previously investigated; Naba et al. [94] demonstrated that there was consistency in ECM proteins between two samples from each patient as well as among three patients. Similar to our results, ECM regulators were the least common identified proteins between the two samples from each patient. In our model, the similarity of identified ECM proteins between CRC-PDX tumors and 3D-eCRC-PDX tissues, as well as the consistency among separately prepared batches, was relatively high; however, the number of ECM proteins identified in our in vitro and in vivo CRC-PDX tissues was lower than that identified in the patient CRC tissues by Naba et al. [94]. For CRC-PDX line employed here, patient tumor tissue for ECM analysis was not available; future work to examine the correlation between the in vitro model established here and the originating patient tumor ECM compositions is warranted.
Cancer progression leads to remodeling of the tumor microenvironment [95, 96]. A driver of microenvironmental remodeling is fibroblast-derived ECM components [97]; fibroblasts also interact with the surrounding ECM proteins and contract the tumor tissue which affects the mechanobiological properties of the tissue [95, 97, 98]. Consistent with prior findings [38, 98–104], we observed that the 3D-eCRC-PDX tissues displayed tissue contraction over time as the mouse stromal cells elongated. We additionally found that the 3D-eCRC-PDX tissue volume decreased temporally, which was not observed in acellular PEG-Fb hydrogels and is, therefore, not intrinsic to the biomaterial properties under our cell culture conditions. This observation suggests that the change in 3D-eCRC-PDX tissue morphology was a result of cell-matrix interactions. Fibroblast-induced contraction and remodeling of ECM, which play roles in matrix stiffening and cell signaling, division, and migration, have been widely investigated [52–55, 98–104]. Marquez et al. [98] demonstrated higher fibroblast density increased ECM contraction and stiffness in collagen I hydrogels. Consistent with the previously reported observation [98–104], our data suggest that the increase in stiffness of the 3D-eCRC-PDX tissue could be partially a result of the increasing number of elongated cells over time.
Tumor tissue stiffness impacts tumor cell proliferation, dormancy, migration and metastasis, and chemotherapeutic response, while simultaneously affecting cell-matrix interactions; thus, stiffness is widely recognized as an important factor for cancer cell behavior [14, 50, 52–55, 105, 106]. In CRC, tumor stiffness correlates to a higher stage, as well as metastatic phenotypes [107]. The bulk elastic modulus was 1.08–12.2 kPa (median = 3.49 kPa) for stage II CRC while the normal tissue presented a stiffness range of 0.37–7.33 kPa (median = 0.936 kPa). Our CRC-PDX tumors fell within the reported stiffness range for the stage II CRC patient tumors. Specifically, we found that CRC-PDX tumor stiffness increased from the core to the periphery, with a regional stiffness ranges of 0.4–0.6 kPa, 1.3–1.9 kPa, 2.4–3.4 kPa at the core, midpoint, and periphery regions, respectively. Interestingly, the time-dependent bulk stiffness of our 3D-eCRC-PDX tissues (ranging from 0.3–3.5 kPa) recapitulated the full range of regional stiffnesses observed in the CRC-PDX tumors.
Prior work has shown that biomaterial matrix composition, stiffness, and structure are important parameters in recapitulating the tumor microenvironment [108]; these parameters, both initially and over time as the PEG-Fb is replaced with cell-secreted ECM, are likely contributing to the maintenance of the originating tumor cell populations and the recapitulation of key molecular features in our study. We have previously reported that PEG-Fb facilitates the co-incorporation of heterogeneous cell types to form 3D engineered breast cancer tissues [50]. Our current findings suggest that PEG-Fb supported culture of heterogeneous cell types from a colon cancer PDX tumor. Tissue stiffness has been well documented to modulate cancer cell proliferation and phenotype, and metastatic properties in a cancer type specific manner [52–57, 109]. Therefore, stiffness of the 3D-eCRC-PDX tissues also may play a role in tumor cell population maintenance; systematic examination of this potential relationship between initial 3DeCRC-PDX tissue stiffness and cell population composition and molecular features is warranted. Finally, unlike physically crosslinked matrices (e.g. collagen and Matrigel, which can rapidly compact as a result of forces exerted by encapsulated cell elongation and migration [110] ), PEG-Fb, as a covalently crosslinked material, maintains its structure until it is enzymatically degraded by the cells. This longer timeframe of structural integrity might also be contributing to originating tumor cell population maintenance in our study. Further studies would be required to determine whether other hydrogel matrices also support maintenance of heterogeneous colon cancer PDX cell types. Consistent with our current findings, it has been observed that stromal cells, primarily fibroblasts, can outgrow cancer cells in 2D culture of primary patient-derived tumors [29].
