Summary
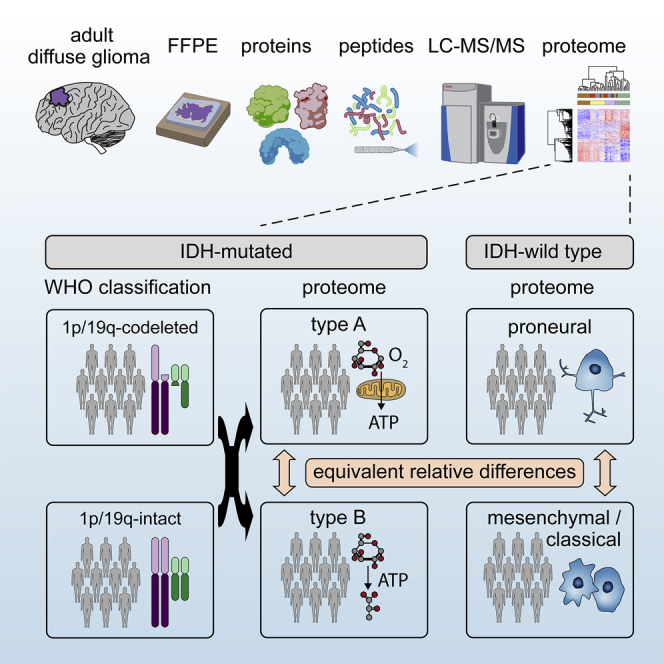
High-grade adult-type diffuse gliomas are malignant neuroepithelial tumors with poor survival rates in combined chemoradiotherapy. The current WHO classification is based on IDH1/2 mutational and 1p/19q codeletion status. Glioma proteome alterations remain undercharacterized despite their promise for a better molecular patient stratification and therapeutic target identification. Here, we use mass spectrometry to characterize 42 formalin-fixed, paraffin-embedded (FFPE) samples from IDH-wild-type (IDHwt) gliomas, IDH-mutant (IDHmut) gliomas with and without 1p/19q codeletion, and non-neoplastic controls. Based on more than 5,500 quantified proteins and 5,000 phosphosites, gliomas separate by IDH1/2 mutational status but not by 1p/19q status. Instead, IDHmut gliomas split into two proteomic subtypes with widespread perturbations, including aerobic/anaerobic energy metabolism. Validations with three independent glioma proteome datasets confirm these subgroups and link the IDHmut subtypes to the established proneural and classic/mesenchymal subtypes in IDHwt glioma. This demonstrates common phenotypic subtypes across the IDH status with potential therapeutic implications for patients with IDHmut gliomas.
Keywords: glioma, glioblastoma, proteomics, isocitrate dehydrogenase, IDH, 1p/19q codeletion
Graphical abstract
Highlights
-
•
The tumor proteomes of IDHmut gliomas do not segregate by 1p/19q codeletion
-
•
IDHmut gliomas show two aerobic/anaerobic energy metabolism subgroups
-
•
Metabolic IDHmut subgroups resemble proneural and mesenchymal/classic IDHwt glioma
-
•
Protein level perturbations of oncogenes and tumor suppressors in glioma
Bader et al. use liquid chromatography-mass spectrometry to characterize the proteomes of diffuse glioma brain tumors. The study reveals that Isocitrate dehydrogenase-mutant gliomas segregate into two metabolic subtypes resembling proneural and classic/mesenchymal subtypes in Isocitrate dehydrogenase wild-type gliomas. This has broad implications for clinical patient stratification and therapy.
Introduction
Gliomas in adulthood account for about 80% of malignant primary brain tumors with an incidence of about 5 per 100,000 people.1 In general, adult diffuse gliomas are classified according to histomorphological and molecular characteristics, including isocitrate dehydrogenase (IDH) mutations and 1p/19q codeletions, and graded (WHO grades 2 to 4) for predicting clinical-biological behavior (WHO Classification of Central Nervous System Tumours, fifth edition2). The category of adult diffuse gliomas comprises astrocytoma, IDH-mutant (IDHmut), oligodendroglioma, IDHmut and 1p/19q codeleted, and glioblastoma, IDH-wild-type (IDHwt). Most IDHwt glioblastoma patients die within 15–18 months after diagnosis and the 5-year survival rate does not exceed 10% despite therapy,3,4 while astrocytoma, IDH-mutant, have a significantly better prognosis. Unfortunately, the overall survival has not been improved markedly in recent years.
Several genetic alterations in the development and progression of diffuse gliomas have been known for years.5 Key genetic events include hotspot mutations in IDH genes 1 and 2 and codeletion of the 1p/19q chromosome arms. The molecular genetics are routinely assessed in clinical practice for diagnostic purposes as both are associated with prognostic and predictive relevance.6,7 Mechanistically, missense mutations alter IDH enzymatic activity such that the oncometabolite D-2-hydroxyglutarate is formed, which leads to epigenetic remodeling via demethylase inhibition and HIF1-dependent survival and angiogenesis.8 Codeletion of 1p/19q occurs in IDHmut oligodendrogliomas but not in astrocytomas; however, its pathological mechanism is less well understood.9,10
Omics technologies have emerged as powerful methods to classify gliomas. Genomic and transcriptomic analyses revealed subtypes of high-grade gliomas with decreasing survival, which are referred to as proneural, classical, and mesenchymal.11,12 Further integrative genomic, transcriptomic, and epigenomic analyses of gliomas WHO grades 2 to 4 indicated at least seven glioma subtypes, with the most prominent classifier being IDH mutational status.13 IDHmut gliomas further separated into a 1p/19q codeletion entity and two non-codeletion entities, namely high and low glioma CpG island methylation phenotype. Notably, the classification of IDHwt gliomas was largely orthogonal to the previous GBM classification, suggesting greater complexity. In addition, methylation-based classification of CNS tumors revealed 82 distinct tumor methylation classes.14
Genome-wide epigenetics and next-generation sequencing of tumor genomes have lately found their way into routine neuropathological diagnostics. Nevertheless, the scientific community has continued to improve the classification of IDHmut gliomas focusing on non-1p/19q codeleted astrocytomas and recently identified CDKN2A/B status as a biomarker.15 Despite these advances in the biological subclassification of CNS tumor entities, there have been no significant breakthroughs in tumor therapy for high-grade gliomas in recent years. Thus, it remains an unmet need to refine the classification of gliomas and to identify cellular targets for future therapies.
In contrast to the advances in the epigenetic, genetic, and transcriptomic characterization of gliomas, an equivalent understanding at the proteome level has not yet been achieved. Mass spectrometry (MS) has emerged as the method of choice for large-scale proteome investigations with many applications in biology and medicine.16 However, previous studies utilizing MS to analyze the human glioma proteome often covered few proteins, were limited to available fresh tumor tissue, or were conducted with immortalized cell lines. We have demonstrated previously that several thousand proteins can be robustly quantified from formalin-fixed, paraffin-embedded (FFPE) tissue, making a plethora of biobank samples amenable to proteomic analysis.17,18 Moreover, the analysis of the phosphoproteome can reveal altered signaling checkpoints of cancerous transformation across all areas of cell biology. Phosphotyrosine signaling has been shown to be preserved in FFPE samples, and the phosphoproteome of FFPE samples, including cancer tissues, has been investigated before.19,20,21 Inspired by recent proteomic and phosphoproteomic studies proving its potential to stratify breast cancer subtypes,22,23,24 we set out to characterize the proteomes of adult-type diffuse high-grade gliomas.
Results
Sample characteristics and proteomic workflow
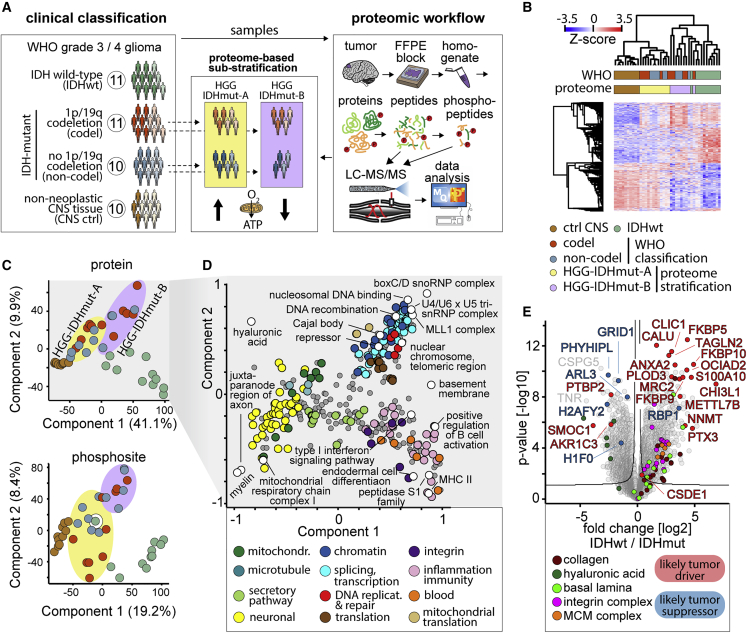
We collected 10 to 11 FFPE samples of glioblastoma, IDHwt; oligodendroglioma IDH-mutant and 1p/19q-codeleted (codel); astrocytoma IDH-mutant (non-codel); and 10 control samples of non-neoplastic CNS tissue (CNS ctrl) (Figure 1A; Table 1). All tumors were classified histomorphologically and immunohistochemically according to the criteria of the WHO classification for CNS tumors (fifth edition). In addition, we characterized all tumors (epi)genetically using genome-wide methylation profiling and copy number variation (CNV) profiling.
Figure 1.
Study and global proteome overview
(A) Cohort overview and schematic proteomic workflow. Sample numbers in circles. Dark colors represent males and light colors females. The icons at the bottom of the proteome-based classification of IDHmut gliomas represents high (arrow up) and low (arrow down) expression levels of mitochondrial respiratory chain proteins.
(B) Unbiased hierarchical clustering of all 42 samples (columns) and proteins (rows), Z-scored protein intensity shown in a heatmap; 3,749 significant proteins included, significance (q < 5% at s0 = 2) according to one-way ANOVA analysis using the WHO-defined entities. Sample clustering based on Euclidian distance and protein clustering based on Pearson correlation.
(C) Principal-component analysis of proteomes of all 42 samples for protein abundance (upper panel) and phosphosite abundance (lower panel). Components 1 and 2 shown, respectively. Sample color code as in (A and B). Colored ellipses highlight the IDHmut HGG-IDHmut-A and HGG-IDHmut-B clusters as defined in (B).
(D) Two-dimensional analysis of protein annotation term enrichment in components 1 and 2 of the principal-component analysis of the proteome dataset linking the components to biological features.
(E) Comparison of IDHwt and IDHmut glioma proteomes. Samples, n = 11 IDHwt and 21 IDHmut.
Table 1.
Cohort composition
| Non-neoplastic controls | Oligodendroglioma IDHmut and 1p/19q codeleted (codel) | Astrocytoma IDHmut (non-codel) | HGG-IDHmut-A | HGG-IDHmut-B | Glioblastoma (IDHwt) | |
|---|---|---|---|---|---|---|
| N | 10 | 11 | 10 | 12 | 9 | 11 |
| Age at diagnosis (mean ± SD) | 39 ± 12 | 48 ± 10 | 36 ± 12 | 38 ± 12 | 47 ± 12 | 73 ± 7 |
| Male:female | 3:7 | 6:5 | 5:5 | 6:6 | 5:4 | 8:3 |
| Histopathology | parahippocampal tissue of patients with hippocampal sclerosis | 11 oligodendroglioma, IDHmut and 1p/19q codeleted (CNS: WHO: 3) | 10 astrocytoma, IDHmut (CNS: WHO 3/4) | 6 oligodendroglioma, IDHmut and 1p/19q codeleted (CNS: WHO: 3), 6 astrocytoma, IDHmut (CNS: WHO 3/4) |
5 oligodendroglioma, IDHmut and 1p/19q codeleted (CNS: WHO: 3), 4 astrocytoma, IDHmut (CNS: WHO 3/4) |
11 glioblastoma, IDHwt (CNS: WHO 4) |
| WHO grade 3:4 | Na | 11:0 | 8:2 | 11:1 | 8:1 | 0:11 |
| IDH status | Na | 11 IDH1 R132H | 10 IDH1 R132H | 12 IDH1 R132H | 9 IDH1 R132H | 11 wt |
| 1p/19q status (LOH microsatellite) | Na | 11 codeletion | 10 intact | 6 codeletion, 6 intact | 5 codeletion, 4 intact | 11 intact |
| CDKN2A/B deletion | Na | 1/11 | 1/10 | 0/12 | 2/9 | 8/11 |
| EGFR amplification | Na | 0/11 | 0/10 | 0/12 | 0/9 | 5/11 |
| Methylation class | Na | 11 methylation class family glioma, IDHmut | 9 methylation class family glioma, IDHmut, 1x non-classifiable |
11 methylation class family glioma, IDHmut, 1 non-classifiable |
9 methylation class family glioma, IDHmut | 11 methylation class family glioma, IDHwt, 1 non-classifiable |
| Methylation subclass | Na | 10 1p/19q-codeleted oligodendroglioma, 1 oligosarcoma |
8 astrocytoma, 1 high-grade astrocytoma, 1 non-classifiable | 6 1p/19q codeleted oligodendroglioma, 5 astrocytoma, 1 non-classifiable | 4 1p/19q codeleted oligodendroglioma, 1 oligosarcoma, 3 astrocytoma, 1 high-grade astrocytoma | 4 RTK II, 3 RTK I, 3 mesenchymal, 1 non-classifiable |
We applied an FFPE proteomics workflow that we established previously and used to analyze ovarian cancer subtypes.17 FFPE slices were deparaffinized, homogenized, and protein extracts were digested by trypsin and LysC. Peptides were subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis or used for phosphopeptide enrichment and subsequent LC-MS/MS analysis (Figure 1A). Our dataset comprised a total of 5,724 proteins and 5,212 high-confidence phosphosites, corresponding to about 5,000 quantified proteins and 3,000 phosphosites per sample (Figure S1A).
Global dataset structure and IDHwt/IDHmut glioma proteome differences
We assessed the molecular relationship of tumor proteomes by unbiased hierarchical clustering and principal-component analysis and linked main proteome differences to biological pathways (Figures 1B–1D).
The CNS ctrls clustered most closely and clearly separated from the gliomas (Figures 1B and 1C). Compared with both IDHwt and IDHmut glioma, the CNS ctrls were enriched in synaptic and myelin-related proteins, reflective of functional brain tissue in good quantitative concordance with CPTAC glioma proteome data (Figures 1D and S1B).25
The IDHwt gliomas in our study were most distinct from CNS ctrl and also segregated from IDHmut gliomas (Figures 1B and 1C). The IDHwt proteome was enriched with proteins linked to inflammation, MCM complex DNA polymerases, an integrin-, collagen-, and laminin-rich “basement membrane-like” extracellular matrix (ECM) profile, low in hyaluronic acid, which is associated with increased malignancy in gliomas (Figures 1C–1E).26 IDHwt/IDHmut differences aligned well with the CPTAC data and were largely unaffected by 1p/19q codeletion status in our data (Figures 1E, S1C, and S1D). In line with a more “aggressive” phenotype of IDHwt gliomas, many outlier proteins with high abundance in IDHwt are cancer drivers, several of them linked to invasion. Outlier proteins associated with IDHmut included tumor suppressors downregulated in IDHwt (Figures 1E and S1D; Table 2). Notably, these tumor suppressors include the histone proteins H1F0 and H2AFY2, which both maintain an epigenetic profile of differentiation and inhibit a return to a proliferative stem cell state through distinct mechanisms.27,28 Known proteome alterations driving progression of the IDHmut were apparent, such as the strong epigenetic downregulation of RBP1, as well as novel ones, such as AKR1C3 overexpression selectively in IDHmut (Figures 1E and S1E; Table 2).
Table 2.
Proteins of interest with differential abundance in various glioma subtypes in the proteomic data of this study
| Sample group | Gene, protein | Role | Tumor entity | Cellular function | Reference |
|---|---|---|---|---|---|
| Selected outlier proteins regulated between IDHwt and IDHmut | |||||
| IDHwt | CHI3L1, chitinase-3-like protein 1 | OG | glioma, breast cancer |
inflammation, angiogenesis | Libreros et al.29; Steponaitis et al.30 |
| FKBP5/FKBP9/FKBP10, peptidyl-prolyl cis-trans isomerase FKBP5/9/10 | OG | glioma, renal cell carcinoma |
FKBP5: NF-κB signalingFKBP9: MAPK signaling FKBP10: proliferation, invasion | Jiang et al.31; Xu et al.32; Ge et al.33 | |
| CALU, calumenin | OG | glioma | EMT | Yang et al.34 | |
| TAGLN2, transgelin-2 | OG | glioma | cell motility | Han et al.35 | |
| PLOD3∗ | OG | glioma | proliferation, invasion, hypoxia | Tsai et al.36 | |
| S100A10, protein S100-A10∗ | OG | glioma, various | ECM remodeling, invasion | Tantyo et al.37 | |
| ANXA2, annexin A2∗ | prevents S100A10 degradation | ||||
| OCIAD2, OCIA domain-containing protein 2 | OG | glioma, various | migration, invasion | Nikas 201638; Sinha et al.39 | |
| MRC2, C-type mannose receptor 2 | OG | hepatocellular carcinoma | migration, invasion, TGF-β signaling | Gai et al.40 | |
| METTL7B, methyltransferase-like protein 7B | OG | lung cancer | cell-cycle progression | Liu et al.41 | |
| CLIC1, chloride intracellular channel protein 1 | OG | glioblastoma | proliferation | Setti et al.42 | |
| PTX3, pentraxin-related protein PTX3 | OG | glioblastoma | autophagy | Wang et al.43 | |
| RBP1, retinol-binding protein 1 | TSG | IDHmut glioma | retinoic acid metabolism | Chou et al.44 | |
| CSDE1, cold-shock domain-containing protein E1 | OG | colorectal cancer | differentiation, EMT | Lee et al., 201745; Martinez-Useros et al.46 | |
| NNMT, nicotinamide N-methyltransferase | OG | ovarian cancer | ECM | Eckert et al.47 | |
| IDHmut | H1F0, histone H1.0 | TSG | various, including glioblastoma | differentiation, epigenome | Torres et al.27 |
| H2AFY2, core histone macro-H2A.2 | TSG | melanoma | differentiation, epigenome | Gaspar-Maia et al.28; Kapoor et al.48 | |
| ARL3, ADP-ribosylation factor-like protein 3 | TSG | glioma | not clear yet | Wang et al.49 | |
| GRID1, glutamate receptor ionotropic, delta-1 | NA | glioma | not clear yet | Wang et al.50 | |
| PHYHIPL, phytanoyl-CoA hydroxylase interacting protein-like | TSG | glioblastoma | TNF signaling | Fu et al.51 | |
| AKR1C3, aldo-keto reductase family1 member C3 | OG | liver cancer, prostate cancer |
metabolism EMT, ERK signaling |
Zhao et al. 52; Wang et al.53 | |
| PTBP2, polypyrimidine tract-binding protein 2 | OG | glioma | proliferation, migration | Cheung et al.54 | |
| SMOC1, SPARC-related modular calcium-binding protein 1 | NA | oligodendroglioma | ECM | Brellier et al.55 | |
| Selected outlier proteins regulated between IDHmut glioma subgroups | |||||
| HGG-IDHmut-B | CCAR1/2, cell division cycle and apoptosis regulator protein ½ | OG, (TSG) | various | Wnt/β-catenin, DNA damage, cell-cycle, cell growth, apoptosis | Johnson et al.56 |
| YBX1, Y-box-binding protein 1 | OG | various, including glioblastoma | proliferation, survival, invasion | Maurya et al. 57; Kuwano et al.58; Gupta et al.59 | |
| PRDX4, periredoxin-4 | OG | various, including glioblastoma | oxidative stress, apoptosis | Jia et al. 60; Kim et al.61 | |
| SUPT5H, transcription elongation factor SPT5 | OG | colorectal carcinoma | telomerase expression | Chen et al.62 | |
| LAMB1, laminin subunit beta-1 | OG | hepatocellular carcinoma | invasion | Govaere et al.63 | |
| LUM, lumican | OG | lung cancer | metastasis | Hsiao et al.64 | |
| ERH, enhancer of rudimentary homolog | OG | breast, ovarian, liver, and bladder urothelial cancer | splicing, cell cycle, DNA replication and repair, EMT and invasion | Graille and Rougemaille65; Pang et al.66; Zhang et al.67 | |
| 1p/19q codel | RPS6K/MSK1, ribosomal protein S6 kinase alpha-5 | OG | breast cancer, glioblastoma | differentiation, metastasis, drug resistance | Gawrzak et al. 68; Wu et al.69 |
| LRP4, low-density lipoprotein receptor-related protein 4 | OG | papillary thyroid cancer | proliferation, invasion | Zhou et al.70 | |
| TRIM67, tripartite motif-containing protein 67 | OG TSG |
lung cancer, colorectal cancer | proliferation, invasion, p53 activation | Jiang et al.71; Wang et al.72 | |
| Non-1p/19q codel | FBLN1, fibulin-1 | TSG | various | apoptosis, cell motility | Kanda et al. 73; Xiao et al.74 |
| TNC, tenascin-C | OG | glioblastoma, various others | invasion, proliferation, EMT | Xia et al.75; Yoshida et al.76 | |
| UniProt annotated tumor suppressors and oncoproteins—analysis across glioma groups | |||||
| Reduced in IDHwt and HGG-IDHmut-B | DMTN, dematin | TSG | colorectal cancer | metastasis | Ye et al.77 |
| CYLD, ubiquitin carboxyl-terminal hydrolase CYLD | TSG | skin cancer, myeloma | NF-κB signaling | Sun78 | |
| BIN1, Myc box-dependent-interacting protein 1 | TSG | lung cancer | c-Myc signaling | Zhang et al.79 | |
| Reduced in IDHwt | NDRG2, protein NDRG2 | TSG | lymphoma | AKT signaling | Nakahata et al.80 |
| CDKN1B/p27Kip1, cyclin-dependent kinase inhibitor 1B | TSG | various | cell cycle | Bencivenga et al.81 | |
| NF1, neurofibromin | TSG | glioma, various | Ras signaling | Lobbous et al.82 | |
| Reduced in HGG-IDHmut-B | PRKCD, protein kinase C delta type | TSG/(OG) | lymphoma, breast cancer |
apoptosis, p53 signaling, ErbB2 signaling |
Dashzeveg and Yoshida 83; Baumann et al.84; Allen-Petersen et al.85 |
| RPS6KA2 | TSG | ovarian cancer | proliferation, apoptosis | Bignone et al.86 | |
| High in IDHwt | PYCARD, apoptosis-associated speck-like protein a CARD | TSG/OG | glioblastoma | inflammasome | Stone et al.87; Sharma et al.88; Martinon et al.89 |
| DAB2, disabled homolog 2 | TSG/other | mesenchymal glioblastoma | tumor microenvironment, inflammation | Behnan et al. 90; Figliuolo da Paz et al.91 | |
| NFKB2, NF-κB p100 subunit | (OG) | glioblastoma | mesenchymal differentiation, EMT | Yamini92 | |
| High in IDHwt and HGG-IDHmut-B | EGFR, epidermal growth factor receptor | OG | glioma | survival, proliferation, invasion | Oprita et al.93 |
| AKT2, protein kinase Akt-2 | OG | glioma | metabolism, proliferation, survival | Pu et al.94; Kim et al.95 | |
| NUP214, nuclear pore complex protein Nup214 DEK, protein DEK SET, protein SET |
OG | leukemia | fusion proteins DEK-NUP214 and SET-NUP214∗: NF-κB signaling | Saito et al.96 | |
| TPM3, tropomyosin alpha-3 chain | OG | glioma | EMT | Tao et al.97 | |
| RBM15, RNA-binding protein 15 | OG | glioma | RNA methylation | Su et al.98 | |
OG, oncogene; TSG, tumor suppressor gene. Sample group denotes the samples with high protein abundance. PLOD3, multifunctional procollagen lysine hydroxylase and glycosyltransferase LH3. ANXA2 and S100A10 strongly correlated (Pearson r = 0.93) in abundance across samples (all glioma and ctrl CNS of this study). Similarly, NUP214, DEK, and SET correlated well (Pearson r = 0.87 NUP214 vs. DEK, r = 0.76 NUP214 vs. SET) across samples (see Figure S5E).
Intriguingly, the IDHmut gliomas did not separate according to the codeletion status in a hierarchical cluster and in the first two principal components, indicating a minor effect of the codeletion status on the proteome (Figures 1B–1C and S1F). Instead, the IDHmut gliomas reproducibly fall into two distinct clusters, which were balanced for the codeletion status and patient gender (Figures 1A–1C and S1G). We refer to them as HGG-IDHmut-A and HGG-IDHmut-B. HGG-IDHmut-A formed a cluster, which was more similar to CNS ctrls than to other gliomas (Figures 1B and 1C). HGG-IDHmut-B formed a more diffuse cluster which was most related to IDHwt glioma.
HGG-IDHmut-A/B sub-stratification is independent of epigenetic profile, CNV alterations, or clinical parameters
We carefully examined potential sources of systematic biases in our study groups. First, we compared the proteome-based sub-stratification with the epigenetic and CNV profile of IDH-mutated gliomas. Both IDHmut-proteome subgroups included gliomas, with astrocytoma and 1p/19q codeleted oligodendroglioma methylation subclasses at a comparable ratio (p > 0.9, Fisher’s exact test; Table 1). Our analysis shows that CNVs did not bias these proteome groups (Figures S1H and S1I). Specifically, an evenly distributed CNV load and comparable chromosome gains or losses were found in both groups. The sub-stratification was also not imbalanced with respect to WHO grade (p > 0.9, Fisher’s exact test; Table 1) and was statistically insignificantly imbalanced in CDKN2A/B status (p = 0.17, Fisher’s exact test; Table 1). Furthermore, there was no imbalance in EGFR amplifications, which were only detected in IDHwt gliomas (5/11). Next, we examined whether the origin of the proteome-based sub-stratification of HGG-IDHmut-A and -B correlated with demographic patient data and histology. HGG-IDHmut-B and HGG-IDHmut-A sample groups, however, were undistinguishable. The sample groups correlated neither with age nor gender of patients, nor was the separation attributable to a different total tumor cell content (Table 1; Figures S2A–S2E; see supplemental information). We did not detect statistically significant differences in progression-free survival between HGG-IDHmut-A and -B in our cohort comprising 21 IDHmut glioma (Figure S2F).
HGG-IDHmut-A/B differences reflect chromosomal and mitochondrial aberrations
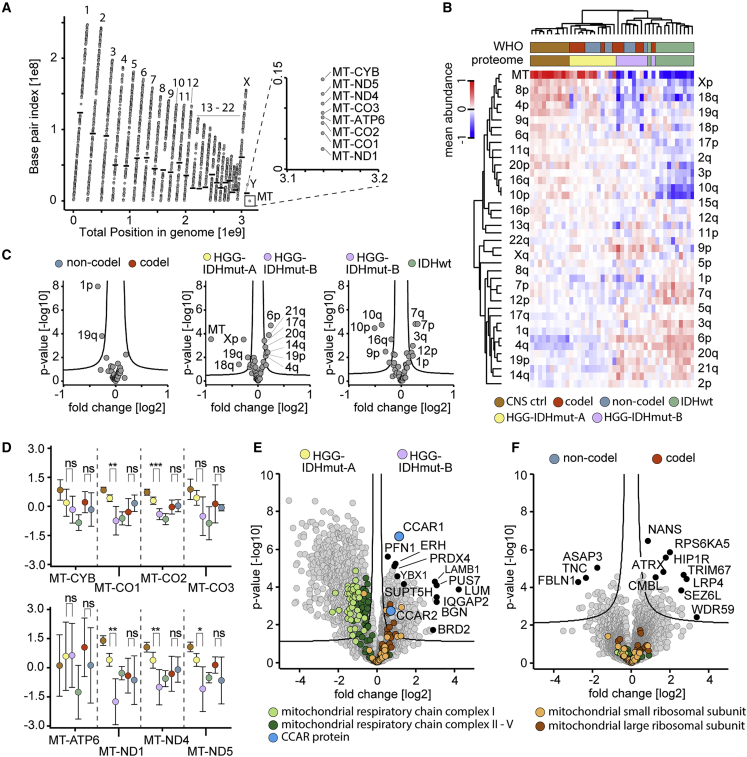
After excluding systematic biases in our study groups, we next assessed whether previously described cancer-associated chromosomal aberrations can be detected in the proteomes of tumor samples. Detected proteins were evenly distributed across almost the entire genome (Figure 2A). Hierarchical clustering across tissue samples revealed a pattern recapitulating the global proteome clustering (Figure 2B). In accordance with the 1p/19q status, codel samples exhibited a significantly reduced abundance of 1p/19q-encoded proteins (Figure 2C). However, the HGG-IDHmut-A/B groups showed distinct chromosome arm-encoded proteomes, with a strikingly reduced abundance of proteins encoded by the mtDNA in HGG-IDHmut-B (Figures 2C and 2D). This trend was also evident for nuclear genome-encoded mitochondrial respiratory chain complex proteins; however, not for mitochondrial ribosomal proteins (Figure 2E). The 1p/19q codeletion status did not exhibit these mitochondrial aberrations (Figures 2D and 2F).
Figure 2.
Chromosomal alterations point at mitochondrial perturbations in the alternatively stratified groups of IDHmut gliomas
(A) Proteome coverage across the human genome in the entire dataset (42 samples). Points indicate quantified proteins, black horizontal bars as boundaries between chromosomal p and q arms; p arms below and q arms above the bar. Uncovered areas included the p arms of chromosomes 13, 14, 15, 21, and 22, and the centromere-proximal quarter of the 9p arm.
(B) Relative abundance of chromosome arm-specific proteomes across samples shown as a heatmap. Abundance as mean intensity of all proteins assigned to a given chromosome arm. Protein intensities normalized by subtraction of median across samples before mean averaging. Samples, n = 10 (ctrl CNS), n = 12 (HGG-IDHmut-A), n = 9 (HGG-IDHmut-B), n = 11 (IDHwt), n = 11 (codel), n = 10 (non-codel).
(C) Abundance difference of chromosome arm-specific proteomes between IDHmut glioma entities. Comparison of codel (n = 11) versus non-codel entity (n = 10) (left), (center), IDHwt (n = 11) versus HGG-IDHmut-B (n = 9) (right).
(D) Abundance of proteins encoded by mitochondrial DNA across the conventional and alternatively defined tumor and control entities of this study. Circles denote means and error bars 95% confidence intervals of the mean. Color code and sample numbers as in (B).
(E and F) Global proteome and abundance differences in mitochondrial proteins between the alternative proteome-defined entities HGG-IDHmut-A (n = 12) and HGG-IDHmut-B (n = 9) (E) and the 1p/19q-codeleted (n = 11) and non-codeleted (n = 10) entities (F) of IDHmut glioma. Mitochondrial respiratory chain complex V refers to ATP synthase.
HGG-IDHmut-A/B stratification is linked to differential expression of cancer driver genes
Several known cancer drivers were enriched in HGG-IDHmut-B over HGG-IDHmut-A (Figure 2E; Table 2). The most significant outlier enriched in HGG-IDHmut-B was CCAR1, the related CCAR2 was less significantly enriched. Both proteins play complex roles in cancer, including gliomas.56,99,100,101 Intriguingly, the best correlating protein to CCAR1 in our dataset was the splicing factor SRSF1 (Figure S3A). Its related protein SRSF5 promotes lung cancer via alternative splicing of CCAR1 upon high availability of glucose.102 In our dataset, SRSF5 was stochastically quantified while SRSF1, 2, 3, 4, 6, 7, 10, and 11 were significantly enriched in HGG-IDHmut-B over HGG-IDHmut-A.
1p/19q codeletion status affects cancer-related proteins
Around 1,900 proteins differed significantly between the HGG-IDHmut-B and HGG-IDHmut-A sample groups in our dataset, whereas only about 100 proteins differed significantly between 1p/19q codel and non-codeleted IDHmut (Figures 2E and 2F). Nevertheless, several of the regulated proteins between codel and non-codel IDHmut have been linked to cancer in various contexts (Figure 2F; Table 2).