Although we demonstrated the ability of CRC PDX cells to be maintained in long-term in vitro culture using tissue engineering tools, the current study had limitations. Our efforts in this study were focused on the engineered CRC-PDX tissue model development, in-depth characterization and comparison between in vitro tissues and in vivo tumors, and batch-to-batch reproducibility. Future work in our laboratory will investigate the applicability of this in vitro 3D model for other CRC PDX lines from multiple patients. Whereas this study employed patient-derived cells, the patient tumor tissue used to initiate the PDX CRC line employed was not available for direct comparison with the in vivo tumors or in vitro tissues. Furthermore, we did not include human stromal cells in our in vitro model as these cells are replaced by mouse stromal cells in PDX tumors. Whereas directly using patient tumor cells to create this engineered CRC tissue model and including patient-specific tumor stromal cells such as cancer-associated fibroblasts and immune cells could potentially result in a better recapitulation of the patient tumor, this approach only allows cells to be obtained once from each patient; given the high level of heterogeneity between patient tumors, using patient tumor cells directly, rather than PDX lines, would not have provided the ability to directly assess and refine model reproducibility. To test the clinical translatability of our model in predicting individual patient drug response, further studies are needed. Furthermore, the use of this in vitro 3D model could be extended to examine the potential to support PDX or patient cells from other types of cancer.
5. Conclusion
In this study, we have developed a new in vitro 3D engineered tissue model (3D-eCRC-PDX tissues) using cells isolated from CRC-PDX tumors. During long-term culture, the 3D-eCRC-PDX tissues maintained the cell populations observed in the originating CRC-PDX tumor, whereas in 2D-CRC-PDX cultures, mouse stromal cells grew at a much faster rate and overtook the human CRC cells, thereby leading to cell population compositions that deviated from the CRC-PDX tumor. The cells within the 3D-eCRC-PDX tissues demonstrated locational heterogeneity in terms of cell subpopulations; elongated mouse cells were primarily present on the surface of the 3D-eCRC-PDX tissues and human colony-forming CRC cells were present throughout the 3D-eCRC-PDX tissues. Furthermore, cellular morphology within the 3D-eCRCPDX tissues was similar to that observed in CRC-PDX tumors. CRC-PDX tumor stiffness varied regionally; 3D-eCRC-PDX tissues increased in stiffness over time in culture, providing recapitulation of the full CRC-PDX tumor stiffness range. Moreover, results from our transcriptomic analysis of CRC-PDX tumors and 3D-eCRC-PDX tissues indicate similarity in enriched gene ontology molecular functions and Hallmark gene sets. Importantly, prognostic Kaplan-Meier plots for overall survival and relapse free survival did not reveal significant differences between CRC-PDX tumors and 3D-eCRC-PDX tissues.
Culturing CRC-PDX tumor-derived cells in vitro through the use of our 3D-eCRC-PDX tissues was found to be mutually beneficial to (1) the advancement of in vitro CRC-PDX tumor-derived cell culture and (2) the capacity of 3D engineered cancer tissue models to recapitulate patient tumors. Introducing the ability to culture PDX cells long-term (29 days) in an in vitro biomimetic environment that preserves native cell behavior overcoming 2D culture and in vivo testing limitations, whereas implementing a more pathophysiologically relevant cell source improves 3D engineered cancer tissue model recapitulation of patient tumors, thus overcoming limitations imposed by the use of cancer cell lines. The cell encapsulation and subsequent tissue formation were robust as no significant differences were observed between multiple batches of 3D-eCRC-PDX tissues prepared from separate CRC-PDX tumors. In conclusion, our novel in vitro CRC model exhibits high potential for the examination of CRC disease progression and tumorigenic mechanisms to mitigate the impact of CRC.
Supplementary Material
Acknowledgments
The authors thank Michelle Aono for kindly performing tissue sectioning and H&E staining, Allison Church Bird from the Flow Cytometry Facility (Auburn University, Auburn, AL, USA) for providing constructive guidance for cell labeling, Dr. Mahmoud Mansour’s laboratory in the Department of Anatomy, Physiology and Pharmacology (Auburn University, Auburn, AL, USA) for facilitating the use of the confocal microscope, and Dr. C. Ross Ethier’s laboratory in the Wallace H. Coulter Department of Biomedical Engineering (Georgia Institute of Technology, Atlanta, GA, USA) for providing access to the MicroSquisher apparatus. We also thank Dr. Sara Cooper (HudsonAlpha Institute for Biotechnology) for assistance with the RNA seq analysis.
The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Funding Sources
The authors acknowledge financial support from the Auburn University Research Initiative in Cancer (AURIC) Seed Grant Program (M.W.G., E.A.L.), AURIC Graduate Fellowships (I.H., N.L.H.), the National Center for Advancing Translational Research of the National Institutes of Health (NIH) (UL1TR003096–01(I.H., B. Anbiah, M.W.G., E.A.L.)), the United States Department of Agriculture, National Institute of Food and Agriculture (NIFA) Hatch Grant (ALA044–1-18037 (M.W.G.)), the UAB Comprehensive Cancer Center, NIH-National Cancer Institute (NCI) (P30CA013148, J.A.M.), and The UAB Institutional Core Funding Mechanism (J.A.M.).
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
The RNA seq data set (GSE151069) is available from the NCBI Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo). Proteomics data will be uploaded to the PRIDE - Proteomics Identification Database - EMBL-EBI upon acceptance of the manuscript. Flow cytometry data are available in the Supplementary Materials. All other data are available from the corresponding authors upon reasonable request.
References
- [1].American Cancer Society 2019 Cancer Facts & Figures 2019. Atlanta: American Cancer Society; ) [Google Scholar]
- [2].SEER Cancer Statistics Review, 1975–2013, National Cancer Institute. Bethesda, MD, https://seer.cancer.gov/archive/csr/1975_2013/, based on November 2015 SEER data submission, posted to the SEER web site, April 2016., ed N Howlader, et alet al. [Google Scholar]
- [3].Risk Lifetime (Percent) of Being Diagnosed with Cancer by Site and Race/Ethnicity Males, 18 SEER Areas, 2011-2013. (Table 1.16).
- [4].Risk Lifetime (Percent) of Being Diagnosed with Cancer by Site and Race/Ethnicity Females, 18 SEER Areas, 2011-2013. (Table 1.17).
- [5].Libutti SK, Saltz LB, Willett CG and Levine R 2015. DeVita, Hellman, and Rosenberg’s Cancer: Principles & Practice of Oncology: Tenth Edition, ed DeVita VT, et al. : Wolters Kluwer Health Adis (ESP)) [Google Scholar]
- [6].Schaeybroeck SV, Lawler M, Johnston B, Salto-Tellez M, Lee J, Loughlin P, Wilson R and Johnston PG 2014. Abeloff’s Clinical Oncology (Fifth Edition), ed Niederhuber JE, et al. (Philadelphia: Elsevier) pp 1278–335.e14 [Google Scholar]
- [7].Mazzoleni G, Di Lorenzo D and Steimberg N 2009. Modelling tissues in 3D: the next future of pharmaco-toxicology and food research? Genes Nutr 4 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Prasetyanti PR and Medema JP 2017. Intra-tumor heterogeneity from a cancer stem cell perspective Mol Cancer 16 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Easwaran H, Tsai HC and Baylin SB 2014. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance Mol Cell 54 716–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nagle RB 2004. Role of the extracellular matrix in prostate carcinogenesis J Cell Biochem 9136–40 [DOI] [PubMed] [Google Scholar]
- [11].Quail DF and Joyce JA 2013. Microenvironmental regulation of tumor progression and metastasis Nat Med 19 1423–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nguyen DX, Bos PD and Massague J 2009. Metastasis: from dissemination to organ-specific colonization Nat Rev Cancer 9274–84 [DOI] [PubMed] [Google Scholar]
- [13].Kraning-Rush CM, Carey SP, Lampi MC and Reinhart-King CA 2013. Microfabricated collagen tracks facilitate single cell metastatic invasion in 3D Integr Biol (Camb) 5 606–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kraning-Rush CM and Reinhart-King CA 2012. Controlling matrix stiffness and topography for the study of tumor cell migration Cell Adh Migr 6 274–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, Martin SS and Konstantopoulos K 2012. Physical confinement alters tumor cell adhesion and migration phenotypes FASEB J 26 4045–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Paul CD, Mistriotis P and Konstantopoulos K 2017. Cancer cell motility: lessons from migration in confined spaces Nat Rev Cancer 17 131–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim DH, Ewald AJ, Park J, Kshitiz, Kwak M, Gray RS, Su CY, Seo J, An SS and Levchenko A 2018. Biomechanical interplay between anisotropic re-organization of cells and the surrounding matrix underlies transition to invasive cancer spread Sci Rep 8 14210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Park J, Kim DH, Kim HN, Wang CJ, Kwak MK, Hur E, Suh KY, An SS and Levchenko A 2016. Directed migration of cancer cells guided by the graded texture of the underlying matrix Nat Mater 15792–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cree IA, Glaysher S and Harvey AL 2010. Efficacy of anti-cancer agents in cell lines versus human primary tumour tissue Curr Opin Pharmacol 10 375–9 [DOI] [PubMed] [Google Scholar]