ATRX was among these differently expressed proteins and had a lower abundance in the non-codel sample group. This is in line with the frequent loss of ATRX in 1p/19q intact but not in 1p/19q codeleted gliomas as ATRX loss activates the “alternative lengthening of telomere”pathway.103,104,105 In contrast, TERT promoter mutations are associated with IDHwt and ATRX expression.106,107,108 TERT, the catalytic subunit of telomerase, was not covered in our dataset, but telomerase-accessory proteins were among the most highly enriched proteins among ATRX-correlating proteins (Figure S3B).
Next, we compared the regulation of proteins affected by common chromosomal alterations in gliomas (1p/19q codeletion, gain of chr. 7, loss of chr. 10; Figure 2C).108,109 In our data, many proteins encoded on chromosome 10 and 7 were significantly regulated (10 reduced, 7 elevated) in IDHwt gliomas, and transcripts of these regulated proteins were expectedly associated with survival according to The Cancer Genome Atlas transcriptomics data (Figure S3C).110 By contrast, proteins encoded on the 1p and particularly the 19q arm exhibited smaller and less statistically significant regulation and their transcripts did not show a clear association with survival (Figure S3D).
In summary, proteins encoded on 1p/19q were less drastically regulated compared with proteins affected by other common chromosomal alterations in glioma. Taken together, the overall glioma proteome showed only minor alterations correlating with the 1p/19q codeletion but reflected known cancer-related proteins, in particular in telomere maintenance pathways.
HGG-IDHmut-A/B stratification correlates with distinct metabolic profiles
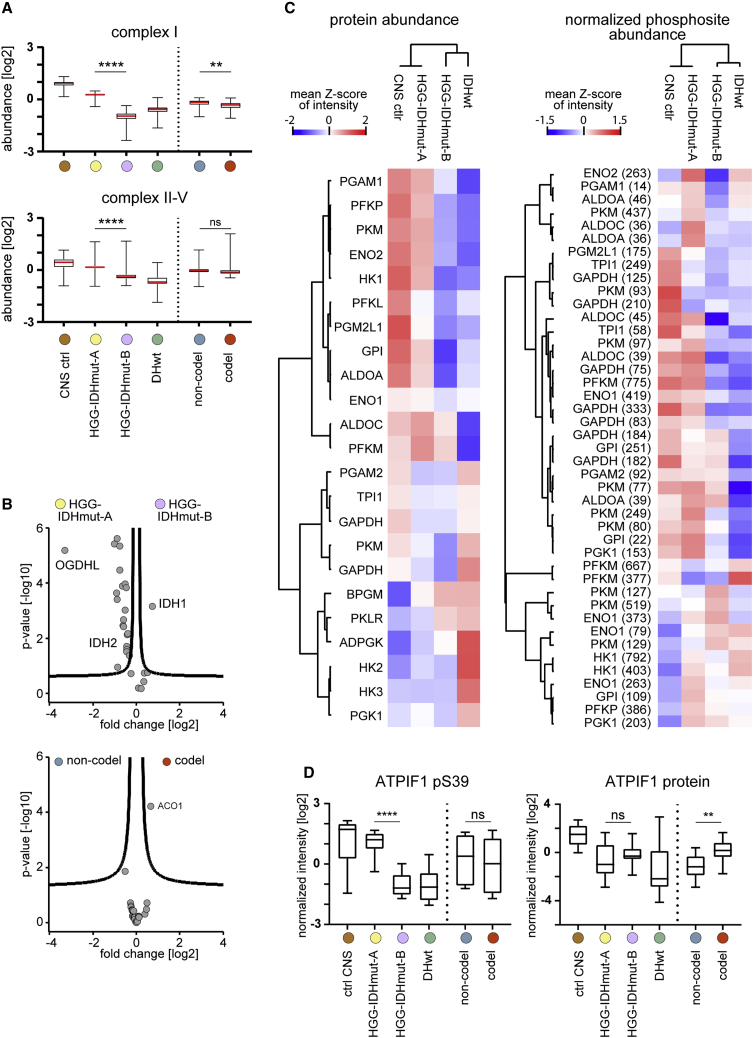
The differential abundance of mitochondrial respiratory chain proteins between HGG-IDHmut-A and -B prompted us to investigate metabolic alterations in greater detail. Respiratory chain complex and tricarboxylic acid (TCA) cycle proteins exhibited a low abundance in IDHwt and HGG-IDHmut-B (Figures 3A, 3B, S4A, and S4B). Intriguingly, mild dysfunction of respiratory chain complex I—the lowest in HGG-IDHmut-B—can promote cancer via the generation of reactive oxygen species (ROS).111 Notably, IDH1 was the only TCA protein with mildly elevated abundance in HGG-IDHmut-B. Conversely, the TCA protein oxoglutarate dehydrogenase L was strongly reduced, which may be linked to its tumor suppressor function.112
Figure 3.
Metabolism-related proteome differences associated with the novel classification of IDHmut tumors
(A) Abundance of mitochondrial respiratory chain complex proteins across the sample entities of this study, split by complex I (upper panel) and complex II-V (lower panel). Samples, n = 10 (ctrl CNS), n = 12 (HGG-IDHmut-A), n = 9 (HGG-IDHmut-B), n = 11 (IDHwt), n = 11 (codel), n = 10 (non-codel).
(B) Regulation of tricarboxylic acid protein abundances between HGG-IDHmut-B and HGG-IDHmut-A entities (upper panel) but not between codeletion-defined entities (lower panel). Sample numbers as in (A).
(C) Glycolysis-related protein profiles (left panel) and protein abundance-normalized phosphosite profiles (right panel) across the proteome-defined entities. Abundances as sample group means of cross-sample Z scores of protein intensities and relative protein-normalized phosphosite intensities. Sample numbers as in (A).
(D) Abundance of the ATPIF1 pS39 phosphosite (left panel) and the protein abundance (right panel) across the entities of this study. Sample numbers as in (A).
Regarding glycolysis, HGG-IDHmut-B again exhibited a protein and phosphosite profile similar to IDHwt (Figures 3C and S4C–S4E). In particular, hexokinase 1 was low abundant in HGG-IDHmut-B and IDHwt. Downregulation of hexokinase 1 promotes glycolysis and induces EMT in several human cancer cell lines and increased malignancy in a xenograft model.113
Likewise, the abundance of the pS39 site on the mitochondrial ATPase inhibitor ATPIF1 was low in both IDHwt and HGG-IDHmut-B compared with HGG-IDHmut-A (Figure 3D). ATPIF1 overexpression promotes a metabolic shift from oxidative phosphorylation to glycolysis in colon cancer,114 while ATPIF1 is inhibited by S39 phosphorylation.115
In summary, HGG-IDHmut-B showed molecular hallmarks of increased glycolytic activity and HGG-IDHmut-A of increased oxidative phosphorylation, independent of 1p/19q codeletion.
Proteome-based sub-stratification correlates better with profiles of annotated tumor suppressors and drivers than 1p/19q codeletion
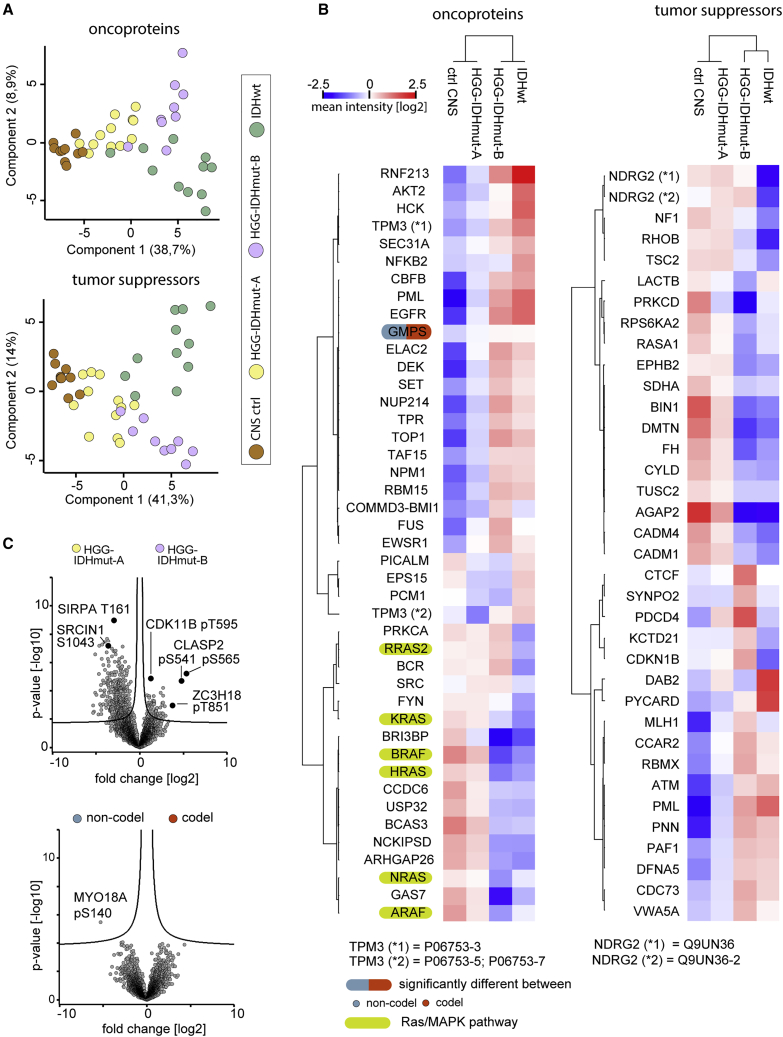
Next, we examined the profiles of annotated tumor suppressors and oncoproteins across the glioma entities. Principal-component analysis separated IDHwt, HGG-IDHmut-A, and HGG-IDHmut-B samples but not the 1p/19q codeletion status (Figures 4A, S5A, and S5B). The HGG-IDHmut-B signature was largely similar to IDHwt, while 1p/19q codeletion status was associated with few statistically significant differences (Figures 4B, S5C, and S5D).
Figure 4.
The alternative sub-stratification of IDHmut gliomas correlates with altered tumor suppressor, onco-proteins, and phosphosite levels
(A) Principal-component analysis of UniProt keyword-annotated proto-oncogenes (upper panel) and tumor suppressor genes (lower panel). Samples, n = 10 (ctrl CNS), n = 12 (HGG-IDHmut-A), n = 9 (HGG-IDHmut-B), n = 11 (IDHwt).
(B) Abundance profiles of oncoprotein (left) and tumor suppressor (right) proteins across the proteomic entities of this study. Protein intensities are first normalized by subtraction of cross-sample median and then averaged by mean sample group abundance. Samples, n = 12 (HGG-IDHmut-A), n = 9 (HGG-IDHmut-B), n = 11 (codel), n = 10 (non-codel).
(C) Phosphosite abundance differences between the proteomic entities of this study (upper panel) and the 1p/19q codeletion-defined entities (lower panel). Sample numbers as in (A).
Some tumor suppressors were specifically downregulated in IDHwt, others only in HGG-IDHmut-B, and some in both (Figure 4B; Table 2). IDHwt and HGG-IDHmut-B sample groups exhibited a downregulation of RAS pathway proteins in line with previous reports for glioblastoma (Figure 4B).116 Conversely, several proto-oncogenes were elevated in IDHwt and HGG-IDHmut-B (Figures 4B and S5E; Table 2). For some regulated proteins, the function in the context of tumorigenesis is unclear or context dependent (Table 2).
HGG-IDHmut-B and HGG-IDHmut-A exhibited strong alterations of tumor suppressors and oncoprotein phosphosite abundances, not associated with 1p/19q codeletion status (Figures 4C and S6A–S6D). For instance, the pT595 site on the cyclin-dependent kinase CDK11B was abundant in both HGG-IDHmut-B and IDHwt compared with HGG-IDHmut-A independent of total protein levels (Figures 4C and S6C). This suggests elevated CDK11B activity due to homology with the activating pT161 site in CDK1,117 linked to cell-cycle progression and altered in various cancers.118 This highlights HGG-IDHmut-A and HGG-IDHmut-B signaling differences vital to cancer.
HGG-IDHmut-A/B differences are evident across studies
To validate major molecular hallmarks supporting a metabolic stratification of IDHmut gliomas independent of 1p/19q status, we reanalyzed three recent proteomics datasets. In one study dataset comprising predominantly IDHmut glioma we identified equivalent IDHmut-A and IDHmut-B subtypes using unbiased hierarchical clustering as in our dataset (Figures S7A and S7B).119 Notably, a small number of samples in that dataset indicate that IDHmut-A/B are also present in WHO grade 2 gliomas and that proteome differences between these are equivalent to those of WHO grade 3 gliomas (Figure S7B). A second study described a similar metabolism-linked proteomic stratification of IDHwt gliomas into a glycolysis and an oxidative phosphorylation subgroup.120 To further validate such a stratification in a larger and multi-omics glioma dataset we integrated a third study, which also comprised predominantly IDHwt gliomas.25
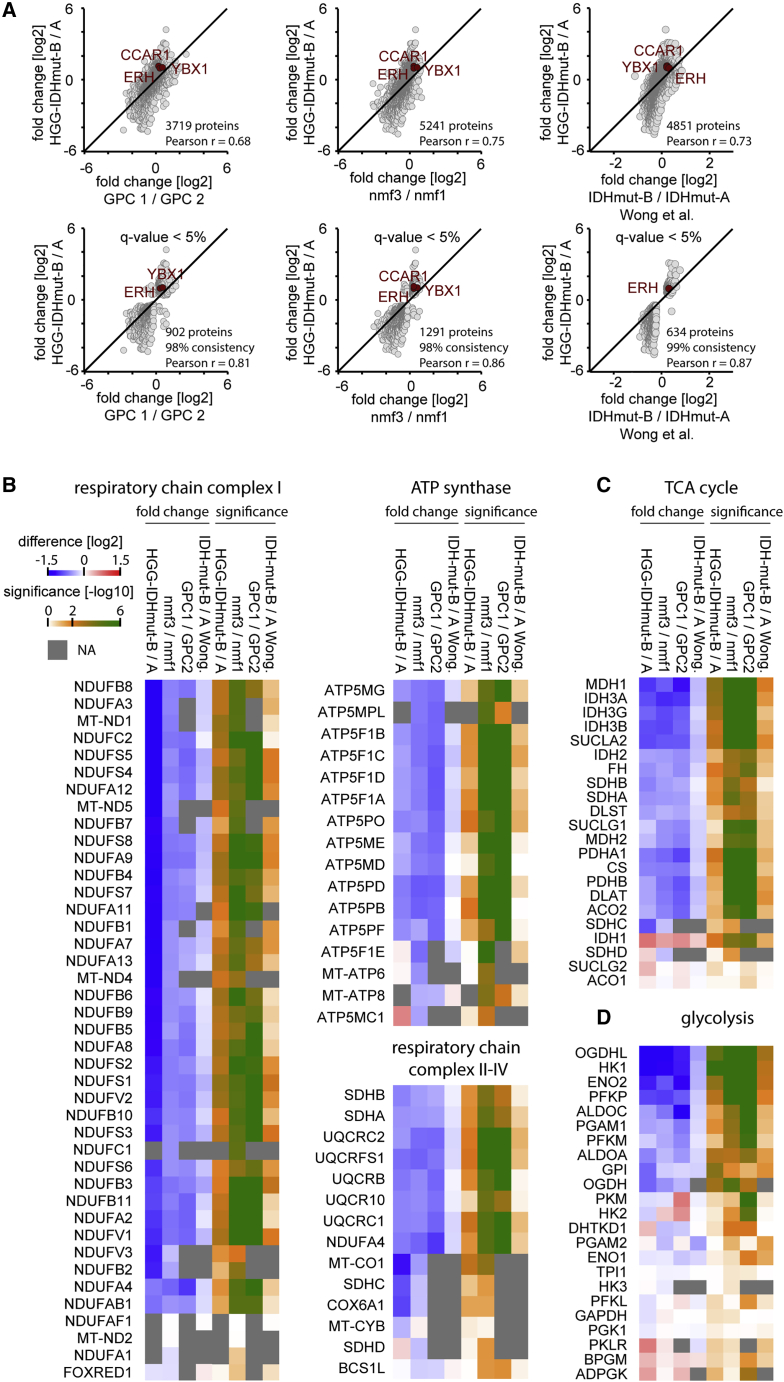
Highly consistent with our study, metabolic proteins involved in glycolysis, TCA cycle, oxidative phosphorylation, and even the global proteome were regulated in great quantitative agreement in the three other studies (Figures 5A–5D and S7C–S7F). Among the metabolic classes, TCA cycle proteins showed the most consistent and quantitatively concordant regulation across studies.