- [20].Fernando A, Glaysher S, Conroy M, Pekalski M, Smith J, Knight LA, Di Nicolantonio F and Cree IA 2006. Effect of culture conditions on the chemosensitivity of ovarian cancer cell lines Anticancer Drugs 17 913–9 [DOI] [PubMed] [Google Scholar]
- [21].Chou J, Fitzgibbon MP, Mortales CL, Towlerton AM, Upton MP, Yeung RS, McIntosh MW and Warren EH 2013. Phenotypic and transcriptional fidelity of patient-derived colon cancer xenografts in immune-deficient mice PLoS One 8 e79874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Brown KM, Xue A, Mittal A, Samra JS, Smith R and Hugh TJ 2016. Patient-derived xenograft models of colorectal cancer in pre-clinical research: a systematic review Oncotarget 7 66212–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee WS, Kim HY, Seok JY, Jang HH, Park YH, Kim SY, Shin DB and Hong S 2014. Genomic profiling of patient-derived colon cancer xenograft models Medicine (Baltimore) 93 e298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cho YB, Hong HK, Choi YL, Oh E, Joo KM, Jin J, Nam DH, Ko YH and Lee WY 2014. Colorectal cancer patient-derived xenografted tumors maintain characteristic features of the original tumors J Surg Res 187 502–9 [DOI] [PubMed] [Google Scholar]
- [25].Day CP, Merlino G and Van Dyke T 2015. Preclinical mouse cancer models: a maze of opportunities and challenges Cell 163 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].McIntyre RE, Buczacki SJ, Arends MJ and Adams DJ 2015. Mouse models of colorectal cancer as preclinical models Bioessays 37 909–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Murayama T and Gotoh N 2019. Patient-Derived Xenograft Models of Breast Cancer and Their Application Cells 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Okada S, Vaeteewoottacharn K and Kariya R 2019. Application of Highly Immunocompromised Mice for the Establishment of Patient-Derived Xenograft (PDX) Models Cells 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kodack DP, Farago AF, Dastur A, Held MA, Dardaei L, Friboulet L, von Flotow F, Damon LJ, Lee D, Parks M, Dicecca R, Greenberg M, Kattermann KE, Riley AK, Fintelmann FJ, Rizzo C, Piotrowska Z, Shaw AT, Gainor JF, Sequist LV, Niederst MJ, Engelman JA and Benes CH 2017. Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care Cell Rep 21 3298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kondo J, Ekawa T, Endo H, Yamazaki K, Tanaka N, Kukita Y, Okuyama H, Okami J, Imamura F, Ohue M, Kato K, Nomura T, Kohara A, Mori S, Dan S and Inoue M 2019. High-throughput screening in colorectal cancer tissue-originated spheroids Cancer Sci 110345–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Boehnke K, Iversen PW, Schumacher D, Lallena MJ, Haro R, Amat J, Haybaeck J, Liebs S, Lange M, Schafer R, Regenbrecht CR, Reinhard C and Velasco JA 2016. Assay Establishment and Validation of a High-Throughput Screening Platform for Three-Dimensional Patient-Derived Colon Cancer Organoid Cultures J Biomol Screen 21 931–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Verissimo CS, Overmeer RM, Ponsioen B, Drost J, Mertens S, Verlaan-Klink I, Gerwen BV, van der Ven M, Wetering MV, Egan DA, Bernards R, Clevers H, Bos JL and Snippert HJ 2016. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cox MC, Reese LM, Bickford LR and Verbridge SS 2015. Toward the Broad Adoption of 3D Tumor Models in the Cancer Drug Pipeline ACS Biomaterials Science & Engineering 1 877–94 [DOI] [PubMed] [Google Scholar]
- [34].Gu L and Mooney DJ 2016. Biomaterials and emerging anticancer therapeutics: engineering the microenvironment Nat Rev Cancer 16 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Carvalho MR, Lima D, Reis RL, Correlo VM and Oliveira JM 2015. Evaluating Biomaterial- and Microfluidic-Based 3D Tumor Models Trends Biotechnol 33 667–78 [DOI] [PubMed] [Google Scholar]
- [36].Pradhan S, Clary JM, Seliktar D and Lipke EA 2017. A three-dimensional spheroidal cancer model based on PEG-fibrinogen hydrogel microspheres Biomaterials 115 141–54 [DOI] [PubMed] [Google Scholar]
- [37].Rios de la Rosa JM, Wubetu J, Tirelli N and Tirella A 2018. Colorectal tumor 3D in vitro models: advantages of biofabrication for the recapitulation of early stages of tumour development Biomedical Physics & Engineering Express 4 [DOI] [PubMed] [Google Scholar]
- [38].Dolznig H, Rupp C, Puri C, Haslinger C, Schweifer N, Wieser E, Kerjaschki D and Garin-Chesa P 2011. Modeling colon adenocarcinomas in vitro a 3D co-culture system induces cancer-relevant pathways upon tumor cell and stromal fibroblast interaction Am J Pathol 179 487–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hoshiba T and Tanaka M 2016 Decellularized matrices as in vitro models of extracellular matrix in tumor tissues at different malignant levels: Mechanism of 5-fluorouracil resistance in colorectal tumor cells Biochim Biophys Acta 1863. 2749–57 [DOI] [PubMed] [Google Scholar]
- [40].Magdeldin T, Lopez-Davila V, Villemant C, Cameron G, Drake R, Cheema U and Loizidou M 2014. The efficacy of cetuximab in a tissue-engineered three-dimensional in vitro model of colorectal cancer J Tissue Eng 5 2041731414544183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dainiak MB, Savina IN, Musolino I, Kumar A, Mattiasson B and Galaev IY 2008. Biomimetic macroporous hydrogel scaffolds in a high-throughput screening format for cell-based assays Biotechnol Prog 24 1373–83 [DOI] [PubMed] [Google Scholar]
- [42].Shakibaei M, Kraehe P, Popper B, Shayan P, Goel A and Buhrmann C 2015. Curcumin potentiates antitumor activity of 5-fluorouracil in a 3D alginate tumor microenvironment of colorectal cancer BMC Cancer 15 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nyga A, Loizidou M, Emberton M and Cheema U 2013. A novel tissue engineered three-dimensional in vitro colorectal cancer model Acta Biomater 9 7917–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Luo X, Fong ELS, Zhu C, Lin QXX, Xiong M, Li A, Li T, Benoukraf T, Yu H and Liu S 2020. Hydrogel-based colorectal cancer organoid co-culture models Acta Biomater [DOI] [PubMed] [Google Scholar]
- [45].Ng S, Tan WJ, Pek MMX, Tan MH and Kurisawa M 2019. Mechanically and chemically defined hydrogel matrices for patient-derived colorectal tumor organoid culture Biomaterials 219 119400 [DOI] [PubMed] [Google Scholar]
- [46].Fong EL, Wan X, Yang J, Morgado M, Mikos AG, Harrington DA, Navone NM and Farach-Carson MC 2016. A 3D in vitro model of patient-derived prostate cancer xenograft for controlled interrogation of in vivo tumor-stromal interactions Biomaterials 77 164–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pedron S, Wolter GL, Chen JE, Laken SE, Sarkaria JN and Harley BAC 2019. Hyaluronic acid-functionalized gelatin hydrogels reveal extracellular matrix signals temper the efficacy of erlotinib against patient-derived glioblastoma specimens Biomaterials 219119371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pedron S, Hanselman JS, Schroeder MA, Sarkaria JN and Harley BAC 2017. Extracellular Hyaluronic Acid Influences the Efficacy of EGFR Tyrosine Kinase Inhibitors in a Biomaterial Model of Glioblastoma Adv Healthc Mater 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pedron S, Polishetty H, Pritchard AM, Mahadik BP, Sarkaria JN and Harley BAC 2017. Spatially graded hydrogels for preclinical testing of glioblastoma anticancer therapeutics MRS Commun 7 442–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pradhan S, Hassani I, Seeto WJ and Lipke EA 2017. PEG-fibrinogen hydrogels for three-dimensional breast cancer cell culture J Biomed Mater Res A 105 236–52 [DOI] [PubMed] [Google Scholar]
- [51].Pradhan S, Smith AM, Garson CJ, Hassani I, Seeto WJ, Pant K, Arnold RD, Prabhakarpandian B and Lipke EA 2018. A Microvascularized Tumor-mimetic Platform for Assessing Anti-cancer Drug Efficacy Sci Rep 8 3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, Walsh S, Benten D, Forbes SJ, Wells RG and Iredale JP 2011. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells Hepatology 53 1192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Park JS, Chu JS, Tsou AD, Diop R, Tang Z, Wang A and Li S 2011. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-beta Biomaterials 32 3921–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Swaminathan V, Mythreye K, O’Brien ET, Berchuck A, Blobe GC and Superfine R 2011. Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines Cancer Res 71 5075–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zaman MH, Trapani LM, Sieminski AL, Mackellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA and Matsudaira P 2006. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis Proc Natl Acad Sci U S A 103 10889–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jabbari E, Sarvestani SK, Daneshian L and Moeinzadeh S 2015. Optimum 3D Matrix Stiffness for Maintenance of Cancer Stem Cells Is Dependent on Tissue Origin of Cancer Cells PLoS One 10 e0132377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yang X, Sarvestani SK, Moeinzadeh S, He X and Jabbari E 2013. Three-dimensional-engineered matrix to study cancer stem cells and tumorsphere formation: effect of matrix modulus Tissue Eng Part A 19 669–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Almany L and Seliktar D 2005. Biosynthetic hydrogel scaffolds made from fibrinogen and polyethylene glycol for 3D cell cultures Biomaterials 26 2467–77 [DOI] [PubMed] [Google Scholar]
- [59].Dikovsky D, Bianco-Peled H and Seliktar D 2006. The effect of structural alterations of PEG-fibrinogen hydrogel scaffolds on 3-D cellular morphology and cellular migration Biomaterials 27 1496–506 [DOI] [PubMed] [Google Scholar]
- [60].Maykel J, Liu JH, Li H, Shultz LD, Greiner DL and Houghton J 2014. NOD-scidIl2rg (tm1Wjl) and NOD-Rag1 (null) Il2rg (tm1Wjl) : a model for stromal cell-tumor cell interaction for human colon cancer Dig Dis Sci 59 1169–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Naba A, Clauser KR, Hoersch S, Liu H, Carr SA and Hynes RO 2012. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices Mol Cell Proteomics 11 M111 014647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Naba A, Clauser KR and Hynes RO 2015. Enrichment of Extracellular Matrix Proteins from Tissues and Digestion into Peptides for Mass Spectrometry Analysis J Vis Exp e53057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ludwig MR, Kojima K, Bowersock GJ, Chen D, Jhala NC, Buchsbaum DJ, Grizzle WE, Klug CA and Mobley JA 2016. Surveying the serologic proteome in a tissue-specific kras(G12D) knockin mouse model of pancreatic cancer Proteomics 16 516–31 [DOI] [PubMed] [Google Scholar]
- [64].Naba A, Pearce OMT, Del Rosario A, Ma D, Ding H, Rajeeve V, Cutillas PR, Balkwill FR and Hynes RO 2017. Characterization of the Extracellular Matrix of Normal and Diseased Tissues Using Proteomics J Proteome Res 16 3083–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR 2013. STAR: ultrafast universal RNA-seq aligner Bioinformatics 29 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Alonso A, Lasseigne BN, Williams K, Nielsen J, Ramaker RC, Hardigan AA, Johnston B, Roberts BS, Cooper SJ and Marsal S 2017. aRNApipe: a balanced, efficient and distributed pipeline for processing RNA-seq data in high-performance computing environments Bioinformatics 33 1727–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Love MI, Huber W and Anders S 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 Genome biology 15 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Eide PW, Bruun J, Lothe RA and Sveen A 2017. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models Scientific reports 7 16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hoshida Y 2010. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment PloS one 5 e15543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Guinney J, Dienstmann R, Wang X, De Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G and Angelino P 2015. The consensus molecular subtypes of colorectal cancer Nature medicine 21 1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM and Castiglioni I 2015. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data Nucleic acids research 44 e71-e [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Peng L, Bian XW, Li DK, Xu C, Wang GM, Xia QY and Xiong Q 2015. Large-scale RNA-Seq Transcriptome Analysis of 4043 Cancers and 548 Normal Tissue Controls across 12 TCGA Cancer Types Scientific Reports 5 13413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES and Mesirov JP 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles Proceedings of the National Academy of Sciences of the United States of America 10215545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Eden E, Navon R, Steinfeld I, Lipson D and Yakhini Z 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists BMC bioinformatics 10 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Supek F, Bošnjak M, Škunca N and Šmuc T 2011. REVIGO summarizes and visualizes long lists of gene ontology terms PloS one 6 e21800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Goswami