Figure 5.
Comparison of HGG-IDHmut-B/HGG-IDHmut-A to the proteomic data of three other glioma studies
HGG-IDHmut-A (n = 12) and HGG-IDHmut-B (n = 9) correlate with the high and low oxidative phosphorylation glioma subgroups reported as GPC2 (n = 13) and GPC1 (n = 26),120 nmf1/proneural-like (n = 29) and nmf3/classical-like (n = 25),25 and IDHmut-A (n = 10) and IDHmut-B (n = 28),119 respectively.
(A) Pairwise comparison of fold changes across all proteins overlapping in both datasets (upper panel) and across all proteins significantly regulated (q < 5%) in both datasets.
(B) Regulation of mitochondrial respiratory chain proteins across datasets.
(C) Regulation of tricarboxylic acid cycle proteins across datasets.
(D) Regulation of glycolysis proteins across datasets.
In the multi-omics dataset, our HGG-IDHmut-A (oxidative phosphorylation high) glioma subgroup corresponded to the proneural-like (nmf1) subgroup (Figure 5A). Our HGG-IDHmut-B (oxidative phosphorylation low) subgroup correlated most strongly with the classical-like (nmf3) glioma subgroup, but also considerably with the mesenchymal-like (nmf2) subgroup (Figures 5A and S7C).
In addition to metabolic changes, similarities across studies extended to other biological pathways, such as mRNA splicing, RNA metabolism, translation, chromatin dynamics, DNA replication, and a collagen-rich ECM, which were enriched in HGG-IDHmut-B and corresponding subgroups across studies (Figures S7G and S7H). Conversely, neural phenotype and mitochondrial proteins were enriched in HGG-IDHmut-A/proneural-like subgroups. Moreover, top outliers and likely cancer drivers CCAR1, YBX1, and ERH enriched in HGG-IDHmut-B were likewise regulated in other datasets (Figures 5A and S7G; Table 2).
Altogether, we show that IDHmut gliomas can be stratified into two proteomic subtypes with diverging energy metabolism and overall stronger differences than those caused by 1p/19q codeletion. Intriguingly, this separation emerges in four independent studies, not only independent of 1p/19q but also across IDHmut status.
Discussion
Classification of CNS tumors has long been based on histology only. In recent years, new molecular methods have significantly improved the diagnosis of diffuse glioma and risk stratification of certain glioma subgroups. At the genetic and epigenetic level, we now know numerous driver mutations and epigenetic profiles that provide information about the emergence of these malignant tumors.
Unfortunately, these successes are not significantly reflected in the treatment of diffuse high-grade adult gliomas. As a rule, adult malignant gliomas have been treated the same way for more than 15 years, mostly independent of the molecular profile, according to the generally applicable STUPP and CATNON regimen with concomitant radiochemotherapy with 6 or 12 cycles of the alkylating agent temozolomide.121,122 A major breakthrough in improving survival, as in other malignancies, such as breast cancer, lung cancer, or colorectal carcinomas, could not be achieved by these therapies. A targeted therapy tailored to the molecular profile, e.g., IDHmut or IDHmut and 1p/19q codeleted gliomas could not be established so far.
Therefore, we hypothesized that, in addition to the known histological-molecular tumor classification (according to the current WHO classification), a proteome-based approach could reveal signaling pathways and metabolic states which pave the way for novel therapeutic strategies.
For this purpose, we characterized the proteome and phospho-proteome of adult-type diffuse high-grade (WHO grades 3 and 4) gliomas from FFPE samples with MS. The genetic subtyping of IDHmut tumors into astrocytoma, IDHmut and oligodendroglioma, IDHmut and 1p/19q codeleted according to the current WHO classification correlated with minor differences in the tumor proteome (WHO Classification of Central Nervous System Tumours, fifth edition). The selective loss of proteins encoded on chromosomal locations 1p and 19q was reflected in the proteomes, albeit to a lower extent than loss of chromosome 10 proteins in IDHwt gliomas. Likewise, the proteomic data showed the expected relationship of 1p/19q codeletion and molecular signatures of telomere maintenance via either the alternative lengthening of telomeres pathway or the telomerase-dependent pathway. These signatures include loss of ATRX in 1p/19q intact gliomas and conversely high abundance of telomerase-associated proteins in ATRX intact (1p/19q codeleted) gliomas. Overall, the proteomics approach discovered expected features associated with the 1p/19q codeletion.
However, the proteomes suggested an alternative sub-stratification of IDHmut gliomas independent of 1p/19q codeletion. This sub-stratification correlated with the loss of mitochondrial DNA-encoded proteins, a loss of mitochondrial respiratory chain proteins, an overall distinct metabolic profile, a basement membrane-like ECM signature, distinct proto-oncogene and tumor suppressor protein signatures, differences in translation, RNA metabolism, DNA replication, and chromatin dynamics, as well as site-specific differences in the phospho-proteomes. In this study cohort, the alternative sub-stratification was not associated with patient gender, age, clinical course, discerning histopathological features, or CNV alterations.
Occurrence of an early 1p/19q codeletion can be a crucial point in cancer development, while both codeleted and non-codeleted gliomas can progress by similar mechanisms. Accordingly, our proteome-based classification does not question the general relevance of the 1p/19q codeletion as an early-event determinant of glioma development. However, in contrast to the more dynamic proteome, 1p/19q status may not be the most appropriate marker for the current state in subsequent cancer progression.
The tumor proteomes in this study showed perturbations of various biological processes, in particular the aerobic/anaerobic energy metabolism. The downregulation of the mitochondrial respiratory chain proteins in the proteomically defined HGG-IDHmut-B subgroup could reflect the Warburg effect, which favors the anaerobic over aerobic metabolism in cancer cells. Notably, complex I was particularly affected. A mild dysfunction of complex I can promote tumor progression via ROS-dependent activation of AKT signaling.111 Moreover, loss of respiratory chain proteins in our dataset was accompanied by a synergistic loss of pS39 phosphorylation on ATPIF1, a rewiring of the TCA cycle and glycolysis enzyme profile, including the loss of HK1, which promotes glycolysis and malignancy in mouse xenograft models.113 Loss of S39 phosphorylation on ATPIF1 promotes a metabolic shift from oxidative phosphorylation to glycolysis and further proliferation and cell death resistance via mitochondrial ROS production in hypoxia and cancer.114,115 Intriguingly, there could also be a link between the metabolic rewiring and CCAR1 dysregulation. In lung cancer, elevated glucose concentrations induce the splicing factor SRSF5 to promote cancer progression via alternative splicing of CCAR1.102 In our dataset, CCAR1 was the top outlier in gliomas with respiratory chain loss and CCAR1’s best correlating protein was the related SRSF1. Overall, our study provides a rich protein and phosphosite resource supporting future mechanism of action and target discovery investigations.
There is increasing evidence in the literature that high-grade gliomas show distinct metabolic subtypes, using glycolysis (Warburg effect) or oxidative phosphorylation as primary energy source.120,123 The proteome differences between HGG-IDHmut-A and -B of our study were strikingly similar to differences of corresponding subtypes we likewise identified in another dataset of IDHmut glioma using unbiased hierarchical clustering.119 Notably, these similarities extended to independently defined IDHwt subtypes in two more studies, both in terms of the global proteome and metabolic classes of proteins, including glycolysis, TCA cycle, and oxidative phosphorylation.25,120
The IDHwt dataset originating from the CPTAC multi-omic glioma study revealed that HGG-IDHmut-A (high oxidative phosphorylation) of our study corresponded to a proneural-like glioma subtype and HGG-IDHmut-B (low oxidative phosphorylation) corresponded best to a classical but also relatively well to a mesenchymal-like glioma subtype.25 Thus, the HGG-IDHmut-B group in our cohort potentially comprises both mesenchymal and classic phenotype-like IDHmut glioma, which have common strong proteome differences to proneural-like IDHmut glioma.
The strong similarity of the proteome differences between IDHmut subtypes in our study, compared with the differences between IDHwt subtypes in the two other studies emerging from our analysis, suggests a shared—but yet to be determined—molecular mechanism for this phenotypic, including metabolic divergence across IDH status. Single-cell transcriptome analysis of gliomas has revealed that tumors comprise cells of four phenotypic subtypes with distinct metabolism in varying overall compositions.123 Intrinsic (e.g., genetic/epigenetic) factors may cause such diversity and extrinsic factors (e.g., tumor microenvironment) could select for a certain cellular or overall tumor type. Further studies based on single-cell characterization, such as by emerging proteomics approaches, will be valuable in addressing these questions by defining the metabolic and neurodevelopmental tumor cell states and their microenvironment.124
Metabolic stratification of gliomas is an emerging approach with broad implications for survival and therapy. Re-analysis of The Cancer Genome Atlas cohort comprising over 300 human IDHwt glioblastoma samples using a pathway-based classification of the transcriptome showed a better survival of oxidative phosphorylation-driven gliomas.123 Patient-derived cells originating from tumors relying on oxidative phosphorylation were uniquely susceptible to compounds targeting mitochondria, including inhibition of respiratory chain complex I (metformin), inhibition of mitochondrial translation (tigecycline), and induction of mitochondrial oxidative stress and apoptosis (menadione). Likewise, these cells also exhibited increased sensitivity to ionizing radiation, which primarily causes mitochondrial rather than nuclear stress.123,125 Similarly, another study screened anticancer agents for efficacy in patient-derived cells originating from IDHwt gliomas with proteomically characterized metabolic state.120 Several agents were selectively cytotoxic in either glycolysis-reliant glioma cells (tandutinib, crizotinib, olaparib, and AZD2014) or in oxidative phosphorylation-reliant cells (erismodegib and canertinib). Moreover, the proteomic/metabolic stratification of IDHmut gliomas may be relevant for treatment with other established drugs. For instance, IDH1 was selectively more abundant in HGG-IDHmut-B than in HGG-IDHmut-A despite the opposite trend for all other regulated TCA cycle proteins, which may correspond to differential sensitivity toward IDH inhibitors, such as ivosidenib, and a benefit in clinical trials.126,127 Similarly, metformin is under discussion as a glioma drug, potentially in synergistic use with temozolomide, and has shown differential efficacy in cellular models of patient-derived cells derived from proteomic-metabolically stratified gliomas.123,128,129 In summary, past studies highlight the promise that metabolic stratification holds for glioma precision medicine and as well as the power of proteomics in discovering such stratification exemplified for IDHwt glioma. By further implicating IDHmut gliomas, our study creates a bigger picture of common phenotypic subtypes independent of the IDH status, which we believe will also benefit patients with IDHmut gliomas by making future proteome/metabolism-directed therapies available to them as well.
Limitations of the study
A limitation of this study is the low number of IDHmut samples (21 IDHmut tissue samples in total). Validation in the three literature datasets provided additional support. Samples in two of the three studies were predominantly IDHwt, but in the third predominantly IDHmut. We conclude that IDHmut gliomas can be classified into metabolic and proteomic subgroups similar to those of IDHwt. However, this study does not ascertain whether proteomic/metabolic status and IDH mutational status are independent and orthogonal classifiers of a continuum of glioma phenotypes. Alternatively, similar metabolic glioma subgroups could exist within both IDHwt and IDHmut. Furthermore, the mechanistic root cause driving subtype divergence and its interplay with the (epi-)genome and tumor microenvironment remains to be elucidated.
On the clinical side, larger and clinically better-characterized study collectives are required to integrate the novel proteomic/metabolic subgroups and the established IDH mutation and 1p/19q codeletion status into a new overall classification scheme and assess the predictive power of these features for survival and treatment response. Moreover, prospective studies are required to assess whether patients with high-grade gliomas benefit from a combination of metabolic/proteomic stratification and precision anticancer agents and radiotherapy.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| FFPE glioma tissue samples | Department of Neuropathology Charité - Universitätsmedizin Berlin | See Data S1 for each sample |
| Chemicals, peptides, and recombinant proteins | ||
| lysyl Endopeptidase®, Mass Spectrometry Grade (Lys-C) | FUJIFILM Wako | WAKO 129-02541 |
| trypsin, porcine, Mass Spectrometry Grade | Sigma Aldrich/Merck | Cat# T6567 |
| styroldivinylbenzol-reversed phase sulfonate (SDB-RPS) | Empore 3M | Cat# 13427427 |
| IDH1 R132H antibody | DIANOVA, clone H09 | Cat# DIA-H09-L |
| Polymerase for IDH1 and IDH2 pyrosequencing | HotStarTag DNA Polymerase, Qiagen | Cat# 203205 |
| Critical commercial assays | ||
| Pierce BCA assay | Thermo Fisher | Cat# 23227 |
| AssayMAP Cartridge Rack Fe(III)-NTA 5μL | Agilent Technologies | Cat# G5496-60085 |
| Qiagen DNeasy blood and tissue DNA extraction kit | Qiagen | Cat# 69581 |
| Maxwell RSC FFPE Plus DNA Purification Kit | Promega | Cat# AS1720 |
| Zymo EZ Methylation Kit | Zymo Research Irvine | Cat# D5004 |
| Infinium HD FFPE DNA Restore Kit | Illumina | Cat# WG-321-1002 |
| Deposited data | ||
| Mass spectrometric raw data | This study | PRIDE repositorium: PXD024427 |
| Methylation and copy number variation raw data | This study | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE212838 |
| Murine brain proteome dataset | Sharma et al., 2015130 | Table S5 |
| CPTAC/Wang et al. glioma proteome dataset: sample annotations | Wang et al., 202125 | Table S1 |
| CPTAC/Wang et al. glioma proteome dataset: protein intensities | Wang et al., 202125 | Table S2, Table 8 “proteome_normalized” |
| Oh et al. glioma proteome dataset: sample annotations | Oh et al., 2020120 | Data S1 |
| Oh et al. glioma proteome dataset: sample annotations | Oh et al., 2020120 | Data S2, Table 2 “Global” |
| Wong et al. glioma proteome dataset: sample annotations | Wong et al., 2022119 | https://github.com/derekwong90/LGG_proteomics/blob/main/data/metadata.txt |
| Wong et al. glioma proteome dataset: protein intensities | Wong et al., 2022119 | https://github.com/derekwong90/LGG_proteomics/blob/main/data/protein_abundance.txt |
| Software and algorithms | ||
| Xcalibur | Thermo Fisher Scientific | OPTON-30965 |
| MaxQuant | Cox and Mann, 2008131 | https://maxquant.org/ |
| Perseus | Tyanova et al., 2016132 | https://maxquant.org/perseus/ |
| R | The R Project for Statistical Computing | https://www.r-project.org/ |
| Brain tumor classifier version v11b4 | Capper et al., 201814 | www.molecularNeuropathology.org; Classifier Version v11b4 |
| Brain tumor classifier version v12.5 | Suwala et al., 2022133 | www.molecularNeuropathology.org; Classifier Version v12.5 |
| R/Bioconductor conumee package version 1.9.0 | Hovestadt and Zapatka134 | https://bioconductor.org/packages/release/bioc/html/conumee.html |
| R package ComplexHeatmaps v2.10.0 | Gu et al., 2016135 | https://www.bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| R package survival v3.1-8 | Therneau et al., 136 | https://github.com/therneau/survival |
| R package survminer v0.4.9 | Kassambra et al., 137 | https://rpkgs.datanovia.com/survminer/index.html |
| Other | ||
| IDH1 and IDH2 pyrosequencing procedure | Radke et al., 2019138 | N/A |
| IDH1 R132H immunohistochemistry procedure | Radke et al., 2019138 | N/A |
| EPIC analyses procedure | Suwala et al., 2022133 | N/A |
| iScan system | Illumina | Cat# SY-101-1001 |
| Bead chip array platform | Illumina | Cat# 20028879 |
| T10 basic Ultra-Turrax blender | IKA | 0003737000 |
| Bioruptor Plus | Diagenode | Cat# B01020001 |
| Concentrator plus | Eppendorf | Cat# 5305000509 |
| Nanodrop 2000 | Thermo Scientific | Cat# ND-2000 |
| AssayMAP Bravo | Agilent | Cat# G5571AA |
| EASY-nLC 1200 | Thermo Fisher Scientific | Cat# LC140 |
| Q Exactive HF | Thermo Fisher Scientific | Cat# 0726041 |
| Q Exactive HF-X | Thermo Fisher Scientific | Cat# 0726042 |
| Fused silica capillaries | Optronis GmbH | TSP 075 375 |
| C18 beads for in-house packed chromatography columns | Dr.Maisch | ReproSil-Pur 120C18-AQ, 1.9 μm |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Felix Meissner (felix.meissner@uni-bonn.de).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects
Formalin fixed paraffin embedded (FFPE) tumor samples were collected from patients who underwent surgery at the Department of Neurosurgery of the Charité, Universitätsmedizin Berlin, and diagnosed at the Department of Neuropathology Charité - Universitätsmedizin Berlin. The diagnoses were made according to the valid WHO classification for tumors of the CNS at the time of diagnosis (revised fourth edition, 2016) and re-evaluated when the WHO classification was updated (fifth edition 2021). Patients’ ages at the time of surgery were 73 ± 7 years (mean ± SD) for IDHwt, 48 ± 10 years for codel, 36 ± 12 years for non-codel, and 39 ± 12 years for the CNS ctrl. The non-neoplastic CNS tissue samples originate from epilepsy patients whose temporal lobe has been resected because of mesial hippocampal sclerosis. Subsequently, the parahippocampal cortical and subcortical tissue was separated from the hippocampus and the white matter tissue in particular was used as controls. Further information can be found in Table S1. All IDH-mutant gliomas included in the study had IDH1 R132H mutations. This study was conducted according to the ethical principles of medical research involving human subjects according to the Declaration of Helsinki. The clinical data were assessed and anonymized for patients’ confidentiality. Ethical approval (EA2/101/08) was granted by the institutional ethics board of the Charité Ethics Committee.
Method details
Clinical diagnostic procedures and sampling
IDH mutation status was determined by IDH1 R132H immunohistochemistry (IHC). In cases with unclear IDH immunohistochemistry results and in patients with glioblastomas under 55 years of age, an IDH1 and IDH2 pyrosequencing analysis was also performed.138,139 The 1p/19q-codeletion status was determined using the EPIC methylation analysis – described below. As current diagnostic criteria for glioma include homozygous deletion of CDKN2A/B, this was inferred from the EPIC analysis later in the study.15 IDHwt glioma exhibited frequent (8/11) loss of CDKN2A/B while only few (2/21) IDHmut glioma showed this. Similarly, EGFR amplification was common in IDHwt (5/11) but absent in IDHmut glioma. The tumor area was marked on an H&E section by a neuropathologist and the corresponding tissue was removed from the paraffin block for the proteome and molecular analyzes. Areas with high tumor cell content (≥50%) were preferably chosen and adjacent sections from the tissue block macro-dissected for experiments. As gliomas are diffusely infiltrating tumors, three pronounced diffuse glioma samples with a tumor cell content of 35-50% were also included. For IDH-mutant samples, tumor content estimation was refined by categorization into solid tumor and infiltration zone.
DNA methylation and copy number variation analysis
For all tumors, DNA methylation and copy number analyses were performed using the EPIC (850k) Bead-Chip array platform (Illumina, USA).133 All analyses were carried out according to the manufacturer's instructions. DNA was extracted from FFPE tumor samples using the Maxwell RSC FFPE Plus DNA Purification Kit (Promega, USA). After bisulfite conversion with the Zymo EZ Methylation Kit (Zymo Research Irvine, USA), the Infinium HD FFPE DNA Restore Kit was used for DNA restoration. The beadchips were scanned on the iScan system (Illumina USA).
Sample preparation for mass spectrometry
FFPE tumor samples were prepared for proteomic analysis as reported previously.17 FFPE biobank specimens with an approximate 1.5 × 1 cm area were cut in 10μm slices with a microtome. Three slices were twice deparaffinized by 1mL of xylene at 50°C for 5min, washed twice with ethanol at RT for 5min and air-dried for 15min. Samples were homogenized in 300μL lysis solution (0.1M Tris/HCl, pH7.5, 10mM dithiothreitol) with an T10 basic Ultra-Turrax blender (IKA, Staufen, Germany). SDS was added to a final concentration of 4% w/v and sample homogenates were incubated at 99°C for 60 min at 600rpm on a Thermomixer (Eppendorf, Germany), cooled to RT, and sonicated on a Bioruptor Plus device for 15 cycles of 15s high power setting and 15s incubation (Diagenode SA, Belgium). After centrifugation for 10 min at 16 000 g at 4°C, proteins in the supernatant were alkylated with 55mM iodoacetamide for 30min in the dark and precipitated with 4-fold excess v/v of −20°C acetone overnight. The pellet was isolated by centrifugation at 16 000 g at 4°C for 10min, washed with 80%–20°C acetone, and resuspended in 100μL 8M urea aided by sonication as before.
The protein concentration was determined by BCA assay and proteins were digested with LysC for 3 hrs and subsequently diluted 3-fold with 50mM ammonium bicarbonate and digested with trypsin overnight. Digestions were performed at an enzyme:protein ratio of 1:50 w/w. Digestion was stopped by addition of trifluoroacetic (TFA) to 1% v/v and peptides were cleaned up according to the iST protocol using styroldivinylbenzol-reversed phase sulfonate material (SDB-RPS; Empore 3M, Germany) on in house-packed spin cartridges.140 In brief, acidified peptides were loaded by centrifugation at 400g, washed twice with 200μL 1% v/v TFA in isopropanol, washed twice with 200μL 0.2% v/v TFA in water, eluted with 150μL 1% ammonia in 80% acetonitrile, and dried in a vacuum concentrator (Concentrator plus, Eppendorf, Germany). Peptides were then resuspended in 150 μL A∗ buffer (2% v/v acetonitrile, 0.1% v/v formic acid) aided by sonication and concentration was measured spectroscopically at 280nm (Nanodrop 2000, Thermo Scientific). One aliquot was kept for protein abundance ‘total’ proteomics while the bulk was used for phosphorylation site proteomics. For that purpose, 80μg of purified peptide were subjected to phosphopeptide enrichment using Fe(III)-NTA cartridges on the AssayMAP Bravo platform (Agilent, USA) according to the manufacturer’s instructions. The eluate was dried and resuspended in 5 μL A∗ buffer.
Ultra-high pressure liquid chromatography and mass spectrometry
Samples were measured using an EASY-nLC 1200 (Thermo Fisher Scientific) coupled to a Q Exactive HF and a Q Exactive HF-X Orbitrap mass spectrometer (Thermo Fisher Scientific) in case of the proteome and phospho proteome samples, respectively. Purified peptides were separated on 50cm UHPLC columns with an inner diameter of 75μm packed in-house with ReproSil-Pur C18-AQ 1.9μm resin (Dr.Maisch GmbH) and ions were generated by a nano-electrospray ion source (Thermo Fisher Scientific). About 500ng of purified un-enriched peptides and the entire phosphopeptide enrichment eluate per sample corresponding to less than 500ng were loaded on the liquid chromatography column. Un-enriched peptides were eluted at a flow rate of 250 nL/min and a temperature of 60°C over a 160min gradient with decreasing concentration of buffer A (0.1% v/v formic acid in water) and concomitantly increasing concentration of buffer B (0.1% v/v formic acid, 80% v/v acetonitrile). The gradient used for separation of un-enriched peptides started at 2% v/v B, followed by several phases of linear increases to 5% at minute 3, to 25% at minute 109, to 35% at minute 136, to 60% at minute 148, to 95% at minute 151, a plateau at 95% up to minute 154, a decrease to 5% reached at minute 157 and further plateau at 5% until minute 160. Phosphopeptide-enriched samples were separated by a gradient starting at 3% B, followed by linear increases to 19% at minute 60, to 41% at minute 90, to 90% at minute 95, and a plateau at 90% until minute 100 at a flow of 300 nL/min at 60°C.
Generally, the mass spectrometers were operated by the Xcalibur software (Thermo Fisher) and MS/MS data were recorded in the data-dependent acquisition (DDA) mode. Survey scan (MS1) settings included an ion target value of 3 × 106 charges in the 300–1650 m/z range with a maximum injection time of 25 ms and a resolution of 60,000 at m/z 200. For non-enriched peptides, up to 15 MS/MS spectra with an ion target value of 105 charges, a maximum injection time of 25 ms, a resolution of 15,000 at m/z 200, a 1.4 m/z precursor isolation window, and a 20s dynamic exclusion list were recorded per DDA cycle. For phospho-enriched peptides, up to 10 MS/MS spectra were acquired per cycle with identical parameters but a maximum injection time of 50ms, a 1.6 m/z isolation window, and a dynamic exclusion window of 30 s. Precursors ions with charges other than 2-5 were not selected for MS/MS events. Fragmentation was performed by higher-energy C-trap dissociation (HCD) with a normalized collision energy of 27eV.
Quantification and statistical analysis
DNA methylation and copy number variation data analysis
DNA based classification was performed for 850k data using the publicly available “brain tumor classifier”, version v11b4, ref. 14. One oligodendroglioma sample with 1p/19q codeletion confirmed by EPIC analyses was surprisingly assigned ‘high grade astrocytoma’ by this algorithm. We thus reanalyzed this sample with the recently improved classifier version v12.5133 which assigned the new class ‘oligosarcoma’ to this sample in line with its oligodendroglial origin.
Copy number variations were calculated from IDAT files using the R/Bioconductor conumee package, and were visualized using ComplexHeatmaps v2.10.0. The Integrative Genomics Viewer (IGV) was used to assess chromosomal gains and losses considering the tumor cell content.134,135,141 In general, changes were considered potentially relevant if the intensity ratio of a segment deviation from the baseline by more than 0.1, ref. 14. In addition, we made summary copy number profiles for both IDH mutated groups (A and B). This analysis was done using an adaption of the conumee script (provided by Dr. Damian Stichel, Neuropathology Heidelberg). This algorithm allowed determining CNV load (CNV-L) for each tumor, resulting in a split for a value of 349.695.798 base pairs.15 This CNV load refers to the sum of all gains and deletions as determined by analysis of the 850k raw data by our performed algorithm.
Progression-free survival analysis
Outcome data were available in 20 of 21 IDHmut A/B patients. Progression free survival analysis was performed using the R v4.1.1 packages survival v3.1-8 and survminer v0.9.9, ref. 136 and 137. We used Kaplan-Meier estimates to investigate differences in progression free survival. The starting point for the analysis of the results was the date of the first histological diagnosis of a glioma. As an “event” for progression analysis, we defined either the performance of another surgical intervention (using the date of surgery as event date) or if available tumor progression on MRI (using the date of the MRI as event date). No patients died during follow up period. Median follow up time was 6.5 years (range 1 year–14 years).
Mass spectrometry data processing
To process MS raw files, we employed the MaxQuant software versions 1.5.8.4 and 1.6.0.15 for the un-enriched peptides and phospho-enriched peptides, respectively.131 Spectra were searched against the UniProtKB human FASTA database of canonical and isoform protein sequences downloaded in March 2018 and comprising 93,786 entries. Default search parameters were utilized unless stated differently. In brief, tryptic peptides with a minimum length of 7 amino acids, a maximum mass of 4600 Da, and two miscleavages at maximum were searched. Carbamidomethlyation was set as a fixed modification and methionine oxidation and protein N-terminal acetylation as variable modifications, for the search of phospho-enriched peptides phosphorylation of serine, threonine and tyrosine was additionally included. A maximum of five modifications per peptide was permitted. A false discovery rate (FDR) cutoff of 1% was applied at the peptide and protein level. The search feature “Match between runs,” which allows the transfer of peptide identifications in the absence of MS/MS-based identification after nonlinear retention time alignment was enabled with a maximum retention time window of 0.7 min. Protein abundances were normalized with the MaxLFQ label-free normalization algorithm incorporated in MaxQuant.
Data preprocessing and bioinformatic analysis
Data analysis was mainly performed in the Perseus environment version 1.6.1.3 and in version 1.6.0.9 for correlation analysis.132 Potential contaminants, proteins only identified by site, and search decoys were excluded from further analysis. Protein abundance was log2-transformed and proteins that were not quantified in at least seven samples of at least one of the three WHO entities and one ctrl (IDHwt, IDHmut, 1p/19q-codel, ctrl CNS) were removed. Missing values for protein abundances were imputed according to Perseus default settings from normal distributions around the detection limit with a SD of 0.3 times that of the observed protein distribution of that sample and a downshift of 1.8 standard deviations. Volcano plots were generated in Perseus and FDR-controlled by q-value calculation using a permutation strategy in conjunction with a SAM-statistic with an s0-parameter of 0.1.142 Proteins above the cutoff line are significant according to q-value <5%. Bar plots and boxplots with boxes and whiskers depicting the inter-quartile and minimum to maximum range, respectively, were created in GraphPad Prism 9. T-tests with Welch’s correction were used for group comparisons in these plots. Significance levels were p < 0.05, <0.01, <0.001, and <0.0001 indicated by one to four stars. Heat maps and hierarchical clusters were generated in Perseus. Protein intensities were Z-Scored or normalized by subtraction of the median intensity across samples as stated in the figure legends. In case of sample groups, mean group intensities were used. Principal component analysis, Pearson correlation analysis, and one-way ANOVA with s0 parameter-modified test statistic were performed using Perseus tools. Analysis of ATRX-correlating proteins was limited to IDHmut tumor samples as ATRX loss is mostly restricted to IDHmut gliomas.143
Phosphoproteome-specific analysis
For phosphosite analysis, the phospho (STY) site table generated by MaxQuant containing multiplicity-level quantification was transformed to phosphosite-level quantification by the Perseus expand site table feature. Like protein data, phosphosite abundances were log2-transformed and filtered for seven quantifications. Moreover, phosphosites were filtered for high localization confidence (p > 0.75), termed class I sites, and the median phosphosite intensity of each sample was subtracted from all phosphosites within that sample. Subsequently, missing values were imputed as above.
For the analysis of the glycolysis enzyme phosphorylation signature, phosphosite abundances were normalized to their matching protein abundance for each sample. To avoid amplification of error due to missing values and imputation on both protein and phosphopeptide level, unimputed protein intensities were subtracted from unimputed phosphosite intensities in log2-space, equivalent to division in linear space. Subsequently, the median was subtracted within samples, normalized phosphosites filtered for at least seven quantifications in at least one of the WHO sample groups and imputation performed as above.
Proteome annotations, chromosomal analysis, and survival associations
Proteins were annotated via the in-built Perseus function with gene ontology terms for biological processes (GOBP), cellular compartments (GOCC), molecular function (GOMF), UniProt Keywords, UniProt protein families, UniProt protein-protein interactions (‘interacts with’), and genomic information including chromosomal and base pair position.
Chromosome arm area abundances were calculated as mean abundance of all proteins with corresponding chromosomal position and normalized by subtracting the median abundance within samples. For heatmap visualization, the median abundance within proteins across samples was subtracted. Genome assembly issues and the Y chromosome for which only one protein was quantified were excluded.
Annotation term enrichment on the PCA loadings, i.e. protein contributions, to the two main principal components was performed with the 2D enrichment tool in Perseus.144 Annotations with a Benjamini-Hochberg-adjusted q-value higher than 0.5% and or less than 5 proteins were excluded. Remaining annotations were manually assigned to the meta-categories such as ‘chromatin’.
For analysis of functional metabolic groups of proteins, proteins were selected according to the terms ‘glycolysis’ (UniProt Keywords) and ‘tricarboxylic acid cycle’ (UniProt Keywords), and the terms for ‘mitochondrial respirator chain complex’ I-IV (GOCC), respectively, and ‘mitochondrial proton-transporting ATP synthase complex’ as complex V (GOCC). Tumor suppressors and oncoproteins were filtered according to UniProt Keywords annotation for ‘tumor suppressor genes’ and ‘proto-oncogenes’.
Survival associations mapped onto outlier proteins regulated on chr. 10, chr. 7, and arms 1p and 19q derived from TCGA glioma Affymetrix human exon 1.0 ST transcriptomics data accessed via betastasis (http://www.betastasis.com). High and low expression were separated by the median expression value. Significance of association calculated by the log -rank test were taken from betastasis.