CP and Nakshatri H 2013. PROGgene: gene expression based survival analysis web application for multiple cancers Journal of clinical bioinformatics 3 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Goswami CP and Nakshatri H 2014. PROGgeneV2: enhancements on the existing database BMC cancer 14 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Petit V, Massonnet G, Maciorowski Z, Touhami J, Thuleau A, Nemati F, Laval J, Chateau-Joubert S, Servely JL, Vallerand D, Fontaine JJ, Taylor N, Battini JL, Sitbon M and Decaudin D 2013. Optimization of tumor xenograft dissociation for the profiling of cell surface markers and nutrient transporters Lab Invest 93 611–21 [DOI] [PubMed] [Google Scholar]
- [79].Truong D, Puleo J, Llave A, Mouneimne G, Kamm RD and Nikkhah M 2016. Breast Cancer Cell Invasion into a Three Dimensional Tumor-Stroma Microenvironment Sci Rep 6 34094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Truong DD, Kratz A, Park JG, Barrientos ES, Saini H, Nguyen T, Pockaj B, Mouneimne G, LaBaer J and Nikkhah M 2019. A Human Organotypic Microfluidic Tumor Model Permits Investigation of the Interplay between Patient-Derived Fibroblasts and Breast Cancer Cells Cancer Res 79 3139–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lyons SM, Alizadeh E, Mannheimer J, Schuamberg K, Castle J, Schroder B, Turk P, Thamm D and Prasad A 2016. Changes in cell shape are correlated with metastatic potential in murine and human osteosarcomas Biol Open 5 289–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kim JH, Rhee YY, Bae JM, Cho NY and Kang GH 2013. Loss of CDX2/CK20 expression is associated with poorly differentiated carcinoma, the CpG island methylator phenotype, and adverse prognosis in microsatellite-unstable colorectal cancer Am J Surg Pathol 37 1532–41 [DOI] [PubMed] [Google Scholar]
- [83].Imai Y, Yamagishi H, Fukuda K, Okamura T, Ono Y, Ban S, Inoue T and Ueda Y 2014. Expression of cytokeratin 20 indicates invasive histological phenotype in poorly differentiated colorectal adenocarcinoma Anticancer Res 34 159–67 [PubMed] [Google Scholar]
- [84].Shin JH, Bae JH, Lee A, Jung CK, Yim HW, Park JS and Lee KY 2010. CK7, CK20, CDX2 and MUC2 Immunohistochemical staining used to distinguish metastatic colorectal carcinoma involving ovary from primary ovarian mucinous adenocarcinoma Jpn J Clin Oncol 40 208–13 [DOI] [PubMed] [Google Scholar]
- [85].Toyoshima M, Momono Y, Makino H, Kudo T, Oka N, Sakurada J, Suzuki H, Kodama H and Yoshinaga K 2016. Cytokeratin 7-positive/cytokeratin 20-negative cecal adenocarcinoma metastatic to the uterine cervix: a case report World J Surg Oncol 14 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bayrak R, Haltas H and Yenidunya S 2012. The value of CDX2 and cytokeratins 7 and 20 expression in differentiating colorectal adenocarcinomas from extraintestinal gastrointestinal adenocarcinomas: cytokeratin 7-/20+ phenotype is more specific than CDX2 antibody Diagn Pathol 7 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Muzny DM et al. 2012. Comprehensive molecular characterization of human colon and rectal cancer Nature 487 330–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Prasetyanti PR, van Hooff S R, van Herwaarden T, de Vries N, Kalloe K, Rodermond H, van Leersum R, de Jong J H, Franitza M and Nürnberg P 2019. Capturing colorectal cancer inter‐tumor heterogeneity in patient‐derived xenograft (PDX) models International journal of cancer 144 366–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov Jill P and Tamayo P 2015. The Molecular Signatures Database Hallmark Gene Set Collection Cell Systems 1 417–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Riedl A, Schlederer M, Pudelko K, Stadler M, Walter S, Unterleuthner D, Unger C, Kramer N, Hengstschlager M, Kenner L, Pfeiffer D, Krupitza G and Dolznig H 2017. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses J Cell Sci 130 203–18 [DOI] [PubMed] [Google Scholar]
- [91].Fong EL, Martinez M, Yang J, Mikos AG, Navone NM, Harrington DA and Farach-Carson MC 2014. Hydrogel-based 3D model of patient-derived prostate xenograft tumors suitable for drug screening Mol Pharm 11 2040–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Vaz MA, Martinez JC, Devesa JM, Trill JD, Abraira V, Riquelme A and Carrato A 2014. Prognostic value of stem cell quantification in stage II colon cancer PLoS One 9 e88480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sachs N and Clevers H 2014. Organoid cultures for the analysis of cancer phenotypes Curr Opin Genet Dev 24 68–73 [DOI] [PubMed] [Google Scholar]