Tumor content analyses
Various controls were performed to check for potential associations of the proteome-based sub-stratification of IDHmut tumors with clinical parameters or tumor content (see supplemental information). For stratifications into ‘high’ and ‘low’ groups for parameters, e.g. solid tumor content, sample assignment was made in the way entailing maximum parameter separation while maintaining balanced composition of the two groups regarding HGG-IDHmut-B and HGG-IDHmut-A. Regression analysis was performed using the lm() linear model in R version 3.6.3, fitting protein abundances based on HGG-IDHmut-A/B status, 1p/19q codeletion status, and two tumor content variables (i: solid tumor area, and ii: infiltration zone area) among IDHmut samples.
Further, proteome differences were compared to cell type differences between isolated astrocytes and neurons using available murine proteomics data.130 Proteins were matched based on identical gene names. Mouse cell data were filtered for astrocytes (n = 3), oligodendrocytes at DIV4 (n = 3), neurons at DIV15, and adult microglia (n = 3). Similar to our proteome data, proteins without at least two out of three quantifications in at least one cell type were removed and subsequently missing values were imputed as above.
Integration with other glioma proteome datasets
Proteome differences between HGG-IDHmut-A and B were compared to published glioma proteome data.25,119,120 The CPTAC/Wang dataset stratified glioma samples by a multi-omics-defined phenotype: 29 nmf1/proneural-like, 37 nmf2/mesenchymal-like, 26 nmf3/classical-like, 6 IDH mutant (IDH1 R132H), and 10 non-neoplastic ctrl CNS. One sample with a non-hotspot IDH-mutation (R222C) in the CPTAC dataset, assigned to nmf3/classical-like, was excluded. The dataset was further filtered for proteins with at least 70% observations in at least 1 ‘multi-omic’ group reducing the dataset from 10,998 to 10,206 proteins. Remaining missing values were imputed analogously as in our dataset. A list of the human proteome with RefSeq and UniProt IDs for each protein was downloaded from UniProt to match proteins identified by RefSeq ID in the CPTAC dataset to proteins specified by UniProt IDs in our dataset, 5195 proteins overlapped. The Oh et al. study comprised 26 GPC1 (GBM proteomic cluster 1, high glycolysis) subtype and 13 GPC2 (high oxidative phosphorylation) subtype IDHwt glioblastoma samples, which were used in their main GPC1/GPC2 clustering and the re-analysis in our study. The proteome dataset contained 3909 proteins without missing values. Proteins were specified by UniProt IDs which we used to match proteins to our data, 3715 proteins overlapped. The Wong dataset comprised 6 control CNS samples, 21 IDH-mutant 1p/19q-codeleted (Wong et al.: ‘type I’), 17 IDH-mutant 1p/19q non-codeleted (‘type II’), 10 IDH-wt (‘type III’) glioma samples and additional reference samples which we removed from further analysis. The dataset comprising 7988 protein groups was filtered for proteins present in all samples reducing the number to 5897 proteins. Proteins were matched to our dataset based on UniProt IDs. Our hierarchical clustering classified the IDHmut glioma into 10 IDHmut-A (comprising 6 ‘type I’ and 4 ‘type II’) and 28 IDHmut-B (comprising 15 ‘type I’ and 13 ‘type II’).
Acknowledgments
We thank Igor Paron for technical assistance and Fabian Coscia for advice regarding the proteomic workflow, as well as Elis Perez and Amos Münch for the evaluation of the CNV data and statistical analyses. Funding was provided by the Max Planck Society for the Advancement of Science and the German Research Foundation DFG under project IDs 360372040 (SFB 1335), 408885537 (TRR 274), and 390873048 (EXC 2151 ImmunoSensation2).
Author contributions
J.M.B. prepared the samples for MS, acquired and analyzed mass spectrometric data, generated figures, and drafted the manuscript. J.M.B. and F.M. analysed and interpreted data. N.D., M. Misch, and A.K. selected and analyzed the samples histopathologically and interpreted the data. A.K. performed the CNV and EPIC analyses, analyzed the data, and generated figures. F.M., N.D., and M. Mann conceived and guided the study. F.M., N.D., and A.K. edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: December 29, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100877.
Contributor Information
Arend Koch, Email: arend.koch@charite.de.
Felix Meissner, Email: felix.meissner@uni-bonn.de.
Supporting citations
The following reference appears in the Supplemental Information: 145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160.
Supplemental information
Data and code availability
-
•
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
The raw methylation data and the CNV profiles calculated from them have been deposited to the Gene Expression Omnibus – NCBI repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
The paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Ostrom Q.T., Bauchet L., Davis F.G., Deltour I., Fisher J.L., Langer C.E., Pekmezci M., Schwartzbaum J.A., Turner M.C., Walsh K.M., et al. The epidemiology of glioma in adults: a state of the science review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Classification of Tumours Editorial Board . 5th ed. International Agency for Research on Cancer; 2021. World Health Organization Classification of Tumours of the Central Nervous System. [Google Scholar]
- 3.Dolecek T.A., Propp J.M., Stroup N.E., Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro Oncol. 2012;14:v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J.B., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 5.Ohgaki H., Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J., et al. Mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu N., Richards R., Jensen R. Role of chromosomal 1p/19q co-deletion on the prognosis of oligodendrogliomas: a systematic review and meta-analysis. Interdiscip. Neurosurg. 2016;5:58–63. doi: 10.1016/j.inat.2016.06.008. [DOI] [Google Scholar]
- 8.Chen J., McKay R.M., Parada L.F. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins R.B., Blair H., Ballman K.V., Giannini C., Arusell R.M., Law M., Flynn H., Passe S., Felten S., Brown P.D., et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 10.Leeper H.E., Caron A.A., Decker P.A., Jenkins R.B., Lachance D.H., Giannini C. IDH mutation, 1p19q codeletion and ATRX loss in WHO grade II gliomas. Oncotarget. 2015;6:30295–30305. doi: 10.18632/oncotarget.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riddick G., Fine H.A. Integration and analysis of genome-scale data from gliomas. Nat. Rev. Neurol. 2011;7:439–450. doi: 10.1038/nrneurol.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin N., Yan W., Gao K., Wang Y., Zhang J., You Y. Prevalence and clinicopathologic characteristics of the molecular subtypes in malignant glioma: a multi-institutional analysis of 941 cases. PLoS One. 2014;9:e94871. doi: 10.1371/journal.pone.0094871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceccarelli M., Barthel F.P., Malta T.M., Sabedot T.S., Salama S.R., Murray B.A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S.M., et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capper D., Jones D.T.W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D.E., et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirahata M., Ono T., Stichel D., Schrimpf D., Reuss D.E., Sahm F., Koelsche C., Wefers A., Reinhardt A., Huang K., et al. Novel, improved grading system(S) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018;136:153–166. doi: 10.1007/s00401-018-1849-4. [DOI] [PubMed] [Google Scholar]
- 16.Aebersold R., Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 17.Coscia F., Lengyel E., Duraiswamy J., Ashcroft B., Bassani-Sternberg M., Wierer M., Johnson A., Wroblewski K., Montag A., Yamada S.D., et al. Multi-level proteomics identifies CT45 as a chemosensitivity mediator and immunotherapy target in ovarian cancer. Cell. 2018;175:159–170.e16. doi: 10.1016/j.cell.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coscia F., Doll S., Bech J.M., Schweizer L., Mund A., Lengyel E., Lindebjerg J., Madsen G.I., Moreira J.M., Mann M. A streamlined mass spectrometry–based proteomics workflow for large-scale FFPE tissue analysis. J. Pathol. 2020;251:100–112. doi: 10.1002/path.5420. [DOI] [PubMed] [Google Scholar]
- 19.Kohale I.N., Burgenske D.M., Mladek A.C., Bakken K.K., Kuang J., Boughey J.C., Wang L., Carter J.M., Haura E.B., Goetz M.P., et al. Quantitative analysis of tyrosine phosphorylation from FFPE tissues reveals patient-specific signaling networks. Cancer Res. 2021;81:3930–3941. doi: 10.1158/0008-5472.CAN-21-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wakabayashi M., Yoshihara H., Masuda T., Tsukahara M., Sugiyama N., Ishihama Y. Phosphoproteome analysis of formalin-fixed and paraffin-embedded tissue sections mounted on microscope slides. J. Proteome Res. 2014;13:915–924. doi: 10.1021/pr400960r. [DOI] [PubMed] [Google Scholar]
- 21.Friedrich C., Schallenberg S., Kirchner M., Ziehm M., Niquet S., Haji M., Beier C., Neudecker J., Klauschen F., Mertins P. Comprehensive micro-scaled proteome and phosphoproteome characterization of archived retrospective cancer repositories. Nat. Commun. 2021;12:3576. doi: 10.1038/s41467-021-23855-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson H.J., Socciarelli F., Vacanti N.M., Haugen M.H., Zhu Y., Siavelis I., Fernandez-Woodbridge A., Aure M.R., Sennblad B., Vesterlund M., et al. Breast cancer quantitative proteome and proteogenomic landscape. Nat. Commun. 2019;10:1600–1614. doi: 10.1038/s41467-019-09018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouchal P., Schubert O.T., Faktor J., Capkova L., Imrichova H., Zoufalova K., Paralova V., Hrstka R., Liu Y., Ebhardt H.A., et al. Breast cancer classification based on proteotypes obtained by SWATH mass spectrometry. Cell Rep. 2019;28:832–843.e7. doi: 10.1016/j.celrep.2019.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zagorac I., Fernandez-Gaitero S., Penning R., Post H., Bueno M.J., Mouron S., Manso L., Morente M.M., Alonso S., Serra V., et al. In vivo phosphoproteomics reveals kinase activity profiles that predict treatment outcome in triple-negative breast cancer. Nat. Commun. 2018;9:3501. doi: 10.1038/s41467-018-05742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L.-B., Karpova A., Gritsenko M.A., Kyle J.E., Cao S., Li Y., Rykunov D., Colaprico A., Rothstein J.H., Hong R., et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39:509–528.e20. doi: 10.1016/j.ccell.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sim H., Hu B., Viapiano M.S. Reduced expression of the hyaluronan and proteoglycan link proteins in malignant gliomas. J. Biol. Chem. 2009;284:26547–26556. doi: 10.1074/jbc.M109.013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres C.M., Biran A., Burney M.J., Patel H., Henser-Brownhill T., Cohen A.H.S., Li Y., Ben-Hamo R., Nye E., Spencer-Dene B., et al. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science. 2016;353:aaf1644. doi: 10.1126/science.aaf1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaspar-Maia A., Qadeer Z.A., Hasson D., Ratnakumar K., Leu N.A., Leroy G., Liu S., Costanzi C., Valle-Garcia D., Schaniel C., et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat. Commun. 2013;4:1565. doi: 10.1038/ncomms2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Libreros S., Garcia-Areas R., Iragavarapu-Charyulu V. CHI3L1 plays a role in cancer through enhanced production of pro-inflammatory/pro-tumorigenic and angiogenic factors. Immunol. Res. 2013;57:99–105. doi: 10.1007/s12026-013-8459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steponaitis G., Skiriutė D., Kazlauskas A., Golubickaitė I., Stakaitis R., Tamašauskas A., Vaitkienė P. High CHI3L1 expression is associated with glioma patient survival. Diagn. Pathol. 2016;11:42. doi: 10.1186/s13000-016-0492-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W., Cazacu S., Xiang C., Zenklusen J.C., Fine H.A., Berens M., Armstrong B., Brodie C., Mikkelsen T. FK506 binding protein mediates glioma cell growth and sensitivity to rapamycin treatment by regulating NF-κB signaling pathway. Neoplasia. 2008;10:235–243. doi: 10.1593/neo.07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu H., Liu P., Yan Y., Fang K., Liang D., Hou X., Zhang X., Wu S., Ma J., Wang R., et al. FKBP9 promotes the malignant behavior of glioblastoma cells and confers resistance to endoplasmic reticulum stress inducers. J. Exp. Clin. Cancer Res. 2020;39:44. doi: 10.1186/s13046-020-1541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge Y., Xu A., Zhang M., Xiong H., Fang L., Zhang X., Liu C., Wu S. FK506 binding protein 10 is overexpressed and promotes renal cell carcinoma. Urol. Int. 2017;98:169–176. doi: 10.1159/000448338. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y., Wang J., Xu S., Shi F., Shan A. Calumenin contributes to epithelial-mesenchymal transition and predicts poor survival in glioma. bioRxiv. 2020 doi: 10.1101/2020.07.05.188318. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han M.Z., Xu R., Xu Y.Y., Zhang X., Ni S.L., Huang B., Chen A.J., Wei Y.Z., Wang S., Li W.J., et al. TAGLN2 is a candidate prognostic biomarker promoting tumorigenesis in human gliomas. J. Exp. Clin. Cancer Res. 2017;36:155. doi: 10.1186/s13046-017-0619-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai C.-K., Huang L.-C., Tsai W.-C., Huang S.-M., Lee J.-T., Hueng D.-Y. Overexpression of PLOD3 promotes tumor progression and poor prognosis in gliomas. Oncotarget. 2018;9:15705–15720. doi: 10.18632/oncotarget.24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tantyo N.A., Karyadi A.S., Rasman S.Z., Salim M.R.G., Devina A., Sumarpo A. The prognostic value of S100A10 expression in cancer. Oncol. Lett. 2019;17:1417–1424. doi: 10.3892/ol.2018.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikas J.B. Independent validation of a mathematical genomic model for survival of glioma patients. Am. J. Cancer Res. 2016;6:1408–1419. [PMC free article] [PubMed] [Google Scholar]
- 39.Sinha S., Bheemsetty V.A., Inamdar M.S. A double helical motif in OCIAD2 is essential for its localization, interactions and STAT3 activation. Sci. Rep. 2018;8:7362. doi: 10.1038/s41598-018-25667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gai X., Tu K., Lu Z., Zheng X. MRC2 expression correlates with TGFβ1 and survival in hepatocellular carcinoma. Int. J. Mol. Sci. 2014;15:15011–15025. doi: 10.3390/ijms150915011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu D., Li W., Zhong F., Yin J., Zhou W., Li S., Sun X., Xu J., Li G., Wen Y., et al. METTL7B is required for cancer cell proliferation and tumorigenesis in non-small cell lung cancer. Front. Pharmacol. 2020;11:178. doi: 10.3389/fphar.2020.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Setti M., Savalli N., Osti D., Richichi C., Angelini M., Brescia P., Fornasari L., Carro M.S., Mazzanti M., Pelicci G. Functional role of CLIC1 ion channel in glioblastoma-derived stem/progenitor cells. J. Natl. Cancer Inst. 2013;105:1644–1655. doi: 10.1093/jnci/djt278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z., Wang X., Zhang N., Zhang H., Dai Z., Zhang M., Feng S., Cheng Q. Pentraxin 3 promotes glioblastoma progression by negative regulating cells autophagy. Front. Cell Dev. Biol. 2020;8:795. doi: 10.3389/fcell.2020.00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chou A.P., Chowdhury R., Li S., Chen W., Kim A.J., Piccioni D.E., Selfridge J.M., Mody R.R., Chang S., Lalezari S., et al. Identification of retinol binding protein 1 promoter hypermethylation in isocitrate dehydrogenase 1 and 2 mutant gliomas. J. Natl. Cancer Inst. 2012;104:1458–1469. doi: 10.1093/jnci/djs357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ju Lee H., Bartsch D., Xiao C., Guerrero S., Ahuja G., Schindler C., Moresco J.J., Yates J.R., Gebauer F., Bazzi H., et al. A post-transcriptional program coordinated by CSDE1 prevents intrinsic neural differentiation of human embryonic stem cells. Nat. Commun. 2017;8:1456. doi: 10.1038/s41467-017-01744-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Useros J., Garcia-Carbonero N., Li W., Fernandez-Aceñero M.J., Cristobal I., Rincon R., Rodriguez-Remirez M., Borrero-Palacios A., Garcia-Foncillas J. UNR/CSDE1 expression is critical to maintain invasive phenotype of colorectal cancer through regulation of c-MYC and epithelial-to-mesenchymal transition. J. Clin. Med. 2019;8:560. doi: 10.3390/jcm8040560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eckert M.A., Coscia F., Chryplewicz A., Chang J.W., Hernandez K.M., Pan S., Tienda S.M., Nahotko D.A., Li G., Blaženović I., et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature. 2019;569:723–728. doi: 10.1038/s41586-019-1173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapoor A., Goldberg M.S., Cumberland L.K., Ratnakumar K., Segura M.F., Emanuel P.O., Menendez S., Vardabasso C., LeRoy G., Vidal C.I., et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y., Zhao W., Liu X., Guan G., Zhuang M. ARL3 is downregulated and acts as a prognostic biomarker in glioma. J. Transl. Med. 2019;17:210–215. doi: 10.1186/s12967-019-1914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang R., Gurguis C.I., Gu W., Ko E.A., Lim I., Bang H., Zhou T., Ko J.H. Ion channel gene expression predicts survival in glioma patients. Sci. Rep. 2015;5:11593. doi: 10.1038/srep11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu H., Ge B., Chen D., Wu Y., Luo Q., Li X., Zheng C., Tang Q. Phytanoyl-CoA 2-hydroxylase-interacting protein-like gene is a therapeutic target gene for glioblastoma multiforme. Med. Sci. Monit. 2019;25:2583–2590. doi: 10.12659/MSM.913895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao S.F., Wang S.G., Zhao Z.Y., Li W.L. AKR1C1-3, notably AKR1C3, are distinct biomarkers for liver cancer diagnosis and prognosis: database mining in malignancies. Oncol. Lett. 2019;18:4515–4522. doi: 10.3892/ol.2019.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang B., Gu Y., Hui K., Huang J., Xu S., Wu S., Li L., Fan J., Wang X., Hsieh J.T., et al. AKR1C3, a crucial androgenic enzyme in prostate cancer, promotes epithelial-mesenchymal transition and metastasis through activating ERK signaling. Urol. Oncol. 2018;36:472.e11–472.e20. doi: 10.1016/j.urolonc.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 54.Cheung H.C., Hai T., Zhu W., Baggerly K.A., Tsavachidis S., Krahe R., Cote G.J. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain. 2009;132:2277–2288. doi: 10.1093/brain/awp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brellier F., Ruggiero S., Zwolanek D., Martina E., Hess D., Brown-Luedi M., Hartmann U., Koch M., Merlo A., Lino M., Chiquet-Ehrismann R. SMOC1 is a tenascin-C interacting protein over-expressed in brain tumors. Matrix Biol. 2011;30:225–233. doi: 10.1016/j.matbio.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 56.Johnson G.S., Rajendran P., Dashwood R.H. CCAR1 and CCAR2 as gene chameleons with antagonistic duality: preclinical, human translational, and mechanistic basis. Cancer Sci. 2020;111:3416–3425. doi: 10.1111/cas.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maurya P.K., Mishra A., Yadav B.S., Singh S., Kumar P., Chaudhary A., Srivastava S., Murugesan S.N., Mani A. Role of Y box protein-1 in cancer: as potential biomarker and novel therapeutic target. J. Cancer. 2017;8:1900–1907. doi: 10.7150/jca.17689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuwano M., Shibata T., Watari K., Ono M. Oncogenic Y-box binding protein-1 as an effective therapeutic target in drug-resistant cancer. Cancer Sci. 2019;110:1536–1543. doi: 10.1111/cas.14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gupta M.K., Polisetty R.V., Sharma R., Ganesh R.A., Gowda H., Purohit A.K., Ankathi P., Prasad K., Mariswamappa K., Lakshmikantha A., et al. Altered transcriptional regulatory proteins in glioblastoma and YBX1 as a potential regulator of tumor invasion. Sci. Rep. 2019;9:10986. doi: 10.1038/s41598-019-47360-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia W., Chen P., Cheng Y. PRDX4 and its roles in various cancers. Technol. Cancer Res. Treat. 2019;18 doi: 10.1177/1533033819864313. 1533033819864313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim T.H., Song J., Alcantara Llaguno S.R., Murnan E., Liyanarachchi S., Palanichamy K., Yi J.Y., Viapiano M.S., Nakano I., Yoon S.O., et al. Suppression of peroxiredoxin 4 in glioblastoma cells increases apoptosis and reduces tumor growth. PLoS One. 2012;7:e42818. doi: 10.1371/journal.pone.0042818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen R., Zhu J., Dong Y., He C., Hu X. Suppressor of Ty homolog-5, a novel tumor-specific human telomerase reverse transcriptase promoter-binding protein and activator in colon cancer cells. Oncotarget. 2015;6:32841–32855. doi: 10.18632/oncotarget.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Govaere O., Petz M., Wouters J., Vandewynckel Y.P., Scott E.J., Topal B., Nevens F., Verslype C., Anstee Q.M., Van Vlierberghe H., et al. The PDGFRα-laminin B1-keratin 19 cascade drives tumor progression at the invasive front of human hepatocellular carcinoma. Oncogene. 2017;36:6605–6616. doi: 10.1038/onc.2017.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsiao K.C., Chu P.Y., Chang G.C., Liu K.J. Elevated expression of lumican in lung cancer cells promotes bone metastasis through an autocrine regulatory mechanism. Cancers. 2020;12:233. doi: 10.3390/cancers12010233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Graille M., Rougemaille M. ERH proteins: connecting RNA processing to tumorigenesis? Curr. Genet. 2020;66:689–692. doi: 10.1007/s00294-020-01065-z. [DOI] [PubMed] [Google Scholar]
- 66.Pang K., Zhang Z., Hao L., Shi Z., Chen B., Zang G., Dong Y., Li R., Liu Y., Wang J., et al. The ERH gene regulates migration and invasion in 5637 and T24 bladder cancer cells. BMC Cancer. 2019;19:225. doi: 10.1186/s12885-019-5423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang D., Chu Y.J., Song K.J., Chen Y.L., Liu W., Lv T., Wang J., Zhao H., Ren Y.Z., Xu J.X., et al. Knockdown of enhancer of rudimentary homolog inhibits proliferation and metastasis in ovarian cancer by regulating epithelial-mesenchymal transition. Biomed. Pharmacother. 2020;125:109974. doi: 10.1016/j.biopha.2020.109974. [DOI] [PubMed] [Google Scholar]
- 68.Gawrzak S., Rinaldi L., Gregorio S., Arenas E.J., Salvador F., Urosevic J., Figueras-Puig C., Rojo F., Del Barco Barrantes I., Cejalvo J.M., et al. MSK1 regulates luminal cell differentiation and metastatic dormancy in ER + breast cancer. Nat. Cell Biol. 2018;20:211–221. doi: 10.1038/s41556-017-0021-z. [DOI] [PubMed] [Google Scholar]
- 69.Wu S., Wang S., Zheng S., Verhaak R., Koul D., Yung W.K.A. MSK1-Mediated βb-catenin phosphorylation confers resistance to PI3K/mTOR inhibitors in glioblastoma. Mol. Cancer Ther. 2016;15:1656–1668. doi: 10.1158/1535-7163.MCT-15-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou X., Xia E., Bhandari A., Zheng C., Xiang J., Guan Y., Zhang X. LRP4 promotes proliferation, migration, and invasion in papillary thyroid cancer. Biochem. Biophys. Res. Commun. 2018;503:257–263. doi: 10.1016/j.bbrc.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 71.Jiang J., Ren H., Xu Y., Wudu M., Wang Q., Liu Z., Su H., Jiang X., Zhang Y., Zhang B., Qiu X. TRIM67 promotes the proliferation, migration, and invasion of non-small-cell lung cancer by positively regulating the Notch pathway. J. Cancer. 2020;11:1240–1249. doi: 10.7150/jca.38286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S., Zhang Y., Huang J., Wong C.C., Zhai J., Li C., Wei G., Zhao L., Wang G., Wei H., et al. TRIM67 activates p53 to suppress colorectal cancer initiation and progression. Cancer Res. 2019;79:4086–4098. doi: 10.1158/0008-5472.CAN-18-3614. [DOI] [PubMed] [Google Scholar]
- 73.Kanda M., Nomoto S., Okamura Y., Hayashi M., Hishida M., Fujii T., Nishikawa Y., Sugimoto H., Takeda S., Nakao A. Promoter hypermethylation of fibulin 1 gene is associated with tumor progression in hepatocellular carcinoma. Mol. Carcinog. 2011;50:571–579. doi: 10.1002/mc.20735. [DOI] [PubMed] [Google Scholar]
- 74.Xiao W., Wang J., Li H., Xia D., Yu G., Yao W., Yang Y., Xiao H., Lang B., Ma X., et al. Fibulin-1 is epigenetically down-regulated and related with bladder cancer recurrence. BMC Cancer. 2014;14:677. doi: 10.1186/1471-2407-14-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xia S., Lal B., Tung B., Wang S., Goodwin C.R., Laterra J. Tumor microenvironment tenascin-C promotes glioblastoma invasion and negatively regulates tumor proliferation. Neuro Oncol. 2016;18:507–517. doi: 10.1093/neuonc/nov171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshida T., Akatsuka T., Imanaka-Yoshida K. Tenascin-C and integrins in cancer. Cell Adh. Migr. 2015;9:96–104. doi: 10.1080/19336918.2015.1008332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye Y.P., Jiao H.L., Wang S.Y., Xiao Z.Y., Zhang D., Qiu J.F., Zhang L.J., Zhao Y.L., Li T.T., Li-Liang, et al. Hypermethylation of DMTN promotes the metastasis of colorectal cancer cells by regulating the actin cytoskeleton through Rac1 signaling activation 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis 06 Biological Sciences 0604 Genetics. J. Exp. Clin. Cancer Res. 2018;37:299. doi: 10.1186/s13046-018-0958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun S.C. CYLD: a tumor suppressor deubiquitinase regulating NF-B activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X., Wang J., Jia Y., Liu T., Wang M., Lv W., Zhang R., Shi J., Liu L. CDK5 neutralizes the tumor suppressing effect of BIN1 via mediating phosphorylation of c-MYC at Ser-62 site in NSCLC. Cancer Cell Int. 2019;19:226. doi: 10.1186/s12935-019-0952-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakahata S., Ichikawa T., Maneesaay P., Saito Y., Nagai K., Tamura T., Manachai N., Yamakawa N., Hamasaki M., Kitabayashi I., et al. Loss of NDRG2 expression activates PI3K-AKT signalling via PTEN phosphorylation in ATLL and other cancers. Nat. Commun. 2014;5:3393. doi: 10.1038/ncomms4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bencivenga D., Caldarelli I., Stampone E., Mancini F.P., Balestrieri M.L., Della Ragione F., Borriello A. p27Kip1 and human cancers: a reappraisal of a still enigmatic protein. Cancer Lett. 2017;403:354–365. doi: 10.1016/j.canlet.2017.06.031. [DOI] [PubMed] [Google Scholar]
- 82.Lobbous M., Bernstock J.D., Coffee E., Friedman G.K., Metrock L.K., Chagoya G., Elsayed G., Nakano I., Hackney J.R., Korf B.R., Nabors L.B. An update on neurofibromatosis type 1-associated gliomas. Cancers. 2020;12:114. doi: 10.3390/cancers12010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dashzeveg N., Yoshida K. Crosstalk between tumor suppressors p53 and PKCδ: execution of the intrinsic apoptotic pathways. Cancer Lett. 2016;377:158–163. doi: 10.1016/j.canlet.2016.04.032. [DOI] [PubMed] [Google Scholar]
- 84.Baumann U., Fern'ndez-S'iz V., Rudelius M., Lemeer S., Rad R., Knorn A.M., Slawska J., Engel K., Jeremias I., Li Z., et al. Disruption of the PRKCD-FBXO25-HAX-1 axis attenuates the apoptotic response and drives lymphomagenesis. Nat. Med. 2014;20:1401–1409. doi: 10.1038/nm.3740. [DOI] [PubMed] [Google Scholar]
- 85.Allen-Petersen B.L., Carter C.J., Ohm A.M., Reyland M.E. Protein kinase Cδ is required for ErbB2-driven mammary gland tumorigenesis and negatively correlates with prognosis in human breast cancer. Oncogene. 2014;33:1306–1315. doi: 10.1038/onc.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bignone P.A., Lee K.Y., Liu Y., Emilion G., Finch J., Soosay A.E.R., Charnock F.M.L., Beck S., Dunham I., Mungall A.J., Ganesan T.S. RPS6KA2, a putative tumour suppressor gene at 6q27 in sporadic epithelial ovarian cancer. Oncogene. 2007;26:683–700. doi: 10.1038/sj.onc.1209827. [DOI] [PubMed] [Google Scholar]
- 87.Stone A.R., Bobo W., Brat D.J., Devi N.S., Van Meir E.G., Vertino P.M. Aberrant methylation and down-regulation of TMS1/ASC in human glioblastoma. Am. J. Pathol. 2004;165:1151–1161. doi: 10.1016/S0002-9440(10)63376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharma N., Saxena S., Agrawal I., Singh S., Srinivasan V., Arvind S., Epari S., Paul S., Jha S. Differential expression profile of NLRs and AIM2 in glioma and implications for NLRP12 in glioblastoma. Sci. Rep. 2019;9:8480. doi: 10.1038/s41598-019-44854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martinon F., Burns K., Tschopp J. The Inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell. 2002;10:417–426. doi: 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 90.Behnan J., Stangeland B., Hosainey S.A.M., Joel M., Olsen T.K., Micci F., Glover J.C., Isakson P., Brinchmann J.E. Differential propagation of stroma and cancer stem cells dictates tumorigenesis and multipotency. Oncogene. 2017;36:570–584. doi: 10.1038/onc.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Figliuolo da Paz V., Ghishan F.K., Kiela P.R. Emerging roles of disabled homolog 2 (DAB2) in immune regulation. Front. Immunol. 2020;11:580302–580311. doi: 10.3389/fimmu.2020.580302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamini B. NF-κB, mesenchymal differentiation and glioblastoma. Cells. 2018;7:125. doi: 10.3390/cells7090125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oprita A., Baloi S.C., Staicu G.A., Alexandru O., Tache D.E., Danoiu S., Micu E.S., Sevastre A.S. Updated insights on EGFR signaling pathways in glioma. Int. J. Mol. Sci. 2021;22:587. doi: 10.3390/ijms22020587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pu P., Kang C., Li J., Jiang H., Cheng J. The effects of antisense AKT2 RNA on the Inhibition of malignant glioma cell growth in vitro and in vivo. J. Neuro Oncol. 2006;76:1–11. doi: 10.1007/s11060-005-3029-3. [DOI] [PubMed] [Google Scholar]
- 95.Kim E.H., Jo Y., Sai S., Park M.J., Kim J.Y., Kim J.S., Lee Y.J., Cho J.M., Kwak S.Y., Baek J.H., et al. Tumor-treating fields induce autophagy by blocking the Akt2/miR29b axis in glioblastoma cells. Oncogene. 2019;38:6630–6646. doi: 10.1038/s41388-019-0882-7. [DOI] [PubMed] [Google Scholar]
- 96.Saito S., Cigdem S., Okuwaki M., Nagata K. Leukemia-associated Nup214 fusion proteins disturb the XPO1-mediated nuclear-cytoplasmic transport pathway and thereby the NF-κB signaling pathway. Mol. Cell Biol. 2016;36:1820–1835. doi: 10.1128/mcb.00158-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tao T., Shi Y., Han D., Luan W., Qian J., Zhang J., Wang Y., You Y., Chinese Glioma Cooperative Group CGCG TPM3, a strong prognosis predictor, is involved in malignant progression through MMP family members and EMT-like activators in gliomas. Tumour Biol. 2014;35:9053–9059. doi: 10.1007/s13277-014-1974-1. [DOI] [PubMed] [Google Scholar]
- 98.Su Y., Huang J., Hu J. M6 a rna methylation regulators contribute to malignant progression and have clinical prognostic impact in gastric cancer. Front. Oncol. 2019;9:1038. doi: 10.3389/fonc.2019.01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee Y., Lee J.K., Ahn S.H., Lee J., Nam D.H. WNT signaling in glioblastoma and therapeutic opportunities. Lab. Invest. 2016;96:137–150. doi: 10.1038/labinvest.2015.140. [DOI] [PubMed] [Google Scholar]