- [94].Naba A, Clauser KR, Whittaker CA, Carr SA, Tanabe KK and Hynes RO 2014. Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver BMC Cancer 14 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Yuan Y, Jiang Y-C, Sun C-K and Chen Q-M 2016. Role of the tumor microenvironment in tumor progression and the clinical applications (Review) Oncology Reports 35 2499–515 [DOI] [PubMed] [Google Scholar]
- [96].Bager CL, Willumsen N, Kehlet SN, Hansen HB, Bay-Jensen AC, Leeming DJ, Dragsbaek K, Neergaard JS, Christiansen C, Hogdall E and Karsdal M 2016. Remodeling of the Tumor Microenvironment Predicts Increased Risk of Cancer in Postmenopausal Women: The Prospective Epidemiologic Risk Factor (PERF I) Study Cancer Epidemiology Biomarkers & Prevention 25 1348–55 [DOI] [PubMed] [Google Scholar]
- [97].Alkasalias T, Moyano-Galceran L, Arsenian-Henriksson M and Lehti K 2018. Fibroblasts in the Tumor Microenvironment: Shield or Spear? International Journal of Molecular Sciences 19 1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Marquez JP, Genin GM, Pryse KM and Elson EL 2006. Cellular and matrix contributions to tissue construct stiffness increase with cellular concentration Ann Biomed Eng 34 1475–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Jin T, Li L, Siow RC and Liu KK 2015. A novel collagen gel-based measurement technique for quantitation of cell contraction force J R Soc Interface 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Bell E, Ivarsson B and Merrill C 1979. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro Proc Natl Acad Sci U S A 76 1274–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Godbout C, Follonier Castella L, Smith EA, Talele N, Chow ML, Garonna A and Hinz B 2013. The mechanical environment modulates intracellular calcium oscillation activities of myofibroblasts PLoS One 8 e64560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Marquez JP, Elson EL and Genin GM 2010. Whole cell mechanics of contractile fibroblasts: relations between effective cellular and extracellular matrix moduli Philos Trans A Math Phys Eng Sci 368 635–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Jansen KA, Bacabac RG, Piechocka IK and Koenderink GH 2013. Cells actively stiffen fibrin networks by generating contractile stress Biophys J 105 2240–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Hinz B, Celetta G, Tomasek JJ, Gabbiani G and Chaponnier C 2001. Alpha-smooth muscle actin expression upregulates fibroblast contractile activity Mol Biol Cell 12 273–041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chen JE, Pedron S and Harley BAC 2017. The Combined Influence of Hydrogel Stiffness and Matrix-Bound Hyaluronic Acid Content on Glioblastoma Invasion Macromol Biosci 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Holt SE, Ward ES, Ober RJ and Alge DL 2017. Shooting for the moon: using tissuemimetic hydrogels to gain new insight on cancer biology and screen therapeutics MRS Communications 7 427–41 [Google Scholar]
- [107].Kawano S, Kojima M, Higuchi Y, Sugimoto M, Ikeda K, Sakuyama N, Takahashi S, Hayashi R, Ochiai A and Saito N 2015. Assessment of elasticity of colorectal cancer tissue, clinical utility, pathological and phenotypical relevance Cancer Sci 106 1232–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Pradhan S, Hassani I, Clary JM and Lipke EA 2016. Polymeric Biomaterials for In Vitro Cancer Tissue Engineering and Drug Testing Applications Tissue Eng Part B Rev 22 470–84 [DOI] [PubMed] [Google Scholar]
- [109].Wang C, Sinha S, Jiang X, Murphy L, Fitch S, Wilson C, Grant G and Yang F 2021. Matrix Stiffness Modulates Patient-Derived Glioblastoma Cell Fates in Three-Dimensional Hydrogels Tissue Eng Part A 27 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Fernandez P and Bausch AR 2009. The compaction of gels by cells: a case of collective mechanical activity Integr Biol (Camb) 1 252-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA seq data set (GSE151069) is available from the NCBI Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo). Proteomics data will be uploaded to the PRIDE - Proteomics Identification Database - EMBL-EBI upon acceptance of the manuscript. Flow cytometry data are available in the Supplementary Materials. All other data are available from the corresponding authors upon reasonable request.