- 100.Zuccarini M., Giuliani P., Ziberi S., Carluccio M., Iorio P.D., Caciagli F., Ciccarelli R. The role of wnt signal in glioblastoma development and progression: a possible new pharmacological target for the therapy of this tumor. Genes. 2018;9:105. doi: 10.3390/genes9020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Manoranjan B., Chokshi C., Venugopal C., Subapanditha M., Savage N., Tatari N., Provias J.P., Murty N.K., Moffat J., Doble B.W., Singh S.K. A CD133-AKT-Wnt signaling axis drives glioblastoma brain tumor-initiating cells. Oncogene. 2020;39:1590–1599. doi: 10.1038/s41388-019-1086-x. [DOI] [PubMed] [Google Scholar]
- 102.Chen Y., Huang Q., Liu W., Zhu Q., Cui C.P., Xu L., Guo X., Wang P., Liu J., Dong G., et al. Mutually exclusive acetylation and ubiquitylation of the splicing factor SRSF5 control tumor growth. Nat. Commun. 2018;9:2464. doi: 10.1038/s41467-018-04815-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nandakumar P., Mansouri A., Das S. The role of ATRX in glioma biology. Front. Oncol. 2017;7:236. doi: 10.3389/fonc.2017.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Clynes D., Jelinska C., Xella B., Ayyub H., Scott C., Mitson M., Taylor S., Higgs D.R., Gibbons R.J. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 2015;6:7538. doi: 10.1038/ncomms8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mukherjee J., Johannessen T.C., Ohba S., Chow T.T., Jones L., Pandita A., Pieper R.O. Mutant IDH1 cooperates with ATRX loss to drive the alternative lengthening of telomere phenotype in glioma. Cancer Res. 2018;78:2966–2977. doi: 10.1158/0008-5472.CAN-17-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Walsh K.M., Wiencke J.K., Lachance D.H., Wiemels J.L., Molinaro A.M., Eckel-Passow J.E., Jenkins R.B., Wrensch M.R. Telomere maintenance and the etiology of adult glioma. Neuro Oncol. 2015;17:1445–1452. doi: 10.1093/neuonc/nov082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ferreira M.S.V., Sørensen M.D., Pusch S., Beier D., Bouillon A.-S., Kristensen B.W., Brümmendorf T.H., Beier C.P., Beier F. Alternative lengthening of telomeres is the major telomere maintenance mechanism in astrocytoma with isocitrate dehydrogenase 1 mutation. J. Neuro Oncol. 2020;147:1–14. doi: 10.1007/s11060-020-03394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Körber V., Yang J., Barah P., Wu Y., Stichel D., Gu Z., Fletcher M.N.C., Jones D., Hentschel B., Lamszus K., et al. Evolutionary trajectories of IDH WT glioblastomas reveal a common path of early tumorigenesis instigated years ahead of initial diagnosis. Cancer Cell. 2019;35:692–704.e12. doi: 10.1016/j.ccell.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 109.Crespo I., Vital A.L., Nieto A.B., Rebelo O., Tão H., Lopes M.C., Oliveira C.R., French P.J., Orfao A., Tabernero M.D. Detailed characterization of alterations of chromosomes 7, 9, and 10 in glioblastomas as assessed by single-nucleotide polymorphism arrays. J. Mol. Diagn. 2011;13:634–647. doi: 10.1016/j.jmoldx.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lopez-Gines C., Cerda-Nicolas M., Gil-Benso R., Pellin A., Lopez-Guerrero J.A., Callaghan R., Benito R., Roldan P., Piquer J., Llacer J., Barbera J. Association of chromosome 7, chromosome 10 and EGFR gene amplification in glioblastoma multiforme. Clin. Neuropathol. 2005;24:209–218. [PubMed] [Google Scholar]
- 111.Leone G., Abla H., Gasparre G., Porcelli A.M., Iommarini L. The oncojanus paradigm of respiratory complex I. Genes. 2018;9:243. doi: 10.3390/genes9050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sen T., Sen N., Noordhuis M.G., Ravi R., Wu T.C., Ha P.K., Sidransky D., Hoque M.O. OGDHL is a modifier of AKT-dependent signaling and NF-κB function. PLoS One. 2012;7:e48770. doi: 10.1371/journal.pone.0048770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tseng P.L., Chen C.W., Hu K.H., Cheng H.C., Lin Y.H., Tsai W.H., Cheng T.J., Wu W.H., Yeh C.W., Lin C.C., et al. The decrease of glycolytic enzyme hexokinase 1 accelerates tumor malignancy via deregulating energy metabolism but sensitizes cancer cells to 2-deoxyglucose inhibition. Oncotarget. 2018;9:18949–18969. doi: 10.18632/oncotarget.24855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Formentini L., S'nchez-Aragó M., S'nchez-Cenizo L., Cuezva J.M. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Mol. Cell. 2012;45:731–742. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 115.García-Bermúdez J., S'nchez-Aragó M., Soldevilla B., Del Arco A., Nuevo-Tapioles C., Cuezva J.M. PKA phosphorylates the ATPase inhibitory factor 1 and inactivates its capacity to bind and inhibit the mitochondrial H(+)-ATP synthase. Cell Rep. 2015;12:2143–2155. doi: 10.1016/j.celrep.2015.08.052. [DOI] [PubMed] [Google Scholar]
- 116.Lymbouridou R., Soufla G., Chatzinikola A.M., Vakis A., Spandidos D.A. Down-regulation of K-ras and H-ras in human brain gliomas. Eur. J. Cancer. 2009;45:1294–1303. doi: 10.1016/j.ejca.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 117.Timofeev O., Cizmecioglu O., Settele F., Kempf T., Hoffmann I. Cdc25 phosphatases are required for timely assembly of CDK1-cyclin B at the G2/M transition. J. Biol. Chem. 2010;285:16978–16990. doi: 10.1074/jbc.M109.096552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou Y., Shen J.K., Hornicek F.J., Kan Q., Duan Z. The emerging roles and therapeutic potential of cyclin-dependent kinase 11 (CDK11) in human cancer. Oncotarget. 2016;7:40846–40859. doi: 10.18632/oncotarget.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wong D., Lee T.H., Lum A., Tao V.L., Yip S. Integrated proteomic analysis of low-grade gliomas reveals contributions of 1p-19q co-deletion to oligodendroglioma. Acta Neuropathol. Commun. 2022;10:70. doi: 10.1186/s40478-022-01372-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oh S., Yeom J., Cho H.J., Kim J.-H., Yoon S.-J., Kim H., Sa J.K., Ju S., Lee H., Oh M.J., et al. Integrated pharmaco-proteogenomics defines two subgroups in isocitrate dehydrogenase wild-type glioblastoma with prognostic and therapeutic opportunities. Nat. Commun. 2020;11:3288. doi: 10.1038/s41467-020-17139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J.B., Belanger K., Brandes A.A., Marosi C., Bogdahn U., et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 122.van den Bent M.J., Tesileanu C.M.S., Wick W., Sanson M., Brandes A.A., Clement P.M., Erridge S., Vogelbaum M.A., Nowak A.K., Baurain J.F., et al. Adjuvant and concurrent temozolomide for 1p/19q non-co-deleted anaplastic glioma (CATNON; EORTC study 26053-22054): second interim analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 2021;22:813–823. doi: 10.1016/S1470-2045(21)00090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Garofano L., Migliozzi S., Oh Y.T., D’Angelo F., Najac R.D., Ko A., Frangaj B., Caruso F.P., Yu K., Yuan J., et al. Pathway-based classification of glioblastoma uncovers a mitochondrial subtype with therapeutic vulnerabilities. Nat. Cancer. 2021;2:141–156. doi: 10.1038/s43018-020-00159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mund A., Coscia F., Hollandi R., Kov'cs F., Kriston A., Brunner A.-D., Bzorek M., Naimy S., Rahbek Gjerdrum L.M., Dyring-Andersen B., et al. AI-driven Deep Visual Proteomics defines cell identity and heterogeneity. bioRxiv. 2021 doi: 10.1101/2021.01.25.427969. Preprint at. [DOI] [Google Scholar]
- 125.Richardson R.B., Harper M.-E. Mitochondrial stress controls the radiosensitivity of the oxygen effect: implications for radiotherapy. Oncotarget. 2016;7:21469–21483. doi: 10.18632/oncotarget.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mellinghoff I.K., Ellingson B.M., Touat M., Maher E., De La Fuente M.I., Holdhoff M., Cote G.M., Burris H., Janku F., Young R.J., et al. Ivosidenib in isocitrate dehydrogenase 1-mutated advanced glioma. J. Clin. Oncol. 2020;38:3398–3406. doi: 10.1200/JCO.19.03327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zarei M., Hue J.J., Hajihassani O., Graor H.J., Katayama E.S., Loftus A.W., Bajor D., Rothermel L.D., Vaziri-Gohar A., Winter J.M. Clinical development of IDH1 inhibitors for cancer therapy. Cancer Treat Rev. 2022;103:102334. doi: 10.1016/j.ctrv.2021.102334. [DOI] [PubMed] [Google Scholar]
- 128.Mazurek M., Litak J., Kamieniak P., Kulesza B., Jonak K., Baj J., Grochowski C. Metformin as potential therapy for high-grade glioma. Cancers. 2020;12:210. doi: 10.3390/cancers12010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Valtorta S., Lo Dico A., Raccagni I., Gaglio D., Belloli S., Politi L.S., Martelli C., Diceglie C., Bonanomi M., Ercoli G., et al. Metformin and temozolomide, a synergic option to overcome resistance in glioblastoma multiforme models. Oncotarget. 2017;8:113090–113104. doi: 10.18632/oncotarget.23028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sharma K., Schmitt S., Bergner C.G., Tyanova S., Kannaiyan N., Manrique-Hoyos N., Kongi K., Cantuti L., Hanisch U.-K., Philips M.-A., et al. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 132.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 133.Suwala A.K., Felix M., Friedel D., Stichel D., Schrimpf D., Hinz F., Hewer E., Schweizer L., Dohmen H., Pohl U., et al. Oligosarcomas, IDH-mutant are distinct and aggressive. Acta Neuropathol. 2022;143:263–281. doi: 10.1007/s00401-021-02395-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.V Hovestadt, M Zapatka. conumee: enhanced copy-number variation analysis using Illumina DNA methylation arrays. [DOI] [PMC free article] [PubMed]
- 135.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 136.Therneau, T M; Lumley, T; Atkinson, E; Crowson, C. Survival (R Package).
- 137.Kassambra, A; Kosinski, M; Biecek, P; Scheipl, F. Survminer (R Package).
- 138.Radke J., Koch A., Pritsch F., Schumann E., Misch M., Hempt C., Lenz K., Löbel F., Paschereit F., Heppner F.L., et al. Predictive MGMT status in a homogeneous cohort of IDH wildtype glioblastoma patients. Acta Neuropathol. Commun. 2019;7:89. doi: 10.1186/s40478-019-0745-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lohkamp L.N., Schinz M., Gehlhaar C., Guse K., Thomale U.W., Vajkoczy P., Heppner F.L., Koch A. MGMT Promoter methylation and BRAF V600E mutations are helpful markers to discriminate pleomorphic xanthoastrocytoma from giant cell glioblastoma. PLoS One. 2016;11:e0156422. doi: 10.1371/journal.pone.0156422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 141.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wiestler B., Capper D., Holland-Letz T., Korshunov A., Von Deimling A., Pfister S.M., Platten M., Weller M., Wick W. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126:443–451. doi: 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 144.Cox J., Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics. 2012;13:S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Alečković M., Wei Y., LeRoy G., Sidoli S., Liu D.D., Garcia B.A., Kang Y. Identification of Nidogen 1 as a lung metastasis protein through secretome analysis. Genes Dev. 2017;31:1439–1455. doi: 10.1101/gad.301937.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Han C., Liao X., Qin W., Yu L., Liu X., Chen G., Liu Z., Lu S., Chen Z., Su H., et al. EGFR and SYNE2 are associated with p21 expression and SYNE2 variants predict post-operative clinical outcomes in HBV-related hepatocellular carcinoma. Sci. Rep. 2016;6:31237. doi: 10.1038/srep31237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gordon-Weeks A., Lim S.Y., Yuzhalin A., Lucotti S. Tumour-derived laminin α5 (LAMA5) promotes colorectal liver metastasis growth, branching angiogenesis and notch pathway inhibition. Cancers. 2019;11 doi: 10.3390/cancers11050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fozzatti L., Park J.W., Zhao L., Willingham M.C., Cheng S.Y. Oncogenic actions of the nuclear receptor corepressor (NCOR1) in a mouse model of thyroid cancer. PLoS One. 2013;8:e67954. doi: 10.1371/journal.pone.0067954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao J., Sun Y., Huang Y., Song F., Huang Z., Bao Y., Zuo J., Saffen D., Shao Z., Liu W., Wang Y. Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing. Sci. Rep. 2017;7:40488. doi: 10.1038/srep40488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Feng T., Liu Y., Li C., Li Z., Cai H. DEK proto-oncogene is highly expressed in astrocytic tumors and regulates glioblastoma cell proliferation and apoptosis. Tumour Biol. 2017;39 doi: 10.1177/1010428317716248. 1010428317716248. [DOI] [PubMed] [Google Scholar]
- 151.Baskin R., Woods N.T., Mendoza-Fandiño G., Forsyth P., Egan K.M., Monteiro A.N.A. Functional analysis of the 11q23.3 glioma susceptibility locus implicates PHLDB1 and DDX6 in glioma susceptibility. Sci. Rep. 2015;5:17367. doi: 10.1038/srep17367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Taniguchi K., Iwatsuki A., Sugito N., Shinohara H., Kuranaga Y., Oshikawa Y., Tajirika T., Futamura M., Yoshida K., Uchiyama K., Akao Y. Oncogene RNA helicase DDX6 promotes the process of c-Myc expression in gastric cancer cells. Mol. Carcinog. 2018;57:579–589. doi: 10.1002/mc.22781. [DOI] [PubMed] [Google Scholar]
- 153.Arentz G., Chataway T., Price T.J., Izwan Z., Hardi G., Cummins A.G., Hardingham J.E. Desmin expression in colorectal cancer stroma correlates with advanced stage disease and marks angiogenic microvessels. Clin. Proteomics. 2011;8:16. doi: 10.1186/1559-0275-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang Z., Kang J., Deng X., Guo B., Wu B., Fan Y. Knockdown of GATAD2A suppresses cell proliferation in thyroid cancer in vitro. Oncol. Rep. 2017;37:2147–2152. doi: 10.3892/or.2017.5436. [DOI] [PubMed] [Google Scholar]
- 155.Anchi T., Tamura K., Furihata M., Satake H., Sakoda H., Kawada C., Kamei M., Shimamoto T., Fukuhara H., Fukata S., et al. SNRPE is involved in cell proliferation and progression of high-grade prostate cancer through the regulation of androgen receptor expression. Oncol. Lett. 2012;3:264–268. doi: 10.3892/ol.2011.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wculek S.K., Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature. 2015;528:413–417. doi: 10.1038/nature16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ivanova A.V., Goparaju C.M.V., Ivanov S.V., Nonaka D., Cruz C., Beck A., Lonardo F., Wali A., Pass H.I. Protumorigenic role of HAPLN1 and its IgV domain in malignant pleural mesothelioma. Clin. Cancer Res. 2009;15:2602–2611. doi: 10.1158/1078-0432.CCR-08-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang J., Yu D., Liu X., Changyong E., Yu S. LINC00641/miR-4262/NRGN axis confines cell proliferation in glioma. Cancer Biol. Ther. 2020;21:758–766. doi: 10.1080/15384047.2020.1776581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Beltran A.S., Russo A., Lara H., Fan C., Lizardi P.M., Blancafort P. Suppression of breast tumor growth and metastasis by an engineered transcription factor. PLoS One. 2011;6:e24595. doi: 10.1371/journal.pone.0024595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhou C., Yu J., Wang M., Yang J., Xiong H., Huang H., Wu D., Hu S., Wang Y., Chen X.Z., Tang J. Identification of glycerol-3-phosphate dehydrogenase 1 as a tumour suppressor in human breast cancer. Oncotarget. 2017;8:101309–101324. doi: 10.18632/oncotarget.21087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
The raw methylation data and the CNV profiles calculated from them have been deposited to the Gene Expression Omnibus – NCBI repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
The paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.